Abstract
The functional significance of naturally occurring variants of human hepatitis B virus (HBV) remains largely unknown. Previously, we reported an immature secretion phenotype caused by a highly frequent mutation at amino acid 97 of the HBV core (capsid) protein (HBcAg). This phenotype is characterized by a nonselective and excessive secretion of virions containing an immature genome of single-stranded viral DNA. To extend our study of virion secretion to other naturally occurring variants, we have characterized mutations at HBcAg codons 5, 38, and 60 via site-directed mutagenesis. Although the phenotype of the mutation at codon 38 is nearly identical to that for the wild-type virus, our study reveals that a single mutation at codon 5 or 60 exhibits a new extracellular phenotype with significantly reduced virion secretion yet maintains normal intracellular viral DNA replication. A complementation study indicates that the mutant core protein alone is sufficient for the “low-secretion” phenotype. Furthermore, the low-secretion phenotype of the codon 5 mutant appears to be induced by the loss of a parental proline residue, rather than by the gain of a new amino acid. Our study underscores the core protein as another crucial determinant in virion secretion, in addition to the known envelope proteins. Our present results suggest that a very precise structure of both α-helical and nonhelical loop regions of the entire HBcAg molecule is important for virion secretion. The low-secretion variants may contribute to the phenomenon of gradually decreasing viremia in chronic carriers during the late phase of persistent infection.
Hepatitis B virus (HBV) is an important human infectious agent. Worldwide, at least 300 million individuals are chronic HBV carriers. Chronic infection with HBV leads to the development of cirrhosis and liver cancer (31, 32). Naturally occurring variants have been found frequently in chronic HBV carriers. The potential role of these variants in chronicity and pathogenesis remains unclear, since no universal assay is available to evaluate these diverse HBV variants. For example, naturally occurring variants of HBV core antigen (HBcAg) have been frequently reported. This 183-amino-acid protein has multiple functions, including interaction with the pregenomic RNA and polymerase during encapsidation, polymerization with itself to form the nucleocapsid, import of relaxed circular DNA to the nucleus, and targeting to the endoplasmic reticulum for envelope formation (3, 7, 16, 25, 26, 37). In addition, HBcAg is structurally related to the secreted e antigen (27), and thus certain mutations in HBcAg should also be present in the e antigen. Because of the highly versatile nature of the HBV core protein, it is difficult to predict which functional assays should be used for the study of HBcAg variants in order to yield more informative results. Therefore, the biological significance of these mutations remains unclear.
Previously, we described several missense mutations in HBcAg which coincide with major histocompatibility complex class II-restricted T-cell epitopes (18). One of these mutations occurs at HBcAg position 38, changing a tyrosine (Y) to a histidine (H). The tyrosine residue at position 38 is highly evolutionarily conserved, present even in woodchuck and ground squirrel hepatitis B viruses. Another mutation occurs at HBcAg position 60, changing a hydrophobic leucine (L) to a hydrophobic valine (V). It is interesting that this L60V mutation is adjacent to a cysteine residue at HBcAg position 61 which is involved in the intermolecular disulfide bonding during the dimerization of HBcAg (24, 45). A close examination of the existing literature revealed that mutations Y38H and L60V often occur together on the same HBV sequence (1, 14, 15, 18, 28, 29, 36). Finally, a very frequent mutation of HBcAg occurs at codon 5, changing a conserved proline (P) to a threonine (T) (1, 2, 6, 11, 13, 14, 18, 22, 33, 38). While the most common mutation found at HBcAg codon 5 is from P to T, other less frequent changes have also been reported, such as proline to serine (S).
Recently, we reported an “immature secretion” phenotype of HBV virions caused by a highly frequent mutation at HBcAg codon 97 (42, 43). Unlike wild-type (WT) HBV (34), this phenotype of mutant 97 is featured by a nonselective and excessive secretion of virions containing immature genomes with single-stranded DNA. Here, we extend our study of HBV virion secretion to six additional core protein variants (Y38H, L60V, double mutant Y38H/L60V, P5T, P5S, and P5A). Surprisingly, we observed that five out of these six different variants display a new extracellular phenotype with significantly reduced levels of secreted virions containing only the mature genome. These “low-secretion” core variants could contribute to the gradually decreasing viremia observed during persistent infection in chronic HBV carriers (4, 9, 10).
MATERIALS AND METHODS
Preparation of HBV intracellular core particles and viral DNA as well as gradient centrifugation analysis of virion secretions were done as detailed elsewhere (41).
Plasmid constructs. (i) Y38H, L60V, Y38H/L60V.
A wild-type HBV monomer of the adr subtype (40) was used as the template to introduce the mutations in a site-specific mutagenesis reaction (Altered Sites II In Vitro Mutagenesis Systems; Promega, Madison, Wis.) as described previously (43). The sequences of oligonucleotides used to create HBcAg mutations at positions 38 (Y to H) and 60 (L to V) are 5′CCTCTGCTCTGCATCGGGAGGCC3′ and 5′GGCAAGCTATTGTGTGTTGGGGT3′, respectively (mutation sites are underlined). HBV mutant Y38H was used to obtain the double mutant Y38H/L60V. HBV monomers were then dimerized in tandem to mimic the circular configuration of HBV. All the mutations were confirmed by DNA sequencing.
(ii) P5T, P5A, and P5S.
Three mutants of subtype ayw origin, P5T, P5S, and P5A, were created by the Kunkel method of mutagenesis using oligonucleotides 5′GACATCGACACTTATAAAGAA3′, 5′GACATCGACTCTTATAAAGAA3′, and 5′GACATCGACGCTTATAAAGAA3′, respectively (18).
(iii) SVC L60V.
The core gene of the L60V HBV mutant was obtained by PCR amplification and subcloned into the expression vector using an SV40 early enhancer and promoter (42). The protein product of HBcAg L60V was confirmed by immunoblot analysis using a rabbit anti-HBcAg polyclonal antibody (21; data not shown).
Double-stranded DNA probe.
The full-length 3.1-kb vector-free HBV DNA fragments (adr and ayw) were purified from pALTER-HBV adr by BamHI digestion and from pWT ayw by EcoRI digestion. Approximately 25 ng of DNA was radiolabeled using a random-primed DNA labeling kit (Roche Co., Indianapolis, Ind.).
Immunoblot analysis of the core protein and enzyme immunoassay for HBeAg and HBsAg.
Core protein expression was detected by Western blot analysis using a rabbit anticore antibody (21). Detection of HBsAg and HBeAg was performed using the Abbott HBsAg kit and HBe (rDNA) kit, respectively (42).
Quantitative measurement of HBV DNA replication intermediates.
Quantitative comparisons of overall replication and the viral secretion levels by Southern blot analysis were performed with the ONE-Dscan computer program (Scanalytics Co., Billerica, Mass.) using images from X-ray film as described elsewhere (42).
RESULTS
No major difference in the levels of intracellular replication between variants Y38H, L60V, and Y38H/L60V and the WT HBV.
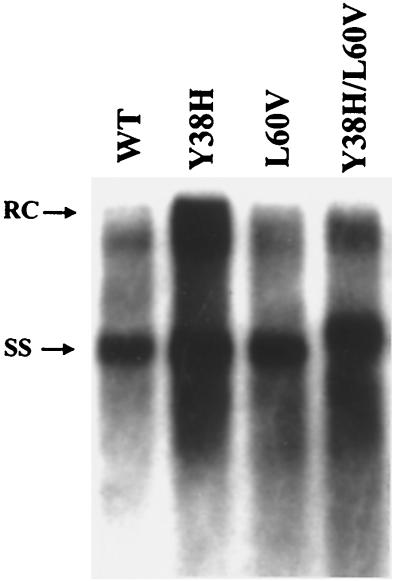
To investigate the functional significance of the naturally occurring mutations at codons 38 and 60 of the core protein, we introduced a tyrosine-to-histidine mutation at position 38 (mutant Y38H) and a leucine-to-valine mutation at position 60 (mutant L60V). These mutants and the double mutant Y38H/L60V were assayed for their levels of intracellular replication in comparison with that of the parental WT HBV. As shown in Fig. 1, the levels of the replicative intermediates of these mutants are comparable to that of the WT HBV. A small, yet reproducible, increase in the level of the replicative intermediates of mutant Y38H was observed (approximately 1.5-fold over that of the WT, based on the average of 10 independent experiments).
FIG. 1.
The distribution of the replicative intermediates of the intracellular HBV DNA shows no major difference between WT HBV (WT) and the core mutants Y38H, L60V, and Y38H/L60V. Ten micrograms of each plasmid DNA was transfected into Huh7 human hepatoma cells. Intracellular core particles were harvested 7 days posttransfection, and the core particle-associated HBV DNA was analyzed by a Southern blot with a 3.1-kb vector-free HBV DNA probe. Full-length relaxed-circle form (RC) DNA at the 4.0-kb position and single-stranded (SS) DNA at the 1.5-kb position are indicated by arrows.
Significant reduction of the amount of secreted virions of HBcAg variant L60V.
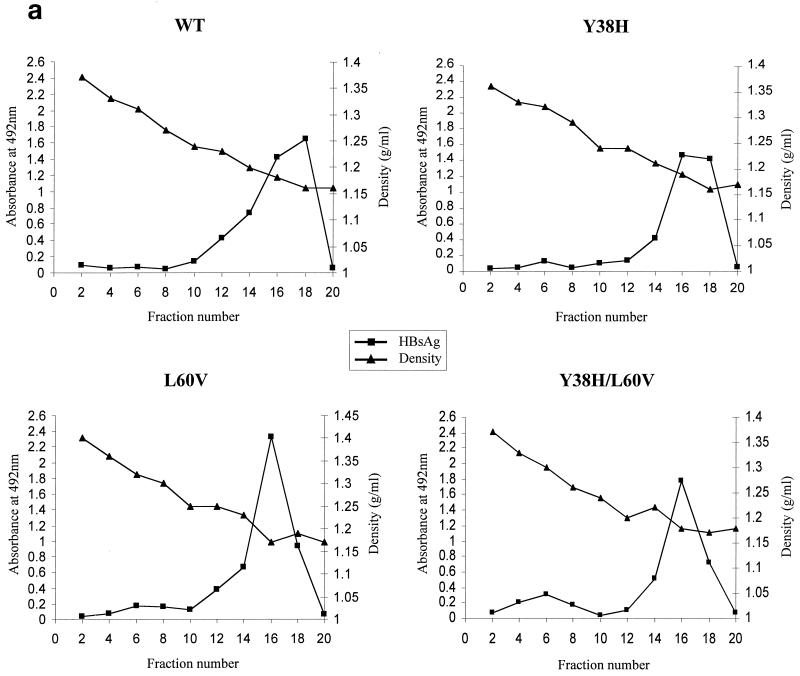
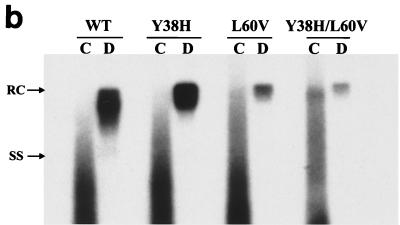
We examined further the secretion profile of these mutant and WT virions by gradient centrifugation and Southern blot procedures. As shown in Fig. 2a, similar levels of HBsAg subviral particles peak at similar positions for both the WT and mutants L60V, Y38H, and Y38/L60V. When viral DNA of the gradient fractions was subsequently analyzed by Southern blotting (Fig. 2b), mutant L60V secreted a significantly reduced amount of virion DNA relative to WT HBV and the Y38H mutant (about a three- to sixfold reduction, based on five independent experiments) in fractions 10 to 16, which correspond to the known density of infectious Dane particles (around 1.24 g/cm3). The nonenveloped core particles (1.35 g/cm3) of hepadnaviruses released into the medium have been observed previously in different tissue culture systems (8, 30, 35, 37); however, they have never been found in vivo. The remote possibility that naked core particles are released due to a small fraction of cell lysis in tissue culture cannot be excluded. Although these naked core particles are not directly relevant to our current study of virion secretion, they are included here as a control to demonstrate the low signals from virions of variant L60V (Fig. 2b).
FIG. 2.
Analysis of the extracellular virion particles by gradient centrifugation reveals the low-secretion phenotype of mutants L60V and Y38H/L60V. Media were collected on days 5 and 7 posttransfection. After centrifugation through a 20% sucrose cushion, the resuspended pellets of HBV particles were separated by isopycnic gradient centrifugation through 20 to 50% (wt/vol) cesium chloride. (a) Viral particles, including naked core particles and virions, were separated according to their buoyant densities (▴), followed by enzyme immunoassay for HBsAg (■) performed on every other fraction after dialysis. The secreted HBsAg, which formed DNA-free subviral particles, was usually several logs more abundant than the DNA-containing virions. (b) Collected fractions corresponding to the naked core particles (pooled from fractions 2 and 4; d = 1.35 g/ml) and to virion particles (pooled from fractions 10, 12, 14, and 16; d = 1.23 g/ml) were dialyzed against TNE buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA [pH 7.4]) before DNA extraction and analyzed further by Southern blotting with a 3.1-kb HBV DNA probe. C, pooled naked core particle fractions (see the text); D, pooled Dane particle (virion) fractions. RC, relaxed-circle form DNA; SS, single-stranded DNA.
The low-secretion phenotype of mutation L60V is dominant over the normal-secretion phenotype of mutation Y38H.
Previously, we demonstrated that a frequent HBcAg mutation, P130T, is compensatory or dominant to the immature secretion phenotype of another frequent mutation, I97L (44). Because of the association between mutations Y38H and L60V, we further investigated the relationship between these two mutations. When the double mutant Y38H/L60V was characterized, the low-secretion phenotype of mutation L60V was clearly dominant over the normal-secretion phenotype of mutation Y38H (Fig. 2b).
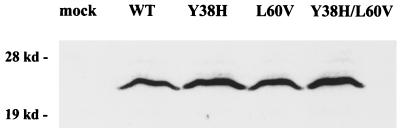
No significant difference in the steady-state levels of mutant L60V and WT core proteins.
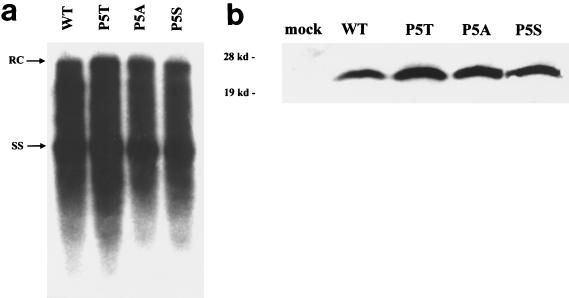
Immunoblot analysis was performed to check the intracellular core protein expression of mutants Y38H, L60V, and Y38H/L60V. As shown in Fig. 3, no major difference was observed. A slight but reproducible increase of the core protein signal was observed for the mutant Y38H, which is consistent with its small increase in replication activity (Fig. 1). These data represent findings from four independent experiments.
FIG. 3.
Similar steady-state levels of HBV core proteins were observed between wild-type and mutant viruses. Briefly, cell lysates harvested 3 days posttransfection were analyzed by Western blotting using a rabbit anticore antibody (21). The same filter was also probed with a monoclonal antibody to the HBV-specific pre-S1 protein as an internal control for equal loading and transfection efficiency (17; data not shown).
The low-secretion phenotype is caused solely by the L60V core protein.
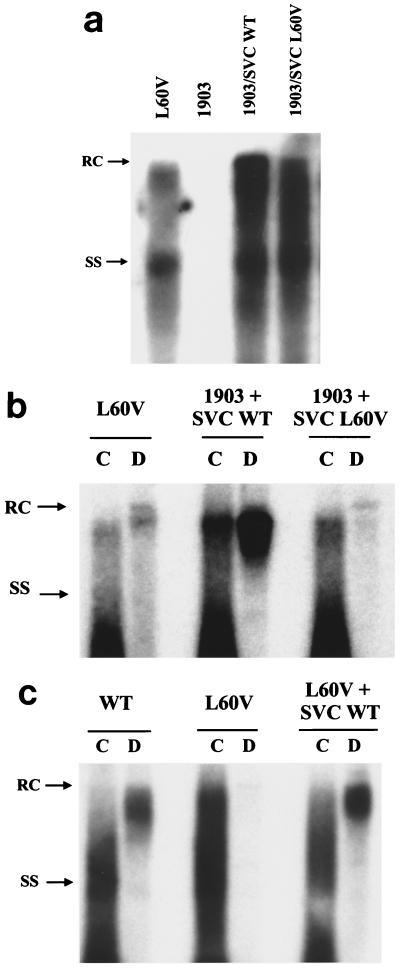
To test the potential trans effect of the mutant L60V core protein, a complementation assay was performed with a core-deficient (and hence replication-defective) mutant HBV, called 1903, and an expression vector producing the L60V core protein (SVC L60V). A WT core protein (subtype adr) cloned in the same expression vector was used as a positive control (SVC WT). As shown in Fig. 4a, the intracellular replication of the core-defective mutant 1903 can be restored efficiently by trans complementation with either the WT or mutant L60V core protein. However, when the virion secretion was analyzed by gradient centrifugation (Fig. 4b), we observed that complementation with the mutant L60V core protein alone can reproduce the low-secretion phenotype. In a separate reciprocal experiment (Fig. 4c), we provided the WT core protein to mutant L60V and observed the successful rescue of the low-secretion phenotype of mutant L60V. Taken together, these experiments indicate that the mutant L60V core protein alone is necessary and sufficient for the low-secretion phenotype.
FIG. 4.
Mutant L60V core protein alone is sufficient to induce a low-secretion phenotype. WT and mutant core proteins expressed from plasmids SVC WT adr and SVC L60V, respectively, were supplied in trans to a core-defective and replication-defective HBV mutant, 1903 (42). Their respective complementation effects on intracellular HBV DNA replication (a) and virion secretion (b) were compared. (c) Furthermore, WT core protein can rescue the low-secretion phenotype of mutant L60V via cotransfection. The analysis of extracellular virion particles was performed by gradient centrifugation as described in the legend for Fig. 2. RC, relaxed-circle form DNA; SS, single-stranded DNA.
No major differences in the level of intracellular replication are seen between mutants P5T, P5S, and P5A and WT HBV.
In addition to mutants 38 and 60 (Fig. 1 and 2), a highly frequent naturally occurring mutation at codon 5 (P5T) was also included in this study (14, 18, 22). In addition to P5T, several other codon 5 mutants (P5S and P5A) were constructed and characterized in parallel. As shown in Fig. 5a, no significant difference in the level of intracellular DNA replication was observed between the codon 5 mutants and WT HBV. As a control, we also show here that there is no significant difference in the steady-state levels of codon 5 mutants and WT core proteins (Fig. 5b).
FIG. 5.
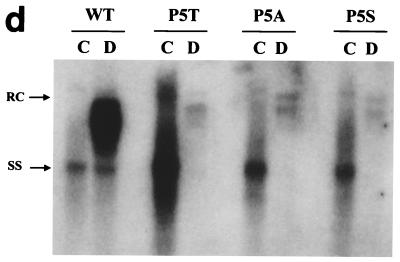
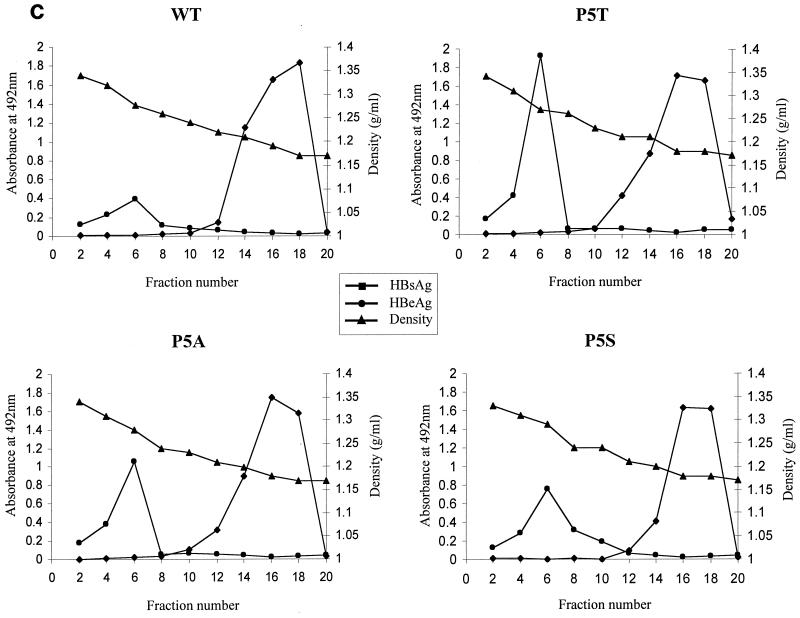
The low-secretion phenotype is also observed for all of the codon 5 mutants: P5T, P5S, and P5A. Viral replication (a) and secretion (c and d) were analyzed as described for Fig. 1 and 2. Detection of HBeAg in each fraction was performed using an enzyme immunoassay kit (Abbott HBe [rDNA]) (panel c [●]). (b) Similar steady-state levels of HBV core proteins were observed between WT and codon 5 mutant viruses by Western blot analysis. The same filter was also probed with a monoclonal antibody to the HBV-specific pre-S1 protein as an internal control for equal loading and transfection efficiency (17; data not shown). RC, relaxed-circle form DNA; SS, single-stranded DNA.
The low-secretion phenotype of the codon 5 mutant virions is caused by the loss of the parental amino acid proline.
Most interestingly, we observed a similar low-secretion phenotype in all three codon 5 mutants (Fig. 5d), ranging from a 7- to 16-fold reduction, based on five independent experiments. In a previous study by members of our group (43), we demonstrated that it is the acquisition of a leucine residue at amino acid position 97 of HBcAg that is important for immature virion secretion. Here, in contrast, the fact that mutants P5T, P5S, and P5A share a common low-secretion phenotype strongly suggests that it is the loss of the WT amino acid proline, rather than the acquisition of a new amino acid (T, S, or A), that is responsible for the aberrant phenotype.
DISCUSSION
In this study, we observed a unique extracellular phenotype associated with naturally occurring mutations of the HBV core protein. The mutation at position 5 or 60 alone had little effect on the intracellular viral DNA replication (Fig. 1; Fig. 5a), the steady-state level of core protein (Fig. 3 and 5b), or the secretion of HBsAg (Fig. 2a and 5c). The only detectable effect of these mutations in the present study was the significant reduction in secreted virions containing normal or mature genomes with relaxed-circle DNA form (Fig. 2b, 4b, and 5d).
The paradox of detecting the low-secretion HBV variants in patients' sera.
The acquisition of a mutation at position 5 or 60 has been reported using serum samples in several longitudinal studies (2, 6, 38). It is interesting to ask how the low-secretion variants, such as mutants P5T and L60V, can be secreted into circulation and eventually predominate over the existing WT HBV population in a natural infection. Several hypotheses can be entertained at this stage. (i) These low-secretion variants are immune escape mutants (18), and their disadvantage in low secretion is outweighed by their advantage in avoiding the immune selective pressure. (ii) An unknown secondary compensatory mutation in the same HBV variants can restore virion secretion. (iii) Coinfection of the same hepatocytes by a mixture of WT and mutant viruses results in the formation of mosaic core particles with intermediate or normal secretion efficiency. So far, we have tested the second hypothesis and demonstrated that mutation Y38H, which often associates with mutation L60V, is not compensatory for the low-secretion phenotype. The third hypothesis is consistent with the presence of both the WT and mutant 5 or 60 in the same individuals at the same time (11, 14). Indeed, we can restore virion secretion to a normal level by providing the wild-type core protein in trans to the low-secretion variants via cotransfection (Fig. 4c).
Difference in the intracellular profiles of viral DNA genome maturity between HBV core and envelope mutants.
It has been shown previously that when virion secretion was blocked by mutations in large S (pre-S1) or small S (HBsAg) envelope proteins, it can result in a higher level of intracellular accumulation of the more fully mature HBV DNA genome, which often migrates at approximately a 4.0-kb position on agarose gel electrophoresis (23, 42). In contrast, despite the significantly reduced level of virion secretion of HBV core mutants 5 and 60, there is no corresponding increase of the steady-state level of intracellular HBV DNA replicative intermediates or more mature forms of HBV DNA (Fig. 1 and 5a). One possibility is that the effect of core mutation 5 or 60 is pleiotropic: the expected increase of the more mature HBV genome intracellularly, due to decreased virion secretion, may be masked by another as yet uncharacterized effect in decreasing the synthesis of HBV replicative intermediates. Alternatively, elongation of the nascent plus-strand DNA may not be able to continue in the core mutants, since their core particles may accumulate in a different subcellular compartment which is not supportive for viral DNA synthesis and genome maturation.
Stringent requirement of a precise conformation of core particles in envelopment or virion secretion.
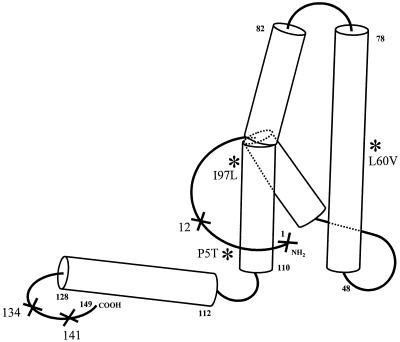
Recently, it has been shown that artificially introduced insertions or deletions in nonhelical regions of the HBV core protein can block virion secretion (Fig. 6) (19, 20). Core proteins with either a 10- or 23-amino-acid extension at the N terminus can allow intracellular viral DNA replication but inhibit virion secretion (19). Koschel et al. also characterized several artificial HBcAg mutants defective in virion secretion, including deletions of amino acids 12 or 134 or insertion at amino acid 141. However, in these cases, all the secretion-defective core mutants are also replication defective by endogenous polymerase assay and Southern blot analysis. In contrast, the naturally occurring variants L60V and P5T display normal intracellular HBV DNA replication, despite their severe defect in virion secretion. In summary, based on the four-helix bundle structure of HBcAg (5, 12, 39) and our current data, both α-helical (L60V and I97L) and nonhelical (P5T) loop regions of the entire HBcAg molecule are important for virion secretion (Fig. 6). Furthermore, the bending created by the proline residue at amino acid 5 of WT HBcAg is critical for productive virion secretion. It is very likely that virion secretion depends on the interactions between the envelopment machinery and capsid particles with a very precise conformation.
FIG. 6.
Distribution of virion secretion-defective mutations on the reported 3D structure of HBV core protein (aa 1 to 149) (5, 12, 39). This cartoon of the HBcAg monomer is drawn as left handed, while the hand of the crystallographic map of HBcAg in reference 39 is right handed. ❉, naturally occurring secretion-defective variants containing substitution mutations; ×, artificial low-secretion or no-secretion mutants containing insertion or deletion mutations (19, 20). Naturally occurring variant 97 is an immature secretion mutant (42).
Our findings of the emergence of secretion-defective HBcAg variants, such as the predominant mutants 5 and 97 (42, 43, 44), and the defective-interfering variants, such as the core internal deletion variants (41), indicate that they are likely mechanisms that contribute to the decreased viremia observed during the late stage of chronic infection in HBV patients (4, 9, 10). Further investigation of these naturally occurring HBV variants will help us understand better the regulation of virion secretion and morphogenesis.
ACKNOWLEDGMENTS
We thank colleagues in C. Shih's laboratory for careful reading of the manuscript. S. Le Pogam is a McLaughlin Postdoctoral Fellow. We thank A. C. Steven for the improvement of the structure of HBcAg in Fig. 6.
This study was mainly supported by NIH grants RO1 CA 70336 and CA84217 to C.S.
REFERENCES
- 1.Akarca U S, Lok A S. Naturally occurring hepatitis B virus core gene mutations. Hepatology. 1995;22:50–60. [PubMed] [Google Scholar]
- 2.Asahina Y, Enomoto N, Ogura Y, Kurosaki M, Sakuma I, Izumi N, Marumo F, Sato C. Sequential changes in full-length genomes of hepatitis B virus accompanying acute exacerbation of chronic hepatitis B. J Hepatol. 1996;25:787–794. doi: 10.1016/s0168-8278(96)80280-7. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg B S, London W T. Hepatitis B virus: pathogenesis and prevention of primary cancer of the liver. Cancer. 1982;50:2657–2665. [PubMed] [Google Scholar]
- 5.Böttcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 6.Bozkaya H, Ayola B, Lok A S. High rate of mutations in the hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology. 1996;24:32–37. doi: 10.1002/hep.510240107. [DOI] [PubMed] [Google Scholar]
- 7.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C M, Jeng K S, Hu C P, Lo S J, Su T S, Ting L P, Chou C K, Han S H, Pfaff E, Salfeld J, Schaller H. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D S. From hepatitis to hepatoma: lessons from type B viral hepatitis. Science. 1993;262:369–370. doi: 10.1126/science.8211155. [DOI] [PubMed] [Google Scholar]
- 10.Chu C M, Karayiannis P, Fowler M J, Monjardino J, Liaw Y F, Thomas H C. Natural history of chronic hepatitis B virus infection in Taiwan: studies of hepatitis B virus DNA in serum. Hepatology. 1985;5:431–434. doi: 10.1002/hep.1840050315. [DOI] [PubMed] [Google Scholar]
- 11.Chuang W L, Omata M, Ehata T, Yokosuka O, Ito Y, Imazeki F, Lu S N, Chang W Y, Ohto M. Precore mutations and core clustering mutations in chronic hepatitis B virus infection. Gastroenterology. 1993;104:263–271. doi: 10.1016/0016-5085(93)90861-6. [DOI] [PubMed] [Google Scholar]
- 12.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 13.Ehata T, Omata M, Chuang W L, Yokosuka O, Ito Y, Hosoda K, Ohto M. Mutations in core nucleotide sequence of hepatitis B virus correlate with fulminant and severe hepatitis. J Clin Investig. 1993;91:1206–1213. doi: 10.1172/JCI116281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther S, Baginski S, Kissel H, Reinke P, Kruger D H, Will H, Meisel H. Accumulation and persistence of hepatitis B virus core gene deletion mutants in renal transplant patients are associated with end-stage liver disease. Hepatology. 1996;24:751–758. doi: 10.1002/hep.510240401. [DOI] [PubMed] [Google Scholar]
- 15.Hannoun C, Horal P, Lindh M. Long-term mutation rates in the Hepatitis B genome. J Gen Virol. 2000;81:75–83. doi: 10.1099/0022-1317-81-1-75. [DOI] [PubMed] [Google Scholar]
- 16.Hatton T, Zhou S, Standring D N. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J Virol. 1992;66:5232–5241. doi: 10.1128/jvi.66.9.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heermann K H, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. Large surface proteins of hepatitis B virus containing the pre-S sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosono S, Tai P C, Wang W, Ambrose M, Hwang D G, Yuan T T, Peng B H, Yang C S, Lee C S, Shih C. Core antigen mutations of human hepatitis B virus in hepatomas accumulate in MHC class II-restricted T cell epitopes. Virology. 1995;212:151–162. doi: 10.1006/viro.1995.1463. [DOI] [PubMed] [Google Scholar]
- 19.Hui E K, Yi Y S, Lo S J. Hepatitis B viral core proteins with an N-terminal extension can assemble into core-like particles but cannot be enveloped. J Gen Virol. 1999;80:2647–2659. doi: 10.1099/0022-1317-80-10-2647. [DOI] [PubMed] [Google Scholar]
- 20.Koschel M, Oed D, Gerelsaikhan T, Thomssen R, Bruss V. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J Virol. 2000;74:1–7. doi: 10.1128/jvi.74.1.1-7.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanford R E, Notvall L M, Dreesman G R, Harrison C R, Lockwood D, Burk K H. Expression and characterization of hepatitis B virus precore-core antigen in E. coli. Viral Immunol. 1987;1:97–109. doi: 10.1089/vim.1987.1.97. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y I, Hur G M, Suh D J, Kim S H. Novel pre-C/C gene mutants of hepatitis B virus in chronic active hepatitis: naturally occurring escape mutants. J Gen Virol. 1996;77:1129–1138. doi: 10.1099/0022-1317-77-6-1129. [DOI] [PubMed] [Google Scholar]
- 23.Melegari M, Scaglioni P P, Wands J R. The small envelope protein is required for secretion of a naturally occurring hepatitis B virus mutant with Pre-S1 deleted. J Virol. 1997;71:5449–5454. doi: 10.1128/jvi.71.7.5449-5454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nassal M, Rieger A, Steinau O. Topological analysis of the hepatitis B virus core particle by cysteine-cysteine cross-linking. J Mol Biol. 1992;225:1013–1025. doi: 10.1016/0022-2836(92)90101-o. [DOI] [PubMed] [Google Scholar]
- 25.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepatitis. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 26.Ou J H, Rutter W J. Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J Virol. 1987;61:782–786. doi: 10.1128/jvi.61.3.782-786.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou J H. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–S187. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 28.Preikschat P, Meisel H, Will H, Gunther S. Hepatitis B virus genomes from long-term immunosuppressed virus carriers are modified by specific mutations in several regions. J Gen Virol. 1999;80:2685–2691. doi: 10.1099/0022-1317-80-10-2685. [DOI] [PubMed] [Google Scholar]
- 29.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum H E. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 30.Sells M A, Chen M L, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih C, Tai P C, Whitehead W, Hosono S, Lee C S, Yang C S. Hepatitis B and C viruses and liver cancer. In: Bertino J R, editor. Encyclopedia of cancer. II. New York, N.Y: Academic Press, Inc.; 1996. pp. 824–834. [Google Scholar]
- 32.Slagle B L, Lee T H, Butel J S. Hepatitis B virus and hepatocellular carcinoma. Prog Med Virol. 1992;39:167. [PubMed] [Google Scholar]
- 33.Sterneck M, Gunther S, Santantonio T, Fischer L, Broelsch C E, Greten H, Will H. Hepatitis B virus genomes of patients with fulminant hepatitis do not share a specific mutation. Hepatology. 1996;24:300–306. doi: 10.1002/hep.510240203. [DOI] [PubMed] [Google Scholar]
- 34.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 35.Sureau C, Romet-Lemonne J L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Akahane Y, Hino K, Ohta Y, Mishiro S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch Virol. 1998;143:2313–2326. doi: 10.1007/s007050050463. [DOI] [PubMed] [Google Scholar]
- 37.Tuttleman J S, Pugh J C, Summers J W. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J Virol. 1986;58:17–25. doi: 10.1128/jvi.58.1.17-25.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida T, Aye T T, Shikata T, Yano M, Yatsuhashi H, Koga M, Mima S. Evolution of the hepatitis B virus gene during chronic infection in seven patients. J Med Virol. 1994;43:148–154. doi: 10.1002/jmv.1890430209. [DOI] [PubMed] [Google Scholar]
- 39.Wynne S A, Crowther R A, Leslie A G. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 40.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan T T, Lin M H, Qiu S M, Shih C. Functional characterization of naturally occurring variants of human hepatitis B virus containing the core internal deletion mutation. J Virol. 1998;72:2168–2176. doi: 10.1128/jvi.72.3.2168-2176.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan T T, Sahu G K, Whitehead W E, Greenberg R, Shih C. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J Virol. 1999;73:5731–5740. doi: 10.1128/jvi.73.7.5731-5740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan T T, Tai P C, Shih C. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human hepatitis B virus core antigen. J Virol. 1999;73:10122–10128. doi: 10.1128/jvi.73.12.10122-10128.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan T T, Shih C. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (I97L) J Virol. 2000;74:4929–4932. doi: 10.1128/jvi.74.10.4929-4932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng J, Schödel F, Peterson D L. The structure of hepadnaviral core antigens. Identification of free thiols and determination of the disulfide bonding pattern. J Biol Chem. 1992;267:9422–9429. [PubMed] [Google Scholar]