Abstract
Objectives
This scoping review aims to present flavonoid compounds’ promising effects and possible mechanisms of action on potential therapeutic targets in the SARS-CoV-2 infection process.
Methods
A search of electronic databases such as PubMed and Scopus was carried out to evaluate the performance of substances from the flavonoid class at different stages of SARS-CoV-2 infection.
Results
The search strategy yielded 382 articles after the exclusion of duplicates. During the screening process, 265 records were deemed as irrelevant. At the end of the full-text appraisal, 37 studies were considered eligible for data extraction and qualitative synthesis. All the studies used virtual molecular docking models to verify the affinity of compounds from the flavonoid class with crucial proteins in the replication cycle of the SARS-CoV-2 virus (Spike protein, PLpro, 3CLpro/ MPro, RdRP, and inhibition of the host’s ACE II receptor). The flavonoids with more targets and lowest binding energies were: orientin, quercetin, epigallocatechin, narcissoside, silymarin, neohesperidin, delphinidin-3,5-diglucoside, and delphinidin-3-sambubioside-5-glucoside.
Conclusion
These studies allow us to provide a basis for in vitro and in vivo assays to assist in developing drugs for the treatment and prevention of COVID-19.
Graphical abstract
Supplementary information
The online version contains supplementary material available at 10.1007/s40199-023-00461-3.
Keywords: Coronavirus, Flavonoids, Respiratory Syndrome, In silico
Introduction
The coronavirus 2019 disease (COVID-19), caused by the SARS-CoV-2 virus, has burdened global healthcare systems in an unprecedented way. Several scientific studies have been produced to reduce the impacts of this disease and side effects by developing new pharmacological treatments, vaccines, and faster and more sustainable diagnostic techniques. Among them, drug-repurposing studies have investigated the use of monoclonal antibodies (tocilizumab), antineoplastics (imatinib), immunosuppressants (mycophenolate mofetil), antiparasitics (niclosamide), and non-steroidal anti-inflammatory drugs steroids (glucocorticoids) [1].
Natural products are common models for synthesizing new antiviral drugs due to their availability in the nature and variability of compounds with therapeutic potential. Around 50% of all approved drugs between 1981–2019 were derivatives from natural products, including active ingredients, such as flavonoids that can be found in several plant species, such as chamomile, mint, orange, lemon, apple, grape, among others [2].
Some flavonoids have important biological activities, such as antivirals (amentoflavone, baicalein, kaempferol, myricitrin, orientin, rutin), anti-inflammatory (hesperidin, phacelianin), antibacterial (naringenin), and immunomodulatory (fisetin, luteolin), and they can be used the prevention and treatment of different kind of diseases [3, 4]. The chemical structures of these natural metabolites are commonly composed of hydrophobic aromatic rings, hydroxyl groups, and sugar moieties (glucosides) that promote molecular interactions observed in silico studies [5].
Computer-Aided Drug Design (CADD) methodologies are used to study and develop new drugs or to reposition old drugs [6–8]. Molecular modeling is defined as the investigation of the structures and molecular properties of the substances of interest employing computational chemistry and graphic visualization techniques. Therefore, the researchers may obtain chemical or biological models, which are then submitted to specific computational programs to visualize, simulate, and interpret interrelated systems, such as those involved in drug-receptor interaction. These combined methods have the advantage of guiding a more assertive development of drugs and the possibility of reducing the wastage of time and investments in molecules with low therapeutic potential [9, 10].
Some recent studies highlight the potential antiviral effects of flavonoids against COVID-19. Santana et al. [11] showed that quercetin, apigenin, vitexin, baicalein, hesperidin, naringin, rutin, luteolin, and myricitrin were effective in reducing some of the main respiratory symptoms caused by COVID-19. An in silico approach using molecular docking to assess the inhibition of the SARS-CoV-2 spike protein revealed that naringin has minimal binding energy [12]. Similarly, Rameshkumar et al. [13] showed that agathisflavone and albireodelphin had high binding energies against RdRP and spike proteins, respectively. Five molecules were identified as potent inhibitors of the COVID-19 virus: albireodelphin, apigenin- 7-(6″-malonylglucoside), cyanidin-3-(p-coumaroyl)-rutinoside-5-glucoside, delphinidin-3,3-O-diglucoside-5-(6-p-coumarylglucoside) and (-)-maackiain-3-O-glucosyl-6″-O-malonate).
In the review study by Khazdair et al. [14], the flavonoid quercetin, when together with apigenin and isorhamnetin, inhibits the life cycle of the hepatitis C virus. In treating SARS-CoV-2, quercetin has been shown to act on the caspase 3, MAPK1, and NF-κB signaling pathways effectively to suppress high levels of cytokines and block binding sites on the surface peaks of SARS-CoV2 and prevent the spread of the virus.
Continuing the study of Khazdair et al. [14], an in silico research, it acts as a potentially highly effective disruptor of the initial infection process by binding to the interface between the SARS-CoV-2 viral spike protein and the ACE2 protein of epithelial cells.
Few other studies on this topic are available, like the study of Kaul et al. [15]. Our study aims to systematically synthesize the available data on the therapeutic potential of this class of natural substances for the treatment of SARS-CoV-2 infection, as well as their mechanisms of action, through a broad scoping review.
Methods
This scoping review was designed according to the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews (PRISMA-ScR) Checklist [16], Cochrane Handbook for Systematic Reviews of Interventions version 6.2, and Joanna Briggs Institute methodology for scoping reviews [17, 18]. This study was registered in the OSF (Open Science Framework), which can be found via the https://doi.org/10.17605/OSF.IO/7QXV8.
The search was performed in PubMed and Scopus electronic databases, on August 27th, 2020, without language or publication date limits. The main descriptors were related to COVID-19 and flavonoids (see the complete search strategy in appendix A provided in the supplemental material). Manual searches on the reference list of included studies were also conducted.
We included in silico studies that evaluated the use of flavonoids (any type and in any dose/regimen), alone or combined with other substances, for the management of SARS-CoV-2 (COVID-19) infections at any stage. During the screening phase (title and abstract) and full-text eligibility phase, articles were excluded if they were: (i) written in non-Roman characters; (ii) designed as case report; (iii) simple reviews; (iv) non-systematic; (v) systematic reviews with meta-analysis; (vi) systematic reviews without meta-analysis; (vii) about analyses of synthetic or; (viii) semi-synthetic flavonoids (see list of inclusion and exclusion criteria for studies in appendix B provided in the supplemental material).
A standardized form was used to extract the following data: study baseline characteristics (author, publication date), interventions/targets, PDB code, computer programs, and main effects/outcomes. These data were extracted from each article and synthetically transformed into the table presented in this study.
Two authors performed all steps in the study selection and data extraction phases independently, with a third author resolving discrepancies during the consensus meetings.
Results
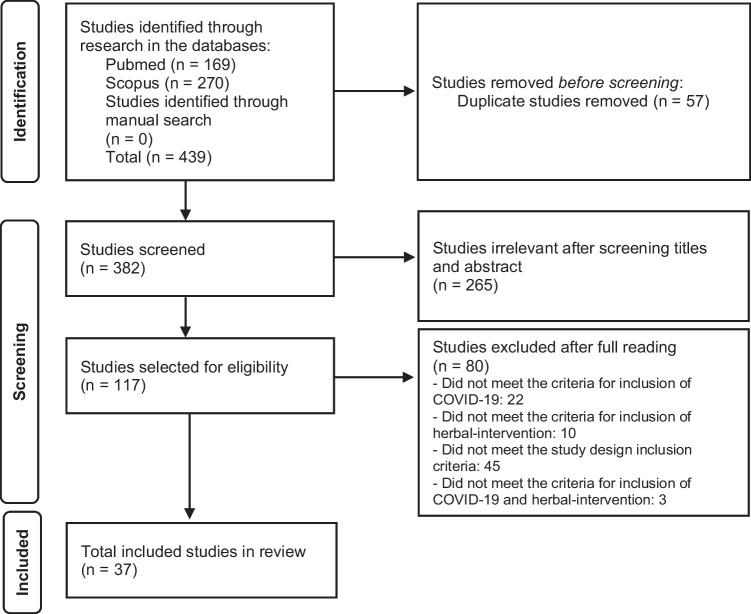
The search strategy yielded 382 articles after the exclusion of duplicates. During the screening process, 265 records were deemed as irrelevant. From the 117 studies read in total, 80 were excluded, and 37 studies were included for qualitative synthesis (see Studies excluded after full reading in appendix C provided in the supplemental material). No additional article was added from the manual search (Fig. 1).
Fig. 1.
Flowchart of the study selection process
Table 1 below shows the main characteristics of these 37 studies included. All of these studies were designed as in silico essays, in which virtual molecular docking models were used to identify SARS-CoV-2 binding potential compounds, such as flavonoids.
Table 1.
Main characteristics and results of the 37 in silico studies
| Author and Year | Country | Flavonoids evaluated | Programs | Target protein and PDB code | Main results |
|---|---|---|---|---|---|
| Chikhale et al. 2020 [91] | United Kingdom | Neohesperidin, myricitrin, quercitrin, naringin, icariin | UniProtKB, Swiss-Model, Schrodinger Package | TMPRSS2 (serine transmembrane protease 2) (1Z8G) | Neohesperidin obtained the best bonding energy (-66.53 kcal/mol for Prime MM-GBSA), (-12,77 kcal/mol for Glide Score and Dock Score) |
| Bhowmik et al. 2020 [92] | India | Rutin | I-TASSER, PyRx, AutoDock, Genbank, Gromacs | Envelope (2MM4), membrane (4f91B) and nucleocapsid (6M3M) | Rutin showed better binding energy for envelope protein (-9.30 kcal/mol) |
| Hamza et al. 2021 [93] | Pakistan | Kaempferol | Mascot Server, Pymol, and BIOVIA Discovery Studio | Viral peptides | Kaempferol showed the best binding energy for viral peptides (-6.20 kcal/mol) |
| Fakhar et al. 2021 [94] | South Africa | Delphinidin-3-sambubioside-5-glucoside, delphinidin 3,30-di-glucoside-5-(6-p-coumarylglucoside), pelargonidin | Package Maestro, Pubchem, AMBER, QikProp Module | Mpro (6LU7) | Delphinidin-3-sambubioside-5-glucoside had the best binding energy (-12.37 kcal/mol) |
| Chitranshi et al. 2020 [29] | Australia | Apigenin, luteolin, quercetin, amentoflavone, bilobetin, ginkgetin | Clustal Omega Server, Interactive Tree of Life (iTOL) online tool, Austin Model-1, Open Babel software, Chimera software, AutoDock, Pubchem | 3CLpro (6Y2G) | Amentoflavone showed the best binding energy (-8.49 kcal/mol) |
| Joshi et al. 2021 [35] | India | Cyanidin, kaempferol, rutin, gallocatechin, epigallocatechin, quercetin, eriodictyol | MEGA software, FigTree software, Cytoscape, AutoDock, AutoGrid, Pubchem, UniProtKB database, SwissADME Server | Mpro (6Y2F), RNA-dependent RNA polymerase (RdRP) (7BTF), ACE II (2AJF) |
Quercetin and eriodictyol presented the best binding energy for Mpro (-9.90 kcal/mol) Epigallocatechin for RdRP (-12.90 kcal/mol) Cyanidin for ACE II (-8.20 kcal/mol) |
| Mahdian et al. 2020 [19] | Iran | Hesperidin | SWISS-MODEL, Drug Bank database, AutoDock, Gromacs package, PyRx tool | 3CLpro, Plpro, TMPRSS2 (serine transmembrane protease 2), spike protein | Hesperidin presented the best binding energy for 3CLpro (-8.00 kcal/mol); for PLpro (-9.40 kcal/mol); for TMPRSS2 (-6.10 kcal/mol) and for protein spike (-7,40 kcal/mol) |
| Meyer-Almes 2020 [33] | Germany | Naringin, epicatechin, Homoorientin, proanthocyanidin, rutin, and quercetin | EOM pharmacophore editor, AutoDock, PyRx, Virtual screening using MOE ZINC15 database, AMBER | 3CLpro (6LU7) | Naringin presented the best bonding energy (-9.70 kcal/mol) |
|
Maiti and Banerjee 2020 [38] |
India | Catechin, Catechin gallate, Epicatechin 3–O-gallate, epigallocatechin, epigallocatechin 3-gallate, gallocatechin, gallocatechin gallate, theaflavin monogallate, and theaflavin digallate | PatchDock web server, AutoDock, RCSB—PDB | ACE II (4APH) | Theaflavin monogallate presented the best binding energy for ACE II (- 6.72 kcal/mol) |
| Wang et al. 2020 [42] | China | Quercetin, formononetin, luteolin | BiocManager, Sybyl package, Software Cytoscape | Not applicable (Interleukins) | Quercetin showed the C-score above 3 for all tested targets |
| Vijayakumar et al. 2020 [22] | India | Luteolin, apigenin, tangeritin, kaempferol, quercetin, myricetin, fisetin, hesperidin, naringenin, eriodictyol, liquiritin, genistein, daidzein, calophyllolide, cyanidin, delphinidin, malvidin, pelargonidin, peonidin, phloridzin | SWISS-MODEL, PROCHECK, AutoDock, PerkinElmer Chem3D, PyRx, the Molinspiration, pkCSM and RCSB | RNA-dependent RNA polymerase (RdRP) (6M71), main protease (M pro) (6YB7) and Spike protein (S) (6LZG) |
Calophyllolide showed the best binding energy for RdRP (-8.70 kcal/mol) and for Mpro (-9,30 kcal/mol Eriodictyol and calophyllolide showed the best binding energy for protein spike (-7.90 kcal/mol) |
| Jo et al. 2020 [5] | Republic of Korea | Baicalin, herbacetin, pectolinarin | Protein Preparation Wizard, Schrodinger Package (Maestro) | 3CLpro (6LU7) | Pectolinarin presented the best bonding energy (-10.97 kcal/mol) |
| Khalifa et al. 2020 [32] | Egypt | Phacelianin, gentiodelphin, cyanidin 3-glucoside, cyanidin 3-rutinoside, pelargonidin 3-glucoside, delphinidin 3-sambudiglucoside | MOE (Molecular Operating Environment Software), PubChem, RCSB, GROMOS Software | 3CLpro (6y84) | Cyanidin 3-rutinoside presented the best bonding energy (-17.02 kcal/mol) |
| Abian et al. 2020 [95] | Spain | Quercetin | AutoDock | 3CLpro (6Y2E) | Quercetin showed the best binding energy (-7.50 kcal/mol) |
| Singh et al. 2020 [102] | India | EGCG (epigallocatechin-3-gallate), theaflavin (TF1), theaflavin-3’-O-gallate (TF2a), theaflavin-3’-gallate (TF2b), theaflavin-3,3’-digallate (TF3), hesperidin, quercetagetin, and myricitrin | AutoDock, Swiss Target Prediction, PubChem, RCSB, AMBER, PkCSM Tool | RNA-dependent RNA polymerase (RdRP) (6M71) | Theaflavin-3,3’-digallate showed the best binding energy (-9.90 kcal/mol) |
| Pandey et al. 2020 [21] | United States | Apigenin, luteolin, quercetin, kaempferol, fisetin, genistein | AutoDock, MGL Tools, PyMol, PubChem, RCSB | ACE II (6VYB), spike protein |
Fisetin and quercetin showed identical and better binding energy for spike protein (-8.50 kcal/mol) Quercetin showed the best binding energy ACE II (-22.17 kcal/mol) |
| Sharma and Shanavas 2020 [39] | India | Delphinidin-3,5-diglucoside, avicularin | Package Schrödinger, RCSB – PDB, Swiss ADME software | Mpro (6lu7), ACE II (1R4L) | Delphinidin-3,5-diglucoside showed the best binding energy for Mpro (-12.20 kcal/mol); ACE II (-13.60 kcal/mol) |
| Fatoki et al. 2021 [26] | Nigeria | Quercetin, kaempferol | PyMol, AutoDock, Intact, Uniprot, DynaMine Server, Expression2Kinases Software, SwissADME | 3CLpro (2XYR), PLpro (3VB6), RNA-dependent RNA polymerase (5Y3E) | Quercetin showed the best binding energy for 3Clpro (-8.20 kcal/mol); for PLpro (-10.20 kcal/mol); for RdRP (-9.20 kcal/mol) |
| Maurya et al. 2020 [20] | India | Quercetin, luteolin, naringenin | Molegro Virtual Docker, Pubchem, swissADME, admetSAR | ACE II (6VXX), Protein S (spike) (1R42) | Quercetin showed the best binding energy for spike protein (-86.22 kcal/mol) and for ACE II (-92.05 kcal/mol) |
|
Tao et al. 2020 [34] |
China | Quercetin, kaempferol, isorhamnetin, baicalein, naringenin, and formononetin | Cytoscape, AutoDock, Pymol, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform | ACE II (6lu7), 3CLpro (1r42) |
Baicalein presented the best binding energy for 3CLpro (- 7.80 kcal/mol) Quercetin showed the best binding energy for ACE II (-8.40 kcal/mol) |
| Kandeel et al. 2021 [27] | Saudi Arabia | Quercetin | Maestro Package, AMBER | PLpro (6w9c) | Quercetin showed the best binding energy (-7.75 kcal/mol) |
| Alagu et al. 2021 [24] | India | Orientin, vitexin | Autodock, Gromacs simulation package | Protein spike (S) (5R82), ACE II (6VYB), Mpro (1R42) | Orientin presented the best binding energy for Mpro (-90.20 kcal/mol); for protein spike (-72.30 kcal/mol); ACE II (-70.60 kcal/mol) |
| Chikhale et al. 2020 [36] | India | Quercetin | Schrodinger Package, glide XP, PubChem, RCSB, AMBER | ACE II (6M0J) | Quercetin showed the best binding energy for ACE II (-4.41 kcal/mol) |
| Narkhede et al. 2020 [96] | India | Hesperetin | AutoDock, Pymol, Discovery Studio Visualizer, PubChem, RCSB, SwissADME | Mpro (6LU7) | Hesperetin showed the best binding energy (-7.90 kcal/mol) |
| Ruan et al. 2020 [41] | China | Kaempferol, quercetin, 2-methyl-7-methoxy-4’-nitro-isoflavone, naringenin, formononetin | AutoDock, Discovery software, PyMOL software, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform, Cytoscape Software | Not applicable (interleukins) | All substances showed energy above -5.00 kcal/mol |
| Fischer et al. 2020 [97] | Switzerland | Rhamnetin | AutoDock, Maestro Package, Protein Data Bank, VirtualToxLab | Mpro (6LU7) | Rhamnetin presented the best binding energy for Mpro (-8.20 kcal/mol) |
| Yu et al. 2020 [23] | China | Luteolin | AutoDock, NCBI, RCSB | 3CLpro (6LU7), PLpro (4OVZ), RdRP (6NUS) and Spike protein (6VSB) | Luteolin showed the best binding energy for 3 CLpro (-5.37 kcal/mol); for RdRP (-7.80 kcal/mol); for protein spike (-7.00 kcal/mol); for PLpro (-6.80 kcal/mol) |
| Das et al. 2020 [98] | India | Rutin, hesperidin, epigallocatechin gallate, epigallocatechin, myricitrin, quercitrin, glabridin, rhoifolin, vitexin | SwissDock server, Pymol | Mpro (6Y84) | Rutin presented the best binding energy (-9.55 kcal/mol) |
| Islam et al. 2020 [99] | Bangladesh | Baicalin, glabridin, cyanidin 3-glucoside, apigenin, quercetin, luteolin | Autodock, Gold software, admetSAR | Mpro (6LU7) | Cyanidin 3-glucoside presented the best binding energy (-8.40 kcal/mol) |
|
Jo et al. 2020 [31] |
Republic of Korea | Herbacetin, rhoifolin, pectolinarin, kaempferol, morin | Schrodinger Package (Maestro), PubChem, RCSB | 3CLpro (4WY3) | Rhoifolin showed the best binding energy (-9.56 kcal/mol) |
| Alamri et al. 2020 [28] | Saudi Arabia | Luteolin and kaempferol | Autodock, PyRx, GROMACS Software, SwissParam | RNA polymerase dependent on viral RNA (RdRP) (6M71), 3CLpro (6W63) and papain as protease (PLpro) (6W9C) | Luteolin showed the best binding energy for RdRP (-9.80 kcal/mol) |
| Dubey and Dubey 2020 [30] | India | Narcissoside | Software Molegro Virtual Docker (MVD), Pubchem | 3CLpro (6W63) | Narcissoside showed the best binding energy (-180.74 kcal/mol) |
| Joshi et al. 2020 [37] | India | Quercetin, Chrysoeriol, delphinidin-3-O-glucoside | PharmaGist web servers, Ligplot Software, PubChem, RCSB, DruLiTo software, admetSar | Mpro (6LU7), ACE II (1R4L) | Quercetin showed the best binding energy for Mpro (-8.30 kcal/mol; for ACE II (-11.30 kcal/mol) |
|
Lung et al. 2020 [100] |
Taiwan | Theaflavin | Modeller; Chimera, SWISS-MODEL, ZINC15 database, Blind Docking server | RNA-dependent RNA polymerase (RdRP) | Theaflavin showed the best binding energy (-9.11 kcal/mol) |
| Owis et al. 2020 [101] | Egypt | Kaempferol | DMP, RCSB | Mpro (6LU7) | Kaempferol showed the best binding energy (-8.12 kcal/mol) |
| Glinsky 2020 [40] | United States | Quercetin | Gene Expression Omnibus (GEO) | ACE II | There was no apply binding energy |
| Gorla et al. 2021 [25] | India | Biochanin A, silibinin (silybin A), silymarin, malvidin, morin, quercetin and diosmetin | Molegro Virtual Docker, NCBI, RCSB, Swiss ADME software | ACE II (6VW1) and protein spike |
Biochanin A showed the best binding energy for protein spike (-78,41 kcal/mol) Silymarin/ silibinin (silybin A) showed the best binding energy for ACE II (- 121.28 kcal/mol) |
DMP: Data Management Plan; MOE: Molecular Operating Environment Software; NCBI: National Center for Biotechnology Information; PDB Code: Protein data bank database code; PKCSM tool: Prediction of pharmacokinetic and toxicity properties of small molecules using chart-based signatures; RCSB: Architectural advances in search for integrated research and efficient access to data from the macromolecular structure of the PDB file
The studies were published in 2020 and 2021, in the following countries: Australia (n = 1), Bangladesh (n = 1), China (n = 4), Egypt (n = 2), Germany (n = 1), India (n = 14), Iran (n = 1), Nigeria (n = 1), Pakistan (n = 1), Republic of Korea (n = 2), Saudi Arabia (n = 2), Spain (n = 1), South Africa (n = 1), Switzerland (n = 1), Taiwan (n = 1), United Kingdom (n = 1), and United States of America (n = 2).
The Schrodinger package, Amber18 software package, I-Tasser, Gromacs, PyRx, AutoDock, PharmaGist web server, Maestro package, MOE pharmacophore editor, Molegro Virtual Docker software, Ligplot software, Gold software, server SwissDock, Pymol, Discovery Studio software, Procheck web server, PerkinElmer, BiocManager Package, PatchDock web server, Austin Model-1, Open Babel software, UCSF Chimera, and Raccoon software were used as the main computer programs.
The main viral proteins analyzed were Glycoprotein S (Spike Protein) (n = 7 studies; 18.91%), Papain-type viral protease (PLpro) (n = 5 studies; 13.51%), Chymotrypsin-type viral protease (3CLpro) (n = 12; studies; 32.43%), Main protein (MPro) (n = 11; 29.73%), RNA-dependent RNA-polymerase (RdRP) (n = 6; 16.22%), and inhibition of the host’s ACE II receptor (n = 11; 29.73%). (Complete information on the main flavonoid chemical structure is available in Appendix D of the supplementary material).
Studies of the Spike protein evaluated a total of 26 different flavonoids: apigenin, biochanin A, calophyllolide, kaempferol, cyanidin, daidzein, delphinidin, diosmetin, eriodictyol, fisetin, genistein, hesperidin, liquiritin, luteolin, tangeritin, malvidin, myricetin, morin, naringenin, orientin, pelargonidin, peonidin, phloridzin, quercetin, silymarin/ silibinin (silybin A), vitexin. With the most promising anti-viral compounds being: biochanin A (-78.41 kcal/mol); calophyllolide and eriodictyol (-7.90 kcal/mol); fisetin (-8.50 kcal/mol); hesperidin (-7.4 kcal/mol); quercetin (-86.22 kcal/mol); luteolin (-7.00 kcal/mol); orientin (-72.30 kcal/mol) [19–25].
Quercetin also presented effects on the inhibition of PLpro, according to Fatoki et al. [26] and Kandeel et al. [27] (binding energies of -10.20 kcal/mol and -7.75 kcal/mol, respectively). These results were similar for hesperidin (binding energy of -9.40 kcal/mol) [19] and luteolin (binding energy of -6.80 kcal/mol) [23].
Among the flavonoids with the potential to inhibit 3CLpro, those that showed the best results were amentoflavone, baicalein, cyanidin 3-rutinoside, hesperidin, kaempferol, luteolin, narcissoside, naringin, pectolinarin, quercetin, and rhoifolin [19, 23, 26, 28–34]. In this scenario Narcissoside has the highest binding energy (-180.74 kcal/mol) [30]. On the other hand, epigallocatechin presented binding energy above -12.90 kcal/mol against RdRP [35]. Finally, the most promising flavonoids inhibiting the host’s ACE II receptor were cyaniding, delphinidin, orientin, quercetin, silymarin/silibinin (silybin A), and theaflavin monogallate with binding energies varying from -4.76 kcal/mol until -121.28 kcal/mol [20, 21, 24, 25, 35–39].
Three of the studies evaluated for data extraction (Glinsky [40], Ruan et al. [41] Wang et al. [42]) used some flavonoids to build molecular maps guided by genomic regulatory elements by analyzing gene silencing and overexpression experiments. Quercetin was a flavonoid identified as a supposed mitigation agent for COVID-19. There is a change in the expression of human genes encoding SARS-CoV-2 target proteins when, by structural similarity, the flavonoid develops as an inhibitor interfering with the functions of SARS-CoV-2 viral proteins in human cells.
The different binding energy values occurred due to the various programs used in the articles since each program has a different algorithm and formula for calculating energy [43]. Despite the heterogeneity between the methodologies used, ranking the most promising flavonoids was suggested, classified according to the software used, lower binding energy, and the number of targets (Table 2).
Table 2.
Classification of energies, programs, and targets
| Target | Flavonoid | Binding energy | Program | Author and Year |
|---|---|---|---|---|
| 3 CLpro | Luteolin | -5.37 kcal/mol | AutoDock vina* | Yu et al. 2020 |
| 3CLpro | Naringin | -9.70 kcal/mol | AutoDock vina* | Meyer-Almes 2020 |
| 3CLpro | Hesperidin | -8.00 kcal/mol | AutoDock vina* | Mahdian et al. 2020 |
| 3CLpro | Quercetin | -7.50 kcal/mol | AutoDock vina* | Abian et al. 2020 |
| 3Clpro | Quercetin | -8.20 kcal/mol | AutoDock vina v1.1.2 | Fatoki et al. 2021 |
| 3CLpro | Baicalein | -7.80 kcal/mol | AutoDock vina v1.1.2 | Tao et al. 2020 |
| 3CLpro | Amentoflavone | -8.49 kcal/mol | AutoDock v4.2 | Chitranshi et al. 2020 |
| 3CLpro | Narcissoside | -180.74 kcal/mol | Molegro Virtual Docker* | Dubey and Dubey 2020 |
| 3CLpro | Rhoifolin | -9.56 kcal/mol | Package Schrodinger software suite (Maestro, version 11.8.012) | Jo et al. 2020 |
| 3CLpro | Pectolinarin | -10.97 kcal/mol | Package Schrodinger software suite (Maestro, version 11.8.012) | Jo et al. 2020 |
| 3CLpro | Cyanidin 3-rutinoside | -17.02 kcal/mol | MOE (Molecular Operating Environment Software)* | Khalifa et al. 2020 |
| ACE II | Quercetin | -11.30 kcal/mol | AutoDock vina* | Joshi et al. 2020 |
| ACE II | Cyanidin | -8.20 kcal/mol | AutoDock vina* | Joshi et al. 2021 |
| ACE II | Quercetin | -8.40 kcal/mol | AutoDock vina v1.1.2 | Tao et al. 2020 |
| ACE II | Orientin | -70.60 kcal/mol | AutoDock v4.2 | Alagu et al. 2021 |
| ACE II | Quercetin | -4.41 kcal/mol | AutoDock v4.2 | Chikhale et al. 2020 |
| ACE II | Quercetin | -22.17 kcal/mol | AutoDock Raccoon | Pandey et al. 2020 |
| ACE II | Quercetin | -92.05 kcal/mol | Molegro Virtual Docker v3.0.0 | Maurya et al. 2020 |
| ACE II | Silymarin | -121.28 kcal/mol | Molegro Virtual Docker v6.0 | Gorla et al. 2021 |
| ACE II | Delphinidin-3,5-diglucoside | -13.60 kcal/mol | Glide package of Schrodinger chemical simulation software* | Sharma and Shanavas 2020 |
| ACE II | Theaflavin monogallate | -6.72 kcal/mol | PatchDock web server* | Maiti and Banerjee 2020 |
| Envelope protein | Rutin | -9.30 kcal/mol | AutoDock 4 and vina | Bhowmik et al. 2020 |
| Mpro | Eriodictyol | -9.90 kcal/mol | AutoDock vina* | Joshi et al. 2021 |
| Mpro | Quercetin | -9.90 kcal/mol | AutoDock vina* | Joshi et al. 2021 |
| Mpro | Cyanidin 3-glucoside | -8.40 kcal/mol | AutoDock vina* | Islam et al. 2020 |
| Mpro | Quercetin | -8.30 kcal/mol | AutoDock vina* | Joshi et al. 2020 |
| Mpro | Calophyllolide | -9,30 kcal/mol | AutoDock vina v1.1.2 | Vijayakumar et al. 2020 |
| Mpro | Rhamnetin | -8.20 kcal/mol | AutoDock vina v1.1.2 | Fischer et al. 2020 |
| Mpro | Hesperetin | -7.90 kcal/mol | AutoDock vina v1.0 | Narkhede et al. 2020 |
| Mpro | Orientin | -90.20 kcal/mol | AutoDock v4.2 | Alagu et al. 2021 |
| Mpro | Delphinidin-3,5-diglucoside | -12.20 kcal/mol | Glide package of Schrodinger chemical simulation software* | Sharma and Shanavas 2020 |
| Mpro | Delphinidin-3-sambubioside-5-glucoside | -12.37 kcal/mol | Package Schrodinger software suite (Maestro, version 11.6) | Fakhar et al. 2021 |
| Mpro | Rutin | -9.55 kcal/mol | Swissdock* | Das et al. 2020 |
| Mpro | Kaempferol | -8.12 kcal/mol | London dG score* | Owis et al. 2020 |
| PLpro | Hesperidin | -9.40 kcal/mol | AutoDock vina* | Mahdian et al. 2020 |
| PLpro | Luteolin | -6.80 kcal/mol | AutoDock vina* | Yu et al. 2020 |
| PLpro | Quercetin | -10.20 kcal/mol | AutoDock vina v1.1.2 | Fatoki et al. 2021 |
| PLpro | Quercetin | -7.75 kcal/mol | Package Schrodinger (Maestro)* | Kandeel et al. 2021 |
| Protein Spike | Quercetin | -8.50 kcal/mol | AutoDock vina* | Pandey et al. 2020 |
| Protein Spike | Hesperidin | -7,40 kcal/mol | AutoDock vina* | Mahdian et al. 2020 |
| Protein Spike | Luteolin | -7.00 kcal/mol | AutoDock vina* | Yu et al. 2020 |
| Protein Spike | Eriodictyol | -7.90 kcal/mol | AutoDock vina v1.1.2 | Vijayakumar et al. 2020 |
| Protein Spike | Calophyllolide | -7.90 kcal/mol | AutoDock vina v1.1.2 | Vijayakumar et al. 2020 |
| Protein Spike | Orientin | -72.30 kcal/mol | AutoDock v4.2 | Alagu et al. 2021 |
| Protein Spike | Quercetin | -86.22 kcal/mol | Molegro Virtual Docker v3.0.0 | Maurya et al. 2020 |
| Protein Spike | Biochanin A | -78.41 kcal/mol | Molegro Virtual Docker v6.0 | Gorla et al. 2021 |
| RdRP | Epigallocatechin | -12.90 kcal/mol | AutoDock vina* | Joshi et al. 2021 |
| RdRP | Luteolin | -7.80 kcal/mol | AutoDock vina* | Yu et al. 2020 |
| RdRP | Theaflavin-3,3’- digallate | -9.90 kcal/mol | AutoDock vina v1.1.2 | Singh et al. 2020 |
| RdRP | Luteolin | -9.80 kcal/mol | AutoDock vina v1.1.2 | Alamri et al. 2020 |
| RdRP | Quercetin | -9.20 kcal/mol | AutoDock vina v1.1.2 | Fatoki et al. 2021 |
| RdRP | Calophyllolide | -8.70 kcal/mol | AutoDock vina v1.1.2 | Vijayakumar et al. 2020 |
| RdRP | Theaflavin | -9.11 kcal/mol | Blind Docking server* | Lung et al. 2020 |
| Spike Protein | Fisetin | -8.50 kcal/mol | AutoDock vina* | Pandey et al. 2020 |
| TMPRSS2 | Hesperidin | -6.10 kcal/mol | AutoDock vina* | Mahdian et al. 2020 |
| TMPRSS2 | Neohesperidin | -66.53 kcal/mol | Glide package of Schrodinger molecular modelling suite (Glide Score)* | Chikhale et al. 2020 |
| TMPRSS2 | Neohesperidin | -12.77 kcal/mol | Glide package of Schrodinger molecular modelling suite (Dock Score)* | Chikhale et al. 2020 |
| TMPRSS2 | Neohesperidin | -12.77 kcal/mol | Glide package of Schrodinger molecular modelling suite (Prime MM-GBSA)* | Chikhale et al. 2020 |
| Viral peptides | Kaempferol | -6.20 kcal/mol | AutoDock vina* | Hamza et al. 2021 |
*Version not specified by the authors
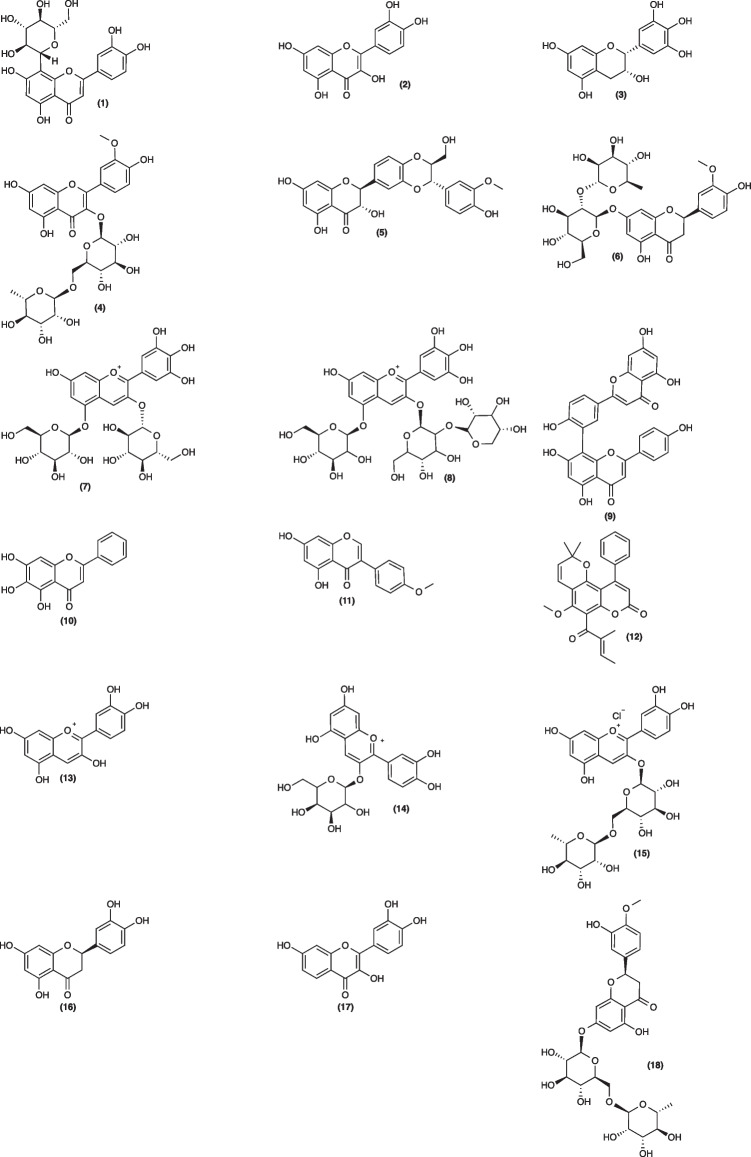
The main chemical structures of flavonoids numbered in sequence (1 to 29), are shown bellow in Fig. 2. Being that the flavonoids that were considered as the most promising according to binding energy, were the sequence 1 to 8, as described below. The sequence 9 to 29 are the other flavonoids highlighted, according to the studies included in this review.
Fig. 2.
Chemical structures described in the most promising results of the 37 in silico studies drawn by ChemDraw version 14.0.0.118. The chemical structures of flavonoids mentioned, correspond to: (1) orientin; (2) quercetin; (3) epigallocatechin; (4) narcissoside; (5) silymarin; (6) neohesperidin; (7) delphinidin-3,5-diglucoside; (8) delphinidin-3-sambubioside-5-glucoside; (9) amentoflavone; (10) baicalein; (11) biochanin A; (12) calophyllolide; (13) cyanidin; (14) cyanidin-3-glucoside; (15) cyaniding-3-rutinoside; (16) eriodictyol; (17) fisetin; (18) hesperidin; (19) hesperitin; (20) kaempferol; (21) luteolin; (22) naringin; (23) pectolinarin; (24) rhamnetin; (25) rhoifolin; (26) rutin; (27) theaflavin; (28) theaflavin-3,3’- digallate; (29) theaflavin monogallate
Within the different versions of the AutoDock program, the most promising flavonoids are orientin (1), whose energy ranged from -90.20 kcal/mol to -70.60 kcal/mol; quercetin (2), ranging from -22.17 kcal/mol to -7.50 kcal/mol; and epigallocatechin (3), with energy of -12.90 kcal/mol. Regarding the number of targets, among the articles included in this review, quercetin (2) presented binding energy in six different targets, orientin in three different targets, and epigallocatechin only one target.
Moreover, with the versions of Molegro Virtual Docker software, narcissoside (4) showed the lowest energy (-180.74 kcal/mol). In this program, the most promising flavonoids are narcissoside (4), with an energy of -180.74 kcal/mol and silymarin (5), with an energy of -121.28 kcal/mol, both of which were bound to only one type of target. While with the Schrodinger Package, neohesperidin (6) had the lowest energy, with -15.83 kcal/mol. In this program, the most promising flavonoids are neohesperidin (6), with -15.83 kcal/mol; delphinidin-3,5-diglucoside (7), with energy ranging from -13.60 kcal/mol to -12.20 kcal/mol; and delphinidin-3-sambubioside-5-glucoside (8), with the lowest energy of -12.37 kcal/mol for only one type of target. In addition to that, appendix D of supplementary material, includes the other chemical structures of the flavonoids (30 to 76) investigated in the studies included in this review.
Discussion
To the best of our knowledge, this is the first comprehensive scoping review to systematically synthesize the available evidence on the potential therapeutic effects of different flavonoids against SARS-CoV-2.
We included 37 articles that refer to in silico models. A recent study design that guides the development of other in vivo and in vitro studies, favoring resource savings and providing insights into the most effective components, speeding up the development process and research direction.
Within molecular mechanics studies, molecules are sets of atoms linked together by harmonic or elastic forces. These have been described as potential energy functions of structural contributions and unbound interactions. When these forces are added together, they form the force field, which can have adjustable parameters to improve the set of properties of the molecule [44, 45].
In this study, we present molecular docking, a molecular modeling method that seeks to predict the structures of receptor-ligand complexes target of interest. The main tools used are the search algorithm and an energy-scoring function. The scoring evaluates the binding energy of interaction between the ligand and the target receptor, classifying the best binding modes interactions between anchored ligands and proteins and predicting possible modes of action or lack thereof. Compounds with lower binding energy may have a higher affinity for target proteins [46, 47].
At the beginning of the pandemic several studies sought to compare the already studied SARS-CoV and the new SARS-CoV-2. Through homology modeling, it was revealed that the Mpro, RdRP, and Spike proteins of SARS-CoV-2 are remarkably similar to SARS-CoV. Thus, anti-coronavirus drug design strategies could be classified through the inhibition of proteins such as Mpro or enzymes that are necessary for replication and synthesis of viral RNA (RdRP) or inhibition of structural proteins such as the spike protein to adhere to host cells by inhibiting domain 2 of the angiotensin-converting enzyme [48–50].
The replicative cycle of SARS-CoV-2 begins with the interaction of the spike glycoprotein (S) located in the viral envelope, responsible for the crown conformation allocated to the Coronaviridae family, with the cell receptor of the angiotensin-converter enzyme 2 (ACE II), located on the surface of the target cell. The link between glycoprotein ACE II is responsible for the tropism of the virus by the host cell [51, 52].
After the adoption and penetration stages, denudation occurs, in which there is the release of the genetic material (RNA) of the virus in the cytoplasm of the host cell. The virus carries the viral proteins that are necessary for its initial survival in the target cell, involved in the process of transcription and viral replication (e. g. nucleocapsid contains papain-type viral proteases (PLpro), chymotrypsin proteases (3CLPro, also called MPro), in addition to RNA-dependent RNA polymerase, helicase, and RNA replicase) [53, 54].
The replication strategies of a ꞵ-coronavirus are based on the initial translation of genomic RNA into a precursor polyprotein, which is processed into non-structural proteins. Thus, genomic RNA is used as a mold by an RNA-dependent viral replication (RdRP) for the complete transcription of a simple negative RNA tape, serving as a template for the transcription of subgenomic messenger Rs RNA used to encode viral structural proteins and transcribing new genomic RNA, originating new viruses [55].
Flavonoids are phenolic compounds subdivided into different groups, including flavonols, flavones, flavanones, catechins, anthocyanins, isoflavones, dihydrophenols, and chalcones. These natural products are known to have antiviral activity, acting mainly on the viral DNA polymerase enzyme and having hydroxyl groups that favor interactions with crucial residues. Additionally, the position and number of hydrogen bonds are essential in the analysis of the inhibitory potential against the SARS-CoV-2 virus due to their binding affinity and amino acid interactions (glycine, alanine, serine, histidine, asparagine, glutamine, cysteine, proline, tyrosine, arginine, aspartic acid, glutamic acid, phenylalanine, valine, tryptophan, threonine, lysine, leucine, isoleucine, and methionine) [56].
Within the viral cycle, the SARS-CoV-2 replication mechanism was primarily led by RdRP, a complex of non-structural proteins [57, 58]. Viral RNA is translated into various polyproteins by the main protease (Mpro) action on SARS-CoV-2. In molecular modeling, the removal/mutation of the amino acids presented in each target, showed the loss of Mpro activation and reversion to the protomer form as well as the spike (S) protein, which attacks human angiotensin-converting enzyme 2 receptors [59–61]. The human equivalent for this particular protease is absent, making this a safe target for anti-SARS-CoV-2 agents.
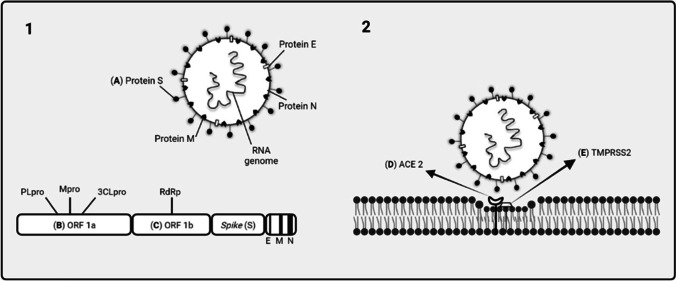
Figure 3 shows the most promising targets and flavonoids obtained from the 37 studies analyzed, according to our research on the reproductive cycle of SARS-CoV-2.
Fig. 3.
SARS-CoV-2 life cycle in the main flavonoids discuss in the scoping review. 1. Shows the main structural and non-structural target proteins of SARS-CoV-2 discussed in the review. The most promising flavonoids analyzed in silico study for (A) protein S were quercetin, eriodictyol, hesperidin, calophyllolide, fisetin, orientin, luteolin, and biochanin A; (B) ORF 1a (PLpro) were quercetin, eriodictyol, hesperidin, and luteolin; (B) ORF 1a (Mpro) were rutin, kaempferol, delphinidin-3-sambubioside-5-glucoside, quercetin, eriodictyol, calophyllolide, delphinidin-3,5-diglucoside, orientin, rhamnetin, and cyanidin-3-glucoside; (B) ORF 1a (3CLpro) were amentoflavone, quercetin, hesperidin, naringin, pectolinarin, cyanidin-3-rutinoside, baicalein, luteolin, rhoifolin, and narcissoside; (C) ORF 1b (RdRP) were quercetin, epigallocatechin, calophyllolide, theaflavin-3,3’-digallate, luteolin, and theaflavin. In 2, the main cellular targets discussed in this study are shown, being (D) ACE 2 and (E) TMPRSS2, and the flavonoids observed for these targets were for (D) quercetin, cyanidin, theaflavin monogallate, delphinidin-3,5-diglucoside, orientin, and silymarin/Silibinin (silybin A); and for (E) neohesperidin, hesperidin
The search for new substances to treat a disease, and even a new condition, is based on the search for articles that present some confirmation of substances that promoted positive effects in similar diseases. Thus, this work shows research and studies with flavonoids that had a positive influence on diseases similar to SARS-CoV-2, influencing the development of in silico studies, and through the promising results, encourage the research of these flavonoids for in vitro and in vivo studies.
Among all the flavonoids, compounds from the flavones class were the first to be associated with antiviral properties. In the late 1940s, quercetin was described as having a “prophylactic effect” against the rabies virus in infected rats [62]. We found this compound with promising activity against SARS-CoV-2 (energies ranging from -4.41 to -92.05 kcal/mol). Previous studies show quercetin can bind with glycoproteins from the viral envelope and cellular receptors by modifying their chemical structure and blocking the virus-binding site. Brum et al. [63] demonstrated a reduction in the virucide activities of some viruses with this substance, while Carvalho et al. [64] confirmed its effects against canine parvovirus in vitro studies.
Other flavonoids, such as rutin, amentoflavone, baicalein, myricitrin, and kaempferol are also related to antiviral activity, respectively, against HIV, herpes simplex, human cytomegalovirus, African swine fever, and influenza A, H1N1 and H9N2 [62, 65]. In the review study by Khazdair et al. [14], it was shown in research results that kaempferol suppresses the activity of influenza viruses, such as H1N1 and H9N2, and shown greats results for in vitro studies for hepatitis B virus, in addition, to presenting other in vitro study, where MH-S cells infected by the H9N2 influenza viruses were treated with kaempferol (50 mM) and this significantly reduced the accumulation of ROS, malondialdehyde, TNF-α, IL-1β, and IL-6. In another dosage, 100 μmol/L,ç completely inhibited the replication of bovine herpesvirus 1 in Madin-Darby bovine renal cells.
Still in the review study by Khazdair et al. [14], in an in vivo study, kaempferol at a dosage of 15 mg/kg reduced pulmonary edema, wet/dry lung weight, myeloperoxidase activity, pulmonary capillary permeability, and the number of inflammatory cells in mice infected with kaempferol. H9N2 influenza virus. For SARS-CoV, he demonstrated that there is potency to block a cation-selective channel that is expressed in the infected cell.
When quercetin and kaempferol are joined, there is a binding to the proteins of SARS-CoV2, which is involved in inflammatory responses and the modulation of the immune system through modification in the expression of cyclooxygenase 2, interleukins, MAPKs, alteration of the signaling cascade [14].
Rutin promotes the normalization of resistance and permeability of the wall of lymphatic and venous vessels. In studies conducted on guinea pig ileum, this compound acted as a non-competitive inhibitor of ACE II and prostaglandin E2 [66], which may justify its antiviral effect, including against SARS-CoV-2. Myricitrin has recently been associated with activities against HIV, influenza, and leukemia [67]. The flavone baicalein from Scutellaria baicalensis Georgi (Lamiaceae) has been described as an anti-HIV compound, as it leads to a dose-dependent inhibition of both the HIV-1 protein and reverse transcriptase. It also interferes with the interaction between viral envelope proteins and CD4 + cells, thus reducing the virus binding capacity to the host cell [68, 69].
Orientin, a flavonoid from the Trollius chinensis Bunge (Ranunculaceae) flowers, is currently used in treating respiratory tract infections in Asian countries [70]. This compound could be a promising alternative against COVID-19 as it inhibits the spike protein. We also found the compound luteolin with high binding energy against several SARS-CoV-2 targets (e.g., PLpro, 3CLpro, RdRP, ACE II), which could be further investigated in future trials. This substance inhibits the activation of T cells and the release of inflammatory cytokines by microglia [71].
Some anthocyanins previously demonstrated antioxidant and anti-inflammatory properties (e.g., inhibition of LDL oxidation) and the potential to decrease the risk of cardiovascular diseases and cancer [72–76]. Compounds such as phacelianin and cyanidin additionally have anti-mutagenic and antiviral activities [77–79].
The delphinidins – i.e., delphinidin-3,30-di-glucoside-5-(6-p-coumarylglucoside) and delphinidin-3,5-diglucoside – act on platelet activity by decreasing the expression of activated αIIβ3 integrin on platelets, inhibiting the platelet aggregation in trials with agonists ADP (Adenosine Diphosphate) and TRAP (Thrombin Receptor Activator Peptide) thrombin agonist, which contributes to the prevention of thrombosis. Additionally, they may improve some endothelial functions by increasing nitric oxide synthesis and reducing platelet aggregation. Other effects of these substances include hepatic protection, antitumor, and anti-inflammatory vascular effects [80, 81].
Compounds such as gallocatechin and neohesperidin (dihydrochalcones class) have proved antioxidant activity similar to vitamin C and vitamin E. Thus, they can strengthen the immune system and help inhibit the action of free radicals, especially in the cardiovascular system [82, 83].
Among the flavanones, hesperidin may play a significant role in the human system as an anti-inflammatory, since it inhibits both the cyclooxygenase (COX) and lipoxygenase pathways [84, 85]. Its anti-inflammatory effects are additionally associated with the inhibition of the synthesis of prostaglandins (PGE2 and PGE2a) [86]. Other substances of this class, such as naringin, also present protective effects on several systems (i.e., renal, cardiovascular, hepatic, intestinal microbiota, and immunological) due to their biological properties as antioxidant, antitumor, antiviral, antibacterial, anti-inflammatory, antiadipogenic, and cardio-protective [87].
Among the isoflavones, formononetin presents an important activity against ACE II and 3CLpro. This substance has anti-inflammatory and antioxidant action through the decrease in the formation of free radicals, preventing lipid peroxidation [88], including in the central nervous system [89].
The study by Kaul et al. [15] shows in their review that several in vitro studies explored the anti-SARS-CoV-2 action of flavonoids through the guidance of in silico studies associated with additional in vitro or in vivo investigations of anti-SARS-CoV and anti-MERS-CoV activities of various flavonoids studied. It is also cited from 2021 studies with clinical trials in patients with COVID-19 showing the promising effect of quercetin.
Computational molecular modeling precedes in vitro and in vivo studies, demonstrating great possibilities of interaction through molecular docking between compounds and the molecular target [90].
In summary, all the studies used virtual molecular docking models to verify the affinity of compounds from the flavonoid class with crucial proteins in the replication cycle of the SARS-CoV-2 virus (Spike protein, PLpro, 3CLpro/ MPro, RdRP, and inhibition of the host’s ACE II receptor). The flavonoids that showed the lowest binding energies and more targets were orientin, quercetin, epigallocatechin, narcissoside, silymarin, neohesperidin, delphinidin-3,5-diglucoside, and delphinidin-3-sambubioside-5-glucoside.
This study has some limitations. No statistical synthesis of the evidence (i.e., employing meta-analysis) was possible since the heterogeneity between studies (e.g., study design and methods used/programs, type of flavonoid, outcome measure) is very high. Thus, other natural compounds (alone or combined) may have some impact in this context and should be better evaluated in other studies. The focus of this review was on the antiviral effects of flavonoids.
Conclusion
In silico models demonstrated that some flavonoids showed the lowest binding energies and most numbers of targets (ranging from three to six), such as orientin, quercetin, epigallocatechin, narcissoside, silymarin, neohesperidin, delphinidin-3,5-diglucoside, and delphinidin-3-sambubioside-5-glucoside, and they showed promising antiviral activities against SARS-CoV-2 through different mechanisms of action.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We would like to thank the Programa de Graduação em Ciências Farmacêuticas at Universidade Estadual do Oeste do Paraná, Universidade Estadual de Maringá, Centro Universitário Ingá, Universidade do Porto and Universidade Federal do Paraná.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest in relation to the data presented in this publication.
Footnotes
In the original publication of the article, the affiliation of Fernando Fernandez Llimos was published incorrectly.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/19/2023
A Correction to this paper has been published: 10.1007/s40199-023-00469-9
References
- 1.Grando RL, Oliveira ACD, Fierro IM. The repositioning of drugs as a potential strategy for the treatment of COVID-19. Observatory on Science, Technology and Innovation in Health at the Oswaldo Cruz Foundation. 2020, p.1–9. Available from: http://observatorio.fiocruz.br/estudos/o-reposicionamento-de-farmacos-como-uma-potencial-estrategia-para-o-tratamento-da-covid-19.
- 2.Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 3.Aviran M, Dornfelt L, Rosenblat M. Pomegranate juice consumption reduces oxidative stress, atherogenic modifications to LDL, and plaquet aggregation: studies in human and in atherosclerotic apolipoprotein E- deficient mice. Am J Clin Nutr. 2000;71(5):1062–1076. doi: 10.1093/ajcn/71.5.1062. [DOI] [PubMed] [Google Scholar]
- 4.Mani JS, Johnson JB, Steel JC, Broszczak DA, Neilsen PM, Walsh KB, Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jo S, Kim S, Kim DY, Kim MS, Shin DH. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J Enzyme Inhib Med Chem. 2020;35(1):1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S, Malik BK, Sharma DK. Molecular drug targets and structure-based drug design: a holistic approach. Bioinformation. 2006;1:314–320. doi: 10.6026/97320630001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekins S, Mestres J, Testa BB. In silico pharmacology for drug discovery: applications to targets and beyond. J Pharmacol. 2007;152:21–37. doi: 10.1038/sj.bjp.0707306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czodrowski P, KriegL JM, Scheuerer S, Fox T. Computational approaches to predict drug metabolism. Expert Opin Drug Metab Toxicol. 2009;5(1):15–27. doi: 10.1517/17425250802568009. [DOI] [PubMed] [Google Scholar]
- 9.Henckel JG, Billings EM. Molecular modeling. In: Foye WO, Lemke TL, Williamns DA (Eds.). Principles of medicinal chemistry (4a ed.). Media: Williams e Wilkins; 1995. p.57–58.
- 10.Sant’Anna CMR. Glossary of terms used in drug planning (IUPAC Recommendations for 1997) New Chem. 2002;25:505–512. doi: 10.1590/S0100-40422002000300027. [DOI] [Google Scholar]
- 11.Santana FPR, Thevenard F, Gomes KS, Taguchi L, Câmara NOS, Stilhano RS, Ureshino RP, Prado CM, Lago JHG. New perspectives on natural flavonoids on COVID-19-induced lung injuries. Phytother Res. 2021;35(9):1–19. doi: 10.1002/ptr.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain AS, Sushma P, Dharmashekar C, Beelagi MS, Prasad SK, Shivamallu C, Prasad KS. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J Biol Sci. 2021;28(1):1040–1051. doi: 10.1016/j.sjbs.2020.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rameshkumar MR, Indu P, Arunagirinathan N, Venkatadri B, El-serehy HA, Ahmad A. Computational selection of flavonoid compounds as inhibitors against SARS-CoV-2 main protease, RNA-dependent RNA polymerase and spike proteins: A molecular docking study. Saudi J Biol Sci. 2021;28(1):448–458. doi: 10.1016/j.sjbs.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khazdair MR, Anaeigoudari A, Agbor GA. Anti-viral and anti-inflammatory effects of kaempferol and quercetin and COVID-2019: A scoping review. Asian Pacific Journal of Tropical Biomedicine. 2021;11:327–34. Available from: https://www.apjtb.org/text.asp?2021/11/8/327/319567
- 15.Kaul R, Paul P, Kumar S, Büsselberg D, Dwivedi VD, Chaari A. Promising antiviral activities of natural flavonoids against SARS-CoV-2 targets: systematic review. Int J Mol Sci. 2021;22(20):11069. doi: 10.3390/ijms222011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, Moher D, Peters MD, Horsley T, Weeks L, Hempel S. PRISMA extension for scope reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169(7):467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 17.Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, Welch VA. Cochrane Handbook for Systematic Reviews of Interventions version 6.2. Cochrane. 2021. Disponível em. www.training.cochrane.org/handbook.
- 18.Joanna Briggs Institute (JBI). Methodology for JBI scoping reviews Joanna Briggs Institute. Manual. 2015. Retrieved 17 June 2021, from. https://joannabriggs.org/assets/docs/sumri/Reviewers-Manual_Methodology-for-JBI-Scoping-Reviews_2015_v2.pdf.
- 19.Mahdian S, Ebrahim-Habibi A, Zarrabi M. Drug repurposing using computational methods to identify therapeutic options for COVID-19. J Diabetes Metab Disord. 2020;19(2):691–699. doi: 10.1007/s40200-020-00546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurya VK, Kumar S, Prasad AK, Bhatt MLB, Saxena SK. Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virus Dis. 2020;31(2):179–193. doi: 10.1007/s13337-020-00598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey P, Rane JS, Chatterjee A, Kumar A, Khan R, Prakash A, Ray S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J Biomol Struct Dyn. 2020;39(16):6306–6316. doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijayakumar BG, Ramesh D, Joji A, Jayachandra PJ, Kannan T. In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS-CoV-2. Eur J Pharmacol. 2020;886:173448. doi: 10.1016/j.ejphar.2020.173448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu R, Chen L, Lan R, Shen R, Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents. 2020;56(2):106012. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alagu Lakshmi S, Shafreen RMB, Priya A, Shunmugiah KP. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J Biomol Struct Dyn. 2021;39(13):459404609. doi: 10.1080/07391102.2020.1778537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorla US, Rao GK, Kulandaivelu US, Alavala RR, Panda SP. Lead finding from selected flavonoids with antiviral (SARS-CoV-2) potentials against COVID-19: an in-silico Evaluation. Comb Chem High Throughput Screen. 2021;24(6):879–890. doi: 10.2174/1386207323999200818162706. [DOI] [PubMed] [Google Scholar]
- 26.Fatoki TH, Ibraheem O, Ogunyemi IO, Akinmoladun AC, Ugboko HU, Adeseko CJ, Awofisayo OA, Olusegun SJ, Enibukun JM. Network analysis, sequence and structure dynamics of key proteins of coronavirus and human host, and molecular docking of selected phytochemicals of nine medicinal plants. J Biomol Struct Dyn. 2021;39(16):6195–6217. doi: 10.1080/07391102.2020.1794971. [DOI] [PubMed] [Google Scholar]
- 27.Kandeel M, Abdelrahman AHM, Oh-Hashi K, Ibrahim A, Venugopala KN, Morsy MA, Ibrahim MAA. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J Biomol Struct Dyn. 2021;39(14):5129–5136. doi: 10.1080/07391102.2020.1784291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alamri MA, Altharawi A, Alabbas AB, Alossaimi MA, Alqahtani SM. Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics. Arab J Chem. 2020;13(9):7224–7234. doi: 10.1016/j.arabjc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chitranshi N, Gupta VK, Rajput R, Godinez A, Pushpitha K, Shen T, Mirzaei M, You Y, Basavarajappa D, Gupta V, Graham SL. Evolving geographic diversity in SARS-CoV2 and in silico analysis of replicating enzyme 3CL(pro) targeting repurposed drug candidates. J Transl Med. 2020;18:278. doi: 10.1186/s12967-020-02448-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubey K, Dubey R. Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomedical Journal. 2020;43:363–367. doi: 10.1016/j.bj.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalifa I, Nawaz A, Sobhy R, Althwab SA, Barakat H. Polyacylated anthocyanins constructively network with catalytic dyad residues of 3CL(pro) of 2019-nCoV than monomeric anthocyanins: A structural-relationship activity study with 10 anthocyanins using in-silico approaches. J Mol Graph Model. 2020;100:107690. doi: 10.1016/j.jmgm.2020.107690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer-Almes FJ. Repurposing approved drugs as potential inhibitors of 3CL-protease of SARS-CoV-2: Virtual screening and structure based drug design. Comput Biol Chem. 2020;88:107351. doi: 10.1016/j.compbiolchem.2020.107351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao Q, Du J, Li X, Zeng J, Tan B, Xu J, Lin W, Chen XL. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharm. 2020;46(8):1345–1353. doi: 10.1080/03639045.2020.1788070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, Pandya VK, Chugh A, Giri AP, Kulkarni MJ. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn. 2021;39(9):3099–3114. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chikhale RV, Gurav SS, Patil RB, Sinha SK, Prasad SK, Shakya A, Shrivastava SK, Gurav NS, Prasad RS. Sars-cov-2 host entry and replication inhibitors from Indian ginseng: an in-silico approach. J Biomol Struct Dyn. 2020;24:1–15. doi: 10.1080/07391102.2020.1784289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi T, Sharma P, Mathpal S, Pundir H, Bhatt V, Chandra S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur Rev Med Pharmacol Sci. 2020;24(8):4529–4536. doi: 10.26355/eurrev_202004_21036. [DOI] [PubMed] [Google Scholar]
- 38.Maiti S, Banerjee A. Epigallocatechin gallate and theaflavin gallate interaction in SARS-CoV-2 spike-protein central channel with reference to the hydroxychloroquine interaction: Bioinformatics and molecular docking study. Drug Dev Res. 2020;7. 10.1002/ddr.21730. 10.1002/ddr.21730. [DOI] [PMC free article] [PubMed]
- 39.Sharma P, Shanavas A. Natural derivatives with dual binding potential against SARS-CoV-2 main protease and human ACE2 possess low oral bioavailability: a brief computational analysis. J Biomol Struct Dyn. 2020;1–12:5819–5830. 10.1080/07391102.2020.1794970. [DOI] [PMC free article] [PubMed]
- 40.Glinsky GV. Tripartite combination of candidate pandemic mitigation agents: vitamin D, quercetin, and estradiol manifest properties of medicinal agents for targeted mitigation of the COVID-19 pandemic defined by genomics-guided tracing of SARS-CoV-2 targets in human cells. Biomedicines. 2020;8(5):129. doi: 10.3390/biomedicines8050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan X, Du P, Zhao K, Huang J, Xia H, Dai D, Huang S, Cui X, Liu L, Zhang J. Mechanism of Dayuanyin in the treatment of coronavirus disease 2019 based on network pharmacology and molecular docking. Chin Med J. 2020;15:62. doi: 10.1186/s13020-020-00346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M, Fu D, Yao L, Li J. Theoretical study of the molecular mechanism of maxingyigan decoction against COVID-19: network pharmacology-based Strategy. Comb Chem High Throughput Screen. 2020 doi: 10.2174/1386207323666200806164635. [DOI] [PubMed] [Google Scholar]
- 43.Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20(7):13384–421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coelho LW, Junqueira GMA, Herrera JOM, Machado SP. Aplicação de mecânica molecular em química inorgânica. Quim Nova. 1999;22(3):396–404. doi: 10.1590/S0100-40421999000300018. [DOI] [Google Scholar]
- 45.Barreiro EJ, Fraga CAM. Química Medicinal, As Bases Moleculares da Ação dos Fármacos. 3. Porto Alegre: Artmed Editora; 2015. p. 608. [Google Scholar]
- 46.Alonso H, Bliznyuk AA, Gready JE. Combining docking and molecular dynamic simulations in drug design. Med Res Rev. 2006;26(5):531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 47.Cheng T, Li Q, Zhou Z. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS PharmSciTech. 2012;14(1):137. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elfiky AA. Ribavirina, remdesivir, sofosbuvir, galidesivir e tenofovir contra RNA polimerase dependente de RNA SARS-CoV-2 (RdRp): um estudo de encaixe molecular. Ciênc Vida. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z. Estrutura da RNA polimerase dependente de RNA do vírus COVID-19. Ciência. 2020 doi: 10.1126/science.abb7498. [DOI] [Google Scholar]
- 50.Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S, Xie YC, Tian G, Jiang HW, Tao SC, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE. Base estrutural para a inibição da RNA polimerase dependente de RNA de SARS-CoV-2 por remdesivir. Ciência. 2020 doi: 10.1126/science.abc1560. [DOI] [Google Scholar]
- 51.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin i to angiotensin 1–9. Circ Res. 2000;87(5):E1–9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 52.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parks JM, Smith JC. How to discover antiviral drugs quickly. N Engl J Medi. 2020;382(23):2261–2264. doi: 10.1056/NEJMcibr2007042. [DOI] [PubMed] [Google Scholar]
- 54.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romanos MTV, de Santos NS, O; Wigg MD, Virologia Humana. 3a. Guanabara Koogan: Rio de Janeiro; 2015. [Google Scholar]
- 56.Middleton EJR, Kandaswam C, Theoharides TC. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease and cancer. Pharmacol Rev. 2000;52(4):673–751. [PubMed] [Google Scholar]
- 57.Jacome R, Becerra A, Ponce de Leon S, Lazcano A. Análise estrutural de polimerases dependentes de RNA monoméricas: implicações evolutivas e terapêuticas. Apr Plos One. 2015;10. 10.1371/journal.pone.0139001.
- 58.Tan YW, Fung TS, Shen H, Huang M, Liu DX. Coronavirus proteínas não estruturais do vírus da bronquite infecciosa 8 e 12 formam um complexo estável independente das regiões não traduzidas do RNA viral e outras proteínas virais. Virologia. 2018;513:75–84. doi: 10.1016/j.virol.2017.10.004. [DOI] [Google Scholar]
- 59.South AM, Tomlinson L, Edmonston D, Hiremath S, Sparks MA. Controvérsias da inibição do sistema renina-angiotensina durante a pandemia de COVID-19. Nat Rev Nephrol. 2020 doi: 10.1038/s41581-020-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q, Wong G, Lu G, Yan J. Proteína de pico Gao GF MERS-CoV: alvos para vacinas e terapêuticas. Antiviral Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Enzima conversora de angiotensina 2 (ACE2) como um receptor SARS-CoV-2: mecanismos moleculares e potencial alvo terapêutico. Ter Intensiva Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang HK, Xia Y, Yang ZY, Jiang S. Recent advances in the Discovery and development of flavonoides and their analogues as antitumor and anti-HIV agentes. Flavonoids in the Living System. Manthey and Buslig Plenum Press, New York. 1998;439:191-22510.1007/978-1-4615-5335-9_15. [DOI] [PubMed]
- 63.Brum LP. Antiviral activity of phenolic compounds (ferulic and transcinnamic acids) and flavonoids (quercetin and kaempferol) on bovine herpesvirus 1, bovine herpesvirus 5 and canine distemper virus. 2006. Doctoral Thesis (Doctorate in Agricultural Biochemistry). Federal University of Viçosa, Viçosa, Minas Gerais; 2006.
- 64.Carvalho OV, Oliveira FS, Saraiva GL, Botelho CV, Ferreira HCC, Santos MR, Silva Júnior A, Almeida MR. Antiviral potential of quercetin on canine parvovirus. Arq Bras Med Vet Zootec. 2013;65(2):353-358, Belo Horizonte. 10.1590/S0102-09352013000200008
- 65.Jo S, Kim S, Shin DH, Kim MS. Inhibition of African swine fever virus protease by myricetin and myricitrin. J Enzyme Inhib Med Chem. 2020;35(1):1045–1049. doi: 10.1080/14756366.2020.1754813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pathak D, Pathak K, Singla AK. Flavonoids as medicinal agents: recent advances. Phytotherapy. 1991;57(5):371–389. [Google Scholar]
- 67.Zakaryan H, Arabyan E, Oo A, Zandi K. Flavonóides: Promising natural compounds against viral infections. Adv Virol. 2017;162:2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li BQ, Tao FU, Dongyan Y, Kang EA. Flavonoid baicalein inhibits HIV-1 Infecction at the level of viral entry. Biochem Biophys Res Commun. 2000;276:534–538. doi: 10.1006/bbrc.2000.3485. [DOI] [PubMed] [Google Scholar]
- 69.Asres K, Seyoum A, Veeresham C, Bucar F, Gibbons S. Naturally derived anti-HIV agents. Phytother Res. 2005;19(7):557–581. doi: 10.1002/ptr.1629. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, Ma S, Yang Y, Ye S, But PP. Antiviral activities of flavonoids and organic acid from Trollius chinensis Bunge. J Ethnopharmacol. 2002;79:365–368. doi: 10.1016/s0378-8741(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 71.Theoharides TC, Stewart JM, Hatziagelaki E, Kolaitis G. Brain "fog," inflammation and obesity: key aspects of neuropsychiatric disorders improved by luteolin. Front Neurosci. 2015;9:225. doi: 10.3389/fnins.2015.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang YC, Huang KX, Huang AC, Ho YC, Wang CJ. Hibiscus anthocyanins-rich extract inhibited LDL oxidation and oxLDL-mediated macrophages apoptosis. Food Chem Toxicol. 2006;44(7):1015–1023. doi: 10.1016/j.fct.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 73.Chen PN, Kuo WH, Chiang CL, Chiou HL, Shou YS, Chuc SC. Black rice anthocyanins inhibit cancer cells invasion via repressions of MMPs and u-PA expression. Chem Biol Interact. 2006;163(3):218–229. doi: 10.1016/j.cbi.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 74.Toufektsian MC, De Lorgeril M, Nagy N, Salen P, Donati MB, Giordano L, Mock HP, Peterek S, Matros A, Petroni K, Pilu R, Rotillo D, Tonelli C, De Leiris J, Boucher F, Martin C. Chronic dietary intake of plant-derived anthocyanins protects the rat heart against ischemia reperfusion injury. J Nutr. 2008;138(4):747–752. doi: 10.1093/jn/138.4.747. [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Alonso M, Minihane AM, Rimbach G, Rivas-Gonzalo JC, Tereza SP. Red wine anthocyanins are rapidly absorbed in humans and affect monocyte chemoattractant protein 1 levels and antioxidant capacity of plasma. J Nutr Biochem. 2009;20(7):521–529. doi: 10.1016/j.jnutbio.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 76.Xia M, Ling W, Zhu H, Ma J, Wang Q, Hou M, Tang Z, Guo H, Liu C, Ye Q. Anthocyanin attenuates CD40-mediated endothelial cell activation and apoptosis by inhibiting CD40-induced MAPK activation. Atherosclerosis. 2009;202(1):41–47. doi: 10.1016/j.atherosclerosis.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 77.Galvano F, La Fauci L, Lazzarino G, Fogliano V, Ritieni A, Ciappellano S, Battistini NC, Tavazzi B, Galvano G. Cyanidins: metabolism and biological properties. J Nutr Biochem. 2004;15(1):2–11. doi: 10.1016/j.jnutbio.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Gerardi C, Frassinetti S, Caltavuturo L, Leone A, Lecci R, Calabriso N. Anti-proliferative, anti-inflammatory and anti-mutagenic activities of a Prunus mahaleb L. anthocyanin-rich fruit extract. J Funct Foods. 2016;27:537–e548. doi: 10.1016/j.jff.2016.09.024. [DOI] [Google Scholar]
- 79.Mohammadi PP, Fakhri SS, Asgary MH, Farzaei J, Echeverría, Signaling Thevias and Therapeutic Targets of Antiviral Agents: Focusing on Antiviral Approaches and Clinical Perspectives of Anthocyanins in Viral Disease Management, Frente. Pharmacology. 2019;10:1207. doi: 10.3389/fphar.2019.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel K, Patel DK. Medicinal importance, pharmacological activities, and analytical aspects of hispidulin: A concise report. J Tradit Complement Med. 2016;7(3):360–366. doi: 10.1016/j.jtcme.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watson RR, Schönlau F. Nutraceutical and antioxidant effects of a delphinidin-rich maqui berry extract Delphinol®: a review. Minerva Cardioangiol. 2015;63(2 Suppl 1):1–12. [PubMed] [Google Scholar]
- 82.Tomonori N, Tadashi H, Tokimitsu I. A green tea extract high in catechins reduces body fat and cardiovascular risks in humans. Obesity. 2007;15:1473–1483. doi: 10.1038/oby.2007.176. [DOI] [PubMed] [Google Scholar]
- 83.Stompor M, Broda D, Bajek-Bil A. Dihydrochalcones: Acquisition methods and pharmacological properties. A first systematic review. Molecules. 2019;24(24):4468. doi: 10.3390/molecules24244468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferrandiz ML, Alcaraz MJ. Antiinflammatory activity and inhibition of arachidonic acid metabolism by flavonoids. Agents Actions. 1991;32(3–4):283–288. doi: 10.1007/BF01980887. [DOI] [PubMed] [Google Scholar]
- 85.Simões CM, Schenkel EP, Bauer L, Langeloh A. Pharmacological investigations on Achyrocline satureioides (LAM.) DC., Compositae. J Ethnopharmacol. 1988;22(3):281–93. doi: 10.1016/0378-8741(88)90239-5. [DOI] [PubMed] [Google Scholar]
- 86.Garg A, et al. Chemistry and pharmacology of the Citrus bioflavonoid hesperidin. Phytother Res. 2001;15(8):655–669. doi: 10.1002/ptr.1074. [DOI] [PubMed] [Google Scholar]
- 87.Salehi B, cols. The therapeutic potential of naringenin: A review of clinical trials. Pharmaceuticals. 2019;12(1):11.10.3390/ph12010011. [DOI] [PMC free article] [PubMed]
- 88.Mu H, Bai YH, Wang ST, Zhu ZM, Zhang YW. Research on antioxidant effects and estrogenic effect of formononetin from Trifolium pratense (red clover) Phytomedicine. 2009;16(4):314–319. doi: 10.1016/j.phymed.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 89.Sun M, Zhou T, Zhou L, Chen Q, Yu Y, Yang H, Zhong K, Zhang X, Xu F, Cai S, Yu A, Zhang H, Xiao R, Xiao D, Chui D. Formononetin protects neurons against hypoxia-induced cytotoxicity through upregulation of ADAM10 and sAβPPα. J Alzheimers Dis. 2012;28(4):795–808. doi: 10.3233/JAD-2011-110506. [DOI] [PubMed] [Google Scholar]
- 90.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA. 2003;100:13190. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chikhale RV, Gupta VK, Eldesoky GE, Wabaidur SM, Patil SA, Islam MA. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. Journal of Biomolecular Structure and Dynamics. 2020;1–16. 10.1080/07391102.2020.1798813. [DOI] [PMC free article] [PubMed]
- 92.Bhowmik D, Nandi R, Jagadeesan R, Kumar N, Prakash A, Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. InfectGenet Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hamza M, Ali A, Khan S, Ahmed S, Attique Z, Ur Rehman S, Khan A, Ali H, Rizwan M, Munir A, Khan AM, Siddique F, Mehmood A, Nouroz F. nCOV-19 peptides mass fingerprinting identification, binding, and blocking of inhibitors flavonoids and anthraquinone of Moringa oleifera and hydroxychloroquine. J Biomol Struct Dyn. 2021;39(11):4089–4099. doi: 10.1080/07391102.2020.1778534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fakhar Z, Faramarzi B, Pacifico S, Faramarzi S. Anthocyanin derivatives as potent inhibitors of SARS-CoV-2 main protease: An in-silico perspective of therapeutic targets against COVID-19 pandemic. J Biomol Struct Dyn. 2021;39(16):6171–6183. doi: 10.1080/07391102.2020.1801510. [DOI] [PubMed] [Google Scholar]
- 95.Abian O, Ortega-Alarcon D, Jimenez-Alesanco A, Ceballos-Laita L, Vega S, Reyburn HT, Rizzuti B, Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int J Biol Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Narkhede RR, Pise AV, Cheke RS, Shinde SD. Recognition of natural products as potential inhibitors of COVID-19 Main Protease (Mpro): In-Silico Evidences. Nat Prod Bioprospect. 2020;10(5):297–306. doi: 10.1007/s13659-020-00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fischer A, Sellner M, Neranjan S, Smieško M, Lill MA. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int J Mol Sci. 2020;21(10):3626. doi: 10.3390/ijms21103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Das S, Sarmah S, Lyndem S, Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2020;39(9):3347–3357. 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed]
- 99.Islam R, Parves MR, Paul AS, Uddin N, Rahman MS, Mamun AA, Hossain MN, Ali MA, Halim MA. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020;1–12. 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed]
- 100.Lung J, Lin YS, Yang YH, Chou YL, Shu LH, Cheng YC, Liu HT, Wu CY. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J Med Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Owis AI, El-Hawary MS, El Amir D, Aly OM, Abdelmohsen UR, Kamel MS. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020;10:19570–19575. doi: 10.1039/D0RA03582C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh S, Sk MF, Sonawane A, Kar P, Sadhukhan S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRP) inhibition: an in-silico analysis. J Biomol Struct Dyn. 2020;39(16):6249-6264.10.1080/07391102.2020.1796810. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.