Summary
Osteoclasts are specialized multinucleated giant cells with unique bone-destroying capacities. A recent study revealed that osteoclasts undergo an alternative cell fate by dividing into daughter cells called osteomorphs. To date, no studies have focused on the mechanisms of osteoclast fission. In this study, we analyzed the alternative cell fate process in vitro and, herein, reported the high expression of mitophagy-related proteins during osteoclast fission. Mitophagy was further confirmed by the colocalization of mitochondria with lysosomes, as observed in fluorescence images and transmission electron microscopy. We investigated the role played by mitophagy in osteoclast fission via drug stimulation experiments. The results showed that mitophagy promoted osteoclast division, and inhibition of mitophagy induced osteoclast apoptosis. In summary, this study reveals the role played by mitophagy as the decisive link in osteoclasts’ fate, providing a new therapeutic target and perspective for the clinical treatment of osteoclast-related diseases.
Subject areas: Molecular interaction, Specialized functions of cells, Functional aspects of cell biology
Graphical abstract
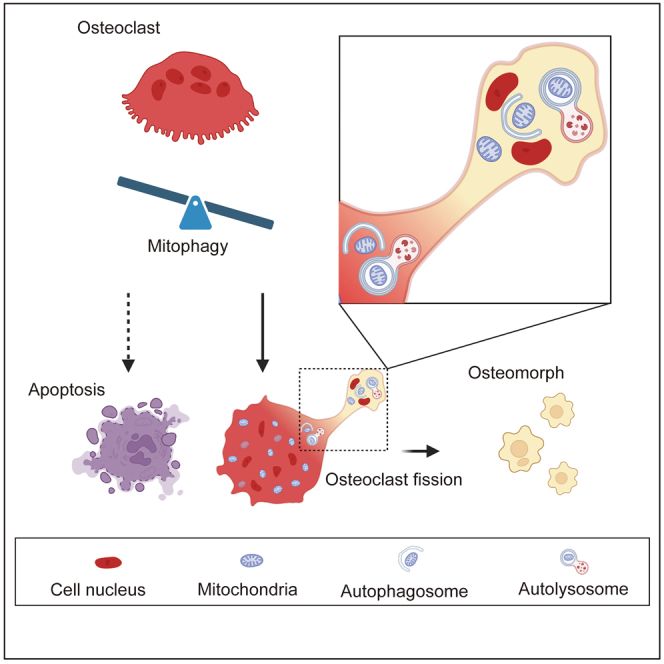
Highlights
-
•
Mitophagy is involved in the osteoclast-osteomorph transformation
-
•
Enhancing mitophagy promotes osteomorph formation
-
•
Mitophagy determines whether osteoclasts undergo apoptosis or fission
Molecular interaction; Specialized functions of cells; Functional aspects of cell biology
Introduction
Bone is a dynamic organ that is formed by osteoblasts and resorbed by osteoclasts continuously throughout life, changing the skeleton structure to maintain optimal function and bone homeostasis. Bone remodeling falls into two classifications. Remodeling-based bone formation (RBF), following osteoclast resorption, plays a significant role in renewing bone tissue. In contrast, another pathway that induces new bone formation is modeling-based bone formation (MBF), which is independent of previous bone resorption and less commonly found in the adult skeleton.1,2 RBF is the main form of bone remodeling in which osteoclasts play a vital role. Osteoclasts are specialized myeloid-derived multinucleated cells formed by the fusion of macrophages and have the unique bone-destroying capacity.3 The significant role played by osteoclasts in bone homeostasis has been established through many disease models such as osteoporosis, rheumatoid arthritis, and Paget’s disease models.4,5,6 Because of the importance of osteoclasts in bone remodeling, gaining an understanding of the biology of this cell type is important.
Osteoclast differentiation is triggered by the stimulation of receptor activators of nuclear factor-κB (NF-κB) ligand (RANKL) and macrophage colony-stimulating factor (M-CSF).7 RANKL associates with its receptor RANK, which activates downstream signaling pathways such as the NF-κB and protein kinase B (AKT) pathways to induce the expression of osteoclast-related genes.8 The nuclear factor of activated T cells 1 (NFATc1) is a master regulator of osteoclastogenesis and is induced by RANKL stimulation,9 and NFATc1 has been reported to induce the expression of genes that are significant to osteoclast differentiation and function.10,11 Under normal physiological conditions, large osteoclasts have a short lifespan and eventually undergo apoptosis.12 Osteoclast apoptosis has been reported to contribute to the treatment of rheumatoid arthritis and osteoporosis.13,14 Osteoclast-derived apoptotic bodies have been shown to exert biological effects in promoting bone defect healing.15 Hence, the fate of large osteoclasts after fusion is as important as their formation process.
However, according to a remarkable in vivo study,16 osteoclasts undergo cell fission instead of apoptosis after formation, which means large polykaryons are separated into smaller daughter cells called osteomorphs. Furthermore, this study revealed that spontaneous osteoclast apoptosis is far less common than fission in vivo.16 Moreover, an in vitro study showed that mature osteoclasts underwent fission and separated into smaller cells.17 These studies shed new light on osteoclast physiology. In addition, single-cell RNA sequencing data showed that osteomorphs differ from osteoclasts and macrophages at the transcriptional level. Daughter cells express a number of noncanonical osteoclast genes, such as Fbxo7, which had not been previously reported to be involved in bone metabolism.16 However, no studies have focused on the specific mechanism of osteoclast fission.
The Fbxo7 protein is a member of the Skp1-Cullin-F-box-type E3 ubiquitin ligases, which play crucial roles in targeting proteins for ubiquitination.18 Previous research revealed that Fbxo7 participates in mitochondrial maintenance by directly interacting with PTEN induced kinase 1 (PINK1) and Parkin, which are key proteins in mitophagy and play important roles in Parkin-mediated mitophagy.19 Another study showed that Parkinson’s disease-associated mutations in Fbxo7 disrupt mitophagy.20 Interestingly, Fbxo7 has also been reported to be overexpressed in osteomorphs,16 suggesting that mitochondrial homeostasis may play a role in osteoclast fission. Mitophagy, which refers to the degradation of mitochondria by autophagy, is a crucial mechanism for mitochondrial quality control.21 There are multiple mitophagy pathways, such as FUN14 domain containing 1 (FUNDC1), PINK1/parkin RBR E3 ubiquitin protein ligase (Parkin), and BCL2/adenovirus E1B interacting protein 3 (BNIP3),22,23 and dysfunctional mitochondria are identified through different autophagic pathways, resulting in them being engulfed by autophagosomes and transported to lysosomes for degradation. Notably, most likely due to the high energy needed for bone resorption, osteoclasts have a dramatically high number of mitochondria.24,25 The fate of these mitochondria during osteoclast division is an interesting question worthy of investigation. Considering the above-mentioned factors, we hypothesize that mitophagy may play a significant role in osteoclast fission.
To test our hypothesis, we demonstrated the physiological process in vitro by first determining whether mitophagy can be observed in osteoclast fission. In addition, to confirm that a change in mitophagy is associated with osteoclast fission, the effect of apoptosis was successfully excluded from the results. Finally, we demonstrated that mitophagy regulates osteoclast fission, which regulates the formation of daughter cells, as evidenced by the results when mitophagy was inhibited or excited.
Results
Osteoclast-osteomorph recycling in vitro
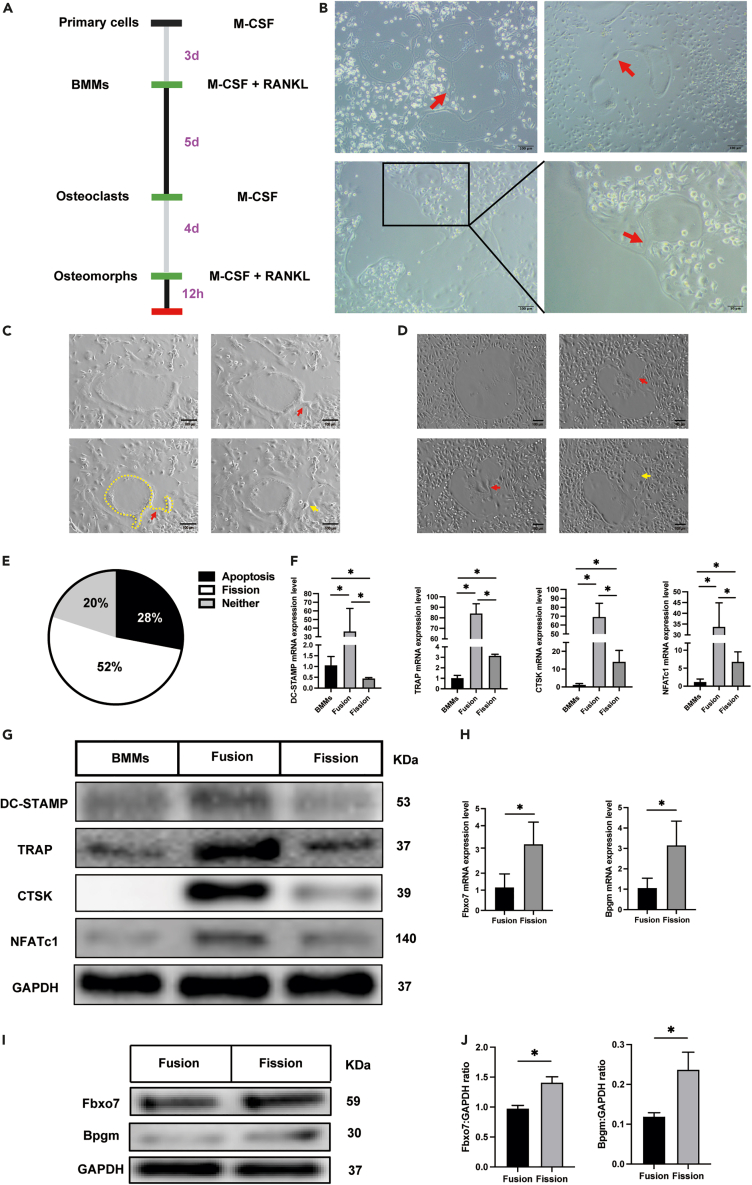
To confirm osteoclast fission in vitro, we monitored the osteoclasts for 4 days after fusion (Figure 1A). We observed that large osteoclasts divided into small cells (Figure 1B, red arrow). Moreover, we ascertained that these small cells were alive by examining their mobility. The time-lapse images confirmed this event and demonstrated that the daughter cells were freely motile (Videos S1 and S2). In the video, we observed that the osteoclasts first formed a bridged process (Figures 1C and 1D, red arrow), and then the process gradually tapered and the daughter cells were released (Figures 1C and 1D, yellow arrow). To analyze the difference between osteoclast fission and apoptosis, we assessed osteoclast apoptosis with time-lapse imaging. As expected, apoptotic cells underwent a totally different process than dividing osteoclasts (Video S3), disintegrating into motionless pieces that were ultimately phagocytosed by other cells. To further confirm the proportion of osteoclasts undergoing apoptosis and division, we randomly selected 25 obvious large osteoclasts in four visual fields and then observed and tracked changes to these cells. Thirteen of these cells underwent division, seven underwent apoptosis, and the remaining five cells showed neither division nor apoptosis at the end of the observation period (Figure 1E). Together, these results indicated that osteoclasts can divide or undergo apoptosis in vitro.
Figure 1.
Osteoclast-osteomorph recycling in vitro
(A) The timeline of the experiment.
(B) Representative images of osteoclast fission. Red arrowheads indicate daughter cells.
(C) Screenshot of the key frame of Video S1. Red arrowheads indicate cell bridged process. Yellow arrowhead indicates daughter cell. Yellow dotted box indicates the cell outline.
(D) Screenshot of the key frame of Video S2. Red arrowheads indicate cell bridged process. Yellow arrowhead indicates daughter cell.
(E) Proportion of osteoclasts with different fates.
(F) The relative mRNA levels of DC-STAMP, TRAP, CTSK, and NFATc1 genes in bone marrow macrophages (BMMs), fused osteoclasts, and osteoclasts undergoing fission (n = 3).
(G) Western blot analysis of the protein levels of DC-STAMP, TRAP, CTSK, NFATc1, and GAPDH in BMMs, fused osteoclasts, and osteoclasts undergoing fission.
(H) The relative mRNA levels of the Fbxo7 and Bpgm genes in fused osteoclasts and osteoclasts undergoing fission (n = 3).
(I) Western blot analysis of the protein levels of Fbxo7, Bpgm, and GAPDH in fused and osteoclasts undergoing fission (n = 3).
(J) Quantification of Fbxo7 and Bpgm by immunoblotting.
∗p < 0.05 by Student’s t test (H and J) or by one-way ANOVA (F).
The large osteoclast divided into small daughter cells, which were mobilizable. These smaller motile cells migrate away from each other.
The large osteoclast divided into small daughter cells, which were mobilizable. These smaller motile cells migrate away from each other.
The large osteoclasts broke up into lots of stationary cell debris, which were finally swallowed by other cells.
We further investigated whether the daughter cells were osteomorphs by measuring the expression levels of several key genes by RT-qPCR. Samples were collected according to the time node of the time-lapse images; when most osteoclast divisions were observed, the dividing phase samples were harvested. As previously reported, the osteomorphs gene expression was similar to that of osteoclasts on one level but different on another level.16 For example, TRAP expression was upregulated in osteoclasts and osteomorphs, but its expression was higher in the osteoclasts. DC-STAMP expression was upregulated in osteoclasts but not in osteomorphs. In contrast, the expression of certain genes, such as Fbxo7 and Bpgm, was significantly upregulated only in osteomorphs, not osteoclasts. In our experiment, expression of both TRAP and DC-STAMP was decreased in cells undergoing fission compared with the level in osteoclasts (Figures 1F and 1G). Compared with that in macrophages, the expression of TRAP was upregulated, and that of DC-STAMP was downregulated, consistent with the aforementioned research (Figures 1F and 1G). Similarly, as markers of osteoclasts, the expressions of CTSK and NFATc1 also decreased significantly after osteoclasts division (Figures 1F and 1G). Additionally, we evaluated the expression of Fbxo7 and Bpgm and found that both showed upregulated expression (Figure 1H). Similarly, the two genes also showed upward trends at the protein level (Figures 1I and 1J). These data suggested that the daughter cells of osteoclasts were osteomorphs.
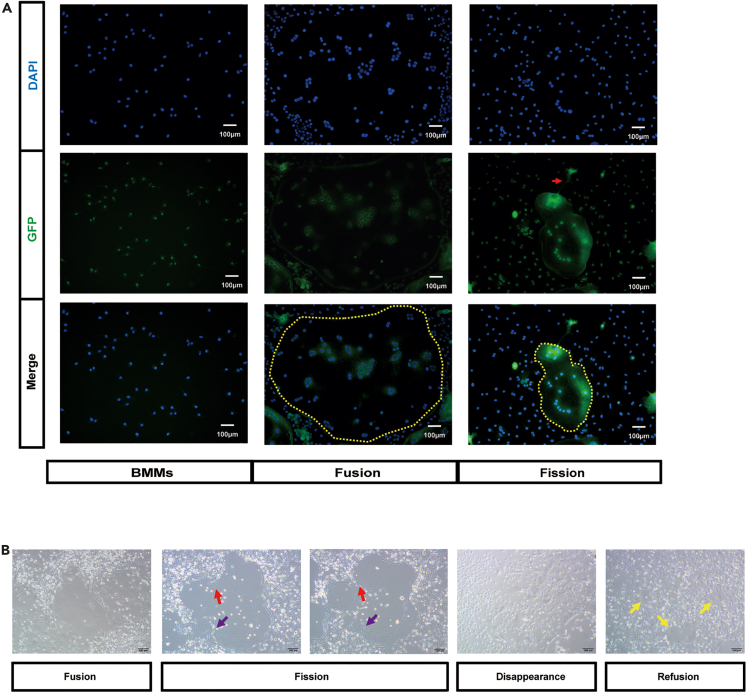
The identity of the daughter cells was further supported by anti-Fbxo7 immunofluorescence showing a marked increase in Fbxo7 expression in the cells undergoing fission (Figure 2A). Fbxo7 was expressed at a low level in bone marrow macrophages (BMMs) and osteoclasts (Figure 2A, yellow circle), even in the main portion of osteoclasts, which were undergoing division. Upregulation of Fbxo7 expression was observed at the site of cell division (Figure 2A, red arrow). Osteoclast protrusions included a nucleus, which demonstrated that the protrusion was destined to be a daughter cell, not a simple protrusion, and that was consistent with the results of time-lapse images. To confirm the function of the osteomorphs, we sought to have the daughter cells re-fuse in vitro. The results showed that large osteoclasts first underwent fission (Figure 2B, red arrow); at the same time, the daughter cell was observed to be fused again (Figure 2B, purple arrow). This phenomenon could be further confirmed by time-lapse images (Video S4). And after 4 days, the osteoclasts completely disappeared (Figure 2B). Then, the osteoclast induction medium was replaced, and new osteoclasts (Figure 2B, yellow arrow) were observed after 12 h, forming much faster than traditional osteoclast induction. Time-lapse images showed this process completely. Osteoclast divided into active daughter cell, which kept in constant motion. When stimulated again with RANKL, it fused to form new osteoclast within a few hours (Video S5). Collectively, these results indicated that osteoclasts underwent fission to produce osteomorphs in vitro and that osteoclast fission differs from apoptosis. Moreover, osteomorphs had the ability to fuse back into osteoclasts.
Figure 2.
Generation and re-fusion of osteomorphs
(A) Representative images of immunofluorescence staining (Fbxo7, green; DAPI, blue) in BMMs, fused osteoclasts, and osteoclasts undergoing fission (n = 5). Yellow dotted circles indicate large osteoclasts. The red arrowhead indicates osteoclasts undergoing fission.
(B) Representative images showing osteoclast division and re-fusion. Red arrowhead indicates osteoclasts undergoing fission. Purple arrowhead indicates osteoclasts undergoing re-fusion. Yellow arrowheads indicate the re-fused osteoclasts generated after treatment with RANKL for 12 h.
The large osteoclast divided into small daughter cells. And these daughter cells fused back into osteoclast.
The large osteoclast divided into small daughter cell, which was mobilizable (yellow arrow). And the daughter cell fused back into osteoclast in a few hours when treated with RANKL (red arrow).
Mitophagy was involved in the osteoclast-osteomorph transformation
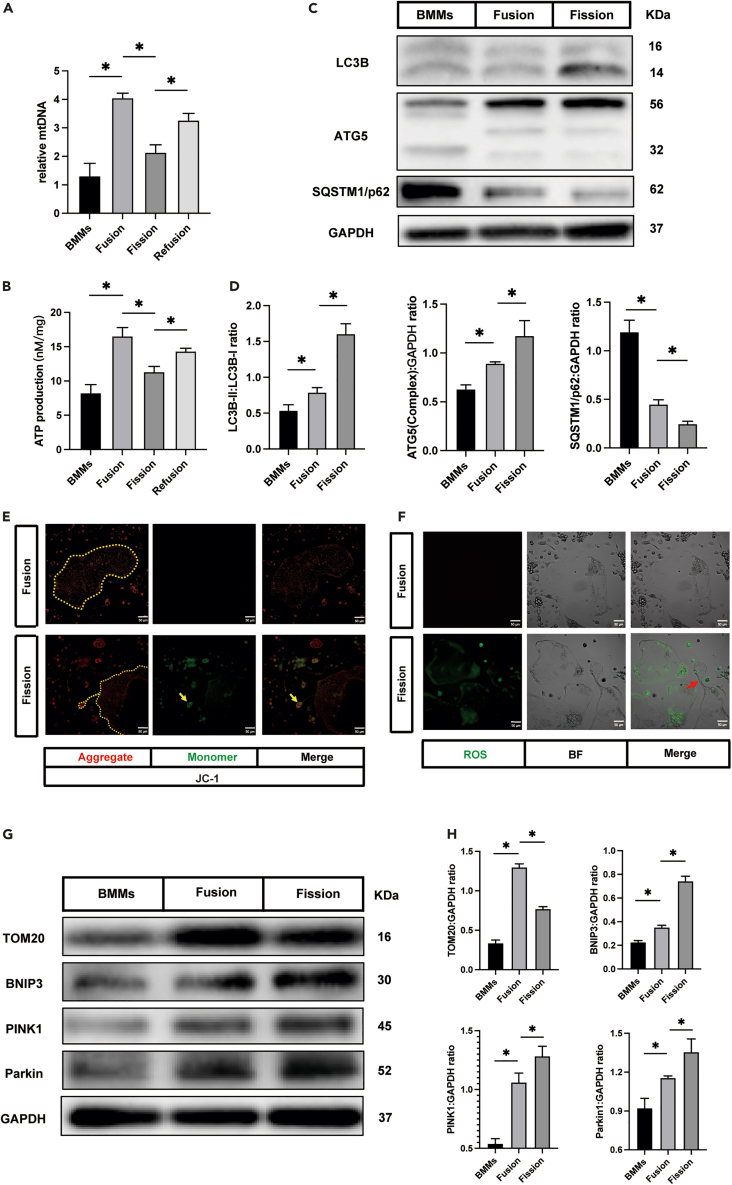
Previous research and the aforementioned results confirmed that Fbxo7 was highly expressed in the osteomorphs.16 Several studies have shown that Fbxo7 is related to Parkinson’s disease by participating in mitophagy.20,26 Therefore, we decided to evaluate certain mitophagy indicators to determine whether mitophagy is associated with osteoclast fission. In addition, we measured the levels of mitochondrial DNA (mtDNA) since the mitochondrial number can be predicted on the basis of the mtDNA copy number. Samples were harvested as described previously. The mtDNA copy number was found to increase when BMMs fused to osteoclasts, a finding that was consistent with that of a previous study (Figure 3A).27 Significantly, osteoclast fission resulted in a decreased mtDNA copy number, which increased again after treatment with fresh osteoclast differentiation medium (Figure 3A). ATP production was analyzed to assess mitochondrial activity. During the differentiation and evolution of osteoclasts, the ATP production level showed a trend similar to that of mtDNA copy number (Figure 3B). To confirm whether mitochondria were removed by mitophagy, we first analyzed the levels of microtubule-associated protein 1 light chain 3 beta (LC3B), autophagy-related protein 5 (ATG5), and sequestosome 1 (SQSTM1/p62). The results showed that LC3B-II and ATG5-ATG12 complex levels were significantly increased and that the level of SQSTM1/p62 was reduced in the osteoclast fission group compared with the BMMs and large osteoclasts (Figures 3C and 3D). In addition, we investigated the mitochondrial membrane potential and reactive oxygen species (ROS) generation rate to determine the function of the mitochondria. 5,5ʹ,6,6ʹ-tetrachloro-1,1ʹ,3,3ʹ-tetraethylbenzimidazolylcarbocyani-ne iodide (JC-1) is a dichromatic dye, which exhibits potential-dependent accumulation in mitochondria. It exists in the form of green fluorescent monomer in depolarizing mitochondria and in the form of red fluorescent aggregation in polarizing mitochondria. The ratio of green to red fluorescence depends only on the mitochondrial membrane potential.28 Compared to the large osteoclasts, the green fluorescence of daughter cells was significantly enhanced, which indicated the loss of mitochondrial membrane potential in these cells (Figure 3E, yellow arrow). Consistently, ROS production was increased in the fission group (Figure 3F). Previous studies had identified that the BNIP3 and PINK1/Parkin pathways are vital for the regulation of mitophagy in mammalian cells.29,30 Therefore, we determined BNIP3, PINK1, and Parkin expression levels in different cell lines. Mitophagy was indicated by the increased levels of these mitophagy protein indicators and the reverse trend of mitochondria protein translocase of outer mitochondrial membrane 20 (TOM20) level (Figures 3G and 3H). These data suggest a role played by mitophagy in osteoclast fission.
Figure 3.
Mitophagy was involved in osteoclast-osteomorph recycling
(A) The relative levels of mitochondrial DNA in BMMs, fused osteoclasts, osteoclasts undergoing fission, and re-fused osteoclasts (n = 3).
(B) The ATP production in BMMs, fused osteoclasts, osteoclasts undergoing fission, and re-fused osteoclasts (n = 3).
(C) Western blot analysis of the protein levels of LC3B, ATG5, p62, and GAPDH in BMMs, fused osteoclasts, and osteoclasts undergoing fission (n = 3).
(D) Quantification of LC3B, ATG5, and p62 by immunoblotting.
(E) Representative JC-1 fluorescence images showing fused osteoclasts and osteoclasts undergoing fission (J-aggregate, red; J-monomer, green) (n = 5). Yellow dotted lines indicate osteoclasts. Yellow arrowheads indicate osteoclasts undergoing fission. The J-monomer signal was significantly enhanced in daughter cells, which demonstrated that the mitochondrial membrane potential had decreased.
(F) Representative ROS fluorescence images showing fused osteoclasts and osteoclasts undergoing fission (ROS, green) (n = 5). The red arrowhead indicates osteoclasts undergoing fission.
(G) Western blot analysis of protein levels of TOM20, BNIP3, PINK1, Parkin, and GAPDH in BMMs, fused osteoclasts, and osteoclasts undergoing fission (n = 3).
(H) Quantification of TOM20, BNIP3, PINK1, and Parkin by immunoblotting.
∗p < 0.05 by one-way ANOVA (A, B, D, and H).
The alterations of mitochondria, lysosomes, and autolysosomes in osteoclast fission
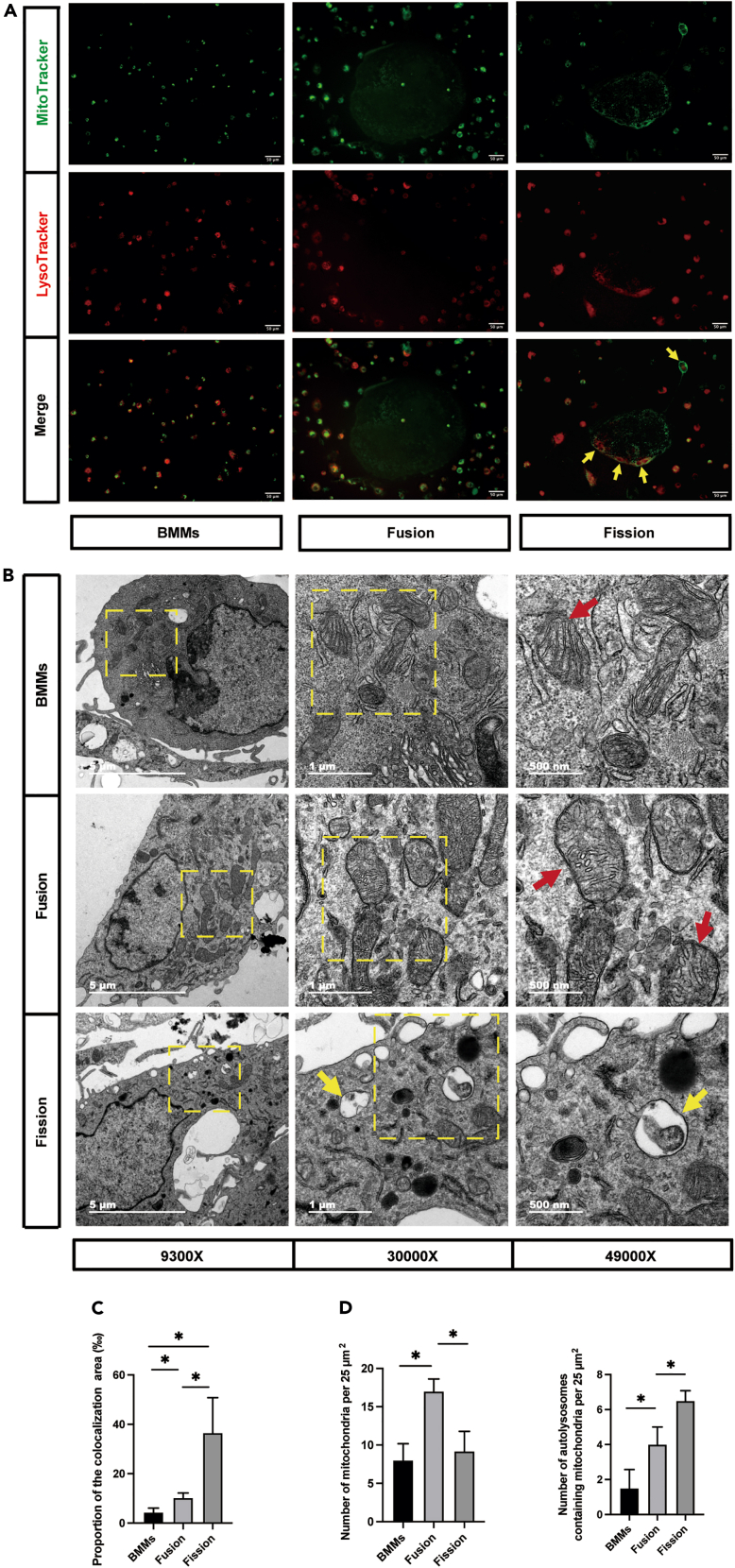
To directly evaluate mitophagy in osteoclast fission, colocalization of MitoTracker Green and LysoTracker Red was performed to assess the mitophagy level. Samples were collected as described previously. The results showed that colocalization of mitochondria with lysosomes was markedly increased in osteoclasts undergoing fission, as evidenced by the merged fluorescence signaling of MitoTracker and LysoTracker (Figure 4A, yellow arrow). The area of colocalization in fission group was significantly increased (Figure 4C). Consistent with the immunofluorescence of the costained mitochondria and lysosomes, autophagosomes were observed in the fission group through transmission electron microscopy (Figure 4B, yellow arrow). We also quantified the mitochondria (Figure 4B, red arrow) and autolysosomes in the images. Similar to the mtDNA copy number and TOM20 results, the number of mitochondria increased and decreased with fusion and division, respectively (Figure 4D). Many more autolysosomes were observed in the fission group than in the BMMs and large osteoclasts (Figure 4D). Taken together, these results confirmed the association of mitophagy with osteoclast fission.
Figure 4.
The alterations of mitochondria, lysosomes, and autolysosomes in osteoclast fission
(A) Representative MitoTracker and LysoTracker staining fluorescence images showing BMMs, fused osteoclasts, and osteoclasts undergoing fission (Mitochondria, green; lysosomes, red) (n = 5). Yellow arrowheads indicate that fluorescent signaling of MitoTracker and LysoTracker were merged in osteoclasts undergoing fission.
(B) Representative transmission electron microscope images showing BMMs, fused osteoclasts, and fission osteoclasts (n = 3). Red arrowheads indicate mitochondria. Yellow arrowheads indicate autolysosomes.
(C) Quantification of the proportion of the colocalization area to the total area in the image (n = 3).
(D) Quantification of mitochondria and autolysosomes by transmission electron microscope.
∗p < 0.05 by one-way ANOVA (C and D).
The role of mitophagy in osteoclast fission and apoptosis
Apoptosis was common during osteoclast culturing in vitro, as Video S3 shows. Hence, confirming whether the upregulation of mitophagy was caused by fission or apoptosis was clearly important. Because the regulatory mechanisms of osteoclast fission have not been revealed, we tried to increase the proportion of apoptotic cells by treating the cultures with staurosporine (STS) and assessing the contribution of apoptosis to mitophagy.
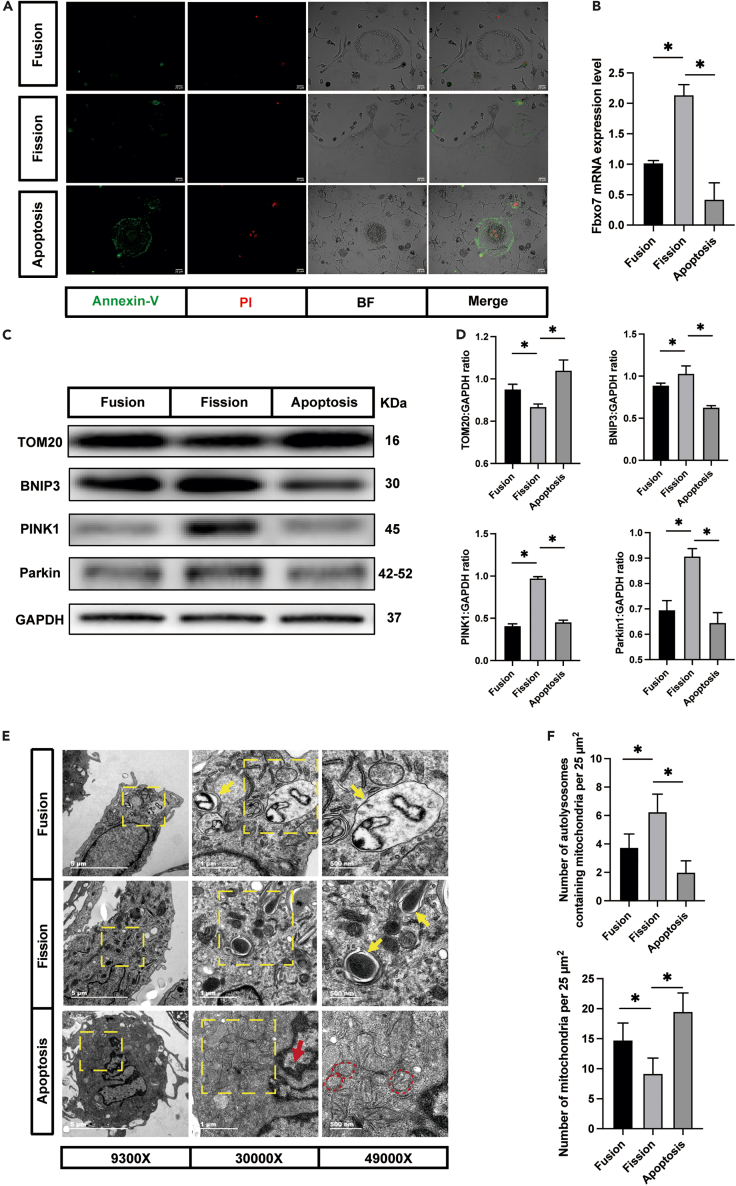
To confirm that apoptosis had been induced successfully, Annexin-V/propidium iodide (PI) staining was performed to assess cell vitality. As expected, Annexin-V- and PI-positive cells were observed after treatment with STS, while the fission cells were negative for both Annexin-V and PI staining (Figure 5A). The images demonstrated that apoptosis induction was realized and proved that the cells undergoing fission were distinct from those undergoing apoptosis.
Figure 5.
The role played by mitophagy in osteoclast fission and apoptosis
(A) Representative Annexin-V/PI staining fluorescence images showing fused osteoclasts, osteoclasts undergoing fission, and apoptosis osteoclasts (Annexin-V, green; PI, red) (n = 5).
(B) The relative mRNA levels of Fbxo7 genes in fused osteoclasts, osteoclasts undergoing fission, and apoptosis osteoclasts (n = 3).
(C) Western blot analysis of the protein levels of TOM20, BNIP3, PINK1, Parkin, and GAPDH in fused osteoclasts, osteoclasts undergoing fission, and apoptosis osteoclasts (n = 3).
(D) Quantification of TOM20, BNIP3, PINK1, and Parkin by immunoblotting.
(E) Representative transmission electron microscope images showing fused osteoclasts, osteoclasts undergoing fission, and apoptosis osteoclasts (n = 3). The red arrowheads indicate the condensation of nuclear chromatin. The red dotted circles indicate mitochondria fracture and decreased density. Yellow arrowheads indicate autolysosomes.
(F) Quantification of mitochondria and autolysosomes by transmission electron microscope.
∗p < 0.05 by one-way ANOVA (B, D and F).
We then asked whether apoptosis contributes to the upregulation of Fbxo7 expression. To this end, we compared the Fbxo7 expression levels in the osteoclast, fission, and apoptosis groups. Notably, Fbxo7 expression was downregulated in apoptotic osteoclasts (Figure 5B). To investigate the contribution of apoptosis to mitophagy, western blotting was performed to evaluate the levels of mitophagy-related proteins. We found that apoptotic osteoclasts expressed much higher levels of TOM20 and lower levels of BNIP3, PINK1, and Parkin than the fission group (Figures 5C and 5D), which suggested that the mitophagy rate was low in the apoptosis group. We also analyzed the mitochondrial membrane potential and ROS generation in apoptotic osteoclasts. Images showed that the mitochondrial membrane potential disappeared and that many more ROS were produced in the apoptotic osteoclasts than in the fission cells (Figures S1A and S1B), which was consistent with previous research on apoptotic cells.31,32 To further probe the relationship between apoptosis and mitophagy in osteoclasts, we took advantage of transmission electron microscopy to observe the apoptotic cells (Figure 5E), and we found that these cells had nuclear chromatin accumulation with uneven distribution (Figure 5E, red arrow). Typical apoptotic features were identified in the mitochondria, such as organelle fracture and decreased density (Figure 5E, red dotted box) caused by changes in mitochondrial outer membrane permeabilization.33,34,35 Moreover, the results demonstrated that the apoptosis group exhibited a lower mitophagic flux, as evidenced by the lack of autolysosomes (Figures 5E and 5F). Collectively, these results indicated that mitophagy was caused by osteoclast fission, not apoptosis.
Inhibiting mitophagy induces osteoclast apoptosis
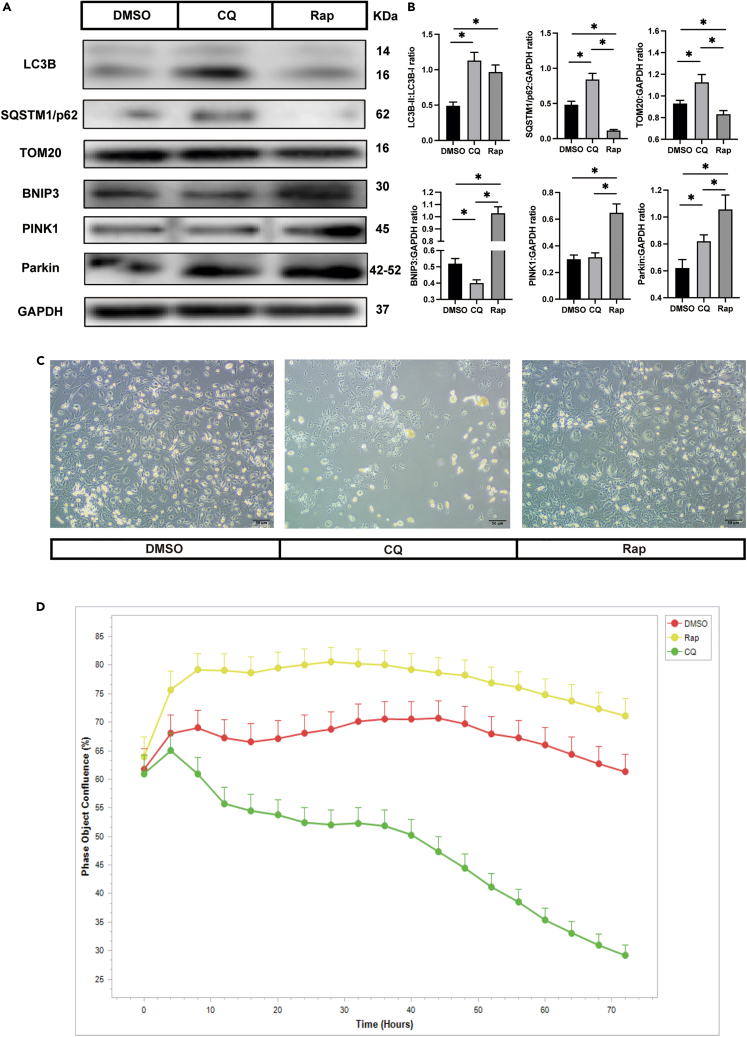
The data presented thus far indicated that osteoclast fission is associated with mitophagy, and therefore we wondered whether mitophagy plays a significant role in osteoclast fission. Chloroquine (CQ) and rapamycin (Rap) are classic mitophagy modulators; the former suppresses mitophagy by preventing the fusion of autophagosomes with lysosomes36; however, Rap promotes mitophagy through the serine/threonine protein kinase mammalian target of rapamycin (mTOR) signaling pathways.37 After treatment with CQ, LC3B-II and SQSTM1/p62 accumulated because of the blockage of autophagosome-lysosome formation. Furthermore, the TOM20 level was reversed by CQ treatment, which indicated mitophagy-related degradation (Figures 6A and 6B). Rap treatment resulted in enhanced LC3B-II conversion and decreased SQSTM1/p62 and TOM20 expression levels. Simultaneously, the levels of BNIP3, PINK1, and Parkin expression were significantly upregulated following treatment with Rap (Figures 6A and 6B). Together, these data demonstrated that mitophagy was successfully regulated.
Figure 6.
Inhibiting mitophagy induces osteoclast apoptosis
(A) Western blot analysis of the protein levels of LC3B, p62, TOM20, BNIP3, PINK1, Parkin, and GAPDH in different osteoclast groups (n = 3). The osteoclasts were treated with DMSO, CQ, and Rap, respectively, after fused.
(B) Quantification of LC3B, p62, TOM20, BNIP3, PINK1, and Parkin by immunoblotting.
(C) Representative images of osteoclasts after treated with drugs 3 days (n = 3).
(D) The cell confluence in different group (n = 5).
∗p < 0.05 by one-way ANOVA (B).
To investigate whether inhibition of mitophagy induces osteoclast apoptosis, large osteoclasts were incubated with CQ and Rap after osteoclasts had fused. Three days later, a large number of apoptotic cells were observed in the CQ group compared with the dimethyl sulfoxide (DMSO) and Rap groups (Figure 6C). To confirm the anti-apoptotic effect of mitophagy in osteoclasts in vitro, live-cell imaging was performed to assess the degree of cell confluence. The results showed that many osteoclasts underwent cell death after treatment with CQ for 72 h, while Rap treatment led to the opposite trend (Figure 6D).
To exclude interference from off-target effects of the drugs, 3-Methyladenine (3-MA) and carbonyl cyanide m-chlorophenylhydrazine (CCCP) were used to repeat the drug stimulation experiment. 3-MA significantly inhibited LC3B-II conversion and increased the content of TOM20. CCCP increased the expression of LC3B-II and decreased the content of TOM20 by promoting the formation of autophagosomes (Figures S2A and S2B). These results suggest that mitophagy was successfully manipulated. The response of osteoclasts to stimulation was similar to that to the stimulation of previous drugs. Osteoclasts underwent apoptosis when mitophagy was suppressed, and promoting mitophagy increased the cell survival curve (Figures S2C and S2D).
Together, these results indicated that mitophagy antagonized the apoptosis of osteoclasts. Here, we hypothesize that mitophagy is a critical process in osteomorph production.
Enhancing mitophagy promotes osteoclast-osteomorph recycle
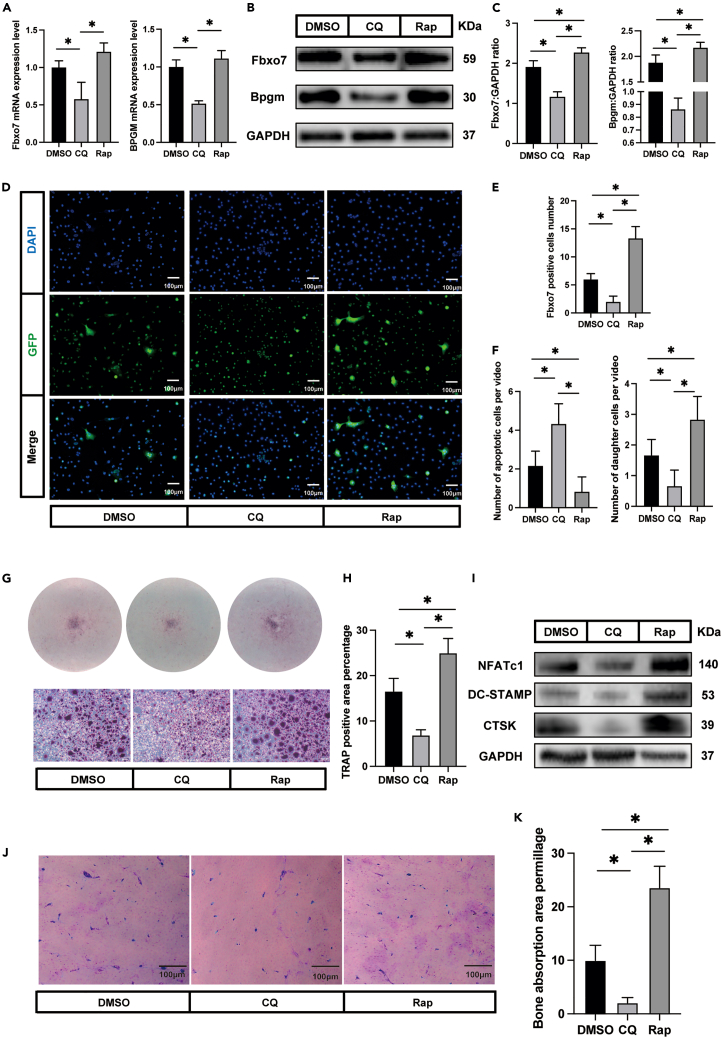
To test our hypothesis that mitophagy is critical to osteomorph generation, we first evaluated the expression of the osteomorph marker genes Fbxo7 and Bpgm after treatment with mitophagy modulators. By comparing their expression in modulator treatment groups with that in the DMSO group, we found that CQ significantly downregulated marker gene expression. In addition, the protein expression levels of Fbxo7 and Bpgm showed the same tendency (Figures 7B and 7C). Rap exerted no significant effect on the expression of Fbxo7 or Bpgm at the mRNA level (Figure 7A). However, the protein expression was significantly increased after Rap treatment. To confirm our initial hypothesis, an Fbxo7 immunofluorescence assay was performed to assess the number of osteomorphs after different treatments. Fluorescence images showed that the osteomorphs were nearly invisible in the CQ group. In contrast, incubation with Rap significantly increased the amount of osteomorphs compared with that in the DMSO group (Figures 7D and 7E).
Figure 7.
Enhancing mitophagy promotes osteoclast-osteomorph recycling
(A) The relative mRNA levels of Fbxo7 and Bpgm genes in DMSO, CQ, and Rap group (n = 3).
(B) Western blot analysis of the protein levels of Fbxo7, Bpgm, and GAPDH in different osteoclast groups (n = 3).
(C) Quantification of Fbxo7 and Bpgm by immunoblotting.
(D) Representative images showing immunofluorescence staining (Fbxo7, green; DAPI, blue) in DMSO, CQ, and Rap group (n = 5).
(E) Number of Fbxo7-positive cells (n = 5).
(F) Number of daughter cells and apoptotic cells in different groups (n = 6).
(G) Representative images showing TRAP staining in DMSO, CQ, and Rap group (n = 3). The cells were treated with RANKL again for 12 h after large osteoclasts disappearing.
(H) Quantification of TRAP-positive area by ImageJ.
(I) Western blot analysis of the protein levels of NFATc1, DC-STAMP, CTSK, and GAPDH in different osteoclast groups.
(J) Representative images showing bone resorption pits in DMSO, CQ, and Rap group (n = 3).
(K) Quantification of bone resorption pits area.
∗p < 0.05 by one-way ANOVA (A, C, E, F, H, and K).
Time-lapse imaging was performed to provide more reliable evidence. Six visual fields were randomly selected from each group, and the morphological changes in these osteoclasts were observed. The number of daughter cells and apoptotic cells were counted. The results indicated that CQ significantly increased the number of apoptotic cells and decreased the number of daughter cells, while Rap led to the opposite effects (Figure 7F).
We ultimately corroborated and extended these findings by examining osteoclast re-fusion. When mitophagy was suppressed, a smaller TRAP-positive area than that in DMSO-treated cells was observed. Rap treatment led to the opposite trend (Figures 7G and 7H). This was also confirmed by the protein expression of osteoclast markers, including NFATc1, DC-STAMP, and CTSK (Figure 7I)). A bone resorption assay was performed to evaluate the function of these re-fusion osteoclasts. Images showed that these cells exhibited the ability to absorb bone tissue (Figure 7J). Furthermore, CQ treatment reduced the formation of resorption pits, but Rap treatment had the opposite effect (Figures 7J and 7K).
Moreover, to mitigate off-target drug effects on the results, we repeated the above-mentioned experiments with 3-MA and CCCP. The results showed that the effect of 3-MA was similar to that of CQ, and the effect of CCCP was similar to that of Rap (Figure S3).
Altogether, these results demonstrated the significant role played by mitophagy in the regulation of osteoclast fission and the osteoclast-osteomorph recycling.
Discussion
Osteoclast fission is a newly discovered phenomenon,16 and the detailed mechanisms have not yet been discerned. Here, we show, for the first time, the specific mechanisms of osteoclast fission and re-fusion in vitro. Our current study clarifies that mitophagy is the key process in the production of osteomorphs and is the decisive link determining whether osteoclasts undergo apoptosis or fission. This research has thrown some new light on the mystery of osteoclasts and presents a new therapeutic target and perspective for the clinical treatment of osteoclast-related diseases.
Previous studies have reported that osteoclasts are terminally differentiated cells that undergo apoptosis after a short lifespan of approximately 2 weeks.12,38 Our study successfully observed the physiological phenomenon of osteoclast division in vitro. Recent research has shown that individual osteoclast syncytia live longer, approximately 6 months, through nucleus renewal.39 According to the most recent study,16 spontaneous osteoclast apoptosis is a rare event in vivo. Osteoclasts divide into daughter cells, which are called osteomorph, to increase the migration rate, revealing a quicker response and higher energy efficiency. Furthermore, osteomorphs are found not only on the surface of bone but also in the bone marrow and blood. Recent discoveries of osteoclast types, such as vessel-associated osteoclasts and osteoclasts derived from erythromyeloid progenitors, also suggested that osteoclasts warrant further exploration.40,41 In this study, we observed that osteoclasts underwent cellular fission in vitro (Videos S1 and S2), which appeared completely different than those undergoing apoptosis (Video S3). The daughter cells acquired superior mobility, allowing rapid osteoclast migration. For this purpose, the ability of osteomorphs to re-fuse into large osteoclasts is vital. As expected, osteomorphs responded rapidly to RANKL treatment. This outcome was consistent with that of a previous study16 and suggested that the fission of osteoclasts and the fusion of osteomorphs contribute to the efficient regulation of bone metabolism.
Mitophagy plays a key role in mitochondrial quality control, which means it maintains the homeostasis of healthy and damaged mitochondria in cells.24,42 Our results show that mitophagy plays an important role not only in the fusion process but also in the fate of osteoclasts after fusion. Mitophagy involves the transport of damaged or excess mitochondria to lysosomes, where these mitochondrial components are degraded and recycled.43,44 Previous studies have revealed that mitophagy plays a vital role in the differentiation of osteoclasts.25,45 For example, upregulated expression of BNIP3, which is a key protein in mitophagy, ultimately leads to osteoclastogenesis.46 Furthermore, the Beclin-1/BECN1 mitophagy pathway is important for osteoclast differentiation.47,48,49 In artificial joint replacement, autophagy was found to promote osteoclastogenesis by increasing the levels of Beclin-1 and soluble RANKL.50 Upregulated mitophagy has been established to protect cells from apoptosis, and suppressing mitophagy induces the apoptosis of osteoclasts.51,52 However, relevant studies have been focused on the process of osteoclast formation because osteoclasts are terminally differentiated cells that undergo apoptosis quickly. This study identified the ability of osteoclasts to divide into osteomorphs and demonstrated that mitophagy plays a crucial role in this biological process.
We confirmed the link between osteoclast fission and mitophagy in three ways. Firstly, we found that the expression of mitophagy indicators in the osteoclast fission group was upregulated, which suggested that mitophagy might be involved in osteoclast division. Recent studies have identified the colocalization of mitochondria and lysosomes as evidence supporting mitophagy.53,54 Therefore, fluorescence images were acquired to assess the colocalization of mitochondria and lysosomes. Furthermore, transmission electron microscopy was performed to directly evaluate mitophagy. Secondly, apoptosis was considered the fate of osteoclasts after bone resorption.55 Recent evidence has indicated that fission is much more common than apoptosis in osteoclasts in vivo.16 However, we still needed to exclude the effect of apoptosis. And we found that the proportion of apoptotic osteoclasts increased when the mitophagy index decreased. The ideal simulation of the spontaneous apoptosis of osteoclasts in vivo is difficult to achieve. STS is a common apoptosis inducer that has been employed in many apoptosis studies.56,57 STS has also been reported to promote mitophagy.58 However, the level of mitophagy in the STS group was still significantly lower than that in the fission group. The suppression of mitophagy in the STS group indicated that mitophagy was induced by osteoclast fission. Finally, we used drugs to regulate mitophagy to observe its effect on osteoclast fate. When mitophagy was inhibited, a large number of osteoclasts underwent apoptosis. And when mitophagy was upregulated, the formation of Fbxo7-positive cells increased. We investigated the formation of osteomorph by observing its re-fusion as we found that the fusion of osteomorph was relatively rapid. However, it is necessary to pay attention to the possible interference of other cells.
Overactivation of osteoclasts has been shown to be associated with a variety of diseases, such as osteoporosis and rheumatoid arthritis.59,60 Osteoporosis therapies fall into two classifications, one of which is antiresorptive drugs, which slow bone resorption by suppressing osteoclast formation and function.61 Nevertheless, treatments, such as bisphosphonates and denosumab, focus on the osteoclast fusion process, and no study has been directed to intervention in the subsequent division process. Furthermore, treatment of osteoporosis with antiresorptive drugs causes side effects. For example, bisphosphonates may lead to medication-related osteonecrosis in the jaw and atypical femoral fractures, and denosumab is likely to result in clusters of rebound-associated vertebral fractures.62 Most previous research has indicated that most osteoclasts undergo division into osteomorphs and remain in the body for a long time,16 which suggests that large osteoclasts in the body may be just a small portion of the cells involved in this process, with more daughter cells in circulation to respond rapidly as needed. Thus, our study was innovatively focused on the fate of large osteoclasts after formation, highlighting the significant role of mitophagy and osteomorphs in bone homeostasis. Interestingly, osteoclast-derived apoptotic bodies have been shown to play a bridging role in osteoclast-osteoblast coupling in bone remodeling to promote bone defect healing.15,63 Different outcomes of osteoclasts may exert distinct effects on bone homeostasis. Altogether, our research provides a promising therapeutic target for osteoclast fusogenesis and osteolytic disease treatment.
This research pioneers the study of the mechanism of osteomorph formation. Mitophagy is more closely related to the physiological activities of osteoclasts, which provide a new understanding of osteoclasts, key cells of bone homeostasis. The limitation of this study is that the specific molecular pathway of mitophagy regulating osteoclast division has not been revealed. And we noted that Fbxo7 was characteristically highly expressed during this process. Here, we try to summarize previous research and combine it with our data to propose possible hypotheses that Fbxo7-mediated PINK1/Parkin pathway may play a crucial role in this process.19,64 We speculate that Fbxo7 induces mitophagy through interaction with PINK1 and Parkin in response to certain physiological signals such as mitochondrial depolarization. And this deep mechanism will be one of the main directions of our subsequent research.
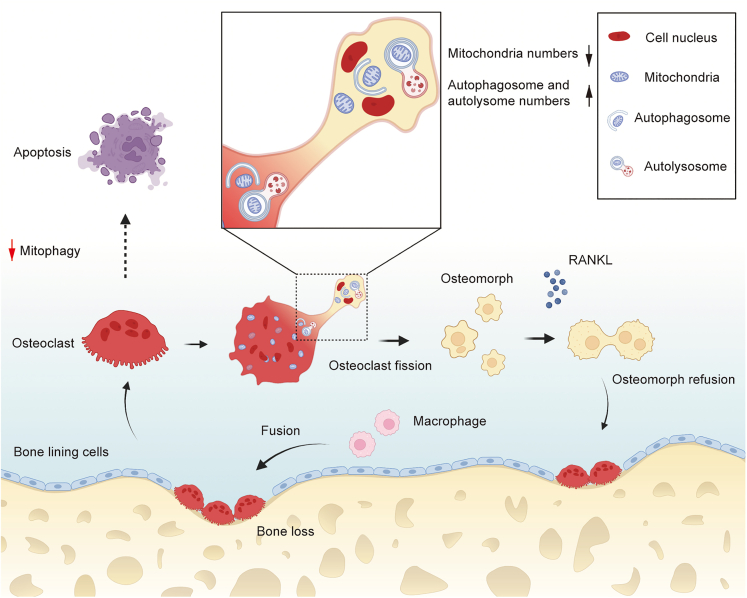
In conclusion, the present work first reveals that mitophagy is an important process for osteoclast fission, which sheds new light on the understanding of osteoclast physiology. After fusion, most osteoclasts undergo cell fission and turn into osteomorphs, which have the ability to re-fuse rapidly into large osteoclasts. Mitophagy is significant for this process; if it is inhibited, osteoclasts will undergo apoptosis (Figure 8). Gaining critical insight into this process will provide clues for the future management of osteoclast-related diseases through the regulation of osteoclast-osteomorph recycling mediated by mitophagy.
Figure 8.
Relationship between mitophagy and osteoclast fate
Large osteoclasts are formed by fusion of macrophagys. After formation, there are two distinct fates for osteoclasts. Most of them undergo fission and transform into osteomorphs, in which mitophagy plays a significant role. And osteomorphs could re-fuse back to osteoclasts to perform a bone resorption function when treated with RANKL again. If mitophagy is suppressed, osteoclasts will undergo apoptosis.
Limitations of the study
It must be pointed out that our study was only conducted in vitro cell experiments, and whether the same mechanism exists in vivo is yet to be verified. Secondly, the subject of the current study is a mixed population of cells, and we try to exclude the interference of other cells, but there is no denying the possibility of other effects. As of yet, osteomorphs and their surface markers have not yet been fully characterized.65 The establishment of in vivo models and the purification of osteomorphs will provide more powerful evidence, which are the focus of our next research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NFATc1 antibody | CST | 8032; RRID:AB_10829466 |
| TRAP antibody | Abcam | ab191406; RRID:AB_2936874 |
| Fbxo7 antibody | GeneTex | GTX65829; RRID:AB_2936875 |
| Bpgm antibody | Santa Cruz | sc-373819; RRID:AB_10989752 |
| TOM antibody | Proteintech | 11802-1-AP; RRID:AB_2207530 |
| CTSK antibody | Proteintech | 11239-1-AP; RRID:AB_2245581 |
| SQSTM1/p62 antibody | Affinity | AF5384; RRID:AB_2837869 |
| BNIP3 antibody | Affinity | DF8188; RRID:AB_2841498 |
| PINK1 antibody | Affinity | DF7742; RRID:AB_2841209 |
| LC3B antibody | Affinity | AF4650; RRID:AB_2844592 |
| APG5L/ATG5 antibody | Affinity | DF6010; RRID:AB_2837987 |
| DC-STAMP antibody | ABclonal | A14630; RRID:AB_2761505 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse M-CSF Protein (M-CSF) | R&D Systems | 416-ML |
| Recombinant Mouse RANKL (RANKL) | R&D Systems | 462-TEC |
| Staurosporine (STS) | MedChemExpress | HY-15141 |
| MitoTracker Green | Beyotime | C1048 |
| LysoTracker Red | Beyotime | C1046 |
| PBS (PBS) | Solarbio | P1020 |
| Methanol | Sangon Biotech | A601617 |
| Triton X-100 | Solarbio | T8200 |
| DAPI reagent | Solarbio | S2110 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Cienry | CR12800-S |
| Fetal Bovine Serum | Gibco | 10270106 |
| RIPA lysis buffer | GenStar | E121-01 |
| Protease inhibitors | Solarbio | P6730 |
| PVDF membranes | Millipore | IPVH00010 |
| SDS-PAGE gels | Fdbio Science Biotech | FD346 |
| Critical commercial assays | ||
| enhanced mitochondrial membrane potential assay kit with JC-1 | Beyotime | C2003S |
| ATP assay kit | Beyotime | S0026 |
| ROS assay kit | Beyotime | S0033S |
| Annexin V-FITC/PI apoptosis detection kit | Solarbio | CA1020 |
| Cells genomic DNA extraction kit | Solarbio | D1700 |
| RT-qPCR kit | Takara | RR047A |
| Experimental models: Organisms/strains | ||
| C57BL/6J mouse | Slaccas | N/A |
| Software and algorithms | ||
| GraphPad Prism Version 8 for Mac | GraphPad Software | https://www.graphpad.com/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/ |
| ImageJ Version 1.53 for Mac | National Institutes of Health | https://www.nih.gov/ |
| IBM SPSS Statistics 13.0 software | IBM lnc | https://www.ibm.com |
| ZEN 3.0 | Zeiss | RRID:SCR_013672 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Guoli Yang (guo_li1214@zju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
C57BL/6J mice were purchased from Slaccas (Shanghai, China). Six weeks old C57BL/6J female mice were the source of bone marrow macrophages in vitro. The cell extraction protocol complied with the regulations of laboratory animal use of Zhejiang University and the National Institutes of Health guide for the care and use of laboratory animals (NIH Publications No. 8023). The influence of gender needs to be studied in the future and is one of our limitations.
Bone marrow macrophages extraction
BMMs were isolated from tibias and femurs of six weeks old C57BL/6J female mice, were the target cells of our study in in vitro experiment. First, mice were sacrificed and disinfected with the help of immersion in 75% alcohol for 10 min. Peel the tibias and femurs from the body, remove the synovium, tendon, adipose and other soft tissues around. Bone marrow cells were flushed from the medullary cavities of tibiae and femurs. Then cells were cultured at 37°C with 5% CO2 in a DMEM medium containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 30 ng/mL M-CSF for 3 days to obtain BMMs.
Method details
Cell culture
Mouse BMMs were obtained as described.66 Briefly, total bone marrow cells were isolated from the femurs and tibias. The cells were plated in plastic petri dishes and cultured in DMEM (supplemented with 10% FBS, 1% penicillin/streptomycin, and 30 ng/mL M-CSF) at 37°C and 5% CO2 for 3 days. Subsequently, adherent cells were harvested and seeded with 1 105 cells per well in a 24-well plate in the presence of 30 ng/mL M-CSF and 50 ng/mL RANKL to induce osteoclast differentiation. The media and cytokines were changed every 3 days. For drug stimulation experiments, cells were treated with vehicle (DMSO 0.005%v/v), CQ (10 μM), Rap (5 μM), 3-MA (2mM) and CCCP (10μM) for 30 min after successful induction of osteoclasts.
Reagents
M-CSF (416-ML) and RANKL (462-TEC) were purchased from R&D Systems. Staurosporine was purchased from MedChemExpress (HY-15141). MitoTracker Green (C1048), LysoTracker Red (C1046), enhanced mitochondrial membrane potential assay kit with JC-1 (C2003S), ATP assay kit (S0026) and a ROS assay kit (S0033S) were purchased from Beyotime. An Annexin V-FITC/PI apoptosis detection kit and cells genomic DNA extraction kit were purchased from Solarbio (CA1020). Following anti-bodies were used in our experiments: anti-Fbxo7 antibody (GeneTex #GTX65829), anti-Bpgm antibody (Santa Cruz #sc-373819), anti-TOM antibody (Proteintech #11802-1-AP), anti-SQSTM1/p62 antibody (Affinity #AF5384), anti-BNIP3 antibody (Affinity #DF8188), anti-PINK1 antibody (Affinity #DF7742), anti-LC3B antibody (Affinity #AF4650), anti-APG5L/ATG5 antibody (Affinity #DF6010), anti-DC-STAMP antibody (ABclonal #A14630), anti-CTSK antibody (Proteintech #11239-1-AP), anti-NFATc1 antibody (CST #8032),anti-TRAP antibody (Abcam #ab191406).
Time lapse microscopy
Time lapse images were taken with a Keyent BZ-X800E fluorescence microscope. The cell plate was placed in a microscope chamber gassed with 5% CO2 and maintained at 37°C. Time-lapse microscopy was performed for 60 h at 30 min/frame intervals. Software-based autofocus was used to eliminate focal plane drift.
Immunofluorescence
Samples were collected according to the time node of the time-lapse images. After washing with PBS, cells were fixed with 4% paraformaldehyde (PFA) for 15 min, followed by permeabilization with 0.5% Triton X-100 in PBS for 30 min. Unspecific binding sites were blocked with 5% BSA for 30 min. Then, the anti-Fbxo7 antibody was applied overnight at 4°C. The next day, the cells were incubated with the secondary antibody for 3 h, followed by 4ʹ,6-diamidino-2-phenylindole (DAPI) staining for 10 min. Finally, fluorescence images were acquired with a fluorescence microscope, and random fields were photographed.
Analysis of the apoptosis rate
Apoptotic cells were examined using Annexin V-FITC/PI staining according to the manufacturer’s protocol. The cells were detected under a fluorescence microscope. Both Annexin V and PI-negative staining indicated live cells; early apoptotic cells were stained only with Annexin V; and both Annexin V- and PI-positive stained cells were in the late stage of apoptosis.67
Bone resorption assay
Bone resorption assays were performed as previously described.68 Briefly, the drug-treated cells were harvested and seeded onto bovine bone slices at 104 cells/well in a 24-well plate. The cells were cultured with 30 ng/mL M-CSF and 50 ng/mL RANKL for 5 days. Then the slices were treated with 1 M NH4OH to remove the cells. Next, the slices were stained with 1% toluidine blue for 1 min. Resorption pits images were captured with a light microscope.
Mitochondrial DNA content
Mitochondrial DNA content was quantified as the ratio of mitochondrial DNA to nuclear DNA. Samples were collected according to the time node of the time-lapse images. Total DNA was isolated with a DNA extraction kit. RT-qPCR was performed using a TB Green kit (TaKaRa, Kyoto, Japan) and a 384 Real-Time PCR system (Bio-Rad, USA). Nuclear DNA B2M or H19 was amplified as internal standards, and mitochondrial DNA ND1 or Cytb was normalized to nuclear DNA. The following primers were used: B2M forward: 5′- ACGTAACACAGTTCCACCC G-3′; B2M reverse: 5′- CGGCCATACTGGCATGCTTA-3′; ND1 forward: 5′- TCTAATCGCCAT AGCCTTCC-3′; ND1 reverse: 5′- GCGTCTGCAAATGGTTGTAA-3′69; H19 forward: 5′- GTCCACGAGACCAATGAC TG-3′; H19 reverse: 5′- GTACCCACCTGTCGTCC-3′; Cytb forward: 5′- ATTCCTTCATGTCGGA CGAG-3′; Cytb reverse: 5′-ACTGAGAAGCCCCCTCAAAT-3′.70
ATP assay
An ATP assay kit was used for the quantification of ATP levels. Briefly, cells were harvested and cleaved according to the manufacturer's manual. The cell lysate was centrifuged to obtain the supernatant (12000g, 4°C, 10min). Then, the supernatant was added to ATP working solution. ATP concentration was measured by measuring the luminescence intensity.
Assessment of mitochondrial membrane potential and ROS generation
JC-1 was used to measure mitochondrial membrane potential according to the kit manufacturer’s manual. In brief, cells were gently washed once with warm PBS, incubated in JC-1 working solution for 20 min at 37°C, and then washed three times. For the detection of ROS, a DCFU-DA probe was used according to the manufacturer’s instructions Briefly, cells were treated with 10 μM DCFU-DA FBS-free medium for 20 min at 37°C after washing with PBS. Images were acquired with a fluorescence microscope.
Transmission electron microscopy and analysis
Transmission electron microscopy was performed according to the protocol in the laboratory.71,72 The cells were first washed with PBS and fixed in 2.5% phosphate-buffered glutaraldehyde overnight at 4°C and then fixed with 1% osmium tetroxide for 1 h followed by 2% uranyl acetate staining. Then, the cells were dehydrated in a graded ethanol series and acetone, and embedded with epoxy resin. Finally, they were cut into approximately 100-nm ultrathin sections. Images were captured with a 120-kV Tecnai G2 Spirit electron microscope.
Identification of mitochondria and lysosomes
For the analysis of mitophagy, mitochondrial and lysosome staining was performed as reported.73 Briefly, two fluorescent selective probes, MitoTracker Green and LysoTracker Red, were used to identify mitochondria and lysosomes, respectively. Cells were incubated with 20 nM MitoTracker and 50 nM LysoTracker at 37°C for 20 min, and then stock solutions were removed. Then cells were observed with a fluorescence microscope.
Cell death assay
For the analysis of cell death, live-cell imaging was performed. In brief, cells were treated as described above. Then, they were photographed every 4 h for 72 h. The cell death test was confirmed by the number of cells, which was determined by the confluence of cells. The experiment was completed using an IncuCyte ZOOM™ live cell Imaging system.
Quantitative real-time PCR analysis
Samples were collected according to the time node of the time-lapse images. Total RNA was isolated and 500 ng of RNA was reversed transcribed to cDNA with a PrimeScriptTM RT reagent kit according to the manufacturer’s instructions. RT-qPCR was performed using a TB Green kit in a 384 Real-Time PCR system. The messenger RNA (mRNA) expression level was calculated using the 2-ΔΔCt method and normalized to the housekeeping gene GAPDH. The following primers were used: Fbxo7 forward: 5′- CGCAGCCAAAGTGTACAAAG-3′; Fbxo7 reverse: 5′- AGGTTCAGTACTTGC CGTGTG-3′; Bpgm forward: 5′- ACCGGAGGTACAAAGTGTGC-3′; Bpgm reverse: 5′- CTCCAGCAGAATCGGAACTC-3′; DC-STAMP forward: 5′- TCCTCCATGAA CAAACAGTTCCAA-3′; DC-STAMP reverse: 5′- AGACGTGGTTTAGGAATGCAGCTC -3′; TRAP forward: 5′- AGCAGCCAAGGAGGACTAC -3′; TRAP reverse: 5′- CAT AGCCCACACCG TTCTC -3′; CTSK forward: 5′- ATGTGGGTGTTCAAGTTTCTGC-3′; CTSK reverse: 5′-CCAC AAGATTCTGGGGACTC-3′; NFATc1 forward: 5′- GGTGCTGTCTGGCCATAACT-3′; NFATc1 reverse: 5′-GAAACGCTGGTACTGGCTTC-3′.
Western blot analysis
Samples were collected according to the time node of the time-lapse images. Cells were harvested and lysed in cell lysis buffer for 30 min on ice. The protein concentration was detected using a bicinchoninic acid kit (Beyotime, China). Equivalent amounts of protein (20 μg) were electrophoresed on 10-12% SDS-PAGE. The proteins were then transferred onto PVDF membranes. Subsequent processes were carried out following standard protocols. A densitometric analysis of the bands was performed with ImageJ software (National Institutes of Health).
Quantification and statistical analysis
All experiments were repeated independently at least three times. The results are expressed as the means ± standard deviation unless otherwise noted. All data were analyzed using the statistical software SPSS 13.0 (SPSS, Chicago, IL, USA). Statistical significance was analyzed using Student’s t test for comparisons between two groups. For comparisons among 3 or more groups, one-way analysis of variance (ANOVA) was performed, followed by Bonferroni's test. P values lower than 0.05 were considered to indicate statistical significance.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No.: 82201008) and Zhejiang Provincial Key Research and Development Program of China (Grant No.: 2021C03113). We thank Yuchen Zhang in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University, for her technical assistance on transmission electron microscopy.
Author contributions
Yang, G.L. and Wang, Y. conceived the ideas. Yang, G.L., Huang, T.B., and Wang, Y.C. designed the study. Wang, Y.C., Yu, Z., Yu, K. Fu, M.D., and Miao, X.Y. conducted the experiments. Wang, Y.C., Huang, T.B., Jiang, Z.W., and Lai, K.C. analyzed the data. Wang, Y.C., Huang, T.B., and Wang, Y. wrote the manuscript. Yang, G.L. led the study and writing. All authors reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: April 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106682.
Contributor Information
Ying Wang, Email: 7314032@zju.edu.cn.
Guoli Yang, Email: guo_li1214@zju.edu.cn.
Supplemental information
The fate of osteoclasts in physiological states.
The fate of osteoclasts under DMSO treatment.
The fate of osteoclasts under CQ treatment.
The fate of osteoclasts under Rap treatment.
The fate of osteoclasts under 3-MA treatment.
The fate of osteoclasts under CCCP treatment.
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wang W., Tseng W.J., Zhao H., Azar T., Pei S., Jiang X., Dyment N., Liu X.S. Activation, development, and attenuation of modeling- and remodeling-based bone formation in adult rats. Biomaterials. 2021;276:121015. doi: 10.1016/j.biomaterials.2021.121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dempster D.W., Zhou H., Recker R.R., Brown J.P., Recknor C.P., Lewiecki E.M., Miller P.D., Rao S.D., Kendler D.L., Lindsay R., et al. Remodeling- and modeling-based bone formation with teriparatide versus denosumab: a longitudinal analysis from baseline to 3 Months in the AVA study. J. Bone Miner. Res. 2018;33:298–306. doi: 10.1002/jbmr.3309. [DOI] [PubMed] [Google Scholar]
- 3.Hasegawa T., Kikuta J., Sudo T., Matsuura Y., Matsui T., Simmons S., Ebina K., Hirao M., Okuzaki D., Yoshida Y., et al. Identification of a novel arthritis-associated osteoclast precursor macrophage regulated by FoxM1. Nat. Immunol. 2019;20:1631–1643. doi: 10.1038/s41590-019-0526-7. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto K., Nakashima T., Shinohara M., Negishi-Koga T., Komatsu N., Terashima A., Sawa S., Nitta T., Takayanagi H. Osteoimmunology: the conceptual framework unifying the immune and skeletal systems. Physiol. Rev. 2017;97:1295–1349. doi: 10.1152/physrev.00036.2016. [DOI] [PubMed] [Google Scholar]
- 5.Wong S.W., Huang B.W., Hu X., Ho Kim E., Kolb J.P., Padilla R.J., Xue P., Wang L., Oguin T.H., 3rd, Miguez P.A., et al. Global deletion of Optineurin results in altered type I IFN signaling and abnormal bone remodeling in a model of Paget's disease. Cell Death Differ. 2020;27:71–84. doi: 10.1038/s41418-019-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rea S.L., Walsh J.P., Layfield R., Ratajczak T., Xu J. New insights into the role of sequestosome 1/p62 mutant proteins in the pathogenesis of Paget's disease of bone. Endocr. Rev. 2013;34:501–524. doi: 10.1210/er.2012-1034. [DOI] [PubMed] [Google Scholar]
- 7.Sun X., Xie Z., Hu B., Zhang B., Ma Y., Pan X., Huang H., Wang J., Zhao X., Jie Z., et al. The Nrf2 activator RTA-408 attenuates osteoclastogenesis by inhibiting STING dependent NF-kappab signaling. Redox Biol. 2020;28:101309. doi: 10.1016/j.redox.2019.101309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang E., Pavlos N.J., Rea S.L., Qi M., Chai T., Walsh J.P., Ratajczak T., Zheng M.H., Xu J. Proteasome inhibitors impair RANKL-induced NF-kappaB activity in osteoclast-like cells via disruption of p62, TRAF6, CYLD, and IkappaBalpha signaling cascades. J. Cell. Physiol. 2009;220:450–459. doi: 10.1002/jcp.21787. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Wang C., Wang G., Sun Y., Deng Z., Chen L., Chen K., Tickner J., Kenny J., Song D., et al. Loureirin B suppresses RANKL-induced osteoclastogenesis and ovariectomized osteoporosis via attenuating NFATc1 and ROS activities. Theranostics. 2019;9:4648–4662. doi: 10.7150/thno.35414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi Y., Sakai E., Okamoto K., Kajiya H., Okabe K., Naito M., Kadowaki T., Tsukuba T. Rab44, a novel large Rab GTPase, negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation. Cell. Mol. Life Sci. 2018;75:33–48. doi: 10.1007/s00018-017-2607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J., Wu H.F., Ang E.S.M., Yip K., Woloszyn M., Zheng M.H., Tan R.X. NF-kappaB modulators in osteolytic bone diseases. Cytokine Growth Factor Rev. 2009;20:7–17. doi: 10.1016/j.cytogfr.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Manolagas S.C. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 13.Deng C., Zhang Q., He P., Zhou B., He K., Sun X., Lei G., Gong T., Zhang Z. Targeted apoptosis of macrophages and osteoclasts in arthritic joints is effective against advanced inflammatory arthritis. Nat. Commun. 2021;12:2174. doi: 10.1038/s41467-021-22454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon Y.J., Zhang Z., Bang I.H., Kwon O.K., Yoon S.J., Kim J.R., Lee S., Bae E.J., Park B.H. Sirtuin 6 in preosteoclasts suppresses age- and estrogen deficiency-related bone loss by stabilizing estrogen receptor alpha. Cell Death Differ. 2019;26:2358–2370. doi: 10.1038/s41418-019-0306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Q., Liang M., Limjunyawong N., Dan Y., Xing J., Li J., Xu J., Dou C. Osteoclast-derived apoptotic bodies show extended biological effects of parental cell in promoting bone defect healing. Theranostics. 2020;10:6825–6838. doi: 10.7150/thno.45170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald M.M., Khoo W.H., Ng P.Y., Xiao Y., Zamerli J., Thatcher P., Kyaw W., Pathmanandavel K., Grootveld A.K., Moran I., et al. Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell. 2021;184:1940. doi: 10.1016/j.cell.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen I.D.C., Vermeer J.A.F., Bloemen V., Stap J., Everts V. Osteoclast fusion and fission. Calcif. Tissue Int. 2012;90:515–522. doi: 10.1007/s00223-012-9600-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng H., Liang H., Jankovic J. F-box only protein 7 gene in parkinsonian-pyramidal disease. JAMA Neurol. 2013;70:20–24. doi: 10.1001/jamaneurol.2013.572. [DOI] [PubMed] [Google Scholar]
- 19.Burchell V.S., Nelson D.E., Sanchez-Martinez A., Delgado-Camprubi M., Ivatt R.M., Pogson J.H., Randle S.J., Wray S., Lewis P.A., Houlden H., et al. The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 2013;16:1257–1265. doi: 10.1038/nn.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado-Camprubi M., Esteras N., Soutar M.P., Plun-Favreau H., Abramov A.Y. Deficiency of Parkinson's disease-related gene Fbxo7 is associated with impaired mitochondrial metabolism by PARP activation. Cell Death Differ. 2017;24:120–131. doi: 10.1038/cdd.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan D.C. Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 2020;15:235–259. doi: 10.1146/annurev-pathmechdis-012419-032711. [DOI] [PubMed] [Google Scholar]
- 22.Lin Q., Li S., Jiang N., Jin H., Shao X., Zhu X., Wu J., Zhang M., Zhang Z., Shen J., et al. Inhibiting NLRP3 inflammasome attenuates apoptosis in contrast-induced acute kidney injury through the upregulation of HIF1A and BNIP3-mediated mitophagy. Autophagy. 2021;17:2975–2990. doi: 10.1080/15548627.2020.1848971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng M.Y.W., Wai T., Simonsen A. Quality control of the mitochondrion. Dev. Cell. 2021;56:881–905. doi: 10.1016/j.devcel.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Ling W., Krager K., Richardson K.K., Warren A.D., Ponte F., Aykin-Burns N., Manolagas S.C., Almeida M., Kim H.N. Mitochondrial Sirt3 contributes to the bone loss caused by aging or estrogen deficiency. JCI Insight. 2021;6:e146728. doi: 10.1172/jci.insight.146728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., Deng Z., Ma Y., Jin J., Qi F., Li S., Liu C., Lyu F.J., Zheng Q. The role of autophagy and mitophagy in bone metabolic disorders. Int. J. Biol. Sci. 2020;16:2675–2691. doi: 10.7150/ijbs.46627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y., Qin L., Pan H., Liu Z., Jiang L., He Y., Zeng Q., Zhou X., Zhou X., Zhou Y., et al. The role of genetics in Parkinson's disease: a large cohort study in Chinese mainland population. Brain. 2020;143:2220–2234. doi: 10.1093/brain/awaa167. [DOI] [PubMed] [Google Scholar]
- 27.Park-Min K.H. Metabolic reprogramming in osteoclasts. Semin. Immunopathol. 2019;41:565–572. doi: 10.1007/s00281-019-00757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livingston M.J., Wang J., Zhou J., Wu G., Ganley I.G., Hill J.A., Yin X.M., Dong Z. Clearance of damaged mitochondria via mitophagy is important to the protective effect of ischemic preconditioning in kidneys. Autophagy. 2019;15:2142–2162. doi: 10.1080/15548627.2019.1615822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen T.N., Padman B.S., Lazarou M. Deciphering the molecular signals of PINK1/parkin mitophagy. Trends Cell Biol. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Choi G.E., Lee H.J., Chae C.W., Cho J.H., Jung Y.H., Kim J.S., Kim S.Y., Lim J.R., Han H.J. BNIP3L/NIX-mediated mitophagy protects against glucocorticoid-induced synapse defects. Nat. Commun. 2021;12:487. doi: 10.1038/s41467-020-20679-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z., Chen X., Liu N., Shi Y., Liu Y., Ouyang L., Tam S., Xiao D., Liu S., Wen F., Tao Y. A nuclear long non-coding RNA LINC00618 accelerates ferroptosis in a manner dependent upon apoptosis. Mol. Ther. 2021;29:263–274. doi: 10.1016/j.ymthe.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baechler B.L., Bloemberg D., Quadrilatero J. Mitophagy regulates mitochondrial network signaling, oxidative stress, and apoptosis during myoblast differentiation. Autophagy. 2019;15:1606–1619. doi: 10.1080/15548627.2019.1591672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie L.L., Shi F., Tan Z., Li Y., Bode A.M., Cao Y. Mitochondrial network structure homeostasis and cell death. Cancer Sci. 2018;109:3686–3694. doi: 10.1111/cas.13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunelle J.K., Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei M.C., Zong W.X., Cheng E.H., Lindsten T., Panoutsakopoulou V., Ross A.J., Roth K.A., MacGregor G.R., Thompson C.B., Korsmeyer S.J. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Bari M.A.A. Co-targeting of lysosome and mitophagy in cancer stem cells with chloroquine analogues and antibiotics. J. Cell Mol. Med. 2020;24:11667–11679. doi: 10.1111/jcmm.15879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang C., Livingston M.J., Liu Z., Dong Z. Autophagy in kidney homeostasis and disease. Nat. Rev. Nephrol. 2020;16:489–508. doi: 10.1038/s41581-020-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krum S.A., Miranda-Carboni G.A., Hauschka P.V., Carroll J.S., Lane T.F., Freedman L.P., Brown M. Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J. 2008;27:535–545. doi: 10.1038/sj.emboj.7601984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacome-Galarza C.E., Percin G.I., Muller J.T., Mass E., Lazarov T., Eitler J., Rauner M., Yadav V.K., Crozet L., Bohm M., et al. Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature. 2019;568:541–545. doi: 10.1038/s41586-019-1105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romeo S.G., Alawi K.M., Rodrigues J., Singh A., Kusumbe A.P., Ramasamy S.K. Endothelial proteolytic activity and interaction with non-resorbing osteoclasts mediate bone elongation. Nat. Cell Biol. 2019;21:430–441. doi: 10.1038/s41556-019-0304-7. [DOI] [PubMed] [Google Scholar]
- 41.Yahara Y., Barrientos T., Tang Y.J., Puviindran V., Nadesan P., Zhang H., Gibson J.R., Gregory S.G., Diao Y., Xiang Y., et al. Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair. Nat. Cell Biol. 2020;22:49–59. doi: 10.1038/s41556-019-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B., Lautrup S., Hasan-Olive M.M., Caponio D., Dan X., et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat. Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palikaras K., Lionaki E., Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T., Liu Q., Gao W., Sehgal S.A., Wu H. The multifaceted regulation of mitophagy by endogenous metabolites. Autophagy. 2022;18:1216–1239. doi: 10.1080/15548627.2021.1975914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin X., Zhou C., Li J., Liu R., Shi B., Yuan Q., Zou S. Autophagy in bone homeostasis and the onset of osteoporosis. Bone Res. 2019;7:28. doi: 10.1038/s41413-019-0058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y., Chen G., Zhang W., Xu N., Zhu J.Y., Jia J., Sun Z.J., Wang Y.N., Zhao Y.F. Autophagy regulates hypoxia-induced osteoclastogenesis through the HIF-1alpha/BNIP3 signaling pathway. J. Cell. Physiol. 2012;227:639–648. doi: 10.1002/jcp.22768. [DOI] [PubMed] [Google Scholar]
- 47.Laha D., Deb M., Das H. KLF2 (kruppel-like factor 2 [lung]) regulates osteoclastogenesis by modulating autophagy. Autophagy. 2019;15:2063–2075. doi: 10.1080/15548627.2019.1596491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao S.J., Kong F.Q., Cai W., Xu T., Zhou Z.M., Wang Z.B., Xu A.D., Yang Y.Q., Chen J., Tang P.Y., et al. GIT1 contributes to autophagy in osteoclast through disruption of the binding of Beclin1 and Bcl2 under starvation condition. Cell Death Dis. 2018;9:1195. doi: 10.1038/s41419-018-1256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terešak P., Lapao A., Subic N., Boya P., Elazar Z., Simonsen A. Regulation of PRKN-independent mitophagy. Autophagy. 2022;18:24–39. doi: 10.1080/15548627.2021.1888244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su B., Li D., Xu J., Zhang Y., Cai Z., Kauther M.D., Ma R. Wear particles enhance autophagy through up-regulation of CD147 to promote osteoclastogenesis. Iran. J. Basic Med. Sci. 2018;21:806–812. doi: 10.22038/IJBMS.2018.29347.7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim C.J., Shin S.H., Kim B.J., Kim C.H., Kim J.H., Kang H.M., Park B.S., Kim I.R. The effects of Kaempferol-inhibited autophagy on osteoclast formation. Int. J. Mol. Sci. 2018;19:125. doi: 10.3390/ijms19010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Morgan M.J., Chen K., Choksi S., Liu Z.G. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood. 2012;119:2895–2905. doi: 10.1182/blood-2011-08-372383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan S., Wu K., Zhao M., Yuan J., Ma S., Zhu E., Chen Y., Ding H., Yi L., Chen J. LDHB inhibition induces mitophagy and facilitates the progression of CSFV infection. Autophagy. 2021;17:2305–2324. doi: 10.1080/15548627.2020.1823123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stathakos P., Jiménez-Moreno N., Crompton L.A., Nistor P.A., Badger J.L., Barbuti P.A., Kerrigan T.L., Randall A.D., Caldwell M.A., Lane J.D. A monolayer hiPSC culture system for autophagy/mitophagy studies in human dopaminergic neurons. Autophagy. 2021;17:855–871. doi: 10.1080/15548627.2020.1739441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyce B.F. Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J. Bone Miner. Res. 2013;28:711–722. doi: 10.1002/jbmr.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An H.K., Chung K.M., Park H., Hong J., Gim J.E., Choi H., Lee Y.W., Choi J., Mun J.Y., Yu S.W. CASP9 (caspase 9) is essential for autophagosome maturation through regulation of mitochondrial homeostasis. Autophagy. 2020;16:1598–1617. doi: 10.1080/15548627.2019.1695398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosa N., Ivanova H., Wagner L.E., 2nd, Kale J., La Rovere R., Welkenhuyzen K., Louros N., Karamanou S., Shabardina V., Lemmens I., et al. Bcl-xL acts as an inhibitor of IP3R channels, thereby antagonizing Ca(2+)-driven apoptosis. Cell Death Differ. 2022;29:788–805. doi: 10.1038/s41418-021-00894-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xian H., Liou Y.C. Loss of MIEF1/MiD51 confers susceptibility to BAX-mediated cell death and PINK1-PRKN-dependent mitophagy. Autophagy. 2019;15:2107–2125. doi: 10.1080/15548627.2019.1596494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komatsu N., Win S., Yan M., Huynh N.C.N., Sawa S., Tsukasaki M., Terashima A., Pluemsakunthai W., Kollias G., Nakashima T., Takayanagi H. Plasma cells promote osteoclastogenesis and periarticular bone loss in autoimmune arthritis. J. Clin. Invest. 2021;131:e143060. doi: 10.1172/JCI143060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chotiyarnwong P., McCloskey E.V. Pathogenesis of glucocorticoid-induced osteoporosis and options for treatment. Nat. Rev. Endocrinol. 2020;16:437–447. doi: 10.1038/s41574-020-0341-0. [DOI] [PubMed] [Google Scholar]
- 61.Rachner T.D., Khosla S., Hofbauer L.C. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reid I.R., Billington E.O. Drug therapy for osteoporosis in older adults. Lancet. 2022;399:1080–1092. doi: 10.1016/S0140-6736(21)02646-5. [DOI] [PubMed] [Google Scholar]
- 63.Ma Q., Liang M., Wu Y., Luo F., Ma Z., Dong S., Xu J., Dou C. Osteoclast-derived apoptotic bodies couple bone resorption and formation in bone remodeling. Bone Res. 2021;9:5. doi: 10.1038/s41413-020-00121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y., Lear T.B., Verma M., Wang K.Z., Otero P.A., McKelvey A.C., Dunn S.R., Steer E., Bateman N.W., Wu C., et al. Chemical inhibition of FBXO7 reduces inflammation and confers neuroprotection by stabilizing the mitochondrial kinase PINK1. JCI Insight. 2020;5:e131834. doi: 10.1172/jci.insight.131834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weivoda M.M., Bradley E.W. Macrophages and bone remodeling. J. Bone Miner. Res. 2023;38:359–369. doi: 10.1002/jbmr.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L., You X., Lotinun S., Zhang L., Wu N., Zou W. Mechanical sensing protein PIEZO1 regulates bone homeostasis via osteoblast-osteoclast crosstalk. Nat. Commun. 2020;11:282. doi: 10.1038/s41467-019-14146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song H., Feng X., Zhang H., Luo Y., Huang J., Lin M., Jin J., Ding X., Wu S., Huang H., et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419–1437. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei L., Chen W., Huang L., Wang H., Su Y., Liang J., Lian H., Xu J., Zhao J., Liu Q. Alpinetin ameliorates bone loss in LPS-induced inflammation osteolysis via ROS mediated P38/PI3K signaling pathway. Pharmacol. Res. 2022;184:106400. doi: 10.1016/j.phrs.2022.106400. [DOI] [PubMed] [Google Scholar]
- 69.Lin Q., Li S., Jiang N., Shao X., Zhang M., Jin H., Zhang Z., Shen J., Zhou Y., Zhou W., et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019;26:101254. doi: 10.1016/j.redox.2019.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H., Wang Y., Li W., Chen H., Du L., Liu D., Wang X., Xu T., Liu L., Chen Q. Deficiency of mitophagy receptor FUNDC1 impairs mitochondrial quality and aggravates dietary-induced obesity and metabolic syndrome. Autophagy. 2019;15:1882–1898. doi: 10.1080/15548627.2019.1596482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu W., Duan X., Fang X., Shang W., Tong C. Mitochondrial protein import regulates cytosolic protein homeostasis and neuronal integrity. Autophagy. 2018;14:1293–1309. doi: 10.1080/15548627.2018.1474991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cloonan S.M., Glass K., Laucho-Contreras M.E., Bhashyam A.R., Cervo M., Pabón M.A., Konrad C., Polverino F., Siempos I.I., Perez E., et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat. Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wan M.C., Tang X.Y., Li J., Gao P., Wang F., Shen M.J., Gu J.T., Tay F., Chen J.H., Niu L.N., et al. Upregulation of mitochondrial dynamics is responsible for osteogenic differentiation of mesenchymal stem cells cultured on self-mineralized collagen membranes. Acta Biomater. 2021;136:137–146. doi: 10.1016/j.actbio.2021.09.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The large osteoclast divided into small daughter cells, which were mobilizable. These smaller motile cells migrate away from each other.
The large osteoclast divided into small daughter cells, which were mobilizable. These smaller motile cells migrate away from each other.
The large osteoclasts broke up into lots of stationary cell debris, which were finally swallowed by other cells.
The large osteoclast divided into small daughter cells. And these daughter cells fused back into osteoclast.
The large osteoclast divided into small daughter cell, which was mobilizable (yellow arrow). And the daughter cell fused back into osteoclast in a few hours when treated with RANKL (red arrow).
The fate of osteoclasts in physiological states.
The fate of osteoclasts under DMSO treatment.
The fate of osteoclasts under CQ treatment.
The fate of osteoclasts under Rap treatment.
The fate of osteoclasts under 3-MA treatment.
The fate of osteoclasts under CCCP treatment.
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.