Abstract
Metastasis and failure of present-day therapies represent the most common causes of mortality in patients with cutaneous melanoma. To identify the underlying genetic and transcriptomic landscapes, in this study we analyzed multi-organ metastases and tumor-adjacent tissues from 11 rapid autopsies after treatment with MAPK inhibitor (MAPKi) and/or immune checkpoint blockade (ICB) and death due to acquired resistance. Either treatment elicits shared genetic alterations that suggest immune-evasive, cross-therapy resistance mechanisms. Large, non-clustered deletions, inversions and inter-chromosomal translocations dominate rearrangements. Analyzing data from separate melanoma cohorts including 345 therapy-naive patients and 35 patients with patient-matched pre-treatment and post-acquired resistance tumor samples, we performed cross-cohort analyses to identify MAPKi and ICB as respective contributors to gene amplifications and deletions enriched in autopsy versus therapy-naive tumors. In the autopsy cohort, private/late mutations and structural variants display shifted mutational and rearrangement signatures, with MAPKi specifically selecting for signatures of defective homologous-recombination, mismatch and base-excision repair. Transcriptomic signatures and crosstalks with tumor-adjacent macroenvironments nominated organ-specific adaptive pathways. An immune-desert, CD8+-macrophage-biased archetype, T-cell exhaustion and type-2 immunity characterized the immune contexture. This multi-organ analysis of therapy-resistant melanoma presents preliminary insights with potential to improve therapeutic strategies.
Subject terms: Melanoma, Translational research, Cancer genomics, Oncogenesis, Metastasis
Genomic analyses in a rapid autopsy cohort study of patients with melanoma identify the genetic and transcriptomic landscape of melanoma with acquired resistance to MAPK inhibitor and immune checkpoint blockade therapies, providing insights for the potential improvement of therapeutic strategies.
Main
Cutaneous melanoma (CM) exhibits a UV-related high mutational burden1,2. Mutually exclusive BRAF and NRAS mutations drive MAPK addiction in ~70% of metastatic CM3. CM genomes also harbor a high burden of structural variants (SVs)4 and chromothripsis5. Current knowledge of this mutational landscape is derived from tumors naive to highly active treatments developed recently and inclusive of earlier-stage disease. How MAPK inhibitor (MAPKi)/immune checkpoint blockade (ICB) therapies alter the mutational landscape and, thereby, cause death remains largely unknown.
Metastases cause most cancer-related deaths6, and CM is among the most metastatic (>60% of autopsies with brain metastases)7,8. Despite experimental metastasis studies, the difficulty of accessing patient-derived metastatic tissues has limited understanding of clinical metastatic and organ-specific evolution. Although the concept of an organ-specific pre-metastatic niche has been demonstrated experimentally9, little is known clinically regarding co-adaptations between metastases and their site-specific macroenvironments.
Although MAPKi/ICB have become standard-of-care therapies for patients with metastatic CM in developed countries, clinical relapse occurs commonly, with multi-therapy resistance being highly lethal. Acquired MAPKi resistance in CM has been evaluated at an omics scale in a few cohorts, but knowledge of clinically acquired ICB resistance is limited10–18. Metastases to accessible anatomic sites overrepresent current datasets on acquired resistance. Monitoring (for example, liquid biopsy) and therapeutic strategies to counter resistance require insights into multi-organ mechanisms and heterogeneity. Whole-exome sequencing (WES) and whole-genome sequencing (WGS) can characterize patterns termed ‘mutational signatures’ that reflect imprints of DNA mutagenic processes and defective DNA damage repair processes19,20. It is unknown whether rare signatures of a particular malignancy, such as UV-related CM, are common with respect to late mutations, potentially due to the influence of a particular therapy. Such signatures that emerge later during tumor evolution might represent targetable pathway defects or synthetic lethalities.
Hence, we assembled a rapid autopsy melanoma (RAM) cohort from patients with BRAFMUT or NRASMUT CM who were treated with and responded initially to MAPKi/ICB therapies but later died because of disease progression. This cohort includes 71 distinct metastatic tumors, 41 tumor-adjacent ‘normal’ (AN) tissues representing organ-specific tumor macroenvironments and 38 tumor-non-adjacent normal (NAN) tissues. We generated and analyzed WES from tumors and patient-matched NANs as well as WGS from a subset. To dissect the contribution of distinct therapies, we comparatively analyzed WES data from longitudinal pre- and post- tumors from patients with CM who had progressed on either MAPKi-only or ICB-only therapy. Moreover, we developed organ-specific metastatic signatures based on tumorcell-enriched transcriptomes and analyzed ligand–receptor signaling between tumor and AN tissues. Finally, we deconvolved tumor, AN and NAN transcriptomes to decipher organ-specific immune contextures.
Results
RAM cohort, omic data and comparative cohorts
From 11 RAM cases, we collected (2012–2019) tumor, AN (≤1 cm from tumor border) and NAN (>1 cm) tissues from deceased patients (five females and six males, all of European ancestry; seven BRAFMUT and four NRASMUT; five of 11 with radiation-treated brain metastasis) (Supplementary Table 1 and Extended Data Fig. 1a). All patients before autopsies progressed on the last therapy with nearly all tumors acquiring resistance, went to hospice and died. Four patients had been treated with only MAPKi; four with only ICB; and three with MAPKi and ICB (in sequence) (Supplementary Table 1). We generated WES (Supplementary Table 2) from 74 tumors (three multi-regional samples) (brain, thyroid, heart or cardio, lung, liver/gallbladder, spleen, adrenal, lymph node (LN), soft tissues (ST) and pleural membrane or the omentum/peritoneum) and 10 patient-matched normals (Extended Data Fig. 1a). We also generated WGS (Supplementary Table 3) from 22 tumors (brain, lung, liver, adrenal, LN and ST) (Extended Data Fig. 1a). Finally, we generated RNA sequencing (RNA-seq) from 93 tumors, 68 ANs and 67 NANs (inclusive of multi-regional samples) from all sites except thyroid (Extended Data Fig. 1a and Supplementary Table 4). We comparatively analyzed (Extended Data Fig. 1b) CM data from (1) TCGA–SKCM2, (2) patient-matched pre and post tumors with MAPKi-only or ICB-only treatments (Supplementary Table 5) and (3) matched vehicle-treated/MAPKi-sensitive and acquired MAPKi-resistant patient-derived xenografts (PDXs) (Supplementary Table 6).
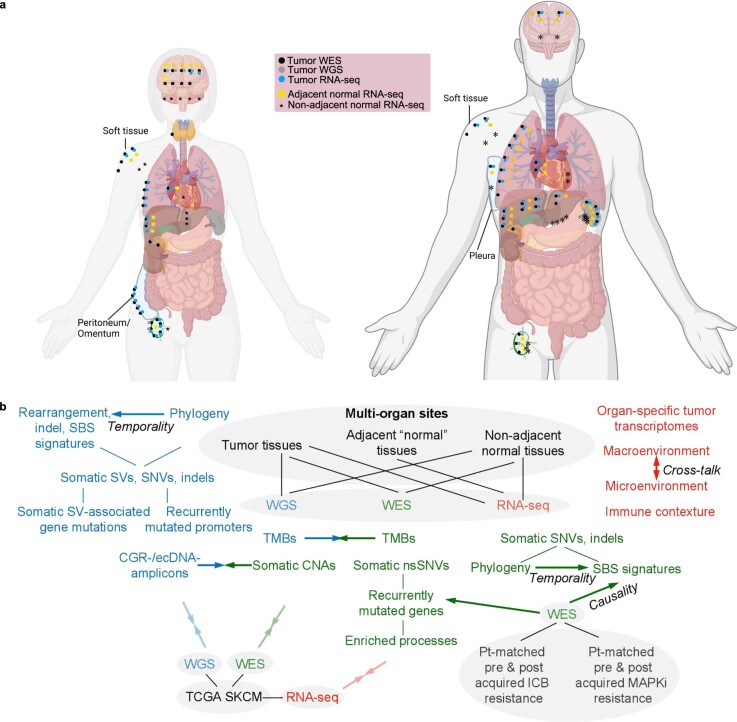
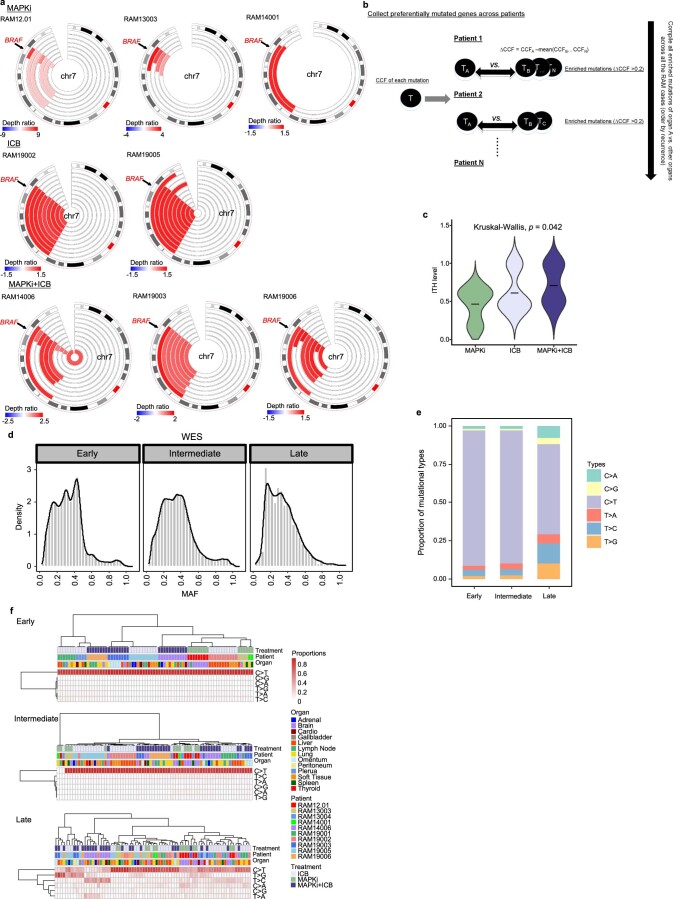
Extended Data Fig. 1. Overview of the tissues sampled at rapid autopsies and the study design.
(a) Numbers and types of tissues, sequencing strategies, and metastatic sites are shown for 11 rapid melanoma autopsy (RAM) cases consisting of five deceased female patients and six deceased male patients with BRAFMUT (n = 7) or NRASMUT (n = 4) cutaneous melanoma. Not represented are RNA-seq derived from triplicates and WES derived from non-adjacent normal tissues. (b) Inclusion of tissue sets and datasets (grey background) and flow chart of analyses performed. RAM is the core tissue or data set, supplemented by comparisons (transparent, adjoined arrows pointing in opposite directions) with two tissue/data sets. RAM cohort (tumor, n = 22 for WGS, n = 74 for WES, n = 93 for RNA-seq; adjacent ‘normal’, n = 68 for RNA-seq; non-adjacent normal, n = 67 for RNA-seq, n = 10 for WES). Pre-and-post cohort consists of patient-matched cutaneous melanoma tumors biopsied pretreatment and post acquired resistance (ICB, n = 7 pairs; MAPKi, n = 59 pairs; both subgroups with patient-matched normal tissues for all patients; n = 102 tumor WES datasets). TCGA–SKCM cohort, only BRAFMUT (n = 233) or NRASMUT (n = 125) tumors included (13 tumors with both BRAF and NRAS mutations; total n = 345 tumors). Bolded arrows, analyses compared to derive insights into temporality or causality.
Somatic exomic alterations
At a median coverage of 217×, the mean number of somatic mutations/tumor and tumor mutational burden (TMB) was 4,401 (range, 167–17,872) and 68.8 mutations per megabase (Mb) (range, 2.6−279.3) (Fig. 1a and Supplementary Table 2). For non-synonymous somatic mutations, a mean of 675 mutations per tumor (range, 24–2,669) corresponded to a TMB of 22.49 mutations per Mb (range, 0.8–88.97) (Supplementary Table 2). These TMBs are higher than or similar to previous estimates (Supplementary Table 7). We detected by Genomic Identification of Significant Targets In Cancer (GISTIC2.0) 48 significantly amplified and 75 significantly deleted regions (Extended Data Fig. 2a and Supplementary Table 8). Amplified regions harbored BRAF (80% of tumors), ACTA1 (66%) and TERT (42%); deleted regions harbored IFN cluster genes and CDKN2A/B (51%), B2M and SPRED1 (45%), BRCA1 (41%) and JAK2 and CD274 (35%) (Extended Data Fig. 2b). Tumors from six of six MAPKi-only and MAPKi+ICB cases harbored BRAF amplification, whereas tumors from two of four ICB-only cases harbored BRAF amplification. Copy number loss of JAK2, previously associated with acquired ICB resistance21,22, was observed in MAPKi-only cases. The findings of mechanisms of acquired MAPKi resistance in tumors with acquired ICB resistance and vice versa suggest cross-resistant mechanisms that converge on immune evasion.
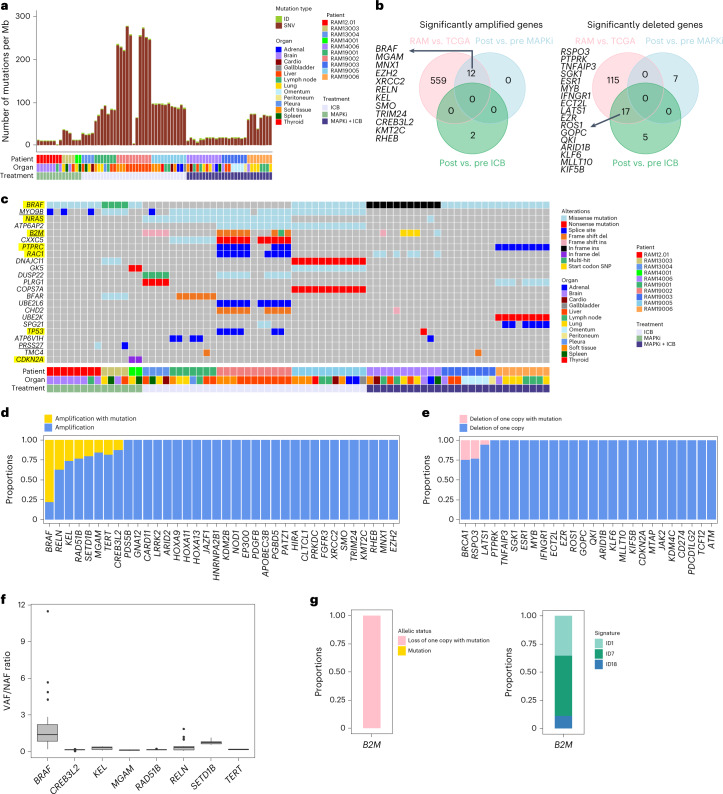
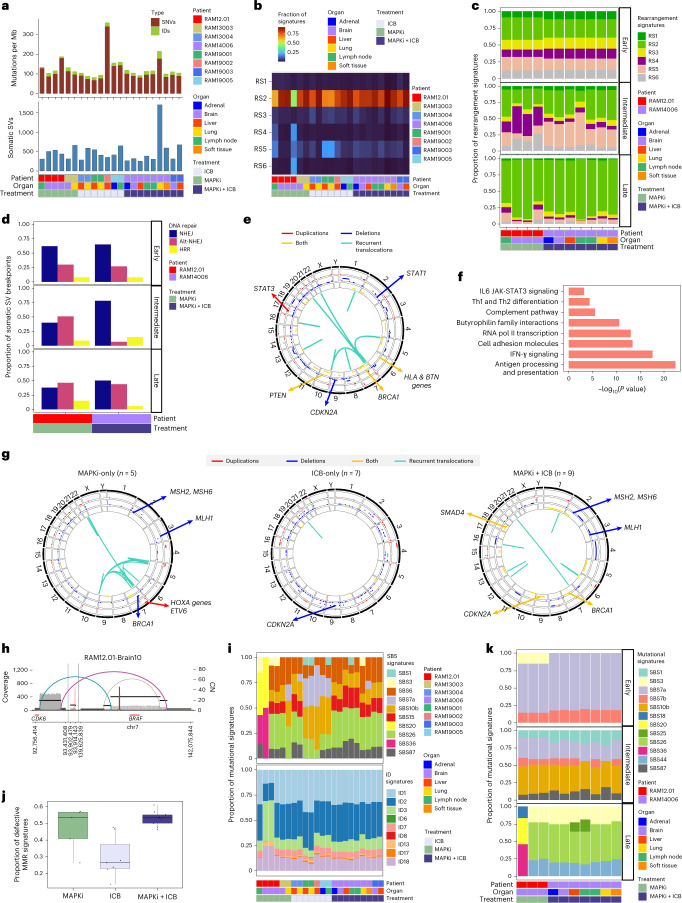
Fig. 1. Whole-exomic landscape of therapy-resistant cutaneous melanoma.
a, Tumor somatic mutational burdens based on synonymous and non-synonymous IDs and SNVs across RAM cases (patients), organ sites and treatment histories. b, Venn diagrams showing overlaps of significantly amplified (left) or deleted (right) genes based on frequency-enriched CNAs observed in three comparative cohorts: (1) RAM tumors versus TCGA–SKCM tumors (BRAF mutant and NRAS mutant only); (2) MAPKi-only post versus pre tumors; and (3) ICB-only post versus pre tumors. c, Highly expressed SMGs in the RAM tumor cohort annotated by mutational types, cases, organ sites and treatment histories. Yellow highlights, SMGs identified by prior published studies (Extended Data Fig. 2g,h). Underlines, SMGs significantly (two-sided Fisher’s exact test, FDR-adjusted P < 0.05) enriched in the RAM tumor cohort (versus the TCGA–SKCM tumor cohort, BRAF mutant and NRAS mutant only). MYO9B, P = 7.97 × 10−5 and adjusted P = 0.004144; B2M, P = 1.75 × 10−3 and adjusted P = 0.044013; PRSS27, P = 2.12 × 10−3 and adjusted P = 0.044013. d,e, Copy number and mutational (missense, nonsense, splice-site, frame-shift IDs) status of overlapping amplified (d) or deleted (e) genes (b and Extended Data Fig. 2c). f, Ratios of variant versus normal allele frequencies in eight overlapping amplified genes (BRAF, n = 39; CREB3L2, n = 7; KEL, n = 15; MGAM, n = 9; RAD51B, n = 5; RELN, n = 26; SETD1B, n = 6; TERT, n = 5 tumors). Central line of each box, median; top and bottom edges of each box, first and third quartiles; whiskers extend 1.5× the interquartile range beyond box edges. g, Copy number and mutational status of the overlapping SMG B2M (Extended Data Fig. 2e) in the RAM cohort (left); ID signatures identified among B2M-mutated RAM tumors (right). del, deletion; ins, insertion; SNP, single-nucleotide polymorphism.
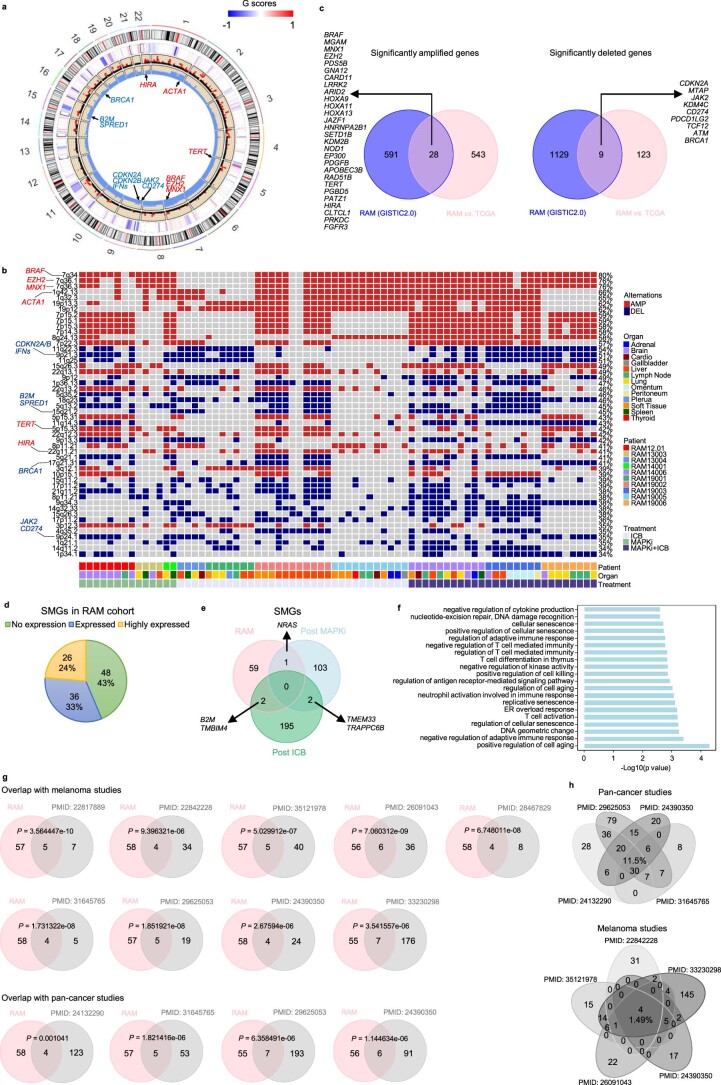
Extended Data Fig. 2. Significant copy-number alterations and mutated genes enriched in the RAM tumor cohort.
(a) Somatic copy-number alterations of the RAM cohort (n = 74 tumors) (left). Outermost layer, chromosomal regions; second layer, heatmap of G scores (GISTIC 2.0); third layer, P values of amplified and deleted regions (red dots indicate significance); innermost layer, frequencies of amplified and deleted regions; arrows, locations of functionally important genes displaying significant amplification (red) or deletion (blue). (b) Top 50 significantly altered exomic regions by GISTIC analysis, listed in decreasing order of recurrence and annotated with associated genes, across RAM cases/patients, organ sites, and treatment histories. AMP, amplification; DEL, deletion. (c) Venn diagrams showing overlaps between (i) significantly amplified (left) or deleted (right) genes based on GISTIC2.0 analysis of the RAM tumors and (ii) frequency-enriched, copy number alterations observed in RAM tumors versus TCGA–SKCM tumors (BRAF- and NRAS-mutant only). (d) The numbers and proportions of significantly mutated genes (SMGs) with no expression (mean log2CPM < 0), expresssion (0 ≤ mean log2CPM < 4), or high expression (mean log2CPM > 4) among RAM tumors. (e) Venn diagram of overlapping SMGs identified in three tumor cohorts: (i) RAM, (ii) clinical post-MAPKi-only (pre-MAPKi SMGs subtracted) and, (iii) clinical post-ICB-only (pre-ICB SMGs subtracted). (f) Top 20 significantly enriched biological processes of SMGs with expression (as defined in d). One-sided Fisher’s exact test without multiple comparisons. (g) Venn diagrams showing overlapping numbers of SMGs identified in the RAM tumor cohort versus SMGs identified in published (PMIDs shown) large-scale melanoma and pan-cancer cohorts. P values by one-sided hypergeometric test. (h) Venn diagrams showing overlapping numbers of SMGs identified in published pan-cancer (top) or melanoma (bottom) cohorts.
To match RAM tumors, we selected TCGA–SKCM tumors driven by BRAF or NRAS mutations. By comparing RAM versus TCGA–SKCM copy number alteration (CNA) frequencies with Fisher’s exact test, we detected 571 significantly amplified and 132 significantly deleted genes in the RAM cohort (Supplementary Table 9). We observed significant overlaps of amplified (but not deleted) genes between findings from GISTIC and RAM-enriched (versus TCGA–SKCM) CNAs (P = 0.0482198, hypergeometric test) (Extended Data Fig. 2c). We tested the hypothesis that a subset of RAM-enriched (versus TCGA–SKCM) CNAs is due to acquired resistance to MAPKi/ICB therapy. We identified significant CNAs in post-treatment (versus pre-treatment) melanoma from patients with either MAPKi-only or ICB-only treatment (between pre and post biopsies) histories. Notably, gene amplifications overlapped significantly between the RAM (versus TCGA–SKCM) and the MAPKi-only post (versus pre) frequency-enriched CNAs (P = 1.205907 × 10−18, hypergeometric test) (Fig. 1b). In contrast, gene deletions overlapped significantly between the RAM (versus TCGA–SKCM) and the ICB-only post (versus pre) frequency-enriched CNAs (P = 6.52371 × 10−33, hypergeometric test) (Fig. 1b). Thus, acquired resistance to ICB versus MAPKi distinctly contributes to the RAM CNA landscape.
We also nominated significantly mutated genes (SMGs) (Fig. 1c and Supplementary Table 10). To circumvent a limited cohort size, we inflated type I error by not performing multiple testing but reduced false positives by enforcing an expression cutoff. MutSig2CV (at a raw P < 0.05 cutoff) called 110 SMGs. Among these, we nominated 62 SMGs (Extended Data Fig. 2d) with RNA (Supplementary Table 10) or high RNA (Fig. 1c and Supplementary Table 10) expression, using cohort-matched transcriptomes. Among high-expression SMGs, we identified resistance driver mutations in BRAF and NRAS and predicted loss-of-function B2M mutations (Fig. 1c). Consistent with resistance causation, B2M was significantly mutated in ICB-only post (but not pre) tumors (Extended Data Fig. 2e). Moreover, the 62 nominated SMGs enriched for immune, cell death and senescence regulation (Extended Data Fig. 2f). We cross-referenced the 62 RAM-nominated SMGs to SMGs reported by large-scale, melanoma-specific or pan-cancer studies and observed highly significant overlaps (Extended Data Fig. 2g). As context, among these publications, the consensus SMGs comprised small fractions (1.49% and 11.5% among melanoma and pan-cancer cohorts, respectively) (Extended Data Fig. 2h). Seven high-expression RAM SMGs (BRAF, NRAS, B2M, PTPRC, RAC1, TP53 and CDKN2A) were previously reported as SMGs (Fig. 1c).
Among genes affected by overlapping CNAs (Fig. 1b and Extended Data Fig. 2c), eight amplified genes harbored mutations in at least one copy (Fig. 1d,e). BRAF displayed a mean ratio of variant-to-normal allelic frequency of 1.87 (Fig. 1f), indicating selective amplification of the mutant allele. Among affected tumors, deleted genes predominantly showed single-copy loss (Fig. 1e). Several genes (BRCA1, RSPO3 and LATS1) were affected by both deletions and mutations. Among B2M-mutated tumors, 100% harbored bi-allelic loss-of-function alterations (deletion of one and mutation of another copy) (Fig. 1g). Because B2M mutations consisted of small insertions and deletions (IDs), we found enrichment of ID1 and ID7 signatures among B2M-mutated tumors (Fig. 1g), suggesting defective DNA mismatch repair (MMR) as a cause.
Phylogeny and heterogeneity
RAM tumors evolved via both linear and branched divergence without any organ-specific pattern (Fig. 2a). Regardless of treatment history, prominent truncal amplifications involved BRAF, MNX1, EZH2, XRCC2 and ACTA, and truncal deletions involved JAK2 and CDKN2A (Fig. 2a and Supplementary Table 1). In contrast, B2M somatic mutations and deletions were exclusive to ICB-only and MAPKi+ICB cases and occurred as truncal, semi-truncal or private events (Fig. 2a). Loss-of-function mutations affecting JAK2, CDKN2A (and other 9p21.3 genes—for example, IFN-induced genes; Extended Data Fig. 2a) and B2M indicated immune evasion as mechanisms of acquired resistance3,10,23. In RAM19002, non-synonymous mutations of MMR genes (MSH6, PMS2 and POLD1) preceded B2M IDs (Fig. 2a). We also observed convergent evolution due to MAPKi/ICB therapies (for example, in RAM14006, distinct, second-hit non-synonymous B2M mutations (Fig. 2a); in multiple cases, distinct BRAF amplicon boundaries (Extended Data Fig. 3a)). By analyzing tumor cell fractions of mutated genes, we failed to detect organ-specific enrichment for specific mutated genes (Extended Data Fig. 3b). Finally, intratumoral heterogeneity (ITH), estimated by the proportion of subclonal mutations, was significantly lower after MAPKi-only treatment (Extended Data Fig. 3c).
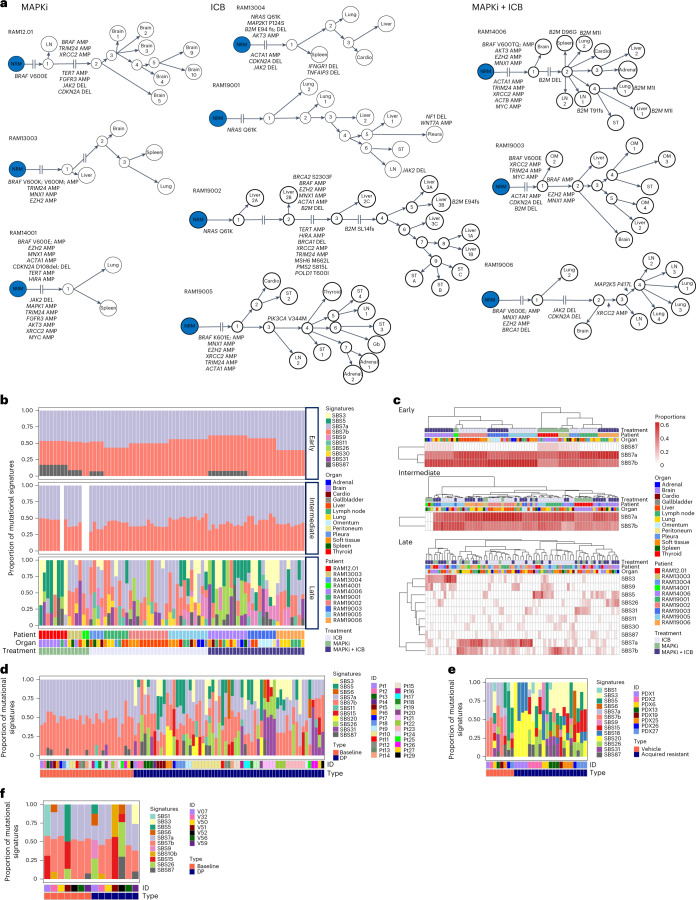
Fig. 2. Multi-organ temporal mutational patterns of RAM tumors.
a, Phylogenetic relationship of multi-organ metastases in 10 RAM cases organized by treatment histories. Each tumorʼs somatic mutations (SNVs and IDs) were used to construct a maximally parsimonious phylogenetic tree. Some branches, as indicated, are not shown to scale owing to extensive lengths. Each evolutionary trajectory is annotated by selected cancer genes and their mutations. AMP, copy number amplification; DEL, copy number deletion; Gb, gallbladder; NRM (blue node), normal tissue; OM, omentum. b, Spectra of mutational signatures among early, intermediate and late mutations, based respectively on shared, semi-private and private SBSs in a across RAM cases, organ sites and treatment histories. With WES of two RAM14001 tumors available, we identified only early and late mutations for signature detection. c, Unsupervised clustering of treatment histories, RAM cases and organ sites based on the proportions or compositions of SBS mutational signatures present in early, intermediate and late mutations in b. d, Analysis of SBS mutational signatures in a clinical cohort of patient-matched MAPKi-sensitive (referred to as baseline) and MAPKi-acquired resistant (referred to as disease progression (DP)) cutaneous BRAFMUT melanoma tumors (n = 88 tumors). MAPKi-sensitive tumors represent ICB-naive, pre-MAPKi-treatment clinical tumors (n = 28 patients). e, As in d, except all samples are PDX tumors from sex-matched NSG mice and consisting of patient-matched (n = 8 models), vehicle-treated and MAPKi-sensitive tumors (n = 8) and acquired MAPKi-resistant BRAF-mutant or NRAS-mutant tumors (n = 21). f, As in d, except all samples are clinical tumors (n = 14) consisting of patient-matched MAPKi-naive (except one), pre-ICB baseline tumors and acquired ICB-resistant tumors (n = 7 patients).
Extended Data Fig. 3. Cancer cell fractions and mutational spectra analysis.
(a) Circos representation of copy number gains harboring BRAF in multiple metastases from eight RAM cases. For each Circos plot (RAM case), the outermost layer represents chromosome 7 regions, and the inner heatmaps indicate copy number status (spanning region and depth ratio) of tumors with BRAF amplification. Arrow indicates the location of BRAF. (b) Schema of sub-clonal analysis of multi-organ metastases to identify organ-specific enrichment of mutations. Each mutation’s cancer cell fraction (CCF) is estimated using PyClone-VI. The ∆CCF of each mutation was calculated between tumors of organ A versus tumors of other organs in each RAM case. Enriched mutations in organ A are defined as those with a ∆CCF difference > 0.2. The recurrence of genes harboring organ A-enriched mutations is computed by combining the enriched mutations of tumors from organ A across all applicable RAM cases. (c) Intratumoral heterogeneity (ITH) levels (fraction of sub-clonal mutations per tumor) of RAM tumors by treatment histories. P values, Kruskal–Wallis test. ICB, n = 33; MAPKi, n = 14; ICB + MAPKi, n = 27. (d) Distribution of mutant allele frequencies (MAF) of early, intermediate, and late mutations. (e) Mutational spectra of early, intermediate, and late mutations. (f) Unsupervised clustering of mutational spectra among early, intermediate, and late mutations across treatment histories, RAM cases/patients, and organ sites.
Altered mutational spectra and signatures
We previously identified altered mutational spectra associated with somatic mutations unique to acquired MAPKi resistance14. Here, we divided somatic mutations into early, intermediate and late mutations and analyzed mutant allelic frequencies (Extended Data Fig. 3d), mutational spectra (Extended Data Fig. 3e) and single-base substitution (SBS) signatures based on COSMIC version 3.3 (Fig. 2b). The mean mutant allelic frequencies of early, intermediate and late somatic single-nucleotide variants (SNVs) were, respectively, 0.39, 0.36 and 0.27. Early and intermediate mutational spectra displayed a case-specific but no site-specific or treatment-specific pattern. However, late mutational spectra clustered independently of case, site or treatment history (Extended Data Fig. 3f), enriching for C>A, T>C and T>G (Extended Data Fig. 3e). Among 10 SBS signatures, UV signatures (SBS7a and SBS7b) dominated early and intermediate somatic mutations (Fig. 2b) in a case-specific pattern (Fig. 2c). Notably, non-UV-related signatures dominated late mutations (Fig. 2b) with extensive intra-patient and inter-patient heterogeneity but treatment-elicited convergence (for example, signatures of defective homologous recombination repair (HHR) and MMR clustering with MAPKi) (Fig. 2c). Notably, SBS3 (defective HRR) was detected among late mutations in nine of 10 patients (Fig. 2b). We also detected SBS5 (clock-like), SBS9 (polymerase eta somatic hypermutation), SBS26 (defective MMR), SBS30 (defective DNA base excision repair (BER) due to NTHL1 mutations), SBS31 (platinum treatment) and SBS87 (thiopurine treatment) among late mutations in most patients. SBS11 (temozolomide) was detected among late somatic mutations in association with ICB treatment (Fig. 2b).
We sought to validate an association between non-UV-related signatures and MAPKi/ICB therapies. We assembled two MAPKi validation cohorts of patient-matched melanoma with WES data: (1) a clinical BRAFMUT cohort (88 tumors from 28 patients) consisting of pre-treatment and acquired-resistant tumors (along with normal genomic DNA (gDNA)) (Supplementary Table 5) and (2) a PDX BRAFMUT or NRASMUT cohort (29 tumors from eight models) consisting of model-matched vehicle-treated and acquired-resistant tumors derived in patient sex-matched NSG mice (information on gender of source patients in Supplementary Table 6) (along with normal gDNAs). All pre-treatment melanomas in both MAPKi cohorts were ICB naive and MAPKi sensitive (Supplementary Tables 5 and 6). We extracted somatic mutations unique to acquired MAPKi resistance and identified the frequencies of SBS signatures, including 12 non-UV signatures of defective HRR, MMR and BER (Fig. 2d,e). In contrast, UV signatures dominated the SNVs in pre/sensitive tumors. We also assembled WES data from patient-matched pre/sensitive and post/acquired ICB-resistant melanoma (14 tumors from seven patients) and normal gDNAs. All pre/sensitive melanoma in this ICB validation cohort, except one (V52), were MAPKi naive (Supplementary Table 5). After extracting SBS signatures from somatic SNVs in pre-treatment tumors and unique to acquired resistance, we detected enrichment of UV signatures in four of seven acquired ICB-resistant tumors (Fig. 2f). Thus, MAPKi (versus ICB) therapy selects for non-UV-related mutational signatures.
Somatic whole-genomic alterations
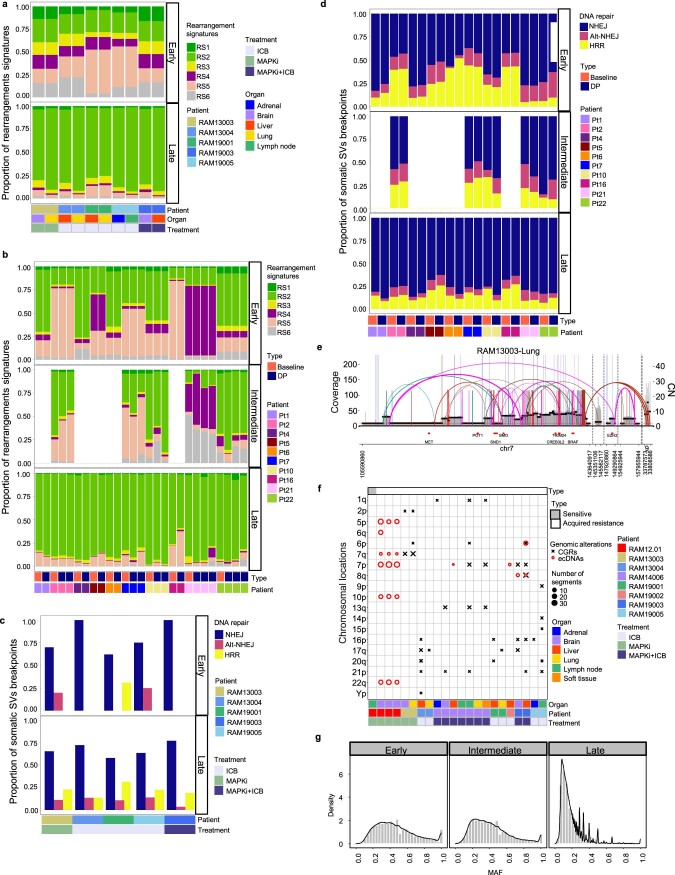
We generated WGS data (median coverage of 23×; range, 10×–43×) from a subset of RAM tumors (one sensitive and 21 acquired- resistant; eight cases; six sites) (Supplementary Table 3). We observed no significant differences in the median SNVs or IDs based on treatment histories. The mean WGS-estimated TMB was 97 mutations per Mb (Fig. 3a), which is higher than WES-estimated TMB (Fig. 1a) and WGS-estimated TMB (39.6 mutations per Mb) in earlier-stage, MAPKi/ICB-naive CM (Supplementary Table 7). We identified a mean of 493 SVs per tumor (range, 254–1,708) (Fig. 3a), in contrast to a mean of 342 or 106 in mucosal or CM cohorts, respectively4. We first classified SV/rearrangements as clustered or non-clustered24,25 and found 67% as non-clustered translocations, 17% as non-clustered deletions and 13% as non-clustered inversions. Rearrangement signature (RS) 2 (ref. 24), defined by large (>100 kb) non-clustered deletions, inversions and inter-chromosomal translocations, was most frequent, regardless of case, site or treatment (Fig. 3b). To dissect the temporality of RSs, we focused on two RAM cases with multiple tumors to reconstruct the phylogeny and determine early (Fig. 3c and Extended Data Fig. 4a), intermediate (Fig. 3c) and late SVs (Fig. 3c and Extended Data Fig. 4a). Analysis of RS1 to RS6 revealed that (1) all six RSs occurred at ≥4% in every tumor among early SVs; (2) the frequencies of RS4 or RS5 (characterized by <100-kb deletions and enriched in BRCA1/2-deficient breast tumors26) increased among intermediate (versus early) SVs; and (3) the frequencies of RS2 increased among late SVs (Fig. 3c and Extended Data Fig. 4a). Moreover, analyzing WGS derived from clinical (pre and post) MAPKi tumors (10 pre and 17 post from 10 patients), we observed that RS2 enrichment dominated late SVs in both pre and post tumors (Extended Data Fig. 4b).
Fig. 3. Whole-genomic landscape of therapy-resistant cutaneous melanoma.
a, TMBs based on somatic IDs, SNVs and SVs across 22 tumors (eight cases), organs and treatment histories. b,c, Spectra of RSs among all SVs (b) or early, intermediate and late SVs (c). Tumors analyzed in c derived from phylogenetic analysis of two RAM cases with multiple tumors. d, Distribution of DNA DSB repair processes (NHEJ, alternative NHEJ and HRR) inferred by breakpoint junctional sequence analysis of early, intermediate and late SVs of RAM cases in c. Homologous sequence lengths at breakpoints of 0–1 bp, 2–6 bp or >6 bp infer NHEJ, alternative NHEJ and HRR, respectively. e, SVs across genomic locations and affected genes. Outer layer, chromosome locations; second layer, large (>1 Mb) duplications (red); third layer, large deletions (blue); inner layer, a combination of both duplications and deletions (yellow); frequencies of occurrence increase from inner to outer circles. Inside the circles, recurrent (≥4 of 21 resistant RAM tumors) intra-chromosomal or inter-chromosomal translocations. f, Pathways enriched in genes overlapping somatic SVs detected in ≥50% (11/21) of RAM tumors (one-sided Fisher’s exact test, adjusted by FDR). g, As in e, except sub-analyses based on treatment histories and recurrent translocation in ≥3 tumors in each treatment history category. h, SV plot indicating extrachromosomal DNA amplicon harboring BRAF in a brain metastasis. Horizontal black and red lines indicate, respectively, genomic segments with similar copy numbers and genes. Each line/arc representing discordant reads is colored based on differences from expected distance or orientation. i, Spectra of SBS and ID signatures based on WGS across 22 tumors (eight cases), organs and treatment histories. j, Proportions of SBS mutational signatures associated with defective MMR, per treatment history (n = 22 tumors). Central line of each box, median; top and bottom edges of each box, first and third quartiles; whiskers extend 1.5× the interquartile range beyond box edges. k, As in i, except for early, intermediate and late SBS mutations in two RAM cases with phylogenetic data.
Extended Data Fig. 4. Structural variations, complex genomic rearrangements (CGRs) and extrachromosomal DNAs (ecDNAs) amplicons, and mutations.
(a) Spectra of rearrangement signatures (RSs) among early or late structural variations (SVs) in RAM cases (n = 5 cases; n = 5 sites). (b) Spectra of RSs among early, intermediate, or late SVs in a clinical MAPKi-treated, patient-matched pre-and-post melanoma cohort (n = 10 patients; n = 10 baseline tumors; n = 17 disease progression or DP tumors). (c) Distribution of double-stranded DNA break (DSB) repair processes (non-homologous end-joining or NHEJ, alternative NHEJ or alt-NHEJ, homologous recombination repair or HRR) inferred by breakpoint-junctional sequence analysis of early or late SVs of RAM cases in (a). (d) Distribution of DSB repair processes inferred by breakpoint-junctional sequence analysis of early, intermediate, or late SVs of the cohort in (b). For cases with multiple DPs, average values are presented. (e) SV plot indicating CGR harboring BRAF in a lung metastasis. Horizontal black and red lines indicate respectively genomic segments with similar copy numbers and genes. Short vertical lines and arcs indicate discordant read pairs linking two amplicons via a structural variant junction. Long vertical lines indicate break ends that map from amplicon into low-complexity regions which cannot be traced further. Each line/arc representing discordant reads is colored based on differences from expected distance or orientation. (f) Chromosomal distribution of ecDNAs and CGRs in RAMs (n = 22; one remained sensitive to treatment; 21 with acquired MAPKi-resistance) across organs, deceased patients, and treatment histories. Circles and Xs represent ecDNAs and CGRs, respectively. The size of circles or Xs indicates the number of copy number variant segments (reference sizes shown). (g) Distribution of mutant (variant) allele frequencies in early, intermediate, or late mutations for RAM12.01 and RAM14006.
To assess DNA double-strand break (DSB) repair mechanisms, we analyzed the breakpoint-junctional sequences of early (Fig. 3d and Extended Data Fig. 4c), intermediate (Fig. 3d) and late SVs (Fig. 3d and Extended Data Fig. 4c). Among early SVs, 76% of breakpoints displayed a homologous sequence (HS) size of 0–1 base pairs (bp), supporting non-homologous end joining (NHEJ) as a key DSB repair mechanism. Among intermediate and late SVs, we inferred either NHEJ or NHEJ + alternative NHEJ. We validated the importance of NHEJ across temporal SVs using WGS data from the clinical pre and post MAPKi cohort (Extended Data Fig. 4d). Thus, NHEJ and alternative NHEJ represent potential targets to blunt SV-driven melanoma progression.
Moreover, we identified pathway enrichments of genes overlapping somatic SVs. First, SV-related deletions occurred in CDKN2A (52%), PTEN (38%) and BRCA1 (~29%) (Fig. 3e). Notably, SVs in chr6p spanning HLA class I and II genes were highly recurrent (62%) (Fig. 3e). Second, we identified recurrent translocations (≥4/21 tumors) involving oncogenes such as PDGFB and JAZF1 (Supplementary Table 11). Third, 491 somatic SV-related genes (Supplementary Table 12) recurrently (>50%) and significantly enriched for immune pathways, such as antigen processing and presentation, IFN-γ, complement and IL-6-mediated JAK-STAT3 signaling (Fig. 3f). Fourth, a higher number of recurrent translocations associated with MAPKi-only or MAPKi+ICB treatment histories (Fig. 3g). MAPKi-only tumors harbored recurrent (60%) duplications of HOXA family genes and ETV1. MAPKi+ICB tumors harbored frequent (43%) SVs involving SMAD4. Notably, MAPKi-only or MAPKi+ICB tumors harbored recurrent deletions in MMR genes (MSH2, MSH6 and MLH1). Finally, we identified BRAF amplification via intrachromosomal complex genomic rearrangements and extrachromosomal DNAs in association with MAPKi treatment (Fig. 3h and Extended Data Fig. 4e,f).
Among ID signatures, ID1 and ID2 were most frequent (Fig. 3i), which suggests mutagenesis via slippage of replicated DNA strands. SBS signatures of defective MMR (SBS6, SBS15, SBS20 and SBS26) were prevalent (Fig. 3i). Their proportions were significantly (Kruskal–Wallis test, P = 0.0082) higher with MAPKi-only and MAPKi+ICB treatments (Fig. 3j). We then temporally ordered WGS-based somatic SNVs. The average mutant allele frequencies of early, intermediate and late SNVs were, respectively, 0.47, 0.41 and 0.20 (Extended Data Fig. 4g). Consistent with WES-based findings (Fig. 2b), UV-related SBS signatures dominated early SNVs (Fig. 3k). Intermediate SNVs enriched for SBS10b (mutations in the polymerase epsilon exonuclease domain, which are associated with hypermutations >100 per Mb) (Fig. 3k). Late mutations enriched for SBS20 and SBS26 (defective MMR) and SBS3 (defective HRR) (Fig. 3k). Moreover, we identified somatic SNVs (3/21 intermediate, 2/21 late) in the TERT promoter (−124C/T and −146C/T). Finally, we assessed the temporality of HRR and MMR alterations. Early nonsense BRCA2 mutations occurred in RAM19005 (ICB-only), and intermediate SV-associated BRCA1 deletions occurred in RAM14006 (MAPKi+ICB), which may explain RS5 enrichment (Fig. 3c). MMR gene deletions occurred as early, intermediate and late somatic SVs, and somatic non-synonymous SNVs occurred late in MMR genes (MLH3, MSH6, MSH2 and PMS2), potentially contributing to MMR SBS signatures (Fig. 3k).
Organ site-specific transcriptomic features
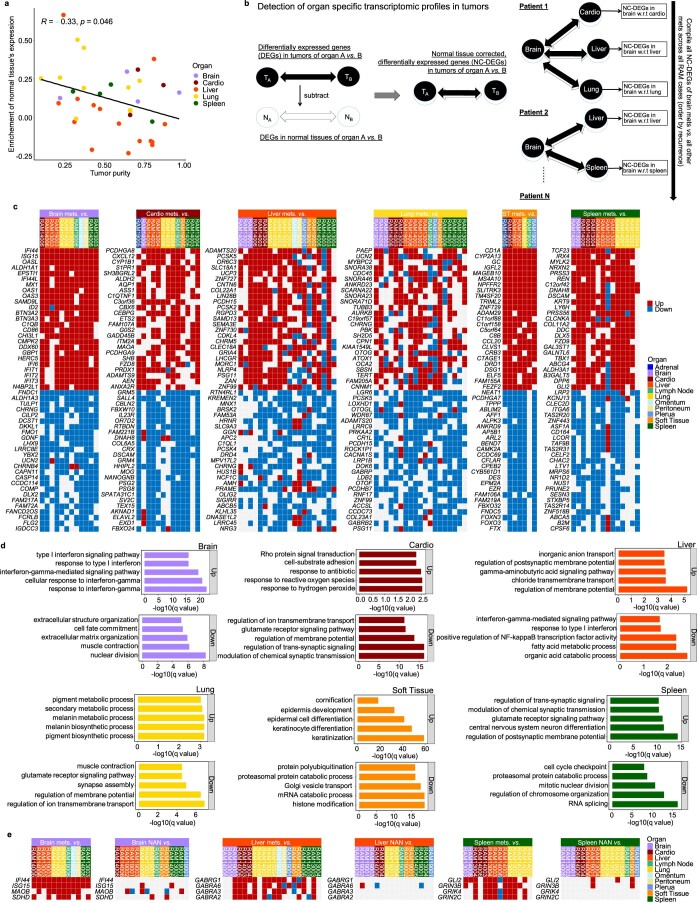
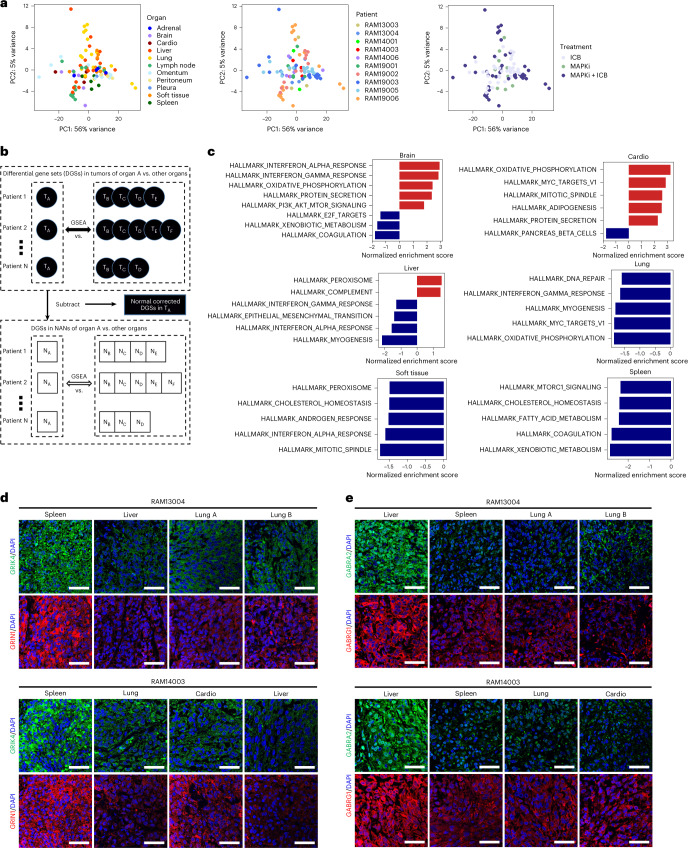
Consistent with ‘contamination’ of bulk tumors by organ-specific cell types, we observed an inverse correlation between WES-based tumor cell purities and enrichment of NAN gene expression (Extended Data Fig. 5a). However, bulk tumor transcriptomes did not segregate by cases, treatment histories or sites, possibly because of wide-ranging tumor purities (<25% to 90%) (Fig. 4a and Supplementary Table 2). We then devised a strategy to identify tumor-cell-enriched signatures by detecting differential gene sets (DGSs) and differentially expressed genes (DEGs), where DGSs and DEGs of organ-specific metastasis are (1) depleted bioinformatically of NAN DGSs and DEGs across case-matched, cross-organ, pair-wise comparisons and (2) consistent across multiple such comparisons in ≥3 cases (Fig. 4b and Extended Data Fig. 5b). We observed upregulated and downregulated DGSs and DEGs specific to brain, cardiac, liver, splenic, lung and ST metastases (Fig. 4c and Extended Data Fig. 5c). We also performed Gene Ontology (GO) enrichment analysis of recurrent DEGs (Extended Data Fig. 5d). Brain metastases organ-specifically upregulated IFN response signatures and associated with oxidative phosphorylation and PI3K-AKT-mTOR signaling27,28. Cardiac metastases upregulated oxidative phosphorylation and response to reactive oxygen species and downregulated neurotransmitter and anabolic hypoxia genes. Liver and splenic metastases both upregulated neural genes/pathways. Liver metastases upregulated the complement pathway but downregulated IFN response genes. Lung metastases upregulated snoRNAs and pigment biosynthesis, whereas ST metastases upregulated epidermal differentiation genes. We confirmed that organ-specific, tumor-cell-enriched transcripts were not differentially expressed by corresponding NANs (Extended Data Fig. 5e). At the protein level, we validated the glutaminergic versus the GABAnergic phenotypes of splenic (Fig. 4d) versus liver (Fig. 4e) metastases.
Extended Data Fig. 5. Organ-specific, normal tissue-depleted expression profiles of metastatic tumors.
(a) Correlation between tumor purity and enrichment of normal organ-tissue expression in tumor samples (n = 36) across the brain, cardio, liver, lung and splenic metastases. Only tumor samples with both WES and RNA-seq data available were included in this analysis. P value, two-sided Pearson correlation. (b) Differentially expressed gene (DEG) analyses of multi-organ metastases. DEGs between tumors from two organs are corrected by removing DEGs between non-adjacent normal tissues from the two respective organs. In the example of the brain, normal corrected-DEGs (NC-DEGs) are identified between brain metastases with respect to (w.r.t.) tumors from all other available organ sites in the same RAM case (patient). Recurrence of organ-specific, NC-DEGs is computed by combining the NC-DEGs of brain metastases across all available cases or deceased patients. (c) Top 25 upregulated (red) and downregulated (blue) genes in tumors metastatic to one specific organ (brain, cardio, liver, lung, soft tissue, or spleen; displayed horizontally) versus other organs (displayed vertically). (d) Top 5 significantly enriched gene ontology (GO) terms (biological processes) for the recurrently upregulated or downregulated genes in tumors metastatic to one specific organ versus other organs. Recurrently upregulated or downregulated genes, ≥ 30% of comparisons (in c). (e) Recurrence of tumor enriched, OXPHOS, IFN-induced (in brain) and neural-related genes (in liver and spleen) in case-matched, cross-organ comparisons for both tumor and non-adjacent normal tissue compartments.
Fig. 4. Normal tissue-depleted transcriptomic signatures of organ-specific metastases.
a, PCA of transcriptional profiles of RAM tumors (n = 93) colored by organ sites, RAM cases and treatment histories. b, DGS analyses of tumor-cell-enriched, organ-specific transcriptomes. DGSs between tumors of organ A versus other organs (that is, B, C, D, E and F) are identified by GSEA. DGSs are similarly identified between NAN tissues (n = 67) of organ A versus other organs. Subtraction of the latter from the former identifies normal-corrected/depleted (or tumor-cell-enriched) DGSs of organ-specific transcriptomes. c, Normalized enrichment scores (NESs) of top five positively and negatively enriched gene sets (hallmark gene sets, Molecular Signatures Database) in organ-specific metastases of therapy-resistant cutaneous melanoma from rapid autopsies. d,e, Immunofluorescent staining of indicated proteins within tumor tissues of distinct organs from two RAM cases (ruler, 50 μm; experiment performed once). PC, principal component.
Organ-specific tumor microenvironmental–macroenvironmental interactions
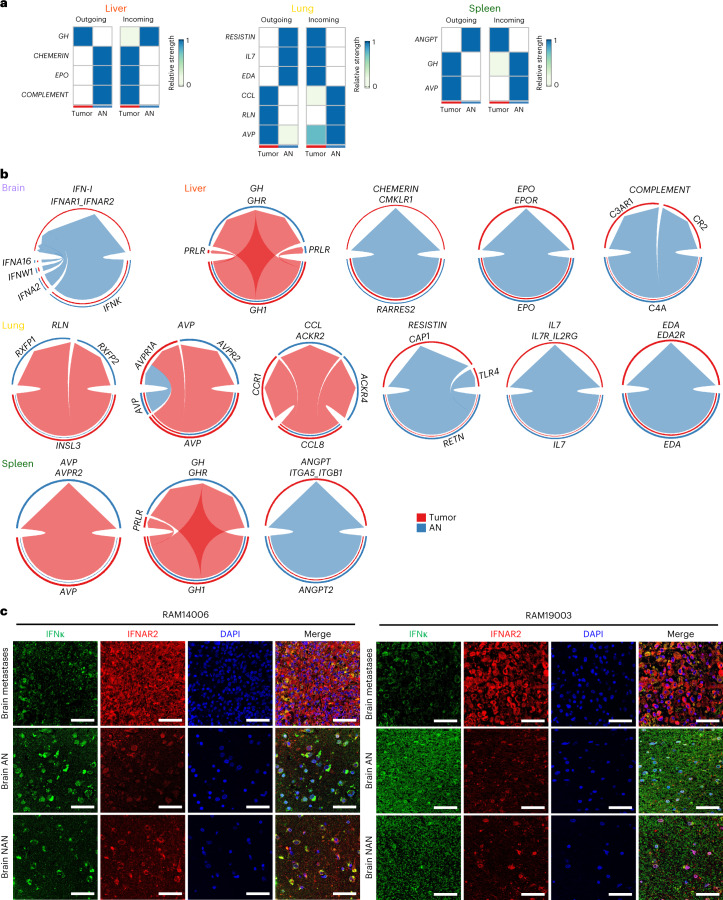
Next, we searched for ligand–receptor signaling across metastases and AN tissues. In four tumor–AN pairs from ≥3 cases, we observed growth factor, inflammation and fibrosis ligand–receptor pairs in the brain, lung, liver and spleen, visualized as outgoing or incoming (Fig. 5a) and with directionality plus connectivity strength (Fig. 5b). Lungtumor-outgoing ligand CCL connected to the tumor and AN as incoming signals via the receptors CCR1 and ACKR2/4, respectively (Fig. 5b). In brain metastases, AN-outgoing type I IFN signaled into tumors via IFNAR1/2 (Fig. 5b), which is consistent with IFN upregulation by brain metastases (Fig. 4 and Extended Data Fig. 5). In lung metastases, AN-to-tumor signals consisted of RESISTIN, IL-7 and EDA; tumor-to-AN signals consisted of CCL, RLN and AVP. In liver metastases, AN-outgoing ligands included complement genes (C4A) (Fig. 4c), EPO and CHEMERIN, and tumor-outgoing ligands included only GH. Moreover, splenic metastases featured both AN-to-tumor (ANGPT2) and tumor-to-AN (GH1 and AVP) axes. Lastly, immunofluorescence studies confirmed IFNκ expression preferentially in the AN of brain metastases, IFNAR2 intratumorally and their overlap intratumorally (Fig. 5c).
Fig. 5. Organ-specific ligand–receptor signaling between metastatic tumors and tumor-adjacent tissue macroenvironments.
a, Analysis of bulk RNA-seq showing signaling strengths of incoming and/or outgoing ligand–receptor pairs between tumor and AN (also known as macroenvironmental) tissues in the liver, lung and spleen. Values are row-scaled. b, As in a, except data visualized as chord diagrams and included the brain. Arrows point to the receivers (receptors). c, Immunofluorescent co-staining of IFNκ and IFNAR2 in RAM case-matched brain tumors, AN and NAN tissues (ruler, 50 μm; experiment performed once).
Organ-specific tumor immune contextures
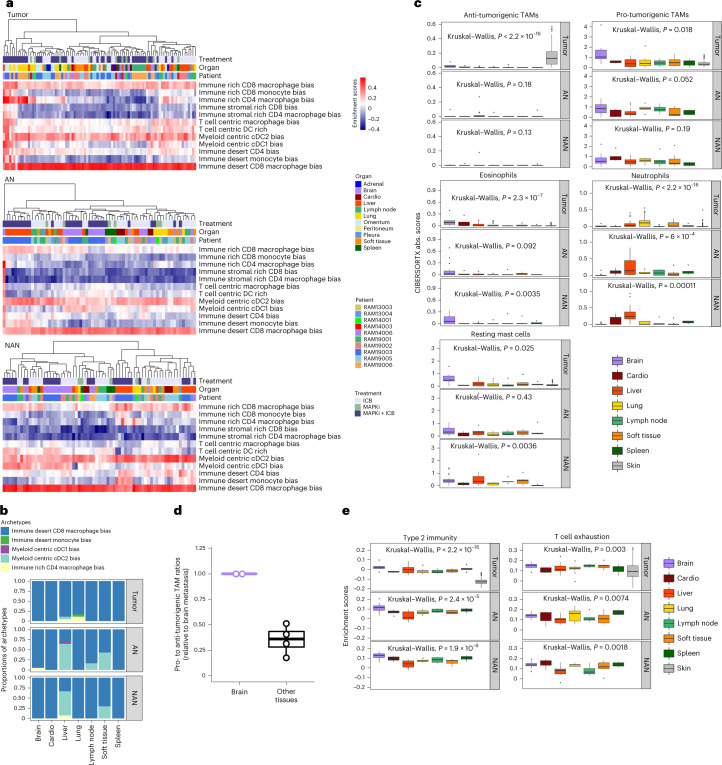
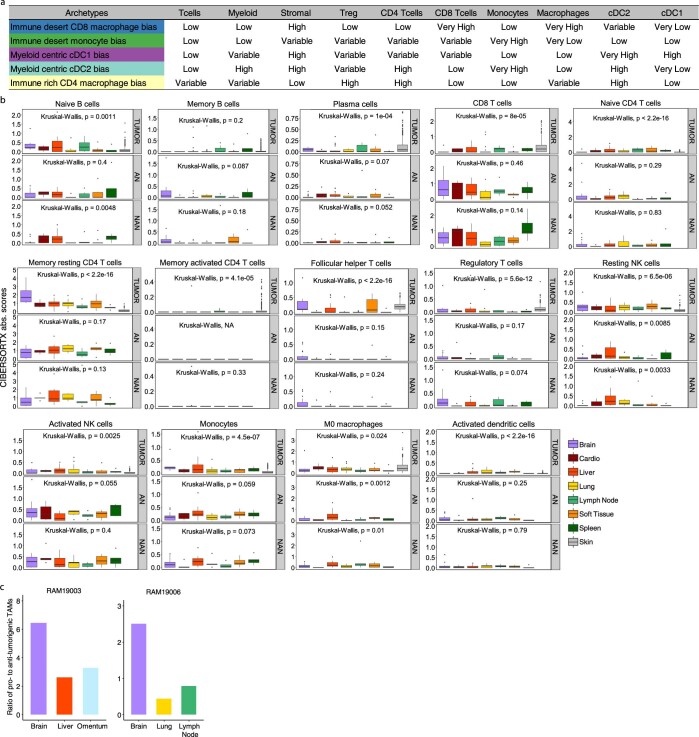
We next evaluated organ-specific (intra)tumor immune microenvironments (TIMEs) and immune macroenvironments (within AN and NAN tissues). Analysis of 12 pan-cancer immune archetypes29 via unsupervised clustering revealed immune macroenvironments, more than TIMEs, as organ-specific or patient-specific (Fig. 6a). We then assigned each tissue sample to the highest enrichment-scoring immune archetype and calculated the distribution of archetypes for each organ (Fig. 6b and Extended Data Fig. 6a). AN and NAN immune archetypes were similar in each organ, and TIMEs uniformly (~100%) enriched for the immune-desert, CD8+-macrophage-biased archetype linked to T-cell exhaustion and the worst survival in patients from the TCGA–SKCM cohort29. This contrasted with a lower frequency (~60%) of the same archetype in TCGA–SKCM tumors29, 30% of which belong to two immune archetypes: T cell-centric macrophage biased and T cell-centric dendritic cell biased29. These and the immune-rich CD8+ or CD4+ archetypes were all but absent in our RAM cohort. Interestingly, ~5% of liver TIMEs comprised the myeloid-centric, cDC2-biased archetype (associated with tumor fibrosis).
Fig. 6. Tumor immune microenvironments and macroenvironments across organ sites.
a, Unsupervised clustering of 12 pan-cancer immune archetypes in the RAM tumor (n = 93), AN (n = 68) and NAN (n = 67) tissue compartments, across RAM cases/patients, organ sites and treatment histories. Each sample’s enrichment scores of immune archetypes were used to generate heat maps. b, The composition of immune archetypes in the tumor, AN and NAN compartments across multiple organ sites. c, Absolute enrichment scores (CIBERSORTx) of anti-tumorigenic (named M1 by CIBERSORTx) or pro-tumorigenic (M2) TAMs, eosinophils, resting mast cells and neutrophils in the tumor, AN and NAN compartments of indicated organ sites, compared against the TCGA–SKCM cohort (labeled ‘skin’ as the organ site) (n = 344 BRAFMUT or NRASMUT melanoma). Comparisons among organs by the Krustal–Wallis test. Central line of each box, median; top and bottom edges of each box, first and third quartiles; whiskers extend 1.5× the interquartile range beyond box edges. Tumor compartment (brain, n = 6; cardio, n = 4; liver, n = 18; lung, n = 17; lymph node, n = 13; spleen, n = 5; soft tissue, n = 10; skin, n = 344). AN (brain, n = 21; cardio, n = 5; liver, n = 17; lung, n = 6; lymph node, n = 6; spleen, n = 5; soft tissue, n = 7). NAN (brain, n = 20; cardio, n = 5; liver, n = 15; lung, n = 5; lymph node, n = 6; spleen, n = 5; soft tissue, n = 10). d, Quantifications by multiplex immunofluorescence of the ratios of pro-tumorigenic (CD68+CD163+CD206+/−) to anti-tumorigenic (CD68+iNOS+/−CD206–CD163−) TAMs in metastatic tumors to visceral organs (n = 4; RAM19003: liver, omentum; RAM19006: lung and LN) relative to ratios in tumors to the brain (n = 2) in two RAM cases. Box plot as defined in c. e, As in c, except enrichment scores of T cell exhaustion and type 2 immunity signatures in the tumor, AN and NAN compartments across organ sites.
Extended Data Fig. 6. Tumor immune microenvironments and macroenvironments across multiple organs.
(a) Summary of characteristics and immune cell features of immune archetypes identified in the RAM tissue cohort. (b) Absolute enrichment scores of 14 immune cell types in the tumor, adjacent normal (AN), and non-adjacent normal (NAN) tissue compartments. Comparisons among organ sites by the Kruskal-Wallis test. Central line of each box, median; top and bottom edges of each box, first and third quartiles; whiskers extend 1.5x the inter-quartile range beyond box edges. TCGA–SKCM labeled as ‘skin’ in the tumor compartment category (n = 344 BRAFMUT or NRASMUT melanoma). Tumor compartment (brain, n = 6; cardio, n = 4; liver, n = 18; lung, n = 17; lymph node, n = 13; spleen, n = 5; soft tissue, n = 10, skin, n = 344). AN (brain, n = 21; cardio, n = 5; liver, n = 17; lung, n = 6; lymph node, n = 6; spleen, n = 5; soft tissue, n = 7). NAN (brain, n = 20; cardio, n = 5; liver, n = 15; lung, n = 5; lymph node, n = 6; spleen, n = 5; soft tissue, n = 10). (c) Quantifications by multiplex immunofluorescence of the ratios of pro-tumorigenic (CD68+CD163+CD206+/−) to anti-tumorigenic (CD68+iNOS+/−CD206−CD163−) TAMs in metastatic tumors to the brain and to indicated visceral organs in two RAM cases.
Using CIBERSORTx, we evaluated RAM versus TCGA–SKCM immune contextures (Fig. 6c and Extended Data Fig. 6b). Across 22 immune cell types, we observed consistent patterns between AN and NAN tissues, except for plasma cells, regulatory T cells and naive/memory B cells. Relative to RAM tumors, TCGA melanomas displayed a higher proportion of anti-tumorigenic macrophages (Fig. 6c). Across RAM organs, brain metastases enriched for pro-tumorigenic macrophages, eosinophils and resting mast cells. Using multi-spectral immunofluorescence, we confirmed a higher pro- to anti-tumorigenic tumor-associated macrophage (TAM) ratio in brain metastases (Fig. 6d and Extended Data Fig. 6c). In addition, lung metastases preferentially comprised neutrophils, potentially related to tumor-derived CCL8 (Fig. 5). Finally, RAM (versus TCGA–SKCM) enriched for T-cell exhaustion and type-2 immunity (Fig. 6e).
Discussion
This RAM study begins to build foundational insights into highly evolved and lethal CMs that resist MAPKi and/or ICB therapies. By comparative analysis of acquired-resistant CM (preceded by only one of the two types of therapies) with patient-matched pre-treatment tumors, we resolved further how each therapy distinctly and convergently shapes the high mutational, CNA and SV burdens of acquired-resistant CM. SMGs and genes altered by CNAs and SVs enrich in immune-evasive processes (for example, BRAFMUT amplification and loss-of-function alterations in B2M, JAK2, CD274/PD-L1 and PTEN; Supplementary Table 13) that may confer cross-therapy resistance, accelerating lethal disease progression. Notably, evolution of MAPKi (versus ICB) resistance shifts the mutational signatures, implicating therapy-elicited DNA damage and/or deficiency in repair pathways (for example, MMR, BER and HRR) as culprits. The evolution of late/private SVs, regardless of treatment history, enriches for RS2. Analysis of breakpoint-junctional sequences of SVs suggests NHEJ as a MAPKi or ICB co-target. Overall, multiple forms of genomic instability may cause and/or result from resistance evolution, with therapeutic implications that warrant mechanistic studies.
Acquired therapy resistance co-evolves with, and may also promote, metastatic progression. However, comparative analysis of RAM versus earlier-stage and MAPKi/ICB-naive CM cohorts is limited by cross-study technical variables (sequencing depth, read lengths, tumor purities and library preparation) and by RAM’s relatively small sample size, which may contribute to false-positive SMGs. We mitigated false positives by requiring SMGs to display gene expression and by analyzing validation cohorts. Future RAM studies should expand the current cohort size and increase representation of CM subtypes, ethnic and ancestral diversities and the treatment-naive landscape.
Our analysis, by computationally depleting bulk metastatic tumor transcriptomes of patient-matched and organ-matched normal tissue-derived transcriptomes, sheds light on organ-specific metastatic signatures. Liver and spleen metastases display neural differentiation, suggesting therapeutic targets30,31. Melanoma brain metastasis (MBM) displays signatures of IFN signaling, oxidative phosphorylation and PI3K-AKT signaling27,28,32. The brain-specific macroenvironment appears to be a predominant source of IFN ligands. Overall, RAM tumors, including MBM, strongly display an immune-desert but CD8+-macrophage-biased archetype with enrichment of T-cell exhaustion. For MBM, loss of antigen presentation and enrichment of type-2 immunity suggest TGFβ blockade and upregulation of cytotoxic natural killer (NK) cell-mediated or CD4+ T cell-mediated anti-tumor immunity33 as potential therapeutic strategies. The pro-tumorigenic TAM phenotype of MBM also suggests therapeutic co-targets. Thus, we have uncovered a preliminary set of organ-specific metastatic signatures, tumor macroenvironment crosstalks and immune contextures that characterize therapy-resistant CM, justifying expanded RAM-based and functional analyses.
Methods
Rapid autopsies and samples
We performed warm autopsies with informed consent at the University of North Carolina at Chapel Hill and the University of California, Los Angeles (UCLA). In brief, patients or persons holding the healthcare power of attorney signed the Autopsy Authorization Form and the institution-specific tumor tissue procurement and banking consent form. Research in this study involving autopsy specimens does not meet the regulatory definition of human subject research. The last tissue sample of each case was excised and stored no longer than 6 hours from death. We collected metastatic tumor, AN (≤1 cm away from tumor border) and NAN (>1 cm) tissues from 11 RAM cases. We collected formalin-fixed, paraffin-embedded (FFPE) tissues from all available sites. If tissue was sufficient, we also collected snap-frozen tissues and stored them at −80 °C. An autopsy pathologist (Leigh B. Thorne) reviewed hematoxylin and eosin tissue sections. Histopathologic analysis selected for high tumor content and against necrosis, AN with ≤10% tumor cell contamination and NAN without tumor cell contamination. We further selected for tumor purity >30% based on Sanger-sequencing-estimated BRAF or NRAS mutant allele frequencies (BRAF forward primer, 5′-GACTCTAAGAGGAAAGATGAAGTAC-3′; BRAF reverse primer, 5′-GTTGAGACCTTCAATGACTTTCTAG-3′; NRAS forward primer, 5′-GGCTTGAATAGTTAGATGCTTATTTAACCTTGGC-3′; and NRAS reverse primer, 5′-GCTCTATCTTCCCTAGTGTGGTAACCTC-3′).
Clinical samples
Tumor tissues from living patients with CM and patient-matched normal tissues were collected with the approval of institutional review boards at UCLA and Vanderbilt Ingram Cancer Center and with informed consent of each patient or the patient’s legal representative. We analyzed by WES 88 tumor samples from 28 patients at UCLA with BRAFMUT CM obtained before treatments with and responses to MAPKi and then at the time of disease progression (that is, acquired resistance), along with patient-matched normal tissues (Supplementary Table 5). From PDX models collected from patients at UCLA, we analyzed by WES 29 tumors and patient-matched normal tissues (from eight patients with BRAFMUT or NRASMUT CM) treated with vehicle or trametinib (at sufficient in vivo doses to induce tumor regression) in NSG mice (Supplementary Table 6). Also, we analyzed by WES 14 tumor samples from seven patients with CM at Vanderbilt obtained before treatments with and responses to ICB and then at the time of disease progression (Supplementary Table 5). Sex/gender was self-reported and not considered in the study design given that each cohort size was small. Participation in research was not compensated.
PDXs and treatments
To develop PDX models, tumor fragments derived from metastatic CM, with approval by UCLA institutional review boards and the UCLA Animal Research Committee, were transplanted subcutaneously in sex-matched NSG mice (4–6 weeks old) from the UCLA vivarium or Jackson Laboratory. We conducted all animal experiments in accordance with approved protocol and regulations (ARC 2016-086). We adhered to the maximal tumor size for experimental endpoints, which was ~1,500–2,000 mm3 without mobility impairment and with body condition score >2. We implanted one tumor fragment in each mouse on the flank and measured tumors with a calliper every 2 days. Tumor volumes were calculated by (length × width2)/2, and we excluded data from occasional animals that died before final analysis. We assigned tumors with volumes ~500 mm3 randomly into experimental groups. A special mouse chow (Test Diet) incorporated trametinib (LC Laboratories) to achieve daily dosing at 5 mg/kg/day.
WES and WGS
We extracted gDNAs from snap-frozen tumors, NANs and ANs using the QIAGEN AllPrep DNA/RNA Mini Kit; from FFPE tumors (clinical ICB cohort) using the QIAGEN QIAamp DNA FFPE Tissue Kit; and from frozen blood using the QIAGEN FlexiGene DNA Kit. Quantification of gDNAs was based on NanoDrop (Thermo Fisher Scientific) and/or Qubit fluorometer using the Qubit dsDNA Broad Range (BR) Assay Kit (Life Technologies). Size and quality of gDNAs were based on TapeStation (Agilent Technologies) and/or agarose gel electrophoresis to ensure gDNA library preparation from equal gDNA input and from intact high-molecular-weight gDNA. We prepared whole-exome libraries using the Roche NimbleGen Exon-Seq Kit, the Roche NimbleGen SeqCap Kit or the Roche KAPA HyperPlus Library Preparation Kit with KAPA HyperCap Workflow version 3.0 for exome hybridization (Supplementary Table 14) and whole-genome libraries using the Roche KAPA HyperPrep Kit. In brief, after enzymatic fragmentation of gDNAs, the libraries were constructed by end-repairing and A-tailing the fragmented DNAs, ligation of adapters and polymerase chain reaction (PCR) amplification. After library construction, we quantified indexed libraries for equal molar pooling and paired-end sequenced with a read length of 2 × 150 bp on the Illumina HiSeq 3000 or Illumina NovaSeq 6000 S4 platform.
RNA-seq
We extracted total RNAs from snap-frozen tissues using the QIAGEN AllPrep DNA/RNA Mini Kit and the Ambion mirVana miRNA Isolation Kit. Total RNA quantification was based on the Qubit RNA High Sensitivity (HS) Assay Kit (Thermo Fisher Scientific) and/or a NanoDrop (Thermo Fisher Scientific). RNA quality was based on TapeStation (Agilent Technologies) and used to calculate the input for RNA library preparation. We prepared RNA libraries using KAPA RNA HyperPrep Kit with RiboErase (Roche) or TruSeq RNA Exome Kit (Illumina), TruSeq RNA Single Indexes Set A Kit (Illumina) and TruSeq RNA Single Indexes Set B Kit (Illumina) following the manufacturers’ protocols (Supplementary Table 15). In brief, RNA libraries, prepared by the KAPA RNA HyperPrep Kit with RiboErase, used 1 μg of total RNA captured by magnetic oligo-dT beads. RNAs were fragmented and cDNAs synthesized using random priming for the first strand. The second strand was synthesized (with dUTP) and marked to convert the cDNA:RNA hybrid to double-stranded cDNA (dscDNA). Then, dscDNA libraries were constructed by adding dAMP to the 3′ ends during A-tailing, and dsDNA adapters with 3′ dTMP overhangs were ligated to library insert fragments during adapter ligation. We carried out amplification of library fragments carrying appropriate adapter sequences at both ends using high-fidelity, low-bias PCR amplification, whereas the strand marked with dUTP was not amplified, to enable strand-specific sequencing. After library construction, we quantified indexed libraries for equal molar pooling and single-end sequenced with a read length of 1 × 50 bp on the Illumina HiSeq 3000 or Illumina NovaSeq 6000 S4 platform. RNA libraries prepared by the TruSeq RNA Exome Kit used 50 ng of RNA for high-quality RNA (DV200 > 70%) and 100 ng of RNA for medium-quality and low-quality RNA (DV200 < 70%). After RNA fragmentation and cDNAs synthesis, we purified dscDNAs using AMPure XP beads (Beckman Coulter). After A-tailed enrichment, we quantified cDNA libraries by the Qubit dsDNA HS Assay Kits (Thermo Fisher Scientific); quality was assessed by TapeStation. We used 200 ng of each DNA library for exome enrichment. After amplification and purification, we removed free adapters by using Illumina Free Adapter Blocking Reagent (Illumina). All libraries were pooled at equal molar and sequenced with a 2 × 150-bp read length on the Illumina NovaSeq 6000 S4 platform.
Somatic SNVs and CNAs
We used BWA for mapping and Picard for removal of duplications. We identified somatic SNVs and IDs of tumors34,35 by using patient-matched normal tissues for germline reference. We called SNVs using the Unified Genotyper tool of GATK, MuTect and VarScan2 and IDs using GATK-UGF, SomaticIndelDetector of GATK (IndelLocator) and VarScan2 (calls made by at least two of three algorithms). SNV/IDs were supported by at least five reads in the tumor samples and none in the patient-matched normal tissues. Somatic SNV/IDs were then annotated by Oncotator36. Finally, we used Sequenza37 to detect tumor purity, ploidy, somatic CNAs and loss-of-heterozygosity regions.
Significant CNA genes in RAM tumors
We applied GISTIC2.0 (ref. 38) to identify the significantly deleted and amplified regions using each RAM tumor’s copy number segmentation file as the input. We generated circos plots with both q values and G-scores representing the amplitude of the aberration. Only regions with q values less than 0.05 were defined as significantly altered regions. We performed Fisher’s exact test to identify differentially amplified or deleted genes in the RAM tumor cohort versus the BRAF or NRAS mutated TCGA–SKCM cohort (n = 345 tumors: seven stage 0, 65 stage 1, 100 stage 2, 130 stage 3, 17 stage 4 and 26 unknown). We downloaded TCGA–SKCM CNA data from cBioPortal and compiled the frequencies of CNA genes (Supplementary Table 16). We applied Fisher’s exact test to individual-level amplified or deleted events, which were counted in a given RAM case if they were identified from at least one tumor of that case. We identified RAM frequency-enriched (versus TCGA–SKCM) CNA genes as amplified or deleted genes with a false discovery rate (FDR)-adjusted P < 0.05 and a higher frequency in RAM (versus TCGA–SKCM) cohort.
CNA genes enriched in post (versus pre) acquired MAPKi resistance
We compared the patient-level frequency of each amplified or deleted gene in post-treatment (acquired-resistant) versus pre-treatment melanoma by using Fisher’s exact test. When multiple post tumors were available from a given patient, they were considered as an entirety, and an amplified or deleted gene was counted in this patient if it was identified from at least one of the post tumors. Multiple pre-treatment tumors from one patient were also counted as an entirety. We nominated a post-enriched amplified or deleted gene if its frequency is higher in post (versus pre) tumors and the FDR-adjusted P value is less than 0.05.
CNA genes enriched in post (versus pre) acquired ICB resistance
An amplified or deleted gene enriched in acquired ICB resistance was defined by its amplification or deletion frequency in the post tumors being >2× that in the pre tumors and by its detection in ≥2 patients’ post tumors.
SMGs
We applied MutSig2CV to identify SMGs using each RAM case’s mutational profile as the input and each RAM case (not each tumor sample) as an identifier. A mutation exists in a given RAM case if it was identified in ≥1 tumor. To circumvent the limitation of a small sample size, we inflated type I error by not performing multiple testing39 and identified by MutSig2CV genes at P < 0.05. To reduce false positives, we nominated SMGs by filtering for RNA expression. Based on the mean values of normalized expression levels (log2 counts per million (CPM)) of RAM tumors, we annotated MutSig2CV SMGs as no expression (mean log2CPM < 0), expressed (0 ≤ mean log2CPM < 4) or highly expressed (mean log2CPM > 4) and excluded those with no expression. We performed Fisher’s exact test to compare the patient-level frequency of each SMG in the RAM versus TCGA–SKCM cohorts and used the FDR approach to adjust the P values. We then performed GO enrichment analysis with the clusterProfiler package to detect the significant biological processes for expressed SMGs. Moreover, we identified SMGs with MutSig2CV for post tumors from patients with MAPKi-only or ICB-only treatments by using each patient’s mutational profile of post tumor(s) as input and each patient (not each post tumor) as an identifier. For patients with multiple post tumors, a mutation was considered to exist in this patient if it was detected in ≥1 tumor. We defined post/acquired-resistant SMGs as genes identified by MutSig2CV at P < 0.05 in the post tumors and not identified in the patient-matched pre tumor(s).
Phylogeny and mutational signatures
We performed phylogenetic analysis using the PHYLIP program with the parsimony algorithm35 and annotated each tree with potential drivers of tumorigenesis and/or resistance. All tumors in each case shared early somatic SNVs; a subset shared intermediate SNVs; and late SNVs were unique to one tumor. The contribution of each mutational type and mutational signature was then determined for early, intermediate and late mutations through the R package deconstructSigs40 with COSMIC SBS signatures version 3.3 as reference. For the pre and post MAPKi-only and ICB-only cohorts, we identified mutations unique to post tumors per patient, which were then subjected to mutational signature analysis. We used the R package MutationalPatterns with COSMIC ID signatures version 3.3 as reference to determine ID signatures underlying all identified B2M IDs.
ITH and preferentially mutated genes
We conducted a subclonal analysis for each RAM case using PyClone-VI41 and assessed each mutation’s cancer cell fraction (CCF). Mutations were clonal if the CCF approaches 1; otherwise, mutations were subclonal. The ratio of subclonal mutations to all mutations determined ITH. We determined preferentially mutated genes for each organ as follows: (1) selected for non-synonymous mutations; (2) calculated ΔCCF (see formula in Extended Data Fig. 3b) of each mutation in the tumor of one specific organ (that is, TA) versus tumors of other organs (that is, TB…TN); (3) identified organ-specific enriched mutations with a ΔCCF of >0.2 for each RAM case; and (4) defined a gene as ‘preferentially mutated genes’ of organ A when organ-A-enriched mutations in this gene occurred in ≥3 patients.
WGS-based SV analysis
We mapped WGS reads to GRCh38/hg38 human reference genome using BWA-MEM42. SAMtools43 sorted alignments and removed PCR duplicates. CNVkit44 with default parameters called CNVs. SVs were concordant by two or more of three methods: SvABA45, TIDDIT46 and DELLY47. For high-coverage data (>15×), we used default parameters. The parameter minimum number of points (-l) was set to 5 in TIDDIT for low-coverage data. SVs detected in matched-normal genomes were removed from the tumor samples to infer somatic SVs. We estimated rearrangement signatures based on a previous classification26. For RAM cases with ≥2 tumors, we computed early, intermediate (only feasible with RAM12.01 and RAM14006) and late somatic SVs. For each case, early and late somatic SVs consisted of SVs common to all tumors and unique to each tumor, respectively, and the rest were classified as intermediate. We carried out breakpoint junctional sequence analysis by examining the presence of homologous sequences. Homologous sequences with 0–1 bp, 2–6 bp and >6 bp were attributed to NHEJ, alternative NHEJ (microhomologous end joining) and HRR, respectively. Genes overlapping with somatic SVs were annotated using AnnotSV48. We performed pathway enrichment analysis using the Molecular Signatures Database (MSigDB)49 with pathway datasets listed in KEGG, Reactome and Pathway Interaction databases.
Reconstruction of focal amplifications
Focal amplicon identification and elucidation of circular extrachromosomal DNAs (ecDNAs) and complex genomic rearrangements (CGRs) using WGS data were carried out by AmpliconArchitect50. In brief, we determined the list of potential intervals for each amplicon, for which copy numbers and SVs were estimated using read depth and discordant read signatures. It then constructed a breakpoint graph. Simple cycles were then decomposed from the breakpoint graphs and amplicons classified into ecDNAs, CGRs and linear amplicons. We used CNVKit to infer the initial set of CNV seed regions. SV view of amplicons was generated using functions available in AmpliconArchitect.
WGS-based mutation analysis
Somatic SNVs and IDs were identified using Strelka2 (ref. 51) with default parameters and then subjected to mutational and ID signature analyses using the R package deconstructSigs40 and MutationalPatterns52, respectively, with COSMIC SBS/ID signatures version 3.3 as reference. Classification of early, intermediate and late somatic SNVs was in accordance with that for WES-based mutations.
RNA-seq analysis
We analyzed single-end and paired-end RNA-seq data3,35 by mapping transcriptome reads to the GRCh38/hg38 human reference genome using HISAT2. Gene-level counts were estimated by the htseqcount program. The normalized expression level of each gene, log2CPM, was calculated by the R package edgeR53 and batch corrected with the ‘removeBatchEffect’ function in the limma R package54. We performed principal component analysis (PCA) using the prcomp function in R package stats to visualize the clustering of samples.
Organ tissue expression in RAM tumors
We first used the DESeq2 package to detect DEGs between NANs from one specific organ versus other organs. We defined organ-specific normal gene signatures as the top five significantly upregulated genes, all of which were confirmed to display organ-specific expression in the human protein atlas55. For RAM tumors, we performed single-sample gene set enrichment analysis (GSEA) to generate the enrichment scores of the organ-matched normal organ-specific gene signature. CPM values were input into the gene set variation analysis (GSVA) program using the default ‘kcdf=Gaussian’ option.
DGS analysis
GSEA via the fgsea package used the human hallmark (H) gene sets (MSigDB). Genes were pre-ordered by the log2-transformed expression fold change metrics (log2FC). We calculated the enrichment nominal P values by permutation test (100,000 permutations), with Benjamini–Hochberg FDR correction for multiple testing. We then performed GSEA to identify the DGSs for both tumors and NANs of one organ versus other organs. We defined normal-corrected DGSs (NC-DGSs) in tumors of organ A as those DGSs detected in the tumors of organ A (versus other organs) but not in the NAN compartment of organ A (versus other organs).
DEG analysis
To filter out normal tissue-specific DEGs from bulk tumor transcriptomes, we used RNA-seq derived from each tumor’s organ-matched NANs and public datasets of normal organ/tissue gene expressions from the Illumina Human Body Map and GTEx. DEGs between a tumor pair from two organs (for example, TA versus TB) were corrected for expression of the same genes between the normal tissue pair of the same two organs (for example, NA versus NB). We calculated normal-corrected fold change (NC-FC) of each gene between TA versus TB as the FC of this gene between TA versus TB divided by the FC of this gene between NA versus NB. We defined the genes with NC-FC > 2 or NC-FC < 0.5 as the upregulated or downregulated NC-DEGs between these two organs. We then computed the recurrence (≥30%) of NC-DEGs by collecting NC-DEGs of tumors from one specific organ against all other tumors from other organs of the same RAM case across all RAM cases. GO enrichment analysis via the ‘clusterProfiler’ package56 identified the top five significant GO biological processes for recurrently upregulated or downregulated NC-DEGs of each organ-specific metastasis.
Analysis of tumor–macroenvironment interactions
Only organ sites with ≥4 tumor–AN pairs and four tumor–NAN pairs available from ≥3 RAM cases were included for this analysis using the CellChat R package57. We culled significant ligand–receptor signaling that was detected from tumor–AN pairs of one specific organ site but not from tumor–NAN pairs of the same organ site. The netAnalysis_signalingRole_heatmap function was used for visualization. The circle plots depicting tumor–AN interactions of each organ were generated by applying netVisual_aggregate with the options layout = ‘circle’.
Analysis of immune contextures
We performed single-sample GSEA to generate the absolute enrichment scores of the 12 immune archetypes for each tissue, using CPM values of all expressed genes as input for GSVA58 in the default ‘kcdf=Gaussian’ option. We then assigned each tissue to the highest enrichment scoring immune archetype. CIBERSORTx59 was used in the ‘absolute mode’ to estimate infiltration levels of 22 immune cell types with CPM values as input. We downloaded the gene-level normalized read counts (RSEM, file name: Batch normalized from Illumina HiSeq_RNASeqV2) of TCGA–SKCM RNA-seq (inclusive of only BRAF or NRAS mutant tumors) from cBioPortal. CIBERSORTx estimated the absolute abundance of 22 immune cell types with the normalized expression level as an input. We calculated the enrichment scores of two signatures, ‘T cell exhaustion’ and ‘type 2 immunity’33, by GSVA using the default ‘kcdf=Gaussian’ option.
Immunofluorescence
FFPE RAM tissue sections were heated at 90 °C for 25 minutes and immersed in xylene and gradient ethanol to achieve deparaffinization and re-hydration. Then, tissue sections were subjected to heat at 95 °C 10 mM citrate buffer (pH 6.0) for 15 minutes to retrieve antigens. After permeabilization and blocking with 0.1% Triton X-100/10% normal goat serum in PBS for 1 hour, tissue sections were incubated with primary antibodies, including anti-GRIK4 (Invitrogen, MA5-31745, 1:200), anti-GRIN1 (Abcam, ab109182, 1:50), anti-GABRG1 (Invitrogen, PA5-99317, 1:100), anti-GABRA2 (Invitrogen, PA5-106894, 1:100), anti-IFNκ (Novus, H00056832-M01, 1:50) and anti-IFNAR2 (Invitrogen, PA5-119915, 1:200), at 4 °C overnight. Visualization was achieved with goat anti-mouse IgG highly cross-absorbed secondary antibody, Alexa Fluor Plus 488 (Invitrogen, A32723, 1:400) or goat anti-rabbit IgG highly cross-absorbed secondary antibody, Alexa Fluor 555 (Invitrogen, A-21429, 1:500), and nuclei were counterstained by DAPI (Sigma-Aldrich, D9542). We captured images on a Leica confocal SP8-STED/FLIM/FCS microscope.
Multi-spectral immunofluorescence
Using Ventana Discovery Ultra (Roche) and Opal fluorophores (Akoya Biosciences), we deparaffinized 5-µm-thick tissue sections using EZ-Prep reagent (Roche) and retrieved antigens in CC1 buffer (pH 9, 95 °C; Roche). Discovery Inhibitor (Roche) was applied to inhibit enzymatic activities, followed by six sequential rounds of staining. Each round included the addition of a primary antibody followed by secondary antibody detection using either OmniMap anti-Ms HRP (Roche, 760-4310, ready-to-use) for mouse or OmniMap anti-Rb-HRP (Roche, 760-4311, ready-to-use) for rabbit following the manufacturer’s specifications. We amplified signals by using Opal fluorophores at 1:400. Between rounds of staining, the tissue sections underwent heat-induced epitope retrieval to remove the primary/secondary-HRP antibody complexes before staining with the subsequent antibody. The primary antibodies and corresponding fluorophores are anti-MRT-1 (Abcam, ab210546, 1:200) in Opal 480; anti-iNOS (Abcam, ab115819, 1:200) in Opal 520; anti-CD68 (Roche, 790-2931, ready-to-use) in Opal 570; anti-CD163 (Abcam, ab182422, 1:200) in Opal 620; anti-CD206 (Cell Signaling Technology, 91992S, 1:200) in Opal 690 and anti-SOX10 (Abcam, ab227680, 1:200) in Opal 780. We counterstained nuclei with Spectral DAPI (Akoya Biosciences, FP1490) and mounted the stained tissues with ProLong Diamond Antifade mounting medium (Thermo Fisher Scientific). Subsequently, we imaged stained tissues (×20) using the Vectra Polaris imaging system (Akoya Biosciences). After image capture, unmixing of the spectral libraries was performed with inForm software (Akoya Biosciences). Unmixed images were then imported into HALO (Indica Labs) for stitching, cell segmentation and cell phenotyping. We analyzed whole tumor regions from each slide. Data were exported and graphed with Prism (GraphPad). Representative images were exported from HALO after spectral unmixing.
Statistical methods
We conducted statistical analyses in R 4.02, Python 3.8.0, Python 2.7.17 and Prism.
Reporting Summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41591-023-02304-9.
Supplementary information
Supplementary Table 1. Clinical information and tissue collection. Table 2. WES data quality, characteristics and availability of sample-matched WGS and/or RNA-seq. Table 3. WGS data quality, characteristics and availability of sample-matched WES and/or RNA-seq. Table 4. RNA-seq data quality and availability of sample-matched WES and/or WGS. Table 5. Clinical information for patient-matched tumors biopsied pre-treatment and at disease progression. Table 6. Clinical information and driver mutations for PDX models. Table 7. TMB of BRAFMUT and NRASMUT melanoma from published cutaneous melanoma studies. Table 8. Significantly amplified and deleted regions and genes in the RAM tumor cohort analyzed by GISTIC2.0. Table 9. Significantly amplified and deleted genes in RAM versus TCGA–SKCM (BRAF and NRAS mutated) cohorts: results of two-sided Fisher’s exact test. P values adjusted by FDR. Table 10. SMGs in the RAM cohort identified by MutSig2CV without multiple testing. Table 11. Recurrent somatic translocations. Table 12. Recurrent somatic structure variation-associated genes. Table 13. Mutational status of genes associated with cross-therapy resistance among RAM cases. Table 14. Summary of WES library preparation kits. Table 15. Summary of RNA-seq library preparation kits. Table 16. CNAs of the BRAF and NRAS mutant TCGA–SKCM cohorts (downloaded from cBioProtal).
Acknowledgements
We thank all members of the Lo laboratory for their critical comments. This research was supported by grants (to R.S.L.) from the National Institutes of Health (NIH) (1R01CA176111A1, 1R21CA215910-01, R21CA255837-01, U54CA274509 and 1P01CA168585), the Melanoma Research Alliance (MRA) (Team Science Awards 2019) and the V Foundation for Cancer Research (Translational Award). Additional funding was provided by the MRA Team Science Award 2022 (to R.S.L. and D.B.J.), the MRA Dermatology Fellows Award (S.L., P.D. and Z.Y.), the MRA Young Investigator Award (G.M.), the Luke and Susan Simons Melanoma Fund (to D.B.J.), the Jonsson Comprehensive Cancer Center (JCCC) Postdoctoral Seed Grant (to Z.Y.) and JCCC Postdoctoral Fellowships (to S.L., P.D., X.Z. and Z.Y.). S.L. is the recipient of a Career Development Award from the Melanoma Research Foundation. Funding for this research was also provided (to S.J.M. and R.S.L.) by Merck Sharp & Dohme, a subsidiary of Merck & Co., Inc. Additional support came from Mary Tanner and Maurizio Grimaldi and the Ressler Family Foundation and the University Cancer Research Fund (Lineberger Comprehensive Cancer Center at the University of North Carolina at Chapel Hill (UNC-CH)). We thank V. J. Moylan, Jr. (Assistant Professor of Pathology and Laboratory Medicine, UNC-CH), Medical Examiner for Orange County, North Carolina, and L. B. Thorne (Professor of Pathology and Laboratory Medicine, UNC-CH) for performing warm autopsies and quality control analysis of tissues used. We also thank X. Li (Director) and the Technology Center for Genomics and Bioinformatics at UCLA for excellent technical support.
Extended data
Source data
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Author contributions
Conceptualization: S.J.M. and R.S.L.; methodology: S.L., P.D., S.H.L., Y.W., X.Z., Z.Y., R.J.L., C.D., P.O.S., G.M., S.J.M. and R.S.L.; formal analysis: S.L., P.D., S.H.L., Y.W., X.Z., R.J.L., C.D., G.M., S.J.M. and R.S.L.; investigation: S.L., P.D., S.H.L., Y.W., X.Z., Z.Y., R.J.L., G.M., S.J.M. and R.S.L.; resources: S.M.D., G.M., D.B.J., S.J.M. and R.S.L.; writing—original draft: S.L., P.D. and R.S.L; writing—review and editing: S.L., P.D., S.H.L., Y.W., X.Z., Z.Y., R.J.L., G.M., D.B.J., S.J.M. and R.S.L; visualization: S.L., P.D., X.Z., R.J.L., G.M. and R.S.L.; supervision: R.S.L.; and funding acquisition: R.S.L.
Peer review
Peer review information
Nature Medicine thanks Carla Daniela Robles-Espinoza, Benjamin Izar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Anna Maria Ranzoni, in collaboration with the Nature Medicine team.
Data availability
The BAM files of WES, WGS and RNA-seq data are deposited in the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/) with accession number EGAS00001006644. Access requires a data sharing agreement. Source data are provided with this paper.
Competing interests
R.S.L. reports research support from Merck and OncoSec, clinical trial support from Pfizer and Bristol Myers Squibb, honorarium from Pfizer and US Patent Application Serial No. 63/488,609. S.J.M. reports research support from Merck, Amgen and SYNDAX and consultant work for EMD Serono and iTeos Therapeutics. D.B.J. reports research support from Bristol Myers Squibb and Incyte and consultant work for Bristol Myers Squibb, Catalyst, Iovance, Jansen, Mallinckrodt, Merck, Mosaic, Novartis, Oncosec, Pfizer and Targovax. S.M.D. reports service on scientific advisory boards for LungLife AI and Early Diagnostics. P.O.S. reports research support from Castle Biosciences, advisory board service for Castle Biosciences and being a co-founder and shareholder of Tempo Therapeutics. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Prashanthi Dharanipragada, Shirley H. Lomeli, Yan Wang, Xiao Zhang.
Extended data
is available for this paper at 10.1038/s41591-023-02304-9.
Supplementary information
The online version contains supplementary material available at 10.1038/s41591-023-02304-9.
References
- 1.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong A, et al. Durable suppression of acquired MEK inhibitor resistance in cancer by sequestering MEK from ERK and promoting antitumor T-cell immunity. Cancer Discov. 2021;11:714–735. doi: 10.1158/2159-8290.CD-20-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 5.Cortes-Ciriano I, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillekas H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8:5574–5576. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budczies J, et al. The landscape of metastatic progression patterns across major human cancers. Oncotarget. 2015;6:570–583. doi: 10.18632/oncotarget.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies MA, et al. Prognostic factors for survival in melanoma patients with brain metastases. Cancer. 2011;117:1687–1696. doi: 10.1002/cncr.25634. [DOI] [PubMed] [Google Scholar]
- 9.Peinado H, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 10.Hugo W, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriceau G, et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 2015;27:240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, et al. A novel AKT1 mutant amplifies an adaptive melanoma response to BRAF inhibition. Cancer Discov. 2014;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, et al. Melanoma whole-exome sequencing identifies V600EB-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagle N, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrov LB, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degasperi, A. et al. Substitution mutational signatures in whole-genome-sequenced cancers in the UK population. Science376, science.abl9283 (2022). [DOI] [PMC free article] [PubMed]
- 21.Liu D, et al. Evolution of delayed resistance to immunotherapy in a melanoma responder. Nat. Med. 2021;27:985–992. doi: 10.1038/s41591-021-01331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Anti-PD-1/L1 lead-in before MAPK inhibitor combination maximizes antitumor immunity and efficacy. Cancer Cell. 2021;39:1375–1387. doi: 10.1016/j.ccell.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, et al. Molecular profiling of patient-matched brain and extracranial melanoma metastases implicates the PI3K pathway as a therapeutic target. Clin. Cancer Res. 2014;20:5537–5546. doi: 10.1158/1078-0432.CCR-13-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho JH, et al. AKT1 activation promotes development of melanoma metastases. Cell Rep. 2015;13:898–905. doi: 10.1016/j.celrep.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Combes AJ, et al. Discovering dominant tumor immune archetypes in a pan-cancer census. Cell. 2022;185:184–203. doi: 10.1016/j.cell.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatesh H, Monje M. Neuronal activity in ontogeny and oncology. Trends Cancer. 2017;3:89–112. doi: 10.1016/j.trecan.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zahalka AH, Frenette PS. Nerves in cancer. Nat. Rev. Cancer. 2020;20:143–157. doi: 10.1038/s41568-019-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M, et al. TGF-β suppresses type 2 immunity to cancer. Nature. 2020;587:115–120. doi: 10.1038/s41586-020-2836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong A, et al. Exploiting drug addiction mechanisms to select against MAPKi-resistant melanoma. Cancer Discov. 2018;8:74–93. doi: 10.1158/2159-8290.CD-17-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S, et al. Response and recurrence correlates in patients treated with neoadjuvant anti-PD-1 therapy for resectable oral-cavity squamous cell carcinoma. Cell Rep. Med. 2021;2:100411. doi: 10.1016/j.xcrm.2021.100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos AH, et al. Oncotator: cancer variant annotation tool. Hum. Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Favero F, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inman GJ, et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat. Commun. 2018;9:3667. doi: 10.1038/s41467-018-06027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gillis S, Roth A. PyClone-VI: scalable inference of clonal population structures using whole genome data. BMC Bioinformatics. 2020;21:571. doi: 10.1186/s12859-020-03919-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wala JA, et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisfeldt J, Vezzi F, Olason P, Nilsson D, Lindstrand A. TIDDIT, an efficient and comprehensive structural variant caller for massive parallel sequencing data. F1000Res. 2017;6:664. doi: 10.12688/f1000research.11168.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geoffroy V, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34:3572–3574. doi: 10.1093/bioinformatics/bty304. [DOI] [PubMed] [Google Scholar]
- 49.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deshpande V, et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat. Commun. 2019;10:392. doi: 10.1038/s41467-018-08200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 52.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 56.Wu T, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb.) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin S, et al. Inference and analysis of cell–cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newman AM, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Clinical information and tissue collection. Table 2. WES data quality, characteristics and availability of sample-matched WGS and/or RNA-seq. Table 3. WGS data quality, characteristics and availability of sample-matched WES and/or RNA-seq. Table 4. RNA-seq data quality and availability of sample-matched WES and/or WGS. Table 5. Clinical information for patient-matched tumors biopsied pre-treatment and at disease progression. Table 6. Clinical information and driver mutations for PDX models. Table 7. TMB of BRAFMUT and NRASMUT melanoma from published cutaneous melanoma studies. Table 8. Significantly amplified and deleted regions and genes in the RAM tumor cohort analyzed by GISTIC2.0. Table 9. Significantly amplified and deleted genes in RAM versus TCGA–SKCM (BRAF and NRAS mutated) cohorts: results of two-sided Fisher’s exact test. P values adjusted by FDR. Table 10. SMGs in the RAM cohort identified by MutSig2CV without multiple testing. Table 11. Recurrent somatic translocations. Table 12. Recurrent somatic structure variation-associated genes. Table 13. Mutational status of genes associated with cross-therapy resistance among RAM cases. Table 14. Summary of WES library preparation kits. Table 15. Summary of RNA-seq library preparation kits. Table 16. CNAs of the BRAF and NRAS mutant TCGA–SKCM cohorts (downloaded from cBioProtal).
Data Availability Statement
The BAM files of WES, WGS and RNA-seq data are deposited in the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/) with accession number EGAS00001006644. Access requires a data sharing agreement. Source data are provided with this paper.