Summary
Genome sequencing (GS) is a powerful test for the diagnosis of rare genetic disorders. Although GS can enumerate most non-coding variation, determining which non-coding variants are disease-causing is challenging. RNA sequencing (RNA-seq) has emerged as an important tool to help address this issue, but its diagnostic utility remains understudied, and the added value of a trio design is unknown. We performed GS plus RNA-seq from blood using an automated clinical-grade high-throughput platform on 97 individuals from 39 families where the proband was a child with unexplained medical complexity. RNA-seq was an effective adjunct test when paired with GS. It enabled clarification of putative splice variants in three families, but it did not reveal variants not already identified by GS analysis. Trio RNA-seq decreased the number of candidates requiring manual review when filtering for de novo dominant disease-causing variants, allowing for the exclusion of 16% of gene-expression outliers and 27% of allele-specific-expression outliers. However, clear diagnostic benefit from the trio design was not observed. Blood-based RNA-seq can facilitate genome analysis in children with suspected undiagnosed genetic disease. In contrast to DNA sequencing, the clinical advantages of a trio RNA-seq design may be more limited.
In this study we applied a trio RNA sequencing approach in a cohort of children with medical complexity who previously underwent genome sequencing. We find that trio analysis, while sometimes helpful in ruling out false-positive RNA-level aberrations, did not increase the diagnostic yield beyond singleton analysis.
Main text
Genome sequencing (GS) is currently the most comprehensive genetic test for the diagnosis of rare Mendelian disorders.1 However, more than half of individuals with a suspected genetic condition remain undiagnosed after GS. One limitation of contemporary genome analysis is the difficulty of filtering, prioritizing, and interpreting relevant non-coding variants beyond those affecting canonical splice sites. RNA sequencing (RNA-seq) has emerged as a promising technology to help address this issue. Initial studies applying RNA-seq in select cohorts of individuals yielded promising results.2,3,4,5,6,7,8,9,10,11,12 By contrast, the yield of RNA-seq as a direct complement to GS and with a family-based design has received limited study.
In this study we performed GS and RNA-seq from blood on 97 total individuals from 39 families: 2 quads, 22 trios, 8 duos, and 7 singletons. All probands were children with medical complexity who had previously undergone GS, and a subset of the GS results were reported previously.13,14 Expression outliers, novel or missing splicing junctions, and putative splicing variants of uncertain significance found by GS were evaluated via RNA-seq in each affected individual. In addition, we utilized the paired GS and RNA-seq data to identify single-nucleotide variants (SNVs) with allele imbalance.
Please see supplemental methods for details on cohort recruitment, GS and RNA-seq methods, and filtering/analysis methods.7,13,15,16,17,18,19,20,21,22 Written consent was provided by each proband’s parents and/or guardians as well as the proband where appropriate. The study was approved by the Research Ethics Board at the Hospital for Sick Children.
We identified gene-expression outliers and aberrant splicing events using blood RNA-seq data of 97 individuals from this cohort and an additional 145 individuals from our internal cohorts (122 individuals with pediatric rare disease and 23 healthy children).15,16 Our internal control cohort was used instead of GTEx given multiple technical differences (supplemental methods) and was expected to result in a lower false-positive rate. For example, when using over 200 selected high-quality blood RNA-seq datasets from GTEx, we identified a median of 3,096 genes with at least one aberrant splicing event, compared to 1,276 using our internal cohort.
Using OUTRIDER, we identified outlier genes with two different cutoffs based on either p value or Z score.23 With an adjusted p value < 0.05, we identified a median of three under-expressed and two over-expressed outlier genes per proband (Figure 1A). To increase the sensitivity of our assay, we expanded our search to genes with an absolute Z score ≥ 3 without a p value cutoff, resulting in a median of 86 under-expressed and 24 over-expressed outliers. Of these, only a median of six and three under- and over-expressed outlier genes, respectively, were associated with Human Phenotype Ontology (HPO) terms relevant to each individual’s phenotypes (Figure 1A).
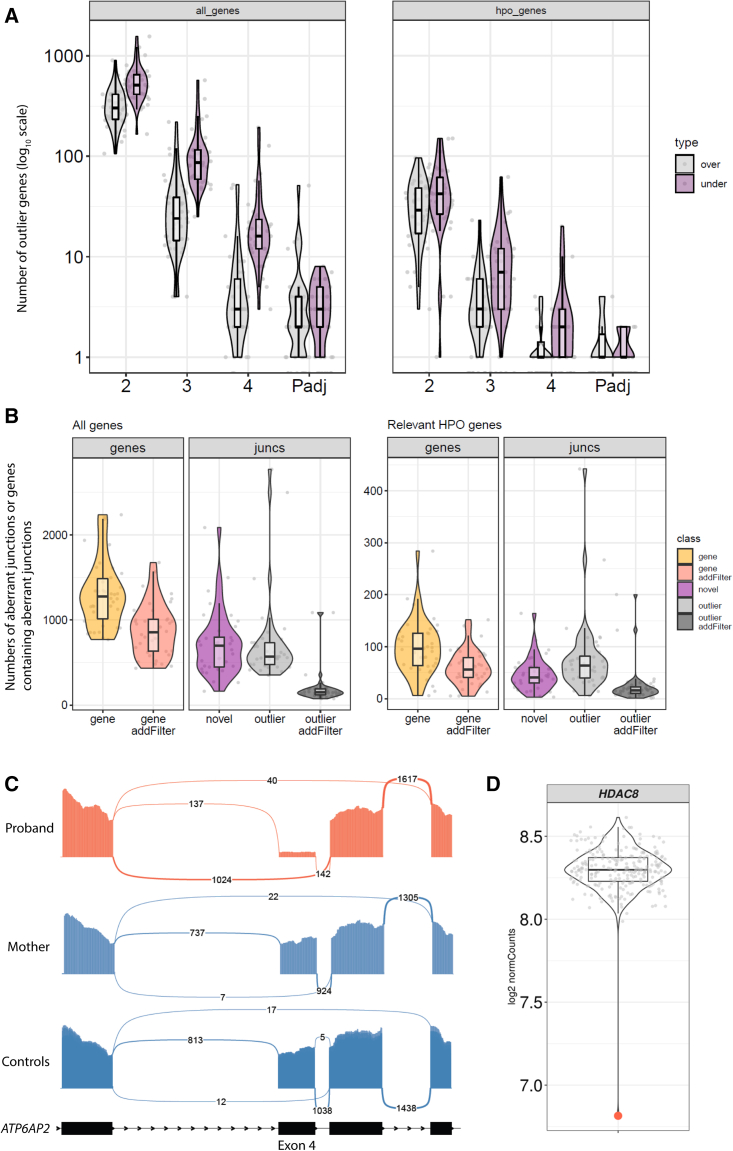
Figure 1.
Identification of gene-expression outliers and aberrant junctions in a cohort of medically complex children
(A) Summary of gene-expression outliers using different cut-offs. Scatterplot showing number of over-expressed (gray) or under-expressed (purple) outlier genes, as compared to our in-house control cohort, at absolute Z score ≤ 2, 3, or 4 or adjusted p value (adjusting across all genes in all samples; adjusted p values [Padj]) < 0.05. Each dot represents one sample. Violin and boxplots summarize the distribution of values in each group. In all boxplots the middle line is the median, the box edges are the 25th and 75th percentiles, and the whiskers represent 1.5× the interquartile range. y axis, numbers of outlier genes (log10 scale). Left panel, all genes; right panel, only genes associated with relevant HPO terms.
(B) Summary of aberrant junctions. Scatterplot showing number of reported genes (yellow), genes after additional filters (red), novel junctions (purple), reported outlier junctions (gray), and reported outlier junctions after additional filters (dark gray). Each dot represents one sample. Violin and boxplots summarize the distribution of values in each group. y axis, numbers of aberrant junctions or genes containing aberrant junctions. Left panel, all genes; right panel, only genes associated with relevant HPO terms.
(C) Sashimi plot of representative aberrant junctions in CMC 46 revealing a predominance of transcripts that skipped exon 4 in ATP6AP2. The skipping of exon 4 is evident in the proband (red) compared to his mother and to 10 randomly selected controls from the cohort (blue). y axis, number of aligned reads. The number of reads supporting each junction is shown between exons. The minimum number of reads to be drawn was set to 5 for this plot, for better visualization.
(D) Expression level of HDAC8 in CMC 6 (red dot) showing decreased expression compared to the rest of the samples in the cohort (gray dots). y axis, log2-normalized counts.
Using a set of relatively lenient cutoffs (see supplemental methods for more details), our bioinformatics pipeline reported a median of 1,276 genes per sample with novel or outlier junctions. This number was reduced to 856 (corresponding to a median of 676 novel junctions and 150 outlier junctions) after applying additional filters to prioritize outlier junctions (supplemental methods). After selecting genes associated with relevant HPO terms, we were left with a median of 16 outlier junctions and 41 novel junctions in 56 genes (Figure 1B).
Using an RNA-first approach (i.e., blind to GS findings) and filtering for splice junctions and expression outliers as described above, we were able to identify putative diagnostic RNA-level aberrations in 3 of the 39 probands (8%). In two individuals, aberrant splicing events were identified, whereas in the third individual, an expression outlier was detected. For the two aberrant splicing events, RNA-seq provided functional supporting evidence facilitating reclassification of the corresponding DNA variants as likely pathogenic (supplemental note).
Several findings are of particular note. Proband CMC 27 had a history of global developmental delay, intellectual disability, and epilepsy. In our initial unbiased filtering approach, missing junctions were identified in SMS (MIM: 300105). Closer examination of the transcriptome revealed multiple splicing abnormalities, including skipping of both exon 4 alone and exons 3 and 4 as well as the inclusion of intron 3 (Figure S1). None of these aberrant splicing events were observed in the parents. GS had previously identified a de novo variant upstream of exon 4 (c.265−5T>A [GenBank: NM_004595.5]); however, there was no consensus between three different in silico splicing prediction tools (MaxEnt,24 −57.6%; NNSPLICE,25 −10.1%; SSF [http://www.umd.be/searchsplicesite.html], −100%), and the pathogenicity of the variant was initially unclear.
Proband CMC 46 had a history of global developmental delay, intellectual disability, epilepsy, and inflammatory bowel disease. Missing/outlier junctions were identified in ATP6AP2 (MIM: 300556), with analysis of the transcript revealing a predominance of transcripts that skipped exon 4 (Figure 1C). A similar skew toward transcripts skipping exon 4 was not seen in either parent. GS had previously identified a de novo intronic indel (c.301−11_301−10delTT [GenBank: NM_005765.3]). There was no consensus between three different in silico splicing prediction tools (MaxEnt, −6.4%; NNSPLICE, −79.9%; SSF, −7.1%), and thus the variant was considered of unknown significance prior to RNA-seq analysis.
Proband CMC 6 had a history of bilateral choanal stenosis, bilateral dysplastic kidneys, dysmorphic features, microcephaly, global developmental delay, chronic lung disease, and intermittent pancytopenia. Gene expression analysis identified decreased HDAC8 (MIM: 300269) expression with a fold change of 0.36, Z score of −10.15, and adjusted p value of 5.82e−12 (Figure 1D). GS had previously identified a de novo frameshift variant, c.134_137del (GenBank: NM_018486.3) (p.Ile45Lysfs∗9).
Selected variants of uncertain diagnostic significance (VUSs) from the cohort were previously described.13 These included five putative splicing variants in a total of four genes: MED23 (MIM: 605042), PLCB1 (MIM: 607120), KIF1A (MIM: 601255), and JAM3 (MIM: 606871). MED23 exhibits measurable expression in blood, thus allowing for the targeted analysis of a homozygous c.3939+5G>A (GenBank: NM_004830.4) variant. This variant was classified as “likely pathogenic” when detected by clinical exome sequencing in CMC 01 (ClinVar: SCV000681305.2). There was no consensus between three different in silico splicing prediction tools (MaxEnt, −55.9%; NNSPLICE, −40.7%; SSF, −13.8%). No aberrant splicing was observed across the transcript, nor any significant difference in overall or allele-specific expression (ASE). Altogether, this downgrades the variant to a VUS with conflicting evidence (Figure S2). JAM3 is not expressed in blood, and KIF1A and PLCB1 are not expressed in any of the typical clinically accessible tissues (lymphoblastoid cell lines, fibroblasts, or blood),17,26 and thus the variants in these genes were not able to be further classified using blood-based RNA-seq.
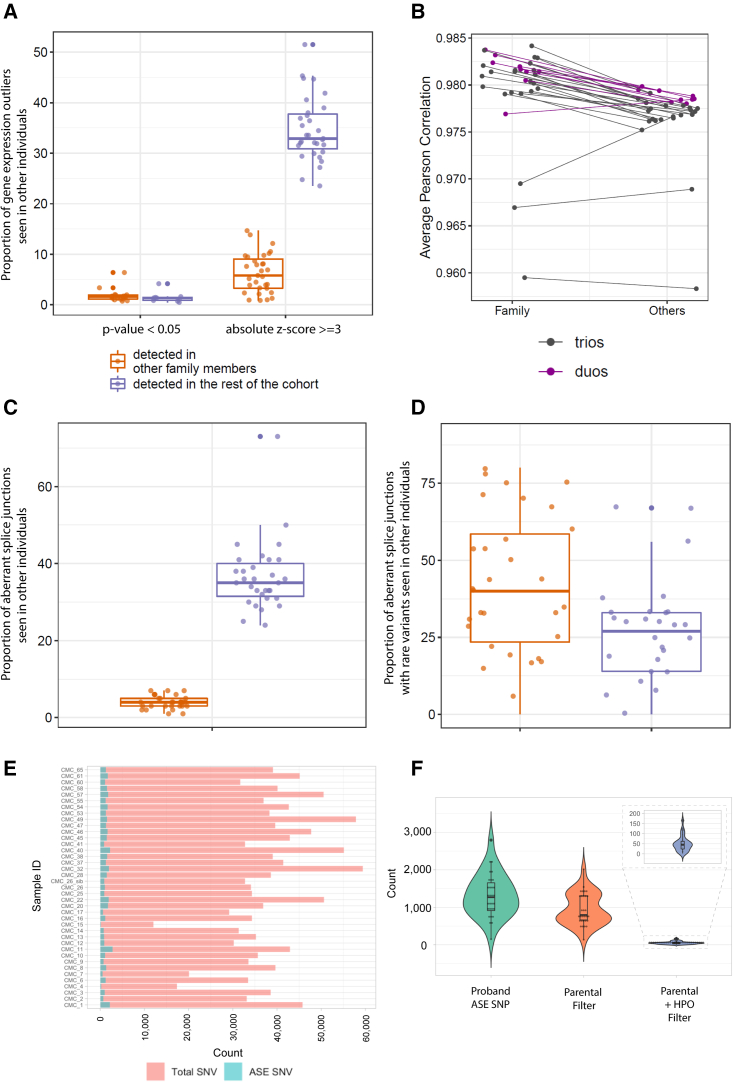
We sought to determine if a family-based RNA-seq design would facilitate filtering and interpretation of results, as is the case for trio genome-wide sequencing compared with singleton testing.27 We performed this filtering based on a de novo dominant model of inheritance where we wanted to prioritize novel events for further study. When examining gene-expression outliers, we found 186 statistically significant gene-expression outlier events (182 unique genes) in the probands with RNA-seq data from at least one parent. Of these, expression outlier events in 30 genes (in 17 probands) were seen in at least one other family member, compared to 14 genes (in 7 probands) that were seen in at least one other individual from the rest of the cohort, resulting in the ability to exclude 16% vs. 7.5% of identified outliers, respectively (Figures 2A and S3). This suggests that the familial samples may be important for prioritizing statistically significant expression outliers likely due to both the genetic and environmental similarities between probands and their family members. Using OUTRIDER-normalized read counts, the average Pearson correlation between a proband and their family members (median = 0.981) was slightly but significantly higher than that between the proband and the rest of the cohort (median = 0.977, p = 1.756e−06, one-sided paired Wilcoxon test) (Figure 2B). Although different RNA-seq normalization methods could have an impact on this analysis, our observation is consistent with a previous study suggesting familial similarity of the blood transcriptome.28 When using a more lenient cut-off for expression outliers (absolute Z score ≥ 3), trio analysis was no longer effective: only 1%–16% of identified genes were seen in at least one other family member, whereas 24%–52% were seen in at least one other internal cohort member (Figure 2A).
Figure 2.
Trio vs. cohort analysis for expression outliers as well as aberrant junctions and allele-specific expression analysis in the cohort
(A) Proportions of gene-expression outliers also detected in other samples showing that trio analysis is effective in filtering out statistically significant expression outliers, but not when using a more lenient cut-off (absolute Z score). Each dot represents a proband with family data available. y axis, proportion of gene-expression outlier defined by statistical significance (adjusted p value < 0.05) or Z scores (absolute Z score ≥ 3) that are also detected in family members (orange) or the rest of the cohort (purple).
(B) Gene expression correlation is consistently higher between probands and their family members than between probands and the rest of the cohort. y axis, average Pearson correlation of gene expression. Each dot represents a proband. Lines connect the same proband in the two columns. Purple, duos; black, trios.
(C) Proportions of total aberrant splicing events also detected in other samples. Each dot represents a proband with family data available. y axis, proportions of genes containing aberrant junctions detected in other family members (orange) or the rest of the cohort (purple).
(D) Proportions of aberrant splicing events with at least one rare variant nearby also detected in other samples. Each dot represents a proband with family data available. y axis, proportions of genes containing aberrant junctions detected in other family members (orange) or the rest of the cohort (purple).
(E) Bar plot of the number of reported SNVs and ASE SNVs for all the affected individuals. Red, total number of rare SNVs; blue, number of ASE events.
(F) Violin plot of the distribution of ASE SNVs after parental filter and HPO term filter.
We next sought to filter splice junctions using the parental data, again based on a de novo dominant model of inheritance. When examining all junction outliers, parental data were not effective in filtering out non-diagnostic splicing events. A median of only 4% of aberrant junctions were seen in at least one family member compared to a median of 35% of aberrant junctions in at least one other individual in the entire cohort (Figures 2C, S4, and S5). This is partially because our pipeline was designed to be highly sensitive at the cost of a higher false-positive rate. In addition, the majority of these aberrant splicing events likely do not have a clear genetic cause and likely have limited biological significance. Indeed, a median of only 10 outlier junctions have at least one rare variant (gnomAD genome and exome allele frequency [AF] < 0.01) nearby (within 10 bp of either end of the junction). We found that 125 of a total of 326 such junctions identified were seen in at least one other family member, compared to 83 seen in other individuals in the cohort (Figures 2D and S6). Neither type of trio analysis resulted in the identification of any additional diagnostic variants.
We used our combined GS and RNA-seq datasets to assess each proband for ASE, where two alleles at the same locus are expressed differently (i.e., skew toward one of the alleles). To do this, we prioritized variants that were rare (gnomAD genome AF < 0.01) and exhibited an imbalanced expression between the two alleles. We reasoned that dominant disorders caused by reduced expression of one allele (i.e., haploinsufficiency) would be detected by our expression outlier analysis above, but wondered if we might be able to identify recessive disorders caused by skewing of expression toward a single pathogenic allele.
ASE analysis was performed on 38 probands and 1 affected sibling. Through our analysis pipeline, a median of 38,308 heterozygous SNV sites were reported per affected individual, and a median of 1,263 (mean of 3.4% of all heterozygous SNV sites) were identified as ASE sites after the filters and QC measures (supplemental methods; Figure 2E). Applying an additional parental filter (for probands with parental samples) that removed sites that were reported as having a significant imbalance in either of the parents lowered the number by 27% to 944 ASE sites. In combination with the relevant HPO term filter, the number is reduced to an average of 50 ASE sites per proband (Figure 2F). Manual review of all filtered ASE sites did not yield any additional diagnoses in our cohort.
Altogether, we find that RNA-seq in blood can facilitate the interpretation of GS data. RNA analysis allowed for the confirmation of two putative diagnostic DNA variants (not including one that was already classified as pathogenic at the DNA level) and the exclusion of one other, thereby resulting in diagnostic utility in 8% (3/39) of the families. Trio RNA-seq analysis did not increase the yield of new diagnoses or candidate variants/genes. Altogether, our data provide further support for the use of singleton RNA-seq as an important diagnostic tool in rare disease.
One caveat of our study is that whole blood was utilized for RNA-seq; this has well-recognized limitations when studying a heterogeneous cohort of individuals with suspected undiagnosed rare genetic diseases.3 Another limitation of our study is that we utilized the trio RNA-seq data in a targeted way: for filtering out false-positive events based on a de novo dominant mode of inheritance. It remains possible that a trio design might facilitate the identification of disease-causing RNA-level aberrations with other modes of inheritance (e.g., inherited dominant-acting variants where there are ASE differences between parent and child causing apparent incomplete penetrance/highly variable expression). Trio RNA-seq may yet show utility if combined with additional novel analysis methods and/or if applied to a different or larger cohort of individuals. One additional caveat of our study is that our bioinformatics pipeline used genome build hg19 and Ensembl v.75, which have both since undergone additional improvements. However, the impact of newer builds on these results is expected to be minimal given that we focused on genes in which variants are known to cause disease, which are expected to already have been well annotated.
In total, these data highlight the value and limitations of blood-based RNA-seq as a clinical diagnostic test for rare genetic disease. Further study is required to better understand its diagnostic utility as a complement to GS in other cohorts of individuals with suspected genetic disorders.
Acknowledgments
We gratefully acknowledge all the individuals and their families who participated in this study. We thank the many healthcare providers involved in the diagnosis and care of these children with medical complexity. Special thanks to all staff affiliated with the Complex Care Program and The Centre for Applied Genomics. M.D.W. was supported by the Canada Research Chairs Program. Funding was provided by Genome Canada (OGI-158, M.D.W., A.S., and J.J.D.), the SickKids Centre for Genetic Medicine, the Sickkids Research Institute, and the University of Toronto McLaughlin Centre.
Declaration of interests
The authors declare no competing interests.
Published: March 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.03.006.
Contributor Information
Gregory Costain, Email: gregory.costain@sickkids.ca.
James J. Dowling, Email: james.dowling@sickkids.ca.
Supplemental information
Data and code availability
The data supporting the current study have not been deposited in a public repository to protect individual confidentiality but are available from the corresponding author on request.
References
- 1.Bick D., Jones M., Taylor S.L., Taft R.J., Belmont J. Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases. J. Med. Genet. 2019;56:783–791. doi: 10.1136/jmedgenet-2019-106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonorazky H.D., Naumenko S., Ramani A.K., Nelakuditi V., Mashouri P., Wang P., Kao D., Ohri K., Viththiyapaskaran S., Tarnopolsky M.A., et al. Expanding the Boundaries of RNA Sequencing as a Diagnostic Tool for Rare Mendelian Disease. Am. J. Hum. Genet. 2019;104:466–483. doi: 10.1016/j.ajhg.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murdock D.R., Dai H., Burrage L.C., Rosenfeld J.A., Ketkar S., Müller M.F., Yépez V.A., Gagneur J., Liu P., Chen S., et al. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J. Clin. Invest. 2021;131:e141500. doi: 10.1172/JCI141500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C., et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yépez V.A., Gusic M., Kopajtich R., Mertes C., Smith N.H., Alston C.L., Ban R., Beblo S., Berutti R., Blessing H., et al. Clinical implementation of RNA sequencing for Mendelian disease diagnostics. Genome Med. 2022;14:38. doi: 10.1186/s13073-022-01019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bournazos A.M., Riley L.G., Bommireddipalli S., Ades L., Akesson L.S., Al-Shinnag M., Alexander S.I., Archibald A.D., Balasubramaniam S., Berman Y., et al. Standardized practices for RNA diagnostics using clinically accessible specimens reclassifies 75% of putative splicing variants. Genet. Med. 2022;24:130–145. doi: 10.1016/j.gim.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Frésard L., Smail C., Ferraro N.M., Teran N.A., Li X., Smith K.S., Bonner D., Kernohan K.D., Marwaha S., Zappala Z., et al. Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med. 2019;25:911–919. doi: 10.1038/s41591-019-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’Grady G.L., et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddirevula S., Kuwahara H., Ewida N., Shamseldin H.E., Patel N., Alzahrani F., AlSheddi T., AlObeid E., Alenazi M., Alsaif H.S., et al. Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics. Genome Biol. 2020;21:145. doi: 10.1186/s13059-020-02053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C., et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wai H.A., Lord J., Lyon M., Gunning A., Kelly H., Cibin P., Seaby E.G., Spiers-Fitzgerald K., Lye J., Ellard S., et al. Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet. Med. 2020;22:1005–1014. doi: 10.1038/s41436-020-0766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truty R., Ouyang K., Rojahn S., Garcia S., Colavin A., Hamlington B., Freivogel M., Nussbaum R.L., Nykamp K., Aradhya S. Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation. Am. J. Hum. Genet. 2021;108:696–708. doi: 10.1016/j.ajhg.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costain G., Walker S., Marano M., Veenma D., Snell M., Curtis M., Luca S., Buera J., Arje D., Reuter M.S., et al. Genome sequencing as a diagnostic test in children with unexplained medical complexity. JAMA Netw. Open. 2020;3:e2018109. doi: 10.1001/jamanetworkopen.2020.18109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haque B., Khan T., Ushcatz I., Curtis M., Pan A., Wu W., Orkin J., Costain G. Contemporary aetiologies of medical complexity in children: a cohort study. Arch. Dis. Child. 2023;108:147–149. doi: 10.1136/archdischild-2022-325094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lionel A.C., Costain G., Monfared N., Walker S., Reuter M.S., Hosseini S.M., Thiruvahindrapuram B., Merico D., Jobling R., Nalpathamkalam T., et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stavropoulos D.J., Merico D., Jobling R., Bowdin S., Monfared N., Thiruvahindrapuram B., Nalpathamkalam T., Pellecchia G., Yuen R.K.C., Szego M.J., et al. Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Pediatric Medicine. NPJ Genom. Med. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker S., Lamoureux S., Khan T., Joynt A.C.M., Bradley M., Branson H.M., Carter M.T., Hayeems R.Z., Jagiello L., Marshall C.R., et al. Genome sequencing for detection of pathogenic deep intronic variation: A clinical case report illustrating opportunities and challenges. Am. J. Med. Genet. 2021;185:3129–3135. doi: 10.1002/ajmg.a.62389. [DOI] [PubMed] [Google Scholar]
- 18.Tan A., Abecasis G.R., Kang H.M. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Geijn B., Mcvicker G., Gilad Y., Pritchard J.K. WASP: Allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo M.A., Banks E., Poplin R., Garimella K. v, Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castel S.E., Levy-Moonshine A., Mohammadi P., Banks E., Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brechtmann F., Mertes C., Matusevičiūtė A., Yépez V.A., Avsec Ž., Herzog M., Bader D.M., Prokisch H., Gagneur J. OUTRIDER: a statistical method for detecting aberrantly expressed genes in RNA sequencing data. Am. J. Hum. Genet. 2018;103:907–917. doi: 10.1016/j.ajhg.2018.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 25.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 26.Aicher J.K., Jewell P., Vaquero-Garcia J., Barash Y., Bhoj E.J. Mapping RNA splicing variations in clinically accessible and nonaccessible tissues to facilitate Mendelian disease diagnosis using RNA-seq. Genet. Med. 2020;22:1181–1190. doi: 10.1038/s41436-020-0780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manickam K., McClain M.R., Demmer L.A., Biswas S., Kearney H.M., Malinowski J., Massingham L.J., Miller D., Yu T.W., Hisama F.M., ACMG Board of Directors Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:2029–2037. doi: 10.1038/s41436-021-01242-6. [DOI] [PubMed] [Google Scholar]
- 28.Tremblay B.L., Guénard F., Lamarche B., Pérusse L., Vohl M.-C. Familial resemblances in human whole blood transcriptome. BMC Genom. 2018;19:300. doi: 10.1186/s12864-018-4698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the current study have not been deposited in a public repository to protect individual confidentiality but are available from the corresponding author on request.