Abstract
REV1 functions in the DNA polymerase ζ mutagenesis pathway. To help understand the role of REV1 in lesion bypass, we have examined activities of purified human REV1 opposite various template bases and several different DNA lesions. Lacking a 3′→5′ proofreading exonuclease activity, purified human REV1 exhibited a DNA polymerase activity on a repeating template G sequence, but catalyzed nucleotide insertion with 6-fold lower efficiency opposite a template A and 19–27-fold lower efficiency opposite a template T or C. Furthermore, dCMP insertion was greatly preferred regardless of the specific template base. Human REV1 inserted a dCMP efficiently opposite a template 8-oxoguanine, (+)-trans-anti-benzo[a]pyrene-N 2-dG, (–)-trans-anti-benzo[a]pyrene-N 2-dG and 1,N 6-ethenoadenine adducts, very inefficiently opposite an acetylaminofluorene-adducted guanine, but was unresponsive to a template TT dimer or TT (6–4) photoproduct. Surprisingly, the REV1 specificity of nucleotide insertion was very similar in response to different DNA lesions with greatly preferred C insertion and least frequent A insertion. By combining the dCMP insertion activity of human REV1 with the extension synthesis activity of human polymerase κ, bypass of the trans-anti-benzo[a]pyrene-N 2 -dG adducts and the 1,N 6-ethenoadenine lesion was achieved by the two-polymerase two-step mechanism. These results suggest that human REV1 is a specialized DNA polymerase that may contribute to dCMP insertion opposite many types of DNA damage during lesion bypass.
INTRODUCTION
Cells contain four complex systems in response to DNA damage: DNA repair, cell cycle checkpoint control, apoptosis and damage tolerance. Damage tolerance enables cells to completely replicate the genome in the presence of DNA damage. Unlike DNA repair, damage tolerance does not remove DNA lesions. Lesion bypass is an important damage tolerance mechanism, which directly copies the damaged DNA template during replication. Conceptually, lesion bypass can be divided into four steps: (i) damage recognition; (ii) nucleotide insertion opposite the lesion; (iii) extension DNA synthesis from opposite the lesion; and (iv) replication restoration. Steps (ii) and (iii) are frequently referred to as translesion synthesis. Two types of lesion bypass are known: error-free lesion bypass and error-prone lesion bypass. The former predominantly incorporates the correct nucleotide opposite the lesion, whereas the latter frequently incorporates an incorrect nucleotide opposite the lesion. Consequently, error-free lesion bypass is a mutation-avoiding mechanism, whereas error-prone lesion bypass is a mutation-generating mechanism. In cells, error-prone lesion bypass constitutes a major mechanism of DNA damage-induced mutagenesis.
Translesion synthesis often requires a specialized DNA polymerase (Pol), as many lesions block replicative polymerases. Recent biochemical studies suggest that DNA polymerases of the newly identified Y (UmuC) family are probably involved in translesion synthesis (1,2). The prototypic members of the Y family consist of the Escherichia coli DNA polymerase IV, the E.coli polymerase V, the yeast Rev1, the yeast Polη and the human Polι (1,2). Yeast Rev1 is a deoxycytidyl (dCMP) transferase, which efficiently inserts a C opposite a template G or apurinic/apyrimidinic (AP) site (3). Lawrence and colleagues observed that yeast cells predominantly incorporate a C opposite a template AP site (4,5). Thus, it was proposed that Rev1 plays a major role in nucleotide insertion opposite the lesion during bypass of a template AP site (3). Supporting this hypothesis, C incorporation opposite a site-specific template AP site was no longer observed in a yeast rev1 deletion mutant strain (5).
In yeast, Rev1 functions in the Polζ mutagenesis pathway (3,6). This pathway is a major error-prone lesion bypass mechanism and is required for mutagenesis induced by UV radiation, acetylaminofluorene (AAF) and AP sites (5,7,8). Rad6, Rad18 and DNA Polζ (the Rev3–Rev7 complex) are additionally required for the Polζ mutagenesis pathway. The Rad6–Rad18 complex is thought to function at an early step of the Polζ mutagenesis pathway (9). Yeast Polζ is able to perform extension DNA synthesis from opposite several lesions examined (10–13). Nucleotide incorporation activity of yeast Polζ has also been observed opposite some DNA lesions (13). Accordingly, Guo et al. (13) proposed a Polζ dual-function model that this polymerase functions at both the translesion synthesis step and the extension synthesis step in response to some DNA damage during lesion bypass. While TT dimer bypass can be efficiently catalyzed by Polη alone (14–16), efficient extension synthesis of many lesions may require Polζ following nucleotide insertion by the Y family polymerases, as suggested by the two-polymerase two-step model of lesion bypass (13).
Human homologs of RAD6, RAD18, REV1, REV3 and REV7 have all been identified and cloned (17–24). Human REV1 protein is also a dCMP transferase capable of C insertion opposite a template G, U and AP site (21). Human REV3 interacts with human REV7 (24), and human RAD18 interacts with the two Rad6 human homologs, HHR6A and HHR6B (18,19). Additionally, human REV1–REV7 interaction has also been observed (25). Human cells with perturbed RAD18 function show sensitivity to several DNA damaging agents (19). Reduced expression of REV1 and REV3 genes in cultured human cells results in significant reduction of UV-induced mutagenesis (22,23). These molecular, biochemical and genetic studies clearly indicate that a Polζ mutagenesis pathway, similar to that in yeast, is operational in humans. Ubiquitous expression of RAD18, REV1 and REV3 genes in various human tissues is consistent with the notion that the Polζ pathway may represent a major mutagenesis mechanism in mammals (18,20,21).
The role of REV1 in the Polζ pathway is poorly understood. Other than a template AP site, it is not known whether REV1 can catalyze nucleotide insertion opposite other DNA lesions. In order to evaluate whether REV1 is responsive to a wider spectrum of DNA lesions, we have analyzed biochemical activities of purified human REV1 protein toward various template bases and DNA lesions. In this report, we show a DNA polymerase activity of human REV1 on repeating template G sequences, demonstrate dCMP insertion activities of human REV1 in response to several types of DNA lesions, and show sequential lesion bypass by the combined activities of human REV1 and Polκ. Our results suggest that human REV1 is a specialized DNA polymerase that may contribute to dCMP insertion opposite many types of DNA damage during lesion bypass.
materials and methods
Materials
A mouse monoclonal antibody against the His6 tag was purchased from Qiagen (Valencia, CA). Alkaline phosphatase conjugated anti-mouse IgG was from Sigma Chemicals (St Louis, MO). All undamaged oligonucleotides were synthesized by Operon (Alameda, CA) unless otherwise indicated. N-Acetoxy-N-2-acetylaminofluorene (the activated form of AAF) was obtained from the Midwest Research Institute (Kansas City, MO). The E.coli strain BL21(DE3) and the E.coli expression vector pET-3d were purchased from Invitrogen (Carlsbad, CA). Human Polκ was purified to near homogeneity as previously described (26).
DNA templates containing a site-specific lesion
The 30mer DNA template, 5′-CCTTCTTCATTCXAACATACTTCTTCTTCC-3′, contained a tetrahydrofuran (AP site analog) at the X position. The 30mer DNA template, 5′-GGATGGACTGCAGGATCCGGAGGCCGCGCG-3′, contained an 8-oxoguanine at the underlined G. The 29mer DNA template, 5′-CCATCGCTACCTACCATCCGAATTCGCCC-3′, contained a 1,N6-ethenoadenine at the underlined A. These three damaged DNA templates were synthesized via automated DNA phosphoramidite methods by Operon. A 33mer DNA template containing either a site-specific (+)-trans-anti-benzo[a]pyrene (BPDE)-N2-dG or a site-specific (–)-trans-anti-BPDE-N2-dG was prepared as previously described (27–29). Its sequence is 5′-CTCGATCGCTAACGCTACCATCCGAATTCGCCC-3′, where the modified guanine is underlined. A 30mer DNA template containing a site-specific AAF-adducted guanine was prepared as previously described (26). Its sequence is 5′-CCTTCTTAATAGCTTCATACTTCTTCTTCC-3′, where the modified guanine is underlined. A 49mer DNA template containing a site-specific cis-syn TT dimer or a TT (6–4) photoproduct was prepared as previously described (30). Its sequence is 5′-AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT-3′, where the modified TT is underlined.
Purification of human REV1 protein
The human REV1 cDNA containing the His6-coding sequence at its 5′ end was cloned into the NcoI and BamHI sites of the E.coli expression plasmid pET-3d, yielding pETh6-hREV1. Escherichia coli BL21(DE3) cells harboring pETh6-hREV1 were grown in 500 ml of LB medium containing 100 µg/ml of ampicillin to 1 OD600. Then, isopropyl β-d-thiogalactopyranoside was added to 1 mM to induce expression of human REV1 for 12 h. Cells were collected by centrifugation, resuspended in 20 ml of Ni buffer A containing 50 mM sodium phosphate, pH 8.0, 0.3 M NaCl, 10% glycerol, 0.5% Igepal CA630, 10 mM imidazole, 5 mM β-mercaptoethanol and protease inhibitors (18). Cells were homogenized by a sonicator (Fisher Scientific, Pittsburgh, PA) with a setting at 30% power, using 30 pulses of 1 s each with a 1 s pause between pulses on ice. The clarified extract (∼20 ml) was loaded onto a NiSO4-charged HiTrap chelating column (1 ml; Amersham Pharmacia Biotech, Piscataway, NJ) at a flow rate of 0.5 ml/min. After washing the column sequentially with 15 ml of Ni buffer A containing 10 mM imidazole and 15 ml of Ni buffer A containing 50 mM imidazole, bound proteins were eluted with a 20 ml linear gradient of 50–500 mM imidazole. The His6-tagged human REV1 protein was identified by western blot analyses using a mouse monoclonal antibody specific to the His6 tag. The pooled human REV1 (10 ml) was concentrated to 1 ml by PEG 8000 in a dialysis bag and loaded onto an FPLC Superdex 200 HR10/30 column (Amersham Pharmacia Biotech). The gel filtration column was eluted with FPLC buffer A (50 mM Tris–HCl, pH 7.5, 10% glycerol, and 5 mM β-mercaptoethanol) containing 300 mM KCl. Silver staining, western blot analyses and dCMP transferase assays were performed to detect and confirm the purified human REV1 protein.
The dCMP transferase assay
Assays of dCMP transferase were performed essentially as previously described (21). This dCMP transferase assay is identical to our standard DNA polymerase assay (31). A standard reaction mixture (10 µl) contained 25 mM KH2PO4 pH 7.0, 5 mM MgCl2, 5 mM dithiothreitol, 100 µg/ml bovine serum albumin, 10% glycerol, 50 µM of dNTPs (dATP, dCTP, dTTP and dGTP individually or together as indicated), 50 fmol of a DNA substrate containing a 5′-32P-labeled primer, and purified human REV1 protein. After incubation at 30°C for 10 min or otherwise indicated, reactions were terminated with 7 µl of a stop solution (20 mM EDTA, 95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol). Reaction products were separated on a 15% polyacrylamide gel containing 8 M urea and visualized by autoradiography.
Kinetic analysis of nucleotide insertion by human REV1
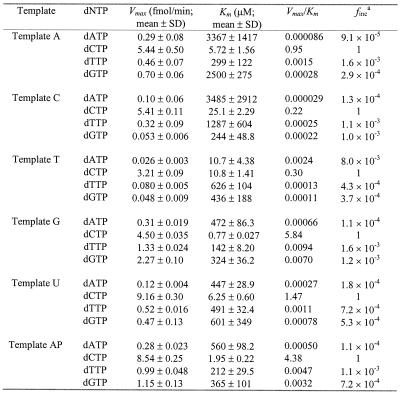
Kinetic analysis of human REV1 was performed as previously described (31,32). Briefly, the assays were performed using 50 fmol of a primed DNA template, 2 ng (14 fmol) of purified human REV1 and increasing concentrations of each dNTP (dATP, dCTP, dTTP or dGTP). Incubations were for 2 min at 30°C under standard DNA polymerase assay conditions. Longer incubations of up to 120 min were required to detect some misincorporations by human REV1. Reaction products were separated by electrophoresis on a 15% denaturing polyacrylamide gel. The fraction (%) of primers extended by human REV1 was calculated following scanning densitometry of the extended DNA band(s) and the remaining primer band on the autoradiogram. Product formed (P) was derived from the calculation P = % primer extension × 50 fmol. Observed velocity (v) was obtained from the calculation v = P/incubation time in min. Then the observed velocity was plotted as a function of dNTP concentration. The plotted data was fitted by a non-linear regression curve to the Michaelis–Menten equation, v = (Vmax × [dNTP])/(Km + [dNTP]), using the SigmaPlot software. Vmax and Km values of nucleotide insertion were obtained from the fitted curves. The rate of A, G and T insertions relative to the C insertion was calculated from the equation finc = (Vmax/Km)A, G or T/(Vmax/Km)C.
RESULTS
DNA polymerase activity of human REV1 protein
Previously, we expressed and purified human REV1 protein from the yeast Saccharomyces cerevisiae (21). To avoid potential interference of REV1 activity by a eukaryotic protein, we expressed human REV1 in E.coli cells and purified the protein again to near homogeneity (Fig. 1A). Human REV1 protein was tagged with six histidine residues at its N-terminus to facilitate its purification and detection. Human REV1 purified from E.coli cells migrated as a 153 kDa protein on a 10% SDS–polyacrylamide gel (Fig. 1A), consistent with its calculated molecular weight of 138 kDa (21). The identity of the tagged human REV1 was confirmed by western blot analysis using a mouse monoclonal antibody specific to the His6 tag (data not shown). Thus, the full-length human REV1 was purified. Three faint protein bands of 140, 107 and 71 kDa, respectively, were additionally detected below the human REV1 band (Fig. 1A). These bands were also detected by western blot analysis using the monoclonal antibody specific to the His6 tag (data not shown), suggesting that they are C-terminal degradation products of human REV1.
Figure 1.
DNA polymerase activity of human REV1 on a repeating template G sequence. (A) The purified human REV1 protein (400 ng) was analyzed by electrophoresis on a 10% SDS–polyacrylamide gel and visualized by silver staining. Protein size markers (lane M) are indicated on the left. (B) DNA polymerase assays were performed in the presence of a single dATP (A), dCTP (C), dTTP (T), dGTP (G) or all four dNTPs (N4) at 30°C for 30 min, using 7 ng (50 fmol) of purified human REV1 and 50 fmol of the indicated template containing a 5′-32P-labeled 20mer primer. DNA size markers in nucleotides are indicated on the left. Quantitation of extended primers is shown below the gel.
Initially, we found that human REV1 possesses a dCMP transferase activity capable of inserting a C opposite a template G (21). To determine whether human REV1 also possess a DNA polymerase activity on repeating template G sequences, we performed standard DNA polymerase assays, using a 33mer DNA template and a 20mer 32P-labeled primer annealed right before a stretch of eight template G residues (Fig. 1B). As shown in Figure 1B (lane 1), a DNA polymerase activity was readily detected with purified human REV1. Various primer extension products were observed that differed in length by 1 nt (Fig. 1B, lane 1), indicating that human REV1 synthesizes DNA in a distributive manner. To identify the nucleotides incorporated, we performed the polymerase assay again in the presence of only one dNTP at a time (Fig. 1B, lanes 2–5). As shown in Figure 1B (lane 3), eight C residues were incorporated by human REV1 opposite the stretch of eight template G residues. Apparently, two C residues were also incorporated opposite the template 3′-AG sequence at the end of the template G8 stretch (Fig. 1B, lane 3). These results show that human REV1 is a DNA polymerase on repeating template G sequences.
Human REV1 does not possess a 3′→5′ proofreading exonuclease activity
To examine whether human REV1 contains a proofreading exonuclease activity, we labeled a 15mer primer at its 5′ end with 32P and annealed it to a 30mer DNA template, yielding an A–C (template–primer) mismatch at the primer 3′ end (Fig. 2). In the absence of dNTPs, a 3′→5′ proofreading exonuclease would recognize a mismatch at the primer 3′ end and subsequently remove one or a few nucleotides from the 3′ end of the primer. Under the conditions used, the proofreading nuclease activity of the Klenow fragment of E.coli DNA polymerase I was readily detected after 2 min incubation (Fig. 2, compare lanes 1 and 2). In contrast, human REV1 did not degrade the mismatched primer even after 30 min incubation in the absence of dNTPs (Fig. 2, lane 3). These results show that human REV1 does not possess a 3′→5′ proofreading exonuclease activity.
Figure 2.
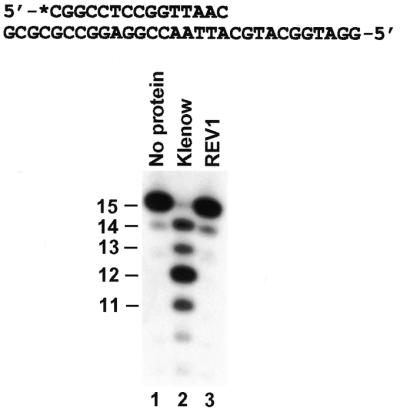
Proofreading exonuclease assays of human REV1 protein. A 15mer 5′-32P-labeled primer was annealed to the indicated DNA template, forming an A–C (template–primer) mismatch at the primer 3′ end. The DNA substrate was incubated with the purified Klenow fragment of E.coli DNA polymerase I (1 U) (lane 2), purified human REV1 protein (10 ng, 72 fmol) (lane 3) or without any protein (lane 1) at 37°C. The reaction was 2 min for the Klenow fragment and 30 min for human REV1. DNA size markers in nucleotides are indicated on the left.
DNA synthesis opposite various template bases
To examine whether human REV1 is capable of utilizing template A, C and T bases for DNA synthesis, we performed DNA polymerase assays with the purified protein (71 fmol), using templates A, T and C containing a 17mer 32P-labeled primer (50 fmol) (Fig. 3A). Templates G, U and AP site (50 fmol) were also included for comparison (Fig. 3A). In the presence of all four dNTPs, human REV1 inserted only one nucleotide opposite the template C and the template AP site (Fig. 3B, lanes 16 and 26), mainly one nucleotide opposite the template A (Fig. 3B, lane 1), and two nucleotides opposite the templates T, G and U (Fig. 3B, lanes 6, 11 and 21). However, nucleotide insertions opposite templates A, T and C were less efficient than those opposite templates G, U and AP site (Fig. 3B, compare lanes 1, 6 and 16 with lanes 11, 21 and 26). To determine what nucleotide was inserted by human REV1, DNA polymerase assays were carried out in the presence of only one dNTP at a time. As shown in Figure 3B, C was predominantly inserted by human REV1 with every template. Furthermore, human REV1 extended the T–C (template–primer) and U–C mismatches by 1 nt with C insertion opposite the next template C (Fig. 3B, lanes 8 and 23), although such extension was significantly less efficient than that from the G–C base pair (Fig. 3B, lane 13).
Figure 3.
Activity of human REV1 opposite various template bases. (A) Templates used for DNA polymerase assays. A 17mer primer was labeled with 32P at its 5′ end as indicated by an asterisk and annealed separately to each DNA template. (B) Standard DNA polymerase assays were performed with 50 fmol DNA substrate and 10 ng (72 fmol) of human REV1 in the presence of a single dATP (A), dCTP (C), dTTP (T), dGTP (G) or all four dNTPs (N4) as indicated. DNA size markers in nucleotides are indicated on the sides.
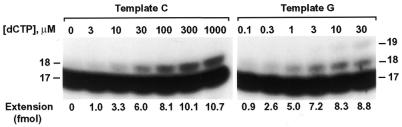
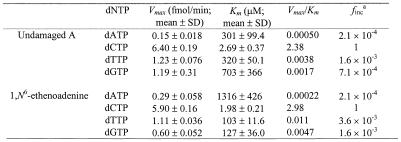
To quantitatively compare the efficiency and specificity of nucleotide insertion, we performed kinetic analysis of nucleotide insertion by human REV1. DNA polymerase assays were performed with human REV1 (14 fmol), using different DNA templates (50 fmol) and increasing concentrations of a single dNTP. Nucleotide insertion products were then quantified from the gel autoradiogram. As an example, results of dCMP insertion opposite the templates C and G are shown in Figure 4. From these assays, the kinetic parameters Vmax and Km were obtained. As indicated by the Vmax/Km values, C insertion opposite templates A, C, T, G, U and AP site was most efficient compared with other nucleotide insertions (Table 1). With a relative rate of 1 for C insertion, other nucleotides were incorporated with rates in the order of 10–3–10–5 (Table 1). Based on the Vmax/Km values (Table 1), relative efficiency of C insertion opposite templates G, AP site, U, A, T and C were 1, 0.8, 0.3, 0.2, 0.05 and 0.04, respectively. These results show that human REV1 recognizes template G efficiently, template A less efficiently, and templates T and C inefficiently, but indiscriminately inserts dCMP opposite all four template bases.
Figure 4.
Kinetic analysis of nucleotide insertion by human REV1. Standard DNA polymerase assays were performed at 30°C for 2 min, using 2 ng (14 fmol) of purified human REV1 and 50 fmol of DNA templates containing a 32P-labeled 17mer primer. Increasing concentrations of a single dNTP were used in the assay. The reaction products were separated by electrophoresis on a 15% denaturing polyacrylamide gel and visualized by autoradiography. After quantifying extended primers as shown below the gels, the kinetic Vmax and Km values were obtained as described in Materials and Methods. Autoradiograms of dCMP insertion opposite templates C and G, respectively, are shown. DNA templates used are shown in Figure 3A. DNA size markers in nucleotides are indicated on the sides.
Table 1. Kinetic measurement of nucleotide insertion by human REV1 opposite various template bases.
afinc = (Vmax/Km)A, G or T/(Vmax/Km)C.
Accurate nucleotide insertion by human REV1 opposite damaged template G
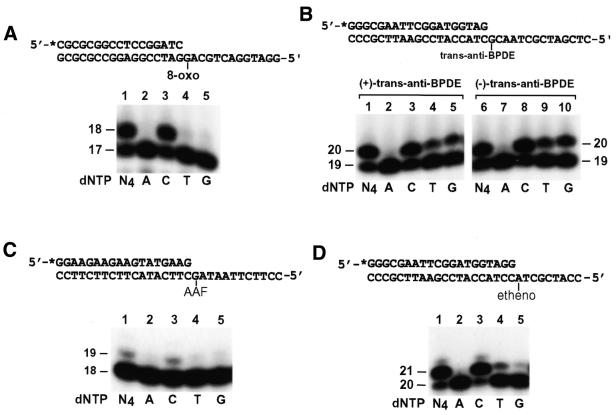
Efficient recognition of template G by human REV1 for DNA synthesis raised the question whether this enzyme is able to perform nucleotide insertion opposite damaged template G. To answer this question, we examined the response of human REV1 to four template G lesions: 8-oxoguanine, (+)-trans-anti-BPDE-N2-dG adduct, (–)-trans-anti-BPDE-N2-dG adduct and AAF-adducted guanine. Opposite the first three lesions, human REV1 effectively inserted 1 nt (Fig. 5A and B, lane 1; Fig. 5B, lane 6). Inefficiently, human REV1 also inserted one nucleotide opposite the AAF-adducted guanine (Fig. 5C, lane 1). Regardless of the specific damage, a C was predominantly inserted by human REV1 opposite the lesion (Fig. 5A–C, lane 3; Fig. 5B, lane 8). Minor G and T insertions were also observed opposite (+)-trans-anti-BPDE-N2-dG adduct and (–)-trans-anti-BPDE-N2-dG adduct (Fig. 5B, lanes 4, 5, 9 and 10).
Figure 5.
Nucleotide insertion by human REV1 opposite various DNA lesions. DNA primers were labeled with 32P (asterisks) at their 5′ ends and separately annealed right before a template lesion as indicated. Standard DNA polymerase assays with the damaged DNA template (50 fmol) were performed in the presence of a single dATP (A), dCTP (C), dTTP (T), dGTP (G) or all four dNTPs (N4) at 30°C for 10 min, using 10 ng (72 fmol) (A, B and D) or 40 ng (290 fmol) (C) of purified human REV1. (A) Template containing an 8-oxoguanine and a 17mer primer. (B) Templates containing a (+)-trans-anti-BPDE-N2-dG adduct or (–)-trans-anti-BPDE-N2-dG adduct, and a 19mer primer. (C) Template containing an AAF-guanine adduct and an 18mer primer. (D) Template containing a 1,N6-ethenoadenine and a 20mer primer. DNA size markers in nucleotides are indicated on the left or on the sides.
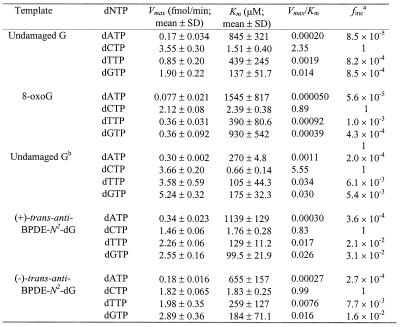
To quantitatively determine if the lesion affects kinetics of human REV1 catalysis, we measured the kinetic values of nucleotide insertion opposite damaged versus undamaged template G. As indicated by the Vmax/Km values (Table 2), 8-oxoguanine, (+)-trans-anti-BPDE-N2-dG and (–)-trans-anti-BPDE-N2-dG inhibited dCMP insertion of human REV1 by 2.6-, 6.7- and 5.6-fold, respectively. The specificity of nucleotide insertion by human REV1, however, was quantitatively very similar with or without the damage (Table 2). The Vmax/Km value of human REV1 opposite a template AAF-guanine was 0.0034, compared with 3.86 opposite the undamaged G in the same sequence context (1135-fold inhibition). Kinetic parameters of other nucleotide insertion opposite the AAF-guanine adduct were not measurable under the experimental conditions used. These results show that human REV1 is capable of accurate nucleotide insertion with varying efficiencies opposite damaged template G, such as 8-oxoguanine, (+)-trans-anti-BPDE-N2-dG adduct, (–)-trans-anti-BPDE-N2-dG adduct and AAF-adducted guanine.
Table 2. Kinetic measurement of nucleotide insertion by human REV1 opposite damaged template G.
afinc = (Vmax/Km)A, G or T/(Vmax/Km)C.
bUndamaged control for the BPDE-modified templates.
Error-prone nucleotide insertion by human REV1
To examine whether human REV1 is capable of nucleotide insertion opposite DNA lesions other than damaged G, we examined the response of this enzyme to a template 1,N6-ethenoadenine. A 29mer DNA template was synthesized, which contained a site-specific 1,N6-ethenoadenine. A 20mer 5′ 32P-labeled primer was then annealed right before the lesion (Fig. 5D). In the presence of all four dNTPs, purified human REV1 efficiently inserted 1 nt opposite the lesion (Fig. 5D, lane 1). To identify the inserted nucleotide, the reaction was performed again in the presence of only one dNTP at a time. As shown in Figure 5D (lane 3), C was predominantly inserted by human REV1 opposite the template 1,N6-ethenoadenine. Much less efficiently, T and G insertions were also detected (Fig. 5D, lanes 4 and 5). Based on kinetic measurements, C insertion by human REV1 opposite 1,N6-ethenoadenine was as efficient as opposite undamaged template A (Table 3). Furthermore, the specificity of nucleotide insertion by human REV1 opposite 1,N6-ethenoadenine was similar to that opposite 8-oxoguanine, (+)-trans-anti-BPDE-N2-dG and (–)-trans-anti-BPDE-N2-dG (Tables 2 and 3). These results show that human REV1 performs efficient and predominant dCMP insertion opposite a template 1,N6-ethenoadenine, leading to an error-prone consequence.
Table 3. Kinetic measurement of nucleotide insertion by human REV1 opposite a template 1,N6-ethenoadenine.
afinc = (Vmax/Km)A, G or T/(Vmax/Km)C.
Lesion bypass by the sequential actions of human REV1 and human Polκ
If dCMP insertion by human REV1 contributes to lesion bypass, extension DNA synthesis from opposite the lesion will have to be carried out by another DNA polymerase. To determine whether such extension synthesis can indeed take place following REV1 action, we performed lesion bypass experiments with the combined activities of purified human REV1 and human Polκ. Following REV1 action (10 min at 30°C) opposite the template (+)-trans-anti-BPDE-N2-dG adduct and (–)-trans-anti-BPDE-N2-dG adduct, purified human Polκ was added and the reaction was continued for another 10 min at 30°C. As shown in Figure 6A (lanes 2 and 4), human Polκ efficiently catalyzed extension DNA synthesis from opposite the lesion following REV1 dCMP insertion. Similar lesion bypass experiments were also performed with the template 1,N6-ethenoadenine. Following REV1 dCMP insertion opposite this lesion (Fig. 6B, lane 1), purified human Polκ again catalyzed extension DNA synthesis from opposite the lesion, resulting in ∼40% of the primers being extended beyond the lesion site in 10 min at 30°C (Fig. 6B, lane 2). These results show that the (+)- and (–)-trans-anti-BPDE-N2-dG adducts and the 1,N6-ethenoadenine can be effectively bypassed by the sequential actions of human REV1 and human Polκ in vitro.
Figure 6.
DNA lesion bypass by the combined activities of human REV1 and Polκ. Standard DNA polymerase assays with the indicated DNA templates (50 fmol) were performed in the presence of all four dNTPs, using 10 ng (72 fmol) (A) or 5 ng (36 fmol) (B) of purified human REV1. After 10 min reaction at 30°C, 5 ng (50 fmol) of purified human Polκ was then added to the reaction and the incubation continued for another 10 min. (A) Bypass of the 1,N6-ethenoadenine lesion. (B) Bypass of the (+)- and (–)-trans-anti-BPDE-N2-dG adducts. Lanes 1 and 3, reactions with REV1 alone for 20 min at 30°C; P, the primer. DNA size markers in nucleotides are indicated on the right.
Human REV1 is unresponsive to a template TT dimer and TT (6–4) photoproduct
In cells, REV1 is required for UV-induced mutagenesis (23). However, it is not known whether human REV1 is able to perform nucleotide insertion opposite UV lesions. Using a template cis–syn TT dimer and a TT (6–4) photoproduct, we did not detect nucleotide insertion with purified human REV1 opposite the 3′ T of either the template TT dimer or the TT (6–4) photoproduct even if excessive amounts of the protein (40 ng, 290 fmol) were used (Fig. 7A, lanes 2 and 3). Human REV1 was also unable to perform nucleotide insertion opposite the 5′ T of these two lesions (Fig. 7B, lanes 2 and 3). In separate experiments, purified human Polη performed translesion synthesis opposite the TT dimer and nucleotide insertion opposite the 5′ T of the TT (6–4) photoproduct in these same DNA templates (33), confirming the integrity of the templates used. These results show that human REV1 is unresponsive to a template TT dimer or a TT (6–4) photoproduct in vitro.
Figure 7.
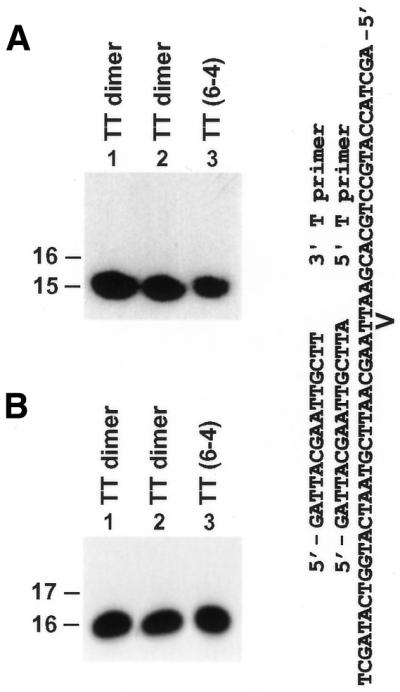
Response of human REV1 to a template TT dimer and TT (6–4) photoproduct. Two primers were labeled with 32P at their 5′ ends and separately annealed to a template containing a site-specific TT dimer or TT (6–4) photoproduct as shown. The 3′ T primer (15mer) and the 5′ T primer (16mer) were annealed right before the 3′ T and the 5′ T of the lesion, respectively. Standard DNA polymerase assays were performed with 50 fmol of the damaged DNA template and 40 ng (290 fmol) of human REV1 in the presence of all four dNTPs. (A) DNA polymerase assays with substrates containing the 3′ T primer. (B) DNA polymerase assays with substrates containing the 5′ T primer. Lane 1, reaction with the TT dimer template without human REV1 protein. DNA size markers in nucleotides are indicated on the left.
DISCUSSION
REV1 is known as a DNA template-dependent dCMP transferase (3,21). We have now demonstrated that human REV1 acts as a DNA polymerase on repeating template G sequences. Accordingly, human REV1 may be considered as a specialized DNA polymerase with a typical polymerase property opposite template G but altered biochemical properties opposite the other three template bases. Surprisingly, regardless of the base identity in the template, human REV1 greatly prefers dCMP insertion, with highest efficiency opposite a template G and lowest efficiency opposite a template C or T. Following dCMP insertion, extension by human REV1 is very inefficient or undetectable from opposite a template T, A and C. Hence, the template base strongly affects the rate of nucleotide insertion and subsequent extension, but not the specificity of nucleotide insertion. This unique property explains our previous observation that dCMP is efficiently inserted by human REV1 opposite a template U (21). Remarkably, human REV1 activity opposite template U is ∼5-fold higher than that opposite template T. Apparently, the 5-methyl group of a template T dramatically decreases recognition by human REV1.
Our results clearly show that human REV1 is not specific to a template AP site, but is catalytically responsive to a wide spectrum of different DNA lesions in vitro. Despite drastic differences in lesion structure, REV1 specificity of nucleotide insertion is surprisingly similar: predominant dCMP insertion and least frequent dAMP insertion opposite the lesion. Thus, even though human REV1 recognizes various damaged or undamaged template bases with very different affinity, this enzyme incorporates nucleotide with a similar specificity and strongly prefers dCMP insertion. These observations indicate that nucleotide selection by REV1 is not determined by hydrogen bonding and geometric fitting that normally rule DNA synthesis. The unique base selection of REV1 has profound biological impact. If REV1 indeed contributes to nucleotide insertion during lesion bypass in cells, dCMP insertion would lead to an error-free outcome opposite damaged G, but an error-prone consequence opposite other damaged bases. REV1 activity is much more efficient opposite templates G and A than opposite templates C and T. Coincidentally, nucleotide insertion by REV1 was observed opposite damaged templates G and A, but not opposite a template TT dimer and a TT (6–4) photoproduct. At present, it is not clear whether dCMP insertion by REV1 is restricted to damaged purines and AP sites in DNA.
The 1,N6-ethenoadenine lesion is an exocyclic DNA adduct induced by the carcinogens vinyl chloride and urethane, or spontaneously formed in cells from lipid peroxidation products (34,35). This lesion is mutagenic in mammalian cells (35–37). In simian kidney cells, 1,N6-ethenoadenine predominantly induces A→G transitions as a result of C insertion opposite the lesion (37). In one study with human cells, it was found that 1,N6-ethenoadenine induces slightly more A→T than A→G mutations and least frequently A→C mutations (36). Based on our in vitro results that human REV1 efficiently inserts a C opposite a template 1,N6-ethenoadenine, we hypothesize that REV1 may play an important role in dCMP insertion during bypass of 1,N6-ethenoadenine in cells. In vitro translesion synthesis of a template 1,N6-ethenoadenine by purified human Polη has also been reported recently (38).
The (+)-trans-anti-BPDE-N2-dG and (–)-trans-anti-BPDE-N2-dG lesions are two stereoisomeric DNA adducts induced by the potent carcinogen benzo[a]pyrene (39–41). Efficient C insertion by human REV1 opposite both lesions raised the possibility that this protein may contribute to error-free lesion bypass of benzo[a]pyrene-induced guanine DNA adducts in cells. Previously, we found that human Polκ is capable of efficient error-free lesion bypass of a template (–)-trans-anti-BPDE-N2-dG adduct, whereas human Polη is able to perform error-prone translesion synthesis opposite a template (+)-trans-anti-BPDE-N2-dG adduct in vitro (26,33). Further genetic studies are needed to definitively prove that REV1 and Polκ contribute to error-free bypass of the trans-anti-BPDE-N2-dG adducts and that Polη is involved in error-prone translesion synthesis of these lesions in cells.
Most recently, we found that purified human Polκ is able to catalyze extension DNA synthesis from opposite a template trans-anti-BPDE-N2-dG adduct, regardless of what nucleotide resides opposite the lesion (Y.Zhang, X.Wu, D.Guo, O.Rechkoblit and Z.Wang, manuscript submitted). Hence, Polκ may serve as the second DNA polymerase acting at the second step (extension DNA synthesis) during bypass of some lesions by the two-polymerase two-step mechanism (Y.Zhang, X.Wu, D.Guo, O.Rechkoblit and Z.Wang, manuscript submitted). Supporting this model, human REV1 and Polκ can indeed sequentially bypass a template trans-anti-BPDE-N2-dG adduct or a template 1,N6-ethenoadenine lesion. These results are consistent with a role of human REV1 in dCMP insertion opposite the lesion during bypass of some DNA damage in cells.
In vitro, human REV1 is unable to perform nucleotide insertion opposite a template TT dimer or TT (6–4) photoproduct, two major DNA lesions induced by UV radiation. Since it is involved in UV-induced mutagenesis (23), REV1 may perform another function different from its dCMP transferase activity in response to UV radiation. This has also been suggested for the yeast Rev1 protein (5). However, this presumed second function of REV1 is presently unknown. Among the Y family DNA polymerases, Polη is the only enzyme in humans that can efficiently perform translesion synthesis opposite a template TT dimer. Since yeast Polζ is completely blocked by a TT dimer in vitro (12,13), human Polζ is likely to be unresponsive to template TT dimers, too. This underscores the pivotal role of Polη in protecting humans from the deleterious effect of UV radiation, as exemplified in xeroderma pigmentosum variant patients who have lost the Polη function (16,42). In the absence of Polη, the very limited nucleotide insertion activity of human Polι opposite TT dimers may become important in response to UV radiation, and may significantly contribute to mutagenesis induced by TT dimers in DNA (43,44). It appears that Drosophila Polι catalyzes efficient error-free bypass of a template TT dimer, and may thus function redundantly with Polη in response to UV-induced TT dimers during replication (45).
The biochemical capability of REV1 to respond to a wide spectrum of different DNA lesions further strengthened the notion that the whole Y family of DNA polymerases is involved in copying damaged DNA templates during replication (2). The involvement of multiple DNA polymerases would form an effective lesion bypass system in response to the many different lesions that can arise in DNA.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Fenghuan Yuan for technical assistance in the purification of human REV1 protein. This work was supported by a New Investigator Award in Toxicology from Burroughs Wellcome Fund (Z.W.) and NIH grants CA40463 (J.-S.T.) and CA20851 (N.E.G.).
REFERENCES
- 1.Ohmori H., Friedberg,E.C., Fuchs,R.P.P., Goodman,M.F., Hanaoka,F., Hinkle,D., Kunkel,T.A., Lawrence,C.W., Livneh,Z., Nohmi,T., Prakash,L., Prakash,S., Todo,T., Walker,G.C., Wang,Z. and Woodgate,R. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z. (2001) Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res., 486, 59–70. [DOI] [PubMed] [Google Scholar]
- 3.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs P.E. and Lawrence,C.W. (1995) Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol., 251, 229–236. [DOI] [PubMed] [Google Scholar]
- 5.Nelson J.R., Gibbs,P.E., Nowicka,A.M., Hinkle,D.C. and Lawrence,C.W. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol., 37, 549–554. [DOI] [PubMed] [Google Scholar]
- 6.Larimer F.W., Perry,J.R. and Hardigree,A.A. (1989) The REV1 gene of Saccharomyces cerevisiae: isolation, sequence and functional analysis. J. Bacteriol., 171, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison A., Christensen,R.B., Alley,J., Beck,A.K., Bernstine,E.G., Lemontt,J.F. and Lawrence,C.W. (1989) REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol., 171, 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baynton K., Bresson-Roy,A. and Fuchs,R.P. (1998) Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol. Cell. Biol., 18, 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailly V., Lamb,J., Sung,P., Prakash,S. and Prakash,L. (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev., 8, 811–820. [DOI] [PubMed] [Google Scholar]
- 10.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- 11.Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.-S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R.E., Washington,M.T., Haracska,L., Prakash,S. and Prakash,L. (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature, 406, 1015–1019. [DOI] [PubMed] [Google Scholar]
- 13.Guo D., Wu,X., Rajpal,D.K., Taylor,J.-S. and Wang,Z. (2001) Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res., 29, 2875–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R.E., Prakash,S. and Prakash,L. (1999) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 15.Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 17.Koken M.H.M., Reynolds,P., Jaspers-Dekker,I., Prakash,L., Prakash,S., Bootsma,D. and Hoeijmakers,J.H.J. (1991) Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl Acad. Sci. USA, 88, 8865–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xin H., Lin,W., Sumanasekera,W., Zhang,Y., Wu,X. and Wang,Z. (2000) The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res., 28, 2847–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tateishi S., Sakuraba,Y., Masuyama,S., Inoue,H. and Yamaizumi,M. (2000) Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl Acad. Sci. USA, 97, 7927–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin W., Wu,X. and Wang,Z. (1999) A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- 21.Lin W., Xin,H., Zhang,Y., Wu,X., Yuan,F. and Wang,Z. (1999) The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res., 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs P.E., Wang,X.D., Li,Z., McManus,T.P., McGregor,W.G., Lawrence,C.W. and Maher,V.M. (2000) The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl Acad. Sci. USA, 97, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakumo Y., Roth,T., Ishii,H., Rasio,D., Numata,S., Croce,C.M. and Fishel,R. (2000) A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem., 275, 4391–4397. [DOI] [PubMed] [Google Scholar]
- 25.Murakumo Y., Ogura,Y., Ishii,H., Numata,S., Ichihara,M., Croce,C.M., Fishel,R. and Takahashi,M. (2001) Interactions in the error-prone postreplicaiton repair proteins, hREV1, hREV3, and hREV7. J. Biol. Chem., 276, 35644–35651. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Yuan,F., Wu,X., Wang,M., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. (2000) Error-free and error-prone lesion bypass by human DNA polymerase κin vitro. Nucleic Acids Res., 28, 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosman M., Ibanez,V., Geacintov,N.E. and Harvey,R.G. (1990) Preparation and isolation of adducts in high yield derived from the binding of two benzo[a]pyrene-7,8-dihydroxy-9,10-oxide stereoisomers to the oligonucleotide d(ATATGTATA). Carcinogenesis, 11, 1667–1672. [DOI] [PubMed] [Google Scholar]
- 28.Geacintov N.E., Cosman,M., Mao,B., Alfano,A., Ibanez,V. and Harvey,R.G. (1991) Spectroscopic characteristics and site I/site II classification of cis and trans benzo[a]pyrene diolepoxide enantiomer-guanosine adducts in oligonucleotides and polynucleotides. Carcinogenesis, 12, 2099–2108. [DOI] [PubMed] [Google Scholar]
- 29.Rechkoblit O., Amin,S. and Geacintov,N.E. (1999) Primer length dependence of binding of DNA polymerase I Klenow fragment to template–primer complexes containing site-specific bulky lesions. Biochemistry, 38, 11834–11843. [DOI] [PubMed] [Google Scholar]
- 30.Smith C.A. and Taylor,J.S. (1993) Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn, trans-syn-I, (6-4) and Dewar photoproducts of thymidylyl(3′→5′)-thymidine. J. Biol. Chem., 268, 11143–11151. [PubMed] [Google Scholar]
- 31.Zhang Y., Yuan,F., Wu,X. and Wang,Z. (2000) Preferential incorporation of G opposite template T by the low fidelity human DNA polymerase ι. Mol. Cell. Biol., 20, 7099–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creighton S., Bloom,L.B. and Goodman,M.F. (1995) Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading and lesion bypass efficiencies. Methods Enzymol., 262, 232–256. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartsch H., Barbin,A., Marion,M.J., Nair,J. and Guichard,Y. (1994) Formation, detection and role in carcinogenesis of ethenobases in DNA. Drug Metab. Rev., 26, 349–371. [DOI] [PubMed] [Google Scholar]
- 35.Nair J., Barbin,A., Velic,I. and Bartsch,H. (1999) Etheno DNA-base adducts from endogenous reactive species. Mutat. Res., 424, 59–69. [DOI] [PubMed] [Google Scholar]
- 36.Levine R.L., Yang,I.Y., Hossain,M., Pandya,G.A., Grollman,A.P. and Moriya,M. (2000) Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res., 60, 4098–4104. [PubMed] [Google Scholar]
- 37.Pandya G.A. and Moriya,M. (1996) 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry, 35, 11487–11492. [DOI] [PubMed] [Google Scholar]
- 38.Levine R.L., Miller,H., Grollman,A., Ohashi,E., Ohmori,H., Masutani,C., Hanaoka,F. and Moriya,M. (2001) Translesion DNA synthesis catalyzed by human pol η and pol κ across 1,N6-ethenodeoxyadenosine. J. Biol. Chem., 276, 18717–18721. [DOI] [PubMed] [Google Scholar]
- 39.Cheng S.C., Hilton,B.D., Roman,J.M. and Dipple,A. (1989) DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem. Res. Toxicol., 2, 334–340. [DOI] [PubMed] [Google Scholar]
- 40.Peltonen K. and Dipple,A. (1995) Polycyclic aromatic hydrocarbons: chemistry of DNA adduct formation. J. Occup. Environ. Med., 37, 52–58. [DOI] [PubMed] [Google Scholar]
- 41.Geacintov N.E., Cosman,M., Hingerty,B.E., Amin,S., Broyde,S. and Patel,D.J. (1997) NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen–DNA adduct: principles, patterns and diversity. Chem. Res. Toxicol., 10, 111–146. [DOI] [PubMed] [Google Scholar]
- 42.Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- 43.Tissier A., Frank,E.G., McDonald,J.P., Iwai,S., Hanaoka,F. and Woodgate,R. (2000) Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase ι. EMBO J., 19, 5259–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Yuan,F., Wu,X., Taylor,J.-S. and Wang,Z. (2001) Response of human DNA polymerase ι to DNA lesions. Nucleic Acids Res., 29, 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa T., Uematsu,N., Mizukoshi,T., Iwai,S., Iwasaki,H., Masutani,C., Hanaoka,F., Ueda,R., Ohmori,H. and Todo,T. (2001) Mutagenic and nonmutagenic bypass of DNA lesions by Drosophila DNA polymerases dpolη and dpolι. J. Biol. Chem., 276, 15155–15163. [DOI] [PubMed] [Google Scholar]