Abstract
The micronuclear DNA of Paramecium tetraurelia is estimated to contain over 50,000 short DNA elements that are precisely removed during the formation of the transcriptionally active macronucleus. Each internal eliminated sequence (IES) is bounded by 5′-TA-3′ dinucleotide repeats, a feature common to some classes of DNA transposons. We have developed an in vivo assay to analyze these highly efficient and precise DNA excision events. The microinjection of a cloned IES into mating cells results in accurately spliced products, and the transformed cells maintain the injected DNA as extrachromosomal molecules. A series of deletions flanking one side of a 28-bp IES were constructed and analyzed with the in vivo assay. Whereas 72 bp of DNA flanking the eliminated region is sufficient for excision, lengths of 31 and 18 bp result in reduced excision and removal of all wild-type sequences adjacent to the TA results in complete failure of excision. In contrast, nucleotide mutations within the middle of the 28-bp IES do not prevent excision. The results are consistent with a functional role for perfect inverted repeats flanking the IES.
Ciliated protozoa provide a unique biological system for the study of DNA rearrangements. During sexual reproduction, a transcriptionally active macronucleus is formed from the germ line DNA in the micronucleus. The precise details of the process vary among different ciliates, but common features include fragmentation of germ line chromosomes, elimination of specific DNA elements, and amplification of the macronucleus-destined linear fragments (reviewed in references 2, 13, and 21). Elimination of relatively small regions of the genome (14 bp to several kilobases) followed by rejoining of the adjacent sequences has been observed in a wide variety of ciliate species. These excised DNA elements are commonly referred to as internal eliminated sequences (IESs) to distinguish them from elimination that results from fragmentation events.
In Paramecium tetraurelia, the micronuclear genome contains relatively short IESs (26 bp to about 1 kb) that always begin and end with a 5′-TA-3′ dinucleotide. Excision results in precise removal of the element, leaving a single TA within the macronuclear DNA (4, 5, 15, 23, 26, 27). No significant open reading frames are encoded by these elements, and comparison of evolutionarily related IESs within the variable surface antigen genes of P. tetraurelia revealed substantial variation in the size and sequence of an IES relative to the adjacent macronuclear DNA (23). The ends of the element generally include a perfect inverted repeat that includes the TA and extends either into the IES or out toward the macronucleus-destined DNA (26). Statistical analysis of 20 IESs from P. tetraurelia identified an 8-bp consensus inverted terminal repeat that includes the invariant TA dinucleotide (12). This consensus repeat is inside the IES and therefore does not necessarily include the perfect inverted repeats. The functional significance of the consensus repeat is supported by the analysis of Paramecium mutant cell lines defective in IES excision. Isolated cell lines that are unable to excise a specific IES contain single nucleotide mutations in the consensus region (16, 17).
The structure of a Paramecium IES can be complex. There are at least two examples in which one IES is located inside a larger IES (6, 16, 17). In this report (which focuses on one of the complex IESs), we will refer to the smaller IES located inside another as an internal IES. The enzymatic machinery responsible for IES excision has not been identified, but analysis of a pleiotropic mutant line has shown that excision of one IES (or a small subclass) is inhibited by a mutation in an unlinked locus (19).
Paramecium is not the only ciliate that contains TA IESs (IESs bounded by TA repeats). Euplotes crassus contains IESs bounded by 5′-TA-3′ direct repeats, and they have similar consensus terminal inverted repeats (reviewed in reference 11). Interestingly, the consensus repeats from both Paramecium and Euplotes have some similarity to the termini of Tc1 transposable elements. These findings combined with a long history of other observations led to models that proposed at least some IESs are ancestral remnants of transposons (reviewed in reference 13). Unfortunately, evaluation of this hypothesis and other investigations of cis-acting functional requirements for TA IES excision have been difficult because of technical limitations.
To date, the most thoroughly investigated DNA elimination events are those in Tetrahymena thermophila (reviewed in reference 28). These eliminated sequences are not flanked by conserved 5′-TA-3′ repeats and generally contain short direct repeats at their boundary. Analysis of the M element revealed a requirement for a 10-bp DNA sequence (A5G5) located about 45 bp outside the left boundary that has a corresponding partner found in inverted orientation about 45 bp outside the right boundary (8). Inserting different lengths of DNA between the 10-bp sequence and the normal splice boundary does not prevent excision but alters the boundary in a corresponding manner so that a distance of about 45 bp is maintained (9). Therefore, the sequence element not only is required but also specifies the deletion boundary.
In this paper we describe an in vivo method for the analysis of Paramecium IES excision. We show that injection of cloned copies of an IES into cells undergoing sexual reproduction (the formation of a new macronucleus) results in accurately spliced DNA products. We used this procedure to analyze the effects of various deletions on excision of an internal 28-bp IES. The results show that flanking sequences are required for IES excision, but the alteration of four nucleotides inside the IES does not prevent excision. A decrease in excision efficiency is correlated with the location of an 8-bp perfect inverted repeat about 50 bp outside the eliminated region. Excision is completely abolished by a deletion that removes all flanking DNA adjacent to the TA dinucleotide. The possible relationship between Paramecium IESs and those in Tetrahymena is discussed.
MATERIALS AND METHODS
Cell lines, media, and culture conditions.
Stock d4-110 from the culture collection of J. Preer (Indiana University, Bloomington, Ind.) was used for all injection experiments. This derived strain of P. tetraurelia, stock 51, contains a mutant allele called high reactor (hrB) that maintains mating reactivity (induced by starvation) longer than wild-type cells (25). This strain was used as a matter of convenience to isolate mating cell pairs. All cells were cultured in a 0.25% wheat grass medium buffered with sodium phosphate (0.45 g/liter) and supplemented with stigmasterol (0.25 mg/liter). The medium was inoculated with a nonpathogenic strain of Klebsiella pneumoniae 1 day prior to use. All cell lines were maintained at 27°C and cultured as described by Sonneborn (24).
Transformation of mating cells.
Mating reactive cells were mixed; after 1 h, conjugating pairs were isolated into a separate depression well. Only those pairs that remained firmly united after forceful passage through a micropipette were used for injection. Microinjection was performed between 18 and 21 h after mixing (27°C), which is after the first postzygotic cell division but prior to formation of the mature macronucleus. Injection was performed as described by Godiska et al. (7) on an inverted microscope. DNAs were dissolved at a final concentration of 1 to 2 mg/ml in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Between 3 and 6 pl of this solution was injected into each cell by using a glass microneedle 1 to 2 μm in diameter at the tip. Most injections in this study used supercoiled plasmid, but transformation was also successful with linear plasmid DNA. Since the developing macronucleus is not visible under phase-contrast optics, injections were directed toward the mid-anterior region of the cell. After injection, individual cells were placed into 1-ml depression wells with fresh wheat grass medium and cultured without selection at 27°C for 24 h. Cell lines were treated with G418 (Sigma Chemical Company, St. Louis, Mo.) at a final concentration of 20 μg/ml for at least 2 days. Those cell lines surviving drug treatment were grown without selection in large cultures for DNA isolation. The transformation efficiency was typically between 1 and 3% of injected cells.
Plasmid construction.
All plasmids used in this study were made by inserting DNA fragments into the unique BamHI site of pPXV-NEO (10). PCR amplifications were used to generate the appropriate fragments, and DNA sequencing was used to confirm the sequence of each insert. The series of flanking deletions were constructed by using one primer located beyond a BglII site on the right flank of the IES (5′GCAGGTTGCTGGAGAGG) and the following primers that contain a BamHI site (underlined): pNEO-IES (5′GCATGGATCCGGCATGTAGAAGTGCAA), pIES-72 (5′CATGGGATCCGCTTTGAATTGTGAAATAATTC), pIES-31 (5′GCTAGGATCCGAAAGTAGGAAAAATTTAAAAAAG), pIES-18 (5′GCTAGGATCCATTTAAAAAAGAGTATGTTATAG), and pIES-1 (5′GTCAGGATCCTATAGTGATTATTAAAATAC). The resulting PCR products were digested with BglII and BamHI and inserted into the BamHI site of pPXV-NEO. All flanking A51 sequences distal to the primer were deleted, and the endpoint of the deletion was attached to the calmodulin promoter region (Fig. 1). The pIES-SacI insert was constructed by ligation of two PCR products. One product used the primer on the right flank of the IES (above) and a second primer (5′CGATGAGCTCTTGATTCATAAGTTTAAAAGC) complementary to the internal 28-bp IES except for a SacI site (underlined). The other PCR product used the pNEO-IES primer on the left flank (see above) with a second primer (5′CGATGAGCTCTAATAATCACTATAACATAC) complementary to the internal 28-bp IES containing a SacI site. Digestion of products with SacI and BamHI or BglII was followed by a three-way ligation into the BamHI site of pPXV-NEO. The inserts from all constructs were sequenced to confirm that no extraneous mutations were induced by PCR.
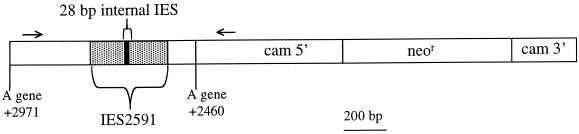
FIG. 1.
Diagram of the pIES-NEO plasmid insert. An 887-bp fragment containing the 370-bp A51 gene IES2591 and flanking DNA was ligated into the unique BamHI site of pPXV-NEO (10). Transformed cell lines were assayed for IES excision by PCR using one primer in the 5′ upstream region of the calmodulin gene (cam) and the other primer in the A51 flanking sequences (primers are represented by arrows). The calmodulin 5′ upstream sequence and 3′ downstream sequence flank the neomycin resistance coding region (neor).
Isolation of genomic DNA.
Total genomic DNA was isolated from 1-liter cultures (≥1,000 cells/ml). The cells were pelleted, resuspended in 0.5 ml of culture medium, and squirted into 4 ml of lysing solution (10 mM Tris-HCl [pH 9.5], 1% sodium dodecyl sulfate [SDS], 50 mM EDTA) at 65°C. After 10 min, the solution was phenol-chloroform (1:1) extracted and then ethanol precipitated. The pellet was dissolved in approximately 0.5 ml of TE (10 mM Tris, 1 mM EDTA [pH 8.0]).
PCR amplification.
Analysis of IES excision from the transformed cell lines was performed by PCR amplification of the region in the plasmid containing the A51 gene IES2591 followed by gel electrophoresis of the resulting products. The amplification reactions used one primer (5′GGCATTAAGCTTGTGTC) in the A51 gene and one primer (5′TGTATATAAAAAGGTTCAGAAGGG) in the calmodulin promoter that drives transcription of the antibiotic resistance gene (neo). Products were separated on 4% agarose gels (NuSieve 3:1 agarose; BioWhittaker Molecular Applications, Rockland, Maine). Independent excision of the internal 28-bp IES was assayed using one primer outside IES2591 (+3021 in the A51 gene, 5′GCAGGTTGCTGGAGAGG) and one primer complementary to the splice junction of the 28-bp internal IES (5′AAAAAAGAGTATGTTAAG). This set of primers can amplify a product from molecules that have excised the 28-bp internal IES but not the entire IES2591. Total genomic DNA isolated from various time points after a mass mating was used as the template for PCR.
Southern hybridization.
Southern blot analyses were performed by the method of Sambrook et al. (22). Gels were blotted to a nitrocellulose filter (Schleicher & Schuell, Keene, N.H.), which was UV cross-linked and washed in a solution of 10× Denhardt's solution 0.2 M phosphate buffer, 0.1% SDS, and 5× SET (1× SET is 0.15 M NaCl, 30 mM Tris, and 2 mM EDTA) at 65°C for 1 h. The filter was incubated in hybridization solution (1× Denhardt's solution, 0.02 M phosphate buffer, 5× SET, 0.25% SDS) for 1 h before the labeled probe was added. IES excision assay blots were probed with a 5′-end-labeled oligonucleotide (5′AAAGAGTATGTTAAGTTTAAAAGCTT) that is complementary to the splice junction of the 28-bp internal IES. The oligonucleotide hybridization filters were washed three times for 30 min each at 45°C with Wash II, containing 1× SET, 1× Denhardt's solution, 0.025 M phosphate buffer, and 0.1% sodium pyrophosphate. The filter containing PCR products to detect independent excision of the 28-bp internal IES (Fig. 4) was probed with a fragment starting at the 28-bp internal IES and ending at nucleotide position +3021 relative to the start of translation. The filter was washed three times for 30 min each at 65°C with Wash III, containing 0.2× SET, 0.025 M phosphate buffer, 0.1% sodium pyrophosphate, and 0.1% SDS.
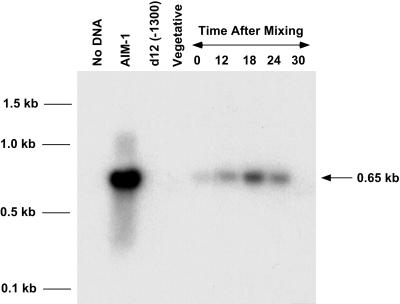
FIG. 4.
The 28-bp internal IES can be independently removed from IES2591. PCR was performed using one primer flanking IES2591 and one primer complementary to the spliced junction of the 28-bp internal IES. Mating reactive cells were mixed, and DNA was isolated 0, 12, 18, 24, and 30 h after mixing. The partially spliced product should be present only during sexual reproduction. Lanes: No DNA, no DNA PCR control; AIM-1, mutant cell line that eliminates the 28-bp internal IES but contains the remaining IES2591 in the macronucleus; d12 (−1300), deletion mutant of the A51 gene; Vegetative, wild-type vegetative cells.
RESULTS
Accurate excision of a cloned IES occurs in mated cells.
Our approach to the development of a DNA excision assay was to introduce a drug resistance gene along with an IES into the developing macronucleus and then select drug-resistant (Neor) transformants and assay for excision of the IES from the plasmid. We constructed a plasmid with an IES from the micronuclear copy of the A51 surface antigen gene (IES2591) inserted into a unique BamHI site in the drug resistance plasmid pPXV-NEO (10). The resulting pIES-NEO construct contains both the IES and a neo gene driven by the Paramecium calmodulin promoter (Fig. 1). Resistance to G418 does not require excision of the IES; therefore, transformation experiments followed by drug selection can identify cells containing the plasmid, but this does not necessarily mean they have excised the IES.
Cells were prepared for injection by mixing mating reactive cultures and isolating tight pairs 1 h later. The optimal time for injection was determined empirically. Injection of cells earlier than 18 h after mixing rarely produced transformants, and cells injected beyond 25 h showed little evidence of DNA excision. Cells between 18 and 20 h after mixing were used for all microinjection experiments reported here unless otherwise stated. Three to four days after injection, drug-resistant cell lines were identified and cultured to at least 200 ml before isolation of genomic DNA. The portion of the plasmid containing the IES was amplified with PCR using one primer in the A51 flanking sequence and one primer in the calmodulin promoter (Fig. 1). Since the A51 gene and the calmodulin gene are unlinked in the Paramecium genome, only the plasmid fragment is amplified.
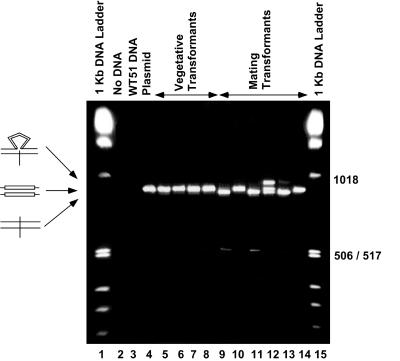
IES2591 is 370 bp in length, but previous studies have shown that it contains an internal 28-bp IES that is removed when a mutation prevents excision of the entire IES (16). Consequently, there are two potential products, one resulting from excision of the 370-bp IES and one resulting from excision of the 28-bp IES. The PCR fragments amplified from selected examples of vegetative and mating transformants and electrophoresed on a 4% agarose gel are shown in Fig. 2. As expected, no DNA is amplified from the negative controls (lanes 2 and 3). A full-length (nonspliced) fragment is amplified from the plasmid control and vegetative transformants. In contrast, at least three new bands are detected in the products from the mated cell transformants. A low-intensity band migrating at approximately 500 bp (lanes 9 and 11) is produced upon excision of the entire 370-bp IES2591 (difficult to detect in this photographic exposure). Purification of the band and direct DNA sequencing confirmed that the product was accurately spliced at the TA dinucleotide (data not shown). Among transformants that showed splicing of the entire 370-bp IES, usually less than 5% of the product was in this form. Lanes 9 and 11 also contain a major band that migrates slightly faster than the full-length PCR product. Sequence analysis of these bands showed that they are products from accurate excision of the 28-bp internal IES. Surprisingly, we also observed bands that migrate more slowly than the full-length product (lane 12). These low-mobility bands are the result of heteroduplex formation between one DNA strand that has excised the 28-bp IES and a second full-length DNA strand (Fig. 3 and data not shown). The resulting heteroduplex with a single-strand loop migrates with lower mobility than the corresponding full-length PCR fragment on a 4% agarose gel. Supporting this hypothesis is our ability to generate the low-mobility band (heteroduplex) by mixing, denaturing, and then annealing pure samples of the two faster-migrating bands (data not shown). Most likely the heteroduplex is formed by the melting and reannealing steps that occur in the final cycles of PCR from transformants that do not completely excise the 28-bp IES.
FIG. 2.
Ethidium bromide-stained gel of PCR products from vegetative and mating cell transformants. Drug-resistant cell lines were isolated after injection of pIES-NEO into vegetative or mated Paramecium cells. PCR products were electrophoresed on a 4% agarose gel that was stained with ethidium bromide. As expected, no product was detected from the no-DNA control or the wild-type (WT) genomic DNA (lanes 2 and 3). Vegetative cells transformed with pIES-NEO shown in lanes 5 to 8 contain a single band that migrates at the same position as the full-length plasmid product (876 bp; lane 4). Lanes 9 to 14 contain products from mating cell transformants. A faint band at the 506/517-bp marker in lanes 9 and 11 is expected upon removal of the entire 370-bp IES (506 bp). Small differences in mobility of the 876-bp fragment are due to removal of the internal 28-bp IES. Since the 28-bp IES is not eliminated from every molecule, heteroduplexes containing unspliced and spliced strands are formed and migrate more slowly than either homoduplex (e.g., lane 12).
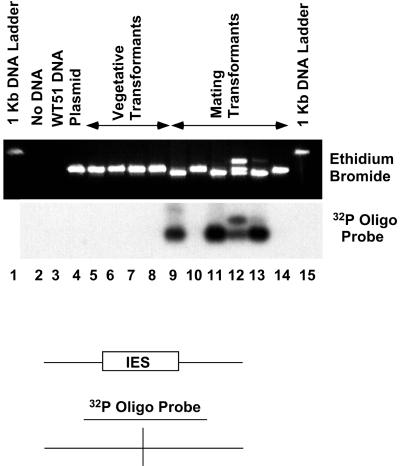
FIG. 3.
Accurate excision revealed by Southern hybridization to the PCR products from the in vivo excision assay. The ethidium bromide-stained gel shown in Fig. 2 was blotted to a nylon membrane and hybridized with a 26-mer oligonucleotide complementary to the junction of the excised 28-bp internal IES. Washing conditions were adjusted so that the probe hybridized to the spliced product but not the unspliced PCR products from the original plasmid (lane 4) or vegetative transformants (lanes 5 to 8). Note the hybridization signal corresponding to the low-mobility (heteroduplex) band in lane 12.
To confirm accurate excision of each product, the agarose gel was blotted to a nylon membrane and probed with a 32P-labeled oligonucleotide complementary to the junction of the spliced 28-bp IES. As seen in Fig. 3, the hybridization signal is present only from products of the mated cells (lanes 9, 11, 12, and 13). Also, the low-mobility heteroduplex as well as the homoduplex of excised DNA hybridizes with the oligonucleotide (lane 12). These data clearly demonstrate that injection of plasmid DNA into cells 18 h after initiation of mating results in the accurate excision of either the entire 370-bp IES or the 28-bp internal IES. It should be noted that a small percentage of transformants (Neor cell lines) show no evidence of IES excision. This population represents about 10 to 20% of transformants from mated cells (Table 1); two examples are shown in Fig. 3. Southern hybridization analysis showed that transformed cell lines contained the plasmid as extrachromosomal molecules (data not shown).
TABLE 1.
Effects of flanking sequence deletions on IES excision
| Injected plasmid | No. of transformed lines showing excision | Total Neor transformed lines | % Excisiona |
|---|---|---|---|
| pIES-NEO | 8 | 10 | 80 |
| pIES-72 | 13 | 15 | 87 |
| pIES-31 | 2 | 10 | 20 |
| pIES-18 | 6 | 14 | 43 |
| pIES-1 | 0 | 15 | 0 |
Calculated by taking the number of transformed cell lines that contain some spliced 28-bp IES and dividing by the total number of transformed lines. Transformed lines showing excision varied in the amount of spliced product (10 to 100%).
The 28-bp internal IES is independently excised in vivo.
The results from our mated cell transformants suggested that the 28-bp internal IES was removed from the plasmid more efficiently than the entire 370-bp IES (Fig. 2). This observation led us to focus on the 28-bp IES. Although excision of the 28-bp IES was observed in a mutant cell line and in cell lines that artificially inhibit excision of IES2591 (6, 16), we wanted to demonstrate independent excision of the 28-bp internal IES in a wild-type cell. We used a PCR-based assay to demonstrate independent excision of the 28-bp internal IES. One oligonucleotide primer complementary to the junction of the excised 28-bp IES was used in combination with a second primer located 428 bp outside of IES2591. A DNA fragment will be amplified from this primer pair only if the 28-bp internal IES is removed prior to excision of the entire IES2591. Since this cannot occur in the micronucleus or mature macronucleus, the PCR product should be specific to macronuclear development. Figure 4 shows the results of Southern hybridization of the PCR amplifications using these primers on cells at 0, 12, 18, 24, and 30 h after mixing. As expected, there was almost no detectable signal at time zero, but the 12- and 18-h time points show substantial amounts of the expected 650-bp product. By 30 h, formation of the macronucleus is complete and little product is detected. The small amount of signal at time zero is most likely due to self-fertilization (autogamy) caused in a small fraction of the cells as a result of the starvation conditions required to induce mating reactivity. The results demonstrate that at least some copies of IES2591 excise the 28-bp IES prior to complete excision. The sequence features and in vivo behavior of the 28-bp internal IES demonstrate that it is a model substrate for studying IES excision.
Flanking sequences are required for IES excision.
Prior to the development of our in vivo assay, there was no direct method to investigate the role of flanking sequences in Paramecium IES excision. To test whether flanking sequences are required for excision, a series of deletions were made on one end of the 28-bp IES such that 72, 31, 18, and 0 bp (a BamHI site) were located adjacent to the TA dinucleotide boundary. All A51 gene sequences distal to those positions were deleted from each construct (see Fig. 6 for a summary of the deletion endpoints), yielding plasmids pIES-72, pIES-31, pIES-18, and pIES-1, respectively. Each plasmid was injected into mated cells; then drug-resistant cell lines were identified and assayed as described previously for pIES-NEO. The results in Table 1 show that pIES-72L is excised as efficiently as the full-length IES (pIES-NEO), but there is a drop in efficiency of excision in pIES-31 and pIES-18. Finally, eliminating all wild-type flanking sequence (pIES-1) results in complete inhibition of splicing. The results clearly show the importance of flanking DNA for excision of the 28-bp IES. Although we cannot claim that this feature is universal among all Paramecium IESs, it shows that the mechanism required for excision in some cases requires sequence features outside the eliminated region. The precise identity of the required flanking sequence is unknown, but our model (presented in Discussion) proposes that two sets of inverted repeats, one approximately 50 bp outside the IES and the other adjacent to the TA, function in the excision process.
FIG. 6.
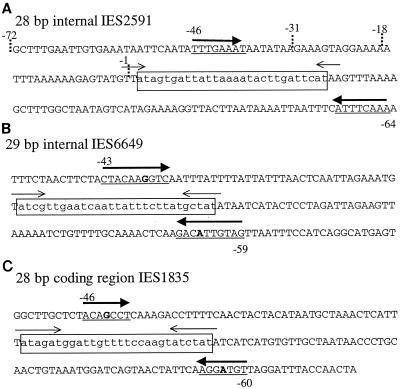
Identification of inverted repeats that flank short Paramecium IESs. (A) Sequence of the 28-bp internal IES analyzed in this study. The 28-bp eliminated sequence is boxed and in lowercase letters. The previously identified inverted repeats that are adjacent to the flanking TA are indicated by thin arrows, and the perfect 8-bp inverted repeat flanking the element (identified in this study) is indicated by thick arrows and underlines. The dotted lines at −1, −18, −31, and −72 mark the deletion endpoints for pIES-1, pIES-18, pIES-31, and pIES-72 in Table 1. Numbers above and below each underlined sequence indicate the distance between the most distal nucleotide in the inverted sequence and the closest boundary (5′-TA-3′) of the IES (also in panels B and C). Consequently the numbers on both flanks of the IES are negative. The first nucleotide prior to the left TA is −1, and the first nucleotide after the right TA is −1. (B) The inverted repeats flanking (underlined) and adjacent to the 29-bp internal IES of IES6649. One A-G mismatch in the 10-bp inverted repeat is shown in boldface; one is seen also in IES1835 (C). (C) The inverted repeats surrounding IES1835, a 28-bp IES located in the coding region of the A51 gene.
Nucleotide mutations inside the 28-bp IES do not prevent excision.
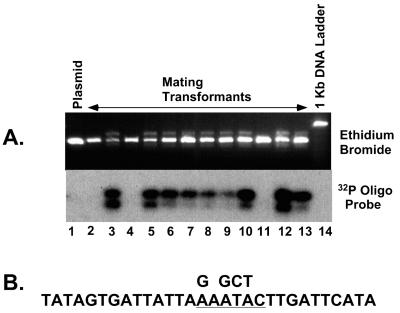
Sequence comparisons between evolutionarily related IESs from variable surface antigen genes show that the internal region (with the exception of the first 8 bp on either end) is generally not conserved in terms of size or primary sequence (23). We sought to take advantage of this internal sequence flexibility in order to manipulate the sequence of the 28-bp IES. A unique SacI restriction site was introduced approximately in the middle of the element by altering four nucleotides (Fig. 5B). The resulting plasmid, called pIES-SacI, was transformed into mated cells, and 12 drug-resistant cell lines were isolated. As shown in Fig. 5A, 9 of the 12 transformed cell lines showed evidence of DNA excision. Although we have not investigated the limits of internal sequence alterations, these results are consistent with the observed flexibility of sequences in the internal region of Paramecium IESs.
FIG. 5.
Alteration of nucleotides inside the 28-bp IES does not prevent excision. Four nucleotides inside the 28-bp IES were altered to create a unique SacI restriction site in pIES-SacI (B). After injection of pIES-SacI into mating cells and isolation of DNA from drug-resistant transformants, amplified PCR products were separated on a 4% agarose gel (A). (A) Agarose gel stained with ethidium bromide (top) and Southern hybridization of the blotted gel using the 28-bp IES junction specific oligonucleotide (see Materials and Methods) (bottom). Lane 1 contains product from pIES-SacI.
DISCUSSION
Role of flanking DNA in Paramecium IES excision.
Developmentally regulated DNA rearrangements have been observed in several different species of ciliated protozoa (reviewed in references 2, 13, and 21). Although the sequence characteristics of these eliminated elements differ among diverse ciliates, it is not clear whether these features are representative of a major difference in the mechanisms of excision or superficial alterations of the same fundamental molecular process. Functional analysis of these DNA elements has been difficult due to the lack of techniques for analyzing eliminated sequences in different organisms.
Previous studies of Paramecium IESs have identified conserved sequence features that include the flanking TA dinucleotide, a perfect inverted repeat adjacent to the TA (26), and an eight-nucleotide consensus terminal inverted repeat sequence that includes the TA dinucleotide (12). Analysis of mutations in the A51 variable surface protein gene showed that single nucleotide mutations located in the consensus sequence can prevent excision of the IES (16, 17). Although the isolation of mutants is useful, it cannot substitute for more extensive manipulations using recombinant DNA. We have demonstrated that microinjection of cloned Paramecium IESs into mated cells can be used to analyze the cis-acting sequence requirements for developmentally controlled DNA excision. Similar methods were previously developed for T. thermophila (29), and studies of eliminated sequences in this organism have shown that flanking DNA plays an important role in these events (3, 8, 14, 20). In the case of the Tetrahymena M element, it is clear that a 10-bp sequence (A5G5) located 50 bp outside the deleted region is sufficient to specify one end of the DNA splice junction (9). Interestingly, our results demonstrate the importance of flanking sequences in elimination of the Paramecium 28-bp internal IES.
Although we do not have direct experimental data to identify the critical cis-acting sequences, examination of the sequences flanking the 28-bp IES and results of the deletion analysis (Table 1) lead us to propose a model that emphasizes the importance of flanking inverted repeats. The drop in excision efficiency between bp −72 and −31 correlates with the presence of an 8-bp sequence (position −46) that is a perfect inverted repeat of a sequence located at −64 bp outside the right flank of the IES (Fig. 6A). Despite the lack of direct experimental data, we note that other short eliminated sequences (28 or 29 bp) also have inverted repeats approximately the same distance outside the element (Fig. 6B and C). In particular, the other known internal IES, a 29-bp element inside IES6649, has a 10-bp inverted repeat with a single A/G mismatch. The length and position of this repeat relative to the internal 28-bp IES make a strong argument for its significance. A third example is an inverted repeat outside the 28-bp IES1835. This IES sits directly in the coding region of the A51 gene; therefore, the repeat (7 bp with one mismatch) is part of the macronuclear A51 gene. Additional examples of inverted repeats flanking Paramecium IESs can be found, but it is difficult to evaluate their significance because we have no information concerning the necessary length or acceptable number of mismatches.
Sequence comparisons between various alleles could be used to evaluate whether the flanking repeats are conserved structural features of Paramecium IESs. Unfortunately, the only available IES2591 sequence is from P. tetraurelia, stock 29 (A29); with the exception of a 12-bp insertion, it is 99% identical to A51 (6). This single highly conserved sequence obviously is not informative, but a comparative sequence analysis might be a useful approach for future studies.
Despite our finding that inverted repeats are located about 50 bp outside the element, only the pIES-1 construct completely prevented excision. One possibility is that a previously undefined sequence element required for IES excision is located between −18 and −1. Alternatively, IES excision may require a combination of distant inverted repeats (50 bp away, as described above) and adjacent inverted repeats that include the flanking TA dinucleotide. In the pIES-1 construct, a BamHI site directly adjacent to the TA dinucleotide eliminates one nucleotide of the four-base inverted repeat. The loss of the distant flanking inverted sequences as well as the inverted repeat adjacent to the TA might completely eliminate excision. In fact, one could imagine that the two sets of inverted repeats are synergistic; a longer inverted sequence adjacent to the TA might compensate for a short flanking inverted sequence. The A51 IES1835 shown in Fig. 6C has the longest inverted repeat adjacent to the TA (seven nucleotides) but the shortest flanking repeat (seven with one mismatch).
Relationship between Paramecium IESs and DNA elimination in other ciliates.
The most thoroughly studied eliminated sequences in ciliates are those from T. thermophila (reviewed in references 2 and 28). Unlike the case for Paramecium, these eliminated DNAs are bounded by short direct repeats and do not have a consensus inverted terminal repeat with similarity to the Tc1 family of transposons. Analysis of the M element revealed that a 10-bp polypurine tract located approximately 45 bp outside the eliminated region is required to specify the deletion boundary. Interestingly, this sequence (A5G5) flanks both ends of the M element but is positioned in inverted orientation, analogous to the inverted orientation of the 8-bp sequence (TTTGAAAT) outside the 28-bp Paramecium IES. A more recent analysis of the eliminated R element in Tetrahymena found that again flanking sequences specify the splice junction (3). Unlike the M element, the required flanking sequence spans roughly a 70-bp region and has no apparent similarity to the A5G5 sequence. It is interesting that both Paramecium and Tetrahymena require flanking sequences for DNA elimination. Drawing from the analysis of the M element in Tetrahymena, there is the added possibility that both systems identify eliminated DNA using signals that include flanking inverted sequences.
The short IESs in E. crassus are structurally similar to Paramecium IESs (reviewed in reference 11). They are bounded by TA dinucleotides and frequently have perfect inverted repeats near their termini. Like Paramecium IESs, the Euplotes IESs are precisely removed, and statistical analysis has revealed a consensus terminal inverted repeat sequence that is similar (but not identical) to the Paramecium consensus. Despite the similarity in IES structure between the two organisms, we were unable to detect excision of a Euplotes IES in our Paramecium in vivo assay (M. Ku and J. Forney, unpublished data). The failure of this first attempt could reflect a real incompatibility between the two systems of excision or merely a technical issue such as the timing of microinjection (see below).
The in vivo assay as a tool for analysis of Paramecium DNA elimination.
Despite our success using the in vivo assay to analyze Paramecium IES excision, some technical points remain unresolved. One is the fact that our transformed cell lines do not show complete processing. Only a fraction of cell lines have excised the entire 370-bp IES2591 and even the 28-bp internal IES is not removed from all copies of the plasmid in all cell lines. The incomplete processing may be a result of the relatively late time of injection. Both our own analysis of IES2591 (Fig. 4) and recently published work by Betermier et al. (1) indicate that Paramecium IES excision begins within 12 h after mixing; therefore, injection at 18 h may not leave enough time for complete processing of the large number of injected copies. Alternatively, the low efficiency of excision may result simply from the large number of plasmid molecules that are injected. Previous studies have shown that a subset of Paramecium IESs (including IES2591) inhibit excision of their micronuclear homolog when they are present in the old macronuclear genome (5, 6, 16, 18). The inhibition of excision is dependent on high-copy-number plasmids in the macronucleus; therefore, it is possible that a similar phenomenon occurs in our injection system. Microinjection with lower concentrations of plasmid may improve the percentage of spliced DNA within transformed cells.
Finally, understanding the relationship between the cis-acting requirements for the 28-bp internal IES and a typical IES will require further investigations. Although we have shown that the 28-bp internal IES is removed as a normal part of DNA processing, it is possible that the regulatory elements controlling excision differ between different types of IESs. Regardless of the differences, it is clear that analysis of the 28-bp internal IES will provide critical insights into the regulation of DNA elimination in Paramecium. Future experiments with additional IES substrates will provide an interesting comparative analysis.
ACKNOWLEDGMENT
This work was supported by National Science Foundation grant MCB-9808285.
Footnotes
Paper number 16335 from the Purdue Agricultural Experiment Station.
REFERENCES
- 1.Betermier M, Duharcourt S, Seitz H, Meyer E. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol Cell Biol. 2000;20:1553–1561. doi: 10.1128/mcb.20.5.1553-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyne R S, Chalker D L, Yao M-C. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 3.Chalker D L, La Terza A, Wilson A, Kroenke C, Yao M-C. Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol Cell Biol. 1999;19:5631–5641. doi: 10.1128/mcb.19.8.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubrana K, Le Mouel A, Amar L. Deletion endpoint allele-specificity in the developmentally regulated elimination of an internal sequence (IES) in Paramecium. Nucleic Acids Res. 1997;25:2448–2454. doi: 10.1093/nar/25.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence in Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- 6.Duharcourt S, Keller A-M, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godiska R, Aufderheide K, Gilley D, Hendrie P, Fitzwater T, Preer L B, Polisky B, Preer J R., Jr Transformation of Paramecium by microinjection of a cloned serotype gene. Proc Natl Acad Sci USA. 1987;84:7590–7594. doi: 10.1073/pnas.84.21.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godiska R, Yao M-C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990;61:1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- 9.Godiska R, James C, Yao M-C. A distant 10 bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 1993;7:2357–2365. doi: 10.1101/gad.7.12a.2357. [DOI] [PubMed] [Google Scholar]
- 10.Haynes J W, Ling K-T, Saimi Y, Kung C. Induction of antibiotic resistance in Paramecium tetraurelia by the bacterial gene APH-3′-II. J Eukaryot Microbiol. 1995;42:83–91. doi: 10.1111/j.1550-7408.1995.tb01545.x. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs M E, Klobutcher L A. The long and the short of developmental DNA deletion in Euplotes crassus. J Eukaryot Microbiol. 1996;43:442–452. doi: 10.1111/j.1550-7408.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- 12.Klobutcher L A, Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klobutcher L A, Herrick G. Developmental genome reorganization in ciliated Protozoa: the transposon link. Prog Nucleic Acid Res. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Pearlman R E. Programmed DNA rearrangement from an intron during nuclear development in Tetrahymena thermophila: molecular analysis and identification of potential cis-acting sequences. Nucleic Acids Res. 1996;24:1943–1949. doi: 10.1093/nar/24.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling K Y, Vaillant B, Haynes W J, Saimi Y, Kung C. A comparison of internal eliminated sequences in the genes that encode two K+ channel isoforms in Paramecium tetraurelia. J Eukaryot Microbiol. 1998;45:459–465. doi: 10.1111/j.1550-7408.1998.tb05100.x. [DOI] [PubMed] [Google Scholar]
- 16.Mayer K M, Mikami K, Forney J D. A mutation in Paramecium tetraurelia reveals functional and structural features of developmentally excised DNA elements. Genetics. 1998;148:139–149. doi: 10.1093/genetics/148.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer K M, Forney J D. A mutation in the flanking 5′-TA-3′ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics. 1999;151:597–604. doi: 10.1093/genetics/151.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer E, Duharcourt S. Epigenetic programming of developmental genome rearrangements in ciliates. Cell. 1996;87:9–12. doi: 10.1016/s0092-8674(00)81317-3. [DOI] [PubMed] [Google Scholar]
- 19.Meyer E, Keller A M. A Mendelian mutation affecting mating-type determination also affects developmental genomic rearrangements in Paramecium tetraurelia. Genetics. 1996;143:191–202. doi: 10.1093/genetics/143.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patil N S, Karrer K M. A developmentally regulated deletion element with long terminal repeats has cis-acting sequences in the flanking DNA. Nucleic Acids Res. 2000;28:1465–1472. doi: 10.1093/nar/28.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott D M. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Scott J, Leeck C, Forney J. Analysis of the micronuclear B type surface protein gene in Paramecium tetraurelia. Nucleic Acids Res. 1994;22:5079–5084. doi: 10.1093/nar/22.23.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonneborn T M. Methods in Paramecium research. Methods Cell Physiol. 1970;4:242–339. [Google Scholar]
- 25.Sonneborn T M. Paramecium aurelia. In: King R, editor. Handbook of genetics. Vol. 2. New York, N.Y: Plenum Press; 1975. pp. 469–594. [Google Scholar]
- 26.Steele C J, Barkocy-Gallagher G A, Preer L B, Preer J R., Jr Developmentally excised sequences in micronuclear DNA of Paramecium. Proc Natl Acad Sci USA. 1994;91:2255–2259. doi: 10.1073/pnas.91.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vayssie L, Sperling L, Madeddu L. Characterization of multigene families in the micronuclear genome of Paramecium tetraurelia reveals a germline specific sequence in an intron of a centrin gene. Nucleic Acids Res. 1997;25:1036–1041. doi: 10.1093/nar/25.5.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao M-C. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet. 1996;12:26–30. doi: 10.1016/0168-9525(96)81385-0. [DOI] [PubMed] [Google Scholar]
- 29.Yao M-C, Yao C-H. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol Cell Biol. 1989;9:1092–1099. doi: 10.1128/mcb.9.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]