Abstract
Acinetobacter baumannii is a common cause of multidrug-resistant (MDR) nosocomial infections around the world. However, little is known about the persistence and dynamics of A. baumannii in a healthy community. This study investigated the role of the community as a prospective reservoir for A. baumannii and explored possible links between hospital and community isolates. A total of 12 independent A. baumannii strains were isolated from human faecal samples from the community in Segamat, Malaysia, in 2018 and 2019. Another 15 were obtained in 2020 from patients at the co-located tertiary public hospital. The antimicrobial resistance profile and biofilm formation ability were analysed, and the relatedness of community and hospital isolates was determined using whole-genome sequencing (WGS). Antibiotic profile analysis revealed that 12 out of 15 hospital isolates were MDR, but none of the community isolates were MDR. However, phylogenetic analysis based on single-nucleotide polymorphisms (SNPs) and a pangenome analysis of core genes showed clustering between four community and two hospital strains. Such clustering of strains from two different settings based on their genomes suggests that these strains could persist in both. WGS revealed 41 potential resistance genes on average in the hospital strains, but fewer (n=32) were detected in the community strains. In contrast, 68 virulence genes were commonly seen in strains from both sources. This study highlights the possible transmission threat to public health posed by virulent A. baumannii present in the gut of asymptomatic individuals in the community.
Keywords: Acinetobacter baumannii, community, hospital, faecal, comparative genomics, antimicrobial resistance, virulence, CRISPR–Cas, Malaysia
Data Summary
All sequence data are available online. The assembled sequencing reads generated in the present study are publicly available at the National Center for Biotechnology Information (NCBI), BioProject Number: PRJNA851747 and PRJNA659865. The authors confirm that all supporting data, code and protocols have been provided within the article or through supplementary data files. All new multilocus sequence typing (MLST) sequences generated were deposited onto the respective databases available on PubMLST (https://pubmlst.org/organisms/Acinetobacter-baumannii). Additional A. baumannii genomes used in this study are available and can be downloaded from GenBank (accession numbers available in File S2).
Impact Statement.
Studies on Acinetobacter baumannii isolated from healthy individuals in the community are quite rare. Hospital-derived isolates are used in the majority of studies currently available. We obtained A. baumannii isolates from the faeces of community members in Malaysia and contrasted them with isolates from local hospitals. According to our findings, low and transient carriage of A. baumannii was deduced in the gut of community individuals. Community isolates were similar to hospital isolates in terms of virulence determinants but not antibiotic resistance. Using core-genome single-nucleotide polymorphism (SNP), pangenome, phylogenetic and CRISPR array analyses from whole-genome sequencing, it was demonstrated that some hospital and community isolates clustered together in all of the analyses. Our findings highlight the significance of A. baumannii carriage in healthy persons as well as the potential transmission risk. If strains of A. baumannii present in the gut acquire antimicrobial resistance (AMR) genes, infections caused by these organisms may become difficult to treat.
Introduction
Antimicrobial resistance (AMR) has been a global threat to public health and a leading cause of long-term hospitalization, morbidity, mortality and costs over the years [1]. Hospital settings were considered to be the primary reservoirs of infections caused by antimicrobial-resistant bacteria, but a more pressing concern now is the spread of antimicrobial-resistant bacteria within the community and the environment [2, 3]. Widespread use of antimicrobials is the principal cause of the development of AMR inside and outside of the hospital [4]. Studies have demonstrated that communities can act as a key reservoir for antimicrobial-resistant bacteria [5–7].
Acinetobacter baumannii, a lactose-non-fermenting Gram-negative pathogen, is one of the most commonly identified multidrug-resistant bacteria. Although it was formerly considered to be a low-category pathogen, it has now emerged as the primary cause of hospital and community-acquired infections [8]. A. baumannii is an opportunistic pathogen that belongs to the ESKAPE group categorized by the Infectious Diseases Society of America (IDSA) [9]. The World Health Organization (WHO) has designated this bacterium as a priority 1 critical pathogen since 2017 [10]. Three major global clones of A. baumannii (IC- I, IC- II and IC- III) emerged as high-risk pandemic lineages with significant persistence in hospital settings [11]. Multidrug resistance is frequently linked to isolates from these global clones [12, 13]. A. baumannii has been related to a wide range of human diseases, including ventilator-associated pneumonia, bloodstream, skin and urinary tract infections, and secondary meningitis [14].
A combination of mechanisms, including drug target modification, the production of hydrolyzing enzymes such as beta-lactamases, alteration of bacteria cell membrane permeability, increased expression of efflux pumps and altered topoisomerases can lead to antibiotic resistance in A. baumannii [1, 15]. In addition, the ability of A. baumannii to develop biofilms on a wide range of surfaces can be associated with its persistence in hospital settings and the emergence of recalcitrant and chronic infections [16].
A. baumannii has been isolated from diverse sources, including hospitalized and non-hospitalized individuals, different environmental sources and slaughtered animals, but the ecology outside hospitals is not well understood [9, 17–20]. The mechanisms of A. baumannii virulence are well characterized in different settings [17, 18]. Although significant genomic differences and differences in phenotypes associated with virulence have been reported between single community-acquired and hospital-acquired strains, data on a larger number of community and hospital isolates from one locality to corroborate these observations are still lacking [19]. Without triggering an infection, A. baumannii can also colonize the skin and respiratory system [17]. In hot and humid climates – notably in the Asia-Pacific region – A. baumannii has emerged as a cause of severe community-acquired infections [21–23]. Studies have found Acinetobacter spp. in the intestinal microbiota of healthy volunteers (12.2 %) [24]. Nevertheless, the origins of colonization and factors influencing it are still unknown [25].
This study aimed to explore the role of the community as a potential reservoir for A. baumannii and possible transmission between the community and the hospital. We characterized A. baumannii isolates from the community in Segamat, a small town in southern peninsular Malaysia, and compared them with isolates obtained from a local government hospital. A longitudinal study was also conducted to detect whether isolates are commensal or transient in the community. The AMR profile and biofilm-forming ability of A. baumannii isolates from the community and the hospital were compared. Whole-genome sequencing (WGS) was used to investigate virulence determinants, as well as genes linked with antibiotic resistance and the presence of CRISPR arrays, followed by single-nucleotide polymorphism (SNP) and pangenome-based phylogenetic correlation between these strains. This study sheds light on the relationship between hospital and community isolates from one district in Malaysia, contributing to a better understanding of the epidemiology and pathogenesis of A. baumannii in the community.
Methods
Study location and sampling
The study was conducted in collaboration with the South East Asia Community Observatory (SEACO), a community-based research platform in Segamat District, Johor, Malaysia [26]. The human faecal samples were sourced from recruited people as described in [27]. Briefly, fresh faecal samples were collected from participants between May and August 2018. Resampling was carried out from the same individuals and other household members in November 2019 to determine if the isolates are transient or commensal and their transmission between other family members. Hospital Segamat provided hospital isolates from June to October 2020. This is the only tertiary government hospital in Segamat District, Johor, Malaysia.
Isolation and identification of A. baumannii
To isolate A. baumannii , approximately 1 g of faecal sample was suspended in 9 ml of buffered peptone water (Oxoid, UK) and vortexed. Following this, a 10-fold serial dilution was carried out using buffered peptone water. From each dilution, 100 µl was spread on Leeds Acinetobacter Agar (HiMedia, India) and subsequently plates were incubated at 37 °C for 24 h. The colony morphology and nature of the strains were observed and recorded. Three colonies with A. baumannii morphology were selected and identified from each potentially positive sample based on aerobic, Gram-negative, catalase-positive, oxidase-negative, nonmotile, nonfermenting coccobacilli nature [28].
PCR amplification of 16S rRNA gene fragments and subsequent sequencing was performed to confirm Acinetobacter spp. at the genus level. The 16S rRNA gene was targeted using the universal primers described in previous studies [29]. Bacterial DNA extraction for PCR was carried out by the boiling extraction method [30]. Details of the extraction method and 16S rRNA PCR reaction can be found in Text S1, Table S7-S9 (available in the online version of this article).
Species identification by phenotypic methods and 16S rRNA is insufficient for unambiguous identification of A. baumannii [31]. Thus, the detection of A. baumannii with species-specific PCR was performed based on [32]. The internal fragment of gyrB gene was targeted for PCR amplification to detect A. baumannii (detailed in Table S10 and S11). A. baumannii ATCC BAA 1605 (ATCC BAA-1605) was used as control.
Antimicrobial resistance profiling
Antibiotic susceptibility of A. baumannii isolates was determined against 13 antibiotics using the Kirby–Bauer disc diffusion method. The different classes of antibiotics used were penicillin (piperacillin, PRL 100); ß-lactamase inhibitor combinations (piperacillin–tazobactam, TZP 110; ampicillin–sulbactam, SAM 20); third- and fourth-generation cephalosporin (ceftazidime, CAZ 30; cefotaxime, CTX 30; cefepime, FEP 30); carbapenem (imipenem, IPM 10; meropenem MEM, 10); aminoglycoside (gentamicin, CN 10; amikacin AK 30); fluoroquinolone (ciprofloxacin, CIP 5) and tetracycline (TET 30). For colistin and polymyxin B, the broth microdilution method was used. Results were interpreted based on the Clinical and Laboratory Standard Institute (CLSI) guidelines [33]. A. baumannii ATCC BAA 1605 (ATCC BAA-1605) and Escherichia coli ATCC BAA 2325 (ATCC BAA-2523) with known resistance patterns were purchased from the American Type Culture Collection (ATCC, USA) and used in the study as controls. Three biological and technical replicates were used for this profiling.
Biofilm formation analysis
Biofilm formation by each isolate was detected using crystal violet (CV) and XTT assays. Biofilm production and quantification using CV assay were performed according to [34] and the XTT assay was performed according to [35] with slight modifications. The results were interpreted according to the criteria suggested in [36]. Detailed descriptions of the XTT and CV assay can be found in Text S1.
WGS
The total genomic DNA of A. baumannii isolates was extracted using the Wizard genomic DNA Extraction kit (Promega, USA) according to the manufacturer’s instructions. Extracted DNA quality and concentration were assessed using a Nanodrop bioanalyser spectrophotometer (Thermo Scientific, USA). Illumina sequencing libraries were prepared using the Nextera XT DNA Preparation Kit (Illumina, USA). All isolates were sequenced using the MiSeq Reagent kit v3 with a 2×251 bp paired-end read configuration.
Illumina reads were processed using Trimmomatic with the following parameters: PE, ILLUMINACLIP: adapters/NexteraPE.fa:2 : 30 : 10 : 8, LEADING:3, TRAILING:3, SLIDINGWINDOW:5 : 20, MINLEN:35 [37]. For each sequenced strain, the trimmed paired-end reads were then de novo assembled using SPAdes 3.15.2 [38]. Functional annotation was performed using Prokka 1.13 [39]. The MLST Oxford scheme was performed by targeting seven housekeeping genes (gltA, gdhB, gyrB, gpi, cpn60, recA and rpoD) from the assembled WGS sequences to obtain the allele profile and sequence types (STs) using the MLST 2.0 online tool at the Center for Genomic Epidemiology (CGE) (https://cge.cbs.dtu.dk/services/MLST/) [40]. Subsequently, goeBURST analysis (http://eburst.mlst.net/) was carried out to determine the clonal complexes present in our isolates and A. baumannii isolates in the PubMLST database (https://pubmlst.org/). AMR genes were identified by Abricate v1.0.1 (https://github.com/tseemann/abricate), using the Comprehensive Antibiotic Resistance Database (CARD) [41] and ResFinder [42]. Virulence-associated genes were identified using the virulence factor database (VFDB4) [43]. Gene content matrices were obtained using Roary 3.13.0 [44], with a minimum of 90 % identity between coding sequences (CDSs) required for a gene to belong to the same family. The CRISPR–Cas array was detected using CRISPRCasFinder 4.2.20 [45]. The number of CRISPR arrays in each genome with an evidence level ≥2 was counted and allocated to the genome. Mobile genetic elements from each isolate were detected using Mobile Element Finder v1.0.3 at the CGEserver. Single-nucleotide variants (SNVs) were detected using the SNIPPY v4.6.0 variant calling tool with multi-SNIPPY default parameters [46]. SNP sites were used to identify core genomic SNPs [47]. Genealogies Unbiased By recomBinations In Nucleotide Sequences (Gubbins) with default parameters was used to detect and analyse SNPs likely introduced simultaneously during a homologous recombination event [48]. Phylogenetic trees were built using Gubbins based on the alignment of the non-recombinant SNPs obtained and using a maximum-likelihood (ML) phylogeny inferred from the alignment of these SNPs. Phylogenetic trees were constructed using FastTree [49]. Phylogenetic clusters (n≥2) were identified using TreeCluster with a bootstrap threshold of 90 % and a genetic distance of 0.045 [50]. The online tool iTOL V6 was used to annotate and visualize the phylogenetic trees [51].
Results
Phenotypic characterization of A. baumannii isolates from the Segamat community and hospital
A total of 233 human faecal samples from 110 households in Segamat, Malaysia were tested in 2018 for the presence of A. baumannii . After screening, only nine A . baumannii were isolated. In 2019, resampling was carried out for all of these carriers and community members, where 3 A. baumannii isolates were detected from 126 faecal samples and all of these were from new individuals (2.4 %) and households. Based on our resampling data in 2019, none of the individuals carried A. baumannii for a year. Thus, overall 12 community strains were characterized.
In addition, 15 A . baumannii isolates were provided by Hospital Segamat from hospitalized patients. These isolates were obtained from blood culture (n=5), tracheal aspirates (n=7), urine (n=1) and sputum (n=1) assessments. One of the isolates lacked information concerning the isolation source. Detailed information on community and hospital isolates can be found in Tables S1 and S2.
Substantial differences in antimicrobial susceptibility profile
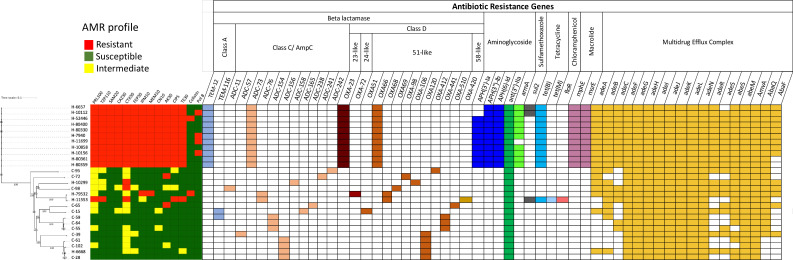
The analysis of the antibiotic susceptibility profile of A. baumannii community and hospital isolates revealed considerable differences. Out of 15 hospital isolates examined, 12 (80 %) were classified as MDR and were resistant to cephalosporin, carbapenems, fluoroquinolones, aminoglycosides, tetracycline and β-lactam combination agents (Fig. 1). In addition, 2/15 and 4/15 of the hospital isolates showed resistance to colistin and polymyxin B, respectively (Fig. 1). In contrast, only three (25 %) of the community isolates showed resistance to one or two antibiotics and none were MDR.
Fig. 1.
Antibiotic susceptibility profile and presence of resistance genes from whole-genome sequences of independent A. baumannii isolated from the Segamat community and hospital. Fourteen antibiotics were tested: PRL100, piperacillin 100 µg; TZP110, piperacillin–tazobactam 110 µg; SAM20, ampicillin–sulbactam; CAZ30, ceftazidime 30 µg; CTX30, cefotaxime 30 µg; FEP30, cefepime 30 µg; IPM10, imipenem 10 µg; MEM10, meropenem 10 µg; CN10, gentamicin 10 µg; AK30, amikacin 30 µg; CIP5, ciprofloxacin 5 µg; colistin (≥4 µg ml−1) and polymixin B (≥4 µg ml−1). The isolates were organized here along with a phylogenetic tree (left) constructed by whole-genome SNP analysis. In this study, isolate names with ‘H’ and ‘C’ represent the hospital and community isolates, respectively. The presence of genes in an isolate is specified by a coloured rectangle, with different genes coloured differently according to the classes to which they confer resistance. The absence of genes is represented by white rectangles.
These data showed that multidrug resistance was more common in the A. baumannii hospital isolates than the community strains (Fisher’s exact test P-value <0.0001).
Similar biofilm formation ability
Crystal violet (CV) and XTT assays were used to measure the biofilm biomass and metabolic activity of A. baumannii isolates. Both community and hospital isolates had variable abilities to form biofilms and the results varied depending on the assay used. However, the hospital strains had somewhat higher biofilm-producing ability compared to the community ones (Fig. S1), but the difference was not statistically significant. (chi-square test, P-value >0.05).
To determine the association between biofilm-forming ability and antibiotic resistance, statistical analysis (chi-square test) was carried out. However, no significant association was found between biofilm-forming ability and antibiotic resistance among these A. baumannii strains (P-value >0.05).
Genomic characterization of A. baumannii isolates from the Segamat community and hospital shows relatedness
All 27 A . baumannii strains were subjected to WGS. Selected features of the sequenced A. baumannii isolates are shown in File S2 (Genomic features). The assembled draft genomes of these strains showed an average cumulative length of 3.6–3.8 Mb and 39 % GC content, consistent with the genome assembly of most published A. baumannii isolates in the GenBank database. The newly sequenced isolates from Segamat, Malaysia reported here showed more than 97 % pairwise average nucleotide identity (ANI) with the A. baumannii reference strain AC30.
MLST
STs of community and hospital A. baumannii isolates were determined using the MLST Oxford scheme from WGS-assembled sequences. The analysis identified 19 different STs from the 27 hospital and community isolates. Among them, nine STs (ST1930, ST2230, ST2232, ST2234 and ST2236 from the community and ST2237, ST2238 and ST2241 from the hospital) are being described for the first time in this study and were deposited in the PubMLST database (Table S3). The existing STs detected here are ST128, ST231, ST503, ST1463 and ST1912 from the community and ST208, ST447, ST547, ST642 and ST684 from the hospital. The remaining isolates were assigned new STs. No identical STs were detected between the community and hospital strains. ST208 was the predominant ST, comprising (46.6 %, 7/15) of the hospital isolates.
Based on the goeBURST analysis, eight clonal complexes (CCs) were detected, summarized in Table S5. They are CC208, CC231, CC474, CC953, CC642, CC1108, CC1171 and CC1178. ST208, ST547 and ST684 belong to the globally distributed clonal complex CC208 (previously known as CC92), which corresponds to IC2 (international clone 2) (Fig. S2). CC208 has been identified as a major epidemic clonal complex of carbapenem-resistant A. baumannii [52]. All CC208 isolates were obtained from the hospital. Interestingly, one community isolate, C-98 (ST231), belonged to CC231 that clustered with the previously identified international clonal lineage IC1.
SNV analysis
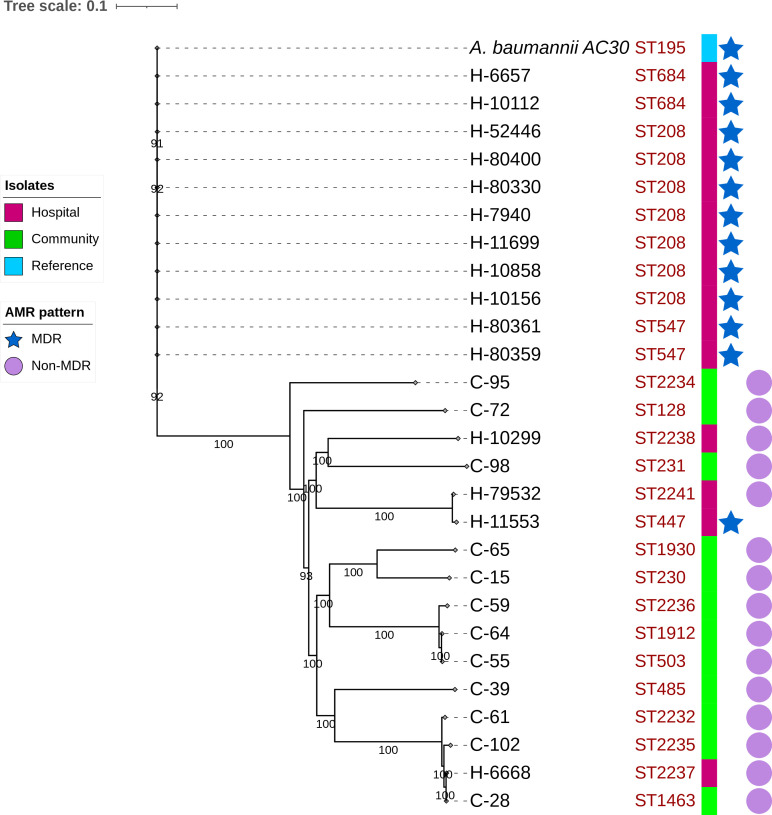
The variation between the genomes of the current study, along with a reference strain A. baumannii AC30, was assessed. Considerable differences between A. baumannii MDR and sensitive isolates were observed. Against the reference strain, 11 MDR hospital isolates showed less nucleotide variation than the other 16 strains (Table S5). The number of nucleotide insertions and deletions was also different within the MDR and non-MDR classes. Total SNPs detected ranged from 447 (H-11699) to 39 120 (H-79532). A phylogenetic tree was constructed based on the core-genome SNP alignment (Fig. 2). From the phylogeny, two clusters were observed between hospital and community strains. In one cluster, three community strains were located in one clade with one hospital strain (C-28, C-102, C-61 and H-6668). The number of SNPs in these isolates was 36 447, 36 205, 36 572 and 37 320, respectively. The other cluster was observed between C-98 and H-10299, with 37 745 and 37 773 SNPs, respectively. Only H-11533 was MDR out of these hospital strains, clustering with the non-MDR strain H-79532.
Fig. 2.
Phylogenetic tree using the maximum-likelihood method based on the core-genome SNP alignment of 27 sequenced community and hospital strains along with A. baumannii strain AC30 as a reference. Isolates are colour coded as purple and green for hospital and community isolates, respectively, and STs for each isolate are shown. The isolates’ sources and AMR patterns are displayed at the right and are represented by coloured squares, an asterisk for MDR isolates and circles for non-MDR, respectively. Bootstrap values are shown on each node.
Antibiotic resistance-related genes
The variable profile of antibiotic resistance identified among the strains led us to analyse the known resistance-associated genes in the sequenced genomes. These included genes encoding intrinsic and acquired β-lactamases, genes that confer resistance to aminoglycosides, fluoroquinolones, chloramphenicol, macrolides and tetracycline, as well as efflux pump-associated genes, even though not all may necessarily be required for resistance (Fig. 1).
A total of 55 antibiotic resistance genes were detected across all 27 isolates. Three types of β-lactamase genes encoding enzymes for class A, class C and class D β-lactamase superfamilies were detected in our isolates. The intrinsic AmpC β-lactamase (bla ADC) and OXA-51 serine-type oxacillinase (bla OXA-51) genes were found in all A. baumannii isolates (n=27) [53]. However, translations of the sequence revealed different STs corresponding to different protein clades for products of both these genes (Figs S3 and S4) [54]. Such differences in sequence may be allied with the differences in the phenotypes seen. Further, the OXA24 type gene (bla OXA72) and OXA58 type gene (bla OXA-420), which confer carbapenem resistance, were found in hospital isolates H-79532 and H-11553, respectively. Another widely distributed carbapenem-resistance gene, bla OXA23, and an extended-spectrum β-lactamase (ESBL) gene (TEM-12) were only detected in hospital strains (n=11/15). Two community isolates also harboured ESBL containing the gene for TEM-116.
The mechanisms by which A. baumannii develops resistance to aminoglycoside agents are varied, but they almost always involve the production of aminoglycoside-modifying enzymes. These enzymes can be categorized as aminoglycoside acetyltransferases (AAC), aminoglycoside phosphotransferases (APH), and/or aminoglycoside nucleotidyltransferases (ANT or AAD), depending on their specific functions [55]. We only found aminoglycoside O-phosphotransferase (APH(3′)-Ia (n=9/27), APH(3″)-Ib (n=11/27) and APH(6)-Id) (n=11/27) in hospital isolates (none of the community isolates). Aminoglycoside nucleotidyltransferases ANT(3″)-IIa, intrinsic in this species, were found in all the isolates (n=27). Another major resistance mechanism to aminoglycoside is the acquisition of a 16S methyltransferase (armA), which confers broad-spectrum resistance to all clinically relevant drugs in this class of antibiotics, was also only found in the hospital strains (n=10/27) [56]. Three isolates (H-11699, C-72 and C-65) showed amikacin resistance despite the lack of the armA gene. The reasons for this would need further investigation. The sulfonamide resistance dihydropteroate synthase gene sul2 was found in three hospital strains. However, tetB was the most frequently identified gene among MDR hospital strains, followed by macrolide resistance genes (mphE and msrE) (n=12/15), which conforms with the phenotype data.
Antimicrobial resistance in A. baumannii has been associated with four families of efflux pumps: the resistance nodulation division (RND) family, the major facilitator superfamily (MFS) family, the multidrug and toxic compound extrusion (MATE) family, and the small multidrug resistance (SMR) family [57]. The adeABC and RND-type efflux pumps are not only associated with aminoglycoside resistance but also with resistance to tigecycline lactams, chloramphenicol, erythromycin and tetracycline [58]. The complete adeABC package was detected in 12 hospital and 1 community A. baumannii isolates, with another 10 isolates carrying either 1 or 2 genes. Other RND-type efflux pumps, including adeFGH and adeIJK, which can contribute to multidrug resistance in A. baumannii , were commonly found in all 27 isolates. Apart from this, MFS efflux pumps, abaQ (n=23) and amvA (n=27) mediate resistance to different types of antibiotics, including fluoroquinolones and macrolides [59, 60]. We found the SMR efflux pump-related gene abeS in 25 isolates, whereas a MATE family pump, abeM, was present in all A. baumannii isolates in our study. The chloramphenicol exporter gene, flor, was detected in a hospital strain, H-11553.
Virulence factors
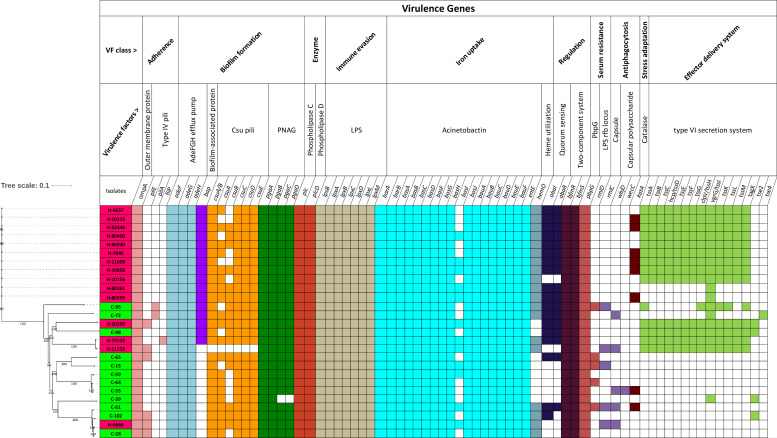
All sequenced strains were examined for genes encoding virulence factors selected from the virulence factor database (VFDB). The results are summarized in Fig. 3. Similar virulence genes were present in most community and hospital isolates, which suggests the pathogenic nature of most strains in the current study.
Fig. 3.
Presence of genes involved in virulence in the community and hospital A. baumannii genomes. The names of the isolates are shown on the left of the figure and are colour coded, as shown in Fig. 2. The isolates were organized here along with a phylogenetic tree (left) constructed by whole-genome SNP analysis. The presence of genes in an isolate is specified by a coloured rectangle, with different genes being coloured differently according to their mechanistic traits. The absence of genes is shown as white rectangles. These genes are grouped in the figure according to the mechanistic trait they are predicted to impart to A. baumannii strains.
However, there was a clear difference in the presence of genes encoding the type VI secretion system, which plays a vital role in the virulence of A. baumannii . The set of genes are present in 13 isolates, with another 5 isolates carrying some genes of this system [61]. Strikingly, the complete set of these genes, including core tss (tssA-M) and tag, was only found in 12 hospital isolates (not found in H-6668, H-80359 and H-80361) and 1 community isolate (C-98). A chi-square test revealed significant associations with presence in hospital strains and absence in community strains for 13 of the 15 genes tested that make up the type VI secretion system (P<0.05) (Table S4). The only genes not associated are tse2 and tse4. Additionally, the gene encoding the biofilm-associated protein (bap), which acts in biofilm formation [62, 63], was found in 13 hospital and 3 community strains. This gene is complex in nature, having a long coding sequence comprising a variable number of repetitive regions [64].
Other virulence genes did not show any significant difference in frequency between hospital and community strains. To persist in iron-limited host habitats, A. baumannii develops high-affinity iron acquisition mechanisms, such as the siderophore acinetobactin [65]. Acinetobactin gene clusters (barAB, basA-J, bauA-F, entE) were present in almost all community and hospital isolates in this study, with the single exception that basI was not present in 15 isolates. Genes involved in biofilm and pili formation, adherence, quorum sensing, lipid A biosynthesis, phospholipase, two-component regulator systems and serum resistance were also detected in most of the A. baumannii isolates. The operon encoding the csu pili chaperone–usher assembly system that contributes to biofilm formation [66] and the pgaABCD operon, required for intercellular adhesin synthesis, were present in most of the isolates. OmpA, which codes for an outer-membrane protein, a key virulence factor that mediates bacterial biofilm formation, eukaryotic cell infection, antibiotic resistance and immunomodulation [67], was present in all isolates. The serum resistance gene pbpG (encoding penicillin-binding protein) was also found in all of our isolates.
Mobile genetic elements (MGEs)
High genetic plasticity in A. baumannii enables the accumulation of resistance determinants and the horizontal transfer of resistance genes through MGEs [68]. The presence of known MGEs, including transposons and insertion sequences in the sequenced community and hospital strains (shown in File S2; MGEs), was investigated. A search for composite transposons revealed that carbapenem resistance gene bla OXA23 was present inside Tn2007 in all strains that carry the gene. Another strain, H-11553, carried bla OXA-58 within a composite transposon of ISPssp2 (IS1 family). Apart from this, the transposon Tn6080 surrounding target site duplications was found to carry the bla OXA-51-like gene in all 27 isolates.
The current study also found 16 insertion sequences (ISs) from 8 different families: IS3, IS5, IS6, IS8, IS30, IS66, IS91 and IS256. Some were widely distributed among the genomes investigated, such as IS17, while others were restricted to a single isolate (e.g. ISAba10, ISAba49, ISAba27, ISEc29).
The CRISPR arrays of community and hospital isolates were found to differ considerably
Out of 27 sequenced strains in this study, CRISPR arrays were detected in 8 community and 2 hospital strains (Table S6). Two CRISPR arrays were detected in three community strains (C-28, C-39, C-61) and one hospital strain (H-79532). No CRISPR arrays were present in any MDR strain. Antibiotic-susceptible strains or those with a lower number of resistance genes had higher instances of CRISPR. The arrays exhibited a series of repeated sequences and spacers associated with type I-F CRISPR systems.
We found identical spacer and repeat sequences between community strain C-61 and hospital strain H-6668. All 20 spacers of the C-61 CRISPR2 array were identical to the spacers of H-6668. This suggests a very close ancestral history between these isolates, consistent with their close placement in the phylogram in Fig. 2. We also detected identical spacers among two community strains, C-61 and C-28. However, no significant association was found between the CRISPR array and antibiotic resistance in our 27 isolates. (P-value >0.05).
Pangenome analysis
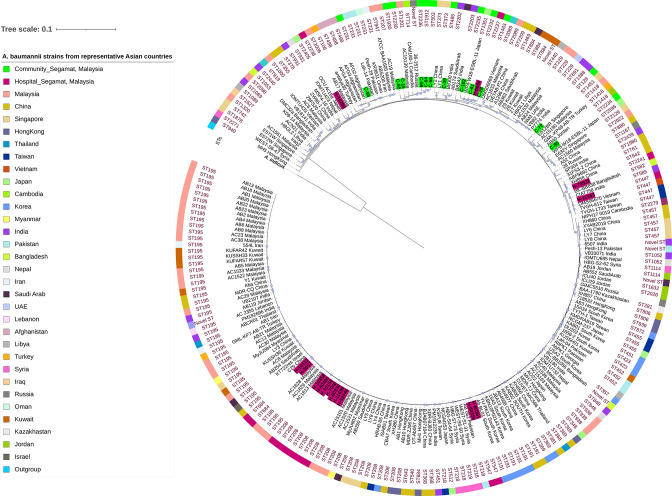
The functional adaptability of bacterial species can be better understood by extensive analysis of their pan-genome [69]. This analysis helps to identify core and accessory genes in the genome of A. baumannii species that might contribute to finding an ancestral relationship based on their genomic variation [70]. To assess the ancestral relationship of A. baumannii strains circulating in the Asian region, the gene content of the 27 genomes in this study was compared with another 191 A . baumannii sequences from Asian countries retrieved from the National Center for Biotechnology Information (NCBI) database (File S2; A. baumannii genomes from NCBI). The pangenome comprised a total of 22 885 genes, of which 18 682 (81.63 %), 1966 (8.59 %), 1481 (6.47 %) and 756 (3.30 %) were identified as cloud, shell, soft-core and core genes shared among the 218 isolates. Cloud (0 %<=strains<15 %), shell (15 %<=strains<95 %) and soft-core genes (95 %<=strains<99 %) are the accessory genes (gene set shared within one or some strains) as opposed to core genes (99 %<=strains<=100 %) that are shared by almost all clade members [71]. In these isolates, the proportion of the pan-genome-containing genes related to transcriptional regulators and transporters was 3.15 % (721) and 3.70 % (847), respectively. Moreover, we discovered genes that code for transposases and bacteriophage proteins (heads, tails and capsids) in 3.39 % (727) and 1.52 % (327) of the accessory genome, respectively. Genes encoding hypothetical proteins were found in 0.61 % (140) of core genomes and 45.64 % (10 446) of accessory genomes. A core gene-based phylogenetic tree was created from the pangenome, which reflects the relationships between the genomes of our study and selected other genomes (Fig. 4). The phylogenetic tree could be divided into numerous clades based on STs. Acinetobacter indicus was used as an outgroup. We observed a similar clustering pattern to that of the SNP-based phylogeny. The hospital strain H-6668 clustered with C-28, C-61 and C-102, consistent with the SNP phylogeny, and again indicating a close relationship between these isolates. Similar clustering was also observed between C-98 and H-10299 in the SNP phylogeny, as we found them in the same branch in pangenome phylogeny. When we made a comparison with other Asian countries, 10 hospital and 2 community isolates from our study clustered with other Malaysian isolates. The remaining 15 hospital and community isolates were clustered with isolates from PR China, Japan, the Republic of Korea, Vietnam, India, Bangladesh, Singapore, the Russian Federation and Afghanistan.
Fig. 4.
Core-genome-derived phylogenetic tree from pangenomes of 191 A . baumannii strains from Asian countries retrieved from the NCBI and the 27 A . baumannii strains from the current study. Terminal branches are labelled with the strain name and representative country names. Sequence types (STs) determined using the Oxford MLST scheme. Samples characterized in this study are shown in purple (hospital strains) and green (community strains). The blue circle symbols near the branches indicate bootstrap values of 100,
Discussion
A. baumannii is one of the most frequently encountered pathogens in human infections and is recognized as a significant reservoir of MDR genes [72]. Most of the previous studies have focused on A. baumannii isolated from hospitals and only a few studies have focused on isolates from the community or environment [73, 74]. Ours is the first study investigating the epidemiology of A. baumannii recovered from the human gut as a potential reservoir in the community and from a tertiary care hospital located in the same district to establish whether there is possible transmission between these two sources. This is of crucial importance in the field of epidemiology due to A. baumannii being a priority pathogen with scarce data on community carriage, especially in healthy individuals in Southeast Asia [75].
The isolation rate of A. baumannii from the human faecal samples in the Segamat community was 3.9 % (n=9/233) and 2.4 % (n=3/126) in 2018 and 2019, respectively, which is consistent with prevalence rates from faecal samples reported in Senegal 5.4 % (n=39/774), the UK and the Netherlands (2/226) [74, 75]. It is possible that a more exhaustive analysis of individual faecal samples would have revealed greater carriage rates, but that was not possible due to our large sample sizes. Several studies have also focused on human skin as a community reservoir of A. baumannii . Various studies have found the prevalence of A. baumannii on human skin to be 0.5, 2.5 % or much higher [74, 76, 77]. Thus, despite A. baumannii not being widespread in non-hospitalized individuals, it can be, and has been, recovered at low frequencies from the skin and faecal flora. The 12 isolates from the community in our study appear not to be commensal. However, these organisms could act as potential reservoirs of genes and transfer antibiotic resistance genes horizontally to other bacterial species in the human gut and impact on community health during their carriage in the gut. Besides community isolates, we obtained 15 A . baumannii isolates from hospitalized individuals and the majority were from tracheal aspirates (7/15).
A comparison of antibiotic resistance profiles between community and hospital isolates revealed that the hospital isolates had an elevated level of resistance. Most community isolates were either susceptible or showed intermediate resistance to the antibiotics tested. Our study found colistin- and polymyxin B-resistant hospital strains. These two antibiotics have become the last-line therapy for antibiotic-resistant A. baumannii infections [78]. Moreover, 80 % of the hospital isolates from Segamat were highly resistant to carbapenem (imipenem and meropenem). Malaysia’s National Surveillance on Antibiotic Resistance (NSAR) reported that countrywide carbapenem resistance rates were ~50–60 % from 2008 to 2016 [79], and even higher rates have been reported in the university hospital in Kuala Lumpur [80]. These studies, along with ours, suggest that carbapenem-resistant A. baumannii strains are widely distributed in hospital settings, with a risk of spreading into the community.
Considerable differences were found between the MDR and non-MDR phenotype and genotype data among the community and hospital A. baumannii isolates, which were expected, as the hospital is known to be a hotspot for MDR bacteria. AMR gene analysis based on whole-genome sequences revealed that all strains carried beta-lactam resistance genes (OXA51, bla ADC). However, most community and three hospital strains were sensitive to beta-lactam antibiotics. We have found several point mutations in beta-lactamase genes in non-resistant isolates leading to non-conservative amino acid changes. These changes may lead to lower functionality of the protein products. However, this needs further testing. In addition, the carbapenem resistance OXA-23 and OXA-24 like genes were identified in 11 and 1 hospital strains, respectively, confirming our AST results indicating that these strains were resistant to carbapenems. Apart from AMR, WGS revealed several virulence factors that were found in all 27 sequenced strains. Genes coding for the type VI secretion system (T6SS), which is known as a major virulence factor in A. baumannii , were found in most of the hospital strains and one community strain. T6SS genes can be exploited to produce toxins that kill other bacteria and even eukaryotic cells [81].
MGEs play an essential role in regulating and disseminating antimicrobial resistance genes [64]. Composite transposons (Tn2006, Tn2007, Tn2008 and Tn2009) flanked by two copies of similar insertion sequence elements are associated with the transfer of bla OXA23 in A. baumannii [82]. Our study also found the presence of OXA23 genes within Tn2007 in MDR hospital strains. Likewise, transposons and different insertion sequences were widely distributed among community strains. The role of MGEs in genetic exchange between hospital and community strains was not explored.
The CRISPR–Cas mechanism is considered to be an adaptive immune system based on identifying past infections in prokaryotes. According to CRISPRCasdb [83], roughly 20 % of genus Acinetobacter representatives and 18 % of isolates of A. baumannii species possess both CRISPR arrays and Cas genes [84]. The current analysis discovered putative CRISPR–Cas systems in eight community and just two hospital strains, and all of these isolates were susceptible to antibiotics. Isolates lacking CRISPR arrays and active Cas genes were demonstrated to contain considerably more antibiotic resistance genes than those lacking either or both, although the numbers analysed were not sufficient to produce a statistically significant result. These results suggest a link between the susceptible and resistant genotype/phenotypic strain of A. baumannii and the type of its CRISPR–Cas system. Thus, CRISPR–Cas systems could also play a critical role in controlling the expression of many genes that determine the resistance pattern and pathogenicity of isolates and the propagation of antibiotic resistance genes in A. baumannii [84, 85].
Comparative whole-genome sequence analysis of the community and hospital A. baumannii strains revealed the possible ancestral relationship of some isolates within these settings. Nearly similar clustering was observed in both phylogenetic trees after whole-genome SNP and pangenome-based core gene analysis. The drug-susceptible isolates in this study were distinct from the MDR strains and there was apparent clustering between the hospital isolate H-6668 and the community strains C-28, C-61 and C-102. Furthermore, C-61 and H-6668 had identical CRISPR arrays. All of these isolates were found to be antibiotic-susceptible. We also observed another cluster between H-10299 and C-98 after pangenome and SNP core genome phylogeny. There was also one cluster containing MDR strain H-11553 and non-MDR strain H-79532. Both of the strains carried similar beta-lactam resistance genes with certain mutations (ADC-76 and OXA-66). They also carried similar type VI secretion-related genes.
These observations are consistent with previous reports of transmission from patient to patient, patient to health care worker, patient to the environment and other related sources [86]. The results demonstrate that there could have been a common ancestor of some of these hospital and community A. baumannii isolates if we disregard the possibility of adventitious isolation.
Conclusion
This study characterized the variation between A. baumannii strains isolated from two different sources in a district in Malaysia as a pilot study. We found that the human gut could potentially act as a reservoir for A. baumannii colonization, even though carriage in the same individuals was not detected after a year. Hospitals have been identified as the primary source of MDR A. baumannii in Malaysia. Less drug resistance was demonstrated by community isolates, indicating that higher antibiotic selection pressure occurred in the hospital settings. Comparative WGS results of A. baumannii strains revealed the possible ancestral relationship between some isolates, suggesting the chances of a single origin for some of the strains in these two different settings. This study also suggests that the community strains could easily turn into pathogenic strains if they gain resistance genes via horizontal gene transfer, perhaps by losing the CRISPR loci. Studies focusing on a larger sample of A. baumannii isolates from the community, hospitals and surrounding environments in different geographical locations would improve our understanding of this relationship. This study sheds light on the public health concerns associated with the dissemination of A. baumannii in a healthy community and may aid in devising public health measures to limit the spread of this pathogen in the future.
Supplementary Data
Funding information
This study was supported by the Fundamental Research Grant Scheme (FRGS) from the Ministry of Education (MOE), Malaysia (grant number FRGS/1/2019/SKK01/MUSM/01/1), the 2017 Monash Malaysia Strategic Large Grant Scheme (LG-2017–01-SCI) and the 2017 Tropical Medicine and Biology Grant for Malaysian Microbiome in health and disease project.
Acknowledgements
The authors would like to express their gratitude to Monash University Malaysia, the Tropical Medicine and Biology Multidisciplinary Platform and the South East Asia Community Observatory (Segamat, State of Johor, Malaysia) for their support. They are also thankful to Associate Professor Dr Lee Sui Mae for her support during sample collection and project initiation.
Author contributions
N.H.M.: conceptualization, methodology, formal analysis, investigation, resources, writing – original draft preparation. M.H.H.: analysis, writing – review and editing. M.A.L.H.: writing – review and editing. J.D. writing – review and editing. T.T.S.: writing – review and editing, funding. D.R.: writing – review and editing, funding. F.M.: sampling, funding. Q.A.: writing – review and editing, supervision. H.S.T.: methodology, writing – review and editing, supervision. S.R.: conceptualization, methodology, writing – review and editing, supervision, funding.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Authorization to conduct the study was attained from the Monash University Human Research Ethics Committee (MUHREC, project number: 1516), which is in accordance with the WMA Declaration of Helsinki (WMA and World Medical Association 2013) and Medical Research and Health Committee, Ministry of Health Malaysia (project number: NMRR-17-3524-36764-IIR).
Footnotes
Abbreviations: AMR, antimicrobial resistance; ATCC, American Type Culture Collection; CLSI, Clinical and Laboratory Standard Institute; IC, international clone; IDSA, Infectious Diseases Society of America; MDR, multidrug resistant; MGE, mobile genetic element; MLST, multilocus sequence typing; SEACO, South East Asia Community Observatory; SNP, single nucleotide polymorphisms; SNV, single nucleotide variants; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary material files are available with the online version of this article.
References
- 1.Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10:1–31. doi: 10.3390/pathogens10030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuya EY, Lowy FD. Antimicrobial-resistant bacteria in the community setting. Nat Rev Microbiol. 2006;4:36–45. doi: 10.1038/nrmicro1325. [DOI] [PubMed] [Google Scholar]
- 3.Cave R, Cole J, Mkrtchyan HV. Surveillance and prevalence of antimicrobial resistant bacteria from public settings within urban built environments: challenges and opportunities for hygiene and infection control. Environ Int. 2021;157:106836. doi: 10.1016/j.envint.2021.106836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldeyab MA, Harbarth S, Vernaz N, Kearney MP, Scott MG, et al. The impact of antibiotic use on the incidence and resistance pattern of extended-spectrum beta-lactamase-producing bacteria in primary and secondary healthcare settings. Br J Clin Pharmacol. 2012;74:171–179. doi: 10.1111/j.1365-2125.2011.04161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeana C, Larson E, Sahni J, Bayuga SJ, Wu F, et al. The epidemiology of multidrug-resistant Acinetobacter baumannii: does the community represent a reservoir? Infect Control Hosp Epidemiol. 2003;24:275–279. doi: 10.1086/502209. [DOI] [PubMed] [Google Scholar]
- 6.Karanika S, Karantanos T, Arvanitis M, Grigoras C, Mylonakis E. Fecal colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin Infect Dis. 2016;63:310–318. doi: 10.1093/cid/ciw283. [DOI] [PubMed] [Google Scholar]
- 7.Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asif M, Alvi IA, Rehman SU. Insight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect Drug Resist. 2018;11:1249–1260. doi: 10.2147/IDR.S166750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekic S, Hrenovic J, Ivankovic T, van Wilpe E. Survival of ESKAPE pathogen Acinetobacter baumannii in water of different temperatures and pH. Water Sci Technol. 2018;78:1370–1376. doi: 10.2166/wst.2018.409. [DOI] [PubMed] [Google Scholar]
- 10.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 11.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemec A, Dijkshoorn L, van der Reijden TJK. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J Med Microbiol. 2004;53:147–153. doi: 10.1099/jmm.0.05445-0. [DOI] [PubMed] [Google Scholar]
- 13.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed army medical center. Antimicrob Agents Chemother. 2006;50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wareth G, Linde J, Nguyen NH, Nguyen TNM, Sprague LD, et al. WGS-based analysis of carbapenem-resistant Acinetobacter baumannii in Vietnam and molecular characterization of antimicrobial determinants and MLST in Southeast Asia. Antibiotics (Basel) 2021;10:1–15. doi: 10.3390/antibiotics10050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh H, Thangaraj P, Chakrabarti A. Acinetobacter baumannii: a brief account of mechanisms of multidrug resistance and current and future therapeutic management. J Clin Diagn Res. 2013;7:2602–2605. doi: 10.7860/JCDR/2013/6337.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii. Virulence. 2012;3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doughari HJ, Ndakidemi PA, Human IS, Benade S. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ. 2011;26:101–112. doi: 10.1264/jsme2.me10179. [DOI] [PubMed] [Google Scholar]
- 18.Rafei R, Hamze M, Pailhoriès H, Eveillard M, Marsollier L, et al. Extrahuman epidemiology of Acinetobacter baumannii in Lebanon. Appl Environ Microbiol. 2015;81:2359–2367. doi: 10.1128/AEM.03824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 20.Hamouda A, Findlay J, Al Hassan L, Amyes SGB. Epidemiology of Acinetobacter baumannii of animal origin. Int J Antimicrob Agents. 2011;38:314–318. doi: 10.1016/j.ijantimicag.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Meumann EM, Anstey NM, Currie BJ, Piera KA, Kenyon JJ, et al. Genomic epidemiology of severe community-onset Acinetobacter baumannii infection. Microb Genom. 2019;5:e000258. doi: 10.1099/mgen.0.000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung WS, Chu CM, Chan VL, Lam JY, Ho PL. Fulminant community acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest. 2005;128:149S. doi: 10.1378/chest.128.4_MeetingAbstracts.149S. [DOI] [PubMed] [Google Scholar]
- 23.Wang JT, McDonald LC, Chang SC, Ho M. Community-acquired Acinetobacter baumannii bacteremia in adult patients in Taiwan. J Clin Microbiol. 2002;40:1526–1529. doi: 10.1128/JCM.40.4.1526-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G, Yang M, Zhou K, Zhang L, Tian L, et al. Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol. 2015;25:1136–1145. doi: 10.4014/jmb.1412.12047. [DOI] [PubMed] [Google Scholar]
- 25.Al Atrouni A, Joly-Guillou M-L, Hamze M, Kempf M. Reservoirs of non-baumannii Acinetobacter species. Front Microbiol. 2016;7:49. doi: 10.3389/fmicb.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partap U, Young EH, Allotey P, Soyiri IN, Jahan N, et al. HDSS profile: The South East Asia community observatory health and demographic surveillance system (SEACO HDSS) Int J Epidemiol. 2017;46:1370–1371G. doi: 10.1093/ije/dyx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huët MAL, Wong LW, Goh CBS, Hussain MH, Muzahid NH, et al. Investigation of culturable human gut mycobiota from the segamat community in Johor, Malaysia. World J Microbiol Biotechnol. 2021;37:1–15. doi: 10.1007/s11274-021-03083-6. [DOI] [PubMed] [Google Scholar]
- 28.Alsan M, Klompas M. Acinetobacter baumannii: an emerging and important pathogen. J Clin Outcomes Manag. 2010;17:27–35. [PMC free article] [PubMed] [Google Scholar]
- 29.Schuurman T, de Boer RF, Kooistra-Smid AMD, van Zwet AA. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol. 2004;42:734–740. doi: 10.1128/JCM.42.2.734-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dashti A, Jadaon M, Abdulsamad A, Dashti H. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J. 2009;41 [Google Scholar]
- 31.Vijayakumar S, Biswas I, Veeraraghavan B. Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci OA. 2019;5:FSO395. doi: 10.2144/fsoa-2018-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins PG, Lehmann M, Wisplinghoff H, Seifert H. gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J Clin Microbiol. 2010;48:4592–4594. doi: 10.1128/JCM.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CLSI CLSI supplement M100: Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 34.Amin M, Navidifar T, Shooshtari FS, Rashno M, Savari M, et al. Association between biofilm formation, structure, and the expression levels of genes related to biofilm formation and biofilm-specific resistance of Acinetobacter baumannii strains isolated from burn infection in Ahvaz, Iran. Infect Drug Resist. 2019;12:3867–3881. doi: 10.2147/IDR.S228981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin MF, Lin YY, Lan CY. Characterization of biofilm production in different strains of Acinetobacter baumannii and the effects of chemical compounds on biofilm formation. PeerJ. 2020;8:e9020. doi: 10.7717/peerj.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Xia J, Xu Y, Gong M, Zhou Y, et al. Biological features of biofilm-forming ability of Acinetobacter baumannii strains derived from 121 elderly patients with hospital-acquired pneumonia. Clin Exp Med. 2016;16:73–80. doi: 10.1007/s10238-014-0333-2. [DOI] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De novo assembler. Curr Protoc Bioinformatics. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 39.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, et al. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii . J Clin Microbiol. 2005;43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Yang J, Yu J, Yao Z, Sun L, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:325–328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitto F, Battistuzzi FU. Estimating pangenomes with roary. Mol Biol Evol. 2020;37:933–939. doi: 10.1093/molbev/msz284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, et al. CRISPRCasFinder, an update of crisrfinder, includes a portable version, enhanced performance and integrates search for cas proteins. Nucleic Acids Res. 2018;46:W246–51. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimura D, Kajitani R, Gotoh Y, Katahira K, Okuno M, et al. Evaluation of SNP calling methods for closely related bacterial isolates and a novel high-accuracy pipeline: BactSNP. Microb genomics. 2019;5:1–8. doi: 10.1099/mgen.0.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balaban M, Moshiri N, Mai U, Jia X, Mirarab S. TreeCluster: clustering biological sequences using phylogenetic trees. PLoS One. 2019;14:e0221068. doi: 10.1371/journal.pone.0221068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bian X, Liu X, Zhang X, Li X, Zhang J, et al. Correction to: epidemiological and genomic characteristics of A. baumannii from different infection sites using comparative genomics (BMC Genomics, (2021), 22, 1, (530), 10.1186/s12864-021-07842-5) BMC Genomics. 2021;22:649. doi: 10.1186/s12864-021-07942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, et al. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans BA, Amyes SGB. OXA β-lactamases. Clin Microbiol Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tahbaz SV, Azimi L, Lari AR. Characterization of aminoglycoside resistance mechanisms in Acinetobacter baumannii isolates from burn wound colonization. Ann Burns Fire Disasters. 2019;32:115–121. [PMC free article] [PubMed] [Google Scholar]
- 56.Hasani A, Sheikhalizadeh V, Ahangarzadeh Rezaee M, Rahmati-Yamchi M, Hasani A, et al. Frequency of aminoglycoside-modifying enzymes and ArmA among different sequence groups of Acinetobacter baumannii in Iran. Microb Drug Resist. 2016;22:347–353. doi: 10.1089/mdr.2015.0254. [DOI] [PubMed] [Google Scholar]
- 57.Lin MF, Lin YY, Tu CC, Lan CY. Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J Microbiol Immunol Infect. 2017;50:224–231. doi: 10.1016/j.jmii.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Abdi SN, Ghotaslou R, Ganbarov K, Mobed A, Tanomand A, et al. Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect Drug Resist. 2020;13:423–434. doi: 10.2147/IDR.S228089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pérez-Varela M, Corral J, Aranda J, Barbé J. Functional characterization of AbaQ, a novel efflux pump mediating quinolone resistance in Acinetobacter baumannii . Antimicrob Agents Chemother. 2018;62:1–4. doi: 10.1128/AAC.00906-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajamohan G, Srinivasan VB, Gebreyes WA. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii . J Antimicrob Chemother. 2010;65:1919–1925. doi: 10.1093/jac/dkq195. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Zhou Z, He F, Ruan Z, Jiang Y, et al. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One. 2018;13:e0192288. doi: 10.1371/journal.pone.0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brossard KA, Campagnari AA. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun. 2012;80:228–233. doi: 10.1128/IAI.05913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Longo F, Vuotto C, Donelli G. Biofilm formation in Acinetobacter baumannii . New Microbiol. 2014;37:119–127. [PubMed] [Google Scholar]
- 64.Leal NC, Campos TL, Rezende AM, Docena C, Mendes-Marques CL, et al. Comparative genomics of Acinetobacter baumannii clinical strains from Brazil reveals polyclonal dissemination and selective exchange of mobile genetic elements associated with resistance genes. Front Microbiol. 2020;11:1176. doi: 10.3389/fmicb.2020.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheldon JR, Skaar EP. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020;16:e1008995. doi: 10.1371/journal.ppat.1008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 67.Nie D, Hu Y, Chen Z, Li M, Hou Z, et al. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J Biomed Sci. 2020;27:26. doi: 10.1186/s12929-020-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Li H, Zhang J, Wang H. Co-Occurrence of bla(OXA-23) in the chromosome and plasmid: increased fitness in carbapenem-resistant Acinetobacter baumannii . Antibiotics (Basel, Switzerland) 2021;10:1196. doi: 10.3390/antibiotics10101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hassan A, Naz A, Obaid A, Paracha RZ, Naz K, et al. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genomics. 2016;17:732. doi: 10.1186/s12864-016-2951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaba S, Kumari A, Medema M, Kaushik R. Pan-genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super-order. Sci Rep. 2020;10:21205. doi: 10.1038/s41598-020-77723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park SC, Lee K, Kim YO, Won S, Chun J. Large-scale genomics reveals the genetic characteristics of seven species and importance of phylogenetic distance for estimating pan-genome size. Front Microbiol. 2019;10:834. doi: 10.3389/fmicb.2019.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo L, Wei D, Zhang X, Wu Y, Li Q, et al. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol. 2019;10:2752. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeana C, Larson E, Sahni J, Bayuga SJ, Wu F, et al. The epidemiology of multidrug-resistant Acinetobacter baumannii: does the community represent a reservoir? Infect Control Hosp Epidemiol. 2003;24:275–279. doi: 10.1086/502209. [DOI] [PubMed] [Google Scholar]
- 74.Dijkshoorn L, van Aken E, Shunburne L, van der Reijden TJK, Bernards AT, et al. Prevalence of Acinetobacter baumannii and other Acinetobacter spp. in faecal samples from non-hospitalised individuals. Clin Microbiol Infect. 2005;11:329–332. doi: 10.1111/j.1469-0691.2005.01093.x. [DOI] [PubMed] [Google Scholar]
- 75.Kempf M, Rolain J-M, Diatta G, Azza S, Samb B, et al. Carbapenem resistance and Acinetobacter baumannii in Senegal: the paradigm of a common phenomenon in natural reservoirs. PLoS One. 2012;7:e39495. doi: 10.1371/journal.pone.0039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berlau J, Aucken H, Malnick H, Pitt T. Distribution of Acinetobacter species on skin of healthy humans. Eur J Clin Microbiol Infect Dis. 1999;18:179–183. doi: 10.1007/s100960050254. [DOI] [PubMed] [Google Scholar]
- 77.Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, et al. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol. 1997;35:2819–2825. doi: 10.1128/jcm.35.11.2819-2825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dortet L, Potron A, Bonnin RA, Plesiat P, Naas T, et al. Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci Rep. 2018;8:16910. doi: 10.1038/s41598-018-35041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmad N, Hashim R, Amran F, Zamri H, Ling L, et al. Inst Med Res Kuala Lumpur Malaysia; 2017. National antimicrobial surveillance report. [Google Scholar]
- 80.Kong BH, Hanifah YA, Yusof MYM, Thong KL. Antimicrobial susceptibility profiling and genomic diversity of multidrug-resistant Acinetobacter baumannii isolates from a teaching hospital in Malaysia. Jpn J Infect Dis. 2011;64:337–340. [PubMed] [Google Scholar]
- 81.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, et al. Genomic and functional analysis of the type VI secretion system in Acinetobacter . PLoS One. 2013;8:e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu L-L, Ji S-J, Ruan Z, Fu Y, Fu Y-Q, et al. Dissemination of blaOXA-23 in Acinetobacter spp. in China: main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob Agents Chemother. 2015;59:1998–2005. doi: 10.1128/AAC.04574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:1–10. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tyumentseva M, Mikhaylova Y, Prelovskaya A, Tyumentsev A, Petrova L, et al. Genomic and phenotypic analysis of multidrug-resistant Acinetobacter baumannii clinical isolates carrying different types of CRISPR/Cas systems. Pathogens. 2021;10:205. doi: 10.3390/pathogens10020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 86.Uwingabiye J, Lemnouer A, Roca I, Alouane T, Frikh M, et al. Clonal diversity and detection of carbapenem resistance encoding genes among multidrug-resistant Acinetobacter baumannii isolates recovered from patients and environment in two intensive care units in a Moroccan hospital. Antimicrob Resist Infect Control. 2017;6:1–9. doi: 10.1186/s13756-017-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.