Abstract
Reactive oxygen species (ROS) posed a risk for the transition of vertebrates from aquatic to terrestrial life. How ancestral organisms adapted to such ROS exposure has remained a mystery. Here, we show that attenuation of the activity of the ubiquitin ligase CRL3Keap1 for the transcription factor Nrf2 during evolution was key to development of an efficient response to ROS exposure. The Keap1 gene was duplicated in fish to give rise to Keap1A and the only remaining mammalian paralog Keap1B, the latter of which shows a lower affinity for Cul3 and contributes to robust Nrf2 induction in response to ROS exposure. Mutation of mammalian Keap1 to resemble zebrafish Keap1A resulted in an attenuated Nrf2 response, and most knock-in mice expressing such a Keap1 mutant died on exposure as neonates to sunlight-level ultraviolet radiation. Our results suggest that molecular evolution of Keap1 was essential for adaptation to terrestrial life.
Molecular evolution of Keap1 allowed vertebrates to transition from an aquatic to a highly oxidizing terrestrial environment.
INTRODUCTION
The transition of organisms from an aquatic to a terrestrial environment was a key event in evolution. The development of aerobic respiration, which produces energy far more efficiently than does anaerobic respiration, is thought to be one adaptation that allowed organisms to migrate to and thrive on land (1). However, oxygen concentrations were higher on land than in the sea, and terrestrial organisms were at risk of exposure to higher levels of reactive oxygen species (ROS). The evolution of defense mechanisms against ROS is therefore thought to have been necessary as organisms moved onto land. It has remained unclear, however, what molecular changes were key to such evolution of ROS defense mechanisms.
NF-E2–related factor 2 (Nrf2) is a basic leucine zipper transcription factor that functions as a key regulator of stress responses by inducing the expression of antioxidant proteins and thereby protecting cells from oxidative damage (2). Under basal conditions, Nrf2 binds to Kelch-like ECH-associating protein 1 (Keap1; also known as KLHL19), a substrate receptor subunit of the Cul3-RING ubiquitin ligase (CRL3), in the cytoplasm and is rapidly degraded in a proteasome-dependent manner. Exposure of cells to oxidative stress or toxic chemicals (many of which are electrophilic) results in attenuation of the ubiquitylation activity of CRL3Keap1 and translocation of synthesized Nrf2 to the nucleus, where it activates the transcription of genes containing an antioxidant response element (2).
Nrf2-deficient mice manifest no apparent abnormalities in their birth, growth, or fertility (3), but they show increased sensitivity to a variety of environmental and pharmacological toxicants including acetaminophen, which induces acute liver damage (4, 5); butylated hydroxytoluene, which induces acute respiratory distress syndrome (6); and bleomycin, which induces pulmonary fibrosis (7). They also show severe phenotypes under various conditions related to oxidative stress, including ischemia-reperfusion injury of various tissues (8–11) as well as exposure to high oxygen concentrations (12, 13) or ionizing radiation such as ultraviolet (UV) (14–17). Nrf2 also evokes marked anti-inflammatory responses, not only indirectly by attenuating the production of damage-associated molecular patterns that trigger inflammation but also directly by inhibiting the induction of proinflammatory genes (18).
Keap1 is the most studied of the 42 members of the Kelch-like (KLHL) protein family (19), which are responsible for substrate recognition by CRL3 complexes. KLHL proteins harbor three key functional domains: a BTB domain, which mediates association with Cul3 and Rbx1; a BACK domain, which also interacts via its 3-box motif with Cul3 (20); and a Kelch-repeat domain, which interacts with substrates (19). In addition to KLHL proteins, some Kelch repeat– and BTB domain–containing (KBTBD) proteins (21), BTB domain–containing (BTBD) proteins (22), speckle-type POZ (SPOP) proteins (20), potassium channel tetramerization domain–containing (KCTD) proteins (23), and RhoBTB proteins (atypical members of the Rho family of small guanosine triphosphatases) (24) have BTB domains that interact with Cul3 and function as substrate receptors of CRL3.
We here found that the interaction of human Keap1 with Cul3 is weaker than that of other human KLHL proteins. The BTB domain of human Keap1 has a Cul3-interacting region (designated C3IR) that differs from that of other human Cul3-interacting BTB domains, and replacement of this region of Keap1 with the corresponding sequence of KLHL3 conferred stronger binding to Cul3. Evolutionary tracing suggests that the Keap1 gene was duplicated during whole-genome duplication in fish to give rise to Keap1A and Keap1B, which bind to Cul3 strongly or weakly, respectively. Keap1A was subsequently lost in reptiles and their descendants, and Keap1B evolved into mammalian Keap1. We found that replacement of C3IR of mammalian Keap1 with the corresponding region of zebrafish Keap1A attenuated Nrf2 activity in cells subjected to stress, and most mouse neonates harboring this mutation were found to die on exposure to sunlight-level UV radiation. Our results suggest that the molecular evolution of Keap1 was essential for adaptation of organisms to the higher ROS levels associated with a terrestrial environment.
RESULTS
Interaction of Keap1 with Cul3 is weaker than that of other KLHL proteins
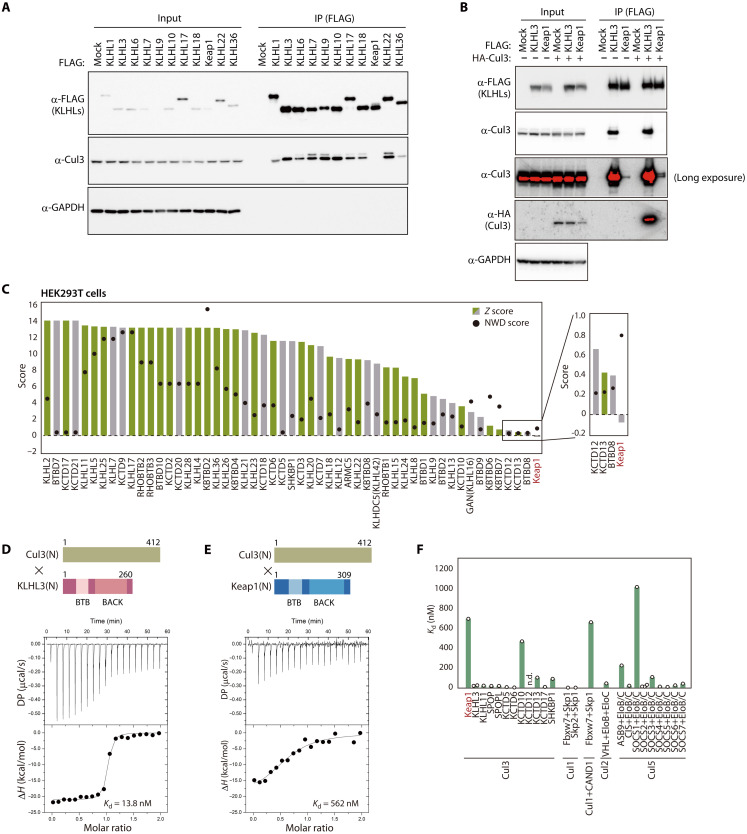
To compare interactions between KLHL proteins and Cul3, we forcibly expressed various FLAG epitope–tagged human KLHL proteins in human embryonic kidney (HEK) 293T cells and examined their binding to endogenous Cul3 by coimmunoprecipitation analysis (Fig. 1A). Unexpectedly, in contrast to the results of previous studies in which Cul3 was overexpressed, the interaction between FLAG-Keap1 and endogenous Cul3 was hardly detectable, whereas that of the other FLAG-tagged KLHL proteins examined was readily apparent. The interaction between FLAG-Keap1 and endogenous or hemagglutinin epitope (HA)–tagged Cul3 was detected by analysis of FLAG immunoprecipitates with antibodies to HA or to Cul3, although the extent of Cul3 binding to Keap1 was markedly smaller than that to KLHL3 (Fig. 1B). Overexpressed Keap1 (but not KLHL3) bound to Nrf2, excluding the possibility that Keap1 aggregation prevented its binding to Cul3 (fig. S1). Reanalysis of mass spectrometry–derived data for Cul3 binding proteins in HEK293T cells (25) revealed that Keap1 had the lowest z score among 53 BTB proteins (Fig. 1C), with its normalized weighted D score (NWD score) (26) being <1. According to the most recent data from BioPlex 3.0 (accessed on 31 August 2022), a project to identify binding proteins in a comprehensive manner (27), KLHL3 and many other BTB proteins, but not Keap1, have been identified as Cul3 binding proteins in HEK293T or HCT116 cells (Fig. 1C and fig. S2).
Fig. 1. Weaker interaction of Keap1 with Cul3 compared with other receptors.
(A) Lysates of HEK293T cells expressing FLAG-tagged human KLHL proteins were subjected to immunoprecipitation (IP) with antibodies to FLAG, and the resulting precipitates as well as the original lysates (Input) were subjected to immunoblot analysis with the indicated antibodies (α-). GAPDH was examined as a loading control. (B) Lysates of HEK293T cells expressing FLAG-tagged human KLHL3 or Keap1 together with HA-tagged human Cul3 were subjected to immunoprecipitation with antibodies to FLAG, and the resulting precipitates as well as the original lysates were subjected to immunoblot analysis with the indicated antibodies. GAPDH was examined as a loading control. (C) Fifty-three BTB proteins previously found to interact with Cul3 in HEK293T cells (25). Z and NWD scores were obtained from CompPASS analysis (26). Cul3 interactants in the BioPlex 3.0 database (27) are shown in green, with others shown in gray. (D and E) Representative ITC titration curves for interaction between human Cul3(N) and either human KLHL3(N) (D) or human Keap1(N) (E) as well as domain organization of the protein fragments. Baseline-corrected differential power (DP) versus time as well as the normalized binding curve, with integrated changes in enthalpy (ΔH) plotted against molar ratio, are shown. Each experiment was performed independently at least twice. (F) Dissociation constants for Cullins and the indicated receptors. Values other than those for Keap1 and KLHL3 were obtained from previous studies (21, 23, 28, 54–62). n.d., not detected.
We therefore attempted to measure the binding affinity of Keap1 and KLHL3 for Cul3 by isothermal titration calorimetry (ITC) with recombinant N-terminal portions of human Keap1 [Keap1(N)] or KLHL3 [KLHL3(N)] that spanned the BTB and BACK domains as well as the recombinant N-terminal domain of human Cul3 [Cul3(N)]. The dissociation constant (Kd) for the interaction between Cul3(N) and KLHL3(N) was 13.8 nM (Fig. 1D), consistent with the results of a previous study (28), whereas the binding affinity between Cul3(N) and Keap1(N) was ~40-fold weaker (Kd = 562 nM) (Fig. 1E). Comparison with previous data revealed that the Kd for Keap1-Cul3 is the highest among BTB proteins that bind to Cul3, being almost identical to that for the interaction between Cul1 and Skp1/Fbxw7 in the presence of CAND1, a receptor dissociation-promoting protein for Cullins (Fig. 1F). SOCS1 showed the highest Kd value among CRL complexes previously studied, consistent with the fact that SOCS1 has an incomplete Cul5-interacting domain (29). Collectively, these results indicated that the binding of Keap1 to Cul3 is weaker than that of other receptor subunits.
C3IR of Keap1 is responsible for weak interaction with Cul3
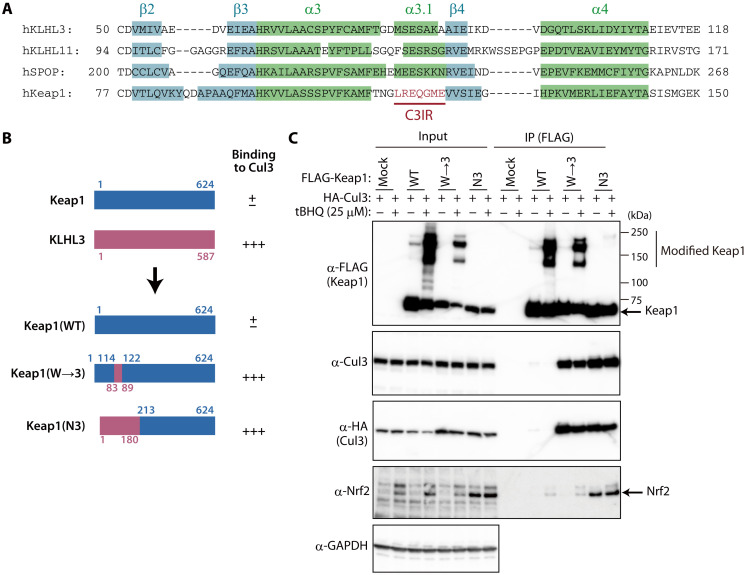
To explore structural features that might account for the differences in Cul3 binding between Keap1 and other BTB proteins, we compared the secondary structures of human Keap1 and three Cul3 binding proteins (KLHL3, KLHL11, and SPOP) whose structures in complex with Cul3 have been solved by x-ray crystallography (21, 30, 31). The four BTB domains showed almost identical secondary structures, with the exception that KLHL3, KLHL11, and SPOP each had an α3.1 helix structure, whereas Keap1 instead had a unique seven–amino acid stretch (C3IR) (Fig. 2A). This finding is consistent with the previous observation that the α3-β4 loop contributes most to Cul3 binding (31). We then superimposed the Cul3-KLHL3 crystal structure on the Keap1 crystal structure and found that, consistent with the secondary structure information, the BTB domains of KLHL3 and Keap1 overlapped in most regions with the exception of C3IR of Keap1 (fig. S3A). Four residues of KLHL3 (Asp82, Met83, Ser84, and Glu85) likely contribute to binding to Cul3, whereas the corresponding amino acids of Keap1 are positioned away from the Cul3 surface (fig. S3B).
Fig. 2. C3IR of Keap1 is responsible for weak binding to Cul3.
(A) Alignment of amino acid sequences for the BTB domains of human (h) KLHL3, KLHL11, SPOP, and Keap1. Information on secondary structure was obtained from PDB (4HXI for KLHL3, 4AP2 for KLHL11, 4EOZ for SPOP, and 4CXI for Keap1). The numbers of α helices and β sheets are in accordance with a previous study (30). (B) Domain organization of human Keap1, human KLHL3, and the exchange mutant proteins Keap1(N3) and Keap1(W→3) as well as a summary of their interaction with Cul3 as determined in (C). (C) HEK293T cells expressing FLAG-tagged wild-type (WT) or mutant forms of human Keap1 together with HA-tagged human Cul3 were incubated in the absence or presence of 25 μM tBHQ for 24 hours, lysed, and subjected to immunoprecipitation with antibodies to FLAG. The resulting precipitates as well as the original cell lysates were subjected to immunoblot analysis with the indicated antibodies.
To examine whether C3IR is responsible for the weak binding of Keap1 to Cul3, we constructed exchange mutants of human Keap1 and KLHL3. Keap1(N3), a Keap1 mutant that contains the BTB domain (but not the BACK domain) of KLHL3 (Fig. 2B), showed robust binding to Cul3 in HEK293T cells (Fig. 2C), indicating that the BTB domain of Keap1 contributes to the weak interaction with Cul3. Binding to Cul3 was also increased for the Keap1(W→3) mutant, in which only C3IR was replaced by the amino acids constituting the α3.1 helix of KLHL3 (Fig. 2, B and C). The interaction of these mutants with Cul3 was largely unaffected by cell treatment with tert-butylhydroquinone (tBHQ), an Nrf2-inducing chemical that modifies Cys151 in the BTB domain of Keap1. tBHQ treatment induced Nrf2 accumulation as well as the appearance of modified (higher molecular weight) forms of Keap1 in the cells forcibly expressing wild-type (WT) or W→3 mutant forms of Keap1, consistent with previous observations (32). Given that the Keap1(N3) mutant lacks the Cys151 residue of Keap1, modified forms of the mutant protein were not detected in tBHQ-treated cells. In the cells expressing Keap1(N3), Nrf2 accumulated even in the absence of tBHQ, suggesting that a conformational change induced by the substitution of the BTB domain of KLHL3 may have a dominant negative effect on Nrf2 degradation. Collectively, these results suggested that C3IR of Keap1 dictates its weak binding to Cul3.
C3IR of Keap1 was changed during evolution
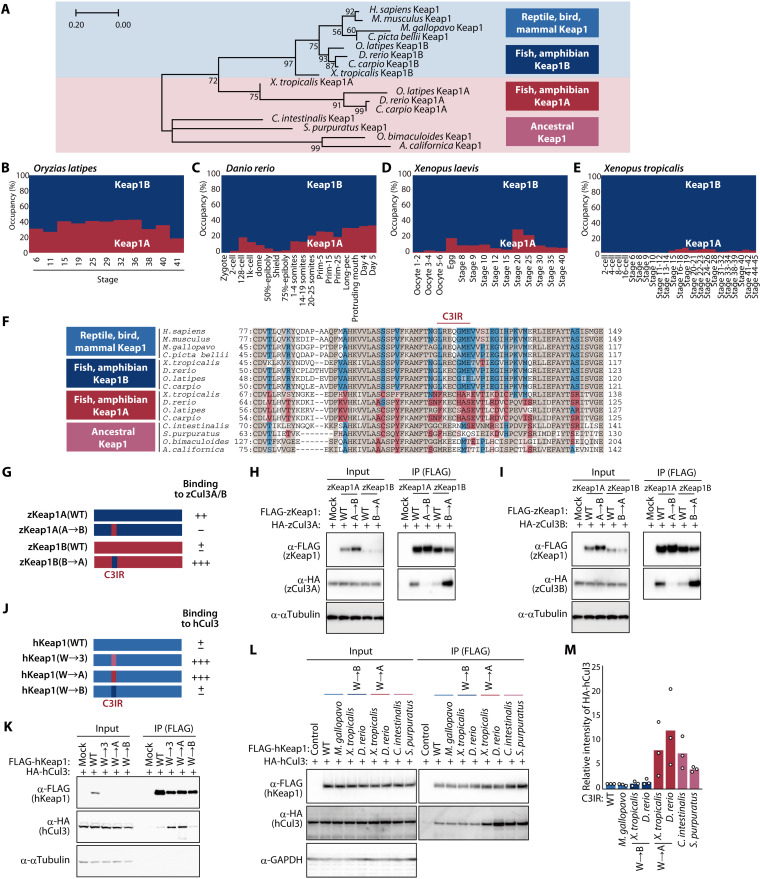
Given that the binding of Keap1 to Cul3 was weaker than that of other KLHL proteins, we speculated that Keap1 might have acquired a mutation that restrains its binding to Cul3 during evolution. Mammalian Keap1 belongs to the Keap1B subfamily of Keap1 proteins, with no reptile, bird, or mammalian species having been found to harbor a Keap1A gene, suggesting that both Keap1A and Keap1B were conserved during evolution from fish to tetrapods and that Keap1A was subsequently lost during further evolution to amniotes (32). Consistent with this scenario, phylogenetic analysis of the BTB domain of Keap1 proteins revealed that this domain in amniotes is similar to that of fish and amphibian Keap1B, whereas the BTB domain of Keap1A in Xenopus tropicalis resembles most closely that of Keap1 in ancestral animals such as mollusks and chordates (Fig. 3A). Previous RNA-sequencing (RNA-seq) analysis revealed that the proportion of Keap1A mRNA among total Keap1 mRNA during development is ~40% in the fish Oryzias latipes (Fig. 3B) and Danio rerio (Fig. 3C), whereas it is only 10 to 20% in the aquatic amphibian Xenopus laevis (Fig. 3D) and was barely detectable in X. tropicalis (Fig. 3E), which is aquatic but can also live in puddles, suggesting that the Keap1A/B mRNA ratio might be correlated with the extent of onshore advance.
Fig. 3. Evolutionary changes in C3IR of Keap1 weakened its interaction with Cul3.
(A) Phylogenetic tree for the BTB domain of Keap1 proteins. The numbers associated with the branches indicate bootstrap values. (B to E) Ratio of Keap1A and Keap1B mRNAs at the indicated developmental stages in O. latipes (B), D. rerio (C), X. laevis (D), and X. tropicalis (E). Data are derived from RNA-seq analysis in previous studies (50–53). (F) Alignment of amino acid sequences for the BTB domain of Keap1 proteins. Keap1A- or Keap1B-specific amino acids are highlighted in red or blue, respectively, with common amino acids shown in brown. (G) Domain organization of zKeap1A, zKeap1B, and the C3IR mutants zKeap1A(A→B) and zKeap1B(B→A) as well as a summary of their interaction with zCul3 as determined in (H) and (I). (H and I) Lysates of HEK293T cells expressing FLAG-tagged zKeap1A, zKeap1B, zKeap1A(A→B), or zKeap1B(B→A) together with HA-tagged zCul3A (H) or zCul3B (I) were subjected to immunoprecipitation with antibodies to FLAG. (J) Domain organization of human Keap1 and the C3IR mutants hKeap1(W→3), hKeap1(W→A), and hKeap1(W→B) as well as a summary of their interaction with human Cul3 as determined in (K). (K) Lysates of HEK293T cells expressing FLAG-tagged hKeap1, hKeap1(W→3), hKeap1(W→A), or hKeap1(W→B) together with HA-tagged hCul3 were subjected to immunoprecipitation with antibodies to FLAG. (L and M) Lysates of HEK293T cells expressing HA-tagged hCul3 together with FLAG-tagged hKeap1 or its exchange mutants containing C3IR of Keap1 proteins from the indicated species were subjected to immunoprecipitation with antibodies to FLAG (L). The relative extent of HA-hCul3 binding to the hKeap1 mutants was also quantitated by densitometry (M). Quantitative data are means from three independent experiments.
C3IR and its surrounding regions show the most amino acid differences within the BTB domain between Keap1A and Keap1B of fish and amphibians (Fig. 3F). To compare the extent of binding to Cul3 between Keap1A and Keap1B, we forcibly expressed zebrafish (D. rerio) Keap1A (zKeap1A), zKeap1B, or their exchange mutants (Fig. 3G) in HEK293T cells and examined their interactions with exogenous zCul3A (Fig. 3H) or zCul3B (Fig. 3I) by coimmunoprecipitation analysis. The extent of interaction with either zCul3A or zCul3B was greater for zKeap1A than for zKeap1B. The interaction of a zKeap1A mutant containing C3IR of zKeap1B [zKeap1A(A→B)] with zCul3A or zCul3B was attenuated compared with that of zKeap1A. On the other hand, the reciprocal mutant in which C3IR of zKeap1B was replaced by C3IR of zKeap1A [zKeap1B(B→A)] interacted to a greater extent with Cul3 than did zKeap1B, suggesting that C3IR of zKeap1 is the major determinant of binding affinity for Cul3.
We also generated human Keap1 mutants in which C3IR was replaced by that of zKeap1A [Keap1(W→A)] or zKeap1B [Keap1(W→B)] (Fig. 3J). Coimmunoprecipitation analysis in HEK293T cells showed that Keap1(W→A) interacted with human Cul3 to the same extent as did the Keap1(W→3) mutant, whereas Keap1(W→B) showed no change in the extent of Cul3 binding compared with the WT protein (Fig. 3K). We next replaced C3IR of human Keap1 with that of various animals. In addition to Keap1(W→A), human Keap1 substitution mutants containing C3IR of X. tropicalis Keap1A or of Ciona intestinalis or Strongylocentrotus purpuratus Keap1 showed stronger binding to Cul3 compared with the WT protein (Fig. 3, L and M). In contrast, the corresponding human substitution mutants containing C3IR of Meleagris gallopavo Keap1 or of X. tropicalis Keap1B bound to Cul3 to the same extent as did WT human Keap1 (Fig. 3, L and M). These results thus suggested that Keap1 of ancestral aquatic organisms binds strongly to Cul3, that whole-genome duplication in fish resulted in the generation of two Keap1 proteins—Keap1A and Keap1B—that bind strongly and weakly to Cul3, respectively, and that only Keap1B was subsequently retained by species that progressed to live on land.
Nrf2-dependent responses were impaired in cells expressing the Keap1(W→A) mutant
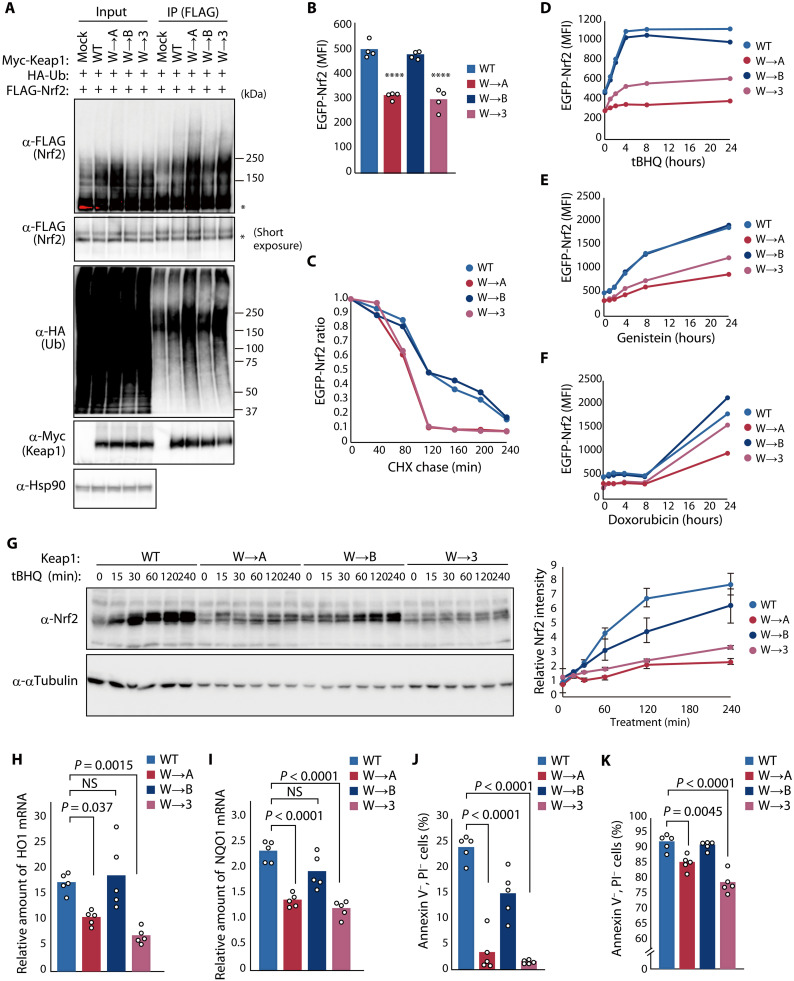
We next examined the effect of the binding affinity between Cul3 and Keap1 on Nrf2 induction and Nrf2-dependent responses to oxidative stress. Analysis of FLAG-tagged human Nrf2 ubiquitylation in HEK293T cells expressing HA-ubiquitin and Myc epitope–tagged human Keap1 revealed that the two Keap1 mutants—Keap1(W→3) and Keap1(W→A)—that showed stronger binding to Cul3 mediated ubiquitylation of Nrf2 to a greater extent than did WT Keap1 or Keap1(W→B) (Fig. 4A). Similar forced expression of Keap1 in HEK293T cells also stably harboring enhanced green fluorescent protein (EGFP)–tagged human Nrf2 revealed that the level of EGFP-Nrf2 fluorescence was reduced in cells expressing Keap1(W→3) or Keap1(W→A) compared with that in cells expressing WT Keap1 or Keap1(W→B) (Fig. 4B). Furthermore, a cycloheximide chase assay revealed that the half-life of EGFP-Nrf2 was decreased substantially in cells expressing Keap1(W→3) or Keap1(W→A) compared with that in cells expressing WT Keap1 or Keap1(W→B) (Fig. 4C). We next treated these various transfected cells with tBHQ (Fig. 4D), genistein (Fig. 4E), or doxorubicin (Fig. 4F), all of which induce Nrf2, and monitored changes in EGFP-Nrf2 fluorescence over time. EGFP-Nrf2 induction by each agent was markedly attenuated in the cells expressing Keap1(W→3) or Keap1(W→A) compared with that in those expressing WT Keap1 or Keap1(W→B).
Fig. 4. Evolutionary changes in C3IR of Keap1 attenuate ubiquitylation activity and promote Nrf2-dependent responses to oxidative stress.
(A) HEK293T cells expressing Myc-Keap1 or its indicated C3IR mutants together with HA-ubiquitin (Ub) and FLAG-Nrf2 were treated with 10 μM MG132 for 6 hours, lysed, and subjected to immunoprecipitation with antibodies to FLAG. Asterisks indicate nonubiquitylated forms of FLAG-Nrf2. (B) Median fluorescence intensity (MFI) determined by flow cytometry for EGFP-Nrf2 expressed in HEK293T cells together with FLAG-Keap1 or its C3IR mutants. Data are means; n = 4. ****P < 0.0001 versus WT. (C) MFI of EGFP-Nrf2 expressed together with FLAG-Keap1 or its C3IR mutants that were treated with 1 μM MLN4924 for 24 hours before washout of MLN4924 and incubation of the cells with cycloheximide (CHX; 50 μg/ml) for the indicated times. Data are expressed relative to the value for time 0. (D to F) MFI of EGFP-Nrf2 expressed together with FLAG-Keap1 or its C3IR mutants during exposure to 25 μM tBHQ (D), 100 μM genistein (E), or 400 nM doxorubicin (F). (G) WT or Keap1 C3IR mutation knock-in HEK293T cells were exposed to 25 μM tBHQ, lysed, and subjected to immunoblot analysis with antibodies to Nrf2 (left). The relative abundance of endogenous Nrf2 was also quantitated (right). Data are means ± SEM; n = 5. (H and I) RT and real-time PCR analysis of HO1 (H) and NQO1 (I) mRNA abundance in WT or Keap1 C3IR mutation knock-in cells at 8 hours after treatment with 25 μM tBHQ. Data are means; n = 5. NS, not significant. (J and K) Flow cytometric analysis of WT or Keap1 C3IR mutation knock-in cells that were negative for annexin V and propidium iodide (PI) staining after treatment with 0.003% H2O2 (J) or 5 μM doxorubicin (K) for 24 hours. Data are means; n = 5.
To avoid potential artifactual effects of protein overexpression, we generated HEK293T cells that express endogenous Keap1 in which C3IR has been replaced with the corresponding region of zKeap1A, zKeap1B, or human KLHL3 with the CRISPR-Cas9 system (fig. S4, A to C). Polymerase chain reaction (PCR) analysis of these C3IR knock-in cells showed that all alleles of five cell clones for each mutation were replaced by the corresponding mutant allele (fig. S4, D to F), whereas immunoblot analysis revealed that Keap1 abundance was essentially unchanged in the mutant cells compared with the parental cells (fig. S4, G to I). The extent of Nrf2 ubiquitylation in W→A or W→3 knock-in cells expressing HA-ubiquitin was increased compared with that in WT or W→B knock-in cells (fig. S5A). Treatment of the C3IR mutant knock-in cells with tBHQ (Fig. 4G) or genistein (fig. S5B) resulted in little Nrf2 accumulation in W→A or W→3 knock-in cells compared with WT or W→B knock-in cells. Treatment with tBHQ also induced transcription of the Nrf2 target genes for heme oxygenase 1 (HO1) and NAD(P)H (reduced form of nicotinamide adenine dinucleotide phosphate) quinone dehydrogenase 1 (NQO1) to a much greater extent in WT and W→B knock-in cells than in W→A or W→3 knock-in cells (Fig. 4, H and I). Treatment with hydrogen peroxide (H2O2) reduced cell viability to a markedly greater extent in W→A and W→3 knock-in cells than in WT or W→B knock-in cells (Fig. 4J). We also found that W→A and W→3 knock-in cells were more sensitive to doxorubicin, another inducer of apoptosis associated with ROS generation and Nrf2-dependent gene transcription, than were WT or W→B knock-in cells (Fig. 4K). Together, these results suggested that human cells expressing a form of Keap1 that is similar to that of ancestral aquatic organisms and that binds more strongly to Cul3 are more sensitive to oxidative stress as a result of weak induction of Nrf2.
Keap1(W→A) knock-in mice are more vulnerable to APAP-induced stress
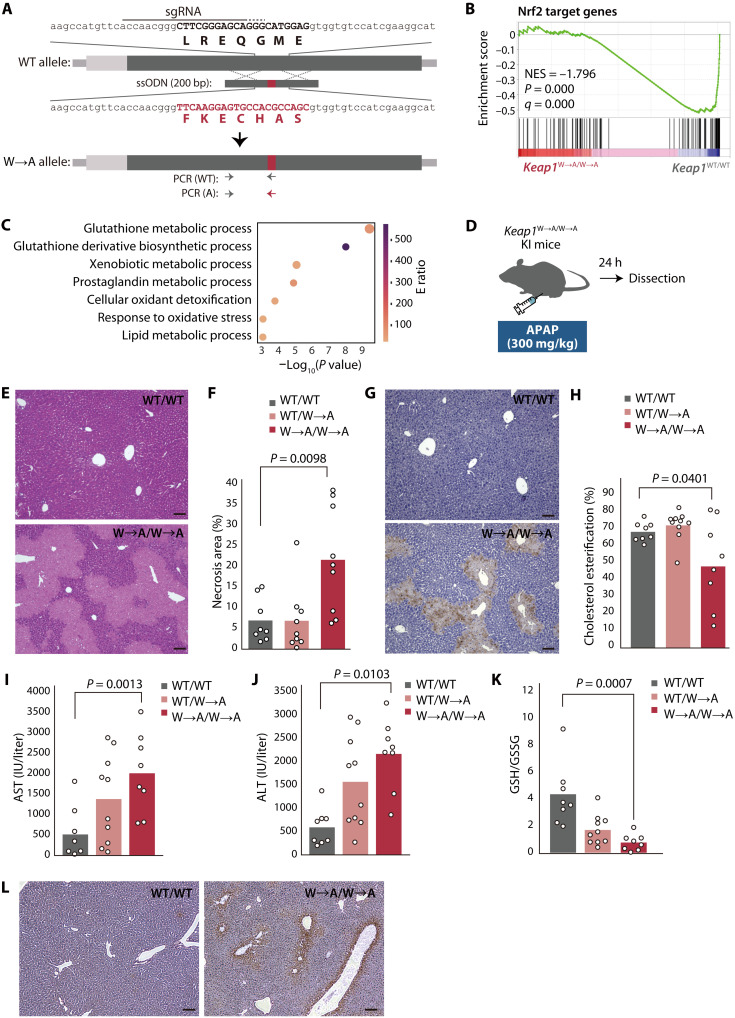
To investigate why Keap1 of terrestrial organisms has a weak affinity for Cul3, we generated mice in which C3IR of endogenous Keap1 has been replaced with that of zKeap1A so as to increase its affinity for Cul3 (Fig. 5A and fig. S6A). These Keap1(W→A) knock-in mice showed no apparent abnormalities in their birth, growth, or reproductive efficiency (fig. S6, B and C). The abundance of Keap1 in the liver was essentially unchanged for Keap1W→A/W→A mice compared with WT mice (fig. S6D). Gene set enrichment analysis (GSEA) of RNA-seq data obtained from the liver of Keap1W→A/W→A and WT mice revealed that the expression of Nrf2 target genes was markedly down-regulated in the mutant animals (Fig. 5B). Gene ontology (GO) term analysis of core enrichment genes in the GSEA indicated that genes related to the glutathione metabolic system were prominently down-regulated in the liver of Keap1W→A/W→A mice (Fig. 5C).
Fig. 5. Keap1W→A/W→A knock-in mice are more sensitive to APAP-induced oxidative stress.
(A) Schematic representation of exon 2 of the WT mouse Keap1 allele, the ssODN, and the W→A (replacement of C3IR of the mouse protein with that of zKeap1A) mutant allele after homologous recombination. The sgRNA and its protospacer adjacent motif are indicated by continuous and dotted overlines, respectively. The 5′ untranslated region and open reading frame of Keap1 are represented by the light and dark gray boxes, respectively. The mutation is shown in red. (B) GSEA plot performed for a gene set consisting of Nrf2 target genes with RNA-seq data obtained from the liver of 8-week-old Keap1W→A/W→A and WT mice. NES, normalized enrichment score. (C) GO analysis of core enrichment genes in (B). The size of the bubbles indicates the number of genes enriched in the corresponding GO term, and their color represents the E ratio (48). (D) Experimental plan for assessment of liver injury induced by an overdose of APAP. (E and F) Representative hematoxylin and eosin staining of liver sections from APAP-treated Keap1W→A/W→A or WT mice (E). The area of necrosis in such sections was quantified with ImageJ/Fiji (F). Quantitative data are means from eight or nine mice of each genotype. (G) Representative TUNEL staining for the liver of APAP-treated Keap1W→A/W→A or WT mice. (H to J) Serum cholesterol ester/total cholesterol ratio (H) as well as AST (I) and ALT (J) activities for APAP-treated WT, Keap1WT/W→A, or Keap1W→A/W→A mice. Data are means for 8 or 10 mice of each genotype. (K) GSH/GSSG ratio for the liver of APAP-treated WT, Keap1WT/W→A, or Keap1W→A/W→A mice. Data are means for 8 or 10 mice of each genotype. (L) Immunohistochemical staining of 4-HNE in liver sections from APAP-treated Keap1W→A/W→A or WT mice. Scale bars, 100 μm.
Whereas Nrf2-deficient mice were previously found not to manifest abnormalities under normal conditions, they were more vulnerable to various stress conditions including an overdose of N-acetyl-p-aminophenol (APAP, or acetaminophen) (2), which induced pronounced liver injury associated with excessive ROS production in the mutant mice (4, 5). To determine whether Keap1W→A/W→A mice are similarly susceptible to APAP-induced hepatotoxicity, we injected them intraperitoneally with APAP (300 mg/kg) and examined them 24 hours later (Fig. 5D). Centrilobular necrosis was pronounced in Keap1W→A/W→A mice but minimal in WT mice (Fig. 5, E and F). The extent of APAP-induced apoptosis, visualized by TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining, was also more pronounced for Keap1W→A/W→A mice than for WT mice (Fig. 5G). The percentage cholesterol esterification in serum, an index of liver function, was lower in Keap1W→A/W→A mice than in WT mice (Fig. 5H and fig. S6, E to G). Serum levels of aspartate transaminase (AST) (Fig. 5I) and alanine transaminase (ALT) (Fig. 5J) were significantly higher in Keap1W→A/W→A mice than in WT mice, indicative of liver damage. Furthermore, the amount of reduced glutathione (GSH) and the ratio of GSH to oxidized glutathione (GSSG) in the liver were decreased for Keap1W→A/W→A mice compared with WT mice (Fig. 5K and fig. S6, H to J), suggestive of a markedly oxidized milieu in the mutant liver. Consistent with this finding, immunohistochemical staining showed that the amount of 4-hydroxy-2-nonenal (4-HNE), a marker of lipid peroxidation, was increased in the Keap1W→A/W→A mouse liver relative to the WT liver (Fig. 5L).
RNA-seq analysis of the liver of APAP-treated mice showed that the difference in expression of genes related to the glutathione metabolism system between Keap1W→A/W→A and WT mice was more pronounced than that apparent for nontreated animals (fig. S7). A similar difference between genotypes in the expression of genes related to the mitochondrial electron transfer system was also more pronounced after APAP administration (fig. S7), suggestive of mitochondrial dysfunction in the liver of the mutant mice treated with APAP (33). The RNA-seq data also suggested that neutrophil chemotaxis was increased to a greater extent in the liver of Keap1W→A/W→A mice relative to that of WT mice after APAP administration, consistent with the results of a previous study showing that an inflammatory response was triggered by the release of cellular contents from necrotic hepatocytes (34). Together, these findings indicated that, similar to Nrf2-deficient mice, Keap1W→A/W→A mice are more sensitive to APAP.
Sunlight-level UV radiation is lethal for Keap1(W→A) knock-in mice
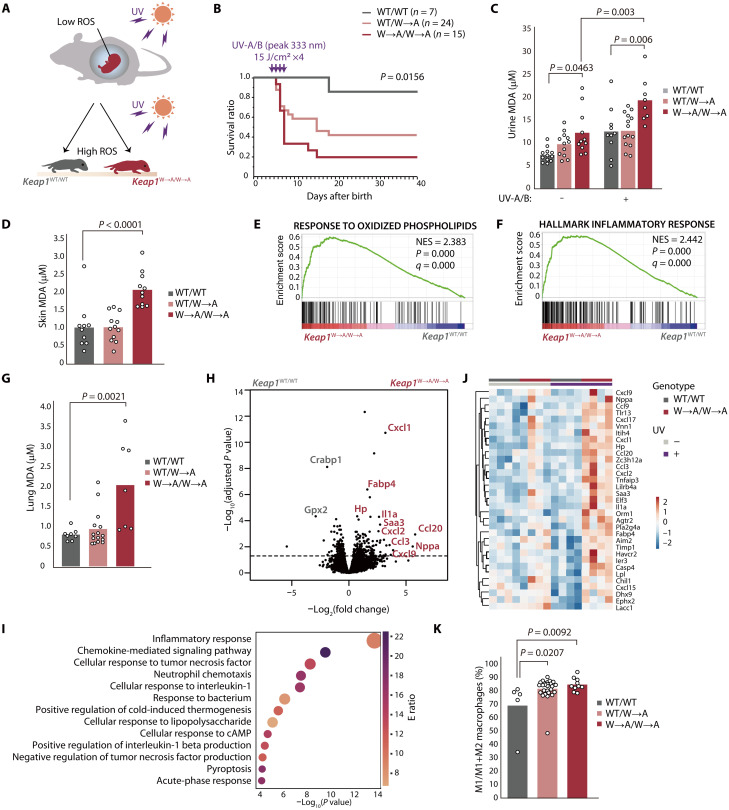
Although fetuses of terrestrial vertebrates experience hypoxia and low ROS levels in the womb or egg, they are exposed to high ROS levels after birth as a result of the increased ambient oxygen concentration and UV radiation of sunlight (Fig. 6A). We exposed neonatal mice at postnatal day (P) 3.5 to UV-A/B radiation (wavelength peak of 333 nm) for 4 days at a dose of 15 J/cm2 per day, which is equivalent to ~2.5 hours of natural sunlight in November at a latitude of 33.5°N (that of Fukuoka). Whereas the survival of most WT mice was not affected by this level of UV radiation, ~80% of Keap1W→A/W→A mice died within 8 days after completion of UV treatment (Fig. 6B). The concentration of malondialdehyde (MDA), an indicator of lipid peroxidation, in both urine (Fig. 6C) and skin (Fig. 6D) was significantly higher for Keap1W→A/W→A mice than for WT mice after UV irradiation for 3 days at 15 J/cm2 per day, suggesting that the mutant animals experienced severe oxidative damage.
Fig. 6. Keap1W→A/W→A knock-in mice are sensitive to oxidative stress induced by sunlight-level UV radiation.
(A) The mouse fetus is protected from oxidative stress in the womb but is exposed to high ROS levels after birth as a result of the high ambient oxygen concentration and UV in sunlight. (B) Kaplan-Meier survival curves for WT, Keap1WT/W→A, and Keap1W→A/W→A mice exposed to UV-A/B radiation. (C) Concentration of MDA in urine of indicated mice that had been subjected (or not) to UV-A/B irradiation. Data are means for 8 to 14 mice of each genotype. (D) Concentration of MDA in skin of indicated mice that had been subjected to UV-A/B irradiation. Data are means for 10 to 12 mice of each genotype. (E and F) GSEA plots for gene sets associated with the response to oxidized phospholipids (E) or the inflammatory response (F) that were constructed from RNA-seq data for skin of Keap1W→A/W→A and WT mice that had been exposed to UV-A/B radiation. (G) Concentration of MDA in lung of indicated mice that had been subjected to UV-A/B irradiation. Data are means for 10 to 12 mice of each genotype. (H) Volcano plot for RNA-seq data from lung of Keap1W→A/W→A and WT mice that had been exposed to UV-A/B radiation. (I) GO analysis of differentially expressed genes up-regulated in Keap1W→A/W→A mouse lung in (H). (J) Heatmap of differentially expressed genes in the gene set related to the inflammatory response (F) that was constructed from the RNA-seq data for lung tissue in (H) as well as for that of mice not subjected to UV irradiation. (K) Proportion of monocyte-derived M1 macrophages in lung of indicated mice that had been subjected to UV-A/B irradiation. Data are means for 5 to 27 mice of each genotype.
GSEA of RNA-seq data obtained from skin of UV-irradiated mice revealed that gene sets for the response to oxidized phospholipids (Fig. 6E) and the inflammatory response (Fig. 6F) were enriched in Keap1W→A/W→A mice relative to WT mice. Such enrichment was not apparent for nonirradiated animals (fig. S8, A and B), suggesting that oxidative stress evokes inflammation in skin of the mutant mice.
The MDA level in lung was also higher for Keap1W→A/W→A mice than for WT mice after exposure to UV for 3 days at 15 J/cm2 per day (Fig. 6G). RNA-seq analysis of lung tissue from irradiated mice showed that expression of many genes related to inflammation was increased in the mutant animals (Fig. 6, H and I). The expression of inflammation-related genes was not substantially increased in lung of Keap1W→A/W→A mice that had not been exposed to UV (Fig. 6J and fig. S8C), suggesting that the lung inflammation apparent in the mutant mice was UV dependent. Although the number of bone marrow–derived macrophages in lung was not affected, the percentage of M1 macrophages was increased in lung of irradiated Keap1W→A/W→A mice compared with that of irradiated WT mice (Fig. 6K), consistent with the previous finding that Nrf2 inhibits transcription of M1-related genes (18). Sustained or excessive activation of M1 macrophages promotes oxidative stress and induces secondary damage to host tissue (35). Collectively, these results indicated that mice expressing a form of Keap1 similar to the ancestral aquatic form are highly sensitive to oxidative stress induced by sunlight-level UV radiation, which elicits a pronounced inflammatory response in the mutant animals. They further suggest that molecular evolution of Keap1 was necessary for vertebrates to be able to tolerate exposure to the level of UV radiation in sunlight.
DISCUSSION
Reliance on oxygen for the efficient production of energy in living organisms is inevitably associated with tissue damage induced by ROS. The Keap1-Nrf2 pathway plays a central role in the response of cells to ROS and has evolved as the partial pressure of oxygen on Earth has increased (36). Orthologs of Nrf2 first appeared in fungi ~1.5 billion years ago, when photosynthetic oxygen was being absorbed into the oceans, and subsequent divergence of Nrf2 occurred both ~1.0 to 1.2 billion years ago, when oceanic ventilation released free oxygen into the atmosphere, and after atmospheric oxygen levels increased in the late Paleozoic (359 to 252 million years ago) (36). Our present results now suggest that molecular evolution of Keap1, resulting in a weakening of its binding to Cul3, may have allowed vertebrates to adapt to the higher concentrations of ROS associated with life on land. Keap1 in a neoavian ancestor was recently shown to harbor a mutation that disrupted its binding to and suppression of Nrf2, suggesting that persistent activation of Nrf2 resulting from this mutation might have been necessary for adaptation to the higher ROS levels associated with active flight (37). The long-lived naked mole rat has also been shown to manifest higher Nrf2 signaling activity associated with increased Nrf2 binding to ARE and increased transcription of the Keap1 gene compared with mice, implicating these changes in life-span extension (38). Changes to the function of the Keap1-Nrf2 system during evolution thus appear to underlie adaptive protection of organisms from ROS-induced tissue damage.
We found that zebrafish Keap1A binds more strongly to Cul3 than does zebrafish Keap1B. Although zebrafish Keap1A and Keap1B were previously shown to similarly suppress Nrf2 (39), the amount of Keap1B mRNA required for a similar level of Nrf2 suppression was greater than that of Keap1A mRNA, suggesting that Keap1A has a higher Nrf2-suppressive activity. Keap1A-deficient zebrafish showed increased transcription of genes related to ROS reduction and glutathione metabolism compared with WT or Keap1B-deficient zebrafish (32). Keap1A-deficient larvae were also resistant to H2O2 under sulforaphane treatment, whereas about half of WT and Keap1B-deficient larvae died under the same condition, suggesting that the expression of Keap1A renders zebrafish vulnerable to oxidative stress.
Keap1 may mediate the ubiquitylation of multiple substrates in addition to Nrf2 (40), so we cannot exclude the possibility that the molecular evolution of Keap1 has had some impact beyond the Nrf2-mediated oxidative stress response. Given that Nrf2 also protects against exposure to diverse phytochemicals present in terrestrial plants (41), the change in Keap1 activity associated with the transition of vertebrates to a terrestrial environment may represent a countermeasure to phytochemicals as well as to increased oxygen pressure and UV-induced ROS stress. Nrf2 also plays important roles in protection against various pathological conditions including cancer, neurodegenerative disease, cardiovascular disease, and acute lung injury (42), and the evolutionary attenuation of Keap1 activity may thus also protect terrestrial vertebrates from these conditions. In conclusion, our results suggest that the molecular evolution of Keap1 was essential for vertebrates to adapt to life on land.
MATERIALS AND METHODS
Antibodies
A list of the antibodies used in the study is provided in table S1.
Construction of expression plasmids and cell transfection
Complementary DNAs (cDNAs) encoding WT or mutant forms of FLAG- or Myc epitope–tagged hKeap1, hKLHL1, hKLHL3, hKLHL6, hKLHL7, hKLHL9, hKLHL10, hKLHL17, hKLHL18, hKLHL22, hKLHL36, zKeap1A, or zKeap1B as well as those encoding Myc epitope– or HA-tagged hCul3, zCul3A, or zCul3B were subcloned into the pcDNA3 vector (Invitrogen). The cDNA for HA-tagged human ubiquitin was subcloned into the pCGN vector. Those encoding EGFP- or FLAG-tagged hNrf2 were subcloned into pCSII–cytomegalovirus (CMV). For transient transfection, vectors were introduced into HEK293T cells with the XtremeGENE 9 reagent (Roche).
Lentiviral infection
HEK293T cells were transfected for 48 hours with the transfer vector together with pCAG-HIVgp and pCMV-VSV-G-RSV-Rev (both provided by H. Miyoshi, RIKEN, Tsukuba, Japan) and with the XtremeGENE 9 reagent (Roche). Culture supernatants containing lentiviruses were then collected, and HEK293T cells (1 × 105 per well of a 12-well plate) were infected with the viruses by culture with 1 ml of the supernatants for 36 hours. Cells expressing EGFP-Nrf2 were then cloned before experiments.
Cell culture
HEK293T cells (American Type Culture Collection, CRL-11268) were cultured under an atmosphere of 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM; Wako) supplemented with 10% fetal bovine serum, sodium pyruvate (1 mM), L-glutamine (2 mM), 2-mercaptoethanol (50 μM), nonessential amino acids (10 ml/liter; Gibco), penicillin (100 U/ml), and streptomycin (100 mg/ml). Cells were treated with tBHQ (25 μM, Sigma-Aldrich), cycloheximide (50 μg/ml; Wako), doxorubicin (400 nM or 5 μM; Sigma-Aldrich), genistein (100 μM; Nacalai Tesque), or H2O2 (0.003%; Wako) as indicated.
Establishment of knock-in cells with the CRISPR-Cas9 system
Complementary oligonucleotides containing the human Keap1 single-guide RNA (sgRNA) target sequences (table S2) were annealed and cloned into the Bbs I site of pX330 (Addgene). The resulting sgRNA vectors and single-stranded oligodeoxynucleotides (ssODNs) (table S2) were introduced into HEK293T cells with the XtremeGENE 9 transfection reagent (Roche). Genotypes of cloned cells were determined by PCR analysis with specific primers (table S2).
Immunoprecipitation and immunoblot analysis
Immunoprecipitation was performed as described previously (43). Cells were incubated for 6 hours in the presence of MG132 (10 μM; Peptide Institute) or MLN4924 (1 μM; LifeSensors) before lysis for the ubiquitylation assay. Protein samples were subjected to SDS-polyacrylamide gel electrophoresis on 5 to 20% SuperSep Ace Precast Gels (Wako), membranes were incubated consecutively with primary antibodies and horseradish peroxidase–conjugated secondary antibodies (Promega), and signals were visualized with SuperSignal West Pico PLUS or Dura reagents (Thermo Fisher Scientific) and a ChemiDoc imaging system (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α-tubulin, and Hsp90 were examined as loading controls.
Protein expression and purification
The N-terminal regions of human KLHL3 (residues 1 to 279), Keap1 (residues 1 to 309), and Cul3 (residues 1 to 417) were expressed in Escherichia coli strain BL21(DE3) pLys(S) with the heat shock method for chemically competent cells. The bacteria were grown at 16°C in 2× YT medium with shaking at 200 rpm to an optical density at 600 nm of 0.6 to 0.8, after which recombinant protein expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 0.1 mM and incubation overnight at 37°C. The cells were isolated by centrifugation at 8000g for 10 min at 4°C and stored at −80°C until further processing. They were subsequently thawed; suspended in a solution containing 50 mM tris-HCl (pH 8.0), 500 mM NaCl, and 1 mM 2-mercaptoethanol; and lysed by ultrasonic treatment (Branson Sonifier). The lysate was centrifuged at 8000g for 10 min at 4°C to remove debris, and the resulting supernatant was mixed with Ni-NTA beads (Invitrogen) in a solution containing 1% Nonidet P-40, 100 mM imidazole (pH 8.0), and 10% glycerol and then incubated overnight at 4°C with rotation. The beads were washed with a solution containing 50 mM tris-HCl (pH 8.0), 500 mM NaCl, 1 mM 2-mercaptoethanol, 100 mM imidazole, and 10% glycerol, and the His10-tagged proteins were eluted with the same solution but containing 400 mM imidazole. Eluted proteins were pooled and further purified by size exclusion chromatography with a Superdex 200 column (GE Healthcare) that had been equilibrated with gel filtration buffer [50 mM Hepes-NaOH (pH 7.5), 150 mM NaCl, 10% glycerol, and 0.5 mM tris(2-carboxyethyl)phosphine]. Purified proteins were concentrated by ultrafiltration (Amicon Ultra, Merck).
Isothermal titration calorimetry
ITC was performed at 25°C with a MicroCal Auto-iTC200 system (Malvern Panalytical). Titrant [100 μM Cul3(N)] was titrated into 385 μl of 10 μM titrate [KLHL3(N) or Keap1(N)] as 19 × 2-μl injections. Data were analyzed with Origin software (OriginLab) and fitted with a one-site binding model.
Structure comparisons
The structures of the KLHL3-Cul3 complex [Protein Data Bank (PDB): 4HXI] and of Keap1 (PDB: 4CXI) were superimposed to obtain structural alignment with the Matchmaker tool and the option of best-aligning pair of chains in ChimeraX 1.4 (University of California San Francisco).
Phylogenetic tree
The amino acid sequence of the BTB domain of each Keap1 protein was obtained from National Center for Biotechnology Information (NCBI) Gene (table S3). Phylogenetic analysis was performed with the MEGA11 tool (44). Alignment was achieved with the default parameters of the MUSCLE algorithm (45). The model that best described the substitution pattern was screened by model selection analysis. The lowest Bayesian information criterion score was 27372.3342, and the model used for maximum likelihood tree construction was that of Jones-Taylor-Thornton (46). The nonuniformity of evolutionary rates among sites was modeled with a discrete gamma distribution (five categories). The bootstrap method, with the number of bootstrap replications set at 1000, was applied to test the inferred phylogeny (47).
Reverse transcription and real-time PCR analysis
Total RNA (1 μg) isolated from cells with Isogen (Nippon Gene) and the PureLink RNA Mini Kit (Thermo Fisher Scientific) was subjected to reverse transcription (RT) with the QuantiTect Reverse Transcription Kit (Qiagen), and the resulting cDNA was subjected to real-time PCR analysis with SYBR Premix Ex Taq (TaKaRa Bio) and specific primers (table S2) in the StepOnePlus Real-Time PCR System (Applied Biosystems). The amounts of target mRNAs were normalized by that of RPS18 mRNA.
Flow cytometry
For detection of apoptosis, HEK293T cells were washed twice with ice-cold Binding Buffer (BD Biosciences) and then stained simultaneously with allophycocyanin-conjugated annexin V and propidium iodide (both from BD Biosciences) in Binding Buffer for 20 min on ice and in the dark. The cells were subsequently analyzed with a FACSVerse flow cytometer (BD Biosciences) or a FACSAria II cell sorter (BD Biosciences) and with FlowJo software (Tree Star).
For analysis of lung leukocytes, a single-cell suspension was obtained by digestion of lung tissue with collagenase (1 mg/ml; Wako) for 60 min at 37°C, passage of the digested material through a 70-μm cell strainer, and lysis of red blood cells (with 0.14 M NH4Cl and 0.01 M tris-HCl at pH 7.5) for 5 min at room temperature. The remaining cells were incubated for 15 min on ice with antibodies to CD16/32 (2.4G2, BD Biosciences) so as to block Fc receptors and were then stained for 30 min on ice with antibodies to CD45.2 (104, BioLegend), to CD11b (M1/70, BioLegend), to F4/80 (BM8, BioLegend), to CD11c (N418, BioLegend), to SiglecF (E50-2440, BioLegend), to CD80 (16-10A1, BioLegend), and to CD206 (MMR) (C068C2, BioLegend). The stained cells were treated with 7-aminoactinomycin D (BD Biosciences) to exclude dead cells and were then analyzed with a FACSVerse flow cytometer (BD Biosciences). Cells with forward scatter–area (FSC-A) or side scatter–area (SSC-A) values that were too small (regarded as debris), those with FSC-W (width) values that were too high relative to FSC-A values (regarded as doublets), those positive for 7- aminoactinomycin D staining (regarded as dead cells), and those negative for CD45 (regarded as nonleukocytes) were discarded. CD11b+F4/80+CD80+CD206− cells were regarded as M1 macrophages, and CD11b+F4/80+CD80−CD206+ cells as M2 macrophages.
Animals
All animal experiments were approved by the animal ethics committee of Kyushu University (approval codes A20-169-3 and A22-240-0) and were conducted in compliance with university guidelines and regulations for animal experimentation. Mice were housed in the specific pathogen–free animal facility at Kyushu University in accordance with institutional guidelines and under the following conditions: ambient temperature of 22°C, humidity of 50 to 60%, 12-hour light/12-hour dark cycle, and free access to water and rodent chow (CA-1, CLEA Japan). All experiments were performed with mice on the C57BL/6J background. For generation of knock-in mice with the C3IR sequence of zKeap1A, embryonic stem cells derived at embryonic day 4 were cultured on a feeder layer of mouse embryonic fibroblasts in Knockout DMEM (Thermo Fisher Scientific) supplemented with KnockOut Serum Replacement (Thermo Fisher Scientific) and ESGRO Recombinant Mouse LIF Protein (Sigma-Aldrich). They were transfected with the ssODN (table S2), CRISPR vector pX330 (Addgene) encoding Cas9 and a Keap1-specific sgRNA (table S2), and the pCAGI-Puro vector. The transfected cells were subjected to selection for 36 hours in the presence of puromycin (1 μg/ml), expanded for 7 days, and picked up as single colonies. Embryonic stem cell clones that had undergone homologous recombination were identified by PCR analysis and confirmed by Sanger sequencing. They were then expanded and injected into C57BL/6J mouse blastocysts to generate chimeric mice. Germline transmission of the W→A allele was achieved by crossing of chimeras with C57BL/6J mice.
APAP administration
Male mice at 8 weeks of age were deprived of food for 16 hours before treatment. APAP (Sigma-Aldrich) was dissolved at a concentration of 10 mg/ml in phosphate-buffered saline that had been warmed to body temperature and was then injected intraperitoneally at a dose of 300 mg/kg. Blood was collected from the tail vein or inferior vena cava for isolation of serum by centrifugation. The activities of AST and ALT in serum were measured with Transaminase CII Test (Wako), and the serum concentrations of total cholesterol and free cholesterol were measured with Cholesterol E Test (Wako) and Free Cholesterol E Test (Wako), respectively. Glutathione concentrations in liver extracts were measured with the GSSG/GSH Quantification Kit (Dojindo).
Histology and immunohistochemical analysis
Tissue was fixed with 4% paraformaldehyde in phosphate-buffered saline, embedded in paraffin, and sectioned with a cryostat at a thickness of 3 μm, and the sections were then depleted of paraffin with xylene, rehydrated with a graded series of ethanol solutions, and stained with hematoxylin and eosin. TUNEL staining was performed with an Apoptosis In Situ Detection Kit (Wako). For 4-HNE staining, antigens were retrieved by microwave irradiation before incubation of sections with antibodies to 4-HNE (JAICA) and POD Conjugate Set Anti Mouse For Mouse Tissue (TaKaRa Bio). Immune complexes were detected with DAB Substrate (TaKaRa Bio), and sections were counterstained with hematoxylin (Sakura Finetek Japan). All sections were examined with a digital microscope (BZ-X800, Keyence), and regions of necrosis were quantified with ImageJ/Fiji software.
UV irradiation
Neonatal mice at P3.5 to P6.5 were exposed to radiation from a UV-A/B spiral lamp (peak wavelength of 333 nm, Vivaria) with a spectral range (280 to 800 nm) corresponding to natural sunlight. Irradiation was performed with mice positioned in a box so that the distance between the light source and target skin was 8 cm. The temperature inside the box was monitored and controlled so as not to exceed 26°C. The UV dose (15 J/cm2 per day) is the equivalent of exposure to ~2.5 hours of natural sunlight in November at a latitude of 33.5°N (that of Fukuoka).
RNA-seq analysis
Frozen skin, lung, or liver of adult or neonatal mice was powdered with a multibead shocker (Yasui Kikai), and total RNA was extracted and purified with Isogen (Nippon Gene) and the PureLink RNA Mini Kit (Thermo Fisher Scientific). The quality of the purified RNA was checked with 2100 Bioanalyzer (Agilent), mRNA was selected with the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs), and libraries were prepared with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs). The cDNAs were sequenced with a NovaSeq 6000 system (Illumina). Single-end reads were mapped against the mouse (mm10) genome with HISAT2, and data normalization was performed with DESeq2. The expression data were analyzed with GSEA v4.2.1 or HemI 2.0 (48). The gene sets for GSEA were obtained from the Molecular Signatures Database v4.0 distributed at the GSEA web site. A gene set designated “Nrf2 target genes” [mouse genes with a binding site for Nrf2 located ±5 kilo–base pairs (kbp) relative to the transcription start site] was generated from the ChIP-Atlas database (49). A heatmap was constructed with ClustVis (https://biit.cs.ut.ee/clustvis). The ratio of Keap1A to Keap1B mRNAs at each stage of development was determined from the TPM (transcripts per million) values calculated from RNA-seq data obtained previously from the indicated animals (50–53).
Lipid peroxidation analysis
Frozen skin or lung tissue was powdered with a multibead shocker (Yasui Kikai), and tissue lysates were prepared with radioimmunoprecipitation assay buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS] and centrifuged three times at 20,400g for 10 min at 4°C to remove debris. The resulting skin and lung extracts as well as urine were assayed for MDA with a TBARS (TCA method) assay kit (Cayman Chemical).
Statistical analysis
All data were analyzed with JMP Pro software (SAS Institute Japan). Comparisons between two or among three or more groups were performed with Student’s unpaired t test or one-way analysis of variance (ANOVA), followed by Tukey-Kramer post hoc test, respectively, with the exception that Kaplan-Meier survival curves were compared with the log-rank test. A P value of <0.05 was considered statistically significant.
Acknowledgments
We thank H. Miyoshi for pCAG-HIVgp and pCMV-VSV-G-RSV-Rev; members of the Department of Molecular and Cellular Biology as well as the Research Promotion Unit, Medical Institute of Bioregulation, Kyushu University, for technical assistance; Y. Okabe, A. Matsukuma, and A. Ohta for help with preparation of the manuscript; and D. Kohda and K. Tamura for helpful discussion. Computations were performed in part on the NIG supercomputer at ROIS National Institute of Genetics.
Funding: This research was supported in part by the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] of the Japan Agency for Medical Research and Development (AMED); by Japan Society for the Promotion of Science KAKENHI grants JP20K07592 and JP19J40055 (to K.Y.) and JP18H05215 (to K.I.N.); and by grant JPMJFR216G (to K.Y.) from the FOREST Program of the Japan Science and Technology Agency (JST).
Author contributions: K.Y. designed and performed most experiments. S.S. performed flow cytometric analysis of lung macrophages. S.M. helped with immunoblot analysis. D.T. helped with ITC. K.I.N. supervised the project. K.Y. and K.I.N. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data generated in this study have been deposited in the DDBJ Sequence Read Archive (DRA) and are publicly available. Accession numbers are DRA014902 and DRA014903. This paper does not report original code. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S3
REFERENCES AND NOTES
- 1.Taverne Y. J., Merkus D., Bogers A. J., Halliwell B., Duncker D. J., Lyons T. W., Reactive oxygen species: Radical factors in the evolution of animal life. Bioessays 40, 1700158 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto M., Kensler T. W., Motohashi H., The KEAP1-NRF2 System: A thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 98, 1169–1203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan K., Lu R., Chang J. C., Kan Y. W., NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. U.S.A. 93, 13943–13948 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan K., Han X. D., Kan Y. W., An important function of Nrf2 in combating oxidative stress: Detoxification of acetaminophen. Proc. Natl. Acad. Sci. U.S.A. 98, 4611–4616 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enomoto A., Itoh K., Nagayoshi E., Haruta J., Kimura T., O’Connor T., Harada T., Yamamoto M., High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59, 169–177 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Chan K., Kan Y. W., Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U.S.A. 96, 12731–12736 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho H.-Y., Reddy S. P. M., Yamamoto M., Kleeberger S. R., The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 18, 1258–1260 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Katsumata Y., Shinmura K., Sugiura Y., Tohyama S., Matsuhashi T., Ito H., Yan X., Ito K., Yuasa S., Ieda M., Urade Y., Suematsu M., Fukuda K., Sano M., Endogenous prostaglandin D2 and its metabolites protect the heart against ischemia-reperfusion injury by activating Nrf2. Hypertension 63, 80–87 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Nezu M., Souma T., Yu L., Suzuki T., Saigusa D., Ito S., Suzuki N., Yamamoto M., Transcription factor Nrf2 hyperactivation in early-phase renal ischemia-reperfusion injury prevents tubular damage progression. Kidney Int. 91, 387–401 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Shah Z. A., Li R. C., Thimmulappa R. K., Kensler T. W., Yamamoto M., Biswal S., Doré S., Role of reactive oxygen species in modulation of Nrf2 following ischemic reperfusion injury. Neuroscience 147, 53–59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Fields J., Zhao C., Langer J., Thimmulappa R. K., Kensler T. W., Yamamoto M., Biswal S., Doré S., Role of Nrf2 in protection against intracerebral hemorrhage injury in mice. Free Radic. Biol. Med. 43, 408–414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho H. Y., Jedlicka A. E., Reddy S. P. M., Kensler T. W., Yamamoto M., Zhang L. Y., Kleeberger S. R., Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Reddy N. M., Suryanaraya V., Yates M. S., Kleeberger S. R., Hassoun P. M., Yamamoto M., Liby K. T., Sporn M. B., Kensler T. W., Reddy S. P., The triterpenoid CDDO-imidazolide confers potent protection against hyperoxic acute lung injury in mice. Am. J. Respir. Crit. Care Med. 180, 867–874 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota A., Kawachi Y., Yamamoto M., Koga T., Hamada K., Otsuka F., Acceleration of UVB-induced photoageing in nrf2 gene-deficient mice. Exp. Dermatol. 20, 664–668 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Saw C. L., Huang M. T., Liu Y., Khor T. O., Conney A. H., Kong A. N., Impact of Nrf2 on UVB-induced skin inflammation/photoprotection and photoprotective effect of sulforaphane. Mol. Carcinog. 50, 479–486 (2011). [DOI] [PubMed] [Google Scholar]
- 16.C. L. L. Saw, Yang A. Y., Huang M.-T., Liu Y., Lee J. H., Khor T. O., Su Z.-Y., Shu L., Lu Y., Conney A. H., Kong Ah-Ng Tony, Nrf2 null enhances UVB-induced skin inflammation and extracellular matrix damages. Cell Biosci. 4, 39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knatko E. V., Ibbotson S. H., Zhang Y., Higgins M., Fahey J. W., Talalay P., Dawe R. S., Ferguson J., Huang J. T. J., Clarke R., Zheng S., Saito A., Kalra S., Benedict A. L., Honda T., Proby C. M., Dinkova-Kostova A. T., Nrf2 activation protects against solar-simulated ultraviolet radiation in mice and humans. Cancer Prev. Res. (Phila.) 8, 475–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi E. H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M., Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 7, 11624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhanoa B. S., Cogliati T., Satish A. G., Bruford E. A., Friedman J. S., Update on the Kelch-like (KLHL) gene family. Hum. Genomics 7, 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang M., Calabrese M. F., Liu J., Waddell M. B., Nourse A., Hammel M., Miller D. J., Walden H., Duda D. M., Seyedin S. N., Hoggard T., Harper J. W., White K. P., Schulman B. A., Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36, 39–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canning P., Cooper C. D. O., Krojer T., Murray J. W., Pike A. C. W., Chaikuad A., Keates T., Thangaratnarajah C., Hojzan V., Marsden B. D., Gileadi O., Knapp S., von Delft F., Bullock A. N., Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 288, 7803–7814 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa M., He Y. J., Borchers C., Xiong Y., Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001–1007 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Pinkas D. M., Sanvitale C. E., Bufton J. C., Sorrell F. J., Solcan N., Chalk R., Doutch J., Bullock A. N., Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem. J. 474, 3747–3761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthold J., Schenková K., Ramos S., Miura Y., Furukawa M., Aspenström P., Rivero F., Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes—Evidence for an autoregulatory mechanism. Exp. Cell Res. 314, 3453–3465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett E. J., Rush J., Gygi S. P., Harper J. W., Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttlin E. L., Bruckner R. J., Navarrete-Perea J., Cannon J. R., Baltier K., Gebreab F., Gygi M. P., Thornock A., Zarraga G., Tam S., Szpyt J., Gassaway B. M., Panov A., Parzen H., Fu S., Golbazi A., Maenpaa E., Stricker K., Guha Thakurta S., Zhang T., Rad R., Pan J., Nusinow D. P., Paulo J. A., Schweppe D. K., Vaites L. P., Harper J. W., Gygi S. P., Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184, 3022–3040.e28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao C., Pallett M. A., Croll T. I., Smith G. L., Graham S. C., Molecular basis of cullin-3 (Cul3) ubiquitin ligase subversion by vaccinia virus protein A55. J. Biol. Chem. 294, 6416–6429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I., VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18, 3055–3065 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji A. X., Prive G. G., Crystal structure of KLHL3 in complex with Cullin3. PLOS ONE 8, e60445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Errington W. J., Khan M. Q., Bueler S. A., Rubinstein J. L., Chakrabartty A., Privé G. G., Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure 20, 1141–1153 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Nguyen V. T., Bian L., Tamaoki J., Otsubo S., Muratani M., Kawahara A., Kobayashi M., Generation and characterization of keap1a- and keap1b-knockout zebrafish. Redox Biol. 36, 101667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran A., Jaeschke H., Acetaminophen hepatotoxicity: A mitochondrial perspective. Adv. Pharmacol. 85, 195–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams C. D., Bajt M. L., Sharpe M. R., McGill M. R., Farhood A., Jaeschke H., Neutrophil activation during acetaminophen hepatotoxicity and repair in mice and humans. Toxicol. Appl. Pharmacol. 275, 122–133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittal M., Siddiqui M. R., Tran K., Reddy S. P., Malik A. B., Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 20, 1126–1167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gacesa R., Dunlap W. C., Barlow D. J., Laskowski R. A., Long P. F., Rising levels of atmospheric oxygen and evolution of Nrf2. Sci. Rep. 6, 27740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castiglione G. M., Xu Z., Zhou L., Duh E. J., Adaptation of the master antioxidant response connects metabolism, lifespan and feather development pathways in birds. Nat. Commun. 11, 2476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis K. N., Wason E., Edrey Y. H., Kristan D. M., Nevo E., Buffenstein R., Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. U.S.A. 112, 3722–3727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L., Kobayashi M., Kaneko H., Nakajima-Takagi Y., Nakayama Y., Yamamoto M., Molecular evolution of Keap1. J. Biol. Chem. 283, 3248–3255 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Kopacz A., Kloska D., Forman H. J., Jozkowicz A., Grochot-Przeczek A., Beyond repression of Nrf2: An update on Keap1. Free Radic. Biol. Med. 157, 63–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin S., Hou D. X., Multiple regulations of Keap1/Nrf2 system by dietary phytochemicals. Mol. Nutr. Food Res. 60, 1731–1755 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Copple I. M., The Keap1-Nrf2 cell defense pathway—A promising therapeutic target? Adv. Pharmacol. 63, 43–79 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Yumimoto K., Matsumoto M., Onoyama I., Imaizumi K., Nakayama K. I., F-box and WD repeat domain-containing-7 (Fbxw7) protein targets endoplasmic reticulum-anchored osteogenic and chondrogenic transcriptional factors for degradation. J. Biol. Chem. 288, 28488–28502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K., Stecher G., Kumar S., MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones D. T., Taylor W. R., Thornton J. M., The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Felsenstein J., Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- 48.Ning W., Wei Y., Gao L., Han C., Gou Y., Fu S., Liu D., Zhang C., Huang X., Wu S., Peng D., Wang C., Xue Y., HemI 2.0: An online service for heatmap illustration. Nucleic Acids Res. 50, W405–W411 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C., ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Liu Y., Yang H., Zhang T., Naruse K., Tu Q., Dynamic transcriptional and chromatin accessibility landscape of medaka embryogenesis. Genome Res. 30, 924–937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White R. J., Collins J. E., Sealy I. M., Wali N., Dooley C. M., Digby Z., Stemple D. L., Murphy D. N., Billis K., Hourlier T., Füllgrabe A., Davis M. P., Enright A. J., Busch-Nentwich E. M., A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 6, e30860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan M. H., Au K. F., Yablonovitch A. L., Wills A. E., Chuang J., Baker J. C., Wong W. H., Li J. B., RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 23, 201–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Session A. M., Uno Y., Kwon T., Chapman J. A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., van Heeringen S. J., Quigley I., Heinz S., Ogino H., Ochi H., Hellsten U., Lyons J. B., Simakov O., Putnam N., Stites J., Kuroki Y., Tanaka T., Michiue T., Watanabe M., Bogdanovic O., Lister R., Georgiou G., Paranjpe S. S., van Kruijsbergen I., Shu S., Carlson J., Kinoshita T., Ohta Y., Mawaribuchi S., Jenkins J., Grimwood J., Schmutz J., Mitros T., Mozaffari S. V., Suzuki Y., Haramoto Y., Yamamoto T. S., Takagi C., Heald R., Miller K., Haudenschild C., Kitzman J., Nakayama T., Izutsu Y., Robert J., Fortriede J., Burns K., Lotay V., Karimi K., Yasuoka Y., Dichmann D. S., Flajnik M. F., Houston D. W., Shendure J., DuPasquier L., Vize P. D., Zorn A. M., Ito M., Marcotte E. M., Wallingford J. B., Ito Y., Asashima M., Ueno N., Matsuda Y., Veenstra G. J. C., Fujiyama A., Harland R. M., Taira M., Rokhsar D. S., Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babon J. J., Sabo J. K., Zhang J. G., Nicola N. A., Norton R. S., The SOCS box encodes a hierarchy of affinities for Cullin5: Implications for ubiquitin ligase formation and cytokine signalling suppression. J. Mol. Biol. 387, 162–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardote T. A. F., Gadd M. S., Ciulli A., Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex. Structure 25, 901–911.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y. K., Kwak M. J., Ku B., Suh H. Y., Joo K., Lee J., Jung J. U., Oh B. H., Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes. Acta Crystallogr. D Biol. Crystallogr. 69, 1587–1597 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Liu X., Reitsma J. M., Mamrosh J. L., Zhang Y., Straube R., Deshaies R. J., Cand1-mediated adaptive exchange mechanism enables variation in F-box protein expression. Mol. Cell 69, 773–786.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maine G. N., Mao X., Komarck C. M., Burstein E., COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 26, 436–447 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L. J., Larimore E. A., Rome M., Zheng N., Clurman B. E., Hess S., Shan S. O., Deshaies R. J., Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153, 206–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salter J. D., Lippa G. M., Belashov I. A., Wedekind J. E., Core-binding factor β increases the affinity between human Cullin 5 and HIV-1 Vif within an E3 ligase complex. Biochemistry 51, 8702–8704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smaldone G., Pirone L., Balasco N., di Gaetano S., Pedone E. M., Vitagliano L., Cullin 3 recognition is not a universal property among KCTD proteins. PLOS ONE 10, e0126808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas J. C., Matak-Vinkovic D., Van Molle I., Ciulli A., Multimeric complexes among ankyrin-repeat and SOCS-box protein 9 (ASB9), ElonginBC, and Cullin 5: Insights into the structure and assembly of ECS-type Cullin-RING E3 ubiquitin ligases. Biochemistry 52, 5236–5246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S3