Abstract
The structural phosphoprotein M2-1 of human respiratory syncytial virus (HRSV) Long strain shows RNA binding capacity in three different assays that detect RNA-protein complexes: cross-linking, gel retardation, and Northern-Western assays. It is able to bind HRSV leader RNA specifically with cooperative kinetics, with an apparent Kd of at least 90 nM. It also binds to long RNAs with no sequence specificity. The RNA binding domain has been located between amino acid residues 59 and 85, at the NH2 terminus of the protein. This region contains the phosphorylatable amino acid residues threonine 56 and serine 58, whose modification decreases the binding capacity of M2-1 protein to long RNAs.
Human respiratory syncytial virus (HRSV), a member of the Pneumovirus genus of the Paramyxoviridae family, is the most common infectious agent in bronchiolitis and pneumonia requiring hospitalization of infants and young children. Immunocompromised and elderly people are also severely affected. No vaccine or specific antiviral treatment is available at present (11). The best way to control these infections, according to the characteristics of the human populations mainly affected, is the use of different agents as live attenuated vaccines (38), humanized neutralizing monoclonal antibodies (21), and antiviral compounds (27).
The HRSV nucleocapsids are, as in all paramyxoviruses, structural components and functional units for replication and transcription processes (11). The nucleocapsid proteins are thus multifunctional, serving as ideal targets for the action of antiviral compounds and for conveying attenuation mutations. The design of both types of reagents would be aided by understanding nucleocapsid protein functions.
HRSV nucleocapsids are composed of the negative-sense RNA genome (15,222 nucleotides long), tightly bound to the N protein, the large polymerase protein (L), the phosphoprotein (P), and probably the M2-1 protein (11, 16). The RNA polymerizing activity of the viral nucleocapsids is due to L protein, for which activity P protein is essential (41). N protein plays a histone-like function, since synthesis of viral RNA always occurs on ribonucleoprotein templates. P protein also acts as a chaperone of N protein, maintaining it competent for encapsidation (30). M2-1 protein enhances the processivity of the viral polymerase (9, 10) and its readthrough of intergenic junctions (17). These properties identified M2-1 protein as an elongation and antiterminator transcription factor, a constituent of the nucleocapsids. The protein is encoded by the M2 gene, present only in the pneumoviruses, which codes for an mRNA containing two open reading frames (ORFs). ORF1 encodes the 22K structural protein, also known as M2-ORF1 or M2-1 protein (194 amino acids). ORF2 encodes M2-2 protein (90 amino acids), detected in HRSV-infected cells, which has a regulatory function in the balance between viral transcription and replication (1, 6, 8).
The M2-1 protein was long ago described as a structural membrane phosphoprotein that could be present as different species in HRSV-infected cells (20, 23). These species could differ in the number of intramolecular disulfide bonds formed among its four cysteine residues (23). Three of the four cysteine residues are at the amino-terminal end, forming a C3H1 motif that is highly conserved among members of the genus. In other proteins, this motif has been shown to bind Zn ions (40). Mutation at the predicted Zn coordinating residues of the C3H1 motif (C7, C15, and H25) abolished M2-1 enhancement of transcriptional readthrough (18). Interaction of M2-1 with the N protein has been also described in coexpression experiments (16).
Here we present evidence that M2-1 is an RNA binding protein, able to bind the RNA leader specifically, whose binding capacity is modulated by phosphorylation. The amino acid residues in contact with the RNA and those modified by phosphorylation have been located.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells, COS1 cells (CV1 cells containing the simian virus 40 T antigen), HRSV Long strain, and the vaccinia virus recombinant vTF-3 were used throughout this study in experimental conditions previously described (35, 37).
Transfection.
cDNAs of different viral proteins, all cloned in pGEM3, were transiently expressed in the vaccinia virus-based expression system under the control of the T7 RNA polymerase promoter (vTF-3). The corresponding soluble proteins were prepared as described elsewhere (37). Briefly, cell cultures were recovered by low-speed centrifugation, twice washed with phosphate-buffered, and then homogenized in 10 mM Tris-HCl (pH 7.5)–140 mM NaCl–5 mM EDTA–1% Triton X-100 and–1% deoxycholate. The supernatant fraction obtained after centrifugation in a minicentrifuge (15 min, 4°C) was considered the soluble protein fraction. For some variants, insoluble proteins were obtained after removal of soluble proteins; they were further solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Plasmids.
The cDNAs corresponding to N, P, and M2 genes of HRSV Long strain, cloned in the corresponding pBSV9 recombinant plasmids (24), were subcloned as HpaI-StuI fragments in the SmaI site of the plasmid vector pGEM3.
Plasmid pL was obtained by cloning, in the EcoRI site of vector pUC19, a cDNA fragment obtained by reverse transcriptase-PCR containing the promoter sequences of the T7 RNA polymerase and the 674 nucleotides of the HRSV Long strain genomic 3′ end. This fragment contains the leader (44 nucleotides plus three extra G's, which increase transcription by the T7 RNA polymerase [9]), together with the NS1 gene and the first 80 nucleotides of the NS2 gene.
The pGEM3 recombinant plasmids containing the cDNAs corresponding to the M2-1 variants were obtained by directed mutagenesis (19) or after cutting the M2 cDNA with restriction enzymes PstI, DpnI, EcoRV, and EcoRV plus NdeI, creating the variants PstI, DpnI, EcoRV, and EcoRV-NdeI, respectively. Variant EcoRVts is similar to EcoRV, but with T56 and S58 replaced by alanine. In the cases of variants PstI and EcoRV-NdeI, after cutting with the corresponding restriction enzyme, the DNA ends generated were blunted by the action of the 3′ exonuclease or polymerase activities of T4 DNA polymerase. In this way, the structure of the mRNA was preserved to facilitate stability and translation.
M2-1 protein purification.
HEp-2 cells were infected with HRSV, and the microfilament-containing protein fraction and extracellular viral particles were obtained as previously described (32). The proteins contained in the different fractions were separated and characterized in SDS-gels (12% acrylamide) containing 0.4% Coomassie blue in the electrophoresis buffer or after negative staining (14). The piece of gel containing the M2-1 protein was excised, and the protein was electroeluted. The protein was then precipitated with acetone and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]) containing 6 M guanidine chloride. The protein was renatured by chromatography through an acrylamide column using TE as the running buffer. The fractions containing the M2-1 protein were detected after SDS-PAGE, pooled, and densitometrically quantified by comparison to known amounts of bovine serum albumin (BSA), separated in the same gel.
Preparation of labeled riboprobes.
The 32P-labeled riboprobes used were prepared by in vitro transcription of the corresponding pGEM3 recombinant plasmids, using T7 or SP6 RNA polymerase and adding [α-32P]CTP to the reaction (25). In this way, 32P-labeled plus- or minus-polarity RNAs were obtained, corresponding to genes P (1,000 nucleotides) and N (1,200 nucleotides). A plus-polarity RNA (700 nucleotides) containing the leader sequence plus three extra G's at the 5′ end was obtained using plasmid pL digested with NcoI in the in vitro transcription reaction.
The short riboprobe (80 nucleotides) containing the leader sequence was obtained using plasmid pL digested with StyI. This short probe contains three extra G's, the 44 nucleotides of the leader, and 33 nucleotides of the NS1 gene at its 5′ end. The nonspecific RNA control was obtained using pGEM4 digested with EcoRI (58 nucleotides).
UV cross-linking assay.
M2-1 protein (10 to 180 ng) was mixed with 1 to 5 ng of the indicated riboprobe (specific activity, 30,000 Cerenkov cpm/ng) and incubated in binding buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 100 μM ZnCl2, 4 mM dithiothreitol). The RNA-protein complex was UV cross-linked as previously described (3). After treatment with bovine pancreas RNase A (60 μg/ml), SDS-PAGE (12% acrylamide gel) was carried out. The gels were stained with Coomassie blue and autoradiographed.
Gel retardation assay.
The protein and riboprobes were incubated as indicated above and then electrophoresed in a gel containing 3.5% acrylamide and 0.5× Tris-borate-EDTA buffer at 100 V. The gels were fixed with 95% ethanol, dried, and autoradiographed. The amount of radioactivity present in the riboprobe position was quantified by a PhosphorImager. This procedure was used for kinetic studies to measure the formation of the protein-RNA complex. The protein concentration at which 50% of the labeled RNA formed an RNA-protein complex was defined as the apparent Kd (dissociation constant).
Northern-Western assay.
Protein (30 to 40 μg) obtained after transfection experiments was separated by SDS-PAGE and electrotransferred to nitrocellulose paper. The paper was washed with 100 mM NaCl–40 mM Tris-HCl (pH 8.0), and the proteins were renatured by incubation in the same buffer containing 0.02% PVP-40 (polyvinylpyrrolidone), 0.02% BSA, 0.02% Ficoll 400, 0.1% Triton X-100, and 50 μM ZnSO4 for 3 h at 20°C. The 32P-labeled riboprobe was added in the same buffer without ZnSO4 and containing yeast RNA (1 μg/ml). After 2 h of incubation, the probe was removed and the paper was washed several times with incubation buffer. The dried paper was exposed in a PhosphorImager.
Chemical and enzymatic treatments of the M2-1 protein.
The cross-linking reaction described above was carried out with 80 ng of purified M2-1 protein and 5 ng of 32P-labeled riboprobe. After RNase digestion and precipitation with acetone, the labeled M2-1 protein with associated radioactivity was mixed with 100 to 300 ng of electroeluted M2-1 protein. The mixture was then treated with 14 or 28 mg of cyanogen bromide (CNBr) per ml in 50% formic acid at 37°C overnight or for 4 h (15). After incubation, the sample was vacuum dried, neutralized by several cycles of resuspension in 0.1 M NH4HCO3 (pH 8.3), and again vacuum dried. When the sample was at neutral pH, it was resuspended in 50 μl of electrophoresis sample buffer. In other cases, the protein was treated with N-bromosuccinimide or with V8 protease as previously described (15, 39).
The peptides generated after treatment were separated by SDS-PAGE as described elsewhere (31) and transferred to an Inmobilon membrane. After peptide staining with amido black, labeled peptides were detected by autoradiography using a PhosphorImager, and their amino-terminal ends were determined by partial sequencing (34).
RESULTS
M2-1 is an RNA binding protein.
To determine whether HRSV nucleocapsid proteins are able to interact with viral RNA, protein fractions from HRSV-infected HEp-2 cells were chromatographed through Sepharose 4B containing covalently bound monoclonal antibodies specific for the different viral proteins. The viral proteins purified in this way were unable to bind 32P-labeled RNAs by cross-linking and by retardation in gel electrophoresis assays. These results could be due to the denaturation of the purified proteins, to the fact that RNA is already bound to them, or to both possibilities. To avoid this difficulty, we first purified the nucleocapsid proteins by SDS-PAGE and then renatured them. As a nucleocapsid source, a microfilament fraction and extracellular viral particles from infected HEp-2 cells were used, because nucleocapsid proteins are in a higher proportion in these fractions than in other subcellular fractions. The protein composition of the protein fractions obtained from HRSV-infected HEp-2 cells and the purity of the viral nucleocapsid proteins after purification and renaturation are shown in Fig. 1A and B.
FIG. 1.
Purified and renatured M2-1 protein binds to RNA, as shown by cross-linking with UV and in electrophoresis gel retardation experiments. (A) SDS-polyacrylamide gel stained by Coomassie blue showing the protein composition of 20 μg of protein contained in extracellular viral particles (V) and soluble (S), microfilament-bound (M), and insoluble (I) protein fractions from HRSV-infected HEp-2 cells. (B) Purity of viral proteins after renaturation and quantification. The BSA slots correspond to 92, 231, 462, 693, and 926 ng of pure BSA. The following slots correspond to different volumes (2 and 4 μl) of purified and renatured N, P, M, and M2-1 proteins. The calculated protein concentrations varied between 0.1 and 0.2 mg/ml. The electrophoretic mobility of HRSV proteins and of molecular weight markers (MW; expressed in kilodaltons) are indicated at the left and right, respectively. (C and D) The indicated amounts (nanograms) of purified M2-1 protein were incubated with 2 ng of RNA (1,000 nucleotides long) labeled with 32P (35,000 Cerenkov cpm/ng) under the indicated conditions. Samples were then UV cross-linked and analyzed by SDS-PAGE following RNase digestion (C) or run in a 5% acrylamide gel (D). Asterisks indicate the electrophoretic mobility of M2-1 and its dimers.
To determine the capacity of M2-1 protein to bind RNA, the purified protein was incubated with 32P-labeled riboprobes and binding was assayed by UV cross-linking (Fig. 1C) and by retardation of the electrophoretic mobility of 32P-labeled riboprobes in gel electrophoresis (Fig. 1D). In both assays, M2-1 protein binding to RNA was detected, but the cross-linking assay did not always yield a linear response with increased amounts of protein. For this reason, kinetic studies of the binding reaction were carried out using gel electrophoresis retardation assays.
The appearance in the cross-linking assays of a labeled protein with an electrophoretic mobility corresponding approximately to a 44-kDa protein, the predicted position for M2-1 dimers, is noteworthy. In addition, N and M but not P proteins were able to bind RNA in assays similar to those described above. The RNA binding capacity of N protein was modulated by P protein, as determined when both proteins were renatured together. This supports the suggested chaperone function of N as described for P protein (30) (data not shown).
M2-1 protein specifically binds to short RNAs containing the 5′-end leader sequence.
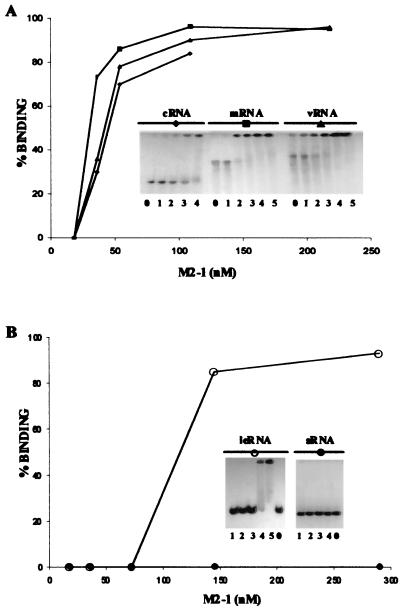
Different amounts of the renatured M2-1 protein were assayed by mobility retardation in gel electrophoresis, using 1 ng of 32P-labeled riboprobes corresponding to plus (mRNA) and to minus (viral RNA) polarities of HRSV P and N genes (1,000 and 1,300 nucleotides, respectively) and to plus-polarity RNA containing the HRSV leader plus three extra G's at the 5′ end (700 nucleotides) and therefore with the structure at the 5′ end of the viral cRNA. Similar kinetics were obtained in all cases tested (Fig. 2A). The protein is also able to bind to other long RNAs unrelated to HRSV (data not shown); M2-1 protein therefore binds without sequence specificity to long RNAs. The apparent Kd, calculated from quantification of the results, is 30 nM for these long RNAs. The retardation of the probes in all cases is due to binding of the protein to a long RNA since binding can be competed for by different unlabeled long RNAs but not by short yeast RNA, which has an average size of 100 nucleotides (data not shown).
FIG. 2.
M2-1 protein binds specifically to HRSV leader RNA. Different quantities of purified M2-1 protein were incubated with 1 ng of 32P-labeled (30,000 Cerenkov cpm/ng) long riboprobes (vRNA [viral RNA] and mRNA) corresponding to the RNAs for the P gene in minus and plus polarities, respectively (cRNA corresponds to 700-nucleotide positive-polarity long RNA containing the leader sequence at its 5′ end) (A) or with 0.1 ng of 32P-labeled (100,000 Cerenkov cpm) short RNAs (leRNA, 90 nucleotides long, containing the leader sequence at its 5′ end; sRNA, a pGEM4-derived RNA, 50 nucleotides long) (B) with increasing amounts of M2-1 protein. Lanes labeled 0, 1, 2, 3, 4, and 5 correspond to the absence of protein or to the addition of M2-1 protein at final concentrations of 18, 36, 54, 109, and 218 (A) and 18, 36, 72, 145, and 290 (B) nM. The amount of radioactivity was quantified by PhosphorImager. The results represent the mean of several experiments with each riboprobe.
To determine the specificity of M2-1 binding to any RNA sequence, shorter RNAs were assayed. A specific 80-nucleotide cRNA, containing three extra G's and the leader sequences (44 nucleotides) at its 5′ end, was also tested. In this case, M2-1 specifically binds to 0.1 ng of the cRNA leader in the presence of 25 ng of yeast RNA (Fig. 2B). The apparent Kd calculated for this RNA binding was 90 nM. There is no binding to the 58-nucleotide nonspecific probe or to 20- to 100-nucleotide-long yeast RNA, as indicated above. This result indicates that M2-1 protein binds specifically to the leader RNA contained in short (80-nucleotide) but not long (700-nucleotide) RNAs.
Mapping of the M2-1 RNA binding domain.
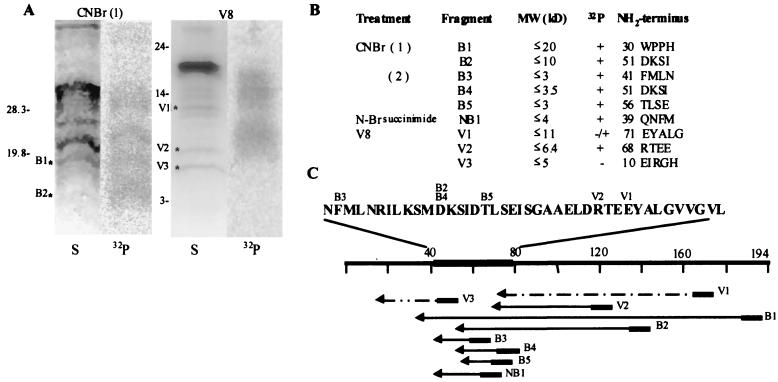
After preparative cross-linking and RNase digestion, the labeled protein was treated with CNBr, N-bromosuccinimide, or V8 protease. The peptides generated after the different treatments were separated by electrophoresis and visualized by staining with amido black; those labeled with 32P were detected by autoradiography. Figure 3A shows the result obtained after treatment with CNBr and V8 protease digestion. The amino-terminal sequence of the peptides generated was determined. The data from several experiments (summarized in Fig. 3B) indicate the calculated sizes for the fragments and the amino acid residues located at their NH2-terminal ends, although the calculated fragment sizes may be overestimated due to their smallness and to the possible presence of oligoribonucleotides bound to them. The approximate locations of these fragments on the M2-1 molecule are shown, although their C-terminal ends are unknown (Fig. 3C). These results indicate that at least some of the residues in contact with the RNA are between positions 55 and 75 but do not rule out the existence of others around this region. The RNA binding region was thus considered to be located between amino acid residues 40 and 80. It also appears that the zinc finger domain does not contain the residues in direct contact with the RNA since the V3 fragment, which starts at the residue at position 10, contains no associated radioactivity.
FIG. 3.
Location of M2-1 protein amino acids residues in contact with RNA. (A) Preparative RNA binding assays were carried out with 150 ng of M2-1 protein and 8 ng of 32P-labeled RNA (1,000 nucleotides long). After UV cross-linking, RNase A digestion, and acetone precipitation, the covalently bound RNA–M2-1 protein was resuspended in TE buffer containing 300 ng of purified M2-1 protein. The mixture was treated with 28 mg of CNBr per ml in 50% formic acid [CNBr(1)] or with 1 μg of protease V8 (V8) in 50 mM NH4HCO3 for 4 or 2 h, respectively, at 37°C. The peptides generated were separated in SDS-polyacrylamide gels containing 6% urea and Tricine in the electrophoresis buffer (31). After electrophoresis, peptides were transferred to Inmobilon membranes and detected by amido black staining (S) or by PhosphorImager (32P). (B) NH2-terminal sequences for the peptides. Sizes of molecular weight (MW) markers are indicated in kilodaltons. (C) Representation of the M2-1 protein molecule and the mapping of unphosphorylated and phosphorylated peptides to it. The dashed lines at the C-terminal ends of fragments indicate that their exact end positions are unknown. The amino acid sequence in the M2-1 protein RNA binding region is indicated. The residues starting the different phosphorylated peptides are also indicated in that sequence.
Deletion variants of M2-1 protein and their RNA binding capacities.
To confirm that between amino acids 40 and 80 of M2-1 protein are those that contact directly RNA and to pinpoint the residues essential for RNA binding, we tested the RNA binding capacity of M2-1 protein and different deletion variants by Northern-Western blotting using M2-1 proteins transiently expressed in the vaccinia virus-based expression system. The cDNA corresponding to the HRSV M2 gene was expressed in this system, and a protein with a mobility slower than that of M2-1 present in the purified virions was observed. This protein has been identified as phosphorylated M2-1 protein (see below).
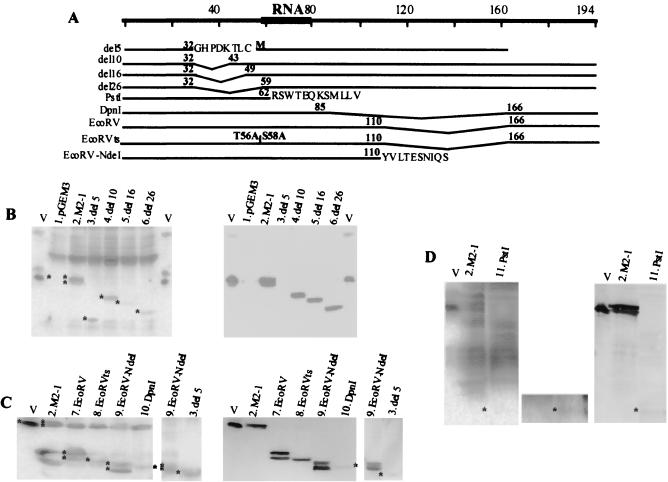
We made sequential and C-terminal amino acid sequence deletions of M2-1 protein. The sequential deletions (del5, del10, del16, and del26) were those in which 5, 10, 16, and 26 amino acids were eliminated between positions 32 and 58. The C-terminal deletions correspond to M2-1 variants PstI, DpnI, EcoRV, EcoRVts, and EcoRV-NdeI. The amino acid residues deleted and/or added up to the first termination codon in each M2-1 variant are indicated in Fig. 4A. All nucleotide sequences of the M2 gene variants were determined by automatic sequencing.
FIG. 4.
M2-1 protein deletion variants and their RNA binding capacities determined by Northern-Western assays. (A) Diagram representation of M2-1 protein indicating the amino acid residues deleted in the different variants as a broken lines. The residues unrelated to M2-1 protein and added as a consequence of the cloning procedures are also shown. Right and left parts of panels B to D correspond to Northern-Western and Western assays, respectively, of 50 μg of total protein. The different M2-1 protein variants are indicated by asterisks in both assays. In panel B, lane V corresponds to extracellular viral particles, and lanes 1, 2, 3, 4, 5, and 6 correspond to soluble proteins from transfections with pGEM3 and pGEM3 recombinants M2-1, del5, del10, del16, and del26, respectively. In panel C, lanes, 2, 3, 7, 8, 9, and 10 correspond to insoluble proteins from transfection with pGEM3 recombinant plasmid M2-1, del5, EcoRV, EcoRVts, EcoRV-NdeI, and DpnI constructs. In panel D, lane 11 corresponds to soluble protein from transfection with the pGEM3 recombinant plasmid PstI. The center portion corresponds to a longer exposure of the Northern-Western blot; the asterisk indicates the PstI variant M2-1 protein position. The Western blots were developed with monospecific anti-M2-1 protein serum except for the insets containing lanes 9 and 3 in panel C, which were developed with anti-HRSV serum.
Assay of the M2-1 protein variants for RNA binding capacity in Northern-Western assays showed that all were able to bind long RNAs (Fig. 4B to D). Comparison of RNA binding capacities of different amounts of unphosphorylated and phosphorylated M2-1 protein indicated that the phosphorylated form has a decreased RNA binding capacity (data not shown). The data from the distinct variants suggested that the RNA binding region is located between amino acid residues 59 and 85. The RNA binding of variant PstI, which contains only the first 62 M2-1 protein amino acid residues, and the fact that the protein containing deletions between residues 32 and 58 does bind to RNA indicate that residues 59 to 62 (EISG) participate in and may be sufficient for RNA binding. The del5 variant (Fig. 4B, lane 3), which is devoid of the zinc finger, is not essential for binding, as suggested by the preparative cross-linking experiments. In this variant, two G insertions occurred at nucleotide 100 of the 5′ end of the mRNA; this produces a frameshift that results in the appearance of a stop codon 24 nucleotides after the insertion. The protein synthesized by this variant probably starts at the third methionine of the M2-1 protein that has a G at position +4, according to the Kozak rules (22), and has a C-terminal truncation, according to its observed size. These results are in agreement with those obtained in the preparative cross-linking assays, which locate the RNA binding region between amino acids 40 and 80, with some of the residues in contact with RNA located between positions 55 and 75. It is also clear that the amino acid residues located between positions 85 and 194 are unnecessary for RNA binding.
M2-1 protein is phosphorylated mainly when expressed in the absence of other viral proteins.
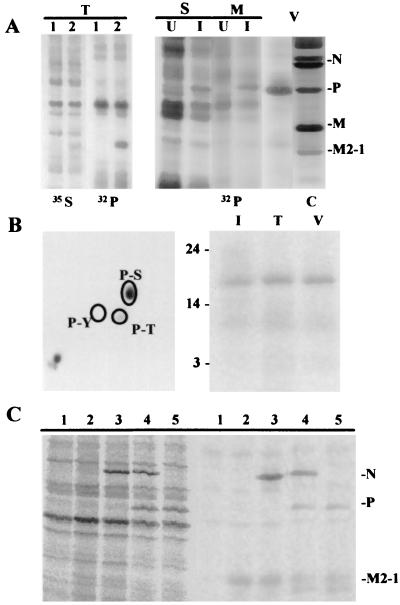
HEp-2 cells infected with the vaccinia virus recombinant vTF-3 were transfected with plasmid pGEM3 or pGEM3-M2. After labeling with [35S]methionine, cell extracts were prepared, and the proteins were analyzed by SDS-PAGE. In pGEM3-M2-transfected cells, we detected a protein that is absent in the cells transfected with pGEM3. This protein showed a mobility slower than that of M2-1 protein present in purified virions (Fig. 5A, lane 2, 35S). Previous results (23) described a 24-kDa structural phosphoprotein, in addition to P protein, which may be M2-1 protein. The change in electrophoretic mobility observed for the transiently expressed M2-1 protein may thus be due to phosphorylation. To test this, the above experiment was repeated but with [32P]orthophosphate labeling. 32P labeling was found in the protein expressed by the recombinant pGEM3-M2 plasmid (Fig. 5A, lane 2, 32P).
FIG. 5.
M2-1 protein is phosphorylated at serine and threonine residues when is expressed in transfected and HRSV-infected HEp-2 cells. Direct or indirect interactions with P protein modulate M2-1 protein phosphorylation. (A) HEp-2 cells were transfected (T) with plasmid pGEM3 (lane 1) or pGem3-M2 (lane 2), mock (U) or HRSV (I) infected, and labeled with [32P]orthophosphate. In the transfection experiment, the protein was also labeled with [35S]methionine. Soluble (S) and microfilament-associated (M) protein fractions were obtained. The proteins were separated by SDS-PAGE and visualized in a PhosphorImager. The viral proteins in extracellular viral particles (V) stained by Coomassie blue (C) are shown at the right. (B) (Left) The piece of gel containing the phosphorylated protein, with electrophoretic mobility slower than that of M2-1 protein of extracellular viral particles expressed by the recombinant plasmid in the transfection experiment (T), was excised from the gel and hydrolyzed with 6 N HCl for 2 h at 110°C. The resulting amino acid residues were separated by two-dimensional electrophoresis in thin layer and characterized by their 32P labels. Unlabeled phosphothreonine (P-T), phosphoserine (P-S), and phosphotyrosine (P-Y) were included as markers and detected with ninhydrin. (Right) Phosphorylated protein expressed by pGEM2-M2 (T), and polypeptides with the same electrophoretic mobility in the microfilament fraction of infected cells (I) and in virions (V), after excision from the gel and CNBr treatment. After renaturation, the peptides were separated in SDS–20% urea acrylamide gels, and those in phosphorylated form were detected by a PhosphorImager. Sizes are indicated in kilodaltons. (C) HEp-2 cells were infected with recombinant vaccinia virus vTF-3 and transfected with pGEM3 (lane 1), with pGEM3-M2 (lanes 2 to 5), and with pGEM3-M2 plus pGEM3-N (lane 3), pGEM3-N and pGEM3-P (lane 4), and pGEM-P (lane 5). The cultures were [35S]methionine labeled, and the soluble proteins were analyzed by SDS-PAGE before (left) or after (right) immunoprecipitation with rabbit anti-HRSV serum.
When 32P-labeled mock- or HRSV-infected HEp-2 cells were fractionated (Fig. 5A) to obtain soluble and microfilament-bound protein fractions and extracellular viral particles, a phosphorylated protein was found with the same electrophoretic mobility as that detected after transfection with the pGEM3 recombinant M2, mainly in the microfilament fraction and extracellular viral particles. The piece of gel containing M2-1 protein, transiently expressed in transfected cells, was excised, and its phosphoamino acid residues were determined (Fig. 5B, left). It was found that M2-1 protein is phosphorylated on serine and to a lesser extent on threonine residues. Similar results were obtained using phosphorylated M2-1 protein from HRSV-infected HEp-2 cells (data not shown). Phosphorylated M2-1 protein from transfected cells, present in the microfilament fraction of HRSV-infected HEp-2 cells and in extracellular viral particles, was treated with CNBr, and the peptides generated were separated by SDS-PAGE; phosphorylated peptides were detected by autoradiography (Fig. 5B, right). The same phosphopeptides were detected in all cases. Also, the two species of M2-1 have been detected at early and late times in HRSV-infected HEp-2 cells (13).
These results, like others recently published (18), indicated that M2-1 protein is phosphorylated and that phosphorylation modifies its electrophoretic mobility. When it is expressed alone, without other viral proteins, all molecules are modified; in the presence of the remaining viral proteins, the bulk of M2-1 protein remains mainly in an unphosphorylated form. It appears that when M2-1 is expressed alone, its phosphorylation sites are exposed to protein kinases and the action of cellular protein phosphatases is prevented. In contrast, in the presence of the rest of viral proteins, M2-1 phosphorylation sites are partially hidden from the protein kinases or the phosphorylated residues are exposed to phosphatase action, probably due to conformational changes in the protein caused by protein-protein interactions.
Coexpression experiments with viral proteins N and P indicate that M2-1 coexpression with P protein results in the lack of phosphorylation of a part of the M2-1 molecules (Fig. 5C). Direct or indirect interactions between P and M2-1 proteins thus result in the prevention of exposure of phosphorylatable M2-1 protein residues to the action of cellular protein kinases or phosphatases, respectively.
Deletions variants of M2-1 protein and their phosphorylation capacity.
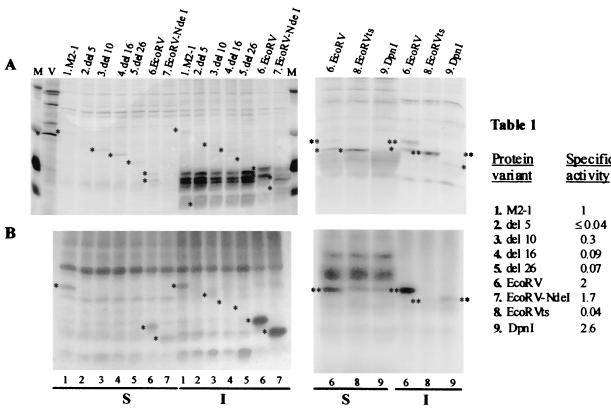
When M2-1 protein and its variants were transiently expressed, differences in solubility and in the presence of two molecular species were observed (Fig. 6A). To determine whether the presence of a second protein species is due to phosphorylation, transfected cells were labeled with [32P]orthophosphate (Fig. 6B), their specific activities were determined, and the results obtained were compared to those for the normal M2-1 protein (Fig. 6; Table 1).
FIG. 6.
Phosphorylation of M2-1 deletion variant proteins. HEp2-cells were infected with vaccinia virus recombinant vTF-3 and transfected with the following or with pGEM3 recombinant plasmids: M2 (lane 1), del5 (lane 2), del10 (lane 3), del16 (lane 4), del26 (lane 5), EcoRV (lane 6), EcoRV-NdeI (lane 7), EcoRVts (lane 8), and DpnI (lane 9). The cultures were labeled with [32P]orthophosphate, and soluble (S) and insoluble (I) protein fractions were isolated, analyzed in SDS–15% polyacrylamide gels, and visualized by Coomassie blue staining (A, left) or by autoradiography (A, right), [35S]methionine-labeled cultures; (B, right and left; [32P]orthophosphate-labeled cultures). M, size markers; V, viral proteins contained in extracellular viral particles. Table 1 shows specific activity (phosphorylations), relative to that of control M2-1, for protein variants.
M2-1 proteins with deletions between amino acids 32 and 42, 32 and 48, and 32 and 58 (del10, del16, and del26) were phosphorylated 30, 9, and 7% with respect to the phosphorylation level of the whole M2-1 protein. Only one protein species was expressed for these variants, except for del10. For the M2-1 variants containing large deletions (EcoRV [amino acids 110 to 166 deleted], DpnI [amino acids 85 to 166 deleted], and EcoRV-NdeI [amino acids 110 to 194 deleted]), higher specific activities (2.0, 2.6, and 1.7, respectively) than those found for the control M2-1 protein were observed. In these variants, two proteins species were detected. These results indicated that the absence of amino acids 1 to 59 or 110 to 162 regulates M2-1 protein phosphorylation in a negative or positive manner.
Identification and localization of phosphorylated residues on the M2-1 molecule.
M2-1 protein transiently expressed in HEp-2 cells with the vaccinia virus-based system was labeled with [32P]orthophosphate or [35S]methionine, fractionated by gel electrophoresis, transferred to Inmobilon membranes, and excised. Proteins were then treated with CNBr and N-bromosuccinimide. Labeled peptides were separated and further identified by autoradiography. In accordance with the cleavage specificity of the chemical agents used, peptide size, and methionine residue distribution in the M2-1 molecule, the phosphorylated residues were tentatively located at the NH2-terminal region between amino acid residues 40 and 100 (data not shown). Concurring with this location, the deletion variants lacking amino acid residues between positions 85 and 166, 110 and 166, and 110 and 194 were phosphorylated at a higher level than the normal protein. The slight phosphorylation for del10 and the lack of phosphorylation found for del16 and del26 indicated that the phosphorylated residues could be located between residues 48 and 85, taking into account that there are no serine and threonine residues between amino acid residues 32 and 48 of M2-1 protein. Previous experiments indicated (36) that purified HRSV particles contain casein kinase II (CKII)-like activity, able to phosphorylate their P protein among others proteins present in the purified viral particles. One of these proteins has the same electrophoretic mobility as the phosphorylated M2-1 protein. The phosphorylated amino acids of M2-1 may thus be modified by CKII protein.
We looked for S and T residues between amino acids 48 and 85 containing the signal recognition sequence for CKII (29) (an acidic or phosphorylated residue at position +3). T56 has this recognition sequence; if it is phosphorylated, S53 could also be a suitable target for phosphorylation. In addition, S58 has the CKI consensus sequence (an acidic or phosphorylated residue at position −3), and if S58 is phosphorylated, S61 also can be phosphorylated by the same protein kinase. This tentative location is in agreement with the lack of phosphorylation observed for del16 and del26, although only del26 has lost the S and T residues present at positions 58 and 56. Changes in protein structure may explain the lack of phosphorylation in the other M2-1 protein variants. We thus replaced T56 and S58 with alanine in deletion variant EcoRV and confirmed the resulting nucleotide sequence by automated sequencing.
This new M2-1 protein variant, EcoRVts, was transiently expressed and labeled with [35S]methionine and [32P]orthophosphate. The protein in the cell extracts was fractionated by SDS-PAGE and visualized by autoradiography (Fig. 6A, lane 8). The EcoRVts variant protein was expressed but was poorly phosphorylated. Calculation of its specific activity showed that it is phosphorylated only to 2 to 4% of the level of the EcoRV variant protein. The same phosphorylation level was obtained in relation to M2-1 when the T56A and S58A substitutions were incorporated into wild-type M2-1 protein (data not shown). Thus, it seems that one or both of the substituted residues are responsible for 96 to 98% of the M2-1 phosphorylation, although it cannot be excluded that substitutions T56A and S58A avoid M2-1 phosphorylation by inducing conformational changes. It is noteworthy that phosphorylatable residues precede amino acid residues 59 to 62, which may be essential for M2-1 protein RNA binding.
DISCUSSION
M2-1 protein has been described as a putative nucleocapsid structural component that acts as a transcription factor by increasing the processivity and readthrough of intergenic sequences by the viral polymerase constituted by the L and P proteins (9, 10, 17). Nevertheless, the increased readthrough of the viral polymerase in the presence of the M2-1 protein does not increase the replication products of HRSV RNA analogs (13). M2-1 incorporation into the viral polymerase complex composed of L and P proteins is thus not related to a switch of polymerase activity from the transcription to the replication mode (10).
In its NH2-terminal sequence, the protein has a Zn finger formed by three cysteine residues and one histidine residue, like several other transcription factors (40). Like them, M2-1 has RNA binding capacity, shown here using three different assays.
By testing long (700- to 1,300-nucleotide), RNAs we found that M2-1 protein may bind to different RNAs with the same affinity, suggesting a histone-like activity for this protein similar to that ascribed to the N protein in paramyxoviruses (12). M2-1 shows also specificity for certain RNA sequences. To determine sequence specificity, we tested shorter RNA molecules and found M2-1 to specifically recognize the leader RNA (44 nucleotides) at the 5′ end of short RNAs (80 nucleotides). The distinct affinities of the M2-1 protein for long and leader RNAs could be due to the binding of different M2-1 oligomers (monomers and trimers or dimers and hexamers) for each type of RNA. It may also be due to the increased number of nonspecific binding sites in the long RNAs. On the other hand, the phosphorylation state of M2-1 may change the affinity and perhaps the specificity of the protein for the different RNAs. The M2-1 RNA binding domain has been mapped outside the zinc finger by preparative cross-linking of the protein to labeled riboprobes. In addition, an M2-1 protein variant (del5) without the Zn finger (located in the first 30 amino acid residues) binds to RNA with greater affinity than does the intact M2-1 protein, according to their expression levels of both proteins (Fig. 4C), indicating the the Zn finger is not essential for M2-1 RNA binding. The amino acid residues in contact with the RNA are located between residues 59 and 85, as indicated by binding capacities of variants lacking residues 32 to 58. In addition, residues at positions 59 to 62 may be sufficient for RNA binding, as indicated by positive RNA binding by the PstI deletion mutant. Nevertheless, the RNA binding capacity of the residues added in the cloning procedure involved in obtaining variant PstI cannot be excluded. Other residues in contact with the RNA, between residues 62 and 85, cannot be ruled out. In addition, the binding capacity of the protein variants devoid of residues 85 to 194 suggests that this part of the molecule is not important for M2-1 binding to RNA. The affinities of the different variants remain to be determined to ensure the importance of different residues in RNA binding.
In contrast, the retroviral nucleoprotein interacts with viral RNA through a Zn finger. In human immunodeficiency virus, basic residues in the first and second Zn fingers (33) are involved in electrostatic contacts with RNA and may be responsible for the specificity of RNA encapsidation. Other Zn fingers are not involved in the binding to nucleic acids; replication protein A (26), for example, binds to single-stranded DNA through molecular regions located outside the Zn finger. These metal binding domains have been identified in several proteins of different origins (5); although they are able to bind nucleic acids in some cases, they also facilitate interactions between proteins and other macromolecules. The metal binding domain can thus also be involved in interactions between protein monomers (28). One can therefore speculate that the metal binding domain of M2-1 plays a role in the dimer formation detected in the UV cross-linking experiments. Its presence could also stabilize the native protein conformation. It appears that the native conformation of the RNA binding domain is facilitated by the presence of Zn ions during the renaturation process, in accordance with the importance of this domain in global structure of the protein. Thus, substitution of residues C7, C15, and H25 predicted at those that coordinate Zn and prevent M2-1 readthrough transcription activity, phosphorylation, and interaction with N protein indicates that the C3H1 motif is essential for maintaining the functional integrity of the protein (18).
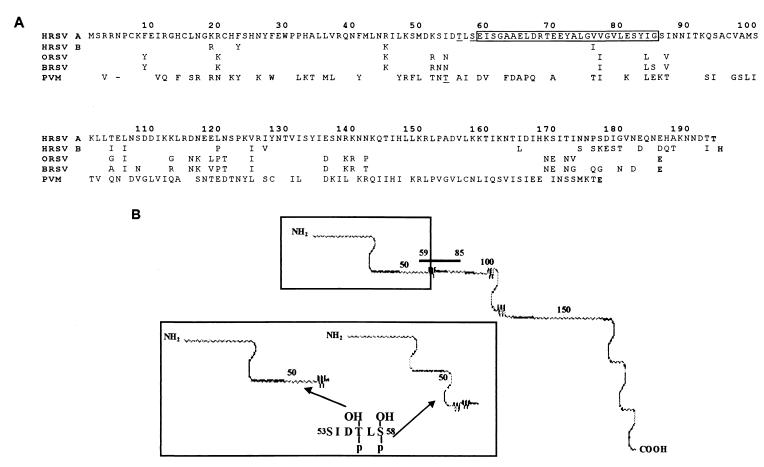
In the M2-1 proteins of human, bovine, and ovine virus origin, amino acid conservation between residues 59 and 80 is 95% (2). When the comparison includes the mouse pneumovirus M2-1 protein (1), there is only 45% similarity between residues 59 and 70 (residues S58, E/D59, I/V60, S61, G62, R68, and T69). Between residues 70 and 80, the conservation is 90% among all strains. The majority of these residues have an aromatic and hydrophobic nature (Fig. 7), similar to those in contact with the RNA in other RNA binding domains (5). The secondary structure of these binding region is a β sheet, according to Chou and Fasman predictions (7). For the amino acid residues at positions 50 to 62, an α-helix structure is predicted (7). These secondary structures have been found in the RNA binding domains of other RNA binding proteins (5).
FIG. 7.
Comparison of pneumovirus M2-1 protein primary structures, showing conservation of the RNA binding domain and phosphorylatable residues. (A) Amino acid sequences of pneumovirus M2-1 proteins of human (A and B subgroup), ovine, bovine, and murine origins (1, 2). The RNA binding domain is boxed; phosphorylated residues are underlined; the last residue from each sequence is indicated in boldface. (B) Secondary structure prediction of M2-1 protein by the Chou and Fasman method (7). The rectangles represent blowups of the M2-1 protein NH2-terminal region showing the effect of substitutions T56E and S58E on its secondary structure.
It is noteworthy that the phosphorylatable residues, whose modification changes the protein RNA binding capacity, are also located in the M2-1 protein RNA binding domain. M2-1 appears to be phosphorylated at T56 and S58, with modification at these residues responsible for 96 to 98% of M2-1 protein phosphorylation.
T56 has the recognition consensus sequence described for the cellular protein kinase CKII (29), and its modification allows the same protein kinase to phosphorylate S53. T56 is not conserved in the mouse pneumovirus M2-1; in this case there is a threonine at position 52, with a consensus recognition sequence for CKII. In this protein, no additional residues could be modified after phosphorylation at T52. The amount of phosphorylated serine is higher than that of phosphorylated threonine; thus, additional serines seem to be modified. We therefore prepared the mutant T56A, S58A, which lacks 96 to 98% of the corresponding phosphorylation found in the EcoRV variant. This indicates that simultaneous phosphorylation at T56 and S58 is responsible for 96 to 98% of M2-1 phosphorylation. Other possibilities nonetheless remain open, as phosphorylation at these residues may be essential for further phosphorylation at a distant site located between amino acid residues 40 and 100, or replacement of these phosphorylatable residues by alanines, like the presence of del10 and del16, may disrupt the protein conformation essential for phosphorylation.
It is nevertheless clear that after removal of these residues, other threonine or serine residues are phosphorylated, since 2 to 4% of the phosphorylation level of the EcoRV mutant was observed. In the M2-1 protein sequence, S and T residues contained in the EcoRV variant with a signal recognition sequence for known kinases are S2 (a residue present in a sequence similar to that of the serine phosphorylated in the nucleoprotein of influenza virus [4]), S108 (CKI), and T180 (CKII). It is likely that this threonine residue or the serines at positions 2 and 108 are also modified, although further experiments are needed to probe this point.
M2-1 protein phosphorylation decreases its RNA binding capacity at least 5- to 10-fold in Northern-Western assays using long RNAs (data not shown). According to this result, modification in the predicted secondary structure of the M2-1 protein may occur when T56 and S58 are replaced by glutamic acid (Fig. 7).
M2-1 protein increases the processitivity of the viral polymerase intragenic (9, 10) and its readthrough capacity on all gene junctions, except at the junction leader-NS1 gene (13). It is tempting to speculate that the M2-1 protein capacity to bind leader RNA, present in short RNAs, should be related to the RNA polymerase ability to start RNA synthesis at the boundary leader-NS1 gene during viral transcription. In this point, it would be important to test the ability of M2-1 variants, mainly new ones with deletions in the RNA binding domain, for transcription processivity and readthrough. These and other intriguing possibilities are now being followed up. Because the direct or mediated interaction of M2-1 with P protein may determine the M2-1 phosphorylation level, the formation of these complexes could also modulate M2-1 protein RNA binding capacity.
ACKNOWLEDGMENTS
We are grateful to R. Serra for preliminary experiments, to J. Avila for many comments and suggestions, to G. Wertz for critical reading of the manuscript, to M. I. García-Albert and R. Martinez for excellent technical assistance, and to K. Mark for editing.
X. Geng and A. Asenjo were fellows of the Agencia Española de Cooperación Internacional and Instituto de Salud Carlos III, respectively. This work was supported by FIS 00/0204.
REFERENCES
- 1.Ahmadian G, Chambers P, Easton A J. Detection and characterization of proteins encoded by the second ORF of the M2 gene of pneumovirus. J Gen Virol. 1999;80:2011–2016. doi: 10.1099/0022-1317-80-8-2011. [DOI] [PubMed] [Google Scholar]
- 2.Alansari H, Potgieter L N D. Molecular cloning and sequence analysis of the phosphoprotein, nucleocapsid protein, matrix protein and 22K (M2) protein of the ovine respiratory syncytial virus. J Gen Virol. 1994;75:3597–3601. doi: 10.1099/0022-1317-75-12-3597. [DOI] [PubMed] [Google Scholar]
- 3.Albó C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrese M, Portela A. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J Virol. 1996;70:3385–3391. doi: 10.1128/jvi.70.6.3385-3391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg J M. Zinc fingers and other metal-binding domains. J Biol Chem. 1990;265:6513–6516. [PubMed] [Google Scholar]
- 6.Bermingham A, Collins P L. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc Natl Acad Sci USA. 1999;96:11259–11264. doi: 10.1073/pnas.96.20.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou P Y, Fasman G D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M, Johnson P R. The two open reading frames of the 22K mRNA of human respiratory syncytial virus: sequence comparison of antigenic subgroups A and B and expression in vitro. J Gen Virol. 1990;71:3015–3020. doi: 10.1099/0022-1317-71-12-3015. [DOI] [PubMed] [Google Scholar]
- 9.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1313–1351. [Google Scholar]
- 12.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 13.Fearns R, Collins P L. Role of the M2-1 transcription antiterminator protein of respiratory syncytial virus in sequential transcription. J Virol. 1999;73:5852–5864. doi: 10.1128/jvi.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Patrón C, Calero M, Rodríguez Collazo P, García J R, Madrazo J, Musachio A, Soriano F, Estrada R, Frank R, Castellano-Serra L R, Méndez E. Protein reverse staining: high efficiency microanalysis of unmodified proteins detected on electrophoresis gels. Anal Biochem. 1995;224:203–211. doi: 10.1006/abio.1995.1031. [DOI] [PubMed] [Google Scholar]
- 15.Fontana A, Gross E. Fragmentation of polypeptides by chemical methods. In: Darbre A, editor. Practical protein chemistry—a handbook. New York, N.Y: Plenum Press; 1986. pp. 67–120. [Google Scholar]
- 16.García J, García-Barreno B, Vivo A, Melero J A. Cytoplasmic inclusions of respiratory syncytial virus infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein and the 22K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 17.Hardy R, Wertz G. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy R, Wertz G. The Cys3-Hys1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J Virol. 2000;74:5880–5885. doi: 10.1128/jvi.74.13.5880-5885.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y T, Collins P L, Wertz G. Characterisation of 10 proteins of human respiratory syncytial virus: identification of a four envelope associated protein. Virus Res. 1985;2:157–173. doi: 10.1016/0168-1702(85)90246-1. [DOI] [PubMed] [Google Scholar]
- 21.Johnson S, Griego S D, Pfarr D S, Doyle M L, Woods R, Carlin D, Prince G A, Koenig S, Young J F, Dillon S B. A direct comparison of the activities of two humanised respiratory syncytial virus monoclonal antibodies: MEDI-493 and RSHZ19. J Infect Dis. 1999;180:35–40. doi: 10.1086/314846. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Effects of intercistronic length on efficiency of reinitiation by eukaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert D M, Hambor J, Diebold M, Galiski B. Kinetics studies and phosphorylation of respiratory syncytial virus polypeptides. J Gen Virol. 1988;69:313–323. doi: 10.1099/0022-1317-69-2-313. [DOI] [PubMed] [Google Scholar]
- 24.López J A, Villanueva N, Melero J A, Portela A. Nucleotide sequences of the fusion and phosphoprotein genes of the human respiratory syncytial (RS) virus Long strain: evidence of subtype genetic heterogeneity. Virus Res. 1988;10:249–262. doi: 10.1016/0168-1702(88)90020-2. [DOI] [PubMed] [Google Scholar]
- 25.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J S, Wang M, Park S J, Lee S H. Zinc finger of replication protein A, a non-DNA binding element, regulates its binding activity through redox. J Biol Chem. 1999;274:29075–29080. doi: 10.1074/jbc.274.41.29075. [DOI] [PubMed] [Google Scholar]
- 27.Pastey M K, Gower T L, Spearman P W, Crowe J E, Jr, Graham B S. A rho A-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat Med. 2000;6:35–40. doi: 10.1038/71503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan T, Giedroc D P, Coleman J E. 1H NMR studies of T4 gene 32 protein: effects of zinc removal and reconstitution. Biochemistry. 1989;28:8828–8832. doi: 10.1021/bi00448a022. [DOI] [PubMed] [Google Scholar]
- 29.Pearson R B, Kemp B E. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 30.Peeples M, Collins P L. Mutations in the 5′ trailer region of respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Ulloa L, Serra R, Asenjo A, Villanueva N. Interactions between cellular actin and human respiratory syncytial virus (HRSV) Virus Res. 1998;53:13–25. doi: 10.1016/s0168-1702(97)00121-4. [DOI] [PubMed] [Google Scholar]
- 33.Urbaneja M A, Kane B P, Johnson D G, Gorelick R J, Henderson L E, Casas-Finet J R. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J Mol Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 34.Vanderkerkhove J, Bauw G, Puype M, Van Damme J, Montagu M. Protein-blotting on polyprene-coated glass-fiber sheets. A basis for acid hydrolysis and gas-phase sequencing of picomole quantities of protein previously separated on sodium dodecyl sulfate polyacrylamide gel. Eur J Biochem. 1985;152:9–19. doi: 10.1111/j.1432-1033.1985.tb09157.x. [DOI] [PubMed] [Google Scholar]
- 35.Villanueva N, Navarro J, Cubero E. Antiviral effects of xanthate D609 on the human respiratory syncytial virus growth cycle. Virology. 1991;181:101–108. doi: 10.1016/0042-6822(91)90474-p. [DOI] [PubMed] [Google Scholar]
- 36.Villanueva N, Navarro J, Méndez E, García-Albert M I. Identification of a protein kinase involved in the phosphorylation of the C-terminal region of human respiratory syncytial virus P protein. J Gen Virol. 1994;75:555–565. doi: 10.1099/0022-1317-75-3-555. [DOI] [PubMed] [Google Scholar]
- 37.Villanueva N, Hardy R, Asenjo A, Qinghong Y, Wertz G. The bulk of human respiratory syncytial virus (HRSV) P phosphoprotein (P protein) is not essential but modulates RNA viral transcription and replication. J Gen Virol. 2000;81:123–133. doi: 10.1099/0022-1317-81-1-129. [DOI] [PubMed] [Google Scholar]
- 38.Whitehead S S, Burreyev A, Teng M N, Firestone C Y, St. Claire M, Elkins W R, Collins P L, Murphy B R. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson J M. Fragmentation of polypeptides by enzymatic methods. In: Darbre A, editor. Practical protein chemistry—a handbook. New York, N.Y: Plenum Press; 1986. pp. 121–148. [Google Scholar]
- 40.Worthington M, Amann B T, Nathans D, Berg J M. Metal binding properties and secondary structure of the Zn binding domain of Nup475. Proc Natl Acad Sci USA. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Q, Hardy R W, Wertz G. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define the minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]