Abstract
Carcinogenesis is a multi-step process by which normal cells acquire genetic and epigenetic changes that result in cancer. In combination with host genetic susceptibility and environmental exposures, a prominent pro-carcinogenic role for the microbiota has recently emerged. In colorectal cancer (CRC), three nefarious microbes have been consistently linked to cancer development: 1) Colibactin-producing Escherichia coli initiates carcinogenic DNA damage, 2) Enterotoxigenic Bacteroides fragilis promotes tumorigenesis via toxin-induced cell proliferation and tumor-promoting inflammation, 3) Fusobacterium nucleatum enhances CRC progression through two adhesins, FadA and Fap2, that promote proliferation, anti-tumor immune evasion, and may contribute to metastases. Herein, we use these three prominent microbes to discuss the experimental evidence linking microbial activities to carcinogenesis and the specific mechanisms driving this stepwise process. Precisely defining mechanisms by which the microbiota impact carcinogenesis at each stage is essential for us to develop microbiota-targeted strategies for the diagnosis, prognosis, and treatment of cancer.
Keywords: Carcinogenesis, Colorectal cancer, Microbiota, Colibactin, B. fragilis toxin, Fusobacterium
Introduction to the microbiota
At birth, humans are seeded with a diverse collection of microbes, including bacteria, viruses, fungi, archaea, protozoa, and helminths (1-3). Humans and microbes are in symbiosis, supporting host physiology, immunological development, and metabolism among other essential functions (4-6). This human-associated microbial consortium is termed the “microbiota”. Bacteria are the best studied gut microbiota members, due to advances in sequencing technology and bioinformatics pipelines that distinguish bacterial taxa by polymorphisms in the ubiquitous bacterial 16S ribosomal RNA genomic sequence (7-9). By scrutinizing the genomic content (“microbiome”) of these microbes, our research community has uncovered some general principles regarding the gut microbiota.
Microbial communities are finely tuned to fit the ecology and function of each body site, likely benefiting from the nutrients in each microenvironment (3, 10). Many of our microbial species evolve with us, acquiring traits through mutation and horizontal gene transfer over our lifetime that may influence health and disease (11). The gut microbiota is the best studied human-associated microbial community, due to its diversity, abundance, and ease of sample collection (feces/stool). The gut microbiota community composition and function differ longitudinally down the gastrointestinal (GI) tract from mouth to anus, depending on the physiological needs of each niche (12). Interestingly, gut microbiota are more similar between individuals than between body sites of one individual (i.e. skin vs. gut) (5). However, the composition of our individual gut microbiota significantly differs (3).
Researchers have not found a “core microbiota”. However, when we consider the capabilities – genes and pathways – harbored in individual microbiomes, we see strong similarities amongst people (3) Thus, it appears that the function of the microbiota may be more important than the presence or absence of species within the community (13). Therefore, individual microbiota hold a potential to impact human health and disease that may be overlooked when simply classifying by taxonomy. The pro-carcinogenic microbes discussed in this review all harbor specific genes and capabilities absent from the core genome of their particular species (i.e. Escherichia coli, Bacteroides fragilis, and Fusobacterium nucleatum) (14-16). Therefore, we have an obligation to look deeper than community structure to evaluate the pro-carcinogenic capability of the microbiota.
We previously explored this body of knowledge using Hanahan and Weinberg’s “Hallmarks of Cancer” (17) as a framework to classify specific mechanisms by which microbes, microbial communities, and microbial metabolites may impact cancer development (15). These ten Hallmarks comprise key biological capabilities acquired by normal cells as they develop traits of cancer cells and progress toward tumor development. Here we will review and expand upon mechanisms by which specific members of the microbiota influence the development of cancer and speculate upon what stages of carcinogenesis they principally impact.
Foundational associations between inflammation, cancer, and the microbiome
In 1984, Drs. Barry Marshall and J. Robin Warren performed an unprecedented experiment in which self-colonization of Dr. Marshall with patient-derived Helicobacter pylori rapidly induced gastritis that was ameliorated by eradication of H. pylori with antibiotics (18, 19). Others in the gastric cancer field had been skeptical that bacteria could survive the acidic environment of the stomach and attributed gastric cancer development to genetic susceptibility or other host physiological causes. Helicobacter is now recognized as a group I carcinogen and the primary cause of gastritis, peptic ulcers, and gastric cancer (20). Helicobacter deploys the cytotoxin-associated gene A (CagA) toxin as the predominant oncoprotein that hijacks multiple epithelial signaling pathways and initiates carcinogenesis with chronic inflammation fueling cancer progression (20). With their infamous experiment, Drs. Marshall and Warren had not simply fulfilled Koch’s postulates for H. pylori and gastritis – they piqued interest in the nuances of this complex relationship between host and microbe(s) and a new field of research emerged in microbial-induced chronic inflammation and carcinogenesis.
With the advent of high-throughput sequencing and an explosion of knowledge about our intestinal microbiota, it is now clear that our gut microbiota influences cancer development, most notably colorectal cancer (CRC) (21-23). Microbiota impact host metabolism, inflammation, immunity, and cellular proliferation, which are all processes that when dysregulated can promote tumorigenesis (24). Furthermore, ample evidence suggests that the microbiota can directly impact tumor formation. Fecal transplants from human patients with CRC promote carcinogenesis in germ-free (sterile mice devoid of any microbiota) and conventional mice administered the colon-specific carcinogen azoxymethane (AOM) (25). Transferring the microbiota of tumor-bearing mice versus non-tumor-bearing mice accelerates the development and severity of tumorigenesis in the AOM/dextran sulphate sodium (DSS) mouse model (26). The structure and physiological state of the microbiota also influences pro-carcinogenic effects, as biofilm-associated communities from both CRC and healthy individuals induce more tumorigenesis than non-biofilm communities in mouse models (27). These studies demonstrate a causal relationship between the microbiota and CRC development and provide rationale for further mechanistic studies.
The microbiota in carcinogenesis: Initiation, promotion, and progression
Over the past decade, pre-clinical and clinical evidence connects the microbiota and their metabolites to carcinogenesis. The conventional paradigm proposes microbial eubiosis (balanced flora) is positively health-associated, while a change in microbial diversity or functionality (dysbiosis, unbalanced flora) can promote disease development, including various cancers (14, 28). Dysbiotic triggers include changes in genetics, environment (e.g. inflammation, medication, diet), or pathogenic infection. However, it is still debated whether microbial community alterations are a cause or effect of carcinogenesis.
Data suggests microbial pathogens drive cancer formation in 15-20% of cancer cases (29). Currently, the International Agency for research on Cancer (IARC) classifies 10 microbial species as group 1 human carcinogens with Helicobacter pylori, Hepatitis B virus (HBV), Hepatitis C virus (HCV), and Human papillomavirus (HPV) driving 90% of infection-associated cancers (21, 29, 30). Despite pathogen-triggered carcinogenesis being the focus for the past 10 years, association studies and studies with selectively colonized (gnotobiotic) mouse models clearly demonstrate the pro-carcinogenic capability of commensal microbes (Table 1).
Table 1.
Host-associated microbes impact carcinogenesis at all stages
| Stage of Carcinogenesis |
Microbe | Microbial Carcinoaenic Agent |
Proposed Host Effect | Site of Cancer | IARC Classification |
References |
|---|---|---|---|---|---|---|
| Initiation | Adherent-invasive Escherichia coli | Colibactin produced from the polyketide synthase (pks) gene cluster | DNA damage | Colon | N.C. | (33, 34, 38-40, 43, 49, 50) |
| Campylobacter jejuni | Cytolethal distending toxin (CDT) | DNA damage | Colon | N.C. | (105-107) | |
| Enterococcus faecalis | Reactive oxygen species (ROS) | DNA damage, genomic instability, cell cycle arrest | Colon | N.C. | (90, 91) | |
| Helicobacter hepaticus | Cytolethal distending toxin (CDT) | DNA damage and impairment of DNA repair | Billiary tract, intestine, breast | N.C. | (108-110) | |
| Initiation and promotion | Chlamydia trachomatis | Unknown | DNA damage, impaired DNA repair, cell survival and proliferation | Urogenital | N.C. | (111-113) |
| Helicobacter pylori | Cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA) | DNA damage, cell survival, cell proliferation | Stomach | 1 | (18-20) | |
| Neisseria gonorrhoeae | Restriction endonucleases | DNA damage, cell survival, cell proliferation | Prostate | N.C. | (114-116) | |
| Salmonella spp. | AvrA and cytolethal distending toxin (CdtB) | DNA damage, cell survival, cell proliferation, cell cycle arrest | Intestine and hepatobiliary | N.C. | (106, 117-119) | |
| Promotion | Clonorchis sinensis | Parasitic excretory-secretory products | Cell proliferation | Bile duct | 1 | (120-122) |
| Enterotoxigenic Bacteroides fragilis (ETBF) | B. fragilis -derived toxin (BFT) | Cell proliferation and tumor-promoting inflammation | Colon | N.C. | (57, 60-62, 64, 106) | |
| Hepatitis B virus (HBV) | Hepatitis B virus X (HBx) and Hepatits B surface Protein (HB) | Genetic instability, cell survival, cell proliferation | Liver | 1 | (123) | |
| Hepatitis C virus (HCV) | Core and Non-structural protein 3 (NS3), and NS5a | Genetic instability, cell survival, cell proliferation | Liver | 1 | (124) | |
| Human Papillomavirus (HPV) types 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 | E6 and E7 proteins | Genetic instability, cell survival and proliferation | Urogenital and oropharynx | 1 | (125-127) | |
| Human T-cell lymphotropic virus 1 (HTLV-1) | HTLV-1 transactivator protein (Tax) and HTLV-1 basic leucine zipper factor (HBZ) | Cell proliferation | Adult T-cell lymphoma | 1 | (128, 129) | |
| Opisthorchis viverrini | Parasitic excretory-secretory products | Cell proliferation and pro-carcinogenic inflammation | Liver | 1 | (122, 130) | |
| Porphyromonas gingivalis | Gingipain | Cell survival | Esophogeal, oral, and pancreas | N.C. | (131, 132) | |
| Schistosoma haematobium | Excretory-secretory products | Cell proliferation and pro-carcinogenic inflammation | Bile duct and Bladder | 1 | (122) | |
| Promotion and progression | Epstein-Barr virus (EBV) | Epstein-Barr virus nuclear antigens (EBNA) and Latent membrane proteins (LMP) | Cell survival, proliferation, differentiation, migration | Burkitt's lymphoma, Nasopharynx, Hodgkin's lymphoma | 1 | (133, 134) |
| Fusobacterium nucleatum | Fusobacterium adhesin A (FadA) and Fusobacterial apoptosis protein 2 (Fap2) | Tumor binding, invasion, cell survival, cell proliferation, immune | Colon and breast | N.C. | (71-73, 76-81, 82) | |
| Kaposi sarcoma-associated herpesvirus (KSHV or HHV8) | K1, K15, and vGPCR | Cell survival, cell proliferation, cell differentiation | Kaposi sarcoma and primary effusion lymphoma | 1 | (135, 136) |
IARC = International Agency for Resarch on Cancer
N.C. = Not classified
Carcinogenesis can be divided into three stages: Initiation, promotion, and progression (31). Initiation is defined by spontaneous or induced genetic alterations, such as exposure to a carcinogenic agent; this alters the responsiveness of cells to their environment and provides a proliferative advantage (31). Promotion is a period of preneoplastic cell proliferation and accumulation, inducing additional genetic damage and amplifying mutations (31). Progression is marked by further neoplastic expansion, with enhanced tumor growth rate, invasiveness, and metastasis. The microbiota has the potential to impact carcinogenesis at all stages (Table 1). Here we provide a detailed discussion of three prominent microbes and the mechanisms by which they initiate, promote, and enhance progression of carcinogenesis in CRC.
Colibactin-producing pks+ Escherichia coli: Initiates carcinogenesis
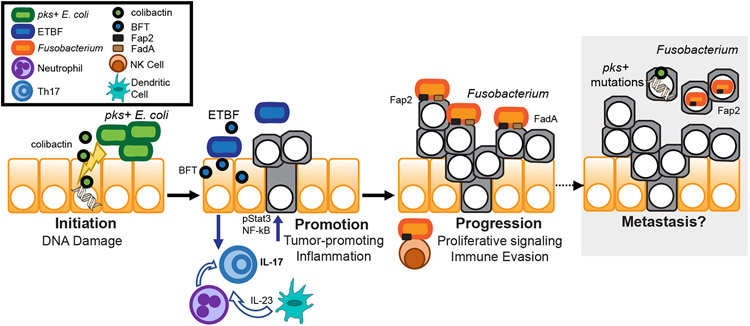
During carcinogenesis, normal host cells acquire mutations that confer growth and survival advantages. Cancer formation is often initiated by a chemical carcinogen, which induces genotoxicity or DNA damage (31). The microbiota is predicted to produce hundreds of unique small molecules and secondary metabolites that may influence host health and disease (32). These metabolites are often synthesized by complex enzymatic assembly lines encoded by biosynthetic gene clusters. One cancer-associated genotoxic molecule is colibactin, produced from the polyketide synthase (pks) gene cluster present among certain strains of E. coli (33, 34). Pks+ E. coli strains are prevalent in the microbiota of CRC patients (34, 35), induce CRC in mouse models (34, 36-38), and leave a distinct mutational fingerprint in human colorectal tumors that signifies former exposure and points to a role in cancer initiation (39, 40) (Figure 1).
Figure 1: Microbiota impact all stages of carcinogenesis.
Initiation (left): first stage of carcinogenesis characterized by DNA alterations to normal cells. Colibactin, a specialized metabolite produced by pks+ E. coli (green), has genotoxic activity that damages DNA and leads to mutations. Promotion (center): second stage of carcinogenesis characterized by proliferation of transformed cells. BFT produced by ETBF (blue) damages the colonic epithelium and barrier integrity. This disruption leads to pro-carcinogenic T helper 17 (Th17) dominant inflammation. Epithelial cells, neutrophils, and dendritic cells produce cytokines that activate T cells to promote Th17 inflammation, including dendritic cell derived IL-23. IL-17-producing T cells signal back to the epithelium and induce epithelial cell proliferation driven by pStat3 and NF-kB pathways. Progression (right): final stage of carcinogenesis characterized by tumor growth and invasion, leading to metastases. Fusobacterium (orange) uses adhesins FadA to bind to E-cadherin and Fap2 to bind to Gal-GalNAc on tumor cells to promote proliferative signaling. Fap2 also binds TIGIT on Natural Killer (NK) cells to enhance immune evasion. Although there is not yet strong evidence that these bacteria promote metastasis, Fusobacterium and a pks+ mutagenic signature have been found in metastases and may contribute to this stage of carcinogenesis.
The pks island was first described in 2006, as a 54kb genomic island of 19 genes (clbA to clbS) that encodes a large and sophisticated non-ribosomal peptide and polyketide synthase assembly line (NRPS-PKS) (33, 41). Not all E. coli harbor the pks island, but those that do are restricted to E. coli phylotype B2 and represent both commensal and pathogenic strains (42). Among human microbiota, pks+ E. coli are highly prevalent in CRC patients (34, 35), with one study estimating carriage among 66.7% of CRC patients, 40.0% of inflammatory bowel disease patients, and only 20.8% of healthy patients (34). These correlative findings suggested pks may play a role in disease promotion.
Early studies demonstrated pks was responsible for inducing cell cycle arrest and activation of DNA repair machinery in mammalian cells exposed to E. coli, suggesting pks products were microbially-derived genotoxins (33, 43). More specifically, epithelial cells that encounter colibactin-producing E. coli exhibit DNA double-strand breaks and are characterized by ɣ-H2AX foci, G2/M cell cycle arrest, megalocytosis, and activation of ATM/CHK/CDC25/CDK1 DNA damage signaling cascades (33, 34, 43). The pro-tumorigenic role of the pks+ island was first demonstrated to enhance tumor multiplicity and invasion in the AOM/interleukin 10-deficient (Il10−/−) colitis-associated CRC mouse model (34). These pro-carcinogenic effects were validated by multiple groups in additional mouse models that also defined the role of various pks genes and proteins required for colibactin’s genotoxic effects (reviewed in (42)).
While the precise chemical identity of bioactive colibactin has remained elusive, chemical and structural analyses have defined inactive precolibactins and stable colibactin-DNA lesions that can lead to mutation and tumorigenesis (44-46). Briefly, inactive precursors are synthesized in the bacterial cytoplasm and then deacetylated in the periplasm by the peptidase ClbP (42, 47, 48). In the mammalian cell nucleus, colibactin alkylates DNA with a ‘double warhead’ comprised of a cyclopropane ring conjugated to an alpha,beta-unsaturated imine, creating adenine-colibactin adducts and DNA crosslinks (49-51). Although bacterial:mammalian cell contact is required (33) for genotoxicity, beyond that it is currently unknown how bioactive colibactin is released from the bacteria and enters the mammalian cell to cause DNA damage.
It was predicted that colibactin-DNA lesions lead to mutations in oncogenes or tumor suppressors that drive cancer. Indeed, two recent studies defined unique mutational signatures caused by colibactin exposure (39, 40). Both studies repeatedly exposed mammalian cells to pks+ E. coli in culture and identified single base pair substitutions contained in specific AT-rich motifs that are structurally and chemically consistent with the effects of previously identified adenine-colibactin adducts. The single base pair substitution (SBS) signature was termed SBS-pks and includes ATA, ATT and TTT with the middle base mutated (39). An additional signature contained single T deletions at T homopolymers, with enrichment of adenines upstream of the insertion/deletion site (termed indels) and was termed ID-pks (39). Importantly, mining established whole-genome sequencing (WGS) datasets revealed that these signatures predominated in CRC tumors and metastases relative to other cancer types (39, 40). SBS-pks and ID-pks signatures were positively correlated, suggesting they derived from a common origin – colibactin exposure (39). Both studies linked the location of these pks signatures to CRC mutational hotspots, with the adenomatous polyposis coli gene APC (the most commonly mutated gene in CRC (52, 53)) harboring the highest amount of mutations with the SBS-pks or ID-pks mutational signatures (39). These signatures can serve as biomarkers of past colibactin exposure and the findings clearly link the mutational signature of colibactin exposure to known CRC driver mutations.
Intriguingly, a separate study examining non-neoplastic colon tissue detected SBS-pks and ID-pks in 29 of 42 healthy individuals and data modeling revealed these signatures were likely acquired before 10 years of age (54). Thus, early exposure to pks+ E. coli and a prominent pks mutational signature found early in life may indicate a greater risk for CRC. In combination with genetic susceptibility and other risk factors, the presence of colibactin-derived mutational signatures may inform new prognostic algorithms for CRC. It will be important to better understand how the genomic location and/or abundance of colibactin-DNA adducts may relate to future cancer risk.
Enterotoxigenic Bacteroides fragilis (ETBF): Promotes carcinogenesis
Almost every neoplastic lesion contains immune cells. Once thought to solely be an anti-tumoral response, inflammation can enhance tumor promotion and progression (30, 55, 56). The close proximity of the microbiota and mucosal immune system provides opportunity for resident microbes to elicit pro-tumorigenic immune responses. Bacteroides spp. are normal inhabitants of the intestinal microbiota, representing approximately 30% of the gut community members, and help shape mucosal immune responses (57).
B. fragilis is a key Bacteroides community member that represents about 0.5-2% of the entire gut microbiota (57). Strain level differences render B. fragilis either beneficial or pro-inflammatory and pro-carcinogenic. Beneficial B. fragilis, referred to as non-toxigenic B. fragilis (NTBF), promotes regulatory T cell development and suppression of inappropriate inflammation through the production of Polysaccharide A (PSA) (58, 59). In contrast, enterotoxigenic B. fragilis (ETBF) produces a proteolytic enterotoxin, termed B. fragilis toxin (BFT) or fragilysin (60). BFT is a heat-labile metalloprotease that is produced as a pro-toxin and activated by the B. fragilis cysteine protease, fragipain (57). ETBF promotes inflammation and CRC predominantly through BFT.
Genetically susceptible mouse models have been instrumental in demonstrating the inflammatory and tumorigenic effects of ETBF. APC is a chief tumor suppressor protein, commonly mutated in CRC patients (52, 53). Apcmin/+ mice and mouse models with truncated Apc spontaneously develop numerous intestinal tumors, mainly localized to the small intestine. However, when colonized with ETBF, tumors now develop in the colon within a month of inoculation (61). ETBF primarily resides in the colon, where it is thought to drive tumorigenic effects via local production of BFT. Colonization with NTBF does not induce colonic tumors, demonstrating the reliance on BFT for B. fragilis pro-carcinogenic activities (61).
Upon exposure to epithelial cells, BFT damages colonic epithelial barrier integrity by inducing cleavage of the zonula adherens protein, E-cadherin (62). Oncogenic beta-catenin is released from E-cadherin and translocates to the nucleus where it acts as a transcription factor and induces epithelial hyperproliferation (63). Normally, cytosolic beta-catenin is restrained by the host APC protein and is continually targeted for proteasomal degradation (63). However, the APC gene is mutated in 70-80% of CRC patients (52, 53), diminishing APC tumor suppressive function. Therefore, beta-catenin oncogenic signaling is likely enhanced by microbial-derived BFT.
BFT-mediated E-cadherin cleavage not only induces proliferative signaling, but also increases gut permeability that enhances translocation of microbial products (57). The disruption of epithelial integrity triggers a pro-inflammatory cascade that leads to rapid and sustained interleukin-17 (IL-17) production by colonic T cells, the defining feature of T helper 17 (Th17) cell immune responses (60, 64). IL-17 production evoked by B. fragilis is a key driver of colon tumorigenesis, which is inhibited by IL-17 neutralization. Thus, ETBF promote cancer development by invoking tumorigenic inflammation, in part through BFT (Figure 1).
Th17 immune responses, induced by microbes and their metabolites, are associated with worsened CRC patient prognosis (55, 65). Under homeostatic conditions and exposure to epithelial-adherent commensals, Th17 immunity is trained as a protective host defense response (66). However, Th17 release can be maladaptive in the context of inflammation and cancer (64). At sites of inflammation and on developing adenomas, epithelial barrier defects and defective mucin production permit microbial sampling by intra-tumoral dendritic cells that then produce IL-23 (55). Neutrophils, other local immune cells, and epithelial cells produce pro-inflammatory cytokines including IL-1β and IL-6 in this microenvironment (56). This intra-tumoral cytokine milieu including IL-23 and IL-6 causes recruitment and expansion of IL-17 producing T cells (IL-17A specifically), which signal to epithelial cells through the receptor IL-17RA (64, 65). Epithelial recognition of IL-6 and IL-17 activates a signaling cascade that involves phosphorylated Stat3, NF-kB, and MAPK (64). This signaling cascade induces anti-apoptotic and pro-proliferative genes that promote cancer development (60). These data suggest that inappropriate exposure to BFT induces a coordinated response between epithelial, myeloid, and lymphoid cells, which establishes a microenvironment of tumor-promoting inflammation that enhances cancer development.
Fusobacterium nucleatum: Enhances cancer progression
Healthy tissues tightly control cellular signals to modulate growth, maintain homeostatic cell densities, tissue architecture, and function. Dysregulated cellular signaling can permit sustained and potentially deleterious cell proliferation. As discussed above in relation to ETBF, microbial-induced dissociation of beta-catenin from E-cadherin drives proliferative pathways that support tumor promotion and progression (63). Fusobacterium nucleatum is a normal inhabitant of the oral microbiota that can cause inflammation in the gingival tissue and infectious inflammatory conditions at multiple body sites (28, 30, 67-69).
Mis-localization of F. nucleatum to the colon is associated with CRC. Although luminal spread seems possible from the oral cavity to the colon, evidence suggests that F. nucleatum reaches sites of inflammation and tumorigenesis via a hematogenous route (70, 71). Fusobacterium is prevalent in CRC patient tissue (72, 73) and its abundance positively correlates with cancer severity (74, 75), supporting a role for Fusobacterium in cancer progression.
As with other pro-carcinogenic bacteria, strain-specific differences in F. nucleatum drive commensal versus pro-carcinogenic behavior (68). An early study demonstrated that daily gastric inoculation of F. nucleatum into CRC-susceptible mice enhanced tumorigenesis, suggesting a causative role (72). Since early associations, the pro-tumorigenic role of F. nucleatum has been supported by ample evidence (67, 72, 73, 76-80). Furthermore, we now understand F. nucleatum carcinogenic effects are primarily mediated by the adhesins Fusobacterial apoptosis protein 2 (Fap2) and Fusobacterium adhesin A (FadA) (Figure 1).
A transposon screen revealed the importance of the adhesin Fap2 in binding microbial and mammalian cells (81). Galactose-inhibited adhesion had been reported previously in studies involving various oral microbes and mammalian cells (67). Fap2 binding was blocked by galactose (81), whose partner was Gal-GalNAc, a disaccharide highly expressed on CRC tumors and metastases (82). Thus F. nucleatum can hone to developing and established tumors, contributing to cancer progression.
For tumorigenesis to progress, neoplastic cells must avoid immune detection and destruction. Natural killer (NK) cells are a key part of immune surveillance by killing non-self cells (i.e. virus-infected and tumor cells) via coordination of activating and inhibitory receptors. F. nucleatum Fap2 binds NK cell inhibitory receptor TIGIT (T cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibitory motif (ITIM) domains), which inhibits NK cell activation and allows tumor cells to evade elimination (78). Fusobacterium also induces immunosuppressive myeloid-derived suppressor cells (MDSCs), which can boost tumor development by interfering with immune surveillance (78).
Another Fusobacterium adhesin, FadA, is implicated in carcinogenesis. FadA binds E-cadherin, activates beta-catenin, and enhances experimental CRC tumor xenograft growth (79). FadA is essential for active invasion of epithelial and endothelial cells (80, 83). A recent study demonstrated that FadA+ F. nucleatum invades HCT116 CRC cells and induces production of CXCL1 and IL-8, chemokines that then promote HCT116 migration (80). These data suggest that colorectal cell invasion by F. nucleatum may enhance metastatic potential. Interestingly, invasion of phagocytic cells – cultured neutrophils and macrophages – was FadA independent (80). Fusobacterium likely harbor additional factors that facilitate invasion and pathogenic interactions with mammalian cells, as invasive strains harbor large genomes predicted to encode multiple FadA-related adhesins and similar surface-associated proteins (84). Thus, FadA promotes proliferative signaling and upon invading CRC cells, may enable cellular migration and metastasis.
Although it is unclear precisely what role the microbiota may play in metastatic growth, some evidence suggests that F. nucleatum plays a role in CRC metastases. One study identified that clonal Fusobacterium strains were found in a majority of primary CRC tumors and paired liver metastases (77). Furthermore, when Fusobacterium was found in CRC metastases, much of the primary tumor microbiome was present as well (77); this suggests that Fusobacterium may be a hub for multi-species pro-carcinogenic activities. Fusobacterium-containing patient-derived xenografts had viable Fusobacterium that appeared to be cancer cell invasive (77). In addition, tumor growth was reduced by treatment with metronidazole, an antibiotic highly effective against Fusobacterium (77). Interestingly, a recent study reported that F. nucleatum could accelerate experimental breast cancer and metastatic progression (71). Similar to findings in CRC, this pro-carcinogenic activity involved Fap2 binding Gal-GalNAc on breast cancer cells and suppression of tumor infiltrating T cells (71). Metastatic progression was inhibited by antibiotic treatment with metronidazole (71). Overall, it is clear that F. nucleatum contributes to cancer progression, in part via FadA and Fap2 adhesins. While additional investigation is needed, evidence suggests a likely role for F. nucleatum in metastasis (Figure 1).
Carcinogenesis is mediated by diverse microbial functions
Our microbiota adapt to an array of microenvironmental shifts during our lifetime, shaping human development, health, and survival. At the same time, these diverse microbial communities can impact chronic disease and diseases of aging, including cancer (24). Carcinogenesis is a multi-step process by which normal host cells acquire genetic and epigenetic changes that result in cancer (31). In combination with host genetic susceptibility and environmental exposures, a prominent pro-carcinogenic role for the microbiota has recently emerged (85).
The microbiota comprises vast communities of microbes that inhabit most body sites. Although we generally exist in a healthy symbiotic relationship with our microbiota, an altered or “dysbiotic” microbial community can contribute to the carcinogenic process. Links between carcinogenesis and ecological alterations to the microbiota are exemplified by CRC, where there is intimate association between host and a rich/diverse community of microbes. Human microbiota studies and experimental animal models of cancer have consistently highlighted several microbes that impact carcinogenesis: E. coli, ETBF, and F. nucleatum (28, 30). In this review, we have described mechanisms driving the pro-carcinogenic effects of these key bacterial species. Furthermore, we proposed that each of these microbes uniquely influence specific stages of carcinogenesis. Colibactin-producing E. coli initiate, ETBF promote, and Fap2+ and FadA+ F. nucleatum enhance progression of carcinogenesis. We use these microbes to give tangible evidence to the stepwise pro-carcinogenic potential of the microbiota.
Outstanding questions and future directions.
Recently in 2019, the International Cancer Microbiome Consortium published a consensus statement on the role of the human microbiome in carcinogenesis stating, “The microbiome is one apex of a tripartite, multidirectional interactome alongside environmental factors and an epigenetically/genetically vulnerable host that combine to cause cancer” (85). As elaborated herein, microbiota have local effects on cancer formation and contribute to systemic effects through biotransformation of chemotherapeutics and immunotherapies (see Sidebar for more information). In this review, we have discussed recent evidence that human-associated microbes can impact each stage of carcinogenesis: initiation, promotion, and progression. However, many important questions remain:
What other microbial factors induce, promote, or progress carcinogenesis?
The microbiota harbors a tremendous capacity for generating novel metabolites (32). Microbial-derived metabolites like short chain fatty acids (SCFAs) and hydrogen sulfide (H2S) can impact CRC (28, 30). The SCFA butyrate provides energy to healthy colonocytes and is less abundant in CRC patients. Administering butyrate or butyrate-producing microbes enhances mitochondrial respiration in healthy colonocytes and is tumor-suppressive in a mouse model of cancer (86, 87). Conversely, H2S is enriched in early stage tumor samples and may promote inflammation/tumorigenesis (88). Various species like B. wadsworthia and Alistipes spp. are abundant in CRC patients and produce H2S that is toxic to epithelial cells and causes DNA damage (28, 30).
Do microbial factors drive specific types of CRC?
One study evaluated 83 patients with a 44 patient validation cohort, in which patients were stratified by mismatch repair (MMR) status (88). This study found MMR status was one of the strongest predictors of microbial community variance and that MMR-deficient patients harbor different microbes and metabolites than MMR-proficient patients (88). Larger cohorts and longitudinal studies are likely to uncover stronger links between the presence of certain microbial signatures and CRC subtypes.
How do the microbiota alter host products to influence cancer development?
Many microbes and their metabolites stimulate reactive oxygen species (ROS) production from host cells, leading to ROS-induced DNA damage that can promote genomic instability and mutations (89-91). In addition, bile acids are notoriously altered by the microbiota and can impact colorectal and hepatocellular carcinoma (92). With the vast amount of metabolites evoked or chemically transformed by the microbiota, carcinogenesis is undoubtedly altered by the milieu of an individual’s microbiota.
Does the physiological state of microbial communities impact their pro-carcinogenic potential?
Biofilms have consistently been found in right-sided (proximal) CRC and can contain ETBF and pks+ E. coli (93, 94). Genetically susceptible mice develop tumors upon inoculation with human colonic biofilms, but rarely with non-biofilm communities, from both healthy individuals and cancer patients (27). However, taxonomy differed between biofilm-positive and biofilm-negative microbial communities (27), making it difficult to discern whether the pro-carcinogenic effects were biofilm-dependent or due to differences in microbial composition. Nonetheless, these findings suggest the biogeographic distribution of the microbiota and inter-microbial interactions play an understudied role in host interactions and possibly cancer.
How does the external environment shape the microbiome to a carcinogenic state?
Chronic inflammation increases cancer risk and severity. A recent manuscript demonstrates that reducing inflammation through TNF-alpha neutralization alters the microbiota and renders it less carcinogenic when transplanted to germ-free cancer-susceptible mice (95). In healthy individuals, the microbiota adapts over a lifetime. B. fragilis and other members of the microbiota continually adapt in the gut via de novo mutations, with the appearance of novel strain variants that could perhaps acquire pro-carcinogenic traits (11).
Significance and therapeutic potential.
Over the past ~250 years, life-changing discoveries have informed cancer research and improved patient prognoses (96). Today, we continue to see an overall decrease in cancer-related deaths amongst men, woman, and children in the United States (96). However, rates of new cancers remain stable or are increasing for some demographics (96). Sustained cancer incidence means the millennia-long fight against cancer persists. Two large limitations to fighting cancer are: 1) modeling the complexity of the host-microbe interactions within the tumor microenvironment and 2) personalization of medicine to optimally treat an individual’s unique disease features, including pro-carcinogenic microbes.
To address these limitations, we need to build longitudinal clinical studies to truly demonstrate microbial-causation among human carcinogenesis. Mechanistic studies and experiments with animal models build the foundation for human-based research. By understanding how and at what stage microbes impact carcinogenesis, we can lessen the gap towards improved clinical intervention. By defining which microbes initiate cancer, we identify biomarkers to predict cancer formation in susceptible individuals. Looking at microbes that promote cancer, we can estimate therapeutic efficiency. Finally, by addressing cancer progressing microbes, we can get a better understanding of prognosis. Furthermore, this information will allow us to best target these carcinogenic microbes and microbial metabolites for elimination, in combination with strategies to enhance beneficial community function. This knowledge can be broadly applied to other microbial-driven conditions of chronic inflammation, infection, and extraintestinal cancers. This should be our focus in the next decade to continue the fight against cancer.
Summary points:
1. Our microbiota influence the initiation, promotion, and progression of carcinogenesis.
2. Currently, microbiota-mediated pro-carcinogenic effects are best exemplified by the gut microbiota in colorectal cancer (CRC).
3. Colibactin-producing polyketide synthase (pks+) E. coli initiate carcinogenesis by inducing DNA damage.
4. Enterotoxigenic Bacteroides fragilis (ETBF) promote carcinogenesis, directly through B.fragilis-derived toxin (BFT) and indirectly through interleukin-17 (IL-17) dominant tumor-promoting inflammation.
5. Fusobacterium nucleatum enhances cancer progression via its adhesins Fusobacterial apoptosis protein 2 (Fap2) and Fusobacterium adhesin A (FadA), which enhance proliferation, promote cellular invasion, and help evade anti-tumor immunity.
6. Defining how and when the microbiota impact carcinogenesis will improve the timing and strategies for risk assessment and personalized cancer treatments.
Sidebar: Biotransformation of chemotherapeutics by the microbiota.
The microbiota can directly impact metabolism of xenobiotics, including chemotherapeutics (97, 98). Pancreatic ductal adenocarcinomas (PDACs) can harbor Gammaproteobacteria able to metabolize gemcitabine, a common chemotherapeutic for PDAC. Gemcitabine inactivation is dependent upon expression of a particular isoform of the bacterial enzyme cytidine deaminase (CDDL), common among Gammaproteobacteria (99). Although the host also produces cytidine deaminases, these results suggest intratumor bacteria may contribute to PDAC resistance to gemcitabine. Irinotecan, a chemotherapeutic used to treat colorectal and pancreatic cancer, has limited efficacy due to GI toxicity caused by reactivation of the drug in the colon by bacterial β-glucuronidases (100). Inhibiting bacterial β-glucuronidases prevents GI toxicity and reduces gut epithelial damage, which may allow administration of higher effective doses (100-102). In future, clinicians may consider individual variations in the microbiome to inform the most effective use of chemotherapeutics, as personalized medicine. Some approaches include building pharmacokinetic models to predict microbiome contributions to the metabolism and absorption of specific drugs and chemotherapeutics (103). Another way to capture the variability in drug metabolism across various patient microbiota is to employ in fimo screening (experimental examination of stool samples), inoculating patient-derived fecal samples with a drug of choice to determine the functional output of an individual’s microbial community (104).
Acknowledgements
We apologize to the authors whose primary literature we could not cite due to space restrictions. This work was supported by funding from NIH R01DK124617 (JCA), pilot funding from NIH P30DK034987 (subcontract JCA), UNC Lineberger Comprehensive Cancer Center Innovation Award (JCA), and SPIRE (NIH IRACDA program) NIH/NIGMS 5K12 GM000678-21 (RMB).
References
- 1.Eckburg PB, Bik EM, Bernstein CN, et al. 2005. Diversity of the human intestinal microbial flora. Science. 308(5728):1635–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, et al. 2005. Host-bacterial mutualism in the human intestine. Science. 307(5717):1915–20 [DOI] [PubMed] [Google Scholar]
- 3.The Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486(7402):207–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lloyd-Price J, Mahurkar A, Rahnavard G, et al. 2017. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 550(7674):61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello EK, Lauber CL, Hamady M, et al. 2009. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 326(5960):1694–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy R, Magis AT, Earls JC, et al. 2020. Longitudinal analysis reveals transition barriers between dominant ecological states in the gut microbiome. Proceedings of the National Academy of Sciences. 117(24):13839–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamady M, Knight R. 2009. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 19(7):1141–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71(12):8228–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporaso JG, Kuczynski J, Stombaugh J, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7(5):335–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone CA, Stombaugh JI, Gordon JI, et al. 2012. Diversity, stability and resilience of the human gut microbiota. Nature. 489(7415):220–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S, Lieberman TD, Poyet M, et al. 2019. Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host and Microbe. 25(5):656–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasapolli R, Schütte K, Schulz C, et al. 2019. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology. 157(4):1081–83 [DOI] [PubMed] [Google Scholar]
- 13.Gilbert JA, Blaser MJ, Caporaso JG, et al. 2018. Current understanding of the human microbiome. Nature Medicine. 24(4):392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dzutsev A, Badger JH, Perez-Chanona E, et al. 2017. Microbes and Cancer. Annu. Rev. Immunol 35(1):199–228 [DOI] [PubMed] [Google Scholar]
- 15.Fulbright LE, Ellermann M, Arthur JC. 2017. The microbiome and the hallmarks of cancer. PLoS Pathog. 13(9):e1006480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong SH, Yu J. 2019. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nature Reviews Gastroenterology & Hepatology, 16(11):690–704. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. 144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 18.Marshall BJ. 1995. The 1995 Albert Lasker Medical Research Award. Helicobacter pylori. The etiologic agent for peptic ulcer. JAMA. 274(13):1064–66 [DOI] [PubMed] [Google Scholar]
- 19.Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1(8390):1311–15 [DOI] [PubMed] [Google Scholar]
- 20.Amieva M, Peek RM Jr. 2016. Pathobiology of Helicobacter pylori–Induced Gastric Cancer. Gastroenterology. 150(1):64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt AP, Redinbo MR, Bultman SJ. 2017. The role of the microbiome in cancer development and therapy. CA: A Cancer Journal for Clinicians. 67(4):326–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Keefe SJD. 2016. Diet, microorganisms and their metabolites, and colon cancer. Nature Reviews Gastroenterology & Hepatology. 13(12):691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsilimigras MCB, Fodor A, Jobin C. 2017. Carcinogenesis and therapeutics: the microbiota perspective. Nature Microbiology. 2:17008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tibbs TN, Lopez LR, Arthur JC. 2019. The influence of the microbiota on immune development, chronic inflammation, and cancer in the context of aging. Microb Cell. 6(8):324–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong SH, Zhao L, Zhang X, et al. 2017. Gavage of Fecal Samples From Patients with Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-free and Conventional Mice. Gastroenterology, 153(6):1621–1633.e6. [DOI] [PubMed] [Google Scholar]
- 26.Zackular JP, Baxter NT, Iverson KD, et al. 2013. The gut microbiome modulates colon tumorigenesis. MBio. 4(6):doi:10.1128-mBio.00692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomkovich S, Dejea CM, Winglee K, et al. 2019. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Invest 129(4):1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwabe RF, Jobin C. 2013. The microbiome and cancer. Nat. Rev. Cancer 13(11):800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. 2012. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 100(Pt B):1–441 [PMC free article] [PubMed] [Google Scholar]
- 30.Dzutsev A, Badger JH, Perez-Chanona E, et al. 2017. Microbes and Cancer. Annu. Rev. Immunol 35(1):199–228 [DOI] [PubMed] [Google Scholar]
- 31.Loeb LA, Harris CC. 2008. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 68(17):6863–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donia MS, Fischbach MA. 2015. Small molecules from the human microbiota. Science. 349(6246):1254766–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nougayrede JP, Homburg S, Taieb F, et al. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 313(5788):848–51 [DOI] [PubMed] [Google Scholar]
- 34.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 338(6103):120–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buc E, Dubois D, Sauvanet P, et al. 2013. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE. 8(2):e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arthur JC, Gharaibeh RZ, Mühlbauer M, et al. 2014. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 5:4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomkovich S, Yang Y, Winglee K, et al. 2017. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 77(10):2620–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cougnoux A, Dalmasso G, Martinez R, et al. 2014. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 63(12):1932–42 [DOI] [PubMed] [Google Scholar]
- 39.Pleguezuelos-Manzano C, Puschhof J, Huber AR, et al. 2020. Mutational signature in colorectal cancer caused by genotoxic pks + E. coli. Nature, 580(7802):269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dziubańska-Kusibab PJ, Berger H, Battistini F, et al. 2020. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nature Medicine. 313(6457):848–7 [DOI] [PubMed] [Google Scholar]
- 41.Homburg S, Oswald E, Hacker J, et al. 2007. Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli. FEMS Microbiol. Lett 275(2):255–62 [DOI] [PubMed] [Google Scholar]
- 42.Faïs T, Delmas J, Barnich N, et al. 2018. Colibactin: More Than a New Bacterial Toxin. Toxins. 10(4):151–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuevas-Ramos G, Petit CR, Marcq I, et al. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 107(25):11537–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balskus EP. 2015. Colibactin: understanding an elusive gut bacterial genotoxin. Natural Product Reports. 32(11):1534–40 [DOI] [PubMed] [Google Scholar]
- 45.Bleich RM, Arthur JC. 2019. Revealing a microbial carcinogen. Science. 363(6428):689–90 [DOI] [PubMed] [Google Scholar]
- 46.Wernke KM, Xue M, Tirla A, et al. 2020. Structure and bioactivity of colibactin. Bioorg. Med. Chem. Lett 30(15):127280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cougnoux A, Gibold L, Robin F, et al. 2012. Analysis of structure-function relationships in the colibactin-maturating enzyme ClbP. J. Mol. Biol 424(3-4):203–14 [DOI] [PubMed] [Google Scholar]
- 48.Brotherton CA, Balskus EP. 2013. A Prodrug Resistance Mechanism Is Involved in Colibactin Biosynthesis and Cytotoxicity. J. Am. Chem. Soc 135(9):3359–62 [DOI] [PubMed] [Google Scholar]
- 49.Wilson MR, Jiang Y, Villalta PW, et al. 2019. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 363(6428):eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue M, Kim Cs, Healy AR, et al. 2019. Structure elucidation of colibactin and its DNA cross-links. Science. 365(6457):eaax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vizcaino MI, Crawford JM. 2015. The colibactin warhead crosslinks DNA. Nature Chem. 7(5):411–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kinzler KW, Vogelstein B. 1996. Lessons from Hereditary Colorectal Cancer. Cell. 87(2):159–70 [DOI] [PubMed] [Google Scholar]
- 53.Rowan AJ, Lamlum H, Ilyas M, et al. 2000. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the ‘two hits’. Proceedings of the National Academy of Sciences. 97(7):3352–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee-Six H, Olafsson S, Ellis P, et al. 2019. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 574(7779):532–37 [DOI] [PubMed] [Google Scholar]
- 55.Grivennikov SI, Wang K, Mucida D, et al. 2012. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 491(7423):254–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greten FR, Grivennikov SI. 2019. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 51(1):27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valguarnera E, Wardenburg JB. 2020. Good Gone Bad: One Toxin Away From Disease for Bacteroides fragilis. J. Mol. Biol 432(4):765–85 [DOI] [PubMed] [Google Scholar]
- 58.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 453(7195):620–25 [DOI] [PubMed] [Google Scholar]
- 59.Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences. 107(27):12204–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sears CL, Geis AL, Housseau F. 2014. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J. Clin. Invest 124(10):4166–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu S, Rhee KJ, Albesiano E, et al. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 15(9):1016–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu S, Lim KC, Huang J, et al. 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proceedings of the National Academy of Sciences. 95(25):14979–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clevers H 2006. Wnt/β-Catenin Signaling in Development and Disease. Cell. 127(3):469–80 [DOI] [PubMed] [Google Scholar]
- 64.Hurtado CG, Wan F, Housseau F, et al. 2018. Roles for Interleukin 17 and Adaptive Immunity in Pathogenesis of Colorectal Cancer. Gastroenterology. 155(6):1706–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K, Kim MK, Di Caro G, et al. 2014. Interleukin-17 Receptor A Signaling in Transformed Enterocytes Promotes Early Colorectal Tumorigenesis. Immunity. 41(6):1052–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atarashi K, Tanoue T, Ando M, et al. 2015. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 163(2):367–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han YW. 2015. Fusobacterium nucleatum: a commensal-turned pathogen. Current Opinion in Microbiology. 23:141–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strauss J, Kaplan GG, Beck PL, et al. 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis 17(9):1971–78 [DOI] [PubMed] [Google Scholar]
- 69.Allen-Vercoe E, Jobin C. 2014. Fusobacterium and Enterobacteriaceae: Important players for CRC? Immunology Letters. 162(2):54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cochrane K, Robinson AV, Holt RA, et al. 2020. A survey of Fusobacterium nucleatum genes modulated by host cell infection. Microbial Genomics. 6(2):1258–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parhi L, Alon-Maimon T, Sol A, et al. 2020. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun, 11(1):3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kostic AD, Chun E, Robertson L, et al. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 14(2):207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castellarin M, Warren RL, Freeman JD, et al. 2011. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 22(2):299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCoy AN, Araujo-Perez F, Azcárate-Peril A, et al. 2013. Fusobacterium Is Associated with Colorectal Adenomas. PLoS ONE. 8(1):e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mima K, Nishihara R, Qian ZR, et al. 2016. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 65(12):1973–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kostic AD, Gevers D, Pedamallu CS, et al. 2011. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 22(2):292–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bullman S, Pedamallu CS, Sicinska E, et al. 2017. Analysis of Fusobacteriumpersistence and antibiotic response in colorectal cancer. Science. 358(6369):1443–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gur C, Ibrahim Y, Isaacson B, et al. 2015. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity. 42(2):344–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubinstein MR, Wang X, Liu W, et al. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 14(2):195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Casasanta MA, Yoo CC, Udayasuryan B, et al. 2020. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. 13(641):eaba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coppenhagen-Glazer S, Sol A, Abed J, et al. 2015. Fap2 of Fusobacterium nucleatum Is a Galactose-Inhibitable Adhesin Involved in Coaggregation, Cell Adhesion, and Preterm Birth. Infection and Immunity. 83(3):1104–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abed J, Emgård JEM, Zamir G, et al. 2016. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host and Microbe. 20(2):215–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu M, Yamada M, Li M, et al. 2007. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem 282(34):25000–25009 [DOI] [PubMed] [Google Scholar]
- 84.Manson McGuire A, Cochrane K, Griggs AD, et al. 2014. Evolution of invasion in a diverse set of Fusobacterium species. MBio. 5(6):e01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scott AJ, Alexander JL, Merrifield CA, et al. 2019. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 68(9):1624–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Donohoe DR, Garge N, Zhang X, et al. 2011. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13(5):517–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Donohoe DR, Holley D, Collins LB, et al. 2014. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 4(12):1387–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hale VL, Jeraldo P, Chen J, et al. 2018. Distinct Microbes, Metabolites, and Ecologies Define the Microbiome in Deficient and Proficient Mismatch Repair Colorectal Cancers, 10(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia J, Chiu L-Y, Nehring RB, et al. 2019. Bacteria-to-Human Protein Networks Reveal Origins of Endogenous DNA Damage. Cell. 176(1–2):127–143.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X, Huycke MM. 2007. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 132(2):551–61 [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Allen TD, May RJ, et al. 2008. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 68(23):9909–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia W, Xie G, Jia W. 2017. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nature Reviews Gastroenterology & Hepatology. 15(2):111–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dejea CM, Fathi P, Craig JM, et al. 2018. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 359(6375):592–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dejea CM, Wick EC, Hechenbleikner EM, et al. 2014. Microbiota organization is a distinct feature of proximal colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 111(51):18321–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Y, Gharaibeh RZ, Newsome RC, et al. 2020. Amending microbiota by targeting intestinal inflammation with TNF blockade attenuates development of colorectal cancer. Nature Cancer, pp. 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.NIH National Cancer Institute (www.cancer.gov): https://www.cancer.gov/research/progress/250-years-milestones, https://www.cancer.gov/research/progress/annual-report-nation.
- 97.Lam KN, Alexander M, Turnbaugh PJ. 2019. Precision Medicine Goes Microscopic: Engineering the Microbiome to Improve Drug Outcomes. Cell Host and Microbe. 26(1):22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 152(1–2):39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Geller LT, Barzily-Rokni M, Danino T, et al. 2017. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 357(6356):1156–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wallace BD, Wang H, Lane KT, et al. 2010. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 330(6005):831–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhatt AP, Pellock SJ, Biernat KA, et al. 2020. Targeted inhibition of gut bacterial β-glucuronidase activity enhances anticancer drug efficacy. Proceedings of the National Academy of Sciences of the United States of America. 117(13):7374–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bhatt AP, Redinbo MR, Bultman SJ. 2017. The role of the microbiome in cancer development and therapy. CA: A Cancer Journal for Clinicians. 67(4):326–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, et al. 2019. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 363(6427):eaat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhatt AP, Grillo L, Redinbo MR. 2019. In Fimo: A Term Proposed for Excrement Examined Experimentally. Gastroenterology. 156(5):1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.He Z, Gharaibeh RZ, Newsome RC, et al. 2019. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 68(2):289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosadi F, Fiorentini C, Fabbri A. 2016. Bacterial protein toxins in human cancers. Pathogens and Disease. 74(1):ftv105. [DOI] [PubMed] [Google Scholar]
- 107.Guidi R, Guerra L, Levi L, et al. 2013. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell. Microbiol 15(1):98–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou D, Wang J-D, Weng M-Z, et al. 2013. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: a meta-analysis. Eur J Gastroenterol Hepatol. 25(4):447–54 [DOI] [PubMed] [Google Scholar]
- 109.Rao VP, Poutahidis T, Ge Z, et al. 2006. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 66(15):7395–7400 [DOI] [PubMed] [Google Scholar]
- 110.Nagamine CM, Sohn JJ, Rickman BH, et al. 2008. Helicobacter hepaticus infection promotes colon tumorigenesis in the BALB/c-Rag2(−/−) Apc(Min/+) mouse. Infect. Immun 76(6):2758–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chumduri C, Gurumurthy RK, Zadora PK, et al. 2013. Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host and Microbe. 13(6):746–58 [DOI] [PubMed] [Google Scholar]
- 112.Siegl C, Prusty BK, Karunakaran K, et al. 2014. Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. CellReports. 9(3):918–29 [DOI] [PubMed] [Google Scholar]
- 113.González E, Rother M, Kerr MC, et al. 2014. Chlamydia infection depends on a functional MDM2-p53 axis. Nat Commun. 5(1):5201–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caini S, Gandini S, Dudas M, et al. 2014. Sexually transmitted infections and prostate cancer risk: a systematic review and meta-analysis. Cancer Epidemiol. 38(4):329–38 [DOI] [PubMed] [Google Scholar]
- 115.Weyler L, Engelbrecht M, Mata Forsberg M, et al. 2014. Restriction endonucleases from invasive Neisseria gonorrhoeae cause double-strand breaks and distort mitosis in epithelial cells during infection. PLoS ONE. 9(12):e114208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vielfort K, Söderholm N, Weyler L, et al. 2013. Neisseria gonorrhoeae infection causes DNA damage and affects the expression of p21, p27 and p53 in non-tumor epithelial cells. J. Cell. Sci 126(Pt 1):339–47 [DOI] [PubMed] [Google Scholar]
- 117.Mughini-Gras L, Schaapveld M, Kramers J, et al. 2018. Increased colon cancer risk after severe Salmonella infection. PLoS ONE. 13(1):e0189721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Di Domenico EG, Cavallo I, Pontone M, et al. 2017. Biofilm Producing Salmonella Typhi: Chronic Colonization and Development of Gallbladder Cancer. Int J Mol Sci. 18(9):1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martin OCB, Bergonzini A, D'Amico F, et al. 2019. Infection with genotoxin-producing Salmonella enterica synergises with loss of the tumour suppressor APC in promoting genomic instability via the PI3K pathway in colonic epithelial cells. Cell. Microbiol 21(12):e13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Won J, Cho Y, Lee D, et al. 2019. Clonorchis sinensis excretory-secretory products increase malignant characteristics of cholangiocarcinoma cells in three-dimensional co-culture with biliary ductal plates. PLoS Pathog. 15(5):e1007818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim T-S, Pak JH, Kim J-B, et al. 2016. Clonorchis sinensis, an oriental liver fluke, as a human biological agent of cholangiocarcinoma: a brief review. BMB Rep. 49(11):590–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arora N, Kaur R, Anjum F, et al. 2019. Neglected Agent Eminent Disease: Linking Human Helminthic Infection, Inflammation, and Malignancy. Front Cell Infect Microbiol. 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tarocchi M, Polvani S, Marroncini G, et al. 2014. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J. Gastroenterol 20(33):11630–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vescovo T, Refolo G, Vitagliano G, et al. 2016. Molecular mechanisms of hepatitis C virus-induced hepatocellular carcinoma. Clin. Microbiol. Infect 22(10):853–61 [DOI] [PubMed] [Google Scholar]
- 125.Wang B, Li X, Liu L, et al. 2020. Β-Catenin: oncogenic role and therapeutic target in cervical cancer. Biol. Res 53(1):33–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Al-Thawadi H, Gupta I, Jabeen A, et al. 2020. Co-presence of human papillomaviruses and Epstein-Barr virus is linked with advanced tumor stage: a tissue microarray study in head and neck cancer patients. Cancer Cell Int. 20:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yim E-K, Park J-S. 2005. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 37(6):319–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mohanty S, Harhaj EW. 2020. Mechanisms of Oncogenesis by HTLV-1 Tax. Pathogens. 9(7):543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Boxus M, Willems L. 2009. Mechanisms of HTLV-1 persistence and transformation. Br. J. Cancer 101(9):1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sripa B, Brindley PJ, Mulvenna J, et al. 2012. The tumorigenic liver fluke Opisthorchis viverrini--multiple pathways to cancer. Trends Parasitol. 28(10):395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gao S, Li S, Ma Z, et al. 2016. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect. Agents Cancer 11(1):3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Malinowski B, Wŝierska A, Zalewska K, et al. 2019. The role of Tannerella forsythia and Porphyromonas gingivalis in pathogenesis of esophageal cancer. Infect. Agents Cancer 14(1):3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nakanishi Y, Wakisaka N, Kondo S, et al. 2017. Progression of understanding for the role of Epstein-Barr virus and management of nasopharyngeal carcinoma. Cancer Metastasis Rev. 36(3):435–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shannon-Lowe C, Rickinson A. 2019. The Global Landscape of EBV-Associated Tumors. Front Oncol. 9:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abere B, Schulz TF. 2016. KSHV non-structural membrane proteins involved in the activation of intracellular signaling pathways and the pathogenesis of Kaposi's sarcoma. Curr Opin Virol. 20:11–19 [DOI] [PubMed] [Google Scholar]
- 136.Broussard G, Damania B. 2019. KSHV: Immune Modulation and Immunotherapy. Front. Immunol 10:3084. [DOI] [PMC free article] [PubMed] [Google Scholar]