Abstract
Nanoparticle transport across tumor blood vessels is a key step in nanoparticle delivery to solid tumors. However, the specific pathways and mechanisms of this nanoparticle delivery process are not fully understood. Here, the biological and physical characteristics of the tumor vasculature and the tumor microenvironment are explored and how these features affect nanoparticle transport across tumor blood vessels is discussed. The biological and physical methods to deliver nanoparticles into tumors are reviewed and paracellular and transcellular nanoparticle transport pathways are explored. Understanding the underlying pathways and mechanisms of nanoparticle tumor delivery will inform the engineering of safer and more effective nanomedicines for clinical translation.
Keywords: cancer nanomedicine, drug delivery, endothelial cells, nanoparticles, transcellular transport, transcytosis, tumor vasculature
1. Introduction
There are over 8 million cancer related deaths worldwide each year with a projected increase in annual new cases.[1] As a result, there is a need for safe and effective treatments. The four cancer treatment strategies that are commonly used in the clinic are: i) cytoreductive surgery; ii) radiation therapy; iii) chemotherapy; and iv) immunotherapy.[2] Nanomedicine can be applied to each of these four treatment regimens at the preclinical and clinical stages. For example, nanoparticles have been applied in imaging guided surgery;[3,4] as agents for localizing heat or radiation to tumors and overcoming radiation resistance;[5–7] as clinically approved chemotherapeutic drugs, such as Doxil and Abraxane (R);[8] and in the development of safer and more effective immunotherapeutics.[9] However, to elicit clinical benefits, all of these strategies have a common need for efficient nanoparticle tumor delivery.
The most direct way to deliver nanoparticles into a solid tumor is by intratumoral injection.[10] While this approach may result in a high number of nanoparticles localized within the tumor, its usefulness and practicality are limited. For example, nanoparticles tend to distribute inhomogeneously throughout the tumor microenvironment upon local administration due to the relatively dense extracellular matrix that limits nanoparticle diffusion.[11] In addition, it may not be feasible to treat metastatic tumors with many neoplastic lesions throughout the body via local injections, meaning that systemic administration is required.[12]
Systemically administered nanoparticles have shown promise at both preclinical and clinical stages for diagnosis and treatment of cancer, however, there are several delivery barriers that nanoparticles need to overcome en route to solid tumors. Each delivery barrier is tied to the distinct phase of the nanoparticle’s journey to reach its destination, outlined in the so-called CAPIR cascade. This five-step cascade describes nanoparticles during: i) circulation throughout the blood stream; ii) accumulation in the tumor microenvironment; iii) penetration and distribution through tumor tissues; iv) internalization into tumor cells; and v) release of nanoparticle payloads.[13]
For systemically administered nanoparicles, the typical nanoparticle tumor delivery efficiency is ≈0.7% (median) of the injected nanoparticle dose.[14] As outlined in Figure 1, there are several reasons for this low nanoparticle delivery efficiency. Upon injection into the blood stream, nanoparticles are subject to proteins adsorbing onto their surfaces, forming what is known as a protein corona. The protein corona changes the nanoparticle physiochemical properties from a synthetic identity to a biological identity, which may affect nanoparticle pharmacokinetics, biodistribution, and toxicity.[15–18] Among these adsorbed proteins are opsonins, which may trigger phagocytosis in macrophages and other cells to swiftly remove circulating nanoparticles from the bloodstream.[19,20] Nanoparticle accumulation in off-target organs greatly reduces the number of nanoparticles in circulation.[21] As a result, there has been much research focused on increasing the nanoparticle blood circulation time by reducing nanoparticle interactions with serum proteins and immune cells. A common method to achieve this goal is coating nanoparticle surfaces with antifouling polymers, such as poly(ethylene glycol) (PEG).[22] Recently, nanoparticle surface modification with cellular membranes, such as membranes from red blood cells, has emerged as an alternative method to camouflage nanoparticles and to increase blood circulation times.[23] In addition, Chan and co-workers have shown that there is a nanoparticle dose threshold for nanoparticle clearance from circulation. By administering nanoparticle doses above the threshold (>1 trillion nanoparticles in mice), the nanoparticle uptake rates of liver phagocytes, such as Kupffer cells, can be overwhelmed to reduce liver clearance. This strategy has been reported to result in nanoparticle tumor delivery efficiencies of up to 12% of the injected dose, with nanoparticles being found within 93% of tumor cells.[24]
Figure 1.
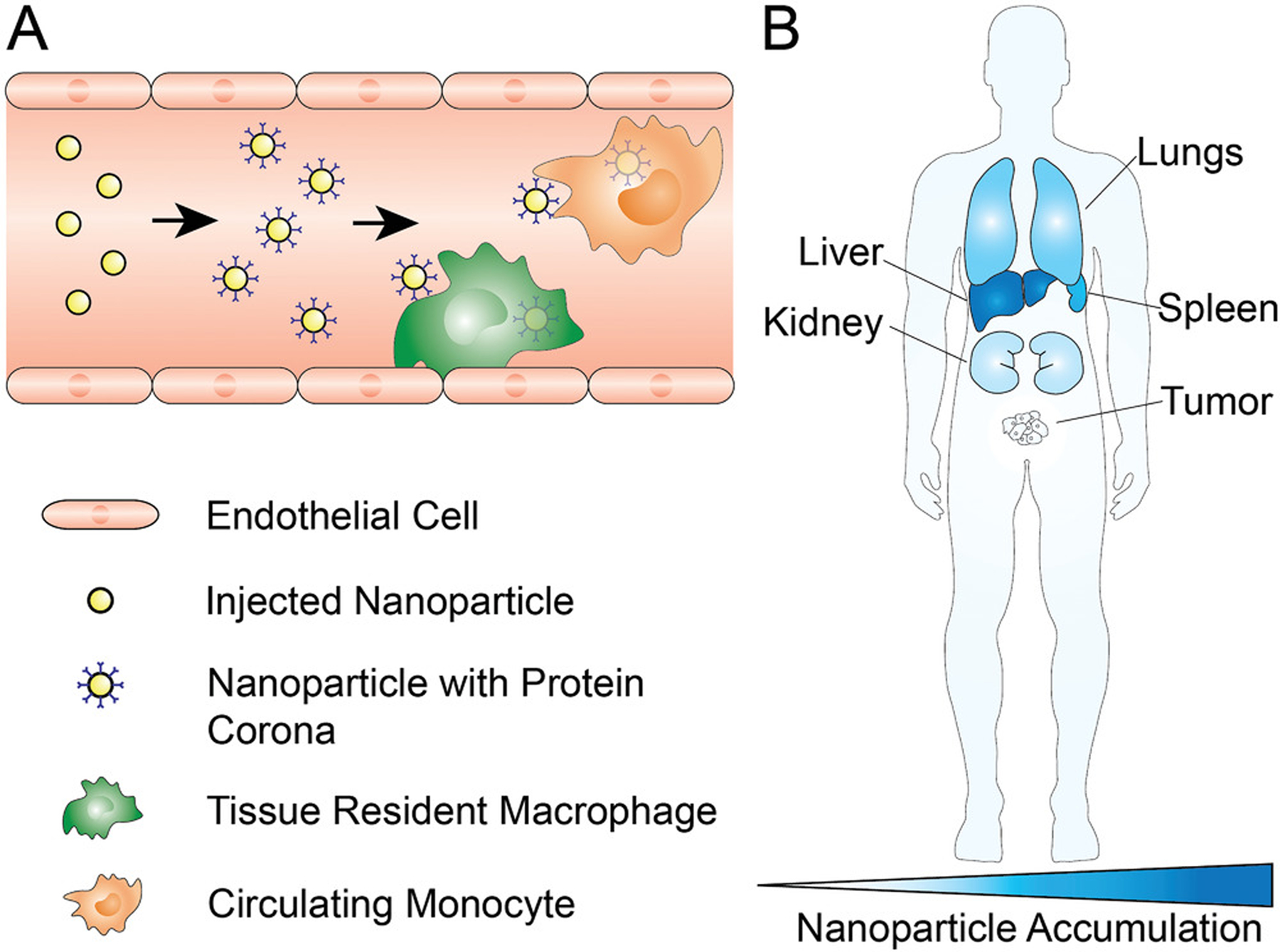
Systemic barriers to nanoparticle tumor delivery. A) After intravenous administration of nanoparticles, various serum proteins adsorb onto the nanoparticle surface and form a protein corona; among these proteins are opsonins, that trigger nanoparticle phagocytosis by immune cells such as circulating or tissue resident macrophages. B) Off target accumulation of nanoparticles in various organs results in fewer nanoparticles reaching the tumor microenvironment. Typically, the liver, spleen, and lungs sequester a large portion of administered nanoparticles. This accumulation is largely dependent on nanoparticle size and surface chemistry. Due to the filtration limit of kidneys being roughly 6 nm, larger nanoparticles do not greatly accumulate in kidneys, however, the kidneys have a much larger role in the accumulation and elimination of sub-6-nm nanoparticles.
The next barrier for nanoparticles is the tumor endothelium, and the transport from the tumor blood vessel lumen across the endothelium into the tumor microenvironment.[11,25] The longstanding paradigm of the enhanced permeability and retention (EPR) effect suggests that nanoparticles passively leak out from tumor vasculature between gaps in endothelial cells, coupled with poor lymphatic drainage of the tumor tissue.[26] Nanoparticle transport through leaky vasculature may occur through convection and diffusion, and may be limited by the increased interstitial fluid pressure observed in solid tumors.[27,28] In contrast to passive nanoparticle transport across tumor blood vessels as suggested by the EPR effect, transcytosis has been proposed as an active nanoparticle transport pathway since as early as the 1990s.[29] However, the contribution of nanoparticle transcytosis to the overall tumor accumulation had not been quantified. Recently, Chan and co-workers reported that only 3–25% of gold nanoparticles reach solid tumors by passive transport, depending on nanoparticle size, indicating that up to 75–97% of nanoparticles undergo active transcytosis transport. Interestingly, these studies were done with gold nanoparticle doses higher than the dose threshold for improved tumor delivery of ≈1 trillion nanoparticles in mice. The nanoparticle doses ranged from 2 × 1012 to 1 × 1014 nanoparticles, depending on size.[24,30] Nanoparticle transport mechanisms across tumor endothelium may be affected by changes in nanoparticle dose. More research is needed to determine how nanoparticle dose may alter extravasation mechanisms, pathways, and nanoparticle tumor delivery efficiency.
In this review, we discuss the specific properties of the tumor microenvironment and vasculature that need be considered for effective nanoparticle transport and tumor delivery. We review the common endocytic pathways that nanoparticles may undergo for transcellular transport across tumor endothelial cells, and how these endocytic pathways may be exploited by specific nanoparticle designs for delivering nanomedicines to solid tumors.
2. The Tumor Microenvironment and Vasculature
2.1. The Tumor Microenvironment
Solid tumors are generally composed of malignant parenchyma, and the surrounding benign tumor stroma.[14] Although the isolated stroma cannot form tumors when planted into host animals, it is essential in supporting tumor growth and architecture of the tumor microenvironment.[31] The tumor stroma is composed of diverse cell types, including caner-associated fibroblasts and immune cells. Cancer-associated fibroblasts produce and remodel the extracellular matrix (ECM), and at the same time secret growth factors that induce angiogenesis or suppress immune cells with the goal to support tumor growth.
Many types of immune cells are found in solid tumors and execute various functions. Briefly, CD8+ cytotoxic T Cells, CD4+ Th1 helper T cells, NK cells, M1 macrophages, and dendritic cells are generally considered as tumor inhibiting, while regulatory T cells (Treg), CD4+ Th2 helper T cells and M2 macrophages are immune suppressing, thus promoting angiogenesis, tumor growth, and metastasis.[32,33] Tumor and stromal cells are embedded in the ECM composed of collagen, fibronectin, fibrin, hyaluronan, and proteoglycans, which provide the mechanical support of the tumor microenvironment. At the same time, plenty of functional cytokines and growth factors, such as vascular endothelial growth factor (VEGF), disperse throughout the tumor forming the noncellular stroma together with the ECM. In addition, solid tumors are characterized by abnormal vasculature, low pH, hypoxia, high interstitial pressure, and crosstalk between individual tumor cell types.[11,34] All these components interplay in forming a complex tumor microenvironment and affect tumor development as well as treatment responses (Figure 2).
Figure 2.
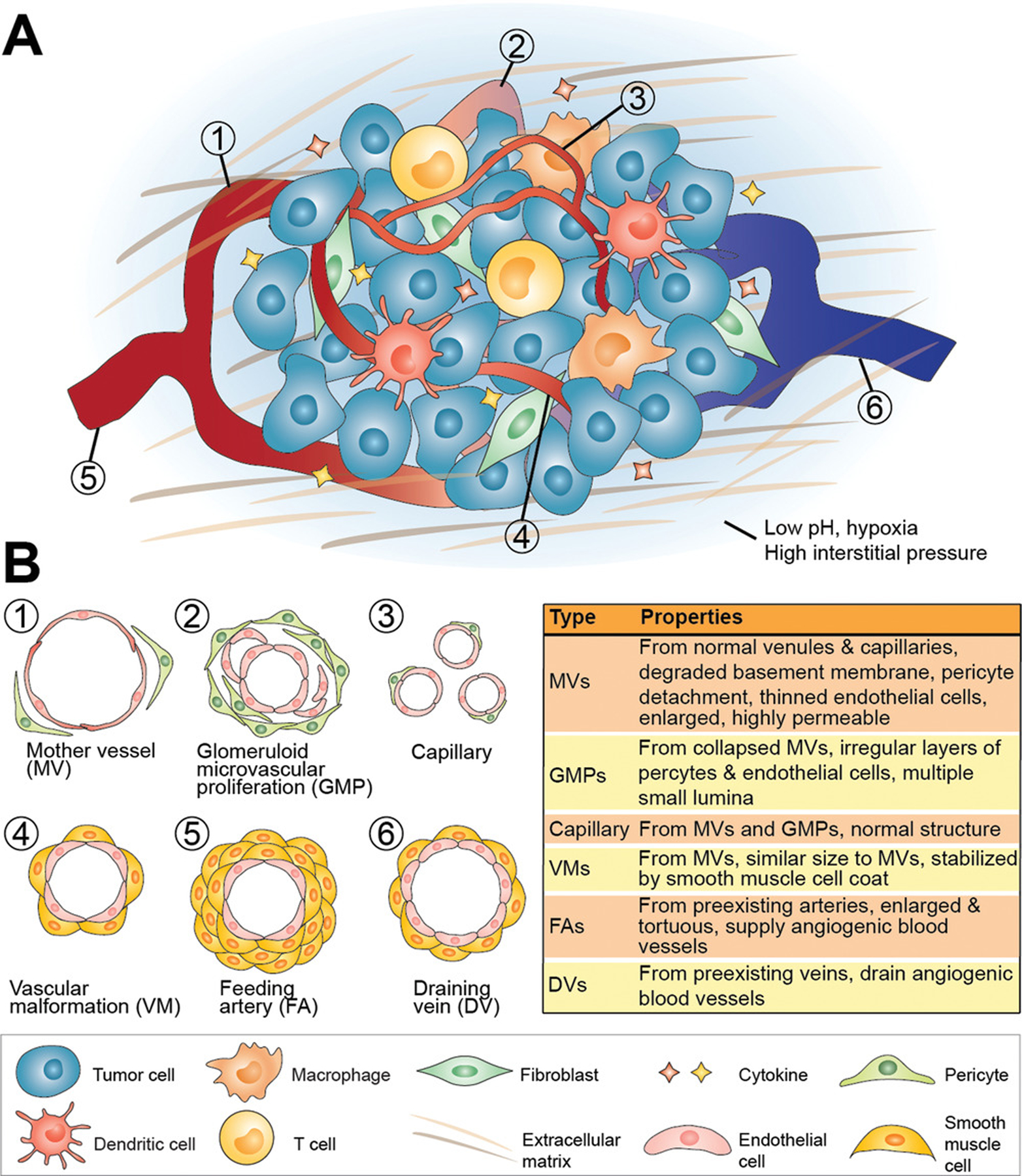
Tumor architecture. A) A solid tumor is composed of malignant parenchyma and benign tumor stroma that supports tumor growth and structure. There are diverse types of stromal cells including cancer-associated fibroblasts, immune cells, and other cells forming the cellular part of tumor stroma. The noncellular parts of the tumor stroma including extracellular matrix and cytokines surround and interact with the embedded cells. An abnormal vascular network is always observed in a solid tumor, which is essential for tumor supply. In addition, low pH, hypoxia, and high interstitial pressure are charateristics of solid tumors. All these tumor components interplay to form a complex microenvironment and drive the tumor development. B) The tumor vasculature is highly abnormal and at least six types of blood vessels with different characteristics have been distinguished.
Besides the complex composition of solid tumors, the phenotype and ratio of both tumor and stroma cells are highly heterogeneous between different patients, different loci within the same patient, and even different sites within the same tumor.[35,36] The tumor development is dynamic and at different development stages, the microenvironment shows variable characteristics.[37] For example, cell plasticity, i.e., the ability of tumor cells to transform and switch their phenotype, is a considerable challenge in the development of cell targeted therapies.[38,39] Characteristics of the tumor microenvironment, as well as its interactions with nanoparticles, have been reviewed in greater detail by Mukherjee and co-workers.[11]
2.2. Angiogenesis and Tumor Vasculature
Angiogenesis and neoangiogenesis are VEGF-dependent processes of forming new blood vessels from preexisting vessels to supply nutrients and oxygen to tumors for development and growth.[40] In some tumors, tissue growth is so fast that tumor cells are located relatively far away from blood vessels, which induces hypoxia, i.e., oxygen deprivation. Hypoxic cells then overexpress VEGF, leading to neoangiogenesis by recruitment of bone marrow derived endothelial progenitor cells to the tumor vascular bed, where these cells mature and release other proangiogenic growth factors.[11,41] The newly formed tumor blood vessels are known to lack some of the structural integrity that is seen in healthy blood vessels. For example, tumor blood vessels may exhibit gaps between endothelial cells, and smooth muscle cells, pericytes, and basement membrane may be missing or exhibit discontinuity as a result of an abnormal expression of certain growth factors, such as angiopoietin-1.[42]
Dvorak and co-workers described six distinct types of tumor blood vessels: i) mother vessels (MVs); ii) glomeruloid microvascular proliferations (GMPs); iii) capillaries; iv) vascular malformations (VMs); v) feeding arteries (FAs); and vi) draining veins (DVs) (Figure 2).[43] Mother vessels are the first angiogenic blood vessels to form from existing venules and capillaries after the degradation of basement membrane, which provides structural support, and the detachment of pericytes, which help control blood flow.[44] This process allows for blood vessel expansion through the intravascular hydrostatic pressure, given that the two aforementioned vessel features that prevent vessel growth are removed, resulting in a thinned and highly permeable endothelium. When MVs collapse, GMPs are formed that accumulate pericytes and macrophages, while also making new basement membrane. Alternatively, MVs can accumulate smooth muscle cells and perivascular collagen to become VMs, which effectively reduces their permeability. Through arteriovenogenesis, FAs and DVs are formed from existing healthy veins and arteries, to supply and drain blood to and from the other types of tumor blood vessels.[31]
These differences in blood vessel structure are important to note for nanomedicine delivery purposes, as nanoparticles may likely interact with each type of tumor blood vessel differently, which could result in varying nanoparticle delivery efficiencies throughout a single solid tumor. Such differences could be potentially exploited, however, by designing nanoparticles that specifically target features that are present in some types of tumor vessels but not others, such as pericytes,[45] for better nanoparticle tumor accumulation and distribution.
The intercellular gaps between endothelial cells in tumor blood vessels form the basis for nanoparticle extravasation according to the EPR effect. The EPR effect suggests that nanoparticles extravasate passively from tumor blood vessels into the tumor microenvironment by convection and diffusion through the leaky vasculature. In addition, it is suggested that the impaired lymphatic system within solid tumors reduces nanoparticle clearance.[26] The EPR effect has been a longstanding paradigm in cancer nanomedicine, and has been exploited as the main tumor delivery mechanism for different types of nanoparticles, including inorganic (such as noble metal, oxide, upconversion, and carbon-based nanoparticles) and organic nanoparticles (such as liposomes or lipid-based nanoparticles, polymeric nanoparticles and dendrimers).[25,46–53] However, it is not the only pathway for nanoparticles to cross tumor blood vessels, as depicted in Figure 3. In general, we can differentiate two main nanoparticle transport pathways: i) paracellular transport by diffusion through intercellular gaps; and ii) transcellular nanoparticle transport through tumor endothelial cells.
Figure 3.
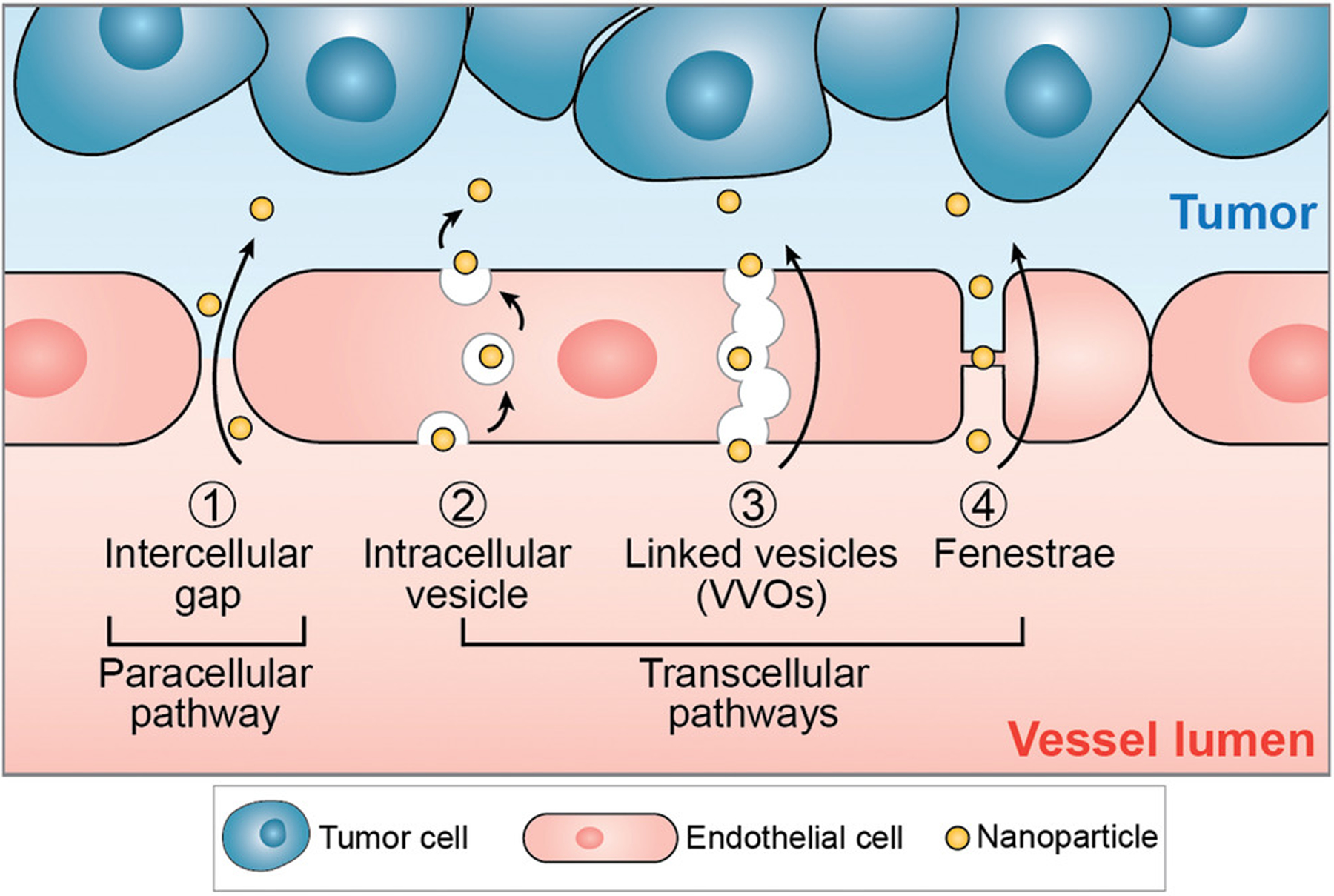
Nanoparticles can extravasate from tumor vascular lumen into the tumor microenvironment by both paracellular 1) and 2–4) transcellular pathways. For the paracellular pathway, nanoparticles transport passively through gaps in the endothelium, i.e., between adjacent endothelial cells. These intercellular gaps (up to 2 μm in size) result from the abnormal vessel structures caused by rapid tumor angiogenesis and are fundamental for the enhanced permeability and retention (EPR) effect. For transcellular pathways, nanoparticles get transported actively into the tumor microenvironment via intracellular vesicles or through transcellular pores. 2) When transported by intracellular vesicles, nanoparticles first enter the cell and locate in vesicles through endocytosis, then get transported across the cytoplasm, and finally exit the cell through exocytosis. 3) VVO and 4) fenestrae are both trans-endothelial pathways for nanoparticle transport. While VVOs are intracellular organelles composed of linked vesicles, fenestrae represent transcellular pores spanned by a fenestral diaphragm.
Transcellular nanoparticle transport is enabled by endocytic vesicles in tumor endothelial cells that deliver nanoparticles from the apical side of the cell to the basal side.[54] While endocytosis is the primary mechanism and pathway for transcellular transport, there are a few alternatives. One of these alternative transcellular pathways may be mediated by vesiculo-vacuolar organelles (VVOs) inside tumor endothelial cells. Little is known about this VVO-mediated transport pathway. However, VVOs have been characterized by Dvorak et al. as membrane-bound, linked vesicles and vacuoles that create a channel for macromolecules to cross the endothelium.[55] VVOs are rarely observed in cultured endothelial cells under standard culture methods and may occur at greater frequency in vivo.[56] Further work is needed to determine if the VVO-mediated pathway is a viable transport route for nanoparticles and nanomedicines. Another potential nanoparticle transport pathway is through fenestrae, i.e., transcellular pores that are typically found in liver sinusoidal and glomerular endothelial cells,[57,58] which have also been observed and documented in tumor vessels, for example in MVs and capillaries (Figure 3).[59] To probe and understand the nanoparticle transport mechanisms across tumor vasculature, the use of 3D microfluidic models that more truthfully recapitulate the tumor microenvironment in vitro may be of value.[60,61]
3. The Entry of Nanoparticles into the Tumor Microenvironment
3.1. Overview of Existing Paradigms
The field of cancer nanomedicine has gone through many advancements over the past decades, as summarized chronologically in Figure 4.[62] The introduction of liposomes and their later conjugation with antibodies for specific, active targeting, known as immunoliposomes, serve as major milestones in the origin of the field.[63–65] In the 1970s and 1980s, methods for improving nanoparticle tumor delivery were already underway, as noted by the discovery that locally heating a tumor can cause an increase in nanoparticle extravasation until blood vessel destruction occurs.[66,67] Reports of receptor-mediated endocytic nanoparticle uptake into tumor cells opened the door to the possibility of controlling cell specific nanoparticle delivery.[68]
Figure 4.
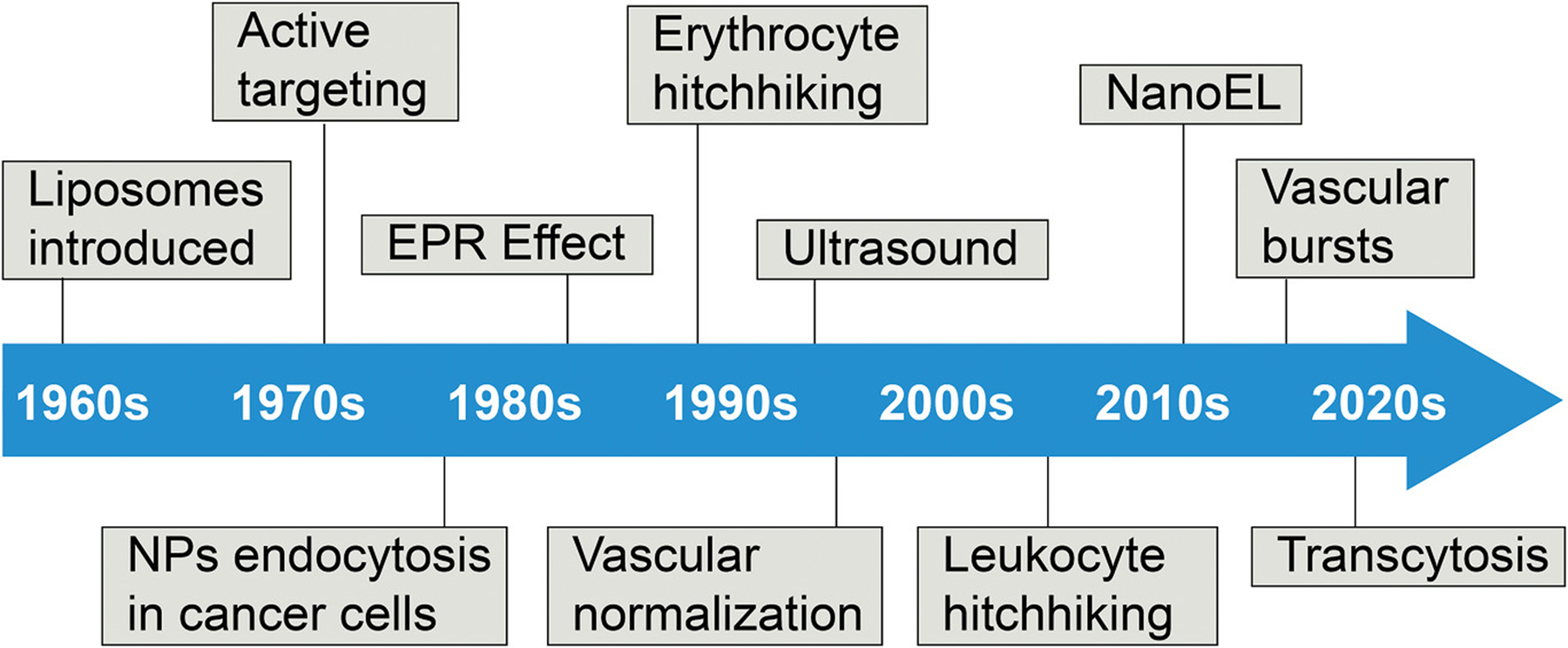
Timeline of different suggested nanoparticle–tumor accumulation pathways and methods. Since the first suggestion of nanotechnology, there have been many different pathways and methods suggested to explain and improve the extravasation of nanoparticles in to the TME.
The longstanding delivery paradigm in cancer nanomedicine, the EPR effect, was introduced in 1986 to explain that nanoparticles accumulate in tumors as a result of vascular leakiness and poor lymphatic drainage.[69] However, the well noted low nanoparticle tumor accumulation has brought the impact of the EPR effect into question.[14] Consequently, a variety of work has been done to find ways that improve nanoparticle tumor delivery and to understand the mechanisms behind it.
One of the earlier methods was erythrocyte hitchhiking, which involved removing erythrocytes from a patient, loading them with drugs, and re-administering them back into the patient.[70] This process has since evolved to have the removed erythrocytes conjugated to drug carrying nanoparticles, so that they would accumulate in the nearest downstream organ from the injection site.[71]
The concept of vascular normalization was then proposed as an extension of typical antiangiogenic treatments. These combined treatments aim to make the tumor vasculature more functionally similar to normal vasculature, resulting in a less constricted delivery of therapeutics to tumors. Antiangiogenic treatments are then applied to constrict the tumor vasculature to starve the tumor of nutrients needed for its survival.[72,73] It was recently shown that gold nanoparticles can accomplish this, along with inhibiting angiogenesis, by disrupting the signaling between tumor cells and endothelial cells.[74,75]
The use of ultrasound has been explored, as the tensile pressure of ultrasonic waves on tumors can cause blood vessel perforation and microconvection in the tumor interstitium, leading to higher nanoparticle extravasation.[76] Later studies have shown that ultrasound waves can be used to release drugs from liposomes[77] and mircobubbles, with the ability to convert the latter into nanobubbles.[78]
Several years later, leukocyte hitchhiking was discussed, and it was shown that antigen-specific T cells can be removed from the body and loaded ex vivo with nanoparticles, which will then target tumors upon re-administration into the body.[79] In addition, it was shown that intravenously administered nanoparticles can be phagocytosed by monocytes, allowing for photothermal therapy to be applied. These cells will then travel to the tumor microenvironment and differentiate into tumor-associated macrophages (TAMs) to migrate into the hypoxic tumor core, where near-IR irradiation can destroy the TAMs by heating the nanoparticles to destroy the surrounding tissue.[7] Recently, it was shown that photothermal therapy can be combined with vascular disruption agents that cause gold nanoparticles to aggregate at targeted locations, for an improved photothermal ablation of tumor cells.[80]
The methods and strategies described here are summarized in Table 1, along with current advances in their applications. Other methods, such as electroporation and the use of magnetic fields, are summarized in greater detail in a recently published review article by Mitragotri and co-workers.[81]
Table 1.
Examples of nanoparticle delivery mechanisms across tumor vasculature.
| Method | Year first described | Classification | Description | References |
|---|---|---|---|---|
| Active targeting liposomes | 1979 | Biological | Involves the attachment of specific antibodies or other molecules to the nanoparticle surface for targeting of complementary receptors on cancer cells. The tumor penetrating peptide, iRGD, and its derivatives, are widely used examples. | [64,65,202,203] |
| Heat treatment | 1979 | Physical | Local heating of tumors several degrees above the average temperature has been shown to significantly increase the extravasation of nanoparticles until vessel destruction occurs. Photosensitive and paramagnetic nanoparticles are being studied to improve these effects. | [66,67,204] |
| Erythrocyte hitchhiking | 1987 | Biological/cell mediated | This process has evolved from erythrocytes being removed from the patient and loaded with drugs and readministered to removed erythrocytes being conjugated with drug carrying nanoparticles and readministered to localize at the nearest downstream organ from the injection site. Work has also been done on coating nanoparticles with intact membranes from erythrocytes containing the typically expressed surface proteins for improved circulation. |
[23,70,71] |
| Vascular normalization | 1996 | Biological | This is an extension of typical antiangiogenic treatments which aim to make tumor vasculature more functionally similar to normal vasculature by removing excessive endothelial cells that make up immature blood vessels, for a less constricted delivery of therapeutics to tumors before cutting off the blood supply to the tumor. Antiangiogenic nanoparticles loaded with anticancer drugs are being explored as a method of simultaneously normalizing tumor vasculature and deliver cytotoxic drugs. | [72,73,205] |
| Ultrasound | 1999 | Physical | The tensile pressure from ultrasonic waves directed at tumors has been shown to cause blood vessel perforation via cavitation, as well as microconvection that results in higher extravasation. Ultrasound has also been used to cause the release of drugs such as doxorubicin from liposomes upon insonation, possibly through the formation of transient pores or other membrane defects |
[76,77] |
| Leukocyte hitchhiking | 2005 | Biological/cell mediated | Antigen-specific T cells can be removed and loaded with viral vectors, that once readministered, will target tumors and transfer the viral vectors. Intravenously administered nanoparticles can be phagocytosed by monocytes, which travel to the tumor microenvironment and differentiate into tumor-associated macrophages (TAMs) to migrate into the hypoxic tumor core. Near-IR irradiation can be directed toward the tumor, which may destroy the TAMs and heat up the nanoparticles to destroy surrounding tissues. The membranes of leukocytes can be coated onto nanoparticles in a similar manner as discussed above with erythrocytes, which could be applied to tumor targeting. |
[7,79,206] |
Significant work has further been done to describe the extravasation of nanoparticles from tumor vasculature based on the biological properties of endothelial cells. Leong and co-workers suggested that certain nanoparticles, such as titanium and gold nanoparticles, can induce the widening of gaps between endothelial cells by disrupting interactions between pairs of vascular endothelial cadherin, allowing for nanoparticles to leak out of the vasculature, in a process they named NanoEL.[82,83] A different paracellular pathway mechanism called vascular bursts was proposed by Kataoka and co-workers, who suggested that dynamic vents open and close at endothelial cell junctions, causing fluid to flow outward into the tumor interstitium and carrying nanoparticles with it.[84]
In a recent paper by Chan and co-workers, the contribution of paracellular nanoparticle transport across tumor blood vessels was quantified using a so-called Zombie model, a fixed tumor-bearing mouse model with blood artificially circulating with a peristaltic pump. Given that fixed cells cannot perform active transport, the only nanoparticles that could accumulate in a solid tumor were those that passively leaked from intercellular gaps. The passive paracellular transport pathway was found to only contribute to 3–25% of the total nanoparticle tumor accumulation seen in living control tumor-bearing mice.[30] Combined with transmission electron micrographs of nanoparticles inside intracellular vesicles within tumor endothelial cells, this study suggests that nanoparticles primarily take active transcellular routes to transport from tumor blood vessels into the tumor microenvironment.
3.2. Endocytosis Mechanisms of Tumor Endothelial Cells
For nanoparticles to transcytose across the tumor endothelium, they first need to endocytose into tumor endothelial cells. There are many different pathways that have been defined for endocytosis, but not all of them may be useful for transcytosis. The most common of these pathways are clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis (Figure 5). While caveolae-mediated endocytosis is the pathway that is most associated with transcytosis across endothelial barriers, clathrin-mediated endocytosis is well-noted as a transcytotic mechanism for crossing the blood–brain barrier. It has further been suggested that both clathrin-mediated and macropinocytosis contribute to blood vessel permeability.[85–87] Several groups have reported the endocytic cell uptake of various nanoparticles, with suggestions that nanoparticles take multiple different uptake routes.[88,89] Understanding the mechanisms behind these endocytosis pathways will allow for targeting of specific transport routes to deliver nanomedicines more efficiently into tumors.
Figure 5.
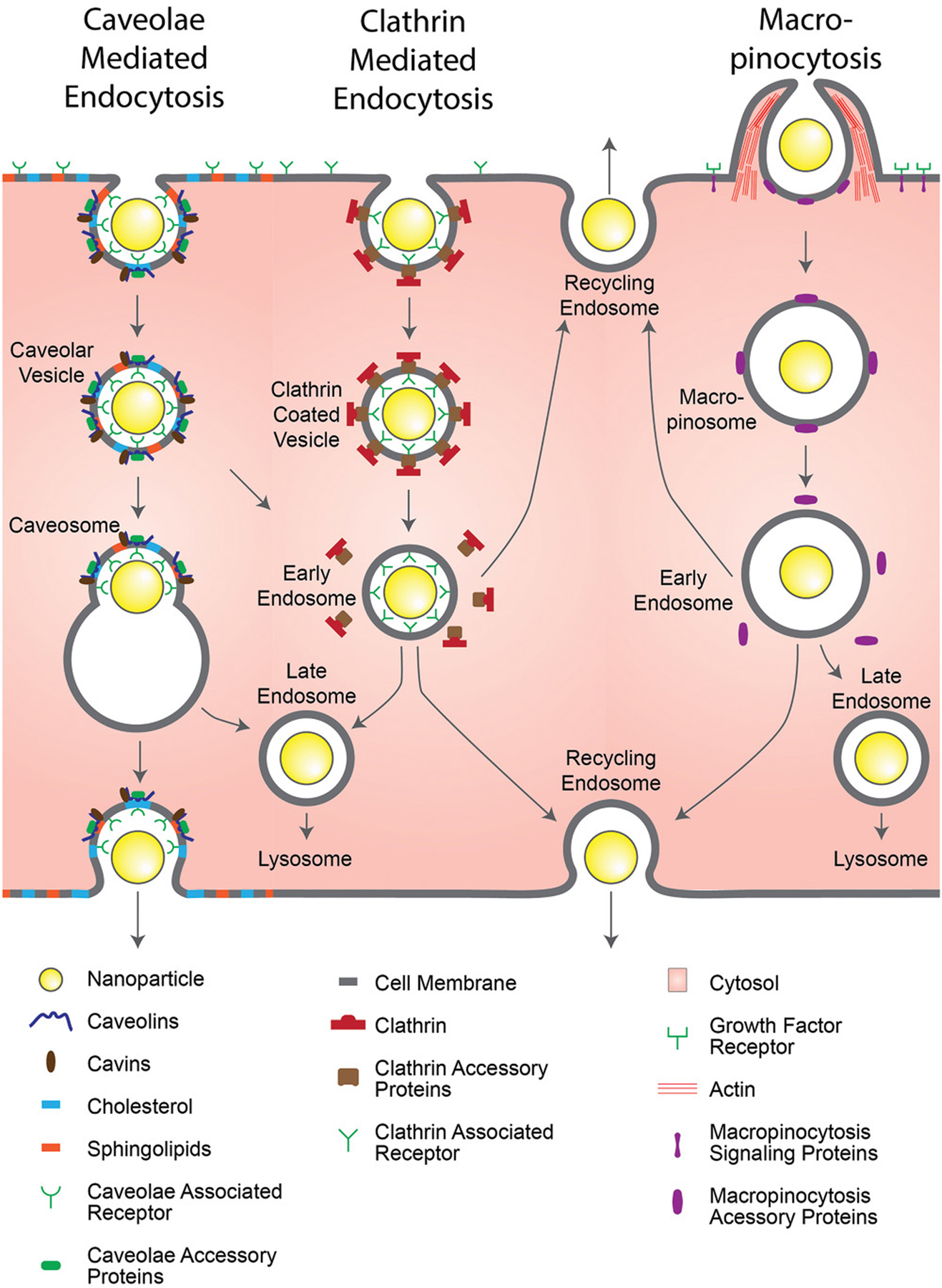
Common endocytic pathway mechanisms in endothelial cells. The specific pathways that nanoparticles take to enter endothelial cells vary, with the receptors that trigger the pathways as well as the nanoparticles’ destination varying with the pathways themselves. Cell membrane invaginations are a typical occurrence for the receptor-mediated endocytic pathways of caveolae-mediated endocytosis and clathrin-mediated endocytosis, however, caveolar vesicles are typically trafficked to the endoplasmic reticulum before being exocytosed, while clathrin coated vesicles are typically trafficked to lysosomes for degradation. The growth factor triggered macropinocytosis involves a heavy actin remodeling to nonspecifically engulf fluid in the area, packaging any contents in to a macropinosomes, which are also typically trafficked to lysosomes for degradation.
3.2.1. Clathrin-Mediated Endocytosis
Clathrin-mediated endocytosis is a receptor specific form of endocytosis that uses vesicles coated with the triskelion protein, clathrin, to internalize materials that bind to its surface receptors.[90] Clathrin does not directly bind to the cell membrane or its specific receptors, and as such, requires several other proteins for binding and vesicle formation.[91] Specific proteins of note are the adaptor protein complex-2 (AP-2) complex, which serves as an intermediate between the cell membrane and clathrin,[92] the clathrin assembly lymphoid myeloid leukemia protein (CALM), which helps control vesicle size,[93] and dynamin, which regulates the maturation of clathrin coated pits and also catalyzes the snipping of the vesicle from the membrane.[94] The vesicles formed in this process are typically sized at around 80–100 nm. However, it has been shown that nanoparticles (522 nm in size) conjugated with transferrin, a clathrin-mediated endocytosis tracer, have been uptaken by clathrin coated vesicles in HeLa cells, indicating that there is a potential variability in the vesicle size.[93,95]
Following the snipping of the vesicles, the clathrin coat is disassembled, allowing the removed proteins to be reused for other clathrin-mediated endocytosis events.[96] At this stage, the vesicles are sorted based on their ligand and receptor contents to early endosomes for trafficking to either late endosomes and are transported to lysosomes for degradation (seen with the epidermal growth factor), or are recycled back to the membrane with the contents exocytosed (seen with the transferrin).[97–99] The recycling endosome has been shown to traffic to either the apical surface or the basal/basolateral surfaces in other cell types such as blood–brain barrier endothelial cells and epithelial cells, which could be useful as a transcytotic pathway for nanoparticle tumor delivery.[100,101] Clathrin-mediated endocytosis has been of particular interest for blood–brain barrier permeability,[102,103] though it may still be relevant for nanoparticle extravasation in tumor vasculature. This concept is evidenced by Bendas and co-workers, who used liposomes conjugated with antibodies against vascular cell adhesion molecule 1 (VCAM-1), which is expressed on activated tumor endothelial cells, to access a clathrin-mediated uptake pathway in a mouse xenograft tumor model (Colo677—human lung cancer).[104] Further research is needed to determine if a basal recycling endosome pathway can be exploited for the transcytotic delivery of nanomedicine across tumor blood vessels.
3.2.2. Caveolae-Mediated Endocytosis
Caveolae-mediated endocytosis is another form of receptor specific endocytosis that is based on caveolae, membrane invaginations that are furnished with cholesterol and sphingolipids.[105] Caveolae are not ubiquitous; most cell types contain caveolae; however, they are more prevalent in endothelial cells, epithelial cells, smooth and striated muscle cells, adipocytes, and fibroblasts.[106] Alongside their endocytic capabilities, they have several other functions, including reducing the tension a cell experiences under mechanical stress,[107] regulating intracellular signal transduction,[108] and mediating neurovascular coupling.[109]
The caveolin family of proteins serves major roles in the functions of caveolae. Caveolin-1 is a cholesterol-binding structural protein that surrounds the invaginations and is necessary for the formation of caveolae.[110] Caveolin-2 has a role in signal regulation and is dependent upon caveolin-1.[111] Caveolin-3 is similar to caveolin-1, however, it is mostly found in muscle cells.[112] The more recently discovered cavin family of proteins also serve essential structural roles for the formation of caveolae.[113] Dynamin has also been shown to be involved in the scission of the endocytic vesicle of caveolae from the plasma membrane, as it does in clathrin-mediated endocytosis, forming vesicles that are typically 50–100 nm wide.[54,114] While caveolin coats do not disassemble before fusing with endosomes unlike clathrin, their vesicles share a similarity in having multiple destinations based on their cargo.[99,115] These endosomes can be trafficked to lysosomes for degradation or trafficked to the Golgi bodies and endoplasmic reticulum for transcytotic purposes.[116,117] This characteristic makes caveolar endocytosis particularly attractive for the delivery of nanoparticles across tumor vasculature.
Malik and co-workers have demonstrated the targeting of caveolae-mediated endocytosis in bovine lung microvessel endothelial cells using polymer nanoparticles coated with fluorescently tagged albumin, a caveolae-mediated endocytosis tracer.[118] Similarly, Astilean and co-workers have shown the specific targeting of caveolae-mediated endocytosis in NIH:OVCAR-3 cells (human ovarian cancer) using nanoparticles made of albumin, conjugated with folic acid for folate receptor alpha targeting, as this marker is overexpressed on these cells.[119,120] In vivo targeting of caveolae has been demonstrated by Schnitzer and co-workers, who used gold nanoparticles conjugated with aminopeptidase P antibodies to target caveolae in the lung endothelium of rats,[121] or gold nanoparticles conjugated with annexin A1 antibodies to target caveolae in the tumorous lung endothelium of rats.[122] These experiments have shown that nanoparticles can be modified in specific ways to target and to exploit transcytosis in tumor endothelial cells using different transport mechanisms, including caveolae-mediated transport.
3.2.3. Macropinocytosis
Macropinocytosis is a nonspecific form of fluid phase endocytosis that involves membrane extensions for relatively large-volume engulfment.[123] This process is triggered and controlled by growth factor signaling, which causes the remodeling of actin in the cytoskeleton to create membrane ruffles that then close back in toward the rest of the membrane.[124,125] The resulting vesicles, known as macropinosomes, vary greatly in size, typically ranging from 500 to 2500 nm, though sizes as low as 200 nm and as high as 5000 nm are also possible.[126,127]
Similar to clathrin coated vesicles, macropinosomes can either mature from early endosomes to late endosomes before trafficking to lysosomes for degradation or can recycle their contents back to the apical or basal/basolateral membrane.[100,128] The visualization of macropinosomes is somewhat less direct than the previously discussed vesicles; while clathrin-coated vesicles and caveolae can be visualized optically with fluorescently tagged antibodies against clathrin heavy/light chain and caveolin-1, macropinosomes have no such marker.[123] Consequently, alternative methods had to be employed, with the most commonly used of them being visualizing the uptake of fluorescently tagged dextran,[129] an established macropinocytosis tracer, or by visualizing the rearrangement of fluorescently tagged f-actin.[130] Receptor tyrosine kinase activation and the oncogene RAS have been established as macropinocytosis triggers, with the process usually being positively regulated by environmental factors, such as nutrient availability through the amino acid activated mammalian target of rapamycin complex 1 (mTORC1).[131,132] Macropinocytosis has been suggested to be highly upregulated in cancers occurring from RAS mutations and serves the cancer cells’ primary method for their increased nutrient collection needs.[133] The increased level of uptaken proteins results in a higher availability of amino acids following lysosomal degradation, leading to higher mTORC1 activity.[134]
3.2.4. Other Endocytic or Transcellular Pathways
The described endocytic pathways are major cell uptake routes, but they are not the only endocytic or transcytotic pathways that occur in endothelial cells. One of these endocytic pathways is phagocytosis, which is typically associated with immune cells, such as macrophages, neutrophils, and dendritic cells, with an endosome formation and intracellular fate that is relatively similar to that of macropinocytosis.[135] While phagocytosis is not a niche for endothelial cells, they are still capable of performing it.[136,137]
Certain clathrin and caveolae independent pathways that are also independent of dynamin and lack a defined protein coat for encapsulating endocytic cargo.[138] One of these pathways is termed the clathrin independent carriers and glycophosphatidylinositol enriched endocytic compartments (CLIC/GEEC) pathway, which is used for the endocytosis of many glycosylphosphatidylinositol anchored proteins, and certain toxins and viruses. CLIC/GEEC endosomes are formed through the activation of the ADP-ribosylation factor 1 (ARF1), where CLICs are formed at the front of migratory cells, and the GEECs that are formed from this fuse with early endosomes.[139,140] A similar pathway is dependent on the ADP-ribosylation factor 6 (ARF6), known as the ARF6-associated pathway. Here, ARF6 is activated and inactivated to control membrane trafficking and recycling. It is currently unknown whether the ARF6-associated pathway and the CLIC/GEEC pathway are truly distinct pathways.[138]
Another endocytic pathway that is potentially prevalent in endothelial cells is lipid raft mediated endocytosis. This pathway is based on cholesterol and sphingolipid rich microdomains on the cell membrane. However, the existence of lipids rafts is a matter of debate in the literature given that they have not been visualized yet in vivo.[141,142]
One pathway that is particularly important for cancer nanomedicine delivery is known as the C-end Rule (CendR) pathway, a neuropilin-1-mediated uptake that is similar to macropinocytosis, and is specific to peptides with a C-terminal arginine or lysine, and is the method that the tumor penetrating peptide, iRGD, takes after it is cleaved by αV integrins on the surface of tumor cells.[143,144] The previously mentioned VVOs and fenestrae are also possible routes, though more work will be needed to determine their feasibilities for nanomedicine delivery purposes.
4. Tools and Techniques to Investigate Nanoparticle Transport Pathways across Tumor Endothelial Cells
The specific targeting of endocytic pathways would be the first step in designing nanoparticles that efficiently and selectively transcytose through tumor blood vessel endothelial cells. A common method of accomplishing this is through the modification of the nanoparticle surfaces with molecular ligands that are specific to endocytic receptors, along with necessary intermediates, as mentioned with the transferrin, albumin, and folic acid conjugations.[95,118,119,145–147] Table 2 lists several common nanoparticle surface ligands, the receptors that these ligands target, and the pathways these ligands are internalized by cells.
Table 2.
Surface receptors for targeting specific endocytic pathways in vitro and in vivo. Abbreviations: AuNP, gold nanoparticle; LDL, low-density lipoprotein; MAL, maleimide; NP, nanoparticle; PEG, poly(ethylene glycol); PLA, polylactic acid; PLGA, poly(lactic-co-glycolic acid).
| Surface receptor target | Targeting molecule | Pathway | Specificity | Nanoparticle examples | References |
|---|---|---|---|---|---|
| Annexin A1 | mAnnA1 | Caveolae | mAnnA1 is specific to Annexin A1 on caveolae of tumor endothelium in humans, rats, and mice, Annexin A1 is not prevalent in caveolae from healthy tissues | -Conjugated to AuNPs for targeting tumor endothelium in rats injected with 13 762 mammary adenocarcinoma cells | [122] |
| GP60 | Albumin | Caveolae | GP60 is specific for albumin across the continuous endothelium | -Conjugated to polymer NPs for targeting BLMVECs -Conjugated to iron oxide NPs for targeting U87MG tumors in mice |
[118,159,207,208] |
| Folate receptor alpha (FRα) | Folic acid | Caveolae | FRα is overexpressed in epithelial malignancies, has limited expression in healthy cells | -Conjugated to albumin NPs for targeting NIH:OVCAR3 cells | [119] |
| Aminopeptidase P (APP) | mAPP | Caveolae | APP is expressed in the endothelium of lungs, kidneys, and livers | -Conjugated to AuNPs for targeting the lung endothelium of athymic, nude mice | [121] |
| CD36—Scavenger receptor class B (SR-B) | A variety of ligands including LDLs and thrombospondin | Caveolae | CD36 is located on many types of cells past endothelial cells, such as macrophages and platelets, and binds to many different ligands | -Phosphatidylcholine conjugated to lipid NPs for targeting macrophages in C57BL/6 mice | [110,209] |
| CD204—Scavenger receptor class A (SR-A) | A variety of ligands including acetylated LDLs and lipopolysaccharide | Caveolae and clathrin | CD204 is located on many types of cells past endothelial cells such as macrophages and epithelial cells, and binds to many different ligands. | -Anti-CD204 conjugated to micelles for targeting macrophages in mice | [210–212] |
| LDL receptor family | A variety of ligands including LDLs and lactoferrin | Caveolae and clathrin | LDL receptors are located on many types of cells past endothelial cells such as macrophages and epithelial cells, overexpressed in liver tumors, and binds to many different ligands. | -Apolipoprotein-B lipid NPs for targeting HepG2 tumors in mice | [213–215] |
| Neonatal FC receptor | IgG FC, albumin | Clathrin | Neonatal FC receptor is specific to epithelial and endothelial cells in humans and mice | -Conjugated to PLA-PEG-MAL NPs for crossing the intestinal epithelium after oral administration in mice | [158,216,217] |
| Transferrin receptor | Transferrin | Clathrin | Highly expressed in solid tumor cells, as well as in blood brain barrier endothelial cells | -Conjugated to PLGA NPs for targeting brain capillary endothelial cells and astrocytes -Conjugated to AuNPs for targeting S.C. Neuro2A tumors in mice |
[218–221] |
| Vascular cell adhesion molecule (VCAM-1) | Anti-VCAM-1 | Clathrin | VCAM-1 is expressed on activated endothelial cells in tumors and during inflammation | -Conjugated to PEG-liposomes for targeting bEnd.3 cells and tumor vasculature in a CD1 nude, human Colo 677 xenograft mouse model | [104] |
| Epidermal growth factor receptor (EGFR) | Epidermal growth factor | Macropinocytosis | Present across many different cell types in the body, but is overexpressed in many tumor cell lines. EGFR also binds to many other types of growth factors | -Conjugated to AuNPs for targeting EMT-6 mammary carcinomas in mice | [222,223] |
| αV Integrins | iRGD | C-end rule (CendR) | Specific to αV integrins on tumor endothelium as well as angiogenic endothelium After binding to αV integrins, iRGD is cleaved in to CRDGK/R and then binds to neuropilin-1 | -Conjugated to micelles for targeting 22Rv1, PC-3, PPC1, MIA PaCa-2, and BT474 tumors in mice | [143,144,224] |
Thorough understanding of how the different pathways function and contribute to nanoparticle cellular uptake is necessary for exploiting them for efficient nanomedicine delivery. Methods for isolating a pathway’s contribution to the total uptake via pathway inhibition, pathway visualization methods, and models for more accurate uptake studies are discussed in this section.
4.1. Inhibiting Endocytic Pathways
Many methods have been employed for studying the pathways that nanoparticles take, both with and without active targeting. The use of small molecule inhibitors has been a popular method for blocking an endocytic pathway and observing changes in cellular uptake, through methods such as fluorescence microscopy,[148] flow cytometry,[149] inductively coupled plasma mass spectrometry,[150–152] and radioactive decay measurements.[153] A common control for these studies is cooling the cells being studied to 4 °C, as this nonspecifically inhibits all endocytosis.[154] Table 3 lists several endocytosis inhibitors that have been studied, the pathways they inhibit, their mechanism of action, and their reported efficiency. However, direct comparisons of the inhibitors listed here are difficult because of the differences in cell types, nanoparticles, and inhibitor concentrations used, leading to the need for further studies that directly test the efficiencies of many different inhibitors on the uptake of multiple tracers for each pathway.
Table 3.
Small molecule endocytosis inhibitors and their effectiveness. Abbreviations: CHO, Chinese hamster ovary cells; NP, nanoparticle; AuNP, gold nanoparticle; HUVECs, human umbilical vein endothelial cells; BLMVECs, bovine lung microvessel endothelial cells; BAEC, bovine aortic endothelial cells; RFCs, rat epididymal fat pad; LacCer, lactosylceramides; WGA, wheat germ agglutinin.
| Inhibitory agent | Pathway inhibited | Inhibition mechanism | Uptake inhibition effectiveness | Considerations | References |
|---|---|---|---|---|---|
| Methyl-β-cyclodextrin (MβCD) | Caveolae-mediated endocytosis and clathrin-mediated endocytosis | Removes cholesterol from the plasma membrane | -53% inhibition of lactosylceramide in HUVECs -56% inhibition of transferrin in HUVECs -80% inhibition of 20 and 40 nm polystyrene NPs in HUVECs -36% inhibition of 100 and 200 nm polystyrene NPs in HUVECs |
Affects multiple pathways | [225–227] |
| N-ethyl-maleimide (NEM) | Caveolae-mediated endocytosis | Inactivating the ATPase NEM-sensitive factor | -75% inhibition of albumin gold complex in BLMVECs and BAECs -<80% inhibition of cationic liposomes in Cos-7 cells |
Stimulates macropinocytosis in various epithelial cell lines | [228–232] |
| Filipin | Caveolae-mediated endocytosis | Removes cholesterol from the plasma membrane | -70% inhibition of albumin-gold complexes in BLMVECs, BAECs, and RFCs -40% inhibition in WGA-PEG-PLA NPs encapsulating CdSe/ZnS quantum dots in CaCo-2 cells |
Sterols and sphingolipids likely play a role in clathrin-mediated endocytosis | [89,153,233,234] |
| Nystatin | Caveolae-mediated endocytosis | Removes cholesterol from the plasma membrane | -80% inhibition of BODIPY-LacCer in rat fibroblasts -No significant inhibition of PEG-PHDCA NPs in rat brain endothelial cells |
Sterols and sphingolipids likely play a role in clathrin-mediated endocytosis | [148,163,233–236] |
| Genistein | Caveolae-mediated endocytosis | Tyrosine kinase inhibitor, and also inhibits the recruitment of dynamin II | -80% inhibition of BODIPY-LacCer in rat fibroblasts -15% inhibition of heparosan based micelles in B16 cells -No significant inhibition of heparosan based micelles in A549 or MGC80-3 cells |
Inhibits clathrin-mediated endocytosis and clathrin and caveolae independent endocytosis pathways that are dependent on tyrosine kinase phosphorylation or dynamin II | [88,148,155,235,237,238] |
| Fumonisin B1 (FB1) | Caveolae-mediated endocytosis | Blocks glycosphin-golipid formation by inhibiting the acetylation of sphingosine and dihydrosphingosine | -60–80% inhibition of BODIPY-LacCer in CHO-K1 cells | Sterols and sphingolipids likely play a role in clathrin-mediated endocytosis | [155,234,239,240] |
| Indometh-acin | Caveolae-mediated endocytosis | Increases the amount of arachidonate in the cell, preventing plasma-lemmal vesicles from being formed, while also causing existing vesicles to resurface | -50% inhibition of[3H] folic acid in monkey kidney epithelial cells -No significant inhibition of heparosan based micelles in A549, MGC80-3, or B16 cells |
May stimulate the caveolae independent endocytosis in certain cell types | [88,157,241–243] |
| Chlorprom-azine | Clathrin-mediated endocytosis | Translocates clathrin and AP-2 into intracellular vesicles from the plasma membrane | -80% inhibition of transferrin in rat fibroblasts -20% inhibition of heparosan based micelles in A549 cells -25% inhibition of heparosan based micelles in MGC80-3 cells -20% inhibition of heparosan based micelles in B16 cells |
Cells may quickly adapt to chlorpromazine in the presence of transferrin to use alternative uptake pathways, limiting the inhibitory effect | [88,148,163,244] |
| Potassium depletion | Clathrin-mediated endocytosis | Dissociates clathrin lattices on the plasma membrane | -80% inhibition of transferrin in rat fibroblasts -50% inhibition of chitosan nanoparticles in A549 cells |
Slight inhibition of clathrin independent pathways | [148,237,245] |
| Dynasore | Clathrin-mediated endocytosis and caveolae-mediatec endocytosis | Inhibits the GTPase activity of dynamin | -<75% inhibition of transferrin in HeLa cells -<60% inhibition of carboxylate-modified polystyrene beads in HeLa cells |
Affects multiple pathways Shown to enhance TGFβ signaling | [114,246–248] |
| Chloroquine | Clathrin-mediated endocytosis | Prevents the transition of type 2 clathrin coated pits to type 3 clathrin coated pits, preventing the formation of clathrin coated vesicles | -85% inhibition of TGFβ1 in Mv1Lu cells -<80% inhibition of cationic liposomes in Cos-7 cells |
Enhances TGFβ signaling | [230,248] |
| Cytocha-lasin D | Macropinocytosis | Binds to actin filaments, preventing the association and disassociation of subunits at the binding sites | -<60 inhibition of FITC-dextran in monocyte derived dendritic cells -50% inhibition of 50–50 PLGA NPs in rabbit conjunctival epithelial cells |
Inhibits phagocytosis and other actin polymerization dependent processes | [149,249–251] |
| 5-(N-Ethyl-N-iso-propyl) amiloride (EIPA) | Macropinocytosis | Inhibits Na+/H+ exchange to disrupt actin polymerization | -90% inhibition of TMR-dextran in T24 cells -60% inhibition of positively charged polystyrene NPs in HeLa cells |
Inhibits phagocytosis and other actin polymerization dependent processes | [133,149,252,253] |
| Wortmannin | Macropinocytosis | Blocks the activity of the phosphoinositide 3-kinase (PI3), which prevents membrane ruffling | -<90% inhibition of fluorescein dextran in murine bone marrow-derived macrophages -60% inhibition of PFBT NPs in J774A.1 cells |
Inhibits phagocytosis, and possibly also clathrin-mediated endocytosis | [125,149,254,255] |
| Imipramine | Macropinocytosis | Inhibits the ruffling of plasma membranes, and potentially inhibits redox signaling | -95% inhibition of FITC-dextran in RAW 264.7 macrophages -50% inhibition of AuNPs in Madin Darby canine kidney cells |
The mechanism of inhibition for this inhibitor is not yet fully understood | [149,256] |
| Colchicine | Macropinocytosis and macrophage endocytosis | Decreases cell motility by preventing tubulin polymerization to block microtubule formation | -37% inhibition of intratracheally delivered gold colloids in macrophages of Syrian golden hamsters -15% inhibition of heparosan based micelles in A549 cells -15% inhibition of heparosan based micelles in MGC80-3 cells -11% inhibition of heparosan based micelles in B16 cells |
Shown to affect multiple pathways | [88,257,258] |
| Rottlerin | Macropinocytosis and fluid phase endocytosis | Inhibits protein kinase C delta (PKCδ) activity | -<90 inhibition of lucifer yellow in monocyte derived dendritic cells | Shown to affect multiple pathways | [249,259] |
| Brefeldin A | Vesicle trafficking | Inhibits the formation of COP1 coats on vesicles to prevent ER-Golgi trafficking | -99.6% inhibition of lunasin in THP-1 macrophages -40% inhibition in WGA-PEG-PLA NPs encapsulating CdSe/ZnS quantum dots in CaCo-2 cells |
Shown to inhibit transcytosis and suggested to additionally inhibit endocytosis | [89,260–262] |
| Dextran sulfate | Scavenger receptor-mediated endocytosis | Competitively binds to scavenger receptors on endothelial cells, such a stabilin-1 and stabilin-2 | -42% inhibition of succinylatedbovine serum albumin in BMECs -66% inhibition of DOPG liposomes and a 75% inhibition of DOPC liposomes in venular endothelial cells of zebrafish embryo compared to arterial endothelial cells |
Dextran sulfate will also bind to the mannose receptor | [263–266] |
It is worth noting that there are certain considerations that must be taken into account for the use of these small molecule inhibitors. First, these cell treatments are not typically 100% specific or efficient in blocking a particular cell uptake pathway, meaning that it must be determined if the remaining uptake can be attributed to either remnants of the pathway being blocked, or to regular uptake from other pathways. Second, close attention must be paid to the mechanism of action of the inhibitors in question, as it is possible that they can affect the uptake of pathways other than the one that is intended; for example, Fumonisin B1 inhibits caveolae-mediated endocytosis by blocking sphingolipid formation through the inhibition of the acylation of sphingosine and dihydrosphingosine.[155] However, it has also been suggested that sphingolipid synthesis could be necessary for clathrin-mediated endocytosis as well.[156] Similarly, a widely used stimulant for macropinocytosis, phorbol-12-myristate-13-acetate (PMA), has been shown to inhibit caveolae-mediated endocytosis.[149,157] There also exists a possibility that the blocking of one pathway increases the uptake in other pathways from what typically occurs, producing results that do not accurately reflect normal physiologic conditions. Third, the specificity of the tracer must be considered, given that there is the possibility for certain tracers to take other pathways as well; for example, albumin, a known tracer for caveolae-mediated endocytosis, has been shown to be uptaken through clathrin-mediated endocytosis when attached to the FC neonatal receptor [158] rather than the typical gp60.[159]
Finally, the size of the nanoparticles being used may be considered, given that there are finite sizes of the endocytic vesicles being studied. Figure 6 demonstrates the typical size ranges of these vesicles, as well as those of their most common tracers and commonly used nanoparticle sizes. While it has been shown that these vesicles can be dynamic and holding larger nanoparticles than what their typical sizes suggest,[95] it is likely that this will need to be determined on a case-by-case basis, taking into account factors such as nanoparticle material, surface charge, shape, and which specific ligands and receptor combos are being used. All of these different considerations imply that further experiments and analysis past just changing uptake with the inhibitor treatment would be required to determine a single pathway’s contribution to the uptake of the tracer or nanoparticle being studied.
Figure 6.
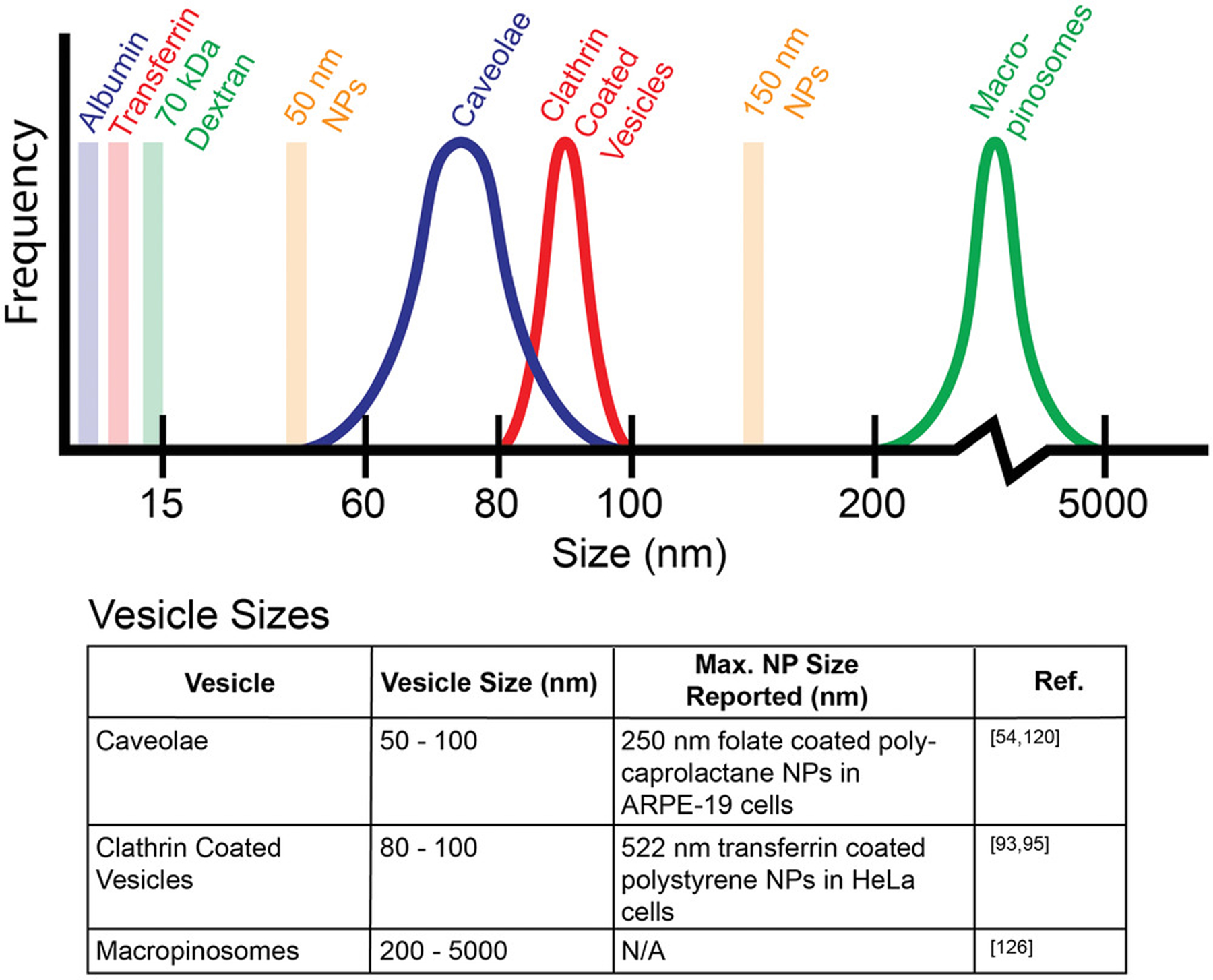
Typical sizes of vesicles formed during endocytosis. The reported size ranges of vesicles formed by the three most common endocytosis pathways are shown, along with typical tracers used in endocytosis studies, albumin for caveolae-mediated pathway, transferrin for clathrin-mediated pathway, and 70 kDa dextran for macropinocytosis. Further studies are needed to compare the sizes of each of these vesicles directly for the same cell type and with the same tracers. While intracellular vesicles exhibit reported size limitations, there are reported cases of caveolae expanding to carry 100 nm nanoparticles. Since all of the tracers are well below the typical sizes of endocytosis vesicles, meaning that each vesicle type can physically accommodate each tracer type, there is a factor of specificity for endocytosis uptake of nanoparticles (and tracers) that is beyond physical size.
An alternative method of studying uptake pathways that has been explored is the genetic alteration of cells to knock down or knock out the expression of relevant proteins. This can be accomplished through the use of small interfering RNA (siRNA) duplexes that cleave its complementary mRNA that codes for target proteins, resulting in the degradation of that mRNA, transiently silencing the expression of the protein in question.[160,161] The inhibition of endocytic pathways has been demonstrated both in vitro and in vivo for various cell types, by targeting proteins such as caveolin-1, clathrin heavy chain, and PAK-1 (a macropinocytosis signaling protein).[162–166] For in vivo systems, the knockdown can be either localized or global.[167,168] While this method is more specific, siRNA is known to be unstable in blood, immunogenic, and cannot easily cross cell membranes.[169] Therefore, for siRNA treatments to be efficient, they need a carrier, with a liposome formulation known as Lipofectamine being a common choice.[170] Alternatively, a permanent, heritable method of gene knockout is achieved through the use of the CRISPR–Cas systems.[171]
4.2. Methods for Studying Endocytic Pathways
Optical microscopy, such as confocal laser scanning microscopy, has been applied for visualizing cell uptake pathways, either by fluorescently tagging associated proteins, tracers, or the nanoparticles themselves. This method is somewhat effective, though there is a considerable limitation in its effectiveness, stemming from the physical limitations of optical microscopy—the diffraction limit of light is roughly 200 nm.[172] While this is sufficient for visualizing whole cells, it is difficult to accurately see certain subcellular structures. Electron microscopy methods have been employed for imaging at sub-nanometer resolution.[173] However, transmission electron microscopy comes at the cost of requiring thin tissue slices for imaging, typically 50–100 nm thick, resulting in a loss of 3D information,[174] which can make differentiating between various types of vesicles and channels difficult or requiring laborious imaging and image processing of multiple sections.
Efforts have been made to surpass the optical diffraction limit without the limitations presented by electron microscopy, and several super resolution microscopy methods have resulted. One such method is near field scanning optical microscopy (NSOM), which surpasses the optical diffraction limit by using a probe that is positioned close to the sample at a distance that is shorter than the excitation wavelength being imaged. However, this method also requires expensive and specialized equipment.[175] Other methods, such as stochastic optical reconstruction microscopy (STORM) and stimulated emission depletion (STED) achieve super resolution by assigning fluorescent and nonfluorescent states to fluorophores, either randomly to create a reconstructed data map, or in a targeted manner that does not require post processing, at the cost of elevated photobleaching concerns.[176] An alternative method called expansion microscopy has been developed that involves anchoring the proteins found in a cell or tissue sample to a superabsorbent hydrogel and allowing it to expand in water, mechanically stretching the sample so that objects smaller than the diffraction limit would be made larger, and therefore, resolvable.[177,178]
Given that blood vessels in living organisms are not static, physical stresses on endothelial cells and nanoparticles must also be taken into account for more informative in vitro studies. One of these stresses that has been studied is shear stress, which is caused by the movement of a fluid across constraining walls (blood through blood vessels, in this case)—it has been found that this can cause cytoskeletal rearrangement in endothelial cells.[179,180] Microfluidic models have been employed to simulate the physiological shear conditions in blood vessels. This was demonstrated in a 2D flow model by Volkov and co-workers, who showed that shear stress is critical for the uptake of cadmium telluride quantum dots and silicon dioxide nanoparticles in human umbilical vein endothelial cells.[181] Lipke and co-workers developed 3D microfluidic chips that model tumor microvascular networks, which are now commercially available prefabricated through the company SynVivo, to test the efficacy of anticancer drugs in metastatic and non-metastatic breast cancer cells.[60] Recently, Chan and co-workers have demonstrated a 3D microfluidic model of entire blood vessel networks that can be coated with endothelial cells for a much closer representation of in vivo conditions in an in vitro system, designed by casting dissolvable 3D printed models of vessel network derived from 3D fluorescent imaging in polydimethylsiloxane.[61] Laser ablation has also been explored as a method for generating highly accurate and precise vascular networks within hydrogels through the degradation of the hydrogel with a pulsed laser on an image-guided control system, so that cells can then be seeded in the newly formed channels.[182,183] These strategies for engineering vasculature for in vitro studies and implantations, along with several others, are discussed in detail in recently published reviews by Vunjak-Novakoiv and co-workers[184] and Slater co-workers[185]
The uptake of nanoparticles in vivo is, more difficult to visualize and study. It is possible that transcytosis rates in tumor blood vessels decrease with age, and that vessels without pericytes have lower transcytosis rates than those with pericytes, given that it has recently been shown that transcytosis through the blood brain barrier is impaired with age, coupled with a loss of pericytes,[186] so this might be considered for in vivo nanoparticle uptake studies. Ex vivo imaging and other quantification methods have been particularly useful for the analysis of nanoparticle accumulation in tumors and organs, i.e., resecting the mass of interest and then imaging and/or quantifying nanoparticle uptake with standard techniques.[187–189] True in vivo imaging to visualize nanoparticle transport is possible through a variety of methods. Intravital microscopy (IVM) is a common method of accomplishing this goal using principles of confocal laser scanning and multiphoton microscopy,[190] as demonstrated by Lo and co-workers, who used IVM to visualize the uptake of mesoporous silica nanoparticles into hepatocytes.[191] This can be further improved with tissue clearing methods such as clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ-hybridization-compatible tissue hydrogel or clear, unobstructed brain imaging cocktails and computational analysis protocols that minimize the effects of light scattering from tissue samples,[192] shown by Chan and co-workers to be effective for imaging of gold nanoparticles and liposomes in whole intact organs and tissues.[193–196] Another tissue clearing method, called vDISCO, works on whole intact mice.[197] Light sheet fluorescence microscopy can then be used for fast, high resolution, optically sectioned imaging, followed by computational 3D reconstruction.[198] These concepts are covered in great detail in a recently published review by Weissleder and co-workers[199] Tissue clearing and light sheet microscopy have been combined with machine learning algorithms to create a framework for quantifying and analyzing brain vasculature, called the vessel segmentation and analysis pipeline (VesSAP), for automatic, unbiased, and scalable vasculature analysis.[200] The use of optical and electron microscopy methods could provide answers to the questions that surround the complex mechanisms behind nanoparticle accumulation in solid tumors.
5. Conclusions
Efficient nanoparticle delivery to tumors requires fundamental understanding of the active and passive transport pathways and mechanisms that nanoparticles use to cross the tumor endothelium.[36] Further knowledge of the different types of tumor blood vessels and how these different vessel types affect nanoparticle transport will be instrumental. The design of tumor targeted nanoparticles that can undergo selective transcellular transport across tumor endothelial cells represents a new frontier in cancer nanomedicine research. Future studies will focus on spatiotemporal characterization of nanoparticle interactions with different tumor blood vessel types and the relationships between nanoparticle physicochemical properties and specific endocytosis and transcytosis pathways in tumor endothelial cells. In addition to ultrastructural imaging approaches, there is a need to characterize nanoparticle physiochemical properties, including changes in the nanoparticle protein corona composition, before and after transport across tumor blood vessels.[201] Such research in combination with genetically engineered and gene knockout animal models may identify pathways, mechanisms, and specific biomolecules involved in trans-endothelial transport and nanoparticle tumor delivery. The successful design of nanoparticles that selectively transport therapeutic and imaging payloads across tumor blood vessels will enable a new generation of safer and more effective cancer nanomedicines for clinical translation.
Acknowledgements
S.W. acknowledges the funding support from the University of Oklahoma IBEST-OUHSC Seed Grant for Interdisciplinary Research, the Oklahoma Tobacco Settlement Endowment Trust awarded to the University of Oklahoma—Stephenson Cancer Center, and the Oklahoma Center for Advancement of Science and Technology (OCAST) Health Research Program. Authors thank R.B. and P.M. acknowledge funding support from the National Institutes of Health (2R01CA136494 and R01CA213278). W. Yang for feedback and discussions and S. Quine for assistance with illustrations.
Biographies

Vinit Sheth is a Ph.D. graduate student in Prof. Stefan Wilhelm’s group at the Stephenson School of Biomedical Engineering (University of Oklahoma). In his research, he applies novel superresolution microscopy methods to study nanomedicine delivery mechanisms.

Lin Wang is a postdoctoral fellow in Prof. Stefan Wilhelm’s group at the Stephenson School of Biomedical Engineering (University of Oklahoma). In her research, she focuses on developing new nanoparticle engineering strategies to improve nanomedicine delivery.

Resham Bhattacharya is an associate professor in the Department of Obstetrics and Gynecology, an adjunct faculty in the Department of Cell Biology, and a full member of the Stephenson Cancer Center at the University of Oklahoma Health Science Center. She received her Ph.D. in 2002 from the Bowling Green State University, Ohio. Her research interests include investigating the basic and clinical translational aspects of molecular signaling in vascular pathologies and gynecologic cancers, primarily ovarian and uterine cancer, and she has also reported seminal observations on the nanomaterial–cellular interface pertaining to mechanisms of cellular uptake, drug delivery, nanodesign, and novel signaling.

Priyabrata Mukherjee is a tenured professor of pathology at the University of Oklahoma Health Sciences Center and holds the Presbyterian health foundation presidential professorship and the Peggy and Charles Stephenson Endowed chair position in Cancer Laboratory Research. He serves as the associate director for translational research and co-director of the Nanomedicine Program at the NCI-designated Stephenson Cancer Center. He has been working in the fields of nanomedicine, cancer biology, angiogenesis, and materials sciences for nearly 20 years.

Stefan Wilhelm is the Stephenson assistant professor at the Stephenson School of Biomedical Engineering (University of Oklahoma) and an associate member of the Stephenson Cancer Center at the University of Oklahoma Health Sciences Center. In 2014, he received his Ph.D. in Chemistry from the University of Regensburg, Germany. For his postdoctoral studies, he joined the Institute of Biomedical Engineering at the University of Toronto, Canada. His research is focused on studying nano–bio interactions for the development of next-generation nanomedicines.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Vinit Sheth, Stephenson School of Biomedical Engineering, University of Oklahoma, 173 Felgar St, Norman, OK 73019, USA.
Lin Wang, Stephenson School of Biomedical Engineering, University of Oklahoma, 173 Felgar St, Norman, OK 73019, USA.
Resham Bhattacharya, Department of Obstetrics and Gynecology, Peggy and Charles Stephenson Cancer Center, University of Oklahoma Health Science Center, 800 NE 10th St, Oklahoma City, OK 73104, USA.
Priyabrata Mukherjee, Department of Pathology, Peggy and Charles Stephenson Cancer Center, University of Oklahoma Health Science Center, 800 NE 10th St, Oklahoma City, OK 73104, USA.
Stefan Wilhelm, Stephenson School of Biomedical Engineering, University of Oklahoma, 173 Felgar St, Norman, OK 73019, USA.
References
- [1].Kowalik A, Kowalewska M, Góźdź S, Transl. Res 2017, 185, 58. [DOI] [PubMed] [Google Scholar]
- [2].Chen HHW, Kuo MT, Oncotarget 2017, 8, 62742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shou K, Tang Y, Chen H, Chen S, Zhang L, Zhang A, Fan Q, Yu A, Cheng Z, Chem. Sci 2018, 9, 3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen F, Madajewski B, Ma K, Zanoni DK, Stambuk H, Turker MZ, Monette S, Zhang L, Yoo B, Chen P, Meester RJC, de Jonge S, Montero P, Phillips E, Quinn TP, Gönen M, Sequeira S, de Stanchina E, Zanzonico P, Wiesner U, Patel SG, Bradbury MS, Sci. Adv 2019, 5, eaax5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alqathami M, Blencowe A, Yeo UJ, Doran SJ, Qiao G, Geso M, Int. J. Radiat. Oncol., Biol., Phys 2012, 84, e549. [DOI] [PubMed] [Google Scholar]
- [6].Kwatra D, Venugopal A, Anant S, Transl. Cancer Res 2013, 2, 330. [Google Scholar]
- [7].Choi M-R, Stanton-Maxey KJ, Stanley JK, Levin CS, Bardhan R, Akin D, Badve S, Sturgis J, P Robinson J, Bashir R, Halas NJ, Clare SE, Nano Lett. 2007, 7, 3759. [DOI] [PubMed] [Google Scholar]
- [8].Anselmo AC, Mitragotri S, Bioeng. Transl. Med 2019, 4, 10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Velpurisiva P, Gad A, Piel B, Jadia R, Rai P, J. Biomed 2017, 2, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Al-Ghananeem AM, Malkawi AH, Muammer YM, Balko JM, Black EP, Mourad W, Romond E, AAPS PharmSciTech 2009, 10, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huai Y, Md. Hossen N, Wilhelm S, Bhattacharya R, Mukherjee P, Bioconjugate Chem. 2019, 30, 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kingston BR, Syed AM, Ngai J, Sindhwani S, Chan WCW, Proc. Natl. Acad. Sci. USA 2019, 116, 14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhou Z, Liu X, Zhu D, Wang Y, Zhang Z, Zhou X, Qiu N, Chen X, Shen Y, Adv. Drug Delivery Rev 2017, 115, 115. [DOI] [PubMed] [Google Scholar]
- [14].Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Nat. Rev. Mater 2016, 1, 16014. [Google Scholar]
- [15].Walkey CD, Chan WCW, Chem. Soc. Rev 2012, 41, 2780. [DOI] [PubMed] [Google Scholar]
- [16].Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DWH, Cohen Y, Emili A, Chan WCW, ACS Nano 2014, 8, 2439. [DOI] [PubMed] [Google Scholar]
- [17].Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW, J. Am. Chem. Soc 2012, 134, 2139. [DOI] [PubMed] [Google Scholar]
- [18].Yang W, Wang L, Mettenbrink EM, Deangelis PL, Wilhelm S, Annu. Rev. Pharmacol. Toxicol 2021, 61, 1. [DOI] [PubMed] [Google Scholar]
- [19].Nguyen VH, Lee B-J, Int. J. Nanomed 2017, 12, 3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lazarovits J, Chen YY, Sykes EA, Chan WCW, Chem. Commun 2015, 51, 2756. [DOI] [PubMed] [Google Scholar]
- [21].Blanco E, Shen H, Ferrari M, Nat. Biotechnol 2015, 33, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Suk JS, Xu Q, Kim N, Hanes J, Ensign LM, Adv. Drug Delivery Rev 2016, 99, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hu C-MJ, Zhang L, Aryal S, Cheung C, Fang RH, Zhang L, Proc. Natl. Acad. Sci. USA 2011, 108, 10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ouyang B, Poon W, Zhang YN, Lin ZP, Kingston BR, Tavares AJ, Zhang Y, Chen J, Valic MS, Syed AM, MacMillan P, Couture-Senécal J, Zheng G, Chan WCW, Nat. Mater 2020, 10.1038/s41563-020-0755-z. [DOI] [PubMed] [Google Scholar]
- [25].Narum SM, Le T, Le DP, Lee JC, Donahue ND, Yang W, Wilhelm S, Nanoparticles for Biomedical Applications, Elsevier, Amsterdam: 2020, pp. 37–53. [Google Scholar]
- [26].Maeda H, Wu J, Sawa T, Matsumura Y, Hori K, J. Controlled Release 2000, 65, 271. [DOI] [PubMed] [Google Scholar]
- [27].Torosean S, Flynn B, Axelsson J, Gunn J, Samkoe KS, Hasan T, Doyley MM, Pogue BW, Nanomedicine 2013, 9, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Engin AB, Nikitovic D, Neagu M, Henrich-Noack P, Docea AO, Shtilman MI, Golokhvast K, Tsatsakis AM, Part. Fibre Toxicol 2017, 14, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang SK, Martin FJ, Jay G, Vogel J, Papahadjopoulos D, Friend DS, Am. J. Pathol 1993, 143, 10. [PMC free article] [PubMed] [Google Scholar]
- [30].Sindhwani S, Syed AM, Ngai J, Kingston BR, Maiorino L, Rothschild J, MacMillan P, Zhang Y, Rajesh NU, Hoang T, Wu JLY, Wilhelm S, Zilman A, Gadde S, Sulaiman A, Ouyang B, Lin Z, Wang L, Egeblad M, Chan WCW, Nat. Mater 2020, 19, 566. [DOI] [PubMed] [Google Scholar]
- [31].Dvorak HF, Cancer J. 2015, 21, 237. [DOI] [PubMed] [Google Scholar]
- [32].De Palma M, Biziato D, Petrova TV, Nat. Rev. Cancer 2017, 17, 457. [DOI] [PubMed] [Google Scholar]
- [33].Wei R, Liu S, Zhang S, Min L, Zhu S, Anal. Cell. Pathol 2020, 2020, 6283796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang Y, Elechalawar CK, Md. Hossen N, Francek ER, Dey A, Wilhelm S, Bhattacharya R, Mukherjee P, Bioact. Mater 2021, 6, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dagogo-Jack I, Shaw AT, Nat. Rev. Clin. Oncol 2018, 15, 81. [DOI] [PubMed] [Google Scholar]
- [36].Dai Q, Wilhelm S, Ding D, Syed AM, Sindhwani S, Zhang Y, Chen YY, Macmillan P, Chan WCW, ACS Nano 2018, 12, 8423. [DOI] [PubMed] [Google Scholar]
- [37].Sykes EA, Dai Q, Sarsons CD, Chen J, Rocheleau JV, Hwang DM, Zheng G, Cramb DT, Rinker KD, Chan WCW, Proc. Natl. Acad. Sci. USA 2016, 113, E1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Boumahdi S, De Sauvage FJ, Nat. Rev. Drug Discovery 2020, 19, 39. [DOI] [PubMed] [Google Scholar]
- [39].Vessoni AT, Filippi-Chiela EC, Lenz G, Batista LFZ, Onco-gene 2020, 39, 2055. [DOI] [PubMed] [Google Scholar]
- [40].Moore MAS, J. Clin. Invest 2002, 109, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhao X, Liu H-Q, Li J, Liu X-L, Oncol. Lett 2016, 12, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Metheny-Barlow LJ, Li LY, Cell Res. 2003, 13, 309. [DOI] [PubMed] [Google Scholar]
- [43].Nagy JA, Chang S-H, Dvorak AM, Dvorak HF, Br. J. Cancer 2009, 100, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bergers G, Song S, Neuro Oncol. 2005, 7, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kang E, Shin JW, Int. J. Nanomed 2016, 11, 2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC, Adv. Drug Delivery Rev 2014, 66, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jain RK, Stylianopoulos T, Nat. Rev. Clin. Oncol 2010, 7, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Golombek SK, May J-N, Theek B, Appold L, Drude N, Kiessling F, Lammers T, Adv. Drug Delivery Rev 2018, 130, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shi J, Kantoff PW, Wooster R, Farokhzad OC, Nat. Rev. Cancer 2017, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kong N, Tao W, Ling X, Wang J, Xiao Y, Shi S, Ji X, Shajii A, Gan ST, Kim NY, Duda DG, Xie T, Farokhzad OC, Shi J, Sci. Transl. Med 2019, 11, eaaw1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Islam MA, Xu Y, Tao W, Ubellacker JM, Lim M, Aum D, Lee GY, Zhou K, Zope H, Yu M, Cao W, Oswald JT, Dinarvand M, Mahmoudi M, Langer R, Kantoff PW, Farokhzad OC, Zetter BR, Shi J, Nat. Biomed. Eng 2018, 2, 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhang C, Zhang X, Zhao W, Zeng C, Li W, Li B, Luo X, Li J, Jiang J, Deng B, Mccomb DW, Dong Y, Nano Res. 2019, 12, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wilhelm S, ACS Nano 2017, 11, 10644. [DOI] [PubMed] [Google Scholar]
- [54].Wang Z, Tiruppathi C, Cho J, Minshall RD, Malik AB, IUBMB Life 2011, 63, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dvorak AM, Kohn S, Morgan ES, Fox P, Nagy JA, Dvorak HF, J. Leukocyte Biol 1996, 59, 100. [PubMed] [Google Scholar]
- [56].Vasile E, Hong Q, Dvorak HF, Dvorak AM, J. Histochem. Cytochem 1999, 47, 159. [DOI] [PubMed] [Google Scholar]
- [57].Cogger VC, Mcnerney GP, Nyunt T, Deleve LD, Mccourt P, Smedsrød B, Le Couteur DG, Huser TR, J. Struct. Biol 2010, 171, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ballermann BJ, Kidney Int. 2005, 67, 1668. [DOI] [PubMed] [Google Scholar]
- [59].Feng D, Nagy JA, Dvorak AM, Dvorak HF, Microvasc. Res 2000, 59, 24. [DOI] [PubMed] [Google Scholar]
- [60].Pradhan S, Smith AM, Garson CJ, Hassani I, Seeto WJ, Pant K, Arnold RD, Prabhakarpandian B, Lipke EA, Sci. Rep 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen YY, Kingston BR, Chan WCW, Adv. Mater. Technol 2020, 5, 2000103. [Google Scholar]
- [62].Claude A, Porter KR, Pickels EG, Cancer Res. 1947, 7, 421. [Google Scholar]
- [63].Bangham AD, Horne RW, J. Mol. Biol 1964, 8, 660. [DOI] [PubMed] [Google Scholar]
- [64].Leserman LD, Weinstein JN, Blumenthal R, Sharrow SO, Terry WD, J. Immunol 1979, 122, 585. [PubMed] [Google Scholar]
- [65].Huang A, Kennel SJ, Huang L, Immunol J. Methods 1981, 46, 141. [DOI] [PubMed] [Google Scholar]
- [66].Weinstein J, Magin R, Yatvin M, Zaharko D, Science 1979, 204, 188. [DOI] [PubMed] [Google Scholar]
- [67].Kong G, Dewhirst MW, Int. J. Hyperthermia 1999, 15, 345. [DOI] [PubMed] [Google Scholar]
- [68].Leserman LD, Weinstein JN, Blumenthal R, Terry WD, Proc. Natl. Acad. Sci. USA 1980, 77, 4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Matsumura Y, Maeda H, Cancer Res. 1986, 46, 6387. [PubMed] [Google Scholar]
- [70].Sprandel U, Lanz D-J, von Hörsten W, Methods Enzymol. 1987, 149, 301. [DOI] [PubMed] [Google Scholar]
- [71].Brenner JS, Pan DC, Myerson JW, Marcos-Contreras OA, Villa CH, Patel P, Hekierski H, Chatterjee S, Tao JQ, Parhiz H, Bhamidipati K, Uhler TG, Hood ED, Kiseleva RY, Shuvaev VS, Shuvaeva T, Khoshnejad M, Johnston I, Gregory JV, Lahann J, Wang T, Cantu E, Armstead WM, Mitragotri S, Muzykantov V, Nat. Commun 2018, 9, 2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yuan F, Chen Y, Dellian M, Safabakhsh N, Ferrara N, Jain RK, Proc. Natl. Acad. Sci. USA 1996, 93, 14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jain RK, Nat. Med 2001, 7, 987. [DOI] [PubMed] [Google Scholar]
- [74].Huang N, Liu Y, Fang Y, Zheng S, Wu J, Wang M, Zhong W, Shi M, Xing M, Liao W, ACS Nano 2020, 14, 7940. [DOI] [PubMed] [Google Scholar]
- [75].Zhang Y, Xiong X, Huai Y, Dey A, Md. Hossen N, Roy RV, Elechalawar CK, Rao G, Bhattacharya R, Mukherjee P, Bioconjugate Chem. 2019, 30, 1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Esenaliev RO, Proc. SPIE 1999, 3601, 166. [Google Scholar]
- [77].Pitt WG, Husseini GA, Roeder BL, Dickinson DJ, Warden DR, Hartley JM, Jones PW, J. Nanosci. Nanotechnol 2011, 11, 1866. [DOI] [PubMed] [Google Scholar]
- [78].Pellow C, O’Reilly MA, Hynynen K, Zheng G, Goertz DE, Nano Lett. 2020, 20, 4512. [DOI] [PubMed] [Google Scholar]
- [79].Cole C, Qiao J, Kottke T, Diaz RM, Ahmed A, Sanchez-Perez L, Brunn G, Thompson J, Chester J, Vile RG, Nat. Med 2005, 11, 1073. [DOI] [PubMed] [Google Scholar]
- [80].Hong S, Zheng D-W, Zhang C, Huang Q-X, Cheng S-X, Zhang X-Z, Sci. Adv 2020, 6, eabb0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sun T, Dasgupta A, Zhao Z, Nurunnabi M, Mitragotri S, Adv. Drug Delivery Rev 2020, S0169–409X, 30056–9, 10.1016/j.addr.2020.06.010. [DOI] [PubMed] [Google Scholar]
- [82].Setyawati MI, Tay CY, Chia SL, Goh SL, Fang W, Neo MJ, Chong HC, Tan SM, Loo SCJ, Ng KW, Xie JP, Ong CN, Tan NS, Leong DT, Nat. Commun 2013, 4, 1673. [DOI] [PubMed] [Google Scholar]
- [83].Setyawati MI, Tay CY, Bay BH, Leong DT, ACS Nano 2017, 11, 5020. [DOI] [PubMed] [Google Scholar]
- [84].Matsumoto Y, Nichols JW, Toh K, Nomoto T, Cabral H, Miura Y, J Christie R, Yamada N, Ogura T, Kano MR, Matsumura Y, Nishiyama N, Yamasoba T, Bae YH, Kataoka K, Nat. Nanotechnol 2016, 11, 533. [DOI] [PubMed] [Google Scholar]
- [85].Villaseñor R, Lampe J, Schwaninger M, Collin L, Cell. Mol. Life Sci 2019, 76, 1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Liu X, Jiang J, Meng H, Theranostics 2019, 9, 8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nel A, Ruoslahti E, Meng H, ACS Nano 2017, 11, 9567. [DOI] [PubMed] [Google Scholar]
- [88].Qiu L, Shan X, Long M, Ahmed KS, Zhao L, Mao J, Zhang H, Sun C, You C, Lv G, Chen J, Int. J. Biol. Macromol 2019, 130, 755. [DOI] [PubMed] [Google Scholar]
- [89].Gao X, Wang T, Wu B, Chen J, Chen J, Yue Y, Dai N, Chen H, Jiang X, Biochem. Biophys. Res. Commun 2008, 377, 35. [DOI] [PubMed] [Google Scholar]
- [90].Mcmahon HT, Boucrot E, Nat. Rev. Mol. Cell Biol 2011, 12, 517. [DOI] [PubMed] [Google Scholar]
- [91].Chang MP, Mallet WG, Mostov KE, Brodsky FM, EMBO J. 1993, 12, 2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kelly BT, Graham SC, Liska N, Dannhauser PN, Honing S, Ungewickell EJ, Owen DJ, Science 2014, 345, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Miller SE, Mathiasen S, Bright NA, Pierre F, Kelly BT, Kladt N, Schauss A, Merrifield CJ, Stamou D, Höning S, Owen DJ, Dev. Cell 2015, 33, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Cocucci E, Gaudin R, Kirchhausen T, Mol. Biol. Cell 2014, 25, 3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tsuji T, Yoshitomi H, Usukura J, J. Electron Microsc 2013, 62, 341. [DOI] [PubMed] [Google Scholar]
- [96].Krantz KC, Puchalla J, Thapa R, Kobayashi C, Bisher M, Viehweg J, Carr CM, Rye HS, J. Biol. Chem 2013, 288, 26721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lakadamyali M, Rust MJ, Zhuang X, Cell 2006, 124, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Donahue ND, Acar H, Wilhelm S, Adv. Drug Delivery Rev 2019, 143, 68. [DOI] [PubMed] [Google Scholar]
- [99].Md. Hossen N, Wang L, Chinthalapally HR, Robertson JD, Fung K-M, Wilhelm S, Bieniasz M, Bhattacharya R, Mukherjee P, Sci. Adv 2020, 6, eaba5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Preston JE, Joan Abbott N, Begley DJ, Physiol. Rev 2014, 83, 147. [DOI] [PubMed] [Google Scholar]
- [101].Tuma PL, Hubbard AL, Physiol. Rev 2003, 83, 871. [DOI] [PubMed] [Google Scholar]
- [102].Xiao G, Gan LS, Int. J. Cell Biol 2013, 2013, 703545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].You L, Wang J, Liu T, Zhang Y, Han X, Wang T, Guo S, Dong T, Xu J, Anderson GJ, Liu Q, Chang Y-Z, Lou X, Nie G, ACS Nano 2018, 12, 4123. [DOI] [PubMed] [Google Scholar]
- [104].Gosk S, Moos T, Gottstein C, Bendas G, Biochim. Biophys. Acta, Biomembr 2008, 1778, 854. [DOI] [PubMed] [Google Scholar]
- [105].Sharma DK, Brown JC, Choudhury A, Peterson TE, Holicky E, Marks DL, Simari R, Parton RG, Pagano RE, Mol. Biol. Cell 2004, 15, 3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Couet J, Belanger MM, Roussel E, Drolet M-C, Adv. Drug Delivery Rev 2001, 49, 223. [DOI] [PubMed] [Google Scholar]
- [107].Sinha B, Köster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C, Nassoy P, Cell 2011, 144, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Fridolfsson HN, Roth DM, Insel PA, Patel HH, FASEB J. 2014, 28, 3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chow BW, Nuñez V, Kaplan L, Granger AJ, Bistrong K, Zucker HL, Kumar P, Sabatini BL, Gu C, Nature 2020, 579, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gerbod-Giannone M-C, Dallet L, Naudin G, Sahin A, Decossas M, Poussard S, Lambert O, Biochim. Biophys. Acta, Gen. Subj 2019, 1863, 830. [DOI] [PubMed] [Google Scholar]
- [111].Sowa G, Biochem. Res. Int 2011, 2011, 809259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Williams TM, Lisanti MP, Genome Biol. 2004, 5, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kovtun O, Tillu VA, Ariotti N, Parton RG, Collins BM, J. Cell Sci 2015, 128, 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Henley JR, Krueger EWA, Oswald BJ, Mcniven MA, J. Cell Biol 1998, 141, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Schnitzer JE, Adv. Drug Delivery Rev 2001, 49, 265. [DOI] [PubMed] [Google Scholar]
- [116].Il Choi S, Maeng YS, Kim TI, Lee Y, Kim YS, Kim EK, PLoS One 2015, 10, e0119561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Le PU, J. Cell Sci 2003, 116, 1059. [DOI] [PubMed] [Google Scholar]
- [118].Wang Z, Tiruppathi C, Minshall RD, Malik AB, ACS Nano 2009, 3, 4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Borlan R, Tatar A-S, Soritau O, Maniu D, Marc G, Florea A, Focsan M, Astilean S, Nanotechnology 2020, 31, 315102. [DOI] [PubMed] [Google Scholar]
- [120].Suen W−LL, Chau Y, J. Pharm. Pharmacol 2014, 66, 564. [DOI] [PubMed] [Google Scholar]
- [121].Oh P, Borgström P, Witkiewicz H, Li Y, Borgström BJ, Chrastina A, Iwata K, Zinn KR, Baldwin R, Testa JE, Schnitzer JE, Nat. Biotechnol 2007, 25, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Oh P, Testa JE, Borgstrom P, Witkiewicz H, Li Y, Schnitzer JE, Nat. Med 2014, 20, 1062. [DOI] [PubMed] [Google Scholar]
- [123].Kerr MC, Teasdale RD, Traffic 2009, 10, 364. [DOI] [PubMed] [Google Scholar]
- [124].Palm W, Philos. Trans. R. Soc., B 2019, 374, 20180285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Araki N, Johnson MT, Swanson JA, J. Cell Biol 1996, 135, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Hewlett L, Prescott A, Watts C, J. Cell Biol 1994, 124, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lim JP, Gleeson PA, Immunol. Cell Biol 2011, 89, 836. [DOI] [PubMed] [Google Scholar]
- [128].Kasahara K, Nakayama Y, Sato I, Ikeda K, Hoshino M, Endo T, Yamaguchi N, J. Cell. Physiol 2007, 211, 220. [DOI] [PubMed] [Google Scholar]
- [129].Commisso C, Flinn RJ, Bar-Sagi D, Nat. Protoc 2014, 9, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lee E, Knecht DA, Traffic 2002, 3, 186. [DOI] [PubMed] [Google Scholar]
- [131].Recouvreux MV, Commisso C, Front. Endocrinol 2017, 8, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Williams TD, Kay RR, J. Cell Sci 2018, 131, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D, Nature 2013, 497, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Charpentier JC, Chen D, Lapinski PE, Turner J, Grigorova I, Swanson JA, King PD, Nat. Commun 2020, 11, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Flannagan RS, Jaumouillé V, Grinstein S, Annu. Rev. Pathol. Mech. Dis 2012, 7, 61. [DOI] [PubMed] [Google Scholar]
- [136].Ji S, Dong W, Qi Y, Gao H, Zhao D, Xu M, Li T, Yu H, Sun Y, Ma R, Shi J, Gao C, Thromb J. Haemostasis 2020, 18, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Xie R, Gao C, Li W, Zhu J, Novakovic V, Wang J, Ma R, Zhou J, Gilbert GE, Shi J, Blood 2012, 119, 2325. [DOI] [PubMed] [Google Scholar]
- [138].Mayor S, Parton RG, Donaldson JG, Cold Spring Harbor Perspect. Biol 2014, 6, a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lundmark R, Doherty GJ, Howes MT, Cortese K, Vallis Y, Parton RG, Mcmahon HT, Curr. Biol 2008, 18, 1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Sathe M, Muthukrishnan G, Rae J, Disanza A, Thattai M, Scita G, Parton RG, Mayor S, Nat. Commun 2018, 9, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Wei X, She G, Wu T, Xue C, Cao Y, Vet. Res 2020, 51, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Sevcsik E, Brameshuber M, Fölser M, Weghuber J, Honigmann A, Schütz GJ, Nat. Commun 2015, 6, 6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E, Cancer Cell 2009, 16, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Pang H-B, Braun GB, Ruoslahti E, Sci. Adv 2015, 1, e1500821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Lee JC, Donahue ND, Mao AS, Karim A, Komarneni M, Thomas EE, Francek ER, Yang W, Wilhelm S, ACS Appl. Nano Mater 2020, 3, 2421. [Google Scholar]
- [146].Wilhelm S, Hirsch T, Patterson WM, Scheucher E, Mayr T, Wolfbeis OS, Theranostics 2013, 3, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lazarovits J, Chen YY, Song F, Ngo W, Tavares AJ, Zhang Y-N, Audet J, Tang B, Lin Q, Tleugabulova MC, Wilhelm S, Krieger JR, Mallevaey T, Chan WCW, Nano Lett. 2019, 19, 116. [DOI] [PubMed] [Google Scholar]
- [148].Singh RD, Puri V, Valiyaveettil JT, Marks DL, Bittman R, Pagano RE, Mol. Biol. Cell 2003, 14, 3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Lin H-P, Singla B, Ghoshal P, Faulkner JL, Cherian-Shaw M, O’connor PM, She J-X, Belin De Chantemele EJ, Csányi G, Br. J. Pharmacol 2018, 175, 3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Li Y, Monteiro-Riviere NA, Nanomedicine 2016, 11, 3185. [DOI] [PubMed] [Google Scholar]
- [151].Donahue ND, Francek ER, Kiyotake E, Thomas EE, Yang W, Wang L, Detamore MS, Wilhelm S, Anal. Bioanal. Chem 2020, 412, 5205. [DOI] [PubMed] [Google Scholar]
- [152].Wilhelm S, Bensen RC, Kothapalli NR, Burgett AWG, Merrifield R, Stephan C, PerkinElmer Application Note 2018.
- [153].Schnitzer JE, Oh P, Pinney E, Allard J, J. Cell Biol 1994, 127, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Chang C-C, Wu M, Yuan F, Mol. Ther.– Methods Clin. Dev 2014, 1, 14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Shu L, J. Glycomics Lipidomics 2012, 01, 3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Meyer SGE, Wendt AE, Scherer M, Liebisch G, Kerkweg U, Schmitz G, De Groot H, Arch. Biochem. Biophys 2012, 526, 60. [DOI] [PubMed] [Google Scholar]
- [157].Smart EJ, Estes K, Anderson RGW, Cold Spring Harbor Symp. Quant. Biol 1995, 60, 243. [DOI] [PubMed] [Google Scholar]
- [158].Pyzik M, Sand KMK, Hubbard JJ, Andersen JT, Sandlie I, Blumberg RS, Front. Immunol 2019, 10, 1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Merlot AM, Kalinowski DS, Richardson DR, Front. Physiol 2014, 5, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ma Y, Jin J, Dong C, Cheng E-C, Lin H, Huang Y, Qiu C, RNA 2010, 16, 2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Scherer LJ, Rossi JJ, Nat. Biotechnol 2003, 21, 1457. [DOI] [PubMed] [Google Scholar]
- [162].Gonzalez E, Nagiel A, Lin AJ, Golan DE, Michel T, J. Biol. Chem 2004, 279, 40659. [DOI] [PubMed] [Google Scholar]
- [163].Zhu X-D, Zhuang Y, Ben J-J, Qian L-L, Huang H-P, Bai H, Sha J-H, He Z-G, Chen Q, J. Biol. Chem 2011, 286, 8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Miyawaki-Shimizu K, Predescu D, Shimizu J, Broman M, Predescu S, Malik AB, Am. J. Physiol.: Lung Cell. Mol. Physiol 2006, 290, L405. [DOI] [PubMed] [Google Scholar]
- [165].Hinrichsen L, Harborth J, Andrees L, Weber K, Ungewickell EJ, J. Biol. Chem 2003, 278, 45160. [DOI] [PubMed] [Google Scholar]
- [166].Maekawa M, Terasaka S, Mochizuki Y, Kawai K, Ikeda Y, Araki N, Skolnik EY, Taguchi T, Arai H, Proc. Natl. Acad. Sci. USA 2014, 278, E978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Razani B, Zhang XL, Bitzer M, Von Gersdorff G, Böttinger EP, Lisanti MP, J. Biol. Chem 2001, 276, 6727. [DOI] [PubMed] [Google Scholar]
- [168].Gao C, Li R, Huan J, Li W, J. Trauma: Inj., Infect., Crit. Care 2011, 70, 210. [DOI] [PubMed] [Google Scholar]
- [169].Tatiparti K, Sau S, Kashaw S, Iyer A, Nanomaterials 2017, 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Dalby B, Methods 2004, 33, 95.15121163 [Google Scholar]
- [171].Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M, Nat. Biotechnol 2013, 31, 681. [DOI] [PubMed] [Google Scholar]
- [172].Betzig E, Trautman JK, Science 1992, 257, 189. [DOI] [PubMed] [Google Scholar]
- [173].Smith DJ, Mater. Today 2008, 11, 30. [Google Scholar]
- [174].Titze B, Genoud C, Biol. Cell 2016, 108, 307. [DOI] [PubMed] [Google Scholar]
- [175].Lereu AL, Passian A, Dumas P, Int. J. Nanotechnol 2012, 9, 488. [Google Scholar]
- [176].Tam J, Merino D, J. Neurochem 2015, 135, 643. [DOI] [PubMed] [Google Scholar]
- [177].Chen F, Tillberg PW, Boyden ES, Science 2015, 347, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Zhao Y, Bucur O, Irshad H, Chen F, Weins A, Stancu AL, Oh E-Y, Distasio M, Torous V, Glass B, Stillman IE, Schnitt SJ, Beck AH, Boyden ES, Nat. Biotechnol 2017, 35, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Dewey CF, Bussolari SR, Gimbrone MA, Davies PF, J. Biomech. Eng 1981, 103, 177. [DOI] [PubMed] [Google Scholar]
- [180].Resnick N, Yahav H, Shay-Salit A, Shushy M, Schubert S, Zilberman LCM, Wofovitz E, Prog. Biophys. Mol. Biol 2003, 81, 177. [DOI] [PubMed] [Google Scholar]
- [181].Samuel SP, Jain N, O’Dowd F, Paul T, Kashanin D, Gerard VA, Gun’ko YK, Prina-Mello A, Volkov Y, Int. J. Nanomed 2012, 7, 2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Brandenberg N, Lutolf MP, Adv. Mater 2016, 28, 7450. [DOI] [PubMed] [Google Scholar]
- [183].Heintz KA, Bregenzer ME, Mantle JL, Lee KH, West JL, Slater JH, Adv. Healthcare Mater 2016, 5, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Fleischer S, Tavakol DN, Vunjak-Novakovic G, Adv. Funct. Mater 2020, 30, 1910811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Pradhan S, Banda OA, Farino CJ, Sperduto JL, Keller KA, Taitano R, Slater JH, Adv. Healthcare Mater 2020, 9, 1901255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Yang AC, Stevens MY, Chen MB, Lee DP, Stähli D, Gate D, Contrepois K, Chen W, Iram T, Zhang L, Vest RT, Chaney A, Lehallier B, Olsson N, du Bois H, Hsieh R, Cropper HC, Berdnik D, Li L, Wang EY, Traber GM, Bertozzi CR, Luo J, Snyder MP, Elias JE, Quake SR, James ML, Wyss-Coray T, Nature 2020, 7, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Moss JI, Barjat H, Emmas S-A, Strittmatter N, Maynard J, Goodwin RJA, Storm G, Lammers T, Puri S, Ashford MB, Barry ST, Theranostics 2020, 10, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Tam AL, Melancon MP, Abdelsalam M, Figueira TA, Dixon K, Mcwatters A, Zhou M, Huang Q, Mawlawi O, Dunner K Jr., Li C, Gupta S, J. Biomed. Nanotechnol 2016, 12, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Sharma A, Cornejo C, Mihalic J, Geyh A, Bordelon DE, Korangath P, Westphal F, Gruettner C, Ivkov R, Sci. Rep 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Fukumura D, Duda DG, Munn LL, Jain RK, Microcirculation 2010, 17, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Cheng S-H, Li F-C, Souris JS, Yang C-S, Tseng F-G, Lee H-S, Chen C-T, Dong C-Y, Lo L-W, ACS Nano 2012, 6, 4122. [DOI] [PubMed] [Google Scholar]
- [192].Richardson DS, Lichtman JW, Cell 2015, 162, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Sindhwani S, Syed AM, Wilhelm S, Glancy DR, Chen YY, Dobosz M, Chan WCW, ACS Nano 2016, 10, 5468. [DOI] [PubMed] [Google Scholar]
- [194].Syed AM, Macmillan P, Ngai J, Wilhelm S, Sindhwani S, Kingston BR, Wu JLY, Llano-Suárez P, Lin ZP, Ouyang B, Kahiel Z, Gadde S, Chan WCW, Nano Lett. 2020, 20, 1362. [DOI] [PubMed] [Google Scholar]
- [195].Sindhwani S, Syed AM, Wilhelm S, Chan WCW, Bioconjugate Chem. 2017, 28, 253. [DOI] [PubMed] [Google Scholar]
- [196].Syed AM, Sindhwani S, Wilhelm S, Kingston BR, Lee DSW, Gommerman JL, Chan WCW, J. Am. Chem. Soc 2017, 139, 9961. [DOI] [PubMed] [Google Scholar]
- [197].Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S, Bhatia HS, Parra-Damas A, Mrowka L, Theodorou D, Rempfler M, Xavier ALR, Kress BT, Benakis C, Steinke H, Liebscher S, Bechmann I, Liesz A, Menze B, Kerschensteiner M, Nedergaard M, Ertürk A, Nat. Neurosci 2019, 22, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Power RM, Huisken J, Nat. Methods 2017, 14, 360. [DOI] [PubMed] [Google Scholar]
- [199].Li R, Ng TSC, Garlin MA, Weissleder R, Miller MA, Adv. Funct. Mater 2020, 17, 1910369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Todorov MI, Paetzold JC, Schoppe O, Tetteh G, Shit S, Efremov V, Todorov-Völgyi K, Düring M, Dichgans M, Piraud M, Menze B, Ertürk A, Nat. Methods 2020, 17, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Stephan MT, Moon JJ, Um SH, Bershteyn A, Irvine DJ, Nat. Med 2010, 16, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Sugahara KN, Braun GB, De Mendoza TH, Kotamraju VR, French RP, Lowy AM, Teesalu T, Ruoslahti E, Mol. Cancer Ther 2015, 14, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Liu X, Lin P, Perrett I, Lin J, Liao Y-P, Chang CH, Jiang J, Wu N, Donahue T, Wainberg Z, Nel AE, Meng H, J. Clin. Invest 2017, 127, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Shah SA, Aslam Khan MU, Arshad M, Awan SU, Hashmi MU, Ahmad N, Colloids Surf., B 2016, 148, 157. [DOI] [PubMed] [Google Scholar]
- [205].Du S, Xiong H, Xu C, Lu Y, Yao J, Biomater. Sci 2019, 7, 1147. [DOI] [PubMed] [Google Scholar]
- [206].Zhang Q, Dehaini D, Zhang Y, Zhou J, Chen X, Zhang L, Fang RH, Gao W, Zhang L, Nat. Nanotechnol 2018, 13, 1182. [DOI] [PubMed] [Google Scholar]
- [207].Tiruppathi C, Finnegan A, Malik AB, Proc. Natl. Acad. Sci. USA 1996, 93, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Xie J, Wang J, Niu G, Huang J, Chen K, Li X, Chen X, Chem. Commun 2010, 46, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Zhang J, Nie S, Zu Y, Abbasi M, Cao J, Li C, Wu D, Labib S, Brackee G, Shen C-L, Wang S, J. Controlled Release 2019, 303, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Kelley JL, Ozment TR, Li C, Schweitzer JB, Williams DL, Crit. Rev. Immunol 2014, 34, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Dunzendorfer S, Lee H-K, Soldau K, Tobias PS, J. Immunol 2004, 173, 1166. [DOI] [PubMed] [Google Scholar]
- [212].Lipinski MJ, Amirbekian V, Frias JC, Aguinaldo JGS, Mani V, Briley-Saebo KC, Fuster V, Fallon JT, Fisher EA, Fayad ZA, Magn. Reson. Med 2006, 56, 601. [DOI] [PubMed] [Google Scholar]
- [213].Zhang X, Sessa WC, Fernández-Hernando C, Front. Cardiovasc. Med 2018, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Strickland DK, Gonias SL, S Argraves W, Trends Endocrinol. Metab 2002, 13, 66. [DOI] [PubMed] [Google Scholar]
- [215].Wang Z, Duan X, Lv Y, Zhao Y, Life Sci. 2019, 239, 117013. [DOI] [PubMed] [Google Scholar]
- [216].Pridgen EM, Alexis F, Kuo TT, Levy-Nissenbaum E, Karnik R, Blumberg RS, Langer R, Farokhzad OC, Sci. Transl. Med 2013, 5, 213ra167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Goebl NA, Babbey CM, Datta-Mannan A, Witcher DR, Wroblewski VJ, Dunn KW, Mol. Biol. Cell 2008, 19, 5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Mayle KM, Le AM, Kamei DT, Biochim. Biophys. Acta, Gen. Subj 2012, 1820, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Li H, Qian ZM, Med. Res. Rev 2002, 22, 225. [DOI] [PubMed] [Google Scholar]
- [220].Chang J, Jallouli Y, Kroubi M, Yuan X-B, Feng W, Kang C-S, Pu P-Y, Betbeder D, Int. J. Pharm 2009, 379, 285. [DOI] [PubMed] [Google Scholar]
- [221].Choi CHJ, Alabi CA, Webster P, Davis ME, Proc. Natl. Acad. Sci. USA 2010, 107, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Bryant DM, Kerr MC, Hammond LA, Joseph SR, Mostov KE, Teasdale RD, Stow JL, J. Cell Sci 2007, 120, 1818. [DOI] [PubMed] [Google Scholar]
- [223].Herbst RS, Int. J. Radiat. Oncol., Biol. Phys 2004, 59, S21. [Google Scholar]
- [224].Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E, Science 2010, 328, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Ho YT, Kamm RD, Kah JCY, Nanoscale 2018, 10, 12386. [DOI] [PubMed] [Google Scholar]
- [226].Ao M, Wu L, Zhou X, Chen Y, Biol. Pharm. Bull 2016, 39, 1029. [DOI] [PubMed] [Google Scholar]
- [227].Rodal SK, Skretting G, Garred Ø, Vilhardt F, Van Deurs B, Sandvig K, Mol. Biol. Cell 1999, 10, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Schnitzer JE, Allard J, Oh P, Am. J. Physiol.: Heart Circ. Physiol 1995, 268, H48. [DOI] [PubMed] [Google Scholar]
- [229].Predescu SA, Predescu DN, Palade GE, Mol. Biol. Cell 2001, 12, 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Huang Y-Z, Gao J-Q, Chen J-L, Liang W-Q, Colloids Surf., B 2006, 49, 158. [DOI] [PubMed] [Google Scholar]
- [231].Sandvig K, Llorente A, Rodal SK, Eker P, Garred Ø, Stahlhut M, Van Deurs B, Eur. J. Cell Biol 2000, 79, 447. [DOI] [PubMed] [Google Scholar]
- [232].Malyukova I, Murray KF, Zhu C, Boedeker E, Kane A, Patterson K, Peterson JR, Donowitz M, Kovbasnjuk O, Am. J. Physiol.: Gastrointest. Liver Physiol 2009, 296, G78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Rothberg KG, Heuser JE, Donzell WC, Ying Y-S, Glenney JR, Anderson RGW, Cell 1992, 68, 673. [DOI] [PubMed] [Google Scholar]
- [234].Kim JH, Singh A, Del Poeta M, Brown DA, London E, J. Cell Sci 2017, 130, 2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Puri V, Watanabe R, Singh RD, Dominguez M, Brown JC, Wheatley CL, Marks DL, Pagano RE, J. Cell Biol 2001, 154, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].R Kim H, Gil S, Andrieux K, Nicolas V, Appel M, Chacun H, Desmaële D, Taran F, Georgin D, Couvreur P, Cell. Mol. Life Sci 2007, 64, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC, Sanders NN, Braeckmans K, Mol. Ther 2010, 18, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Sandvig K, Kavaliauskiene S, Skotland T, Histochem. Cell Biol 2018, 150, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Cheng Z-J, Singh RD, Sharma DK, Holicky EL, Hanada K, Marks DL, Pagano RE, Mol. Biol. Cell 2006, 17, 3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Bhattacharyya S, Singh RD, Pagano R, D Robertson J, Bhattacharya R, Mukherjee P, Angew. Chem., Int. Ed 2012, 51, 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Smart EJ, Ying YS, Anderson RG, J. Cell Biol 1995, 131, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Kamen BA, Johnson CA, Wang MT, Anderson RG, J. Clin. Invest 1989, 84, 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Yumoto R, Nishikawa H, Okamoto M, Katayama H, Nagai J, Takano M, Am. J. Physiol.: Lung Cell. Mol. Physiol 2006, 290, L946. [DOI] [PubMed] [Google Scholar]
- [244].Francia V, Reker-Smit C, Boel G, Salvati A, Nanomedicine 2019, 14, 1533. [DOI] [PubMed] [Google Scholar]
- [245].Huang M, Ma Z, Khor E, Lim L-Y, Pharm. Res 2002, 19, 1488. [DOI] [PubMed] [Google Scholar]
- [246].Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T, Dev. Cell 2006, 10, 839. [DOI] [PubMed] [Google Scholar]
- [247].Smith PJ, Giroud M, Wiggins HL, Gower F, Thorley JA, Stolpe B, Mazzolini J, Dyson RJ, Rappoport JZ, Int. J. Nanomed 2012, 7, 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Chen C-L, Hou W-H, Liu I-H, Hsiao G, Huang SS, Huang JS, J. Cell Sci 2009, 122, 1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Sarkar K, Kruhlak MJ, Erlandsen SL, Shaw S, Immunology 2005, 116, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Cooper JA, J. Cell Biol 1987, 105, 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Qaddoumi MG, Ueda H, Yang J, Davda J, Labhasetwar V, Lee VHL, Pharm. Res 2004, 21, 641. [DOI] [PubMed] [Google Scholar]
- [252].Koivusalo M, Welch C, Hayashi H, Scott CC, Kim M, Alexander T, Touret N, Hahn KM, Grinstein S, J. Cell Biol 2010, 188, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Dausend J, Musyanovych A, Dass M, Walther P, Schrezenmeier H, Landfester K, Mailänder V, Macromol. Biosci 2008, 8, 1135. [DOI] [PubMed] [Google Scholar]
- [254].Fernando LP, Kandel PK, Yu J, Mcneill J, C Ackroyd P, Christensen KA, Biomacromolecules 2010, 11, 2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Kjeken R, Mousavi SA, Brech A, Griffiths G, Berg T, Biochem. J 2001, 357, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Van Der Hoeven D, Cho K-J, Zhou Y, Ma X, Chen W, Naji A, Montufar-Solis D, Zuo Y, Kovar SE, Levental KR, Frost JA, Van Der Hoeven R, Hancock JF, Mol. Cell. Biol 2017, 38, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Valberg PA, Brain JD, Kane D, J. Appl. Physiol.: Respir., Environ. Exercise Physiol 1981, 50, 621. [DOI] [PubMed] [Google Scholar]
- [258].Zhang J, Yu J, Jiang J, Chen X, Sun Y, Yang Z, Yang T, Cai C, Zhao X, Ding P, J. Cell. Biochem 2017, 118, 903. [DOI] [PubMed] [Google Scholar]
- [259].Singla B, Ghoshal P, Lin H, Wei Q, Dong Z, Csányi G, Front. Immunol 2018, 13, e0193737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Alvarez C, Sztul ES, Eur. J. Cell Biol 1999, 78, 1. [DOI] [PubMed] [Google Scholar]
- [261].Cam A, Sivaguru M, Gonzalez de Mejia E, PLoS One 2013, 8, e72115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Hunziker W, Andrew Whitney J, Mellman I, Cell 1991, 67, 617. [DOI] [PubMed] [Google Scholar]
- [263].Tokuda H, Masuda S, Takakura Y, Sezaki H, Hashida M, Biochem. Biophys. Res. Commun 1993, 196, 18. [DOI] [PubMed] [Google Scholar]
- [264].Campbell F, Bos FL, Sieber S, Arias-Alpizar G, Koch BE, Huwyler J, Kros A, Bussmann J, ACS Nano 2018, 12, 2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Jang S, Ohtani K, Fukuoh A, Yoshizaki T, Fukuda M, Motomura W, Mori K, Fukuzawa J, Kitamoto N, Yoshida I, Suzuki Y, Wakamiya N, J. Biol. Chem 2009, 284, 3956. [DOI] [PubMed] [Google Scholar]
- [266].Jansen RW, Molema G, Ching TL, Oosting R, Harms G, Moolenaar F, Hardonk MJ, Meijer DKF, J. Biol. Chem 1991, 266, 3343. [PubMed] [Google Scholar]


