Abstract
Cdc6 performs an essential role in the initiation of eukaryotic DNA replication by recruiting the minichromosome maintenance (MCM) complex onto DNA. Using immunodepletion/add-back experiments in Xenopus egg extracts, we have determined that both Walker A (ATP binding) and Walker B (ATP hydrolysis) motifs of Xenopus Cdc6 (Xcdc6) are essential, but have distinct functional roles. Although Walker B mutant protein binds chromatin well, Walker A mutant protein binds chromatin poorly. Neither Walker A nor Walker B mutant protein, however, load appreciable MCM onto DNA. Herein, we provide evidence that Cdc6 functions as a multimer: 1) mutant and wild-type Xcdc6 form multimers; 2) either mutant protein is dominant negative when added before wild-type Xcdc6, but stimulates DNA replication when added simultaneously with wild-type Xcdc6; and 3) the two mutants restore DNA replication when added together, in the absence of wild-type Xcdc6. Our findings suggest that ATP may play a key regulatory role within this multimer: its binding to Cdc6 promotes chromatin association and its hydrolysis facilitates MCM loading. Moreover, ATP binding and hydrolysis may occur in trans between Cdc6 subunits within the complex.
INTRODUCTION
The initiation of DNA replication constitutes one of the major control points in cell cycle progression. The DNA replication machinery is activated when the cell has reached a critical mass and contains sufficient levels of components, such as nucleoside triphosphates, that are necessary for DNA synthesis. The assembly of eukaryotic trans-acting initiator elements on cis-acting origins of replication is best understood in budding yeast. The origin recognition complex (ORC), consisting of six subunits, binds to origins throughout the cell cycle. Additional factors associate with the DNA-bound ORC during G1 phase to form the prereplication complex. These factors include Cdc6, members of the minichromosome maintenance (MCM) family, and Cdc45 (reviewed in Kelly and Brown, 2000). Once DNA has been licensed to replicate, S phase promoting factor (consisting of various regulatory kinases, e.g., Cdc7/Dbf4 and cyclin-dependent kinase Cdc28) triggers prereplication complexes to fire. Significantly, homologs for many of these proteins have been found in multicellular eukaryotes. For example, ORC, Cdc6, MCMs, Cdc45, Cdc7, and cyclin-dependent kinase (Cdk) 2 proteins are required for DNA replication in Xenopus (Chong et al., 1995; Kubota et al., 1995; Carpenter et al., 1996; Coleman et al., 1996; Sato et al., 1997; Findeisen et al., 1999; Jares and Blow, 2000; Mimura et al., 2000) and mammals (Jiang and Hunter, 1997; Sato et al., 1997; Williams et al., 1997; Saha et al., 1998; Petersen et al., 1999; Mendez and Stillman, 2000; Natale et al., 2000). Thus, both the regulatory system and functional downstream components governing the initiation of DNA replication are likely conserved in all eukaryotes.
One function of Cdc6 homologs is to load the MCMs onto chromatin, a critical step in licensing the DNA for replication (Coleman et al., 1996; Donovan et al., 1997; Tada et al., 1999). A purine nucleotide binding site, composed of two motifs of the type identified by Walker et al. (1982), is found in all Cdc6 homologs. Structural analysis of a number of nucleoside triphosphate-binding proteins containing such motifs indicates that the Walker A motif (GxxGxGKT) or P-loop contains an invariant lysine residue, which interacts directly with the γ-phosphate of ATP and is critical for ATP binding. In contrast, the acidic residues within the Walker B motif or DExD box coordinate a magnesium ion and are essential for ATP hydrolysis (Story and Steitz, 1992). To date, some work has probed the requirement for these sequences in yeast and human Cdc6. Mutant Walker A Cdc6 is nonfunctional in yeast (Perkins and Diffley, 1998; DeRyckere et al., 1999; Wang et al., 1999; Weinreich et al., 1999), suggesting that Cdc6 requires ATP binding to form a productive and stable interaction with ORC and MCMs at replication origins. Microinjection of Walker A mutant human Cdc6 (HsCdc6) into HeLa cells, however, results in a dominant negative phenotype characterized by a block in replication (Herbig et al., 1999), suggesting that the human Walker A mutant protein disrupts the ability of endogenous wild-type Cdc6 to load MCMs stably onto chromatin. In budding yeast, alanine substitution within the Walker B motif (DE223, 224AA) produces a fully functional Cdc6 protein (Weinreich et al., 1999), whereas a glutamic acid-to-glycine change in the Walker B motif (E224G) is dominant negative, blocking cells in late G1 or early S phase (Perkins and Diffley, 1998). In human cells, microinjecting mutant Walker B (E285Q) HsCdc6 also results in a dominant negative phenotype (Herbig et al., 1999). Together, these findings argue that Cdc6 ATP binding and ATP hydrolysis have separable roles, but the precise details remain unclear.
The Xenopus early embryo cell cycles are characterized by rapid oscillations between S and M phases and thus impose constraints on DNA replication that differ from those characterized in yeast and mammalian tissue culture. The egg achieves these rapid cleavages, in part, by using large maternally derived stores of DNA replication machinery, including a surplus of Xcdc6. For example, phosphorylation appears to control the subcellular localization of both HsCdc6 and Xcdc6 but the activities of these proteins vary dramatically. Overexpression (Jiang et al., 1999) or microinjection (Herbig et al., 2000) of unphosphorylatable HsCdc6 inhibits DNA replication. In contrast, unphosphorylatable Xcdc6 does not act as dominant negative inhibitor, but is actually fully functional (Pelizon et al., 2000). These interesting observations suggest that Xcdc6 may be differentially regulated in the early embryo in other respects, such as the requirement of nucleotide binding/hydrolysis.
In this report, we have assessed the activity of mutant forms of Xenopus Cdc6 (Xcdc6). We find that recombinant Xcdc6 harboring point mutations within either the Walker A or Walker B motif is unable to restore DNA replication to an Xcdc6-depleted extract. Immunoblot analyses revealed that, when added alone, the Walker A mutant bound chromatin poorly and failed to recruit MCM to the DNA. In contrast, the Walker B mutant bound chromatin well but recruited little MCM to the DNA. A dominant negative phenotype is observed if either Walker A or Walker B mutant Xcdc6 is added to extracts before wild-type protein. Remarkably, however, both Walker A and Walker B mutant proteins can stimulate wild-type Xcdc6 when added simultaneously with wild-type protein. Under similar conditions, Walker A mutant Xcdc6 can also stimulate Walker B mutant protein. Furthermore, the presence of wild-type Xcdc6 stimulated binding of Walker A mutant Xcdc6 to DNA. Together, these results are consistent with a model in which ATP binding facilitates Cdc6 association with chromatin, whereas ATP hydrolysis is required for MCM loading. Moreover, Cdc6 appears to function as an oligomer in which only a subset of the subunits must be competent to bind and/or hydrolyze ATP for the oligomer to function.
MATERIALS AND METHODS
Construction of Expression Vectors
All Xcdc6 alterations were constructed by polymerase chain reaction (PCR). For N-terminal Xcdc6 (amino acids 1–165), the 5′ primer was used as previously (Coleman et al., 1996), wherein the initiation codon was altered to an NdeI site. The 3′ primer GGTCCGAATTCTCAGCGCTCTGGTATAGCCGTATTCAAAGC contains an additional 10 base pairs that encode a stop codon and an EcoRI site immediately downstream of amino acid 165. For C-terminal Xcdc6 (amino acids 166–554), the 5′ primer GGTCCGAATTCCTGTTGGCTCGTGAGAGTGAG incorporates an EcoRI site immediately upstream of amino acid 166; the 3′ primer GGTCCTCTAGAAATCCCTGAATTGAGAACATTCCC incorporates an XbaI site immediately downstream of amino acid 554. We targeted this truncated protein to the nucleus by fusing the C-terminal portion with simian virus 40 large T antigen nuclear localization signal (NLS). This NLS was made by annealing the following oligonucleotides: GATCCATGGGTGCTCCTCCAAAAAAGAAGAGAAAGGTAGCTCCAG- and AATTCTGGAGCTACCTTTCTCTTCTTTTTTGGAGGAGCAC-CCATG to produce a double-stranded oligonucleotide, which contains 5′ BamHI and 3′ EcoRI restriction sites. PCR reactions contained template (50 ng), primers (0.5 pmol of each), Pfu DNA polymerase (Stratagene, La Jolla, CA), and the manufacturer's buffer. The reactions were cycled 30 times as follows: 94°C, 1 min; 60°C, 3 min; and 72°C, 5 min. The product of the PCR amplification was used directly for subcloning into TAII (Invitrogen, Carlsbad, CA) and the nucleotide sequence was confirmed. For expression in Sf9 insect cells, we modified the pFastBac1 (Invitrogen) to contain a six-histidine tag (His6) and an NdeI site at its N-terminal end by subcloning nim1 coding sequences (containing an N-terminal His6 tag) from pVL1393N-His6 (Coleman et al., 1993) as a BamHI-EcoRI fragment into pFastBacI to produce pFastBac1-His6. N-terminal Xcdc6 was subcloned into pFastBac1-His6 as an NdeI-EcoRI fragment. C-Terminal Xcdc6 was assembled in a trimolecular ligation of BamHI-EcoRI (NLS), EcoRI-XbaI (C terminus), and BamHI-XbaI pFastBac1-His6 (vector). The Walker A and Walker B point mutations were made using the QuikChange site-directed mutagenesis kit (Stratagene) and the following primers: GGTGCTCCTGGTACCGGCGAAACTGCGTGC and GCACGCAGTTTCGCCGGTACCAGGAGCACC (Walker A, K202E) and GGTGTTGGATGGGATGGATCA-GCTGG and CCAGCTGATCCATCCCATCCAACACC (Walker B, E277G). We verified the correct reading frame by sequencing and subcloned BglII-Nsi I fragments encompassing these mutations into pGEX 4T3-Xcdc6 (kindly provided by P. Jackson, Stanford University School of Medicine, Palo Alto, CA).
Expression and Purification of Recombinant Xcdc6
Recombinant baculoviruses encoding N-terminal and C-terminal Xcdc6 were isolated by standard procedures (Invitrogen). Sf9 insect cell lysates containing either full-length (Coleman et al., 1996), N-terminal, or C-terminal His6-Xcdc6 were prepared (Desai et al., 1992) and the histidine-tagged proteins were purified on Ni-IDA (iminodiacetic acid) (Sigma Chemical, St. Louis, MO) as described (Kumagai and Dunphy, 1995) and washed three times each with high salt buffer (10 mM HEPES, pH 7.4, 1.5 M NaCl, 0.1% Triton X-100, 15 mM imidazole) followed by HBS (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.1 mM dithiothreitol) containing 0.2 mM phenylmethylsulfonyl fluoride. Fractions were eluted in HBS made 500 mM in imidazole, dialyzed against HBS, drop-frozen in liquid nitrogen, and stored at −80°C.
Because the mutated forms of Xcdc6 were poorly expressed using a baculovirus system (our unpublished data), mutant and wild-type Xcdc6 were produced as glutathione S-transferase (GST) fusion proteins in bacteria (Furstenthal et al., 2001). GST fusion proteins were expressed in BL21-CodonPlus (Stratagene) following a protocol kindly provided by Furstenthal and Jackson (Stanford University School of Medicine). Cells were grown at 37°C, induced with 0.3 mM isopropyl β-d-thiogalactoside for 4 h, harvested by centrifugation, resuspended in lysis buffer (10 mM sodium phosphate, pH 7.2, 0.5 M NaCl, 1 mM EGTA, 1 mM dithiothreitol, 0.25% Tween 20) containing 0.2 mM phenylmethylsulfonyl fluoride and 10 μg/ml each of pepstatin, chymostatin, and leupeptin, drop-frozen in liquid nitrogen, and stored at −80°C. To purify GST-Xcdc6, the thawed lysate was sonicated and centrifuged in an F18S rotor (Sorvall, Newton, CT) at 12,500 rpm for 30 min. The supernatant was mixed with glutathione-Sepharose (Amersham Biosciences, Piscataway, NJ), prewashed in lysis buffer, and incubated with gentle agitation at 4°C for 1 h to facilitate binding to the beads. The GST-fusion protein/glutathione-Sepharose bead complexes were isolated by centrifugation and transferred to 4 ml of Econocolumn (Bio-Rad, Hercules, CA). After washing with 20 column volumes of lysis buffer, GST fusion proteins were eluted from the beads in lysis buffer (pH adjusted to 8.0) containing 10 mM glutathione. The proteins were eluted in fractions of 300 μl. The peak protein fractions were pooled, dialyzed against HBS, drop-frozen in liquid nitrogen, and stored at −80°C. Protein concentrations were determined using the Bio-Rad protein assay kit.
Xenopus Egg Extracts and Immunodepletions
Xenopus cytostatic factor (CSF)-arrested egg extracts were prepared from unactivated eggs as described (Murray, 1991). Xenopus cytostatic factor-arrested extracts were supplemented with 100 μg/ml cycloheximide and 0.4 mM CaCl2. Immunodepletions were performed on interphase extracts (15-min postactivation) by using protein A-agarose containing either affinity-purified anti-Xcdc6 or control rabbit anti-mouse IgG (Zymed Laboratories, South San Francisco, CA) antibodies (10 μg of antibody/130 μl of extract). All experiments were performed using double-depleted extracts and efficient removal confirmed by immunoblot analyses (our unpublished data). For most experiments, interphase extracts were supplemented with demembranated sperm chromatin (600 nuclei/μl for replication assays, 3000 nuclei/μl for chromatin blots).
DNA Replication Assays
Replication of sperm chromatin was monitored by agarose gel electrophoresis after labeling with [α-32P]dCTP (Dasso and Newport, 1990). Typically, labeling was performed in two time windows (e.g., amount of replication that occurred in 0–45 and 45–90 min) for each sample, as indicated. Quantitation of replication assays was performed using a phosphorimager (Fuji Instruments, Tokyo, Japan). The rate of DNA replication for the Walker B mutant GST-Xcdc6 (Figure 2) was estimated by comparing the level of replication in the first time window (∼30% of total) relative to the amount of replication in the first time window for either the mock depleted (∼75%) or Xcdc6 depleted reconstituted with wild-type GST-Xcdc6 (∼75%), as normalized by protein concentration.
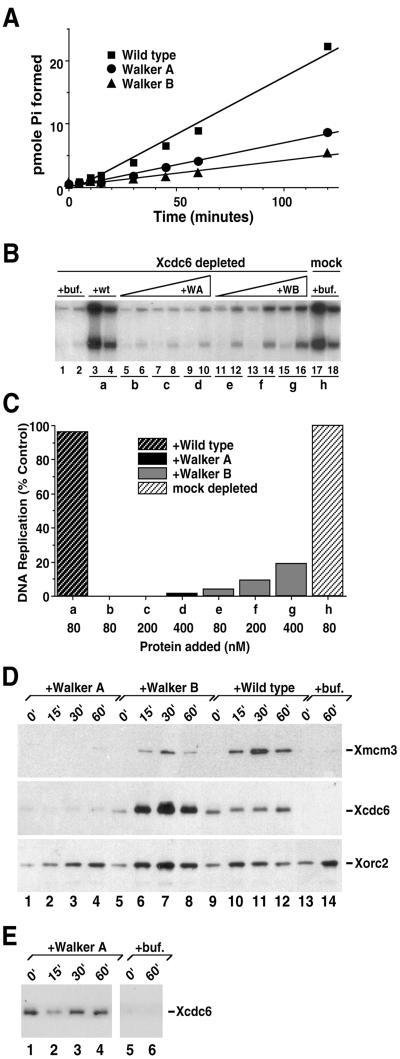
Figure 2.
Intact Walker A and Walker B motifs within the Xcdc6 nucleotide binding site are required for Xcdc6 function. (A) Wild-type GST-Xcdc6 (▪), GST-Xcdc6WA (K202E; ●), and GST-Xcdc6WB (E227G; ▴) were incubated with [γ-32P]ATP for the indicated times at 37°C. Hydrolysis products were separated by thin layer chromatography and the amount of phosphate formed was quantitated. (B) Replication of sperm chromatin was determined in Xcdc6-depleted (lanes 1–16) or mock-depleted (lanes 17 and 18) interphase extracts in the presence of control buffer (lanes 1 and 2; 17 and 18), purified wild-type GST-Xcdc6 (80 nM, lanes 3 and 4), GST-Xcdc6WA (80 nM, lanes 5 and 6; 200 nM, lanes 7 and 8; 400 nM, lanes 9 and 10), or GST-Xcdc6WB (80 nM, lanes 11 and 12; 200 nM, lanes 13 and 14; 400 nM, lanes 15 and 16). Each lane depicts the 32P incorporation into chromosomal DNA that occurred during the following times: 1–60 min (odd-numbered lanes) and 60–120 min (even-numbered lanes). (C) Quantitation of the total chromosomal DNA replication for the indicated samples shown in B over a 120-min period in either a mock-depleted extract in the presence of control buffer alone (h) or Xcdc6-depleted extract in the presence of wild-type GST-Xcdc6 (a), GST-Xcdc6WA (b–d), or GST-Xcdc6WB (e–g) at the indicated protein concentrations. Background was subtracted and histograms were normalized as described in Figure 1. (D) Immunoblots were performed on detergent-insoluble fractions isolated from Xcdc6-depleted interphase extracts containing chromatin incubated in the presence of control buffer (lanes 13 and 14), 400 nM GST-Xcdc6WA (lanes 1–4), 400 nM GST-Xcdc6WB (lanes 5–8), or 400 nM wild-type GST-Xcdc6 (lanes 9–12). Samples were incubated at 23°C for the indicated times and prepared for immunoblot analyses by using anti-Xmcm3 (top), anti-Xcdc6 (middle), or anti-Xorc2 (bottom) antibodies. (E) Longer exposure of portions of the anti-Xcdc6 immunoblot shown in D to demonstrate the weaker but easily detectable GST-Xcdc6WA chromatin association (lanes 1–4) relative to the control buffer treated samples (lanes 5 and 6).
Chromatin Blots
Chromatin blots were performed as described for Xenopus low-speed supernatants (Furstenthal et al., 2001), substituting a Beckman Coulter Microfuge E for the Beckman Coulter 152 microfuge. In some cases, endogenous wild-type Xcdc6 from undepleted egg extracts (where it is present at ∼80 nM; Coleman et al., 1996) was added to Xcdc6-depleted extracts to achieve intermediate Xcdc6 concentrations, as indicated. Immunoblotting was performed using anti-Xorc2 (Carpenter et al., 1996), anti-Xcdc6 (Coleman et al., 1996), and anti-Xmcm3 (Hua and Newport, 1998) antibodies and developed using Renaissance chemiluminescence reagent (PerkinElmer Life Sciences, Boston, MA).
Cell Synchronization and Microinjections
HeLa cells were grown as monolayers in DMEM supplemented with antibiotics and 10% fetal bovine serum in a humidified incubator at 37°C and 5% CO2. To determine the optimal G1 phase bromodeoxyuridine (BrdU) staining schedule, we synchronized cells by mitotic shake off and examined BrdU uptake at different times after replating. G1 phase lasted 7–8 h, and S phase peaked at 9–10 h post shake off and was complete by 12–13 h, as indicated by the appearance of mitotic cells (our unpublished data). Mitotic shake-off cells were incubated for ∼6 h to ensure secure reattachment and then, in view of the above-mentioned cell cycle timing, microinjected (>200 cells for each protein). For the G1/S phase nuclear injections, cells on coverslips were synchronized by double thymidine block composed of sequential growth in media supplemented with 2 mM thymidine (14 h), plain media (8 h), and media supplemented with 2 mM thymidine (14 h). Cells were subsequently released from thymidine block in plain media and immediately microinjected. After microinjection, the media was supplemented with 10 μM BrdU and 1 μM fluorodeoxyuridine (1:1000 dilution, cell proliferation labeling reagent; Amersham Biosciences). Each protein sample for microinjection contained GST at a concentration of 4 mg/ml to facilitate detection of injected cells. GST fusion proteins were injected at a concentration of ∼0.5 mg/ml. Immediately before microinjection, each protein sample was filtered by microfuge centrifugation at 8000 rpm for 1–2 min at 4°C through a 0.22-μm filter unit (Ultrafree-MC; Millipore, Bedford, MA) and cleared by microfuge centrifugation at 14,000 rpm for 5 min at 4°C. Proteins were injected into the nuclei of cells by using purchased needles (Femtotips; Eppendorf - 5 Prime, Boulder, CO) with a microinjector (model 5242; Eppendorf - 5 Prime) and a manipulator (model 5171; Eppendorf - 5 Prime) mounted on an inverted microscope (Nikon TE300).
Immunofluorescence
Immunostaining was performed as described (Herbig et al., 1999). Briefly, cells were washed three times with PBS, fixed for 20 min (3.5% formaldehyde in PBS), permeabilized with 0.2% Triton X-100 for 20 min, and incubated with 10% fetal bovine serum in PBS for 1 h. GST was detected by staining with a rabbit polyclonal anti-GST antibody (Santa Cruz Biotechnology) at a dilution of 1:200 in PBS supplemented with 10% fetal bovine serum (F-PBS) for 2 h, followed by staining with AlexaFluor 488 goat anti-rabbit IgG conjugate (Molecular Probes, Eugene, OR) at a dilution of 1:1000 in F-PBS for 1 h. BrdU incorporated into DNA was detected by staining with a mouse monoclonal anti-BrdU antibody (BD PharMingen, San Diego, CA) at a dilution of 1:500 in F-PBS containing 125 U/ml benzon nuclease (Novagen, Madison, WI) for 2 h, followed by staining with AlexaFluor 594 goat anti-mouse IgG conjugate (Molecular Probes) at a dilution of 1:500 in F-PBS. The cells were then washed two times with PBS for 5 min each, and once with PBS containing 0.25 μg/ml Hoechst (Sigma Chemical) for 5 min. All incubations with antibody were done at room temperature. The coverslips were mounted in 90% glycerol and viewed on a Nikon Eclipse E800 equipped with a 60× objective. All images were captured at identical magnification and exposure times using a Quantix cooled charge-coupled device camera (Photometrics, Tucson, AZ) with Isee software (Inovision, Raleigh, NC).
Protein Binding Assays
35S-Labeled untagged wild-type Xcdc6 was translated using a TNT reticulocyte (Promega, Madison, WI) system and incubated with GST fusion proteins immobilized on glutathione-agarose in the presence of 2% nonfat dry milk and washed essentially as described (Herbig et al., 1999) without any added nucleotides. The bound proteins were eluted by boiling in 2 volumes of SDS sample buffer, separated by SDS-PAGE, and detected by fluorography with enhance (PerkinElmer Life Sciences).
ATPase Reactions
ATPase reactions were performed as previously described (Herbig et al., 1999) by using 25 μM [γ-32P]ATP with 0.5 pmol of GST fusion protein per 10-μl reaction volume. The rates of ATP hydrolysis were determined in the linear range of protein concentration dependence. Aliquots of the reaction mixtures were placed in stop buffer and the equivalent of 1/30 of the reaction was spotted on a polyethyleneimine-cellulose thin layer plate (EM Scientific, Gibbstown, NJ), which was developed in 0.9 M LiCl, 0.05 M EDTA, pH 6.1. The amount of [γ-32P]ATP hydrolyzed to [32P]orthophosphate was quantitated using a phosphorimager (Fuji Instruments).
RESULTS
C-Terminal Two-Thirds of Xcdc6 Is Necessary and Sufficient for DNA Replication
To delineate the functional domains of Xcdc6, deletion mutants were constructed that contained either the C-terminal two-thirds or the N-terminal one-third of the Xenopus protein. The C-terminal domain shows significant homology to the ATPases associated with various cellular activities (AAA+) family of ATPases (Perkins and Diffley, 1998; Liu et al., 2000) and encompasses a characteristic purine nucleotide binding site composed of Walker A and Walker B motifs (Figure 1A), which regulate the conformation and activity of these proteins. In contrast, the N-terminal one-third of Xcdc6 contains a putative NLS and a majority of the S/P or T/P motifs, which comprise potential recognition sites for Cdks and other cell cycle-regulated kinases (Figure 1A). These features suggest that the N-terminal domain might play a regulatory role governing the activity of a C-terminal catalytic domain.
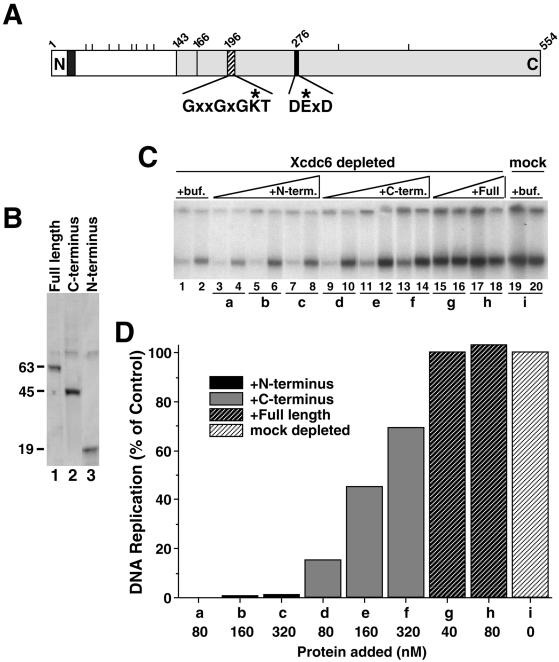
Figure 1.
The C-terminal two-thirds of Xcdc6 is necessary and sufficient to reconstitute DNA replication in an Xcdc6-depleted egg extract. (A) Schematic representation of Xcdc6 showing the approximate locations of a potential nuclear localization signal (dark-shaded box), multiple potential phosphorylation sites (short perpendicular lines above schematic), the region with significant homology (29% identity) to the archael Cdc6 (light-shaded box), Walker A motif (hatched box), Walker B motif (black box), and amino acid number (numbers above schematic). The consensus Walker A and B sequences (asterisks indicate residues mutated in this study) are indicated below the schematic. (B) Purified His6-Xcdc6 proteins (full length, lane 1; C terminus, lane 2; N terminus, lane 3) were stained with Coomassie blue. Approximate molecular weights (in Daltons, × 10−3) are indicated to the left of the gel. (C) Replication of sperm chromatin was determined in Xcdc6-depleted (lanes 1–18) or mock-depleted (lanes 19 and 20) interphase extract in the presence of control buffer (lanes 1 and 2; 19 and 20), purified N-terminal (80 nM, lanes 3 and 4; 160 nM, lanes 5 and 6; 320 nM, lanes 7 and 8), C-terminal (80 nM, lanes 9 and 10; 160 nM, lanes 11 and 12; 320 nM, lanes 13 and 14), or full length (40 nM, lanes 15 and 16; 80 nM, lanes 17 and 18) His6-Xcdc6. Each lane depicts the 32P incorporation into chromosomal DNA that occurred during the following times: 1–45 min (odd-numbered lanes) and 45–90 min (even-numbered lanes). (D) Quantitation of the total chromosomal DNA replication for the indicated samples shown in C over a 90-min period in either a mock-depleted extract in the presence of buffer alone (i) or Xcdc6-depleted extract in the presence of N-terminal (a–c), C-terminal (bars d–f), or full-length (g and h) His6-Xcdc6 at the indicated protein concentrations. In this analysis, we have subtracted the background (replication that occurred in an Xcdc6-depleted extract plus control buffer) and defined 100% as the amount of replication that occurred in a mock-depleted extract in the presence of control buffer.
To characterize the role of the Xcdc6 domains in DNA replication, recombinant histidine-tagged (His6) fusion proteins containing full-length wild-type or deletion mutants of Xcdc6 were purified from baculovirus-infected Sf9 cells (Figure 1B). The ability of these Xcdc6 domains to restore DNA replication in Xcdc6-immunodepleted extracts was evaluated. In the DNA replication assays presented in this study, we have used sperm chromatin as a template and examined the replication that occurred during two sequential time windows (typically, 0–45 and 45–90 min) for each condition. Depending on the quality of the extract, the majority of replication is usually complete in the first time point but examining multiple time points yields useful kinetic information. In this assay, DNA replication was severely diminished in the Xcdc6-depleted extract relative to a mock-depleted extract assessed in parallel (Figure 1C, compare lanes 1 and 2 with 19 and 20), but was fully restored by addition of recombinant full-length His6-Xcdc6 protein (Figure 1, C and D, g and h). The C-terminal two-thirds of Xcdc6 was also able to restore DNA replication to ∼20% of wild-type Xcdc6 activity (Figure 1, C and D, d). In contrast, the N-terminal one-third of Xcdc6 failed to induce any replication, even when present at concentrations fourfold that of wild-type Xcdc6 (Figure 1, C and D, a–c). These findings indicate that the C-terminal domain is necessary and sufficient to act in this assay.
A somewhat larger Xenopus C-terminal domain (amino acids 126–554) has full activity (Pelizon et al., 2000), suggesting that critical elements lie between amino acids 125–165. Indeed, by analogy with the recently solved crystal structure of an archael Cdc6 homolog (Liu et al., 2000), it is likely that Xenopus Cdc6 residues 143–164 form an 32 α-helix that stabilizes the interface of the two domains that comprise the nucleotide binding site (Figure 1A). Addition of a mix of both the N- and C-terminal domains used in this study to an Xcdc6-depleted extract did not result in an increase of DNA replication relative to that observed with the C-terminal domain alone. Furthermore, neither of these Xcdc6 truncations interfered with the ability of an undepleted egg extract to replicate chromosomal DNA (our unpublished data). Together, these findings support the idea of a C-terminal catalytic domain, an N-terminal function that is regulatory but dispensable, and inability of either separate domain to compete effectively with the function of the wild-type protein.
Intact Walker A and Walker B Motifs Are Required for Xcdc6 Activity
Because the C-terminal two-thirds of Cdc6 is sufficient to replicate DNA, we next investigated the role of the highly conserved purine nucleotide binding site within this domain. Accordingly, we mutated the critical lysine residue within the Walker A site (K202E, hereafter referred to as Xcdc6WA) and one of the conserved acidic residues within the Walker B site (E277G, hereafter referred to as Xcdc6WB) in the context of full-length Xcdc6 (Figure 1A).
The ATPase activity of purified wild-type and mutant GST-Xcdc6 was assessed using [γ-32P]ATP as a substrate. Like its yeast (Zwerschke et al., 1994) and human (Herbig et al., 1999) homologs, wild-type GST-Xcdc6 consistently displayed weak but detectable ATPase activity (Figure 2A, squares). The hydrolysis rate for wild-type Xcdc6 (1 pmol of GST Xcdc6 is able to hydrolyze ∼3.2 pmol of ATP/min) is nearly identical to that reported for the human GST-Xcdc6 protein (Herbig et al., 1999). In contrast, GST-Xcdc6WA (Figure 2A, circles) and GST-Xcdc6WB (Figure 2A, triangles) had reduced ATPase activity. In multiple ATPase assays, we consistently observed slightly higher hydrolysis for GST-Xcdc6WA than for GST-Xcdc6WB. Because unfused GST protein purified in parallel had no ATPase activity (our unpublished data), we feel this residual activity of mutant Xcdc6 is likely due to the fact that replacement of a single residue may not completely abolish hydrolysis in this in vitro assay. Even with this caveat, it is clear that mutations in either the Walker A or B motif interfere with ATP hydrolysis.
We next tested whether the replication defect in Xcdc6-depleted extracts was complemented by the addition of recombinant mutant Xcdc6 (Figure 2, B and C). In addition, because Cdc6 is known to bind chromatin in an ORC-dependent manner and recruit MCMs to DNA, we also performed chromatin immunoblot analyses (Figure 2, D and E). As expected, in the absence of Xcdc6, little DNA replication occurs (Figure 2B, lanes 1 and 2) and no Xmcm3 is loaded onto DNA (Figure 2D, lanes 13 and 14). Like the wild-type His6-Xcdc6 (Figure 1), wild-type GST-Xcdc6 completely reconstituted DNA replication in an Xcdc6-depleted extract (Figure 2, B and C, a). As anticipated, recombinant wild-type GST-Xcdc6 binds chromatin very rapidly, stays on the chromatin throughout interphase, and loads Xmcm3 efficiently (Figure 2D, lanes 9–12) in a manner that is indistinguishable from endogenous Xcdc6 (our unpublished data). In contrast, GST-Xcdc6WA could not restore DNA replication to a depleted extract (Figure 2, B and C, b–d). Consistent with this mutant's lack of function in a DNA replication assay, GST-Xcdc6WA bound chromatin poorly and loaded no MCM on the DNA (Figure 2D, lanes 1–4). A longer exposure, however, detected GST-Xcdc6WA on chromatin (Figure 2E, compare lanes 1–4 with lanes 5 and 6). Importantly, this weak GST-Xcdc6WA signal is chromatin dependent (our unpublished data), raising the possibility of a low affinity or transient chromatin interaction. Finally, GST-Xcdc6WB exhibited ∼4% of the activity and replicated DNA at ∼8% of the rate (see MATERIALS AND METHODS) observed for wild-type GST-Xcdc6 (Figure 2, B and C, e–g). Strikingly, GST-Xcdc6WB binds chromatin well, yet loads Xmcm3 poorly (Figure 2D, lanes 5–8). This weak but detectable Xmcm3 loading in the presence of GST-Xcdc6WB correlates well with the modest DNA replication induced by high concentrations of this mutant and likely reflects an incomplete loss of ATP hydrolysis in this single amino acid substitution. Significantly, Xorc2 binds chromatin independent of wild-type or mutant Xcdc6 (Figure 2D) and provides a useful control. From these data, we conclude that an intact Walker A motif is necessary for efficient Xcdc6 chromatin association, whereas an intact Walker B motif is critical for MCM loading and/or Xcdc6 dissociation from chromatin.
Walker A and Walker B Mutant Xcdc6 Block Chromosomal DNA Replication In Vitro and In Vivo
The effect of mutations in Cdc6 Walker A and Walker B motifs has been studied in other systems but the findings have not been consistent. The activities of these mutant proteins range from dominant negative to completely nonfunctional to fully functional. For example, glutamic acid substitution of the Walker A critical lysine in budding yeast (K114E) results in a nonfunctional protein (Perkins and Diffley, 1998), whereas alanine substitution of the analogous lysine in human Cdc6 (K208A) results in a dominant negative protein (Herbig et al., 1999). In budding yeast, alanine substitution within the Walker B motif (DE223, 224AA) is fully functional (Weinreich et al., 1999), whereas a glutamic acid-to-glycine substitution (E224G) is dominant negative (Perkins and Diffley, 1998). It is of interest, therefore, to characterize Cdc6 Walker mutants in a system such as the Xenopus egg extract in which the amounts, combinations, and timing of mutant/wild type protein addition can be precisely controlled. DNA replication was not inhibited in extracts that contained up to fivefold molar excess (vs. endogenous Xcdc6) of either GST-Xcdc6WA or GST-Xcdc6WB (our unpublished data). We reasoned that the failure of these mutants in inhibiting DNA replication might be due to the much higher concentration of endogenous Xcdc6 protein present in the extracts compared with the levels present in yeast and human somatic cells.
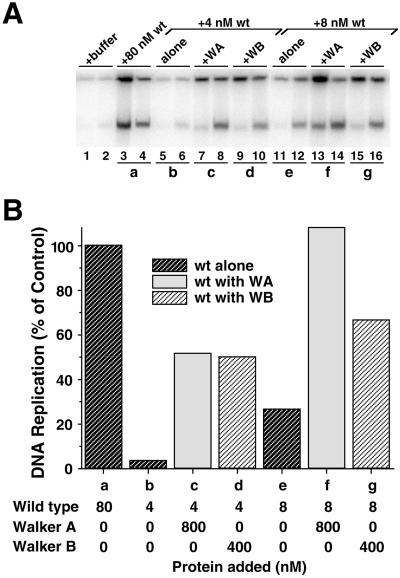
To test this hypothesis, we depleted an egg extract of endogenous Xcdc6 and added back low concentrations of wild-type GST-Xcdc6, either alone or in combination with mutant GST-Xcdc6. Although the absolute value of DNA replication varied between experiments, we found that reconstituting an Xcdc6-depleted extract with either 4 or 8 nM wild-type GST-Xcdc6 (Figure 3, A and B, b and e, respectively) reproducibly gave intermediate values of DNA replication relative to that which occurred in the presence of physiological concentrations (80 nM) of wild-type GST-Xcdc6 (Figure 3, A and B, h). This variability in absolute amount of DNA replication between experiments is likely due to a number of factors, including quality of extract, extent of Xcdc6 immunodepletion, and activity of recombinant protein. Importantly, within any given experiment, the amount of replication was reproducible and between experiments, the trends were fully consistent. Preincubation of GST-Xcdc6WA in an Xcdc6-depleted egg extract containing chromatin blocked the ability of subsequently added wild-type GST-Xcdc6 to replicate chromatin. Similar results were obtained at both 4 nM (Figure 3, A and B, compare b and d) and 8 nM (Figure 3, A and B, compare e and g) wild-type GST-Xcdc6. At >40 nM wild-type Xcdc6, however, the inhibitory activity was not apparent (Figure 3, A and B, compare h and i), arguing that high concentrations of wild-type protein can overwhelm the dominant negative phenotype. A similar dominant negative effect was observed for GST-Xcdc6WB at 8 nM (Figure 3, C and D, compare e and g) wild-type GST-Xcdc6. We did not always observe this GST-Xcdc6WB dominant negative effect at 4 nM wild-type protein (Figure 3, C and D, compare b and d), probably due to the fact that GST-Xcdc6WB alone imparts limited DNA replication (Figures 2 and 3, C and D, a). Together, these data argue that both Walker A and Walker B mutant proteins can manifest a dominant negative phenotype, provided that they are added to the extract before prereplication complex formation and the concentration of wild-type Xcdc6 is <40 mM (our unpublished data). Moreover, these dominant negative phenotypes strongly argue that the recombinant mutant proteins have retained some functional properties of wild-type Xcdc6.
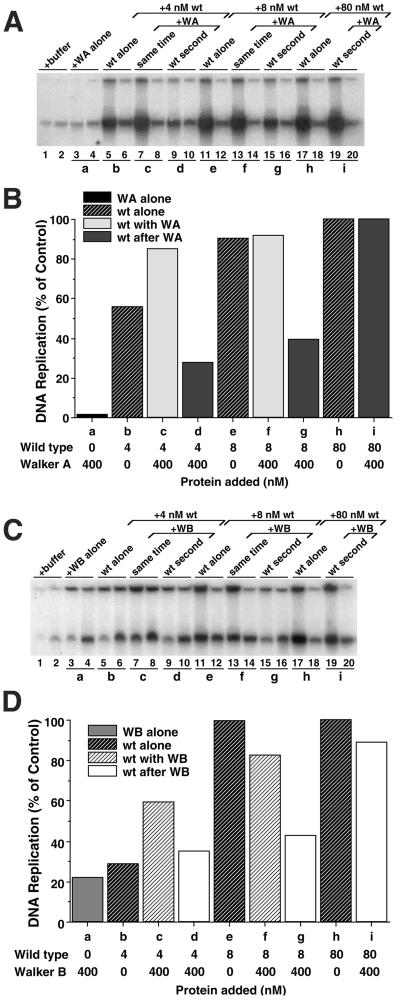
Figure 3.
Walker A and Walker B Xcdc6 mutants are dominant negative in Xenopus egg extracts when they are added to chromatin before addition of wild-type Xcdc6 but stimulate wild-type Xcdc6 if added at the same time. (A) Replication of sperm chromatin was determined in an Xcdc6-depleted interphase egg extract in the presence of buffer (lanes 1 and 2), GST-Xcdc6WA alone (400 nM, lanes 3 and 4), wild-type GST-Xcdc6 alone (4 nM, lanes 5 and 6; 8 nM, lanes 11 and 12; 80 nM, lanes 17 and 18), or 400 nM GST-Xcdc6WA plus various concentrations of wild-type GST-Xcdc6 (4 nM, lanes 7–10; 8 nM, lanes 13–16; 80 nM, lanes 19–20). To assess the importance of the order in which mutant and/or wild-type Xcdc6 bound to chromatin, we either mixed GST-Xcdc6WA with wild-type GST-Xcdc6 before adding it to an Xcdc6-depleted extract containing chromatin (lanes 7 and 8 and 13 and 14) or preincubated GST-Xcdc6WA in an Xcdc6-depleted extract containing chromatin for 5 min at 23°C before adding wild-type GST-Xcdc6 (lanes 9 and 10, 15 and 16, and 19 and 20). Each lane depicts the 32P incorporation into chromosomal DNA that occurred during the following times: 1–45 min (odd-numbered lanes) and 45–90 min (even-numbered lanes). (B) Quantitation of the total chromosomal DNA replication for the indicated samples shown in A over a 90-min period after addition of GST-Xcdc6WA alone (WA alone; a), wild-type GST-Xcdc6 alone (wt alone; b, e, and h), GST-Xcdc6WA with wild-type added together (wt with WA; c and f), or wild-type added 5 min after GST-Xcdc6WA (wt after WA; d, g, and i) at the indicated protein concentrations. In this analysis, we have subtracted the background (replication that occurred in an Xcdc6-depleted extract plus control buffer) and defined 100% as the amount of replication that occurred in an Xcdc6-depleted extract reconstituted with 80 nM wild-type GST-Xcdc6. (C and D) Identical experiment as depicted in A and B, only using 400 nM GST-Xcdc6WB instead of GST-Xcdc6WA, as indicated.
As outlined in INTRODUCTION, it is possible that mutations in the Xcdc6 Walker A and Walker B motifs may result in different phenotypes, depending on whether the protein is present in an embryonic or somatic cell background. To pursue this issue, we sought to characterize the Xenopus Cdc6 mutants in an established somatic system by using in vivo microinjection. Accordingly, we microinjected purified wild type and the two mutant forms of Xcdc6 into HeLa cells and monitored DNA replication. We used HeLa cells for these experiments for two reasons. First, Xenopus tissue culture cells grow very slowly, making synchronization problematic. Second, the Xenopus and human Cdc6 proteins share nearly 80% identity at the amino acid level, and in vitro studies indicate that Xcdc6 can function in human nuclear extracts (Stoeber et al., 1998).
HeLa cells were microinjected with proteins during early-to-mid-G1, before prereplication complex formation and, therefore, before Cdc6 activity. We tried a variety of labeling times with BrdU, from brief pulses at various times after injection to continuous labeling (6–17 h). In all cases, GST-Xcdc6WA and GST-Xcdc6WB inhibited BrdU incorporation, whereas GST alone or wild-type Xcdc6 had no effect (Figure 4). We determined that 60–70% of cells microinjected with either control protein (GST or wild-type GST-Xcdc6; Figure 4A, first and second row, respectively) exhibited intense BrdU staining (BrdU +/+; Figure 4B), whereas only ∼40% of the cells microinjected with either GST-Xcdc6WA or GST-Xcdc6WB (Figure 4A, row 3 and 4, respectively) exhibited intense BrdU staining (Figure 4B). In some cases, the cells had detectable BrdU incorporation but the level of incorporation was lower (Figure 4A, row 3, asterisk). These intermediately stained (BrdU +/−) nuclei may result from either slowed progression through S phase or a delayed entry into S phase and premature arrest of progression upon fixation. Consistent with the hypothesis that GST-Xcdc6WA or GST-Xcdc6WB delay S phase, the proportion of BrdU +/− nuclei was reproducibly higher in the G1 phase cells microinjected with either mutant protein relative to those microinjected with either wild-type GST-Xcdc6 or GST alone (Figure 4B, white stacked histograms).
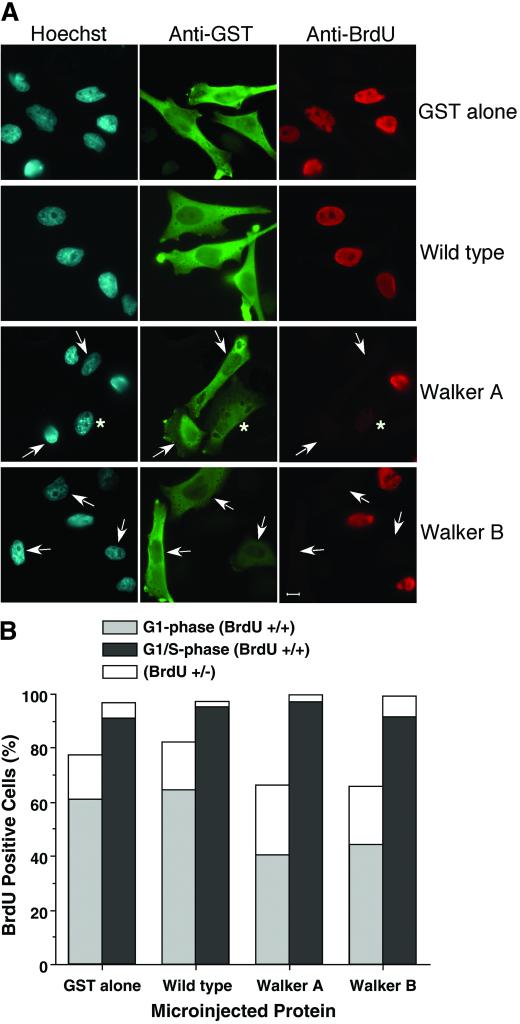
Figure 4.
Walker A and Walker B Xcdc6 mutants are dominant negative when microinjected into G1 phase but not G1/S phase human somatic cells. (A) Immunofluorescence analysis of HeLa cells that were microinjected in early-to-mid-G1 and stained for total DNA (Hoechst, left), microinjection status (anti-GST, middle), or entry into S phase (anti-BrdU, right). Control experiments microinjecting GST alone (row 1) or with wild-type GST-Xcdc6 (row 2) show no effect on S phase entry. In contrast, microinjecting GST with either GST-Xcdc6WA (row 3) or GST-Xcdc6WB (row 4) results in delayed entry into S phase as indicated by absence of BrdU staining (arrowheads). Although we performed nuclear injections, GST staining was found throughout the cell. This cytoplasmic staining is likely due to the high concentration of GST (4 mg/ml) microinjected coupled with the fact that GST is sufficiently small to traverse the nuclear pore, and the microinjector uses a continuous flow, resulting in some protein necessarily entering the cytoplasm. (B) GST was coinjected with buffer, wild-type GST-Xcdc6, GST-Xcdc6WA, or GST-Xcdc6WB into either early to mid-G1 cells (light-shaded bars) or G1/S cells (dark-shaded bars). After injection, cells were grown in medium containing BrdU for 9 h and processed for indirect immunofluorescence by using anti-GST and anti-BrdU antibodies. Microinjected cells (GST positive) were evaluated for entry into S phase (BrdU positive) by scoring as either bright staining (+/+), intermediate staining (+/−), or nonstaining (−/−). Significantly, the proportion of nuclei displaying intermediate BrdU staining (e.g., asterisk, A; white histograms, B) was reproducibly higher in G1 phase cells microinjected with either GST-Xcdc6WA or GST-Xcdc6WB. Each bar represents the average of three experiments where the number of microinjected cells was ∼250.
To investigate the difference in response of the cells to the microinjection, we performed correlation analysis of the outcomes of G1-phase experiments. We compared the percentage of BrdU positive, intermediate, and negative nuclei in injected and uninjected cells in the three experiments. This analysis revealed strong correlation (R2 > 0.9) between the GST alone and wild-type GST-Xcdc6, indicating no statistical difference in these results. In contrast, the outcomes for either of the mutant GST-Xcdc6 proteins were not correlated (R2 < 0.5), suggesting that there were significant changes in the cells induced by injection. These results support the hypothesis that the Xenopus Walker A and Walker B mutant Xcdc6 proteins are dominant negative in human cells. These results are similar to those of Fanning and coworkers who microinjected HsCdc6 Walker A and B mutants into HeLa cells (Herbig et al., 1999). The primary difference between our studies is that we used a heterologous system, Xenopus protein into human cells, whereas they used an entirely homologous system. It is likely that this heterologous system accounts for the reduced magnitude in our dominant negative effect. That is, Xcdc6 may not interact efficiently with human DNA replication machinery. Alternatively, Xcdc6 may not function optimally at 37°C, as is known to be the case for the Xenopus wee1 kinase (Kumagai and Dunphy, 1995).
An important negative control was to microinject into cells synchronized at the boundary when they will have already formed the prereplication complex and thus be refractory to the presence of mutant Cdc6. Accordingly, HeLa cells were synchronized in G1/S phase, microinjected, labeled with BrdU for 9 h, and processed for immunofluorescence. As predicted, microinjection of any of the four proteins, including the GST-Xcdc6WA and GST-Xcdc6WB, had no effect on DNA replication (Figure 4B, right histograms). Together, these data indicate that the mutant Xcdc6 proteins inhibit DNA replication in both embryonic and somatic cells, provided they are introduced before prereplication complex formation.
Walker A and Walker B Mutant Xcdc6 Act Synergistically with Wild-Type Protein If Added Together
In the process of demonstrating the dominant negative effect of Walker A and Walker B mutants in Xenopus egg extracts, we made the surprising observation that mutant Xcdc6 can stimulate the activity of low concentrations of wild-type Xcdc6 when the proteins are added simultaneously. For example, when 400 nM GST-Xcdc6WA was added to extracts with 4 nM wild-type GST-Xcdc6, DNA replication levels increased relative to those with wild-type Xcdc6 alone (Figure 3, A and B, compare b with c). Given that 400 nM GST-Xcdc6WA alone accounts for essentially no DNA replication (Figure 3, A and B, a), this effect cannot be merely additive. Subsequent experiments established that this stimulatory effect of GST-Xcdc6WA required >20 nM protein and saturated at ∼100 nM GST-Xcdc6WA (our unpublished data). Qualitatively similar stimulation of low concentrations of wild-type Xcdc6 to replicate DNA was observed with GST-Xcdc6WB (Figure 3, C and D, compare b with c), with the caveat that GST-Xcdc6WB alone imparts limited DNA replication to an Xcdc6-depleted extract (Figure 3, C and D, a).
We performed additional experiments to compare the ability of GST-Xcdc6WA and GST-Xcdc6WB to stimulate the activity of low concentrations of wild-type Xcdc6. Endogenous Xcdc6 was immunodepleted from egg extracts, resulting in the virtual absence of DNA replication in the presence of buffer alone (Figure 5A, lanes 1 and 2). As a positive control, the extract was reconstituted with physiological concentrations of wild-type GST-Xcdc6 (Figure 5A, lanes 3 and 4), producing a level of replication defined as 100% (Figure 5B, a). As demonstrated in Figure 3, reconstituting an Xcdc6-depleted extract with either 4 or 8 nM wild-type GST-Xcdc6 yielded intermediate DNA replication (Figure 5, A and B, b and e, respectively). When added together with either GST-Xcdc6WA or GST-Xcdc6WB, however, DNA replication was dramatically increased (2.5–14-fold) (Figure 5, A and B, compare b–d; e–g). These results indicate that mutant forms of Xcdc6, which are severely impaired at replicating DNA by themselves, can nonetheless stimulate the ability of wild-type Xcdc6 to replicate DNA.
Figure 5.
Walker A and Walker B Xcdc6 mutants stimulate the activity of low concentrations of wild-type Xcdc6 in Xenopus egg extracts. (A) Replication of sperm chromatin was determined in an Xcdc6-depleted interphase egg extract in the presence of buffer (lanes 1 and 2), wild-type GST-Xcdc6 (80 nM, lanes 3 and 4; 4 nM, lanes 5–10; 8 nM, lanes 11–16), in the presence of 800 nM GST-Xcdc6WA (lanes 7 and 8; lanes 13 and 14), or 400 nM GST-Xcdc6WB (lanes 9 and 10; lanes 15 and 16). Each lane depicts the 32P incorporation into chromosomal DNA that occurred during the following times: 1–60 min (odd-numbered lanes) and 60–120 min (even-numbered lanes). (B) Quantitation of the total chromosomal DNA replication for the indicated samples shown in A over a 120-min period after addition of wild-type GST-Xcdc6 (wt alone; a, b, and e), wild-type with GST-Xcdc6WA (wt with WA; c and f), or wild-type with GST-Xcdc6WB (wt with WB; d and g) at the indicated protein concentrations. Background was subtracted and histograms were normalized as described in Figure 3.
Xcdc6 Forms Multimers
Two models would be consistent with the mutant's ability to stimulate wild-type Xcdc6. An excess of mutant Xcdc6 might act as an antagonist to inactivate a hypothetical negative regulator of DNA synthesis. In this scenario, low concentrations of wild-type Xcdc6 would be sequestered in an inhibited complex. In the presence of excess mutant Xcdc6, however, the wild-type Xcdc6 would be released from inhibition. We do not favor this model because the Xenopus early embryonic cell cycles appear to be free of such negative regulation and, unless this negative regulator has a low affinity for Xcdc6 in solution, it would likely coimmunoprecipitate and be removed with the endogenous Xcdc6 during immunodepletion. Moreover, this model is difficult to reconcile with the observed dominant negative phenotype when mutant Xcdc6 is preincubated with chromatin before addition of wild-type protein (see DISCUSSION). A second model invokes Cdc6 acting as an oligomer comprising at least two activities, only one of which is ATP dependent. In this model, low concentrations of wild-type Xcdc6 might be unable to form a sufficient level of oligomer required for efficient DNA replication. In the presence of excess mutant Xcdc6, a functional heteroligomer could form. In this scenario, the mutant subunit(s) might contribute in a structural way (e.g., ORC/MCM binding), whereas the wild-type subunit(s) would catalyze an ATP-driven process (e.g., complex disassembly or MCM loading).
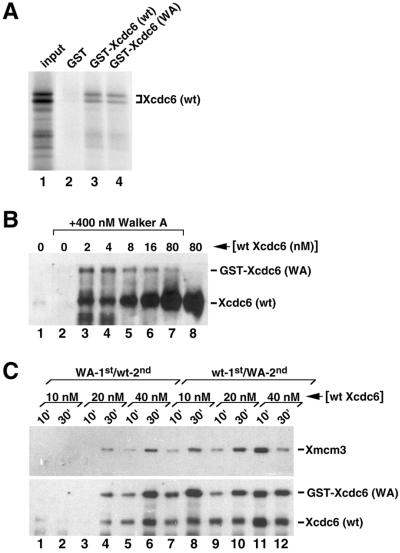
To test this model, we investigated whether purified mutant and wild-type Xcdc6 might form a complex. Significantly, human Cdc6 has been shown to form complexes in vitro (Saha et al., 1998; Herbig et al., 1999) and we used a similar approach to demonstrate Xcdc6 interactions. Briefly, we incubated 35S-radiolabeled untagged wild-type Xcdc6 with glutathione-agarose beads bound to either control GST alone, wild-type GST-Xcdc6, or Walker A GST-Xcdc6 in the presence of 2% nonfat milk. In this assay, the beads were washed extensively and probed for the presence of 35S-radiolabeled untagged Xcdc6 by fluorography (Figure 6A). Radiolabeled Xcdc6 bound both wild-type GST-Xcdc6 and GST-Xcdc6WA (Figure 6A, lanes 3 and 4, respectively), but not GST alone (Figure 6A, lane 2). Significantly, only trace amounts of 35S-radiolabeled control luciferase protein bound to fused or unfused GST (our unpublished data). Together, these results suggest that Xcdc6 monomers can form a complex.
Figure 6.
Wild-type Xcdc6 forms oligomers and recruits Walker A mutant Xcdc6 to chromatin. (A) Glutathione-agarose beads containing equal amounts of GST alone (lane 2), wild-type GST-Xcdc6 (lane 3), or GST-Xcdc6WA (lane 4) were incubated with 35S-radiolabeled untagged wild-type Xcdc6 in the presence of 2% nonfat dry milk. After washing, the beads were prepared for SDS-PAGE and fluorography. Lane 1 contains approximately one-fifth of the labeled input protein as a size marker. (B) Immunoblots were performed on detergent-insoluble fractions isolated from Xcdc6-depleted (lanes 1–6) or undepleted (lanes 7 and 8) interphase extracts incubated in the presence of buffer (lanes 1 and 8) or 400 nM Walker A GST-Xcdc6 (lanes 2–7). GST-Xcdc6WA containing extracts were supplemented with undepleted extract to achieve the indicated concentrations of endogenous wild-type Xcdc6 (lanes 3–6). Samples were incubated for 30 min at 23°C to allow Xcdc6 binding and prepared for immunoblot analysis by using anti-Xcdc6 antibodies. The relative mobilities of the GST-Xcdc6WA and endogenous Xcdc6 are indicated. (C) Immunoblots were preformed on detergent-insoluble fractions isolated from interphase extracts that contain 400 nM GST-Xcdc6WA along with various concentrations of endogenous Xcdc6 (10 nM: lanes 1 and 2, 7 and 8; 20 nM: lanes 3 and 4, 9 and 10; 40 nM: lanes 5 and 6, 11 and 12). In half of the samples, the GST-Xcdc6WA was preincubated in an Xcdc6-depleted interphase extract containing chromatin in advance of adding wild-type Xcdc6 (WA-1st/wt-2nd, lanes 1–6). In parallel samples, the undepleted extract was diluted with Xcdc6 depleted extract to achieve the indicated concentrations of endogenous Xcdc6 and preincubated with chromatin in advance of adding 400 nM GST-Xcdc6WA (wt-1st/WA-2nd, lanes 7–12). All samples were further incubated at 23°C for the indicated times and prepared for immunoblot analyses by using anti-Xmcm3 (top) or anti-Xcdc6 (bottom) antibodies. The relative mobilities of the Xmcm3, GST-Xcdc6WA, and endogenous Xcdc6 are indicated.
Wild-Type Xcdc6 Facilitates Walker A Mutant Xcdc6 Chromatin Binding
Because GST-Xcdc6WA binds chromatin poorly (Figure 2, D and E), we investigated whether conditions that favor synergy (presence of low concentrations of wild-type Xcdc6) might promote Walker A chromatin binding. That is, if Walker A mutant Xcdc6 stimulates low concentrations of wild-type Xcdc6 through formation of functional heteroligomers then we would anticipate increased chromatin association of GST-Xcdc6WA under these conditions. Accordingly, we supplemented an Xcdc6-depleted extract with 400 nM GST-Xcdc6WA and an undepleted extract (known to contain 80 nM Xcdc6; Coleman et al., 1996) to achieve low concentrations (2–16 nM) of endogenous wild-type Xcdc6, thereby reproducing conditions known to promote synergy between mutant and wild-type Xcdc6 (Figures 3 and 5). We then incubated these chromatin-containing extracts at 23°C for 30 min, a time during which we observe maximal Xcdc6 chromatin binding. Finally, chromatin fractions were isolated and probed for the presence of Xcdc6. When incubated in the absence of wild-type Xcdc6, GST-Xcdc6WA fails to bind chromatin at appreciable levels (Figure 6B, lane 2). Strikingly, the addition of low concentrations of endogenous wild-type Xcdc6 facilitates GST-Xcdc6WA chromatin binding (Figure 6B, lanes 3–6). At higher concentrations of wild-type Xcdc6, however, the GST-Xcdc6WA binding to chromatin is greatly reduced (Figure 6B, lane 7). Moreover, in similar experiments, we also observe increased wild-type Xcdc6 chromatin binding in the presence of Walker A mutant Xcdc6, relative to that observed with wild-type Xcdc6 alone (our unpublished data). Under these conditions, therefore, both Walker A mutant and additional wild-type Xcdc6 are recruited to chromatin. Together, this enhanced wild-type and mutant Xcdc6 chromatin binding provides a molecular basis for the observed stimulation of DNA replication.
To establish further the mechanism by which the mutants collectively synergize with or inhibit the activity of wild-type Xcdc6, we performed additional GST-Xcdc6WA and wild-type Xcdc6 mixing experiments and assessed prereplication complex formation by probing chromatin fractions for the presence of both Xcdc6 and Xmcm3. In one experiment, we preincubated chromatin with GST-Xcdc6WA before adding wild-type Xcdc6 (Figure 6C, lanes 1–6). In parallel, a complementary experiment was performed in which wild-type Xcdc6 was preincubated before GST-Xcdc6WA was added (Figure 6C, lanes 7–12). All samples were subsequently incubated and processed for immunoblot analyses at the indicated times. Because preincubating GST-Xcdc6WA with chromatin inhibits the action of low concentrations of wild-type Xcdc6 (Figure 3B), we reasoned that we should observe less wild-type Xcdc6 and less Xmcm3 bound to chromatin under these conditions. Strikingly, significantly less Xcdc6 (wild type and mutant) and Xmcm3 are bound to chromatin when GST-Xcdc6WA is preincubated with chromatin compared with the amounts bound when wild-type Xcdc6 is preincubated with chromatin (Figure 6C, compare lanes 1–6 with lanes 7–12). These experiments provide support for a mechanism whereby the mutant Xcdc6 imparts its dominant negative effect by establishing a nonproductive prereplication complex. Moreover, in view of the increased GST-Xcdc6WA chromatin binding, preincubation of wild-type Xcdc6 with chromatin may facilitate heteroligomer formation. Together, our data are consistent with Cdc6 functioning as an oligomer with multiple functional domains. One activity is ATP dependent; a separate activity may be structural in nature and can be fulfilled by a catalytically dead subunit(s).
Walker A Mutant Xcdc6 Stimulates Activity of Walker B Mutant Xcdc6
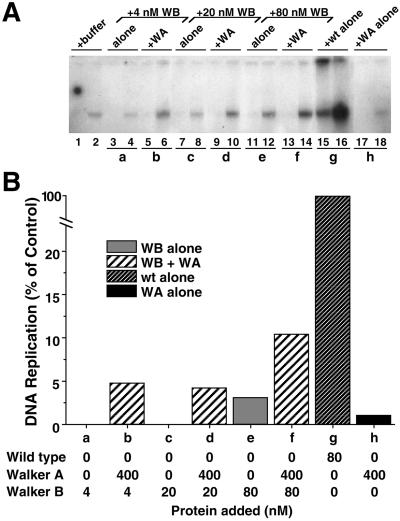
Collectively, the above-mentioned experiments indicate that GST-Xcdc6WA binds chromatin poorly and cannot restore DNA replication to a depleted extract yet can stimulate the activity of wild-type GST-Xcdc6, possibly through heterologimer formation on chromatin. To examine further the stimulatory activity of GST-Xcdc6WA, we investigated whether GST-Xcdc6WA might stimulate the activity of GST-Xcdc6WB. As expected, after immunodepletion of Xcdc6, little DNA replication occurs (Figure 7A, lanes 1 and 2), whereas addition of wild-type GST-Xcdc6 fully reconstituted DNA replication (Figure 7, A and B, g). As shown previously (Figure 2), GST-Xcdc6WB alone restored very limited DNA replication (Figure 7, A and B, a, c, and e). Strikingly, the addition of 400 nM GST-Xcdc6WA, which is essentially devoid of activity in this assay (Figure 7, A and B, h), with GST-Xcdc6WB resulted in increased DNA replication relative to GST-Xcdc6WB alone (Figure 7, A and B; compare a and b, c and d, and e and f). From these data, we conclude that catalytically inactive GST-Xcdc6WA can stimulate catalytically impaired GST-Xcdc6WB. The mechanism underling this GST-Xcdc6WB stimulation is likely to be the same as that for the stimulation of low concentrations of wild-type Cdc6.
Figure 7.
Walker A mutant Xcdc6 stimulates the activity of Walker B mutant Xcdc6. Replication of sperm chromatin was determined in an Xcdc6-depleted interphase egg extract in the presence of buffer (lanes 1 and 2), GST-Xcdc6WB alone (4 nM, lanes 3 and 4; 20 nM, lanes 7 and 8; 80 nM, lanes 11 and 12), 400 nM GST-Xcdc6WA with GST-Xcdc6WB (4 nM, lanes 5 and 6; 20 nM, lanes 9 and 10; 80 nM, lanes 13 and 14), wild-type GST-Xcdc6 alone (80 nM, lanes 15 and 16), or GST-Xcdc6WA alone (400 nM, lanes 17 and 18). Each lane depicts the 32P incorporation into chromosomal DNA that occurred during the following times: 1–45 min (odd-numbered lanes) and 45–90 min (even-numbered lanes). (B) Quantitation of the total chromosomal DNA replication for the indicated samples shown in A over a 90-min period after addition of GST-Xcdc6WB alone (WB alone; a, c, and e), GST-Xcdc6WB with GST-Xcdc6WA (WB + WA; b, d, and f), wild-type GST-Xcdc6 alone (wt alone; g), or GST-Xcdc6WA alone (WA alone; h) at the indicated protein concentrations. Background was subtracted and histograms are normalized as described in Figure 3.
DISCUSSION
The data presented herein suggest that Xcdc6 possesses two distinct activities and may act as an oligomer. One essential activity appears to be ATP dependent and involves Cdc6 acting as a switch that couples key events during the initiation of DNA replication. A point mutation in the Walker A motif, which likely inhibits ATP binding, results in decreased chromatin association and inability to restore DNA replication to an Xcdc6-depleted extract. In contrast, a point mutation in the Walker B motif, which inhibits ATP hydrolysis, binds chromatin well but loads MCMs poorly and is nearly nonfunctional. We demonstrate that both GST-Xcdc6WA and GST-Xcdc6WB can function as dominant negative mutants when either microinjected into HeLa cell nuclei or incubated in Xenopus egg extracts, provided they are added before prereplication complex formation. These results argue that the Xenopus Cdc6 protein can interact with human DNA replication machinery, underscoring the conserved nature of eukaryotic DNA replication machinery. A second, non-ATP–dependent activity of Xcdc6 was also uncovered. We found that GST-Xcdc6WA, which is nonfunctional by itself, can stimulate the activity of both wild-type and catalytically impaired GST-Xcdc6WB. We present evidence that Xcdc6 can form multimers and speculate that this stimulatory activity is imparted via heteroligomer formation on chromatin.
Cdc6 May Function As a Clamp Loader
Clamp loaders are members of the AAA+ family of ATPases that function as multimeric ATP-dependent machines to assemble their respective clamps around DNA (Mossi and Hubscher, 1998; Neuwald et al., 1999; Lee and Bell, 2000). Despite having limited sequence similarity, the three-dimensional structures of clamp loaders are remarkably similar among archaeal, prokaryotic, and eukaryotic organisms (Mossi and Hubscher, 1998; Hingorani and O'Donnell, 2000). Extrapolating from several well-studied examples, it is likely that all clamp loaders use ATP binding and hydrolysis as molecular switches to couple key events in DNA metabolism (Lee and Bell, 2000). ATP binding appears to promote stable clamp loader DNA interactions. The exact requirement for ATP hydrolysis, however, remains unclear. Although hydrolysis is not required for subsequent clamp binding, the DNA clamp complexes formed in the absence of hydrolysis are unstable (Mossi and Hubscher, 1998). The energy from ATP hydrolysis, therefore, may be used to close the clamp around DNA or dissociate the clamp loader from the clamp and DNA (Hingorani et al., 1999).
In our in vitro hydrolysis assays, GST-Xcdc6WA reproducibly had higher activity than GST-Xcdc6WB; yet, in the DNA replication assays, GST-Xcdc6WB was more active. Thus, it seems unlikely that our mutants' intrinsic level of ATPase activity accounts for their different biochemical phenotypes. Indeed, given the low ATPase activity of wild-type Xenopus (this study) and human (Herbig et al., 1999) Cdc6, one wonders about the physiological relevance of this in vitro hydrolysis assay. In vivo, Cdc6 likely functions in a large macromolecular machine that hydrolyzes ATP much faster than this in vitro assay would indicate.
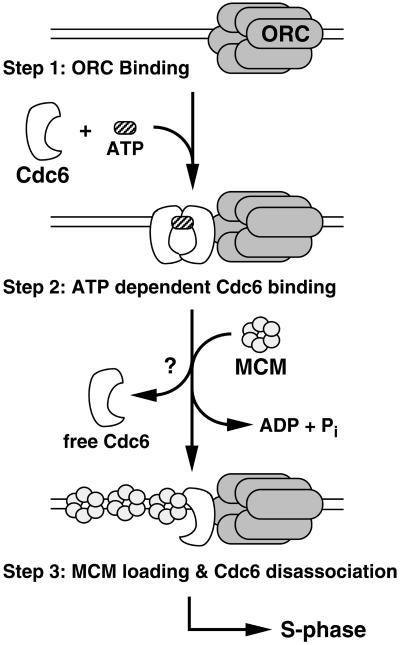
The results presented herein, coupled with previous work in yeast (Perkins and Diffley, 1998; DeRyckere et al., 1999; Wang et al., 1999; Weinreich et al., 1999) and human cells (Herbig et al., 1999), suggest that Cdc6 and Cdt1 (Maiorano et al., 2000; Nishitani et al., 2000) collaborate with the ORC to load the MCM complex onto chromatin. In view of the sequence similarity and biochemical data, it is tempting to speculate that Cdc6 may function as a clamp loader in a multistep process resulting in MCM (a potential ring-shaped clamp) recruitment around DNA. In the first step, ORC binds to DNA independent of Cdc6 (Figure 8, step 1). In a subsequent step, Cdc6 forms a productive interaction with ORC and DNA in a process that likely requires ATP binding (Figure 8, step 2). On the basis of studies with AAA+ family members, GST-Xcdc6WA is expected to have reduced ATP binding (and, therefore, hydrolysis), whereas GST-Xcdc6WB would maintain ATP binding but have reduced hydrolysis (Story and Steitz, 1992). Because GST-Xcdc6WA is severely impaired with respect to chromatin binding and ability to load Xmcm3 (Figure 2D, lanes 1–4), the data presented herein correlate well with the clamp loader model (Figure 8, arrest at step 1 in the absence of ATP binding). Next, Cdc6-mediated ATP hydrolysis may either drive MCM loading and/or disassociation of Cdc6 from the DNA (Figure 8, step 3). Our chromatin immunoblots (Figure 2D, lanes 5–8) are also consistent with this step, because we observe increased GST-Xcdc6WB chromatin binding and reduced Xmcm3 loading in the presence of this mutant (Figure 8, arrest at step 2 in the absence of ATP hydrolysis). Finally, the inability of either GST-Xcdc6WA or GST-Xcdc6WB to restore DNA replication to an Xcdc6-depleted egg extract (Figure 2, B and C) is also consistent with this model, because neither mutant would be expected to promote DNA replication. Although it remains unclear whether the MCM complex is opened by Cdc6 and then topologically linked to DNA, our results are consistent with this clamp loader model. That is, like known clamp loaders, Cdc6 appears to act as a molecular switch in which ATP binding and hydrolysis regulate separate functions that serve in combination to load a complex onto DNA.
Figure 8.
Model summarizing how ATP binding and hydrolysis may regulate Cdc6 activity. At least three steps are involved in the Cdc6-dependent loading of MCM complex. First, the ORC complex must assemble on the DNA (top). Second, Cdc6 associates with ATP, likely eliciting a conformational change to promote Xcdc6 chromatin binding (middle). Finally, the MCM complex is recruited and Cdc6 dissociates in a process that requires ATP hydrolysis. Our data are consistent with Cdc6 acting as a multimer (represented by a dimer) that can be composed of some subunits that do not bind/hydrolyze ATP. Neither the precise oligomeric state nor the stoichiometry of ATP hydrolysis is known. For clarity, Cdt1 has been omitted from this model.
Xcdc6 Walker A and Walker B Mutants Are Dominant Negative in Embryonic and Somatic Cell Cycles
Two models would be consistent with the mutants' ability to inhibit wild-type Xenopus (Figure 3) or human (Figure 4) Cdc6. For example, an excess of the mutant protein may interact with a component of the replication complex and sequester it away from the origin of replication, resulting in a dominant negative phenotype. However, we can detect no significant interaction between Xcdc6 and members of either ORC or MCM complexes in solution (our unpublished data). We present evidence that GST-Xcdc6WA can interact with wild-type Xcdc6 (Figure 6) but do not feel this interaction inhibits wild-type Xcdc6 activity (see below). More importantly, this model is difficult to reconcile with the observation that adding an excess of the mutant Xcdc6 simultaneously with wild-type Xcdc6 results in stimulation rather than inhibition of DNA replication. That is, we observe the dominant negative phenotype only when we preincubate mutant Xcdc6 with chromatin then add wild-type Xcdc6 but not when equivalent concentrations of mutant and wild-type Xcdc6 are added simultaneously (Figure 3). A second model postulates that mutant Xcdc6 forms an unproductive prereplication complex (Figure 8, arrest at step 2). In this scenario, a nonfunctional prereplication complex may preclude subsequent wild-type Xcdc6 chromatin interactions and would explain the observed requirement of mutant protein addition before prereplication complex formation. This model is especially compelling for GST-Xcdc6WB, which binds chromatin well but loads MCM poorly (corresponding to Figure 8, arrest at step 2), but harder to reconcile with the reduced GST-Xcdc6WA chromatin binding. As noted, however, long exposures of chromatin immunoblots do detect significant GST-Xcdc6WA chromatin interaction (Figure 2E). Importantly, this GST-Xcdc6WA signal is dependent on the presence of both chromatin and recombinant GST-Xcdc6WA (Figure 2E). It is very possible, therefore, that Cdc6 can bind chromatin in the absence of ATP but that ATP association stabilizes this interaction. A transient or low-affinity GST-Xcdc6WA chromatin interaction may be disrupted during chromatin isolation but sufficiently strong to interfere with wild-type Xcdc6 chromatin binding. Thus, we suggest that both Walker A and Walker B mutant Xcdc6 form unproductive prereplication complexes, thereby inhibiting the activity of subsequently added wild-type Xcdc6.
Does Cdc6 Function As an Oligomer?
In marked contrast to the inhibitory activity when added before wild-type Xcdc6, both GST-Xcdc6WA and GST-Xcdc6WB stimulate DNA replication when added to chromatin simultaneously with low concentrations of wild-type Xcdc6 (Figures 3 and 5). Our data are consistent with Cdc6 functioning as an oligomer, wherein low concentrations of wild-type Xcdc6 that are insufficient to replicate DNA efficiently can form functional heteroligomers when added to chromatin in the presence of excess mutant Xcdc6. Although it is formally possible that wild-type Xcdc6 may mediate the chromatin association of GST-Xcdc6WA in a mechanism that does not involve heteroligomer formation, we favor this oligomer model for several reasons. First, Xenopus (Figure 6A) and human (Saha et al., 1998; Herbig et al., 1999) Cdc6 form complexes in solution. Second, not only do low concentrations of wild-type Xcdc6 promote GST-Xcdc6WA chromatin binding (Figure 6B) but also high concentrations of GST-Xcdc6WA facilitate additional wild-type Xcdc6 chromatin association (our unpublished data). This mutual stimulation is suggestive of heteroligomer formation and is not an artifact of GST-mediated dimerization because we observe augmented GST-Xcdc6WA binding by using untagged wild-type Xcdc6. Third, the formation of a functional Xcdc6 heteroligomer provides a mechanism to explain the observed stimulation of DNA replication under these conditions. Our bias is that oligomerization is greatly stabilized in the presence of DNA and other DNA replication components. Indeed, we present evidence that such a complex likely forms on chromatin (represented by the dimer in Figure 8, step 2), wherein the mutant subunit(s) provide some activity (e.g., ORC/MCM binding) and the wild-type subunit(s) catalyze an essential ATP driven process (e.g., MCM loading and/or complex disassembly).
This oligomerization model is strengthened by the observation that GST-Xcdc6WA can stimulate not only wild-type Xcdc6 but also GST-Xcdc6WB (Figure 7). In view of the recently described crystal structures of several ATP-binding cassette family members, which reveal the composite nature of ATPase active sites, the current data provide grounds for the speculation that Cdc6 may also form a dimer with a composite ATPase domain. In one example illustrating the composite nature of an ATPase active site, the two ATPase domains of the MutS mismatch repair form an interleaved ATPase interface, in which a conserved flexible loop from one monomer forms part of the opposing monomer's catalytic site (Lamers et al., 2000; Obmolova et al., 2000; Junop et al., 2001). A composite ATPase active site may be a general feature of the ATP binding cassette superfamily because a similar arrangement of two complementary ATPase domains is also observed in the DNA repair protein Rad50 (Hopfner et al., 2000). The data present herein are consistent with Cdc6 forming a dimer with a similar composite ATPase active site (Figure 8, step 2). Although speculative, in such a composite ATPase active site, the GST-Xcdc6WB subunit might bind ATP and impart stable DNA chromatin interactions to the GST-Xcdc6WA subunit, whereas GST-Xcdc6WA might promote ATP hydrolysis and MCM loading.
In conclusion, the experiments presented herein provide further evidence that Walker A and Walker B are essential motifs of Cdc6 that are required for proper prereplication complex formation. Our experiments support a mechanism to explain how ATP binding and hydrolysis contribute in a complementary manner to enable Cdc6 function. Our data are consistent with Cdc6 acting as a molecular switch, wherein ATP binding promotes stable chromatin association and ATP hydrolysis induces MCM loading. Disruption of either of these activities results in dominant negative Cdc6 mutants, most likely due to the assembly of a nonproductive prereplication complex. Finally, we present the first evidence in support of Cdc6 acting as an oligomer, wherein catalytically inactive mutant Cdc6 likely contributes a structural role (e.g., ORC/MCM binding) to the heteroligomer. These findings provide a biochemical framework with which to dissect further the role of Cdc6 in initiating DNA replication.
ACKNOWLEDGMENTS
We thank Jim Hittle for performing the microinjections; Drs. Chernoff, Furstenthal (Stanford University), Herbig (Brown University, Providence, RI), Jackson (Stanford University), Markham, Moss, and Stoyanova, and the members of the Yen laboratory for many insightful discussions; Dr. Newport (University of San Diego, San Diego, CA) for graciously providing the anti-Xmcm3 antibodies; and Drs. Fashena, Golemis, Moss, Strich, and Yen for critical comments on the manuscript. We acknowledge Research Secretarial Services, and the Fox Chase Cancer Center's Glasswashing, Cell Culture, and Fannie E. Rippel Biotechnology Facilities. Awards from the American Cancer Society and the National Institutes of Health (CA-06927 and GM-58924) supported this work. We also gratefully acknowledge support from The V Foundation and an appropriation from the Commonwealth of Pennsylvania.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–08–0382. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–08–0382.
REFERENCES
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Mahbubani HM, Khoo C-Y, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Tang Z, Dunphy WG. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- DeRyckere D, Smith CL, Martin GS. The role of nucleotide binding and hydrolysis in the function of the fission yeast cdc18(+) gene product. Genetics. 1999;151:1445–1457. doi: 10.1093/genetics/151.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai D, Gu Y, Morgan DO. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen M, El-Denary M, Kapitza T, Graf R, Strausfeld U. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur J Biochem. 1999;264:415–426. doi: 10.1046/j.1432-1327.1999.00613.x. [DOI] [PubMed] [Google Scholar]
- Furstenthal L, Kaiser BK, Swanson C, Jackson PK. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J Cell Biol. 2001;152:1267–1278. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Griffith JW, Fanning E. Mutation of Cyclin/cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells. Mol Biol Cell. 2000;11:4117–4130. doi: 10.1091/mbc.11.12.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Marlar CA, Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol Biol Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani MM, Bloom LB, Goodman MF, O'Donnell M. Division of labor–sequential ATP hydrolysis drives assembly of a DNA polymerase sliding clamp around DNA. EMBO J. 1999;18:5131–5144. doi: 10.1093/emboj/18.18.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani MM, O'Donnell M. Sliding clamps: a (tail)ored fit. Curr Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Blow JJ. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wells NJ, Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc Natl Acad Sci USA. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Brown GW. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S-I, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Control of the cdc2/cyclin B complex in Xenopus egg extracts arrested at a G2/M checkpoint with DNA synthesis inhibitors. Mol Biol Cell. 1995;6:199–213. doi: 10.1091/mbc.6.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Lee DG, Bell SP. ATPase switches controlling DNA replication initiation. Curr Opin Cell Biol. 2000;12:280–285. doi: 10.1016/s0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Mossi R, Hubscher U. Clamping down on clamps and clamp loaders–the eukaryotic replication factor C. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- Murray AW. Cell-cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Natale DA, Li CJ, Sun WH, DePamphilis ML. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 2000;19:2728–2738. doi: 10.1093/emboj/19.11.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Pelizon C, Madine MA, Romanowski P, Laskey RA. Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 2000;14:2526–2533. doi: 10.1101/gad.176300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Diffley JFX. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, Lawlis SJ, Hou ZH, Hendricks M, Parvin JD, Dutta A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Arai K-I, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Tada S, Chong JP, Mahbubani HM, Blow JJ. The RLF-B component of the replication licensing system is distinct from Cdc6 and functions after Cdc6 binds to chromatin. Curr Biol. 1999;9:211–214. doi: 10.1016/s0960-9822(99)80092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Feng L, Hu Y, Huang SH, Reynolds CP, Wu L, Jong AY. The essential role of Saccharomyces cerevisiae CDC6 nucleotide-binding site in cell growth, DNA synthesis, and Orc1 association. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W, Rottjakob H-W, Küntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]