Abstract
Mitochondria are a culprit in the onset of Parkinson’s disease, but their role during disease progression is unclear. Here we used Cox proportional hazards models to exam the effect of variation in the mitochondrial genome on longitudinal cognitive and motor progression over time in 4064 patients with Parkinson’s disease. Mitochondrial macro-haplogroup was associated with reduced risk of cognitive disease progression in the discovery and replication population. In the combined analysis, patients with the super macro-haplogroup J, T, U# had a 41% lower risk of cognitive progression with P = 2.42 × 10−6 compared to those with macro-haplogroup H. Exploratory analysis indicated that the common mitochondrial DNA variant, m.2706A>G, was associated with slower cognitive decline with a hazard ratio of 0.68 (95% confidence interval 0.56–0.81) and P = 2.46 × 10−5. Mitochondrial haplogroups were not appreciably linked to motor progression. This initial genetic survival study of the mitochondrial genome suggests that mitochondrial haplogroups may be associated with the pace of cognitive progression in Parkinson’s disease over time.
Keywords: Parkinson’s disease, mitochondrial haplogroups, cognitive progression
Liu et al. report that specific mitochondrial haplogroups are associated with the progression of cognitive decline in patients with Parkinson's disease, but not with the progression of motor impairment. Mitochondrial haplotypes may thus be useful for stratifying patients according to their risk of cognitive decline.
Introduction
Disability and quality of life of patients with Parkinson’s disease (PD) is affected by progressive cognitive impairment.1 Increasing numbers of cognitively impaired patients with PD pose a medical and socio-economic challenge in many countries.2 The pace of cognitive changes during the disease course, however, varies substantially from patient to patient3 and the genetic architecture accounting for this heterogeneity in disease progression has not been well established.
Genome-wide association studies (GWAS) during the past decade have delineated the genetic architecture of disease susceptibility with 90 association signals in 78 common autosomal loci in PD patients of European ancestry.4 Our recent genome-wide survival study identified associations with longitudinal progression from PD to Lewy body dementia in five loci, RIMS2, GBA, and APOE, WWOX and TMEM108.5 This extends and confirms longitudinal studies implicating GBA variants6,7 and APOE ε48 in cognitive decline in PD. These genome-wide and targeted sequencing efforts have paved the way for unravelling the genetic architecture of disease progression in PD, but have not yet investigated the second critical source of human DNA—the mitochondrial genome (mtDNA).
MtDNA mutations contribute to a spectrum of human diseases,9 and in PD there is accumulating genetic and environmental evidence that mitochondrial dysfunction may play a key role in the pathogenesis of the disease.10 There are high level of somatic mtDNA mutations in substantia nigra neurons in early PD11 and dysregulation of mtDNA homeostasis in sporadic PD.12 Mutations in the nuclear-encoded PINK1 and PRKN cause autosomal recessive PD and disrupt mitophagy.13 Moreover, there is a pervasive defect in PGC-1alpha-regulated mitochondrial bioenergetics gene expression in nigral dopamine neurons and substantia nigra even in prodromal, subclinical Lewy body neuropathology.14
The diversity of modern human mtDNA haplogroups (variants) has provided valuable information to trace the history of human evolution, and many studies in recent years have reported links between specific mtDNA haplogroups and susceptibility for PD,15 however, the impact of mtDNA haplogroups or variants on progression in PD has not been defined. To characterize whether genetic variation in the mitochondrial genome influences the progression of PD, we performed a longitudinal, multi-cohort analysis, and identified specific mitochondrial haplogroups linked to cognitive decline in PD. Further exploratory analysis indicated two single nucleotide polymorphisms (SNPs) in mtDNA specifically associated with cognitive progression.
Materials and methods
Study participants
The cohorts included in this study were described in previous work from the International Genetics of Parkinson Disease Progression Consortium.5–7 In brief, 4491 patients with PD (with available genotyping data and quality control) were longitudinally assessed with 33 406 study visits in 15 cohorts from North America and Europe between 1986 and 2017 (Supplementary material). Written informed consent for DNA collection and phenotypic data collection for secondary research use for each cohort was obtained from the participants with approval from the local ethics committees. The Institutional Review Board of Mass General Brigham and the Institutional Review Board of the School of Medicine, Sun Yat-sen University approved the current analyses. Patients whose longitudinal follow-up evaluations were not consistent with a diagnosis of PD were excluded. Fifteen cohorts were a priori assigned to discovery or replication cohorts as we previously described5 (Supplementary Fig. 2). This achieves an approximately two-thirds to one-third split among the two stages and a balanced distribution of the distinct types of cohorts (for example, purpose-designed biomarkers studies, phase 3 clinical trials, population-based cohorts) across stages.
Polymorphism identification and haplogroup classification
We analysed 763 mitochondrial SNPs in 4491 patients with PD and predicted their mitochondrial haplogroup using Haplogrep2.016 with default parameters using the mitochondrial Revised Cambridge Reference Sequence (Supplementary Fig. 1). We next simplified the sub-haplogroups (455 sub-haplogroups) to the 34 haplogroups (Supplementary Table 1). After quality control (Supplementary material), 4064 subjects with 30 515 study visits were used for haplogroup analysis [including H, HV* (excluding H, V), I, J, K, T and U# (excluding K) haplogroups]. Out of 763 mitochondrial SNPs, 102 SNPs with allele frequency >1% were used for single SNP Cox regression analysis.
Statistical analysis
The Cox proportional hazards (Cox PH) analysis was used to estimate the influence of different mitochondrial haplogroups on time (years from onset of PD) to reaching the endpoint of global cognitive impairment (GCI) as indicated by a Serial Mini Mental State Exam (MMSE) ≤ 25 according to the recommendation the International Parkinson and Movement Disorder Society (MDS) Task Force17 and adjusting for the covariates of age at onset, gender, years of education and polygenic hazard score (PHS) as fixed effects, and for a cohort term as a random effect. A second endpoint was time to motor disability with postural instability as indicated by Hoehn and Yahr stage 3 adjusting for age at onset, gender, GBA carrier status and the cohort term similar to Liu et al.6 (see Supplementary material for details). For the single nucleotide variants, a similar Cox PH analysis was used (using the same co-variants as mentioned above) to investigate the effect of each SNP on time to cognitive impairment.
Generalized longitudinal mixed fixed and random effects analysis of cognitive decline was performed with MMSE scores longitudinally assessed at varying times (enrollment visit and multiple longitudinal follow-up visits) in the combined data set (Supplementary material). All analyses were conducted in the R statistical environment version 4.0.2.
Data availability
The genotype and clinical data for the Parkinson’s progression markers initiative (PPMI) included in this study are publicly available upon request to ppmi@loni.usc.edu through a PPMI Whole Genome Sequencing Data Agreement. Clinical data for the Parkinson’s disease biomarker program (PDBP) included in this study are publicly available through https://pdbp.ninds.nih.gov. Clinical longitudinal data and genotyping data for the other cohorts included are accessible through appropriate data sharing agreements that protect patient privacy with the institutions that conducted or are conducting study consents and clinical assessments under local institutional review board approvals.
Results
Mitochondrial haplogroup is associated with cognitive decline in patients with Parkinson's disease
The genotyped data of 4491 patients with PD across 15 cohorts from North America and Europe were used to estimate their mitochondrial haplogroups. 4447 patients with 33 068 longitudinal study visits passed quality control (Supplementary Fig. 1A) and were classified into eight groups: seven macro-haplogroups (H, HV*, I, J, T, K, U#) and a group comprising various other haplogroups (Supplementary Fig. 1B and Supplementary Table 1). Overall, 41.13%, (1829) patients belonged to macro-haplogroup H, which is a common mtDNA clade in Europe and found in approximately 43.10% of UK Biobank individuals.18 There were no significant differences in demographic and clinical characteristics of the patients in the various macro-haplogroups (Supplementary Table 2). The proportion of the seven macro-haplogroups was consistent with a previous survey in various European countries (Supplementary Table 3) and did not differ between the 15 cohorts (P ≈ 1, Fisher’s exact test, Supplementary Fig. 2). For 4064 patients within seven macro-haplogroups, we assigned 2811 patients and 12 605 longitudinal visits to the discovery population. 1253 patients and 17 910 visits comprised the replication population.
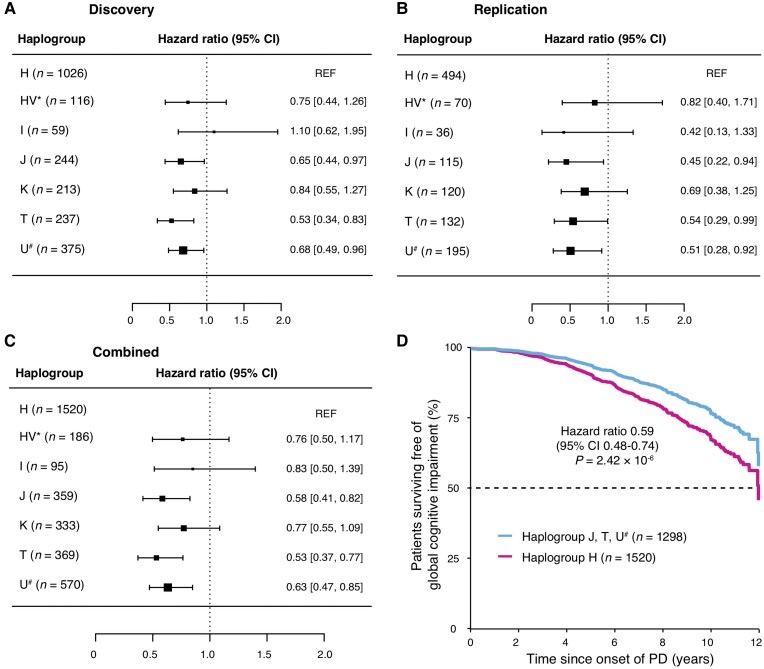
We then investigated the effect of seven macro-haplogroups on the risk of cognitive and motor impairment during the progression of Parkinson’s disease in discovery and replication populations. ‘Haplogroup’ was an unordered categorical variable in our Cox PH model. An omnibus test for haplogroup variation with six degrees of freedom showed that the seven haplogroups in general were differed from each other in their association with cognitive progression (the null hypothesis is that the haplogroups have the same effect) with an ‘omnibus’ test P-value < 0.001 in the discovery stage. We followed up this omnibus test with pertinent post hoc likelihood ratio tests which are the pairwise comparisons of each of the haplogroups against the ‘reference’ haplogroup H. J, T and U# haplogroups were associated with a reduced risk for GCI (MMSE ≤ 25) compared to the common haplogroup H with a hazard ratio (HR) of 0.65 [95% confidence interval (CI) 0.44–0.97] and P = 0.033, HR of 0.53 (95% CI 0.34–0.83) and P = 0.0052 and HR of 0.68 (95% CI 0.49–0.96) with P = 0.028 in the discovery stage, respectively (Fig. 1A). We further confirmed these associations in a replication population, where the HRs were 0.45 (95% CI 0.22–0.94), 0.54 (95% CI 0.29–0.99) and 0.51 (95% CI 0.28–0.92) with P values of 0.033, 0.047 and 0.025 for J, T and U# compared with Haplogroup H, respectively (Fig. 1B). Consistently, in the combined analysis, HR were 0.58 (95% CI 0.41–0.82) with P = 0.0023, 0.53 (95% CI 0.37–0.77) with P = 0.0007, and 0.63 (95% CI 0.47–0.85) with P = 0.0023, respectively (Fig. 1C). For each haplogroup compared to haplogroup H, the Cochran's Q-test and the I2 index showed that HRs across studies were homogeneous (Supplementary Table 4).
Figure 1.
Mitochondrial haplogroups and risk for GCI over time in patients with PD. The forest plot shows HRs for global cognitive impairment in specific types of macro-haplogroups compared to macro-haplogroup H in patients with PD from the discovery (A), replication (B) and combined (C) populations. The squares represent point estimates, with the sides of the square inversely proportional to the standard error of the estimates. The horizontal lines indicate 95% CIs of the estimates. (D) Covariate-adjusted survival curves for patients with PD in macro-haplogroups J, T and U# (cyan line) and those in macro-haplogroups H (magenta line).
There was no difference in HR for GCI among sub-haplogroups of H (Supplementary Fig. 3). There was no difference in HR for motor progression to Hoehn and Yahr stage 3 (motor disability with postural instability in PD) for each of the seven macro-haplotypes in discovery, replication or combined populations (Supplementary Fig. 4). A PHS based on five nuclear genetic loci exhibited a substantial aggregate association with progression to PD dementia in our recent study.5 Here, we calculated the PHS for each patient and found no association between PHS and mtDNA haplogroups (Kruskal–Wallis rank sum test P = 0.59; Supplementary Fig. 5). This suggests that mitochondrial and nuclear genome variants may play independent roles in the cognitive progression of PD.
Since subjects with macro-haplogroups J, T and U# showed a protective effect compared to haplogroup H, we combined these subjects into a super-group (n = 1298) and showed reduced risk for GCI with HR = 0.59 (95% CI 0.48–0.74) and P = 2.42 × 10−6 (Fig. 1D) (macro-haplogroup H as reference) after adjusting for covariates. A linear mixed model analysis indicated that serial MMSE scores in patients with macro-haplogroups J, T and U# declined more slowly over time compared to patients in the common macro-haplogroup H (P = 0.018).
Exploratory analysis of single nucleotide polymorphisms in mtDNA and cognitive decline in PD
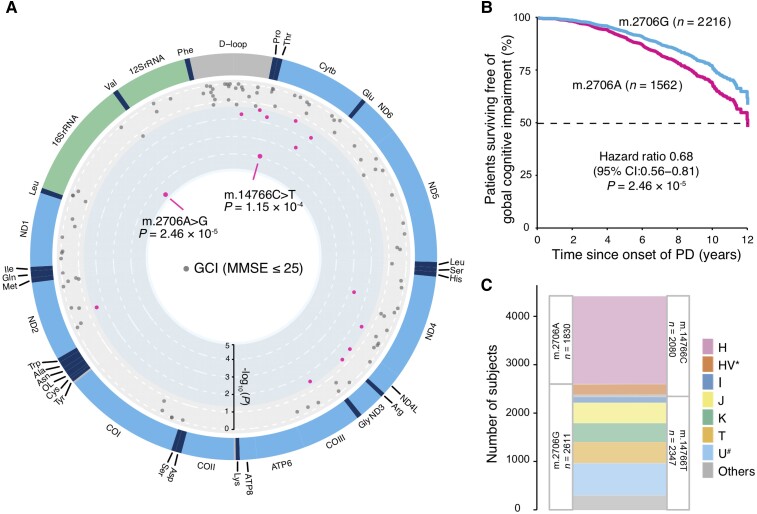
We next carried out an exploratory analysis to investigate the effect of single nucleotide polymorphisms in mtDNA on cognitive impairment during the progression of PD in the combined population (see ‘Methods’ section). We observed that two variants, m.2706A>G and m.14766C>T, were associated with cognitive decline (Fig. 2A). The common m.2706A>G variant (G allele carriers, 58.3% in our cohorts) is located in the 16S rRNA locus. Patients with the m.2706G allele had a reduced risk of developing GCI with an HR = 0.68 (95% CI 0.56–0.81) and P = 2.46 × 10−5 compared to patients with the A allele (Fig. 2B). The common variant m.14766C>T (C allele carrier, 47.5% in our cohorts) codes for an amino acid substitution of an isoleucine for threonine at amino acid site 7 in CYTB. Patients with PD and m.14766T had a reduced risk of developing GCI with a HR = 0.70 (95% CI 0.58–0.84) and P = 1.15 × 10−4 compared to patients carrying the C allele. For m.2706A>G and m.14766C>T, proportional HRs across studies were homogeneous with P = 0.46 (I2 = 0%) and P = 0.44 (I2 = 0.96%), respectively, according to a Cochran’s Q-test for heterogeneity. Associations of these two variants remained significant after considering multiple-testing with both P values lower than the Bonferroni-corrected significance threshold (0.05/102 variants tested = 4.9 × 10−4). Twelve additional variants were associated with cognitive decline during the course of PD with P < 0.05 (Fig. 2A and Table 1).
Figure 2.
mtSNPs associated with cognitive progression in patients with PD. (A) Association plot of SNPs in mtDNA associated with risk of developing global cognitive impairment (dot) in the combined population. The outside labels indicate mitochondrial genes; circular axis from outside to inside represents the value of -log10(P) from 0 to 5; SNPs with P < 0.05 are shown in magenta, while SNPs with P ≥ 0.05 are shown in grey. (B) Covariate-adjusted survival curves for patients with PD carrying mtDNA m.2706G (cyan line) and those with m.2706A (magenta line). m.2706A was used as the reference allele to calculate the HR from the Cox PH analysis; P values from two-sided Wald tests. (C) Overlap between carriers of the m.2706A>G and the m.14766C>T variant. Out of 2611 m.2706G allele carriers and 2347 m.14766T allele carriers, 2342 individuals carried both alleles. Out of 1830 m.2706A allele carriers and 2080 m.14766C allele carriers, 1819 individuals carried both alleles.
Table 1.
Association of mtDNA SNPs with global cognitive impairment during the progression of PD
| rCRS | Effect allele | Alternative allele | P | P a | HR (95% CI) | EAFb | EAF in European | EAF in East Asian | EAF in African |
|---|---|---|---|---|---|---|---|---|---|
| m.2706A>G | G | A | 2.46 × 10−5 | 0.003 | 0.68 (0.56–0.81) | 0.5826 | 0.5746 | 0.9960 | 0.9970 |
| m.14766C>T | T | C | 1.15 × 10−4 | 0.012 | 0.70 (0.58–0.84) | 0.5249 | 0.5169 | 0.9960 | 0.9985 |
| m.11251A>G | G | A | 0.002 | 0.204 | 0.67 (0.52–0.86) | 0.1942 | 0.1610 | 0.0000 | 0.0015 |
| m.15452C>A | A | C | 0.002 | 0.204 | 0.67 (0.52–0.87) | 0.1951 | 0.1610 | 0.0000 | 0.0000 |
| m.15607A>G | G | A | 0.017 | 1 | 0.65 (0.46–0.93) | 0.0984 | 0.0875 | 0.0000 | 0.0015 |
| m.16162A>G | G | A | 0.019 | 1 | 1.95 (1.12–3.40) | 0.0235 | 0.0199 | 0.0417 | 0.0015 |
| m.15928G>A | A | G | 0.021 | 1 | 0.66 (0.47–0.94) | 0.0998 | 0.0875 | 0.0080 | 0.0000 |
| m.11812A>G | G | A | 0.029 | 1 | 0.65 (0.44–0.96) | 0.0787 | 0.0696 | 0.0000 | 0.0045 |
| m.4917A>G | G | A | 0.030 | 1 | 0.66 (0.46–0.96) | 0.0971 | 0.0875 | 0.0000 | 0.0000 |
| m.9477G>A | A | G | 0.031 | 1 | 0.66 (0.46–0.96) | 0.0926 | 0.1392 | 0.0000 | 0.0061 |
| m.10589G>A | A | G | 0.041 | 1 | 1.89 (1.03–3.47) | 0.0110 | 0.0060 | 0.0020 | 0.0530 |
| m.16482A>G | G | A | 0.043 | 1 | 1.62 (1.02–2.59) | 0.0200 | 0.0139 | 0.0020 | 0.0000 |
| m.15218A>G | G | A | 0.045 | 1 | 0.55 (0.31–0.99) | 0.0453 | 0.0437 | 0.0119 | 0.0000 |
| m.10463T>C | C | T | 0.048 | 1 | 0.71 (0.51–1.00) | 0.1030 | 0.0875 | 0.0040 | 0.0000 |
P from the Cox proportional hazards statistic used to estimate the influence of SNP on time (years from onset of PD) to reaching the endpoint of GCI as indicated by a MMSE ≤ 25 in exploratory analyses using the combined population; age at onset of PD, sex, years of education and PHS (including GBA mutation status, APOE ε4 allele haplotype, and rs182987047, rs138073281 and rs8050111) were included as covariates in the Cox analyses. A ‘cohort’ term was included as a random effect. rCRS = revised Cambridge Reference Sequence.
Bonferroni correction based on the result of 102 mtDNA SNPs from combined analysis was performed using the p.adjust function with the ‘Bonferroni’ method in R.
Based on 4491 patients with PD across 15 cohorts. EAF in 503 European, 503 East Asian or 661 African was calculated based on dataset of Phase 1 and 3 of the 1000 Genome Project mitochondrial variants calling by the MToolBox pipeline. EAF = Effect allele frequency.
Both m.2706A and m.14766C are largely specific to the H or HV* haplogroup. The alternative alleles m.2706G and m.14766T occur in other haplogroups (Fig. 2C). These results are consistent with our haplogroup analysis as patients within haplogroups J, T and U# have a lower risk for cognitive progression compared to those with haplogroup H. We found high correlation (r2 = 0.78) of these two common variants in our cohorts and 94.1% of patients carried the same risk/protective alleles (m.2706A/m.14766C or m.2706G/m.14766T). After correcting for the effect of m.2706A>G, conditional Cox PH analysis no longer showed an association of m.14766C>T with cognitive decline [HR = 0.92 (95% CI 0.62–1.38), P = 0.7]. Thus, m.14766C>T was dependent with m.2706A>G in our cohorts.
Age at disease onset, years of education, sex, MMSE at enrollment, Movement Disorder Society Revision of the Unified Parkinson's Disease Rating Scale (MDS-UPDRS III) score at enrollment and depression at enrollment are clinical variables associated with cognitive decline in PD.7 A total of 2629 patients were included in both our previous7 and current studies, and we used these 2376 patients (253 left censored patients were removed) for further analyses (Supplementary material). m.2706A and m.14766C carriers showed significant HRs of 1.48 (95% CI 1.18–1.86, P = 8.21 × 10−4) and 1.38 (95% CI 1.09–1.74, P = 7.23 × 10−3) for risk of progression to GCI, respectively, adjusting for all six clinical predictors (Supplementary Fig. 6).
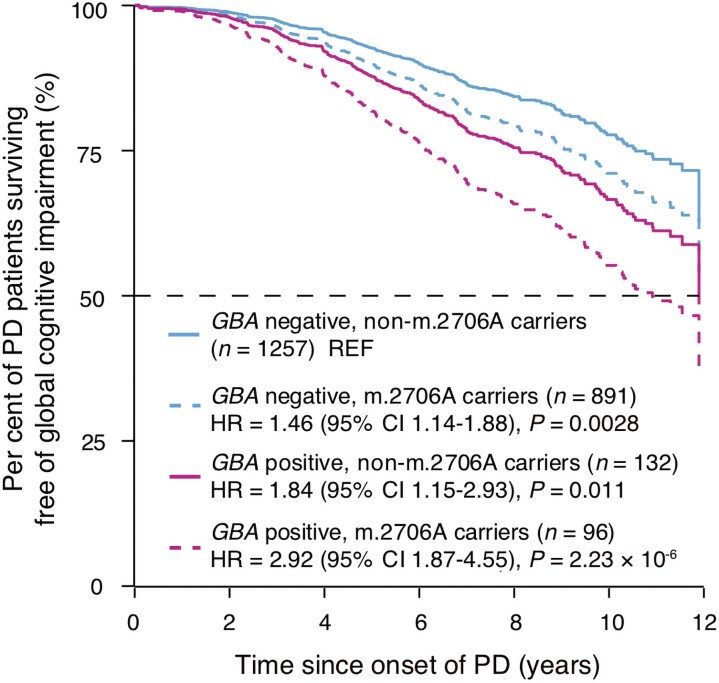
Consistent with our previous genome-wide survival analysis for progression from PD to PD dementia, GBA carriers had an HR of 1.91 (95% 1.39–2.64) with P = 7.76 × 10−5 and APOE ɛ4 carriers had an HR of 1.29 (95% 1.03–1.62) with P = 0.028 for cognitive decline (without accounting for mitochondrial variants; Supplementary Fig. 6). GBA carriers who carried the mitochondrial m.2706A allele (linked to relatively more ‘rapid’ progression compared to the m.2706G allele) had an HR of 2.92 (95% CI, 1.87–4.55, P = 2.23 × 10−6). GBA-positive non-m.2706A carriers had the second highest HR of 1.84 (95% CI 1.15–2.93, P = 0.011), and GBA-negative m.2706A carriers had an HR of 1.46 (95% CI 1.14–1.88, P = 0.0028) compared to patients carrying neither GBA variants nor the m.2706A variant (Fig. 3). Thus, m.2706A>G and GBA variants may have additive effects. Moreover, patients homozygous for the APOE ɛ4 allele and carrying m.2706A had a substantially elevated risk for longitudinal cognitive decline with HR = 5.09 (95% CI 2.04–12.56 P = 0.0005) compared to patients carrying neither the APOE ɛ4 allele nor the m.2706A variant (Supplementary Fig. 7).
Figure 3.
Effects of GBA variants and mtSNPs on global cognitive impairment in patients with PD. Covariate-adjusted survival curves for patients with PD stratified into four subgroups: GBA-negative and non-m.2706A carriers (n = 1257), GBA-negative and m.2706A carriers (n = 891), GBA-positive and non-m.2706G carriers (n = 132) and GBA-positive and m.2706A carriers (n = 96). HR and P-values were calculated adjusting for clinical covariates and study cohort as a random term. The group of GBA-negative and non-m.2706A carriers is denoted as reference group (REF) in this Cox PH analysis.
Discussion
This genetic survival study overall indicates that mitochondrial macro-haplogroups are associated with reduced risk of cognitive disease progression in PD. Post hoc analyses identified the haplogroups J, T and U# as the haplogroups associated with reduced risk compared to the macro-haplogroup H in Parkinson’s patients, but further research is required to definitively identify the contribution and statistical significance of each individual haplogroup. Previous meta-analyses found that the haplogroups J, K and T are associated with reduced susceptibility for PD and the haplogroup H is linked to elevated susceptibility for PD.15
About 41% of patients with PD in this study belong to the macro-haplogroup H, the most common genotype in Europeans. The European mtDNA haplogroup H is associated with a higher survival ratio after sepsis,19 but is linked to higher risk of developing PD in late life.15 On the flip side, our findings are consistent with a relatively more deleterious effect of haplogroup H on the progression of PD compared to haplogroups J, T and U#. This may represent an evolutionary trade-off,20 whereby genetic variants that increase the chance of surviving early-life illness such as sepsis might contribute to pathogenic events later in life.20
Alzheimer's disease-associated plaques and tangles are found in a substantial proportion of brains with of patients with PD dementia in addition to Lewy bodies.21 H and HV are risk haplogroups for Alzheimer’s disease,22 while the JT haplogroup was protective in a prior study23; evidence for the other haplogroups (K, J, T, U) is limited and controversial (e.g. J22,24; Supplementary Table 5). This is also consistent with our study, where H carriers had a relatively more rapid cognitive progression compared to the protective haplogroups J, T and U#.
Two common significant mtSNPs showed effects on the risk of global cognitive impairment and are related to the haplogroups (Fig. 2C). The common m.2706A>G variant, located at 16S rRNA gene, is close to the ribosomal peptidyl transferase center, and might be relevant to many diseases, such as mitochondrial encephalopathy, lactic acidosis and stroke-like episodes (MELAS), Alzheimer’s disease and PD.25 This variant can induce substantial alterations in the mitochondrial 16S rRNA secondary structure.26 The m.14766C variant might increase the risk for late-onset Alzheimer’s disease23 consistent with our findings. Interestingly, contrary to our data, m.2706G was associated with faster cognitive ageing in a large longitudinal cohort of African Americans but not Caucasian Americans.27
Our study is limited in sample size and statistical power. P values for individual haplogroups were not adjusted for multiple testing. Another limitation of this study is that we evaluated the effects of mitochondrial genetic variants in patients with European ancestry only. The mtSNPs (m.2706A>G or m.14766C>T) are rare in populations from East Asia or Africa (Table 1). Further studies in other populations are urgently needed because of differences in mtDNA haplogroups, considering that more than 60% of PD patients are expected to live in the Western Pacific Region by 2030,28 most of them belonging to haplogroups A, B, C, D, F and G. Moreover, replication of our exploratory findings in additional longitudinal patient populations of European ancestry is needed.
This study suggests that mitochondrial genotypes may not be innocent bystanders in the progression of PD, but might play a role in modulating disease progression. Our study provides evidence for the role of mitochondrial haplogroups in the progression of PD towards Lewy body dementia, and this association appears independent of GBA and APOE.
Mitochondrial dysfunction14 and alpha-synuclein accumulation are two pathologically and biologically linked culprits of PD. Alpha-synuclein triplication causes mitochondrial bioenergetics dysfunction.29 Conversely, the mitochondrial toxin rotenone leads to alpha-synuclein accumulation.30 Taken together with our new findings, this body of evidence suggests that mitochondria might play a role not only in the onset, but also in the progression of Parkinson’s disease.
Supplementary Material
Acknowledgements
We thank Mr Ofer Nemirovsky for his invaluable support, encouragement, insights and dedication to accelerating PD research. We thank all study participants, their families, and friends for their support and participation, and our study coordinators for making this work possible. For each individual cohorts, acknowledgements and funding are listed in the Supplementary material. For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising from this submission.
Appendix 1
International Genetics of Parkinson Disease Progression (IGPP) Consortium
Ganqiang Liu, PhD, Rebecca R. Valentino, PhD, Jiajie Peng, PhD, Zhixiang Liao, MS, Joseph J. Locascio, PhD, Jean-Christophe Corvol, MD, Xianjun Dong, PhD, Jodi Maple-Grødem, PhD, Meghan C. Campbell, PhD, Alexis Elbaz, MD, Suzanne Lesage, PhD, Alexis Brice, MD, Graziella Mangone, MD, John H. Growdon, MD, Albert Y. Hung, MD, PhD, Michael A. Schwarzchild, MD, PhD, Michael T. Hayes, MD, Anne-Marie Wills, MD, Todd M. Herrington, MD, Bernard Ravian, MD, Ira Shoulson, MD, Pille Taba, MD, PhD, Sulev Kõks, MD, PhD, Thomas G. Beach, MD, PhD, Florence Cormier-Dequaire, MD, Guido Alves, PhD, Ole-Bjørn Tysnes, MD, Joel S. Perlmutter, MD, Peter Heutink, PhD, Jacobus J. van Hilten, MD, Meike Kasten, MD, Brit Mollenhauer, MD, Claudia Trenkwalder, MD, Christine Klein, MD, Roger A. Barker, PhD, Caroline H. Williams-Gray, PhD, Johan Marinus, PhD and Clemens R. Scherzer, MD
Contributor Information
Ganqiang Liu, Neurobiology Research Center, School of Medicine, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, China.
Chunming Ni, Neurobiology Research Center, School of Medicine, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, China.
Jiamin Zhan, Neurobiology Research Center, School of Medicine, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, China.
Weimin Li, Neurobiology Research Center, School of Medicine, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, China.
Junfeng Luo, Neurobiology Research Center, School of Medicine, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, China.
Zhixiang Liao, APDA Center for Advanced Parkinson Research, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115, USA; Neurogenomics Lab, Harvard Medical School, Brigham and Women's Hospital, Boston, MA 02115, USA.
Joseph J Locascio, APDA Center for Advanced Parkinson Research, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115, USA; Neurogenomics Lab, Harvard Medical School, Brigham and Women's Hospital, Boston, MA 02115, USA; Department of Neurology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Wenbiao Xian, Department of Neurology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong 510080, China.
Ling Chen, Department of Neurology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong 510080, China.
Zhong Pei, Department of Neurology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong 510080, China.
Jean-Christophe Corvol, Sorbonne Université, Institut du Cerveau – Paris Brain Institute - ICM, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Assistance Publique Hôpitaux de Paris, Département de Neurologie et de Génétique, Hôpital Pitié-Salpêtrière, F-75013 Paris, France.
Jodi Maple-Grødem, The Norwegian Centre for Movement Disorders, Stavanger University Hospital, 4068 Stavanger, Norway; Department of Chemistry, Bioscience and Environmental Engineering, University of Stavanger, 4021 Stavanger, Norway.
Meghan C Campbell, Department of Neurology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Alexis Elbaz, Paris-Saclay University, UVSQ, Inserm, Gustave Roussy, ‘Exposome and Heredity’ Team, CESP, F94805 Villejuif, France.
Suzanne Lesage, Sorbonne Université, Institut du Cerveau – Paris Brain Institute - ICM, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Assistance Publique Hôpitaux de Paris, Département de Neurologie et de Génétique, Hôpital Pitié-Salpêtrière, F-75013 Paris, France.
Alexis Brice, Sorbonne Université, Institut du Cerveau – Paris Brain Institute - ICM, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Assistance Publique Hôpitaux de Paris, Département de Neurologie et de Génétique, Hôpital Pitié-Salpêtrière, F-75013 Paris, France.
Albert Y Hung, Department of Neurology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Michael A Schwarzschild, Department of Neurology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Michael T Hayes, Department of Neurology, Brigham and Women’s Hospital, Boston, MA 02115, USA.
Anne-Marie Wills, Department of Neurology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Bernard Ravina, Praxis Precision Medicines, Cambridge, MA 02142, USA.
Ira Shoulson, Department of Neurology, Center for Health and Technology, University of Rochester, Rochester, NY 14642, USA.
Pille Taba, Department of Neurology and Neurosurgery, Institute of Clinical Medicine, University of Tartu, Tartu 50406, Estonia; Neurology Clinic, Tartu University Hospital, Tartu 50406, Estonia.
Sulev Kõks, Centre for Molecular Medicine and Innovative Therapeutics, Murdoch University, Murdoch, Perth, WA 6150, Australia; Perron Institute for Neurological and Translational Science, Nedlands, WA 6009, Australia.
Thomas G Beach, Banner Sun Health Research Institute, Sun City, AZ 85351, USA.
Florence Cormier-Dequaire, Sorbonne Université, Institut du Cerveau – Paris Brain Institute - ICM, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Assistance Publique Hôpitaux de Paris, Département de Neurologie et de Génétique, Hôpital Pitié-Salpêtrière, F-75013 Paris, France.
Guido Alves, The Norwegian Centre for Movement Disorders, Stavanger University Hospital, 4068 Stavanger, Norway; Department of Chemistry, Bioscience and Environmental Engineering, University of Stavanger, 4021 Stavanger, Norway; Department of Neurology, Stavanger University Hospital, 4068 Stavanger, Norway.
Ole-Bjørn Tysnes, Department of Neurology, Haukeland University Hospital, 5020 Bergen, Norway; Department of Clinical Medicine, University of Bergen, 5020 Bergen, Norway.
Joel S Perlmutter, Department of Neurology, Washington University School of Medicine, St. Louis, MO 63110, USA; Departments of Radiology and Neuroscience, Washington University School of Medicine, St. Louis, MO 63110, USA; Program of Physical Therapy and Program of Occupational Therapy, Washington University School of Medicine, St. Louis, MO 63110, USA.
Peter Heutink, German Center for Neurodegenerative diseases (DZNE), 72076 Tübingen, Germany.
Jacobus J van Hilten, Department of Neurology, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.
Roger A Barker, John Van Geest Centre for Brain Repair, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0PY, UK; Wellcome—MRC Cambridge Stem Cell Institute, University of Cambridge, Cambridge CB2 0AW, UK.
Caroline H Williams-Gray, John Van Geest Centre for Brain Repair, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0PY, UK.
Clemens R Scherzer, APDA Center for Advanced Parkinson Research, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115, USA; Neurogenomics Lab, Harvard Medical School, Brigham and Women's Hospital, Boston, MA 02115, USA; Department of Neurology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA; Department of Neurology, Brigham and Women’s Hospital, Boston, MA 02115, USA.
International Genetics of Parkinson Disease Progression (IGPP) Consortium:
Ganqiang Liu, Rebecca R Valentino, Jiajie Peng, Zhixiang Liao, Joseph J Locascio, Jean-Christophe Corvol, Xianjun Dong, Jodi Maple-Grødem, Meghan C Campbell, Alexis Elbaz, Suzanne Lesage, Alexis Brice, Graziella Mangone, John H Growdon, Albert Y Hung, Michael A Schwarzchild, Michael T Hayes, Anne-Marie Wills, Todd M Herrington, Bernard Ravian, Ira Shoulson, Pille Taba, Sulev Kõks, Thomas G Beach, Florence Cormier-Dequaire, Guido Alves, Ole-Bjørn Tysnes, Joel S Perlmutter, Peter Heutink, Jacobus J van Hilten, Meike Kasten, Brit Mollenhauer, Claudia Trenkwalder, Christine Klein, Roger A Barker, Caroline H Williams-Gray, Johan Marinus, and Clemens R Scherzer
Funding
G.L. is supported by the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (22ykqb07), National Natural Science Foundation of China (31900475), Guangdong Basic and Applied Basic Research Foundation (2022A1515011440), Shenzhen Fundamental Research Program (JCYJ20190807161601692) and The Pearl River Talent Recruitment Program (2019QN01Y139). C.R.S. is supported by NIH grants NINDS/NIA R01NS115144, U01NS095736, U01NS100603, and the American Parkinson Disease Association Center for Advanced Parkinson Research. The study was made possible in part by philanthropic support for Illumina MEGA chip genotyping (to Brigham and Women's Hospital and C.R.S.).
Competing interests
Outside this work, C.R.S. has served as consultant, scientific collaborator or on scientific advisory boards for Sanofi, Berg Health, Pfizer and Biogen and has received grants from NIH, U.S. Department of Defense, American Parkinson Disease Association, and the Michael J Fox Foundation (MJFF). C.R.S. is named as co-inventor on U.S. patent applications held by Brigham and Women’s Hospital relating to therapeutics; biomarkers; and polygenic scores for neurodegenerative diseases. M.A.S. has no conflict of interest related to this work. Outside this work, M.A.S. has received grants from NINDS, DoD, MJFF, the Parkinson’s Foundation and Farmer Family Foundation and has served as a consultant to commercial programs: Eli Lilly & Co (data monitoring committee), Prevail Therapeutics (scientific advisory board) and Denali Therapeutics (scientific advisory board); and via the Parkinson Study Group to nQ Medical (scientific advisory board), Chase Therapeutics (scientific advisory board) and Partner Therapeutics (scientific advisory board). A.-M.W. has received research funding from the ALS Association, the Parkinson's Foundation, has participated in clinical trials funded by Acorda, Biogen, Bristol-Myers Squibb, Sanofi/Genzyme, Pfizer and Abbvie and received consultant payments from Mitsubishi Tanabe and Accordant. J.-C.C. has no conflict of interest related to this work. Outside this work, J.C.C. has received honoraria for consulting in advisory boards for Abbvie, Actelion, Air Liquide, Biogen, BMS, BrainEver, Clevexel, Denali, Pfizer, Theranexus and Zambon. B.R. is an employee of and holds equity in Praxis Precision Medicines and is an advisor for Caraway Therapeutics and Brain Neurotherapy Bio. I.S. is the Principal Investigator of a MJFF Computational Science Grant (2017–19). S.K. is supported by Multiple Sclerosis of Western-Australia (MSWA) and the Perron Institute. P.H. is a Scientific Advisor of Neuron23. T.G.B has no conflict of interest related to this work. Outside this work, T.G.B. has received grants from NIA, NINDS, MJFF and the State of Arizona, has served as a scientific advisory board member (with stock options) and consultant to Vivid Genomics, Inc. and has received honoraria from the World PD Coalition. J.J.v.H. has no conflict of interest related to this work. Outside this work, J.J.v.H. has received grants from the Alkemade-Keuls Foundation, Stichting Parkinson Fonds, Parkinson Vereniging, The Netherlands Organisation for Health Research and Development, The Netherlands Organisation for Scientific Research, Hersenstichting, AbbVie, MJFF and research support from Hoffmann-La-Roche, Lundbeck and the Centre of Human Drug Research. R.A.B. has no conflict of interest related to this work. Outside this work, R.A.B. received consultancy monies from LCT, FCDI, Novo Nordisk, Cellino, Sana, UCB; received royalties from Wiley and Springer-Nature; grant funding from CPT, NIHR Cambridge Biomedical Research Centre (146281), MRC, Wellcome (203151/Z/16/Z) and Rosetrees Trust (A1519 M654). C.H.W.-G. has no conflict of interest related to this work. C.H.W.-G. is supported by a RCUK/UKRI Research Innovation Fellowship awarded by the Medical Research Council (MR/R007446/1) and the NIHR Cambridge Biomedical Research Centre and received grant support from MJFF, the Evelyn Trust, the Cure Parkinson’s Trust, Parkinson’s UK, the Rosetrees Trust and the Cambridge Centre for Parkinson-Plus. C.H.W.-G. has received honoraria from Lundbeck and Profile Pharma Ltd and consultancy payments from Modus Outcomes and Evidera. The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Poewe W, Seppi K, Tanner CM, et al. Parkinson disease. Nat Rev Dis Primers. 2017;3:17013. [DOI] [PubMed] [Google Scholar]
- 2. Yang W, Hamilton JL, Kopil C, et al. Current and projected future economic burden of Parkinson's disease in the U.S. NPJ Parkinsons Dis. 2020;6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greenland JC, Williams-Gray CH, Barker RA. The clinical heterogeneity of Parkinson's disease and its therapeutic implications. Eur J Neurosci. 2019;49:328–338. [DOI] [PubMed] [Google Scholar]
- 4. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu G, Peng J, Liao Z, et al. Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson's disease. Nat Genet. 2021;53:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu G, Boot B, Locascio JJ, et al. Specifically neuropathic gaucher's mutations accelerate cognitive decline in Parkinson's. Annals of neurology. 2016;80:674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu G, Locascio JJ, Corvol JC, et al. Prediction of cognition in Parkinson's disease with a clinical-genetic score: a longitudinal analysis of nine cohorts. Lancet Neurol. 2017;16:620–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iwaki H, Blauwendraat C, Leonard HL, et al. Genomewide association study of Parkinson's disease clinical biomarkers in 12 longitudinal patients’ cohorts. Mov Disord. 2019;34:1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. [DOI] [PubMed] [Google Scholar]
- 10. Giannoccaro MP, La Morgia C, Rizzo G, Carelli V. Mitochondrial DNA and primary mitochondrial dysfunction in Parkinson's disease. Mov Disord. 2017;32:346–363. [DOI] [PubMed] [Google Scholar]
- 11. Lin MT, Cantuti-Castelvetri I, Zheng K, et al. Somatic mitochondrial DNA mutations in early Parkinson and incidental Lewy body disease. Annals of neurology. 2012;71:850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dölle C, Flønes I, Nido GS, et al. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat Commun. 2016;7:13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng B, Liao Z, Locascio JJ, et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med. 2010;2:52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hudson G, Nalls M, Evans JR, et al. Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease. Neurology. 2013;80:2042–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weissensteiner H, Pacher D, Kloss-Brandstätter A, et al. Haplogrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–W63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dubois B, Burn D, Goetz C, et al. Diagnostic procedures for Parkinson's disease dementia: recommendations from the movement disorder society task force. Mov Disord. 2007;22:2314–2324. [DOI] [PubMed] [Google Scholar]
- 18. Yonova-Doing E, Calabrese C, Gomez-Duran A, et al. An atlas of mitochondrial DNA genotype-phenotype associations in the UK biobank. Nat Genet. 2021;53:982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baudouin SV, Saunders D, Tiangyou W, et al. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. [DOI] [PubMed] [Google Scholar]
- 20. Benton ML, Abraham A, LaBella AL, Abbot P, Rokas A, Capra JA. The influence of evolutionary history on human health and disease. Nat Rev Genet. 2021;22:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aarsland D, Batzu L, Halliday GM, et al. Parkinson disease-associated cognitive impairment. Nat Rev Dis Primers. 2021;7:47. [DOI] [PubMed] [Google Scholar]
- 22. Maruszak A, Canter JA, Styczyńska M, Zekanowski C, Barcikowska M. Mitochondrial haplogroup H and Alzheimer's disease–is there a connection? Neurobiol Aging. 2009;30:1749–1755. [DOI] [PubMed] [Google Scholar]
- 23. Maruszak A, Safranow K, Branicki W, et al. The impact of mitochondrial and nuclear DNA variants on late-onset Alzheimer's disease risk. J Alzheimers Dis. 2011;27:197–210. [DOI] [PubMed] [Google Scholar]
- 24. Swerdlow RH, Hui D, Chalise P, et al. Exploratory analysis of mtDNA haplogroups in two Alzheimer's longitudinal cohorts. Alzheimer's & Dementia. 2020;16:1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katzman SM, Strotmeyer ES, Nalls MA, et al. Mitochondrial DNA sequence variation associated with peripheral nerve function in the elderly. J Gerontol A Biol Sci Med Sci. 2015;70:1400–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rovcanin B, Jancic J, Samardzic J, et al. In silico model of mtDNA mutations effect on secondary and 3D structure of mitochondrial rRNA and tRNA in leber's hereditary optic neuropathy. Exp Eye Res. 2020;201:108277. [DOI] [PubMed] [Google Scholar]
- 27. Yen K, Wan J, Mehta HH, et al. Humanin prevents age-related cognitive decline in mice and is associated with improved cognitive age in humans. Sci Rep. 2018;8:14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim SY, Tan AH, Ahmad-Annuar A, et al. Parkinson's disease in the western pacific region. Lancet Neurol. 2019;18:865–879. [DOI] [PubMed] [Google Scholar]
- 29. Zambon F, Cherubini M, Fernandes HJR, et al. Cellular alpha-synuclein pathology is associated with bioenergetic dysfunction in Parkinson's iPSC-derived dopamine neurons. Hum Mol Genet. 2019;28:2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotype and clinical data for the Parkinson’s progression markers initiative (PPMI) included in this study are publicly available upon request to ppmi@loni.usc.edu through a PPMI Whole Genome Sequencing Data Agreement. Clinical data for the Parkinson’s disease biomarker program (PDBP) included in this study are publicly available through https://pdbp.ninds.nih.gov. Clinical longitudinal data and genotyping data for the other cohorts included are accessible through appropriate data sharing agreements that protect patient privacy with the institutions that conducted or are conducting study consents and clinical assessments under local institutional review board approvals.