Abstract
The class II transactivator (CIITA), the master regulator of the tissue-specific and interferon gamma-inducible expression of major histocompatibility complex class II genes, synergizes with the histone acetylase coactivator CBP to activate gene transcription. Here we demonstrate that in addition to CBP, PCAF binds to CIITA both in vivo and in vitro and enhances CIITA-dependent transcriptional activation of class II promoters. Accordingly, E1A mutants defective for PCAF or CBP interaction show reduced ability in suppressing CIITA activity. Interestingly, CBP and PCAF acetylate CIITA at lysine residues within a nuclear localization signal. We show that CIITA is shuttling between the nucleus and cytoplasm. The shuttling behavior and activity of the protein are regulated by acetylation: overexpression of PCAF or inhibition of cellular deacetylases by trichostatin A increases the nuclear accumulation of CIITA in a manner determined by the presence of the acetylation target lysines. Furthermore, mutagenesis of the acetylated residues reduces the transactivation ability of CIITA. These results support a novel function for acetylation, i.e., to regulate gene expression by stimulating the nuclear accumulation of an activator.
Major histocompatibility complex (MHC) class II genes encode heterodimeric cell surface molecules that are essential for the presentation of foreign antigenic peptides to helper T cells. Human and mouse genes are expressed in antigen-presenting cells as well as in various cell types upon gamma interferon (IFN-γ) stimulation (16, 34, 52). The expression of these genes occurs mainly at the transcriptional level and is regulated by an array of functional cis elements (H/W, X, and Y) that are conserved among all class II genes (16). Transcription of class II genes is orchestrated by the assembly of a higher-order multiprotein complex on the promoter and requires recruitment of the class II transactivator, CIITA (6, 34). Both constitutive and IFN-γ-inducible expression of class II genes are determined by the presence of CIITA in a variety of cell types (8, 34, 49). Functional analysis of the structure of CIITA revealed the presence of a C-terminal region required for promoter recruitment (44, 59) and an N-terminal acidic transactivation domain that can contact the basic transcriptional machinery (13, 35).
Recently, we and others have demonstrated that the histone acetylase CREB binding protein (CBP) interacts with CIITA and functions as a coactivator for both B-cell-specific and IFN-γ-induced transcription of MHC class II genes (13, 30). Consequently, expression of MHC class II genes was suppressed by the adenovirus E1A protein (30), which is known to strongly bind to and inhibit CBP action (1, 33).
The discovery that transcriptional coactivators have histone acetylase activity (4, 41) provided important insights into the process that links chromatin acetylation to transcriptional activation (20, 31, 50). CBP/p300 and the associated factor PCAF collaborate with many transcription factors as well as with other coactivators, such as SRC-1 and ACTR, to regulate cell proliferation and differentiation (29).
In addition to histones, CBP/p300 and PCAF can acetylate nonhistone proteins such as TFIIE, TFIIF (25), p53 (17, 32, 45), EKLF (58), GATA-1 (7, 23), HMG I (Y) (38), HMG17 (21), ACTR (9), Tat (27), MyoD (46), and E2F1 (36).
In this paper we demonstrate that, similarly to CBP, PCAF binds to and enhances the action of CIITA as an MHC class II gene coactivator. The weak inhibitory activity of E1A mutants defective for binding to either coactivator shows that the action of CIITA depends on the independent and redundant recruitment of either CBP or PCAF. Furthermore, we demonstrate that PCAF and CBP acetylate specific lysine residues within a novel nuclear localization sequence (NLS) of CIITA. We show that CIITA exits from the nucleus in a CRM-1-dependent manner. Acetylation leads to an increase of CIITA accumulation in the nucleus. Mutations of acetylation-target lysines reduce the nuclear levels and transcriptional ability of CIITA. Based on these data, we propose a novel role of acetylation to regulate class II gene expression by affecting the nuclear accumulation of CIITA.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa and COS-1 cell lines were maintained in Dulbecco's modified Eagle's medium and transfected as previously described (51). Luciferase and chloramphenicol acetyltransferase assays were performed at 24 and 36 h posttransfection, respectively. When indicated, cells were treated with 50 U of IFN-γ (R&D) per ml for 12 to 20 h before being harvested.
Plasmids.
The class II −353 Eα chloramphenicol acetyltransferase construct has been described previously (51) and used to generate an equivalent luciferase reporter. Full-length CIITA or its derivatives were expressed from the pCDNA3 expression vector (30). pRSV5E1A expressing the 13S product and pRSVmCR1 and pRSVmCR2, which express molecules with deletions in these domains (from amino acids 38 to 65, 125 to 133, and 140 to 185, respectively), were provided by A. van der Eb (40). Rous sarcoma virus E1A expression plasmids with mutations within the CR1 domain that affect binding to pRb (TK496, amino acids 38 to 44 mutated to alanine [3]), CBP (TK460, amino acids 64 to 68 deleted [3]), and PCAF (E55, amino acids 55 to 60 mutated to alanine [43]) were provided by T. Kouzarides. CBP expression plasmids have been described previously (30). PCAF-expressing construct was kindly provided by Y. Nakatani. The CIITA lysine mutants were constructed with the Gene Editor in vitro site-directed mutagenesis system from Promega. The mutagenic primers were K141,144R (5′-GTTGGGCAGAGAAGTCAGAGAAGACCCTTC) and K156,159R (5′-GCAGACCTGAGGCACTGGAGGCCAGCTGAG). All constructions were verified by sequencing.
In vitro protein-protein interaction experiments.
Fragments of PCAF and CIITA were subcloned into pGEX vectors (Pharmacia) in frame with glutathione S-transferase. Approximately 2 μg of fusion proteins was immobilized to glutathione-Sepharose beads and incubated with in vitro-translated and 35S-labeled (TNT; Promega) CIITA or PCAF protein in a buffer containing 150 mM KCl, 20 mM HEPES (pH 7.9), 0.1% NP-40, 5 mM MgCl2, and 0.2% bovine serum albumin and supplemented with protease inhibitors. Reactions were carried out at 4°C for 5 h, and the mixtures were washed three times in the same buffer without bovine serum albumin. Bound proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by autoradiography.
Immunoprecipitation (IP) and Western blot analysis.
For in vivo protein-protein interactions, COS-1 cells in 100-mm-diameter dishes were transfected with 5 μg of each plasmid using the calcium phosphate method. Whole-cell extracts were prepared in lysis buffer containing 10 mM Tris-HCl (pH 8), 170 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol, and protease inhibitors. Extracts equivalent to about 5 × 106 cells were incubated for 16 h at 4°C with anti-Flag M2 agarose (Sigma). The immunoprecipitated samples were washed four times with lysis buffer containing 250 mM NaCl and subjected to SDS-PAGE. Western blot analysis was performed using monoclonal anti-HA (Santa Cruz), anti-Flag (Kodak), or anti-green fluorescent protein (GFP) (Clontech) antibodies.
In vitro acetylation assays.
Substrate proteins (1 to 2 μg) (glutathione S-transferase [GST] fusions of CIITA fragments) were incubated with 0.2 μg of GST-PCAF protein in 30 μl of acetylation buffer containing 50 mM HEPES (pH 7.9), 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 1 μl of [3H]acetyl coenzyme A (41). For IP-histone acetyltransferase (HAT) assays (4) whole-cell extracts from COS-1 cells transfected with CBP or PCAF were immunoprecipitated with anti-CBP (A22 and C20 [Santa Cruz]) or anti-Flag antibodies.
In vivo labeling.
COS-1 cells were transfected with constructs expressing tagged wild-type or mutant GFP-CIITA from amino acids 1 to 114 and 1 to 408. At 36 h after transfection, the cells were incubated for 1 h in Dulbecco's modified Eagle's medium containing sodium [3H]acetate (1 mCi/ml). Whole-cell extracts were immunoprecipitated with anti-GFP antibody. Immunopurified proteins were subjected to SDS-PAGE and detected by autoradiography. An identical SDS-PAGE gel was transferred to nitrocellulose, and proteins were detected by immunoblotting with anti-GFP monoclonal antibody.
GFP analysis.
Fusions with GFP were constructed in the vector pEGFP-C1 (Clontech). The GFP-p65 construct was provided by D. Thanos. Localization of transfected proteins was detected by using an Olympus IMT2 fluorescence microscope on living or fixed (PBS-acetone, 2:3) cells. When required, cells were counterstained with Hoecht 33342 stain. Quantitative protein expression was determined by Western blot analysis with an anti-GFP monoclonal antibody (Clontech).
RESULTS
The acidic activation domain of CIITA binds to PCAF in vitro and in vivo.
Previous studies have shown that CBP, a protein acetylase, synergizes with CIITA for maximal expression of MHC class II genes (30). In the present study, we decided to investigate the role of PCAF, an acetylase that associates with CBP (56) and forms a higher-order multiprotein complex (PCAF complex [42]), in MHC class II gene transcription.
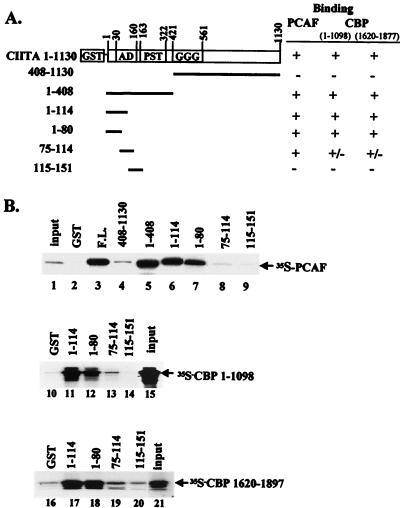
To identify the CIITA region involved in PCAF binding, we used different CIITA fragments fused to GST (Fig. 1A). CIITA fragments containing the amino-terminal transcription activation domain of CIITA (fragments 1–408 and 1–114 [lanes 5 and 6, Fig. 1B]) interacted with PCAF. Interestingly, the same amino-terminal region (amino acids 1 to 114) was also responsible for binding to CBP (30). In an attempt to map more precisely the CBP and PCAF interaction sites within the N-terminal domain of CIITA, the region extending between amino acids 1 to 151 was divided into three smaller parts. As shown in Fig. 1B, both PCAF and CBP bind predominantly to the first 80 amino acids of CIITA (Fig. 1B lanes 7, 12, and 18), which contain an α-helix required for full activity of CIITA (13).
FIG. 1.
The activation domain of CIITA binds to both CBP and PCAF. (A) GST fusions of different parts of the CIITA protein used for interaction with PCAF and CBP are shown schematically. (B) In vitro-translated and 35S-labeled PCAF or two regions of CBP (CBP 1–1098 and CBP 1620–1877) were used in a GST pull-down assay with equal amounts (1 μg) of GST alone (lanes 2, 10, and 16) or fused with the indicated fragments of CIITA. Input was 5% in lane 1, 30% in lane 15, and 20% in lane 21.
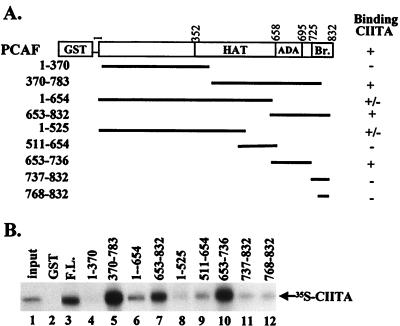
To map the region of PCAF that binds CIITA, we used the PCAF deletion constructs depicted in Fig. 2A. As shown in Fig. 2B, the C-terminal region (amino acids 370 to 783) of PCAF is sufficient for CIITA binding (lane 5) whereas the N-terminal region (amino acids 1 to 370) is not (lane 4). Further analysis showed that the HAT domain (contained in fragments 1–654 and 511–654 in lanes 6 and 9) interacted weakly with CIITA whereas the region that harbors the ADA binding domain (fragment 653–736) showed a strong interaction (lane 10). The Bromo domain was devoid of the ability to interact (lanes 11 and 12). Taken together, these results show that the region of PCAF that binds to CIITA is distinct from the region required for binding to CBP and to nuclear receptors and coactivators.
FIG. 2.
Regions of PCAF that interact with CIITA. (A) Scheme of the GST-PCAF proteins used for interaction with CIITA. Br., bromodomain. (B) In vitro 35S-labeled CIITA was used in a GST pull-down assay with equal amounts of GST alone (lane 2) or GST fused to the indicated regions of PCAF (lanes 3 to 12).
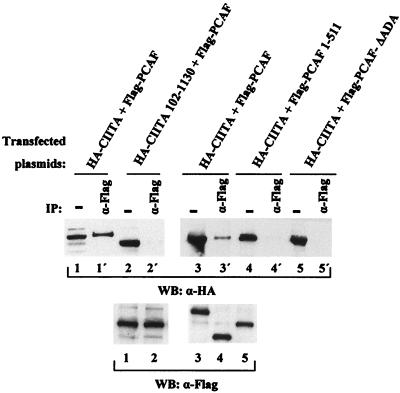
To examine whether PCAF and CIITA associate in vivo, we cotransfected COS-1 cells with expression vectors encoding Flag-tagged PCAF and HA-tagged CIITA and performed coimmunoprecipitation experiments. Figure 3 shows that the intact CIITA protein interacts with PCAF (lane 1') whereas a truncation of the first 102 amino acids (which are required for binding in vitro to PCAF) does not (lane 2'). We also determined which regions of PCAF are important for in vivo association with CIITA. A PCAF protein that contains only the first 511 amino acids (PCAF 1–511) and a PCAF that lacks the ADA binding region (amino acids 653 to 736) (PCAF ΔADA) cannot interact with CIITA (lanes 4' and 5', respectively), in contrast to the wild-type protein (lane 3'). Therefore, the in vivo interaction analysis correlates well with the in vitro data presented previously.
FIG. 3.
In vivo interaction between CIITA and PCAF. Whole-cell extracts from COS-1 cells transfected with the indicated plasmids (5 μg each) were immunoblotted (WB) with anti-HA antibody (α-HA) before (lanes 1 to 5) or after (lanes 1' to 5') immunoprecipitation (IP) with anti-Flag M2 agarose. In lanes 1 to 5 inputs are 10% of the extract used in immunoprecipitation. Equivalent amounts of inputs were also checked for expression of PCAF derivatives using anti-Flag antibody (α-Flag).
PCAF coactivates MHC class II genes.
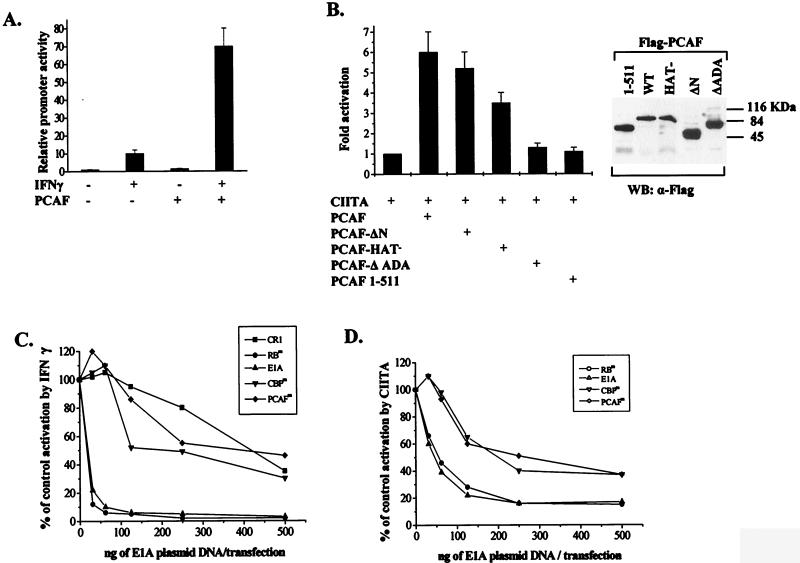
We next asked if PCAF was able to coactivate MHC class II genes. HeLa cells were transiently transfected with the Eα MHC class II promoter construct along with a PCAF-expressing plasmid. The activity of the promoter was determined from untreated cells or cells after a 20-h treatment with IFN-γ, which is known to induce CIITA expression, which in turn induces class II gene transcription. Although the presence of PCAF did not significantly affect the basal promoter activity, it strongly enhanced (up to sevenfold) the IFN-γ-induced expression (Fig. 4A). Since the effect of IFN-γ on class II gene expression is mediated by CIITA, we examined the ability of PCAF to directly coactivate exogenously provided CIITA. Transfection of a CIITA expression vector resulted in activation of the Eα class II promoter in HeLa cells, whereas PCAF alone did not produce any effect (data not shown). Coexpression of PCAF augmented the CIITA-mediated activation (Fig. 4B). Therefore, PCAF and CIITA synergistically activate class II promoter transcription. Deletion of the CBP interaction domain of PCAF (amino acids 1 to 372) did not abolish the ability of PCAF to stimulate CIITA-dependent class II transcription (PCAF-ΔN; Fig. 4B). Thus, the ability of PCAF to stimulate CIITA activity is mainly CBP independent. A PCAF protein bearing mutations in the HAT domain (PCAF HAT− [28]) has a lowered capacity to coactivate CIITA compared to that of the wild-type protein. This suggests that histone or protein acetylation contributes to the synergy observed between PCAF and CIITA. Two PCAF deletion mutants that do not interact with CIITA (PCAF 1–511 and PCAF ΔADA [Fig. 4B]) were unable to coactivate CIITA. This result underlines the importance of strong PCAF binding to CIITA for the coactivation of class II genes.
FIG. 4.
PCAF coactivates MHC class II gene expression. (A) Plasmids encoding PCAF or vector control (0.5 to 1 μg) were cotransfected with 1 μg of an MHC class II promoter-CAT reporter plasmid into HeLa cells. Uninduced and IFN-γ (50 U/ml)-induced activities were assayed 24 h after IFN-γ addition. The CAT activity of vector transfected-uninduced cells was set to 1. Bars represent the standard error of the mean (SEM) from four experiments. (B) HeLa cells were cotransfected with a class II-luciferase reporter (1 μg) a CIITA-expressing plasmid (20 ng), and 0.5 to 1 μg of plasmids expressing wild-type PCAF, a deletion lacking its first 352 amino acids (PCAF-ΔN), a mutant that has no HAT activity (PCAF-HAT−) a mutant containing only the first 511 amino acids (PCAF 1–511), and a mutant that lacks amino acids 653 to 736 (PCAF-Δ ADA). Luc activity was measured 24 h after transfection. The activity of the class II-Luc reporter in the presence of CIITA protein was set to 1 and represents an induction range between 7- and 15-fold. Bars represent SEM from four experiments. The expression levels of the PCAF molecules were analyzed by Western blotting (WB) using an anti-Flag antibody (α-Flag). (C) HeLa cells were transfected with 1 μg of class II-CAT plasmid along with vector control or increasing amounts of plasmids expressing the indicated E1A products. Results are means of three experiments, with standard deviation less than 25% of the mean value. IFN-γ induction ranged between 15- and 45-fold. (D) HeLa cells were cotransfected with 1 μg of class II-CAT and 100 ng of a CIITA-expressing plasmid and vector control plasmid or increasing amounts of plasmids expressing the indicated E1A derivatives. Results are expressed as a percentage of the vector control activation by CIITA and are averages of three experiments, with standard deviation less than 30% of the mean value. Activation by CIITA ranged between 75- and 120-fold.
To evaluate the relative importance of CBP and PCAF in MHC class II regulation, we took advantage of E1A mutants (Fig. 4C and D) that specifically impair the binding to either PCAF or CBP. Mutant E55 (PCAFm) shows impaired binding to PCAF (43), and mutant TK460 (CBPm), in which amino acids 64 to 68 are deleted, cannot bind to CBP (3). Both types of mutations strongly reduced the ability of E1A to suppress IFN-γ induction as well as the direct activation by CIITA of a class II promoter (Fig. 4C and D). Cotransfection of the 13s E1A or a mutant defective for pRb binding, Rbm (mutant TK496 in which amino acids 38 to 44 are mutated to alanine [3]), showed strong suppression, in contrast to a CR1 deletion, which was a very weak inhibitor (Fig. 4C and D). Thus, sequestration of either coactivator by E1A is not sufficient to reduce promoter activation. Taken together, these results demonstrate the functional redundancy of PCAF and CBP, which can independently enhance the action of CIITA.
PCAF and CBP acetylate CIITA.
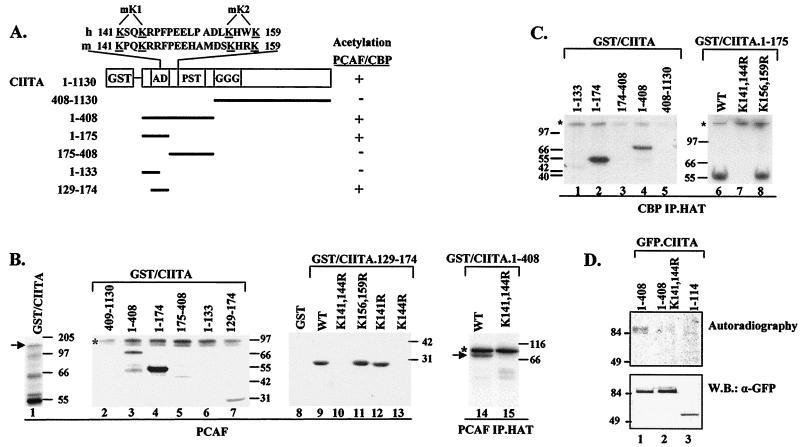
PCAF and CBP are protein acetyltransferases that use as substrates both histones and other proteins (reviewed in references 5 and 29). Therefore, we investigated whether CIITA could be acetylated by PCAF and CBP. Figure 5B shows that PCAF can acetylate CIITA in vitro (lane 1). Deletion analysis indicated that acetylation of CIITA is restricted to amino acids 129 to 174 (lane 7). This region contains two pairs of closely spaced lysines, which are well conserved between human and mouse CIITA (Fig. 5A). Mutation of the first pair, K141,144 (designated mK1), abolished acetylation (lane 10), whereas mutation of the second pair, K156,159 (designated mK2), had no effect (lane 11). Therefore, PCAF acetylates the first lysine pair, K141,144. Further mutagenesis indicated that lysine 144 is the acetylation target of PCAF (lane 13). A similar result was obtained when acetylation was performed using PCAF immunoprecipitated from transfected COS-1 cells (lanes 14 and 15).
FIG. 5.
CIITA is acetylated in vitro and in vivo. (A) GST-CIITA fusion proteins that were assayed for in vitro acetylation. The aligned amino acid sequence of the human and murine CIITA region spanning amino acids 141 to 159 is shown, and critical lysine residues are underlined. mK1 and mK2 designate mutations of lysine pairs K141,144 and K156,159, respectively. (B) PCAF acetylation assays. Portions (1 to 2 μg) of the indicated CIITA fragments were subjected to in vitro acetylation using 500 ng of GST-PCAF (lanes 1 to 13) or PCAF immunoprecipitated on anti-Flag M2 agarose from COS-1 cells (lanes 14 and 15). ∗, PCAF autoacetylation. WT, wild type. (C) CBP immunoprecipitation-acetylation assay. CBP was immunoprecipitated (IP) from COS-1 cell extracts with anti-CBP antibodies and used for acetylation of the indicated CIITA fragments. ∗, CBP autoacetylation. (D) In vivo acetylation. COS-1 cells transfected with the indicated plasmids were labeled with [3H]acetate for 1 h. The resulting whole-cell extracts were immunoprecipitated with anti-GFP monoclonal antibody, subjected to SDS-PAGE (10% polyacrylamide), and subjected to autoradiography. Identical nonradiolabeled samples were immunoblotted (W.B.) with anti-GFP antibody (α-GFP).
We next checked the ability of CBP to acetylate CIITA. Since a bacterially produced CBP HAT domain was unable to acetylate CIITA, we used CBP immunoprecipitated from transfected COS cells. As shown in Fig. 5C, CBP can acetylate CIITA. Deletion analysis limited the area that was acetylated to the first 174 residues (lane 2). Using mutant versions of the region from amino acids 1 to 174, we determined that CBP acetylates the lysine pair K141,144 of CIITA (lane 7) but does not acetylate the lysine pair K156,159 (lane 8). It is unlikely that acetylation of CIITA is due to some other acetylase that coimmunoprecipitates with CBP, since no CIITA acetylation was obtained when extracts prepared from cells transfected with a CBP HAT− expression vector were immunoprecipitated (not shown).
To test whether the in vitro-detected acetylation sites are also targets for acetylation in vivo, we performed metabolic labeling of cells expressing GFP-tagged CIITA proteins. Figure 5D shows that a truncated protein that contains the first 114 amino acids of CIITA and is devoid of any lysine is not labeled (lane 3), excluding the possibility that we detected amino-terminal acetylations. The wild-type 1–408 CIITA protein was efficiently labeled (lane 1), whereas mutant K141,144R showed severely reduced acetylation (lane 2). The intact CIITA protein was also acetylated in vivo (not shown), although the effect of mutating lysines 141 and 144 was less clear, presumably because additional lysine targets might exist in the protein. The conclusion of our acetylation analysis is that the lysine pair K141,144 of CIITA is acetylated both in vitro and in vivo. From now on, mutants K141,144R and K156,159R will be called mK1 and mK2, respectively.
CIITA is acetylated on an NLS.
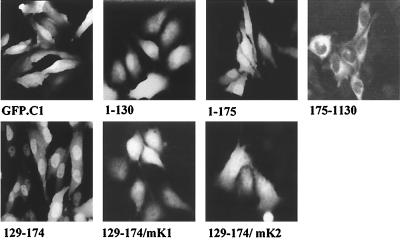
The CIITA region which contains the acetylated lysine residues (Fig. 5A) is highly conserved between human and murine CIITA (47) and has similarity to a bipartite NLS. To test the ability of this region to mediate CIITA import in the nucleus, we used fusions of CIITA with GFP. Fusion of GFP with the first 130 amino acids of CIITA (1–130) produced a protein that was evenly distributed, like the GFP protein itself, whereas a fusion of the first 175 amino acids (1–175) showed nuclear localization (Fig. 6), suggesting that NLSs may reside in this fragment. Accordingly, an amino-terminal deletion of 174 amino acids (175–1130) rendered CIITA cytoplasmic. The NLS was further restricted between amino acids 129 to 174, as indicated by the corresponding GFP fusion, which showed a prominent nuclear fluorescence (Fig. 6). Furthermore, replacement of lysines 141 and 144 (mK1) or 156 and 159 (mK2) with arginines in the context of GFP-CIITA/129–174 generated proteins that were homogeneously distributed in both the nucleus and cytoplasm. Therefore, the region extending from lysines 141 to 159 is a bipartite NLS that requires both motifs for its function.
FIG. 6.
An NLS is contained between amino acids 141 and 159 of CIITA. GFP fusions of the indicated regions of CIITA were transfected in HeLa cells. Green fluorescence was observed 24 h later.
Acetylation increases CIITA accumulation in the nucleus.
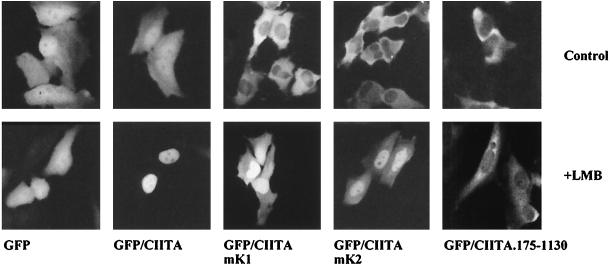
Since acetylation target lysines reside within an NLS, we tested whether acetylation might affect the nuclear localization of the protein. To do this we used GFP fusions to study the subcellular distribution of the acetylation-deficient mutant mK1 in comparison to mK2 and the wild-type protein. The GFP-CIITA fusion protein was distributed in both the nucleus and the cytoplasm, whereas mK1 and mK2 mutants showed a predominantly cytoplasmic localization (Fig. 7, upper panel). More specifically, as shown in Table 1, wild-type CIITA showed stronger nuclear than cytoplasmic (N > C) distribution in 33% of cells, in contrast to only 0.5% of cells expressing either mK1 or mK2 mutant protein. Furthermore, mK2 was homogeneously (N = C) distributed in 14.4% of cells, compared to only 3.5% of mK1 protein. Interestingly, treatment with leptomycin B (LMB) (54), a well-characterized inhibitor of CRM-1-mediated nuclear export (14, 15), led to strong nuclear localization of CIITA (Fig. 7, LMB). Mutants mK1 and mK2 also responded to LMB (Fig. 7, LMB) to a lesser extent, suggesting that they retain residual import activity and are able to accumulate in the nucleus when export is inhibited. As a control, we showed that an N-terminally truncated protein that lacks the NLS and contains amino acids 175 to 1130 was also cytoplasmic but did not respond to LMB (Fig. 7). Thus, the observed cellular distribution of CIITA is determined by both nuclear import and a CRM-1-dependent export.
FIG. 7.
CIITA exits from the nucleus via CRM-1. The nucleocytoplasmic distribution of wild-type and mutant GFP-CIITA fusion proteins in HeLa cells is shown. LMB indicates a 2-h treatment with 20 nM LMB.
TABLE 1.
Subcellular distribution of GFP fusion proteins after TSA treatment
| Distribution | % of cellsa with fluorescence due to:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild-type CIITA
|
mK1
|
mK2
|
p65
|
|||||
| −TSA | +TSA | −TSA | +TSA | −TSA | +TSA | −TSA | +TSA | |
| N > C | 33 ± 3 | 77 ± 8.5 | 0.5 ± 0.2 | 0.6 ± 0.2 | 0.4 ± 0.2 | 2.2 ± 0.7 | 13.3 ± 3.1 | 14.2 ± 4 |
| N = C | 54.4 ± 4.2 | 19.5 ± 3 | 3.5 ± 1 | 5.5 ± 2 | 14.4 ± 2.7 | 60.4 ± 8.1 | 14.7 ± 4 | 13.1 ± 3.5 |
| C > N | 12.6 ± 2 | 3.5 ± 0.8 | 96 ± 1.2 | 93.4 ± 3 | 85.2 ± 2.8 | 37.4 ± 7.2 | 72 ± 6.7 | 72.7 ± 6.5 |
The percentage of cells with clearly visible nuclear over cytoplasmic fluorescence (N > C), fluorescence homogeneously distributed in both nucleus and the cytoplasm (N = C), or predominantly cytoplasmic fluorescence (C > N) is shown 24 h posttransfection without (−) or in the presence of (+) 1 μM TSA. The results shown are the average percentages together with their standard deviation derived from five experiments. During each experiment, more than 1,000 cells were scored.
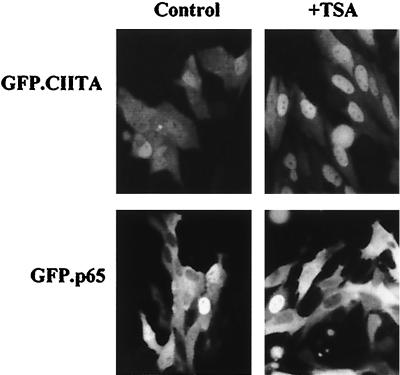
To correlate the subcellular distribution of CIITA with its acetylation state, we used the deacetylase inhibitor trichostatin A (TSA) (57). HeLa cells were transfected with GFP fusions of wild-type or mutant CIITA and GFP-p65, and the cytoplasmic-nuclear distribution of these proteins was determined by fluorescence microscopy. Figure 8 shows that the addition of TSA strongly enhanced the nuclear accumulation of GFP-CIITA but did not significantly affect the subcellular distribution of GFP-p65, which is not acetylated by CBP or PCAF (38) and shuttles between the nucleus and cytoplasm in a regulated fashion (2, 18). Table 1 quantitates the nuclear and cytoplasmic distribution of wild-type, mutant CIITA, and p65 before and after addition of TSA. A 24-h treatment with TSA led to strong redistribution of GFP-CIITA toward the nuclear compartment (N > C cells increased from 33 to 77%) with a concomitant decrease of the homogeneously (N = C cells decreased from 54 to 19%) and cytoplasmically (C > N) expressed protein. Interestingly, TSA treatment significantly affected the localization of mK2, by increasing its homogenous distribution to 60%, but did not affect the distribution of the acetylation mutant mK1. As a control, we showed that TSA did not affect the subcellular distribution of p65.
FIG. 8.
Inhibition of deacetylases by TSA enhances nuclear accumulation of CIITA. HeLa cells were transfected with GFP-CIITA or GFP-p65 expression plasmids and further cultivated in the absence (Control) or presence of 1 μM TSA (+TSA). The picture shows representative fields of fluorescent cells 24 h after the addition of TSA.
Similar results were obtained in parallel experiments with COS-1 cells (data not shown). Thus, inhibition of deacetylation shifts the equilibrium of CIITA from the cytoplasm to the nucleus in a manner dependent on the presence of the lysine pair K141,144.
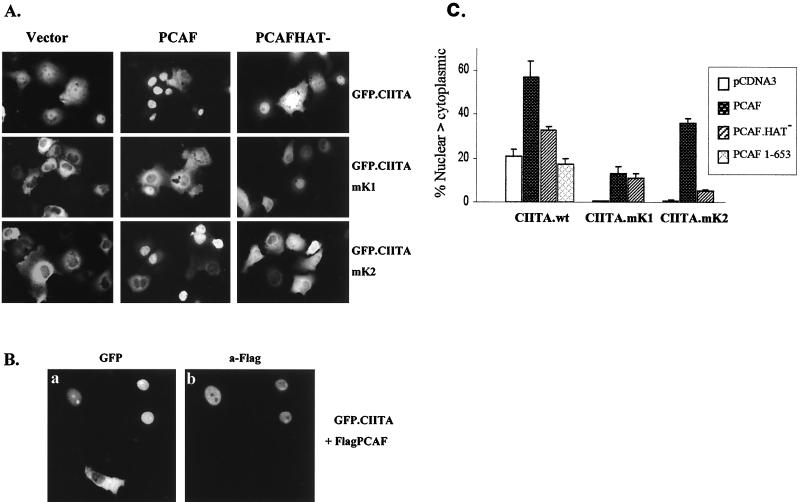
To obtain direct proof that CIITA acetylation by PCAF affects its nuclear and cytoplasmic distribution, we cotransfected GFP-CIITA and PCAF expressing plasmids in COS-1 cells. Exogenously expressed PCAF was able to increase the nuclear levels of CIITA. As shown in Fig. 9A and C, PCAF coexpression promoted the nuclear localization of GFP-CIITA whereas a PCAF HAT− protein was significantly less effective. Cells expressing high levels of PCAF almost invariably presented strong nuclear localization of CIITA. Figure 9B exemplifies this by showing that nuclear GFP-CIITA localization coincides with PCAF expression. The subcellular distribution of the GFP vector alone was not affected by coexpressing PCAF (data not shown). The NLS mutant mK2 also responded strongly to PCAF (the percentage of cells bearing nuclear fluorescence was raised from 0.7 to 33%), and this effect was dependent on HAT activity (Fig. 9C). However, the response of the NLS acetylation mutant mK1 was significantly lower and did not depend on the presence of an intact HAT domain (Fig. 9C). Deletions of PCAF that do not retain strong interaction with CIITA, such as PCAF 1–653 (Fig. 9C) or PCAFΔADA (data not shown), were not able to relocate CIITA to the nucleus. Thus, the effect of PCAF on CIITA localization depends on its ability to both bind and acetylate. Taken together, these results show that acetylation by PCAF or inhibition of deacetylation leads to increased nuclear levels of CIITA by targeting lysines 141 and 144.
FIG. 9.
PCAF promotes nuclear localization of CIITA. (A) Cells were transfected with 1 μg of GFP-CIITA fusion plasmid and 3 μg of Flag-PCAF or Flag-PCAF-HAT− expressing plasmids or empty vector alone. The cells were analyzed 20 to 24 h later. (B) Coexpression of PCAF produces high levels of nuclear CIITA. Cells transfected with GFP-CIITA and Flag-PCAF were stained with an anti-Flag antibody followed by rhodamine-conjugated secondary antibody and analyzed for either GFP fluorescence (a) or rhodamine stain (b). (C) Quantitative analysis of the above results. The percentage of cells expressing predominantly or fully nuclear CIITA (N > C) in the presence of vector or the indicated PCAF expression plasmids is shown. Results presented were derived from the analysis of more than 300 cells 24 h after transfection and are averages from four independent experiments.
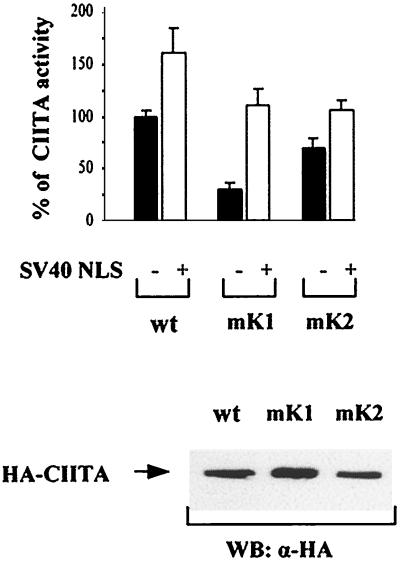
We next investigated the consequences of CIITA acetylation on MHC class II gene transcription. For this purpose, we examined the abilities of wild-type and mutant CIITA proteins to activate a class II promoter. Figure 10 shows that the acetylation NLS mutant mK1 had a strongly reduced ability to transactivate a class II promoter (25% of the wild-type level). In contrast, the NLS mutant mK2 retained 70% of wild-type activity (Fig. 10). The differential activity of the two mutant proteins correlated with their difference in subcellular CIITA distribution (Table 1). To determine whether we could influence the activity of wild-type and mutant CIITA by increasing their nuclear content, we produced fusions carrying at their N terminus the NLS of simian virus 40 (SV40). The addition of the SV40 NLS rendered all these proteins nuclear (data not shown), leading to an increase of their activity (Fig. 10). More specifically, the activity of the acetylation mutant mK1 was restored and reached a level comparable to that of mK2. Therefore, enhanced nuclear accumulation of CIITA leads to higher transactivation of class II genes.
FIG. 10.
Transactivation by CIITA acetylation mutants. COS-1 cells were cotransfected with 1 μg of Eα class II-Luc reporter and 25 ng of the indicated wild-type (wt) or mutant CIITA expression plasmids. Luciferase activity was measured 24 h posttransfection. The results are presented as the percentage of promoter activation obtained by the wild-type CIITA, which is set to 100%, and they are average values of four experiments. The solid and open bars indicate the activities of CIITA derivatives in the absence or presence of the SV40 NLS, respectively. The expression pattern of CIITA proteins was analyzed by immunoblotting (WB) with an anti-HA antibody.
DISCUSSION
In this study we examined the role of PCAF in the transcriptional regulation of MHC class II genes. We showed that in addition to CBP (13, 30), PCAF binds to the class II transactivator and is able to augment the IFN-γ- or CIITA-mediated transcriptional activation of class II genes. Binding of PCAF or CBP leads to acetylation of CIITA at lysines that reside within a NLS. We further demonstrate that CIITA exits the nucleus via a CRM-1-dependent pathway. Based on the effect of the deacetylase inhibitor TSA and the action of PCAF to increase the nuclear levels of CIITA through acetylation of these lysines, we propose that a novel function of acetylation is to regulate the activity of an activator by increasing nuclear accumulation.
Both CBP and PCAF are essential for MHC class II gene transcription.
We demonstrate here that PCAF binds to and synergizes with CIITA to induce class II gene transcription. This effect requires a strong interaction between the two proteins and also the acetyltransferase activity of PCAF. The full effect of PCAF on a class II promoter is derived from its combined action on the recruitment of the transcriptional machinery and acetylation of chromatin and, interestingly, CIITA itself. We have observed that CBP and PCAF cannot bind simultaneously to CIITA in vitro (data not shown). This is not unexpected considering the short CIITA region—the first 80 amino-terminal amino acids—with which they both interact. In this study we show that E1A oncoprotein, which inhibits the synergy between CIITA and CBP (30), also interferes with the interaction between CIITA and PCAF. This interference might result from the prevention of CIITA binding to PCAF, since E1A (43) and CIITA bind to the same region of PCAF. E1A mutants that cannot bind to either CBP or PCAF (3, 43) have equally reduced ability to inhibit the activity of CIITA on an MHC class II promoter. Taken together, the physical interaction properties of CIITA with those coactivators and the pattern of suppression by various E1A mutants are in support of their redundant and independent recruitment by CIITA.
CBP and PCAF are histone and factor acetyltransferases that participate in the formation of diverse multisubunit complexes. CBP is a component of the RNA polymerase II holoenzyme (39), whereas PCAF is found in human cells in a complex that is the equivalent of the yeast SAGA and includes human counterparts of yADA2, yADA3, ySpt3, and histone-like TAFs (42). Thus, the ability of CIITA to recruit CBP and PCAF acetyltransferases independently provides it with the capability to modulate chromatin structure by alternative interactions with multiple complexes. CIITA is required for the establishment of an “occupied” class II promoter configuration in IFN-γ-induced fibroblasts but not in B lymphocytes (53, 55). This difference may be due to cell-type-specific chromatin-modifying complexes that interact with CIITA. There is little information regarding the regulation of activity of CBP and PCAF themselves in different cells and in response to different signals (37). Therefore, we assume that conditions that regulate the availability and stoichiometry of each of the two coactivators may also affect the function of CIITA.
CIITA acetylation as a regulatory mechanism for its subcellular distribution.
We show that acetylation can regulate the subcellular localization of CIITA, which shuttles between the nucleus and the cytoplasm. CIITA shuttling is achieved by the combined action of nuclear import and CRM-1-dependent nuclear export. The latter involves nuclear export sequences recently characterized in CIITA (C. Spilianakis et al., unpublished data). We identify here a bipartite NLS in the amino-terminal region of CIITA. Another NLS, which was deleted in a bare lymphocyte syndrome patient, was recently identified in the carboxy-terminal region (amino acids 955 to 960) (11). It seems that both NLSs are required for efficient nuclear import and function of CIITA. The first half of the amino-terminally located NLS contains lysines 141 and 144, which are targeted for acetylation by CBP and PCAF. We demonstrate that conditions favoring acetylation, such as coexpression of PCAF or inhibition of deacetylases, lead to increased nuclear levels of CIITA. This effect requires (i) direct physical interaction between PCAF and CIITA and (ii) lysine pair K141,144. When nuclear import of CIITA is compromised by the NLS mutation mK2, the action of acetylation to control nuclear levels becomes even more apparent. The NLS acetylation mutant mK1 is not affected by the acetylase activity of PCAF, although it is relocated to a small extent in the nucleus by PCAF in a HAT-independent manner. This effect underlines the ability of simple binding to PCAF to provoke sequestration in the nucleus of mK1 and, to a lesser extent, of mK2 and the wild-type molecule. The deacetylase inhibitor TSA mimicked, although to a lesser extent, the effect of PCAF on the mK2 mutant. A plausible interpretation of this could be that TSA is acting indirectly and thus with slower kinetics than PCAF. Increased nuclear levels of CIITA can be attributed to either increased import or decreased export. Since acetylases such as PCAF are nuclear proteins, we favor the second hypothesis, although the first cannot be formally excluded. These alternative mechanisms are currently under investigation.
The acetylation state of CIITA is important for its activity. The ability of a HAT-deficient PCAF to coactivate CIITA is lowered in comparison to that of the wild-type molecule but is not abolished. This lack of abolition may be attributed to the ability of a HAT− PCAF to recruit the acetylase CBP. The importance of CIITA acetylation on the transcriptional activation of a class II promoter is shown by the reduced transactivation capability of the acetylation mutant mK1 compared to the wild type and the NLS mutant mK2. Since both mutations retain a residual ability for nuclear import, as judged by the action of LMB (Fig. 7), the difference in transactivation capability between mK1 and mK2 underlines the importance of acetylation in CIITA function.
Acetylation is believed to induce structural alterations that could affect protein-DNA or protein-protein interactions. For CIITA, acetylation might induce a conformational change, which alters the way in which CIITA interacts with the nuclear export machinery. Alternatively, it is possible that acetylated CIITA is retained in the nucleus due to association with an architectural nuclear structure (the nucleoskeleton) (10, 22, 24, 26), possibly via intermediary molecules like PCAF. Acetylation might also facilitate or stabilize CIITA recruitment on MHC class II promoters. Conversely, deacetylation could reduce nuclear levels and promoter complex assembly, thus limiting the ability of CIITA for transcriptional activation.
It was recently reported that GTP binding is required for nuclear localization and activation of target genes by CIITA (19). The possibility of a functional connection of these two posttranslational modifications—acetylation and GTP binding—to regulate the subcellular distribution and activity of CIITA is under investigation.
Control of intracellular protein trafficking is a novel mechanism through which acetylation may regulate the action of a transcriptional activator or coactivator such as CIITA. Remarkably, in two other cases, acetylated residues reside within NLSs. Both acetylation sites of p53, by CBP and PCAF, reside in NLSs (45), whereas acetylated lysines of EKLF also overlap an NLS (58). It will be interesting to investigate whether in these cases the acetylation state of the proteins affects their subcellular distribution.
In conclusion, our data show that PCAF activates the transcription of MHC class II genes through both acetylation-dependent and -independent mechanisms. We demonstrate that a novel function of acetylation is to shift the nucleocytoplasmic equilibrium of an activator toward the nucleus.
In a paper published prior to this submission, it is shown that acetylation by CBP is required for nuclear retention of hepatocyte nuclear factor 4 (48).
ACKNOWLEDGMENTS
We thank T. Makatounakis and G. Vretzos for providing excellent technical assistance and T. Kouzarides and Y. Nakatani for providing the indicated reagents. We are indebted to D. Thanos for helpful discussions and critical reading of the manuscript. We also thank C. Mamalaki, I. Talianidis, and S. Georgatos for critical reading; Nektaria Kelaidi for help with secretarial work; L. Kalogeraki for photographic work; and T. Makatounakis for assistance with the presentation.
The present work was supported by the Greek Secretariat General for Research through Institutional funds and European Union (EPET II) grant 236.234.603 and National (PENED) grant 2016.
REFERENCES
- 1.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A S. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Berger S L. Gene activation by histone and factor acetyltransferases. Curr Opin Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 6.Boss J M. Regulation of transcriptional of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 7.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 8.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Lin R, Xie W, Wilpitz D, Evans R. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 10.Cook P R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 11.Cressman D E, Chin K C, Taxman D J, Ting J P Y. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 12.Fontes J D, Jiang B, Peterlin B M. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 16.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Roeder R. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Harhaj E W, Sun S C. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol Cell Biol. 1999;19:7088–7095. doi: 10.1128/mcb.19.10.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harton J, Cressman D, Chin K, Der C, Ting J. GTP binding by class II transactivator. Role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 20.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrera J, Sakagushi K, Berger M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosome protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hozak P. The nucleoskeleton and attached activities. Exp Cell Res. 1996;229:267–271. doi: 10.1006/excr.1996.0370. [DOI] [PubMed] [Google Scholar]
- 23.Hung H L, Lau J, Kim A, Weiss M, Blobel G. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iborra F J, Pombo A, Jackson D A, Cook P R. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 25.Imhof A, Yang X J, Ogryzko V, Nakatani Y, Wolffe A, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 26.Jackson D A, Hassan A B, Errington R J, Cook P R. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiernan R, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang K-T, Benkirane M, Van Lint C. HIV-1 Tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korzus E, Torchia J, Rose D, Xu L, Kurokawa R, McInerney E, Mullen T-M, Glass C, Rosenfeld M. Transcription factor-specific requirements for coactivators and their associated acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 30.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo M-H, Zhou J, Jampeck P, Churchill M, Allis D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Scolnick D, Trievel R, Zhang H B, Marmorstein R, Halazonetis T, Berger S. p53 acetylated in vitro by pCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 34.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 35.Mahanta S K, Scholl T, Yang F C, Strominger J L. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci USA. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Balbas M, Bauer U-M, Nielsen S, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masumi A, Wang I M, Lefebvre B, Yang X J, Nakatani Y, Ozato K. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol Cell Biol. 1999;19:1810–1820. doi: 10.1128/mcb.19.3.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I (Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima T, Ushida C, Anderson S, Parvin J, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 40.Offringa R, Gebel S, van Dam H, Timmers M, Smits A, Zwart R, Stein B, Boss J L, van der Eb A, Herrlich P. A novel function of the transforming domain of E1A: repression of AP-1 activity. Cell. 1990;62:527–538. doi: 10.1016/0092-8674(90)90017-9. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko V, Schiltz L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Ogryzko V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X J, Howard B, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 43.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferases. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 45.Sakaguchi K, Herrera J, Saito S, Miki T, Bustin M, Vassilev A, Anderson C, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang J, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 47.Sims T, Elliot J, Ramassar V, Denney D, Halloran P. Mouse class II transactivator: cDNA sequence and amino acid comparison with the human class II transactivator. Immunogenetics. 1997;45:220–222. doi: 10.1007/s002510050193. [DOI] [PubMed] [Google Scholar]
- 48.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 49.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 50.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 51.Thanos D, Mavrothalassitis G, Papamatheakis J. Multiple regulatory regions on the 5′ side of the mouse E alpha gene. Proc Natl Acad Sci USA. 1988;85:3075–3079. doi: 10.1073/pnas.85.9.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ting J P, Baldwin A S. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 53.Villard J, Muhtethaler-Mottet A, Bontron S, Mach B, Reith W. CIITA-induced occupation of MHC class II promoters is independent of the cooperative stabilization of the promoter-bound multi-protein complexes. Int Immunol. 1999;11:461–469. doi: 10.1093/intimm/11.3.461. [DOI] [PubMed] [Google Scholar]
- 54.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 55.Wright K, Chin K, Linhoff M, Skinner C, Brown J, Boss J, Stark G, Ting J. CIITA stimulation of transcription factor binding to major histocompatibility complex class II and associated promoters in vivo. Proc Natl Acad Sci USA. 1998;95:6267–6272. doi: 10.1073/pnas.95.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X J, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 58.Zhang W, Bieker J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou H, Glimcher L H. Human class II gene transcription directed by the carboxy terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]