Abstract
Yeast cells contain a family of three monothiol glutaredoxins: Grx3, 4, and 5. Absence of Grx5 leads to constitutive oxidative damage, exacerbating that caused by external oxidants. Phenotypic defects associated with the absence of Grx5 are suppressed by overexpression of SSQ1 and ISA2, two genes involved in the synthesis and assembly of iron/sulfur clusters into proteins. Grx5 localizes at the mitochondrial matrix, like other proteins involved in the synthesis of these clusters, and the mature form lacks the first 29 amino acids of the translation product. Absence of Grx5 causes: 1) iron accumulation in the cell, which in turn could promote oxidative damage, and 2) inactivation of enzymes requiring iron/sulfur clusters for their activity. Reduction of iron levels in grx5 null mutants does not restore the activity of iron/sulfur enzymes, and cell growth defects are not suppressed in anaerobiosis or in the presence of disulfide reductants. Hence, Grx5 forms part of the mitochondrial machinery involved in the synthesis and assembly of iron/sulfur centers.
INTRODUCTION
Glutaredoxins and thioredoxins are thioloxidoreductases required for maintaining thiol/disulfide equilibrium in cell proteins (Holmgren, 1989; Carmel-Harel and Storz, 2000; Grant, 2001) and also for the activity of specific enzymes (Aslund et al., 1994; Lillig et al., 1999). Glutaredoxin requires the reduced form of glutathione (GSH) as an electron donor (Holmgren and Aslund, 1995). In Saccharomyces cerevisiae, five glutaredoxins have been identified. Grx1 and Grx2 have two cysteine residues each at their active sites and play different roles in protecting cells against oxidants such as hydrogen peroxide and menadione (Luikenhuis et al., 1998). The defensive roles of Grx1 and Grx2 may overlap with those of the cytosolic Trx1 and Trx2 thioredoxins: at least one Grx1/Grx2 glutaredoxin or Trx1/Trx2 thioredoxin is required for yeast cell viability (Draculic et al., 2000). Another mitochondrial thioredoxin, Trx3, has been described in S. cerevisiae (Pedrajas et al., 1999). Besides Grx1 and Grx2, yeast cells have three monocysteine glutaredoxins: Grx3, Grx4, and Grx5 (Rodríguez-Manzaneque et al., 1999). The absence of Grx3 or Grx4 does not have a dramatic effect on sensitivity to oxidants. In contrast, the absence of Grx5 results in high sensitivity to hydrogen peroxide and menadione, increased protein oxidative damage, growth defects in minimal medium, and inability for respiratory growth (Rodríguez-Manzaneque et al., 1999). The Grx3, Grx4, and Grx5 proteins have been included in a large protein superfamily that contains a conserved structural domain (from amino acids 46–132 in Grx5) defined after the human PICOT protein (Isakov et al., 2000). PICOT may play a negative regulatory role in protein kinase Cθ-mediated activation of the transcription factors AP-1 and NF-κB in human cells (Witte et al., 2000). The N-terminal extension of Grx3 and Grx4 (not present in Grx5) shares additional sequence homology with the N-terminal moiety of human PICOT and other members of the superfamily (Isakov et al., 2000; Rahlfs et al., 2001).
Respiratory growth defects of grx5 mutant cells suggest impairment of mitochondrial functions. One of the essential processes occurring in the mitochondrial matrix of yeast cells is the generation of iron/sulfur (Fe/S) clusters, which are assembled in proteins destined to mitochondrial, cytosolic, or nuclear compartments (Craig et al., 1999; Lill et al., 1999; Lill and Kispal, 2000). These clusters are especially sensitive to oxidants (Keyer and Imlay, 1996) and liberate free iron that could further reactive oxygen species (ROS) production (Cadenas, 1989). Biogenesis of Fe/S clusters is a conserved process from bacteria to higher eukaryotes (Lill et al., 1999; Lill and Kispal, 2000), although in humans biosynthetic complexes for Fe/S cluster assembly are also found in the cytosol (Tong and Rouault, 2000). In S. cerevisiae, synthesis and assembly of Fe/S clusters involves Nfs1 cysteine desulfurase (Kispal et al., 1999; Li et al., 1999), the ferric ion-binding proteins Isu1 and Isu2 (Garland et al., 1999; Schilke et al., 1999), the Yah1 ferrodoxin (Barros and Nobrega, 1999; Lange et al., 2000), the Arh1 ferrodoxin reductase (Lacour et al., 1998; Manzella et al., 1998), the molecular chaperones Ssq1 (Hsp70-type) and Jac1 (Hsp40 or J-type; Schilke et al., 1996; Strain et al., 1998; Schilke et al., 1999; Lutz et al., 2001; Voisine et al., 2001), the homologous proteins Isa1 and Isa2 (Jensen and Culotta, 2000; Kaut et al., 2000; Pelzer et al., 2000), and the functionally uncharacterized protein Nfu1 (Schilke et al., 1999). Exporting Fe/S for extramitochondrial proteins requires the mitochondrial ABC transporter Atm1 (Kispal et al., 1999). Mutations in yeast genes involved in Fe/S cluster assembly cause iron accumulation in the mitochondria, mitochondrial DNA damage, and respiratory metabolism failure (Craig et al., 1999; Lill and Kispal, 2000). Similar phenotypes are observed in null mutants for YFH1 (Babcock et al., 1997; Foury and Cazzalini, 1997), the yeast homologue for the human frataxin gene, with a possible role in mitochondrial iron homeostasis (Foury, 1999; Radisky et al., 1999; although other functions have also been suggested for YFH1 [Ristow et al., 2000]). All of these observations suggest a relationship among Fe/S cluster biogenesis, mitochondrial metal homeostasis, and oxidative damage.
In this work we demonstrate that Grx5 is a mitochondrial glutaredoxin required for the activity of Fe/S-containing enzymes and that its absence affects iron homeostasis and causes osmotic stress at the cell.
MATERIALS AND METHODS
Yeast Strains and Plasmids
The yeast strains are described in Table 1. The following plasmids contain the genes indicated in parenthesis plus their own promoter and terminator sequences (without adjacent complete open reading frames [ORFs]) cloned in the multicopy vector Yeplac181 (Gietz and Sugino, 1988): pMM62 (GRX5), pMM70 (SSQ1), pMM72 (ISA1), and pMM74 (ISA2). Plasmids pCM316, pCM317, and pCM318 contain the complete GRX4, GRX3, and GRX5 ORFs, respectively (without further sequences), cloned in the multiple cloning site of pCM190 (Garíet al., 1997) under the control of the doxycycline-regulated tetO7 promoter. pMM54 is a YIplac128 (Gietz and Sugino, 1988) derivative with GRX5 under its own promoter, tagged at the 3′-end with three hemagglutinin (HA) epitopes in tandem. pMM117 is a derivative of YIplac211 (Gietz and Sugino, 1988) containing the doxycycline-regulated tTA activator (Garíet al., 1997) and the GRX5 ORF (plus terminator sequences) under the control of the tetO7 promoter.
Table 1.
Strains used in this work
| Strain | Relevant phenotype | Comments |
|---|---|---|
| CML235 | MATa ura3-52 leu2Δ1 his3Δ200 | Wild type (Rodriguez-Manzaneque et al., 1999) |
| CML236 | As CML235 but MATα | Wild type |
| MML19 | MATa grx5∷kanMX4 | Deletion of GRX5 in CML235 |
| MML246 | MATa/MATα GRX5/grx5∷kanMX4 | Diploid from a cross CML236 × MML19 |
| MML248 | MATa/MATα GRX5/grx5∷kanMX4 trx3∷CaURA3 | Deletion of TRX3 in MML246 |
| MML264 | MATa grx5∷kanMX4 trx3∷CaURA3 | Spore from MML248 |
| W303-1A | MATa ura3-1 ade2-1 leu2-3,112 trp1-1 his3-11,15 | Wild type |
| W303-1B | As W303-1B but MATα | Wild type |
| MML100 | MATa grx5∷kanMX4 | Deletion of GRX5 in W303-1A |
| MML235 | MATα [pMM54(GRX5-3HA)]∷LEU2 | Integration of linear pMM54 in W303-1B |
| MML240 | MATa grx5∷kanMX4 [pMM54(GRX5-3HA)]∷LEU2 | Spore from a cross MML100 × MML235 |
| MML266 | MATα [pMM96(grx5-Δ8-3HA)]∷LEU2 | Integration of linear pMM96 in W303-1B |
| MML271 | MATα (pMM98[grx5-Δ23-3HA])∷LEU2 | Integration of linear pMM98 in W303-1B |
| MML289 | MATα grx5∷kanMX4 | Spore from a cross W303-1B × MML100 |
| MML290 | MATa grx5∷kanMX4 (pMM96 [grx5-Δ8-3HA])∷LEU2 | Spore from a cross MML100 × MML266 |
| MML298 | MATa yfh∷kanMX4 | Spore from a cross MML100 × MML281 |
| MML300 | MATa yfh∷kanMX4 grx5∷kanMX4 | Spore from a cross MML100 × MML281 |
| MML312 | MATa (pMM117[tTA tetO-GRX5])∷URA3 tetR′-Ssn6∷LEU2 | Integration of linear pMM117 in CML240 (Belli et al., 1998) |
| MML313 | MATa grx5∷kanMX4 (pMM117[tTA tetO-GRX5])∷URA3 tetR′-Ssn6∷LEU2 | Spore from a cross MML289 × MML312 |
| MML345 | MATa aft1-Δ5∷URA3 grx5∷kanMX4 | Spore from a cross W303-1B × MML348 |
| MML348 | MATa aft1-Δ5∷URA3 | Several backcrosses from CML126 (Casas et al., 1997) to introduce the aft1-Δ5 mutation in the W303 background |
Growth Conditions and Determination of Cell Parameters
Cells were grown at 30°C in YPD, YPG (as YPD but with 3% glycerol instead of dextrose), SD medium (0.67% yeast nitrogen base, 2% glucose, and auxotrophic requirements), or SC medium (the same as SD plus drop-out mixture [Kaiser et al., 1994 ]). Specific supplements were omitted from the SC medium when required. For growth in anaerobic conditions, inoculated plates were incubated in an anaerobiosis chamber. Cell numbers (in formaldehyde-fixed samples) and mean cell volumes were determined in a Coulter Z2 counter (Beckman Coulter, Fullerton, CA). 4′,6-Diamidino-2-phenylindole staining was done as described by Kaiser et al. (1994).
Gene Disruptions and Other Genetic Methods
Standard methods were used for DNA manipulation, transformation, crosses between yeast strains, sporulation, and tetrad analyses. SC medium with appropriate supplements was used to select transformants on grx5 mutant cells. The wild-type GRX5 allele was disrupted in the W303 background using the kanMX4 cassette, as previously described (Rodríguez-Manzaneque et al., 1999). A similar approach was used to construct a null mutant in TRX3, using a polymerase chain reaction-amplified cassette with the CaURA3 marker from pAG60 (Goldstein et al., 1999). Oligonucleotides for amplification of the disruptant cassettes were designed to disrupt most of the targeted gene upon transformation with the amplified DNA.
Isolation of grx5Δ Suppressors
Exponentially growing MML19 cells were transformed with a yeast genomic DNA library in the multicopy plasmid YEp13. The transformation mixture was then plated on SD agar plates, which were incubated for 6 d at 30°C. Wild-type CML235 cells were transformed in parallel as a control to quantify transformation efficiency. Two independent transformation experiments were carried out with ∼60,000 transformants on wild-type cells. Plasmids were recovered from growing grx5 transformants, amplified in Escherichia coli, and retransformed on MML19 cells to confirm their ability to suppress the transformation defect of grx5 mutants. Finally, we recovered nine plasmids that gave stable transformation on grx5 cells. Restriction fragment analysis and hybridization with a GRX5 probe demonstrated that the plasmids corresponded to four different clones. One (three separate isolates) contained GRX5, and the other three clones contained inserts from other chromosomal locations: pMM44 (one isolate), pMM45 (two isolates), and pMM46 (three isolates). Partial sequencing of insert ends was carried out to reveal the genes included in each suppressor plasmid.
Construction of GRX5 Derivatives with Deletions at the Mitochondrial Targeting Sequence
Plasmid pMM54 (GRX5-3HA) was used as a template to generate two constructions with deletions in the GRX5 coding sequence, using the ExSite approach (Weiner and Costa, 1995). The resulting pMM96 plasmid contains a deletion that spans from base pairs +4 to +27 (grx5-Δ8), and pMM98 has a deletion spanning from base pairs +4 to +72 (grx5-Δ23). Linearized (EcoRV digestion) plasmids pMM96 and pMM97 were stably integrated at the chromosomal LEU2 locus of W303–1B cells and generated strains MML266 and MML271, respectively.
Sensitivity to Menadione
Cells growing exponentially in YPD medium at 30°C (2 × 107 cells/ml) were added with menadione. Plasmid-transformed cultures were grown in SD medium in selective conditions. In this case, cells were transferred to YPD medium for 4 h before sensitivity analyses. After various treatment times, 1:5 serial dilutions were made, and drops were spotted onto YPD plates. Growth was recorded after 2 d of incubation at 30°C.
Western Blot Analyses
Western blot analyses were done according to the method of Bellíet al. (1998). 12CA5 anti-HA mAb (Roche Diagnostics, Mannheim, Germany) was used at a 1:5000 dilution. An anti-lipoic acid antibody was used at a 1:50,000 dilution to detect protein-bound lipoic acid (Cabiscol et al., 2000). Anti-Aco1 aconitase (1:2000 dilution, from R. Lill), anti-succinate dehydrogenase (1:1000 dilution, from B. Lemire), and anti-cytochrome b2 (1:1000 dilution, from E. Valentín) antibodies were also used.
Isolation of Mitochondrial Fractions
Mitochondria were purified as described by Luttik et al. (1998). Zymolyase 20T (ICN Biochemicals, Cleveland, OH) was used at 3 mg/g of cells (dry weight). Spheroplasts were broken (eight strokes) with a Dounce homogenizer. Mitochondria (pellet) were separated from the postmitochondrial (supernatant) fraction, resuspended in a hypotonic solution (20 mM HEPES plus 1 mM phenylmethylsulfonyl fluoride), and centrifuged in a microfuge (12,000 rpm, 10 min) at 0°C. The resulting pellet and supernatant were respectively considered as the intermembrane space and matrix fractions.
Identification of the Signal Peptide Cleavage Site for Grx5
Four grams of cells grown in YPG medium were resuspended in 50 mM Tris buffer, pH 7.5, plus 20 mM NaCl and disrupted in a French Press (SLM Aminco). After centrifugation (12,000 rpm, 30 min), the crude supernatant was applied to an ionic exchange column (DEAE 15HR, Waters, Milford, MA). Proteins were eluted with a linear gradient (20–400 mM NaCl in Tris-HCl buffer, pH 7.5), and fractions containing Grx5 were identified by Western blot using specific polyclonal antibodies. Proteins in the fractions were separated by two-dimensional electrophoresis. First dimension was performed in a Protean Isoelectric Focusing Cell (Bio-Rad, Hercules, CA) using 17-cm IPG strips (Bio-Rad Ready Strip, pH range 3–11). Second dimension was performed according to the denaturing discontinuous buffer system of Laemmli. Proteins were transferred to a polyvinylidene difluoride membrane using a semidry system, and the spot corresponding to Grx5 (identified by Western blot in a duplicate membrane with the above antibodies) was N-terminal sequenced by Edman degradation using a Beckman LF3000 sequencer equipped with a phenylthiohydantoin derivative analyzer (System Gold, Beckman).
Other Methods
Analysis of protein carbonylation after derivatization of carbonyl groups with dinitrophenylhydrazine was carried out as described by Cabiscol et al. (2000). Enzymatic activities were assayed by the following standard methods: aconitase, citrate synthase, malate dehydrogenase (Robinson et al., 1987), and succinate dehydrogenase (Munujos et al., 1993). Activities were expressed in units (nanomoles per minute) per milligram of cell protein. Extracts were prepared in 0.1 M Tris buffer, pH 8.1, plus 2 mM EDTA using glass beads to disrupt the cells. Nonmitochondrial citrate synthase is not stable at this pH (Liao et al., 1991). Whole cell and mitochondrial iron were determined under reducing conditions (Fish, 1988), with bathophenanthroline sulfonate (BPS) as chelator and after acid digestion of cells or mitochondria with 3% nitric acid. Mitochondrial and postmitochondrial iron were also determined according to the method of Tangeras et al. (1980) after incubation of the samples with 10 mM 2-(N-morpholino)ethanesulfonic acid-KOH buffer, pH 4.5, plus 1% SDS (1 h, 95°C). No significant differences were observed among results with both methods. Heme covalently bound to cytochrome c was detected as described by Vargas et al. (1993), using the Supersignal detection system (Pierce Chemical, Indianapolis, IN). To purify yeast Grx5 glutaredoxin, the entire GRX5 ORF was cloned in-frame in pGEX-4T1 (Amersham Pharmacia Biotech, Piscataway, NJ) to generate a GSH S-transferase (GST)-Grx5 fusion protein. The construct was expressed in E. coli cells. GST-Grx5 was purified from bacterial cell extracts using GSH-Sepharose 4B columns (Amersham Pharmacia). After thrombin cleavage, Grx5 was separated from GST and contaminants by preparative electrophoresis, using a Bio-Rad 491 PrepCell. Polyclonal anti-Grx5 antibodies were raised in rabbits and purified from rabbit serum in a protein A-Sepharose CL-4B column (Amersham Pharmacia).
RESULTS
Defects in grx5 Mutants Are Suppressed by Genes Involved in Fe/S Cluster Assembly
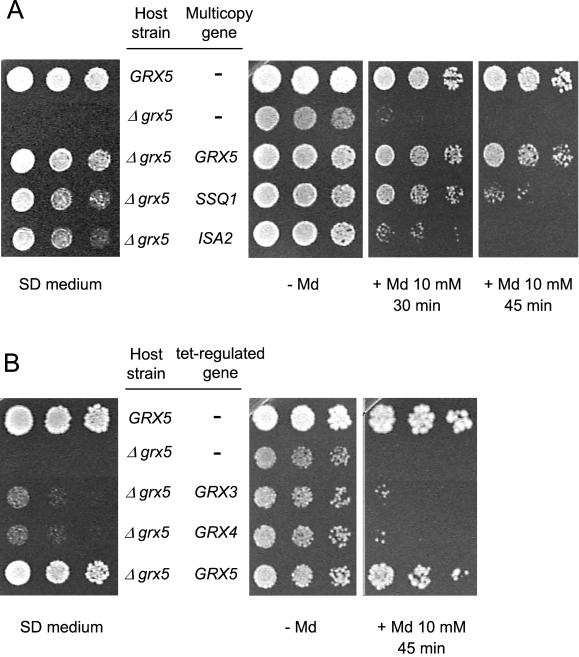
Given the defective growth phenotype of null grx5 mutants in minimal SD medium, we isolated transformants (on MML19 cells) from a multicopy genomic DNA library that were able to grow in such conditions. Three different clones, whose inserts were characterized by DNA sequencing, were redundantly isolated in two independent experiments. Plasmid pMM44 contains two complete ORFs, SSQ1 and ARC18. Plasmid pMM45 contains ROX1, UBA3, ISA2, HOS1, and SPE3, and pMM46 contains RPS22B, YLR368w, SSQ1, and ARC18. We focused our attention on SSQ1 and ISA2, two genes involved in Fe/S cluster assembly at the mitochondria (see INTRODUCTION). To confirm that SSQ1 and ISA2 were really suppressors of grx5 defects, both genes and their respective promoter and terminator sequences were cloned in the multicopy plasmid YEplac181 and then used to transform a grx5 null mutant. This mutant was also transformed with a construction carrying GRX5 under its own promoter in YEplac181. Transformants were obtained in all three cases (∼50% transformation efficiency for SSQ1 and 30% for ISA2 relative to GRX5). Multicopy plasmids containing SSQ1 or ISA2 allowed grx5 cell growth in SD medium (Figure 1A, left). In addition, the SSQ1 plasmid suppressed better the sensitivity of grx5 cells to menadione than did the ISA2 plasmid (Figure 1A, right). Although ISA2 is highly homologous in sequence to ISA1, overexpression of the latter was not capable of suppressing the grx5 phenotypes (Rodríguez-Manzaneque, Tamarit, Bellí, Ros, and Herrero, unpublished results). We therefore conclude that, when overexpressed, some (but not all) of the genes involved in the Fe/S cluster assembly are capable of counteracting the absence of GRX5. This suggests a functional relationship between Grx5 glutaredoxin and Fe/S cluster assembly at the mitochondria. As with mutants in the Fe/S assembly machinery and with yeast frataxin mutants (Babcock et al., 1997; Jensen and Culotta, 2000; Kaut et al., 2000), grx5 cells are unable to use glycerol as a sole carbon source (Rodríguez-Manzaneque et al., 1999). In contrast with other phenotypes shown in Figure 1A, grx5 cell respiratory ability was not directly rescued by transformation with the GRX5 gene. To the contrary, a plasmid carrying GRX5 only rescued growth ability on glycerol in a chromosomal grx5 background after this mutant had been crossed with wild-type cells transformed with the GRX5 plasmid, followed by sporulation of the resulting diploid. This is consistent with grx5 cells having extensively accumulated mutations in mitochondrial DNA, thereby causing the respiratory defect.
Figure 1.
Suppression of phenotypic defects of null grx5 mutants. (A) CML235 wild-type (GRX5) cells and MML19 (Δgrx5) cells nontransformed or transformed with the multicopy plasmids pMM62 (GRX5), pMM70 (SSQ1), or pMM74 (ISA2) were tested for growth on SD medium plates (left) and for sensitivity to 10 mM menadione (Md) treatment at 30°C (right). Serial dilutions of exponential cultures (treated with menadione for the indicated times) were plated on YPD plates. (B) CML235 and MML19 cells transformed with the pCM190 (tetO7 promoter)-derived plasmids pCM317 (GRX3), pCM316 (GRX4), or pCM318 (GRX5) were tested for growth on SD medium plates (left) and for menadione sensitivity (right) in overexpression conditions (minus doxycycline).
We also tested whether GRX3 or GRX4 (the other two members of the same gene family) suppressed the phenotypes of a grx5 mutant when overexpressed from the doxycycline-regulatable tet promoter (Garíet al., 1997). Grx3 and Grx4 only slightly overcame the sensitivity to menadione or the growth defect in SD medium of cells deficient in Grx5 (Figure 1B), suggesting that Grx5 performs different functions from the other two members of the family.
Grx5 Is a Mitochondrial Glutaredoxin
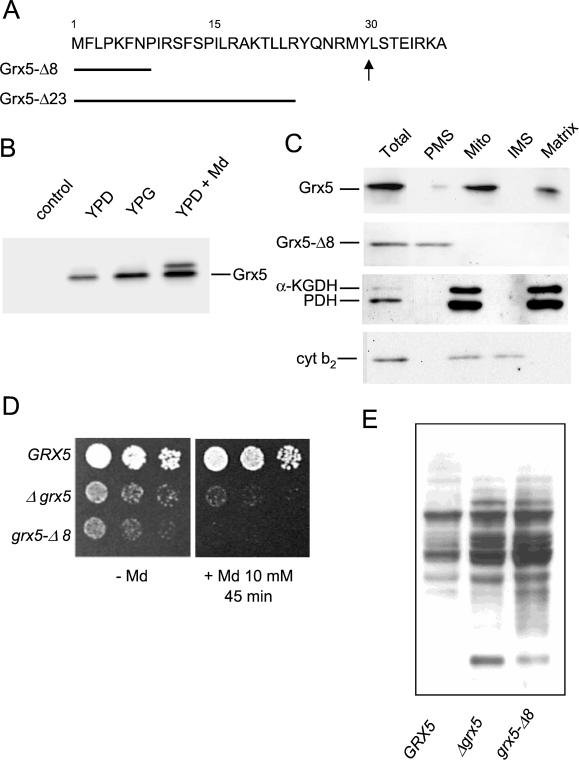
Genetic interactions between Grx5 and mitochondrial proteins implicated in the biogenesis and protein assembly of Fe/S clusters suggested that Grx5 could be a mitochondrial glutaredoxin. PSORT analysis predicts a mitochondrial location for Grx5 due to mitochondrial targeting signatures at its N-terminal region (Pon and Schatz, 1991). To confirm this, we raised antibodies against Grx5 protein that had been expressed in E. coli and then purified. We used these antibodies to purify the mature form of the overexpressed (from the tet promoter) Grx5 protein in S. cerevisiae. N terminus sequencing of the protein spot isolated from a two-dimensional gel showed that mature Grx5 begins with the sequence LSTEIRKA. Hence, the mature product lacks the first 29 amino acids predicted from the proposed GRX5 ORF (Figure 2A). Remarkably, GRX3 and GRX4 sequences have no homology with these N-terminal Grx5 residues (Rodríguez-Manzaneque et al., 1999), and PSORT analysis predicts no mitochondrial location for either Grx3 or Grx4.
Figure 2.
Grx5 is a mitochondrial glutaredoxin. (A) N terminus Grx5 amino acid sequence. Arrow marks where the precursor form is processed, as determined by N terminus sequencing of mature Grx5. The length of the two constructed signal peptide deletions (Grx5-Δ8 and Grx5-Δ23) is indicated. (B) Western blot immunodetection of 3HA-tagged Grx5 in extracts from exponential cultures of MML240 cells in YPD and YPG medium at 30°C and in YPD medium plus menadione (Md, 30 min). Left lane (control) corresponds to W303–1A cell extracts. The same amount of total cell protein (40 μg) was run in each lane. (C) MML235 and MML266 (Grx5-Δ8) cells grown exponentially in YPG medium at 30°C were fractionated, and the resulting fractions were analyzed by Western blot. Anti-HA antibodies were used to detect Grx5 (MML235 fractions) and Grx5-Δ8 (MML266 fractions), and anti-lipoic acid antibodies were used to detect the matrix markers pyruvate dehydrogenase (PDH) and α-ketoglutarate dehydrogenase (α-KGDH). Cytochrome b2 (cyt b2) was used as an intermembrane space (IMS) marker. Results for pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, and cytochrome b2 are shown only for MML266; similar results were observed for MML235. Ten micrograms of protein were loaded in each lane for “Total” cell extracts and postmitochondrial supernatant (PMS) fractions, and 3 μg were loaded for the mitochondrial (Mito), intermembrane space, and matrix fractions. (D) Sensitivity to menadione in strains MML240 (GRX5), MML100 (Δgrx5), and MML290 (grx5-Δ8) growing exponentially at 30°C in YPD medium. (E) Protein carbonylation in extracts from exponentially growing MML240, MML100, and MML290 cells in YPD medium at 30°C.
We tested whether it was possible to detect a 3HA-tagged version of Grx5 expressed under its own promoter. This tagged form fully complemented all the grx5 mutant defects in the MML240 strain, obtained after crossing the grx5 mutant with a wild strain carrying an integrative plasmid with the GRX5–3HA construction. A major form of the expected mobility in Western blots was detected in extracts from exponential cells grown on both glucose and glycerol (Figure 2B). A larger minor form was also observed, especially in cells grown in YPD medium after menadione treatment. Although this treatment did not up-regulate GRX5 expression (Rodríguez-Manzaneque et al., 1999), the total amount of immunodetectable Grx5 protein almost doubled compared with untreated cells (Figure 2B). This increase points to some kind of posttranscriptional regulation of Grx5 levels in oxidative conditions. We then used the tagged version of Grx5 to determine its cellular location. Cell fractionation studies demonstrated that wild-type Grx5 is located at the mitochondria (Figure 2C). After causing outer membrane disruption under hypotonic conditions, mitochondrial subfractionation showed that Grx5 entirely colocalized with two matrix lipoic acid-modified proteins, pyruvate dehydrogenase and α-ketoglutarate dehydrogenase (Figure 2C). From these results, we hypothesized that the deletion of a number of amino acids at the N terminus would cause Grx5 to remain at the cytoplasm. Two shorter forms of Grx5 were constructed, one lacking the first eight amino acids (Grx5-Δ8) and the other lacking the first 23 residues (Grx5-Δ23; Figure 2A). We obtained the same results with both, although here we only show results corresponding to the eight-residue deletion. The shorter version of Grx5 remained exclusively at the cytoplasm (Figure 2C), thus confirming the importance of N terminus residues for adequate targeting of Grx5 at the mitochondria.
We then addressed the possibility that this version of Grx5, present at the cytosol, could also protect cells against oxidative stress. We therefore constructed a strain that produced only the cytosolic Grx5-Δ8 version. This strain was hypersensitive to menadione (Figure 2D) and showed constitutive carbonylation levels even higher than those of cells containing no Grx5 (Figure 2E). We conclude that Grx5 is located in the mitochondria and that the abnormal presence of Grx5 at the cytosol does not complement the phenotypes that result from the absence of mitochondrial Grx5. The moderately dominant negative effect of the grx5-Δ8 allele leaves open the possibility that the mutant protein interferes with some cytosolic mechanism involved in oxidative stress defense.
The Absence of Grx5 Causes Inactivation of Mitochondrial Fe/S Enzymes
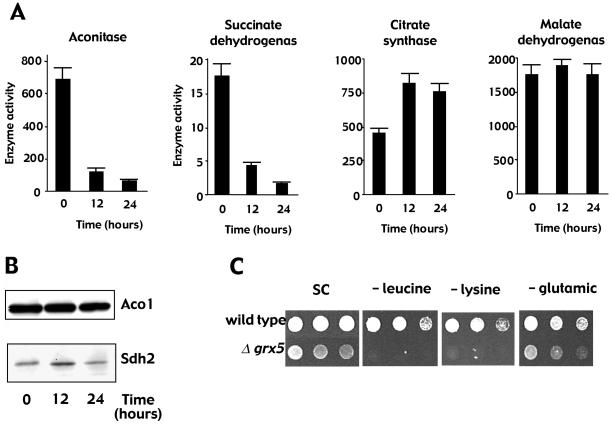
The defects associated with the absence of GRX5 are common to mutants in mitochondrial proteins involved in the biosynthesis/assembly of Fe/S clusters (Lill and Kispal, 2000, and references therein). These defects and the genetic interactions between GRX5 and genes responsible for the biogenesis of Fe/S clusters led us to investigate the involvement of Grx5 in the biogenesis and/or repair of such clusters. We therefore measured the activity of mitochondrial enzymes containing Fe/S clusters in a conditional mutant in which GRX5 expression under the tet promoter was doxycycline regulated. For this purpose, the wild-type GRX5 allele was deleted and a tet-GRX5 construction was integrated at the LEU2 locus. The resulting strain (MML313) grew on glycerol in the absence of doxycycline (GRX5 expressed), but upon addition of the antibiotic (GRX5 transcription immediately repressed), growth became arrested in ∼12 h. 4′,6-Diamidino-2-phenylindole staining confirmed that mitochondria retained the DNA after 24 h of inhibition of GRX5 expression, which is in accordance with a [rho−] phenotype. The activity of two mitochondrial enzymes with Fe/S clusters (aconitase and succinate dehydrogenase) decreased dramatically when GRX5 was not expressed (<15% activity after 24 h; Figure 3A). In contrast, the activity of two mitochondrial enzymes not depending on Fe/S clusters (citrate synthase and malate dehydrogenase) remained unchanged. Western blot analyses of aconitase and succinate dehydrogenase demonstrated that the amounts of the two proteins were not affected by inhibition of GRX5 expression (Figure 3B), indicating that the changes in activity can be attributed to impairment of formation of mature molecules.
Figure 3.
Lack of Grx5 negatively affects Fe/S enzyme activity. (A) MML313 cells growing exponentially in YPG medium at 30°C (1 × 107 cells/ml) were added at time 0 with doxycycline (2 μg/ml) to inhibit GRX5 expression. Enzyme activity in cell extracts was determined at the indicated times. (B) Western blot analysis of aconitase (Aco1) and succinate dehydrogenase (Sdh2) in the same samples as in A. (C) Growth of wild-type CML235 and mutant MML19 (Δgrx5) cells containing the integrative LEU2 plasmid YIplac128 (Gietz and Sugino, 1988) in SC medium with all the supplements or deprived of leucine, lysine, or glutamic acid.
The inhibition of Fe/S enzyme activities could give a clue to the grx5 mutant growth defects in minimal SD medium. When comparing growth of wild-type and grx5 cells in defined SC medium that lacked each of the 20 amino acids, we observed that grx5 cells were unable to grow (or grew poorly) when deprived of leucine, lysine, or glutamic acid (Figure 3C). However, the absence of any other amino acid did not affect growth. The growth defect was rescued when the mutant cells were transformed with a centromeric plasmid expressing GRX5 (Rodríguez-Manzaneque, Tamarit, Bellí, Ros, and Herrero, unpublished results). Thus, auxotrophy for these three amino acids could explain defective growth of the mutant in minimal medium. All three amino acids require Fe/S enzymes for their biosynthesis. Glutamate biosynthesis requires mitochondrial aconitase (Gangloff et al., 1990), and the inactivation of this Fe/S enzyme also explains the glutamate requirement of isa1 and isa2 mutants (Jensen and Culotta, 2000). Lysine auxotrophy probably results from the inactivation of the mitochondrial Fe/S enzyme homoaconitase, which is involved in its synthesis (Bhattacharjee, 1985; De Freitas et al., 2000). Lysine auxotrophy also occurs in isa mutants (Jensen and Culotta, 2000). Leucine biosynthesis requires the cytoplasmic Fe/S enzyme isopropyl malate isomerase, Leu1 (Kohlhaw, 1988). Inactivation of this enzyme has been described for a number of mutants altered in the assembly of Fe/S cluster proteins (Kispal et al., 1999; Kaut et al., 2000; Lange et al., 2000). All of these data support the relationship between the growth defects observed in grx5 cells in minimal medium and the inactivation of Fe/S cluster-containing enzymes implicated in amino acid biosynthesis.
Cells without Grx5 Are Not Defective in the Holoforms of Heme-containing Proteins
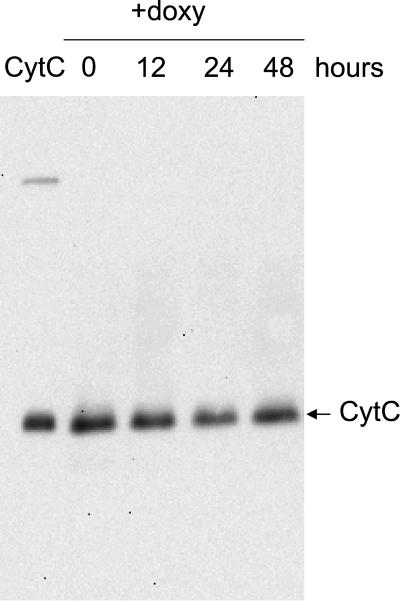
Although heme groups and Fe/S clusters have different biosynthetic pathways, both are also synthesized within the mitochondria, require a source of reduced iron, and are sensitive to oxidative stress (Kranz et al., 1998; Lange et al., 1999). Heme deficiency in yeast has also been found associated with mutations such as atm1 (Kispal et al., 1997), jac1 (Voisine et al., 2001), and yfh1 (Foury, 1999), although this could be the result of alterations in iron homeostasis. Therefore, we analyzed the presence of the mature form of cytochrome c in cell extracts from MML313 cells (tetO7-GRX5) grown in YPG medium before and after the addition of doxycycline. Holo-cytochrome c was detected through the peroxidase activity displayed by the heme groups. The content of holo-cytochrome c did not decrease even after 48 h of repression of GRX5 expression (Figure 4). Although cytochrome c is rather stable in normal growth conditions (half-life of ∼7 h [Pearce and Sherman, 1995]), a moderate to strong effect of Grx5 absence on heme synthesis would have been detected during the time course of the experiment. Thus, Grx5 is not essential for the biosynthesis of heme-containing proteins in mitochondria.
Figure 4.
Effect of Grx5 depletion on the amount of heme covalently bound to cytochrome c. Expression of GRX5 was interrupted (time 0) by the addition of doxycycline (2 μg/ml) to MML313 cells growing exponentially in YPG medium at 30°C. Proteins from whole cell lysates (samples taken at the indicated times after antibiotic addition) were separated by nonreducing SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and analyzed for heme-carrying proteins. In these conditions, heme bound to cytochrome c was the most prominent band detected (marked with an arrow). As a standard, 0.1 μg of cytochrome c from bovine heart was loaded on the left-most line.
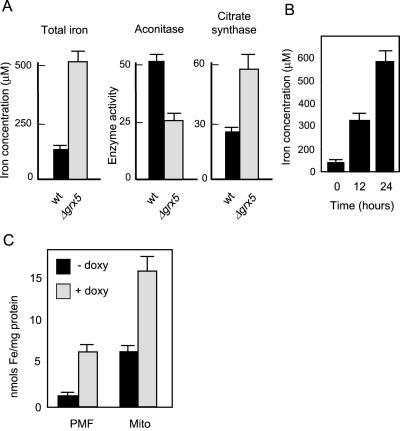
Iron Accumulates in the Cells in the Absence of Grx5
We next analyzed whether, as with other mutants in Fe/S cluster biogenesis, the absence of Grx5 caused iron accumulation in the cells. In the grx5 mutant growing exponentially in YPD medium, an almost sixfold increase in total cell iron was observed with respect to wild-type cells (Figure 5A). This was paralleled by a decrease in aconitase activity but not in the activity of the non-Fe/S enzyme citrate synthase (Figure 5A). Inhibition of aconitase in the grx5 cells grown on glucose was not as dramatic as that observed in cells conditionally expressing GRX5 on glycerol (Figure 3A). The iron accumulation was attributable to the absence of Grx5. In fact, in MML313 (tetO7-GRX5) cells cultured in YPG medium under the same conditions as in Figure 3A, inhibition of GRX5 expression by doxycycline induced iron accumulation in the cell (Figure 5B).
Figure 5.
Iron accumulates in the cell in the absence of Grx5. (A) W303–1A (wt) and MML100 (Δgrx5) cells were grown exponentially in YPD medium at 30°C to determine iron concentration in the cell and also aconitase and citrate synthase activities in total cell extracts. (B) MML313 cells growing exponentially in YPG medium at 30°C were added (time 0) with doxycycline (2 μg/ml), and total cell iron concentration was determined at different intervals after antibiotic addition. (C) Distribution of iron (relative to total protein in the fraction) between postmitochondrial fraction (PMF) and mitochondria (Mito), in MML313 cells untreated (−doxy) or treated for 24 h with doxycycline at 2 μg/ml (+doxy).
A yeast frataxin mutant (yfh1) preferentially accumulates iron at the mitochondria, whereas cytosolic iron becomes depleted (Babcock et al., 1997; Radisky et al., 1999). On the contrary, when non-GRX5–expressing cells were subfractionated into mitochondrial and postmitochondrial fractions, iron was shown to hyperaccumulate in both fractions (Figure 5C). Control analyses showed that the postmitochondrial fraction was not contaminated by mitochondrial enzymes (citrate synthase and α-ketoglutarate dehydrogenase; Rodríguez-Manzaneque, Tamarit, Bellí, Ros, and Herrero, unpublished results). Under our conditions, mitochondrial iron represents 23–28% of total cell iron in both the absence and presence of GRX5 expression (as calculated from the data in Figure 5C).
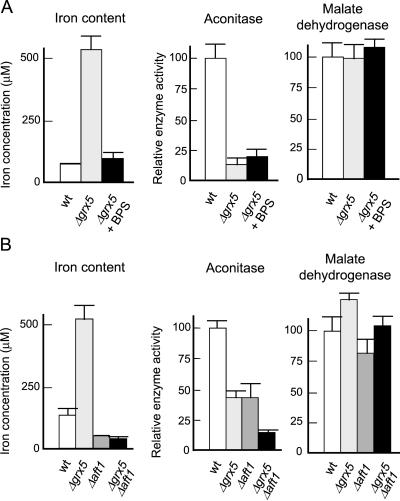
Inactivation of Fe/S Enzymes in the Absence of Grx5 Is Not a Consequence of Iron Accumulation in the Cell
Grx5 could be directly responsible for maintaining iron homeostasis. In that case, inactivation of Fe/S enzymes in grx5 cells could be caused by the generation of ROS due to the presence of high levels of iron. Alternatively, Grx5 could be directly implicated in the biogenesis of Fe/S-protein complexes. In the latter case, increased iron levels in both cytosol and mitochondria would be a consequence of impairment of the formation of such complexes in the absence of the glutaredoxin. We used two approaches to test the direct involvement of Grx5 in Fe/S-enzyme biogenesis independently of the iron levels existing in the cell. First, we used the iron chelator BPS to create conditions in which internal iron levels in grx5 cells were similar to those in wild-type cells. In such conditions, aconitase activity remained greatly diminished in the mutant, in contrast to the non-Fe/S enzyme malate dehydrogenase (Figure 6A). It should be noted that, in this particular genetic background, reduction of aconitase activity in grx5 cells compared with wild-type cells is even higher than in the W303 background in the same growth conditions (compare Figures 6A and 5A).
Figure 6.
Activity of Fe/S enzymes is inhibited in grx5 cells independently of intracellular iron concentration. (A) CML235 (wt) or MML19 (Δgrx5) cells were grown exponentially in YPD medium (also in the presence of 80 μM BPS in the case of MML19 cells), and total cellular iron concentration and aconitase and malate dehydrogenase activity were determined. (B) The same parameters as in A were determined in exponential YPD cultures at 30°C of W303–1A (wt), MML100 (Δgrx5), MML348 (Δaft1), and MML345 (Δgrx5 Δaft1) cells.
Second, based on the fact that Aft1 is a transcriptional factor involved in the expression of genes responsible for the high-affinity iron transport system (Yamaguchi-Iwai et al., 1995; Casas et al., 1997), we constructed a single aft1 and a double aft1 grx5 mutant. As expected, aft1 cells had reduced intracellular iron levels, and, probably as a consequence of this, activity of iron-dependent enzymes such as aconitase was also reduced (Figure 6B). The absence of Grx5 did not lead to iron accumulation in the aft1 grx5 cells, indicating that increased levels of the metal in grx5 cells requires Aft1-dependent iron transport. Importantly, the double mutant displayed additional reduction of aconitase activity compared with aft1 cells, in conditions where intracellular iron remained low (Figure 6B), thus confirming that the primary consequence of the absence of Grx5 is not the accumulation of iron.
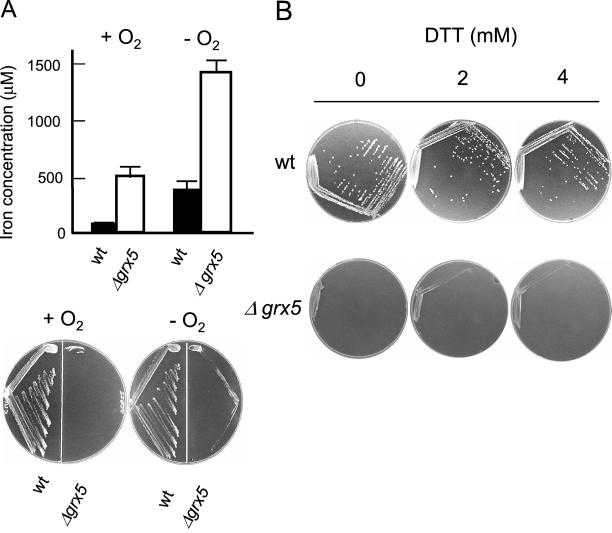
Modification of the Intracellular Redox Potential Does Not Suppress the grx5 Growth Defects
Glutaredoxins have been assigned a role as general reductants of disulfide bonds in cell proteins (Prinz et al., 1997; Carmel-Harel and Storz, 2000). In E. coli, inactivation of the glutaredoxin and/or thioredoxin systems alters the thiol-disulfide equilibrium, which can be reversed in anaerobic conditions or by external reductants such as dithiothreitol (DTT; Prinz et al., 1997). We hypothesized that the growth defects in the absence of Grx5 could be caused by the alteration of the redox potential at the mitochondria and, consequently, by the inhibition of oxidation-sensitive Fe/S-containing proteins. To address this problem, wild-type and grx5 cells were cultured in SD minimal medium in anaerobic conditions, a situation that should compensate, at least in part, for the effect of the grx5 mutation on the thiol-disulfide equilibrium. However, the mutant was unable to grow on SD plates in anaerobiosis (Figure 7A). While growing in anaerobiosis in SC medium, grx5 cells still accumulated high amounts of iron compared with wild-type cells (Figure 7A). As a second approach, cells were cultured on SD plates to which with different amounts of DTT were added, in conditions where this reductant has been shown to be active on yeast cells in vivo (Holst et al., 1997). DTT was unable to suppress the growth defects of the grx5 mutant at concentrations up to 4 mM (Figure 7B). Higher DTT concentrations were partially inhibitory of growth of wild-type cells in SD medium, whereas in complete medium grx5 cells did not show higher sensitivity to DTT than wild-type cells (Rodríguez-Manzaneque, Tamarit, Bellí, Ros, and Herrero, unpublished results). We conclude that the grx5 defective growth is not primarily due to the alteration of the intracellular redox potential, thus supporting the direct participation of Grx5 glutaredoxin in Fe/S cluster biogenesis.
Figure 7.
Growth of grx5 cells is affected in anaerobiosis or in the presence of external reductants. (A) CML235 (wt) and MML19 (Δgrx5) cells were grown exponentially in SC liquid medium at 30°C to exponential phase, in anaerobic or aerobic conditions, and total iron concentration in the cell was determined (top). The same strains were inoculated on SD agar plates, and growth was recorded after 3 d (+ O2) or 4 d (−O2) of incubation at 30°C. (B) CML235 (wt) and MML19 (Δgrx5) cells were inoculated on SD agar plates containing different DTT concentrations, and growth was recorded after 3 d (wt) or 4 d (Δgrx5) of incubation at 30°C.
DISCUSSION
Yeast cells contain both dithiol (Grx1, Grx2) and monothiol (Grx3, Grx4, Grx5) glutaredoxins. The two types of glutaredoxins coexist in many species from bacteria to humans (Rodríguez-Manzaneque et al., 1999), but specific roles for monothiol glutaredoxins have not previously been established. Grx5 is the yeast glutaredoxin whose absence causes the most dramatic effects on the oxidative damage to cell proteins, sensitivity to external oxidants, and general growth defects (Rodríguez-Manzaneque et al., 1999). We have shown that Grx5 is located at the mitochondrial matrix and that its absence has a negative effect on the activity of mitochondrial proteins with Fe/S clusters but not on heme-containing proteins. Mitochondrial matrix location has also been demonstrated for other proteins that participate in Fe/S center protein assembly (reviewed by Lill and Kispal, 2000). The functional relationship between Grx5 and Fe/S cluster assembly was confirmed by the fact that overexpression of genes participating in Fe/S cluster assembly partially suppressed various grx5 cell phenotypes. Of those, SSQ1 codes for a Hsp70-type chaperone that might stabilize apoproteins for Fe/S cluster coordination (Lill and Kispal, 2000) or even participate directly in the recognition/transfer step of clusters from Isu proteins to receptor polypeptides (Silberg et al., 2001). The other partial suppressor of grx5 mutants is ISA2. IscA, its product homologue in E. coli, complexes with ferrodoxin to transfer iron and sulfide to form [2Fe-2S]-ferrodoxin (Ollagnier-de-Choudens et al., 2001). The fact that ferrodoxin is also a component of the Fe/S cluster synthesis machinery in yeast suggests the hypothesis that multiprotein complexes form part of such a machinery.
The function of Grx5 in the assembly of Fe/S centers is not apparently related to that proposed for the human homologue PICOT, which would regulate signaling through the protein kinase Cθ pathway (Isakov et al., 2000; Witte et al., 2000). Although Grx5 contains the PICOT homology domain, it lacks the N-terminal extension (with a thioredoxin or glutaredoxin-like module) present in Grx3 and Grx4 and also in other members of the PICOT superfamily (Isakov et al., 2000). This, coupled with the differential compartmentalization, supports the hypothesis that the PICOT domain could be shared by various oxidoreductases, which contain a CKSF motif in the domain (Rodríguez-Manzaneque et al., 1999) but serve different biological functions.
Besides the inactivation of enzymes with Fe/S clusters such as aconitase or succinate dehydrogenase, mutants deficient in GRX5 share a number of phenotypes with others affected in the Fe/S cluster synthesis. These deficiencies include inability to grow in respiratory conditions and iron accumulation in their mitochondria. The last of these may be responsible for the oxidative damage observed in various cellular macromolecules when Fe/S cluster assembly is disrupted because of iron-mediated ROS formation. We observed additive effects on growth rate and on protein oxidative damage between mutants in GRX5 and in other glutaredoxin genes (Rodríguez-Manzaneque et al., 1999) and also between grx5 and sod1 mutations (Rodríguez-Manzaneque, Tamarit, Bellí, Ros, and Herrero, unpublished observations). This could be the consequence of the inability to repair iron-mediated macromolecular damage (both in mitochondria and cytosol) in Grx5-depleted cells in the absence of other glutaredoxins or of cytosolic superoxide dismutase. In either case, compartmentalization studies and sequence analysis indicate that Grx5 is the only dithiol plus monothiol glutaredoxin that is mitochondrially located and suggests that it does not directly share functions with Grx1–4. Inability to suppress grx5 defects in conditions of GRX3/4 overexpression confirms this suggestion. Similarly, a double grx5 trx3 mutant is no more sensitive to oxidants or growth defects than a single grx5 mutant (Rodríguez-Manzaneque, Tamarit, Bellí, Ros, and Herrero, unpublished observations), thus arguing in favor of completely separate functions for Grx5 and the mitochondrial Trx3 thioredoxin. However, the relatively mild phenotypes of grx5 cells compared with some other mutants in Fe/S cluster synthesis point to partial functional redundancy between Grx5 and other thiol oxidoreductases in the cell.
In Grx5-deficient cells, iron accumulation occurs at similar levels in mitochondria and extramitochondrial fractions, whereas in frataxin mutants iron accumulates exclusively at the mitochondria at the expense of cytosolic iron (Babcock et al., 1997; Foury and Cazzalini, 1997; Radisky et al., 1999). Although these differences argue against Grx5 acting in parallel with Yfh1 frataxin in the maintenance of iron homeostasis in the cell, it was still possible that extensive iron accumulation in grx5 cells was a direct consequence of the absence of Grx5. Consequently, alterations in Fe/S enzyme activity could have been the result of the sensitivity of Fe/S clusters to high levels of ROS generated at increased iron concentrations. However, we can discard this possibility because the reduction of intracellular iron levels to almost normal or even lower than normal concentrations does not suppress the inactivation of Fe/S enzymes in grx5 cells. Similar conclusions have been reached for Jac1 function (Voisine et al., 2001). These facts, together with the inability to suppress the grx5 growth phenotypes in anaerobiosis or by the addition of external reductants, support a direct participation of Grx5 in Fe/S cluster assembly. The question remains as to whether the disruption of Fe/S cluster assembly could cause such high levels of mitochondrial iron and, in the case of grx5 cells, cytosolic iron. Although the process of iron transport across the cytoplasmic membrane of yeast cells has been elucidated (Askwith and Kaplan, 1998; Eide, 1998), the mechanism of iron entry into the mitochondria remains uncertain (Lange et al., 1999). A protein containing Fe/S clusters might participate in the regulation of mitochondrial iron assimilation, perhaps acting as an iron sensor that could act upstream of the nuclear transcription factor Aft1. This would explain the high levels of this metal found at the mitochondria in the absence of normal Fe/S cluster assembly at the organelle.
Although knowledge of the gene products involved in the maturation of Fe/S proteins at the yeast mitochondria has improved in recent years, the specific role of individual proteins and the biochemistry of the process remain obscure. Assembly of Fe/S centers in the apoprotein requires reduction of disulfide bridges between cysteine residues for coordination of Fe atoms (Beinert et al., 1997). In the case of SoxR (a transcriptional regulator of E. coli involved in oxidative stress response whose activity depends on the redox state of a Fe/S cluster present in it [Hidalgo et al., 1997]), GSH reductase and thioredoxins are required for the in vivo response of SoxR to oxidants (Ding and Demple, 1998). Grx5 could play a similar role in yeast mitochondria, although in this case a monothiol mechanism for disulfide bridge reduction should be postulated. This would require a mixed disulfide intermediary between one of the cysteine residues and GSH that would be attacked by the monothiol glutaredoxin (Bushweller et al., 1992). A variant of this hypothesis may be formulated from the observation that GSH and other monothiols are able to disassemble Fe/S clusters through GSH-derived reactive free radicals. The latter could form inactivating mixed disulfides with the cysteine residues responsible for iron chelation (Ding and Demple, 1996). Grx5 could be required to repair such toxic disulfides, therefore restoring the ability to assemble the clusters on the sulfhydryl groups of the apoproteins. Finally, Grx5 could play a role during the Nfs1-catalized desulfuration of cysteine. This reaction has been studied in the Azotobacter vinelandii homologue NiFS and involves formation of a persulfide between sulfur and Cys329 of NiFS (Zheng et al., 1994). In yeast, Grx5 could be involved in the cleavage of this persulfide, leading to the release of sulfur and regeneration of reduced Nfs1. More studies are needed to determine the biochemical role of Grx5 in the formation of Fe/S clusters and to confirm whether Grx5 is part of a mitochondrial matrix multiprotein complex responsible for such a process.
ACKNOWLEDGMENTS
We thank Lidia Piedrafita for her excellent technical assistance and María Angeles de la Torre, Elisa Cabiscol, Pedro Echave, and Jordi Torres for their comments. The gifts of strains and antibodies by Gyula Kispal, Roland Lill, Bernard Lemire, and Eulogio Valentín are also acknowledged. This work was supported by grants (to E.H. and J.R.) from the Ministerio de Educación y Ciencia and the Generalitat de Catalunya. J.T. received a postdoctoral grant from the Generalitat de Catalunya.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0517. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0517.
REFERENCES
- Askwith C, Kaplan J. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem Sci. 1998;23:135–138. doi: 10.1016/s0968-0004(98)01192-x. [DOI] [PubMed] [Google Scholar]
- Aslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 as a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M, De Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- Barros MH, Nobrega FG. YAH1 of Saccharomyces cerevisiae: a new essential gene that codes for a protein homologous to human adrenodoxin. Gene. 1999;233:197–203. doi: 10.1016/s0378-1119(99)00137-7. [DOI] [PubMed] [Google Scholar]
- Beinert H, Holm RH, Münck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- Bellí G, Garí E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee JK. Alpha-aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit Rev Microbiol. 1985;12:131–151. doi: 10.3109/10408418509104427. [DOI] [PubMed] [Google Scholar]
- Bushweller JH, Aslund F, Wuthrich K, Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14-S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- Cadenas E. Biochemistry of oxygen toxicity. Annu Rev Biochem. 1989;58:79–110. doi: 10.1146/annurev.bi.58.070189.000455. [DOI] [PubMed] [Google Scholar]
- Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000;34:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- Casas C, Aldea M, Espinet C, Gallego C, Gil R, Herrero E. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast. 1997;13:621–637. doi: 10.1002/(SICI)1097-0061(19970615)13:7<621::AID-YEA121>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Craig EA, Voisine C, Schilke B. Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol Chem. 1999;380:1167–1173. doi: 10.1515/BC.1999.148. [DOI] [PubMed] [Google Scholar]
- De Freitas JM, Liba A, Meneghini R, Valentine JS, Gralla EB. Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. J Biol Chem. 2000;275:11645–11649. doi: 10.1074/jbc.275.16.11645. [DOI] [PubMed] [Google Scholar]
- Ding H, Demple B. Glutathione-mediated destabilization in vitro of [2Fe-2S] centers in the SoxR regulatory protein. Proc Natl Acad Sci USA. 1996;93:9449–9453. doi: 10.1073/pnas.93.18.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Demple B. Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry. 1998;37:17280–17286. doi: 10.1021/bi980532g. [DOI] [PubMed] [Google Scholar]
- Draculic T, Dawes IW, Grant CM. A single glutaredoxin or thioredoxin gene is essential for viability in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2000;36:1167–1174. doi: 10.1046/j.1365-2958.2000.01948.x. [DOI] [PubMed] [Google Scholar]
- Eide DJ. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu Rev Nutr. 1998;18:441–469. doi: 10.1146/annurev.nutr.18.1.441. [DOI] [PubMed] [Google Scholar]
- Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- Foury F. Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Lett. 1999;456:281–284. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Gangloff SP, Marguet D, Lauquin GJ-M. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergic regulation of expression by glucose plus glutamate. Mol Cell Biol. 1990;10:3551–3561. doi: 10.1128/mcb.10.7.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Garland SA, Hoff K, Vickery LE, Culotta VC. Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J Mol Biol. 1999;294:897–907. doi: 10.1006/jmbi.1999.3294. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:3065–3073. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, Pan X, McCusker JH. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol. 2001;39:533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo E, Ding H, Demple B. Redox signal transduction: mutations shifting [2Fe-2S] centers of the SoxR sensor-regulator to the oxidized form. Cell. 1997;88:121–129. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;254:13963–13966. [PubMed] [Google Scholar]
- Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- Holst B, Tachibana C, Winther JR. Active site mutations in yeast protein disulfide isomerase cause dithiothreitol sensitivity and a reduced rate of protein folding in the endoplasmic reticulum. J Cell Biol. 1997;138:1229–1238. doi: 10.1083/jcb.138.6.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakov N, Witte S, Altman A. PICOT-HD: a highly conserved oritein domain that is often associated with thioredoxin and glutaredoxin modules. Trends Biochem Sci. 2000;25:537–539. doi: 10.1016/s0968-0004(00)01685-6. [DOI] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC. Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol Cell Biol. 2000;20:3918–3927. doi: 10.1128/mcb.20.11.3918-3927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kaut A, Lange H, Diekert K, Kispal G, Lill R. Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J Biol Chem. 2000;275:15955–15961. doi: 10.1074/jbc.M909502199. [DOI] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Csere P, Guiard B, Lill R. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 1997;418:346–350. doi: 10.1016/s0014-5793(97)01414-2. [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are required for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw GB. Isopropylmalate dehydratase from yeast. Methods Enzymol. 1988;166:429–435. doi: 10.1016/s0076-6879(88)66055-1. [DOI] [PubMed] [Google Scholar]
- Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- Lacour T, Achstetter T, Dumas B. Characterization of recombinant adrenodoxin reductase homologue (Arh1p) from yeast: implication in in vitro cytochrome p45011 beta monooxygenase system. J Biol Chem. 1998;273:23984–23992. doi: 10.1074/jbc.273.37.23984. [DOI] [PubMed] [Google Scholar]
- Lange H, Kispal G, Lill R. Mechanism of iron transport to the site of heme synthesis inside yeast mitochondria. J Biol Chem. 1999;274:18989–18996. doi: 10.1074/jbc.274.27.18989. [DOI] [PubMed] [Google Scholar]
- Lange H, Kaut A, Kispal G, Lill R. A mitochondrial ferrodoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc Natl Acad Sci USA. 2000;97:1050–1055. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, Dancis A. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem. 1999;274:33025–33034. doi: 10.1074/jbc.274.46.33025. [DOI] [PubMed] [Google Scholar]
- Liao X, Samall WC, Srere PA, Butow RA. Intramitochondrial functions regulate non-mitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Diekert K, Kaut A, Lange H, Pelzer W, Prohl C, Kispal G. The essential role of mitochondria in the biogenesis of cellular iron-sulfur proteins. Biol Chem. 1999;380:1157–1166. doi: 10.1515/BC.1999.147. [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- Lillig CH, Prior A, Schwenn JD, Aslund F, Ritz D, Vlamis-Gardikas A, Holmgren A. New thioredoxins and glutaredoxins as electron donors of 3′-phosphoadenylylsulfate reductase. J Biol Chem. 1999;274:7695–7698. doi: 10.1074/jbc.274.12.7695. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Dawes IW, Grant CM. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol Biol Cell. 1998;9:1081–1091. doi: 10.1091/mbc.9.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik MAH, Overkamp KM, Kötter P, de Vries S, van Dijken JP, Pronk JT. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- Lutz T, Westermann B, Neupert W, Herrmann JM. The mitochondrial proteins ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol. 2001;307:815–825. doi: 10.1006/jmbi.2001.4527. [DOI] [PubMed] [Google Scholar]
- Manzella L, Barros MH, Nobrega FG. ARH1 of Saccharomyces cerevisiae: a new essential gene that codes for a protein homologous to the human adrenodoxin reductase. Yeast. 1998;14:839–846. doi: 10.1002/(SICI)1097-0061(19980630)14:9<839::AID-YEA283>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Munujos P, Coll-Canti J, González-Sastre F, Gella FJ. Assay of succinate dehydrogenase activity by a colorimetric continuous using iodonitrotetrazolium chloride as electron acceptor. Anal Biochem. 1993;212:506–509. doi: 10.1006/abio.1993.1360. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M. Iron sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferrodoxin. J Biol Chem. 2001;276:22604–22607. doi: 10.1074/jbc.M102902200. [DOI] [PubMed] [Google Scholar]
- Pearce DA, Sherman F. Diminished degradation of yeast cytochrome c by interactions with its physiological partners. Proc Natl Acad Sci USA. 1995;92:3735–3739. doi: 10.1073/pnas.92.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrajas J R, Kosmidou E, Miranda-Vizuete A, Gustafsson JA, Wright APH, Spyrou G. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J Biol Chem. 1999;274:6366–6373. doi: 10.1074/jbc.274.10.6366. [DOI] [PubMed] [Google Scholar]
- Pelzer W, Múhlenhoff U, Diekert K, Siegmund K, Kispal G, Lill R. Mitochondrial Isa2p plays a crucial role in the maturation of cellular iron-sulfur proteins. FEBS Lett. 2000;476:134–139. doi: 10.1016/s0014-5793(00)01711-7. [DOI] [PubMed] [Google Scholar]
- Pon L, Schatz G. Biogenesis of yeast mitochondria. In: Broach JR, Pringle JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 333–406. [Google Scholar]
- Prinz WA, Aslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Babcock MC, Kaplan J. The yeast frataxin homologue mediates mitochondrial iron efflux. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- Rahlfs S, Fischer M, Becker K. Plasmodium falciparum possesses a classical glutaredoxin and a second, glutaredoxin-like protein with a PICOT homology domain. J Biol Chem. 2001;276:37133–37140. doi: 10.1074/jbc.M105524200. [DOI] [PubMed] [Google Scholar]
- Ristow M, Pfister MF, Yee AJ, Schubert M, Michael L, Zhang CY, Ueki K, Michael MD, II, Lowell BB, Kahn CR. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc Natl Acad Sci USA. 2000;22:12239–12243. doi: 10.1073/pnas.220403797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JB, Jr, Brent LG, Sumegi B, Srere PA. An enzymatic approach to the study of the Krebs tricarboxylic acid cycle. In: Darley-Usmar WM, Rickwood D, Wilson MT, editors. Mitochondria: A Practical Approach. Oxford, UK: IRL Press; 1987. pp. 153–170. [Google Scholar]
- Rodríguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke B, Forster J, Davis J, James P, Walter W, Laloraya S, Johnson J, Miao B, Craig E. The cold sensitivity of a mutant of Saccharomyces cerevisiae lacking a mitochondrial heat shock protein 70 is suppressed by loss of mitochondrial DNA. J Cell Biol. 1996;134:603–613. doi: 10.1083/jcb.134.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg JJ, Hoff KG, Tapley Tl, Vickery LE. The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem. 2001;276:1696–1700. doi: 10.1074/jbc.M009542200. [DOI] [PubMed] [Google Scholar]
- Strain J, Lorenz CR, Bode J, Garland S, Smolen GA, Ta DT, Vickery LE, Culotta VC. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae: identification of proteins predicted to mediate iron-sulfur cluster assembly. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- Tangeras A, Flatmark T, Bäcström D, Ehrenberg A. Mitochondrial iron not bound in heme and iron-sulfur centers: estimation, compartmentation and redox state. Biochim Biophys Acta. 1980;589:162–175. doi: 10.1016/0005-2728(80)90035-3. [DOI] [PubMed] [Google Scholar]
- Tong WH, Rouault T. Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 2000;19:5692–5700. doi: 10.1093/emboj/19.21.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas C, McEwan AG, Downie JA. Detection of c-type cytochromes using enhanced chemiluminescence. Anal Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- Voisine C, Cheng YC, Ohlson M, Schilke B, Hoff K, Beinert H, Marszalek J, Craig EA. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:1483–1488. doi: 10.1073/pnas.98.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MP, Costa GL. Rapid PCR site-directed mutagenesis. In: Dieffenbach CW, Dveksler GS, editors. PCR Primer: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laborarory Press; 1995. pp. 613–621. [Google Scholar]
- Witte S, Villalba M, Bi K, Liu Y, Isakov N, Altman A. Inhibition of the c-jun N-terminal kinase/AP-1 and NF-κB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J Biol Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. AFT1: a mediator of iron-regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Dean DR. Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]