SUMMARY
The bacterial cell wall is composed of a highly crosslinked matrix of glycopeptide polymers known as peptidoglycan that dictates bacterial cell morphology and protects against environmental stresses. Regulation of peptidoglycan turnover is therefore crucial for bacterial survival and growth and is mediated by key protein complexes and enzyme families. Here, we review the prevalence, structure, and activity of NlpC/P60 peptidases, a family of peptidoglycan hydrolases that are crucial for cell wall turnover and division as well as interactions with antibiotics and different hosts. Understanding the molecular functions of NlpC/P60 peptidases should provide important insight into bacterial physiology, their interactions with different kingdoms of life and the development of new therapeutic approaches.
eTOC
Enzymes that remodel the bacterial cell wall, peptidoglycan, are critical for both cell division and their interactions with host organisms, making them potentially useful targets for therapeutic applications. In this review, Griffin M et al. provide an overview of NlpC/P60 peptidoglycan hydrolases and summarize their unique functions across different microbes.
INTRODUCTION
Bacteria live in a variety of biomes from the natural environment to cohabitation with other creatures, each with their own harsh and ever-changing physiological conditions. To combat these environmental stresses, bacteria produce protective glycopeptide polymers within their cell walls known as peptidoglycan (PG).1,2 PG consists of a repeating disaccharide unit of N-acetylglucosamine (NAG, GlcNAc) and N-acetylmuramic acid (NAM, MurNAc), with a stem peptide attached to the 3-O-lactoyl group of NAM (Figure 1). Stem peptide structures are highly heterogeneous within the cell wall of individual bacterial cells. In newly incorporated PG monomers, the stem peptide generally exists as a pentapeptide chain, which can be modified and crosslinked with other stem peptides during PG maturation. Across bacterial species, PG also exhibits significant structural diversity, with substitutions at multiple amino acid residues of the stem peptide and crossbridge as well as modifications to the carbohydrate backbone.
Figure 1. Peptidoglycan structure and remodeling.
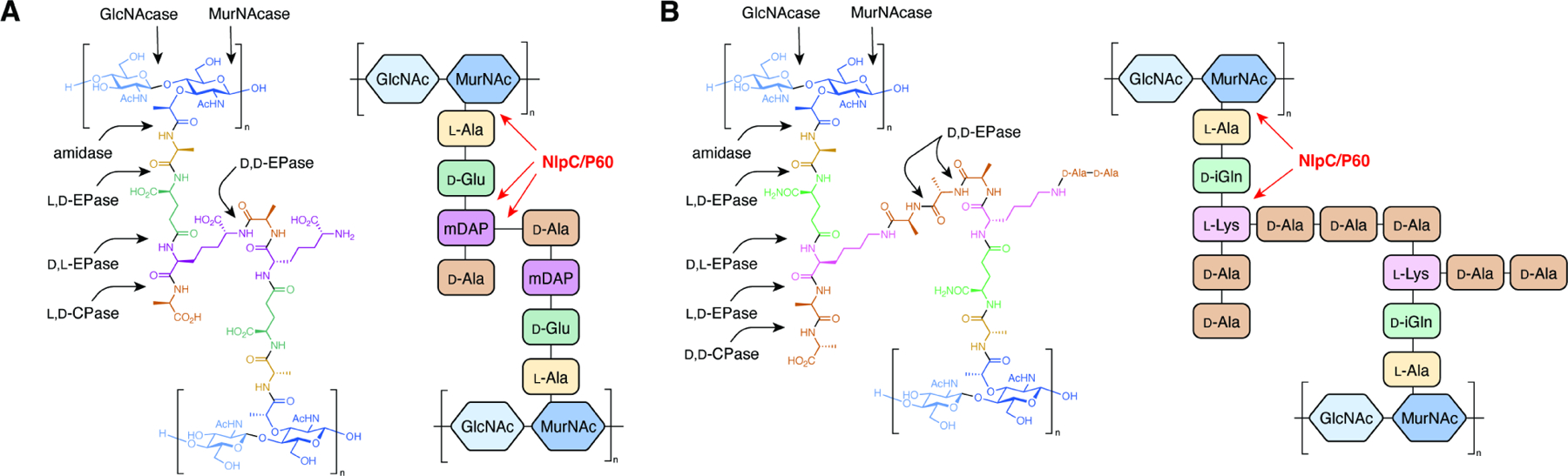
Generalized chemical structure of a crosslinked peptidoglycan monomer with the potential bonds cleaved by different classes of peptidoglycan remodeling enzymes (arrows) for (A) Gram-negative E. coli and (B) Gram-positive E. faecalis.
Although the rigid structure of PG can prevent cell lysis under osmotic shock, PG must be degraded and reformed to allow for cell growth and division.3 Bacteria have developed highly regulated systems of enzymes to remodel PG.4,5 The nomenclature of these proteins is often complicated due to historical precedence and overlapping enzyme classifications.6 In accordance with previous literature, we will refer to remodeling enzymes as three separate groups based on their substrate specificity: glycosidases, which act on the glycosidic bonds between NAG and NAM residues; amidases, which degrade the amide bond between NAM and the stem peptide; and peptidases, which cleave amide bonds between two amino acid residues of the stem peptides or crosslinking bridges. Within the peptidase family, enzymes can be further classified by the stereochemistries of the two amino acid residues they hydrolyze (L,D vs. D,L vs. D,D) and their relative location within the stem peptide (endo- vs. carboxypeptidase). In addition to substrate- or function-based classifications, these proteins have also historically been categorized by structural homology of their catalytic domains. For example, PG peptidase activity is accomplished by at least 10 distinct domain types.6 In this review, we will focus on efforts to understand the activities and biological functions of the “new lipoprotein C from Escherchia coli / protein of 60 kDa of Listeria monocytogenes” (NlpC/P60) superfamily of cysteine peptidases (Figure 1). Through the biochemical and functional characterization of these enzymes, we will highlight how this widespread family of PG hydrolases are critical mediators of both bacterial physiology and microbial-host interactions. Together, the broad roles of NlpC/P60-containing proteins underscore their untapped potential as novel targets for antibiotics and modulators of host immunity.
The NlpC/P60 family of cysteine peptidases
The NlpC/P60 domain is approximately 150 amino acid residues in length and 15 kDa in size (Figure 2A).7 Nearly all of these domains fold akin to other papain-like proteases, with a series of three N-terminal ɑ-helices followed by a five-stranded β-barrel. This tertiary structural motif is similar to other cysteine peptidase families, which aided in initial predictions of the catalytic core of this domain. NlpC/P60 domains are unified by the presence of conserved cysteine and histidine residues, which are found at the N-terminus of the second, conserved ɑ-helix and within the first two β-strands, respectively (Figure 2A). Like other cysteine proteases, the cysteine residue acts as a nucleophile to break the targeted PG amide bond, whereas the histidine residue functions as a general acid-base. In addition to the invariant catalytic diad, some NlpC/P60 domains contain a third polar amino acid residue (His, Glu, or less frequently Gln or Asn) that may act as a coordinating residue for the catalytic histidine through hydrogen bonding.
Figure 2. Biochemical structures of NlpC/P60 proteins.
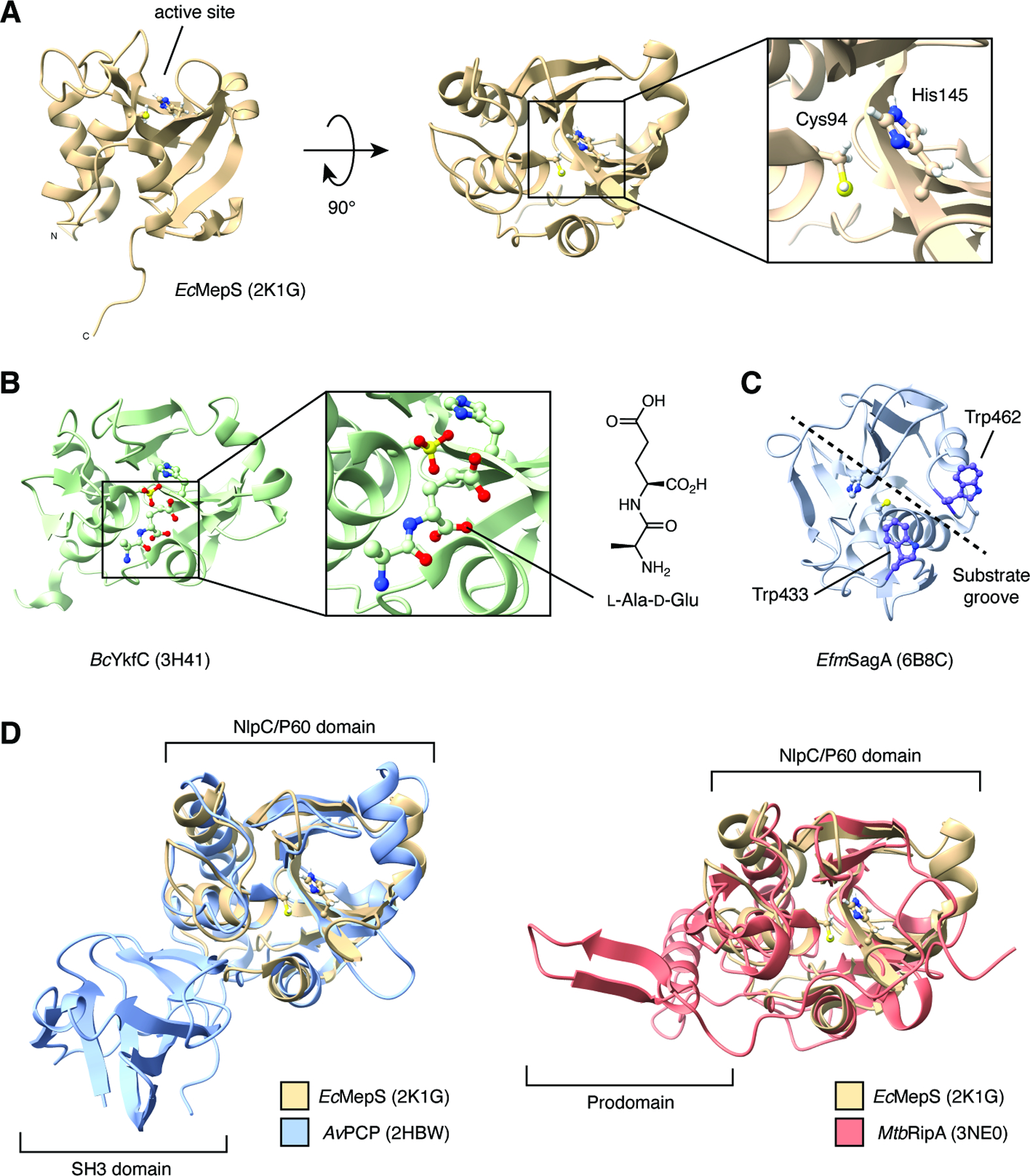
(A) NMR structure and catalytic dyad of E. coli MepS (Spr).
(B) Crystal structure of B. cereus YkfC with dipeptide product of peptidoglycan hydrolysis bound to the active site.
(C) Crystal structure of E. faecium SagA with catalytic dyad and putative substrate-binding Trp residues highlighted.
(D) Overlay of E. coli MepS and A. variabilis AvPCP and M. tuberculosis RipA showing additional regulatory domains.
Peptidase activity of NlpC/P60 domains is most frequently specific for the amide bond between the second and third amino acid residues of the stem peptide (Figure 1).6,7 These residues differ between bacterial species (ɣ-D-Glu vs. ɣ-D-Gln at the second position and L-Lys vs. meso-DAP vs. amidated meso-DAP at the third position), leading to peptidases with differential molecular specificities. In addition, these enzymes often have differential specificities towards substrates depending on chemical structures distal from the targeted amide bond, including the length and crosslinking of the stem peptide, the presence of the glycan residues and the extent of polysaccharide polymerization. NlpC/P60 activity has also been demonstrated on other peptide linkages including between meso-DAP and the fourth stem peptide residue, D-Ala, or between meso-DAP and a crosslinked D-Ala residue from a separate stem peptide 6,7. Beyond strict peptidase activity, some NlpC/P60-containing proteins possess amidase activity to separate MurNAc residues from the stem peptide. Although the protein of unknown function DUF1460 family (IPR010846) is classified separately by InterPro, recent evidence has indicated that DUF1460 represents a new group of evolutionarily and structurally related NlpC/P60 amidases 8. Together, bacteria take advantage of these diverse substrate preferences by often expressing multiple, differentially regulated, and localized NlpC/P60 proteins, allowing them to coordinate the complex process of PG remodeling.
NlpC/P60 domains have been extensively examined through structural studies, which date back to the first solution NMR structure of E. coli MepS (Spr) reported in 2008 (Figure 2A) 9. To date, there are over 25 structures currently deposited in the Protein Data Bank, allowing for direct comparisons of protein architecture across the bacterial kingdom. Crucially, these studies have highlighted conserved mechanisms of substrate recognition. The NlpC/P60 substrate-binding pocket was first mapped for YkfC from Bacillus cereus (Figure 2B).10 X-ray crystallography of the purified enzyme revealed unexpected electron density within the putative enzyme active site. Although no substrate was added in the purification process, this density was assigned to the L-Ala-D-Glu dipeptide that may have been derived from PG the recombinant E. coli host. The bound dipeptide was found to be stabilized by multiple hydrogen bonds within the substrate binding groove, including Glu83, Tyr118 and Asp256, which are highly conserved in the YkfC subfamily of NlpC/P60 enzymes. Guided by this initial structure, multiple computational docking studies of muropeptides with solved structures also aided in defining the substrate-binding cleft of NlpC/P60 proteins. Together, these data uncovered additional regions and residues that may be critical for substrate recognition. For example, docking of an mDAP-type GlcNAc-MurNAc (GM)-tetrapeptide fragment to CwlT of Staphylococcus aureus found that conformational rearrangement of the flexible β4/β5 loop is required for enzymatic function and may dictate substrate specificity.11 Docking of Lys-type GM-tetrapeptide and GM-pentapeptide to SagA from Enterococcus faecium revealed that two aromatic residues, Trp433 and Trp462, interact with the glycan headgroup of the muropeptide and may function as a clamp for substrate binding (Figure 2C).12 Further in silico alanine screening and in vitro validation showed that multiple aromatic and hydrophilic residues along an extended substrate-binding groove were also crucial for enzymatic turnover, likely through stabilization of the muropeptide-bound structure.13 Iterative superimposition and rigid-body fitting of PG fragments to Acd24020 from Clostridium difficile was used to generate a model of the enzyme bound to a crosslinked PG substrate.14 This model revealed that crosslinked PG forms an H-like structure, with the stem peptide accommodated within the substrate-binding groove and the polysaccharide backbone extending perpendicular to the groove along the surface of the domain. The bound structure and subsequent in vitro characterization demonstrated that the unique hourglass shape of the Acd24020 groove enabled recognition of the direct mDAP-D-Ala crosslinks found in PG from C. difficile, underscoring that the composition and orientation of the substrate groove within NlpC/P60 proteins may regulate its specificity for distinct PG chemical structures.
As an additional layer of regulation, NlpC/P60 domains are frequently found in larger proteins that can contain other structural regions or defined domains (Figure 2D). For example, many NlpC/P60-containing proteins including AvPCP from Anabaena variabilis possess one or more bacterial Src homology 3 (SH3) domains, which have been predicted to mediate protein-protein interactions.15 Other common coinciding features include LysM domains, which are thought to be involved in direct PG or cell wall binding,16 as well as prodomains that may regulate catalytic activity as seen in RipA from Mycobacterium tuberculosis.17 Therefore, the overall structure of NlpC/P60 domain-containing proteins may also serve as an additional determinant of biological specificity. Together with substrate preference and endogenous mechanisms of gene regulation, these differentiating factors across bacterial peptidases allow NlpC/P60 domains to exert potentially non-overlapping functions.
The NlpC/P60 domain is found broadly throughout bacteria as well as within viruses, archaea, and even eukaryotes.7 The InterPro classification of protein families categorizes the NlpC/P60 domain (IPR000064) along with the structurally similar cysteine, histidine-dependent amidohydrolases and peptidases (CHAP, IPR007921) domain as part of the papain-like cysteine peptidase superfamily (IPR038765).18 Via sequence analysis, NlpC/P60 domains have been separated into four major groups: (1) P60-like, (2) AcmB/LytN-like, (3) YaeF/Poxvirus G6R-like, and (4) lecithin retinol acyltransferase-like (LRAT-like). The P60-like family proper encompasses most PG-degrading D,L-endopeptidases within bacteria, whereas the AcmB/LytN-like group is a divergent and likely monophyletic clade found in a subset of Gram-positive bacteria arising from rapid sequence divergence. The YaeF/Poxvirus G6R type domain is distributed across taxonomic kingdoms in bacteria, archaea, C. elegans, and poxviruses, whereas the LRAT-like family is also found in viruses, eukaryotes, and rarely within bacterial species. NlpC/P60 domain organization is circularly permuted in the YaeF/Poxvirus G6R-like groups and LRAT-like groups, in which the histidine-containing β-barrel is found prior to the cysteine-containing ɑ-helix in the primary amino acid sequence while maintaining the overall orientation of the catalytic residues.
Functions of NlpC/P60 hydrolases in bacterial growth and division
NlpC/P60 proteins have been identified and characterized in diverse bacteria, including Gram-negative, Gram-positive, and Actinobacteria taxa. These proteins are involved in numerous cellular processes, including growth, division, competition, defense, virulence, sporulation, and biofilm formation. Their expression is controlled by regulatory elements such as two-component systems, alternative sigma factors and riboswitches, and their functionality is modulated by protein-protein interactions, post-translational modifications, and proteolysis. This information is summarized in Table 1.
Table 1:
Summary of bacterial NlpC/P60 proteins
Summary of bacterial NlpC/P60 proteins that have been biochemically or biologically characterized beyond sequence homology.
| Organism | NlpC/P60 Protein | Specific Function(s)* | Notes | Reference(s) |
|---|---|---|---|---|
| Gram-Negative Bacteria | ||||
| Anabaena variabilis | AvPCP | Structure determined (PDB 2HBW) | 15 | |
| Bacteriodes ovatus | YkfC | Structure determined (PDB 3NPF) | 19 | |
| Bacteriodes thetaiotaomicron | YkfC | Structure determined (PDB 3PVQ & 4R0K) | 19 | |
| Bacteriodes uniformis | AmiA | Structure determined (PDB 4H4J [AmiA alone], 4Q68 [AmiA/GlcNAc], and 4Q5K [AmiA/GlcNAc-AnhMurNAc]) | 8 | |
| Desulfovibrio vulgaris | DvLysin | Structure determined (PDB 3M1U) | 19 | |
| Enterobacter cloacae | Tae4 | Cellular defense | Secreted by a T6SS; self-protection is accomplished by the immunity protein Tai4 | 20 |
| Escherechia coli | NlpC | 21 | ||
| MepS (Spr) | Virulence | DD-endopeptidase; regulated by proteolysis (degraded by NlpI/Prc system); synthetic lethality with MepM (a non-NlpC/P60 hydrolase) in rich media but not minimal; contributes to kidney infections; deletion confers sensitivity to EDTA; structure determined (PDB 2K1G) |
22
21 9 23 24 25 26 27 28 29 30 |
|
| MepH (YdhO) | Potentially regulated by nitric oxide |
21
24 26 28 |
||
| YafL | 21 | |||
| Nostoc punctiforme | NpPCP | Structure determined (PDB 2EVR & 2FG0) | 15 | |
| Pseudomonas aeruginosa | MagC | PG binding | Anchors Mag complex to PG; NlpC/P60 domain is inactive | 31 |
| Tse1 | Cellular defense | Secreted by a T6SS; self-protection is accomplished by the immunity protein Tsi1; structures determined for Tse1 (PDB 3VPI) and Tse1/Tsi1 complex (PDB 3VPJ) |
32
33 34 |
|
| PA1198 | Homologous to E. coli MepS (a DD-endopeptidase); regulated by proteolysis (degraded by LbcA/CtpA system) |
35
36 37 38 |
||
| PA1199 | Also homologous to E. coli MepS; C-terminal end plays a role in degradation | |||
| Photobacterium damselae subspecies piscicida | PnpA | Cellular defense | Secreted by a T2SS; Is specific for peptidoglycan of competing bacteria, does not degrade P. damselae peptidoglycan | 39 |
| Salmonella enterica serovar Typhimurium | EcgA | Virulence | Regulated by PhoP; induced in non-growing cells |
40
41 |
| Tae4 | Cellular defense | Secreted by a T6SS; self-protection is accomplished by the immunity protein Tai4 |
42
20 |
|
| MepS | Regulated by ScwA |
43
44 |
||
| MepH | Regulated by ScwA |
43
44 |
||
| NlpC | Regulated by ScwA |
43
44 |
||
| Thermus thermophilus | P60_tth | Structure determined (PDB 4XCM, 4UZ2, and 4UZ3) | 45 | |
| Vibrio cholerae | NlpC | 46 | ||
| TseH | Cellular defense | Structure determined (PDB 6V98) |
47
48 |
|
| Gram-Positive Bacteria | ||||
| Bacillus anthracis | BAS1812 (BA1952) | Sporulation | Seropositive/immunogenic; contains three SH3b domains; mutant has impared endospore formation and germination, and increased susceptibility to detergent; nearly identical to B. cereus CwpFM |
49
50 51 |
| Bacillus cereus | CwpFM (EntFM) | Adhesion, biofilm formation, and virulence |
52
53 54 55 |
|
| YkfC | Structure determined (PDB 3H41) | 10 | ||
| BcPPNE | 56 | |||
| Bacillus sphaericus | DPP VI | Sporulation | Upregulated during sporulation, but no direct evidence of participation |
57
15 |
| Bacillus subtilis | CwlO (YvcE) | Cell elongation | Synthetic lethality with LytE; regulated by WalRK; localizes to lateral cell wall; highly unstable transcript; binds to and depends on FtsEX |
58
59 60 61 62 63 64 65 66 67 62 68 69 70 71 |
| LytE (CwlF) | Cell elongation and cell separation | Synthetic lethality with CwlO; regulated by WalRK; localizes to poles, septa, and lateral cell wall; upregulated when cwlO is deleted |
59
60 61 72 73 74 75 76 62 64 67 62 77 78 68 69 65 71 |
|
| LytF (CwlE) | Cell separation | Non-essential; localizes to septa and poles; can be used to overcome the synthetic lethality of LytE/CwlO if expressed from the cwlO promoter; regulated by sigma factor D. |
59
61 75 79 80 81 82 83 76 68 69 84 |
|
| CwlS (YojL) | Cell separaton | Non-essential; localizes to septa and poles; cannot be used to overcome the synthetic lethality of LytE/CwlO even if expressed from the cwlO promoter |
59
61 85 68 16 69 |
|
| YkfC | Non-essential | 61 | ||
| CwlT (YddH) | Bifunctional (has an NlpC/P60 domain as well as a lysozyme domain); part of an integrative and conjugative element (ICE) |
86
87 |
||
| PgdS | Poly-γ-glutamic acid degradation | Does not degrade peptidoglycan |
88
89 61 90 |
|
| Tgl | Sporulation | Acts by crosslinking proteins to the spore coat |
91
92 93 |
|
| Clostridiodes difficile | CwlA | Cell separation | Phosphorylation dependent | 94 |
| CwlT | 11 | |||
| Cwl0971 | Virulence, sporulation | 95 | ||
| Acd24020 | Structure determined (PDB 7CFL) | 14 | ||
| Enterococcus faecium | SagA | Septum formation and host immune modulation | Essential; localizes to septum; generates muropeptides that enhance host immunity to other bacteria (PDB 6B8C) |
96
13 12 97 98 |
| Lactobacillus casei | P40 |
99
100 |
||
| P75 |
99
100 |
|||
| Lactobacillus paracasei | P40 | 100 | ||
| P75 | 100 | |||
| Lactobacillus plantarum | LytA | Cell elongation | 101 | |
| LytB | Septum maturation | 101 | ||
| LytC | 101 | |||
| LytD | 101 | |||
| Lactobacillus rhamnosus GG | Msp1 (P75, LGG_00324) | Cell separation | Accumulates at septum; is O-glycosylated |
102
103 |
| LGG_02016 | Strongly upregulated in acidic conditions | 104 | ||
| Lactobacillus salivarius | UC118 | Host immune modulation | 105 | |
| Lactococcus lactis | PgpA (YjgB) | Non-essential, mutants have no phenotype | 106 | |
| Listeria monocytogenes | P45 (Spl) |
107
108 |
||
| P60 | Motility and virulence |
107
109 110 111 |
||
| Nocardia seriolae | NlpC/P60 | Virulence | Induces apoptosis | 112 |
| Staphylococcus aureus | CwlT | Structure determined (PDB 4FDY) | 11 | |
| LnsA | Lipoprotein acylation | 113 | ||
| Streptococcus pneumoniae phage Dp-1 | Pal | Phage virulence |
114
115 |
|
| Streptomyces coelicolor | SwlA | Sporulation |
116
117 |
|
| Actinobacteria | ||||
| Corynebacterium diptheriae | DIP1281 | Virulence |
118
119 |
|
| DIP1621 |
120
119 |
|||
| Corynebacterium glutamicum | RipC (CgR_1596, CgP_1735) | Cell separation | Nomenclature note: In C. glutamicum R, this protein is referred to as CgR_1596. In C. glutamicum MB001, it is CgP_1735, which was named RipC by Bernhardt et al. |
121
122 123 124 |
| CgR_2070 | Cell separation | 121 | ||
| Mycobacterium avium subspecies paratuberculosis | MAP_1272c | NlpC/P60 domain is inactive |
125
126 127 128 |
|
| MAP_1203 | 125 | |||
| MAP_1204 | 125 | |||
| MAP_1928c | 125 | |||
| MAP_0036 | 125 | |||
| Mycobacterium marinum | IipA | Virulence | Ortholog of M. tuberculosis RipA | 129 |
| IipB | Virulence | Ortholog of M. tuberculosis RipB | 129 | |
| Mycobacterium smegmatis | RipA | Synthetic lethality with RipB | 130 | |
| RipB | Synthetic lethality with RipA | 130 | ||
| Mycobacterium tuberculosis | RipA (Rv1477) | Septum formation | Synthetic lethality with RipB; localizes to septum and occasionally poles; interacts with RpfB and PBP1 (via PonA1); processed by MarP; structure determined (PDB 6EWY & 3NE0) |
131
132 133 134 135 136 137 17 138 139 140 141 142 143 |
| RipB (Rv1478) | Synthetic lethality with RipA; structure determined (PDB 3PBI) |
139
138 |
||
| RipC (Rv2190c) | Growth and virulence | Activated by FtsX |
144
145 146 |
|
| RipD (1566c) | NlpC/P60 domain is inactive | 147 | ||
| Rv0024 | Biofilm formation and drug resistance | Expression in Mycobacterium smegmatis causes increased biofilm formation and antibiotic resistance | 148 | |
Specific functions beyond growth and division are noted in the table.
A fundamental role of PG hydrolases in bacterial physiology is to aid in growth and division (Figure 3). If cells did not have the ability to degrade and reassemble their own PG, they would become trapped by their own cell walls, unable to expand, elongate, or separate during binary fission. The essential roles of PG remodeling can be seen using the antibiotics complestatin or corbomycin, which inhibit PG hydrolysis by binding to PG and occluding access by hydrolases. Treatment of Bacillus subtilis with either molecule results in a thickening of the cell wall, and ultimately cell death.149 Unsurprisingly, NlpC/P60-containing enzymes have been broadly implicated in bacterial proliferation across multiple genera.
Figure 3. Roles of NlpC/P60 proteins in growth and division.
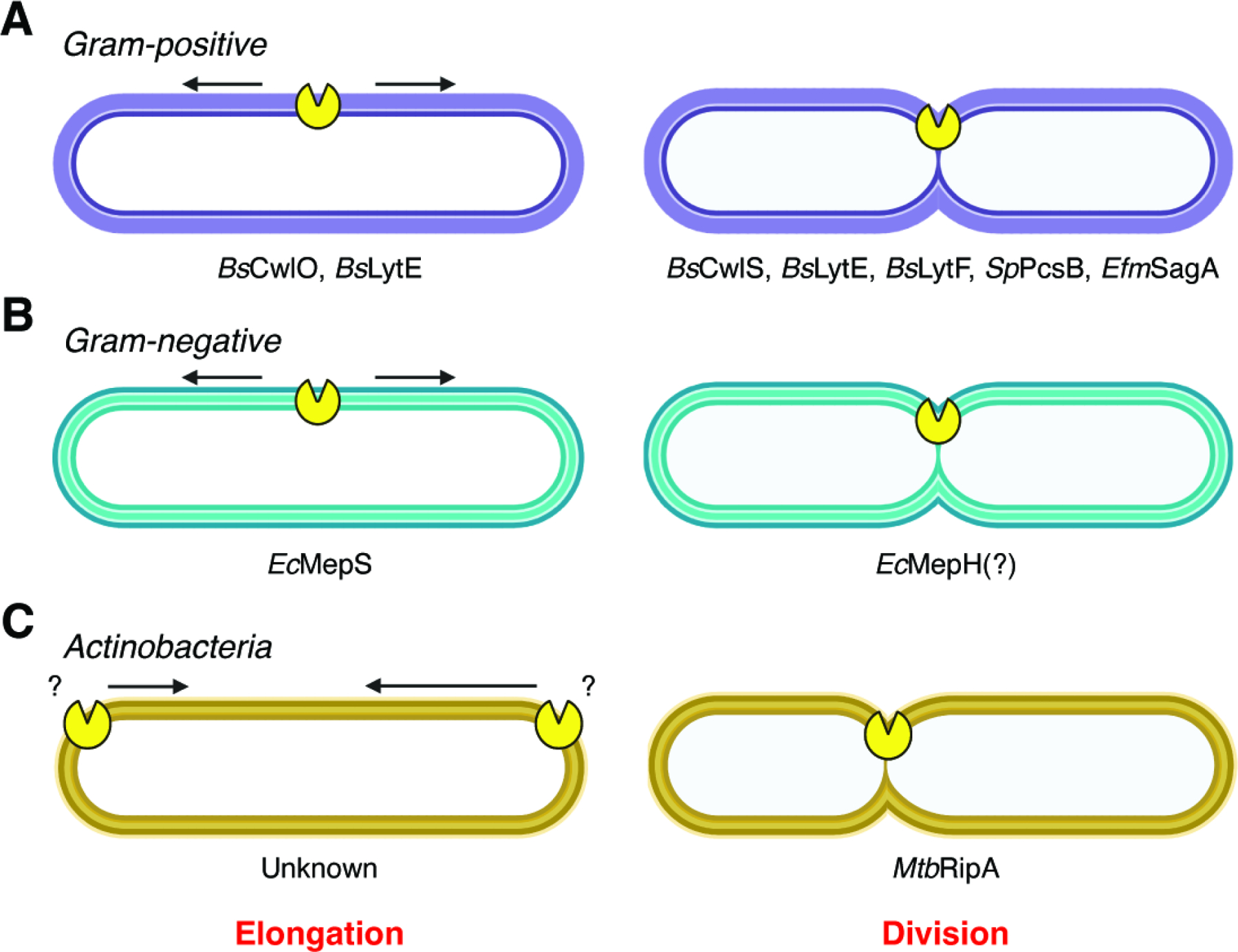
Characterized NlpC/P60 proteins involved in the elongation and division of (A) Gram-positive bacteria, (B) Gram-negative bacteria, and (C) Actinobacteria.
In Gram-positive bacteria, the specific roles of NlpC/P60 hydrolases in growth and division have been well studied (Figure 3A). The Gram-positive model organism B. subtilis contains at least four active NlpC/P60 D,L-endopeptidases: CwlO, LytF, CwlS, and LytE.59 CwlO is localized to the cylindrical part of the rod-shaped cell and is specifically involved in elongation. LytF and CwlS are localized proximal to the septum and poles of dividing cells and are implicated in cell separation. LytE is present along the cell rod, as well as the septum and poles, and this enzyme has roles in both elongation and cell separation. Double knockout of cwlO and lytE is synthetically lethal due to the necessity for D,L-endopeptidase activity within the cell rod during growth.59,69 This essential activity is underscored by recent results showing that all other 40 annotated PG hydrolases in B. subtilis can be simultaneously knocked out in combination with either cwlO or lytE without affecting survival, but loss of only these two enzymes is lethal.61 Both CwlO and LytE are thought to be recruited by homologs of eukaryotic actin. The actin homolog Mbl directs the subcellular localization of CwlO by facilitating its interaction with FtsX in the FtsEX complex, which both activates CwlO and anchors the protein to the membrane-proximal portion of the cell wall.65 Similarly, the actin homolog MreBH recruits LytE to the periphery of the cell, where the protein is secreted and noncovalently bound to the growing sacculus in a helical pattern.72 As the sacculus grows, the newly incorporated LytE becomes dispersed throughout both the membrane-proximal and membrane-distal portions of the cell wall. LytF has been shown to be both necessary and sufficient for complete or near-complete cell separation, with most lytF deficient cells found in long septate chains.84 However, loss of lytE, lytF, and cwlS completely prevents separation and causes cells to chain.85 Interestingly, cell separation can be restored in this triple mutant strain by expression of various PG hydrolases, including non-D,L-endopeptidases and non-NlpC/P60 hydrolases, that are targeted to the septum by fusion with the PG-binding domain of LytE.69 This shows that unlike elongation, which specifically requires D,L-endopeptidase activity, separation can be facilitated by any type of PG hydrolysis.
Several other NlpC/P60 hydrolases from Gram-positive bacteria have also been assigned specific roles in growth and division, such as Lactobacillus plantarum LytA (elongation and septum positioning) and LytB (septum maturation);101 Lactobacillus rhamnosus Msp1 (cell separation);102 Clostriodiodes difficile CwlA (cell separation);94 and Enterococcus faceium SagA (cell separation).13 The Streptococcus pneumoniae CHAP protein PcsB has also been implicated in cell separation.150,151 Notably, SagA and PcsB have the same domain architecture as B. subtilis CwlO, containing an N-terminal coiled-coil domain and a C-terminal NlpC/P60 or CHAP domain. The coiled-coil domain has been shown to facilitate interaction with FtsX for CwlO and PcsB.152 Although the function of the SagA coiled-coil domain remains unclear, E. faecium strains encode FtsEX homologs that warrant further investigation with SagA. Interestingly, SagA, like either CwlO or LytE, has also been reported to be essential.96
Much less is known about the specific functions of NlpC/P60 hydrolases in Gram-negative bacteria (Figure 3B). The Gram-negative model organism Escherichia coli contains at least four NlpC/P60 proteins: MepS (Spr), MepH (YdhO), YafL, and NlpC.21 Knocking out all four of these genes simultaneously inhibits growth on Nutrient Agar but not on standard Luria Broth (LB) or minimal medium. Of the four NlpC/P60 proteins in E. coli, only MepS has been studied thoroughly. MepS is a D,D-endopeptidase that promotes cell elongation and indirectly inhibits division.30 Simultaneous knockout of MepS along with another non-NlpC/P60 PG hydrolase, MepM, is synthetically lethal in LB but not in minimal medium, indicating that the cell’s growth program in standard conditions requires these endopeptidases in a way that growth in nutrient-poor conditions does not. Differential growth under these two conditions was found to be regulated by amino acid availability; however, the amino acids that regulate each enzyme are distinct.24
Five NlpC/P60 proteins have been identified in the model mycobacterium Mycobacterium tuberculosis: RipA, RipB, RipC, RipD, and Rv0024 (Figure 3C).153 The first four are named “resuscitation-promoting factor (rpf)-interacting proteins” due to their complexation with proteins that were originally thought to enable latent M. tuberculosis infections to become virulent.131 RipA localizes to the septum of dividing cells and is involved in cell separation. ripA can be deleted as long as ripB (which is immediately downstream of ripA in the same operon) is present, indicating that there is some degree of functional redundancy between the two.130 However, the phenotypes of ripA and ripB knockout strains are different. The ripA knockout strain produces elongated cells that have more septa than wild-type cells, whereas the ripB knockout strain does not produce either of these phenotypes, indicating that RipA is more influential on overall cell morphology. RipC has been implicated in growth but not cell septation. Mutation of ripC results in a significant decrease in growth as measured by optical density, which is partially reversed by complementation. However, the mutant cells are still capable of forming normal septa, as observed by electron microscopy. RipA and RipC both contain an N-terminal coiled-coil domain. For RipC, this domain allows for binding to FtsX,145 similar to the coiled-coil domains of B. subtilis CwlO and S. pneumoniae PcsB. Conversely, this domain is cleaved off of RipA during activation, so its function in protein activity or regulation remains unknown. The importance of RipD and Rv0024 in mycobacterium biology are less well studied. The NlpC/P60 domain of RipD possesses mutations in the predicted catalytic triad and has been confirmed to be inactive against PG.147 Rv0024 lacks a canonical signal sequence,153 so it is unclear if and how this enzyme is directed to the extracytosolic environment. Nevertheless, ectopic expression of Rv0024 in Mycobacterium smegmatis results in increased biofilm formation and antibiotic tolerance due to its effects on the hydrophobicity of the cell surface,148 suggesting that the protein is exported through an unknown mechanism.
Additional roles of bacterial NlpC/P60 proteins
Competition and cellular defense –
While PG remodeling is crucial for survival and homeostasis, the ability to cleave PG of other species presents an opportunity for niche competition. Several Gram-negative bacterial species use Type VI Secretion Systems (T6SS) to deploy NlpC/P60 hydrolases that degrade the peptidoglycan of competing bacteria (Figure 4A). For example, Pseudomonas aeruginosa injects the Tse1 hydrolase into its competitors’ periplasm. Because this mechanism can also deliver Tse1 to other P. aeruginosa cells, an immunity protein called Tsi1 is employed for self-protection.32–34 In Salmonella enterica subspecies Typhimurium and Enterobacter cloacae, the secreted hydrolase Tae4 and its immunity protein Tai4 serve a similar role, though the mechanism by which the hydrolase is inhibited by its immunity protein is distinct from that of Tse1/Tsi1.20,42 Vibrio cholerae employs the secreted hydrolase TseH for competition with bacteria that occupy similar ecological niches.47,48 The cell killing activity of TseH was inhibited by the putative V. cholerae immunity protein TsiH when over-expressed in E. coli.47 Interestingly, V. cholerae with all other anti-bacterial effectors inactivated except TseH was unable to kill a ΔtsiH V. cholerae strain.48 Further assays demonstrated that the WigKR envelope stress response element in V. cholerae can also protect against the hydrolase in the absence of TsiH, providing multiple routes to prevent T6SS-mediated cell killing. In addition to these T6SS-secreted hydrolases, the Gram-negative bacterium Photobacterium damselae subspecies piscicida has been shown to use a Type II Secretion System (T2SS) to inject the protein PnpA into the extracellular environment. PnpA does not hydrolyze P. damselae peptidoglycan, but it does degrade PG of two competing bacteria, Vibrio anguillarum and Vibrio vulnificus.39 However, how PnpA accesses the cell wall of these Gram-negative organisms remains unclear and requires further investigation. These examples highlight how NlpC/P60-mediated competition can be accomplished through distinct strategies using either differential substrate specificities or effector/immunity pairs.
Figure 4. Functions of prokaryotic NlpC/P60 proteins in survival and infection.
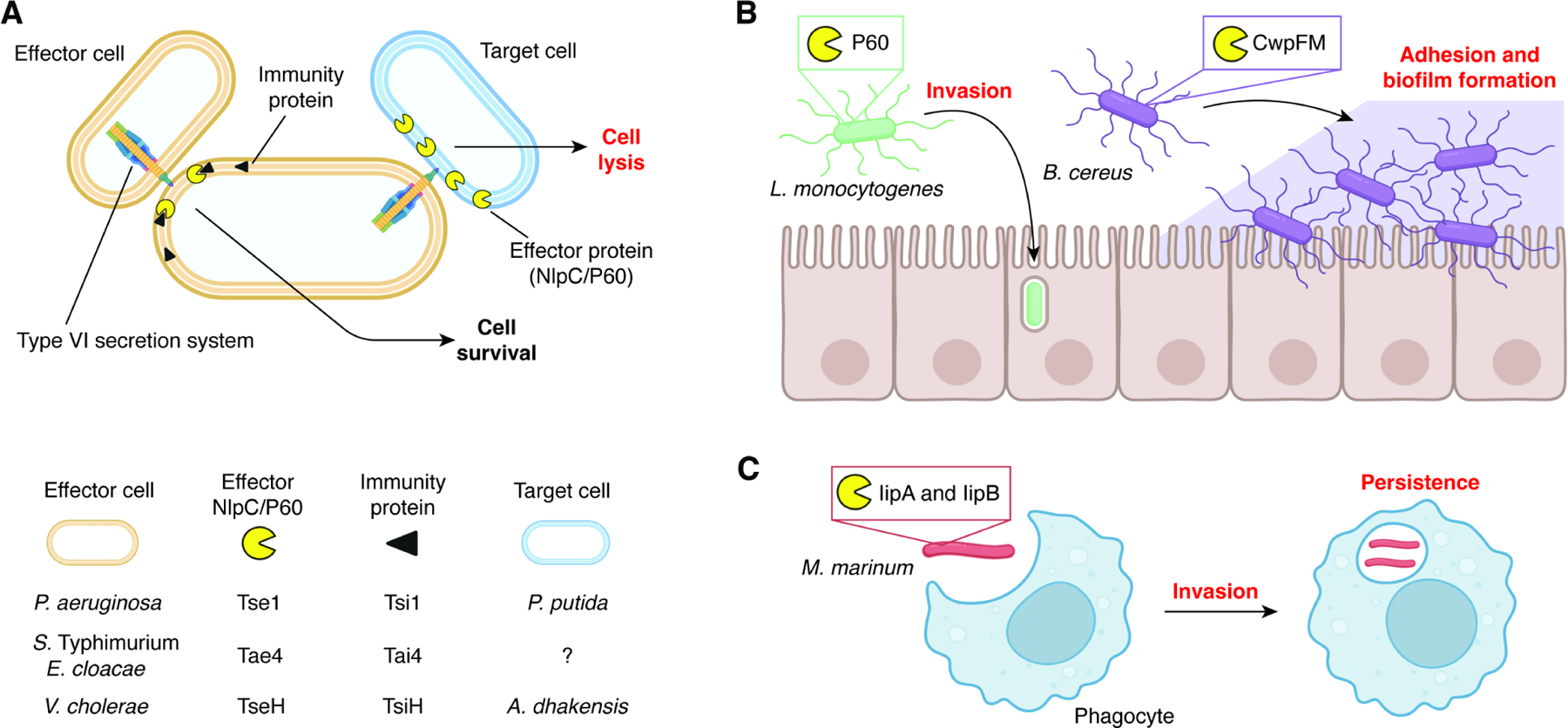
(A) Multiple bacterial species use NlpC/P60 proteins as secretion system effectors to induce cell lysis of other competing microbes.
(B) Expression of bacterial NlpC/P60s enable host colonization at and within cells at the epithelial interface in the gut.
(C) NlpC/P60s produced by mycobacteria improve both invasion and persistence during infection in phagocytes.
Virulence –
Several NlpC/P60 hydrolases have been implicated in virulence (Figure 4B). The founding member of this protein superfamily, P60 from the Gram-positive bacterium Listeria monocytogenes, was originally identified as an invasion-related protein that aided in inducing phagocytosis by host cells,109 a key step in the organism’s pathogenic life cycle. This biological activity was subsequently linked to the PG hydrolase activity of P60.111 A second protein in this organism, P45, was also shown to have a similar role and function in L. monocytogenes virulence.108
The Bacillus cereus and Bacillus thurengiensis protein CwpFM was originally identified as an enterotoxin named EntFM and was shown to be highly prevalent in outbreak-causing strains.54,55 Subsequent analysis showed that this protein is an NlpC/P60 peptidase52,53 that modulates cell motility, morphology, adherence to epithelial cells, biofilm formation, and macrophage vacuolization. Mortality of insect larvae after infection with B. thurengiensis was significantly reduced in a cwpFM mutant compared to wild-type.52
In Corynebacterium diptheriae, DIP1281 has been shown to be critical for adhesion and invasion into epithelial cells.119 dip1281 mutant cells do not separate properly and instead form chains; however, this phenotype is not related to PG hydrolysis but rather to rearrangements of the protein layer on the cell surface.118
The zebrafish pathogen Mycobacterium marinum expresses IipA and IipB, orthologs of M. tuberculosis RipA and RipB, respectively.129 These proteins have been shown to be essential for bacterial invasion and intracellular persistence (iip) within macrophages (Figure 4C). Although zebrafish infection with wild-type M. marinum showed complete lethality, a mutant strain with a disrupted iip locus was completely tolerated. Interestingly, the authors in that study found that complementation with M. tuberculosis RipA (Rv1477) completely restored the lethal phenotype, while complementation with RipB (Rv1478) did not.129 Similarly, the lethality of M. tuberculosis in mouse infection models was also attenuated by ripC mutation, and this activity was partially restored by complementation,144 suggesting NlpC/P60 hydrolases contribute to mycobacteria virulence in different hosts.
Sporulation and germination –
Certain NlpC/P60 enzymes aid in sporulation and germination. Sporulation is a complex process in which certain bacterial species (usually Gram-positive) respond to nutrient deprivation by differentiating into a resistant endospore within a so-called mother cell. Lysis of the mother cell releases the mature spore into the environment. When nutrients improved conditions are detected, the spore undergoes germination to restore the cell to a functional state. As expected, these processes are known to require extensive PG remodeling.154–158 For example, the Streptomyces coelicolor NlpC/P60 protein SwlA has been shown to be involved in both sporulation and germination. A swlA mutant produces defective spores that have abnormal morphologies and are hypersentisive to heat.116 Importantly, single mutations of three other, non-NlpC/P60 hydrolase genes (rpfA, swlB, and swlC) also confer these phenotypes, suggesting that SwlA is just one of several enzymes responsible for proper peptidoglycan remodeling during spore formation in this organism. Spores from the swlA and rpfA mutants are also slower to germinate than wild-type spores, although they are capable of full resuscitation.116
The Bacillus anthracis protein BAS1812 was originally identified as BA1952 from a reverse-vaccinology screen of seropositive antigens51 and was later shown to have activity in sporulation and germination. Identification and characterization of proteins related to B. anthracis sporulation are particularly important, as the spores of this organism are the cause of anthrax disease. Like mutation of swlA in S. coelicolor, deletion of BAS1812 in B. anthracis resulted in impaired endospore formation and delayed germination.49 Both of these phenotypes were partially reversed by complementation with a second copy of BAS1812. However, the BAS1812 deletion also conferred detergent sensitivity that the S. coelicolor swlA mutant did not exhibit. BAS1812 is nearly identical to the previously discussed CwpFM protein of the spore-forming organism B. cereus, suggesting that this protein may play a sporulation/germination role in that organism as well. In Bacillus sphaericus, the protein DPP VI has been identified as an NlpC/P60 hydrolase and is highly upregulated during sporulation.57 However, the direct roles of CwpFM and DPP VI in endospore formation have not been demonstrated, indicating the need for further examination of NlpC/P60 proteins in these processes.
The mechanisms underlying the sporulation and germination-related phenotypes of the mutants listed above are not known and will require further study to elucidate. However, there is one sporulation-related NlpC/P60 enzyme in B. subtilis, the transglutaminase Tgl, that has been studied mechanistically. Instead of acting as a PG hydrolase, Tgl covalently crosslinks proteins that make up the spore coat.91–93 Thus, this protein is one of several NlpC/P60 proteins that serve non-hydrolytic functions.
Other non-PG hydrolytic functions –
In some cases, NlpC/P60-containing proteins have evolved to perform additional cellular functions that are unrelated to PG hydrolysis. Some examples are the P. aeruginosa MagC protein, which serves to anchor the Mag complex to PG, but has not been reported to hydrolyze PG.31 Additionally, the Staphylococcus aureus protein LnsA has been suggested to mediate acylation of lipoproteins113 and the B. subtilis protein PgdS has been shown to degrade poly-γ-glutamic acid.88,89
Regulation of bacterial NlpC/P60 proteins
In general, PG hydrolysis is a highly regulated process.4,5 Overactive PG hydrolysis can compromise the integrity of the cell wall and leave the cell vulnerable to physical stresses, whereas hypoactive PG hydrolysis can prevent cell growth and division. Moreover, the proper amount of PG hydrolysis changes over the lifetime of a cell based on factors such as nutrient availability, temperature, pH, and antibiotic stress. Cells must be able to respond to these types of environmental cues in order to ensure that the balance between peptidoglycan synthesis and hydrolysis is maintained at all times.
The mechanisms by which NlpC/P60 hydrolases are regulated are diverse (Figure 5). At the transcriptional level, two-component systems and alternative sigma factors play key roles. For example, the B. subtilis WalRK two-component system (previously known as YycFG) controls expression of cwlO and lytE (Figure 5A).62 The sensor histidine kinase WalK senses the cleavage products produced by CwlO and LytE67 and relays this information to the response regulator WalR, which in turn binds to the cwlO and lytE promoters and modulates their expression. When the cleavage products are below the proper concentration, WalRK upregulates transcription of cwlO and lytE and vice-versa. The WalRK regulon also includes iseA, which encodes an inhibitor of CwlO and LytE.159 When the cleavage product concentration is too high, iseA transcription is upregulated and vice-versa. Another two-component system that regulates an NlpC/P60 enzyme is the PhoPQ system in S. Typhimurium. This system regulates many genes involved in virulence 160 including the ecgA, which encodes an NlpC/P60 family member.40 Here, the sensor histidine kinase PhoQ senses various extracellular signals, such as pH, osmolarity, and magnesium ion concentration and relays the signal to the response regulator PhoP, which then modulates expression of ecgA. In addition to transcription factors like WalR and PhoP, the use of alternative sigma factors allows a cell to selectively transcribe certain genes based on the cell’s progression through the cell cycle. In B. subtilis, lytE is trancsribed by RNA polymerase with σI,77 while lytF is transcribed by RNA polymerase with σD.81 At least one NlpC/P60 hydrolase is regulated at the post-transcriptional level as well (Figure 5B). The RNA transcript of cwlO is extremely unstable,63 and CwlO protein also has a very short half-life (~5 min),60 which allows for tighter control of CwlO regulation through the need for constant regeneration of cwlO mRNA. Bioinformatic analysis has predicted that some NlpC/P60 enzymes may be regulated by riboswitches;161 however, experiments will be needed to confirm this and to determine the mechanism of action.
Figure 5. Diverse mechanisms of NlpC/P60 regulation in prokaryotes.
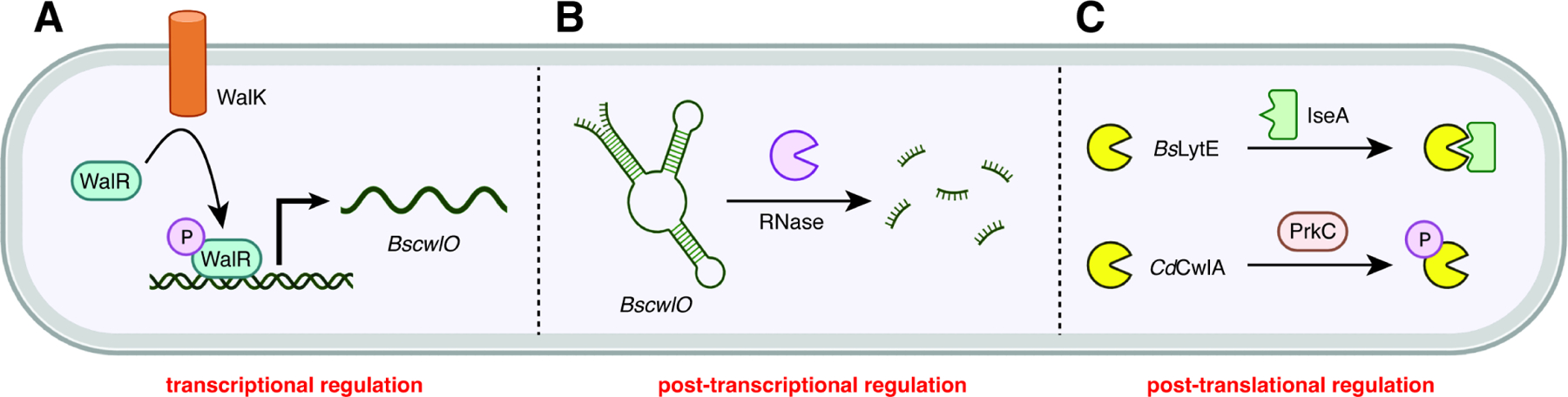
(A) The two-component WalRK signal transduction system in B. subtilis controls expression of the NlpC/P60 cwlO gene.
(B) The cwlO leader sequence forms a secondary structure that is cleaved by RNase Y, leading to degradation of transcript.
(C) Post-translational regulation can occur through protein-protein interactions including immunity proteins and posttranslational modifications like phosphorylation.
The timing and amount of NlpC/P60 hydrolase production are key to their ability to perform their functions properly. This complex form of transcriptional and translational regulation is highlighted by work showing that the synthetic lethality of cwlO and lytE in B. subtilis64 can be overcome by lytF expressed from the cwlO promoter (PcwlO-lytF).68 However, the PcwlO-cwlS construct is unable to rescue the cwlO and lytE double knockout strain, despite CwlS being very similar to LytF.68 Although the exact molecular mechanism underlying this partial redundancy between NlpC/P60 proteins is unknown, this example underscores the importance of examining both protein activity and regulation to determine their roles in bacterial physiology.
NlpC/P60 enzymes are also regulated at the post-translational level (Figure 5C). Protein-protein interactions such as those described above for hydrolases and actin homologs, FtsEX, inhibitors like the immunity proteins for T6SS-secreted hydrolases or IseA, and putative tuberculoidal resuscitation factors, constitute one mechanism of post-translational regulation. A second mechanism is post-translational modification. For example, C. difficile CwlA is differentially localized based on whether it is phosphorylated. Unphosphorylated CwlA is exported to the extracytosolic environment where it can hydrolyze PG, whereas phosphorylated CwlA is retained in the cytoplasm and is thus unable to hydrolyze PG.94 The L. rhamnosus GG protein Msp1 undergoes O-glycosylation at two serine residues. This modification allows for proper localization of the protein and confers protection against proteases, though it does not directly affect hydrolase activity.103 Two hydrolases in M. tuberculosis, RipA and RipB, are post-translationally modified by cleavage of a pro-domain. Only after this cleavage do the NlpC/P60 domains of these enzymes become active. Finally, some NlpC/P60 hydrolases are degraded through regulated proteolysis. E. coli MepS, for example, is degraded by the NlpI/Prc system,22 while P. aeruginosa PA1198 is degraded by the LbcA/CtpA system.35–38
Functions of bacterial NlpC/P60 domains in eukaryotic biology
NlpC/P60 enzymes can also play a more complex role in communication across kingdoms through the generation of immune active molecules (Figure 6). The hydrolysis of PG via NlpC/P60 domains yields small muramyl peptides.162 These fragments can in turn be sensed by eukaryotic pattern recognition receptors to elicit nonspecific immune responses.163 This central pillar of innate immunity allows animals to rapidly respond to a variety of microbial threats. PG fragments are detected by the nucleotide-binding domain, leucine-rich containing (NLR) proteins, NOD1 and NOD2.164–166 NOD1 recognizes fragments containing the dipeptide isoglutamate-meso-DAP (iE-DAP), which is present primarily in Gram-negative bacteria. Conversely, NOD2 binds to fragments containing the glycopeptide MurNAc-alanine-isoglutamate/isoglutamine or muramyl dipeptide (MDP). This structure is present in both Gram-negative and Gram-positive bacteria and can be selectively generated from an intact stem peptide through the D,L-endopeptidase activity of NlpC/P60 enzymes. Therefore, NlpC/P60 proteins may uniquely be able to activate host immunity via their PG remodeling capacity.
Figure 6. Functions of prokaryotic NlpC/P60 proteins in host-microbial interactions.
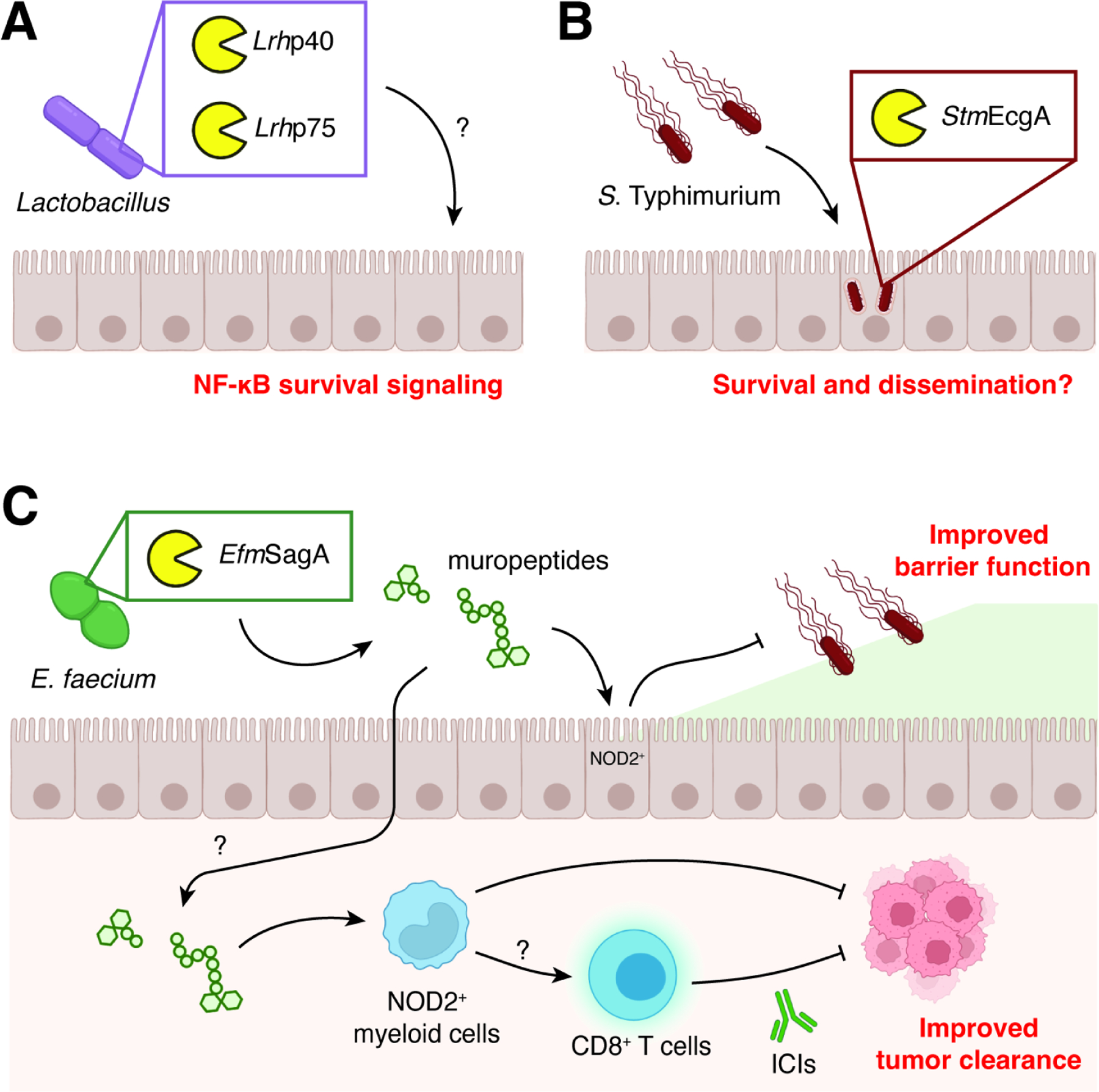
(A) Lactobacillus species produce secreted PG degrading enzymes that induce host immune signaling through NF-κB.
(B) S. Typhimurium selectively expresses EcgA while residing in intracellular vesicles, which increases overall survival and dissemination.
(C) Enterococcus species like E. faecium secrete SagA, which can stimulate NOD2-dependent responses in the host during infection and cancer immunotherapy through the production of muropeptides.
The activity of probiotic bacteria may be mediated by NlpC/P60 catalytic activity. Species of Lactobacillus including L. rhamnosus and L. casei express and secrete two proteins (p40 and p75) that contain CHAP and NlpC/P60 domains, respectively (Figure 6A).99,167 Both of these proteins possess hydrolytic activity against PG, and the p75 protein is specific for the amide bond between isoglutamine and lysine as expected for other NlpC/P60 D,L-endopeptidases.102 Interestingly, the presence of these two hydrolytic enzymes also had significant effects on intestinal epithelial cell (IEC) biology. Treatment of IECs with conditioned medium or purified proteins activated survival signaling via Akt, inhibited cytokine-induced apoptosis, and improved cell proliferation. Nevertheless, it remains unknown whether the probiotic effects of these enzymes were due to direct generation of immune active muramyl peptides or other mechanisms, such as their ability to bind to mammalian extracellular matrix proteins.
PG remodeling by NlpC/P60s may be an adaptive advantage used by infectious bacteria. For example, Salmonella enterica serovar Typhimurium selectively expresses the NlpC/P60 protein EcgA in nutrient-limiting conditions such as intracellular vesicles during infection of mammalian cells (Figure 6B).40,41 In biochemical assays, EcgA was shown to have specialized D,L-endopeptidase activity only on uncrosslinked PG fragments. The exact function of the specific modification remains unknown; however, in vivo experiments revealed that a ΔecgA mutant strain showed a significant competitive disadvantage using an infection model with intraperitoneal injection of Salmonella but not with oral infection. Together, these data suggest that NlpC/P60 enzymes may play a role in pathogen colonization of and dissemination within the host.
Beyond its bacteria-intrinsic activities, the E. faecium enzyme SagA has profound effects on host physiology (Figure 6C).97,168 Colonization of both C. elegans and mice with SagA-expressing E. faecium led to protection against infection by S. Typhimurium. The pro-survival activity of this NlpC/P60 enzyme was confirmed by direct administration of the enzyme itself, as well as colonization with other, nonprotective species, E. faecalis and L. plantarum, that had been engineered to stably express SagA. Using a mutant variant of SagA where its putative catalytic residues were converted to alanine, catalytically dead SagA was no longer able to rescue worms from Salmonella pathogenesis. In mice, colonization with SagA-expressing bacteria showed an increase in the expression of the barrier protein mucin 2 as well as antimicrobial peptides cryptdin 2 and REGIIIɣ. Survival upon Salmonella challenge required the host PG receptor NOD2 as well as epithelial cell-specific MYD88, a signaling adaptor for Toll-like receptors. Finally, the pro-survival activity of SagA was found to protect against Clostridium difficile pathogenesis, highlighting the broad host effects of SagA against enteric infections.
The genus Enterococcus has also been linked to a variety of systemic immune disease phenotypes, including increased susceptibility to graft-versus-host disease169 and increased efficacy of cancer chemo- and immunotherapies.170,171 In addition to E. faecium, other species such as E. durans, E. hirae, and E. mundtii were recovered from cancer patients who responded to anti-PD-1 immune checkpoint inhibitor therapy.172,173 Monocolonization with these bacteria using a murine model of melanoma confirmed the causal link between Enterococcus and immunotherapy response.98 Bioinformatic and biochemical analyses revealed that the active Enterococcus species expressed and secreted orthologs of E. faecium SagA. In vitro enzyme assays showed that the SagA orthologs all possessed D,L-endopeptidase activity with no discernable differences in substrate specificity. Importantly, colonization with L. lactis strains engineered to express wild-type or catalytically dead SagA demonstrated that the PG remodeling activity of SagA was required for anti-tumor activity. Responsiveness to immunotherapy was lost in animals lacking the NOD2 receptor, indicating that PG sensing was necessary for improved drug efficacy. Finally, colonization with SagA-expressing bacteria led to increased immune cell infiltration into tumors, with higher levels of effector and tumor antigen-specific CD8 T cells, providing a cellular basis for the observed anti-tumor activity of SagA.
More broadly, PG remodeling by other NlpC/p60 enzymes from gut microbiota with D,L-endopeptidase activity may function as a general mechanism to modulate host immunity. Recent evidence has found that overall D,L-endopeptidase activity in fecal samples was anti-correlated with Crohn’s disease in humans and led to predisposition to colitis in mice upon fecal microbiota transplantation.105 Oral teatment of animals with either the purified NlpC/p60 protein UC118 or the UC118-expressing bacterium Lactobacillus salivarius ameliorated DSS-induced colitis. However, these beneficial effects were not restored in mice treated with a catalytically dead mutant of UC118, suggesting that the phenotype was driven by the catalytic activity of UC118. Screening of almost 50 commercially available bacterial species found that only a subset were able to strongly activate NOD2, and importantly, all NOD2-activating isolates expressed D,L-endopeptidases. Thus, NlpC/p60 enzymes with this specific activity may serve as both a prognostic biomarker and therapeutic treatment for colitis in NOD2-sufficient patients.
Functions of eukaryotic NlpC/P60 domains
Although NlpC/P60 domains are most frequently found in prokaryotes, rare instances of NlpC/P60 proteins are found within Eukarya.7 Their evolutionary origin(s) remains unclear; however, it has been suggested that some of these enzymes were acquired by horizontal gene transfer from bacteria. For example, the human parasite responsible for trichomoniasis, Trichomonas vaginalis, possesses at least two NlpC/P60 domain-containing proteins (TVAG_457240 and TVAG_209010) with hydrolytic activity against bacterial PG (Figure 7A).174 Like bacterial enzymes, the proteins are exported to the cell surface of the pathogen, and their catalytic activity required the active site cysteine residue. Interestingly, the NlpC/P60 enzymes were active against intact E. coli PG, and co-incubation of the pathogen over-expressing one NlpC/P60 enzyme with E. coli led to bacterial lysis. Since NlpC/P60 enzymes do not typically penetrate the outer membrane of Gram-negative bacteria, further investigation of how these Trichomonas vaginalis NlpC/P60 domain-containing proteins gain access to the PG layer will be important. Expression levels of the two enzymes was also responsive to environmental conditions. If these genes were acquired by horizontal gene transfer, then their genomic incorporation was followed by the evolution of functional secretion signals and regulated expression to best utilize the adaptive advantage provided by NlpC/p60 activity during mucosal colonization.
Figure 7. Functions of eukaryotic NlpC/P60s.
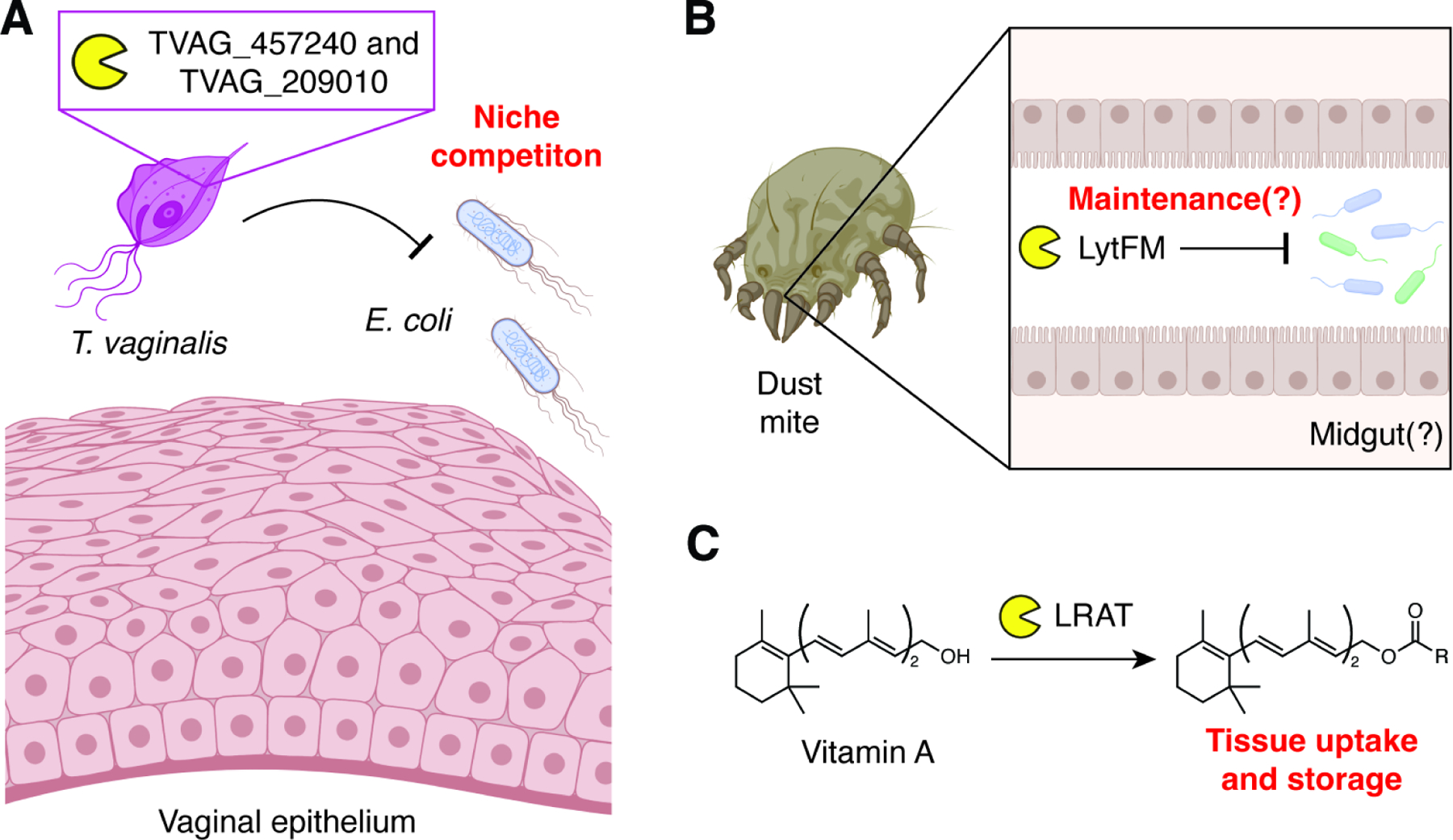
(A) The protozoan T. vaginalis expresses two NlpC/P60s that may improve niche competition at the vaginal epithelium.
(B) Dust mites express LytFM, which may be involved in maintenance of its midgut microbiota.
(C) Mammalian LRAT-like NlpC/P60s like LRAT itself exhibit acyltransferase activities on non-peptidoglycan substrates.
Another potential instance of horizontal gene transfer occurred in the evolutionary ancestors of the house dust mite, genus Dermatophagoides. First discovered in the clinically relevant species D. pteronyssinus and D. farinae, multiple species of dust mite express and secrete a 14 kDa protein that shows homology to bacterial NlpC/P60 enzymes (Figure 7B).175 Termed lytFM, this protein is capable of lysing Gram-positive bacteria, and further biochemical characterization by zymography indicated that the protein is capable of hydrolyzing PG. The enzyme is expressed under the control of a bacterial promoter, suggesting that lytFM may derive from a prokaryotic endosymbiont. The presence of highly similar NlpC/P60-containing proteins in dust mite-associated Bacillus and Staphylococcus species further support the endosymbiont gene transfer hypothesis.176 Although the bacteriocidal activity of lytFM may help the dust mite to maintain its associated microbiota, the biological importance of this eukaryotic NlpC/P60 protein has not been fully elucidated.
Other eukaryotic NlpC/P60 proteins are often members of the LRAT-like group (Figure 7C). LRAT-like NlpC/P60 domains do not hydrolyze PG; instead, they often act as phospholipases/acyltransferases (PLA/ATs).177 For example, the mammalian protein and founding member of this group, lecithin retinol acyltransferase (LRAT), catalyzes the esterification of Vitamin A (all-trans-retinol) via acyl transfer from phosphatidylcholine, which enables retention of Vitamin A in peripheral tissues.178 Mammals also express H-REV107, which functions as a tumor suppressor through negative regulation of the proto-oncogene H-RAS.179 Similar to LRAT, this protein acts as a PLA/AT using phopsholipid substrates. H-REV107 inhibits H-RAS palmitoylation and its downstream signaling, which can be reversed via chemical inhibition of H-REV107 acyltransferase activity. However, whether H-REV107 directly modifies H-RAS or an associated palmitoyl transferase to mediate this activity remains unclear. LRAT-like enzymes are also found in lower organisms, including the protein EGL-26 from the nematode Caenorhabditis elegans.180 Although the molecular function of EGL-26 catalysis is unknown, loss of this protein or mutagenesis of its putative active site residues led to atypical vulval cell morphogensis and decreased egg laying, highlighting the broad biological functions of eukaryotic LRAT-like NlpC/p60 proteins.
Outlook and future directions
The effects of microbial biology on their resident hosts will remain a key concern for human health in the foreseeable future. The battlefronts of host-microbial interactions are quite diverse, from the ongoing emergence of antibiotic-resistant microbial strains181 to the intimate roles of microbes on immune development and homeostasis.182 Nevertheless, these myriad effects are often difficult to target due to a lack of information regarding exact molecular connections between microbial activity and host outcomes. The biosynthesis, remodeling, and degradation of PG is an essential function of virtually all bacteria, and these crucial and bacterial-specific processes have been targeted throughout the history of antibiotic development from its birth with penicillin-based beta lactams to today’s glycopeptide derivatives such as vancomycin and corbomycin.183 Nevertheless, much remains unknown about the molecular intricacies of PG turnover, including the roles of many seemingly redundant remodeling proteins.
As summarized in this review, NlpC/P60 enzymes are widespread components of the PG remodeling machinery that are often essential for the survival and growth of many bacterial species. Thus, we propose that these near ubiquitous cysteine proteases may be a functional target for next-generation antibiotics. Importantly, the development of enzyme-specific inhibitors may allow for precise control of individual species and strains within complex communities, avoiding broad changes to microbiota communities often associated with wide-spectrum antibiotics. Moreover, the presence of catalytic cysteine residues within the active sites of these enzymes opens the door for the development of covalent-based inhibitors, which have recently seen widespread success in the targeting of mammalian proteins.184 Alternatively, compounds that activate endogenous NlpC/P60 activity may serve as bacterial lysis agents.
Finally, the intrinsic endopeptidase activity and substrate specificity of NlpC/P60 enzymes may also offer avenues to direct host immune modulation. The ability of NlpC/P60 proteins like SagA to generate muropeptides may be exploited to activate innate immune pathways using engineered probiotics.98 Rather than relying on the black box of existing, poorly defined strains, the development of live biotherapeutic products with known enzymatic activity and small molecule output may be a feasible strategy for next-generation anti-infectives and immunotherapies. Together, these studies and potential applications highlight the necessity for a better molecular-level understanding of host-microbial interactions through the further study of PG remodeling NlpC/P60 enzymes.
SIGNIFICANCE
Microbes residing upon or within larger organisms can be beneficial or antagonistic for host biology. Understanding the molecular determinants of these interactions may provide new targets to control microbial activity and their effects on the host. This review summarizes how the broad family of NlpC/P60 enzymes play critical roles in bacterial cell-intrinsic and -extrinsic phenomena. Multiple studies have found that the ability to degrade peptidoglycan by NlpC/P60 proteins is essential for bacterial survival, growth, and division. The generation of fragmented peptidoglycan metabolites by these enzymes can also initiate host immune signaling to improve intestinal barrier function and elicit adaptive immune responses. Therefore, a better fundamental understanding of NlpC/P60 structure, activity, and function may provide targets to control prokaryotic growth via next-generation anti-infectives and new routes to improve host immunity through defined, biotherapeutic adjuvants.
Highlights.
NlpC/P60 hydrolases degrade bacterial peptidoglycan and generate muropeptides.
These enzymes are important for proper bacterial growth and cell division.
NlpC/P60 proteins influence bacterial-host interactions including immune stimulation.
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute (5R01CA245292, H.C.H.) and National Institute of Allergy and Infectious Diseases (5R21AI156674-02, H.C.H.). We also acknowledge support from the Hope Funds for Cancer Research (HCFR-19-03-02, M.E.G.), Melanoma Research Foundation (career development award, M.E.G.), and the National Institutes of Health (5T32AI070084-13, J.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
M.E.G. and H.C.H. have filed a patent application (PCT/US2020/019038) for the commercial use of SagA-expressing bacteria to improve checkpoint blockade immunotherapy.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Vollmer W, Blanot D, and de Pedro MA (2008). Peptidoglycan structure and architecture. FEMS Microbiol Rev 32, 149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Lovering AL, Safadi SS, and Strynadka NC (2012). Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem 81, 451–478. 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 3.Rohs PDA, and Bernhardt TG (2021). Growth and division of the peptidoglycan matrix. Annu Rev Microbiol 75, 315–336. 10.1146/annurev-micro-020518-120056. [DOI] [PubMed] [Google Scholar]
- 4.Typas A, Banzhaf M, Gross CA, and Vollmer W (2011). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10, 123–136. 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan AJF, Errington J, and Vollmer W (2020). Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 18, 446–460. 10.1038/s41579-020-0366-3. [DOI] [PubMed] [Google Scholar]
- 6.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, and Desvaux M (2019). Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10, 331. 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anantharaman V, and Aravind L (2003). Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol 4, R11. 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Q, Mengin-Lecreulx D, Patin D, Grant JC, Chiu HJ, Jaroszewski L, Knuth MW, Godzik A, Lesley SA, Elsliger MA, et al. (2014). Structure-guided functional characterization of DUF1460 reveals a highly specific NlpC/P60 amidase family. Structure 22, 1799–1809. 10.1016/j.str.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aramini JM, Rossi P, Huang YJ, Zhao L, Jiang M, Maglaqui M, Xiao R, Locke J, Nair R, Rost B, et al. (2008). Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry 47, 9715–9717. 10.1021/bi8010779. [DOI] [PubMed] [Google Scholar]
- 10.Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Chiu M, et al. (2010). Structure of the gamma-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-Ala-gamma-D-Glu: insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr Sect F Struct Biol Cryst Commun 66, 1354–1364. 10.1107/S1744309110021214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Q, Chiu HJ, Farr CL, Jaroszewski L, Knuth MW, Miller MD, Lesley SA, Godzik A, Elsliger MA, Deacon AM, and Wilson IA (2014). Structures of a bifunctional cell wall hydrolase CwlT containing a novel bacterial lysozyme and an NlpC/P60 DL-endopeptidase. J Mol Biol 426, 169–184. 10.1016/j.jmb.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim B, Wang YC, Hespen CW, Espinosa J, Salje J, Rangan KJ, Oren DA, Kang JY, Pedicord VA, and Hang HC (2019). Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. Elife 8. 10.7554/eLife.45343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinosa J, Lin TY, Estrella Y, Kim B, Molina H, and Hang HC (2020). Enterococcus NlpC/p60 Peptidoglycan Hydrolase SagA Localizes to Sites of Cell Division and Requires Only a Catalytic Dyad for Protease Activity. Biochemistry 59, 4470–4480. 10.1021/acs.biochem.0c00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekiya H, Tamai E, Kawasaki J, Murakami K, and Kamitori S (2021). Structural and biochemical characterizations of the novel autolysin Acd24020 from Clostridioides difficile and its full-function catalytic domain as a lytic enzyme. Mol Microbiol 115, 684–698. 10.1111/mmi.14636. [DOI] [PubMed] [Google Scholar]
- 15.Xu Q, Sudek S, McMullan D, Miller MD, Geierstanger B, Jones DH, Krishna SS, Spraggon G, Bursalay B, Abdubek P, et al. (2009). Structural basis of murein peptide specificity of a gamma-D-glutamyl-l-diamino acid endopeptidase. Structure 17, 303–313. 10.1016/j.str.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong JE, Alsarraf HM, Kaspersen JD, Pedersen JS, Stougaard J, Thirup S, and Blaise M (2014). Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J 281, 1196–1208. 10.1111/febs.12698. [DOI] [PubMed] [Google Scholar]
- 17.Ruggiero A, Marasco D, Squeglia F, Soldini S, Pedone E, Pedone C, and Berisio R (2010). Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure 18, 1184–1190. 10.1016/j.str.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A, and Rawlings ND (2003). The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci 28, 234–237. 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q, Mengin-Lecreulx D, Liu XW, Patin D, Farr CL, Grant JC, Chiu HJ, Jaroszewski L, Knuth MW, Godzik A, et al. (2015). Insights into Substrate Specificity of NlpC/P60 Cell Wall Hydrolases Containing Bacterial SH3 Domains. mBio 6, e02327–02314. 10.1128/mBio.02327-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Zhang H, Gao ZQ, Wang WJ, Liu GF, Xu JH, Su XD, and Dong YH (2013). Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. J Biol Chem 288, 5928–5939. 10.1074/jbc.M112.434357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh SK, SaiSree L, Amrutha RN, and Reddy M (2012). Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86, 1036–1051. 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 22.Singh SK, Parveen S, SaiSree L, and Reddy M (2015). Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci U S A 112, 10956–10961. 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang WC, Hashimoto M, Shih YL, Wu CC, Lee MF, Chen YL, Wu JJ, Wang MC, Lin WH, Hong MY, and Teng CH (2020). Peptidoglycan Endopeptidase Spr of Uropathogenic Escherichia coli Contributes to Kidney Infections and Competitive Fitness During Bladder Colonization. Front Microbiol 11, 586214. 10.3389/fmicb.2020.586214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YJ, Choi BJ, Park SH, Lee HB, Son JE, Choi U, Chi WJ, and Lee CR (2021). Distinct Amino Acid Availability-Dependent Regulatory Mechanisms of MepS and MepM Levels in Escherichia coli. Front Microbiol 12, 677739. 10.3389/fmicb.2021.677739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su MY, Som N, Wu CY, Su SC, Kuo YT, Ke LC, Ho MR, Tzeng SR, Teng CH, Mengin-Lecreulx D, et al. (2017). Structural basis of adaptor-mediated protein degradation by the tail-specific PDZ-protease Prc. Nat Commun 8, 1516. 10.1038/s41467-017-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voedts H, Dorchene D, Lodge A, Vollmer W, Arthur M, and Hugonnet JE (2021). Role of endopeptidases in peptidoglycan synthesis mediated by alternative cross-linking enzymes in Escherichia coli. EMBO J 40, e108126. 10.15252/embj.2021108126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai GC, Cho H, and Bernhardt TG (2017). The mecillinam resistome reveals a role for peptidoglycan endopeptidases in stimulating cell wall synthesis in Escherichia coli. PLoS Genet 13, e1006934. 10.1371/journal.pgen.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SH, Kim YJ, Lee HB, Seok YJ, and Lee CR (2020). Genetic Evidence for Distinct Functions of Peptidoglycan Endopeptidases in Escherichia coli. Front Microbiol 11, 565767. 10.3389/fmicb.2020.565767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banzhaf M, Yau HC, Verheul J, Lodge A, Kritikos G, Mateus A, Cordier B, Hov AK, Stein F, Wartel M, et al. (2020). Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J 39, e102246. 10.15252/embj.2019102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truong TT, Vettiger A, and Bernhardt TG (2020). Cell division is antagonized by the activity of peptidoglycan endopeptidases that promote cell elongation. Mol Microbiol 114, 966–978. 10.1111/mmi.14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zouhir S, Contreras-Martel C, Maragno Trindade D, Attree I, Dessen A, and Macheboeuf P (2021). MagC is a NplC/P60-like member of the alpha-2-macroglobulin Mag complex of Pseudomonas aeruginosa that interacts with peptidoglycan. FEBS Lett 595, 2034–2046. 10.1002/1873-3468.14148. [DOI] [PubMed] [Google Scholar]
- 32.Ding J, Wang W, Feng H, Zhang Y, and Wang DC (2012). Structural insights into the Pseudomonas aeruginosa type VI virulence effector Tse1 bacteriolysis and self-protection mechanisms. J Biol Chem 287, 26911–26920. 10.1074/jbc.M112.368043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, and Mougous JD (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava D, Seo J, Rimal B, Kim SJ, Zhen S, and Darwin AJ (2018). A Proteolytic Complex Targets Multiple Cell Wall Hydrolases in Pseudomonas aeruginosa. mBio 9. 10.1128/mBio.00972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung S, and Darwin AJ (2020). The C-terminus of substrates is critical but not sufficient for their degradation by the Pseudomonas aeruginosa CtpA protease. J Bacteriol 10.1128/JB.00174-20. [DOI] [PMC free article] [PubMed]
- 37.Hsu HC, Wang M, Kovach A, Darwin AJ, and Li H (2022). Pseudomonas aeruginosa C-Terminal Processing Protease CtpA Assembles into a Hexameric Structure That Requires Activation by a Spiral-Shaped Lipoprotein-Binding Partner. mBio, e0368021. 10.1128/mbio.03680-21. [DOI] [PMC free article] [PubMed]
- 38.Chakraborty D, and Darwin AJ (2021). Direct and Indirect Interactions Promote Complexes of the Lipoprotein LbcA, the CtpA Protease and Its Substrates, and Other Cell Wall Proteins in Pseudomonas aeruginosa. J Bacteriol 203, e0039321. 10.1128/JB.00393-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lisboa J, Pereira C, Rifflet A, Ayala J, Terceti MS, Barca AV, Rodrigues I, Pereira PJB, Osorio CR, Garcia-Del Portillo F, et al. (2021). A Secreted NlpC/P60 Endopeptidase from Photobacterium damselae subsp. piscicida Cleaves the Peptidoglycan of Potentially Competing Bacteria. mSphere 6. 10.1128/mSphere.00736-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rico-Perez G, Pezza A, Pucciarelli MG, de Pedro MA, Soncini FC, and Garcia-del Portillo F (2016). A novel peptidoglycan D,L-endopeptidase induced by Salmonella inside eukaryotic cells contributes to virulence. Mol Microbiol 99, 546–556. 10.1111/mmi.13248. [DOI] [PubMed] [Google Scholar]
- 41.Hernandez SB, Castanheira S, Pucciarelli MG, Cestero JJ, Rico-Perez G, Paradela A, Ayala JA, Velazquez S, San-Felix A, Cava F, and Garcia-Del Portillo F (2022). Peptidoglycan editing in non-proliferating intracellular Salmonella as source of interference with immune signaling. PLoS Pathog 18, e1010241. 10.1371/journal.ppat.1010241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benz J, Reinstein J, and Meinhart A (2013). Structural Insights into the Effector - Immunity System Tae4/Tai4 from Salmonella typhimurium. PLoS One 8, e67362. 10.1371/journal.pone.0067362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vesto K, Huseby DL, Snygg I, Wang H, Hughes D, and Rhen M (2018). Muramyl Endopeptidase Spr Contributes to Intrinsic Vancomycin Resistance in Salmonella enterica Serovar Typhimurium. Front Microbiol 9, 2941. 10.3389/fmicb.2018.02941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cestero JJ, Castanheira S, Pucciarelli MG, and Garcia-Del Portillo F (2021). A Novel Salmonella Periplasmic Protein Controlling Cell Wall Homeostasis and Virulence. Front Microbiol 12, 633701. 10.3389/fmicb.2021.633701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong JE, Midtgaard SR, Gysel K, Thygesen MB, Sorensen KK, Jensen KJ, Stougaard J, Thirup S, and Blaise M (2015). An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase. Acta Crystallogr D Biol Crystallogr 71, 592–605. 10.1107/S139900471402793X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorr T, Davis BM, and Waldor MK (2015). Endopeptidase-mediated beta lactam tolerance. PLoS Pathog 11, e1004850. 10.1371/journal.ppat.1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altindis E, Dong T, Catalano C, and Mekalanos J (2015). Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. mBio 6, e00075. 10.1128/mBio.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hersch SJ, Watanabe N, Stietz MS, Manera K, Kamal F, Burkinshaw B, Lam L, Pun A, Li M, Savchenko A, and Dong TG (2020). Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat Microbiol 5, 706–714. 10.1038/s41564-020-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SK, Park YM, Jung KH, and Chai YG (2017). Deletion of a putative NlpC/P60 endopeptidase BAS1812 affects germination, long-term survival and endospore formation in Bacillus anthracis. Microbiology (Reading) 163, 144–152. 10.1099/mic.0.000416. [DOI] [PubMed] [Google Scholar]
- 50.Gat O, Grosfeld H, Ariel N, Inbar I, Zaide G, Broder Y, Zvi A, Chitlaru T, Altboum Z, Stein D, et al. (2006). Search for Bacillus anthracis potential vaccine candidates by a functional genomic-serologic screen. Infect Immun 74, 3987–4001. 10.1128/IAI.00174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sela-Abramovich S, Chitlaru T, Gat O, Grosfeld H, Cohen O, and Shafferman A (2009). Novel and unique diagnostic biomarkers for Bacillus anthracis infection. Appl Environ Microbiol 75, 6157–6167. 10.1128/AEM.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran SL, Guillemet E, Gohar M, Lereclus D, and Ramarao N (2010). CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J Bacteriol 192, 2638–2642. 10.1128/JB.01315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran SL, Cormontagne D, Vidic J, Andre-Leroux G, and Ramarao N (2020). Structural Modeling of Cell Wall Peptidase CwpFM (EntFM) Reveals Distinct Intrinsically Disordered Extensions Specific to Pathogenic Bacillus cereus Strains. Toxins (Basel) 12. 10.3390/toxins12090593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh YM, Sheu SJ, Chen YL, and Tsen HY (1999). Enterotoxigenic profiles and polymerase chain reaction detection of Bacillus cereus group cells and B. cereus strains from foods and food-borne outbreaks. J Appl Microbiol 87, 481–490. 10.1046/j.1365-2672.1999.00837.x. [DOI] [PubMed] [Google Scholar]
- 55.Ngamwongsatit P, Buasri W, Pianariyanon P, Pulsrikarn C, Ohba M, Assavanig A, and Panbangred W (2008). Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int J Food Microbiol 121, 352–356. 10.1016/j.ijfoodmicro.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Xu Q, Rawlings ND, Chiu HJ, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Elsliger MA, Deacon AM, Godzik A, et al. (2011). Structural analysis of papain-like NlpC/P60 superfamily enzymes with a circularly permuted topology reveals potential lipid binding sites. PLoS One 6, e22013. 10.1371/journal.pone.0022013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guinand M (2013). Chapter 554 - Dipeptidyl-Peptidase VI. In Handbook of Proteolytic Enzymes (Third Edition), Rawlings ND, and Salvesen G, eds. (Academic Press; ), pp. 2477–2478. 10.1016/B978-0-12-382219-2.00553-6. [DOI] [Google Scholar]
- 58.Yamaguchi H, Furuhata K, Fukushima T, Yamamoto H, and Sekiguchi J (2004). Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein. J Biosci Bioeng 98, 174–181. 10.1016/S1389-1723(04)00262-2. [DOI] [PubMed] [Google Scholar]
- 59.Hashimoto M, Ooiwa S, and Sekiguchi J (2012). Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall. J Bacteriol 194, 796–803. 10.1128/JB.05569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobihal GS, Flores-Kim J, Roney IJ, Wang X, and Rudner DZ (2022). The WalR-WalK signaling pathway modulates the activities of both CwlO and LytE through control of the peptidoglycan deacetylase PdaC in Bacillus subtilis. J Bacteriol 204, e0053321. 10.1128/JB.00533-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson S, and Garner E (2021). An Exhaustive Multiple Knockout Approach to Understanding Cell Wall Hydrolase Function in Bacillus subtilis. bioRxiv, 2021.2002.2018.431929. 10.1101/2021.02.18.431929. [DOI] [PMC free article] [PubMed]
- 62.Salzberg LI, Powell L, Hokamp K, Botella E, Noone D, and Devine KM (2013). The WalRK (YycFG) and sigma(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol Microbiol 87, 180–195. 10.1111/mmi.12092. [DOI] [PubMed] [Google Scholar]
- 63.Noone D, Salzberg LI, Botella E, Basell K, Becher D, Antelmann H, and Devine KM (2014). A highly unstable transcript makes CwlO D,L-endopeptidase expression responsive to growth conditions in Bacillus subtilis. J Bacteriol 196, 237–247. 10.1128/JB.00986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, and Devine KM (2007). The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 65, 180–200. 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 65.Dominguez-Cuevas P, Porcelli I, Daniel RA, and Errington J (2013). Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol 89, 1084–1098. 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, and Rudner DZ (2013). FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89, 1069–1083. 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobihal GS, Brunet YR, Flores-Kim J, and Rudner DZ (2019). Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. Elife 8. 10.7554/eLife.52088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uelze L (2022). An analysis of LysM domain function in LytE when fulfilling the D,L-endopeptidase requirement for viability in Bacillus subtilis. bioRxiv, 2022.2001.2012.475998. 10.1101/2022.01.12.475998. [DOI]
- 69.Hashimoto M, Matsushima H, Suparthana IP, Ogasawara H, Yamamoto H, Teng C, and Sekiguchi J (2018). Digestion of peptidoglycan near the cross-link is necessary for the growth of Bacillus subtilis. Microbiology (Reading) 164, 299–307. 10.1099/mic.0.000614. [DOI] [PubMed] [Google Scholar]
- 70.Brunet YR, Wang X, and Rudner DZ (2019). SweC and SweD are essential co-factors of the FtsEX-CwlO cell wall hydrolase complex in Bacillus subtilis. PLoS Genet 15, e1008296. 10.1371/journal.pgen.1008296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sassine J, Sousa J, Lalk M, Daniel RA, and Vollmer W (2020). Cell morphology maintenance in Bacillus subtilis through balanced peptidoglycan synthesis and hydrolysis. Sci Rep 10, 17910. 10.1038/s41598-020-74609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, and Errington J (2006). Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell 11, 399–409. 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 73.Patel Y, Zhao H, and Helmann JD (2020). A regulatory pathway that selectively up-regulates elongasome function in the absence of class A PBPs. Elife 9. 10.7554/eLife.57902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kasahara J, Kiriyama Y, Miyashita M, Kondo T, Yamada T, Yazawa K, Yoshikawa R, and Yamamoto H (2016). Teichoic Acid Polymers Affect Expression and Localization of dl-Endopeptidase LytE Required for Lateral Cell Wall Hydrolysis in Bacillus subtilis. J Bacteriol 198, 1585–1594. 10.1128/JB.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamamoto H, Kurosawa S, and Sekiguchi J (2003). Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J Bacteriol 185, 6666–6677. 10.1128/JB.185.22.6666-6677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mamou G, Fiyaksel O, Sinai L, and Ben-Yehuda S (2017). Deficiency in Lipoteichoic Acid Synthesis Causes a Failure in Executing the Colony Developmental Program in Bacillus subtilis. Front Microbiol 8, 1991. 10.3389/fmicb.2017.01991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tseng CL, Chen JT, Lin JH, Huang WZ, and Shaw GC (2011). Genetic evidence for involvement of the alternative sigma factor SigI in controlling expression of the cell wall hydrolase gene lytE and contribution of LytE to heat survival of Bacillus subtilis. Arch Microbiol 193, 677–685. 10.1007/s00203-011-0710-0. [DOI] [PubMed] [Google Scholar]
- 78.Tseng CL, and Shaw GC (2008). Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis. J Bacteriol 190, 1561–1567. 10.1128/JB.01497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, and Sekiguchi J (2008). The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70, 297–310. 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- 80.Kiriyama Y, Yazawa K, Tanaka T, Yoshikawa R, Yamane H, Hashimoto M, Sekiguchi J, and Yamamoto H (2014). Localization and expression of the Bacillus subtilis DL-endopeptidase LytF are influenced by mutations in LTA synthases and glycolipid anchor synthetic enzymes. Microbiology (Reading) 160, 2639–2649. 10.1099/mic.0.080366-0. [DOI] [PubMed] [Google Scholar]
- 81.Margot P, Pagni M, and Karamata D (1999). Bacillus subtilis 168 gene lytF encodes a gamma-D-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, sigmaD. Microbiology (Reading) 145 (Pt 1), 57–65. 10.1099/13500872-145-1-57. [DOI] [PubMed] [Google Scholar]
- 82.Ohnishi R, Ishikawa S, and Sekiguchi J (1999). Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis. J Bacteriol 181, 3178–3184. 10.1128/JB.181.10.3178-3184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dergham Y, Sanchez-Vizuete P, Le Coq D, Deschamps J, Bridier A, Hamze K, and Briandet R (2021). Comparison of the Genetic Features Involved in Bacillus subtilis Biofilm Formation Using Multi-Culturing Approaches. Microorganisms 9. 10.3390/microorganisms9030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen R, Guttenplan SB, Blair KM, and Kearns DB (2009). Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity. J Bacteriol 191, 5775–5784. 10.1128/JB.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fukushima T, Afkham A, Kurosawa S, Tanabe T, Yamamoto H, and Sekiguchi J (2006). A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis. J Bacteriol 188, 5541–5550. 10.1128/JB.00188-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fukushima T, Kitajima T, Yamaguchi H, Ouyang Q, Furuhata K, Yamamoto H, Shida T, and Sekiguchi J (2008). Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting n-acetylmuramidase and DL-endopeptidase activities. J Biol Chem 283, 11117–11125. 10.1074/jbc.M706626200. [DOI] [PubMed] [Google Scholar]
- 87.DeWitt T, and Grossman AD (2014). The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol 196, 1588–1596. 10.1128/JB.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeng J, Jin Y, and Liu Z (2018). Solution scattering study of the Bacillus subtilis PgdS enzyme involved in poly-gamma-glutamic acids degradation. PLoS One 13, e0195355. 10.1371/journal.pone.0195355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scoffone V, Dondi D, Biino G, Borghese G, Pasini D, Galizzi A, and Calvio C (2013). Knockout of pgdS and ggt genes improves gamma-PGA yield in B. subtilis. Biotechnol Bioeng 110, 2006–2012. 10.1002/bit.24846. [DOI] [PubMed] [Google Scholar]
- 90.Fukushima T, Uchida N, Ide M, Kodama T, and Sekiguchi J (2018). DL-endopeptidases function as both cell wall hydrolases and poly-gamma-glutamic acid hydrolases. Microbiology (Reading) 164, 277–286. 10.1099/mic.0.000609. [DOI] [PubMed] [Google Scholar]
- 91.Fernandes CG, Placido D, Lousa D, Brito JA, Isidro A, Soares CM, Pohl J, Carrondo MA, Archer M, and Henriques AO (2015). Structural and Functional Characterization of an Ancient Bacterial Transglutaminase Sheds Light on the Minimal Requirements for Protein Cross-Linking. Biochemistry 54, 5723–5734. 10.1021/acs.biochem.5b00661. [DOI] [PubMed] [Google Scholar]
- 92.Placido D, Fernandes CG, Isidro A, Carrondo MA, Henriques AO, and Archer M (2008). Auto-induction and purification of a Bacillus subtilis transglutaminase (Tgl) and its preliminary crystallographic characterization. Protein Expr Purif 59, 1–8. 10.1016/j.pep.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 93.Fernandes CG, Martins D, Hernandez G, Sousa AL, Freitas C, Tranfield EM, Cordeiro TN, Serrano M, Moran CP Jr., and Henriques AO (2019). Temporal and spatial regulation of protein cross-linking by the pre-assembled substrates of a Bacillus subtilis spore coat transglutaminase. PLoS Genet 15, e1007912. 10.1371/journal.pgen.1007912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garcia-Garcia T, Poncet S, Cuenot E, Douche T, Giai Gianetto Q, Peltier J, Courtin P, Chapot-Chartier MP, Matondo M, Dupuy B, et al. (2021). Ser/Thr Kinase-Dependent Phosphorylation of the Peptidoglycan Hydrolase CwlA Controls Its Export and Modulates Cell Division in Clostridioides difficile. mBio 12. 10.1128/mBio.00519-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu D, Patabendige H, Tomlinson BR, Wang S, Hussain S, Flores D, He Y, Shaw LN, and Sun X (2021). Cwl0971, a novel peptidoglycan hydrolase, plays pleiotropic roles in Clostridioides difficile R20291. Environ Microbiol 23, 5222–5238. 10.1111/1462-2920.15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teng F, Kawalec M, Weinstock GM, Hryniewicz W, and Murray BE (2003). An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect Immun 71, 5033–5041. 10.1128/IAI.71.9.5033-5041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, Mucida D, and Hang HC (2016). A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 353, 1434–1437. 10.1126/science.aaf3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Griffin ME, Espinosa J, Becker JL, Luo JD, Carroll TS, Jha JK, Fanger GR, and Hang HC (2021). Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 373, 1040–1046. 10.1126/science.abc9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bauerl C, Perez-Martinez G, Yan F, Polk DB, and Monedero V (2010). Functional analysis of the p40 and p75 proteins from Lactobacillus casei BL23. J Mol Microbiol Biotechnol 19, 231–241. 10.1159/000322233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bauerl C, Abitayeva G, Sosa-Carrillo S, Mencher-Beltran A, Navarro-Lleo N, Coll-Marques JM, Zuniga-Cabrera M, Shaikhin S, and Perez-Martinez G (2019). P40 and P75 Are Singular Functional Muramidases Present in the Lactobacillus casei /paracasei/rhamnosus Taxon. Front Microbiol 10, 1420. 10.3389/fmicb.2019.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duchene MC, Rolain T, Knoops A, Courtin P, Chapot-Chartier MP, Dufrene YF, Hallet BF, and Hols P (2019). Distinct and Specific Role of NlpC/P60 Endopeptidases LytA and LytB in Cell Elongation and Division of Lactobacillus plantarum. Front Microbiol 10, 713. 10.3389/fmicb.2019.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Claes IJ, Schoofs G, Regulski K, Courtin P, Chapot-Chartier MP, Rolain T, Hols P, von Ossowski I, Reunanen J, de Vos WM, et al. (2012). Genetic and biochemical characterization of the cell wall hydrolase activity of the major secreted protein of Lactobacillus rhamnosus GG. PLoS One 7, e31588. 10.1371/journal.pone.0031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lebeer S, Claes IJ, Balog CI, Schoofs G, Verhoeven TL, Nys K, von Ossowski I, de Vos WM, Tytgat HL, Agostinis P, et al. (2012). The major secreted protein Msp1/p75 is O-glycosylated in Lactobacillus rhamnosus GG. Microb Cell Fact 11, 15. 10.1186/1475-2859-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koponen J, Laakso K, Koskenniemi K, Kankainen M, Savijoki K, Nyman TA, de Vos WM, Tynkkynen S, Kalkkinen N, and Varmanen P (2012). Effect of acid stress on protein expression and phosphorylation in Lactobacillus rhamnosus GG. J Proteomics 75, 1357–1374. 10.1016/j.jprot.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 105.Gao J, Zhao X, Hu S, Huang Z, Hu M, Jin S, Lu B, Sun K, Wang Z, Fu J, et al. (2022). Gut microbial DL-endopeptidase alleviates Crohn’s disease via the NOD2 pathway. Cell Host & Microbe 30, 1–15. 10.1016/j.chom.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 106.Redko Y, Courtin P, Mezange C, Huard C, and Chapot-Chartier MP (2007). Lactococcus lactis gene yjgB encodes a gamma-D-glutaminyl-L-lysyl-endopeptidase which hydrolyzes peptidoglycan. Appl Environ Microbiol 73, 5825–5831. 10.1128/AEM.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carvalho F, Sousa S, and Cabanes D (2014). How Listeria monocytogenes organizes its surface for virulence. Front Cell Infect Microbiol 4, 48. 10.3389/fcimb.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schubert K, Bichlmaier AM, Mager E, Wolff K, Ruhland G, and Fiedler F (2000). P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch Microbiol 173, 21–28. 10.1007/s002030050003. [DOI] [PubMed] [Google Scholar]
- 109.Kuhn M, and Goebel W (1989). Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun 57, 55–61. 10.1128/iai.57.1.55-61.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pilgrim S, Kolb-Maurer A, Gentschev I, Goebel W, and Kuhn M (2003). Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect Immun 71, 3473–3484. 10.1128/IAI.71.6.3473-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wuenscher MD, Kohler S, Bubert A, Gerike U, and Goebel W (1993). The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol 175, 3491–3501. 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hou S, Chen G, Wang W, Xia L, Wang Z, and Lu Y (2020). Identification of a cell-wall peptidase (NlpC/P60) from Nocardia seriolae which induces apoptosis in fathead minnow cells. J Fish Dis 43, 571–581. 10.1111/jfd.13154. [DOI] [PubMed] [Google Scholar]
- 113.Gardiner J.H.t., Komazin G, Matsuo M, Cole K, Gotz F, and Meredith TC (2020). Lipoprotein N-Acylation in Staphylococcus aureus Is Catalyzed by a Two-Component Acyl Transferase System. mBio 11. 10.1128/mBio.01619-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garcia P, Mendez E, Garcia E, Ronda C, and Lopez R (1984). Biochemical characterization of a murein hydrolase induced by bacteriophage Dp-1 in Streptococcus pneumoniae: comparative study between bacteriophage-associated lysin and the host amidase. J Bacteriol 159, 793–796. 10.1128/jb.159.2.793-796.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Loeffler JM, Nelson D, and Fischetti VA (2001). Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294, 2170–2172. 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 116.Haiser HJ, Yousef MR, and Elliot MA (2009). Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J Bacteriol 191, 6501–6512. 10.1128/JB.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bobek J, Strakova E, Zikova A, and Vohradsky J (2014). Changes in activity of metabolic and regulatory pathways during germination of S. coelicolor. BMC Genomics 15, 1173. 10.1186/1471-2164-15-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ott L, Holler M, Gerlach RG, Hensel M, Rheinlaender J, Schaffer TE, and Burkovski A (2010). Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells. BMC Microbiol 10, 2. 10.1186/1471-2180-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ott L (2018). Adhesion properties of toxigenic corynebacteria. AIMS Microbiol 4, 85–103. 10.3934/microbiol.2018.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kolodkina V, Denisevich T, and Titov L (2011). Identification of Corynebacterium diphtheriae gene involved in adherence to epithelial cells. Infect Genet Evol 11, 518–521. 10.1016/j.meegid.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 121.Tsuge Y, Ogino H, Teramoto H, Inui M, and Yukawa H (2008). Deletion of cgR_1596 and cgR_2070, encoding NlpC/P60 proteins, causes a defect in cell separation in Corynebacterium glutamicum R. J Bacteriol 190, 8204–8214. 10.1128/JB.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou X, Rodriguez-Rivera FP, Lim HC, Bell JC, Bernhardt TG, Bertozzi CR, and Theriot JA (2019). Sequential assembly of the septal cell envelope prior to V snapping in Corynebacterium glutamicum. Nat Chem Biol 15, 221–231. 10.1038/s41589-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lim HC, Sher JW, Rodriguez-Rivera FP, Fumeaux C, Bertozzi CR, and Bernhardt TG (2019). Identification of new components of the RipC-FtsEX cell separation pathway of Corynebacterineae. PLoS Genet 15, e1008284. 10.1371/journal.pgen.1008284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maeda T, Tanaka Y, Takemoto N, Hamamoto N, and Inui M (2016). RNase III mediated cleavage of the coding region of mraZ mRNA is required for efficient cell division in Corynebacterium glutamicum. Mol Microbiol 99, 1149–1166. 10.1111/mmi.13295. [DOI] [PubMed] [Google Scholar]
- 125.Bannantine JP, Lingle CK, Adam PR, Ramyar KX, McWhorter WJ, Stabel JR, Picking WD, and Geisbrecht BV (2016). NlpC/P60 domain-containing proteins of Mycobacterium avium subspecies paratuberculosis that differentially bind and hydrolyze peptidoglycan. Protein Sci 25, 840–851. 10.1002/pro.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bannantine JP, Lingle CK, Stabel JR, Ramyar KX, Garcia BL, Raeber AJ, Schacher P, Kapur V, and Geisbrecht BV (2012). MAP1272c encodes an NlpC/P60 protein, an antigen detected in cattle with Johne’s disease. Clin Vaccine Immunol 19, 1083–1092. 10.1128/CVI.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bannantine JP, Stabel JR, Lippolis JD, and Reinhardt TA (2018). Membrane and Cytoplasmic Proteins of Mycobacterium avium subspecies paratuberculosis that Bind to Novel Monoclonal Antibodies. Microorganisms 6. 10.3390/microorganisms6040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eckelt E, Meissner T, Meens J, Laarmann K, Nerlich A, Jarek M, Weiss S, Gerlach GF, and Goethe R (2015). FurA contributes to the oxidative stress response regulation of Mycobacterium avium ssp. paratuberculosis. Front Microbiol 6, 16. 10.3389/fmicb.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao LY, Pak M, Kish R, Kajihara K, and Brown EJ (2006). A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect Immun 74, 1757–1767. 10.1128/IAI.74.3.1757-1767.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martinelli DJ, and Pavelka MS Jr. (2016). The RipA and RipB Peptidoglycan Endopeptidases Are Individually Nonessential to Mycobacterium smegmatis. J Bacteriol 198, 1464–1475. 10.1128/JB.00059-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, and Rubin EJ (2007). A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol 66, 658–668. 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 132.Hett EC, Chao MC, and Rubin EJ (2010). Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog 6, e1001020. 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Botella H, Vaubourgeix J, Lee MH, Song N, Xu W, Makinoshima H, Glickman MS, and Ehrt S (2017). Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress. EMBO J 36, 536–548. 10.15252/embj.201695028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gustine JN, Au MB, Haserick JR, Hett EC, Rubin EJ, Gibson FC 3rd, and Deng LL (2019). Cell Wall Hydrolytic Enzymes Enhance Antimicrobial Drug Activity Against Mycobacterium. Curr Microbiol 76, 398–409. 10.1007/s00284-018-1620-z. [DOI] [PubMed] [Google Scholar]
- 135.Bhuwan M, Arora N, Sharma A, Khubaib M, Pandey S, Chaudhuri TK, Hasnain SE, and Ehtesham NZ (2016). Interaction of Mycobacterium tuberculosis Virulence Factor RipA with Chaperone MoxR1 Is Required for Transport through the TAT Secretion System. mBio 7, e02259. 10.1128/mBio.02259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hett EC, Chao MC, Deng LL, and Rubin EJ (2008). A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog 4, e1000001. 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Santos-Beneit F (2021). Expression of the Mycobacterium tuberculosis RipA cell wall hydrolase in Streptomyces coelicolor hampers vancomycin resistance. J Glob Antimicrob Resist 27, 115–117. 10.1016/j.jgar.2021.08.013. [DOI] [PubMed] [Google Scholar]
- 138.Healy C, Gouzy A, and Ehrt S (2020). Peptidoglycan Hydrolases RipA and Ami1 Are Critical for Replication and Persistence of Mycobacterium tuberculosis in the Host. mBio 11. 10.1128/mBio.03315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Both D, Schneider G, and Schnell R (2011). Peptidoglycan remodeling in Mycobacterium tuberculosis: comparison of structures and catalytic activities of RipA and RipB. J Mol Biol 413, 247–260. 10.1016/j.jmb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 140.Steiner EM, Lyngso J, Guy JE, Bourenkov G, Lindqvist Y, Schneider TR, Pedersen JS, Schneider G, and Schnell R (2018). The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity. Proteins 86, 912–923. 10.1002/prot.25523. [DOI] [PubMed] [Google Scholar]
- 141.Gao B, Wang J, Huang J, Huang X, Sha W, and Qin L (2019). The dynamic region of the peptidoglycan synthase gene, Rv0050, induces the growth rate and morphologic heterogeneity in Mycobacteria. Infect Genet Evol 72, 86–92. 10.1016/j.meegid.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 142.Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, Chao MC, Rego EH, Sassetti CM, Fortune SM, and Rubin EJ (2015). Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria. PLoS Pathog 11, e1005010. 10.1371/journal.ppat.1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chao MC, Kieser KJ, Minami S, Mavrici D, Aldridge BB, Fortune SM, Alber T, and Rubin EJ (2013). Protein complexes and proteolytic activation of the cell wall hydrolase RipA regulate septal resolution in mycobacteria. PLoS Pathog 9, e1003197. 10.1371/journal.ppat.1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Parthasarathy G, Lun S, Guo H, Ammerman NC, Geiman DE, and Bishai WR (2012). Rv2190c, an NlpC/P60 family protein, is required for full virulence of Mycobacterium tuberculosis. PLoS One 7, e43429. 10.1371/journal.pone.0043429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mavrici D, Marakalala MJ, Holton JM, Prigozhin DM, Gee CL, Zhang YJ, Rubin EJ, and Alber T (2014). Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc Natl Acad Sci U S A 111, 8037–8042. 10.1073/pnas.1321812111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liang Y, Zhang X, Bai X, Xiao L, Wang X, Zhang J, Yang Y, Song J, Wang L, and Wu X (2017). Immunogenicity and therapeutic effects of a Mycobacterium tuberculosis rv2190c DNA vaccine in mice. BMC Immunol 18, 11. 10.1186/s12865-017-0196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Both D, Steiner EM, Izumi A, Schneider G, and Schnell R (2014). RipD (Rv1566c) from Mycobacterium tuberculosis: adaptation of an NlpC/p60 domain to a non-catalytic peptidoglycan-binding function. Biochem J 457, 33–41. 10.1042/BJ20131227. [DOI] [PubMed] [Google Scholar]
- 148.Padhi A, Naik SK, Sengupta S, Ganguli G, and Sonawane A (2016). Expression of Mycobacterium tuberculosis NLPC/p60 family protein Rv0024 induce biofilm formation and resistance against cell wall acting anti-tuberculosis drugs in Mycobacterium smegmatis. Microbes Infect 18, 224–236. 10.1016/j.micinf.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 149.Culp EJ, Waglechner N, Wang W, Fiebig-Comyn AA, Hsu YP, Koteva K, Sychantha D, Coombes BK, Van Nieuwenhze MS, Brun YV, and Wright GD (2020). Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 578, 582–587. 10.1038/s41586-020-1990-9. [DOI] [PubMed] [Google Scholar]
- 150.Reinscheid DJ, Gottschalk B, Schubert A, Eikmanns BJ, and Chhatwal GS (2001). Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J Bacteriol 183, 1175–1183. 10.1128/JB.183.4.1175-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bartual SG, Straume D, Stamsas GA, Munoz IG, Alfonso C, Martinez-Ripoll M, Havarstein LS, and Hermoso JA (2014). Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat Commun 5, 3842. 10.1038/ncomms4842. [DOI] [PubMed] [Google Scholar]
- 152.Rued BE, Alcorlo M, Edmonds KA, Martinez-Caballero S, Straume D, Fu Y, Bruce KE, Wu H, Havarstein LS, Hermoso JA, et al. (2019). Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae. mBio 10. 10.1128/mBio.02622-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Squeglia F, Moreira M, Ruggiero A, and Berisio R (2019). The Cell Wall Hydrolytic NlpC/P60 Endopeptidases in Mycobacterial Cytokinesis: A Structural Perspective. Cells 8. 10.3390/cells8060609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, Brun YV, Pogliano K, and Jensen GJ (2013). Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol 88, 673–686. 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, and Jensen GJ (2011). Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146, 799–812. 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Khanna K, Lopez-Garrido J, Zhao Z, Watanabe R, Yuan Y, Sugie J, Pogliano K, and Villa E (2019). The molecular architecture of engulfment during Bacillus subtilis sporulation. Elife 8. 10.7554/eLife.45257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ojkic N, Lopez-Garrido J, Pogliano K, and Endres RG (2016). Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. Elife 5. 10.7554/eLife.18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Muchova K, Chromikova Z, and Barak I (2020). Linking the Peptidoglycan Synthesis Protein Complex with Asymmetric Cell Division during Bacillus subtilis Sporulation. Int J Mol Sci 21. 10.3390/ijms21124513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Arai R, Fukui S, Kobayashi N, and Sekiguchi J (2012). Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis, reveals a novel fold with a characteristic inhibitory loop. J Biol Chem 287, 44736–44748. 10.1074/jbc.M112.414763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Groisman EA, Duprey A, and Choi J (2021). How the PhoP/PhoQ System Controls Virulence and Mg(2+) Homeostasis: Lessons in Signal Transduction, Pathogenesis, Physiology, and Evolution. Microbiol Mol Biol Rev 85, e0017620. 10.1128/MMBR.00176-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Harris KA, Zhou Z, Peters ML, Wilkins SG, and Breaker RR (2018). A second RNA-binding protein is essential for ethanol tolerance provided by the bacterial OLE ribonucleoprotein complex. Proc Natl Acad Sci U S A 115, E6319–E6328. 10.1073/pnas.1803191115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Irazoki O, Hernandez SB, and Cava F (2019). Peptidoglycan muropeptides: release, perception, and functions as signaling molecules. Front Microbiol 10, 500. 10.3389/fmicb.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bastos PAD, Wheeler R, and Boneca IG (2021). Uptake, recognition and responses to peptidoglycan in the mammalian host. FEMS Microbiol Rev 45. 10.1093/femsre/fuaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Caruso R, Warner N, Inohara N, and Nunez G (2014). NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41, 898–908. 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, and Girardin SE (2014). NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol 14, 9–23. 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 166.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, and Girardin SE (2019). NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys 670, 69–81. 10.1016/j.abb.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 167.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, and Polk DB (2007). Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132, 562–575. 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, and Mucida D (2016). Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci Immunol 1. 10.1126/sciimmunol.aai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, Clurman AG, Armijo G, Gomes ALC, Shono Y, et al. (2019). Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science 366, 1143–1149. 10.1126/science.aax3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP, et al. (2016). Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 45, 931–943. 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 171.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, and Wargo JA (2018). The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33, 570–580. 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 359, 91–97. 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 174.Pinheiro J, Biboy J, Vollmer W, Hirt RP, Keown JR, Artuyants A, Black MM, Goldstone DC, and Simoes-Barbosa A (2018). The protozoan Trichomonas vaginalis targets bacteria with laterally acquired NlpC/P60 peptidoglycan hydrolases. mBio 9. 10.1128/mBio.01784-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tang VH, Stewart GA, and Chang BJ (2015). House dust mites possess a polymorphic, single domain putative peptidoglycan d,l endopeptidase belonging to the NlpC/P60 Superfamily. FEBS Open Bio 5, 813–823. 10.1016/j.fob.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tang VH, Stewart GA, and Chang BJ (2017). Dermatophagoides pteronyssinus lytFM encoding an NlpC/P60 endopeptidase is also present in mite-associated bacteria that express LytFM variants. FEBS Open Bio 7, 1267–1280. 10.1002/2211-5463.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Golczak M, Kiser PD, Sears AE, Lodowski DT, Blaner WS, and Palczewski K (2012). Structural basis for the acyltransferase activity of lecithin:retinol acyltransferase-like proteins. J Biol Chem 287, 23790–23807. 10.1074/jbc.M112.361550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, and Palczewski K (2004). Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem 279, 10422–10432. 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang CH, Shyu RY, Wu CC, Tsai TC, Wang LK, Chen ML, Jiang SY, and Tsai FM (2014). Phospholipase A/Acyltransferase enzyme activity of H-rev107 inhibits the H-RAS signaling pathway. J Biomed Sci 21, 36. 10.1186/1423-0127-21-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Estes KA, Kalamegham R, and Hanna-Rose W (2007). Membrane localization of the NlpC/P60 family protein EGL-26 correlates with regulation of vulval cell morphogenesis in Caenorhabditis elegans. Dev Biol 308, 196–205. 10.1016/j.ydbio.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 181.Antimicrobial Resistance C (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ansaldo E, Farley TK, and Belkaid Y (2021). Control of immunity by the microbiota. Annu Rev Immunol 39, 449–479. 10.1146/annurev-immunol-093019-112348. [DOI] [PubMed] [Google Scholar]
- 183.Aminov RI (2010). A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1, 134. 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, González-Páez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, et al. (2016). Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574. 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]


