ABSTRACT
Hox genes encode evolutionarily conserved transcription factors that are essential for the proper development of bilaterian organisms. Hox genes are unique because they are spatially and temporally regulated during development in a manner that is dictated by their tightly linked genomic organization. Although their genetic function during embryonic development has been interrogated, less is known about how these transcription factors regulate downstream genes to direct morphogenetic events. Moreover, the continued expression and function of Hox genes at postnatal and adult stages highlights crucial roles for these genes throughout the life of an organism. Here, we provide an overview of Hox genes, highlighting their evolutionary history, their unique genomic organization and how this impacts the regulation of their expression, what is known about their protein structure, and their deployment in development and beyond.
Keywords: Hox gene regulation, Embryonic development, Patterning, Transcription factor
Summary: This Development at a Glance article provides an overview of the genomic organization, protein structure and regulation of Hox genes and our current understanding of their roles both during and after embryogenesis.
Introduction
Homeobox transcription factors are defined by a conserved 180 bp region of DNA, the homeobox sequence, that encodes a DNA-binding homeodomain. Phylogenetically, there are 11 groups of homeodomain-containing genes with Hox genes belonging to the ANTP group (Holland et al., 2007; reviewed by Ferrier, 2016). These genes were first described in the fruit fly, Drosophila (Lewis, 1978; Nüsslein-Volhard and Wieschaus, 1980), with subsequent studies identifying and characterizing them in a variety of other species, ranging from planaria to zebrafish to mouse and humans (reviewed by Feng et al., 2021; Ghosh and Sagerström, 2018; Mallo and Alonso, 2013; Olson, 2008; Quinonez and Innis, 2014). Distinguishing features of Hox genes in bilaterians are their linked clustering on the chromosome and their temporally and spatially directed expression (Duboule and Dollé, 1989; Izpisúa-Belmonte et al., 1991). Although we know that the temporal and spatial activation of Hox genes is essential for organismal development, many genomic targets of these transcription factors remain a mystery. In addition, how Hox protein structure, beyond the conserved homeodomain, and associated YWPM domain contributes to the control of gene expression is understudied. Here, we provide a brief overview of this important and interesting group of genes. We discuss the evolutionary history and the genomic organization of these genes and how this impacts cluster expression, Hox protein structure, and some key genetic functions described for Hox genes, with an emphasis on mammalian/vertebrate studies.
The evolution and genomic organization of Hox gene families
Homeobox-containing genes often encode transcription factors that play crucial roles in development and patterning. Significant work has been done to categorize the hundreds of homeobox genes into families and classes. The result of this work has established 11 homeobox-containing gene classes in animals (ANTP, PRD, LIM, POU, HNF, SINE, TALE, CUT, PROS, ZF, CERS) and 14 in plants (HD-ZIP I-IV, KNOX, BEL, PLINC, WOX, DDT, PHD, NDX, LD, PINTOX, SAWADEE) (Holland et al., 2007; Mukherjee et al., 2009). Hox genes are members of the ANTP class (Holland et al., 2007; reviewed by Holland, 2013).
Homeobox-containing genes are present in the three kingdoms of Eukarya (plants, animals and fungi), but Hox genes did not appear until after the divergence of the Porifera (sponges), evidenced by their presence in Cnidaria (corals, hydra, jellyfish, etc.) (Burke et al., 1995; Chourrout et al., 2006; Larroux et al., 2007; reviewed by Holland, 2013; Hrycaj and Wellik, 2016; Lemons and McGinnis, 2006). Cnidaria, however, lack a clustered arrangement of Hox genes; instead, Hox genes are randomly distributed throughout the genome, perhaps influencing the radial symmetry of Cnidaria (He et al., 2018; Kamm et al., 2006). Cnidaria and Bilateria likely diverged from a common ancestor, placing them on different evolutionary trajectories. Unlike Cnidaria, Bilaterian Hox genes are clustered on the chromosome and their expression patterns are under both spatial and temporal control. The order and developmental timing of expression of Hox genes along the chromosome mirrors their anterior-to-posterior expression, a property that has been referred to as colinearity (Duboule, 1994, 1998; Duboule and Dollé, 1989; Graham et al., 1989).
Although both invertebrates and vertebrates share colinear and spatiotemporal features of Hox gene expression, there are subtle differences in the genomic organization. In Drosophila, for example, eight Hox genes (lab, pb, Dfd, Scr, Antp, Ubx, abd-A and Abd-B) are linked, but the single Hox gene cluster is split across two different chromosomes (Von Allmen et al., 1996). In vertebrates, it is hypothesized that, in early vertebrate evolution, Hox genes were consolidated to form a compact, organized Hox cluster on the same strand of DNA, without non-Hox genes or repeat sequences. Global, regulatory control of the expression of these genes then evolved such that their chromosomal arrangement and structure became fixed (reviewed by Duboule, 2007). The most posterior member of the original cluster, Abd-B, expanded prior to duplication events, which resulted in additional Hox genes in vertebrates (39 in mammals and 47 in teleost), that are organized into four chromosomal clusters in mammals (HoxA, HoxB, HoxC and HoxD) and seven in zebrafish (Amores et al., 1998; Izpisúa-Belmonte et al., 1991; reviewed by Holland, 2013; Meyer and Málaga-Trillo, 1999; Singh and Krumlauf, 2022; Wagner et al., 2003). Each duplicated Hox gene in vertebrates is referred to as a paralog and this term refers to orthologous Hox genes within a species that have been classified based on genomic location, similarity of DNA sequence, and expression. For example, in mice, there are three members of the Hox1 paralogous group that reside on clusters A, B and D (Hoxa1, Hoxb1 and Hoxd1).
Initiation and regulation of Hox gene expression
Spatiotemporal initiation of Hox gene expression
The initiation of Hox gene expression and the spatiotemporally controlled onset of expression relies on strict epigenetic regulation of Hox gene clusters. In vertebrates, from the early zygote stage until the initiation of gastrulation, Hox gene clusters are tightly packed and inaccessible, and are they are not expressed. Histone modifier complexes, such as the Trithorax (TrxG) and Polycomb (PcG) group complexes are important for regulating this chromatin environment (reviewed by Casaca et al., 2014; Gentile and Kmita, 2020; Jambhekar et al., 2020; Kassis et al., 2017; Mallo and Alonso, 2013; Noordermeer and Duboule, 2013; Schuettengruber et al., 2017; Schwartz and Pirrotta, 2007). As gastrulation commences, Hox expression progressively initiates, starting with the Hox1 and Hox2 paralogs in the newly forming anterior regions of the embryo (Forlani et al., 2003; Wacker et al., 2004). As gastrulation proceeds, the Hox gene complex becomes progressively available, promoting deployment of the next set of paralogs (Holland and Hogan, 1988; reviewed by Deschamps and Duboule, 2017; Deschamps and van Nes, 2005). Hox13 paralogs are turned on last, and this set of paralogs plays crucial roles in ending gastrulation along with sister homeobox genes from the Cdx family (Economides et al., 2003; Neijts et al., 2017; van de Ven et al., 2011; Young et al., 2009; reviewed by Mallo et al., 2010). Similarly, TrxG and PcG complexes regulate chromatin accessibility and segmentation in the developing fly (Bantignies et al., 2011; Bi et al., 2022; Cao et al., 2002; Loker et al., 2021; reviewed by Kassis et al., 2017).
Hox gene regulation
Global enhancer sequences located outside the consolidated Hox gene clusters and the colinear arrangement of genes within the cluster control the pace of Hox gene deployment during axial segmentation (Crocker et al., 2015; Gould et al., 1997; Kmita et al., 2000; Miller and Posakony, 2020; Sharpe et al., 1998; Spitz et al., 2003; van der Hoeven et al., 1996; Zákány et al., 2001; reviewed by Afzal and Krumlauf, 2022; Mallo, 2018; Soshnikova, 2014). Genomes are organized into topologically associated domains (TADs); sequences within a given TAD will interact with higher frequency compared with those outside the TAD (Dixon et al., 2012). Both the HoxA and HoxD clusters contain TAD boundaries, which influence chromatin accessibility. During limb development, enhancers on either side of the TAD work to coordinate two transcriptional waves that permit limb patterning; the early wave patterns the stylopod and zeugopod, whereas the late wave patterns the digits (Montavon et al., 2011; Tarchini and Duboule, 2006; reviewed by Woltering and Duboule, 2010). The switch from early to late transcriptional waves for Hoxd13 is facilitated by enhancers posited in telomeric gene deserts in two TADs that reside outside the Hox gene clusters (Andrey et al., 2013). Single-cell assay for transposase-accessible chromatin (ATAC), chromatin immunoprecipitation sequencing (ChIP) and RNA-sequencing studies further supported the role of Hox13 in this transition, by demonstrating that Hoxa13 and Hoxd13 are required to permit an open chromatin conformation and, thus, the transition from early/proximal to late/distal limb patterning (Desanlis et al., 2020a,b; Sheth et al., 2016).
Although zebrafish and amniotes have similar TAD features, zebrafish TADs lack some enhancers, suggesting that these elements are important for the evolution of fins to limbs (Acemel et al., 2016; Freitas et al., 2012; Gehrke et al., 2015; Woltering et al., 2014; reviewed by Mallo, 2018). In Drosophila, recent work identified tethering elements, which function in conjunction with TADs to regulate developmental processes. Tethering elements facilitate the interactions between long-range enhancers and promoters, whereas TADs function to prevent unintentional interactions with enhancers (Batut et al., 2022). Enhancer sharing has also been documented in Drosophila, where one enhancer regulates the expression of two Hox genes (Eksi et al., 2018; Miller and Posakony, 2020).
Hox protein structure and transcriptional specificity
Hox protein structure
Hox proteins share a 60 amino acid homeodomain that contains a helix-turn-helix DNA-binding domain, which binds to -TAAT- motifs (Desplan et al., 1985; Gehring et al., 1994; Passner et al., 1999). All Hox proteins bind this same consensus site with approximately equal affinity; thus, although the homeodomain is a defining and crucial feature of Hox transcriptional regulation, it does not impart transcriptional specificity (reviewed by Mann et al., 2009). Some Hox proteins additionally contain a hexapeptide (Hx) motif, comprising a highly conserved YPWM motif and variable linker region, immediately N-terminal to the homeodomain sequence. Hexapeptide domains are present in Hox paralogous groups 1-8, and paralogous groups 9 and 10 proteins have a similar motif that retains some structural and functional roles, but the hexapeptide motif is absent from paralogous groups 11-13 proteins. This motif plays crucial roles in TALE (three amino acid loop extension) co-factor selectivity to regulate a subset of Hox targets (Chang et al., 1995; Dard et al., 2019; LaRonde-LeBlanc and Wolberger, 2003; Merabet and Galliot, 2015; Merabet et al., 2003; reviewed by Casaca et al., 2014; In der Rieden et al., 2004; Merabet and Mann, 2016). Additionally, there are substantial differences in the size of N- and C-terminal regions of Hox proteins that are hypothesized to play roles in Hox specificity (Joshi et al., 2007; Liu et al., 2008; Papadopoulos et al., 2010).
Mechanisms of Hox specificity
Despite binding to the same DNA consensus site with equal affinity, Hox proteins have transcriptional specificity. One example of how Hox proteins achieve specificity is through interaction of the YPWM motif with TALE domain co-factors. Like Hox proteins, TALE proteins are highly conserved throughout evolution, and their interaction with Hox proteins increases Hox target selectivity (Bridoux et al., 2020; Dard et al., 2019; Slattery et al., 2011; reviewed by Kassis et al., 2017; Merabet and Mann, 2016; Moens and Selleri, 2006; Rezsohazy et al., 2015). There are two classes of TALE proteins: PBC proteins and MEIS proteins. Drosophila Extradenticle (Exd) and vertebrate Pbx (1-4) proteins make up the PBC class, whereas Drosophila Homothorax (Hth) and vertebrate Meis (1-3) and Prep (1 and 2; Pknox1 and 2) are MEIS class proteins (reviewed by Moens and Selleri, 2006). Although PBC/MEIS proteins also have non-Hox-related activities, they function together with Hox proteins to regulate the development of the hindbrain, limb, axial skeleton, hematopoietic system and other organs (Capellini et al., 2006; Dassé et al., 2012; Deflorian et al., 2004; Delgado et al., 2021; Di Rosa et al., 2007; Erickson et al., 2007; Khandelwal et al., 2017; Machon et al., 2015; Peifer and Wieschaus, 1990; reviewed by Moens and Selleri, 2006; Schulte and Frank, 2014). In zebrafish, for instance, pbx4 (Lazarus) is required for Hox1 and Hox2 paralog function in the control of motor neuron migration in the hindbrain (Cooper et al., 2003). During branchial arch development, Hoxa2 and Meis cooperatively bind DNA to specify the identity of the second branchial arch (Amin et al., 2015; Hunter and Prince, 2002). In Drosophila limb regulation, Exd and Hth enhance abdominal Hox protein Ubx, but not thoracic Hox protein Antp, to bind to the Dll leg selector regulatory sequence (Gebelein et al., 2002). In the mouse proximodistal limb axis, Meis1/2 serve as a co-factors for Hox proteins, responding to both FGF and retinoic acid gradients to pattern the developing limb and specify anterior-posterior identity, respectively (López-Delgado et al., 2021; Mercader et al., 2000). Accordingly, genetic elimination of Meis1/2 leads to limb defects with proximalization of the skeletal elements (Delgado et al., 2021, 2020).
In addition to the conserved homeobox region and the YPWM motifs, Hox proteins contain substantial protein sequences both N-terminal and C-terminal to these domains. These regions are of variable sizes, with the N-terminal region ranging from 96 amino acids (in Hoxa2) to 339 amino acids (in Hoxa10), and the C-terminal domain varying from eight amino acids (in Hoxb9) to 194 amino acids (in Hoxa3) in mice. In general, the N- and C-terminal regions are highly conserved among paralogs, but more divergent between paralogs. There is, however, remarkably little known about the function of these domains, although they have been shown to be important for transcriptional regulation in some systems (reviewed by Mann et al., 2009). For example, Six2 is a downstream target of both Hox11 and Hoxa2; loss of the Hox11 paralogous group genes results in loss of Six2 expression, whereas loss of Hoxa2 leads to increased Six2 expression (Kutejova et al., 2008; Wellik et al., 2002). These transcriptional activities rely on interactions with the N- and C-terminal domains of Hox11 and Hoxa2 (Yallowitz et al., 2009). In addition, it has been shown that the N-terminal region of Hoxa10 is necessary for rib suppression, and that mutations in the N-terminal regions of Hoxa13 and Hoxd13 result in limb and genital defects (Fantini et al., 2009; Guerreiro et al., 2012; reviewed by Brison et al., 2012). Recent work suggests that the C-terminal region and homeodomain also influence interactions with previously inaccessible chromatin (Bulajić et al., 2020). Given the lack of specificity provided by the homeodomain, the N- and C-terminal domains are likely responsible for much of the region- and tissue-specific transcriptional activities of Hox proteins. Interrogation of these domains and identification of additional binding partners with Hox proteins is understudied.
Regional specificity and function of Hox genes during embryonic development
One of the key features of Hox transcription factors is their ability to impart different identities or morphologies along the axes in which they are expressed. During embryogenesis, Hox gene expression becomes spatially restricted along the anterior-posterior (AP) axis of the body (i.e. along the neural tube, axial skeleton and internal organs) and Hox9-13 genes additionally show spatial restriction along the proximodistal (PD) axis of the limbs (reviewed by Deschamps and Duboule, 2017; Mallo, 2018; Mallo et al., 2010; Wellik, 2009).
Neurectoderm
During gastrulation, the ectoderm forms and divides into the surface ectoderm (which gives rise to hair, nails and epidermis) and the neuroectoderm (which gives rise to the neural tube). As development proceeds, the neural tube becomes subdivided into forebrain, hindbrain, midbrain and spinal cord. Neural precursor cells in these developing areas differentiate to form nerve and glial cells specific to each functional region of the brain and spinal cord (reviewed by Jessell, 2000).
The developing hindbrain is divided into eight segments called rhombomeres, which give rise to cranial nerves. Hox1-4 paralogous group genes are expressed in the hindbrain and regulate rhombomere-specific development of hindbrain motor and cranial neurons; defects in paralogous groups 1-4 affect hindbrain segmentation and boundary formation (Barrow et al., 2000; Carpenter et al., 1993; Chisaka et al., 1992; Davenne et al., 1999; Gaufo et al., 2003; Gavalas et al., 1997, 2003; Mark et al., 1993; Prin et al., 2014; Rossel and Capecchi, 1999; Studer et al., 1998, 1996; reviewed by Ghosh and Sagerström, 2018; Krumlauf and Wilkinson, 2021; Parker et al., 2016; Parker and Krumlauf, 2020; Tümpel et al., 2009). Hox5-11 paralogous group genes are colinearly expressed in the spinal cord and play roles in defining motor neuron types derived from the phrenic, lateral, hypaxial, preganglionic and medial motor columns, whereas Hox13 is thought to act as a repressor of neuronal growth along the AP axis (Economides et al., 2003; Holstege et al., 2008; Hostikka et al., 2009; Jung et al., 2010; Lacombe et al., 2013; Lin and Carpenter, 2003; Philippidou et al., 2012; Tiret et al., 1998; van den Akker et al., 1999; Vermot et al., 2005; Wahba et al., 2001; Wu et al., 2008; reviewed by Joshi et al., 2021; Philippidou and Dasen, 2013).
Axial skeleton
In vertebrates, the mesodermally derived somites form alongside the neural tube during gastrulation and these important structures give rise to the dermis, skeletal muscles, tendons and axial vertebrae (reviewed by Williams et al., 2019). The vertebrate axial skeleton attains region-specific morphology as a result of Hox expression and function, subdividing regions from anterior to posterior into cervical, thoracic, lumbar, sacral and caudal domains. Within hours to days of the initiation of expression of each Hox paralogous group, the anterior boundary of expression for each Hox gene is fixed and vertebral axial fate and morphology are irreversibly established (reviewed by Mallo et al., 2010; Wellik, 2007).
Each Hox paralog group exhibits a high level of functional redundancy in patterning the axial vertebrae. It is only when the function of all alleles in a paralog group is lost that dramatic regional homeotic transformations occur (Horan et al., 1995a,b; McIntyre et al., 2007; van den Akker et al., 1999; Wellik and Capecchi, 2003). Loss of paralog function in the axial skeleton results in homeotic transformations. For example, mice with mutations in all three members of the Hox10 paralogous group do not develop normal lumbar vertebrae. Instead, the Hox10 paralogous mutants present with ribs that abnormally extend from the thoracic into lumbar and sacral regions (Wellik and Capecchi, 2003). Recent work demonstrated homeotic transformations in zebrafish for the first time when CRISPR-Cas9 was utilized to create loss-of-function alleles in each of the seven Hox clusters, highlighting a role for Hox genes in axial patterning in fish as well (Yamada et al., 2021).
Elegant support for the extent to which paralogs maintain redundant function has been demonstrated by the exact swapping of Hoxa3 and Hoxd3 coding sequences. Mouse lines were engineered such that the two exons of Hoxd3 exactly replace the exons of Hoxa3 (and the compliment replacement of Hoxd3 exons with those from Hoxa3). Hoxa3 and Hoxd3 single loss-of-function mutant mice displayed remarkably different phenotypes, yet the ‘swapped’ animals displayed no phenotypes, providing strong evidence that Hoxd3 and Hoxa3 are functionally equivalent when expressed appropriately (Chisaka and Capecchi, 1991; Condie and Capecchi, 1993; Greer et al., 2000). Similar results were subsequently obtained by swapping Hoxa1 and Hoxb1 coding sequences (Tvrdik and Capecchi, 2006).
Organ development along the anterior-posterior axis
Hox paralogs also redundantly participate in patterning many organs along the AP axis. Hox3 paralogous genes have been shown to be required for thymus, thyroid and parathyroid development (Condie and Capecchi, 1994; Manley and Capecchi, 1995). Hox5 genes play early roles in the growth and branching of the embryonic lung, and Hox6 genes are important in pancreas development (Hrycaj et al., 2015, 2018; Larsen et al., 2015). Hox10 and Hox11 genes are both important for proper development of the kidney, with loss of Hox11 leading to kidney agenesis (Wellik et al., 2002; Yallowitz et al., 2011). Hox9-13 paralogous groups impact the patterning of the reproductive tract in both males and females, and mutations in these genes result in dramatic transformations of the reproductive tract and lead to infertility (Cermik et al., 2001; Dollé et al., 1991; Ma et al., 1998; Raines et al., 2013; Taylor et al., 1999, 1997).
In humans, 11 syndrome-causing germline Hox mutations have been characterized. The known human Hox syndromes tend to present in the most distal areas (syndactyly of hands and feet, microtias, hypospadias and craniofacial abnormalities), likely because other Hox mutation-causing abnormalities are not easily visualized (i.e. internal abnormalities) (reviewed by Quinonez and Innis, 2014).
Limb development along the proximal-distal axis
Limb buds emerge as mesodermal bulges adjacent from the lateral plate mesoderm. In vertebrates, the forelimb bud arises at the cervicothoracic transition and the hindlimbs develop at the lumbosacral transition, linking axial patterning to limb development (reviewed by Petit et al., 2017; Song et al., 2019). Three main limb skeletal domains form within the limb bud soon after its emergence. From proximal to distal, these are: the stylopod (humerus/femur), the zeugopod (radius and ulna/tibia and fibula) and the autopod (hand/forepaw and foot/hindpaw bones). Abd-B-related Hox genes, groups 9-13, pattern the morphology of each of these segments, with Hox9 and 10 paralogous group genes responsible for establishing stylopod morphology, Hox11 paralog group genes patterning the zeugopod, and Hox13 paralogs (specifically Hoxa13 and Hoxd13) patterning the autopod (Davis et al., 1995; Fromental-Ramain et al., 1996a,b; Wellik and Capecchi, 2003).
Continuing roles for Hox genes after embryogenesis
There is significant evidence accumulating in the literature of continued Hox gene expression in postnatal and adult animals. Hox expression in adults has been reported in the female reproductive tract, the dermis, lung, muscles, the hematopoietic system, synovial cartilage, nervous system, skeleton, and other tissues (Giampaolo et al., 1994; Hutlet et al., 2021; Kawagoe et al., 1999; Li et al., 2021; Pineault et al., 2002; Rux et al., 2021; Song et al., 2020; Taylor et al., 1997; Yoshioka et al., 2021; Yu et al., 2018). Moreover, the stromal cells within many organ systems show continued Hox expression from embryonic to adult stages (examples include the gut, lung, bone, pancreas, muscle and reproductive tract) (Garcia et al., 2020; Hrycaj et al., 2018; Larsen et al., 2015; Song et al., 2020; Taylor et al., 1997; Yahagi et al., 2004; Yoshioka et al., 2021; reviewed by Rux and Wellik, 2017). Intriguingly, Hox genes maintain their strict, regionally restricted expression that is established at embryonic stages.
Continued expression and function in the skeletal system
In many cases, the function of Hox expression in adult tissues has not been explored, but in examples for which post-embryonic function has been assessed, work has shown that Hox genes continue to be important. The mouse skeletal system is one system in which adult Hox expression and function has been rigorously examined in recent years. The expression of Hox genes (visualized with a Hoxa11eGFP endogenous reporter, in one example) is absent from the cartilage condensations and osteoblasts that establish the radial and ulnar bones (Swinehart et al., 2013). Instead, Hoxa11eGFP expression is observed in the outer perichondral/periosteal layer that surrounds these bones and continues to be regionally expressed in the periosteal layer of the radius and ulna throughout life and is also visualized in the endosteum (inner bone lining) and in bone marrow stromal cells (Swinehart et al., 2013; Rux et al., 2016). This expression is maintained regionally in the zeugopod through postnatal and adult stages. The isolation of Hoxa11eGFP-expressing cells from adult bone revealed that these cells are progenitor-enriched mesenchymal stem/stromal cells, with progenitor activity in vitro (Rux et al., 2016).
The broader significance of this finding is supported by the demonstration that region-specific Hox expression in progenitor-enriched mesenchymal stem/stromal cells of other skeletal elements, including in human, continues throughout life (Bradaschia-Correa et al., 2020; Rux et al., 2016). Genetic lineage analyses in mice further supports this finding. Using Hoxa11-CreERT2 to induce lineage labeling at embryonic, postnatal or adult stages, Hoxa11-expressing cells were demonstrated to serve as a stable stem/progenitor population, contributing to chondrocytes, osteoblasts and bone marrow adipose throughout life (Pineault et al., 2019). Conditional loss of Hox11 function at adult stages resulted in progressive and severe defects in the remodeling of the radius and ulna during homeostasis, highlighting a continued role for Hox genes in the vertebrate skeleton (Song et al., 2020).
Continued expression and function in other tissues
Extensive work supports the role for Hox genes in the development of the hindbrain and spinal cord (reviewed by Parker and Krumlauf, 2020; Philippidou and Dasen, 2013). However, additional recent studies document continued expression of Hox genes in neurons after birth (Coughlan et al., 2019; Karmakar et al., 2017; Lizen et al., 2017). Conditional loss of Hox2 genes in post-mitotic glutaminergic bushy cells of the anterior ventral nucleus permits proper tonography and sound transmission that is lost in null mutants, but results in defects in sound frequency discrimination, demonstrating a continuing role for Hox2 genes in shaping the neural circuitry of the mammalian auditory system during postnatal stages (Karmakar et al., 2017). Mis-regulation of factors in glutamatergic/GABAergic synapses and calcium signaling results from loss of Hoxa5 in postnatal stages (Lizen et al., 2017). As many Hox family genes have continued expression after birth in the spinal cord, it will be of great interest to explore their functional roles at postnatal and adult stages (Coughlan et al., 2019).
In the lung, Hox5 gene expression peaks 1-2 weeks after birth and conditional loss of Hox5 during postnatal development results in dramatic defects in alveologenesis, with expanded airways and bronchopulmonary dysplasia (Hrycaj et al., 2018). Although Hox5 expression decreases at adult stages, it remains present, and conditional loss of function in adult mice results in the rapid onset of an emphysema-like phenotype (Li et al., 2021). Potential post-embryonic roles for Hox genes in other organs and tissues are even less explored, but continued Hox gene expression supports this possibility. Substantial literature documents strong, posterior Hox gene expression (Hox9-13) in stromal cells of the adult female and male reproductive tracts and mutations in Hox9-13 lead to infertility (Ma et al., 1998; Mucenski et al., 2019; Oefelein et al., 1996; Taylor et al., 1998, 1997). Moreover, the mammary gland is developmentally regulated by the Hox9 paralog group, and expression of these genes continues in adult tissue (Chen and Capecchi, 1999). Pancreatic mesoderm strongly expresses Hox6 genes, and a subset of stromal cells maintain Hox6 gene expression throughout life, although the functional significance of this is unknown (Garcia et al., 2020; Larsen et al., 2015).
Although the hematopoietic system does not obey simple positional Hox gene expression, there are many publications documenting Hox expression and function in hematopoiesis and hematopoietic cancer; whether roles in this system are developmental or continuing is less clear (reviewed by Alsayegh et al., 2019; Bhatlekar et al., 2018; Feng et al., 2021). The association of Hoxa9 with acute myeloid leukemia is perhaps the most studied, but Hox mutations are associated with several other cancers, including breast, pancreatic, lung, liver and ovarian (Calvo et al., 2001; Kroon et al., 1998; reviewed by Feng et al., 2021; Morgan et al., 2017).
Continued expression and function in invertebrates
Post-metamorphic and adult expression of Hox genes have received less attention in organisms other than mammals, but recent work highlights emerging roles for Hox proteins in homeostatic processes. In Drosophila, changes in post-developmental Hox gene expression affect behavior, such as locomotion, the ability to self-right, and flight (Enriquez et al., 2015; Issa et al., 2022, 2019; reviewed by Buffry and McGregor, 2022; Joshi et al., 2021). Lin-39 (Hox4-5), in Caenorhabditis elegans, is required to maintain the terminal identity of cholinergic motor neurons (Feng et al., 2020). Finally, several Hox genes (hox1, lox5b, hox3a, hox3b and post2b) regulate different aspects of tissue segmentation and fission behavior during asexual reproduction in planaria (Arnold et al., 2021).
Conclusions and perspectives
Despite crucial roles for Hox function in so many aspects of development, tissue homeostasis and regeneration/repair, there are fundamental gaps in our existing knowledge. How these transcription factors achieve specificity in transcriptional regulation represents important unanswered questions in the field. Our understanding of what proteins serve as co-regulators with Hox proteins in addition to the PBC-class proteins to potentially achieve this specificity remains limited. The regions N- and C-terminal to the homeodomain are sure to harbor important interacting domains and it will be essential to identify additional Hox partners. During development, what does positional information mean? What are the mechanisms by which Hox-regulated ‘region-specific’ information is translated in vivo?
Another underexplored area is how Hox genes continue to play important functional roles after embryogenesis. The recent identification of Hox-expressing cells as adult stem/progenitor cells in at least some organ systems emphasizes the importance of examining later functions in other tissues. With sequencing costs rapidly decreasing, obtaining ‘-omic’ data is becoming feasible and has the power to address at least some of these questions. The challenge of testing and providing evidence for new hypotheses that emerge will be more difficult – and more important. Elucidating the mechanisms of Hox gene regulation of the many morphogenetic processes they control will no doubt further our understanding of development and evolution and could provide important opportunities for novel regenerative therapies.
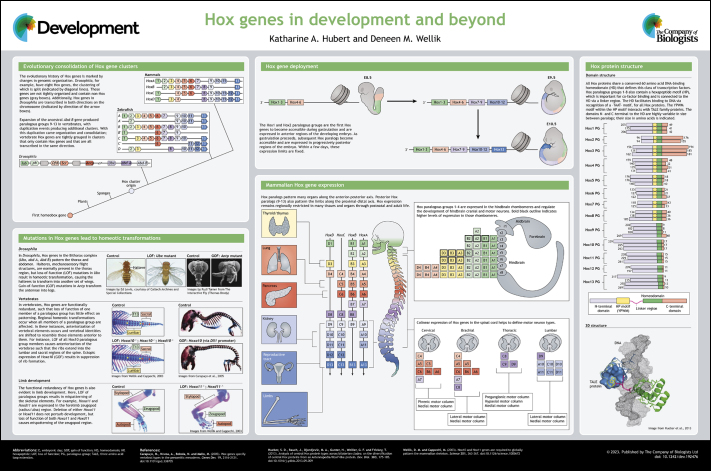
Poster
Footnotes
Funding
This work was funded by the National Institutes of Health (F31 AR079866 to K.A.H. and R37 AR061402 to D.M.W.). Deposited in PMC for release after 12 months.
Development at a glance
A high-resolution version of the poster is available for downloading in the online version of this article at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.192476#supplementary-data.
References
- Acemel, R. D., Tena, J. J., Irastorza-Azcarate, I., Marlétaz, F., Gómez-Marín, C., De La Calle-Mustienes, E., Bertrand, S., Diaz, S. G., Aldea, D., Aury, J. M.et al. (2016). A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 48, 336-341. 10.1038/ng.3497 [DOI] [PubMed] [Google Scholar]
- Afzal, Z. and Krumlauf, R. (2022). Transcriptional regulation and implications for controlling Hox gene expression. J. Dev. Biol. 10, 4. 10.3390/jdb10010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsayegh, K., Cortés-Medina, L. V., Ramos-Mandujano, G., Badraiq, H. and Li, M. (2019). Hematopoietic differentiation of human pluripotent stem cells: HOX and GATA transcription factors as master regulators. Curr. Genomics 20, 438-452. 10.2174/1389202920666191017163837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, S., Donaldson, I. J., Zannino, D. A., Hensman, J., Rattray, M., Losa, M., Spitz, F., Ladam, F., Sagerström, C. and Bobola, N. (2015). Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 32, 265-277. 10.1016/j.devcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores, A., Force, A., Yan, Y. L., Joly, L., Amemiya, C., Fritz, A., Ho, R. K., Langeland, J., Prince, V., Wang, Y. L.et al. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711-1714. 10.1126/science.282.5394.1711 [DOI] [PubMed] [Google Scholar]
- Andrey, G., Montavon, T., Mascrez, B., Gonzalez, F., Noordermeer, D., Leleu, M., Trono, D., Spitz, F. and Duboule, D. (2013). A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167. 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]
- Arnold, C. P., Lozano, A. M., Mann, F. G., Nowotarski, S. H., Haug, J. O., Lange, J. J., Seidel, C. W. and Alvarado, A. S. (2021). Hox genes regulate asexual reproductive behavior and tissue segmentation in adult animals. Nat. Commun. 12, 6706. 10.1038/s41467-021-26986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies, F., Roure, V., Comet, I., Leblanc, B., Schuettengruber, B., Bonnet, J., Tixier, V., Mas, A. and Cavalli, G. (2011). Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144, 214-226. 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Barrow, J. R., Stadler, H. S. and Capecchi, M. R. (2000). Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development 127, 933-944. 10.1242/dev.127.5.933 [DOI] [PubMed] [Google Scholar]
- Batut, P. J., Bing, X. Y., Sisco, Z., Raimundo, J., Levo, M. and Levine, M. S. (2022). Genome organization controls transcriptional dynamics during development. Science 375, 566-570. 10.1126/science.abi7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatlekar, S., Fields, J. Z. and Boman, B. M. (2018). Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018, 3569493. 10.1155/2018/3569493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, C.-L., Cheng, Q., Yan, L.-Y., Wu, H.-Y., Wang, Q., Wang, P., Cheng, L., Wang, R., Yang, L., Li, J.et al. (2022). A prominent gene activation role for C-terminal binding protein in mediating PcG/trxG proteins through Hox gene regulation. Development 149, dev200153. 10.1242/dev.200153 [DOI] [PubMed] [Google Scholar]
- Bradaschia-Correa, V., Leclerc, K., Josephson, A. M., Lee, S., Palma, L., Litwa, H. P., Neibart, S. S., Huo, J. C. and Leucht, P. (2020). Author Correction: Hox gene expression determines cell fate of adult periosteal stem/progenitor cells. Sci. Rep. 10, 3220. 10.1038/s41598-020-59764-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridoux, L., Zarrineh, P., Mallen, J., Phuycharoen, M., Latorre, V., Ladam, F., Losa, M., Baker, S. M., Sagerstrom, C., Mace, K. A.et al. (2020). HOX paralogs selectively convert binding of ubiquitous transcription factors into tissue-specific patterns of enhancer activation. PLoS Genet. 16, e1009162. 10.1371/journal.pgen.1009162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brison, N., Tylzanowski, P. and Debeer, P. (2012). Limb skeletal malformations - what the HOX is going on? Eur. J. Med. Genet. 55, 1-7. 10.1016/j.ejmg.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Buffry, A. D. and Mcgregor, A. P. (2022). Micromanagement of micromanagement of Drosophila post-embryonic development by Hox genes. J. Dev. Biol. 10, 13. 10.3390/jdb10010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulajić, M., Srivastava, D., Dasen, J. S., Wichterle, H., Mahony, S. and Mazzoni, E. O. (2020). Differential abilities to engage inaccessible chromatin diversify vertebrate Hox binding patterns. Development 147, dev194761. 10.1242/dev.194761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, A. C., Nelson, C. E., Morgan, B. A. and Tabin, C. (1995). Hox genes and the evolution of vertebrate axial morphology. Development 121, 333-346. 10.1242/dev.121.2.333 [DOI] [PubMed] [Google Scholar]
- Calvo, K. R., Knoepfler, P. S., Sykes, D. B., Pasillas, M. P. and Kamps, M. P. (2001). Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc. Natl. Acad. Sci. USA 98, 13120-13125. 10.1073/pnas.231115398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R. S. and Zhang, Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039-1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Capellini, T. D., Di Giacomo, G., Salsi, V., Brendolan, A., Ferretti, E., Srivastava, D., Zappavigna, V. and Selleri, L. (2006). Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 133, 2263-2273. 10.1242/dev.02395 [DOI] [PubMed] [Google Scholar]
- Carpenter, E. M., Goddard, J. M., Chisaka, O., Manley, N. R. and Capecchi, M. R. (1993). Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development 118, 1063-1075. 10.1242/dev.118.4.1063 [DOI] [PubMed] [Google Scholar]
- Casaca, A., Santos, A. C. and Mallo, M. (2014). Controlling Hox gene expression and activity to build the vertebrate axial skeleton. Dev. Dyn. 243, 24-36. 10.1002/dvdy.24007 [DOI] [PubMed] [Google Scholar]
- Cermik, D., Karaca, M. and Taylor, H. S. (2001). HOXA10 expression is repressed by progesterone in the myometrium: differential tissue-specific regulation of HOX gene expression in the reproductive tract. J. Clin. Endocrinol. Metab. 86, 3387-3392. [DOI] [PubMed] [Google Scholar]
- Chang, C. P., Shen, W. F., Rozenfeld, S., Lawrence, H. J., Largman, C. and Cleary, M. L. (1995). Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 9, 663-674. 10.1101/gad.9.6.663 [DOI] [PubMed] [Google Scholar]
- Chen, F. and Capecchi, M. R. (1999). Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc. Natl. Acad. Sci. USA 96, 541-546. 10.1073/pnas.96.2.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisaka, O. and Capecchi, M. R. (1991). Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature 350, 473-479. 10.1038/350473a0 [DOI] [PubMed] [Google Scholar]
- Chisaka, O., Musci, T. S. and Capecchi, M. R. (1992). Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature 355, 516-520. 10.1038/355516a0 [DOI] [PubMed] [Google Scholar]
- Chourrout, D., Delsuc, F., Chourrout, P., Edvardsen, R. B., Rentzsch, F., Renfer, E., Jensen, M. F., Zhu, B., De Jong, P., Steele, R. E.et al. (2006). Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature 442, 684-687. 10.1038/nature04863 [DOI] [PubMed] [Google Scholar]
- Condie, B. G. and Capecchi, M. R. (1993). Mice homozygous for a targeted disruption of Hoxd-3 (Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and the axis. Development 119, 579-595. 10.1242/dev.119.3.579 [DOI] [PubMed] [Google Scholar]
- Condie, B. G. and Capecchi, M. R. (1994). Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature 370, 304-307. 10.1038/370304a0 [DOI] [PubMed] [Google Scholar]
- Cooper, K. L., Leisenring, W. M. and Moens, C. B. (2003). Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev. Biol. 253, 200-213. 10.1016/S0012-1606(02)00018-0 [DOI] [PubMed] [Google Scholar]
- Coughlan, E., Garside, V. C., Wong, S. F. L., Liang, H., Kraus, D., Karmakar, K., Maheshwari, U., Rijli, F. M., Bourne, J. and Mcglinn, E. (2019). A Hox code defines spinocerebellar neuron subtype regionalization. Cell Rep. 29, 2408-2421.e4. 10.1016/j.celrep.2019.10.048 [DOI] [PubMed] [Google Scholar]
- Crocker, J., Abe, N., Rinaldi, L., Mcgregor, A. P., Frankel, N., Wang, S., Alsawadi, A., Valenti, P., Plaza, S., Payre, F.et al. (2015). Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160, 191-203. 10.1016/j.cell.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard, A., Jia, Y., Reboulet, J., Bleicher, F., Lavau, C. and Merabet, S. (2019). The human HOXA9 protein uses paralog-specific residues of the homeodomain to interact with TALE-class cofactors. Sci. Rep. 9, 5664. 10.1038/s41598-019-42096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassé, E., Volpe, G., Walton, D. S., Wilson, N., Del Pozzo, W., O'Neill, L. P., Slany, R. K., Frampton, J. and Dumon, S. (2012). Distinct regulation of c-myb gene expression by HoxA9, Meis1 and Pbx proteins in normal hematopoietic progenitors and transformed myeloid cells. Blood Cancer J. 2, e76. 10.1038/bcj.2012.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenne, M., Maconochie, M. K., Neun, R., Pattyn, A., Chambon, P., Krumlauf, R. and Rijli, F. M. (1999). Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron 22, 677-691. 10.1016/S0896-6273(00)80728-X [DOI] [PubMed] [Google Scholar]
- Davis, A. P., Witte, D. P., Hsieh-Li, H. M., Potter, S. S. and Capecchi, M. R. (1995). Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375, 791-795. 10.1038/375791a0 [DOI] [PubMed] [Google Scholar]
- Deflorian, G., Tiso, N., Ferretti, E., Meyer, D., Blasi, F., Bortolussi, M. and Argenton, F. (2004). Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development 131, 613-627. 10.1242/dev.00948 [DOI] [PubMed] [Google Scholar]
- Delgado, I., López-Delgado, A. C., Roselló-Díez, A., Giovinazzo, G., Cadenas, V., Fernández-De-Manuel, L., Sánchez-Cabo, F., Anderson, M. J., Lewandoski, M. and Torres, M. (2020). Proximo-distal positional information encoded by an Fgf-regulated gradient of homeodomain transcription factors in the vertebrate limb. Sci. Adv. 6, eaaz0742. 10.1126/sciadv.aaz0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado, I., Giovinazzo, G., Temiño, S., Gauthier, Y., Balsalobre, A., Drouin, J. and Torres, M. (2021). Control of mouse limb initiation and antero-posterior patterning by Meis transcription factors. Nat. Commun. 12, 3086. 10.1038/s41467-021-23373-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desanlis, I., Kherdjemil, Y., Mayran, A., Bouklouch, Y., Gentile, C., Sheth, R., Zeller, R., Drouin, J. and Kmita, M. (2020a). HOX13-dependent chromatin accessibility underlies the transition towards the digit development program. Nat. Commun. 11, 2491. 10.1038/s41467-020-16317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desanlis, I., Paul, R. and Kmita, M. (2020b). Transcriptional trajectories in mouse limb buds reveal the transition from anterior-posterior to proximal-distal patterning at early limb bud stage. J. Dev. Biol. 8, 31. 10.3390/jdb8040031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps, J. and Duboule, D. (2017). Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 31, 1406-1416. 10.1101/gad.303123.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps, J. and Van Nes, J. (2005). Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 132, 2931-2942. 10.1242/dev.01897 [DOI] [PubMed] [Google Scholar]
- Desplan, C., Theis, J. and O'Farrell, P. H. (1985). The Drosophila developmental gene, engrailed, encodes a sequence-specific DNA binding activity. Nature 318, 630-635. 10.1038/318630a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rosa, P., Villaescusa, J. C., Longobardi, E., Iotti, G., Ferretti, E., Diaz, V. M., Miccio, A., Ferrari, G. and Blasi, F. (2007). The homeodomain transcription factor Prep1 (pKnox1) is required for hematopoietic stem and progenitor cell activity. Dev. Biol. 311, 324-334. 10.1016/j.ydbio.2007.08.031 [DOI] [PubMed] [Google Scholar]
- Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., Hu, M., Liu, J. S. and Ren, B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé, P., Izpisúa-Belmonte, J. C., Brown, J. M., Tickle, C. and Duboule, D. (1991). HOX-4 genes and the morphogenesis of mammalian genitalia. Genes Dev. 5, 1767-1776. 10.1101/gad.5.10.1767 [DOI] [PubMed] [Google Scholar]
- Duboule, D. (1994). Temporal colinearity and the phylotypic progression: a basis for the stability of avertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 135-142. [PubMed] [Google Scholar]
- Duboule, D. (1998). Vertebrate hox gene regulation: clustering and/or colinearity? Curr. Opin. Genet. Dev. 8, 514-518. 10.1016/S0959-437X(98)80004-X [DOI] [PubMed] [Google Scholar]
- Duboule, D. (2007). The rise and fall of Hox gene clusters. Development 134, 2549-2560. 10.1242/dev.001065 [DOI] [PubMed] [Google Scholar]
- Duboule, D. and Dollé, P. (1989). The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8, 1497-1505. 10.1002/j.1460-2075.1989.tb03534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economides, K. D., Zeltser, L. and Capecchi, M. R. (2003). Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev. Biol. 256, 317-330. 10.1016/S0012-1606(02)00137-9 [DOI] [PubMed] [Google Scholar]
- Eksi, S. E., Barmina, O., Mccallough, C. L., Kopp, A. and Orenic, T. V. (2018). A Distalless-responsive enhancer of the Hox gene Sex combs reduced is required for segment- and sex-specific sensory organ development in Drosophila. PLoS Genet. 14, e1007320. 10.1371/journal.pgen.1007320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez, J., Venkatasubramanian, L., Baek, M., Peterson, M., Aghayeva, U. and Mann, R. S. (2015). Specification of individual adult motor neuron morphologies by combinatorial transcription factor codes. Neuron 86, 955-970. 10.1016/j.neuron.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, T., Scholpp, S., Brand, M., Moens, C. B. and Waskiewicz, A. J. (2007). Pbx proteins cooperate with Engrailed to pattern the midbrain-hindbrain and diencephalic-mesencephalic boundaries. Dev. Biol. 301, 504-517. 10.1016/j.ydbio.2006.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini, S., Vaccari, G., Brison, N., Debeer, P., Tylzanowski, P. and Zappavigna, V. (2009). A G220V substitution within the N-terminal transcription regulating domain of HOXD13 causes a variant synpolydactyly phenotype. Hum. Mol. Genet. 18, 847-860. [DOI] [PubMed] [Google Scholar]
- Feng, W., Li, Y., Dao, P., Aburas, J., Islam, P., Elbaz, B., Kolarzyk, A., Brown, A. E. and Kratsios, P. (2020). A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life. Elife 9, e50065. 10.7554/eLife.50065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y., Zhang, T., Wang, Y., Xie, M., Ji, X., Luo, X., Huang, W. and Xia, L. (2021). Homeobox genes in cancers: from carcinogenesis to recent therapeutic intervention. Front. Oncol. 11, 770428. 10.3389/fonc.2021.770428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier, D. E. K. (2016). Evolution of Homeobox gene clusters in animals:the giga-cluster and primary vs.secondary clustering. Front. Ecol. Evol. 4, 1-13. [Google Scholar]
- Forlani, S., Lawson, K. A. and Deschamps, J. (2003). Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development 130, 3807-3819. 10.1242/dev.00573 [DOI] [PubMed] [Google Scholar]
- Freitas, R., Gómez-Marín, C., Wilson, J. M., Casares, F. and Gómez-Skarmeta, J. L. (2012). Hoxd13 contribution to the evolution of vertebrate appendages. Dev. Cell 23, 1219-1229. 10.1016/j.devcel.2012.10.015 [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain, C., Warot, X., Lakkaraju, S., Favier, B., Haack, H., Birling, C., Dierich, A., Doll E, P. and Chambon, P. (1996a). Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development 122, 461-472. 10.1242/dev.122.2.461 [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain, C., Warot, X., Messadecq, N., Lemeur, M., Dollé, P. and Chambon, P. (1996b). Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997-3011. 10.1242/dev.122.10.2997 [DOI] [PubMed] [Google Scholar]
- Garcia, P. E., Adoumie, M., Kim, E. C., Zhang, Y., Scales, M. K., El-Tawil, Y. S., Shaikh, A. Z., Wen, H. J., Bednar, F., Allen, B. L.et al. (2020). Differential contribution of pancreatic fibroblast subsets to the pancreatic cancer stroma. Cell Mol. Gastroenterol. Hepatol. 10, 581-599. 10.1016/j.jcmgh.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaufo, G. O., Thomas, K. R. and Capecchi, M. R. (2003). Hox3 genes coordinate mechanisms of genetic suppression and activation in the generation of branchial and somatic motoneurons. Development 130, 5191-5201. 10.1242/dev.00730 [DOI] [PubMed] [Google Scholar]
- Gavalas, A., Davenne, M., Lumsden, A., Chambon, P. and Rijli, F. M. (1997). Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development 124, 3693-3702. 10.1242/dev.124.19.3693 [DOI] [PubMed] [Google Scholar]
- Gavalas, A., Ruhrberg, C., Livet, J., Henderson, C. E. and Krumlauf, R. (2003). Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development 130, 5663-5679. 10.1242/dev.00802 [DOI] [PubMed] [Google Scholar]
- Gebelein, B., Culi, J., Ryoo, H. D., Zhang, W. and Mann, R. S. (2002). Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev. Cell 3, 487-498. 10.1016/S1534-5807(02)00257-5 [DOI] [PubMed] [Google Scholar]
- Gehring, W. J., Qian, Y. Q., Billeter, M., Furukubo-Tokunaga, K., Schier, A. F., Resendez-Perez, D., Affolter, M., Otting, G. and Wüthrich, K. (1994). Homeodomain-DNA recognition. Cell 78, 211-223. 10.1016/0092-8674(94)90292-5 [DOI] [PubMed] [Google Scholar]
- Gehrke, A. R., Schneider, I., De La Calle-Mustienes, E., Tena, J. J., Gomez-Marin, C., Chandran, M., Nakamura, T., Braasch, I., Postlethwait, J. H., Gómez-Skarmeta, J. L.et al. (2015). Deep conservation of wrist and digit enhancers in fish. Proc. Natl. Acad. Sci. USA 112, 803-808. 10.1073/pnas.1420208112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile, C. and Kmita, M. (2020). Polycomb repressive complexes in Hox gene regulation: silencing and beyond: the functional dynamics of polycomb repressive complexes in Hox gene regulation. BioEssays 42, e1900249. 10.1002/bies.201900249 [DOI] [PubMed] [Google Scholar]
- Ghosh, P. and Sagerström, C. G. (2018). Developing roles for Hox proteins in hindbrain gene regulatory networks. Int. J. Dev. Biol. 62, 767-774. 10.1387/ijdb.180141cs [DOI] [PubMed] [Google Scholar]
- Giampaolo, A., Sterpetti, P., Bulgarini, D., Samoggia, P., Pelosi, E., Valtieri, M. and Peschle, C. (1994). Key functional role and lineage-specific expression of selected HOXB genes in purified hematopoietic progenitor differentiation. Blood 84, 3637-3647. 10.1182/blood.V84.11.3637.bloodjournal84113637 [DOI] [PubMed] [Google Scholar]
- Gould, A., Morrison, A., Sproat, G., White, R. A. and Krumlauf, R. (1997). Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11, 900-913. 10.1101/gad.11.7.900 [DOI] [PubMed] [Google Scholar]
- Graham, A., Papalopulu, N. and Krumlauf, R. (1989). The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57, 367-378. 10.1016/0092-8674(89)90912-4 [DOI] [PubMed] [Google Scholar]
- Greer, J. M., Puetz, J., Thomas, K. R. and Capecchi, M. R. (2000). Maintenance of functional equivalence during paralogous Hox gene evolution. Nature 403, 661-665. 10.1038/35001077 [DOI] [PubMed] [Google Scholar]
- Guerreiro, I., Casaca, A., Nunes, A., Monteiro, S., Nóvoa, A., Ferreira, R. B., Bom, J. and Mallo, M. (2012). Regulatory role for a conserved motif adjacent to the homeodomain of Hox10 proteins. Development 139, 2703-2710. 10.1242/dev.081448 [DOI] [PubMed] [Google Scholar]
- He, S., Del Viso, F., Chen, C. Y., Ikmi, A., Kroesen, A. E. and Gibson, M. C. (2018). An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 361, 1377-1380. 10.1126/science.aar8384 [DOI] [PubMed] [Google Scholar]
- Holland, P. W. (2013). Evolution of homeobox genes. Wiley Interdiscip Rev. Dev. Biol. 2, 31-45. 10.1002/wdev.78 [DOI] [PubMed] [Google Scholar]
- Holland, P. W. and Hogan, B. L. (1988). Expression of homeo box genes during mouse development: a review. Genes Dev. 2, 773-782. 10.1101/gad.2.7.773 [DOI] [PubMed] [Google Scholar]
- Holland, P. W., Booth, H. A. and Bruford, E. A. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5, 47. 10.1186/1741-7007-5-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege, J. C., De Graaff, W., Hossaini, M., Cardona Cano, S., Jaarsma, D., Van Den Akker, E. and Deschamps, J. (2008). Loss of Hoxb8 alters spinal dorsal laminae and sensory responses in mice. Proc. Natl. Acad. Sci. USA 105, 6338-6343. 10.1073/pnas.0802176105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan, G. S., Kovàcs, E. N., Behringer, R. R. and Featherstone, M. S. (1995a). Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function. Dev. Biol. 169, 359-372. 10.1006/dbio.1995.1150 [DOI] [PubMed] [Google Scholar]
- Horan, G. S., Ramírez-Solis, R., Featherstone, M. S., Wolgemuth, D. J., Bradley, A. and Behringer, R. R. (1995b). Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 9, 1667-1677. 10.1101/gad.9.13.1667 [DOI] [PubMed] [Google Scholar]
- Hostikka, S. L., Gong, J. and Carpenter, E. M. (2009). Axial and appendicular skeletal transformations, ligament alterations, and motor neuron loss in Hoxc10 mutants. Int. J. Biol. Sci. 5, 397-410. 10.7150/ijbs.5.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycaj, S. M. and Wellik, D. M. (2016). Hox genes and evolution. F1000Res 5, F1000 Faculty Rev-859. 10.12688/f1000research.7663.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycaj, S. M., Dye, B. R., Baker, N. C., Larsen, B. M., Burke, A. C., Spence, J. R. and Wellik, D. M. (2015). Hox5 genes regulate the Wnt2/2b-Bmp4-signaling axis during lung development. Cell Rep. 12, 903-912. 10.1016/j.celrep.2015.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycaj, S. M., Marty-Santos, L., Cebrian, C., Rasky, A. J., Ptaschinski, C., Lukacs, N. W. and Wellik, D. M. (2018). Hox5 genes direct elastin network formation during alveologenesis by regulating myofibroblast adhesion. Proc. Natl. Acad. Sci. USA 115, E10605-E10614. 10.1073/pnas.1807067115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, M. P. and Prince, V. E. (2002). Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev. Biol. 247, 367-389. 10.1006/dbio.2002.0701 [DOI] [PubMed] [Google Scholar]
- Hutlet, B., Theys, N., Coste, C., Ahn, M. T., Doshishti-Agolli, K., Lizen, B. and Gofflot, F. (2021). Correction to: Systematic expression analysis of Hox genes at adulthood reveals novel patterns in the central nervous system. Brain Struct. Funct. 226, 939-940. 10.1007/s00429-021-02252-5 [DOI] [PubMed] [Google Scholar]
- In Der Rieden, P. M., Mainguy, G., Woltering, J. M. and Durston, A. J. (2004). Homeodomain to hexapeptide or PBC-interaction-domain distance: size apparently matters. Trends Genet. 20, 76-79. 10.1016/j.tig.2003.12.001 [DOI] [PubMed] [Google Scholar]
- Issa, A. R., Picao-Osorio, J., Rito, N., Chiappe, M. E. and Alonso, C. R. (2019). A single MicroRNA-Hox gene module controls equivalent movements in biomechanically distinct forms of Drosophila. Curr. Biol. 29, 2665-2675.e4. 10.1016/j.cub.2019.06.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, A. R., Menzies, J. A. C., Padmanabhan, A. and Alonso, C. R. (2022). A novel post-developmental role of the Hox genes underlies normal adult behaviour. Proc. Natl. Acad. Sci. U.S.A. 119, e2209531119. 10.1073/pnas.2209531119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izpisúa-Belmonte, J. C., Falkenstein, H., Dollé, P., Renucci, A. and Duboule, D. (1991). Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 10, 2279-2289. 10.1002/j.1460-2075.1991.tb07764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar, A., Dhall, A. and Shi, Y. (2020). Author Correction: Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 21, 59. 10.1038/s41580-019-0192-5 [DOI] [PubMed] [Google Scholar]
- Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20-29. 10.1038/35049541 [DOI] [PubMed] [Google Scholar]
- Joshi, R., Passner, J. M., Rohs, R., Jain, R., Sosinsky, A., Crickmore, M. A., Jacob, V., Aggarwal, A. K., Honig, B. and Mann, R. S. (2007). Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131, 530-543. 10.1016/j.cell.2007.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, R., Sipani, R. and Bakshi, A. (2021). Roles of Drosophila Hox genes in the assembly of neuromuscular networks and behavior. Front. Cell Dev. Biol. 9, 786993. 10.3389/fcell.2021.786993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H., Lacombe, J., Mazzoni, E. O., Liem, K. F., Grinstein, J., Mahony, S., Mukhopadhyay, D., Gifford, D. K., Young, R. A., Anderson, K. V.et al. (2010). Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67, 781-796. 10.1016/j.neuron.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm, K., Schierwater, B., Jakob, W., Dellaporta, S. L. and Miller, D. J. (2006). Axial patterning and diversification in the cnidaria predate the Hox system. Curr. Biol. 16, 920-926. 10.1016/j.cub.2006.03.036 [DOI] [PubMed] [Google Scholar]
- Karmakar, K., Narita, Y., Fadok, J., Ducret, S., Loche, A., Kitazawa, T., Genoud, C., Di Meglio, T., Thierry, R., Bacelo, J.et al. (2017). Hox2 genes are required for tonotopic map precision and sound discrimination in the mouse auditory brainstem. Cell Rep. 18, 185-197. 10.1016/j.celrep.2016.12.021 [DOI] [PubMed] [Google Scholar]
- Kassis, J. A., Kennison, J. A. and Tamkun, J. W. (2017). Polycomb and trithorax group genes in Drosophila. Genetics 206, 1699-1725. 10.1534/genetics.115.185116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe, H., Humphries, R. K., Blair, A., Sutherland, H. J. and Hogge, D. E. (1999). Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia 13, 687-698. 10.1038/sj.leu.2401410 [DOI] [PubMed] [Google Scholar]
- Khandelwal, R., Sipani, R., Govinda Rajan, S., Kumar, R. and Joshi, R. (2017). Combinatorial action of Grainyhead, Extradenticle and Notch in regulating Hox mediated apoptosis in Drosophila larval CNS. PLoS Genet. 13, e1007043. 10.1371/journal.pgen.1007043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita, M., Kondo, T. and Duboule, D. (2000). Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing. Nat. Genet. 26, 451-454. 10.1038/82593 [DOI] [PubMed] [Google Scholar]
- Kroon, E., Krosl, J., Thorsteinsdottir, U., Baban, S., Buchberg, A. M. and Sauvageau, G. (1998). Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17, 3714-3725. 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf, R. and Wilkinson, D. G. (2021). Segmentation and patterning of the vertebrate hindbrain. Development 148, dev186460. 10.1242/dev.186460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutejova, E., Engist, B., Self, M., Oliver, G., Kirilenko, P. and Bobola, N. (2008). Six2 functions redundantly immediately downstream of Hoxa2. Development 135, 1463-1470. 10.1242/dev.017624 [DOI] [PubMed] [Google Scholar]
- Lacombe, J., Hanley, O., Jung, H., Philippidou, P., Surmeli, G., Grinstein, J. and Dasen, J. S. (2013). Genetic and functional modularity of Hox activities in the specification of limb-innervating motor neurons. PLoS Genet. 9, e1003184. 10.1371/journal.pgen.1003184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laronde-Leblanc, N. A. and Wolberger, C. (2003). Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17, 2060-2072. 10.1101/gad.1103303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroux, C., Fahey, B., Degnan, S. M., Adamski, M., Rokhsar, D. S. and Degnan, B. M. (2007). The NK homeobox gene cluster predates the origin of Hox genes. Curr. Biol. 17, 706-710. 10.1016/j.cub.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Larsen, B. M., Hrycaj, S. M., Newman, M., Li, Y. and Wellik, D. M. (2015). Mesenchymal Hox6 function is required for mouse pancreatic endocrine cell differentiation. Development 142, 3859-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons, D. and Mcginnis, W. (2006). Genomic evolution of Hox gene clusters. Science 313, 1918-1922. 10.1126/science.1132040 [DOI] [PubMed] [Google Scholar]
- Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565-570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Li, M. H., Marty-Santos, L. M., Van Ginkel, P. R., Mcdermott, A. E., Rasky, A. J., Lukacs, N. W. and Wellik, D. M. (2021). The lung elastin matrix undergoes rapid degradation upon adult loss of. Front. Cell Dev. Biol. 9, 767454. 10.3389/fcell.2021.767454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, A. W. and Carpenter, E. M. (2003). Hoxa10 and Hoxd10 coordinately regulate lumbar motor neuron patterning. J. Neurobiol. 56, 328-337. 10.1002/neu.10239 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Matthews, K. S. and Bondos, S. E. (2008). Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J. Biol. Chem. 283, 20874-20887. 10.1074/jbc.M800375200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizen, B., Moens, C., Mouheiche, J., Sacré, T., Ahn, M. T., Jeannotte, L., Salti, A. and Gofflot, F. (2017). Conditional loss of Hoxa5 function early after birth impacts on expression of genes with synaptic function. Front. Mol. Neurosci. 10, 369. 10.3389/fnmol.2017.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker, R., Sanner, J. E. and Mann, R. S. (2021). Cell-type-specific Hox regulatory strategies orchestrate tissue identity. Curr. Biol. 31, 4246-4255.e4. 10.1016/j.cub.2021.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Delgado, A. C., Delgado, I., Cadenas, V., Sánchez-Cabo, F. and Torres, M. (2021). Axial skeleton anterior-posterior patterning is regulated through feedback regulation between Meis transcription factors and retinoic acid. Development 148, dev193813. 10.1242/dev.193813 [DOI] [PubMed] [Google Scholar]
- Ma, L., Benson, G. V., Lim, H., Dey, S. K. and Maas, R. L. (1998). Abdominal B (AbdB) Hoxa genes: regulation in adult uterus by estrogen and progesterone and repression in Müllerian duct by the synthetic estrogen diethylstilbestrol (DES). Dev. Biol. 197, 141-154. 10.1006/dbio.1998.8907 [DOI] [PubMed] [Google Scholar]
- Machon, O., Masek, J., Machonova, O., Krauss, S. and Kozmik, Z. (2015). Meis2 is essential for cranial and cardiac neural crest development. BMC Dev. Biol. 15, 40. 10.1186/s12861-015-0093-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo, M. (2018). Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet. 34, 209-217. 10.1016/j.tig.2017.11.007 [DOI] [PubMed] [Google Scholar]
- Mallo, M. and Alonso, C. R. (2013). The regulation of Hox gene expression during animal development. Development 140, 3951-3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Mallo, M., Wellik, D. M. and Deschamps, J. (2010). Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344, 7-15. 10.1016/j.ydbio.2010.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley, N. R. and Capecchi, M. R. (1995). The role of Hoxa-3 in mouse thymus and thyroid development. Development 121, 1989-2003. 10.1242/dev.121.7.1989 [DOI] [PubMed] [Google Scholar]
- Mann, R. S., Lelli, K. M. and Joshi, R. (2009). Hox Specificity: Unique roles for factors and collaborators. Curr. Top. Dev. Biol. 88, 63-101. 10.1016/S0070-2153(09)88003-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark, M., Lufkin, T., Vonesch, J. L., Ruberte, E., Olivo, J. C., Dollé, P., Gorry, P., Lumsden, A. and Chambon, P. (1993). Two rhombomeres are altered in Hoxa-1 mutant mice. Development 119, 319-338. 10.1242/dev.119.2.319 [DOI] [PubMed] [Google Scholar]
- Mcintyre, D. C., Rakshit, S., Yallowitz, A. R., Loken, L., Jeannotte, L., Capecchi, M. R. and Wellik, D. M. (2007). Hox patterning of the vertebrate rib cage. Development 134, 2981-2989. 10.1242/dev.007567 [DOI] [PubMed] [Google Scholar]
- Merabet, S. and Galliot, B. (2015). The TALE face of Hox proteins in animal evolution. Front. Genet. 6, 267. 10.3389/fgene.2015.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet, S. and Mann, R. S. (2016). To be specific or not: the critical relationship between Hox and TALE proteins. Trends Genet. 32, 334-347. 10.1016/j.tig.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet, S., Kambris, Z., Capovilla, M., Bérenger, H., Pradel, J. and Graba, Y. (2003). The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev. Cell 4, 761-768. 10.1016/S1534-5807(03)00126-6 [DOI] [PubMed] [Google Scholar]
- Mercader, N., Leonardo, E., Piedra, M. E., Martínez-A, C., Ros, M. A. and Torres, M. (2000). Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127, 3961-3970. 10.1242/dev.127.18.3961 [DOI] [PubMed] [Google Scholar]
- Meyer, A. and Málaga-Trillo, E. (1999). Vertebrate genomics: more fishy tales about Hox genes. Curr. Biol. 9, R210-R213. 10.1016/S0960-9822(99)80131-6 [DOI] [PubMed] [Google Scholar]
- Miller, S. W. and Posakony, J. W. (2020). Disparate expression specificities coded by a shared Hox-C enhancer. Elife 9, e39876. 10.7554/eLife.39876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, C. B. and Selleri, L. (2006). Hox cofactors in vertebrate development. Dev. Biol. 291, 193-206. 10.1016/j.ydbio.2005.10.032 [DOI] [PubMed] [Google Scholar]
- Montavon, T., Soshnikova, N., Mascrez, B., Joye, E., Thevenet, L., Splinter, E., De Laat, W., Spitz, F. and Duboule, D. (2011). A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132-1145. 10.1016/j.cell.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Morgan, R., El-Tanani, M., Hunter, K. D., Harrington, K. J. and Pandha, H. S. (2017). Targeting HOX/PBX dimers in cancer. Oncotarget 8, 32322-32331. 10.18632/oncotarget.15971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski, M. L., Mahoney, R., Adam, M., Potter, A. S. and Potter, S. S. (2019). Single cell RNA-seq study of wild type and Hox9,10,11 mutant developing uterus. Sci. Rep. 9, 4557. 10.1038/s41598-019-40923-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, K., Brocchieri, L. and Bürglin, T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26, 2775-2794. 10.1093/molbev/msp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts, R., Amin, S., Van Rooijen, C. and Deschamps, J. (2017). Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev. Biol. 422, 146-154. 10.1016/j.ydbio.2016.12.024 [DOI] [PubMed] [Google Scholar]
- Noordermeer, D. and Duboule, D. (2013). Chromatin architectures and Hox gene collinearity. Curr. Top. Dev. Biol. 104, 113-148. 10.1016/B978-0-12-416027-9.00004-8 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard, C. and Wieschaus, E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795-801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Oefelein, M., Chin-Chance, C. and Bushman, W. (1996). Expression of the homeotic gene Hox-d13 in the developing and adult mouse prostate. J. Urol. 155, 342-346. 10.1016/S0022-5347(01)66657-6 [DOI] [PubMed] [Google Scholar]
- Olson, P. D. (2008). Hox genes and the parasitic flatworms: new opportunities, challenges and lessons from the free-living. Parasitol. Int. 57, 8-17. 10.1016/j.parint.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Papadopoulos, D. K., Vukojevic, V., Adachi, Y., Terenius, L., Rigler, R. and Gehring, W. J. (2010). Function and specificity of synthetic Hox transcription factors in vivo. Proc. Natl. Acad. Sci. USA 107, 4087-4092. 10.1073/pnas.0914595107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, H. J. and Krumlauf, R. (2020). A Hox gene regulatory network for hindbrain segmentation. Curr. Top. Dev. Biol. 139, 169-203. 10.1016/bs.ctdb.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Parker, H. J., Bronner, M. E. and Krumlauf, R. (2016). The vertebrate Hox gene regulatory network for hindbrain segmentation: Evolution and diversification: Coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. BioEssays 38, 526-538. 10.1002/bies.201600010 [DOI] [PubMed] [Google Scholar]
- Passner, J. M., Ryoo, H. D., Shen, L., Mann, R. S. and Aggarwal, A. K. (1999). Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397, 714-719. 10.1038/17833 [DOI] [PubMed] [Google Scholar]
- Peifer, M. and Wieschaus, E. (1990). Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 4, 1209-1223. 10.1101/gad.4.7.1209 [DOI] [PubMed] [Google Scholar]
- Petit, F., Sears, K. E. and Ahituv, N. (2017). Limb development: a paradigm of gene regulation. Nat. Rev. Genet. 18, 245-258. 10.1038/nrg.2016.167 [DOI] [PubMed] [Google Scholar]
- Philippidou, P. and Dasen, J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80, 12-34. 10.1016/j.neuron.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou, P., Walsh, C. M., Aubin, J., Jeannotte, L. and Dasen, J. S. (2012). Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat. Neurosci. 15, 1636-1644. 10.1038/nn.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault, K. M., Song, J. Y., Kozloff, K. M., Lucas, D. and Wellik, D. M. (2019). Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life. Nat. Commun. 10, 3168. 10.1038/s41467-019-11100-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault, N., Helgason, C. D., Lawrence, H. J. and Humphries, R. K. (2002). Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 30, 49-57. 10.1016/S0301-472X(01)00757-3 [DOI] [PubMed] [Google Scholar]
- Prin, F., Serpente, P., Itasaki, N. and Gould, A. P. (2014). Hox proteins drive cell segregation and non-autonomous apical remodelling during hindbrain segmentation. Development 141, 1492-1502. 10.1242/dev.098954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinonez, S. C. and Innis, J. W. (2014). Human HOX gene disorders. Mol. Genet. Metab. 111, 4-15. 10.1016/j.ymgme.2013.10.012 [DOI] [PubMed] [Google Scholar]
- Raines, A. M., Adam, M., Magella, B., Meyer, S. E., Grimes, H. L., Dey, S. K. and Potter, S. S. (2013). Recombineering-based dissection of flanking and paralogous Hox gene functions in mouse reproductive tracts. Development 140, 2942-2952. 10.1242/dev.092569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezsohazy, R., Saurin, A. J., Maurel-Zaffran, C. and Graba, Y. (2015). Cellular and molecular insights into Hox protein action. Development 142, 1212-1227. 10.1242/dev.109785 [DOI] [PubMed] [Google Scholar]
- Rossel, M. and Capecchi, M. R. (1999). Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 126, 5027-5040. 10.1242/dev.126.22.5027 [DOI] [PubMed] [Google Scholar]
- Rux, D. R. and Wellik, D. M. (2017). Hox genes in the adult skeleton: novel functions beyond embryonic development. Dev. Dyn. 246, 310-317. 10.1002/dvdy.24482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rux, D. R., Song, J. Y., Swinehart, I. T., Pineault, K. M., Schlientz, A. J., Trulik, K. G., Goldstein, S. A., Kozloff, K. M., Lucas, D. and Wellik, D. M. (2016). Regionally restricted Hox function in adult bone marrow multipotent mesenchymal stem/stromal cells. Dev. Cell 39, 653-666. 10.1016/j.devcel.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rux, D., Helbig, K., Koyama, E. and Pacifici, M. (2021). Hox11 expression characterizes developing zeugopod synovial joints and is coupled to postnatal articular cartilage morphogenesis into functional zones in mice. Dev. Biol. 477, 49-63. 10.1016/j.ydbio.2021.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber, B., Bourbon, H. M., Di Croce, L. and Cavalli, G. (2017). Genome regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34-57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Schulte, D. and Frank, D. (2014). TALE transcription factors during early development of the vertebrate brain and eye. Dev. Dyn. 243, 99-116. 10.1002/dvdy.24030 [DOI] [PubMed] [Google Scholar]
- Schwartz, Y. B. and Pirrotta, V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9-22. 10.1038/nrg1981 [DOI] [PubMed] [Google Scholar]
- Sharpe, J., Nonchev, S., Gould, A., Whiting, J. and Krumlauf, R. (1998). Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 17, 1788-1798. 10.1093/emboj/17.6.1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, R., Barozzi, I., Langlais, D., Osterwalder, M., Nemec, S., Carlson, H. L., Stadler, H. S., Visel, A., Drouin, J. and Kmita, M. (2016). Distal limb patterning requires modulation of cis-regulatory activities by HOX13. Cell Rep. 17, 2913-2926. 10.1016/j.celrep.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. P. and Krumlauf, R. (2022). Diversification and functional evolution of HOX proteins. Front. Cell Dev. Biol. 10, 798812. 10.3389/fcell.2022.798812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery, M., Riley, T., Liu, P., Abe, N., Gomez-Alcala, P., Dror, I., Zhou, T., Rohs, R., Honig, B., Bussemaker, H. J.et al. (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270-1282. 10.1016/j.cell.2011.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. Y., Pineault, K. M. and Wellik, D. M. (2019). Development, repair, and regeneration of the limb musculoskeletal system. Curr. Top. Dev. Biol. 132, 451-486. 10.1016/bs.ctdb.2018.12.011 [DOI] [PubMed] [Google Scholar]
- Song, J. Y., Pineault, K. M., Dones, J. M., Raines, R. T. and Wellik, D. M. (2020). Hox genes maintain critical roles in the adult skeleton. Proc. Natl. Acad. Sci. USA 117, 7296-7304. 10.1073/pnas.1920860117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova, N. (2014). Hox genes regulation in vertebrates. Dev. Dyn. 243, 49-58. 10.1002/dvdy.24014 [DOI] [PubMed] [Google Scholar]
- Spitz, F., Gonzalez, F. and Duboule, D. (2003). A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405-417. 10.1016/S0092-8674(03)00310-6 [DOI] [PubMed] [Google Scholar]
- Studer, M., Lumsden, A., Ariza-Mcnaughton, L., Bradley, A. and Krumlauf, R. (1996). Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 384, 630-634. 10.1038/384630a0 [DOI] [PubMed] [Google Scholar]
- Studer, M., Gavalas, A., Marshall, H., Ariza-Mcnaughton, L., Rijli, F. M., Chambon, P. and Krumlauf, R. (1998). Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125, 1025-1036. 10.1242/dev.125.6.1025 [DOI] [PubMed] [Google Scholar]
- Swinehart, I. T., Schlientz, A. J., Quintanilla, C. A., Mortlock, D. P. and Wellik, D. M. (2013). Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development 140, 4574-4582. 10.1242/dev.096693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini, B. and Duboule, D. (2006). Control of Hoxd genes’ collinearity during early limb development. Dev. Cell 10, 93-103. 10.1016/j.devcel.2005.11.014 [DOI] [PubMed] [Google Scholar]
- Taylor, H. S., Vanden Heuvel, G. B. and Igarashi, P. (1997). A conserved Hox axis in the mouse and human female reproductive system: late establishment and persistent adult expression of the Hoxa cluster genes. Biol. Reprod. 57, 1338-1345. 10.1095/biolreprod57.6.1338 [DOI] [PubMed] [Google Scholar]
- Taylor, H. S., Arici, A., Olive, D. and Igarashi, P. (1998). HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J. Clin. Invest. 101, 1379-1384. 10.1172/JCI1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, H. S., Igarashi, P., Olive, D. L. and Arici, A. (1999). Sex steroids mediate HOXA11 expression in the human peri-implantation endometrium. J. Clin. Endocrinol. Metab. 84, 1129-1135. 10.1210/jcem.84.3.5573 [DOI] [PubMed] [Google Scholar]
- Tiret, L., Le Mouellic, H., Maury, M. and Brûlet, P. (1998). Increased apoptosis of motoneurons and altered somatotopic maps in the brachial spinal cord of Hoxc-8-deficient mice. Development 125, 279-291. 10.1242/dev.125.2.279 [DOI] [PubMed] [Google Scholar]
- Tvrdik, P. and Capecchi, M. R. (2006). Reversal of Hox1 gene subfunctionalization in the mouse. Dev. Cell 11, 239-250. 10.1016/j.devcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Tümpel, S., Wiedemann, L. M. and Krumlauf, R. (2009). Hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 88, 103-137. 10.1016/S0070-2153(09)88004-6 [DOI] [PubMed] [Google Scholar]
- Van De Ven, C., Bialecka, M., Neijts, R., Young, T., Rowland, J. E., Stringer, E. J., Van Rooijen, C., Meijlink, F., Nóvoa, A., Freund, J. N.et al. (2011). Concerted involvement of Cdx/Hox genes and Wnt signaling in morphogenesis of the caudal neural tube and cloacal derivatives from the posterior growth zone. Development 138, 3451-3462. 10.1242/dev.066118 [DOI] [PubMed] [Google Scholar]
- Van Den Akker, E., Reijnen, M., Korving, J., Brouwer, A., Meijlink, F. and Deschamps, J. (1999). Targeted inactivation of Hoxb8 affects survival of a spinal ganglion and causes aberrant limb reflexes. Mech. Dev. 89, 103-114. 10.1016/S0925-4773(99)00212-9 [DOI] [PubMed] [Google Scholar]
- Van Der Hoeven, F., Zákány, J. and Duboule, D. (1996). Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell 85, 1025-1035. 10.1016/S0092-8674(00)81303-3 [DOI] [PubMed] [Google Scholar]
- Vermot, J., Schuhbaur, B., Le Mouellic, H., Mccaffery, P., Garnier, J. M., Hentsch, D., Brûlet, P., Niederreither, K., Chambon, P., Dollé, P.et al. (2005). Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development 132, 1611-1621. 10.1242/dev.01718 [DOI] [PubMed] [Google Scholar]
- Von Allmen, G., Hogga, I., Spierer, A., Karch, F., Bender, W., Gyurkovics, H. and Lewis, E. (1996). Splits in fruitfly Hox gene complexes. Nature 380, 116. 10.1038/380116a0 [DOI] [PubMed] [Google Scholar]
- Wacker, S. A., Jansen, H. J., Mcnulty, C. L., Houtzager, E. and Durston, A. J. (2004). Timed interactions between the Hox expressing non-organiser mesoderm and the Spemann organiser generate positional information during vertebrate gastrulation. Dev. Biol. 268, 207-219. 10.1016/j.ydbio.2003.12.022 [DOI] [PubMed] [Google Scholar]
- Wagner, G. P., Amemiya, C. and Ruddle, F. (2003). Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl. Acad. Sci. USA 100, 14603-14606. 10.1073/pnas.2536656100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba, G. M., Hostikka, S. L. and Carpenter, E. M. (2001). The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev. Biol. 231, 87-102. 10.1006/dbio.2000.0130 [DOI] [PubMed] [Google Scholar]
- Wellik, D. M. (2007). Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 236, 2454-2463. 10.1002/dvdy.21286 [DOI] [PubMed] [Google Scholar]
- Wellik, D. M. (2009). Hox genes and vertebrate axial pattern. Curr. Top. Dev. Biol. 88, 257-278. 10.1016/S0070-2153(09)88009-5 [DOI] [PubMed] [Google Scholar]
- Wellik, D. M. and Capecchi, M. R. (2003). Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301, 363-367. 10.1126/science.1085672 [DOI] [PubMed] [Google Scholar]
- Wellik, D. M., Hawkes, P. J. and Capecchi, M. R. (2002). Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 16, 1423-1432. 10.1101/gad.993302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S., Alkhatib, B. and Serra, R. (2019). Development of the axial skeleton and intervertebral disc. Curr. Top. Dev. Biol. 133, 49-90. 10.1016/bs.ctdb.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering, J. M. and Duboule, D. (2010). The origin of digits: expression patterns versus regulatory mechanisms. Dev. Cell 18, 526-532. 10.1016/j.devcel.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Woltering, J. M., Noordermeer, D., Leleu, M. and Duboule, D. (2014). Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 12, e1001773. 10.1371/journal.pbio.1001773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Wang, G., Scott, S. A. and Capecchi, M. R. (2008). Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development 135, 171-182. 10.1242/dev.009225 [DOI] [PubMed] [Google Scholar]
- Yahagi, N., Kosaki, R., Ito, T., Mitsuhashi, T., Shimada, H., Tomita, M., Takahashi, T. and Kosaki, K. (2004). Position-specific expression of Hox genes along the gastrointestinal tract. Congenit Anom (Kyoto) 44, 18-26. 10.1111/j.1741-4520.2003.00004.x [DOI] [PubMed] [Google Scholar]
- Yallowitz, A. R., Gong, K. Q., Swinehart, I. T., Nelson, L. T. and Wellik, D. M. (2009). Non-homeodomain regions of Hox proteins mediate activation versus repression of Six2 via a single enhancer site in vivo. Dev. Biol. 335, 156-165. 10.1016/j.ydbio.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yallowitz, A. R., Hrycaj, S. M., Short, K. M., Smyth, I. M. and Wellik, D. M. (2011). Hox10 genes function in kidney development in the differentiation and integration of the cortical stroma. PLoS One 6, e23410. 10.1371/journal.pone.0023410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., Maeno, A., Araki, S., Kikuchi, M., Suzuki, M., Ishizaka, M., Satoh, K., Akama, K., Kawabe, Y., Suzuki, K.et al. (2021). An atlas of seven zebrafish hox cluster mutants provides insights into sub/neofunctionalization of vertebrate Hox clusters. Development 148, dev198325. 10.1242/dev.198325 [DOI] [PubMed] [Google Scholar]
- Yoshioka, K., Nagahisa, H., Miura, F., Araki, H., Kamei, Y., Kitajima, Y., Seko, D., Nogami, J., Tsuchiya, Y., Okazaki, N.et al. (2021). Hoxa10 mediates positional memory to govern stem cell function in adult skeletal muscle. Sci. Adv. 7, eabd7924. 10.1126/sciadv.abd7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, T., Rowland, J. E., Van De Ven, C., Bialecka, M., Novoa, A., Carapuco, M., Van Nes, J., De Graaff, W., Duluc, I., Freund, J. N.et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516-526. 10.1016/j.devcel.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Yu, Z., Jiang, K., Xu, Z., Huang, H., Qian, N., Lu, Z., Chen, D., Di, R., Yuan, T., Du, Z.et al. (2018). Hoxc-dependent mesenchymal niche heterogeneity drives regional hair follicle regeneration. Cell Stem Cell 23, 487-500.e6. 10.1016/j.stem.2018.07.016 [DOI] [PubMed] [Google Scholar]
- Zákány, J., Kmita, M., Alarcon, P., De La Pompa, J. L. and Duboule, D. (2001). Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell 106, 207-217. 10.1016/S0092-8674(01)00436-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.