Significance
In mammals, lymph nodes play a critical role in the initiation of adaptive immune responses by providing a dedicated place for T cells to scan antigen-presenting cells. Birds, reptiles, amphibians, and fish all maintain diverse repertoires of T cells but lack lymph nodes, raising questions about how adaptive immunity functions in lower jawed vertebrates. Here, we describe a network of lymphocytes in zebrafish that supports whole-body T cell trafficking and provides a site for antigen search, mirroring the function of mammalian lymph nodes. Within this network, T cells can prioritize large-scale trafficking or antigen scanning by toggling between two distinct modes of migration. This network provides valuable insights into the evolution of adaptive immunity.
Keywords: antigen surveillance, motility, collective cell migration, streaming, lymphoid tissue
Abstract
Homeostatic trafficking to lymph nodes allows T cells to efficiently survey the host for cognate antigen. Nonmammalian jawed vertebrates lack lymph nodes but maintain diverse T cell pools. Here, we exploit in vivo imaging of transparent zebrafish to investigate how T cells organize and survey for antigen in an animal devoid of lymph nodes. We find that naïve-like T cells in zebrafish organize into a previously undescribed whole-body lymphoid network that supports streaming migration and coordinated trafficking through the host. This network has the cellular hallmarks of a mammalian lymph node, including naïve T cells and CCR7-ligand expressing nonhematopoietic cells, and facilitates rapid collective migration. During infection, T cells transition to a random walk that supports antigen-presenting cell interactions and subsequent activation. Our results reveal that T cells can toggle between collective migration and individual random walks to prioritize either large-scale trafficking or antigen search in situ. This lymphoid network thus facilitates whole-body T cell trafficking and antigen surveillance in the absence of a lymph node system.
Adaptive immunity relies on rare, naïve T cells finding cognate antigen that arise in the body’s many diverse tissues during an infection (1). In mammals, antigen discovery and subsequent T cell activation occur primarily in lymph nodes, regional organs that provide a specialized space for T cells to efficiently migrate and scan antigen-presenting cells (APCs) (1, 2). In this lymph node system, naïve T cells serially enter, search within, and exit lymph nodes that are distributed strategically throughout the host, allowing individual T cell clones to perform whole-body antigen surveillance on the timescale of days (1, 3).
While lymph nodes are largely restricted to mammals, lymphocytes with diverse antigen receptors are present in all jawed vertebrates (4, 5). It remains unclear how T cells in fish, reptiles, amphibians, and birds effectively survey for antigen in a host devoid of lymph nodes. Further, since diverse lymphocyte repertoires emerged before lymph nodes, adaptive immunity necessarily evolved a different surveillance strategy than the one operating in mammals (4, 5). While lymphocytes typically populate and even form lymphoid tissues around the respiratory and digestive organs of lower jawed vertebrates (6, 7), these animals produce robust adaptive immune responses when these barrier sites are bypassed by intraperitoneal or intramuscular immunization (8, 9). These data indicate that nonmammalian jawed vertebrates also perform whole-body antigen surveillance, but how T cells effectively search for antigen in the absence of lymph nodes remains unknown.
Studies in transparent zebrafish have previously uncovered new mechanisms of immune cell migration and behavior in situ (10–14), and this model organism provides a unique opportunity to investigate T cell surveillance in an organism lacking lymph nodes. Utilizing a combination of microscopy and single-cell RNA-sequencing (scRNA-seq), we identified a whole-body lymphoid tissue that encompasses the major cellular hallmarks of a mammalian lymph node. Live imaging revealed that T cells can toggle between two major modes of motility within this lymphoid tissue: one that uses collective T cell streaming to facilitate long-distance coordinated movement and a second more individual random walk that facilitates antigen-presenting (APC) cell scanning. We find that infection drives a switch from directed streaming to a diffusive random walk and provides evidence that T cells are detecting antigen, activating, and differentiating within this lymphoid network. Collectively, we show that zebrafish T cells utilize distinct modes of motility within this lymphoid network to perform whole-body antigen surveillance in the absence of lymph nodes.
Results
T Cells Organize into a Tessellated Lymphoid Network in Juvenile and Adult Zebrafish.
Prior studies have used larval-stage animals to study T cell motility; however, a functional adaptive immune system does not emerge until 4 to 6 wk postfertilization (wpf) (15, 16). We therefore investigated T cell distribution and trafficking in 10 wpf transparent zebrafish when the adaptive immune system is well established (16–18). Organism-scale imaging of tg(lck:GFP) revealed that T cells organize into a regularly repeating hexagonal pattern across the body of the animal (Fig. 1A). This hexagonal pattern was unique to the body, with T cells in the fins and head assuming a more diffuse distribution (Fig. 1A and SI Appendix, Fig. S1 A and B). Within this hexagonal pattern, GFP-labeled cells were particularly abundant at the dorsal and ventral poles, giving the network a zigzag appearance (Fig. 1A). This pattern is completely absent in the more well-studied larval zebrafish but becomes progressively apparent throughout the juvenile stages of development and is maintained into adulthood (Fig. 1B). Individual T cells are apparent in larval fish (SI Appendix, Fig. S1C), but they do not organize into this higher-order pattern (Fig. 1B). Notably, the emergence of this T cell pattern corresponds temporally with the maturation of adaptive immunity (16).
Fig. 1.
A tessellated lymphoid network supports coordinated whole-body T cell trafficking in juvenile and adult zebrafish. (A) The organism-scale organization of T cells in a 10 wpf tg(lck:GFP) zebrafish, with the inset showing the hexagonal "zigzag" pattern. (B) The emergence of the T cell network is shown with representative images of the caudal peduncle of tg(lck:GFP) zebrafish at 3, 5, 7, 10, and 15 wpf. (C) Imaging of the caudal peduncle shows the organization of T cells (lck:GFP), macrophages (mpeg1:mCherryCAAX), and neutrophils (lyzc:BFP) in triple transgenic zebrafish. (D and E) Imaging of the caudal peduncle at low (d) or high (e) magnification shows the presence of dual labeled cells in tg(lck:GFP;cd4-1:mCherry) fish. MΦ = macrophage, L.E. = leading edge, Ur. = uropod. (F) Timelapse imaging of T cells migrating through the TLN over 20 min, with the cell tracks and directions of eight representative GFP+ cells shown in magenta. (Scale bar = 100 microns.) (G) Plots show the cell tracks of GFP+ cells migrating in the TLN of tg(lck:GFP;cd4-1:mCherry) in the indicated regions of five individual fish pooled from three independent experiments. Each dot = average per fish, asterisks = average total movement. Scale is in microns. (H) A summary of the movement of T cells in the TLN.
To our knowledge, a network that organizes T cells into a repeating pattern has never been observed in any organism. To determine whether other leukocyte populations assume a similar distribution, we generated triple-transgenic lck:GFP;mpeg1:mCherryCAAX;lyzc:BFP zebrafish to visualize T cells, macrophages, and neutrophils simultaneously (18–20). These animals revealed that this zigzag pattern was unique to T cells, with both macrophages and neutrophils showing a more diffuse organization (Fig. 1C). Using double-transgenic lck:GFP;cd4-1:mCherry fish (21), we found mCherry-expressing T cells to be common within the network (Fig. 1 D and E) but infrequent in larval zebrafish (SI Appendix, Fig. S1C). GFP-negative, mCherry-expressing macrophages, described previously (21), are also apparent in proximity to the network (Fig. 1E). Reflecting its repeating pattern, we named this network the tessellated lymphoid network (TLN).
T Cells Traffic through the Host in a Coordinated Ventral-to-Dorsal Loop.
TLN T cells commonly assume the “hand-mirror” morphology typical of motile lymphocytes, prompting us to perform live imaging experiments (Fig. 1E, Inset). TLN T cells migrated in a remarkably coordinated and directional manner, with cells maintaining directional persistence over hundreds of microns as they zigzagged through the network (Fig. 1F and Movie S1). To understand this movement on the organism scale, we performed live imaging in four discrete positions: dorsal anterior, dorsal posterior, ventral anterior, and ventral posterior (Fig. 1G). Remarkably, T cells showed clear rostral movement on the dorsal side and clear caudal movement on the ventral side (Fig. 1 G and H). The TLN is punctuated on the rostral and caudal ends of the body by large reservoirs of T cells (SI Appendix, Fig. S1 D and E). Live imaging is technically challenging at the rostral reservoir due to the presence of the operculum and pectoral fin, but within the caudal reservoir, we observed clear T cell movement from the ventral TLN to the dorsal TLN (SI Appendix, Fig. S1 F–I). These data collectively suggest that the ventral and dorsal portions of the TLN are continuous, allowing T cells to traffic through the host in a coordinated ventral-to-dorsal loop (Fig. 1H).
The TLN Localizes to a Pocket Formed by Overlapping Scales and Is the Largest Reservoir of T Cells in Zebrafish.
Fish scales organize in a hexagonal pattern, like the TLN (22). To determine whether the TLN associates with fish scales, we imaged adult tg(lck:GFP) zebrafish stained with alizarin red and found the TLN and scale pattern to be interrelated (Fig. 2A). Single z-section images showed that the TLN localizes to narrow regions formed between adjacent scales (Fig. 2B), positioning this network just underneath the barrier surface of the animal (Fig. 2C). Given this localization, we next investigated whether TLN T cells could be isolated by descaling adult zebrafish (Fig. 2 D and E). Descaling resulted in an almost complete loss of T cells from the TLN without causing loss of T cells from the nearby fins (Fig. 2 D and E). When descaling was performed on adult tg(lck:GFP) zebrafish in isotonic buffer, GFP-expressing cells from the TLN appeared as a suspension with GFP-negative cells (Fig. 2F), and flow cytometric analysis of this suspension revealed that the GFP-positive cells were live and localized to the lymphocyte gate as expected (SI Appendix, Fig. S2 A and B). We characterized the TLN by flow cytometry and compared it to other known reservoirs of T cells in fish: the gills, gut, spleen, and kidney (a major hematopoietic organ in fish) (Fig. 2G) (6, 7, 21). The TLN, gills, and gut all had a similar proportion of GFP-positive cells (Fig. 2G), but the TLN harbored more total live cells than all sites tested besides the kidney (Fig. 2H). In total, we harvested nearly 1x105 GFP-positive cells from the TLN of each 15 wpf tg(lck:GFP) fish, more than all other sites tested (Fig. 2I). While the spleen is considered the primordial secondary lymphoid organ (23), the presence and organization of white pulp is highly variable between teleost species and in zebrafish appears primarily composed of macrophages and reticular cells (24, 25). In accordance, we found a paucity of GFP-expressing cells in the spleen of adult tg(lck-GFP) zebrafish (Fig. 2I). Using tg(lck:GFP;cd4-1:mCherry) zebrafish, we found that approximately 66% of the TLN T cells also expressed cd4-1 (SI Appendix, Fig. S2C). Other groups have estimated that adult zebrafish have 2 × 105 total T cells (26), suggesting that this lymphoid network is the largest reservoir of T cells in the host.
Fig. 2.

The TLN is the largest reservoir of zebrafish T cells and shares the cellular hallmarks of a mammalian lymph node. (A) The relationship between the T cell pattern and scales shown by imaging the caudal peduncle of tg(lck:GFP) fish stained with Alizarin red. (B) The tissue localization of the T cell network shown by imaging a single z-plane of the caudal peduncle of a tg(lck:GFP) fish, with the yellow transparent overlays emphasizing the scales. (C) Imaging of a tg(lck:GFP) zebrafish stained with Alizarin red shows T cells (white arrows) in a pocket formed by overlapping scales. (D and E) Imaging of control and descaled tg(lck:GFP) adult zebrafish stained with alizarin shows the loss of the TLN T cells upon descaling (yellow inset) but not fin T cells (blue inset). (F) After descaling tg(lck:GFP) zebrafish, the buffer contains a single-cell suspension of GFP-expressing (arrows) and GFP-negative cells. (Scale bar = 10 microns.) (G–I) Representative flow plots (G) and enumeration of live (H) and GFP-expressing (I) cells harvested from the indicated organ of tg(lck:GFP;cd4-1:mCherry) 15 wpf zebrafish, pregated on live singlets, with the frequency of GFP-expressing cells shown. (E) Each dot corresponds to a single fish showing five to seven individuals pooled from four separate experiments. The mean of the TLN was compared to the mean of every other group with an ordinary one-way ANOVA. (J) RNA-seq profiling for all cells collected by descaling adult zebrafish, with leukocyte populations outlined in gray. (K and L) Representative gene expression in each of the major cell types as a dot plot showing select chemokine receptor (K) and chemokine (L) expression.
The TLN Harbors Naïve-Like T Cells and Antigen-Presenting Cells.
We reasoned that if the TLN was supporting antigen surveillance, it must harbor naïve-like T cells as well as antigen-presenting cells. To determine whether these cell populations exist within the TLN, we performed scRNA-seq on the cells harvested by descaling. Leukocytes made up the majority of identified cells, with T cells making up the most abundant cell type (Fig. 2J and SI Appendix, Fig. S2D). Based on the expression pattern of lineage-defining genes, we also identified all three of the major APC populations (macrophages, dendritic-like cells, and B cells) as well as a variety of other leukocytes (Fig. 2J and SI Appendix, Fig. S2 D and E). Further clustering of the T cells revealed subpopulations, including two large clusters of naïve-like T cells that expressed homeostatic chemokine receptors (ccr7, cxcr4a) but showed little-to-no expression of various cytokines (il4, il13, il11b), effector proteins (gzma, nkl1.2), or markers of proliferation (mki67, top2a) (SI Appendix, Fig. S2 F and G). We also identified a regulatory foxp3a-expressing T cell population, a small population of T cells exhibiting constitutive expression of type-2 cytokines (il4, il13, il11b; Th2/ILC2 cell), two discrete populations with gene signatures corresponding to cytotoxic T cells (cytotoxic T cell-1 and 2), a population expressing proliferation markers (mki67, top2a; cycling T cell), and various other minor T cell populations (SI Appendix, Fig. S2 F and G). The TLN thus harbors both naïve-like T cells and APCs, matching the cellular hallmarks of a lymph node.
A Chemokine-Expressing Intermediate Epithelial Cell Population Likely Represents the Putative Cellular Scaffold for TLN T Cells.
We next probed our scRNA-seq dataset to investigate how TLN T cell organization and motility is regulated. Given their coordinated movement, we initially considered that these T cells might be within or associated with either lymphatic or blood vessels. However, we did not identify any blood or lymphatic endothelial cell populations in our scRNA-seq dataset (Fig. 2J and SI Appendix, Fig. S2E). By generating double-transgenic cd4-1:mCherry;mrc1a:GFP fish, we also found no obvious relationship between the TLN and lymphatic vessels (SI Appendix, Fig. S3A)(27). Even in regions where the TLN and lymphatics appeared to overlap in z-projections, we found that the vessels were deeper in the tissue (SI Appendix, Fig. S3A). Using double-transgenic lck:GFP;kdrl:mCherry fish (28), we also found no clear relationship between the TLN and blood vasculature (SI Appendix, Fig. S3B). Some blood vessels are in the same z-region as the TLN, but TLN T cell organization and motility do not appear to be associated with these vessels (SI Appendix, Fig. S3B). These data suggest that TLN T cells are not associated with blood or lymphatic vessels; however, we cannot rule out their association with a different vessel-like structure.
In mammalian lymph nodes, T cell organization and migration is largely regulated by their expression of CCR7 and the expression of the corresponding chemokines (ccl19, ccl21) by fibroblastic reticular cells (29, 30). We found that the chemokine receptor expression profile for leukocytes in the TLN was highly similar to lymph nodes (Fig. 2K) (30). All leukocytes expressed at least one of the two CXCR4 orthologs (cxcr4a, cxcr4b) (Fig. 2K). Expression of ccr7 was only detected in T cells, and this expression was concentrated in the naïve-like populations (Fig. 2K and SI Appendix, Fig. S2 G and H). Zebrafish lack a ccl21 ortholog, but ccl25a and ccl25b signal through both ccr7 and ccr9 in fish (31). We found that one of the two zebrafish orthologs of CCR9, ccr9a, was broadly expressed by all leukocytes, including most of the T cell subsets (Fig. 2K and SI Appendix, Fig. S2 I and J). We next probed our scRNA-seq dataset for expression for the corresponding ligands of CXCR4 (cxc12a, cxcl12b), CCR7 (ccl19a.1, ccl19a.2, ccl19b, ccl25a, ccl25b), and CCR9 (ccl25a, ccl25b). As expected, expression of the relevant chemokines was virtually undetectable in all the hematopoietic cell populations (Fig. 2L). Intermediate epithelial cells, a poorly understood population of cells in the fish skin, expressed high levels of cxcl12a as well as all the known CCR7 and CCR9 ligands except for ccl19a.1 (Fig. 2L). Although basal epithelial cells and mesenchymal cells also showed appreciable expression of ccl19a.2 and ccl25b respectively, intermediate epithelial cells appeared to be the primary source of CXCR4, CCR7, and CCR9 ligands in the TLN (Fig. 2L). If intermediate epithelial cells serve as an organizational and migratory scaffold for TLN T cells, they should assume a similar whole-body scale-associated pattern. While no reporter fish presently exist for these cells or for any CCR7 or CCR9 ligand, our data indicate that these intermediate epithelial cells express cxcl12a (Fig. 2L), for which a reporter exists. Adult tg(cxcl12a:DsRed2) zebrafish show a body-spanning fluorescence pattern that appears interrelated with the scale pattern, similar to the TLN (32). While further research is needed, these data suggest that intermediate epithelial cells are a key regulatory cell population for TLN T cells, mirroring the function of lymph node fibroblastic reticular cells.
TLN T Cells Use Directed Collective Migration.
The TLN maintains all the cellular components that permit effective antigen surveillance in mammalian lymph nodes, so we next investigated TLN T cell search strategy and migration using real-time imaging with tg(lck:GFP;cd4-1:mCherry) fish. In contrast to the random walk exhibited by T cells in lymph nodes, TLN T cells moved in a remarkably coordinated manner, with nearly every T cell migrating in the same direction (Fig. 3A and Movie S2). TLN T cells migrated at an average velocity of 14.2 microns/min and with consistently high directionality (Fig. 3 B and C). We additionally found that TLN T cell motility shows a linear relationship with displacement rather than mean squared displacement (MSD), indicating that these cells are utilizing a superdiffusive mode of migration (Fig. 3D and SI Appendix, Fig. S3C). To further characterize this behavior, we fit curves of MSD versus time to a power law function (see fit statistics in SI Appendix, Fig. S3D). On log–log axes, the slope (α) can be used to differentiate between Brownian random walks (α = 1), superdiffusive motility (1 2), and straight-line directed (ballistic) motility (α = 2) (33). Across multiple fish, we found that α = 1.9, indicating that TLN T cell motility is superdiffusive and highly directed, approaching the ballistic mode (Fig. 3E and SI Appendix, Fig. S3 D and E). An α value this high suggests that TLN T cell motility is different from T cells that utilize Lévy walks for antigen search in vivo (where α ~ 1.4) (34). This motility is also significantly more directional than how T cells migrate in larval zebrafish prior to the development of the TLN (15).
Fig. 3.

TLN T cells use ballistic multicellular streaming to migrate. (A) Timelapse imaging of the TLN in adult tg(lck:GFP;cd4-1:mCherry) zebrafish shows the directional migration of 3 outlined T cells. (B and C) Average T cell velocity (B) and directionality (C) of T cells within the TLN, with each dot corresponding to the average of all T cells from an individual fish pooled from three independent experiments. (D) A plot shows the average displacement vs time of T cells migrating in the TLN. (E) Cell motility is characterized with a log–log plot of mean squared displacement (MSD) versus time where MSD ∝ tα. The mean α for the 5 fish of 1.9022 indicates superdiffusive motion near the ballistic regime. (F) Imaging shows the head-to-tail interaction between two T cells. mm:ss, scale = 10 microns. (G) Timelapse imaging shows five outlined T cells in a tg(lck:GFP;cd4-1:mCherry) zebrafish forming, breaking, and reforming head-to-tail interactions as they migrate through the TLN of a 6-WPF juvenile fish, with the final frame showing the cell tracks. (H and I) Imaging of the TLN in tg(lck:GFP;mpeg1:mCherry) shows T cells organizing into a fibrous network with embedded macrophages as TLN goes from maturing (H) to fully mature (I) in late juvenile or adult zebrafish.
The coordinated and directed nature of this movement may be explained by the prominent head-to-tail interactions of TLN T cells, as observed in the contacts between the leading edge and uropod of mCherry-positive and mCherry-negative T cells in tg(lck:GFP;cd4-1:mCherry) fish (Fig. 3F). In younger juvenile fish where the TLN is sparsely populated, T cells can be seen forming, breaking, and reforming these associations multiple times as they migrate in multicellular units (Fig. 3G). As the TLN matures, these interactions become stable and T cells form a continuum that resembles a flowing fibrous network (Fig. 3 H and I and Movies S1 and S3). This stable multicellular streaming resembles Dictyostelium discoideum streaming motility (35) and allows these T cells to migrate collectively, which explains how T cells maintain directional persistence over hundreds of microns within the TLN (Fig. 1F and Movie S1).
A Switch from Directed Collective Migration to Individual Random Walks Occurs during Infection.
While directed streaming motility is well suited for trafficking through the host, it is less appropriate for antigen search. Macrophages are stably embedded within the TLN, but streaming T cells contact these APCs only transiently (Fig. 3 H and I and Movie S3). We hypothesized that this behavior may change during infections. To this end, we developed a localized bacterial infection model by puncturing the dorsal side of the caudal peduncle with a needle and incubating the wounded fish for 2 h in a bath of mCherry-expressing L. monocytogenes (SI Appendix, Fig. S4A). The infection expands and resolves over a 7-d timespan in most fish, with TLN T cells accumulating in regions proximal to the infected wound between 1 and 5 d postinfection (dpi; SI Appendix, Fig. S4 B and C and Fig. 4 A and B).
Fig. 4.
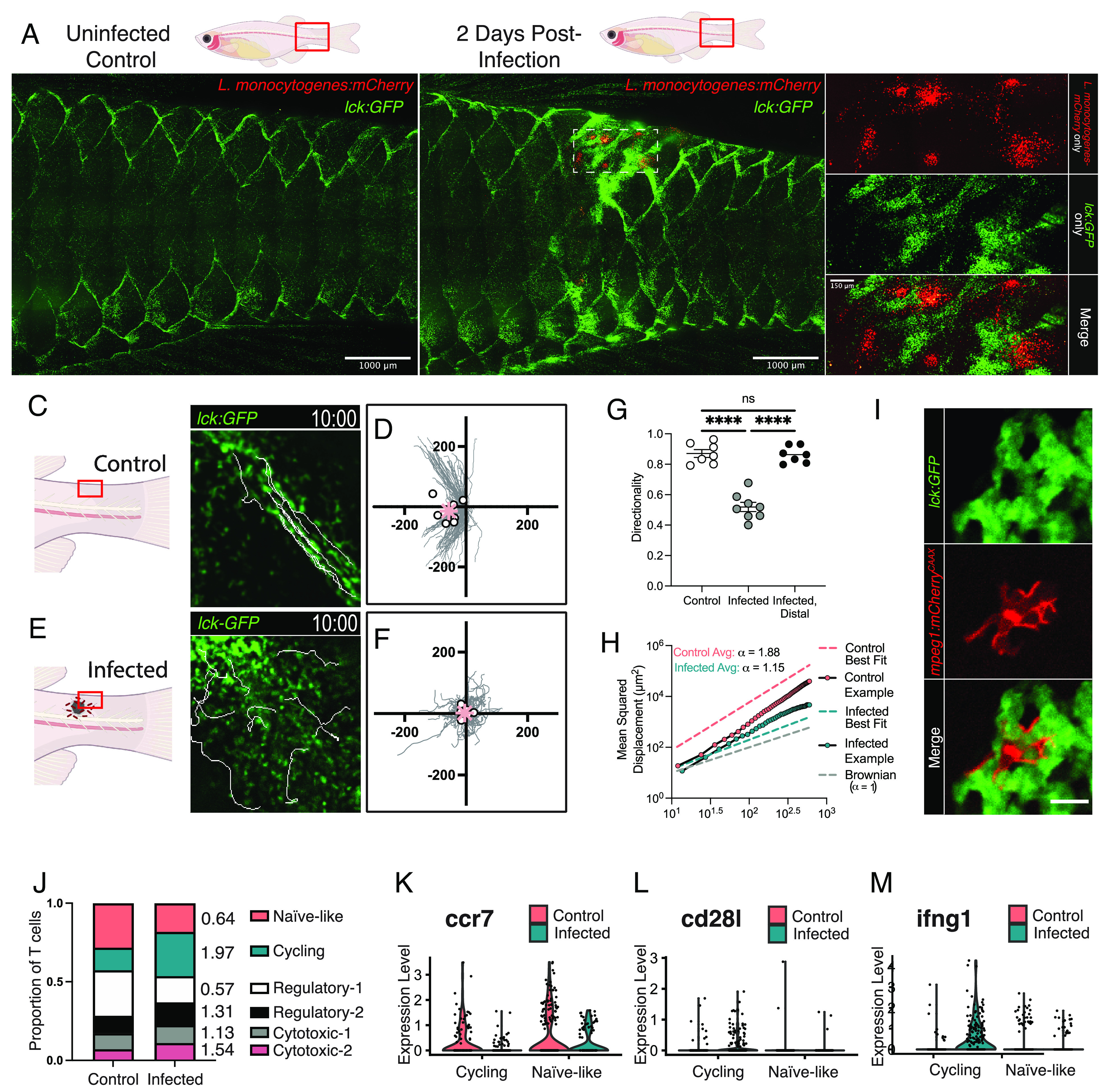
TLN T cells transition to a random walk during infection to scan APCs. (A and B) T cells accumulate around infections in tg(lck:GFP) zebrafish at 2 dpi with mCherry-expressing L. monocytogenes. (C and E) Altered T cell motility at 2 dpi (E) compared to control (C), with 10 overlaid cell tracks. (D and F) Ten-minute cell tracks from 7 to 8 uninfected (d) or 2 dpi (f) tg(lck:GFP;cd4-1:mCherry) fish pooled from three independent experiments. Dots = average movement per fish, asterisk = average total movement. The scale is in microns. (G) Infected-proximal but not infected-distal T cells exhibit reduced directionality, with each dot corresponding to the average of an individual fish pooled from three independent experiments. Means compared with an ordinary one-way ANOVA. (H) Representative plots of MSD versus time indicate departure from the superdiffusive motion characteristic of noninfected fish toward more Brownian-like diffusion in proximal T cells of infected fish at 2 dpi. Values of α were determined as an average of five fish for each condition (control vs. infection); however, plots only depict representative examples from a single control and single infected fish. Model-fitting statistics for all fish is in SI Appendix, Fig. S4E. (I) T cells associate with infection-proximal macrophages in tg(lck:GFP;mpeg1-mcherryCAAX) fish. (Scale bar = 10 microns (J) Proportions of T cell subtypes in control and infected conditions with fold-change noted. (K–M) Violin plots show the expression levels of ccr7 (K), cd28l (L), and ifng1 (M), in cycling and naïve-like T cells harvested by descaling control or infected fish.
While TLN T cells in control fish are highly motile, T cells around the infected wound showed both motile and sessile behaviors (Movie S4). Motile T cells proximal to the infection did not exhibit directed streaming and instead migrated as single cells in apparently random directions with reduced directionality (Fig. 4 C–G and Movie S5). This effect was limited to infection-proximal T cells, as TLN T cells distal to the infection (>500 microns) maintained a highly directional mode of migration (Fig. 4G). Infection-proximal TLN T cells also showed a sublinear relationship between displacement and time, indicating a more diffusive mode of migration (SI Appendix, Fig. S4D). We further characterized this behavior utilizing α values from MSD log–log plots as in Fig. 3D. Control cells continued to exhibit nearly ballistic movement (α = 1.88), but this value dropped considerably for infection-proximal TLN T cells (α = 1.15) and was not statistically different from α = 1 (Fig. 4H and SI Appendix, Fig. S4 E and F). These data indicate that infection-proximal T cells are now utilizing a diffusive random walk. Infection-proximal T cells often interacted sequentially with multiple macrophages over the course of these walks, similar to how T cells search for antigen in mammalian lymph nodes (Movie S6)(2, 36).
T Cell Activation and Differentiation within the TLN during Infections.
We noticed that the sessile population of T cells proximal to the infection were often in intimate, stable contact with regional macrophages (Fig. 4I). Signaling through the T cell receptor (TCR) acts as a migratory stop signal, which facilitates a stable interaction with APCs that is important for T cell proliferation and differentiation (37). To determine whether TLN T cells were activated, we performed single-cell analysis of the TLN harvested from infected fish. We observed several transcriptional changes consistent with T cell activation after infection. In our integrated analysis, T cells segregated into 6 discrete subsets: a naïve-like population, two foxp3a-expressing regulatory T cell populations, two cytotoxic T cell populations, and a cycling T cell population enriched for proliferation markers (mki67, top2a; SI Appendix, Fig. S4 G and H and Fig. 4J). Consistent with T cells becoming activated within the TLN, the proportion of naïve-like T cells decreased, while the cycling T cells and cytotoxic T cell-2 population increased (Fig. 4J). Mirroring the TCR-dependent transcriptional changes found in mammalian T cells, ccr7 expression was decreased and cd28l (ortholog of CTLA-4) was increased in T cells harvested from infected fish, especially within the cycling population (Fig. 4 K and L and SI Appendix, Fig. S4H) (38, 39). In mice, L. monocytogenes is a Th1-skewing infection, with activated T cells up-regulating type 1 effector genes such as ifng (40–42). Likewise, cycling T cells harvested from infected fish had higher expression of ifng compared to cycling T cells from control fish or naïve-like T cells from either group (Fig. 4M and SI Appendix, Fig. S4H). These data indicate that T cells are both activating and differentiating within the TLN during infection.
Discussion
In summary, we have identified a regularly repeating pattern of T cells in zebrafish that we have termed the tessellated lymphoid network. Akin to mammalian lymph nodes, the TLN harbors ccr7-expressing naïve-like T cells, antigen-presenting cells, and a nonhematopoietic epithelial cell population producing CCR7 and other chemokine receptor ligands. Within the TLN, T cells use a directional streaming mode of motility that allows them to traffic through the TLN in an efficient and coordinated ventral-to-dorsal loop. In the context of infection, TLN T cells transition to a diffusive random walk that facilitates interactions with infection-proximal APCs. Finally, scRNA-seq data provide evidence that T cells are detecting antigen, activating, and differentiating within this network. The TLN thus utilizes a different approach to solve the same problem addressed by the lymph node system in mammals: Antigen may arise in distant tissues during an infection, and adaptive immune responses rely upon antigen-specific T cells finding it. By toggling between two modes of migration, directional multicellular streaming or individual random walks, TLN T cells can prioritize either large-scale trafficking or local antigen search.
The degree to which multicellular streaming or collective migration may occur in mammalian lymph nodes is unclear, but it is supported by modeling and direct observation (43). Collective migration is difficult to study with intravital imaging in mammals because only a small fraction of T cells are labeled in these experiments. However, several reports have demonstrated that lymphocytes clearly possess the ability to alternate between Brownian motion and directed migration (44–46). Our directed T cell migration strongly resembles that described by Chauveau et al. for T cells undergoing unidirectional migration on splenic stromal cells to enter the T cell zones in a CCR7-dependent manner (47). The emerging consensus between this report and that of previous literature is that random walks are useful for scanning for antigen, and collective directed migration is well suited for traveling long distances within a tissue.
Our data are consistent with CCR7 playing a central role in directional streaming, as leukocytes lacking CCR7 expression (macrophages, neutrophils) do not organize into the TLN and do not stream when within the network (Fig. 1C and Movie S3). The notable expression of ccr9a by TLN T cells and the corresponding chemokines by intermediate epithelial cells is surprising because in mammals this receptor is primarily expressed in thymocytes and T cells in the small intestine where the corresponding ligands are produced by epithelial cells (29, 30, 48). It will therefore be important to dissect the relative importance of both CCR7 and CCR9a in TLN organization and migration.
The widespread use of directed streaming under homeostatic conditions indicates that TLN T cells are not thoroughly searching for antigen unless there is active inflammation. Lymph node T cells also alter their search strategy depending on inflammation, with some T cells exiting uninflamed lymph nodes within minutes of entering, searching for antigen minimally if at all (49). Grigorova et al. (49) have proposed a hierarchical model of antigen surveillance in lymph nodes where T cells initially survey for regional inflammation and then secondarily search for antigen. This model appears to also describe antigen surveillance in the TLN. Within this framework, streaming may help limit T cell reactivity to self-antigens by restricting antigen surveillance to scenarios where cognate antigen is likely to be found. This model also suggests that T cell search in most settings is deliberately suboptimal and warrants careful comparisons of T cell motility in control and inflamed settings.
While we anticipate that a TLN-like system is present in all teleost fish, the ventral-to-dorsal loop that we describe here would likely be inefficient for larger fish, potentially occurring on the timescale of months rather than days. This trafficking program would be consistent with the reduced diversity of the T cell repertoire found in fish as compared to mammals (26, 50). While further research is warranted on the subject, we hypothesize that increased T cell receptor diversity evolved in jawed vertebrates as more efficient antigen surveillance strategies developed in the lymph node system.
How far a TLN-like system extends through jawed vertebrates is a question of considerable interest. All amphibians and most reptiles and birds lack lymph nodes, but it is presently unknown whether they utilize a TLN-like system for antigen surveillance (4, 5). We favor a model where the TLN gradually evolves over time and represents the precursor to the modern mammalian lymph node. To this end, a better understanding of the molecular and genetic regulators of TLN formation will facilitate phylogenetic analysis across species. We anticipate that a deeper understanding of the evolution of adaptive immunity will provide unexpected insights into human immunology. Indeed, the zebrafish tessellated lymphoid network provides a way to study T cell antigen search without surgical manipulation, multiphoton microscopy, or having to limit analysis to a small fraction of T cells. The TLN shares a number of characteristics with tertiary lymphoid organs, ectopic lymphoid structures that form in peripheral tissues under conditions of chronic inflammation, and may therefore be useful for the study of their formation (51). From a cell biology perspective, this report provides unambiguous evidence that lymphocytes can migrate collectively in vivo, providing a model to further investigate this behavior.
Materials and Methods
Zebrafish Husbandry and Maintenance.
All protocols using zebrafish in this study were approved by the University of Wisconsin-Madison Research Animals Resource Center (protocol M005405-A02). Adult AB strain fish and transgenic zebrafish lines including the Casper WT line (17) and previously published transgenic reporter lines lck:GFP (18), cd4-1:mCherry (21), mpeg1.1:mCherryCAAX (19), lyzC:BFP (20), mrc1a:GFP, (27), and kdrl:mCherry (28) were used in this study. Double- or triple-transgenic reporters were generated by crossing individual lines and screening for fluorescence. For most experiments, these fish were then in-crossed to generate fish utilized for experiments. After breeding, fertilized embryos were placed into E3 media (5 mM NaCl, 0.17 mM KCl, 0.44 mM CaCl2, 0.33 mM MgSO4, 0.025 mM NaOH, and 0.0003% methylene blue) and maintained at 28.5 °C in a Petri dish in a laboratory incubator. Lck:GFP and cd4-1:mCherry larvae were screened at 6 to 7 d post fertilization (dpf) for fluorescence signal emanating from the paired thymus in the head. mpeg1.1:mCherryCAAXand lyzC:BFP reporter fish were screened looking for fluorescent macrophages and neutrophils in the caudal hematopoietic tissue anytime from 3 to 7 dpf. At 7 dpf, screened larvae were transferred to a fish tank with still water where they were fed twice daily and maintained on a 14-h/10-h light/dark schedule. At 10 dpf, a drip of fresh water was applied to the tank, and at 21 dpf, a gentle stream of water was applied. The fish were maintained at a density of approximately 20 fish per 3-L tank to maintain experiment-by-experiment growth consistency. The fish were removed from this environment as needed for experiments. When taking whole-animal confocal tiling images, the fish were fasted overnight to reduce nonspecific fluorescence coming from the intestinal tract.
Fish Handling for Imaging.
For live imaging experiments, juvenile or adult zebrafish were added to water containing 0.08 mg/mL tricaine (MS222/ethyl 3-aminobenzoate; Sigma-Aldrich) until the fish became nonresponsive to touch (<5 min). In some cases, additional tricaine was added dropwise to achieve timely nonresponsiveness. The anesthetized fish were moved to an imaging chamber with a transfer pipet (Fisher 13-711-7M) cut halfway to the base to increase the hole size. Depending on the size of the specimen, the fish were either maintained in 4-well (Ibidi 80427), 2-well (Ibidi 80287), or 1-well (Nunc Lab-Tek II 155360) glass-bottom imaging chambers. The smallest possible chamber was always used and enough water with tricaine was added to the chamber until the fish began to float. Drift was then limited by adding soaked sponge cloth to the chamber around the fish, and an additional soaked sponge cloth was draped over the fish head and upward-facing gills to maintain viability. In all cases, the fish were imaged for ≤20 min. Following imaging, the fish were killed by keeping them in 0.16 mg/mL tricaine for 20 min. For nonterminal experiments, the fish were quickly transferred to a tank of fresh RO water to promote recovery following imaging experiment. We found the fish could tolerate <10 min in 0.08 to 0.16 tricaine and reliably recover.
For whole-fish confocal tiling images, the fish were first killed and then immobilized in a 1-well glass-bottom chamber (Nunc Lab-Tek II 155360) with low gelling temperature agarose (Sigma Aldrich 19045). To visualize scales, we immersed the euthanized fish into 0.04% solution of Alizarin red powder (Sigma Aldrich 130-22-3) dissolved in RO water in a 6-well plate for 20 min. The fish were then washed for three times for 5 min in fresh RO water with gentle rocking prior to imaging.
Puncture Wound and Listeria Infection Model.
A streak plate from L. monocytogenes strain 10403S frozen stock was grown at 37 °C. A fresh colony was picked and grown statically in 1 mL brain–heart infusion (BHI) medium overnight at 30 °C to reach stationary phase. Bacteria were subcultured for ~1.5 to 2 h in fresh BHI (4:1, BHI:overnight culture) to achieve growth to mid-logarithmic phase (OD600 ≈ 0.6 to 0.8). Three milliliters of the midlogarithmic phase bacterial culture was spun down and washed three times in phosphate-buffered saline (PBS; Gibco 14190-144) and resuspended in 100 µL PBS for infection. This bacterial suspension was then added to 10 mL water in a six-well plate. Adult zebrafish were anesthetized in 0.16 mg/mL tricaine and transferred to a damp sponge cloth. A 30G½ needle (Becton Dickinson PrecisionGlide) was used to puncture the caudal peduncle on the dorsal side of the animal. Pressure was applied until the needle came out the other side. The wounded fish were placed in a tank of water until movement was observed (0 to 3 min) at which point they were added to the six-well plate with bacteria. This plate was gently rocked for 2 h in the dark at which point the fish were moved to a 1-L tank with fresh water and maintained in a laboratory 28.5 °C incubator until ready for imaging. Water was exchanged daily and the fish were fed one to two times daily.
Microscopy and Image Preparation.
For imaging the TLN over development (Fig. 1B), fish were imaged on a Zeiss Zoomscope (EMS3/SyCoP3; 1× Plan-NeoFluar Z objective; Zeiss) with an Axiocam Mrm charge-coupled device camera using ZenPro 2012 software (Zeiss). For most other fish imaging, we used a spinning disc confocal microscope (CSU-X, Yokogawa, Sugar Land, TX) with a confocal scanhead on a Zeiss Observer Z.1 inverted microscope, either an EC Plan-Neofluar 10×/0.30 or EX Plan-Neofluar 40×/0.75 object, a Photometrics Evolve EMCCD camera, and Zen software (Zeiss). To stitch tiled images in ZenPro 2012 software (Zeiss), we first performed a max intensity z-projection and then stitched with a 10% maximal shift. All images were prepared for publication with ImageJ (v. 2.1.0/1.53c)
Cell Tracking and Motility Analysis.
Cell movies to be tracked were imported into ImageJ (v. 2.1.0/1.53c), and individual cells were tracked using the Manual Tracking plugin. Some movies were rotated prior to tracking to ensure that dorsal side of the fish was always pointing straight upward. Even when the mCherry channel is not shown, all cell tracking was performed using dual labeled tg(lck:GFP;cd4-1:mCherry) fish as differentiating and tracking individual T cells was prohibitively difficult otherwise. For Fig. 3, every trackable CD4+Lck+ cell was tracked. Cells were only excluded if they left the field of view during the imaging window. CD4-Lck+ cells appeared to move indistinguishably from CD4+Lck+ cells, although we did not attempt to track them. For Fig. 4, the cell density around the infected wounds was too high to track every cell. Instead, we tracked 20 randomly chosen (CD4+ or CD4-) motile T cells from every video. Cell tracking XY coordinates were copied into Microsoft Excel (v. 16.65), which was used to calculate velocity, directionality, displacement, and mean squared displacement. Additionally, Excel was used to generate the cell track rose plots, which were then overlaid in Illustrator (Adobe Inc.; v. 26.02) with graphs of the fish-by-fish cell averages to generate complete figures.
Cell motility within the TLN was characterized by plotting MSD versus time where so that is the slope when plotted on a log–log scale. Values of thus correspond to confined diffusion, to random walk Brownian diffusion, super diffusion, and ballistic motion (e.g., straight line without any turns). Data were fit to with corresponding model parameters, degrees of freedom, and assessment of fit by by shown in SI Appendix, Fig. S3D (control only) and SI Appendix, Fig. S4E (control and infected). Values of were determined by averaging over n = 5 fish for each condition.
Dissection and Cell Isolation from the Gut, Gills, Spleen, and Kidney.
We dissected adult zebrafish as previously described (52). Briefly, we used dissecting scissors to cut along the belly of killed adult zebrafish to expose the internal organs. The spleen was separated from the gut with curved tweezers and added to a 40-micron filter in a six-well plate containing 8 mL of cell isolation media (L-15 + 1 mM EDTA, penicillin–streptomycin, and 2% FBS). The gut was then added to a 1.5-mL Eppendorf tubed filled with 1 mL prewarmed 37 °C collagenase/dispase solution (Roche 10 269 638 001) and incubated for 90 s before adding to a 40-micron filter as above. The operculum, pectoral fin, and pectoral girdle were then all cut out, and the intact gills were removed from with curved tweezers and added to a dish of PBS. Nongill tissue was then carefully removed under a dissecting microscope. In three of six fish, only half the gill arches were cleanly isolated, so final cell counts were doubled in these cases to get estimates of the gill totals. Finally, curved tweezers were used to isolate the entirety of the kidney, which was then added to a 40-micron filter in a 6-well plate filled with 8 mL of cell isolation buffer. All organs were then mechanically disrupted with the back end of a plastic syringe, and the resulting filtered cell suspension was used for further analysis.
Descaling, TLN Cell Isolation, and Flow Cytometry.
Euthanized adult zebrafish were briefly dunked in room-temperature PBS and then placed in a 60-mm Petri dish (Fisher FB0875713A) containing 8 mL of room-temperature PBS. One (gloved) hand was used to hold the fish in place by firmly grasping the head, and the second hand was used to scrape rostrally along the body of the fish with an angled dissecting knife (Fine Science Tools 10056-12) to descale. This was done under a dissecting microscope and scale loss was visually monitored. Scales at the base of the caudal, dorsal, and anal fins typically had to be individually removed by plucking with thin-tip tweezers (Dumont). PBS from the Petri dsh was then washed over the scaled fish for approximately 30 s with a transfer pipette (Fisher 13-711-7M) to promote cells entering the suspension, and the buffer was then passed through a 40-micron filter. The pressure from descaling often caused males and females to release their eggs and milt, respectively. For this reason, only females were used for analysis, as their eggs could be readily filtered out. This cell suspension was then mixed 1:1 with cell isolation media in a 15-mL Falcon tube, washed, resuspended in 500 µL ACK lysis buffer (Cold Spring Harbor Protocols) for 90 s to lyse red blood cells, and washed once more with cell isolation media. For Fig. 2C, these cells were added to an eight-well chamber glass-bottom chamber (Ibidi 80807) and imaged at room temperature on the Zeiss Zoomscope (EMS3/SyCoP3; 1× Plan-NeoFluar Z objective; Zeiss) with an Axiocam Mrm charge-coupled device camera using ZenPro 2012 software (Zeiss). For flow cytometry, cells isolated from tg(lck:GFP;cd4-1:mCherry) by descaling were stained with Ghost Dye™ Red 780 (Tonbo) per manufacturer’s instructions, washed with twice with FACS buffer (PBS + 1 mM EDTA + 5% FBS + penicillin/streptomycin), and analyzed by flow cytometry. A ThermoFisher Attune was used for all experiments, allowing quantitation of total cell numbers of interest in the sample.
Single-Cell RNA-Sequencing.
For scRNA-seq, cells were isolated from uninfected control and 4 d postinfection zebrafish exactly as described above except that all buffers were prepared without EDTA. To enrich for cells involved in the immune response, we only descaled the caudal peduncle rather than the entire fish to harvest cells for scRNA-seq. Libraries were constructed from these cells by the University of Wisconsin-Madison Biotechnology & Gene Expression Center (RRID: SCR_017757) using the Chromium Single Cell Gene Expression Solution 3′ v2 (10x Genomics) and sequenced by the DNA Sequencing Facility (RRID: SCR_017759) using NovaSeq6000 (Illumina) with read lengths of 29-bp + 90-bp (Read1 + Read2). Raw reads were processed by cellranger count (v6.1.2, 10x Genomics) with default parameters for read tagging, alignment to zebrafish reference genome (GRCz11), and feature counting based on an improved zebrafish transcriptome annotation (53). In total, 386M and 486M reads were sequenced for the control and infected samples, respectively, both with valid barcode rates over 95% and a valid UMI rate of 99.9%.
Unsupervised Clustering and Cell Type Identification.
Filtered cell-by-gene count matrices generated were used for graph-based clustering with Seurat v4.1.1 (54). We filtered out genes that are detected in less than four cells and cells with less than 200 genes detected or a mitochondrial ratio >5%. Doublets were also removed using DoubletFinder with an estimated rate of 3% (55). Post doublet removal, the control sample has 3,141 cells with a median of 1,271 captured genes, while the infected sample has 4,309 cells with a median of 2,125 captured genes.
Normalization (SCTransform) and dimensional reduction were done with default parameters in Seurat package prior to clustering and UMAP projection (56). Clustering and visualizations were either performed on the control sample only, or with both samples integrated via canonical correlation analysis (CCA)-based method from Seurat (57). To identify cell types and states at chosen clustering resolution (0.6 for control only, 0.5 for control and infected integrated), we performed differential expression analysis using Wilcoxon rank sum-based method (log2 fold change > 0.25, % of expressing cells > 0.25). Known markers were used to identify T cells (cd247l, lck, il7r, and cxcr4a) (58–60), dendritic cell-like populations (ctsbb, tlr7) (61, 62), B cells (cd37, pax5) (63, 64), macrophages (mpeg1.1, grn1) (65, 66), neutrophils (mpx, il6r) (11, 67), erythrocytes (hbba2, hemgn) (68), thrombocytes (thbs1b, fn1b) (62), superficial epithelial cells (krt1-19d, cldne) (69), ionocytes (trpv6, foxi3b) (70), intermediate epithelial cells (cldna, tp63) (69), basal epithelial cells (cldn1, cldni) (69), mesenchymal cells (vcana, clu) (71), lateral line-like cells (prox1a, prr15la) (72), and epidermal mucous cells (agr2, cldnh) (69). We found one population with some cells locating closer to the T cell populations but without clear enrichment for known signatures and included this population for a subclustering analysis with other T cells. We identified one subcluster with epithelial features but without known T cell features, thus labeled them as others-epithelial cells and removed from the T cell-focused analysis. T cell subsets were identified using known marker gene expression patterns: naïve T cells (ccr7, cxcr4a) (73), cycling T cells (mki67, top2a) (74), regulatory-like T cells (foxp3a, ccr6a, ccl20a.3) (75, 76), cytotoxic T cells (cxcr3.1, gzma, prf1.1, nkl.2, ccr2, prf1.9) (77), ccl38.6-high T cells (il2rb, tnfsf14, ccl38.6), Th2 cell/ILC2 (il11b, il4, il13), rag1/2+ T cells (bcl11ba, rag1, rag2) (62), and lymphocyte-like cells (ccl33.3, lcp2b). For a more stringent T cell-focused analysis in the control and infected integrated sample, we extracted T cells using the expression of both cd247l and lck (log2FC > 0.3, % of expressing cells > 0.3).
Statistical Analysis.
Statistical tests were all performed using GraphPad Prism (version 9.4.1), with specific tests indicated in each figure legend. P values were denoted on graphs as follows: n.s., P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Supplementary Material
Appendix 01 (PDF)
Live confocal imaging of the TLN of an adult tg(lck:GFP;cd4-1:mCherry) shows the characteristic streaming motility with a 10x objective to highlight T cell movement through the TLN network over hundreds of microns. This movie corresponds to Figure 1F, where individual cell tracks are shown. Time in mm:ss.
Live confocal imaging of the TLN of an adult tg(lck:GFP;cd4-1:mCherry) shows the directional movement of T cells. Three representative cells are tracked (white dot and lines) corresponding to the outlined cells in Figure 3A. Time in mm:ss.
Live confocal imaging of the TLN of an adult tg(lck:GFP;mpeg1::mCherryCAAX) display the streaming motility behavior of T cells. Two side-by-side representative movies are show, with the left movie corresponding to figure 3I. Macrophages are distributed throughout the network. Individual T cells are difficult to track by just the GFP channel, but the movie demonstrates T cells streaming past TLN-localized macrophages under homeostatic conditions. Time in mm:ss.
The left portion of this video shows a still image of a region proximal to the puncture wound l. monocytogenes infection of a transgenic lck:GFP adult zebrafish. The two boxed regions in the still image are shown as movies on the right. The ‘motile’ region shows a group T cells that mostly migratory, and the ‘sessile’ region shows a group that is mostly non-migratory. Time in mm:ss.
Live imaging of the region proximal to the puncture wound L. monocytogenes infection of an adult transgenic lck:GFP;cd4-1:mCherry zebrafish. The video first shows still frames with the relevant macrophages labelled (M) and a white dot over the GFP+mCherry+ T cells. Once the movie starts, the white dot follows the T cell as it contacts all three of these infection-proximal macrophages. Time in mm:ss.
Acknowledgments
We would like to thank A. Horn, V. Miskolci, J. Squirrell, D. Bennin, T. Schoen, M. Giese, A. Fister, G. Ramakrishnan, A. Peterson, and N. Mercado-Soto for their helpful discussion throughout the design and execution of this project. We would like to thank the Gene Expression Center at the University of Wisconsin-Madison for sequencing assistance, and the Carbone Center Flow lab for assistance in training and guidance of flow cytometry experiments. The mrc1a:GFP transgenic zebrafish were a kind gift from the Weinstein lab (NIH). Figures throughout this paper were made in part with biorender.com. This study was funded by NIH grant 1 F32 GM146398-01 (T.F.R), NIH grant T32 HL07899 (T.F.R.), and NIH grant R35 GM118027 (A.H.)
Author contributions
T.F.R., and A.H. designed research; T.F.R. and S.S. performed research; S.S., J.R., and J.-D.S. contributed new reagents/analytic tools; T.F.R., Y.H., J.S., and H.Q.D. analyzed data; A.H. oversaw the project; and T.F.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All data are available in the main text or the supplementary materials. scRNA-seq data are available in the NCBI Expression Omnibus (GSE215189). scRNA-seq analysis codes are available at: https://github.com/k326xh/TLN-Tcell-scRNA-analysis.
Supporting Information
References
- 1.Masopust D., Schenkel J. M., The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 13, 309–320 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Krummel M. F., Bartumeus F., Gérard A., T cell migration, search strategies and mechanisms. Nat. Rev. Immunol. 16, 193–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obar J. J., Khanna K. M., Lefrançois L., Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28, 859–869 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neely H. R., Flajnik M. F., Emergence and evolution of secondary lymphoid organs. Annu. Rev. Cell Dev. Biol. 32, 693–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehm T., Hess I., Swann J. B., Evolution of lymphoid tissues. Trends Immunol. 33, 315–321 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Dalum A. S., et al. , High-resolution, 3D imaging of the zebrafish gill-associated lymphoid tissue (GIALT) reveals a novel lymphoid structure, the amphibranchial lymphoid tissue. Front. Immunol. 12, 769901 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abelli L., Picchietti S., Romano N., Mastrolia L., Scapigliati G., Immunohistochemistry of gut-associated lymphoid tissue of the sea bassDicentrarchus labrax(L.). Fish Shellfish Immunol. 7, 235–245 (1997). [Google Scholar]
- 8.Heppell J., et al. , Development of DNA vaccines for fish: Vector design, intramuscular injection and antigen expression using viral haemorrhagic septicaemia virus genes as model. Fish Shellfish Immunol. 8, 271–286 (1998). [Google Scholar]
- 9.Ma J., Bruce T. J., Jones E. M., Cain K. D., A review of fish vaccine development strategies: Conventional methods and modern biotechnological approaches. Microorganisms 7, 569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson T. F., Huttenlocher A., Real-time imaging of inflammation and its resolution: It’s apparent because it’s transparent*. Immunol. Rev. 306, 258–270 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathias J. R., et al. , Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J. Leukoc Biol. 80, 1281–1288 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Wang J., et al. , Visualizing the function and fate of neutrophils in sterile injury and repair. Science 358, 111–116 (2017). [DOI] [PubMed] [Google Scholar]
- 13.de Oliveira S., Rosowski E. E., Huttenlocher A., Neutrophil migration in infection and wound repair: Going forward in reverse. Nat. Rev. Immunol. 16, 378–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson A. L., et al. , A zebrafish compound screen reveals modulation of neutrophil reverse migration as an anti-inflammatory mechanism. Sci. Transl. Med. 6, 225ra229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jerison E. R., Quake S. R., Heterogeneous T cell motility behaviors emerge from a coupling between speed and turning in vivo. Elife 9, e53933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam S. H., Chua H. L., Gong Z., Lam T. J., Sin Y. M., Development and maturation of the immune system in zebrafish, Danio rerio: A gene expression profiling, in situ hybridization and immunological study. Dev. Comparative Immunol. 28, 9–28 (2004). [DOI] [PubMed] [Google Scholar]
- 17.White R. M., et al. , Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langenau D. M., et al. , In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 101, 7369–7374 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellett F., Pase L., Hayman J. W., Andrianopoulos A., Lieschke G. J., mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Oliveira S., et al. , Metformin modulates innate immune-mediated inflammation and early progression of NAFLD-associated hepatocellular carcinoma in zebrafish. J. Hepatol. 70, 710–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dee C. T., et al. , CD4-transgenic zebrafish reveal tissue-resident Th2- and regulatory T cell-like populations and diverse mononuclear phagocytes. J. Immunol. 197, 3520–3530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aman A. J., Fulbright A. N., Parichy D. M., Wnt/β-catenin regulates an ancient signaling network during zebrafish scale development. Elife 7, e37001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flajnik M. F., A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 18, 438–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sales C. F., et al. , Comparative histology in the liver and spleen of three species of freshwater teleost. Neotrop. Ichthyol. 15 (2017). [Google Scholar]
- 25.Menke A. L., Spitsbergen J. M., Wolterbeek A. P. M., Woutersen R. A., Normal anatomy and histology of the adult zebrafish. Toxicol. Pathol. 39, 759–775 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Covacu R., et al. , System-wide analysis of the T cell response. Cell Rep. 14, 2733–2744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung H. M., et al. , Development of the larval lymphatic system in zebrafish. Development 144, 2070–2081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S.-W., Beis D., Mitchell T., Chen J.-N., Stainier D. Y. R., Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Fletcher A. L., Acton S. E., Knoblich K., Lymph node fibroblastic reticular cells in health and disease. Nat. Rev. Immunol. 15, 350–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller S. N., Germain R. N., Stromal cell contributions to the homeostasis and functionality of the immune system. Nat. Rev. Immunol. 9, 618–629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bussmann J., Raz E., Chemokine-guided cell migration and motility in zebrafish development. Embo J. 34, 1309–1318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glass T. J., et al. , Stromal cell-derived factor-1 and hematopoietic cell homing in an adult zebrafish model of hematopoietic cell transplantation. Blood 118, 766–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzler R., Klafter J., The random walk’s guide to anomalous diffusion: A fractional dynamics approach. Phys. Rep. 339, 1–77 (2000). [Google Scholar]
- 34.Harris T. H., et al. , Generalized Lévy walks and the role of chemokines in migration of effector CD8+ T cells. Nature 486, 545–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriebel P. W., Barr V. A., Parent C. A., Adenylyl cyclase localization regulates streaming during chemotaxis. Cell 112, 549–560 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Miller M. J., Safrina O., Parker I., Cahalan M. D., Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J. Exp. Med. 200, 847–856 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dustin M. L., Bromley S. K., Kan Z., Peterson D. A., Unanue E. R., Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 94, 3909–3913 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorkdahl O., et al. , Characterization of CC-chemokine receptor 7 expression on murine T cells in lymphoid tissues. Immunology 110, 170–179 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindsten T., et al. , Characterization of CTLA-4 structure and expression on human T cells. J. Immunol. 151, 3489–3499 (1993). [PubMed] [Google Scholar]
- 40.Orgun N. N., Mathis M. A., Wilson C. B., Way S. S., Deviation from a strong Th1-dominated to a modest Th17-dominated CD4 T cell response in the absence of IL-12p40 and type I IFNs sustains protective CD8 T cells. J. Immunol. 180, 4109–4115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Way S. S., Havenar-Daughton C., Kolumam G. A., Orgun N. N., Murali-Krishna K., IL-12 and type-I IFN synergize for IFN-gamma production by CD4 T cells, whereas neither are required for IFN-gamma production by CD8 T cells after Listeria monocytogenes infection. J. Immunol. 178, 4498–4505 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Z., Khairallah C., Sheridan B. S., Listeria monocytogenes: A model pathogen continues to refine our knowledge of the CD8 T cell response. Pathogens 7, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltman J. B., Marée A. F. M., Lynch J. N., Miller M. J., de Boer R. J., Lymph node topology dictates T cell migration behavior. J. Exp. Med. 204, 771–780 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witt C. M., Raychaudhuri S., Schaefer B., Chakraborty A. K., Robey E. A., Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 3, e160 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okada T., et al. , Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 3, e150 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen C. D., Okada T., Tang H. L., Cyster J. G., Imaging of germinal center selection events during affinity maturation. Science 315, 528–531 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Chauveau A., et al. , Visualization of T cell migration in the spleen reveals a network of perivascular pathways that guide entry into T zones. Immunity 52, 794–807.e797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X., et al. , The roles of CCR9/CCL25 in inflammation and inflammation-associated diseases. Front. Dev. Biol. 9, 686548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grigorova I. L., Panteleev M., Cyster J. G., Lymph node cortical sinus organization and relationship to lymphocyte egress dynamics and antigen exposure. Proc. Natl. Acad. Sci. U.S.A. 107, 20447–20452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sunyer J. O., Fishing for mammalian paradigms in the teleost immune system. Nat. Immunol. 14, 320–326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bery A. I., et al. , Role of tertiary lymphoid organs in the regulation of immune responses in the periphery. Cell Mol. Life Sci. 79, 359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta T., Mullins M. C., Dissection of organs from the adult zebrafish. J. Vis. Exp. 4, 1717 (2010), 10.3791/1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawson N. D., et al. , An improved zebrafish transcriptome annotation for sensitive and comprehensive detection of cell type-specific genes. ELife 9, e55792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao Y., et al. , Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 8, 329–337.e324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carrette F., Surh C. D., IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol. 24, 209–217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palacios E. H., Weiss A., Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene 23, 7990–8000 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Schabath R., et al. , The murine chemokine receptor CXCR4 is tightly regulated during T cell development and activation. J. Leukoc Biol. 66, 996–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Ito T., et al. , Interferon-alpha and interleukin-12 are induced differentially by Toll-like receptor 7 ligands in human blood dendritic cell subsets. J. Exp. Med. 195, 1507–1512 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubin S. A., et al. , Single-cell analyses reveal early thymic progenitors and pre-B cells in zebrafish. J. Exp. Med. 219, e20220038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nutt S. L., Heavey B., Rolink A. G., Busslinger M., Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556–562 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Schwartz-Albiez R., Dörken B., Hofmann W., Moldenhauer G., The B cell-associated CD37 antigen (gp40-52). Structure and subcellular expression of an extensively glycosylated glycoprotein. J. Immunol. 140, 905–914 (1988). [PubMed] [Google Scholar]
- 65.Rougeot J., et al. , RNAseq profiling of leukocyte populations in zebrafish larvae reveals a cxcl11 chemokine gene as a marker of macrophage polarization during mycobacterial infection. Front. Immunol. 10, 832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zakrzewska A., et al. , Macrophage-specific gene functions in Spi1-directed innate immunity. Blood 116, e1–11 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Wright H. L., Cross A. L., Edwards S. W., Moots R. J., Effects of IL-6 and IL-6 blockade on neutrophil function in vitro and in vivo. Rheumatology 53, 1321–1331 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Chan F. Y., et al. , Characterization of adult alpha- and beta-globin genes in the zebrafish. Blood 89, 688–700 (1997). [PubMed] [Google Scholar]
- 69.Hou Y., et al. , Cellular diversity of the regenerating caudal fin. Sci. Adv. 6, eaba2084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jänicke M., Carney T. J., Hammerschmidt M., Foxi3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Dev. Biol. 307, 258–271 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Kimata K., et al. , A large chondroitin sulfate proteoglycan (PG-M) synthesized before chondrogenesis in the limb bud of chick embryo. J. Biol. Chem. 261, 13517–13525 (1986). [PubMed] [Google Scholar]
- 72.Pistocchi A., et al. , The zebrafish prospero homolog prox1 is required for mechanosensory hair cell differentiation and functionality in the lateral line. BMC Dev. Biol. 9, 58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sallusto F., Mackay C. R., Lanzavecchia A., The role of chemokine receptors in primary, effector, and memory immune responses. Annu. Rev. Immunol. 18, 593–620 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Kowalczyk M. S., et al. , Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hui S. P., et al. , Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659–672.e655 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Lee A. Y. S., et al. , Expression of membrane-bound CC chemokine ligand 20 on follicular T helper cells in T-B-cell conjugates. Front. Immunol. 8, 1871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lieberman J., Granzyme A activates another way to die. Immunol. Rev. 235, 93–104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Live confocal imaging of the TLN of an adult tg(lck:GFP;cd4-1:mCherry) shows the characteristic streaming motility with a 10x objective to highlight T cell movement through the TLN network over hundreds of microns. This movie corresponds to Figure 1F, where individual cell tracks are shown. Time in mm:ss.
Live confocal imaging of the TLN of an adult tg(lck:GFP;cd4-1:mCherry) shows the directional movement of T cells. Three representative cells are tracked (white dot and lines) corresponding to the outlined cells in Figure 3A. Time in mm:ss.
Live confocal imaging of the TLN of an adult tg(lck:GFP;mpeg1::mCherryCAAX) display the streaming motility behavior of T cells. Two side-by-side representative movies are show, with the left movie corresponding to figure 3I. Macrophages are distributed throughout the network. Individual T cells are difficult to track by just the GFP channel, but the movie demonstrates T cells streaming past TLN-localized macrophages under homeostatic conditions. Time in mm:ss.
The left portion of this video shows a still image of a region proximal to the puncture wound l. monocytogenes infection of a transgenic lck:GFP adult zebrafish. The two boxed regions in the still image are shown as movies on the right. The ‘motile’ region shows a group T cells that mostly migratory, and the ‘sessile’ region shows a group that is mostly non-migratory. Time in mm:ss.
Live imaging of the region proximal to the puncture wound L. monocytogenes infection of an adult transgenic lck:GFP;cd4-1:mCherry zebrafish. The video first shows still frames with the relevant macrophages labelled (M) and a white dot over the GFP+mCherry+ T cells. Once the movie starts, the white dot follows the T cell as it contacts all three of these infection-proximal macrophages. Time in mm:ss.
Data Availability Statement
All data are available in the main text or the supplementary materials. scRNA-seq data are available in the NCBI Expression Omnibus (GSE215189). scRNA-seq analysis codes are available at: https://github.com/k326xh/TLN-Tcell-scRNA-analysis.



