Significance
Dissimilatory sulfate reduction (DSR) is one of the oldest and most prominent microbial metabolic pathways on Earth. It is generally accompanied by zero-valent sulfur (ZVS) that is involved in several cryptic pathways in marine and terrestrial environments. In this study, we identified a to-date unknown DSR pathway or sulfate-to-ZVS conversion mediated by sulfate-reducing microorganisms. This finding provides further insights into the sulfur cycle, which may help reveal details about cryptic element cycling pathways and improve our understanding of the sulfur metabolism in early Archaean microorganisms.
Keywords: zero-valent sulfur, dissimilatory sulfate reduction, sulfate-reducing microorganism, sulfate reduction pathway, sulfur cycle
Abstract
Dissimilatory sulfate reduction (DSR) mediated by sulfate-reducing microorganisms (SRMs) plays a pivotal role in global sulfur, carbon, oxygen, and iron cycles since at least 3.5 billion y ago. The canonical DSR pathway is believed to be sulfate reduction to sulfide. Herein, we report a DSR pathway in phylogenetically diverse SRMs through which zero-valent sulfur (ZVS) is directly generated. We identified that approximately 9% of sulfate reduction was directed toward ZVS with S8 as a predominant product, and the ratio of sulfate-to-ZVS could be changed with SRMs’ growth conditions, particularly the medium salinity. Further coculturing experiments and metadata analyses revealed that DSR-derived ZVS supported the growth of various ZVS-metabolizing microorganisms, highlighting this pathway as an essential component of the sulfur biogeochemical cycle.
Dissimilatory sulfate reduction (DSR) mediated by sulfate-reducing microorganisms (SRMs) is a ubiquitous microbial process that occurs in both natural and engineered ecosystems (1–3), such as the global anaerobic oxidation of methane (AOM) mediated by microbial consortia of archaea (ANME) (4–7) and the remediation of acid mine drainage (AMD) and sites contaminated by organic pollutants and heavy metals (1, 3, 8, 9). Isotopic pieces of evidence in sedimentary rocks further indicate that SRMs emerged in the early Archaean era (at least 3.5 Gyr) (10) and have played a crucial role in the evolution of Earth’s surface redox state and climate by altering global sulfur, carbon, oxygen, and iron cycles (11). To date, phylogenetically diverse SRMs have been identified from global terrestrial and marine environments (1, 5, 12, 13). Although trace amounts of thionates were observed as intermediates in enzymes or in pretreated SRM cells (14–16), the canonical DSR pathway is thought to reduce sulfate to sulfide as a sole metabolic end product, of which enzymatic mechanisms remain partially understood (17, 18). Zero-valent sulfur (ZVS) has been widely detected in anaerobic conditions along with the DSR-derived sulfide (3, 19–21), which is generally proposed to be a sulfide reoxidation product (12, 21). Nonetheless, whether massive sulfide-to-ZVS reoxidation can occur in anaerobic natural environments with limited source of oxidants remains debatable (6, 21). In contrast, ZVS is proposed as a key intermediate in ANME-mediated anaerobic methane oxidation by sulfate, which is present in ANME intracellularly (4). This could be further disproportionated by cocultured SRMs in the AOM microbial consortia (4). However, a different relation between ANME and SRMs for AOM was documented very recently (5, 6), in which ANME lacking genetic capacity for DSR assimilated inorganic sulfur compounds being more oxidized than sulfide. Therefore, the mechanism through which ZVS is generated under these anaerobic conditions remains poorly understood.
Results and Discussion
Characterization of ZVS Generated from SRM-Mediated DSR.
Recently, we identified ZVS generation in a sulfate-reducing and methanogenic bioreactor, in which known sulfide-oxidizing microorganisms and available sources of oxidants were excluded (20). We hypothesized that ZVS could be derived from the DSR rather than through the reoxidation of sulfide under anaerobic conditions. To test this hypothesis, we examined ZVS generation in six typical strains of phylogenetically diverse SRMs isolated from varied niches (SI Appendix, Fig. S1), including Desulfovibrio vulgaris Hildenborough (DvH) as a model SRM. In these pure cultures, ZVS generation occurred along with sulfate-to-sulfide reduction and peaked when sulfate reduction terminated, resulting in 6.9 to 12.9% (a mean value of 8.9%) sulfate reduction to ZVS, e.g., 8.4 ± 1.5% in DvH (Fig. 1A and SI Appendix, Fig. S2). Thereafter, ZVS levels decreased and stabilized in the late stationary phase (Fig. 1A and SI Appendix, Fig. S2). The ZVS generation was further corroborated by the appearance of yellowish color (because of elemental sulfur) in SRM cultures without amendment of Fe(II/III) ions (Fig. 1B). Such yellowish color could be over-shadowed by black FeS precipitates in the SRM-cultivating LS4D medium. Quantification of the total and extracellular ZVS showed that 8.5 to 38.1% of the ZVS was intracellular sulfur (Fig. 1C and SI Appendix, Fig. S3). Desulforamulus ruminis DL (DLT) as an SRM with a gram-positive-type cell wall structure (22) generated comparatively more abundant intracellular sulfur relative to that of gram-negative DvH, i.e., 38.1 ± 3.7% and 11.3 ± 1.8% intracellular sulfur in DLT and DvH cultures, respectively, fed with a molar ratio of lactate (C) to sulfate (S) of 1:1 (Fig. 1C). This difference in intracellular sulfur accumulation could be attributed to the difficulty in crossing-cell-wall transportation of ZVS in DLT.
Fig. 1.
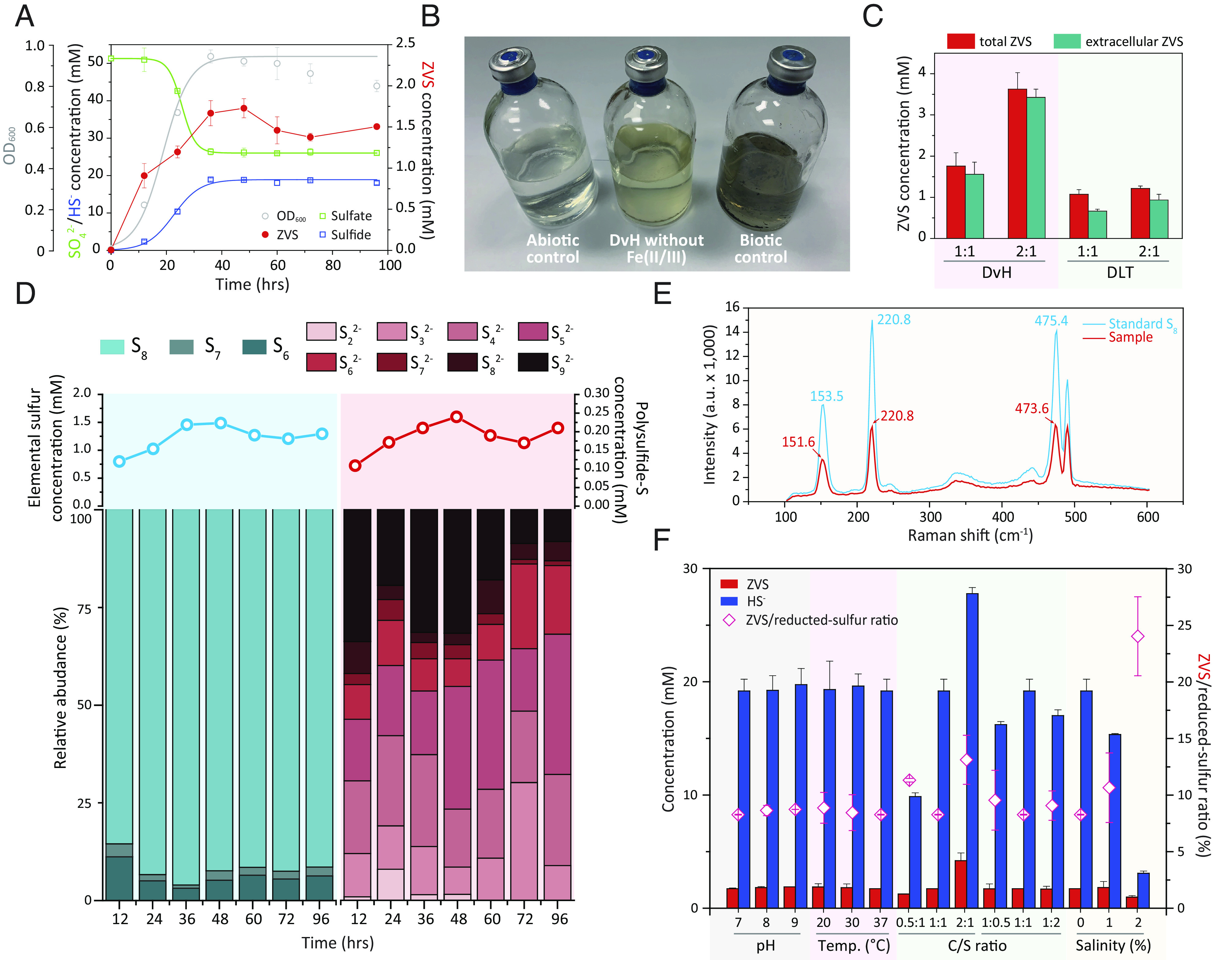
Generation of zero-valent sulfur (ZVS) from DSR. (A) Dissimilatory sulfate reduction (DSR) and ZVS generation in Desulfovibrio vulgaris Hildenborough (DvH). (B) Observed yellowish color in DvH cultures without amendment of Fe(II/III). DvH without Fe(II/III), DvH in the LS4D medium without the amendment of Fe(II/III); Abiotic control, LS4D medium with sulfate- and sulfide-amendment but without DvH inoculation; Biotic control, DvH in the LS4D medium. (C) The total/extracellular ZVS generation of DvH and DLT (Desulforamulus rumins DL) in cultures amended with two different molar ratios of lactate (C) to sulfate (S), i.e., C/S ratios of 1:1 and 2:1. (D) The temporal patterns of ZVS compositional profiles in DvH. Data are presented as mean value of triplicate cultures. (E) Representative Raman spectrum of elemental sulfur in DvH, containing prominent peaks of the elemental sulfur (S8) at 151.6, 220.8, and 473.6 cm−1; a.u., arbitrary units. (F) DSR-derived ZVS and sulfide, as well as the ratio of ZVS to reduced sulfur (i.e., ZVS and sulfide), in DvH cultures with varied pH, temperature (Temp.), C/S ratio, and salinity (%, w/v). Error bars represent SDs of triplicate cultures.
ZVS could be present as elemental sulfur ( ) and/or polysulfide ( ), qualitative and quantitative analyses of which are challenging (20, 21). In this study, multiple complementary methods were used to identify and quantify total ZVS (23, 24), elemental sulfur (25, 26), and polysulfide (27) under anaerobic conditions. The elemental sulfur accounted for 84.1 to 90.4% (a mean value of 87.2%) of the total ZVS at ZVS peak time (Fig.1D and SI Appendix, Fig. S3) and mainly constituted of S8, S7, and S6 (Fig. 1D and SI Appendix, Fig. S4A). The high ratio of S8 (>84.0% of the elemental sulfur) was confirmed by the strong Raman peaks for S8 (Fig. 1E), which could be further converted into S6 and S7 (SI Appendix, Fig. S4 A and B) (28). In contrast, a diverse range of polysulfide species (from di- to nona-sulfide) were identified to be generated in DSR (SI Appendix, Fig. S4C), of which the total amount only accounted for 9.6 to 15.9% (a mean value of 12.8%) of the total ZVS (Fig. 1D and SI Appendix, Fig. S3). The temporal changes of the elemental sulfur and polysulfide followed a similar trend in DvH (Fig. 1D and SI Appendix, Fig. S4D). The nona-sulfide could be derived from the biogenetic and pH-dependent reaction of S8 and sulfide, which was further transferred into di- to octa-sulfide species (29). Notably, extracellular ZVS shared a similar composition profile with that of the total ZVS in the DvH culture (SI Appendix, Fig. S4D).
Environmental stresses (e.g., limited reducing equivalent, high pH, low temperature, and high salinity) could change the metabolism and, consequently, sulfate reduction of SRMs (12). We compared the molar ratio of ZVS to reduced sulfur (ZVS and sulfide) and ZVS compositions in DvH cultures under growth conditions with varied pH, temperature, C/S ratio, and salinity (Fig. 1F). Adjustment of C/S ratios by changing molar concentrations of lactate (C) and sulfate (S) altered the ratios of ZVS to reduced sulfur in the range of 8.2 to 13.1% (Fig. 1F and SI Appendix, Fig. S5). In contrast, high pH (pH = 9.0) and low temperature (20 °C) slightly changed the ratio of ZVS to reduced sulfur, compared to control cultures (Fig. 1F). Strikingly, high salinity (2%, w/v) remarkably increased the ratio of ZVS to reduced sulfur, e.g., 8.2 ± 0.3% in cultures with no salinity vs. 24.0 ± 3.5% in cultures with 2% (w/v) salinity (Fig. 1F). S8 was generated as the predominant ZVS species under these saline conditions (SI Appendix, Fig. S5). The comparatively higher ratio of ZVS to reduced sulfur under saline conditions hint a higher flow ratio of sulfate-to-ZVS in marine environments relative to fresh waters, being consistent with observations of a large amount of ZVS in marine sediments (21, 30).
ZVS Generation from DSR rather than Reoxidation of Sulfide.
To eliminate the possibility that ZVS could have been generated from the oxidation of sulfide under anaerobic conditions (3, 31), we carried out experiments using radiolabeled sulfate (35S-sulfate) or sulfide (35S-sulfide) in DvH cultures. If sulfide-to-ZVS re-oxidation occurred, one would expect i) a lag time between sulfide- and ZVS-generation curves in the 35S-sulfate-fed cultures and ii) an accumulation of radiolabeled ZVS (35S-ZVS) along with decreases in 35S-sulfide in the 35S-sulfide-amended cultures (32, 33). This prediction was contradicted by our observations of synchronous generation of sulfide and 35S-ZVS in 35S-sulfate-fed cultures (Fig. 2A and SI Appendix, Fig. S6) and by the absence of 35S-ZVS accumulation or 35S-sulfide consumption in 35S-sulfide-amended cultures (Fig. 2B). Additional evidence of the negligible role of sulfide-to-ZVS oxidation in our experiments comes from transcription and translation analyses. The biotic sulfide-to-ZVS oxidation can be catalyzed by sulfide-quinone reductase (Sqr) and flavocytochrome c sulfide dehydrogenase (3, 31, 33), potentially harbored by SRMs (31). Although DvH contains a sqr-like gene (SI Appendix, Fig. S7), transcriptome and proteome analyses of DvH cultures fed with lactate and sulfate at their molar ratios of 1:1 and 2:1 showed high and comparable transcriptional/translational levels of dissimilatory sulfite reductase-encoding genes (dsrABC), especially the highly transcribed and translated dsrC gene, while the transcription and translation of sqr-like gene were negligible (Fig. 2C and SI Appendix, Fig. S8 and Table S1). The qPCR quantification of temporally transcribed dsrC and sqr-like genes demonstrated no correlation between the sqr-like gene transcription and ZVS generation (Fig. 2D), further ascertaining the negligible role of Sqr in mediating reoxidation of sulfide-to-ZVS in DvH cultures. Collectively, 35S-radiosulfur-isotope-labeled experiments and transcription/translation analyses unambiguously eliminate the possibility of sulfide-to-ZVS oxidation and reinforce our discovery of synchronous generation of elemental sulfur S8, along with sulfide, from DSR in SRMs and subsequent partial conversion to polysulfide (Fig. 2E). In this pathway, DsrC-trisulfide as an important intermediate from DsrAB-mediated sulfite reduction (17) could be critical to its subsequent conversion to both ZVS and sulfide, which awaits future studies.
Fig. 2.
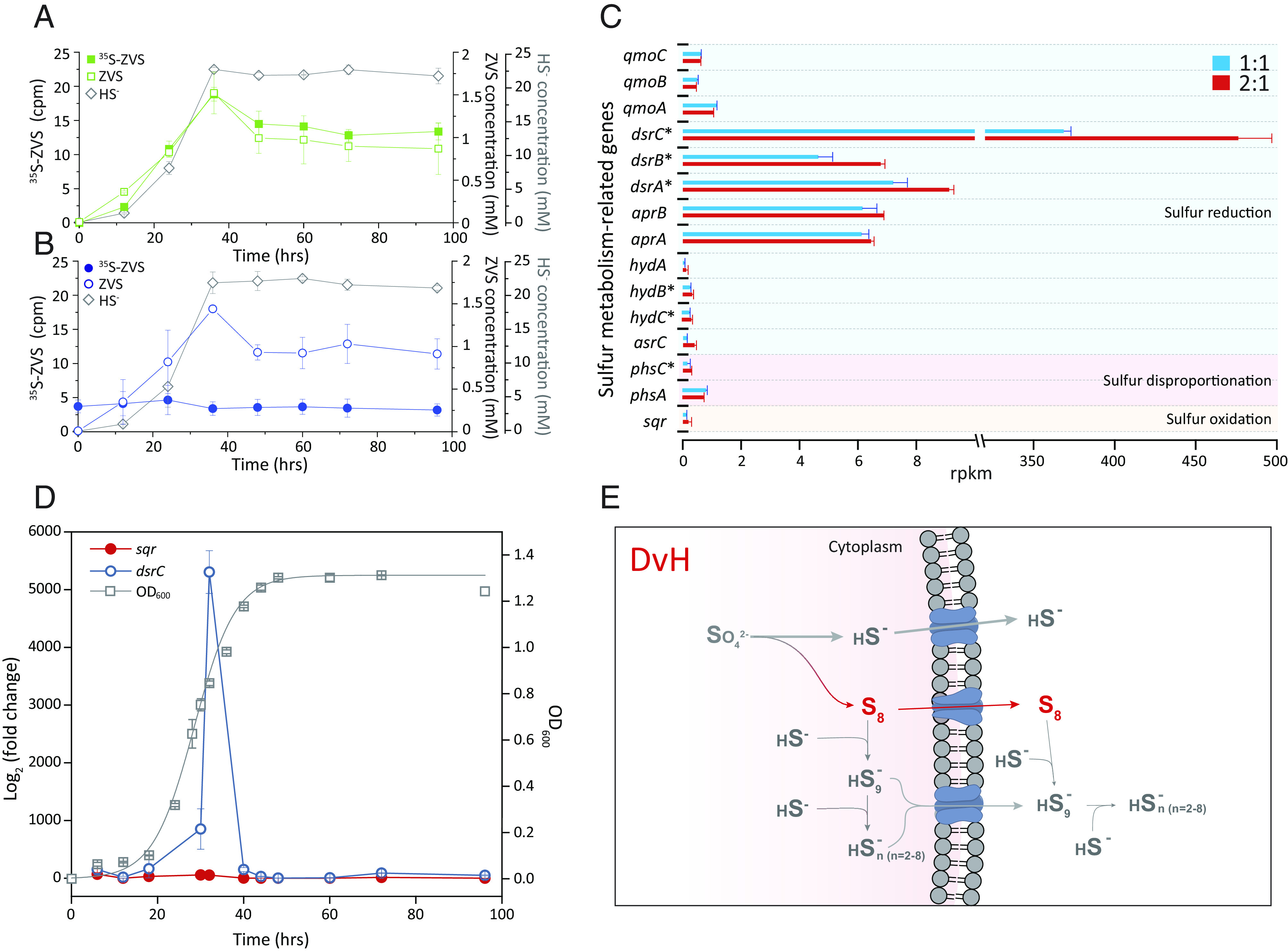
Isotopic and transcriptomic analyses of ZVS generation from DSR. (A) 35S-ZVS accumulation in DvH cultures fed with 35S-sulfate. (B) 35S-ZVS in DvH cultures fed with both sulfate and 35S-sulfide. (C) Transcription of key sulfur metabolism-related genes in DvH cultures fed with different molar ratios of lactate to sulfate, i.e., C/S ratios of 1:1 and 2:1. Statistical significance is based on the T test (n = 3), which is denoted by asterisks (*), P < 0.05. (D) qPCR-quantified transcription of dsrC and sqr genes in DvH. (E) A proposed model for the ZVS generation from DSR in SRB. Error bars represent SDs of triplicate cultures.
Biogeochemical Implications of DSR-Derived ZVS.
The SRMs-mediated DSR plays a key role in the past and present global cycle of sulfur with profound influence on the global cycles of other elements as well (1, 2, 34). In the modern marine environment, about 11.3 teramoles of sulfate are estimated to be reduced yearly, which accounts for the oxidation of 12 to 29% of the global organic carbon flux to the sea floor (2). If 8.9% of the sulfate reduction is channeled to ZVS, there will be around 1.01 teramoles of DSR-derived ZVS that may support a wide range of ZVS-metabolizing microorganisms, e.g., Geobacter, Pelobacter, and Dethiobacter (SI Appendix, Table S2) under thermodynamically favorable conditions (SI Appendix, Table S3). In our previous studies, we isolated a ZVS-reducing Geobacter (Geo) with acetate as both a carbon source and electron donor (35), which was used to form a DvH-Geo coculture fed with lactate and sulfate in this study. Compared to the DvH pure culture, lower concentrations of sulfide and slightly higher concentrations of ZVS were generated in the DvH-Geo coculture (Fig. 3A), suggesting an improved sulfate reduction in the coculture relative to the DvH pure culture. In contrast to no cell growth in the pure Geo culture fed with lactate and sulfate, both Geo and DvH in the coculture grew with the sulfate reduction and ZVS generation at the highest cell concentrations of 2.22 × 105 and 3.97 × 108 16S rRNA gene copies per ml, respectively, at the stationary phase (Fig. 3B). The low cell concentration of Geo in the coculture could be due to the limited amount of ZVS and low energy yield derived from ZVS reduction (SI Appendix, Table S2) as well as trace amount of acetate generated by DvH. The two populations may form a network through carbon- and sulfur-metabolisms, suggesting the DvH-derived ZVS-dependent cell growth of Geo (Fig. 3C). The analysis of 16S rRNA gene amplicon sequencing-based metadata of worldwide marine and terrestrial samples showed that similar metabolic networks between varied lineages of SRMs and ZVS-metabolizing microorganisms (S0MMs, including ZVS-reducing-microorganisms/S0RMs and ZVS-disproportionating-microorganisms/S0DMs) were ubiquitous in natural environments (Fig. 3D and SI Appendix, Table S4). Notably, many DSR sites being in the absence of taxonomically characterized S0MMs had both polysulfide reductase-encoding gene (psr) and dsr genes in their metagenomic data (Fig. 3D and SI Appendix, Table S5), indicating the coexistence of taxonomically uncharacterized S0MMs with SRMs. Therefore, both the 16S rRNA and metagenomic data analyses indicated the global distribution of coexisted SRMs and S0MMs. The multicomponent interaction between sulfur-based metabolizing microorganisms is also essential for us to precisely decipher geological records toward a deeper understanding of our Earth’s earliest microbial ecosystems (10, 36).
Fig. 3.
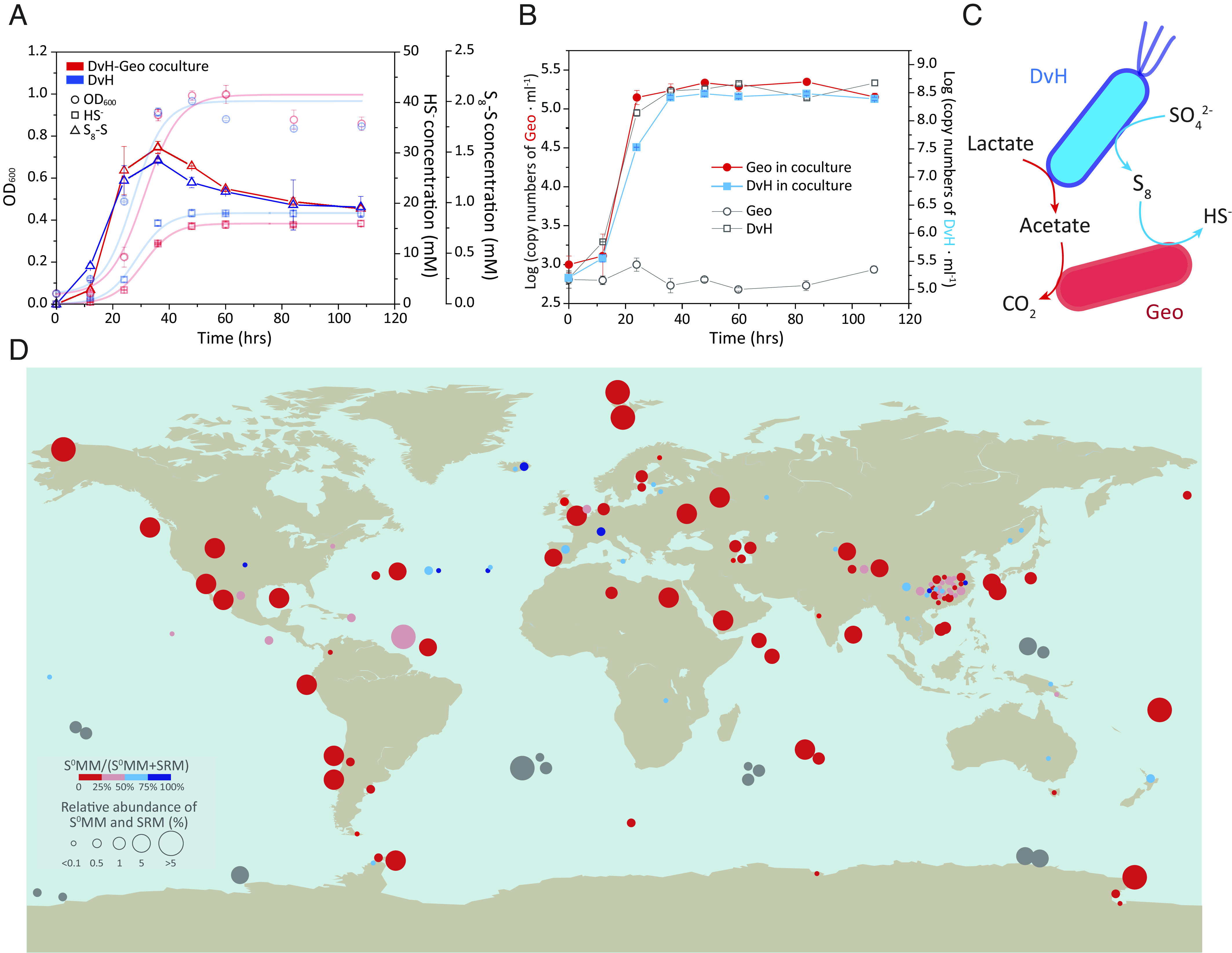
DSR-derived ZVS supported cell growth of ZVS-metabolizing microorganisms (S0MMs). (A) DSR and ZVS generation in a coculture of DvH and Geobacter lovelyi LYY (Geo). (B) Cell growth of DvH and Geo in the coculture with pure cultures as the controls. Pure culture controls of DvH and Geo were prepared under the same growth conditions with the DvH-Geo coculture. (C) Proposed syntrophic interactions between DvH and Geo in the coculture. (D) The global distribution of cooccurred SRMs and ZVS-metabolizing microorganisms (S0MMs, i.e., ZVS-reducing-microorganisms/S0RMs and ZVS-disproportionating-microorganisms/S0DMs) in marine and terrestrial environments by meta-analysis. The metadata were retrieved from previous studies based on 16S rRNA gene amplicon sequences (colorized bubbles) and metagenome sequencing data (gray bubbles). See SI Appendix, Tables S4 and S5 for the detailed metadata information. Error bars represent SDs of triplicate cultures.
In summary, we unraveled a thus far unrecognized pathway of sulfate-to-ZVS in the SRMs-mediated DSR process. This pathway may lay the foundation for better understanding of the fate and role of ZVS in the cryptic sulfur cycle in oxygen minimum zones (37), ANME-SRMs-based methane oxidation (4–6, 34, 38), sedimentary pyrite synthesis (16), and even the sulfur metabolism of early Archaean microorganisms (36). The identification of DSR-derived ZVS also fuels future exploration of the underappreciated sulfur metabolism in worldwide marine and terrestrial environments for the complete understanding of how sulfur biogeochemical cycling shapes our planet’s surface, atmosphere, and climate.
Materials and Methods
Cultivation of SRMs and S0RMs.
Seven SRMs/S0RMs strains and their growth conditions used in this study are listed in SI Appendix, Fig. S1, i.e., Desulfovibrio vulgaris Hildenborough (DvH), Desulforamulus ruminis DL (DLT), Desulfomicrobium baculatum DSM4028 (DSM), Pseudodesulfovibrio indicus J2 (PDI), Thermodesulfatator autotrophicus S606 (TDA), and Halodesulfovibrio marinisediminis C/L 2 (HDM) were selected as SRMs, and Geobacter lovleyi LYY (Geo) was chosen as S0RM. These SRMs from 3 phyla were selected based on their taxonomy and source derivation from both marine and terrestrial environments (SI Appendix, Fig. S1). Five strains of the SRMs (i.e., DvH, DLT, DSM, PDI, and HDM) were grown in an LS4D medium amended with sulfate (50 mM) as the electron acceptor and lactate (60 mM) as the carbon source and reducing equivalent (39). The chemolithoautotrophic and thermophilic SRM strain—TDA—was cultivated in SO4PN salts medium fed with hydrogen (160 kPa, gas phase), carbon dioxide (40 kPa, gas phase), and sulfate (10 mM) as the primary electron donor, carbon source, and electron acceptor, respectively (40). The ZVS-reducing Geo was isolated and maintained in a defined mineral salt medium amended with acetate as a carbon source and electron donor and tetrachloroethene as an electron acceptor, which could also couple cell growth with the reduction of ZVS but not sulfate (35). DvH-Geo coculture was transferred twice in the LS4D medium prior to the coculture experiments, with DvH and Geo pure cultures as biotic controls. For the culturing of SRMs and S0RMs, the completely defined, neutral mineral salts medium was prepared under strict anaerobic conditions as previously described (39–41), in which resazurin (20 μM) was added as a redox indicator. Cultures were transferred in 160-mL serum bottles containing a 98-mL sterilized medium with 2% (v/v) inoculum. Three sets of abiotic controls were prepared with different sulfur species (i.e., 50 mM sulfate, 50 mM sulfide, or 50 mM of both sulfate and sulfide) and without culture inoculation to test ZVS generation under abiotic conditions. Batch experiments were setup with DvH to test the impact of varied C/S ratio, pH, temperature, and salinity on ZVS generation: two experimental sets of C/S molar ratios (lactate:sulfate) including one with a fixed concentration of 50 mM lactate (i.e., 50:25, 50:50, and 50:100) and the other one with a fixed concentration of 50 mM sulfate (i.e., 25:50, 50:50, and 100:50); a gradient of temperature (i.e., 20 °C, 30 °C, and 37 °C), pH (i.e., 7, 8, and 9), and salinity (i.e., 0, 1%, and 2% NaCl, w/v) were prepared individually with the DvH-growing LS4D medium to investigate ZVS generation under different growth conditions. To investigate the color change in the culture medium during ZVS generation, DvH was transferred to no-Fe(II/III)-amendment LS4D medium with an inoculation ratio of 2%. Unless stated otherwise, cultures were incubated at 37 °C (65 °C for the TDA) in the dark without shaking.
Analytical Methods.
ZVS could be present as elemental sulfurs ( ) or polysulfides ( ). The total amount of ZVS was analyzed with ion chromatography (IC; ICS-600; Thermo Fisher Scientific; Carlsbad, CA, USA) via conversion of ZVS to thiosulfate as previously described (25, 42). First, 1-mL samples were collected from cultures using nitrogen-flushed syringes and then injected into 10-mL sealed vials containing 0.1 mL formaldehyde, 1 mL Na2SO3 (1 M) solution and 0.1 mL NaOH solution (1 M), and 2.8 mL deoxygenated ultrapure water. The deoxygenated ultrapure water, Na2SO3, and NaOH stock solutions were prepared and equilibrated in an anaerobic chamber to keep them under strict anaerobic conditions. Then, the mixed solution bottles were incubated in a 60 °C water bath for 24 h prior to subsequent IC test. To prevent the influence of SRM-generated thiosulfate, we also quantify the thiosulfate directly and set it as a control in testing ZVS. To test the total amount of intracellular and extracellular ZVS, mechanical cell disruption was conducted as described earlier (43, 44). Briefly, samples were collected and treated with FastPrep-24 5G Instrument (MP Biochemicals; Santa Ana, CA, USA) under anaerobic conditions. Then, the treated samples were subjected to the test of total ZVS. The extracellular ZVS was analyzed similarly without disrupting cells.
The elemental sulfurs were analyzed based on both a gas chromatograph-mass spectrometer (GC-MS) and a UV–Vis spectrometer: 1) For the GC-MS method (25, 42), 1 mL sample was extracted with 5 mL carbon tetrachloride, and then the solvent phase of 1 mL was transferred to a 2-mL amber vial for subsequent analysis with GC-MS (TQ8040; Shimazu; Kyoto, Japan) equipped with a DB-5 capillary column (30 m × 0.32 mm × 0.25 μm film thickness; Agilent; Folsom, CA, USA). The mass spectrometer was operated in electron impact mode (EI) at an ionization potential of 70 eV. Following mass-to-ion (m/z) ratios were used to detect different elemental sulfur species, i.e., 64, 96, 128, 160, 192, 224, 256, 257, 258, and 259. Temperatures of the injector, ion source, and column were all set at 180 °C, while the interface temperature was adjusted to 220 °C. Helium was used as a carrier gas at a flow rate of 1.2 mL·min−1. 2) For the S8-specific UV–Vis spectrometer method (26), 1 mL sample was extracted with 5 mL dichloromethane, and then, the solvent phase was collected for subsequent measurement with the UV–Vis spectrometer at 270 nm. Standard solutions for elemental sulfur quantification were prepared by dissolving S8 (sulfur powder; Alfa Aesar; Ward Hill, MA, USA) in carbon tetrachloride or dichloromethane. To prevent the potential impact of sulfide on the elemental sulfur test, sulfate reduction–derived sulfide in samples was removed with zinc acetate as described before (25, 42) prior to subsequent elemental sulfur analyses. Raman spectroscopy (inVia Qontor, Renishaw; Gloucestershire, UK) was used to further confirm the DSR-derived S8 (4, 45). In the first step, samples for the Raman spectroscopy analysis were extracted with carbon tetrachloride and then evaporated on calcium fluoride (CaF2) slides. The Raman spectra were recorded between 100 and 600 cm−1 with a 50-mW 532-nm laser and acquired with around 1% laser power and an acquisition time of 150 s.
Polysulfides were determined through high-performance liquid chromatography (HPLC) as described earlier (27). Briefly, 800 μL of deoxygenated methanol was placed in a 2-mL septum-closed HPLC vial, to which 200 μL sample and 5 μL methyl triflate were consecutively added. Prior to the sample injection to the mixture, samples were methylated using methyl triflate to form methyl polysulfides ( ). The mixture was first fortified with an internal standard (dibenz[a,h]anthracene in 1,4-dioxane) and then subjected to HPLC analysis. The HPLC (1290 Infinity II; Agilent; Santa Clara, CA, USA) was equipped with a C18 reverse phase column (Zorbax; Agilent) and a UV-detector at 220 and 230 nm wavelengths. Methanol was used as the mobile phase. A polysulfide standard for qualitative determination was prepared with potassium polysulfide (Sigma-Aldrich; Burlington, MA, USA) based on the methyl triflate derivatization (27). The dimethyl disulfide and dimethyl trisulfide were used as standards for the quantification of disulfide and trisulfide, respectively. Remaining polysulfides ( , n = 4 to 9) were quantified as described previously (46). To exclude the impact of S8 on polysulfide analyses, elemental sulfur extraction with carbon tetrachloride was used to pretreat samples. The total concentrations of the GC-MS-analyzed elemental sulfur and HPLC-analyzed polysulfide account for 78.6 to 120.3% of the IC-analyzed total ZVS concentrations.
Sulfate and thiosulfate were measured using the IC equipped with an IonPac AS23 analytical column (Thermo Fisher Scientific) as previously described (47). Carbonate buffer (0.8 mM NaHCO3 and 4.5 mM Na2CO3) was used as the eluent, and the flow rate was set at 1 mL min−1. Prior to the IC analysis, samples were filtered through a Fast & Low Binding Millipore filter (0.22 μM; Merck; Darmstadt, Germany) with zinc acetate (50 g L−1) for sulfide removal. Consequent sulfide measurement was conducted with a UV–Vis spectrometer (UV-2100; Shimadzu; Kyoto, Japan) at 675 nm using the classical methylene blue method (48, 49). The methylene blue method was based on the reaction of sulfide with N, N-dimethylphenyl-1,4-diamin in the presence of ferric ions. Unless stated otherwise, chemicals were purchased from Sigma-Aldrich at the highest purity available. For these analytical tests, sampling and solution preparation were conducted under anaerobic conditions to prevent sulfur oxidation.
Confirmation of DSR-Derived ZVS Using Radiotracers.
Radiolabeled sulfate (35SO42—) was purchased from PerkinElmer (NEX042001MC; Waltham, MA, USA). The 35S-labeled sulfide was obtained by reduction of diluent 35SO42— based on the modified strongly reducing hydriodic-hypophosphorous-hydrochloric acid (STRIP) method as described previously (50, 51). The reducing mixture constituting of 200 mL of concentrated HCl (37%, w/w), 330 mL hydriodic acid (57%, w/w), and 100 mL hypophosphorous acid (50%, w/w) was prepared in a 1-L round-bottom flask and boiled gently with N2 flush for 4 h to remove any sulfur contamination (51, 52). The reducing agent was stored in a brown glass bottle to avoid photooxidation after it was cooled under an inert atmosphere (N2 flush). Then, 30 mL of the reducing agent was added to a reaction flask, which was amended with 35SO42— containing approximately 5,200 Bq mL−1 radioactivity and boiled gently for 3 h. The evolved H2S was transported using N2 gas stream first into a water trap to remove acid fumes and then into a 10 mL ZnCl2 (0.4 mol L−1) trap to generate ZnS (53). The collected ZnS was further thoroughly washed, dried, and acidified with H3PO4 (1 mol L−1) in an anaerobic vial with N2 flush to release labeled sulfide (53).
Two experimental sets were established with DvH in an LS4D medium amended with 35S-sulfate and 35S-sulfide, as follows: 1) Cultures fed with 35S-sulfate (~16 Bq mL−1) were utilized to compare kinetic curves of ZVS- and sulfide-generation; we assumed that sulfide-to-ZVS reoxidation after sulfate-to-sulfide reduction would result in a lag time between the kinetic curves of ZVS- and sulfide-generation; 2) cultures supplemented with unlabeled sulfate and 35S-sulfide (~12.5 Bq mL−1) were used to test whether ZVS could be generated from the 35S-sulfide via oxidation in sulfate-reducing cultures. Abiotic controls were prepared under the same conditions without an inoculum. The time series of 35S activities in three sulfur pools (sulfate, ZVS, and sulfide) were determined in all experiments. Samples were collected and mixed with zinc acetate (0.9 mol L−1) under anaerobic conditions to trap sulfide. 35SO42— was determined after thorough removal of ZnS by centrifugation (15 min, 2,500× g) and ultrafiltration (Anotop® 25 syringe filter; Whatman, USA). The ZnS removal was further verified in filtrate aliquots by acidification with HCl (7 mol L−1; 0.5 mL per ml) and purging with N2 (53). Since ZVS was mainly present as elemental sulfurs (S0), we treated the radiolabeled elemental sulfurs as radiolabeled ZVS (35S-ZVS) that was extracted with carbon tetrachloride for quantitative analysis. Then, 0.5 mL sample containing different sulfur species was mixed with 2 mL of 30% H2O2, 2.5 mL of deionized water, and 10 mL scintillation cocktail (Ultima Gold LLT; PerkinElmer) for subsequent scintillation counting on a Quantulus GCT 6220 liquid scintillation analyzer (PerkinElmer) following the high-sensitivity radiosulfur analytical technique developed for natural abundance radiosulfur measurements (54, 55). Given that the aim of these radiosulfur-labeled experiments was to semiquantitatively identify the source of ZVS, we reported our data in the unit of cpm (counts per minute) and did not correct for the background or counting efficiency to obtain activities at units of disintegration per minute or Bq. Because we used the same amount of solution (0.5 mL) to do all 35S measurements and the chemical matrix was the same, the intercomparison between different experiments in this study could be valid. The background counts of these experiments were 1 to 2 cpm. Comparable relative abundances of accumulated S0 were identified with the isotopic and chromatographic methods. An initial and slight 35S-ZVS enrichment (<5 cpm) was observed in cultures amended with the unlabeled sulfate and 35S-sulfide (Fig. 2B). Given that the 35S activity in ZVS was not changed across the 35S-sulfide experiments, this observation could be explained by the isotope exchange between 35S-sulfide and ZVS (from inocula), which likely reached equilibrium at the very beginning of the experiments (56, 57). In addition, a low amount of 35S-sulfate (<10 cpm) was observed in the 35S-sulfide-fed cultures, which could be due to the sulfur back flux during sulfate reduction (SI Appendix, Fig. S6B) (53). The ZV35S enrichments in 35S-sulfide-fed cultures were significantly smaller than those observed in 35S-sulfate-fed cultures (Fig. 2A) and therefore would not affect our interpretation or change our conclusion.
Cell Growth of Sulfate- and Sulfur-Reducing Bacteria.
The biomass and growth phase of DvH were determined based on the optical density at a wavelength of 600 nm (OD600) as previously described (58). To measure the cell growth of DvH and Geo in their cocultures, quantitative real-time PCR (qPCR; CFX96 Touch System, Bio-Rad; Hercules, CA, USA) was performed with QuantiTect SYBR Green PCR kit (Qiagen; Hilden, Germany) and primers (SI Appendix, Table S6) as previously described (59, 60). Genomic DNA (gDNA) for the qPCR measurement was extracted from the DvH-Geo cocultures and control cultures using the FastDNA Spin Kit for Soil (MP Biomedicals; Carlsbad, CA, USA).
Transcription and Translation of Sulfur Metabolism Genes.
The DvH genome contained multiple gene sets for sulfur metabolism, including sulfate-reducing dsr genes and sulfide-to-ZVS oxidizing sqr-like gene (61, 62). Both transcriptomic and proteomic analyses were conducted to measure the transcription and translation of these sulfur metabolism genes in DvH cultivated using two different molar ratios of lactate to sulfate, i.e., 1:1 and 2:1, as previously described (41). Briefly, samples for total RNA and protein extraction were collected by centrifugation (5 min, 10,000× g, 4 °C) from triplicate cultures at the mid-exponential phase (OD600 = ~0.4). The RNA samples were then mixed with TRIzolTM reagent (Invitrogen; Waltham, MA, USA). Total RNA was extracted with an RNeasy Mini kit (Qiagen), from which fragmented DNA was removed using DNase I. The highly transcribed ribosomal RNAs (rRNAs) were eliminated using the RiBoCop rRNA depletion Kit (Lexogen; Vienna, Austria). The RNA-Seq libraries were constructed from the rRNA-depleted RNA samples using NEB Next® Ultra™ Directional RNA Library Prep Kit (New England Biolabs; Ipswich, MA, USA) and then sequenced on an Illumina Novaseq6000 platform. The RNA-Seq library construction and sequencing services were provided by Magigene (Shenzhen, China). RNA-Seq raw data in fastq format were processed by Trimmomatic (v.0.35) (63) to acquire the clean reads. The clean reads were first mapped to DvH’s rRNA gene to remove rRNA sequences by Bowtie2 (v2.33) (64). Then, the remaining mRNA sequences were mapped to the reference genome of DvH (NC_002937.3) by the Bowtie2. The HTSeq-count (v0.9.1) was used to obtain the read count and function information of each gene according to the mapping results (65). RPKM (reads per kilobase of transcript, per million mapped reads) was calculated to compare the expression levels of different genes (66). Normalized counts of transcription level in DvH were calculated against the expression of the housekeeping gene recA (SI Appendix, Table S1). Samples for proteome analysis were resuspended in 10 mM phosphate buffer solution (pH 7.4) and centrifuged again prior to their storage at −80 °C. Further protein extraction, digestion, and quantification services were provided by Beijing Genomics Institute (BGI; Shenzhen, China) as described (67).
To monitor the temporal transcription of dsrC and sqr-like genes, qPCR enumeration with dsrC/sqr-like gene-specific primers (SI Appendix, Table S6) was performed with a QuantiTect SYBR Green PCR kit (Qiagen). RNA for the qPCR analyses was reverse transcribed into cDNA using a two-step RT-PCR Sensiscript kit as previously described (41). The recA was used as an internal standard (68). Gene expression changes were calculated on the basis of mean change in the cycle threshold (ΔCt) compared to the recA (69). Each sample was analyzed with at least three technical replicates.
Metadata Analyses of the Coexisting SRMs and ZVS-Metabolizing Microorganisms in Natural Environments.
To explore the global coexistence of SRMs and ZVS-metabolizing microorganisms in various natural environments, published 16S rRNA gene amplicon sequencing data of marine (marine sediment and hydrothermal vent) and terrestrial (soil, river sediment, hot springs, and acid mine drainage) samples (SI Appendix, Table S4) were retrieved from NCBI’s Sequence Read Archive (SRA) database. Raw data containing paired-end reads were merged and filtered to remove low-quality sequences or ambiguous sequences using mothur (v1.39) (70). Unique sequences were identified and denoised using usearch (v11.0.667). The unoise3 algorithm (71) was used to construct zero-radius operational taxonomic units (zOTUs) and to filter chimeras. Taxonomies of the zOTUs were assigned with RDP Classifier (72) with an 80% confidence cut-off against the SILVA (v138) reference database. Then, the zOTUs were divided by total sample reads and collapsed to the genus level to generate their relative abundance.
For the sulfate-reducing sites having SRMs but without known S0MMs, metagenomic data were collected from the NCBI’s SRA database to retrieve both dsr and psr gene sequences (SI Appendix, Table S5). The metagenomic sequencing raw data were filtered to remove low-quality reads using Sickle (73) with parameters set to “−q = 20”. The clean paired-end reads were merged by PEAR (74). The merged sequences were then searched against the SCycDB databases (75) using the DIAMOND program with following parameters: −k 1 −e 0.0001 (76). Sequences matched with psr and dsrC genes were counted, and the relative abundance of dsrC-containing SRMs and psr-containing S0MMs was calculated as described earlier (66).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Inês A.C. Pereira for discussions and Xiaomin Lin for the isotopic measurements. This study was financially supported by the National Natural Science Foundation of China (42161160306 and 41922049) and the Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (No. SML2021SP317). M.L. was supported by the Key Research Program of Frontier Sciences from the Chinese Academy of Sciences (ZDBS-LY-DQC035) and the Guangdong Pearl River Talents Program (2019QN01L150).
Author contributions
S.W. conceived the idea and designed the study. Q.L., Z.L. and X.Y. performed laboratory experiments. Q.L., Z.L. and S.W. analyzed the data. All authors discussed and interpreted the data. S.W. wrote the manuscript with inputs from all authors.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The RNA-seq and mass-spectrometry proteomics data have been deposited in the EMBL BioStudies (E-MTAB-12239) (77) and the Proteomics Identifications Database (PRIDE, PXD040825) (78), respectively. All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Muyzer G., Stams A. J., The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Bowles M. W., Mogollón J. M., Kasten S., Zabel M., Hinrichs K. U., Global rates of marine sulfate reduction and implications for subsea-floor metabolic activities. Science 344, 889–891 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Wu B., et al. , Microbial sulfur metabolism and environmental implications. Sci. Total Environ. 778, 146085 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Milucka J., et al. , Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491, 541–546 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Yu H., et al. , Comparative genomics and proteomic analysis of assimilatory sulfate reduction pathways in anaerobic methanotrophic archaea. Front. Microbiol. 9, 2917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H., et al. , Sulfate differentially stimulates but is not respired by diverse anaerobic methanotrophic archaea. ISME J. 16, 168–177 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vigneron A., et al. , Contrasting pathways for anaerobic methane oxidation in gulf of mexico cold seep sediments. mSystems 4, e00091–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott P., Ragusa S., Catcheside D., Growth of sulfate-reducing bacteria under acidic conditions in an upflow anaerobic bioreactor as a treatment system for acid mine drainage. Water Res. 32, 3724–3730 (1998). [Google Scholar]
- 9.Qian Z., Tianwei H., Mackey H. R., van Loosdrecht M. C. M., Guanghao C., Recent advances in dissimilatory sulfate reduction: From metabolic study to application. Water Res. 150, 162–181 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Shen Y., Buick R., Canfield D. E., Isotopic evidence for microbial sulphate reduction in the early archaean era. Nature 410, 77–81 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Fike D. A., Bradley A. S., Rose C. V., Rethinking the ancient sulfur cycle. Annu. Rev. Earth Planet. Sci. 43, 593–622 (2015). [Google Scholar]
- 12.Zhou J., et al. , How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat. Rev. Microbiol. 9, 452–66 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Wunder L. C., et al. , Iron and sulfate reduction structure microbial communities in (sub-)antarctic sediments. ISME J. 15, 3587–3604 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi K., Tachibana S., Ishimoto M., Intermediary formation of trithionate in sulfite reduction by a sulfate-reducing bacterium. J. Biochem. 65, 155 (1969). [PubMed] [Google Scholar]
- 15.Sass H., Steuber J., Kroder M., Kroneck P. M., Cypionka H., Formation of thionates by fresh-water and marine strains of sulphate-reducing bacteria. Arch. Microbiol. 158, 418–421 (1992). [Google Scholar]
- 16.Fitz R. M., Cypionka H., Formation of thiosulfate and trithionate during sulfite reduction by washed cells of desulfovibrio desulfuricans. Arch. Microbiol. 154, 400–406 (1990). [Google Scholar]
- 17.Santos A. A., et al. , A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350, 1541–1545 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Ferreira D., et al. , The DsrD functional marker protein is an allosteric activator of the DsrAB dissimilatory sulfite reductase. Proc. Natl. Acad. Sci. U.S.A. 119, e2118880119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luther G. W., Pyrite synthesis via polysulfide compounds. Geochim. Cosmochim. Acta 55, 2839–2849 (1991). [Google Scholar]
- 20.Fang W., Gu M., Liang D., Chen G. H., Wang S., Generation of zero valent sulfur from dissimilatory sulfate reduction under methanogenic conditions. J. Hazard. Mater. 10, 121197 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Labrado A. L., Brunner B., Bernasconi S. M., Peckmann J., Formation of large native sulfur deposits does not require molecular oxygen. Front. Microbiol. 10, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe M., Fukui M., Kuever J., "Desulforamulus gen. nov" in Bergey’s Manual of Systematics of Archaea and Bacteria, Anonymous N. N., Ed. (John Wiley & Sons, New York, 2020), pp. 2–8. [Google Scholar]
- 23.Kamyshny A., Borkenstein C. G., Ferdelman T., Protocol for quantitative detection of elemental sulfur and polysulfide zero-valent sulfur distribution in natural aquatic samples. Geostand. Geoanal. Res. 33, 415–435 (2009). [Google Scholar]
- 24.Wu D., et al. , Simultaneous nitrogen and phosphorus removal in the sulfur cycle-associated enhanced biological phosphorus removal (EBPR) process. Water Res. 49, 251–264 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Joly H. A., Belzile N., Determination of elemental sulfur in environmental samples by gas chromatography-mass spectrometry. Chem. Geo. 137, 195–200 (1997). [Google Scholar]
- 26.Houghton J. L., et al. , Thiosulfate oxidation by Thiomicrospira thermophila: Metabolic flexibility in response to ambient geochemistry. Environ. Microbiol. 18, 3057–3072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamyshny A., Ekeltchik I., Gun J., Lev O., Method for the determination of inorganic polysulfide distribution in aquatic systems. Anal. Chem. 78, 2631–2639 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Tebbe F. N., Wasserman E., Peet W. G., Vatvars A., Hayman C., Composition of elemental sulfur in solution: Equilibrium of S6, S7, and S8 at ambient temperatures. J. Am. Chem. Soc. 104, 4972–4974 (1982). [Google Scholar]
- 29.Jarmakka M., et al. , Molecular mechanism of energy conservation in polysulfide respiration. Nat. Struct. Mol. Biol. 15, 730–737 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Tessier A., Zero-valent sulfur and metal speciation in sediment porewaters of freshwater lakes. Environ. Sci. Technol. 43, 7252–7257 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Dahl C., "Bacterial intracellular sulphur globules" in Bacterial Organelles and Organelle-Like Inclusions Microbiology Monographs, Jendrossek D., Ed. (Springer, Cham, 2020), vol. 34, 10.1007/978-3-030-60173-7_2. [DOI] [Google Scholar]
- 32.Roy H., Weber H. S., Tarpgaard I. H., Ferdelman T. G., Jorgensen B. B., Detemination of dissimilatory sulfate reduction rates in marine sediment via radioactive 35S tracer. Limnol. Oceanogr. Methods 12, 196–211 (2014). [Google Scholar]
- 33.Guo W., Cecchetti R. A., Wen Y., Zhou Q., Sedlak D. L., Sulfur cycle in a wetland microcosm: Extended 34S-stable isotope analysis and mass balance. Environ. Sci. Technol. 54, 5498–5508 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Vigneron A., et al. , Beyond the tip of the iceberg; A new view of the diversity of sulfite- and sulfate-reducing microorganisms. ISME J. 12, 2096–2099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Y., et al. , Substrate-dependent competition and cooperation relationships between Geobacter and Dehalococcoides for their organohalide respiration. ISME Commun. 1, 23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philippot P., et al. , Early archaean microorganisms preferred elemental sulfur, not sulfate. Science 317, 1534–1537 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Canfield D. E., et al. , A cryptic sulfur cycle in oxygen-minimum–zone waters off the chilean coast. Science 330, 1375–1378 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Bell E., et al. , Active anaerobic methane oxidation and sulfur disproportionation in the deep terrestrial subsurface. ISME J. 16, 1583–1593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay A., et al. , Salt stress in Desulfovibrio vulgaris Hildenborough: An integrated genomics approach. J. Bacteriol. 188, 4068–4078 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alain K., et al. , Thermodesulfatator atlanticus sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a mid-atlantic ridge hydrothermal vent. Int. J. Sys. Evo. Microbiol. 60, 33–38 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Wang S., et al. , Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc. Natl. Acad. Sci. U.S.A. 111, 12103–12108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogowska J., Sychowska J., Cieszynska-Semenowicz M., Wolska L., Elemental sulfur in sediments: Analytical problems. Environ. Sci. Pollut. Res. 23, 24871–24879 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Whyte J. D., Advances in flavivirus research applications: New techniques using the FastPrep-24 5G sample preparation system. Nat. Method. 13, i–iii (2016). [Google Scholar]
- 44.Nagler M., Podmirseg S. M., Mayr M., The masking effect of extracellular DNA and robustness of intracellular DNA in anaerobic digester NGS studies: A discriminatory study of the total DNA pool. Mol. Ecol. 30, 438–450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J., et al. , A novel bacterial thiosulfate oxidation pathway provides a new clue about the formation of zero-valent sulfur in deep sea. ISME J. 14, 2261–2274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizkov D., Lev O., Gun J., Development of in-house reference materials for determination of inorganic polysulfides in water. Accred. Qual. Assur. 9, 399–403 (2004). [Google Scholar]
- 47.Karmarkar S. V., Quick ion chromatographic determination of sulfate alone in soil extracts and natural waters. Commun. Soil Sci. Plan. 27, 843–852 (1996). [Google Scholar]
- 48.Mousavi M. F., Sarlack N., Spectrophotometric determination of trace amounts of sulfide ion based on its catalytic reduction reaction with methylene blue in the presence of Te(IV). Anal. Lett. 30, 1567–1578 (1997). [Google Scholar]
- 49.Lawrence N. S., Davis J., Compton R. G., Analytical strategies for the detection of sulfide: A review. Talanta 52, 771–784 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Luke C. L., Determination of total sulfur in rubber. Ind. Eng. Chem. 15, 602–604 (1943). [Google Scholar]
- 51.Arnold G. L., Brunner B., Muller I. A., Roy H., Modern applications for a total sulfur reduction distillation method-what’s old is new again. Geochem. Trans. 15, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin M., Thiemens M. H., A simple elemental sulfur reduction method for isotopic analysis and pilot experimental tests of sysmmetry-dependent sulfur isotope effects in planetary processes. Geochem. Geophy. Geosystem. 21, e2020GC009051 (2020). [Google Scholar]
- 53.Holler T., et al. , Carbon and sulfur back flux during anaerobic microbial oxidation of methane and coupled sulfate reduction. Proc. Natl. Acad. Sci. U.S.A. 108, E1484–E1490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin M., Thiemens M. H., Accurate quantification of radiosulfur in chemically complex atmospheric samples. Anal. Chem. 90, 2884–2890 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Lin X., Yu X., Lin M., Analysis of atmospheric radiosulfur at natural abundance by a new-type liquid scintillation counter equipped with guard compensation technology. ACS Earth Space Chem. 6, 1868–1875 (2022), 10.1021/acsearthspacechem.2c00104. [DOI] [Google Scholar]
- 56.Jørgensen B. B., Kuenen J. G., Cohen Y., Microbial transformations of sulfur compounds in a stratified lake (Solar Lake, Sinai). Limnol. Oceanogr. 24, 799–822 (1979). [Google Scholar]
- 57.Fossing H., Jørgensen B. B., Isotope exchange reactions with radiolabeled sulfur compounds in anoxic seawater. Biogeochemistry 9, 223–245 (1990). [Google Scholar]
- 58.Zhou A., et al. , Characterization of NaCl tolerance in Desulfovibrio vulgaris Hildenborough through experimental evolution. ISME J. 7, 1790–1802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fite A., Macfarlane G. T., Cummings J. H., Identification and quantitation of mucosal and faecal Desulfovibrio using real time polymerase chain reaction. Gut 53, 523–529 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amos B. K., et al. , Detection and quantification of Geobacter lovleyi strain SZ: Implications for bioremediation at tetrachloroethene- and uranium-impacted sites. Appl. Environ. Microbiol. 73, 6898–904 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langwig M. V., et al. , Large-scale protein level comparison of detaproteobacteria reveals cohesive metabolic groups. ISME J. 16, 307–320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heidelberg J. F., et al. , The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B., Salzberg S., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S., Pyl P. T., Huber W., HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Zheng H., et al. , General quantitative relations linking cell growth and the cell cycle in escherichia coli. Nat. Microbiol. 5, 995–1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bose A., Gardel E. J., Vidoudez C., Parra E. A., Girguis P. R., Electron uptake by iron-oxidizing phototrophic bacteria. Nat. Commun. 5, 3391 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Schloss P. D., et al. , Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R. C., Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 34, 2371–2375 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Wang Q., Garrity G. M., Tiedje J. M., Cole J. R., Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi N. A., Fass J. N., JSickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files, Version 1.33. https://github.com/najoshi/sickle. Accessed 20 May 2021.
- 74.Zhang J., Kobert K., Flouri T., Stamatakis A., PEAR: A fast and accurate illumina paired-end reAd mergeR. Bioinformatics 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu X., et al. , SCycDB: A curated functional gene database for metagenomic profiling of sulphur cycling pathways. Mol. Ecol. Resour. 21, 924–940 (2021). [Google Scholar]
- 76.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Liang Z., Lu Q., Wang S., Generation of zero-valent sulfur from dissimilatory sulfate reduction in sulfate-reducing bacteria. BioStudies Database. https://www.ebi.ac.uk/biostudies/studies/E-MTAB-12239. Deposited 23 September 2022.
- 78.Liang Z., Lu Q., Wang S., Generation of zero-valent sulfur from dissimilatory sulfate reduction in sulfate-reducing microorganisms. PRIDE Database. http://www.ebi.ac.uk/pride/archive/projects/PXD040825. Deposited 13 March 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The RNA-seq and mass-spectrometry proteomics data have been deposited in the EMBL BioStudies (E-MTAB-12239) (77) and the Proteomics Identifications Database (PRIDE, PXD040825) (78), respectively. All other study data are included in the article and/or SI Appendix.


