Abstract
Activation of mitogen-activated protein kinase (MAP kinase) cascades is essential for plant immunity. Upon activation by surface-localized immune receptors, receptor-like cytoplasmic kinases (RLCKs) in the cytoplasm phosphorylate MAP kinase kinase kinases (MAPKKKs) to initiate MAP kinase activation. Surprisingly, we found that both the phosphorylation of Arabidopsis (Arabidopsis thaliana) MAPKKKs and the subsequent activation of MAP kinase cascades require the λ and κ isoforms of 14-3-3 proteins, which directly interact with multiple RLCKs and MAPKKKs. The N- and C-termini of MAPKKK5 interact intramolecularly to inhibit the access to the C terminus by RLCKs, whereas the 14-3-3 proteins relieve this inhibition and facilitate the interaction of RLCKs with the C-terminus of MAPKKK5. This enables the phosphorylation of MAPKK5 at Ser599 and Ser682, thus promoting MAP kinase activation and enhancing plant disease resistance. Our study reveals a role of 14-3-3 proteins as scaffolds and activators in the regulation of the RLCK-MAPKKK5 module and provides insight into the mechanism of plant immune signaling.
Specific 14-3-3 proteins enable activation of MAPK cascades by surface immune receptors.
IN A NUTSHELL.
Background: The plant innate immune system detects the presence of microbial pathogens and triggers defense responses to terminate or restrict pathogen progression. The BIK1 family receptor-like cytoplasmic kinases (RLCKs) play a crucial role in immune signal transduction and regulate multiple substrate proteins. Much less is known concerning how these kinases and their substrates are organized in a way for robust signaling.
Question: We were interested in identifying interactors of RLCKs and understanding how they regulate immunity.
Findings: We found that the 14-3-3 proteins GRF6 and GRF8 play a key role in both anti-bacterial and anti-fungal immunity in Arabidopsis. We show that these 14-3-3 proteins directly interact with MAP kinase kinase kinase 5 (MAPKKK5) to enable their activation by upstream RLCKs. Our study reveals how GRF6 and GRF8 act as a scaffold to regulate the RLCK-MAPKKK5 module to facilitate the activation of MAP kinase cascades during immune signaling.
Next steps: It would be of interest to investigate whether different 14-3-3 members regulate different immune responses and how.
Introduction
Pattern recognition receptors (PRRs) on the cell surface perceive immunogenic patterns from intercellular spaces and transduce extracellular signals into the cell to activate an initial level of immunity called pattern-triggered immunity (PTI) (DeFalco and Zipfel 2021; Tang et al. 2017). PRRs are primarily receptor kinases and receptor-like proteins and include FLAGELLIN SENSITIVE2 (FLS2), EF-TU RECEPTOR, LYSIN MOTIF RECEPTOR KINASE5, and PLANT ELICITOR PEPTIDE RECEPTORs (PEPRs). These PRRs recognize the bacterial flagellin epitope flg22, the bacterial translation elongation factor EF-Tu epitope elf18, the fungal cell wall component chitin, and plant elicitor peptides (Peps), respectively (Cao et al. 2014; Chinchilla et al. 2006; Kaku et al. 2006; Miya et al. 2007; Yamaguchi et al. 2010; Zipfel et al. 2006). Plants also contain a large repertoire of nucleotide-binding, leucine-rich-repeat receptors (NLRs) that recognize pathogen effectors, which initially evolved as virulence factors but also activate effector-triggered immunity (ETI) (Jones et al. 2016). Recent advances indicate that PTI and ETI potentiate each other to form a robust immune system (Ngou et al. 2021; Yuan et al. 2021).
Advances in the last 2 decades have identified key components linking cell surface PRR recognition of extracellular signals to downstream intracellular responses. A number of receptor-like cytoplasmic kinases (RLCKs) belonging to subfamilies VII and XII play pivotal roles in defenses by linking PRRs to key downstream responses (Liang and Zhou 2018). These RLCKs phosphorylate key immune signaling components and can trigger an influx of calcium ions, a burst of reactive oxygen species (ROS), activation of mitogen-activated protein kinase (MAP kinase) cascades, and activation of heterotrimeric G proteins (Kadota et al. 2014; Li et al. 2014a; Liang et al. 2016; Yamada et al. 2016; Bi et al. 2018; Yan et al. 2018; Tian et al. 2019; Thor et al. 2020; Shi et al. 2022; Ma et al. 2022).
Two well-known MAP kinase cascades are activated during PTI in Arabidopsis thaliana (Arabidopsis). One cascade contains MAP kinase or ERK kinase (MEK) kinase 1 (MEKK1), that activates MAP kinase MPK4 and MPK11. Another cascade has 3 redundant MAP kinase kinase kinases (MAPKKKs), MAPKKK3, MAPKKK5, and YODA, which activate MPK3 and MPK6 (Asai et al. 2002; Bi et al. 2018; Gao et al. 2008; Suarez-Rodriguez et al. 2007; Sun et al. 2018; Yamada et al. 2016; Yan et al. 2018; Liu et al. 2022; Wang et al. 2022). The latter cascade plays a more significant role in defense gene expression and stomatal immunity in plants (Bi et al. 2018; Li et al. 2012; Liu and Zhang 2004; Su et al. 2017; Xu et al. 2016). Multiple RLCK subfamily VII members have been shown to phosphorylate the C-terminus of MAPKKK5 and MEKK1, thus activating the cascades (Bi et al. 2018; Yamada et al. 2016). In addition, the RLCK subfamily XII member BSK1 can phosphorylate the N-terminus of MAPKKK5, and the phosphorylation site is important for the function of MAPKKK5 in disease resistance (Yan et al. 2018). A recent study shows that MPK15 is also activated during and required for resistance to the powdery mildew fungus Golovinomyces cichoracearum (Shi et al. 2022).
14-3-3 proteins are ubiquitous eukaryotic signaling components and are associated with multiple physiological processes, including primary metabolism, cell division, biotic and abiotic stresses, apoptosis, and cancer (Aitken 2006; Denison et al. 2011; Freeman and Morrison 2011; Paul et al. 2012; Bridges and Moorhead 2005). They form homodimers or heterodimers via their N-termini, with each monomer possessing a binding pocket for a target protein. This allows the dimer not only to act as a scaffold but also to regulate catalytic activity, enhance stability, or alter subcellular localization of their target proteins (Muslin et al. 1996; Yaffe et al. 1997; Yang et al. 2006). The Arabidopsis genome encodes 13 functional 14-3-3 proteins, which are known to regulate activities of a variety of target proteins, such as plasma membrane H + -ATPases (Fuglsang et al. 2007), the SALT OVERLY SENSITIVE2 kinase (Yang et al. 2019; Zhou et al. 2014), and NITRATE REDUCTASE (Huber et al. 2002).
Increasing evidence indicates that 14-3-3 proteins also play a crucial role in plant immunity (Lozano-Durán and Robatzek 2015). Arabidopsis GRF6 (a 14-3-3λ protein) interacts with the resistance protein RPW8.2 and positively regulates disease resistance (Yang et al. 2009). Pharmacological experiments indicate that 14-3-3 proteins play a crucial role in PTI responses in Arabidopsis (Lozano-Durán et al. 2014). Multiple pathogen effectors have been shown to target 14-3-3 proteins in Arabidopsis and tomato (Solanum lycopersicum) plants (Giska et al. 2013; Dubrow et al. 2018; Li et al. 2013; Teper et al. 2014; Gao et al. 2022; Chaudhary et al. 2019), further supporting an important role of 14-3-3 proteins in plant immunity.
In addition, the tomato 14-3-3 protein TFT7 has been shown to interact with the tomato MAP kinase kinase kinase (SlMAPKKKα) and the MAP kinase kinase (SlMKK2) to positively regulate cell death (Oh and Martin 2011; Oh et al. 2010). Multiple tomato 14-3-3 proteins have been shown to contribute to cell death triggered by various effectors produced by Xanthomonas bacteria (Giska et al. 2013), and a recent report showed that 14-3-3 proteins play an antiviral role in Nicotiana benthamiana (Gao et al. 2022). Nevertheless, we have limited knowledge concerning how 14-3-3 proteins regulate immune signaling components in plants at the biochemical level.
In this study, we show that GRF6 and GRF8 (14-3-3κ) positively regulate resistance to bacterial and fungal pathogens in Arabidopsis. We show not only that these 14-3-3 proteins directly interact with MAPKKK5, MEKK1, and multiple RLCK subfamily VII members but also that they are required for the activation of MAP kinase cascades downstream of PRRs. Although access of the RLCK subfamily VII member AVRPPHB SUSCEPTIBLE 1 LIKE 19 (PBL19) to the C-terminus of MAPKKK5 is autoinhibited by the MAPKKK5 N-terminus, GRF6 can bind to the MAPKKK5 C-terminus to enhance accessibility by PBL19 and counteract this inhibition. This promotes the phosphorylation of the MAPKKK5 C-terminus by PBL19 and is required for the activation of the MAP kinase cascade during initiation of the immune response. Together, our data reveal a key mechanism by which 14-3-3 proteins regulate the RLCK-MAP kinase module in plant immune signaling.
Results
14-3-3 isoforms GRF6 and GRF8 are required to activate the expression of FRK1 and PR1 triggered by flg22 and chitin
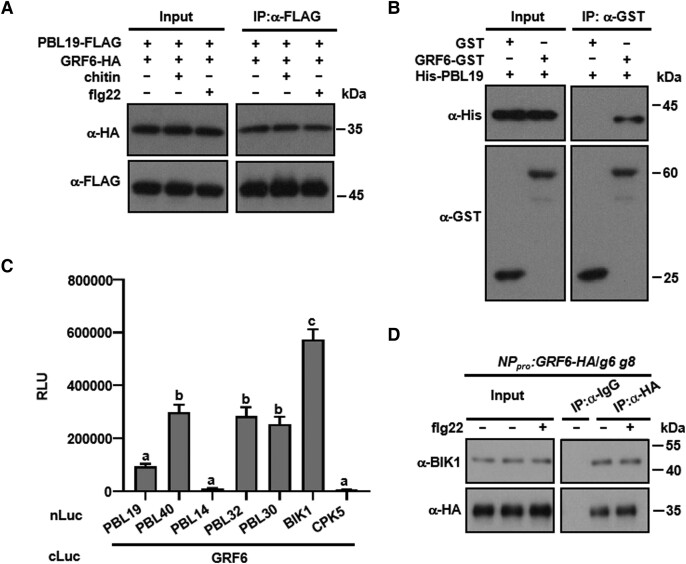
We previously showed that PBL19, PBL20, PBL37, PBL38, PBL39, and PBL40 redundantly regulate both chitin-triggered MAP kinase activation (for antifungal defense) and anti-bacterial immunity (Rao et al. 2018). In an effort to identify proteins involved in immune signaling, FLAG-tagged PBL19 and PBL20 proteins were expressed in Arabidopsis protoplasts, treated with flg22, and total protein extracts were subjected to anti-FLAG immunoprecipitation followed by LC-MS/MS analysis. Strikingly, 10 different 14-3-3 isoforms were pulled-down by both PBL19 and PBL20, each with numerous tryptic fragments identified (Supplemental Table S1). Additional candidate interacting proteins include SOLUBLE N-ETHYLMALEIMIDE-SENSITIVE FACTOR ADAPTOR PROTEIN 33 (SNAP33), PLANT U-BOX 25 (PUB25), PUMILIO 5 (PUM5), E. COLI RAS-LIKE PROTEIN-RELATED GTPASE 1 (ERA1), and ZYGOTIC ARREST 1 (ORPK1). We decided to focus on 14-3-3 proteins in this study. Co-immunoprecipitation (Co-IP) and glutathione-S-transferase (GST) pull-down experiments verified the PBL19 interaction with GRF6 (Fig. 1, A and B; Supplemental Fig. S1A), and we found that this interaction was not affected by either flg22 or chitin treatment (Fig. 1A). We further used luciferase complementation assays to test a variety of RLCK VII members, including BIK1, PBL14, PBL19, PBL30, PBL32, and PBL40 for potential interactions with GRF6. The majority of the RLCKs tested, except for PBL14, interacted with GRF6 in N. benthamiana leaves (Fig. 1C; Supplemental Fig. S1B). The results suggested that the interaction between RLCKs and the 14-3-3 protein is common. To test whether an RLCK interacts with GRF6 in the native configuration, we generated a stable transgenic line carrying GRF6-HA under the control of its native promoter (NPpro) in the grf6 grf8 background. Co-IP assays indicated that BIK1 indeed interacted with GRF6 in Arabidopsis seedlings (Fig. 1D). A GST pull-down experiment further showed that the BIK1-GRF6 interaction was direct (Supplemental Fig. S1C). GRF6 also interacted with FLS2 and BAK1 in protoplasts (Supplemental Fig. S1, D and E), a result consistent with the notion that BIK1 exists in the receptor complex.
Figure 1.
GRFs interact with PBL19 and other RLCKs. A) GRF6 interacts with PBL19 in Arabidopsis protoplasts. Col-0 protoplasts expressing the indicated constructs were treated with chitin or flg22 or neither, and Co-IP assays were performed and detected with anti-FLAG and anti-HA immunoblots. B) GRF6 interacts with PBL19 in vitro. His-tagged PBL19 was incubated with either GST-tagged GRF6 recombinant protein or GST. GST pull-down assays were performed, and input and immunoprecipitated proteins were detected using anti-GST and anti-His antibodies. C) GRF6 interacts with multiple RLCK VII members. The indicated Nluc and Cluc constructs were transiently expressed in N. benthamiana for luciferase complementation assays. Data are presented as mean relative luminescence units (RLUs) ± se. Different letters indicate significant differences at P < 0.05 (n = 12, one-way ANOVA, Tukey post-test). CPK5 was used as a negative control. D) Co-IP assays showing that BIK1 interacts with GRF6 in vivo. Ten-day-old seedlings were treated with or without flg22 and then samples were harvested. Total proteins were extracted and subsequently incubated with anti-IgG Affinity Gel and the HA antibody. The total and precipitated proteins were examined by immunoblotting using antibodies against BIK1 and HA, respectively. Each experiment was repeated at least 3 times with similar results, and results from one representative experiment are shown.
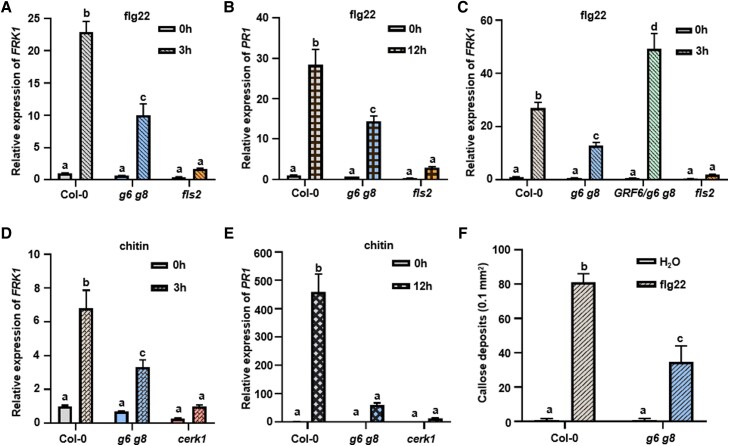
We focused on GRF6 and its closely related isoform GRF8, which are 93% identical at the amino acid level, because GRF8 has been suggested to play a role in PTI (Lozano-Durán et al. 2014). We systematically examined PTI responses in the grf6 grf8 (g6 g8) double mutant, which has been characterized previously due to its involvement plant responses to cold stress (Liu et al. 2017). Upon flg22 and chitin treatments, the expression of FLG22-INDUCED RECEPTOR KINASE1 (FRK1) and PATHOGENESIS-RELATED GENE1 (PR1) were strongly induced in Col-0 plants (Fig. 2, A to E), a result that was consistent with previous reports (Bjornson et al. 2021 and references therein).
Figure 2.
14-3-3 are required for specific PTI responses. A–C) GRF6 is essential for flg22-induced FRK1 expression. Ten-day-old seedlings were sprayed with flg22 or water, and samples were harvested 3 h later. ACTIN1 was used as the internal standard for normalization. The fls2 mutant in Fig. 2, A to C was used as a negative control. D and E) Chitin-induced expression of FRK1 (D) and PR1 (E) was impaired in g6 g8 mutant plants. Ten-day-old seedlings were sprayed with chitin, and samples were harvested 0, 3, or 12 h later. ACTIN1 was used as the internal standard for normalization. The cerk1 mutant was used as a negative control of chitin treatment. F) Flg22-triggered callose deposition is impaired in g6 g8 mutant plants. Error bars are presented as means ± Sd (n = 4). Each experiment was repeated 3 times with similar results. Each experiment was repeated 3 times with similar results, and data from one representative experiment are shown. Values are means ± Sd. Different letters indicate significant differences at P < 0.05 (n = 3 for A to E, n = 4 for F, 1-way ANOVA, Tukey post-test). Statistical analysis results are presented in Supplemental Data Set S1.
In g6 g8 double mutant plants, flg22- and chitin-induced expression of FRK1 and PR1 was reduced by 50% to 80%. In g6 g8 transgenic plants complemented with GRF6, flg22-induced FRK1 gene expression was 2-fold higher compared to Col-0 (Fig. 2C). The flg22- and chitin-induced expression of FRK1 and PR1 were completely abolished in fls2 and cerk1 mutants, respectively, with the latter lacks the chitin co-receptor LYSM DOMAIN RECEPTOR-LIKE KINASE 1 (CERK1) (Fig. 2, D and E; Miya et al. 2007). An examination of grf6 and grf8 single mutant plants showed that flg22-induced FRK1 expression was slightly reduced, whereas chitin-induced FRK1 expression was unchanged (Supplemental Fig. S2, A and B), indicating that GRF6 and GRF8 act redundantly.
We also tested flg22-induced callose deposition. Contrary to Col-0 seedlings, which showed strong callose deposition upon flg22 treatment, the g6 g8 mutant seedlings had diminished callose deposition (Fig. 2F). However, both the g6 g8 mutant plants complemented with GRF6, and plants of a GRF6 overexpression line (GRF6-OE) were indistinguishable from Col-0 plants in ROS burst activity regardless of the immunogenic epitopes used (Supplemental Fig. S3, A to D).
GRF6 and GRF8 positively regulate basal stomatal immunity against bacterial and fungal pathogens
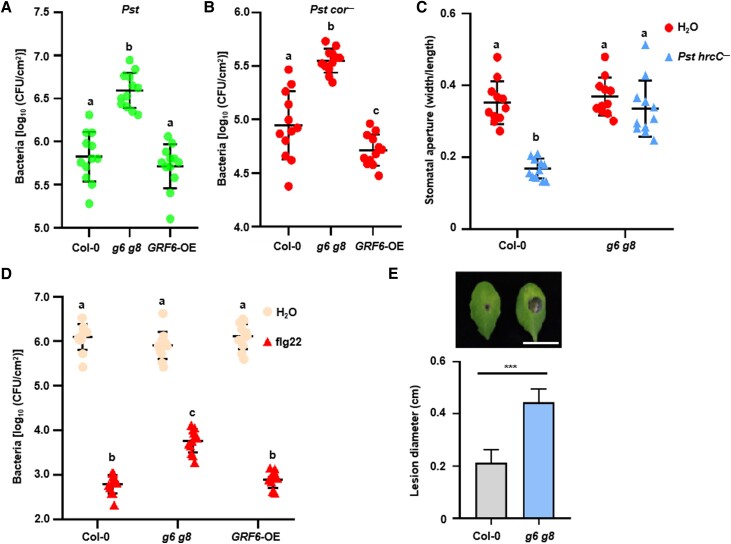
We next asked whether the GRF6 and GRF8 isoforms are required for antibacterial immunity. Perception of immunogenic patterns is known to induce stomatal closure and restrict bacterial entry, and this process is referred to as stomatal immunity (Melotto et al. 2006). Spray-inoculation of Arabidopsis Col-0 leaves with wild-type Pseudomonas syringae pv. tomato (Pst) DC3000 bacteria showed that g6 g8 double mutant plants supported significantly more bacterial growth, whereas the GRF6 overexpression plants had similar bacterial titers compared to Col-0 (Fig. 3A). Bacterial entry assays with the Pst cor− mutant strain, which lacks coronatine known to reopen stomata (Mittal and Davis 1995), showed that, in the g6 g8 double mutant plants, there was significantly more bacterial entry into leaves compared to Col-0 plants (Fig. 3B). The GRF6-OE plants had slightly fewer bacteria entering compared to Col-0, but the difference was statistically insignificant. Spraying of Col-0 plants with Pst hrc− mutant bacteria, which are defective in the secretion of effector proteins, led to reduced stomatal apertures (Fig. 3C), confirming that stomatal immunity can be induced by bacterial immunogenic patterns. The same treatment of g6 g8 mutant plants had no effect on the stomatal apertures, indicating that GRF6 and GRF8 are 14-3-3 isoforms that are required for stomatal immunity.
Figure 3.
GRF6 and GRF8 are required for basal immunity against bacterial and fungal pathogens. A)g6 g8 mutant plants display increased susceptibility to Pst bacteria. Plants were sprayed with wild-type Pst DC3000, and bacterial populations in the leaf were counted 3 d later. B) The g6 g8 mutant displays increased bacterial entry into leaf tissues. Bacterial populations in the leaf were measured 1.5 h following soaking with Pst DC3000 cor−, and the bacterial number was then determined. C) The g6 g8 mutant is impaired in bacterium-induced stomatal closure. Leaf discs were soaked with Pst DC3000 hrcC−, and stomatal aperture was determined 1 h later. Stomatal aperture images were recorded by a microscope, and sizes were then calculated. D) GRF6 and GRF8 are required for flg22-induced resistance to Pst DC3000. Plants were pretreated with H2O or flg22 for 1 d and then infiltrated with Pst DC3000. Bacterial populations in the leaf were determined 2 d later. E) *** in E indicates a significant difference at P < 0.001 (n = 16, 2-tailed Student's t-test). Statistical analysis results are presented in Supplemental Data Set S1.
Flg22 protection assays were performed to determine whether these 2 isoforms are also required for PTI in mesophyll cells (Zipfel et al. 2004). In this assay, plants were pretreated with flg22 for 1 d before infiltrating Pst bacteria, which bypasses natural stomatal entry. Col-0 plants pretreated with flg22 had significantly fewer bacteria in leaves (approximately 0.1%) compared to the H2O control treatment (Fig. 3D). In g6 g8 mutant plants, bacterial were approximately 10-fold higher than in Col-0, indicating that the 2 14-3-3 isoforms are required for flg22-induced mesophyll immunity. GRF6-OE plants had indistinguishable amount of bacteria compared to Col-0 (Fig. 3D), suggesting that GRF6 is not rate-limiting in Col-0 plants. We then tested whether these 2 isoforms are also required for antifungal immunity. When g6 g8 mutant plants were inoculated with Botrytis cinerea, they developed significantly larger disease lesions compared to Col-0 (Fig. 3E), indicating that GRF6 and GRF8 are indeed required for antifungal immunity.
Because recent studies have indicated that PTI and ETI signaling are linked through multiple nodes (Ngou et al. 2021; Yuan et al. 2021), we tested whether GRF6 and GRF8 are required for ETI. Pst strains carrying hopZ1a, avrB, avrRpt2, or avrRps4 avirulence genes are recognized by NLRs ZAR1, RPM1, RPS2, or RPS4, respectively, triggering ETI on Col-0 plants. Both Col-0 and g6 g8 mutant plants showed complete resistance to these strains (Supplemental Fig. S4, A to D), suggesting that the 2 14-3-3 isoforms were not required for ETI. We further subjected grf6 and grf8 single mutant plants to the aforementioned disease resistance assays. Neither of the mutants displayed any defects in antibacterial nor antifungal immunity (Supplemental Fig. S5, A to D), indicating that GRF6 and GRF8 are functionally redundant.
GRF6 and GRF8 are required for pattern-triggered activation of MAP kinases
We next sought to dissect the PTI defect in the g6 g8 mutant at the biochemical level using the FLS2 pathway as a proxy, as different PRRs often share similar downstream components. Considering that GRF6 interacted with RLCKs, FLS2, and BAK1, we systematically examined early molecular events of PTI signaling. To this end, we first tested accumulation of FLS2 and its co-receptor BAK1 and found that levels of these were unaltered in g6 g8 mutant seedlings (Supplemental Fig. S6A). Perception of flg22 induces FLS2-BAK1 interaction (Chinchilla et al. 2007; Heese et al. 2007; Sun et al. 2013), this induced interaction occurred similarly in Col-0 and g6 g8 mutant protoplasts (Supplemental Fig. S6B). Perception of flg22 also induces BIK1 phosphorylation and subsequent dissociation from the FLS2 complex (Lu et al. 2010; Zhang et al. 2010). We found that both BIK1 phosphorylation and the dissociation from FLS2 occurred normally in g6 g8 mutant protoplasts (Supplemental Fig. S6C). Although flg22 induces phosphorylation of the heterotrimeric G protein, XLG2 (Liang et al. 2016), we found via immunoblotting analysis that XLG2 phosphorylation was not affected in g6 g8 mutant protoplasts (Supplemental Fig. S6D).
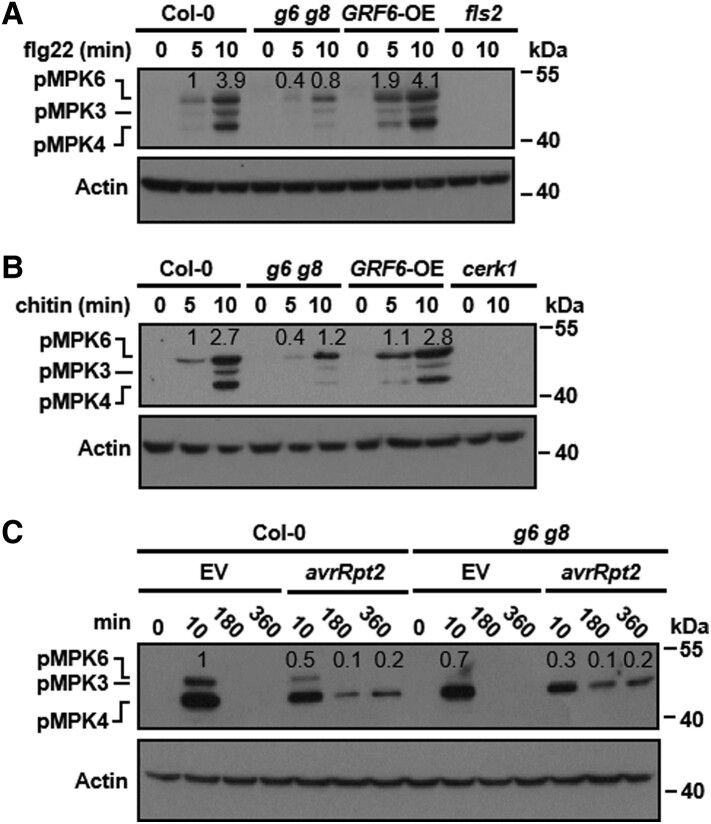
We next tested whether the 2 14-3-3 isoforms are required for flg22-triggered MAP kinase activation in seedlings. Flg22 treatment strongly induced phosphorylation of MPKs indicative of MAP kinase activation within 5–10 min in Col-0 plants, but not in fls2 mutant plants (Fig. 4A). This effect was greatly diminished in g6 g8 mutant plants and was slightly enhanced in the GRF6-OE plants. Similarly, chitin-triggered MAP kinase activation was also diminished in the g6 g8 mutant plants (Fig. 4B). In contrast to the g6 g8 double mutant plants, flg22- and chitin-induced MAP kinase activation was completely normal in the grf6 and grf8 single mutant plants (Supplemental Fig. S7, A and B). This indicated that GRF6 and GRF8 redundantly regulate MAP kinase activation during PTI. Although PTI is associated with only transient MAP kinase activation, ETI is known to be associated with prolonged MAP kinase activation (Tsuda et al. 2013). We therefore inoculated plants with Pst carrying an either empty vector (EV) control or avrRpt2 and examined possible subsequent MAP kinase activation. As expected, Pst EV-induced MAP kinase activation in Col-0 at the 10-min time point, but not later at the 180- and 360-min time points (Fig. 4C). Whereas the early induction was reduced in the g6 g8 mutant, the prolonged MAP kinase activation at the 180 and 360 min time points was indistinguishable in Col-0 vs. g6 g8 plants following inoculation with Pst avrRpt2. Together, these results indicated that GRF6 and GRF8 are specifically required for MAP kinase activation, but not for the other early PTI signaling events we tested.
Figure 4.
GRF6 and GRF8 are required for pattern-triggered MAP kinase activation. A and B) Pattern-triggered MAP kinase activation is impaired in g6 g8 double mutant plants. Seedlings were sprayed with flg22 (A) or chitin (B), and samples were harvested at the indicated times for immunoblot analysis using an anti-pERK antibody. MAP kinase activation is indicated by the accumulation of phosphorylated MPK3, MPK4, and MPK6 (abbreviated as pMPK3, pMPK4, and pMPK6). Anti-ACTIN immunoblotting was used as a loading control. C) MAP kinase activation in Col-0 and g6 g8 plants at different time points following inoculation with the indicated bacterial strains. Seedlings were infiltrated with Pst DC3000 carrying avrRpt2, and samples were harvested at the indicated times for immunoblotting analysis using an anti-pERK antibody. Anti-ACTIN immunoblotting was used as a loading control. Numbers above the lanes in the blots show relative band intensity quantified by Image J software and normalized to the level of actin. Each experiment was repeated 3 times with similar results, and results from one representative experiment are shown.
The 14-3-3 isoforms GRF6 and GRF8 are required for MAPKKK5 phosphorylation and its interaction with PBL19
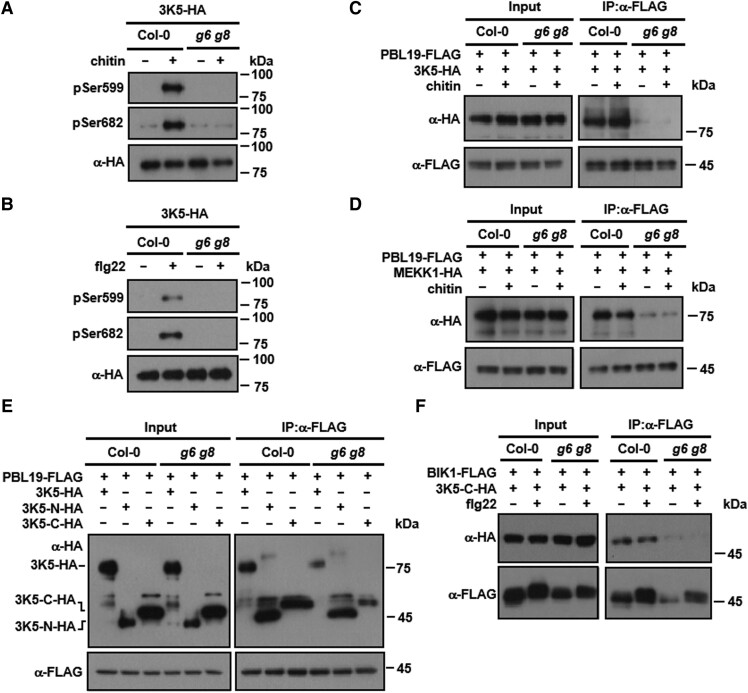
We previously showed that pattern-triggered MAP kinase activation is mediated by RLCKs, that directly phosphorylate MAPKKKs in their C-termini, and that this phosphorylation could be detected by using antibodies that recognize specific phosphorylated Ser residues (anti-pSer599 and anti-pSer682) in MAPKKK5-HA (Bi et al. 2018). As expected, chitin treatment induced strong phosphorylation of MAPKKK5 at Ser599 and Ser682 in MAPKKK5-HA transgenic seedlings in the Col-0 background (Fig. 5A). Strikingly, this induced phosphorylation was abolished in MAPKKK5-HA transgenic plants in the g6 g8 mutant background (Fig. 5A). Likewise, flg22 also induced MAPKKK5 phosphorylation at Ser599 and Ser682, and these phosphorylation events were also abolished in the g6 g8 mutant plants (Fig. 5B).
Figure 5.
GRF6 and GRF8 are required for PBL19-MAPKKK5 interaction and MAPKKK5 phosphorylation. A and B) Pattern-triggered MAPKKK5 phosphorylation is impaired in g6 g8 mutant plants. Seedlings of Col-0 and g6 g8 transgenic lines (T3 generation) expressing MAPKKK5-HA were sprayed with solutions containing chitin (A) or flg22 (B), and proteins were affinity purified using an anti-HA antibody and detected by immunoblot analysis using anti-phosphoSer599 (pSer599) and anti-phosphoSer682 (pSer682) antibodies. 3K5: MAPKKK5. C and D) GRF6 and GRF8 are required for the interaction of PBL19 with MAPKKK5 (C) and MEKK1 (D). Protoplasts from Col-0 or g6 g8 plants expressing the indicated proteins were treated with chitin, and Co-IP was performed using an anti-FLAG antibody. E and F) GRF6 enhances the interaction of the MAPKKK5 C-terminus with PBL19 in Arabidopsis protoplasts. Protoplasts of the indicated genotypes were transfected with indicated constructs, and Co-IP was performed using an anti-FLAG antibody. Each experiment was repeated at least 3 times with similar results (3 times for A to E and 4 times for F), and results from one representative experiment are shown.
The requirement of GRF6 and GRF8 for pattern-triggered MAPKKK5 phosphorylation was unexpected, as MAPKKK5 Ser599 and Ser682 are directly phosphorylated by RLCKs. We therefore tested whether GRF6 and GRF8 are required for interactions between RLCKs and MAPKKKs. Co-IP assays showed that PBL19 strongly interacted with MAPKKK5 in Col-0 protoplasts in the presence or absence of chitin treatment (Fig. 5C; Supplemental Fig. S8A). This interaction was nearly abolished in g6 g8 mutant plants. Similarly, the PBL19-MEKK1 interaction was impaired in g6 g8 mutant protoplasts (Fig. 5D; Supplemental Fig. S8B). These results indicated that the GRF6 and GRF8 isoforms play an important role in the regulation of MAPKKK phosphorylation, likely by promoting interactions between MAPKKKs and RLCKs.
We next asked how GRF6 and GRF8 promote interactions between MAPKKKs and RLCKs. Co-IP assays showed that PBL19 interacted with full-length MAPKKK5 and both its N-terminus and C-terminus in Col-0 protoplasts (Fig. 5E; Supplemental Fig. S8C). In g6 g8 mutant protoplasts, the interactions of PBL19 with both full-length MAPKKK5 and its C-terminal region were clearly diminished, whereas the interaction with the MAPKKK5 N-terminus was not affected. Furthermore, Co-IP assays showed that BIK1 similarly interacted with the C-terminus of MAPKKK5 in a manner dependent on GRF6 and GRF8 (Fig. 5F; Supplemental Fig. S8D). Together, these results suggested that these 14-3-3 proteins are required for proper interaction between the C-terminus of MAPKKK5 and RLCKs.
GRF6 promotes PBL19 access to the MAPKKK5 C-terminus
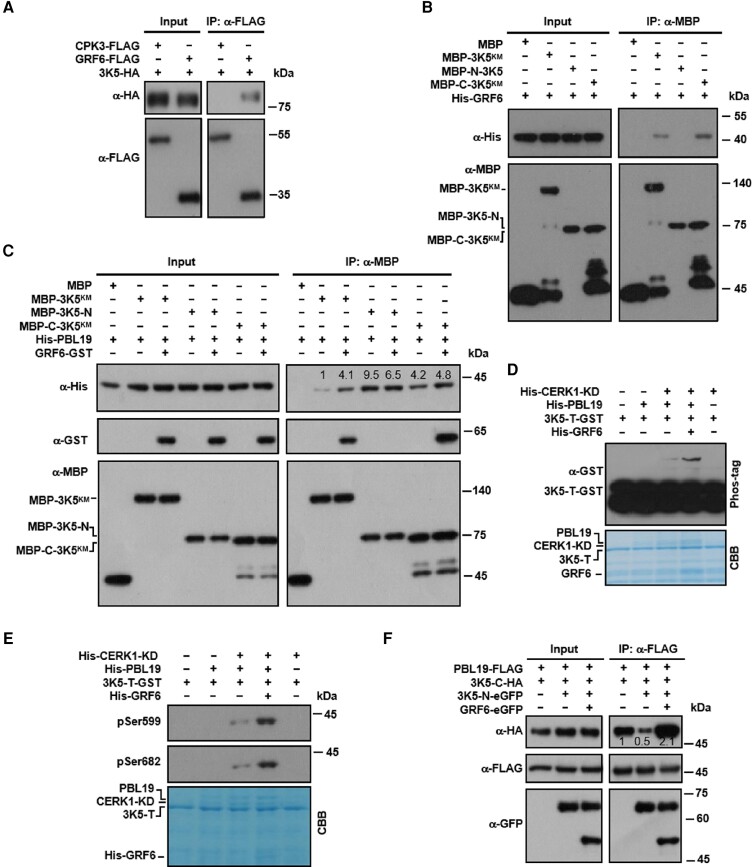
To elucidate the underlying mechanism by which GRF6 regulates MAPKKK5 phosphorylation, we tested whether and how GRF6 interacted with MAPKKK5. Co-IP assays showed that GRF6 indeed interacted with MAPKKK5 in protoplasts independent of flg22 treatment (Fig. 6A; Supplemental Fig. S9A). In vitro pull-down assays showed that GRF6 directly interacted with the C-terminus, but not with the N-terminus, of MAPKKK5 (Fig. 6B).
Figure 6.
GRF6 promotes the access to and phosphorylation of the C-terminus of MAPKKK5 by PBL19. A) GRF6 interacts with MAPKKK5 in Arabidopsis protoplasts. Protoplasts from Col-0 were transfected with the indicated constructs, and Co-IP was performed using an anti-FLAG antibody. CPK3 was used as a negative control. B) GRF6 interacts with the C-terminus of MAPKKK5. His-tagged GRF6 recombinant protein was incubated with MBP-tagged kinase-dead (K375M; KM) variants of full-length or truncated MAPKKK5 proteins, and the interaction was tested in a MBP pull-down assay. C) GRF6 directly enhances the interaction of PBL19 with the MAPKKK5 C-terminus in vitro. GST-tagged GRF6 and His-tagged PBL19 recombinant proteins were incubated with MBP-tagged kinase-dead (KM) variants of full-length or truncated MAPKKK5. MBP pull-down assays were performed, and the pulled-down proteins were analyzed by immunoblotting using anti-His or anti-GST antibodies. D) GRF6 promotes the phosphorylation of the C-terminal tail of MAPKKK5 by PBL19. In the presence of the CERK1 kinase domain (CERK1-KD), MAPKKK5-T-GST was incubated with His-PBL19 with or without His-GRF6 protein. Total protein was analyzed by SDS-PAGE containing Phos-tag acrylamide, and proteins were detected by immunoblotting using an anti-GST antibody. E) GRF6 promotes the PBL19-mediated phosphorylation of MAPKKK5 at Ser599 and Ser682 in vitro. His-GRF6 was incubated with HIS-CERK1-KD, His-PBL19, and/or GST-MAPKKKT-T in kinase reaction buffer. Protein phosphorylation was detected by anti-pSer599 and pSer682 immunoblotting. F) GRF6 promotes the interaction between PBL19 and the C-terminus of MAPKKK5 in the presence of the added N-terminus. Protoplasts from Col-0 were transfected with the indicated constructs, and Co-IP was performed using an anti-FLAG antibody. Numbers above the lanes in the blots (C and F) show relative band intensities quantified by Image J software and normalized to the MBP signal (C) or the FLAG signal (F) in the IP products. The experiment of (A) was repeated 4 times, and the experiments of (B to F) were repeated 3 times. All of the experiments were with similar results, and results from one representative experiment are shown.
We next studied GRF6, PBL19, and MAPKKK5 recombinant proteins in vitro to ask whether GRF6 directly regulated the interaction between PBL19 and the C-terminus of MAPKKK5. In vitro pull-down assays showed that, in the absence of GRF6, PBL19 interacted strongly with the MAPKKK5 N-terminus and modestly with its C-terminus and only weakly with full-length MAPKKK5 (Fig. 6C). In the presence of GRF6, the PBL19 interactions with both full-length MAPKKK5 and its C-terminus were enhanced, whereas the interaction with the MAPKKK5 N-terminus was not affected. Similarly, the presence of GRF6 also enhanced the interaction between BIK1 and the C-terminus of MAPKKK5 (Supplemental Fig. S10A). These results indicated that GRF6 can promote the interactions of the MAPKKK5 C-terminus with both PBL19 and BIK1.
We next tested the CERK1 kinase domain, PBL19, the C-terminal tail of MAPKKK5, and GRF6 in vitro to test whether GRF6 could promote phosphorylation of the MAPKKK5 C-terminal tail by PBL19. As we have previously reported, phos-tag assays showed that PBL19 was able to phosphorylate the MAPKKK5 C-terminal region only in the presence of the CERK1 kinase domain, and this was greatly enhanced in the presence of GRF6 (Fig. 6D). These findings were confirmed by immunoblot analysis using specific anti-pSer599 and anti-pSer682 antibodies (Fig. 6E). These results indicated that GRF6 can directly enhance PBL19-mediated phosphorylation of the MAPKKK5 C-terminus.
We noted that the PBL19 interaction with the MAPKKK5 C-terminus was stronger than its interaction with full-length MAPKKK5, suggesting an inhibitory role of the N-terminus on the MAPKKK5-PBL19 interaction. Therefore, we tested for a potential intramolecular interaction between the N-terminus and C-terminus of MAPKKK5. Indeed, Co-IP assays showed that the N-terminus and C-terminus of MAPKKK5 did interact with each other in protoplasts (Supplemental Fig. S10B). Overexpression of the N-terminus of MAPKKK5 greatly reduced the interaction between PBL19 and the C-terminus of MAPKKK5 in protoplasts (Fig. 6F), probably by competing with PBL19 for access to the MAPKKK5 C-terminus. This suggested that the intramolecular interaction between the N-terminus and C-terminus of MAPKKK5 normally impedes the accessibility of the C-terminus to PBL19. The ability of PBL19 to interact with the MAPKKK5 C-terminus in the presence of MAPKKK5 N-terminus was not affected by flg22 and chitin treatments (Supplemental Fig. S9, B and C), suggesting that pattern perception does not increase the accessibility. Simultaneously overexpressing GRF6 restored the strong interaction between PBL19 and the C-terminus of MAPKKK5 in the presence of the additional N-termini, suggesting that GRF6 can relieve intramolecular inhibition caused by the MAPKKK5 N-terminus.
Discussion
In this study, we demonstrated that the Arabidopsis 14-3-3 proteins GRF6 and GRF8 play a key role in the regulation of PTI. These 2 isoforms interacted with RLCKs (Fig. 1) and were required for expression of pattern-induced defense genes, callose deposition (Fig. 2), and immunity against both bacterial (P. syringae pv. tomato) and fungal (B. cinerea) pathogens (Fig. 3). Further characterization indicated that the 2 isoforms were specifically required for pattern-triggered activation of MAP kinases (Fig. 4, A and B), but not for other early signaling events such as an ROS burst (Supplemental Fig. S3) and phosphorylation of BIK1 and RGS1 (Supplemental Fig. S6D). We showed that GRF6 directly interacted with the C-terminus of MAPKKK5 to promote its accessibility by immune-activated PBL19 (Fig. 6C), revealing a unique mechanism underlying pattern-triggered MAP kinase activation and immunity.
An previous report showed that Arabidopsis GRF8 is targeted by the Pst effector HopM1, which suppresses PTI (Lozano-Durán et al. 2014). Inhibition of 14-3-3 proteins in Arabidopsis and N. benthamiana plants leads to defects in PTI including ROS burst and stomatal closure, although the specific isoforms required remained unknown. Here, we showed that 14-3-3 isoforms GRF6 and GRF8 are redundantly required for flg22- and chitin-induced MAP kinase activation, defense gene expression, and resistance against both bacterial and fungal pathogens. A role of these 2 isoforms in basal resistance is also consistent with a previous report that GRF6 interacts with RPW8 and is required for basal resistance to powdery mildew (Yang et al. 2009). Interestingly, g6 g8 double mutant plants showed a normal ROS burst, suggesting that other isoforms may play a role in ROS regulation during PTI. Consistent with this possibility, we found that at least ten different 14-3-3 members can interact with both PBL19 and PBL20, and that multiple RLCK members can interact with GRF6 when overexpressed in Arabidopsis protoplasts or N. benthamiana plants (Fig. 1). Although the BIK1-GRF6 interaction was confirmed in the native context, future work is needed to systematically determine whether interactions of specific General Regulatory Factors (GRFs) and RLCKs occur in the native in planta context and whether they regulate different signaling networks in PTI. More importantly, future work should address not only whether different GRF members prefer specific target proteins in different signaling networks but also whether this contributes to different signal outputs.
It has been shown that the tomato 14-3-3 protein TFT7, which is similar to human 14-3-3ε, interacts with both SlMAPKKKα, an ortholog of Arabidopsis MAPKKK3, and SlMKK2 to positively regulate cell death during ETI (Oh and Martin 2011; Oh et al. 2010). Overexpression of TFT7 enhances SlMAPKKKα stability and enhances MAP kinase activation. In addition, multiple tomato 14-3-3 proteins have been shown to contribute to ETI triggered by AvrXv3, although the underlying mechanisms remain unclear (Giska et al. 2013). GRF6 and GRF8, however, are dispensable for ETI triggered by multiple effectors. Whereas g6 g8 double mutant plants were impaired in PTI-triggered MAP kinase activation, they displayed normal extended MAP kinase activation triggered by Pst avrRpt2. It remains to be tested whether 14-3-3 isoforms other than GRF6 and GRF8 are involved in ETI in Arabidopsis.
Although numerous reports have indicated that 14-3-3 proteins play an important role in plant immunity by interacting with various target proteins such as Arabidopsis protein RPW8.2 and tomato SlMAPKKKα, the underlying biochemical mechanisms remain poorly understood. Of note, TFT7 has been proposed to act as scaffold to recruit and bridge SlMAPKKKα and SlMKK2 to promote MAP kinase activation during ETI (Oh and Martin 2011). However, SlMAPKKKα and SlMKK2 do not seem to directly interact. Furthermore, TFT7 can also enhance the stability of SlMAPKKKα (Oh et al. 2010). The exact mechanism by which TFT7 enhances MAP kinase activation during ETI is not fully understood. Our study revealed an important mechanism underlying PTI signaling. We showed that the N-terminus of MAPKKK5 interacted with its own C-terminus to prevent access by PBL19 and subsequent PBL19-induced phosphorylation. GRF6 interacted with both PBL19 and the MAPKKK5 C-terminus to enable optimum interaction between PBL19 and the MAPKKK5 C-terminus. Thus, GRF6 acts as a scaffold to assemble PBL19 and MAPKKK5 into a competent complex that is necessary for the phosphorylation of MAPKKK5 C-terminal tail by PBL19. This mechanism likely applies also to GRF8, as GRF6 and GRF8 isoforms are redundantly required both for the interaction between PBL19 and MAPKKK5 and for pattern-triggered MAP kinase activation. The PBL19-MEKK1 interaction similarly required both of the GRF6 and GRF8 isoforms. Furthermore, multiple RLCKs can interact with GRF6, suggesting that this mechanism is used by a variety of PRRs in the activation of MAP kinase cascades.
In summary, our findings have elucidated a unique, detailed mechanism by which 14-3-3 proteins regulate plant immunity (Supplemental Fig. S10C). The 14-3-3 isoforms GRF6 and GRF8 act as scaffold proteins that recruit RLCKs and MAPKKK5 and facilitate rapid activation of the MAP kinase cascade. The interaction between these 14-3-3 proteins with the C-terminus of MAPKKK5 exposes its C-terminus, which is otherwise blocked by an intramolecular interaction with its own N-terminus. This enables better access to and phosphorylation by RLCKs, which is required for the subsequent activation of the MAP kinase cascade.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) plants used in this study include Col-0, grf6 (Liu et al. 2017), grf8 (Liu et al. 2017), grf6 grf8 (Liu et al. 2017), grf6 grf8 complemented with GRF6 (Liu et al. 2017), and the GRF6 overexpression line GRF6-OE (Liu et al. 2017), plus Col-0 and grf6 grf8 transgenic plant lines carrying MAPKKK5-HA under the control of its native promoter (NPpro:MAPKKK5-HA), and a grf6 grf8 transgenic line carrying the GRF6-HA transgene under the control of the native promoter (NPpro:GRF6-HA; this study). The N. benthamiana plants used for the luciferase complementation assays and the Arabidopsis plants used for protoplast transfection and pathogen inoculation were grown in soil with a 2:1 ratio of vermiculite to nutrient soil (Pindstrup, Denmark) at 23 °C with a photoperiod of 10 h white light and 14 h darkness for 4 to 5 wk. The light source was white fluorescent bulbs (YZ28RR16; TOPSTAR) providing an intensity of 90 μE m−2 s−1.
Bacterial strains and growth condition
The bacterial strains used in this study include Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) (Zhou et al. 2015), Pst DC3000 cor− (Zhou et al. 2015), Pst DC3000 hrcC− (Li et al. 2014a), Pst DC3000 carrying hopZ1a (Hu et al. 2020), avrB (Feng et al. 2012), avrRpt2 (Li et al. 2014b), and avrRps4 (Nie et al. 2012).
These strains were grown on King's B medium [2.9% (w/v) Bacto proteose peptone (1264614, Gibco), 0.2% (w/v) K2HPO4 (60353, Sigma), 0.8% (v/v) glycerin (0854, Amresco), 0.15% (w/v) Mg2SO4 (7487-88-9, Sinopharm), and 1.5% (w/v) agar (A038, Caisson)] containing the appropriate antibiotics, and the culture was placed at 28 °C for 2 d.
Plasmid construction and transgenic plants
Constructs used to produce BAK1, BIK1, FLS2, XLG2-NT, MAPKKK5, MAPKKK5-N, MAPKKK5-C, and MEKK1-HA/FLAG proteins under the control of the cauliflower mosaic virus 35S promoter (35Spro) have been described previously (Bi et al. 2018; Feng et al. 2012; Li et al. 2014; Liang et al. 2016; Wang et al. 2015). To generate GRF6-FLAG, CPK5-FLAG, GRF6-HA, MAPKKK5-N-HA, and MAPKKK5-C-HA constructs for transient expression in protoplasts, the appropriate PCR fragments were cloned into pUC19-35S-HA-RBS or pUC19-35S-Flag-RBS vector (New England BioLabs, Li et al. 2005) using a ClonExpress II One Step Cloning Kit (no. C112; Vazyme Biotech). To generate GRF6-eGFP and MAPKKK5-N-eGFP constructs for transient expression in protoplasts, the eGFP fragment was fisrt cloned into pUC19-35S-HA-RBS vector to replace its HA-tag (abbreviated as pUC19-35S-eGFP), and then the GRF6 or MAPKKK5-N PCR fragments was cloned into pUC19-35S-eGFP.
Constructs used to produce BIK1-GST, MAPKKK5-T-GST, His-PBL19, and His-CERK1-KD have been described previously (Bi et al. 2018). To generate GRF6-GST, MBP-MAPKKK5-N, MBP-MAPKKK5-C-K375M, and His-GRF6, the appropriate PCR fragments were cloned into the pGEX-6p-1 vector (GE Healthcare) encoding GST tag, pMAL-c2 vector (New England BioLabs) encoding a maltose-binding protein (MBP) tag, and pET28a (Novagen) encoding a His tag using a ClonExpress II One Step Cloning Kit (no. C112; Vazyme Biotech), respectively. PBL19, PBL40, PBL14, PBL32, PBL30, and BIK1 constructs with HA-Nluc-double tag for luciferase complementation assays have been described previously (Bi et al. 2018). To generate GRF6-Cluc and CPK5-Nluc for luciferase complementation assays, the appropriate PCR fragments were cloned into pCAMBIA1300-Cluc and pCAMBIA1300-HA-Nluc, respectively (Liang et al. 2016). All mutant constructs were generated by site-directed mutagenesis. Primers used in this study are listed in Supplemental Table S2.
To generate NPpro:GRF6-HA transgenic plants, a 4,435 bp genomic fragment from Col-0 including 2,625 bp upstream of the start codon was PCR-amplified and cloned into pCAMBIA1300 (Cambia; Brisbane, QLD, Australia). To generate the NPpro:MAPKKK5-HA transgenic plants, a genomic fragment from MAPKKK5 and the entire coding region was PCR-amplified and cloned into pCAMBIA1300 (Bi et al. 2018). These constructs were introduced into either g6 g8 double mutant plants or Col-0 plants by Agrobacterium tumefaciens (Agrobacterium)-mediated transformation (Clough and Bent 1998). T3 lines were used for experiments described in this study.
Antibodies
Commercial monoclonal antibodies used in this study were: anti-HA (CWBIO, CW0092m), anti-FLAG (Sigma, F3165-5MG), anti-His (CWBIO´CW0286M)´anti-GST (abcam, ab9085), anti-GFP (RM1008-T), anti-MBP (BE2020-100), anti-pERK (4370L, Cell Signaling), and anti-ACTIN (BE0028-100, EASYBIO). Polyclonal antibodies used include anti-FLS2, anti-BIK1, and anti-BAK1 antibodies, which were generated in rabbits (JV222-RB35) by immunizing rabbits with recombinant proteins. Specific anti-pS599 and anti-pS682 antibodies were described previously (Bi et al. 2018).
In planta bacterial growth assays
For spray inoculations, leaves of 4-week-old Arabidopsis plants were sprayed with Pseudomonas syringae pv. tomato (Pst) DC3000 bacteria at 5× 108 colony-forming units/mL, and the number of bacterial in leaves was determined 3 d later. Briefly, leaf discs of 5 mm in diameter were punched from leaves with a cork borer and ground in microfuge tubes with a pestle placed on a handrill. The bacterial suspension was serial-diluted and plated on TSA agar medium [1% (w/v) BactoTM tryptone (211705, BD), 1% (w/v) sucrose (0335, Amresco), 8% (v/v) glutamic acid (G1626, Sigma), 0.1% (w/v) Mg2SO4, and 1.5% (w/v) agar] with appropriate antibiotics. The plates were incubated at 28 °C for 2 d, the colony-forming units (CFUs) were counted.
For bacterial entry assays, leaves of 4-week-old plants were incubated in water overnight and then soaked in a Pst DC3000 cor− bacterial suspension (2 × 108 CFU/mL) containing 0.02% (v/v) Silwet L-77 (Coolaber, CS9791-10 mL) for 1.5 h. The leaves were then washed in sterile H2O containing 0.02% (v/v) Silwet L-77 in a 3 L beaker on a magnetic stir for 30 s to remove surface bacteria, and the number of bacteria in leaves was determined as above.
For flg22 (synthesized by Sangon Biotech and dissoveled in H2O)-protection assays, leaves of 4-week-old Arabidopsis plants were syringe-infiltrated with 1 μM flg22 or H2O 1 d before syringe infiltration of Pst DC3000 at 1 × 106 CFU/mL. The number of bacterial in leaves was determined 2 d later.
To perform bacterial growth assays for ETI, leaves of 4-week-old Arabidopsis plants were infiltrated using a needleless syringe with bacterial strains at 1 × 106 colony-forming units/mL using a needleless syringe. The number of bacteria in leaves was determined 3 d later. The bacterial strain Pst DC3000 and Pst DC3000 strains carrying either hopZ1a, avrB, avrRpt2, or avrPS4 were used in this work.
For Botrytis cinerea inoculation, the central vein of leaves of 4-week old Arabidopsis plants was punctured with a needle, and a 5-μL droplet of conidial suspension (5 × 105 conidia/mL) was placed on the wound. Lesion sizes were measured 3 d later.
Stomatal aperture measurements
Four-week-old plants were kept under light for 3 h to fully open the stomata. Epidermal peels were collected with forceps from the abaxial side of rosette leaves and floated either in mock solution (suspension buffer without bacteria) or in suspension buffer [10 mM MES (E169, Amresco), pH 6.15, 10 mM KCl (0395, Amresco), 10 mM CaCl2 (0556, Amresco)] containing Pst DC3000 hrcC− at 5 × 108 CFU/mL for 1 h. Stomata were examined with a microscope (Axio Imager.A2; Zeiss) under bright field using the 403 objective lens, and images were captured by using the ZEN lite software. Stomatal aperture was calculated using Image J.
Callose deposition
The procedure to measure callose induction was adapted from Clay et al. (2009). Briefly, Arabidopsis seeds were germinated in 1/2-strength MS medium containing 0.23% (w/v) MS Salt (M519, Phytotech), 1.5% (w/v) Sucrose (0335, Amresco), and 0.3% (w/v) Phytagel (P8169, Sigma) with the pH adjusted to 5.8 with 1 M KOH (1310-58-3, Sinopharm) (Zhang et al. 2010) and grown in growth chamber (CU-36L5, Percival) at 22 °C with a photoperiod of 16 h light and 8 h darkness for 8 d. Seedlings were treated with H2O or 1 mM flg22 for 18 h. Epifluorescence images were then captured under ultraviolet light with a fluorescence microscope (Axio Imager.A2; Zeiss). The number of callose deposits per 0.1 mm2 was calculated by using Image J software.
Mass spectrometric analysis of PBL19- and PBL20-interacting proteins
IP-coupled mass spectrometric analysis was performed as previously described (Li et al. 2014). Briefly, Arabidopsis protoplasts expressing PBL19-FLAG or PBL20-FLAG were treated with flg22, lysed in 20 mL IP buffer [50 mM HEPES (H3375, Sigma), pH 7.5, 50 mM NaCl (X190, Amresco), 0.2% Triton X-100 (AP093, GPC), 1 mM DTT (0281, Amresco), 1× protease inhibitor cocktail (04693116001, Roche)]. After IP with anti-FLAG agarose beads, the eluted proteins were run on a 4% to 10% SDS-PAGE gel (Invitrogen), stained with colloidal, and protein bands were digested in-gel with trypsin. After extraction, the peptides were separated by an analytical capillary column (50 μm × 10 cm) packed with 5 μm spherical C18 reversed phase material (YMC, Kyoyo, Japan). The eluted peptides were sprayed into a LTQ mass spectrometer (Thermo Fisher Scientific; San Jose, CA, USA) equipped with a nano-ESI ion source. Database searches were performed on an in-house Mascot server (Matrix Science Ltd., London, UK) against International Protein Index Arabidopsis protein database.
Reverse transcription quantitative PCR
Ten-day-old seedlings grown on 1/2-strength MS were sprayed with 1 μM flg22 or 200 μg/mL chitin (C9752, Sigma) dissolved in H2O and harvested at the indicated time points. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized with the first-strand cDNA synthesis kit (Thermo), quantitative PCR was performed in triplicate for each cDNA sample, and the relative expression levels were normalized to the ACTIN1 gene.
MAP kinase activation assay
Ten-day-old Arabidopsis seedlings grown on 1/2-strength MS medium were sprayed with 1 μM flg22 or 200 μg/mL chitin. For bacterium-triggered MAP kinase activation, leaves of 4-week-old Arabidopsis plants were infiltrated with Pst DC3000 or Pst DC3000 AvrRpt2 at 1 × 106 colony-forming units/mL, and samples were harvested at the indicated time points. Leaves were ground in liquid nitrogen with a grinding rod and extracted using IP buffer (50 mM HEPES-KOH, pH 7.5, 150 mM KCl, 1 mM EDTA (BG292, Dogesce), 0.3% Triton X-100, 1 mM DTT, and 1× complete protease inhibitors and analyzed by immunoblotting with an anti-pERK antibody (4370; Cell Signaling Technology) at 1:4000 dilution. An anti-ACTIN immunoblot was used as a loading control at 1:3000 dilution. The accumulation of phosphorylated MPKs (pMPKs) indicates MAP kinase activation.
Oxidative burst assay
Leaves of 4-week-old plant leaves were sliced into approximately 1 mm strips, incubated overnight in water in a 96-well plate, and then treated with 1 μM flg22, 1 μM pep2, which is a peptide recognized by PEPTIDE 1 RECEPTOR 2 (PEPR2), 1 μM elf18, or 200 μg/mL chitin in 100 μL of buffer [20 mM luminol (A4685, Sigma) and 10 mg/mL horseradish peroxidase (P1709, Sigma)]. Luminescence was recorded using the EnSpire Multimode plate reader (Perkin Elmer).
Co-immunoprecipitation assays
Co-IP in protoplasts was performed as previously described (Li et al. 2014). Briefly, protoplasts of the desired genotypes were transfected with the indicated plasmids and incubated overnight. Protoplasts were then lysed in IP buffer and total protein was incubated with 40 μL agarose-conjugated anti-FLAG antibody (A2220, Sigma) for 2 h to enrich FLAG-tagged proteins. The beads were washed 3 times with IP buffer, and the bound protein was eluted with IP buffer containing 0.5 mg/ml 3 × FLAG peptide (F4799, Sigma).
For anti-HA IP in Arabidopsis plants, 10-day-old plants were used for protein extraction. The protein was precleared with protein A agarose (83219, Sigma) for 1 h and then incubated with an anti-HA antibody (CW0092m, CWBIO) and protein A agarose for 2 h. The agarose beads were washed as in co-IP in protoplasts and boiled with SDS loading buffer to elute the bound protein [1 M Tris-HCl (pH 6.8), 6% (w/v) SDS (11667289001, Roche), 40% (v/v) Glycerin (0854, Amresco), 0.032% (w/v) bromophenol blue (BES-0490BR, BIOSEN), and 1.7% (v/v) β-mercaptoethanol (B0452, New England BioLabs)].
Eluted proteins from co-IP described above were separated in 10% SDS-PAGE gels and detected by immunoblotting using anti-FLAG and anti-HA antibodies (1:3,000 dilution), anti-FLS2 and anti-BAK1 antibodies (1:1,000 dilution), and anti-BIK1 antibodies (1:500 dilution).
Split-luciferase complementation assays
Split-luciferase complementation assays were performed as described previously (Chen et al. 2008). Briefly, Agrobacterium strain GV3101 containing the indicated construct was syringe-infiltrated into expanded leaves of 6-week-old N. benthamiana plants. Leaf discs with of 5 mm in diameter were harvested 2 d after inoculation with a cork borer and placed in a 96-well plate with the luciferase (10476498001, Roche). The luc activities were recorded using the EnSpire Multimode plate reader (Perkin Elmer).
In vitro pull-down assays
Recombinant proteins were expressed in E. coli and affinity-purified using glutathione-Sepharose for GST-tagged proteins (17-0756-01, GE Healthcare), Ni-Sepharose (30210, Qiagen) for His-tagged proteins, or amylose resin (E8021L, New England BioLabs) for MBP-tagged proteins, respectively. For GST pull-down assays, 2.5 μg of the appropriate tagged protein was incubated with 25 μL glutathione agarose beads in 200 μL GST binding buffer [25 mM Tris-HCl (10812846001, Roche), pH 7.5, 100 mM NaCl, 1 mM DTT] for 2 h at 4 °C, washed 3 times with GST wash buffer (25 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 mM DTT), and then eluted with GST elution buffer containing 15 mM GSH (G4251, Sigma). MBP pull-down assays were similarly performed with different binding buffer (20 mM Tris-HCl, pH 7.4, 1.17% (w/v) NaCl and 1 mM EDTA) and elution buffer [20 mM Tris-HCl, pH 7.4, 1.17% (w/v) NaCl, 1 mM EDTA and 0.004% meltose (BES-0798BR, BIOESN)]. The presence of GST-tagged, His-tagged, and MBP-tagged proteins was examined by immunoblotting using anti-GST (1:2,000 dilution), anti-His (1:2,000 dilution), or anti-MBP antibodies (1:3,000 dilution).
In vitro kinase assay and detection of MAPKKK5 phosphorylation
For in vitro kinase assays, 200 ng His-CERK1-KD, 200 ng His-PBL19, or 200 ng His-GRF6 protein was incubated with 2 μg MAPKKK5-T-GST in 20 μL reaction buffer [25 mM Tris-HCl, pH 7.5, 10 mM MgCl2 (7791-18-6, HARVEYBIO), 1 mM DTT, and 100 mM ATP (18330019, Invitrogen)] for 30 min at 30 °C. The reaction was stopped by boiling in the SDS loading buffer. The phosphorylation state of proteins was detected by phos-tag or immunoblotting using antibodies recognizing specific phosphorylated peptides.
Protein samples from the in vitro kinase assay were subjected to phos-tag assays to detect protein phosphorylation. Briefly, samples were electrophoresed in 10% SDS-polyacrylamide gels containing 100 μM MnCl2 and 50 μM Phos-tag Acrylamide AAL-107 (NARD Institute). The gel was then incubated in transfer buffer lacking methanol (20 mM Tris, pH 7.4, and 150 mM glycine) containing 1 mM EDTA for 10 min and subsequently incubated in the transfer buffer lacking EDTA for 10 min. Proteins were then transferred to PVDF membranes and detected by immunoblotting using an anti-GST monoclonal antibody (AB101-02; Tiangen) at 1:2000 dilution.
Phosphorylation of specific MAPKKK5 residues in in vitro phosphorylated protein and seedling proteins was detected by immunoblotting using anti-pS599 and anti-pS682 antibodies at 1:500 dilution (Bi et al. 2018).
Statistical analyses
Statistical analyses were performed using GraphPad Prism version 8, GraphPad Software. Statistical data are provided in Supplemental Data Set S1, and source data are provided in Supplemental Data Set S2.
Accession numbers
Sequence data for genes described in this article can be found in the TAIR database (https://www.arabidopsis.org) and GenBank data libraries under the following accession numbers: AT5G61210 for SNAP33, AT3G19380 for PUB25, AT3G20250 for PUM5, AT5G66470 for ERA1, AT2G01210 for ORPK1, AT4G09000 for GRF1, AT1G78300 for GRF2, AT5G38480 for GRF3, AT1G35160 for GRF4, AT5G16050 for GRF5, AT5G10450 for GRF6, AT3G02520 for GRF7, AT5G65430 for GRF8, AT2G42590 for GRF9, AT1G22300 for GRF10, AT5G46330 for FLS2, AT4G33430 for BAK1, AT4G34390 for XLG2, AT2G19190 for FRK1, AT2G14610 for PR1, AT3G45640 for MPK3, AT4G01370 for MPK4, AT2G43790 for MPK6, AT5G66850 for MAPKKK5, AT4G08500 for MEKK1, AT2G39660 for BIK1, AT5G47070 for PBL19, AT5G03320 for PBL40, AT2G05940 for PBL14, AT2G17220 for PBL32, AT4G35600 for PBL30, AT4G23650 for CPK3, AT4G35310 for CPK5, At1G17750 for PEPR2, At3G21630 for CERK1, At2G37620 for ACTIN1, AAR02168 for hopZ1a, M21965 for avrB, L11355 for avrRpt2, and L43559 for avrRps4.
Supplementary Material
Acknowledgments
We thank Shuhua Yang for 14-3-3-related seeds including grf6 grf8 double mutant, GRF6-OE transgenic seeds and NPpro:GRF6 complemention line in grf6 grf8 double mutant background and Dingzhong Tang for the MBP-MAPKKK5-K375M plasmid.
Contributor Information
Xiaojing Dong, State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Feng Feng, Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK 74078, USA.
Yangjun Li, State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Lin Li, National Institute of Biological Sciences, Beijing 102206, China.
She Chen, National Institute of Biological Sciences, Beijing 102206, China.
Jian-Min Zhou, State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China; Chinese Academy of Sciences Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing 100049, China; Hainan Yazhou Bay Seed Laboratory, Sanya, Hainan 572025, China.
Author contributions
J.-M.Z. and X.D. designed the research and wrote the article. X.D., F.F., and Y.L. performed the experiments. L.L. and S.C. performed mass spectrometry analyses.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. GRF6 interacts with FLS2, BAK1 and RLCKs.
Supplemental Figure S2. grf6 and grf8 single mutants display minor defects in PTI signaling.
Supplemental Figure S3. GRF6 and GRF8 are not required for pattern-triggered ROS production.
Supplemental Figure S4. GRF6 and GRF8 are not required for ETI.
Supplemental Figure S5. The grf6 and grf8 single mutants display normal basal resistance.
Supplemental Figure S6. GRF6 and GRF8 are not required for the FLS2-BAK1 and FLS2-BIK1 interactions, nor are they required for flg22-induced XLG2 phosphorylation.
Supplemental Figure S7. The grf6 and grf8 single mutants display normal MAP kinase activation.
Supplemental Figure S8. PBL19 and BIK1 interact with MAPKKK5 and MEKK1.
Supplemental Figure S9. GRF6 promotes the access to the MAPKKK5 C-terminus by PBL19 independent of pattern perception.
Supplemental Figure S10. GRF6 interacts with C-terminus of MAPKKK5 to promote MAP kinase activation.
Supplemental Table S1. The candidate proteins of LC-MS.
Supplemental Table S2. The primers used in this study.
Supplemental Data Set S1. Statistical data.
Supplemental Data Set S2. Source data of reverse transcription quantitative PCR assays in this study.
Funding
The work was supported by grants from National Key Research and Development Program of China (2021YFA1300701), the National Science Foundation of China (32120103004, 31830019), the Hainan Excellent Talent Team, and the State Key Laboratory of Plant Genomics (SKLPG2016B-2), to J.-M.Z.
Data availability
All data to support the conclusions of this manuscript are provided in the main figures and supplementary information. Source data are also provided in supplementary information.
References
- Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006:16(3):162–172. 10.1016/j.semcancer.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002:415(6875):977–983. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S, Zhou JM. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell. 2018:30(7):1543–1561. 10.1105/tpc.17.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M, Pimprikar P, Nurnberger T, Zipfel C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat Plants. 2021:7(5):579–586. 10.1038/s41477-021-00874-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB. 14-3-3 Proteins: a number of functions for a numbered protein. Sci STKE. 2005:2005(296):10. 10.1126/stke.2962005re10 [DOI] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife. 2014:3:e03766. 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Peng HC, He J, MacWilliams J, Teixeira M, Tsuchiya T, Chesnais Q, Mudgett MB, Kaloshian I. Aphid effector Me10 interacts with tomato TFT7, a 14-3-3 isoform involved in aphid resistance. New Phytol. 2019:221(3):1518–1528. 10.1111/nph.15475 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):323–324. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006:18(2):465–476. 10.1105/tpc.105.036574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007:448(7152):497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009:323(5910):95–101. 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for agrobacteriummediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Zipfel C. Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell. 2021:81(17):3449–3467. 10.1016/j.molcel.2021.07.029 [DOI] [PubMed] [Google Scholar]
- Denison FC, Paul AL, Zupanska AK, Ferl RJ. 14-3-3 Proteins in plant physiology. Semin. Cell Dev. Biol. 2011:22(7):720–727. 10.1016/j.semcdb.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Dubrow Z, Sunitha S, Kim JG, Aakre CD, Girija AM, Sobol G, Teper D, Chen YC, Ozbaki-Yagan N, Vance H, et al. Tomato 14-3-3 proteins are required for Xv3 disease resistance and interact with a subset of Xanthomonas euvesicatoria effectors. Mol. Plant Microbe Interact. 2018:31(12):1301–1311. 10.1094/MPMI-02-18-0048-R [DOI] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012:485(7396):114–118. 10.1038/nature10962 [DOI] [PubMed] [Google Scholar]
- Freeman AK, Morrison DK. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011:22(7):681–687. 10.1016/j.semcdb.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007:19(5):1617–1634. 10.1105/tpc.105.035626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008:18(12):1190–1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang D, Wang X, Zhang X, Wen Z, Zhang Q, Li D, Dinesh-Kumar SP, Zhang Y. Coat proteins of necroviruses target 14-3-3a to subvert MAPKKKα-mediated antiviral immunity in plants. Nat Commun. 2022:13(1):716. 10.1038/s41467-022-28395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giska F, Lichocka M, Piechocki M, Dadlez M, Schmelzer E, Hennig J, Krzymowska M. Phosphorylation of HopQ1, a type III effector from Pseudomonas syringae, creates a binding site for host 14-3-3 proteins. Plant Physiol. 2013:161(4):2049–2061. 10.1104/pp.112.209023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Ibanez SG, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci U S A. 2007:104(29):12217–12222. 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Qi J, Bi G, Zhou JM. Bacterial effectors induce oligomerization of immune receptor ZAR1 in vivo. Mol Plant. 2020:13(5):793–801. 10.1016/j.molp.2020.03.004 [DOI] [PubMed] [Google Scholar]
- Huber SC, MacKintosh C, Kaiser WM. Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol. Biol. 2002:50(6):1053–1063. 10.1023/A:1021284002779 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science. 2016:354(6316):aaf6395. 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 2014:54(1):43–55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Minami NI, Tomiyama CA, Dohmae N, Takio K, Minami E, Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U S A. 2006:103(29):11086. 10.1073/pnas.0508882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014a:15(3):329–338. 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA. 2005:102(36):12990–12995. 10.1073/pnas.0502425102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ma X, Chiang YH, Yadeta KA, Ding P, Dong L, Zhao Y, Li X, Yu F, Zhang L, et al. Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in Arabidopsis. Cell Host Microbe. 2014b:16(4):473–483. 10.1016/j.chom.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 2012:8(6):e1002767. 10.1371/journal.pgen.1002767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yadeta KA, Elmore JM, Coaker G. The Pseudomonas syringae effector HopQ1 promotes bacterial virulence and interacts with tomato 14-3-3 proteins in a phosphorylation-dependent manner. Plant Physiol. 2013:161(4):2062–2074. 10.1104/pp.112.211748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife. 2016:5:e13568. 10.7554/eLife.13568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou JM. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 2018:69(1):267–299. 10.1146/annurev-arplant-042817-040540 [DOI] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell. 2017:66(1):117–128.e5. 10.1016/j.molcel.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Liu Y, Leary E, Saffaf O, Baker RF, Zhang S. Overlapping functions of YDA and MAPKKK3/MAPKKK5 upstream of MPK3/MPK6 in plant immunity and growth/development. J. Integr. Plant Biol. 2022:64(8):1531–1542. 10.1111/jipb.13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004:16(12):3386–3399. 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Duran R, Bourdais G, He SY, Robatzek S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014:202(1):259–269. 10.1111/nph.12651 [DOI] [PubMed] [Google Scholar]
- Lozano-Duran R, Robatzek S. 14-3-3 proteins in plant-pathogen interactions. Mol. Plant Microbe Interact. 2015:28(5):511–518. 10.1094/MPMI-10-14-0322-CR [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci U S A. 2010:107(1):496–501. 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Wang W, Fei Y, Cheng H-Y, Song B, Zhou Z, Zhao Y, Zhang X, Li L, Chen S, et al. A surface-receptor-coupled G protein regulates plant immunity through nuclear kinases. Cell Host Microbe. 2022:30(11):1602–1614.e5. 10.1016/j.chom.2022.09.012 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006:126(5):969–980. 10.1016/j.cell.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Mittal S, Davis KR. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant-Microbe Interact. 1995:8(1):165–171. 10.1094/MPMI-8-0165 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2007:104(49):19613–19618. 10.1073/pnas.0705147104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996:84(6):889–897. 10.1016/S0092-8674(00)81067-3 [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 2021:592(7852):110–115. 10.1038/s41586-021-03315-7 [DOI] [PubMed] [Google Scholar]
- Nie H, Zhao C, Wu G, Wu Y, Chen Y, Tang D. SR1, A calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 2012:158(4):1847–1859. 10.1104/pp.111.192310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Martin GB. Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J Biol Chem. 2011:286(16):14129–14136. 10.1074/jbc.M111.225086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKKα. Plant Cell. 2010:22(1):260–272. 10.1105/tpc.109.070664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Denison FC, Schultz ER, Zupanska AK, Ferl RJ. 14-3-3 phosphoprotein interaction networks-does isoform diversity present functional interaction specification? Front. Plant Sci. 2012:3:190. 10.3389/fpls.2012.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou JM. Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol. 2018:177(4):1679–1690. 10.1104/pp.18.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Li Q, Luo M, Yan H, Xie B, Li X, Zhong G, Chen D, Tang D. Brassinosteroid-signaling kinase1 modulates MAP kinase15 phosphorylation to confer powdery mildew resistance in Arabidopsis. Plant Cell. 2022:34(5):1768–1783. 10.1093/plcell/koac027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Zhang M, Zhang L, Sun T, Liu Y, Lukowitz W, Xu J, Zhang S. Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell. 2017:29(3):526–542. 10.1105/tpc.16.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007:143(2):661–669. 10.1104/pp.106.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2- BAK1 immune complex. Science. 2013:342(6158):624–628. 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- Sun T, Nitta Y, Zhang Q, Wu D, Tian H, Lee JS, Zhang Y. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 2018:19(7):e45324. 10.15252/embr.201745324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell. 2017:29(4):618–637. 10.1105/tpc.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper D, Salomon D, Sunitha S, Kim JG, Mudgett MB, Sessa G. Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector-triggered immunity. Plant J. 2014:77(2):297–309. 10.1111/tpj.12391 [DOI] [PubMed] [Google Scholar]
- Thor K, Jiang S, Michard E, George J, Scherzer S, Huang S, Dindas J, Derbyshire P, Leitão N, DeFalco TA, et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature. 2020:585(7826):569–573. 10.1038/s41586-020-2702-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature. 2019:572(7767):131–135. 10.1038/s41586-019-1413-y [DOI] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 2013:9(12):e1004015. 10.1371/journal.pgen.1004015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud MF, Chabannes M, et al. The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe. 2015:18(3):285–295. 10.1016/j.chom.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu Y, Zhang H, Wang P, Xia Y. Arabidopsis MAPKK kinases YODA, MAPKKK3, and MAPKKK5 are functionally redundant in development and immunity. Plant Physiol. 2022:190(1):206–210. 10.1093/plphys/kiac270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meng J, Meng X, Zhao Y, Liu J, Sun T, Liu Y, Wang Q, Zhang S. Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell. 2016:28(5):1144–1162. 10.1105/tpc.15.00871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Volinia S, Leffers H, Rittinger K, Caron PR, Aitken A, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997:91(7):961–971. 10.1016/S0092-8674(00)80487-0 [DOI] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016:35(22):2468–2483. 10.15252/embj.201694248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. PEPR2 is a second receptor for the pep1 and pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell. 2010:22(2):508–522. 10.1105/tpc.109.068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhao Y, Shi H, Li J, Wang Y, Tang D. BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 2018:176(4):2991–3002. 10.1104/pp.17.01757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Lee WH, Sobott F, Papagrigoriou E, Robinson CV, Grossmann JG, Sundström M, Doyle DA, Elkins JM. Structural basis for protein–protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U S A. 2006:103(46):17237–17242. 10.1073/pnas.0605779103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 2009:60(3):539–550. 10.1111/j.1365-313X.2009.03978.x [DOI] [PubMed] [Google Scholar]
- Yang Z, Wang C, Xue Y, Liu X, Chen S, Song C, Yang Y, Guo Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat Commun. 2019:10(1):1199. 10.1038/s41467-019-09181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021:592(7852):105–109. 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010:7(4):290–301. 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell. 2014:26(3):1166–1182. 10.1105/tpc.113.117069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wu Y, Yang Y, Du M, Zhang X, Guo Y, Li C, Zhou JM. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell. 2015:27(7):2032–2041. 10.1105/tpc.15.00466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-tu by the receptor EFR restricts agrobacterium-mediated transformation. Cell. 2006:125(4):749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004:428(6984):764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to support the conclusions of this manuscript are provided in the main figures and supplementary information. Source data are also provided in supplementary information.