Abstract
Ig VDJ genes in rabbit somatically diversify by both hyperpointmutation and gene conversion. To elucidate the mechanism of gene conversion of IgH genes, we cloned a rabbit homologue of RAD51, a gene involved in gene conversion in Saccharomyces cerevisiae (yeast), and tested whether it could complement a yeast rad51 mutant deficient in recombination repair. We found that rabbit RAD51 partially complemented the defect in switching mating types by gene conversion as well as in DNA double-strand break repair after γ-irradiation. Further, by Western blot analysis, we found that levels of Rad51 were higher in appendix-derived B lymphocytes of 6-wk-old rabbits, a time at which IgH genes diversify by somatic gene conversion. We suggest that Rad51 is involved in somatic gene conversion of rabbit Ig genes.
Antibody diversity is generated by somatic rearrangement of V, D, and J gene segments during maturation of B lymphocytes. These rearrangements result from site-specific DNA recombination that is mediated by several trans-acting factors, including RAG1 (1), RAG2 (2), Ku 70/86 (3–5), XRCC4 (6) and DNA-PKCS (7, 8). In response to antigenic stimulation, the rearranged V(D)J genes in mouse and humans undergo further somatic diversification by hyperpointmutation (9–18). In other species, including chickens and rabbits, further somatic diversification occurs by gene conversion as well as by hyperpointmutation (19–24). Although several cis-acting factors are known to regulate hyperpointmutation (25–31), little is known about trans-acting factors for either somatic gene conversion or hyperpointmutation of Ig genes.
Gene conversion is the nonreciprocal exchange of genetic information that results in an unequal recovery of genetic alleles (32, 33). In Saccharomyces cerevisiae (yeast), gene conversion events may result from the recombinational repair of DNA double-strand breaks that is mediated by the RAD52 epistasis group (34). It is believed that homologous recombination pathways that generate gene conversion events are conserved as a result of the discovery of recombinases that have significant amino acid similarities. Accumulating biochemical and molecular evidence that S. cerevisiae RAD51 is a structural and functional eukaryotic homologue of bacterial RecA (35–40), a gene long known to be critical for recombination (41), has stimulated intensive efforts to identify mammalian equivalents of RAD51. Eukaryotic homologues to RecA/RAD51 are now known in several higher species, including chicken (42), mouse (43), and human (44). Biochemical activities of the purified human Rad51, including formation of nucleoprotein filaments with both single-stranded and double-stranded DNA, DNA-dependent ATPase activity (45, 46), pairing homologous DNA molecules (47), and catalyzing strand exchange with a polarity similar to yeast Rad51 (39, 48), suggest that Rad51 may play a role in homologous recombination. However, it is difficult to demonstrate that RAD51 is involved in recombination in vivo because disruption of the RAD51 gene leads to lethality of both cell lines and embryos (49, 50).
DNA sequence analysis of diversified rabbit VDJ gene rearrangements and nonrearranged VH gene segments has shown that gene conversion events occur with the VDJ gene as a recipient and nonrearranged VH gene segments as donors (22). This process occurs in gut-associated lymphoid tissue, beginning at ~4 wk of age (51–53). We have begun to search for trans-acting factors involved in somatic gene conversion of rabbit Ig genes, and, as part of this effort, we have cloned a rabbit RAD51 homologue. Because of the difficulty in determining, in vivo, the function of RAD51 in vertebrates, we tested whether rabbit RAD51 could function in homologous recombination using rad51 mutant yeast, which are defective in homologous recombination and therefore cannot repair DNA damage caused by ionizing radiation and cannot switch mating types by gene conversion (54–56). Further, to investigate the role of RAD51 in the generation of Ig gene diversity, we determined whether its expression correlated with the timing and location of somatic gene conversion of IgH genes.
Materials and Methods
Cloning and nucleotide sequence analysis of rabbit RAD51
Thymic cDNA was synthesized and amplified by PCR using as a 5′ primer (5′-ATGGCAATGCAGATGCAGCT-3′) and as a 3′ primer (5′-TCAGTCTTTGGCATCTCCCA-3′), sequences that are homologous to the 5′ and 3′ ends of human RAD51, respectively (44). The PCR product was cloned into M13 mp18/mp19 and the nucleotide sequence was determined (57).
Genomic and Northern blot analysis
Genomic DNA was prepared from rabbit liver as described (58), and Southern blot analysis of restricted DNA (10 μg) was performed using the rabbit RAD51 PCR product as a probe (59). RNA was isolated by density gradient centrifugation in CsCl, and poly(A)+ RNA was purified using a Quick Prepmicro mRNA purification kit (Pharmacia, Piscataway, NJ). For Northern blot analysis, ~3 μg of poly(A)+ RNA was separated by electrophoresis in 1.5% agarose and 1 M formaldehyde gels, and the gels were blotted onto nylon membrane.
Genomic library
A partial MboI genomic DNA library in EMBL4 (60), prepared from DNA of a VHa2 allotype rabbit, was screened using the rabbit RAD51 cDNA as probe. RAD51+ phage were isolated and characterized by restriction mapping and Southern blot analysis. Individual exons were cloned into M13/mp18 and the nucleotide sequences were determined. A size-selected library of 8 to 10-kb EcoRI fragments isolated from liver genomic DNA was constructed in pGEM3 (60) and probed with an exon 1 and exon 2 PCR product by using 5′-ATGGCAATGCAGATGCAGCT-3′ as a 5′ primer and 5′-CAGAATTTTATCAGCTTTTG-3′ as a 3′ primer.
Anti-Rad51 serum
Anti-Rad51 serum was prepared by immunizing rabbits with histidinetagged human Rad51 purified from Escherichia coli by ion-exchange chromatography (Qiagen, Santa Clarita, CA). The plasmid encoding the histidine-HsRad51 fusion protein was kindly provided by Charles Radding (Yale University). Serum from an immunized rabbit reacted with a 38-kDa protein (the expected size of Rad51) present in lysates from rabbit, mouse, and chicken cell lines as determined by Western blot analysis, whereas preimmunization serum from the same rabbit did not recognize this protein.
Western blot analysis
IgM+ and IgL+ cells were isolated from the appendix by flow cytometry after reaction with biotinylated goat anti-rabbit L-chain followed by FITC-conjugated avidin or with mouse monoclonal anti-rabbit μ-chain followed by FITC-conjugated goat anti-mouse Ig. By reanalysis, the sorted cells were shown to be >95% Ig+ (FACStarPlus, Becton Dickinson, San Jose, CA). Lysates from 1 × 106 purified IgM+ or IgL+ cells were electrophoresed on an SDS/10% polyacrylamide gel and blotted to a 0.1 μm nitro-cellulose membrane. Rabbit Rad51 was detected by using rabbit anti-human Rad51 polyclonal antiserum as primary reagent, biotinylated goat anti-rabbit L-chain as secondary reagent, followed by avidin-HRP and enhanced chemiluminescence substrate (Amersham, Buckinghamshire, U.K.).
γ-Irradiation of cell lines
B lineage cells, 55D1 (61), were irradiated with 6 Gy of ionizing radiation using a Nordion 1.8 kCi 137Cs irradiator as a γ-ray source at 7.8 krad/h. At each time point tested after irradiation, cell viability was determined by trypan blue dye exclusion. The cells were pelleted and resuspended in guanidine isothiocyanate, and RNA was prepared. Cell cycle analysis of irradiated and unirradiated cells was performed by flow cytometry after the cells were stained with Hoechst 33342 dye (62).
Yeast complementation
The mutant rad51 strain (g896–7Dα) containing the rad51–1 allele was obtained from John Game (Lawrence Berkeley Lab; Berkeley, CA) (63). Meiotic segregants were obtained from a diploid cross of g896–7Da with w303–1A (MATa ura3–1 trp1–1 his3–11, 15 ade2–1 leu2–3, 112 can1–100) by selecting on yeast peptone dextrose supplemented with canavanine (64). A meiotic segregant containing MATa rad51–1 ura3–1 trp1–1 leu2–3, 112 was used for subsequent studies.
Rabbit RAD51 cDNA was cloned into the pYES2.0 shuttle vector under the GAL1 promoter. This vector was introduced by transformation into the yeast rad51 strain, and transformants were selected in synthetic media without uracil. RAD51 expression was induced with 5% galactose. After 2 days, transformant yeast cells were plated on YpGal (5%). For DNA double-strand break repair studies, the plates were exposed to ionizing radiation as described above at 7.8 krad/h and incubated at 30°C. The percentage of viable cells after 3 days, relative to time zero, was determined. The percentage of viable cells after 1 wk was similar to that determined after 3 days. Each sample was determined in duplicate, with typically <10% variation.
For mating type-switch complementation, rad51 S. cerevisiae pYES2.0 transformants were additionally transformed with pGHOT-GAL3 (Trp+) (65), containing the HO endonuclease gene controlled by the GAL1 promoter. pYES2.0, pGHOT-GAL3 double transformants were selected by growing them in synthetic media lacking both uracil and tryptophan but containing galactose (5%) to induce switch through expression of HO endonuclease. After 40–48 h of induction, double-transformant cells were plated onto synthetic dextrose media without uracil and tryptophan. Individual colonies were tested for switch by mating with both MATa and MATα tester strains. The genotype of the MATa tester is his7 ura1 cdc4–1, whereas the genotype of the MATα is his7 cdc4–1 hom3 can1–100 sap3. Importantly, because the genetic backgrounds of rad51 and tester S. cerevisiae are heteroallelic at multiple alleles, one can test for switch by genetic cross-complementation. For example, neither rad51 nor tester haploid strains are able to grow in media lacking histidine; however, if mating has occurred, diploid cells are able to grow on media deficient in histidine. Because MATa (rad51–1) cells can only mate with the MATα tester, all unswitched cells will only grow when spotted onto synthetic dextrose plates containing a lawn of the MATα tester. Conversely, rad51 cells that have switched can grow on synthetic dextrose plates containing the MATa tester. S. cerevisiae rad51 cells containing rabbit RAD51 that had switched mating types from MATa to MATα were tested for their ability to switch back to MATa by mating with the MATα tester strain.
Results
Cloning and sequence analysis of rabbit RAD51
We PCR-amplified RAD51 from rabbit thymus cDNA using oligomers derived from human RAD51 and determined the nucleotide sequence (Fig. 1A). Genomic RAD51 was cloned from rabbit genomic phage and plasmid libraries. By restriction mapping and Southern blot analysis, we identified four overlapping RAD51+ phage clones and one nonoverlapping RAD51+ plasmid clone (Fig. 1B). We determined the nucleotide sequence of the fragments that hybridized with RAD51 and identified 10 RAD51 exons ranging in size between 60 bp and 123 bp. The exons were separated by as little as 200 bp and as much as 10 kb. To confirm the restriction map, and to localize the nonoverlapping plasmid clone containing exons 1 and 2, we performed Southern blot analysis of XbaI-digested genomic DNA using exon-specific probes. The probes for exons 3 to 10 hybridized with XbaI fragments identical in size to those cloned in the phage (Fig. 2A). The probe for exons 1 and 2 hybridized with the 9-kb XbaI fragment. On the basis of Southern hybridization of HindIII-digested rabbit genomic DNA with probes for exons 2 and 3 (data not shown), we estimate that this plasmid fragment lies 6 kb upstream of the DNA in phage 11.1 (Fig. 1B). Based on the restriction map of the phage and plasmid clones, we conclude that the entire RAD51 gene spans a region >35 kb.
FIGURE 1.
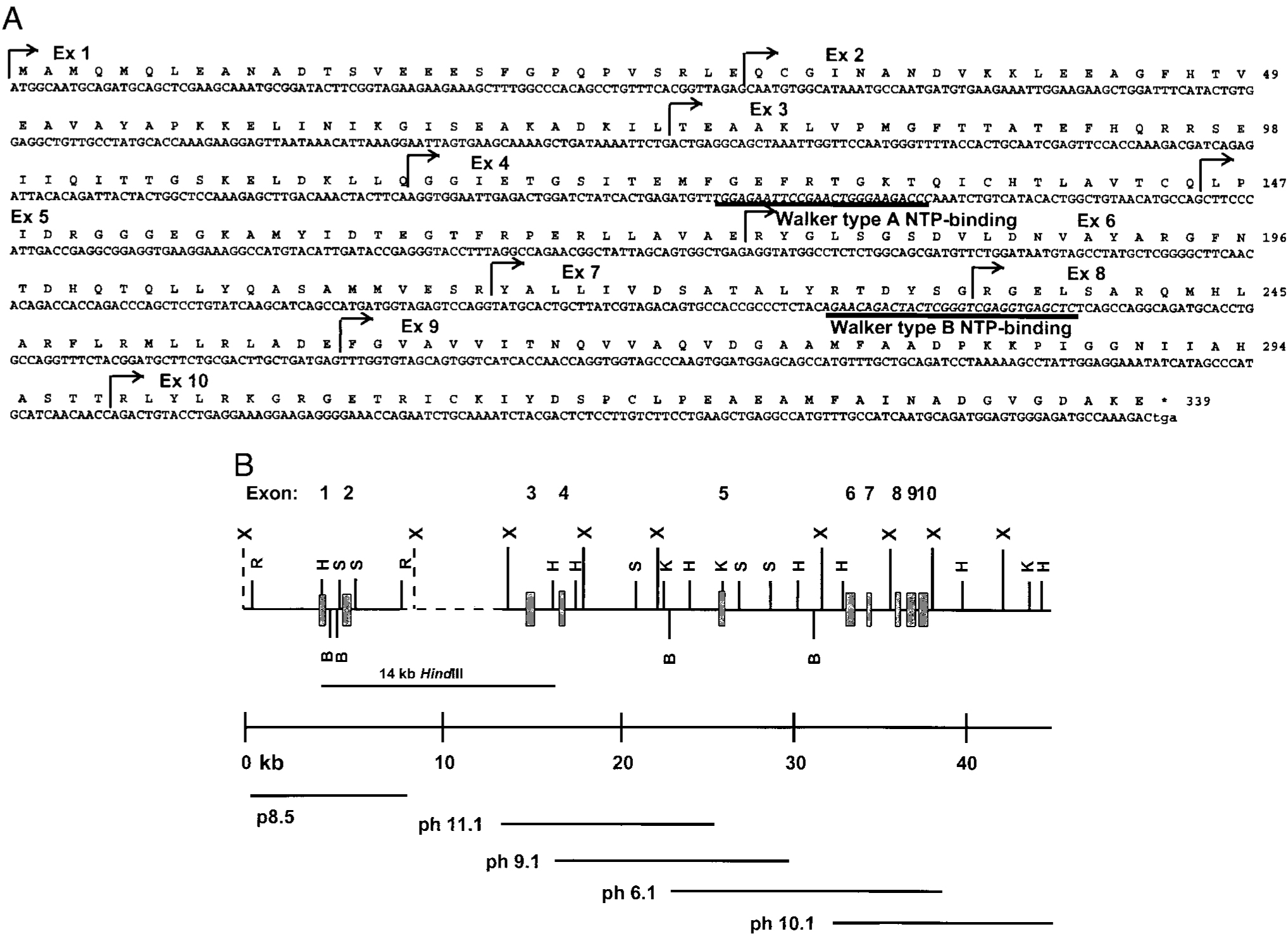
Rabbit RAD51 cDNA and genomic locus. A, Nucleotide and deduced amino acid sequence of rabbit RAD51 cDNA. Amino acid numbering is shown at the right. Boundaries of exons are marked by arrowed lines. Consensus Walker type-A and type-B nucleotide binding sites are underlined. B, Partial restriction map of RAD51 genomic phage (ph) and plasmid (p) clones. The locations of exons 1–10 (vertical boxes) were determined by restriction mapping and nucleotide sequence analysis. Restriction sites: X, XbaI; B, BamHI; H, HindIII; S, SacII; K, KpnI; R, EcoRI (not all EcoRI sites are shown). Dashed line indicates region of genomic DNA not cloned. ph11.1 and p8.5 are separated by 6 kb as determined in genomic Southern blot analysis by the hybridization of a 14-kb HindIII fragment with both exon 2 and exon 3 probes.
FIGURE 2.
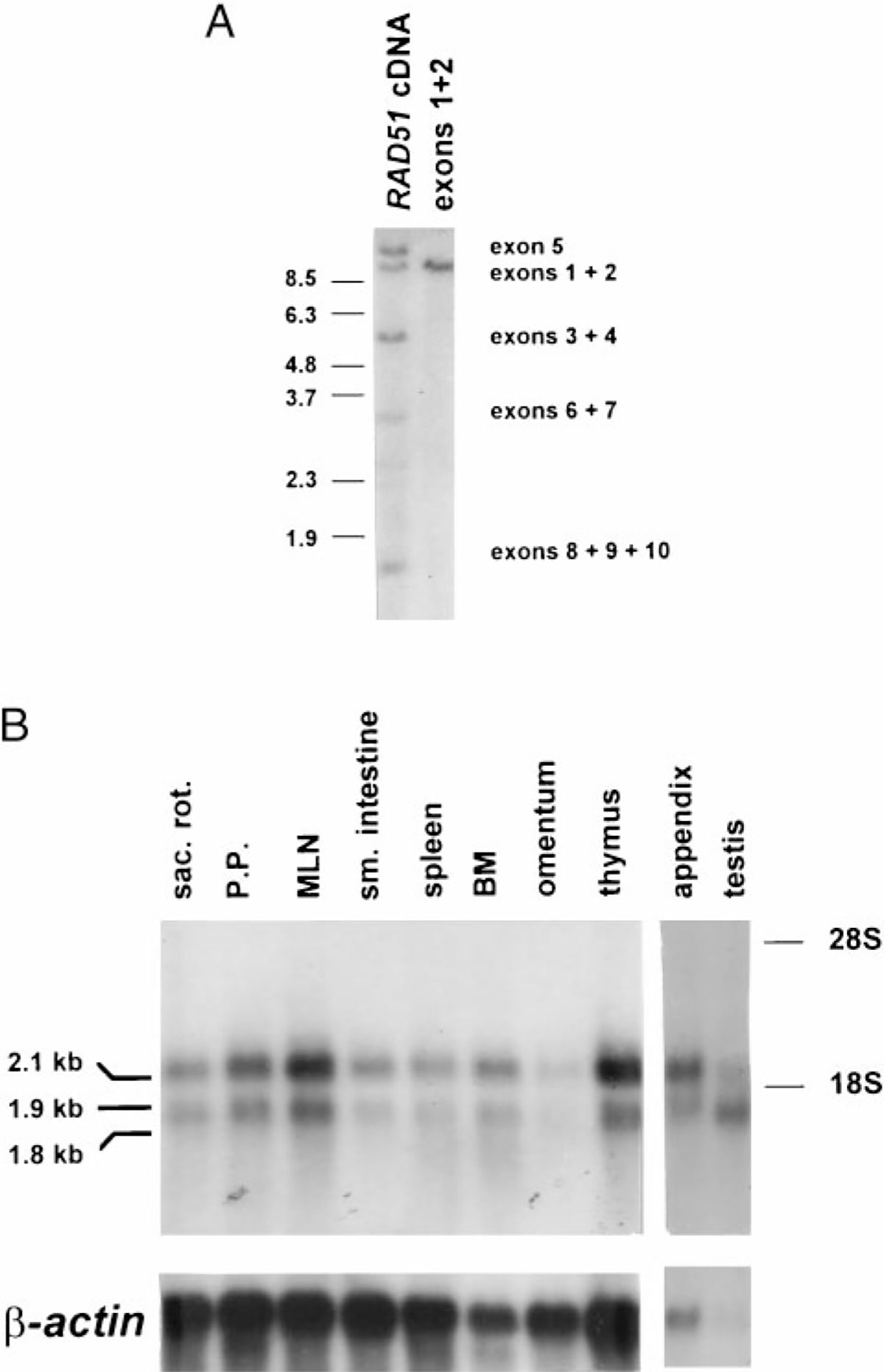
Southern and Northern blot analyses of rabbit RAD51. A, Southern blot analysis of XbaI-digested liver genomic DNA. The blot was probed with a PCR-generated fragment spanning exons 1 and 2 (right) and subsequently with full-length RAD51 cDNA (left). Bands corresponding to each RAD51 exon were similarly confirmed by using exon-specific probes as indicated at the right. The size standard is BstEII-restricted λ phage DNA, and the sizes are indicated on the left. B, Northern blot analysis of RAD51 mRNA expression in selected tissues of 6-wk-old rabbits. Each lane was loaded with 3 μg of poly(A)+ RNA. RAD51 and β-actin cDNAs were used as probes. Positions of the 28S and 18S ribosomal bands are shown. Sizes of mRNA species are based on HindIII-digested λ DNA run in formaldehyde buffer.
Tissue expression
We examined the expression of RAD51 in various tissues by poly(A)+ Northern blot analysis and found that it was expressed in nine lymphoid tissues, including sacculus rotundus, Peyer’s patches, mesenteric lymph node, small intestine, spleen, bone marrow, omentum, thymus, and appendix (Fig. 2B). RAD51 expression was undetectable in brain but was found at low levels in lung, liver, and kidney (data not shown).
We detected three different mRNA species, two in mitotic tissues (2.1 kb and 1.9 kb) and a third in testis (1.8 kb) (Fig. 2B). To determine whether all the RAD51 exons are represented in each mRNA species, we hybridized poly(A)+ Northern blots of appendix and testis RNA with exon-specific probes. We found that, although both the 2.1-kb and 1.9-kb mRNA species hybridized to each exon probe, the 1.8-kb mRNA species from testis did not hybridize to the exon 1–2 probe (data not shown). These data suggest that the 1.8-kb mRNA species lacks the first two exons of RAD51 and that the 1.9-kb and 2.1-kb mRNA species result from either differential transcription start sites or differential polyadenylation.
Function of RAD51
Recombinational DNA repair.
In S. cerevisiae, DNA double-strand breaks, such as those caused by ionizing radiation, are frequently repaired by homologous recombination involving RAD51 (34, 54, 55). Accordingly, RAD51 mRNA levels are up-regulated after exposure to DNA-damaging agents (35, 36, 66). In mammals, it also appears that homology-directed repair is an important DNA repair pathway (67). We determined whether rabbit RAD51 was up-regulated in a manner similar to that of yeast RAD51 in cell lines in response to irradiation. We irradiated rabbit B cell lines with 6 or 7 Gy of ionizing radiation and measured RAD51 expression by Northern blot analysis of poly(A)+ RNA (Fig. 3A). By analyzing RAD51 mRNA levels before and at 1, 3, 4, 6, 12, and 18 h after irradiation, we found that RAD51 expression increased significantly soon after irradiation. After normalizing RAD51 mRNA to the amount of β-actin mRNA, we found, in each of four experiments at 6 Gy and two experiments at 7 Gy, a 2- to 15-fold increase in RAD51 mRNA, with peak induction occurring at 6 h after irradiation (t test = p < 0.1; Fig. 3B). We conclude that RAD51 mRNA is induced in B cell lines after exposure to ionizing radiation, a finding consistent with the potential involvement of RAD51 in homology-directed DNA double-strand break repair.
FIGURE 3.
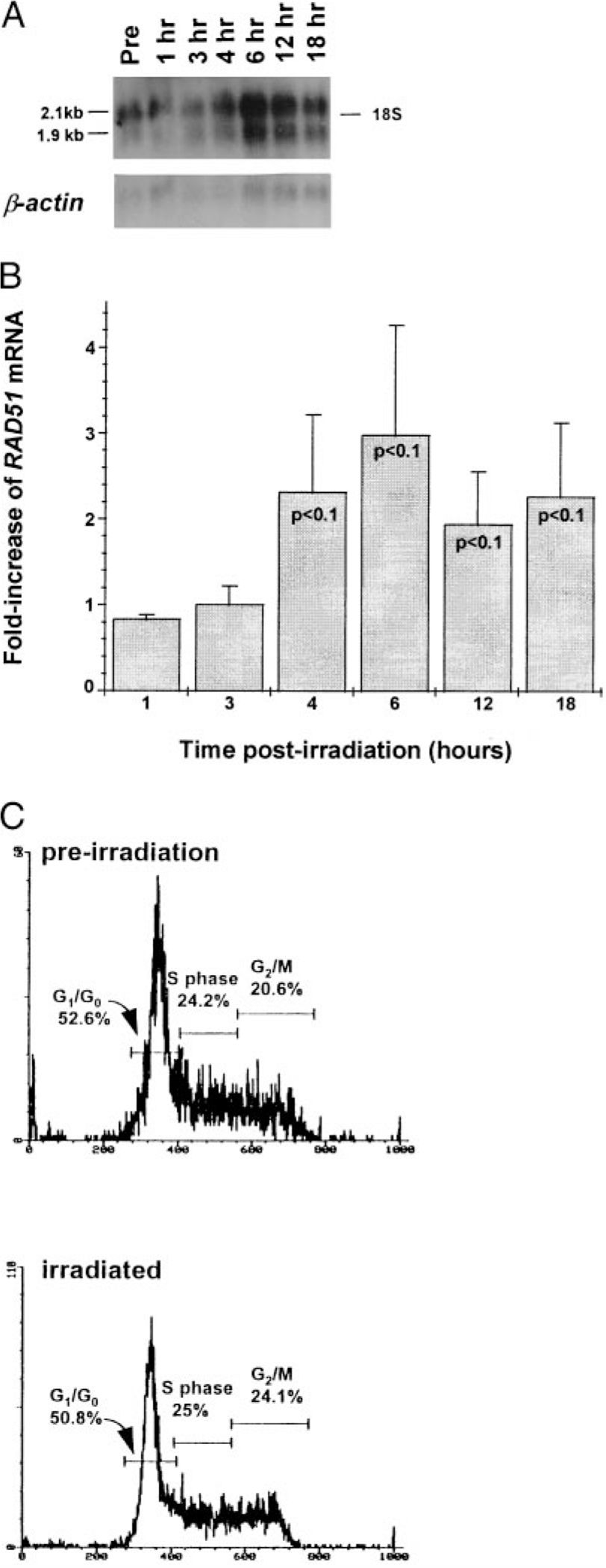
Effects of γ-irradiation on expression of RAD51 mRNA and on cell cycle of the rabbit cell line, 55D1. A, Northern blot analysis of RAD51 mRNA expression after exposure to 6 Gy of γ-irradiation. Each lane contains 3 μg of poly(A)+ RNA isolated at the given times before and after irradiation. RAD51 and β-actin cDNAs were used as probes. Position of the 18S ribosomal bands is shown. In this experiment, after normalizing to the amount of β-actin, the level of RAD51 mRNA at 4 h was 2-fold greater than the level in preirradiated control cells. B, Summary graph of four independent experiments measuring RAD51 mRNA level following exposure of 55D1 cells to 6 Gy ionizing radiation relative to the RAD51 mRNA level before irradiation (fold increase). RAD51 levels for each time point were normalized to the amount of β-actin mRNA by densitometry and the fold increase was determined as the ratio of RAD51 mRNA level after irradiation to the level before irradiation; p values (t test) for 4, 6, 12, and 18 h time points are given. When data from two additional experiments performed with 7 Gy ionizing radiation are included in the analysis, p values were p < 0.025 for the 6 and 12 h time points; p < 0.05 for the 18-h time point. C, FACS profiles of 55D1 cells before (top) and 4 h after (bottom) exposure to 6 Gy of γ-irradiation. The cells were stained with Hoechst 33342 dye 1 h before analysis by flow cytometry.
Ionizing radiation can cause DNA damage that arrests the cell cycle (68). Because it appears that Rad51 protein is regulated by the cell cycle in mammals (69, 70), we tested whether the increase in RAD51 mRNA in 55D1 cells after irradiation resulted from cell cycle arrest at a stage when RAD51 mRNA accumulates. We analyzed the cell cycle profiles of irradiated (6 Gy) and unirradiated cells by staining them with Hoechst 33342 dye and found that the profiles were nearly identical (Fig. 3C). We conclude that the increase in RAD51 mRNA after exposure to ionizing radiation does not result from cell cycle arrest at a phase where RAD51 mRNA accumulates but, instead, is likely due to other biological effects resulting from ionizing radiation.
To examine whether rabbit RAD51 can function in homologous recombination, we tested whether it could complement the homologous recombination defect in S. cerevisiae rad51 mutants. Because of this defect, rad51 mutants are hypersensitive to ionizing radiation. We cloned rabbit RAD51 into a yeast expression vector, introduced this vector into rad51 mutant yeast, and determined whether the yeast survived exposure to ionizing radiation. As shown in Fig. 4, we found that rad51 yeast that expressed rabbit RAD51 were less sensitive to ionizing radiation than were rad51 mutant yeast. For example, rad51 cells expressing rabbit RAD51 that were exposed to 23.4 krad of γ-irradiation were ~10 times more viable than rad51 cells expressing either vector alone or rabbit RAD51 in the antisense orientation. These data demonstrate that rabbit RAD51 can partially complement the γ-ray sensitivity of the rad51 mutant. Because yeast use homologous recombination to repair DNA damage caused by ionizing radiation, we conclude that rabbit RAD51 can function in recombinational repair of DNA.
FIGURE 4.
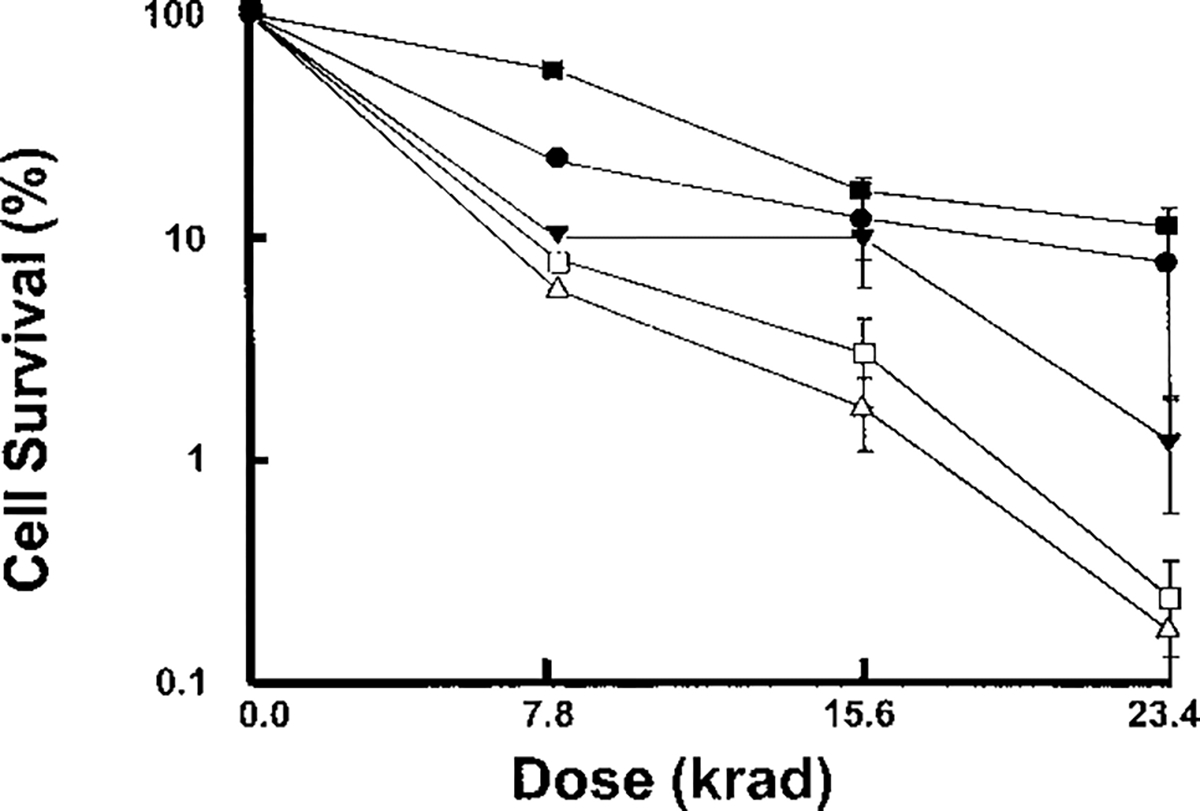
Complementation of rad51 S. cerevisiae by rabbit RAD51 after exposure to γ-irradiation. Fraction of surviving rad51 yeast containing plasmid with rabbit RAD51 (▼), rabbit RAD51 in the antisense orientation (△), vector alone (□), or S. cerevisiae RAD51 (●) is shown. RAD51 yeast (■) were also tested.
Gene conversion.
Because ionizing radiation primarily stimulates recombinational repair that results in gene conversion (71), we decided to determine whether rabbit RAD51 can function in sitespecific repair of a DNA double-strand break that results in gene conversion. Yeast mating-type switch occurs by gene conversion, which is initiated by HO endonuclease through a site-specific DNA double-strand break at the MAT locus. When one of two flanking loci, HMRa or HMLα, is used as a donor, the recipient MAT gene with the double-strand break is converted to the opposite mating type (72). We tested whether rabbit RAD51 could complement a rad51 (MATa) yeast strain that is unable to switch mating types. Cells were tested for their ability to switch by measuring whether they could mate with cells of the original type after induction of switch. For example, rad51 cells begin as MATa and can only mate with MATα strains. After they switch, these cells become MATα and can then mate with MATa strains. We expressed both rabbit RAD51 and HO endonuclease in MATa rad51 cells and assayed for mating type switch to MATα by testing whether the switched cells could mate when mixed with a MATa tester strain. As a positive control, we expressed yeast RAD51 in rad51 cells and determined that >20% of the cells undergo mating type switch (Table I). Six of 2851 rad51 yeast isolates expressing rabbit RAD51 mated with the MATa tester strain, indicating that they had switched mating types. In contrast, none of >5000 rad51 isolates expressing either vector alone or the rabbit gene in the antisense direction grew on MATa-coated minimal media plates. Statistical analysis using Fisher’s exact test showed a significant difference (p < 0.0015) in mating type-switch frequencies between rad51 cells containing rabbit RAD51 and those containing vector alone and rabbit RAD51 in the antisense orientation.
Table I.
Complementation of mating-type switch in rad51 S. cerevisiae
| rad51 Transformant | Total Screened | Total Switcheda | Switch Frequency (%) |
|---|---|---|---|
|
| |||
| S. cerevisiae RAD51 | 886 | 198 | 22.3 |
| Rabbit RAD51 (5′–3′) | 2851 | 6b | 0.2 |
| Rabbit RAD51 (3′–5′) | 2574 | 0 | 0 |
| Vector | 2998 | 0 | 0 |
Only patches with more than one papillae were counted.
Statistical analysis using the Fisher’s exact test showed a difference between switch in rabbit RAD51 (5′–3′) and lack of switch in rabbit RAD51 (3′–5′) and vector (p < 0.0015).
To rule out the possibility that the cells that had switched mating type were RAD51+ revertants, two of the six MATα isolates containing rabbit RAD51 were retested for their ability to switch mating type. If RAD51+ revertants were responsible for the observed mating-type switch, then we would expect the frequency of switching to MATa to be >20%. In both isolates, the frequency of mating type switching remained very low (<5%), indicating that the rad51–1 allele had not reverted back to wild-type (data not shown). These data indicate that rad51 yeast expressing rabbit RAD51 can switch mating types, suggesting that rabbit RAD51 functions in gene conversion.
RAD51 expression and ongoing somatic gene conversion
RAD51 mRNA levels during an immune response.
Recently, Winstead and Knight (unpublished data) identified somatic gene conversion-like events within clonally related VDJ sequences isolated from regional popliteal lymph node germinal center lymphocytes after local immunization. To test whether RAD51 expression is up-regulated in similar circumstances, we immunized rabbits with keyhole limpet hemocyanin and performed Northern blot analysis on poly(A)+ RNA isolated from the regional popliteal lymph nodes (PLN)4 (Fig. 5). We found a striking increase in RAD51 expression. By densitometric analysis of six independent experiments, the level of RAD51 expression in the PLN from the immunized leg was 13-fold (p < 0.025) greater than RAD51 expression in the PLN from the unimmunized leg.
FIGURE 5.
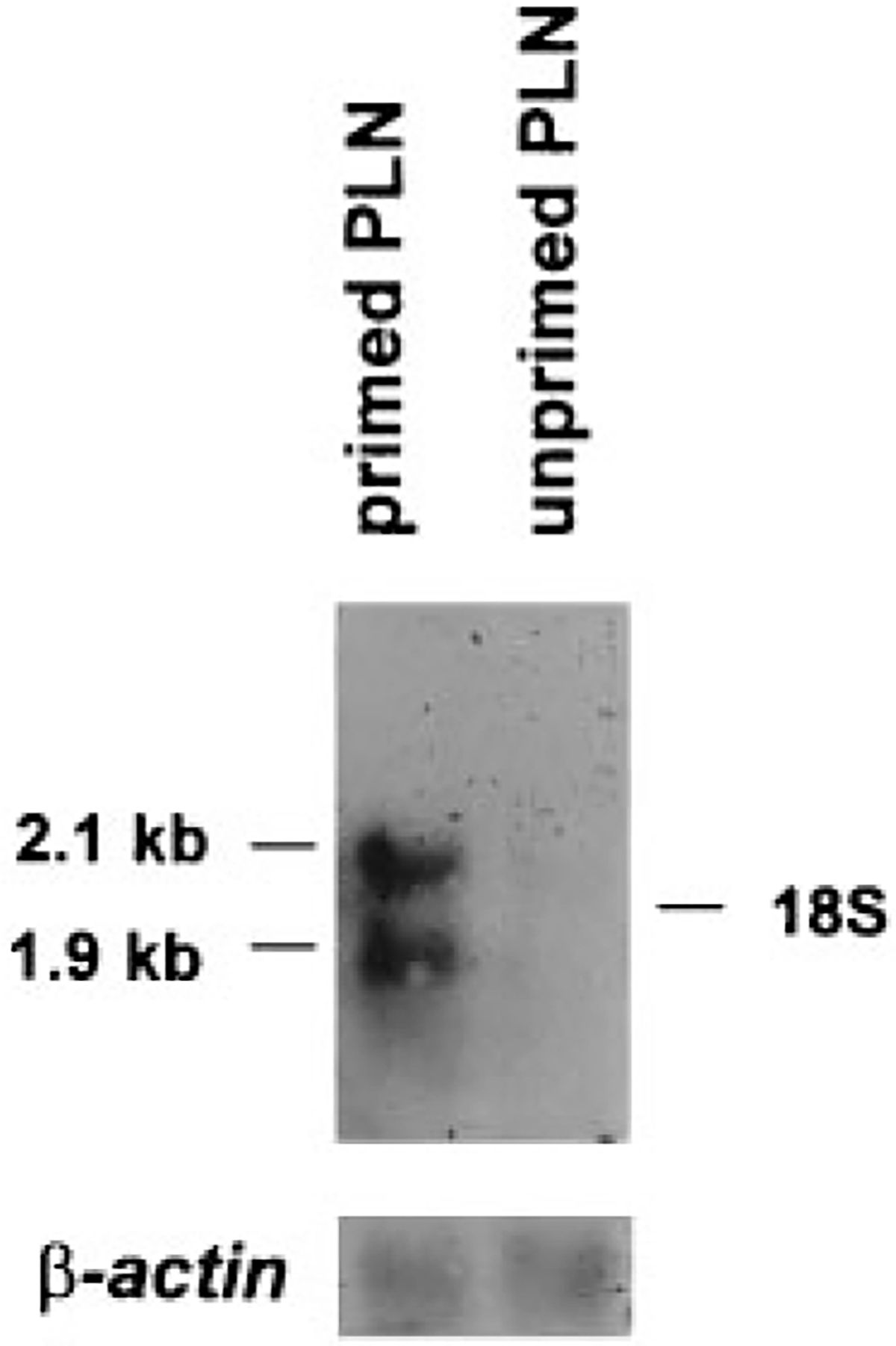
Northern blot analysis of RAD51 mRNA expression after local immunization. Poly(A)+ RNA (3 μg/lane) was isolated from regional PLN from the immunized leg (primed PLN) and the unimmunized leg (unprimed PLN) of keyhole limpet hemocyanin-immunized rabbits. RAD51 and β-actin cDNAs were used as probes. Position of the 18S ribosomal bands is shown. After normalizing levels of RAD51 mRNA to β-actin, the level of RAD51 mRNA detected in the primed PLN was greater than the level of RAD51 mRNA in the unprimed PLN (p < 0.025 as determined by t test).
RAD51 mRNA and protein levels in appendix during ontogeny.
We analyzed the expression of RAD51 in rabbit appendix because somatic gene conversion occurs there starting at ~4 wk of age. Although RAD51 is expressed in appendix of 2-wk-old animals, much higher levels of both the 2.1-kb and the 1.9-kb RAD51 mRNAs are evident at 6 wk of age (Fig. 6A). By densitometric analysis, normalizing both mRNA species to β-actin mRNA, we found that in each of four independent experiments, RAD51 mRNA was 2- to 5-fold higher in 6-wk-old rabbits relative to the level observed in 2-wk-old or adult rabbits (t test = p < 0.1; Fig. 6B). These data show that RAD51 expression increases at the same time at which Ig genes undergo somatic gene conversion.
FIGURE 6.
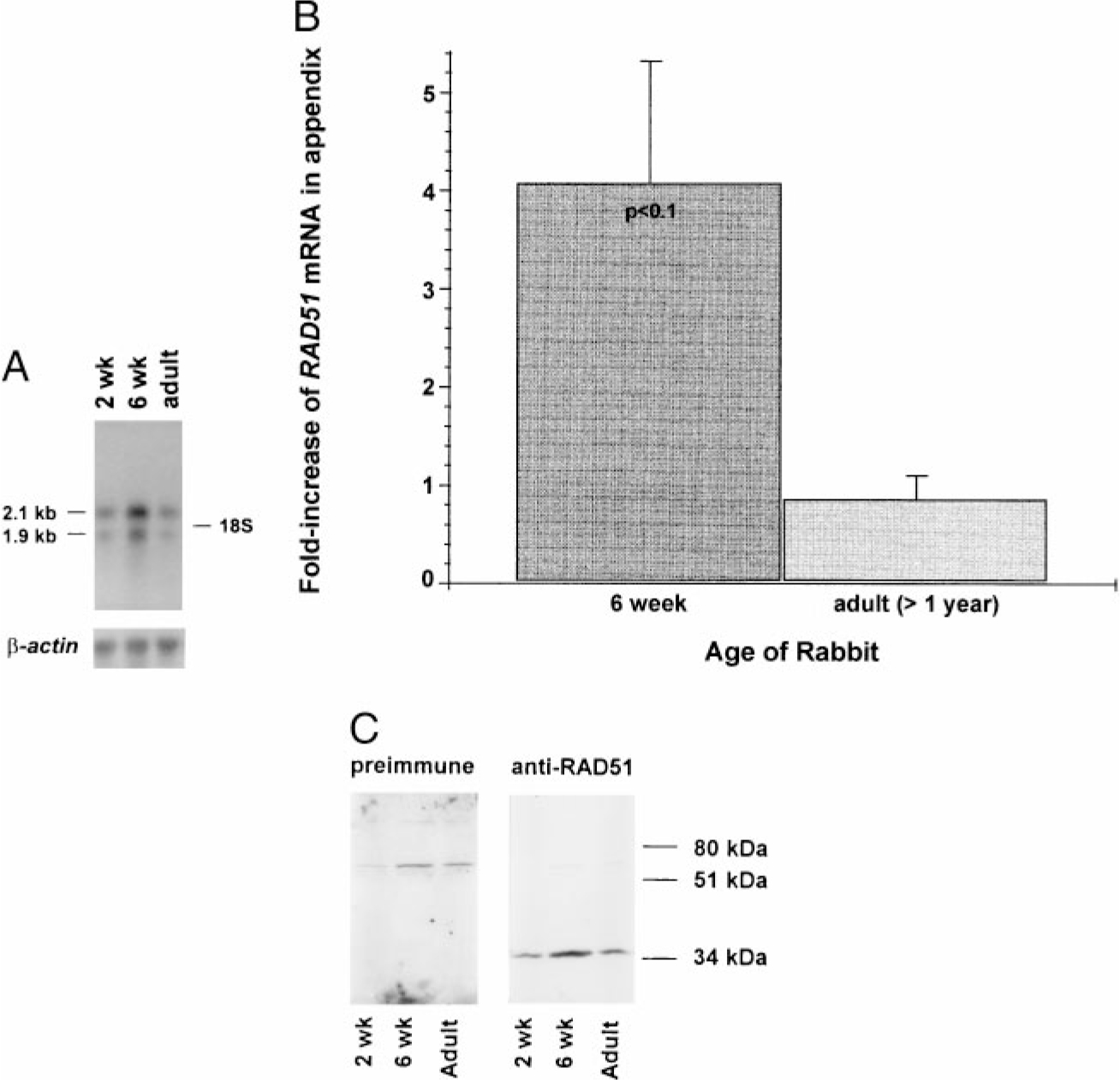
RAD51 mRNA and protein in rabbit appendix. A, Northern blot analysis of RAD51 mRNA expression in appendix from 2-wk-old, 6-wk-old, and adult (>1-yr-old) rabbits. Poly(A)+ RNA (3 μg/lane) from appendices of three different rabbits were pooled for each time point to control for individual animal variation. RAD51 and β-actin cDNAs were used as probes. Position of the 18S ribosomal bands is shown. In this experiment, the level of RAD51 mRNA increased 5-fold in appendix from 6-wk-old rabbits relative to that found in 2-wk-old and adult rabbits. B, Summary graph of four independent experiments measuring RAD51 mRNA levels in 6-wk-old and adult rabbits relative to that found in 2-wk-old rabbits. The p value (t test) for the 6-wk-old rabbits is given. C, Western blot analysis of Rad51 levels in appendix from 2-wk-old, 6-wk-old, and adult rabbits. Lysates (1 × 106 cells/lane) from FACS-sorted IgM+ appendix cells were immunoblotted with preimmune (left) and with anti-human Rad51 (right) serum. Positions of the m.w. (kDa) standards are included for comparison. The m.w. of Rad51 is 38 kDa. In this experiment, the level of Rad51 increased 2-fold in appendix from 6-wk-old rabbits relative to that found in 2-wk-old and adult rabbits. Similar results were obtained using IgL+ cells.
To ascertain whether the increase in RAD51 mRNA is reflected at the protein level and whether it is found in B lymphocytes, we performed Western blot analysis on FACS-sorted appendix B lymphocytes from rabbits of different ages. We generated antiserum against Rad51 by immunizing rabbits with a His-tagged fusion protein and used this antiserum for Western blot analysis (Fig. 6C). Using densitometric analysis we found that, in each of six experiments, B lymphocytes from appendix of 6-wk-old rabbits had from 2- to 12-fold more Rad51 (p < 0.005) than B lymphocytes from 2-wk-old or adult rabbits. These data show that Rad51 protein levels are increased in B lymphocytes from appendix of 6-wk-old rabbits. The timing of this increase correlates with the timing of Ig gene diversification by somatic gene conversion, suggesting that Rad51 may mediate IgH diversification by gene conversion in rabbit.
Discussion
To date, little progress has been made in identifying trans-acting factors involved in somatic diversification of Ig genes by either somatic hyperpoint mutation or somatic gene conversion. Because gene conversion occurs via homologous recombination, protein homologues to those involved in homologous recombination pathways in lower eukaryotes are ideal candidates for studying somatic gene conversion of Ig genes. One such protein is yeast Rad51, a functional homologue of bacterial RecA. In both yeast and bacteria, these molecules are known to mediate homologous recombination (38, 39, 73–75). In the present study, we demonstrated that expression of a rabbit RAD51 homologue in a yeast rad51 mutant restores, in part, DNA double-strand break repair and mating-type switching, both of which occur by homologous recombination. The ability of rabbit RAD51 to partially restore mating-type switching in the rad51 strain that has a switch defect demonstrates clearly that rabbit RAD51 can function in homologous recombination leading to gene conversion. Further, we found that Rad51 levels increase in B lymphocytes from appendix at a time when IgH genes diversify by somatic gene conversion-like events, indicating that Rad51 may mediate IgH gene diversity in rabbits.
Rad51-like molecules have been highly conserved throughout evolution (76), presumably because of their critical role in DNA repair and recombination. Rabbit Rad51 is no exception; it shares a high level of sequence identity with Rad51 homologues from species as distant as Schizosaccharomyces pombe (77) and Xenopus laevis (78) (Fig. 7). One of the most conserved regions of these genes includes consensus Walker type-A and type-B nucleotide binding domains (79), which are important for RAD51-dependent recombination in S. cerevisiae (35, 80, 81). Based on the high degree of sequence identity, it seems likely that rabbit Rad51 functions similarly to yeast Rad51.
FIGURE 7.

Comparison of deduced amino acid sequence of rabbit Rad51 to other Rad51-like molecules. Identity is depicted as dots, and slashes indicate spaces introduced to maximize homology. Walker type-A and type-B nucleotide binding regions are boxed. The N-terminal 52 amino acids from S. cerevisiae and the 18 amino acids from S. pombe are not included. Ra, rabbit; Hs, Homo sapien (accession no. D14134); Mm, Mus musculus (accession no. D13473); Ch, chicken (accession no. D09655); Xe, X. laevis (accession no. D38489); Dm, Drosophila melanogaster (accession no. D17726); Um, Ustilago maydis (accession no. L18882); Sc, S. cerevisiae (accession no. D10023); Sp, S. pombe (accession no. Z22691).
Yeast RAD51 is important for repair of DNA double-strand breaks induced by DNA-damaging agents such as γ-irradiation or the radiomimetic drug methyl methanesulfonate (MMS). Repair of DNA double-strand breaks in yeast after damage by these agents occurs by homologous recombination, frequently resulting in gene conversion (82). Experiments from several laboratories have implicated mammalian Rad51 involvement in recombination. Baumann et al. (47) showed that, similar to yeast Rad51, purified human Rad51 pairs homologous DNA molecules and catalyzes strand exchange in vitro. In addition, Xia et al. (83) demonstrated that increased recombination frequencies in immortalized human cell lines correlated with increases in RAD51 mRNA. Direct evidence that mammalian Rad51 can function in homologous recombination has been obtained through complementation of recombination-deficient yeast. In the present study, we showed that rabbit RAD51 partially complemented the sensitivity of yeast rad51 mutants to ionizing radiation. These data are consistent with the studies of Morita et al. (43) and Knight and Barrington (84), which demonstrated that expression of mouse and rabbit RAD51 in rad51 S. cerevisiae could complement sensitivity to methyl methanesulfonate (MMS).
Like yeast, mammalian cells can repair DNA double-strand breaks by homology-directed recombination (67). Consistent with a role in double-strand break repair in mammals, rabbit RAD51 mRNA is up-regulated in lymphoid cell lines after exposure to ionizing radiation. Although we do not know the exact role of increased levels of RAD51 mRNA after irradiation, Liu et al. (85) described two CHO cell lines that are mildly sensitive to ionizing radiation, XRCC2 and XRCC3, and showed that they are defective in proteins similar to yeast Rad51. Thus, we suggest that, in rabbit, Rad51 may participate in DNA double-strand break repair of radiation-induced damage. This idea needs to be tested directly.
The inability of rabbit RAD51 to fully complement the recombination defects in DNA double-strand break repair or mating-type switch in rad51 yeast could result from its inability to interact optimally with other yeast proteins that are involved in homologous recombination. In yeast, Rad51 interacts with several other RAD52 epistatic group molecules such as Rad52, Rad54, Rad55, and Rad57 (35, 86–90), and these interactions facilitate Rad51-mediated recombination in vitro (40, 91–95). Milne et al. (86) showed that interactions between Rad51 and other molecules involved in homologous recombination are important in vivo by demonstrating that overexpression of a dominant negative homologue of Kluyveromyces lactis RAD52 in wild-type S. cerevisiae interferes with RAD51-mediated repair of damage caused by MMS.
We found that Rad51 levels increase in appendix-derived B lymphocytes of 6-wk-old rabbits, a time at which somatic diversification of IgH genes occurs. Although we suggest that this increase in Rad51 is due to its involvement in gene conversion, we cannot exclude the possibility that the increase results from an increase in cell proliferation. From our data, at least two pieces of evidence suggest that RAD51 has a role beyond that of cell proliferation. First, RAD51 mRNA is up-regulated in rabbit cell lines after exposure to ionizing radiation. Because the cell cycle is relatively unaffected by the γ-ray dose used in our studies, the increase in RAD51 mRNA is not a result of increased proliferation or cell cycle arrest. Second, Rad51 levels in B lymphocytes from appendix of 6-wk-old rabbits are higher than the levels in adult rabbits. By 6 wk of age, the follicular structure of rabbit appendix is fully developed (96, 97), and the high number of germinal centers within the appendix is maintained throughout life as the appendix does not involute in adult rabbits (98). If it is assumed that the percentage of cycling B lymphocytes is similar in these tissues, then the increase in Rad51 is likely to be independent of cell proliferation. It has been difficult to discriminate the role of RAD51 in recombination in contrast to its role in proliferation because RAD51 knockout mutations are lethal in vertebrate cells (49, 50). Despite the lethality of RAD51 mutations in vertebrates, both Tsuzuki et al. (49) and Sonoda et al. (50) showed that RAD51−/− cells undergo one to two rounds of cell division before dying, implying that RAD51 is not essential for proliferation per se. Why do the cells die if RAD51 is not required for proliferation? Sonoda et al. (50) showed that the RAD51−/− cells that did undergo one to two rounds of cell division had increased DNA damage, presumably because DNA damage occurs during replication (99), and RAD51-mediated homologous recombination is required to repair this damage. If this model is correct, then any mutation affecting Rad51 recombination activity would also compromise cell viability, making it difficult to separate a role for RAD51 in homologous recombination from a role in cell proliferation by using traditional mutational analysis. If RAD51 is involved in somatic diversification by gene conversion, then we would expect to find detectable levels in B lymphocytes within germinal centers in rabbit. In mouse, there are conflicting reports regarding the presence of Rad51 within splenic germinal centers. Li et al. (100) detected Rad51 in the periarteriolar lymphoid sheath (PALS), but not within germinal centers, consistent with a function of murine Rad51 or a Rad51-like molecule in isotype switching. In contrast, Yamamoto et al. (101) found Rad51 within germinal centers and rarely in the PALS. More detailed analysis of Rad51 levels in germinal center cell subpopulations are needed to resolve this issue.
We also found that RAD51 mRNA levels increased in the PLN of immunized rabbits. This increase presumably reflects ongoing IgH gene diversification as well as increased proliferation. Rabbits diversify their IgH genes by both somatic gene conversion as well as somatic hyperpointmutation (22–24). Winstead and Knight (unpublished data) recently showed that both of these processes contribute to IgH gene diversification in PLN of immunized rabbits. Although our data do not exclude the involvement of Rad51 in somatic hyperpointmutation of IgH genes, we believe that it is not involved because homologous recombination does not appear to be a major mechanism of hyperpoint mutation (27). Instead, we propose that the increase in RAD51 mRNA in the PLN of immunized rabbits partially results from increased gene conversion.
In this study, we proposed that RAD51 is involved in somatic gene conversion of IgH genes in rabbit. Mammalian homologues to other yeast molecules involved in homologous recombination will also likely be involved in IgH gene diversification. For example, another RAD52 epistasis group member, RAD54, has been implicated in somatic gene conversion of chicken IgH genes. Bezzubova et al. (102) showed that a RAD54−/− mutant of an actively gene converting chicken cell line underwent IgH gene conversion at a lower frequency. Genes involved in other DNA repair pathways, such as mismatch repair (103, 104), may also contribute to IgH gene diversity. An understanding of the regulation of DNA repair genes and their pathways within B lymphocytes will help both in elucidating the mechanisms of Ig diversity and in understanding how different species utilize these mechanisms to generate a diverse Ab repertoire.
Acknowledgments
We thank Dr. Susan Fisher for performing the Fisher’s exact analysis on the mating type switch data.
This work was supported by Grant AI 16611 from the National Institutes of Health.
Footnotes
Abbreviations used in this paper: PLN, popliteal lymph nodes; MMS, methyl methanesulfonate.
References
- 1.Schatz DG, Oettinger MA, and Baltimore D. 1989. The V(D)J recombination activating gene (RAG-1). Cell 59:1035. [DOI] [PubMed] [Google Scholar]
- 2.Oettinger MA, Schatz DG, Gorka C, and Baltimore D. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517. [DOI] [PubMed] [Google Scholar]
- 3.Lee SE, Pulaski CR, He DM, Benjamin DM, Voss MJ, Um J, and Hendrickson EA. 1995. Isolation of mammalian cell mutants that are x-ray sensitive, impaired in DNA double-strand break repair and defective for V(D)J recombination. Mutat. Res. 336:279. [DOI] [PubMed] [Google Scholar]
- 4.Pergola F, Zdzienicka MZ, and Lieber MR. 1993. V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol. Cell. Biol. 13:3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, and Alt FW. 1993. Impairment of V(D)J recombination in double-strand break repair mutants. Science 260:207. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Otevrei T, Gao Y, Cheng H-L, Seed B, Stamato TD, Taccioli GE, and Alt FW. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 83:1079. [DOI] [PubMed] [Google Scholar]
- 7.Kirchgessner CU, Patil CK, Evans JW, Cuomo CA, Fried LM, Carter T, Oettinger MA, and Brown JM. 1995. DNA-dependent kinase (p350) as a candidate gene for the murine scid defect. Science 267:1178. [DOI] [PubMed] [Google Scholar]
- 8.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day III RS, Barron GM, and Allalunis-Turner J. 1995. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 267:1183. [DOI] [PubMed] [Google Scholar]
- 9.Reth M, Hammerling GL, and Rajewsky K. 1978. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. Eur. J. Immunol. 8:393. [DOI] [PubMed] [Google Scholar]
- 10.Bothwell ALM, Paskind M, Reth M, Imanishi-Kari T, Rajewsky K, and Baltimore D. 1981. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a γ−2a variable region. Cell 24:625. [DOI] [PubMed] [Google Scholar]
- 11.Gearhart PJ, Johnson ND, Douglas R, and Hood L. 1981. IgG antibodies to phosphorylcholine exhibit more diversity than their IgM counterparts. Nature 291:29. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Davis M, Sinn E, Patten P, and Hood L. 1981. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell 27:573. [DOI] [PubMed] [Google Scholar]
- 13.Kaartinen M, Griffiths GM, Hamlyn PH, Markham AF, Karjalainen K, Pelkonen JLT, Kakela O, and Milstein C. 1983. Anti-oxazolone light and heavy chain mRNA sequences. J. Immunol. 130:937. [PubMed] [Google Scholar]
- 14.Griffiths GM, Berek C, Kaartinen M, and Milstein C. 1984. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature 312:271. [DOI] [PubMed] [Google Scholar]
- 15.Wysocki L, Manser T, and Gefter ML. 1986. Somatic evolution of variable region structures during an immune response. Proc. Natl. Acad. Sci. USA 83:1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berek C, and Milstein C. 1987. Mutation drift and repertoire shift in the maturation of the immune response. Immunol. Rev. 96:23. [DOI] [PubMed] [Google Scholar]
- 17.Chien NC, Pollock RR, Desaymard C, and Scharff MD. 1988. Point mutations cause the somatic diversification of IgM and IgG2a anti-phosphorylcholine antibodies. J. Exp. Med. 167:954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manser T 1989. Evolution of antibody structure during the immune response. J. Exp. Med. 170:1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynaud C-A, Anquez V, Grimal H, and Weill J-C. 1987. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48:379. [DOI] [PubMed] [Google Scholar]
- 20.Reynaud C-A, Dahan A, Anquez V, and Weill J-C. 1989. Somatic hyperconversion diversifies the single VH gene of the chicken with a high incidence in the D region. Cell 59:171. [DOI] [PubMed] [Google Scholar]
- 21.Thompson CB, and Neiman PE. 1987. Somatic diversification of the chicken immunoglobulin light chain gene is limited to the rearranged variable gene segment. Cell 48:369. [DOI] [PubMed] [Google Scholar]
- 22.Becker RS, and Knight KL. 1990. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell 63:987. [DOI] [PubMed] [Google Scholar]
- 23.Short JA, Sethupathi P, Zhai SK, and Knight KL. 1991. VDJ genes in VHa2 allotype-suppressed rabbits: limited germline VH gene usage and accumulation of somatic mutations in D regions. J. Immunol. 147:4014. [PubMed] [Google Scholar]
- 24.Lanning DK, and Knight KL. 1997. Somatic hypermutation: mutations 3′ of rabbit VDJ H-chain genes. J. Immunol. 159:4403. [PubMed] [Google Scholar]
- 25.Betz AG, Milstein C, González-Fernández A, Pannell R, Larson T, and Neuberger MS. 1994. Elements regulating somatic hypermutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell 77:239. [DOI] [PubMed] [Google Scholar]
- 26.Weber JS, Berry J, Manser T, and Claflin JL. 1994. Mutations in Ig V(D)J genes are distributed asymmetrically and independently of the position of V(D)J. J. Immunol. 153:3594. [PubMed] [Google Scholar]
- 27.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez-Fernandez A, Pannell R, Neuberger MS, and Milstein C. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Science 376:225. [DOI] [PubMed] [Google Scholar]
- 28.Peters A, and Storb U. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4:57. [DOI] [PubMed] [Google Scholar]
- 29.Green NS, Rabinowitz JL, Zhu M, Kobrin BJ, and Scharff MD. 1995. Immunoglobulin variable region hypermutation in hybrids derived from a pre-B and a myeloma cell line. Proc. Natl. Acad. Sci. USA 92:6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachl J, and Wabl M. 1996. An immunoglobulin mutator that targets G-C base pairs. Proc. Natl. Acad. Sci. USA 93:851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachl J, and Wabl M. 1996. Enhancers of hypermutation. Immunogenetics 45:59. [DOI] [PubMed] [Google Scholar]
- 32.Winkler H 1930. Die konversion der gene. Gustav Fisher, Jena. [Google Scholar]
- 33.Lindegren C 1949. The Yeast Cell, its Genetics, and Cytology. Educational Publishing, St. Louis, MO. [Google Scholar]
- 34.Petes TD, Malone RE, and Symington LS. 1991. Recombination in yeast. In The Molecular Biology and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Broach JR, Pringle J, and Jones E, eds. Cold Spring Harbor Laboratory Press, Plainview, NY, p. 407. [Google Scholar]
- 35.Shinohara A, Ogawa H, and Ogawa T. 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69:457. [DOI] [PubMed] [Google Scholar]
- 36.Aboussekhra A, Chanet R, Adjiri A, and Fabre F. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa T, Yu X, Shinohara A, and Egelman EH. 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259:1896. [DOI] [PubMed] [Google Scholar]
- 38.Sung P 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241. [DOI] [PubMed] [Google Scholar]
- 39.Sung P, and Robberson DL. 1995. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82:453. [DOI] [PubMed] [Google Scholar]
- 40.Sung P 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111. [DOI] [PubMed] [Google Scholar]
- 41.Clark AJ, and Margulies AD. 1965. Isolation and characterization of recombination-deficient mutants of Escherichia coli k12. Genetics 53:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bezzubova O, Shinohara A, Mueller RG, Ogawa H, and Buerstedde J-M. 1993. A chicken RAD51 homologue is expressed at high levels in lymphoid and reproductive organs. Nucleic Acids Res. 21:1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita T, Yoshimura Y, Yamamoto A, Murata K, Mori M, Yamamoto H, and Matsushiro A. 1993. A mouse homolog of the Escherichia coli recA and Saccharomyces cerevisiae RAD51 genes. Proc. Natl. Acad. Sci. USA 90:6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura Y, Morita T, Yamamoto A, and Matsushiro A. 1993. Cloning and sequence of the human RecA-like gene cDNA. Nucleic Acids Res. 21:1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson FE, Stasiak A, and West SC. 1994. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 13:5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta RC, Bazemore LR, Golub EI, and Radding CM. 1997. Activities of human recombination protein Rad51. Proc. Natl. Acad. Sci. USA 94:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumann P, Benson FE, and West SC. 1996. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87:757. [DOI] [PubMed] [Google Scholar]
- 48.Baumann P, and West SC. 1997. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 16:5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuzuki T, Yoshimitsu F, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yasuhide Y, and Morita T. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93:6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, and Takeda S. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinstein PD, Mage RG, and Anderson AO. 1994. The appendix functions as a mammalian bursal equivalent in the developing rabbit. Adv. Exp. Med. Biol. 355:249. [DOI] [PubMed] [Google Scholar]
- 52.Crane MA, Kingzette M, and Knight KL. 1996. Evidence for limited B-lymphopoiesis in adult rabbits. J. Exp. Med. 183:2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vajdy M, Sethupathi P and Knight KL. 1998. Dependence of antibody somatic diversification on gut-associated lymphoid tissue in rabbit. J. Immunol. 160:2725. [PubMed] [Google Scholar]
- 54.Morrison DP, and Hastings PJ. 1979. Characterization of the mutation mut5–1. Mol. Gen. Genet. 175:57. [DOI] [PubMed] [Google Scholar]
- 55.Saeki T, Machida I, and Nakai S. 1980. Genetic control of diploid recovery after γ-irradiation in the yeast Saccharomyces cerevisiae. Mutat. Res. 73:251. [DOI] [PubMed] [Google Scholar]
- 56.Sugawara N, Ivanov EL, Fishman-Lobell J, Ray BL, Wu X, and Haber JE. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84. [DOI] [PubMed] [Google Scholar]
- 57.Sanger F, Nicklen S, and Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blin N, and Stafford DW. 1976. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 3:2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Southern E 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503. [DOI] [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch EF, and Maniatis T. 1989. Molecular Cloning: A Laboratory Manual, 2nd Ed. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 61.Knight KL, Spieker-Polet H, Kazdin DS, and Oi VT. 1988. Transgenic rabbits with lymphocytic leukemia induced by the c-myc oncogene fused with the immunoglobulin heavy chain enhancer. Proc. Natl. Acad. Sci. USA 85:3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller W, and Gautier F. 1975. Interactions of heteroaromatic compounds with nucleic acids: A-T-specific non-intercalating DNA ligands. Eur. J. Biochem. 54:385. [DOI] [PubMed] [Google Scholar]
- 63.Roth RM, Game JC, and Peak MJ. 1987. Sensitivities to monochromatic 254-nm and 365-nm radiation of closely related strains of Saccharomyces cerevisiae with differing repair capabilities. Photochem. Photobiol. 45:479. [DOI] [PubMed] [Google Scholar]
- 64.Sherman F, Fink GR, and Hicks JB. 1982. Methods of Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 65.Fasullo M, Bennett T, AhChing P, and Koudelik J. 1998. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol. Cell. Biol. 18:1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Basile G, Aker M, and Mortimer RK. 1992. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol. 12:3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang F, Han M, Romanienko PJ, and Jasin M. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada M, and Puck TT. 1961. Action of radiation on mammalian cells, IV. Reversible mitotic lag in the S3 HeLa cells produced by low doses of X-rays. Proc. Natl. Acad. Sci. USA 47:1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto A, Taki T, Yagi H, Habu T, Yoshida K, Yoshimura Y, Yamamoto K, Matsushiro A, Nishimune Y, and Morita T. 1996. Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol. Gen. Genet. 251:1. [DOI] [PubMed] [Google Scholar]
- 70.Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, and Kamada N. 1996. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene 12:2165. [PubMed] [Google Scholar]
- 71.Davies PJ, Evans WE, and Parry JM. 1975. Mitotic recombination induced by chemical and physical agents in the yeast Saccharomyces cerevisiae. Mutat. Res. 51:327. [DOI] [PubMed] [Google Scholar]
- 72.Haber JE 1992. Mating-type gene switching in Saccharomyces cerevisiae. Trends Genet. 8:446. [DOI] [PubMed] [Google Scholar]
- 73.Shibata T, DasGupta C, Cunningham RP, and Radding CM. 1979. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl. Acad. Sci. USA 76:1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weinstock GM, McEntee K, and Lehman IR. 1979. ATP-dependent renaturation of DNA catalyzed by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 76:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cassuto E, West SC, Mursalim J, Conlon S, and Howard-Flanders P. 1980. Initiation of gentic recombination: homologous pairing between duplex DNA molecules promoted by recA protein. Proc. Natl. Acad. Sci. USA 77:3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Story RM, Bishop DK, Kleckner N, and Steitz TA. 1993. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science 259:1892. [DOI] [PubMed] [Google Scholar]
- 77.Muris DF, Vreeken K, Carr AM, Broughton BC, Lehman AR, Lohman PH, and Pastink A. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maeshima K, Morimatsu K, Shinohara A, and Horii T. 1995. RAD51 homologues in Xenopus laevis: two distinct genes are highly expressed in ovary and testis. Gene 160:195. [DOI] [PubMed] [Google Scholar]
- 79.Walker JE, Saraste M, Runswick MJ, and Gay NJ. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sung P, and Stratton SA. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983. [DOI] [PubMed] [Google Scholar]
- 81.Chanet R, Heude M, Adjiri A, Moloisel L, and Fabre F. 1996. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 16:4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Szostak JW, Orr-Weaver TL, Rothstein RJ, and Stahl FW. 1983. The double-strand-break repair model for recombination. Cell 33:25. [DOI] [PubMed] [Google Scholar]
- 83.Xia SJ, Shammas MA, and Reis RJ. 1997. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol. Cell. Biol. 17:7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Knight KL, and Barrington RA. 1998. Somatic diversification of IgH genes in rabbit. Immunol. Rev. 162:37. [DOI] [PubMed] [Google Scholar]
- 85.Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, et al. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damage. Mol. Cell 1:783. [DOI] [PubMed] [Google Scholar]
- 86.Milne GT, and Weaver DT. 1993. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 7:1755. [DOI] [PubMed] [Google Scholar]
- 87.Donovan JW, Milne GT, and Weaver DT. 1994. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 8:2552. [DOI] [PubMed] [Google Scholar]
- 88.Hays SL, Firmenich AA, and Berg P. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson RD, and Symington LS. 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 15:4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, and Heyer W-D. 1997. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 16:2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sung P 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194. [DOI] [PubMed] [Google Scholar]
- 92.Benson FE, Baumann P, and West SC. 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391:401. [DOI] [PubMed] [Google Scholar]
- 93.New JH, Sugiyama T, Zaitseva E, and Kowalczykowski SC. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407. [DOI] [PubMed] [Google Scholar]
- 94.Shinohara A, and Ogawa T. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404. [DOI] [PubMed] [Google Scholar]
- 95.Petukhova G, Stratton S, and Sung P. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393:91. [DOI] [PubMed] [Google Scholar]
- 96.Cooper MD, Perey DY, Gabrielsen AE, Sutherland EER, McKneally MF, and Good RA. 1968. Production of an antibody deficiency syndrome in rabbits by neonatal removal of organized intestinal lymphoid tissues. Int. Arch. Allergy 33:65. [DOI] [PubMed] [Google Scholar]
- 97.Thorbecke GJ 1960. γ Globulin and antibody formation in vitro. I. γ Globulin formation in tissues from immature and normal adult rabbits. J. Exp. Med. 112:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Archer OK, Sutherland DER, and Good RA. 1964. The developmental biology of lymphoid tissue in the rabbit: consideration of the role of thymus and appendix. Lab. Invest. 13:259. [PubMed] [Google Scholar]
- 99.Barnes G, and Rio D. 1997. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li MJ, Peakman MC, Golub EI, Reddy G, Ward DC, Radding CM, and Maizels N. 1996. Rad51 expression and localization in b cells carrying out class switch recombination. Proc. Natl. Acad. Sci. USA 93:10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamamoto A, Taki T, Yagi H, Habu T, Yoshida K, Yoshimura Y, Yamamoto K, Matsushiro A, Nishimune Y, and Morita T. 1996. Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol. Gen. Genet. 251:1. [DOI] [PubMed] [Google Scholar]
- 102.Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, and Buerstedde JM. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185. [DOI] [PubMed] [Google Scholar]
- 103.Cascalho M, Wong J, Steinberg C, and Wabl M. 1998. Mismatch repair co-opted by hypermutation. Science 279:1207. [DOI] [PubMed] [Google Scholar]
- 104.Storb U, Peters A, Klotz E, Kim N, Shen HM, Hackett J, Rogerson B, and Martin TE. 1998. Cis-acting sequences that affect somatic hypermutation of Ig genes. Immunol. Rev. 162:153. [DOI] [PubMed] [Google Scholar]


