Abstract
Cold atmospheric-pressure plasma (CAP) has emerged as a potential alternative or adjuvant to conventional antibiotics for the treatment of bacterial infections, including those caused by antibiotic-resistant pathogens. The potential of sub-lethal CAP exposures to synergise conventional antimicrobials for the eradication of Pseudomonas aeruginosa biofilms is investigated in this study. The efficacy of antimicrobials following or in the absence of sub-lethal CAP pre-treatment in P. aeruginosa biofilms was assessed. CAP pre-treatment resulted in an increase in both planktonic and biofilm antimicrobial sensitivity for all three strains tested (PAO1, PA14, and PA10548), with both minimum inhibitory concentrations (MICs) and minimum biofilm eradication concentrations (MBECs) of individual antimicrobials, being significantly reduced following CAP pre-treatment of the biofilm (512-fold reduction with ciprofloxacin/gentamicin; and a 256-fold reduction with tobramycin). At all concentrations of antimicrobial used, the combination of sub-lethal CAP exposure and antimicrobials was effective at increasing time-to-peak metabolism, as measured by isothermal microcalorimetry, again indicating enhanced susceptibility. CAP is known to damage bacterial cell membranes and DNA by causing oxidative stress through the in situ generation of reactive oxygen and nitrogen species (RONS). While the exact mechanism is not clear, oxidative stress on outer membrane proteins is thought to damage/perturb cell membranes, confirmed by ATP and LDH leakage, allowing antimicrobials to penetrate the bacterial cell more effectively, thus increasing bacterial susceptibility. Transcriptomic analysis, reveals that cold-plasma mediated oxidative stress caused upregulation of P. aeruginosa superoxide dismutase, cbb3 oxidases, catalases, and peroxidases, and upregulation in denitrification genes, suggesting that P. aeruginosa uses these enzymes to degrade RONS and mitigate the effects of cold plasma mediated oxidative stress. CAP treatment also led to an increased production of the signalling molecule ppGpp in P. aeruginosa, indicative of a stringent response being established. Although we did not directly measure persister cell formation, this stringent response may potentially be associated with the formation of persister cells in biofilm cultures. The production of ppGpp and polyphosphate may be associated with protein synthesis inhibition and increase efflux pump activity, factors which can result in antimicrobial tolerance. The transcriptomic analysis also showed that by 6 h post-treatment, there was downregulation in ribosome modulation factor, which is involved in the formation of persister cells, suggesting that the cells had begun to resuscitate/recover. In addition, CAP treatment at 4 h post-exposure caused downregulation of the virulence factors pyoverdine and pyocyanin; by 6 h post-exposure, virulence factor production was increasing. Transcriptomic analysis provides valuable insights into the mechanisms by which P. aeruginosa biofilms exhibits enhanced susceptibility to antimicrobials. Overall, these findings suggest, for the first time, that short CAP sub-lethal pre-treatment can be an effective strategy for enhancing the susceptibility of P. aeruginosa biofilms to antimicrobials and provides important mechanistic insights into cold plasma-antimicrobial synergy. Transcriptomic analysis of the response to, and recovery from, sub-lethal cold plasma exposures in P. aeruginosa biofilms improves our current understanding of cold plasma biofilm interactions.
Keywords: Pseudomonas aeruginosa, Biofilm, Plasma Medicine, Cold plasma, Antimicrobial synergy, Transcriptomics, ppGpp, Persister cells
1. Introduction
Cold atmospheric-pressure plasma (CAP) has emerged as a promising potential alternative to traditional antibiotic therapy for the treatment of bacterial infections including those caused by antibiotic-resistant pathogens [1]. CAP is a type of ionised gas that inactivates bacteria through a combination of physical and chemical effects produced by the plasma plume. The plasma plume generates a diverse and complex mixture of reactive oxygen species (ROS) and reactive nitrogen species (RNS) that damage the bacteria's cell membrane and DNA with antimicrobial efficacy against a variety of bacterial species including the ESKAPE pathogens [2]. In response to this oxidative stress, bacterial cells may produce enzymes such as superoxide dismutase and catalase, which help to detoxify ROS and RNS. However, this oxidative stress may also induce the formation of persister cells, which are a subpopulation of bacterial cells that are highly tolerant to antibiotics, as described by Lewis, 2010 [3]. Recent studies have revealed that the level of ROS in the environment can impact the dormancy and the subsequent capacity of intracellular Staphylococcus aureus persisters to resuscitate from this resting state, which highlights the importance of oxidative stress in regulating bacterial persistence [4]. Overall, while ROS and RNS are effective antimicrobials, the ability of bacteria to detoxify themselves through enzyme production or, worse still, evade killing through persister cell formation highlights the limitations of using ROS/RNS alone as a treatment strategy against bacterial infections. Therefore, alternative or combination treatment regimens, such as the use of CAP and traditional antimicrobials, should be explored to more effectively target and eradicate bacteria.
Pseudomonas aeruginosa is a common cause of chronic lung infections due to its prevalence in nosocomial infections and has high morbidity rates [5], particularly in cystic fibrosis (CF) patients. P. aeruginosa exhibits both intrinsic and acquired multidrug resistance mechanisms to multiple classes of antibiotics, resistance of which is significantly elevated via the formation of complex, protective biofilms. Biofilms are complex communities of bacteria that produce extensive extracellular polymeric substances (EPS). This EPS contributes to phenotypic and transcriptomic changes, which in turn can result in elevated tolerance to antimicrobial challenge, often requiring eradication concentrations up to 1000 times higher than their planktonic counterparts [6]. The P. aeruginosa biofilm matrix can reduce the efficacy of antibiotics for example, by binding positively charged tobramycin to biofilm matrix components to hinder antibiotic diffusion [7]. Similarly, components of biofilm EPS such as eDNA and alginate provide a possible protective effect and tolerance against CAP [8]. Here we demonstrate that P. aeruginosa biofilms establish a stress response to CAP through the production of the signalling molecule (p)ppGpp (guanosine pentaphosphate). This molecule upregulates a variety of pathways, including those involved in the production of EPS and the detoxification of ROS and RNS.
This study provides detailed mechanistic insights into these potential mechanisms using both culture-based antimicrobial synergy studies and transcriptomic analysis. The study also examines the effect of sub-lethal CAP exposure on P. aeruginosa biofilm to investigate the mechanisms underlying the enhanced susceptibility of P. aeruginosa biofilms to antibiotics after CAP treatment. The use of sub-lethal exposures allows for the assessment of changes in bacterial physiology, gene expression, and other cellular processes induced by CAP exposure, without completely eradicating the bacterial population. By characterising the oxidative stress response and antimicrobial synergy of sub-lethal CAP exposure, improved understanding of the mechanisms of action of CAP and how to effectively harness CAP to eliminate drug-resistant and phenotypically tolerant bacteria in both infection and the environment in the healthcare setting. Although longer exposure times, would decrease the viable bacterial cell population without the use of antibiotics, sub-lethal cold plasma exposures on P. aeruginosa biofilms with transcriptomic analysis improves our current understanding of cold plasma biofilm interactions. Based on these findings, we propose that a combination treatment of CAP followed by antimicrobial treatment may be an effective approach to synergise antibiotic activity against bacterial biofilms in acute and chronic infections.
2. Materials and methods
2.1. Plasma jet configuration
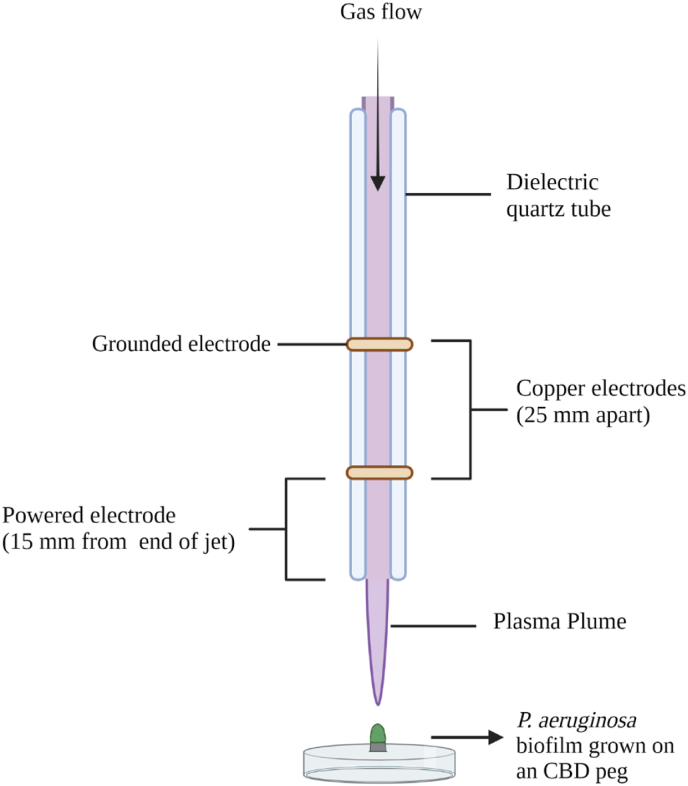
The plasma jet used for this study is an atmospheric-pressure, kHz-driven DBD (dielectric barrier discharge) plasma jet, as described in detail previously by Flynn et al., [2], Alkawareek et al. [9], and Barakat et al., [10]. The jet is powered by a high-voltage AC power supply (Haiden PHK-2k, Haiden Laboratory Inc., Japan) and constitutes a dielectric quartz tube with an outer diameter of 6 mm and an inner diameter of 4 mm, around which two copper electrodes are secured. The 2 mm copper electrodes are 25 mm apart, with the powered electrode positioned 20 mm from the open end of the jet. This investigation used a helium plasma containing 0.5% oxygen, generated at a voltage amplitude of 6 kV and a repetition frequency of 20 kHz. The gaseous components are fed into the jet by a mass flow controller (MKS 247D, MKS Instruments, USA) and the addition of power induces the generation of a glowing plasma between the two electrodes. This plasma becomes more diffuse as it travels down the quartz tube of the jet and forms the plasma plume.
2.2. Microbial strains, growth conditions, and antimicrobial agents
Three P. aeruginosa strains were used in this study: the reference organism P. aeruginosa PAO1, and clinical isolates P. aeruginosa UCBPP-PA14, and P. aeruginosa NCIMB 10548. It is worth noting that NCIMB 10548 is a strain derived from PAO1 [11]. We included both strains in our study as they have been shown to exhibit different phenotypic and genotypic characteristics, which may influence their response to CAP treatment. Moreover, previous studies have shown that strain-specific differences in bacterial response to CAP treatment can occur [12]. Therefore, including both strains in our study allowed us to investigate potential strain-specific differences in response to CAP treatment. Strains were sub-cultured from frozen 50% glycerol stocks (stored at −80 °C in Microbank cryovials (Pro-Lab Diagnostics, Cheshire, UK)). For planktonic experiments, strains were inoculated onto Mueller-Hinton agar (MHA) from the glycerol stocks and incubated in a static incubator at 37 °C overnight before use.
For biofilm experiments, strains were sub-cultured in Mueller-Hinton broth (MHB) and incubated in an orbital incubator at 37 °C and 100 rpm overnight. Cultures were then diluted with fresh sterile MHB to achieve an optical density at 550 nm (OD550) equivalent to 1 × 107 CFU/mL, and 200 μL was used to inoculate the wells of the Calgary Biofilm Device (CBD) (Innovotech, Edmonton, Canada). The pegs of the device were immersed in the bacterial suspension and the CBD was incubated for 24 h at 37 °C and 100 rpm. Biofilm-coated pegs were rinsed in 200 μL phosphate-buffered saline (PBS) before either removal of pegs for CAP exposure, or transfer to an antimicrobial challenge plate. Standard plate counts for these strains was carried out on MHA using the serial dilution method of Miles and Misra and incubated overnight at 37 °C. The antimicrobial agents employed for this study included gentamicin sulfate, tobramycin sulfate, ciprofloxacin, and chlorhexidine digluconate. These were obtained from Sigma-Aldrich (Gillingham, UK), except for tobramycin sulfate, purchased from Alfa Aesar (Lancashire, UK).
2.3. CAP exposure and cell culturability determination
Planktonic and biofilm samples were CAP-exposed across a range of treatment times to obtain survival curves for the selection of a suitable sub-lethal exposure time for further study. Planktonic cells were grown overnight on MHA and transferred to PBS to produce a bacterial solution with an OD550 equivalent to 5 × 108 CFU/mL 100 μL of bacterial suspension was then treated under the plume of the plasma jet, 15 mm from the end of the jet as measured from the end of the quartz tube to the surface of the sample. Samples were treated in triplicate for each time point. After exposure, PBS dilutions of each sample were used to determine microbial cell viability.
Biofilm treatments were also carried out at the same distance from the jet, using 24 h biofilms grown on the pegs of the CBD as previously described (illustrated in Fig. 1). Pegs were rinsed in 200 μL PBS after incubation, snapped off from the lid of the device and fixed in an upright position in a Petri dish for CAP exposure. Samples were treated in triplicate for each time point, with the tip of the biofilm peg 15 mm from the end of the plasma jet. After exposure, pegs were added to 200 μL PBS in a 96-well plate and biofilms were loosened from the pegs into solution by sonication in an Elmasonic S30 ultrasonic device (Elma Schmidbauer GmbH, Singen, Germany). Samples were sonicated for 10 min at a frequency of 37 kHz, employing the sweep function for even sound field distribution. Biofilm pegs were then removed from the wells and PBS dilutions of the resultant liquid bacterial culture were inoculated onto MHA to determine microbial cell culturability. Sub-lethal treatment times were selected to achieve a 1-log reduction of bacterial cells to∼106 CFU/mL for following antimicrobial susceptibility testing. CAP exposures selected from this were 45 s and 90 s respectively for planktonic and biofilm samples.
Fig. 1.
Diagram of the cold plasma jet configuration depicting the electrodes, plasma plume, and the electrode geometry used for the treatment of a P. aeruginosa biofilm grown on a CBD peg.
2.4. Antimicrobial susceptibility investigation
The effects of CAP pre-treatment on the antimicrobial susceptibility of three P. aeruginosa strains were tested against planktonic or biofilm-forming bacteria. Susceptibility endpoints were obtained for untreated planktonic samples of the following: P. aeruginosa PAO1, P. aeruginosa UCBPP-PA14, and P. aeruginosa NCIMB 10548, according to standard protocols for determination of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). Changes to MIC and MBC in CAP-treated bacterial cultures were then assessed after 45 s exposure to the plasma jet, using the same conditions and jet distance as described previously. For each sample, CAP treated bacterial suspension was used to inoculate a prepared antimicrobial challenge 96-well plate (10 μL per well). Each MIC assay was incubated for 16 h at 37 °C, and MIC and MBC values were recorded according to standard protocols. As a control, colony counts were carried out using PBS dilutions of untreated and CAP-treated samples alone to confirm a ∼1-log reduction in cell numbers and ensure that any improved susceptibility was not due to reduced bacterial cell counts after 45 s CAP exposure.
For biofilm samples, susceptibility endpoints were determined for biofilms of each of the three strains according to standard protocols for the determination of minimum biofilm inhibitory concentration (MBIC) and minimum biofilm eradication concentration (MBEC) [13]. When recording MBIC, wells with inhibited growth were also inoculated onto MHA to obtain an MBC value for bacterial cells released from the biofilm peg into the media. Changes to MBIC, MBC, and MBEC in CAP-treated biofilms were then assessed after 90 s exposure to the plasma jet, by immediate addition to an antimicrobial challenge plate. MBIC, MBC, and MBEC values were recorded. Colony counts were also carried out using additional untreated and CAP-treated biofilm pegs to confirm an effect at 90 s CAP exposure, as described previously.
2.5. Measurement of metabolic activity with isothermal microcalorimetry
To measure heat production as a result of metabolic activity in biofilms, the calScreener microcalorimeter (Symcel, Stockholm, Sweden) was used. To grow biofilms, an overnight culture of P. aeruginosa strains (PAO1, PA14, and PA10548) were centrifuged (5000 rpm, 10 min) and resuspended in MHB to a density equivalent to 5 × 107 CFU/mL. Bacterial suspensions were then added (200 μL) to plastic inserts that fit in the titanium cups of the calScreener and incubated at 37 °C for 24 h. Plastic inserts were then removed, spent media was removed and wells were gently rinsed twice with 0.5 mL PBS, and wells with biofilms were then exposed to CAP at a distance of 15 mm from the bottom of the well plate to the end of the quartz tube. Biofilms were exposed for a total of 90 s, and then fresh sterile MHB was immediately added to each plastic insert and transferred back into the titanium cups, which were placed in a 48-well plate and inserted in the instrument. The heat flow was measured at 37 °C, and the resulting data were analysed using the CalView software (Symcel).
2.6. RNA extraction
RNA extraction from PAO1 was performed using the RiboPure – Bacteria kit (Thermo Fisher Scientific, UK) 4, 6, and 24 h post-CAP treatment. We focused on P. aeruginosa strain PA01 for our transcriptomic analysis as it is a well-studied strain that is commonly used as a model organism for biofilm research.
Spent media from unexposed and post-CAP exposed biofilms at 4, 6, and 24 h were removed. The wells of the 24-well plate were rinsed with 0.5 mL sterile PBS to remove non-adherent cells, followed by the addition of 0.5 mL of the RNAWIZ solution from the RiboPure – Bacteria kit to each well. The biofilm-containing wells were scrapped using the end of a 1 mL pipette tip and transferred to an RNAase-free 1.5 mL microtube containing zirconia beads. The biofilm-RNAWIZ solution was beaten for 10 min at maximum speed using a vortex, after which samples were centrifuged at 4 °C for 2 min. Each biofilm lysate was then transferred to a new 1.5 mL microtube wherein chloroform was added for extraction of RNA. The manufacturer's instructions were followed with minor modifications, which included extending the lysis time to 15 min and increasing the amount of chloroform used for RNA extraction. RNA quantification was performed using the Quant-iT RNA assay kit (Thermo Fisher Scientific, UK). The aqueous layers were treated with DNAse for 1 h at 37 °C and eluted in 30 μL of buffer and centrifuged for 5 min using a spin column. RNA quantification was again performed to determine the final RNA concentration.
2.7. Transcriptomic data processing and analysis
Before RNA sequencing, RNA samples were assessed for quality using a 5400 Fragment analyser (Agilent) and for purity using a Nanodrop™ 2000/2000c spectrophotometer. RNA-Seq was performed in the Genomics Core Technology Unit at Queens’ University Belfast, following standard protocols of the KAPA (Stranded) RNA hYPERprep without Riboerase kit (Roche Diagnostics Ltd, UK) after rRNA removal using the Ribo-Zero Plus rRNA Depletion Kit (Illumina). 200 ng RNA was used as input, and generated libraries were quantified using Kapa Quantification before being normalized and pooled in equimolar amounts. Sequencing was performed on the Illumina NextSeq 500 platform, with 75-bp strand-specific paired-end reads. Sequencing data was returned as trimmed FASTQ files. These reads were mapped onto the P. aeruginosa genome PAO1 [14] using Bowtie 2 with default settings [15]. Mapped reads were counted using featurecounts from the Rsubread package [16].
2.8. Differential gene expression analysis
Statistical analysis of the RNA-Seq dataset was performed in the R environment (4.1.0) [17]. Differential gene expression analysis was performed using DESeq2 [18]. Genes with significant expression were defined based on ± log2 fold change of 1 and an FDR of 0.05 using the Benjamini & Hochberg procedure. GO enrichment of either up or down differentially expressed genes was completed using GOseq [19] using the GO terms from Pseudomonas Genome DB [20]. Heatmaps depicting pyoverdine and pyocyanin transcription were produced in GraphPad Prism 9. Data analyses were performed using R Studio 1.2.5033 platform [21], and bar graphs were generated using ggplot2 library [22].
2.9. Hyperphosphorylated guanosine [(p)ppGpp] determination assay
Here, the presence of the signalling molcule (p)ppGpp was detected to assess the stress response in P. aeruginosa biofilms. To do this, a modified colourimetric assay using a Fenton-like reagent was used [23]. This assay exploited the inhibitory effect of (p)ppGpp on the reaction between 2,2’ -azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) and hydrogen peroxide in the presence of Fe3+. As (p)ppGpp has a greater binding affinity to Fe3+, a positive result for the presence of (p)ppGpp was indicated by inhibition of the blue-green colour resulting from the oxidation of ABTS. For this assay, 50 μL of HAc-NaAC buffer was added to the wells of a 96-well plate, followed by 50 μL of supernatant from CAP-treated bacterial suspension at increasing plasma exposures. 50 μL of each of the following was then added; 5 μM Fe3+ solution, 5 mM ABTS solution, 4 mM H2O2 and sterile water. The absorbance at 414 nm was then measured using a BMG FLUOstar Optima Fluorescence plate reader (BMG Labtech Ltd., Aylesbury, UK) for 30 min as the reaction progressed.
2.10. Pyoverdine and pyocyanin determination
To confirm the transcriptomic changes in pyoverdine and pyocyanin production, the production of both virulence factors were measured experimentally. Pyocyanin and pyoverdine values were normalized according to growth at OD550 following CAP treatment at 4 h, 6 h, and 24 h incubation. Culture supernatants were then collected after centrifugation at 10,000 rpm for 10 min. Pyoverdine production was measured from the culture supernatant using its innate fluorescence at ex. 400 nm, and em. 460 nm using a plate reader (BMG Labtech Ltd., Aylesbury, UK). Pyocyanin absorbance in the supernatant was measured at 550 nm using a plate reader.
2.11. Detection of extracellular ATP and LDH
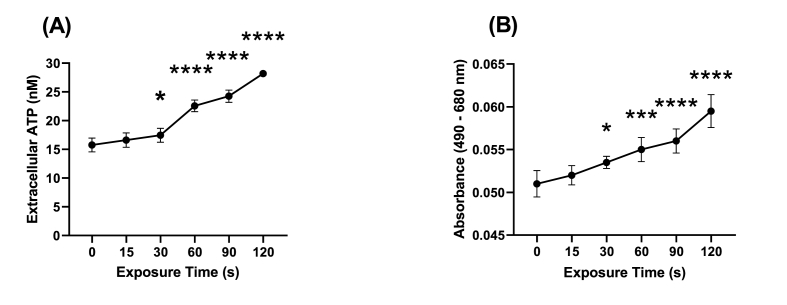
Extracellular ATP and LDH (lactate dehydrogenase) were detected in order to assess the effects of CAP treatment on the membrane integrity and cell viability of P. aeruginosa PAO1 cells. ATP is a marker of cellular damage and release, while LDH is an enzyme that is released from cells during damage or death. By measuring the concentrations of extracellular ATP and LDH in CAP-treated bacterial suspensions at various time points, the study aimed to determine the degree of cellular damage and cytotoxicity induced by CAP treatment. To measure the concentration of extracellular ATP, a firefly luciferase-based bioluminescent assay was used (ATP determination kit; Molecular Probes Inc.). To quantify only the extracellular ATP from bacterial suspension, the procedure was altered to omit the cell-lysing step. A standard curve was generated by measuring the luminescence of standard ATP solutions of varying concentrations with luminescence recorded using a BMG FLUOstar Optima Fluorescence plate reader (BMG Labtech Ltd., Aylesbury, UK). The procedure was then repeated using 50 μL of CAP-treated bacterial suspension from each time point and the amount of ATP was determined by comparison to the standard curve.
Following various durations of CAP treatment of P. aeruginosa PAO1 cells, were quantified LDH concentrations were quantified using the Invitrogen CyQUANT LDH Cytotoxicity Assay kit as per the manufacturer's instructions (Thermo Fisher Scientific Inc.). To prepare the CAP-treated cells, the absorbance obtained from the maximum LDH release control was subtracted from the spontaneous LDH release control and plotted against the cell number to determine the linear range and optimal number of cells. Next, the LDH activity was measured by adding the reaction mixture, incubating for 30 min, adding the stop solution, and calculating the difference between the absorbance at 680 nm and the absorbance at 480 nm.
2.12. Statistical analysis
The results for Fig. 5 the average means and standard deviation from biological triplicates was compared using a two-way ANOVA and Bonferroni's test. For Fig. 2, Fig. 10, Fig. 11, Fig. 12, a one-way ANOVA, with Dunnett's test against the untreated control were used. P values of * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001), and **** (P ≤ 0.0001) indicate statistical significance. GraphPad Prism 9.5.4 was used for all analysis.
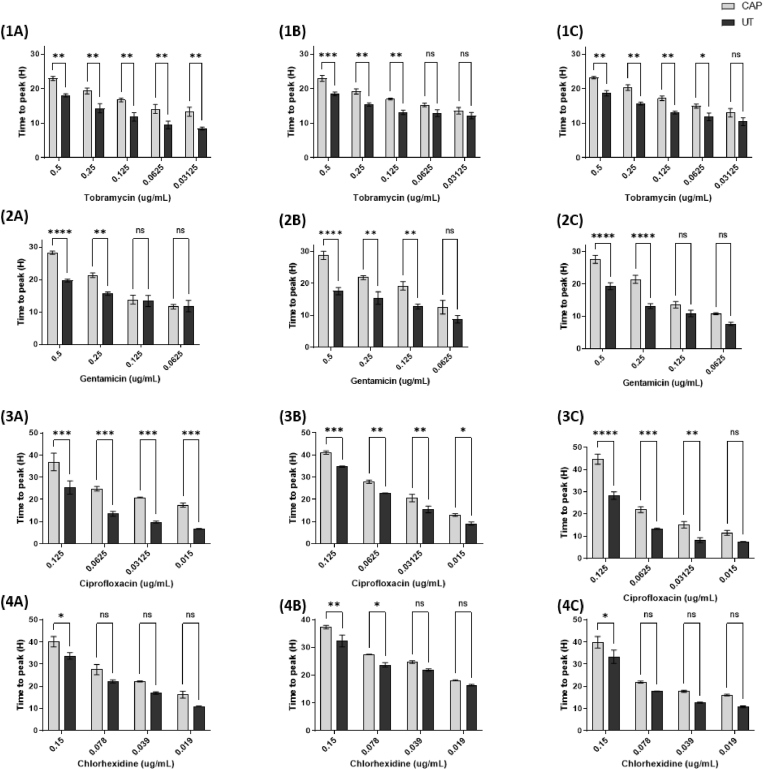
Fig. 5.
Time to peak metabolism (h) of the three P. aeruginosa biofilms - PAO1 (A), PA14 (B), and PA10548 (C) against tobramycin [1], gentamicin [2], ciprofloxacin [3], and chlorhexidine [4]. Biofilms were either untreated or CAP treated for 90 s followed by the addition of antimicrobial. Error bars indicate the mean ± standard deviation determined from biological replicates, asterisks indicate significant differences between the untreated and CAP-treated groups (p < 0.05, p < 0.01, p < 0.001, and p < 0.0001) using one-way ANOVA and Dunnett's post-test analysis (n = 3).
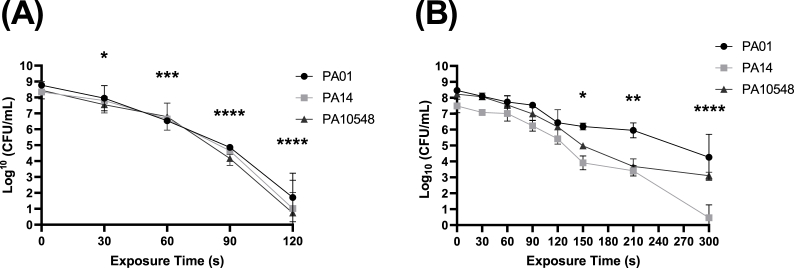
Fig. 2.
Reductions (log10 CFU/mL) in P. aeruginosa culturability over time (s) (A) planktonic (B) biofilm (strains PAO1, PA14, and PA10548) after CAP treatment assessed by colony count assay. Error bars indicate the mean ± standard deviation determined from biological replicates (n = 3). The significance levels were the same for each strain and therefore only one set is displayed. Asterisks denote significant differences between the relevant exposure times compared to baseline (t = 0).
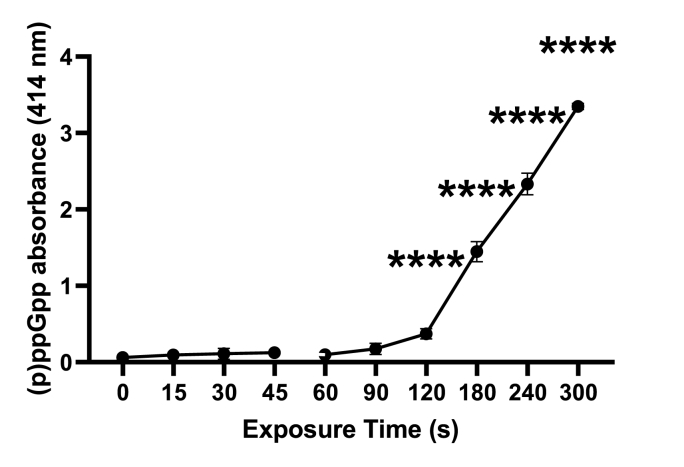
Fig. 10.
Absorbance of (p)ppGpp measured at 414 nm from PAO1 cells following different CAP exposure times (s). Error bars indicate the mean ± standard deviation determined from biological replicates (n = 3). Asterisks denote significant differences between the relevant exposure times compared to baseline (t = 0).
Fig. 11.
Measurement of time-dependant (A) extracellular ATP and (B) LDH leakage following CAP treatment of PAO1 cells. Error bars indicate the mean ± standard deviation determined from biological replicates (n = 3). Asterisks denote significant differences between the relevant exposure times compared to baseline (t = 0).
Fig. 12.
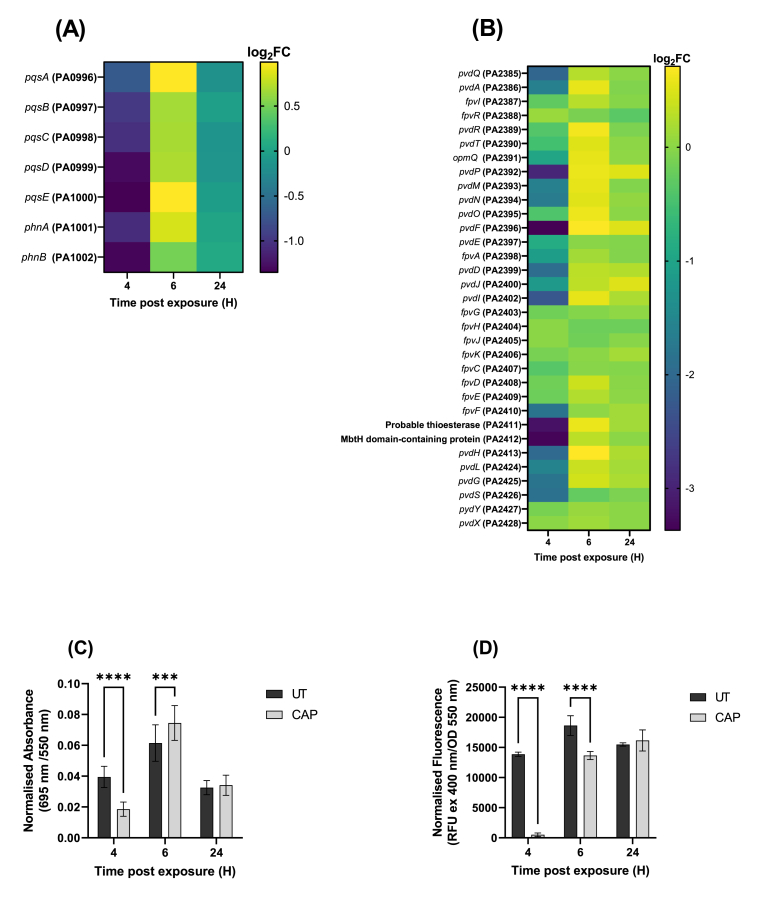
Pyoverdine and pyocyanin production by PAO1 at 4 h, 6 h, and 24 h post CAP treatment. Heatmap depicting changes in the transcript expression with genes related to (A) pyocyanin expression (B) and pyoverdine expression. To confirm these phenotypes, pyocyanin and pyoverdine concentrations were measured using absorbance and fluorescence for (C) pyocyanin (D) and pyoverdine. The error bars indicate the mean ± standard deviation determined from biological replicates (n = 3).
3. Results
3.1. Effect of CAP on P. aeruginosa planktonic cells and biofilms
To assess the antimicrobial activity of CAP on P. aeruginosa strains in both planktonic and biofilms phenotypes, cultures of each strain were exposed to increasing doses of CAP as a function of time. As shown in Fig. 2A, all three strains of P. aeruginosa exhibited comparable susceptibility to CAP treatment when in planktonic form, with near-complete eradication (8–9 logs) after 120 s of treatment. When grown in biofilms (shown in Fig. 2B), the bacteria displayed an elevated tolerance towards CAP treatment with a 4-log reduction in PAO1, a 5-log reduction in P10548, and up to 7-log reduction in PA14 biofilm after 300 s of treatment. These results suggest that CAP is able to kill P. aeruginosa in planktonic form but may be less effective against biofilms.
3.2. Enhanced antimicrobial efficacy with sub-lethal CAP pre-treatment
In order to determine whether a combination of antimicrobials with CAP would result in increased bacterial killing we conducted cell culturability assays. Using the data obtained in Fig. 2, the sub-lethal treatment time (the CAP treatment duration that results in a 1-log or less reduction in colony culturability (CFU/mL)), for planktonic cells this was determined to be 45 s, and for biofilms, 90 s. The MIC and MBC values for tobramycin, gentamicin, ciprofloxacin, and chlorhexidine were initially determined for each strain of P. aeruginosa, and compared to those obtained for planktonic bacteria that were pre-treated with sub-lethal CAP exposure (Fig. 3).
Fig. 3.
MIC and MBC determinations (μg/mL) of tobramycin (A), gentamicin (B), ciprofloxacin (C), and chlorhexidine (D) against P. aeruginosa strains PAO1, PA14, and PA10548. Strains were either untreated (UT) or CAP treated for 45 s, immediately followed by the addition of one of the antimicrobial compounds for 16 h. These results are based on endpoint visual assessment (no error bars) based on three biological replicates (n = 3).
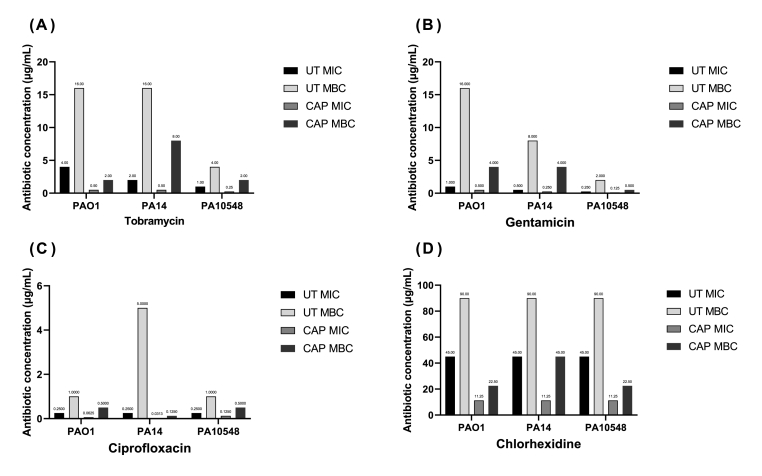
The results from Fig. 3 show that CAP pre-treatment enhanced the susceptibility of planktonic P. aeruginosa to antimicrobial agents. For all three strains tested, tobramycin exhibited the greatest enhancement in antimicrobial activity, with its MIC decreasing eightfold and MBC decreasing similarly. Gentamicin was less affected by CAP pre-exposure, but still showed moderate reduction in MIC and MBC values. Ciprofloxacin in combination with CAP also displayed a reduction in susceptibility similar to gentamicin. Finally, pre-treatment with CAP followed by chlorhexidine was the least pronounced in improving the susceptibility of the P. aeruginosa strains. Comparison of MIC and MBC values for untreated and sub-lethal CAP pre-treated P. aeruginosa are summarised in Table 1.
Table 1.
Comparison of MIC and MBC values of PAO1, PA14, and PA10548, untreated and following sub-lethal CAP treatment.
| Concentration (μg/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tobramycin |
Gentamicin |
Ciprofloxacin |
Chlorhexidine |
||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | ||
| PAO1 | UT | 4 | 16 | 1 | 16 | 0.25 | 1 | 45 | 90 |
| CAP | 0.5 | 2 | 0.5 | 4 | 0.0625 | 0.5 | 11.25 | 22.5 | |
| PA14 | UT | 2 | 16 | 0.5 | 8 | 0.25 | 5 | 45 | 90 |
| CAP | 0.5 | 8 | 0.25 | 4 | 0.0313 | 0.125 | 11.25 | 45 | |
| PA10548 | UT | 1 | 4 | 0.25 | 2 | 0.25 | 1 | 45 | 90 |
| CAP | 0.25 | 2 | 0.125 | 0.5 | 0.125 | 0.5 | 11.25 | 22.5 | |
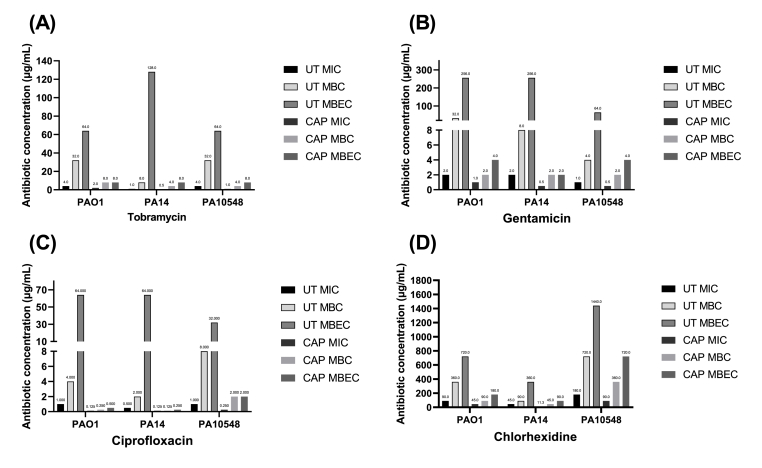
Pre-treatment with CAP also enhanced the susceptibility of P. aeruginosa biofilms to antimicrobial agents, reducing the MIC, MBC, and MBEC values for all four antimicrobials and all three strains of bacteria tested (Fig. 4). The specific changes in antibiotic concentrations are shown in Table 2. The most effective combinational therapy was tobramycin against PA14, with the MIC reduced from 2 μg/mL to 0.5 μg/mL, the MBC reduced from 8 μg/mL to 2 μg/mL, and the MBEC reduced from 256 μg/mL to 2 μg/mL. The least effective therapy was chlorhexidine against PA10458, with the MIC reduced from 180 μg/mL to 90 μg/mL, the MBC reduced from 720 μg/mL to 360 μg/mL, and the MBEC reduced from 1440 μg/mL to 720 μg/mL. These results suggest that pre-treatment with CAP may significantly enhance the effectiveness of certain antimicrobial treatments against P. aeruginosa biofilms.
Fig. 4.
MIC, MBC, and MBEC determinations (μg/mL) of tobramycin (A), gentamicin (B), ciprofloxacin (C), and chlorhexidine (D) for biofilms of P. aeruginosa strains PAO1, PA14, and PA10548. Strains were either untreated (UT) or CAP treated for 90 s followed by the addition of the antimicrobial for 16 h. Results are based on endpoint visual assessment (no error bars) based on three biological replicates (n = 3).
Table 2.
Comparison of MIC, MBC, and MBEC values of PAO1, PA14, and PA10548 biofilms, untreated and following sub-lethal CAP treatment.
| Concentration (μg/mL) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tobramycin |
Gentamicin |
Ciprofloxacin |
Chlorhexidine |
||||||||||
| MIC | MBC | MBEC | MIC | MBC | MBEC | MIC | MBC | MBEC | MIC | MBC | MBEC | ||
| PAO1 | UT | 4 | 32 | 64 | 2 | 32 | 256 | 1 | 4 | 64 | 90 | 360 | 720 |
| CAP | 2 | 8 | 8 | 1 | 2 | 4 | 0.125 | 0.25 | 0.5 | 45 | 90 | 180 | |
| PA14 | UT | 1 | 8 | 128 | 2 | 8 | 256 | 0.5 | 2 | 64 | 45 | 90 | 360 |
| CAP | 0.5 | 4 | 8 | 0.5 | 2 | 2 | 0.125 | 0.125 | 0.25 | 11.25 | 45 | 90 | |
| PA10548 | UT | 4 | 32 | 64 | 1 | 4 | 64 | 1 | 8 | 32 | 180 | 720 | 1440 |
| CAP | 1 | 4 | 8 | 0.5 | 2 | 4 | 0.25 | 2 | 2 | 90 | 360 | 720 | |
3.3. Metabolic changes to P. aeruginosa biofilms following sub-lethal CAP exposures
To determine whether observed increase in antimicrobial susceptibility after sub-lethal CAP treatment was due to changes in bacterial metabolism isothermal microcalorimetry was used to determine the time-to-peak (TTP) metabolism. TTP data is interpreted from thermograms that are unique to each bacteria and the environmental condition in which they grow (Fig. S-1 for representative thermograms). TTP metabolism is an important indicator that reflects the time required for adaptation to stress and is therefore an indirect measure of cell recovery. TTP metabolism allows us to quantify the time for bacterial cells to recover following antimicrobial treatment alone compared CAP and antimicrobial treatment. As shown in Fig. 5, pre-treatment with CAP increased the TTP metabolism for all four antimicrobial agents against all three strains of P. aeruginosa and confirms enhanced susceptibility of P. aeruginosa biofilms to antimicrobial agents after CAP treatment.
With tobramycin alone, there was a dose-dependent effect with an increased time-to-peak that was proportional to the concentration of antimicrobial used (Fig. 5). A CAP pre-treatment further prolonged the time-to-peak metabolism in each strain of P. aeruginosa tested. This effect is also observed at lower concentrations of antibiotic such as 0.0625 μg/mL and 0.03125 μg/mL, the TTP metabolism increased from 9.49 – 12.93 h to 14.11–16.3 h and 8.45–12.17 h to 13.34–13.73 h for each antimicrobial respectively. Gentamicin also increased the time-to-peak metabolism, particularly at higher concentrations of 0.5 μg/mL, which saw an increase from 17.63 - 19.73 h to 26.62–28.79 h across the three strains. In the case with ciprofloxacin and chlorhexidine, the results showed that the TTP metabolism can be detected for longer than 40 h after exposure to the highest concentrations of these antimicrobials - 0.125 μg/mL and 0.15 μg/mL respectively. This suggests that P. aeruginosa is able to recover from exposure to these antimicrobials, and CAP pre-treatment prolongs this recovery time. Overall, these results provide insight into how P. aeruginosa metabolically responds to different antimicrobials and highlights the potential for biofilm recovery.
3.4. Transcriptomic analysis P. aeruginosa biofilms following sub-lethal CAP treatment
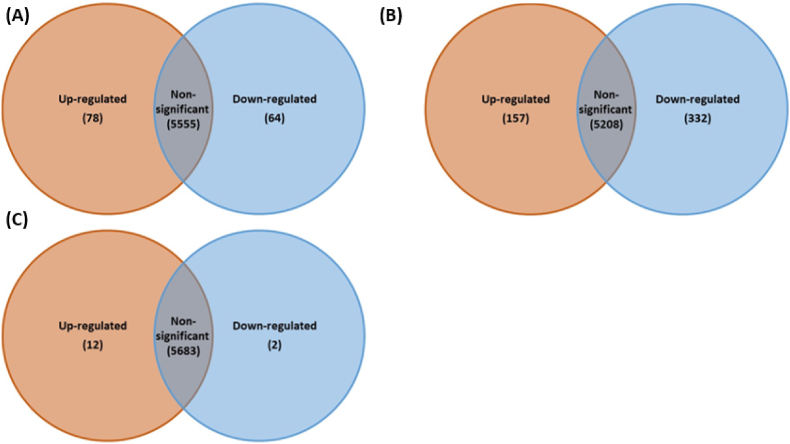
We conducted transcriptomic analysis to investigate changes in gene expression that may account for the enhanced antimicrobial susceptibility observed in P. aeruginosa biofilms following CAP treatment for 90 s. Our analysis (Fig. 6) showed significant changes in gene expression at different time points after treatment, with the number of differentially expressed genes peaking at 6 h post-treatment. At 4 h post CAP treatment, 78 genes were upregulated (>1 log2FC) and 64 genes were downregulated (<-1 log2FC) compared to control untreated biofilm cells as shown in volcano plots (Fig. 6). After 6 h, the number of upregulated genes increased to 157, and the number of downregulated genes increased to 322. At 24 h post-treatment, only 12 upregulated genes and 2 downregulated genes, a time at which cellular transcription had essentially returned to ‘normal’ or non-exposed control, indicating recovery from the cold plasma. These results indicate that CAP treatment causes significant changes in gene expression in P. aeruginosa biofilms during the times at which the bacteria exhibit increased antimicrobial susceptibility. These results are also displayed as a volcano plots with the overall spread of gene expression following CAP exposure (Fig. S-2).
Fig. 6.
Differential gene expression in P. aeruginosa (PA01) biofilms after sub-lethal CAP exposure at different time points post-treatment. (A) 4 h post-exposure, (B) 6 h post-exposure, (C) 24 h post-exposure.
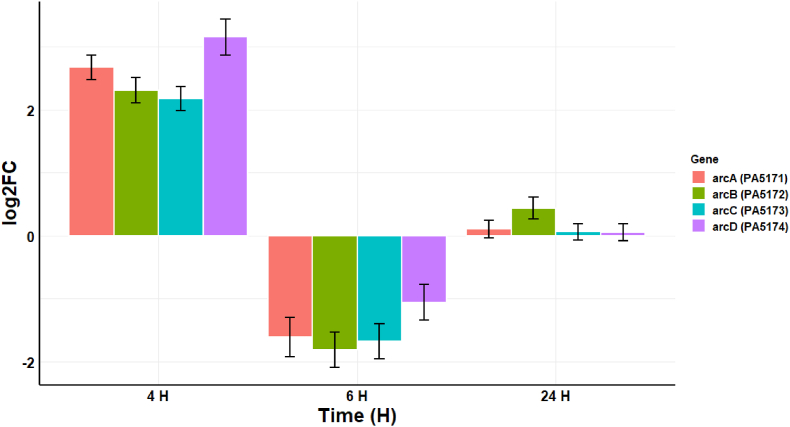
Transcriptomic analysis of the 4 h post-CAP treatment conditions revealed an upregulation in the arginine deaminase system (ADS) evidenced by the increased transcription of PA5170 (log2FC 3.15), PA5171 (log2FC 2.67), PA5172 (log2FC 2.31), and PA5173 (log2FC 2.17) (Fig. 7). The system has three main enzymes: arginine deiminase (PA5171) (arcA), ornithine carbamoyltransferase (PA5172) (arcB), and carbamate kinase (PA5173) (arcC), and an inducible transport system involving ornithine-arginine antiport across the cell membrane (PA5171) (arcD) [24]. By 6 h, there is a down-regulation in the ADS pathway, and this may be an indicator that the P. aeruginosa biofilm has transitioned away from an initial acute stress response to CAP, and by 24 h there are no significant difference in gene expression in this pathway. This would be consistent with a response to the acute oxidative insult of cold plasma exposure, whereby the biofilm is exposed to the primarily short-lived RONS generated by the plasma jet used in this study and described elsewhere [1].
Fig. 7.
Changes in the transcription (log2FC) of four genes related to the arginine deaminase system at 4 h, 6 h, and 24 h post-exposure to CAP. The error bars indicate the mean ± standard deviation determined from biological replicates (n = 3).
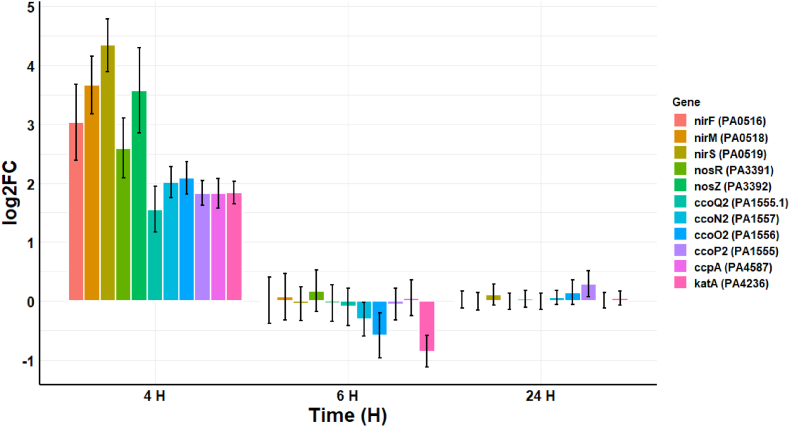
The antibacterial effects of CAP are primarily mediated via short- and longer-lived RONS [25]. These data indicate that P. aeruginosa counteracts the effects of RONS by transitioning to anaerobic respiration, and upregulation of genes related to nitrate and nitrite denitrification at 4 h post-CAP treatment are shown in Fig. 8. These genes include NosR (PA3391) (log2FC 2.60), which is a transcriptional regulator for nitrous-oxide reductase, and nitrous-oxide reductase (PA3392) (log2FC 3.57). There is also upregulation of genes involved in nitrite denitrification, such as protein NirF (PA0516) (log2FC 3.03), responsible for biosynthesis of heme d1 of nitrite reductase, cytochrome c551 (PA0518) (log2FC 3.66), which functions as an electron donor for cytochrome cd1 in nitrite and nitrate respiration, and nitrite reductase (PA0519) (log2FC 4.34). Bacteria also employ a variety of ROS-degrading enzymes to combat oxidative stress, including superoxide dismutases, catalases, alkylhydroperoxidases, superoxide dismutases, superoxide reductases, and peroxidases [[26], [27], [28], [29]]. In the case of P. aeruginosa, several genes that encode for cbb3-type cytochrome c oxidase (PA1555, PA1555.1, PA1556, and PA1557) were upregulated at 4 h post-treatment (log2FC 1.56–2.08). There was also an increase in cytochrome c551 peroxidase (PA4587) (log2FC 1.83) and catalase (PA4236) (log2FC 1.84), both of which protect against toxic peroxides and decompose hydrogen peroxide, respectively. The impact of RONS on P. aeruginosa is reduced at 6 h and 24 h post-treatment, indicating that the bacteria have successfully mitigated the effects of these longer-lived species.
Fig. 8.
Log2FC in the transcription of 11 genes related to CAP generated reactive oxygen and reactive nitrogen species defence at 4 h, 6 h, and 24 h post-exposure. The error bars indicate the mean ± standard deviation determined from biological replicates (n = 3).
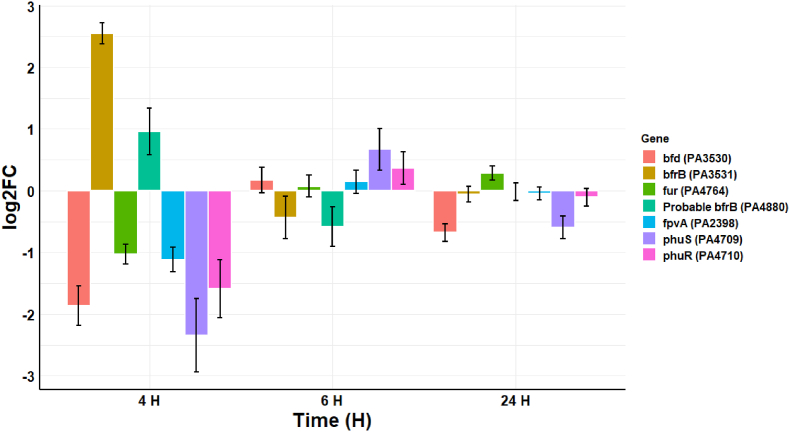
The transcriptome of P. aeruginosa in response to CAP-produced RONS shows a shift in iron acquisition proteins (Fig. 9), probably as a protective mechanism to prevent the generation of toxic hydroxyl radicals (OH•) through Fenton-like biochemical reactions. The increased transcription of bacterioferritin (bfrB) (PA3531) (log2FC 2.55) and another probable bacterioferritin (PA4880) (log2FC 0.96), suggests that there is increased storage and scavenge of iron in response to RONS. This is further supported by the decrease in the transcription of genes involved in the uptake of iron, such as bacterioferritin-associated ferredoxin (bfd) (PA3530) (log2FC −1.87), Ferric uptake regulator (fur) (PA4764) (log2FC −1.03), and ferripyoverdine receptor (fpvA) (PA2398) (log2FC −1.12). In addition, there is downregulation of the TonB-dependent outer membrane receptor that transports heme to the periplasm (phuR) (PA4709) (log2FC −2.35) and the phuS gene (PA4709) (log2FC −2.35), which are part of the phu system responsible for the intracellular trafficking of iron. The phuS gene has been described as acting as a control valve to regulate the influx of heme into the cell [30]. Overall, these results suggest that P. aeruginosa responds to RONS by reducing uptake and increasing storage of iron to protect itself from the toxic effects of RONS.
Fig. 9.
Log2FC in the transcription of 7 genes related to iron storage at 4 h, 6 h, and 24 h post-exposure. The error bars indicate the mean ± standard deviation determined from biological replicates (n = 3).
Several genes related to cell stress and cell morphology also had significant changes in their transcription at 4 h post-CAP exposure (Table 3).
Table 3.
Transcriptional changes of fourteen genes at 4 h post CAP exposure in PAO1 biofilms. The table lists the genes, along with their log2FC values.
| Gene ID | log2FC | Standard error | Annotation |
|---|---|---|---|
| PA4067 | 2.208755 | 0.172204 | Outer membrane protein OprG |
| PA3692 | 1.068882 | 0.237976 | Lipotoxin F, LptF |
| PA4481 | 1.100758 | 0.201629 | Rod shape-determining protein MreB |
| PA0141 | 1.495912 | 0.256964 | Polyphosphate kinase PPK2 1 |
| PA3724 | 1.475389 | 0.211961 | LasB, Elastase |
| PA0962 | 1.458929 | 0.195674 | Probable DNA-binding stress protein |
| PA1092 | 1.206224 | 0.151888 | B-type flagellin |
| PA1871 | 1.11844 | 0.504509 | LasA, protease |
| PA2685 | −1.07631 | 0.22685 | Type VI secretion system spike protein VgrG1c |
| PA2426 | −1.84735 | 0.532096 | Sigma factor PvdS |
| PA5255 | −1.41887 | 0.180495 | Transcriptional regulatory protein AlgQ (Alginate regulatory protein AlgR2) |
The upregulation of MreB (PA4481) (log2FC 1.10) and outer membrane proteins such as Lipotoxin F, LptF (PA3692) (log2FC 1.07), suggests that the cells may be attempting to repair or strengthen their cell membranes in response to the CAP treatment. Another major outer membrane protein, OprG (PA4067) which acts as a porin was also upregulated (log2FC 2.21). The upregulation of outer membrane proteins serves as a resistance mechanism for cells against hydrogen peroxide as previously demonstrated by Damron et al., 2009 [31]. B-type flagellin are important adhesion components of the outer membrane, so their upregulation post-CAP treatment suggests increased motility towards fresh nutrients or repair of damaged flagella. Regarding cell stress, the upregulation of a probable DNA-binding stress protein (PA0962) (log2FC 1.45) is a strong indicator of stress response of CAP to cells. The gene (PA2685) which codes for the Type VI secretion system (T6SS) spike protein VgrG1c was downregulated (log2FC −1.85) following CAP treatment and suggests that this may play a role in preventing ROS-dependent lethality in P. aeruginosa cells. Additionally, two proteases, LasB elastase (PA3724) and LasA protease (PA1871) were upregulated with log2FC values of 1.48 and 1.12 respectively. Previous research has shown that CAP treatment can result in the downregulation of protease activity, and so the upregulation of these may be a sign of cells returning to their normal state [32]. At an earlier time point, the protease activity could still be downregulated, indicating that the cells are still under stress. LasB and LasA proteases are virulence factors that play a role in the pathogenesis of P. aeruginosa infections, so understanding their regulation in response to treatment is important consideration when using CAP for the management of such infections.
3.5. Ribosomal inactivation and persister cell resuscitation
The gene PA0141, also known as polyphosphate kinase (PPK2), performs a variety of functions in P. aeruginosa including stress survival [33]. To study the potential role of PPK2 in persister cell resuscitation, a (p)ppGpp stringent response assay was conducted. The results of this assay, shown in Fig. 10, indicate for the first time that PAO1 cells undergo a stringent response, as indicated by increased production of (p)ppGpp when exposed to CAP, which suggests the upregulation of PPK2 class 1 in response to stress. The increase in ppGpp leakage after 120 s may be a sign of CAP-induced stress, potentially via damage to the cell membrane, which could lead to the release of ATP while the cells remain viable [34]. Overall, these results support the idea that PPK2 plays a role in the stress response and persister cell resuscitation in P. aeruginosa.
Based on transcriptomic analysis, we focused on the upregulation of 15 genes annotated as 30S and 50S ribosomal proteins, which we observed 6 h after CAP treatment (supplementary data Fig. S3), with a log2FC ranging from 1.01 to 1.38. While the exact function of these genes is unclear, six are within a 10-gene operon (PA4250 - PA4259) that was previously thought to be negatively regulated during the stringent response [35]. This suggests that the upregulation of these genes may play a role in the resuscitation of persister cells in P. aeruginosa.
3.6. Changes in permeability of P. aeruginosa cells using ATP and LDH assays
To further investigate potential changes in the cell membrane, indicated by the upregulation of MreB (PA4481), outer membrane proteins and (p)ppGpp, leakage of ATP and LDH following CAP treatment in P. aeruginosa cells was investigated (Fig. 11). The increase in ATP and LDH concentrations in the extracellular milieu indicates time-dependant loss of integrity of the cell membrane of P. aeruginosa following CAP exposure.
3.7. Changes in the production of virulence factors following CAP exposure
The genes PA2385 - PA2410 [36] within the genome of PAO1 are related to pyoverdine production, and the genes PA0996 - PA1002 are associated with to pyocyanin and PQS production [37]. Pyoverdine is a fluorescent siderophore produced by P. aeruginosa involved in iron uptake [38], while pyocyanin is a redox-active pigment that has antimicrobial activity and can enhance the production of ROS in the bacteria [39]. Following CAP treatment an upregulation in a number of genes related to the biosynthesis of pyocyanin and pyoverdine and to genes related to relevant pyocyanin and pyoverdine quorum sensing molecules at 4 h post-exposure (see Fig. 12) was observed. After 6 h, there was a decrease in many of these genes, but these were not considered significant. At 24 h, there is no transcriptomic difference in the plasma treated and untreated biofilm, again indicating recovery from initial sub-lethal cold plasma insult. To confirm these transcriptomic changes in pyoverdine and pyocyanin production P. aeruginosa biofilms were exposed to 90 s CAP treatment and pyoverdine and pyocyanin concentrations were measured. Experimental data (Fig. 12C – D) confirmed these transcriptomic expressions and quantify the extent of reduction of each virulence factor with pyoverdine production nearly absent at 4 h post CAP treatment.
4. Discussion
Our results illustrate that P. aeruginosa biofilms vary in their tolerance to CAP treatment, with PAO1 displaying the greatest tolerance, followed by P10548, and then up to 7-log reduction with PA14 after 300 s exposure. This strain variation in CAP susceptibility is consistent with previous research demonstrating differences in biofilm-formation and EPS production among different strains of P. aeruginosa leading to variation in response to CAP exposure [8,40]. Our original hypothesis that the increased efficacy of cationic antimicrobials following CAP exposure was due to disruption of molecular interactions between the cationic antimicrobials and anionic biofilm components, including alginate, was partially supported by isothermal titration calorimetry data (data not shown). However, transcriptomic analysis suggests a potentially more complex mechanism of cold plasma-antimicrobial synergy, and disruption of such interactions by CAP exposure alone is likely to contribute to a much broader mechanism of synergy. The differences in the efficacy of antimicrobials may be due to their specific mechanisms of action. The combination of CAP and antibiotics was effective at altering the metabolic activity of the biofilms, as demonstrated by increased TTP metabolism at every concentration of antimicrobial used, indicating enhanced susceptibility against the three strains of P. aeruginosa tested. Ciprofloxacin and gentamicin caused the greatest increase in the TTP metabolism when used in higher concentrations, while tobramycin was consistently able to extend the TTP metabolism almost all concentrations tested. Chlorhexidine was not as effective in significantly increasing the TTP metabolism. These findings suggest that the observed increased susceptibility induced by CAP exposure may be due to improved penetration of antibiotics inside the cell, ultimately leading to decreased MBEC values.
The upregulation of ADS in P. aeruginosa biofilms neutralises the acidification caused by CAP-generated RONS, potentially protecting cells superoxide or hydrogen peroxide toxicity normally associated with aerobic respiration. In the oral microbiome, ADS regulates pH homeostasis at neutrality via ammonia production, protecting less acid-tolerant organisms [41]. ADS also produces ATP by converting arginine into ornithine, ammonia, and carbon dioxide [42]. Arginine was previously reported to increase metabolic activity in anaerobic P. aeruginosa biofilms increased ciprofloxacin and tobramycin susceptibility potentially, while a combination of zinc and arginine was effective at reducing dental plaque biofilms. Nitrate displayed a similar effect in the absence of oxygen [43]. It would be interesting to investigate whether plasma devices that produce high quantities of nitrates display a similar effect on P. aeruginosa biofilms [44].
Bacteria produce enzymes to degrade RONS, and their precursors, maintaining stress levels within a tolerable range. Transcriptomic analysis of P. aeruginosa exposed to CAP demonstrated an upregulation of cbb3 oxidases, catalases, and peroxidases, and denitrification to mitigate oxidative stress, degrade antibacterial RONS, while upregulating energy production via anaerobic respiration. Cbb3 oxidases in P. aeruginosa are highly expressed under anoxic conditions [45], altering anaerobic growth, denitrification process, and cell morphology [46]. CAP treatment shifts P. aeruginosa to anaerobic respiration via the glyoxylate shunt pathway and nitrate reduction, evident by the downregulation of fumarate hydratase class II 2 (PA4470) (log2FC −1.03), aconitate hydratase A (PA1562) (log2FC −1.04), citrate synthase (PA0795) (log2FC −1.06), and isocitrate lyase (PA2634) (log2FC −1.09) (data not shown). Additionally, differential expression at 6 h post-treatment suggest recovery from an anoxic state. The glyoxylate cycle allows P. aeruginosa to use acetate and fatty acids as carbon sources, which are inducible to oxidative stress [47], to avoid ROS production. Iron is crucial for P. aeruginosa growth, and critical in mitigating damage from RONS during oxidative stress. CAP treatment causes P. aeruginosa to oxidise soluble iron (Fe2+ to Fe3+) and store it as ferrihydrite or ferric phosphate, while also increasing bacterioferritin to further reduce free iron concentrations [48]. While iron is important for promoting twitching motility and a reduced biofilm thickness [49], a high concentration of iron during the lag phase can make cells transiently sensitive to oxidative stress [50].
Data from this study demonstrates for the first time that CAP treatment triggers a stringent response in P. aeruginosa cells, potentially leading to the development of persister cells within the biofilm, which can subsequently resuscitate. A CAP-induced stringent response has not previously been described in bacteria. Persister cells form within regions of the biofilm with low nutrients, antibiotic insult, and oxidative stress, and play a role in chronic bacterial infections [51]. Indeed, the second phase of the biphasic kill curve (Fig. 2 (B)), and widely reported in the literature, is suggestive of the development of a persister cell phenotype [52]. CAP treatment upregulates genes involved in the production of (p)ppGpp (confirmed in vitro), and polyphosphate (polyP), which are known to inhibit protein synthesis, increase efflux pump activity, and increase cells tolerance to antimicrobials [53] [54]. Surviving persister cells downregulate the TCA and activate the glyoxylate shunt [55]. The upregulation of expression of ribosome modulation factor at 4 h (PA3049) (log2FC 1.73), induced by ppGpp, alters ribosomal RNA structures, impairs protein synthesis, and upregulates stress-related genes, leading to persister cell formation [56]. At 6 h post-treatment, gene expression of ribosome modulation factor decreases (log2FC −2.73), indicating persister cell resuscitation. The transcriptional response plays a crucial role in persister cell development and resuscitation after CAP treatment. Liao et al. previously demonstrated that CAP-induced oxidative stress in Staphylococcus aureus led to a viable but non-culturable state [57], which represents the same dormant phenotype as the persister term [58]. The upregulation of PPK2 class I (PA0141) (log2FC 1.5) at 4 h post CAP treatment suggests that polyP granules are metabolised to provide GTP as an energy reserve in the recovering biofilm. The downregulation of AlgQ (log2FC −1.42) at 4 h, which regulates alginate biosynthesis and other virulence factors indicates that alginate biosynthesis did not occur [53] and confirmed by lack of differential expression of other alginate biosynthesis genes (Supplementary Tables S–1). The downregulation of AlgQ may result in the downregulation of the extracellular proteases LasA and LasB, and pyoverdine and pyocyanin, as well as their sigma factor PvdS (log2FC −1.85). Interestingly, LasB has been shown to play a role in alginate biosynthesis and biofilm formation in P. aeruginosa [59,60].
CAP treatment resulted in downregulation of pyoverdine and pyocyanin genes at 4 h post-exposure, further decreasing at 6 h post-exposure, with full recovery of regular production observed by 24 h. The reduction in pyoverdine biosynthesis may be linked to the increased bacterioferritin production, which lowers free iron concentration in the P. aeruginosa microenvironment, limiting cellular toxicity. Pyocyanin causes oxidative stress in host cells by disrupting host catalase and mitochondrial electron transport, and therefore its decrease may be to prevent oxidative stress in bacterial cells. Experimental studies conducted here and previously by Wang et al. (2020) confirmed these results [61]. Kašparová et al., found that Las-B elastase activity was reduced up to 82% after 15 min CAP treatment, and protease activity and pyocyanin production were completely inhibited after 60 min treatment [32].
Unsurprisingly, following CAP treatment, P. aeruginosa upregulates stress genes, including DNA-binding stress proteins and the spike protein VgrG1c, which is part of the T6SS used to translocate effector proteins into adjacent cells. VgrG1c has been shown to have ROS-dependent lethality in the K. pneumoniae T6SS [62], and its’ the downregulation (log 2FC −1.85) following CAP treatment may prevent ROS-dependent lethality in P. aeruginosa cells, although the exact mechanism remains unclear.
Transcriptomic analysis revealed that CAP upregulates genes involved in the determination of cell shape such as MreB, Lipotoxin F, OprG, and B-type flagellin. Furthermore the increased concentration of extracellular ATP and LDH following CAP treatment suggest membrane damage/perturbations. CAP may cause damage or temporary perturbations of cell membranes, making bacteria more susceptible to antimicrobial agents, facilitating their penetration or translocation into bacterial cells. The increased density of cells within biofilms may increase this effect, resulting in the observed higher reductions in MIC, MBC, and MBEC values compared to planktonic cells. While PA10548 is likely to display a similar transcriptomic profile due to its high genomic similarity to PAO1, PA14 possesses over 800 unique genes that may affect its biological processes and gene expression differently against CAP [11,40].
This study highlights the potential of using CAP treatment alongside conventional antibiotics as a synergistic approach to effectively control biofilms, as it may be possible to eradicate the biofilm completely, including persister cells, and prevent the recurrence of infection. These results have potential clinical implications for biofilm control, since CAP pre-treatment has been shown to reduce the concentration of antimicrobial needed to effectively eradicate a biofilm, thereby potentially reducing side effects, toxicity, and antimicrobial use. CAP treatment may also prolong the time for biofilm maturation/recovery, making it a promising alternative to conventional antibiotics alone, for the treatment of chronic wounds or persistent and recurrent infections in, for example, CF patients, implant or indwelling device infections. The study provides, for the first time, novel mechanistic insights into the mode of antibacterial action of CAP and P. aeruginosa response to and recovery from CAP exposure, as well as potential mechanisms of CAP-biofilm antimicrobial synergy.
Funding
Research reported in this publication was supported by the National Institutes of Health (NIH) under award numbers RO1 AR076941 (NH and TF) and the Northern Ireland HSC Research & Development Division, Public Health Agency award STL/5350/17, as part of a Tripartite Grant to BG and NH and TF. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or HSC/PHA.
CRediT authorship contribution statement
Jordanne-Amee Maybin: and performed experimental work. Thomas P. Thompson: and performed experimental work. Padrig B. Flynn: completed the RNA extraction. Timofey Skvortsov: provided support with transcriptomic analysis. Noreen J. Hickok: Writing – review & editing, guided experimental work and contributed to writing and reviewing the manuscript. Theresa A. Freeman: Writing – review & editing, guided experimental work and contributed to writing and reviewing the manuscript. Brendan F. Gilmore: Writing – review & editing, guided experimental work and contributed to writing and reviewing the manuscript.
Declaration of competing interest
The authors of this manuscript declare that they have no financial or personal relationships that may be perceived as influencing their work. We confirm that all authors have contributed significantly to the planning, execution, and writing of the manuscript. We also confirm that the content of this manuscript has not been published or submitted for publication elsewhere.
Acknowledgements
We are thankful to Symcel for initial loan of the CalScreener device and their assistance regarding technical support and data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100122.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Gilmore B.F., Flynn P.B., O'Brien S., Hickok N., Freeman T., Bourke P. Cold plasmas for biofilm control: opportunities and challenges. Trends Biotechnol. 2018;36(6):627–638. doi: 10.1016/j.tibtech.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Flynn P.B., Higginbotham S., Nid’a H.A., Gorman S.P., Graham W.G., Gilmore B.F. Bactericidal efficacy of atmospheric pressure non-thermal plasma (APNTP) against the ESKAPE pathogens. Int J Antimicrob Agents. 2015;46(1):101–107. doi: 10.1016/j.ijantimicag.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 4.Peyrusson F., Nguyen T.K., Najdovski T., Van Bambeke F. Host cell oxidative stress induces dormant Staphylococcus aureus persisters. Microbiol Spectr. 2022;10(1):e02313–e02321. doi: 10.1128/spectrum.02313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sousa A.M., Pereira M.O. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens. 2014;3(3):680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anwar H., Costerton J. Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicro. Agents Chemother. 1990;34(9):1666–1671. doi: 10.1128/aac.34.9.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng B.S., Zhang W., Harrison J.J., Quach T.P., Song J.L., Penterman J., et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15(10):2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alshraiedeh N.A.H., Kelly S.A., Thompson T.P., Flynn P.B., Tunney M.M., Gilmore B.F. Extracellular polymeric substance‐mediated tolerance of Pseudomonas aeruginosa biofilms to atmospheric pressure nonthermal plasma treatment. Plasma Process Polym. 2020;17(12) [Google Scholar]
- 9.Alkawareek M.Y., Algwari Q.T., Laverty G., Gorman S.P., Graham W.G., O’Connell D., et al. Eradication of Pseudomonas aeruginosa biofilms by atmospheric pressure non-thermal plasma. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0044289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barakat M.M., Dallal Bashi Y.H., Carson L., Graham W.G., Gilmore B.F., Flynn P.B. Atmospheric pressure non-thermal plasma exposure reduces Pseudomonas aeruginosa lipopolysaccharide toxicity in vitro and in vivo. Microb Pathog. 2019;136:103679. doi: 10.1016/j.micpath.2019.103679. [DOI] [PubMed] [Google Scholar]
- 11.De Soyza A., Hall A.J., Mahenthiralingam E., Drevinek P., Kaca W., Drulis‐Kawa Z., et al. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen. 2013;2(6):1010–1023. doi: 10.1002/mbo3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourke P., Ziuzina D., Han L., Cullen P., Gilmore B.F. Microbiological interactions with cold plasma. J Appl Microbiol. 2017;123(2):308–324. doi: 10.1111/jam.13429. [DOI] [PubMed] [Google Scholar]
- 13.Olson M.E., Ceri H., Morck D.W., Buret A.G., Read R.R. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66(2):86. [PMC free article] [PubMed] [Google Scholar]
- 14.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 15.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8):e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Team R.C.R. 2013. A language and environment for statistical computing. [Google Scholar]
- 18.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allaire J. vol. 770. 2012. pp. 165–171. (RStudio: integrated development environment for R). Boston, MA. 394. [Google Scholar]
- 22.Villanueva R.A.M., Chen Z.J. Taylor & Francis; 2019. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 23.Zheng L.L., Huang C.Z. Selective and sensitive colorimetric detection of stringent alarmone ppGpp with Fenton-like reagent. Analyst. 2014;139(23):6284–6289. doi: 10.1039/c4an01632g. [DOI] [PubMed] [Google Scholar]
- 24.Ferro K.J., Bender G.R., Marquis R.E. Coordinately repressible arginine deiminase system inStreptococcus sanguis. Curr Microbiol. 1983;9(3):145–149. [Google Scholar]
- 25.Nicol M.J., Brubaker T.R., Honish B.J., Simmons A.N., Kazemi A., Geissel M.A., et al. Antibacterial effects of low-temperature plasma generated by atmospheric-pressure plasma jet are mediated by reactive oxygen species. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-59652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett D.J., Schweizer H.P., Ohman D.E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177(22):6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan H.M., Schrum L.W. Roles of manganese and iron in the regulation of the biosynthesis of manganese-superoxide dismutase inEscherichia coli. FEMS (Fed Eur Microbiol Soc) Microbiol Rev. 1994;14(4):315–323. doi: 10.1111/j.1574-6976.1994.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 28.Jakopitsch C., Regelsberger G., Georg Furtmüller P., Rüker F., Peschek G.A., Obinger C. Engineering the proximal heme cavity of catalase-peroxidase. J Inorg Biochem. 2002;91(1):78–86. doi: 10.1016/s0162-0134(02)00374-4. [DOI] [PubMed] [Google Scholar]
- 29.Ueda M., Kinoshita H., Maeda S.I., Zou W., Tanaka A. Structure-function study of the amino-terminal stretch of the catalase subunit molecule in oligomerization, heme binding, and activity expression. Appl Microbiol Biotechnol. 2003;61(5–6):488–494. doi: 10.1007/s00253-003-1251-5. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill M.J., Wilks A. The P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem Biol. 2013;8(8):1794–1802. doi: 10.1021/cb400165b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damron F.H., Napper J., Teter M.A., Yu H.D. Lipotoxin F of Pseudomonas aeruginosa is an AlgU-dependent and alginate-independent outer membrane protein involved in resistance to oxidative stress and adhesion to A549 human lung epithelia. Microbiology (Read) 2009;155(Pt 4):1028–1038. doi: 10.1099/mic.0.025833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasparova P., Vankova E., Paldrychova M., Svobodova A., Hadravova R., Jarosova Kolouchova I., et al. Non-thermal plasma causes Pseudomonas aeruginosa biofilm release to planktonic form and inhibits production of Las-B elastase, protease and pyocyanin. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.993029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishige K., Zhang H., Kornberg A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc Natl Acad Sci U S A. 2002;99(26):16684–16688. doi: 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alkawareek M.Y., Algwari Q.T., Laverty G., Gorman S.P., Graham W.G., O'Connell D., et al. Eradication of Pseudomonas aeruginosa biofilms by atmospheric pressure non-thermal plasma. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0044289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eymann C., Homuth G., Scharf C., Hecker M. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol. 2002;184(9):2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith E.E., Sims E.H., Spencer D.H., Kaul R., Olson M.V. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J Bacteriol. 2005;187(6):2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong L., Pang J., Wang X., Zhang Y., Li G., Hu X., et al. Mechanism of pyocyanin abolishment caused by mvaT mvaU double knockout in Pseudomonas aeruginosa PAO1. Virulence. 2019;11(1):57–67. doi: 10.1080/21505594.2019.1708052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravel J., Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11(5):195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 39.O'Malley Y.Q., Reszka K.J., Spitz D.R., Denning G.M., Britigan B.E. Pseudomonas aeruginosa pyocyanin directly oxidizes glutathione and decreases its levels in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2004;287(1):L94–L103. doi: 10.1152/ajplung.00025.2004. [DOI] [PubMed] [Google Scholar]
- 40.Grace A., Sahu R., Owen D.R., Dennis V.A. Pseudomonas aeruginosa reference strains PAO1 and PA14: a genomic, phenotypic, and therapeutic review. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casiano-Colón A., Marquis R.E. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 1988;54(6):1318–1324. doi: 10.1128/aem.54.6.1318-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griswold A., Chen Y.-Y.M., Snyder J.A., Burne R.A. Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl Environ Microbiol. 2004;70(3):1321–1327. doi: 10.1128/AEM.70.3.1321-1327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borriello G., Richards L., Ehrlich G.D., Stewart P.S. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicro. Agents Chemother. 2006;50(1):382–384. doi: 10.1128/AAC.50.1.382-384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gloag E.S., Khosravi Y., Masters J.G., Wozniak D.J., Amorin Daep C., Stoodley P. A combination of zinc and arginine disrupt the mechanical integrity of dental biofilms. Microbiol Spectr. 2022 doi: 10.1128/spectrum.03351-22. e03351-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawakami T., Kuroki M., Ishii M., Igarashi Y., Arai H. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol. 2010;12(6):1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 46.Hamada M., Toyofuku M., Miyano T., Nomura N. Cbb 3-type cytochrome c oxidases, aerobic respiratory enzymes, impact the anaerobic life of Pseudomonas aeruginosa PAO1. J Bacteriol. 2014;196(22):3881–3889. doi: 10.1128/JB.01978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahn S., Jung J., Jang I.-A., Madsen E.L., Park W. Role of glyoxylate shunt in oxidative stress response. J Biol Chem. 2016;291(22):11928–11938. doi: 10.1074/jbc.M115.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yau K.P.S., Murphy A.B., Zhong L., Mai-Prochnow A. Cold plasma effect on the proteome of Pseudomonas aeruginosa – role for bacterioferritin. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0206530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh P.K., Parsek M.R., Greenberg E.P., Welsh M.J. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417(6888):552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 50.Rolfe M.D., Rice C.J., Lucchini S., Pin C., Thompson A., Cameron A.D., et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J Bacteriol. 2012;194(3):686–701. doi: 10.1128/JB.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balaban N.Q., Merrin J., Chait R., Kowalik L., Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 52.Mai-Prochnow A., Bradbury M., Ostrikov K., Murphy A.B. Pseudomonas aeruginosa biofilm response and resistance to cold atmospheric pressure plasma is linked to the redox-active molecule phenazine. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0130373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ledgham F., Soscia C., Chakrabarty A., Lazdunski A., Foglino M. Global regulation in Pseudomonas aeruginosa: the regulatory protein AlgR2 (AlgQ) acts as a modulator of quorum sensing. Res Microbiol. 2003;154(3):207–213. doi: 10.1016/S0923-2508(03)00024-X. [DOI] [PubMed] [Google Scholar]
- 54.Thayil S.M., Morrison N., Schechter N., Rubin H., Karakousis P.C. The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Acker H., Sass A., Bazzini S., De Roy K., Udine C., Messiaen T., et al. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prossliner T., Skovbo Winther K., Sørensen M.A., Gerdes K. Ribosome hibernation. Annu Rev Genet. 2018;52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 57.Liao X., Liu D., Ding T. Nonthermal plasma induces the viable-but-nonculturable state in Staphylococcus aureus via metabolic suppression and the oxidative stress response. Appl Environ Microbiol. 2020;86(5):e02216–e02219. doi: 10.1128/AEM.02216-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J.S., Chowdhury N., Yamasaki R., Wood T.K. Viable but non‐culturable and persistence describe the same bacterial stress state. Environ Microbiol. 2018;20(6):2038–2048. doi: 10.1111/1462-2920.14075. [DOI] [PubMed] [Google Scholar]
- 59.Kamath S., Kapatral V., Chakrabarty A. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol Microbiol. 1998;30(5):933–941. doi: 10.1046/j.1365-2958.1998.01121.x. [DOI] [PubMed] [Google Scholar]
- 60.Cathcart G.R., Quinn D., Greer B., Harriott P., Lynas J.F., Gilmore B.F., et al. Novel inhibitors of the Pseudomonas aeruginosa virulence factor LasB: a potential therapeutic approach for the attenuation of virulence mechanisms in pseudomonal infection. Antimicro. Agents Chemother. 2011;55(6):2670–2678. doi: 10.1128/AAC.00776-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L., Xia C., Guo Y., Yang C., Cheng C., Zhao J., et al. Bactericidal efficacy of cold atmospheric plasma treatment against multidrug-resistant Pseudomonas aeruginosa. Future Microbiol. 2020;15:115–125. doi: 10.2217/fmb-2019-0265. [DOI] [PubMed] [Google Scholar]
- 62.Storey D., McNally A., Astrand M., Sa-Pessoa Graca Santos J., Rodriguez-Escudero I., Elmore B., et al. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 2020;16(3) doi: 10.1371/journal.ppat.1007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.