Summary
Cytosolic innate immune sensing is critical for protecting barrier tissues. NOD1 and NOD2 are cytosolic sensors of small peptidoglycan fragments (muropeptides) derived from the bacterial cell wall. These muropeptides enter cells, especially epithelial cells, through unclear mechanisms. We previously implicated SLC46 transporters in muropeptide transport in Drosophila immunity. Here we focused on Slc46a2, which was highly expressed in mammalian epidermal keratinocytes, and showed that it was critical for the delivery of diaminopimelic acid (DAP)-muropeptides and activation of NOD1 in keratinocytes, while the related transporter Slc46a3 was critical for delivering the NOD2 ligand MDP to keratinocytes. In a mouse model, Slc46a2- and Nod1-deficiency strongly suppressed psoriatic inflammation, while methotrexate, a commonly used psoriasis therapeutic, inhibited Slc46a2-dependent transport of DAP-muropeptides. Collectively these studies define SLC46A2 as a transporter of NOD1activating muropeptides, with critical roles in the skin barrier, and identify this transporter as an important target for anti-inflammatory intervention.
eTOC Blurb
Innate immune receptors NOD1/2 detect bacterial peptidoglycans that access the cytosol. However, the mechanisms by which peptidoglycan fragments reach the cytosol are unclear. Bharadwaj et al. reveal that Slc46a2 is critical for transporting DAP-type peptidoglycans into keratinocytes, that it is inhibited by the anti-inflammatory methotrexate, and that it works with the NOD1 pathway to incite psoriatic inflammation.
Graphical Abstract
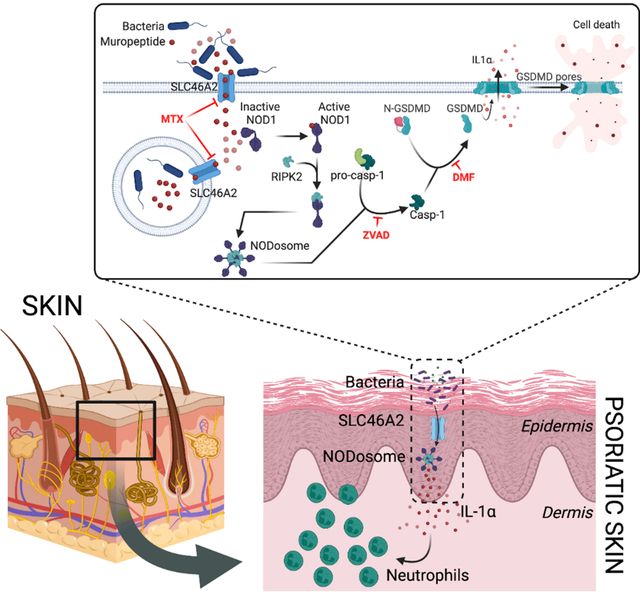
Introduction
Cytosolic innate immune receptors play critical roles in host defense by sensing microbial products that access the cell interior and activating potent immune and inflammatory responses.1,2 These receptors include several nucleic acids sensors, such as cGAS, RIG-I, NLRP1, and AIM2, as well as others that respond to bacterial products (NOD1, NOD2, NAIPs and NLRC4) or danger signals (NLRP1 or 3).2–7 In some cell types, ligands for these cytosolic sensors can be imported into the cytosol.8–12 For instance, fragments of the bacterial cell wall, or muropeptides, enter the cytosol and drive potent inflammatory responses by activating NOD1 or NOD2 and NF-κB.13,14 Muropeptides that contain the amino acid diaminopimelic acid (DAP), common to gram-negative bacteria and gram-positive bacilli, are potent NOD1 agonists, while muramyl-dipeptide (MDP), derived from nearly all bacterial peptidoglycan, activates NOD2.
While muropeptides can enter into some cell types, the underlying mechanisms of entry are unclear.15 Several reports have linked the solute carrier 15 (SLC15) family of peptide transporters to cytosolic muropeptide delivery.16–21 However, these solute carriers have not been directly demonstrated to transport muropeptides and are not specifically required for NOD signaling, but instead have recently been linked to IRF5 activation following stimulation of TLRs as well as NODs.22,23 On the other hand, we recently identified the SLC46 family as candidate muropeptide transporters in Drosophila and mammalian cells.24 In particular, human or mouse SLC46A2 or SLC46A3, but not SLC46A1 (encoding the Proton Coupled Folate Transporter), strongly enhance muropeptide-triggered NOD-dependent NF-κB reporter activity in cell lines. SLC46A2 is highly effective in facilitating the DAP-containing muropeptide tracheal cytotoxin (TCT) activation of the cytosolic innate immune receptor NOD1 in these reporter assays, yet it is expressed in only a limited set of tissues. Analysis of publicly available databases and previous publications reveals that Slc46a2 is predominantly expressed in the skin epidermis as well as cortical epithelial cells of the thymus.25–27 Skin is a critical barrier defense against micro-organisms in the environment and an important immune-responsive organ.28,29 NOD-mediated bacterial recognition plays a critical role in the interaction between gut microbiota and the intestinal epithelia30, yet much less is known about the role of the NOD receptors in other barrier tissues, like the skin.31 Similar to the gut microbiome, the skin microbiome constantly interacts with the epidermis, modulating local and systemic immune responses, and is implicated in inflammatory skin diseases such as psoriasis.32–35 However, the role of NOD1/2 sensing in this tissue has not been evaluated.
While SLC46A1 has little effect on NOD1 or NOD2 sensing of muropeptides in NF-κB reporter assays, it is an established transporter of folates and anti-folates.24,36 The anti-folate Methotrexate (MTX) is both an anti-proliferative agent, used to treat blood cancers, and a potent anti-inflammatory drug commonly used as a first-line treatment for psoriasis, rheumatoid arthritis, and other immune-mediated inflammatory disorders.37,38 For cancer therapy, MTX inhibits cell proliferation by interfering with dihydrofolate folate reductase and other folate-dependent enzymes required for thymidine and purine synthesis. However, the anti-inflammatory mechanism of action for MTX is less clear and is not linked to its antiproliferative activity. While several mechanisms have been suggested, one leading hypothesis argues that MTX, by inhibiting the folate-dependent enzyme 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) transformylase (ATIC), indirectly leads to elevated extracellular adenosine which in turns inhibits inflammatory signaling via the adenosine receptors A2a and A3.39 However, other mechanisms have also been proposed, including effects on NF-κB, lincRNA-p21, and NO synthase. Yet, demonstrating any of these mechanisms of action as central to the response of patients to MTX has been challenging. Nonetheless, it is clear that MTX accesses the cytosol where it is often polyglutamated and potently inhibits several folate-dependent enzymes. The role of SLC46A2 or SLC46A3, which are ~30% identical to SLC46A1, in MTX uptake has not been extensively examined.
Here we characterized a mouse deficient in Slc46a2, demonstrating the essential function of SLC46A2 for delivery of DAP-muropeptides and NOD1 activation in keratinocytes, and identifying a DAP-muropeptide-triggered epidermal inflammatory response. This response involved caspase-1- and gasdermin D-dependent plasma membrane permeabilization of epidermal keratinocytes and the release of IL-1α. In the mouse, this response drove the rapid recruitment of neutrophils to muropeptide-challenged skin and was essential for the development of psoriatic inflammation. Moreover, we also show that this pathway was inhibited by methotrexate, indicating an additional mechanism of action for this commonly used anti-inflammatory drug.37 Human keratinocytes, in the context of 3D skin organoids, also similarly respond to DAP-muropeptides driving an IL-1-dependent response.
Results
NOD1 and NOD2 responses in vivo require Slc46a2 and Slc46a3
Previously, we implicated the SLC46 family of transporters in the delivery of muropeptides to cytosolic innate immune receptors in Drosophila and human cell lines.24 To determine whether SLC46 family members contribute to mammalian responses to muropeptides in vivo, we used a classic assay to monitor NOD1 and NOD2 activaton in Slc46a2−/− and Slc46a3−/− mice (Fig. S1A–E).40–42 iE-DAP or MDP was injected intraperitoneally (IP) and neutrophil recruitment to the peritoneum measured after 3 h (Fig. S1F). Like Nod1−/− mice, Slc46a2−/− mice did not respond to iE-DAP but responded like wild-type to MDP. By contrast, Slc46a3−/− mice phenocopied Nod2−/− mice, responding normally to iE- DAP but failing to respond to MDP (Fig. 1A).
Fig. 1. Slc46a2 is required for neutrophil recruitment in response to NOD1 stimulation in the mouse peritoneum and skin.
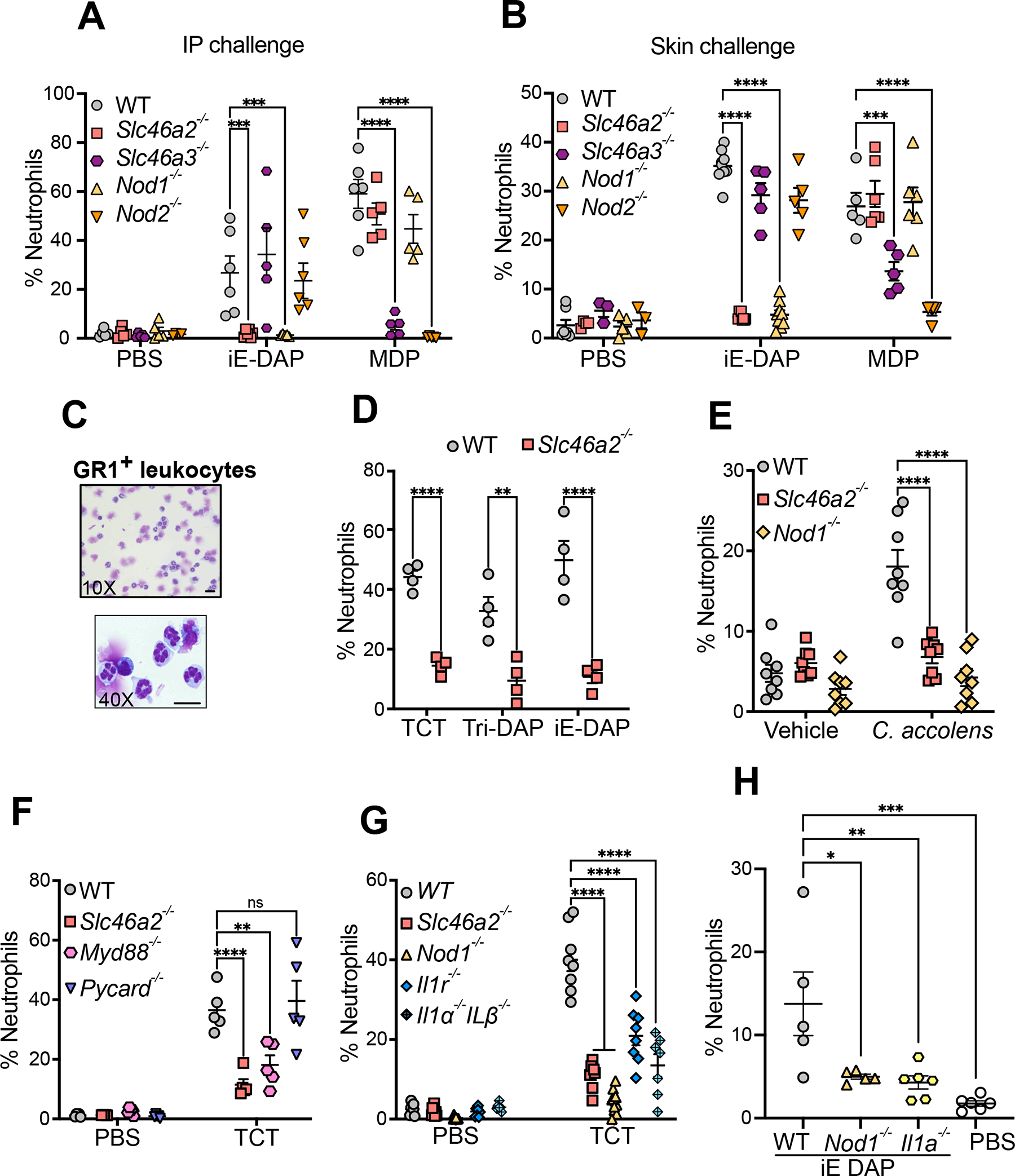
(A) Neutrophil recruitment in the peritoneum 3 h after intraperitoneal injection of 100 μl iE-DAP (30 μM) or MDP (10 μM), shown as a percent of CD45+ cells, in wild type, Slc46a2−/−, Nod1−/−, Slc46a3−/− or Nod2−/− mice. See also Figure S1A–F.
(B) Neutrophil recruitment to the skin 3 h after intradermal injection of 10 μl iE-DAP (30 μM) or MDP (10 μM), shown as a percent of CD45+ cells, in wild type, Slc46a2−/−, Nod1−/−, Slc46a3−/− or Nod2−/− mice. See also Figure S1G–H.
(C) Images of FACS sorted GR1+ neutrophils 3 h after iE-DAP challenge from WT mouse skin. Cells prepared using cytospin and stained with Giemsa stain show multilobed nuclei.
(D) Neutrophil recruitment to the skin shown as a percent of CD45+ cells, 3 h after intradermal challenged with 10 μl different DAP-type muropeptides, TCT (8 μM), iE-DAP (30 μM), or Tri-DAP (25 μM), comparing wild type and Slc46a2−/− animals.
(E) Neutrophil recruitment measured 3 h after topical association of tape-stripped pinnae skin with C. accolens, shown as a percent of CD45+ cells, in wild type, Slc46a2−/−, or Nod1−/− animals. See also Figure S2A.
(F) Recruitment of neutrophils to pinnae 3 h after intradermal injection of 10 μl of 8 μM TCT or PBS in WT, Slc46a2−/−, Myd88−/− or Pycard−/− (Asc-deficient) mice, shown as a percent of CD45+ cells.
(G) Recruitment of neutrophils to pinnae 3 h after intradermal injection of 10 μl of 8 μM TCT or PBS in WT, Slc46a2−/−, Il1r1−/−, or Il1a−/− & Il1b−/−, shown as a percent of CD45+ cells. See also Figure S2B and S2C.
(H) Recruitment of neutrophils after 3 h of intradermal injection of 10 μl of 30 μM iE-DAP in WT, Il1a−/−, or Nod1−/− mice, shown as a percent of CD45+ cells. See also Figure S2D.
Genotypes are indicated on all panels. Comparisons with two-way ANOVA with Tukey’s multiple comparisons test to determine significance. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant. Each dot represents an individual animal, data pooled from two to four separate trials, except for panel C which shows a representative image from three independent experiments. The scale bar is 10μM. See also Figures S1 & S2
Slc46a2 is highly expressed in skin epidermis but not found in many other tissues (Fig. S1G and 43,44). As the activity of NOD1 agonists in skin has not been extensively characterized31, the DAP-muropeptide tracheal cytotoxin (TCT), or LPS as a control, was injected intradermally (ID) in the pinnae and Gr-1+ leukocyte recruitment to the ear was monitored, between 1 and 24 h (Fig. S1H); leukocyte recruitment following DAP-muropeptide was rapid and robust, peaking at 3 h. Using this intradermal challenge assay, Slc46a2−/− mice failed to respond to iE-DAP, but responded normally to MDP, phenocopying Nod1−/− mice (Fig. 1B). Moreover, Slc46a3−/− mice were significantly defective in responding to MDP, like Nod2−/− animals. Gr-1+ leukocytes recruited to the pinnae by iE-DAP challenge were multilobed, consistent with neutrophils, and parallel analysis with Ly6C and Ly6G antibodies similarly showed robust, Slc46a2-dependent recruitment of Ly6Ghi neutrophils (Figure S1I). At this 3 h timepoint, recruitment of T-cells (CD3+) or Ly6Chi macrophages was trending but did not reach significance. H&E staining of pinnae sections confirmed that neutrophils, as well as other immune cells, were recruited to the skin in response to DAP-muropeptide challenge in a Nod1- and Slc46a2-dependent manner (Fig. S1K, multi-lobed neutrophil nuclei marked with arrows). These results, with IP or ID challenge assays, demonstrate that Slc46a2 is selectively required for the NOD1 pathway while Slc46a3 is critical for the NOD2 pathway, consistent with the idea that they each transport NOD1 or NOD2 ligands, respectively, into the cytosol.
To further probe the specificity of SLC46A2 in the response to DAP-muropeptides, we compared three different NOD1 agonists with increasing sizes (iE-DAP, Tri-DAP, and TCT) with ID challenge in the pinnae. Slc46a2-deficient mice displayed significantly reduced neutrophil recruitment in response to TCT, iE-DAP, and Tri-DAP challenge when compared to WT mice (Fig.1D). Further, topical application of DAP-muropeptides on tape-stripped mouse skin also triggered robust Slc46a2- and Nod1-dependent neutrophil recruitment (Fig. S2A). Similarly, tape-stripped skin treated with live Corynebacterium accolens, a common commensal with DAP-type peptidoglycan known to modulate local and systemic immunity when associated with the skin32, resulted in robust neutrophil recruitment in WT skin that was significantly decreased in either the Slc46a2 or Nod1 mutants (Fig. 1E). These results support the hypothesis that SLC46A2 functions in the skin epidermis to deliver DAP-muropeptides to cytosolic NOD1, triggering a rapid neutrophilic influx.
DAP-muropeptide response in vivo requires IL-1α
To investigate the pathways involved in DAP-muropeptide-triggered Slc46a2 and Nod1-dependent neutrophil recruitment to the skin, Pycard- (encoding ASC) and Myd88-deficient mice were analyzed. WT and Pycard-deficient mice showed similar responses to ID DAP-muropeptide challenge, whereas the response was significantly reduced in Myd88-deficient mice, like the Slc46a2-deficient mice (Fig. 1F). Given that the DAP-muropeptide (TCT) used in these assays is LPS and lipopeptide free45, the MyD88 dependence suggests that DAP-muropeptide challenge might trigger an IL-1 response. In fact, mice lacking a functional IL-1 Receptor (Il1r1−/−) or deficient for both IL-1α and IL-1β encoding genes (Il1a and Il1b) showed significantly reduced responses to intradermal TCT challenge, similar to Nod1−/− or Slc46a2−/− mice (Fig. 1G). Importantly, the absence of Myd88 or Il1r1 did not significantly reduce the expression of Slc46a2, Slc46a3, Nod1, or Nod2 in primary mouse keratinocytes (Fig. S2B). We further dissected the individual role of these cytokines (IL-1α and IL-1β); Il1b-deficient mice did not show any defect in responding to DAP-muropeptide (Fig. S2C). By contrast, Il1a-deficient animals failed to respond to intradermal DAP-muropeptide challenge (Fig. 1H). This phenotype was further confirmed by inhibiting lL-1α using a neutralizing antibody (Fig. S2D). Keratinocytes are known for their robust IL-1α expression46 and this cytokine is known for its potent neutrophil recruiting activity.47 These results suggest that SLC46A2-transported DAP-muropeptide and ensuing NOD1 activation induces IL-1α release from keratinocytes and subsequent recruitment of neutrophils in skin.
DAP-muropeptide-stimulated primary keratinocytes release IL-1α through a pyroptotic-like process
To examine IL-1α production, primary mouse keratinocytes, which express Slc46a2 and Nod1 (Fig. S2E), were isolated and cultured ex vivo, stimulated with DAP-muropeptide, and supernatants assayed for cytokine production. As predicted, WT keratinocytes released significant IL-1α protein 24 h post-challenge, whereas IL-1α released by Slc46a2- or Nod1-deficient keratinocytes was significantly reduced compared to WT, and not significantly increased compared to unstimulated cells (Fig. 2A). By contrast, primary dermal fibroblasts did not respond to iE-DAP (Fig. S2F), consistent with no detectable expression of Slc46a2 and Nod1 in these cells (Fig. S2E). Further, conditioned media from DAP-muropeptide stimulated WT keratinocytes, but not control conditioned media from unstimulated cells, triggered significant neutrophil recruitment when injected IP into naïve mice, while the activity of supernatants from IL1a-, Slc46a2- or Nod1-deficient keratinocytes was significantly reduced (Fig. S2G and 2B). Likewise, conditioned media from stimulated WT keratinocytes depleted with anti-IL-1α displayed significantly less neutrophil-attracting activity when injected IP (S2H). Other cytokines, including TNF, IL-6, IL-1β, and IL-17 were not induced in iE-DAP-challenged keratinocytes and were unchanged in the absence of Slc46a2 or Nod1 (Fig. S2I), while IL-23 was not detected in any condition.
Fig. 2. Primary mouse epidermal keratinocytes respond to DAP-muropeptides via Slc46a2 and Nod1.
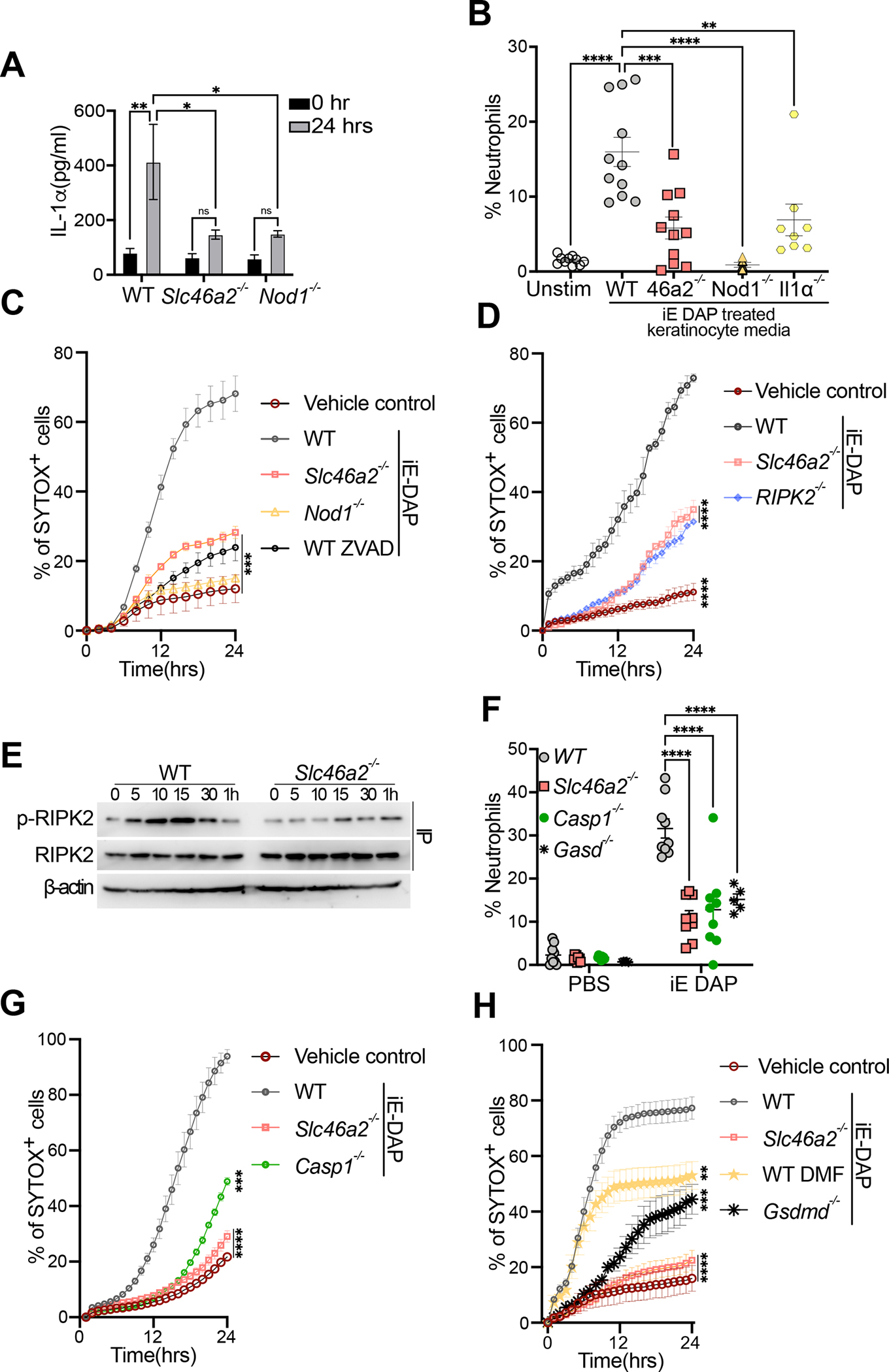
(A) Primary keratinocytes from wild type, Slc46a2−/−, or Nod1−/− mice were isolated and cultured ex vivo, stimulated with 30 μM iE DAP for 24 h, and supernatants were assayed for IL-1α cytokine production by ELISA. See also Figure S2E–F and S2I, where other cytokines were similarly analyzed.
(B) Neutrophil recruitment to the peritoneum of naïve wild type mice after IP injection of conditioned media from WT, Slc46a2−/−, Nod1−/− or Il1a−/− keratinocytes stimulated with 30 μM iE-DAP for 24 h, or unstimulated as a control, shown as a percent of CD45+ cells. See also Figure S2G and S2H.
(C) Using live cell imaging, WT, Slc46a2−/− and Nod1−/− keratinocyte permeabilization was measured with a Sytox dye uptake assay over a 24 h time course following 30 μM iE-DAP treatment. Additionally, WT keratinocytes were pre-treated with pan-caspase inhibitor zVAD-fmk (10μm). An equal volume of DMSO was added to the media as vehicle control.
(D) Similar to (C) using live cell imaging, WT, Slc46a2−/− and Ripk2−/− keratinocyte permeabilization was measured with a Sytox dye uptake assay over a 24 h time course following 30 μM iE-DAP treatment. Vehicle control was media alone.
(E) Immunoprecipitation-immunoblot of lysates from iE-DAP challenged keratinocytes probed for phospho- RIPK2, total RIPK2, and β-actin from whole cell lysates.
(F) Neutrophil recruitment to pinnae 3 h after intradermal injection of 10μl of 30 μM iE-DAP in WT, Slc46a2−/−, Casp1−/− and Gasdmd−/−, shown as a percent of CD45+ cells.
(G) Similar to (C), Sytox uptake was measured in WT, Slc46a2−/−, Nod1−/−, or Casp1−/− keratinocytes for 24 hours after challenge with 30 μM iE-DAP. Vehicle control was media alone.
(H) Sytox dye uptake by keratinocytes over 24h following 30 μM iE-DAP challenge in WT, Slc46a2−/−, and Gasdmd−/−. WT keratinocytes were also treated with the Gasdermin inhibitor DMF (50 μM). An equal volume of DMSO was added to the media as vehicle control.
Genotypes are indicated on all panels. Panels A displays data from 3 independent measurements and is representative of at least 3 separate trials, error bars represent standard error of the mean (SEM) and analyzed by two-way ANOVA and Tukey’s multiple comparisons tests. For panels B & F each dot represents an individual mouse, data pooled from 2 separate trials, analyzed by two-way ANOVA and Tukey’s multiple comparisons tests. In panels C, D, G, & H each data point represents the mean with SEM of at least three independent measurements, and are representative of 2 to 4 separate trials, analyzed by one-way ANOVA and Tukey’s multiple comparisons tests. Panel E shows a representative image from three independent experiments. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant. See also Figures S2.
Alarmins like IL-1α are released by damaged or dying cells, triggering inflammatory responses in nearby cells.48 To monitor cell damage or death, we next examined DAP-muropeptide-triggered keratinocyte permeabilization, quantifying the uptake of the membrane-impermeable Sytox in a live cell imaging assay.49,50 WT keratinocytes were markedly permeabilized in response to DAP-muropeptide, while Slc46a2- and Nod1-deficient keratinocytes were largely protected (Fig. 2C). Given the role of RIPK2 in the NOD1 pathway, we also probed the role of this kinase in iE-DAP triggered cell permeabilization. Keratinocytes from RIPK2 mutant mice were also protected from iE-DAP-induced cell permeabilization, similar to Slc46a2−/− keratinocytes (Fig. 2D). Consistent with these results, iE-DAP stimulation triggered RIPK2 phosphorylation in WT but not Slc46a2−/− keratinocytes (Figure 2E). Thus, the iE-DAP triggered pathway in keratinocytes likely diverges from the canonical NOD1 pathway downstream of RIPK2, leading to cell permeabilization and IL-1α release but not cytokine gene induction.
In order to further probe this iE-DAP-triggered cell permeabilization, WT keratinocytes were treated with the pan-caspase inhibitor zVAD-FMK which prevented DAP-muropeptide-triggered permeabilization of WT keratinocytes (Fig. 2C). The role of caspases and pyroptosis in this process was then analyzed with Casp1- and Gsdmd-deficient mice that were challenged with ID iE-DAP injection, where they exhibited a strong defect in neutrophil recruitment, similar to Slc46a2−/− animals (Fig. 2F). Keratinocytes from these knockouts also showed significantly decreased permeabilization in response to iE-DAP (Fig. 2G & H). A similar phenotype was also observed following treatment with dimethyl fumarate (DMF), a potent inhibitor of gasdermin pore formation (Fig. 2H).51 Altogether, these data show that DAP-muropeptide stimulation of primary keratinocytes drives cell permeabilization through a pathway requiring Slc46a2, Nod1 and RIPK2, involving a caspase-1 and gasdermin D pyroptosis-like process, leading to the release of IL-1α.
Slc46a2 delivers DAP-muropeptide into keratinocytes
To directly evaluate the role of SLC46A2 in DAP-muropeptide transport, we used two complementary chemical biological approaches. First, a modified, biologically active alkyne derivative of iE-DAP was synthesized and utilized to visualize uptake of this muropeptide into primary keratinocytes using “click-chemistry.”52 After 60 minutes, iE-DAP was clearly detected within both WT and Nod1−/− cells, but not in Slc46a2-deficient keratinocytes, by confocal microscopy or in cell lysates (Fig. 3A and S3A, respectively). By contrast, Slc46a2 did not affect the intracellular delivery of click-modified MDP, which was instead dependent on Slc46a3 (Fig. S3A & B). In an orthogonal approach, fluorescently labeled lipid nanoparticles (NP) were loaded with iE-DAP and used to deliver the iE-DAP into keratinocytes, which bypassed the requirement for Slc46a2, but not Nod1, in inducing membrane permeability (Fig. 3B and S3C). Together, these results, visualizing with click-modified muropeptides or delivering muropeptides with lipid nanoparticles, are consistent with data in Figure 1 that implicated SLC46A2 in the NOD1 pathway and SLC46A3 for NOD2 responses. Moreover, they directly demonstrate a role for the SLC46A2 and A3 transporters in transporting DAP-type or MDP muropeptides, respectively, to the cytosol.
Fig. 3. DAP-muropeptide transport requires Slc46a2 and is blocked by methotrexate.
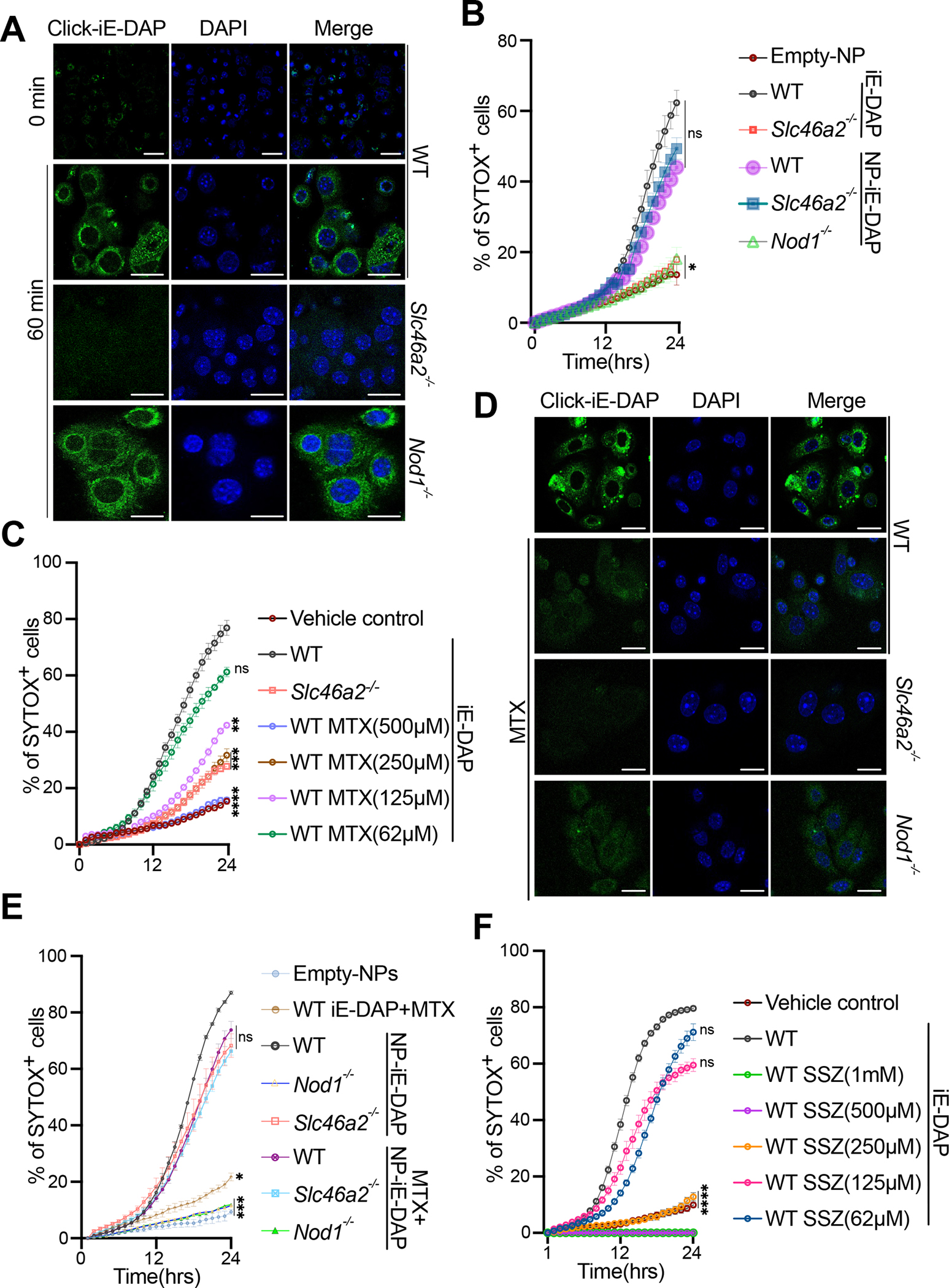
(A) Confocal images of WT, Slc46a2−/− or Nod1−/− keratinocytes after 1 h treatment with 30 μM “click-iE-DAP” or left untreated. Cells were fixed and then visualized with click-reacted Calflour488-azide. See also Figure S3A and S3B.
(B) Using live cell imaging, WT, Slc46a2−/− and Nod1−/− keratinocyte permeabilization was measured with a Sytox dye uptake assay over a 24 h time course following challenge with either 30 μM naked iE-DAP or lipid nanoparticles (NP) loaded with iE-DAP. See also Figure S3C.
(C) Similar to (B), Sytox uptake was measured in WT keratinocytes stimulated with 30 μM iE-DAP plus increasing concentrations of MTX, or vehicle, for 24hrs time course. Slc46a2−/− keratinocytes, without MTX, were included as an additional control. An equal volume of DMSO was added to the media as vehicle control. See also Figure S3D.
(D) Confocal images of WT, Slc46a2−/− or Nod1−/− keratinocytes after 1 h challenge with 30 μM “click-iE-DAP” or with 30μM “click-iE DAP” plus 250 μM methotrexate (MTX). Cells were fixed and then visualized with click-reacted Calflour488-azide. See also Figure S3E.
(E) Similar to (B), Sytox dye uptake using WT, Slc46a2−/− and Nod1−/−keratinocytes over 24h following stimulation with naked iE-DAP plus 250 μM MTX, or lipid nanoparticles (NP) loaded with iE-DAP, with or without 250μM MTX. See also Figure S3F.
(F) Similar to (C), Sytox uptake was measured in WT keratinocytes treated with increasing concentrations of SSZ and challenged with 30 μM iE-DAP. See also Figure S3G.
Genotypes are indicated on all panels. In panels B, C, E, and F each data point shown as a mean plus SEM of at least three independent results, representative of at least 2 to 4 separate trials, analyzed by one-way ANOVA and Tukey’s multiple comparisons tests. Panels A and D are representative images from at least three independent experiments. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant. The scale bar is 10μM. See also Figure S3 and Figure S4 for compound structures.
Common anti-inflammatory drugs inhibit SLC46A2-mediated delivery of muropeptides
SLC46A2 is a paralog of the proton-coupled folate transporter SLC46A1 (~30% identity), which suggests folates and anti-folates could be common cargo for all SLC46 family transporters. 36,53 Methotrexate (MTX) is a potent anti-inflammatory drug commonly used to treat psoriasis and rheumatoid arthritis, although its mechanism(s) of action as an anti-inflammatory are not completely clear. 37–39 Anti-folates like MTX are known to compete with folate for transport through SLC46A1.36 Given the similarity between SLC46A1 and SLC46A2, we hypothesized that MTX may also compete with DAP-muropeptides for docking to and transport by SLC46A2. To test this idea, increasing concentrations of MTX were added to the keratinocyte-based iE-DAP cell permeabilization assays. In a dose-dependent manner, MTX inhibited DAP-muropeptide triggered keratinocyte permeabilization and IL-1α release, similar to the Slc46a2-deficient keratinocytes (Fig. 3C and S3D). Further, MTX blocked the cytosolic accumulation of “click”-iE-DAP, similar to competition with unlabeled iE-DAP, as well as the cytosolic delivery of “click” MDP (Fig. 3D and S3E) but failed to interfere with NP-mediated iE-DAP delivery, assayed by cell permeabilization (Fig. 3E and S3F). Together these results demonstrate that MTX inhibits SLC46A2-mediated DAP-muropeptide transport as well as SLC46A3-mediated MDP delivery in mouse keratinocytes.
Sulfasalazine (SSZ), another common anti-inflammatory drug, has recently been reported to block SLC46A2-mediated cGAMP transport in human monocytes.54 Therefore, we have tested the effect of SSZ on iE-DAP responses in keratinocytes. High concentrations of SSZ (1 mM or 500 μM), similar to what was reported in the cGAMP study, caused Sytox staining to remain undetectable, below that of untreated cells, perhaps indicative of nonspecific inhibition of this dye (Fig 3F). However, lower concentrations of SSZ significantly blocked the cell permeabilization and IL-1α release in WT keratinocytes in a dose-dependent manner (Fig. 3F, S3G). Together, these findings suggest SSZ may also interfere with SLC46A2-mediated iE-DAP transport, similar to MTX.
Overexpression of SLC46s is sufficient for muropeptide delivery
To further characterize SLC46-mediated DAP-muropeptide transport, we used a gain-of-function approach in transfected HEK293T cells, similar to our previous study.24 Cells were transfected with plasmids expressing Slc46a1, Slc46a2, or Slc46a3, challenged with click-iE-DAP, and visualized with reaction to CalFluor 488-Azide. Slc46a2 expression resulted in robust cellular transport of iE-DAP, while Slc46a3 expression provided observable iE-DAP uptake that is significantly less robust than Slc46a2 supported uptake (Fig 4A & quantified in 4B). By contrast, Slc46a1 expression did not support the import of iE-DAP above what was observed in empty vector (EV) control transfected cells.
Fig. 4. In SLC46-transfected cells, fand anti-folates inhibit iE-DAP transport to varying degrees.
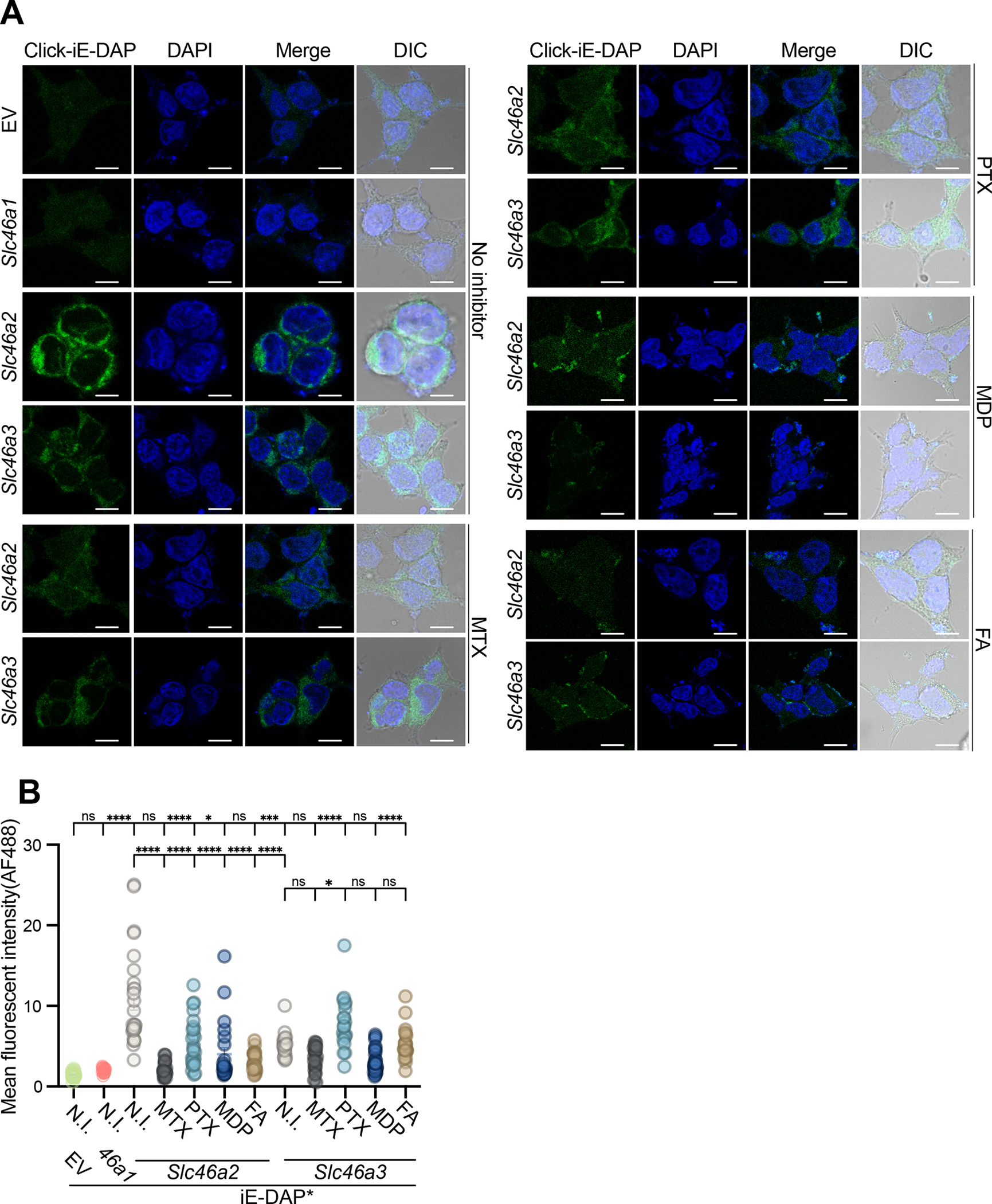
(A) HEK293T cells were transiently transfected with empty vector (EV), Slc46a1, Slc46a2, or Slc46a3 expression plasmids, and challenged with 30 μM click-iE-DAP with or without folates/antifolates. 1 hr after the challenge, cells were fixed, and then iE-DAP entry was visualized by confocal microscopy following reaction with CalFluor488-Azide. SLC46A2 more effectively transported iE-DAP, compared to SLC46A3, while SLC46A1 was inactive. MTX (250 μM) blocked iE-DAP transport through SLC46A2 or SLC46A3 whereas another antifolate, pemetrexed (PTX, 250 μM) and folic acid (FA) (250 μM) interfered with SLC46A2 mediated transport while MDP (30 μM) more potently inhibited SLC46A3 transport.
(B) Mean fluorescence intensity computed from images similar to panel (A) across three independent replicates. Each dot in the graph represents fluorescence from a cell. N.I. indicates no inhibitor.
In panel A, representative images are shown from at least three independent experiments. Panel B uses a two-way ANOVA test and Tukey’s multiple comparisons test to determine significance. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant. The scale bar is 10μM. For inhibitor structures, see Figure S4.
Using this gain-of-function assay, the effect of folates and anti-folates on muropeptide transport was further analyzed. MTX significantly blocked the iE-DAP import in Slc46a2-expressing cells, to near background fluorescence, and also prevented the modest iE-DAP import observed in Slc46a3-expressing cells (Fig. 4A and B). Pemetrexed (PTX), another anti-folate used in cancer therapy, significantly but incompletely interfered with SLC46A2-supported iE-DAP transport but did not affect (even increased) SLC46A3-mediated iE-DAP uptake. Similarly, folic acid blocked iE-DAP transport by SLC46A2, but not by SLC46A3, while MDP competed with SLC46A3-mediated iE-DAP transport more robustly than SLC46A2-mediated transport, consistent with the selectivity of SLC46A3 for MDP. Figure S4 provides the chemical structures of the compounds used in these studies and throughout. These results argue that SLC46A2 and SLC46A3 prefer as cargo iE-DAP or MDP, respectively. However, this selectivity for distinct muropeptides is not completely strict, as revealed in these over-expression transfection-based assays. Similarly, different folates and antifolates competed for muropeptide transport with distinct selectivity, with MTX effectively inhibiting both SLC46A2- and A3-mediated transport while PTX preferentially interfered with transport by SLC46A2, but not SLC46A3. Likewise, folic acid also preferentially inhibited SLC46A2-mediated transport.
Slc46a2 and Nod1 are required for psoriatic inflammation in a mouse model
The above results show that SLC46A2 is inhibited by folates/anti-folates, like MTX, as well as by SSZ. As shown in Figure 1, Slc46a2 is necessary for an inflammatory response in the skin to C. accolens, while Corynebacterium spp. are increased in psoriatic lesions33,55–60, and C. accolens is known to exacerbate psoriasis-like phenotypes in a mouse model.32 In fact, the imiquimod (IMQ) model of psoriatic inflammation requires an intact microbiome.34,35,61 Therefore, we used the IMQ model to probe the role of Slc46a2 and Nod1 in psoriatic-like inflammation. With a 7-day course of topical IMQ application, WT mice displayed the expected psoriatic-like inflammation, assayed by enhanced ear thickness and H&E histology, while Slc46a2- and Nod1-deficient mice were markedly resistant (Fig. 5A, B, and S5A). Application of IMQ for only 3 days followed by topical application C. accolens similarly drove psoriatic-like inflammation in WT mice while both Slc46a2- and Nod1-deficient animals presented reduced inflammation (Fig. 5C and S5B, C). Association of C. accolens was confirmed by counting colony forming unit (CFU) 24 h after bacterial application to otherwise untreated mouse pinnae skin (Fig S5D). Further, topical application of MTX reduced the psoriatic-like inflammation in WT skin, as assayed by ear thickness and histology, while MTX had no observable effect on the residual inflammation in Slc46a2−/− and Nod1−/− skin (Fig. 5D and S5E, F). These results show that SLC46A2 is required for the progression of psoriatic inflammation in the IMQ mouse model, by transporting bacterial-derived DAP-muropeptides and activating NOD1 signaling, while MTX suppresses this psoriatic inflammation by interfering with the SLC46A2-NOD1 response.
Fig. 5. Slc46a2−/− and Nod1−/− mice are resistant to IMQ-induced psoriatic inflammation.
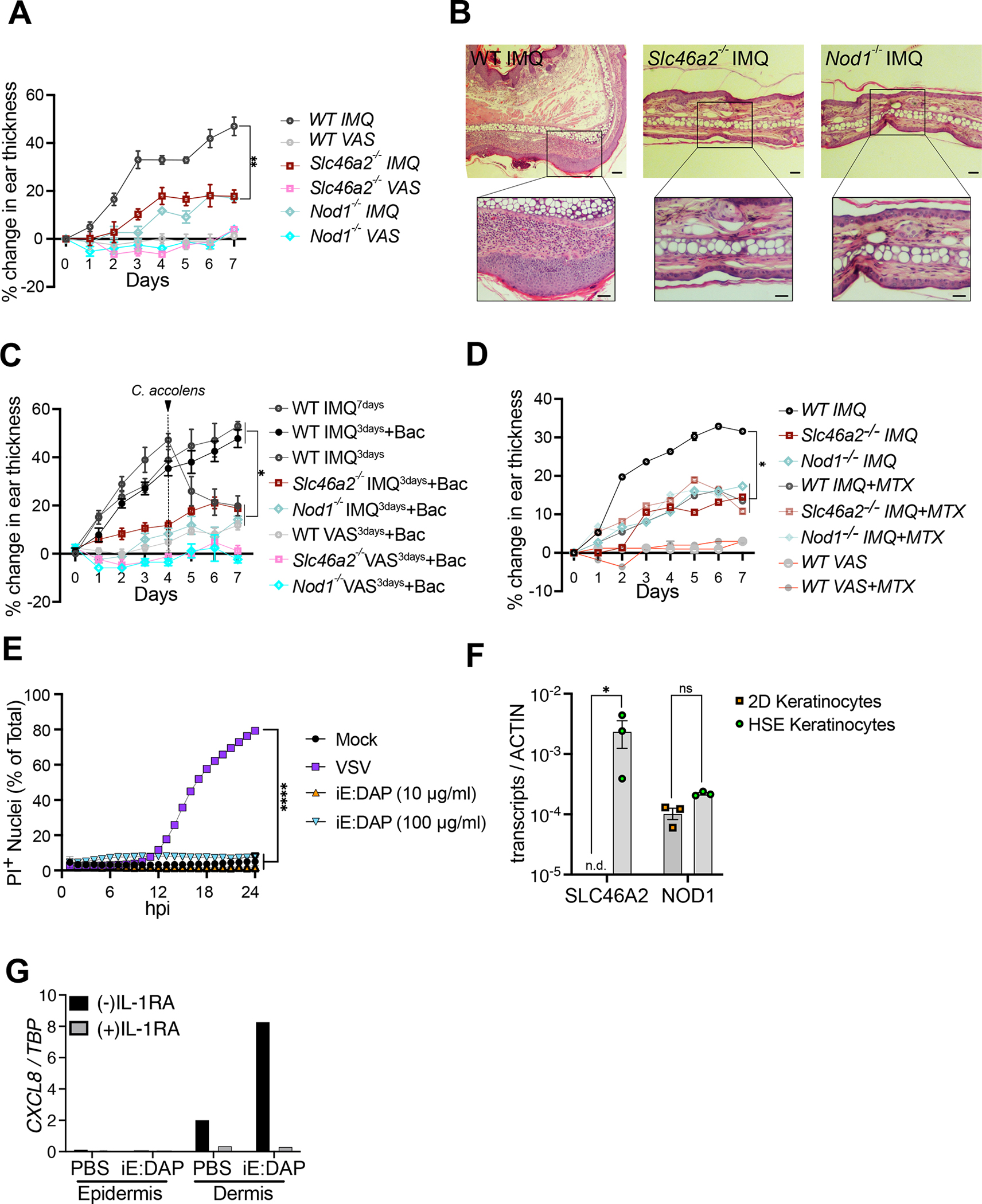
(A) 5% imiquimod (IMQ) was topically applied with daily for 7 days to pinnae to induce psoriasis and ear inflammation was quantified daily by caliper measurement. Contralateral pinnae were treated with Vaseline (VAS) as vehicle control. The mean thickness of the pinnae and SEM are plotted.
(B) H&E-stained histology images of IMQ-treated pinnae on day 7 from the experiment in (A). Genotypes are indicated on all panels. Vaseline control treated pinnae shown in Figure S5A.
(C) IMQ was applied for only 3 days, and then pinnae were treated daily for 3 days with topical application of live C. accolens (107 CFU/ml), except for controls with either a full 7 days or just 3 days of IMQ treatment. Contralateral pinnae were treated with VAS as vehicle control or VAS with C. accolens. The mean thickness and SEM of the pinnae is plotted. See also Figure S5B–S5D.
(D) is similar to (A) except IMQ was applied daily along with 50 μl of 250 μM MTX. The mean thickness and SEM of the pinnae is plotted. See also Figure S5E and S5F.
(E) Propidium Iodide (PI) uptake assay using primary human foreskin keratinocytes challenged with indicated iE-DAP doses or VSV infection (MOI 10) as a positive control. iE-DAP treatment did not induce cell permeabilization in human keratinocytes.
(F) Expression analysis of SLC46A2 and NOD1 in human keratinocytes grown in 2D culture and 3D skin organoids (HSE).
(G) Induction of CXCL8 in skin organoid epidermal and dermal layers after PBS or iE-DAP challenge of the epidermis in the presence and absence of IL-1 receptor inhibitor (IL-1RA). High expression of CXCL8 was observed in the dermis following the iE-DAP challenge of the epidermis, and this was blocked by IL-1RA. See also Figure S5G and S5H.
In panels A, C, D, and E each data point displays the mean and SEM from 5 animals in each group, data representative of 2 to 4 separate trials. Significance was analyzed by one-way ANOVA with Tukey’s multiple comparisons tests. Panels B and G are representatives of three independent experimental results. For panel F, each dot represents an independent experiment result, with bar displaying mean and SEM indicated, analyzed by a two-way ANOVA and Tukey’s multiple comparisons tests. **** P < 0.0001; *** P < 0.001; ** P < 0.01; * P < 0.05; ns, not significant. The scale bar is 100μM. See also Figure S5.
Human keratinocytes in the context of a 3D squamous epithelium respond to iE-DAP
To determine if iE-DAP-triggers inflammatory responses occur in human skin, we first analyzed primary foreskin-derived human keratinocytes. For this experiment, keratinocytes were infected with vesicular stomatitis virus (VSV) as a positive control, as it is known to induce cell permeabilization via a GSDME-dependent pyroptotic pathway.50,62 By contrast, iE-DAP treatment failed to induce membrane permeabilization in these cells (Fig. 5E). Consistent with this lack of response, primary human keratinocytes, in standard tissue culture conditions, did not express SLC46A2 (Fig 5F). When these same keratinocytes are cultured at an air-liquid interface on a bed of collagen-embedded dermal fibroblasts, they differentiate into a multilayered squamous epithelium (known as Human Skin Equivalents (HSE)), with many similarities to human skin.63 In the context of the HSE, differentiated human keratinocytes robustly expressed SLC46A2 and these organoids responded to iE-DAP. Upon treatment of the HSE epidermis, but not the dermis, with 30 μM iE-DAP for 24 h, we observed the induction of CXCL8 in underlying dermal fibroblasts (Fig. 5G and S5G). Moreover, this response was prevented by concomitant treatment with recombinant IL-1R antagonist (IL-1Ra), strongly implicating IL-1 release by keratinocytes in driving CXCL8 induction in HSE dermal fibroblasts. Likewise, in the mouse system, supernatants from iE-DAP-stimulated primary keratinocytes triggered, via Slc46a2- and Nod1-dependent signaling, CXCL1 (KC) production in dermal fibroblasts (Fig. S5H). These findings in human skin organoids argue that in human differentiating keratinocytes, like their mouse counterpart, transport iE-DAP through SLC46A2 to stimulate NOD1 and induce IL-1 signaling.
Discussion
While it has been clear for many years that some cell types import muropeptides and activate the cytosolic innate immune receptors NOD1 or NOD2,64 the underlying cellular and molecular mechanisms that mediate the import of these molecules have remained unclear. Initially, the SLC15 family of oligopeptide transporters was implicated in this process 16–20, but they have never been demonstrated to bind or transport muropeptides while more recent studies instead have shown SLC15A4 functions as a scaffold involved in IRF5 activation downstream of TLR as well as NOD activation.20,22 By contrast, our initial work in Drosophila implicated SLC46s in this process,24 and here we show that SLC46A2 has the properties of a transporter selective for delivering DAP-type muropeptides for NOD1 activation in murine tissues, while SLC46A3 is critical for MDP delivery and NOD2 activation. Using in vivo approaches in two different tissues, as well as ex vivo studies with primary mouse keratinocytes, Slc46a2 was required for the response to multiple NOD1 activating DAP-muropeptides, while Slc46a3 was critical for the response to the NOD2 activating muropeptide MDP. Moreover, two orthogonal approaches were used to demonstrate that intracellular delivery of DAP-muropeptide requires Slc46a2. Intracellular iE-DAP, directly detected via click chemistry dye labeling, required Slc46a2 but not Nod1, while intracellular MDP detection similarly relied on Slc46a3. When packaged in a lipo-nanoparticle for direct intracellular delivery, iE-DAP activation of NOD1 no longer required Slc46a2. Together, these immunological and cell biological data strongly argue for the direct transport of DAP-muropeptides by SLC46A2 and implicate SLC46A3 in the selective delivery of MDP. Primary human keratinocytes, in standard tissue culture conditions, do not express SLC46A2 and do not respond to iE-DAP, while after differentiating into a squamous epithelium in the HSE organoids, SLC46A2 is expressed, and the epidermis responds to DAP-muropeptide.
The data presented here show that Slc46a2 is essential for Nod1 responses, while the Slc46a3 phenotypes with MDP and NOD2 activation in the skin/keratinocytes are robust but partial. For example, with intradermal challenge, neutrophil recruitment following iE-DAP challenge is near background in the Slc46a2 mutant, similar to Nod1, while the response to MDP in Slc46a3 is significantly reduced but is not as low as the Nod2 mutants. Likewise, using click-MDP, a reduction in cellular MDP delivery was observed in the Slc46a3 mutant, but it was not reduced to the background signal in these assays; in fact, MTX treatment was a more complete inhibitor of MDP uptake. From these data, we speculate that SLC46A2, while selective for DAP-muropeptides, may to a lesser degree also transport MDP. This lower MDP transporting activity could be immunologically relevant in the tissues where Slc46a2 is robustly expressed, like keratinocytes, but irrelevant in other cell types. Likewise, in the HEK293T cell transfection assays, overexpression of Slc46a3 can support the modest import of iE-DAP, suggesting selectivity, but not complete specificity, for distinct muropeptides by these two SLC46 transporters. Analysis of a double mutant, Slc46a2−/−, Slc46a3−/− strain, is required to address the possible redundancy of these transporters in muropeptide delivery. Alternatively, the SLC15 family could be involved, to a lesser degree, in MDP transport. Future studies will address these possibilities and the potential interplay between transporters.
In both humans and mice, Slc46a2 is expressed in only a limited set of tissues, including the epidermis.43,44 By contrast, Slc46a3 is expressed widely including in the skin, many myeloid cells, and the gut epithelium.65 While the role of Slc46a3 in these other tissues awaits future study, the limited expression pattern of Slc46a2 led us to explore, in more detail, the role of SLC46A2 and NOD1-dependent responses in the skin. We found a robust neutrophilic response to intradermal DAP-muropeptide challenge that required Slc46a2 and Nod1. Similarly, we also found that murine skin, once the outer waxy stratum corneum was removed, responded to topical application of either iE-DAP or the DAP-producing commensal C. accolens through Slc46a2 and Nod1. Likewise, primary mouse keratinocytes ex vivo responded to DAP-muropeptides with cell permeabilization and the release of active IL-1α. Similar to the in vivo neutrophilic response, the cell permeabilization pathway in keratinocytes involved Casp1 and Gsdmd in addition to Slc46a2 and Nod1. Together these results demonstrate that skin keratinocytes respond to DAP-muropeptides via intracellular delivery by SL46A2 and activation of NOD1, which in turn drives IL-1α release via a pathway that involves caspase-1 and gasdermin D cell permeabilization, and perhaps cell death. The rapidity of the response in vivo suggests this is not a transcription-dependent response, although the response is slower in isolated keratinocytes. Future studies are necessary to more fully characterize the molecular mechanisms linking NOD1 and RIPK2, to this pyroptosis-like response.
Given the established link between C. accolens and psoriasis in the mouse model,32 and the increased Corynebacterium spp. in psoriasis lesions,33,55–60 the role of the SLC46A2-NOD1 pathway in psoriatic inflammation was also examined in the IMQ mouse model. Animals deficient for either Slc46a2 or Nod1 were resistant to psoriatic inflammation and unresponsive to C. accolens-triggered pathology. Given the previous work linking C. accolens to the activation of IL-17-producing γδT cells in the skin32, it will be exciting to learn if the SLC46A2-NOD1-pathway in the skin also drives this IL-17-producing γδ response, in addition to the strong neutrophilic infiltration. Corynebacterium have also been linked to IL-17-producing γδ T-cells responses in the eye66, and it will also be exciting to determine if NOD1 and SLC46A2 are also involved in that model.
Immune-mediated inflammatory diseases, like psoriasis, are often treated with the first-line drug methotrexate. MTX is a historically complicated drug, originally developed as an antiproliferative due to its interference with folate-dependent enzymes required for nucleotide biosynthesis and DNA replication. Although more than 50 years old, MTX is still used as a cancer therapy. Subsequently, MTX was found to be effective in psoriasis, rheumatoid arthritis, and other immune-mediated inflammatory diseases, but at much lower doses that do not impact cell proliferation.37,39,67 The mechanisms of action of MTX as an anti-inflammatory drug are controversial. The leading model argues that MTX functions as a traditional anti-folate interfering with the enzyme AICAR transformylase, leading to increased intracellular AICAR and eventually increased extracellular Adenosine, a known anti-inflammatory molecule that acts via adenosine receptors, such as A2A.39 In addition, MTX has been argued to interfere with NF-κB activation through unknown mechanisms. Here we present another mechanism of action for the anti-inflammatory activity of MTX that does not involve interfering with folate-dependent enzymes. Instead, the data here show that MTX competes with muropeptides for delivery into the cytosol, phenocopying Slc46a2-deficiency, in three separate assays, ex vivo in keratinocytes assays as well as with an in vivo model of psoriatic inflammation. In vivo, MTX application in the Slc46a2 and Nod1 mutants had no effect on the residual inflammation still observed in the psoriasis model, while it strongly suppressed psoriatic-like inflammation in WT mice, exhibiting reduced inflammation similar to that observed in the Slc46a2 or Nod1 mutants. In addition, using a gain-of-function approach in a transfected cell line, MTX potently inhibited iE-DAP import by both SLC46A2 and SLC46A3, while the anti-folate PTX and folic acid more selectively inhibited SLC46A2. Consistent with the selectivity for MDP, SLC46A3-mediated transport, but not SLC46A2-mediated transport, was competed with the excess MDP. Together, these in vivo, ex vivo, and in vitro findings argue that MTX competes with muropeptides as cargo for the transporters SLC46A2 and SLC46A3, and thereby directly interferes with two central inflammatory responses, the NOD1 and NOD2 pathways.
In the future, biophysical approaches will be necessary to probe the cargo binding activity and selectivity of SLC46A2 and SLC46A3, as well as the related transporter SLC46A1 (the established proton-coupled folate/anti-folate transporter); all three members of the SLC46 family share approximately 30% amino acid identity. These approaches can also be used to determine if the glutamic acid moiety shared between muropeptides and folates underlie their connectivity as cargo for the SLC46 transporters. Future work will also focus on precisely mapping the anatomical location(s) of these responses within the epidermis and probe the connection between SLC46A2-NOD1-mediated response in keratinocytes to activation of IL-17 producing γδ T cells.32,68
Through a combination of immunology, chemical biology, and biomedical engineering approaches, we demonstrate that SLC46A2 and SLC46A3 function to deliver DAP-muropeptides or MDP, respectively, to the cytosol of keratinocytes, driving a robust IL-1α-dependent inflammatory response and neutrophil recruitment in mouse skin. This response is critical for psoriatic-like inflammation in a mouse model, while human skin organoids respond similarly although the connection to psoriasis awaits future analysis. Additionally, the anti-inflammatory drug MTX phenocopies Slc46a2 deficiency in vivo, ex vivo in primary mouse keratinocytes, and in transfected cells, arguing that SLC46A2 is a direct anti-inflammatory target of this drug. More generally, these findings identify disease mechanisms and therapeutic targets that could be applicable to a broad number of inflammatory conditions and highlight the role of SLC46 family in host-microbiome interactions.
Limitations of the study
As mentioned above, the possibility of redundancy between Slc46a2 and Slc46a3, especially in regard to MDP delivery in tissues with high levels of Slc46a2 expression, requires further study. Similarly, the connection between SLC46 muropeptide delivery and role of SLC15s in these pathways was not examined here. The response kinetics to DAP-muropeptides was discrepant between the in vivo and ex vivo assays. In vivo skin challenge triggered a rapid neutrophilic influx, readily detectable as early as 1 hr post injection, while in vitro the response in primary keratinocytes was noticeably slower; muropeptide-triggered keratinocyte permeabilization was detectable only starting at ~12 hr. While the underlying cause of this differential timing is unknown, we speculate that the involvement of a full of array of differentiating keratinocytes, immune cells, and underlying dermal fibroblasts in the intact tissue supports the rapid in vivo response, while in vitro the undifferentiated primary keratinocyte in isolation may not be optimal for responding to NOD1 activation. Future work is necessary to define which epidermal layer(s) express Slc46a2 and respond via NOD1 activation. Another limitation is the partial phenotype observed in cell permeabilization assays with Caspase-1 and Gasdermin-D mutant keratinocytes after DAP-muropeptide challenge, compared to Nod1 or Slc46a2 mutant keratinocytes, which opens the possibility of involvement of additional cell permeabilization or cell death factors, such as Caspase-3 or Gasdermin-E, in SLC46A2-NOD1-mediated keratinocyte pyroptosis. The molecular connection between NOD1 and Caspase-1 is also unknown; both proteins include Death Domains and they could interact directly, perhaps in a multi-subunit complex along with RIP2K (also a Death Domain protein). However, these Death Domains, when examined in isolation do not interact in a binary fashion.69 Future studies are required to address these limitations and untangle these complex immunological signaling responses.
RESOURCE AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Neal Silverman (neal.silverman@umassmed.edu)
Materials availability
Reagents generated or used in this study are available on request from the lead contact with a completed Materials Transfer Agreement (MTA). Click-modified muropeptides are available from Catherine Leimkuhler Grimes at the University of Delaware, via an MTA; however, these chemical reagents are available in very limited supply. Information on reagents used in this study is available in the key resources table. Mouse strains and other reagents will be shared with a simple MTA.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| RIPK2 antibody | Invitrogen, Carlsbad, CA | cat# MA5–17221 |
| pRIPK2 antibody | Sigma | cat# SAB4504642 |
| Phospho p65 antibody | Cell Signaling | cat# 3033 |
| Total p65 antibody | Cell Signaling | cat# 8242 |
| Beta-actin-HPR antibody | Sigma | cat# A3854 |
| Mouse IL-1α Neutralizing antibody - Monoclonal Mouse IgG1 (6H7) | invivogen | cat# mabg-mil1a |
| Mouse IgG1 isotype control | invivogen | cat# mabg1-ctrlm |
| Anti-mouse-HRP | GE | cat# NA931V |
| Anti-rabbit-HRP | Biorad | cat# 1705046 |
| PE anti-mouse Ly-6G/Ly-6C (Gr-1) Antibody (RB6–8C5) | Biolegend | cat# 108408 |
| APC anti-mouse/human CD11b Antibody (M1/70) | Biolegend | cat# 101212 |
| Alexa Fluor® 700 anti-mouse CD45 Antibody (30-F11) | Biolegend | cat# 103128 |
| PE/Cyanine7 anti-mouse F4/80 Antibody | Biolegend | cat# 123114 |
| Bacterial and virus strains | ||
| WT VSV | Whelan et al, 1995(62) | N/A |
| Corynebacterium accolens | ATCC via J. Kang | strain# 49725 |
| Biological samples | ||
| Normal human foreskin | UMass Memorial Hospital | N/A |
| NIH/3T3 | ATCC | ATCC# CRL-1658 |
| Chemicals, peptides, and recombinant proteins | ||
| KC basal medium (EpiLife) | Invitrogen, Carlsbad, CA | cat# MEPICF500 |
| Defined Growth Supplement (dGS) | Invitrogen, Carlsbad, CA | cat# S0125 |
| Dispase powder | Invitrogen, Carlsbad, CA | cat# 17105041 |
| Coating Matrix | Invitrogen, Carlsbad, CA | cat# R011K |
| TrypLE | Invitrogen, Carlsbad, CA | cat# 12604–013 |
| CCK-8 cell viability Kit | Dojindo Molecular Technologies | cat# CK04–11 |
| RPMI-1640 | HyClone | cat# SH30027.01 |
| Fetal Calf Serum | HyClone | cat# SV30160.03 |
| 2-mercaptoethanol | Sigma-Aldrich | cat# M3148 |
| Asparagine | Sigma-Aldrich | cat# A4159 |
| Glutamine | Sigma-Aldrich | cat# G8540 |
| Penicillin/Streptomycin | HyClone | cat# SV30010 |
| Ethanol | Merck Millipore | cat# 107017 |
| Collagenase D | Roche Diagnostics | cat# 11088866001 |
| Pronase protease | Merck Millipore | cat# 53702 |
| GeneJuice® Transfection Reagent | Millipore | cat# 70967 |
| Tris buffer (pH 8) | 1st BASE | cat# 1415 |
| 0.5M EDTA (pH 8) | 1st BASE | cat# BUF-1053 |
| 10X Phosphate Buffered Saline (PBS) | 1st BASE | cat# BUF-2040 |
| Trypsin-EDTA solution 10X | Sigma-Aldrich | cat# 59418C |
| Amphotericin B | Sigma-Aldrich | cat# A2492 |
| Brewer thioglycollate medium | Remel | cat# R064702 |
| DMEM F12 media | Thermo Scientific | cat# 12430112 |
| Recombinant Murine M-CSF | Peprotech | cat# 315–02 |
| DMEM (No Calcium) | ThermoFisher | cat# 21068028 |
| Ham’s F-12 Nutrient Mix | ThermoFisher | cat# 11765054 |
| Bovine Calf Serum | HyClone | SH30073.03 |
| L-glutamine | ThermoFisher | cat# 25030081 |
| Adenine | Sigma | cat# A8626–1G |
| Hydrocortisone | Sigma | cat# H-4881 |
| Triiodothyronine | Sigma | cat# T5516 |
| ITS-X | Invitrogen | cat# 51500–056 |
| O-phosphorylethanolamine | Sigma | cat# p0503 |
| Progesterone | Sigma | cat# P8783 |
| Epidermal Growth Factor | Peprotech | cat# AF-100–15 |
| Calcium Chloride | Sigma | cat# C7902 |
| Sodium Ascorbate | Sigma | cat# A4034 |
| Chelex 100 | Sigma | cat# C7901 |
| T-PER | Thermo fisher | cat# 78510 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail (100X) | Thermo fisher | cat# 78440 |
| PVDF membrane | Sigma-milipore | cat# IPVH00010 |
| Protein-A agarose | Thermo fisher | cat# 20333 |
| SDS-gel | biorad | cat# 3450418 |
| Sytox | Thermo fisher | cat# S34859 |
| 1.5M Tris PH 8.8 | Biorad | cat# 1610798 |
| 0.5M Tris PH 6.8 | Biorad | cat# 1610799 |
| TRIzol™ Reagent | Invitrogen | cat# 15596026 |
| Chloroform | Fisher Scientific | cat# C607SK-1 |
| Isopropanol | Fisher Scientific | cat# BP2618500 |
| Ethanol | Fisher Scientific | cat# BP28184 |
| Water | Fisher Scientific | cat# W9–1 |
| iE DAP | Invivogen | cat# tlrl-dap |
| Tri-DAP | Invivogen | cat# tlrl-tdap |
| C12-iE-DAP | Invivogen | cat# tlrl-c12dap |
| MDP | Invivogen | cat# tlrl-mdp |
| TCT | T. Kaneko et al., 2004(45) | N/A |
| Alkyne-iE-DAP |
R. Bharadwaj et al. 2023 (81) | N/A |
| Alkyne-MDP | R. Bharadwaj et al. 2023 (81) | N/A |
| 1X PBS | Sigma | cat# D8537 |
| Protenase K | Sigma | cat# P2308 |
| hoechst | Thermo fisher | cat# 62249 |
| Imiquimod cream | perrigo | NDC#45802–368-62 |
| Histopaque 1119 | Sigma-Aldrich | cat# 11191 |
| Histopaque 1077 | Sigma-Aldrich | cat# 10771 |
| laemmli sample buffer | BioRad | cat# 161–0737 |
| Copper Sulfate | Sigma | cat# PHR1477 |
| BTTAA | Click Chemistry Tools | cat# 1236 |
| sodium ascorbate | Sigma | cat# A4034 |
| CalFluor 488 Azide | Click Chemistry Tools | cat# 1369 |
| Recombinant human IL-1Ra | Biolegend | cat# 553902 |
| Propidium Iodide | ThermoFisher | cat# P1304MP |
| Monarch Total RNA Miniprep Kit | NEB | cat# T2010S |
| Taqman RNA-to-CT 1-Step Kit | ThermoFisher | cat# 4392653 |
| 18:1 (Δ9-Cis) PC (DOPC) | Avanti Polar Lipids | cat# 850375 |
| 18:0 PC (DSPC) | Avanti Polar Lipids | cat# 850365 |
| 18:1 (Δ9-Cis) PG (DOPG) | Avanti Polar Lipids | cat# 840475 |
| Cholesterol (Ovine) | Avanti Polar Lipids | cat# 700000 |
| DiO’; DiOC18(3) (3,3’-Dioctadecyloxacarbocyanine Perchlorate) | ThermoFisher Scientific | cat# D275 |
| DSPE-mPEG 2000 | Laysan Bio | cat# MPEG-DSPE-2000 |
| Pierce Quantitative Peptide Assay and Standards (QuantIT) | ThermoFisher Scientific | cat# 23290 |
| Critical commercial assays | ||
| Pan Dendritic Cell Isolation Kit | Miltenyi Biotech | cat# 130–100-875 |
| IL-1α ELISA kit | R&D systems | cat# DY400 |
| CXCL1 (KC) ELISA kit | R&D systems | cat# DY453 |
| IL-6 | R&D systems | cat# DY406 |
| IL-1B | R&D systems | cat# DY401 |
| TNF-α | R&D systems | cat# DY410 |
| iScript™ gDNA Clear cDNA Synthesis Kit | Biorad | cat# 1725034 |
| Deposited data | ||
| None | ||
| Experimental models: Cell lines | ||
| HEK293T | ATCC | ATCC#CRL-3216 |
| Experimental models: Organisms/strains | ||
| ES cells for making Slc46a2−/− mice | KOMP Repository, University of California | RRID:MMRRC_062453-UCD |
| Slc46a2−/− mice | This study | N/A |
| Slc46a3−/− mice | Jackson Labs | RRID:MMRRC_049738-UCD |
| Nod1−/− mice | M. Chamaillard et al., 2003(71) | N/A |
| Nod2−/− mice | K. S. Kobayashi et al., 2005(30) | N/A |
| IL1a−/− mice | R. K. S. Malireddi et al., 2022(77) | N/A |
| IL1b−/− Mice | M. B. Glaccum et al., 1997(75) | N/A |
| Pycard−/− mice | V. A. K. Rathinam et al., 2010(76) | N/A |
| MyD88−/− mice | T. Kawai, et al., 1999(73) | N/A |
| Casp1−/− mice | S. M. Man et al., 2017(74) | N/A |
| Gasmd−/− mice | N. Kayagaki et al., 2015(72) | N/A |
| Oligonucleotides | ||
| NOD1 FP | GAAGGCACCCCATTGGGTT | N/A |
| NOD1 RP | AATCTCTGCATCTTCGGCTGAC | N/A |
| GAPDH FP | TCACCACCATGGAGAAGGC | N/A |
| GAPDH RP | GCTAAGCAGTTGGTGGTGCA | N/A |
| Slc46a2 FP | CGCTGGACAAGTTCATTGGC | N/A |
| Slc46a2 RP | GCCACAACACCAATTGGGAC | N/A |
| SLC46A3 FP | CCGACTCACTTGGGTTAGGA | N/A |
| SLC46A3 RP | CGGTCGCTACCATTACCAGT | N/A |
| Slc46a2 TDF | GGAGAATGATTTGGGACCATGAAG | N/A |
| Slc46a2 TDR | GTGCTGAAGACCTGACTCGTATC | N/A |
| Slc46a2 LacINF | GGTAAACTGGCTCGGATTAGGG | N/A |
| Slc46a2 LacINR | TTGACTGTAGCGGCTGATGTTG | N/A |
| Human GAPDH Taqman primer/probe | ThermoFisher | cat# Hs02786624_g1 |
| Human CXCL8 Taqman primer/probe | ThermoFisher | cat# Hs00174103_m1 |
| Recombinant DNA | ||
| pEFV5 | D. Paik et al., 2017(24) | N/A |
| pEF-mSlc46a1-V5 | D. Paik et al., 2017(24) | N/A |
| pEF-mSlc46a2-V5 | D. Paik et al., 2017(24) | N/A |
| pEF-mSlc46a3-V5 | D. Paik et al., 2017(24) | N/A |
| Software and algorithms | ||
| Gen5 Software | BioTek | N/A |
| Graphpad Prism 9 | GraphPad Software | N/A |
| Other | ||
| Water bath | GFL | cat# 1002 |
| Centrifuge | Eppendorf | cat# 5810R |
| Digital caliper | Mitutoyo | cat# 500–196-30 |
| Transpore surgical tape | 3M Transpore | cat# 1527–0 |
| Semi-Prep, ZORBAX SB-C18 | Agilent | cat# AG880975–202 |
| 96 well plate | Denville scientific inc. | cat# T1006 |
| centrifugal filters | Milipore | cat# UFC501024 |
| Scissors | Fisher Scientific | cat# 08–940 |
| Forcep | Fisher Scientific | cat# 13–812-211 |
| 0.2 μM syringe filter | Sartorius Stedim | cat# 16534 |
| 1.7 ml microcentrifuge tube | Axygen | cat# MCT-175-C |
| 10 cm cell culture dish | Greiner | cat# 664160 |
| 15 ml conical bottom tube | Greiner | cat# 188271 |
| 50 ml conical bottom tube | Greiner | cat# 227261 |
| 70 μm Cell Strainer Nylon mesh | Corning | cat# 352360 |
| Insulin Syringe | BD | cat# 305937 |
| 6-well Deep Well TC-treated polystyrerene Plates | Corning | cat# 355467 |
| 6 well plate inserts | Corning | cat# 353091 |
Data and code availability
All data are available in the main text or the supplementary materials.
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal studies were performed in compliance with the federal regulations set forth in the Animal Welfare Act (AWA), the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and the guidelines of the UMass Chan Medical School Institutional Animal Use and Care Committee. All protocols used in this study were approved by the Institutional Animal Care and Use Committee at the UMass Chan Medical School (protocol 2056). All mice used in this study were maintained and bred in a pathogen-free environment. All mouse strains were backcrossed and maintained on a C57Bl/6J background. If not otherwise indicated, mice of the same sex and age were paired and assigned into experimental groups.
For generating Slc46a2−/−, ES cells harboring Slc46a2 locus targeted with Zen-Ub1 cassette were purchased from Mutant Mouse Resource and Research Center (MMRRC) at the University of California at Davis, an NIH-funded strain repository, and were donated to the MMRRC by The KOMP Repository, University of California, Davis; originating from David Valenzuela, George Yancopoulos, Regeneron Pharmaceuticals, Inc (RRID: MMRRC_062453-UCD) 70. ES cells were used to generate chimeric founder mice by standard microinjection into albino C57BL/6J blastocysts, by the UMMS transgenic mouse core. Chimeras were mated to albino C57Bl/6J mice to identify germline transmission. Heterozygous animals were identified using PCR (see Key Resources Table for oligos). Wildtype (Slc46a2+/+) and mutant (Slc46a2−/−) lines were established from these heterozygous and used in all experiments. Slc46a3−/− mice (Slc46a3tm1.1(KOMP)Vlcg) were generated by KOMP-Regeneron and acquired from Jackson Labs. For in vivo analysis of NOD1/2 responses in mouse skin, mixed male and female mice aged 4–6 weeks were used, while for IP NOD1/2 challenge assays male mice aged 6–12 weeks were used.
For genotyping, mice were anesthetized, and ear-punch or tail samples were collected in a sterile blade in a PCR tube. 75 μl of DNA extraction buffer (25 mM NaOH and 0.2 mM EDTA) was added to the sample, and it was heated at 95°C for 1 h in a thermocycler. Samples were then treated with 75 μl of neutralizing buffer (40mM Tris Ph-5.5) and stored at analyzed by PCR. Primer sequences are reported in the KRT.
Nod1−/−, Nod2−/−, IL1a−/−, IL1b−/−, IL1R−/−, Pycard−/−, and MyD88−/−, Casp1−/−, and Gasmd−/− mice were reported previously 30,71–77. Note that these Caspase-1 knockouts are wild-type for Caspase-11.
HEK293T cell
HEK293T cells are female and were maintained and cultured in DMEM high glucose (Gibco) with 10% fetal bovine serum (FBS) and 1X penicillin and streptomycin (Corning). Cells were maintained at 37°C with 5% CO2 and 100% humidity. For passaging, cells were washed with 1X PBS (Sigma) and incubated in TrypLE (Thermo) for 5 min at 37°C. Cells were rinsed with complete DMEM and after counting, a known number of cells were plated.
Bacteria culture
C. accolens was cultured in brain-heart infusion (BHI) broth supplemented with 1% Tween-80 overnight at 37°C shaking at 225 RPM. Bacteria were quantified by standard bacteriology techniques, counting CFU and measuring OD at 600 nm.
METHOD DETAILS
Keratinocyte isolation, culture, and analysis
Adult (6–8 weeks) mouse keratinocytes were isolated from male or female animals using protocols described previously 78. In brief, tail skin was incubated in 4mg/ml Dispase II (Roche) overnight at 4°C. The epidermis was physically removed from the dermis with tweezers and subsequently digested with TrypLE (Thermo) for 20 min at room temperature, and keratinocytes were detached by vigorously shaking in culture medium and filtered with a 70 μm strainer (Fisher scientific). Isolated cells were seeded at a density of 105cells/cm2 cultured with EpiLife (Gibco) in 12-well plates precoated with coating matrix (Gibco) at 37°C with 5% CO2 and 100% humidity and used between 3 and 5 days after isolation.
Conditioned media preparation
For the preparation of conditioned media, keratinocytes were first stimulated with 8 μM TCT 45, 30 μM iE-DAP (Invivogen), or 20 ng/ml LPS (Invivogen) for 1 hour (or left unstimulated, control). Then cells were washed 3 times with culture medium to remove agonists and cultured further in complete media for 24 hours when the media was collected and centrifuged. 200 μl of conditioned media was intraperitoneally injected into naïve mice and leukocyte recruitment was assayed by using flow cytometry.
ELISA
For cytokine ELISAs, primary keratinocytes were challenged, or not, with 8 μM TCT or 30 μM iE-DAP for 24 h and culture medium was harvested. Media was centrifuged at 500 g for 5 mins and used for analyzing cytokine release by ELISA (R&D systems) using the manufacturer’s protocol. In brief, 96-well ELISA plates were coated with capture antibody overnight at room temperature. The next day, the plate was washed three times with wash buffer and incubated with media (100 μl) for 2 h followed by three washes and then probed with a biotin-labeled detection antibody for 2 h. After three washes plates were incubated with HRP-streptavidin, developed with TMB (3,3’, 5,5”-tetramethylbenzidine) substrate for 20 min at room temperature, and then stopped with 1N sulfuric acid. Then plates were analyzed at 450 nm with wavelength correction at 540 nm.
Cell permeabilization assay
For cell permeabilization assays, 1X105 keratinocytes/well were seeded in a 96-well plate (Denville scientific inc.). After reaching 80% confluency 30 μM iE-DAP was added with 1nM Sytox red dye (Thermo) and 0.2nM Hoechst stain (Thermo). The plate was then cultured in the Cytation5 (BioTek) live-imaging microscope with quantitative imaging of 1 field of view in each well every 60 min for 24 hours. For analysis, a total number of intact cell nuclei were quantified via Hoechst positive nuclei and the total number of permeabilized cells was quantified by Sytox positive nuclei. The percentage of Sytox+ cells was then calculated.
Pinnae processing
Pinnae were cut into small pieces and incubated for 30 min at 37°C in a solution of 1mg/ml collagenase (Sigma) in DMEM media containing 10% FBS (Gibco). Ice-cold FACS buffer (2% FBS in PBS) was added to skin samples to stop the reaction. Then samples were placed on 70μm cell strainers and ground using a cell strainer pestle to create a single-cell suspension. Cells were spun at 180g for 5min at 4°C and resuspended in FACS buffer.
Flow cytometry
For pinnae,1×106 cells were incubated in an Fc receptor block (anti-CD16) for 10 min and stained with live/dead stain-aqua (Invitrogen) (1:5000), F4/80-PE/Cy7 (1:200), CD45-alexaflour700 (1:200) and Gr1-PE (1:200), or Ly6C & Ly6G, or CD3-FITC for 30 min in ice in the dark. Samples were washed thrice in FACS buffer and analyzed with a Cytek Aurora cytometer. For peritoneal cells, 1×106 cells were incubated in an Fc receptor block (anti-CD16) for 10 min and stained with CD45R, CD11c, CD11b, F4/80, Ly6C, Ly6G, and live/dead stain for 30 min in ice. Samples were washed thrice in FACS buffer and analyzed with a Cytek Aurora cytometer. All flow cytometry data were analyzed with FlowJo software (Tree Star, USA).
Cytospin
Gr-1+ FACS-sorted cells (BD FACS Aria Fusion) were centrifuged in 200 μL media onto a microscope slide using a Cytospin Universal 320 (Hettich, Germany) and stained with H&E stain. Images were acquired with a Nikon microscope equipped with a Canon A620 camera.
Histology
3h after DAP-muropeptide administration or 7 d after topical application of IMQ, pinnae were isolated and fixed in PBS containing 10% formalin. Paraffin-embedded sections were cut at 0.5 mm, stained with hematoxylin and eosin, and imaged by light microscopy.
Isolation of RNA from mouse tissues and cells
Lungs: the whole lung was isolated from euthanized mice and 1mg lung tissue was homogenized in Trizol using a homogenizer and cDNA was prepared as below.
Spleen: the whole spleen was isolated from euthanized mice and single-cell suspension was prepared by meshing through a 70 μm cell strainer with a plunger. These splenocytes were resuspended in Trizol and cDNA was prepared as below.
Gut: Gut was isolated from euthanized mice and 1 mg gut tissue was homogenized in Trizol using homogenizer and cDNA was prepared as below.
Dermis and epidermis: whole skin was extracted from euthanized mice and incubated in 4mg/ml Dispase II (Roche) overnight at 4°C. The epidermis was separated from the dermis with forceps. These tissues were homogenized in Trizol and cDNA was prepared as below.
Bone marrow macrophages: tibia and femur bones were extracted from euthanized mice and crushed using mortar and pestle in DMEM medium. This suspension of cells was passed through a 70 μm cell strainer and 2 × 106 cells/mL were plated in DMEM F12 media with recombinant MCSF (25 ng/ml). After 7 days, cells were harvested in Trizol and cDNA was prepared as below.
Peritoneal macrophages: 2ml of 3% Brewer thioglycollate medium was injected to elicit peritoneal macrophages. 4 days later, peritoneal cells were harvested by injecting 5ml FACS buffer in peritoneal cavity of euthanized mice and collected. These cells were resuspended in Trizol and cDNA was prepared as below.
Dendritic cells: As per manufacturer protocol, Pan dendritic cells were isolated from euthanized mice using Pan Dendritic Cell Isolation Kit from Miltenyi Biotec Inc. cDNA was prepared as below.
Neutrophils: neutrophils were isolated from bone marrow cells. Marrow cells were isolated as described and neutrophils separated from them using histopaque density gradient (Histopaque 1119: Histopaque 1077 in 1:5(Sigma)). The middle layer containing neutrophils was separated, cells resuspended in Trizol and cDNA was prepared as below.
qRT-PCR
Cells or tissue were lysed in TRIzol followed by RNA extraction per manufacture protocol. In brief, TRIzol lysed samples were mixed with chloroform and spun at 12000g for 15min at 4C. RNA from aqueous phase was precipitated with isopropanol and the RNA pellet was washed with 80% ethanol and resuspended in water.
cDNA was prepared from 1μg RNA using iScript gDNA Clear cDNA Synthesis Kit per manufacturer protocol. In brief, 1μg RNA was incubated with DNase mastermix at 25°C for 5 min and followed by reverse transcription for 20 min at 46°C. cDNA was diluted in 1:5 and used directly in qPCR reaction.
Real-time quantitative PCR was performed with 0.4mM primer, 10 μL iQ SYBR Green Supermix (BioRAD), in a final volume of 10 μL on the CFX96 real-time system (BioRAD). All samples were run in triplicate. GAPDH was used to normalize. Primer sequences are reported in KRT.
IMQ model of psoriasis-like dermatitis
Psoriasis-like inflammation on mouse pinnae skin was induced as previously described by 79. In brief, mice were treated daily for up to 6 d with 5 mg 5% IMQ cream topically applied on one pinna, while the other pinnae were treated with a similar amount of petroleum jelly (VAS), as vehicle control. Ear-skin thickness was measured daily using a digital caliper. The change in ear-skin thickness over time was reported as the difference related to measurement immediately prior to the initial IMQ or VAS application. For bacterial association, 107 CFU/ml in 50% glycerol was applied daily by swab to one pinna and 50% glycerol was applied to other pinnae.
To quantify C. accolens association, bacterial-associated and control ear pinnae were swapped with a sterile cotton swab and soaked into 500μl BHI broth with 1% tween 80 and plated onto BHI agar plates. These plates were incubated overnight at 37°C and CFUs were counted the next day.
Tape stripping
The upper most layers of the epidermal barrier, the stratum corneum, of the dorsal pinnae was disrupted by tape stripping: manually applying a small piece of surgical tape (Transpore surgical tape) to the skin and removing, repeating 5 times with a fresh piece of tape for each round 80. Then 107 cfu bacteria or 30μM iE-DAP in 20μl of 50% glycerol was applied to one pinna and only 50% glycerol was applied to another pinna. 3 h later, mice were euthanized, and pinnae were processed as described above.
Click-iE DAP preparation and imaging
iE-DAP and MDP with an alkyne handle for click-chemistry reaction were synthesized using a standard 8-step chemical synthesis, for complete experimental details, see (Bharadwaj et al, 2023)81.
Prior to use in experiments, the final click-muropeptides were purified by semi-prep C18 HPLC and analyzed by NMR & MS. (Thermo Q-Exactive Orbitrap at the Mass Spectroscopy Facility and Bruker AV 400 MHz, AV III 600 MHz NMR at the NMR laboratory, Department of Chemistry and Biochemistry, University of Delaware).
For visualization of muropeptide intracellular delivery, mouse keratinocytes or HEK 293T cells (transfected overnight with pEF-V5, pEF-Slc46a1-V5, pEF-Slc46a2-V5, and pEF-Slc46a3-V5) were challenged with either click-iE-DAP (30 μM) or click-MDP (20 μM) 37°C for 30–60 min in presence or absence of different inhibitors (250 μM MTX, 250 μM PTX, 30 μM MDP, or 250 μM FA). Cells were washed 2X with 1xPBS to remove access of click-muropeptides and fixed with 4% paraformaldehyde in PBS at RT for 10 min. Fixed cells were permeabilized with 1% Triton-X in PBS for 10 min at RT and blocked with 1% BSA in PBS. These permeabilized cells were then incubated in click-reaction conditions (250 μM CuSO4, 35 μM BTAA, 60 μM sodium ascorbate) with 2.5 μM CalFluor 488 Azide at RT for 30 mins. Cells were then washed and mounted on slides, with DAPI containing mounting media. Slides were imaged with a Leica SP8 confocal microscope. For spectrometric quantitation, cells were lysed (T-PER buffer, Thermo) and lysates were cleared by spinning at 13000 rpm for 30 min at 4°C. Click reaction was performed as stated above and fluorescence was measured at 488 nM using a spectrophotometer (Biotek).
Nano-particle preparation and challenge
A lipid/cholesterol matrix consisting of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC, Avanti), 1,2-dioleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (DOPG, Avanti), cholesterol (Avanti), and 1,2-distearoyl-sn-glycero3-phosphoethanolamine-methoxyl poly(ethylene glycol) 2000 (DSPE-mPEG 2000, Laysan Bio) was used to formulate iE-DAP-encapsulated nanoparticles (NPs). We adapted our previously published methods 82 to load iE-DAP into lipid-based NPs. Briefly, a matrix of DOPC/DSPC/DOPG/cholesterol/DSPE-mPEG at 33.5/33.5/20/10/3 mol% was prepared in chloroform and dry lipid/cholesterol films were allowed to form. Films were rehydrated with iE-DAP (Invivogen) prepared in PBS at 1.2 mg/mL and these samples were vortexed for 30 sec every 10 min for 1 hr at 56°C to complete this process. Following rehydration, samples were ultrasonicated in alternating 20 sec pulse/10 sec off cycles for 5 min at an amplitude of 20% to form iE-DAP-encapsulated lipid-based NPs. Samples were dialyzed for 1 hr following ultrasonication. Dynamic light scattering (DLS) and zeta potential measurements using a Malvern Zetasizer were used to characterize NP size and surface charge, respectively. NPs had an average 43.97 nm hydrodynamic diameter, a polydispersity index (PDI) of 0.158, and a zeta potential of −9.90 mV. Quant-IT assay (Thermo Fisher Scientific) measurements were used to measure average iE-DAP encapsulation at 998.1 μg/mL and an average encapsulation efficiency of 57.1%. To visualize iE-DAP-encapsulated NPs, 0.1 mol% of the fluorescent lipid tracer dye 3,3’-dioctadecyloxacarbocyanine perchlorate (DiO) were added to the lipid/cholesterol films.
Mouse keratinocytes were challenged iE-DAP loaded NPs, or control empty NPs, at 37°C at 10μg/ml final concentration (~30 μM iE-DAP), and then monitored for Sytox uptake assay for 24 h, as described above. To visualize NP delivery, cells were washed after 30–60 mins 2X with PBS to remove access NPs and fixed with 4% paraformaldehyde in PBS at RT for 10 min. Cells were blocked with 1% BSA in PBS, washed 3X with PBS and mounted on slides with DAPI containing mounting media. Slides were then visualized with a Leica SP8 confocal microscope.
Human skin equivalent construction and infections
HSEs were constructed as previously described in Carlson et al. 2008.63 In brief, human foreskin fibroblasts (HFFs) were embedded in collagen for 7 days at 37°C with 5% CO2 and 100% humidity. Keratinocytes were added on top of the HFF-collagen matrix and left to grow for another 6–8 days at an air-liquid interface at 37°C until full morphologic differentiation and stratification were observed. For muropeptide challenge assays, 30μM iE-DAP diluted in PBS was added to the epidermal side of the HSE. For IL-1R inhibition studies, 500 ng/ml of IL-1Ra (Biolegend) was added to 8 mL of HSE cornification media at 1 h post-iE-DAP challenge. Keratinocyte and fibroblast layers were separated by 5–10 min incubation in a dispase II containing buffer (150 mM NaCl, 10mM HEPES, 2mM CaCl2, 2.4 U/ml dispase II) followed by gentle peeling of the two layers. Supernatants or individual tissue layers were harvested at 24 h for transcriptional analysis.
Plasmids
pEFV5, pEF-Slc46a1-V5, pEF-Slc46a2-V5, and pEF-Slc46a3-V5 plasmids used in this study were generated previously by D. Paik et al., 2017.24
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed by GraphPad Prism software and enumerated as mean ± standard error of the mean (SEM). The number of independent replicates (n) and methods used to analyze statical significance (P-values) are reported in individual figure legends.
Supplementary Material
Highlights.
SLC46A2 is critical for cytosolic transport of DAP-muropeptides and NOD1 activation
NOD1 activation triggers epidermal keratinocyte pyroptosis through caspase-1 and GSDMD
SLC46A2-NOD1 signaling axis is critical for the progression of psoriatic inflammation
Methotrexate blocks SLC46A2-mediated DAP-muropeptide transport
Acknowledgments
The authors wish to thank Douglas Golenbock, Kate Fitzgerald, Egil Lien and Gabriel Nunez for sharing mouse strains, and Egil Lien for comments on the manuscript, Gail German and Zhaozhao Jiang for technical assistance.
Funding:
National Institutes of Health grant ROAI060025 (NS)
Kenneth Rainin Foundation (NS)
National Institutes of Health grant 5T32GM133395 (KW)
National Institutes of health grant R01GM138599 (CLG)
Pew Biomedical scholars Program (CLG)
National Institutes of Health grant K22CA262355 (PUA)
National Institutes of Health grant R00AI130258 (MO)
Smith Family Award for Excellence in Biomedical Research from the Richard and Susan Smith Family Foundation (MO)
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of interests
A provisional patent on targeting SLC46s to inhibit inflammation in psoriasis and other auto-inflammatory diseases has been filed by some of the authors (NS, RB, & MO). TDK consults for Pfizer.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Ragland SA, and Kagan JC (2021). Cytosolic detection of phagosomal bacteria-Mechanisms underlying PAMP exodus from the phagosome into the cytosol. Mol Microbiol 116, 1420–1432. 10.1111/mmi.14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rathinam VA, and Fitzgerald KA (2011). Cytosolic surveillance and antiviral immunity. Curr Opin Virol 1, 455–462. 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okude H, Ori D, and Kawai T (2020). Signaling Through Nucleic Acid Sensors and Their Roles in Inflammatory Diseases. Front Immunol 11, 625833. 10.3389/fimmu.2020.625833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanson KV, Deng M, and Ting JP (2019). The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19, 477–489. 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston EL, Heras B, Kufer TA, and Kaparakis-Liaskos M (2021). Detection of Bacterial Membrane Vesicles by NOD-Like Receptors. Int J Mol Sci 22. 10.3390/ijms22031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sundaram B, and Kanneganti TD (2021). Advances in Understanding Activation and Function of the NLRC4 Inflammasome. Int J Mol Sci 22. 10.3390/ijms22031048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauernfried S, Scherr MJ, Pichlmair A, Duderstadt KE, and Hornung V (2021). Human NLRP1 is a sensor for double-stranded RNA. Science 371. 10.1126/science.abd0811. [DOI] [PubMed] [Google Scholar]
- 8.Kudo M, Kobayashi-Nakamura K, Kitajima N, and Tsuji-Naito K (2020). Alternate expression of PEPT1 and PEPT2 in epidermal differentiation is required for NOD2 immune responses by bacteria-derived muramyl dipeptide. Biochem Biophys Res Commun 522, 151–156. 10.1016/j.bbrc.2019.11.044. [DOI] [PubMed] [Google Scholar]
- 9.Luteijn RD, Zaver SA, Gowen BG, Wyman SK, Garelis NE, Onia L, McWhirter SM, Katibah GE, Corn JE, Woodward JJ, and Raulet DH (2019). SLC19A1 transports immunoreactive cyclic dinucleotides. Nature 573, 434–438. 10.1038/s41586-019-1553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie C, Cordova AF, Hess GT, Bassik MC, and Li L (2019). SLC19A1 Is an Importer of the Immunotransmitter cGAMP. Mol Cell 75, 372–381 e375. 10.1016/j.molcel.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strober W, and Watanabe T (2011). NOD2, an intracellular innate immune sensor involved in host defense and Crohn’s disease. Mucosal Immunol 4, 484–495. 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al Nabhani Z, Dietrich G, Hugot JP, and Barreau F (2017). Nod2: The intestinal gate keeper. PLoS Pathog 13, e1006177. 10.1371/journal.ppat.1006177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schleifer KH, and Kandler O (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36, 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, and Girardin SE (2019). NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys 670, 69–81. 10.1016/j.abb.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Irazoki O, Hernandez SB, and Cava F (2019). Peptidoglycan Muropeptides: Release, Perception, and Functions as Signaling Molecules. Front Microbiol 10, 500. 10.3389/fmicb.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismair MG, Vavricka SR, Kullak-Ublick GA, Fried M, Mengin-Lecreulx D, and Girardin SE (2006). hPepT1 selectively transports muramyl dipeptide but not Nod1-activating muramyl peptides. Can J Physiol Pharmacol 84, 1313–1319. 10.1139/y06-076. [DOI] [PubMed] [Google Scholar]
- 17.Charriere GM, Ip WE, Dejardin S, Boyer L, Sokolovska A, Cappillino MP, Cherayil BJ, Podolsky DK, Kobayashi KS, Silverman N, et al. (2010). Identification of Drosophila Yin and PEPT2 as evolutionarily conserved phagosome-associated muramyl dipeptide transporters. J Biol Chem 285, 20147–20154. 10.1074/jbc.M110.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Tattoli I, Wojtal KA, Vavricka SR, Philpott DJ, and Girardin SE (2009). pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J Biol Chem 284, 23818–23829. 10.1074/jbc.M109.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, de Maziere A, Klumperman J, Schlatter M, Delamarre L, and Mellman I (2014). Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509, 240–244. 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 20.Sasawatari S, Okamura T, Kasumi E, Tanaka-Furuyama K, Yanobu-Takanashi R, Shirasawa S, Kato N, and Toyama-Sorimachi N (2011). The solute carrier family 15A4 regulates TLR9 and NOD1 functions in the innate immune system and promotes colitis in mice. Gastroenterology 140, 1513–1525. 10.1053/j.gastro.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Song F, Jiang H, Nunez G, and Smith DE (2018). SLC15A2 and SLC15A4 Mediate the Transport of Bacterially Derived Di/Tripeptides To Enhance the Nucleotide-Binding Oligomerization Domain-Dependent Immune Response in Mouse Bone Marrow-Derived Macrophages. J Immunol 201, 652–662. 10.4049/jimmunol.1800210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz LX, Lee J, Kapoor U, Kartnig F, Sedlyarov V, Papakostas K, Cesar-Razquin A, Essletzbichler P, Goldmann U, Stefanovic A, et al. (2020). TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7–9. Nature 581, 316–322. 10.1038/s41586-020-2282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimann I, Gonzalez-Quintial R, Baccala R, Kiosses WB, Teijaro JR, Parker CG, Li X, Beutler B, Kono DH, and Theofilopoulos AN (2022). The solute carrier SLC15A4 is required for optimal trafficking of nucleic acid-sensing TLRs and ligands to endolysosomes. Proc Natl Acad Sci U S A 119, e2200544119. 10.1073/pnas.2200544119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paik D, Monahan A, Caffrey DR, Elling R, Goldman WE, and Silverman N (2017). SLC46 Family Transporters Facilitate Cytosolic Innate Immune Recognition of Monomeric Peptidoglycans. J Immunol 199, 263–270. 10.4049/jimmunol.1600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Kim MG, Soo Lyu M, Kozak CA, Schwartz RH, and Flomerfelt FA (2000). Characterization of the mouse gene, human promoter and human cDNA of TSCOT reveals strong interspecies homology. Biochim Biophys Acta 1493, 159–169. 10.1016/s0167-4781(00)00177-9. [DOI] [PubMed] [Google Scholar]
- 26.Kim KY, Lee G, Yoon M, Cho EH, Park CS, and Kim MG (2015). Expression Analyses Revealed Thymic Stromal Co-Transporter/Slc46A2 Is in Stem Cell Populations and Is a Putative Tumor Suppressor. Mol Cells 38, 548–561. 10.14348/molcells.2015.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 28.Elias PM (2008). Skin barrier function. Curr Allergy Asthma Rep 8, 299–305. 10.1007/s11882-008-0048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouwstra JA, and Ponec M (2006). The skin barrier in healthy and diseased state. Biochim Biophys Acta 1758, 2080–2095. 10.1016/j.bbamem.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, and Flavell RA (2005). Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734. 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 31.Harder J, and Nunez G (2009). Functional expression of the intracellular pattern recognition receptor NOD1 in human keratinocytes. J Invest Dermatol 129, 1299–1302. 10.1038/jid.2008.395. [DOI] [PubMed] [Google Scholar]
- 32.Ridaura VK, Bouladoux N, Claesen J, Chen YE, Byrd AL, Constantinides MG, Merrill ED, Tamoutounour S, Fischbach MA, and Belkaid Y (2018). Contextual control of skin immunity and inflammation by Corynebacterium. J Exp Med 215, 785–799. 10.1084/jem.20171079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balato A, Cacciapuoti S, Di Caprio R, Marasca C, Masara A, Raimondo A, and Fabbrocini G (2019). Human Microbiome: Composition and Role in Inflammatory Skin Diseases. Arch Immunol Ther Exp (Warsz) 67, 1–18. 10.1007/s00005-018-0528-4. [DOI] [PubMed] [Google Scholar]
- 34.Stehlikova Z, Kostovcikova K, Kverka M, Rossmann P, Dvorak J, Novosadova I, Kostovcik M, Coufal S, Srutkova D, Prochazkova P, et al. (2019). Crucial Role of Microbiota in Experimental Psoriasis Revealed by a Gnotobiotic Mouse Model. Front Microbiol 10, 236. 10.3389/fmicb.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanvit P, Konkel JE, Jiao X, Kasagi S, Zhang D, Wu R, Chia C, Ajami NJ, Smith DP, Petrosino JF, et al. (2015). Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat Commun 6, 8424. 10.1038/ncomms9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker JL, Deme JC, Kuteyi G, Wu Z, Huo J, Goldman ID, Owens RJ, Biggin PC, Lea SM, and Newstead S (2021). Structural basis of antifolate recognition and transport by PCFT. Nature 595, 130–134. 10.1038/s41586-021-03579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinblatt A.K.J.a.M.E. (2018). Methotrexate. In Rheumatology, Weisman MHASEGJSMWM, ed. (Elsevier; ). [Google Scholar]
- 38.Alqarni AM, and Zeidler MP (2020). How does methotrexate work? Biochem Soc Trans 48, 559–567. 10.1042/BST20190803. [DOI] [PubMed] [Google Scholar]
- 39.Cronstein BN, and Aune TM (2020). Methotrexate and its mechanisms of action in inflammatory arthritis. Nat Rev Rheumatol 16, 145–154. 10.1038/s41584-020-0373-9. [DOI] [PubMed] [Google Scholar]
- 40.Masumoto J, Yang K, Varambally S, Hasegawa M, Tomlins SA, Qiu S, Fujimoto Y, Kawasaki A, Foster SJ, Horie Y, et al. (2006). Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med 203, 203–213. 10.1084/jem.20051229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenzweig HL, Martin TM, Planck SR, Galster K, Jann MM, Davey MP, Kobayashi K, Flavell RA, and Rosenbaum JT (2008). Activation of NOD2 in vivo induces IL-1beta production in the eye via caspase-1 but results in ocular inflammation independently of IL-1 signaling. J Leukoc Biol 84, 529–536. 10.1189/jlb.0108015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magalhaes JG, Philpott DJ, Nahori MA, Jehanno M, Fritz J, Le Bourhis L, Viala J, Hugot JP, Giovannini M, Bertin J, et al. (2005). Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep 6, 1201–1207. 10.1038/sj.embor.7400552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Jin X, Tsueng G, Afrasiabi C, and Su AI (2016). BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res 44, D313–316. 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlsson M, Zhang C, Mear L, Zhong W, Digre A, Katona B, Sjostedt E, Butler L, Odeberg J, Dusart P, et al. (2021). A single-cell type transcriptomics map of human tissues. Sci Adv 7. 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, and Silverman N (2004). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649. 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 46.Orzalli MH, Smith A, Jurado KA, Iwasaki A, Garlick JA, and Kagan JC (2018). An Antiviral Branch of the IL-1 Signaling Pathway Restricts Immune-Evasive Virus Replication. Mol Cell 71, 825–840 e826. 10.1016/j.molcel.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rider P, Carmi Y, Guttman O, Braiman A, Cohen I, Voronov E, White MR, Dinarello CA, and Apte RN (2011). IL-1alpha and IL-1beta recruit different myeloid cells and promote different stages of sterile inflammation. J Immunol 187, 4835–4843. 10.4049/jimmunol.1102048. [DOI] [PubMed] [Google Scholar]
- 48.Di Paolo NC, and Shayakhmetov DM (2016). Interleukin 1alpha and the inflammatory process. Nat Immunol 17, 906–913. 10.1038/ni.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, et al. (2018). Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069. 10.1126/science.aau2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orzalli MH, Prochera A, Payne L, Smith A, Garlick JA, and Kagan JC (2021). Virus-mediated inactivation of anti-apoptotic Bcl-2 family members promotes Gasdermin-E-dependent pyroptosis in barrier epithelial cells. Immunity 54, 1447–1462 e1445. 10.1016/j.immuni.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphries F, Shmuel-Galia L, Ketelut-Carneiro N, Li S, Wang B, Nemmara VV, Wilson R, Jiang Z, Khalighinejad F, Muneeruddin K, et al. (2020). Succination inactivates gasdermin D and blocks pyroptosis. Science 369, 1633–1637. 10.1126/science.abb9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shieh P, Dien VT, Beahm BJ, Castellano JM, Wyss-Coray T, and Bertozzi CR (2015). CalFluors: A Universal Motif for Fluorogenic Azide Probes across the Visible Spectrum. J Am Chem Soc 137, 7145–7151. 10.1021/jacs.5b02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao R, Aluri S, and Goldman ID (2017). The proton-coupled folate transporter (PCFT-SLC46A1) and the syndrome of systemic and cerebral folate deficiency of infancy: Hereditary folate malabsorption. Mol Aspects Med 53, 57–72. 10.1016/j.mam.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cordova AF, Ritchie C, Böhnert V, and Li L (2021). Human SLC46A2 Is the Dominant cGAMP Importer in Extracellular cGAMP-Sensing Macrophages and Monocytes. ACS Central Science 7, 1073–1088. 10.1021/acscentsci.1c00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quan C, Chen XY, Li X, Xue F, Chen LH, Liu N, Wang B, Wang LQ, Wang XP, Yang H, and Zheng J (2020). Psoriatic lesions are characterized by higher bacterial load and imbalance between Cutibacterium and Corynebacterium. J Am Acad Dermatol 82, 955–961. 10.1016/j.jaad.2019.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Gao Z, Tseng CH, Strober BE, Pei Z, and Blaser MJ (2008). Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One 3, e2719. 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fyhrquist N, Muirhead G, Prast-Nielsen S, Jeanmougin M, Olah P, Skoog T, Jules-Clement G, Feld M, Barrientos-Somarribas M, Sinkko H, et al. (2019). Microbe-host interplay in atopic dermatitis and psoriasis. Nature Communications 10, 4703. 10.1038/s41467-019-12253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yerushalmi M, Ofir E, Melanie A, and Vinod C (2019). The skin microbiome in psoriatic disease: A systematic review and critical appraisal. Journal of Translational Autoimmunity 2, 100009. 10.1016/j.jtauto.2019.100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olejniczak-Staruch I, Ciążyńska M, Sobolewska-Sztychny D, Narbutt J, Skibińska M, and Lesiak A (2021). Alterations of the Skin and Gut Microbiome in Psoriasis and Psoriatic Arthritis. International Journal of Molecular Sciences 22, 3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olunoiki E, Rehner J, Bischoff M, Koshel E, Vogt T, Reichrath J, and Becker SL (2022). Characteristics of the Skin Microbiome in Selected Dermatological Conditions: A Narrative Review. Life 12, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zákostelská Z, Málková J, Klimešová K, Rossmann P, Hornová M, Novosádová I, Stehlíková Z, Kostovčík M, Hudcovic T, Štepánková R, et al. (2016). Intestinal Microbiota Promotes Psoriasis-Like Skin Inflammation by Enhancing Th17 Response. PLoS One 11, e0159539. 10.1371/journal.pone.0159539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whelan SP, Ball LA, Barr JN, and Wertz GT (1995). Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A 92, 8388–8392. 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carlson MW, Alt-Holland A, Egles C, and Garlick JA (2008). Three-dimensional tissue models of normal and diseased skin. Curr Protoc Cell Biol Chapter 19, Unit 19 19. 10.1002/0471143030.cb1909s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bastos PAD, Wheeler R, and Boneca IG (2021). Uptake, recognition and responses to peptidoglycan in the mammalian host. FEMS Microbiol Rev 45. 10.1093/femsre/fuaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, and Sweet MJ (2008). Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4, 5. 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.St Leger AJ, Desai JV, Drummond RA, Kugadas A, Almaghrabi F, Silver P, Raychaudhuri K, Gadjeva M, Iwakura Y, Lionakis MS, and Caspi RR (2017). An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal γδ T Cells. Immunity 47, 148–158.e145. 10.1016/j.immuni.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schrezenmeier E, and Dorner T (2020). Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol 16, 155–166. 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 68.Anthony J, Jigar VD, Rebecca AD, Abirami K, Fatimah A, Phyllis S, Kumarkrishna R, Mihaela G, Yoichiro I, Michail SL, and Rachel RC (2017). An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal γδ T Cells. Immunity 47, 148–158.e145. 10.1016/j.immuni.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou W, Kaneko N, Nakagita T, Takeda H, and Masumoto J (2021). A comprehensive interaction study provides a potential domain interaction network of human death domain superfamily proteins. Cell Death Differ 28, 2991–3008. 10.1038/s41418-021-00796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. (2003). High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21, 652–659. 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 71.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, et al. (2003). An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 4, 702–707. 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 72.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671. 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 73.Kawai T, Adachi O, Ogawa T, Takeda K, and Akira S (1999). Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11, 115–122. 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 74.Man SM, Karki R, Briard B, Burton A, Gingras S, Pelletier S, and Kanneganti TD (2017). Differential roles of caspase-1 and caspase-11 in infection and inflammation. Sci Rep 7, 45126. 10.1038/srep45126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glaccum MB, Stocking KL, Charrier K, Smith JL, Willis CR, Maliszewski C, Livingston DJ, Peschon JJ, and Morrissey PJ (1997). Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J Immunol 159, 3364–3371. [PubMed] [Google Scholar]
- 76.Rathinam VAK, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. (2010). The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature Immunology 11, 395–402. 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malireddi RKS, Bynigeri R, Kancharana B, Sharma BR, Burton AR, Pelletier S, and Kanneganti T-D (2022). Determining distinct roles of IL-1α through generation of an IL-1α knockout mouse with no defect in IL-1β expression. bioRxiv, 2022.2009.2021.508892. 10.1101/2022.09.21.508892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li F, Adase CA, and Zhang LJ (2017). Isolation and Culture of Primary Mouse Keratinocytes from Neonatal and Adult Mouse Skin. J Vis Exp 10.3791/56027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gray EE, Ramirez-Valle F, Xu Y, Wu S, Wu Z, Karjalainen KE, and Cyster JG (2013). Deficiency in IL-17-committed Vgamma4(+) gammadelta T cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nat Immunol 14, 584–592. 10.1038/ni.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Au - Sparber F, and Au - LeibundGut-Landmann S (2019). Infecting Mice with Malassezia spp. to Study the Fungus-Host Interaction. JoVE, e60175. doi: 10.3791/60175. [DOI] [PubMed] [Google Scholar]
- 81.Bharadwaj R, Anonick MV, Mashayekh S, Brown A, Wodzanowski KA, Okuda K, Silverman N, and Grimes CL (2023). Synthesis and validation of click-modified of NOD1/2 agonists. bioRxiv, 2023.2003.2028.534546. 10.1101/2023.03.28.534546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Atukorale PU, Raghunathan SP, Raguveer V, Moon TJ, Zheng C, Bielecki PA, Wiese ML, Goldberg AL, Covarrubias G, Hoimes CJ, and Karathanasis E (2019). Nanoparticle Encapsulation of Synergistic Immune Agonists Enables Systemic Codelivery to Tumor Sites and IFNbeta-Driven Antitumor Immunity. Cancer Res 79, 5394–5406. 10.1158/0008-5472.CAN-19-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


