Abstract
A gene, badH, whose predicted product is a member of the short-chain dehydrogenase/reductase family of enzymes, was recently discovered during studies of anaerobic benzoate degradation by the photoheterotrophic bacterium Rhodopseudomonas palustris. Purified histidine-tagged BadH protein catalyzed the oxidation of 2-hydroxycyclohexanecarboxyl coenzyme A (2-hydroxychc-CoA) to 2-ketocyclohexanecarboxyl-CoA. These compounds are proposed intermediates of a series of three reactions that are shared by the pathways of cyclohexanecarboxylate and benzoate degradation used by R. palustris. The 2-hydroxychc-CoA dehydrogenase activity encoded by badH was dependent on the presence of NAD+; no activity was detected with NADP+ as a cofactor. The dehydrogenase activity was not sensitive to oxygen. The enzyme has apparent Km values of 10 and 200 μM for 2-hydroxychc-CoA and NAD+, respectively. Western blot analysis with antisera raised against purified His-BadH identified a 27-kDa protein that was present in benzoate- and cyclohexanecarboxylate-grown but not in succinate-grown R. palustris cell extracts. The active form of the enzyme is a homotetramer. badH was determined to be the first gene in an operon, termed the cyclohexanecarboxylate degradation operon, containing genes required for both benzoate and cyclohexanecarboxylate degradation. A nonpolar R. palustris badH mutant was unable to grow on benzoate or cyclohexanecarboxylate but had wild-type growth rates on succinate. Cells blocked in expression of the entire cyclohexanecarboxylate degradation operon excreted cyclohex-1-ene-1-carboxylate into the growth medium when given benzoate. This confirms that cyclohex-1-ene-1-carboxyl-CoA is an intermediate of anaerobic benzoate degradation by R. palustris. This compound had previously been shown not to be formed by Thauera aromatica, a denitrifying bacterium that degrades benzoate by a pathway that is slightly different from the R. palustris pathway. 2-Hydroxychc-CoA dehydrogenase does not participate in anaerobic benzoate degradation by T. aromatica and thus may serve as a useful indicator of an R. palustris-type benzoate degradation pathway.
Aromatic compounds in the form of plant phenolics, plant-derived lignin monomers, and aromatic hydrocarbons are among the most abundant compounds on earth (22, 39). Despite the intrinsic resonance stability of the aromatic ring, many microbes can degrade aromatic compounds for use as carbon and energy. Two different biochemical strategies are used by microbes to accomplish the degradation of aromatic molecules, depending on the availability of oxygen. Under aerobic conditions, oxygenases hydroxylate aromatic rings to initiate the degradation process (13, 15). Under anaerobic conditions, reductive reactions relieve the resonance of the aromatic ring. In recent years, research has established that structurally diverse aromatic compounds are converted to benzoate or benzoyl-coenzyme A (CoA) as a starting intermediate for a central anaerobic pathway of aromatic ring reduction culminating in ring cleavage (16, 17).
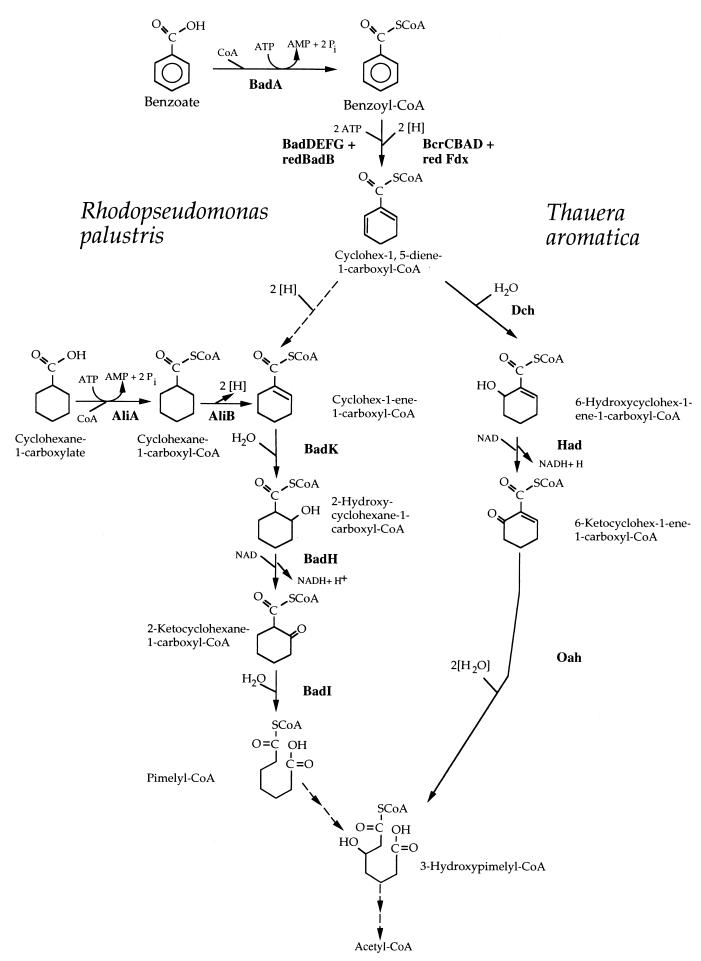
The purple nonsulfur phototrophic bacterium Rhodopseudomonas palustris and the denitrifying bacterium Thauera aromatica have served as model systems for the study of anaerobic benzoate degradation. Initial physiological studies with these two organisms indicated that the pathway of anaerobic benzoate degradation can be divided into several segments that include (i) CoA thioester formation, (ii) ring reduction, (iii) introduction of a hydroxyl group, and (iv) ring fission. Based on this, it was assumed that R. palustris and T. aromatica used identical pathways to degrade benzoate. However, subsequent biochemical and molecular studies have made it clear that the benzoate degradation pathways used by these two bacteria are not exactly the same (16). The two organisms have similar benzoyl-CoA reductase enzymes, but once benzoyl-CoA is reduced to a cyclohexadienecarboxyl-CoA intermediate, the next step in T. aromatica is a hydration of the ring, whereas the next probable step in R. palustris is a two-electron reduction to give cyclohex-1-ene-1-carboxyl-CoA as an intermediate. Two slightly different sets of reactions follow which lead to the formation of two different ring fission products: pimelyl-CoA in the case of R. palustris and 3-hydroxypimelyl-CoA in the case of T. aromatica (Fig. 1).
FIG. 1.
Comparison of anaerobic benzoate degradation by R. palustris and T. aromatica. Reactions involved in funneling cyclohexanecarboxylate into the anaerobic benzoate degradation pathway in R. palustris are shown. Solid arrows indicate enzymatic activities that have been purified from either R. palustris or T. aromatica (1, 6, 11, 23, 25, 26, 33). Dashed arrows indicate hypothetical enzymatic reactions. Assignment of gene products from R. palustris (Fig. 2) and T. aromatica (7) to specific steps is indicated. redBadB, reduced BadB; redFdx, reduced ferredoxin.
Despite differences in the structures of the intermediates, a dehydrogenation reaction is required in each pathway to accomplish the conversion of a ring hydroxyl to a carbonyl. This reaction has not yet been studied for R. palustris, but we have proposed that it is catalyzed by the BadH protein (10). The badH gene is present in a cluster of benzoate degradation genes, and the predicted BadH protein has similarity to short-chain dehydrogenases (10). BadH does not have any significant amino acid sequence identity to the hydroxyacyl-CoA dehydrogenase (Had) used by T. aromatica to degrade benzoate (7, 16). Here we report the purification and characterization of BadH from R. palustris. We also demonstrate that badH is the first gene in an operon involved in cyclohexanecarboxylate utilization by this organism and is an essential gene for anaerobic benzoate degradation. A possible advantage of the R. palustris-type benzoate pathway as opposed to the T. aromatica-type pathway is that it provides a route for the utilization of the alicyclic acids cyclohexanecarboxylate and cyclohex-1-ene-1-carboxylate as growth substrates.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. R. palustris was grown anaerobically in defined mineral medium at 30°C as described previously (11). Carbon sources were added to a final concentration of 3 mM, except succinate, which was used at a final concentration of 10 mM. Growth was monitored spectrophotometrically at 660 nm. Cultures were illuminated with 40- or 100-W incandescent light bulbs. Cells were harvested in the mid- to late logarithmic phase of growth by centrifugation, washed once in 20 mM Tris hydrochloride (pH 7.2) (Tris buffer), and stored at −70°C until used. Escherichia coli strains used for protein expression were grown with shaking at 30°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80dlacZΔM15 | Gibco-BRL |
| S17-1 | thi pro hdsR hdsM+ recA; chromosomal insertion of RP4-2 (Tc::Mu Km::Tn7) | 38 |
| R. palustris strains | ||
| CGA009 | Wild-type strain; spontaneous Cmr derivative | 21 |
| CGA702 | badH::Kmr (polar) | This study |
| CGA720 | badH::Kmr (nonpolar) | This study |
| Plasmids | ||
| pUC18 | High-copy-number cloning vector; ColE1 lacZ Apr | 45 |
| pJQ200KS | Mobilizable suicide vector; sacB Gmr | 35 |
| pUC18K2 | pUC18 containing nonpolar Kmr cassette | 29 |
| pTrcHisA | Six-histidine fusion protein expression vector; ColE1 lacIq Apr | Invitrogen |
| pPE304 | pHC79; cosmid cloning vector containing 35-kb insert including badH; Apr | 10 |
| pDP101 | pUC18 with 2.6-kb fragment from pPE304 containing badH; Apr | This study |
| pDP203 | pDP101 with Kmr gene at the StuI site in badH | This study |
| pDP204 | pJQ200KS; sacB suicide vector containing Kmr cassette at the StuI site of badH | This study |
| pDP212 | pDP101 with nonpolar Kmr gene at the StuI site in badH | This study |
| pDP213 | pJQ200KS; sacB suicide vector with nonpolar Kmr cassette at the StuI site of badH | This study |
| pDPHisH1 | pTrcHisA vector containing badH; Apr | This study |
Apr, ampicillin resistance; Gmr, gentamicin resistance; Cmr, chloramphenicol resistance.
Synthesis of acyl-CoA thioesters.
Cyclohex-1-ene-1-carboxyl-CoA and pimelyl-CoA were synthesized from mixed anhydrides as described by Merkel et al. (30), except that the final alkali treatment step was omitted. The procedure for pimelyl-CoA synthesis yielded mainly pimelyl-CoA and only small amounts of pimelyldi-CoA. 2-Hydroxycyclohexanecarboxyl-CoA (2-hydroxychc-CoA) was synthesized enzymatically from cyclohex-1-ene-1-carboxyl-CoA using crotonase (EC 4.2.1.17) purchased from Sigma (St. Louis, Mo.). 3-Hydroxypimelyl-CoA was synthesized enzymatically using pimelyl-CoA as the starting substrate and partially purified pimelyl-CoA dehydrogenase and dehydropimelyl-CoA hydratase activities from benzoate-grown cells of R. palustris (M. Emig, unpublished data). Acetoacetyl-CoA and 3-hydroxybutyryl-CoA were purchased from Sigma. Acyl-CoA thioesters were purified using C18 reverse-phase Sep-Pack cartridges (Millipore Corp., Milford, Mass.) as described previously (33). High-pressure liquid chromatography (HPLC) analysis using an Ultrasphere octyldecyl silane (C18) reverse-phase column (4.6 mm by 25 cm) (Beckman Instruments, Fullerton, Calif.) was used to confirm that the purified CoA thioester substrates were free of contaminating CoASH. The solvent system used was a mixture of 20 mM ammonium acetate (pH 6.0) and methanol. The column was equilibrated with 20% methanol, and elution was done by a linear gradient of 20 to 80% methanol in 30 min. The absorbance of the effluent was monitored by scanning the region from 210 to 260 nm using a model 996 photodiode array detector (Waters Associates, Milford, Mass.). The products of the synthesis reactions were analyzed by electrospray ionization mass spectrometry at the University of Iowa High-Resolution Mass Spectrometry Facility to confirm that they had the expected molecular mass. Since CoASH and acyl-CoA thioesters have essentially the same extinction coefficients at 254 nm, the acyl-CoA thioester substrates were quantitated spectrophotometrically at this wavelength according to a standard curve of known quantities of CoASH.
Cloning and DNA and RNA manipulations.
Standard protocols were used for DNA cloning and transformation (4). Plasmids were purified using QIAprep spin columns (Qiagen Inc., Chatsworth, Calif.), and restriction fragments used for cloning were extracted from agarose gels with GeneClean spin (Bio 101, La Jolla, Calif.). R. palustris chromosomal DNA was isolated as described previously (9). The access reverse transcription-PCR system (Promega Corp., Madison, Wis.) was used to determine the transcriptional organization of the badHIaliBA and badK genes. Total RNA was isolated from benzoate-grown R. palustris cells using the SV total RNA isolation system (Promega Corp.).
Construction of R. palustris chromosomal mutants.
A badH::Kmr (kanamycin resistant) nonpolar mutant (strain CGA720) was constructed by inserting the 0.85-kb Kmr cassette from pUC18K2 (29) into a unique StuI site on the plasmid pDP101 to yield the plasmid pDP212. The plasmid pDP101 contains the entire badH gene cloned into pUC18. The badH::Kmr nonpolar construct was subcloned into the suicide vector pJQ200KS (35) to generate the plasmid pDP213. The badH::Kmr nonpolar construct (pDP213) was then introduced into R. palustris by conjugation from E. coli S17-1 (38). A badH::Kmr polar mutant (strain CGA702) was constructed by inserting the 1.3-kb Kmr GeneBlock cassette (Pharmacia) into a unique StuI site on the plasmid pDP101 to yield the plasmid pDP203. The construct was cloned into the suicide vector pJQ200KS (35) to generate the plasmid pDP204. The badH::Kmr construct (pDP204) was then introduced into R. palustris by conjugation from E. coli S17-1 (38). Recombinants were selected as described previously (9). The insertion of the nonpolar Kmr cassette into the badH open reading frame was confirmed by PCR amplification of the badH region of the R. palustris chromosome from wild-type (0.8-kb product) and mutant (1.6-kb product) chromosomal DNA. The insertion of the polar Kmr GeneBlock cassette into the badH open reading frame was confirmed by Southern blot hybridization.
Enzyme activities.
Standard assay conditions for hydroxyacyl-CoA dehydrogenase activity were 20 mM Tris-HCl (pH 9.0), 1.5 mM NAD, 0.5 mM hydroxyacyl-CoA substrate, 60 mM hydrazine, and appropriate amounts of protein. Activity was also monitored in the reductase direction using a solution containing 20 mM Tris-HCl (pH 7.0), 100 μM NADH, and 50 μM acetoacetyl-CoA, because 2-hydroxychc-CoA dehydrogenase is active with this substrate. Activity was measured spectrophotometrically by monitoring absorbance at 340 nm, and a molar extinction coefficient of 6.22 mM−1 cm−1 for NADH was used to calculate activities. The standard assay was used to estimate Km values for hydroxyacyl-CoA substrates from double-reciprocal plots with substrates from 1.25 μM to 2 mM. Inhibition experiments were performed using the standard assay conditions except that enzyme was incubated on ice with the test compound for 15 min prior to addition to the assay mixture. 2-Ketocyclohexanecarboxyl-CoA hydrolase activity was measured as described previously (33).
Analysis of R. palustris badH::Km (CGA702) culture supernatants.
CGA702 was grown in the light in defined mineral medium containing 10 mM succinate and 1.5 mM benzoate at 30°C. Aliquots (1.0 ml) were removed at various time points, cells were removed by centrifugation, and 0.5 ml of the resulting supernatant was analyzed by HPLC using an Ultrasphere octyldecyl silane (C18) reverse-phase column (4.6 mm by 25 cm) (Beckman Instruments). The solvent system used was a mixture of 20 mM ammonium acetate (pH 6.0) and methanol. The column was equilibrated with 1% methanol, and elution was done by a linear gradient of 1 to 50% methanol in 30 min. The absorbance of the effluent was monitored by scanning the region from 210 to 260 nm using a model 996 photodiode array detector (Waters Associates). Peaks were identified by spectral properties and comigration by HPLC with authentic standards.
Preparation of R. palustris cell extracts.
Cells were grown to late logarithmic phase, harvested by centrifugation, washed, and suspended in approximately 2 volumes of 20 mM Tris-HCl (pH 7.4) buffer containing 1 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 5 μg of DNase/ml, 5 μg of RNase/ml, and 0.5 mM phenylmethylsulfonyl fluoride. Cells were lysed by sonication, and cell debris was removed by centrifugation at 10,000 × g for 30 min at 4°C. The resulting supernatant was then centrifuged at high speed (100,000 × g) for 1 h at 4°C. The supernatant from the second centrifugation was termed crude cell extract.
Construction of a histidine-BadH fusion expression strain.
A His-BadH fusion was constructed by PCR amplification of badH from pDP101 using primers containing EcoRI and BamHI restriction sites. The EcoRI- and BamHI-digested PCR product was ligated into the EcoRI and BamHI restriction sites of the pTrcHisA vector (Invitrogen, Carlsbad, Calif.) to generate the plasmid pDPHisH1, resulting in an N-terminal six-histidine tag sequence and a factor Xa cleavage site in the recombinant BadH fusion protein. The plasmid pDPHisH1 was introduced into E. coli DH5α by transformation according to standard protocols (4).
Purification of histidine-tagged BadH by affinity chromatography.
The His-BadH fusion protein was expressed in E. coli DH5α Cells were grown to an optical density at 660 nm of 0.6 and were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 to 4 h. Cells from 1 liter of culture were harvested by centrifugation, washed, and resuspended in binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl). Cells were lysed by passage through a French pressure cell at 85 MPa, and cell debris was removed by centrifugation at 10,000 × g for 30 min at 4°C. The supernatant from the centrifugation was termed crude cell extract. All subsequent steps were carried out at 4°C. Crude cell extract (typically 300 to 500 mg of protein) was loaded onto a 5-ml Hitrap chelating column (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) that had been charged with NiSO4 and equilibrated with binding buffer. The column was washed extensively with binding buffer and then was washed with binding buffer containing 5% elution buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 1 M imidazole) for 30 min at 1 ml/min. A linear gradient of 5 to 50% elution buffer in binding buffer was then passed through the column at a flow rate of 1 ml/min over a period of 25 min. Fractions (2 ml) were collected and assayed spectrophotometrically for 2-hydroxychc-CoA dehydrogenase activity. The enzyme activity was eluted at approximately 0.5 M imidazole. The activity-containing fractions from the affinity column were pooled and exchanged into 20 mM Tris-HCl (pH 7.4) using a 5-ml Hitrap desalting column at a flow rate of 1 ml/min. The activity-containing fractions were adjusted to 50% glycerol and stored at −20°C. Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Other analytical procedures.
The native apparent molecular mass of His-BadH was determined by gel filtration chromatography using a Progel TSK G3000SW HPLC column (0.75 by 7.5 mm) (Supelco, Bellefonte, Pa.) equilibrated with 20 mM Tris-HCl (pH 7.5)–30 mM NaCl. The following protein molecular weight standards were used to calibrate the column: chymotrypsinogen A, 25,000; bovine serum albumin, 66,000; aldolase, 158,000; and ferritin, 440,000. SDS-PAGE was carried out with 12% acrylamide gels by standard procedures (24). Separated proteins were visualized by staining with Coomassie blue R-250. Molecular weight standards were from Bio-Rad Laboratories (Richmond, Calif.). The protein concentration was estimated by the dye-binding assay (Bio-Rad) using bovine serum albumin as a standard.
Immunoblotting.
Polyclonal antiserum was prepared from a rabbit inoculated with purified His-BadH at Covance Research Products Inc. (Denver, Pa.). Cell extracts of R. palustris were typically separated on SDS–12% polyacrylamide gels and electroblotted onto an Immobilon polyvinylidene difluoride membrane (Millipore), and antigens were visualized using alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad) together with nitroblue tetrazolium and bromochloroindolyl phosphate as chromogenic substrates (4).
Computer analysis of DNA sequences.
The BLAST network services at the National Center for Biotechnology Information (Bethesda, Md.) were used to search protein databases for similar sequences (3). Amino acid sequence similarities were calculated using the GAP program from the University of Wisconsin Genetics Computer Group software package (version 9.0) (8). The multiple sequence alignment was constructed using the CLUSTALW multiple sequence alignment program at the Baylor College of Medicine Human Genome Center (42). The program BOXSHADE (version 3.21) was used to shade aligned sequences.
RESULTS
badH is the first gene in an operon that contains genes for cyclohexanecarboxylic acid degradation.
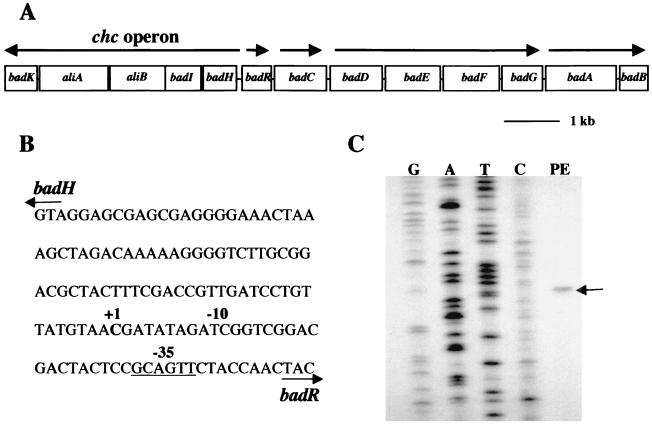
The badH gene is the first gene in a cluster of five genes (badH, badI, aliB [formerly badJ], aliA, and badK) that are predicted to be transcribed in the same direction (Fig. 2). These genes are adjacent to genes known to be required for anaerobic benzoyl-CoA reduction (10). The genes badI, aliB, aliA, and badK have been assigned functions in either benzoate or cyclohexanecarboxylate degradation (Fig. 1). Reverse transcription-PCR amplification of total RNA isolated from benzoate-grown R. palustris cells showed that the five genes are organized as an operon. The transcriptional start site of the badHIaliBAbadK operon (henceforth called the chc degradation operon) was located 81 bp upstream of the badH initiation codon. The badH promoter region has a −35 sequence similar to the consensus RNA polymerase sigma 70 subunit recognition element but no obvious −10 consensus sequence.
FIG. 2.
(A) Map of bad (benzoic acid degradation) gene cluster from R. palustris. Arrows indicate transcriptional units. The genes encode enzymes of anaerobic benzoate or cyclohexanecarboxylate degradation, as indicated in Fig. 1. (B) Nucleotide sequence of the badH promoter region, with numbering indicating the start site of badH transcription (+1) and the −10 and −35 (underlined) nucleotides. (C) Mapping of the badH transcriptional start site by primer extension. The position of the primer extension product is indicated by the arrow (lane PE). RNA for the primer extension reaction was isolated from benzoate-grown cells. A sequence ladder generated with the same primer is shown.
Molecular characteristics of badH.
The badH gene is similar in deduced amino acid sequence to members of the short-chain dehydrogenase/reductase (SDR) family of enzymes (19, 20). Most members of the SDR family of enzymes are NAD(P)-dependent oxidoreductases of 250 to 300 amino acids that contain no metal cofactors and have a wide range of substrate specificities that include alcohols, sugars, prostaglandins, and hydroxysteroids (19, 20, 34). BadH is most similar to eubacterial acetoacetyl-CoA reductases involved in polyhydroxyalkanoate and fatty acid biosynthesis (about 40% amino acid identity) (36, 43). It is 37% identical to 7α-hydroxysteroid dehydrogenase from E. coli and 36% identical to 20β-hydroxysteroid dehydrogenase from Streptomyces exfoliatus. The badH open reading frame is predicted to encode a protein of 255 amino acids with a molecular mass of 26 kDa and a pI of 6.3. Signature features of members of the SDR family include a GxxxGxG motif near the N terminus, proposed to be a nucleotide-binding domain, and three highly conserved amino acids (a serine, a tyrosine, and a lysine) near the middle of the protein that have been implicated in catalysis (12, 28, 40, 41). BadH has these features (Fig. 3).
FIG. 3.
Amino acid sequence alignment of BadH with selected members of the SDR family. The abbreviation, enzyme, number of amino acids, organism, and accession number, respectively, for each family member follow: BadH, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase, 255, R. palustris, and U75363 (10); PhbB, acetoacetyl-CoA reductase, 241, Rhizobium meliloti, and P50205 (43); FabG, 3-ketoacyl-acyl carrier protein reductase, 244, E. coli, and P25716 (36); NodG, nodulation protein G, 246, Azospirillum brasilense, and P17611 (44); 20HSD, 20β-hydroxysteroid dehydrogenase, 255, S. exfoliatus, and P19992 (28); and 7αHSD, 7α-hydroxysteroid dehydrogenase, 255, E. coli, and P25529 (46). Alignment was created using CLUSTALW (version 1.74), and shading was done with the program BOXSHADE. Black shading indicates 100% amino acid identity, and gray shading indicates 100% similarity. ∗, active-site amino acids. The boxed sequence is the proposed pyridine nucleotide-binding site, and the solid black line denotes the SDR family signature (20).
BadH is a 2-hydroxychc-CoA dehydrogenase.
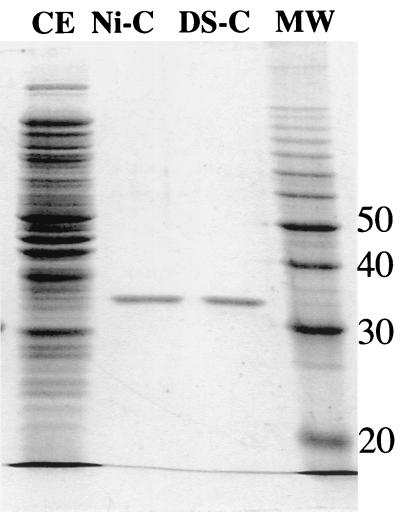
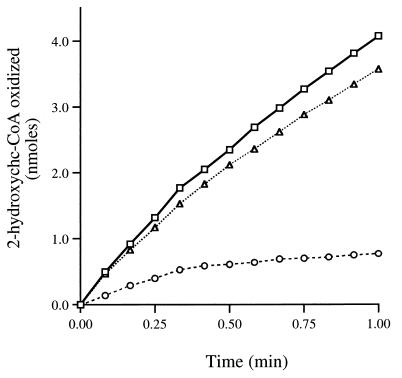
The sequence of reactions that we had proposed for the benzoate degradation pathway included the oxidation of 2-hydroxychc-CoA to 2-ketocyclohexanecarboxyl-CoA. We were able to measure 2-hydroxychc-CoA dehydrogenase activities of 45 and 35 nmol of 2-hydroxychc-CoA oxidized min−1 mg of protein−1 in cell extracts of R. palustris that had been grown on benzoate and cyclohexanecarboxylate, respectively. The level of activity detected in succinate-grown cells was fivefold lower (7 nmol min−1 mg of protein−1). 2-Hydroxychc-CoA dehydrogenase activity was not sensitive to oxygen. Based on its amino acid sequence, we hypothesized that BadH catalyzed this activity. The badH gene was therefore cloned into a histidine fusion expression vector to create the plasmid pDPHisH1. When the histidine-tagged BadH protein was expressed in E. coli, 2-hydroxychc-CoA dehydrogenase activity was detected, with a specific activity of 350 nmol min−1 mg of protein−1. This activity was purified 18-fold to homogeneity by affinity chromatography (Fig. 4). Typical yields of purified enzyme per liter of culture were 5 to 10 mg, with a final specific activity of 6,500 nmol min−1 mg of protein−1. The enzyme could be stored at −20°C in 50% glycerol for several months without significant loss of activity, but enzyme stored without glycerol became inactive within several days. Purified His-BadH had NAD+-dependent dehydrogenase activity with 3-hydroxybutyryl-CoA as well as with 2-hydroxychc-CoA, but no activity was detected with 3-hydroxypimelyl-CoA as a substrate. Activity could be detected in the reverse direction with acetoacetyl-CoA when NADH was supplied as a cofactor. The rate of activity of the purified His-BadH protein with 2-hydroxychc-CoA as a substrate fell off rapidly (Fig. 5). This inhibition could be relieved by addition of hydrazine, which can react with the keto group of the reaction product to form a hydrazone, irreversibly removing the product (32). Similar results were obtained with an addition of purified 2-ketocyclohexanecarboxyl-CoA hydrolase. These data indicate that the equilibrium of the dehydrogenase reaction lies on the side of the hydroxyacyl-CoA substrate. In view of these findings, 60 mM hydrazine was routinely added to 2-hydroxychc-CoA assay mixtures.
FIG. 4.
SDS-PAGE analysis of active protein fractions obtained during purification of His-BadH. Lanes: CE, crude cell extract (20 μg); Ni-C, nickel column pooled fractions (1 μg); DS-C, Hitrap desalting column pooled fractions (1 μg); MW, protein ladder (Bio-Rad). Numbers on the right are molecular weights, in thousands.
FIG. 5.
Time course of 2-hydroxychc-CoA dehydrogenase activity. Activity was measured spectrophotometrically by monitoring the reduction of NAD+ at 340 nm using standard assay conditions as described in Materials and Methods. Reaction mixtures contained purified His-BadH (0.8 μg) plus purified 2-ketocyclohexanecarboxyl-CoA hydrolase (10 μg) (triangles), hydrazine (60 mM) (squares), or neither hydrazine nor 2-ketocyclohexanecarboxyl-CoA hydrolase (circles).
The apparent Kms for 2-hydroxychc-CoA and for NAD+ were 10 μM (± 5 μM) and 200 μM, respectively. The Km for 2-hydroxybutyryl-CoA was 70 μM. No activity was detected with NADP+ as a cofactor. The pH optimum for the 2-hydroxychc-CoA dehydrogenase reaction with purified His-BadH was about pH 9.5 in 20 mM Tris-HCl buffer. Activity at pH 10.0 was the same as that at pH 9.5, while activities at pHs 9.0, 8.0, and 7.0 were 86, 46, and 19%, respectively, of that at pH 9.5.
Active His-BadH had a native molecular mass of 115 kDa as determined by gel filtration chromatography and a subunit molecular mass of 32 kDa as determined by SDS-PAGE analysis. The predicted molecular mass of the histidine-tagged form of BadH is 30 kDa. This indicates that the active enzyme is a homotetramer.
Construction and analysis of a badH mutant.
To determine if BadH is required for growth of R. palustris on aromatic or alicyclic acids, a nonpolar mutation was made in the badH gene by insertion of a kanamycin resistance gene cassette. This cassette contained translational stop codons in all three reading frames at the 5′ end of the kanamycin resistance gene and a ribosome binding site and translational start sites at the 3′ end to ensure translation of downstream genes. The badH mutant (strain CGA720) that was generated in this way was unaffected in its expression of badI, the gene immediately downstream of badH, as determined by Western blot analysis with BadI antiserum. Also, extracts of badH mutant cells grown on succinate plus benzoate had BadI (2-ketocyclohexanecarboxyl-CoA hydrolase) enzymatic activity.
The badH mutant was unable to grow anaerobically on benzoate, cyclohexanecarboxylate, or cyclohex-1-ene-1-carboxylate, but it had wild-type doubling times on succinate. It also failed to grow aerobically on cyclohexanecarboxylate or cyclohex-1-ene-1-carboxylate. Wild-type R. palustris cells grow well on these two alicyclic acids under aerobic conditions, whereas benzoate degradation is sensitive to oxygen. No 2-hydroxychc-CoA dehydrogenase activity was detected in extracts of the badH mutant grown on any of the carbon sources tested. Polyclonal antiserum that was prepared against purified His-BadH reacted with a protein of 27 kDa (the predicted size of BadH) that was present in extracts of benzoate-grown wild-type cells of R. palustris. No BadH protein was detected in extracts of succinate-grown cells or in extracts of badH mutant cells grown on benzoate plus succinate (data not shown).
A mutant blocked in expression of the chc degradation operon excretes cyclohex-1-ene-1-carboxylate when given benzoate.
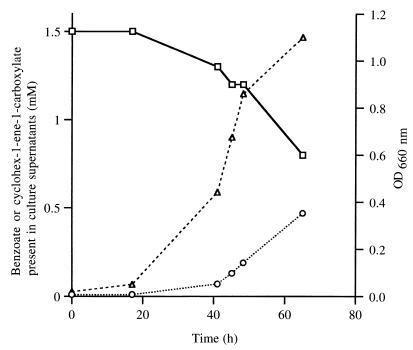
In addition to creating the badH nonpolar mutation described above, we also inserted an unaltered kanamycin resistance gene into badH to generate a mutant that, due to polar effects of the insertion, was defective in expression of all genes of the chc degradation operon. This polar badH mutant was, as expected, unable to grow on benzoate, cyclohexanecarboxylate, or cyclohex-1-ene-1-carboxylate. We also found that mutant cells growing on succinate in the presence of 1.5 mM benzoate excreted substantial amounts of the alicyclic acid cyclohex-1-ene-1-carboxylate (Fig. 6). This was probably derived from cyclohex-1-ene-1-carboxyl-CoA that was formed from benzoate and that accumulated intracellularly in the blocked mutant.
FIG. 6.
Time course of anaerobic benzoate transformation by cultures of CGA702 (badH::Kmr). Cells were grown anaerobically in mineral medium containing 10 mM succinate and 1.5 mM benzoate. Aliquots were removed at various time points, cells were removed by centrifugation, and supernatants were analyzed by C18 reverse-phase HPLC for the presence of benzoate (squares) and cyclohex-1-ene-carboxylate (circles). Growth (represented by triangles) was monitored spectrophotometrically by monitoring optical density at 660 nm (OD660 nm). Data points are averages of duplicates.
DISCUSSION
The ability to degrade benzoate and various aromatic compounds that are processed through benzoyl-CoA is widespread among taxonomically diverse bacteria, including phototrophs, denitrifiers, fermentative bacteria, sulfate reducers, and iron-reducing bacteria (3, 5, 14, 18, 27). To date, however, detailed studies of anaerobic benzoyl-CoA degradation have been carried out with just two species, the denitrifier T. aromatica and the phototroph R. palustris. From this work, it is evident that at least two variants of the benzoyl-CoA degradation pathway exist (Fig. 1) (16). Work presented here provides further evidence in favor of the pathway that we have proposed for R. palustris. Previously we reported the cloning and sequencing of a cluster of five genes, badK, aliA, aliB, badI, and badH, that we hypothesized to have a role in the degradation of both benzoate and the alicyclic acid cyclohexanecarboxylate (10). Here we demonstrate that these genes are organized as an operon. We also provide evidence that cyclohex-1-ene-1-carboxyl-CoA itself, and not a hydroxylated derivative, is an intermediate in anaerobic benzoate degradation by showing that a mutant blocked in the transcription of the chc degradation operon accumulates cyclohex-1-ene-1-carboxylate in the growth medium when it metabolizes benzoate.
Each of the two known variants of the anaerobic benzoate degradation pathways may have advantages. The T. aromatica pathway accomplishes the conversion of benzoyl-CoA to 3-hydroxypimelyl-CoA in just four enzymatic steps, compared to seven steps for R. palustris (16). On this basis, the T. aromatica pathway is more efficient. The R. palustris pathway, on the other hand, provides a point of entry for alicyclic acids, like cyclohexanecarboxylate, to be degraded. At this point, it is not known whether additional variants of the benzoyl-CoA degradation pathway will be found in other bacteria. The fermentative bacteria deserve special consideration, however, because of the severe energetic constraints under which they operate. Bacteria of the genus Syntrophus can grow with benzoate as a sole carbon and energy source when they are in coculture with hydrogen-consuming bacteria, such as methanogens or sulfate reducers, but energy calculations show that they cannot possibly generate sufficient ATP to support growth by using either of the benzoyl-CoA degradation pathways shown in Fig. 1 (31, 37). Thermodynamic calculations suggest that a four-electron rather than a two-electron reduction of benzoyl-CoA would not necessarily require the hydrolysis of ATP. A modification of the pathway, such that benzoyl-CoA is reduced directly to cyclohex-1-ene-1-carboxyl-CoA, would allow sufficient ATP to be generated for growth. This postulated pathway for fermenters would be very similar to the proposed R. palustris pathway, but with the omission of a cyclohexadienecarboxyl-CoA as an intermediate.
With this report, the functions of each of the five gene products of the chc degradation operon have now been confirmed either by N-terminal amino acid sequencing of purified enzymes or by heterologous expression of recombinant enzyme activities in E. coli. The badI gene product has been purified from R. palustris and shown to catalyze the hydrolytic cleavage of 2-ketocyclohexanecarboxyl-CoA (33). The gene aliB encodes an acyl-CoA dehydrogenase that has been heterologously expressed, purified, and shown to catalyze the oxidation of cyclohexanecarboxyl-CoA to cyclohex-1-ene-1-carboxyl-CoA (Emig, unpublished data). The gene aliB was formerly named badJ (10). We propose that this gene be renamed aliB because we have now determined that it functions to funnel the alicyclic compound cyclohexanecarboxylate into the benzoate pathway and does not appear to have a direct role in anaerobic benzoate degradation (Emig, unpublished data). The gene aliA encodes cyclohexanecarboxylate-CoA ligase. This enzyme has been purified from R. palustris, and the N-terminal amino acid sequence was determined (23). The gene badK encodes an enoyl-CoA hydratase that has previously been demonstrated to have activity with cyclohex-1-ene-1-carboxyl-CoA (10). Finally, the badH gene described here was expressed and shown to encode a 2-hydroxychc-CoA dehydrogenase that is required for anaerobic growth on benzoate as well as for growth on cyclohexanecarboxylate under either aerobic or anaerobic conditions.
2-Hydroxychc-CoA dehydrogenase activity may prove to be a useful indicator of an R. palustris-type, as opposed to a T. aromatica-type, benzoyl-CoA degradation pathway. The substrate for this enzyme can be prepared comparatively easily. Cyclohex-1-ene-1-carboxyl-CoA is easily prepared from its commercially available free acid, and it can then be hydrated to form 2-hydroxychc-CoA using commercially available crotonase. The benzoate degradation pathway used by T. aromatica includes a 6-hydroxycyclohex-1-ene-carboxyl-CoA dehydrogenase that is an NAD+-dependent alcohol dehydrogenase rather than an SDR, as is BadH (7, 26). This enzyme has been purified and found not to be active with 2-hydroxychc-CoA as a substrate (26). T. aromatica cell extracts do have a 2-hydroxychc-CoA dehydrogenase activity, but in contrast to R. palustris, the activity is not inducible by anaerobic growth on benzoate (26).
ACKNOWLEDGMENTS
This work was supported by the Division of Energy Biosciences, U.S. Department of Energy (grant DE-FG02-95ER20184), and by the U.S. Army Research Office (grants DAAG55-98-0083 and -0188).
REFERENCES
- 1.Altenschmidt U, Oswald B, Fuchs G. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J Bacteriol. 1991;173:5494–5501. doi: 10.1128/jb.173.17.5494-5501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Anders H-J, Kaetzke A, Kämpfer P, Ludwig W, Fuchs G. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int J Syst Bacteriol. 1995;45:327–333. doi: 10.1099/00207713-45-2-327. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1990. [Google Scholar]
- 5.Beller H R, Spormann A M, Sharma P K, Cole J R, Reinhard M. Isolation and characterization of a novel toluene-degrading, sulfate-reducing bacterium. Appl Environ Microbiol. 1996;62:1188–1196. doi: 10.1128/aem.62.4.1188-1196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boll M, Fuchs G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 7.Breese K, Boll M, Alt-Morbe J, Schägger H, Fuchs G. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur J Biochem. 1998;256:148–154. doi: 10.1046/j.1432-1327.1998.2560148.x. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egland P G, Gibson J, Harwood C S. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J Bacteriol. 1995;177:6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egland P G, Pelletier D A, Dispensa M, Gibson J, Harwood C S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissler J F, Harwood C S, Gibson J. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol. 1988;170:1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh D, Wawrzak Z, Weeks C M, Duax W L, Erman M. The refined three-dimensional structure of 3α,20β-hydroxysteroid dehydrogenase and possible roles of the residues conserved in short-chain dehydrogenases. Structure. 1994;2:629–640. doi: 10.1016/s0969-2126(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 13.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 14.Gorny N, Schink B. Anaerobic degradation of catechol by Desulfobacterium sp. strain Cat2 proceeds via carboxylation to protocatechuate. Appl Environ Microbiol. 1994;60:3396–3400. doi: 10.1128/aem.60.9.3396-3400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 16.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1999;22:439–458. [Google Scholar]
- 17.Heider J, Fuchs G. Anaerobic metabolism of aromatic compounds. Eur J Biochem. 1997;243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 18.Hopkins B T, McInerney M J, Warikoo V. Evidence for an anaerobic syntrophic benzoate degradation threshold and isolation of the syntrophic benzoate degrader. Appl Environ Microbiol. 1995;61:526–530. doi: 10.1128/aem.61.2.526-530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jornvall H, Hoog J-O, Persson B. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999;445:261–264. doi: 10.1016/s0014-5793(99)00130-1. [DOI] [PubMed] [Google Scholar]
- 20.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenase/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 21.Kim M-K, Harwood C S. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett. 1991;83:199–204. [Google Scholar]
- 22.Kirk T K. Degradation of lignin. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 399–437. [Google Scholar]
- 23.Küver J, Xue J Y, Gibson J. Metabolism of cyclohexane carboxylic acid by the photosynthetic bacterium Rhodopseudomonas palustris. Arch Microbiol. 1995;164:337–345. doi: 10.1007/BF02529980. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Laempe D, Eisenreich W, Bacher A, Fuchs G. Cyclohexa-1,5-diene-1-carboxyl-CoA hydratase, an enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1998;255:618–627. doi: 10.1046/j.1432-1327.1998.2550618.x. [DOI] [PubMed] [Google Scholar]
- 26.Laempe D, Jahn M, Fuchs G. 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1999;263:420–429. doi: 10.1046/j.1432-1327.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 27.Lovley D R, Giovannoni S J, White D C, Champine J E, Phillips E J P, Gorby Y A, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 28.Marekov L, Krook M, Jornvall H. Prokaryotic 20β-hydroxysteroid dehydrogenase is an enzyme of the 'short-chain, non-metalloenzyme' alcohol dehydrogenase type. FEBS Lett. 1990;266:51–54. doi: 10.1016/0014-5793(90)81504-h. [DOI] [PubMed] [Google Scholar]
- 29.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merkel S M, Eberhard A E, Gibson J, Harwood C S. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J Bacteriol. 1989;171:1–7. doi: 10.1128/jb.171.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mountfort D O, Brulla W J, Krumholz L R, Bryant M P. Syntrophus buswellii gen. nov., sp. nov.: a benzoate catabolizer from methanogenic ecosystems. Int J Syst Bacteriol. 1984;34:216–217. [Google Scholar]
- 32.Novikov D K, Vanhove G F, Carchon H, Asselberghs S, Eyssen H J, Van Veldhoven P P, Mannaerts G P. Peroxisomal beta-oxidation. Purification of four novel 3-hydroxyacyl-CoA dehydrogenases from rat liver peroxisomes. J Biol Chem. 1994;269:27125–27135. [PubMed] [Google Scholar]
- 33.Pelletier D A, Harwood C S. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J Bacteriol. 1998;180:2330–2336. doi: 10.1128/jb.180.9.2330-2336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persson B, Krook M, Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 35.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 36.Rawlings M, Cronan J J E. The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J Biol Chem. 1992;267:5751–5754. [PubMed] [Google Scholar]
- 37.Schöcke B, Schink B. Energetics of methanogenic benzoate degradation by Syntrophus gentianae in syntrophic coculture. Microbiology. 1997;143:2345–2351. doi: 10.1099/00221287-143-7-2345. [DOI] [PubMed] [Google Scholar]
- 38.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 39.Swoboda-Colberg N G. Chemical contamination of the environment: sources, types and fate of synthetic organic chemicals. In: Young L Y, Cerniglia C E, editors. Microbial transformations and degradation of toxic organic chemicals. New York, N.Y: Wiley-Liss, Inc.; 1995. pp. 27–74. [Google Scholar]
- 40.Tanabe T, Tanaka N, Uchikawa K, Kabashima T, Ito K, Nonaka T, Mitsui Y, Tsuru M, Yoshimoto T. Roles of the ser146, tyr159, and lys163 residues in the catalytic action of 7α-hydroxysteroid dehydrogenase from Escherichia coli. J Biochem. 1998;124:634–641. doi: 10.1093/oxfordjournals.jbchem.a022159. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. Crystal structures of the binary and ternary complexes of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry. 1996;35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 42.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tombolini R, Povolo S, Buson A, Squartini A, Nuti M P. Poly-β-hydroxybutyrate (PHB) biosynthetic genes in Rhizobium meliloti 41. Microbiology. 1995;141:2553–2559. doi: 10.1099/13500872-141-10-2553. [DOI] [PubMed] [Google Scholar]
- 44.Vieille C, Elmerich C. Characterization of an Azospirillum brasilense Sp7 gene homologous to Alcaligenes eutrophus phbB and to Rhizobium meliloti nodG. Mol Gen Genet. 1992;231:375–384. doi: 10.1007/BF00292706. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Yoshimoto T, Higashi H, Kanatani A, Lin X S, Nagai H, Oyama H, Kurazono K, Tsuru D. Cloning and sequencing of the 7α-hydroxysteroid dehydrogenase gene from Escherichia coli HB101 and characterization of the expressed enzyme. J Bacteriol. 1991;173:2173–2179. doi: 10.1128/jb.173.7.2173-2179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]