Abstract
HilA activates the expression of Salmonella enterica serovar Typhimurium invasion genes. To learn more about regulation of hilA, we isolated Tn5 mutants exhibiting reduced hilA and/or invasion gene expression. In addition to expected mutations, we identified Tn5 insertions in pstS, fadD, flhD, flhC, and fliA. Analysis of the pstS mutant indicates that hilA and invasion genes are repressed by the response regulator PhoB in the absence of the Pst high-affinity inorganic phosphate uptake system. This system is required for negative control of the PhoR-PhoB two-component regulatory system, suggesting that hilA expression may be repressed by PhoR-PhoB under low extracellular inorganic phosphate conditions. FadD is required for uptake and degradation of long-chain fatty acids, and our analysis of the fadD mutant indicates that hilA is regulated by a FadD-dependent, FadR-independent mechanism. Thus, fatty acid derivatives may act as intracellular signals to regulate hilA expression. flhDC and fliA encode transcription factors required for flagellum production, motility, and chemotaxis. Complementation studies with flhC and fliA mutants indicate that FliZ, which is encoded in an operon with fliA, activates expression of hilA, linking regulation of hilA with motility. Finally, epistasis tests showed that PhoB, FadD, FliZ, SirA, and EnvZ act independently to regulate hilA expression and invasion. In summary, our screen has identified several distinct pathways that can modulate S. enterica serovar Typhimurium's ability to express hilA and invade host cells. Integration of signals from these different pathways may help restrict invasion gene expression during infection.
Salmonella enterica serovar Typhimurium is a facultative intracellular pathogen that causes gastroenteritis in humans and a typhoid-like disease in mice. Some S. enterica serovar Typhimurium virulence factors are encoded on the 40-kb Salmonella pathogenicity island 1 (SPI1) located at centisome 63 (60). Many SPI1 genes were first identified for their roles in invasion, a process whereby bacteria induce their own uptake into normally nonphagocytic cells (34). These invasion genes have also been implicated in other processes that may contribute to S. enterica serovar Typhimurium virulence, including intestinal colonization (R. A. Murray, unpublished observations), destruction of M cells in Peyer's patches (49, 66), activation of cytokine expression in epithelial cells (45), induction of neutrophil migration across the intestinal epithelium (36, 58), and stimulation of apoptosis in macrophages (18, 44, 61).
SPI1 invasion genes encode a type III secretory apparatus and several secreted factors that are translocated by the secretion system directly into the cytosol of cultured epithelial cells (21, 33, 39) (Fig. 1). During invasion, the secreted effectors are thought to interact with eukaryotic proteins to activate signal transduction pathways, rearrange the actin cytoskeleton, and cause membrane ruffling and macropinocytosis in the host cell, ultimately inducing uptake of bacteria (17, 32, 33, 40, 45). Oxygen, osmolarity, bacterial growth state, and certain mutations affect Salmonella's ability to invade cultured epithelial cells (10, 31, 53, 74, 79). Many of these conditions and mutations also affect expression of invasion genes (9, 65), suggesting that invasiveness may be modulated by regulating invasion gene expression. This regulation is thought to be mediated by several transcriptional regulators on SPI1, including InvF and HilA (9, 23, 29).
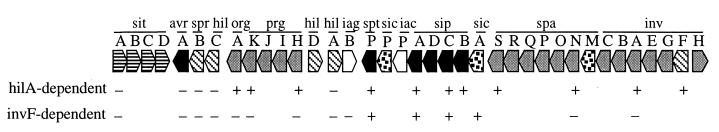
FIG. 1.
Regulation of SPI1 genes by HilA and InvF. The proposed functions of SPI1 gene products are as follows: black boxes, secreted effectors; gray boxes, type III secretory apparatus; dotted boxes, chaperones involved in type III secretion; diagonally striped boxes, transcriptional regulators; horizontally striped boxes, iron uptake system; white boxes, function unknown (73). −, expression of the SPI1 gene is not affected by a mutation in hilA or invF; +, expression of the SPI1 gene is severely reduced by a mutation in hilA or invF (8, 9, 23, 29, 30; this work; S. M. Damrauer, unpublished observations).
InvF is an AraC-like transcriptional regulator required for expression of secreted effectors encoded on SPI1 (Fig. 1), SPI5, and SopEΦ (23, 29). HilA is an OmpR-ToxR family member that appears to directly activate transcription of SPI1 genes encoding components of the type III secretory apparatus (8, 9). HilA also appears to directly activate invF expression, thereby indirectly activating expression of several secreted effectors. Interestingly, two SPI1 effectors may be directly activated by both HilA and InvF (23, 29). One effector, sipC, appears to have two promoters: a HilA-dependent promoter thought to be upstream of spaS and an InvF-dependent promoter located downstream of spaS (23). Thus, HilA directly or indirectly regulates expression of the type III secretion system and its secreted effectors, and this regulation is thought to be mediated by modulating hilA expression.
Changes in oxygen tension, osmolarity, and pH alter expression of hilA and SPI1 invasion genes (9), but the sensors and transcription factors involved in this regulation have not yet been identified. A point mutation (pho-24) in the extracellular cation sensor phoQ drastically reduces hilA and invasion gene expression (9, 65). Normally, the PhoQ sensor kinase is activated and phosphorylates its cognate transcriptional regulator, PhoP, when extracellular cation levels are low (82). In the pho-24 mutant, PhoQ is active even when extracellular cation levels are relatively high, resulting in net phosphorylation of PhoP (37). The strong repressing effect of the pho-24 mutation on hilA and invasion gene expression suggests that PhoP∼P might repress hilA directly or indirectly in response to low extracellular cation levels. Expression of hilA and invasion genes is also reduced by disruptions in sirA or barA (1, 4, 48). The products of these genes resemble GacA and LemA, respectively. GacA and LemA are two-component regulatory factors implicated in Pseudomonas pathogenesis. It is thought that BarA regulates the activity of SirA in response to an unknown environmental cue, and SirA goes on to directly or indirectly activate expression of hilA and invasion genes.
Other mutations causing decreased hilA and invasion gene expression have been identified in csrB and in a second pathogenicity island called SPI2. In Escherichia coli, CsrB is an RNA thought to sequester CsrA, a protein that binds to and accelerates degradation of particular mRNAs (55, 84). Chromosomal disruptions in S. enterica serovar Typhimurium csrB or expression of E. coli csrA from a plasmid reduces hilA expression (4), suggesting that CsrA may degrade an mRNA whose product is involved in regulation of hilA. hilA expression is also reduced by insertions in SPI2 genes that encode structural components of a type III secretion system required by S. enterica serovar Typhimurium to survive inside macrophages (25). Thus, regulation of the SPI1 and SPI2 secretion systems may be interrelated. The mechanisms whereby CsrA-CsrB and SPI2 genes affect hilA expression are unknown.
Recent studies of the hilA promoter suggest that inhibition of hilA expression by high oxygen, low osmolarity, pho-24, or disruptions in sirA or barA requires region −39 to −314 upstream of the hilA transcriptional start site (72). An unknown repressor is thought to act at this site to inhibit hilA expression. hilC, also referred to as sirC (69) or sprA (30), and hilD are SPI1 genes predicted to encode AraC-like transcriptional regulators that may inhibit the repressor under appropriate conditions, allowing expression of hilA (72). hilC mutants exhibit a mild decrease in hilA expression and invasiveness, while a disruption in hilD drastically reduces hilA expression and significantly inhibits S. enterica serovar Typhimurium's ability to invade HEp-2 cells (69, 72). Thus, hilD seems to play a more important role than hilC in regulation of hilA expression and S. enterica serovar Typhimurium invasiveness in vitro. HilC may be capable of activating invF and invA expression directly, since high-level expression of hilC from a plasmid allows expression of these genes in the absence of hilA (30, 69).
Although many genes have been implicated in hilA regulation, only one screen designed to isolate mutations reducing invasion gene expression has been reported (48), and this screen was not saturating. Identification of such mutations might help uncover novel regulatory pathways or elucidate mechanisms involved in regulation of hilA and invasion genes by environmental conditions. Here we report the identification of Tn5 insertions causing decreased hilA and/or invasion gene expression. Our results suggest that several pathways not previously implicated in regulation of hilA work independently of each other to modulate hilA expression and invasion of HEp-2 cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cultures were grown at 37°C in Luria-Bertani (LB) medium composed of 0.5% Bacto-yeast extract, 1% Bacto-tryptone, and 1% NaCl. When appropriate, the medium was supplemented with antibiotics as follows: 100 to 200 μg of ampicillin per ml, 50 to 100 μg of kanamycin per ml, 10 μg of chloramphenicol per ml, and/or 10 μg of tetracycline per ml. Expression of fliA, fliZY, fliZ, and fliY was induced by growing cultures in medium containing 0.01 to 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). β-Galactosidase assays were performed with bacterial cultures grown under low-oxygen conditions as previously described (9, 54), and activities were quantified by the Miller method (59).
TABLE 1.
S. enterica serovar Typhimurium strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 derivatives | ||
| EE633 | sipA4::Tn5lacZY (Tetr) | 9 |
| EE636 | orgA::Tn5lacZY (Tetr) | 9 |
| EE637 | invF11-5::Tn5lacZY (Tetr) | 9 |
| EE638 | sipC11-6::Tn5lacZY (Tetr) | 9 |
| EE656 | prgH020::Tn5lacZY (Tetr) | 9 |
| EE658 | hilA080::Tn5lacZY (Tetr) | 9 |
| EE659 | prgK100::Tn5lacZY (Tetr) | 9 |
| ST54 | fadF103::MudJ (lac Kanr) | 78 |
| CL87 | iagB87::lacZY | This work |
| CL87 derivatives | ||
| RL353 | pst-4::Tn10 (Tetr) | This work and reference 47 |
| RL414 | fadD1::Tn5 (Kanr) | This work |
| RL291 | pstS55::Tn5 (Kanr) | This work |
| RL415 | fliA51::Tn5 (Kanr) | This work |
| RL446 | fliA36::Tn5B50 (Tetr) | This work |
| RL424 | flhC4::Tn5 (Kanr) | This work |
| RL60 | flhC36::Tn5 (Kanr) | This work |
| RL61 | flhD37::Tn5 (Kanr) | This work |
| EE710 | envZ182::cam (Camr) from BA708 | This work; S. Lindgren and B. A. Ahmer, unpublished data |
| EE719 | sirA2::kan (Kanr) from BA732 | This work; B. A. Ahmer, unpublished data |
| RL516 | fadD1::Tn5 envZ182::cam (Kanr Camr) | This work |
| RL421 | fadD1::Tn5 pst-4::Tn10 (Kanr Tetr) | This work |
| RL451 | fadD1::Tn5 fliA36::Tn5B50 (Kanr Tetr) | This work |
| RL423 | pst-4::Tn10 fliA51::Tn5 (Tetr Kanr) | This work |
| RL419 | pst-4::Tn10 envZ182::cam (Tetr Camr) | This work |
| RL462 | pst-4::Tn10 sirA2::kan (Tetr Kanr) | This work |
| RL448 | fliA36::Tn5B50 envZ182::cam (Tetr Camr) | This work |
| RL524 | fliA36::Tn5B50 sirA2::kan (Tetr Kanr) | This work |
| RL449 | flhC4::Tn5 fliA36::Tn5B50 (Kanr Tetr) | This work |
| LT2 derivatives | ||
| TBW19812 | ΔphoB1::cat (Camr) | 47 |
| TR6583 | metE205 ara-9 | 78 |
| LS1860 | TR6583 fadR101 | 78 |
| TBW19912 | pst-4::Tn10 | 47 |
| EE251 | trpA8 hisC527 rpsL | 54 |
| Plasmids | ||
| pTrc99c | Ampr, tac expression vector containing lacIq | 5 |
| pSIIA1 | Ampr, pTrc99c with S. enterica serovar Typhimurium fliA under tac promoter | 52 |
| pN300 | Ampr, pACYC177 containing E. coli fadD | 13 |
| pSN507 | Ampr, pBR322 containing E. coli pstSCAB-phoU operon | 6 |
| pKK223-3 | Ampr, pBR322-derived tac expression vector | 62 |
| pDSM29 | Ampr, pKK223-3 with E. coli fliZ under tac promoter | 62 |
| pDSM30 | Ampr, pKK223-3 with E. coli fliY under tac promoter | 62 |
| pDSM32 | Ampr, pKK223-3 with E. coli fliZY under tac promoter | 62 |
| pAID325 | Camr, pACYC184 containing lacIq | 26 |
Tissue culture growth conditions and invasion assays.
HEp-2 cells (ATCC CCL23) were maintained in Dulbecco's modified Eagle medium supplemented with 5% fetal bovine serum. Invasion assays were performed on HEp-2 monolayers with oxygen-limited bacterial cultures as previously described (9, 54). To verify results from invasion assays, coverslip assays were performed on HEp-2 monolayers that were obtained by seeding approximately 5 × 104 cells per coverslip in a 24-well plate and allowing growth overnight at 37°C with 5% CO2. After inoculation of coverslips with 100 μl of oxygen-limited bacterial cultures, plates were centrifuged at 100 × g for 10 min and incubated at 37°C in a 5% CO2 incubator for 1 h. Cells were then rinsed two times with phosphate-buffered saline (PBS), fixed with methanol for 5 min, and treated with Giemsa stain for 45 min. Following four rinses with distilled water, coverslips were dried, mounted, and examined by bright-field microscopy.
DNA methods.
Restriction enzymes were purchased from New England Biolabs. Chromosomal DNA was isolated with Invitrogen's Easy-DNA kit. PCR was performed with rTaq polymerase from TaKaRa Shuzo Co. PCR products were cloned with a TA-cloning kit from Invitrogen. Plasmid DNA was purified on Qiagen columns. Enzymes and kits were used according to the manufacturers' instructions.
Bacterial strain construction.
Marked mutations were moved into different strain backgrounds and tested for linkage to chromosomal markers by P22 transduction. Plasmids were passaged through r−m+ LT2 strain LB5000 (15, 70) before being electroporated into SL1344 derivatives by standard methods.
CL87 was constructed by generating a chromosomal iagB87::lacZY fusion in SL1344. A promoterless lacZY fragment was isolated by digesting pRS415 (76) with SmaI and StuI. This 5.2-kb fragment was then ligated into an SspI site 90 bp downstream of the iagB translational start site. iagB87::lacZY was crossed into the SL1344 chromosome by allelic exchange with pMAK705 (38). The location and orientation of lacZY were confirmed by PCR with primers in lacZ and hilA.
Motility assays and flagellar staining.
Motility was tested by assessing swarming phenotypes on motility agar (2.5% nutrient broth and 0.5% Bacto-agar) supplemented with antibiotics and IPTG when appropriate. To visualize flagella, cells were grown in tubes on a roller at 37°C to mid-log phase, and 1 drop of each culture was mixed with 5 to 10 μl of RYU Flagella stain (Remel). Cells were then examined by bright-field microscopy.
Tn5 mutagenesis and screening of mutants.
Pools of EE251::Tn5 mutants were generated as previously described (54). To identify mutations that affect hilA and invasion gene expression, Tn5 mutations were moved from EE251 pools into CL87 and EE633 by P22 phage transduction. Transductants were initially selected on LB agar supplemented with 100 μg of kanamycin per ml and 10 mM EGTA. Transductants were scraped off the plates, suspended in LB medium containing 10 mM EGTA, and replated on MacConkey lactose (MacLac) agar supplemented with 100 μg of kanamycin per ml and 10 mM EGTA. Colonies exhibiting a loss-of-red phenotype compared to their wild-type parents were restreaked on MacLac agar containing 10 mM EGTA to purify the bacteria from phage and to confirm the loss-of-red phenotype.
Identification of Tn5 mutations.
RL20, RL119, and RL224 chromosomal DNAs were digested with SalI or EcoRI, ligated into pUC19, and transformed into DH5α. Plasmids from transformants which were resistant to both ampicillin and kanamycin were isolated and sequenced with primer IS50RL (GGTCACATGGAAGTCAGATC), corresponding to a sequence internal to Tn5, or primer M13F (GTAAAACGACGGCCAG). Chromosomal DNAs from RL60, RL61, and RL134 were subjected to PCR with primers IS50RL and tar1 (CTGGCGGAAGCATAACGGTG). PCR products were TA cloned into DH5α and sequenced with primers M13F and M13R (CAGGAAACAGCTATGAC). Sequencing was performed with the PRISM ready reaction dideoxy terminator cycle sequencing kit and the model 373A DNA sequencing system (Applied Biosystems).
Isolation of fliA36::Tn5B50.
Pools of EE251::Tn5B50 mutants were generated as previously described (54). Tn5B50 mutations were transduced into CL87, and transductants were selected on LB agar supplemented with 10 μg of tetracycline per ml and 10 mM EGTA. Mutants were screened on motility agar supplemented with 10 μg of tetracycline per ml and 10 mM EGTA. Nonmotile mutants were examined for flagella and tested for β-galactosidase activity. Tn5B50 insertions in nonmotile mutants with decreased iagB87::lacZY expression were tested for linkage to fliA51::Tn5 by P22 transduction. RL241 was confirmed to have a Tn5B50 insertion in fliA by PCR with primers at the 5′ and 3′ ends of fliA, fliA1 (GGCGCTACAGGTTACATAAG) and fliA2 (TAGTCTATACGTTGTGCGGC), respectively.
RESULTS
Reporter fusion strains.
To monitor the effects of mutations on hilA and invasion gene expression, we utilized the iagB87::lacZY fusion strain CL87 and the sipA4::Tn5lacZY fusion strain EE633. iagB is a gene downstream of hilA encoding a product of unknown function, and sipA encodes a secreted factor whose expression is dependent on HilA (Fig. 1). iagB is thought to be cotranscribed with hilA, since hilA339::kan abolishes iagB87::lacZY expression, even in the presence of plasmids expressing hilA or hilA and iagB (data not shown). Thus, iagB87::lacZY and sipA4::Tn5lacZY are used as reporters of hilA and hilA-dependent invasion gene expression, respectively (9, 48). Both CL87 and EE633 are able to invade HEp-2 cells as efficiently as their wild-type parent, SL1344 (C. A. Lee, unpublished observations).
Isolation of mutants.
To isolate mutants with reduced hilA and invasion gene expression, we transduced Tn5 insertions from our pools of Tn5-mutagenized EE251 into CL87 and EE633. Among approximately 9,000 transductants, we chose 36 exhibiting a lactose-negative (pink or white) phenotype on MacLac agar. Of these, we confirmed that 3 CL87::Tn5 mutants had decreased iagB87::lacZY expression, and 25 EE633::Tn5 mutants had decreased sipA4::Tn5lacZY expression by β-galactosidase assays. Two mutants completely lacking β-galactosidase activity had lost the sipA4::Tn5lacZY reporter fusion during transduction of the Tn5 mutations into EE633 due to linkage between the Tn5 and sipA. We examined the remaining 26 mutants for their ability to invade HEp-2 cells, and, since loss of motility results in decreased invasiveness (50), we tested noninvasive mutants for motility by microscopy and motility agar assays. To determine linkage of the mutations to SPI1 and to test their effects on other invasion genes, we transduced the kanamycin-resistant Tn5 insertions into strains carrying various invasion gene Tn5lacZY reporter fusions encoding tetracycline resistance. In two cases, the phenotypes of the mutants did not cotransduce with the Tn5 insertions, suggesting that the Tn5 mutations are not responsible for decreased sipA4::Tn5lacZY expression in those mutants.
Based on these data, we have classified 3 CL87::Tn5 mutants with decreased iagB expression and 16 EE633::Tn5 mutants with decreased sipA expression into three different groups: (i) 7 motile mutants with Tn5 insertions linked to SPI1, (ii) 7 motile mutants containing Tn5 insertions unlinked to SPI1, and (iii) 5 nonmotile mutants with Tn5 insertions unlinked to SPI1.
Characterization of SPI1-linked Tn5 insertions.
Seven EE633::Tn5 mutants exhibiting 2- to 7-fold decreases in sipA4::Tn5lacZY expression and 50- to >100-fold reductions in invasion of HEp-2 cells contained Tn5 insertions linked to SPI1. One of these insertions was closely linked to sipA4::Tn5lacZY and was not further characterized. Six other insertions were found to be >69% linked to invG, a gene immediately downstream of invF. One of them, invF-29::Tn5, was also tested for linkage to invF11-5::Tn5lacZY and was found to be 100% linked. Since disruptions in invF are known to cause strong defects in invasion and reduced expression of certain hilA-dependent genes (23, 29), the invG-linked Tn5 insertions isolated in our screen are likely to decrease expression of sipA by reducing or abolishing invF expression. Such mutations are expected to decrease expression of sipC, but not hilA or other hilA-dependent genes (Fig. 1). In support of this, we found that invF-29::Tn5 reduced expression of sipA and sipC, but not hilA, orgA, prgH, or prgK (data not shown). Other invG-linked insertions were not further analyzed.
Identification of motile mutants with Tn5 insertions unlinked to SPI1.
One motile CL87::Tn5 mutant with a fivefold decrease in iagB87::lacZY expression and six motile EE633::Tn5 mutants with two- to fourfold decreases in sipA4::Tn5lacZY expression exhibited less-than-sevenfold reductions in invasion of HEp-2 cells and contained Tn5 insertions unlinked to SPI1. Based on sequencing of the Tn5 insertion site junctions, two of these appear to be in the S. enterica serovar Typhimurium pstS and fadD genes. The region flanking pstS55::Tn5 is predicted to encode amino acids that are 93% identical to PstS from E. coli. The insertion site corresponds to a location 798 bp downstream of the translational start site for E. coli pstS. The region flanking fadD1::Tn5 is predicted to encode amino acids that are 75% identical to E. coli FadD. Its insertion site corresponds to a location 125 bp downstream of the translational start site for E. coli fadD. The locations of the other five Tn5 insertions in this group have not yet been determined.
Identification of nonmotile mutants.
Two CL87::Tn5 mutants with 2- to 4-fold decreases in iagB87::lacZY expression and three EE633::Tn5 mutants with 2- to 4-fold decreases in sipA4::Tn5lacZY expression exhibited 50- to 100-fold reductions in invasion of HEp-2 cells and had Tn5 insertions unlinked to SPI1. These mutants were nonmotile, as determined by swarming phenotypes on motility agar plates, and staining results indicated that they lacked flagella (data not shown). Thus, the strong invasion defects of these mutants are most likely due to loss of motility. Based on DNA sequencing, the fliA51::Tn5 mutant has an insertion in S. enterica serovar Typhimurium fliA 63 bp downstream of the translational start site.
The four remaining nonmotile mutants had Tn5 insertions >57% linked to tar::Tn10. For three of these mutants, the regions between tar and Tn5 were amplified by PCR, cloned, and sequenced. Two lie in flhC, and one lies in flhD. The flhC4::Tn5 and flhC36::Tn5 insertions lie 119 and 101 bp downstream of the S. enterica serovar Typhimurium flhC translational start site, respectively. The flhD37::Tn5 insertion lies 303 bp downstream of the S. enterica serovar Typhimurium flhD translational start site. zec57::Tn5 is 57.5% linked to tar, but its insertion site has not yet been identified.
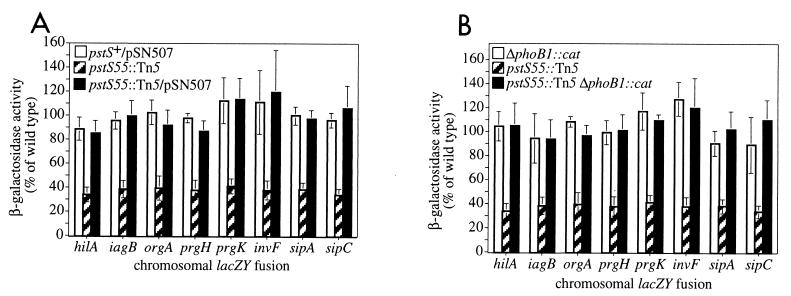
Characterization of pstS55::Tn5 mutant.
The pstS55::Tn5 mutation reduced expression of hilA and invasion genes two- to threefold, and this defect was complemented by pSN507, a pBR322 derivative containing the E. coli pstSCAB-phoU operon (Fig. 2A). pBR322 had no effect on hilA or invasion gene expression (data not shown). In E. coli, the pstSCAB-phoU operon encodes a high-affinity inorganic phosphate (Pi) uptake system (83). In addition to its role in importing Pi, the Pst system is required for negative control of the PhoR-PhoB two-component regulatory system. When extracellular Pi levels are low (<4 μM), PhoR phosphorylates the transcriptional regulator PhoB, activating expression of the phosphate (Pho) regulon (see Fig. 6). When Pi levels are high (>4 μM), PhoR acts as a phosphatase and dephosphorylates PhoB∼P, inhibiting expression of the Pho regulon. In the absence of the Pst system, PhoB∼P accumulates even in the presence of high Pi levels. Thus, the pstS55::Tn5 mutation is expected both to abolish the high-affinity Pst system and lead to accumulation of PhoB∼P.
FIG. 2.
pstS55::Tn5 reduces hilA and invasion gene expression in a phoB-dependent manner. (A) Complementation of pstS55::Tn5 by pSN507, a plasmid containing E. coli pstSCAB-phoU. (B) ΔphoB1::cat suppresses effects of pstS55::Tn5 on hilA and invasion gene expression. β-Galactosidase activity for each fusion is expressed as a percentage of its activity in a wild-type SL1344 background. Average percentages were calculated by using three or more values from at least two different experiments. Error bars represent the standard deviation of normalized values. Typical β-galactosidase activities (Miller units [U]) for fusions in a wild-type background were as follows: hilA080::Tn5lacZY, 710 U; iagB87::lacZY, 637 U; orgA::Tn5lacZY, 2,331 U; prgH020::Tn5lacZY, 1,577 U; prgK100::Tn5lacZY, 3,157 U; invF11-5::Tn5lacZY, 2,663 U; sipA4::Tn5lacZY, 1,836 U; sipC11-6::Tn5lacZY, 5,233 U.
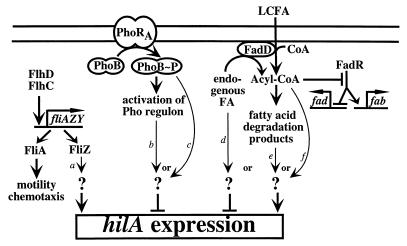
FIG. 6.
Model for regulation of hilA expression by three distinct pathways identified in this study. Thin lines represent potential mechanisms for regulation of hilA by each pathway: a, FliZ induces the expression or activity of an activator of hilA expression; b or c, a gene product in the Pho regulon (b) or PhoB∼P (c) activates expression or activity of a repressor of hilA expression; d, accumulation of an endogenous precursor of fatty acid metabolism activates expression or activity of a repressor of hilA expression; e or f, alternatively, a fatty acid degradation product (e) or acyl-CoA (f) activates the expression or activity of an activator of hilA expression. Other models for regulation of hilA by these pathways are possible.
To distinguish whether loss of the Pst transporter or accumulation of PhoB∼P is responsible for reduced hilA expression in the pstS55::Tn5 mutant, we tested the effect of ΔphoB1::cat. In the presence of pstS55::Tn5, ΔphoB1::cat restored hilA and invasion gene expression to wild-type levels (Fig. 2B). These data suggest that repression of hilA and invasion genes by pstS55::Tn5 is mediated by PhoB and that the PhoR-PhoB two-component regulatory system has the ability to directly or indirectly regulate hilA and invasion gene expression. As expected, ΔphoB1::cat had no effect on hilA or invasion gene expression when the Pst system was intact (Fig. 2B). Under our growth conditions (i.e., rich medium), Pi levels are not limiting and PhoR is able to dephosphorylate PhoB in the presence of an intact Pst system, preventing any accumulation of PhoB∼P.
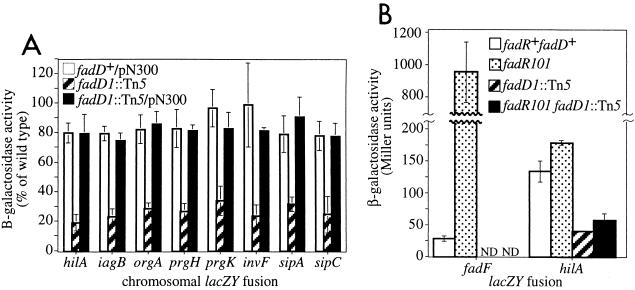
Characterization of fadD1::Tn5 mutant.
fadD1::Tn5 reduced expression of hilA and invasion genes three- to fivefold, and this defect was complemented by pN300, a pACYC177 derivative containing E. coli fadD (Fig. 3A). pACYC177 had no effect on invasion gene expression (data not shown). In E. coli, fadD encodes acyl coenzyme A (CoA) synthetase, a protein required for import of long-chain fatty acids (LCFA) and for activation of LCFA with CoA prior to the first step in β-oxidation (13). As expected, fadD1::Tn5 mutants were unable to grow on LCFA as the sole carbon source (data not shown).
FIG. 3.
fadD1::Tn5 reduces hilA and invasion gene expression in a fadR-independent manner. (A) Complementation of fadD1::Tn5 by pN300, a plasmid containing E. coli fadD. β-Galactosidase activity for each fusion is expressed as a percentage of its activity in a wild-type SL1344 background. Average percentages were calculated by using three or more values from at least two different experiments. Error bars represent the standard deviation of normalized values. (B) fadR101 allows expression of fadF103::MudJ but does not suppress the effect of fadD1::Tn5 on hilA080::Tn5lacZY expression. Reporter fusions and fadD1::Tn5 were P22 transduced into TR6583 and LS1860. β-Galactosidase assays were performed on cultures grown in LB medium under oxygen-limiting conditions. β-Galactosidase activity is expressed as Miller units. Averages were calculated by using four or more values from at least two different experiments. Error bars represent the standard deviations. ND, not determined.
In E. coli, acyl-CoA modulates the activity of the transcriptional regulator, FadR (27, 28). In the absence of LCFA, FadR activates expression of fatty acid biosynthesis (fab) genes and represses expression of fatty acid degradation (fad) genes. In the presence of LCFA, however, long-chain fatty acyl-CoA (LCFACoA) is produced in a fadD-dependent manner and binds to FadR, preventing it from binding to the fab and fad promoters (Fig. 6). In fadD mutants, LCFA cannot be imported and LCFACoA is no longer produced, allowing FadR to bind to the fab and fad promoters even in the presence of exogenous LCFA.
Recent studies suggest that FadR activity is also inhibited by LCFACoA in S. enterica serovar Typhimurium (78). Since loss of fadD abolishes production of LCFACoA, we speculated that decreased hilA and invasion gene expression in the fadD mutant might be due to repression of hilA by FadR. If so, a mutation in fadR should suppress the effect of the fadD mutation on hilA expression. To test this, we transduced hilA080::Tn5lacZY and fadD1::Tn5 into wild-type or fadR101 LT2 strains. As a control, we used fadF103::MudJ, a lacZY fusion known to be repressed by FadR (78). fadF was repressed under our conditions, and, as expected, this repression was alleviated by the mutation in fadR (Fig. 3B). In contrast, hilA expression was relatively high under the same conditions. fadD1::Tn5 reduced hilA expression in the LT2 strain, but this decrease was not suppressed by the fadR mutation. These results indicate that fadD1::Tn5 decreases hilA and invasion gene expression by a fadR-independent mechanism.
Characterization of fliA51::Tn5 and flhC4::Tn5.
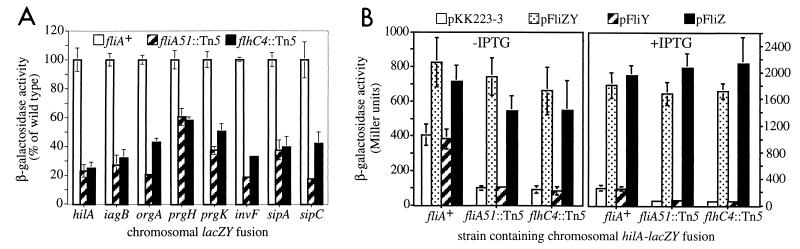
In E. coli and S. enterica serovar Typhimurium flhC and flhD encode master regulators of flagellar genes. FlhC and FlhD form heterotetramers to activate expression of operons encoding the flagellar basal body and FliA (56). FliA is an alternate sigma factor required for the expression of operons encoding flagellin, chemotaxis machinery, and the flagellar motor (64). flhC4::Tn5 and fliA51::Tn5 had comparable effects on invasion gene expression (Fig. 4A), and both abolished swarming phenotypes on motility agar plates (Table 2). As expected, addition of pSIIA1, a plasmid expressing fliA from the tac promoter, restored motility to the fliA51::Tn5 mutant but not the flhC4::Tn5 mutant in the presence of IPTG (Table 2). However, pSIIA1 did not complement the effect of fliA51::Tn5 on hilA expression (data not shown), suggesting that loss of FliA itself is not responsible for decreased hilA and invasion gene expression in this mutant.
FIG. 4.
(A) fliA51::Tn5 and flhC4::Tn5 reduce hilA and invasion gene expression. β-Galactosidase activity for each fusion is expressed as a percentage of its activity in a wild-type SL1344 background. Average percentages were calculated by using at least two values. Error bars represent the standard deviation of normalized values. (B) Effects of fliA51::Tn5 and flhC4::Tn5 on hilA expression are complemented by expression of E. coli fliZ. pFliZ is pDSM29, pFliY is pDSM30, and pFliZY is pDSM32. fliZ, fliY, or fliZY expression was induced by growing cultures in LB medium containing 10 mM IPTG under oxygen-limiting conditions. β-Galactosidase activity is expressed as Miller units. Averages were calculated by using four or more values from at least two different experiments. Error bars represent the standard deviations.
TABLE 2.
Swarming phenotypes on motility agar
| Plasmid | Plasmid genotype | Strain genotype
|
||
|---|---|---|---|---|
| fliA+ | fliA51::Tn5 | flhC4::Tn5 | ||
| pKK223-3 | Vector | + | − | − |
| pDSM29 | fliZ+ | + | − | − |
| pTrc99c | Vector | + | − | − |
| pSIIA1 | fliA+ | + | + | − |
In both E. coli and S. typhimurium, fliA is in an operon with two downstream genes of unknown function, fliZ and fliY (46, 62). Since flhDC is required for expression of fliA and since fliZ and fliY are cotranscribed with fliA (Fig. 6), we postulated that the effects of flhC, flhD, and fliA mutations on hilA and invasion gene expression could be due to reduced or abolished expression of fliZ and/or fliY. We therefore examined the effects of E. coli fliZY, fliZ, or fliY on hilA expression. Even in the absence of IPTG, addition of plasmids expressing E. coli fliZY (pDSM32) or fliZ (pDSM29) from the tac promoter increased expression of hilA in a wild-type background and complemented the effects of fliA51::Tn5 and flhC4::Tn5 on hilA expression (Fig. 4B). In the presence of IPTG, these plasmids induced high-level expression of hilA, even in the fliA and flhC mutants. In contrast, the parent plasmid (pKK223-3) and a plasmid expressing E. coli fliY from the tac promoter (pDSM30) did not increase hilA expression in wild-type or mutant strains in the presence or absence of IPTG. As expected, the plasmid expressing E. coli fliZ from the tac promoter (pDSM29) did not restore motility to the fliA or flhC mutant in the presence of IPTG (Table 2). These results suggest that the effects of flhC4::Tn5 and fliA51::Tn5 on motility are due to reduced or abolished fliA expression, while their effects on hilA and invasion gene expression are due to decreased expression of fliZ.
Effects of double mutations on iagB87::lacZY expression and invasion of HEp-2 cells.
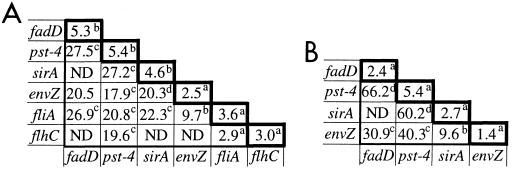
The results presented above show that multiple regulators appear to control hilA and invasion gene expression. We therefore tested whether they operate via common or different regulatory pathways. We predicted that mutations repressing hilA through the same pathway would not reduce hilA expression more when combined than when acting alone. In contrast, we expected that mutations reducing hilA expression by distinct regulatory pathways would decrease hilA expression much more when combined than when acting alone. We therefore used P22 transduction to combine mutations that reduce hilA expression and compared the effects of single and double mutations on iagB87::lacZY expression and invasion of HEp-2 cells.
In addition to the mutations identified in our screen, low osmolarity (9) and disruptions in sirA (1, 48) are known to reduce hilA expression. In E. coli and S. enterica serovar Typhimurium, the EnvZ-OmpR two-component regulatory system is thought to modulate expression of certain genes in response to changes in osmolarity (67). Thus, we also tested mutants containing envZ182::cam or sirA2::kan alone and in combination with mutations identified by our screen. Mutants with insertions in fliA and flhC were not tested in invasion assays, since nonmotility per se is known to greatly reduce invasiveness, even in strains that express hilA (50).
In order to construct certain double mutants, we obtained mutations with alternate antibiotic resistance markers in the pstSCAB-phoU operon and fliA. pst-4::Tn10 disrupts the pstSCAB-phoU operon and encodes tetracycline resistance (47). To obtain a mutation in fliA with a different marker, we transduced Tn5B50 insertions encoding tetracycline resistance into CL87 and screened for nonmotile mutants with decreased iagB87::lacZY expression. We then mapped these mutations relative to fliA51::Tn5 and identified a Tn5B50 insertion in fliA (data not shown).
Compared to pstS55::Tn5 (Fig. 2A), pst-4::Tn10 had a somewhat stronger effect on iagB expression (Fig. 5A). fliA36::Tn5B50's effect on iagB expression was comparable to that of fliA51::Tn5 and was complemented by pDSM29 (data not shown). Other single mutations, including envZ182::cam, mildly reduced iagB expression. However, with the exception of the fliA36::Tn5B50 flhC4::Tn5 mutant, double mutants exhibited much greater reductions in expression of iagB (Fig. 5A) than their single-mutant parents. Similarly, the single mutants invaded HEp-2 cells almost as well as CL87, while the double mutants exhibited strong invasion defects (Fig. 5B). Invasion assay results were confirmed by coverslip assays as described in Materials and Methods (data not shown). These data indicate that most of the mutations act independently of each other to reduce hilA expression and invasion of HEp-2 cells. For example, the mutation in sirA does not appear to reduce hilA expression by the same pathway as the mutation in envZ. In contrast, fliA36::Tn5B50 and flhC4::Tn5 appear to work in the same pathway to reduce hilA expression. The latter result is consistent with our model, in which flhDC is required for expression of the fliAZY operon and fliZ expression is required for maximal hilA and invasion gene expression.
FIG. 5.
Effects of single and double mutations on iagB87::lacZY expression and invasion of HEp-2 cells. The following mutations were used in these experiments: fadD1::Tn5, pst-4::Tn10, sirA2::kan, envZ182::cam, and flhC4::Tn5. fliA51::Tn5 was used in combination with the pst-4::Tn10 mutation, but all other fliA mutants contain fliA36::Tn5B50. (A) Fold reduction in iagB87::lacZY expression. Each value was calculated by dividing the β-galactosidase activity of CL87 (613 ± 100 Miller units) by the β-galactosidase activity (Miller units) of each mutant (n ≥ 4). a, standard deviation (SD) ≤ 0.6; b, SD < 2; c, SD ≤ 3.4; d, SD = 4.9. (B) Fold reduction in invasion indices. Values were calculated by dividing the invasion index of CL87 by the invasion index of each mutant (n ≥ 3). The invasion index for each strain was determined by measuring the percentage of bacterial inoculum surviving gentamicin treatment. a, SD ≤ 1.5; b, SD = 2.8; c, SD ≤ 18.1; d, SD ≤ 31.5. ND, not determined.
DISCUSSION
S. enterica serovar Typhimurium encounters many different environmental conditions and barriers at various sites during infection (73). After being ingested, the bacteria must survive the acidity of the stomach to reach the low-oxygen, hyperosmotic environment of the small intestine. There they must overcome the deleterious effects of defensins, secretory immunoglobulin A, bile salts, and pancreatic enzymes. To cause systemic infection, the bacteria must penetrate the intestinal epithelium and withstand the host's immune system. The latter process may be achieved in part by S. enterica serovar Typhimurium's ability to grow inside macrophages, where the bacteria must overcome exposure to cationic peptides, acid pH, and nutrient deprivation.
Although some genes may be important for S. enterica serovar Typhimurium's survival throughout infection, particular genes seem to be required to overcome unique challenges that bacteria face at specific sites. For example, SPI1 invasion genes are thought to play an important role in colonization and persistence in the intestine (R. A. Murray, unpublished observations), while certain genes on SPI2 (19, 43, 63) and SPI3 (14) appear to be important for survival in macrophages. In order to correlate expression of appropriate sets of genes with specific locations, S. enterica serovar Typhimurium may regulate expression of virulence genes in response to environmental signals found at critical sites during infection.
Fatty acids and virulence.
Genes involved in uptake and degradation of fatty acids may be important at various stages of infection, and many of these genes seem to be induced in vivo (42). For example, in a screen for in vivo-induced genes, Mahan et al. recovered a fadB fusion from the spleens of mice infected intraperitoneally with S. enterica serovar Typhimurium (57). fadB encodes an enzyme required for β-oxidation of LCFA (20). In a similar screen, a fadL fusion was isolated from a mouse intestine 24 h after intragastric inoculation with S. enterica serovar Typhimurium (D. M. Heithoff, C. P. Conner, and M. J. Mahan, unpublished results). fadL is an outer membrane transporter required for uptake of LCFA (11, 12). In addition, a fusion to fadF, a gene required for metabolism of medium-chain fatty acids and LCFA (20), was induced in cultured epithelial cells (78). Finally, a Fad− mutant has been shown to exhibit reduced virulence after oral inoculation in mice (80). Together, these results suggest that fatty acid degradation may be important for S. enterica serovar Typhimurium's survival throughout infection. However, the availability of fatty acids and the effects of these fatty acids on the bacteria's nutritional status, membrane composition, and levels of fatty acid intermediates may vary at different sites during infection. In turn, S. enterica serovar Typhimurium may utilize these variations to evaluate its environment and alter virulence gene expression accordingly.
Our studies with the fadD mutant suggest that loss of acyl-CoA synthetase represses hilA expression by a FadR-independent mechanism (Fig. 6). One explanation may be that LCFACoA regulates the activity or expression of some other transcriptional regulator that, in turn, modulates hilA expression. Another fatty acid-responsive transcriptional regulator called FarR has been identified in E. coli (68). FarR represses its own expression and the expression of genes encoding enzymes of the tricarboxylic acid cycle. Its ability to bind DNA in vitro is inhibited by saturated LCFA and their CoA derivatives. However, an S. enterica serovar Typhimurium FarR homolog is probably not responsible for repression of hilA in the fadD mutant. In our experiments, we found that fadF was repressed in a fadR-dependent manner, implying that LCFACoA levels were quite low under our conditions. In fact, no LCFA were added to the medium. hilA expression, however, was relatively high under these conditions in a fadD+ strain, suggesting that very small amounts of LCFACoA are sufficient for hilA expression. If the reduced expression of hilA in the fadD mutant is due to loss of LCFACoA itself, any fatty acid-responsive transcriptional regulator(s) involved would have to be more sensitive to changes in minute quantities of LCFACoA than FadR. This would seem to eliminate a FarR homolog as a candidate, since E. coli FarR is thought to be much less sensitive to LCFACoA than FadR (27, 28, 68). No fatty acid-responsive regulator with greater sensitivity to LCFACoA than FadR has yet been identified in E. coli or S. enterica serovar Typhimurium. However, it remains possible that another fatty acid-responsive regulator is expressed or activated only in the presence of our low-oxygen, hyperosmotic conditions and that this regulator is responsible for fadD-dependent regulation of hilA.
Another possibility is that some endogenous precursor or product of fatty acid metabolism regulates expression of hilA. Endogenous fatty acids produced by membrane turnover and fatty acid biosynthesis in E. coli are thought to be converted to acyl-CoA for degradation (27). Thus, even without exogenous LCFA, a low level of β-oxidation may occur under the growth conditions used in our study. The fadD mutation blocks degradation of fatty acids and may lead to accumulation of endogenous precursors that might somehow repress hilA expression. One candidate for such an activity is long-chain acyl-acyl carrier protein (acyl-ACP), an important intermediate of fatty acid biosynthesis responsible for feedback inhibition of fatty acid biosynthetic enzymes (27). Long-chain acyl-ACP is a substrate for synthesis of lipid A and phospholipids, and changes in its levels may affect the phospholipid composition of the cellular membrane. Such alterations are known to affect expression of several genes (27). Acyl-ACP has also been implicated in production of autoinducers (71), suggesting another link between this intermediate and gene regulation. Alternatively, a product of β-oxidation may activate hilA expression. However, in the absence of exogenous LCFA, the levels of intermediates in this pathway are extremely low (27). Thus, any transcriptional regulator modulating hilA expression in response to fatty acid degradation products would have to be extremely sensitive to such metabolites.
Finally, it may be that FadD produces short- or medium-chain fatty acyl-CoA derivatives which do not affect FadR but modulate a regulator of hilA. Significant quantities of short-chain fatty acids have been found in the intestinal lumen (22), and their CoA derivatives may act as signals inducing hilA expression. Thus, production of short-chain acyl-CoA may help to activate hilA and invasion gene expression in the intestinal lumen. Whatever the mechanism, our results link fatty acid metabolism with regulation of hilA, suggesting that S. enterica serovar Typhimurium may somehow monitor this pathway to coordinate expression of invasion genes during infection.
Motility and regulation of invasion genes.
Our results indicate that hilA and invasion genes are also regulated by expression of fliZ (Fig. 6). In addition, Eichelberg et al. have reported that invF expression is reduced by a fliA::Tn10 insertion in Salmonella enterica serovar Typhi (K. Eichelberg, K. Kaniga, and J. E. Galan, Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995, abstr. B-319, p. 221, 1995), and our results would suggest that this effect is due to reduced or abolished expression of fliZ. Although an in-frame deletion of fliZ is reported to have a mild effect on motility (46), fliZ's function is unknown, and its predicted product bears no significant homology to any known protein or structural motif. It seems unlikely that FliZ itself acts as a transcriptional regulator, since it contains no apparent DNA binding domain. A mutation in fliA that presumably abolishes fliZ expression does not significantly affect S. enterica serovar Typhimurium's ability to cause disease in mice (75), suggesting that FliZ is not required for virulence.
Because fliZ is cotranscribed with fliA in S. enterica serovar Typhimurium (46), the regulation of hilA by FliZ implies a link between motility and invasion gene expression. Interestingly, many other pathogenic organisms also appear to coordinate motility with expression of virulence factors. In Bordetella bronchiseptica, BvgAS activates expression of certain virulence factors, while inhibiting expression of flagella and motility (2, 3), and in Yersinia enterocolitica, both motility and expression of inv, which encodes an invasion factor, are maximal at 23°C (51). Temperature-dependent expression of inv and flagellin genes seems to be coordinately regulated at the level of transcription (7). In Vibrio cholerae, mutations resulting in nonmotile or hypermotile phenotypes also affect expression of virulence factors, including the toxin-coregulated pilus and cholera toxin (35). Our results indicate that S. enterica serovar Typhimurium motility per se is not required for hilA expression, and fliZ is not necessary for motility. However, environmental signals modulating fliAZY expression may coordinately regulate motility via FliA and expression of invasion genes via FliZ. The significance of this link between motility and invasion gene expression in S. enterica serovar Typhimurium virulence is unknown.
PhoPQ and PhoR-PhoB: differential regulation of virulence genes.
While this study implicates fadD and fliZ in regulating expression of only one set of virulence factors (i.e., invasion genes), evidence suggests that some regulatory pathways modulate expression of more than one set of virulence factors. One example of this is the model for regulation of virulence genes by PhoPQ. When extracellular cation levels are low, PhoPQ activates expression of PhoP-activated genes (pag), including genes on SPI2 (24) and SPI3 (14). Although it is unclear whether cation levels are the true signals for PhoPQ activation in vivo, many PhoP-activated genes are known to be induced in macrophages (81), implying that PhoPQ is active in that environment. However, PhoPQ is thought to inhibit hilA and invasion gene expression (9, 65), suggesting that these genes are repressed in macrophages. In turn, when exposed to conditions that inactivate PhoPQ, S. enterica serovar Typhimurium presumably expresses invasion genes while repressing PhoP-activated genes. Thus, PhoPQ may be inactive in the small intestine where invasion gene expression is thought to be important. PhoPQ's differential regulation of PhoP-activated gene and invasion gene expression evokes a model in which S. enterica serovar Typhimurium uses the same regulatory system to activate the expression of genes whose products are required at a particular site (such as the intestinal lumen or inside macrophages) while simultaneously turning off expression of genes whose products are not needed at that specific location.
An interesting parallel can be drawn between this model and the regulation of virulence genes by the PhoR-PhoB two-component regulatory system. Our results indicate that PhoB∼P directly or indirectly represses hilA and invasion genes, suggesting that low extracellular Pi levels may be capable of repressing hilA and invasion genes via PhoR-PhoB (Fig. 6). This leads to speculation that Pi levels are high in the intestinal lumen, allowing expression of invasion genes at that site. In contrast, Pi levels may be low in the macrophage. gfp fusion studies indicate that the pstSCAB-phoU operon is induced in macrophages (81), implying that Pi uptake and P assimilation are important for survival of S. enterica serovar Typhimurium in this environment. In addition, the pstSCAB-phoU operon is a member of the Pho regulon, and its induction in the macrophage is therefore probably mediated by PhoR-PhoB (83). This implies that Pi levels in the macrophage are low enough (<4 μM) to activate the PhoR sensor kinase, resulting in the accumulation of PhoB∼P. Thus, in response to low levels of Pi in the macrophage, PhoR-PhoB may reduce expression of genes whose products are no longer needed, including invasion genes, while turning on expression of genes that help S. enterica serovar Typhimurium survive the harsh conditions inside macrophages, including pstSCAB-phoU. In support of this model, Deiwick et al. have shown that SPI2 genes are activated by low Pi levels in vitro (24).
As with PhoPQ, it is unclear how PhoR-PhoB might regulate hilA expression. The E. coli Pho regulon consists of at least 31 genes involved in Pi uptake and P assimilation (83). PhoB∼P might repress hilA by modulating the expression of any one of these genes. Also, PhoB∼P may regulate SPI2 genes in response to Pi levels, and since mutations in certain SPI2 genes reduce hilA expression, the regulation of hilA by PhoB∼P may be an indirect effect resulting from a change in expression of those SPI2 genes. Alternatively, PhoB∼P might repress hilA expression through some unknown gene not yet identified as part of the Pho regulon, or it may repress hilA directly. Finally, it could be that an increase in PhoB∼P affects accumulation of an intracellular signal such as ppGpp, which may somehow reduce hilA expression (16, 77). Future studies will aim to distinguish between these possibilities.
Restricted gene expression and complex regulation.
The activation and repression of different virulence genes by PhoPQ and PhoR-PhoB raise an important point about S. enterica serovar Typhimurium pathogenesis. While expression of a particular set of genes at a specific site may be crucial for virulence, repression of a different set of genes at the same site may be equally important for survival of bacteria in the host. Mutations causing ectopic expression of virulence genes can greatly reduce S. enterica serovar Typhimurium's ability to cause disease in mice (41). The attenuation of such mutants may be due in part to premature host immune responses to antigens from inappropriately expressed virulence factors. Some ectopically produced proteins may interfere with other processes whose functions are required at certain sites during infection. Also, it may be important for S. enterica serovar Typhimurium's survival in the host to minimize the expenditure of energy and resources required for producing certain virulence determinants. For example, type III secretion systems are relatively complex structures, and it may be energetically unfavorable to produce them continuously. Thus, it may be necessary to restrict virulence gene expression to one or a few specific locations during infection.
Our results suggest that FliZ, FadD, PhoB, EnvZ, and SirA each affect hilA expression independently and act through distinct pathways. PhoPQ, SPI2 genes, and CsrA-CsrB may represent other independent pathways regulating hilA expression, and many more such pathways may await discovery. The regulation of hilA expression by multiple pathways may be necessary to ensure that SPI1-mediated type III secretion occurs only in the presence of specific environmental conditions representing the precise location(s) where this process is necessary. Several candidate regulators that may integrate signals from various regulatory pathways include HilC, HilD, the unknown repressor, and/or HilA itself.
Prior studies have demonstrated that hilA and invasion genes are severely repressed by high oxygen, low osmolarity, or the pho-24 mutation (9), suggesting that the expression of hilA (and therefore the expression of the secretion apparatus and its secreted effectors) is similar to an all-or-nothing response. However, mutations identified in this study have milder effects than the previously identified repressing environmental signals, and these mutations result in intermediate expression levels of hilA and invasion genes. This suggests that instead of simply being turned on or off, hilA can be modulated incrementally by different regulatory inputs. However, it is possible that the pathways implicated by these mutations really have much stronger effects on hilA expression under certain appropriate conditions.
Another possibility is that the strong repression of hilA by certain conditions involves two or more regulatory pathways. For example, the effect of the envZ mutation on hilA expression is fairly mild, suggesting that this regulatory system cannot fully account for the strong repression of hilA by low osmolarity. However, changes in osmolarity might regulate hilA expression through both EnvZ and some other independent pathway, and it may be necessary to abolish both pathways in order to observe the level of repression seen under low-osmolarity conditions. Thus, the regulation of hilA and invasion genes in response to individual environmental conditions may be more complex than previously thought.
Finally, it may be that hilA and invasion genes are incrementally regulated at certain sites within the host, as suggested by the effects of mutations identified in this screen. This type of regulation may be important if particular environmental conditions are the same at several critical locations during infection. By requiring more than one inhibiting signal, S. enterica serovar Typhimurium may have more flexibility, permitting intermediate levels of hilA and invasion gene expression at sites where one inhibiting condition is present. At the same time, such regulation would allow for inhibition of hilA and invasion gene expression when particular combinations of repressing conditions are encountered. In contrast, signals like high oxygen or low osmolarity that completely repress hilA and invasion genes may represent a very unique environment encountered by the bacteria at a specific site during infection. As previously suggested, a condition at one site in the host may be precisely the opposite of a condition found at another site, allowing differential regulation of distinct sets of virulence factors by a common pathway, such as PhoPQ. In such situations, an all-or-nothing response may be more efficient than incremental regulation. Whether incremental or absolute, the repression of hilA and invasion genes by mutations identified in this study adds to the growing list of regulatory pathways implicated in modulating hilA and invasion gene expression. Future studies will aim to determine the mechanisms whereby these pathways affect expression of hilA and may provide insights into how various signals are integrated to ultimately influence S. enterica serovar Typhimurium invasiveness.
ACKNOWLEDGMENTS
We are grateful to R. Langdon and S. Pardo-Reoyo for preliminary studies of the fadD and envZ mutants; K. Kutsukake, S. Lindgren, and B. Ahmer for sharing strains; and M. Mahan for sharing unpublished data. We also thank L. Schechter, R. Murray, and S. Akbar for critical reading of the manuscript.
This work was supported by American Heart Association grant-in-aid 96006780 (C.A.L.), the Albany Medical College Strategic Research Initiative (C.C.D.), NIH grant no. GM47628-01 (M.P.S.), and NIH Senior Fellowship F33 AI10093 (B.L.W.).
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B J, Miller J F. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol. 1993;175:3468–3479. doi: 10.1128/jb.175.11.3468-3479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akerley B J, Monack D M, Falkow S, Miller J F. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol. 1992;174:980–990. doi: 10.1128/jb.174.3.980-990.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol., in press. [DOI] [PubMed]
- 5.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 6.Amemura M, Shinagawa H, Makino K, Otsuji N, Nakata A. Cloning of and complementation tests with alkaline phosphatase regulatory genes (phoS and phoT) of Escherichia coli. J Bacteriol. 1982;152:692–701. doi: 10.1128/jb.152.2.692-701.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badger J L, Miller V L. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J Bacteriol. 1998;180:793–800. doi: 10.1128/jb.180.4.793-800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 9.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 10.Behlau I, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black P N. Characterization of FadL-specific fatty acid binding in Escherichia coli. Biochim Biophys Acta. 1990;1046:97–105. doi: 10.1016/0005-2760(90)90099-j. [DOI] [PubMed] [Google Scholar]
- 12.Black P N. The fadL gene product of Escherichia coli is an outer membrane protein required for uptake of long-chain fatty acids and involved in sensitivity to bacteriophage T2. J Bacteriol. 1988;170:2850–2854. doi: 10.1128/jb.170.6.2850-2854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black P N, DiRusso C C, Metzger A K, Heimert T L. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem. 1992;267:25513–25520. [PubMed] [Google Scholar]
- 14.Blanc-Potard A-B, Solomon F, Kayser J, Groisman E A. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullas L R, Ryu J-I. Salmonella typhimurium LT2 strains which are r−m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 17.Chen L M, Hobbie S, Galan J E. Requirement for CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 18.Chen L M, Kaniga K, Galan J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 19.Cirillo D, Valdivia R H, Monack D, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 20.Clark D P, Cronan J E., Jr . Two-carbon compounds and fatty acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 343–357. [Google Scholar]
- 21.Collazo C M, Galán J E. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 22.Cummings J H, Pomare E W, Branch W J, Naylor C P, Macfarlane G T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darwin K H, Miller V L. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol. 1999;181:4949–4954. doi: 10.1128/jb.181.16.4949-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 25.Deiwick J, Nikolaus T, Shea J E, Gleeson C, Holden D W, Hensel M. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol. 1998;180:4775–4780. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derman A I, Puziss J W, Bassford P J, Jr, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiRusso C C, Black P N, Weimar J D. Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog Lipid Res. 1999;38:129–197. doi: 10.1016/s0163-7827(98)00022-8. [DOI] [PubMed] [Google Scholar]
- 28.DiRusso C C, Heimert T L, Metzger A K. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. J Biol Chem. 1992;267:8685–8691. [PubMed] [Google Scholar]
- 29.Eichelberg K, Galán J E. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eichelberg K, Hardt W D, Galán J E. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol Microbiol. 1999;33:139–152. doi: 10.1046/j.1365-2958.1999.01458.x. [DOI] [PubMed] [Google Scholar]
- 31.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature. 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y, Galan J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 34.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gewirtz A T, Siber A M, Madara J L, McCormick B A. Orchestration of neutrophil movement by intestinal epithelial cells in response to Salmonella typhimurium can be uncoupled from bacterial internalization. Infect Immun. 1999;67:608–617. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunn J S, Hohmann E L, Miller S I. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardt W D, Galan J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardt W D, Urlaub H, Galan J E. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 42.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan.In vivo gene expression and the adaptive response: from pathogenesis to vaccines and antimicrobials. In H. Smith, C. J. Dorman, G. Dougan, D. W. Holden, and P. Williams (ed.), Royal Society Philosophical Transactions: Biological sciences, in press. The Royal Society, London, United Kingdom. [DOI] [PMC free article] [PubMed]
- 43.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F, Holden D W. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 44.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hobbie S, Chen L M, Davis R J, Galan J E. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 46.Ikebe T, Iyoda S, Kutsukake K. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology. 1999;145:1389–1396. doi: 10.1099/13500872-145-6-1389. [DOI] [PubMed] [Google Scholar]
- 47.Jiang W, Metcalf W W, Lee K-S, Wanner B L. Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J Bacteriol. 1995;177:6411–6421. doi: 10.1128/jb.177.22.6411-6421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnston C, Pegues D A, Hueck C J, Lee C A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 49.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones B D, Lee C A, Falkow S. Invasion of Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapatral V, Olson J W, Pepe J C, Miller V L, Minnich S A. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol. 1996;19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 52.Kutsukake K, Iino T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol. 1994;176:3598–3605. doi: 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu M Y, Yang H, Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol. 1995;177:2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCormick B A, Hofman P M, Kim J, Carnes D K, Miller S I, Madara J L. Surface attachment of Salmonella typhimurium to intestinal epithelia imprints the subepithelial matrix with gradients chemotactic for neutrophils. J Cell Biol. 1995;131:1599–1608. doi: 10.1083/jcb.131.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 60.Mills D M, Bajaj V, Lee C A. A 40kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 61.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mytelka D S, Chamberlin M J. Escherichia coli fliAZY operon. J Bacteriol. 1996;178:24–34. doi: 10.1128/jb.178.1.24-34.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 65.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 66.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 67.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 68.Quail M A, Dempsey C E, Guest J R. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 1994;356:183–187. doi: 10.1016/0014-5793(94)01264-4. [DOI] [PubMed] [Google Scholar]
- 69.Rakeman J L, Bonifield H R, Miller S I. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol. 1999;181:3096–3104. doi: 10.1128/jb.181.10.3096-3104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanderson K E, Stocker B A D. Salmonella typhimurium strains used in genetic analysis. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1220–1224. [Google Scholar]
- 71.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schechter L M, Damrauer S M, Lee C A. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 73.Schechter L M, Lee C A. Salmonella invasion of non-phagocytic cells. In: Oelschlaeger T A, Hacker J H, editors. Subcellular biochemistry. 33. Bacterial invasion into eukaryotic cells. New York, N.Y: Kluwer Academic/Plenum Publishers; 2000. pp. 289–320. [DOI] [PubMed] [Google Scholar]
- 74.Schiemann D A, Shope S R. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437–440. doi: 10.1128/iai.59.1.437-440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitt C K, Darnell S C, Tesh V L, Stocker B A D, O'Brien A D. Mutation of flgM attenuates virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J Bacteriol. 1994;176:368–377. doi: 10.1128/jb.176.2.368-377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 77.Spector M P. The starvation-stress response (SSR) of Salmonella. Adv Microb Physiol. 1998;40:233–279. doi: 10.1016/s0065-2911(08)60133-2. [DOI] [PubMed] [Google Scholar]
- 78.Spector M P, DiRusso C C, Pallen M J, Portillo F G D, Dougan G, Finlay B B. The medium/long-chain fatty acyl-CoA dehydrogenase (fadF) gene of Salmonella typhimurium is a phase 1 starvation-stress response (SSR) locus. Microbiology. 1999;145:15–31. doi: 10.1099/13500872-145-1-15. [DOI] [PubMed] [Google Scholar]
- 79.Tartera C, Metcalf E S. Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal epithelial cells. Infect Immun. 1993;61:3084–3089. doi: 10.1128/iai.61.7.3084-3089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Utley M, Franklin D P, Krogfelt K A, Laux D C, Cohen P S. A Salmonella typhimurium mutant unable to utilize fatty acids and citrate is avirulent and immunogenic in mice. FEMS Microbiol Lett. 1998;163:129–134. doi: 10.1111/j.1574-6968.1998.tb13036.x. [DOI] [PubMed] [Google Scholar]
- 81.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 82.Véscovi E G, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 83.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1357–1381. [Google Scholar]
- 84.Yang H, Liu M Y, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]