Keywords: lncRNA, proteomics, RNA-sequencing, satellite cells, skeletal muscle
Abstract
MicroRNAs (miRs) control stem cell biology and fate. Ubiquitously expressed and conserved miR-16 was the first miR implicated in tumorigenesis. miR-16 is low in muscle during developmental hypertrophy and regeneration. It is enriched in proliferating myogenic progenitor cells but is repressed during differentiation. The induction of miR-16 blocks myoblast differentiation and myotube formation, whereas knockdown enhances these processes. Despite a central role for miR-16 in myogenic cell biology, how it mediates its potent effects is incompletely defined. In this investigation, global transcriptomic and proteomic analyses after miR-16 knockdown in proliferating C2C12 myoblasts revealed how miR-16 influences myogenic cell fate. Eighteen hours after miR-16 inhibition, ribosomal protein gene expression levels were higher relative to control myoblasts and p53 pathway-related gene abundance was lower. At the protein level at this same time point, miR-16 knockdown globally upregulated tricarboxylic acid (TCA) cycle proteins while downregulating RNA metabolism-related proteins. miR-16 inhibition induced specific proteins associated with myogenic differentiation such as ACTA2, EEF1A2, and OPA1. We extend prior work in hypertrophic muscle tissue and show that miR-16 is lower in mechanically overloaded muscle in vivo. Our data collectively point to how miR-16 is implicated in aspects of myogenic cell differentiation. A deeper understanding of the role of miR-16 in myogenic cells has consequences for muscle developmental growth, exercise-induced hypertrophy, and regenerative repair after injury, all of which involve myogenic progenitors.
INTRODUCTION
MicroRNAs (miRNAs) destabilize mRNAs and/or prevent their translation and are key regulators of myogenic stem cell (satellite cell) biology (1). These small noncoding RNAs control nearly every aspect of myogenic cell function including maintenance of quiescence (2–5), activation (6, 7), proliferation (8, 9), self-renewal (10), migration (11), and differentiation (8, 12, 13). The depletion of miRNAs from myogenic cells results in swift cell death (2, 8, 14). Dysregulation of miRNAs in satellite cells in vivo may negatively affect muscle development, adaptability to exercise, the muscle microenvironment with aging, and regenerative capacity after injury; all these processes depend on satellite cells (15–24). Furthermore, miRNAs are released from satellite cells in extracellular vesicles (EVs) and taken up by recipient cells throughout muscle in vivo (14, 25, 26). Satellite cell communication throughout muscle via miRNAs in EVs contributes to muscle adaptation during loading (27, 28). Understanding the role of miRNAs in myogenic progenitors may accelerate the discovery of miRNA-based therapeutics that affect muscle plasticity via fusion-dependent and/or -independent mechanisms (27, 29).
miR-16 is highly conserved and ubiquitously expressed (30–32). It was the first miRNA implicated in tumorigenesis and is a highly influential miRNA in numerous cellular processes, specifically as it relates to tumor growth (32, 33). In skeletal muscle tissue that contains both mature muscle fibers and myogenic satellite cells, miR-16 content rises during avian embryonic development but begins to wane at birth concomitant with secondary myogenesis (34). Low miR-16 levels during development are associated with muscle hypertrophy (35, 36). We reported that muscle miR-16 levels decline after resistance exercise following a week of acclimation to training and that knockdown in myotubes increases muscle protein synthesis (37). Skeletal muscle miR-16 is also lower in regenerating relative to uninjured muscle (6). The repression of miR-16 in muscle is therefore associated with postnatal myofiber growth and regeneration as well as the exercise-induced hypertrophic response. In myogenic precursor cells, miR-16 is induced upon activation (2, 6, 38) and enriched during proliferation (2, 14) but gradually declines in culture (2) and with differentiation (34, 36, 39). Some evidence suggests that miR-16-5p is high in quiescent satellite cells but still drops precipitously during differentiation (1). miR-16 overexpression prevents myogenic cell differentiation and myotube formation in vitro (34), whereas knockdown enhances these processes (34, 40). MyoD contributes to myogenic cell differentiation (41–48). miR-16 knockdown in murine myofiber-associated myogenic cell culture may increase the proportion of MyoD+ satellite cells by 3 days but reduces it by 5 days, pointing to an effect on myogenic cell behavior and fate (6). The literature collectively points to miR-16 having a key function in myogenic cells and in determining skeletal muscle mass. Despite its importance, how miR-16 exerts its effects in myogenic cells is incompletely defined.
The purpose of this investigation was to provide detailed information on how miR-16 influences myogenic cell biology. First, we evaluated miR-16 levels in mechanically overloaded muscle tissue in vivo, which is a well-established model of rapid growth associated with satellite cell differentiation and fusion (49). We then inhibited miR-16-5p in proliferating C2C12 myoblasts and performed RNA-sequencing (RNA-seq) and discovery proteomics. C2C12s express the satellite cell marker Pax7 (50) and can model the behaviors of primary myogenic progenitors in vitro (51–53). Our experiments point to a multifaceted role for miR-16 repression in myogenic cell differentiation. We thus shed light on potential mechanisms whereby miR-16 influences satellite cell-mediated muscle growth and regenerative potential in vivo (6, 36). We also provide general information on miR-16 targets, which may be informative in nonmuscle cell types where miR-16 is enriched (30).
MATERIALS AND METHODS
Murine Experiment
Female C57BL6/J mice (2–3 mo of age) were utilized for in vivo overload experiments. These experiments were performed for a prior publication from our laboratory (54), and the mice were injected with 5-ethenyl uridine 5 h before being euthanized. Experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Kentucky. Mice were housed in a temperature- and humidity-controlled room maintained on a 14:10-h light-dark cycle, and food and water were provided ad libitum throughout experimentation. Briefly, synergist ablation surgery to overload the plantaris muscle was performed as described by Murach et al. (54). Under isoflurane anesthesia, a portion of the gastrocnemius and soleus was removed, leaving the plantaris to be mechanically overloaded during reambulation (n = 4). Sham-operated mice (no removal of muscle) were controls (n = 3). Seventy-two hours after surgery, animals were euthanized in the morning via a lethal dosage of pentobarbital sodium injected intraperitoneally followed by cervical dislocation. Plantaris muscle tissue was harvested and frozen in liquid nitrogen and stored for downstream analyses. miR-16 level in tissue was evaluated with an unpaired directional t test in GraphPad Prism (Boston, MA).
Cell Culture Experiments
C2C12 murine myoblasts (CRL-1772, ATCC, Manassas, VA) were plated in six-well plates (5 × 104 cells/well) with 2 mL of Dulbecco’s modified Eagle medium (DMEM) combined with 20% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S). Cells were incubated at 37°C with 5% CO2, and medium was changed every 48 h. Upon 40–50% confluence, cells were washed with PBS for transfection (37). For proteomics five technical replicates were utilized, and for transcriptomics three control and two knockdown wells were employed. miR-16 levels in cells were evaluated in Prism with an unpaired directional t test with n = 5 technical replicates.
Transfection of C2C12 Myoblasts
Plasmids encoding either an empty vector control (pCMVmiR; PCMVMIR, Origene, Rockville, MD) or miR-16-1 (miR-16-5p) inhibitor were transferred into DH5-a Escherichia coli as we have previously described (37). Plasmid DNA was amplified and isolated from bacteria with the PureLink HiPure Plasmid Filter Maxiprep kit (K211017, Life Technologies, Carlsbad, CA). For miR-16 inhibition, we utilized anti-miR inhibitor (AM17000, Ambion, Austin, TX). Plasmid DNA (1 μg) was diluted in 50 μL of Opti-MEM reduced serum medium (31985088, Life Technologies), combined with 4 μL of Lipofectamine 2000 (11668019, Life Technologies) diluted in 50 μL of Opti-MEM, and incubated for 20 min to allow lipid/DNA complexes to form. Medium was replaced with Opti-MEM, and lipid/DNA complexes were added and incubated for 5 h at 37°C with 5% CO2. After 5-h incubation, medium was replaced with normal growth medium for 18 h. The efficiency of the inhibition of miR-16 was tested and validated previously (37).
RNA Isolation, Quality Check, and qPCR for miR-16
Myoblast and muscle tissue RNA were isolated with TRIzol reagent, which was then homogenized with a Polytron or bullet homogenizer. After homogenization, RNA was isolated with the phase separation method by addition of chloroform or bromochloropropane followed by centrifugation. The aqueous phase was transferred to a new 1.5-mL tube, and an equal amount of 70% of diethyl pyrocarbonate (DEPC)-treated ethanol was added. RNA isolation was further processed with the Invitrogen RNA isolation kit (K145002, Invitrogen, Carlsbad, CA) or the Zymo Direct-zol kit (Zymo Research, Irvine, CA). RNA concentration was determined with a BioTek Take3 microplate with a BioTek PowerWave XS microplate reader as previously described (55) or NanoDrop (ThermoFisher Scientific). RNA samples were only accepted if 260 nm-to-280 nm ratio was >2.0. Samples were stored at −80°C until further use.
Reverse transcription (RT) of miR-16 was performed with the TaqMan MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Waltham, MA). Briefly, 200 ng of total miRNA was added to a master mix comprised of 1 µL of a 10× TaqMan probe (PN4427975, Applied Biosystems) for U6 and miR-16 each (RT:001973 and RT:000391, respectively, Applied Biosystems), 0.3 µL of dNTPs, 3 µL of MultiScribe RTase, 1.5 µL of 10× RTase buffer, 0.19 µL of RNase inhibitor, and nuclease-free water added up to 15 µL of total volume. RT reaction occurred with a hold step of 30 min at 16°C, followed by 30 min at 42°C and finally 5 min at 85°C. Samples were held at 4°C until further analysis. RT-qPCR was performed with the QuantStudio 3 Real-Time PCR system (Applied Biosystems). A 20-µL reaction composed of an adequate amount of TaqMan probes plus TaqMan Fast Advanced Master Mix (4444556, Applied Biosystems) was used to amplify cDNA. Samples followed a protocol consisting of incubation at 95°C for 4 min, followed by 45 cycles of denaturation, annealing, and extension at 95°C and 60°C. TaqMan probes were measured at the end of the extension step of each cycle. Fluorescence-labeled 20× TaqMan probes (PN4427975, Applied Biosystems) included U6 and miR-16 (TM:001973 and TM:000391, respectively, Applied Biosystems). Results were analyzed with QuantStudio Software. Cycle threshold (CT) was determined, and the ΔCT value was calculated as the difference between CT value and U6 CT value. U6 CT values were not different between experimental conditions. Final quantification of gene expression was calculated with the ΔΔCT method. Relative quantification was calculated as and expressed as arbitrary units.
RNA-Sequencing and Transcriptomic Analyses
RNA-sequencing was performed by NovoGene as previously described by us (54). Standard 150-bp paired-end sequencing was performed, and read counts were >20 million. Raw counts from RNA-sequencing were used as inputs into Partek Flow. Alignment was performed with STAR with mmu39. After low-expressed genes were filtered, DESeq2 (version 1.34.0) was used for normalization and differential analyses to identify differentially expressed genes (DEGs) with pairwise comparisons (56). DEGs were identified with a false discovery rate (FDR, step-up procedure) adjusted P value < 0.05. DEGs with adjusted P value < 0.05 were used for downstream functional analysis using ConsensusPath DB (57). Pathway analysis was conducted using the mouse overrepresentation feature, up- or downregulated DEGs, and the Reactome database with default settings. For pathway analysis, q values were used to determine significance based on DEGs with adjusted P < 0.05.
Proteomics: FASP bHPLC-Orbitrap Fusion
Proteomics was performed at the University of Arkansas for Medical Sciences Proteomics Core. Protein samples were reduced, alkylated, and digested by filter-aided sample preparation (58) with sequencing-grade modified porcine trypsin (Promega). Tryptic peptides were separated into 46 fractions on a 100 × 1.0-mm Acquity BEH C18 column (Waters) with an UltiMate 3000 UHPLC system (Thermo) with a 50-min gradient from 99:1 to 60:40 buffer A-to-B ratio under basic pH conditions and then consolidated into 12 superfractions; buffer A = 0.1% formic acid, 0.5% acetonitrile, and buffer B = 0.1% formic acid, 99.9% acetonitrile. Each superfraction was then further separated by reverse-phase XSelect CSH C18 2.5-μm resin (Waters) on an in-line 150 × 0.075-mm column with an UltiMate 3000 RSLCnano system (Thermo). Peptides were eluted with a 45-min gradient from 98:2 to 65:35 buffer A-to-B ratio. Eluted peptides were ionized by electrospray (2.4 kV) followed by mass spectrometric analysis on an Orbitrap Fusion Tribrid mass spectrometer (Thermo). MS data were acquired with the FTMS analyzer in profile mode at a resolution of 240,000 over a range of 375 to 1,500 m/z. After HCD activation, MS/MS data were acquired with the ion trap analyzer in centroid mode and normal mass range with normalized collision energy of 28–31% depending on charge state and precursor selection range. Proteins were identified by database search using MaxQuant (Max Planck Institute) label-free quantification with a parent ion tolerance of 2.5 ppm and a fragment ion tolerance of 0.5 Da. Scaffold Q + S (Proteome Software) was used to verify MS/MS-based peptide and protein identifications. Protein identifications were accepted if they could be established with <1.0% false discovery and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (59). Protein abundance is presented with intensity-based absolute quantification (iBAQ) values. To determine differential expression and pathway analysis between conditions, P values based on unpaired nondirectional t tests were employed with a cutoff of P < 0.05. Benjamini–Hochberg-adjusted P values were subsequently calculated from these P values. Untranslated region (UTR) sequences for genes that resulted in proteins of interest were downloaded from UCSC Genome Browser and used for prediction of miR-16 binding sites. The potential binding site was identified with RNAhybrid software (60, 61), which combines thermodynamic and seed sequence information.
RESULTS
miR-16 Is Lower after 72-h Synergist Ablation Mechanical Overload of the Plantaris Muscle
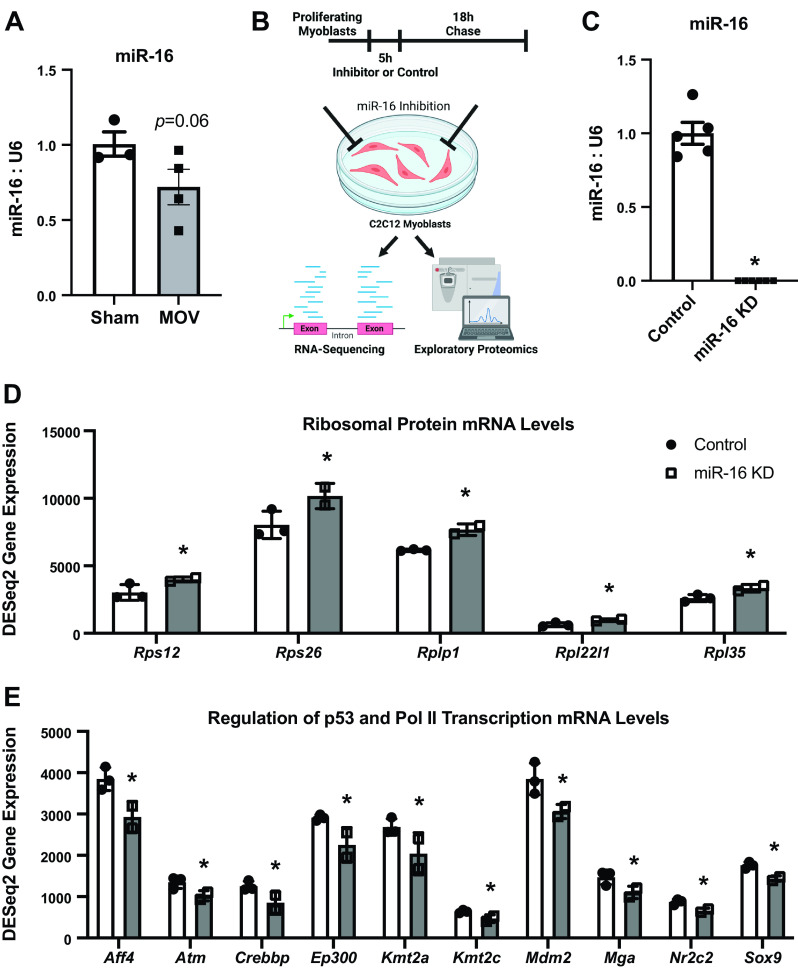
Previous work suggests that muscle miR-16 levels are inversely related to postnatal muscle growth in chickens (34, 36) and are lower during regeneration in mice (24). We showed that miR-16 is lower in muscle tissue after a bout of resistance exercise in rats (37). Previous work showed significant changes in global muscle miRNA levels in the early phase of mechanical overload-induced hypertrophy (62). To determine whether miR-16 declines during the early phase of a well-characterized model of rapid loading-induced hypertrophy in mice (49, 63, 64), we compared miR-16-5p levels in 72-h mechanically overloaded (MOV) plantaris muscle to sham-operated muscles. miR-16 was 28% lower in muscle tissue during MOV (P = 0.06) (Fig. 1A). Overall, low miR-16 appears characteristic of growing muscle in several settings and species (6, 35–37).
Figure 1.
miR-16 levels in overloaded muscle in vivo and RNA-sequencing (RNA-seq) analysis in proliferating C2C12 myoblasts 18 h after miR-16 knockdown (KD). A: miR-16 levels in plantaris skeletal muscle after 72 h of synergist ablation mechanical overload of the plantaris (MOV) relative to sham operation. B: in vitro study design. C: miR-16 levels in control and KD myoblasts. D: levels of ribosomal protein genes in control and KD myoblasts. E: levels of p53- and Pol II transcription-related genes in control and KD myoblasts. For RNA-seq experiments, n = 3 control and n = 2 KD technical replicates. *adj. P < 0.05.
miR-16 Influences Ribosomal-, p53-, and Pol II Transcription-Related mRNA Abundance
The in vitro study design is found in Fig. 1B. The transfection of a miR-16 inhibitor was effective at knocking down miR-16 (miR-16 KD) in proliferating C2C12 myoblasts (Fig. 1C). Inhibition of miR-16 in myoblasts upregulated mRNA levels of Rps12, Rps26, Rplp1, Rpl22l1, and Rpl35 (adj. P < 0.05), which encode ribosomal proteins (Fig. 1D). Additional ribosomal protein genes tended to be higher after miR-16 knockdown (Rpl27 and Rpl27a, adj. P < 0.10) (Supplemental Table S1). The major processes downregulated by miR-16 inhibition were Regulation of TP53 Activity through Methylation (Reactome, q = 0.0014) and RNA Polymerase II Transcription (Reactome, q = 0.0026) (Supplemental Table S2). In the former pathway, Atm, Ep300, and Mdm2 were lower (adj. P < 0.05). In addition to those three genes, Aff4, Crebbp, Kmt2a, Kmt2c, Mga, Nr2c2, and Sox9 were lower after miR-16 knockdown in the latter pathway (adj. P < 0.05) (Fig. 1E). The long noncoding RNA (lncRNA) H19 was elevated with miR-16 inhibition (adj. P = 0.0035), and the lncRNAs Malat1 (adj. P = 0.059) and Xist (adj. P = 0.009) were lower (Supplemental Table S1).
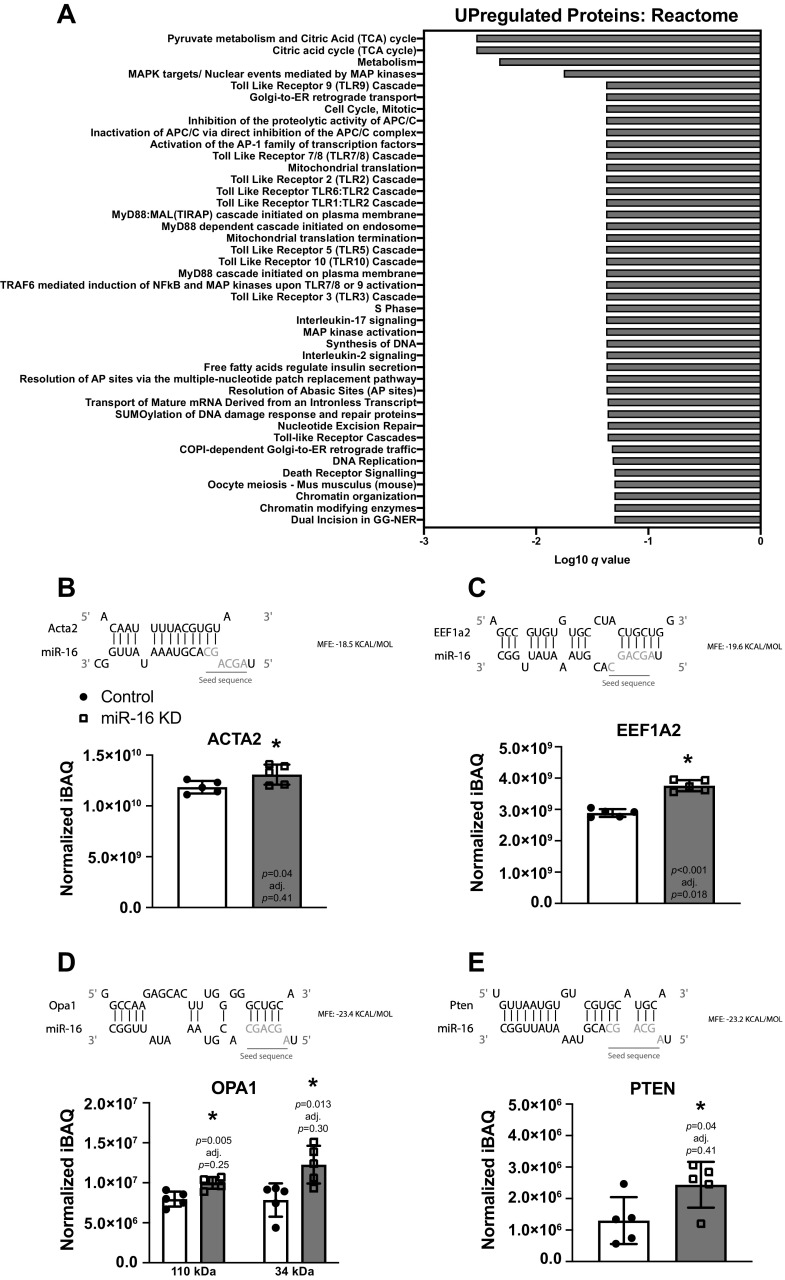
miR-16 Knockdown Increases Metabolism-Related Proteins and Specific Markers of Myogenic Cell Differentiation
Proteomics was performed in five technical replicates per condition. After miR-16 knockdown, five proteins were upregulated (EEF1A2, HIST1H1B, NPC2, PRPH, and TSEN54) and seven proteins were downregulated (GLRX2, GOLPH3, GRK6, LAPTM4A, NDC1, THAP4, and ZC3H11A) with an adjusted P value <0.05, and 899 proteins achieved significance at P < 0.05 (Supplemental Table S3). Proteins involved in different aspects of Metabolism (Reactome, q = 0.0178) were upregulated (assessed using proteins with P < 0.05) (Fig. 2A) (Supplemental Tables S3 and S4). Within the broad Metabolism pathway, various tricarboxylic acid (TCA) cycle proteins were selectively enriched (Reactome, q = 0.00293). Proteins related to Metabolism of RNA (Reactome, q = 0.000456) were most downregulated by miR-16 inhibition (Supplemental Table S4). Markers of myogenic cell maturation, including myosin light chains (MYL6B, MYL9, MYL12A, P < 0.05) as well as smooth muscle actin (ACTA2, P = 0.04, adj. P = 0.41) (Fig. 2B), were upregulated with miR-16 KD. Specific proteins that have a defined role in satellite cell behavior were also altered. In addition to muscle-enriched EEF1A2 (P = 0.0000164, adj. P = 0.018) (Fig. 2C) (65–67), OPA1 (P < 0.05, adj. P = 0.25 and 0.30) (Fig. 2D) (68, 69) and PTEN (P < 0.05, adj. P = 0.41) (Fig. 2E) (70–72) were elevated by miR-16 inhibition; the latter two proteins are of interest but did not achieve significance according to adjusted P values. OPA1 was detected twice (canonical 111 kDa and truncated 34 kDa), and the levels of both isoforms were higher with knockdown (P < 0.05) but not according to adjusted P values.
Figure 2.
Proteomic analysis in proliferating C2C12 myoblasts 18 h after miR-16 knockdown (KD). A: Reactome pathway analysis using upregulated differentially expressed proteins (P < 0.05). B–E: thermodynamic and seed sequence target prediction for miR-16 and protein levels of ACTA2 (B), EEF1A2 (C), OPA1 (D), and PTEN (E) in control and KD myoblasts. iBAQ, intensity-based absolute quantification. For all experiments, n = 5 technical replicates were utilized. *P < 0.05. Created with BioRender.com.
An overlap in gene and protein expression would not necessarily be expected at a discrete time point since protein translation lags behind changes in mRNA expression. Nevertheless, comparison of gene to protein levels revealed agreement for Bmpr2, Cltc, Lama5, Lpp, Mga, Nptx1, and Sema5a, all of which were lowered by miR-16 knockdown (P < 0.05). Rpl27a, which was trending to increase at the gene level, was elevated at the protein level (P = 0.01, adj. P = 0.28). Since we observed upregulation of proteins with miR-16 knockdown, and miRNAs can prevent protein translation without altering transcript levels (14, 73), we performed a 3′ UTR binding affinity analysis of miR-16 for ACTA2, EEF1A2, OPA1, and PTEN (Fig. 2, B–E), using RNAhybrid (60, 61). Based on free energy, the results collectively suggested that miR-16 could regulate the levels of these proteins. The highest complementarity between the miR-16 seed sequence and 3′ UTR sequence was for EEF1A2, OPA1, and PTEN (Fig. 2. C–E).
DISCUSSION
Satellite cells differentiate and begin fusing appreciably to muscle fibers between days 4 and 5 of mechanical overload in adult mice (25) and ∼5 days after transplantation into muscle of mice (74). In our 72-h overload experiments, the contributions of miR-16 from satellite cells versus other cell types in muscle tissue (e.g., muscle fibers or immune cells) (75) cannot be discerned. The influence of hypertrophy per se versus a degeneration/regeneration response that can occur with synergist ablation (19, 76) is also unclear. Nevertheless, our observation of lower miR-16 in murine muscle tissue after 72 h of MOV corresponds with reduced muscle miR-16 levels during early in vivo regeneration after injury (6) and during recovery from a bout of resistance exercise (37). To model miR-16 regulation of myogenic cell behavior, we repressed it in proliferating myoblasts and performed transcriptomic and proteomic profiling. These data provide a framework for understanding miR-16’s role in myogenic cell fate.
miR-16-5p can induce p53 signaling in myogenic cells (34). Repression of p53-related gene expression with miR-16 knockdown dovetails with this observation. In C2C12s in vitro, the expression of ribosomal proteins is upregulated during early differentiation (77, 78). Enrichment of ribosomal protein genes is associated with satellite cell fusion into the myofiber syncytium during hypertrophy (79). The induction of ribosomal protein genes, specifically Rps26, by miR-16 inhibition may characterize myogenic cells that are primed for fusion (79). The lncRNA H19 was elevated with miR-16 knockdown, whereas Malat1 and Xist were lower. lncRNAs are not typically targeted for decay by miRNAs since translation is seemingly required for RNA destabilization (80). H19 induction (81–84) and Malat1 reduction (85) are strongly implicated in myogenic cell differentiation, but how miR-16 could affect lncRNA levels in myogenic cells deserves further investigation. In concert with evidence suggesting that miR-16 can target Myomaker, the gatekeeper of myogenic cell fusion that gradually increases during differentiation (86, 87), our RNA-seq data collectively point to declining miR-16 levels facilitating lineage progression toward differentiation.
At the protein level, repressing miR-16 in proliferating myoblasts leads to several alterations that are indicative of miR-16’s roles in the regulation of differentiation. The enrichment of myosin light chains, noted in our data, is a sign of myogenic cell maturation (88, 89). Upregulation of smooth muscle actin (Acta2) is also strongly associated with myoblast differentiation (90–93). Satellite cell differentiation is controlled by a metabolic shift and progression from glycolysis to the TCA cycle (94). Elevated TCA cycle proteins with miR-16 repression therefore seems intuitive from a metabolic perspective. Induction and phosphorylation of EEF1A2 is linked to myogenic cell differentiation (65–67). EEF1A2 is muscle enriched (95) and highly regulated during muscle development (96), protects myotubes from cell death (65), and controls Utrophin levels in skeletal muscle (97, 98). EEF1A2 is also a core component of cardiomyocyte differentiation (99). The causal functions of EEF1A2 control by miR-16 in myoblast differentiation deserve further study. Higher PTEN and OPA1 with miR-16 inhibition is noteworthy since both are implicated in reinforcing satellite cell quiescence (68, 70, 71). PTEN can repress the satellite cell identity gene Pax7 (72, 100) but may also facilitate a return to quiescence (70, 71) that could ensue if differentiation and fusion does not progress. Whereas OPA1 supports quiescence by maintaining mitochondrial integrity (68), OPA1 induction and mitophagy is an essential component of successful C2C12 myoblast differentiation (69).
The inhibition of miR-16 in proliferating myoblasts, which occurs naturally during myogenic differentiation, reveals its contributions to this process. Specifically, we uncover miR-16’s regulation of ribosomal, p53-related, and lncRNA gene expression as well as metabolic- and muscle maturation-related protein abundance. A more detailed understanding of miR-16 dynamics, specifically in satellite cells in vivo during different stages of myogenesis, regeneration, and hypertrophy, will inform how miR-16 controls muscle mass in varying circumstances. The present study is limited to a single time point with an immortalized cell line. We also do not report on myogenic cell behavior after miR-16 inhibition, although this has been documented in detail elsewhere (6, 34, 40). Limitations aside, we provide insights from two -omic layers to expand our understanding of miR-16 regulation in myogenic cells.
DATA AVAILABILITY
Processed data are provided in Supplemental Tables S1–S4 and were deposited in GEO (GSE229134).
SUPPLEMENTAL DATA
Supplemental Tables S1–S4: https://doi.org/10.6084/m9.figshare.22151624.
GRANTS
This work was supported by NIH Grant R00 AG063994 to K.A.M. Support for proteomics was provided by an Arkansas Biosciences Institute grant to N.P.G.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
N.P.G. and K.A.M. conceived and designed research; S.L., D.L., F.M., P.J.K., and F.v. performed experiments; S.L., F.M., P.J.K., I.J.V., and K.A.M. analyzed data; S.L., F.M., I.J.V., and K.A.M. interpreted results of experiments; S.L., I.J.V., and K.A.M. prepared figures; S.L., D.L., and K.A.M. drafted manuscript; S.L., D.L., F.M., P.J.K., I.J.V., F.v., N.P.G., and K.A.M. edited and revised manuscript; S.L., D.L., F.M., P.J.K., I.J.V., F.v., N.P.G., and K.A.M. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. John J. McCarthy of the University of Kentucky and Dr. Vandre C. Figueiredo of Oakland University for critical feedback on this manuscript. The authors also thank the University of Arkansas for Medical Sciences Proteomics Core for conducting the proteomics and analysis.
The Graphical Abstract was generated with BioRender.
REFERENCES
- 1. Castel D, Baghdadi MB, Mella S, Gayraud-Morel B, Marty V, Cavaillé J, Antoniewski C, Tajbakhsh S. Small-RNA sequencing identifies dynamic microRNA deregulation during skeletal muscle lineage progression. Sci Rep 8: 4208, 2018. doi: 10.1038/s41598-018-21991-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482: 524–528, 2012. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11: 118–126, 2012. [Erratum in Cell Stem Cell 11: 279, 2012]. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 4. Koning M, Werker PM, van Luyn MJ, Krenning G, Harmsen MC. A global downregulation of microRNAs occurs in human quiescent satellite cells during myogenesis. Differentiation 84: 314–321, 2012. doi: 10.1016/j.diff.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 5. de Morree A, Klein JD, Gan Q, Farup J, Urtasun A, Kanugovi A, Bilen B, van Velthoven CT, Quarta M, Rando TA. Alternative polyadenylation of Pax3 controls muscle stem cell fate and muscle function. Science 366: 734–738, 2019. doi: 10.1126/science.aax1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farina NH, Hausburg M, Betta N, Pulliam C, Srivastava D, Cornelison D, Olwin BB. A role for RNA post-transcriptional regulation in satellite cell activation. Skelet Muscle 2: 21, 2012. doi: 10.1186/2044-5040-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell 10: 327–336, 2012. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang D-Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol 190: 867–879, 2010. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lozano-Velasco E, Vallejo D, Esteban FJ, Doherty C, Hernández-Torres F, Franco D, Aránega AE. A Pitx2-MicroRNA pathway modulates cell proliferation in myoblasts and skeletal-muscle satellite cells and promotes their commitment to a myogenic cell fate. Mol Cell Biol 35: 2892–2909, 2015. doi: 10.1128/MCB.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su Y, Yu Y, Liu C, Zhang Y, Liu C, Ge M, Li L, Lan M, Wang T, Li M, Liu F, Xiong L, Wang K, He T, Shi J, Song Y, Zhao Y, Li N, Yu Z, Meng Q. Fate decision of satellite cell differentiation and self-renewal by miR-31-IL34 axis. Cell Death Differ 27: 949–965, 2020. doi: 10.1038/s41418-019-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baghdadi MB, Firmino J, Soni K, Evano B, Di Girolamo D, Mourikis P, Castel D, Tajbakhsh S. Notch-induced miR-708 antagonizes satellite cell migration and maintains quiescence. Cell Stem Cell 23: 859–868.e5, 2018. doi: 10.1016/j.stem.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 12. Goljanek-Whysall K, Sweetman D, Abu-Elmagd M, Chapnik E, Dalmay T, Hornstein E, Münsterberg A. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc Natl Acad Sci USA 108: 11936–11941, 2011. doi: 10.1073/pnas.1105362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goljanek-Whysall K, Pais H, Rathjen T, Sweetman D, Dalmay T, Münsterberg A. Regulation of multiple target genes by miR-1 and miR-206 is pivotal for C2C12 myoblast differentiation. J Cell Sci 125: 3590–3600, 2012. doi: 10.1242/jcs.101758. [DOI] [PubMed] [Google Scholar]
- 14. Fry CS, Kirby TJ, Kosmac K, McCarthy JJ, Peterson CA. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 20: 56–69, 2017. doi: 10.1016/j.stem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bachman JF, Klose A, Liu W, Paris ND, Blanc RS, Schmalz M, Knapp E, Chakkalakal JV. Prepubertal skeletal muscle growth requires Pax7-expressing satellite cell-derived myonuclear contribution. Development 145: dev167197, 2018. doi: 10.1242/dev.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cramer AA, Prasad V, Eftestøl E, Song T, Hansson KA, Dugdale HF, Sadayappan S, Ochala J, Gundersen K, Millay DP. Nuclear numbers in syncytial muscle fibers promote size but limit the development of larger myonuclear domains. Nat Commun 11: 6287, 2020. doi: 10.1038/s41467-020-20058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hansson KA, Eftestøl E, Bruusgaard JC, Juvkam I, Cramer AW, Malthe-Sørenssen A, Millay DP, Gundersen K. Myonuclear content regulates cell size with similar scaling properties in mice and humans. Nat Commun 11: 6288, 2020. doi: 10.1038/s41467-020-20057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fry CS, Lee JD, Mula J, Kirby TJ, Jackson JR, Liu F, Yang L, Mendias CL, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat Med 21: 76–80, 2015. doi: 10.1038/nm.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, Van Zant G, Campbell KS, Esser KA, Dupont-Versteegden EE, Peterson CA. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138: 3657–3666, 2011. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138: 3639–3646, 2011. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138: 3647–3656, 2011. [Erratum in Development 138: 4333, 2011]. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 22. Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138: 3625–3637, 2011. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Englund D, Figueiredo V, Dungan C, Murach K, Peck B, Petrosino J, Brightwell C, Dupont A, Neal A, Fry C, Accornero F, McCarthy J, Peterson C. Satellite cell depletion disrupts transcriptional coordination and muscle adaptation to exercise. Function (Oxf) 2: zqaa033, 2021. doi: 10.1093/function/zqaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Englund DA, Murach KA, Dungan CM, Figueiredo VC, Vechetti IJ Jr, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Depletion of resident muscle stem cells negatively impacts running volume, physical function and muscle hypertrophy in response to lifelong physical activity. Am J Physiol Cell Physiol 318: C1178–C1188, 2020. doi: 10.1152/ajpcell.00090.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murach KA, Peck BD, Policastro RA, Vechetti IJ, Van Pelt DW, Dungan CM, Denes LT, Fu X, Brightwell CR, Zentner GE, Dupont-Versteegden EE, Richards CI, Smith JJ, Fry CS, McCarthy JJ, Peterson CA. Early satellite cell communication creates a permissive environment for long-term muscle growth. iScience 24: 102372, 2021. doi: 10.1016/j.isci.2021.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murach KA, Vechetti IJ Jr, Van Pelt DW, Crow SE, Dungan CM, Figueiredo VC, Kosmac K, Fu X, Richards CI, Fry CS, McCarthy JJ, Peterson CA. Fusion-independent satellite cell communication to muscle fibers during load-induced hypertrophy. Function (Oxf) 1: zqaa009, 2020. doi: 10.1093/function/zqaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murach KA, Fry CS, Dupont‐Versteegden EE, McCarthy JJ, Peterson CA. Fusion and beyond: Satellite cell contributions to loading‐induced skeletal muscle adaptation. FASEB J 35: e21893, 2021. doi: 10.1096/fj.202101096R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murach KA, Fry CS, Kirby TJ, Jackson JR, Lee JD, White SH, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Starring or supporting role? Satellite cells and skeletal muscle fiber size regulation. Physiology (Bethesda) 33: 26–38, 2018. doi: 10.1152/physiol.00019.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bagley JR, Denes LT, McCarthy JJ, Wang ET, Murach KA. The myonuclear domain in adult skeletal muscle fibres: past, present, and future. J Physiol 601: 723–741, 2023. doi: 10.1113/JP283658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yue J, Tigyi G. Conservation of miR-15a/16-1 and miR-15b/16-2 clusters. Mamm Genome 21: 88–94, 2010. doi: 10.1007/s00335-009-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang H, Fu Z, Jiang X, Wang N, Wang F, Wang X, Zhang S, Wang Y, Yan X, Guan WX, Zhang CY, Zen K, Zhang Y, Chen X, Zhou G. miR-16 promotes the apoptosis of human cancer cells by targeting FEAT. BMC Cancer 15: 448, 2015. doi: 10.1186/s12885-015-1458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aqeilan R, Calin G, Croce C. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ 17: 215–220, 2010. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 33. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529, 2002. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cai B, Ma M, Chen B, Li Z, Abdalla BA, Nie Q, Zhang X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis 9: 367, 2018. doi: 10.1038/s41419-018-0403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jia X, Lin H, Nie Q, Zhang X, Lamont SJ. A short insertion mutation disrupts genesis of miR-16 and causes increased body weight in domesticated chicken. Sci Rep 6: 36433, 2016. doi: 10.1038/srep36433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jia X, Ouyang H, Abdalla BA, Xu H, Nie Q, Zhang X. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities. Biochim Biophys Acta Gene Regul Mech 1860: 674–684, 2017. doi: 10.1016/j.bbagrm.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 37. Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA Jr, Wiggs MP, Nilsson MI, Crouse SF, Fluckey JD, Washington TA, Greene NP. microRNA-16 is downregulated during insulin resistance and controls skeletal muscle protein accretion. J Cell Biochem 117: 1775–1787, 2016. doi: 10.1002/jcb.25476. [DOI] [PubMed] [Google Scholar]
- 38. Aguilar CA, Pop R, Shcherbina A, Watts A, Matheny RW Jr, Cacchiarelli D, Han WM, Shin E, Nakhai SA, Jang YC, Carrigan CT, Gifford CA, Kottke MA, Cesana M, Lee J, Urso ML, Meissner A. Transcriptional and chromatin dynamics of muscle regeneration after severe trauma. Stem Cell Reports 7: 983–997, 2016. doi: 10.1016/j.stemcr.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siengdee P, Trakooljul N, Murani E, Schwerin M, Wimmers K, Ponsuksili S. MicroRNAs regulate cellular ATP levels by targeting mitochondrial energy metabolism genes during C2C12 myoblast differentiation. PLoS One 10: e0127850, 2015. doi: 10.1371/journal.pone.0127850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harding RL, Velleman SG. MicroRNA regulation of myogenic satellite cell proliferation and differentiation. Mol Cell Biochem 412: 181–195, 2016. doi: 10.1007/s11010-015-2625-6. [DOI] [PubMed] [Google Scholar]
- 41. Cornelison D, Olwin BB, Rudnicki MA, Wold BJ. MyoD−/− satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol 224: 122–137, 2000. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- 42. Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol 144: 631–643, 1999. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2-and COOH-terminal regions. J Cell Biol 171: 471–482, 2005. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 18: 2348–2353, 2004. doi: 10.1101/gad.1234304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol 210: 440–455, 1999. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Di Carlo A, De Mori R, Martelli F, Pompilio G, Capogrossi MC, Germani A. Hypoxia inhibits myogenic differentiation through accelerated MyoD degradation. J Biol Chem 279: 16332–16338, 2004. doi: 10.1074/jbc.M313931200. [DOI] [PubMed] [Google Scholar]
- 47. Zhang H, Wen J, Bigot A, Chen J, Shang R, Mouly V, Bi P. Human myotube formation is determined by MyoD–Myomixer/Myomaker axis. Sci Adv 6: eabc4062, 2020. doi: 10.1126/sciadv.abc4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamamoto M, Legendre NP, Biswas AA, Lawton A, Yamamoto S, Tajbakhsh S, Kardon G, Goldhamer DJ. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Reports 10: 956–969, 2018. doi: 10.1016/j.stemcr.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kirby TJ, McCarthy JJ, Peterson CA, Fry CS. Synergist ablation as a rodent model to study satellite cell dynamics in adult skeletal muscle. Methods Mol Biol 1460: 43–52, 2016. doi: 10.1007/978-1-4939-3810-0_4. [DOI] [PubMed] [Google Scholar]
- 50. Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell 102: 777–786, 2000. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 51. Asakura A, Rudnicki MA, Komaki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 68: 245–253, 2001. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- 52. Lamon S, Zacharewicz E, Stephens AN, Russell AP. EPO‐receptor is present in mouse C2C12 and human primary skeletal muscle cells but EPO does not influence myogenesis. Physiol Rep 2: e00256, 2014. doi: 10.1002/phy2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270: 725–727, 1977. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 54. Murach KA, Liu Z, Jude B, Figueiredo VC, Wen Y, Khadgi S, Lim S, Morena da Silva F, Greene NP, Lanner JT, McCarthy JJ, Vechetti IJ Jr, von Walden F. Multi-transcriptome analysis following an acute skeletal muscle growth stimulus yields tools for discerning global and MYC regulatory networks. J Biol Chem 298: 102515, 2022. doi: 10.1016/j.jbc.2022.102515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lim S, Deaver JW, Rosa-Caldwell ME, Haynie WS, Morena da Silva F, Cabrera AR, Schrems ER, Saling LW, Jansen LT, Dunlap KR, Wiggs MP, Washington TA, Greene NP. Development of metabolic and contractile alterations in development of cancer cachexia in female tumor-bearing mice. J Appl Physiol (1985) 132: 58–72, 2022. doi: 10.1152/japplphysiol.00660.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kamburov A, Herwig R. ConsensusPathDB 2022: molecular interactions update as a resource for network biology. Nucleic Acids Res 50: D587–D595, 2022. doi: 10.1093/nar/gkab1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362, 2009. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 59. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658, 2003. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 60. Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34: W451–W454, 2006. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517, 2004. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vechetti IJ Jr, Peck BD, Wen Y, Walton RG, Valentino TR, Alimov AP, Dungan CM, Van Pelt DW, von Walden F, Alkner B, Peterson CA, McCarthy JJ. Mechanical overload‐induced muscle‐derived extracellular vesicles promote adipose tissue lipolysis. FASEB J 35: e21644, 2021. doi: 10.1096/fj.202100242R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roy RR, Edgerton V. Response of mouse plantaris muscle to functional overload: comparison with rat and cat. Comp Biochem Physiol A Physiol 111: 569–575, 1995. doi: 10.1016/0300-9629(95)00062-c. [DOI] [PubMed] [Google Scholar]
- 64. Murach KA, McCarthy JJ, Peterson CA, Dungan CM. Making mice mighty: recent advances in translational models of load-induced muscle hypertrophy. J Appl Physiol (1985) 129: 516–521, 2020. doi: 10.1152/japplphysiol.00319.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem 277: 5418–5425, 2002. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Piazzi M, Bavelloni A, Faenza I, Blalock W, Urbani A, D’Aguanno S, Fiume R, Ramazzotti G, Maraldi NM, Cocco L. eEF1A phosphorylation in the nucleus of insulin-stimulated C2C12 myoblasts. Mol Cell Proteomics 9: 2719–2728, 2010. doi: 10.1074/mcp.M110.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vislovukh AA, Groisman IS, El’skaya AV, Negrutskii BS, Polesskaya AN. Transcriptional and post-transcriptional control of eEF1A2 expression during myoblast differentiation. Вiopolymers Cell 28: 456–460, 2012. doi: 10.7124/bc.000136. [DOI] [Google Scholar]
- 68. Baker N, Wade S, Triolo M, Girgis J, Chwastek D, Larrigan S, Feige P, Fujita R, Crist C, Rudnicki MA, Burelle Y, Khacho M. The mitochondrial protein OPA1 regulates the quiescent state of adult muscle stem cells. Cell Stem Cell 29: 1315–1332.e9, 2022. doi: 10.1016/j.stem.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sin J, Andres AM, Taylor DJ, Weston T, Hiraumi Y, Stotland A, Kim BJ, Huang C, Doran KS, Gottlieb RA. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 12: 369–380, 2016. doi: 10.1080/15548627.2015.1115172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yue F, Bi P, Wang C, Shan T, Nie Y, Ratliff TL, Gavin TP, Kuang S. Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat Commun 8: 14328, 2017. doi: 10.1038/ncomms14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yue F, Bi P, Wang C, Li J, Liu X, Kuang S. Conditional loss of Pten in myogenic progenitors leads to postnatal skeletal muscle hypertrophy but age-dependent exhaustion of satellite cells. Cell Rep 17: 2340–2353, 2016. doi: 10.1016/j.celrep.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Langdon CG, Gadek KE, Garcia MR, Evans MK, Reed KB, Bush M, Hanna JA, Drummond CJ, Maguire MC, Leavey PJ, Finkelstein D, Jin H, Schreiner PA, Rehg JE, Hatley ME. Synthetic essentiality between PTEN and core dependency factor PAX7 dictates rhabdomyosarcoma identity. Nat Commun 12: 5520, 2021. doi: 10.1038/s41467-021-25829-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114, 2008. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 74. Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol 125: 1275–1287, 1994. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. von Walden F, Rea M, Mobley CB, Fondufe-Mittendorf Y, McCarthy JJ, Peterson CA, Murach KA. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics 15: 1151–1162, 2020. doi: 10.1080/15592294.2020.1755581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Murach KA, White SH, Wen Y, Ho A, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Differential requirement for satellite cells during overload-induced muscle hypertrophy in growing versus mature mice. Skelet Muscle 7: 14, 2017. doi: 10.1186/s13395-017-0132-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stansfield BN, Brown AD, Stewart CE, Burniston JG. Dynamic profiling of protein mole synthesis rates during C2C12 myoblast differentiation. Proteomics 21: 2000071, 2021. doi: 10.1002/pmic.202000071. [DOI] [PubMed] [Google Scholar]
- 78. Brown AD, Stewart CE, Burniston JG. Degradation of ribosomal and chaperone proteins is attenuated during the differentiation of replicatively aged C2C12 myoblasts. J Cachexia Sarcopenia Muscle 13: 2562–2575, 2022. doi: 10.1002/jcsm.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Murach KA, Dungan CM, von Walden F, Wen Y. Epigenetic evidence for distinct contributions of resident and acquired myonuclei during long-term exercise adaptation using timed in vivo myonuclear labeling. Am J Physiol Cell Physiol 322: C86–C93, 2022. doi: 10.1152/ajpcell.00358.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Biasini A, Abdulkarim B, de Pretis S, Tan JY, Arora R, Wischnewski H, Dreos R, Pelizzola M, Ciaudo C, Marques AC. Translation is required for miRNA‐dependent decay of endogenous transcripts. EMBO J 40: e104569, 2021. doi: 10.15252/embj.2020104569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu X, Ji S, Li W, Yi B, Li H, Zhang H, Ma W. LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/FoxO1. Cell Mol Biol Lett 22: 10, 2017. doi: 10.1186/s11658-017-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li J, Su T, Zou C, Luo W, Shi G, Chen L, Fang C, Li C. Long non-coding RNA H19 regulates porcine satellite cell differentiation through miR-140-5p/SOX4 and DBN1. Front Cell Dev Biol 8: 518724, 2020. doi: 10.3389/fcell.2020.518724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li J, Zhao W, Li Q, Huang Z, Shi G, Li C. Long non-coding RNA H19 Promotes porcine satellite cell differentiation by interacting with TDP43. Genes 11: 259, 2020. doi: 10.3390/genes11030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28: 491–501, 2014. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen X, He L, Zhao Y, Li Y, Zhang S, Sun K, So K, Chen F, Zhou L, Lu L, Wang L, Zhu X, Bao X, Esteban MA, Nakagawa S, Prasanth KV, Wu Z, Sun H, Wang H. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov 3: 17002, 2017. doi: 10.1038/celldisc.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499: 301–305, 2013. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. McKellar DW, Walter LD, Song LT, Mantri M, Wang MF, De Vlaminck I, Cosgrove BD. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun Biol 4: 1280, 2021. doi: 10.1038/s42003-021-02810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xue Q, Zhang G, Li T, Ling J, Zhang X, Wang J. Transcriptomic profile of leg muscle during early growth in chicken. PLoS One 12: e0173824, 2017. doi: 10.1371/journal.pone.0173824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maire P, Dos Santos M, Madani R, Sakakibara I, Viaut C, Wurmser M. Myogenesis control by SIX transcriptional complexes. Semin Cell Dev Biol 104: 51–64, 2020. doi: 10.1016/j.semcdb.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 90. Randrianarison-Huetz V, Papaefthymiou A, Herledan G, Noviello C, Faradova U, Collard L, Pincini A, Schol E, Decaux JF, Maire P, Vassilopoulos S, Sotiropoulos A. Srf controls satellite cell fusion through the maintenance of actin architecture. J Cell Biol 217: 685–700, 2018. doi: 10.1083/jcb.201705130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Al Tanoury Z, Rao J, Tassy O, Gobert B, Gapon S, Garnier JM, Wagner E, Hick A, Hall A, Gussoni E, Pourquié O. Differentiation of the human PAX7-positive myogenic precursors/satellite cell lineage in vitro. Development 147: dev187344, 2020. doi: 10.1242/dev.187344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang J, Broer T, Chavez T, Zhou CJ, Tran S, Xiang Y, Khodabukus A, Diao Y, Bursac N. Myoblast deactivation within engineered human skeletal muscle creates a transcriptionally heterogeneous population of quiescent satellite-like cells. Biomaterials 284: 121508, 2022. doi: 10.1016/j.biomaterials.2022.121508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14: 309–315, 2017. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hori S, Hiramuki Y, Nishimura D, Sato F, Sehara-Fujisawa A. PDH‐mediated metabolic flow is critical for skeletal muscle stem cell differentiation and myotube formation during regeneration in mice. FASEB J 33: 8094–8109, 2019. doi: 10.1096/fj.201802479R. [DOI] [PubMed] [Google Scholar]
- 95. Kahns S, Knudsen C, Clark B, Lund A, Kristensen P, Cavallius J, Merrick W. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res 26: 1884–1890, 1998. doi: 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hudson NJ, Lyons RE, Reverter A, Greenwood PL, Dalrymple BP. Inferring the in vivo cellular program of developing bovine skeletal muscle from expression data. Gene Expr Patterns 13: 109–125, 2013. doi: 10.1016/j.gep.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 97. Péladeau C, Adam N, Bronicki LM, Coriati A, Thabet M, Al-Rewashdy H, Vanstone J, Mears A, Renaud JM, Holcik M, Jasmin BJ. Identification of therapeutics that target eEF1A2 and upregulate utrophin A translation in dystrophic muscles. Nat Commun 11: 1990, 2020. doi: 10.1038/s41467-020-15971-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Miura P, Coriati A, Bélanger G, De Repentigny Y, Lee J, Kothary R, Holcik M, Jasmin B. The utrophin A 5′-UTR drives cap-independent translation exclusively in skeletal muscles of transgenic mice and interacts with eEF1A2. Hum Mol Genet 19: 1211–1220, 2010. doi: 10.1093/hmg/ddp591. [DOI] [PubMed] [Google Scholar]
- 99. Lyu Y, Jia W, Wu Y, Zhao X, Xia Y, Guo X, Kang J. Cpmer: a new conserved eEF1A2-binding partner that regulates Eomes translation and cardiomyocyte differentiation. Stem Cell Reports 17: 1154–1169, 2022. doi: 10.1016/j.stemcr.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Duan S, Yuan G, Liu X, Ren R, Li J, Zhang W, Wu J, Xu X, Fu L, Li Y, Yang J, Zhang W, Bai R, Yi F, Suzuki K, Gao H, Esteban CR, Zhang C, Izpisua Belmonte JC, Chen Z, Wang X, Jiang T, Qu J, Tang F, Liu G-H. PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nat Commun 6: 10068, 2015. doi: 10.1038/ncomms10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables S1–S4: https://doi.org/10.6084/m9.figshare.22151624.
Data Availability Statement
Processed data are provided in Supplemental Tables S1–S4 and were deposited in GEO (GSE229134).