Abstract
A mutation in the Arabidopsis gene STARIK leads to dwarfism and chlorosis of plants with an altered morphology of leaf and cell nuclei. We show that the STARIK gene encodes the mitochondrial ABC transporter Sta1 that belongs to a subfamily of Arabidopsis half-ABC transporters. The severity of the starik phenotype is suppressed by the ectopic expression of the STA2 homolog; thus, Sta1 function is partially redundant. Sta1 supports the maturation of cytosolic Fe/S protein in Δatm1 yeast, substituting for the ABC transporter Atm1p. Similar to Atm1p-deficient yeast, mitochondria of the starik mutant accumulated more nonheme, nonprotein iron than did wild-type organelles. We further show that plant mitochondria contain a putative l-cysteine desulfurase. Taken together, our results suggest that plant mitochondria possess an evolutionarily conserved Fe/S cluster biosynthesis pathway, which is linked to the intracellular iron homeostasis by the function of Atm1p-like ABC transporters.
INTRODUCTION
ABC transporters constitute one of the largest protein families with diverse functions in membrane transport. The designation of ABC transporter recognizes a highly conserved ATP binding cassette, which is the most characteristic feature of this superfamily. These proteins mediate the relatively specific, active transmembrane transport of molecules that can range in size from small ions to proteins (reviewed in Higgins, 1992) and utilize the energy of ATP hydrolysis to pump substrates across membranes, usually against a concentration gradient. The typical ABC transporter consists of four domains. Two of these domains are hydrophobic, and each comprises six membrane-spanning segments. The transmembrane domains are believed to form a channel and to determine the specificity of the transporter. The other two domains are located at the periphery of the membrane and couple ATP hydrolysis to the transport process.
The ABC transporter Atm1p of budding yeast mitochondria represents a “half-transporter” in which one transmembrane and one ATP binding domain are expressed in a single polypeptide (Leighton and Schatz, 1995). Atm1p is localized in the mitochondrial inner membrane and is believed to function as an exporter, because its ABC domains face the mitochondrial matrix. The ATM1 loss-of-function results in respiration-deficient mitochondria that lack cytochromes (Leighton and Schatz, 1995; Kispal et al., 1997) and that tend to loose their DNA (Leighton and Schatz, 1995). These pleiotropic effects of the Atm1p deficiency are generally attributed to the fact that in Δatm1 cells, mitochondria accumulate up to 30-fold higher levels of iron than do wild-type organelles (Kispal et al., 1997; Mitsuhashi et al., 2000). The Atm1p transport function appears to be conserved in eukaryotes, and the human mitochondrial ABC transporters Abc7 and MTABC3 have been shown to be true functional orthologs of the Atm1p (Csere et al., 1998; Mitsuhashi et al., 2000). The human ABC7 gene has been implicated in hereditary X-linked sideroblastic anemia and ataxia, an iron storage disease in which cells contain high deposits of nonheme iron within their mitochondria (Allikmets et al., 1999). The human MTABC3 gene is a candidate gene for lethal neonatal metabolic syndrome, a disorder of the mitochondrial function associated with iron metabolism (Mitsuhashi et al., 2000).
The substrates for Atm1p are unknown, although a function of Atm1p in the maturation of cytosolic Fe/S proteins has been recently demonstrated (Kispal et al., 1999). Fe/S clusters are prosthetic groups whose chemical characteristics define the redox, catalytic, and regulatory properties of many proteins (Beinert et al., 1997). In budding yeast, a complex machinery, composed of some 10 mitochondrial proteins, mediates Fe/S biogenesis (reviewed in Lill and Kispal, 2000) and resembles the well-characterized prokaryotic iron-sulfur-cluster pathway from Azotobacter vinelandii (Zheng et al., 1998). This process can be divided into three steps: formation of elemental sulfur, assembly of sulfur and iron into a cluster, and insertion of the clusters into apoproteins. The functions of Nfs1p, Isu1p and Isu2p, Yah1p, and Jac1p are all essential for yeast. These recent findings demonstrate that in yeast, (i) mitochondria are the primary site of Fe/S cluster formation, and (ii) mitochondria not only produce their own set of Fe/S proteins but, importantly, are also required for the maturation of cytosolic Fe/S proteins.
Here, we describe an Arabidopsis mutant starik (Russian for “old man”) that is deficient in the function of the mitochondrial ABC transporter Sta1. Complementation experiments in yeast indicate that Sta1 is a functional ortholog of the Atm1p. These results emphasize the importance of ABC transporters for the integration of mitochondria within plant cellular iron homeostasis.
RESULTS
The Arabidopsis Mutant starik Has an Altered Leaf Morphology and Structural Changes in Mesophyll Cell Nuclei
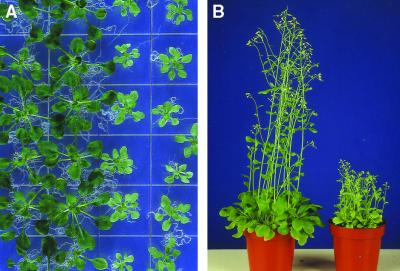
The starik mutation segregated out in the progeny of the Arabidopsis exon–trap line SK3-3 (Babiychuk et al., 1997). This recessive, Mendelian mutation results in dwarfism and chlorosis of plants. The degree of chlorosis is positively correlated with light intensity and depends on growth conditions. As presented in Figure 1A, the most severe phenotype was observed when plants were grown in vitro on synthetic media. Nevertheless, mutant plants were photoautotrophic and fertile, and they set fully viable seeds (Figure 1B). Figure 2 shows that leaves of the mutant plants were thicker, that they had enlarged cells with more air space in between them, and that the palisade and the spongy parenchyma in mutant leaves could not be clearly distinguished (Figures 2A to 2C and 2E).
Figure 1.
Chlorotic Phenotype and Stunted Growth Tendency of the starik Mutant.
(A) Three-week-old Arabidopsis plants grown in vitro on synthetic media; starik plants are in the two rows at right.
(B) starik plants (right) at the early flowering stage; for comparison, at left are wild-type Arabidopsis Col5 plants of the same age.
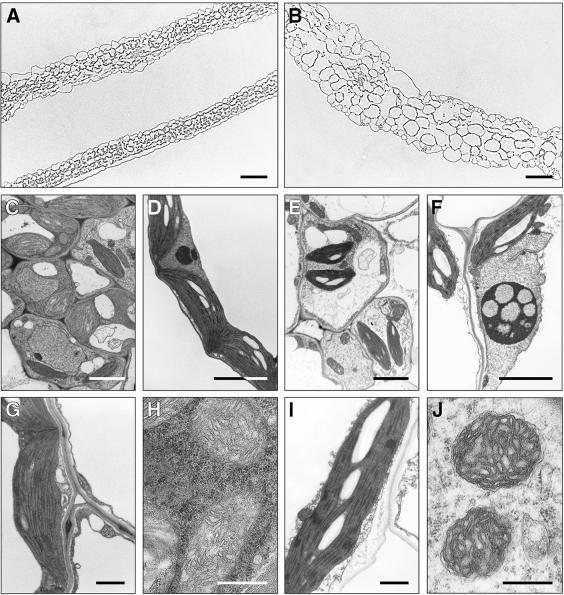
Figure 2.
Cellular Phenotype of the starik Mutant.
(A) Typical cross-section of wild-type leaves.
(B) Cross-section of a starik mutant leaf shown at the same magnification as in (A). Leaves are thicker and cells are larger in sta1.
(C) Cells near vascular bundles in the wild type as seen by transmission electron microscopy.
(D) Part of a wild-type mesophyll cell, including a wild-type nucleus.
(E) Cells near vascular bundles of the starik mutant.
(F) A typical nucleus in mesophyll cells of the starik mutant. Nuclei are larger, often lobe shaped, and develop large nucleoli with electron-transparent, vacuoli-like inclusions (cf. with [D]).
(G) Wild-type chloroplast.
(H) Wild-type mitochondria.
(I) Chloroplast from sta1 cells. These organelles have starch granules and a developed thylakoid membrane system similar to that of wild-type chloroplasts.
(J) Mitochondria observed in sta1 cells. Development, size, and shape are similar to those of wild-type mitochondria.
 ;
;  ;
; 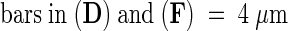 ;
; 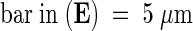 ;
; 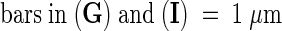 ;
; 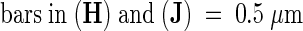 .
.
Transmission electron microscopy was used to visualize organelle development in sta1 cells. The starik mutation had no significant effect on plastid differentiation, and chloroplasts in sta1 cells had extensively developed thylakoid membranes stacked into granas (Figures 2G and 2I). Furthermore, the internal membrane development in mitochondria appeared normal (Figures 2H and 2J). Rather, the most prominent change in mutant cells was found in the nuclei. In starik, the nuclei in many leaf cells both were larger and contained nucleoli with typical vacuole-like electron-transparent inclusions (cf. that of the wild type and a mutant nucleus in Figures 2D and 2F, respectively).
The STARIK Gene Encodes a Mitochondrial ABC Transporter
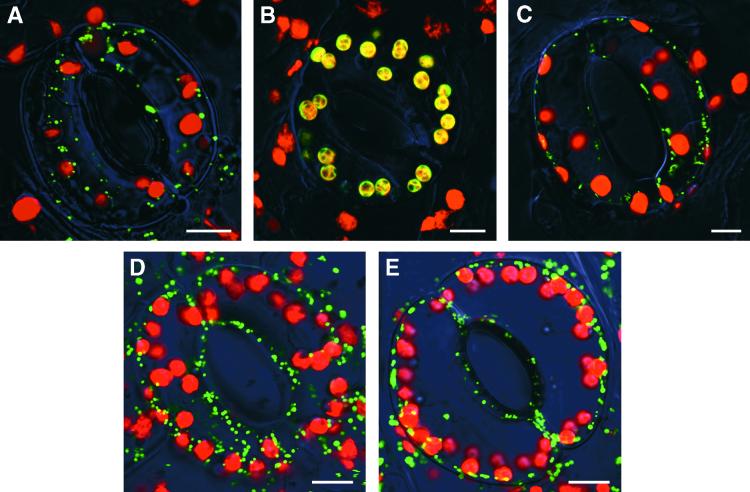
The starik mutation is a T-DNA insertion allele. The 35S promoter-driven expression of the complete STA1 cDNA-coding sequence isolated in this study (EMBL accession number AJ272202) was sufficient to reverse the sta1 mutant phenotype to wild type in a genetic complementation experiment. Overexpression of STA1 had no obvious effects on plants grown under standard conditions (data not shown). The STA1 gene has been sequenced within the Arabidopsis genome sequencing projects. The gene-coding region is split into 18 exons, as found by comparing genomic sequences (GenBank accession number AB019228 of the P1 clone MCK7, chromosome 5) with our cDNA sequence. The Sta1 polypeptide encompasses 728 amino acid residues and is 51.9 and 56.6% identical with the mitochondrial ABC transporter Atm1p from yeast and ABC7 from human cells, respectively. The homology between these proteins extends along the entire sequence, and respective transmembrane and ABC domains could be recognized in Sta1 (data not shown). The N-terminal sequence of the Sta1 polypeptide resembled mitochondrial targeting signals. To verify this conclusion, we prepared a translational gene fusion between the first 104 N-terminal amino acid residues of the Sta1 and the green fluorescent protein (GFP). Figure 3 illustrates that Sta11–104-GFP was localized exclusively in mitochondria. Thus, the STARIK gene encodes a mitochondrial half-ABC transporter.
Figure 3.
Intracellular Targeting of the GFP Fusion Proteins in Tobacco Cells.
(A) The N-terminal 104–amino acid residue of Sta1, Sta11–104-GFP.
(B) Residues 1 to 56 of the pea small subunit of ribulose-1,5-bisphosphate carboxylase, RbcS1–56-GFP. Because both red fluorescence signal from chlorophyll and green fluorescence signal from RbcS1–56-GFP are emitted from chloroplasts, they appear as yellowish green.
(C) The mitochondrial targeting peptide (residues 1 to 31) of the tobacco Mn-superoxide dismutase, MnSOD1–31-GFP.
(D) Residues 1 to 80 of Sta2, Sta21–80-GFP.
(E) The N-terminal 54–amino acid residue of the Arabidopsis Nfs1p homolog, Nfs1–54-GFP.
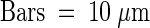 .
.
The sta1 mutant allele was generated with an exon trap T-DNA. Mutant cells express the translational fusion between Sta11–486 and neomycin phosphotransferase. Such a chimeric protein lacks the entire ATP binding cassette of Sta1, but in principle it could heterodimerize with homologous half-transporters (see below). However, in ABC transporters, both ATP binding domains are required for function (Higgins, 1992); therefore, the mutant holo-transporter(s) is (are) probably not functional.
Sta1 Transport Function Is Partially Redundant
Two genes encoding proteins with high similarity to Sta1 (on average, 83% similarity at the amino acid level) were identified by screening Arabidopsis genomic sequences. These genes were provisionally designated STA2 and STA3. STA2 and STA3 genes are positioned in tandem on chromosome 4 (Arabidopsis clone ATT5F17; GenBank accession number AL049917) and therefore might represent a gene duplication event. Unlike STA1, both STA2 and STA3 genes have a single intron. The presence of STA3-derived expressed sequence tags in a database and the detection of STA2 transcript by RNA gel blot hybridization indicated that both genes are expressed; in addition, our polymerase chain reaction (PCR) analysis showed that the intron was correctly spliced out in the STA2 mRNA (data not shown).
The fluorescent image of transgenic tobacco cells presented in Figure 3D shows that the Sta21–80-GFP translational fusion is targeted into mitochondria. To verify that the Sta1 function could be redundant and that the starik phenotype might develop because of tissue specificity in gene expression, we transformed the starik mutant with a chimeric gene cassette in which the STA2 cDNA was placed under the control of the 35S promoter. The sta1(35S::STA2) plants did not revert to wild type, but Figure 4 shows that they grew more vigorously and that they had larger leaves that were obviously not chlorotic and siliques that were more elongated. These results suggest that Sta2 and maybe Sta3 ABC transporters have a function similar to that of Sta1.
Figure 4.
Partial Complementation of the starik Mutant by Overexpression of Sta2.
(A) Plants at the rosette stage. The full-length cDNA of STA2 was placed under the control of the 35S promoter, and the resulting expression cassette was introduced into the sta1 plants. The starik mutant phenotype was assessed in a progeny of 12 independent transformants. Wild type (C24), starik mutant (sta1), and progeny from two independent lines of the starik mutant transformed with the 35S::STA2 gene expression cassette were planted together as indicated. Yellow dots mark sta1 mutant plants segregating out in the progeny of line STA2-9. Transgenic plants are larger than the mutant and show little chlorosis, yet they differ from the wild type in size and are lighter green.
(B) Plants at flowering stage. Shown are the same plants as in (A) but 2 weeks later. Transgenic plants never reached the height of the wild-type plants, but they were significantly more vigorous than the mutant.
(C) Fully elongated siliques. Three siliques collected from wild-type (left) and starik mutant (right) plants are compared with siliques collected from three independent transgenic lines of sta1 (35S::STA2) plants (middle). 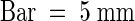 .
.
Sta1 Is a Functional Ortholog of the Yeast Mitochondrial ABC Transporter Atm1p
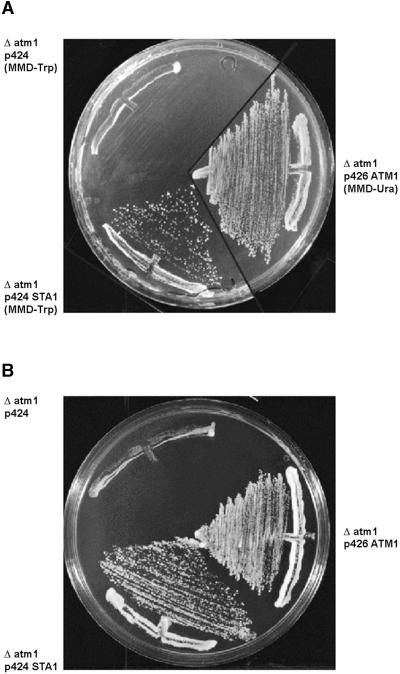
The Sta1 sequence and the mitochondrial localization of the Sta11–104-GFP fusion protein indicated that the Sta1 protein might be a functional ortholog of yeast Atm1p. Mutant yeast strains lacking Atm1p are not viable on media containing nonfermentable carbon sources and grow poorly on media containing fermentable carbon sources (Leighton and Schatz, 1995; Kispal et al., 1997). To test the functional replacement of Atm1p by Sta1, we tested the influence of the plant protein on the growth of Δatm1 yeast cells. As shown in Figure 5, expression of STA1 restored growth of Δatm1 cells on glucose-containing minimal and rich media to the same extent as did the expression of the yeast ATM1, indicating that both proteins performed a similar function.
Figure 5.
Effects of STA1 Expression on the Growth of Δatm1 Yeast.
(A) Cells grown on agar plates with minimal medium containing glucose for 3 days at 30°C. The full-length sta1 cDNA or the yeast ATM1 gene was inserted into the multicopy yeast expression vector pRS424-GPD (Mumberg et al., 1995). Δatm1 cells were transformed with the plasmids or the vector without DNA insert. To apply selective pressure for maintenance of the vectors in yeast cells, minimal medium was depleted of either tryptophan (-Trp) or uracil (-Ura).
(B) Cells grown on YPD medium (1% yeast extract, 2% bactopeptone, 2% glucose, and 1.5% agar) under the same conditions as given for (A).
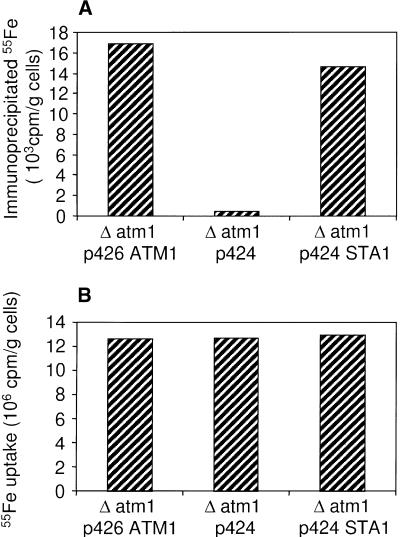
To verify this conclusion in more detail, we investigated the potential of Sta1 to mediate the maturation of cytosolic Fe/S proteins by measuring the de novo assembly of an Fe/S cluster into cytosolic isopropyl malate isomerase, Leu1p (Kispal et al., 1999). Cells were radiolabeled with 55Fe for 1 hr, and a cell extract was prepared. The incorporation of an Fe/S cluster into Leu1p was estimated from the radioactive iron that was coimmunoprecipitated with anti-Leu1p-specific antibodies. Figure 6 summarizes the experimental data. In the presence of Sta1 or Atm1p, similar amounts of radioactive iron were coimmunoprecipitated, indicating that Sta1 was as efficient as Atm1p in promoting the maturation of cytosolic Leu1p (Figure 6A). In contrast, hardly any radioactivity was detected in extracts of Δatm1 cells. The amount of radioactive iron in the three cell extracts was similar (Figure 6B), excluding the possibility that the various cells differed in their capability to accumulate iron into the cell. From these observations, we conclude that Sta1 is a functional ortholog of Atm1p and can support the maturation of Fe/S proteins in the yeast cytosol.
Figure 6.
Sta1 Influence on the Assembly of the Cytosolic Fe/S Protein Leu1p and Iron Uptake in Δatm1 Yeast Cells.
(A) Coimmunoprecipitated 55Fe estimated by liquid scintillation counting. Δatm1 cells transformed with pRS424-GPD containing no DNA insert, the STA1 cDNA, or the ATM1 gene were grown in glucose-containing minimal medium and were radiolabeled for 1 hr with 55Fe. Cells were harvested, an extract was prepared by lysis with glass beads, and Leu1p polypeptide was immunoprecipitated with anti-Leu1p antibody. A background signal obtained for immunoprecipitation with antibodies derived from preimmune serum was subtracted (∼2.2 × 103 cpm/g cells).
(B) Uptake of radioactive 55Fe into the cells measured from the content of 55Fe in the extracts by liquid scintillation counting.
The Classical Fe/S Cluster Biosynthesis Pathway Is Present in Plant Mitochondria
To answer the important question of whether plant mitochondria possess the molecular machinery for Fe/S cluster biogenesis as found in yeast, we performed a screening of the available Arabidopsis sequences in the databases to identify plant homologs.
We found that the Arabidopsis genome encoded at least (i) one Nfs1p-like protein (67% similarity/57% identity at the amino acid sequence level); (ii) four proteins highly similar in size and sequence to Isu1p (on average 70% identity); (iii) three proteins similar in size and sequence to Isa1p (45% average identity); (iv) three proteins resembling yeast Nfu1p (40% average identity, but over a small domain of ∼70 amino acid residues); and (v) one Yah1p homolog (45% identity). All the Arabidopsis polypeptides identified possessed a putative organellar targeting sequence. However, it is difficult to distinguish between chloroplast and mitochondrial transit peptides, and in fact, bifunctional transit peptides have been described for some plant proteins (Menand et al., 1998).
Therefore, we analyzed the intracellular targeting of the putative Arabidopsis l-cysteine desulfurase, which could be a functional ortholog of Nsf1p, an enzyme initiating Fe/S cluster assembly in yeast mitochondria (Kispal et al., 1999). The N-terminal 56–amino acid residue of the Nfs1p homolog was fused to GFP, and the intracellular localization of the resulting Nfs1–56-GFP fusion protein was determined. Figure 3E shows that the Nfs1–56-GFP fusion was exclusively targeted to mitochondria. Taken together, these data strongly suggest that plant cells, like yeast and mammals (Lill and Kispal, 2000), possess a mitochondrial Fe/S cluster biosynthesis machinery resembling that of bacteria (Zheng et al., 1998).
Biochemical and Gene Expression Analysis of the starik Mutant
The phenotype of the starik mutant is most probably pleiotropic. By analogy with known yeast mutants, the phenotype development could be related either directly to the loss of activity of Fe/S cluster proteins in extramitochondrial plant cell compartments or indirectly to a misbalance in iron homeostasis. We analyzed the total activities of two Fe/S cluster-dependent enzymes: (i) cytosolic and mitochondrial isoforms of aconitase (De Bellis et al., 1993) and (ii) mitochondrial and plastidic ferrochelatases (Papenbrock et al., 1999). No significant differences between the sta1 mutant and wild type were found (data not shown). The total content of iron in sta1 plants did not differ significantly from that in the wild type, indicating that this mutation did not result in gross aberrations in iron availability to plant cells, as has been described for several other chlorotic plant mutants (Briat and Lobréaux, 1997). Further analysis of the iron status in mitochondria purified from achlorophyllous tissues has shown that mitochondria purified from sta1 cells contained 1.5- to 1.8-fold higher levels of the “free” (nonheme, nonprotein) iron than did wild-type mitochondria. Although the iron overload of mitochondria in sta1 cells was not very high compared with that of mitochondria of Δatm1 yeast (Kispal et al., 1997), this finding again indicates a similarity of functions between Sta1 and Atm1p.
Changes in the pool of free iron in mitochondria suggested that sta1 cells might be experiencing increased rates of iron-mediated oxidative damage (Halliwell and Gutteridge, 1989). We found that genes encoding ascorbate peroxidase (APX) and copper/zinc superoxide dismutases (Cu/ZnSODs) were upregulated in the starik mutant (Table 1), thus supporting this notion. Alteration in copper or zinc availability as a cause of the Cu/ZnSOD gene responses was ruled out by an in-gel isoenzyme analysis. However, as summarized in Table 1, the overall response of the antioxidant defense system was low, indicating that sta1 cells did not experience severe oxidative stress. This conclusion was further supported by biochemical measurements: (i) the content and the redox status of low molecular weight antioxidants, such as ascorbic acid and glutathione, were not altered in sta1; (ii) mutant plants had wild-type levels of nicotinamine, which is a specific iron chelator found in plant cells; (iii) mitochondrial proteins with covalently attached heme groups (Kispal et al., 1997) were not affected in sta1 mitochondria; and (iv) total activities of catalases and APXs were not significantly altered in the starik mutant. Additionally, as shown in Figure 7A, the starik mutant responded normally to experimentally induced oxidative stress.
Table 1.
Gene Expression Analysis on the sta1 Mutant Backgrounda
| Processb | Protein/Genec | Proven/Assumed Function | Protein Localization |
Reference | Transcript Levels in sta1 vs. Wild Typed |
|---|---|---|---|---|---|
| Oxidative stress | Apx1 | H2O2 scavenging | Cytosol | Kubo et al. (1993) | ⩽2↑ |
| Cu/ZnSOD | O2·− scavenging | Cytosol | Kliebenstein et al. (1998) | 2↑ | |
| Cu/ZnSOD | O2·− scavenging | Plastids | Kliebenstein et al. (1998) | 2↑ | |
| FeSOD | O2·− scavenging | Plastids | Kliebenstein et al. (1998) | WT | |
| MnSOD | O2·− scavenging | Mitochondria | Kliebenstein et al. (1998) | WT | |
| P1 | NADPH oxidoreductase | Cytosol | Babiychuk et al. (1995) | WT | |
| Senescence | Cat3 | H2O2 scavenging | Peroxisomes | Park et al. (1998) | WT |
| Sag12 | Proteolysis (?) | (?) | Lohman et al. (1994) | ND | |
| Iron homeostasis | Ferritin | Iron storage | Plastids | Gaymard et al. (1996) | WT |
| Pathogen response | PR1 | (?) | (?) | Chen et al. (1993) | WT |
| Oxidative stress/ thermogenesis |
Alternative oxidase Aox1 Alternative oxidase Aox2a |
CN-resistant respiration CN-resistant respiration |
Mitochondria Mitochondria |
Saisho et al. (1997) Saisho et al. (1997) |
WT ND |
| Fe/S cluster biogenesis | Nfs1p-like | l-cysteine desulfurase (?) | Mitochondria | This work | 2↑ |
| Sta2 | ABC transporter | Mitochondria | This work | WT | |
| Branched-chain amino acid synthesis (?) |
Bat1p-like | Transaminase; suppressor of atm1 mutation |
(?) | Kispal et al. (1996) | 2↑ |
| DNA repair/recombination | Zap | Poly(ADP-ribos)ylation | Nucleus | Babiychuk et al. (1998) | 2–3↑ |
| App | Poly(ADP-ribos)ylation | Nucleus | Babiychuk et al. (1998) | 6–10↑ | |
| AtRad51 | DNA strand exchange | Nucleus | Doutriaux et al. (1998) | 4–6↑ |
Transcript levels were measured in the wild type and sta1 by using RNA gel blot hybridization with radioactively labeled DNA probes and quantification of the hybridization signal on a phosphorimaging device. Total RNA was extracted from in vitro–grown plants; developmental defects of sta1 are most pronounced under these conditions (see Methods for details). (?) indicates cases in which function, intracellular localization of protein, or the process in which it is involved is not known or proven formally.
Although the process in which the analyzed genes are involved is indicated, this grouping does not mean necessarily that genes belong to the same regulon.
DNA probes were prepared from cDNAs. APX1, all of SODs, P1, ZAP, and APP cDNAs were from a collection of the Department of Plant Genetics (Ghent University, Belgium); Nfs and STA2 were amplified by PCR in this study; CAT3, ferritin, Bat1p-like, and PR1 cDNAs reported as expressed sequence tag markers in public databases were provided by the Ohio Arabidopsis Stock Center (Columbus); all clones were confirmed by partial DNA sequencing. Clones SAG12, AOX1 and AOX2a, and AtRAD51 were generous gifts of Richard Amasino, Mikio Nakazono, and Florence Couteau, respectively.
Relative to the wild type, the upregulation (↑) of the gene is indicated as fold; a steady state level of expression that did not exceed a twofold difference with the wild type is indicated by WT; ND, no transcript was detected both in the wild type and sta1.
Figure 7.
Gene Expression Analysis.
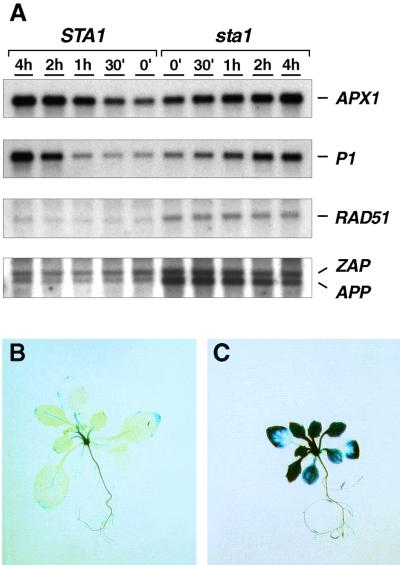
(A) RNA gel blot analysis. Three-week-old in vitro–grown wild-type (STA1) and mutant (sta1) plants were infiltrated with 10−6 M methyl viologen and sampled at the time intervals indicated above the lanes. To analyze steady state mRNA levels and gene responses to experimental oxidative stress, we extracted RNA and analyzed it by RNA gel blot hybridization. The same nylon membrane was consequently hybridized with probes made from cDNAs that encoded cytosolic ascorbate peroxidase (APX1), ζ-crystallin-like NADP(H)oxidoreductase P1 (P1), Arabidopsis homolog of Rad51 (RAD51), and probes of Arabidopsis poly(ADP-ribose) polymerases App (APP) and Zap (ZAP) mixed together. For summary of the gene expression analysis, see Table 1. h, hour; (′), minutes.
(B) GUS staining of an in vitro–grown wild-type plant. The activity of the promoter APP-uidA gene fusion (Babiychuk et al., 1998) was analyzed in STA1 and sta1 plants segregating in F2-segregating populations of a respective cross.
(C) GUS staining of a sta1 plant.
A misbalance in iron homeostasis is well known to cause DNA damage (Nunoshiba et al., 1999). Therefore, we analyzed the expression of genes encoding plant poly(ADP-ribose) polymerases (APP and ZAP) and Rad51-like protein (AtRAD51), which might constitute the components of the base excision repair (Babiychuk et al., 1998) and recombination repair pathways (Doutriaux et al., 1998; Ries et al., 2000), respectively. The genes analyzed might represent part of a genotoxic response regulon in plants because they are known to be induced by the ionizing radiation; the UV-B light also induces at least AtRAD51. Figure 7 illustrates that all three genes were induced in the sta1 plants. The expression of the APP promoter–uidA gene fusion on the sta1 mutant background was further analyzed. Figure 7C shows that all sta1 shoot tissues were highly β-glucuronidase (GUS) positive. The GUS activity pattern in root tissues of the mutant did not differ significantly from that of the wild type. Although these data suggested that Sta1 deficiency eventually led to substantial DNA damage in photosynthetic tissues of plants, we found no increase in steady state levels of abasic sites (AP) in DNA of the mutant (data not shown). Thus, the specificity of the gene response found remains to be determined.
DISCUSSION
We have characterized a nuclear-encoded mitochondrial ABC transporter Sta1 and demonstrated that it represents a functional ortholog of the yeast ABC transporter Atm1p. The Arabidopsis genome codes for Sta2 and Sta3 ABC transporters that are highly homologous to Sta1. The ectopic expression of the mitochondrial Sta2 homolog partially suppresses the starik mutant phenotype caused by the mutation in Sta1. The results of this experiment indicate that Sta1, Sta2, and possibly Sta3 have overlapping functions. In addition, the analysis of Arabidopsis genomic sequences suggests the presence of several genes encoding half-transporters that might be targeted to mitochondria but that are less homologous to Sta1 (unpublished data). Although the amino acid sequences of human Abc7 and MTABC3 share only 42.9% similarity, they both suppress the loss of Atm1p function in yeast and are candidate genes for human genetic diseases caused by the misbalance of iron homeostasis (Csere et al., 1998; Allikmets et al., 1999; Mitsuhashi et al., 2000). These data suggest that the larger families of Atm1p orthologous ABC transporters might have more diverse functions in higher eukaryotes than in yeast.
The sta1 mutation results in an increased expression of Arabidopsis genes encoding a putative Nfs1p-like cysteine desulfurase and a branched-chain amino acid transaminase (Table 1), homologous to the yeast Bat1p (Kispal et al., 1997). In yeast, both Nsf1p and Bat1p/Bat2p are required for the biogenesis of the cytosolic Fe/S isopropyl malate isomerase Leu1p (Kispal et al., 1999; Prohl et al., 2000). A compensatory response of plant cells toward a misbalance in the biogenesis of Fe/S cluster proteins may provide a simple explanation for the observed changes in steady state mRNA levels. The STA1 gene deficiency leads to the 1.5- to 1.8-fold increase in the pool of the free (nonprotein, nonheme) iron within mitochondria, which is a general consequence of the deficiency in genes involved in Fe/S cluster biogenesis (Lill and Kispal, 2000).
The possible role of plant mitochondria in Fe/S cluster biogenesis has not been documented in the literature. Although chloroplasts have been shown to promote the formation of endogeneous Fe/S ferredoxin in vitro (Takahashi et al., 1991), additional proofs of chloroplast self-sufficiency in Fe/S biosynthesis are necessary. Here, we demonstrate that plant mitochondria contain at least one putative l-cysteine desulfurase, Nfs. In Fe/S biosynthesis, the pyridoxal phosphate-dependent l-cysteine desulfurases initiate Fe/S assembly by producing and carrying elemental sulfur (Zheng et al., 1998; Kispal et al., 1999; Yuvaniyama et al., 2000). In addition, a number of protein homologs, which constitute the Fe/S cluster biosynthesis machinery of yeast mitochondria, have been identified in the Arabidopsis genome. Taken together, these data strongly suggest that plant mitochondria can support (i) the maturation of their own Fe/S proteins and (ii) the maturation of Fe/S proteins in extramitochondrial plant cell compartments, the latter function being mediated by Atm1p-like plant mitochondrial ABC transporters.
A mutation in the STA1 gene results in a light-dependent chlorosis and dwarfism of plants as well as alterations in the structure of leaf and nuclei. By analogy with Δatm1 yeast (Kispal et al., 1999), the development of the starik mutant phenotype could be related to a defect in the maturation of Fe/S proteins. Here, no significant differences were found in the combined enzymatic activities of plastidic and mitochondrial ferrochelatases and in cytosolic and mitochondrial aconitases between the wild type and the starik mutant. The mitochondrial isoforms are apparently unaffected in sta1 cells, which is consistent with data on the biogenesis of mitochondrial Fe/S proteins in Δatm1 yeast (Kispal et al., 1999). The partially redundant function of Sta2 and Sta3 proteins might therefore be sufficient to support the biogenesis of the cytosolic aconitase in plants. The plastidic ferrochelatase most probably depends on Fe/S clusters synthesized within chloroplasts (Takahashi et al., 1991).
The available data strongly suggest that overall, the starik phenotype is probably stress related and arises as a secondary effect because of a misbalance in intracellular iron homeostasis. The light dependence of chlorotic symptoms in the sta1 plants indicates photooxidative damage (Lindahl et al., 2000). Interestingly, the mitochondrial gene copy number in sta1 cells is increased at least threefold (unpublished data). The plant mitochondriome shows a similar response toward photooxidative stress in barley seedlings bleached with norfluorazon, but also in the barley albostrians albino mutant (Hedtke et al., 1999). We found a strong activation of genes encoding plant poly(ADP-ribose)polymerases and Rad51-like protein. Considering that the nucleus undergoes the most apparent morphological changes in the sta1 mutant, the sta1-dependent activation of genes encoding nuclear proteins with a function in DNA repair seems to be all the more significant. However, the starik mutant had wild-type steady state levels of abasic sites in DNA; thus, the specificity of this gene response remains to be proven. The analysis of retrograde signaling in yeast, that is, modification of nuclear gene expression by multiple mitochondrial signals, has revealed the complex nature of the physiological processes that are induced by mitochondrial dysfunction (Hallstrom and Moye-Rowley, 2000). The compromise of mitochondrial function in plants is also known to result in several pleiotropic phenotypes, including alterations in plastid differentiation, although the underlying molecular mechanisms and the physiological significance behind them are not fully understood (Mackenzie and McIntosh, 1999, and references therein). Assessing the gene responses characterized in this study in other plant mutants together with additional analyses of the starik mutant may facilitate our understanding of the essential cellular signals that couple mitochondrial, chloroplastic, and nuclear functions throughout development.
METHODS
Handling of Laboratory Strains
The starik (sta1) mutation was isolated on a genetic background of Arabidopsis thaliana ecotype Col5, also known as C24 (Valvekens et al., 1988). To prepare achlorophyllous Arabidopsis tissues for mitochondria isolation, we cultured roots from plants grown in liquid Murashige and Skoog medium in a liquid callus-inducing medium (Valvekens et al., 1988) for 2 weeks. Transgenic Arabidopsis and Nicotiana tabacum SR1 plants were generated by standard in vitro plant transformation protocols (Valvekens et al., 1988). To select sta1/sta1;papp-uidA plants, plants from three independent transgenic lines (Sal5, Sal21, and Sal33) of Arabidopsis Col1 transformed with the APP gene promoter–uidA fusion (Babiychuk et al., 1998) were crossed with the mutant, and the progeny from kanamycin+BASTA–resistant F1 plants were analyzed for β-glucuronidase (GUS) activity.
Agrobacterium tumefaciens GV3101(pMP90) was used for the delivery of T-DNA from binary vectors into plant cells. Escherichia coli JM109 (Promega, Madison, WI) was used for molecular cloning. Bacterial strains were grown on a standard Luria-Bertani medium (Gibco BRL, Gaithersburg, MD).
The following Saccharomyces cerevisiae strain was used as wild type: YPH500 (MATα, ura3-52, lys2-801, ade2-101, trp1-63, his3-200, leu2-1). The Δatm1 cells (MATα, ura3-52, lys2-801, ade2-101, trp1-63, his3-200, leu2-1, atm1::LEU2) have previously been described (Kispal et al., 1997) as well as techniques for growth and transformation of yeast cells (Kispal et al., 1997).
Molecular Cloning
STA1 full-length cDNA was reconstructed by polymerase chain reaction (PCR) methods. Based on the sequence of the chimeric exon from the line SK3-3, the 5′ end sequences of the STA1 cDNA were amplified with a nested set of primers positioned ∼300 bp from the 5′ end of the messenger by using a DNA template prepared with a Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA). The same template was used to amplify downstream sequences in a 3′ rapid amplification of cDNA ends configuration. The reconstructed cDNA sequence was used to design primers for the amplification of a complete coding region using a proofreading Pfu DNA polymerase (Stratagene, La Jolla, CA). STA2 cDNA was amplified by PCR with a pair of primers, the sequence of which was designed on the basis of the annotated genomic sequence of the STA2 gene. PCR mutagenesis was used to prepare translational fusions with the green fluorescent protein (GFP). The GFP-coding region adapted for the expression in plant cells was derived from pBIN-m-gfp5-ER, which was a generous gift of Jim Haseloff (University of Cambridge, UK). An expression cassette used for driving the expression of chimeric genes was derived from pE2113-GUS (Mitsuhara et al., 1996). All expression cassettes were carried within a T-DNA region of the binary vector pGSV6, which was kindly provided by Plant Genetics Systems N.V. (Gent, Belgium). Yeast shuttle vector pRS423-GDP (Mumberg et al., 1995) was used for the expression of cDNAs in yeast. pBluescript KS+ (Stratagene) was used for routine subcloning.
Mitochondria Isolation and Analysis
Mitochondria were isolated from Arabidopsis achlorophyllous tissues by the Percoll gradient method essentially as described (Klein et al., 1998). To assess the intactness of mitochondria, we measured the activity of aconitase by following the decrease in absorbance of cis-aconitate at 240 nm (De Bellis et al., 1993). Total mitochondrial proteins were measured with a modified Bradford reagent (Coomassie Plus Protein Assay Reagent; Pierce, Rockford, IL). Analyses of proteins with covalently attached heme group and mitochondrial pools of non–Fe/S and of nonheme iron were performed as described (Kispal et al., 1997).
Microscopical Analysis
Light and transmission electron microscopy was performed according to standard protocols (Hayat, 1986). Microscopic analysis data shown were performed on tissues harvested from soil-grown plants of the same age (3 to 4 weeks old). Fully expanded leaves of plants were fixed, embedded in London Resin White, and sectioned. For confocal microscopy analysis, epidermis peels were mounted in water under cover slips. Specimens were examined with a ×63/1.2-Wcorr water immersion lens on an LSM 510 laser scanning confocal microscope (Karl Zeiss Inc., Thornwood, NY). Fluorescence images were obtained at 2048 × 2048 pixel resolution (with 2.24 μsec pixel time) by using the 488-nm excitation line of an argon ion laser (458, 488, and 514 nm) with the appropriate emission filters 505 to 530 BP and 650 LP for GFP and chlorophyll, respectively. For differential interference contrast (DIC) reference images, laser was used as a transmission light source. Optical sections were taken at different channels and were overlayed onto DIC images with the LSM510 software package supplied with the microscope. Images were exported as TIFF files and further processed with Adobe Photoshop (version 5.02; Adobe Systems, San Jose, CA).
Other Methods
The 55Fe incorporation into Leu1p yeast polypeptide and iron uptake experiments were performed according to Kispal et al. (1999). Methods described previously were used to analyze the total iron content in leaf tissues (Schmidke and Stephan, 1995); nicotinamine (Schmidke and Stephan 1995); ascorbic acid and glutathione (Davey et al., 1999); activities of catalase, ascorbate peroxidase, and aconitase (De Bellis et al., 1993; Kampfenkel et al., 1995); superoxide dismutase isoforms by PAGE under native conditions (Kliebenstein et al., 1998); Fe-chelatase activity (Papenbrock et al., 1999); and abasic sites in DNA after modification with an aldehyde-reactive probe, ARP (Dojindo Laboratories, Kumamoto, Japan) (Nakamura et al. 1998). RNA was extracted from 3-week-old seedlings grown in vitro and analyzed by gel blot hybridization as described (Babiychuk et al., 1998). RNA was quantified spectrophotometrically, and the equal loading as well as the RNA transfer efficiency were additionally verified by staining of RNA gel blots with methylene blue. Hybridization signals were visualized and quantified on a PhosphorImager model 445SI (Amersham Pharmacia Biotech, Little Chalfont, UK), using a software provided with the instrument. 32P-α-dCTP-labeled DNA probes were prepared with the Rediprime DNA labeling system (Amersham Pharmacia Biotech). DNA sequencing analysis was performed with the dideoxy chain termination method (BigDye Terminator Sequencing kit; PE Applied Biosystems, Foster City, CA) on an ABI377 automatic DNA sequencer (PE Applied Biosystems).
Acknowledgments
We thank Freya De Winter and Els Van Lerberge for technical assistance and Martine De Cock, Karel Spruyt, and Rebecca Verbanck for help in preparing the manuscript and artwork. The help of Wally Wendt (Gatersleben, Germany) for performing the nicotinamine measurements is greatly appreciated. We are grateful to our colleagues who shared materials and ideas, to Jim Haseloff for the mGFP5 clone, to Yoo-Sun Noh and Richard Amasino for the SAG12 cDNA, to Mikio Nakazono for alternative oxidase cDNA clones, to Florence Couteau for AtRAD51, to Peter Hedden for advice and discussion on experiments with gibberellic acid, to Nicole Mang, Jörg Kuddla, and Axel Brennicke for the mitochondrial and chloroplast DNA probes, to Masanobu Shiga and Jun Nakamura for advice on AP site determination, and to Kim Hanson and Joanne Chory for seeds of the cue1-2 mutant. The Arabidopsis Biological Resource Center (Ohio State University, Columbus) is acknowledged for making Arabidopsis cDNA clones freely available. This research was supported in part by grants from the Diensten van de Eerste Minister, Wetenschappelijke, Technische en Culturele Aangelegenheden (DWTC T960226), the Sonderforschungsbereich 286 of the Deutsche Forschungsgemeinschaft, the Volkswagenstiftung, the Fonds der Chemischen Industrie, and the Hungarian Funds OKTA.
References
- Allikmets, R., Raskind, W.H., Hutchinson, A., Schueck, N.D., Dean, M., and Koeller, D.M. (1999). Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8 743–749. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Kushnir, S., Belles-Boix, E., Van Montagu, M., and Inzé, D. (1995). Arabidopsis thaliana NADPH oxidoreductase homologs confer tolerance of yeasts toward the thiol-oxidizing drug diamide. J. Biol. Chem. 270 26224–26231. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Fuangthong, M., Van Montagu, M., Inzé, D., and Kushnir, S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk, E., Cottrill, P.B., Storozhenko, S., Fuangthong, M., Chen, Y., O'Farrell, M.K., Van Montagu, M., Inzé, D., and Kushnir, S. (1998). Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J. 15 635–645. [DOI] [PubMed] [Google Scholar]
- Beinert, H., Holm, R.H., and Münck, E. (1997). Iron-sulfur clusters: Nature's modular, multipurpose structures. Science 277 653–659. [DOI] [PubMed] [Google Scholar]
- Briat, J.-F., and Lobréaux, S. (1997). Iron transport and storage in plants. Trends Plant Sci. 2 187–193. [Google Scholar]
- Chen, Z., Silva, H., and Klessig, D.F. (1993). Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262 1883–1886. [DOI] [PubMed] [Google Scholar]
- Csere, P., Lill, R., and Kispal, G. (1998). Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Lett. 441 266–270. [DOI] [PubMed] [Google Scholar]
- Davey, M.W., Gilot, C., Persiau, G., Østergaard, J., Han, Y., Bauw, G.C., and Van Montagu, M.C. (1999). Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol. 121 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bellis, L., Tsugeki, R., Alpi, A., and Nishimura, M. (1993). Purification and characterization of aconitase isoforms from etiolated pumpkin cotyledons. Physiol. Plant. 88 485–492. [Google Scholar]
- Doutriaux, M.-P., Couteau, F., Bergounioux, C., and White, C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257 283–291. [DOI] [PubMed] [Google Scholar]
- Gaymard, F., Boucherez, J., and Briat, J.-F. (1996). Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem. J. 318 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J.M.C. (1989). Free Radicals in Biology and Medicine. (Oxford: Clarendon Press).
- Hallstrom, T.C., and Moye-Rowley, W.S. (2000). Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Chem. Biol. 275 37347–37356. [DOI] [PubMed] [Google Scholar]
- Hayat, M.A. (1986). Basic Techniques for Transmission Electron Microscopy. (San Diego, CA: Academic Press).
- Hedtke, B., Wagner, I., Börner, T., and Hess, W.R. (1999). Inter-organellar crosstalk in higher plants: Impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 19 635–643. [DOI] [PubMed] [Google Scholar]
- Higgins, C.F. (1992). ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 8 67–113. [DOI] [PubMed] [Google Scholar]
- Kampfenkel, K., Van Montagu, M., and Inzé, D. (1995). Effects of iron excess on Nicotiana plumbaginifolia plants. Implications to oxidative stress. Plant Physiol. 107 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal, G., Steiner, H., Court, D.A., Rolinski, B., and Lill, R. (1996). Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Ec39 protein. J. Biol. Chem. 271 24458–24464. [DOI] [PubMed] [Google Scholar]
- Kispal, G., Csere, P., Guiard, B., and Lill, R. (1997). The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418 346–350. [DOI] [PubMed] [Google Scholar]
- Kispal, G., Csere, P., Prohl, C., and Lill, R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, M., Binder, S., and Brennicke, A. (1998). Purification of mitochondria from Arabidopsis. In Arabidopsis Protocols, Methods in Molecular Biology, Vol. 82, J.M. Martínez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 49–53. [DOI] [PubMed]
- Kliebenstein, D.J., Monde, R.-A., and Last, R.L. (1998). Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A., Saji, H., Tanaka, K., and Kondo, N. (1993). Genomic DNA structure of a gene encoding cytosolic ascorbate peroxidase from Arabidopsis thaliana. FEBS Lett. 315 313–317. [DOI] [PubMed] [Google Scholar]
- Leighton, J., and Schatz, G. (1995). An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 14 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill, R., and Kispal, G. (2000). Maturation of cellular Fe-S proteins: An essential function of mitochondria. Trends Biochem. Sci. 25 352–356. [DOI] [PubMed] [Google Scholar]
- Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A.B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman, K.N., Gan, S., John, M.C., and Amasino, R.M. (1994). Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 92 322–328. [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand, B., Maréchal-Drouard, L., Sakamoto, W., Dietrich, A., and Wintz, H. (1998). A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl–tRNA synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara, I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37 49–59. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi, N., Miki, T., Senbongi, H., Yokoi, N., Yano, H., Miyazaki, M., Nakajima, N., Iwanaga, T., Yokoyama, Y., Shibata, T., and Seino, S. (2000). MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis. J. Biol. Chem. 275 17536–17540. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Müller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119–122. [DOI] [PubMed] [Google Scholar]
- Nakamura, J., Walker, V.E., Upton, P.B., Chiang, S.-Y., Kow, Y.W., and Swenberg, J.A. (1998). Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 58 222–225. [PubMed] [Google Scholar]
- Nunoshiba, T., Obata, F., Boss, A.C., Oikawa, S., Mori, T., Kawanishi, S., and Yamamoto, K. (1999). Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J. Biol. Chem. 274 34832–34837. [DOI] [PubMed] [Google Scholar]
- Papenbrock, J., Mock, H.-P., Kruse, E., and Grimm, B. (1999). Expression studies in tetrapyrrole biosynthesis: Inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208 264–273. [Google Scholar]
- Park, J.-H., Oh, S.A., Kim, Y.H., Woo, H.R., and Nam, H.G. (1998). Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol. Biol. 37 445–454. [DOI] [PubMed] [Google Scholar]
- Prohl, C., Kispal, G., and Lill, R. (2000). Branched-chain amino acid transaminases of yeast Saccharomyces cerevisiae. In Branched Chain Amino Acids, Part B, Methods in Enzymology, Vol. 324, J.R. Sokatch and R.A. Harris, eds (San Diego, CA: Academic Press), pp. 365–375. [DOI] [PubMed]
- Ries, G., Heller, W., Puchta, H., Sandermann, H., Seidlitz, H.K., and Hohn, B. (2000). Elevated UV-B radiation reduces genome stability in plants. Nature 406 98–101. [DOI] [PubMed] [Google Scholar]
- Saisho, D., Nambara, E., Naito, S., Tsutsumi, N., Hirai, A., and Nakazono, M. (1997). Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol. Biol. 35 585–596. [DOI] [PubMed] [Google Scholar]
- Schmidke, I., and Stephan, U.W. (1995). Transport of metal micronutrients in the phloem of castor bean (Ricinus communis) seedlings. Physiol. Plant. 95 147–153. [Google Scholar]
- Takahashi, Y., Mitsui, A., and Matsubara, H. (1991). Formation of the Fe-S cluster of ferredoxin in lysed spinach chloroplasts. Plant Physiol. 95 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens–mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama, P., Agar, J.N., Cash, V.L., Johnson, M.K., and Dean, D.R. (2000). NifS-directed assembly of a transient [2Fe-2S] cluster with the NifU protein. Proc. Natl. Acad. Sci. USA 97 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Cash, V.L., Flint, D.H., and Dean, D.R. (1998). Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273 13264–13272. [DOI] [PubMed] [Google Scholar]