Abstract
The matrix (M) protein of vesicular stomatitis virus (VSV) is a potent inhibitor of bidirectional nuclear transport. Here we demonstrate that inhibition occurs when M protein is in the nucleus of Xenopus laevis oocytes and that M activity is readily reversed by a monoclonal antibody (αM). We identify a region of M protein, amino acids 51 to 59, that is required both for inhibition of transport and for efficient recognition by αM. When expressed in transfected HeLa cells, M protein colocalizes with nuclear pore complexes (NPCs) at the nuclear rim. Moreover, mutation of a single amino acid, methionine 51, eliminates both transport inhibition and targeting to NPCs. We propose that M protein inhibits bidirectional transport by interacting with a component of the NPC or an NPC-associated factor that participates in nucleocytoplasmic transport.
Trafficking of large macromolecules (more than 50 kDa) between the nucleus and the cytoplasm occurs through nuclear pore complexes (NPCs) via signal-dependent, carrier-mediated processes (reviewed in references 39 and 54). This transport is subject to control in response to a variety of stimuli such as progression through the cell cycle, exposure to stress, and infection by viruses (reviewed in reference 39). Thus, control of nucleocytoplasmic transport is an important element in the regulation of gene expression.
Much of the carrier-mediated movement through NPCs requires cargo-specific transport receptors called importins and exportins (or karyopherins), which are members of the importin β superfamily of proteins (16, 21, 45, 61). Transport receptors can bind their cargoes either directly or via specialized adapter proteins. For example, importin β mediates import of proteins containing basic nuclear localization sequences and small nuclear ribonucleoproteins (snRNPs) using the adapter proteins importin α (1) and snurportin (28, 43), respectively. Importin β can also interact directly with import cargoes, such as cyclin B (40, 53) and certain ribosomal proteins (30). CRM1 (Exportin1) mediates the export of proteins containing leucine-rich nuclear export signals (NES) as well as unspliced viral mRNAs and pre-snRNAs that are bound to specific NES-containing adapter proteins (14, 17, 51, 57). Exportin-t binds directly to its RNA export cargo, tRNA (3, 33).
Directionality of nuclear transport appears to be governed largely by Ran, a small GTPase that is a central component of most known nucleocytoplasmic transport pathways (reviewed in references 9 and 41). Owing to the asymmetric localization of the Ran effector proteins RanGAP (the GTPase activating protein in the cytoplasm) and RCC1 (the guanine nucleotide exchange factor in the nucleus), a steep concentration gradient of RanGTP is presumed to exist across the nuclear envelope (29). This gradient plays a pivotal role in nucleocytoplasmic transport by triggering both assembly and disassembly of receptor-cargo complexes in the appropriate subcellular compartment (60). Thus, import complexes assemble in the cytoplasm in the absence of RanGTP and disassemble in the nucleus in the presence of RanGTP, whereas export complexes form upon binding to RanGTP in the nucleus and dissociate upon removal and hydrolysis of RanGTP in the cytoplasm. Consequently, collapse of the RanGTP-GDP gradient leads to a block of most nucleocytoplasmic transport (29).
Nucleocytoplasmic transport is subject to regulation during infection by many types of viruses. For example, the NS1 protein of influenza virus inhibits export of cellular poly(A)+ mRNA (7). Expression of the Rev protein of human immunodeficiency virus type 1, which functions as an adapter for CRM1, allows export of incompletely spliced viral mRNAs (14, 17, 25, 38). The E1B oncoprotein of adenovirus type 5 promotes export of viral mRNAs and inhibits export of most cellular mRNA species (11). The matrix (M) protein of vesicular stomatitis virus (VSV) inhibits bidirectional nuclear transport of both RNAs and proteins (27).
Infection of cells by VSV, a negative-strand RNA virus that replicates in the cytoplasm, results in rapid shutoff of cellular gene expression (59) and snRNA processing (18). The M protein, a major structural component of the VSV virion, plays a central role both in the inhibition of host cell gene expression (5, 42) and in viral assembly (59). These two properties are genetically separable from each other (6, 8, 37) in that methionine 51 (Met-51) of the M protein is required for inhibition of host cell gene expression, but not for viral assembly, whereas amino acids 4 to 21 are needed for viral assembly but not for inhibition of host cell gene expression.
Previously, we showed that M protein synthesized in Xenopus laevis oocytes inhibits the import of snRNPs and karyophilic proteins, as well as the export of snRNAs and mRNAs, but not tRNAs (27). Here we use a monoclonal antibody that recognizes M protein (αM) to investigate the mechanism of this inhibition of bidirectional nuclear transport. We demonstrate that inhibition of both export and import is readily reversed by this antibody and that M protein works from within the nucleus. Furthermore, we show that Met-51 and the adjacent residues 52 to 59 are necessary both for efficient recognition by αM and for the inhibition of nuclear transport. Met-51 is also important for colocalization of M protein and NPCs, indicating that the interaction between M protein and an NPC-associated factor(s) is necessary for M activity.
MATERIALS AND METHODS
DNA templates and in vitro RNA synthesis.
DNA templates for transcription of RNA import or export substrates were generated either by PCR amplification (U1, U1Sm−, U5, and U6 snRNAs, U3 snoRNA, and NL-15 RNA) or by linearization of plasmid DNAs (AdML pre-mRNAs and tRNA) as described previously (22, 44, 55). In vitro synthesis of [α-32P]GTP-labeled RNAs was performed in 20-μl reactions using SP6 polymerase (U1, U1Sm−, U5, U3, and AdML RNAs) or T7 polymerase (tRNA and NL-15 and U6 RNAs) as detailed elsewhere (44). U1, U1Sm−, U5, U3, and AdML RNAs were synthesized with m7GpppG caps, whereas U6 received a γmepppG cap. All RNAs were purified as described previously (55).
DNA templates and in vitro synthesis of M protein mRNA.
DNA templates for synthesis of polyadenylated mRNAs encoding histidine-tagged wild-type, Δ2–46, Δ4–21, and Δ199–229 M proteins were generated by PCR amplification (see Table 1). Following PCR, the DNA products were cleaved with the appropriate restriction enzymes and cloned into the pSP64poly(A) vector (Promega). DNA templates for synthesis of mRNAs encoding histidine-tagged internal deletions Δ47–75, Δ116–175, and Δ155–185 of M protein were constructed using site-directed mutagenesis (Clontech). A mutagenic primer (Table 1) complementary to regions flanking the deletion and a selection primer were annealed to the DNA template, pSP64.OMHis6 (Table 1). The primers were extended using the Klenow fragment of DNA polymerase, and the DNAs were ligated and transformed into Escherichia coli BMH 71-18 muts cells. A DNA template for synthesis of mRNA encoding a histidine-tagged internal deletion (Δ75–106) of M protein was obtained by digestion of pSP64.OMHis6 DNA (Table 1) with BglII, which removed a fragment encoding a region within the M protein corresponding to amino acids 75 to 106. The remaining large DNA fragment was blunt ended with deoxynucleoside triphosphates using T4 DNA polymerase, gel purified, and recircularized using T4 DNA ligase; the ligated DNA was transformed into E. coli JM109 cells. The pSV2-O82M plasmid, encoding M protein with the M51R mutation (6), was kindly provided by Douglas Lyles (Wake Forest University). The HindIII fragment of this DNA containing the O82M coding region was ligated to HindIII-cut pSP64poly(A)+ vector DNA and transformed into E. coli DH5α cells. The nucleotide sequences of all clones were confirmed by DNA sequencing.
TABLE 1.
Plasmid constructions
| Clone | Oligonucleotide(s)a | Template |
|---|---|---|
| pSP64.OMHis6 | 5′-GATTTAGGTGACACTATAG-3′; OMHis-3′ (5′-TAAGGGATCCCTGCAGTCATCAGTGGTGGTGGTGGTGGTGCTCGAGTTTGAAGTGGCTGACAG-3′) | pSP64.OM(22) |
| pSP64.OM2-46His6 | 5′-ATTTCTAGATTCATCATGGGAGTTGACGAGATGGAC-3′; OMHis-3′ | pSP64.OMHis6 |
| pSP64.OM4-21His6 | 5′-GTGTTATCCCAATCCATTCATCATATGAGTTCCATCGCACCACCCCCTTATGAAGAGG-3′; OMHis-3′ | pSP64.OMHis6 |
| pSP64.OM199-229His6 | 5′-GATTTAGGTGACACTATAG-3′; 5′-GAATTCGGATCCTCATCAAAATTTGGAAGAATTGAAATGATCC-3′ | pSP64.OMHis6 |
| pSP64.OM23-46His6 | Selection primer (5′-GTGCCACCTGATATCTAAGAAACC-3′); 5′-GAAATCTAAGAAATTAGGGATCGGAGTTGACGAGATGGAC-3′ | pSP64.OMHis6 |
| pSP64.OM47-75His6 | Selection primer; 5′-CAAAATTGACAAATCCTATTTTCGTCCGTTCAGAACATACTCAG-3′ | pSP64.OMHis6 |
| pSP64.OM116-146His6 | Selection primer; 5′-GGCTTTTTTGGGTTCTTCTAATAAGACCCCTCCCATGCTCATG-3′ | pSP64.OMHis6 |
| pSP64.OM155-185His6 | Selection primer; 5′-GACCCCTCCCATGCTCAATGTAGCTCCTATGATCTGGGATC-3′ | pSP64.OMHis6 |
| pGEX-2T-OM | GEX-OM1 (5′-CATCATGGGATCCTTAAAGAAGATTCTCGG-3′); SP64-3′ (5′-GGGAGCTCGCCCGGGGATCC-3′) | pSP64.OM(22) |
| pGEX-2T-OM(M51A) | 5′-GAGTTGACGAGGCTGACACTCATGATCCGCATC-3′; 5′-CATGAGTGTCAGCCTCGTCAACTCCAAAATAGG-3′ | pSP64.OM(22) |
| pGEX-2T-OM(M51L) | 5′-GAGTTGACGAGCTGGACACTCATGATCCGCATC-3′; 5′-CATGAGTGTCCAGCTCGTCAACTCCAAAATAGG-3′ | pSP64.OM(22) |
| pGEX-2T-OMAAA48 | 5′-CTATTTTGGAGCGGCCGCTATGGACACTCATGATCCGCATC-3′; 5′-GTCCATAGCGGCCGCTCCAAAATAGGATTTGTCAATTGG-3′ | pSP64.OM(22) |
| pGEX-2T-OMAAA51 | 5′-CACGAGCGGCCGCTCATGATCCGCATCAATTAAG-3′; 5′-GGATCATGAGCGGCCGCCTCGTCAACTCCAAAATAGGATTTG-3′ | pSP64.OM(22) |
| pGEX-2T-OMAAA54 | 5′-CACTGCGGCCGCTCATCAATTAAGATATGAGAAATTC-3′; 5′-CTTAATTGATGAGCGGCCGCAGTGTCCATCTCGTCA-3′ | pSP64.OM(22) |
| pGEX-2T-OMAAA57 | 5′-GATCCGGCGGCCGCTAGATATGAGAAATTCTTCTTTAC-3′; 5′-CTCATATCTAGCGGCCGCCGGATCATGAGTGTCCATCTCG-3′ | pSP64.OM(22) |
| pGEX-2T-OMAAA60 | 5′-CATCAATTAGCGGCCGCAAAATTCTTCTTTACAGTGAAAATG-3′; 5′-GAATTTTGCGGGCGCTAATTGATGCGGATCATGAGTG-3′ | pSP64.OM(22) |
| pGEX-2T-OMAAA63 | 5′-GATATGAGGCGGCCGCTTTTACAGTGAAAATGACGGTTAG-3′; 5′-CTGTAAAAGCGGCCGCCTCATATCTTAATTGATGCGGATC-3′ | pSP64.OM(22) |
For each pair of oligonucleotides, the first sequence shown is the forward primer and the second sequence shown is the reverse primer. The sequences of OMHis-3′, selection primer, GEX-OM1, and SP64-3′ are shown in rows 1, 5, and 9. For the last seven clones listed, the second set of primers was GEX-OM1 and SP64-3′.
For in vitro synthesis of poly(A)+ mRNAs encoding the various M proteins, plasmid DNAs were linearized with EcoRI and used in large-scale transcription reactions with SP6 polymerase according to the protocol of Promega, Inc.
Construction of GFP-M DNAs.
To make plasmids encoding chimeric green fluorescent protein (GFP)-M fusion proteins [pEGFP-C1-OM and pEGFP-C1-OM(51R)], the BamHI fragment containing the M protein gene from either the pGEX-2T-OM or the pGEX-2T-OM(51R) plasmid (see below) was ligated to BamHI-cut pEGFP-C1 (Clontech) DNA and transformed into E. coli cells. The correct orientations of the M protein genes were confirmed by digestion of the resulting plasmid DNAs with BglII.
DNA transfections.
For transient transfections of GFP-M DNAs into cells, a six-well tissue culture plate containing coverslips was seeded with 4 × 105 HeLa cells per well 24 h before use. Transfections were carried out according to the protocol of Life Technologies, Inc., using 1 μg of pEGFP-C1-OM or of pEGFP-C1-OM(51R) DNA and 10 μl of Lipofectamine (Life Technologies, Inc.). After 24 h the cells were processed for immunofluorescence.
Construction of GST-M protein DNAs and purification of GST-M proteins.
The pGEX-2T-OM vector encoding a glutathione S-transferase (GST)–M fusion protein, GST-M, was generated by PCR amplification (see Table 1). The upstream primer contained two nucleotide changes at positions 4 and 6 with respect to the nucleotide sequence of M protein (Orsay strain). These changes, which generated a BamHI restriction site, resulted in a serine-to-glycine substitution at amino acid 2. The PCR product and the GST encoding expression vector, pGEX-2T (Pharmacia), were digested with BamHI, ligated together, and transformed into JM109 cells.
To generate pGEX-2T-OM(51R), the pSP64-O82Mpoly(A) and pGEX-2T-OM plasmid DNAs were digested with MfeI and StuI. The fragment released from the region of pSP64-O82Mpoly(A) encoding M protein was gel purified and ligated into the cut pGEX-2T-OM vector, which was then transformed into JM109 cells. The plasmids pGEX-2T-OM(AAA48 through AAA63), pGEX-2T-OM(51A), and pGEX-2T-OM(51L) were generated by two-step PCR (Table 1). PCR products were digested with MfeI and StuI and ligated into the similarly cut vector, pGEX-2T-OM. pGEX-OM(48–62), encoding amino acids 48 to 62 of M protein fused to the C terminus of GST, was made by ligation of complementary oligonucleotides containing the appropriate ends with pGEX-2T plasmid DNA linearized with the same restriction enzymes.
For production of recombinant proteins, all plasmids were transformed into E. coli BL21 cells. Cells were grown overnight at 37°C in Luria-Bertani medium containing ampicillin and then induced for 3 hours with 1 mM IPTG. Cells were harvested by centrifugation, resuspended in one-tenth volume of 1× phosphate buffered saline (PBS; pH 7.4)–1 mM dithiothreitol–1% Triton X-100–1 mM phenylmethylsulfonyl fluoride and immediately lysed by passage through a French press. The lysates were clarified by centrifugation, made 10% with respect to glycerol, and frozen at −70°C until further purification.
For affinity purification of GST-M proteins, lysates (10 ml) were quick-thawed and loaded directly onto a 1-ml glutathione-Sepharose column (Sigma). Columns were washed with 10 ml of wash buffer (PBS, pH 7.4; 1 M NaCl; 1% Triton X-100) and eluted with 50 mM Tris-HCl (pH 8.0)–5 mM glutathione–150 mM KCl–0.01% Triton X-100. Then, 0.5-ml fractions were collected, and aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing GST-M protein were pooled, made 10% with respect to glycerol, and stored at 4°C or frozen at −70°C until use.
Analysis of RNA transport in X. laevis oocytes.
Stage VI oocytes were manually dissected from X. laevis ovaries and maintained in MBS-H medium at 18°C (24). Nuclei or cytoplasms were injected with 12 nl of H2O containing ∼5 fmol each of 32P-labeled RNAs, along with blue dextran (44, 55). After incubation at 18°C for the indicated times, oocytes were manually dissected and analyzed as previously described (22, 35, 44).
For injection of mRNA encoding the wild-type VSV M protein, 25 nl (∼20 fmol) was injected into the cytoplasm ∼18 h prior to the injection of import or export RNAs. For injections of mRNAs encoding M deletion proteins, 25 nl (∼200 fmol) was injected. For injection of purified GST-M proteins (∼100 μg/ml), 15 nl was injected into the nucleus or 25 nl was injected into the cytoplasm. For injection of GST-M(48–62) protein, the concentration of protein in the injection mix was 3.6 mg/ml. When GST-M protein and antibodies were coinjected, the concentration of GST-M protein in the injection mixes was ∼25 μg/ml. For inhibition of protein synthesis, cycloheximide was added directly to the MBS-H medium at a final concentration of 200 μg/ml.
Analysis of protein synthesis in X. laevis oocytes.
Stage VI oocytes were injected into the cytoplasm with mRNAs for M proteins and incubated for 16 to 24 h in MBS-H containing 0.5 to 1.0 μCi of [35S]methionine (Amersham) (24). The nuclear and cytoplasmic fractions from such oocytes were analyzed as previously described (27, 46).
Antibodies.
The hybridoma cell line synthesizing the αM monoclonal antibody (23H12) (36) was kindly provided by Douglas Lyles (Wake Forest University). Antibodies were purified from hybridoma supernatants on a protein G-Sepharose (Pharmacia) column using standard procedures (26). Antibodies were stored at −20°C until use. For use in oocytes, αM was concentrated to 27 mg/ml (Amicon), and 15 nl (nucleus) or 25 nl (cytoplasm) was injected. The nonimmune rabbit anti-mouse immunoglobulin G (IgG) antibody (Cappell) was concentrated to 22 mg/ml (Amicon) and either 15 nl (nucleus) or 25 nl (cytoplasm) was injected into oocytes. The monoclonal αGST antibody (Pharmacia) was dialyzed against PBS at 0.2 mg/ml. For coinjection of αM or αGST antibody and GST-M protein, the concentration of antibody in the injection mixtures was ∼0.18 mg/ml.
Western blotting and immunoprecipitations.
For Western blot analysis, purified GST-M proteins or oocyte extracts were fractionated by SDS-PAGE and the proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). Membranes were probed with antibodies in TBS-T (10 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1 mM EDTA; 0.25% Tween 20) containing 5% powdered milk (Carnation) and developed with LumiGLO (Kirkegaard and Perry).
For immunoprecipitations, 9 μg of αM was coupled to 20 μl of protein A-Sepharose beads (Sigma) in TBS-T for 2 h at 4°C. Either 40 ng of recombinant M protein or 0.5 oocyte equivalent of [35S]methionine-labeled cytoplasmic extract, prepared from oocytes expressing M protein, was added to the αM beads. After overnight rotation at 4°C, the supernatant was collected and the pellets were washed five times with 500 μl of TBS-T. The supernatant and pellet fractions were visualized by SDS-PAGE, followed by autoradiography.
Immunofluorescence.
To process cells for immunofluorescence, cells were either fixed with 2% paraformaldehyde for 15 min followed by permeabilization with 0.5% Triton X-100 or else extracted first with 0.5% Triton X-100 for 3 min, followed by paraformaldehyde fixation. GFP-M proteins were visualized using the ×100 objective of an Axioplan 2 fluorescence microscope (Zeiss). For double-labeling experiments, GFP-M protein expressing cells were extracted with Triton X-100, fixed with formaldehyde, and incubated with 5 μg of mAb414 (kindly provided by Laura Davis, Brandeis University). The subsequent incubation was with Alexa 568-conjugated goat anti-mouse secondary antibody (Molecular Probes).
Phage display.
Isolation of peptides that could be recognized by the αM antibody (23H12) was done by phage display as described previously (49). The CW1 M13 phage library, a 12-mer random peptide library, was kindly provided by Brian Kay (University of Wisconsin—Madison). After three rounds of selection, the resulting phage were plaque purified. Binding activities of the cloned phage were confirmed by enzyme-linked immunosorbent assay, and the peptide sequences of isolated αM binding phage were deduced from DNA sequence analysis.
RESULTS
αM antibody in either the nucleus or the cytoplasm reverses the inhibitory activity of M protein.
The M protein of VSV is a potent inhibitor of bidirectional nuclear transport (27). As we reported previously, export of RNAs (snRNA and mRNA but not tRNA) from oocyte nuclei is selectively blocked upon the synthesis of M protein following cytoplasmic injection of in vitro-transcribed M protein mRNA (Fig. 1A, panel c). M protein also inhibits the import of karyophilic proteins (27; data not shown) and several RNAs, such as U1, U5, and U6 snRNAs and NL-15 RNA (Fig. 1C, panel d), which use different import pathways (22). U3 snoRNA, which normally is not exported (56) or imported (50), was included here as a control for accuracy of injection and dissection of the oocytes.
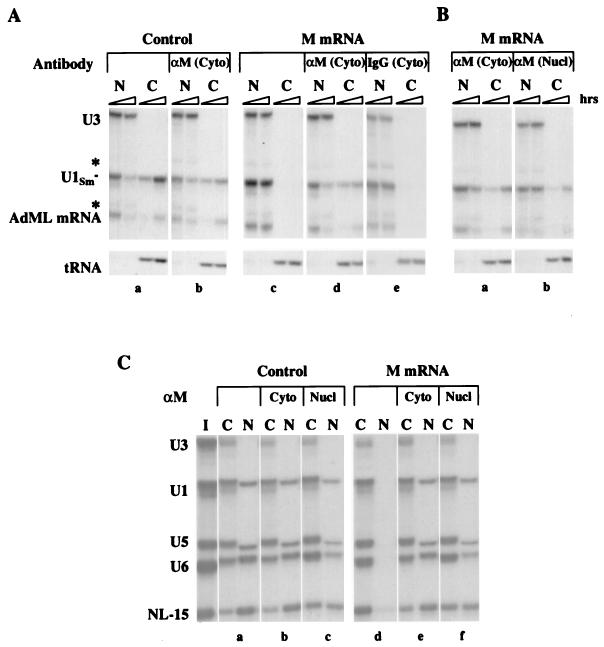
FIG. 1.
Inhibition of bidirectional nuclear transport by M protein is reversed by αM antibody. (A) Neutralization of M activity. A mixture of 32P-labeled U3 and U1Sm− snRNAs, AdML pre-mRNA, and tRNA was injected into the nuclei of oocytes that had been preinjected with M protein mRNA (M mRNA) and antibody in the cytoplasm. αM or nonimmune rabbit anti-mouse IgG was injected 0.5 h prior to the injection of M protein mRNA, and oocytes were incubated for an additional 16 h prior to the injection of export RNAs. At 1 and 3 h after RNA injection, oocytes were dissected into nuclear (N) and cytoplasmic (C) fractions; RNAs were isolated and 0.5 oocyte equivalents were resolved on 8% denaturing polyacrylamide gels and detected by autoradiography. AdML mRNA is generated by splicing of the injected pre-mRNA (not shown). Asterisks indicate degradation products arising from AdML pre-mRNA and U3 snoRNA. RNA export was monitored in control oocytes in the absence (a) and in the presence (b) of cytoplasmic αM antibody and in oocytes expressing M protein (M oocytes) in the absence (c) and in the presence of cytoplasmic αM antibody (d) or nonimmune rabbit anti-mouse IgG (e). (B) Reversal of inhibition of RNA export. αM was injected into the nucleus (Nucl) or cytoplasm (Cyto) 16 h after the injection of M protein mRNA and 2 h prior to the injection of export RNAs. RNA export was monitored as in panel A in M oocytes in the presence of cytoplasmic (a) or nuclear (b) αM antibody. (C) Reversal of inhibition of RNA import. A mixture of 32P-labeled U3, U1, U5, and U6 snRNAs and NL-15 RNA was injected into the cytoplasms of control and M oocytes in the absence or presence of αM antibody. αM was injected into the nucleus (Nucl) or cytoplasm (Cyto) 16 h after the injection of M protein mRNA and 2 h prior to injection of import RNAs. At 28 h after RNA injection, the nucleocytoplasmic distributions of RNAs were analyzed as in panel A. The RNAs in the injection mixture (I) are shown. Import was monitored in control and M oocytes in the absence (a and d) or in the presence of cytoplasmic (b and e) or nuclear (c and f) αM antibody.
Here, we refer to the ability of M protein to block nucleocytoplasmic transport as M activity. To probe the mechanism of M activity, we utilized a mouse monoclonal IgG antibody, 23H12, which is specific for M protein (αM) (36). We first showed that αM can neutralize M activity by introducing αM into the cytoplasm of oocytes prior to the injection of M mRNA (Fig. 1A). The presence of this antibody in the cytoplasm eliminated the inhibitory activity of the newly synthesized M protein (panel d). As expected, RNA export in oocytes containing no M mRNA was unaffected by cytoplasmic (panel b) or nuclear αM antibody (data not shown). The ability to neutralize M activity was specific to αM since injection of nonspecific IgGs into the cytoplasm (panel e) was without effect.
We then asked if αM could reverse M activity once the inhibition of transport had been established in oocytes expressing M protein (Fig. 1B and C). Inhibition of export of both mRNA and snRNA (Fig. 1B) was reversed regardless of whether αM was injected into the cytoplasm (panel a) or the nucleus (panel b). RNA import (Fig. 1C) also was restored by αM, independent of where the antibody was introduced (panels d to f); again, the antibody had little or no effect on RNA import in the absence of M protein (panels b and c). Likewise, protein import was restored by αM that was injected into either the nucleus or the cytoplasm (data not shown). Thus, αM in either the nucleus or the cytoplasm can reverse M activity, raising the possibility that M protein functions in the nucleus.
αM antibody eliminates M activity by nuclear depletion or neutralization of M protein.
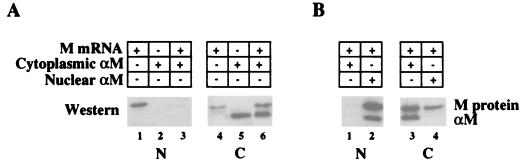
To determine how αM neutralizes M activity, we monitored the intracellular distribution of M protein in the presence and absence of αM (Fig. 2A). Because of its small size (∼28 kDa), M protein might be able to enter and exit the nucleus either by diffusion or by carrier-mediated transport. In the absence of the antibody, M protein was present in both the nucleus and the cytoplasm (27) (Fig. 2A, lanes 1 and 4). Independent of the expression of M protein, αM antibody remained in the cell compartment into which it was introduced (Fig. 2A, lanes 5 and 6; see also Fig. 2B, lanes 2 and 3). Injection of αM into the cytoplasm prior to injection of M mRNA led to sequestration of M protein in the cytoplasm (Fig. 2A, lanes 3 and 6), showing that complexes of M protein plus αM do not transit the NPCs (Fig. 2A, lane 6; Fig. 2B, lanes 2 and 3). Thus, neutralization of M activity by cytoplasmic αM (Fig. 1A) is due either to the formation of antibody-M protein complexes in the cytoplasm or to the lack of M protein in the nucleus.
FIG. 2.
Nucleocytoplasmic distribution of M protein in the absence or presence of αM. One oocyte equivalent of nuclear (N) or cytoplasmic (C) extract was analyzed by SDS-PAGE, followed by Western blotting with αM. (A) αM antibody was injected 0.5 h prior to the injection of M protein mRNA, and 21 h later extracts were prepared. The distribution of M protein was monitored in the absence (lanes 1 and 4) or presence (lanes 3 and 6) of cytoplasmic αM antibody. (B) αM antibody was injected 16 h after the injection of M protein mRNA, and 5 h later extracts were prepared. The distribution of M protein was monitored in the presence of cytoplasmic (lanes 1 and 3) or nuclear (lanes 2 and 4) αM antibody. In all cases, the antibody remained in the compartment into which it was injected, as shown by the location of the αM light chain (panel A, lanes 5 and 6; panel B, lanes 2 and 3).
Because αM also reversed M activity after transport had already been established (Fig. 1B and C), we analyzed the distribution of M protein in such oocytes (Fig. 2B). Injection of αM into the cytoplasm resulted in the nuclear depletion of M protein (lanes 1 and 3), indicating that the nuclear M protein either exits to the cytoplasm or turns over. Injection of αM into the nucleus led to increased levels of nuclear M protein but did not deplete the cytoplasmic pool (lanes 2 and 4), presumably because of continued synthesis of M protein. The fact that transport in these oocytes was normal indicates either that this cytoplasmic M protein is in an inactive form or that the target of M protein resides in the nucleus. In any case, the elimination of M activity (Fig. 1) correlates with the lack of free M protein in the nucleus, due to binding of αM to M protein in the nucleus, nuclear turnover, or sequestration of the protein in the cytoplasm. Therefore, we propose that M protein inhibits nuclear transport from within the nucleus.
Inhibition of nuclear transport occurs from within the nucleus.
To introduce M protein directly into the nucleus (or the cytoplasm), we generated a fusion protein containing GST and M protein (GST-M protein). This recombinant protein had a molecular weight of about 56,000, thereby reducing its rate of passive diffusion through NPCs (54). Upon injection into either the nucleus or the cytoplasm, GST-M protein was stable, and it gradually distributed between both compartments over a 20-h period (Fig. 3A). Injected recombinant GST-M protein inhibited both import of RNAs (Fig. 3B) and export of U1Sm− RNA (Fig. 3C, panels b and d) and AdML mRNA (Fig. 4b). Moreover, the inhibitory activity was independent of whether the protein was injected into the nucleus (Fig. 3B, panel b; Fig. 3C, panel b) or the cytoplasm (Fig. 3B, panel c; Fig. 3C, panel d).
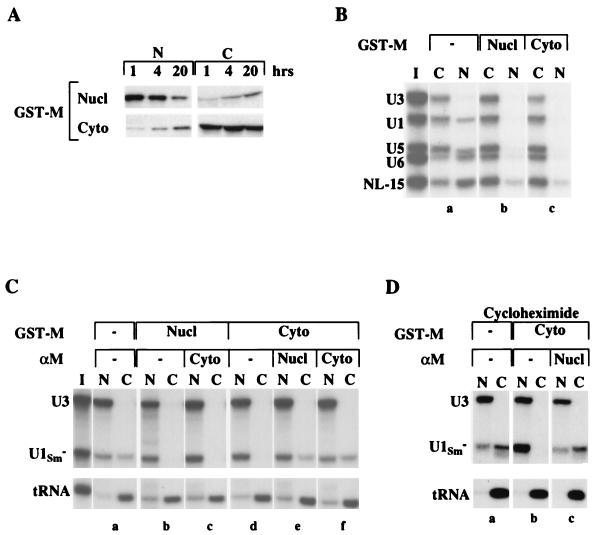
FIG. 3.
Inhibition of transport by GST-M protein occurs from within the nucleus. (A) Stability and distribution of GST-M protein. The nucleocytoplasmic distribution of GST-M protein was monitored by Western blotting with αGST antibody. Extracts were prepared 1, 4, and 20 h after nuclear (top panels) or cytoplasmic (bottom panels) injection of GST-M protein; one oocyte equivalent of the nuclear (N) or cytoplasmic (C) extracts was analyzed. (B) Inhibitory activity of GST-M protein. Import of 32P-labeled U1, U5, and U6 snRNAs and NL-15 RNA (import RNAs) was analyzed in control oocytes (a) and in oocytes preinjected with GST-M protein in the nucleus (b) or the cytoplasm (c). GST-M protein was injected 2 h prior to injection of import RNAs. Import was monitored 28 h after injection of the RNA mixture (I). (C) Nuclear function of GST-M protein. Export of 32P-labeled U1Sm− snRNA and tRNA was analyzed in control oocytes (a) and in oocytes preinjected with GST-M protein (b and d) or with both GST-M protein and αM antibody (c, e, and f). αM was injected 1 h prior to the injection of GST-M protein, which was followed 1 h later by the injection of the export RNAs. Export was monitored 1 h after injection of the RNA mixture (I). (D) Reversal of transport inhibition in the absence of protein synthesis. Export of 32P-labeled U1Sm− snRNA and tRNA was analyzed in the presence of cycloheximide in control oocytes (a) and in oocytes preinjected with GST-M protein (b) or with GST-M protein and nuclear αM antibody (c). GST-M protein was injected into the cytoplasm, and 1.5 h later cycloheximide (200 μg/ml) was added to the oocyte incubation medium. αM was injected 1 h after the addition of cycloheximide and 1 h prior to the injection of export RNAs. This amount of cycloheximide was sufficient to block protein synthesis as monitored by [35S]methionine labeling (data not shown).
FIG. 4.
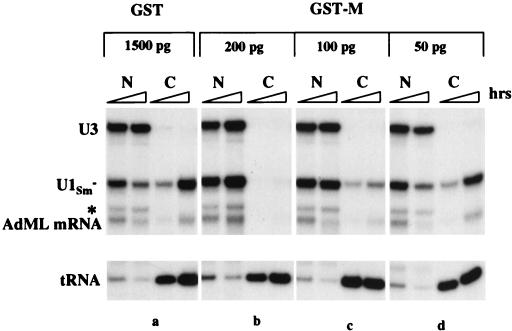
Titration of the amount of nuclear GST-M protein required for M activity. Export of 32P-labeled U1Sm− snRNA, AdML mRNA and tRNA was analyzed in oocytes preinjected 20 min earlier in the nucleus with 1,500 pg of recombinant GST protein (a) and with the indicated amounts (b to d) of GST-M protein. RNA export was monitored 1 and 3 h after injection of export substrates. The asterisk indicates a degradation product of U3 snoRNA.
As we observed with M protein synthesized in oocytes, the activity of cytoplasmically injected M protein was eliminated when the neutralizing antibody αM was present in either the nucleus or the cytoplasm (Fig. 3C, panels e and f). Therefore, we tested our model that M protein acts from within the nucleus by injecting GST-M protein into the nucleus and αM into the cytoplasm. When transport was monitored 1 h after injection of RNA substrates, GST-M protein inhibited RNA export (panel c), demonstrating that nuclear M protein is sufficient for M activity. At later times, this GST-M activity was attenuated (data not shown), consistent with our previous finding that cytoplasmic αM eliminates M activity by sequestering M protein as it exits the nucleus (Fig. 1B and 2B). We conclude that GST-M protein functions from within the nucleus to inhibit nuclear transport.
Nuclear M protein operates efficiently.
Only a low level of cytoplasmically injected GST-M protein appeared in the nucleus within 1 h (Fig. 3A), but this was sufficient for M activity (Fig. 3C, panel d). To determine the amount of nuclear GST-M protein required for inhibition of U1Sm− RNA export, we varied the amount of protein injected. About 2 × 109 molecules (∼200 pg) per oocyte nucleus sufficed (Fig. 4b), but 100 pg did not (panel c). Because it is unlikely that all of the recombinant protein molecules were active, 200 pg of GST-M protein represents an upper limit of the amount required.
The rate at which nuclear GST-M protein inhibits export of RNA was estimated by coinjection of GST-M protein and RNA export substrates into the nucleus (data not shown). In this case, <10% of the U1Sm− RNA was exported within the first hour compared to 50% of the RNA in the absence of GST-M protein. Thus, M protein functions very quickly within the nucleus to inhibit transport.
Nuclear αM antibody reverses inhibition by shielding a region necessary for M activity.
Since reversal of transport inhibition by nuclear αM, in principle, could be due to the synthesis of new protein(s) required for transport, we tested if αM could restore transport in the absence of protein synthesis. Inhibition of RNA export by GST-M protein was reversed by nuclear αM, even in the presence of cycloheximide (Fig. 3D), demonstrating that M activity does not involve the irreversible modification or destruction of a component that participates in nuclear transport. Also, the neutralization of GST-M protein by αM was dependent on binding of the monoclonal antibody to its specific epitope, since a GST-specific monoclonal antibody (αGST) did not interfere with the activity of GST-M protein (data not shown). Thus, αM may reverse transport inhibition by binding to a region of M protein necessary for M activity, thereby disrupting interactions between the protein and its target(s).
Met-51 of M protein contributes to recognition by αM antibody.
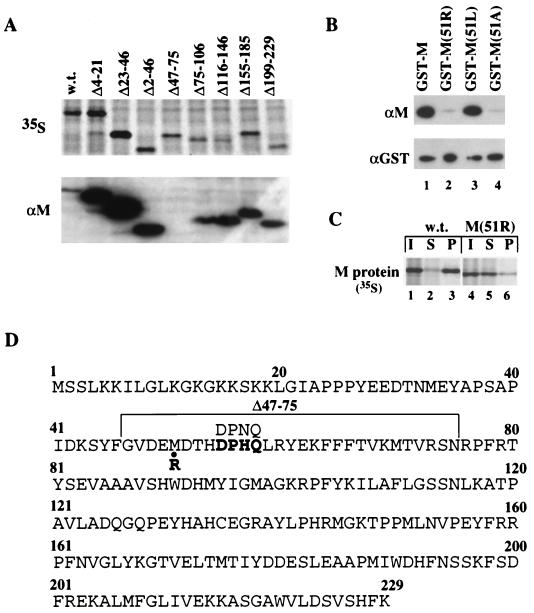
The region of M protein recognized by αM was identified by Western blot analysis of mutant M proteins. Several mRNAs encoding deletion mutants of M protein were generated, and the proteins were expressed in oocytes, as demonstrated by labeling with [35S]methionine (Fig. 5A, top panel). All of these mutant M proteins, except for the Δ47–75 deletion protein, were recognized by αM (bottom panel). In addition, an M13 phage library displaying random peptides 12 amino acids in length was screened for the ability to be recognized by αM. A consensus sequence present in seven of the nine selected peptides, DPNQ, is present once in the immunizing antigen (M protein of the San Juan strain) (data not shown). This sequence is very similar to the amino acid sequence, DPHQ, found at the same position (amino acids 55 to 58) of the M protein (Orsay strain) used here (Fig. 5D). Thus, the αM epitope is contained within amino acids 47 to 75 of M protein and includes residues 55 to 58.
FIG. 5.
Met-51 of M protein contributes to recognition by αM antibody. (A) Mapping of the αM epitope region. mRNAs encoding the wild type (w.t.) and the indicated deletion mutants of M protein were injected into the cytoplasm of oocytes, and the newly synthesized proteins were labeled by the addition of [35S]methionine to the incubation medium. After 18 h, whole-oocyte extracts were prepared and analyzed by SDS-PAGE, followed by autoradiography (top panel) or Western blotting with the αM antibody (bottom panel). (B) αM reactivity of wild-type and mutant M proteins. Western blot analysis was performed with 10 ng each of GST-M (lane 1), GST-M(51R) (lane 2), GST-M(51L) (lane 3), and GST-M(51A) (lane 4) proteins using αM (top panel) or αGST (bottom panel) antibody. (C) Inefficient recognition by αM of native M(51R) protein. [35S]methionine-labeled M proteins were synthesized in oocytes as in panel A and 0.5 oocyte equivalents of cytoplasmic extracts (I) were immunoprecipitated with αM antibody. Both the supernatant (S) and the pellet (P) fractions from immunoprecipitations of wild-type M (lanes 2 and 3) or M(51R) (lanes 5 and 6) proteins were analyzed by SDS-PAGE, followed by autoradiography. (D) Amino acid sequence of the VSV M protein of the Orsay strain. Amino acids 47 to 75 are bracketed. The DPHQ sequence (amino acids 55 to 58) is shown in boldface. The corresponding sequence (DPNQ) in the immunizing antigen, VSV M protein of the San Juan strain, is shown above the DPHQ sequence. The substitution of Met-51 to Arg-51, found in the M protein of the temperature-sensitive VSV mutant, tsO82 (8), is indicated.
A spontaneous temperature-sensitive mutant of VSV (tsO82) that is defective in blocking host cell gene expression has an altered M protein in which Met-51 is changed to arginine [M(51R) protein] (6, 8). Because Met-51 lies within the region of M protein containing the αM epitope, we tested if this residue is important for recognition by αM. GST-M(51R) mutant protein reacted poorly with αM in Western blots (Fig. 5B, top panel, lane 2), a result consistent with the unpublished results of others analyzing tsO82 mutant M protein synthesized in mammalian cells (D. Lyles, personal communication). Also, the native M(51R) protein was recognized inefficiently by αM in an immunoprecipitation assay (Fig. 5C, compare lanes 2 and 3 with lanes 5 and 6).
Since the M51R mutation introduces a positively charged residue at position 51, we asked if a conservative substitution of Met-51 with the hydrophobic residues leucine or alanine could maintain αM recognition. In Western blots, the GST-M(51L) but not the GST-M(51A) protein was recognized like wild-type GST-M protein (Fig. 5B, top panel, lanes 3 and 4). Thus, an amino acid with a long aliphatic side chain is required at position 51 of M protein for efficient binding to αM.
Met-51 is essential for M activity.
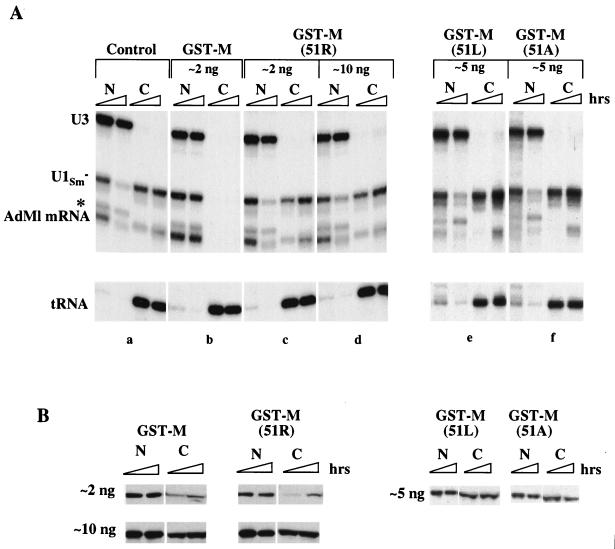
To test if the αM epitope region is also required for inhibition of nuclear transport, we assayed the effect of the M51R mutation on M activity. GST-M(51R) protein injected into the nucleus was unable to inhibit RNA export (Fig. 6A), showing that an arginine at position 51 of M protein eliminates M activity. Inhibition of transport was not observed even when the amount of injected mutant protein was 50 times that needed for inhibition by wild-type GST-M protein (cf. Fig. 4). Also, M(51R) protein was inactive for inhibition of protein import (data not shown). Western blot analysis, using αGST antibody, demonstrated that the protein was stable in oocytes and that its distribution was similar to that of GST-M protein (Fig. 6B). Thus, the Met-51 to Arg-51 mutation abolishes the ability of M protein to inhibit both nuclear transport and gene expression (6, 13, 52), showing that the mechanisms of inhibition of these processes are closely linked.
FIG. 6.
Met-51 of M protein is essential for inhibition of nuclear export. (A) RNA export. Wild-type and mutant GST-M proteins were injected into the nucleus 1 h prior to the injection of RNA export substrates. Export of 32P-labeled U1Sm− snRNA, AdML mRNA, and tRNA was monitored 1 and 3 h after RNA injection. RNA export was analyzed in the absence (a) or in the presence of the indicated amounts of GST-M (b), GST-M(51R) (c and d), GST-M(51L) (e), or GST-M(51A) (f) proteins. (B) GST-M protein stability and distributions. The nucleocytoplasmic distributions of GST-M proteins were monitored 2 and 5 h after nuclear injection of the indicated amounts of GST-M, GST-M(51R), GST-M(51L), or GST-M(51A) proteins.
We tested if the inactivity of GST-M(51R) protein was due to the introduction of a positive charge by assaying the activities of mutant M proteins having the conservative amino acid substitutions M51L and M51A. Neither of these proteins (which were stable in oocytes; Fig. 6B) was able to inhibit RNA export (Fig. 6A, panels e and f) even when injected into the nucleus at 30 times the amount needed for inhibition by wild-type protein (data not shown). Thus, although leucine at position 51 allows for recognition by αM, it does not suffice for M activity, showing that a methionine at position 51 is essential for the inhibition of nuclear transport.
Amino acids 51 to 59 are necessary, but not sufficient, for inhibition of transport by M protein.
Secondary structure predictions by the PhD and PREDATOR algorithms (19, 20, 47) suggest that the αM epitope is contained within a region of M protein (50-EMDTHDPHQ-58) that is likely to be in either a loop or a turn structure. This region probably is exposed on the surface of the protein because most of these amino acids are hydrophilic and αM can bind and inactivate M protein in vivo. This model is in agreement with previous findings by others demonstrating that a major V8 protease cleavage site in M protein occurs at position 50 (31).
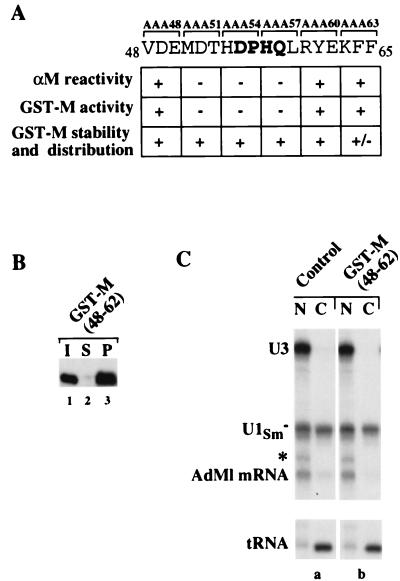
To determine if amino acids in the loop region surrounding Met-51 also are important for M activity, a series of templates was constructed encoding mutant GST-M proteins with triple alanine substitutions from positions 48 through 65 (three amino acids substituted in each mutant). As indicated, three of these proteins, mutated in amino acids 51 through 59, were not recognized by αM (Fig. 7A), a result consistent with our αM epitope mapping (Fig. 5). When tested for their abilities to inhibit transport, the same three mutant GST-M proteins were inactive, even though they were stable and distributed similarly to the GST-M protein in injected oocytes (Fig. 7A). Thus, in addition to Met-51, amino acids 52 to 59 are necessary for both αM recognition and GST-M activity.
FIG. 7.
Amino acids 51 to 59 of M protein are necessary, but not sufficient, for the inhibition of nuclear transport. (A) Activity and αM reactivity of triple alanine scanning mutants of M protein. The αM reactivities of the mutant M proteins were analyzed as in Fig. 5A and B, and the inhibitory activities, stabilities, and intracellular distributions were determined as in Fig. 6. +, Behavior indistinguishable from that of wild-type protein; −, behavior similar to that of GST-M(51R) mutant protein. (B) αM reactivity of native GST-M(48–62) protein. GST-M(48–62) protein (I) was immunoprecipitated with αM antibody and the supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE, followed by Western blotting with the αGST antibody. (C) RNA export. Export of 32P-labeled U1Sm− snRNA, AdML mRNA, and tRNA was analyzed in the absence (a) or in the presence (b) of GST-M(48–62) protein. GST-M(48–62) protein was injected into the nucleus 1 h prior to the injection of export substrates. RNA export was monitored 1 h after RNA injection. The molar concentration of the injected GST-M(48–62) protein was ∼75 times that of wild-type GST-M protein used in comparable experiments (e.g., Fig. 6A, panel b).
To test if the amino acids of this loop region were sufficient for M activity, we generated a fusion protein composed of GST and amino acids 48 to 62 of M protein. The chimeric protein was recognized by αM in immunoprecipitations (Fig. 7B), showing that the αM epitope region was presented in a context resembling that of the native M protein. Although a large nuclear pool of this protein remained for at least 18 h after injection into oocytes (data not shown), it had no effect on the export of mRNA or snRNAs (Fig. 7C). Thus, amino acids 51 to 59 are necessary, but not sufficient, for M activity.
Met-51 is necessary for targeting M protein to the nuclear rim.
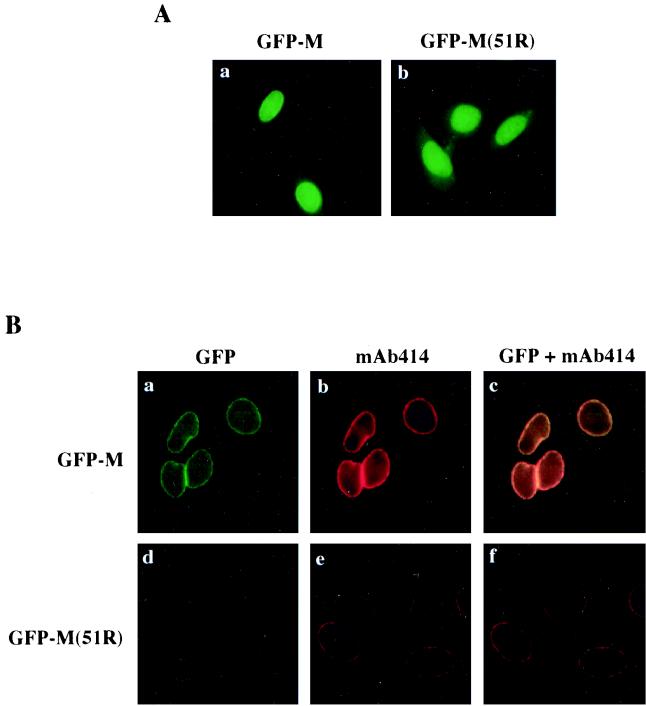
To determine where within the nucleus M protein might function, GFP-tagged wild-type and mutant [M and M(51R)] proteins were expressed in transiently transfected HeLa cells. A comparable recombinant GFP-tagged M protein inhibited nuclear transport upon injection into oocytes (data not shown). The wild-type GFP-M protein localized to the nucleoplasm, the cytoplasm, and the nuclear rim (Fig. 8A, panel a). In contrast, the M(51R) mutant protein showed only nucleoplasmic and cytoplasmic localization; no nuclear rim association was readily observable (panel b), indicating that the intracellular distribution of the inactive M protein differs from that of wild-type M protein.
FIG. 8.
Wild-type GFP-M protein, but not mutant GFP-M(51R) protein, colocalizes with NPCs. Plasmid DNAs encoding GFP-tagged wild-type and M(51R) mutant M proteins were transfected into HeLa cells, and 24 h later the cells were either fixed with formaldehyde (A) or extracted with 0.5% Triton X-100 prior to formaldehyde fixation and visualization by fluorescence microscopy (B). (A) Intracellular localization of GFP-M proteins in mammalian cells. The localization of GFP-M (a) and GFP-M(51R) (b) proteins in transiently transfected HeLa cells was monitored by direct fluorescence. (B) Colocalization of GFP-M protein and NPCs. The association of GFP-M protein with the nuclear rim was monitored in Triton-extracted cells that were expressing either GFP-M (a to c) or GFP-M(51R) (d to f) proteins. GFP-M (a) and GFP-M(51R) (d) proteins were detected by direct fluorescence, and NPCs (b and e) were visualized by immunostaining with mAb414 antibody and a rhodamine-labeled secondary antibody. Colocalization of NPCs and GFP-M (c) or GFP-M(51R) (f) proteins is indicated by the overlap (yellow) of the rhodamine (red) and the GFP (green) fluorescent signals.
To visualize the nuclear-rim-associated fraction of GFP-M proteins, the transfected HeLa cells were treated with Triton X-100 prior to fixation with formaldehyde, thereby releasing soluble M protein (4). M protein associated with the nuclear rim was resistant to this treatment, whereas the nucleoplasmic and cytoplasmic fractions of GFP-M protein were extracted (Fig. 8B, panel a). As expected, in GFP-M(51R)-transfected cells, no nuclear-rim-associated protein was detectable (panel d). Thus, the elimination of M activity correlates with the loss of nuclear rim association, suggesting that targeting of M protein to the nuclear rim is essential for M activity.
We asked if a putative target for M protein might be contained within NPCs. To visualize the NPCs (Fig. 8B, panels b and e), we used monoclonal antibody mAb414, which recognizes FG repeat containing nucleoporins (10). As shown by the overlap of the fluorescence signals from GFP and mAb414 (panels c and f), GFP-M protein and NPCs colocalize at the nuclear rim (panel c). This suggests that wild-type, but not M(51R) mutant (panel f), M protein interacts with a nuclear component of NPCs. We propose that M protein acts as an inhibitor of bidirectional nuclear transport when associated with the intranuclear side of the NPC.
DISCUSSION
We have demonstrated here that the M protein of VSV must be in the nucleus to inhibit bidirectional nuclear transport and that the target of M protein is likely to be a component of the NPC. Our results show that inhibition of transport is readily reversible, since a monoclonal antibody (αM) can restore transport, even after inhibition has been established. Amino acids 51 to 59 of M protein are important both for the inhibition of nuclear transport and for recognition by αM. Moreover, we have identified a single amino acid, Met-51, that is necessary for the association of M protein with the nuclear rim. We propose that interaction of M protein with a nuclear component(s) of the NPC and/or an NPC-associated factor(s) is responsible for its inhibition of nuclear transport.
Our results show that M protein must be in the nucleus, and not complexed with αM, in order to inhibit nucleocytoplasmic transport. Transport is restored either when the activity of M protein is neutralized by nuclear αM or when M protein is sequestered and neutralized in the cytoplasm by αM (Fig. 1 and 2). Moreover, GST-M protein introduced into the nucleus inhibits transport, even when neutralizing αM antibody is present in the cytoplasm (Fig. 3C). Consistent with the need for a pool of nuclear GST-M protein, GST-M activity dissipates with time as the protein is depleted from the nucleus and sequestered by αM in the cytoplasm. Thus, M protein exerts its inhibitory effect from within the nuclear compartment.
The interaction of M protein with a nuclear target(s) must be rapid, since inhibition of transport occurs soon after delivery of M protein to the nucleus. Also, this association must be dynamic, since αM can restore transport even when protein synthesis is blocked (Fig. 3D), ruling out resynthesis of targets as a way to restore transport. Thus, M protein may modulate the activity of a nuclear transport factor through reversible modification (e.g., phosphorylation, ADP ribosylation, etc.). Alternatively, M protein could disrupt associations between factors that participate in nuclear transport by competitively binding to one of them.
The M(51R) mutant protein, originally identified in a spontaneous temperature-sensitive mutant of VSV (tsO82) (8), inactivates the ability of M protein to inhibit transport (Fig. 6A) and affects binding to αM (Fig. 5B). Conservative substitution of Met-51 with either leucine or alanine also eliminates M activity (Fig. 6B), demonstrating that a methionine at this position is essential for the inhibitory effect of M protein. In the case of GST-M(51L) protein, its inactivity cannot be attributed to gross misfolding of the epitope region, since αM recognition is not altered by this mutation (Fig. 5B). We propose that Met-51 of M protein is necessary for the interaction between M protein and its target(s) and that binding of αM to the epitope region of M protein disrupts this association.
Met-51 lies within a region of M protein (amino acids 50 to 58) predicted to form an exposed loop or turn structure. Triple alanine scanning mutations of the adjacent amino acids in this presumptive loop region (amino acids 52 to 59) abolished both M activity and recognition by αM (Fig. 7A). However, substitution of amino acids 53 to 59 of M protein with a sequence composed of both hydrophilic and hydrophobic amino acids, which are less likely than consecutive alanine residues to distort the structure of this region, did not eliminate M activity (unpublished data). This raises the possibility that amino acids 53 to 59 within the loop region do not play a direct role in the interaction of M protein and its target. We are currently testing precisely which amino acids in this region are involved in essential contacts between M protein and its nuclear target(s).
Our analyses of the intracellular localization of wild-type and mutant M proteins (Fig. 8) indicate that the nuclear components of the NPC and associated transport factors are likely targets for inactivation by M protein. In transiently transfected HeLa cells, GFP-M protein associated with the nuclear rim, and this localization was coincident with that of FG-repeat containing nucleoporins. In contrast, the nonfunctional mutant GFP-M(51R) protein did not colocalize with NPCs, leading to the model that M protein is associated with the NPC when it inhibits transport.
In support of this model, the pattern of inhibition by M protein resembles that observed when the activities of specific nucleoporins or transport factors are inhibited by antibodies or dominant-negative mutant proteins. Antibodies to Nup98 or Nup153, two intranuclear components of the NPC, block the export of mRNA and snRNA, but not tRNA (46, 58). Likewise, the isolated nucleoporin binding domains of the transport factors, importin β and TAP, act as dominant-negative mutants to inhibit the export of mRNA and snRNA and, to a much lesser extent, tRNA (4, 32). The dominant-negative form of importin β is also an efficient inhibitor of protein import (32). Both the antibodies and dominant-negative mutants are proposed to inhibit NPC function by blocking docking sites for transporters and their respective cargoes. We propose that M protein functions in a similar manner to inhibit bidirectional nuclear transport. Consistent with this proposal, mAb414, which inhibits the export of most classes of RNAs (23), does not block the export of ET202, an RNA that was selected solely for its ability to be exported in the presence of M protein (23).
VSV replicates its genome in the cytoplasm and does not require nuclear factors for virus production (15); nonetheless, M protein can be detected in the nuclei of VSV-infected cells (36). In agreement with that observation, we have shown that M protein distributes to both the cytoplasm and the nucleus in Xenopus oocytes and in transfected HeLa cells (27) (Fig. 2 and 8) and that the protein is actively imported in an in vitro system (J. M. Petersen, unpublished data). The mutant M(51R) and M(51L) proteins also enter the nucleus both in vivo and in vitro (Fig. 8) (37; unpublished data), showing that Met-51 is not required for nuclear uptake of M protein. It is unclear whether M protein exits the nucleus by diffusion or active transport.
In addition to inhibiting nucleocytoplasmic transport, wild-type M protein was previously reported to inhibit gene expression in cultured cells (5, 6, 13, 42) and transcription in vitro (2, 62). We have not observed an inhibition of transcription by M protein in Xenopus oocytes, but we recognize that oocyte nuclei may be unusual in that they contain large stockpiles of transcription and replication factors (24). In any case, the loss of inhibition of both gene expression and transport upon mutation of Met-51 indicates that a common mechanism is involved. We have proposed (27) that M protein may affect gene expression directly by blocking mRNA export. In addition, inhibition of transport might secondarily affect the import or function of factors that are essential for the assembly of active transcription complexes. However, M protein may use the same mechanism to inactivate different targets that affect either transport or transcription independently. Recent evidence suggests that inhibition of RNA polymerase II transcription by M protein alters the activity of the transcription factor TFIID (62).
Inhibition of transport by M protein, presumably via association with a nuclear component of NPCs, may recapitulate control systems that normally modulate nucleocytoplasmic transport in uninfected cells. For example, the target(s) of M protein could be a component of the NPC, the function of which could be altered during induction of a stress response, reduction of cell growth rate, or progression through the cell cycle (12, 34, 48). Thus, M protein might provide a potent tool for investigation of cellular mechanisms that regulate nuclear transport and transcription. Our current efforts are focused on the identification of the intranuclear components of the NPC that interact with M protein and on elucidation of how inhibition of transport and transcription are linked.
ACKNOWLEDGMENTS
We thank Douglas Lyles, Jen Bachorik, and Laura Davis for kindly supplying reagents and Brian Kay for assistance with phage display selection. We also thank Susanne Blaser-Imboden, Matt Bohlman, and Thomas Jensen for technical help and Christopher Trotta and Doreen Glodowski for critical comments on the manuscript.
This work was supported by NIH grant GM30220 and by a DARPA grant (MDA972-97-1-0005) to J.E.D. V.V. was supported by an NIH training grant (GM08349-08). J.M.P. is a Burroughs Wellcome Fund Fellow of the Life Sciences Research Foundation.
REFERENCES
- 1.Adam E J, Adam S A. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed M, Lyles D S. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J Virol. 1998;72:8413–8419. doi: 10.1128/jvi.72.10.8413-8419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts G J, Fornerod M, Mattaj I W. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 4.Bachi A, Braun I C, Rodrigues J P, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black B L, Lyles D S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black B L, Rhodes R B, McKenzie M, Lyles D S. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J Virol. 1993;67:4814–4821. doi: 10.1128/jvi.67.8.4814-4821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Li Y, Krug R M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulon P, Deutsch V, Lafay F, Martinet-Edelist C, Wyers F, Herman R C, Flamand A. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J Gen Virol. 1990;71:991–996. doi: 10.1099/0022-1317-71-4-991. [DOI] [PubMed] [Google Scholar]
- 9.Dahlberg J E, Lund E. Functions of the GTPase Ran in RNA export from the nucleus. Curr Opin Cell Biol. 1998;10:400–408. doi: 10.1016/s0955-0674(98)80017-3. [DOI] [PubMed] [Google Scholar]
- 10.Davis L I, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 11.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldherr C M, Akin D. Signal-mediated nuclear transport in proliferating and growth-arrested BALB/c 3T3 cells. J Cell Biol. 1991;115:933–939. doi: 10.1083/jcb.115.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferran M C, Lucas-Lenard J M. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 15.Follett E A, Pringle C R, Wunner W H, Skehel J J. Virus replication in enucleate cells: vesicular stomatitis virus and influenza virus. J Virol. 1974;13:394–399. doi: 10.1128/jvi.13.2.394-399.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 18.Fresco L D, Kurilla M G, Keene J D. Rapid inhibition of processing and assembly of small nuclear ribonucleoproteins after infection with vesicular stomatitis virus. Mol Cell Biol. 1987;7:1148–1155. doi: 10.1128/mcb.7.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 20.Frishman D, Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimm C, Lund E, Dahlberg J E. In vivo selection of RNAs that localize in the nucleus. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimm C, Lund E, Dahlberg J E. Selection and nuclear immobilization of exportable RNAs. Proc Natl Acad Sci USA. 1997;94:10122–10127. doi: 10.1073/pnas.94.19.10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurdon J B, Wickens W P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- 25.Hammarskjöld M L, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Storing and purifying antibodies. In: Harlow E, Lanes D, editors. Antibodies: a laboratory manual. New York, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 285–318. [Google Scholar]
- 27.Her L-S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 28.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäkel S, Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaptur P E, Rhodes R B, Lyles D S. Sequences of the vesicular stomatitis virus matrix protein involved in binding to nucleocapsids. J Virol. 1991;65:1057–1065. doi: 10.1128/jvi.65.3.1057-1065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Liang S, Tartakoff A M. Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)+ RNA export. EMBO J. 1996;15:6750–6757. [PMC free article] [PubMed] [Google Scholar]
- 35.Lund E, Paine P L. Nonaqueous isolation of transcriptionally active nuclei from Xenopus oocytes. Methods Enzymol. 1990;181:36–43. doi: 10.1016/0076-6879(90)81110-g. [DOI] [PubMed] [Google Scholar]
- 36.Lyles D S, Puddington L, McCreedy B J., Jr Vesicular stomatitis virus M protein in the nuclei of infected cells. J Virol. 1988;62:4387–4392. doi: 10.1128/jvi.62.11.4387-4392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyles D S, McKenzie M O. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology. 1997;229:77–89. doi: 10.1006/viro.1996.8415. [DOI] [PubMed] [Google Scholar]
- 38.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 40.Moore J D, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore M S. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 42.Paik S Y, Banerjee A C, Harmison G G, Chen S J, Schubert M. Inducible and conditional inhibition of human immunodeficiency virus proviral expression by vesicular stomatitis virus matrix protein. J Virol. 1995;69:3529–3537. doi: 10.1128/jvi.69.6.3529-3537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palacios I, Hetzer M, Adam S A, Mattaj I W. Nuclear import of U snRNPs requires importin beta. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasquinelli A E, Dahlberg J E, Lund E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- 45.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 46.Powers M A, Forbes D J, Dahlberg J E, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 48.Saavedra C, Tung K S, Amberg D C, Hopper A K, Cole C N. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- 49.Sparks A B, Adey N B, Cwirla S, Kay B K. Screening phage-displayed random peptide libraries. In: Kay B K, Winter J, McCafferty J, editors. Phage display of peptides and proteins. San Diego, Calif: Academic Press; 1996. pp. 227–252. [Google Scholar]
- 50.Speckmann W, Narayanan A, Terns R, Terns M P. Nuclear retention elements of U3 small nucleolar RNA. Mol Cell Biol. 1999;19:8412–8421. doi: 10.1128/mcb.19.12.8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 52.Stanners C P, Francoeur A M, Lam T. Analysis of VSV mutant with attenuated cytopathogenicity: mutation in viral function, P, for inhibition of protein synthesis. Cell. 1977;11:273–281. doi: 10.1016/0092-8674(77)90044-7. [DOI] [PubMed] [Google Scholar]
- 53.Takizawa C G, Weis K, Morgan D O. Ran-independent nuclear import of cyclin B1-Cdc2 by importin beta. Proc Natl Acad Sci USA. 1999;96:7938–7943. doi: 10.1073/pnas.96.14.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talcott B, Moore M S. Getting across the nuclear pore complex. Trends Cell Biol. 1999;8:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- 55.Terns M P, Dahlberg J E, Lund E. Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- 56.Terns M P, Dahlberg J E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- 57.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 58.Ullman K S, Shah S, Powers M A, Forbes D J. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner R R. Rhabdovirus biology and infection: an overview. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum Publishing Corp.; 1987. pp. 9–74. [Google Scholar]
- 60.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 61.Wozniak R W, Rout M P, Aitchison J D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 62.Yuan H, Yoza B K, Lyles D S. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology. 1998;251:383–392. doi: 10.1006/viro.1998.9413. [DOI] [PubMed] [Google Scholar]