Abstract
Background
Cholinergic hypofunction and sleep disturbance are hallmarks of Alzheimer’s disease (AD), a progressive disorder leading to neuronal deterioration. Muscarinic acetylcholine receptors (M1-5 or mAChRs), expressed in hippocampus and cerebral cortex, play a pivotal role in the aberrant alterations of cognitive processing, memory, and learning, observed in AD. Recent evidence shows that two mAChRs, M1 and M3, encoded by CHRM1 and CHRM3 genes, respectively, are involved in sleep functions and, peculiarly, in rapid eye movement (REM) sleep.
Methods
We used twenty microarray datasets extrapolated from post-mortem brain tissue of non-demented healthy controls (NDHC) and AD patients to examine the expression profile of CHRM1 and CHRM3 genes. Samples were from eight brain regions and stratified according to age and sex.
Results
CHRM1 and CHRM3 expression levels were significantly reduced in AD compared with age- and sex-matched NDHC brains. A negative correlation with age emerged for both CHRM1 and CHRM3 in NDHC but not in AD brains. Notably, a marked positive correlation was also revealed between the neurogranin (NRGN) and both CHRM1 and CHRM3 genes. These associations were modulated by sex. Accordingly, in the temporal and occipital regions of NDHC subjects, males expressed higher levels of CHRM1 and CHRM3, respectively, than females. In AD patients, males expressed higher levels of CHRM1 and CHRM3 in the temporal and frontal regions, respectively, than females.
Conclusion
Thus, substantial differences, all strictly linked to the brain region analyzed, age, and sex, exist in CHRM1 and CHRM3 brain levels both in NDHC subjects and in AD patients.
Keywords: Alzheimer’s disease, REM-Sleep, sleep disturbance, bioinformatics, CHRM1, CHRM3
1. INTRODUCTION
Dementia is a syndrome characterized by a decline in cognitive skills, behavior, and in ability to perform simple daily activities. The drastic reduction of cognitive functions is accompanied and sometimes preceded, by altered emotional control, social behavior or motivation. To date, dementia is a major cause of disability and dependence among elderly people around the world [1]. Major causes of dementia include diseases and injuries that primarily or secondarily affect the brain, such as Alzheimer's disease (AD) or stroke [2].
AD is a progressive, neurodegenerative disorder, finally resulting in a continuous decline in thinking, behavior, and social interactions [3]. Typically, AD progresses from mild cognitive impairment to severe dementia over several years [4]. About fifteen years before symptoms of cognitive impairment appear, amyloid plaques begin to accumulate in the extracellular matrix of the adult brain. In cerebrospinal fluid (CSF), this latter scenario is coincidently reflected by reduced levels of the less soluble form of amyloid beta (Aβ), Aβ42, thus allowing a preclinical diagnosis [5-7]. As for the neurofibrillary tangles - aggregates of hyperphosphorylated tau protein- also used as a pathological biomarker of AD, develop in the second phase of the disease [8]. Currently, the diagnosis of AD depends on clinical assessments and post-mortem neuropathology, which are unbenefited for early diagnosis and progressive monitoring. Studies have reported that the levels of cerebrospinal fluid (CSF) and blood neurogranin (NRGN) are related to the occurrence and the subsequent progression of AD [9].
As AD disease progresses, amyloid plaques and neurofibrillary tangles tend to accumulate and compromise several areas of the brain, including regions critical for the sleep-wake cycle, such as the cerebral cortex, basal anterior brain, the locus coeruleus, and the hypothalamus [10]. Sleep-wake cycle alteration appears to precede the onset of cognitive symptoms in AD patients. Besides, previously published work showed a strong association between interrupted sleep and the onset of AD [11]. The use of techniques such as polysomnography has highlighted modifications in the sleep cycle of AD patients. In particular, AD patients have a decrease in slow wave sleep and rapid eye movement (REM), prolonged REM latency, an increase in stages N1 and N2 non-REM (NREM), and higher sleep fragmentation; finally leading to a general reduction in sleep duration [11, 12]. Accordingly, early sleep assessment with polysomnography (PSG) can provide more sensitive diagnostic tools for preclinical detection of AD rather than the more traditional cognitive tests [13]. In addition, to facing the devastating effects of the illness on daily life, AD patients suffer from circadian rhythm disturbances, epiphenomena linked to insomnia, sleep fragmentation and excessive daytime sleepiness [14-16]. Sleep and circadian disturbances have been proved to worsen along with the progression of the disease. [12, 17]. Recently, a correlation has been established between the sleep-wake cycle and AD [18], suggesting relevant implications for patient clinical management and potential treatment strategies.
Indeed, there is increasing evidence that sleep is critical for cognitive processing, despite a causal relationship between sleep and the development of dementia is difficult to establish since the two phenomena are bi-directionally linked. Nevertheless, sleep is now widely recognized as crucial for memory consolidation and removal of exceeding Aβ and hyper-phosphorylated tau protein that accumulate in AD patients’ brains [19]. Usually, it is possible to distinguish between declarative memories (i.e., memory for conscious events), enhanced by NREM or slow wave sleep, and non-declarative memories (i.e., procedural memories), consolidated during REM sleep [20, 21]. Moreover, sleep deprivation results in increased deficits in plasticity and synaptic memory processes [22-24].
The presynaptic cholinergic hypofunction is one of the major consequences of AD and the cholinergic replacement therapy could prove beneficial effects in alleviating cognitive dysfunction [25]. Pharmacological restoration of acetylcholine levels using acetylcholinesterase (AChE) inhibitors and N‐methyl‐d‐aspartate (NMDA) antagonists has been proven successful; acetylcholine receptor (AChR) agonists might be considered effective treatment of AD in the future [26].
AChRs are classified as muscarinic acetylcholine receptors (mAChRs) and nicotinic acetylcholine receptors (nAChRs) according to pharmacological affinities and sensitivities to endogenous ligands [26]. Muscarinic receptors are involved in memory, motor control, and learning [27, 28]. They are classified into M1 (CHRM1), M2 (CHRM2), M3 (CHRM3), M4 (CHRM4), and M5 (CHRM5) [29]. The M1, M3, and M5 mAChRs are coupled with Gq proteins [30]. M1 receptors are responsible for cholinergic functions and are involved in synaptic plasticity, learning and memory (cognition), neuronal differentiation during development and neuronal excitability [29]. M3 receptors are highly expressed in the hypothalamus, and to a less extent in the hippocampus and play major roles in food intake and body growth [31].
Alteration of mAChRs has been widely linked to AD [32-36]. Prior studies proved that both M1 and M4 subtype muscarinic AchRs were significantly reduced in the cerebral cortex and AD patients' hippocampus [37]. Due to the salient role played by these two areas in memory, cognitive processing and learning and considering the largely attenuated cholinergic signal, the most significant feature in the AD brain, it is clear the importance of restoring this altered scenario via a cholinergic activation. In the central nervous system, mAChRs can be divided into two groups, M1/M3 and M2/M4, based on their transduction pathways and activities. Intriguingly, γ‐secretase activity increases and thus less β‐amyloid peptide is formed upon stimulation of M1/M3 receptors [26]. It is reported that G‐protein coupled M1 muscarinic receptor is impaired in the neocortex region of the patients with AD and that severity of cognitive symptoms in the AD is considerably related to the degree of M1/G‐protein uncoupling [38]. Experiments conducted on M1 mAChR knockout mice displayed increased amyloid processing of APP, a critical step in the progression of the AD, thus suggesting a role of mAChRs in processing APP [39].
To date, no exhaustive explanation has been given on the mechanisms underlying poor sleep and circadian rhythm disturbances in AD. Recently, both M1 and M3 receptors have been shown to be essential for REM sleep [40]. Acetylcholine per se drives the REM sleep regulation [41]. A potential function of acetylcholine is likely the regulation of cellular properties involved in theta oscillation rather than the switching of neural circuits. Consistently with this, isolated hippocampal neurons exhibit oscillations in the theta frequency band under pharmacological stimulation with an acetylcholine receptor agonist [42-44]. Throughout mutagenization of acethylcholine receptors, Niwa and his associates revealed that DKO mice of muscarinic acetylcholine receptors coupled with Gq proteins (M1 and M3) abolished REM sleep and that this phenomenon was associated with an increase in theta waves during the sleep phase, leaving the oscillation of the theta largely unaffected during wakefulness [40]. To confirm these results, pharmacological studies have shown that the use of muscarinic receptor inhibitors, such as atropine or scopolamine, reduced the oscillation of theta electroencephalogram (EEG) in anesthetized animals [45, 46].
In the light of these data, we set out to verify the possible relationship between CHRM1 and CHRM3 gene expression levels and AD in human specimens. With this aim, we investigated the expression of these two genes in post-mortem brain tissue of not demented healthy control subjects and AD patients using a public online microarray dataset. The data were downloaded from GEO Database and statistically reprocessed.
2. MATERIALS AND METHODS
2.1. Data Selection
In order to test our hypotheses, we have collected several microarray datasets to obtain the largest possible number of brain samples of non-demented subjects who died from causes not attributable to neurodegenerative diseases and of deceased patients suffering from AD. As regards to the transcriptomes datasets, we downloaded them from NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) [47]. Mesh terms “Brain region”, “Human”, and “Alzheimer's disease”, were used to identify human potential datasets of interest. Twenty datasets were selected following the criteria exposed in the section “Clinical and neuropathological criteria”. The datasets selected are shown in Table 1 [48-67].
Table 1.
The datasets selected.
| - | Dataset | Organism | Individuals in the Dataset | Platform | Samples Origin |
NDHC
Samples |
AD
Samples |
References |
|---|---|---|---|---|---|---|---|---|
| 1 | GSE33000 | HS | 624 | GPL4372 | HBTRC | 156 | 310 | [48] |
| 2 | GSE28894 | HS | 114 | GPL6104 | NIA | 29 | 0 | [49] |
| 3 | GSE35978 | HS | 50 | GPL6244 | SMRI | 24 | 0 | [50] |
| 4 | GSE15745 | HS | 150 | GPL6104 | UMBB | 189 | 0 | [51] |
| 5 | GSE44772 | HS | 1098 | GPL4372 | HBTRC | 297 | 387 | [52] |
| 6 | GSE36192 | HS | 396 | GPL6947 | NIA | 481 | 0 | [53] |
| 7 | GSE60862 | HS | 134 | GPL5175 | UKBEC | 364 | 0 | [54] |
| 8 | GSE118553 | HS | 112 | GPL10558 | MRC-LBB | 88 | 167 | [55] |
| 9 | GSE25219 | HS | 57 | GPL5175 | BTBDDUM | 33 | 0 | [56] |
| 10 | GSE71620 | HS | 210 | GPL11532 | PBTDP | 292 | 0 | [57] |
| 11 | GSE36980 | HS | 88 | GPL6244 | KU | 47 | 32 | [58] |
| 12 | GSE26927 | HS | 113 | GPL6255 | BNEN | 6 | 11 | [59] |
| 13 | GSE84422 | HS | 125 | GPL570 | MSBB | 28 | 74 | [60] |
| 14 | GSE5281 | HS | 150 | GPL570 | ADRCs | 74 | 87 | [61] |
| 15 | GSE48350 | HS | 131 | GPL570 | ADRC | 105 | 80 | [62] |
| 16 | GSE11882 | HS | 63 | GPL570 | MSBB | 105 | 0 | [63] |
| 17 | GSE35864 | HS | 72 | GPL570 | NNATCB | 4 | 0 | [64] |
| 18 | GSE28146 | HS | 30 | GPL570 | BBADRCUK | 8 | 22 | [65] |
| 19 | GSE132903 | HS | 195 | GPL10558 | TGRI | 98 | 97 | [66] |
| 20 | GSE39420 | HS | 21 | GPL11532 | IDIBAPS | 7 | 14 | [67] |
Abbreviations: Harvard Brain tissue resource center (HBTRC); Brain Donor Program at the University of California (BDP); Mount Sinai Medical Center Brain Bank (MSBB); National Institute on Aging Alzheimer’s Disease Center (ADRC); National Institute on Aging (NIA); Stanley Medical Research Institute (SMRI); University of Maryland Brain Bank (UMBB); UK Brain Expression Consortium (UKBEC); Medical Research Council (MRC) London Neurodegenerative Diseases Brain Bank (from now on referred to as MRC-LBB); University of Pittsburgh’s Brain Tissue Donation Program (PBTDP); Pritzker Consortium (PC); Neuropathology Consortium of the Stanley brain collection (Stanley Medical Research Institute, US) (SMRIC); Translational Genomic Research Institute (TGRI); Kyushu University (KU); BrainNet Europe network (BNEN); Mount Sinai/JJ Peters VA Medical Center Brain Bank (MSBB); Alzheimer’s Disease Centers (ADCs); The National NeuroAIDS Tissue Consortium Brain (NNATCB); Brain Bank of the Alzheimer's Disease Research Center at the University of Kentucky (BBADRCUK); Brain and Tissue Bank for Developmental Disorders at the University of Maryland (BTBDDUM); Neurological Tissue Bank of the Biobank-Hospital Clínic-IDIBAPS and the Neuropathology Institute from the Hospital Universitari de Bellvitge; NDHC= non demented healthy controls subjects; AD= Alzheimer’s Disease patients; HS= homo sapiens.
2.2. Clinical and Neuropathological Criteria
The brain of each subject with Alzheimer’s disease was age-matched to a healthy brain. Furthermore, we stratified the samples according to sex and age, as shown in Table 2.
Table 2.
Sample stratification according to sex and age.
| S. No. | Age Stage | NDHC | AD |
|---|---|---|---|
| 1 | 50–65 middle-age | 1176 = 903 ♂+273 ♀ | 73 = 36 ♂+37 ♀ |
| 2 | 65–75 senior | 450 = 307 ♂+143 ♀ | 229 = 151 ♂+78 ♀ |
| 3 | 76-89 elderly | 578 = 374 ♂+204 ♀ | 699 = 310 ♂+389 ♀ |
| 4 | 90-99 nonagenarian | 206 = 71 ♂+135 ♀ | 262 = 58 ♂+204 ♀ |
| 5 | >100 centenarian | 25 = 5 ♂+20 ♀ | 18 = 5 ♂+13 ♀ |
| Total sample | 2435 (1660 ♂+775 ♀) | 1281 (560 ♂+721 ♀) |
Abbreviations: NDHC = non-demented healthy controls subjects; AD = Alzheimer’s disease patients.
Five groups were obtained: middle-age (50-65 years), senior (66-75 years), elderly (76-89 years), nonagenarian (90-99 years), and centenarian (>100 years) [68].
All brain samples analyzed were grouped into eight main brain regions (pre-frontal, frontal, occipital, cerebellum, temporal, cingulate, diencephalon, and limbic system). Complete details of examined brain regions are shown in Table 3.
Table 3.
Sample stratification according to brain regions.
| N° | Brain Regions | Abs | Brain Portions | n° of NDHC Samples |
n° of AD
Samples |
|
|---|---|---|---|---|---|---|
| 1 | Pre-frontal | PFC | Pre frontal cortex ; dorso-lateral pre frontal cortex; medial prefrontal cortex; ofc (orbitofrontal cortex); orbital prefrontal cortex; ventral forebrain; ventrolateral cortex; ventrolateral prefrontal cortex. | 560 (438♂+122♀) | 439 (197♂+242♀) | |
| 2 | Frontal | FC | Frontal cortex frontal pole (Brodmann area 9, 10); medial frontal cortex. | 455 (305♂+150♀) | 55 (21♂+34♀) |
|
| 3 | Occipital | OC | Occipital cortex; primary visual cortex; visual cortex. | 118 (92♂+26♀) |
148 (73♂+75♀) |
|
| 4 | Cerebellum | CB | Cerebellar cortex; cerebellum; upper (rostral) rhombic lip. | 460 (320♂+140♀) | 167 (80♂+87♀) |
|
| 5 | Temporal | TP | Inferior temporal cortex; primary auditory cortex; superior temporal cortex; superior temporal cortex (Brodman area 22); temporal cortex; ventral head of the caudate nucleus. | 199 (132♂+67♀) | 62 (28♂+34♀) |
|
| 6 | Cingulate | CYN | Anterior cingulate; caudal ganglionic eminence; lateral ganglionic eminence; medial ganglionic eminence; medial temporal gyrus; post central gyrus; posterior cingulate; posterior cingulate cortex; subpial grey matter lesions from the frontal gyri; superior frontal gyrus. | 246 (124♂+122♀) |
191 (95♂+96♀) |
|
| 7 | Diencephalon | DIE | Basal ganglia; dorsal thalamus; putamen; striatum; nucleus accumbens; substantia nigra; thalamus; mediodorsal nucleus of the thalamus. | 111 (77♂+34♀) |
52 (11♂+41♀) |
|
| 8 | Limbic System | LS | Amygdala; entorhinal cortex; hippocampus. | 286 (172♂+114♀) |
167 (55♂+112♀) |
|
Note: All brain samples analyzed were divided into 58 portions, subsequently grouped into the eight main brain regions (pre-frontal, frontal, occipital, cerebellum, temporal, cingulate, diencephalon, and limbic system).
Abbreviations: NDHC = non demented healthy controls subjects; AD = Alzheimer’s Disease patients.
Among data deriving from frozen tissue samples of subjects who did not die from causes related to neurological diseases, 2435 were selected and identified as non-demented healthy controls (NDHC) (68.24 ± 14.57 years), whereas 1281 samples were from AD patients (82.03 ± 9.29 years). In order to limit the distortion effect due to age differences, we compared the brain of subjects with AD to age-matched NDHC brains.
Most of the samples analyzed were obtained from public brain databases (Table 1). Postmortem interval (PMI), sample pH, and RNA integrity number (RIN) were elements of pre-selection by the authors of the reference microarray datasets and, subsequently object to our further exclusion analysis. For the diagnosis of AD, we took into consideration the investigations carried out by the authors of the individual datasets (Consortium to Establish a Registry for Alzheimer's Disease (CERAD) guidelines, progressive decline in memory, cognitive deficits in two or more areas, MMSE, CDR, Braak stage, general and regional atrophy, gray and white matter atrophy, ventricular enlargement, and cataloged neuropathological diagnosis by pathologists based on, e.g., Neurofibrillary Tangle (NFT) counts). With regard to the cognitive integrity of the healthy subjects included in our analysis, we considered the selection criteria and cognitive tests carried out by the authors of the microarray datasets listed in Table 1 (memory complaints, history of memory complaints, normal cognitive function documented by scoring age and education adjusted, Mini-mental status examination (MMSE), and global clinical dementia rating, CDR).
Unfortunately, the available datasets did not provide information regarding sleep disorders. The data used to classify patients were unavailable for individual patients inside the matrix datasets analyzed or in the relative publications associated with the dataset.
2.3. Data Processing and Experimental Design
In order to process and identify Significantly Different Expressed Genes (SDEG) in all selected datasets, we used the MultiExperiment Viewer (MeV) software (The Institute for Genomic Research (TIGR), J. Craig Venter Institute, USA). In cases where multiple gene probes insisted on the same GeneID, we used those with the highest variance. The significance threshold level for all data sets was p < 0.05. Statistically significant genes were selected for further analysis. For all datasets, we performed a Benjamini & Hochberg FDR (False discovery rate) to adjust P values for multiple comparisons [69-72].
2.4. Statistical Analysis
For statistical analysis, Prism 9 software (GraphPad Software, USA) was used. Based on the Shapiro-Wilk test, almost all data were normal, so parametric tests were used. Significant differences between groups were assessed using the Ordinary one-way ANOVA test, and Tukey’s multiple comparisons test was performed to compare data between all groups. Correlations were determined using Pearson correlation. All tests were two-sided, and significance was determined at P < 0.05. All MD selected were transformed for the analysis in the Z-score intensity signal. Z score is constructed by taking the ratio of weighted mean difference and combined standard deviation according to Box and Tiao (1992) [73]. The application of a classical method of data normalization, z-score transformation, provides a way of standardizing data across a wide range of experiments and allows the comparison of microarray data independent of the original hybridization intensities. The z-score is considered a reliable procedure for this type of analysis and can be considered a state-of-the-art method, as demonstrated by numerous bibliographies [74-78]. Z scores are calculated by subtracting the overall average gene intensity (within a single experiment) from the raw intensity data for each gene, and dividing that result by the SD of all the measured intensities, according to the formula:
Z score (intensity G - mean intensity G1. . .Gn)/SDG1. . .Gn
where G is any gene on the microarray and G1. . . Gn represents the aggregate measure of all of the genes.
Diagnostic accuracies were tested in logistic regression models separately for NDHC versus AD patients. All models were evaluated for the significance of the included biomarkers and overall diagnostic accuracy. The area under the ROC curve (AUC) and its 95% confidence interval (95% CI) indicates diagnostic efficiency. The accuracy of the test with the percent error is reported.
3. RESULTS
3.1. CHRM1 and CHRM3 Expression Levels are Downregulated in AD Brains
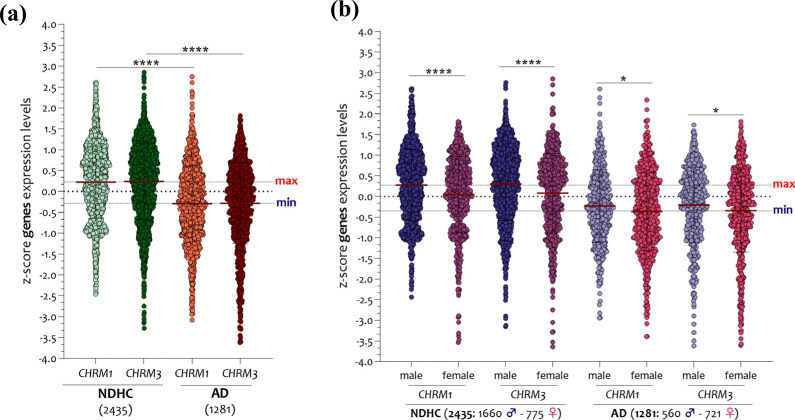
In a preliminary analysis, we first documented a downregulation of the brain expression levels of CHRM1 and CHRM3 in the whole brain of AD patients compared to NDHC subjects (Fig. 1a). Grouping the samples according to sex, we found that both in the brain of NDHC subjects and in those of AD patients, the expression levels of CHRM1 and CHRM3 were significantly higher in males than in females and that these differences were attenuated in AD (Fig. 1b).
Fig. (1).
Sex-dependent difference in CHRM1 and CHRM3 brain expression levels of NDHC subjects and AD patients. Analysis of CHRM1 and CHRM3 in brains of 2435 NDHC subjects and 1281 AD patients (a), sorted according to sex (b). The dashed lines indicate the maximum and minimum mean values. Data are expressed as z-score intensity expression levels (means and SD) and presented as dot plots. P values <0.05 were considered as statistically significant (*p < 0.01; ****p < 0.00001).
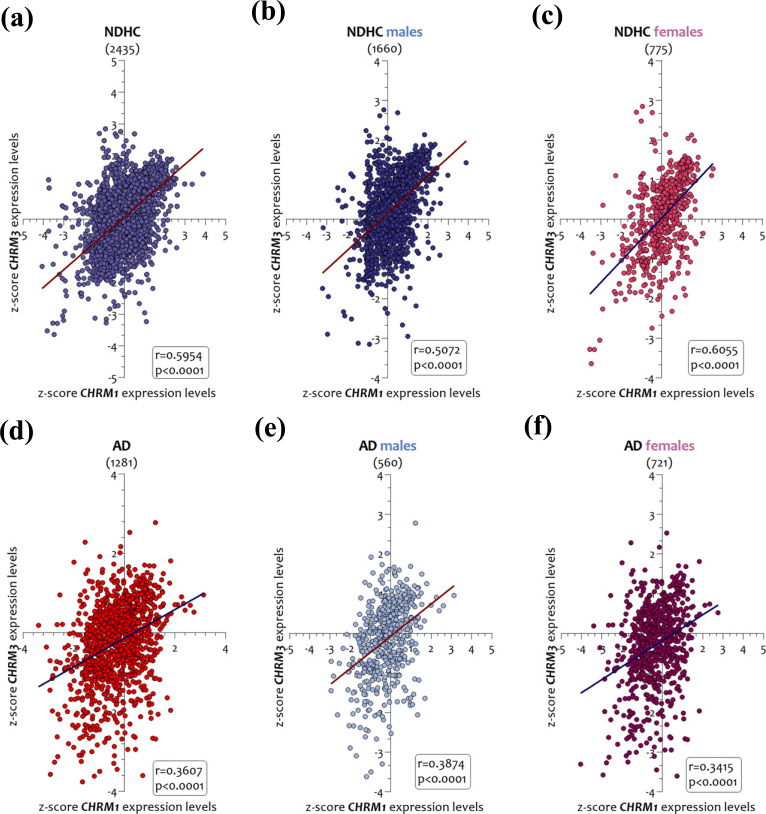
We next observed that the brain CHRM1 and CHRM3 expression levels were significantly correlated both in AD patients and in NDHC subjects (Fig. 2). Particularly, in the brain of NDHC subjects, the CHRM1 and CHRM3 expression levels were strongly correlated (r = 0.5954, p < 0.0001) (Fig. 2a). This correlation was maintained when we decided to separate the samples according to sex. Notably, males (r= 0.5072, p < 0.0001) (Fig. 2b) had a lower R-squared than females (r = 0.6055, p < 0.0001) (Fig. 2c). In the brains of AD patients, we observed that the CHRM1 and CHRM3 expression levels had a marked correlation with each other but showed a lower trend compared to NDHC (r = 0.3607, p < 0.0001) (Figs. 2d). This correlation also emerged when we grouped the samples according to sex. Indeed, both in brains of male AD patients (r = 0.3874, p < 0.0001) (Fig. 2e) and in those of females (r = 0.3415, p < 0.0001) (Fig. 2f), the expression levels of CHRM1 and CHRM3 were significantly correlated.
Fig. (2).
Correlation analysis between CHRM1 and CHRM3 brain expression levels in NDHC subjects and AD patients. Correlation analysis between brain CHRM1 and CHRM3 expression levels in NDHC subjects and AD patients sorted according to sex. CHRM1 and CHRM3 expression levels in 2435 NDHC whole brains (a) and stratified in 1660 males (b) and in 775 female brains (c). CHRM1 and CHRM3 expression levels in 1281 brains of AD patients (d) and stratified according to sex in 560 males (e) and 721 females (f). Data are expressed as z-score intensity expression levels. P values < 0.05 were considered statistically significant.
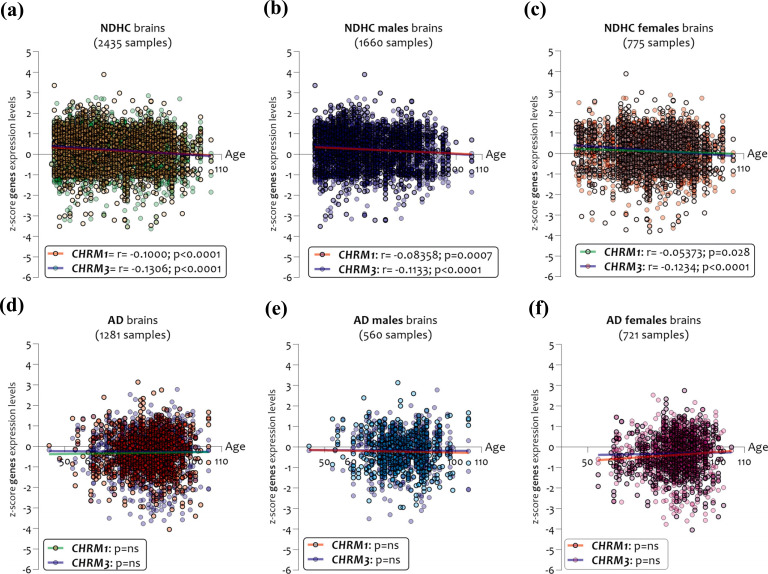
By stratifying the samples according to age, sex, and disease, we found significant differences between NDHC subjects and AD patients (Fig. 3). In brains of NDHCs subjects, we observed that the expression levels of CHRM1 and CHRM3 were significantly and inversely correlated with age, both in the whole sample (2435 samples) (CHRM1 r = -0.1000 and p < 0.0001, CHRM3 r = - 0.1306 and p < 0.0001) (Fig. 3a), and when stratifying the samples according to sex, in the 1660 males (CHRM1 r = -0.083 and p = 0.0007, CHRM3 r = -0.1133 and p < 0.0001) (Fig. 3b) and in 775 females (CHRM1 r = -0.053 and p = 0.028, CHRM3 r = -0.1234 and p < 0.0001) (Fig. 3c). No significant correlations between CHRMs expression and sex were observed in the AD samples analyzed (Figs. 3d, e, and f).
Fig. (3).
Age correlation analysis between CHRM1 and CHRM3 expression levels in NDHC subjects and AD patients according to sex. CHRM1 and CHRM3 expression levels in 2435 NDHC whole brains (a) and stratified in 1660 males (b) and in 775 female brains (c) sorted by age. CHRM1 and CHRM3 expression levels in 1281 brains of AD patients (d) and stratified according to sex in 560 males (e) and 721 females (f) sorted by age. Data are expressed as z-score intensity expression levels. P values <0.05 were considered statistically significant.
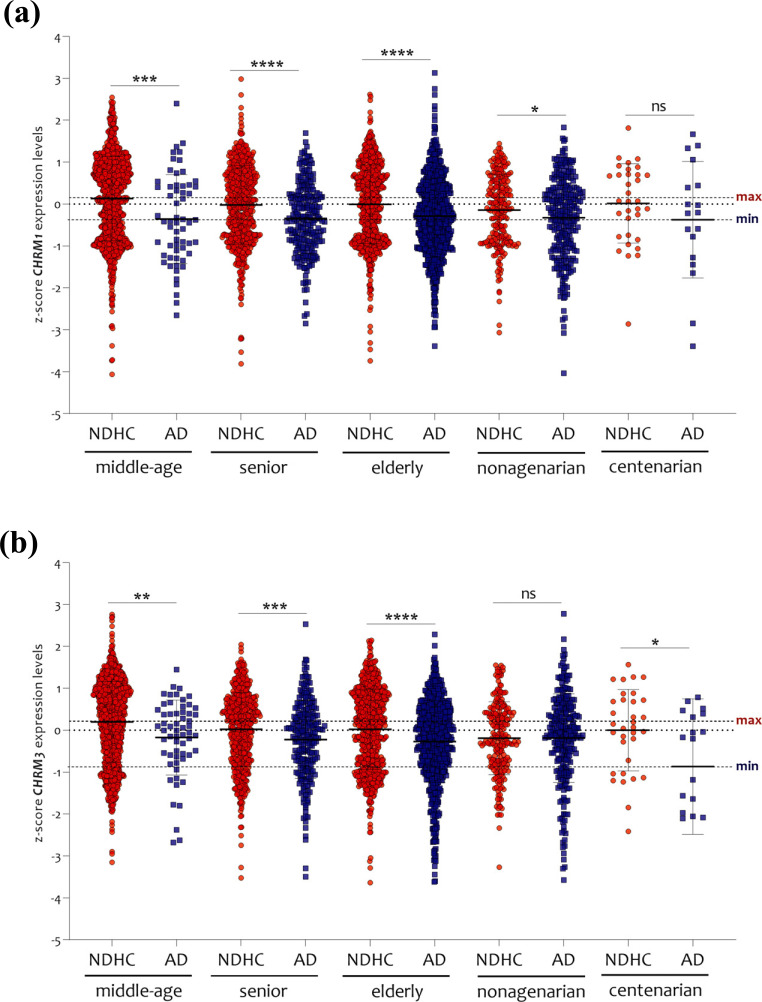
Expression analysis according to the age-matched showed a significant reduction in CHRM1 (Fig. 4a) and CHRM3 (Fig. 4b) in AD patients in comparison with NDHC subjects. No significant changes in CHRM1 expression levels emerged in AD centenarian patients (Fig. 4a) and in CHRM3 in nonagenarian patients compared to age-matched NDHC subjects (Fig. 4b). Among the age-matched groups, only the NDHC subjects presented different levels of expression of CHRM1 and CHRM3 (Supplementary Fig. 1 (1.3MB, pdf) ). Indeed, in the brain of NDHC subjects, CHRM1 expression levels were significantly reduced in nonagenarian subjects compared to middle-aged subjects (p < 0.001). Significant reductions of CHRM3 expression were present in senior (p < 0.01), elderly (p < 0.001), and nonagenarians (p < 0.00001), compared to middle-ages (Supplementary Fig. 1 (1.3MB, pdf) ) (Supplementary Table 1 (1.3MB, pdf) ). No significant changes emerged by comparing the different age ranges in AD patients (Supplementary Fig. 1 (1.3MB, pdf) ) (Supplementary Table 1 (1.3MB, pdf) ), thus indicating that age per se does not affect CHRM1 and CHRM3 expression levels in AD patients.
Fig. (4).
CHRM1 e CHRM3 expression analysis in middle-age, senior, elderly, nonagenarian and centenarian. The expression levels of CHRM1 (a) and CHRM3 (b) were significantly decreased in AD patients compared to NDHC in all age groups selected, except for CHRM1 in centenarians and CHRM3 in nonagenarians. The dashed lines indicate the maximum and minimum mean values. Data are expressed as z-score intensity expression levels (means and SD) and presented as dot plots. P values < 0.05 were considered as statistically significant (*p < 0.01; **p < 0.001, ***p < 0.0001 and ****p < 0.00001).
3.2. Brain Region-specific Changes in CHRM1 and CHRM3 Expression
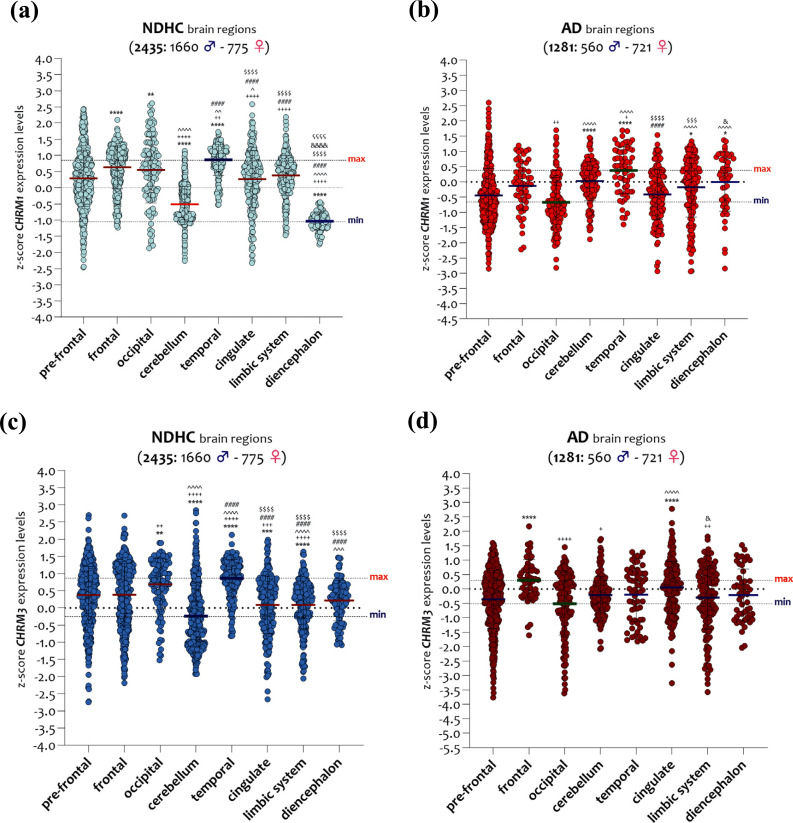
According to brain regions and sex, we analyzed the transcript expressions of CHRM1 and CHRM3 and found that in NDHC subjects, CHRM1 was highly expressed in the temporal region, while the lowest levels were present in the diencephalon region (Fig. 5a). Significant differences were observed between the temporal region and the cingulate (p < 0.0001), the limbic system (p < 0.0001), and the diencephalon (p < 0.0001), and between the diencephalon and all the other brain regions analyzed (p < 0.0001) (Fig. 5a).
Fig. (5).
CHRM1 and CHRM3 expression levels in brain regions of NDHC subjects and AD patients. CHRM1 expression levels in eight brain regions of 2435 NDHC subjects (a) and 1281 AD patients (b). CHRM3 expression levels in eight brain regions of 2435 NDHC subjects (c) and 1281 AD patients (d). Assignment of symbols of statistical significance: (*) pre-frontal vs. all, (+) frontal vs. all, (^) occipital vs. all, (#) cerebellum vs. all, ($) temporal vs. all, (&) cingulate vs. all, (ç) limbic system vs. all. The dashed lines indicate the maximum and minimum mean values. Data are expressed as z-score intensity expression levels (means and SD) and presented as violin dot plots. P values <0.05 were considered as statistically significant (*p < 0.01, **p < 0.001, ***p < 0.0001, and ****p < 0.00001).
Regarding the CHRM1 expression levels in the brain of AD patients, we observed a reduction in the differences between the various brain regions examined compared to NDHC subjects (Fig. 5a). The temporal region expressed the highest levels of CHRM1, in contrast to the lowest ones expressed in the occipital region. Notably, the occipital region showed significantly lower expression levels of CHRM1 than the cerebellum (p < 0.0001), the temporal region (p < 0.0001), the limbic system region (p < 0.0001), and the diencephalon (p < 0.0001) (Fig. 5b). The temporal region exhibited significantly higher levels of CHRM1 than the cingulate (p < 0.0001) and limbic system (p < 0.0001) (Fig. 5b).
A partially overlapping trend was observed when we analyzed the CHRM3 transcript in different brain regions of NDHC subjects and AD patients. We found that in brain regions of NDHC subjects, the CHRM3 transcript was highly expressed in the temporal region, while the lowest levels were expressed in the cerebellum (Fig. 5c). Relevant differences were observed comparing the temporal region to the cingulate (p < 0.0001), the limbic system (p < 0.0001), and the diencephalon (p < 0.0001) (Fig. 5c). Cerebellar expression levels of CHRM3 were significantly lower than the temporal region (p < 0.0001), cingulate, limbic system (p < 0.0001), and diencephalon (p < 0.0001) (Fig. 5c).
CHRM3 expression analysis in brain regions of AD patients unveiled smaller region-specific differences than observed among the controls. Indeed, we observed that the frontal cortex exhibited the highest levels of CHRM3 while the occipital cortex had the lowest (Fig. 5d). CHRM3 levels expressed in the occipital region were significantly lower than those detected in the cingulate (p < 0.001), while in the frontal cortex, we found significantly higher levels of expression than in the occipital region (p < 0.0001), in the cerebellum (p < 0.01), and in the limbic system (p < 0.001) (Fig. 5d).
3.3. Augmented Levels of CHRM1 in Cerebellum and Diencephalon of AD Patients
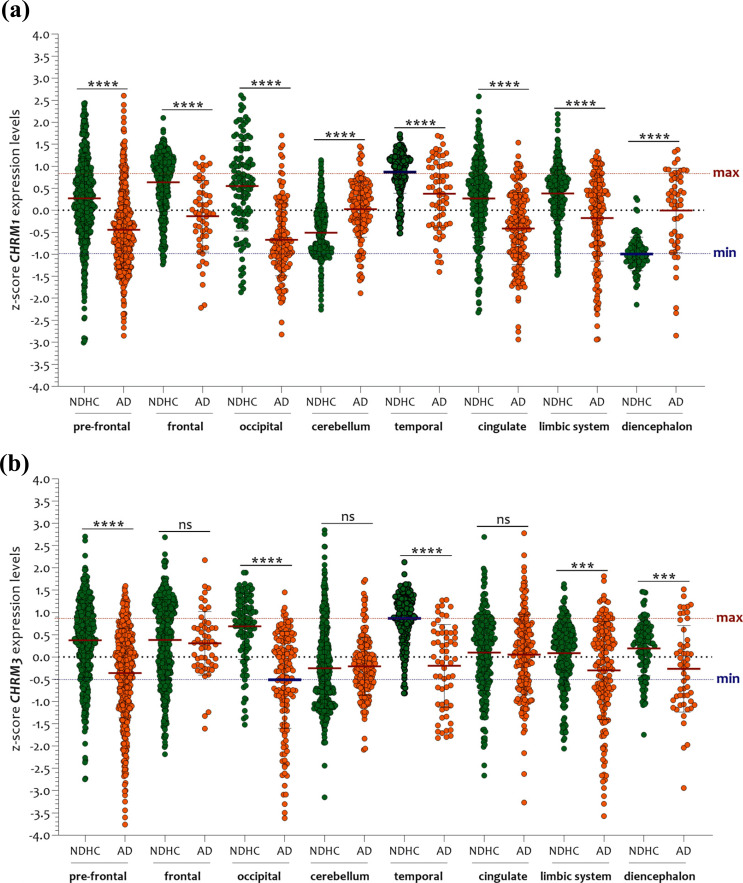
Compared with NDHC subjects, CHRM1 transcription levels of AD patients were markedly reduced in: pre-frontal (p < 0.0001), frontal (p < 0.0001), occipital (p < 0.0001), and temporal (p < 0.0001) cortices, as well as in the cingulate (p < 0.0001) and limbic system (p < 0.0001) (Fig. 6a). By contrast, in the NDHC group, the cerebellum (p < 0.0001) and diencephalon (p < 0.00001) presented lower expression levels of CHRM1 than those measured in the AD group (Fig. 6a).
Fig. (6).
Comparative analysis of CHRM1 and CHRM3 expression levels between NDHC subjects and AD patients. (a) Compared to NDHC, the AD group exhibits largely decreased levels of CHRM1 in: pre-frontal (p < 0.0001), frontal (p < 0.0001), occipital (p < 0.0001), and temporal cortices (p < 0.0001), in cingulate (p < 0.0001), and in limbic system (p < 0.0001). The sole exceptions are of cerebellum (p < 0.0001) and diencephalon (p < 0.00001). (b) In comparison to NDHC subjects, transcription levels of CHRM3 were significantly attenuated in pre-frontal (p < 0.0001), occipital (p < 0.0001), and temporal (p < 0.0001) cortices, in the limbic system (p < 0.0001), and in the diencephalon of AD patients. No significant variation was registered by comparing the expression levels of CHRM3 in the frontal cortex, cerebellum, and cingulate. The dashed lines indicate the maximum and minimum mean values. Data are expressed as z-score intensity expression levels (means and SD) and presented as dot plots. P values <0.05 were considered as statistically significant (***p < 0.0001, and ****p < 0.00001).
These results were also confirmed in males and females when we stratified the samples according to sex (Supplementary Figs. 2a (1.3MB, pdf) and 2b (1.3MB, pdf) ). Apropos CHRM3 expression levels, we showed that it was significantly reduced in the pre-frontal (p < 0.0001), occipital (p < 0.0001), and temporal (p < 0.0001) cortices, as well as in the limbic system (p < 0.0001) and diencephalon (p < 0.001) of AD patients compared with NDHC subjects (Fig. 6b). No significant variation was observed comparing the expression levels of CHRM3 in the frontal cortex, cerebellum, and cingulate (Fig. 6b). These results overlapped with those obtained by stratifying the samples according to sex (Supplementary Figs. 3a (1.3MB, pdf) and 3b (1.3MB, pdf) ).
3.4. Sex Influence in Brain CHRM1 and CHRM3 Expression
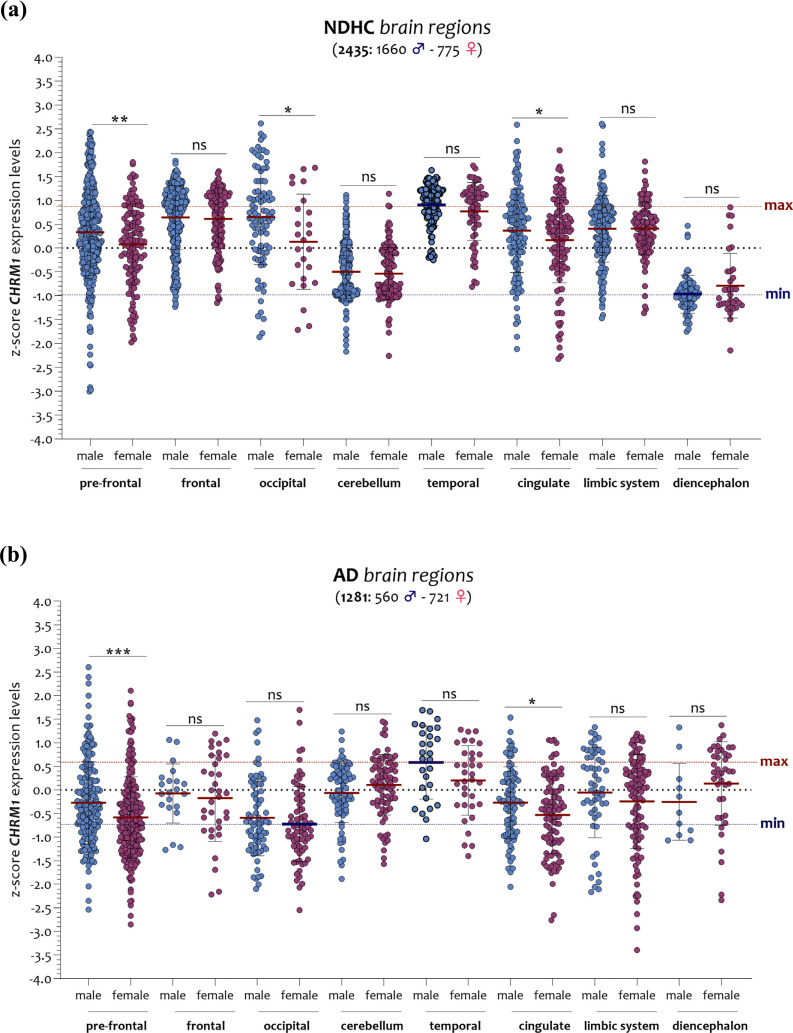
In NDHC subjects, the diencephalon of males presented the lowest levels of CHRM1, while in contrast, the temporal region showed the highest. As for the brain of AD patients, the occipital region of females was characterized by the lowest levels of CHRM1, while the highest levels were detected in the temporal region of males (Fig. 7a and b). Among NDHC subjects, we observed that females had significantly lower CHRM1 expression levels than males in the pre-frontal cortex (p < 0.001), in the occipital region (p < 0.01), and in the cingulate region (p < 0.01) (Fig. 7a). These differences were also present in the brain of AD patients (pre-frontal p < 0.0001 and cingulate p < 0.01), with the exception of the occipital region (p = ns) (Fig. 7b).
Fig. (7).
Sex-matched analysis of the CHRM1 expression levels in brain regions of NDHC subjects and AD patients. In NDHC males, diencephalon expressed the lowest levels of CHRM1, whereas the temporal region had the highest (a). As for the brain of AD patients, the occipital region of females expressed the lowest levels of CHRM1, in contrast with the temporal region of males showing the highest (b). The dashed lines indicate the maximum and minimum mean values. Data are expressed as z-score intensity expression levels (means and SD) and presented as dot plots. P values < 0.05 were considered as statistically significant (p = ns, (*p < 0.01, **p < 0.001, ***p < 0.0001).
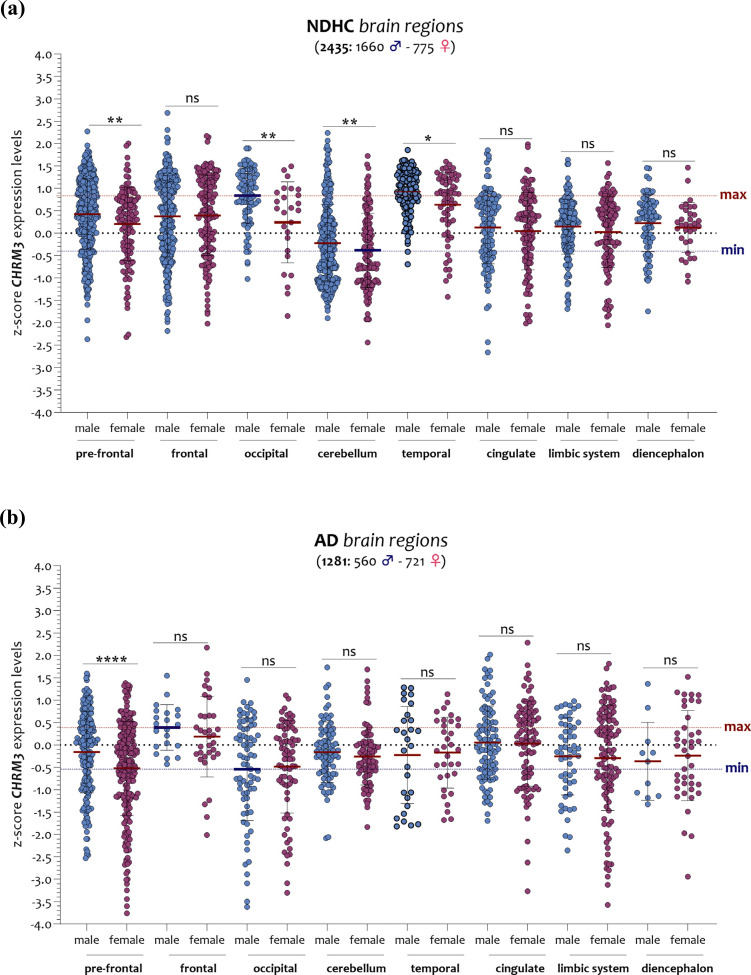
In NDHC female subjects, we observed the lowest levels of CHRM3 in the cerebellum, in contrast with the highest in the occipital region. The AD brain analysis showed that the lowest levels of CHRM3 were expressed by males in the occipital region and the highest by males in the frontal region (Fig. 8a and b). In particular, in the brain of NDHC subjects, we observed that females had significantly lower CHRM3 expression levels than males in the pre-frontal cortex (p < 0.001), in the occipital region (p < 0.001), in the cerebellum (p < 0.001), and in the temporal region (p < 0.01) (Fig. 8a). In AD, these differences were present only in the pre-frontal cortex (p < 0.0001) (Fig. 8b).
Fig. (8).
Analysis of the CHRM3 expression levels in the brain regions of NDHC subjects and AD patients according to sex. The brain of NDHC females’ subjects expresses the lowest levels of CHRM3 in the cerebellum region while highest in the males’ occipital region (a). As for the brain of AD patients, the lowest levels of CHRM3 were expressed by males in the occipital region and the highest by males in the frontal region (b). The dashed lines indicate the maximum and minimum mean values. Data are expressed as z-score intensity expression levels (means and SD) and presented as violin dot plots. P values <0.05 were considered as statistically significant (p=ns, (*p < 0.01, **p < 0.001, ****p < 0.00001).
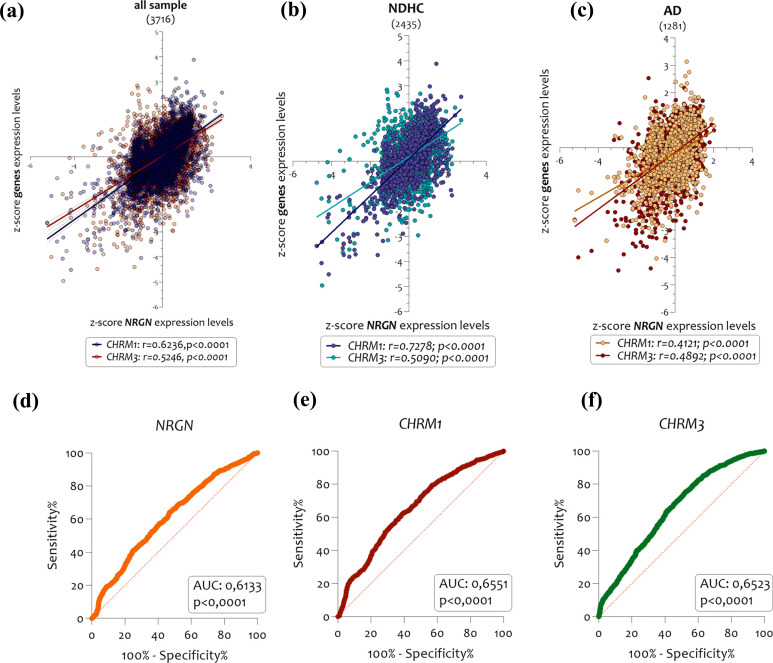
3.5. CHRM1 and CHRM3 Expression Levels are Correlated to NRGN
In order to investigate possible relationships between the CHRM1 and CHRM3 expression levels with the Alzheimer's disease progression, we compared their levels of expression with those of neurogranin (NRGN), a gene coding for a synaptic protein related to AD progression. The use of NRGN protein as a biomarker of Alzheimer's disease progression is well established [9, 79, 80]. Our analysis demonstrated the existence of a significant positive correlation between the expression of NRGN and both CHRM1 and CHRM3 (Fig. 9a) in the brain of NDHC subjects (Fig. 9b) and in those of AD patients as well (Fig. 9c). Particularly, we reported that the expression of CHRM1 and CHRM3 compared to NRGN, showed a positive correlation in all sample selected (CHRM1 r = 0.6236, p < 0.0001; CHRM3 r = 0.5246, p < 0.0001) (Fig. 9a), in NDHC subjects (CHRM1 r = 0.7278, p < 0.0001; CHRM3 r = 0.5090, p < 0.0001) (Fig. 9b), and in AD patients (CHRM1 r = 0.4121, p < 0.0001; CHRM3 r = 0.4892, p < 0.0001) (Fig. 9c). Overlapping results were obtained by stratifying samples according to sex (Supplementary Fig. 4 (1.3MB, pdf) ). We found a stronger linear relationship in females than males in the expression levels of CHRM1, CHRM3, and NRGN, both in the brain of NDHC subjects (Supplementary Fig. 4a (1.3MB, pdf) ) and in AD patients (Supplementary Fig. 4b (1.3MB, pdf) ).
Fig. (9).
Correlation analysis between CHRM1, CHRM3, and NRGN expression levels in NDHC subjects and AD patients’ brains. CHRM1 and CHRM3 expression levels positively correlated with NRGN in the overall sample (a), NDHC subjects (b) and AD patients (c). Moreover, NRGN (AUC = 0.6133, p < 0.0001) (d), CHRM1 (AUC=0.6551, p < 0.0001) (e), and CHRM3 (AUC = 0.6523, p < 0.0001) (f) were all significant predictors of AD. Data are expressed as z-score intensity expression levels and presented as dot plots. P values <0.05 were considered statistically significant.
Furthermore, we tested the diagnostic accuracy of NRGN, CHRM1, and CHRM3 mRNA for AD patients using logistic regression models. NRGN (AUC=0.6133, p < 0.0001), CHRM1 (AUC=0.6551, p < 0.0001), and CHRM3 (AUC = 0.6523, p < 0.0001) were all significant predictors of AD (Fig. 9d, e, f). Surprisingly, for single predictors, CHRM1 had the highest accuracy, followed by CHRM3 and NRGN.
4. DISCUSSION
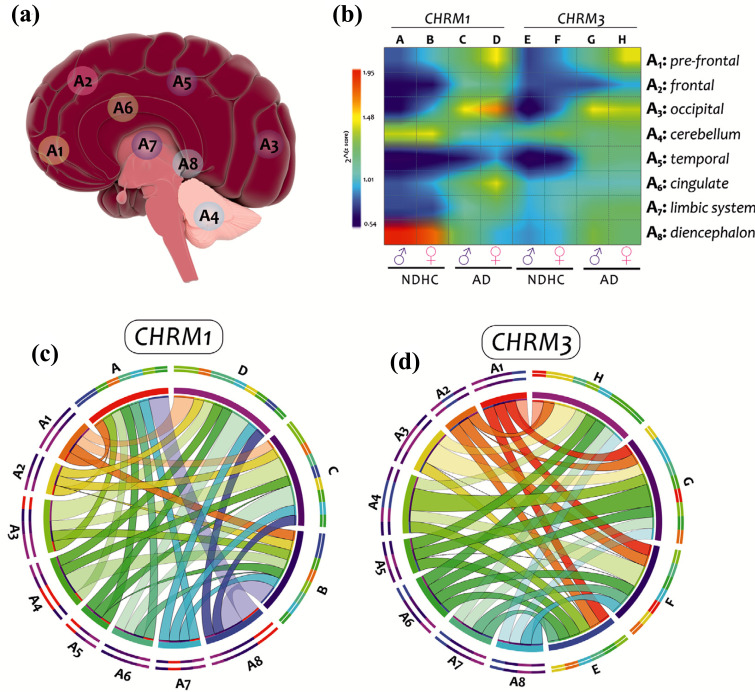
In our investigation, in order to examine the expression profile of CHRM1 and CHMR3 genes, we used twenty microarray datasets generated from brain biopsies of NDHC subjects and AD patients. We reported that CHRM1 and CHRM3 expression levels were significantly reduced in AD brains compared with NDHC. The expression levels of the two genes were positively correlated both in NDHC and in AD brain patients. Older NDHC subjects showed lower expression than the younger ones. In addition, there was a significant correlation between CHRM1, CHRM3, and NRGN expression, and these correlations were potentially a function of both disease state and sex (Fig. 10).
Fig. (10).
Analysis results graphical summary. The eight brain regions analyzed in this study (a). Plot matrix Heat map colored by the interpolate-cold hot. A, C, E, and G indicate the results of the brain regions analysis in males with NDHC and AD. B, D, F, and H indicate the results of the brain regions analysis in females NDHC and AD. The gradation color from blue (0.54 value) to red (1.95 value) indicates the z-score levels. As an interpretation example, the highest levels of CHRM1 were expressed by NDHC males (column A) in the diencephalic region (line A8), indicated by the red color (b). CHRM1 (c) and CHRM3 (d) expression levels in males and females brain regions of NDHCs subjects and AD patients were expressed as z-score and depicted as cord diagrams using the online tool CIRCOS. Ribbon sizes indicate the z-score value. The eight brain regions are indicated as follow: A1 (pre-frontal), A2 (frontal), A3 (occipital), A4 (cerebellum), A5 (temporal), A6 (cingulate), A7 (diencephalon), A8 (limbic system).
In recent years, the use of datasets available in public databases has grown exponentially. Several research groups - including ours - have extensively used the analysis of public transcriptome datasets for the identification of novel pathogenic pathways and therapeutic targets in several human pathologies [81, 82], such as neurodegenerative disease [83-89] and cancer [90]. Through a meta-analysis of public array datasets, it is possible to increase the statistical power to obtain a more precise estimate of gene expression differentials and assess the heterogeneity of the overall estimate. Meta-analysis is relatively inexpensive since it makes comprehensive use of already available data and represents a vast source of information that could make a difference in setting up highly targeted experimental strategies.
One of the important hallmarks of AD is cholinergic hypo-function [91]. It has been shown that in AD brains, there are reduced choline acetyltransferase levels accompanied by decreased ACh synthesis and significant loss of cholinergic neurons. Recent evidence indicates that cholinergic hypofunction is closely related to Aβ and tau pathologies [92]. Members of the muscarinic acetylcholine receptors family are expressed in several brain regions and play crucial roles in a variety of physiological processes such as memory, attention, nociception, motor control, and sleep-wake cycle [93]. Among the five members of the mAChR family (M1-M5), the M1 (CHRM1) subtype makes up 50-60% of the total and is predominantly expressed in the hippocampus, cerebral cortex, corpus striatum, and thalamus [94]. CHRM1-knockout mice show a series of cognitive deficits and impairments in long-term potentiation, indicating that the CHRM1 subtype is physiologically linked to multiple functions such as synaptic plasticity, neuronal excitability, and neuronal differentiation during early development, learning, and memory [95]. M3 mAChR (CHRM3) subtypes are widely distributed in the CNS, although at a lower level than other mAChR subtypes. CHRM3 is expressed at a relatively high level in the hypothalamus but it is also found in many other regions, including the hippocampus [95]. Our results appear to be in line with existing evidence on the role of the CHRM1 and CHRM3 genes in AD pathophysiology. Indeed, alterations in learning and memory skills and disorders in food intake are peculiar characteristics of AD [96, 97]. Interestingly, here we demonstrate how the brain of AD females had lower levels of CHRM1 and CHRM3 than age-matched males. Differences in the incidence and prevalence of AD between female and male subjects are well established; notably, two thirds of AD patients are women [98]. Our findings partly confirmed this trend by comparing the CHRM1 and CHRM3 expression levels with the neurogranin (NRGN) gene expression. Indeed, we showed that in the brain of NDHC females, there was the strongest linear relationship between the expression levels of NRGN and both CHRM1 and CHRM3, compared to males NDHC. Overlapping data were obtained when we performed the analysis in AD patients. Furthermore, ROC curve analysis showed that for single predictors, CHRM1 had the highest accuracy, followed by CHRM3 and NRGN. These results confirm the potential role played by CHRM1 and CHRM3 receptors in AD. Moreover, since neurogranin is a marker of the disease progression, CHRM1 and CHRM3 receptors could likely be causally linked to the cognitive impairment associated with AD.
A limitation of our manuscript derives from the absence of information regarding the clinical data of the selected samples. Unfortunately, the analyzed datasets did not provide any information on concurrent sleep disorders. Such information would have allowed us a deeper analysis and could have finally unveiled the link between muscarinic receptors and sleep disturbances both in AD patients and in potentially healthy subjects during ageing. To support this, we have been able to provide our results and the bibliography published up to this moment. Sleep disturbances and sleep-wake rhythm disturbances are established symptoms of AD and may precede the other clinical symptoms of this neurodegenerative disease [99]. Researchers speculate that cognitive decline distinguishing AD might originally affect brain areas responsible for sleep, thus explaining the strict association between sleep disorders and the disease [100]. Recent evidence indicates that people with sleep fragmentation have a higher risk of developing AD in the following years [101], suggesting that sleep disturbances, as a potential early biomarker of AD, may occur several years before the appearance of cognitive decline. Nevertheless, the causal relationship between altered sleep and AD has not been proved yet [19]. In 2014, Peever J and colleagues from the University of Toronto in Canada found that rapid eye movement sleep is the best current predictor of brain diseases like Parkinson's and Alzheimer's [100]. A recent genetic study revealed that the G protein-coupled muscarinic acetylcholine receptors, CHRM1 and CHRM3, are essential for REM sleep, as REM sleep and its associated enrichment of EEG theta oscillation could be hardly detected in CHRM1 and CHRM3 double-knockout (DKO) mice during sleep [40]. REM sleep phase is characterized by rapid side-to-side eye movements, muscle atony and a mixture of high-frequency, low-amplitude brain waves similar to those in the waking state. Several areas in the brain have been recognized as linked to the REM sleep, such as the hippocampus, temporal-occipital areas, cortex, and basal forebrain. The limbic system, including the amygdala, is an active region during REM sleep. Our results showed a significant reduction in CHRM1 and CHRM3 expression levels in the whole brain of AD patients compared to age-matched NDHC subjects. These results have also been confirmed in regions that regulate REM sleep activity, such as the pre-frontal, frontal, occipital, and limbic system. These findings suggest an active role of the CHRM1 and CHRM3 genes in sleep disorders present in AD patients. Only in the diencephalon and cerebellum of AD patients we have observed an increase in CHRM1 expression levels compared to NDHC subjects. Recently, it has been shown that rather than being ‘spared’, the cerebellum is affected in a different way than other brain regions and that, since it shows few pathological signs, this scenario may reflect a plausible protective capability of this region [102]. The increase of CHRM1 would appear to be in antithesis with the other brain regions analyzed. Such a discrepancy could be related to a possible disproportion in the deposition of the Ab plaques with a consequent compensatory CHRM1 gene transcription.
Furthermore, the sex-related differences in CHRM1 and CHRM3 expression in brains of both NDHC subjects and AD patients highlighted by our analysis agree with the evidence that females develop sleep disturbances more frequently than males. Women across a wide range of ages report more sleep complaints. In subjective studies with self-assessments, women report disrupted and insufficient sleep more commonly than men [103]. They report poorer sleep quality, difficulties falling asleep, frequent night awakenings, and longer periods of time awake throughout the night [104]. Consistently, sex differences related to insomnia, which emerges after puberty and tends to increase with ageing, have been reported, suggesting the involvement of hormonal influence [105]. Along with the development of Alzheimer’s disease, sex-related differences in the CHRM1 and CHRM3 brain expression levels were significantly attenuated in comparison with NDHC subjects. Most likely, once the disease takes over, the resulting neuronal death tends to reduce the differences between sexes, manifesting a common phenotype, the AD phenotype. Despite this, however, in AD patients, sex-related differences related to sleep disturbances are more present in females than in males [106].
Our results unveiled a significant variation in CHRM1 and CHRM3 expression levels among the different brain areas analyzed, both in healthy subjects and in AD patients. Expression of the CHRM1 receptor is well-characterized in all major areas of the forebrain, including the hippocampus, cerebral cortex, corpus striatum, and thalamus [95]. As regards, CHRM1 distribution has relatively high levels in the hypothalamus but is also found in several other regions including the the hippocampus [94]. Our data showed the highest levels of both CHRM1 and CHRM3 in the temporal lobe region in healthy subjects, while, in AD patients, at the temporal lobe level for CHRM1 and at the frontal area level for CHRM3. The data showed compatibility both with bibliographic findings and with the function performed. The temporal lobe communicates with the hippocampus and is crucial in forming long-term memory. In AD patients’ brains, the temporal lobe levels of both CHRM1 and CHRM3 were significantly reduced, likely suggesting a potential reduction in neurotransmission function. It has been shown that, in the early stages, Alzheimer's disease typically affects short-term memory. However, as the disease progresses, people gradually experience a higher long-term memory loss, also called amnesia [107].
Regarding the changes in brain CHRM1 and CHRM3 expression as a function of sex found during our analysis, the cellular distribution of these two receptors must be taken into consideration. Neuronal pyramidal cells express CHRM1 and CHRM3[108]. Loss of large pyramidal cells, along with plaques and tangles, is fundamental to the neuropathology of Alzheimer's disease [109]. Studies reported sex differences in the morphology of pyramidal neurons. The degree of clinical dementia well correlates with the extent of pyramidal cell loss. The triggering cause of this cell loss remains unknown but probably, relates to neurofibrillary degeneration through oxidative stress and neuroinflammation. When affected by AD, females progress more often to severe cognitive dysfunction due to more severe neurofibrillary degeneration and greater loss of brain parenchyma [110]. Thus, we found sex-related differences in AD brains CHRM1 and CHRM3 expression could be attributable to a more extensive loss of pyramidal neurons and more severe neurofibrillary degeneration.
In line with our finding and considering a likely clinical relevance for our presented results, we need to mention that a positive allosteric modulator (PAM) of M1 muscarinic acetylcholine receptors has been successfully tested in preclinical studies on Alzheimer's disease [111-113]. Certainly, the use of PAMs leads to reduced side-effects in comparison to agonist drugs, being the receptor exclusively activated under physiological stimuli, nevertheless, a selective M1 muscarinic acetylcholine receptors agonist is in clinical development for the treatment of AD [112]. Furthermore, the data reported could open to future gendered therapeutic approaches since muscarinic receptors are more expressed in AD patients in males than in females.
CONCLUSION
Our study unveils substantial differences in CHRM1 and CHRM3 brain transcripts of AD patients compared to NDHCs subjects, differences that are strictly linked to the brain regions and the sex (Fig. 10).
Taken together, these results partially confirm the clinical findings that emerged from AD patients in the last years. As an exploratory study, further investigations are needed in order to corroborate our findings.
ACKNOWLEDGEMENTS
We would like to show our gratitude to the authors of microarray datasets made available online for consultation and re-analysis. We also want to thank Hans Zimmer for realizing the songs “Dream Is Collapsing” and “Dream Within a Dream” inserted in the “Inception” movie as a soundtrack, an inspiration during the writing of this manuscript.
LIST OF ABBREVIATIONS
- AD
Alzheimer’s Disease
- CHRM1
Muscarinic Acetylcholine Receptor M1
- CHRM3
Muscarinic Acetylcholine Receptor M3
- CNS
Central Nervous System
- CSF
Cerebrospinal Fluid
- EEG
Electroencephalogram
- FDR
False Discovery Rate
- GEO
Gene Expression Omnibus
- MD
Microarray Datasets
- MeV
MultiExperiment Viewer
- N_REM
Non-REM
- NDHC
Non-demented Healthy Controls
- REM
Rapid Eye Movement
- SDEG
Significantly Different Expressed Genes
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
All clinical investigations have been conducted according to the Declaration of Helsinki principles, as stated by the original authors of the deposited datasets.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets analysed during the current study are available from the corresponding author on reasonable request.
FUNDING
Henrik Zetterberg is a Wallenberg Scholar supported by grants from the Swedish Research Council (Grant no. 2018-02532), the European Research Council (Grant no. 681712), Swedish State Support for Clinical Research (Grant no. ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (Grant no. 201809-2016862), and the UK Dementia Research Institute at UCL. Michelino Di Rosa was supported by the University Research Project Grant (Grant no. PIACERI 2020-2022), Department of Biomedical and Biotechnological Sciences (BIOMETEC), University of Catania, Italy, for the project NEURAGITION (NEURological AGIng DeconvoluTION).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
REFERENCES
- 1.Ahmed R.M., Paterson R.W., Warren J.D., Zetterberg H., O’Brien J.T., Fox N.C., Halliday G.M., Schott J.M. Biomarkers in dementia: clinical utility and new directions. J. Neurol. Neurosurg. Psychiatry. 2014;85(12):1426–1434. doi: 10.1136/jnnp-2014-307662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blennow K., Zetterberg H. Biomarkers for Alzheimer’s disease: current status and prospects for the future. J. Intern. Med. 2018;284(6):643–663. doi: 10.1111/joim.12816. [DOI] [PubMed] [Google Scholar]
- 3.Blennow K., de Leon M.J., Zetterberg H. Alzheimer’s disease. Lancet. 2006;368(9533):387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 4.Quiroz Y.T., Zetterberg H., Reiman E.M., Chen Y., Su Y., Fox-Fuller J.T., Garcia G., Villegas A., Sepulveda-Falla D., Villada M., Arboleda-Velasquez J.F., Guzmán-Vélez E., Vila-Castelar C., Gordon B.A., Schultz S.A., Protas H.D., Ghisays V., Giraldo M., Tirado V., Baena A., Munoz C., Rios-Romenets S., Tariot P.N., Blennow K., Lopera F. Plasma neurofilament light chain in the presenilin 1 E280A autosomal dominant Alzheimer’s disease kindred: a cross-sectional and longitudinal cohort study. Lancet Neurol. 2020;19(6):513–521. doi: 10.1016/S1474-4422(20)30137-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sperling R.A., Aisen P.S., Beckett L.A., Bennett D.A., Craft S., Fagan A.M., Iwatsubo T., Jack C.R., Jr, Kaye J., Montine T.J., Park D.C., Reiman E.M., Rowe C.C., Siemers E., Stern Y., Yaffe K., Carrillo M.C., Thies B., Morrison-Bogorad M., Wagster M.V., Phelps C.H. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman R.J., Xiong C., Benzinger T.L., Fagan A.M., Goate A., Fox N.C., Marcus D.S., Cairns N.J., Xie X., Blazey T.M., Holtzman D.M., Santacruz A., Buckles V., Oliver A., Moulder K., Aisen P.S., Ghetti B., Klunk W.E., McDade E., Martins R.N., Masters C.L., Mayeux R., Ringman J.M., Rossor M.N., Schofield P.R., Sperling R.A., Salloway S., Morris J.C., Dominantly Inherited Alzheimer N. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 2012;367(9):795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagan A.M., Xiong C., Jasielec M.S., Bateman R.J., Goate A.M., Benzinger T.L., Ghetti B., Martins R.N., Masters C.L., Mayeux R., Ringman J.M., Rossor M.N., Salloway S., Schofield P.R., Sperling R.A., Marcus D., Cairns N.J., Buckles V.D., Ladenson J.H., Morris J.C., Holtzman D.M., Dominantly Inherited Alzheimer N. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci. Transl. Med. 2014;6(226):226ra30. doi: 10.1126/scitranslmed.3007901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández F., Avila J. Tauopathies. Cell. Mol. Life Sci. 2007;64(17):2219–2233. doi: 10.1007/s00018-007-7220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Lin H., He X., Chen L., Dai Y., Jia W., Xue X., Tao J., Chen L. Neurogranin as a cognitive biomarker in cerebrospinal fluid and blood exosomes for Alzheimer’s disease and mild cognitive impairment. Transl. Psychiatry. 2020;10(1):125. doi: 10.1038/s41398-020-0801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudelli R.D., Ambler M.W., Wisniewski H.M. Morphology and distribution of Alzheimer neuritic (senile) and amyloid plaques in striatum and diencephalon. Acta Neuropathol. 1984;64(4):273–281. doi: 10.1007/BF00690393. [DOI] [PubMed] [Google Scholar]
- 11.Ju Y.E., Lucey B.P., Holtzman D.M. Sleep and Alzheimer disease pathology--a bidirectional relationship. Nat. Rev. Neurol. 2014;10(2):115–119. doi: 10.1038/nrneurol.2013.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moe K.E., Vitiello M.V., Larsen L.H., Prinz P.N. Symposium: Cognitive processes and sleep disturbances: Sleep/wake patterns in Alzheimer’s disease: relationships with cognition and function. J. Sleep Res. 1995;4(1):15–20. doi: 10.1111/j.1365-2869.1995.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 13.Jackson C.E., Snyder P.J. Electroencephalography and event-related potentials as biomarkers of mild cognitive impairment and mild Alzheimer’s disease. Alzheimers Dement. 2008;4(1) Suppl. 1:S137–S143. doi: 10.1016/j.jalz.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Moran M., Lynch C.A., Walsh C., Coen R., Coakley D., Lawlor B.A. Sleep disturbance in mild to moderate Alzheimer’s disease. Sleep Med. 2005;6(4):347–352. doi: 10.1016/j.sleep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Beaulieu-Bonneau S., Hudon C. Sleep disturbances in older adults with mild cognitive impairment. Int. Psychogeriatr. 2009;21(4):654–666. doi: 10.1017/S1041610209009120. [DOI] [PubMed] [Google Scholar]
- 16.Craig D., Hart D.J., Passmore A.P. Genetically increased risk of sleep disruption in Alzheimer’s disease. Sleep. 2006;29(8):1003–1007. doi: 10.1093/sleep/29.8.1003. [DOI] [PubMed] [Google Scholar]
- 17.Pace-Schott E.F., Spencer R.M. Age-related changes in the cognitive function of sleep. Prog. Brain Res. 2011. p. 191, 75-89. [DOI] [PubMed] [Google Scholar]
- 18.Lim M.M., Gerstner J.R., Holtzman D.M. The sleep-wake cycle and Alzheimer’s disease: what do we know? Neurodegener. Dis. Manag. 2014;4(5):351–362. doi: 10.2217/nmt.14.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloret M.A., Cervera-Ferri A., Nepomuceno M., Monllor P., Esteve D., Lloret A. Is sleep disruption a cause or consequence of Alzheimer’s disease? reviewing its possible role as a biomarker. Int. J. Mol. Sci. 2020;21(3):21. doi: 10.3390/ijms21031168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abel T., Havekes R., Saletin J.M., Walker M.P. Sleep, plasticity and memory from molecules to whole-brain networks. Curr. Biol. 2013;23(17):R774–R788. doi: 10.1016/j.cub.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maquet P. The role of sleep in learning and memory. Science. 2001;294(5544):1048–1052. doi: 10.1126/science.1062856. [DOI] [PubMed] [Google Scholar]
- 22.Casement M.D., Broussard J.L., Mullington J.M., Press D.Z. The contribution of sleep to improvements in working memory scanning speed: a study of prolonged sleep restriction. Biol. Psychol. 2006;72(2):208–212. doi: 10.1016/j.biopsycho.2005.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graves L.A., Heller E.A., Pack A.I., Abel T. Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learn. Mem. 2003;10(3):168–176. doi: 10.1101/lm.48803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prince T.M., Wimmer M., Choi J., Havekes R., Aton S., Abel T. Sleep deprivation during a specific 3-hour time window post-training impairs hippocampal synaptic plasticity and memory. Neurobiol. Learn. Mem. 2014;109:122–130. doi: 10.1016/j.nlm.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swetha R., Kumar D., Gupta S.K., Ganeshpurkar A., Singh R., Gutti G., Kumar D., Jana S., Krishnamurthy S., Singh S.K. Multifunctional hybrid sulfonamides as novel therapeutic agents for Alzheimer’s disease. Future Med. Chem. 2019;11(24):3161–3178. doi: 10.4155/fmc-2019-0106. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira-Vieira T.H., Guimaraes I.M., Silva F.R., Ribeiro F.M. Alzheimer’s disease: Targeting the cholinergic system. Curr. Neuropharmacol. 2016;14(1):101–115. doi: 10.2174/1570159X13666150716165726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veeraragavan S., Bui N., Perkins J.R., Yuva-Paylor L.A., Carpenter R.L., Paylor R. Modulation of behavioral phenotypes by a muscarinic M1 antagonist in a mouse model of fragile X syndrome. Psychopharmacology (Berl.) 2011;217(1):143–151. doi: 10.1007/s00213-011-2276-6. [DOI] [PubMed] [Google Scholar]
- 28.Verma S., Kumar A., Tripathi T., Kumar A. Muscarinic and nicotinic acetylcholine receptor agonists: current scenario in Alzheimer’s disease therapy. J. Pharm. Pharmacol. 2018;70(8):985–993. doi: 10.1111/jphp.12919. [DOI] [PubMed] [Google Scholar]
- 29.Roeren T., LeVeen R.F., Nugent L. Photoplethysmographic documentation of improved microcirculation after pentoxifylline therapy. Angiology. 1988;39(11):929–933. doi: 10.1177/000331978803901101. [DOI] [PubMed] [Google Scholar]
- 30.Caulfield M.P., Birdsall N.J. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol. Rev. 1998;50(2):279–290. [PubMed] [Google Scholar]
- 31.Weston-Green K., Huang X.F., Lian J., Deng C. Effects of olanzapine on muscarinic M3 receptor binding density in the brain relates to weight gain, plasma insulin and metabolic hormone levels. Eur. Neuropsychopharmacol. 2012;22(5):364–373. doi: 10.1016/j.euroneuro.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Lang W., Henke H. Cholinergic receptor binding and autoradiography in brains of non-neurological and senile dementia of Alzheimer-type patients. Brain Res. 1983;267(2):271–280. doi: 10.1016/0006-8993(83)90879-X. [DOI] [PubMed] [Google Scholar]
- 33.Piggott M.A., Owens J., O’Brien J., Colloby S., Fenwick J., Wyper D., Jaros E., Johnson M., Perry R.H., Perry E.K. Muscarinic receptors in basal ganglia in dementia with Lewy bodies, Parkinson’s disease and Alzheimer’s disease. J. Chem. Neuroanat. 2003;25(3):161–173. doi: 10.1016/S0891-0618(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 34.Scarr E., McLean C., Dean B. Higher levels of different muscarinic receptors in the cortex and hippocampus from subjects with Alzheimer’s disease. J. Neural Transm. (Vienna) 2017;124(3):273–284. doi: 10.1007/s00702-016-1625-3. [DOI] [PubMed] [Google Scholar]
- 35.Colloby S.J., McKeith I.G., Wyper D.J., O’Brien J.T., Taylor J.P. Regional covariance of muscarinic acetylcholine receptors in Alzheimer’s disease using (R, R) [(123)I]-QNB SPECT. J. Neurol. 2015;262(9):2144–2153. doi: 10.1007/s00415-015-7827-z. [DOI] [PubMed] [Google Scholar]
- 36.Holman B.L., Gibson R.E., Hill T.C., Eckelman W.C., Albert M., Reba R.C. Muscarinic acetylcholine receptors in Alzheimer’s disease. In vivo imaging with iodine 123-labeled 3-quinuclidinyl-4-iodobenzilate and emission tomography. JAMA. 1985;254(21):3063–3066. doi: 10.1001/jama.1985.03360210079035. [DOI] [PubMed] [Google Scholar]
- 37.Jiang S., Li Y., Zhang C., Zhao Y., Bu G., Xu H., Zhang Y.W. M1 muscarinic acetylcholine receptor in Alzheimer’s disease. Neurosci. Bull. 2014;30(2):295–307. doi: 10.1007/s12264-013-1406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conn P.J., Jones C.K., Lindsley C.W. Subtype-selective allosteric modulators of muscarinic receptors for the treatment of CNS disorders. Trends Pharmacol. Sci. 2009;30(3):148–155. doi: 10.1016/j.tips.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis A.A., Fritz J.J., Wess J., Lah J.J., Levey A.I. Deletion of M1 muscarinic acetylcholine receptors increases amyloid pathology in vitro and in vivo. J. Neurosci. 2010;30(12):4190–4196. doi: 10.1523/JNEUROSCI.6393-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa Y., Kanda G.N., Yamada R.G., Shi S., Sunagawa G.A., Ukai-Tadenuma M., Fujishima H., Matsumoto N., Masumoto K.H., Nagano M., Kasukawa T., Galloway J., Perrin D., Shigeyoshi Y., Ukai H., Kiyonari H., Sumiyama K., Ueda H.R. Muscarinic acetylcholine receptors Chrm1 and Chrm3 are essential for REM sleep. Cell Rep. 2018;24(9):2231–2247.e7. doi: 10.1016/j.celrep.2018.07.082. [DOI] [PubMed] [Google Scholar]
- 41.Murillo-Rodriguez E., Arias-Carrion O., Zavala-Garcia A., Sarro-Ramirez A., Huitron-Resendiz S., Arankowsky-Sandoval G. Basic sleep mechanisms: an integrative review. Cent. Nerv. Syst. Agents Med. Chem. 2012;12(1):38–54. doi: 10.2174/187152412800229107. [DOI] [PubMed] [Google Scholar]
- 42.Williams J.H., Kauer J.A. Properties of carbachol-induced oscillatory activity in rat hippocampus. J. Neurophysiol. 1997;78(5):2631–2640. doi: 10.1152/jn.1997.78.5.2631. [DOI] [PubMed] [Google Scholar]
- 43.Fellous J.M., Sejnowski T.J. Cholinergic induction of oscillations in the hippocampal slice in the slow (0.5-2 Hz), theta (5-12 Hz), and gamma (35-70 Hz) bands. Hippocampus. 2000;10(2):187–197. doi: 10.1002/(SICI)1098-1063(2000)10:2<187:AID-HIPO8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 44.Manseau F., Danik M., Williams S. A functional glutamatergic neurone network in the medial septum and diagonal band area. J. Physiol. 2005;566(Pt 3):865–884. doi: 10.1113/jphysiol.2005.089664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim E.J., Jeong D.U. Transdermal scopolamine alters phasic REM activity in normal young adults. Sleep. 1999;22(4):515–520. doi: 10.1093/sleep/22.4.515. [DOI] [PubMed] [Google Scholar]
- 46.Buzsáki G. Theta oscillations in the hippocampus. Neuron. 2002;33(3):325–340. doi: 10.1016/S0896-6273(02)00586-X. [DOI] [PubMed] [Google Scholar]
- 47.Sjöstedt E., Zhong W., Fagerberg L., Karlsson M., Mitsios N., Adori C., Oksvold P., Edfors F., Limiszewska A., Hikmet F., Huang J., Du Y., Lin L., Dong Z., Yang L., Liu X., Jiang H., Xu X., Wang J., Yang H., Bolund L., Mardinoglu A., Zhang C., von Feilitzen K., Lindskog C., Pontén F., Luo Y., Hökfelt T., Uhlén M., Mulder J. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367(6482):367. doi: 10.1126/science.aay5947. [DOI] [PubMed] [Google Scholar]
- 48.Narayanan M., Huynh J.L., Wang K., Yang X., Yoo S., McElwee J., Zhang B., Zhang C., Lamb J.R., Xie T., Suver C., Molony C., Melquist S., Johnson A.D., Fan G., Stone D.J., Schadt E.E., Casaccia P., Emilsson V., Zhu J. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014;10:743. doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumaran R., Cookson M.R. Pathways to Parkinsonism Redux: convergent pathobiological mechanisms in genetics of Parkinson’s disease. Hum. Mol. Genet. 2015;24(R1):R32–R44. doi: 10.1093/hmg/ddv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., Meng Q., Xia Y., Ding C., Wang L., Dai R., Cheng L., Gunaratne P., Gibbs R.A., Min S., Coarfa C., Reid J.G., Zhang C., Jiao C., Jiang Y., Giase G., Thomas A., Fitzgerald D., Brunetti T., Shieh A., Xia C., Wang Y., Wang Y., Badner J.A., Gershon E.S., White K.P., Liu C. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci. Transl. Med. 2018;10(472):10. doi: 10.1126/scitranslmed.aat8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., Johnson R., Zielke H.R., Ferrucci L., Longo D.L., Cookson M.R., Singleton A.B. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., MacDonald M.E., Lamb J.R., Bennett D.A., Molony C., Stone D.J., Gudnason V., Myers A.J., Schadt E.E., Neumann H., Zhu J., Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153(3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernandez D.G., Nalls M.A., Moore M., Chong S., Dillman A., Trabzuni D., Gibbs J.R., Ryten M., Arepalli S., Weale M.E., Zonderman A.B., Troncoso J., O’Brien R., Walker R., Smith C., Bandinelli S., Traynor B.J., Hardy J., Singleton A.B., Cookson M.R. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol. Dis. 2012;47(1):20–28. doi: 10.1016/j.nbd.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trabzuni D., Ramasamy A., Imran S., Walker R., Smith C., Weale M.E., Hardy J., Ryten M. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 2013;4:2771. doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel H., Hodges A.K., Curtis C., Lee S.H., Troakes C., Dobson R.J.B., Newhouse S.J. Transcriptomic analysis of probable asymptomatic and symptomatic alzheimer brains. Brain Behav. Immun. 2019;80:644–656. doi: 10.1016/j.bbi.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M., Pletikos M., Meyer K.A., Sedmak G., Guennel T., Shin Y., Johnson M.B., Krsnik Z., Mayer S., Fertuzinhos S., Umlauf S., Lisgo S.N., Vortmeyer A., Weinberger D.R., Mane S., Hyde T.M., Huttner A., Reimers M., Kleinman J.E., Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.French L., Ma T., Oh H., Tseng G.C., Sibille E. Age-related gene expression in the frontal cortex suggests synaptic function changes in specific inhibitory neuron subtypes. Front. Aging Neurosci. 2017;9:162. doi: 10.3389/fnagi.2017.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hokama M., Oka S., Leon J., Ninomiya T., Honda H., Sasaki K., Iwaki T., Ohara T., Sasaki T., LaFerla F.M., Kiyohara Y., Nakabeppu Y. Altered expression of diabetes-related genes in Alzheimer’s disease brains: the Hisayama study. Cereb. Cortex. 2014;24(9):2476–2488. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durrenberger P.F., Fernando F.S., Kashefi S.N., Bonnert T.P., Seilhean D., Nait-Oumesmar B., Schmitt A., Gebicke-Haerter P.J., Falkai P., Grünblatt E., Palkovits M., Arzberger T., Kretzschmar H., Dexter D.T., Reynolds R. Common mechanisms in neurodegeneration and neuroinflammation: a BrainNet Europe gene expression microarray study. J. Neural Transm. (Vienna) 2015;122(7):1055–1068. doi: 10.1007/s00702-014-1293-0. [DOI] [PubMed] [Google Scholar]
- 60.Wang M., Roussos P., McKenzie A., Zhou X., Kajiwara Y., Brennand K.J., De Luca G.C., Crary J.F., Casaccia P., Buxbaum J.D., Ehrlich M., Gandy S., Goate A., Katsel P., Schadt E., Haroutunian V., Zhang B. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer’s disease. Genome Med. 2016;8(1):104. doi: 10.1186/s13073-016-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang W.S., Dunckley T., Beach T.G., Grover A., Mastroeni D., Walker D.G., Caselli R.J., Kukull W.A., McKeel D., Morris J.C., Hulette C., Schmechel D., Alexander G.E., Reiman E.M., Rogers J., Stephan D.A. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol. Genomics. 2007;28(3):311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berchtold N.C., Cribbs D.H., Coleman P.D., Rogers J., Head E., Kim R., Beach T., Miller C., Troncoso J., Trojanowski J.Q., Zielke H.R., Cotman C.W. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. USA. 2008;105(40):15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berchtold N.C., Coleman P.D., Cribbs D.H., Rogers J., Gillen D.L., Cotman C.W. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol. Aging. 2013;34(6):1653–1661. doi: 10.1016/j.neurobiolaging.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gelman B.B., Chen T., Lisinicchia J.G., Soukup V.M., Carmical J.R., Starkey J.M., Masliah E., Commins D.L., Brandt D., Grant I., Singer E.J., Levine A.J., Miller J., Winkler J.M., Fox H.S., Luxon B.A., Morgello S. The National NeuroAIDS Tissue Consortium brain gene array: two types of HIV-associated neurocognitive impairment. PLoS One. 2012;7(9):e46178. doi: 10.1371/journal.pone.0046178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blalock E.M., Buechel H.M., Popovic J., Geddes J.W., Landfield P.W. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J. Chem. Neuroanat. 2011;42(2):118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Piras I.S., Krate J., Delvaux E., Nolz J., Mastroeni D.F., Persico A.M., Jepsen W.M., Beach T.G., Huentelman M.J., Coleman P.D. Transcriptome changes in the Alzheimer’s disease middle temporal gyrus: Importance of RNA metabolism and mitochondria-associated membrane genes. J. Alzheimers Dis. 2019;70(3):691–713. doi: 10.3233/JAD-181113. [DOI] [PubMed] [Google Scholar]
- 67.Antonell A., Lladó A., Altirriba J., Botta-Orfila T., Balasa M., Fernández M., Ferrer I., Sánchez-Valle R., Molinuevo J.L. A preliminary study of the whole-genome expression profile of sporadic and monogenic early-onset Alzheimer’s disease. Neurobiol. Aging. 2013;34(7):1772–1778. doi: 10.1016/j.neurobiolaging.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 68.Sanfilippo C., Castrogiovanni P., Imbesi R., Kazakowa M., Musumeci G., Blennow K., Zetterberg H., Di Rosa M. Sex difference in CHI3L1 expression levels in human brain aging and in Alzheimer’s disease. Brain Res. 2019;1720:146305. doi: 10.1016/j.brainres.2019.146305. [DOI] [PubMed] [Google Scholar]
- 69.Xiao J., Cao H., Chen J. False discovery rate control incorporating phylogenetic tree increases detection power in microbiome-wide multiple testing. Bioinformatics. 2017;33(18):2873–2881. doi: 10.1093/bioinformatics/btx311. [DOI] [PubMed] [Google Scholar]
- 70.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 71.Davis S., Meltzer P.S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 72.Mauri E., Sacchetti A., Vicario N., Peruzzotti-Jametti L., Rossi F., Pluchino S. Evaluation of RGD functionalization in hybrid hydrogels as 3D neural stem cell culture systems. Biomater. Sci. 2018;6(3):501–510. doi: 10.1039/C7BM01056G. [DOI] [PubMed] [Google Scholar]
- 73.Tiao, GEPBGC Bayesian Inference in Statistical Analysis; , 1992.
- 74.Cheadle C., Vawter M.P., Freed W.J., Becker K.G. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003;5(2):73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J., Coombes K.R., Highsmith W.E., Keating M.J., Abruzzo L.V. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: a meta-analysis of three microarray studies. Bioinformatics. 2004;20(17):3166–3178. doi: 10.1093/bioinformatics/bth381. [DOI] [PubMed] [Google Scholar]
- 76.Reddy T.B., Riley R., Wymore F., Montgomery P., DeCaprio D., Engels R., Gellesch M., Hubble J., Jen D., Jin H., Koehrsen M., Larson L., Mao M., Nitzberg M., Sisk P., Stolte C., Weiner B., White J., Zachariah Z.K., Sherlock G., Galagan J.E., Ball C.A., Schoolnik G.K. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37(Database issue):D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mehmood R., El-Ashram S., Bie R., Dawood H., Kos A. Clustering by fast search and merge of local density peaks for gene expression microarray data. Sci. Rep. 2017;7:45602. doi: 10.1038/srep45602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang C., Huo Y., Xin L., Tian B., Yu B. Feature selection and tumor classification for microarray data using relaxed Lasso and generalized multi-class support vector machine. J. Theor. Biol. 2019;463:77–91. doi: 10.1016/j.jtbi.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 79.De Vos A., Jacobs D., Struyfs H., Fransen E., Andersson K., Portelius E., Andreasson U., De Surgeloose D., Hernalsteen D., Sleegers K., Robberecht C., Van Broeckhoven C., Zetterberg H., Blennow K., Engelborghs S., Vanmechelen E. C-terminal neurogranin is increased in cerebrospinal fluid but unchanged in plasma in Alzheimer’s disease. Alzheimers Dement. 2015;11(12):1461–1469. doi: 10.1016/j.jalz.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 80.Kvartsberg H., Lashley T., Murray C.E., Brinkmalm G., Cullen N.C., Höglund K., Zetterberg H., Blennow K., Portelius E. The intact postsynaptic protein neurogranin is reduced in brain tissue from patients with familial and sporadic Alzheimer’s disease. Acta Neuropathol. 2019;137(1):89–102. doi: 10.1007/s00401-018-1910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caltabiano R., Castrogiovanni P., Barbagallo I., Ravalli S., Szychlinska M.A., Favilla V., Schiavo L., Imbesi R., Musumeci G., Di Rosa M. Identification of novel markers of prostate cancer progression, potentially modulated by vitamin D. Appl. Sci. 2019;9(22):4923. doi: 10.3390/app9224923. [DOI] [Google Scholar]
- 82.Fagone P., Nunnari G., Lazzara F., Longo A., Cambria D., Distefano G., Palumbo M., Nicoletti F., Malaguarnera L., Di Rosa M. Induction of OAS gene family in HIV monocyte infected patients with high and low viral load. Antiviral Res. 2016;131:66–73. doi: 10.1016/j.antiviral.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 83.Sanfilippo C., Castrogiovanni P., Imbesi R., Tibullo D., Li Volti G., Barbagallo I., Vicario N., Musumeci G., Di Rosa M. Middle-aged healthy women and Alzheimer’s disease patients present an overlapping of brain cell transcriptional profile. Neuroscience. 2019;406:333–344. doi: 10.1016/j.neuroscience.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Castrogiovanni P., Li Volti G., Sanfilippo C., Tibullo D., Galvano F., Vecchio M., Avola R., Barbagallo I., Malaguarnera L., Castorina S., Musumeci G., Imbesi R., Di Rosa M. Fasting and fast food diet play an opposite role in mice brain aging. Mol. Neurobiol. 2018;55(8):6881–6893. doi: 10.1007/s12035-018-0891-5. [DOI] [PubMed] [Google Scholar]
- 85.Sanfilippo C., Nunnari G., Calcagno A., Malaguarnera L., Blennow K., Zetterberg H., Di Rosa M. The chitinases expression is related to simian immunodeficiency virus encephalitis (SIVE) and in HIV encephalitis (HIVE). Virus Res. 2017;227:220–230. doi: 10.1016/j.virusres.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 86.Sanfilippo C., Malaguarnera L., Di Rosa M. Chitinase expression in Alzheimer’s disease and non-demented brains regions. J. Neurol. Sci. 2016;369:242–249. doi: 10.1016/j.jns.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 87.Sanfilippo C., Longo A., Lazzara F., Cambria D., Distefano G., Palumbo M., Cantarella A., Malaguarnera L., Di Rosa M. CHI3L1 and CHI3L2 overexpression in motor cortex and spinal cord of sALS patients. Mol. Cell. Neurosci. 2017;85:162–169. doi: 10.1016/j.mcn.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 88.Sanfilippo C., Pinzone M.R., Cambria D., Longo A., Palumbo M., Di Marco R., Condorelli F., Nunnari G., Malaguarnera L., Di Rosa M. OAS gene family expression is associated with HIV-related neurocognitive disorders. Mol. Neurobiol. 2018;55(3):1905–1914. doi: 10.1007/s12035-017-0460-3. [DOI] [PubMed] [Google Scholar]
- 89.Sanfilippo C., Castrogiovanni P., Imbesi R., Di Rosa M. CHI3L2 expression levels are correlated with AIF1, PECAM1, and CALB1 in the brains of Alzheimer’s disease patients. J. Mol. Neurosci. 2020;70(10):1598–1610. doi: 10.1007/s12031-020-01667-9. [DOI] [PubMed] [Google Scholar]
- 90.Di Rosa M., Sanfilippo C., Libra M., Musumeci G., Malaguarnera L. Different pediatric brain tumors are associated with different gene expression profiling. Acta Histochem. 2015;117(4-5):477–485. doi: 10.1016/j.acthis.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 91.Richter J.A., Perry E.K., Tomlinson B.E. Acetylcholine and choline levels in post-mortem human brain tissue: preliminary observations in Alzheimer’s disease. Life Sci. 1980;26(20):1683–1689. doi: 10.1016/0024-3205(80)90176-9. [DOI] [PubMed] [Google Scholar]
- 92.Pákáski M., Kálmán J. Interactions between the amyloid and cholinergic mechanisms in Alzheimer’s disease. Neurochem. Int. 2008;53(5):103–111. doi: 10.1016/j.neuint.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 93.Bonner T.I., Buckley N.J., Young A.C., Brann M.R. Identification of a family of muscarinic acetylcholine receptor genes. Science. 1987;237(4814):527–532. doi: 10.1126/science.3037705. [DOI] [PubMed] [Google Scholar]
- 94.Levey A.I. Immunological localization of m1-m5 muscarinic acetylcholine receptors in peripheral tissues and brain. Life Sci. 1993;52(5-6):441–448. doi: 10.1016/0024-3205(93)90300-R. [DOI] [PubMed] [Google Scholar]
- 95.Hamilton S.E., Loose M.D., Qi M., Levey A.I., Hille B., McKnight G.S., Idzerda R.L., Nathanson N.M. Disruption of the m1 receptor gene ablates muscarinic receptor-dependent M current regulation and seizure activity in mice. Proc. Natl. Acad. Sci. USA. 1997;94(24):13311–13316. doi: 10.1073/pnas.94.24.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang C.C., Lin Y.F., Chiu C.H., Liao Y.M., Ho M.H., Lin Y.K., Chou K.R., Liu M.F. Prevalence and factors associated with food intake difficulties among residents with dementia. PLoS One. 2017;12(2):e0171770. doi: 10.1371/journal.pone.0171770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wong S., Irish M., Savage G., Hodges J.R., Piguet O., Hornberger M. Strategic value-directed learning and memory in Alzheimer’s disease and behavioural-variant frontotemporal dementia. J. Neuropsychol. 2019;13(2):328–353. doi: 10.1111/jnp.12152. [DOI] [PubMed] [Google Scholar]
- 98.Mazure C.M., Swendsen J. Sex differences in Alzheimer’s disease and other dementias. Lancet Neurol. 2016;15(5):451–452. doi: 10.1016/S1474-4422(16)00067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brzecka A., Leszek J., Ashraf G.M., Ejma M., Ávila-Rodriguez M.F., Yarla N.S., Tarasov V.V., Chubarev V.N., Samsonova A.N., Barreto G.E., Aliev G. Sleep disorders associated with Alzheimer’s disease: A perspective. Front. Neurosci. 2018;12:330. doi: 10.3389/fnins.2018.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peever J., Luppi P.H., Montplaisir J. Breakdown in REM sleep circuitry underlies REM sleep behavior disorder. Trends Neurosci. 2014;37(5):279–288. doi: 10.1016/j.tins.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 101.Lim A.S., Kowgier M., Yu L., Buchman A.S., Bennett D.A. Sleep Fragmentation and the Risk of Incident Alzheimer’s Disease and Cognitive Decline in Older Persons. Sleep. 2013;36(7):1027–1032. doi: 10.5665/sleep.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu J., Patassini S., Rustogi N., Riba-Garcia I., Hale B.D., Phillips A.M., Waldvogel H., Haines R., Bradbury P., Stevens A., Faull R.L.M., Dowsey A.W., Cooper G.J.S., Unwin R.D. Regional protein expression in human Alzheimer’s brain correlates with disease severity. Commun. Biol. 2019;2:43. doi: 10.1038/s42003-018-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lindberg E., Janson C., Gislason T., Björnsson E., Hetta J., Boman G. Sleep disturbances in a young adult population: can gender differences be explained by differences in psychological status? Sleep. 1997;20(6):381–387. doi: 10.1093/sleep/20.6.381. [DOI] [PubMed] [Google Scholar]
- 104.Zhang B., Wing Y.K. Sex differences in insomnia: a meta-analysis. Sleep. 2006;29(1):85–93. doi: 10.1093/sleep/29.1.85. [DOI] [PubMed] [Google Scholar]
- 105.Mong J.A., Cusmano D.M. Sex differences in sleep: impact of biological sex and sex steroids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371(1688):20150110. doi: 10.1098/rstb.2015.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jee H.J., Shin W., Jung H.J., Kim B., Lee B.K., Jung Y.S. Impact of Sleep Disorder as a Risk Factor for Dementia in Men and Women. Biomol. Ther. (Seoul) 2020;28(1):58–73. doi: 10.4062/biomolther.2019.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jahn H. Memory loss in Alzheimer’s disease. Dialogues Clin. Neurosci. 2013;15(4):445–454. doi: 10.31887/DCNS.2013.15.4/hjahn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurowski P., Gawlak M., Szulczyk P. Muscarinic receptor control of pyramidal neuron membrane potential in the medial prefrontal cortex (mPFC) in rats. Neuroscience. 2015;303:474–488. doi: 10.1016/j.neuroscience.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 109.Balhorn R., Gledhill B.L., Wyrobek A.J. Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry. 1977;16(18):4074–4080. doi: 10.1021/bi00637a021. [DOI] [PubMed] [Google Scholar]
- 110.Filon J.R., Intorcia A.J., Sue L.I., Vazquez Arreola E., Wilson J., Davis K.J., Sabbagh M.N., Belden C.M., Caselli R.J., Adler C.H., Woodruff B.K., Rapscak S.Z., Ahern G.L., Burke A.D., Jacobson S., Shill H.A., Driver-Dunckley E., Chen K., Reiman E.M., Beach T.G., Serrano G.E. Gender differences in Alzheimer disease: Brain atrophy, histopathology burden, and cognition. J. Neuropathol. Exp. Neurol. 2016;75(8):748–754. doi: 10.1093/jnen/nlw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Caccamo A., Fisher A., LaFerla F.M. M1 agonists as a potential disease-modifying therapy for Alzheimer’s disease. Curr. Alzheimer Res. 2009;6(2):112–117. doi: 10.2174/156720509787602915. [DOI] [PubMed] [Google Scholar]
- 112.Nathan P.J., Millais S.B., Godwood A., Dewit O., Cross D.M., Liptrot J., Ruparelia B., Jones S.P., Bakker G., Maruff P.T., Light G.A., Brown A.J.H., Weir M.P., Congreve M., Tasker T. A phase 1b/2a multicenter study of the safety and preliminary pharmacodynamic effects of selective muscarinic M1 receptor agonist HTL0018318 in patients with mild-to-moderate Alzheimer’s disease. Alzheimers Dement. (N. Y.) 2022;8(1):e12273. doi: 10.1002/trc2.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abd-Elrahman K.S., Sarasija S., Colson T.L., Ferguson S.S.G. A positive allosteric modulator for the muscarinic receptor (M1 mAChR) improves pathology and cognitive deficits in female APPswe/PSEN1ΔE9 mice. Br. J. Pharmacol. 2022;179(8):1769–1783. doi: 10.1111/bph.15750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
The datasets analysed during the current study are available from the corresponding author on reasonable request.