Abstract
tmRNA has a dual function as a tRNA and an mRNA to facilitate trans-translation, in which a ribosome can switch between translation of a truncated mRNA and the tmRNA’s tag sequence. SmpB is a tmRNA binding protein that has been identified to be essential for trans-translation in vivo. To further study the function of SmpB, an S30 fraction from an Escherichia coli strain, in which the set of genes for SmpB and tmRNA has been deleted from the genome, and His-tagged SmpB active in trans-translation were prepared. The SmpB-depleted S30 fraction had an ability to facilitate poly(U)-dependent tag-peptide synthesis in vitro when purified His-tagged SmpB was exogenously added together with tmRNA, although SmpB was not required for in vitro poly(U)-dependent poly(Phe) synthesis. It was also found that depletion of SmpB leads to a decrease in the level of tmRNA in the cell. In addition, SmpB considerably enhanced the aminoacylation of tmRNA by alanyl-tRNA synthetase in vitro. The aminoacylation enhancement by SmpB, the binding of SmpB to tmRNA and the effect of depletion of SmpB on the expression level of tmRNA in the cell were all affected by some mutations in the tRNA-like domain which cause a defect in ribosome binding leading to a trans-translation deficiency. These results demonstrate that, via binding to the tRNA-like domain of tmRNA, SmpB plays various roles: rescuing the tmRNA molecule from degradation in the cell, enhancing the aminoacylation of tmRNA and mediating the binding of tmRNA to ribosome.
INTRODUCTION
tmRNA (transfer-messenger RNA, also known as 10Sa RNA or SsrA RNA) is widely distributed among eubacteria, and has also been found in some chloroplasts (1–3). It is a unique molecule in that it possesses both tRNA and mRNA properties to facilitate an unusual translation reaction, trans-translation.
Both terminal regions of this molecule can be folded into a partial tRNA-like structure comprising a TΨC-arm and an amino acid acceptor stem leading to the universal 3′-terminal CCA sequence (4,5) with two tRNA-specific modified nucleotides in the putative TΨC-loop (6). This tRNA-like structure is supported by chemical and enzymatic probing studies as well as comparative studies (7–10). It contains major identity determinants of tRNAAla, and can be recognized by RNase P (5,11), EF-Tu (12,13) and alanyl-tRNA synthetase (4,5).
The mRNA domain located in the middle of this molecule encodes the last 10 residues of the 11 amino acid tag-sequence (AANDENYALAA), which was first found on the truncated C-terminus of a fraction of mouse interleukin-6 expressed in Escherichia coli (14), and was also found on other polypeptides when they were translated from mRNAs lacking a termination codon (15), possessing a cluster of rare codons (16) or from mRNA for E.coli LacI (17) and on an in vitro translation product in the presence of tmRNA and poly(U) mimicking a truncated mRNA (18). It has recently been shown that the trans-translation can also occur at the termination codon on a normal mRNA (19).
It has been proposed that the molecular interplay between these two functions of this molecule facilitates an unusual translation reaction—trans-translation—in which a ribosome can switch from the translation of a truncated mRNA to the tag-encoded sequence of tmRNA. This would relieve stalled translation from mRNAs lacking a stop codon or possessing a cluster of rare codons with the addition of a specific tag-peptide as a degradation signal to the truncated C-termini of polypeptides decoded (15,16,20,21). A series of these processes may promote recycling of ribosomes as well as prevent accumulation of abortively synthesized polypeptides, providing some advantage to the cell for survival (22,23). The trans-translation model has also been supported by several other findings showing that the function as a tRNA is a prerequisite for the function as an mRNA in vitro (18), that an amino acid aminoacylated to tmRNA is actually incorporated into the tag-peptide (24) and that tmRNA binds predominantly to 70S ribosome (4,25) but scarcely to polysome (26).
According to the trans-translation, the first GCA codon for tag-peptide should be settled on the A-site after the first translocation event. It has been shown in vivo and in vitro that several nucleotides upstream of the tag-encoding region are important for efficient and correct initiation of tag-translation (27,28). In addition, the first pseudoknot (PK1) 12 nt upstream of the tag-initiation point, but not the other three pseudoknots (PK2–PK4) downstream of the tag-encoding region, has been shown to be important for efficient tag-translation (29,30). However, any intermolecular or intramolecular interactions with these cis-acting sequences and structures on tmRNA should be required to reasonably explain the mechanism underlying resumption of translation at the definite position on tmRNA.
Recently, the small basic protein SmpB, the gene of which is located immediately upstream of the tmRNA gene on the genome, has been identified in E.coli as a tmRNA binding protein (31), and it is included in the complex involving tmRNA and several other proteins (32). Among these protein factors, SmpB is the only one that has been shown to be essential for trans-translation. The depletion of SmpB causes an unsuccessful binding of tmRNA to ribosome leading to the failure of trans-translation in vivo, although the mode of interaction between SmpB and tmRNA has yet to be identified.
In the present study, we focused on the role of SmpB in the processes of trans-translation. The results showed that tmRNA, via binding to the tRNA-like domain of tmRNA, plays various roles: rescuing the tmRNA molecule from degradation in the cell, enhancing the aminoacylation of tmRNA and mediating the binding of tmRNA to ribosome.
MATERIALS AND METHODS
Construction of the strain W3110 ΔsmpBΔssrA::cat
The ScaI–SacI fragment including the smpB and ssrA genes derived from a Kohara lambda phage clone, number 439 (33) was cloned into pUC119. The NspV–NcoI region of this plasmid covering the C-terminal six-sevenths of smpB and the whole of ssrA was then replaced by a gene conferring chloramphenicol resistance (Cmr GenBlock in pHSG398). The ScaI–SacI fragment excised from the resulting plasmid was transformed into E.coli JC7623. In the transformants of chloramphenicol-resistance obtained, the expected gene substitution in the genome was confirmed by northern hybridization using oligodeoxynucleotide probes for SmpB and tmRNA. The disrupted set of the smpB and ssrA genes in JC7623 was then transferred to W3110 by phage P1 using chloramphenicol resistance as a selection marker. We confirmed that the final W3110 ΔsmpBΔssrA (smpB-ssrA::cat) failed to produce tmRNA by northern hybridization using an oligodeoxynucleotide probe for tmRNA, and by PCR for both genes.
Construction of the plasmids encoding wild-type and His-tagged SmpB
The gene for wild-type SmpB, as well as for N- and C-terminally His-tagged SmpB in which a 6× His sequence was immediately fused to the N- and C-termini, respectively, of the coding region of SmpB, was amplified by primer-directed PCR, and ligated downstream of native SmpB promoter sequence. The amplified DNA fragment was cloned into pGEMEX-2 (Promega). The resulting plasmids were named pSmpB, pNHisSmpB and pCHisSmpB, respectively.
Purification of His-tagged SmpB protein
The strain W3110 ΔsmpBΔssrA/pNHisSmpB or ΔsmpBΔssrA/pCHisSmpB was grown in 6 l of LB broth at 37°C to an A600 of 1.0. The cells were harvested by centrifugation, resuspended in 30 ml of buffer A [50 mM HEPES (pH 7.5), 100 mM potassium chloride, 10% glycerol (w/v) and 1 mM dithiothreitol] and lysed by sonication. The S100 fraction prepared from the lysate was loaded on an SP Sepharose Fast Flow column (Amersham Pharmacia Biotech). The column was washed with buffer A and eluted with a linear gradient from 100 to 1000 mM potassium chloride in buffer A (300 ml). SmpB was eluted from the column in the fractions of 580–700 mM potassium chloride. Fractions containing SmpB were dialyzed with buffer B [50 mM HEPES (pH 7.5), 200 mM potassium chloride, 10% glycerol and 1 mM dithiothreitol] and loaded onto Ni-NTA agarose (Qiagen). The column was washed with buffer C [50 mM HEPES (pH 7.5), 200 mM potassium chloride, 10% glycerol, 20 mM imidazole and 1 mM dithiothreitol] and eluted with a linear gradient from 20 to 1000 mM imidazole in buffer C. Typically, 0.2 mg of SmpB with >95% purity was yielded from a liter of cell culture. The concentration of protein was measured by the Bradford method.
Preparations of tmRNA variants and purification of tRNAAla from E.coli cells
This plasmid encoding each of tmRNA variants was cotransformed with pACYC184 encoding the T7 RNA polymerase gene under the lac-promoter sequence into E.coli strain W3110 ΔssrA, as described (18,28,29). tmRNA induced by the addition of 1.0 mM isopropyl-1-thio-β-d-galactopyranoside was purified as described (4).
The nucleic acid fraction was extracted with phenol from mid-log phase cells followed by ethanol precipitation. After 2× phenol extraction and ethanol precipitation, the resulting fraction was subjected to differential isopropylalcohol precipitations to roughly remove DNA, followed by incubation with RNase-free DNase I (Amersham Pharmacia Biotech). tmRNA was purified by electrophoresis on a 5% polyacrylamide gel containing 7 M urea. Spectrophotometric measurements were made to determine the concentration of RNA.
The preparation of solid-phase DNA probes and the preparative hybrid selection of tRNAs were carried out as described earlier (34). The sequence of biotinylated DNA probe is 5′-biotin-TGGTGGAGCTATGCGGGATC-3′, which is complementary to the region from the variable arm to the 3′-CCA end of E.coli tRNAAla. The resulting tRNA fraction was further purified by 15% polyacrylamide gel electrophoresis (35).
Aminoacylation
Alanyl-tRNA synthetase was purified from an alanyl-tRNA synthetase overproducing strain (5) with DEAE-Toyopearl 650 (Tosoh, Tokyo) and subsequent hydroxyapatite column chromatography (Gigapite, Seikagaku Corporation, Tokyo) (35). The final fraction of alanyl-tRNA synthetase had a specific activity of 16.78 U/mg, when 1 U of enzyme catalyzed the formation of 1 nmol of aminoacyl-tRNA per 10 min under the reaction conditions described below.
The aminoacylation reaction proceeded at 37°C, in a 50 µl reaction mixture containing 60 mM Tris–HCl (pH 7.5), 10 mM magnesium chloride, 30 mM potassium chloride, 5.0 mM dithiothreitol, 2.5 mM ATP and 25 µM l-[U-14C]alanine (6.5 GBq/mmol; NEN Life Science Products), 1.0 µM tmRNA variants (when 1 A260 unit corresponds to 325 pmol) or tRNAAla transcript, 8.5 × 10–2 or 8.5 × 10–3 U of alanyl-tRNA synthetase and varying amounts (0–4 µM) of SmpB. At the times specified, a 12 µl aliquot was withdrawn and spotted on Whatman 3MM filter paper, and radioactivity in the trichloroacetic acid-insoluble fraction was measured by a liquid scintillation counter.
SmpB-dependent trans-translation in vitro
S30 fraction was prepared from E.coli cells, as described previously (18). The reaction mixture (100 µl) contained 80 mM Tris–HCl (pH 7.8), 7 mM magnesium acetate, 150 mM ammonium chloride, 2.5 mM dithiothreitol, 5 mM phosphoenolpyruvate, 1 mM ATP, 0.2 mM GTP, 20 µM l-[U-14C]alanine (6.5 GBq/mmol), and 0.05 mM each of the remaining unlabeled 19 amino acids, 0.1 µM tmRNA, 20 µl of the S30 fraction, 250 µg of poly(U) (50–100mer; Sigma) and SmpB (0–4 µM). tmRNA variants were used in the reaction without any refolding procedure after the purification from the gel containing 7 M urea. The reaction mixture was incubated at 37°C. At intervals, a 23.5 µl aliquot was withdrawn from a 100 µl reaction mixture and spotted onto Whatman 3MM filter paper, and radioactivity in the hot trichloroacetic acid-insoluble fraction was measured by a liquid scintillation counter.
SmpB-dependent interaction of tmRNA with ribosome
After 5 min preincubation at 37°C, 80 µl of reaction mixture containing 100 mM Tris–HCl (pH 7.8), 8.75 mM magnesium acetate, 187.5 mM ammonium chloride, 375 mM potassium chloride, 3.125 mM dithiothreitol, 1.25 mM ATP, 0.25 mM GTP, 6.25 mM phosphoenol pyruvate, 0.0625 mM each of 20 amino acids, 25 pmol tmRNA and 250 µg poly(U) with or without 2.5 µM SmpB, 20 µl of S30 fraction from ΔsmpBΔssrA cells were added and incubated at 37°C for 10 min. The reaction mixture was immediately loaded on a 5–20% linear sucrose density gradient containing 10 mM Tris–HCl (pH 7.8), 10 mM magnesium chloride and 300 mM potassium chloride, and was centrifuged at 25 000 r.p.m. with a Hitachi P28S rotor for 5 h at 4°C. Nucleic acids prepared from each fraction by phenol extraction and ethanol precipitation were separated by electrophoresis on a 1.5% agarose gel containing 6.3% formaldehyde, and were then blotted onto a nylon membrane. tmRNA was detected by northern hybridization using a 3′-digoxygenin-labeled oligodeoxyribonucleotide (4) complementary to a portion of the tmRNA sequence (nucleotides 251–280).
Glycerol density gradient centrifugation
tmRNA (25 µg, 1 µM) and His-tagged SmpB (16 µg, 4 µM) were mixed in 200 µl 80 mM Tris–HCl (pH 7.8) buffer containing 7 mM magnesium acetate, 150 mM ammonium cloride and 2.5 mM dithiothreitol, and incubated at 37°C for 10 min. The mixture was loaded on a 10–30% linear glycerol density gradient (12 ml) made in the same buffer, and was centrifuged at 40 000 r.p.m. for 20 h at 4°C in a Hitachi P28S rotor. Gradients were fractionated into 25 fractions. Each fraction was divided into two portions: 80% was used for detection of SmpB and 20% was used for detection of tmRNA.
Proteins in each fraction were precipitated with one-third vol of 100% trichloroacetic acid and then were resuspended in 150 mM Tris (pH 6.8), 30% glycerol, 6% SDS, 300 mM dithiothreitol and 0.3% bromophenol blue. After electrophoresis, the SDS-containing 15% polyacrylamide gel was electroblotted onto a nitrocellulose membrane and was then probed with an antibody raised against a peptide having an amino acid sequence of positions 130–149 of E.coli SmpB. Antibody was visualized by ECL plus chemiluminescence (Amersham Pharmacia Biotech) after binding a horseradish peroxidase-conjugated secondary antibody.
Nucleic acids prepared from each fraction by phenol extraction and ethanol precipitation were separated by electrophoresis on a 1.5% agarose gel containing 6.3% formaldehyde, and were then blotted onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech). tmRNA was detected by northern hybridization using a 3′-digoxygenin-labeled oligodeoxyribonucleotide complementary to a portion of the tmRNA sequence (nucleotides 331–360) (4).
Gel-mobility shift assay
tmRNA was dephosphorylated by T4 alkaline phophatase (Takara) and was then rephosphorylated with [γ-32P]ATP by T4 polynucleotide kinase (Takara). Binding reaction mixtures (20 µl) contained 50 mM MES (pH 6.5), 200 mM potassium chloride, 5% glycerol, 5 mM 2-mercaptoethanol, 0.01% NP-40, 0.1 mg/ml bovine serum albumin, labeled tmRNA (100 pM) and different amounts of His-tagged SmpB. After incubation at room temperature for 10 min, mixtures were electrophoresed on a 5% polyacrylamide gel in TBE buffer [50 mM Tris (pH 8.3), 25 mM borate and 2 mM EDTA]. Radioactivity on a gel was monitored by Bio-Image Analyzer FLA3000 (Fuji Film).
Detection of RNA in the cell by northern hybridization
Total RNA from cells was prepared by phenol extraction as described (4). Varying amounts of total RNA were electrophoresed on a 1.5% agarose gel containing 6.3% formaldehyde, and were then blotted onto a nylon membrane. tmRNA was detected by northern hybridization using a 3′-digoxygenin-labeled oligodeoxyribonucleotide complementary to a portion of the tmRNA sequence (nucleotides 331–360) or a portion of 5S rRNA (nucleotides 71–100).
RESULTS
Disruption of the set of genes for SmpB and tmRNA
The SmpB gene (smpB) is located immediately upstream of the tmRNA gene (ssrA). We first constructed a strain in which the region of the E.coli genome covering the C-terminal 85% of smpB and the whole of ssrA has been replaced by a chloramphenicol-resistant gene (ΔsmpBΔssrA or smpB-ssrA::cat). The parental strain (W3110), the otherwise isogenic ΔssrA (ssrA::kan) (5) and ΔsmpBΔssrA cells displayed no difference in growth rate at either 37 or 42°C in rich medium (data not shown).
SmpB is required for poly(U)-dependent tag-peptide synthesis but not for poly(U)-dependent poly(Phe) synthesis
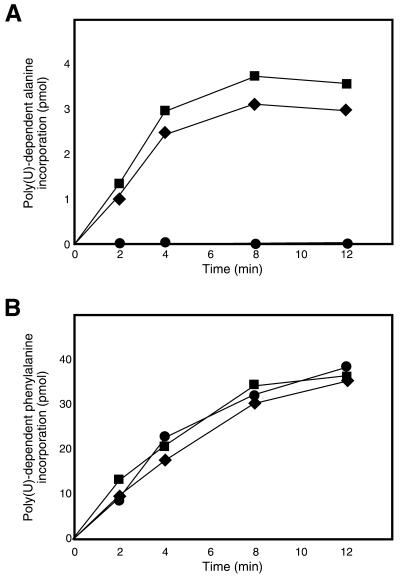
It has been shown that the ability of tag-peptide synthesis directed by tmRNA can be assessed by monitoring the poly(U)-dependent alanine incorporation into the polypeptide fraction using the S30 fractions from ΔssrA cells (18). Using this method, the ability of the SmpB-depleted S30 fraction from ΔsmpBΔssrA cells to facilitate tag-peptide synthesis was examined. The SmpB-containing S30 fraction from ΔssrA cells had an ability to facilitate poly(U)-dependent alanine incorporation in the presence of exogenous tmRNA, whereas the SmpB-depleted S30 fraction from ΔsmpBΔssrA cells did not (Fig. 1A). These results confirm in vitro that SmpB is required for trans-translation. On the other hand, poly(U)-dependent poly(Phe) synthesis in the presence of exogenous tmRNA proceeded similarly with the S30 fractions from two strains (Fig. 1B), indicating that SmpB is not a prerequisite for canonical translation.
Figure 1.
(A) Poly(U)-dependent alanine incorporation using the S30 fractions from ΔsmpBΔssrA cells with (squares) and without (circles) plasmid-encoded SmpB and from ΔssrA cells (diamonds) in the presence of exogenous tmRNA. (B) Poly(U)-dependent phenylalanine incorporation using the S30 fractions from ΔsmpBΔssrA cells with (squares) and without (circles) plasmid-encoded SmpB and from ΔssrA cells (diamonds) in the presence of exogenous tmRNA.
Next, the S30 fraction was prepared from the ΔsmpBΔssrA cells into which a pMW119-based low copy number plasmid carrying the SmpB gene with or without the 6× His (His-tag) sequence at the N- or C-terminus under the native promoter was introduced. This S30 fraction supplemented with plasmid-encoded SmpB without the His-tag sequence had an ability to facilitate poly(U)-dependent tag-peptide synthesis in the presence of exogenous tmRNA with an efficiency almost equivalent to that for the S30 fraction from the ΔssrA cells (Fig. 1A). The S30 fraction supplemented with plasmid-encoded N- or C-terminally His-tagged SmpB also had an ability to facilitate poly(U)-dependent tag-peptide synthesis in the presence of exogenous tmRNA with a similar efficiency (Fig. 1A).
In vitro trans-translation using the SmpB-depleted S30 fraction was facilitated by an exogenous addition of purified His-tagged SmpB
We prepared SmpB possessing a His-tag at the C-terminus in E.coli. His-tagged SmpB was successfully expressed and solubilized. We purified it by cation exchange chromatography followed by nickel-chelate affinity chromatography to a single band of ∼19 kDa using SDS–polyacrylamide gel electrophoresis.
A gel-mobility shift assay was performed for this His-tagged SmpB. The Kd was ∼30 nM, only slightly higher than the value (10 nM) reported for the wild-type SmpB purified from an inclusion body via a procedure of denaturation by 3 M guanidium hydrochloride (31).
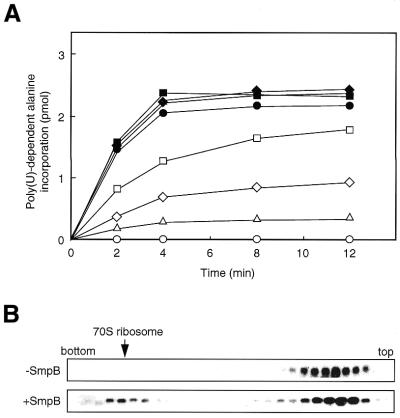
Then we examined the activity of this purified His-tagged SmpB in in vitro poly(U)-dependent tag-peptide synthesis in the presence of exogenous tmRNA using the SmpB-depleted S30 fractions from ΔsmpBΔssrA cells. As shown in Figure 2A, tag-peptide synthesis occurred when His-tagged SmpB was exogenously added in the reaction. The level of tag-peptide synthesis increased with increasing amount of SmpB, and it was substantially saturated at a SmpB concentration >1.6 µM. These results indicate that purified His-tagged SmpB is active in trans-translation.
Figure 2.
(A) The effect of the addition of purified His-tagged SmpB into the S30 fractions from ΔsmpBΔssrA cells on poly(U)-dependent alanine incorporation in vitro. 0 (open circles), 0.05 (open triangles), 0.1 (open diamonds), 0.2 (open squares), 0.4 (closed circles), 0.8 (closed triangles), 1.6 (closed diamonds) and 3.2 µM (closed squares) of His-tagged SmpB were added to the reaction. (B) The effect of purified His-tagged SmpB on the ribosome binding of tmRNA in vitro.
We also examined the effect of His-tagged SmpB on the ribosome binding property of tmRNA in vitro. The mixture of the trans-translation reaction in the presence or absence of 2 µM His-tagged SmpB was subjected to a sucrose density gradient centrifugation. In the absence of SmpB, tmRNA appears exclusively in the soluble fraction (Fig. 2B). In the presence of SmpB, the signal of tmRNA appears in the 70S ribosome fraction as well as in the soluble fraction. This is in agreement with an earlier finding that depletion of SmpB seriously affects the ribosome binding capacity of tmRNA in vivo (31).
SmpB binding property for tmRNA variants analyzed by glycerol density gradient centrifugation
Next we examined the binding property of SmpB to tmRNA by glycerol density gradient centrifugation. The mixture of tmRNA and purified His-tagged SmpB was centrifuged through a glycerol density gradient, and the fractions were subjected to western blotting analysis using anti-SmpB antibody as well as northern blotting analysis using a DNA probe complementary to a portion of tmRNA.
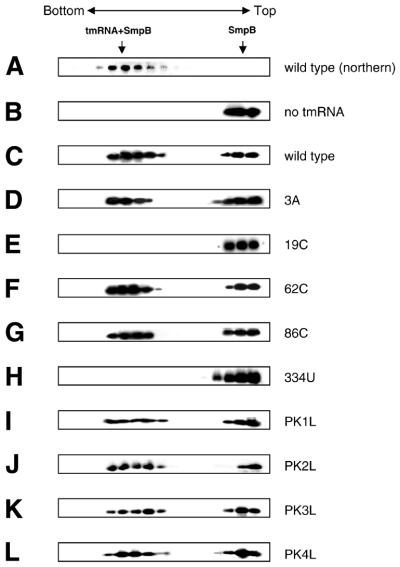
As shown in Figure 3B, in the absence of tmRNA, the signal for SmpB was observed in the slower sedimenting fractions around the top of the tube. In the presence of tmRNA, the signal was shifted to the lower fractions (Fig. 3C) in which the tmRNA signal was also detected by northern hybridization (Fig. 3A). Essentially the same patterns were obtained, when SmpB was detected using the anti-His-tag antibody (data not shown). These results indicate that the His-tagged SmpB made a complex with tmRNA in the mixture.
Figure 3.
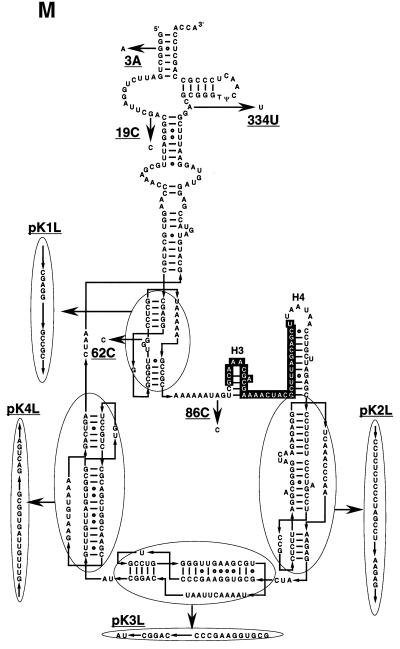
Interaction of SmpB with tmRNA variants. (A) tmRNA fractionated by centrifugation on a glycerol density gradient was detected by northern hybridization. His-tagged SmpB alone (B) or mixed with wild-type tmRNA (C), 3A (D), 19C (E), 62C (F) 86C (G), 334U (H), pK1L (I), pK2L (J), pK3L (K) or pK4L (L) was fractionated by glycerol density gradient centrifugation and was detected by western blotting using an antibody raised against SmpB. (M) Mutations designated on a secondary structure model of E.coli tmRNA. The tag-encoded sequence highlighted by white with a black background is surrounded by four pseudoknots (PK1–PK4). Non-Watson–Crick base pairs are shown by open circles. This RNA has two tRNA-specific modified nucleotides, 5-methyl U and pseudouridine in the T-loop (6), indicated as T and Ψ, respectively. Arrows indicate the mutations used in this study.
Using this system, we examined the SmpB binding property of 3A mutant having a G→A mutation at position 3 which has no capacity of aminoacylation due to the lack of the identity determinant for alanyl-tRNA synthetase. This mutant successfully made a complex with SmpB (Fig. 3D), suggesting that aminoacylation with alanine is not a prerequisite for SmpB binding to tmRNA.
In earlier studies, several tmRNA mutants, which cause serious damage to trans-translation in vitro, have been obtained. Among them, we selected four single-point mutants named 19C, 62C, 86C and 334U, to examine the effects of mutations on the complex formation with SmpB (Fig. 3M). Each of 19C and 334U having a G→C mutation at 19 and an A→U mutation at 334, respectively, completely damages the trans-translation in vitro (36). 62C having a G→C mutation at position 62 in PK1 decreases the in vitro trans-translation efficiency to less than half (29), and 86C having an A→C mutation at 86, 4 nt upstream of the tag-initiation point, completely damages the trans-translation in vitro (28). Each mutation causes serious damage to trans-translation in vitro, but does not affect the aminoacylation. We examined the complex formation with SmpB for these mutants by glycerol density gradient centrifugation. It was found that both 62C and 86C mutants made a complex with SmpB (Fig. 3F and G), whereas 19C and 334U mutants did not (Fig. 3E and H). These results suggest that SmpB contacts with positions G19 and A334 and/or their close proximities, but not with the bases at 62 or 86.
We also examined the effects for the pseudoknot mutants pK1L, pK2L, pK3L and pK4L, in which psudoknot structure is replaced by single-stranded RNA (Fig. 3M) (37). Each of these four pseudoknot mutants made a complex with SmpB (Fig. 3I–L).
SmpB enhances aminoacylation of tmRNA
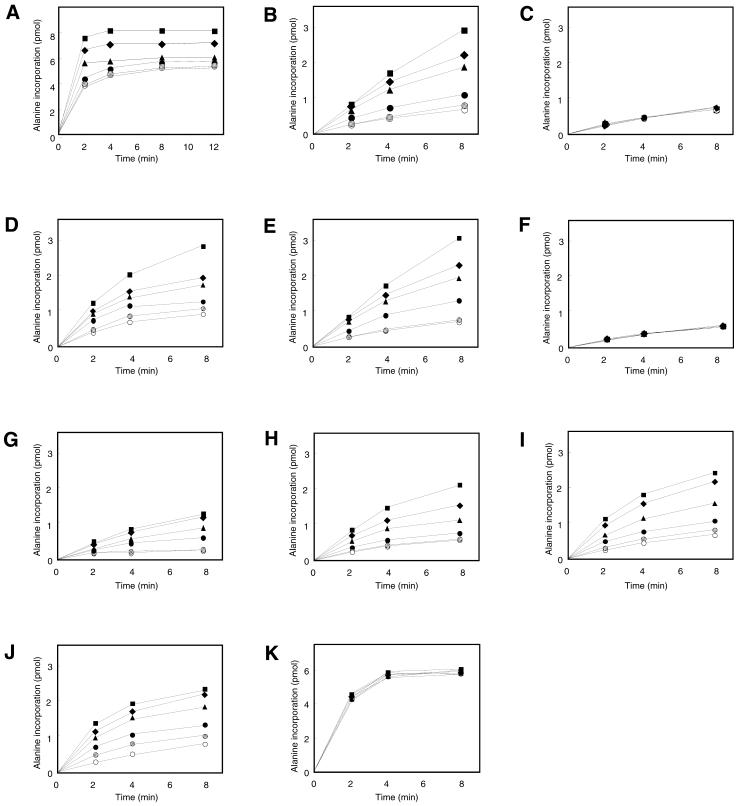
Next the effect of His-tagged SmpB on the aminoacylation reaction of tmRNA by alanyl-tRNA synthetase was examined. Under excess alanyl-tRNA synthetase (8.5 × 10–2 U), the plateau level of aminoacylation reached only ∼50% in the absence of SmpB (Fig. 4A). The low level of aminoacylation plateau has also been reported earlier (6,13), and it was not attributed to the contamination of nucleases in alanyl-tRNA synthetase, because of no detectable degradation of the tmRNA band on the gel after prolonged incubation of the reaction and an unchanged level of aminoacylation plateau after prolonged preincubation without amino acid (data not shown). Interestingly, the plateau level was increased with increasing amount of SmpB and it reached 75% in the presence of 4 µM SmpB. Under the limited concentration of alanyl-tRNA synthetase (8.5 × 10–3 U), the effect of SmpB was more obvious (Fig. 4B). The aminoacylation efficiency at 12 min in the presence of 4 µM SmpB was ∼5-fold higher than that in the absence of SmpB. As a control molecule aminoacylatable with alanine, tRNAAla from E.coli was examined. In contrast to the case for tmRNA, the aminoacylation efficiency of tRNAAla was unchanged by the addition of SmpB (Fig. 4K).
Figure 4.
The effect of SmpB on aminoacylation of tmRNA by 8.5 × 10–2 U (A) and 8.5 × 10–3 U (B) of alanyl-tRNA synthetase. The effect of SmpB on aminoacylation of tmRNA mutants 19C (C), 62C (D) 86C (E), 334U (F), pK1L (G), pK2L (H), pK3L (I), pK4L (J) and E.coli tRNAAla (K), using 8.5 × 10–3 U of alanyl-tRNA synthetase. 0 µM (open circles), 0.1 µM (hatched circles), 0.5 µM (closed circles), 1 µM (closed triangles), 2 µM (closed diamonds) and 4 µM (closed squares) SmpB were added to the reaction of aminoacylation.
We examined the effect of SmpB on aminoacylation for the trans-translation deficient mutants described above. As already reported (28,29), aminoacylation of 62C and 86C mutants occurs to a similar extent as that for wild-type tmRNA in the absence of SmpB (Fig. 4A, D and E). The aminoacylation for 62C and 86C mutants was considerably enhanced by the addition of SmpB (Fig. 4D and E) in a manner similar to that for wild-type tmRNA. pK2L, pK3L and pK4L mutants are efficiently aminoacylated, whereas pK1L has a significantly reduced aminoacylation ability (37). The enhancement of aminoacyation by SmpB was observed in these pseudoknot mutant tmRNAs, even in pK1L (Fig. 4G–I). In contrast, no significant enhancement was observed for 19C and 334U mutants (Fig. 4C and F), which does not bind to SmpB (Fig. 3E and H). Thus, the enhancement of aminoacylation by SmpB was observed only in the tmRNA mutants which can make a complex with SmpB (Fig. 3E–H). These results indicate that SmpB has a role in the enhancement of aminoacylation of tmRNA via binding to G19 and A334 and/or their close proximities, and that a defect in the normal binding between tmRNA and SmpB abolishes the enhancement of the aminoacylation.
Binding to SmpB increases the level of tmRNA in the cell
We compared the expression level of tmRNA between the SmpB-depleted cells (ΔsmpBΔssrA) and the SmpB-containing cells (ΔssrA). A pMW119-based low copy number plasmid carrying the tmRNA gene under the native promoter sequence was transformed into these two strains. Northern blot analysis confirmed that the expression level of tmRNA in the ΔssrA cells was comparable to that in the cell of parental strain (data not shown). In contrast, the amount of tmRNA in the ΔsmpBΔssrA cells reduced ∼4-fold (Fig. 5A and B). As a control, 5S ribosomal RNA was tested, and no significant difference in the amount between the two strains was observed (Fig. 5E).
Figure 5.
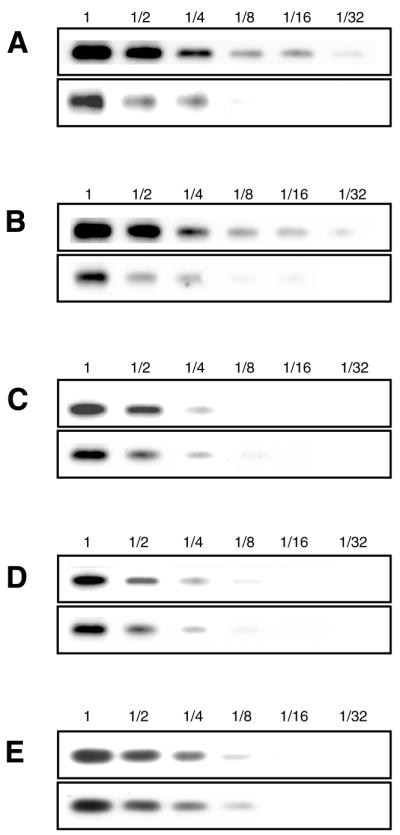
The expression levels of tmRNA in the cells of ΔssrA (top) and ΔsmpBΔssrA (bottom) strains carrying the low-copy number plasmid-encoded (A) wild-type tmRNA, (B) 3A, (C) 19C and (D) 334U mutants. (E) The expression levels of 5S ribosomal RNA in the cells of two strains carrying the low-copy number plasmid-encoded wild-type tmRNA. Varying dilutions of total RNA from the cell extract of each strain were electrophoresed on a 1.5% agarose gel containing 6.3% formaldehyde, and were then blotted onto a nylon membrane. tmRNA was detected by northern hybridization using a 3′-digoxygenin-labeled oligodeoxyribonucleotide. No degradation products of the tmRNA molecule were detected in the lower part of the gel irrespective of the presence or absence of SmpB. Presumably the primary degradation induces further degradation in the cell.
Next we examined the effect of depletion of SmpB on the expression levels of tmRNA mutants 3A, 19C and 334U. We prepared pMW119-based low copy number plasmids each carrying tmRNA mutant gene under the native promoter sequence, and then transformed them into ΔsmpBΔssrA and ΔssrA cells. The difference between the two strains was similarly observed for 3A mutant (Fig. 5B), which has no aminoacylation ability (18). The expression level of 19C or 334U mutants in the presence of SmpB was apparently lower than that of wild-type or 3A mutant (Fig. 5A–D). Furthermore, the levels of these two mutant tmRNAs were not affected by SmpB, in contrast to the wild-type and 3A tmRNAs (Fig. 5A–D). Thus, the difference in expression levels between the two strains was observed only in the tmRNA mutants capable of binding SmpB. The results strongly suggest that the observed effect is not a result of a regulation at the transcription level and that SmpB functions to rescue tmRNA from degradation by nucleases via binding to tmRNA in the cell.
DISCUSSION
Karzai et al. (31) have shown that SmpB is a tmRNA binding protein that has been identified to be essential for trans-translation in vivo, and its depletion affects the binding of tmRNA to ribosome. Their overproduced SmpB has a high affinity to tmRNA, although with no information about the trans-translation activity. In the present study, we made an SmpB- and tmRNA-depleted strain ΔsmpBΔssrA. Using this strain, we first confirmed that SmpB is essential for trans-translation, but not for canonical translation in vitro. The strain ΔsmpBΔssrA in addition to another tmRNA-depleted strain ΔssrA allowed us to evaluate the trans-translation activity of SmpB in vitro as well as the effect of SmpB in combination with the mutation of tmRNA in vivo and in vitro. Using this system, we confirmed that the overproduced His-tagged SmpB is active in both tmRNA binding and trans-translation. This active SmpB considerably enhanced the aminoacylation activity of tmRNA. It was also found that the expression level of tmRNA is significantly reduced in the SmpB-depleted cell. In addition, the present in vitro study supports an earlier in vivo study (31) indicating that SmpB has a role to facilitate the binding to the ribosome. SmpB might be a direct mediator to recruit tmRNA into the ribosome. Alternatively, SmpB only induces a conformation change of tmRNA so that tmRNA can enter the ribosome.
Here, three functions of SmpB have been established: SmpB rescues the tmRNA molecule from degradation in the cell, enhances aminoacylation by alanyl-tRNA synthetase and facilitates the binding of tmRNA to the ribosome. All of these functions were found to be affected by some single-point mutations in the tRNA-like domain of tmRNA which also affect the binding of SmpB to tmRNA (Table 1). Conversely, the binding of SmpB was still observed in any mutant in which SmpB protects from degradation in the cell or enhances aminoacylation. This clearly indicates that these functions are mediated by the binding of SmpB to the lower portion of the tRNA-like domain of tmRNA.
Table 1. The effects of mutations on aminoacylation, alanine incorporation, SmpB binding stabilization of tmRNA by SmpB in the cell and ribosome binding capacity.
| Aminoacylationa | Alanine incorporationb | SmpB bindingh | Stabilization of tmRNA by SmpB in the celli | Ribosome binding capacity | ||
|---|---|---|---|---|---|---|
| – SmpB | + SmpB (4 µM) | |||||
| Wild-type tmRNA | 1 | 4.15 | 1 | + | + | + |
| 3A | <0.01 | <0.01 | <0.01c | + | + | –c |
| 19C | 0.94 | 1.0 | <0.01d | – | – | –c |
| 334U | 1.0 | 1.1 | <0.01d | – | – | –d |
| 62C | 1.7 | 4.9 | 0.43e | + | ndj | ndj |
| 86C | 1.1 | 4.1 | 0.05f | + | ndj | +f |
| PK1L | 0.43 | 2.1 | 0.06g | + | ndj | ndj |
| PK2L | 0.91 | 3.6 | 0.36g | + | ndj | ndj |
| PK3L | 1.1 | 4.2 | 0.76g | + | ndj | ndj |
| PK4L | 1.1 | 4.3 | 0.90g | + | ndj | ndj |
aThe values relative to that for wild-type tmRNA are those after 4 min incubation in the absence of SmpB using 8.5 × 10–3 U of alanyl-tRNA synthetase in Figure 4.
bThe values are those after 12 min incubation relative to that for wild-type tmRNA.
cData are from Himeno et al. (18).
dData are from Hanawa-Suetsugu et al. (36).
eData are from Nameki et al. (29).
fData are from Lee et al. (28).
gData are from Nameki et al. (37).
hData are from Figure 3.
iData are from Figure 5.
jNot determined.
Earlier studies have shown that the aminoacylation capacity of tmRNA is significantly poor as compared with that of tRNAAla (13,24). The present study demonstrates that pre-aminoacylated tmRNA is not optimal for recognition by alanyl-tRNA synthetase and that SmpB has a high affinity to pre-aminoacylated tmRNA to elevate the aminoacylation efficiency to a considerable extent. A large population of tmRNA in the cell may form a complex with SmpB prior to aminoacylation (Fig. 6): this is supported by the present findings that the non-aminoacylatable 3A mutant can form a complex with SmpB and that it is protected from degradation by SmpB in the cell to an extent comparable with the case for wild-type tmRNA. SmpB may interact with the tRNA-like region of non-aminoacylated tmRNA to modulate the conformation of the tRNA-like domain more suitable for recognition by alanyl-tRNA synthetase (Fig. 6). An earlier study has shown that His-tagged SmpB has no effect on aminoacylation of the transcript of tmRNA (31), which is in sharp contrast with the present results. We have no idea whether this discrepancy can be attributed to the fact that their SmpB was purified from an inclusion body via a procedure of denaturation by 3 M guanidium hydrochloride. We can at least rule out the possibility that the observed aminoacylation enhancement in vitro was due to the protection of tmRNA by SmpB from degradation by nucleases in the alanyl-tRNA synthetase of our preparation.
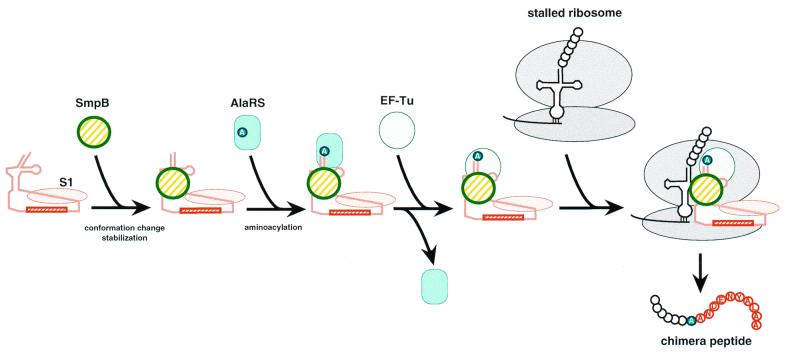
Figure 6.
A possible pathway of the tmRNA complex formation for trans-translation. Judging from the dissociation constant with tmRNA of 10 nM or lower (36,44), the association of S1 might occur in the early stage of the complex formation. This model adopts a likely complex of SmpB, EF-Tu, S1 and tmRNA as a complex just prior to entrance to the stalled ribosome, as mentioned by Wower et al. (44). Some other possible factors such as phosphoribosyl pyrophosphate synthase, RNase R, YfbG (32) and tRNAAla (46) that have not yet been identified to be essential for trans-translation are not shown here.
The lower half of the tRNA-like domain encompassing 13–21 and 332–335 are highly conserved, and our recent study has shown that several base substitutions in this region affect the trans-translation in vitro (36). In particular, 19C and 334U, single-point mutations at the strictly conserved positions at 19 and 334, respectively, severely damaged the binding to the ribosome leading to a defect in trans-translation, whereas it did not affect aminoacylation by alanyl-tRNA synthetase at all. Note that both positions are in the tRNA-like domain, but apart from the internal coding region. The present study reveals that the defect in the ribosome binding caused by each mutation is attributed to the unsuccessful interaction with SmpB. It has been proposed that G19 and A334 together with A20 and G333 form an AG:GA sheared base pair, a characteristic RNA structure found in hammerhead ribozyme and in ribosomal RNA (36,38). Considering that either mutation at 19 or 334 induces only a minor change of the local conformation of the tmRNA molecule, SmpB may interact with these nucleotides directly or in close proximity of them. These positions are close to the area of interaction with alanyl-tRNA synthetase, which primarily covers the acceptor helix leading to the 3′-CCA terminal (39–41), and is extended to the D-loop (42,43) and the anticodon stem in tRNAAla (43). The present results indicate that SmpB and alanyl-tRNA synthetase bind tmRNA cooperatively rather than in a mutually exclusive manner.
It has recently been shown that SmpB can form a complex with tmRNA and other proteins including a small subunit ribosomal protein S1, phosphoribosyl pyrophosphate synthase, RNase R and YfbG (32). Among them, phosphoribosyl pyrophosphate synthase and RNase R are not essential for trans-translation. S1 can interact with PK2, PK3 and PK4 (44), although S1 is not universal among prokaryotes (45). The present results indicate that SmpB does not bind any of PK1, PK2, PK3 and PK4. Therefore, SmpB and S1 would be capable of binding to a tmRNA molecule simultaneously (Fig. 6), as mentioned in Wower et al. (44).
The 19C or 334U mutation, which seriously affects binding to SmpB (Fig. 3), has no longer ribosome binding capacity (36). In contrast, the 86C mutation 4 nt upstream of the tag-initiation point, which does not affect binding to SmpB (Fig. 3), retains the capability of binding to the ribosome (28). These findings, together with an earlier in vivo (31) and the present in vitro results showing that depletion of SmpB seriously affects the ribosome binding capacity of tmRNA, support the idea that SmpB has a crucial role in recruitment of tmRNA to the ribosome. However, the behavior of SmpB during the ribosomal processes has yet to be characterized. The 86C mutation seriously damages the trans-translation due to some defect in the ribosomal processes (28). U86, together with A87 and G88, has been shown in vivo and in vitro to serve as a core nucleotide to determine the tag-resuming position, and thus it might be a recognition site of some trans-acting factor (27,28). Although the present results indicate that SmpB does not interact with this core nucleotide at least in solution, it is possible that a conformation change of the tmRNA–SmpB complex in the ribosome induces the contact between the core nucleotides and SmpB. Further studies about such a conformation change in the ribosome or another molecule possibly interacting with SmpB as well as the conformation of tmRNA complexed with SmpB are required to rationalize the elaborate interplay between the two apparently distant domains during the ribosomal processes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Nobukazu Nameki, Gota Kawai, Brice Felden and Chie Takemoto for helpful advice on this study, and Miss Katsura Mizushima for technical assistance. We also thank the Gene Research Center of Hirosaki University for the use of the facility. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan, to A.M. and H.H., grants (‘Research for the Future’ Program, JSPS-RFTF96100305 and JSPS-RFTF97L00593) from the Japan Society for the Promotion of Science to A.M. and H.H., a Human Frontier Science Program research grant (RG0291/2000-M 100) to H.H., and Research Fellowships of Japan Society for the Promotion of Science for Young Scientists to K.H.-S.
REFERENCES
- 1.Williams K.P. (2000). The tmRNA website. Nucleic Acids Res., 28, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwieb C. and Wower,J. (2000). (tmRDB) tmRNA database. Nucleic Acids Res., 28, 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The SsrA–SmpB system for protein tagging, directed degradation and ribosome rescue. NatureStruct. Biol., 7, 449–455. [DOI] [PubMed] [Google Scholar]
- 4.Ushida C., Himeno,H., Watanabe,T. and Muto,A. (1994) tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res., 22, 3392–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komine Y., Kitabatake,M., Yokogawa,T., Nishikawa,K. and Inokuchi,H. (1994) A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felden B., Hanawa,K., Atkins,J.F., Himeno,H., Muto,A., Gesteland,R.F., McCloskey,J.A. and Crain,P.F. (1998) Presence and location of modified nucleotides in Escherichia coli tmRNA: structural mimicry with tRNA acceptor branches. EMBO J., 17, 3188–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felden B., Himeno,H., Muto,A., Atkins,J.F. and Gesteland,R.F. (1996) Structural organization of Escherichia coli tmRNA. Biochimie, 78, 979–983. [DOI] [PubMed] [Google Scholar]
- 8.Felden B., Himeno,H., Muto,A., McCutcheon,J.P., Atkins,J.F. and Gesteland,R.F. (1997) Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA, 3, 89–104. [PMC free article] [PubMed] [Google Scholar]
- 9.Williams K.P. and Bartel,D.P. (1996) Phylogenetic analysis of tmRNA secondary structures. RNA, 2, 1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 10.Hickerson R., Watkins-Sims,C.D., Burrows,C.J., Atkins,J.F., Gesteland,R.F. and Felden,B. (1998) A nickel complex cleaves uridines in folded RNA structures: application to E. coli tmRNA and related engineered molecules. J. Mol. Biol., 279, 577–587. [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Pandit,S. and Deutscher,M.P. (1998) 3′ Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudinger-Thirion J., Giegé,R. and Felden,B. (1999) Aminoacylated tmRNA from Escherichia coli interacts with procaryotic elongation factor Tu. RNA, 5, 989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barends S., Wower,J. and Kraal,B. (2000) Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry, 39, 2652–2658. [DOI] [PubMed] [Google Scholar]
- 14.Tu G.-F., Reid,G.E., Zhang,J.-G., Moritz,R.L. and Simpson,R.J. (1995) C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem., 270, 9322–9326. [DOI] [PubMed] [Google Scholar]
- 15.Keiler K.C., Waller,P.R.H. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of protein synthesized from messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- 16.Roche E.D. and Sauer,R.T. (1999) SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J., 18, 4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abo T., Inada,T., Ogawa,K. and Aiba,H. (2000) SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J., 19, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himeno H., Sato,M., Tadaki,T., Fukushima,M., Ushida,C. and Muto,A. (1997) In vitro trans translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol., 268, 803–808. [DOI] [PubMed] [Google Scholar]
- 19.Roche E.D. and Sauer,R.T. (2001) Identification of endogenous SsrA-tagging proteins reveals tagging at positions corresponding to stop codons. J. Biol. Chem., 276, 28509–28515. [DOI] [PubMed] [Google Scholar]
- 20.Muto A., Sato,M., Tadaki,T., Fukushima,M., Ushida,C. and Himeno,H. (1996) Structure and function of bacterial 10Sa RNA: trans-translation system. Biochimie, 78, 985–991. [DOI] [PubMed] [Google Scholar]
- 21.Muto A., Ushida,C. and Himeno,H. (1998) A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci., 23, 25–29. [DOI] [PubMed] [Google Scholar]
- 22.Huang C., Wolfgang,M.C., Withey,J., Koomey,M. and Friedman,D.I. (2000) Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J., 19, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muto A., Fujihara,A., Ito,K., Matsuno,J., Ushida C. and Himeno,H. (2000). Requirement of transfer-messenger RNA (tmRNA) for the growth of Bacillus subtilis under stresses. Genes Cells, 5, 627–636. [DOI] [PubMed] [Google Scholar]
- 24.Nameki N., Tadaki,T., Muto,A. and Himeno,H. (1999) Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. J. Mol. Biol., 289, 1–7. [DOI] [PubMed] [Google Scholar]
- 25.Komine Y., Kitabatake,M. and Inokuchi,H. (1996) 10Sa RNA is associated with 70S ribosome particles in Escherichia coli. J. Biochem., 119, 463–467. [DOI] [PubMed] [Google Scholar]
- 26.Tadaki T., Fukushima,M., Ushida,C., Himeno,H. and Muto,A. (1996) Interaction of 10Sa RNA with ribosomes in Escherichia coli. FEBS Lett., 399, 223–226. [DOI] [PubMed] [Google Scholar]
- 27.Williams K.P., Martindale,K.A. and Bartel,D.P. (1999) Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J., 18, 5423–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S., Ishii,M., Tadaki,T., Muto,A. and Himeno,H. (2001) Determinants on tmRNA for initiating efficient and precise trans-translation: Some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA, 7, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nameki N., Felden,B., Atkins,J.F., Gesteland,R.F., Himeno,H. and Muto,A. (1999) Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J. Mol. Biol., 286, 733–744. [DOI] [PubMed] [Google Scholar]
- 30.Nameki N., Chattopadhyay,P., Himeno,H., Muto,A. and Kawai,G. (1999) An NMR and mutational analysis of an RNA pseudoknot of E. coli tmRNA involved in trans-translation. Nucleic Acids Res., 27, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karzai A.W., Susskind,M.M. and Sauer,R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karzai A.W. and Sauer,R.T. (2001) Protein factors associated with the SsrA-SmpB tagging and ribosome rescue complex. Proc. Natl Acad. Sci. USA, 98, 3040–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohara Y., Akiyama,K. and Isono,K. (1987) The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell, 50, 495–508. [DOI] [PubMed] [Google Scholar]
- 34.Tsurui H., Kumazawa,Y., Sanokawa,R., Watanabe,Y., Kuroda,T., Wada,A., Watanabe,K. and Shirai,T. (1994) Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem., 221, 166–172. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Asahara,H., Himeno,H., Hasegawa,T. and Shimizu,M. (1991) Identity determinants of Escherichia coli tRNAAla. J. Mol. Recognit., 4, 129–132. [DOI] [PubMed] [Google Scholar]
- 36.Hanawa-Suetsugu K., Bordeau,V., Himeno,H., Muto,A. and Felden,B. (2001) Importance of the conserved nucleotides around the tRNA-like structure of Escherichia coli transfer-messenger RNA for protein tagging. Nucleic Acids Res., 29, 4663–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nameki N., Tadaki,T., Himeno,H. and Muto,A. (2000) Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett., 470, 345–349. [DOI] [PubMed] [Google Scholar]
- 38.Zwieb C., Guven,S.A., Wower,I.K. and Wower,J. (2001) Three-dimensional folding of the tRNA-like domain of Escherichia coli tmRNA. Biochemistry, 40, 9587–9595. [DOI] [PubMed] [Google Scholar]
- 39.McClain W.H. and Foss,K. (1988) Changing the identity of a tRNA by introducing a G–U wobble pair near the 3′ acceptor end. Science, 240, 793–796. [DOI] [PubMed] [Google Scholar]
- 40.Hou Y.-M. and Schimmel,P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature, 333, 140–145. [DOI] [PubMed] [Google Scholar]
- 41.Shi J.-P. and Schimmel,P. (1991) Aminoacylation of alanine minihelices. ‘Discriminator’ base modulates transition state of single turnover reaction. J. Biol. Chem., 266, 2705–2708. [PubMed] [Google Scholar]
- 42.Pleiss J.A., Wolfson,A.D. and Ulenbeck,O.C. (2000) Mapping contacts between Escherichia coli alanyl-tRNA synthetase and 2′ hydroxyls using a complete tRNA molecule. Biochemistry, 39, 8250–8258. [DOI] [PubMed] [Google Scholar]
- 43.Park S.J. and Schimmel,P. (1988) Evidence for interaction of an aminoacyl transfer RNA synthetase with a region important for the identity of its cognate transfer RNA. J. Biol. Chem., 263, 16527–16530. [PubMed] [Google Scholar]
- 44.Wower J., Zwieb,C., Guven,S.A. and Wower,I. (2000) Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J., 19, 6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert M.W. and Rabinowitz,J.C. (1989) The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J. Biol. Chem., 264, 2228–2235. [PubMed] [Google Scholar]
- 46.Gillet R. and Felden,B. (2001) Transfer RNAAla recognizes transfer-messenger RNA with specificity; a functional complex prior to entering the ribosome? EMBO J., 20, 2966–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]