Abstract
Stimulation of the NF-κB pathway often causes p65-p50 and p50-p50 dimers to be simultaneously present in the cell nucleus. A natural polymorphism at nucleotide −863 in the human TNF promoter (encoding tumor necrosis factor [TNF]) region provides an opportunity to dissect the functional interaction of p65-p50 and p50-p50 at a single NF-κB binding site. We found that this site normally binds both p65-p50 and p50-p50, but a single base change specifically inhibits p50-p50 binding. Reporter gene analysis in COS-7 cells expressing both p65-p50 and p50-p50 shows that the ability to bind p50-p50 reduces the enhancer effect of this NF-κB site. Using an adenoviral reporter assay, we found that the variant which binds p50-p50 results in a reduction of lipopolysaccharide-inducible gene expression in primary human monocytes. This finding adds to a growing body of experimental evidence that p50-p50 can inhibit the transactivating effects of p65-p50 and illustrates the potential for genetic modulation of inflammatory gene regulation in humans by subtle nucleotide changes that alter the relative binding affinities of different forms of the NF-κB complex.
The NF-κB/Rel family of transcription factors is involved in many physiological processes, including regulation of a wide range of inflammatory mediators (2). Inflammation is a critical component of host defense, but it is also responsible for many of the clinical symptoms of infection and injury and can be fatal if elicited in excess. This raises the fundamental question of how the level of response to NF-κB is optimized across a large number of different inflammatory genes. Often this may involve functional interactions between NF-κB and other transcription factors (24). This paper considers another mechanism, which has received relatively little attention, namely, through variation in the composition of NF-κB dimers that bind to a specific regulatory site. The canonical form of NF-κB is a heterodimer comprising a p65 subunit, containing both a DNA binding domain and a domain that is essential for transcriptional activation, plus a p50 subunit which has a DNA binding domain but no activation domain. The biological role of p50 was initially thought to relate solely to its DNA binding properties within the active p65-p50 complex, but more recent evidence suggests that p50-p50 homodimers are capable of acting as transcriptional repressors. For example, artificial overexpression of p50 acts to suppress the transactivating effects of p65 at some NF-κB sites (15, 17), and this mechanism has been implicated in the downregulation of major histocompatibility complex expression (16) and in viral postinduction repression of the beta interferon gene (23). Since the optimal binding sequences for p65 and p50 are similar but not identical (12, 15, 23), it is possible that NF-κB-induced responses might be fine-tuned by minor sequence variations that alter the relative binding of p65-p50 and p50-p50 to different regulatory elements. To examine this question, we have investigated the functional properties of a naturally occurring, single nucleotide polymorphism that specifically affects the binding of p50-p50 to an NF-κB site in the human TNF promoter (encoding tumor necrosis factor [TNF]) region.
MATERIALS AND METHODS
Plasmids.
Human p50- and p65-expressing constructs in the Rc/CMV vector (Invitrogen) were previously described (13). The human TNF promoter construct (−1173)-pGL3 (TNF −863C) was described previously (25). The corresponding fragment containing the C-to-A substitution at nucleotide (nt) −863 (TNF −863A) was generated by site-directed mutagenesis in the construct −1173-pGL3 using oligonucleotides bearing nucleotide substitution −863A (forward primer, GGACCCCCaCTTAACGAAG; reverse primer, CTTCGTTAAGtGGGGGTCC), along with vector-specific primers HindIII (AATGCCAAGCTTGGAAGAG) as the forward primer and KpnI (TCGATAGGTACCGAGCTCTT) as the reverse primer, as previously described (22). (Lowercase letters indicate polymorphic mutations, and sequences of restriction sites are underlined). Final PCR products were cloned into HindIII/KpnI sites of modified pGL3-basic vector. Two sets of constructs were generated in pAdTrack vector (7): with (pAdTrack-TNF-luc-3′UTR) and without (pAdTrack-TNF-luc) the 3′ untranslated region (UTR) of the human TNF gene. For later ones, KpnI/SalI fragments containing the human TNF promoter, luciferase reporter gene, and simian virus 40 (SV40) late poly(A) signal were derived from TNF −863C or TNF −863A constructs and cloned into KpnI/SalI sites of the pAdTrack vector. 3′ UTR constructs were obtained by substituting the XbaI/BamHI fragment containing the SV40 late poly(A) signal in TNF −863C or TNF −863A plasmids for a 1,041-bp fragment of 3′ UTR amplified by PCR with corresponding primers 3′ UTR-F(XbaI) (aattctagaGGAGGACGAACATCCAAC) and 3′ UTR-R(BamHI) (aatGgATcCCC CAGAGTTGGAAATTC). KpnI/SalI fragments were subsequently cloned into pAdTrack vector. All constructs were verified by DNA sequencing.
Protein extracts and electrophoretic mobility shift assay (EMSA).
The following oligonucleotide probes were radiolabeled with [α-32P]dCTP (Amersham Pharmacia Biotech): κB1−863A (forward primer, agctGAGTATGGGGACCCCCCCTTAA; reverse primer, agctTTAA; reverse primer, agctTTAA GtGGGGGTCCCCATACTC). Mono Mac 6 cells (10 × 106 to 20 × 106) were stimulated with 100 ng of lipopolysaccharide (LPS) per ml for 1 h, and nuclear extracts were prepared as previously described (18). COS-7 cells were transfected with cytomegalovirus (CMV) p50- and CMV p65-expressing constructs, and total protein extracts were prepared by lysing cells in lysis buffer (20 mM Tris-Cl [pH 8.0], 300 mM NaCl, 0.1% NP-40, 10% glycerol) supplemented with protease inhibitors (Boehringer Mannheim). The binding reaction mixture contained 12 mM HEPES [pH 7.8], 80 to 100 mM KCl, 1 mM EDTA, 1 mM EGTA, 12% glycerol, and 0.5 μg of poly(dI-dC) (Amersham Pharmacia Biotech). Protein extracts (1 to 4 μg) were mixed in an 8-μl reaction mixture with 0.2 to 0.5 ng of radiolabeled probe (1 × 104 to 5 × 104 cpm) and incubated at room temperature for 10 min. Where indicated below, a competitive cold probe or corresponding antibodies (all from Santa Cruz) were included with the radiolabeled probe. The reaction was analyzed by electrophoresis in a nondenaturing 5% polyacrylamide gel at 4°C in 0.5× Tris-borate-EDTA buffer. Where indicated, gels were quantified using a PhosphorImager (Molecular Dynamics).
Cell culture, transfections, and luciferase assay.
Mono Mac 6 cells were maintained as previously described (27). COS-7 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, 0.2 mM l-glutamine, and 0.1% glucose. Transient transfections were performed on COS-7 cells with TNF promoter luciferase reporter constructs along with p50- and p65-expressing constructs by using Fugene 6 nonliposomal reagent according to the manufacturer's instructions (Boehringer Mannheim). After transfection, cells were incubated for 24 h prior to harvesting. Luciferase assay was performed using the luciferase assay system (Promega) and the Turner Designs model 20 luminometer (Promega) according to the protocol supplied.
Purification of human monocytes and adenoviral infection.
Mononuclear cells were isolated from single-donor plateletpheresis residues from the North London Blood Transfusion Centre (London, United Kingdom) as described previously (5). Monocytes were treated with macrophage colony-stimulating factor (100 ng/ml) for 48 h. Synovium from rheumatoid patients undergoing joint replacement surgery at the Rheumatology Clinic, Charing Cross Hospital (London, United Kingdom), was dissociated by cutting it into small pieces and digested in medium containing 0.15 mg of DNase I (Sigma, Poole, United Kingdom) per ml and 5 mg of collagenase (Roche, Lewes, United Kingdom) per ml for 1 to 2 h at 37°C. After passing cells through nylon mesh to exclude cell debris, the total cell mixture was cultured at 106 cells/ml as described previously (5). The pAdEasy-1 adenoviral plasmid was provided by B. Vogelstein (The Howard Hughes Medical Institute, Baltimore, Md.). Recombinant viruses were generated in BJ5183 bacterial cells transformed with 1 μg of linearized pAdTrack-TNF-luc-3′UTR or pAdTrack-TNF-luc constructs and 100 ng of pAdEasy-1 vector by the heat shock method. After selection, DNA extracted from recombinant clones was used for virus propagation in 293 human embryonic kidney cells and purified by ultracentrifugation through two cesium chloride gradients essentially as described previously (7). Titers of viral stocks were determined by plaque assay in 293 cells after exposure to virus for 1 h in serum-free DMEM (Gibco BRL) and subsequently overlaid with an agarose-DMEM mixture and incubated for 10 to 14 days. After macrophage colony-stimulating factor treatment, macrophages were exposed to virus for 1 h in serum-free RPMI 1640 medium followed by washing and reculturing in RPMI medium with 2% fetal bovine serum for 48 h. Cells were stimulated with LPS (10 ng/ml) for 4 h and assayed for green Fluorescent protein (GFP) and luciferase production.
RESULTS
Reduction of p50-p50 binding by a single nucleotide polymorphism.
Automated sequencing of the human TNF gene revealed a C-to-A transition at nt −863 relative to the transcription start site. Genotyping of DNA from 200 European and 300 West African adults by PCR with TNF promoter sequence-specific primers gave TNF −863A gene frequencies of 15 and 6%, respectively. The same single nucleotide polymorphism has been identified by others, with estimated gene frequencies of 17% in North Americans of Caucasian origin, 30% in Cambodians, and 14% in the Japanese (8, 26).
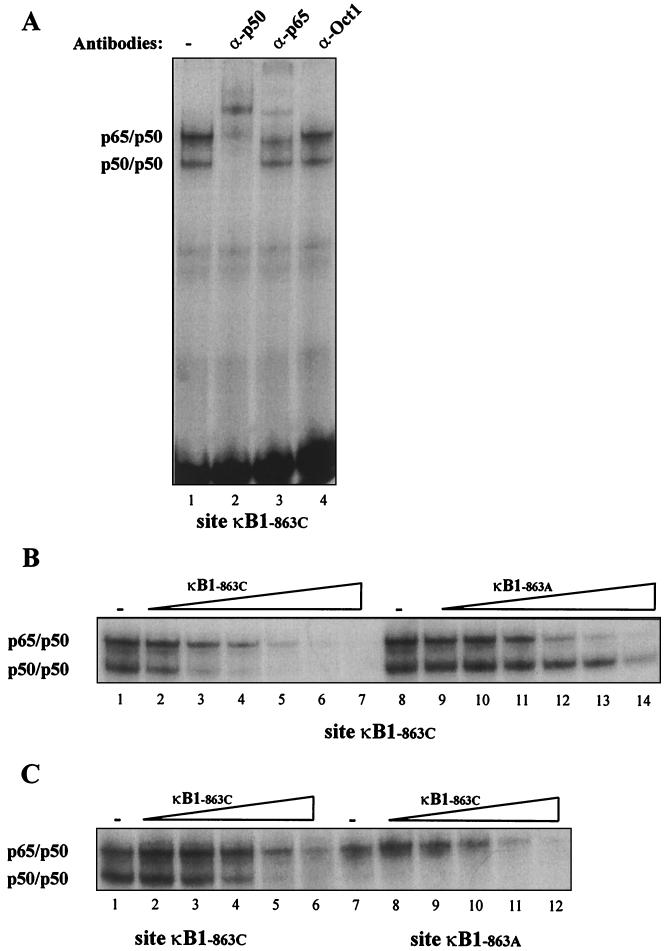
This single nucleotide polymorphism changes the sequence from −873- to −863 from GGGGACCCCCC to GGGGACCCCCA. Both sequences have NF-κB binding features, and it has been previously noted that nuclear extracts from activated human monocytes can form NF-κB complexes with the GGGGACCCCC sequence located at nt −873 of the TNF promoter region (25). To investigate how the polymorphism might affect DNA-protein interactions, we performed EMSA with the oligonucleotide sequence from nt −879 to −858 using nuclear extracts obtained from cells of the human monocyte line Mono Mac 6 after LPS stimulation. The oligoduplex containing −863C formed two complexes that were identified by supershift assay as p65-p50 and p50-p50 (Fig. 1A). In contrast, the oligoduplex containing −863A formed a complex with p65-p50 but not with p50-p50 (Fig. 1C, lanes 1 and 7).
FIG. 1.
Nuclear factors binding to sites κB1−863C and κB1−863A. Nuclear extracts from Mono Mac 6 cells after 1 h of stimulation with LPS were used in an EMSA with a radioactive probe corresponding to site κB1−863C. (A) Supershift experiment using no antibodies (lane 1) or antibodies against p50 (lane 2), p65 (lane 3), or Oct-1 (lane 4). (B) Competition with the same amount (lanes 2 and 9) or a 3-fold excess (lanes 3 and 10), 9-fold excess (lanes 4 and 11), 27-fold excess (lanes 5 and 12), 81-fold excess (lanes 6 and 13), or 243-fold excess (lanes 7 and 14) of unlabeled site κB1−863C or site κB1−863A. The upper complex consists of the p65-p50 heterodimer, and the lower complex consists of the p50-p50 homodimer. (C) Nuclear extracts from Mono Mac 6 cells after 1 h of stimulation with LPS were used in an EMSA with a probe corresponding to sites κB1−863C (lanes 1 through 6) or κB1−863C (lanes 7 through 12). Competition was performed with the same amount (lanes 2 and 8) or a 3-fold excess (lanes 3 and 9), 9-fold excess (lanes 4 and 10), 27-fold excess (lanes 5 and 11), or 81-fold excess (lanes 6 and 12) of unlabeled site κB1−863C.
To compare the abilities of the two different sequences to form NF-κB complexes, we performed competition experiments. The p65-p50 complex formed by the radiolabeled oligoduplex containing −863C was markedly reduced by a 27-fold excess of unlabeled oligoduplex containing either −863C or −863A (Fig. 1B, lanes 5 and 12). Similarly, the p65-p50 complex formed by the radiolabeled oligoduplex containing −863A was markedly reduced by a 27-fold excess of unlabeled oligoduplex containing −863A (Fig. 1C, lane 5). In contrast, the p50-p50 complex formed by the radiolabeled oligoduplex containing −863C was almost abolished by a 9-fold excess of unlabeled oligoduplex containing −863C (Fig. 1B, lane 4) but was only slightly reduced by an 81-fold excess of unlabeled oligonucleotide containing −863A (Fig. 1B, lane 13). These findings confirm that the sequence containing −863A has a markedly reduced ability to bind p50-p50, whereas p65-p50 binding does not differ greatly between the two sequences.
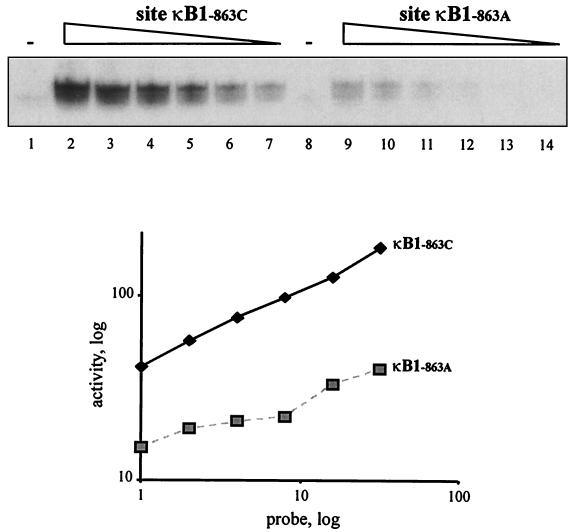
To estimate p50-p50 binding affinity with greater specificity, we obtained p50-enriched protein extracts by transiently expressing a construct containing the p50 gene in COS-7 cells. When these p50-enriched protein extracts were incubated with different concentrations of the oligoduplex probes described above, it was found that at least 30 times the amount of probe containing −863A was required to bind the same amount of p50-p50 as the probe containing −863C (Fig. 2, lanes 7 and 9). We concluded that binding affinity for p50-p50 is reduced more than 10-fold as a result of the single nucleotide polymorphism at −863.
FIG. 2.
Binding affinity of the p50-p50 homodimer to sites κB1−863C and κB1−863A. Standard amounts of protein extracts from COS-7 cells overexpressing p50 protein were used in an EMSA with radioactive probes (diluted as described below) corresponding to site κB1−863C (lanes 1 through 7) or κB1−863A (lanes 8 through 14). Lanes 2 and 9, nondiluted probe; lanes 3 and 10, 1:2 dilution; lanes 4 and 11, 1:4 dilution; lanes 5 and 12, 1:8 dilution; lanes 6 and 13, 1:16 dilution; lanes 7 and 14, 1:32 dilution. Lanes 1 and 8 show nondiluted probes incubated with extracts from mock-transfected cells. The graph represents quantitative analysis of the autoradiograms.
Effect on gene expression depends on NF-κB dimer ratio.
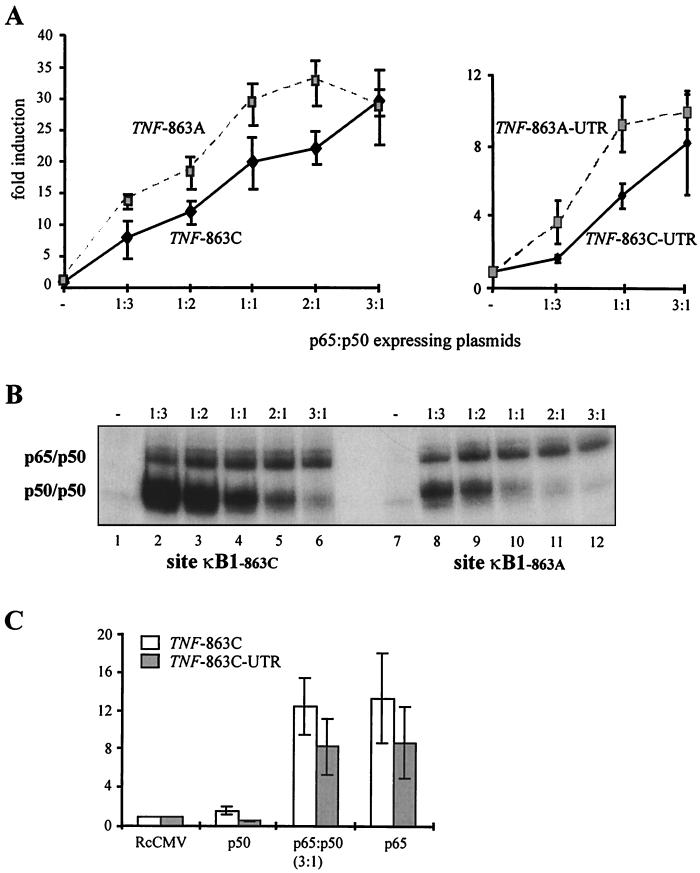
The above findings suggested that the effect of this polymorphism on gene expression might depend on the types of NF-κB dimer that are present. To test this hypothesis we cotransfected COS-7 cells with p65- and p50-expressing plasmids plus a reporter construct containing the human TNF promoter linked to a luciferase reporter gene, with or without a downstream segment of the TNF 3′ UTR. Each experiment compared reporter gene expression for the −863C and −863A forms of the TNF promoter region, using different ratios of p65- and p50-expressing plasmids but keeping the total amount of plasmid constant. As shown in Fig. 3A, reporter gene activity increased with the amount of p65-expressing plasmid. When the amount of p65-expressing plasmid exceeded the amount of p50-expressing plasmid threefold, the two forms of the TNF promoter gave similar levels of reporter gene expression, but at lower ratios the −863A promoter variant gave higher levels of reporter gene expression than the −863C promoter variant. For a p65-to-p50 plasmid ratio of 1:1, this difference between the two allelic forms was statistically significant in independent sets of experiments with the 3′ UTR (n = 3, P < 0.05 by paired t test) and without the 3′ UTR (n = 6, P < 0.05).
FIG. 3.
Effect of the −863A polymorphism on NF-κB-dependent TNF promoter activity. (A) TNF promoter constructs (TNF −863C or TNF −863A, left panel; TNF −863C-3′UTR or TNF −863A-3′UTR, right panel) were expressed in COS-7 cells along with different ratios of CMV -p65- to CMV p50-expressing constructs. Results are shown as means and standard errors of six (left panel) and three (right panel) independent experiments. (B) Protein extracts from COS-7 cells overexpressing p65 and p50 proteins were used in an EMSA with radioactive probes corresponding to site κB1−863C (lanes 1 through 6) or κB1−863A (lanes 7 through 12). Lanes 2 and 8, ratio of CMV p65 to CMV p50 plasmids of 1:3; lanes 3 and 9, ratio of 1:2; lanes 4 and 10, ratio of 1:1; lanes 5 and 11, ratio of 2:1; lanes 6 and 12, ratio of 3:1. Lanes 1 and 7 show probes incubated with extracts from mock-transfected cells. The composition of the complexes was confirmed by supershift analysis (data not shown). (C) TNF −863C promoter constructs (with and without the 3′ UTR) were expressed in COS-7 cells with CMV p50 alone, with CMV p65 alone, or with a mixture of the two plasmids in a ratio of CMV p65 to CMV p50 of 3:1. RcCMV, cells transfected with an empty expression vector. Results are shown as means and standard errors of three independent experiments.
We sought to relate the allelic differences in transcriptional activation to levels of p65-p50 and p50-p50 binding. Protein extracts from p65- and p50-transfected COS-7 cells were analyzed by EMSA using the radiolabeled oligoduplexes described for Fig. 1. As shown in Fig. 3B, the amount of p65-p50 binding to both −863C and −863A oligoduplexes was relatively constant across the range of plasmid concentrations used in experiments described above. For the −863C oligoduplex, p50-p50 binding equaled or exceeded p65-p50 binding for most plasmid concentrations, although p50-p50 disappeared when the p65 plasmid was in a threefold excess. The −863A oligoduplex bound much less p50-p50 than the −863C oligoduplex at all plasmid concentrations. Taken with the reporter gene data, these results indicate that the −863A polymorphism acts to increase gene expression under conditions in which a significant amount of p50-p50 is present.
A question raised by these observations is how the transcriptional activity of p50-p50 alone compares with that of p65-p50 or p65-p65 in this experimental system. We therefore conducted reporter gene experiments using the TNF −863C allele, in which p65-p50 (based on the data in Fig. 3B we used a p65-to-p50 plasmid ratio of 3:1) was compared with p50-p50 or p65-p65 (i.e., using p50 or p65 alone but keeping the total plasmid amount constant). As shown in Fig. 3C, p65-p65 was found to have transcriptional activity similar to that of p65-p50, while p50-p50 entirely lacked activity (Fig. 3C).
Adenovirus-based reporter analysis in primary human monocytes.
Previously reported investigations of the effects on the TNF −863 polymorphism in different systems have yielded conflicting results; these include measurements of TNF levels in human plasma, TNF production by peripheral blood mononuclear cells, and reporter gene expression by cell lines of lymphocytic or hepatic origin (8, 21, 26). At least two biological variables might explain such differences: firstly, the data in Fig. 3 indicate that the functional effects of the TNF −863 polymorphism depend on the ratio of p65-p50 to p50-p50, which is likely to vary with stimulus, time, and cell type; secondly, this polymorphism will undoubtedly be in linkage disequilibrium with neighboring polymorphisms, which may confound comparisons between individuals of different TNF −863 genotypes. We therefore sought to examine the effect of the polymorphism in the specific context of LPS-stimulated peripheral blood monocytes, using a reporter assay method to avoid the confounding effects of other genetic factors. By directly comparing the two alleles in the same host cell preparation, reporter assays also remove the confounding effect of experimental variability arising from the process of monocyte purification. Since primary human monocytes are refractory to conventional transfection techniques, we adapted an adenoviral delivery system (5, 7) to analyze TNF promoter function in these cells.
We generated a set of recombinant viruses in which a luciferase reporter gene was placed immediately downstream of the TNF promoter region, containing either −863C or −863A, and immediately upstream of the TNF 3′ UTR. The gene for GFP, located downstream of a CMV promoter in a separate part of the construct, was used to normalize the luciferase reporter data. Monocytes were infected with recombinant virus for 48 h prior to LPS stimulation.
The results of independent experiments performed in duplicate on elutriated monocytes from unrelated donors are summarized in Table 1. In unstimulated monocytes, all constructs gave low levels of reporter gene expression, with no significant difference between the TNF −863C and TNF −863A promoters. Following LPS stimulation, the level of reporter gene expression increased by about a factor of 80 for the TNF −863A promoter, compared to about a factor of 20 for the TNF −863C promoter.
TABLE 1.
Effect of −863A polymorphism on TNF promoter activity in human monocytesa
| Promoter construct | Luciferase activity
|
|||
|---|---|---|---|---|
| NA | LPS | GFP normalized
|
||
| NA | LPS | |||
| With 3′ UTR | ||||
| TNF −863C | 17 (3–85) | 693 (435–1,107) | 22 (5–96) | 868 (485–1,554) |
| TNF −863A | 52 (35–73) | 4,632 (3,289–6,523) | 39 (22–70) | 3,508 (2,226–5,528) |
| Without 3′ UTR | ||||
| TNF −863C | 31 (16–61) | 2,910 (1,698–4,987) | 43 (20–91) | 3,896 (1,612–9,418) |
| TNF −863A | 28 (16–47) | 2,590 (1,748–3,835) | 43 (23–81) | 4,036 (1,982–8,220) |
Purified human monocytes were infected with adenoviral particles carrying variants of TNF promoters and luciferase reporter genes. Luciferase activity, in arbitrary units, was normalized by the protein concentration of cellular extract and, where indicated, by GFP activity. Results are shown as geometric means (with 95% confidence intervals) of duplicate estimates in three independent experiments. NA, nonactivated cells; LPS, cells activated with 10 ng of LPS per ml.
The TNF 3′ UTR contains a UA-rich sequence that acts to destabilize mRNA and thereby limits gene expression. To investigate functional interactions between the promoter and the 3′ UTR, we also tested reporter constructs containing the same TNF promoter sequences but lacking the TNF 3′ UTR, which was replaced by the SV40 late poly(A) signal. As expected these constructs gave a significantly higher level of inducible gene expression. However, the difference between TNF −863C and TNF −863A promoter variants was not seen in constructs lacking the TNF 3′ UTR, suggesting that the functional manifestations of this polymorphism may depend on a high rate of mRNA turnover. A similar result was obtained when recombinant adenovirus was used to introduce TNF promoter 3′ UTR constructs into primary macrophages from inflamed synovial tissue obtained from a patient undergoing knee replacement for osteoarthritis. The level of luciferase expression for the TNF −863A allele was 2,127 ± 126 U (mean ± standard deviation of triplicate measurements), compared to 492 ± 65 U for the TNF −863C allele. In these synovial macrophages, stimulation in vitro with LPS caused no significant increment in gene expression (TNF −863A, 1,983 ± 92 U; TNF −863C, 482 ± 28 U).
DISCUSSION
Previous studies of this polymorphism in vivo have yielded a confusing picture. The TNF −863A allele has been reported to be associated with elevated TNF production by peripheral blood mononuclear cells stimulated with concanavalin A (8), while others have reported that this allele is associated with a lowering of resting plasma TNF levels (21), but such data must be interpreted with caution because of the potential confounding effect of other strongly linked polymorphisms within the TNF promoter region (8). In reporter gene analysis of T- and B-cell lines, the TNF −863A polymorphism has been reported to have no detectable effect (26), while it has been associated with a reduction of basal reporter gene expression in a hepatoblastoma cell line (21). We have no simple explanation for these differences, except that we would expect the functional effects of the TNF −863A allele to be highly context dependent, depending on the exact proportion of the p65-p50 and p50-p50 complexes within a specific cell type and possibly on other cell-specific factors with which they may interact.
In this study we have used a COS cell model to reduce the complexity of the system and address the question of how differences in p65-p50 and p50-p50 binding affinity may affect transcriptional regulation in the absence of other confounding factors. Our findings are consistent with a growing body of evidence that p50-p50 may exert inhibitory effects on transcriptional activation. Schmitz and Baeuerle (17) observed that the transactivating effects of p65 at a canonical NF-κB domain can be suppressed by overexpression of p50, and they postulated that certain κB motifs may be more susceptible to negative regulation by p50-p50 than others. Overexpression of p50 blocked LPS-induced transcription from a TNF promoter reporter construct, showing that this transcription factor is an inhibitor of the TNF gene (1). It has also been observed that expression of beta interferon, which is transcriptionally regulated by NF-κB, is increased in mice with disruptions of the p50 gene (19), while in mice with elevated constitutive levels of p50 due to p105 gene disruption, NF-κB-regulated cytokine production is increased in some cell types and suppressed in others (9). One possible explanation for the inhibitory effect is that p50-p50 reduces transcriptional activation by competing for binding with p65-p50, since p65 carries a transactivating domain and p50 does not. An alternative explanation is that p50-p50 interacts with a transcriptional repressor, as suggested by the observation that Drosophila dorsal switch protein 1 converts NF-κB from a transcriptional activator to a repressor only in the presence of p50 (14). As for the mechanism that operates in our experimental system, two observations may be relevant. Firstly, p50-p50 alone fails to induce gene expression, excluding the possibility that it is simply a weaker stimulator than p65-p50. Secondly, if the inhibitory effect of p50-p50 was primarily due to binding competition with p65-p50, then we might expect the TNF −863A allele to show increased p65-p50 binding, but this was not evident in these experiments. These observations raise the possibility that p50-p50 may actively repress transactivation by p65-p50, but further work is needed to resolve this issue with confidence.
The structural reasons why the TNF −863A allele reduces p50-p50 binding more than 10-fold, but has little effect on p65-p50 binding, may be complex. Optimal binding sequences for p65-p50 and p50-p50 are known to be similar but not identical (12, 15, 23). The sequence from nt −872 to −863 sequence matches an optimal p65-p50 binding motif but has three mismatches with an optimal p50-p50 motif, and the variant allele adds an extra mismatch for both motifs. The sequence from nt −873 to −864 matches an optimal p65-p50 motif and has one mismatch with an optimal p50-p50 motif. Although the variant allele is just outside the latter sequence, it may be functionally relevant in view of crystallographic data suggesting that interactions with flanking nucleotides are more critical for p50-p50 than for p65-p50 (3). It is also conceivable that the variant allele reduces p50-p50 binding by altering other protein interactions in this region. In general terms, the effects of this polymorphism are consistent with the structural observation that p50-p50 has more stringent binding requirements than p65-p50.
We have described an adenovirus-based method for introducing promoter-reporter gene constructs into primary human cells. One advantage of this method is that it permits examination of allelic effects on a specific cell type—such as purified monocytes from peripheral blood or synovial macrophages from a diseased joint—free of the experimental and biological noise that invariably accompanies comparisons of purified primary cell fractions from individuals of different genotypes. In this model we found that the TNF −863A allele results in three- to fourfold increases in LPS-induced gene expression in primary human monocytes, and although the data are preliminary, there seems to be a comparable effect on basal gene expression in synovial macrophages from a diseased joint. Intriguingly, in both of these primary cell types the allelic difference depends on the presence of the TNF 3′ UTR, whereas in COS-7 cells it is independent of the 3′ UTR. This is not the first TNF promoter polymorphism whose functional effect in cells of the macrophage lineage has been reported to depend on the 3′ UTR (11). We do not currently understand why this is so. It might reflect a functional interaction between 5′ and 3′ enhancer elements; alternatively, since the TNF 3′ UTR contains a UA-rich motif that destabilizes mRNA and may suppress translation (6, 20), it is conceivable that subtle effects of a promoter variant on the transcription rate may be evident only under conditions of rapid mRNA turnover. Whatever the explanation, this result highlights the importance of taking all components of gene regulation into consideration when designing experimental systems for functional analysis.
Regulation of TNF is of clinical importance because of its potentially damaging proinflammatory effects. The TNF response to LPS in human monocytes is remarkably transient, with a significant amount of p65-p50 in the nucleus during the initial phase of this response, while TNF mRNA levels fall as the amount of p50-p50 increases (10). These observations raise the possibility that a site which binds both p65-p50 and p50-p50 might function as a transcriptional activator in the initial phase but as a repressor in the later phase of the response. Such regulatory processes might be of considerable importance in an inflammatory disease such as rheumatoid arthritis, where there is strong evidence that TNF has a causal role in pathogenesis (4) and that NF-κB is critical for TNF production by synovial macrophages taken from diseased joints (5). Our present data would suggest that individuals with the TNF −863A allele might have increased susceptibility to severe rheumatoid arthritis, and this is supported by preliminary case control data for United Kingdom patients with accelerated erosive joint disease (I. A. Udalova et al., unpublished data). More detailed genetic investigation of the TNF −863 polymorphism in different infectious and inflammatory diseases may help to resolve its functional significance and the evolutionary question of whether its frequency of 27% in Europeans, compared to 12% in Africans, is the result of a specific selection pressure.
ACKNOWLEDGMENTS
We thank Vincent Vidal and Meike Hensmann for technical help and critical comments and Scott Silverman for assistance with manuscript preparation.
This work was supported by the Medical Research Council (I.A.U., A.R., and D.K.) and by the Arthritis Research Campaign (C.S. and B.F.) A.D. was supported by an EU Training and Mobility of Researchers Award.
REFERENCES
- 1.Baer M, Dillner A, Schwartz R C, Sedon C, Nedospasov S, Johnson P F. Tumor necrosis factor alpha transcription in macrophages is attenuated by an autocrine factor that preferentially induces NF-κB p50. Mol Cell Biol. 1998;18:5678–5689. doi: 10.1128/mcb.18.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin A S., Jr The NF-κB and IB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Chen F E, Huang D B, Chen Y Q, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 4.Elliott M J, Maini R N, Feldmann M, Long-Fox A, Charles P, Bijl H, Woody J N. Repeated therapy with monoclonal antibody to tumour necrosis factor alpha (cA2) in patients with rheumatoid arthritis. Lancet. 1994;344:1125–1127. doi: 10.1016/s0140-6736(94)90632-7. [DOI] [PubMed] [Google Scholar]
- 5.Foxwell B, Browne K, Bondeson J, Clarke C, de Martin R, Brennan F, Feldmann M. Efficient adenoviral infection with IB alpha reveals that macrophage tumor necrosis factor alpha production in rheumatoid arthritis is NF-κB dependent. Proc Natl Acad Sci USA. 1998;95:8211–8215. doi: 10.1073/pnas.95.14.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachetin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171:465–475. doi: 10.1084/jem.171.2.465. . (Erratum, 171:971–972.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, Itoh K. Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens. 1998;51:605–612. doi: 10.1111/j.1399-0039.1998.tb03002.x. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa H, Claudio E, Dambach D, Raventos-Suarez C, Ryan C, Bravo R. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p)105 precursor (NF-κB1) but expressing p50. J Exp Med. 1998;187:985–996. doi: 10.1084/jem.187.7.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastenbauer S, Ziegler-Heitbrock H W L. NF-κB1 (p50) is upregulated in lipopolysaccharide tolerance and can block tumor necrosis factor gene expression. Infect Immun. 1999;67:1553–1559. doi: 10.1128/iai.67.4.1553-1559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroeger K M, Carville K S, Abraham L J. The −308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol. 1997;34:391–399. doi: 10.1016/s0161-5890(97)00052-7. [DOI] [PubMed] [Google Scholar]
- 12.Kunsch C, Ruben S M, Rosen C A. Selection of optimal κB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuprash D V, Udalova I A, Turetskaya R L, Rice N R, Nedospasov S A. Conserved kappa B element located downstream of the tumor necrosis factor alpha gene: distinct NF-κB binding pattern and enhancer activity in LPS activated murine macrophages. Oncogene. 1995;11:97–106. [PubMed] [Google Scholar]
- 14.Lehming N, Thanos D, Brickman J M, Ma J, Maniatis T, Ptashne M. An HMG-like protein that can switch a transcriptional activator to a repressor. Nature. 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 15.Perkins N D, Schmid R M, Duckett C S, Leung K, Rice N R, Nabel G J. Distinct combinations of NF-κB subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci USA. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaksin D, Baeuerle P A, Eisenbach L. KBF1 (p50 NF-κB homodimer) acts as a repressor of H-2Kb gene expression in metastatic tumor cells. J Exp Med. 1993;177:1651–1662. doi: 10.1084/jem.177.6.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz M L, Baeuerle P A. The p65 subunit is responsible for the strong transcription activating potential of NF-κB. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sha W C, Liou H C, Tuomanen E I, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 20.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 21.Skoog T, Hooft F M, Kallin B, Jovinge S, Boquist S, Nilsson J, Eriksson P, Hamsten A. A common functional polymorphism (C→A substitution at position −863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. 1999;8:1443–1449. doi: 10.1093/hmg/8.8.1443. [DOI] [PubMed] [Google Scholar]
- 22.Stuber F, Udalova I A, Book M, Drutskaya L N, Kuprash D V, Turetskaya R L, Schade F U, Nedospasov S A. −308 tumor necrosis factor (TNF) polymorphism is not associated with survival in severe sepsis and is unrelated to lipopolysaccharide inducibility of the human TNF promoter. J Inflamm. 1995;46:42–50. [PubMed] [Google Scholar]
- 23.Thanos D, Maniatis T. Identification of the rel family members required for virus induction of the human beta interferon gene. Mol Cell Biol. 1995;15:152–164. doi: 10.1128/mcb.15.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 25.Udalova I A, Knight J C, Vidal V, Nedospasov S A, Kwiatkowski D. Complex NF-κB interactions at the distal tumor necrosis factor promoter region in human monocytes. J Biol Chem. 1998;273:21178–21186. doi: 10.1074/jbc.273.33.21178. [DOI] [PubMed] [Google Scholar]
- 26.Uglialoro A M, Turbay D, Pesavento P A, Delgado J C, McKenzie F E, Gribben J G, Hartl D, Yunis E J, Goldfeld A E. Identification of three new single nucleotide polymorphisms in the human tumor necrosis factor-alpha gene promoter. Tissue Antigens. 1998;52:359–367. doi: 10.1111/j.1399-0039.1998.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler-Heitbrock H W, Thiel E, Futterer A, Herzog V, Wirtz A, Riethmuller G. Establishment of a human cell line (Mono Mac 6) with characteristics of mature monocytes. Int J Cancer. 1988;41:456–461. doi: 10.1002/ijc.2910410324. [DOI] [PubMed] [Google Scholar]