Abstract
The multiprotein human SWI-SNF (hSWI-SNF) complex is a chromatin-remodeling machine that facilitates transcription by overcoming chromatin-mediated gene repression. We had previously shown that hSNF5/INI1, an intrinsic, consistent component of the hSWI/SNF complex, is associated with Epstein-Barr nuclear antigen 2 (EBNA2) and have proposed that EBNA2 directs this complex to key EBNA2-responsive viral and cellular genes. Using chromatin immunoprecipitation and quantitative PCR, we show that antibodies directed against components of the hSWI-SNF complex preferentially precipitate chromatin-associated DNA that contains a targeted EBNA2-responsive element in the context of both episomal and cellular chromatin. This enrichment does not occur in EBNA2-negative cells or when the EBNA2-responsive element is mutated. The stable association of the hSWI-SNF complex with the EBNA2-responsive promoter can also be disrupted by deletion of the TATA element, suggesting that EBNA2 in itself is insufficient to mediate stable targeting of the hSWI-SNF complex. These results demonstrate that recruitment of the hSWI-SNF complex to selected promoters can occur in vivo through its interaction with site-specific activator proteins and that stable targeting may require the presence of basal transcription factors.
Epstein-Barr virus (EBV) immortalizes B lymphocytes, with attendant expression of a small subset of viral genes including Epstein-Barr nuclear protein 2 (EBNA2), which is required for immortalization (14). EBNA2 contains an acidic transcriptional activation domain (8) and activates several viral and cellular genes that are involved in the immortalization process (9, 23, 51, 52, 66). The activation domain of EBNA2 binds to TFIIH, TATA binding protein (TBP)-associated factor TAF40, and TFIIB and to a protein that binds TFIIE (46, 47) and can interact both functionally and in vitro with transcription coactivators p300/CBP and P/CAF (4, 18, 53). EBNA2 is tethered to a subset of EBNA2-responsive promoters through its association with RBP-Jκ/CBF1 (13, 15, 49, 65). This interaction is mediated by a domain of EBNA2 that is conserved among various gammaherpesviruses (CR6) that is distinct from the transactivation domain (27, 37). Another factor, SKIP, binds to a distinct conserved region of EBNA2 (CR5) to direct transcription repressor complexes to sites of EBNA2 action (67). Through these interactions, EBNA2 serves the pivotal function of acting as an adapter molecule that can direct various multiprotein complexes to its sites of action to integrate both transcriptional activation and repression functions that are required to maintain the growth-transformed state. We had shown that a phosphorylated fraction of nuclear EBNA2 in lymphocytes is associated with an invariant member of the human SWI-SNF (hSWI-SNF) complex, hSNF5/INI1, the human homologue of yeast SNF5 protein (60). The association with hSNF5/INI1 is also mediated through a region of EBNA2 that is distinct from its transactivation domain.
The hSWI-SNF complex is one of a family of multiprotein complexes that are conserved in evolution and serve to remodel chromatin in a catalytic, energy-dependent fashion. Overcoming the barrier to transcription imposed by chromatin is a pivotal aspect of the control of gene expression. Derepression can occur by perturbing the interaction between nucleosomes and DNA through histone acetylation by the coactivators p300/CBP and P/CAF (5, 34), by activator binding, or by remodeling by multiprotein complexes (reviewed in reference 66). The chromatin-remodeling complexes of both yeast and higher eukaryotes share several features. Each includes a homologue of the yeast SNF2-SWI2 protein that contains DNA-dependent ATPase and a helicase motif (19, 30, 54) and encodes the core catalytic activity of the complex (38) and is invariably associated with three other proteins in all of the SWI-SNF homologous complexes studied to date: SNF5 or its human homologue hSNF5/INI1 (29), SWP73 or its human homologue BAF60, and SWI3 or its homologues BAF170 and BAF155 (54).
The SWI-SNF complex in yeast is required for the induced expression of a small number of genes but is dispensable for growth under normal condition (57). The relatively low abundance (100 to 2,000 copies per cell) of SWI-SNF complexes suggests that some form of targeting is required to bring the complex to specific genes or families of genes. It has been proposed that the SWI-SNF complex is recruited to promoters as a part of the RNA polymerase II (Pol II) holoenzyme (7, 56), although some biochemical data and in vitro analyses of SWI-SNF complexes have been at variance with this model (31, 64). The complex might also be targeted through its association with site-specific DNA binding proteins. The latter hypothesis is supported by the requirement of SWI-SNF proteins for glucocorticoid receptor function in yeast cells (30, 63), by its activation of several nuclear receptors in mammalian cells (6, 55), and by recruitment of yeast SWI-SNF in vitro by a peptide in the glucocorticoid receptor (48). Targeting of SWI-SNF has recently been substantiated by several reports demonstrating that components of the yeast and human SWI-SNF complexes can be targeted by their association with the activation domains of general (32, 33, 64) or gene-specific (1, 10, 20, 25, 36) transcription factors. In all examples to date, targeting is mediated through SWI-SNF interaction with the activation domain of the associated transcription factor.
To determine whether the hSWI-SNF complex is targeted to EBNA2-responsive promoters through its association with EBNA2, we have generated multicopy episomal chromatin templates that contain the EBNA2-responsive element of the EBV terminal protein 1 (TP1/LMP2A) gene. EBNA2 activates TP1/LMP2A through a promoter that contains two RBP-Jκ/CBF1 binding sites (GTGGGAA) (24, 65). Using quantitative PCR, we show that antibodies directed against hSWI-SNF components preferentially immunoprecipitate chromatin-associated DNA that is at least 5- to 10-fold enriched for the targeted EBNA2-responsive sequence. We also show that the hSWI-SNF complex is targeted to cellular chromatin of the CD23 gene, which is induced by EBNA2. The region of EBNA2 that mediates this interaction is distinct from the activation domain. These data show that EBNA2 acts as an adapter protein that can target the hSWI-SNF complex to a specific region in chromatin. Targeting of hSWI-SNF by EBNA2 is consistent with its playing a significant role in regulating gene expression from viral or cellular chromatin.
MATERIALS AND METHODS
Cloning of plasmids and generation of Raji cell sublines.
The reporter gene TpTATAMG was generated within plasmid pSp1TATAMG by removing the 12 tandem Sp1 binding sites with HpaI and BglII, blunt ending, and ligating with the double-stranded 54-bp oligodeoxyribonucleotide derived from TP1/LMP2A promoter with the sequence CTCGCGACTCGTGGGAAAATGGGCGGAAGGGCACCGTGGGAAAATAGTTCCAGG. The TpTATAMG insert was then removed from this vector by digestion with NruI and NdeI, blunt ended, and ligated into the HindIII site of pHeBO, creating plasmid pTpTATAMG. Plasmid pTpmutTATAMG, containing mutated CBF1/RBP-Jκ binding sites, was made in the same manner using the oligonucleotide CTCGCGACTCGTTTTAAAATGGGCGGAAGGGCACCGTTTTAAAATAGTTCCAGG. Plasmid pTATAMG lacks the TP1/LMP2A sequence and was generated without the oligonucleotide insert. Plasmid pTpMG lacks the TATA element and was made by digesting pSp1TATAMG with HpaI and EcoRI to remove the Sp1 sites and the TATA element and ligating with the TP1/LMP2A oligonucleotide. All plasmids were transferred into Raji cells by electroporation and then selected and maintained in hygromycin B. The episome copy number in each cell line was determined by Southern blot analysis.
Generation and purification of chromatin fragments.
The procedures to generate purified chromatin fragments were modified from the method of Grandori et al. (12). Episome-containing Raji cells (1.5 × 108 to 2 × 108 cells) washed with phosphate-buffered saline (PBS) were lysed with 10 ml of reticulocyte standard buffer (RSB) (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2) with 0.25% NP-40, and nuclei were collected by centrifugation at 300 × g and 4°C for 5 min. The nuclear pellet was resuspended in 5 ml of RSB–10% glycerol, adjusted to 10 mM MgCl2 and CaCl2, and digested with micrococcal nuclease (50 to 100 U/107 nuclei) for 10 min at 30°C. Reactions were terminated with an equal volume of ice-cold TEE (20 mM Tris [pH 7.5], 60 mM EDTA, 30 mM EGTA) and centrifuged at 450 × g for 5 min. The nuclear pellet was then lysed with TEP (12 mM Tris [pH 7.7], 3 mM sodium-free EDTA) containing 1 μM leupeptin and 0.2 μM phenylmethylsulfonyl fluoride at 107 nuclei/5 ml for 5 min and homogenized in a Dounce homogenizer. Nuclear debris was removed by centrifugation at 3,000 × g for 15 min, and the supernatant was adjusted to final concentrations of 0.1 mg of bovine serum albumin per ml, 20 mM Tris (pH 7.5), 125 mM KCl, 5% glycerol, and 0.1% NP-40. The suspension was then concentrated to 1 ml in a Millipore Ultrafree filtration concentrator, and the optical density at 260 nm was determined. Equal amounts of DNA were layered onto 10-ml 5 to 30% sucrose gradients (20 mM Tris [pH 7.5], 125 mM KCl, 1 mM EDTA) and centrifuged at 25,000 × g and 4°C for 16 to 20 h. Fractions of 0.5 ml, each containing 2 to 10 μg of DNA, were collected for immunoprecipitation.
Immunoprecipitation of chromatin fragments and PCR analysis.
Sucrose gradient-purified chromatin preparations containing equal amounts (±10%) of DNA were incubated with antibodies directed against hSNF5/INI1 (Rb2464), BRG1, TFIID (SI-1; Santa Cruz Biotechnology), or Pol II (8WG16; Research Diagnostics, Inc.) or with control antibodies (preimmunized rabbit serum or anti-FLAG antibody M2) at 1:25 dilution for 2 h at 4°C. The immune complexes were precipitated with protein A-Sepharose, washed three times with wash buffer (20 mM Tris [pH 7.5], 125 mM KCl, 1 mM EDTA, 0.1% NP-40), and digested with proteinase K in sodium dodecyl sulfate (SDS) stop buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 5 mM EDTA, 0.5% SDS) overnight at 37°C. DNA was isolated by phenol-chloroform extraction and ethanol precipitation and then resuspended in Tris-EDTA.
Quantitative PCR analysis.
Quantitative PCR was performed with appropriate primer pairs on 2 to 5 μl of the DNA sample and appropriate plasmid controls (10−14 to 10−12 M). PCRs were carried out for 26 to 29 cycles (for episomal analysis) or 30 to 34 cycles (for genomic analysis) in the presence of 0.5 μM primer, 100 μM deoxynucleoside triphosphate (with or without [α-32P] dATP [1 μCi/20 nmol]), 1.25 mM MgCl2, and either 0.5 U of Taq (Perkin-Elmer) or 1.0 U of Platinum-Taq (Gibco-BRL) polymerase in a final volume of 100 μl. Each cycle consisted of 1 min at 65°C, 2 min at 72°C, and 1 min at 92°C. PCR primers sequences are listed in Table 1. To determine the abundance of the Myc-globin sequence, we used primers 5′MycGb1 and 3′MycGb1; for the β-lactamase gene, we used primers 5′ AmpR and 3′ AmpR. The three PCR primer sets used to analyze the promoter region of the CD23 gene, CD23A, CD23B, and CD23C, are shown in Table 1 and Fig. 5. Quantitation of the PCR products was performed by radionucleotide incorporation or by Southern blot analysis with radiolabeled DNA fragments derived from pTpTATAMG, pTpMG, pTATAMG, pH5, or pFE1. pH5 is a plasmid containing a 5-kb HindIII fragment of the CD23 (−1.3 kb through exon 6), and pFE1 contains a 3.5-kb SalI-HindIII fragment (−4.8 to −1.3) upstream from CD23. Radioactivity signals were quantitated by PhosphorImager analysis.
TABLE 1.
Sequences of PCR primers used in targeting studies
| PCR primer set | Sequence | Size (bp) of PCR product |
|---|---|---|
| 3′MycGb1/5′MycGb1 | 5′-GTGCCTGACTGCGTTAGCAA/5′-CCTATTCGCTCCGGATCTCC | 267 |
| 5′AmpR/3′AmpR | 5′-GCTGCGCCTTATCCGGTAAC/5′-TCTGCGCGTAATCTGCTGCT | 312 |
| 5′CD23A/3′CD23A | 5′-GCCACTATGCCACCTAGACTTC/5′-CAGCTGTTCTGGAGTGCAGG | 310 |
| 5′CD23B/3′CD23B | 5′-CAGTGTGCAGTAACAGTGGTTC/5′-GAAAGTAGAGCCACTGACAGC | 241 |
| 5′CD23C/3′CD23C | 5′-GCAGTGTGGACAGAATCTCGAG/5′-CAACTCCAGGCCGTCCTTCTAA | 177 |
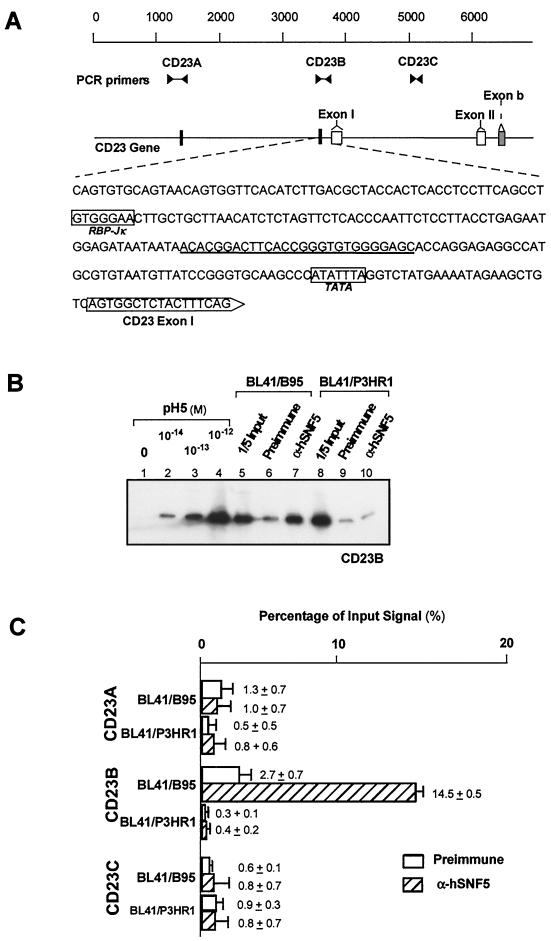
FIG. 5.
Targeting of the hSWI-SNF complex to the CD23 gene. (A) Schematic of the upstream region of the CD23 gene containing exons I, II, and b. The two RBP-Jκ/CBF1 binding sites present in this region are indicated by solid rectangles. The upstream sequence from exon I is shown with the RPB-Jκ/CBF1 and TATA elements. Positions of the PCR primers (sequences shown in Table 1) and their product lengths are also shown. (B) Targeting of the hSWI-SNF complex to three regions on the CD23 gene in BL41/B95 and BL41/P3HR1 cells were measured by immunoprecipitation and quantitative PCR with specific primer sets. The autoradiogram of quantitative PCR using the CD23B primer set is depicted. Lanes 1 to 4, PCR of template controls; lanes 5 and 8, PCR results from the preimmunoprecipitated input (1/5 of total) chromatin; lanes 6, 7, 9, and 10, PCR results from the immunoprecipitated samples. (C) Results of PhosphorImager analysis of PCR performed with CD23A, CD23B, and CD23C primer sets on chromatin immunoprecipitation samples. Results are reported as the percentage of signal obtained from chromatin prior to immunoprecipitation.
Metabolic labeling of nuclear proteins.
Raji cells (108) harboring pTpTATAMG were washed in PBS plus methionine-cysteine-free RPMI 1640 with 20% dialyzed fetal calf serum and incubated for 2 h in the same medium. The cells were transferred to 5 ml of fresh medium containing 1 mCi of Tran35S-label (1,175 Ci/mmol; ICN) and grown overnight. Nuclear isolation, micrococcal nuclease digestion, and sucrose gradient purification were carried out as described above. Then 300 μl of each fraction was immunoprecipitated with 3 μl of Rb2464–protein A-Sepharose, washed, and eluted with 20 μl of 2× SDS sample buffer. The labeled proteins were separated by polyacrylamide gel electrophoresis (PAGE) on an SDS–10% polyacrylamide gel and visualized by autoradiography.
Micrococcal nuclease analysis.
Nuclei were prepared as described above, resuspended at 2 × 107 nuclei/ml in RSB–10% glycerol–10 mM CaCl2–10 mM MgCl2, and digested with increasing amounts of micrococcal nuclease (0 to 180 U/5 × 106 nuclei) for 5 min at 37°C. The reactions were terminated with SDS stop buffer and digested with proteinase K overnight. DNA was phenol-chloroform extracted and ethanol precipitated; 20 μg of each sample was digested with NotI and NsiI, Southern blotted, and probed with a 237-bp NsiI/BamHI fragment probe of pTpTATAMG.
S1 nuclease assay.
Transcription from the Myc-globin reporter gene was determined by measuring the steady-state mRNA by S1 assay (21, 22). The S1 nuclease assay was performed as reported previously with a 162-nucleotide single-stranded DNA probe corresponding to positions −36 to +126 of the Myc-globin transcription start site. Myc-globin mRNA transcribed from the proper initiation site results in the protection of a 126-nucleotide 5′-32P-labeled probe fragment. The samples from the S1 nuclease assay were subjected to 6% neutral polyacrylamide gel electrophoresis. The results of the S1 nuclease assays were then normalized with respect to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA present in each sample and the relative copy number of the episomal reporter construct present in the Raji cell sublines.
RESULTS
Incorporation of reporter episomes in Raji cells.
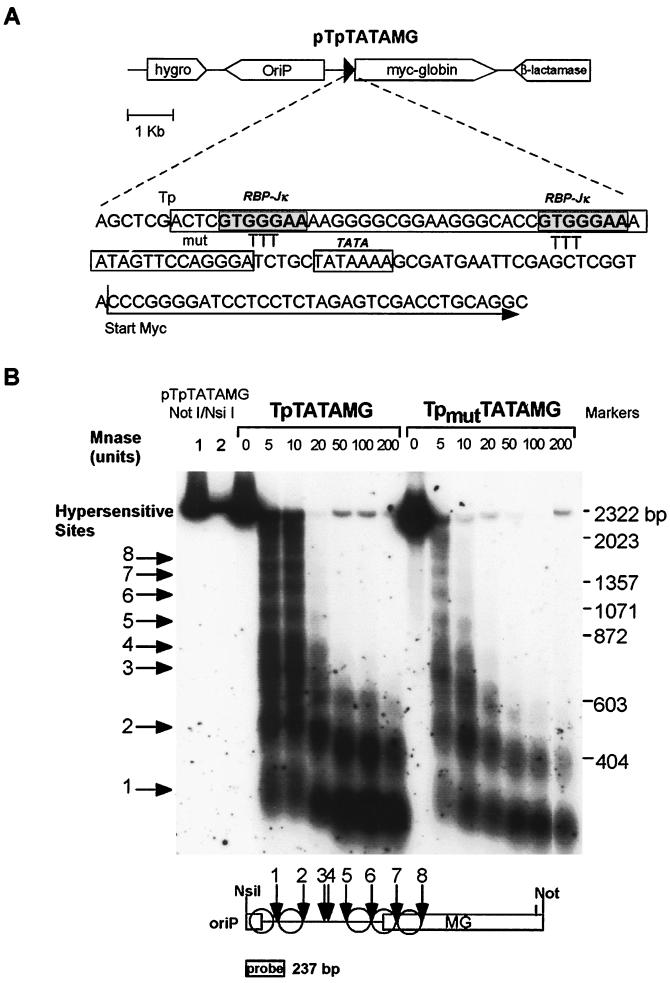
To study targeting of the hSWI-SNF complex by EBNA2 to an EBNA2-responsive element in vivo, we used an artificial reporter gene consisting of multicopy episomal chromatin templates that contain sequences derived from a well-characterized EBNA2-responsive element (24, 66). The episomal templates contain EBV Ori-P (44) and are maintained in Raji Burkitt lymphoma cells at 50 to 200 copies per cell (data not shown). The reporter gene, TpTATAMG, contains a 54-bp sequence from the EBV TP1/LMP2A promoter (24) placed directly upstream from the adenovirus major late promoter TATA element and a Myc-globin fusion gene (Fig. 1A) (21). The promoter contains two RBP-Jκ/CBF1 binding sites (GTGGGAA), which are mutated by triplet substitutions (GGG→TTT) in the pTpmutTATAMG construct. Micrococcal nuclease digestion and indirect end-labeling analysis (59) of Raji cells harboring these constructs showed that the episomes are assembled into ordered arrays of positioned nucleosomes (Fig. 1B). Mutations at the RBP-Jκ/CBF1 binding sites did not result in detectable alterations in the micrococcal nuclease sensitivity patterns. The inferred positions of nucleosomes in the upstream region of the reporter gene are depicted in Fig. 1B.
FIG. 1.
Schematic representation of pTpTATAMG and micrococcal nuclease digestion of pTpTATAMG and pTpmutTATAMG. (A) Main features of plasmid pTpTATAMG. The sequence of the EBV TP1/LMP2A promoter, TATA box, upstream Myc-globin reporter gene, and heptameric RBP-Jκ/CBF1 binding sites within the TP1/LMP2A promoter are indicated. Substitutions in plasmid pTpmutTATAMG (mut) replace the GGG sequence within the RPB-Jκ site with TTT. (B) Micrococcal nuclease (Mnase) digestion and indirect end labeling of the upstream region of the Myc-globin reporter gene. The probe for hybridization was derived from the 237-bp NsiI/BamHI fragment of pTpTATAMG (position indicated). The eight micrococcal nuclease-hypersensitive sites identified are numbered 1 to 8. Proposed positions of the nucleosomes (circles) are depicted in the lower panel. Marker lanes (1 and 2) contain 10 and 2 pg of pTpTATAMG digested with NotI and NsiI.
Targeting analysis of the hSWI-SNF proteins.
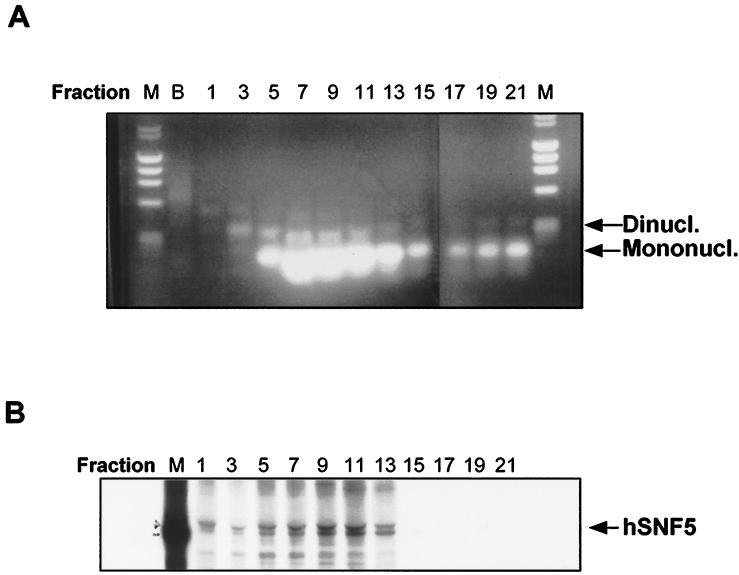
Chromatin fragments from Raji cells were prepared by digestion with micrococcal nuclease, lysis with hypotonic buffer, and separation of chromatin fragments on a sucrose gradient. This procedure generated a predominance of mono-, di-, and trinucleosomes (Fig. 2A). To ensure that hSNF5/INI1 remained associated with the oligonucleosomes through the purification, we metabolically labeled the Raji cells with [35S]methionine-cysteine prior to these procedures. The fractions (7 to 11) containing hSNF5/INI1 (47- to 49-kDa doublet) were identified by immunoprecipitation with an anti-hSNF5/INI1 antibody (Rb2464) as previously described (60) and corresponded to the highest concentration of oligonucleosomes (Fig. 2B).
FIG. 2.
Purification of chromatin fragments. (A) Aliquots of DNA extracted from chromatin fractions derived from sucrose gradient purification were separated on a 1.5% agarose gel and stained with ethidium bromide. Migration of mono- and dinucleosomes is indicated at the right. Lane B designates the bottom gradient fraction, which includes chromatin fragment sediments; lanes M contain size markers. (B) Immunoprecipitation of sucrose gradient fractions with anti-hSNF5/INI1 antibodies. 35S-labeled chromatin fragments were prepared and purified on the sucrose gradient as described for panel A. Each sucrose gradient fraction was then immunoprecipitated with Rb2464 (anti-hSNF5 antibody), separated by SDS-PAGE, and visualized by autoradiography.
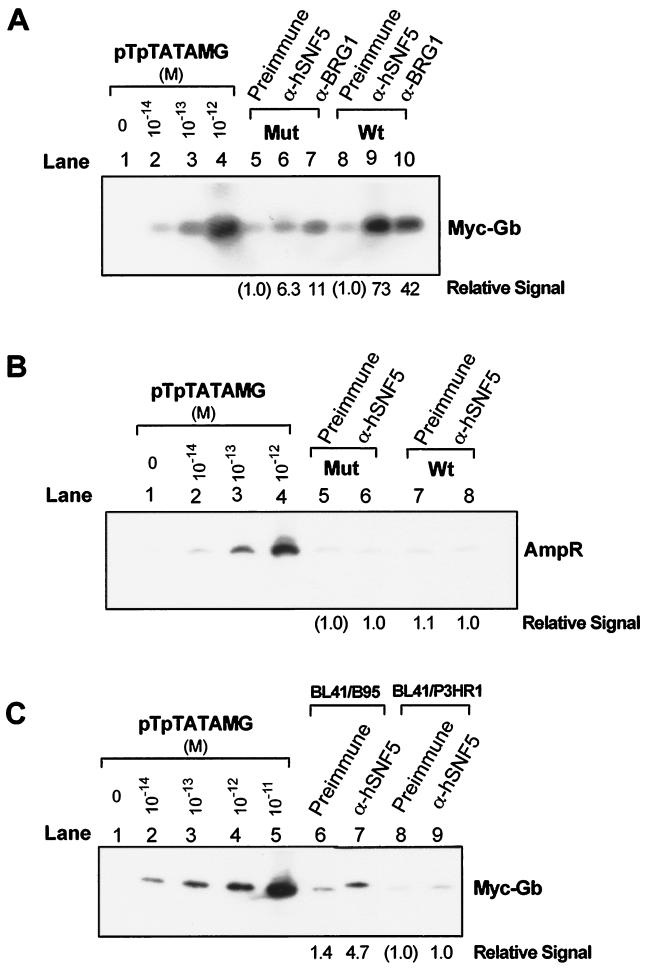
To determine if the hSWI-SNF complex is preferentially associated with EBNA2-responsive promoter sequences, we carried out chromatin immunoprecipitation with antibodies directed against two components of the hSWI-SNF complex, hSNF5/INI1 and BRG1, on the pooled sucrose gradient fractions 8 to 13. Quantitative PCR was then performed on DNA purified from the immune complexes and analyzed on Southern blots with sequence-specific probes. Figure 3A shows a blot of the PCR products obtained with these primers on the naked pTpTATAMG plasmid (lanes 1 to 4) and immunoprecipitated sample DNA (lanes 5 to 10) derived from Raji cells harboring either pTpmutTATAMG or pTpTATAMG. The PCR assay is quantitative over 2 orders of magnitude (10−14 to 10−12 M), and the control lanes were used to generate a standard curve for PhosphorImager analysis of the relative signals in lanes 5 to 10. In Raji cells harboring the unaltered pTpTATAMG episomes, anti-hSNF5/INI1 antibody preferentially enriched the Tp1/LMP2A promoter sequence (10- to 70-fold in four independent experiments) compared to the preimmune serum (compare lanes 8 and 9 in Fig. 3A). Anti-BRG1 antibody also enriched the Tp1/LMP2A sequence (5- to 40-fold in four independent experiments) relative to the preimmune serum (compare lanes 10 and 8). When the EBNA2-responsive element within the Tp1/LMP2A sequence was mutated, the antibody-specific enrichment of Tp1/LMP2A sequence was reduced (lanes 5 to 7). The small but persistent enrichment of the promoter target sequence by both anti-hSNF5/INI1 and anti-BRG1 antibodies in the mutated RBP-Jκ/CBF1 construct suggests that some association of hSWI-SNF complex with this DNA region may occur independently of EBNA2 and RBP-Jκ/CBF1. As an additional control, we determined whether the β-lactamase gene sequences that lie 3 kb from the Tp1/LMP2A sequence on the episome were enriched in immunoprecipitates. We found no enrichment with anti-hSNF5/INI1 antibody (Fig. 3B). In these experiments, equal amounts of DNA were subjected to sucrose gradient purification prior to immunoprecipitation. The episome copy number was found to be 1.5-fold higher in Raji/pTpTATAMG cells than in Raji/pTpmutTATAMG cells (data not shown). This disparity does not account for the observed differences obtained from the PCR analysis.
FIG. 3.
Quantitative PCR of DNA from immunoprecipitated chromatin fractions. Sucrose gradient-purified chromatin fragments pooled from fractions 8 to 13 were immunoprecipitated with preimmune serum (Preimmune), anti-hSNF5 antibody (α-hSNF5), or anti-BRG1 antibody (α-BRG1). DNA isolated from immunoprecipitates was subjected to quantitative PCR to determine relative abundance of the TP1/LMP2A promoter and β-lactamase sequences. Control PCR series were performed with plasmid pTpTATAMG (10−14 to 10−11 M) as the template. The results were analyzed on a PhosphorImager and are reported as the relative amount of PCR product compared with preimmune control (indicated below of each lane). (A) Autoradiogram of quantitative PCR of Myc-globin sequences from Raji/pTpTATAMG (Wt [wild type]; lanes 8 to 10) and Raji/pTpmutTATAMG (Mut [mutant]; lanes 5 to 7) chromatin fragments immunopurified with preimmune or hSNF5- or BRG1-specific antibodies. (B) Autoradiogram of PCR of β-lactamase sequence from immunopurified Raji/pTpTATAMG (Wt; lanes 7 and 8) and Raji/ pTpmutTATAMG (Mut; lanes 5 and 6) chromatin fragments using the AmpR primer set. (C) Autoradiogram of PCR of Myc-globin sequence from chromatin immunoprecipitation of BL41/B95 (EBNA2-positive) or BL41/P3HR1 (EBNA2-negative) cells that harbor the episomal pTpTATAMG.
To determine whether the association of hSNF5/INI1 with the TP1/LMP2 promoter is EBNA2 dependent, we performed the same analysis on the Burkitt lymphoma (BL41) sublines that had been converted with either the EBNA2-negative (P3HR1) or EBNA2-positive (B95) EBV strain (Fig. 3C). These cells support lower episome copy numbers (5 to 20 copies per cell) than Raji cells (50 to 200 copies per cell). Quantitative PCR performed on chromatin immunoprecipitation samples derived from the BL41 cells shows that anti-hSNF5/INI1 antibody (Fig. 3C, lane 7) enriched the Tp1/LMP2A target sequence fivefold relative to the preimmune serum (lane 6) in the EBNA2-positive BL41/B95 cells but not in the EBNA2-negative BL41/P3HR1 cells (lanes 8 and 9). We consistently found a more pronounced targeting effect in Raji cells than in BL41/B95 cells. We presume that this disparity results from the differences in episome copy number between Raji and BL41 cells. The experiment shown in Fig. 3 demonstrates that the hSNF5/INI1 and BRG1 proteins are preferentially associated with the promoter region of the pTpTATAMG reporter gene. This association can be disrupted by mutations introduced into the RBP-Jκ/CBF1 elements within the TP1/LMP2A promoter and is dependent on the expression of EBNA2.
The TATA element is necessary for hSWI-SNF targeting.
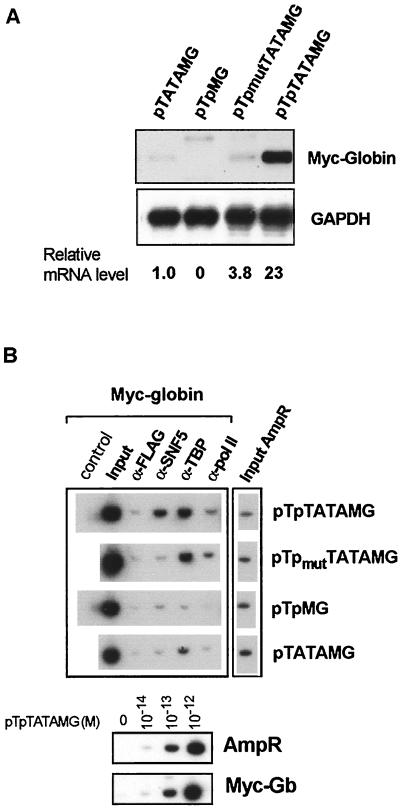
The yeast SWI-SNF complex has been shown to alter chromatin structure at the TATA element in the Saccharomyces cerevisiae SUC2 promoter (61). Furthermore, both RBP-Jκ/CBF1 and EBNA2 can associate with TFIID (35, 47). Because of these observations, we determined whether binding of TBP at the TATA element is required for stable targeting of the hSWI-SNF complex. To test this hypothesis, we constructed episomes lacking either the TP1/LMP2A promoter (pTATAMG) or the TATA sequence (pTpMG) and compared these to the unaltered pTpTATAMG episome with respect to both transcriptional activation of the Myc-globin reporter gene and the presence of hSNF5/INI1, TBP, and RNA Pol II at the promoter region. The steady-state levels of properly initiated mRNA from the Myc-globin reporter gene of these constructs were measured by a quantitative S1 nuclease protection assay (21). Myc-globin mRNA from the normal transcription initiation site of pTpTATAMG results in the protection of a 126-nucleotide fragment (Fig. 4A). The S1 nuclease assays were normalized for the relative copy number of each reporter episome. The relative steady-state levels of properly initiated Myc-globin mRNA adjusted for the episome copy number are depicted below the autoradiogram. The results in Fig. 4A demonstrate that TP1/LMP2A is a functional promoter in the pTpTATAMG reporter episome and that low levels of reporter gene expression persist even from a promoterless episome, pTATAMG.
FIG. 4.
Analysis of the contribution of TP1/LMP2A promoter and TATA elements to reporter expression and targeting of the hSWI-SNF complex. (A) The steady-state levels of Myc-globin and GAPDH mRNA of Raji/pTpTATAMG, Raji/pTpmutTATAMG, Raji/pTATAMG, and Raji/pTpMG cells were determined by the S1 nuclease assay. The results were normalized for the relative copy number of reporter episomes present in these cells (4.8 for pTATAMG, 4.0 for pTpMG, 0.5 for pTpmutTATAMG, and 1.0 for pTpTATAMG) and reported as relative mRNA levels with respect to the mRNA in the promoterless Raji/pTATAMG cells. (B) Targeting of hSNF5/INI1, TBP, and Pol II was determined by immunoprecipitation and PCR incorporation of [α-32P]dATP. Input DNA prior to immunoprecipitation was quantitated by PCR for both Myc-globin and β-lactamase (AmpR) sequences, showing nearly equal input DNA in cells harboring the four episomal constructs. Quantitative incorporation of [α-32P]dATP is linear over 2 orders of magnitude of template concentrations for both AmpR and MycGb primer sets (lower panel).
In an effort to correlate components of the transcription complex with gene expression, we performed chromatin immunoprecipitation assays with antibodies directed against hSNF5/INI1, TBP, and RNA Pol II. In this experiment, quantitation was done by radionucleotide incorporation of PCR products. The lower panel of Fig. 4B shows that this assay is quantitative over 2 orders of magnitude using a plasmid template.
As shown in Fig. 4B, mutation (pTpmutTATAMG) or deletion (pTATAMG) of the Tp promoter sequence does not influence the binding of TBP to the promoter region but does disrupt the targeting of the hSNF5/INI1 protein to this region. By contrast, when the TATA element is removed from the promoter (pTpMG), neither TBP nor hSNF5/INI1 is found to be associated with the promoter DNA. These observations suggest that binding of TBP may stabilize the association between hSWI-SNF and the target site in the promoter, possibly through an indirect association with the basal transcription complex. RNA Pol II was not preferentially bound in the promoter region, and it is not required for the targeting of the hSWI-SNF complex. Since transcription persists from the TP1/LMP2A promoterless construct (pTATAMG) (Fig. 4A), this result implies that the hSWI-SNF complex is not required for basal transcription of the Myc-globin reporter gene. Taken together, these results demonstrate that EBNA2 is necessary but not sufficient for targeting of hSWI-SNF complexes to the EBNA2-responsive promoter.
Targeting of the hSWI-SNF complex to the cellular CD23 promoter.
CD23 is an EBNA2-responsive cellular gene (9) whose product can act as an autocrine growth factor for EBV-immortalized B lymphocytes (45). It contains an RBP-Jκ/CBF1 site 171 bp upstream from the CD23 transcription start site (Fig. 5A). Another RBP-Jκ/CBF1 site lies in the opposite orientation at position −2471. To test whether the hSWI-SNF complex is targeted to these sequences by EBNA2, we performed the same targeting analysis directed at these two sites and at a third sequence in intron I that does not contain the RBP-Jκ/CBF1 site. We performed this analysis in both EBNA2-positive (BL41/B95) and EBNA2-negative (BL41/P3HR1) cells. CD23 mRNA expression was found to be 50-fold greater in BL41/B95 cells than in BL41/P3HR1 cells (data not shown). Three sets of PCR primers, CD23A, CD23B, and CD23C (Table 1), were used for these analyses; their positions are depicted in Fig. 5A. The chromatin immunoprecipitation analysis shows that anti-hSNF5/INI1 antibody specifically enriched the exon I (CD23B locus) sequence fivefold compared to the preimmune serum in BL41/B95 but not in BL41/P3HR1 cells (Fig. 5B and C). PCR analyses using the CD23A and CD23C primers show that neither the upstream RBP-Jκ/CBF1 site nor the intron I sequence was enriched by anti-hSNF5/INI1 antibody regardless of EBNA2 expression (Fig. 5C). The quantitative results from triplicate immunoprecipitations (Fig. 5C) show that the hSWI-SNF complex is targeted to a specific region of the CD23 gene in an EBNA2-dependent manner. Because there was no targeting to the RBP-Jκ/CBF1 sequence residing 2,471 bp upstream from the exon I start site (Fig. 5C), these results further demonstrate that other cis-acting elements are required for stable hSWI-SNF complex targeting.
DISCUSSION
The hSWI-SNF complex is targeted to EBNA2-responsive promoters through EBNA2 and RBP-Jκ/CBF1.
The low abundance and catalytic rate of remodeling of nucleosomal arrays by hSWI-SNF suggest that some mechanism exists for directing the complex to its targets. Because continuous action of the SWI-SNF is required to maintain transcription of SWI-SNF-inducible genes, the complex must be retained beyond the point of assembly of the initiation complex (3, 43). Here we show that the hSWI-SNF complex is targeted to the EBNA2-responsive sequences of both reporter episomes and the lymphocyte CD23 gene in vivo by demonstrating that antibodies directed against components of the hSWI-SNF complex selectively precipitate chromatin fragments containing RBP-Jκ/CBF1 binding sites of EBNA2-responsive promoters. This preferential association of the hSWI-SNF complex with these DNA sequences was not observed in an EBNA2-negative cell line and was diminished by mutations at the RBP-Jκ/CBF1 binding sites that disrupt binding of this factor. These results extend our previous observation that a phosphorylated fraction of nuclear EBNA2 is associated the hSNF5/INI1 subunit of the hSWI-SNF complex in lymphocytes (60) and suggest the participation of the hSWI-SNF complex in the transcriptional activation function of EBNA2. While this work was being completed, several reports appeared demonstrating targeting of SWI-SNF complexes by binding to acidic activation domains through binding to the SWI2-SNF2 or SNF5 homologues of either yeast or mammalian SWI-SNF complexes (20, 33, 48).
The role of hSWI-SNF complex in EBNA2-mediated transactivation.
The control of transcription by local chromatin structure has been shown to be an essential feature of many eukaryotic genes. In this respect, the EBV transcriptional program in latently infected B cells must proceed in the same chromatin-repressed promoter environment. If recruitment of the hSWI-SNF complex by EBNA2 to responsive promoter chromatin is one strategy that EBV has adopted to overcome chromatin-mediated transcriptional repression, this would explain the marked effects of EBNA2 expression on cellular and viral gene expression compared with its modest effects on transiently transfected reporter constructs. Also, the acidic activation domain of EBNA2 interacts with p300/CBP and P/CAF histone acetyltransferases (HATs) to enhance the transactivation of the viral LMP1 promoter (53). It is unclear if hSWI-SNF and HAT proteins are selectively used by EBNA2 at a subset of EBNA2-responsive genes or if they are functionally overlapping. Thus, EBNA2 emerges as a pivotal mediator of coordinated assembly of factors in a combinatorial fashion acting at its target promoters. Through its interaction with RBP-Jκ/CBF1, it mediates derepression (16). Interactions between the activation domain of EBNA2 and basal transcription factors act to assemble the transcription initiation complex (46, 47). The acidic activation domain also interacts with accessory factors such as p300/CBP and P/CAF to transduce HAT activity (53). The present work shows that a distinct region of EBNA2 recruits a chromatin-remodeling complex to EBNA2 targets. EBNA2 also appears to compete with the interaction between histone deacetylase and the SKIP protein that is tethered to RBP-Jκ/CBF1 (67). Presumably additional promoter-specific factors determine which of these EBNA2-mediated functions predominates at distinct EBNA2-regulated promoters.
Stable targeting of the hSWI-SNF complex by EBNA2 requires TBP.
Deletion of the TATA element from the episomal promoter abolishes both transcription and targeting of hSNF5/INI1 (Fig. 4). This finding suggests that binding of TBP to the TATA element is a prerequisite both for transcription and for stabilization of hSNF5/INI1 binding to chromatin in this region. Although EBNA2 binds weakly to TBP, it interacts with TAF40 through its acidic activation domain (47). RBP-Jκ/CBF1 has also been reported to bind the TAFII110 component of TFIID complex (35). In addition, partially purified hSWI-SNF has been shown to facilitate in vitro binding of TBP to the TATA element in nucleosomes (17) and yeast SWI-SNF complex alters the chromatin structure over both the enhancer and the TATA elements of the SUC2 gene (61). These reports and our results suggest that TBP may be associated with both EBNA2 and hSWI-SNF in a complex over the EBNA2-responsive promoters. The presence of both hSNF5/INI1 and TBP at the transcriptionally active episomal promoter also suggests that both the hSWI-SNF complex and TBP are retained beyond the point of transcription initiation.
Influence of the hSWI-SNF complex on transcription of the reporter gene in artificial episomes.
Despite the apparent targeting of the hSWI-SNF complex to the promoter region in our reporter episome, we did not find dramatic enhancement in the steady-state level of transcription of the linked reporter gene that was similar to that seen for EBNA2-responsive genes in vivo (Fig. 4). For instance, the steady-state transcription of the CD23 gene in EBNA-positive BL41/B95 cells is at least 50-fold greater than the level in EBNA2-negative BL41/P3HR1 cells (data not shown). The relatively meager transcriptional enhancement by the EBNA2-responsive TP1/LMP2A promoter in episomes could be due to a basal chromatin structure that is unusually permissive to transcription and is relatively independent of hSWI-SNF complex remodeling. The permissive nature of the episomal chromatin is supported by three observations. First, restriction endonuclease digestion of nuclei shows that these sites in plasmids are far more sensitive to digestion than the level of nuclease sensitivity of chromosomal sites or reconstituted nucleosomal sites (data not shown). We also could not detect significant differences in the restriction endonuclease sensitivity of the TP1/LMP2 region of the episomes that distinguish normal or mutated RBP-Jκ/CBF1 sites (data not shown). The episomes in these Raji cells may thus reside in an open chromatin conformation that is minimally influenced by hSWI-SNF remodeling. Second, we found that the promoterless pTATAMG episomes can direct a low level of reporter gene transcription. This construct contains a TATA element but no other enhancer or promoter element to drive transcription of the reporter gene. The finding of reporter transcription in this context suggests that the episomal chromatin may lack elements that normally repress transcription. Third, DNase I digestion of the episomes in Raji cells showed that they lack hypersensitive sites in the region of the reporter gene (data not shown). This suggests that despite being packaged into an ordered array of nucleosomes, the DNA in these nucleosomes is uniformly accessible to nuclease and is in this regard structurally distinct from chromosomal DNA.
Although previous studies showed minimal differences that distinguish the transcriptional regulation and structure of cellular chromatin from those properties of amplified episomal chromatin (41), two features of the cellular environment of our experimental system could result a relatively open chromatin conformation in the episomal constructs: deletion of the EBNA3C gene from Raji cells and the OriP enhancer residing in the episomes. EBNA3C is a viral protein that also binds indirectly to DNA through RBP-Jκ/CBF1 (2, 39) and acts as an expression silencer by opposing the activating effects of EBNA2 (26, 42, 50) in part through its ability to associate with histone deacetylase I (40). Binding of EBNA1 to OriP, the plasmid origin of replication, permits stable plasmid replication and segregation (62) but also activates an enhancer in OriP (11). Thus, the binding of EBNA1 to OriP may exert a widespread effect resulting in derepression of the plasmid chromatin. Although our work clearly demonstrates targeting, the functional consequences of high-level transcriptional activation and chromatin remodeling may be inapparent due to these effects. Nonetheless, these episomes do not reside in an unrestricted open conformation since prior studies using OriP episomes in Raji cells showed that addition of a potent enhancer could activate transcription, increase nuclease sensitivity, and cause histone hyperacetylation (28). As discussed above, targeting of the hSWI-SNF complex and HAT activity may be functionally redundant at this episomal promoter, and this could also result in inapparent chromatin structural alteration in this experimental system.
Role of the activation domain in recruitment of SWI-SNF complexes.
The region of EBNA2 that mediates its interaction with hSWI5/INI1 is distinct from its transcription activation domain (60). In this regard, EBNA2 acts as an adapter protein that may recruit the complex to sites of action similar to the recruitment of SWI-SNF by Swi5p in yeast cells (10). By contrast, several in vitro and in vivo studies of recruitment of SWI-SNF components showed strict dependence on the activation domain (20, 33, 48, 64). These results suggest that EBNA2 may activate transcription both by recruiting basal transcription factors and cofactors through the activation domain and by association with hSNF5/INI1 and recruitment of the hSWI-SNF complex. Analysis of targeting and expression in cells expressing EBNA2 mutants that separately disrupt the activation and hSNF5/INI1 binding domains will define the relative importance of these separate means of recruitment.
ACKNOWLEDGMENTS
We thank Gerald Crabtree for the anti-BRG1 antibody, Carla Grandori for helpful suggestions regarding the experimental procedures, and Sarah Gimmestad and William Tuttle for expert technical assistance.
This work was supported by grants from the Department of Veterans Affairs (W.H.S.) and Public Health Service grants (CA719829 [D.Y.W.], CA54337 [A.K.], and CA82459 [W.H.S.]) from the National Cancer Institute.
REFERENCES
- 1.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 2.Bain M, Watson R J, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70:2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggar S R, Crabtree G R. Continuous and widespread roles for Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibiting p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J I, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordier M, Calendar A, Billaud M, Zimber U, Rousselet G, Pavlish O, Banchereau J, Tursz T, Bornkamm G W, Lenoir G M. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces the expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 11.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 15.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RPB Jκ-repressed genes by a mechanism resembling Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 18.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 20.Kowenz-Leutz E, Leutz A. A C-EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 21.Krumm A, Hickey L B, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 22.Krumm A, Madisen L, Yang X J, Goodman R, Nakatani Y, Groudine M. Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc Natl Acad Sci USA. 1998;95:13501–13506. doi: 10.1073/pnas.95.23.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larcher C, Kempkes B, Kremmer E, Prodinger W M, Pawlita M, Bornkamm G W, Dierich M P. Expression of Epstein-Barr virus nuclear antigen-2 (EBNA2) induces CD21/CR2 on B and T cell lines and shedding of soluble CD21. Eur J Immunol. 1995;25:1713–1719. doi: 10.1002/eji.1830250634. [DOI] [PubMed] [Google Scholar]
- 24.Laux G, Dugrillon F, Eckert C, Adam B, Zimber-Strobl U, Bornkamm G W. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C H, Murphy M R, Lee J S, Chung J H. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc Natl Acad Sci USA. 1999;96:12311–12315. doi: 10.1073/pnas.96.22.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Roux A, Kerdiles B, Walls D, Dedieu J F, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 27.Ling P D, Hayward S D. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBP Jκ. J Virol. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madisen L, Krumm A, Hebbes T R, Groudine M. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natarajan K, Jackson B M, Zhou H, Winston F, Hinnebusch A G. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 33.Neely K E, Hassan A H, Wallberg A E, Steger D J, Cairns B R, Wright A P, Workman J L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 34.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 35.Olave I, Reinberg D, Vales L D. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc Natl Acad Sci USA. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng R, Gordadze A V, Fuentes-Penana E M, Wang F, Zong J, Hayward G S, Tan J, Ling P D. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J Virol. 2000;74:379–389. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 39.Radkov S A, Bain M, Farrell P J, West M, Rowe M, Allday M J. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radkov S A, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday M J. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richard-Foy H, Hager G L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saudarsanam P, Cao Y, Wu L, Laurent B C, Winston R. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with histone acetyltransferase GCN5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swendeman S, Thorley-Lawson D A. The activation antigen BLAST-2, when shed, is an autocrine growth BCGF for normal and transformed B-lymphoblasts. EMBO J. 1987;6:1637–1642. doi: 10.1002/j.1460-2075.1987.tb02412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallberg A E, Neely K E, Hassan A H, Gustafsson J A, Workman J L, Wright A P. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol Cell Biol. 2000;20:2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F, Kikutani H, Tsang S F, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F, Tsang S F, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 56.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 57.Winston F, Carlson M. Yeast SNF-SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 58.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 59.Wu C. The 5′ end of Drosophila heat shock genes in chromatin are hypersensitive to DNAse I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 60.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu L, Winston F. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4230–4234. doi: 10.1093/nar/25.21.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 63.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 64.Yudkovsky N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou S, Fujimuro M, Hsieh J J, Chen L, Hayward S D. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74:1939–1947. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]