Abstract
We have used a quantitative RNase protection assay to characterize the relative accumulation and abundance of individual adeno-associated virus type 2 (AAV) RNAs throughout the course of AAV-adenovirus coinfections and preinfections. We have demonstrated that there is a previously unrecognized temporal order to the appearance of AAV RNAs. First, unspliced P5-generated transcripts, which encode Rep78, were detectable prior to the significant accumulation of other AAV RNAs. Ultimately, as previously demonstrated, P19-generated products accumulated to levels greater than those generated from P5, and P40-generated transcripts predominated in the total RNA pool. Second, the percentage of each class of AAV RNA that was spliced increased during infection, and the degree of this increase was different for the P5/P19 products than for those generated by P40. At late times postcoinfection, approximately 90% of P40 products, but only approximately 50% of RNAs generated by P5 and P19, were seen to be spliced; thus, the AAV intron was removed to different final levels from these different RNA species. We have shown that each of the AAV RNAs is quite stable; the majority of each RNA species persisted 6 h after treatment with actinomycin D. Quantification of the accumulation of individual AAV RNAs, over intervals during which degradation was negligible, allowed us to infer that at late times during infection the relative strength of P5, P19, and P40 was approximately 1:3:18, respectively, consistent with the steady-state accumulated levels of the RNAs generated by each promoter. All AAV RNAs exited to the cytoplasm with similar efficiencies in the presence or absence of adenovirus; however, adenovirus coinfection appeared to stimulate total splicing of AAV RNAs and the relative use of the downstream intron acceptor. Our results confirm and extend previous observations concerning the appearance and processing of AAV-generated RNAs.
The human parvovirus adeno-associated virus type 2 (AAV-2) is a single-stranded DNA virus which requires coinfection of helper virus for establishment of a productive infection (see reference 1 for a review). The genome of approximately 4.7 kb is flanked by palindromic inverted terminal repeats that are required for viral replication and site-specific integration into human chromosome 19 in the absence of helper virus or cellular stress (2). RNAs generated from the three viral promoters (P5, P19, and P40) give rise to the two large replication proteins (Rep78 and Rep68), two small replication proteins (Rep52 and Rep40), and three capsid proteins (VP1, VP2, and VP3), respectively (1, 2). Each AAV pre-mRNA class contains an intron which, when excised, utilizes a single splice donor site and either one of two splice acceptor sites. Rep78 and Rep52 are encoded by intron-containing RNAs, while Rep68 and Rep40 are derived from spliced transcripts. Although use of the different splice acceptor sites would be predicted to result in Rep68 and Rep40 proteins with different carboxyl termini, these variants have not been demonstrated, and so it is unclear whether they have distinct roles during infection (1, 26). All AAV capsid proteins are apparently translated from spliced templates (26).
Expression and replication of the AAV genome is controlled by both the AAV-encoded Rep proteins and coinfecting helper virus (commonly adenovirus [Ad] or herpesvirus) (1, 2, 3, 16, 29). Rep78 and Rep68 have been demonstrated to both positively and negatively regulate the three AAV promoters (11, 12, 19, 27, 28). They have also been suggested to influence AAV gene expression posttranscriptionally by specifically down-regulating translation of AAV RNAs by an unknown mechanism (27).
A number of Ad proteins that stimulate AAV replication during coinfection have been identified, and in most cases their roles have been partially elucidated (see references 1 and 2 for reviews). The Ad E1A protein, required for the transcription of other Ad early genes, also induces the expression of the AAV genome. E1A participates in transactivation of the AAV P5 promoter through its interaction with at least two cellular proteins, MLTF and YY1 (24). The Ad E2A protein (the Ad DNA-binding protein), although not absolutely required for AAV infection, has been shown to stimulate AAV particle formation and AAV promoter activity (8, 22, 30). The requirement for the Ad VA1 gene product seems to be in facilitating AAV protein synthesis during Ad infection (1). The Ad E1B55k and E4orf6 proteins are involved in multiple aspects of Ad-cell interactions, including (i) stimulating cellular transformation by binding and inactivation of P53 and (ii) promoting the transport of late Ad transcripts from the nucleus while concomitantly blocking the export of cellular mRNA (7, 9, 13, 31). The E1B55k-E4orf6 complex has been shown to influence the timely cytoplasmic accumulation of AAV RNAs during coinfection with Ad (22). The E4orf6 protein has also been shown to be necessary for efficient processive extension of the single-strand AAV recombinant genome to its duplex form (8).
Previous work has addressed the general relative steady-state levels of the major classes of AAV RNAs (12, 26), the relative usage of the different intron splice acceptor sites (26), and the general stability of AAV RNAs (5) and has suggested that Ad coinfection has stimulating effects on the splicing and export of AAV RNA (22, 26, 27). Together these studies have generated our current view of AAV RNA biogenesis and processing. To and more precisely characterize these important aspects of AAV infection, we have developed and applied an RNase protection assay (RPA) for the quantitative detection of individual AAV RNA species throughout infection. In this report we have characterized the relative individual transcript abundance throughout the course of AAV-Ad coinfections and preinfections. We have found a previously unrecognized temporal order to the appearance of AAV RNAs: P5-generated transcripts were detectable prior to the significant accumulation of other AAV RNAs. In addition, the relative ratio of spliced to unspliced AAV RNAs increases during infection, and to different degrees for each class of RNA. Unspliced RNAs generated by the P5 and P19 promoters accumulate to greater relative levels, in both total and cytoplasmic RNA pools, than do those generated from P40. We have shown that each of the AAV RNAs are quite stable and thereby have inferred the relative strength of the three AAV promoters during infection. We have also shown that Ad stimulates both the total levels of AAV splicing and the relative usage of the downstream acceptor (A2). Our results have confirmed and extended previous observations concerning the biogenesis and processing of AAV RNA in a more quantified way as a prelude for further investigation.
MATERIALS AND METHODS
Plasmids, cells, and viruses.
293-rAAV cells were isolated from a clonal 293 cell population with increased sensitivity to AAV infection and were a generous gift from R. J. Samulski (University of North Carolina—Chapel Hill). HeLa cells were purchased from the American Type Culture Collection (Manassas, Va.). AAV-2 and Ad type 5 (dl309) were also generous gifts of R. J. Samulski. Herpes simplex virus type 1F (HSV-1F) was a kind gift from J. Mitchell (University of Missouri—Columbia). Infections were consistently performed at multiplicities of infection (MOIs) of 10 for AAV and 2 to 5 for helper virus. SSV9 (pSub201) was a gift from R. J. Samulski (21). CMV (cytomegalovirus)-AAV was generated by cloning the 4,306-nucleotide (nt) XbaI fragment (nt 184 to 4490) from the infectious clone of AAV-2 DNA, SSV9, into the pCI cloning vector (Promega, Madison, Wis.). Transient transfections were performed using Lipofectamine and Plus reagents as recommended by the supplier (Gibco BRL, Grand Island, N.Y.). AAV and Ad stocks were prepared by freeze-thaw lysis of virus-containing cells and purified by cesium chloride ultracentrifugation, and fractions were pooled by dot-blot identification as described elsewhere (14).
RNA isolation and characterization.
Total RNA was prepared 24 to 48 h after infection or transfection, as noted, using guanidine isothiocyanate lysis and cesium chloride ultracentrifugation purification as described elsewhere (17). Cytoplasmic RNA was prepared by depleting cytosol of intact nuclei by centrifugation as described elsewhere (23). Cytoplasmic RNA was shown to be free of nuclear precursor rRNA by gel electrophoresis as described previously (17). Poly(A)+ RNA was prepared from total RNA stocks using a Dynabeads mRNA purification kit as recommended by the supplier (Dynal, Lake Success, N.Y.). The RP, SB, and DH probes were generated by cloning AAV restriction fragments from nt 1767 to 1958, 810 to 965, and 2194 to 2395, respectively, into the T7-driven pGEM-3Z transcription vector (Promega) RPAs were performed on 10 μg of total or cytoplasmic RNA, in substantial probe excess, as described elsewhere (23). RPA analysis was quantified using a Molecular Imager FX and Quantity One imaging software (Bio-Rad, Hercules, Calif.). Relative molar ratios of individual RNAs were determined after adjusting for the number of labeled uridines in each protected fragment as previously described (23).
RNA stability and AAV promoter strength.
AAV RNA stability was determined by coinfecting 293 cells with AAV and Ad for 12 h, adding actinomycin D (40 mM) (31), and isolating total RNA at approximately 0, 0.5, 3, and 6 h posttreatment. The efficiency of actinomycin D to terminate nascent RNA synthesis was determined following addition of 32P in phosphate-free medium immediately after actinomycin D treatment. Actinomycin D treatment reduced 32P incorporation more than 200-fold (data not shown). AAV RNA stability over 6 h was determined as a percentage of 0-h AAV-specific RNA remaining at each time point. For assays of relative AAV promoter strength, 293 cells preinfected with Ad for 12 h were infected with AAV, and total RNA was isolated at hourly intervals. Data were plotted using the Axum6 graph software; lines were drawn using the program's best-fit algorithm (Mathsoft, Seattle, Wash.). Relative promoter rates were calculated by averaging the slopes determined over 3-h intervals where RNA degradation was demonstrated to be negligible.
RESULTS
Temporal accumulation of AAV RNA during helper virus coinfection and preinfection.
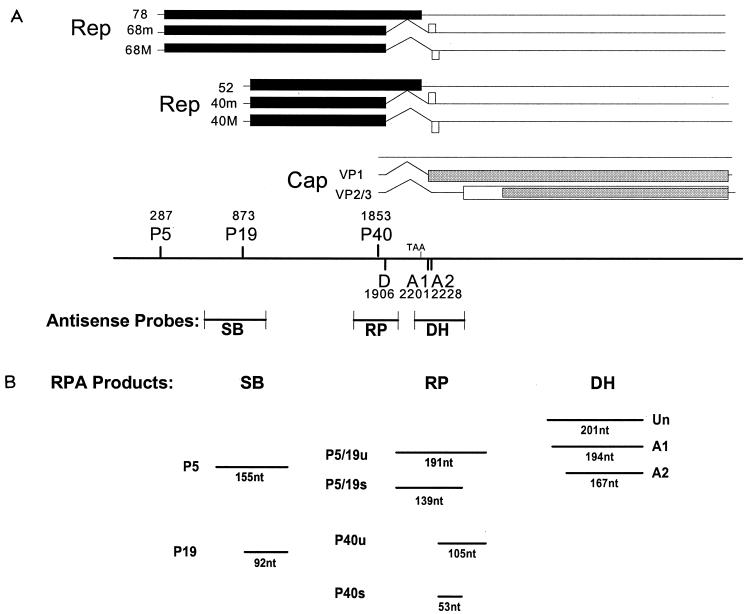
Our first objective was to determine a detailed profile of the temporal accumulation of AAV RNA during AAV-Ad coinfection. For this purpose, we developed a quantitative RPA which could distinguish the various spliced and unspliced RNAs generated from the three viral promoters. Although no single RPA probe could be developed to usefully compare all AAV RNAs simultaneously, the use of three probes allowed comparison between almost all AAV RNA species at various points during infection. (The relative ratio of spliced to unspliced P5- and P19-generated RNAs could not be individually determined in our assay.) Figure 1A depicts the three probes used and their positions relative to the AAV genome. The SB probe spans P19 and is protected differentially by transcripts generated from P5 versus those derived from P19. The RP probe spans a region from upstream of P40 to within the intron and allows comparative quantification of spliced and unspliced transcripts from P40 and from P5 and P19 (P5/P19). The DH probe spans the splice acceptor sites and detects both unspliced RNA and splice acceptor usage for the total AAV spliced RNA population. The various expected nuclease-resistant probe fragments are diagramed in Fig. 1B.
FIG. 1.
(A) Genetic map of AAV. Transcripts and protein-encoding regions for the three AAV promoters are shown. Promoters and intron donor and acceptor sites are mapped with their AAV nucleotide designations. Antisense AAV RNA probes used in our assays are positioned relative to the AAV genome and are described in Materials and Methods. (B) Schematic representation of probe fragments protected by AAV transcripts. Individual fragments are diagramed in their approximate relative positions when separated on a 6% denaturing polyacrylamide gel. U, unspliced; s, sliced.
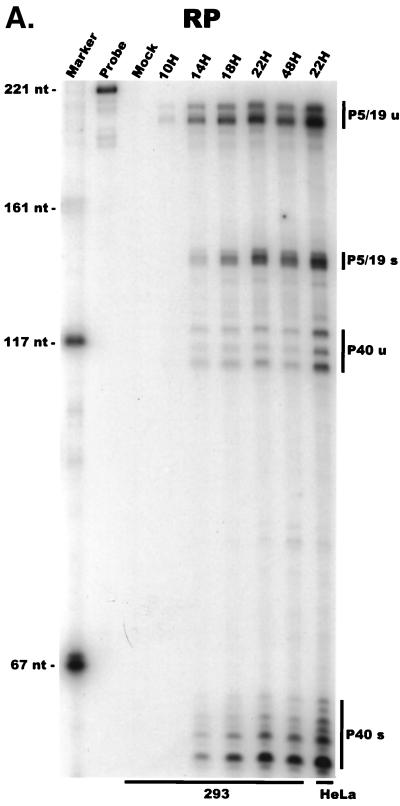
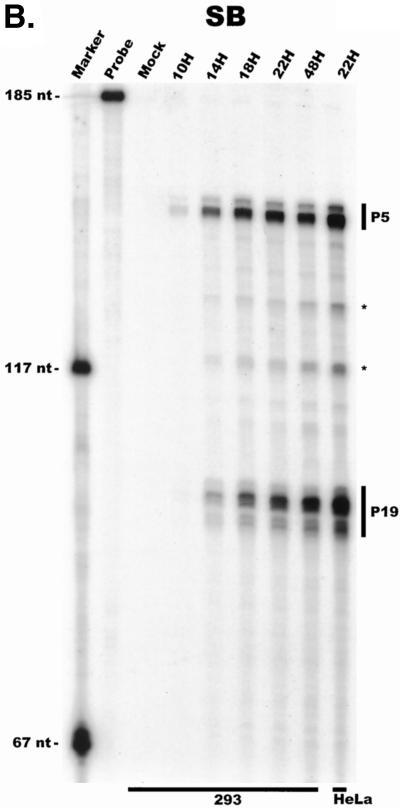
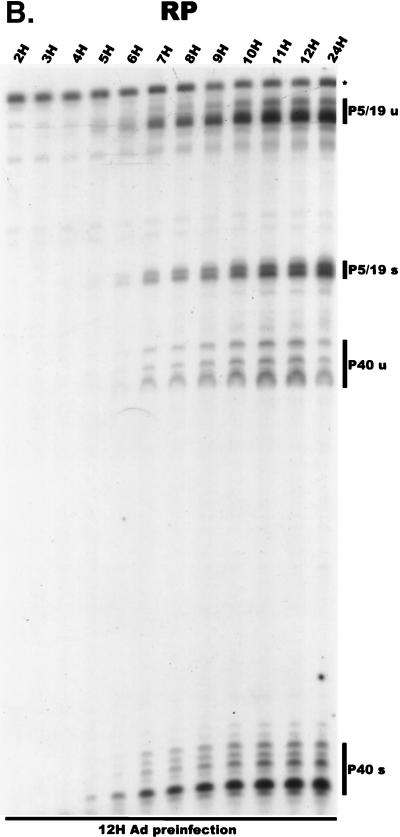
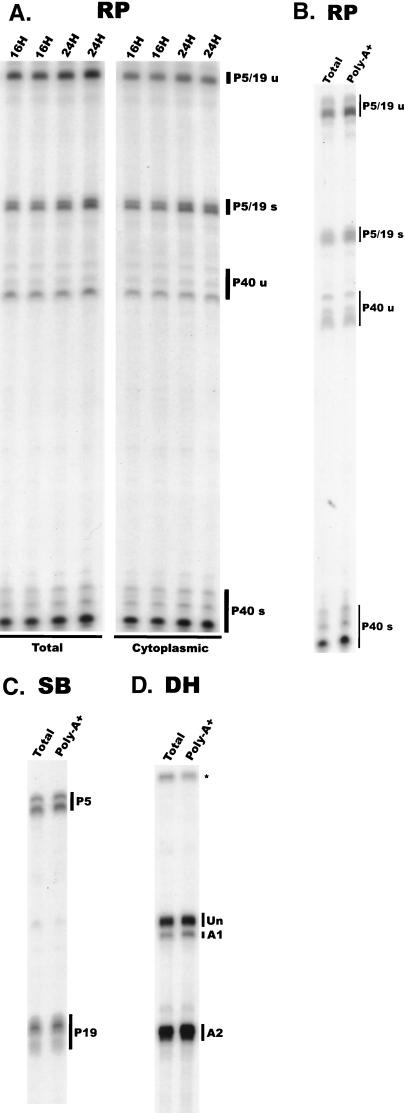
For our initial analysis, total RNA harvested at various times following coinfection of 293 cells with AAV and Ad was subjected to quantitative RPA using the individual probes described above. All predicted RNA species were readily detectable in these experiments (Fig. 2A to C; multiple bands protected by P40- and P19-generated products using the RP [Fig. 2A] and SB [Fig. 2B] probes, respectively, likely reflect multiple sites of RNA initiation from these promoters). Our results indicated the following points. First, although the characteristics of temporal accumulation varied slightly between experiments, low levels of accumulated P5 transcripts could be detected prior to the significant accumulation of products of either the P19 or P40 promoter (Fig. 2A and B). In the experiment shown in Fig. 2A, RPA using the RP probe demonstrated that at 10 h postinfection, unspliced P5/19 products could be detected prior to those generated from P40. Analysis of the same RNA sample using the SB probe demonstrated that at this early time point P5 products were preferentially detected (Fig. 2B). Thus, unspliced products of the P5 promoter (which encode Rep78) accumulate to significant levels at early times during coinfection with Ad prior to other AAV RNAs. As infection progressed, P19-generated transcripts eventually accumulated to approximately two- to threefold-greater steady-state levels than P5 products (Fig. 2B), and as expected (27), P40 transcripts accumulated to predominate in the total AAV RNA pool (Fig. 2A). Combined data generated from multiple experiments with the RP and SB probes demonstrated that at late times during infection the relative steady-state accumulated level of viral transcripts generated from P5, P19, and P40 was approximately 1:3:20, respectively, which is in good agreement with previous results obtained using other methods (27).
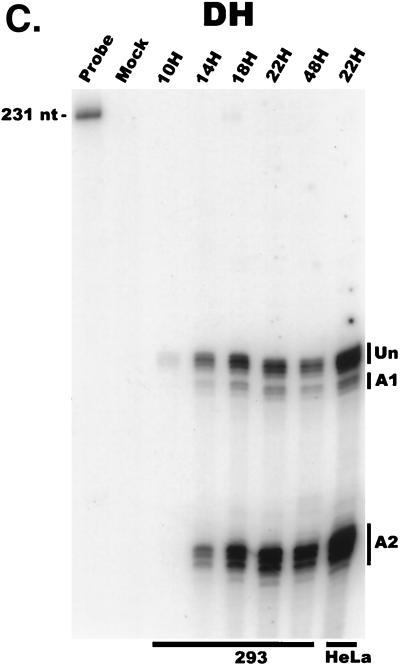
FIG. 2.
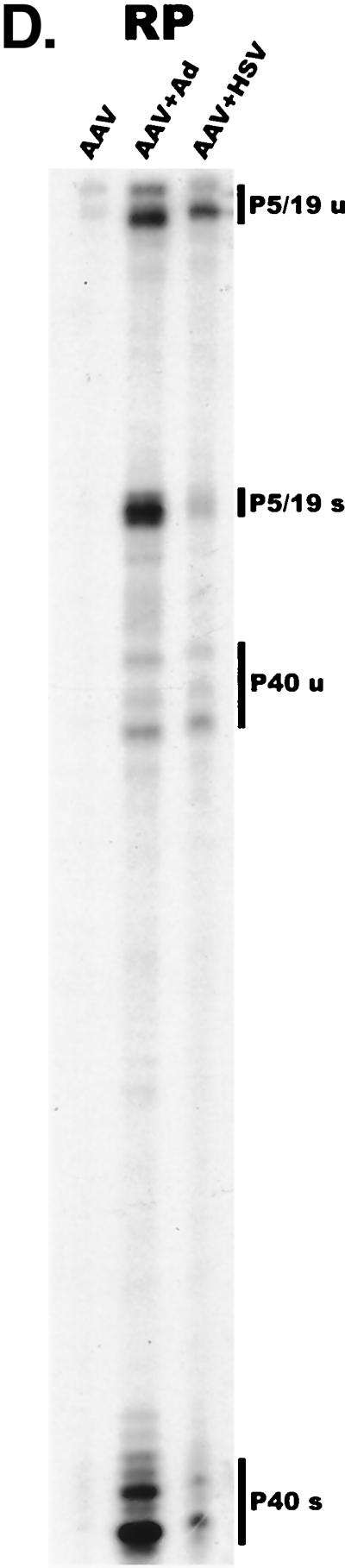
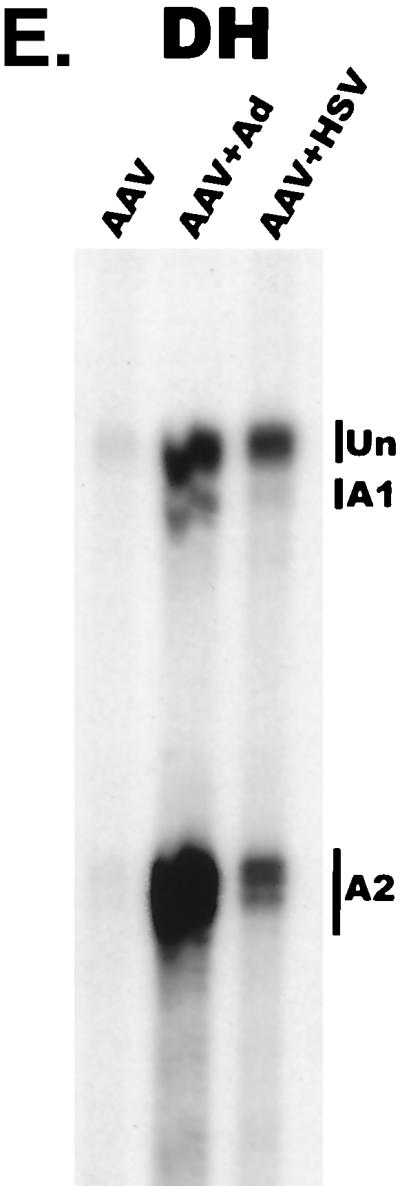
AAV RNA accumulates in a temporal manner during helper virus coinfection. (A) P5/19 unspliced transcripts are detectable prior to significant accumulation of other AAV RNAs. The RP probe was used to protect 10 μg of total RNA isolated at indicated times from infected 293 or HeLa cells (indicated at the bottom). Cells were coinfected with AAV (MOI = 10) and Ad (MOI = 2 to 5) at time zero. AAV-specific products are indicated on the right (u, unspliced; s, spliced). Marker and Probe lanes display reference size standards which are indicated on the left. Lanes labeled Mock display protections of uninfected 293 cell RNA. Multiple bands seen for the probes spanning P19 and P40 likely reflect the use of multiple initiation sites within these regions; additional bands may include incomplete RNase digestion of probe-RNA complexes. (B) P5-generated transcripts are detectable before significant accumulation of those generated by P19. The SB probe was used to protect the same RNA and exactly as described for panel A. At 10 h postcoinfection, P5 transcripts predominate. Later in infection (18 to 221 h), P19 products accumulate to approximately two- to threefold-greater levels than P5 products. Asterisks indicate likely breakdown products of P5 protected fragments and were excluded from quantification. (C) Levels of spliced versus unspliced RNAs increase over the course of infection. The DH probe was used to protect the same RNA samples as above exactly as described for panel A, except that the nt 161 marker had just run off the bottom of this gel. (D) Coinfection of AAV with HSV yields AAV steady-state RNA ratios similar to those found after AAV-Ad coinfection. The RP probe was used to protect 10 μg of total RNA isolated from 293 cells approximately 24 h after infection with AAV (MOI = 10) alone, coinfection with Ad (MOI = 2 to 10), or coinfection with HSV (MOI = 2 to 10). In this experiment, likely due to infection conditions, the level of AAV expression in response to HSV coinfection was atypically low. The ratio of the various RNA species, however, remains the same. (E) HSV stimulates AAV splice acceptor usage similarly to Ad. The DH probe was used to protect the same RNA samples as used for the experiments shown in panel D.
At 14 h postinfection, although some P5/19-generated spliced products were detectable, the majority of P5/19-generated products that had accumulated were unspliced (Fig. 2A to C). In contrast, even when first detected, spliced P40 products were more abundant than unspliced P40 products (Fig. 2A). In our assays, P40 transcripts were never detectable in an unspliced form in the absence of spliced P40 products. Furthermore, during the course of infection, the ratio of spliced to unspliced P5/19- and P40-generated products increased to different final steady-state ratios (Fig. 2A). Ultimately, late in infection, the ratio of spliced to unspliced P5/19 products reached approximately 1:1, while approximately 90% of P40 RNAs accumulated as spliced products. In addition to the obvious implications for the generation of AAV proteins, these results suggest that a mechanism must exist to ensure that the same intron is removed to different final levels from the individual classes of viral RNAs.
Examination and quantification of AAV RNAs with the DH probe (Fig. 2C) confirmed that the accumulated ratio of spliced to unspliced AAV RNA increased over the course of infection, to levels consistent with those seen using the RP probe. Quantification showed that the splice site acceptor usage (A1 or A2 [Fig. 1]) remained relatively constant over this time frame; approximately 10% of all spliced transcripts utilized the upstream acceptor (A1), and 90% utilized the downstream acceptor (A2).
Coinfection of HeLa cells by Ad and AAV showed essentially the same temporal pattern of AAV RNA accumulation (data not shown). A representative analysis of steady-state RNA accumulated levels in HeLa cells is shown for each probe (Fig. 2A to C, lanes 22H). Similar RPA analysis of AAV RNA following HSV coinfection demonstrated that the steady-state RNA ratios and splice acceptor usage are similar to those reached late during coinfection with Ad (Fig. 2D and E).
Temporal accumulation of AAV RNA during helper virus preinfection.
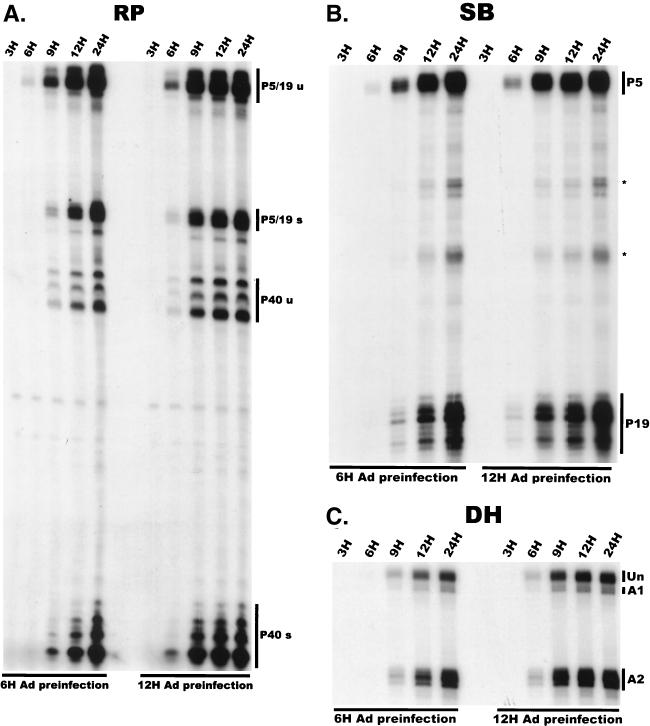
To address whether the temporal order of AAV RNA expression merely reflected the temporal expression of Ad gene products, we examined the accumulation of AAV RNA in 293 cells preinfected with Ad for various times (Fig. 3). Under these conditions, the temporal accumulation of spliced and unspliced AAV RNAs occurred in a manner very similar to that seen during simultaneous coinfection, except that the lag time between AAV infection and the appearance of detectable AAV gene products was significantly reduced (compare Fig. 2 and 3). Following a 6-h preinfection, unspliced P5 products preceded those generated by P19 and were clearly detectable prior to those generated by P40 (Fig. 3A and B, left panels). Preinfection with Ad for 12 h reduced the interval prior to the appearance of AAV RNAs to a greater extent than did a 6-h preinfection (Fig. 3A). Although under these compacted conditions a 3-h sampling period did not reveal a point where P5 products significantly preceded those of P19 and P40, P5, P19, and P40 RNAs still accumulated in a relative temporal manner (Fig. 3A and B). These observations are consistent with the suggestion that there is a temporal regulation of AAV RNAs independent of the temporal expression of Ad genes and that the initial appearance of AAV transcripts was dependent on limiting factors supplied by, or activated by, coinfected Ad. Results using the DH probe confirmed that in the presence of Ad preinfection there was a shift to the relative predominance of spliced species and that the ratio of A2 to A1 usage was also relatively constant under these conditions (Fig. 3C).
FIG. 3.
AAV RNA accumulation following helper virus preinfection. (A) P5/19 products accumulate prior to other AAV RNAs in 293 cells preinfected with Ad. The RP probe was used to protect 10 μg of total RNA exactly as for Fig. 2 except that 293 cells were preinfected with Ad (MOI = 2 to 5) for 6 or 12 h (indicated at the bottom). Total RNA was isolated at the times indicated after AAV infection at time zero. u, unspliced; s, spliced. (B) P5 products accumulate prior to P19 products in 293 cells preinfected with Ad. The SB probe was used to protect the same RNA samples used for experiments shown in panel A. Asterisks indicate likely breakdown products of P5 protected fragments and were excluded from quantification. (C) There is a shift to the relative predominance of spliced AAV RNA species in 293 cells preinfected with Ad. The DH probe was used to protect the same RNA samples as used for experiments shown in panels A and B. Unspliced RNAs and spliced RNAs utilizing either A1 or A2 are indicated.
Stability and promoter strength.
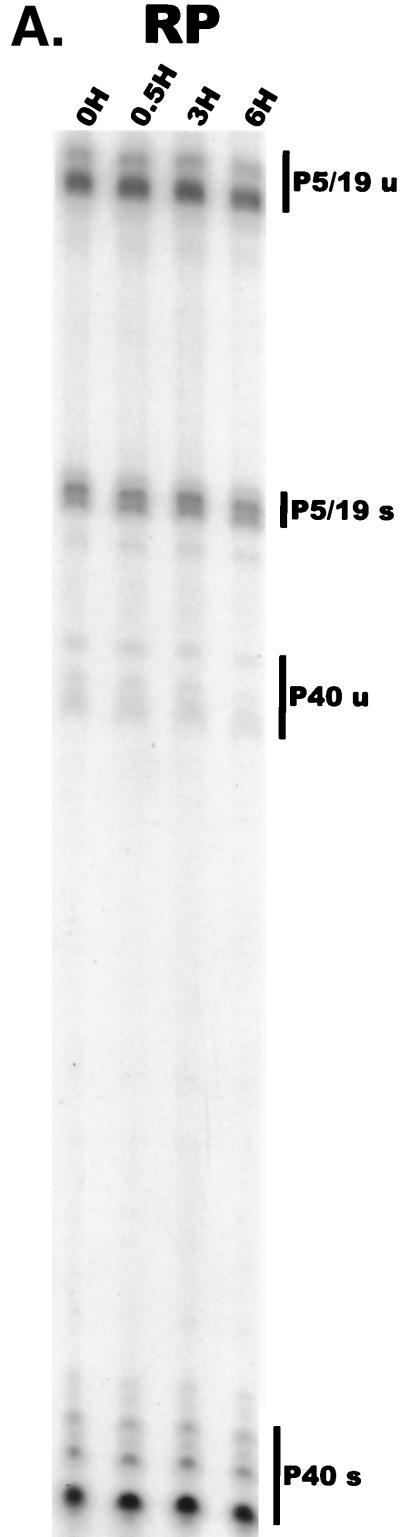
AAV RNAs are very stable. As shown in Fig. 4A and summarized in Table 1, under conditions at which the incorporation of 32P into all nascent transcripts in infected cells was reduced more than 200-fold (data not shown), beginning at 12 h post AAV-Ad coinfection, all AAV-generated RNAs were seen to be generally stable over a 6-h period in total RNA pools. Interestingly, even unspliced RNAs were quite stable over this period, suggesting that by our first time point, they had efficiently localized to the cytoplasm, where they could not be spliced. (The small but detectable decrease during this time in the levels of P5/19 and P40 unspliced RNAs [Table 1] most likely reflects the further processing of a small residual nuclear pool of unspliced AAV RNAs to their spliced counterparts.)
FIG. 4.
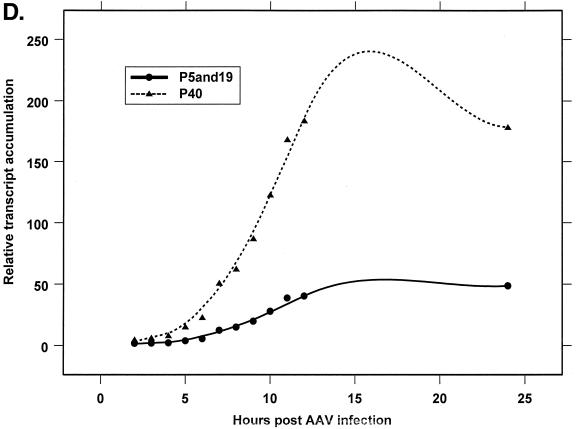
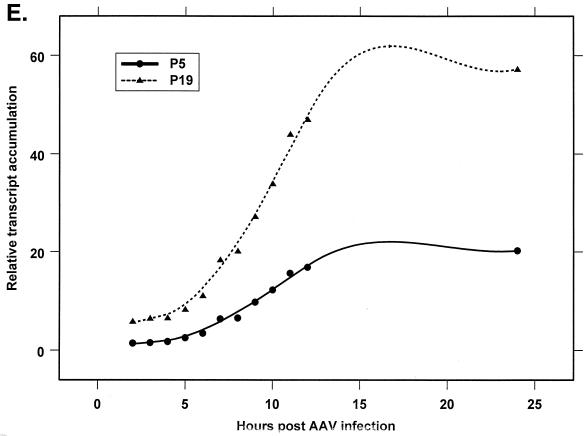
Analysis of AAV RNA stability and accumulation. (A) AAV RNAs are stable. The RP probe was used to protect AAV RNA exactly as described for Fig. 2A except that at 12 h after AAV-Ad coinfection 40 mM actinomycin D was added at time zero and RNA was isolated at indicated times after actinomycin D treatment (described in Materials and Methods). All AAV RNAs are present at high levels at the 6-h time point (Table 1). Comparison between lanes relied on equivalent loading conditions and multiple repetition of the experiment, since unpredictable effects of Ad on cellular RNAs precluded their use as an internal standard. u, unspliced; s, spliced. (B) AAV transcript accumulation over 1-h intervals during a productive AAV infection (RP probe). The RP probe was used to protect RNA exactly as for Fig. 2D except that Ad was preinfected for 12 h and total RNA was isolated at indicated time points after AAV infection at time zero. The asterisk indicates excess undigested RP probe. See panels D and E and Table 2 for quantification. (C) AAV transcript accumulation over 1-h intervals during a productive AAV infection (SB probe). The SB probe was used to protect the same RNA samples exactly as described for panel B. Asterisks indicate likely breakdown products pf P5 protected fragments and were excluded from quantification. See panels D and E and Table 2 for quantification. (D) P40-generated transcripts accumulate at a greater rate than P5- and P19-generated transcripts. P40 unspliced and spliced RNAs were quantified; values were combined and plotted versus combined quantified levels of P5 and P19 unspliced and spliced RNAs for each time point from the experiment shown in panel B. Rates were derived from these curves as described in Materials and Methods and are reported in Table 2. (E) P19-generated transcripts accumulate at a greater rate than P5-generated transcripts. Data from panel C are plotted exactly as in panel D and reported in Table 2.
TABLE 1.
Measure of AAV RNA stability during a productive AAV-Ad coinfectiona
| Time (h) posttreatment | AAV RNA stability (mean % remaining after treatment) ± SD)
|
|||
|---|---|---|---|---|
| P5/19
|
P40
|
|||
| Unspliced | Spliced | Unspliced | Spliced | |
| 0.5 | 102 ± 13 | 108 ± 15 | 91 ± 10 | 102 ± 15 |
| 3 | 92b | 97b | 79b | 94b |
| 5–6 | 84 ± 14 | 102 ± 17 | 71 ± 6 | 97 ± 22 |
At 12 h after AAV-Ad coinfection, cells were treated with 40 mM actinomycin D for the periods indicated (see Materials and Methods for details).
n = 2.
Quantitative RPAs can be used to measure the increase in specific RNA accumulation over a short time interval. Furthermore, because AAV RNAs are stable, transcript accumulation over a period of time where AAV RNA degradation was negligible (3-h intervals) should be proportional to promoter strength. A determination of the increase in individual full-length RNA abundance over such short intervals as the infection progressed was, therefore, used to indirectly determine the relative rate of AAV promoter activity during normal infection. This indirect measure of promoter strength does not take into account possible attenuation of transcripts prior to the region of each RNA protected in our assays.
Figure 4B and C show the accumulation of AAV RNAs over hourly time intervals, using the RP and SB probes, respectively, in experiments in which cells were infected with Ad 12 h prior to infection with AAV. These results are shown in a graphical form in Fig. 4D and E, and results representative of a number of such experiments are summarized in Table 2. The slopes of the curves in Fig. 4D indicate that total P40 products accumulated at a relative rate approximately 4.5-fold greater than generated by P5/19, and this rate was relatively constant at all points assayed (Table 2). Data generated with the SB probe (Fig. 4E) indicated that P19 was approximately 2.5-fold more active than P5 (Table 2). At the earliest time point examined, the ratio of P19 to P5 activity measured in this way was less than that seen at later time points, consistent with the results seen in Fig. 2B. This analysis yielded an average relative ratio of promoter activity of approximately 1:3:18 from P5, P19, and P40, respectively, which is in good agreement with the relative steady-state ratios of accumulated AAV transcripts calculated late in AAV-Ad coinfection (Fig. 2A and B).
TABLE 2.
Relative AAV promoter strengtha
| Time interval (h) during infection | P40:P5/19b | P19:P5c |
|---|---|---|
| 3–5 | 4.79 | 1.80 |
| 4–6 | 4.52 | 2.65 |
| 5–7 | 4.14 | 2.66 |
| 6–8 | 4.14 | 2.94 |
| 7–9 | 4.89 | 2.62 |
| 8–10 | 4.65 | 2.42 |
| 9–11 | 4.29 | 2.85 |
| 10–12 | 4.93 | 2.85 |
| Mean ± SD | 4.54 ± 0.32 | 2.60 ± 0.36 |
Derived from the slopes of the curves in Fig. 4D and E over short (3-h) time intervals where AAV RNAs were shown to be stable.
The rate (increase/time) of P40 accumulation divided by the rate of P5/19 accumulation (both taken from Fig. 4D and E) for each time interval shown; the RP probe was used.
The rate (increase/time) of P19 accumulation divided by the rate of P5 accumulation (both taken from Fig. 4D and E) for each time interval shown; the SB probe was used.
Accumulation of cytoplasmic and polyadenylated RNAs.
Both spliced and unspliced RNAs generated from P5 and P19 encode proteins critical for AAV replication. Therefore, we next determined the levels of accumulation of the individual AAV RNA species in the cytoplasm of 293 cells. Our results demonstrated that at two time points in an established AAV infection (16 and 24 h), the relative steady-state accumulated levels in the cytoplasm of all AAV RNA species were similar to those seen in total RNA pools (Fig. 5A). This is in contrast to accumulation of RNA generated by the autonomous parvovirus minute virus of mice (MVM), for which only spliced RNAs are detectable in the cytoplasm (6).
FIG. 5.
All AAV RNAs are transported efficiently to the cytoplasm. (A) Total and cytoplasmic RNAs purified at 16 and 24 h after AAV-Ad coinfection were examined using the RP probe in duplicate to control for experimental error. Each species of AAV RNA (indicated on the right [u, unspliced; s, spliced]) accumulates in the cytoplasm in a ratio similar to that found in total RNA preparations. Each species of AAV RNA is efficiently polyadenylated. The RP (B), SB (C), and DH (D) probes were used to protect 10 μg of total RNA purified from AAV-Ad-coinfected 293 cells and poly(A)+-containing RNA purified from 20 μg of the same total RNA preparations, as described in Materials and Methods. The ratio of AAV RNA species (indicated on the right of each panel) in poly(A)+-purified RNA is indistinguishable from that of total RNA pools for each probe. The asterisk indicates excess undigested DH probe.
The ratios of spliced to unspliced polyadenylated AAV RNAs were indistinguishable to the accumulation of the individual RNAs in total RNA pools (Fig. 5B and C), suggesting that splicing of AAV RNA is not a prerequisite for their polyadenylation. Further, RPA using the DH probe shows that splice acceptor usage for the pool of polyadenylated RNA is essentially the same as for total RNA pools (Fig. 5D).
Ad stimulates total splicing of AAV RNA and the relative use of the downstream acceptor A2.
Infection by AAV in the absence of helper Ad did not yield detectable levels of AAV-specific RNAs in either HeLa or 293 cells (data not shown). Significant levels of AAV RNA were detectable, however, in 293 cells transfected with the infectious AAV plasmid SSV9, either in the absence or in the presence of Ad, which allowed the following observations.
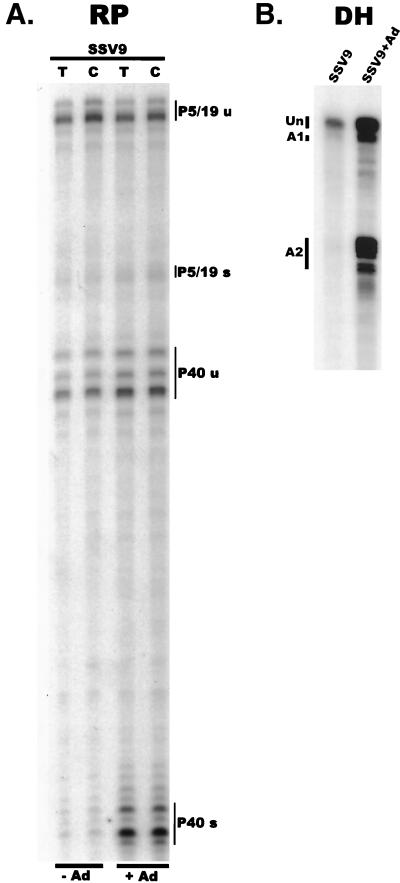
First, although present at reduced total levels, the majority of AAV RNA generated from SSV9 in 293 cells in the absence of Ad was found to be unspliced (Fig. 6). Ad coinfection both increased the amount of AAV RNA that was generated and significantly increased (>10-fold) the percentage of these RNAs that accumulated as spliced RNAs so that spliced species predominated (Fig. 6). Ad infection stimulated the splicing of P40-generated RNAs more dramatically so than RNAs generated from P5 or P19 (Fig. 6A). Analysis of a number of experiments has demonstrated that although the increase in the ratio of spliced to unspliced AAV RNA generated from transfected SSV9 was substantially increased in the presence of Ad, this ratio did not reach total spliced levels seen during coinfection (compare levels in Fig. 6 and Fig. 2). Splice acceptor site usage was also significantly influenced by Ad. In the absence of Ad, rare spliced AAV RNA molecules generated from SSV9 transfection of 293 cells utilized acceptors A1 and A2 at approximately equal frequencies (Fig. 6B). Ad infection increased the ratio of A2 usage relative to A1 at least 4-fold, which remained somewhat less than the relative 10-fold ratio (A2:A1) seen during coinfection. Thus, infection with Ad increased total SSV9-generated AAV RNA levels, the ratio of spliced to unspliced AAV RNA, and the relative utilization of acceptor A2.
FIG. 6.
(A) Plasmids expressing a recombinant AAV genome from different promoters are differentially effected by Ad. The RP probe was used to protect either total or cytoplasmic RNA (indicated at the top) isolated from 293 cells transfected with the SSV9 plasmid and either not infected or infected with Ad (MOI = 2 to 5) (indicated at the bottom). AAV RNA species are indicated at the right (u, unspliced; s, spliced). (B) In the presence of Ad, the ratio of spliced to unspliced AAV RNAs dramatically increases. The DH probe was used to protect total RNA isolated from 293 cells transfected with SSV9 and either not infected or infected with Ad (MOI = 2 to 5).
AAV RNAs generated by SSV9 were able to accumulate in the cytoplasm of 293 cells at similar efficiencies in the presence or absence of Ad (Fig. 6A). Similar results were obtained in HeLa cells following transfection of an AAV-expressing construct driven by the CMV immediate-early promoter cloned upstream of P5 (data not shown). Notably, these results suggest that unspliced AAV RNAs (at least those generated from transfected templates) do not require helper virus for efficient export to the cytoplasm.
DISCUSSION
In this report we describe the characterization of various aspects of the accumulation and abundance of AAV RNA during infection. First, our results demonstrate a temporal order of accumulation of AAV RNAs during AAV-Ad coinfection. Using a quantitative RPA, we have demonstrated that the P5-generated RNAs accumulate to detectable levels prior to other AAV RNAs in coinfected 293 and HeLa cells. At subsequent time points, P19-generated products accumulated to levels greater than those generated from P5, and P40-generated transcripts soon predominated. This temporal order of expression is consistent with the generally accepted model that in the presence of helper virus, the AAV P19 and P40 promoters are stimulated by the large Rep proteins (12, 19, 27, 28) and is so analogous to the ordered expression of the autonomous parvovirus MVM (6). Our results suggest that very early in infection Rep78 may be present in infected cells prior to expression of any other AAV-specific proteins; however, previous studies have not corroborated this. The appearance of spliced P5-generated RNAs coincides with the accumulation of P19 and P40 transcripts, consistent with previous studies that have implicated Rep68 as the primary P19- and P40-transactivating protein (1, 2). The steady-state transcript ratio achieved late in infection for P5, P19, and P40 is approximately 1:3:20, respectively. The same relative accumulated levels of AAV transcripts were detected in AAV-Ad-coinfected HeLa cells and in HeLa cells coinfected with HSV and are present within the poly(A)+ RNA population.
When 293 cells were preinfected with Ad for 6 or 12 h, there was a shift in AAV gene expression to earlier times after AAV infection. In effect, in the presence of Ad early gene products, the lag time between AAV infection and subsequent RNA accumulation was reduced proportionally to the length of Ad preinfection; however, a temporal order of AAV RNA expression was still evident. Following a 6-h Ad preinfection, it was possible to detect P5 unspliced transcripts prior to other AAV RNAs. Following a 12-h Ad preinfection, although our 3-h sampling period did not reveal a point where only P5 products were detected, there was still a detectable shift in the relative abundance of AAV P5, P19, and P40 RNAs. These results suggest that there is a temporal regulation of the appearance of AAV RNAs that is independent of any temporal appearance of Ad gene products and, further, that Ad preinfection renders the cellular environment competent for rapid and efficient AAV infection either by directly supplying or by stimulating cellular factors required for full AAV RNA expression and processing.
In contrast to the autonomous parvovirus MVM (20), the relative ratio of accumulated spliced to unspliced AAV RNAs changes temporally during infection. Unspliced P5/19 products could be detected prior to spliced P5/19 products, but P40 unspliced RNAs were not detected in the absence of their spliced counterparts. The percentage of each class of AAV RNA that was spliced increased during infection, and the rate of increase was different for the P5/19 products than for those generated by P40. In total RNA pools at steady-state levels, the P5 and P19 spliced and unspliced RNAs, each of which encode required replication proteins, were present at approximately equimolar ratios, while P40 unspliced RNAs, which do not encode functional polypeptides, were the most slowly accumulating and least abundant transcripts found at steady-state levels. Excision of a common intron to different final levels therefore paralleled the requirement for functional polypeptides. While the different levels of excision for these classes of RNA may reflect actual splicing differences governed by cis-acting sequences within these RNAs or differential promoter architecture (4), other effects such as selective, active export from the nucleus (7, 13, 18, 25) could also affect the ultimate ratio of accumulated unspliced and spliced RNAs.
Examination of total RNA after treatment of coinfected cells with actinomycin D demonstrated that AAV RNAs are, for the most part, very stable. Both spliced and unspliced AAV RNAs are stable for long periods of time, further suggesting that most unspliced RNAs probably have exited from the nucleus and are no longer accessible to the splicing machinery. Almost equimolar amounts of spliced and unspliced P5 and P19 RNAs, which each encode viable Rep proteins, apparently exit the nucleus as stable molecules prior to splicing, while more than 90% of the P40 products are spliced in the nucleus prior to transport to the cytoplasm.
Our examination of accumulating levels of stable AAV RNAs allowed calculation of the relative steady-state promoter strength of P5, P19, and P40 to be approximately 1:3:18, respectively. Our experiments were conducted in 293 cells preinfected with Ad for approximately 12 h to help ensure that the cellular environment was staged to support maximal levels of AAV gene expression. As expected if RNA degradation was not a factor, these calculated values are similar to the steady-state RNA levels reached during coinfection. For reasons that are not clear, these values differ from those obtained previously by nuclear run-on assays (12), which are perhaps more qualitative in nature.
Examination of total and cytoplasmic RNA preparations from identical time points during coinfection showed that both spliced and unspliced RNAs accumulate efficiently in the cytoplasm in the presence of Ad (although as described above, a much larger percentage of P5- and P19-generated unspliced RNAs accumulate in both total and cytoplasmic pools). Further, transfection of the AAV-expressing plasmids showed that both spliced and unspliced AAV RNAs are efficiently transported to the cytoplasm in the absence of Ad in both 293 and HeLa cells. Transport of unspliced, intron-containing eukaryotic RNAs to the cytoplasm is generally tightly regulated, and the export of these RNAs is often facilitated by regulatory RNA-binding proteins (10, 15, 18, 25). Since helper virus is not required for the transport of unspliced AAV RNAs, if such regulation exists it may be controlled by cellular factors, AAV factors (although no evidence for this exists), or a combination both and would likely involve cis-acting AAV regulatory sequences.
Experiments using transfected AAV-expressing plasmids allowed initial examination of the processing of AAV RNA in the presence and absence of helper virus. Our results confirmed previous observations that when expressed alone, AAV RNAs remained primarily unspliced and that coinfection with Ad increased the ratio of spliced to unspliced AAV RNA, primarily the P40-generated RNAs (26). Further, we demonstrated that helper virus coinfection altered AAV splice site acceptor usage. When expressed in 293 cells in the absence of Ad, rare spliced AAV RNAs utilized A1 and A2 at approximately the same frequency; however, coinfection with Ad stimulated A2 usage to levels approximately fourfold that of A1 (which is still less than seen during coinfection), in agreement with previous observations (26). The stimulation, following infection with Ad, of total splicing and of relative A2 usage of RNAs generated from transfected plasmids never reaches the final levels seen in coinfection. While excision and replication of AAV from the transfected SSV9 plasmid would be expected in our experiments, it is not known what percentage of the RNA that we detect is derived from replicating templates versus from the input transfected DNA templates. Perhaps the inability following transfection to achieve the spliced levels seen during coinfection may somehow reflect a difference between RNA generated from these two different transcription templates.
In summary, our results show that during infection the AAV genome is transcribed in a temporal order, that the relative ratio of spliced versus unspliced AAV RNAs increases during infection, that P5- and P19-generated unspliced RNAs accumulate to greater relative ratios than unspliced RNAs generated by P40, and that Ad stimulates total splicing of AAV RNAs and the relative use of the intron acceptor (A2). Others have shown that Ad gene products influence AAV RNA transport during infection (22), and certain Ad gene products have been shown to have effects on the processing of Ad RNAs (13). It will be interesting to determine whether the effects of Ad on AAV processing are direct or indirect.
ACKNOWLEDGMENTS
We are particularly grateful to R. Jude Samulski and the Vector Core Facility at the University of North Carolina—Chapel Hill for valuable reagents, methods, discussion, hospitality, and generous help in beginning to work with AAV. We thank Greg Tullis (Avigen, Inc.) for plasmids and advice and Jianming Qiu of our laboratory for helpful discussions. We also thank Lisa Burger for excellent technical assistance.
This work was supported by PHS grants ROI A146458 and ROI A121302 from NIAID to D.J.P.
REFERENCES
- 1.Berns K. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1017–1041. [Google Scholar]
- 2.Berns K I, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Buller R M, Janik J E, Sebring E D, Rose J A. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmer P, Pesce C G, Baralle F E, Kornblihtt A R. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter B J. Transcription. In: Ward D, Tattersall P, editors. Replication of mammalian proviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1978. pp. 33–52. [Google Scholar]
- 6.Clemens K E, Pintel D J. The two transcription units of the autonomous parvovirus minute virus of mice are transcribed in a temporal order. J Virol. 1988;62:1448–1451. doi: 10.1128/jvi.62.4.1448-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrun F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Wimler K M, Carmichael G G. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyöstiö S R M, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 Rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leppard K N. Regulated RNA processing and RNA transport during adenovirus infection. Semin Virol. 1998;8:301–307. [Google Scholar]
- 14.Li J, Samulski R J, Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo M J, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra L, Rose J A. Adeno-associated virus DNA synthesis. Virology. 1990;179:632–638. doi: 10.1016/0042-6822(90)90130-j. [DOI] [PubMed] [Google Scholar]
- 17.Naeger L K, Schoborg R V, Zhao Q, Tullis G E, Pintel D J. Nonsense mutation inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1111. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- 18.Nakielny S, Fischer U, Michael W M, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 19.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pintel D J, Gersappe A, Haut D, Pearson J. Determinants that govern alternative splicing of parvovirus pre-mRNAs. Semin Virol. 1995;6:283–290. [Google Scholar]
- 21.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samulski R J, Shenk T. Adenovirus E1B 55-Mr polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J Virol. 1988;62:206–210. doi: 10.1128/jvi.62.1.206-210.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 24.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 25.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 26.Trempe J P, Carter B J. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J Virol. 1988;62:3356–3363. doi: 10.1128/jvi.62.9.3356-3363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trempe J P, Carter B J. Regulation of adeno-associated virus gene expression in 293 cells: control of mRNA abundance and translation. J Virol. 1988;62:68–74. doi: 10.1128/jvi.62.1.68-74.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weger S, Wistuba A, Grimm D, Kleinschmidt J A. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J Virol. 1997;71:8437–8447. doi: 10.1128/jvi.71.11.8437-8447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weindler F W, Heilbronn R. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J Virol. 1991;65:2476–2482. doi: 10.1128/jvi.65.5.2476-2483.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang U-C, Huang W, Flint S J. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J Virol. 1996;70:4071–4080. doi: 10.1128/jvi.70.6.4071-4080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]