Abstract
Background & Aims
Around 20% of patients with non-alcoholic fatty liver disease (NAFLD) are lean. Increasing evidence suggests that lean NAFLD is a unique subtype of the disease. We aimed to explore the metabolic profile, genetic basis, causal risk factors, and clinical sequelae underlying lean NAFLD.
Methods
NAFLD was diagnosed by whole liver proton density fat fraction ≥5%. Whole liver proton density fat fraction and hepatic iron were quantified using magnetic resonance imaging in the UK Biobank. Individuals in this study were stratified according to the World Health Organization criteria of obesity, into lean, overweight, and obese. Mediation analysis, Mendelian randomisation analysis, and Bayesian networks were used to identify a risk factor or a clinical sequela of lean/obese NAFLD.
Results
Lean NAFLD manifested a distinct metabolic profile, featured by elevated hepatic iron and fasting glucose. Four loci, namely, HFE rs1800562, SLC17A3-SLC17A2-TRIM38 rs9348697, PNPLA3 rs738409, and TM6SF2 rs58542926, were associated with lean NAFLD (p <5 × 10-8). HFE rs1800562 was specifically associated with lean NAFLD and demonstrated a significant mediation effect through elevating hepatic iron. Type 2 diabetes was the most pronounced clinical sequela of lean NAFLD, followed by liver cirrhosis.
Conclusions
Our study suggested that HFE plays a potential steatogenic role rather than regulating iron homoeostasis in patients with lean NAFLD. The increased liver iron deposition is associated with lean NAFLD, whereas obese NAFLD is not related to hepatic iron. The clinical management of patients with lean NAFLD shall be concerned with the prevention and treatment of type 2 diabetes and liver cirrhosis.
Impact and implications
Lean NAFLD has a distinct natural history from obese NAFLD. This study underscored liver iron content and the genetic variant of the iron homoeostasis gene HFE as major risks of lean NAFLD, in addition to the unique metabolic profile. The development of type 2 diabetes or liver cirrhosis shall be closely monitored and prevented in patients with lean NAFLD.
Keywords: Magnetic resonance imaging, Non-alcoholic fatty liver disease, BMI, Genome-wide association study, Hepatic iron
Graphical abstract
Highlights
-
•
Lean NAFLD is genetically and metabolically different from obese NAFLD.
-
•
Variants of the iron homoeostasis gene HFE are specifically associated with lean NAFLD.
-
•
Patients with lean NAFLD are susceptible to type 2 diabetes and liver fibrosis.
Introduction
Non-alcoholic fatty liver disease (NAFLD), characterised by excessive liver fat deposition, was estimated to affect 24% of the world population.1 NAFLD is highly correlated with obesity, which is a criterion of clinically suspected fatty liver. However, nearly 40% of patients with NAFLD are not obese (BMI ≤30 kg/m2), and ∼20% are lean (BMI ≤25 kg/m2).2 Therefore, lean NAFLD may be underdiagnosed in regular clinical screening. Lean NAFLD was found to be associated with milder metabolic symptoms. However, the histological severity of lean NAFLD can be comparable with that of obese individuals.3 Compared with obese NAFLD, lean NAFLD is likely a unique subtype of metabolic (dysfunction)-associated fatty liver disease.4,5 Previous studies have found that the genetic variant TM6SF2 rs58542926 was more frequent in patient with lean NAFLD than in patients with obese NAFLD,6 whereas PNPLA3 rs738409 has a similar trend among Asians.7 Unfortunately, the current understanding of the genetic and pathobiological basis underlying lean NAFLD remains largely unexplored. A comprehensive genome-wide assessment for the genetic basis of lean NAFLD has not been reported. Meanwhile, it remains largely unclear what risk factors and long-term clinical sequelae for lean NAFLD are. Addressing these questions will not only significantly further our understanding of the aetiology of lean NAFLD but will also help develop effective and evidence-based strategies for both prevention and long-term management of this disease subtype.
Liver iron could exert a synergic effect together with steatosis in promoting hepatic lipid deposition.8 However, mounting evidence also implied that altered hepatic iron level is associated with insulin resistance and inflammation in patients NAFLD,9 whereas animal and cell model studies also revealed the pathogenic role of increased adipose tissue iron in the occurrence and development of NAFLD.10 The human study linking hepatic iron to NAFLD is less conclusive.
In this study, we performed a genome-wide association study (GWAS) and multiple causal inferences in a large cohort, aiming to identify genetic polymorphisms, manifestations, and clinical sequelae of lean NAFLD. Our results implicated a causal relationship between the homoeostatic iron regulator gene (HFE), increased hepatic iron content, and the onset of lean NAFLD.
Materials and methods
Details about materials and methods were included in the Supplementary methods.
UK Biobank participants and sample exclusion
Genotyping, data collection, and quality control were conducted by the UK Biobank team according to the protocols published previously.11 The details of genetic data quality control are provided in the Supplementary methods. Participants with alcoholic disease, alcohol harmful use, diagnosed haemochromatosis, viral hepatitis, Wilson disease, and liver damage drug use were excluded from the analyses (Table S1).
Image processing and WL-PDFF and liver iron calculation
The whole-liver proton density fat fraction (WL-PDFF) was calculated based on the magnetic resonance images of ∼45,000 individuals of the UK Biobank. Liver iron images of ∼37,000 individuals were generated using the pipeline published elsewhere (Fig. S1).12 Regions of interest marked by a team of experienced radiologists were used to train the model. The deep learning model nnUnet13 was constructed with a two-dimensional model for liver iron and a three-dimensional model for WL-PDFF to segment the liver and further calculated WL-PDFF/liver iron content for each individual.
NAFLD cases and controls in different obesity strata
NAFLD was diagnosed by WL-PDFF ≥5%, whereas the healthy controls were defined as WL-PDFF <5%. The cohort was stratified according to the World Health Organization criteria of obesity, into lean (BMI <25 kg/m2, n = 13,614), overweight (25 kg/m2 ≤BMI <30 kg/m2, n = 13,710), and obese (BMI ≥30 kg/m2, n = 5,617).
GWAS and heritability calculation
We undertook genome-wide association analyses in three BMI strata, as well as a combined GWAS that included all participants in the cohort. The cohort consisted of participants with European ancestry. GWAS, conditional analysis, and sensitivity analysis were conducted using BOLT-LMM.14 Diplotype assignments were conducted using PLINK v1.07.15 LDSC16 was used to estimate single-nucleotide polymorphism (SNP) heritability.
Mendelian randomisation
Two-stage least-squares linear regression was used to perform one-sample Mendelian randomisation (MR).17 Polygenic risk score (PRS) of lean NAFLD and liver iron in different BMI strata was calculated using genetic variables obtained from GWAS results using PRSice2.18 MR p value <0.05 was used to indicate significant causal associations between lean NAFLD and clinical outcomes.
Mediation model
The mediation model was used to identify the potential mechanism in which the genetic variant played a role in lean NAFLD. Our mediation analysis comprises three schemes, that is, X→Y, X→M, and X + M→Y, where M is a mediator that explains the underlying mechanism of the relationship between X and Y. We estimated CIs and tested whether the mediation effect was statistically significant via bootstrap with 1,000 resampling using the R package ‘mediation’ (R Foundation for Statistical Computing, Vienna, Austria; https://www.r-project.org/). Covariates used in mediation analysis included sex and age.
Bayesian networks
We used Bayesian networks (BNs) to validate the clinical outcomes of lean NAFLD inferred by MR. A BN uses a graphical model, directed acyclic graph, to illustrate the conditional dependencies of variables.19 We selected the R package ‘bnlearn’ as our analysis tool. We calculated average networks by bootstrapping the data with the replacement for 1,000 times in which the best network is recorded in each iteration. Using this approach, we computed the joint strength of bidirectional causal relationships among related factors, that is, genetic variants, lean NAFLD, and potential clinical outcomes. We included only the significant lean NAFLD variants in this analysis so that BN results could be comparable with MR results.20 Only the causal relationships with a possibility >50% were considered significant and showed in the network plot.
Statistical analysis
Two-tailed Student’s t test and ANOVA were applied to test the differences between lean healthy, lean NAFLD, obese healthy, and obese NAFLD in metabolic profiling. The t test, ANOVA, linear/logistic regression, and the estimation of the Pearson correlation test were all performed using R (R Foundation for Statistical Computing).
Results
Estimating liver fat content and liver iron content from magnetic resonance imaging data
The UK Biobank imaging cohort involved participants with abdominal magnetic resonance images of the Dixon technique and IDEAL protocol. WL-PDFF and liver iron were quantified using a trained deep learning pipeline. Using this pipeline, WL-PDFFs of 42,005 participants were successfully estimated and liver iron contents were calculated in 34,625 participants (Fig. 1, Phase 1).
Fig. 1.
Study flowchart.
GWAS, genome-wide association study; HFE, homoeostatic iron regulator; MR, magnetic resonance; NAFLD, non-alcoholic fatty liver disease; WL-PDFF, whole-liver proton density fat fraction.
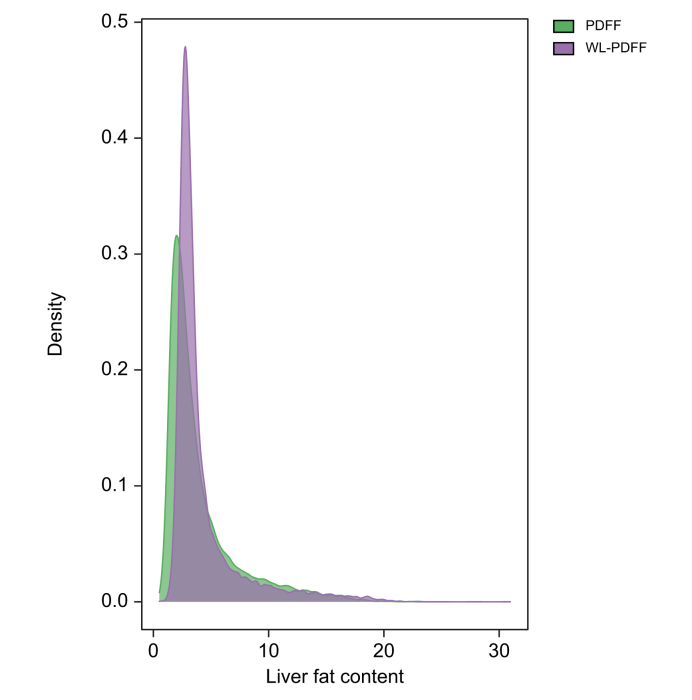
The mean of WL-PDFF was 4.27 (SD 3.06). We further correlated WL-PDFF with proton density fat fraction (PDFF) that were previously calculated based on 3–9 points in a single slice of the images and were now available for a subset of the imaging cohort (n = 15,750, UK Biobank field 24352) to confirm the accuracy of WL-PDFF. WL-PDFF was highly correlated with PDFF, with a correlation coefficient r = 0.893 (95% CI 0.889–0.896, p <2.2 × 10-16). On average, the liver fat content estimated with WL-PDFF was slightly higher than that quantified with PDFF, especially in those individuals with lower liver fat content (Fig. 2). The mean of liver iron was 1.22 (SD 0.21), which is consistent with a previous report.12
Fig. 2.
Distribution plot showing the agreement between WL-PDFF and PDFF.
PDFF, proton density fat fraction in the UK Biobank; WL-PDFF, whole-liver proton density fat fraction inferred by us.
Identification of patients with lean NAFLD and their metabolic profiling
Although liver fat and iron contents as well as BMI are all continuous variables, we primarily conducted our study based on a case–control design. This is because such a design allows us to evaluate lean NAFLD as a clinically relevant disease rather than a biological trait. As such, our findings associated with lean NAFLD would be more clinically interpretable. We identified 810 NAFLD cases and 12,804 healthy controls in lean participants, 3,069 NAFLD cases and 10,641 healthy controls in overweight participants, as well as 2,744 NAFLD cases and 2,873 healthy controls in obese individuals.
To better characterise the clinical profile of patients with lean NAFLD, we compared the metabolic biomarkers and characteristics between lean healthy participants, participants with lean NAFLD, participants with obese non-NAFLD, and participants with obese NAFLD (Fig. 1, Phase 2). Considering that the age and sex distribution were different in these four groups, we also conducted metabolic profiling adjusted for age and sex (Fig. 3).
Fig. 3.
Dot plots showing the metabolic profiling of participants in four groups.
Levels of biomarkers were normalised by adjusting for age and sex in linear regression models. Two-tailed Student’s t tests were applied to test the differences between lean healthy and lean NAFLD, obese healthy and obese NAFLD, and lean NAFLD and obese NAFLD. Values of p were shown between different groups. ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CRP, C-reactive protein; diastolicBP, diastolic blood pressure; FastingGlucose, glucose levels under fasting time >8 h; GGT, gamma glutamyltransferase; HDL, HDL cholesterol; iron, liver iron; LDL, LDL (directly measured); systolicBP, systolic blood pressure; TG, triglycerides; VAT, visceral adipose tissue; WHR, waist-to-hip ratio.
Most of these clinical markers showed a similar pattern (detrimental level: obese NAFLD > obese ≈ lean NAFLD > lean), including serum levels of liver enzymes, C-reactive protein, urate, HDL cholesterol, LDL cholesterol, triglycerides, abdominal obesity (as indicated by waist-to-hip ratio and visceral adipose tissue), and diastolic/systolic blood pressure. However, total cholesterol did not differ among the groups. It is also noteworthy that patients with lean NAFLD manifested the highest fasting glucose level among all four groups (ANOVA, p = 1.9 × 10-5), although the difference between lean NAFLD and obese NAFLD was not significant. This also implied that both individuals with lean NAFLD and those with obese NAFLD may experience a similar abnormal glucose metabolism. Interestingly, liver iron content was also the highest in patients with lean NAFLD (ANOVA, p = 5.5 × 10-21). Moreover, both obese NAFLD and lean NAFLD affected more males than females as compared with controls with obese and lean non-NAFLD, respectively (obese: 57 vs. 46%; lean: 58 vs. 39%). Furthermore, the individuals with lean NAFLD were 2.7 years older (p = 4.8 × 10-18) than the lean healthy people and 2.2 years older (p = 4.5 × 10-21) than the individuals with obese NAFLD. There was no age difference between the obese NAFLD and obese groups (p = 0.2). Other clinical characteristics are shown in Table S2.
Genetic susceptibility of NAFLD stratified by BMI
We next sought to identify genetic variants conferring risks for lean NAFLD, with also an aim to compare them with those for overweight and obese NAFLD (Fig. 1, Phase 3). We also conducted an overall GWAS on general NAFLD without BMI stratification. GWAS analyses were performed on a total of 32,941 participants with European ancestry.
Our analyses demonstrated that the well-established SNPs PNPLA3 rs738409 and TM6SF2 rs58542926 were significantly associated with NAFLD in all strata (Table 1 and Fig. 4), although stratum-specific associations were also observed.
Table 1.
NAFLD genetic association in multiple BMI strata.
| SNP | CHR:BP | Gene | A0 | A1 | OR | SE | p value | MAF |
|---|---|---|---|---|---|---|---|---|
| Lean (BMI <25 kg/m2) | ||||||||
| rs9348697 | 6:25890834 | SLC17A3-SLC17A2-TRIM38 | C | T | 1.34 | 0.05 | 1.40 × 10-8 | 0.37 |
| rs1800562 | 6:26093141 | HFE | G | A | 2.19 | 0.10 | 2.70 × 10-16 | 0.08 |
| rs58542926 | 19:19379549 | TM6SF2 | C | T | 2.06 | 0.10 | 4·70 × 10-14 | 0.08 |
| rs738409 | 22:44324727 | PNPLA3 | C | G | 1.54 | 0.06 | 8.60 × 10-13 | 0.22 |
| Overweight (25 kg/m2 ≤ BMI <30 kg/m2) | ||||||||
| rs882551 | 5:155015814 | SGCD | A | G | 1.21 | 0.03 | 1.60 × 10-08 | 0.02 |
| rs58542926 | 19:19379549 | TM6SF2 | C | T | 1.84 | 0.06 | 2.10 × 10-28 | 0.07 |
| rs429358 | 19:45411941 | APOE | T | C | 0.76 | 0.04 | 1.10 × 10-11 | 0.15 |
| rs738409 | 22:44324727 | PNPLA3 | C | G | 1.60 | 0.04 | 2.70 × 10-41 | 0.21 |
| Obese (BMI ≥30 kg/m2) | ||||||||
| rs58542926 | 19:19379549 | TM6SF2 | C | T | 1.76 | 0.07 | 4.90 × 10-15 | 0.07 |
| rs738409 | 22:44324727 | PNPLA3 | C | G | 1.48 | 0.05 | 1.00 × 10-17 | 0.2 |
| Combined (lean + overweight + obese) | ||||||||
| rs1800562 | 6:26093141 | HFE | G | A | 1.25 | 0.03 | 3.10 × 10-11 | 0.08 |
| rs429358 | 19:45411941 | APOE | T | C | 0.88 | 0.02 | 6.30 × 10-16 | 0.15 |
| rs58542926 | 19:19379549 | TM6SF2 | C | T | 1.4 | 0.02 | 6.70 × 10-53 | 0.07 |
| rs738409 | 22:44324727 | PNPLA3 | C | G | 1.27 | 0.01 | 1.20 × 10-66 | 0.21 |
The association tests were conducted in lean, overweight, or obese people using linear mixed models. An additional association test was also conducted in a combined cohort without BMI stratification (combined). OR, SE, and p were calculated between phenotype and genotype using linear mixed models.
CHR, chromosome; BP, base pair; A0, the other allele; A1, effect allele; MAF, minor allele frequency; NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; SE, standard error of OR; SNP, single-nucleotide polymorphism.
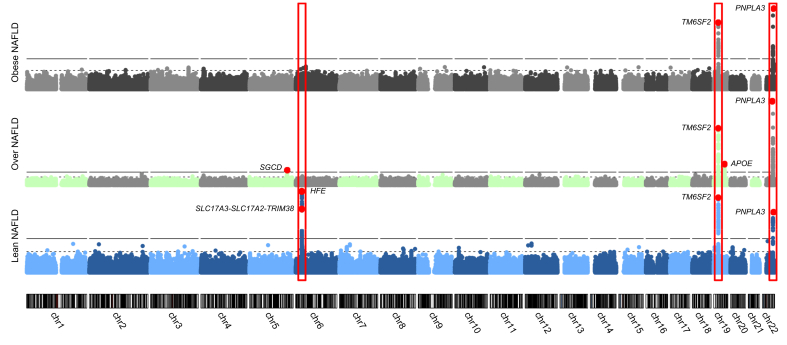
Fig. 4.
Manhattans plots showing the genetic associations with NAFLD in three BMI strata.
The x-axis is the genomic position. The y-axis is the significance of association in -log10(p values). Red points indicate the lead variants in each stratum. The significance of associations between genotype and lean/overweight/obese NAFLD were calculated using linear mixed models, adjusting for age, sex, and BMI. NAFLD, non-alcoholic fatty liver disease.
Among lean participants, in addition to the PNPLA3 and TM6SF2 loci, the 6p22.2 locus was uniquely shown to be associated with NAFLD. Two lead SNPs, rs1800562 in HFE (homoeostatic iron regulator; odds ratio [OR] 2.19, p = 2.7 × 10-16) and rs9348697 in the locus of SLC17A3-SLC17A2-TRIM38 (OR 1.34, p = 1.4 × 10-8), were significantly associated with NAFLD in this lean group. The HFE rs1800562 is also known as the non-synonymous variant C282Y, which is primarily identified in the European population with a minor allele frequency of 5.88% (<2% among other ethnic populations) and is known to be associated with the autosomal recessive hereditary haemochromatosis (HH), which features excessive iron absorption and iron overload.21,22 Another lead SNP, rs9348697, is located in an intergenic region between SLC17A3 (solute carrier family 17 member 3) and SLC17A2 (solute carrier family 17 member 2), and in the upstream of TRIM38 (tripartite motif containing 38) (Fig. S2). This SNP was demonstrated as an expression quantitative trait locus (eQTL) of TRIM38 (r = 0.195, p = 6.9 × 10-125) in the blood.23 According to the Genotype-Tissue Expression (https://gtexportal.org/) database,24 rs9348697 is a significant eQTL for SLC17A4 (p = 8.6 × 10-6) and a splicing QTL for SLC17A2 in liver (p = 8.8 × 10-13). A conditional analysis by adjusting for HFE rs1800562 demonstrated that SLC17A3-SLC17A2-TRIM38 locus SNP rs9348697 remained to be statistically significant (adjusted p = 2.6 × 10-3). No other independent association signals in this genomic locus were observed (Fig. S3).
The effect of HFE rs1800562 C282Y on lean NAFLD was likely imposed in a recessive model (OR 22.95, p = 7.9 × 10-33) (Table S3). Either the C282Y homozygous Y/Y genotype or a compounded heterozygous genotype of C282Y and H63D (rs1799945) was associated with HH.25 Moreover, the participants carrying the diplotype 282Y–63H/282C-63D were significantly susceptible to lean NAFLD (OR 2.39, p = 7.2 × 10-6) (Table S4). These lean 282Y–63H/282C-63D compound heterozygotes also had significantly higher WL-PDFF levels than C282Y heterozygotes, H63D heterozygotes, and H63D homozygotes (ANOVA, p = 1.4 × 10-83) (Fig. S4).
In the overweight participants, in addition to the PNPLA3 and TM6SF2 variants, we also identified rs882551, which is located in an intergenic region close to the SGCD (sarcoglycan delta) gene (Fig. S5) and rs429358 of the APOE (apolipoprotein E) gene, to be associated with NAFLD (p <5 × 10-8) (Table 1). The lead SNP rs882551 is an eQTL of FADXC2 (fatty acid hydroxylase domain containing 2), a positive regulator of protein phosphorylation that plays a role in oxidoreductase activity and iron ion binding.23 The lead SNP rs429358 (OR 0.76, p = 1.1 × 10-11) in APOE, the gene encoding apolipoprotein E, has been associated with fatty liver disease in a previous study.26
Among obese cases, the PNPLA3 and TM6SF2 variants were the only two reaching the genome-wide significance level (Table 1). In the GWAS (n = 32,941) combined all obesity categories, no additional association signals were observed (Table 1 and Fig. S6).
SNP-based heritability (h2SNP) of lean, overweight, and obese NAFLD was estimated to be 6.5, 14.1, and 10.0%, respectively.
We also checked the known variants that are associated with NAFLD and/or PDFF-quantified liver fat content.12,[27], [28], [29] We found that the majority of these variants were replicated (p <0.05), including those located near or in MTARC1, GCKR, MBOAT7, TRIB1, GPAM, C2orf16, ZNF512, SUGP1, and PBX4, in at least one of the three BMI strata cohorts as well as in the combined GWAS with the same direction and even similar ORs. Especially in the lean group, variants in TRIB1, APOE, GPAM, SUGP1, and PBX4 were replicated (p <0.05). However, the well-known variants in HSD17B13 that are associated with NAFLD were not replicated in either separate or the combined cohort (Table S5).
Sensitivity analysis for lean NAFLD GWAS
The HFE 282Y/Y homozygous is a cause of HH, an autosomal recessive genetic disorder that featured iron overload in the liver and blood.30 However, HH had a low penetrance possibly attributed to genetic variants in other genes that regulate iron metabolism31 or obesity.32 Thus, International Classification of Diseases 10th Revision (ICD-10) code for disorders of iron metabolism (E831) could not be used to thoroughly exclude patients with HH. We performed a sensitivity analysis that excluded patients with potential HH based on individual genotypes (homozygous pC282Y). We identified in total 2,889 HFE rs1800562 homozygous individuals in the UK Biobank, of which 61 lean individuals had WL-PDFF measurements. After excluding the potential HH, we confirmed that HFE rs1800562 was still associated with lean NAFLD with a reduced effect (OR 1.31, p = 9.0 × 10-3), whereas SLC17A3-SLC17A2-TRIM38 rs9348697 shared a similar pattern (OR 1.18, p = 1.7 × 10-3).
Alcohol intake is another relevant issue when determining genetic susceptibility of NAFLD, especially in the lean cohort. The ICD-10 records may not be able to exclude heavy drinkers. According to the AASLD guidelines, we identified 369 heavy drinkers in lean participants (Supplementary method). After further eliminating heavy drinkers in lean participants, along with excluding potential HH, HFE rs1800562 (OR 1.31, p = 6.8 × 10-3) and SLC17A3-SLC17A2-TRIM38 rs9348697 remained significant (OR 1.17, p = 0.03).
Other sensitivity analyses by controlling for additional covariates also did not change the significance of these two lead SNPs. Well-established SNPs PNPLA3 rs738409 and TM6SF2 rs58542926 were significant in all sensitivity analyses (p <5 × 10-8) (Table S6).
The causal role of liver iron in lean/obese NAFLD
We set out to explore whether there is a causal relationship between lean/obese NAFLD, HFE rs1800562, and liver iron content (Fig. 1, Phase 4). We first confirmed the association between lean NAFLD and liver iron content in 10,892 lean participants (β = 0.12, p = 8.1 × 10-29). As alcohol intake may increase iron absorption, we further examined this association in 1,887 lean teetotallers (Supplementary method) as a sensitivity analysis. After adjusting for sex and age, the association between lean NAFLD and liver iron in lean teetotallers persists (β = 0.12, p = 2.0 × 10-6). We also performed a sensitivity analysis by controlling for gamma glutamyltransferase (GGT), a marker of excessive alcohol intake, to further validate that the association between liver iron and lean NAFLD was not biased by alcohol intake. After controlling for the GGT level in 10,892 lean participants, the association between liver iron and lean NAFLD remains significant (β = 0.12, p = 8.2 × 10-27).
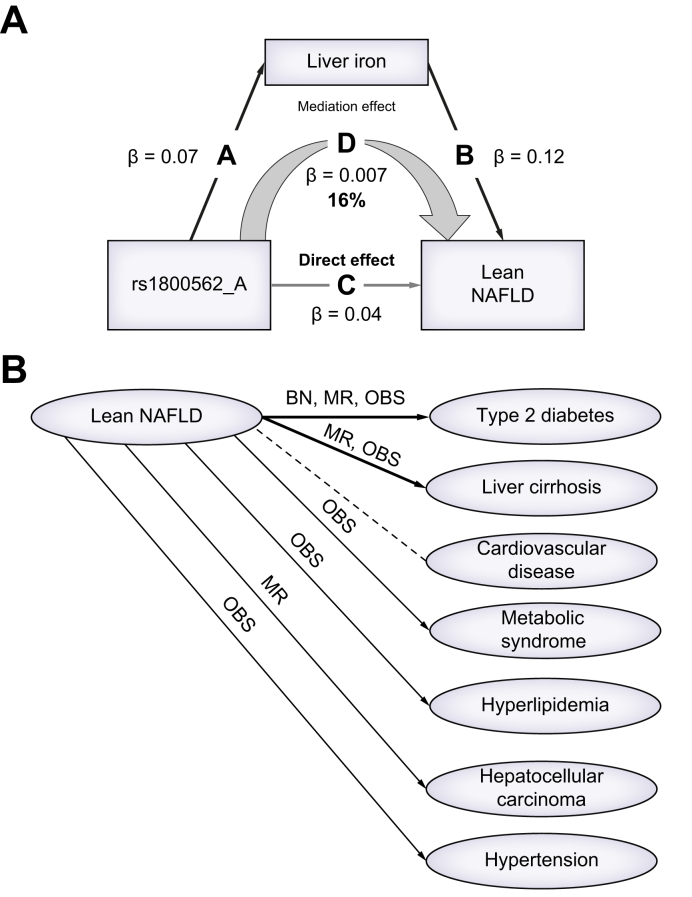
A mediation model was used to explore the liver iron impact on lean/obese NAFLD and determine the extent to which the effect of HFE rs1800562 on lean NAFLD is exerted through elevated liver iron content. A total of 10,892 lean participants and 3,765 obese participants were used in this analysis. Three regression models were constructed to estimate the mediation effect. The first regression revealed that HFE rs1800562 was associated with lean NAFLD, which was estimated in our GWAS. We further estimated the second regression showing that HFE rs1800562 was positively correlated with liver iron in lean NAFLD (β = 0.07, p = 1.1 × 10-35), and the last regression identified that liver iron (β = 0.11, p = 1.5 × 10-25) and HFE rs1800562 (β = 0.04, p = 1.2 × 10-9) would share the effect of increasing the risk of lean NAFLD. As a result, 17% effect of HFE rs1800562 on lean NAFLD was mediated by liver iron content (β = 0.007, p <2.2 × 10-16), whereas the direct effect remains significant (β = 0.038, p <2.2 × 10-16) (Fig. 5A). Similar mediation models were constructed for obese NAFLD, but no significant mediation effect was observed (p = 0.22).
Fig. 5.
Association models for lean NAFLD.
(A) Mediation model showing the causal relationship between HFE rs1800562, liver iron, and lean NAFLD. Every edge represents a significant association with different effect size calculated using linear regression models(p <0.05). In total, 16% effect of HFE rs1800562 on lean NAFLD was mediated by liver iron content. (B) Association between lean NAFLD and all potential clinical outcomes. An arrow indicates that the association between lean NAFLD and a clinical outcome was significant (p <0.05). The dotted arrow represents no association was observed. The abbreviations on the arrows represent the models used to identify the association. BN, Bayesian network; NAFLD, non-alcoholic fatty liver disease; MR, Mendelian randomisation; OBS, observation test.
MR was used to confirm this nonexisting causal relationship as obese NAFLD was not associated with HFE. We first constructed PRS for liver iron in obese individuals, using 86 linkage disequilibrium-clumped SNPs at the significance level of p <1 × 10-5 (R2 = 0.18, p = 1.1 × 10-188) to obtain instrumental variables. MR revealed that there was no causal relationship between liver iron and NAFLD in obese individuals (β = 0.12, p = 0.20).
We further explored the causal effect of lean/obese NAFLD on hepatic iron levels using MR. By using two well-established variants PNPLA3 rs738409 and TM6SF2 rs58542926 as instrumental variants for lean/obese NAFLD, we found that NAFLD in both lean and obese individuals do not causally result in increased hepatic iron content (causal effect β = 0.09, p = 0.82, and β = 0.09, p = 0.55, respectively). Details of the MR results are in Table S7.
Clinical sequelae of lean NAFLD
We aimed to further examine the potential clinical sequelae of lean NAFLD (Table S8) using observational analysis and MR analysis. Four SNPs associated with lean NAFLD, namely, SLC17A3-SLC17A2-TRIM38 rs9348697, HFE rs1800562, TM6SF2 rs58542926, and PNPLA3 rs738409, were used to construct lean NAFLD PRS. Using this lean NAFLD PRS as the instrumental variable, we inferred the causal effects of lean NAFLD on clinical outcomes with MR. Seven well-known NAFLD comorbidities were investigated, namely, type 2 diabetes, metabolic syndrome, cardiovascular disease, hyperlipidaemia, liver cirrhosis, hepatocellular carcinoma, and hypertension. Lean NAFLD causally increased the risk for type 2 diabetes (β = 0.10, p = 1.77 × 10-2) and non-alcoholic liver cirrhosis (β = 0.02, p = 7.19 × 10-3), which agreed with observational analysis results (Table S9). BN analysis also showed that lean NAFLD had a 62% probability of leading to type 2 diabetes, whereas liver fibrosis was not significant possibly because of the limited number of cases. Fig. 5B summarised all the associations between lean NAFLD and comorbidities.
As diabetes was a well-known complication of haemochromatosis, we also conducted sensitivity analysis by excluding potential HH. After the exclusion of homozygous individuals, the causal relationship between lean NAFLD and type 2 diabetes remains significant (β = 0.14, p = 0.01). This result was also confirmed by BN analysis (probability = 0.62).
Discussion
The aetiology and clinical profile of NAFLD in lean individuals remain under investigated. In this study, we generated the WL-PDFF and liver iron content of over 37,000 individuals from magnetic resonance data using a noninterventional deep learning approach. With these data, we for the first time explored the genetic basis underlying lean NAFLD in a European population at the genome-wide level. By leveraging the existing rich data accumulated in the UK Biobank, we further demonstrated the causal role of increased liver iron as well as the potentially causal clinical sequelae of lean NAFLD. Our study provides important data to accelerate the research and clinical management for lean NAFLD.
Patients with lean NAFLD manifest a distinctive metabolic profile as compared with lean healthy individuals, individuals with obese NAFLD, or individuals with obese non-NAFLD. We observed that the liver iron content of lean NAFLD cases is higher than that of obese NAFLD and lean healthy cases. Although individuals with lean NAFLD generally exert a milder metabolic profile than NAFLD cases in the obese group, they demonstrated significantly unhealthy levels of the majority of metabolic traits when compared with the lean healthy group, which is consistent with the previous observation.5 Although this may indicate a logical correlation between the severity of liver histology and overall metabolic health, it should be noted that these profile data in the UK Biobank cohort are 8–10 years before the NAFLD is defined, suggesting that (1) these comparisons may underestimate the health condition of lean NAFLD and (2) deviations of metabolic profiles from a healthy condition may occur earlier before the development of NAFLD in lean individuals.
It is also important to know what long-term clinical sequelae might be associated with lean NAFLD. Using MR analysis, we found that lean NAFLD causally increased risks for type 2 diabetes and liver cirrhosis, suggesting (1) a potential contribution of lean NAFLD to other metabolic diseases risks and (2) that excessive fat accumulation in the liver of lean individuals also increases risks for more severe liver damages. More large-scale studies are needed to further investigate the natural history of lean NAFLD.
Our study revealed the genetic bases underlying NAFLD among individuals with different BMI strata. Interestingly, the well-established29,33 PNPLA3 and TM6SF2 variants were found to be top risk factors for NAFLD among all BMI-stratified groups, highlighting the fundamental role of these two genes in the aetiology of NAFLD, although adiposity may further exacerbate the disease severity. In addition, we found that variants of two candidate genes SGCD and APOE were more specifically associated with NAFLD among overweight individuals, as opposed to the lean or obese groups. The reason underlying these specific associations in the overweight group remains unclear and thus should be further investigated.
More specifically, iron homoeostasis genes may be involved in the aetiology of lean NAFLD. Our study identified that the iron homoeostasis variant HFE C282Y is associated with lean NAFLD but not with NAFLD in obese individuals. Before our study, whether this HFE variant is associated with NAFLD in humans is inconclusive.34,35 A few studies have reported the association between this variant and NAFLD,36,37 whereas others did not confirm this association.35,38 Furthermore, in one Italian study, the patients with NAFLD carrying the HFE C282Y variant were reported to be leaner than the non-carriers.39 Our study for the first time demonstrated that this variant is associated with NAFLD only in lean individuals, which may explain the mixed results of the aforementioned association studies.
HFE is a membrane protein acting to regulate cellular iron intake by competing with transferrin for binding to the transferrin receptor, whereas transferrin is the transporter for ferric iron. The missense variant C282Y in HFE has been demonstrated to lead to hepcidin deficiency and subsequently increase iron absorption by enterocytes and accumulation in the liver.40 We observed in our mediation analysis that the impact of HFE C282Y mutation on the development of lean NAFLD is only moderately mediated via increased liver iron (about 17%), indicating that non-iron modulation function of HFE may also contribute to the development of lean NAFLD. Indeed, a recent study has identified HFE as a negative regulator of LDL receptor expression in hepatocytes,41 indicating a potential steatogenic role of HFE. Steatogenic mechanisms of HFE C282Y rs1800562-A on lean NAFLD may be attributed to oxidative stress,42 macrophage activation,43 stellate cell activation,44 endoplasmic reticulum stress,45 and increased cholesterol synthesis.46
Hepatic iron overload has been associated with NAFLD, which was deemed to be attributable to the increased oxidative stress that further induces dysfunction of mitochondria47 and autolysosomes,48 both resulting in insulin resistance49 in the liver, and thus leads to cellular damage.50 The production of excessive reactive oxygen species induced by elevated liver iron content could also attack phospholipids with polyunsaturated acyl tails to produce peroxided lipids and enhance ferroptosis, thus causing steatohepatitis.51 Other studies also found that perturbations of iron homoeostasis were frequently observed in patients with NAFLD.39,52 In our study, the causal association between liver iron content and lean NAFLD was inferred using an observational study and causal statistical models. We found that liver iron has a causal effect on lean NAFLD, although lean NAFLD does not causally lead to increased liver iron accumulation, suggesting that liver iron is a pathogenic factor (although it may not be necessarily a direct factor) for lean NAFLD.
The moderate effect of liver iron on lean NAFLD leads to a small difference in liver iron content between patients with lean NAFLD and lean healthy patients. Therefore, we compared the degree of steatosis between lean patients with haemochromatosis (patients with exact high liver iron content) and lean individuals without HH to further validate the causal association between steatosis and liver iron. In a smaller available cohort consisting of 24 lean patients with haemochromatosis and 10,897 lean individuals without HH, both the average hepatic fat fraction (5.01 vs. 3.11%, t test, p = 0.01) and NAFLD prevalence (33.33 vs. 5.79%, Fisher’s exact test, p = 4.18 × 10-5) were higher in lean patients with haemochromatosis than in lean individuals without HH. Taken together, our study suggests that increased hepatic iron level is a causal factor for NAFLD in lean individuals.
The mechanism by which liver iron specifically affects NAFLD in lean participants also remained to be investigated. However, current works of literature together with our results suggest that excessive adipose tissue accumulation may reduce iron absorption. Our BMI-stratified analysis indicated that the effect of the HFE C282Y variant on NAFLD risk was attenuated by increased BMI. Meanwhile, the liver iron content of obese participants was lower than that of lean participants and played no causal role in NAFLD. This implicates that liver iron load itself and effect on liver fat deposition are possibly reduced by adiposity. At the same time, it is known that the serum iron load in obese individuals was lower than that in lean healthy controls,53,54 which is likely the result of impaired absorption of supplemented iron.55 Increasing studies over the past decades suggested that accumulated adiposity reduces iron absorption from enterocytes of the duodenum,[56], [57], [58] which is at least in part attributed to increased inflammatory cytokine production from adipose tissue. These elevated inflammatory cytokines can induce the hepatic production of the iron-regulatory hormone hepcidin, which is also associated with reduced intestinal absorption of iron.59 However, weight loss in overweight and obese individuals decreased inflammatory and hepcidin levels60 and restored the serum iron level.61 Therefore, it is plausible that increased BMI associated with obesity may reduce intestinal iron absorption, thus attenuating the effect of increased iron uptake of hepatocytes caused by HFE deficiency. Collectively, adiposity and HFE deficiency may reciprocally interact with each other, which leads to the specific association between the HFE variant and NAFLD in lean individuals. Notably, unknown causality other than liver iron was also indicated in our mediation analysis, with the detailed underlying mechanism remaining to be further elucidated. Our study further underscores the notion that NAFLD in lean individuals may be a disease subtype with unique genetic and pathobiological basis.
There are limitations in our study. First, the definition of NAFLD in our study is just based on the fat content quantified by WL-PDFF, which may not coincide with the disease diagnosed based on liver histology. Future studies should validate our findings in clinically and histologically characterised individuals. Second, because the participants in this study are primarily from the UK Biobank, the causal relationship between hepatic iron and lean NAFLD needs to be further studied in non-European cohorts, especially in ancestry with a low frequency of HFE C282Y. Third, this study lacked independent cell/animal experiments to confirm this causality. Fourth, alcohol intake is known to be associated with increased hepatic iron load62 and steatosis. Unfortunately, the unavailability of more specific alcohol biomarkers, such as phosphatidyl ethanol or carbohydrate-deficient transferrin63 limited our chance to further validate the impact of alcohol intake on confounding the association between liver iron and lean NAFLD. We attempted to use self-report alcohol consumption combined with GGT measurements to define teetotallers, moderate drinkers, and heavy drinkers and discovered that the association between liver iron and lean NAFLD is independent of alcohol intake. However, self-report alcohol consumption used in this study is based on 1-day recall, which may give false-negative results as patients tend not to drink the day before the hospital visit.
Conclusion
Lean NAFLD is a subtype of fatty liver disease with a distinct metabolic profile and pathobiological process. The HFE C282Y variant and increased liver iron content are associated with NAFLD in lean people, rather than in obese people. With the high prevalence and potential underdiagnosis, it is necessary to screen people with normal body weight for lean NAFLD. Our findings generated novel scientific hypotheses warranting further investigations.
Financial support
This work was supported by the ‘Changbai Mountain Scholar’ Distinguished Professor Awarding Program of the Department of Education of Jilin Province, China.
Authors’ contributions
Designed the research: PC, WL.
Contributed to the funding acquisition and data acquisition: PC.
Drew the liver regions of interest and conducted phantom study: XP, JS, XT, DT.
Processed images, trained the nnU-Net model, and calculated WL-PDFF and liver iron content: XJ, SH, TW, ZS, YZ, PC.
Analysed the data: ZS, PC, AT, IS.
Interpreted the results: PC, WL.
Wrote the first draft of the manuscript: ZS, PC, WL, AT.
Revised the manuscript and approved the submission: all authors.
Managed the project: PC, YZ.
Data availability statement
The individual-level genetic data and phenotype data are available through application at the UK Biobank (https://www.ukbiobank.ac.uk/). Whole liver proton density fat fraction (WL-PDFF) and liver iron content data generated in this study will be available through application after publication of this study. GWAS summary statistics will be shared on GWAS catalogue (https://www.ebi.ac.uk/gwas/).
Conflicts of interest
The authors declare no competing interests.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
This study used the UK Biobank resources under application numbers 53562 and 71668.
Footnotes
Author names in bold designate shared co-first authorship.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100744.
Contributor Information
Yonggang Zhang, Email: zhangyg@jlu.edu.cn.
Wanqing Liu, Email: wliu@wayne.edu.
Peng Chen, Email: pchen@jlu.edu.cn.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Younossi Z., Anstee Q.M., Marietti M., Hardy T., Henry L., Eslam M., et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Ye Q., Zou B., Yeo Y.H., Li J., Huang D.Q., Wu Y., et al. Global prevalence, incidence, and outcomes of non-obese or lean non-alcoholic fatty liver disease: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:739–752. doi: 10.1016/S2468-1253(20)30077-7. [DOI] [PubMed] [Google Scholar]
- 3.Hagström H., Nasr P., Ekstedt M., Hammar U., Stål P., Hultcrantz R., et al. Risk for development of severe liver disease in lean patients with nonalcoholic fatty liver disease: a long-term follow-up study. Hepatol Commun. 2018;2:48–57. doi: 10.1002/hep4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eslam M., Sanyal A.J., George J., International Consensus Panel MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999–2014 e1991. doi: 10.1053/j.gastro.2019.11.312. [DOI] [PubMed] [Google Scholar]
- 5.Eslam M., El-Serag H.B., Francque S., Sarin S.K., Wei L., Bugianesi E., et al. Metabolic (dysfunction)-associated fatty liver disease in individuals of normal weight. Nat Rev Gastroenterol Hepatol. 2022;19:638–651. doi: 10.1038/s41575-022-00635-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen F., Esmaili S., Rogers G.B., Bugianesi E., Petta S., Marchesini G., et al. Lean NAFLD: a distinct entity shaped by differential metabolic adaptation. Hepatology. 2020;71:1213–1227. doi: 10.1002/hep.30908. [DOI] [PubMed] [Google Scholar]
- 7.Honda Y., Yoneda M., Kessoku T., Ogawa Y., Tomeno W., Imajo K., et al. Characteristics of non-obese non-alcoholic fatty liver disease: effect of genetic and environmental factors. Hepatol Res. 2016;46:1011–1018. doi: 10.1111/hepr.12648. [DOI] [PubMed] [Google Scholar]
- 8.Powell E.E., Ali A., Clouston A.D., Dixon J.L., Lincoln D.J., Purdie D.M., et al. Steatosis is a cofactor in liver injury in hemochromatosis. Gastroenterology. 2005;129:1937–1943. doi: 10.1053/j.gastro.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Dongiovanni P., Fracanzani A.L., Fargion S., Valenti L. Iron in fatty liver and in the metabolic syndrome: a promising therapeutic target. J Hepatol. 2011;55:920–932. doi: 10.1016/j.jhep.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Britton L.J., Subramaniam V.N., Crawford D.H. Iron and non-alcoholic fatty liver disease. World J Gastroenterol. 2016;22:8112–8122. doi: 10.3748/wjg.v22.i36.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Basty N., Whitcher B., Bell J.D., Sorokin E.P., van Bruggen N., et al. Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. Elife. 2021;10 doi: 10.7554/eLife.65554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Maier-Hein K.H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18:203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- 14.Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, et al. Efficient Bayesian mixed model analysis increases association power in large cohorts. Nature Genetics. 2015 doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J. Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess S., Small D.S., Thompson S.G. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333–2355. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi S.W., O'Reilly P.F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8:giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearl J. An introduction to causal inference. Int J Biostat. 2010;6:7. doi: 10.2202/1557-4679.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howey R., Shin S.Y., Relton C., Davey Smith G., Cordell H.J. Bayesian network analysis incorporating genetic anchors complements conventional Mendelian randomization approaches for exploratory analysis of causal relationships in complex data. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C.Y., Babitt J.L. Liver iron sensing and body iron homeostasis. Blood. 2019;133:18–29. doi: 10.1182/blood-2018-06-815894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corradini E., Buzzetti E., Pietrangelo A. Genetic iron overload disorders. Mol Aspects Med. 2020;75 doi: 10.1016/j.mam.2020.100896. [DOI] [PubMed] [Google Scholar]
- 23.Võsa U., Claringbould A., Westra H.J., Bonder M.J., Deelen P., Zeng B., et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53:1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurrin L.C., Bertalli N.A., Dalton G.W., Osborne N.J., Constantine C.C., McLaren C.E., et al. HFE C282Y/H63D compound heterozygotes are at low risk of hemochromatosis-related morbidity. Hepatology. 2009;50:94–101. doi: 10.1002/hep.22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamialahmadi O., Mancina R.M., Ciociola E., Tavaglione F., Luukkonen P.K., Baselli G., et al. Exome-wide association study on alanine aminotransferase identifies sequence variants in the GPAM and APOE associated with fatty liver disease. Gastroenterology. 2021;160:1634–1646 e1637. doi: 10.1053/j.gastro.2020.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Ghodsian N., Abner E., Emdin C.A., Gobeil E., Taba N., Haas M.E., et al. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teo K., Abeysekera K.W.M., Adams L., Aigner E., Anstee Q.M., Banales J.M., et al. rs641738C>T near MBOAT7 is associated with liver fat, ALT and fibrosis in NAFLD: a meta-analysis. J Hepatol. 2021;74:20–30. doi: 10.1016/j.jhep.2020.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anstee Q.M., Darlay R., Cockell S., Meroni M., Govaere O., Tiniakos D., et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol. 2020;73:505–515. doi: 10.1016/j.jhep.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Kowdley K.V., Brown K.E., Ahn J., Sundaram V. Correction: ACG clinical guideline: hereditary hemochromatosis. Am J Gastroenterol. 2019;114:1927. doi: 10.14309/ajg.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 31.Milet J., Dehais V., Bourgain C., Jouanolle A.M., Mosser A., Perrin M., et al. Common variants in the BMP2, BMP4, and HJV genes of the hepcidin regulation pathway modulate HFE hemochromatosis penetrance. Am J Hum Genet. 2007;81:799–807. doi: 10.1086/520001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbas M.A., Abraham D., Kushner J.P., McClain D.A. Anti-obesity and pro-diabetic effects of hemochromatosis. Obesity. 2014;22:2120–2122. doi: 10.1002/oby.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisinos C.A., Wilman H.R., Thomas E.L., Kelly M., Nicholls R.C., McGonigle J., et al. Genome-wide and Mendelian randomisation studies of liver MRI yield insights into the pathogenesis of steatohepatitis. J Hepatol. 2020;73:241–251. doi: 10.1016/j.jhep.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellervik C., Birgens H., Tybjaerg-Hansen A., Nordestgaard B.G. Hemochromatosis genotypes and risk of 31 disease endpoints: meta-analyses including 66,000 cases and 226,000 controls. Hepatology. 2007;46:1071–1080. doi: 10.1002/hep.21885. [DOI] [PubMed] [Google Scholar]
- 35.Hernaez R., Yeung E., Clark J.M., Kowdley K.V., Brancati F.L., Kao W.H. Hemochromatosis gene and nonalcoholic fatty liver disease: a systematic review and meta-analysis. J Hepatol. 2011;55:1079–1085. doi: 10.1016/j.jhep.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saremi L., Lotfipanah S., Mohammadi M., Hosseinzadeh H., Sayad A., Saltanatpour Z. Association of HFE gene mutations with nonalcoholic fatty liver disease in the Iranian population. Cell Mol Biol. 2016;62:123–128. doi: 10.14715/cmb/2016.62.12.21. [DOI] [PubMed] [Google Scholar]
- 37.Nelson J.E., Brunt E.M., Kowdley K.V., Nonalcoholic Steatohepatitis Clinical Research Network Lower serum hepcidin and greater parenchymal iron in nonalcoholic fatty liver disease patients with C282Y HFE mutations. Hepatology. 2012;56:1730–1740. doi: 10.1002/hep.25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raszeja-Wyszomirska J., Kurzawski G., Lawniczak M., Miezynska-Kurtycz J., Lubinski J. Nonalcoholic fatty liver disease and HFE gene mutations: a Polish study. World J Gastroenterol. 2010;16:2531–2536. doi: 10.3748/wjg.v16.i20.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valenti L., Dongiovanni P., Fracanzani A.L., Santorelli G., Fatta E., Bertelli C., et al. Increased susceptibility to nonalcoholic fatty liver disease in heterozygotes for the mutation responsible for hereditary hemochromatosis. Dig Liver Dis. 2003;35:172–178. doi: 10.1016/s1590-8658(03)00025-2. [DOI] [PubMed] [Google Scholar]
- 40.Barton J.C., Edwards C.Q., Acton R.T. HFE gene: structure, function, mutations, and associated iron abnormalities. Gene. 2015;574:179–192. doi: 10.1016/j.gene.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demetz E., Tymoszuk P., Hilbe R., Volani C., Haschka D., Heim C., et al. The haemochromatosis gene Hfe and Kupffer cells control LDL cholesterol homeostasis and impact on atherosclerosis development. Eur Heart J. 2020;41:3949–3959. doi: 10.1093/eurheartj/ehaa140. [DOI] [PubMed] [Google Scholar]
- 42.Maliken B.D., Nelson J.E., Klintworth H.M., Beauchamp M., Yeh M.M., Kowdley K.V. Hepatic reticuloendothelial system cell iron deposition is associated with increased apoptosis in nonalcoholic fatty liver disease. Hepatology. 2013;57:1806–1813. doi: 10.1002/hep.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handa P., Morgan-Stevenson V., Maliken B.D., Nelson J.E., Washington S., Westerman M., et al. Iron overload results in hepatic oxidative stress, immune cell activation, and hepatocellular ballooning injury, leading to nonalcoholic steatohepatitis in genetically obese mice. Am J Physiol Gastrointest Liver Physiol. 2016;310:G117–G127. doi: 10.1152/ajpgi.00246.2015. [DOI] [PubMed] [Google Scholar]
- 44.Gao H., Jin Z., Bandyopadhyay G., Wang G., Zhang D., Rocha K.C.E., et al. Aberrant iron distribution via hepatocyte-stellate cell axis drives liver lipogenesis and fibrosis. Cell Metab. 2022;34:1201–1213 e1205. doi: 10.1016/j.cmet.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan T.C., Crawford D.H., Jaskowski L.A., Subramaniam V.N., Clouston A.D., Crane D.I., et al. Excess iron modulates endoplasmic reticulum stress-associated pathways in a mouse model of alcohol and high-fat diet-induced liver injury. Lab Invest. 2013;93:1295–1312. doi: 10.1038/labinvest.2013.121. [DOI] [PubMed] [Google Scholar]
- 46.Graham R.M., Chua A.C., Carter K.W., Delima R.D., Johnstone D., Herbison C.E., et al. Hepatic iron loading in mice increases cholesterol biosynthesis. Hepatology. 2010;52:462–471. doi: 10.1002/hep.23712. [DOI] [PubMed] [Google Scholar]
- 47.Jadhav S., Protchenko O., Li F., Baratz E., Shakoury-Elizeh M., Maschek A., et al. Mitochondrial dysfunction in mouse livers depleted of iron chaperone PCBP1. Free Radic Biol Med. 2021;175:18–27. doi: 10.1016/j.freeradbiomed.2021.08.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jahng J.W.S., Alsaadi R.M., Palanivel R., Song E., Hipolito V.E.B., Sung H.K., et al. Iron overload inhibits late stage autophagic flux leading to insulin resistance. EMBO Rep. 2019;20 doi: 10.15252/embr.201947911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das M., Sauceda C., Webster N.J.G. Mitochondrial dysfunction in obesity and reproduction. Endocrinology. 2021;162:bqaa158. doi: 10.1210/endocr/bqaa158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muckenthaler M.U., Rivella S., Hentze M.W., Galy B. A red carpet for iron metabolism. Cell. 2017;168:344–361. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cel Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendler M.H., Turlin B., Moirand R., Jouanolle A.M., Sapey T., Guyader D., et al. Insulin resistance-associated hepatic iron overload. Gastroenterology. 1999;117:1155–1163. doi: 10.1016/s0016-5085(99)70401-4. [DOI] [PubMed] [Google Scholar]
- 53.Zhao L., Zhang X., Shen Y., Fang X., Wang Y., Wang F. Obesity and iron deficiency: a quantitative meta-analysis. Obes Rev. 2015;16:1081–1093. doi: 10.1111/obr.12323. [DOI] [PubMed] [Google Scholar]
- 54.Pinhas-Hamiel O., Newfield R.S., Koren I., Agmon A., Lilos P., Phillip M. Greater prevalence of iron deficiency in overweight and obese children and adolescents. Int J Obes Relat Metab Disord. 2003;27:416–418. doi: 10.1038/sj.ijo.0802224. [DOI] [PubMed] [Google Scholar]
- 55.Baumgartner J., Smuts C.M., Aeberli I., Malan L., Tjalsma H., Zimmermann M.B. Overweight impairs efficacy of iron supplementation in iron-deficient South African children: a randomized controlled intervention. Int J Obes. 2013;37:24–30. doi: 10.1038/ijo.2012.145. [DOI] [PubMed] [Google Scholar]
- 56.Cepeda-Lopez A.C., Melse-Boonstra A., Zimmermann M.B., Herter-Aeberli I. In overweight and obese women, dietary iron absorption is reduced and the enhancement of iron absorption by ascorbic acid is one-half that in normal-weight women. Am J Clin Nutr. 2015;102:1389–1397. doi: 10.3945/ajcn.114.099218. [DOI] [PubMed] [Google Scholar]
- 57.Aigner E., Feldman A., Datz C. Obesity as an emerging risk factor for iron deficiency. Nutrients. 2014;6:3587–3600. doi: 10.3390/nu6093587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnweber T., Ress C., Nairz M., Theurl I., Schroll A., Murphy A.T., et al. High-fat diet causes iron deficiency via hepcidin-independent reduction of duodenal iron absorption. J Nutr Biochem. 2012;23:1600–1608. doi: 10.1016/j.jnutbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Stoffel N.U., El-Mallah C., Herter-Aeberli I., Bissani N., Wehbe N., Obeid O., et al. The effect of central obesity on inflammation, hepcidin, and iron metabolism in young women. Int J Obes. 2020;44:1291–1300. doi: 10.1038/s41366-020-0522-x. [DOI] [PubMed] [Google Scholar]
- 60.Chang J.S., Li Y.L., Lu C.H., Owaga E., Chen W.Y., Chiou H.Y. Interleukin-10 as a potential regulator of hepcidin homeostasis in overweight and obese children: a cross-sectional study in Taiwan. Nutrition. 2014;30:1165–1170. doi: 10.1016/j.nut.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 61.Teng I.C., Tseng S.H., Aulia B., Shih C.K., Bai C.H., Chang J.S. Can diet-induced weight loss improve iron homoeostasis in patients with obesity: a systematic review and meta-analysis. Obes Rev. 2020;21 doi: 10.1111/obr.13080. [DOI] [PubMed] [Google Scholar]
- 62.Harrison-Findik D.D. Role of alcohol in the regulation of iron metabolism. World J Gastroenterol. 2007;13:4925–4930. doi: 10.3748/wjg.v13.i37.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cabezas J., Lucey M.R., Bataller R. Biomarkers for monitoring alcohol use. Clin Liver Dis. 2016;8:59–63. doi: 10.1002/cld.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual-level genetic data and phenotype data are available through application at the UK Biobank (https://www.ukbiobank.ac.uk/). Whole liver proton density fat fraction (WL-PDFF) and liver iron content data generated in this study will be available through application after publication of this study. GWAS summary statistics will be shared on GWAS catalogue (https://www.ebi.ac.uk/gwas/).