Abstract
The lysine demethylase LSD1 (also called KDM1A) plays important roles in promoting multiple malignancies including both hematologic cancers and solid tumors. LSD1 targets histone and non-histone proteins and can function as a transcriptional corepressor and coactivator. LSD1 has been reported to act as a coactivator of androgen receptor (AR) in prostate cancer (PCa) and to regulate the AR cistrome via demethylation of its pioneer factor FOXA1. A deeper understanding of the key oncogenic programs targeted by LSD1 could help stratify PCa patients for treatment with LSD1 inhibitors, which are currently under clinical investigation. In this study, we performed transcriptomic profiling in an array of castration-resistant PCa (CRPC) xenograft models that are sensitive to LSD1 inhibitor treatment. Impaired tumor growth by LSD1 inhibition was attributed to significantly decreased MYC signaling, and MYC was found to be a consistent target of LSD1. Moreover, LSD1 formed a network with BRD4 and FOXA1 and was enriched at super-enhancer (SE) regions exhibiting liquid-liquid phase separation. Combining LSD1 inhibitors with BET inhibitors exhibited strong synergy in disrupting the activities of multiple drivers in CRPC, thereby inducing significant growth repression of tumors. Importantly, the combination treatment showed superior effects than either inhibitor alone in disrupting a subset of newly identified CRPC-specific SEs. These results provide mechanistic and therapeutic insights for co-targeting two key epigenetic factors and could be rapidly translated in the clinic for CRPC patients.
Keywords: LSD1, BRD4, MYC, Super Enhancer, prostate cancer, castration-resistant prostate cancer, FOXA1
INTRODUCTION
Although the androgen receptor (AR) pathway is the major factor to drive prostate cancer (PCa) tumorigenesis, patients only benefit from the AR signaling inhibitors (ARSi) for a limited time and eventually develop resistance, and PCa tumors enter a stage called castration-resistant PCa (CRPC) leading to lethality (1). Multiple molecular pathways promote the survival and the progression of PCa cells under therapeutic pressure, including aberrant AR genetic alteration (amplification, splicing variant), AR cofactor mutations, and AR-independent mechanisms (2–5). Therefore, developing novel therapeutic regimens to improve clinical outcomes for CRPC patients is of paramount importance.
Mounting evidence has suggested that alterations of epigenetic regulators may play an essential role in conferring PCa growth advantage during drug resistance evolution (6). LSD1/KDM1A (Lysine-Specific Demethylase 1), a histone lysine demethylase, was initially established as a transcriptional repressor, via removing methyl groups of mono- and di-methylated histone 3 lysine 4 (H3K4me1/2) (7). However, LSD1 was later shown to function as a transcriptional activator in PCa and breast cancer (BCa), possibly via switching its histone substrate to H3K9me1/2, a mark of repressive gene transcription (8–10). While our early integrative genome-wide studies confirm that LSD1 functions as a coactivator of AR, the data also indicate that LSD1 can still maintain its H3K4 demethylase activity at AR binding sites (11). Significantly, our recent findings demonstrate that LSD1 chromatin binding enriches at active enhancers, and inhibition of LSD1 drastically disrupts the global binding of FOXA1, a critical pioneer factor for AR (12), via blocking K270-demethylation of FOXA1, and leads to impaired enhancer accessibility (13). Moreover, our studies using preclinical models in PCa suggest that FOXA1 expression level may predict the efficacy of LSD1 inhibitors (LSD1-i) in suppressing tumor growth (13). However, whether LSD1-i can target other key oncogenic pathways in PCa is still largely unknown.
Multiple clinical trials are currently investigating the therapeutic potential of targeting LSD1 in various cancers (14). The LSD1 inhibitor treatment has been moving forward in malignancies such as hematologic cancers (15), lung cancer (16), and Ewing Sarcomas (17), but has shown limited advancement in the clinical investigation of PCa. Therefore, it is critical to interrogate the function of LSD1 in various models of PCa. In this study, we elucidated the transcriptomic profiles of LSD1-i in an array of CRPC xenograft models that are sensitive to LSD1-i treatments. In agreement with the impaired tumor growth, inhibition of LSD1 led to decreased MYC signaling including the MYC gene itself. It is well established that MYC is highly regulated by its associated Super-Enhancers (SEs), which are defined as a cluster of enhancers that are in close proximity (~12.5kb) (18). A high density of master transcriptional activators (e.g., Oct4) and mediators (e.g., Med1) assemble on the SEs, establishing a powerful transcription apparatus (19,20). Recent findings have shown that the intrinsically disordered regions within the key transcriptional regulators mediate the liquid-liquid phase separation (LLPS), which can facilitate the formation of large complexes at SEs and promotes robust gene transcription (21–23). Therefore, the transcription of SE-driven oncogenes, such as MYC, is exceptionally sensitive to inhibitors targeting BRD4, a member of the bromodomain and extraterminal (BET) family that serves as a master regulator of SE organization (20). By examining the molecular mechanism of LSD1 regulation of MYC, we showed that LSD1-mediated FOXA1 demethylation could facilitate the BRD4 chromatin binding. We further demonstrated that LSD1 global chromatin occupancy enriched at the SEs and formed biomolecular condensation in the nucleus. Inhibition of LSD1 attenuated the expression of SE-driven genes and could synergize with BET inhibitors in decreasing the CRPC tumor burden.
MATERIALS AND METHODS
Cell Culture:
CWR-22RV1 (22RV1) and LNCaP cell lines were purchased from ATCC, authenticated every six months using short tandem repeat (STR) profiling, and frequently tested for mycoplasma contamination using the MycoAlert mycoplasma detection kit (Lonza). LNCaP95 cells were derived from LNCaP cells and cultured with 10% phenol red-free charcoal-stripped serum (CSS). 22RV1, LNCaP, and LNCaP-derived stable cell lines were generally cultured in RPMI-1640 medium with 10% FBS, which was replaced by the hormone-depleted medium (5% CSS) for 2–3 days before the subsequent assays.
Chromatin Immunoprecipitation (ChIP) and ChIP-seq Analysis:
For the preparation of ChIP, dispensed cells were formalin-fixed, lysed, and sonicated to break the chromatin into 500–800 bp fragments, followed by immunoprecipitation with ChIP grade antibodies: anti-BRD4 (A301–985A100, Bethyl Laboratories), anti-V5 (R960, Thermo Fisher Scientific), or Rabbit/Mouse IgG (Millipore). The qPCR analysis was carried out using the SYBR Green method on the QuantStudio 3 Real-time PCR system (Thermo Fisher Scientific). Primers for KLK3-Enh and ZBTB16-Enh were previously listed (13). Sequences for MYC super-enhancers - SE1, Forward: 5’-GGTTTTACGCATTCATGGGGG-3’, Reverse: 5’-GCTCAAGCAATTGAGGCTGC-3’; SE2, Forward:5’- CAGCTCTCCTTGGAGTCTCTC-3’, Reverse: 5’-ATAAACCTCTCGTGTGAGCCC-3’.
ChIP-seq libraries were constructed using the SMARTer ThruPLEX DNA-Seq Prep Kit (Takara Bio USA). Next-generation sequencing (51nt, single-end) was performed using Illumina HiSeq2500. ChIP-sequencing reads were mapped to the hg19 human genome using bwa (version 0.7.17) with aln and samse sub-commands (24). Samtools (version 1.10) was used to convert sam files to bam format. The significance of enriched ChIP regions was evaluated by using MACS2 (version 2.2.7) (25). The R package IRanges (version 2.30.0) was used to analyze peak intervals and determine the overlapped regions. Venn diagrams were generated using VennDiagram (version 1.7.3) R package. The signals associated with genomic regions were visualized by using computeMatrix and plotHeatmap tools from deepTools (version 3.5.0). computeMatrix with reference-point mode was used to calculate scores for each genomic region, and plotHeatmap was used to create a heatmap for scores associated with genomic regions. Motif enrichment analysis was performed by using SeqPos with the default setting in Galaxy/Cistrome (26).
Immunoprecipitation and Immunoblotting:
For immunoprecipitation assays (IP), CWR-22RV1 cells or LuCaP PDX tumor tissues were lysed in Triton Lysis buffer and treated with protein inhibitor cocktails (Thermo Fisher Scientific), followed by a brief sonication, and then the lysates were immunoprecipitated with anti-LSD1 (ab17721, Abcam), anti-BRD4 (A301–985A100, Bethyl Laboratories), or anti-FOXA1 (ab23738, Abcam). For immunoblotting, cells were lysed with RIPA buffer containing protease inhibitor cocktail (Thermo Fisher Scientific) and anti-MYC (9402, CellSig), anti-FOXA1 (ab23738, Abcam), anti-BRD4 (ab128874, Abcam), anti-LSD1 (ab41969, Abcam), anti-CoREST (ab183711, Abcam), anti-GAPDH (ab8245, Abcam) were used.
Quantitative Real-time PCR and RNA-Seq Analysis:
RNA from cell lines was extracted by TRIzol reagent (Invitrogen). The expression of mRNA was measured using real-time RT-PCR (qRT-PCR) analyses with Taqman Fast one-step Mix RT-PCR reagents (Thermo Fisher Scientific) on the QuantStudio 3 Real-time PCR system. The results were normalized to co-amplified GAPDH. The primer and probe sets for ZBTB16, MBOAT2, ELOVL7, COL23A1, CDH2, MYC, and GAPDH were purchased as an inventoried mix from Applied Biosystems (Thermo Fisher Scientific).
RNA from tumor tissue samples was extracted by using TissueLyser LT (QIAGEN) and RNeasy Kit (QIAGEN). RNA-Seq library was prepared using TruSeq Stranded RNA LT Kit (Illumina). Sequencing was performed on NextSeq 2000 Illumina Genome Analyzer. The single-end reads were processed by FastQC and aligned by STAR (version 2.7.9a) to the human Ensemble genome (Ensembl, GRCh37) with all default parameters (27). featureCounts (version 2.0.2) from Subread package was used to assign sequence reads to the genomic features. All gene counts were processed with R package limma (3.52.0) to evaluate the differential expression using the Benjamini–Hochberg false discovery rate (FDR)-adjusted P value (28). The expression values were centered and scaled across samples and then displayed using the ComplexHeatmap (version 2.12.0) R package. The pre-ranked gene lists were used to conduct Gene Set Enrichment Analysis (GSEA) by using R package fgsea (version 1.22.0) (29). The top pathways with normalized enrichment scores (NES) ranked by P value were plotted for visualization.
Single-cell RNA-seq Analysis:
Single cell solution of tissue samples was obtained following a published protocol (30). Briefly, tissue fragments were incubated in 3 ml Accumax (Innovative Cell Technologies, AM105) for 10–20 minutes at room temperature on a rocking shaker. Cell suspensions were then filtered with a 70μm cell strainer and spun at 580g for 5 min at 4 °C. Red blood cells were lysed with ACK Lysing Buffer (Thermo Fisher Scientific, A1049201) on ice for 1 min, followed by quenching with PBS and then filtered again with a 40-μm cell strainer. Cell suspensions were washed three times and resuspended with PBS plus 0.04% BSA. The final cell viability of suspensions was determined by Countess Automated Cell Counter (Thermo Fisher Scientific).
For scRNA-seq data processing and quality control, scRNA sequencing reads were processed into FASTQ format. Then single-cell feature counts were quantified into the form of an h5 filtered matrix using Cell Ranger (version 6.0.1). We used the human genome (GRCh37) as a reference. The Cell Ranger output was imported into Seurat (version 4.1.1) R package for further analysis (31). After removing unwanted cells from the dataset, we employed “LogNormalize” to normalize the feature expression measurements for each cell by the total expression. Then the Seurat FindVariableFeatures() function was used to select 2000 highly variable features and the Seurat ScaleData function was used to perform scaling on the previously identified variable features. For Single-cell RNA clustering, the scaled data was performed by principal component analysis (Seruat RunPCA). We determined the k-nearest neighbors of each cell and constructed a shared nearest-neighbor (SNN) graph (Seurat FindNeighbors). Subsequently, we identified clusters by applying the modularity optimization technique and using the top ten PCs (Seurat FindClusters). Uniform manifold approximation and projection (UMAP) nonlinear dimensionality reduction was performed on the first ten PCs to generate the two-dimensional plots accordingly.
Super Enhancer Identification:
To define enhancers, we collected publicly available ChIP-seq of H3K27ac from the GEO dataset (GSE130408) (32), including 37 normal prostate epithelium samples, 32 primary prostate tumors, and 17 metastatic prostate cancer human specimens. H3K27ac ChIP-seq reads were mapped to hg19 reference genome using bwa. MACS2 was used to identify enhancer-enriched regions. Then the ROSE (Rank Ordering of Super-Enhancers) algorithm was used to identify super-enhancers (19,20). ROSE was run by allowing enhancers within 12,500 bp to be stitched together. Stitched enhancers were ranked according to the intensity of H3K27ac ChIP-seq signal. A threshold was then determined according to the geometric inflection point to distinguish between enhancers and super-enhancers. The identified super-enhancer regions were further annotated by annotatePeaks. The common super-enhancers of each group were defined when these regions occur in over 85% of samples.
Confocal Immunofluorescence Microscopy:
Cells grown on glass coverslips were fixed in 4% paraformaldehyde and then permeabilized in 0.5% triton X100 (FisherSci). For blocking, cells were incubated in 10% goat serum (Cellsig) and followed by primary antibodies incubation for overnight (anti-BRD4, Abcam ab128874; anti-LSD1, CellSig 2139) and then secondary antibodies incubation (Goat anti-Rabbit IgG Alexa Fluor 488, Goat anti-Rabbit IgG Alexa Fluor 594, Life Technologies). Nuclei were stained using DAPI (Life Technologies). Images were taken by 63X objective and post-processed using Fiji Is Just ImageJ.
Cell Proliferation Assay:
Cells were seeded in 96-well plates and treated with inhibitors for 4 days. Cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega) per the manufacturer’s protocol.
Migration Assay:
Transwell migration assays were performed with Corning® FluoroBlok™ Inserts (351152, Corning). Per the manufacturer’s protocol, the same number of 22RV1 cells were seeded in the pre-moisturized upper chamber with serum-free medium and the lower chamber was filled with medium containing 20% FBS as a chemoattractant. After two days of drug treatments, migrated cells were stained by Corning Calcein AM Fluorescent Dye (354217, Corning). All experiments were done in biological triplicates and images were taken by the EVOS auto fluorescence microscope.
Mouse Xenografts:
All animal experiments were approved by the University of Massachusetts, Boston Institutional Animal Care and Use Committee and conducted following institutional and national (USA) guidelines. 22RV1 cells were resuspended in serum-free RPMI 1640 medium and mixed in a 1:1 ratio with Matrigel (BD Biosciences) prior to subcutaneous implantation (2×106 cells per injection) on flanks of castrated SCID mice (4 to 6 weeks old; Taconic). Xenograft tumors, including PDXs, were further passaged in castrated male SCID mice. Tumor length (L) and width (W) were measured by caliper at the indicated time, and tumor volumes were calculated (LxW2/2). The housing conditions were ambient temperatures of 65–75°F with 40–60% humidity and 12h light/12h dark cycle.
Statistical Analysis:
Data in bar graphs represent the mean±SD of at least 3 biological repeats. For most studies, statistical analysis was performed using Student’s t-test by comparing treatment versus vehicle or otherwise as indicated: * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001. The results for immunoblotting are representative of at least three experiments. Boxplots of signature scores and gene expression were compared using the Wilcoxon test for comparison between two groups of samples. All other statistical analyses and visualization were performed with R (version 4.2.0) unless otherwise specified.
Data availability:
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE209889. All other raw data generated in this study is available from the corresponding author upon request. The data analyzed in this study were obtained from GEO at GSE94013 and GSE130408.
RESULTS
LSD1 inhibition in CRPC models consistently targets MYC signaling
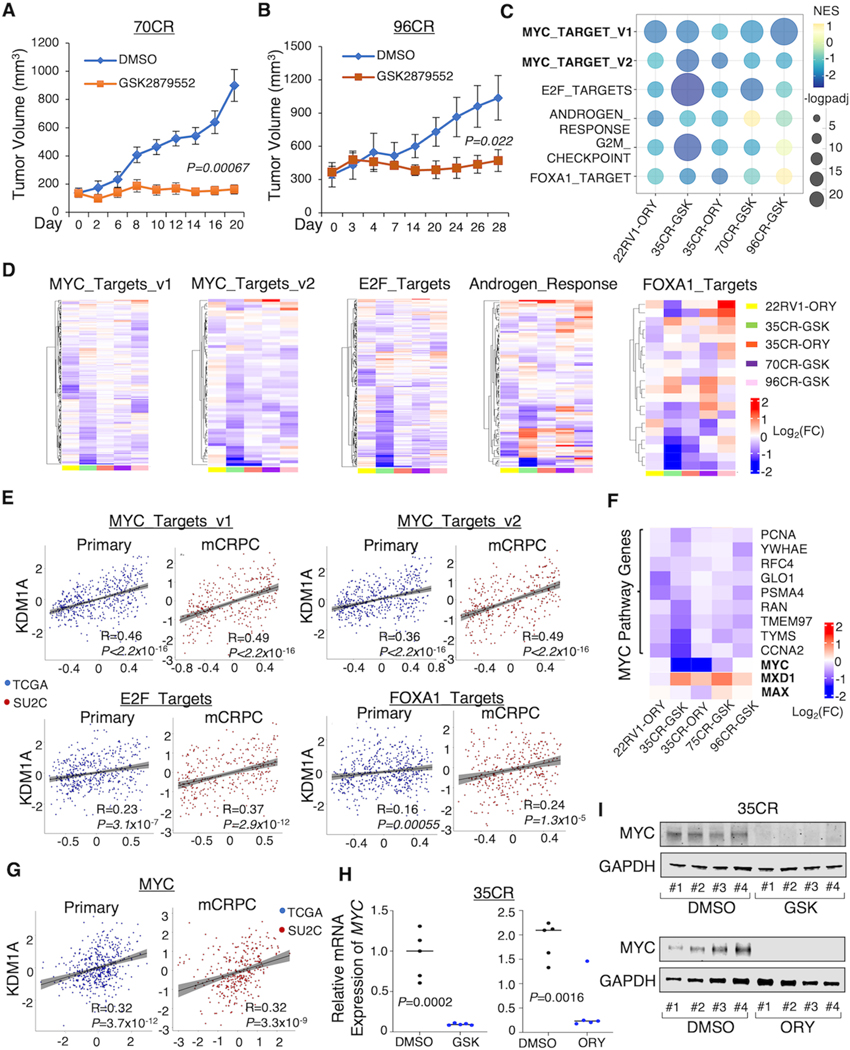
We have previously found that CWR-22RV1 and LuCaP 35CR models are strong responders to LSD1-i treatment and LuCaP 77CR is a modest responder (13). To further understand the actions of LSD1-i in CRPC, we tested two additional CRPC patient-derived xenograft (PDX) models, LuCaP 70CR and 96CR (33). As shown in Fig. 1A, B and Supplementary Fig. S1A, LSD1-i treatment (GSK2879552) significantly decreased the xenograft tumor growth in both models and increased cellular levels of H3K4me2. We then performed a RNA-seq analysis in all the strong responders (22RV1, 35CR, 70CR, and 96CR) to further determine the molecular underpinnings of how LSD1-i represses tumor growth by collecting a total of 34 tumor samples from CRPC xenografts treated with LSD1-i: GSK2879552 (16) or ORY-1001 (34). Principal Component Analysis (PCA) revealed that 70CR and 96CR share a more similar transcriptomic profile, which is distinct from 35CR and 22RV1 xenograft (Supplementary Fig. S1B). Using Gene Set Enrichment Analysis (GSEA) on the RNA-seq data, we found that the expression levels of MYC and E2F transcriptional targets were consistently downregulated by LSD1-i (Fig. 1C and D). Decreased expression of E2F targets by LSD1-i is consistent with our recent finding that LSD1 can increase E2F1 activity by enhancing its chromatin binding (35). AR activity and G2/M cell cycle-related genes were also inhibited in most of the models, consistent with previous reports (8,11). Since our recent study has suggested that a major molecular function of LSD1 is to stabilize FOXA1 chromatin binding and thus increase enhancer accessibility (13), we next examined whether LSD1-i targets the FOXA1 activity in these CRPC models. To test that, we developed a FOXA1-target signature (upregulated genes) that was derived from FOXA1 silencing in LNCaP and 22RV1 cells (FOXA1_TARGETS) (36,37) and found that LSD1-i treatment decreased the FOXA1 activity in all models but 96CR. In addition, we also observed upregulated immune response hallmark genes which were activated in all the models (Supplementary Fig. S1C and D), consistent with the previous finding showing that LSD1 suppresses tumor immunogenicity (38). To further corroborate the above findings, we interrogated the association between LSD1 and the identified LSD1-i targeted pathways in the clinical samples. There was a significant positive correlation between LSD1 expression level and MYC, E2F, or FOXA1 targets in primary PCa (TCGA dataset, (39)) and CRPC (SU2C dataset,(40)) patient cohorts, and the correlation appeared to be stronger in CRPC samples (Fig. 1E).
Figure 1. LSD1 inhibition represses MYC signaling in various CRPC models.
(A, B) Castrated SCID mice bearing LuCaP 70CR (A) and 96CR (B) PDX xenografts were treated daily with DMSO or LSD1 inhibitor GSK2879552 (33 mg/kg) via intraperitoneal injection, and the tumor volume was measured by caliper at indicated time points. (C, D) Tumor samples from each xenograft model were subjected to RNA-seq studies. Gene set enrichment analysis (GSEA) was done by comparing the LSD1 inhibitor treatment with vehicle treatment. The bubble plot (C) and heatmap views (D) show the top ranked LSD1-i-repressed pathways that are most common among all models. (E) Correlations between KDM1A level and LSD1-i repressed pathways in TCGA and SU2C patient cohorts. (F) The heatmap view of MYC, MYC cofactors, and MYC targets. (G) The correlation between KDM1A level and MYC in TCGA and SU2C patient cohorts. (H, I) MYC expression levels in the 35CR model treated by LSD1-i were measured by RT-qPCR (H) and western blot (I).
Next, we focused on the transcriptional regulation of LSD1 on MYC signaling. As shown in Fig. 1F, the expressions of the MYC gene and a panel of MYC targets, including important cell cycle regulators such as PCNA and CCNA2, were significantly decreased by LSD1-i. On the contrary, MYC antagonist protein, such as MXD1 was upregulated by LSD1-i, suggesting that LSD1 could activate MYC signaling via multiple mechanisms. Importantly, LSD1 was also highly correlated with MYC expression in patient PCa samples (Fig. 1G). Examining the tumor samples from 35CR and 22RV1 xenograft models, we confirmed that LSD1-i markedly decreased both mRNA and protein expression of MYC (Fig. 1H and I; Supplementary Fig. S1E). Taken together, our transcriptomic profiling study in CRPC xenograft models revealed that the tumor-suppressive effect of LSD1-i in CRPC is mediated via targeting multiple oncogenic pathways, including MYC signaling.
Heterogenous response to LSD1 inhibition in the 96CR model
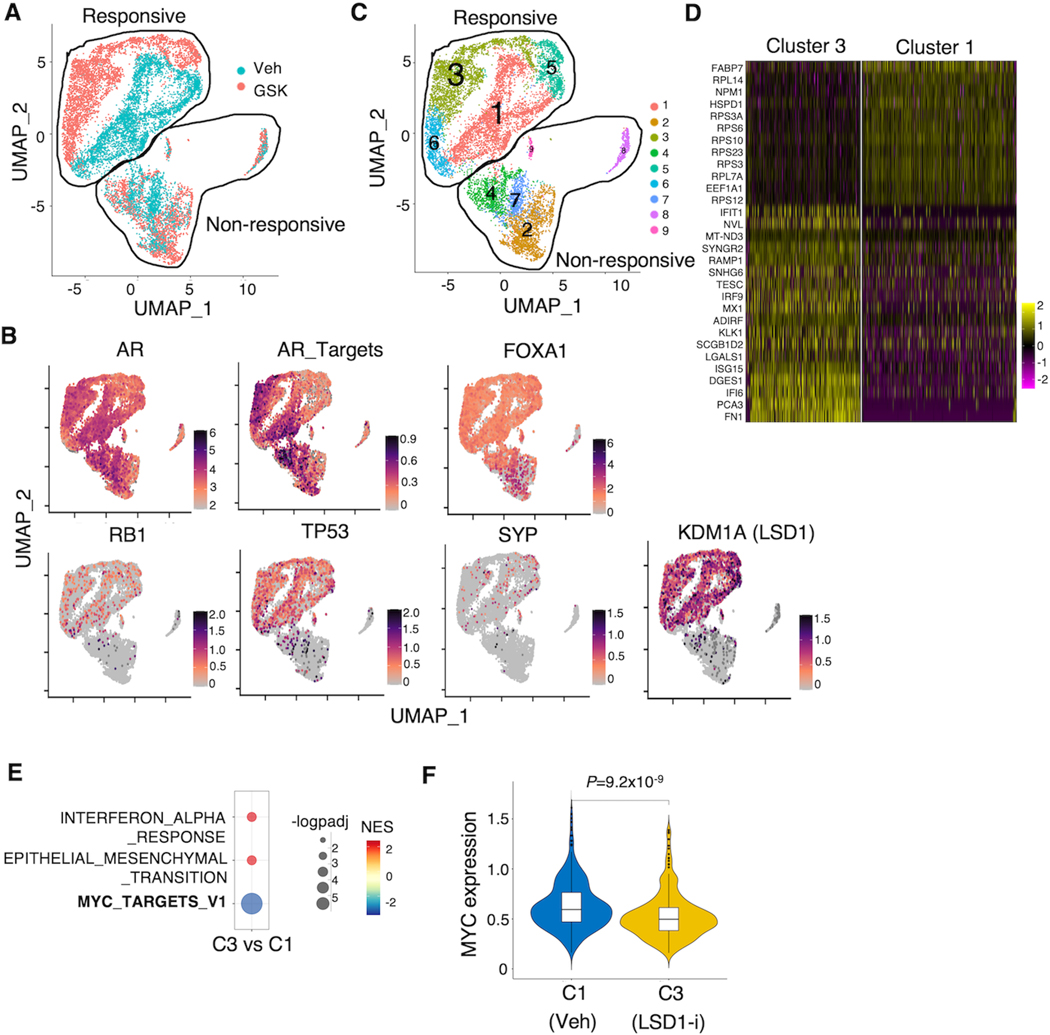
PCa is a highly heterogeneous disease and sub-populations carrying distinct genomic backgrounds can contribute to the disease progression differently. Since LSD1-i appeared to be effective in the 96CR model, whereas some of the transcriptional output responses were different from other models (see Fig. 1D), we hypothesized that the intrinsic heterogeneity of 96CR may contribute to this effect. Interestingly, 96CR was originally derived from the hormone-dependent LuCaP96 PDX which was characterized as heterozygous RB1 deletion. However, homozygous RB1 deletion was detected in tumor cells when LuCaP96 progressed to 96CR after castration (33). Our recent finding in the 96CR model still indicates the detection of Rb-positive tumor populations (41), suggesting a possible mixed population of tumor cells with distinct genomic backgrounds. In addition to alterations of RB1, 96CR tumors also contain AR and MYC amplification and heterozygous deletion of TP53 (33). Therefore, we sought to determine the cellular responses to LSD1-i on the single-cell level in 96CR tumors. As shown in Fig. 2A, there appeared to be two distinct cell populations that showed different transcriptional responses to LSD1-i: Responsive (clear separation of transcriptomes) and Non-responsive (overlapped transcriptomes) clusters. While AR, AR targets, and FOXA1 were highly expressed in both populations, the non-responsive population clearly lacked expression of RB1 and had low TP53 expression, which are well-known molecular characteristics of Neuroendocrine PCa (NEPC) (Fig. 2B). However, this non-responsive population appeared to be largely comprised by non-NEPC cells as they barely expressed NEPC marker SYP. Importantly, the overall LSD1 expression was much lower in the non-responsive population. Therefore, these ARhigh/FOXA1high/RB1−/TP53low/KDM1Alow tumor cells may represent a previously undefined molecular subtype that is intrinsically resistant to LSD1 inhibitor treatment. Nonetheless, via unsupervised clustering, we compared the transcriptomes between Cluster 1 (vehicle-treated) and Cluster 3 (LSD1-i treated) within the responsive population (Fig. 2C and D) and found that MYC targets were significantly decreased by LSD1-i (Fig. 2E), which was consistent with the bulk RNA-seq data. In addition, MYC gene expression was also decreased by the treatment of LSD1-i (Fig. 2F). On the contrary, the comparison of Cluster 7 (vehicle-treated) and 2 (LSD1-i treated) within the non-responsive population showed neither the enrichment of MYC signaling nor the change of MYC expression (Supplementary Fig. S2A and B).
Figure 2. Heterogenous response to LSD1 inhibition in LuCaP 96CR model.
(A) Single-cell RNA-seq analysis demonstrated by the Uniform Manifold Approximation and Projection (UMAP) plot was generated (blue: vehicle, red: LSD1-i). (B) Expression levels of indicated genes were plotted onto the UMAP. (C) Cluster 1 (Vehicle treated) and cluster 3 (GSK treated) are highlighted. (D) Top-ranked genes that are differentially expressed between cluster 3 and cluster 1. (E) The GSEA showing the hallmark pathways that were significantly changed by GSK treatment (cutoff: P-adj<0.05). (F) The violin plot of MYC gene expression in Vehicle (cluster 1) vs. GSK2879552 (cluster 3).
To further elucidate the molecular characteristics of the non-responsive subpopulation, we probed the transcriptomic profiling between Cluster 7 (non-responsive, vehicle-treated) and Cluster 1 (responsive, vehicle-treated). These non-responsive tumor cells showed enrichment of E2F targets and cell cycle pathways and de-enrichment of Oxidative Phosphorylation pathway (metabolic pathways) (Supplementary Fig. S2C–E). Taken together, using this single-cell RNA-seq analysis in the 96CR model, we confirmed that LSD1-i targets MYC signaling in LSD1-high CRPC cells and identified a molecular subtype of LSD1-low CRPC cells, which have higher cell cycle signatures but lower metabolic signatures and are resistant to LSD1-i.
LSD1 enriches at super-enhancers and forms phase separation
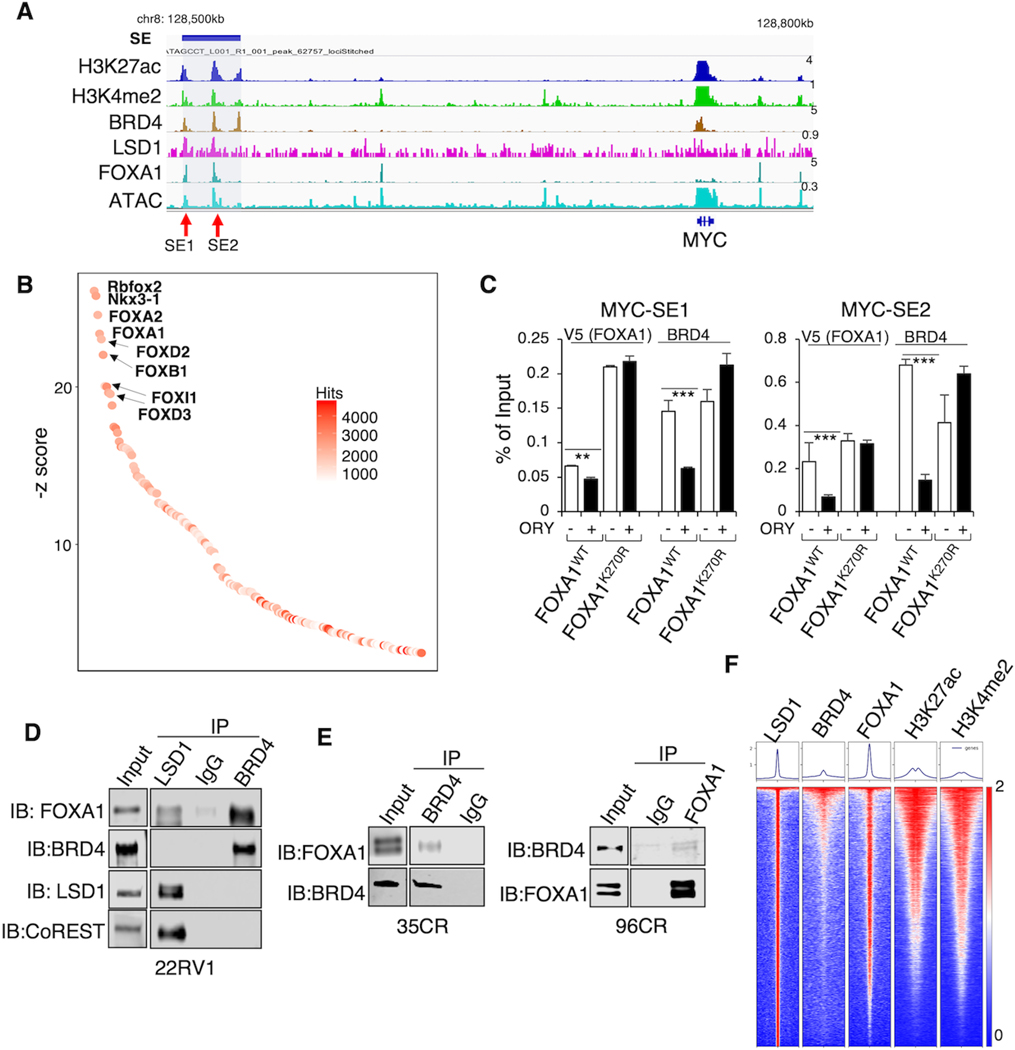
MYC gene expression is well-known for its regulation by Super-Enhancers (SEs) occupied by master regulators such as BRD4 (20). Since LSD1 can function to increase chromatin accessibility by promoting the binding of pioneer factor FOXA1 (13), we hypothesize that LSD1 may be critical for maintaining SEs in PCa cells. At the region ~250kb upstream of the MYC gene, we identified a SE which is marked by a cluster of H3K27ac and H3K4me2 peaks and co-occupied by BRD4, LSD1, and FOXA1 (Fig. 3A). Interestingly, the motif enrichment analysis of published BRD4 ChIP-seq peaks revealed strong enrichment of Forkhead DNA binding motifs (42) (Fig. 3B). To determine whether LSD1-mediated FOXA1 chromatin binding affects BRD4 recruitment, we performed ChIP-V5 and ChIP-BRD4 in previously established 22RV1 stable cell lines expressing doxycycline-inducible V5-tagged wildtype (WT) or mutant (K270R) FOXA1, the latter of which binds to chromatin more tightly and is resistant to LSD1 inhibition (13) (Fig. 3C). BRD4 binding was markedly decreased by LSD1-i in the WT cell line but was not affected in the K270R mutant cell line. These results suggest that unmethylated FOXA1 may recruit BRD4 to the chromatin. Next, we sought to investigate the interaction between FOXA1 and BRD4. In 22RV1 cells cultured under hormone-depleted conditions, LSD1 interacted with FOXA1, consistent with the previous study (11) (Fig. 3D). However, LSD1 did not seem to directly interact with BRD4, whereas FOXA1 strongly interacted with BRD4, and this interaction does not seem to be affected by the LSD1 demethylase activity (Supplementary Fig. S3A). Moreover, we can also detect the interaction between BRD4 and FOXA1 in the tumor samples of 35CR and 96CR PDXs (Fig. 3E). To further examine whether BRD4 binding is globally associated with LSD1 and FOXA1, we performed BRD4 ChIP-seq in LNCaP cells under hormone-depleted conditions, and BRD4 binding was associated with LSD1, FOXA1, and active enhancer marks (Fig. 3F). Together, these results suggest a framework of LSD1, FOXA1, and BRD4, in which LSD1-demethylated FOXA1 can recruit BRD4 to chromatin, and the association between FOXA1 and BRD4 may be important for enhancer activation.
Figure 3. LSD1-mediated FOXA1 chromatin binding promotes BRD4 recruitment.
(A) The genome view of indicated ChIP-seq peaks within an identified super-enhancer (SE) upstream of MYC locus (hg19), which contains two genomic regions, SE1 and SE2, based on strong enhancer markers. (B) Top-ranked motifs from published BRD4 ChIP-seq in 22RV1 cells (GSE94013). (C) Stable 22RV1 cells expressing V5-tagged WT FOXA1 or K270R mutant (grown in hormone-depleted medium) were pretreated with doxycycline to induce FOXA1 expression, and then treated with DMSO or ORY-1001 (5μM) for 1 day. (D) Immunoprecipitation (IP) of LSD1 or BRD4 in 22RV1 cells (grown in hormone-depleted medium) followed by immunoblotting of FOXA1, BRD4, and LSD1. (E) IP of BRD4 followed by immunoblotting of FOXA1 in 35CR and 96CR xenograft tissue. (F) Heatmap view of ChIP-seq intensity of LSD1, BRD4, FOXA1, H3K27ac, and H3K4me2 in LNCaP cells (grown in medium with charcoal-striped serum, CSS).
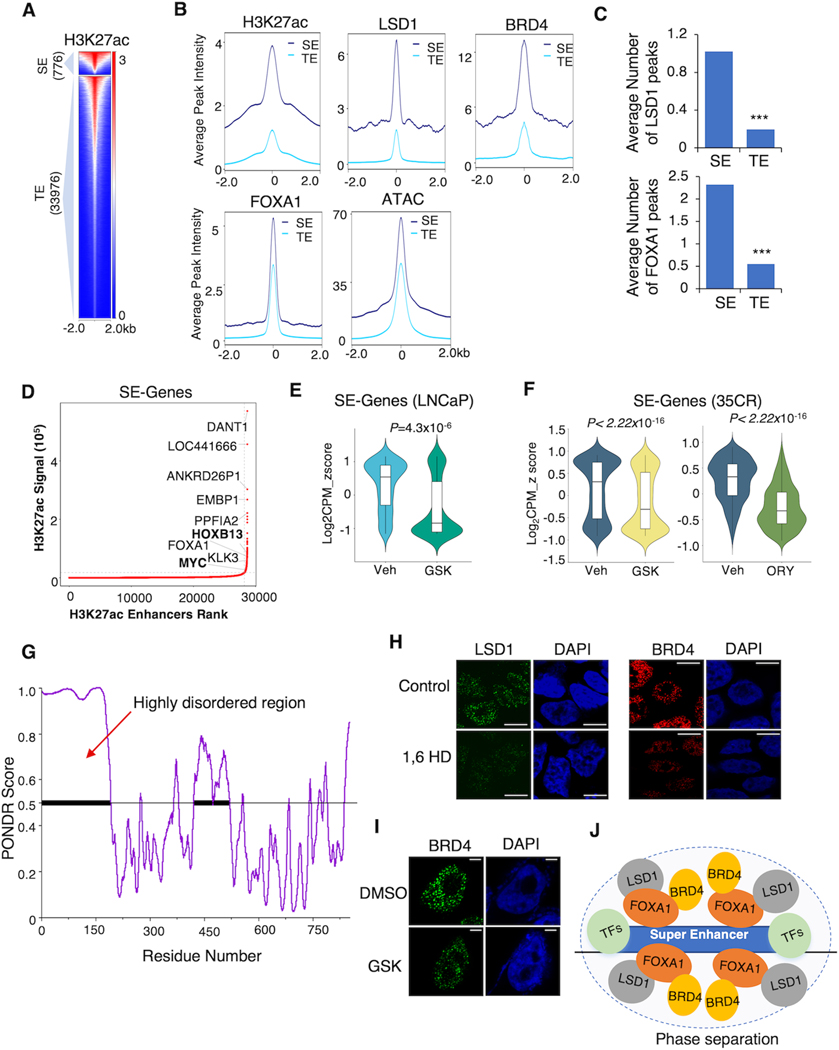
Since BRD4 is well-known for its enrichment at SEs, we hypothesized that the chromatin occupancy of LSD1 may also concentrate at the SEs. To test this hypothesis, we performed the ROSE analysis (Ranking of Super Enhancers) using the H3K27ac ChIP-seq in LNCaP cells (under hormone-depleted conditions) and identified 776 SEs and 33,976 typical enhancers (TEs) (Fig. 4A). Consistent with previous findings, H3K27ac and BRD4 levels were significantly higher at SEs than TEs. Similar to BRD4 binding levels, LSD1 exhibited much stronger ChIP-seq signals on SEs than TEs. The peak intensities of FOXA1 and ATAC signals were also higher at SEs than TEs (Fig. 4B) and can be quickly decreased by LSD1-i (Supplementary Fig. S3B). Concordantly, the average number of LSD1 or FOXA1 peaks distributed on SEs was higher than TEs (Fig. 4C). We then identified a list of SE-associated genes, including MYC and HOXB13, and then examined whether targeting LSD1 can impair the transcription of these genes using results from RNA-seq analyses (Fig. 4D). As shown in Fig. 4E and F, LSD1-i broadly decreased the expression of the SE-associated genes in LNCaP cells under castrated conditions and in 35CR tumors, indicating a critical role of LSD1 in stabilizing SEs in PCa cells.
Figure 4. LSD1 enriches at SEs and forms LLPS.
(A) The heatmap view of H3K27ac ChIP-seq intensity at SEs and typical enhancers (TEs) defined in LNCaP cells. (B) Average ChIP-seq signals of H3K27ac, LSD1, BRD4, FOXA1, and ATAC at SEs and TEs. (C) The average number of LSD1 and FOXA1 peaks distributed on the SEs and TEs. (D) Top-ranked SE-associated genes identified in the LNCaP cells (in CSS). (E) The violin plot showing the expression levels of SE-associated genes in LNCaP cells treated with Vehicle or GSK2879552 for 2 days. (F) The violin plot showing the expression levels of SE-associated genes in 35CR xenograft tissue samples that received vehicle or LSD1 inhibitor treatment. (G) PONDR (VSL2) scores of LSD1 protein for its disordered regions. (H) Representative immunofluorescent images of LSD1 or BRD4 in 22RV1 cells (in CSS), before and after treatment with 3% hexanediol for 30 seconds. Scale bar represents 5 μm. (I) Representative immunofluorescent images of BRD4 in 22RV1 cells (in CSS) treated with DMSO or GSK2879552 (1μM, 4 hours) Scale bar represents 2 μm. (J) Proposed working model.
It has been well characterized that phase separation, via recruiting a high density of transcriptional apparatus, occurs on SEs to drive robust transcription (21), and the intrinsically disordered regions of proteins are attributed to the formation of biomolecular condensates (22). Indeed, the N-terminus of LSD1 (aa 1–165) was predicted to be highly disordered and may contribute to the condensate formation (Fig. 4G). Under high-resolution confocal microscopy in 22RV1 cells, we observed that LSD1 exhibited similar puncta-like staining as BRD4, which is a characteristic of LLPS. Treating cells with 1,6-hexanediol, a commonly used compound for disrupting biomolecular condensates, reduced the puncta-like formation of LSD1 and BRD4 (Fig. 4H). Deleting the N-terminal disordered region of LSD1 could also disrupt the puncta formation (Supplementary Fig. S3C and D), confirming that the N-terminal region of LSD1 protein is critical for forming the phase separation. More importantly, LSD1-i treatment in 22RV1 cells disrupted the puncta-like formation of BRD4 (Fig. 4I), suggesting that LSD1 may function to stabilize the BRD4-enriched nuclear condensates which potentially interact with SEs (Fig. 4J).
LSD1-i and BET-i can synergistically suppress CRPC tumor growth in vitro and in vivo
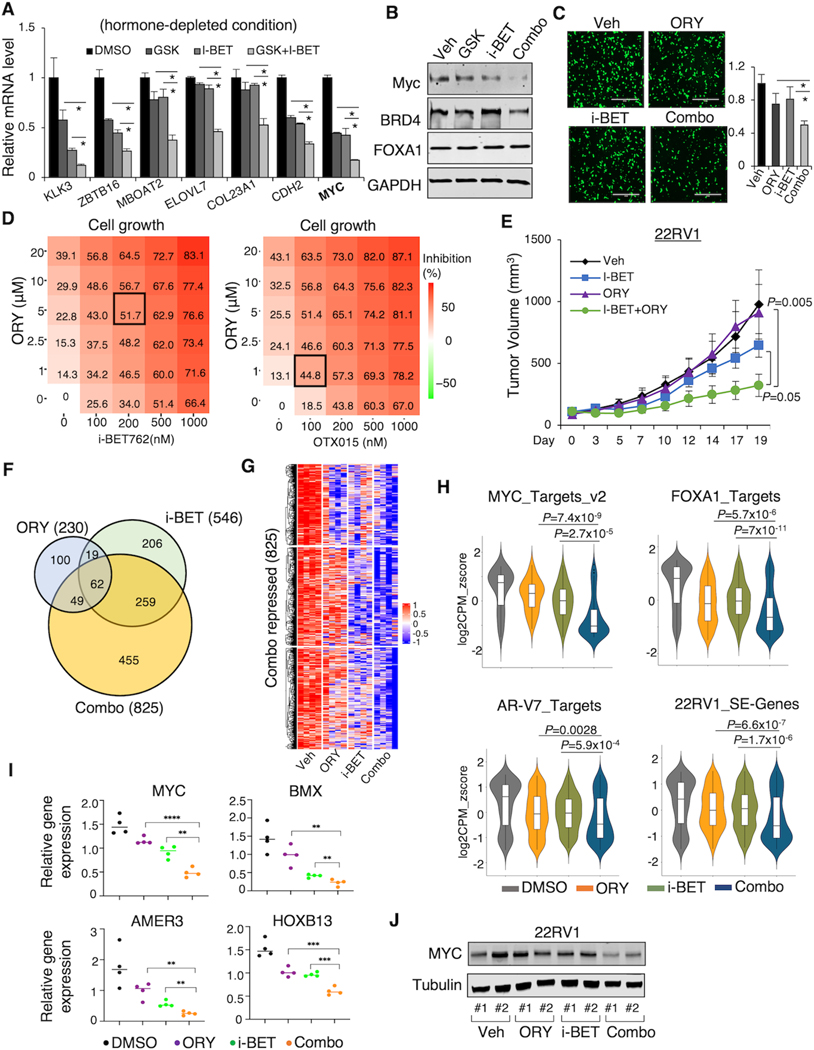
The above results rationalized a therapeutic strategy by disrupting oncogenic SEs using LSD1 and BET inhibitors which could confer a synergistic anti-tumor effect in CRPC. To test the efficacy of the combinational treatment of LSD1-i and BET-i in CRPC, we first treated 22RV1 cells (express high levels of AR splice variants) with GSK2879552 and i-BET762 (also known as GSK525762 (43)) under the hormone-depleted condition. We first assessed the effects of the combinational treatment on AR/AR-V signaling. As shown in Supplementary Fig. S4A, BRD4 chromatin bindings at AR/AR-V7-regulated enhancers were decreased by both LSD1-i and BET-i. Significantly, a clear additive effect on a panel of previously reported AR/AR-V7 target genes (KLK3, ZBTB16, MBOAT2, ELOVL7) (44) or AR-V7-specific target genes (COL23A1, CDH2) was observed (Fig. 5A). Furthermore, MYC mRNA and protein levels were also synergistically decreased by the combination treatment (Fig. 5B). Next, we examined how the combination treatment affect tumor cell migration and growth by using another potent LSD1-i, ORY-1001, which is currently tested in multiple clinical trials (45). As shown in Fig. 5C, the combination treatment had a stronger effect on PCa cell migration than the single agent treatment. Moreover, the combination treatment of LSD1-i with two BET inhibitors all synergistically repressed the growth of 22RV1 cells (Fig. 5D and Supplementary Fig. S4B and C). Similar results on AR/AR-V7 pathway, MYC expression, and cell growth were also obtained in the LNCaP95 model, a LNCaP derived CRPC cell line expressing high levels of AR splice variants (Supplementary Fig. S4D–F), and the parental LNCaP cells (under full serum conditions) (Supplementary Fig. S4G–I).
Figure 5. LSD1-i and BET-i synergistically suppress CRPC tumor growth.
(A, B, C) 22RV1 cells (in CSS) were treated with 1μM GSK2879552, 1μM i-BET762, or the combination for 1 day, and subjected to RT-qPCR analysis. Relative gene expression of AR/AR-V7 targets (A), protein expression of Myc, BRD4, and FOXA1 (B), and representative image and quantitative results of transwell migration assay (C) are shown. (D) The heatmap view of the percentage of inhibition of 22RV1 cells (in CSS) treated with the combination of indicated doses of ORY-1001 and BET inhibitors for 4 days. The dosages that achieve the highest synergy are highlighted. (E) Castrated SCID mice bearing 22RV1 xenografts were treated with vehicle, i-BET762 (16mg/kg, daily), ORY-1001 (0.03mg/kg, every other day), or the combination via intraperitoneal injection (for ORY-1001) or oral gavage (for i-BET762). Tumor volume was measured by caliper. (F) RNA-seq analyses were performed using the tumor samples collected at the end of the experiment. The Venn diagram for ORY-1001-repressed genes, i-BET762-repressed genes, and the combination treatment-repressed genes is shown. (G) The heatmap view for the combination treatment-repressed genes in all tumor samples. (H) The violin plots showing the indicated gene signatures in the 22RV1 xenograft tumor samples. (I) RT-qPCR analysis of indicated gene expression in the tumor samples. (J) Immunoblotting for c-Myc expression in the tumor samples. Data in bar graphs represent the mean ± SD. Data in growth curves represent the mean ± SD.
To evaluate the effects of co-targeting LSD1 and BRD4 on tumor growth in vivo, we treated the castrated mice bearing 22RV1 CRPC xenograft tumors with ORY-1001 (at a lower dosage than used as a single treatment in previous experiments) and i-BET762 alone or in combination. As shown in Fig. 5E, while the single agent treatment (at a lower dosage) did not significantly repress the tumor growth, the combination treatment induced strong tumor regression. More importantly, there was no overt toxicity observed in mice (Supplementary Fig. S4J). We next conducted an RNA-seq analysis using the tumor tissue samples from these 22RV1 xenografts. The combination treatment showed more potent repression than any single agent (Fig. 5F and 5G). We then interrogated the transcriptional outputs exerted by the combined treatment: MYC targets, FOXA1 targets, and AR-V7 targets. As expected, in concordance with the synergistic effect on tumor regression, co-targeting LSD1 and BRD4 can more effectively downregulate these targets. More importantly, the expression levels of SE-associated genes (defined by H3K27ac) in 22RV1 were also synergistically impaired by the combination treatment (Fig. 5H). RT-qPCR validation of the RNA-seq analysis showed that MYC and several other SE-associated genes were synergistically downregulated by the combined treatment (Fig. 5I and J). These data indicated that co-targeting LSD1 and BRD4 may achieve a more favorable clinical output in CRPC.
Co-targeting LSD1 and BRD4 synergistically impairs clinically identified CRPC-specific SEs
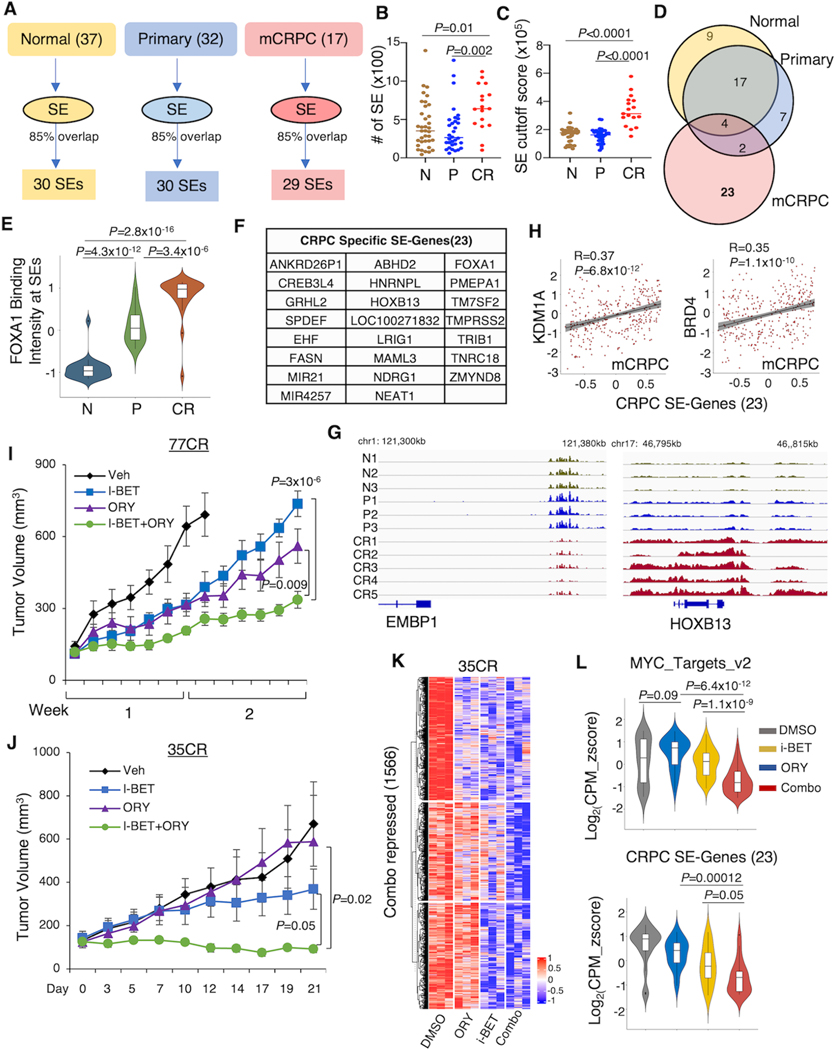
The above results strongly suggested that targeting LSD1 and BRD4 could converge on disrupting oncogenic SEs that drive CRPC progression. Using ROSE algorithm and public H3K27ac ChIP-seq performed in samples representing different stages of PCa development (prostate epithelium: n=37, primary PCa tissue: n=32, and CRPC PDX tumor tissue: n=17) (32), we identified common SEs genes that are generally shared among samples within each group (at least 85% of samples shared) and identified 30, 30, and 29 SEs in normal (N), primary PCa (P), and CRPC (CR) group (Fig. 6A). Interestingly, we found that CRPC samples have larger numbers of SEs than normal prostate or primary PCa (Fig. 6B). The SEs in the CRPC also showed much stronger H3K27ac signals, suggesting more robust enhancer activities (Fig. 6C). Interestingly, there was a major overlap between the normal and primary groups, whereas CRPC group had a distinct list of SEs (n=23) (Fig. 6D). To gain more insights into the epigenomic profiles of these CRPC-specific SEs, we investigated the FOXA1 chromatin binding on these genomic regions using published FOXA1 ChIP-seq (32). Interestingly, FOXA1 showed stronger binding signals in the CPRC samples than others, suggesting that FOXA1 may play a critical role in activating these CRPC-specific SEs (Fig. 6E), although this effect could be contributed by the differences in ChIP materials (biopsies versus PDXs). These CRPC-specific SEs-associated genes include FOXA1, HOXB13, FASN (a key lipogenic regulator), and non-coding RNAs NEAT1 (Fig. 6F, 6G). More importantly, these 23 CRPC-specific SE-associated genes were significantly correlated with KDM1A and BRD4 expression levels in the CRPC SU2C patient cohort (Fig. 6H). To further determine whether the combination treatment can target these CRPC-specific SEs, we assessed the effects of the combination treatment in two LuCaP CRPC PDX models, 77CR (a modest responder to LSD1-i) and 35CR (a strong responder to LSD1-i), both of which shared these CRPC-specific SEs. Even though 77CR tumors grew much more robustly, the combination treatment showed a significant advantage in repressing the tumor growth than any single agent, although tumors may seem to eventually relapse (Fig. 6I). More significantly, the combo treatment achieved almost a complete blockade of tumor growth in the 35CR model (Fig. 6J). We then performed RNA-seq analysis in the 35CR model to examine the transcriptome changes by the treatments. As shown in Fig. 6K and Supplementary Fig. S5A, the combination treatment can more broadly repress gene expression. More importantly, co-targeting LSD1 and BRD4 achieved synergism in repressing MYC signaling and CRPC-specific SE-associated genes (Fig. 6L). Together, these data demonstrated a strong clinical potential of combining LSD1-i and BET-i in treating CRPC via disrupting SE-driven oncogenic transcription networks.
Figure 6. The combination of LSD1-i and BET-i disrupts CRPC-specific SEs.
(A) Schematic view of SE identification using published H3K37ac ChIP-seq data in normal, primary PCa, and CRPC tissue samples (GSE130408). (B, C) The number of SEs (B) and SE cutoff scores (C) in these samples. (D) The Venn diagram showing the overlap of SEs in different tissues. (E) The violin plot showing the FOXA1 chromatin binding signal on the identified 23 CRPC-specific SEs. (F) A list of CRPC-specific SE-associated genes. (G) The genome view of the H3K27ac tracks on EMBP1 and HOXB13. (H) Correlations between KDM1A or BRD4 with CRPC-specific SE-associated genes in SU2C mCRPC patient dataset. (I, J) Castrated SCID mice bearing 77CR (I) and 35CR (J) xenografts were treated with vehicle, i-BET762 (16mg/kg, daily), ORY-1001 (0.03mg/kg, every two days), or the combination. Tumor volume was measured by caliper. (K) RNA-seq analyses were performed using the tissue samples from 35CR collected at the end of the experiment. The heatmap view for the combination treatment-repressed genes in all tumor samples is shown. (L) The violin plot showing the expression changes of MYC targets (upper panel) and CRPC-specific SE-associated genes (lower panel) in these tumor samples.
DISCUSSION
Recent preclinical studies of inhibiting key epigenetic regulators in various malignancies have shed light on developing epigenetic therapies in the clinic. However, the most common hurdle of moving forward with these therapies in clinical trials is the adverse side effects due to the high dosage treatment of inhibitors. In this study, we aim to fully understand the molecular consequences of targeting LSD1 in the CRPC models and provide novel insights for developing more optimal strategies to further improve the anti-tumoral effect of LSD1-i. Our RNA-seq analysis using tumor samples from CRPC xenograft tumors treated with LSD1-i indicates that multiple oncogenic pathways can be targeted by LSD1-i, including transcription pathways of AR, FOXA1, E2F, and MYC. In agreement with previous studies (38), we also observed that LSD1-i consistently activated hallmark immunogenic pathways despite that the mice in our studies are immunocompromised strains, suggesting that LSD1-i may potentially improve the clinical output of PD-(L)1 blockade in CRPC by turning the immune ‘cold’ microenvironment into immune ‘hot’. While AR signaling has been well recognized as a target of LSD1-i (8,9,11,13), our recent work also uncovered an additional oncogenic function of LSD1 in stabilizing E2F chromatin binding (35). This is of critical clinical significance, as Rb-deficient cells (~10–15% of CRPC) may be more susceptible to LSD1-i. Our data here shows that LSD1-i consistently suppressed E2F signaling in all the tested CRPC models. Interestingly, in our single-cell RNA-seq analysis performed in the 96CR model, the apparent Rb-deficient cells express very low LSD1, thereby are resistant to LSD1-i. We determined this non-responsive population as ARhigh/FOXA1high/RB1−/TP53low/LSD1low, which does not express typical NE markers and retains the AR-FOXA1 axis. Importantly, this sub-population appeared to be less metabolic than the responsive counterpart. These data highlight the intrinsic heterogeneity of CRPC, and underscore the importance of determining the survival dependency within each subpopulation of CRPC tumors.
One major mechanism for cancer cells to acquire drug resistance is the oncogenic reprogramming driven by SEs that confer growth advantages under therapeutic pressure (46–48). One of the prominent examples of SE-driven oncogenes is MYC (49), which is highly expressed in PCa and can drive disease progression (50,51). In our study, we identified the association between LSD1, FOXA1, and BRD4 on the MYC SEs. Since aberrant SE reprogramming has been demonstrated as a mechanism for driving CRPC (47,52,53), we systematically compared the SE profiles in normal, primary PCa, and CRPC samples. This method led us to uncover a major SE reprogramming in CRPC and identify 23 SE-associated genes. Although we did not include the MYC gene due to the use of high stringency cutoff rate (85%), these 23 SE-associated genes appear to be involved in various tumor-promoting pathways. For example, we found well-characterized pioneer transcription factors such as FOXA1 and HOXB13, which have been shown to collaborate with AR in determining oncogenic reprogramming (54). We also found AR coregulators such as GRHL2 (Grainyhead-like protein 2), which has been shown to collaborate with FOXA1 to drive drug resistance in BCa and can enhance AR activity in PCa (55,56), and SPDEF (SAM pointed domain containing ETS transcription factor), whose expression levels are elevated in the tumors samples and has been linked to poor prognosis in PCa (57). Previous studies from us and others have shown that the lipid synthesis pathway is highly activated in CRPC and promotes tumor metastasis (44,58). The identification of FASN (fatty acid synthase) as one CRPC-specific SE gene further highlights that targeting lipogenic pathways could benefit a subset of CRPC patients. It should be noted that among the list of CRPC-specific SE-associated genes, several are non-coding RNAs. In particular, the long non-coding RNA NEAT1 was recently reported to promote bone metastasis in PCa (59). Importantly, in our attempt to understand the epigenomic landscape of CRPC SEs, we observed that the FOXA1 chromatin binding on these 23 SEs is stronger in CRPC than in primary PCa, indicating that there is potential positive feedback to promote the disease progression. It also alludes that the observed framework of LSD1, FOXA1, and BRD4 may be essential for mediating the SEs activities in CRPC. Overall, our analysis suggests that SEs in CRPC may activate a multifaceted oncogenic program.
Previous reports in the embryonic stem cell models have demonstrated that LSD1 in association with the repressive complex members HDAC1 and 2 enriches at the SEs, where LSD1 still demethylates H3K4me and may function to restrain the enhancer activation during the differentiation process (60). In that model, LSD1 demethylase activity is under check by a feedback mechanism involving histone acetyltransferase-mediated acetylation. A more recent study in BCa cells reports that LSD1 also enriches at SEs, and collaborates with BRD4 to repress a list of therapeutic resistance genes (61). While these studies demonstrated that LSD1 is tightly associated with SEs, our results in conjunction with the previous report on LSD1-mediated regulation of FOXA1 reveal a new model for the LSD1 involvement in a transcription activator network of FOXA1 and BRD4 on the CRPC SEs. Interestingly, another recent report by Sehrawat et al., showed that LSD1 drives PCa development via interaction with ZNF217 and this activity does not require its demethylase activity (62). However, in our model, LSD1 enzymatic activity is clearly required for SE activation. Therefore, whether ZNF217 is involved in the LSD1/FOXA1/BRD4 complex still requires further investigation. Our model suggests that LSD1 demethylates FOXA1 to stabilize FOXA1 binding, and that the unmethylated FOXA1 can more strongly bind to SEs and thus increases the recruitment of BRD4 to maintain SE activity (although we cannot exclude the possibility that other LSD1-interacting partners, such as CoREST and HDAC1/2, are also involved in the complex). BRD4 is recruited to chromatin by recognizing histone acetylation through its bromodomain. Our data suggest that FOXA1 DNA binding, which is mediated through the high-affinity interaction of its Forkhead domain with specific DNA sequences, may function as an anchor for BRD4 residency on chromatin. Interestingly, previous proteomic profiling studies suggest that BRD4 can recognize non-histone proteins that contain a Kac-XX-Kac motif (63) and multiple K-XX-K motifs can be found within the Forkhead DNA binding domain and the C terminal transactivation domain of FOXA1, although our previous mass-spectrometry studies did not find any strong signal of acetylation in these regions (13). Future investigation will be needed to determine the specific domains that mediate the physical interaction between FOXA1 and BRD4.
Furthermore, we present for the first time that LSD1 forms LLPS in CRPC cells, a property that is contributed by the highly disordered N-terminus of the protein, and that inhibition of LSD1 by the irreversible covalent inhibitors can lead to the disruption of BRD4-enriched nuclear condensates, which is further supported by a recent study showing that small molecule drugs are preferentially concentrated in the condensate where their target proteins are enriched in (23). This observation corroborates the important role of LSD1 in maintaining SE activities, where a high concentration of LSD1 could be associated with a multitude of transcriptional factors (e.g., FOXA1 and BRD4) and drive robust oncogenic transcription. Further investigation is required to fully understand the molecular composition and the biophysical principles of condensate formation by LSD1.
Although both synthetic and natural LSD1-i are in rapid development (64), the lethal hematological toxicity from targeting LSD1 may still prevent the clinical application of LSD1-i (14). A regimen that deploys lower dosages of inhibitors without compromising the anti-tumor efficacy is of paramount importance. Our results from in vitro and in vivo studies strongly suggest that co-targeting LSD1 and BRD4 using lower dosages could outperform the high-dose single agent treatment, without inducing overt toxicity in vivo. Interestingly, BET-i has also been shown to block AR/AR-V7 signaling in PCa cells (65–67) and BET inhibitors in conjunction with other targeted therapies are currently tested in multiple clinical trials for CRPC patients (68,69). Therefore, our results demonstrate a strong therapeutic potential of combining LSD1-i and BET-i.
In conclusion, our transcriptomic profiling study using the xenograft CRPC models has revealed that LSD1 inhibition can target multiple oncogenic pathways in CRPC, and that disrupting MYC signaling is a major consequence of LSD1-i. This study also further advances our understanding of the molecular mechanism of LSD1 activation function at enhancers, particularly SEs, in CRPC, which allows LSD1 to form a network with FOXA1 and BRD4 to maintain SE activity. Moreover, our study provides important mechanistic and therapeutic insights into developing the combination treatment of LSD1-i and BET-i as a novel therapeutic strategy for CRPC patients.
Supplementary Material
SIGNIFICANCE.
LSD1 drives prostate cancer progression by activating super-enhancer-mediated oncogenic programs, which can be targeted with the combination of LSD1 and BRD4 inhibitors to suppress growth of castration-resistant prostate cancer.
ACKNOWLEDGMENTS
This work is supported by grants from NIH (R01CA211350 to C Cai, U54CA156734 to JA Macoska, P01CA163227 to SP Balk), DOD (W81XWH-19-1-0361 and W81XWH-21-1-0267 to C Cai, W81XWH-19-1-0777 to S Gao), and CIHR (142246 and 15967 to H He). M Liu was supported by the graduate fellowship from Integrative Biosciences Program at University of Massachusetts Boston. W Han, Z Wang, and A Besschetnova were supported by CSM (College of Science and Mathematics) Dean’s Doctoral Research Fellowship from University of Massachusetts Boston. C.C. is supported by Proposal Development Grant Program from University of Massachusetts Boston. The establishment, characterization, and maintenance of the LuCaP PDXs are funded by NIH (P50CA097186 and P01CA163227).
Footnotes
The authors declare no potential conflict of interest.
REFERENCES
- 1.Nuhn P, De Bono JS, Fizazi K, Freedland SJ, Grilli M, Kantoff PW, et al. Update on Systemic Prostate Cancer Therapies: Management of Metastatic Castration-resistant Prostate Cancer in the Era of Precision Oncology. Eur Urol 2019;75:88–99 [DOI] [PubMed] [Google Scholar]
- 2.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS, et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res 2014;20:1590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda DY, Spisak S, Seo JH, Bell C, O’Connor E, Korthauer K, et al. A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell 2018;174:422–32 e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao S, Chen S, Han D, Barrett D, Han W, Ahmed M, et al. Forkhead domain mutations in FOXA1 drive prostate cancer progression. Cell Res 2019;29:770–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015;15:701–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conteduca V, Hess J, Yamada Y, Ku SY, Beltran H. Epigenetics in prostate cancer: clinical implications. Transl Androl Urol 2021;10:3104–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941–53 [DOI] [PubMed] [Google Scholar]
- 8.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005;437:436–9 [DOI] [PubMed] [Google Scholar]
- 9.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 2007;9:347–53 [DOI] [PubMed] [Google Scholar]
- 10.Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N, et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol 2008;10:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai C, He HH, Gao S, Chen S, Yu Z, Gao Y, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep 2014;9:1618–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer 2012;12:381–5 [DOI] [PubMed] [Google Scholar]
- 13.Gao S, Chen S, Han D, Wang Z, Li M, Han W, et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat Genet 2020;52:1011–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol 2019;12:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wass M, Gollner S, Besenbeck B, Schlenk RF, Mundmann P, Gothert JR, et al. A proof of concept phase I/II pilot trial of LSD1 inhibition by tranylcypromine combined with ATRA in refractory/relapsed AML patients not eligible for intensive therapy. Leukemia 2021;35:701–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015;28:57–69 [DOI] [PubMed] [Google Scholar]
- 17.Reed DR, Mascarenhas L, Meyers PA, Chawla SP, Harrison DJ, Setty B, et al. A phase I/II clinical trial of the reversible LSD1 inhibitor, seclidemstat, in patients with relapsed/refractory Ewing sarcoma. Journal of Clinical Oncology 2020;38:TPS11567-TPS [Google Scholar]
- 18.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153:320–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018;175:1842–55 e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018;361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall’Agnese A, et al. Partitioning of cancer therapeutics in nuclear condensates. Science 2020;368:1386–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 2011;12:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He MX, Cuoco MS, Crowdis J, Bosma-Moody A, Zhang Z, Bi K, et al. Transcriptional mediators of treatment resistance in lethal prostate cancer. Nat Med 2021;27:426–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87 e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomerantz MM, Qiu X, Zhu Y, Takeda DY, Pan W, Baca SC, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet 2020;52:790–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate 2017;77:654–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maes T, Mascaro C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, et al. ORY-1001, a Potent and Selective Covalent KDM1A Inhibitor, for the Treatment of Acute Leukemia. Cancer Cell 2018;33:495–511 e12 [DOI] [PubMed] [Google Scholar]
- 35.Han W, Liu M, Han D, Li M, Toure AA, Wang Z, et al. RB1 loss in castration-resistant prostate cancer confers vulnerability to LSD1 inhibition. Oncogene 2022;41:852–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin HJ, Zhao JC, Ogden I, Bergan RC, Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res 2013;73:3725–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song B, Park SH, Zhao JC, Fong KW, Li S, Lee Y, et al. Targeting FOXA1-mediated repression of TGF-beta signaling suppresses castration-resistant prostate cancer progression. J Clin Invest 2019;129:569–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 2018;174:549–63 e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han W, Liu M, Han D, Toure AA, Li M, Besschetnova A, et al. Exploiting the tumor-suppressive activity of the androgen receptor by CDK4/6 inhibition in castration-resistant prostate cancer. Mol Ther 2022;30:1628–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai L, Tsai YH, Wang P, Wang J, Li D, Fan H, et al. ZFX Mediates Non-canonical Oncogenic Functions of the Androgen Receptor Splice Variant 7 in Castrate-Resistant Prostate Cancer. Mol Cell 2018;72:341–54 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirguet O, Gosmini R, Toum J, Clement CA, Barnathan M, Brusq JM, et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem 2013;56:7501–15 [DOI] [PubMed] [Google Scholar]
- 44.Han W, Gao S, Barrett D, Ahmed M, Han D, Macoska JA, et al. Reactivation of androgen receptor-regulated lipid biosynthesis drives the progression of castration-resistant prostate cancer. Oncogene 2018;37:710–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salamero O, Montesinos P, Willekens C, Perez-Simon JA, Pigneux A, Recher C, et al. First-in-Human Phase I Study of Iadademstat (ORY-1001): A First-in-Class Lysine-Specific Histone Demethylase 1A Inhibitor, in Relapsed or Refractory Acute Myeloid Leukemia. J Clin Oncol 2020;38:4260–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Ma Q, Shang Z, Niu Y. Super-enhancer in prostate cancer: transcriptional disorders and therapeutic targets. NPJ Precis Oncol 2020;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen S, He Y, Wang L, Zhang J, Quan C, Niu Y, et al. Aberrant activation of super enhancer and choline metabolism drive antiandrogen therapy resistance in prostate cancer. Oncogene 2020;39:6556–71 [DOI] [PubMed] [Google Scholar]
- 48.Li GH, Qu Q, Qi TT, Teng XQ, Zhu HH, Wang JJ, et al. Super-enhancers: a new frontier for epigenetic modifiers in cancer chemoresistance. J Exp Clin Cancer Res 2021;40:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuijers J, Manteiga JC, Weintraub AS, Day DS, Zamudio AV, Hnisz D, et al. Transcriptional Dysregulation of MYC Reveals Common Enhancer-Docking Mechanism. Cell Rep 2018;23:349–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corey E, Quinn JE, Buhler KR, Nelson PS, Macoska JA, True LD, et al. LuCaP 35: a new model of prostate cancer progression to androgen independence. Prostate 2003;55:239–46 [DOI] [PubMed] [Google Scholar]
- 51.Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, et al. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol 2013;44:1617–23 [DOI] [PubMed] [Google Scholar]
- 52.Guo H, Wu Y, Nouri M, Spisak S, Russo JW, Sowalsky AG, et al. Androgen receptor and MYC equilibration centralizes on developmental super-enhancer. Nat Commun 2021;12:7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgart SJ, Nevedomskaya E, Lesche R, Newman R, Mumberg D, Haendler B. Darolutamide antagonizes androgen signaling by blocking enhancer and super-enhancer activation. Mol Oncol 2020;14:2022–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015;47:1346–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paltoglou S, Das R, Townley SL, Hickey TE, Tarulli GA, Coutinho I, et al. Novel Androgen Receptor Coregulator GRHL2 Exerts Both Oncogenic and Antimetastatic Functions in Prostate Cancer. Cancer Res 2017;77:3417–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cocce KJ, Jasper JS, Desautels TK, Everett L, Wardell S, Westerling T, et al. The Lineage Determining Factor GRHL2 Collaborates with FOXA1 to Establish a Targetable Pathway in Endocrine Therapy-Resistant Breast Cancer. Cell Rep 2019;29:889–903 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meiners J, Schulz K, Moller K, Hoflmayer D, Burdelski C, Hube-Magg C, et al. Upregulation of SPDEF is associated with poor prognosis in prostate cancer. Oncol Lett 2019;18:5107–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu X, Fong KW, Gritsina G, Wang F, Baca SC, Brea LT, et al. HOXB13 suppresses de novo lipogenesis through HDAC3-mediated epigenetic reprogramming in prostate cancer. Nat Genet 2022;54:670–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer 2020;19:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012;482:221–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu B, Liu X, Han L, Chen X, Wu X, Wu J, et al. BRD4-directed super-enhancer organization of transcription repression programs links to chemotherapeutic efficacy in breast cancer. Proc Natl Acad Sci U S A 2022;119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sehrawat A, Gao L, Wang Y, Bankhead A 3rd, McWeeney SK, King CJ, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci U S A 2018;115:E4179–E88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uuskula-Reimand L, et al. Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains. Mol Cell 2019;73:621–38 e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong J, Pervaiz W, Tayyab B, Li D, Kang L, Zhang H, et al. A comprehensive comparative study on LSD1 in different cancers and tumor specific LSD1 inhibitors. Eur J Med Chem 2022;240:114564 [DOI] [PubMed] [Google Scholar]
- 65.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014;510:278–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welti J, Sharp A, Yuan W, Dolling D, Nava Rodrigues D, Figueiredo I, et al. Targeting Bromodomain and Extra-Terminal (BET) Family Proteins in Castration-Resistant Prostate Cancer (CRPC). Clin Cancer Res 2018;24:3149–62 [DOI] [PubMed] [Google Scholar]
- 67.Kim DH, Sun D, Storck WK, Welker Leng K, Jenkins C, Coleman DJ, et al. BET Bromodomain Inhibition Blocks an AR-Repressed, E2F1-Activated Treatment-Emergent Neuroendocrine Prostate Cancer Lineage Plasticity Program. Clin Cancer Res 2021;27:4923–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017;12:323–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aggarwal RR, Schweizer MT, Nanus DM, Pantuck AJ, Heath EI, Campeau E, et al. A Phase Ib/IIa Study of the Pan-BET Inhibitor ZEN-3694 in Combination with Enzalutamide in Patients with Metastatic Castration-resistant Prostate Cancer. Clin Cancer Res 2020;26:5338–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE209889. All other raw data generated in this study is available from the corresponding author upon request. The data analyzed in this study were obtained from GEO at GSE94013 and GSE130408.