Abstract
Plant cells can exhibit highly complex nuclear organization. Through dye-labeling experiments in untransformed onion epidermal and tobacco culture cells and through the expression of green fluorescent protein targeted to either the nucleus or the lumen of the endoplasmic reticulum/nuclear envelope in these cells, we have visualized deep grooves and invaginations into the large nuclei of these cells. In onion, these structures, which are similar to invaginations seen in some animal cells, form tubular or planelike infoldings of the nuclear envelope. Both grooves and invaginations are stable structures, and both have cytoplasmic cores containing actin bundles that can support cytoplasmic streaming. In dividing tobacco cells, invaginations seem to form during cell division, possibly from strands of the endoplasmic reticulum trapped in the reforming nucleus. The substantial increase in nuclear surface area resulting from these grooves and invaginations, their apparent preference for association with nucleoli, and the presence in them of actin bundles that support vesicle motility suggest that the structures might function both in mRNA export from the nucleus and in protein import from the cytoplasm to the nucleus.
INTRODUCTION
Although cellular function often requires maximization of surface area relative to volume, notably in organelles such as the endoplasmic reticulum (ER) and Golgi apparatus, traditional representations of the nucleus depict a rounded structure with little internal organization. Recently, however, the nuclei of animal cells have been found to show considerable spatial and structural organization at the chromosomal level. The discrete and comparatively stable territories that chromosomes occupy within the nucleus are separated by interchromosomal domains through which transcribed RNA and other macromolecules can diffuse (reviewed in Lamond and Earnshaw, 1998). Animal nuclei also sometimes deviate from the characteristic rounded shape. Deviations include a folded, grooved, or notched surface (Majno et al., 1969) and tubular invaginations. Electron microscopy has shown that these nuclear invaginations can penetrate deep into the nucleus and confirmed the presence of the double membrane of the nuclear envelope (Ellenberg et al., 1997; Fricker et al., 1997; Clubb and Locke, 1998). However, revelation of the full nature of nuclear invaginations in living animal cells has required confocal microscopy of fluorescent dyes (Fricker et al., 1997; Lui et al., 1998a, 1998b) or green fluorescent protein (GFP) fusion proteins targeted to the nuclear envelope (Ellenberg et al., 1997; Broers et al., 1999). The number and nature of invaginations vary from cell type to cell type, ranging from simple invaginations to intricate branched structures that can penetrate into and through the nucleus. Invaginations often associate with nucleoli (Fricker et al., 1997) and appear to be stable (Ellenberg et al., 1997; Broers et al., 1999). Nuclear grooves and invaginations substantially increase the surface area of the nucleus and have been suggested to function in signaling from the cytoplasm to the nucleus (Lui et al., 1998a, 1998b) or to coordinate transport processes between the nucleus and the cytoplasm (Fricker et al., 1997). However, whether any relationship exists between the distribution of nuclear invaginations and chromosome domains within the animal nucleus remains to be determined.
Plant nuclei show numerous structural and organizational features that are similar to those of animal cells. Chromosomes occupy specific territories within the nucleus (Heslop-Harrison et al., 1990; Abranches et al., 1998; Gonzalez-Melendi et al., 2000; reviewed in Heslop-Harrison and Bennett, 1990; Franklin and Cande, 1999) and can be dynamic, having the ability to change shape (Heslop-Harrison and Heslop-Harrison, 1989; Chytilova et al., 2000).
Plant nuclei, too, can deviate from the traditional rounded structures shown in textbooks in various ways. For example, placental cells in Lilium ovaries contain a nuclear reticulum in which the inner nuclear membrane forms extensive tubular invaginations that penetrate through the center of the nucleus, the lumen of this reticulum being contiguous with the lumen of the ER (Singh and Walles, 1995; Singh et al., 1998). This type of structure differs from the shallow and narrow invaginations of both membranes of the nuclear envelope found in developing microspores of various species (Aldrich and Vasil, 1970; Dickinson and Bell, 1970, 1972; Li and Dickinson, 1987) and also differs from nuclear vacuoles, membrane-bound inclusions within the nucleus that are also generally found in meiotic cells (Sheffield et al., 1979; Karasawa and Ueda, 1983; Sangwan, 1986; Yi et al., 1994). Additional published examples document deep cytoplasmic channels penetrating into the nuclei of nonreproductive cells of various species, including Narcissus (Gunning and Steer, 1996), Pisum (Bowes, 1996), and onion epidermis (Kartusch et al., 2000). In fact, the large nuclei of onion epidermal cells show marked irregularities in nuclear shape, having large groovelike infoldings that contain rapid cytoplasmic movement (Lichtscheidl and Url, 1988; N.S. Allen, personal observation). However, whether these nuclear infoldings represent invaginations or slices through nuclear grooves cannot be determined without optical sectioning techniques. Furthermore, the function (if any) of these structures remains unknown; as with animal cells, however, the great increase in the contact area between the nucleus and the cytoplasm may be important.
The current study evolved from confocal investigations with fluorescently labeled probes that nonspecifically labeled the interior membranes of onion epidermal cells, revealing not only ER organization and dynamics but also extensive grooves that scored the nucleus. Subsequently, we have transiently expressed GFP targeted either to the nucleus (N-GFP) or to the ER (ER-GFP) to characterize these grooves, along with tubular nuclear invaginations, in living onion nuclei. We have furthermore observed similar structures in the nuclei of suspension-cultured tobacco cells. These structures, similar to the grooves and invaginations described by Ellenberg et al. (1997), Fricker et al. (1997), Lui et al. (1998a)(1998b), and Broers et al. (1999), substantially increase the surface area of the nucleus. Importantly, investigating nuclear grooves and invaginations in GFP-expressing cells should further our understanding of communication between the nucleus and the cytoplasm.
RESULTS
Untransformed Onion Epidermal Nuclei Are Highly Convoluted and Contain Deep Grooves and Invaginations
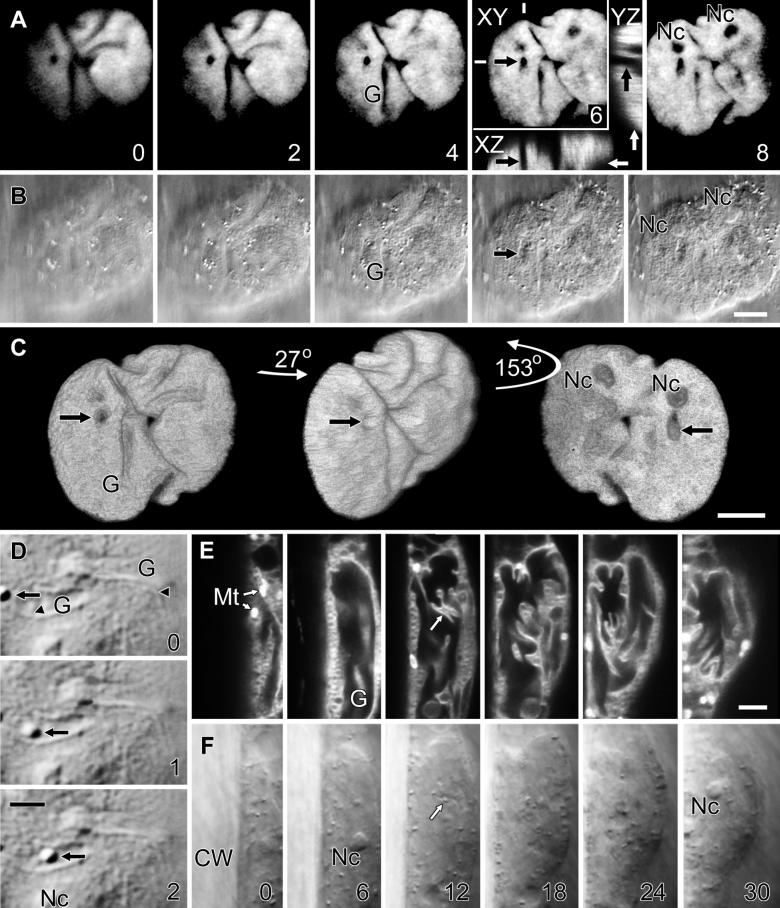
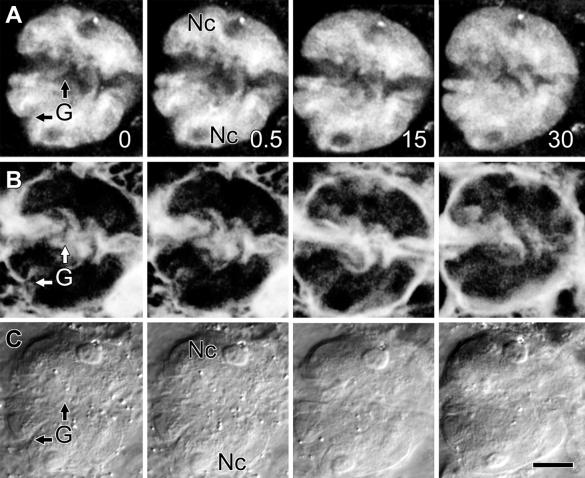
Untransformed onion epidermal nuclei are lens-shaped, are as large as 30 μm in diameter, and as shown by 4′,6-diamidino-2-phenylindole (DAPI) labeling of nuclear DNA in Figure 1 (and the QuickTime movie Fig1.mov included in the online version of this article), contain deep grooves that score their surface and from which DAPI labeling was absent (G in Figure 1A [and in Fig1.mov, sequence 1]). These grooves penetrated as much as 6 μm into the nucleus, deeper than previous differential interference contrast (DIC) video microscopy might suggest (Lichtscheidl and Url, 1988). DAPI labeling was also absent from tubular invaginations that projected as far as 8 μm into the center of the nucleus (Figure 1A, arrow [Fig1.mov, sequence 1, arrow]). Furthermore, image reconstructions in orthogonal planes (Figure 1A, planes XZ and YZ) confirmed that DAPI-labeled DNA surrounded these nuclear invaginations. DAPI labeling was likewise absent from nucleoli (Nc in Figures 1A and 1B [and in Fig1.mov, sequence 1]), which in this case were separated by a nuclear groove. Interestingly, one nucleolus lay close to the nuclear invagination (Figure 1A, arrow). Three-dimensional image reconstructions with a surface shadowing algorithm emphasized the highly convoluted shape of this onion nucleus, showing dramatically how the deep grooves score the nuclear surface and how the invagination penetrates through the nucleus (Figure 1C [Fig1.mov, sequence 2]). Concurrent DIC imaging clearly visualized nuclear grooves and invaginations (G and arrow in Figure 1B) and demonstrated that these structures lay outside the nucleus and were not simply areas within the nucleus that lacked DNA.
Figure 1.
Untransformed Onion Epidermal Nuclei Are Highly Lobed and Grooved.
(A) and (B) Optical sections shown at 2.0-μm intervals from the surface of an untransformed onion nucleus, as indicated by numbers in the lower right of sections in (A). (A) Confocal fluorescence images of DAPI-labeled DNA, revealing numerous nuclear grooves (G), two nucleoli (Nc) in separate nuclear lobes, and an invagination (black arrows) that could be followed through multiple focal planes. White bars in the section at 6 μm from the nuclear surface show the locations at which Metamorph software was used to reconstruct orthogonal slices in the XZ and YZ planes from the entire data set. These reconstructed slices are shown in the corresponding inset, where white arrows indicate the plane of the original optical section. These reconstructions confirm that the nuclear invagination is surrounded entirely by DNA. (B) Concurrent DIC images showing that grooves (G), nucleoli (Nc), and the nuclear invagination (black arrow) are all visible with transmitted light.
(C) Three-dimensional reconstruction with surface shadowing of DAPI fluorescence from (A), revealing the convoluted shape of the nucleus. The nucleus was rotated 27° in the direction of the arrow between the left and central projections and shows the grooves (G) that lay across the nuclear surface. Rotation through a further 153° shows the nucleus in cross-section roughly at its center, where two nucleoli (Nc) and the nuclear invagination (arrows) are seen.
(D) Polarization modulation DIC images (Holzwarth et al., 1997) of an onion nucleus showing the nuclear envelope (arrowheads), nucleolus (Nc), and two nuclear grooves (G). A single vesicle (arrows) moved rapidly through one of these grooves at ∼3 μm sec−1. Times in seconds are shown at the lower right of each image.
(E) and (F) Optical sections shown at 6-μm intervals from the surface of an untransformed onion nucleus, as indicated by numbers in the lower right of sections in (F). The nucleus is seen in cross-section and lies adjacent to the cell wall. (E) Confocal fluorescence images of DIOC6(3) labeling of internal membranes show bright mitochondria (Mt) adjacent to the nucleus and extensive nuclear invaginations (arrow) and grooves (G) inside the nucleus. Note that the nuclear groove lies very close to the position of the nucleolus. (F) Concurrent DIC images showing that nucleoli (Nc) and the nuclear invagination (arrow) are visible with transmitted light. CW, cell wall.
 ;
;  (
( )
) 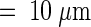 ;
; 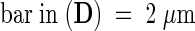 ;
; 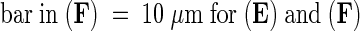 .
.
That nuclear grooves and invaginations lie outside the onion nucleus was further confirmed by high-resolution polarization-modulation DIC video microscopy (Holzwarth et al., 1997). Two grooves that penetrated several micrometers into the nucleus were visible, one of which lay adjacent to a nucleolus (Figure 1D, Nc) and showed rapid cytoplasmic streaming (∼3 μm sec−1) of spherosomes and other small vesicles (Figure 1D, arrow). Vesicle movement was also seen in nuclear invaginations (data not shown).
Confocal imaging of 3,3′-dihexyloxacarbocyanine iodide (DIOC6(3)), a membrane-permeant carbocyanine dye that labels both ER and mitochondria, further demonstrated the presence in onion nuclei of deep grooves and invaginations (Figure 1E), although this technique presented several problems, including rapid fading of signal and labeling of other particles such as mitochondria. In both fluorescence (Figure 1E) and DIC images (Figure 1F), extensive nuclear grooves and invaginations (arrow) were visible, often close to the nucleoli.
Nuclear-Targeted GFP Confirms the Irregular Shape of Onion Nuclei
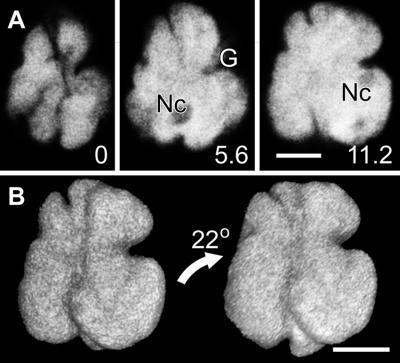
Confocal optical sections through the nuclei of living onion cells transiently expressing N-GFP confirmed observations made with DAPI labeling. Because the N-GFP is not a DNA stain and is free to diffuse throughout the nucleus (Chytilova et al., 2000), the extensive nuclear grooves (Figure 2A, G) that slice deeply into the nuclear surface near to nucleoli (Figure 2A, Nc) must lie outside the nucleus, rather than being DNA-free compartments inside the nucleus. The resulting highly convoluted shape of the nucleus is visible in three-dimensional reconstructions (Figure 2B).
Figure 2.
N-GFP Confirms the Presence of Nuclear Grooves in Onion Cells.
(A) Optical sections at different depths into the nucleus of an N-GFP–expressing onion cell (relative distances in micrometers are shown at the lower right of each image) reveal the complex nuclear organization typical of onion epidermal cells. Nuclear grooves (G) exclude N-GFP, which lie outside the nucleus, although nucleoli (Nc) within the nucleus also exclude the N-GFP.
(B) A three-dimensional reconstruction of N-GFP in the nucleus from (A) confirms the convoluted shape of the nucleus. The nucleus was rotated 22° in the direction of the arrow between projections.
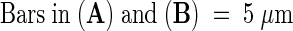 .
.
ER-GFP Localizes to the Nuclear Envelope That Bounds Invaginations and Grooves
Imaging of ER-GFP demonstrates dynamic subcortical ER and stable polygonal arrays of cortical ER (Haseloff et al., 1997; Ridge et al., 1999; Scott et al., 1999), which we used, along with the presence of cytoplasmic streaming, to confirm that the cells remained healthy during the experiments. However, as shown in Figure 3, ER-GFP also reveals nuclear structure because the lumen of the ER is continuous with the nuclear envelope (Herman et al., 1990).
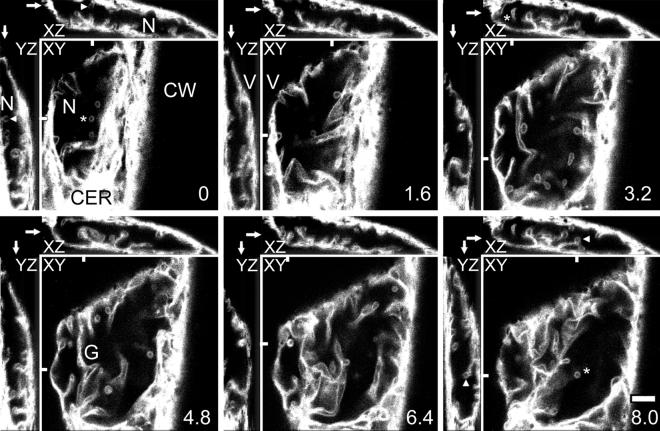
Figure 3.
Three-Dimensional Reconstructions of ER-GFP Fluorescence Reveal the Highly Complex Organization of Onion Nuclear Grooves and Invaginations.
Confocal optical sections were taken at 0.4-μm intervals through an ER-GFP–expressing onion cell but are presented here at 1.6-μm intervals in sections labeled XY, with relative depths in micrometers given at the lower right of each section. Orthogonal slices in the XZ and YZ planes were calculated from the entire data set at the locations marked with bars in the XY sections and are presented in the insets labeled XZ and YZ, above and at the left of the XY images. Arrows in the corresponding XZ and YZ orthogonal sections indicate the planes of the original optical sections. The lens-shaped nucleus in this cell is typical of nuclei in the onion epidermis. It is squeezed between the central vacuole (V) and the cell wall (CW). A thin layer of cortical ER (CER) is present between the plasma membrane and the nucleus. Well-defined but branched nuclear grooves (G) wind through the nucleus, whereas invaginations are visible as projections in longitudinal section or as rings in cross-section (asterisks). Such rings appear as projections in the orthogonal reconstructions (arrowheads). Grooves and invaginations often show nonfluorescent centers, which suggests that they contain cytoplasmic cores. 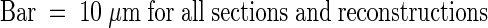 .
.
Confocal optical sections (Figure 3, XY planes) through an ER-GFP–expressing onion cell, and three-dimensional reconstructions of the orthogonal planes derived from these sections (XZ and YZ), show how the lens-shaped nucleus was squeezed between the central vacuole and the cortical ER layer adjacent to the cell wall. Consistent with DAPI and DIOC6(3) observations, two distinct variations from a rounded nuclear structure were apparent. First, the nucleus contained numerous grooves—planes of cytoplasm, often narrow (one to several micrometers wide) and branched—that extended many micrometers into the core of the nucleus and split the nucleus into several lobes. The nucleus shown in Figure 3 also contained nuclear invaginations, linear projections of ER that also can extend several micrometers into the nucleus. When seen in longitudinal section, the invaginations looked similar to nuclear grooves, but when cross-sectioned they were visualized as rings. However, invaginations seen as rings in the XY plane appeared as linear or curved structures in the reconstructed perpendicular XZ and YZ planes (arrowheads). Neither nuclear grooves nor nuclear invaginations were found in any preferred orientation.
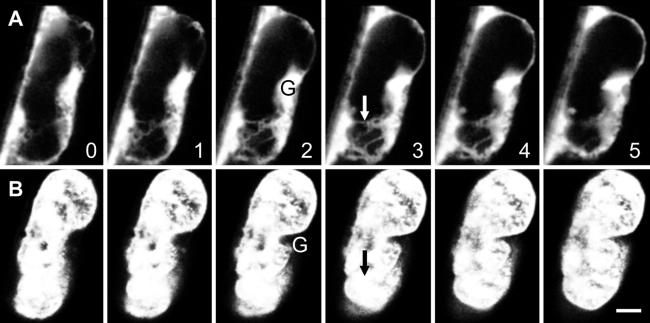
As shown in Figure 4 (and the online movie Fig4.mov), some nuclei showed variations in nuclear invaginations. Faint and branched channels of ER-GFP traversed the entire nucleus (Figure 4A, arrow [Fig4.mov, arrows]) but were not visible in transmitted light (data not shown), nor were they visible with DAPI counterstaining. However, DAPI staining confirmed that the location of these ER channels was entirely surrounded by DNA, indicating that they were tubular in nature rather than deep grooves (Figure 4B). ER-GFP was also prominent in the nuclear grooves found between nuclear lobes (Figure 4A, G). The saturation of the fluorescence signal apparent in these locations, and the lack of discernable structure because of this, resulted from the need to detect faint fluorescent signals from the nuclear channels.
Figure 4.
ER-GFP Confirms That Invaginations Can Pass Entirely through the Onion Nucleus.
(A) ER-GFP accumulates in nuclear grooves (G) with the fluorescence signal here becoming saturated so that several fainter ER strands can be imaged. These strands (arrow) represent an invagination that passes completely through the nucleus.
(B) DAPI labeling confirms that the location of the ER strands, marked by the arrow in the identical location as that shown in (A), occurs in the center of the nucleus.
Shown are simultaneous confocal optical sections of ER-GFP and DAPI-labeled DNA in a lens-shaped onion nucleus, seen in cross-section. Relative depths from the approximate center of the nucleus are indicated in micrometers at the lower right side of each section in (A). 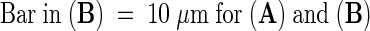 .
.
Actin Is Present in Grooves and Invaginations
The presence of actin bundles around the nucleus of permeabilized onion epidermal cells is demonstrated in Figure 5. Actin is also prominent in nuclear grooves visualized with DAPI-labeling of the DNA (Figure 5A, arrows) and DIC optics (Figure 5D, arrows). Actin bundles are also present in the highly convoluted nucleus, within invagination-like structures (Figures 5B, 5C, and 5F). These observations are consistent with the presence of cytoplasmic streaming within grooves (Figure 1D).
Figure 5.
F-Actin Is Present in Onion Nuclear Grooves and Surrounds the Nucleus.
Optical sectioning of a glycerol-permeabilized onion epidermal cell. Relative depths from the top of the nucleus are given in micrometers and are indicated at the bottom right of sections shown in (A). Arrows placed in identical locations in (A) to (D) show the locations of actin-containing nuclear grooves.
(A) A highly convoluted nuclear shape is defined by DAPI labeling of DNA.
(B) Rhodamine–phalloidin—labeled actin is present in several nuclear grooves and also apparently within the nucleus (F).
(C) False color images showing both DAPI (cyan) and actin (red) indicate the presence of actin in grooves and in other sites within the nucleus (Nc) that are presumed to be invaginations.
(D) Nuclear grooves (arrows) and nucleoli (Nc) were visible by transmitted light in DIC optics.
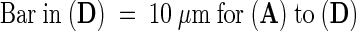 .
.
Onion Nuclear Grooves and Invaginations Are Stable Structures
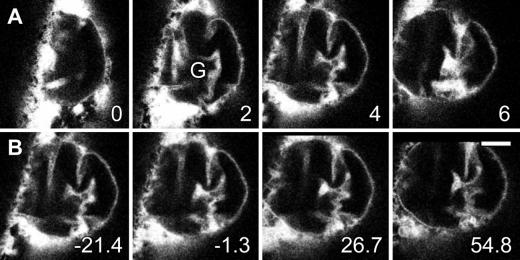
The plant nucleus is a motile structure, showing actin-dependent rotation and translocation within the cell (Chytilova et al., 2000). Although this dynamism partially confounded our analyses, time-lapse studies of ER-GFP–expressing and DAPI-labeled cells confirmed that the nuclear lobes, grooves, and invaginations of onion epidermal cells were stable, as shown in Figures 6 and 7 (online movie Fig6.mov). A DAPI-labeled nucleus of an ER-GFP–expressing cell having a deep groove between two lobes, with each lobe containing a single nucleolus, was visualized at a single focal plane by time-lapse confocal microscopy for 1 hr. Although the distribution of perinuclear ER showed rapid changes, groove structure as visualized with DAPI, ER-GFP, and DIC optics (Figures 6A to 6C, respectively) remained constant. Rendering of the full time-lapse data set into a movie sequence confirmed the dynamism of the perinuclear ER and the stability of the prominent nuclear groove; moreover, it demonstrated that the nucleus slowly rocked backward and forward (Fig6.mov). Observations of N-GFP–expressing onion epidermal cells demonstrated similar stability (data not shown).
Figure 6.
Structures of Invaginations and Grooves in Onion Nuclei Are Stable.
This confocal time-lapse sequence shows an ER-GFP–expressing cell at a single focal plane. Times in minutes are given at the lower left of time-lapse images shown in (A).
(A) DAPI fluorescence revealed the presence of two nucleoli (Nc) that were present in different nuclear lobes separated by one of several prominent nuclear grooves (G).
(B) ER-GFP fluorescence showed little change of nuclear-associated ER but substantial changes in ER patterns around the nucleus.
(C) Concurrent DIC images showed that the location of the two nucleoli (Nc) remained constant, demonstrating that the nucleus moved little through the focal plane.
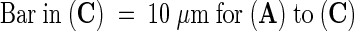 .
.
Figure 7.
Onion Nuclear Structure Is Stable, Even after Disruption of the Actin Cytoskeleton.
(A) Optical sections through an ER-GFP–expressing onion cell demonstrate numerous nuclear grooves (G). Relative depths from the top of the nucleus are shown in micrometers at the lower right of sections.
(B) Time-lapsed confocal sections show that grooves remain unchanged both before and after actin disruption. Times in minutes before (minus sign) and after the addition of 1 μM cytochalasin D are shown at the lower right of the time-lapse images. Because the nucleus moved slightly within the cell, and because of movement of the onion epidermal layer on the microscope stage, this sequence comprises images from optical stacks that were manually aligned.
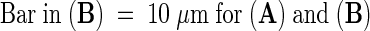 .
.
Disruption of actin with cytochalasin D (1.0 μM) does not destabilize the nuclear grooves, although such treatment inhibits cytoplasmic streaming (Figure 7). As Figure 7A shows, confocal optical sectioning revealed the presence of extensive grooving within a nucleus. The addition to that nucleus of 1 μM cytochalasin D (at 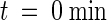 ), a concentration that disrupts actin organization in onion epidermal cells (data not shown), did not affect nuclear shape or organization of the nuclear grooves (Figure 7B); the cytochalasin did, however, stop cytoplasmic streaming (data not shown). Similarly, microtubule depolymerization with oryzalin (10 μM) did not modify nuclear structure (data not shown).
), a concentration that disrupts actin organization in onion epidermal cells (data not shown), did not affect nuclear shape or organization of the nuclear grooves (Figure 7B); the cytochalasin did, however, stop cytoplasmic streaming (data not shown). Similarly, microtubule depolymerization with oryzalin (10 μM) did not modify nuclear structure (data not shown).
Tobacco NT1 Nuclei Contain Invaginations and Grooves That May Originate during Cell Division
The nuclei of tobacco NT1 suspension-cultured cells are large, as large as 25 μm in diameter; appear to have a smooth surface, unlike the convoluted shape of onion nuclei; and contain a single, prominent nucleolus. DIOC6(3) labeling of NT1 cells showed the dynamic arrays of cortical and subcortical ER normally found in plant cells but also revealed membranous inclusions into many nuclei in the form of narrow invaginations or channels through the nucleus (data not shown). However, these inclusions were difficult to image because of image fading and a high background.
Stable transformants of the NT1 cell line that express ER-GFP have proved ideal for investigating nuclear invaginations, because at least 20% of the cells from cultures undergoing logarithmic growth and a high percentage of cells from a stationary-phase culture demonstrate various forms of nuclear channels and invaginations (S.L. Gupton, D.A. Collings, and N.S. Allen, unpublished data). As shown in Figure 8, ER-GFP–expressing cells showed normal patterns of dynamic ER organization in the cell cortex (Figure 8A, 0 μm). Figure 8A also shows two different types of ER-bounded nuclear inclusions: a long ER strand arched through the nucleus in multiple optical sections (arrows), whereas several smaller invaginations were present in the nearly spherical nuclear surface (arrowheads). When the fluorescence image was overlaid on the DIC image, the ER strand was clearly shown to be wrapped around the nucleolus (Figure 8C, arrows), and many of the invaginations projected in toward the nucleolus (arrowheads). However, neither channels nor invaginations were visible with transmitted light (Figure 8B). Furthermore, in most tobacco NT1 cells in which nuclear invaginations and channels were observed, they clearly associated with the nucleoli.
Figure 8.
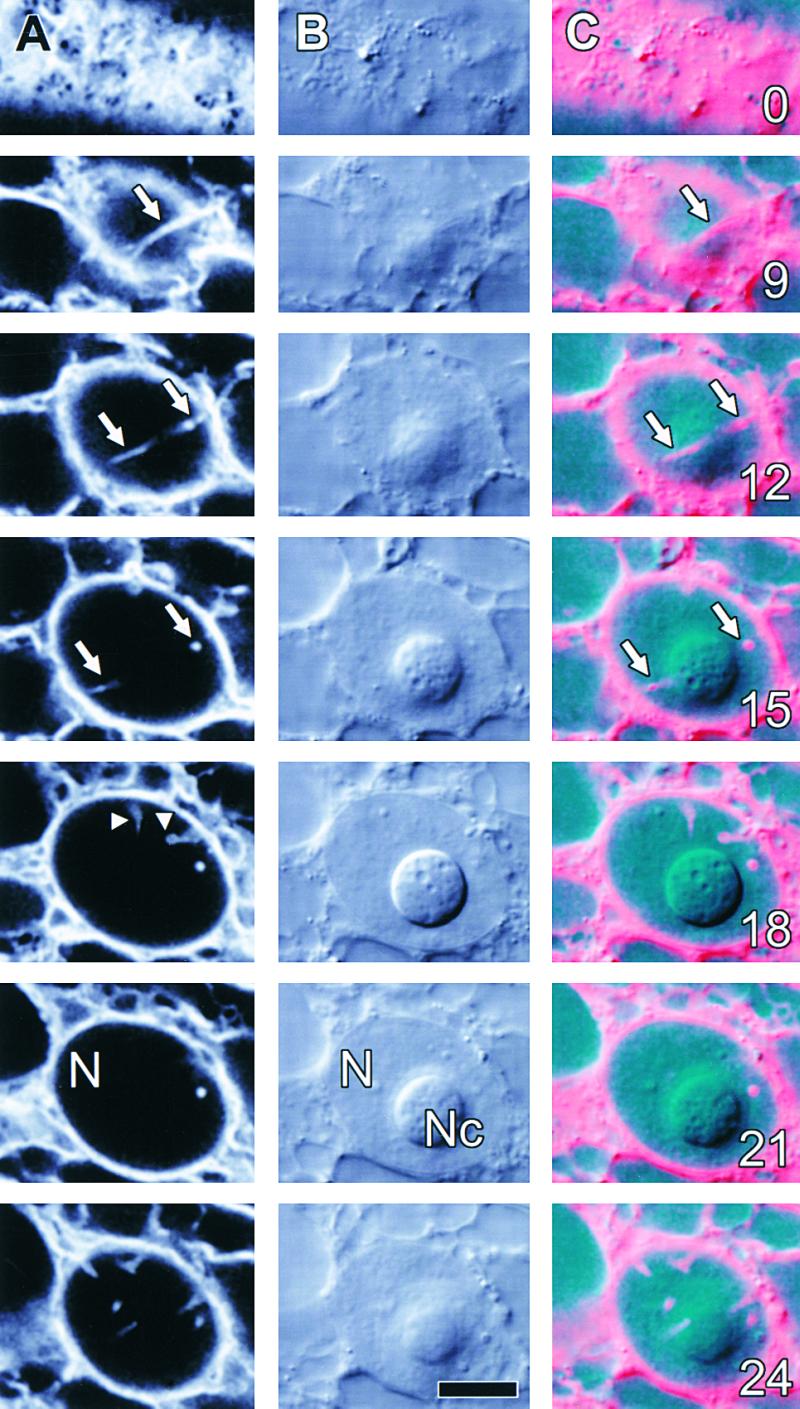
Nuclear Grooves and Invaginations Occur in Tobacco NT1 Suspension-Cultured Cells.
Selected optical sections of a tobacco NT1 cell stably expressing ER-GFP are shown at various intervals from the cell surface. The depths in micrometers are indicated at the lower right of sections in (C).
(A) ER-GFP demonstrates a single groove-like structure (arrows) and several invaginations (arrowheads) within the nucleus (N).
(B) Concurrent DIC images show a well-defined nucleolus (Nc) at the center of the nucleus (N). Grooves and invaginations could not be visualized in transmitted light.
(C) A composite image showing GFP (red) and transmitted light (cyan) demonstrates that the nuclear groove (arrows) wraps around the nucleolus.
 .
.
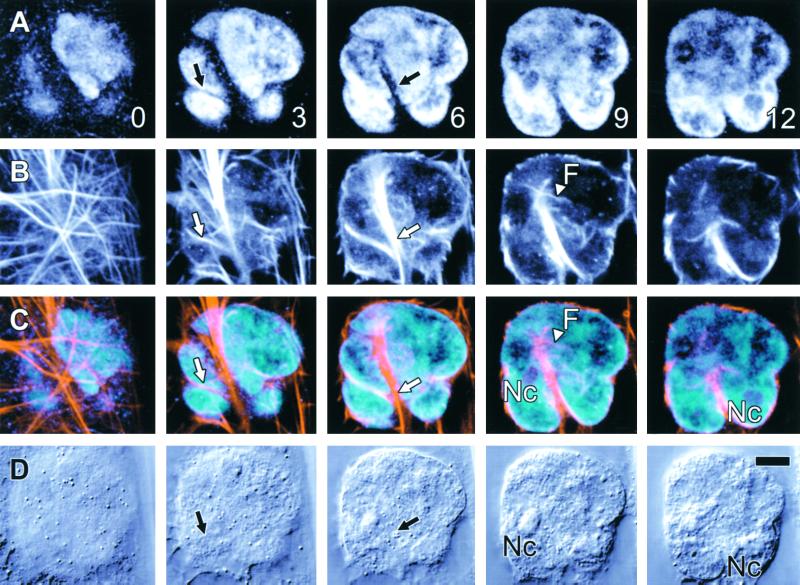
The presence of nuclear invaginations and channels in tobacco NT1 cells immediately after cytokinesis suggests that such structures form during cell division. Figure 9 shows that nuclear channels and invaginations occurred in cells immediately after reformation of the nuclear envelope, even before the cell plate had fully fused to the parent cell wall. Importantly, in both daughter nuclei, there was a close association of the nuclear channels and invaginations with the redeveloping nucleoli that had yet to return to their normal spherical shape (Figures 9A and 9C). Furthermore, in the majority of cases observed, the nuclear channels ran parallel to the presumed orientation of the spindle axis, as judged by the position of the expanding cell plate (Figure 9B).
Figure 9.
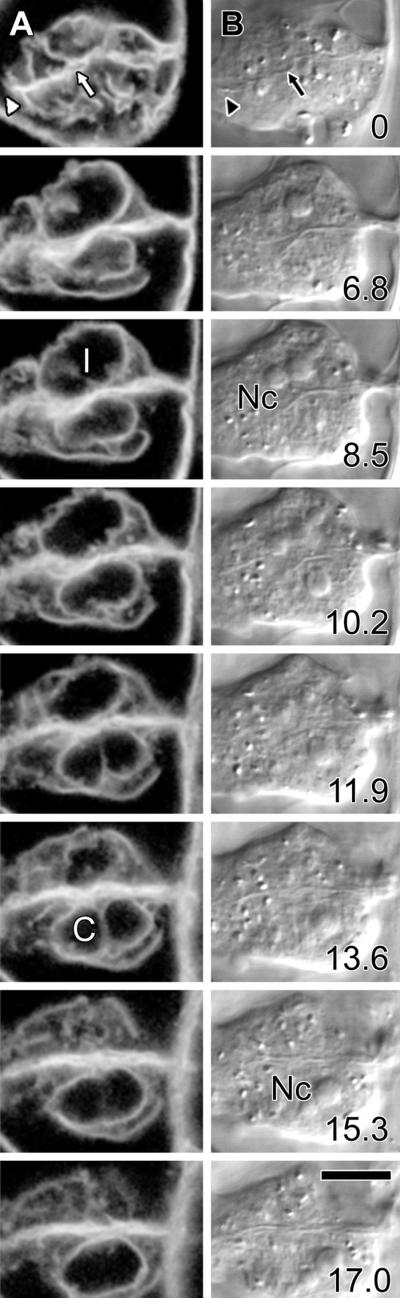
Tobacco NT1 Nuclear Invaginations and Grooves May Form during Cytokinesis.
Two tobacco cells undergoing cytokinesis demonstrate that transnuclear ER strands may form during cell division.
(A) ER-GFP fluorescence shows that cytokinesis is incomplete, because the cell plate has yet to fuse completely with the parent cell walls (arrowhead) and remains wavy (arrow), although the nuclear envelopes have reformed around the daughter nuclei. Two types of irregularities occur in these nuclei, with the upper nucleus showing an invagination (I), whereas the lower nucleus shows a channel that passes completely through the nucleus and is surrounded both above and below by the nucleus (C). Note that this channel lies parallel to the presumed spindle axis.
(B) Concurrent DIC images confirm that the cell plate has yet to fuse with the parent cell wall (arrow and arrowhead) and that neither channels nor invaginations are visible by transmitted light. However, comparison of fluorescence and transmitted light images indicates that both the channel and the invagination show close association with the redeveloping nucleoli (Nc) of the daughter cells.
Optical sections at different depths from the surface of the upper cell are shown in micrometers at the lower right of sections in (B).  .
.
As found with onion epidermal nuclear grooves and invaginations, the channels and invaginations present in tobacco NT1 cells, including the cell shown in Figure 8, were stable and remained unchanged for as long as 1 hr (data not shown).
DISCUSSION
Through dye-labeling experiments in untransformed cells, and through the expression of GFP targeted to either the nucleus or the lumen of the ER/nuclear envelope, we have demonstrated that the nuclei of several plants contain various structures such as grooves, invaginations, and channels that greatly increase the surface area of the nucleus. Although no function can yet be ascribed to these structures, their apparent association with nucleoli suggests that they could function in increasing nucleocytoplasmic trafficking of proteins and RNA.
Transient Expression of GFP Visualizes Grooves and Invaginations in Onion Nuclei
The nuclei of onion epidermal cells deviate from a lenslike shape in two ways—through the formation of nuclear grooves and nuclear invaginations. Invaginations are linear projections of cytoplasm into the nucleus, surrounded on all sides by the nucleus, whereas grooves are sheets of cytoplasm folded or trapped between lobes of the nucleus. (Nuclear channels were also observed on occasions, but we consider these a special case of nuclear invagination that extends entirely through the nucleus.) These structures share several common features, including being bounded by the nuclear envelope/ER, and three pieces of evidence confirming that they both contain cytoplasmic cores. First, the centers of both grooves and invaginations lack fluorescence when seen in cross-section; second, both grooves and invaginations contain actin, which supports vesicle motility and cytoplasmic streaming; and third, ER dynamics sometimes are observed within grooves and invaginations.
Overnight incubation of epidermal peels on agar plates, along with the ensuing transient expression of GFP constructs, does not induce the formation of nuclear grooves and invaginations in onion epidermal cells. These structures were also found in untransformed tissue and could be observed by DIC video microscopy, by DAPI and DIOC6(3) labeling, and by the microinjection of fluorescently labeled 70-kD dextrans, the molecular mass of which exceeded the exclusion limit for passive diffusion through nuclear pores (data not shown). Furthermore, we can see no immediately apparent way in which N-GFP, a β-glucuronidase (GUS)–GFP fusion protein with a nuclear localization sequence (Grebenok et al., 1997), or ER-GFP, a fusion of GFP with ER targeting and retention sequences (Haseloff et al., 1997), might induce the formation of nuclear grooves and invaginations. This is not necessarily the case for all GFP constructs. For example, although Ellenberg et al. (1997) could observe nuclear invaginations in untransformed cells, the frequency of invaginations was greater in cells expressing GFP fused to the lamin B receptor, a protein normally found on the nuclear face of the nuclear envelope, indicating that expression of this transgene might induce the formation of invaginations.
Nuclear grooves and invaginations appear to associate preferentially with nucleoli in onion epidermal cells. This was apparent with transmitted light (Figure 1D), DAPI-labeling of DNA (Figures 1A, 1B, 5C, and 6A), transmitted light in combination with DIOC6(3) labeling of internal membrane (Figures 1E and 1F), N-GFP localized to the nucleus (Figure 2A), and transmitted light in combination with ER-GFP (Figures 6B and 6C). Detailed measurements of this phenomenon, and an accompanying statistical analysis to determine whether the apparent association is indeed significant, lie beyond the scope of this article. However, because of the functional implications of such a localization (see below), further investigation is required.
If nuclear grooves and invaginations occur in untransformed onion epidermal cells, why have they not been characterized previously? Previous research has demonstrated the presence of groovelike structures in onion epidermal cells by DIC (Lichtscheidl and Url, 1988; N.S. Allen, personal observation) and electron microscopy (Kartusch et al., 2000). These methods, however, are limited in their ability to resolve structures in three dimensions. Confocal microscopy of DIOC6(3) labeling partially overcame such problems but was limited by rapid fading, the labeling of other organelles (including mitochondria), and the possibility that long-term treatment with DIOC6(3) might itself induce changes in ER organization. GFP is thus an ideal way to visualize invaginations and grooves, but even the approach used in this study has its limitations. Unlike the studies of Ellenberg et al. (1997) and Broers et al. (1999), in which fusion to specific proteins (lamin B receptor or lamin A, respectively) restricted GFP to the nuclear envelope, our ER-GFP construct diffused freely throughout the ER. This made the imaging of faint intranuclear structures against a background of ER accumulation around the nucleus difficult and often required the fluorescence signal from the ER to be saturated, as is apparent in Figure 4.
Nuclear Invaginations in Onion Are Similar to Structures Seen in Animal Cells
The nuclear invaginations found in onion epidermal cells are unlike the various small nuclear invaginations and nuclear vacuoles primarily associated with meiotic plant cells (Sassen, 1964; Aldrich and Vasil, 1970; Dickinson and Bell, 1970, 1972; Sheffield et al., 1979; Karasawa and Ueda, 1983; Sangwan, 1986; Li and Dickinson, 1987). Furthermore, because the structures in onion contain a cytoplasmic core, they are unlike the nuclear reticulum found in Lilium ovary placental cells, in which only the inner membrane of the nuclear envelope invaginates, so that the centers of these invaginations are continuous with the lumen of the ER rather than the cytoplasm (Singh and Walles, 1995; Singh et al., 1998).
Grooves and invaginations in onion may be similar to the deep cytoplasmic grooves occasionally reported in electron micrographs of various different plant nuclei, including Narcissus (Gunning and Steer, 1996, see plate 29) and Pisum (Bowes, 1996, see plate 2.23). Interestingly, extensive infolding has also been shown in onion epidermal nuclei by electron (Kartusch et al., 2000, Figure 3B) and DIC video microscopy (Lichtscheidl and Url, 1988). However, the true nature of these structures, and whether they represent nuclear grooves or invaginations, has not been determined.
Instead, the invaginations seen in onion nuclei seem most similar to those found in living nuclei of various animals. Animal nuclear invaginations are stable structures as large as 1 or 2 μm in diameter and can branch or traverse the nucleus, although the number and form of the invaginations vary from cell type to cell type (Ellenberg et al., 1997; Fricker et al., 1997; Lui et al., 1998a, 1998b; Broers et al., 1999). Electron microscopy has shown these invaginations to be bounded by the double nuclear membrane and to contain nuclear pores (Fricker et al., 1997). Importantly, animal nuclear invaginations often show a preferred orientation—perpendicular to the substrate—although once again the degree of orientation varies from cell type to cell type (Fricker et al., 1997; Broers et al., 1999). Animal nuclear invaginations can also associate with nucleoli, which is believed to have functional implications (Fricker et al., 1997).
Nuclear invaginations visualized with ER-GFP in plant cells were stable for extended times (as long as 1 hr) in both onion and tobacco, consistent with observations in various animal cell types (Ellenberg et al., 1997; Broers et al., 1999), although these observations stand in contrast to the apparently dynamic invaginations reported in various animal cells by Fricker et al. (1997). The factors controlling the stability of invaginations remain unknown. However, stability appears to reside within the nucleus itself, or more likely, within the nuclear envelope and the accompanying nuclear matrix (Yu and de la Espina, 1999; reviewed in de la Espina, 1995; Smith, 1999). This is because the cytoplasmic cytoskeleton is highly unlikely to contribute to nuclear invagination stability in either plant or animal nuclei. Although onion nuclear grooves and invaginations contain actin bundles, actin destabilization with cytochalasin neither modifies nor destabilizes the nuclear groove or invagination structure. Depolymerization of microtubules with oryzalin also did not modify grooves or invaginations, although this is less surprising because the interphase plant microtubules are primarily cortical rather than associated with the nucleus (Hepler and Hush, 1996). Similarly, although nuclear invaginations in animal cells have been observed to contain actin bundles (Suarez-Quian and Dym, 1992; Clubb and Locke, 1998), they also remain stable when either actin microfilaments or microtubules are depolymerized (Suarez-Quian and Dym, 1992).
Transient Expression of GFP Visualizes Grooves, Channels, and Invaginations in Tobacco Nuclei
The nuclei of tobacco NT1 cells deviate from spherical through the formation of nuclear grooves and nuclear invaginations, structures similar to those seen in onion nuclei. Although nuclear grooves were rare, extensive invaginations were seen that, when extending fully through the nucleus, formed nuclear channels. Nuclear grooves and invaginations were best studied in stably transformed cell lines expressing ER-GFP; however, the fact that these structures were also visible in untransformed cells labeled with DIOC6(3) indicates that they were not induced by the transformation process or by GFP expression.
The nuclear grooves and invaginations present in tobacco NT1 cells have considerable similarities to the grooves and invaginations seen in onion epidermal nuclei. For example, the association of channels and invaginations with the nucleoli of tobacco NT1 cells was pronounced and persisted throughout interphase, consistent with our observations of grooves and invaginations in onion and with others' observations of invaginations in various animal cells (Fricker et al., 1997). Rhodamine–phalloidin labeling visualizes actin bundles traversing the nucleus (Collings et al., 1998) and present in nuclear inclusions (Kengen et al., 1993) in tobacco BY-2 cells, which are closely related to the NT1 cell line. This not only suggests the presence of channels and invaginations but also indicates that such structures have cytoplasmic centers, similar to those found in onion nuclei. However, several differences exist between tobacco and onion cells. Tobacco NT1 nuclei are rounded, lacking the extensive grooves found in onion nuclei, and the invaginations, grooves, and channels seen with confocal microscopy were not visible by transmitted light. Such differences may explain why these nuclear structures have not previously been visualized in tobacco NT1 cells.
Nuclear invaginations and channels were regularly observed to have a preferred orientation in recently divided cells—adjacent to the reformed nucleoli and running parallel to the presumed axis of the spindle—although no preferred orientation was observed for the channels and invaginations at later stages of interphase. On the basis of dye studies, Fricker et al. (1997) proposed that the nuclear invaginations found in various living animal cells form during mitosis and remain in the nucleus through interphase. Subsequent experiments by Ellenberg et al. (1997) and Broers et al. (1999), using GFP fused to the lamin B receptor and lamin A, respectively, confirmed this by showing nuclear invaginations reappearing as the nuclear envelope reformed after mitosis. We are currently investigating the nature of invagination formation in synchronized NT1 cell cultures and determining whether their formation occurs through ER strands being trapped in the nucleus during chromatin condensation, as seems likely. However, the origin of grooves and invaginations in onion bulb epidermal cells, a storage tissue where cells remain undivided for many months, remains unknown.
Are Grooves and Invaginations Present in All Plant Nuclei?
The distribution of nuclear grooves and invaginations, and related structures, among different higher plants has not been determined. Using ER-targeted GFP, we visualized these structures in tobacco NT1 and onion epidermal cells and observed similar structures in the epidermal nuclei of cells in whole Nicotiana benthamiana plants (seed lines courtesy of Dr. David Baulcombe, John Innes Institute, Norwich, UK). Other studies have shown similar nuclear infoldings in electron micrographs of petals from Narcissus (Gunning and Steer, 1996), root cells from Pisum (Bowes, 1996), and radical and hyphal cells from the parasitic plant Cuscuta (dodder) (K. Vaughn, personal communication). Given the many examples of cells that lack nuclear invaginations, presumably grooves and invaginations are limited to specific cell types, possibly those with larger nuclei or those that have undergone multiple rounds of nuclear endoreduplication. Such specialization strongly hints at a functional significance for the presence of these structures in particular cells.
The Functions of Grooves and Invaginations
What, if any, functional significance do grooves and invaginations have in plant nuclei? Various roles have been proposed for the invaginations present in animal nuclei, including suggestions that increased nuclear surface area resulting from grooves and invaginations leads to increases in rates of nucleocytoplasmic transport and signaling. Fricker et al. (1997) suggested, partially on the preferential distribution of invaginations near nucleoli, that these structures might facilitate mRNA export to the cytoplasm. Alternatively, Lui et al. (1998a, 1998b) proposed that nuclear invaginations might contribute to the transmission of calcium signals from the cytoplasm to the nucleus. Calcium within the nuclear envelope can also regulate the functioning of nuclear pores (Perez-Terzic et al., 1997).
The structural properties of nuclear invaginations in plant cells and their similarities to the structures seen in animal cells suggest that similar functions might occur. A role in calcium signaling is possible: the plant ER acts as a calcium store (Malhó et al., 1998), and plant nuclei respond to calcium signals (Tähtiharju et al., 1997). However, the extensive DNA endoreduplication present in differentiated plant tissues, notably in epidermal cells, where grooves and invaginations are often found, and the accompanying increase in nuclear volume (Traas et al., 1999) make it more likely that grooves and invaginations are involved in the efficient trafficking of RNA out of the nucleus and of proteins into the nucleus. Although such transport could occur by diffusion, the movement of some mRNA in animal cells is an actin-dependent process (Bassell and Singer, 1997). Evidence also indicates that mRNA localization in plant cells is cytoskeleton dependent (Muench et al., 1998), as is the import of proteins and other signaling molecules from the cytoplasm into the plant nucleus (Smith and Raikhel, 1998, 1999). Thus, the presence of cytoplasmic streaming and actin bundles within grooves and invaginations would be consistent with a role in mRNA transport and protein import. Consistent with this hypothesis, nuclear pores are seen in the infoldings present in Narcissus nuclei (Gunning and Steer, 1996), and we are now determining the distribution of nuclear pores in onion epidermal cells to determine how grooves might function in trafficking between the nucleus and the cytoplasm.
Although the processes described above might be aided by the presence of grooves and invaginations, these structures might not be necessary, for mRNA export or protein/signal import. However, a function for these structures in nucleocytoplasmic trafficking would take on further significance if a relationship between chromosome positioning and the orientation of nuclear grooves could be established. In animal cells, fluorescent in situ hybridization of fixed nuclei (Lamond and Earnshaw, 1998) and GFP techniques in living cells (Marshall et al., 1997) have demonstrated that chromosomes localize into discrete and reproducible zones that remain constant throughout interphase and within different cells. Although the application of such chromosome painting methods to plant nuclei has been limited to systems in which chromosomes derive from different parent species, a similar extent of chromosome organization is apparent (Heslop-Harrison et al., 1990; Abranches et al., 1998; Gonzalez-Melendi et al., 2000; reviewed in Heslop-Harrison and Bennett, 1990; Franklin and Cande, 1999). Because onion epidermal cells show a consistent distribution of nucleoli with respect to nuclear lobes and grooves, with nucleoli generally occurring in separate lobes, and because invaginations in tobacco nuclei show a marked preference for localizing near nucleoli, some regular chromosome zonation might exist relative to the position of nuclear grooves and invaginations. The potential for the functional significance of such organization, with respect to the export of mRNA or signaling, suggests that further investigation of chromosome organization in onion and tobacco nuclei is warranted.
METHODS
Labeling of Onion and Tobacco NT1 Cells
To label their DNA, onion (Allium cepa cv Tango; Martin Produce, Greeley, CO) and tobacco (Nicotiana tabacum NT1 suspension culture line) cells were incubated for 60 min with 1.0 μg mL−1 4′,6-diamidino-2-phenylindole (DAPI), rinsed briefly, and viewed immediately. Internal membranes of onion and tobacco cells were labeled for 1 to 2 min with 20 to 40 μg mL−1 3,3′-dihexyloxacarbocyanine iodide (DIOC6(3)) and viewed immediately.
Rhodamine–Phalloidin Labeling of Actin in Onion 24 Epidermal Cells
Actin was labeled in permeabilized epidermal peels by the methods of Olyslaegers and Verbelen (1998). Inner epidermal peels were floated for 20 min on cytoskeleton stabilization buffer (100 mM Pipes, pH 6.9, 10 mM EGTA, 4 mM MgSO4, and 0.2 M mannitol) containing 2% (w/v) glycerol and 0.2 μM rhodamine–phalloidin (Molecular Probes, Eugene, OR), then washed with and mounted in cytoskeleton stabilization buffer containing 0.1% p-phenylenediamine and 1 μg mL−1 DAPI. Attempts to colocalize actin and endoplasmic reticulum (ER)–targeted green fluorescent protein (GFP) in permeabilized cells failed because the glycerol treatment compromised the integrity of the ER.
Transient Expression of GFP in Onion Epidermal Cells
Epidermal cells of onion bulbs were transformed according to Scott et al. (1999). Epidermal peels from the inner epidermis of onion scales were placed mesophyll-side down on 2.0% (w/v) agar plates (Murashige and Skoog [1962] media supplemented with 3% sucrose) and bombarded under vacuum at 1100 mm Hg (Biolistic PDS-1000/He; Bio-Rad) with 1-μm-diameter gold particles coated with DNA. Two constructs were used. Nuclear-targeted GFP (N-GFP) consisted of a nuclear localization signal–GFP–β-glucuronidase (GUS) fusion protein, behind the cauliflower mosaic virus 35S promoter inserted into a pUCAP vector (Grebenok et al., 1997); GFP targeted to the ER (ER-GFP) used a signal sequence coupled to GFP and the HDEL ER-retention sequence (mGFP5; Haseloff et al., 1997) behind the 35S promoter inserted into a pUCAP vector. Before observation, peels were incubated for 18 to 24 hr at 22°C in the dark. Some preparations were also counterstained with DAPI, as described above.
Tobacco NT1 Cell Lines Expressing ER-Targeted GFP
Protoplasts of the tobacco NT1 suspension culture line were transformed by electroporation with a pUC-based vector containing the cauliflower mosaic virus 35S promoter, signal sequence, mGFP5 construct, HDEL retention sequence, and a nopaline synthase terminator. Callus cells were regenerated, and after 3 weeks, a stable transformant was selected on the basis of high GFP expression. The selected cell line was grown in suspension culture with weekly subculturing in Murashige and Skoog medium supplemented with 3% sucrose, 1 μg mL−1 thiamine, 100 μg mL−1 inositol, 0.2 μg mL−1 2,4-dichlorophenoxy acetic acid, and 255 μg mL−1 KH2PO4, in full darkness at 24°C, with shaking at 120 rpm.
Microscopy
Confocal fluorescence and concurrent differential interference contrast (DIC) images were recorded in optical section and time series modes with Leica TCS NT and SP confocal systems with 20 × NA 0.8 dry, 40 × NA 1.25 oil-immersion, and 63 × NA 1.2 water-immersion lenses (Leica, Wetzlar, Germany). Excitation sources were 351 and 361 nm for DAPI, 488 nm for GFP and DIOC6(3), and 568 nm for rhodamine–phalloidin. Fluorescence was recorded either with band-pass filters (420 to 460 nm for DAPI, 515 to 545 nm for GFP and DIOC6(3), and 585 to 615 nm for rhodamine–phalloidin) or with the SP-scanning head set to similar wavelengths. DIC images were recorded concurrently with a red helium/neon laser (633 nm) and a red filter placed in front of the transmitted light detector, taking advantage of the stable helium/neon laser. For a single experiment (see Figure 3), a Zeiss (Carl Zeiss, Inc., Thornwood, NY) confocal microscope with a 40 × NA 1.2 water-immersion lens was used, with excitation from an argon/krypton laser at 488 nm and emission recorded with a 515- to 545-nm band-pass filter. Confocal optical stacks were transformed into three-dimensional reconstructions with a surface shadowing algorithm in the Leica software; orthogonal slices were produced by using Metamorph (Universal Imaging, West Chester, PA). All images were processed with Adobe Photoshop (Grand Prairie, TX).
Supplementary Material
Acknowledgments
We thank George Allen, Eric Davies, and Timothy Oliver (North Carolina State University [NCSU]) for technical assistance and comments; Dana Moxley (NCSU) for culturing tobacco NT1 cells and conducting the DIOC6(3) experiments; and Kristi de Courcy (Virginia Tech, Blacksburg, VA) for assistance with the Zeiss confocal microscope. We also thank George Allen (NCSU) for his generosity in providing the tobacco NT1 cell line transformed with mGFP5 (originally constructed by Jim Haseloff, Medical Research Council, Cambridge, UK), Jim Haseloff (MRC) for the ER-GFP (mGFP5) clone, and David Galbraith (University of Arizona, Tucson, AZ) for the N-GFP construct. Ingeborg Pauluzzi (University of Vienna, Austria) kindly provided a video copy of Lichtscheidl and Url's 1988 film. Early studies were conducted at the Marine Biological Laboratory, Woods Hole, MA. This work was supported by the North Carolina State University (NCSU) NASA Specialized Center of Research and Training Grant No. NAGW-4984 and by NCSU North Carolina Agricultural Research Service grants.
Footnotes
Online version contains Web-only data
References
- Abranches, R., Beven, A.F., Aragón-Alcaide, L., and Shaw, P.J. (1998). Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 143 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich, H.C., and Vasil, I.K. (1970). Ultrastructure of the postmeiotic nuclear envelope in microspores of Podocarpus macrophyllus. J. Ultrastruct. Res. 32 307–315. [DOI] [PubMed] [Google Scholar]
- Bassell, G., and Singer, R.H. (1997). mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 9 109–115. [DOI] [PubMed] [Google Scholar]
- Bowes, B.G. (1996). A Colour Atlas of Plant Structure. (London: Manson Publishing).
- Broers, J.L.V., Machiels, B.M., van Eys, G.J.J.M., Kuijpers, H.J.H., Manders, E.M.M., van Driel, R., and Ramaekers, F.C.S. (1999). Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 112 3463–3475. [DOI] [PubMed] [Google Scholar]
- Chytilova, E., Macas, J., Sliwinska, E., Rafelski, S.M., Lambert, G.M., and Galbraith, D.W. (2000). Nuclear dynamics in Arabidopsis thaliana. Mol. Biol. Cell 11 2733–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clubb, B.H., and Locke, M. (1998). 3T3 cells have nuclear invaginations containing F-actin. Tissue Cell 30 684–691. [DOI] [PubMed] [Google Scholar]
- Collings, D.A., Asada, T., Allen, N.S., and Shibaoka, H. (1998). Plasma membrane–associated actin in Bright Yellow 2 tobacco cells: Evidence for interaction with microtubules. Plant Physiol. 118 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Espina, S.M.P. (1995). Nuclear matrix isolated from plant cells. Int. Rev. Cytol. 162B 75–139. [DOI] [PubMed] [Google Scholar]
- Dickinson, H.G., and Bell, P.R. (1970). Nucleocytoplasmic interaction at the nuclear envelope in post-mitotic microspores of Pinus banksiana. J. Ultrastruct. Res. 33 356–359. [DOI] [PubMed] [Google Scholar]
- Dickinson, H.G., and Bell, P.R. (1972). Structures resembling nuclear pores at the orifice of nuclear invaginations in developing microspores of Pinus banksiana. Dev. Biol. 27 425–429. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E.D., Moriera, J.E., Smith, C.L., Presley, J.F., Worman, H.J., and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: Targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, A.E., and Cande, W.Z. (1999). Nuclear organization and chromosome segregation. Plant Cell 11 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker, M., Hollinshead, M., White, N., and Vaux, D. (1997). Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane–bound invaginations of the nuclear envelope. J. Cell Biol. 136 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Melendi, P., Beven, A., Boudonck, K., Abranches, R., Wells, B., Dolan, L., and Shaw, P. (2000). The nucleus: A highly organized but dynamic structure. J. Microsc. 198 199–207. [DOI] [PubMed] [Google Scholar]
- Grebenok, R.J., Pierson, E., Lambert, G.M., Gong, F.C., Afonso, C.L., Haldemann, C.R., Carrington, J.C., and Galbraith, D.W. (1997). Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11 573–586. [DOI] [PubMed] [Google Scholar]
- Gunning, B.E.S., and Steer, M.W. (1996). Plant Cell Biology: Structure and Function. (Boston: Jones and Bartlett Publishers).
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., and Hush, J.M. (1996). Behavior of microtubules in living plant cells. Plant Physiol. 112 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, E.J., Tague, B.W., Hoffman, L.M., Kjemtrup, S.E., and Chrispeels, M.J. (1990). Retention of phytohemagglutinin with carboxyterminal tetrapeptide KDEL in the nuclear envelope and the endoplasmic reticulum. Planta 182 305–312. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J., and Heslop-Harrison, Y. (1989). Conformation and movement of the vegetative nucleus of the angiosperm pollen tube: Association with the actin cytoskeleton. J. Cell Sci. 93 299–308. [Google Scholar]
- Heslop-Harrison, J.S., and Bennett, M.D. (1990). Nuclear architecture in plants. Trends Cell Biol. 12 401–405. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S., Leitch, A.R., Schwarzacher, T., and Anamthawat-Jónsson, K. (1990). Detection and characterization of 1B/1R translocations in hexaploid wheat. Heredity 65 385–392. [Google Scholar]
- Holzwarth, G., Webb, S.C., Kubinski, D.J., and Allen, N.S. (1997). Improving DIC microscopy with polarization modulation. J. Microsc. 188 249–254. [Google Scholar]
- Karasawa, K., and Ueda, K. (1983). Occurrence of nuclear vacuole in meiotic prophase nuclei in compositae. Caryologia 36 145–153. [Google Scholar]
- Kartusch, R., Lichtscheidl, I.K., and Weidinger, M.-L. (2000). Brefeldin A induces callose formation in onion inner epidermal cells. Protoplasma 212 250–261. [Google Scholar]
- Kengen, H.M.P., van Amstel, T., and Knuiman, B. (1993). Basket-shaped structures formed by F-actin in the nuclei of elongating cells of Nicotiana tabacum. Can. J. Bot. 71 725–731. [Google Scholar]
- Lamond, A.I., and Earnshaw, W.C. (1998). Structure and function in the nucleus. Science 280 547–553. [DOI] [PubMed] [Google Scholar]
- Li, F.L., and Dickinson, H.G. (1987). The structure and function of nuclear invaginations characteristic of microsporogenesis in Pinus banksiana. Ann. Bot. 60 321–330. [Google Scholar]
- Lichtscheidl, I.K., and Url, W.G. (1988). The cytoplasm of the plant cell: Allium cepa inner epidermis. Film C2209. (Vienna, Austria: Österreichisches Bundesinstitut für den Wissenschaftlichen Film).
- Lui, P.P.Y., Kong, S.K., Kwok, T.T., and Lee, C.Y. (1998. a). The nucleus of HeLa cell contains tubular structures for Ca2+ signalling. Biochem. Biophys. Res. Commun. 247 88–93. [DOI] [PubMed] [Google Scholar]
- Lui, P.P.Y., Lee, C.Y., Tsand, D., and Kong, S.K. (1998. b). Ca2+ is released from the nuclear tubular structure into nucleoplasm in C6 glioma cells after stimulation with phorbol ester. FEBS Lett. 432 82–87. [DOI] [PubMed] [Google Scholar]
- Majno, M., Shea, S.M., and Leventhal, M. (1969). Endothelial contraction induced by histamine-type mediators. J. Cell Biol. 42 647–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhó, R., Moutinho, A., van der Luit, A., and Trewavas, A.J. (1998). Spatial characteristics of calcium signalling: The calcium wave as a basic unit in plant cell calcium signalling. Philos. Trans. R. Soc. Lond. Ser. B 353 1463–1473. [Google Scholar]
- Marshall, W.F., Straight, A., Marko, J.F., Swedlow, J., Dernburg, A., Belmont, A., Murray, A.W., Agard, D.A., and Sedat, J.W. (1997). Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7 930–939. [DOI] [PubMed] [Google Scholar]
- Muench, D.G., Wu, Y., Coughlan, S.J., and Okita, T.W. (1998). Evidence for a cytoskeleton-associated binding site involved in prolamine mRNA localization to the protein bodies in rice endosperm tissue. Plant Physiol. 116 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Olyslaegers, G., and Verbelen, J. (1998). Improved staining of F-actin and co-localization of mitochondria in plant cells. J. Microsc. 192 73–77. [Google Scholar]
- Perez-Terzic, C., Jaconi, M., and Clapham, D.E. (1997). Nuclear calcium and the regulation of the nuclear pore complex. Bioessays 19 787–792. [DOI] [PubMed] [Google Scholar]
- Ridge, R.W., Uozumi, I., Plazinski, J., Hurley, U.A., and Williamson, R.E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40 1253–1261. [DOI] [PubMed] [Google Scholar]
- Sangwan, R.S. (1986). Formation and cytohistochemistry of nuclear vacuoles during meiosis in Datura. Eur. J. Cell Biol. 40 210–218. [Google Scholar]
- Sassen, M.M.A. (1964). Fine structure of Petunia pollen grain and pollen tube. Acta Bot. Neerl. 13 175–181. [Google Scholar]
- Scott, A.C., Wyatt, S., Tsou, P., Robertson, D., and Allen, N.S. (1999). Model system for plant cell biology: GFP imaging in living onion epidermal cells. BioTechniques 26 1125–1132. [DOI] [PubMed] [Google Scholar]
- Sheffield, E., Cawood, A.H., Bell, P.R., and Dickinson, H.G. (1979). The development of nuclear vacuoles during meiosis in plants. Planta 146 597–601. [DOI] [PubMed] [Google Scholar]
- Singh, S., and Walles, B. (1995). Ultrastructural differentiation of the ovarian transmitting tissue in Lilium regale. Ann. Bot. 75 455–462. [Google Scholar]
- Singh, S., Lazzaro, M.D., and Walles, B. (1998). The nuclear reticulum in placental cells of Lilium regale is a part of the endomembrane system. Protoplasma 203 144–152. [Google Scholar]
- Smith, H.B. (1999). The nucleocytoplasmic continuum: Pushing the (nuclear) envelope. Plant Cell 11 989–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., and Raikhel, N.V. (1998). Nuclear localization signal receptor importin associates with the cytoskeleton. Plant Cell 10 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., and Raikhel, N.V. (1999). Protein targeting to the nuclear pore. What can we learn from plants? Plant Physiol. 119 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Quian, C.A., and Dym, M. (1992). Characterization of Sertoli cell perinuclear filaments. Microsc. Res. Tech. 20 219–231. [DOI] [PubMed] [Google Scholar]
- Tähtiharju, S., Sangwan, V., Monroy, A.F., Dhindsa, R.S., and Borg, M. (1997). The induction of kin genes in cold-acclimating Arabidopsis thaliana. Evidence of a role for calcium. Planta 203 442–447. [DOI] [PubMed] [Google Scholar]
- Traas, J., Hülskamp, M., Gendreau, E., and Höfte, H. (1999). Endoreduplication and development: Rule without dividing? Curr. Opin. Plant Biol. 1 498–503. [DOI] [PubMed] [Google Scholar]
- Yi, W., Shi-Jie, Y., Ming-Yi, L., and Cheng-Hou, L. (1994). Nuclear invaginations and nuclear vacuole formation in several plants. Acta Bot. Sin. 36 963–966. [Google Scholar]
- Yu, W., and de la Espina, S.M.P. (1999). The plant nucleoskeleton: Ultrastructural organization and identification of NuMA homologues in the nuclear matrix and mitotic spindle of plant cells. Exp. Cell Res. 246 516–526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.