Abstract
Disease resistance proteins containing a nucleotide binding site (NBS) and a leucine-rich repeat (LRR) region compose the largest class of disease resistance proteins. These so-called NBS-LRR proteins confer resistance against a wide variety of phytopathogens. To help elucidate the mechanism by which NBS-LRR proteins recognize and transmit pathogen-derived signals, we analyzed mutant versions of the Arabidopsis NBS-LRR protein RPS2. The RPS2 gene confers resistance against Pseudomonas syringae strains carrying the avirulence gene avrRpt2. The activity of RPS2 derivatives in response to AvrRpt2 was measured by using a functional transient expression assay or by expressing the mutant proteins in transgenic plants. Directed mutagenesis revealed that the NBS and an N-terminal leucine zipper (LZ) motif were critical for RPS2 function. Mutations near the N terminus, including an LZ mutation, resulted in proteins that exhibited a dominant negative effect on wild-type RPS2. Scanning the RPS2 molecule with a small in-frame internal deletion demonstrated that RPS2 does not have a large dispensable region. Overexpression of RPS2 in the transient assay in the absence of avrRpt2 also led to an apparent resistant response, presumably a consequence of a low basal activity of RPS2. The NBS and LZ were essential for this overdose effect, whereas the entire LRR was dispensable. RPS2 interaction with a 75-kD protein (p75) required an N-terminal portion of RPS2 that is smaller than the region required for the overdose effect. These findings illuminate the pathogen recognition mechanisms common among NBS-LRR proteins.
INTRODUCTION
Plants have evolved a surveillance system to recognize attacking pathogens and to induce appropriate defense responses. The specificity of pathogen recognition often is determined by a pathogen avirulence (avr) gene and a corresponding plant resistance (R) gene (Hammond-Kosack and Jones, 1997). Accumulating evidence indicates that R gene products function either directly or indirectly as receptors for the products of avr genes, thereby providing an early indication of pathogen attack (Scofield et al., 1996; Tang et al., 1996; Thomas et al., 1997; Ellis et al., 1999; Leister and Katagiri, 2000). Many avr genes appear to encode virulence factors, which in the case of bacterial pathogens, are transported directly from the pathogen into plant cells by way of a pathogen-encoded type III secretion system (Mudgett and Staskawicz, 1998; Galan and Collmer, 1999). Avr–R interactions lead to activation of various host defense responses, including a specialized type of programmed cell death known as the hypersensitive response (HR; Hammond-Kosack and Jones, 1997). According to their encoded protein structures, R genes can be divided into four major classes, among which the largest is the nucleotide binding site (NBS)–leucine-rich repeat (LRR) class (Hammond-Kosack and Jones, 1997). NBS-LRR proteins are numerous—the Arabidopsis genome is estimated to have ∼200 members in this gene family (Meyers et al., 1999)—and confer resistance against a wide variety of pathogens and pests, including viruses, bacteria, fungi, oomycetes, nematodes, and insects (Hammond-Kosack and Jones, 1997; Milligan et al., 1998; Rossi et al., 1998). Although many NBS-LRR resistance proteins have been identified in a wide variety of plant species, the molecular mechanism by which NBS-LRR proteins recognize pathogens and interact with downstream signal transduction components is poorly understood.
The Arabidopsis NBS-LRR R gene RPS2 confers resistance to Pseudomonas syringae strains that express the avr gene avrRpt2 (Bent et al., 1994; Mindrinos et al., 1994). Several lines of evidence suggest that AvrRpt2 protein is transported into plant cells, in which it interacts with and is recognized by the RPS2 protein–based surveillance system (Leister et al., 1996; McNellis et al., 1998; Mudgett and Staskawicz, 1999; Leister and Katagiri, 2000): (1) the ability of AvrRpt2 to elicit an HR and other plant defense responses depends on the function of the hrp type III secretion system; (2) direct expression of avrRpt2 in plant cells is sufficient to induce RPS2-dependent responses; (3) AvrRpt2 appears to be processed by a plant intracellular protease; (4) RPS2 probably is localized in the plant cytoplasm; and (5) AvrRpt2 and RPS2 can form an immunoprecipitable complex in the plant cell. Observations similar to (1) and (2) have been made for several bacterial avr genes (Bonas and van den Ackervaken, 1997). Moreover, similar to (3), several bacterial Avr proteins apparently are acylated in the plant cell (Nimchuk et al., 2000). Observations equivalent to (2) also have been made for viral avr genes (Bendahmane et al., 1999; Erickson et al., 1999), which correspond to NBS-LRR R genes. Regarding (4), not only RPS2 but also other NBS-LRR proteins are predicted to be cytoplasmic, based on their primary structures (Hammond-Kosack and Jones, 1997). Peripheral association with the plasma membrane has been demonstrated for the Arabidopsis NBS-LRR R protein RPM1 (Boyes et al., 1998). In summary, NBS-LRR proteins as a group appear to constitute a surveillance system that functions inside plant cells to detect pathogen-derived molecules. A recent report (Jia et al., 2000) that the rice Pi-ta R protein, which is closely related to NBS-LRR proteins but does not have the consensus repeat structure in the leucine-rich domain, can directly bind the corresponding fungal Avr protein suggests that NBS-LRR proteins may function as receptors of pathogen-derived molecules.
Here, we report structure–function analysis of the Arabidopsis RPS2 protein. RPS2 does not have a large dispensable region. The N-terminal portion, which includes the NBS and a leucine zipper (LZ) motif, appears to be crucial for downstream signaling. A 75-kD protein (p75) that interacts with RPS2 in vivo interacts with the N-terminal portion of RPS2 and may be involved in downstream signaling functions.
RESULTS
NBS and LZ Motifs Are Required for RPS2 Function
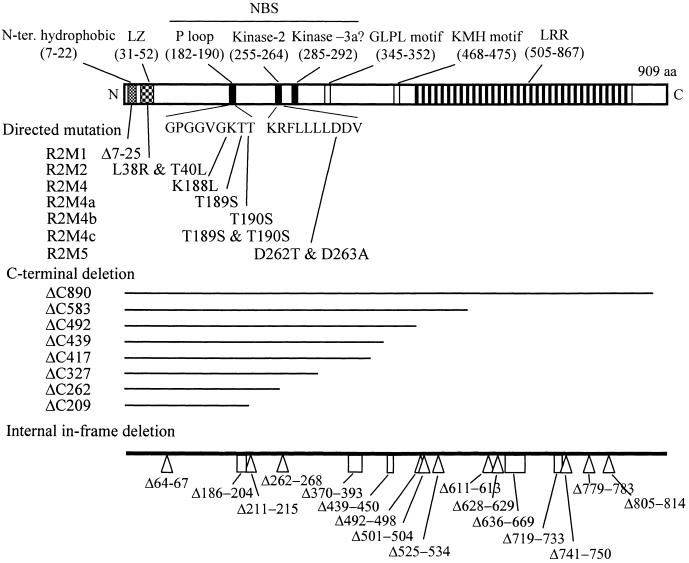
RPS2 contains several possible functional peptide motifs, including an N-terminal hydrophobic region, an LZ region, and an NBS (Bent et al., 1994; Mindrinos et al., 1994). Figure 1 indicates directed mutations made in these motifs. The 19–amino acid residue N-terminal hydrophobic region and the adjacent arginine residues at positions 24 and 25 are deleted (Δ7-25) in R2M1. R2M2 carries two amino acid substitution mutations in the LZ (L38R and T40L), which should disrupt presumed ability of the LZ to engage in protein–protein interactions by way of coiled-coil formation. Two submotifs of the NBS, the P-loop and the kinase-2 motif (Traut, 1994), also were mutagenized. In R2M4, a lysine residue that is highly conserved in P-loops from various proteins is replaced with a leucine residue (K188L). In R2M5, the highly conserved tandem aspartate residues in the kinase-2 motifs of NBS-LRR proteins (Meyers et al., 1999) are replaced with a threonine and an alanine (D262T and D263A). For a negative control, we used R2M7, in which W235 is replaced with a stop codon (Leister et al., 1996)—the same mutation as in the rps2-101C allele.
Figure 1.
RPS2 Derivatives Used in the Study.
All the RPS2 derivatives used in the study are depicted in a schematic representation of the RPS2 primary structure (909 amino acid [aa] residues). The N-terminal (N-ter.) hydrophobic region, LZ, NBS motifs, LRR, and two of the motifs (GLPL and KMH) that are highly conserved among NBS-LRR proteins are shown. Amino acid substitution mutants are indicated. The lines for the C-terminal deletion mutants represent the amino acid sequence regions that remain in the mutants. The number in each C-terminal deletion mutant name represents the position of the first deleted amino acid residue. The positions of small in-frame internal deletions are indicated along the bottom line. The numbers in each in-frame internal deletion name represent the positions of the first and the last deleted amino acid residues. The mutations corresponding to deletions of <11 amino acid residues are represented by triangles; deletions of >10 amino acid residues are represented by rectangles, the widths of which correspond to the sizes of the deletions.
rps2-101C plants were transformed with the RPS2 mutant derivatives ligated (as described in Methods) to a 1.4-kb Arabidopsis genomic sequence upstream of the RPS2 coding sequence. As previously reported, this region was sufficient to express a wild-type RPS2 cDNA clone and complement the rps2-101C mutation (Leister et al., 1996). For each mutant, at least six independent transformants were tested for the ability to mount an HR in response to avrRpt2. Leaves of the transgenic plants were hand-inoculated with Pseudomonas syringae pv phaseolicola strain NPS3121 expressing avrRpt2 on the plasmid vector pLAFR3 (3121/avrRpt2) or with the same P. syringae strain carrying a vector control (3121/−) at a dose of 108 colony-forming units mL−1. The typical resistance response to this interaction is development of a confluent HR within 1 day after infiltration. As shown in Table 1, none of the rps2-101C plants carrying any of the mutant RPS2 transgenes developed such an HR in response to 3121/avrRpt2, whereas rps2-101C plants carrying the wild-type transgene did. In this particular experiment, only one transgenic plant for the RPS2 wild-type transgene was tested, but we have analyzed many independent lines transformed with the wild-type RPS2 transgene in other experiments with similar results (e.g., Leister et al., 1996). None of the plants showed an HR in response to 3121/−.
Table 1.
HR in Response to Infection by P. syringae Expressing avrRpt2
| Transgenesa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Recipient | No transgene | RPS2 WTb | R2M1 | R2M2 | R2M4 | R2M5 | R2M7 | Δ501–504 | Δ611–613 | Δ805–814 |
| rps2-101C | 0/2 | 1/1 | 0/6 | 0/8 | 0/8 | 0/6 | 0/6 | 0/8 | 0/8 | 0/8 |
| RPS2 wild type | 2/2 | 5/5 | 1/7 | 1/7 | 8/8 | 3/3 | 5/5 | 8/8 | 8/8 | 8/8 |
The numbers indicate the number of HR-positive plants per the number of independent transgenic lines tested.
WT, wild type.
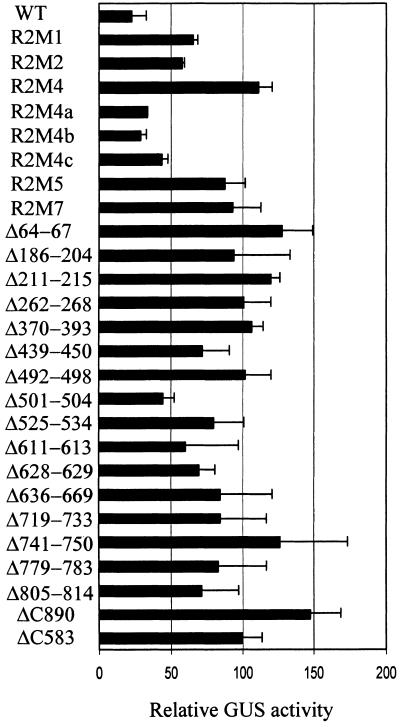
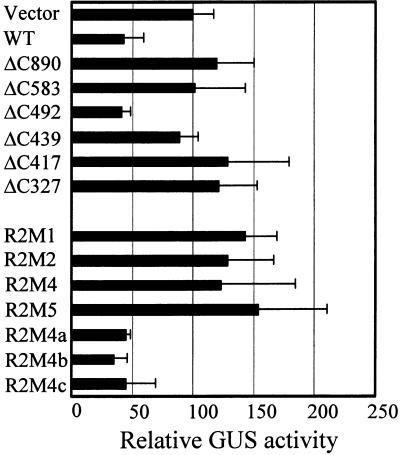
Figure 2 shows the results for the RPS2 derivatives analyzed with the biolistic bombardment transient expression assay (Leister et al., 1996). In this assay, the response resulting from the AvrRpt2–RPS2 interaction is detected as a decrease in β-glucuronidase (GUS) activity from the cotransformed GUS reporter gene (uidA), which is presumably associated with RPS2-mediated elicitation of cell death. In Figure 2, the GUS activity obtained with the indicated RPS2 derivative cobombarded with avrRpt2 is expressed as a percentage of the GUS activity obtained with the same RPS2 derivative cobombarded with the vector control. The value for an inactive RPS2 derivative (which does not respond to AvrRpt2) is ∼100, whereas that for an active one, such as wild-type RPS2, is <50. All four RPS2 mutants described above—R2M1, R2M2, R2M4, and R2M5—were judged to be inactive in the transient expression assay. Although R2M1 (N-terminal hydrophobic region deletion) and R2M2 (LZ mutant) reproducibly showed less GUS activity than did R2M4 (P-loop mutant), they were clearly more active than was wild-type RPS2. Because the results obtained by analysis of transgenic plants and by the transient expression assay correlated well, we used the transient expression assay for most subsequent analyses.
Figure 2.
RPS2 Activity of RPS2 Derivatives Measured by a Transient Expression Assay.
RPS2 activity of RPS2 derivatives in response to avrRpt2 was measured by using a transient expression assay, with a decrease in GUS activity being a measure of the RPS2 activity and with luciferase activity as a normalization factor for the transformation efficiency, as previously described (Leister et al., 1996). Similar assays were performed with RPS2 derivatives in the absence of avrRpt2. For each derivative, the data are expressed as the percentage of the normalized GUS activity observed in the presence of avrRpt2 divided by that in the absence of avrRpt2. The more active a derivative, the less its relative GUS activity. Error bars represent the standard deviation. WT, wild type.
In the P-loop region, two threonine residues (positions 189 and 190 in RPS2) are highly conserved among the NBS-LRR proteins (Meyers et al., 1999); in the general P-loop consensus sequence, however, the residues for these positions are either serine or threonine (Traut, 1994). Three mutants, in which serines were substituted for these threonines [R2M4a (T189S), R2M4b (T190S), and R2M4c (T189S and T190S)], were made and tested for their activity in the transient expression assay. R2M4a and R2M4b were as active as wild type whereas R2M4c activity was slightly weaker (Figure 2). Conceivably, this slight decrease in the activity of R2M4c could be important, such that R2M4c might appear to be inactive if transgenic plants carrying R2M4c were tested by inoculation of a P. syringae strain expressing avrRpt2 (see below the result with Δ501-504, which shows an activity similar to that of R2M4c in Figure 2). Thus, replacing either of the conserved threonine residues with a serine residue does not seem to affect RPS2 activity, whereas replacing both of them with serines may affect the activity.
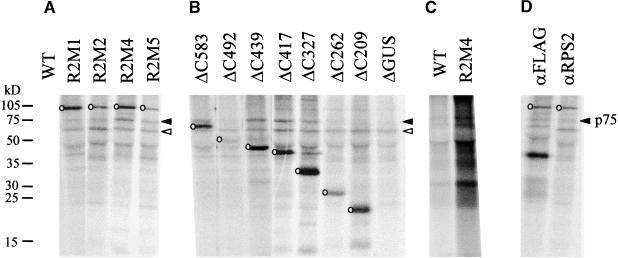
To exclude the possibility that the loss of R gene function seen in most RPS2 mutants is due to instability of the mutant proteins, we examined the quantities of the mutant proteins in Arabidopsis protoplasts. The RPS2 derivatives were transiently expressed and radiolabeled in Arabidopsis rps2-101C protoplasts and then immunoprecipitated with a polyclonal anti-RPS2 antibody. After SDS-PAGE, the precipitated RPS2 proteins were detected by using a phosphorimager. Interestingly, as shown in Figure 3A, the inactive RPS2 mutant proteins accumulated in much greater amounts than did wild-type RPS2 protein; the overall incorporation of label in the wild-type lane, including incorporation into nonspecific protein bands, was much less than in the other lanes (see below for an explanation). Because the mutant proteins are not unstable, we conclude that the reason the mutations in the N-terminal hydrophobic region, the LZ, and the NBS cause loss of RPS2 function is that all of these motifs are required for RPS2 function.
Figure 3.
In Vivo Stability of RPS2 Derivative Proteins and Their Ability to Interact with p75.
RPS2 and its derivative proteins were transiently expressed in Arabidopsis protoplasts. The RPS2 derivatives and other cellular proteins were radiolabeled with 35S-methionine, and total proteins or immunoprecipitated proteins were resolved by SDS-PAGE and visualized with a phosphorimager.
(A) and (B) RPS2 and its derivatives were immunoprecipitated with an anti-RPS2 antiserum; shown are composites of the results from four independent experiments.
(C) Labeled total proteins from protoplasts transfected with either RPS2 or R2M4 were precipitated with trichloroacetic acid. The amount of extract used in each lane in (C) is equivalent to ∼5% of the extract used in each lane in (A), (B), and (D).
(D) A C-terminal FLAG-tagged R2M4 (left lane) and R2M4 (right lane) were immunoprecipitated with an anti-FLAG antibody and an anti-RPS2 antiserum, respectively.
We attempted to normalize the results from different experiments by adjusting the imaging brightness to make comparable between experiments the intensity of the 65-kD nonspecific band (labeled by open arrowheads) for R2M4, which was included in every experiment as a positive control. The bands corresponding to the RPS2 derivatives are marked by open circles. The position of p75 is indicated by filled arrowheads. The positions of molecular mass markers are indicated at left in kilodaltons. WT, wild type.
Mutations Close to the N Terminus Result in Dominant Negative Phenotypes
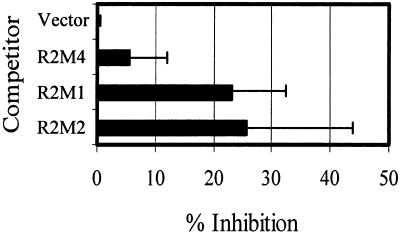
Table 1 shows that expression of R2M1 (N-terminal hydrophobic deletion) and R2M2 (LZ amino acid residue substitutions) in RPS2/RPS2 wild-type plants resulted in suppression of the RPS2 wild-type phenotype, whereas expression of wild-type RPS2 or of R2M4, R2M5, R2M7, and some of the deletion mutants described below did not. Although we cannot exclude the possibility that cosuppression occurred only with R2M1 and R2M2, it seems unlikely considering the minimal level of alteration in the RNA structures of these two RPS2 derivatives. Thus, R2M1 and R2M2 appear to have a dominant negative effect. We confirmed this conclusion using the transient expression assay. The wild-type RPS2 construct was mixed with a 10-fold excess of the construct for R2M1, R2M2 or R2M4, and the mixture was subjected to the transient expression assay. As shown in Figure 4, R2M4 showed little effect on RPS2 activity, but R2M1 and R2M2 markedly inhibited the RPS2 response. The dominant negative effects of R2M1 and R2M2 observed in the transient assay were relatively small (31 and 33% inhibition, respectively), which may reflect residual activity of R2M1 and R2M2 (Figure 2) or greater sensitivity of the transient expression assay than the transgenic plant HR assay.
Figure 4.
Dominant Negative Effect of R2M1 and R2M2 Measured by a Transient Expression Assay.
The RPS2 wild-type construct was cobombarded with a 10-fold excess of the indicated RPS2 derivatives or the vector as competitors in a transient expression assay to measure the response to avrRpt2. The decrease in GUS activity with the vector competitor control was set to 0% inhibition, and the GUS activity observed in the absence of RPS2 (no response) was set to 100% inhibition. The error bars indicate the standard deviation.
RPS2 Does Not Have a Large Dispensable Region
To determine whether RPS2 has a relatively large dispensable region in which the protein could be readily manipulated without affecting activity, we scanned the protein with a series of small in-frame deletions and analyzed the deletion mutants for RPS2 activity. Because these internal deletion mutants have linker sequences equivalent to eight or nine amino acid residues inserted where the deletions were made (Figure 1), it is possible that the observed effect of some of the mutations may not be due to the deletion but rather to the insertion of the short unrelated sequence. Two additional mutants with relatively small C-terminal deletions were included in this analysis (Figure 1). In the transient assay, most of these deletion mutants exhibited no RPS2 activity (Figure 2). Even a small C-terminal 20–amino acid residue deletion was inactive (ΔC890). However, the C terminus per se is not required because, as we previously reported, RPS2 is tolerant to a C-terminal fusion of a relatively large polypeptide (209 amino acid residues; Leister and Katagiri, 2000). A few internal deletions, such as Δ501–504, exhibited weak but detectable activities. These latter deletions—Δ501–504, Δ611–613, and Δ805–814—also were analyzed by the HR assay in transgenic rps2-101C plants. In contrast with the results obtained in the transient assay, none of the transgenic plants developed an HR in response to avrRpt2 (Table 1). We speculate that the transient assay may have a greater sensitivity than does the HR assay. The fact that most of the deletion mutants tested exhibited loss of activity suggests that RPS2 does not have a large dispensable region.
RPS2 Exhibits an Overdose Effect
In optimizing the transient expression assay, we observed that bombardment of leaves with an above-optimal amount of the RPS2 construct decreased the GUS reporter activity in the absence of avrRpt2 expression (Figure 5). Although we cannot exclude other possible mechanisms, we interpret these results as demonstrating that RPS2 has a low basal activity in the absence of AvrRpt2 and that overexpression of RPS2 activates a downstream defense response pathway. We call this phenomenon the RPS2 “overdose effect.”
Figure 5.
Overdose Effect of RPS2 and Its Derivatives.
In the absence of the AvrRpt2 construct, we used 20-fold more of the wild-type (WT) RPS2 construct than is normally used in the transient expression assay. The same type of assay was performed with the indicated RPS2 derivatives. Relative GUS activity (normalized for transformation efficiency, as measured by luciferase activity) for the vector control was set to 100. The error bars indicate the standard deviation.
When testing a series of C-terminal RPS2 deletion mutants for the overdose effect (Figure 5), we found that a region near the C terminus (see ΔC890, Figures 1 and 5) was required. Interestingly, however, when the entire LRR region and the C-terminal region were deleted (ΔC492), the overdose effect was similar to wild-type RPS2. Thus, when the LRR is entirely removed, the C-terminal region is not required for the overdose effect. Further deletions (from the C terminus) into the N-terminal portion of RPS2, which contains peptide sequences that are highly conserved among NBS-LRR proteins, such as the KMH motif around position 470, resulted in loss of the overdose effect (ΔC439, ΔC417, and ΔC327).
RPS2 mutants R2M1, R2M2, R2M4, R2M5, R2M4a, R2M4b, and R2M4c also were tested for the overdose effect (Figure 5). R2M1, R2M2, R2M4, and R2M5, which do not exhibit RPS2 activity (i.e., do not respond to AvrRpt2), also do not exhibit the overdose effect. R2M4a, R2M4b, and R2M4c, however, were active in both responding to AvrRpt2 and exhibiting the overdose effect. As shown in Figures 3A and 3B, when RPS2 derivatives were overexpressed in Arabidopsis protoplasts, all of the RPS2 derivatives that did not show the overdose effect accumulated in greater amounts than did the ones that showed the effect (wild type and ΔC492). This observation rules out the possibility that the RPS2 derivatives not showing the overdose effect are unstable in plant cells. Therefore, the N-terminal half of RPS2, including the N-terminal hydrophobic region, the LZ region, and the NBS, is functionally important for the overdose effect.
When wild-type RPS2 protein was overexpressed in protoplasts, not only was incorporation of radioactivity into RPS2 diminished compared with that of R2M4 (which does not show the overdose effect), but also incorporation of radioactivity into other endogenous proteins was greatly reduced (Figure 3C). This observation was highly reproducible and explains why signals are barely detectable in the wild-type lane in Figure 3A. R2M1, R2M2, R2M5, ΔC583, ΔC439, ΔC417, ΔC327, ΔC262, and ΔC209 (as well as the unrelated control ΔGUS), which are inactive for the overdose effect, showed as much incorporation of radioactivity into general proteins as R2M4, whereas ΔC492, which is active in the overdose effect, showed much less incorporation of radioactivity (data not shown). Therefore, there is a strong correlation between the overdose effect activity and decreased incorporation of radioactivity into proteins in general. These results indicate that the RPS2 overdose effect is associated with nonspecific inhibition of de novo protein synthesis, induction of massive, nonspecific protein degradation, or both. Whatever the molecular mechanism involved, we believe this nonspecific inhibition of protein accumulation is the basis on which we can measure both RPS2 activity and the overdose effect in the transient expression assays.
RPS2 Activity Measured by Transient Expression and the Overdose Effect Are Not Strongly Affected by ndr1-1 Mutation
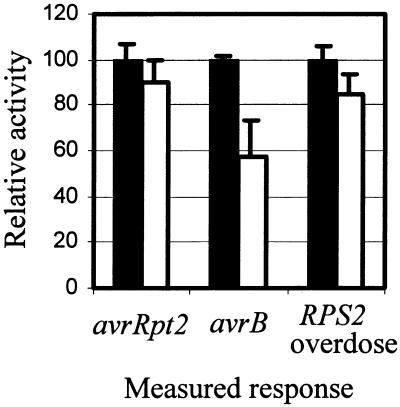
Arabidopsis NDR1 gene function is required for both the HR and the limitation of bacterial growth resulting from the avrRpt2–RPS2 interaction (Century et al., 1995, 1997). In contrast, although NDR1 is also required for the limitation of bacterial growth in an avrB–RPM1 interaction, it is not required for the HR (Century et al., 1995). We therefore tested whether NDR1 is required in the transient expression assay for RPS2 and RPM1 activities and in the RPS2 overdose effect. (RPM1 did not exhibit an overdose effect at the concentration of plasmid DNA at which RPS2 shows the effect; results not shown.) As shown in Figure 6, comparing ndr1-1 leaves with wild-type leaves in the transient expression assay indicated no substantial difference in the response to avrRpt2. The response to avrB in ndr1-1 plants was ∼60% of the wild-type control. The RPS2 overdose effect was only slightly reduced in ndr1-1.
Figure 6.
Effect of the ndr1-1 Mutation on the Transient Expression Assay.
Columbia-0 (Col-0) wild-type (solid bars) and ndr1-1 plants (open bars) were tested in the transient expression assays for the response to avrRpt2, the response to avrB, and the RPS2 overdose effect. Both wild-type and ndr1-1 plants are RPS2 and RPM1 wild type. To represent the relative activity, we set the decrease of the normalized GUS activity with wild-type plants to 100, and the normalized GUS activity with vector controls was set to 0. The error bars indicate the standard deviation.
p75 Interacts with an N-Terminal Region of RPS2
In a previous study, we showed that when a FLAG-tagged version of RPS2 derivative R2M4 was expressed in Arabidopsis protoplasts, immunoprecipitation with an anti-FLAG antibody coimmunoprecipitated a 75-kD plant protein, p75 (Leister and Katagiri, 2000). Figure 3D shows that p75 is also coimmunoprecipitated with R2M4 by an anti-RPS2 antibody. Because the anti-RPS2 antibody recognizes residues close to the N terminus (amino acid residues 23 to 36), whereas the FLAG tag is fused to the C terminus of R2M4 (909 amino acid residues), p75 cannot be a truncated version of RPS2.
Figures 3A and 3B show that p75 was coimmunoprecipitated with various RPS2 derivatives. Examination of the RPS2 deletion series shows that when the deletion from the C terminus extended past residue 326, no p75 interaction was detected (ΔC262 and ΔC209). Thus, the portion of RPS2 that is sufficient for p75 interaction was delimited to the N-terminal 326 amino acids. p75 interaction was not affected by any of the inactive directed mutants (R2M2, R2M4, and R2M5), except for R2M1, which reproducibly reduced the amount of p75 coimmunoprecipitated. Because of the inhibition of radiolabeled protein accumulation as described above, we were unable to determine whether wild-type RPS2 or ΔC492 can interact with p75.
DISCUSSION
To gain insight into the molecular mechanisms involved in gene-for-gene interactions, we performed structure–function analyses of the Arabidopsis RPS2 protein. The major results of this analysis can be summarized as follows: RPS2 does not have a large region that is dispensable for its activity in response to AvrRpt2. Overexpression of RPS2 apparently results in activation of downstream responses in the absence of AvrRpt2, and an N-terminal portion that includes the LZ and NBS regions is important for this activity (overdose effect) whereas the LRR is not. RPS2 activity and the overdose effect that we measure by the transient expression assay are barely affected by a presumptive null mutation in the NDR1 gene. The region of RPS2 involved in interaction with p75 is located within the first 326 amino acids.
An Integrated Three-Dimensional Structure for RPS2?
RPS2 apparently does not have a large region that is dispensable for its activity in response to AvrRpt2. This observation suggests that RPS2 protein has an integrated tertiary structure rather than a modular structure composed of well-defined domains. An integrated three-dimensional structure might explain the difficulty of constructing functionally active chimeras between RPS2 and RPM1, even though both of these Arabidopsis R proteins belong to the LZ subclass of NBS-LRR proteins (F. Katagiri, unpublished data). The observation that most RPS2 mutants with small in-frame deletions are inactive calls for caution in interpreting the results obtained with single amino acid substitution mutants. Although the amino acid substitutions probably did disrupt the function of the motif in which the mutation was created, these single amino acid changes conceivably could affect the overall conformation of the integrated protein structure. On the other hand, as the data in Figure 3 show, we can exclude the trivial possibility that the amino acid substitution mutations affected the stability of RPS2 protein.
RPS2 Dominant Negative Mutants
Two of the amino acid substitution mutants, R2M1 (N-terminal hydrophobic region) and R2M2 (LZ), exhibited a dominant negative effect over the wild-type protein. These mutations are located close to the N terminus. Because no other inactive mutants showed this effect, we conclude that the way these two mutants disrupt RPS2 function differs at least partly from the others. The simplest model for a dominant negative mutation is that the mutant protein sequesters limiting molecules (which can be the corresponding wild-type protein) into an unproductive complex. Because R2M4 and R2M5, as well as R2M1 and R2M2, can interact with p75 (Figures 3A and 3B), p75 is unlikely to be such a limiting factor. RPS5 is an NBS-LRR R gene conferring resistance against P. syringae strains carrying avrPphB (Warren et al., 1998). Although rps5-1 is a recessive mutation in the third repeat of the LRR, it nevertheless interferes partially with the function of other NBS-LRR proteins. Specifically, it has been postulated that RPS5-1 protein sequesters a factor shared by many R gene products. It will be of interest to determine whether R2M1 and R2M2 also can interfere with other R protein functions.
RPS2 Overdose Effect
Overexpression of RPS2 apparently leads to activation of the defense response. This is reminiscent of an often-observed phenomenon in signal transduction studies: overexpression of a single signal transduction component leads to constitutive activation of the signal transduction pathway in the absence of the corresponding stimulus. Similar observations also have been made with other R genes, including Pto (Tang et al., 1999) and Prf (Oldroyd and Staskawicz, 1998), the overexpression of which results in constitutive expression of defense-related responses. We speculate that RPS2 has low basal activity in the absence of AvrRpt2 and that overexpression exceeds the threshold needed to turn on downstream responses. This interpretation provides a simple explanation for our previous report that transgenic plants containing an RPS2 transgene under the control of the strong 35S* promoter (see Methods) all exhibit cosuppression of RPS2 gene function (Mindrinos et al., 1994). Given that overexpression of RPS2 even in the absence of AvrRpt2 leads to HR cell death, only cosuppressed transgenic lines can be recovered. In the transient expression assay for RPS2 function involving relatively small doses of RPS2, the specificity of the assay is assured by the specificity of the stimulus, that is, avrRpt2. However, in the overdose effect assay, the amount of GUS activity measured is a consequence of the inhibition of accumulation of newly synthesized proteins (see Figure 3C). This inhibition may, in turn, be the consequence of RPS2-mediated signal transduction, but it also could be caused by a mechanism other than the one directly related to the responses initiated by the RPS2–AvrRpt2 interaction.
Assuming that our interpretation is correct and that the overdose effect is a direct reflection of the downstream signaling function of RPS2, loss of the overdose effect in a mutant implies that the downstream signaling process has been impaired. The overdose effect is probably a consequence of the inhibition of de novo protein synthesis or massive nonspecific protein degradation (or both). Boyes et al. (1998) reported that RPM1 protein was rapidly degraded when the resistance response was activated. Despite an apparent similarity between the observation by Boyes et al. and the RPS2 overdose effect, there are clear differences between them. Protein degradation observed by Boyes et al. is relatively specific to RPM1, whereas the RPS2 overdose affects proteins nonspecifically (Figure 3C). Given a lack of obvious changes in the appearance of protoplasts 1 day after transfection with wild-type RPS2 (data not shown), we prefer the idea that the overdose effect is a consequence of protein synthesis inhibition rather than massive nonspecific protein degradation. That the transfected protoplasts appear normal also suggests that the decrease in reporter activity measured by the transient expression assays is not a consequence of cell death per se, which is commonly defined by loss of the plasma membrane integrity; nonetheless, general inhibition of protein accumulation indicates that a fundamental cell function is impaired.
In the smallest RPS2 derivative that is active in the overdose effect, the C-terminal 418 amino acid residues, including the entire LRR region, have been deleted (ΔC492). Not only is the LRR therefore unnecessary for the basic downstream signaling function, but also the LRR may have an inhibitory effect on the overdose effect. This latter speculation is based on the observations that ΔC890 and ΔC583, which retain at least part of the LRR, did not exhibit the overdose effect and that the overdose effect can be restored by deleting the entire LRR (ΔC492). Given that the LRR or corresponding leucine-rich domain is the major avr-specificity determinant in NBS-LRR proteins or a related protein, respectively (Ellis et al., 1999; Jia et al., 2000), the LRR might function as an inhibitory domain, and recognition of the avr-based signal by the LRR might release the N-terminal downstream signaling portion of the protein from inhibition. In the overdose effect, as in the standard transient assay for RPS2 function, the N-terminal hydrophobic region, the LZ region, and the NBS are all required. However, we tested mutations in these motifs only in the context of the full-length RPS2 protein. Some of these motifs may be involved in mediating an interaction with the C-terminal positively regulating region of RPS2, and possibly some of these motifs may not be required for the overdose effect in the ΔC492 deletion mutant. Additional highly conserved motifs among NBS-LRR proteins probably are also required for downstream signaling because the C-terminal deletion mutant that removes the highly conserved KMH motif (near amino acid residue 470) did not show the overdose effect (ΔC417). In all cases, the mutant proteins that did not exhibit the overdose effect were accumulated in Arabidopsis protoplasts, eliminating protein instability as the cause for loss of activity.
NDR1 Function Not Required in the Transient Assay
We found that both RPS2 function in response to AvrRpt2 and the RPS2 overdose effect as measured in the transient expression assay are barely affected by the ndr1-1 mutation. Our results from the transient assay represent the first reported case in which an avrRpt2-elicited response is not strongly inhibited by the ndr1-1 mutation. One explanation is that the inhibition of accumulation of newly synthesized proteins, which we measured in the overdose assay, may branch from the signal transduction pathway before the step involving NDR1. This explanation includes the following possibility: Because the activities observed with the biolistic transient expression assays using leaf tissue are restricted to the cells in which R protein–based signals are generated, the effect of the ndr1-1 mutation would not be detected if the function of NDR1 in the resistance response were crucial only in the adjacent cells. Alternatively, NDR1 may be a quantitative factor, and the transient expression assay may be more sensitive for detecting RPS2 activity than is the HR assay by using transgenic plants. If so, the greater sensitivity of the transient assay may be attributable to expression of greater amounts of AvrRpt2 and RPS2 in the cell, which might cause a low saturation point for the activity in the assay. Assuming that NDR1 is not absolutely required for RPS2 function but rather serves as an ancillary stimulatory factor, the effect of its absence may be difficult to measure in the transient assay.
Perspectives
Our structure–function analysis of RPS2 has yielded insight into how NBS-LRR proteins function in pathogen recognition signal cascades. Our results are consistent with a model in which the LRR region of the protein is involved in signal perception, and the N-terminal portion of the protein, including the N-terminal hydrophobic domain, the LZ, the NBS and other conserved domains, functions in signal transduction. One practical way to test these hypotheses is to examine whether other NBS-LRR R proteins, such as RPM1, behave similarly in our transient expression systems. For example, testing whether other NBS-LRR proteins can exhibit an overdose effect when their LRR regions are deleted and whether mutations near the N terminus cause dominant negative effects should lead to interesting findings. Identifying common features of NBS-LRR proteins will help elucidate how the gene-for-gene resistance system functions to activate host defense responses.
METHODS
Plant and Bacterial Strains
The Arabidopsis thaliana ecotype Columbia (Col-0) plants used in the study were RPS2/RPS2 (wild type), rps2-101C/rps2-101C (rsp2 mutant; Yu et al., 1993), and ndr1-1/ndr1-1 (ndr1 mutant; Century et al., 1995). The rps2-101C Δrpm1 double mutant (rps2-101C/rps2-101C; Δrpm1/Δrpm1) is a Col-0 × Nd-0 hybrid (Mindrinos et al., 1994). Plants were grown at 22°C with 80% relative humidity and a 12-hr-light/12-hr-dark cycle in environmentally controlled chambers. Escherichia coli strains DH5α and DH10B (Life Technologies, Bethesda, MD) were used for plasmid construction procedures. The uidA mutant (E. coli strain PK803) was a gift of E. Signer (Jefferson et al., 1986). Pseudomonas syringae pv phaseolicola NPS3121 strains carrying pLAFR3 or pLH12 (avrRpt2) were used for the confluent hypersensitive response (HR) assay as described by Yu et al. (1993).
Construction of Plant Transient Expression Vector pKEx4tr
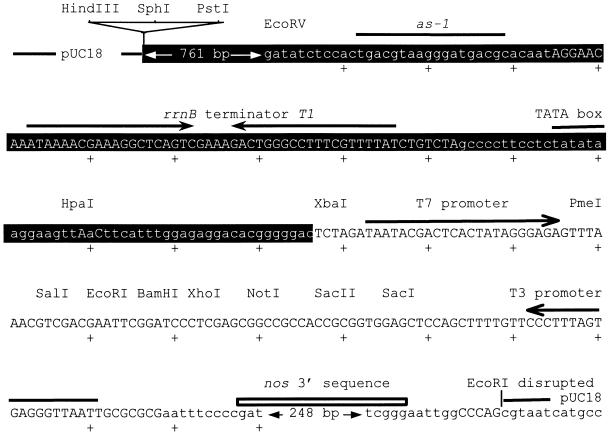
We constructed pKEx4tr, a plasmid vector for transient expression analysis in plants. It originally was designed to be a cDNA cloning vector in which the function of a cDNA insert could be readily analyzed (Mindrinos et al., 1994) or in which functionally active cDNA inserts could be screened in plant cells by using a transient expression assay. For use as a general cDNA cloning vector, we considered it crucial to have very low expression of cDNA inserts in E. coli because some plant proteins might have a deleterious effect on E. coli when expressed. At the same time, to facilitate analysis in plant cells, the cDNA inserts should be under the control of a strong plant promoter. Figure 7 illustrates features of pKEx4tr that differ from the parent plasmid pBI221 (Clonetech, Palo Alto, CA). pBI221, derived from pUC19, has a wild-type 35S promoter (Odell et al., 1985), the E. coli reporter gene for β-glucuronidase (GUS) activity (uidA) (Jefferson et al., 1986, 1987), and the nos 3′ sequence (Bevan et al., 1983). To obtain a high expression of cDNA inserts in plants and a low expression in E. coli, we made two changes in pBI221. First, on the basis of a report about transcription initiation within the 35S promoter sequence in E. coli (Assaad and Signer, 1990), which showed that E. coli RNA polymerase appears to use the 35S TATA box as a −10 sequence, we inserted the E. coli rrnB terminator T1 sequence (Brosius et al., 1981; Brosius, 1984) upstream of the TATA box. This disrupts transcription from the most downstream cryptic E. coli promoter and also disrupts transcription from additional upstream cryptic transcription initiation sites in the 35S promoter. Importantly, the rrnB terminator T1 insertion does not disrupt crucial elements of the 35S promoter for transcription in plants, such as the as-1 element (Lam et al., 1989) and the TATA box; rather, this insertion merely increases the distance between the as-1 element and the TATA box by 42 bp. We refer to the altered 35S promoter as 35S*. Second, the entire region spanning the 35S* promoter, the multiple cloning site, and the nos 3′ sequence was transferred from pBI221 into pUC18 because in E. coli we observed substantial read-through from the lacZ promoter, located upstream of the 35S promoter (data not shown). In the pUC18 version, the orientation of the insert was opposite that of the lacZ promoter; that is, in pKEx4tr, the lacZ promoter is located downstream from the nos 3′ sequence.
Figure 7.
Structure of pKEx4tr.
The top strand of the sequence surrounding the multiple cloning site in pKEx4tr is shown. Nucleotides that differ from the corresponding nucleotides in pBI221 are shown in uppercase letters. The 35S* promoter region is indicated by white letters in black background boxes. The 5′ region of the 35S* promoter, which is indicated as a 761-bp region, is identical to that of the wild-type 35S promoter in pBI221. The regions derived from pUC18 are depicted schematically. The as-1 element and the TATA box of the 35S promoter, the E. coli rrnB terminator T1, some of the unique restriction sites in the vector, the T7 and T3 promoters, and the nos 3′ sequence are indicated. Plus signs indicate positions for every 10 nucleotides.
The entire sequence of pKEx4tr has been deposited in GenBank (accession number AF044029). Because pKEx4tr was constructed through several trial-and-error steps, the construction steps are not described here. Details of the construction are available from F.K. on request. Other useful features of pKEx4tr are that (1) the multiple cloning site located between the 35S* promoter and the nos 3′ sequence is compatible with commercially available unidirectional cDNA cloning kits; (2) the multiple cloning site contains restriction sites for PmeI and NotI, which recognize 8-bp sites and can be used to excise cDNA inserts cloned by using one of the commercial cDNA cloning kits; and (3) the multiple cloning site is flanked by T7 and T3 promoters, which are convenient for RNA probe synthesis and for in vitro transcription/translation studies (e.g., Leister et al., 1996).
The amount of expression of the 35S::uidA construct in pBI221 was compared with that of the 35S*::uidA construct in pKEx4tr (pKEx4tr-G). First, the amounts of GUS expression in Arabidopsis leaves were compared by using a biolistic transient expression system. pKEx4tr (vector control), pKEx4tr-G, or pBI221 DNA was cobombarded into leaves with an internal reference construct, p35S-LUC DNA (Chern et al., 1996), in which the uidA sequence of pBI221 had been replaced with the firefly luciferase gene. After a 17-hr incubation at room temperature in the dark, leaf extracts were made, and the GUS and luciferase (LUC) enzyme activities in the extracts were measured. As shown in Table 2, pKEx4tr-G expressed approximately threefold more GUS activity than did pBI221. This greater expression of the insert in pKEx4tr-G than in pBI221 was fortuitous.
Table 2.
Expression of 35S*-GUS in Arabidopsis Plants and in E. coli
| Plasmid Construct
|
|||
|---|---|---|---|
| Activity | pKEx4tr | pBI221 | pKEx4tr-G |
| Relative GUS activity (mean ± se) in biolistically bombarded plants | −0.2 ± 0.2a | 100 ± 6a | 286 ± 29a |
| Relative GUS activity (mean ± se) in E. coli | 0.02 ± 0.01b | 100 ± 22b | 0.07 ± 0.02b |
The measured GUS activities were normalized by the corresponding luciferase activities. All values were renormalized, setting the mean value for pBI221 to 100. Four replicates were measured for each construct.
The measured GUS activities were normalized by the relative copy numbers of the plasmids. All values were renormalized, setting the mean value for pBI221 to 100. Four independent transformants were analyzed for each construct.
Next, the amount of expression of GUS in pKEx4tr-G and pBI221 was compared in E. coli by transforming strain PK803, which contains a uidA deletion (Jefferson et al., 1986), with pKEx4tr, pKEx4tr-G, and pBI221. Table 2 shows that pKEx4tr-G had 1400-fold less GUS activity than did pBI221. Previously, we reported that the 35S* promoter was ∼50-fold less active than was the 35S promoter in E. coli (Mindrinos et al., 1994). However, we realized that the strains derived from PK803 in the previous experiment had not been properly calibrated; we had overlooked that PK803 is temperature sensitive and that it is RecA+, which resulted in variable copy numbers of multimerized plasmids. When we repeated the experiment, we correctly calibrated the PK803-derived strains and normalized the GUS activity, obtaining the results shown in Table 2.
The method we used to reduce the activity of the 35S promoter in E. coli should be applicable to other plant promoters as well as eukaryotic promoters in general. Such modified promoters are useful for a variety of biotechnology applications.
Other Plasmid Constructs
As described in detail below, plasmid constructs for transient expression assays were made in pKEx4tr using the 35S* promoter unless specified, and constructs for generating transgenic plants were made in pBI1.Rpro11 by using a 1.4-kb region upstream of the RPS2 coding region as a promoter.
A series of C-terminal deletions of RPS2 was generated as follows. First, to obtain pX11C, the LS12 linker (see below) was inserted into the NotI-SacI site in pRPS2 (RPS2 cDNA clone 11 in pKEx4tr) (Leister et al., 1996), which is located at the 3′ end of the cDNA insert. LS12 contains a stop codon in each of the three reading frames.
Second, pX11C was digested with KpnI and a 5′ overhang restriction enzyme (NotI or other enzymes with sites internal to the RPS2 coding region), and a deletion series was made by an ExoIII deletion method (Ausubel et al., 1998). The break points of the deletions were determined by sequencing.
To generate a series of small in-frame internal deletions of RPS2, a series of N-terminal deletions were generated (which became the C-terminal portion of the internal deletions). The N-terminal deletions were then combined with appropriate C-terminal deletions (which became the N-terminal portion of the deletions). To generate N-terminal deletions, first we inserted linker LS34 (see below) into the SphI-SalI site in pRPS2, which is located at the 5′ end of the cDNA insert, to obtain pX11N.
Second, pX11N was digested with SphI and a restriction enzyme that generates a 5′ overhanging end (AscI or other enzymes with sites internal to the RPS2 coding region), and a deletion series was made by the ExoIII deletion method. The break points of the deletions were determined by sequencing. To create small in-frame internal deletions, we chose pairs of N-terminal and C-terminal deletions. The insert of an N-terminal deletion construct was excised by a KpnI and SacI digestion, and the insert was cloned into the ClaI-SacI site of the corresponding C-terminal deletion construct, together with the appropriate ADx (AD0, AD1, AD2; see below) adaptor linkers to adjust the reading frames between the deletions. Each adaptor linker had an XhoI site to facilitate detection and was compatible with ClaI and KpnI sites.
The junction sequences of the internal deletions were confirmed to be in-frame by sequencing.
The amino acid substitution mutants were generated by a polymerase chain reaction (PCR) soeing–based method, in which two amplified fragments with an intended mutation in the overlapping region are joined together by PCR (Horton et al., 1990). Pfu DNA polymerase was used for amplification. The primer pairs used to create mutations were as follows (the complementary parts used for soeing are aligned):
To clone the RPS2 promoter region, pBI1.R2 (Mindrinos et al., 1994) was digested with BamHI and EcoRI, and a 1.9-kb DNA fragment was cloned into the BamHI-EcoRI site of pBluescript II SK+ (Stratagene) to obtain pK2-5. The 1.7-kb DNA region of the insert, which was bounded by a KpnI site (originating from the vector) at the upstream end and by the base pair that was 22 bp upstream of the beginning of the RPS2 coding sequence at the downstream end (where a PstI site was created by using the PCR primer 5′-CGCGGATCCTGCAGCTTTTACTTGTCTGAGC-3′), was recloned into the KpnI-PstI site of pBluescript II SK+ to obtain pK2-5X1; this added an appropriate multiple cloning site from the vector to the 3′ end of the RPS2 promoter region. The 1.5-kb HindIII (an internal site)-SacI (the 3′ end of the multiple cloning site) fragment of pK2-5X1, which contained a 1397-bp RPS2 upstream region, was cloned into the HindIII-SacI site of the plant transformation vector pBI1.4t (a pBIN19 derivative) to replace the 35S* promoter and the multiple cloning site of pBI1.4t. The resulting construct, which was named pBI1.Rpro11, is a plant transformation vector containing the RPS2 promoter (corresponding to nucleotide numbers 60,955 to 62,352 in the Arabidopsis bacterial artificial chromosome clone with GenBank accession number AL049483) followed by a multiple cloning site (containing unique SmaI, BamHI, XbaI, and SacI sites) and the nos 3′ sequence. For the plant transformation constructs with RPS2 derivatives, the RPS2 derivative genes were excised from the appropriate pKEx4tr-based constructs with PmeI (blunt end) and SacI and then were cloned into the SmaI (blunt end)-SacI site of pBI1.Rpro11.
Transient Expression in Arabidopsis
Biolistic bombardment was performed as described (Mindrinos et al., 1994). For analysis of the 35S* promoter, 0.8 μg of pKEx4tr, pKEx4tr-G, or pBI221; 0.2 μg of p35S-LUC; and 0.5 mg of 1-μm-diameter gold particles (Bio-Rad) were used for each bombardment. The transient expression assay for RPS2 activity was performed as described (Leister et al., 1996). For analysis of the dominant negative effect, the amount of the competitor plasmid DNA exceeded by 10-fold that of the wild-type plasmid DNA. For analysis of the RPS2 overdose effect, the amount of the RPS2 derivative plasmid DNA used was 20-fold more than the amount used in the ordinary RPS2 activity assay.
Generation of Transgenic Arabidopsis
Transgenic Arabidopsis plants were generated as described previously (Mindrinos et al., 1994; Clough and Bent, 1998). Multiple independent lines of first-generation transgenic plants were used for analysis.
Reporter Assays
The GUS and LUC activities in extracts were measured by using a fluorogenic assay with 4-methylumbelliferyl-β-d-glucuronide as a GUS substrate (Jefferson, 1987) and by a luciferase assay kit (Promega), respectively. E. coli extracts for measurement of GUS activity were prepared as follows. The bacteria were harvested from 400 μL of LB medium culture by centrifugation, suspended in 400 μL of GUS assay buffer (50 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% sodium N-lauroylsarcosine, 0.1% Triton X-100, and 10 mM 2-mercaptoethanol), and sonicated briefly. The lysate was cleared by centrifugation. The GUS activities in E. coli extracts were normalized according to the relative plasmid amount of each bacterial sample used for extract preparation to cancel any difference in plasmid copy number (although the difference was small).
Anti-RPS2 Antibody
A peptide corresponding to amino acid residues 23 to 36 (ERRGHKTDLROAIT) of RPS2 was synthesized by using multiple antigen peptide technology (Tam, 1988). Two rabbits were immunized with the peptide, and antisera were collected from them. Peptide synthesis and antiserum production were performed by Research Genetics (Huntsville, AL). One of two antisera showed better efficiency in immunoprecipitation of in vitro–translated RPS2 and was chosen for use.
In Vivo Labeling and Immunoprecipitation of RPS2 Derivatives
In vivo labeling of RPS2 derivatives with 35S-methionine by using a transient expression in Arabidopsis protoplasts was performed as described by Leister and Katagiri (2000). Immunoprecipitation also was performed as described (Leister and Katagiri, 2000) except that 10 μL of anti-RPS2 antiserum was used per sample.
GenBank Accession Numbers
The sequence of pKEx4tr (accession number AF044029) was submitted to GenBank.
Acknowledgments
We thank Ethan Signer for PK803, Brian Staskawicz for the ndr1-1 mutant, and Jane Glazebrook for critical reading of the manuscript. This work was supported by National Science Foundation Grant MCB-9604830 to F.K. and National Institutes of Health Grant GM48707 to F.M.A. F.Y. was partly supported by the Chinese National Foundation of Natural Sciences.
References
- Assaad, F.F., and Signer, E.R. (1990). Cauliflower mosaic virus P35S promoter activity in Escherichia coli. Mol. Gen. Genet. 223 517–520. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1998). Current Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bevan, M., Barnes, W.M., and Chilton, M.D. (1983). Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 11 369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas, U., and van den Ackervaken, G. (1997). Recognition of bacterial avirulence proteins occurs inside the plant cell: A general phenomenon in resistance to bacterial diseases? Plant J. 12 1–7. [DOI] [PubMed] [Google Scholar]
- Boyes, D.C., Nam, J., and Dangl, J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hy-persensitive response. Proc. Natl. Acad. Sci. USA 95 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius, J. (1984). Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene 27 162–172. [DOI] [PubMed] [Google Scholar]
- Brosius, J., Dull, T.J., Sleeter, D.D., and Noller, H.F. (1981). Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148 107–127. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chern, M.-S., Bobb, A.J., and Bustos, M.M. (1996). The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 8 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, F.L., Holzberg, S., Calderon-Urrea, A., Handley, V., Axtell, M., Corr, C., and Baker, B. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defense response in tobacco. Plant J. 18 67–75. [DOI] [PubMed] [Google Scholar]
- Galan, J.E., and Collmer, A. (1999). Type III secretion machines: Bacterial devices for protein delivery into host cells. Science 284 1322–1328. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 575–607. [DOI] [PubMed] [Google Scholar]
- Horton, R.M., Cai, Z.L., Ho, S.N., and Pease, L.R. (1990). Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. BioTechniques 8 528–535. [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Jefferson, R.A., Burgess, S.M., and Hirsh, D. (1986). β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 13 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Benfey, P.N., Gilmartin, P.M., Fang, R.-X., and Chua, N.-H. (1989). Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc. Natl. Acad. Sci. USA 86 7890–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, R.T., and Katagiri, F. (2000). A resistance gene product of the nucleotide binding site—Leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 22 345–354. [DOI] [PubMed] [Google Scholar]
- Leister, R.T., Ausubel, F.M., and Katagiri, F. (1996). Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA 93 15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., Mudgett, M.B., Li, K., Aoyama, T., Horvath, D., and Chua, N.-H. (1998). Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 14 247–257. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Dickerman, A.W., Michelmore, R.W., Sivaramakrishnan, S., Sobral, B.W., and Young, N.D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20 317–332. [DOI] [PubMed] [Google Scholar]
- Milligan, S.B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V.M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F.M. (1994). The Arabidopsis thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78 1089–1099. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B., and Staskawicz, B. (1998). Protein signaling via type III secretion pathways in phytopathogenic bacteria. Curr. Opin. Microbiol. 1 109–114. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B., and Staskawicz, B.J. (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: Demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32 927–941. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R.T., Katagiri, F., and Dangl, J.L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101 353–363. [DOI] [PubMed] [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.-H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Staskawicz, B.J. (1998). Genetically engineered broad-spectrum disease resistance in tomato. Proc. Natl. Acad. Sci. USA 95 10300–10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, M., Goggin, F.L., Milligan, S.B., Kaloshian, I., Ullman, D.E., and Williamson, V.M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065. [DOI] [PubMed] [Google Scholar]
- Tam, J.P. (1988). Synthetic peptide vaccine design: Synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85 5409–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Jones, D.A., Parniske, M., Harrison, K., Balint-Kurti, P.J., Hatzinxanthis, K., and Jones, J.D. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut, T.W. (1994). The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem. 229 9–19. [DOI] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R.W. (1998). A mutation in the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G.-L., Katagiri, F., and Ausubel, F.M. (1993). Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol. Plant-Microbe Interact. 6 434–443. [DOI] [PubMed] [Google Scholar]