Abstract
We have developed a strategy to introduce in vitro-methylated DNA into defined chromosomal locations. Using this system, we examined the effects of methylation on transcription, chromatin structure, histone acetylation, and replication timing by targeting methylated and unmethylated constructs to marked genomic sites. At two sites, which support stable expression from an unmethylated enhancer-reporter construct, introduction of an in vitro-methylated but otherwise identical construct results in specific changes in transgene conformation and activity, including loss of the promoter DNase I-hypersensitive site, localized hypoacetylation of histones H3 and H4 within the reporter gene, and a block to transcriptional initiation. Insertion of methylated constructs does not alter the early replication timing of the loci and does not result in de novo methylation of flanking genomic sequences. Methylation at the promoter and gene is stable over time, as is the repression of transcription. Surprisingly, sequences within the enhancer are demethylated, the hypersensitive site forms, and the enhancer is hyperacetylated. Nevertheless, the enhancer is unable to activate the methylated and hypoacetylated reporter. Our findings suggest that CpG methylation represses transcription by interfering with RNA polymerase initiation via a mechanism that involves localized histone deacetylation. This repression is dominant over a remodeled enhancer but neither results in nor requires region-wide changes in DNA replication or chromatin structure.
In vertebrates, methylation of DNA occurs predominantly at the cytosine of CpG dinucleotides. This reversible modification is required for mouse development (33), plays an active role in X-chromosome inactivation and imprinting (25), and may be involved in tissue-specific gene repression (4) and in the silencing of parasitic sequences (52). Dynamic changes in methylation have been implicated in malignant transformation (26), and thus far two genetic disorders have been correlated to defects in genes involved in maintenance of methylation and methylation-induced repression (18).
The predominant consequence of methylation is transcriptional repression, which can be mediated either directly, by blocking the binding of transcription factors to CpG containing binding sites (23), or indirectly by proteins that specifically bind to methylated DNA via a methyl-CpG-binding domain (MDB) (37). Recently, several MBD-containing proteins have been described (19), of which four have been implicated in transcriptional repression. These proteins are thought to modify chromatin structure by recruiting histone deacetylase (HDAC) activity to methylated DNA, resulting in a repressive nucleosomal structure (reviewed in references 1 and 43).
The repressive effect of methylation on a given gene depends on the nature of its control elements (such as enhancer and promoter) (2), the density of methylated CpGs (21), the protein environment of a given cell type, and the chromosomal context of the gene, which can support or repress transcription. Thus, to determine the consequences of methylation on gene activity, it is important to compare unmethylated and methylated DNAs in the same cellular system and at the same position in the genome. The availability of methylases from bacteria permits the methylation of plasmid DNA in vitro prior to transfer into vertebrate cells. Thus far, standard techniques of gene transfer involving injection or transfection have been used to introduce such in vitro-methylated DNA into cells to determine the effects of DNA methylation on expression and/or chromatin structure. Studies using this experimental approach have contributed much information to our current understanding of methylation-induced repression. However, this approach is limited by the non-chromosomal-chromatin structure and the absence of replication in the case of the nonintegrated DNA and by the influences of copy number and different integration site(s) on transcription of the transgene(s) in the case of the stable transfections.
Here we show that in vitro-methylated DNA can be efficiently targeted into defined genomic sites using Cre recombinase. In order to analyze the mechanism of methylation-induced repression, as well as the dynamics of the methylation pattern, we used this approach to compare methylated and unmethylated DNA after insertion into the same chromosomal position. We targeted two genomic loci, both of which support expression from an unmethylated transgene (9), with either a fully methylated construct or an unmethylated, but otherwise identical control.
Our results suggest that DNA methylation at a genomic site permissive for transcription is stably propagated and is sufficient to repress transcription. This repression occurs in the absence of de novo methylation of adjacent DNA and without a change in the early timing of replication, suggesting that methylation does not result in a widespread change in the structure of the locus. Furthermore, we show that the enhancer becomes demethylated and remodeled but is not sufficient to overcome the repression, which occurs at the level of transcriptional initiation. Consistent with the model of HDAC recruitment by methylated DNA (1), we observe hypoacetylation of histones H3 and H4 at the methylated regions of the transgene, implicating a localized histone deacetylation as the cause of repression.
MATERIALS AND METHODS
Vectors and in vitro methylation.
The targeting plasmid pL1HS2EGFP1L was constructed by standard methods; the complete sequence is available on request. It contains L1 and 1L loxP sites as defined previously (10) flanking a HS2 fragment of the human β-globin LCR (GenBank HUMHBB file 7764 to 9218) linked to the human β-globin promoter (fragment −374 to +44 relative to the cap site) driving an enhanced green fluorescent protein (EGFP) reporter gene. The EGFP reporter consist of the simian virus 40 (SV40) 16S-19S splicing sites fused to the EGFP coding sequences (fragment NcoI-NotI, positions 677 to 1401 of Clontech [Palo Alto, Calif.] plasmid pEGFP-N1) and to SV40 polyadenylation sites. The SV40 16S-19S splicing sites and the poly(A) signal were derived from Clontech plasmid pCMVBeta. In vitro methylation was performed with SssI methylase (NEB) according to the protocol supplied by the manufacturer, followed by organic extraction and ethanol precipitation. Completeness of reaction was verified by full resistance to digestion with the methylation-sensitive enzymes HpaII and HhaI.
Cell lines and gene targeting.
MEL cell clones RL5 and RL6 contain a HYTK fusion gene flanked by inverted loxP sites (9). These cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% calf serum and split every 4 days. Prior to Cre-mediated targeting, cells were cultured in medium supplemented with 750 μg of hygromycin (Roche) per ml to select cells expressing the HYTK fusion gene. After selection, 4 × 106 cells were cotransfected with 25 μg of pL1HS2EGFP1L, 20 μg of Cre expression plasmid (CMV-Cre) (17), and 200 μg of sonicated salmon sperm DNA as a carrier in a BTX electroporator set to 250 V and 1,100 μF. Cells were plated in nonselective media and split after 3 days into media containing 10 μM ganciclovir to select against HYTK-expressing cells. After 1 week in selection, dilutions were plated to obtain single clones, which were then expanded and analyzed by genomic DNA Southern blot.
FACS analysis.
For GFP expression analysis, a single-cell suspension was harvested and washed with staining media (phosphate-buffered saline supplemented with 3% calf serum). Cells were resuspended in staining media supplemented with 1 μg of propidium iodide (PI) per ml for live-dead discrimination. Fluorescence-activated cell sorter (FACS) analysis was carried out on a FACSCalibur cytometer (Becton Dickinson) equipped with the standard fluorescein filter set. Data on a minimum of 10,000 live cells were collected and analyzed with the software CellQuest (Becton Dickinson).
Nuclease sensitivity analysis.
DNase I digestion of nuclei and subsequent Southern blot analyses were performed as described previously (11). The complete GFP coding region was used as a probe.
Replication timing analysis.
Replication timing was analyzed essentially as described elsewhere (7). Exponentially growing cells were pulse-labeled with bromodeoxyuridine (BrdU) and fixed. After being stained with PI, cells were sorted into different phases of the cell cycle according to DNA content, and BrdU-containing nascent DNA was purified by immunoprecipitation with an antibody against BrdU-DNA (Becton Dickinson). PCR (23 cycles) was performed using 2 μl (500 cell equivalents) of each nascent strand sample as a template. Southern blots were prepared and probed with radiolabeled probes synthesized by random priming the equivalent PCR product, amplified separately from a clone containing the transgene. In each experiment, genomic DNA from the same clone was included as a control for the strength and specificity of the PCR. All primers were specific and yielded a single primary product.
Analysis of histone acetylation.
Chromatin fixation and purification were performed as described earlier (46). Exponentially growing cells (2 × 108) were fixed in 150 ml of Dulbecco's modified Eagle's medium with 1% formaldehyde for 3 min at room temperature. After sonication, protein-DNA complexes were purified by isopycnic centrifugation (40). DNA content of cross-linked chromatin was quantified using a Hoefer Instruments fluorometer. Polyclonal antibodies against all acetylated isoforms of histone H4 (αH4-Ac) and against histone H3 acetylated at lysines 9 and 14 (αH3-Ac) were purchased from Upstate Biotechnology. Immunoprecipitation conditions for both antisera were as described elsewhere (46). Quantitative PCR of input and antibody-bound chromatin was performed with 1 to 2 ng of DNA as a template in a total volume of 25 μl with the appropriate primer pairs. Primers for transgene sequences were designed and tested to be specific and to give a product size ranging from 340 to 380 bp. The primer pair for the mouse amylase gene (amy4+6) gives a product of 400 bp, allowing us to perform duplex PCR with any of the transgene primer sets. A total of 0.1 μl of [α32P]dCTP (NEN) was added to each reaction. For each sequence, PCR reactions were performed in parallel under conditions of linear amplification (see Fig. 2 in reference 46; also data not shown) in a Perkin-Elmer 9600 thermocycler, for 27 cycles, using identical temperature profiles for all primer pairs. One-third of the reaction was subjected to electrophoresis on a 5% nondenaturing polyacrylamide gel, and products were quantified with a PhosphorImager and the ImageQuant software (Molecular Dynamics).
Nuclear run-on analysis.
Nuclear run-on assays were performed as described earlier (31) using [α-32P]CTP as the label. A 369-bp fragment, starting 47 bp downstream of the cap site and ending 170 bp into the GFP reading frame was used as a promoter-proximal probe, generated by PCR using the primer pair roGFP1+2. The distal probe was generated with the primer pair GFP1+2 and corresponds to the 3′ half of the GFP gene (bp 232 to 611 of the reading frame).
Methylation analysis.
Southern blot analysis to detect the methylation state of HpaII sites was carried out using standard procedures. Bisulfite conversion was conducted as described previously (34). To obtain the methylation status of the enhancer, nested PCR of converted genomic DNA was carried out with primer pairs +bisHS2-1 and −bisHS2-1 in the first round (30 cycles; annealing temperature, 50°C) and +bisHS2-2 and −bisHS2-2 in the second round of PCR (29 cycles; annealing temperature, 50°C). For the β promoter, primer pairs +bisβpr1 and −bisβpr1 (30 cycles; annealing temperature, 50°C) and +bisβpr2 and −bisβpr1 (29 cycles; annealing temperature, 50°C) were used in the first and second rounds, respectively. PCR products were cloned using the TA Cloning Kit (Invitrogen, Carlsbad, Calif.), and individual clones were sequenced with an ABI PRISM 377 DNA sequencer (Perkin-Elmer) as described earlier (34).
Primer sequences.
Listed are the names, product sizes, and sequences (in parentheses) of primers used in this study. The sequences in the transgene were as follows: GFP1+2, 377 bp, GFP-1 (ACATGAAGCAGCACGACTTC) and GFP-2 (TGCTCAGGTAGTGGTTGTC); roGFP1+2, 369 bp, roGFP-1 (ACCGGTGGTCGAGGAACTGA) and roGFP-2 (AGGGCACGGGCAGCTTGC); hubPr1+5, 342 bp, hubPr-1 (TGCTTACCAAGCTGTGATTCC) and hubPr-5 (GTGTCTGTTTGAGGTTGCTAG); huHS21+4, 343 bp, huHS2-1 (TTCCAGCATCCTCATCTCTGA) and huHS2-4 (TTTAGTCAGGTGGTCAGCTTCTC); mouse amylase 2.1y gene amy4+6, 401 bp, Amy4 (TCAGTTGTAATTCTCCTTGTACGG) and Amy6 (CATTCCTTGGCAATATCAACC); amyl1+2, 370 bp, mAmyl1 (AGCACTGAGGATTCAGTCTATG) and mAmyl2, (CCCGTACAAGGAGAATTACAAC); and mouse β-globin 5′Ey (located 1.1 kb 5′ of the Ey start codon), 376 bp, 5Ey-3 (GCACATGGATGCAGTTAAACAC) and 5Ey-4 (GAGTGACAGTGTAGAGAAGATG). The primers for bisulfite converted DNA of the transgene were as follows: +bisHS2-1 (GTTATATTTTTGTGTGTTTTTATTAGTGAT), +bisHS2-2 (TATAGTTTAAGTATGAGTAGTTTTGGTTAG), +bisHS2-2 (TATAGTTTAAGTATGAGTAGTTTTGGTTAG), −bisHS2-2 (TACACATATATTAATAAAACCTAATTCTAC), +bisβpr-1 (ATATGAAATAAGGATATGGAAGAGGAAGGT), +bisβpr-2 (TTTTAAGGTATTTTTGGATAGTTAGGTGGT), and −bisβpr-1 (CAAACCTAAAAATAAAAACAACATCCACTA).
RESULTS
Experimental strategy.
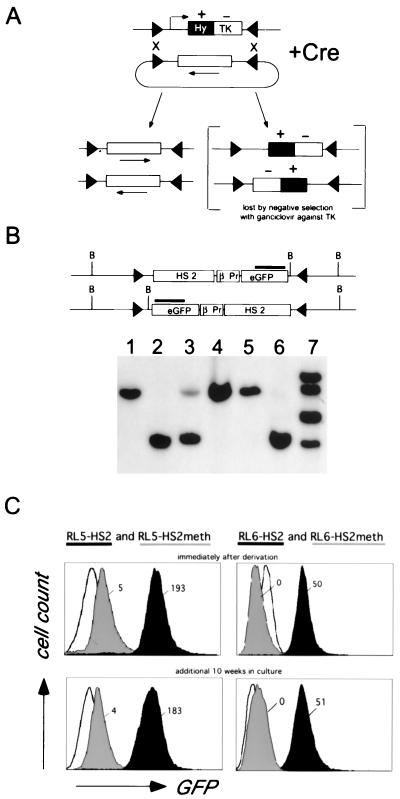
Our goal was to define the effects of DNA methylation in a defined chromosomal position. To accomplish this, we targeted control and in vitro-methylated DNA to the same sites in the genome. The likely repressive effect of methylation on transcription precluded the use of homologous or site-specific recombination targeting strategies, which depend upon the expression of a marker gene on the DNA molecule to be inserted. Instead, we made use of recombinase-mediated cassette exchange (RMCE), which allows the targeted insertion of a DNA cassette by selection against the HYTK fusion gene introduced during the original derivation of the targeting sites (10, 47).
We chose two genomic sites (RL5 and RL6) in mouse erythroleukemia (MEL) cells which can be targeted with Cre-recombinase using RMCE (Fig. 1A [9, 10]). A DNA construct containing the HS2 enhancer element from the human β-globin LCR, the human β-globin gene promoter, and the GFP reporter gene was either left unmodified or in vitro-methylated with SssI methylase (which methylates every CpG) and subsequently inserted into these sites.
FIG. 1.
(A) Principle of Cre-RMCE with inverted loxP sites. First, a stable cell line is generated with a construct encoding the positive-negative selectable marker gene HYTK (a fusion of hygromycin B phosphotransferase and herpes simplex virus thymidine kinase) flanked by inverted loxP sites. For the replacement reaction, a construct containing a similar set of loxP sites flanking the cassette to be recombined is transfected together with a Cre recombinase expression plasmid. Recombination between the loxP sites in the two constructs results in exchange of the cassettes and loss of the TK-negative selectable marker. The inverted loxP sites on the same DNA molecule can also recombine, resulting in the inversion of the intervening DNA (10). Ganciclovir is used to select against cells that still express the HYTK gene, allowing isolation of cells that have undergone the targeting reaction. (B) Representative Southern blot analysis of clones derived from RL6 using a restriction enzyme and probe combination that allows determination of the correct integration and orientation. Clones containing the transgene exclusively in one orientation (lanes 1, 2, 4, 5, and 6) were further analyzed, a mixture of both orientations (lane 3), or additional random insertions of the targeting construct (lane 7) were discarded. (C) Expression of the reporter gene, as measured by flow cytometry, is independent of the time in culture and is repressed by in vitro methylation. After targeted insertion into RL5 and RL6, clones containing the unmethylated transgene (RL5-HS2 and RL6-HS2 [black]) or the in vitro-methylated transgene (RL5-HS2meth and RL6-HS2meth [grey]) were analyzed by FACS either immediately after derivation (upper profile) or after an additional 10 weeks in culture (lower profile). The original RL5 and RL6 clones (containing only the HYTK marker) served as a negative control (white). The fluorescence values shown reflect the difference between the median of the transgene containing clone and that of the GFP negative parental clone.
Clones were derived and analyzed by Southern blotting for legitimate exchange of the cassette as shown in Fig. 1B. RL5-HS2 and RL6-HS2 refer to the unmethylated construct inserted into RL5 and RL6, respectively, and RL5-HS2meth and RL6-HS2meth refer to the in vitro-methylated constructs at these sites. The targeting efficiency, measured as the percentage of ganciclovir-resistant clones that have been correctly targeted, varied between 40 and 80% (data not shown). In vitro methylation of the plasmid did not decrease the targeting efficiency or the total number of clones, suggesting that CpG methylation does not interfere with recombinase activity. Thus, Cre-RMCE is suitable to target in vitro-methylated DNA into previously marked genomic sites.
Methylation-induced repression in permissive genomic sites.
To measure the effect of methylation on reporter gene expression, GFP fluorescence was analyzed by flow cytometry. As shown in Fig. 1C, both RL5 and RL6 insertion sites support pancellular GFP expression from the unmethylated transgene at high levels. In contrast, at both genomic sites, methylation of the reporter construct represses GFP expression in all cells analyzed. At RL5, GFP expression from the methylated construct is reduced to just above the background fluorescence level. Analysis of steady-state RNA using Northern blot analysis revealed that the residual GFP fluorescence reflects low-level transcription (data not shown). A similar expression pattern for the unmethylated and in vitro-methylated construct was observed at RL5 and RL6 when the transgene was integrated in the opposite orientation, indicating that in both genomic sites these expression characteristics are not orientation dependent (data not shown). The repressed and active expression states of the methylated and unmethylated transgenes, respectively, are stable over at least 12 weeks in culture, corresponding to ca. 100 cell divisions (Fig. 1C). The expression status at the RL5 integration site did not change even after 10 months in culture, whereas at RL6 noticeable silencing of the unmethylated transgene was observed in one orientation by the fourth month of culture, as described elsewhere (9).
Maintenance of methylation.
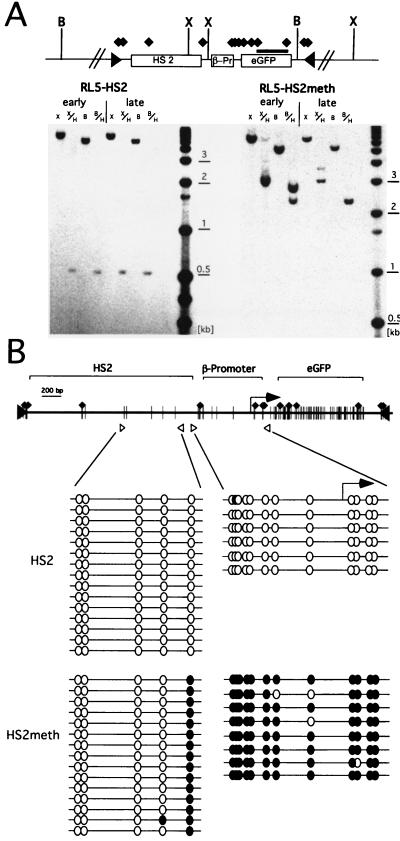
The stable expression states of the unmethylated and premethylated transgenes at RL5, even after extended periods in culture, suggest that the original methylation state is propagated in vivo. To determine if the methylation state of the introduced cassette is faithfully maintained, genomic DNA was isolated immediately after clone derivation (day 14) and after an additional 10 weeks in culture (day 90). First, the extent of methylation of the transgene and the adjacent genomic sequence was characterized by Southern blotting using the methylation-sensitive restriction enzyme HpaII. The position of each restriction site and the DNA fragment used as a probe are shown in Fig. 2A. The HpaII sites in the GFP gene of the unmethylated construct at RL5 (RL5-HS2) are susceptible to digestion at the early and late time points, suggesting that no de novo methylation has occurred. In contrast, digestion of the RL5-HS2meth clone yields a larger fragment, indicating that these sites are methylated. The XbaI/HpaII digest reveals that all nine HpaII sites in the promoter and the GFP gene are blocked, suggesting that this part of the transgene remains completely methylated. However, the resulting fragment is smaller than a fragment obtained with the XbaI digest alone, indicating that digestion occurs at an endogenous, unmethylated HpaII site outside of the transgene. Thus methylation is stably maintained in this part of the transgene, and we find no evidence for de novo methylation at one CpG in the flanking genomic DNA.
FIG. 2.
Maintenance of the methylation status. (A) Map of the L1-HS2GFP-1L transgene, including locations of the HpaII (black diamonds) XbaI (X), BglII (B), and loxP sites (black triangles). For Southern blot analyses, genomic DNA from the early and late time points was digested with either XbaI or BglII, in combination with the methylation-sensitive enzyme HpaII, and hybridized with a GFP probe (black bar). The unmethylated clone RL5-HS2 yields a 600-bp fragment with both HpaII-containing digests at both time points, indicating that no de novo methylation of the GFP gene has occurred. The in vitro-methylated clone RL5-HS2meth shows methylation of all HpaII sites in the transgene, with the exception of the three HpaII sites at the 5′ end of the transgene, as indicated by the 2.4-kb fragment obtained with a BglII/HpaII digest (see the text). (B) Detailed mapping of the methylation status of the enhancer and promoter region. Genomic DNA from the late time point was bisulfite converted, and the sequences of interest were PCR amplified, subcloned, and sequenced (see Materials and Methods). The positions of primers are indicated by open triangles. Open or filled circles correspond to unmethylated or methylated CpGs, respectively.
The BglII/HpaII restriction digest reveals the methylation status of the GFP gene, promoter, enhancer, and 5′-flanking genomic DNA (Fig. 2A). This digest yields a 2.4-kb fragment in case of RL5-HS2meth, which is indicative of methylation of all eight HpaII sites in the coding region and promoter but demethylation of a HpaII site in the enhancer. This site is partially demethylated at the early time point and fully demethylated after 10 weeks in culture.
To further characterize the extent of demethylation in this region, we mapped the methylation state of all CpGs in the enhancer and promoter region using bisulfite conversion and sequencing (34; see also Materials and Methods). This technique allows the analysis of the methylation state of any cytosine, independent of its sequence context. Primers to PCR amplify the bisulfite-converted genomic DNA were chosen to be specific for the core of HS2 or the promoter; the resulting methylation data are shown in Fig. 2B. Consistent with the Southern blot analysis, the promoter is methylated in the RL5-HS2meth clone, indicating that methylation is maintained in this region. However, the CpGs present in the HS2 enhancer are unmethylated in both RL5-HS2 and RL5-HS2meth, the latter suggesting that demethylation of the enhancer has occurred in vivo (Fig. 2B).
In summary, the introduced methylation is stably maintained at the promoter and coding region and no spreading of methylation into adjacent genomic DNA occurs. However, the enhancer is demethylated in the in vitro-methylated clone. Despite this demethylation, the enhancer is unable to overcome the methylation-induced repression.
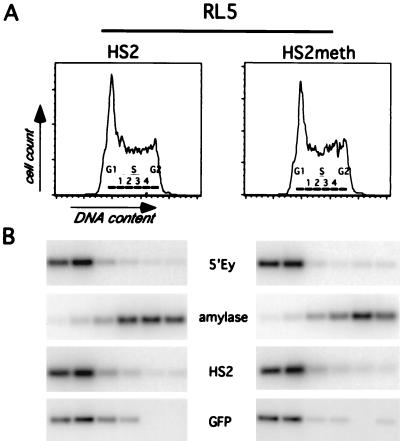
Early timing of replication in the methylated state.
Early timing of DNA replication has been associated in many systems with active transcription, open chromatin structure, and hypomethylation of the DNA, whereas late replication has been correlated with transcriptional inactivity, closed chromatin structure, and hypermethylation (48). Thus, we sought to determine whether the targeted introduction of methylation affects the replication timing at both integration sites (RL5 and RL6) by determining the relative abundance of specific genomic sequences in nascent DNA synthesized during different windows of the cell cycle (Fig. 3 and reference 7). Exponentially growing cells were pulse-labeled with BrdU and sorted by FACS into different fractions of the cell cycle based on their DNA content. BrdU-labeled DNA was enriched by immunoprecipitation and analyzed by PCR, using primers specific for the transgene or endogenous loci with a known timing of replication. As a control for early replication, we used the endogenous mouse β-globin locus and as a control for late replication we used the mouse amylase 2.1y gene, which we have shown previously to be late replicating in erythroid cells (7). The unmethylated transgene in the clone RL5-HS2 replicates early during S phase in comparison to the late control and as early as the mouse β-globin locus (Fig. 3). The replication timing of the RL5 locus is also early in the methylated and transcriptionally repressed clone RL5-HS2meth (Fig. 3), which is indistinguishable from the unmethylated clone. At the RL6 locus we find the same result: early timing of replication with both the unmethylated and the methylated constructs (data not shown). Thus, at both genomic sites, methylation of the transgene does not interfere with its early replication. We conclude that the establishment of methylation-induced repression neither requires nor results in late replication of these genomic regions.
FIG. 3.
Replication timing of the RL5 insertion site containing the unmethylated (RL5-HS2) and methylated transgene (RL5-HS2meth). (A) Histograms of PI staining intensity (DNA content) of cells for timing analysis are shown. The gates used to sort cells into fractions corresponding approximately to G1, S phase (S1 to S4), and G2 are labeled. (B) PCR-Southern analysis of replication timing of the transgene and control loci. Analysis was performed as described in Materials and Methods, using primers for the early-replicating control loci (endogenous murine β-globin [5′Ey3+4]), a late-replicating control locus (murine amylase [mAmyl1+2]), and the transgene enhancer (huHS21+2) and reporter gene (roGFP1+2). In RL5-HS2 and RL5-HS2meth, the transgene replicates as early as the endogenous murine β-globin locus, indicating that methylation of the transgene does not delay its replication timing.
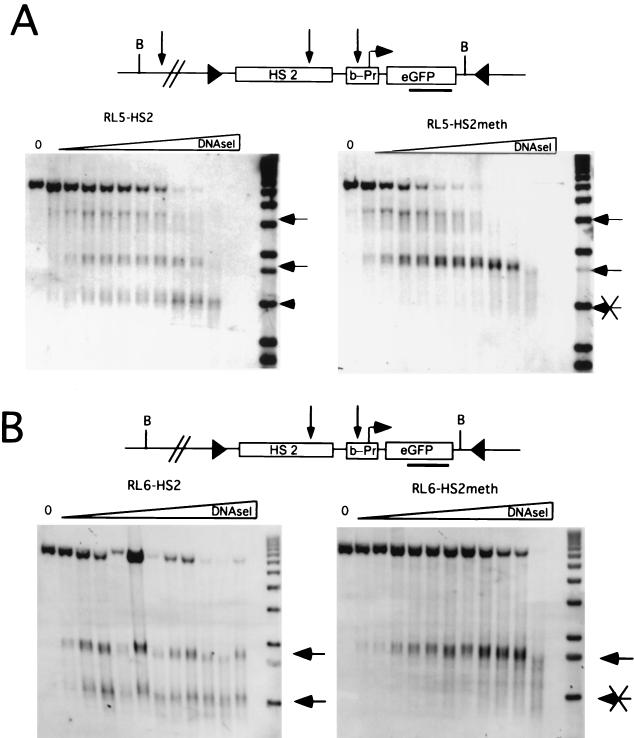
Remodeling of the enhancer is not influenced by the local methylation state.
Despite the localized demethylation of HS2 observed in the RL5-HS2meth transgene, the enhancer is unable to activate transcription. Thus, we asked whether methylation of the promoter and gene interferes with remodeling of the enhancer. Nuclei were isolated and incubated with increasing amount of DNase I, and the resulting genomic DNA was analyzed on a Southern blot. In the unmethylated RL5-HS2 clone, we detect hypersensitive-site (HS) formation at the β-promoter, at the HS2 enhancer and in addition, at an endogenous site 5′ of the transgene (Fig. 4A). In the transcriptionally repressed and methylated clone RL5-HS2meth, no HS forms at the promoter, while the enhancer and the endogenous sites form as in the unmethylated control.
FIG. 4.
Analysis of enhancer and promoter remodeling. Nuclei were isolated and digested with increasing concentrations of DNase I. Subsequently, genomic DNA was isolated, digested with Bgl II, and hybridized with a probe corresponding to the GFP gene. (A) Analysis of integration site RL5 with the unmethylated (RL5-HS2) and methylated (RL5-HS2meth) transgene. (B) Analysis of integration site RL6. Each hypersensitive site detected is marked with an arrow.
To test whether enhancer HS formation at the RL5 integration site is influenced by the presence of the endogenous HS, a similar DNase I series was performed with the corresponding constructs integrated at RL6 which does not show an endogenous HS in proximity to the 5′ end of the transgene. At this locus, both the promoter and the enhancer HSs form in the unmethylated and expressing clone, whereas only the enhancer HS is present in the methylated clone (Fig. 4B). Thus, in both genomic loci, the enhancer HS forms regardless of the methylation state of the downstream region. Since the formation of an HS is a consequence of non-histone protein binding (3), we conclude that the enhancer sequence is accessible to transactivators, even in close proximity to the methylated promoter and gene.
Methylation represses initiation of transcription.
Several mechanisms have been proposed to explain how DNA methylation interferes with the process of transcription. A study in the fungus Neurospora crassa suggested that DNA methylation inhibits RNA-polymerase elongation, whereas the loading of polymerases is not disturbed (45). On the other hand, experiments in Xenopus oocytes with nonreplicating plasmids suggested that methylation blocks the loading of RNA-polymerase (29). Genomic targeting of in vitro-methylated DNA allowed us to examine this question in a mammalian system, with the advantage that the methylated and unmethylated constructs reside in the same chromosomal locus and in a single copy.
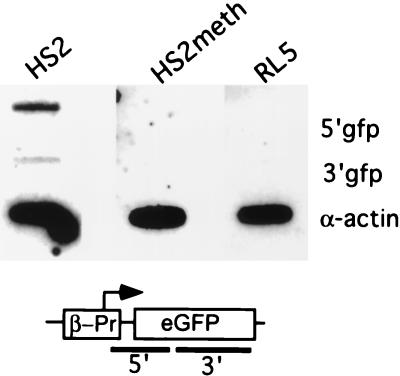
To determine whether the methylated and unmethylated transgenes show equivalent loading of polymerase, nuclear run-on assays were performed with nuclei isolated from RL5-HS2, RL5-HS2meth, and the parental clone RL5. In the run-on assay, elongation of already-initiated transcript proceeds in the presence of radiolabeled CTP under conditions which dissociate from DNA any nonpolymerase protein which could interfere with transcriptional elongation (16) (see Materials and Methods). The resulting nuclear run-on RNA(s) were analyzed by hybridization to an endogenous control and to sequences corresponding to a promoter-proximal and distal sequence of the reporter gene. As shown in Fig. 5, the unmethylated and transcribing RL5-HS2 clone shows a strong promoter-proximal signal and a weaker promoter-distal signal. Both signals indicate active transcription, the difference in intensity between promoter-proximal and distal probe suggests a higher density of polymerases at the promoter than at the gene. This indicates a high degree of polymerase loading and possible promoter-proximal pausing, as we and others have described previously for a number of endogenous genes and synthetic constructs (reviewed in reference 32). However, the nuclear RNA from RL5-HS2meth shows no signal above background for both reporter gene probes, indicating that polymerase loading is significantly reduced in the methylated construct. Thus, in a chromosomal context in mammalian cells, methylation interferes with polymerase loading, as described previously in Xenopus oocytes.
FIG. 5.
Nuclear run-on analysis to determine the density of polymerases in the unmethylated and methylated transgenes at the RL5 integration site. Nuclei from the RL5-HS2 clone, containing the unmethylated transgene, the RL5-HS2meth clone, containing the methylated transgene and the control parental clone RL5 without the transgene were isolated. Nuclear run-on assays were performed in the presence of radioactively labeled CTP, and nascent RNA was hybridized to three different DNA probes: α-actin as an endogenous control; 5′GFP, a promoter-proximal fragment containing the 3′ end of the β-globin promoter and the 5′ half of the GFP gene (a PCR product generated with the primer pair roGFP1+2); and 3′GFP, a promoter-distal fragment containing most of the 3′ half of the GFP coding region (generated with the primer pair GFP1+2). The actively expressing clone RL5-HS2 yields a higher signal for the proximal than for the distal probe, indicating that promoter-proximal pausing of polymerases occurs (see the text). In contrast, RL5-HS2meth gives no signal above background for either probes, suggesting a strong reduction of polymerase loading in the methylated state.
Methylation density defines the level of histone acetylation.
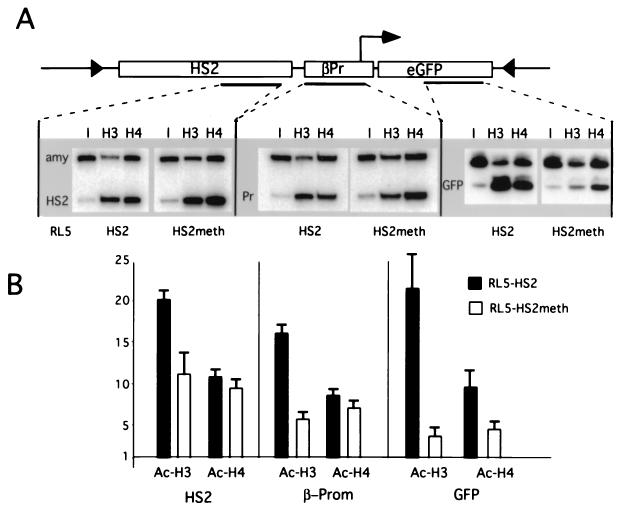
Hyperacetylation of histones has been shown to mark open chromatin and to be required for transcriptional activation (49). The recent finding that MBD proteins interact with HDACs suggests that methylation represses transcription by recruiting HDAC activity, resulting in hypoacetylation of histones residing in methylated DNA (1). We measured the relative level of histone acetylation of the methylated and unmethylated transgenes integrated at the RL5 genomic site. Formaldehyde cross-linked chromatin was purified and immunoprecipitated with antisera against acetylated isoforms of histone H3 and H4, as described previously (46). The antibody-bound DNA was analyzed with a duplex PCR approach using one primer pair specific for a transgenic sequence and a second pair specific for the endogenous amylase 2.1y gene, under conditions of linear amplification (46; see also Materials and Methods). This amylase gene is in a closed chromatin conformation and is characterized by relative hypoacetylation of histones H3 and H4 in MEL cells (46). The ratio of the two PCR products was determined for the antibody-bound fraction and normalized to the ratio obtained from the input material prior to immunoprecipitation. Three different regions in the RL5-HS2 and RL5-HS2meth transgenes were analyzed: the enhancer, the promoter, and the coding region of the reporter gene. A representative set of PCR products and the resulting enrichments relative to amylase are shown in Fig. 6A.
FIG. 6.
Chromatin immunoprecipitation analysis of histone H3 and H4 acetylation in different regions of the methylated and unmethylated transgene. Antibodies recognizing all acetylated isoforms of H4 (H4) or histone H3 acetylated at lysines 9 and 14 (H3) were used for immunoprecipitation. PCRs were performed on the input and antibody-bound chromatin fractions in the presence of a radiolabeled nucleotide under conditions of linear amplifications, as we have shown previously (see reference 46 and Materials and Methods). One primer pair amplifies a sequence from the transgene and the other amplifies a sequence from the endogenous mouse amylase 2.1y gene. The PCR products from the input (I) and antibody-bound DNA (H3 and H4) were electrophoresed on a nondenaturing acrylamide gel; a representative gel is shown. (B) Quantification of duplex PCR products from three independent immunoprecipitation experiments. The transgene/amylase ratio from each bound fraction was standardized by dividing by the transgene/amylase ratio from the input material to determine the relative enrichment of transgenic sequences during the immunoprecipitation. The mean value and standard error of the mean for the enrichment are plotted (see the text). The x axis is drawn at 1, which reflects no enrichment.
In this analysis, the unmethylated and expressing clone RL5-HS2 shows strong and comparable enrichment (17- to 22-fold) for all three sequences in the transgene with the antibody against acetylated histone H3. The antibody against acetylated histone H4 also showed strong enrichment (8- to 11-fold), with no detectable difference between the three sequences, suggesting uniform hyperacetylation of both H4 and H3 throughout the transgene in this clone. In contrast, the level of enrichment for H4 and H3 in the methylated clone RL5-HS2meth varies among the three sequences, with the highest level of enrichment at HS2, an intermediate level at the promoter, and the lowest level at the reporter gene (Fig. 6A). A direct comparison of the H3 acetylation between methylated and unmethylated constructs shows that the methylated clone is almost twofold less acetylated at HS2, threefold less acetylated at the promoter, and over sixfold less acetylated at the gene. The extent of this localized deacetylation directly correlates with the CpG density, which is highest in the GFP gene (see Fig. 2A), indicating that methylation density defines the degree of local hypoacetylation. These results are consistent with the recruitment of HDAC-containing complexes by MBDs and suggest that this recruitment results in a very localized deacetylation.
DISCUSSION
Genomic targeting of methylated DNA results in stable transcriptional repression.
We have targeted in vitro-methylated DNA into the genome to analyze methylation-induced repression at defined genomic insertion sites. DNA methylation studies have typically utilized either nonchromosomal templates, such as transiently transfected plasmids (2, 29), drug-selectable episomal constructs (21), or stably integrated transgenes (12, 30). While these experiments have been informative, nonchromosomal templates do not necessarily resemble the chromatin structure of chromosomal DNA and thus may not accurately reflect the effect of CpG methylation on transcription and chromatin structure. While stable transfection results in chromosomal integration, current protocols do not allow for control of the copy number or integration site, and thus different constructs are analyzed in different chromosomal contexts. Variable effects of different integration sites on transcription and chromatin structure are well documented (13, 20), complicating the analysis of cell lines harboring stably transfected reporter constructs. The Cre-RMCE targeting strategy described here circumvents these limitations, permitting the stable introduction of unmethylated and in vitro-methylated DNA at the same integration site.
The construct we analyzed contains the GFP reporter gene driven by the human β-globin promoter and the HS2 enhancer element from the human β-globin locus control region. This plasmid was either unmethylated or methylated in vitro at all CpGs using the bacterial methyltransferase SssI and introduced with similar high efficiency into two defined genomic integration sites that support stable expression from an unmethylated transgene. While the unmethylated construct is stably expressed even after long-term passage in culture, in vitro methylation of the reporter results in strongly reduced expression at either integration site (Fig. 1C), suggesting that the methylation is maintained and is responsible for this repression.
Methylation of the transgene does not alter the methylation state, chromatin structure, or replication timing of flanking DNA.
Consistent with the active expression state of the unmethylated transgene over time, we find no de novo methylation of this construct. On the other hand repression of the in vitro-methylated transgene is stable, a result consistent with the maintenance of its methylation at the promoter and the reporter gene. It has been proposed that spreading of methylation in cis into nonmethylated DNA is one mechanism by which de novo methylation occurs (50); however, we do not observe de novo methylation in the genomic DNA adjacent to the methylated construct. In addition, a DNase I-hypersensitive site flanking one of the insertion sites is present independent of the methylation state of the transgene. These observations suggest that the introduced patch of methylated DNA at the promoter and gene is propagated through cell division but is not sufficient to cause de novo methylation or to alter the chromatin structure of flanking DNA.
At many loci, replication timing is correlated with transcriptional activity; expressed loci are early replicating and silent loci are late replicating (15). Since transcriptional activators may be limited in late S phase, late replication itself may play a role in gene repression (reviewed in reference 48). As would be predicted, the active unmethylated constructs are early replicating. Surprisingly, at both genomic sites, the silent, in vitro-methylated constructs are also early replicating, indicating that a change in replication timing to late S phase is neither a requirement for, nor a consequence of, methylation-induced repression at these genomic loci. The maintenance methylase DNMT1, which preferentially binds to hemimethylated DNA, was recently reported to be associated with HDACs (14, 44) at replication foci. This interaction may ensure that, independent of the timing of replication, even hemimethylated DNA is in a repressive chromatin state.
Taken together, the lack of methylation spreading, the preservation of early replication timing, and the presence of a flanking HS after integration of the methylated construct suggest that transcriptional repression is not due to widespread changes in the activity or structure of the locus per se but rather to the local effects of methylation on the transgene itself.
Methylation-induced repression of transcriptional initiation is dominant over a demethylated and remodeled enhancer.
Analysis of the methylation status of the transgenes by bisulfite sequencing reveals that methylation at the promoter and gene is maintained over time, whereas the enhancer is demethylated after integration of the in vitro-methylated construct. Previous reports suggest that the binding of transactivators to DNA can interfere with the maintenance of methylation, probably by masking the CpG dinucleotide after DNA replication (22, 35). Thus, the observed demethylation at the enhancer could be a consequence of transcription factor binding to the enhancer.
Given the demethylated state of the enhancer, it is perhaps not surprising that the enhancer HS still forms (Fig. 3). Since HS formation requires non-histone protein binding (3), the enhancer of the methylated construct is occupied despite its close proximity to a high density of methylated CpGs. Nevertheless, this is not sufficient to overcome methylation-induced repression, and we conclude that methylation-induced repression does not result from inhibition of transcription factor binding at the enhancer. In contrast, the methylation is maintained over the promoter and gene and the promoter HS does not form. Consequently, the promoter and/or gene are the sites at which the methylation-induced repression mechanism operates.
To directly address whether polymerase loading or elongation are effected by DNA methylation, we used the nuclear run-on assay and measured the density of polymerases on the unmethylated and methylated transgenes. Previous studies using in vitro-methylated DNA containing mammalian promoters injected into Xenopus oocytes (29) suggest that methylation results in a block to transcription initiation. In contrast, experiments in N. crassa suggest that a block to transcriptional elongation is the major mechanism for methylation-induced repression in this organism (45). Our results indicate that, on a chromosomal template in mammalian cells, methylation interferes with transcriptional initiation. However, we cannot rule out that methylation has an additional effect on transcriptional elongation, since such an effect would be masked by the repressed initiation.
Is localized deacetylation of histones sufficient for repression?
Two mechanisms of methylation-induced transcriptional repression have been proposed. The binding of a subset of transcription factors is sensitive to methylation of their cognate binding sites (23), suggesting that CpG methylation of a promoter could directly block transactivator binding. However, a direct block of binding is unlikely to be responsible for the repression of the β-globin promoter used in this study, as the 120-bp element upstream of the initiation site does not contain any CpGs (Fig. 2B), yet is sufficient for promoter activity (reference 36 and references therein). An alternative mechanism of transcriptional repression involving the MBD family of proteins has been proposed (reviewed in reference 1). These proteins interact with, or are integral components of, complexes which include HDACs (27, 38, 39, 51, 53), suggesting that recruitment of HDAC activity, resulting in a modified nucleosomal structure, is a common motif in methylation-induced repression. Accordingly, it has been shown that methylated transgenes are hypoacetylated (6, 8, 12, 42). Here, we observe a reduction of histone acetylation at the promoter and gene of the in vitro-methylated transgene. The degree of deacetylation is more prominent for histone H3 than for H4 and correlates with the density of methylated CpGs: the GFP gene, which has a high density of CpGs, is the most hypoacetylated, while the promoter and enhancer, with lower densities, are acetylated to a lesser extent. This localized deacetylation suggests that the recruited HDACs act only on nearby nucleosomes, a result consistent with that reported for the HDAC activity of the yeast Sin3A complex (28).
In several studies treatment with the HDAC inhibitor trichostatin A (TSA) partially relieved the transcriptional repression of in vitro-methylated constructs (6, 8), whereas in others no reactivation was observed (34, 41). Here, TSA treatment of cells containing the inactive, methylated transgene did not result in reactivation of reporter gene transcription (data not shown), a result consistent with our previous study in MEL cells showing that a densely methylated provirus containing the same reporter gene could not be reactivated by TSA alone (34). Since the HDACs currently known to be involved in methylation-induced repression are at least partially sensitive to TSA treatment, we speculated that a lack of reactivation indicates an additional HDAC independent mode of repression (34). However, the recent finding that the HDAC activity of yeast SIR2 (24) and yeast HOS3 (5) is not inhibited by TSA indicates that TSA does not inhibit all HDAC activity, and it remains to be determined if methylation-induced repression involves a TSA-resistant HDAC activity.
Together, our experiments show that DNA methylation results in a localized histone deacetylation without affecting the chromatin structure and replication timing of the insertion site. Previously, we have shown that the transcriptionally active β-globin promoter in its native location is hyperacetylated (46), and we speculated that this hyperacetylation is required for activation. Thus, a localized hypoacetylation mediated by CpG methylation may be sufficient to account for the observed repression.
ACKNOWLEDGMENTS
This work was supported by a fellowship from the Deutsche Forschungsgemeinschaft to D.S.; NIH fellowship GM 19767/01 to M.C.L.; a fellowship from the American Cancer Society to D.M.C.; and NIH grants HL38655, DK56845, and HL554350 to E.E.B. and grants DK44746, HL57620, and CA54337 to M.G.
We thank Jürgen Bode, Steven Fiering, Ross Hardison, Anton Krumm, and the members of the Groudine lab for helpful suggestions; Claire Francastel for helpful comments on the manuscript; and Joan Hamilton, David Scalzo, Jennifer Stout, and Urszula Maliszewski for technical assistance.
REFERENCES
- 1.Bird A P, Wolffe A P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 2.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 4.Brandeis M, Ariel M, Cedar H. Dynamics of DNA methylation during development. Bioessays. 1993;15:709–713. doi: 10.1002/bies.950151103. [DOI] [PubMed] [Google Scholar]
- 5.Carmen A A, Griffin P R, Calaycay J R, Rundlett S E, Suka Y, Grunstein M. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc Natl Acad Sci USA. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W Y, Townes T M. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc Natl Acad Sci USA. 2000;97:377–382. doi: 10.1073/pnas.97.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimbora D M, Schübeler D, Reik A, Hamilton J, Francastel C, Epner E M, Groudine M. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol Cell Biol. 2000;20:5581–5591. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne A W. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Y. Q., M. C. Lorincz, S. Fiering, J. M. Greally, and E. Bouhassira. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 10.Feng Y Q, Seibler J, Alami R, Eisen A, Westerman K A, Leboulch P, Fiering S, Bouhassira E E. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 11.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 12.Forrester W C, Fernandez L A, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–3014. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francastel C, Walters M C, Groudine M, Martin D I. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- 14.Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 15.Goldman M A, Holmquist G P, Gray M C, Caston L A, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- 16.Groudine M, Peretz M, Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981;1:281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu H, Zou Y R, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich B. Methylation moves into medicine. Curr Biol. 2000;10:R60–R63. doi: 10.1016/s0960-9822(00)00286-4. [DOI] [PubMed] [Google Scholar]
- 19.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henikoff S. Position effect and related phenomena. Curr Opin Genet Dev. 1992;2:907–912. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh C L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh C L. Evidence that protein binding specifies sites of DNA demethylation. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iguchi-Ariga S M, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 24.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 25.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 26.Jones P A, Laird P W. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 27.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 28.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kass S U, Landsberger N, Wolffe A P. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 30.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 31.Krumm A, Hickey L B, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 32.Krumm A, Meulia T, Groudine M. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays. 1993;15:659–665. doi: 10.1002/bies.950151005. [DOI] [PubMed] [Google Scholar]
- 33.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 34.Lorincz M C, Schübeler D, Goeke S C, Walters M, Groudine M, Martin D I. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol Cell Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers R M, Tilly K, Maniatis T. Fine structure genetic analysis of a beta-globin promoter. Science. 1986;232:613–618. doi: 10.1126/science.3457470. [DOI] [PubMed] [Google Scholar]
- 37.Nan X, Meehan R R, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 39.Ng H H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 40.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 41.Osborne C S, Pasceri P, Singal R, Sukonnik T, Ginder G D, Ellis J. Amelioration of retroviral vector silencing in locus control region beta-globin-transgenic mice and transduced F9 embryonic cells. J Virol. 1999;73:5490–5496. doi: 10.1128/jvi.73.7.5490-5496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razin A. CpG methylation, chromatin structure, and gene silencing—a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rountree M R, Bachman K E, Baylin S B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 45.Rountree M R, Selker E U. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schübeler D, Francastel C, Cimbora D M, Reik A, Martin D I, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 47.Seibler J, Schübeler D, Fiering S, Groudine M, Bode J. DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- 48.Simon I, Cedar H. Temporal order of DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 387–408. [Google Scholar]
- 49.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 50.Tollefsbol T O, Hutchison C A., III Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J Mol Biol. 1997;269:494–504. doi: 10.1006/jmbi.1997.1064. [DOI] [PubMed] [Google Scholar]
- 51.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 52.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]