Abstract
Herpesvirus DNA packaging involves binding and cleavage of DNA containing the specific DNA-packaging motifs. Here we report a first characterization of the terminase subunits pUL56 and pUL89 of human cytomegalovirus (HCMV). Both gene products were shown to have comparable nuclease activities in vitro. Under limiting protein concentrations the nuclease activity is enhanced by interaction of pUL56 and pUL89. High amounts of 2-bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole partially inhibited the pUL89-associated nuclease activity. It was demonstrated that pUL56 is able to bind to nucleocapsids in vivo. Electron microscopy (EM) and image analysis of purified pUL56 revealed that the molecules occurred as a distinct ring-shaped structure with a pronounced cleft. EM analysis of purified pUL89 demonstrated that this protein is also a toroidal DNA-metabolizing protein. Upon interaction of pUL56 with linearized DNA, the DNA remains uncut while the cutting event itself is mediated by pUL89. Using biochemical assays in conjunction with EM pUL56 was shown to (i) bind to DNA and (ii) associate with the capsid. In contrast to this, EM analysis implied that pUL89 is required to effect DNA cleavage. The data provide the first insights into the terminase-dependent viral DNA-packaging mechanism of HCMV.
INTRODUCTION
Herpesvirus DNA replication results in the formation of large head-to-tail DNA concatemers, and subsequent maturation into unit length molecules involves site-specific cleavage at sequence motifs (pac motifs) located within the a sequence (1–3). Unit length DNA is then encapsidated into preformed capsids. Herpesviruses share common features with double-stranded (ds)DNA bacteriophages, such as DNA replication into high molecular mass concatenated DNA and procapsid assembly around a scaffold (4,5). Initiation signals for DNA packaging as well as termination sequences vary in different phages. One group, e.g. phage λ and T7, initiates and terminates packaging at specific sites called cos or pac sites (6–8). However, phages P1, P22 and SPP1 initiate packaging at specific sites (pac) but termination is achieved by a ‘headful packaging’ mechanism, in which cleavage occurs as soon as a certain amount of DNA has entered the preformed capsid. Two different models have been proposed for the initiation of DNA packaging: one proposes a non-specific initiation point at partially single-stranded regions together with ‘headful packaging’ of 102–103% of the genome (9) while the other model (in T4) favors initiation by a gene 16-dependent mechanism at a specific pac site near the end of gene 16 (10). Initiation as well as termination of DNA packaging requires the introduction of endonuclease-induced dsDNA cuts. The generation of unit length genomes during DNA packaging is catalyzed by a class of proteins known as terminases, which catalyze the ATP-dependent translocation of genomic DNA into the bacteriophage procapsid and bind and cleave concatenated DNA (4,11). Most bacteriophage terminases are hetero-oligomers with each subunit carrying a different function (12–15). Mutations in any of the encoding genes lead to the accumulation of empty procapsids (proheads) and DNA concatemers (4).
Recently it was demonstrated that the human cytomegalovirus (HCMV) pUL56 gene product (pUL56), the homolog of the herpesvirus simplex type-1 (HSV-1) ORF UL28, is associated with specific binding of DNA containing packaging motifs, leading to the suggestion that pUL56 plays a key role in DNA packaging (16–18). Comparable observations were reported for the HSV-1 homolog of pUL56 (UL28), demonstrating a direct interaction of pUL28 with DNA containing the pac1 motif (19).
Here we present the characterization of the function of the purified terminase subunits pUL56 and pUL89. Furthermore, we show that pUL56 is associated with capsids. Electron microscopic analysis of pUL56 and pUL89 established a toroidal architecture, commonly found for other DNA-metabolizing proteins (20). Nuclease activity together with EM analysis demonstrated that pUL89 is necessary for the cleavage of viral DNA.
MATERIALS AND METHODS
Cells and viruses
5B1-4 (High Five) or SF9 insect cells were grown in TC-100 medium supplemented with 10% fetal calf serum, glutamine and gentamicin (60 µg/ml) as described previously (21). For generation of recombinant baculovirus, transposition into the bacmid bMON14272 was used, as described by Luckow et al. (22).
Plasmid construction and oligonucleotides
Restriction enzymes were purchased from New England Biolabs (Beverly, MA) and used as instructed by the manufacturer. The UL89 ORF was generated by cDNA construction and PCR amplification. PCR primers amplified DNA segments from –17 to + 2306 bp of the HCMV (strain AD169) UL89 ORF sequence except that a single nucleotide 5′ of the A of the initiation codon was deleted to generate a NdeI site. This was cloned into the pCRII vector (Invitrogen) to generate construct pUB8. A BamHI–XbaI PCR fragment from pUB8 was cloned into the same sites of the pcDNA3.1/His C vector (Invitrogen) to generate pcDNA-UL89. A EcoRI–XbaI PCR fragment from pcDNA-UL89 containing the Anti-Xpress antibody epitope was inserted into the baculovirus vector pFastBac HTa (Life Technologies) to generate pBac-UL89. Recombinant baculovirus expressing pUL89 was generated by site-specific transposition of the expression cassette into the bacmid bMON14272, as described by Life Technology.
Protein purification
High Five cells (7 × 107) expressing recombinant pUL89 or pUL56 were harvested 48 h post-infection. Purification protocols were identical for both proteins. Sedimented cells were lysed in 4 ml of harvest buffer (10 mM Tris–HCl pH 7.4, 50 mM NaCl and protease inhibitors) and sonicated on ice. Undissolved material was sedimented at 5000 g at 4°C and passed through a 0.2 µm filter. Purification was performed in two steps using an Äkta FPLC column (Amersham Pharmacia Biotech) at 4°C. The first step entailed anion exchange with a 6 ml Resource Q column (Amersham Pharmacia Biotech). The column was washed with three bed volumes of harvest buffer prior to loading the proteins. Elution was achieved using a linear salt gradient of 50 mM–1 M NaCl. Twenty fractions were collected and analyzed by SDS–PAGE. The second chromatography step was carried out on a HiPrep 16/60 Sephacryl S-300 HR gel permeation column (Amersham Pharmacia Biotech). After equilibrating the column with three bed volumes of 20 mM HEPES, pH 7.4, 150 mM NaCl, the pooled fractions from the previous step were loaded. Forty fractions were collected prior to analysis by SDS–PAGE. Interactions between the proteins and the column matrix prevent molecular mass determination, because they skew the elution profile towards smaller apparent Stokes radii. Those fractions containing the purified protein were pooled and spin concentrated on a PES membrane (Spin-MICRO) with a 30 kDa cut-off (membraPure, Bodenheim, Germany), aliquoted and stored at –80°C.
In vitro translation
Plasmids pcDNA-UL89 (0.5 µg) and pcDNA-UL56 (0.5 µg) were incubated with [35S]methionine (10 mCi/ml) and 20 µl of TNT T7 Quick Master Mix (Promega, Madison, WI) in a final volume of 30 µl for 1 h at 30°C. Translation products were analyzed by SDS–PAGE.
PAGE and western blot analysis
High Five cells were infected at a multiplicity of infection of 2 with either wild-type virus or recombinant baculovirus pUL89 (rpUL89). The cells were harvested 48 h after infection, sonicated and centrifuged at 2000 g for 5 min. The supernatants were separated on 8% (w/v) polyacrylamide gels, transferred to nitrocellulose sheets and subjected to western blot analysis as described previously (16). The primary antibody used was the Anti-Xpress antibody (1:1000; Invitrogen) specific for rpUL89. With the separated capsid/tegument fractions, affinity-purified human pAbUL56 (1:10), mAb58-15 (1:10) specific for the major tegument protein pp65 and mAb28-4 specific for the major capsid protein (MCP) (1:10), respectively, served as primary antibodies.
Nuclease activity
Supercoiled plasmid DNA containing the a sequence between the L-S junction (pUC-aseq) or without the a sequence (pBR322) was amplified in Escherichia coli XL1blue and purified by Qiagen midipreparation (Qiagen, Hilden, Germany) as specified by the supplier. Extracts from purified proteins (3 or 1.2 µg/ml) were incubated with 1 µg plasmid DNA in a final volume of 50 µl in nuclease buffer (10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 50 mM NaCl) for 1 h at 37°C. The samples were incubated with proteinase K (final concentration 1 µg/µl) for a further 1 h at 37°C. DNA was fractionated by electrophoresis in a 1% agarose gel (17).
Purification of HCMV capsids
Seventy-two hours after infection extracellular virions were separated from dense bodies and non-infectious particles by sedimentation through a sodium tartrate gradient according to Talbot and Almeida (23). Isolated virions were incubated with non-ionic detergent (1% v/v NP-40) for 10 min on ice prior to separation through a 15% (w/v) sucrose cushion in TE buffer. The sediment, which constitutes the capsid/tegument fraction, was treated with 1% (v/v) β-mercaptoethanol for 10 min on ice. Following this treatment, the supernatant contains the tegument and the sediment the capsid fraction. The capsid fraction was subjected to SDS–PAGE prior to immunostaining.
Electron microscopy and image processing
Negatively stained specimens were prepared essentially according to Valentine et al. (24) using an aqueous solution of uranyl acetate (4% w/v, pH 4.3) and omitting any fixation steps. Specimens were mounted on 400 mesh copper grids and observed in a Zeiss EM T109 electron microscope operated at an acceleration voltage of 80 kV. Electron micrographs were recorded at a calibrated magnification (29 600×). Selected micrographs were digitized using a Microtex Scan Maker E6. Scanning increments were adjusted so that the final scan step corresponded to 4.76 Å/pixel at the specimen level. Single particle analysis was carried out using the image analysis software programs of IMAGIC-5 (25) and EMAN (26) running on a Pentium PC under Linux.
DNA–protein complexes were prepared by incubating the purified proteins (pUL56 and pUL89, 1–2 µg) with linearized DNA (0.2 µg) containing the a sequence (30 min at 30°C) in a final volume of 50 µl of 20 mM HEPES buffer, pH 7.4. The formed complexes were fixed with glutaraldehyde by adding 1.25 µl of an 8% (v/v) solution made up fresh in distilled water and incubation for 10 min. Specimens were mounted onto freshly glow-discharged carbon-coated copper grids (G400) and incubated for 60 s prior to staining with uranyl acetate (see above) for 60 s and blotting dry.
For immunoelectron microscopy purified capsids were labeled with pAbUL56 (16,27). Unambiguous detection of pAbUL56 was achieved using protein A–gold. Stability tests at high ionic strength (28) ensured that the gold particles had been quantitatively covered by protein A, thus minimizing the possibility of non-specific binding. The samples were subjected to labeling prior to fixation with 0.1% (v/v) glutaraldehyde and negative staining using 4% (w/v) uranyl acetate following published procedures (28).
Thin sections
HFF cells were infected with HCMV at a multiplicity of infection of 4 and harvested in Dulbecco’s modified Eagle’s medium (DMEM) 72 h post-infection. Cells were fixed with DMEM containing 4% paraformaldehyde, dehydrated, post-stained with 0.2% (w/v) uranyl acetate and embedded in LR-Gold and 0.1% (w/v) benzoylperoxide. Polymerization was performed at –20°C under UV irradiation. Immunoelectron microscopy was performed according to Kolesnikova et al. (29).
RESULTS
Purification of pUL56 and pUL89
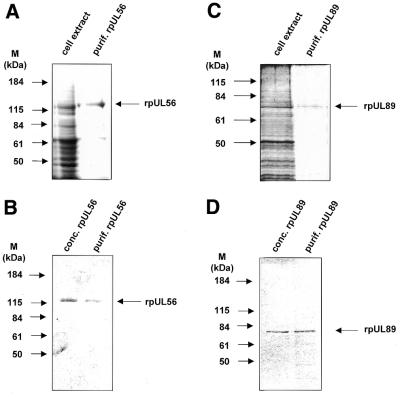
Recombinant baculovirus pUL89- or pUL56-infected cell extracts were harvested and subjected to anion exchange chromatography. Aliquots of every fraction were assayed for protein content by SDS–PAGE. Fractions containing rpUL56 or rpUL89 were pooled and loaded onto a gel permeation chromatography column. Both proteins were essentially homogeneous after this second purification step (Fig. 1A and C) and ready for further analysis after spin concentration (Fig. 1B and D).
Figure 1.
SDS–PAGE of cell lysates and purified proteins. (A) Aliquots of extracts from High Five cells infected with baculovirus UL56 (cell extract) and rpUL56 obtained after a two-step purification using anion exchange followed by gel permeation chromatography (purif. rpUL56) were subjected to SDS–PAGE prior to staining with silver. (B) Aliquots after purification (purif. rpUL56) and following spin concentration (conc. rpUl56) were subjected to SDS–PAGE prior to staining with Coomassie brilliant blue. (C) Aliquots of extracts from High Five cells infected with baculovirus UL89 (cell extract) and rpUL89 obtained after two-step purification using anion exchange followed by gel permeation chromatography (purif. rpUL89) were subjected to SDS–PAGE prior to staining with silver. (D) Aliquots after purification (purif. rpUL89) and following spin concentration (conc. rpUL89) were subjected to SDS–PAGE prior to staining with Coomassie brilliant blue. Molecular weight markers (M) are indicated on the left; the positions of the proteins are indicated on the right.
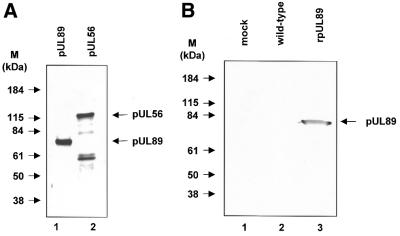
Identification of in vitro translated pUL89 and recombinant baculovirus-expressed epitopically tagged rpUL89
Products of in vitro translation of pcDNA-UL89 were subjected to SDS–PAGE prior to visualization by autoradiography. Only one specific protein was observed after in vitro translation of UL89 (Fig. 2A). Mock- and wild-type baculovirus-infected cells and purified rpUL89 were separated on a denaturing polyacrylamide gel followed by transfer to nitrocellulose. Nitrocellulose filters reacted with the Anti-Xpress antibody detected only a protein with an Mr of ∼70 000–75 000 (Fig. 2B, lane 3). No staining was observed with mock- or wild-type baculovirus-infected cells (Fig. 2B, lanes 1 and 2). Since the molecular mass of the detected protein corresponded to the expected size of the amino acid sequence, we concluded that the 75 000 Mr protein is the product of the UL89 gene.
Figure 2.
Identification of HCMV pUL89 in vitro translated or synthesized in insect cells infected with recombinant baculovirus UL89 or baculoviurs UL56. (A) Aliquots of pUL89 in vitro translation products were subjected to SDS–PAGE (8% polyacrylamide) prior to detection by autoradiography. (B) Aliquots of extracts from mock-transfected High Five cells (lane 1) or High Five cells infected with wild-type baculovirus (lane 2) or purified recombinant rpUL89 (lane 3) were separated by SDS–PAGE followed by immunoblotting with the specific anti-Xpress antibody against Xpress-tagged pUL89. Molecular weight standards (M) are indicated on the left; the positions of pUL89 and pUL56 are indicated by arrows.
Association of pUL56 with capsids
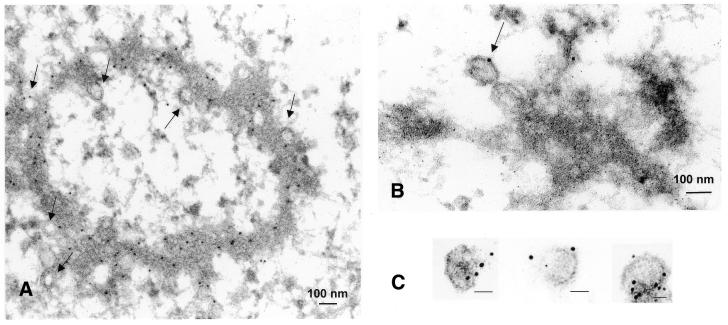
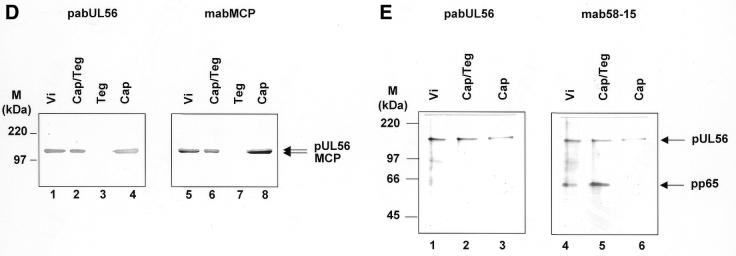
For immunoelectron microscopy of thin sections an affinity-purified human antibody against pUL56 (pAbUL56) (27) and the pUL44-specific antibody RG1202 (Rumbaugh-Goodwin Institute for Cancer Research) and secondary antibodies conjugated with colloidal gold were used. In the case of purified capsids only pAbUL56 was used. Immunostaining of ultrathin sections of infected cells showed an electron-dense material, the viral DNA replication centers, which were heavily labeled with gold particles (Fig. 3A). Labeling was also observed on some forming capsids, demonstrating an association of pUL56 with capsids (Fig. 3A and B). Furthermore, background staining of uninfected cells was not detectable (data not shown). Electron microscopic observation of purified capsids revealed that the protein A–gold particles were almost exclusively found attached to capsids (Fig. 3C). Purification of the capsids was analyzed by immunoblots with antibodies against capsid or tegument proteins (Fig. 3D and E). The pUL56 polypeptide separated with the nucleocapsid fraction. The control protein, pp65, the major tegument protein, was detected in every fraction that contained tegument (Fig. 3E), while the MCP served as the indicator for purification of the capsid fraction (Fig. 3D). These data strongly suggest that HCMV pUL56 is closely associated with the nucleocapsid.
Figure 3.
Association of pUL56 with viral capsids. (A) Immunoelectron microscopy (IEM) analysis of an ultrathin section of HCMV-infected HFF cells. Immunogold labeling was carried out using pAbUL56 (dilution 1:10) and a goat anti-human antibody conjugated with colloidal gold (bead diameter 5 nm, dilution 1:50). (B) IEM analysis with double labeling was performed with (i) pAbUL56 and goat anti-human antibody conjugated with colloidal gold (bead diameter 10 nm) and (ii) mAb against pUL44 (dilution 1:10) and a goat anti-mouse antibody conjugated with colloidal gold (bead diameter 5 nm). (C) Immunogold labeling of purified capsids. Capsids were labeled with pAbUL56 and protein A conjugated with 5 nm colloidal gold. Arrows indicated capsids. The scale bars represent 100 nm. (D and E) Extracellular virions (vi) were separated into nucleocapsid/tegument (Cap/Teg), tegument (Teg) and nucleocapsid (Cap) components and analyzed by immunoblotting with pAbUL56 and an antibody against MCP (D) or with pAbUL56 and an antibody against pp65 (E). The arrows indicate the positions of pUL56, MCP and pp65 and the molecular weight standards (M) are indicated on the left.
Nuclease activity of rpUL89
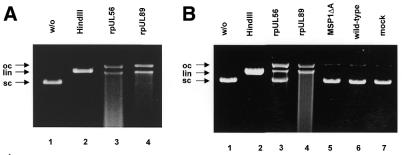
In order to determine whether HCMV pUL89 has the capacity to cleave DNA containing the packaging motifs, purified rpUL89 or rpUL56 (3.0 µg/ml) was incubated with circular plasmid DNA (pUC-aseq) bearing a single a sequence or one without an a sequence (pBR322) as substrate. Separation by agarose gel electrophoresis showed that plasmid DNA molecules incubated in the absence of protein were present as a fast migrating supercoiled (sc) form (Fig. 4, lane 1). After treatment with the restriction enzyme HindIII the fast migrating forms were converted to linear (lin) molecules (Fig. 4, lane 2). In the presence of extracts containing rpUL56, sc plasmid DNA was converted to open circular (oc) as well as linear (lin) molecules (Fig. 4A, lane 3). The same observation was made with purified rpUL56 (Fig. 4A, lane 3). Incubation of circular plasmid DNA in the presence of rpUL89 also led to the formation of oc and lin molecules (Fig. 4A, lane 4). As terminases are able to cleave non-specifically in vitro, experiments were carried out using circular plasmid pBR322 devoid of pac sites, confirming that both proteins have a nuclease activity when non-specific DNA is used (Fig. 4B, lanes 3 and 4). A control protein (the purified Toxoplasma gondii protein MSP1ΔA) does not have nuclease activity (Fig. 4B, lane 5). Taken together, these results indicate that HCMV pUL89 has the capacity to cleave DNA in vitro and that this enzymatic activity is, in vitro, a random reaction.
Figure 4.
Nuclease activity of HCMV rpUL56 and rpUL89. (A) Nuclease assay with purified proteins. Lane 1, pUC-aseq; lane 2, incubation in the presence of HindIII; lane 3, pUC-aseq incubated with purified rpUL56; lane 4, pUC-aseq incubated with purified rpUL89. (B) Nuclease assay with pBR322 as substrate. Lane 1, plasmid pBR322 in the absence of protein; lane 2, pBR322 with the restriction endonuclease HindIII; lane 3, pBR322 incubated with purified rpUL56; lane 4, pBR322 incubated with purified rpUL89; lane 5, incubation with purified MSP1ΔA; lane 6, pBR322 incubated with wild-type baculovirus-infected extracts; lane 7, pBR322 incubated in the presence of mock-infected extracts. The arrows indicate three different plasmid DNA forms: open circular molecules (oc), linear forms (lin) and supercoiled DNA (sc).
Influence of 2-bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (BDCRB) on nuclease activity
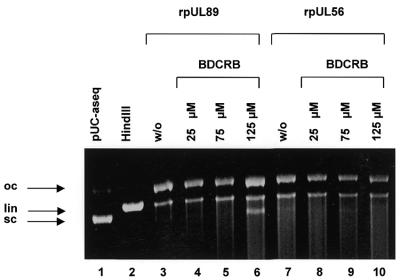
Experiments in the presence of BDCRB were carried out to reveal whether this drug targeted to pUL89 has an effect on cleavage of DNA. The results shown in Figure 5 demonstrate that even at high concentrations, BDCRB had no effect on the enzyme activity of rpUL56 (Fig. 5, lanes 8–10). However, the nuclease activity of rpUL89 was reduced in the presence of 125 µM BDCRB, as indicated by the appearance of sc DNA (Fig. 5, lane 4). In contrast, high amounts of ganciclovir had no effect on the enzyme reaction (data not shown). These results suggest that high concentrations of BDCRB could reduce the nuclease activity of rpUL89.
Figure 5.
Influence of BDCRB on nuclease activity. Lane 1, pUC-aseq alone; lane 2, pUC-aseq treated with HindIII; lane 3, pUC-aseq plus purified rpUL56; lane 4, pUC-aseq incubated with rpUL89 and 25 µM BDCRB; lane 5, pUC-aseq incubated with rpUL89 and 75 µM BDCRB; lane 6, pUC-aseq incubated with rpUL89 and 125 µM BDCRB; lane 7, incubation with rpUL56; lane 8, incubation with rpUL56 and 25 µm BDCRB; lane 9, incubation with rpUL56 and 75 µM BDCRB; lane 10, incubation with rpUL56 and 125 µM BDCRB. The arrows indicate three different plasmid forms: open circular (oc), linear (lin) and supercoiled molecules (sc).
rpUL56 could enhance rpUL89 nuclease activity
To further investigate whether rpUL56 could enhance the nuclease activity of rpUL89, experiments were performed under limiting concentrations (1.2 µg/ml). In the presence of a limited amount of purified rpUL56 (1.2 µg/ml) or rpUL89 (1.2 µg/ml), only ∼30 and 60% of ccc plasmid DNA was converted to oc as well as lin molecules (Fig. 6, lanes 3 and 4). However, incubation with both proteins resulted in an enhancement of ccc cleavage (Fig. 6, lane 5). This finding implies that the enhancement of nuclease activity in vitro is based on an interaction of the two proteins.
Figure 6.

Nuclease activity with limited amounts of rpUL56 and rpUL89. Nuclease assay with purified proteins. Lane 1, plasmid pUC-aseq in the absence of protein; lane 2, pUC-aseq treated with the restriction endonuclease HindIII; lane 3, pUC-aseq incubated with rpUL56 (1.2 µg/ml); lane 4, pUC-aseq incubated with rpUL89 (1.2 µg/ml); lane 5, pUC-aseq incubated with rpUL56 and rpUL89 (1.2 µg/ml each). The arrows indicate three different plasmid DNA forms: open circular molecules (oc), linear forms (lin) and supercoiled DNA (sc).
rpUL56 has a toroidal structure
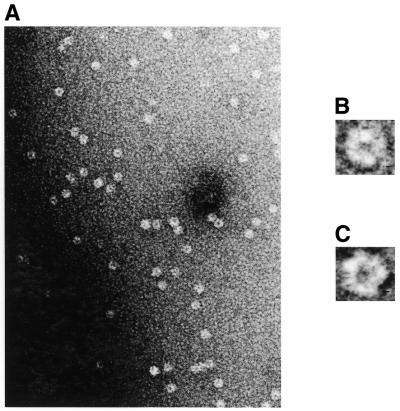
In order to obtain the first direct structural data for purified pUL56, electron microscopic studies were carried out in conjunction with digital image analysis (Fig. 7). The overview presented in Figure 7A shows that the protein solution is homogeneous and monodisperse, with very little tendency for aggregation. All protein molecules display a ring-shaped structure clearly discernible prior to image processing. Images of single pUL56 molecules such as those shown in Figure 7A were subjected to multivariate statistical analysis together with hierarchical ascendant classification, revealing two major projections, i.e. >95% of the molecules adopted one of two orientations relative to the support film (Fig. 7B and C), either of them corresponding to what we defined as face-on views. However, while face-on view 2 shows a distinct stain-filled cleft originating at the central torus and opening into the surrounding medium, view 1 does not. Furthermore, the diameter of the torus is slightly larger in view 2 (4 nm) compared with view 1 (3 nm). This trend is also echoed by the slight decrease in the overall diameter, i.e. 11 nm for view 2 versus 9 nm for view 1. Views 1 and 2 could either correspond to differences in top and bottom face-on projections, thereby endowing pUL56 with the 3-dimensional shape of a truncated cone (assuming a monomer) or, alternatively, these two projections could represent different functional states. The latter possibility is supported by the observation that in the presence of DNA, view 2 occurs more frequently (>50% of the entire population of face-on views) than in the absence of DNA (<20% of the entire population), where view 1 is the dominant projection. It was also noted that pUL56 shows a more pronounced cleft/side opening in the presence of DNA (data not shown). While these observations favor the presence of at least two functional states, the exact molecular basis of these differences remains to be established. It is also interesting to note that amongst all molecules observed, not a single side-on view, i.e. a projection orthogonal to the face-on views, could be detected. This implies that the molecule must be rather flat and unable to assume a stable orientation when standing on its side. This idea is reinforced by the fact that a spherical protein of calculated mass 93.5 kDa is ∼6 nm in diameter based on the findings of Zipper et al. (30) that a volume of 1.37 Å3 ≈ 1 Da. Since the diameter of pUL56 in the electron microscope was measured to be 9 or 11 nm, the molecule must adopt a toroidal structure with a calculated height of only 2–3 nm (again, assuming a monomer). It is also noticeable that the contour lines corresponding to higher protein densities in the average of view 2 (Fig. 7E) seem to delineate three separate domains giving rise to a roughly pentagonal outline (see Discussion).
Figure 7.
Electron microscopic analysis of purified rpUL56 after negative staining. (A) An overview. (B and C) Representative molecules belonging to the two different projections (views 1 and 2, respectively). Note the toroidal structure of both projections and the pronounced cleft at the 9 o’clock position in (C). Averaged projections after digital image analysis are depicted in (D) (view 1) and (E) (view 2). The scale bars correspond to 50 (A), 10 (B and C) and 5 nm (D and E).
pUL89 is a ring-shaped protein
The baculovirus expressed purified rpUL89 protein was used to prepare specimens for electron microscopic analysis. The overview in Figure 8A shows that the protein has little tendency to aggregate and occurs as monomers. It has, like pUL56, a ring-shaped structure with, however, a more irregular outline (Fig. 8B and C). The rings have a diameter of ∼8 nm and the central protein cleft is ∼2 nm across. This toroidal structure demonstrates that pUL89 is in line with a number of DNA-metabolizing proteins.
Figure 8.
Electron microscopy of purified rpUL89. (A) An overview of rpUL89 negative stained with uranyl acetate. (B and C) Representative molecules. The scale bars correspond to 50 (A) and 1 nm (B and C).
DNA binding and cutting is a concerted action
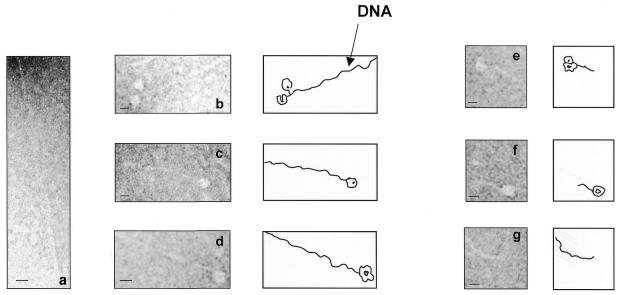
Electron microscopic observations of pUL56 incubated with linearized DNA (2.8 kb pUC-aseq containing one a sequence) showed DNA strands with contour lengths of ∼1 µm, which is in line with the DNA size (Fig. 9a–d). Upon addition of pUL89 only very short DNA fragments were observed (Fig. 9e–g). This experiment demonstrates that after binding of pUL56 to the DNA, the DNA remains uncut, while the cutting event itself is mediated by pUL89.
Figure 9.
Representative electron micrographic projections of linearized pUC-aseq after incubation with rpUL56 (a–d) and rpUL56 + rpUL89 (e–g). The sections show an ∼1 µM DNA molecule. The scale bars correspond to 10 nm.
DISCUSSION
dsDNA packaging into the capsid shell of bacteriophage is a complex biological process which is mediated by two non-structural proteins, the terminase subunits (4,7,11). It has been postulated that an analogous process occurs in herpesviruses. Recently we demonstrated that the gene product of ORF UL89 interacts with the terminase subunit pUL56, indicating that this highly conserved protein is the other terminase subunit (31). Abbots et al. (32) reported that the homologous proteins in HSV-1, pUL28 and pUL15 could also interact with each other. Here we have characterized pUL89, the homolog of HSV-1 pUL15 as well as the bacteriophage T4 gp17 (33). Both the in vitro translation product and the recombinant protein (rpUL89) have a molecular mass of ∼70–75 kDa. This observation is in line with previous reports on HSV-1 (34).
Using circular plasmid DNA containing a single a sequence as substrate, purified baculovirus-expressed HCMV pUL89 (rpUL89) exhibited enzymatic activity that converted sc plasmid DNA into oc as well as lin molecules. In addition, the ability of both proteins to cleave DNA without packaging signals demonstrated that the enzymatic activity is random under in vitro conditions. Comparable observations were made with the T4 homolog gp17, which cleaves plasmid DNA as well as E.coli genomic DNA in a non-specific manner (35,36). Limiting amounts of rpUL89 exhibited a reduced enzymatic activity that converted ∼60% of sc plasmid DNA into oc as well as lin molecules. This activity can be enhanced by incubation of the plasmid DNA in the presence of both proteins. While we were able to show in the nuclease assay that there is positive cooperativity between the two proteins converting sc DNA to linear form, it was not possible to directly differentiate between the pUL56 and pUL89 activities. Electron microscopic analysis demonstrated that rpUL56 is necessary for DNA binding, whereas rpUL89 mediates the cleavage of linearized DNA, i.e. rpUL56 is unable to exert specific cleavage by itself. However, it is important to point out that UL56 and UL89 are both able to randomly nick DNA, which will also lead to DNA cleavage if the nicking sites are <10 bp apart. Furthermore, the production of smaller linearized fragments of many different sizes will result in a dilution of the DNA in the gel and could thereby render them invisible or produce smeared bands, while in the electron microscope one is able to observe even single DNA molecules.
It is reported that bacteriophage T4 gp17 alone can catalyze a number of functions involved in packaging the unit length genome (4,35–38). This bacteriophage terminase subunit also exhibits an in vitro packaging activity, an ATPase activity, and contains a portal vertex-binding site for linkage of DNA to the procapsid. Sheaffer et al. (39) reported that in the case of HSV-1 the gene product of ORF UL6 could function as a portal protein and may be integrated into the capsid shell. Our results demonstrate that pUL56 binds to mature and immature capsids and is found to be associated on one side of the capsid shell. This finding may suggest that in HCMV pUL56 could also function as a portal protein or contains a portal protein-binding site. However, it is known that at least six other proteins, the gene products of UL6, UL15, UL17, UL25, UL32 and UL33, are required for packaging of HSV-1 DNA into the procapsid (40–44). Homologous proteins are found in HCMV, but the mode of action of each protein is not known to date.
Recently Lin et al. (45) described a bacteriophage gp16 subunit essential for recognition of DNA and initiation of packaging. Although HCMV rpUL56 also binds to DNA containing the packaging motifs (17) pac1 and pac2 and the observed ATPase activity (31), it is more likely that pUL56 is analogous to T4 gp17. We therefore propose that DNA packaging, entailing DNA binding, cleavage, translocation and attachment of the DNA to the procapsids, constitutes a general process in nucleocapsid-containing DNA viruses.
Based on our observations we postulate that the initiation of DNA packaging takes place in discrete stages: attachment of pUL56 to pac sites of concatemeric DNA (stage 1) and specific cleavage by the concerted action of pUL89 and pUL56 (stage 2). The provision of the energy for translocation of concatemeric DNA to the procapsids is mediated by pUL56. The association of pUL56 with the capsid furthermore implies that the third stage, i.e. attachment of DNA–protein complexes to the procapsid, is also catalyzed by pUL56. Finally, in the light of all data presented herein, the recent studies by Giesen et al. (27) demonstrating that pUL56 is associated with viral replication centers seem to suggest that DNA replication is coupled with packaging.
Figure 10 presents a preliminary working model for pUL56 as derived from electron microscopic data. The overall distribution of densities and the variations therein suggest that the protein consists of several discrete domains. This in itself could endow the molecule with a certain amount of flexibility. The apex of the pentagonal outline in Figure 7E coincides with the location of the cleft, again suggesting that conformational flexibility could well be within the remit of, if not a prerequisite for, the biological activity of pUL56. It is noticeable that in the averaged projection the cleft (view 2) is less pronounced than in individual particles (see for example Fig. 7C) where this discontinuity seems to extend throughout the molecule. This could be due to variations in stain depth as much as structural fluctuations. It is therefore possible that the two structural states observed correspond to different functional states. At present it can only be speculated that the cleft, together with the central torus, is intimately involved in the biological function of pUL56 by facilitating protein–DNA interaction and increasing the surface at the protein–DNA interface. In line with this is the observation of larger scale structural changes upon interaction of DNA with pUL56. The model in Figure 10 therefore shows a ‘silent’ (A) and an active state (B) for pUL56. In the active state the diameter of the molecule itself, as well as the central torus, has increased and the whole molecule opens up between two flanking regions. These regions would correspond to the protein domains on either site of the cleft (Fig. 7C and E) and they may be involved in DNA binding. Electron microscopic analysis of pUL89 demonstrated that this molecule also has a ring-shaped structure (Fig. 8). This structure is comparable with that shown for the small terminase subunit gp16 of bacteriophage T4 (46). The toroids of bacteriophages are formed by oligomers. In the case of HCMV, GST pull-down assays demonstrated that only pUL56 could interact with itself, implying an ability to form oligomers. At the moment we cannot exclude the possibility that the toroids are monomeric, but a dimeric quaternary structure with two toroids on top of each other, for instance, would strongly support the discussed function-dependent structural changes. While a full characterization of these interactions must await further studies, the overall toroidal shapes of pUL56 and pUL89 are in line with a large body of data suggesting that ring-shaped proteins play a major role in DNA metabolism (20).
Figure 10.
Preliminary, schematic model of HCMV pUL56 depicting the proposed silent (A) and active (B) states. In the active state, DNA is thought to be encased by the protein. The proposed conformational changes might be a prerequisite for DNA binding of pUL56. HCMV pUL56 drawn to show the different forms as top and bottom views of pUL56 (C and D).
Recent genetic analysis has demonstrated that a new class of anti-HCMV agents, benzimidazole ribonucleosides (47), inhibits HCMV DNA maturation via involvement of the viral UL89 (48) and UL56 (49) gene products. Our experiments show that UL89-associated nuclease activity is reduced in the presence of high concentrations of BDCRB. However, since there is no 100% reduction, it is obvious that the effect of BDCRB on pUL89 could not solely be explained by a decrease in nuclease activity. Further analyses need to be done to elucidate the effect. Based on the limited efficacy of the current drugs, ganciclovir, cidofovir and foscarnet, new anti-viral therapeutics appear to be in demand. Inhibitors targeting pUL56 and/or pUL89 may offer an attractive alternative since mammalian cell DNA replication does not involve cleavage of concatemeric DNA. Drugs targeted to terminase-like proteins would therefore be safe and highly selective. To this end, further studies will need to be undertaken aimed at a deepening of our understanding of viral DNA packaging processes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Claudia Grosskurth and Sonja Heck (Institute of Virology, Marburg) for expert technical assistance, and especially Johannes Gottanka and Marco Gösswein (Institute of Anatomy II, Erlangen) for helpful assistance concerning the electron microscopy. We are grateful to Mark Underwood (GlaxoWellcome) for kindly providing plasmid pUB8, William J. Britt for the monoclonal antibodies mAb28-4 and mAb58-15, and Alexandro Ripalti for the mAb RG1202. We also acknowledge Leroy Townsend for providing BDCRB. We thank Mark Stinski and Fred Homa for helpful discussions. This study was supported by the Johannes und Frieda Marohn-Stiftung. E.B. is a recipient of a Habilitationstipendium from the Deutsche Forschungsgemeinschaft (DFG) and thanks B. Fleckenstein for kind support.
REFERENCES
- 1.Chou J. and Roizman,B. (1985) The isomerization of the herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell, 41, 803–811. [DOI] [PubMed] [Google Scholar]
- 2.Deiss L.P., Chou,J. and Frenkel,N. (1986) Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol., 59, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spaete R.R. and Mocarski,E.S. (1985) The a sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J. Virol., 54, 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black L.W. (1988) DNA packaging in dsDNA bacteriophages. In Calendar,R. (ed.), The Bacteriophages. Plenum, New York, NY, pp. 321–373.
- 5.Murialdo H. and Becker,A. (1978) Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol. Rev., 42, 529–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker A. and Murialdo,H. (1990) Bacteriophage lambda DNA: the beginning of the end. J. Bacteriol., 172, 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins R.R. and Becker,A. (1994) Chromosome end formation in phage lambda, catalyzed by terminases, is controlled by two DNA elements of cos, cosN and R3 and by ATP. EMBO J., 13, 6152–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith M.P. and Feiss,M. (1993) Sites and gene products involved in lambdoid phage DNA packaging. J. Bacteriol., 175, 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosig G. (1994) In Karm,J., Drake,J.W., Kreuzer,K.N., Mosig,G., Hall,D.H., Eiserling,F.A., Black,L.W., Spicer,E.K., Kutter,E., Carlson,K. and Miller,E.S. (eds), Molecular Biology of Bacteriophage T4. American Society for Microbiology, Washington, DC, pp. 54–82.
- 10.Black L.W. (1995) DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays, 17, 1025–1030. [DOI] [PubMed] [Google Scholar]
- 11.Feiss M. and Becker,A. (1983) DNA packaging and cutting. In Hendrix,R.W., Roberts,J.W., Stahl,F.W. and Weisberg,R.A. (eds), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 305–330.
- 12.Black L.W., Showe,M.K. and Steven,A.C. (1994) Morphogenesis of the T4 head. In Karam,J., Drake,J.W., Kreuzer,K.N., Mosig,G., Hall,D.H., Eiserling,F.A., Black,L.W., Spicer,E.K., Kutter,E., Carlson,K. and Miller,E.S. (eds), Molecular Biology of Bacteriophage T4. American Society for Microbiology, Wahington, DC, pp. 218–258.
- 13.Casjens S. and Hendrix,R. (1988) Control mechanisms in dsDNA bacteriophage assembly. In Calendar,R. (ed.), The Bacteriophages. Plenum Press, New York, NY, Vol. 1, pp. 15–91.
- 14.Eppler K., Wyckoff,E., Goates,J., Parr,R. and Casjens,S. (1991) Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology, 183, 519–538. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa H. and Hearin,P. (1994) Structure, function and specificity of the DNA packaging signals in double-stranded DNA viruses. Semin. Virol., 5, 5–13. [Google Scholar]
- 16.Bogner E., Reschke,M., Reis,B., Mockenhaupt,T. and Radsak,K. (1993) Identification of the gene product encoded by ORF UL56 of human cytomegalovirus genome. Virology, 196, 290–293. [DOI] [PubMed] [Google Scholar]
- 17.Bogner E., Radsak,K. and Stinski,M.F. (1998) The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol., 72, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogner E. (1999) Human cytomegalovirus (HCMV) nuclease: implications for new strategies in gene therapy. Gene Ther. Mol. Biol., 3, 75–78. [Google Scholar]
- 19.Adelman K., Salmon,B. and Baines,J.D. (2001) Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl Acad. Sci. USA, 98, 3086–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hingorani M.M. and O’Donnell,M. (1998) Toroidal proteins: running rings around DNA. Curr. Biol., 8, 83–86. [DOI] [PubMed] [Google Scholar]
- 21.Summers M.D. and Smith,G.E. (1987) A Manual of Methods for Baculovirus Vectors and Insect Cell Culture Procedures. Texas Agriculture Experiment Station, College Station, TX.
- 22.Luckow V.A., Lee,S.C., Barry,G.F. and Olins,P.O. (1993) Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol., 67, 4566–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talbot P. and Almeida,J.D. (1977) Human cytomegalovirus: purification of enveloped virions and dense bodies. J. Gen. Virol., 36, 345–349. [DOI] [PubMed] [Google Scholar]
- 24.Valentine R., Shapiro,C. and Stadtman,E.R. (1968) Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry, 7, 2143–2152. [DOI] [PubMed] [Google Scholar]
- 25.van Heel M., Harauz,G. and Orlova,E.V. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke S.J., Baldwin,P.R. and Chiu,W. (1999) EMAN: semiautomated software for high-resolution single-particles reconstructions. J. Struct. Biol., 128, 82–97. [DOI] [PubMed] [Google Scholar]
- 27.Giesen K., Radsak,K. and Bogner,E. (2000) The potential terminase subunit pUL56 of human cytomegalovirus is translocated into the nucleus by its own NLS and interacts with importin α. J. Gen. Virol., 81, 2231–2244. [DOI] [PubMed] [Google Scholar]
- 28.Hoppert M. and Holzenburg,A. (1998) Electron Microscopy in Microbiology, Royal Microscopical Society Handbook Vol. 43. Bios Scientific Publishers, Oxford, UK.
- 29.Kolesnikova L., Mühlberger,E., Ryabchikova,E. and Becker,S. (2000) Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol., 74, 3899–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zipper P., Kratky,O., Herrmann,R. and Hohn,T. (1971) An X-ray small angle study of the bacteriophage fr and R17. Eur. J. Biochem., 18, 1–9. [DOI] [PubMed] [Google Scholar]
- 31.Hwang J.-S. and Bogner,E. (2002) ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem., 277, 6943–6948. [DOI] [PubMed] [Google Scholar]
- 32.Abbots A.P., Preston,V.G., Hughes,M., Patel,A.H. and Stow,N.D. (2000) Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol., 81, 2999–3009. [DOI] [PubMed] [Google Scholar]
- 33.Davison A.J. (1992) Channel catfish virus: a new type of herpesvirus. Virology, 186, 9–14. [DOI] [PubMed] [Google Scholar]
- 34.Baines J.D., Poon,A.P.W., Rovnak,J. and Roizman,B. (1994) The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol., 68, 8118–8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya S.P. and Rao,V.B. (1993) A novel terminase activity associated with the DNA-packaging protein gp17 of bacteriophage T4. Virology, 196, 34–44. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya S.P. and Rao,V.B. (1994) Structural analysis of DNA cleaved in vivo by bacteriophage T4 terminase. Gene, 146, 67–72. [DOI] [PubMed] [Google Scholar]
- 37.Powell D., Franklin,J., Arisaka,F. and Mosig,G. (1990) Bacteriophage T4 DNA-packaging genes 16 and 17. Nucleic Acids Res., 18, 4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao V.B., Leffers,G. and Kuebler,D. (1996) Molecular genetic and biochemical analysis of the DNA packaging and terminase functions of gp17 from bacteriophage T4. In The 11th Evergrenn International Bacteriophage T4 Meeting, Olympia, WA, p. 28.
- 39.Sheaffer A.K., Newcomb,W.W., Gao,M., Yu,D., Weller,S.K., Brown,J.C. and Tenney,D.J. (2001) Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol., 75, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamberti C. and Weller,S.K. (1996). The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology, 226, 403–407. [DOI] [PubMed] [Google Scholar]
- 41.McNab A.R., Desai,P., Person,S., Roof,L.L., Thomson,D.R., Newcomb,W.W., Brown,J.C. and Homa,F.L. (1998) The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol., 72, 1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds A.E., Fan,Y. and Baines,J.D. (2000) Characterization of the U(L) 33 gene product of herpes simplex virus 1. Virology, 266, 310–318. [DOI] [PubMed] [Google Scholar]
- 43.Salmon B., Cunningham,C., Davison,J.A., Harris,W.J. and Baines,J.D. (1998) The herpes simplex virus type 1U(L)17 gene encodes virion tegument proteins that are required for cleavage packaging of viral DNA. J. Virol., 72, 3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu D. and Weller,S.K. (1998) Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol., 72, 7428–74309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H., Simon,M.N. and Black,L.W. (1997) Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA-packaging. J. Biol. Chem., 272, 3495–3501. [DOI] [PubMed] [Google Scholar]
- 46.Kuebler D. and Rao,V.B. (1998) Functional analysis of the DNA-packaging/terminase protein gp17 from bacteriophage T4. J. Mol. Biol., 281, 803–814. [DOI] [PubMed] [Google Scholar]
- 47.Townsend L.B., Devivar,R.V., Turk,S.R., Nassiri,M.R. and Drach,J.C. (1995) Design, synthesis and antiviral activity of certain 2,5,6-trihalo-1-(beta-D-ribofuranosyl) benzimidazoles. J. Med. Chem., 38, 4098–4150. [DOI] [PubMed] [Google Scholar]
- 48.Underwood M.R., Harvey,R.J., Stanat,S.C., Hemphill,M.L., Miller,T., Drach,J.C., Townsend,L.B. and Biron,K.K. (1998) Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol., 72, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krosky P.M., Underwood,M.R., Turk,S.R., Feng,K.W.-H, Jain,R.K., Ptak,R.G., Westerman,A.C., Biron,K.K., Townsend,L.B. and Drach,J.C. (1998) Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol., 72, 4721–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]