Highlights
-
•
The piRNA expression profiles of HSV-1-infected human fibroblasts identified 69 differentially expressed piRNAs.
-
•
Functional enrichment analysis revealed putative biological function of dysregulated piRNAs associated with antiviral immunity.
-
•
Two piRNAs, piRNA-hsa-28,382 and piRNA-hsa-28,190, potentially regulate HSV-1 replication.
Keywords: HSV-1, piRNA, RNA-seq, Expression pattern, Viral replication
Abstract
Piwi-interacting RNAs (piRNAs) are a class of non-coding RNAs that play a key role in spermatogenesis. However, little is known about their expression characterization and role in somatic cells infected with herpes simplex virus type 1 (HSV-1). In this study, we systematically investigated the cellular piRNA expression profiles of HSV-1-infected human lung fibroblasts. Compared with the control group, 69 differentially expressed piRNAs were identified in the infection group, among which 52 were up-regulated and 17 were down-regulated. The changes in the expression of 8 piRNAs were further verified by RT-qPCR with a similar expression trend. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that the target genes of piRNAs were mainly involved in antiviral immunity and various human disease-related signaling pathways. Furthermore, we tested the effects of four up-regulated piRNAs on viral replication by transfecting piRNA mimics. The results showed that the virus titers of the group transfected with piRNA-hsa-28,382 (alias piR-36,233) mimic decreased significantly, and that of the group transfected piRNA-hsa-28,190 (alias piR-36,041) mimic significantly increased. Overall, our results revealed the expression characteristics of piRNAs in HSV-1-infected cells. We also screened two piRNAs that potentially regulate HSV-1 replication. These results may promote a better understanding of the regulatory mechanism of pathophysiological changes induced by HSV-1 infection.
1. Introduction
Herpes simplex virus type 1 (HSV-1) is a highly infectious virus that is primarily transmitted by oral-oral contact (saliva). It belongs to the herpes α virus subfamily and its seroprevalence rate in the population can even reach 80% (James et al., 2020; Whitley and Roizman, 2001). Unlike HSV-2, which belongs to the same alphaherpesvirinae, HSV-1 rarely causes genital area infection, but up to 40% of infected patients may develop clinical symptoms such as skin lesions and even central nervous system complications (Duarte et al., 2019). HSV-1 can cause lifelong latent infection. When the host is subjected to various nonspecific stimuli, it can periodically reactivate the latently infected virus, leading to recurrent disease (Elbers et al., 2007). At present, there is no effective vaccine against HSV-1, and antiviral therapy is the main clinical control strategy for HSV infection (Johnston et al., 2016). Yet, high drug resistance has emerged in people with low immune function (Greeley et al., 2020). Therefore, it is of urgent importance to deeply understand the infection of HSV-1 and its interaction with the host.
PIWI-interacting RNAs (piRNAs) are a class of small silent RNAs containing 24 to 31 nucleotides (nt) (Aravin et al., 2001). The piRNAs were first identified in the testis of Drosophila, which can silence Stellate (the multi-copy gene) on the X chromosome of Drosophila (Aravin et al., 2001). PIWI proteins belong to the argonaute/PIWI family (Carmell et al., 2002) and contain domains of endonuclease activity that enable them to cleave RNA. In humans, there are four PIWI homologues, namely PIWIL1, PIWIL2, PIWIL3, and PIWIL4 (Kim, 2019). As a member of non-coding RNAs, piRNAs usually interact with PIWI proteins to induce target gene silencing by forming an RNA-induced silencing complex (RISC) (Girard et al., 2006). Compared with well-known microRNAs, piRNAs are different in length, quantity, and expression pattern (Aravin et al., 2006). The roles of piRNAs are very conservative in animal germ cells and can protect the vast majority of animal germ line genomes from transposon expression (Lewis et al., 2018). The piRNAs can also function outside the germ line. Abnormal regulation of piRNAs has been found in different tumor tissues and may be closely related to the malignant phenotype and clinical stage of tumor cells (Ozata et al., 2019).
Previously, the process of achieving antiviral defense based on RNA interference mechanism was usually attributed to siRNAs, especially in plant and insect virus infection (Berkhout, 2018). With an increasing number of studies on piRNAs in recent years, the role of piRNAs in the process of virus infection has also gained new understanding. For example, studies have found that mosquitoes can fight RNA virus infections through the ping-pong pathway (Miesen et al., 2015, 2016). However, there is still a big gap in the research on the expression pattern and function of piRNAs in virus-infected mammalian cells. Previously, the expression of piRNAs induced by coxsackievirus B3 (CVB3) and human rhinovirus (HRV) infections was analyzed (Li et al., 2021; Yao et al., 2020). The possible mechanisms of piRNAs involved in the regulation of pathophysiological changes in human cells infected with viruses were initially explored. However, the expression pattern and function of piRNAs during HSV-1 infection remains unknown.
In this study, we sought to investigate the effect of HSV-1 infection on piRNA expression and explore the possible role of differentially expressed piRNAs in the process of HSV-1 infection. We investigated host piRNAs' expression patterns and function during HSV-1 infection using high-throughput RNA sequencing and functional analysis in HSV-1 infected and mock-infected human lung fibroblasts. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyzed differentially expressed piRNAs to reveal the potential roles of piRNAs in the process of HSV-1 infection. Furthermore, we tested the effects of four up-regulated piRNAs on viral replication by transfecting piRNA mimics. Overall, our results revealed the expression patterns of piRNAs induced by HSV-1 infection and identified two piRNAs that potentially regulate HSV-1 replication, which furthers our understanding of the regulatory mechanism of pathophysiological changes induced by HSV-1 infection.
2. Materials and methods
2.1. Cells culture, virus infection, and piRNAs transfection
Human embryonic lung diploid fibroblast (KMB17) cells are susceptible to HSV-1 and have revealed the expression pattern of circular RNAs in our previous study (Shi et al., 2018). Human embryonic lung diploid fibroblast (KMB17) cells are susceptible to HSV-1, and the expression pattern of circular RNAs has been revealed in our previous study. KMB17 cells were cultured in MEM medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin, and streptomycin in a humidified atmosphere containing 5% CO2/ 95% air at 37 °C. The HSV-1 strain 17 was used to infect KMB17 cells at a multiplicity of infection (MOI) of 1 for sample preparation. piRNA mimics, which were used to overexpress piRNAs, were synthesized by the GenePharma (Shanghai, China). For piRNA mimic transfection, KMB17 cells were seeded uniformly in 6-well plates at 50,000 cells per well; after 12 h, piRNA mimics transfection was performed using Lipofectamine 3000 (Cat no. L3000008, Thermo Fisher, USA) according to the manufacturer's protocol.
2.2. Immunofluorescence
At 48 h after infection with HSV-1 at a MOI of 1, KMB17 cells in cell climbing slices in 12-well plates were fixed with 4% paraformaldehyde for 20 min at room temperature (RT) , permeated with 0.1% Triton X-100, blocked with 5% BSA, and incubated with HSV-1 ICP5 antibody (Cat no. ab6508, 1:500, Abcam, UK) overnight at 4 °C. Fluorescein (FITC)-Conjugated Affinipure Goat Anti-Mouse IgG (Cat No. SA00003–1, 1:200, Proteintech Group, USA) was used as a secondary antibody. Finally, the nuclei were stained with DAPI at RT for 10 min. The images were captured using a panoramic MIDI digital scanner (3D HISTECH, Hungary).
2.3. Data processing and analysis
In order to analyze the expression patterns of piRNAs during HSV-1 infection, we used the RNA-seq dataset from our previous study (Shi et al., 2018), numbered GSE102470, which has been uploaded to the public database GEO (https://www.ncbi.nlm.nih.gov/geo/). There were six samples and three biological replicates for each of the HSV-1 infection and the control groups. The Cutadapt (version 1.14) (Marcel, 2011) software was used to remove the connector sequences from the original data and filter the sequence length (sequences lengths 〈 15 nt and 〉 41 nt were removed). Fastx-toolkit (version 0.0.13) (Gordon and Hannon, 2010) software was used to perform Q20 quality control on sequences, and the sequences whose Q20 reached 80% or more were retained. The reads containing N bases were then filtered out by using NGS QC Toolkit (version 2.3.3) (Patel and Jain, 2012)software. High-quality clean reads were finally obtained and used for subsequent analysis. Fastx-toolkit (Version 0.0.13) (Gordon and Hannon, 2010) software was used to count the types of clean reads and obtain unique reads.
2.4. Alignment of small RNAs and identification of piRNAs
Bowtie (Langmead, 2010) software was used to annotate and identify small RNAs, while clean reads were compared with the Rfam (version 10.0) (Griffiths-Jones et al., 2003)database. The rRNA, snRNA, Cis-reg, tRNA, and other sequences were annotated and removed as far as possible and were not used for subsequent known piRNAs alignment. The filtered reads were matched with the transcript sequences without error, and the transcript sequences could be completely aligned; the sequences longer than 26 nt were considered degraded fragments of mRNA, and this part of the sequences was filtered. Then, the miRBase (version 22.0) (Griffiths-Jones et al., 2006, 2008) database was used to make zero base mismatch matching between the sequences without aligned transcripts, and the miRBase database was used to filter out the miRNAs. Finally, the sequences with 18–34 nt in the filtered reads were matched with piRNAs of this species in piRBase (version 1.0) (Wang et al., 2019) database using Bowtie software. The aligned sequences were considered as known piRNAs. Based on this, the expression of piRNAs was counted, and the subsequent differential expression analysis was carried out.
2.5. Differential expression analysis of piRNAs
Transcripts Per million (TPM) (Sun et al., 2014) was used to quantify known piRNAs. The R package DESEq2 (Anders, 2010) was applied to screen the differential piRNAs among different samples. For differential expression analysis of piRNAs, the default conditions for filtering differences had p-value < 0.05 and |log2 foldchange| ≥ 1. The expression level of piRNAs was used to conduct principal component analysis (PCA) to investigate the distribution and correlation between biological samples.
2.6. Target gene prediction of piRNAs
The miRanda (John et al., 2004) algorithm was used to predict host target genes of differentially expressed piRNAs. The principles of miRanda algorithm are as follows: (1) piRNAs and mRNAs sequences matching; (2) according to the energy stability of piRNA and mRNA binding, the target relationship between piRNA and mRNA is comprehensively predicted. The interaction between the top 10 up-regulated differentially expressed piRNAs and HSV-1 major genes was analyzed by RNAInter (version 4.0) (Kang et al., 2022). The piRNA-target gene network was constructed using Cytoscape software (3.9.1) (Shannon et al., 2003).
2.7. GO and KEGG pathway analysis
The Gene Ontology (GO) functional analysis was performed to annotate and classify all genes of species and target genes of differentially expressed piRNAs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is the main public database for biological pathways. Pathways analysis of target genes of differentially expressed piRNAs was performed using the KEGG database. A hypergeometric distribution test was used to calculate the significance of enrichment of target genes of differentially expressed piRNAs in each Pathway entry.
2.8. RT-qPCR analysis of piRNAs
To verify the expression patterns of piRNAs, 8 piRNAs (4 up-regulated and 4 down-regulated) were randomly selected from the top 20 differentially expressed piRNAs (Tables 1 and 2) for RT-qPCR analysis. KMB17 cells were infected with HSV-1 at a MOI of 1 in 6-well cell culture plates for 48 h. Then infected or uninfected cells were harvested from 3 wells using TRIzol Reagent, and total RNA was extracted. cDNA was synthesized using specific stem-loop RT primers (Table S1) and GoScript™ Reverse Transcription System (Cat no. A5001, Promega, USA). RT-qPCR was performed by designing specific forward and universal reverse primers for each piRNA using GoTaq® qPCR Master Mix reagent (Cat no. A6002, Promega, USA) on Bio-Rad CFX96 real-time detection system. The RT-qPCR programs were as follow: 95 °C for 5 min, 40 circles at 95 °C for 30 s, 50 °C for 30 s. All samples were normalized by U6 small nuclear RNA. Finally, 2−ΔΔCt method (Livak and Schmittgen, 2001) was used for data analysis.
Table 1.
Characteristics of top 10 up-regulated piRNAs induced by HSV-1 infection in KMB17 cells.
| piRNA id | Chromosome | Strand | Start | End | Sequence |
|---|---|---|---|---|---|
| piR-hsa-28,634 | chr21 | + | 8,218,733 | 8,218,764 | GUCCGGUGCGGAGUGCCCUUCGUCCUGGGAA |
| piR-hsa-24,085 | chrUn | + | 157,608 | 157,637 | CUGGGAAUGCAGCCCAAAGCGGGUGGUAA |
| piR-hsa-23,326 | chr21 | + | 8,255,104 | 8,255,134 | CCGGCCCGGACACGGACAGGAUUGACAGAU |
| piR-hsa-23,248 | chr21 | + | 8,210,895 | 8,210,927 | CCCGGCCCGGACACGGACAGGAUUGACAGAUU |
| piR-hsa-28,382 | chr12 | – | 127,166,399 | 127,166,425 | GGGGCGAAGCUACCAUCUGUGGGAUU |
| piR-hsa-18,905 | chr21 | + | 8,441,435 | 8,441,462 | UGGGAAUGCAGCCCAAAGCGGGUGGUA |
| piR-hsa-28,190 | chr11 | + | 109,603,196 | 109,603,226 | GGCCGUGAUCGUAUAGUGGUUAGUACUCUG |
| piR-hsa-26,399 | chrUn | + | 157,612 | 157,640 | GAAUGCAGCCCAAAGCGGGUGGUAAACU |
| piR-hsa-11,080 | chr12 | – | 127,166,408 | 127,166,437 | UGAGGAGCCAAUGGGGCGAAGCUACCAUC |
| piR-hsa-26,946 | chrUn | + | 159,995 | 160,024 | GAGGGGCCCGUGCCUUGGAAAGCGUCGCG |
Table 2.
Characteristics of top 10 down-regulated piRNAs induced by HSV-1 infection in KMB17 cells.
| piRNA id | Chromosome | Strand | Start | End | Sequence |
|---|---|---|---|---|---|
| piR-hsa-21,509 | chrM | – | 248 | 278 | UGUCUGUGUGGAAAGCGGCUGUGCAGACAU |
| piR-hsa-21,508 | chrM | – | 249 | 278 | UGUCUGUGUGGAAAGCGGCUGUGCAGACA |
| piR-hsa-28,391 | chr6 | – | 172,971 | 173,002 | GGGGGGUGUAGCUCAGUGGUAGAGCGCGUGC |
| piR-hsa-27,222 | chr17 | – | 18,253,912 | 18,253,939 | GAUUAUGAUGAUGCCUUAACACUGACU |
| piR-hsa-27,400 | chr6 | – | 27,592,859 | 27,592,891 | GCAGAGUGGCGCAGCGGAAGCGUGCUGGGCCC |
| piR-hsa-27,138 | chr6 | – | 27,335,032 | 27,335,062 | GCCUGGGUAGCUCAGUCGGUAGAGCAUCAG |
| piR-hsa-1245 | chr9 | – | 137,013,749 | 137,013,775 | AGCCCUGAUGAUGCCCACUCCUGAGC |
| piR-hsa-28,183 | chrX | – | 3,915,277 | 3,915,303 | GGCCGGUUAGCUCAGUUGGUAAGAGC |
| piR-hsa-23,230 | chr2 | + | 205,354,229 | 205,354,259 | CCCCUGGUCGUCUAGUGGUUAGGAUUCGGC |
| piR-hsa-28,845 | chr1 | – | 30,968,167 | 30,968,195 | GUUCACUGAUGAGAGCAUUGUUCUGAGC |
2.9. Determination of HSV-1 titer
To investigate the roles of differentially expressed piRNAs, piRNA mimics were transfected into KMB17 cells. At 12 h post-transfection, the cells were infected with HSV-1. At 36 h post-infection, the supernatant of infected cells was collected and titrated with TCID50 (tissue culture infective dose 50%) assay.
2.10. Statistical analysis
Data represent the mean ± SD from at least three independent experiments. The differences between the two groups were compared using a two-tailed unpaired Student's t-test. P-value <0.05 was considered a statistically significant difference.
3. Results
3.1. Identification and characterization of piRNAs in HSV-1 infected KMB17 cells
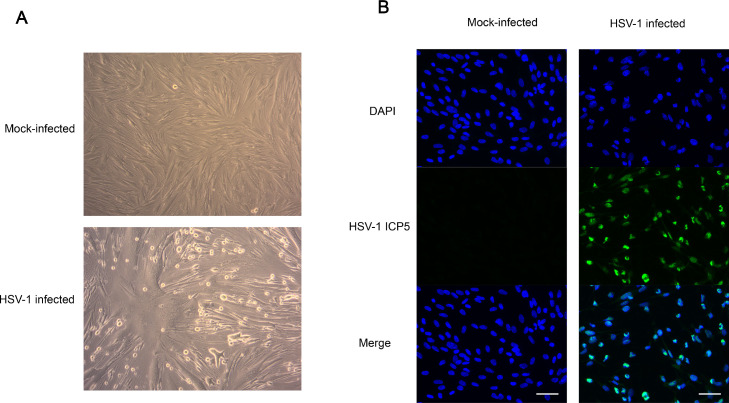
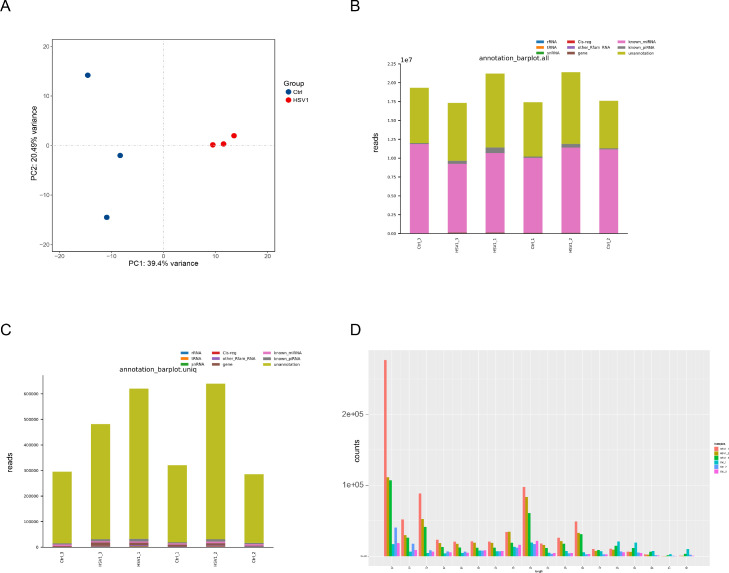
In order to ensure the infection efficiency of HSV-1 in KMB17 cells, we evaluated HSV-1-infected and uninfected cells with ordinary light microscopy (Fig. 1A) and immunofluorescence of HSV-1-specific protein ICP5 (Fig. 1B). Microscope images showed obvious cytopathic effects 48 h after infection compared to control cells. Immunofluorescence further showed that HSV-1-specific green fluorescence appeared in infected cells. These results indicated that HSV-1 could efficiently infect KMB17 cells. Next, we annotated and identified piRNAs. The high-throughput sequencing results showed that the clean reads of each sample ranged from 17.29 M to 21.36 M. The genome alignment rate ranged from 95.37 to 97.85%, and the known piRNAs alignment rate ranged from 0.65 to 3.59%. Principal component analysis (PCA) showed high repeatability and correlation among six biological samples (Fig. 2A). Through sRNA comparison and piRNA identification, the distribution of total sRNA and unique sRNA in each sample was analyzed (Fig. 2B, C). Among them, 849, 849, and 737 piRNAs were identified in the HSV-1 infection group. The control group identified 519, 628 and 606 piRNAs. Next, we analyzed the bases preference of the first site and each site of the detected piRNAs. However, the piRNAs in each sample did not show a strong preference for specific bases of the first position (1 U) and 10 position (10A) (Fig. S1, S2). At the same time, the sequence length statistics of all piRNAs showed obvious peaks at 17 nt, 23 nt, and 26 nt (Fig. 2D), which is consistent with the length characteristics of piRNAs and consistent with previous studies (Li et al., 2021; Yao et al., 2020).
Fig. 1.
At 48 h after infection with HSV-1 at a MOI of 1, HSV-1 replicated and proliferated in KMB17 cells and led to morphological changes in KMB17 cells. (A) KMB17 cells seen under a 200-magnification normal light microscope. Compared with normal KMB17 cells without infection, the morphology of KMB17 cells after HSV-1 infection was altered. (B) HSV-1 ICP5 (green) specific fluorescence under fluorescence microscopy. The blue color is DAPI. The scale bar is 50 μm.
Fig. 2.
Identification and annotation of piRNAs in KMB17 cells. (A) Principal component analysis (PCA) diagram of piRNAs expressed among samples. (B) Histogram of total RNA classification annotation in each sample, with the horizontal axis showing samples and the vertical axis showing the number of reads of each type of small RNA. (C) Bar graph of unique RNA classification annotation in each sample. The horizontal axis is the sample, and the vertical axis is the number of reads of each type of small RNA. (D) Global nucleotide length distribution of piRNA sequencing results.
3.2. Differential expression analysis of piRNAs
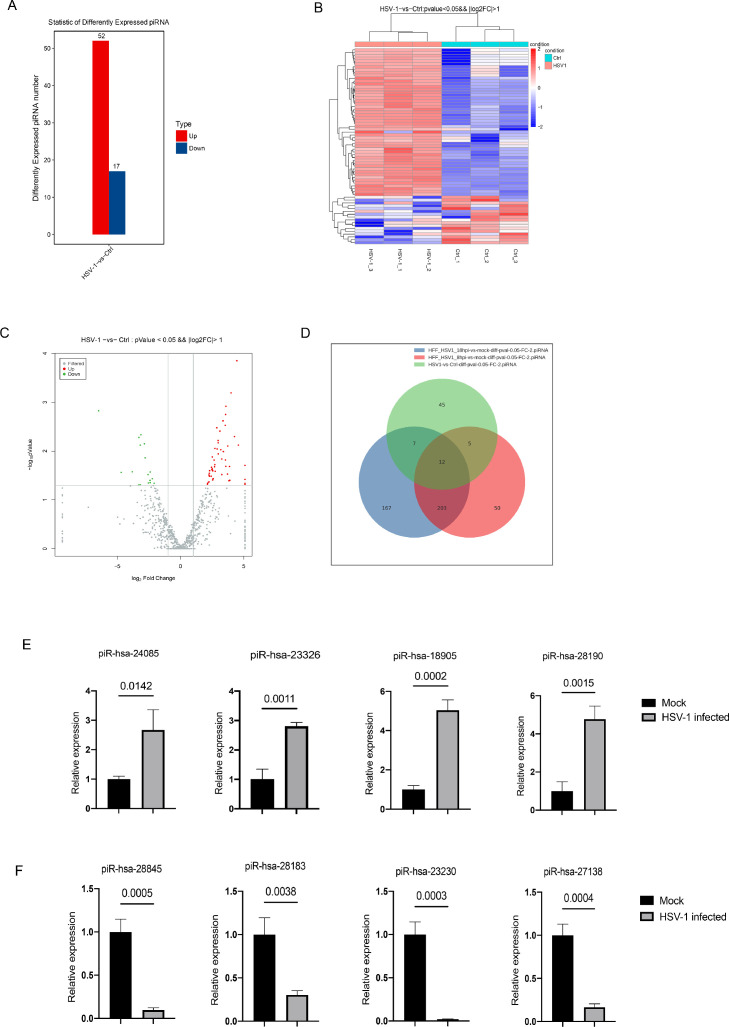
To analyze the differences of piRNA expression between the two groups during HSV-1 infection, we used the package R DESeq2 to screen the differential piRNAs between HSV-1 infected and mock-infected group. As a results, we identified 69 piRNAs that were significantly differentially expressed between two groups. Among them, 52 piRNAs were up-regulated and 17 were down-regulated (Fig. 3A). And the|log2 foldchange| of up regulation range from 2.10785922 to 4.56268419, the log2 foldchange of up regulation range from 2.110090519 to 6.497873947. The heat map shows the piRNAs cluster analysis results, indicating significant expression differences among samples (Fig. 3B). In addition, the volcano map was drawn to show the overall distribution of differential piRNAs (Fig. 3C). To check the reproducibility of the deregulation of piRNAs, we analyzed the sequences of 12 publicly available data (SRA accession number: PRJNA924274) of HSV-1 infected and mock-infected primary human foreskin fibroblast (HFF) from Zubković et al. (2022). The results showed that a large number of differentially expressed piRNAs were identified at 8 and 18 h after HSV-1 infected HFF cells, among which, a total of 12 piRNAs were reproducibly deregulated in HFF and KMB17 cell lines infected with HSV-1 and to show their biological relevance in infection (Fig. 3D and Table S2). Furthermore, to confirm these differentially expressed piRNAs identified by RNA-seq analysis, we randomly selected 8 of the top 20 differentially expressed piRNAs (4 up-regulated and 4 down-regulated) for stem-loop RT-qPCR analysis. The result showed that, compared with the control group, the selected piR-hsa-24,085, piR-hsa-23,326, piR-hsa-18,905, and piR-hsa-28,190 were significantly up-regulated in the HSV-1 infected group (Fig. 3E). At the same time, the selected piR-hsa-28,845, piR-hsa-28,138, piR-hsa-23,230, and piR-hsa-27,138 were significantly down-regulated in the HSV-1 infected group (Fig. 3F). The RT-qPCR result of the selected piRNAs is consistent with the RNA-seq result. To sum up, these changes of piRNAs expression in host cells during HSV-1 infection may suggest that host cells have complex molecular behavior mediated by piRNAs in viral infection.
Fig. 3.
piRNAs are differentially expressed during HSV-1 infection. (A) Statistical histogram of differentially expressed piRNAs. (B) Heatmap of cluster analysis of differentially expressed piRNAs. (C) Volcano map of differentially expressed piRNAs. Red dots represent up-regulated piRNAs and green dots represent down-regulated piRNAs. (D) Venn diagram of the common differentially expressed piRNAs in HFF and KMB17 cells. (E, F) Randomly selected up-regulated piRNAs and down-regulated piRNAs were verified by Stem-loop RT-qPCR. Data were presented as the mean ± SD of three independent experiments.
3.3. Functional prediction of differentially expressed piRNAs during HSV-1 infection
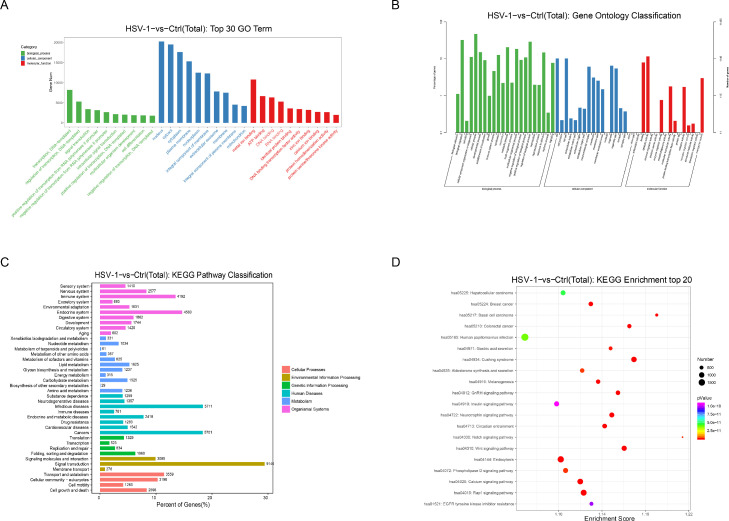
To better understand the potential roles of differentially expressed piRNAs during HSV-1 infection, we analyzed the host target genes of these piRNAs using the GO and KEGG databases (Table S3). Enrichment analysis showed that the parental genes of differentially expressed piRNAs are related to transcription, regulation of transcription, and signal transduction (Fig. 4A). The GO level 2 also indicated that differentially expressed piRNAs might be involved in biological processes, such as the immune and stimulus-response (Fig. 4B, Fig. S3). Next, we analyzed the functions of differentially expressed piRNAs by KEGG ernichment analysis. KEGG level 2 distribution showed that differentially expressed piRNAs were enriched in the nervous system, immune system, infectious diseases, and immune disease signaling pathways (Fig. 4C). At the same time, bubble plots showed that differentially expressed piRNAs are involved in the regulation of a wide range of signaling pathways, such as calcium signaling pathway (hsa04020), human papillomavirus infection (hsa05165), and basal cell carcinoma (hsa05217) (Fig. 4D). These results suggested that the differentially expressed piRNAs involved in various biological functions, including viral infection and immune response.
Fig. 4.
GO and KEGG analysis of differentially expressed piRNAs. (A) Top 30 GO terms of differentially expressed piRNAs in biological process, cellular component, and molecular function. (B) GO Level2 level distribution map of differentially expressed piRNAs target genes. (C) KEGG Level2 level distribution map of differentially expressed piRNAs target genes. The horizontal axis is the total number of differential piRNAs target genes annotated to each Level2 pathway and the corresponding ratio. The vertical axis represents the name of the Level2 pathway, and the number on the right side of the column represents the number of differential piRNAs target genes annotated to the Level 2 pathway. (D) KEGG enrichment analysis top20 (sorted by-log10Pvalue corresponding to each entry) bubble chart.
3.4. piRNAs induced by HSV-1 potentially affect viral replication
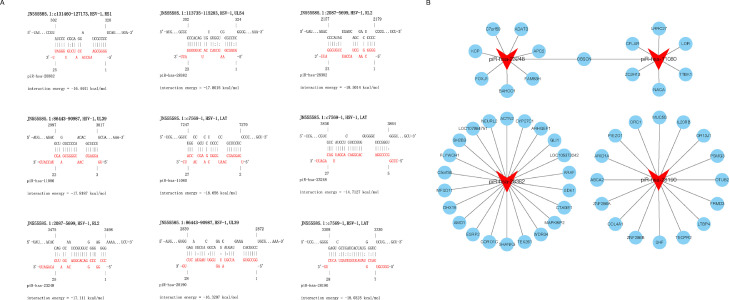
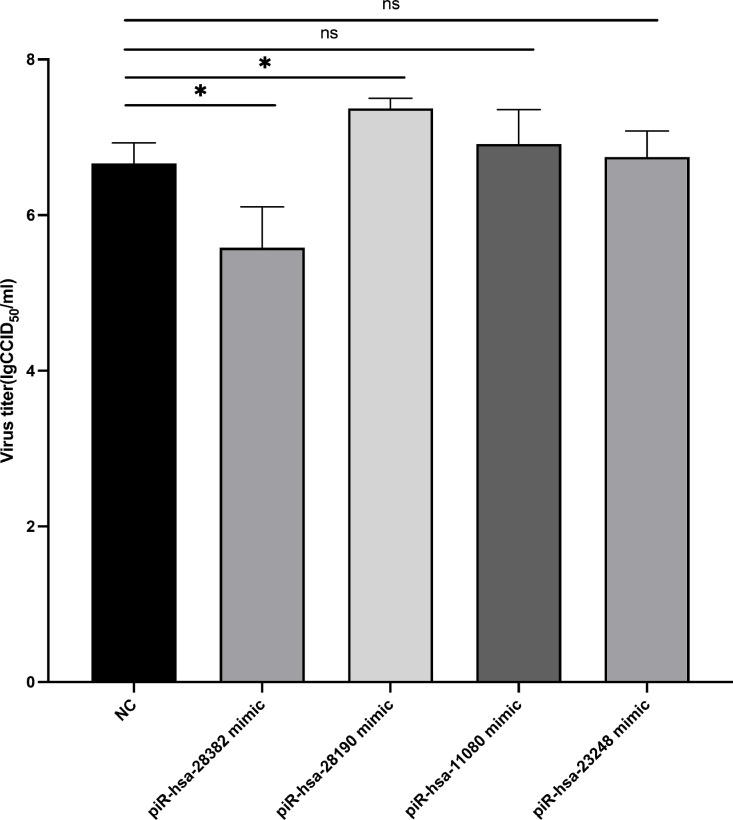
To test whether differential piRNAs could affect HSV-1 replication, the RNAInter V4.0 (Kang et al., 2022) was used to predict the targeting relationship between the top 10 up-regulated piRNAs and HSV-1 major viral genes. Finally, we selected four piRNAs that are most likely to interact with HSV-1, namely, piR-hsa-28,382 (alias piR-36,233), piR-hsa-11,080 (alias piR-48,966), piR-hsa-23,248 (alias piR-33,082), and piR-hsa-28,190 (alias piR-36,041). All these four piRNAs were predicted to interact with at least two HSV-1 genes (Fig. 5A). After that, Cytoscape software (3.9.1) (Shannon et al., 2003) was used to predict the host target genes of these four piRNAs. The results showed that each piRNA could target at least 7 host genes (Fig. 5B). These target genes are potentially involved in several important biological processes, including cell growth and development, humoral immunity and protein transportation. Then, the corresponding piRNA mimics were transfected into KMB17 cells for HSV-1 infection, and the supernatant was collected 36 h after HSV-1 infection at a MOI of 1. The titer of HSV-1 in the supernatant of the cells was measured, respectively. The result showed that compared with the negative control group, the viral titer of the piRNA-hsa-28,382 mimic group significantly decreased, while the titer of the piRNA-has-28,190 mimic group significantly increased, and there was no significant difference in the other two groups (Fig. 6). These results suggest that these up-regulated piRNAs can affect HSV-1 replication and proliferation, which may be achieved by piRNAs targeting the viral genes or the host cellular genes.
Fig. 5.
Functional interaction analysis of differentially expressed piRNAs and their target genes. (A) piR-hsa-28,382 interacts with HSV-1 RL2, RS1, and UL54. piR-hsa-11,080 interacts with HSV-1 LAT and UL39. piR-hsa-23,248 interacts with HSV-1 RL2 and LAT. piR-hsa-28,190 interacts with HSV-1 LAT and UL39. (B) Interaction network of differentially expressed piRNAs and their cellular target genes.
Fig. 6.
After piRNA mimics transfection, the HSV-1 titers in the supernatant of the cells shown by histogram. Data are presented as the mean ± SD of three independent experiments.
4. Discussion
In this study, we systematically analyzed the piRNA expression profiles of KMB17 cells in HSV-1 infected and control groups, and analyzed the functions of differentially expressed piRNAs. After ensuring the good quality of samples and sequencing data, we found that the expression of piRNAs in KMB17 cells was significantly altered upon HSV-1 infection. Sixty-nine differentially expressed piRNAs were identified, among which 52 were up-regulated and 17 were down-regulated (Fig. 3A). Interestingly, there were more up-regulated piRNAs than down-regulated piRNAs, which is consistent with the study of piRNA expression profiles in Coxsackievirus B3 infection (Yao et al., 2020). This indicates that viral infection is likely to induce the high expression of piRNAs in the host. In this study, a total of 12 differentially expressed piRNAs are the same as the previous study of Zubković et al., which to a certain extent reflects the universality of piRNA expression in the host cells during HSV-1 infection. Herein, we further selected 4 up-regulated piRNA mimics to transfect KMB17 cells and measureed the virus titer in the supernatant 36 h after HSV-1 infection. The results showed that piRNA-hsa-28,382 mimic could decrease the titer of HSV-1 in the supernatant of infected cells, while piRNA-hsa-28,190 mimic could increase the titer of HSV-1 (Fig. 6). This suggests that up-regulated piRNA-hsa-28,382 likely has an important role in fighting HSV-1 infection. Moreover, combined with our previous analysis of the interaction between piRNAs and major genes of HSV-1, piRNA-hsa-28,382 participates in anti-virus proliferation by silencing the expression of virus genes. At the same time, piRNA-hsa-28,190 may promote HSV-1 infection, which is not in line with our previous conjecture. It may also partcipate in promoting HSV-1 infection in other ways.
The GO annotation analysis showed that the differentially expressed piRNAs in HSV-1 infected cells might be involved in biological functions, such as biological adhesion, immune system process, and response to stimulus, which are very common in many viral infections and may be the key to the interaction with the host in some viral infections (Aoshi et al., 2011; Xu and Shaw, 2016). These results suggest that these differentially expressed piRNAs involve in the process of HSV-1 infecting host cells. In addition, protein binding, transcription, and regulation of transcription were also specifically enriched in infected cells. Some studies have shown that protein binding is crucial for virus infection, and may be involved in virus entering host cells (Qiao and Olvera de la Cruz, 2020; Sathiyamoorthy et al., 2017). Moreover, the differentially expressed piRNAs may have an important role in the process of viral genome replication (Dremel and DeLuca, 2019) based on the GO term transcription and regulation of transcription. The multiple human disease-related signaling pathways were enriched in the HSV-1 infected group compared with the uninfected group, including neurodegenerative diseases, infectious diseases, and immune diseases. In particular, it should be noted that related genes were also enriched in the human papillomavirus infection signal pathway, suggesting that the process of HSV-1 infection may be similar to that of human papillomavirus infection.
KEGG level 2 distribution showed that differentially expressed piRNAs were enriched in the nervous system, immune system, infectious diseases, and immune disease signaling pathways. In addition, the Wnt signaling pathway was enriched in KEGG analysis. A previous study showed that the Wnt signaling pathway is related to the toxicity of influenza virus (Forero et al., 2015) and participates in virus proliferation and cytomegalovirus infection (Baasch et al., 2021).
The importance of endocytosis in the process of virus infection is self-evident. Endocytic vesicles are conducive to transporting virus particles into the deep cytoplasm without being squeezed by cytoplasm and hindered by cytoskeleton (Cossart and Helenius, 2014). Furthermore, the calcium signaling pathway has been proved to play an important role in virus entry, virus gene expression, virus protein post-translation processing, and virus particle maturation and release (Zhou et al., 2009). In this study, we found that differentially expressed piRNAs in HSV-1 infected cells may affect the viral life cycle by altering these signaling pathways. In addition to viral infection-related pathways, cancer-related pathways were enriched, including hepatocellular carcinoma, breast cancer, basal cell carcinoma, and mammalian cancer. Furthermore, studies have shown that Notch signaling pathway roles in cancer are very extensive; in cetin cancers, Notch signaling may act as a carcinogenic factor while in others as a tumor inhibitor (Aster et al., 2017). On the other hand, in recent years, oncolytic HSV obtained by genetic modification of HSV has made some progress in the research of tumor immunotherapy (Jahan et al., 2021; Mondal et al., 2020). At the same time, the differentially expressed piRNAs found in this study during HSV-1 infection may provide some new clues for anticancer therapy.
In summary, we used a high-throughput RNA sequencing technique to analyze piRNA expression patterns and their roles in KMB17 cells infected by HSV-1. Our results revealed the expression characteristics of piRNA in HSV-1 infected cells and screened two piRNAs that potentially regulate HSV-1 replication. The target genes of differentially expressed piRNAs were predicted and their function were analyzed by GO and KEGG enrichment analysis. These results will help reveal the roles and mechanisms of piRNAs in the process of HSV-1 infection.
Author contributions statement
JD. S, J.S, and YZ. Y. conceptualized and supervised the study; XW, PH, MY, L, YM, and HL, C, performed the experiments, analyzed the data, and participated in the experimental design. XW wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-036), Natural Science Foundation of Yunnan Province (202001AS070046), the Yunnan Provinal Key Research and Development Program (202102AA100017), the Innovation Team Project of Yunnan Science and Technology Department (202105AE160020), and the Fund for Reserve Talents of Young and Middle-aged Academic and Technical Leaders of Yunnan Province (2019HB043).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199087.
Contributor Information
Jing Sun, Email: jingzikm@imbcams.com.cn.
Yunzhang Hu, Email: huyunzhang@imbcams.com.cn.
Jiandong Shi, Email: shijiandong@imbcams.com.cn.
Appendix. Supplementary materials
Fig. S1. First base bias statistics of piRNAs detected in each sample.
Fig. S2. The bias statistics of each base of piRNAs detected in each sample.
Fig. S3. GO analysis of directed acyclic graph of differentially expressed piRNAs.
Table S1. Stem-loop RT-qPCR primers and programs for differentially expressed piRNAs.
Table S2. Twelve common differentially expressed piRNAs.
Table S3. Prediction of host target genes of differentially expressed piRNAs.
Data availability
Data will be made available on request.
References
- Anders S. Analysing RNA-Seq data with the DESeq package. Mol. Biol. (N.Y.) 2010 [Google Scholar]
- Aoshi T., Koyama S., Kobiyama K., Akira S., Ishii K.J. Innate and adaptive immune responses to viral infection and vaccination. Curr. Opin. Virol. 2011;1(4):226–232. doi: 10.1016/j.coviro.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Chien M., Russo J.J., Ju J., Sheridan R., Sander C., Zavolan M., Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Naumova N.M., Tulin A.V., Vagin V.V., Rozovsky Y.M., Gvozdev V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11(13):1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aster J.C., Pear W.S., Blacklow S.C. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baasch S., Giansanti P., Kolter J., Riedl A., Forde A.J., Runge S., Zenke S., Elling R., Halenius A., Brabletz S., Hengel H., Kuster B., Brabletz T., Cicin-Sain L., Arens R., Vlachos A., Rohr J.C., Stemmler M.P., Kopf M., Ruzsics Z., Henneke P. Cytomegalovirus subverts macrophage identity. Cell. 2021;184(14):3774–3793. doi: 10.1016/j.cell.2021.05.009. e3725. [DOI] [PubMed] [Google Scholar]
- Berkhout B. RNAi-mediated antiviral immunity in mammals. Curr. Opin. Virol. 2018;32:9–14. doi: 10.1016/j.coviro.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16(21):2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Cossart P., Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 2014;6(8) doi: 10.1101/cshperspect.a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dremel S.E., DeLuca N.A. Genome replication affects transcription factor binding mediating the cascade of herpes simplex virus transcription. Proc. Natl. Acad. Sci. U. S. A. 2019;116(9):3734–3739. doi: 10.1073/pnas.1818463116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte L.F., Farías M.A., Álvarez D.M., Bueno S.M., Riedel C.A., González P.A. Herpes simplex virus Type 1 infection of the central nervous system: insights into proposed interrelationships with neurodegenerative disorders. Front. Cell Neurosci. 2019;13:46. doi: 10.3389/fncel.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers J.M., Bitnun A., Richardson S.E., Ford-Jones E.L., Tellier R., Wald R.M., Petric M., Kolski H., Heurter H., MacGregor D. A 12-year prospective study of childhood herpes simplex encephalitis: is there a broader spectrum of disease? Pediatrics. 2007;119(2):e399–e407. doi: 10.1542/peds.2006-1494. [DOI] [PubMed] [Google Scholar]
- Forero A., Tisoncik-Go J., Watanabe T., Zhong G., Hatta M., Tchitchek N., Selinger C., Chang J., Barker K., Morrison J., Berndt J.D., Moon R.T., Josset L., Kawaoka Y., Katze M.G. The 1918 influenza virus PB2 protein enhances virulence through the disruption of inflammatory and Wnt-mediated signaling in Mice. J. Virol. 2015;90(5):2240–2253. doi: 10.1128/JVI.02974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gordon, A., Hannon, G., 2010. Fastx-toolkit. FASTQ/A short-reads preprocessing tools (unpublished) http://hannonlab.cshl.edu/fastx_toolkit5.
- Greeley Z.W., Giannasca N.J., Porter M.J., Margulies B.J. Acyclovir, cidofovir, and amenamevir have additive antiviral effects on herpes simplex virus TYPE 1. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104754. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic. Acids. Res. 2003;31(1):439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic. Acids. Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic. Acids. Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan N., Ghouse S.M., Martuza R.L., Rabkin S.D. In Situ cancer vaccination and immunovirotherapy using oncolytic HSV. Viruses. 2021;13(9) doi: 10.3390/v13091740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C., Harfouche M., Welton N.J., Turner K.M., Abu-Raddad L.J., Gottlieb S.L., Looker K.J. Herpes simplex virus: global infection prevalence and incidence estimates, 2016. Bull. World Health Organ. 2020;98(5):315–329. doi: 10.2471/BLT.19.237149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C., Gottlieb S.L., Wald A. Status of vaccine research and development of vaccines for herpes simplex virus. Vaccine. 2016;34(26):2948–2952. doi: 10.1016/j.vaccine.2015.12.076. [DOI] [PubMed] [Google Scholar]
- Kang J., Tang Q., He J., Li L., Yang N., Yu S., Wang M., Zhang Y., Lin J., Cui T., Hu Y., Tan P., Cheng J., Zheng H., Wang D., Su X., Chen W., Huang Y. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic. Acids. Res. 2022;50(D1):D326–d332. doi: 10.1093/nar/gkab997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.W. PIWI proteins and piRNAs in the nervous system. Mol. Cells. 2019;42(12):828–835. doi: 10.14348/molcells.2019.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics Chapter. 2010;11 doi: 10.1002/0471250953.bi1107s32. Unit 11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.H., Quarles K.A., Yang Y., Tanguy M., Frézal L., Smith S.A., Sharma P.P., Cordaux R., Gilbert C., Giraud I., Collins D.H., Zamore P.D., Miska E.A., Sarkies P., Jiggins F.M. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2018;2(1):174–181. doi: 10.1038/s41559-017-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X., Wang Y., Song J., Song Q., Wang Y., Han J. HRV16 infection induces changes in the expression of multiple piRNAs. Virol. Sin. 2021;36(4):736–745. doi: 10.1007/s12250-021-00344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marcel M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17(1) [Google Scholar]
- Miesen P., Girardi E., van Rij R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic. Acids. Res. 2015;43(13):6545–6556. doi: 10.1093/nar/gkv590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesen P., Joosten J., van Rij R.P. PIWIs Go Viral: arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog. 2016;12(12) doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M., Guo J., He P., Zhou D. Recent advances of oncolytic virus in cancer therapy. Hum. Vaccin. Immunother. 2020;16(10):2389–2402. doi: 10.1080/21645515.2020.1723363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata D.M., Gainetdinov I., Zoch A., O'Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 2019;20(2):89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao B., Olvera de la Cruz M. Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano. 2020;14(8):10616–10623. doi: 10.1021/acsnano.0c04798. [DOI] [PubMed] [Google Scholar]
- Sathiyamoorthy K., Chen J., Longnecker R., Jardetzky T.S. The COMPLEXity in herpesvirus entry. Curr. Opin. Virol. 2017;24:97–104. doi: 10.1016/j.coviro.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Hu N., Mo L., Zeng Z., Sun J., Hu Y. Deep RNA sequencing reveals a repertoire of human fibroblast circular RNAs associated with cellular responses to herpes simplex virus 1 infection. Cell. Physiol. Biochem. 2018;47(5):2031–2045. doi: 10.1159/000491471. [DOI] [PubMed] [Google Scholar]
- Sun J., Wang S., Li C., Ren Y., Wang J. Novel expression profiles of microRNAs suggest that specific miRNAs regulate gene expression for the sexual maturation of female Schistosoma japonicum after pairing. Parasit. Vect. 2014;7:177. doi: 10.1186/1756-3305-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang P., Lu Y., Li Y., Zheng Y., Kan Y., Chen R., He S. piRBase: a comprehensive database of piRNA sequences. Nucleic. Acids. Res. 2019;47(D1):D175–d180. doi: 10.1093/nar/gky1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R.J., Roizman B. Herpes simplex virus infections. Lancet. 2001;357(9267):1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- Xu H., Shaw D.E. A simple model of multivalent adhesion and its application to influenza infection. Biophys. J. 2016;110(1):218–233. doi: 10.1016/j.bpj.2015.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Wang X., Song J., Wang Y., Song Q., Han J. Coxsackievirus B3 infection induces changes in the expression of numerous piRNAs. Arch. Virol. 2020;165(1):105–114. doi: 10.1007/s00705-019-04451-2. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Frey T.K., Yang J.J. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium. 2009;46(1):1–17. doi: 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubković A., Žarak I., Ratkaj I., Rokić F., Jekić M., Pribanić Matešić M., Lebrón R., Gómez-Martín C., Lisnić B., Lisnić V.J., Jonjić S., Pan D., Vugrek O., Hackenberg M., Jurak I. The virus-induced upregulation of the miR-183/96/182 cluster and the FoxO Family Protein Members Are Not Required For Efficient Replication of HSV-1. Viruses. 2022;14(8) doi: 10.3390/v14081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. First base bias statistics of piRNAs detected in each sample.
Fig. S2. The bias statistics of each base of piRNAs detected in each sample.
Fig. S3. GO analysis of directed acyclic graph of differentially expressed piRNAs.
Table S1. Stem-loop RT-qPCR primers and programs for differentially expressed piRNAs.
Table S2. Twelve common differentially expressed piRNAs.
Table S3. Prediction of host target genes of differentially expressed piRNAs.
Data Availability Statement
Data will be made available on request.