Abstract
Type IV secretion systems direct transport of protein or nucleoprotein complexes across the cell envelopes of prokaryotic donor and eukaryotic or prokaryotic recipient cells. The process is mediated by a membrane-spanning multiprotein assembly. Potential NTPases belonging to the VirB11 family are an essential part of the membrane-spanning complex. Three representatives of these NTPases originating from the conjugative transfer regions of plasmids RP4 (TrbB) and R388 (TrwD) and from the cag pathogenicity island of Helicobacter pylori (HP0525) were overproduced and purified in native form. The proteins display NTPase activity with distinct substrate specificities in vitro. TrbB shows its highest specific hydrolase activity with dATP, and the preferred substrate for HP0525 is ATP. Analysis of defined TrbB mutations altered in motifs conserved within the VirB11 protein family shows that there is a correlation between the loss or reduction of NTPase activity and transfer frequency. Tryptophan fluorescence spectroscopy of TrbB and HP0525 suggests that both interact with phospholipid membranes, changing their conformation. NTPase activity of both proteins was stimulated by the addition of certain phospholipids. According to our results, Virb11-like proteins seem to most likely be involved in the assembly of the membrane-spanning multiprotein complex.
Translocation of macromolecules across the cell envelope of gram-negative bacteria is facilitated by multiprotein complexes classified in four major families and designated as type I to type IV secretion systems (for recent reviews, see references 4, 8, 20, and 34). Type IV systems are characterized by their unique ability to transfer both DNA and protein molecules unidirectionally to recipient cells. Interestingly, type IV transfer of DNA is apparently always coupled to protein secretion, whereas protein transfer takes place independently and thus is not coupled to DNA transfer. In fact, several type IV systems that are apparently exclusively dedicated to protein transfer have been recently discovered (9, 38, 39a, 43).
Type IV systems comprise the DNA transfer systems specified by conjugative plasmids (29), the agrobacterial T-DNA transfer system (14), and the pertussis toxin secretion system encoded by Bordetella pertussis (Ptl) (43). In the course of the last 2 years, other pathogenicity-related type IV secretion systems have been discovered: Legionella pneumophila Dot/Mrc, which is required for intracellular propagation of the bacterium but also catalyzes the transfer of DNA (39, 42), and the Helicobacter pylori cag pathogenicity island (9), which is involved in the induction of the inflammatory response of infected epithelial cells. Recently, the genome sequence of the obligate intracellular pathogen Rickettsia prowazekii (1) and Actinobacillus actinomycetemcomitans revealed the presence of a gene cluster that evidently encodes another representative of type IV secretion systems. In addition, a type IV secretion system homologue essential for intracellular multiplication has been found in Brucella suis (11).
The presence of gene products with nucleoside triphosphate (NTP) binding motifs in all four families of secretion systems reflects a common requirement for an energy source to drive the macromolecular transport reaction or to assemble the membrane-spanning transport complex. That also underscores the general importance of these proteins for understanding macromolecule secretion. Whereas traffic nucleoside triphosphate hydrolases (NTPases) of type I systems are integral inner membrane proteins, those from types II, III, and IV have been found in peripheral association at the cytoplasmatic side of the inner membrane or, at least partially, in the cytoplasm (10, 16, 26, 31–34, 36, 41).
Interestingly, NTPases encoded by type II and type IV systems are evolutionary related, as suggested by a high degree of sequence relationship (31, 33).
Here, we report the purification of three potential NTPases from type IV systems: the TrbB and TrwD proteins, encoded by the conjugative plasmids RP4 (IncPα) and R388 (IncW), respectively, and the cag HP0525 protein specified by the cag pathogenicity island of H. pylori. All three proteins were overproduced in soluble form in Escherichia coli and were purified by conventional column chromatography. Purification procedures were carried out under native conditions and with the unmodified proteins.
This study was organized by using three prototype proteins of well-defined systems in order to settle some controversies raised in previous publications. For instance, VirB11 solubilized under denaturing conditions (10) and GST (glutathione S-transferase)(226-230)-TrwD (33) were reported as active ATPases while MBP (maltose binding protein)-VirB11 (40) was reported not to hydrolyze ATP (10, 40). Here, we report on the enzymology of the purified proteins, all three displaying NTPase activity in vitro. cag HP0525 and RP4 TrbB interact with phospholipid membranes as demonstrated by fluorescence spectroscopy. Possible functions of transport NTPases within the process of assembly of the transport complex or in energizing the translocation reaction are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and medium.
E. coli K-12 strains SCS1 (18) and HB101 (6) were used as hosts for plasmids. HB101 Nxr is a spontaneous nalidixic acid-resistant derivative of HB101 that served as a recipient in conjugative experiments. Cells were grown in YT medium (27) buffered with 3-(N-morpholino)propanesulfonic acid (sodium salt, pH 8.0) and supplemented with 0.1% glucose and 25 μg of thiamin hydrochloride per ml. When appropriate, antibiotics were added as follows: ampicillin (sodium salt), 100 μg/ml; chloramphenicol, 10 μg/ml; and nalidixic acid, 30 μg/ml. Plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Relevant genotype | Selective markera | Replicon | Reference or source |
|---|---|---|---|---|---|
| pDB126 | pML123Ω(BamHI; RP4 BfaI 45.893-53.462 kbb) | trbB-trbM+ | Cm | ColD | 2 |
| traF-traM+ | |||||
| oriT+ | |||||
| pDB179 | pMS54Δ(NdeI-HindIII)Ω(pDB778 NdeI-HindIII, 0.4 kb) | trbB+ trbC+ | Ap | pMB1 | 17 |
| pMS470Δ8 | pMS119EHΔ(XbaI-PstI)Ω(pT7-7 XbaI-NdeI 40-bp fragment, R751 traC AvaI-SphI 1.4 kb) | Ap | pMB1 | 3 | |
| pMS54 | pMS470Δ8Δ(NdeI-HindIII)Ω(RP4 18.825-19.931 kb) | trbB+ | Ap | pMB1 | 28 |
| pMS55.1 | pMS470Δ8Δ(NdeI-HindIII)Ω(pSU4116 8.353-9.465 kb) | trwD+ | Ap | pMB1 | This work |
| pMS55.2 | pMS470Δ8Δ(NdeI-HindIII)Ω(pSU4116 8.419-9.465 kb) | trwD+ | Ap | pMB1 | This work |
| pSK126 | pDB126Δ(EcoRI-NdeI)Ω(EcoRI-BsrI linker)Ω(pML123 0.960-1.130 kb) | trbB mutant | Cm | ColD | This work |
| trbC-trbM+ | |||||
| traF-traM+ | |||||
| oriT+ | |||||
| pSU4116 | Traw+ | 33 | |||
| pWP4760 | pMS470Δ8Δ(NdeI-HindIII)Ω(NCTC1163 28.052-29.065 kbc) | HP0525+ | Ap | pMB1 | This work |
Reagents and buffers.
Heparin-Sepharose CL-6B, HiTrap heparin, HiTrap Q, and Superose-12 columns were obtained from Pharmacia Biotech. Hydroxylapatite HT gel was from Bio-Rad. Unlabeled nucleoside triphosphates were from Roche Molecular Biochemicals. Radioactive nucleotides were obtained from Amersham Corp. All enzymes were obtained from New England Biolabs and used under the conditions recommended by the manufacturer. Buffer A consisted of 20 mM Tris-HCl (pH 7.6), 0.1 M NaCl, 1 mM dithiothreitol, 0.1 mM EDTA, and 10% glycerol. Buffer B, used for hydroxylapatite chromatography, was made of 20 mM potassium phosphate (pH 6.9), 50 mM KCl, 1 mM dithiothreitol, and 10% glycerol. Buffer C was used for NTPase assays and contained 50 mM 2-(cyclohexylamino)ethanesulfonic acid (CHES) (pH 9.5), 2 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol and 50 μg of bovine serum albumin per ml.
DNA techniques.
Standard molecular cloning techniques were performed by the methods described in Sambrook et al. (35). PCR fragments were generated by using DeepVentR DNA polymerase under the condition recommended by the manufacturer. Plasmid pSK126 is an in-frame deletion mutant of trbB based on plasmid pDB126 (2). For its construction, pDB126 was digested with EcoRI and NdeI, and the resulting 1,130-bp fragment was isolated and digested with BsrI. The 22.6-kb fragment from pDB126, the 171-bp BsrI-NdeI fragment from the BsrI digestion, and an EcoRI-BsrI linker (AATTCGTAGGGGTTACTGAAAAGTGAGC [18,825 to 18,830 bp]) and TCACTTTTCAGTAACCCCTACG [18,825 to 18,828 bp]) were used for a ligation reaction, yielding pSK126. The RP4 sequence is shown in italics; the numbers in brackets refer to sequence coordinates of RP4 (accession no. M93696). The construction was verified by sequencing. DNA fragments containing trwD were generated by the PCR with pSU4116 plasmid DNA (33) as the template by applying two different primer pairs (GGGAATTCATATGTCTACAGTCTCGAAAGC [8,355 to 8,374 bp] [ATG start] or GGGAATTCATATGGCGCAACTCCTGCG [8,419 to 8,434 bp] [GTG start] and GGGTTTAAGCTTCCCCGATACAGCCG [9,449 to 9,464 bp] [R388, accession no. X81123]). The resulting PCR fragments were treated with NdeI and HindIII and were inserted into the multicloning site of pMS470Δ8 (3), yielding pMS55.1 (ATG start) and pMS55.2 (GTG start). Application of the PCR with a primer pair (GGAATAAGCATATGACTGAAGACAGATTGAG [complementary to the sequence with accession no. AC1000108] [28,041 to 28,071 bp]; nucleotides in italics deviate from those in the AC1000108 sequence] and GGTGTTATACAAAAAAGCTTCCATTGGCC [29,047 to 29,076 bp]) and H. pylori NCTC11638 chromosomal DNA (generously provided by Antonello Covacci, Siena, Italy) resulted in the generation of a PCR fragment (1,036 bp) containing the complete HP0525 reading frame. Restriction sites NdeI and HindIII were introduced with the primers so that the NdeI-HindIII-treated PCR fragment could be inserted into the multicloning site of the expression vector pMS470Δ8, resulting in pWP4760.
Mutagenesis of the RP4 trbB coding region.
Three trbB point mutations were generated by using the QuickChange site-directed mutagenesis kit (Stratagene) under the conditions recommended by the manufacturer. pMS54 (28) was used as the template. Oligonucleotide primers applied for K161A were GGTACTGGCTCGGGCGCGACCACGCTCGTC (19,290 to 19,320 bp) and GACGAGCGTGGTCGCGCCCGAGCCAGTACC (19,290 to 19,320 bp), primers applied for D186A were CGTCATCATCGAGGGCCTCGATGATGACG (19,367 to 19,395 bp]), and primers applied for R217T were CAAGACAACGCTGACTATGCGCCCCGACC (19,460 to 19,489 bp) and GGTCGGGGCGCATAGTCAGCGTTGTCTTG (19,460 to 19,489 bp). Deviations from the wild-type (wt) trbB sequence are underlined. Plasmids were digested with KpnI and HindIII, and fragments containing the mutations were subcloned into pMS470Δ8 (3), resulting in the plasmids pMS54K161A, pMS54D186A, and pMS54R217T. Base exchanges introduced in trbB were verified by nucleotide sequencing. For the complementation assay, the SphI and KpnI fragments of the plasmids pMS54K161A, pMS54D186A, and pMS54R217T containing the mutation were used to replace the SphI and KpnI fragment in pDB179.
Complementation assay.
Conjugation experiments employed E. coli HB101 Nxr as the recipient and E. coli HB101 carrying pSK126 and pDB179 as the donor. With a donor-to-recipient cell ratio of 1:10, mixed cultures were filtered on Millipore type HA filters (0.45-μm pore size) and the filters were incubated on YT agar plates without selection for 45 min at 37°C and were washed in 10 mM MgSO4. Serial dilutions were plated onto selective media. Transfer frequencies were expressed as the number of transconjugants per donor cell. Conjugation experiments involving the R388 trwD overexpression plasmids pMS55.1 and pMS55.2 were performed on solid media as described previously (33).
Protein purification.
All operations were carried out at 4°C unless noted otherwise. Protein concentration was analyzed by using the Bradford reagent (7) obtained commercially from Bio-Rad. The results from typical preparations of RP4 TrbB, cag HP0525, and R388 TrwD are summarized in Table 2. Cultures (1.2 liters in 5-liter flasks) of SCS1 (pMS54), SCS1 (pWP4760), or SCS1 (pMS55.1) were grown by shaking at 37°C to an A600 of 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and growth was continued for 5 h. Cells were harvested by centrifugation at 20°C, were resuspended in 5 ml of spermidine solution (100 mM spermidine · 3 HCl, 200 mM NaCl, 2 mM EDTA) per g of [wet weight] of cells, and were stored in liquid nitrogen at −70°C.
TABLE 2.
Purification of TrbB, HP0525, and TrwD
| Protein | Fraction | Purification step | Protein (mg) | Yield (%) | Purity (%) |
|---|---|---|---|---|---|
| TrbB | I | Crude cell extract | 912 | 100 | 27.1 |
| II | (NH4)2SO4 precipitation | 600 | 65.8 | 32.3 | |
| III | Heparin-Sepharose CL-6B chromatography | 262 | 28.7 | 47.6 | |
| IV | Hydroxylapatite chromatography | 85 | 9.3 | 99.8 | |
| HP0525 | I | (NH4)2SO4 precipitation | 918 | 100 | 10.7 |
| II | HiTrap Q chromatography | 146 | 38 | 25.6 | |
| III | Hydroxylapatite chromatography | 19 | 17 | 89.7 | |
| IV | Superose 12 gel filtration | 6.1 | 6 | 98.3 | |
| TrwD | I | Crude cell extract | 1075 | 100 | 31.9 |
| II | (NH4)2SO4 precipitation | 280 | 26.1 | 32.5 | |
| III | Heparin-Sepharose CL-6B chromatography | 135 | 12.6 | 35.4 | |
| IV | Hydroxylapatite chromatography | 23 | 2.1 | 93.4 |
Purification of RP4 TrbB.
Cells (wet weight, 7 g) were thawed, supplemented with 40 ml of a solution containing 10% sucrose, 100 mM Tris-HCl [pH 7.6], 0.6 ml of lysozyme (50 mg/ml), and 1.4 ml of 10% Brij-58 and 29 ml of a solution containing 5% sucrose, 100 mM Tris-HCl [pH 7.6], and 100 mM NaCl. The mixture was stirred for 90 min. After centrifugation at 100,000 × g for 90 min, supernatant I was kept, the pellet was resuspended in 100 ml of buffer A supplemented with 1 M NaCl (final concentration), and the mixture was stirred for 90 min. The resulting lysate was centrifuged at 100,000 × g for 90 min. Supernatant II was combined with supernatant I (fraction I, 164 ml). Fraction I was saturated with (NH4)2SO4 to 60% and was stirred for 2 h. Precipitated proteins were collected by centrifugation at 100,000 × g for 45 min and were resuspended in 100 ml of buffer A with 500 mM NaCl (fraction II, 100 ml). Fraction II was dialyzed against buffer A until a conductivity equivalent to that of buffer A was reached and was applied at a flow rate of 50 ml/h to a heparin-Sepharose column (15 by 110 mm) equilibrated with buffer A and 50 mM NaCl. Proteins were eluted with 400 ml of a linear gradient from 50 mM to 600 mM NaCl in buffer A. TrbB eluted at 375 mM NaCl. The peak fractions were pooled (fraction III, 45 ml). Fraction III was applied directly, at a flow rate of 50 ml/h, to a hydroxylapatite column (15 by 100 mm) equilibrated with buffer B. The column was washed with 60 ml of buffer B and then with 60 ml of buffer B containing 250 mM potassium phosphate. Proteins were eluted with 60 ml of buffer B with 500 mM potassium phosphate, and fractions containing TrbB were pooled (fraction IV, 29 ml). To concentrate fraction IV, it was dialyzed against buffer A with 200 mM NaCl and 20% polyethylene glycol (PEG) 20,000. TrbB has been stored in 50% glycerol in buffer A with 200 mM NaCl at −20°C without loss of activity for 3 years.
Purification of H. pylori cag HP0525.
Cells (13 g [wet weight] in spermidine solution, 80 ml) were lysed at 0°C in a solution containing 5% sucrose, 100 mM Tris-HCl [pH 7.6], 100 mM NaCl, 0.25% Brij-35, and 0.2 mg of lysozyme per ml (final concentrations are given) (final volume, 325 ml). After 90 min, the highly viscous lysate was centrifuged at 100,000 × g for 90 min. Proteins in the supernatant (300 ml) were precipitated by the addition of (NH4)2SO4 (60% saturation at 0°C). Following stirring at 0°C for 2 h, the precipitate was collected by centrifugation for 30 min at 10,000 × g. Pellets were resuspended in 80 ml of buffer A containing 0.01% Brij-35 and were dialyzed against several changes of the same buffer (fraction I, 80 ml). Fraction I (two aliquots of 40 ml each) was applied to a 5-ml HiTrap Q column equilibrated in buffer A–0.01% Brij-35. Using Pharmacia fast protein liquid chromatography equipment, proteins were eluted with a 40-ml linear gradient of 100 to 600 mM NaCl in buffer A–0.01% Brij-35, and 5-ml fractions were collected. HP0525 eluted at approximately 400 mM NaCl (fraction II, 10 ml). Fraction II (two aliquots of 5 ml each) was applied to a 5-ml hydroxylapatite column equilibrated in buffer B. Proteins were eluted with a 30-ml linear gradient from 20 to 350 mM potassium phosphate in buffer B, and 2.5-ml fractions were collected. HP0525 eluted at 190 mM potassium phosphate (fraction III, 15 ml). Prior to gel filtration, fraction III was concentrated by dialysis against 20% PEG 20,000 in buffer A–0.01% Brij-35. When a final volume of 3.5 ml was reached, several 200-μl aliquots, each containing 1 mg of protein, were applied to a Superose 12 column (10 by 300 mm) preequilibrated in buffer A–0.01% Brij-35. Proteins were eluted at a flow rate of 0.5 ml/min, and 0.5-ml fractions were collected. Fractions containing HP0525 were pooled, were concentrated by dialysis against 20% PEG 20,000 in buffer A–0.01% Brij-35, and were finally dialyzed against 50% glycerol in buffer A–0.01% Brij-35 (fraction IV, 2 ml). Fraction IV was stored at −20°C without loss of activity for 1 year.
Purification of R388 TrwD.
Cells (10 g [wet weight] of bacterial pellet) were thawed and supplemented with 52 ml of a solution containing 10% sucrose, 100 mM Tris-HCl [pH 7.6], 1.75 ml of lysozyme (50 mg/ml), 3.92 ml of 10% Brij-58, 20 ml of 5 M NaCl, and 123 ml of 5% sucrose-100 mM Tris-HCl [pH 7.6]-100 mM NaCl and were stirred for 90 min. Following centrifugation at 100,000 × g for 90 min, supernatant I was kept and the pellet was resuspended in 40 ml of buffer A supplemented with 1 M NaCl (final concentration) and was stirred for 90 min. The resulting lysate was centrifuged at 100,000 × g for 90 min, and supernatant II was combined with supernatant I (fraction I, 212 ml). Fraction I was saturated with (NH4)2SO4 to 60% and was stirred for 2 h. Precipitated proteins were collected by centrifugation at 100,000 × g for 45 min and were resuspended in 35 ml of buffer A with 500 mM NaCl (fraction II, 35 ml). Fraction II was dialyzed against buffer A-150 mM NaCl to a conductivity equivalent to that of buffer A-150 mM NaCl and was applied, at a flow rate of 50 ml/h, to a heparin-Sepharose column (15 by 280 mm) equilibrated with buffer A-150 mM NaCl. Proteins were eluted with 500 ml of a linear gradient of 150 to 600 mM NaCl in buffer A. TrwD eluted at 220 mM NaCl. The peak fractions were pooled (fraction III, 150 ml). Fraction III was applied directly, at a flow rate of 50 ml/h, to a hydroxylapatite column (15 by 140 mm) equilibrated with buffer B. The column was washed with 150 ml of buffer B. Proteins were eluted with 250 ml of a linear gradient of 20 to 500 mM potassium phosphate in buffer B, and fractions containing TrwD were pooled (fraction IV, 120 ml). To concentrate fraction IV, it was dialyzed against buffer A with 500 mM NaCl and 20% PEG 20,000. TrwD has been stored in 50% glycerol in buffer A with 500 mM NaCl at −20°C without loss of activity for 1 year.
NTPase Assay.
NTP hydrolysis reactions (20 μl) were performed with 1.5 μM purified protein at 30°C for 20 min in buffer C containing 0.1 μCi of [α-32P]- or [γ-32P]NTP and 0.2 mM unlabeled NTP. Reaction products were separated by thin-layer chromatography on cellulose MN300 polyethyleneimine from Macherey-Nagel (37), and radioactive NTP and nucleoside diphosphate (NDP) or Pi were quantified by using storage phosphor technology (22) and the ImageQuant software (Molecular Dynamics).
Preparation of phospholipid membranes.
Phospholipids were purchased from Sigma as 10-mg/ml solutions in chloroform-methanol (9:1). The required amount was dried under a nitrogen stream and was suspended at a concentration of 1 mg/ml in the appropriate volume of 20 mM Tris-HCl [pH 7.6] and 100 mM NaCl. Samples were sonicated for 15 min at 10°C and were used immediately for further experiments.
RESULTS
Heterologous overexpression of RP4 trbB, cag HP0525, and R388 trwD in E. coli results in overproduction of a soluble protein.
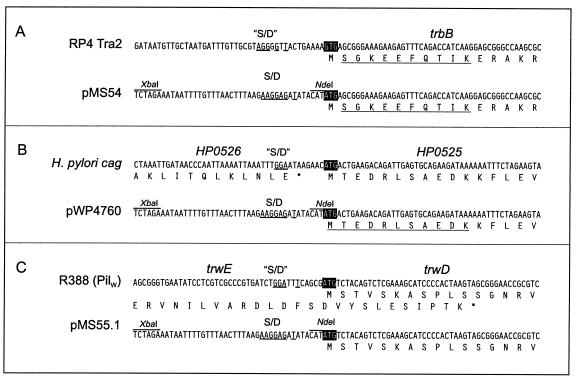
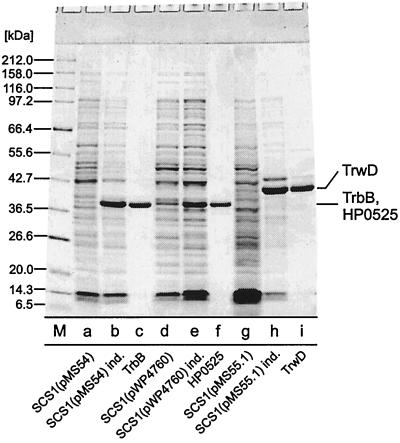
The trbB structural gene was obtained via NdeI restriction of RP4 (28). HP0525 and trwD structural genes were obtained from PCR fragments. The trwD open reading frame contains two potential start codons: an ATG initiation codon and a GTG located 22 codons downstream. Rivas et al. (33) have demonstrated that an N-terminal GST fusion to Ser-2 of TrwD was fully active in vivo. To test whether trwD starts with the ATG or the GTG initiation codon, two corresponding PCR fragments were generated, resulting in pMS55.1 (ATG start) and pMS55.2 (GTG start). Genes were placed under transcriptional control by the tac promoter and under translational control by the T7 gene 10 Shine-Dalgarno sequence by insertion into the vector pMS470Δ8 (Fig. 1). The insertions were sequenced after molecular cloning. Induction of the resulting strains SCS1(pMS54), SCS1(pWP4760), SCS1(pMS55.1), and SCS1(pMS55.2) with 1 mM IPTG for 5 h resulted in overproduction of additional polypeptides with apparent molecular masses of 32, 35, 44, and 41 kDa, respectively. These values are in good agreement with the calculated masses for TrbB (34,995.1 Da) and HP0525 (37,579.5 Da), respectively. The mass of the protein produced by SCS1(pMS55.1) differs significantly from the calculated molecular mass of TrwD (38,002.4 Da), but the mass of the protein produced in SCS1(pMS55.2) is in good agreement with the calculated mass. However, the latter protein was not functional in vivo (vide infra). Given the rather acidic calculated pI of TrwD of 5.9, the difference of the mass of TrwD and the calculated molecular mass is not unexpected. All three proteins could be identified in Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels representing approximately 52% of the SDS-soluble proteins in SCS1(pMS54), approximately 12% of the SDS-soluble proteins in SCS1(pWP4760), and approximately 62% of the SDS-soluble proteins in SCS1(pMS55.1) (Fig. 2). Solubility studies indicated that at least 50% of the overproduced proteins were soluble under low-salt conditions (50 mM NaCl) in buffers containing 0.1% of the nonionic detergent Brij-35 or Brij-58. These soluble protein fractions served as the starting material for purification of TrbB, HP0525, and TrwD.
FIG. 1.
Construction of overexpression plasmids for RP4 trbB, cag HP0525, and R388 trwD. Upper panels, nucleotide sequences flanking the initiation codons of trbB (A), HP0525 (B), and trwD (C) in their native environments. Nucleotide sequences resembling Shine-Dalgarno sequences (“S/D”) are underlined. Initiation codons are marked by black boxes. Partial amino acid sequences of gene products are shown below the nucleotide sequences. Lower panels, genes were placed under translational control by the T7 gene 10 ribosomal binding site (S/D) by insertion into the expression vector pMS470Δ8. Underlined parts of the amino acid sequences were confirmed by N-terminal sequencing.
FIG. 2.
Purification of NTPases of type IV secretion pathways. Lane a, SCS1 (pMS54), no IPTG added; lane b, SCS1(pMS54), cells were induced for 5 h with 1 mM IPTG; lane c, TrbB, fraction V, 5 μg; lane d, SCS1(pWP4760), no IPTG added; lane e, SCS1(pWP4760), cells were induced for 5 h with 1 mM IPTG; lane f, HP0525, fraction IV, 5 μg; lane g, SCS1(pMS55.1), no IPTG added; lane h, SCS1(pMS55.1) cells were induced for 5 h with 1 mM IPTG; lane i, TrwD, fraction V, 5 μg; M, molecular mass standards, masses are given in kDa. Samples were electrophoresed in an SDS–15% polyacrylamide gel. The gel was stained with Serva Blue R.
Overproduced proteins are functional in complementing corresponding mutants in vivo.
pDB179 (trbB, trbC) was able to complement an in-frame deletion mutation of trbB in vivo as shown by a complementation assay. TrbB alone was not able to restore the wt transfer efficiency of the trbB deletion mutant (data not shown). This phenomenon was also observed for the complementation of a multiple reading frame insertion linker insertion mutation in trbB (17, 25). pMS55.1 (trwD, ATG start) was shown to be active in vivo by applying a complementation assay. pMS55.2 (trwD, GTG start) could not complement a trwD mutant (data not shown), indicating that the trwD gene starts with an ATG. A corresponding complementation assay for the overexpressed HP0525 gene was not performed. However, it is known that nonpolar knockout mutants in HP0525 are impaired in eliciting interleukin-8 production, and hence the inflammatory response in infected mammalian cells, demonstrating the functional importance of HP0525 for the H. pylori cag region (A. Covacci, personal communication).
Purification of RP4 TrbB.
The purification of TrbB from an extract of the overproducing strain SCS1 (pMS54) is summarized in Table 2. TrbB was purified by heparin-Sepharose chromatography followed by hydroxylapatite chromatography. With a sodium chloride gradient, TrbB eluted as a single peak at 375 mM NaCl from heparin-Sepharose (data not shown). This protein solution consisted mainly of TrbB and lysozyme (the latter was used to break up the cells). Lysozyme was separated from TrbB by hydroxylapatite chromatography. TrbB eluted as a single peak when the column was washed with 500 mM potassium phosphate. The preparation of purified TrbB (fraction IV) was 99.8% pure as judged by Coomassie blue staining of an SDS–15% polyacrylamide gel (Fig. 2, lane c).
Purification of cag HP0525.
HP0525 was purified to near homogeneity by a fast protein liquid chromatography-based three-step procedure (Table 2). Surprisingly, HP0525 did not bind to heparin columns under low-salt conditions, as did TrbB or TrwD. Therefore, anion-exchange chromatography on HiTrap Q was chosen as the first purification step. A second chromatography step on hydroxylapatite yielded an almost pure protein fraction. Remaining impurities were removed by gel filtration on Superose 12, resulting in a 98.3% pure preparation of HP0525 (Table 2 and Fig. 2, lane f).
Purification of R388 TrwD.
The purification of TrwD from an extract of the overproducing strain SCS1(pMS55.1) is summarized in Table 2. TrwD was purified by applying the same two-step procedure as for TrbB. However, TrwD was found to bind less tightly than TrbB to both heparin-Sepharose and hydroxylapatite. Elution from heparin-Sepharose took place at 220 mM NaCl and from hydroxylapatite at 160 mM potassium phosphate when a linear potassium phosphate gradient was applied. The preparation of TrwD (fraction IV) was 93.4% pure as judged by Coomassie blue staining of an SDS–15% polyacrylamide gel (Fig. 2, lane i).
R388 TrwD forms ring-shaped oligomers.
Image processing of TrbB and HP0525 micrographs showed that both proteins form hexameric ring-shaped assemblies (23). Electron microscopy was applied to purified TrwD to verify that the oligomeric structure most likely hexameric is also common to TrwD. Experiments were carried out as described previously (23).
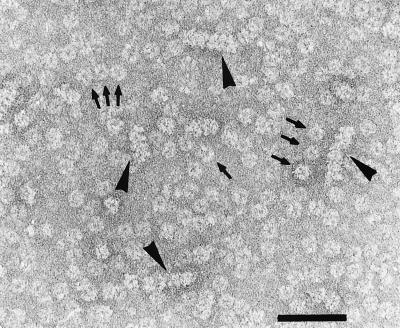
As already demonstrated for TrbB, TrwD also formed ring-shaped complexes in the presence of ATP and Mg2+. Similar to TrbB, TrwD oligomers were not very stable, resulting in partially disassembled protein complexes such as open rings and lower oligomers that were detectable among the familiar ring-shaped assemblies (Fig. 3). We were not able to stabilize the TrwD structure by using glutaraldehyde, as it was possible for TrbB since TrwD tends to aggregate under these conditions. Interestingly, tube-like structures were also observed (Fig. 3).
FIG. 3.
R388 TrwD forms hexameric rings. TrwD (50 ng/μl) was preincubated in 20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM ATP, and 10 mM MgCl2 for 10 min at room temperature. Samples were stained with 1% uranyl acetate (12) and were observed in a Phillips CM 100 electron microscope. Bar, 100 nm. Arrows indicate ring-shaped molecules and arrow heads indicate tube-like structures.
RP4 TrbB, cag HP0525, and R388 TrwD display NTPase activity in vitro.
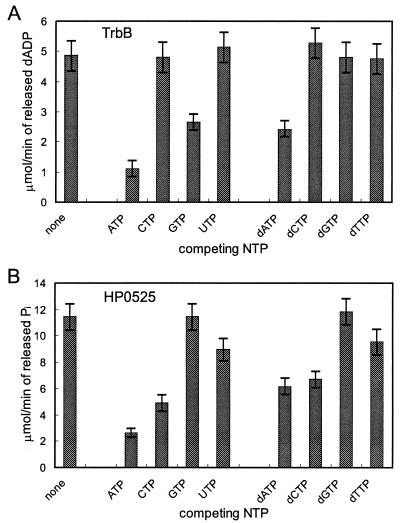
The most prominent feature in the amino acid sequences of PulE family proteins is the presence of a conserved NTP binding motif. To explore the functional significance of this element, we tested purified preparations of TrbB and HP0525 for NTPase activity in vitro. TrbB NTPase activity was characterized under standard conditions (see Materials and Methods) when not indicated otherwise. Purified TrbB protein hydrolyzed [α-32P]dATP, [γ-32P]GTP, and [γ-32P]ATP to the corresponding nucleoside diphosphate and Pi as shown by thin-layer chromatography. The dATPase activity coincided with the protein peak when purified TrbB was subjected to glycerol gradient centrifugation, indicating that the detected activity is not due to impurities (23). At pH 7.5 and 2 mM Mg2+, GTP and ATP were found to be poorer substrates than dATP since the rate of hydrolysis was one-third and 1/10, respectively, that of dATP. dATPase activity was assayed in the presence of 200 μM dATP, and the other ribo- or deoxyribonucleoside triphosphates were added to the assay to determine whether they can compete for binding to TrbB (Fig. 4A). None of the tested nucleotides was found to be a better substrate for this enzyme. dATPase, GTPase, and ATPase activities were also determined when each of the two other nucleotides was competing for binding to TrbB. Thus, a decrease of the hydrolyzing activity compared to the activity with the labeled nucleotide alone would indicate a better binding of the added unlabeled nucleotide to the active site of TrbB. This assay showed that TrbB bound ATP more tightly than GTP and dATP; however, it hydrolyzed dATP with the highest efficiency. Native single-stranded DNA of bacteriophage M13 mp19 did not stimulate the dATPase, ATPase, or GTPase activity.
FIG. 4.
TrbB and HP0525 display distinct substrate affinities. (A) TrbB was incubated under standard conditions with labeled [α-32P]dATP (final concentration, 200 μM). Unlabeled NTPs or dNTPs were added to the reaction in fivefold molar excess, and dATP hydrolase activity was determined as described in Materials and Methods. (B) HP0525 was incubated under standard conditions with labeled [γ-32P]ATP (final concentration, 500 μM). Unlabeled NTPs or dNTPs were added to the reaction in fivefold molar excess, and ATP hydrolase activity was determined as described in Materials and Methods.
Optimal conditions for the TrbB dATPase, GTPase, and ATPase activities were pH 9.5 and 2 mM Mg2+. A second pH maximum of approximately 7.0 was observed for the GTPase and ATPase activity. When Mg2+ was replaced by 1 mM Mn2+, activities with the three substrates were reduced by 50% (dATPase), 70% (GTPase), or 30% (ATPase), whereas no activity was observed (<2%) when Ca+2 was substituted for Mg+2.
Characteristics of the HP0525 NTP hydrolase activity are very similar to those of TrbB. The pH optimum of the ATPase locates at around 9.5, and divalent cations are required for the reaction. Mg2+ supports ATP hydrolysis best, followed by Mn2+ (80%) and Ca2+ (35%). Thus, the only difference with respect to TrbB is that Ca2+ supports the reaction, although with low efficiency. More pronounced differences between TrbB and HP0525 are seen when the substrate requirements are compared: HP0525 hydrolyzes ATP with the highest efficiency, followed by dATP. By using the competition assay described above for detecting ATP hydrolysis in the presence of various concentrations of other (d)NTPs, relative affinities were determined to be in the order ATP, dATP > CTP, dCTP > TTP, UTP (Fig. 4). GTP and dGTP apparently are not at all accepted as substrates by HP0525 (Fig. 4B). This is a clear difference from TrbB, which hydrolyzes ATP with only 10% efficiency but GTP with 30% efficiency compared to that of its preferred substrate dATP.
For GST(226-230)-TrwD, it was reported that its preferred substrate is ATP over GTP (33). The purified TrwD was able to hydrolyze ATP (data not shown). Furthermore, it was found that dATP is also a substrate for TrwD, but dATP was hydrolyzed to a lesser extent than ATP (data not shown). Furthermore, the dATPase activity of TrwD was lower than the one of TrbB and HP0525.
Phospholipids stimulate NTP hydrolase activity of RP4 TrbB and cag HP0525.
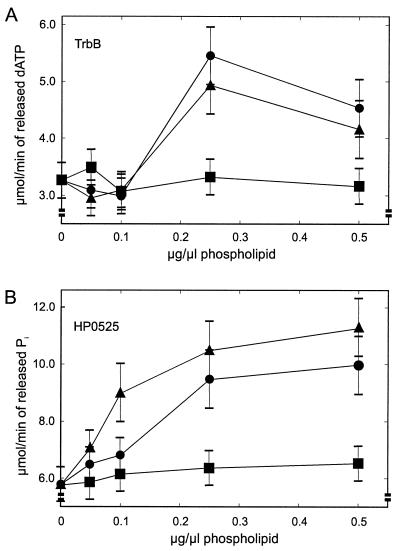
Neither TrbB nor HP0525 or TrwD contain apparent membrane-spanning domains. However, several VirB11-like proteins colocalize with the inner membrane fraction in sedimentation experiments (10, 16, 32, 33). Linked to this observation, it was found that the ATP hydrolase activity of GST(226-230)-TrwD was enhanced by phospholipids (33). Therefore, the influence of phospholipids, the main constituents of the inner bacterial membrane, on the NTP hydrolase activity of both TrbB and HP0525 was analyzed. Phospholipid membrane suspensions were incubated at various concentrations with TrbB and HP0525, and the amount of hydrolyzed input (d)ATP was determined after incubation for 30 min at 30°C (Fig. 5). Both TrbB and HP0525 are stimulated up to about twofold at 0.25 μg of cardiolipin (CL) and phosphatidylglycerol (PG) per μl. In both proteins, higher concentrations (0.5 μg/μl) do not lead to a significantly different stimulatory effect. Phosphatidylethanolamine (PE) had no significant effect on either protein (Fig. 5).
FIG. 5.
Phospholipids stimulate NTP hydrolase activities of transport NTPases. (A) TrbB dATP hydrolase activity in the presence of various phospholipids; (B) ATP hydrolase activity of HP0525 in the presence of various phospholipids. NTPase assays were performed in 50 mM Tris-HCl [pH 7.5], 50 mM KCl, 2 mM MgCl2, 0.05 mg of bovine serum albumin, 1 mM dithiothreitol, and 0.25 mM EDTA. Phospholipids were added to the reactions as sonicated suspensions (see Materials and Methods), and after a preincubation period of 10 min at 30°C, 0.1 μCi of radiolabeled (d)ATP was added (final concentration, 0.2 mM). Incubation was continued for 30 min at 30°C, and the reactions were stopped by the addition of EDTA (final concentration, 50 mM). Products were analyzed by thin-layer chromatography and were quantified as described in Materials and Methods. Symbols: ▴, CL; ●, PG; ■, PE.
RP4 TrbB and cag HP0525 undergo conformational changes in the presence of phospholipids.
Tryptophan fluorescence spectroscopy was applied to see whether the stimulation of NTP-hydrolase activity that is observed with HP0525 and TrbB in the presence of phospholipids is reflected by conformational changes of the proteins. Both TrbB and HP0525 contain only two tryptophan residues each, which served as convenient probes to monitor polarity changes in the environment of the corresponding protein domains. The tryptophan residues within TrbB are well separated (Trp-47 to Trp-240), thus occupying different domains of the protein, whereas the tryptophan residues in HP0525 are separated by only six amino acids (Trp-57 to Trp-64).
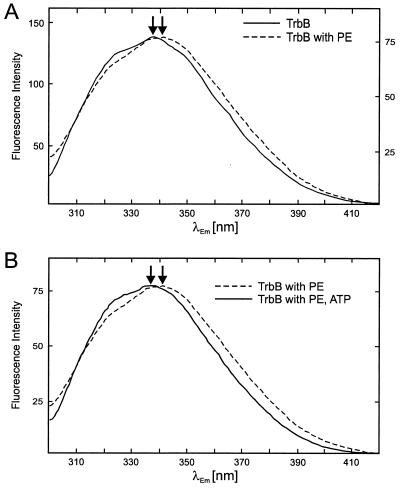
The fluorescence spectrum of TrbB shows a peak with a maximum at 336 nm and a shoulder that corresponds to an estimated peak maximum around 325 nm (Fig. 6). Addition of PE resulted in a moderate quench of fluorescence intensity by about 50%. The maximum of fluorescence intensity in the presence of PE was shifted to 342 nm (Fig. 6A), indicating an increase of the polarity in the environment of the respective tryptophan residue (24). The position of the shoulder remained unaffected. Hexameric rings of TrbB are known to be stabilized by nucleotides and Mg2+ (23); therefore, the influence of ATP and Mg2+ on the fluorescence spectrum of TrbB was tested. Subsequent addition of ATP in the presence of Mg2+ had no effect on the TrbB fluorescence spectrum (data not shown). However, the shift of the peak maximum that was caused by addition of PE was not observed when TrbB was preincubated with ATP and Mg2+ (Fig. 6B). Incubation of TrbB with CL or PG under the same conditions gave virtually identical results (data not shown). Therefore, Fig. 6 illustrates only the data obtained with PE, the most abundant phospholipid in the E. coli cytoplasmic membrane (15).
FIG. 6.
Fluorescence spectroscopy of TrbB. (A) TrbB undergoes conformational changes in the presence of PE. Continuous line, the fluorescence spectrum of TrbB was measured at a protein concentration of 100 μg/ml in 20 mM Tris-HCl [pH 7.6], 50 mM NaCl, 5 mM MgCl2, and 0.1 mM EDTA. Fluorescence was measured at an excitation wavelength of 288 nm with a Perkin-Elmer LS50B spectrofluorimeter. Emission was recorded between 300 and 420 nm. Excitation and emission slits were set to 2.5 and 4 nm, respectively. Dotted line, PE was added as a sonicated suspension to a final concentration of 100 μg/ml. The TrbB/PE mixture was incubated for 30 min at 37°C, and the fluorescence spectrum was recorded. Peak intensities of the spectra were equalized. Arbitrary units for fluorescence intensities in the absence and presence of PE are given on the left- and right-hand side of the spectrum, respectively. The TrbB emission maxima in the absence and presence of PE are marked by arrows. (B) Dotted line, same spectrum as indicated by the dotted line in panel A; continuous line, TrbB was preincubated for 15 min at 37°C in the presence of 1 mM ATP. PE was added as described above, and after incubation of the TrbB-PE-ATP mixture for 30 min at 37°C, the fluorescence spectrum was recorded as described. Emission maxima with and without preincubation of TrbB with ATP are marked by arrows.
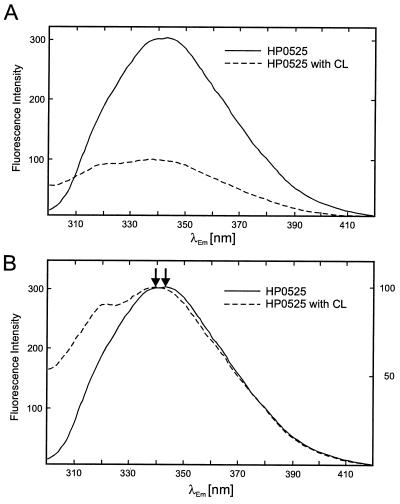
The fluorescence spectrum of HP0525 shows a single maximum at 343 nm, consistent with the assumption that its two closely spaced tryptophan residues face very similar environments (Fig. 7). Addition of CL resulted in a strong quench (Fig. 7A), and the fluorescence maximum shifted to 339 nm (Fig. 7B). Both are indications for a conformational change that renders the environment of the HP0525 domain containing the two tryptophan residues less polar. Preincubation of HP0525 with ATP in the presence of Mg2+ had no effect on the spectra (data not shown). CL had the strongest effect on the peak position of the HP0525 spectrum. Addition of PE or PG resulted in a comparable quenching effect; however, the peak position remained largely unaffected with PE and a lesser shift to shorter wavelength than with CL was observed with PG (data not shown).
FIG. 7.
Fluorescence spectroscopy of cag HP0525. (A) Quenching of HP0525 fluorescence in the presence of CL; continuous line, the fluorescence spectrum of HP0525 was measured at a protein concentration of 25 μg/ml in 20 mM Tris-HCl [pH 7.6], 50 mM NaCl, and 0.1 mM EDTA. Parameters for fluorescence measurement were as described in the legend to Fig. 6. Dotted line, CL was added as a sonicated suspension to a final concentration of 100 μg/ml. The HP0525-CL mixture was incubated for 30 min at 37°C, and the fluorescence spectrum was recorded as described above. (B) Same as for panel A; however, the peak intensities of the spectra are equalized. The HP0525 emission maxima in the absence and presence of CL are marked by arrows. Arbitrary units for fluorescence intensities in the absence and presence of CL are given on the left- and right-hand side of the spectrum, respectively.
The nucleotide binding site type A of RP4 TrbB is essential for conjugative transfer.
Four highly conserved motifs are common among the members of the VirB11 family: a nucleotide binding site type A (Walker A Box), an aspartate box (Asp Box), a nucleotide binding site type B (Walker B Box), and a histidine box (His Box) (23, 33). For functional and structural studies of TrbB, we chose lysine 161 in the Walker A Box, aspartate 186, and arginine 217 in the Walker Box as targets for site-directed mutagenesis. Rules for amino acid replacements were followed to minimize structural distortions in the protein (5).
For measuring conjugative plasmid transfer, the complementation assay system employed strains carrying pSK126(ΔtrbB) and pDB179 or deviates of pDB179 (see Materials and Methods). A mutation in the Walker A Box of TrbB (K161A) abolished the capacity to restore conjugation in the complementation assay, whereas the other two mutations in the Asp Box (D186A) and in the Walker B Box (R217T) only slightly affected transfer (Table 3).
TABLE 3.
Properties of trbB mutants
| trbB mutant | Transfer frequencya | Activity ofb:
|
Assembly to hexameric rings | Trans- dominancec | ||||
|---|---|---|---|---|---|---|---|---|
| dATPase (pH 9.5) | GTPase
|
ATPase
|
||||||
| pH 7.5 | pH 9.5 | pH 7.5 | pH 9.5 | |||||
| wt | 2.0 × 100 | 4.48 ± 0.5 | 0.9 ± 0.1 | 1.88 ± 0.3 | 0.55 ± 0.1 | 0.91 ± 0.1 | + | 4.2 × 10−1 |
| wtd | 2.27 ± 0.4 | 0.26 ± 0.03 | 0.34 ± 0.03 | 0.28 ± 0.03 | 0.31 ± 0.03 | + | ||
| K161A | <10−7 | <0.03 | <0.03 | <0.03 | <0.03 | <0.03 | + | 1.4 × 10−4 |
| D186A | 4.8 × 10−1 | <0.03 | 0.11 ± 0.03 | <0.07 | <0.07 | 0.22 ± 0.03 | + | NDe |
| R217T | 9.6 × 10−1 | <0.07 | <0.09 | <0.03 | <0.03 | <0.03 | + | ND |
Transfer frequencies are expressed as transconjugants per donor cell.
The NTPase activities were analyzed with a 1.5 μM concentration of the respective protein at 30°C for 20 min. Activity values are given in micromoles of released NDP per minute.
For determination of trans-dominance of trbB mutants, merodiploid strains HB101(pSK126 trbBwt, pDB179 trbBmut) expressing both wt and mutant trbB genes were employed as donor strains. Transfer frequencies are expressed as transconjugants per donor cell.
Purified using a 6 M urea extract as starting material.
ND, not determined.
Mutant proteins TrbB K161A and TrbB R217T were as soluble under native conditions as wt TrbB. The corresponding cell extracts were used as starting material for purification. Mutation D186A resulted in a protein which was insoluble under native conditions but which could be solubilized with 6 M urea. Thus, a urea extract which was dialyzed against buffer A to renature the protein was used as starting material for purification. As a control, wt TrbB was also purified from a renatured urea extract to verify that TrbB retained its activity upon de- and renaturation.
For determination of their NTPase activities, the three mutant proteins were purified by applying the same purification procedure as used for wt TrbB. Mutant proteins were assayed as described in Materials and Methods. Since the GTPase and ATPase activity of wt TrbB contained two pH maxima, at pH 7.0 and 9.5, mutant proteins were additionally assayed at pH 7.5 for GTPase and ATPase activity. All three mutant proteins were found to be deficient in hydrolyzing dATP. TrbB K161A was inactive in all tested NTPase activities. TrbB D186A retained 40% of the wt GTPase at pH 7.5 and 60% of the wt ATPase at pH 9.5. Interestingly, the highest GTPase activity of TrbB D186A was not detected at pH 9.5 as for wt TrbB but at pH 7.5 (Table 3). TrbB R217T did not hydrolyze ATP but retained a very weak GTPase activity at pH 7.5.
Electron microscopy showed that all three mutant proteins were still as able as wt TrbB to form hexameric rings when they were incubated in the presence of ATP and Mg2+ (data not shown). The transdominant effect of trbB K161A on wt trbB indicates that the mutant protein is able to form hetero-oligomers with the wt protein or that mutant protein competes with the wt protein for interaction with other components of the secretion complex. Since TrbB K161A and TrbB R217T did not hydrolyze ATP, it is the binding of the nucleotide and not the hydrolysis that seems to stabilize the ring form.
DISCUSSION
We have overproduced and purified three representatives from the VirB11 family of transport NTPases. All three proteins were obtained in substantial amounts in E. coli, and the majority of the material appeared as soluble protein in the cytoplasmatic fraction, allowing purification under native conditions. Thus, a comparative study of the enzymatic properties of three related NTPases encoded by type IV secretion systems was possible.
TrbB, GST(226-230)-TrwD (33), TrwD (data not shown), and HP0525 display a rather weak NTPase activity in vitro within an order of magnitude, similar to chaperons like DnaK (44) and ClpA (21). It is therefore very suggestive that NTP hydrolysis is used by these proteins to energize a chaperone-like process involved in the assembly of the membrane-spanning multiprotein complex. Interestingly, the substrate spectra of TrbB and HP0525 differ considerably in vitro. Substrates for TrbB are dATP, GTP, and ATP, whereas HP0525 has a much wider spectrum with a preference for ATP and dATP. In contrast to TrbB, GTP does not compete for ATP hydrolysis by HP0525. The biological significance of these differences remains to be elucidated.
Mutant protein TrbB K161A was deficient in restoring wt transfer frequency in a trbB0 mutant strain and did not hydrolyze any of the nucleotide substrates used by TrbB. The analyses of the NTPase activity of TrbB D186A and TrbB R217T, which were both able to mediate conjugation, did not allow a clear-cut conclusion because some of the NTPase activities of the mutant proteins were close to the detection level of the method. There is a correlation between loss or reduction of NTPase activity and transfer frequency, but, surprisingly, transfer has decreased only to levels of 25% (TrbB D186A) or 50% (TrbB R217T) of the wt level. This might indicate that the conserved aspartate and arginine residues in E. coli do not seem to play an important role for the structure or function of TrbB in vivo. Another explanation could be that TrbB and its homologues are indeed involved only in the assembly of the type IV transporter, and once this complex has formed it is fully functional without requiring the activity of TrbB anymore. Thus, a very low residual activity of the mutant TrbB could still account for rather high levels of DNA transfer.
Incubation of HP0525 and TrbB with the phospholipids CL and PG resulted in a significant stimulation of the NTPase activity. Stimulation of NTPase activity by phospholipids has also been reported earlier for GST(226-230)-TrwD (33). These findings are in accordance with the described association of VirB11-like proteins with the inner membrane (10, 16, 32, 33). An association with the inner membrane was also indicated by fluorescence spectroscopy, since the conformations of TrbB and HP0525 are changed in the presence of certain phospholipids.
Incubation of HP0525 with CL, and to a lesser extent with other phospholipids, resulted in a strong quench of tryptophan fluorescence and a shift of the emission maximum to shorter wavelengths. The shift is indicative of a less polar environment of the tryptophan residues in the presence of phospholipid (24). A tight interaction of the N-terminal domain harboring the two HP0525 tryptophan residues (Trp-57 and Trp-64) with the phospholipid membranes, possibly by integration or peripheral association, might explain this polarity change.
In contrast to HP0525, different fluorescence spectra were observed when TrbB was first incubated with ATP and Mg2+ followed by incubation with PE versus when the incubation order was reversed. A much lesser quenching of tryptophan fluorescence was observed with HP0525 and no shift of the emission maximum was detected with TrbB when preincubated with ATP and Mg2+, conditions known to stabilize the hexameric structure of TrbB. Thus, neither of the TrbB tryptophan residues (Trp-47 and Trp-240) in a TrbB hexamer seem to interact with phospholipid membranes as shown for HP0525. This is not surprising, since the positions of the tryptophan residues in TrbB differ from those in HP0525.
On the other hand, when TrbB was incubated with PE first, a shift of the emission maximum to longer wavelengths, indicative of a more polar environment, was observed. The exposure of at least one of the two TrbB tryptophan residues to a more polar environment might be due to a conformational change of TrbB, e.g., dissociation into single subunits. Subsequent addition of ATP plus Mg2+ had no further effect on the fluorescence spectrum, indicating that once the PE-induced change of the TrbB conformation has occurred it cannot be influenced or reversed by ATP plus Mg2+.
In contrast to the phospholipid-induced changes in the HP0525 fluorescence spectrum, changes observed in TrbB obviously do not reflect the interactions responsible for phospholipid-induced NTPase stimulation. Changes in the TrbB spectrum in the absence of ATP plus Mg2+ are independent of the type of phospholipid applied. However, NTPase is only stimulated by CL and PG. Moreover, in the presence of ATP, no changes in the emission maximum of the TrbB spectrum are observed. The opposite is true for HP0525, where only those phospholipid species which stimulate NTPase shift the emission maximum, and changes in the fluorescence spectrum are observed both in the presence and absence of ATP plus Mg2+.
Nevertheless, the interaction of TrbB with the inner membrane in vivo might be dependent on the ATP concentration in the cell. In this context, it is interesting to mention that no functional receptor for donor-specific phages is detectable after arsenate addition (13). This indicates an energy requirement in the form of ATP to keep the Mpf complex active since the entire Mpf complex is needed to present the receptor on the cell surface.
As it was already shown for TrbB and HP0525, TrwD forms ring-like structures which are stabilized by NTPs and Mg2+, similar to TrbB (23). Additionally, TrwD tends to form tube-like aggregates, but the significance of these structures for the function of TrwD remains to be elucidated.
The existence of ring-shaped oligomers is a very common feature in NTP-hydrolyzing enzymes that catalyze repetitive reactions or need a high concentration of active sites, allowing a simultaneous interaction at different places of a substrate. Examples include DNA helicases, chaperones like GroEL or ClpX, and motor-like proteins such as the F0F1-ATPase (19).
Proteins belonging to the VirB11 family are essential components of type IV secretion systems. Thus, they are involved in the assembly of the membrane-spanning transport complex and in the transfer of the cognate substrate of the transport complex which could be a protein, a nucleoprotein complex, or DNA. Since the activity of neither NTPase described here is stimulated by the addition of single-stranded DNA (data not shown), it is rather likely that a protein and not DNA or a nucleoprotein complex is the substrate for the VirB11-like proteins. Since the VirB11-like proteins are mostly localized in the cytoplasm associated with the inner membrane, it is difficult to imagine that they energize the transport process of a protein across the entire cell envelope. Furthermore, the type IV secretion systems encode another class of potential NTPases, e.g., RP4 TrbE, which is tightly membrane associated and thus an additional candidate for a traffic NTPase. Thus, it seems more likely that VirB11-like proteins energize the assembly of the membrane-spanning multiprotein complex. The similarities in the structures and NTPase activities of VirB11-like proteins to those of chaperones suggest that the binding of VirB11-like proteins to components of the transport complex might aid the translocation of the component across the inner membrane and/or the binding of the component to further components to form the entire membrane-spanning complex.
ACKNOWLEDGMENTS
S.K. and E.L. are grateful to Hans Lehrach for his generous support. The expert technical assistance of Marianne Schlicht is greatly appreciated. W.P. thanks Paul Hooykaas for generous support and stimulating discussions.
The work of S.K. was financially supported by Sonderforschungsbereich grant 344/A8 to E.L. from the Deutsche Forschungsgemeinschaft. The work of W.P. was financially supported by Biotech grant ERB4001GT963065 from the European Union. F.C. was supported by grant PB95-1269 from the DGICYT (Ministry of Education, Spain).
REFERENCES
- 1.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Pontén T, Alsmark U C M, Podowski R M, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 2.Balzer D, Pansegrau W, Lanka E. Essential motifs of relaxase (TraI) and TraG protein involved in conjugative transfer of plasmid RP4. J Bacteriol. 1994;176:4285–4295. doi: 10.1128/jb.176.14.4285-4295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzer D, Ziegelin G, Pansegrau W, Kruft V, Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992;20:1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binet R, Letoffe S, Ghigo J M, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 5.Bordo D, Argos P. Suggestions for “safe” residue substitutions in site-directed mutageneses. J Mol Biol. 1991;217:721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Burns D L. Biochemistry of type IV secretion. Curr Opin Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 9.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factor. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie P J, Ward J E, Jr, Gordon M P, Nester E W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covacci A, Telford J L, Giudice G D, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 12.Crowther R A, Amos L A. Harmonic analysis of electron microscope images with rotational symmetry. J Mol Biol. 1971;60:123–130. doi: 10.1016/0022-2836(71)90452-9. [DOI] [PubMed] [Google Scholar]
- 13.Daugelavičius R, Bamford J K H, Grahn A M, Lanka E, Bamford D H. IncP plasmid encoded cell envelope-associated DNA transfer complex increases the cell permeability. J Bacteriol. 1997;179:5195–5202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Cruz F, Lanka E. Function of the Ti-plasmid Vir proteins: T-complex formation and transfer to the plant cell. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Boston, Mass: Kluwer Academic Publishers; 1998. pp. 281–301. [Google Scholar]
- 15.Dowhan W. CDP-diacylglycerol synthase of microorganisms. Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 16.Grahn A M, Haase J, Bamford D H, Lanka E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Hingorani M M, O'Donnell M. Toroidal proteins: running rings around DNA. Curr Biol. 1998;8:R83–R86. doi: 10.1016/s0960-9822(98)70052-1. [DOI] [PubMed] [Google Scholar]
- 20.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang B J, Woo K M, Goldberg A L, Chung C H. Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J Biol Chem. 1988;263:8727–8734. [PubMed] [Google Scholar]
- 22.Johnston R F, Pickett S C, Barker D L. Autoradiography using storage phosphor technology. Electrophoresis. 1990;11:355–360. doi: 10.1002/elps.1150110503. [DOI] [PubMed] [Google Scholar]
- 23.Krause, S., M. Bárcena, W. Pansegrau, R. Lurz, J. M. Carazo, and E. Lanka. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 24.Lakowicz J R. Principles of fluorescence spectroscopy. New York, N.Y: Plenum Press; 1983. [Google Scholar]
- 25.Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lory S. Secretion of proteins and assembly of bacterial surface organelles: shared pathways of extracellular protein targeting. Curr Opin Microbiol. 1998;1:27–35. doi: 10.1016/s1369-5274(98)80139-2. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–433. [Google Scholar]
- 28.Motallebi-Veshareh M, Balzer D, Lanka E, Jagura-Burdzy G, Thomas C M. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with vegetative replication. Mol Microbiol. 1992;6:907–920. doi: 10.1111/j.1365-2958.1992.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 29.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 30.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 31.Possot O, Pugsley A P. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 32.Rashkova S, Spudich G M, Christie P J. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J Bacteriol. 1998;179:583–591. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivas S, Bolland S, Cabezón E, Goni F M, de la Cruz F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- 34.Salmond G P C. Secretion of extracellular virulence factors by plant pathogenetic bacteria. Annu Rev Phytopathol. 1994;32:181–200. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sandkvist M, Bagdasarin M, Howard S P, DiRita V J. Interaction between the autokinase EspE and EspL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherzinger E, Ziegelin G, Bárcena M, Carazo J M, Lurz R, Lanka E. The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J Biol Chem. 1997;272:30228–30236. doi: 10.1074/jbc.272.48.30228. [DOI] [PubMed] [Google Scholar]
- 38.Segal E D, Cha J, Falkow J L S, Tompkins L S. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal G, Purcell M, Shuman H A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of genes of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens K M, Roush C, Nester E. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner L R, Olson J W, Lory S. The XcpR protein of Pseudomonas aeruginosa dimerizes via its N-terminus. Mol Microbiol. 1997;26:877–887. doi: 10.1046/j.1365-2958.1997.6201986.x. [DOI] [PubMed] [Google Scholar]
- 42.Vogel P J, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–875. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 43.Winans S C, Burns D L, Christie P J. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zylicz M, LeBowitz J H, McMacken R, Georgopoulos C. The DnaK-protein of Escherichia coli possesses an ATPase and autophosphorylating activity and is essential in an in vitro DNA replication system. Proc Natl Acad Sci USA. 1983;80:6431–6435. doi: 10.1073/pnas.80.21.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]