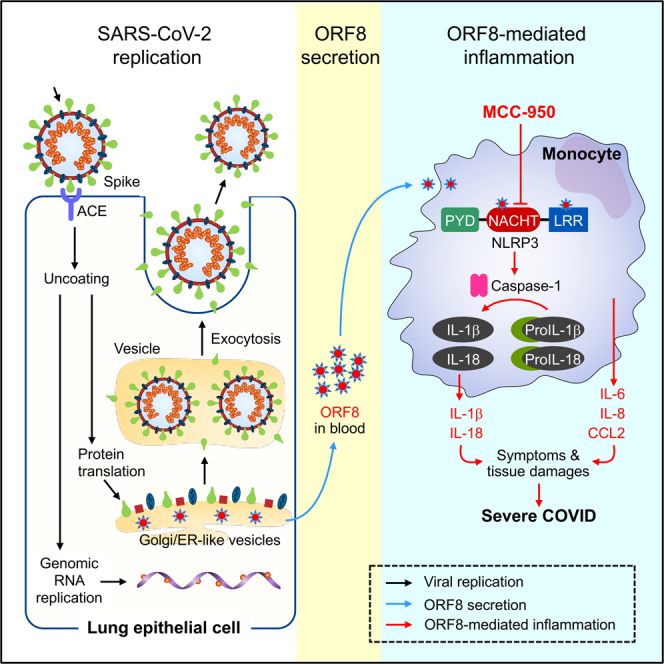
Summary
Despite extensive research, the specific factor associated with SARS-CoV-2 infection that mediates the life-threatening inflammatory cytokine response in patients with severe COVID-19 remains unidentified. Herein we demonstrate that the virus-encoded Open Reading Frame 8 (ORF8) protein is abundantly secreted as a glycoprotein in vitro and in symptomatic patients with COVID-19. ORF8 specifically binds to the NOD-like receptor family pyrin domain-containing 3 (NLRP3) in CD14+ monocytes to induce inflammasomal cytokine/chemokine responses including IL1β, IL8, and CCL2. Levels of ORF8 protein in the blood correlate with severity and disease-specific mortality in patients with acute SARS-CoV-2 infection. Furthermore, the ORF8-induced inflammasome response was readily inhibited by the NLRP3 inhibitor MCC950 in vitro. Our study identifies a dominant cause of pathogenesis, its underlying mechanism, and a potential new treatment strategy for severe COVID-19.
Subject areas: Immunity, Virology
Graphical abstract
Highlights
-
•
ORF8 and select NSPs of the SARS-CoV-2 virus are secreted from human cells
-
•
ORF8 induces monocytic pro-inflammatory cytokines via NLPR3 inflammasome pathways
-
•
Blood ORF8 levels in symptomatic covid-19 patients predict prognosis
-
•
NLRP3 inhibitor MCC950 blocks ORF8 induced pro-inflammatory cytokine production
Immunity; Virology
Introduction
COVID-19, the global pandemic caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected more than 750 million people and caused nearly seven million deaths worldwide as of April 5, 2023.1 Numerous studies have shown that the production of pro-inflammatory cytokines/chemokines including IL1β, IL6, IL8, and CCL2 is responsible for life-threatening symptoms.2,3 It is also known that the viral load, cytokine levels, and disease severity are tightly associated,4,5,6 and that virus-neutralizing antibodies and IL1β pathway antagonists can readily mitigate symptoms and improve clinical outcomes.7,8 However, the intermediate viral factor that directly causes the inflammatory cytokine responses remains unidentified. It has been demonstrated that SARS-CoV-2 infection localizes to nasal and pulmonary epithelial cells,9,10 whereas the cytokine response is more systemic. It seems irreconcilable how this cytokine response is initiated given that no live virus has been reported in the blood based on transfusion medicine studies.11,12,13,14 We hypothesized that an inflammatory byproduct of SARS-CoV-2 replication is released into the bloodstream resulting in a systematic cytokine response in severe COVID-19 patients.
On infection of human cells, the SARS-CoV-2 virus replicates its 29.9 kb RNA genome and produces up to 29 possible viral proteins, including 16 non-structural proteins (NSP1-16), four structural proteins Spike (SPK), Membrane (MEM), Envelope (ENV) and Nucleocapsid (NUC) and nine accessory proteins (ORF3A, 3B, 6, 7A, 7B, 8, 9b, 9c, and 10). Although only four structural proteins with the RNA genome are assembled into new viral particles, the other viral proteins are thought to be left behind15 which may disrupt host cell functions.16,17 Herein, we demonstrate that the SARS-CoV-2 encoded ORF8 is abundantly secreted as a glycoprotein into culture supernatant in vitro and into the bloodstream in patients with COVID-19. In contrast to other secreted NSPs, only glycosylated ORF8 stimulates CD14+ monocytes to produce a group of pro-inflammatory cytokines/chemokines through an NLRP3-mediated inflammasome response. In addition, the ORF8/NLRP3 axis was readily inhibited by MCC950, a specific NLRP3 inhibitor that blocks the production of the master cytokine IL1β.
Results
Secretion of SARS-CoV-2 proteins from human cells
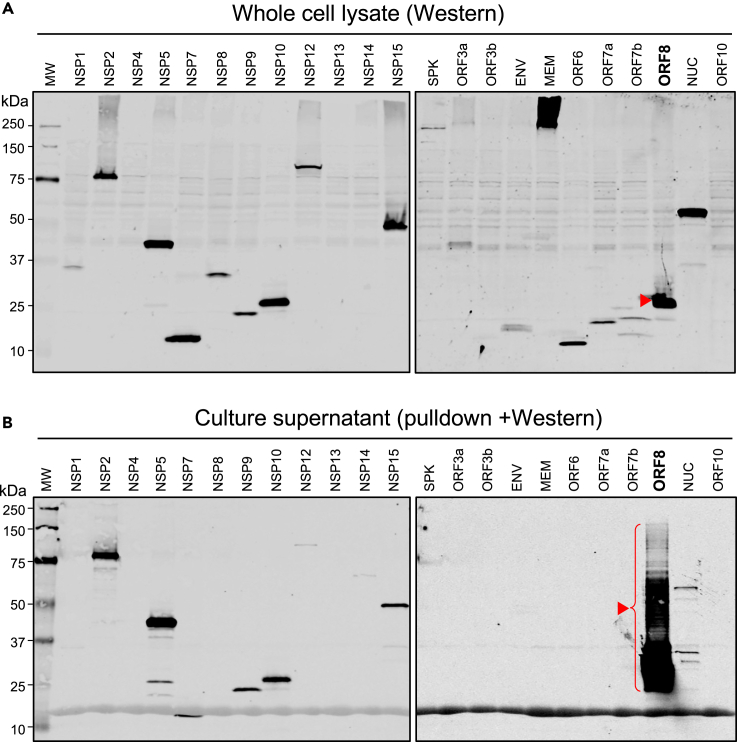
To determine the secretion potential of SARS-CoV-2 encoded proteins from human cells, we transduced HEK293 cells, a human cell line commonly used for the study of protein expression and secretion,18 with one of 22 available lentiviral constructs each expressing a SARS-CoV-2 protein tagged Strep II (key resources table) except the SPK was tagged with hemagglutinin (HA) epitope tag. The secreted viral proteins in the culture supernatants were enriched and analyzed by Western blotting, and their levels were compared to their total cellular expression in whole cell lysates. As shown in Figure 1A, most viral proteins were robustly expressed with the expected molecular sizes except for NSP13, ORF10, MEM, and NSP4 because of their small sizes or insolubility issues. Although NSP2, 5, 7, 9, 10, 12, 14, 15, ORF8, and NUC proteins were all detected as secreted proteins, ORF8 was the single most robustly secreted protein (Figure 1B). This is consistent with in silico analysis (Figure S1) showing that all secreted NSPs possess an unconventional protein secretion signal (UPS)19 whereas ORF8 carries a classical protein signal sequence (SS) and an N-link glycosylation sites (78NYTV). Consistent with its secretory nature, immunofluorescence staining of ORF8 showed that the protein was localized to cytoplasmic vesicular structures (Figure S2). These results suggest that ORF8 is efficiently secreted through the classical ER/Golgi protein secretion pathway, and its glycosylation explains the up-shifting and smearing of the ORF8 protein band (Figure 1B).
Figure 1.
Expression and secretion property of SARS-CoV-2 proteins
(A) Western blot image of total cell lysates for validating SARS-CoV-2 protein expression in HEK293 cells. The blots were probed with anti-Strep II tag and anti-HA (for SPK protein) antibodies.
(B) Western image of SARS-CoV-2 protein pulldown using Strep-Tactin beads and anti-HA-beads (for SPK) from the culture supernatants for validating SARS-CoV-2 protein secretion from HEK293 cells. The blots were also probed with anti-Strep II tag and anti-HA (for SPK) antibodies.
Secreted ORF8 protein induces the production of pro-inflammatory cytokines
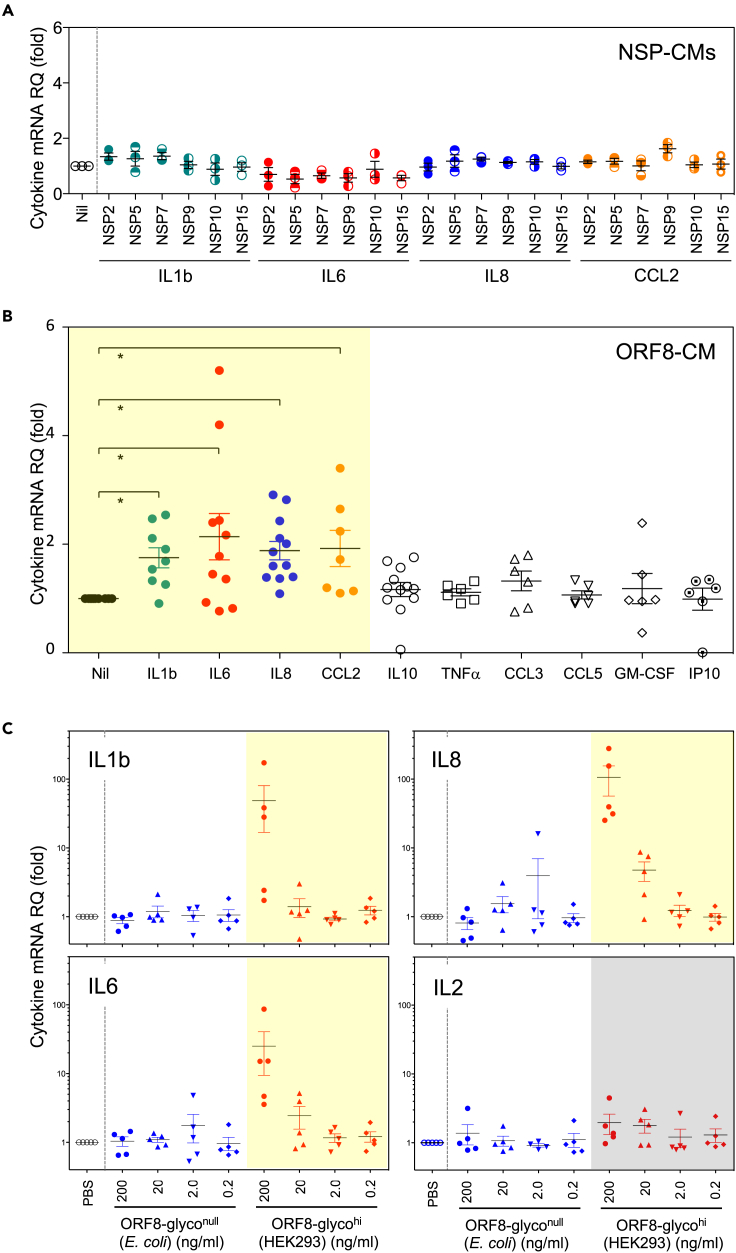
We then queried if any of the secreted SARS-CoV-2 proteins could induce the expression of pro-inflammatory cytokines seen in patients with COVID-19. We treated human peripheral blood mononuclear cells (PBMCs) from healthy donors with conditioned media (CM) containing major secreted NSPs or ORF8 proteins followed by an assessment of cytokine expression. We found that none of the secreted NSP proteins induced cytokine expression (Figure 2A) whereas ORF8-containing CM (ORF8-CM) induced the expression of IL1β, IL6, IL8, and CCL2 up to 5-fold in PBMCs from select donors (Figure 2B). Because IL1β, IL6, IL8, and CCL2 are among the key cytokines elevated in patients with severe COVID-19,20 we postulated that secreted ORF8 is the viral factor responsible for inflammatory cytokine response in patients with severe COVID-19.
Figure 2.
Secreted glycosylated ORF8 specifically induces the expression of pro-inflammatory cytokines
(A) Conditioned media containing major secreted NSP proteins did not induce pro-inflammatory cytokines in PBMCs of healthy donors.
(B) Conditioned media containing secreted ORF8 specifically induced pro-inflammatory cytokines (highlighted in yellow) IL1β, IL6, IL8, and CCL2 in PBMCs from unselected healthy donors. ∗p≤0.05.
(C) Purified highly glycosylated ORF8 (ORF8-glycolhi) from HEK293 but not unglycosylated ORF8 (ORF8-glycolnull) from E. coli stimulates PBMCs to produce pro-inflammatory cytokines IL1β, IL6, IL8 but not T cell cytokine IL2.
To evaluate whether the glycosylation of ORF8 affected the cytokine induction, we purified ORF8 from the HEK293 culture supernatant and from ORF8-expressing Escherichia coli and designated them as ORF8-glycohi and ORF8-glyconull, respectively. As shown in Figure 2C, IL1β, IL-6, and IL-8 but not IL-2 were robustly induced by the pure ORF8-glycohi in a dose-dependent manner confirming that secreted ORF8 is capable of inducing pro-inflammatory cytokines seen in COVID-19 patients. Of interest, E. coli expressed ORF8-glyconull, had no cytokine induction activity even at high doses, suggesting that proper ORF8 glycosylation is necessary for cytokine induction.
We then tested the cytokine induction activities of ORF8 from HEK293 cells treated with the Golgi inhibitor cocktail Brefeldin A and Monensin (BFA/M) and found that when the glycosylation level was reduced (Figure S3) the cytokine induction activity of BFA/M-treated ORF8 was indeed altered (actually increased), suggesting that the cytokine induction activity of ORF8 is governed by its glycosylation at the Golgi complex. Our protein localization data (Figures S4A–S4AC) indicate that several SARS-CoV-2 proteins are localized to Golgi membranes and may potentially modify the glycosylation and the cytokine induction activity of ORF8. Indeed, co-expressing MEM, ENV, ORF3A, or ORF7A was able to alter the molecule weight and the cytokine-inducing activities of ORF8 (Figures S4D–S4E).
ORF8 stimulates CD14+ monocytes to produce pro-inflammatory cytokines
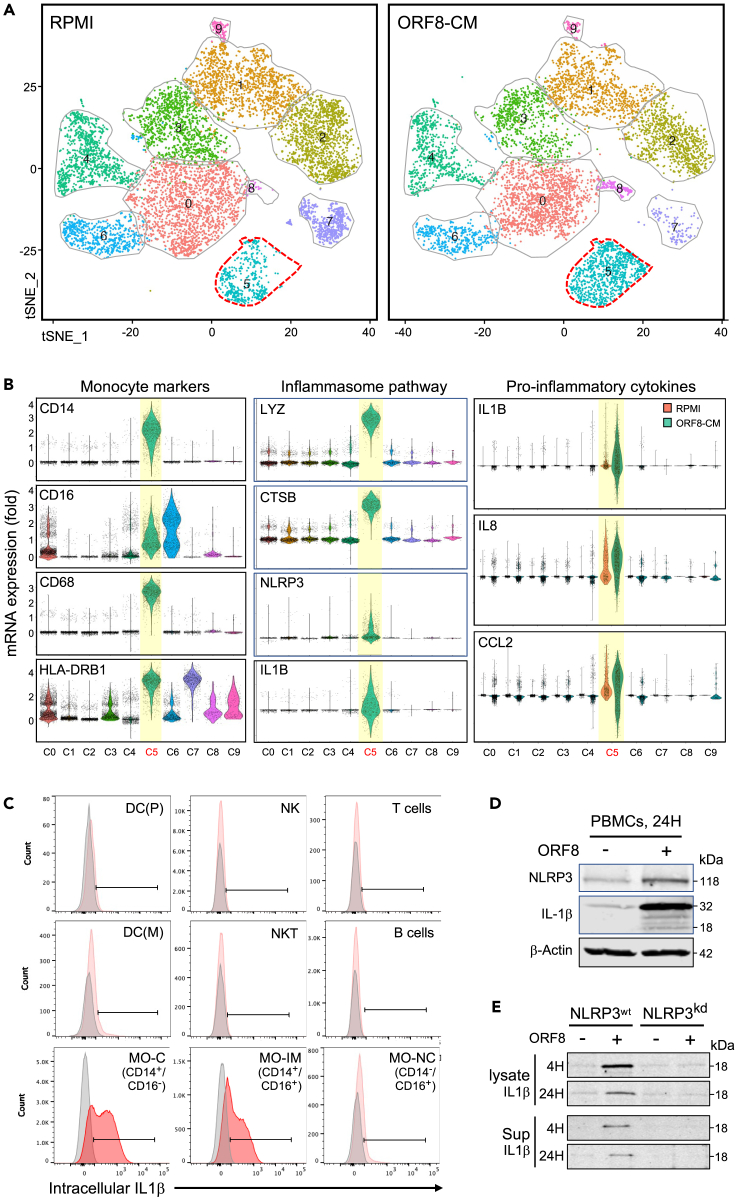
Next, we asked which specific cell subsets in PBMCs are the primary targets of ORF8, and what is the resulting transcriptional signatures in those cells. We performed single-cell RNA sequencing (scRNA-Seq) analysis on three PBMC samples treated with ORF8-CM or control-CM. Single-cell transcriptome-based cell clustering showed that cells in cluster 5 (C5) on the t-SNE plot (Figure 3A) were induced to express cytokines including IL1β, IL8, and CCL2 and inflammasome pathway components on ORF8 treatment (Figure 3B, Table S1). The same cytokines are known to be elevated in COVID-19 patients.21 The expression of CD14, CD16, CD68, and HLA-DR identifies those cytokine-secreting cells as activated monocytes.22,23
Figure 3.
ORF8 targets CD14+ monocytes to induce pro-inflammatory cytokines by activating the NLRP3-mediated inflammasome pathway
(A) tSNE maps of cell clusters using concatenated scRNA-Seq data from two ORF8-responsive PBMC samples. Cells in cluster 5 were responsive to ORF8 treatment.
(B) Violin plots show the monocyte markers CD14, CD16, CD68, and HLA-DRB1, key inflammasome pathway components LYZ, CTSB, NLRP3, IL1β, IL8, and CCL2 mRNA expression in cells of cluster 5.
(C) Flow cytometry analysis of intracellular IL1β protein expression in different PBMC subsets in response to ORF8 treatment. The plots show that IL1β was exclusively produced by CD14+CD16− (classical) and CD14+CD16+ (intermediate) monocyte subsets.
(D) Western blot images validate the induction of NLRP3 and IL1β in PBMCs on stimulation with ORF8 at the protein level.
(E) Western images showing in response to ORF8 treatment, the IL1β induction in parental THP-1 cells became diminished in NLRP3 knockdown (NLRP3kd) THP-1 cells. The data shown are representative of at least two independent experiments except for the scRNA-Seq.
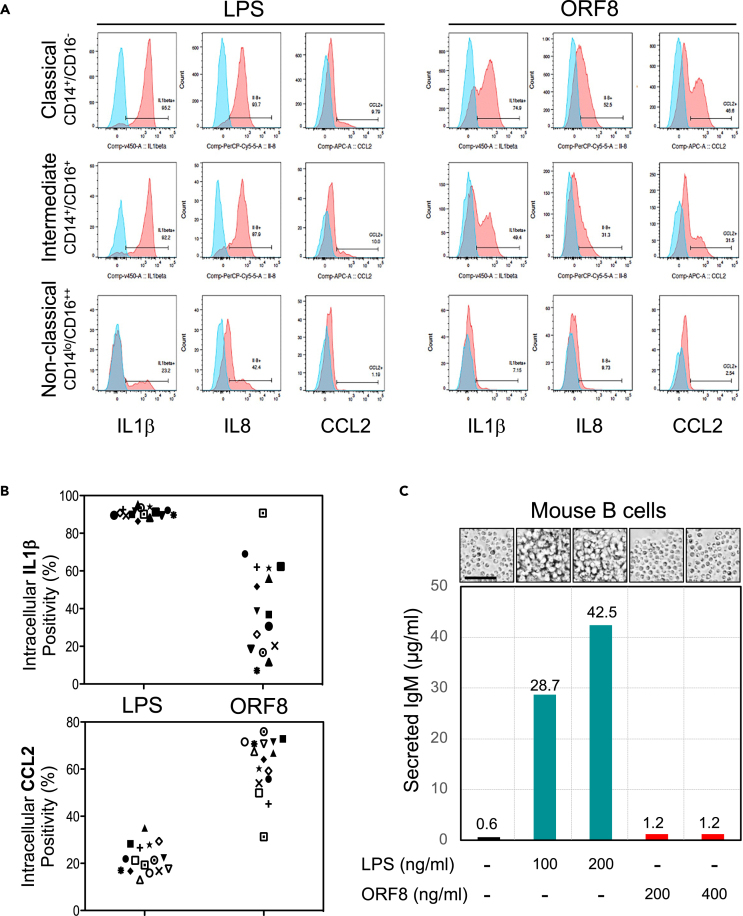
To identify which monocyte subsets are the targets of ORF8, we analyzed intracellular cytokines in ORF8 stimulated PBMCs from 15 donors by flow cytometry. Significant IL1β expression was detected in classical (CD14+/CD16-) and intermediate (CD14+/CD16+) subsets but not non-classical (CD14low/CD16+) monocytes nor cells of other lineages (B or T-lymphocytes, dendritic, NK cells) in all 15 donors (Figure 3C). Similar results were observed for IL8 and CCL2 expression as well (Figure 5A). Our data demonstrate that the CD14+ monocyte subsets are the producer of the pro-inflammatory cytokines in response to ORF8 stimulation.
Figure 5.
ORF8 is functionally distinct from LPS
(A) Representative flow data showing LPS activates multiple cytokine responses in all three monocyte subsets of PMBCs from all 15 healthy donors examined whereas ORF8 only activates CD14+ Monocytes but not CD14low/CD16++ non-classical monocytes (red boxed) suggesting LPS and ORF8 may target monocytes through different mechanisms.
(B) Intracellular cytokine flow cytometry data summary on 15 healthy donors on treatment with ORF8 or LPS, suggesting that ORF8 and LPS are different in cytokine induction specificities.
(C) Mouse B cells responded differently to the treatment of ORF8 and LPS as measured by IgM production and the morphological features of cell proliferation status. Scale bar: 100 μm. The dissimilar characteristics of ORF8 and LPS dismiss the possibility that the cytokine-inducing function of ORF8 resulted from the contamination of LPS.
ORF8 activates the inflammasome responses is NLRP3 dependent
To decipher the mechanism by which ORF8 induces pro-inflammatory cytokines in CD14+ monocytes, using the Gene Set Enrichment Analysis (GSEA) and Kyoto Encyclopedia of Gene and Genome (KEGG) tools, we found that the SARS-CoV-2 infection, NOD-like receptor signaling, and the NF-κB pathways were among the top enriched, with mRNAs of lysosomal enzymes (LYZ, CTSB, CTSD, CTSS), inflammasomal protein NLPR3, and cytokines IL1β, IL8, CCL2 being among the top expressed (Figure 3B, Table S1).
We then examined the role of the NLRP3-mediated inflammasome pathway in ORF8-mediated cytokine production. Figure 3D shows that both NLPR3 and IL1β proteins were induced in PBMCs on ORF8 treatment, and such induction was diminished in NLPR3 knockdown cells (THP1-defNLRP3, InvivoGen) compared to the parental THP-1 cells, demonstrating the NLRP3 dependency of ORF8 mediated cytokine response in human monocytes (Figure 3E).
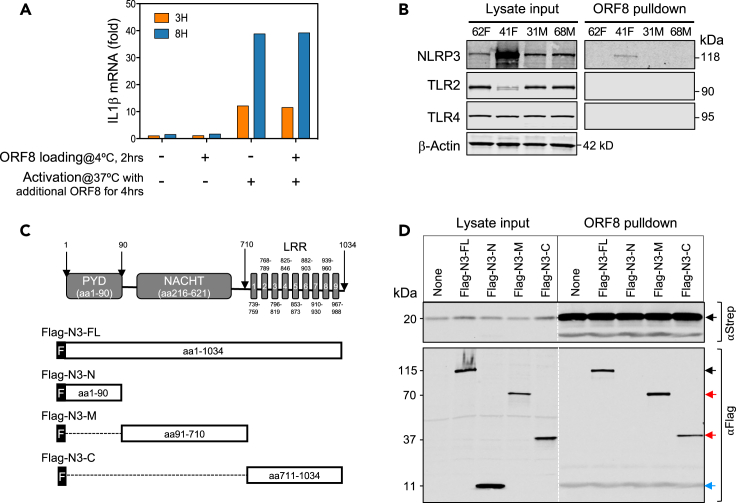
To determine how ORF8 molecules enter CD14+ monocytes, we first saturated ORF8 binding receptors (if any) on PBMCs with purified ORF8 protein on ice and then washed the cells to remove unbound ORF8 protein. The resulting cells were then incubated at 37°C to initiate ORF8-mediated cell activation in the presence or absence of additional ORF8 protein. Figure 4A shows robust IL1β mRNA expression was observed in cells only exposed to additional ORF8 but not those only pre-incubated with ORF8, suggesting that ORF8 likely enters monocytes through a non-receptor-mediated process, such as phagocytosis.
Figure 4.
ORF8 induces pro-inflammatory cytokines through binding to NLRP3
(A) ORF8 induces IL1β production through a non-surface-receptor-mediated process. PBMCs were pre-incubated on ice with ORF8 (500 ng/mL) for 2 h to allow ORF8 to bind to its “surface receptors” followed by washing and incubating at 37°C to activate the cytokine production in the presence or absence of additional ORF8 (200 ng/mL). IL1β expression was measured by qPCR.
(B) ORF8 directly binds to NLRP3 but not TLR2 nor TRL4 in primary monocytes shown by affinity pulldown assay.
(C) Schematic drawing of NLRP3 deletion constructs for mapping ORF8 binding domains in NLPR3.
(D) Mapping NLRP3 domains that bind to ORF8 by affinity pulldown assay using ORF8-Strep-Tacin beads. Western images of total cell lysates (left) were used as expression controls and Western images of precipitated proteins (right) showing ORF8 efficiently binds the full-length as well as two NLRP3 deletion mutant proteins.
In complementary experiments, we tested if ORF8 would bind to TLR2, TLR4, CD14, or NLRP3, receptors known to be involved in inflammasome activation. We incubated ORF8 protein-coated beads with monocyte lysates from two ORF8 responding and two ORF8 non-responding healthy donors. Figure 4B shows that neither TLR2, TLR4, nor CD14 were co-precipitated with ORF8 (CD14 data not shown). However, NLRP3 was readily detected in one of the ORF8-responders, suggesting that indeed ORF8 binds to NLRP3 in primary human monocytes.
To further dissect how NLRP3 binds to ORF8, we transiently transfected ORF8-expressing HEK293 cells with Flag-tagged NLRP3 constructs Flag-N3-FL (full-length NLRP3), or one of the three deletion mutants Flag-N3-N, Flag-N3-M, or Flag-N3-C (Figure 4C).24 Cell lysates were then incubated with Strep-Tactin beads to pull down Strep-tagged ORF8 and its binding proteins. In addition to NLRP3-FL (Figure 4D), both Flag-N3-M, and Flag-N3-C but not Flag-N3-N also strongly co-precipitated with ORF8 suggesting both the middle NACHT domain and C-terminal LRR domain of NLRP3 each can independently bind to ORF8. These results clearly demonstrate inflammasome protein NLRP3 directly interacts with ORF8.
ORF8 activates the NLRP3-mediated inflammasome pathways
The effect of LPS in inflammasomal activation has been well established; therefore, it was important to demonstrate that our purified ORF8 protein was free of LPS contamination. ORF8 and LPS were compared for their effects on IL1β, IL8, and CCL2 induction in three monocyte subsets (classical, intermediate, and non-classical) from 15 healthy donors. Figure 5A shows that all three monocyte subsets responded to LPS whereas only CD14+ (classical and intermediate) responded to ORF8 with variable induction of IL1β, IL8, and CCL2. These results show that ORF8 and LPS target monocytes with different cell type specificities. We then examined the responsiveness of CD14+ monocytes to LPS or ORF8 treatment using IL1β, IL8, and CCL2 expression as the readout, and we found that intracellular IL1β was detected in the majority (90.1%) of LPS-treated CD14+ monocytes but only in a subpopulation (mean = 38%) of ORF8 treated CD14+ monocytes. In contrast, CCL2 expression was detected only in a minor population (mean = 22.1%) of LPS-treated CD14+ monocytes but in a larger population of ORF8-treated CD14+ monocytes (mean = 63.7%) (Figure 5B). These data clearly demonstrate that ORF8 and LPS have different cytokine induction specificities. In addition, we examined the cell proliferation and the IgM induction of mouse splenic B cells on ORF8 and LPS treatment and found that the mouse splenic B cells did not proliferate, nor produce any polyclonal IgM 96 h after ORF8 treatment, whereas LPS-treated cells showed robust proliferation morphology and massive production of IgM (Figure 5C). In the aggregate, our data clearly demonstrate that the cytokine inducing activity of ORF8 is not because of LPS contamination.
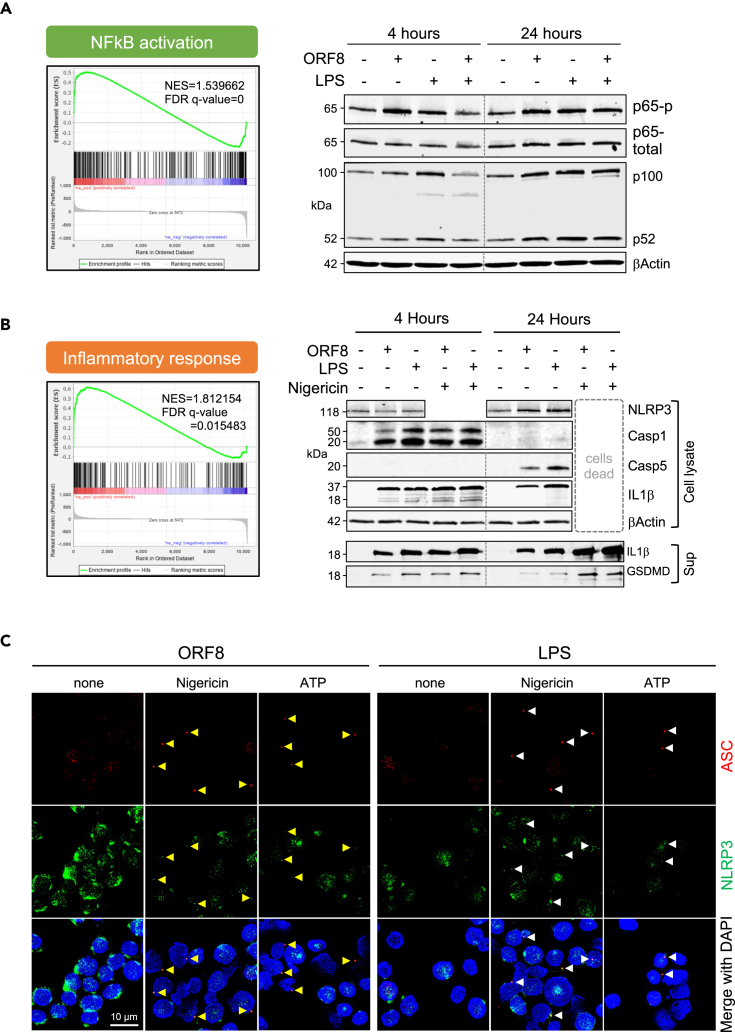
To examine the activation status of the canonical NF-κB pathway necessary for the induction of inflammasomal proteins including NLPR3, Casp-1, and IL1β in ORF8 treated monocytes, we examined the GSEA analysis results and found that NF-κB pathway components were transcriptionally enriched (NES = 1.539662, FDR q-value = 0) in ORF8 treated monocytes in cluster 5 (Figure 6A, left panel). To validate these at the protein level, we treated THP-1 cells with ORF8 for 4 and 24 h and determined their NF-κB pathway protein expression. Figure 6A (right panels) shows that the increased level of phospho-p65 and the conversion of p100 to p52 were readily detected 4 h after ORF8 treatment. Similar results were also observed in LPS-treated cells. These data demonstrate that ORF8 stimulation activates the NF-κB pathway triggering the activation of the inflammasome pathway in human monocytes.
Figure 6.
Profiles of ORF8-mediated signaling pathways
(A) Gene enrichment analysis of scRNA-Seq (left panel) and Western confirmation of key NFkB pathway molecules (right panels) on stimulation of ORF8 for 4 or 24 h.
(B) Gene enrichment analysis of scRNA-Seq (left panel) and Western confirmation of NLRP3, IL1β, Casp-1, and Casp-5 in THP-1 cell lysates and IL1β and GSDMD in the culture supernatants (right panels) on stimulation of ORF8 for 4 or 24 h.
(C) Immunofluorescence images show that LPS and ORF8 activate ASC speck (arrowhead) formation only when a second signal (nigericin or extracellular ATP) was present. Cells were first treated with 200 ng/mL ORF8 or 100 ng/mL LPS for 2.5 h followed by 90 min treatment with 5 μM Nigericin or 5 μM ATP before cell harvest for immunofluorescence staining. Scale bar: 10 μm.
Similarly, the GSEA analysis data revealed the mRNAs of inflammasome pathway components were also significantly enriched (NES = 1.812154, FDR q-value =0.015483) (Figure 6B, left panel). We then treated THP-1 cells for 4 or 24 h with ORF8 or LPS alone or in combination with nigericin, a potent agent that provides a second signal for the canonical inflammasomal pathway activation. The cell lysates and culture supernatants were analyzed by Western blotting for the expression of NLPR3, Caspase-1, Caspase-5, IL1β, and the pore-forming fragment of gasdermin D (GSDMD) proteins. We found that ORF8 induces the inflammasome pathway by 1) activating the production of IL1β without requiring a separate priming step; 2) signaling through the non-canonical pathway using Casp-1 (early hours) followed by switching to Casp-4/5; 3) maintaining cell viability without triggering pyroptosis unless the second signal such as nigericin is present which rapidly initiates pyroptosis (Figure 6B, right panel). Our data suggest that ORF8 and LPS may represent a family of pyrogens by triggering a non-canonical inflammasomal pathway leading to the production and release of IL1β without activating GSDMD to the level necessary for pyroptosis, a process called hyperactivation for LPS.25,26 However, ORF8 can also serve as a priming agent for the canonical inflammasome pathway to evoke a prompt pyroptosis in the presence of second signal molecules such as extracellular ATP from damaged cells.
To validate the dual roles of ORF8 in the activation of canonical and non-canonical inflammasome pathways, we examined ASC speck formation in ORF8- or LPS-treated THP-1 cells by immunofluorescence. As shown in Figure 6C, ORF8 alone was insufficient to induce ASC speck formation. However, distinct pre-nuclear ASC specks were readily detected when nigericin or extracellular ATP was also present. In addition, the NLPR3 protein was also co-localized with ASC protein to the specks (Figure 6C), ASC speck formation always precedes cell pyroptosis in our system, and ORF8 is slightly more robust than LPS in mediating ASC formation in the presence of nigericin or extracellular ATP. The results confirm the dual roles of ORF8 in the activation of the non-canonical inflammasome pathway when the second signal is absent, and the canonical inflammasome pathway by priming the cells when the second signal is present. These data further suggest that the mode of action of ORF8 is dependent on the presence or absence of a second signal such as extracellular ATP released from damaged cells/tissues nearby.
Blood ORF8 protein levels correlate with the mortality and disease course of severe patients with COVID-19
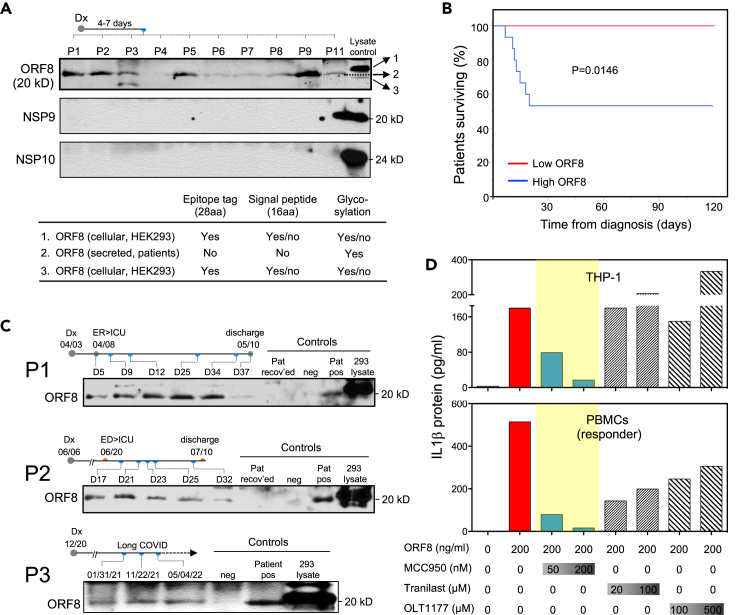
Having demonstrated that ORF8 is secreted as a glycoprotein and capable of inducing inflammatory cytokine responses in vitro, we then asked if ORF8 is also secreted and glycosylated in blood from patients infected with SARS-CoV-2. By analyzing serum samples collected from patients within 4–7 days of COVID-19 diagnosis using Western blotting, we found that ORF8 protein was readily detected at various levels in 92% (23/25) of newly infected patients (Figure 7A). Of interest, the same samples showed no detectable NSP9 and NSP10 which are also secreted suggesting not all the secreted SARS-CoV-2 proteins are present at detectable levels in blood circulation. To determine the glycosylation status of the serum ORF8, we compared the sizes of ORF8 from patients and HEK293 lysate. Our results (Figure 7A) show that patient serum ORF8 is indeed glycosylated when compared to the ORF8 from HEK293 lysate which has an extra Strep-II tag (28 aa) and possibly an uncleaved signal sequence (16 aa). Owing to the lack of a quantitative assay, we were unable to determine the absolute quantity of ORF8 in patient samples; however, our semi-quantitative Western blot could readily detect ORF8 in as little as four microliters of patient serum suggesting the protein is present at significant levels in patient blood at the onset of COVID-19.
Figure 7.
The blood levels of ORF8 protein in COVID-19 patients correlate with the disease outcome, and the ORF8/NLRP3 axis is targetable with NLRP3 inhibitor MCC950
(A) Representative western blot images showing ORF8 protein but not NSP9 or NSP10 were detectable in as little as 4.0 μL of serum samples from newly infected COVID-19 patients. ORF8 protein in patient sera shown as glycosylated by size estimation.
(B) Kaplan-Meier curve showing fatality in hospitalized patients is associated with higher serum level of ORF8. p=0.0146.
(C) Western images showing ORF8 levels at different time points correlate with disease course in two patients with prolonged clinical courses, and in one patient with long-covid.
(D) Targetability of ORF8 mediated IL1β expression with various NLRP3 inhibitors in THP1 cells and human primary PBMCs. The data shown are representative of three independent experiments. Abbreviations: Dx = diagnosis; ED = emergency department; ICU = intensive care unit.
Given its activity in inducing inflammatory cytokines in vitro, and its significant presence in the blood of newly infected patients, we then asked if the levels of ORF8 protein would correlate with disease severity and outcome. To that end, we examined the correlation between blood ORF8 levels and patients’ survival outcomes in our cohort of 25 hospitalized patients. Figure 7B shows that after 120 days of follow-up, all seven fatalities were exclusively associated with the ORF8-high group (scored 2+ or 3+ on Western blot) whereas all patients in the ORF8-low group (scored 0 or 1+) had mild symptoms and quick recovery without any events of death. These results demonstrate a clear correlation between the ORF8 load and disease severity in newly infected patients. We then monitored the ORF8 levels at various time points during the disease course in two patients with prolonged COVID-19, and one patient with long covid lasting more than 17 months. As demonstrated in Figure 7C, two patients with high blood ORF8 levels had persistent severe symptoms requiring treatment in the intensive care unit, and their blood ORF8 levels diminished by the time they were recovered and discharged from the hospitals. The long covid patient (Pat 3) with lingering symptoms throughout remained ORF8 positive in all three serial blood samples collected over a 16-month period (Figure 7C). These results suggest that the ORF8 level is prognostic for COVID-19 outcome, further supporting our hypothesis that the ORF8 protein is a pathogenic cause of severe COVID-19 in patients.
ORF8-mediated cytokine induction is targetable by select NLRP3 inhibitors
Given its causative role in COVID-19 pathogenesis, we then tested the targetability of ORF8-mediated cytokine response using three investigational NLRP3 inhibitors - MCC950, Tranilast, and OLT117 (Dapansutrile). As shown in Figure 7D, the NLRP3 inhibitor MCC950 showed effective inhibition on the production of IL1β in both THP-1 cells and PBMCs (blue bars), while Tranilast and OLT117 (hatched bars) exhibited moderate to marginal effect in PBMCs but no effect in THP-1 cells. Our results suggest that ORF8-mediated IL1β production can be effectively inhibited by the NLRP3 inhibitor MCC950 at a nanomolar dose range, demonstrating that targeting the ORF8/NLRP3 axis is a promising strategy for treating patients with symptomatic COVID-19.
Discussion
Despite the success in the development and implementation of effective vaccines to prevent SARS-CoV-2 infection, COVID-19 remains a major challenge for many reasons. These include, (1) a significant population remains unvaccinated, (2) new variants keep emerging and evading current vaccine protection, (3) patients with compromised immunity including those with blood cancers on immunosuppressive medications are poorly protected,27,28 and more importantly, (4) many previously infected patients have developed lingering symptoms or long covid. Therefore, better biomarkers, therapies and clinical management tools for COVID-19 are urgently needed.
SARS-CoV-2 accessory protein ORF8, a 121 amino acid protein, is the least conserved protein in the beta-coronavirus family.29,30 Here we report that ORF8 is a major secreted viral glycoprotein in vitro and in COVID-19 patients. Our data, summarized in Graphical Abstract, support the concept that the ORF8 protein secreted from locally infected cells in the lung and released to the bloodstream then stimulates circulating CD14+ monocytes to initiate systematic cytokine responses. This is consistent with findings by others that monocytes are the single most affected WBC subset in the blood by SARS-CoV-2 infection.31 It has been reported that about 6% of monocytes were infected with the virus in COVID-19 patients through CD16-mediated uptake of antibody-opsonized SARS-CoV-2 virus. Whether these cells were truly infected or simply engulfed with infected cells/debris remains to be seen because no live virus was detected.32 Nevertheless, these monocytes may represent a parallel and complementary process to the ORF8-mediated inflammatory pathway described herein. Our intracellular IL1β data showed a mean of 43% (range, 7.2–90.7%) of monocytes that are ORF8 responsive, suggesting that the ORF8-mediated process dominates. We conclude that secreted ORF8 is a key disease-causing viral factor and can be targeted by NLRP3 inhibitors, offering a potential new treatment option.
NLRP3-mediated inflammasome activation is a critical part of the innate immune response. However, the precise mechanisms for different pathogens have not been fully delineated. Our study indicated that ORF8 may enter monocytes through a non-receptor- mediated ORF8 internalization, followed by lysosomal action involving lysozyme (LYZ) and cathepsin proteases (CTSB, CTSD). ORF8 then may bind to the NACHT and/or LRR domain to activate NLRP3 and recruit Casp-1 (early) and Casp-4/5 (later). It is not known what the roles of these two different bindings play, and if they are important in pathway choice (canonical versus non-canonical). It is also interesting that, like LPS, ORF8 induces the expression/activation of Caspase-1 early (hour 4) but switches to Casp-4/5 later (at hour 24) even when nigericin is present (data not shown). Our data shows that the mode of action of ORF8 is readily changed by the presence of the second signal leading to the switch from non-canonical to canonical inflammasomal response. Therefore, it is conceivable that, in patients with COVID-19, the inflammation may be aggravated by the additional tissue/cell damages that release ATP, leading to the activation of an ORF8-mediated canonical inflammasome pathway and causing pyroptosis.
Owing to the lack of effective COVID-19 animal models, it is challenging to genetically test the pathogenic role and targetability of the ORF8 protein in vivo. However, a SARS-CoV-2 variant (Δ382) found in Taiwan and Singapore with a complete loss of the ORF8 gene serves as a tailor-made natural genetic model that validates our findings.33,34 Young et al. analyzed a cohort of 92 patients infected with the wildtype virus and 29 patients infected with the Δ382 variant and found that patients infected with the wildtype SARS-CoV-2 exhibited much higher levels of pro-inflammatory cytokines than those infected with the Δ382 variant. Furthermore, no patients with the Δ382 variant required supplemental oxygen whereas it was required for 26 (28%) patients infected with wildtype virus. These observations provide yet another line of evidence supporting the link between ORF8 and COVID-19 disease severity, and an in vivo genetic proof-of-principle for targeting ORF8 to prevent severe COVID-19 infection. In addition, various clinical studies and trials have shown that the IL1 receptor antagonist anakinra significantly reduced the mortality risk in hospitalized patients with moderate to severe inflammation symptoms.35,36,37 The data from our present study suggests that targeting NLRP3 or ORF8 upstream of the IL1 receptor would provide greater therapeutic benefit to patients with severe COVID-19 symptoms, and could also provide prophylactic benefit to patients at risk of developing severe COVID-19 infection.
MCC950, also known as CRID3 or CP-456773, is a sulfonylurea-containing compound developed by Pfizer and first disclosed in 1998.38 Multiple studies have demonstrated that MCC950 inhibits both canonical and non-canonical activation of NLRP3 mediated inflammation pathways and is effective in specifically blocking the induction of IL1β in vitro and in various mouse models. However, testing MCC950 in phase II clinical trials for patients with rheumatoid arthritis was terminated early because of hepatotoxicity.39 This trial has not been fully published; therefore, MCC950 and other drugs of the class hold great promise for symptomatic Covid-19 patients.
Limitations of the study
The bulk of this study was carried out in ex vivo systems using expression constructs for the SARS-CoV-2 coding genes and serum samples obtained from patients with Covid-19. It would be valuable to validate our findings in vivo in either SARS-CoV-2 infected or ORF8 transgenic animal models. In addition, our work provides evidence that ORF8-mediated inflammasome pathways can be targeted by the NLRP3 inhibitor MCC950 in vitro; however, the true clinical benefits of this intervention must be rigorously tested in human patients through clinical trials.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-ORF8, SARS-Cov2 | MyBioResource | Cat# MBS3014575 |
| Rabbit anti-NSP9, SARS-Cov2 | Abcam | Cat#ab284037 |
| Rabbit anti-NSP10, SARS-Cov2 | R&D | Cat#MAB11049-SP |
| Rabbit anti-human IL1beta | Abcam | Cat#Ab216995, RRID:AB_2894877 |
| Mouse anti-human ASC (B-3) | Santa Cruz | Sc-514414, RRID:AB_2737351 |
| Rabbit anti-human NLRP3 | Cell Signaling | Cat#15101S, RRID:AB_2722591 |
| Rabbit anti-human Cleaved Caspase-1 (asp297) | Cell Signaling | Cat#4199S, RRID:AB_1903916 |
| Rabbit anti-human Caspase-5 | Cell Signaling | Cat#46680, RRID:AB_2799306 |
| Rabbit anti-human Gasdermin D | Cell Signaling | Cat#97558, RRID:AB_2864253 |
| Mouse anti-NFkB p65 | Cell Signaling | Cat#6956, RRID:AB_10828935 |
| Rabbit anti-pNFkB | Cell Signaling | Cat#3033, RRID:AB_331284 |
| Rabbit anti-NFkB2, p105/p50 | Cell Signaling | Cat#12540, RRID:AB_2687614 |
| Rabbit anti-IkB alpha | Abcam | Cat#9242, RRID:AB_331623 |
| Mouse anti-Strep Tag II, | Millipore | Cat#71590-3, RRID:AB_10807650 |
| Mouse anti-HA, clone 12CA5 | Sigma | Cat#11583816001, RRID:AB_514505 |
| Mouse anti-Flag M2 | Sigma | Cat#3165, RRID:AB_259529 |
| Mouse anti-human IL1beta-BV421, clone AS10 | BD Pharmingen | Cat#567791, RRID:AB_2916736 |
| Rat Anti-Human IL-6-PE, clone MQ2-6A3 | BD Pharmingen | Cat# 559331, RRID:AB_397228 |
| Mouse anti-human CCL2-APC, clone 5D3-F7 | Invitrogen | Cat#17-7099-81, RRID:AB_469500 |
| Mouse anti-human IL8-PerCP-eFluor 710, clone 8CH | Invitrogen | Cat#46-8088-42, RRID:AB_10805748 |
| CD3-FITC, clone OKT3 | Biolegend | Cat# 317306, RRID:AB_571907 |
| CD19-FITC, clone SJ25-C1 | Biolegend | Cat# 363025, RRID:AB_2564254 |
| CD14-APC-Cy7, clone M5E2 | Biolegend | Cat# 301820, RRID:AB_493695 |
| CD16-PE-Cy7, clone 3G8 | Biolegend | Cat# 302016, RRID:AB_314216 |
| CD56-PE-Dazzle, clone HCD56 | Biolegend | Cat# 318347, RRID:AB_2563563 |
| CD16-BUV496, clone 3G8 | BD | Cat# 564653, RRID:AB_2744294 |
| CD11c-PE-Cy7, clone B-ly6 | BD | Cat# 561356, RRID:AB_10611859 |
| CD5-BV605, clone L17F12 | BD | Cat# 742550, RRID:AB_2740860 |
| HLA-DR-BV785, clone GF6-6 | BD | Cat# 564041, RRID:AB_2738559 |
| Ghost Dye Violet 510 | Tonbo | Cat# 13-0870-T100 |
| Bacterial and virus strains | ||
| Stbl3 | ThermoFisher | Cat# C7373-03 |
| Biological samples | ||
| Plasma samples, patients with Covid-19 | Mayo Clinic | N/A |
| PBMCs, healthy donors | Mayo Clinic | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ORF8, SARS-CoV2, purified from HEK293 | This paper | N/A |
| ORF8, SARS-CoV2, purified from E. coli | This paper | N/A |
| Lipopolysaccharides (LPS) | Sigma | Cat# L2630 |
| Nigericin | Sigma | Cat# N7143 |
| MCC950 | Sigma | Cat# S7809 |
| Tranilast (SB 252218) | Selleckchem | Cat# S1439 |
| OLT1177 (Dapansutrile) | Tocris | Cat# 6902 |
| Deposited data | ||
| Single-cell RNA-seq | This paper | SRA: PRJNA971080 |
| Experimental models: Cell lines | ||
| HEK293 | ATCC | RRID:CVCL_0045 |
| HEK293-F | ATCC | RRID:CVCL_6642 |
| THP1 | ATCC | RRID:CVCL_0006 |
| THP-1, NLRP3 Knocked down | Invivogen | Cat code: Thp-dnlp |
| Oligonucleotides | ||
| Forward primer for IL1β qPCR: CCACAGACCTTCCAGGAGAATG | This paper | N/A |
| Reverse primer for IL1β qPCR: GTGCAGTTCAGTGATCGTACAGG | This paper | N/A |
| Forward primer for IL6 qPCR: AGACAGCCACTCACCTCTTCAG | This paper | N/A |
| Reverse primer for IL6 qPCR: TTCTGCCAGTGCCTCTTTGCTG | This paper | N/A |
| Forward primer for IL8 qPCR: GAGAGTGATTGAGAGTGGACCAC | This paper | N/A |
| Reverse primer for IL8 qPCR: CACAACCCTCTGCACCCAGTTT | This paper | N/A |
| Forward primer for CCL2 qPCR: AGAATCACCAGCAGCAAGTGTCC | This paper | N/A |
| Reverse primer for CCL2 qPCR: TCCTGAACCCACTTCTGCTTGG | This paper | N/A |
| Forward primer for beta-Actin qPCR: CACCATTGGCAATGAGCGGTTC | This paper | N/A |
| Reverse primer for beta-Actin qPCR: AGGTCTTTGCGGATGTCCACGT | This paper | N/A |
| Forward primer for GAPDH qPCR: GTCTCCTCTGACTTCAACAGCG | This paper | N/A |
| Reverse primer for GAPDH qPCR: ACCACCCTGTTGCTGTAGCCAA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-EF1alpha-SARS-CoV-2-NSP1-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141367 |
| pLVX-EF1alpha-SARS-CoV-2-NSP2-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141368 |
| pLVX-EF1alpha-SARS-CoV-2-NSP4-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141369 |
| pLVX-EF1alpha-SARS-CoV-2-NSP5-C145A-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141371 |
| pLVX-EF1alpha-SARS-CoV-2-NSP7-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141373 |
| pLVX-EF1alpha-SARS-CoV-2-NSP8-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141374 |
| pLVX-EF1alpha-SARS-CoV-2-NSP9-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141375 |
| pLVX-EF1alpha-SARS-CoV-2-NSP10-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141376 |
| pLVX-EF1alpha-SARS-CoV-2-NSP12-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141378 |
| pLVX-EF1alpha-SARS-CoV-2-NSP13-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141379 |
| pLXV-EF1alpha-2xStrep-SARS-CoV-2-NSP14-IRES-Puro | Addgene | RRID:Addgene_141380 |
| pLVX-EF1alpha-SARS-CoV-2-NSP15-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141381 |
| pBOB-CAG-SARS-CoV2-SPK-HA | Addgene | RRID:Addgene_141347 |
| pLVX-EF1alpha-SARS-CoV-2-ORF3a-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141383 |
| pLXV-EF1alpha-2xStrep-SARS-CoV-2-ORF3b-IRES-Puro | Addgene | RRID:Addgene_141384 |
| pLVX-EF1alpha-SARS-CoV-2-ENV-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141385 |
| pLVX-EF1alpha-SARS-CoV-2-MEM-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141386 |
| pLVX-EF1alpha-SARS-CoV-2-ORF6-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141387 |
| pLVX-EF1alpha-SARS-CoV-2-ORF7a-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141388 |
| pLXV-EF1alpha-2xStrep-SARS-CoV-2-ORF7b-IRES-Puro | Addgene | RRID:Addgene_141389 |
| pLVX-EF1alpha-SARS-CoV-2-ORF8-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141390 |
| pLVX-EF1alpha-SARS-CoV-2-NUC-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141391 |
| pLVX-EF1alpha-SARS-CoV-2-ORF10-2xStrep-IRES-Puro | Addgene | RRID:Addgene_141394 |
| pcDNA3-N-Flag-NLRP3 | Addgene | RRID:Addgene_75127 |
| pcDNA3-N-Flag-NLRP3 1-90 | Addgene | RRID:Addgene_75137 |
| pcDNA3-N-Flag-NLRP3 91-710 | Addgene | RRID:Addgene_75140 |
| pcDNA3-N-Flag-NLRP3 711-1034 | Addgene | RRID:Addgene_75141 |
| Software and algorithms | ||
| Outcyte 1.0 | http://www.outcyte.com/analyse/ | |
| SignaIP 5.0 | https://services.healthtech.dtu.dk/services/SignalP-5.0/ | |
| NetNGlyc 1.0 | https://services.healthtech.dtu.dk/services/NetNGlyc-1.0/ | |
| NetOGlyc 4.0 | https://services.healthtech.dtu.dk/services/NetOGlyc-4.0/ | |
| TMHMM 2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ | |
| Flowjo version 10.8.1 | https://www.flowjo.com/ | |
| GraphPad Prism version 8.0 | https://www.graphpad.com/ | |
| JMP version 14.0 | https://www.jmp.com/ | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Xiaosheng Wu (wu.xiaosheng@mayo.edu).
Materials availability
In this study, we produced ORF8 protein in both HEK293 cells and in E. coli, and purified it to homogeneity for assessing its cytokine induction potential. The availability of these unique reagents is very limited. However, we are more than happy to provide technical support if you need assistance with creating the proteins yourself.
Experimental model and subject details
Study samples
Human serum/plasma samples were collected from Mayo Clinic patients with a documented diagnosis of COVID-19 infection and consented to COVID-19 Research Task Force Specimen Biobank. Samples-used in this study were requested and approved by the Task Force Review Committee and the Mayo Clinic Institutional Review Board (IRB). Deidentified fresh leukocyte cones from healthy donors were obtained from the Mayo Clinic Blood Bank. The use of mouse splenic B cells from C57/B6 mice was approved by the Institutional Animal Care and Use Committee (IACUC) of the Mayo Clinic.
Method details
SARS-CoV-2 protein expression in human cells
Lentiviral constructs expressing SARS-CoV-2 proteins originally described by Gordon, et al.16 were purchased from Addgene (key resources table) and used to make stable expression cell lines in HEK293 cells by lentiviral transduction. Transient expression of ORF8 construct DNA in HEK293F cells with the HyCell TransFx media (Cytiva) method for ORF8 protein purification. Initially, we used the conditioned media (CM) (culture supernatants dialyzed twice against sterile PBS and once against RPMI) for PBMC stimulation at 20% (v/v) for 72 hours. It was later replaced with purified ORF8 glycoprotein at 200 ng/ml for 24 hours of incubation based on our titration data (Figure S5).
Detection of SARS-CoV-2 protein secretion
To detect protein secretion, HEK293 cells expressing the protein of interest were seeded at 70% confluency in a T25 flask in 4.0 ml of DMEM/10% FCS. After 48 hours of culture, the supernatants were collected and cleared for cells and debris by centrifugation at 3000 rpm for 5 min. Four mL of cleared supernatants were then incubated overnight with 40μl of 50% slurry of StrepTactin (or anti-HA for SPK protein) beads. The beads were then washed twice with PBS, then resuspended in 40 μl Laemmli sample buffer with 5% BME. The amount of sample equivalent to 1ml of culture supernatant was loaded into a 4-20% gradient gel followed by western blotting per routine probing with anti-Strep II antibody at 1:1000 dilution (#71590, Millipore) and imaged on a Li-COR scanner.
Cell stimulation and cytokine detection
Fresh PBMCs (3x106) isolated from healthy donors were stimulated with either 20% (v/v) of CM-ORF8 or 200ng/ml of pure ORF8 for 24 hours in a total volume of 1.5ml RPMI/10% FCS in a 12-well plate. Cytokine mRNA expression was determined by SYBR-green-based qPCR method using the primers listed in the key resources table, and housekeeping gene β-Actin or GAPDH was used for normalization. By comparing with cytokine protein detection in the culture supernatants by Luminex assay using premade Luminex detection kit for IL1β, IL6, IL8, and TNFa) (R&D systems) or a custom Luminex cytokine detection kit for IL1β, IL6, IL8, IL18, CCL2, and TNFα (ThermoFisher), we found that the qPCR method was more sensitive (Figure S6). We also performed intracellular cytokines detection by flow cytometry (see below for method details).
Intracellular cytokine detection by flowcytometry
Two million PBMCs were activated with ORF8 or LPS for 18.5 hours, and Golgi-Plug (Brefeldin A, Cat # 51-2301KZ, BD Pharmingen) was added at 1:1000 for additional 5.5 hours of incubation. The cells were then collected and stained with cell lineage surface markers (CD3, CD19, CD14, CD16, CD11c, CD56, HLA-Dr) and FVD for live/dead gating followed by staining intracellular IL1β, IL8, IL6, and CCL2 upon fix and permeabilization using a BD Cytofix/Cytoperm fixation/Permeabilization Kit (Cat # 554714). All antibodies used are listed in key resources table. The flow cytometry 13-color detections were run on an LSRFortessa X-20 (BD). The Data were analyzed using Flowjo software V10.
Single-cell RNA sequencing of ORF8 treated PBMCs
Three million PMBCs were treated with either media control- or ORF8-CM for three days; cells were collected and enriched for viable cells to greater than 90% using the dead cell removing kit (Miltenyi Biotec, Cat#130-090-101), then submitted for cell capturing, and single-cell RNA sequencing. The cell suspension and Chromium Single Cell 3’ v2 library master mix (10x Genomics), gel beads, and partitioning oil were added to a Chromium Single Cell A chip which was then loaded into the Chromium Controller for capturing 4000 single cells and subsequent cDNA synthesis. The resulting single-cell cDNA libraries were pooled and sequenced as 100x2 paired-end reads on an Illumina HiSeq 4000 using HiSeq 3000/4000 sequencing kit.
Single-cell RNA sequencing data analysis
The Seurat package (v4.0.1) was used to perform integrated analyses of single cells. Genes expressed in less than 3 cells, cells expressed less than 200 genes, and more than 40% of the genes were mitochondrial were excluded for downstream analysis in each sample. For scRNA-Seq analysis, we followed the Seurat integration workflow and the comparative analysis workflow. Each dataset was normalized and the top 2000 Highly Variable Genes (HVGs) across cells were selected. The datasets were integrated based on “anchors” identified between datasets before Principal Component Analysis was performed to do a linear dimensional reduction. Shared Nearest Neighbor Graph was constructed to identify clusters in the low-dimensional space (top 50 statistically significant principal components). Enriched marker genes in each cluster conserved across all samples were identified, and differentially expressed genes between two selected conditions were detected using the default Wilcoxon Rank Sum test at the cluster level. Both GSEA and KEGG pathway enrichment analyses were performed.
ORF8 expression and purification from HEK293 cells
Transfected HEK293F cells were grown for 3-4 days at a cell density below 6x106 cells per ml. Cells were pelleted by centrifugation (200 x g for 5 min, 23°C) and the supernatant containing secreted ORF8 was supplemented with a Complete-EDTA free protease inhibitor cocktail (Roche), 1mM BME, and 1 mL of biotin blocking solution (BioLock). The media were loaded on a 5 mL StrepTactin Sepharose High-Performance column (GE Healthcare) in PBS buffer and eluted with a linear gradient of 0-50% 5 mM Biotin in PBS. Fractions containing ORF8 were pooled, concentrated by ultrafiltration (Amicon), and then purified using a Superdex 200 Increase 10/300 GL (Cytiva) in PBS, followed by anion exchange chromatography using a Source 15Q 4.6/10 column (Cytiva) with a gradient of 0–500 mM NaCl in 20mM Tris pH 7.5. Fractions containing ORF8 were pooled and concentrated by ultrafiltration and stored at 4°C.
ORF8 expression and purification from E. Coli
DNA encoding SARS CoV-2 ORF8 codon-optimized for expression in E. coli was cloned into pMCSG740 using Stbl3 cells (EMD) and transformed into Rosetta2 cells (EMD), and protein expression was induced with 100 μΜ IPTG overnight at 15°C. E. coli culture was pelleted, lysed, and protein purified as described previously,41 except that cells were lysed by sonication with a Branson sonicator in place of Dounce homogenization. The concentrated, refolded protein was loaded on a Superdex S75 size-exclusion column equilibrated in PBS. Fractions containing ORF8 were pooled and concentrated by ultrafiltration (Amicon). The His-tag was removed by incubation with TEV protease. Untagged ORF8 was run on a Source 15Q 4.6/10 anion exchange column (Cytiva) equilibrated in wash buffer (20 mM Tris pH 7.5) and eluted with a linear gradient of 0-100% 500 mM NaCl in 20 mM Tris pH 7.5. Monomer and dimer peaks were pooled and concentrated.
Immunofluorescence staining
For intracellular localization of SARS-CoV2 proteins (Figure S1), HeLa cells, seeded on a 12-well chamber slide (Erie Scientific) at 70% confluency, transiently transfected with various SARS-CoV2 protein constructs using lipofectamine 2000 for 36 hours. The cells were then fixed with 4% paraformaldehyde (PFA) at room temperature for 10 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes and blocked for 2 hours with the blocking buffer (3% fat-free milk in PBS supplemented with 0.02% Triton X-100, insoluble debris was cleared by centrifugation at 14,000 rpm for 15 minutes) at room temperature. The cells were then incubated with mouse monoclonal anti-HA antibody at 1:100 in the blocking buffer for the HA-tagged Spike protein, or anti-Strep Tag II antibody at 1:100 in the blocking buffer for all other Strep II-tagged SARS-Cov2 proteins at 4°C overnight. After washing with the blocking buffer five times, the cells were incubated with anti-mouse Ig conjugated with Alexa 565 at 1:500 in the blocking buffer for 1 hour at room temperature, again washed twice with the blocking buffer and three times with PBS, then refixed with 4% PFA for 5 minutes and washed twice with water. The slides were mounted with DAPI-containing vectorshield and imaged on a Zeiss 780 confocal microscope.
For detection of apoptosis-associated speck-like protein containing CARD (ASC) specks (Figure 5C) in THP-1 cells, we first treated Thp-1 cells with ORF8 (200 ng/ml) or LPS (100 ng/ml) for 2.5 hours followed by adding 0 or 5 μg/ml Nigericin for an additional 1.5 hours, the cells amounted to slides using cytospin, fixed with 4% PFA for 10 min, and stained for ASC and NLRP3 with respective antibodies both at 1:100 dilution and performed as described above.
Blood ORF8 detection
For detection of ORF8 protein in serum samples from patients newly infected with SARS-CoV-2, four microliters of serum were directly mixed with 36 microliters of 2× Laemmli buffer with 5% of BME, boiled for 3 min before load to 4-15% SDS-PAGE gels, the blots were probed with 1:5000 diluted rabbit anti-ORF8 antibody (MyBioSource, Cat# MBS3014575) at 4°C overnight. The images were developed using an ECL reagent kit. The same positive and negative controls were included in each gel for batch normalization.
Quantification and statistical analysis
Data analysis
Cytokine data were analyzed using GraphPad prism version 8.0. Flowcytometry data analysis was done with Flowjo version 10.0, Immunofluorescence images were processed in Adobe Photoshop version 2021. Overall survival was calculated from the time of COVID-19 diagnosis, using the Kaplan-Meir method on JMP 14.0 software.
Acknowledgments
We thank The Mayo Clinic Genome Analysis Core, Bioinformatics Core, Microscopy and Cell Analysis Core, and the CMM Electron Microscopy Facility of the University of California San Diego (supported by NIH equipment grant 1S10OD023527-01) for their expert service, Mayo Clinical Blood Bank for providing fresh leukocyte cones of healthy donors, and the Mayo Clinic Center for Individualized Medicine for funding and access to their biobank data. We also thank the University of Iowa/Mayo Clinic Lymphoma SPORE (NIH/NCI grant P50 CA97274) and the Predolin Foundation Biobank for sharing some of their resources for this project.
Author contributions
X.W. and T.E.W. designed the experiments, analyzed, and interpreted results, and wrote the manuscript; X.W., M.K.M., G.J.R., K.E.N., K.A.G., X.T., T.L.W., V.T., A.W., and M.J.S. conducted experiments and analyzed data; X.W., J.P.A., Y.Y., X.T., S.D., and M.J.S. analyzed data and generated figures; A.D.B., J.P.A., G.R., J.P., S.M.A., V.B., H.D., and T.E.W. provided experiment samples; J.P.A., G.J.R., J.P., T.L.W., S.M.A., and M.J.S. discussed and interpreted results; all authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106929.
Contributor Information
Xiaosheng Wu, Email: wu.xiaosheng@mayo.edu.
Thomas E. Witzig, Email: witzig.thomas@mayo.edu.
Supplemental information
Data and code availability
-
•
All raw data in this publication will be shared upon request. The single cell RNA-seq data (SRA: PRJNA971080) presented in Figure 3 is available for public access.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.WHO World Health Organization - Covid Dashboard. 2023. https://covid19.who.int/ Last update: April 5, 2023.
- 2.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) isclosely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin R. COVID-19 and blood donation. JAMA. 2022;327:615. doi: 10.1001/jama.2022.0763. [DOI] [PubMed] [Google Scholar]
- 12.Andersson M.I., Arancibia-Carcamo C.V., Auckland K., Baillie J.K., Barnes E., Beneke T., Bibi S., Brooks T., Carroll M., Crook D., et al. SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus. Wellcome Open Res. 2020;5:181. doi: 10.12688/wellcomeopenres.16002.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappy P., Candotti D., Sauvage V., Lucas Q., Boizeau L., Gomez J., Enouf V., Chabli L., Pillonel J., Tiberghien P., et al. No evidence of SARS-CoV-2 transfusion transmission despite RNA detection in blood donors showing symptoms after donation. Blood. 2020;136:1888–1891. doi: 10.1182/blood.2020008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owusu M., Sylverken A.A., El-Duah P., Ayisi-Boateng N.K., Yeboah R., Adu E., Asamoah J., Frimpong M., Senyo J., Acheampong G., et al. Low risk of SARS-CoV-2 in blood transfusion. PLoS One. 2021;16:e0249069. doi: 10.1371/journal.pone.0249069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural characterization of SARS-CoV-2: where we are, and where we need to Be. Front. Mol. Biosci. 2020;7:605236. doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bäck N., Kanerva K., Kurutihalli V., Yanik A., Ikonen E., Mains R.E., Eipper B.A. The endocytic pathways of a secretory granule membrane protein in HEK293 cells: PAM and EGF traverse a dynamic multivesicular body network together. Eur. J. Cell Biol. 2017;96:407–417. doi: 10.1016/j.ejcb.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27:230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin K., Bardes E.E., Mitelpunkt A., Wang J.Y., Bhatnagar S., Sengupta S., Krummel D.P., Rothenberg M.E., Aronow B.J. Implicating gene and cell networks responsible for differential COVID-19 host responses via an interactive single cell web portal. bioRxiv. 2021 doi: 10.1101/2021.06.07.447287. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert C., Preijers F., Yanikkaya Demirel G., Sack U. Monocytes and macrophages in flow: an ESCCA initiative on advanced analyses of monocyte lineage using flow cytometry. Cytometry B Clin. Cytometry. 2017;92:180–188. doi: 10.1002/cyto.b.21280. [DOI] [PubMed] [Google Scholar]
- 23.Kapellos T.S., Bonaguro L., Gemund I., Reusch N., Saglam A., Hinkley E.R., Schultze J.L. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front. Immunol. 2019;10:2035. doi: 10.3389/fimmu.2019.02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H., Wang Y., Li X., Zhan X., Tang M., Fina M., Su L., Pratt D., Bu C.H., Hildebrand S., et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016;17:250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanoni I., Tan Y., Di Gioia M., Springstead J.R., Kagan J.C. By capturing inflammatory lipids released from dying cells, the receptor CD14 induces inflammasome-dependent phagocyte hyperactivation. Immunity. 2017;47:697–709.e3. doi: 10.1016/j.immuni.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evavold C.L., Ruan J., Tan Y., Xia S., Wu H., Kagan J.C. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity. 2018;48:35–44.e36. doi: 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roeker L.E., Knorr D.A., Thompson M.C., Nivar M., Lebowitz S., Peters N., Deonarine I., Jr., Momotaj S., Sharan S., Chanlatte V., et al. COVID-19 vaccine efficacy in patients with chronic lymphocytic leukemia. Leukemia. 2021;35:2703–2705. doi: 10.1038/s41375-021-01270-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberger L.M., Saltzman L.A., Senefeld J.W., Johnson P.W., DeGennaro L.J., Nichols G.L. Antibody response to SARS-CoV-2 vaccines in patients with hematologic malignancies. Cancer Cell. 2021;39:1031–1033. doi: 10.1016/j.ccell.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y., Schneider T., Leong M., Aravind L., Zhang D. Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of SARS-CoV-2-related viruses. mBio. 2020;11:e00760-20. doi: 10.1128/mBio.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notarbartolo S., Ranzani V., Bandera A., Gruarin P., Bevilacqua V., Putignano A.R., Gobbini A., Galeota E., Manara C., Bombaci M., et al. Integrated longitudinal immunophenotypic, transcriptional and repertoire analyses delineate immune responses in COVID-19 patients. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg5021. [DOI] [PubMed] [Google Scholar]
- 32.Junqueira C., Crespo Â., Ranjbar S., de Lacerda L.B., Lewandrowski M., Ingber J., Parry B., Ravid S., Clark S., Schrimpf M.R., et al. FcgammaR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;606:576–584. doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong Y.N., Tsao K.C., Hsiao M.J., Huang C.G., Huang P.N., Huang P.W., Lee K.M., Liu Y.C., Yang S.L., Kuo R.L., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microb. Infect. 2020;9:1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young B.E., Fong S.W., Chan Y.H., Mak T.M., Ang L.W., Anderson D.E., Lee C.Y.P., Amrun S.N., Lee B., Goh Y.S., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caricchio R., Abbate A., Gordeev I., Meng J., Hsue P.Y., Neogi T., Arduino R., Fomina D., Bogdanov R., Stepanenko T., et al. Effect of canakinumab vs placebo on survival without invasive mechanical ventilation in patients hospitalized with severe COVID-19: a randomized clinical trial. JAMA. 2021;326:230–239. doi: 10.1001/jama.2021.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyriazopoulou E., Huet T., Cavalli G., Gori A., Kyprianou M., Pickkers P., Eugen-Olsen J., Clerici M., Veas F., Chatellier G., et al. Effect of anakinra on mortality in patients with COVID-19: a systematic review and patient-level meta-analysis. Lancet Rheumatol. 2021;3:e690–e697. doi: 10.1016/S2665-9913(21)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyriazopoulou E., Poulakou G., Milionis H., Metallidis S., Adamis G., Tsiakos K., Fragkou A., Rapti A., Damoulari C., Fantoni M., et al. Early treatment of COVID-19 with anakinra guided by soluble urokinase plasminogen receptor plasma levels: a double-blind, randomized controlled phase 3 trial. Nat. Med. 2021;27:1752–1760. doi: 10.1038/s41591-021-01499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urban F.J. 1998. Efficent synthesis of furan sulfonamide compounds used in the synthesis of new IL-1 inhibitors. US6022984A. [Google Scholar]
- 39.Li H., Guan Y., Liang B., Ding P., Hou X., Wei W., Ma Y. Therapeutic potential of MCC950, a specific inhibitor of NLRP3 inflammasome. Eur. J. Pharmacol. 2022;928:175091. doi: 10.1016/j.ejphar.2022.175091. [DOI] [PubMed] [Google Scholar]
- 40.Stols L., Gu M., Dieckman L., Raffen R., Collart F.R., Donnelly M.I. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 41.Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X., Hurley J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021785118. e2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw data in this publication will be shared upon request. The single cell RNA-seq data (SRA: PRJNA971080) presented in Figure 3 is available for public access.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.