Abstract
Alzheimer’s disease biomarkers are widely accepted as surrogate markers of underlying neuropathological changes. However, few studies have evaluated whether preclinical Alzheimer’s disease biomarkers predict Alzheimer’s neuropathology at autopsy. We sought to determine whether amyloid PET imaging or CSF biomarkers accurately predict cognitive outcomes and Alzheimer’s disease neuropathological findings.
This study included 720 participants, 42–91 years of age, who were enrolled in longitudinal studies of memory and aging in the Washington University Knight Alzheimer Disease Research Center and were cognitively normal at baseline, underwent amyloid PET imaging and/or CSF collection within 1 year of baseline clinical assessment, and had subsequent clinical follow-up. Cognitive status was assessed longitudinally by Clinical Dementia Rating®. Biomarker status was assessed using predefined cut-offs for amyloid PET imaging or CSF p-tau181/amyloid-β42. Subsequently, 57 participants died and underwent neuropathologic examination. Alzheimer’s disease neuropathological changes were assessed using standard criteria. We assessed the predictive value of Alzheimer’s disease biomarker status on progression to cognitive impairment and for presence of Alzheimer’s disease neuropathological changes.
Among cognitively normal participants with positive biomarkers, 34.4% developed cognitive impairment (Clinical Dementia Rating > 0) as compared to 8.4% of those with negative biomarkers. Cox proportional hazards modelling indicated that preclinical Alzheimer's disease biomarker status, APOE ɛ4 carrier status, polygenic risk score and centred age influenced risk of developing cognitive impairment. Among autopsied participants, 90.9% of biomarker-positive participants and 8.6% of biomarker-negative participants had Alzheimer's disease neuropathological changes. Sensitivity was 87.0%, specificity 94.1%, positive predictive value 90.9% and negative predictive value 91.4% for detection of Alzheimer's disease neuropathological changes by preclinical biomarkers. Single CSF and amyloid PET baseline biomarkers were also predictive of Alzheimer’s disease neuropathological changes, as well as Thal phase and Braak stage of pathology at autopsy. Biomarker-negative participants who developed cognitive impairment were more likely to exhibit non-Alzheimer's disease pathology at autopsy.
The detection of preclinical Alzheimer's disease biomarkers is strongly predictive of future cognitive impairment and accurately predicts presence of Alzheimer's disease neuropathology at autopsy.
Keywords: PET scan, CSF, neuropathology, biomarker, validation study
Long et al. show that in participants who are cognitively normal at baseline, CSF and amyloid PET biomarkers strongly predict progression to cognitive impairment and presence of Alzheimer’s disease neuropathology at autopsy. These results support the use of biomarkers for detecting preclinical Alzheimer’s disease.
Introduction
Alzheimer’s disease is the most common cause of dementia in older adults and is characterized neuropathologically by the presence of extracellular plaques consisting of aggregated amyloid-β (Aβ) peptide and neurofibrillary tangles consisting of intraneuronal inclusions of aggregated, hyperphosphorylated tau protein.1 Substantial evidence supports the observation that Alzheimer’s disease neuropathologic changes (ADNC) accrue in the brain for years before the onset of symptomatic disease, representing a preclinical phase. This has been supported by: post-mortem studies demonstrating presence of ADNC in a substantial number of cognitively normal older adults2–4; detection of intracerebral accumulation of amyloid over time by PET imaging, which is associated with increased risk of progression to symptomatic Alzheimer’s disease5,6; inverse correlation of CSF levels of Aβ42 with amyloid imaging markers in cognitively normal individuals,7 which is predictive of progression to symptomatic Alzheimer’s disease and whole brain atrophy8; and reductions in Aβ42 and elevated levels of total tau (t-tau) and tau phosphorylated at position 181 (p-tau181) that appear decades before the expected time of onset of symptomatic disease in autosomal dominant Alzheimer’s disease cohorts.9,10 Several longitudinal studies have assessed outcomes in older adults with preclinical Alzheimer’s disease and demonstrated that risk of progression from preclinical to symptomatic Alzheimer’s disease increases over time.11–15
Amyloid PET imaging and CSF p-tau181/Aβ42 are accepted methods for diagnosing Alzheimer’s disease in patients with established cognitive impairment and are widely viewed as valid modalities for detecting the presence of ante-mortem Alzheimer’s disease neuropathology. However, the early neuropathologic correlation studies that validated these methods used cohorts with small participant numbers16–19 or were based on post-mortem biomarker assessment.20 Larger follow-up studies in autopsy-confirmed cohorts have either focused on terminally ill participants with short life expectancy21 or cohorts primarily consisting of cognitively impaired participants.22–25 Most studies evaluating performance of CSF or amyloid PET biomarkers in the preclinical phase of the disease have evaluated longitudinal changes in biomarkers over time,26,27 or assessed the performance of such tests relative to one another15,28,29 or in relation to the onset of cognitive impairment.8,14 Few studies of preclinical Alzheimer’s disease have validated these tests in relation to gold-standard neuropathologic diagnosis and have generally been limited by small cohort sample size prohibiting the assessment of diagnostic accuracy.11 With the development of disease-modifying therapies, clinical trials are now moving towards primary prevention paradigms, enrolling cognitively normal participants based on the presence of preclinical Alzheimer’s disease biomarkers.30,31 Therefore, it is crucial to validate that biomarker testing in this population reliably detects individuals with confirmed Alzheimer’s disease neuropathological change.
In this study, we have evaluated whether the aggregate use of amyloid PET imaging or CSF biomarker testing in cognitively normal participants in a large study of community-dwelling participants accurately predicts cognitive outcomes and ADNC at autopsy.
Methods
Participants
Participants were cognitively normal community-dwelling individuals, aged 42.5–91.2 years at baseline, who were enrolled in longitudinal studies of memory and aging at the Knight Alzheimer Disease Research Center (ADRC) at the Washington University School of Medicine in St Louis. All participants underwent annual clinical assessments, including Clinical Dementia Rating® (CDR®)32 and Mini-Mental State Examination (MMSE).33 The CDR is a well-validated instrument consisting of semi-structured interviews with the participant and their collateral source as conducted by an experienced clinician to identify intra-individual changes in participant cognitive and functional abilities. Assigned CDR scores describe dementia severity (CDR 0 = cognitively normal, CDR 0.5 = very mild dementia, CDR 1 = mild dementia, CDR 2 = moderate dementia, CDR 3 = severe dementia). Participants were included in this study if they were CDR 0 at baseline clinical assessment, underwent CSF collection by lumbar puncture (LP) and/or amyloid PET imaging [either 11C Pittsburgh compound B (PIB) or 18F-AV-45 (AV45)] within 12 months of baseline clinical assessment, and had at least one additional clinical assessment after their baseline visit.
For participants who progressed to CDR > 0 during longitudinal follow-up, aetiologic diagnoses were assigned by experienced clinicians within the Knight ADRC following review of clinical history obtained at time of CDR assessment along with limited cognitive testing and based on standard research criteria for diagnosis of various dementia syndromes. Clinical diagnoses were determined independently of knowledge of biomarker status. Participant clinical assessments in this study were performed between 6 March 2002 and 1 January 2019.
Standard protocol approvals, registrations and patient consents
The Washington University Human Research Protection Office approved all study procedures. Written informed consent was obtained according to the Declaration of Helsinki from all participants before study enrolment.
Amyloid imaging
Participants underwent a PET scan with either PIB or AV45. PIB PET imaging was performed with a Siemens 962 HR + ECAT PET or Biograph mCT scanner (Siemens/CTI). AV45 PET imaging was performed with a Siemens Biograph mMR scanner (Siemens/CTI). Data were processed using regions of interest derived from T1-weighted MRIs using Freesufer.34 Data from the 30–60 min post-injection window for PIB and the 50–70 min post-injection window for AV45 were converted to standardized uptake value ratios (SUVRs) relative to the cerebellar cortex. Data were partial volume corrected using a geometric transfer matrix.35 Values from regions known to be involved in Alzheimer's diseaseee were averaged together to provide a summary measure of amyloid deposition. Amyloid PET positivity was defined by previously established SUVR cut-offs for PIB (>1.42)26 and AV45 (>1.22).36 For some analyses, PIB and AV45 tracer data were converted to centiloid units37–39 and combined using previously established cut-offs (centiloid units > 16.4).
CSF collection and processing
Participants underwent CSF collection by LP. CSF was collected as previously described.7 Participants underwent LP at ∼8 AM following overnight fasting. CSF (20–30 ml) was collected in a 50-ml polypropylene tube via gravity drip using an atraumatic Sprotte 22-G spinal needle. The tube was inverted gently to disrupt potential gradient effects and centrifuged at low speed to pellet any cellular debris. The CSF was then aliquoted into polypropylene tubes and stored at −80°C. To limit issues related to assay drift40 and lot-to-lot variability41 associated with commercial enzyme-linked immunosorbent assays, only those participants whose CSF was processed using next-generation automated Roche Elecsys assays were included for analysis. CSF Aβ42, t-tau and p-tau181 were measured with corresponding Elecsys immunoassays on the Roche cobas e601 analyser. CSF biomarker positivity was defined by the ratio of CSF p-tau181/Aβ42 using a cut-off of >0.0198. This biomarker ratio and this cut-off were chosen to yield high concordance and positive percent and negative percentage agreement with PIB status.28
APOE ɛ genotyping and computation of polygenic risk score
APOE ɛ genotype status of each participant was obtained using previously described methods42 and provided by the Genetics core of the Knight ADRC. Polygenic risk score (PRS) for each participant derived using genotype status across 24 single nucleotide polymorphisms known to be associated with Alzheimer’s disease risk were computed and provided by the Genetics core. PRS are presented as summed log2-transformed odds ratio (OR). Methods describing the computation of PRS have been described elsewhere.43,44
Neuropathological assessment
Eighty-two participants died during the study period. Of these, 57 (70%) provided autopsy consent and the brains of those participants were evaluated by experienced neuropathologists within the Knight ADRC Neuropathology Core. Autopsies in this study were performed between 6 September 2004 and 10 July 2020. Standard procedures45 and consensus criteria46–60 were used to establish neuropathological diagnoses. For purposes of this study, Alzheimer’s disease pathology was scored as present or absent based on specimen categorization by either National Institute on Aging (NIA)-Reagan61 or NIA-Alzheimer’s Association (AA) criteria,62 depending on when each autopsy was completed. For those specimens categorized using NIA-Reagan criteria, Alzheimer’s disease pathology was considered ‘present’ if specimens were classified as having either intermediate or high likelihood that Alzheimer’s disease pathological changes underlie dementia. For those specimens categorized by NIA-AA criteria, Alzheimer’s disease pathology was considered ‘present’ if specimens were classified as having either intermediate or high ADNC; by NIA-AA criteria, intermediate and high ADNC (but not low ADNC) are considered sufficient to account for observed dementia. Braak neurofibrillary tangle stage46 and Thal amyloid phase48 were recorded for each participant, when available. Comorbid neuropathological diagnoses, including Lewy body pathology, hippocampal sclerosis, TDP-43 inclusions, frontotemporal lobar degeneration, other primary tauopathies, cerebrovascular disease, infarctions and haemorrhages were also recorded.
Study design
In participants who were cognitively normal at the time of biomarker assessment, we assessed whether a ‘positive’ Alzheimer’s disease molecular biomarker value predicted progression to cognitive impairment (CDR > 0) and/or the presence of ADNC at autopsy. For participants who completed amyloid PET imaging as described previously, this modality was used to classify biomarker positivity. For participants who underwent CSF collection, CSF p-tau181/Aβ42 ratio was used to establish biomarker positivity. For participants assessed by both modalities, amyloid PET was used to define biomarker status to maximize the number of longitudinal biomarker data points available for assessment. ADNC was defined as described previously.
Statistical analyses
Summary statistics for continuous variables (means/medians and standard deviations) and categorical variables (percentages) were calculated for all participants as well as the biomarker-negative and -positive subgroups. For biomarker-negative versus -positive comparisons, two-sample t-tests were used for continuous variables and z-score tests for two proportions were used for categorical variables. Wilcoxon rank sum tests for continuous variables and Chi-square tests and binomial tests for proportions were used if the parametric test assumptions were not met. Cox proportional hazards models with death as a competing risk were used to compare the probability of progression to CDR > 0 for biomarker-negative and -positive participants. Biomarker positivity was used as the main effect and centred age (normalized age used for statistical modelling), APOE ε4 carrier status (0 = non-carrier, 1 = carrier), gender, self-reported race, centred education (years), PRS, the time interval from baseline to last clinical assessment (years) and all two-way interaction terms for these variables were included as covariates. Centred age and centred education were calculated by subtracting overall means of each metric across the entire dataset from individual values. For these models, hazard ratios (HR) and their 95% confidence intervals with P-values were reported. Logistic regression models were used to assess the effect of baseline positive biomarkers on the odds of intermediate-high ADNC at autopsy for those participants with neuropathological data (n = 46). As with the Cox models, biomarker positivity, centred age, APOE ε4 carrier status, gender, race, centred education, PRS and the time interval from baseline to death were included in the models. Univariate logistic regression models were used to assess the effect of each baseline individual biomarker (CSF Aβ42, p-tau181, t-tau, p-tau181/Aβ42, amyloid PET centiloid) on the odds of intermediate-high ADNC at autopsy. Univariate ordinal logistic regression models were used to assess the effect of each baseline individual biomarker on the cumulative odds of less advanced Braak stage and Thal phase. For each ordinal regression model, assumption of proportional odds was met. For binomial and ordinal logistical regression models, the ORs and their 95% confidence intervals with P-values were reported.
Data availability
All data from analyses in this paper will be deposited in the Washington University Knight ADRC dataset and made available to qualified investigators by request following approval by the Knight ADRC data request committee.
Results
Participants
Table 1 summarizes baseline characteristics and cognitive outcomes for participants in this study. Race was self-reported by participants. A total of 720 participants met criteria for inclusion as described previously; 117 (16.3%) underwent CSF collection only, 252 (35.0%) underwent amyloid PET imaging only and 351 (48.7%) completed both CSF collection and amyloid PET (in these participants, amyloid PET was used for biomarker status determination). 22.5% of biomarker-positive and 14.4% of biomarker-negative participants were defined on the basis of CSF assessment, whereas 77.5% of biomarker-positive and 85.6% of biomarker-negative participants were defined on the basis of amyloid PET assessment.
Table 1.
Demographics and clinical characteristics for cognitively normal (CDR = 0) participants at baseline
| All participants | Biomarker negative | Biomarker positive | P-valuea | ||||
|---|---|---|---|---|---|---|---|
| n | Mean (SD), or % | n | Mean (SD), or % | n | Mean (SD), or % | ||
| Age, mean (SD), years | 720 | 69.2 (9.2) | 560 | 67.9 (9.3) | 160 | 73.9 (6.8) | <0.0001 |
| Male sex, % | 302 | 41.9 | 231 | 41.2 | 71 | 44.4 | 0.4778 |
| Race, % | |||||||
| Non-Hispanic White | 621 | 86.3 | 470 | 83.9 | 151 | 94.4 | 0.0007 |
| African-American | 96 | 13.3 | 87 | 15.5 | 9 | 0.6 | 0.0012 |
| Asian | 2 | 0.3 | 2 | 0.4 | 0 | 0 | 0.4237 |
| More than one | 1 | 0.1 | 1 | 0.2 | 0 | 0 | 0.5687 |
| APOE ɛ4 carriers, % | 241 | 33.5 | 147 | 26.2 | 94 | 58.7 | <0.0001 |
| PRS, mean (SD) | 683 | −0.00355 (0.00729) | 531 | −0.00405 (0.00711) | 152 | −0.00181 (0.00762) | 0.0007 |
| Education, mean (SD), years | 720 | 16.0 (2.6) | 560 | 16.0 (2.5) | 160 | 16.0 (3.0) | 0.9890 |
| Baseline MMSE, mean (SD) | 720 | 29.1 (1.2) | 560 | 29.1 (1.1) | 160 | 29.0 (1.3) | 0.1954 |
| Final MMSE, mean (SD) | 718 | 28.7 (2.1) | 560 | 28.9 (1.5) | 158 | 27.7 (3.3) | <0.0001 |
| Interval from baseline assessment to final assessment, mean (SD) [median], years | 720 | 5.8 (3.4) [5.2] | 560 | 5.8 (3.5) [5.2] | 160 | 5.7 (3.1) [5.3] | 0.8415 |
| Interval from baseline biomarker to final assessment, mean (SD) [median], years | 720 | 5.6 (3.4) [5.1] | 560 | 5.6 (3.5) [5.1] | 160 | 5.6 (3.1) [5.1] | 0.8509 |
| Interval from baseline biomarker to study conclusionb or death, mean (SD) [median], years | 720 | 7.6 (3.7) [7.5] | 560 | 7.7 (3.8) [7.3] | 160 | 7.6 (3.4) [7.5] | 0.9424 |
| Mortality, % | 82 | 11.4 | 54 | 9.6 | 28 | 17.5 | 0.0058 |
| Progression to CDR > 0, %c | 102 | 14.17 | 47 | 8.4 | 55 | 34.4 | <0.0001 |
| CDR at final assessment | |||||||
| 0 | 633 | 87.9 | 521 | 93.0 | 112 | 70.0 | <0.0001 |
| 0.5 | 60 | 8.3 | 31 | 5.6 | 29 | 18.1 | <0.0001 |
| 1 | 24 | 3.3 | 8 | 1.4 | 16 | 10.0 | <0.0001 |
| 2 | 3 | 0.4 | 0 | 0 | 3 | 1.9 | 0.0012 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | – |
| Progression to CDR > 0 by time interval of follow-up from initial biomarker to final assessment, % | |||||||
| 0–5 years | 39 | 11.0 | 17 | 6.1 | 22 | 28.6 | <0.0001 |
| 5–10 years | 45 | 16.2 | 20 | 9.5 | 25 | 36.8 | <0.0001 |
| 10–15 years | 17 | 19.5 | 9 | 12.5 | 8 | 53.3 | 0.0003 |
| 15–20 years | 1 | 50.0 | 1 | 50.0 | 0 | – | – |
| Clinical diagnoses assigned at time of final assessment for progressors (CDR > 0), % | |||||||
| AD dementiad | 70 | 68.6 | 26 | 55.3 | 44 | 80.0 | 0.0073 |
| Parkinson's disease dementia | 2 | 1.0 | 0 | 0 | 2 | 3.6 | 0.1868 |
| DLB | 1 | 1.0 | 1 | 2.1 | 0 | 0 | 0.2757 |
| Vascular dementia | 3 | 3.0 | 2 | 4.2 | 1 | 1.8 | 0.4654 |
| Non-AD dementia | 2 | 2.0 | 1 | 2.1 | 1 | 1.8 | 0.9124 |
| Uncertain dementiae | 24 | 23.5 | 17 | 36.1 | 7 | 12.7 | 0.0054 |
| No. defined by CSF biomarker, % | 117 | 16.3 | 81 | 14.4 | 36 | 22.5 | 0.0151 |
| t-tau, mean (SD), pg/ml | 117 | 234.8 (104.7) | 81 | 194.0 (57.0) | 36 | 326.8 (127.9) | <0.0001 |
| p-tau181 mean (SD), pg/ml | 117 | 21.5 (11.3) | 81 | 16.6 (4.6) | 36 | 32.6 (13.8) | <0.0001 |
| Aβ42 mean (SD), pg/ml | 117 | 1382.5 (623.6) | 81 | 1622.4 (578.3) | 36 | 842.8 (302.7) | <0.0001 |
| p-tau181/Aβ42, mean (SD) | 117 | 0.020 (0.018) | 81 | 0.011 (0.003) | 36 | 0.041 (0.020) | <0.0001 |
| No. defined by amyloid PET biomarker | 603 | 83.7 | 479 | 85.5 | 124 | 77.5 | 0.0151 |
| PIB SUVR, mean (SD) | 406 | 1.300 (0.610) | 318 | 1.021 (0.095) | 88 | 2.307 (0.624) | <0.0001 |
| AV45 SUVR, mean (SD) | 197 | 1.135 (0.459) | 161 | 0.951 (0.160) | 36 | 1.958 (0.457) | <0.0001 |
AD = Alzheimer’s disease; DLB = dementia with Lewy bodies.
P-values derived from tests comparing mean or % values between biomarker-negative and -positive subgroups. P-values < 0.05 are highlighted in bold.
Study conclusion date 19 April 2019.
Row highlighted in bold indicates main finding from this table.
AD dementia category includes the following diagnostic subgroups containing only a single participant: Biomarker negative = CDR 0.5 in memory only; Biomarker-positive = AD dementia with other contributing aetiology, AD dementia with depression contributing, AD dementia with disturbed social comportment, AD dementia with language dysfunction, CDR 0.5 in memory only.
Uncertain dementia category includes the following diagnostic subgroups: Uncertain dementia, Uncertain—possible non-AD dementia, Uncertain—questionable impairment.
A total of 160 participants (22.2%) were designated biomarker-positive at baseline assessment (91.9% age ≥ 65 years), while 560 participants (78.8%) were designated biomarker-negative (68.0% age ≥ 65 years). Participants defined as biomarker-positive tended to be older (P < 0.0001), were more likely APOE ε4 allele carriers (P < 0.0001), and were more likely to self-report their race as White (P = 0.0007) as compared to biomarker-negative participants. The mean interval from baseline to final clinical assessment was 5.8 years (median 5.3 years; range 0.8–15.9 years) and this did not differ based on biomarker designation. Biomarker-positive participants had higher overall mortality compared to biomarker-negative participants (P = 0.0058).
Cognitive outcomes
A significantly higher proportion of biomarker-positive participants (34.4%) progressed from normal cognition (CDR = 0) to impaired cognition (CDR > 0) at last assessment as compared to biomarker-negative participants (8.4%) during the period of evaluation (P < 0.0001). This was reflected by a lower average MMSE at the time of last clinical assessment in biomarker-positive versus -negative participants (P < 0.0001). When evaluated by time interval of follow-up from initial biomarker to final assessment, a higher proportion of biomarker-positive (53.3%) compared to biomarker-negative (12.5%) participants progressed to CDR > 0 when followed for 10–15 years from baseline assessment (P = 0.0003). Among all biomarker-positive participants who progressed to CDR > 0 (n = 55) at last assessment, the average time interval from detection of positive biomarkers to development of cognitive impairment was 3.9 years (SD 2.5 years, range 0.9–10.9 years). There were no observed differences in these outcomes between African-American/Black participants and White participants, however the small number of biomarker-positive African-American/Black participants limited detection of differences due to lack of power.
To better assess the underlying clinical phenotypes associated with progression to CDR > 0 among biomarker-positive and biomarker-negative participants, the clinical diagnoses assigned at final assessment are listed in Table 1. Alzheimer’s disease dementia was the assigned diagnosis in 80% of biomarker-positive CDR progressors compared to 55.3% of biomarker-negative CDR progressors (P = 0.0073). Uncertain dementia, reflecting either atypical pattern or questionable level of impairment, was the assigned diagnosis in 12.7% of biomarker-positive CDR progressors compared to 36.1% of biomarker-negative CDR progressors (P = 0.0054).
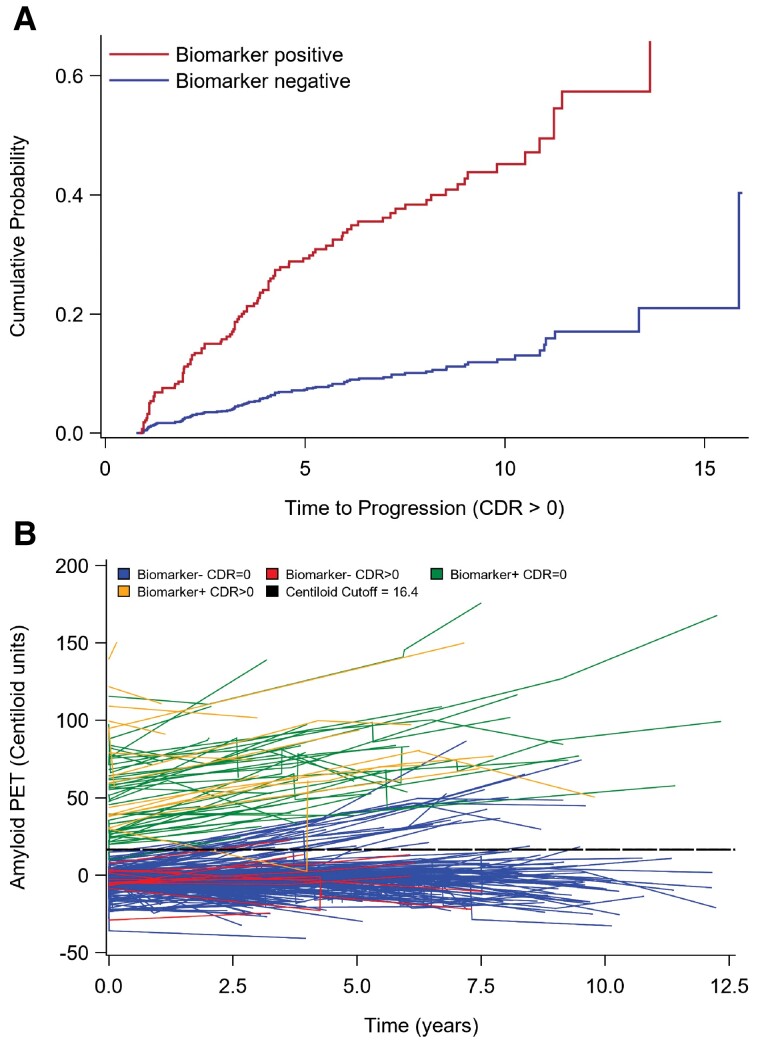
To determine the effect of biomarker status, age, sex, self-reported race, years of education, APOE ɛ4 carrier status, PRS and interval from baseline assessment to final assessment on risk of progression to CDR > 0, multivariable Cox proportional hazard regression with death as a competing risk was used to model the effects of these covariates on CDR progression (Supplementary Table 1). Due to missing covariates, only 680 of 720 available participants were included in this analysis. In this model, preclinical Alzheimer’s disease biomarker positivity (HR = 2.794; P < 0.0001) and APOE ɛ4 + carrier status (HR = 1.941; P = 0.0266) had the strongest independent effect on risk of developing cognitive impairment. Age at baseline (independent of APOE ɛ4 carrier status) and PRS also significantly contributed to risk of developing cognitive impairment. There was a statistically significant interaction between APOE ɛ4 carrier status and age (P = 0.0045), highlighting a reduced effect of age on risk of CDR progression among APOE ɛ4 carriers as compared to APOE ɛ4 non-carriers. All other tested covariates in the model did not significantly contribute to increased risk of developing cognitive impairment. Cumulative incidence function curves derived from this model (Fig. 1A) demonstrate the increased probability of CDR progression over time in biomarker-positive compared to biomarker-negative individuals.
Figure 1.
Survival analysis and longitudinal biomarker trends in biomarker-negative and -positive groups. (A) Model-based cumulative incidence function curves for probability of progression to CDR > 0 in biomarker-negative (blue; lower curve) and biomarker-positive (red; upper curve) populations. (B) Spaghetti plots of longitudinal amyloid PET biomarker trends in CDR progressors (CDR > 0) and non-progressors (CDR = 0). Amyloid PET PIB and AV-45 SUVR measurements were converted to Centiloid units to combine tracer data. Data were plotted and subcategorized by biomarker-positive and -negative assignment. Biomarker category assignment was based on SUVR cut-offs. The comparable converted Centiloid cut-off value (16.4) is presented for visualization but was not used in determining biomarker status. Fitted lines obtained by simple linear regression with 95% CI are also shown. Only participants with at least one additional biomarker assessment of the same modality used to define original biomarker status were included.
There was a surprisingly large number of biomarker-negative individuals with CDR progression (n = 47 of total n = 560), among which 55.3% were assigned a clinical diagnosis of Alzheimer’s disease dementia (described previously). To better understand the relationship between longitudinal biomarkers, biomarker status and cognitive outcomes, participants from the original cohort with at least one additional biomarker assessment of the same modality used to define original biomarker status were separately analysed and presented in Supplementary Table 2. Nearly all of these participants with longitudinal biomarkers were assigned biomarker status based on amyloid PET imaging. The biomarker-positive cohort had significantly higher baseline (P = 0.0448) and final (P = 0.0337) amyloid PET centiloid units among CDR progressors compared to non-progressors, whereas there was no significant difference in baseline or final amyloid PET centiloid units between the CDR progressors and non-progressors in the biomarker-negative cohort. A total of 28 biomarker-negative participants in this cohort converted to biomarker-positive status over the course of longitudinal biomarker assessment, however only one of these progressed to CDR > 0. This is supported by plots of longitudinal amyloid PET trends over time (Fig. 1B). Among biomarker-negative CDR progressors, there is no trend towards increased amyloid accumulation.
Neuropathological outcomes
During the course of follow-up, 82 participants died and 57 (70%) came to autopsy. Clinical characteristics and neuropathological outcomes are detailed in Table 2. Average age at death was 85.5 years and was not significantly different between biomarker-negative and -positive participants. The average interval from time of initial biomarker assessment to death was 7.0 years (median 7.0 years; range 2.2–11.9 years) and also was not different between biomarker-negative or -positive participants. Amyloid PET imaging was used to categorize biomarker status for 37 participants, while 20 participants were categorized by CSF. 22 participants (38.6%) were biomarker positive at baseline assessment and 35 (61.4%) were biomarker negative. 68.2% of biomarker-positive participants who underwent autopsy had cognitive impairment at time of expiration compared to 48.6% of biomarker-negative participants (P = 0.1471).
Table 2.
Clinical characteristics and neuropathological diagnoses of deceased participants with completed autopsy
| Total | Biomarker negative | Biomarker positive | P-valuea | ||||
|---|---|---|---|---|---|---|---|
| n | Mean (SD) or % | n | Mean (SD) or % | n | Mean (SD) or % | ||
| Age at baseline, mean (SD), years | 57 | 78.8 (6.9) | 35 | 79.0 (7.3) | 22 | 78.6 (6.3) | 0.8310 |
| Age at death, mean (SD), years | 57 | 85.5 (7.5) | 34 | 85.9 (8.3) | 19 | 86.3 (6.9) | 0.8546 |
| Male sex, % | 25 | 43.9 | 17 | 48.6 | 8 | 36.4 | 0.3681 |
| Race, % | |||||||
| Non-Hispanic White | 53 | 93.0 | 32 | 91.4 | 21 | 95.4 | 0.5619 |
| African-American | 4 | 7.0 | 3 | 8.6 | 1 | 4.6 | 0.5619 |
| Asian | 0 | 0 | 0 | 0 | 0 | 0 | – |
| More than one | 0 | 0 | 0 | 0 | 0 | 0 | – |
| APOE ɛ4 carriers, % | 14 | 24.6 | 4 | 11.4 | 10 | 45.4 | 0.0036 |
| Education, mean (SD), years | 57 | 16.2 (3.1) | 35 | 15.8 (2.7) | 22 | 16.7 (3.6) | 0.3158 |
| Interval from baseline assessment to death, mean (SD) [median], years | 57 | 7.2 (3.3) [7.2] | 35 | 6.9 (3.3) [7.2] | 22 | 7.6 (3.3) [7.2] | 0.4434 |
| Interval from baseline biomarker to death, mean (SD) [median], years | 57 | 7.0 (3.2) [7.0] | 35 | 6.8 (3.2) [7.3] | 22 | 7.4 (3.2) [6.9] | 0.4657 |
| Progression to CDR > 0 at expiration, % | 32 | 56.1 | 17 | 48.6 | 15 | 68.2 | 0.1471 |
| CDR at expiration, % | |||||||
| 0 | 25 | 43.9 | 18 | 51.4 | 7 | 31.8 | 0.1471 |
| 0.5 | 11 | 19.3 | 9 | 25.7 | 2 | 9.1 | 0.1211 |
| 1 | 4 | 7.0 | 3 | 8.6 | 1 | 4.6 | 0.5619 |
| 2 | 5 | 8.8 | 2 | 5.7 | 3 | 13.6 | 0.3030 |
| 3 | 12 | 21.0 | 3 | 8.6 | 9 | 40.9 | 0.0035 |
| Intermediate-high ADNC b , % | 23 | 40.3 | 3 | 8.6 | 20 | 90.9 | <0.0001 |
| ADNC by severity, % | |||||||
| None | 7 | 12.3 | 7 | 20.0 | 0 | 0 | 0.0251 |
| Low | 27 | 47.4 | 25 | 71.4 | 2 | 9.1 | <0.0001 |
| Intermediate | 9 | 15.8 | 1 | 2.9 | 8 | 36.4 | 0.0007 |
| High | 14 | 24.5 | 2 | 5.7 | 12 | 54.5 | <0.0001 |
| No. defined by CSF biomarker, % | 20 | 35.1 | 12 | 60.0 | 8 | 40.0 | 0.2077 |
| Intermediate-high ADNC, % CSF group | 9 | 45.0 | 1 | 8.3 | 8 | 100.0 | <0.0001 |
| Elecsys t-tau, mean (SD), pg/ml | 20 | 253.556 (135.095) | 12 | 188.152 (66.145) | 8 | 351.662 (155.978) | 0.0213 |
| Elecsys p-tau181, mean (SD), pg/ml | 20 | 24.238 (15.961) | 12 | 15.579 (5.263) | 8 | 37.227 (18.071) | 0.0113 |
| Elecsys Aβ42, mean (SD), pg/ml | 20 | 1277.65 (619.144) | 12 | 1622.28 (552.519) | 8 | 760.700 (227.481) | 0.0002 |
| p-tau181/Aβ42 Ratio, mean (SD) | 20 | 0.0264 (0.0262) | 12 | 0.0099 (0.0024) | 8 | 0.0512 (0.0262) | 0.0029 |
| No. defined by amyloid PET biomarker, % | 37 | 64.9 | 23 | 62.2 | 14 | 37.8 | 0.0366 |
| Intermediate-high ADNC, % PET group | 14 | 37.8 | 2 | 8.7 | 12 | 85.7 | <0.0001 |
| PIB SUVR, mean (SD) | 32 | 1.605 (0.935) | 20 | 1.025 (0.122) | 12 | 2.572 (0.899) | <0.0001 |
| AV45 SUVR, mean (SD) | 5 | 1.393 (0.589) | 3 | 1.109 (0.196) | 2 | 1.818 (0.840) | 0.2258 |
P-values derived from tests comparing mean or % values between biomarker-negative and -positive subgroups. P-values < 0.05 are highlighted in bold.
Defined by NIA-Regan intermediate-to-high likelihood of Alzheimer's diseaseee or NIA-AA intermediate-to-high ADNC. Row highlighted in bold indicates main finding from this table.
Preclinical Alzheimer’s disease biomarker positivity was a very strong predictor of underlying intermediate-to-high ADNC at autopsy. 90.9% of participants with positive preclinical Alzheimer’s disease biomarkers had intermediate-to-high ADNC on autopsy, whereas only 8.6% of participants negative for preclinical Alzheimer’s disease biomarkers had intermediate-to-high ADNC (P < 0.0001). Preclinical Alzheimer’s disease biomarker positivity was associated with high sensitivity (87.0%), specificity (94.1%), positive predictive value (PPV) (90.9%) and negative predictive value (NPV) (91.4%) for predicting intermediate-to-high ADNC at autopsy (Table 3). When participant cohorts categorized by either CSF or PET were evaluated separately, sensitivity, specificity, PPV and NPV remained high. Although CSF testing seemed to marginally outperform amyloid PET imaging for predicting ADNC at autopsy, this was not statistically significant when directly comparing diagnostic metrics between CSF and PET (sensitivity P = 0.824, specificity P = 0.3134, PPV P = 0.2622, NPV P = 0.971).
Table 3.
Diagnostic performance of preclinical Alzheimer's disease biomarkers for predicting intermediate-high ADNC at autopsy
| n | Sensitivity | 95% CI | Specificity | 95% CI | PPV | 95% CI | NPV | 95% CI | |
|---|---|---|---|---|---|---|---|---|---|
| Total, % | 57 | 87.0 | 73.2–100.0 | 94.1 | 86.2–100.0 | 90.9 | 78.9–100.0 | 91.4 | 82.1–100.0 |
| CSF p-tau181/Aβ42, % | 20 | 88.9 | 68.4–100.0 | 100.0 | 100.0–100.0 | 100.0 | 100.0–100.0 | 91.7 | 76.0–100.0 |
| Amyloid PET, % | 37 | 85.7 | 67.4–100.0 | 91.3 | 79.8–100.0 | 85.7 | 67.4–100.0 | 91.3 | 79.8–100.0 |
To evaluate the effect of preclinical biomarkers, centred age, APOE ɛ4 carrier status, sex, race, education, PRS and interval from baseline biomarker assessment to death on odds of harbouring ADNC, a logistic regression model was employed using these covariates. The only variable significantly associated with an effect on odds of harbouring ADNC in this cohort was preclinical Alzheimer’s disease biomarker positivity, which exhibited a very large effect size because so few individuals with positive biomarkers had absent or low ADNC (OR 886.15, 95% CI 23.85 >999.9, P = 0.0002). No other covariates demonstrated independent significant associations with diagnosis of ADNC.
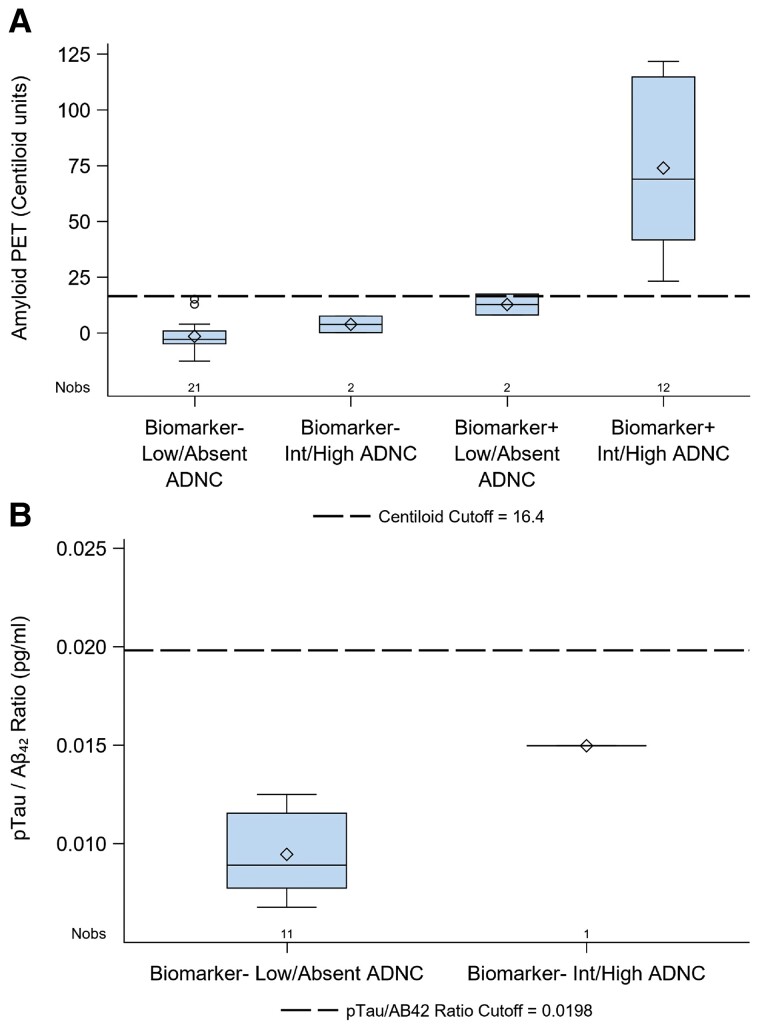
In this cohort, baseline preclinical Alzheimer’s disease biomarker evaluation resulted in three ‘false-negative’ and two ‘false-positive’ assignments when compared to gold-standard neuropathological assessment. To better evaluate these discordant biomarker assignments, baseline biomarker values were plotted after stratifying by baseline biomarker designation and final Alzheimer’s disease neuropathological outcome (Fig. 2). In all cases, those participants with false-positive or -negative baseline biomarker assignments had baseline biomarker values that were closer to the threshold value compared to those participants who were correctly assigned.
Figure 2.
Baseline biomarker levels in participants stratified by biomarker status and final Alzheimer's disease neuropathological diagnosis. (A) Boxplots of baseline amyloid PET biomarker levels stratified by biomarker positivity and low/absent or intermediate/high ADNC at autopsy. Amyloid PET PIB and AV-45 SUVR measurements were converted to Centiloid units to combine tracer data. Biomarker category assignment was based on SUVR cut-offs. The comparable converted Centiloid cut-off value (16.4) is presented for ease of visualization but was not used as a threshold for biomarker positivity. (B) Boxplots of baseline CSF p-tau181/Aβ42 levels for biomarker-negative participants stratified by low/absent or intermediate/high ADNC at autopsy. Among the participants whose biomarker status was based on CSF, there were no biomarker-positive participants without ADNC in the autopsy cohort. So, biomarker-positive participants were excluded from this plot. In both A and B, the box size defines the interquartile range, the horizontal line indicates the median, the diamond indicates the mean, the whiskers indicate maximum and minimum range of data points and open circles indicate outliers.
To investigate this further, individual Alzheimer’s disease neuropathological scores were evaluated in these discordant cases. Among the three biomarker-negative participants with underlying intermediate-high ADNC (false negatives), two cases were clearly discordant with substantial amyloid (Thal phase 4, CERAD score 3) and tau (Braak stage V) burden qualifying for a high ADNC designation despite negative biomarker assignment. The third ‘false-negative’ case had moderate amyloid (Thal phase 3, CERAD 1) and tau (Braak stage III) burden, qualifying for intermediate ADNC designation. There were two biomarker-positive participants lacking intermediate-high ADNC (false positives) and each had moderate amyloid burden (Thal phase 3–4, CERAD score 0) and low tau burden (Braak stage I-II), qualifying for low ADNC designation.
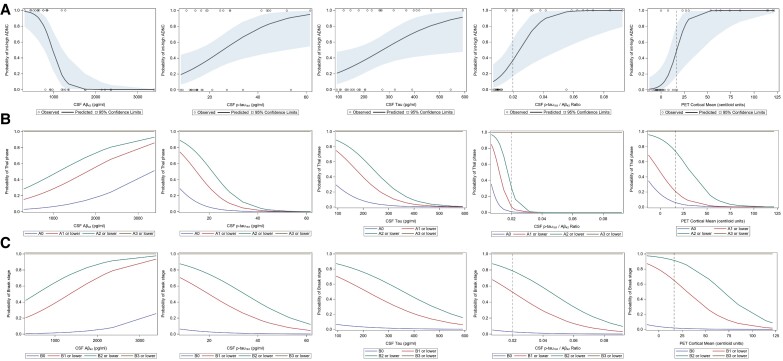
The association between biomarker levels and neuropathological outcomes was further explored by using univariate logistical regression models with baseline single biomarker levels as independent variable and probability of intermediate-high ADNC as dependent variable. OR from these analyses are summarized in Table 4. These models demonstrated that increased CSF Aβ42 levels were associated with significantly lower odds of intermediate-high ADNC per unit change, whereas increased CSF p-tau181, t-tau, p-tau181/Aβ42 and amyloid PET centiloid values were associated with significantly higher odds of intermediate-high ADNC per unit change. CSF Aβ40 levels were not available with this specific Elecsys dataset, so Aβ42/Aβ40 ratios could not be calculated. Probability curves produced by these models are shown in Fig. 3A. Similarly, the effect of baseline single biomarker levels on the probability of having more advanced Braak stage and Thal phase pathology at autopsy was assessed using univariate ordinal logistic regression models. OR from these analyses are summarized in Table 5. The models similarly demonstrated that increased Aβ42 levels were associated with significantly lower odds, whereas p-tau-181, t-tau, p-tau-181/Aβ42 and amyloid PET centiloid values were associated with significantly higher odds of more advanced Braak stage and Thal phase pathology at autopsy per biomarker unit change. Cumulative probability curves from the ordinal logistic regression models are shown in Fig. 3B and C.
Table 4.
Effect of baseline individual biomarkers on odds of intermediate-high ADNC present at autopsy
| Biomarker | Odds ratio | 95% CI | P-valuea |
|---|---|---|---|
| Elecsys Aβ42b (per 100 pg/ml increase) | 0.513 | 0.326–0.807 | 0.0039 |
| Elecsys p-tau181b (per 1 pg/ml increase) | 1.086 | 1.015–1.161 | 0.0161 |
| Elecsys t-Taub (per 10 pg/ml increase) | 1.077 | 1.007–1.152 | 0.0297 |
| p-tau181/Aβ42 ratiob (per 0.001 unit increase) | 1.134 | 1.031–1.248 | 0.0099 |
| Centiloidc (per 5 unit increase) | 2.248 | 1.219–4.145 | 0.0095 |
P-values < 0.05 are highlighted in bold
Total n limited to 33 of 57 due to missing CSF values.
Total n limited to 37 of 57 due to missing amyloid PET Centiloid values.
Figure 3.
Probability curves of ADNC pathology as a function of baseline single biomarker values. (A) Probability curves (with shaded 95% confidence intervals) of intermediate-high ADNC at autopsy as a function of baseline levels of Aβ42, p-tau181, t-tau, p-tau181/Aβ42 ratio and amyloid PET Centiloid units. Probability curves are generated from univariate logistic regression models with biomarker level as independent variable and probability of intermediate-high ADNC as dependent variable. (B) Cumulative probability curves of Thal phase pathology at autopsy as a function of baseline levels Aβ42, p-tau181, t-tau, p-tau181/Aβ42 ratio and amyloid PET Centiloid units. Thal phases are grouped in this model consistent with NIA-AA criteria62: A0 = Thal phase 0, A1 = Thal phase 1 or 2, A2 = Thal phase 3, A3 = Thal phase 4 or 5. (C) Cumulative probability curves of Braak stage pathology at autopsy as a function of baseline levels Aβ42, p-tau181, t-tau, p-tau181/Aβ42 ratio and amyloid PET Centiloid units. Braak stages are grouped in this model consistent with NIA-AA criteria62: B0 = Braak stage 0, B1 = Braak stage I or II, B2 = Braak stage III or IV, B3 = Braak stage V or VI. (B and C) Probability curves were generated from univariate ordinal logistic regression models with biomarker level as independent variable and cumulative probability of pathologic group as dependent variable. Modelled probabilities are cumulated over lower pathology groups, such that each curve delineates the probability of a given pathologic group or lower. Vertical dashed lines highlight p-tau181/Aβ42 ratio and Centiloid cut-off thresholds for reference.
Table 5.
Effect of baseline individual biomarkers on cumulative odds of less advanced Braak stage or Thal phase pathology
| Biomarker | Thal phasea | Braak stageb | ||||
|---|---|---|---|---|---|---|
| Odds ratioh | 95% CI | P-value | Odds ratioh | 95% CI | P-valuec | |
| Elecsys Aβ42a (per 100 pg/ml increase) | 1.121d | 1.010–1.245 | 0.0321 | 1.140e | 1.019–1.277 | 0.0227 |
| Elecsys p-tau181a (per 1 pg/ml increase) | 0.854d | 0.763–0.956 | 0.0061 | 0.929e | 0.873–0.978 | 0.0083 |
| Elecsys t-Taua (per 10 pg/ml increase) | 0.867d | 0.787–0.955 | 0.0040 | 0.930e | 0.877–0.986 | 0.0152 |
| p-tau181/Aβ42 Ratioa (per 0.001 unit increase) | 0.721d | 0.536–0.873 | 0.0081 | 0.952e | 0.917–0.980 | 0.0033 |
| Centiloidb (per 5 unit increase) | 0.799f | 0.707–0.903 | 0.0003 | 0.691g | 0.506–0.848 | 0.0039 |
Thal phases grouped as follows for ordinal logistic regression model: phase 0 = A0, phase 1–2 = A1, phase 3 = A2, phase 4–5 = A3.
Braak stages grouped as follows for ordinal logistic regression model: stage 0 = B0, stage I–II = B1, stage III–IV = B2, stage V–VI = B3.
P-values < 0.05 are highlighted in bold.
Total n limited to 31 of 57 due to missing CSF values or missing Thal phase.
Total n limited to 33 of 57 due to missing CSF values or missing Braak stage.
Total n limited to 34 of 57 due to missing Centiloid values or missing Thal phase.
Total n limited to 36 of 57 due to missing Centiloid values or missing Braak stage.
For the ordinal logistic regression, probabilities modelled are cumulated over lower Thal and Braak stages such that the OR reflect change in odds of being at a lower stage or phase of pathology per unit change of biomarker.
As described previously, there were a large number of biomarker-negative CDR progressors (48.6%) in this autopsy cohort but only a low percentage with underlying ADNC (8.6%). To better understand the underlying neuropathologies associated with each subcategory, the cohort was analysed separately by biomarker status and CDR progressor status (Supplementary Table 3). Cerebrovascular disease was highly prevalent across all subcategories, with only two participants lacking any vascular pathology. When vascular disease was limited to pathology more likely to influence cognition (infarcts, haemorrhages, severe arteriolosclerosis or severe white matter changes), pathological changes remained prevalent (∼40% across the whole cohort) but were significantly less prevalent amongst biomarker-positive CDR non-progressors (n = 0; 0%) as compared to biomarker-positive CDR progressors (n = 9; 60%). Primary tau pathologies, which include typical frontotemporal lobar degeneration pathologies as well as common age-related primary tauopathies such as argyrophilic grain disease, primary age-related tauopathy and ageing-related tau astrogliopathy, were commonly observed across the cohort (43.8%) but noticeably absent in any biomarker-positive CDR non-progressor. Overall, 12 of 17 biomarker-negative CDR progressors had neuropathological diagnoses expected to influence cognition, including discordantly assigned ADNC, cerebral Lewy bodies, hippocampal sclerosis, frontotemporal lobar degeneration-TDP43 and prion disease. In comparison, only 3 of 18 biomarker-negative CDR non-progressors had similar diagnoses, including cerebral Lewy bodies and medial temporal lobe TDP43 pathology.
Discussion
This prospective study evaluated clinical and neuropathological outcomes in a community-dwelling cohort of cognitively normal participants stratified by the presence or absence of preclinical Alzheimer’s disease. Preclinical Alzheimer’s disease biomarker positivity, as defined by either CSF p-tau181/Aβ42 or amyloid PET imaging in individuals with normal cognition, was associated with significantly increased risk of developing cognitive impairment over time and was highly predictive of intermediate-high ADNC at autopsy. This study adds to the base of evidence supporting the use of these biomarkers to define Alzheimer’s disease in its preclinical phase by providing crucial neuropathological validation. Indeed, in this study, 20 out of 22 participants with positive preclinical Alzheimer’s disease biomarkers who went to autopsy were found to have intermediate-high ADNC.
Most recent Alzheimer’s disease clinical trials have added detection of either PET- or CSF-based Alzheimer’s disease biomarkers as inclusion criteria to enroll only those cognitively impaired participants with anticipated ADNC. With the development of Alzheimer’s disease disease-modifying therapies,1 treatment trials are beginning to focus on prevention paradigms in preclinical Alzheimer’s disease populations.30,31 In studies focused on prevention of sporadic Alzheimer’s disease, this requires screening strategies to identify possible participants. These data support the use of these modalities for screening cognitively normal individuals in the community. The recent discovery of promising plasma-based Alzheimer’s disease biomarkers63,64 should enable even broader screening of the general population. Neuropathological correlation will be required for thorough validation of plasma-based preclinical Alzheimer’s disease biomarkers.
CSF- and PET-based preclinical Alzheimer’s disease diagnosis in this study was associated with high sensitivity, specificity, PPV and NPV for detection of intermediate-high ADNC. Although there were a significant number of biomarker-negative participants who developed cognitive impairment prior to death, 53% of these participants had underlying non-Alzheimer’s disease neuropathological diagnoses that could influence cognition, whereas only 17% had ‘false-negative’ ADNC.
The strong association of preclinical Alzheimer’s disease biomarker positivity with subsequent development of cognitive impairment reported here is well-established and has been presented previously in a number of prior studies, both from the Washington University Knight ADRC cohort5,8,11,14 as well as other cohorts of ageing.12,65–68 A unique feature of this cohort is the size and duration of prospective longitudinal cognitive follow-up, with 87 participants (12.1% of total cohort) being followed for 10–15 years from time of initial biomarker assessment. In this study, though the proportion of biomarker-positive individuals with progression to CDR > 0 increased significantly with interval of follow-up, only 53.3% of biomarker-positive participants progressed when followed for 10–15 years from baseline assessment, suggesting an extended period between development of ADNC and onset of cognitive impairment. This finding is consistent with previous studies that have evaluated time to incident cognitive impairment in relation to biomarker positivity and found that a proportion of biomarker-positive participants persist beyond 15 years without progression to CDR > 0, supporting a preclinical stage of Alzheimer’s disease that can extend beyond 15 years.14
In this study, there were two ‘false-positive’ and three ‘false-negative’ cases, as determined by discordance between biomarker status and presence or absence of intermediate-high ADNC. One possible interpretation is that these participants were falsely assigned at baseline due to the imprecision of the biomarker cut-off threshold, supported by the fact that all discordant cases had biomarker levels near the cut-off threshold. An alternative explanation is that the false-positive participants had low levels of ADNC below the threshold for intermediate-to-high ADNC designation on autopsy. Indeed, the two false-positive cases showed low ADNC at autopsy with moderate amyloid burden but insufficient tau pathology to qualify as intermediate-to-high ADNC. By analogy, the false-negative participants could have relatively modest amyloid burden at autopsy with ADNC stage driven primarily by high levels of tau pathology. However, in reality, two of the false-negative cases harboured substantial amyloid and tau pathology consistent with high ADNC, whereas the third case had moderate amyloid and tau burden consistent with intermediate ADNC. Therefore, it is also possible these false-negative participants may have developed more substantial ADNC in the interval between baseline biomarker assessment and autopsy.
Tau PET imaging was not assessed in this cohort, as its inclusion would have significantly limited cohort size. However, tau PET has been shown to accurately predict high ADNC in a terminally ill population69 and seems to reflect underlying Alzheimer’s disease tau pathology.70,71 Although the biomarker false-positive rate was low in this study, it is likely that assessment of tau PET status in a preclinical population would further lower this false-positive rate, potentially at the expense of a higher false-negative rate.
Previous studies have demonstrated an association between Thal amyloid phase, but not Braak stage, on the level of ante-mortem PIB PET binding.72,73 However, these prior studies have not evaluated whether baseline Alzheimer’s disease biomarker levels in cognitively normal participants influence the level of Thal phase and Braak stage pathology at autopsy. In this study, baseline individual biomarker levels (CSF Aβ42, p-tau181, t-tau, p-tau181/Aβ42 and amyloid PET centiloid) were all significantly associated with the probability of harbouring intermediate-high ADNC and having more or less advanced Thal phase and Braak stage pathology at autopsy
Neuropathological assessment also demonstrated frequent occurrence of non-Alzheimer’s disease comorbid pathological diagnoses, although these did not appear evenly distributed across groups when stratified by progression to CDR > 0. These data indicate that among biomarker-positive CDR non-progressors, non-Alzheimer’s disease neuropathological diagnoses are infrequent, whereas among biomarker-negative CDR progressors, non-Alzheimer’s disease neuropathologic diagnoses more commonly underlie cognitive impairment, with some contribution from limited numbers of false-negative ADNC diagnoses.
One concern about a cross-sectional screening approach for detecting preclinical Alzheimer’s disease in the general population is the possibility for progression from negative to positive biomarkers after the initial screening test and the theoretical risk for developing Alzheimer’s disease dementia after a negative screening test. Indeed, in this cohort there was a surprisingly large number of biomarker-negative individuals who progressed to CDR > 0, most of which were assigned a clinical diagnosis of Alzheimer’s disease dementia. In the longitudinal biomarker clinical cohort of this study, conversion from biomarker negative to positive was observed among ∼10% of biomarker-negative participants. However, the rate of progression to CDR > 0 among this group was 3.5%, significantly less than the base rate of CDR progression in the full biomarker-negative cohort (8.4%). Therefore, in this cohort with a mean interval follow-up of ∼6 years, conversion from negative to positive preclinical Alzheimer’s disease biomarkers was not highly predictive of onset of cognitive impairment, and, it seems unlikely that CDR progression among biomarker-negative participants in this dataset can be attributed to rapid development of substantial Alzheimer’s disease neuropathologic change. While future strategies for screening populations for preclinical Alzheimer’s disease may require longitudinal screening tests performed at regular intervals, the appropriate time interval for screening will need to be determined on the basis of more in-depth longitudinal analyses of cohorts of biomarker converters.
This study has a number of strengths. First, unlike most published biomarker-autopsy validation studies, this study by design evaluated a community-based cohort of cognitively normal participants, thereby exclusively focusing on the validity of these markers in the preclinical phase of disease. Since this cohort consists of participants living in the local community and not recruited from tertiary referral memory clinics with potential referral biases, the spectrum of comorbid neuropathology observed on autopsy is less likely to be skewed compared to that observed in other studies.22 This is supported by the near universal presence of cerebrovascular disease noted across all categories of brain specimens in the autopsy cohort, a highly prevalent diagnosis in the general population of older adults.74
This study also has important limitations. The participant population included individuals highly motivated to participate in research, with many agreeing to serial LP. This population is unlikely to be representative of the general community. The total number of autopsied participants with positive preclinical Alzheimer’s disease biomarkers was relatively low (38.6%). This limited sample size restricts the extent of neuropathological assessment. Also, a significant proportion of participants at autopsy with positive preclinical Alzheimer’s disease biomarkers had not yet developed cognitive impairment prior to autopsy (31.8%), meaning they did not meet clinical criteria for Alzheimer’s disease dementia. However, this limitation would not be expected to influence the accuracy of detecting ADNC. Another limitation of this study is that it included participants defined by either CSF or amyloid PET in combination and did not limit analysis to only one method for biomarker detection. This choice was made to broaden the number of participants with longitudinal assessments and the number available for inclusion in the autopsy cohort. There is risk that discordance in positivity between these two methods might bias the data. We tried to limit this risk by using predefined cut-offs to establish positivity that have been shown previously by Knight ADRC investigators to demonstrate high concordance between CSF and PET modalities.28 One final limitation of this study is that it does not definitively demonstrate whether detection of preclinical Alzheimer’s disease biomarkers or ADNC inevitably leads to cognitive impairment in affected participants. Although the data in this study and many others strongly support the association between detection of preclinical Alzheimer’s disease biomarkers and onset of cognitive impairment, it remains an open question as to what proportion of these participants, with sufficient longitudinal follow-up, will ultimately develop cognitive impairment.
In summary, preclinical detection of Alzheimer’s disease biomarkers by CSF or amyloid PET is associated with significantly elevated risk of developing cognitive impairment and is highly predictive of intermediate-to-high ADNC (considered sufficient to account for dementia) at autopsy. This study provides neuropathological validation supporting the use of these modalities for preclinical Alzheimer’s disease screening. Further longitudinal follow-up will be required to establish the long-term clinical outcomes among individuals with positive preclinical Alzheimer’s disease biomarkers and among biomarker converters.
Supplementary Material
Acknowledgements
The authors thank the many research participants enrolled in studies at the Knight ADRC. The authors also thank the staff members of the Administration, Clinical, Genetics, Imaging, Biostatistics and Neuropathology Cores of the Knight ADRC who performed participant assessments, and collected and processed specimens and data used in this study.
Contributor Information
Justin M Long, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Neurology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Dean W Coble, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Division of Biostatistics, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Chengjie Xiong, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Division of Biostatistics, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Suzanne E Schindler, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Neurology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Richard J Perrin, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Neurology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Pathology and Immunology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Brian A Gordon, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Mallinckrodt Institute of Radiology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Tammie L S Benzinger, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Mallinckrodt Institute of Radiology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Elizabeth Grant, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Division of Biostatistics, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Anne M Fagan, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Neurology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Oscar Harari, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Psychiatry, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Carlos Cruchaga, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Psychiatry, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
David M Holtzman, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Neurology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
John C Morris, Charles F. and Joanne Knight Alzheimer Disease Research Center, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Neurology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA; Department of Pathology and Immunology, Washington University School of Medicine in St Louis, St Louis, MO 63110, USA.
Funding
This work was supported by awards from the NIH K08AG068611, the Alzheimer’s Association AACSF-18-564776 and the Mary E. Groff Charitable Trust to J.M.L. This work was also supported by NIH P30AG066444, P01AG03991 and P01AG026276 grants to J.C.M. The Research Education Component through NIH grant P30AG066444 provided individual support to J.M.L.
Competing interests
J.M.L., D.W.C., C.X., S.E.S., R.J.P., B.A.G., T.L.B., E.G., O.H. and C.C. report no competing interests. A.M.F. has received research funding from the National Institute on Aging of the National Institutes of Health, Biogen, Centene, Fujirebio and Roche Diagnostics. She is a member of the scientific advisory boards for Roche Diagnostics, Genentech and Diadem and also consults for DiamiR and Siemens Healthcare Diagnostics Inc. There are no competing interests. D.M.H. reports being a cofounder with equity in C2N Diagnostics, LLC. He is on the scientific advisory boards of Genentech, Denali, C2N Diagnostics and Cajal Neurosciences and consults for Takeda, Casma and Eli Lilly. He is an inventor of a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. This antibody program was licensed to AbbVie. He is an inventor on a patent licensed by Washington University to Eli Lilly on a humanized anti-Aβ antibody. His laboratory receives research grants from the NIH, Cure Alzheimer’s Fund, the Rainwater Foundation, the JPB Foundation, Good Ventures, C2N Diagnostics, NextCure, Denali and Novartis. J.C.M. does not own stock nor has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Long JM, Holtzman DM. Alzheimer disease: An update on pathobiology and treatment strategies. Cell. 2019;179:312–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Price JL, McKeel DW, Buckles VD, et al. . Neuropathology of nondemented aging: Presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging. 2009;30:1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Price JL, Morris JC. Tangles and plaques in nondemented aging and “preclinical” Alzheimer’s disease. Ann Neurol. 1999;45:358–368. [DOI] [PubMed] [Google Scholar]
- 4. Knopman DS, Parisi JE, Salviati A, et al. . Neuropathology of cognitively normal elderly. J Neuropathol Exp Neurol. 2003;62:1087–1095. [DOI] [PubMed] [Google Scholar]
- 5. Morris JC, Roe CM, Grant EA, et al. . Pittsburgh Compound B imaging and prediction of progression from cognitive normality to symptomatic Alzheimer disease. Arch Neurol. 2009;66:1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mintun MA, Larossa GN, Sheline YI, et al. . [11c]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. [DOI] [PubMed] [Google Scholar]
- 7. Fagan AM, Mintun MA, Mach RH, et al. . Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–519. [DOI] [PubMed] [Google Scholar]
- 8. Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64:343–349. [DOI] [PubMed] [Google Scholar]
- 9. Bateman RJ, Xiong C, Benzinger TLS, et al. . Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fagan AM, Xiong C, Jasielec MS, et al. . Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci Transl Med. 2014;6:226ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vos SJ, Xiong C, Visser PJ, et al. . Preclinical Alzheimer’s disease and its outcome: A longitudinal cohort study. Lancet Neurol. 2013;12:957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donohue MC, Sperling RA, Petersen R, et al. . Association between elevated brain amyloid and subsequent cognitive decline among cognitively normal persons. JAMA. 2017;317:2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dumurgier J, Hanseeuw BJ, Hatling FB, et al. . Alzheimer’s disease biomarkers and future decline in cognitive normal older adults. J Alzheimers Dis. 2017;60:1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roe CM, Ances BM, Head D, et al. . Incident cognitive impairment: Longitudinal changes in molecular, structural and cognitive biomarkers. Brain. 2018;141:3233–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vos SJB, Gordon BA, Su Y, et al. . NIA-AA staging of preclinical Alzheimer disease: Discordance and concordance of CSF and imaging biomarkers. Neurobiol Aging. 2016;44:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leinonen V, Alafuzoff I, Aalto S, et al. . Assessment of beta-amyloid in a frontal cortical brain biopsy specimen and by positron emission tomography with carbon 11-labeled Pittsburgh compound B. Arch Neurol. 2008;65:1304–1309. [DOI] [PubMed] [Google Scholar]
- 17. Wolk DA, Grachev ID, Buckley C, et al. . Association between in vivo fluorine 18-labeled flutemetamol amyloid positron emission tomography imaging and in vivo cerebral cortical histopathology. Arch Neurol. 2011;68:1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sojkova J, Driscoll I, Iacono D, et al. . In vivo fibrillar beta-amyloid detected using [11C]PiB positron emission tomography and neuropathologic assessment in older adults. Arch Neurol. 2011;68:232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikonomovic MD, Klunk WE, Abrahamson EE, et al. . Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer’s disease. Brain. 2008; 131(Pt 6):1630–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strozyk D, Blennow K, White LR, Launer LJ. CSF Abeta 42 levels correlate with amyloid-neuropathology in a population-based autopsy study. Neurology. 2003;60:652–656. [DOI] [PubMed] [Google Scholar]
- 21. Clark CM, Pontecorvo MJ, Beach TG, et al. . Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-β plaques: A prospective cohort study. Lancet Neurol. 2012;11:669–678. [DOI] [PubMed] [Google Scholar]
- 22. Lesman-Segev OH, La Joie R, Iaccarino L, et al. . Diagnostic accuracy of amyloid versus 18 F-fluorodeoxyglucose positron emission tomography in autopsy-confirmed dementia. Ann Neurol. 2021;89:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reimand J, Boon BDC, Collij LE, et al. . Amyloid-β PET and CSF in an autopsy-confirmed cohort. Ann Clin Transl Neurol. 2020;7:2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen CD, Joseph-Mathurin N, Sinha N, et al. . Comparing amyloid-β plaque burden with antemortem PiB PET in autosomal dominant and late-onset Alzheimer disease. Acta Neuropathol. 2021;142:689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grothe MJ, Moscoso A, Ashton NJ, et al. . Associations of fully automated CSF and novel plasma biomarkers with Alzheimer disease neuropathology at autopsy. Neurology. 2021;97:e1229–e1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vlassenko AG, McCue L, Jasielec MS, et al. . Imaging and cerebrospinal fluid biomarkers in early preclinical Alzheimer disease. Ann Neurol. 2016;80:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanseeuw BJ, Betensky RA, Jacobs HIL, et al. . Association of amyloid and tau with cognition in preclinical Alzheimer disease: A longitudinal study. JAMA Neurol. 2019;76:915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schindler SE, Gray JD, Gordon BA, et al. . Cerebrospinal fluid biomarkers measured by Elecsys assays compared to amyloid imaging. Alzheimers Dement. 2018;14:1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palmqvist S, Mattsson N, Hansson O, Alzheimer’s disease neuroimaging initiative . Cerebrospinal fluid analysis detects cerebral amyloid-β accumulation earlier than positron emission tomography. Brain. 2016;139:1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sperling RA, Rentz DM, Johnson KA, et al. . The A4 study: Stopping AD before symptoms begin? Sci Transl Med. 2014;6:228fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bateman RJ, Benzinger TL, Berry S, et al. . The DIAN-TU next generation Alzheimer’s prevention trial: Adaptive design and disease progression model. Alzheimers Dement. 2017;13:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morris JC. The clinical dementia rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. [DOI] [PubMed] [Google Scholar]
- 33. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 34. Su Y, D’Angelo GM, Vlassenko AG, et al. . Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS ONE. 2013;8:e73377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su Y, Blazey TM, Snyder AZ, et al. . Partial volume correction in quantitative amyloid imaging. Neuroimage. 2015;107:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mishra S, Gordon BA, Su Y, et al. . AV-1451 PET Imaging of tau pathology in preclinical Alzheimer disease: Defining a summary measure. Neuroimage. 2017;161:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klunk WE, Koeppe RA, Price JC, et al. . The centiloid project: Standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11:1–15.e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Su Y, Flores S, Hornbeck RC, et al. . Utilizing the centiloid scale in cross-sectional and longitudinal PiB PET studies. Neuroimage Clin. 2018;19:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su Y, Flores S, Wang G, et al. . Comparison of Pittsburgh compound B and florbetapir in cross-sectional and longitudinal studies. Alzheimers Dement (Amst). 2019; 11:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schindler SE, Sutphen CL, Teunissen C, et al. . Upward drift in cerebrospinal fluid amyloid β 42 assay values for more than 10 years. Alzheimers Dement. 2018;14:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vos SJB, Visser PJ, Verhey F, et al. . Variability of CSF Alzheimer’s disease biomarkers: Implications for clinical practice. PLoS ONE. 2014;9:e100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cruchaga C, Kauwe JSK, Nowotny P, et al. . Cerebrospinal fluid APOE levels: An endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet. 2012;21:4558–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cruchaga C, Del-Aguila JL, Saef B, et al. . Polygenic risk score of sporadic late-onset Alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimers Dement. 2018;14:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Del-Aguila JL, Fernández MV, Schindler S, et al. . Assessment of the genetic architecture of Alzheimer’s disease risk in rate of memory decline. J Alzheimers Dis. 2018;62:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cairns NJ, Taylor-Reinwald L, Morris JC. Alzheimer’s disease neuroimaging initiative. Autopsy consent, brain collection, and standardized neuropathologic assessment of ADNI participants: The essential role of the neuropathology core. Alzheimers Dement. 2010;6:274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. [DOI] [PubMed] [Google Scholar]
- 47. Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thal DR, Rüb U, Orantes M, Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58:1791–1800. [DOI] [PubMed] [Google Scholar]
- 49. Khachaturian ZS. Diagnosis of Alzheimer’s disease. Arch Neurol. 1985;42:1097–1105. [DOI] [PubMed] [Google Scholar]
- 50. Braak H, Del Tredici K, Rüb U, de Vos RAI, Jansen Steur ENH, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. [DOI] [PubMed] [Google Scholar]
- 51. McKeith IG, Galasko D, Kosaka K, et al. . Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): Report of the Consortium on DLB International Workshop. Neurology. 1996;47:1113–1124. [DOI] [PubMed] [Google Scholar]
- 52. McKeith IG, Dickson DW, Lowe J, et al. . Diagnosis and management of dementia with Lewy bodies: Third report of the DLB Consortium. Neurology. 2005;65:1863–1872. [DOI] [PubMed] [Google Scholar]
- 53. McKeith IG, Boeve BF, Dickson DW, et al. . Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kovacs GG, Ferrer I, Grinberg LT, et al. . Aging-related tau astrogliopathy (ARTAG): Harmonized evaluation strategy. Acta Neuropathol. 2016;131:87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kovacs GG, Robinson JL, Xie SX, et al. . Evaluating the patterns of aging-related tau astrogliopathy unravels novel insights into brain aging and neurodegenerative diseases. J Neuropathol Exp Neurol. 2017;76:270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Josephs KA, Murray ME, Whitwell JL, et al. . Staging TDP-43 pathology in Alzheimer’s disease. Acta Neuropathol. 2014;127:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Josephs KA, Murray ME, Whitwell JL, et al. . Updated TDP-43 in Alzheimer’s disease staging scheme. Acta Neuropathol. 2016;131:571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nelson PT, Dickson DW, Trojanowski JQ, et al. . Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142:1503–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Montine TJ, Phelps CH, Beach TG, et al. . National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 2012;123:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease . The National Institute on Aging, and Reagan Institute Working Group on Diagnostic criteria for the Neuropathological Assessment of Alzheimer’s Disease. Neurobiol Aging. 1997;18(4 Suppl):S1–S2. [PubMed] [Google Scholar]
- 61. Hyman BT, Trojanowski JQ. Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer Disease. J Neuropathol Exp Neurol. 1997;56:1095–1097. [DOI] [PubMed] [Google Scholar]
- 62. Hyman BT, Phelps CH, Beach TG, et al. . National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schindler SE, Bollinger JG, Ovod V, et al. . High-precision plasma β-amyloid 42/40 predicts current and future brain amyloidosis. Neurology. 2019;93:e1647–e1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakamura A, Kaneko N, Villemagne VL, et al. . High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554:249–254. [DOI] [PubMed] [Google Scholar]
- 65. Knopman DS, Jack CR, Wiste HJ, et al. . Short-term clinical outcomes for stages of NIA-AA preclinical Alzheimer disease. Neurology. 2012;78:1576–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moghekar A, Li S, Lu Y, et al. . CSF Biomarker changes precede symptom onset of mild cognitive impairment. Neurology. 2013;81:1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stomrud E, Minthon L, Zetterberg H, Blennow K, Hansson O. Longitudinal cerebrospinal fluid biomarker measurements in preclinical sporadic Alzheimer’s disease: A prospective 9-year study. Alzheimers Dement (Amst). 2015; 1:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Clark LR, Racine AM, Koscik RL, et al. . Beta-amyloid and cognitive decline in late middle age: Findings from the WRAP study. Alzheimers Dement. 2016;12:805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fleisher AS, Pontecorvo MJ, Devous MD, et al. . Positron emission tomography imaging with [18F]flortaucipir and postmortem assessment of Alzheimer disease neuropathologic changes. JAMA Neurol. 2020;77:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen CD, Holden TR, Gordon BA, et al. . Ante- and postmortem tau in autosomal dominant and late-onset Alzheimer’s disease. Ann Clin Transl Neurol. 2020;7:2475–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Soleimani-Meigooni DN, Iaccarino L, La Joie R, et al. . 18F-flortaucipir PET to autopsy comparisons in Alzheimer’s disease and other neurodegenerative diseases. Brain. 2020;143:3477–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Murray ME, Lowe VJ, Graff-Radford NR, et al. . Clinicopathologic and 11C-Pittsburgh compound B implications of thal amyloid phase across the Alzheimer’s disease spectrum. Brain. 2015;138(Pt 5):1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lowe VJ, Lundt ES, Albertson SM, et al. . Neuroimaging correlates with neuropathologic schemes in neurodegenerative disease. Alzheimers Dement. 2019;15:927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vermeer SE, Longstreth WT, Koudstaal PJ. Silent brain infarcts: a systematic review. Lancet Neurol. 2007;6:611–619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from analyses in this paper will be deposited in the Washington University Knight ADRC dataset and made available to qualified investigators by request following approval by the Knight ADRC data request committee.