ABSTRACT
Objective: Sepsis is a complex disease characterized by an inflammatory response and tissue hypoxia. Hypoxia-inducible factor 1α (HIF-1α) expression level is regulated by hypoxia and inflammation. This study aimed to explore the correlation between HIF-1α expression level and sepsis by bioinformatics analysis and clinical investigation. Methods: Bioinformatics tools were used to identify differentially expressed genes between sepsis and nonsepsis groups using the Gene Expression Omnibus data set. A clinical investigation was carried out to validate HIF-1α protein level in 54 nonseptic patients and 173 septic patients who were followed up for 28 days. Results: Bioinformatics analysis revealed that HIF-1α messenger RNA level was significantly different between septic and nonseptic patients (P < 0.05). Consistent with the study hypothesis, higher HIF-1α levels in plasma were found in septic patients compared with those in nonseptic patients. The diagnostic accuracy for sepsis, as quantified by the area under the curve, was 0.926 (0.885–0.968) for HIF-1α expression level combined with oxygen saturation to fraction of inspired oxygen (SpO2/FiO2), white blood cell, and blood urea nitrogen. The HIF-1α expression level was also significantly correlated with the severity of the disease. The results of the restricted cubic splines model indicated a U-shaped relationship between HIF-1α expression level and intensive care unit (ICU) mortality. Univariate and multivariate linear regression analyses indicated that septic patients with the elevated HIF-1α expression levels had shorter length of ICU stay versus those with the lower HIF-1α expression levels. Conclusion: Hypoxia-inducible factor 1α expression level can be used for diagnosing disease, assessing severity, and predicting length of ICU stay in septic patients.
KEYWORDS/ABBREVIATIONS: Hypoxia-inducible factor 1α, sepsis, mortality, biomarker, intensive care unit, HIF-1α—hypoxia-inducible factor 1α, DEG—differentially expressed gene, GEO—Gene Expression Omnibus, WBC—white blood cell, BUN—blood urea nitrogen, RCS—restricted cubic spline, ICU—intensive care unit, WGCNA—weighted gene co-expression network analysis, PPI–protein-protein interaction, GO—Gene Ontology, KEGG—Kyoto Encylopedia of Genes and Genomes, HRG—hypoxia-related gene, PO2/FiO2—arterial oxygen partial pressure/fractional inspired oxygen, aCCI—age-adjusted Charlson Comorbidity Index, SOFA—sequential organ failure assessment, qSOFA—Quick SOFA, ELISA—enzyme-linked immunosorbent assay, ROC—receiver operating characteristic, IMV—invasive mechanical ventilation, AUC—area under the curve, RR—respiratory rate, bpm—beats/breaths per minute, WBC—white blood cell, ALT—alanine aminotransferase, AST—aspartate aminotransferase, Cr—creatinine, Lac—lactic acid, PHD—prolyl hydroxylase domain, SpO2—pulse oxygen saturation, FiO2—fraction of inspiration O2, FPR—false positive rate, TPR—true positive rate
INTRODUCTION
Sepsis was defined as life-threatening organ dysfunction caused by a dysregulated body response to infection (1). Despite advances in biomedicine and clinical medicine and the fact that the global age-standardized incidence of sepsis per 1,000 of the population gradually decreased from 1.075 in 1990 to 0.678 in 2017 (2), sepsis still accounts for 19.7% of all deaths worldwide (2). Thus, further research on sepsis is warranted.
Hemodynamics in sepsis changes as sepsis develops and includes macrocirculatory derangement, such as relative hypovolemia, decreased vascular tone, and microcirculatory dysfunction, including tissue hypoperfusion and insufficient oxygen level (3), which have also been considered as potent causes of the development of secondary organ dysfunction (4). At the cellular level, sepsis is characterized by an impairment of oxygen uptake and utilization. Therefore, an in-depth understanding of the molecular changes during hypoxia in sepsis and their clinical significance may facilitate management of critically ill patients. Furthermore, exploration of the underlying molecular mechanism may improve the understanding of the pathophysiological processes associated with sepsis and lead to the development of personalized medicine by selectively targeting the molecular target.
The hypoxic response is mainly regulated by the heterodimeric transcription factor hypoxia-inducible factor (HIF) family, including HIF-1α, HIF-2α, HIF-3α, and HIF-1β (5). In contrast to HIF-2α–mediated long-term acute hypoxia, HIF-1α is generally accepted to play a role in short-term hypoxia. Over the past decade, cumulative evidence demonstrated that HIF-1α expression level was correlated with clinical phenotype in sepsis, such as sepsis-induced lung injury and septic shock (6–9). However, there is a paucity of research assessing the correlation between circulating HIF-1α expression level and clinical characteristics in patients with sepsis. Because HIF-1α expression level generally degrades rapidly under aerobic conditions, it has shown limited and variable clinical value.
To evaluate the diagnostic and prognostic value of HIF in patients with sepsis, bioinformatics analysis and clinical investigation were conducted in this study. This study may contribute to promote understanding of the pathophysiological process underlying sepsis and lead to the development of more efficient targeted therapeutic strategies.
MATERIALS AND METHODS
Data sets collection and processing
Gene expression profiling data that investigated the gene expression levels in septic patients and nonseptic subjects were downloaded from the Gene Expression Omnibus database, which included GSE100159 and GSE80496 data sets (10,11). Two data sets were collected from adults and children with sepsis, respectively. The differentially expressed genes (DEGs) obtained by taking the intersection of the two gene sets mentioned previously were better representatives. The whole blood samples were collected within the first 24 hours after the diagnosis of sepsis. The corresponding expression matrix and clinical data were downloaded and matched. The study design is shown in Figure 1.
FIG. 1.
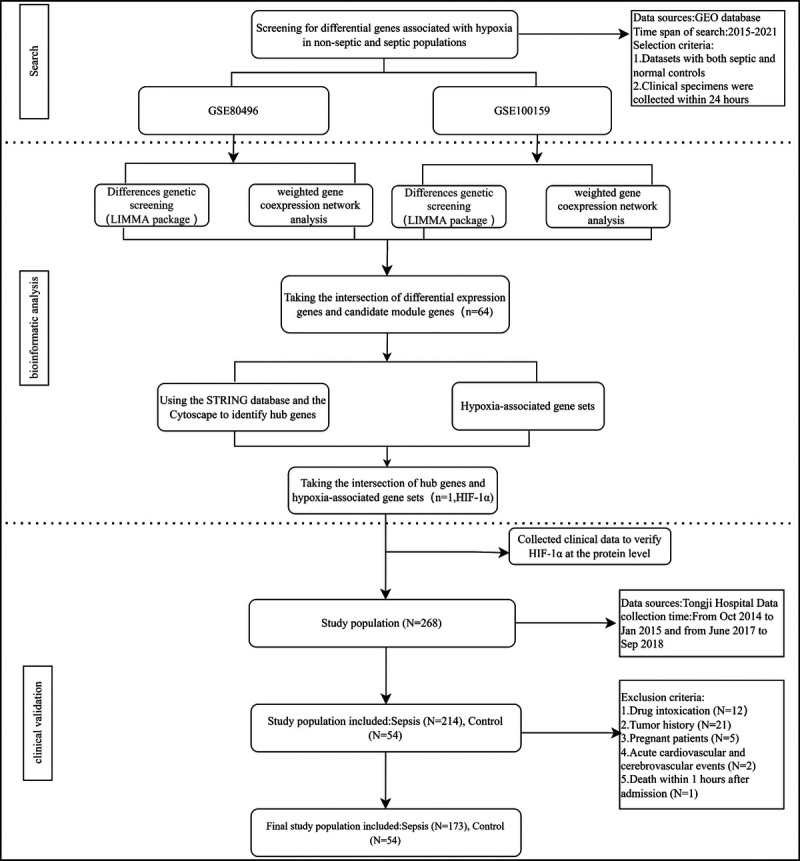
The flowchart of study design.
Bioinformatic analysis
To identify DEGs, the “limma” R package was used, and DEGs were screened using the following criteria (12,13): |Log2FoldChange| > 1 and P < 0.05. Then, weighted gene co-expression network analysis was used to construct gene co-expression networks, find co-expression modules, and promote clinical gene biomarker screening (14). Subsequently, “pickSoftThreshold” function was used as the proper soft-thresholding power for the network construction, identification of gene modules was performed via an average linkage clustering algorithm, and Pearson correlation analysis of gene and clinical traits was carried out. According to the screening conditions (|Module Membership| > 0.8; gene significance, >0.1), modules that were highly correlated with the clinically significant modules (correlation, >0.8) were identified as candidate genes. A Venn diagram was used to draw the intersection of genes, which overlapped between DEGs and candidate genes. The protein-protein interaction (PPI) networks were constructed via the STRING online database (string-db.org) with a high confidence (0.700) (15). Top 10 hub genes were confirmed via topological data analysis using Cytohubba plugin in Cytoscape 3.8.0 software (Institute for Systems Biology at the University of California, San Diego, CA) (16). Gene Ontology (GO) enrichment analysis and Kyoto Encylopedia of Genes and Genomes (KEGG) pathway enrichment analysis were used to exhibit the molecular function and critical pathways associated with intersection genes via the “org.Hs.eg.db” and “clusterProfiler” R packages (17,18). P value <0.05 was set as the cutoff criterion for the enriched terms.
To identify hypoxia-related hub genes, hypoxia-related genes (HRGs) from HALLMARK_HYPOXIA and BIOCARTA_HIF_PATHWAY gene sets were downloaded from the MSigDB database (http://www.gsea-msigdb.org/gsea/msigdb/help.jsp). To explore more specific target genes associated with the hypoxia pathway, hub genes and HRGs were intersected.
Clinical study design and subjects
The primary study population has been described previously (19). According to the “Sepsis-3.0 criteria” (1), it was attempted to regroup the study population into two groups: nonseptic group and septic group. All patients with sepsis who met diagnostic criteria within 24 h, who were admitted to the intensive care unit (ICU) of three medical centers (Tongji Guanggu Hospital, Zhongfa Hospital, and Hankou Hospital, which all were located in Wuhan, China) from October 2014 to January 2015 and from June 2017 to September 2018, were enrolled in this study. In the same period, healthy individuals who were attended in medical examinations were assigned to the control group. The study population was divided into sepsis group and nonsepsis group based on the aforementioned criteria. For assessing oxygenation status in septic patients, the arterial oxygen partial pressure/fractional inspired oxygen (PO2/FiO2) ratio has been the criterion standard. Patients in the sepsis group were further divided into three subgroups according to PO2/FiO2 ratio. The exclusion criteria were as follows: paraquat poisoning, age at the time of diagnosis was younger than 18 years, death within 1 h after admission, and acute cardiocerebrovascular events, such as acute coronary syndrome and stroke (20,21).
Age, sex, age-adjusted Charlson Comorbidity Index (aCCI), sequential organ failure assessment (SOFA) score, vital signs, routine blood test, liver function, renal function, and other laboratory parameters were analyzed. This scoring system was used to evaluate comorbidity in the study population. Quick SOFA (qSOFA) is known as a precious measure for phenotypic screening, and it was therefore selected as a reference index for evaluating the performance of the diagnostic tools (22).
Ethical approval was obtained from the Ethics Committee of Tongji Hospital Affiliated to Tongji Medical College (Wuhan, China; approval no. TJ-IRB20150318). All subjects signed the informed consent form before enrollment.
Detection of HIF-1α level and follow-up
Whole blood samples for HIF-1α measurement were collected on day 1 of ICU hospitalization and placed into tubes containing ethylenediaminetetraacetic acid (EDTA), and a cocktail of protease inhibitors was added immediately (Sigma-Aldrich, MO). Plasma was separated from the aforementioned blood samples via centrifugation at 3,000g for 15 min at 4°C. The remaining blood samples were centrifuged and immediately stored in the −80°C super cold refrigerator as described previously (19). The sample collection, storage, and testing methods were proved to be effective in preventing protein degradation (23–25).
Hypoxia-inducible factor 1α expression level was measured using enzyme-linked immunosorbent assay (ELISA) kits (HM10162; BioSwamp, Wuhan, China) (25). The following steps were performed for ELISA experiment: (1) Diluting standards were implemented according to the manufacturer's instructions. (2) Addition to samples: blank hole, standard hole, and sample hole were set on a 96-well enzyme-labeled plate. A volume of 50 μL/well standard sample was added to standard holes into a 96-well plate. Next, 40 μL of sample and 10 μL of antibody were added to sample holes. (3) Enzyme addition and incubation: Each well, except for the blank hole, was added to 50 μL enzymatic labeling solution and incubated at 30°C for 30 min. (4) Washing: The 30-fold washing solution of configured liquid was diluted for 30 times with double-distilled water. Every hole was rinsed with washing solution, which allowed to stand for 30 s and was then discarded (five replicates). (5) Color development: Chromogenic agent A (50 μL) and chromogen B (50 μL) were added to each well in sequence and gently mixed and incubated for 10 min at 37°C. (6) Termination and measurement: A 50-μL stop solution was added to each hole. Blank well was taken as zero, and the optical density was measured at the wavelength of 450 nm after adding.
The primary outcome measure was the ICU mortality, and the secondary outcome measures were length of ICU stay and 28-day survival. All patients were followed-up for 28 days by telephone after the onset of the disease.
Statistical analysis
The statistical analysis was performed using R 3.6.3 and STATA 14.0 (Stata Corp, College Station, TX) software. Normally distributed continuous data were presented as mean ± standard deviation and compared using the Student t test or one-way analysis of variance, whereas nonparametric data were presented as median (interquartile range) and compared using the Kruskal-Wallis test or Mann-Whitney U test. The categorical variables were tested by the chi-square test. Correlations were evaluated by the Spearman correlation coefficient. Receiver operating characteristic (ROC) curves were plotted to evaluate the diagnostic value of HIF-1α for sepsis. DeLong test was performed to compare the accuracy of different indicators using ROC curves. Logistic/Cox regression models were used to investigate the association between HIF-1α expression level and adverse outcomes, such as invasive mechanical ventilation (IMV), intrahospital mortality, and 28-day all-cause mortality. Potential nonlinear relationships between HIF-1α expression level and in-hospital mortality were tested with restricted cubic splines (RCSs). Testing of a U-shaped relationship was carried out by including squared terms in the models and “utest” method (26). Stepwise multiple regression analysis was used to assess the relationship between HIF-1α expression level and length of ICU stay. A two-sided P < 0.05 was considered statistically significant.
RESULTS
HIF-1α signaling pathway was enriched in Sepsis patients
Differential gene expression analysis showed 2,132 DEGs in the GSE100159 data set, including 891 upregulated and 1,241 downregulated DEGs (Supplementary Fig. 1, A–D, http://links.lww.com/SHK/B672). In the GSE80496 data set, 947 DEGs were obtained, including 481 upregulated and 466 downregulated DEGs (Supplementary Fig. 1, E–H, http://links.lww.com/SHK/B672). Weighted gene co-expression network analysis identified 2,143 and 1,244 sepsis-associated candidate genes in the GSE80496 and GSE10095 data sets, respectively (Supplementary Fig. 2 and 3, http://links.lww.com/SHK/B672). The intersection between upregulated DEGs and sepsis-associated candidate gene data sets revealed 177 genes (Fig. 2A). Furthermore, the GO molecular function enrichment analysis showed that HIF-1α genes were involved in protein dimerization activity (Fig. 2B). The KEGG pathway enrichment analysis indicated that the intersection of gene sets was enriched in the pathway of HIF-1α signaling pathway (Fig. 2C).
FIG. 2.
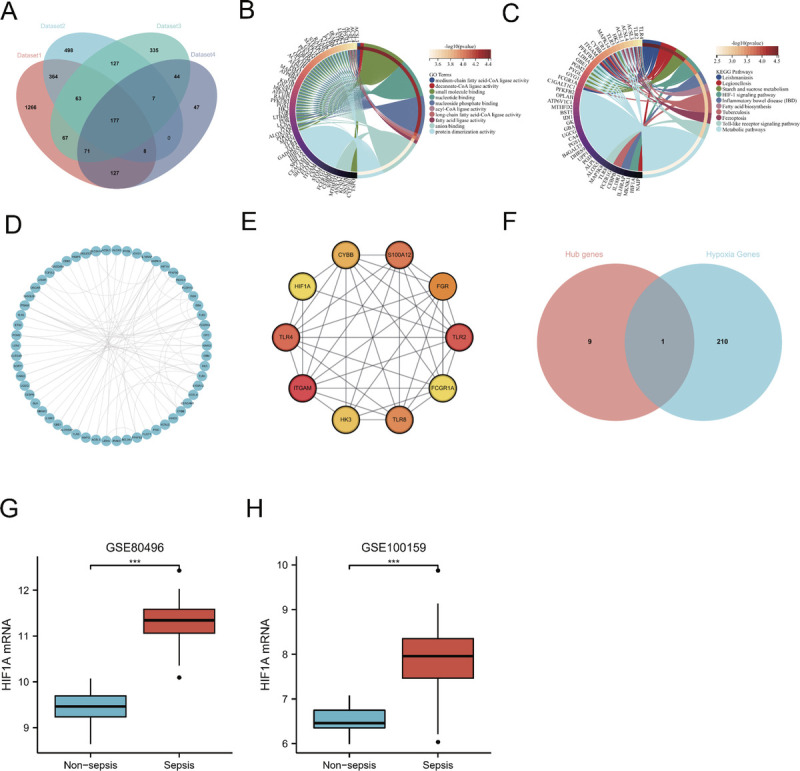
Identification and validation of hypoxia-related hub genes. A, A Venn diagram program was used to reflect the intersection between WGCNA candidate genes and upregulated DEGs in GSE100159 and GSE80496 data sets. B, The GO enrichment analysis of intersection genes. C, The KEGG enrichment analysis of intersected genes. D, The PPI network of intersected genes, including ITGAM, TLR2, TLR4, S100A12, TLR8, FGR, CYBB, HK3, HIF-1α, and FCGR1A. E, The PPI network of hub genes. F, The intersections of hub genes and hypoxia gene set. G and H, The HIF-1α mRNA expression level in the sepsis is significantly higher than that in the nonsepsis group (P < 0.001) (GSE80496, GSE100159).
HIF-1α was identified as one of the hub genes in septic patients
The top 10 hub genes were identified using the PPI network and the connectivity degree method among 177 intersection genes (Fig. 2D, E). To further identify hypoxia-related hub genes, an analysis was performed between the top 10 hub genes and HRGs using a Venn diagram. As a result, the hypoxia-related hub gene HIF-1α was obtained (Fig. 2F).
HIF-1α expression level significantly increased during sepsis
There were significant differences in the level of expression of HIF-1α expression level between the sepsis and nonsepsis groups (GSE80496, 9.440 ± 0.387 vs. 11.328 ± 0.505, P < 0.001, Fig. 2G; GSE100159, 6.540 ± 0.314 vs. 7.860 ± 0.848, P < 0.001, Fig. 2H).
The baseline characteristics of the study population
The study ultimately included 54 nonseptic patients and a total of 173 septic patients. Table 1 summarizes the baseline characteristics of the nonseptic and septic groups. Vital signs, biochemical markers, and ICU parameters were measured within the first 4 h after ICU admission. Patients in the septic group had significantly higher temperature, heart rate, and mean arterial pressure compared with those in the nonseptic group (all, P < 0.001). Furthermore, patients in the septic group had higher values of laboratory parameters, including infection-related indicators such as white blood cell (WBC) count, and liver function indicators such as alanine aminotransferase and aspartate aminotransferase, as well as renal function indicators such as creatinine (Cr) and blood urea nitrogen (BUN), when compared with those in the nonseptic group (all, P < 0.05). In addition, patients in the septic group also had higher SOFA scores on admission to the hospital, longer duration of undergoing IMV, and higher 28-day all-cause mortality rates compared with patients in the nonseptic group (all P < 0.05). According to the intragroup comparison, there was an increase in HIF-1α expression level with the reduced PO2/FiO2 ratio in the septic group, although the differences were not statistically significant (P > 0.05).
TABLE 1.
Clinical characteristics and laboratory tests of the study population
| Characteristic | Control (n = 54) | Sepsis (n = 173, PO2/FiO2, mm Hg) | |||
|---|---|---|---|---|---|
| >300 (n = 46) | 100–300 (n = 108) | <100 (n = 19) | P | ||
| Female (n, %) | 31 (13.7) | 22 (12.7) | 46 (26.6) | 5 (2.9) | 0.072,* 0.277† |
| Age <60 y (n, %) | 39 (17.2) | 33 (19.1) | 60 (34.7) | 12 (6.9) | 0.170,* 0.695† |
| Vital signs | |||||
| Temperature (°C) | 36.5 (36.32–36.9) | 37.15 (36.8–37.6) | 37.3 (36.5–38.2) | 37.8 (36.95–38.4) | <0.001,* 0.563† |
| Heart rate (bpm) | 83 (76.5, 90.25) | 112.2 ± 28.91 | 116.44 ± 28.77 | 125.26 ± 19.89 | <0.001,* 0.233† |
| Respiratory rate (bpm) | 20 (19–20) | 20.5 (10.25–26) | 20 (0–30.25) | 18 (0–33) | 0.806,* 0.654† |
| MAP (mm Hg) | 89 (82.67–98.61) | 81.22 ± 22.5 | 81.63 ± 20.98 | 78.23 ± 20.28 | <0.001,* 0.814† |
| HIF-1α (ng/mL) | 2.19 (1.59–3.32) | 2.74 (1.7–3.32) | 3 (2.04–3.91) | 3.25 (2.32–3.97) | 0.011,* 0.078† |
| Laboratory index | |||||
| WBC (109/L) | 6.56 (5.07–8.76) | 12.43 (8.83–18.57) | 15.23 (8.81–19.34) | 21.8 (14.47–23.1) | <0.001,* 0.058† |
| ALT (U/L) | 17.5 (11–28) | 23.5 (15–74.75) | 28.5 (14–82.5) | 35 (12–58.5) | <0.001,* 0.905† |
| AST (U/L) | 20 (15–28.75) | 48 (23–84) | 47.5 (24–119.25) | 61 (20–136.5) | <0.001,* 0.821† |
| BUN (mmol/L) | 4.71 (4.03–5.91) | 11.65 (6.96–19) | 10.35 (6.74–15.95) | 13 (7.77–21.38) | <0.001,* 0.437† |
| Cr (μmol/L) | 64 (54–73) | 123.5 (79–287.75) | 117.5 (74.75–269) | 162 (122–247) | <0.001,* 0.551† |
| Critical illness score | |||||
| aCCI | 2 (0–3) | 2 (1–3) | 2 (0.75–4) | 2 (0.5–4) | 0.919,* 0.919† |
| qSOFA | 0 (0–1) | 2 (1–2) | 2 (1–2) | 2 (2–2) | <0.001,* 0.695† |
| SOFA | 1 (0–2.75) | 6 (4–10.5) | 8 (5.75–10) | 10 (9–12) | 0.001,* 0.001† |
| Arterial blood gas | |||||
| PCO2 (mm Hg) | — | 36.45 (29.02–40.98) | 36.4 (29.67–42.23) | 37 (29.1–50.35) | 0.616† |
| PO2 (mm Hg) | — | 143.5 (119.25–181.75) | 84 (69.68–99.25) | 57 (50.5–60.5) | <0.001† |
| SaO2 (%) | 100 (96.25–100) | 98.5 (94.75–100) | 93 (90.5–97) | <0.001† | |
| Lac (mmol/L) | — | 3.45 (2.35–5.08) | 3.88 (2.83–6.29) | 5.06 (2.84–8.88) | 0.191† |
| PO2/FiO2 (mm Hg) | — | 365.43 (330.49–466.43) | 201.35 (151.5–241.53) | 66.25 (60–85.53) | <0.001† |
| Clinical outcomes | |||||
| IMV (n, %) | — | 20 (11.6) | 62 (35.8) | 16 (9.2) | 0.010† |
| IMV time (h) | — | 0 (0–33.75) | 11 (0–87) | 31 (15.5–74) | 0.033† |
| In-ICU mortality (n, %) | — | 4 (2.3) | 13 (7.5) | 1 (0.6) | 0.808† |
| ICU stay time (h) | — | 86 (41.5–161.25) | 90.5 (39.75–214.5) | 60 (26–159.5) | 0.387† |
| 28-d Mortality (n, %) | — | 9 (5.2) | 33 (19.1) | 9 (5.2) | 0.077† |
| 28-d Survival (d) | — | 28 (28–28) | 28 (20–28) | 28 (13–28) | 0.147† |
*Comparisons between nonseptic and septic group.
†Within septic subgroup comparisons.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; bpm, beats/breaths per minute; Lac, lactic acid; PCO2, partial pressure of carbon dioxide; SaO2, arterial oxygen saturation.
Elevated HIF-1α expression level might be a potential biomarker for adjuvant diagnosis of sepsis
On the first day of admission, HIF-1α expression level was significantly higher in the septic group than that in the nonseptic group (2.980 [1.990–3.857] ng/mL vs. 2.194 [1.589–3.316] ng/mL, P < 0.05). Area under the ROC curve values of clinical indicators were calculated to compare the diagnostic performance obtained by the ROC curve analysis (Fig. 3). A poorer diagnostic performance of HIF-1α was found in the diagnosis of septic and nonseptic participants compared with qSOFA (Table 2). To enhance the diagnostic performance, an indicator, namely, HIF-1αadj, which could combine clinical parameters and HIF-1α expression level, was proposed. The HIF-1αadj index was formulated as follows: HIF-1αadj = HIF-1α × FiO2/SpO2 (Equation 1). According to the ROC curve analysis, the diagnostic accuracy in differentiating the septic group from the nonseptic group using HIF-1αadj was moderate, which was similar to that of qSOFA. White blood cell and BUN showed a similar diagnostic performance. However, a significant increase in the diagnostic performance was found when HIF-1αadj was combined with WBC and BUN. The combination of HIF-1αadj and BUN decreased the diagnostic performance, whereas this phenomenon was not found in the combination of HIF-1αadj and WBC.
FIG. 3.
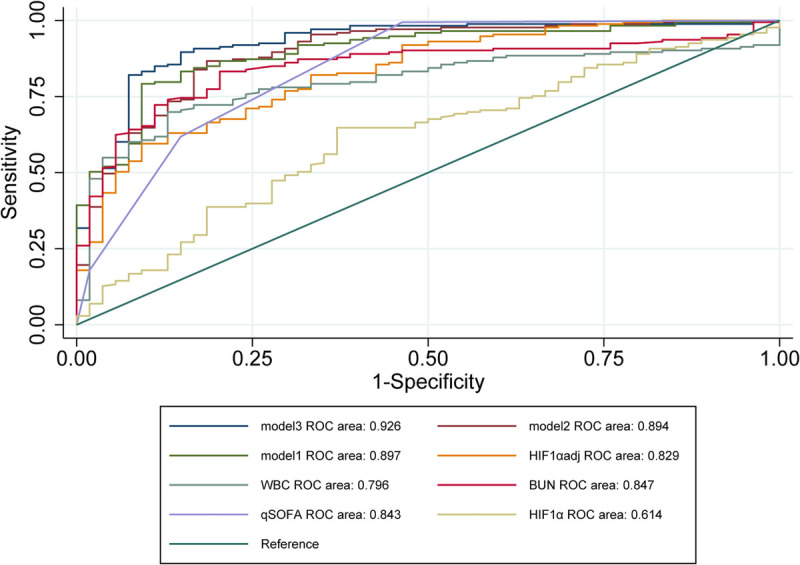
The ROC curve for the diagnosis of sepsis. Receiver operating characteristic curves were compared between the nonseptic and septic groups. FPR indicates false positive rate; TPR, true positive rate.
TABLE 2.
ROC curves in assessing the statue of sepsis
| Variables | Cutoff values | Sensitivity (%) | Specificity (%) | AUC (95% CI) | Youden index | P |
|---|---|---|---|---|---|---|
| Model 3 | 0.418 | 0.896 | 0.852 | 0.926 (0.885–0.968) | 0.748 | 0.001 |
| Model 2 | 0.569 | 0.867 | 0.815 | 0.894 (0.844–0.945) | 0.682 | 0.051 |
| Model 1 | 1.268 | 0.792 | 0.907 | 0.897 (0.851–0.942) | 0.699 | 0.049 |
| HIF-1αadj | 1.220 | 0.595 | 0.907 | 0.829 (0.769–0.890) | 0.503 | 0.686 |
| WBC | 10.045 | 0.870 | 0.699 | 0.796 (0.736–0.856) | 0.570 | 0.174 |
| BUN | 6.035 | 0.796 | 0.832 | 0.847 (0.794–0.901) | 0.629 | 0.889 |
| HIF-1α | 2.429 | 0.630 | 0.647 | 0.614 (0.530–0.699) | 0.277 | <0.001 |
| qSOFA | 0.500 | 0.994 | 0.537 | 0.843 (0.780–0.905) | 0.531 | Ref. |
HIF-1αadj = HIF-1α × Fio2/Spo2. Model 1, HIF-1αadj + BUN; model 2, HIF-1αadj + WBC; model 3, HIF-1αadj + WBC + BUN; P values: DeLong test was used for the comparison of different AUCs.
AUC, area under the curve; Ref., reference.
Associations of HIF-1α expression level with different parameters
Heat map showing HIF-1α expression level was positively correlated with a variety of factors, such as laboratory indicators, hypoxic indicators, and scoring systems (Fig. 4A). Hypoxia-inducible factor 1α expression level was found to be correlated with the severity of the disease. In the beginning, all patients were divided into four groups according to qSOFA score. A statistically significant increase in HIF-1α expression level was found in both groups qSOFA (1), qSOFA (2), and qSOFA (3) groups compared with qSOFA (0) (Fig. 4B). However, there was no significant difference in HIF-1α expression level among qSOFA (1), qSOFA (2), and qSOFA (3) groups. Hypoxia-inducible factor 1α was also noted to be closely associated with the SOFA score and aCCI in our population studies (Fig. 4, C and D). Thus, HIF-1α could be used to assess the severity of the disease. Moreover, HIF-1α expression level was associated with various clinical indicators in the present study. Spearman correlation analysis revealed that FIO2, Cr, and BUN (Fig. 4, E–G) were positively correlated with the HIF-1α expression level, and oxygen saturation to fraction of inspired oxygen (SpO2/FiO2) ratio (Fig. 4H) exhibited a negatively correlation.
FIG. 4.
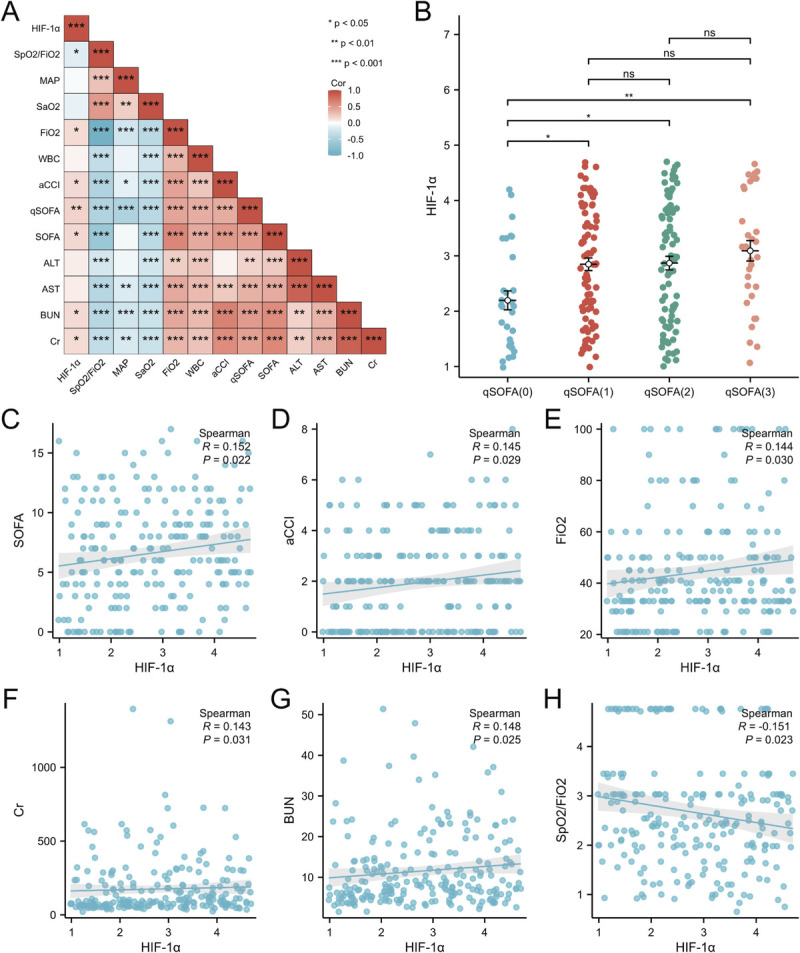
The association of HIF-1α protein levels, clinical characteristics, and clinical prognosis. A, The Spearman rank correlation test was used for exploration of the correlations. Positive correlations are illustrated by red squares; negative correlations are presented by blue squares. B, Hypoxia-inducible factor 1α distributions for different qSOFA score-based subgroups (2.09 [1.40–2.83] vs. 2.84 [2.04–3.77] vs. 2.94 [1.83–3.84] vs. 3.13 [2.40–4.22], P < 0.01, P [overall] = 0.0063) (*P < 0.05, **P < 0.01, ***P < 0.001). C, The relationship between SOFA score and HIF-1α level. D, The relationship between aCCI score and HIF-1α level. D, The relationship between FIO2 and HIF-1α level. F, The relationship between Cr level and HIF-1α level. G, The relationship between BUN level and HIF-1α level. H, The relationship between SpO2/FIO2 and HIF-1α level.
In addition, associations between HIF-1α expression level and different parameters were further validated using univariate and multivariate linear regression models. As shown in Table 3, the univariate linear regression model indicated that HIF-1α was correlated with SOFA, qSOFA, and SpO2/FiO2. The aforementioned results were confirmed by the age- and sex-adjusted multivariate linear regression model. However, the associations between HIF-1α expression level and aCCI were unapparent in the multivariate linear regression model aCCI model. The remaining indicators did not show a significant correlation.
TABLE 3.
Correlations between HIF-1α and different parameters in septic patients
| Variables | Univariate (95% CI) | P | Multivariate (95% CI)* | P |
|---|---|---|---|---|
| SOFA | 0.038 (0.005–0.071) | 0.023 | 0.035 (0.001–0.069) | 0.046 |
| qSOFA | 0.232 (0.077–0.387) | 0.003 | 0.211 (0.051–0.371) | 0.01 |
| aCCI | 0.085 (0.009–0.161) | 0.029 | 0.077 (−0.035 to 0.189) | 0.176 |
| FIO2 | 0.007 (−0.0001 to 0.013) | 0.055 | 0.005 (−0.002 to 0.012) | 0.126 |
| Cr | 0.0002 (−0.0005 to 0.0009) | 0.532 | 0.0002 (−0.0005 to 0.001) | 0.563 |
| BUN | 0.013 (−0.002 to 0.029) | 0.095 | 0.011 (−0.005 to 0.027) | 0.190 |
| SpO2/FiO2 | −0.157 (−0.279 to −0.034) | 0.012 | −0.146 (−0.282 to −0.009) | 0.037 |
*Adjusted for age and sex.
Association between HIF-1α expression level and in-ICU mortality in septic patients
The RCS method was used to examine the dose-response relationship between the levels of HIF-1α expression level and in-ICU mortality in septic patients. The risk of mortality corresponding to minimum value of HIF-1α expression level was used as a reference point. The uncorrected RCS showed a nonlinear relationship between HIF-1α expression level and in-ICU mortality (Fig. 5). The quadratic regression model showed HIF-1α and HIF-1α (2) (Supplementary Table 1, http://links.lww.com/SHK/B672). The aforementioned results indicate a possible regional variance in a U-shaped relationship, and the relationship was further examined using the U test method. The results indicated a U-shaped relationship between the levels of HIF-1α expression level and ICU mortality (HIF-1α extreme point, 2.558; U test, P < 0.05).
FIG. 5.
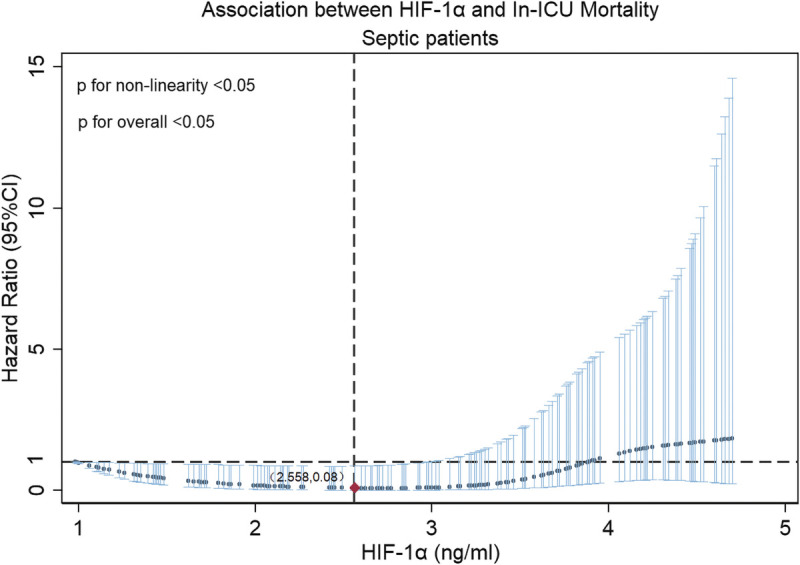
Restricted cubic spline regression model. Unadjusted, RCSs with four knots were used to model the association of HIF-1α expression level with ICU mortality in septic patients.
The relationship between HIF-1α expression level and time-to-treatment was explored using univariate and multivariate linear regression analysis. It was found that HIF-1α expression level was negatively correlated with the length of ICU stay time in univariate model and in multivariate model (Table 4). No correlation was found between the HIF-1α expression level and 28-day survival.
TABLE 4.
Association between HIF-1α and the treatment time among septic patients
| Variables | Univariate (95% CI) | P | Multivariate (95% CI)* | P |
|---|---|---|---|---|
| 28-d Survival | −0.359 (−1.496 to 0.778) | 0.534 | −0.145 (−1.248 to 0.958) | 0.796 |
| ICU stay time | −32.152 (−54.809 to 9.496) | 0.006 | −30.111 (−52.780 to 7.443) | 0.01 |
*Adjusted for age and sex.
DISCUSSION
In the present study, the value of HIF-1α in diagnosing sepsis was investigated, the clinical relevance of HIF-1α was elucidated, and the ability of HIF-1α was further assessed to predict clinical outcomes. The bioinformatics analysis was first used to identify DEGs, and the differential expression of HIF-1α in the septic and nonseptic groups was further verified. Subsequently, HIF-1α expression levels on the first day of admission were significantly different in septic patients compared with that in nonseptic patients; it was positively correlated with SOFA and qSOFA. This indicates that HIF-1α expression level could also be used as a marker of the severity of the disease, particularly in septic patients. Second, the significance of the HIF-1α–based indicators as a diagnostic and prognostic biomarker for sepsis was assessed and verified. Third, a U-shaped association was found between HIF-1α expression levels and ICU mortality, indicating that HIF-1α expression level is highly closely correlated with ICU mortality in septic patients. Finally, septic patients with high HIF-1α levels had a shorter ICU stay compared with patients with low HIF-1α levels. Thus, high HIF-1α levels on the first day of admission can be used as a diagnostic and prognostic indicator for sepsis.
The present study primarily concentrated on investigating the messenger RNA (mRNA) levels of HIF-1α and assessed the effects of HIF-1α protein. Some results of the present study were consistent with those reported previously. Bacterial LPS-induced HIF-1α activation in human monocytes upregulated HIF-1α mRNA expression and HIF-1α protein accumulation in vitro (27). An in vivo prospective clinical study compared HIF-1α mRNA level in the blood of healthy volunteers and shock patients (including septic shock, hemorrhagic shock, and cardiogenic shock) and found that HIF-1α mRNA level was significantly higher in patients with shock than that in healthy volunteers (9). Using bioinformatics analysis, Ferreira et al. (8) analyzed public databases of septic patients and healthy volunteers and found the elevated HIF-1α mRNA level in blood specimens of septic patients. However, there were some discrepancies between the results of the present study and those achieved previously; for instance, a significant decrease in HIF-1α expression level was detected between patients with sepsis and healthy volunteers, rather than an increase, as reported by Schafer et al. (28). In another in vitro study, HIF-1α mRNA level increased after a 6-h LPS stimulation, whereas it decreased after a 48-h LPS stimulation (28).
Two possible reasons may be responsible for these discrepancies. The first cause could be LPS tolerance, a phenomenon that the expression levels of inflammatory genes are upregulated with the onset of an inflammatory or hypoxic stimulus, whereas repetitive stimuli can inhibit the expression levels of inflammatory genes. A class of time-dose-response models incorporating LPS stimulation and hypoxic conditions in zebrafish larvae showed that HIF-1α mRNA level reached the peak after 8 h (29). Another study of LPS-stimulated neutrophils experiment indicated that 4-h LPS stimulation upregulated HIF-1α protein level and then significantly decreased gradually (30). Second, the differences in therapeutic alternatives between ventilation and oxygen therapies may explain some discrepancies. Studies demonstrated that HIF-1α is hydroxylated by oxygen-dependent prolyl hydroxylase domain (PHD), ubiquitinated by the E3 ubiquitin ligase Von Hippel-Lindau protein, and it is then rapidly degraded by the proteasome under normoxic conditions (31). A previous study described that long-term exposure to hypoxia could enhance HIF-α prolyl-4-hydroxylase capacity of cytoplasmic and nuclear protein extracts, which accelerated decomposition of HIF-1α after reoxygenation (32). The PHD activity decreased under hypoxic conditions, blocking hydroxylation and stabilizing HIF-1α expression level (33). Oxygen therapy, including nasal catheter oxygen or mask oxygen therapy, IMV, and other methods, was applied to patients with sepsis at admission, whereas improving hypoxic conditions might remarkably reduce HIF-1α expression level. However, in the present study, septic patients were classified into three groups according to their PO2/FiO2 (group 1, >300 mm Hg; group 2, 100–300 mm Hg; group 3, <100 mm Hg), and no significant differences were detected among these groups. Considering all the aforementioned factors, the timing of sepsis episodes is a major risk factor for elevated HIF-1α expression level.
As a strength of this study, HIF-1α expression level was measured after patients' admission to ICU (within the first 4 h of admission) and was evaluated in the different PO2/FiO2 subgroup of sepsis. This avoids the interference caused by administering anti-infective medications to treat and the duration of disease. Second, previous studies have mainly concentrated on HIF-1α mRNA level in different diseases. In the present study, HIF1a protein level in septic patients was examined for the first time. This is one of the novelties of this research. The present study also has some limitations. First, sepsis is a heterogeneous disease characterized by dysregulated systemic responses resulting from infections caused by bacteria, fungi, or viruses (34). Comorbidities, age, genetic predispositions, and infection sources can complicate HIF-1α expression level. Although several factors were considered in the analysis and assessment, it was still difficult to avoid confounding factors and different clinical manifestations of septic patients. Second, as circulating HIF-1α expression level was measured after sepsis, it was infeasible to determine whether the elevated HIF-1α expression level could be causally related to sepsis. Third, because of the lack of in vitro cellular experiments, the differences in mRNA and protein expression levels of HIF-1α between sepsis and normal culture conditions were not analyzed. Fourth, HIF-1α is not actively released into the circulation, and it was speculated that circulation HIF-1α protein is a marker for cell damage in sepsis. The hypothesis will be verified in the future studies. Further large-scale study is advantageous to dynamically evaluate the influences of HIF-1α expression level on sepsis development and prognosis and to explore its underlying mechanisms. In the future study, cellular and animal experiments will be conducted to determine how the elevated HIF-1α expression level can affect septic patients' prognosis. Infection and other influential factors will also be profoundly studied.
CONCLUSION
In conclusion, a new indicator was proposed for sepsis, HIF-1α, and it was revealed that HIF-1α could serve as a robust diagnostic and prognostic biomarker for sepsis. The HIF-1α expression level increased as the severity of the sepsis was enhanced. However, it is essential to conduct additional, comprehensive clinical studies on HIF-1α and sepsis, to confirm the aforementioned findings. Nonetheless, these findings have substantial clinical and public health implications.
Supplementary Material
Footnotes
H.R. and Y.-z.L. contributed equally to this work.
The data sets used and analyzed during the current study are available from the corresponding author upon reasonable request.
H.R., Y.-z.L., and Q.Z. participated in writing the manuscript. B.-r.W. and R.W. participated in the collection and interpretation of data. S.-s.L. and X.R. were involved in the review of the manuscript.
This study was financially supported by funding from the National Natural Science Foundation of China (grant no. 82271358) and the Scientific Research Foundation for Returned Overseas Chinese Scholars of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, and Talents Project of Public Health in Hubei Province (grant no. 2022SCZ048).
The authors report no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citation appears in the printed text and is provided in the HTML and PDF versions of this article on the journal’s Web site (www.shockjournal.com).
Contributor Information
Hang Ruan, Email: 17843102798@139.com.
Yao-zhuo Li, Email: yaozhuo_li@outlook.com.
Qin Zhang, Email: qzhang8@tjh.tjmu.edu.cn.
Bin-ran Wang, Email: wangbr15@outlook.com.
Rongxue Wu, Email: Rwu3@uchicago.edu.
Shu-sheng Li, Email: Shushengli16@sina.com.
REFERENCES
- 1.Singer M Deutschman CS Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd KE Johnson SC Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rovas A Seidel LM Vink H, et al. Association of sublingual microcirculation parameters and endothelial glycocalyx dimensions in resuscitated sepsis. Crit Care. 2019;23:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tadros T Traber DL Heggers JP, et al. Angiotensin II inhibitor DuP753 attenuates burn- and endotoxin-induced gut ischemia, lipid peroxidation, mucosal permeability, and bacterial translocation. Ann Surg. 2000;231(4):566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lv QW Zheng ZQ Zhang H, et al. Serum hypoxia-inducible factor 1alpha emerges as a prognostic factor for severe traumatic brain injury. Clin Chim Acta. 2021;522:77–82. [DOI] [PubMed] [Google Scholar]
- 6.Sun HD Liu YJ Chen J, et al. The pivotal role of HIF-1α in lung inflammatory injury induced by septic mesenteric lymph. Biomed Pharmacother. 2017;91:476–484. [DOI] [PubMed] [Google Scholar]
- 7.Shalova IN Lim JY Chittezhath M, et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. 2015;42(3):484–498. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira BL Leite GGF Brunialti MKC, et al. HIF-1α and hypoxia responsive genes are differentially expressed in leukocytes from survivors and non-survivors patients during clinical sepsis. Shock. 2021;56(1):80–91. [DOI] [PubMed] [Google Scholar]
- 9.Textoris J Beaufils N Quintana G, et al. Hypoxia-inducible factor (HIF1 alpha) gene expression in human shock states. Crit Care. 2012;16(4):R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman MC Rinchai D Baldwin N, et al. Development of a fixed module repertoire for the analysis and interpretation of blood transcriptome data. Nat Commun. 2021;12(1):4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herberg JA Kaforou M Wright VJ, et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA. 2016;316(8):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME Phipson B Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muenchen RA, Hilbe JM: Graphics with ggplot2. In: R for Stata Users. New York, NY: Springer New York, 2010:385–452. [Google Scholar]
- 14.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D Gable AL Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P Markiel A Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbon S Douglass E Good BM, et al. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu GC Wang LG Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran X Zhang Q Li S, et al. Tissue kallikrein exacerbating sepsis-induced endothelial hyperpermeability is highly predictive of severity and mortality in sepsis. J Inflamm Res. 2021;14:3321–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y Tan J Xie H, et al. HIF-1α regulates EMT via the snail and β-catenin pathways in paraquat poisoning-induced early pulmonary fibrosis. J Cell Mol Med. 2016;20(4):688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsao CC Baumann J Huang SF, et al. Pericyte hypoxia-inducible factor-1 (HIF-1) drives blood-brain barrier disruption and impacts acute ischemic stroke outcome. Angiogenesis. 2021;24(4):823–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans L Rhodes A Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit Care Med. 2021;49(11):E1063–E1143. [DOI] [PubMed] [Google Scholar]
- 23.Heikal L Ghezzi P Mengozzi M, et al. Assessment of HIF-1α expression and release following endothelial injury in-vitro and in-vivo. Mol Med. 2018;24:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W Sun Y Wang H, et al. HIF-1α enhances vascular endothelial cell permeability through degradation and translocation of vascular endothelial cadherin and claudin-5 in rats with burn Injury. J Burn Care Res. 2021;42(2):258–268. [DOI] [PubMed] [Google Scholar]
- 25.Li J Li SX Gao XH, et al. HIF1A and VEGF regulate each other by competing endogenous RNA mechanism and involve in the pathogenesis of peritoneal fibrosis. Pathol Res Pract. 2019;215(4):644–652. [DOI] [PubMed] [Google Scholar]
- 26.Lind JT, Mehlum H. With or without U? The appropriate test for a U-shaped relationship. Oxford Bull Econ Stat. 2010;72(1):109–118. [Google Scholar]
- 27.Frede S Stockmann C Freitag P, et al. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-kappaB. Biochem J. 2006;396:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer ST Frede S Winning S, et al. Hypoxia-inducible factor and target gene expression are decreased in patients with sepsis prospective observational clinical and cellular studies. Anesthesiology. 2013;118(6):1426–1436. [DOI] [PubMed] [Google Scholar]
- 29.Liu S Zhu K Chen N, et al. Identification of HIF-1 alpha promoter and expression regulation of HIF-1 alpha gene by LPS and hypoxia in zebrafish. Fish Physiol Biochem. 2013;39(5):1153–1163. [DOI] [PubMed] [Google Scholar]
- 30.Pan T Sun S Chen Y, et al. Immune effects of PI3K/Akt/HIF-1α–regulated glycolysis in polymorphonuclear neutrophils during sepsis. Crit Care. 2022;26(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lima B Lam GKW Xie L, et al. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci U S A. 2009;106(15):6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marxsen JH Stengel P Doege K, et al. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381:761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu K Jiao HL Li S, et al. ATF4 promotes bone angiogenesis by increasing vegf expression and release in the bone environment. J Bone Miner Res. 2013;28(9):1870–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu JY Li Q Wu ZM, et al. Two gene set variation indexes as potential diagnostic tool for sepsis. Am J Transl Res. 2020;12(6):2749–2759. [PMC free article] [PubMed] [Google Scholar]


