ABSTRACT
Asian lineage Zika virus (ZIKV) strains emerged globally, causing outbreaks linked with critical clinical disease outcomes unless the virus is effectively restricted by host immunity. We have previously shown that retinoic acid-inducible gene-I (RIG-I) senses ZIKV to trigger innate immunity to direct interferon (IFN) production and antiviral responses that can control ZIKV infection. However, ZIKV proteins have been demonstrated to antagonize IFN. Here, we conducted in vitro analyses to assess how divergent prototypic ZIKV variants differ in virologic properties, innate immune regulation, and infection outcome. We comparatively assessed African lineage ZIKV/Dakar/1984/ArD41519 (ZIKV/Dakar) and Asian lineage ZIKV/Malaysia/1966/P6740 (ZIKV/Malaysia) in a human epithelial cell infection model. De novo viral sequence determination identified amino acid changes within the ZIKV/Dakar genome compared to ZIKV/Malaysia. Viral growth analyses revealed that ZIKV/Malaysia accumulated viral proteins and genome copies earlier and to higher levels than ZIKV/Dakar. Both ZIKV strains activated RIG-I/IFN regulatory factor (IRF3) and NF-κB pathways to induce inflammatory cytokine expression and types I and III IFNs. However, ZIKV/Malaysia, but not ZIKV/Dakar, potently blocked downstream IFN signaling. Remarkably, ZIKV/Dakar protein accumulation and genome replication were rescued in RIG-I knockout (KO) cells late in acute infection, resulting in ZIKV/Dakar-mediated blockade of IFN signaling. We found that RIG-I signaling specifically restricts viral protein accumulation late in acute infection where early accumulation of viral proteins in infected cells confers enhanced ability to limit IFN signaling, promoting viral replication and spread. Our results demonstrate that RIG-I-mediated innate immune signaling imparts restriction of ZIKV protein accumulation, which permits IFN signaling and antiviral actions controlling ZIKV infection.
IMPORTANCE ZIKV isolates are classified under African or Asian lineages. Infection with emerging Asian lineage-derived ZIKV strains is associated with increased incidence of neurological symptoms that were not previously reported during infection with African or preemergent Asian lineage viruses. In this study, we utilized in vitro models to compare the virologic properties of and innate immune responses to two prototypic ZIKV strains from distinct lineages: African lineage ZIKV/Dakar and Asian lineage ZIKV/Malaysia. Compared to ZIKV/Dakar, ZIKV/Malaysia accumulates viral proteins earlier, replicates to higher levels, and robustly blocks IFN signaling during acute infection. Early accumulation of ZIKV/Malaysia NS5 protein confers enhanced ability to antagonize IFN signaling, dampening innate immune responses to promote viral spread. Our data identify the kinetics of viral protein accumulation as a major regulator of host innate immunity, influencing host-mediated control of ZIKV replication and spread. Importantly, these findings provide a novel framework for evaluating the virulence of emerging variants.
KEYWORDS: RIG-I, Zika virus, innate immunity
INTRODUCTION
Zika virus (ZIKV) remains an emerging threat of public health importance. Originally isolated in Uganda in 1947, ZIKV circulated mostly undetected in Africa and southeast Asia with limited mosquito to human transmission until 2007, when the first large-scale outbreak was reported in the Yap State of the Federated States of Micronesia (1–3). Prior to this point, ZIKV infection was largely asymptomatic and occasionally resulted in mild human disease with flu-like symptoms (4–6). However, in 2013 and 2015, French Polynesia and the Americas, respectively, reported widespread ZIKV outbreaks characterized by more severe clinical outcomes. Retrospective studies of the 2013 French Polynesian outbreak showed that adult infection with ZIKV was associated with an increased incidence of Guillain-Barré syndrome, while in utero infections during the 2015 outbreak in the Americas were linked to a series of developmental impairments described as congenital Zika syndrome (CZS) (7–10). Thus, the recent ZIKV epidemics marked the emergence of severe clinical manifestations for adult and in utero infections, the latter etiologically linked with CZS, including altered fetal development, microcephaly, and fetal demise (11–13). Moreover, novel routes of transmission were documented, including through blood and sexual contact and via vertical transmission at the maternal-fetal interface (14–20). The reemergence of ZIKV as a global health threat has revealed important gaps in our understanding of ZIKV biology, including the host innate immune response to acute ZIKV infection (21, 22).
ZIKV is a positive-sense single-stranded RNA virus that is genetically clustered into two distinct lineages, African and Asian (23, 24). The Asian lineage encompasses the emerging strains associated with recent epidemics (23–25). The recent large-scale ZIKV epidemics highlight the global threat ZIKV poses to immunologically naive populations, spurring research to elucidate how ZIKV has evolved to cause severe disease. Several studies have performed comparative virologic analyses of African and Asian lineage ZIKV variants and revealed differences in cytopathic effect and replication kinetics (26–30). Other studies have conducted comparative analyses of pre-epidemic and epidemic ZIKV strains and identified differences in viral proteins that enhance fetal brain damage, viral infectivity, or viral transmissibility (31–37). These findings implicate viral genetics and interactions with host innate immune programs as major determinants of ZIKV replication, transmission, and infection outcome.
Host innate immunity serves as the first line of defense against RNA virus infection (38–41). The retinoic acid-inducible gene-I (RIG-I) protein serves as the main sensor of ZIKV in human cells and, upon recognition of ZIKV pathogen-associated molecular pattern (PAMP), signals through mitochondrial antiviral signaling (MAVS) adaptor protein to activate interferon regulatory factor (IRF) 3 and NF-κB transcription factors (27, 38, 41–45). These factors translocate to the nucleus to induce the expression of target genes that encode antiviral, immunomodulatory, and inflammatory factors, including types I and III interferons (IFNs), inflammatory cytokines, and chemokines (38, 41, 46). IFN-β is an immediate product of this response and is secreted from infected cells, signaling through the IFN receptor (IFNAR) in an autocrine and paracrine manner to activate the JAK/signal transducer and activator of transcription (STAT) signaling pathway (38, 46–48). Activation of JAK/STAT signaling induces the expression of hundreds of interferon-stimulated genes (ISGs) that exert antiviral and immunomodulatory functions to restrict viral replication and spread (49–52). In vivo and in vitro studies utilizing knockouts of different components of the RIG-I-like receptor (RLR) and IFN signaling pathways have demonstrated the critical role that innate immune activation and signaling play in restricting flavivirus infection in general and in suppressing ZIKV replication and virulence (27, 53–60). To counteract the antiviral activity of RLR and IFN signaling, ZIKV has evolved various mechanisms to disrupt these signaling pathways. During replication, ZIKV nonstructural proteins induce the formation of invaginations along the endoplasmic reticulum membrane that house the viral replication machinery and envelop nascent viral RNA, thereby preventing or delaying RIG-I detection of ZIKV PAMPs (61–64). In addition, ZIKV proteins can further block host innate immunity at the initial RLR or downstream IFN signaling stages. Previous studies have demonstrated that ZIKV differentially activates or suppresses RIG-I signaling, differentially modulating or significantly inhibiting IFN induction (27, 65–73). Studies evaluating the interaction between ZIKV and host IFN signaling have identified host proteins that are targeted by ZIKV structural and nonstructural proteins to dampen ISG expression (74–79). These features highlight the delicate balance between the accumulation of viral proteins that serve as innate immune antagonists and the induction and actions of the host antiviral response that determines infection outcome. However, the impact of ZIKV replication kinetics on the extent of host innate immune activation and antiviral actions across distinct stages of the ZIKV life cycle remains poorly understood.
Here, we present a comprehensive virologic and host innate immune analysis of two prototypic ZIKV variants from different lineages, African lineage ZIKV/Dakar/1984/ArD41519 (ZIKV/Dakar) and Asian lineage ZIKV/Malaysia/1966/P6740 (ZIKV/Malaysia). Our infection studies utilize immunocompetent human epithelial cells to assess ZIKV replication and examine the innate immune response induced by infection. De novo assembly of viral sequences revealed amino acid changes within the ZIKV/Dakar genome compared to ZIKV/Malaysia. One-step viral growth analyses demonstrated that while ZIKV/Malaysia replicated the viral genome more efficiently, it produced fewer infectious virions than ZIKV/Dakar. During the acute infection time course, ZIKV/Malaysia accumulated viral proteins and RNA earlier and to a greater abundance and exhibited a higher infection efficiency than ZIKV/Dakar. Viral growth analyses in control and RIG-I knockout (KO) cells revealed that RIG-I is critical for restricting ZIKV spread late in acute infection, wherein early accumulation of viral proteins in infected cells links with an enhanced ability to restrict IFN signaling. Thus, RIG-I directs an innate immune response that imparts control of viral replicative fitness impacting the kinetics of viral protein accumulation and IFN signaling to restrict ZIKV genome replication and spread. Our data implicate RIG-I signaling in limiting the epidemic potential of ZIKV variants and identify the window prior to the accumulation of viral innate immune antagonists as critical for viral sensitivity to restriction by the innate immune response.
RESULTS
Prototypic ZIKV variants encode genomic differences.
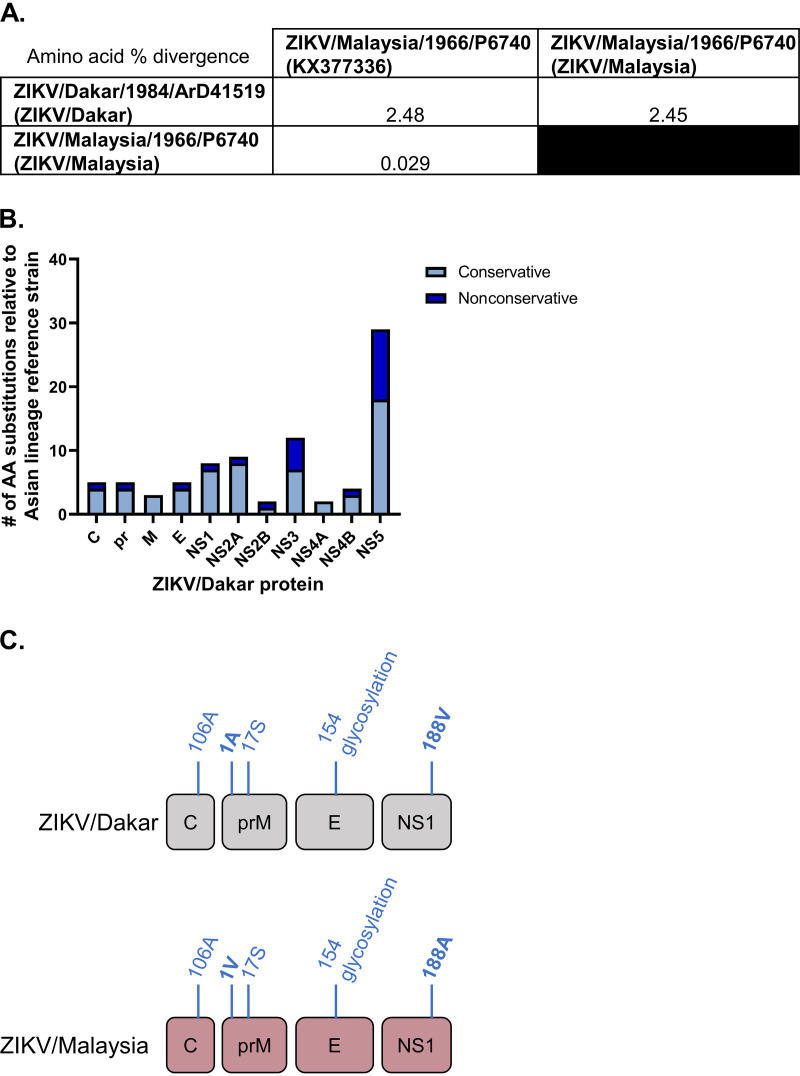
To evaluate how changes in ZIKV genome sequences contribute to viral replicative fitness, we first sequenced two prototypic ZIKV strains from distinct lineages, African lineage ZIKV/Dakar (GenBank OQ180929) and Asian lineage ZIKV/Malaysia (GenBank OQ165285). Though the original ZIKV strain isolated in Uganda in 1947 (MR766) is the prototypic African lineage ZIKV strain, it was passaged extensively in mice prior to its global distribution for research applications, whereas ZIKV/Dakar has undergone only limited cell culture passaging since its original isolation (1, 23, 80). We note that ZIKV/Malaysia is the prototypic virus of the Asian lineage and was thus included in our comparative analyses with African lineage ZIKV/Dakar (81). We also included the sequence of a reference Asian lineage strain in our genomic analysis (GenBank KX377336) (82). We determined the percent amino acid divergence of our ZIKV variants compared to the reference Asian lineage strain and to one another. Compared to the ZIKV Asian lineage reference genome, ZIKV/Malaysia displays low (0.029%) amino acid sequence divergence. Importantly, ZIKV/Dakar exhibits increased amino acid sequence divergence from the Asian lineage reference strain compared to ZIKV/Malaysia with 2.48% amino acid divergence (Fig. 1A).
FIG 1.
ZIKV variants are genetically distinct, encoding sequence differences. Viral stocks were deep sequenced, and overlapping contigs were generated de novo to assemble amino acid sequences of the open reading frame of ZIKV strains. (A) The matrix presents the percent divergence of ZIKV amino acid sequences compared to an Asian lineage reference genome obtained from GenBank (KX377336). (B) The total number of amino acid substitutions in each ZIKV/Dakar protein compared to an Asian lineage reference strain is depicted. The substitutions are categorized as conservative or nonconservative. (C) Amino acid residues that have previously been demonstrated to impact viral fitness. Residues in bold are not shared between ZIKV/Dakar and ZIKV/Malaysia.
We identified several nonconservative and conservative amino acid substitutions in the ZIKV/Dakar structural and nonstructural proteins compared to the Asian lineage reference strain with the greatest number of substitutions located within the NS5 protein (Fig. 1B; Table 1). We found that five of the amino acid substitutions identified in our genomic analyses have been previously demonstrated to impart phenotypic differences in viral infectivity, virulence, and modulation of IFN production in mosquito and mammalian hosts and cell lines (Fig. 1C; Table 2) (31, 32, 35–37, 83–85). However, our sequencing revealed that ZIKV/Dakar and ZIKV/Malaysia carry different amino acid residues at two of these sites: position 1 of prM protein and position 188 of NS1. This mutation in prM has been shown to impact viral fitness and virulence in mammalian hosts, while the mutation in NS1 has been demonstrated to influence both viral infectivity in the mosquito vector and regulation of IFN induction in vitro and in vivo, suggesting that differences at these sites may contribute to distinct phenotypes between ZIKV/Dakar and ZIKV/Malaysia in mosquitoes and mammalian hosts and in IFN production during infection (31, 36, 85). We also identified several amino acid substitutions within the remaining nonstructural proteins that have not previously been directly demonstrated to contribute to differences in viral fitness or host interactions.
TABLE 1.
All amino acid differences between ZIKV/Dakar and an Asian lineage reference straina
| Position | Substitution | Type of substitution | Protein |
|---|---|---|---|
| 7 | K to N | Nonconservative | Capsid |
| 25 | S to N | Conservative | Capsid |
| 27 | F to L | Conservative | Capsid |
| 101 | K to R | Conservative | Capsid |
| 110 | V to I | Conservative | Capsid |
| 123 | V to A | Conservative | prb |
| 125 | V to I | Conservative | pr |
| 148 | P to A | Conservative | pr |
| 157 | Y to H | Nonconservative | pr |
| 158 | I to V | Conservative | pr |
| 246 | R to K | Conservative | Mb |
| 260 | A to L | Conservative | M |
| 262 | A to V | Conservative | M |
| 575 | S to F | Nonconservative | Eb |
| 607 | I to V | Conservative | E |
| 727 | A to V | Conservative | E |
| 728 | L to F | Conservative | E |
| 785 | L to M | Conservative | E |
| 846 | D to E | Conservative | NS1 |
| 863 | R to K | Conservative | NS1 |
| 926 | S to R | Nonconservative | NS1 |
| 956 | V to I | Conservative | NS1 |
| 982 | A to V | Conservative | NS1 |
| 988 | V to A | Conservative | NS1 |
| 1007 | K to R | Conservative | NS1 |
| 1030 | I to V | Conservative | NS1 |
| 1180 | I to M | Conservative | NS2A |
| 1191 | I to V | Conservative | NS2A |
| 1204 | A to V | Conservative | NS2A |
| 1226 | I to V | Conservative | NS2A |
| 1263 | V to A | Conservative | NS2A |
| 1270 | D to E | Conservative | NS2A |
| 1275 | I to V | Conservative | NS2A |
| 1297 | T to A | Nonconservative | NS2A |
| 1354 | L to I | Conservative | NS2A |
| 1461 | D to E | Conservative | NS2B |
| 1477 | T to A | Nonconservative | NS2B |
| 1553 | H to Q | Nonconservative | NS3 |
| 1558 | S to A | Nonconservative | NS3 |
| 1594 | H to L | Nonconservative | NS3 |
| 1620 | T to K | Nonconservative | NS3 |
| 1671 | R to K | Conservative | NS3 |
| 1676 | T to A | Conservative | NS3 |
| 1687 | K to R | Conservative | NS3 |
| 1717 | T to K | Nonconservative | NS3 |
| 1862 | V to I | Conservative | NS3 |
| 1909 | V to I | Conservative | NS3 |
| 1962 | I to V | Conservative | NS3 |
| 2085 | R to K | Conservative | NS3 |
| 2123 | F to L | Conservative | NS4A |
| 2127 | E to D | Conservative | NS4A |
| 2282 | L to I | Conservative | NS4B |
| 2283 | G to A | Conservative | NS4B |
| 2289 | R to K | Conservative | NS4B |
| 2293 | A to T | Nonconservative | NS4B |
| 2594 | Y to H | Nonconservative | NS5 |
| 2598 | I to V | Conservative | NS5 |
| 2621 | K to R | Conservative | NS5 |
| 2634 | T to M | Nonconservative | NS5 |
| 2659 | S to P | Nonconservative | NS5 |
| 2679 | A to T | Nonconservative | NS5 |
| 2715 | L to M | Conservative | NS5 |
| 2722 | Y to H | Nonconservative | NS5 |
| 2765 | D to E | Conservative | NS5 |
| 2795 | L to M | Conservative | NS5 |
| 2800 | N to R | Nonconservative | NS5 |
| 2802 | V to I | Conservative | NS5 |
| 2807 | S to N | Conservative | NS5 |
| 2842 | I to V | Conservative | NS5 |
| 2909 | H to R | Conservative | NS5 |
| 2969 | Q to H | Nonconservative | NS5 |
| 3039 | V to I | Conservative | NS5 |
| 3044 | S to N | Conservative | NS5 |
| 3046 | T to A | Nonconservative | NS5 |
| 3154 | Q to H | Nonconservative | NS5 |
| 3161 | R to K | Conservative | NS5 |
| 3167 | S to R | Nonconservative | NS5 |
| 3239 | Y to H | Nonconservative | NS5 |
| 3257 | I to F | Conservative | NS5 |
| 3304 | S to A | Nonconservative | NS5 |
| 3307 | V to I | Conservative | NS5 |
| 3333 | V to M | Conservative | NS5 |
| 3335 | W to C | Conservative | NS5 |
| 3402 | Y to F | Nonconservative | NS5 |
Column 1 indicates the position of amino acid residue relative to the polyprotein. Column 3 shows whether the amino acid substitution retains (conservative) or changes (nonconservative) the biochemical properties of the original amino acid residue.
pr, peptide; M, membrane; E, envelope.
TABLE 2.
Previously characterized ZIKV amino acid substitutions and their phenotypesa
| Protein | Position | Residue | Phenotype(s) |
|---|---|---|---|
| Capsid | 106 | Ala | Enhanced infectivity and fitness in mosquitoes and IFNAR KO mice (32, 35) |
| prM | 123 | Val | Enhanced infectivity in mosquito cells; increased frequency of fetal demise in IFNAR KO mice (36) |
| prM | 139 | Asn | Enhanced infectivity in human and murine neural progenitor cells; increased microcephaly severity in BALB/c mice (37) |
| Envelope | 443 | Asn | Enhanced infectivity and virulence in IFNAR KO mice and mosquitoes; reduced infectivity and entry into mosquito cells; reduced neurovirulence in BALB/c mice (83, 84, 115, 116) |
| NS1 | 982 | Val | Increased NS1 antigenemia and infectivity in mosquitoes; enhanced ability to block IFN induction (31, 85) |
Columns 1 to 3 indicate the viral protein, position relative to the viral polyprotein, and amino acid residue, respectively. Column 4 describes the phenotype associated with the specific amino acid residue as previously demonstrated.
ZIKV strains exhibit replication differences in immunocompetent cells.
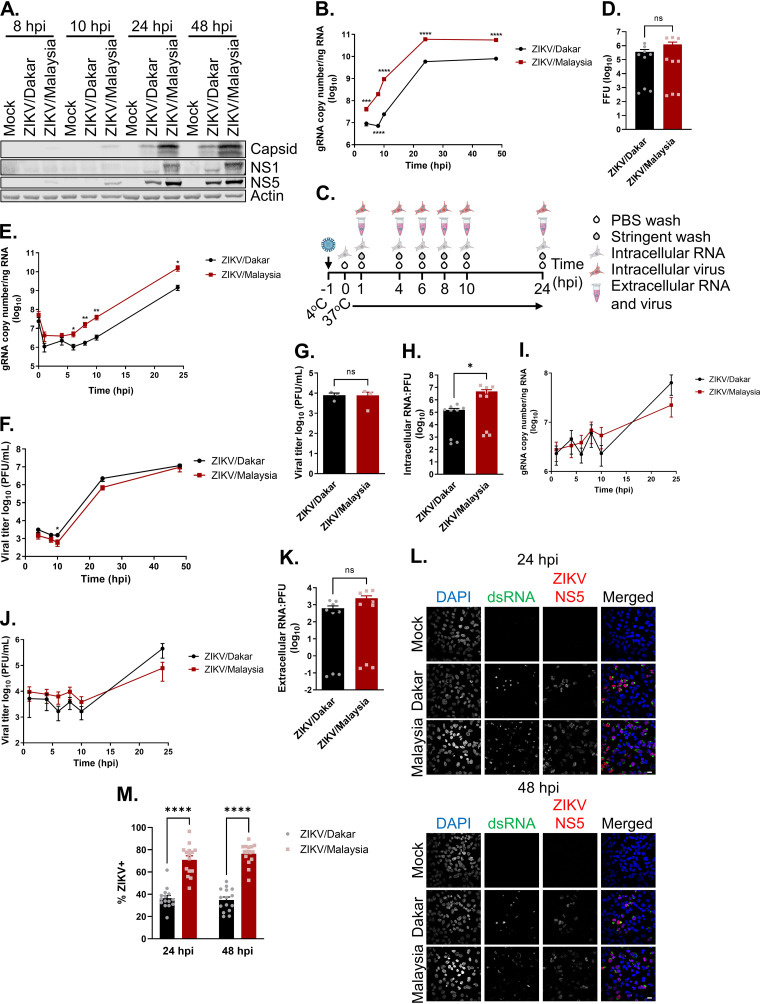
To evaluate strain-specific differences in viral replication, we carried out analyses to examine viral protein accumulation, genome replication, virion secretion, and spread of our ZIKV variants in an established immunocompetent human epithelial cell (A549) infection model (27, 43, 77). We carried out immunoblot analyses to evaluate the accumulation of viral proteins over a 48-h acute infection time course. These analyses revealed that ZIKV/Malaysia-infected samples accumulated detectable levels of the capsid protein, NS1, and NS5 by 8 h postinfection (hpi), while these viral proteins were not detectable in ZIKV/Dakar-infected samples until 24 hpi (Fig. 2A). We also observed that ZIKV/Malaysia-infected samples had greater abundance of the capsid protein, NS1, and NS5 throughout the infection time course than ZIKV/Dakar-infected samples. Next, we conducted viral growth analyses to characterize one-step growth kinetics of these variants during acute infection at a high multiplicity of infection (MOI). We observed that ZIKV/Malaysia replicated to significantly higher levels, with viral RNA peaking at 24 hpi and plateauing over the remainder of the time course (Fig. 2B). In comparison, ZIKV/Dakar did not reach the exponential phase of genome replication until 8 hpi and accumulated significantly fewer copies of genomic RNA (Fig. 2B). Given the stark differences in the accumulation of viral proteins and intracellular viral RNA between cells infected with ZIKV/Dakar and ZIKV/Malaysia, we investigated whether our ZIKV variants exhibited differences in viral attachment and entry that could possibly be linked to these outcomes (Fig. 2C). Viral attachment analyses demonstrated that ZIKV/Dakar and ZIKV/Malaysia exhibited similar efficiency of cell binding. Though we observed a trend towards increased viral attachment by ZIKV/Malaysia compared to ZIKV/Dakar, this difference was not statistically significant, indicating that the differences in the accumulation of viral proteins and intracellular genome copies are not due to differences in viral attachment to cells (Fig. 2D). We then assessed viral entry and observed that the intracellular viral RNA levels at 1 hpi were comparable between the ZIKV variants, indicating that these variants did not exhibit differences in viral entry (Fig. 2E). Analysis of genome replication revealed differences in viral RNA abundance at early time points during the time course, with higher levels exhibited by ZIKV/Malaysia than ZIKV/Dakar, which set the stage for higher viral RNA levels late in the time course (Fig. 2E). Indeed, we observed that ZIKV/Malaysia accumulated higher levels of intracellular viral RNA than ZIKV/Dakar at 24 hpi. Together, our findings demonstrate that ZIKV/Dakar and ZIKV/Malaysia differ in their genome replication efficiencies.
FIG 2.
ZIKV variants exhibit replication differences in immunocompetent cells. (A) A549 cells were mock infected or infected with ZIKV/Dakar or ZIKV/Malaysia over a 48-h time course at an MOI of 5. Cell lysates were analyzed via immunoblotting. One immunoblot representative of three independent experiments (n = 3) is presented. (B and F) A549 cells were infected with ZIKV/Dakar or ZIKV/Malaysia at an MOI of 5 over a 48-h time course. Results are representative of three independent experiments (n = 3). (B) Viral copy numbers were determined from RNA lysates by qRT-PCR. Depicted are the means from three biological and technical replicates that were pooled for statistical analysis. (C to E and G to K) A549 cells were mock-infected or infected with ZIKV variants over a 24-h time course at an MOI of 5. Results are representative of three independent experiments (n = 3). (C) Schematic of experimental setup. Synchronous infection was initiated by exposing virus to cells at 4°C followed by moving cultures to 37°C to initiate synchronous viral entry. (D) Virus/cell binding analysis. RNA lysates were harvested at 0 hpi, and viral copy numbers were determined by qRT-PCR and converted to FFUs. Results depict the means from three biological and technical replicates that were pooled for statistical analysis. (E) Viral RNA levels during synchronous infection. Intracellular viral copy numbers were quantified from RNA lysates by qRT-PCR. Results depict the means from three biological and technical replicates that were pooled for statistical analysis. (F) Viral titers of the cell culture supernatants were determined by plaque assay. Results are depicted as means. (G) Intracellular viral titers were determined from cell pellets from 24 hpi by plaque assay and depicted as means. (H) Intracellular RNA:PFU ratios. Ratios were calculated for ZIKV variants. Depicted are the means from three biological and technical replicates. (I and J) Cell culture supernatants were collected at the indicated time points. (I) The extracellular viral copy number was quantified via qRT-PCR. Results depict the means from three biological and technical replicates that were pooled for statistical analysis. (J) Extracellular viral titers were determined by plaque assay and depicted as means. (K) Extracellular RNA:PFU ratios. Ratios were calculated for ZIKV variants. Depicted are the means from three biological and technical replicates. (L and M) A549 cells were infected with ZIKV strains at an MOI of 5 to quantify the percentage of infected cells at each time point by immunofluorescence analysis. For each sample, five randomly selected fields of view representing at least 190 cells total were analyzed. Images were manually analyzed using the multipoint tool in Fiji. Scale bar = 10 μm. The percentage of ZIKV+ cells (defined as dsRNA+ or NS5+) is presented after normalizing to the total number of cells as determined by DAPI staining. Depicted are the means of five randomly selected fields from three independent experiments that were pooled for statistical analysis. For all figures, error bars represent the standard error of the mean (SEM). All statistical analyses were performed with a two-tailed unpaired t test at each time point (ns, not significant; *, P < 0.05; **, P < 0.005; ****, P < 0.0001).
We then evaluated virion production and found that cells infected with the ZIKV variants released similar amounts of infectious particles over the 48-h infection time course despite the significant differences in the accumulation of viral genome copies (Fig. 2F). To determine whether differences in virion assembly or secretion kinetics could account for this discrepancy, we employed a synchronous infection analysis as outlined in Fig. 2C. We observed that cells synchronously infected with the ZIKV variants had similar amounts of intracellular infectious particles at 24 hpi despite significant differences observed in the levels of intracellular viral RNA (Fig. 2E and G). Given this observation, we calculated the intracellular RNA:PFU ratios to evaluate the virion assembly efficiency of these variants, revealing that ZIKV/Dakar was significantly more efficient at virion assembly than ZIKV/Malaysia (Fig. 2H). When we quantified the numbers of extracellular infectious virions over the 24-h time course, we observed no differences in the levels of extracellular viral RNA and infectious particles (Fig. 2I and J). Comparisons of extracellular RNA:PFU ratios to measure the infectivity of progeny virions revealed no statistically significant differences in the infectivity of progeny produced by the ZIKV variants, though ZIKV/Malaysia trended toward a higher extracellular RNA:PFU ratio (Fig. 2K). These findings demonstrate that ZIKV/Dakar and ZIKV/Malaysia do not differ in their kinetics of virion secretion or infectivity of progeny virus; instead, they differed in virion assembly efficiency, with ZIKV/Dakar exhibiting enhanced virion assembly efficiency.
To evaluate the spread of ZIKV/Dakar and ZIKV/Malaysia over a 48-h acute infection time course, we carried out immunofluorescence analyses of cells stained with antibodies specific to double-stranded RNA (dsRNA; a marker for viral replication) and ZIKV NS5 protein to compare the percentage of infected cells during high-MOI infection with these variants. We observed that at both 24 and 48 hpi, ZIKV/Malaysia infected a significantly higher percentage of cells than ZIKV/Dakar as indicated by dsRNA and ZIKV NS5 staining (Fig. 2L and M). There was no significant increase in the percentage of infected cells for either ZIKV strain between 24 and 48 hpi, though there was a nonsignificant trend of increased ZIKV/Malaysia infection between these time points, suggesting that there was limited viral spread between these time points (Fig. 2M). As the percentage of infected cells between ZIKV variants exhibited only a 2-fold difference, it is likely that this difference contributes only minimally to the 10-fold difference in viral genome copies between ZIKV/Dakar and ZIKV/Malaysia (see Fig. 2B). Since we did not observe any significant differences in cell binding or viral entry between ZIKV/Dakar and ZIKV/Malaysia, it is possible that discrepancies in the interactions between ZIKV variants and host factors of viral control may differentially induce early antiviral responses that restrict viral infection. Together, our data clearly demonstrate that ZIKV/Dakar and ZIKV/Malaysia exhibit unique replication differences, specifically at the stages of viral protein accumulation, genome replication, and virion assembly that together impact viral spread.
ZIKV variants induce robust innate immune activation.
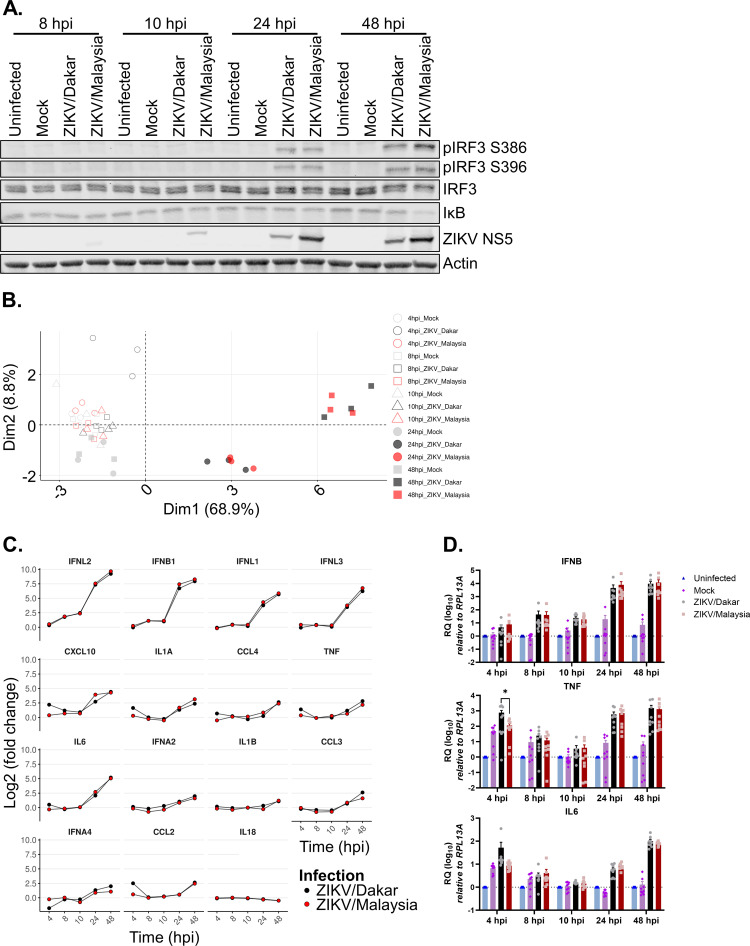
To investigate the contribution of innate immunity to replication differences of the ZIKV variants, we performed immunoblot analyses to evaluate activation of RLR signaling and response over a 48-h acute infection time course. We probed for phospho-IRF3 (pIRF3) S386 and S396 and IκB abundance as markers of IRF3 and NF-κB activation, respectively. At 24 and 48 hpi, both ZIKV/Dakar and ZIKV/Malaysia induced IRF3 and NF-κB activation to a similar extent, with ZIKV/Malaysia inducing RLR activation to a slightly greater extent at 48 hpi as indicated by the increased abundance of pIRF3 S386 and S396 and the decreased abundance of IκB (Fig. 3A; Table 3). Thus, both ZIKV variants activate RLR signaling, with ZIKV/Malaysia inducing moderately increased RLR signaling late in acute infection.
FIG 3.
ZIKV variants activate RLR signaling. (A and D) A549 cells were uninfected, mock-infected, or infected with ZIKV variants over a 48-h time course at an MOI of 5. (A) Cell lysates were analyzed via immunoblotting. Depicted is an immunoblot representative of three independent experiments (n = 3). (B and C) A549 cells were mock infected or infected with either ZIKV/Dakar or ZIKV/Malaysia over a 48-h time course at an MOI of 5. RNA lysates were collected, and transcript levels of cytokines and IFNs were quantified on the NanoString nCounter. Raw and processed expression data can be found in Table S1. Results are representative of three independent experiments (n = 3). (B) The PCA plot depicts changes in cytokine transcriptional profiles over the infection time course. (C) Log2 fold changes over mock infection in the transcript levels of GOIs are depicted individually for each gene. Statistical analysis was performed with a two-tailed t test followed by Benjamini-Hochberg (BH) correction for each time point. (D) RNA lysates were collected for qRT-PCR analysis. IFNB, TNF, and IL-6 transcript levels were normalized to the RPL13A housekeeping gene of uninfected samples at each time point. Results depict the means from three biological and technical replicates, which were pooled for statistical analysis; error bars represent SEMs. Statistical analysis was performed with a one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test for each time point. RQ, relative quantification.
TABLE 3.
Quantification of relevant immunoblot protein abundancea
| Figure | Protein | Mean abundance ± SEM at 24 hpib | Mean abundance ± SEM at 48 hpib | P value (24 hpi) | P value (48 hpi) |
|---|---|---|---|---|---|
| Fig. 3A | pIRF3 S386 | 0.347 ± 0.004 (ZIKV/Dakar) 0.294 ± 0.035 (ZIKV/Malaysia) |
0.812 ± 0.137 (ZIKV/Dakar) 1.128 ± 0.159 (ZIKV/Malaysia) |
0.2639 | 0.2080 |
| Fig. 3A | pIRF3 S396 | 0.378 ± 0.103 (ZIKV/Dakar) 0.282 ± 0.071 (ZIKV/Malaysia) |
0.985 ± 0.387 (ZIKV/Dakar) 1.033 ± 0.458 (ZIKV/Malaysia) |
0.4926 | 0.9407 |
| Fig. 3A | IκB | 0.236 ± 0.060 (ZIKV/Dakar) 0.176 ± 0.053 (ZIKV/Malaysia) |
0.231 ± 0.084 (ZIKV/Dakar) 0.088 ± 0.037 (ZIKV/Malaysia) |
0.4991 | 0.2268 |
| Fig. 4A | pSTAT1 | 0.805 ± 0.011 (ZIKV/Dakar) 0.764 ± 0.080 (ZIKV/Malaysia) |
1.517 ± 0.147 (ZIKV/Dakar) 0.366 ± 0.128 (ZIKV/Malaysia) |
0.6562 | 0.0044 |
| Fig. 4A | STAT1 | 0.588 ± 0.286 (ZIKV/Dakar) 0.393 ± 0.195 (ZIKV/Malaysia) |
1.460 ± 0.810 (ZIKV/Dakar) 0.358 ± 0.174 (ZIKV/Malaysia) |
0.6060 | 0.3052 |
| Fig. 4A | STAT2 | 0.260 ± 0.052 (ZIKV/Dakar) 0.094 ± 0.030 (ZIKV/Malaysia) |
0.490 ± 0.089 (ZIKV/Dakar) 0.037 ± 0.017 (ZIKV/Malaysia) |
0.0649 | 0.0328 |
| Fig. 4A | RIG-I | 0.659 ± 0.128 (ZIKV/Dakar) 0.602 ± 0.203 (ZIKV/Malaysia) |
1.607 ± 0.555 (ZIKV/Dakar) 0.640 ± 0.210 (ZIKV/Malaysia) |
0.8238 | 0.2171 |
| Fig. 4A | IFITM1 | 0.016 ± 0.008 (ZIKV/Dakar) 0.007 ± 0.001 (ZIKV/Malaysia) |
0.613 ± 0.099 (ZIKV/Dakar) 0.024 ± 0.009 (ZIKV/Malaysia) |
0.2845 | 0.0262 |
| Fig. 4A | OAS1 | 0.190 ± 0.128 (ZIKV/Dakar) 0.088 ± 0.045 (ZIKV/Malaysia) |
0.294 ± 0.192 (ZIKV/Dakar) 0.024 ± 0.019 (ZIKV/Malaysia) |
0.5193 | 0.2935 |
| Fig. 4A | Mx1 | 0.073 ± 0.045 (ZIKV/Dakar) 0.044 ± 0.034 (ZIKV/Malaysia) |
0.835 ± 0.274 (ZIKV/Dakar) 0.042 ± 0.023 (ZIKV/Malaysia) |
0.4866 | 0.1008 |
| Fig. 4A | IFIT1 | 0.380 ± 0.079 (ZIKV/Dakar) 0.180 ± 0.063 (ZIKV/Malaysia) |
1.223 ± 0.263 (ZIKV/Dakar) 0.228 ± 0.058 (ZIKV/Malaysia) |
0.1219 | 0.0575 |
The mean abundance of relevant proteins at 24 and 48 hpi from three independent experiments were quantified by densitometry and are depicted. The corresponding P-values are presented in columns 5 and 6.
Mean protein abundance derived from three independent experiments (n = 3). Statistical analysis was performed with a two-tailed unpaired t test with Welch’s correction at each time point.
As RLR activation and signaling stimulate the expression of IFN-β and other cytokines, we next analyzed a select cytokine mRNA expression profile induced in response to infection with ZIKV/Dakar or ZIKV/Malaysia at the transcriptional level using a custom NanoString innate immunity gene panel, which includes target genes of IRF3, NF-κB, IFN, and inflammatory cytokine signaling marking activation of these pathways (38, 41, 44, 45). We note that NanoString analysis provides a direct measure of the absolute abundance of the target mRNAs noted (86). We compared transcript levels from each infection condition to mock infection at each time point to calculate the fold change in gene expression. Principal-component analysis (PCA) revealed that infected samples clustered with mock-infected samples prior to 24 hpi (Fig. 3B). At 24 and 48 hpi, ZIKV/Dakar- and ZIKV/Malaysia-infected samples clustered away from the mock-infected samples, with all infected samples clustering close together at each of these time points, indicating that our samples clustered by infection condition and time postinfection. To gain insight into the cytokine-specific transcriptional profiles induced by ZIKV infection, we visualized the fold change in the expression levels of cytokines over our acute infection time course (Fig. 3C). Compared to mock infection, infection with ZIKV variants similarly induced the expression of several cytokines, especially types I and III IFNs, demonstrating innate immune activation of IRF3 and NF-κB signaling. To validate our NanoString results, we quantified transcript levels of IFNB, tumor necrosis factor (TNF), and interleukin-6 (IL-6) genes via reverse transcription-quantitative PCR (qRT-PCR) (Fig. 3D). Consistent with the results from our NanoString analysis, infection with ZIKV/Dakar and ZIKV/Malaysia induced the expression of these cytokines to a similar extent. Hence, our analyses of RLR signaling in response to infection with the ZIKV variants indicate that ZIKV/Dakar and ZIKV/Malaysia activate RLR signaling to the same extent to induce similar innate immune activation gene expression profiles.
ZIKV/Malaysia blocks IFN signaling to suppress ISG expression.
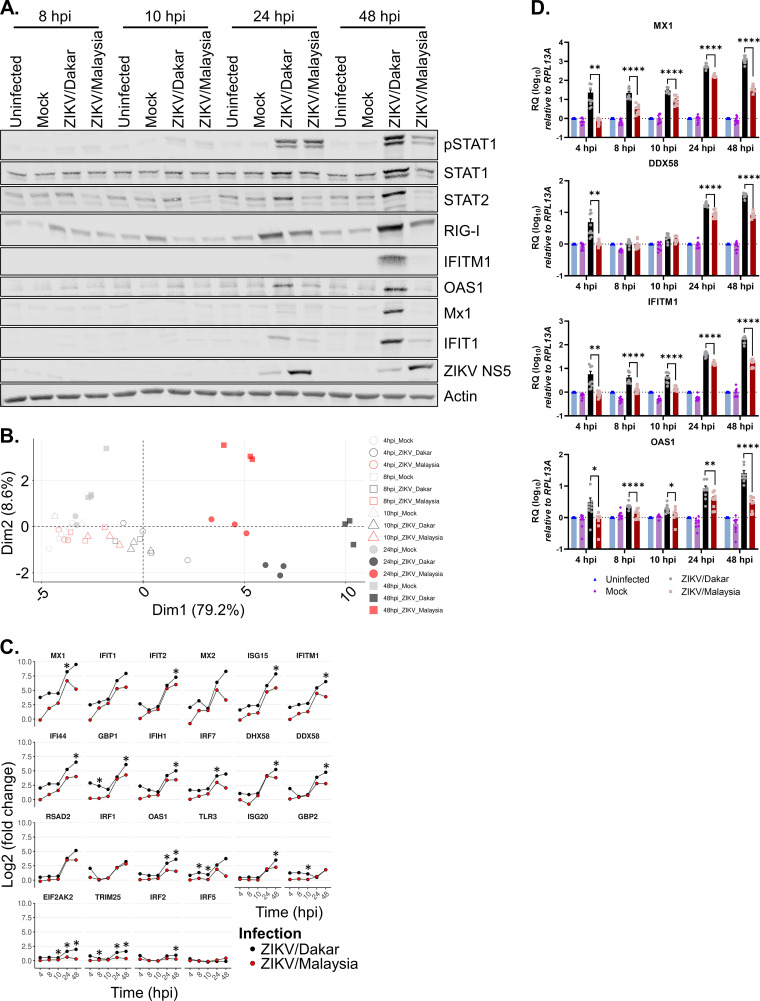
Given the robust induction of IFNs in response to acute ZIKV infection, we sought to examine the activation status of the JAK/STAT signaling pathway and the expression of ISGs at the protein level utilizing an immunoblot assay. At 24 hpi, ZIKV variants induced STAT1 phosphorylation (pSTAT1) to similar levels (Fig. 4A; Table 3). However, by 48 hpi, the levels of pSTAT1 induced by ZIKV/Dakar and ZIKV/Malaysia infections diverged, with lower pSTAT1 levels detected during ZIKV/Malaysia infection and higher pSTAT1 levels detected during ZIKV/Dakar infection. To evaluate whether the differential phosphorylation of STAT1 at 48 hpi resulted in differential downstream induction of ISGs, we examined the protein abundance for several ISGs, including STAT1, RIG-I, interferon-induced transmembrane protein 1 (IFITM1), 2′,5′-oligoadenylate synthetase 1 (OAS1), myxoma resistance protein 1 (Mx1), and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1). We observed that the protein abundance of these ISGs was lower during ZIKV/Malaysia infection than during ZIKV/Dakar infection, demonstrating that a reduction in pSTAT1 levels links with a reduction in ISG induction (Fig. 4A; Table 3). Additionally, we assessed STAT2 abundance, as ZIKV has been shown to target STAT2 for proteasomal degradation as an additional mechanism to antagonize JAK/STAT signaling (74, 75). We observed that STAT2 levels were consistently lower during ZIKV/Malaysia infection throughout the acute infection time course (Fig. 4A; Table 3). Indeed, we observed STAT2 reduction as early as 8 hpi, which coincided with detection of ZIKV/Malaysia NS5. Thus, despite the robust acute induction of IFNs during infection with both ZIKV variants, we observed a lower level of IFN signaling during infection with ZIKV/Malaysia than during ZIKV/Dakar infection, indicating that ZIKV/Malaysia can block IFN signaling more efficiently than ZIKV/Dakar.
FIG 4.
ZIKV/Malaysia robustly blocks IFN signaling to suppress ISG expression. (A and D) A549 cells were uninfected, mock infected, or infected with ZIKV variants over a 48-h time course at an MOI of 5. (A) Cell lysates were analyzed by immunoblotting. Depicted is one immunoblot that is representative of three independent experiments (n = 3). (B and C) A549 cells were mock infected or infected with either ZIKV/Dakar or ZIKV/Malaysia over a 48-h time course at an MOI of 5, and RNA lysates were collected. RNA transcript levels of a panel of ISGs were quantified on the NanoString nCounter. Raw and processed expression data can be found in Table S1. Results are representative of three independent experiments (n = 3). (B) The PCA plot shows the impact of each infection condition on the ISG transcriptome over the infection time course. (C) The log2 fold change in the transcript levels of ISGs is depicted for each gene. Statistical analysis was performed with a two-tailed t test followed by BH correction for each time point. (D) RNA lysates were collected for qRT-PCR analysis. MX1, IFITM1, DDX58, and OAS1 transcript levels were normalized to the RPL13A housekeeping gene of uninfected samples at each time point. Results depict the mean values from three biological and technical replicates that were pooled for statistical analysis; error bars represent the SEMs. Statistical analysis was performed with a one-way ANOVA followed by Tukey’s multiple-comparison test (*, P < 0.05; **, P < 0.005; ****, P < 0.0001).
To further determine the effects of innate immune activation and JAK/STAT signaling regulation on downstream gene expression during infection, we evaluated the IRF3-target gene and ISG profiles induced by infection with the ZIKV variants at the transcriptional level using our custom NanoString innate immunity gene panel. We compared transcript levels of ISGs from each infection condition to mock infection at each time point to derive the fold change in gene expression. PCA revealed that IRF3-target gene and ISG expression profiles were similar between mock-infected and infected samples up to 24 hpi (Fig. 4B). At 24 and 48 hpi, samples infected with ZIKV/Dakar and ZIKV/Malaysia exhibited distinct clustering patterns, with samples clustered based on ZIKV variant and time. Compared to mock infection, infection with ZIKV/Dakar and ZIKV/Malaysia induced initial transcript levels of IRF3-target genes and ISGs in our panel at early time points postinfection (Fig. 4C). In accordance with our immunoblot results, we observed that infection with ZIKV/Malaysia suppressed the transcriptional induction of a majority of ISGs in our panel by 24 hpi compared to infection with ZIKV/Dakar, which induced ISGs to higher levels overall. We utilized qRT-PCR to quantify transcript levels of the canonical ISGs, MX1, IFITM1, DDX58, and OAS1, as a means of validating our results from our NanoString analysis (Fig. 4D). We observed that ZIKV/Dakar infection induced the expression of these ISGs to significantly higher levels, confirming our findings from our NanoString analysis. Altogether, our data reveal a striking and unique immunophenotype wherein two prototypic ZIKV strains of different lineages induce similar IRF3 and NF-κB signaling profiles but differentially modulate IFN signaling to regulate downstream expression of ISGs, driving distinct innate immune signatures.
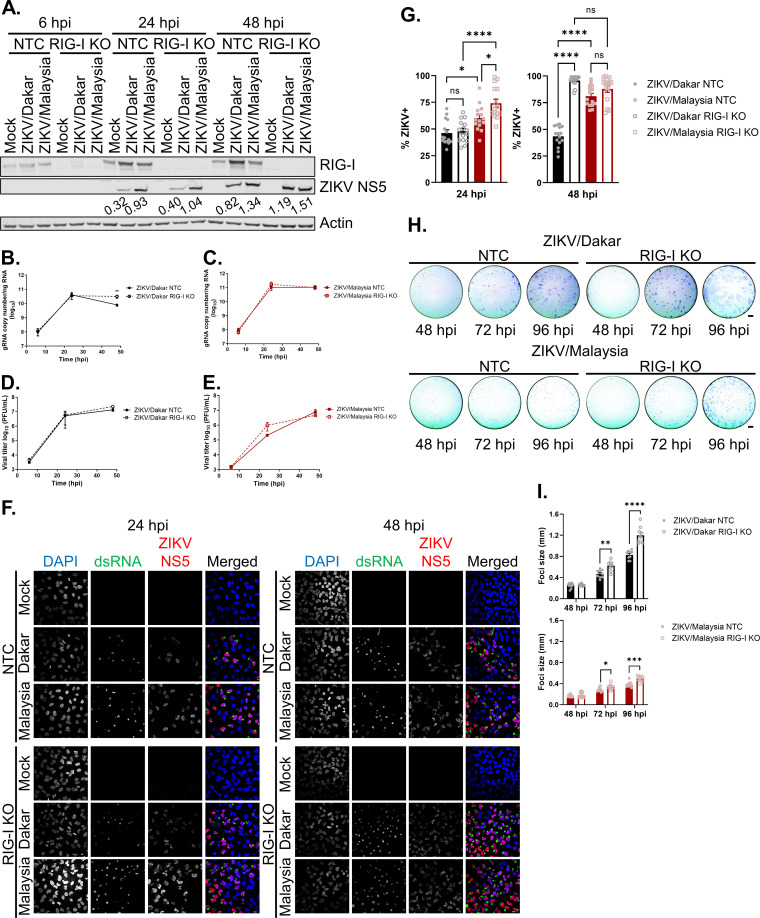
RIG-I-mediated signaling differentially restricts the accumulation of ZIKV/Dakar proteins and viral genome and suppresses spread of ZIKV variants late in acute infection.
To evaluate the role of RIG-I in innate immune control of ZIKV variants, we utilized A549 RIG-I KO cells and assessed the contribution of the antiviral actions of RIG-I signaling on different stages of the ZIKV life cycle. A549 RIG-I KO cells are defective in RLR signaling and do not produce IFN in response to ZIKV infection (27). As a control, we assessed ZIKV infection in A549 nontargeting control (NTC) cells. We infected NTC and RIG-I KO cells with the ZIKV variants over a 48-h infection time course at a high MOI to evaluate the following stages of the viral life cycle: viral protein accumulation, genome replication, infectious virion release, and viral spread. Immunoblot analyses revealed that ZIKV/Malaysia-infected samples accumulated NS5 to a greater abundance in both NTC and RIG-I KO cells at 24 hpi than ZIKV/Dakar-infected samples indicating intrinsic differences in the rate of viral protein accumulation between samples infected with ZIKV/Dakar and ZIKV/Malaysia (Fig. 5A). However, at 48 hpi, levels of NS5 across our ZIKV variants were comparable in RIG-I KO cells, whereas in NTC cells, the levels of ZIKV/Dakar NS5 were lower than in cells infected with ZIKV/Malaysia. Our findings reveal that RIG-I-mediated antiviral signaling directs a response that restricts viral protein accumulation by ZIKV/Dakar, but not ZIKV/Malaysia, at later time points of acute infection.
FIG 5.
RIG-I-mediated signaling restricts the accumulation of ZIKV/Dakar proteins and viral genome and universally suppresses ZIKV spread. (A) A549 NTC and RIG-I KO cells were mock infected or infected with ZIKV/Dakar or ZIKV/Malaysia over a 48-h time course at an MOI of 5. Cell lysates were analyzed via immunoblotting. Depicted is one immunoblot that is representative of three independent experiments (n = 3). ZIKV NS5 immunoblot band intensities were quantified in Fiji and are indicated beneath the corresponding bands. (B to E) A549 NTC and RIG-I KO cells were infected with (B and D) ZIKV/Dakar or (C and E) ZIKV/Malaysia over a 48-h time course at an MOI of 5. (B and C) RNA lysates were analyzed by qRT-PCR to quantify viral copy numbers. Depicted are the means from three biological and technical replicates that were pooled for statistical analysis. Statistical analysis was performed with a two-tailed unpaired t test at each time point (**, P < 0.005). (D and E) Cell culture supernatants were analyzed by plaque assay to quantify the viral titers. Depicted are the means from two to three independent experiments (n = 2 to 3). Statistical analysis was performed with a two-tailed unpaired t test for each time point. (F and G) A549 NTC and RIG-I KO cells were infected with ZIKV/Dakar or ZIKV/Malaysia at an MOI of 5. Samples were analyzed by immunofluorescence staining for dsRNA and ZIKV NS5. For each sample, five randomly selected fields of view representing at least 300 cells total were analyzed. Images were manually analyzed using the multipoint tool in Fiji. Scale bar = 10 μm. The percentage of ZIKV+ cells (defined as dsRNA+ or NS5+) is presented after normalizing to the total number of cells as defined by DAPI staining. Depicted are the means of five randomly selected fields from each of the three independent experiments (n = 3). Data from five randomly selected fields from each of the three independent experiments were pooled for statistical analysis. Statistical analysis was performed with a one-way ANOVA followed by Tukey’s multiple-comparison test (ns, not significant; *, P < 0.05; ****, P < 0.0001). (H and I) A549 NTC and RIG-I KO cells were infected with serial dilutions of ZIKV/Dakar or ZIKV/Malaysia over a 96-h time course. Cells were fixed and stained for viral envelope protein to detect foci of infection. Depicted are representative images of foci from three independent experiments (n = 3). For each of the three technical replicates, the diameter of 10 randomly selected foci was measured. Scale bar = 1 mm. Graphs depict the means of 10 randomly selected foci from each of the three technical replicates across three independent experiments. Statistical analysis was performed with a two-tailed unpaired t test for each time point (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001). For all figures, error bars represent the SEM.
Next, we carried out paired virus growth analyses of the ZIKV variants at a high MOI in NTC and RIG-I KO cells to examine the impact of RIG-I signaling on viral genome replication and virion release. We observed that levels of intracellular viral RNA for each ZIKV strain were similar between NTC and RIG-I KO cells at 6 and 24 hpi (Fig. 5B and C). At 48 hpi, however, we found significantly more ZIKV/Dakar RNA in RIG-I KO cells than in NTC cells, demonstrating that RIG-I signaling is critical in restricting ZIKV/Dakar genome replication as observed previously with ZIKV and other flaviviruses (Fig. 5B) (27, 43, 60). On the other hand, we did not observe any differences in viral RNA levels at 48 hpi between NTC and RIG-I KO cells infected with ZIKV/Malaysia, suggesting that RIG-I signaling cannot restrict replication of this variant at the MOI used (Fig. 5C). Quantification of infectious particles revealed no differences in infectious viral titers between ZIKV variants across cell types (Fig. 5D and E). Our findings indicate that ZIKV/Dakar, but not ZIKV/Malaysia, genome replication is restricted by RIG-I-mediated antiviral signaling at late acute infection time points and reveal that infectious virion release by cells infected with ZIKV/Dakar or ZIKV/Malaysia is insensitive to RIG-I signaling.
To evaluate the impact of RIG-I signaling on virus spread, we infected NTC and RIG-I KO cells at a high MOI and quantified the percentage of infected cells at 24 and 48 hpi. Similar to what we observed in wild-type cells, ZIKV/Malaysia infected a significantly higher percentage of cells at 24 hpi in both NTC and RIG-I KO cultures than ZIKV/Dakar (Fig. 5F and G). ZIKV/Malaysia also infected a significantly higher percentage of RIG-I KO cells than NTC cells at 24 hpi, indicating that RIG-I-mediated antiviral signaling can restrict ZIKV/Malaysia spread early in acute infection under these high-MOI conditions. We observed no differences in the percentages of ZIKV/Malaysia-infected cells at 48 hpi between NTC and RIG-I KO cultures during acute infection at a high MOI. We also found that ZIKV/Malaysia trended towards increased viral spread at 48 hpi in NTC cells, similar to our observations in wild-type cells, though this trend is more prominent in NTC cells. We attribute this difference between wild-type and NTC cells to experimental variation. Remarkably, however, ZIKV/Dakar infected significantly more cells at 48 hpi in RIG-I KO cultures compared to NTC cultures, reaching a similar percentage of infected cells as ZIKV/Malaysia in RIG-I KO cultures. This observation underscores the importance of RIG-I-mediated signaling in restricting ZIKV/Dakar spread late in acute infection. To further evaluate the impact of RIG-I-mediated antiviral signaling on ZIKV spread, we infected NTC and RIG-I KO cells using serial virus dilutions that yielded isolated foci and measured viral spread in the presence of a methylcellulose overlay over a 96-h infection time course. Interestingly, the foci formed during ZIKV/Dakar infection were larger than those formed during ZIKV/Malaysia infection in both NTC and RIG-I KO cells even though ZIKV/Malaysia exhibits enhanced viral spread compared to ZIKV/Dakar in the absence of an overlay (Fig. 5F to I). As each ZIKV/Dakar-infected cell produces more infectious virions than ZIKV/Malaysia-infected cells, the methylcellulose overlay likely concentrates more ZIKV/Dakar virions than ZIKV/Malaysia virions on the surfaces of neighboring uninfected cells to promote infection, thereby resulting in formation of larger foci during ZIKV/Dakar infection. We observed that the foci of infection formed by ZIKV/Dakar and ZIKV/Malaysia were similar in diameter between NTC and RIG-I KO cells at 48 hpi (Fig. 5H and I). However, at 72 and 96 hpi, the diameters of foci for ZIKV/Dakar and ZIKV/Malaysia were increased in RIG-I KO cells compared to NTC cells, indicating that viral spread was enhanced in the absence of RIG-I, particularly at late time points of infection. Thus, our results clearly demonstrate that RIG-I-mediated innate immune activation is critical in restricting ZIKV infection by restricting protein accumulation and genome replication late in acute infection to control viral spread.
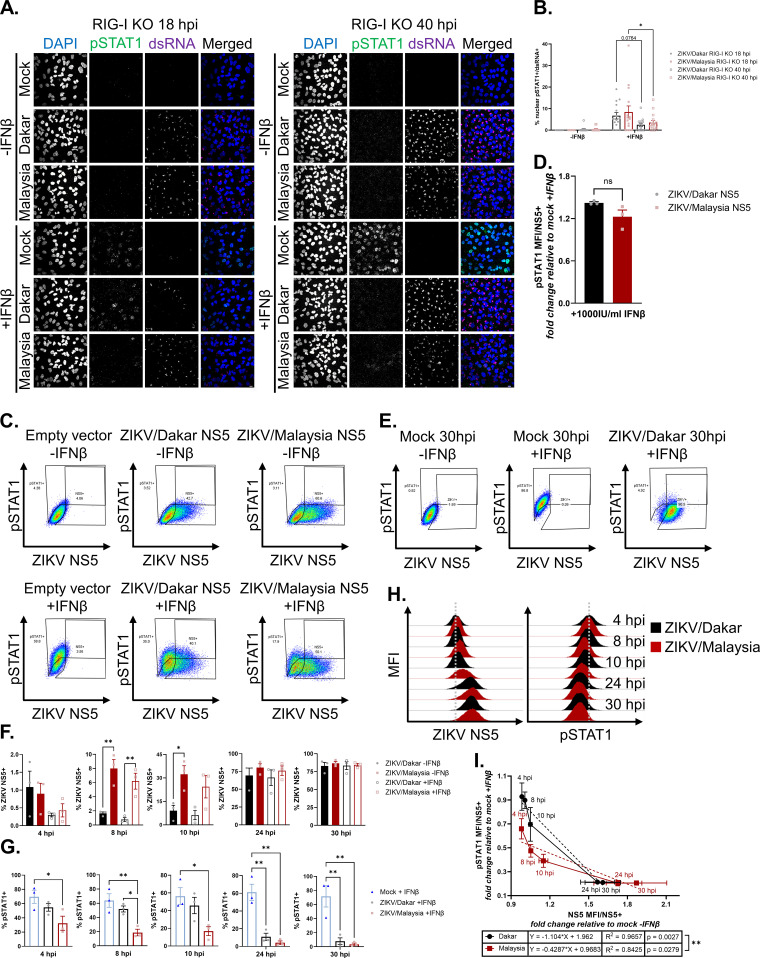
Early accumulation of ZIKV NS5 during ZIKV/Malaysia infection confers enhanced ability to block IFN-β signaling.
To determine how differences in the kinetics of viral protein accumulation impact the response to IFN in cells infected with ZIKV/Dakar and ZIKV/Malaysia, we assessed the IFN response in infected RIG-I KO cells, as they lack the ability to produce IFN-β in response to ZIKV infection. This approach allowed us to exogenously introduce IFN-β to evaluate ZIKV-mediated JAK/STAT blockade comparatively across the ZIKV variants independent of RIG-I-mediated innate immune activation. We infected RIG-I KO cells with either ZIKV/Dakar or ZIKV/Malaysia, subsequently mock-treated or IFN-β-treated cells early (at 18 hpi) or late (at 40 hpi) in infection, and analyzed the percentage of IFN-responsive cells via immunofluorescence assays. We quantified only the percentage of ZIKV-infected cells (defined as dsRNA+ cells) that had nuclear pSTAT1 signal to exclude any possible IFN response from uninfected or bystander cells within the cultures. At both 18 and 40 hpi, we observed no differences in the percentages of ZIKV/Dakar- and ZIKV/Malaysia-infected cells that were responsive to exogenous IFN-β, indicating that the ZIKV variants did not differ in their ability to antagonize IFN-β signaling (Fig. 6A and B). We found that the percentages of infected cells responsive to IFN-β stimulus were significantly reduced at 40 hpi compared to 18 hpi. This reduction in IFN responsiveness coincided with an increase in viral proteins (see Fig. 5A), suggesting that the extent and ability of ZIKV-mediated blockade of IFN signaling are dependent on viral protein accumulation over the course of an acute infection.
FIG 6.
Early accumulation of ZIKV/Malaysia NS5 confers enhanced ability to antagonize IFN-β signaling. (A and B) A549 RIG-I KO cells were mock infected or infected with ZIKV/Dakar or ZIKV/Malaysia at an MOI of 5 and then mock treated or treated with 100 IU/mL of IFN-β for 1 h for immunofluorescence analysis. For each sample, five randomly selected fields of view representing at least 310 cells total were analyzed. Z-stacks with a 2.5 μm step size were acquired and maximum Z-projections were generated in Fiji for manual analysis with the multipoint tool in Fiji. Scale bar represents 10 μm. The percentage of nuclear pSTAT1+ cells is presented after normalizing to the number of infected cells (defined as dsRNA+). Depicted are means of five randomly selected fields from three independent experiments (n = 3), which were pooled for statistical analysis with a two-way ANOVA followed by Tukey’s multiple-comparison test (*, P < 0.05). (C, D) HEK293T cells were transfected with an empty vector, ZIKV/Dakar NS5 construct, or ZIKV/Malaysia NS5 construct and treated with 1000 IU/mL of IFN-β for 8 h. (C) Gating strategy to identify transfected cells (defined as NS5+) and IFN responsive cells (defined as pSTAT1+). (D) pSTAT1 MFIs of transfected cells treated with IFN-β are normalized to pSTAT1 MFI of transfected, mock-treated cells. Depicted are means from three independent experiments (n = 3). Statistical analysis was performed with a two-tailed unpaired t test (ns: not significant). (E – I) A549 RIG-I KO cells were mock infected or infected with ZIKV/Dakar or ZIKV/Malaysia at an MOI of 5 over a 30-h time course and mock-treated or treated with 100 IU/mL of IFN-β for 30 min. Results are representative of three independent experiments (n = 3). (E) Gating strategy to identify infected cells (defined as NS5+) and IFN-responsive cells (defined as pSTAT1+). (F) Depicted is the mean percentage of infected cells of the total cell population. Statistical analysis was performed with a one-way ANOVA followed by Tukey’s multiple-comparison test (*, P < 0.05; **, P < 0.005). (G) Depicted is the mean percentage of pSTAT1+ cells of the total cell population. Statistical analysis was performed with a one-way ANOVA followed by Tukey’s multiple-comparison test (*, P < 0.05; **, P < 0.005). (H) Representative histograms of ZIKV NS5 and pSTAT1 MFIs of the total cell population during each infection time course. Gray dotted lines represent the centers of the histograms for ZIKV/Dakar-infected samples at 4 hpi as a reference point for visualizing the shift in NS5 and pSTAT1 histograms. (I) Depicted are the mean NS5 and pSTAT1 MFIs of the infected cell population over each infection time course. Statistical analysis was performed with linear regressions (**, P < 0.005). For all figures, error bars represent the SEM.
ZIKV NS5 has been demonstrated to be a potent IFN antagonist (74, 77, 78, 87). Since we identified 29 substitutions between ZIKV/Dakar and ZIKV/Malaysia NS5 proteins (see Fig. 1B), we evaluated the ability of these divergent NS5 proteins to modulate IFN signaling outside the context of infection. HEK293T cells expressing either ZIKV/Dakar NS5, ZIKV/Malaysia NS5, or the empty pcDNA3.1+ vector were mock-treated or treated with a high dose of IFN-β. We stained cells for NS5 and pSTAT1 to identify transfected cells and IFN-responsive cells, respectively (Fig. 6C). To account for differences in transfection efficiency between samples, we measured pSTAT1 median fluorescence intensities (MFIs) only within the NS5+ population. Compared to each other, cells expressing ZIKV/Dakar NS5 or ZIKV/Malaysia NS5 and treated with IFN-β both suppressed pSTAT1 accumulation. We observed a trend towards greater suppression of pSTAT1 accumulation by ZIKV/Malaysia NS5 than ZIKV/Dakar NS5, but this difference was not statistically significant (Fig. 6D). Thus, the NS5 protein from ZIKV variants of distinct lineages can suppress the JAK/STAT pathway. Together, our results indicate that the differential modulation of IFN signaling observed during ZIKV/Dakar and ZIKV/Malaysia infections in immunocompetent cells is not due to their divergent NS5 proteins (see Fig. 4).
Since we observed that cells infected with ZIKV/Dakar and ZIKV/Malaysia and cells expressing ZIKV/Dakar and ZIKV/Malaysia NS5 proteins antagonized IFN signaling equally, we hypothesized that the IFN responsiveness of cells is dependent on the rate of ZIKV protein accumulation. To test this hypothesis, we mock infected or infected A549 RIG-I KO cells to exclude endogenous IFN production during infection that otherwise occurs in immunocompetent cells. At various time points over a 30-h infection time course, we mock treated or IFN-β-treated the RIG-I KO cells to evaluate the IFN responsiveness of both the total cell population and the infected cell population via flow cytometry analysis. We stained cells for NS5 and pSTAT1 to identify infected cells and IFN-responsive cells, respectively (Fig. 6E). Over the course of infection, we observed that the percentage of infected RIG-I KO cells increased for both ZIKV variants, with ZIKV/Malaysia infecting a significantly higher percentage of cells than ZIKV/Dakar prior to 24 hpi (Fig. 6F). However, at 24 and 30 hpi, the percentages of infected cells were similar between ZIKV/Dakar- and ZIKV/Malaysia-infected samples. Quantification of the percentage of IFN-responsive cells revealed a decrease in the percentage of pSTAT1+ cells in infected RIG-I KO cultures over time compared to mock-infected samples (Fig. 6G). Remarkably, we observed a reduction in the percentage of pSTAT1+ cells in ZIKV/Malaysia-infected RIG-I KO cultures as early as 4 hpi compared to mock- and ZIKV/Dakar-infected cultures, despite ZIKV/Malaysia-infected samples expressing levels of NS5 below the limit of detection at this time point. Compared to ZIKV/Dakar, ZIKV/Malaysia-infected RIG-I KO cultures had fewer pSTAT1+ cells through 24 hpi, with only statistically significant differences in the percentage of pSTAT1+ cells between ZIKV/Dakar and ZIKV/Malaysia infections observed at 8 hpi. At 24 and 30 hpi, when the percentages of infected cells were comparable between the two variants, both ZIKV/Dakar- and ZIKV/Malaysia-infected samples had significantly lower percentages of pSTAT1+ cells following IFN-β treatment than mock-infected samples. We did not observe differences in the percentages of pSTAT1+ cells between ZIKV/Dakar- and ZIKV/Malaysia-infected cultures at these time points. We observed similar trends when we plotted NS5 and pSTAT1 MFIs of the total cell population (Fig. 6H). NS5 MFI increased over time during infection with either ZIKV strain, while pSTAT1 MFI decreased over the course of both ZIKV/Dakar and ZIKV/Malaysia infections in the RIG-I KO cultures. Of note, compared to ZIKV/Malaysia-infected samples, ZIKV/Dakar-infected samples displayed a delayed increase in NS5 MFI at 4, 8, and 10 hpi, which coincided with a delayed decrease in pSTAT1 MFI at these time points. We observed that NS5 and pSTAT1 MFIs between samples infected with either ZIKV variant were similar at 24 and 30 hpi, suggesting that ZIKV/Dakar and ZIKV/Malaysia can block IFN signaling in the absence of RIG-I when levels of NS5 are equivalent. These findings demonstrate that the IFN responsiveness of ZIKV-infected cultures is inversely correlated with ZIKV NS5 levels.
We next evaluated whether ZIKV NS5 levels significantly predict the IFN responsiveness of infected cells. We plotted NS5 MFI values against pSTAT1 MFI values of infected cells that were treated with IFN-β (Fig. 6I). NS5 and pSTAT1 MFIs were normalized to those of mock-infected IFN-β-treated samples for each time point. NS5 MFIs between ZIKV/Dakar- and ZIKV/Malaysia-infected cells were similar at 4 hpi but differed between 8 and 24 hpi, with ZIKV/Malaysia-infected cells having higher NS5 MFI values between these time points. At 24 and 30 hpi, NS5 MFIs were similar between RIG-I KO cells infected with either ZIKV variant, indicating that levels of NS5 within ZIKV/Dakar- and ZIKV/Malaysia-infected cells were comparable. We observed that pSTAT1 MFIs were different between cells infected with the ZIKV strains as early as 4 hpi, with ZIKV/Dakar-infected cells having higher pSTAT1 MFI values prior to 24 hpi. These observations demonstrate that the differential ability to block IFN-β signaling early in infection is due to differences in NS5 levels in infected cells during these early time points. At 24 and 30 hpi, when NS5 levels were similar between ZIKV/Dakar- and ZIKV/Malaysia-infected RIG-I KO cells, pSTAT1 MFI values were the same between cells infected with either variant. Linear regression analyses confirmed that NS5 MFI of infected cells was inversely correlated with pSTAT1 MFI of infected cells and revealed that NS5 MFI of infected cells significantly predicted pSTAT1 MFI of cells infected with the ZIKV variants. Comparison of the regression slopes revealed that they were significantly different between the ZIKV variants, indicating that ZIKV/Dakar and ZIKV/Malaysia differ in their rate of NS5 accumulation and modulation of IFN-β signaling (Fig. 6I). Taken together, our results clearly demonstrate that the extent of ZIKV-mediated IFN blockade is predominantly dependent on the rate and levels of ZIKV NS5 accumulation in infected cells, such that ZIKV/Dakar and ZIKV/Malaysia do not differ in their ability to block IFN signaling when levels of NS5 are comparable in infected cells. Thus, ZIKV NS5 expression and accumulation kinetics are major determinants of viral control mediated by RIG-I-driven IFN production and signaling.
DISCUSSION
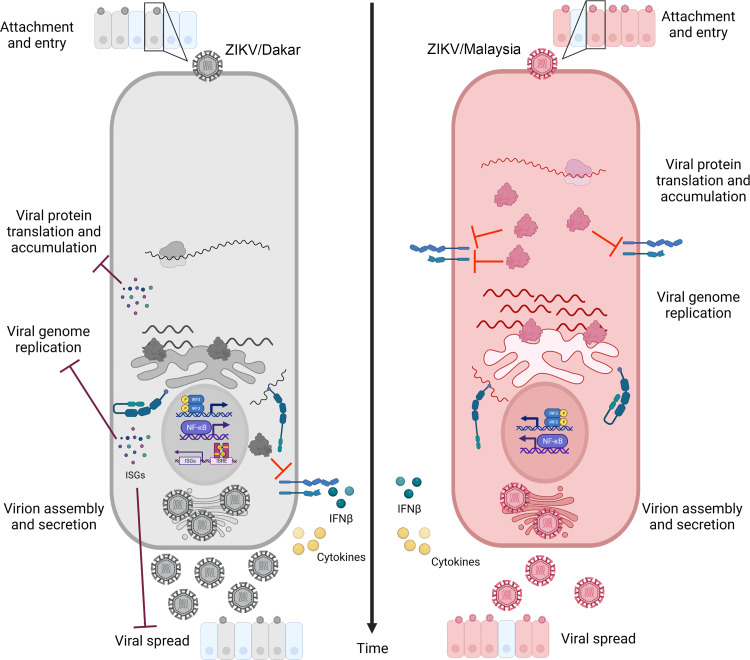
To provide a deeper understanding of differences between ZIKV variants that might impart disease, we conducted virologic and host innate immune analyses between African lineage ZIKV/Dakar and Asian lineage ZIKV/Malaysia. We found that immunocompetent cells infected with ZIKV/Malaysia accumulate viral proteins and genome copies earlier in infection and to higher levels than cells infected with ZIKV/Dakar. Examination of the innate immune response profiles initiated upon infection demonstrated that both variants induced innate immune activation marked by expression of IRF3 and NF-κB target genes, including high levels of IFN-β. Despite the robust IFN induction, ZIKV/Malaysia blocked downstream induction of ISGs, while ZIKV/Dakar did not. Our subsequent analyses in NTC and RIG-I KO cells revealed that RIG-I drives an innate immune response that restricts ZIKV protein accumulation, wherein early accumulation of viral proteins confers enhanced antagonism of IFN signaling, supporting efficient genome replication and viral spread late in acute infection. Based on our findings, we propose a model of RIG-I action during ZIKV infection in which RIG-I recognition of ZIKV PAMPs induces IRF3 and NF-κB activation, leading to innate immune activation with IFN-β production and response (42). The IFN response is controlled by the viral NS5 protein and subjected to differential regulation across viral variants through the kinetics and levels of NS5 accumulation. RIG-I and rapid accumulation of NS5 are key features of this model that, respectively, direct acute innate immune activation and a response that can restrict ZIKV replication and cell-to-cell spread and mediate suppression of IFN signaling to support overall infection progression (Fig. 7). This model highlights the “arms race” between the virus and host to control the outcome of infection. Our comparative analyses show that ZIKV/Malaysia wins the “arms race” over ZIKV/Dakar because it can quickly accumulate NS5 to levels for suppression of IFN signaling. Thus, infection with ZIKV/Malaysia may be linked to worse disease outcome than infection with ZIKV/Dakar.
FIG 7.
Early accumulation of ZIKV proteins impacts the host innate immune response and control of viral replication and spread; model figure demonstrating how early accumulation of viral proteins impacts the host innate immune response to ZIKV/Dakar (left) and ZIKV/Malaysia (right) infections. Early accumulation of ZIKV/Malaysia proteins confers enhanced ability to block IFN-β signaling and ISG induction compared to ZIKV/Dakar. Activation of RIG-I-mediated antiviral signaling restricts ZIKV/Dakar protein accumulation, thereby limiting viral genome accumulation and spread.
Other studies have previously demonstrated that African lineage ZIKV strains replicate to higher levels and produce higher viral titers than Asian lineage variants, but these studies primarily compared African or pre-epidemic Asian lineage viruses with epidemic Asian lineage strains (26–28, 30, 88–93). Moreover, comparative analyses of a pre-epidemic Asian lineage ZIKV variant (ZIKV/Cambodia/2010/FSS13025; ZIKV/Cambodia) with African lineage viruses and epidemic strains in mosquitoes and mammalian models have demonstrated that Asian lineage-derived emerging variants and African lineage viruses exhibit higher replicative fitness than ZIKV/Cambodia (27, 35). Liu et al. identified four amino acid substitutions (T106A in capsid, V1A in prM, A188V in NS1, and M872V in NS5) present in epidemic ZIKV strains but absent in ZIKV/Cambodia that represent direct reversions to residues encoded by African lineage viruses, suggesting that mutations that likely enabled adaptation to the mosquito vectors and mammalian hosts in Asia occurred following the isolation of the prototypic ZIKV/Malaysia isolate and spread of ZIKV throughout Asia (35). Reversion of these four amino acid residues in emerging ZIKV strains was sufficient to restore viral fitness in mosquitoes and mammalian hosts (35). Our findings, however, indicate that the prototypic Asian lineage virus, ZIKV/Malaysia, exhibits enhanced replicative fitness in A549 cells compared to a prototypic African lineage virus, ZIKV/Dakar. As ZIKV/Malaysia was isolated several decades prior to ZIKV/Cambodia, there are likely genetic differences between these two Asian lineage variants. Indeed, we revealed in our sequencing analysis that ZIKV/Malaysia encodes an alanine at position 106 of the capsid protein and a valine at position 872 of NS5, suggesting that the presence of these residues shared with African lineage viruses contributes to the enhanced replicative fitness of ZIKV/Malaysia observed in our study. However, we found that ZIKV/Malaysia encodes the alanine mutation at position 188 of NS1 associated with reduced replicative fitness (35). As the capsid T106A mutation is known to exert a strong effect on viral fitness, it is possible that the effect of this alanine in the capsid protein on viral fitness overcomes the impairment linked with the V188A mutation in NS1, increasing the overall replicative fitness of ZIKV/Malaysia (35). As ZIKV/Malaysia exhibited greater replication efficiency than ZIKV/Dakar, it is likely that there are other uncharacterized genetic differences between these two lineages that contribute to the replication discrepancies observed here. NS5 serves as the viral RNA-dependent RNA polymerase (RdRp), and it is possible that specific NS5 sequence differences identified in this study along with differences in the rate of NS5 accumulation between ZIKV/Dakar and ZIKV/Malaysia could direct differential RdRp activity and confer replication differences between the two viruses (94, 95). Future studies assessing the impact of the genomic differences between African and Asian lineage prototypic strains on viral replicative fitness should define these genetic determinants of viral replicative fitness.
Evaluation of the interplay between ZIKV replication kinetics and host innate immunity revealed that infection with ZIKV/Dakar and ZIKV/Malaysia acutely induced robust RLR activation and induction of antiviral and inflammatory cytokine expression, including IFN-β, to a similar extent. This outcome was unexpected, as previous studies have demonstrated that ZIKV variants of both lineages block IFN induction during acute infection, suggesting that this discrepancy may be ZIKV strain dependent (65, 67, 68, 71, 96). Furthermore, ZIKV/Dakar encodes a valine at position 188 of NS1, which has additionally been shown to confer enhanced ability to block IFN-β induction compared to NS1 proteins encoding an alanine at this position (85). As we observed similar induction of IFN-β by ZIKV/Dakar and ZIKV/Malaysia, which encodes an alanine at this position, our results suggest that the valine at position 188 of NS1 is not sufficient to block IFN induction during in vitro infection of A549 cells. Despite the induction of IFNs during infection, ZIKV/Malaysia robustly blocked IFN signaling and downstream expression of ISGs, showing that ZIKV/Malaysia exhibits enhanced ability to restrict IFN signaling compared to ZIKV/Dakar. However, overexpression of ZIKV/Dakar and ZIKV/Malaysia NS5 proteins indicated that these divergent viral proteins blocked IFN signaling to the same extent, though there was a trend for ZIKV/Malaysia to impart greater suppression of pSTAT1 accumulation. We attribute the robust blockade of IFN signaling imposed by ZIKV/Malaysia to the early accumulation of viral proteins in infected cells, where early accumulation of NS5 in ZIKV/Malaysia-infected cells confers enhanced ability to antagonize IFN signaling. Studies of other flaviviruses as well as SARS-CoV-2 have also demonstrated a clear relationship between the rate of accumulation of viral innate immune antagonists and host innate immune responses (97–100). We demonstrated that the correlations between pSTAT1 and ZIKV NS5 levels in infected cells were different between ZIKV variants, which is likely driven by differences in the rate of NS5 accumulation. Genomic differences in the 3′ untranslated region (UTR) of these variants may impact the rate of NS5 accumulation, as 3′-UTR-derived subgenomic flavivirus RNA has been demonstrated to stabilize NS5 to efficiently inhibit STAT1 phosphorylation (101). Differences in the rate of NS5 accumulation may also alter the kinetics of NS5 interactions with host factors critical for IFN signaling. ZIKV NS5 has been shown to antagonize IFN signaling by targeting STAT2 for proteasomal degradation and by sequestering host chaperone HSP90 to destabilize JAK kinases (74, 77, 87). As such, early accumulation of ZIKV NS5 can drive interactions with STAT2 and HSP90 early in acute infection to blunt the IFN response in infected cells to promote viral replication. Moreover, other ZIKV nonstructural proteins have been demonstrated to impart regulation of the host IFN response (75, 76). As we observed differential accumulation of multiple ZIKV proteins during acute infection with the ZIKV variants, it is possible that early accumulation of other ZIKV proteins during ZIKV/Malaysia infection contributes to the robust blockade of the IFN response imposed by this variant compared to ZIKV/Dakar (see Fig. 2A). Our findings identify the rate of accumulation of ZIKV proteins as an important virologic feature for evaluating the IFN sensitivity of emerging variants.
ZIKV infection had not been previously linked to severe clinical disease until the outbreaks in French Polynesia and the Americas, raising the question as to why infection with ZIKV strains that circulated for decades in Africa and Southeast Asia was not associated with more severe disease, such as congenital abnormalities (4, 102). Comparative in vitro and in vivo studies of African and Asian lineage viruses have shown that African strains are more virulent and more cytopathic, suggesting that in utero infection with an African lineage virus would lead to fetal demise or resorption rather than congenital abnormalities (29, 34, 88). We observed that ZIKV/Malaysia replicated to higher levels than ZIKV/Dakar, suggesting that in utero infection with ZIKV/Malaysia may similarly cause severe fetal damage, whereas ZIKV/Dakar would be acutely restricted by the innate immune response and likely not confer congenital infection. On the other hand, other in vivo studies have shown that high levels of IFNs produced during in utero infection with African and Asian lineage ZIKV strains can result in fetal demise or resorption (103, 104). Our study raises new questions regarding the mechanisms underlying lineage-specific differences in fetal pathology and warrants further examination of prototypic African and Asian lineage ZIKV strains in an established pregnant pigtail macaque model to understand lineage-dependent differences in fetal pathology (12).
The ability of ZIKV/Malaysia to block IFN signaling more potently than ZIKV/Dakar serves as a potential mechanism for persistence in infected hosts, as dampened IFN responses can promote viral dissemination to and persistence in immune-privileged sites, such as the reproductive tissues and brain (105–108). ZIKV strains circulating in southeast Asia have been detected in the placenta and fetal brain several weeks after the onset of symptoms in the infected pregnant woman (109, 110). The detection of ZIKV in maternal reproductive and fetal tissues several weeks following infection highlights the ability of nonemergent ZIKV strains to disseminate to and persist in these tissues. Given that ZIKV/Malaysia robustly blocks IFN signaling compared to ZIKV/Dakar, it is possible that antagonism of IFN signaling may facilitate ZIKV/Malaysia spread to and persistence in the reproductive tissues and brain. Thus, the unique immunophenotypes uncovered in our study emphasize the need for surveillance of endemic ZIKV variants.
Overall, our comparative analysis of African lineage ZIKV/Dakar and Asian lineage ZIKV/Malaysia revealed greater abundance of viral proteins and genome in ZIKV/Malaysia-infected cells than in ZIKV/Dakar-infected cells during acute infection. Early accumulation of viral proteins in ZIKV/Malaysia-infected cells confers enhanced antagonism of IFN signaling, overcoming robust RIG-I activation initiated by infection, to suppress the induction of ISGs. Our findings demonstrate that RIG-I mediates an innate immune response that imparts control of viral fitness by restricting ZIKV protein accumulation to enhance IFN signaling, thereby limiting viral genome replication and spread. RIG-I-targeted therapeutics could be useful to direct innate immune suppression of ZIKV during acute infection.
MATERIALS AND METHODS
Maintenance of cell lines.
A549 (ATCC, Manassas, VA, USA), Vero (WHO, Geneva, Switzerland), and HEK293T cells (ATCC) were cultured in complete Dulbecco’s modified Eagle medium (cDMEM) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA), 10 mM l-glutamine, 5 mM sodium pyruvate, 0.5× nonessential amino acids, 10 mM HEPES, and 1× antibiotics/antimycotics (all from Corning, Corning, NY, USA). A549 NTC and RIG-I KO cells were generated by CRISPR-Cas9 as previously described and were maintained in cDMEM containing 10 μg/mL puromycin (Invivogen, San Diego, CA, USA) (27). ZV-13 8A3.C1 hybridomas were a kind gift from Michael Diamond (Washington University in St. Louis, St. Louis, MO, USA) and were maintained in Iscove’s modified Dulbecco’s medium (IMDM; Thermo Fisher-Life Technologies, Waltham, MA, USA) supplemented with 20% FBS, 1 mM sodium pyruvate, and 1× penicillin/streptomycin (Fisher Scientific, Hampton, NH, USA). All cell lines tested negative for mycoplasma contamination.
Generation of ZIKV working stocks.
ZIKV/Dakar/1984/ArD41519 (ZIKV/Dakar) was provided by Michael Diamond (Washington University in St. Louis), and ZIKV/Malaysia/1966/P6740 (ZIKV/Malaysia) was provided by Robert Tesh (University of Texas Medical Branch, Galveston, TX, USA). Plaque-picked isolates were passaged twice in Vero cells and subsequently used to infect Vero cells at an MOI of 0.05 at 37°C for 2 h with rocking to generate ZIKV working stocks. Following adsorption, inocula were removed and fresh DMEM containing 5% FBS (5% DMEM) was added. At 48 hpi, medium was replaced with fresh 5% DMEM. Virus stocks were harvested 48 h after the medium change, and the titers of working stocks were determined on A549 cells via focus forming unit (FFU) assays.
Deep sequencing of ZIKV stocks and sequence alignment.
ZIKV RNA was extracted from virus working stocks using the QIAmp viral RNA minikit (Qiagen, Hilden, Germany). Isolated RNA was then digested with DNase I and purified using the Qiagen RNeasy kit. RNA quality was analyzed on the 2100 bioanalyzer (Agilent, Santa Clara, CA, USA) using the Agilent RNA 6000 Pico assay and quantified on a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) prior to rRNA depletion and RNA library prep using the KAPA HyperPrep kit (Roche Diagnostics Corporation, Indianapolis, IN, USA). cDNA library qualities were evaluated on the Agilent 2100 bioanalyzer and quantified with the Qubit fluorometer before sequencing on a NextSeq 500 sequencing platform (Illumina, San Diego, CA, USA). 2 × 76-nucleotide stranded paired-end reads were generated. Raw RNA sequencing (RNA seq) data were depleted of adapters, low quality bases, and human reads. Sequencing files were then loaded onto the de novo assembler Trinity (version 2.9.0) and assembled and aligned as previously described (27, 111). Each sample had approximately 25 million raw reads. Viral sequence alignments were loaded into the JALView software (version 2.11.0) to identify amino acid differences between ZIKV strains (112).
ZV-13 antibody purification and quantification.
Hybridoma supernatant was harvested as the source of ZV-13 antibody once ~95% cell death was observed. The supernatant was clarified by centrifuging for 10 min at 400 × g at 4°C and then filtered with a 0.22-μm filter prior to antibody purification via protein A affinity chromatography using the Pierce Protein A IgG purification kit and NAb Protein A Plus spin columns (all from Thermo Fisher-Life Technologies). Purified ZV-13 antibody was concentrated and buffer-exchanged into 1× phosphate-buffered saline (PBS; Fisher Scientific) using Amicon Ultra-0.5 100-kDa centrifugal filter units for quantification by enzyme-linked immunosorbent assay (ELISA; Sigma-Aldrich, St. Louis, MO, USA). 96-well Nunc ELISA plates (Fisher Scientific) were coated with goat anti-mouse IgG2c (Jackson ImmunoResearch, West Grove, PA, USA) diluted to 500 ng/mL in 1× PBS and incubated overnight at 4°C. Coated plates were washed with PBS plus Tween 20 (PBST; Fisher Scientific) and blocked for 1 h at room temperature with PBS plus 1% bovine serum albumin (PBSA; Sigma-Aldrich) supplemented with 2% normal goat serum (NGS; Jackson ImmunoResearch). Following blocking, serial dilutions of hybridoma supernatant and purified ZV-13 antibody were added and incubated for 1 h at 37°C. Plates were washed, and biotin-conjugated anti-mouse IgG2c secondary antibody (Jackson ImmunoResearch) diluted 1:5,000 in PBSA was added and incubated for 1 h at room temperature. Plates were then washed and incubated with streptavidin-horseradish peroxidase (HRP) (Thermo Fisher-Life Technologies) diluted 1:20,000 in PBSA for 30 min at room temperature. Plates were washed, developed by adding 3,3′,5,5′-tetramethylbenzinide (TMB; Surmodics, Eden Prairie, MN, USA), and incubated in the dark. The reaction was stopped with the addition of 1 M H2SO4, and absorbance was measured at 450 nm in a microplate reader (BioLegend, San Diego, CA, USA). Serial dilutions of known concentrations of mouse IgG2c (Southern Biotech, Birmingham, AL, USA) were included to generate a standard curve with optical density (OD) values plotted against concentration in GraphPad Prism (version 9, GraphPad, La Jolla, CA, USA). The linear portion of the standard curve was used to derive a least-squares equation to calculate the concentration of purified ZV-13 antibody.
FFU assay for determining the titers of ZIKV stocks.
A549 cells were seeded in 96-well plates at a density of 2 × 104 cells per well. The following day, virus stocks were serially diluted in DMEM containing 2% FBS (2% DMEM), and cells were inoculated with dilutions in triplicate at 37°C for 2 h. After the incubation, cells were overlaid with DMEM supplemented with 1% carboxymethylcellulose (Fisher Scientific), 2% FBS, 10 mM HEPES, and 1× penicillin/streptomycin. At 48 hpi, the methylcellulose overlay and medium were removed, and cells were fixed with 4% paraformaldehyde (Fisher Scientific) for 20 min at room temperature. Following fixation, cells were washed with 1× PBS, and anti-ZIKV envelope antibody, ZV-13, diluted 1:250 in permeabilization/wash/block buffer (2.5% NGS, 2.5% normal donkey serum [Sigma-Aldrich], 0.2% BSA, and 0.1% Triton X-100 [Fisher Scientific] in 1× PBS) was added. Primary antibody was incubated for 2 h with rocking at room temperature. Plates were then washed, and donkey anti-mouse HRP secondary antibody (Jackson ImmunoResearch) diluted 1:3,000 in permeabilization/wash/block buffer was added and incubated with rocking for 1 h in the dark. Following washes, TrueBlue peroxidase substrate (VWR, Radnor, PA, USA) was added, and the mixture was incubated until foci of infected cells were observed. Distilled water (dH2O) was added to stop the enzymatic reaction and removed prior to imaging. Foci of infected cells were detected via a BioSpot plate reader (Cellular Technology Limited, Cleveland, OH, USA) and manually counted using the multipoint tool in Fiji (version 2.1.0, NIH, USA) to determine viral titers (113).
FFU assay for measuring viral spread.
A549 cells were seeded in 6-well plates at a density of 6 × 105 cells per well. The following day, virus stocks were serially diluted in 2% DMEM, and cells were inoculated and fixed as described above. Following fixation, cells were washed with 1× PBS, and an anti-flavivirus envelope antibody, 4G2 (Ab00230-2.0; Absolute Antibody, Oxford, UK), diluted 1:500 in permeabilization/wash/block buffer (see recipe above) was added to the cells. Following incubation with primary antibody, donkey anti-mouse HRP secondary antibody diluted 1:1,000 in permeabilization/wash/block buffer was added and incubated with rocking for 1 h in the dark. Foci were developed and detected as described above. The diameters of the foci were measured in Fiji.
ZIKV infection.
A549 cells were seeded at a density of 3 × 105 cells per well in 12-well plates. Cells were uninfected, mock-infected, or infected with ZIKV variants the next day at an MOI of 5 FFU/cell, and virus inocula prepared in DMEM were allowed to adsorb at 37°C for 2 h with rocking. Uninfected samples are seeded cells that remained untouched throughout the infection time course, whereas mock-infected samples were inoculated with DMEM. Following adsorption, the inocula were removed, and cells were washed with 1× PBS before fresh cDMEM was added and returned to 37°C until the appropriate time points.
ZIKV attachment and entry assay.
A549 cells were seeded at a density of 3 × 105 cells per well in 12-well plates. Cells were precooled at 4°C prior to the addition of ice-cold virus inoculum. Inocula were allowed to adsorb to cells at 4°C for 1 h. Inocula were then removed, and cells were washed with ice-cold 1× PBS before fresh prewarmed cDMEM was added to cells. Cells were returned to 37°C until the appropriate time points to evaluate viral entry, genome replication, and virion production. For viral attachment assays, intracellular viral RNA was harvested using the Qiagen RNeasy kit with DNase I digestion following ice-cold 1× PBS washes.
Quantification of extracellular and intracellular RNA.
A549 cells were infected as described above. At the appropriate time points, 1 mL of cell culture supernatants was harvested and centrifuged at 2,000 rpm for 10 min at 4°C. Extracellular RNA was extracted from the clarified supernatants using the QIAmp viral RNA minikit to quantify extracellular viral copy numbers. Cells were washed with ice-cold stringent wash buffer (1 M NaCl, 50 mM sodium bicarbonate, pH 9.5) for 3 min to remove cell surface-associated viruses. After ice-cold 1× PBS washes, cells were harvested using the Qiagen RNeasy kit to extract intracellular RNA to quantify intracellular viral copy numbers.
Quantification of extracellular and intracellular virions.
A549 cells were infected as described above. At the appropriate time points, 1 mL of cell culture supernatants was harvested and centrifuged at 2,000 rpm for 10 min at 4°C. The clarified supernatants were used for plaque assays to quantify extracellular virions. Cells were washed with ice-cold stringent wash buffer as described above and then detached with 0.05% trypsin-EDTA (Fisher Scientific). Cells were resuspended in 1 mL cDMEM and centrifuged at 300 × g for 5 min at 4°C. Cell pellets were resuspended in 0.5 to 1 mL cDMEM and stored at −80°C. To quantify intracellular virions, cell pellets were thawed at 37°C to lyse cells, and cellular debris removed by centrifuging at 3,200 × g for 5 min at 4°C. The clarified supernatants were used for plaque assays.
Plaque assay.
Vero cells were seeded at a density of 1.85 × 105 cells per well in 6-well plates. The next day, supernatants from infected samples were serially diluted in DMEM containing 1% FBS. Vero cells were incubated with dilutions in duplicate at 37°C for 1 h with rocking. Following incubation, cells were overlaid with 1% low-melt agarose (Fisher Scientific), 5% sodium bicarbonate (Fisher Scientific), and 5% FBS in 2× DMEM (Fisher Scientific). Plaques were visualized 4 to 5 days later with a 1% low-melt agarose overlay containing 3% neutral red (Sigma-Aldrich), 10% 10× PBS (Fisher Scientific), and dH2O.
Viral copy number quantification by TaqMan qRT-PCR.
cDNA was synthesized from isolated RNA using the iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). cDNA was diluted to a final concentration of 2.5 ng/μL for TaqMan qRT-PCR on the QuantStudio 5 real-time PCR system (Thermo Fisher) using TaqMan universal PCR master mix (Fisher Scientific) and ZIKV NS3-specific primers and probe (Table 4; IDT, Coralville, IA). The ZIKV NS3-specific probe has a 5′ 6-carboxyfluorescein (FAM) reporter dye and a 3′ Iowa Black FQ quencher. ZIKV NS3-specific primers target a conserved 88 nucleotide region in NS3. The ZIKV NS3 amplicon was amplified from ZIKV cDNA and cloned into the multiple cloning site in the pEF mammalian expression plasmid containing an N-terminal FLAG tag using the In-Fusion cloning kit (Table 4; TaKaRa Bio, Kusatsu, Japan) following the manufacturer’s instructions. Each sample was run in triplicate, and dilutions of known concentrations of plasmid containing the NS3 amplicon were included to generate a standard curve with copy number values plotted against threshold cycle (CT) values to derive a semi-log equation in GraphPad Prism to determine the viral copy number of each sample. Dilutions of cDNA synthesized from RNA extracted from ZIKV stocks were used to derive a log-log equation to convert viral copy numbers to their FFU equivalent in GraphPad Prism.
TABLE 4.
Primer sequences for cloning and qRT-PCRa
| Primerb | Sequence (5′–3′) | Source |
|---|---|---|
| Primers for cloning | ||
| pEF N-FLAG vector F | AAACCCGCTGATCAGCCT | This paper |
| pEF N-FLAG vector R | GCTGAATTCCTTGTCATCGTCATCC | This paper |
| ZIKV NS3 F | GACAAGGAATTCAGCGAGGGAGAGTTCAAGCTTAGGACG | This paper |
| ZIKV NS3 R | CTGATCAGCGGGTTTGATAGGCCAGCCAAACAGGAAGA | This paper |
| pcDNA3.1+ ZIKV/Dakar and ZIKV/Malaysia NS5 vector F | GTTTAAACCCGCTGATCAGCCT | This paper |
| pcDNA3.1+ ZIKV/Dakar and ZIKV/Malaysia NS5 vector R | CAGCTTGGGTCTCCCTATAGTGAG | This paper |
| pcDNA3.1+ ZIKV/Dakar NS5 and ZIKV/Malaysia NS5 F | GGGAGACCCAAGCTGATGGGAGGTGGAACGGGAGAG | This paper |
| pcDNA3.1+ ZIKV/Dakar NS5 R | TCAGCGGGTTTAAACTTACAGCACTCCAGGTGTGGAC | This paper |
| pcDNA3.1+ ZIKV/Malaysia NS5 R | TCAGCGGGTTTAAACTTATAACACTCCAGGTGTGGACCC | This paper |
| Primers for TaqMan qRT-PCR | ||
| ZIKV NS3 F | GAGGGAGAGTTCAAGCT | This paper |
| ZIKV NS3 R | GATAGGCCAGCCAAAC | This paper |
| ZIKV NS3 probe (with quencher) | 56-FAM/TTCATGAGTTCCACAAAGGTCTTCCTTTGC/3IABkFQ | This paper |
| Primers for SYBR green qRT-PCR | ||
| IFNB | NA | PPH00384F; Qiagen |
| TNF | NA | PPH00341F; Qiagen |
| IL-6 | NA | PPH00560C-200; Qiagen |
| MX1 | NA | PPH01325A; Qiagen |
| IFITM1 | NA | PPH05981C; Qiagen |
| DDX58 | NA | PPH20774A-200; Qiagen |
| OAS1 | NA | PPH01324A-200; Qiagen |
| RPL13A F | GCCCTACGACAAGAAAAAGCG | 27 |
| RPL13A R | TACTTCCAGCCAACCTCGTGA | 27 |
All sequences used for SYBR green qRT-PCR are of human origin.
F, forward; R, reverse; NA, not available.
Transcript analysis by SYBR green qRT-PCR.
cDNA was synthesized as described above. For each sample, cDNA was diluted to a final concentration of 2.5 ng/μL for SYBR green qRT-PCR on the QuantStudio 5 real-time PCR system using the SYBR green master mix (Thermo Fisher) and gene-specific primers (Table 4; Qiagen; IDT). Each sample was run in triplicate.
Transcript analysis by NanoString.
RNA samples were diluted to a final concentration of 20 ng/μL and hybridized to reporters and capture probes from a custom-generated innate immunity probe set (NanoString, Seattle, WA, USA). Following hybridization for 16 h at 65°C, sample preparation was completed on an nCounter MAX prep station (NanoString) according to the manufacturer’s protocol. RNA transcript counts were quantified on a NanoString nCounter (NanoString) according to the manufacturer’s instructions.
Bioinformatics analyses of NanoString data.
Raw reads were assessed for technical quality control flags using the nSolver software (NanoString) and transferred into RStudio (Boston, MA, USA). As the design of our probe set was nonrandom, we utilized a custom approach for data analysis as previously described to account for large differences in the number of observed counts between mock-infected and infected samples (114). Briefly, we used count data within each sample vector as an internal reference, against which we compared genes of interest (GOIs). We identified reference genes by first measuring the coefficient of variation (CV) for all genes in the probe set, which included GOI, housekeeping (HK) genes, and positive and negative controls. We then used the calculated CVs to select the least variable HK genes. To obtain our within-sample reference value, we calculated the geometric mean of the selected HK genes. We then “anchored” all count data from each sample to its within-sample reference in ratio form to derive anchored gene counts (AGCs). To calculate the log2 fold change, we formed a ratio using the AGC for each GOI against its values from mock infection and took the log2 of these values, which were used to generate PCA plots and line graphs.
Protein lysate quantification.
Cells were washed with 1× PBS and lysed on ice with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.6, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate) containing freshly added phosphatase inhibitors (1:100; VWR), protease inhibitors (1:100; Sigma-Aldrich), and 250 nM okadaic acid (1:1,000; Fisher Scientific). Cell lysates were collected and stored at −80°C. Lysates were subsequently thawed on ice and sonicated in a water bath containing ice slurry for three 30-s bursts on the highest setting with two 20- s pauses in between. Lysates were then centrifuged at 14,000 rpm for 15 min at 4°C to pellet the cellular debris. Clarified lysates were collected and quantified using a Bio-Rad protein assay kit (Bio-Rad).
Immunoblot analysis.
For immunoblot analysis, protein lysates were incubated with 4× Laemmli buffer (Bio-Rad) containing 10% β-mercaptoethanol (Thermo Fisher-Life Technologies) at 95°C for 3 min. Approximately 7 to 10 μg of total protein was loaded per lane onto 4 to 20% Criterion TGX gradient gels (Bio-Rad) and electrophoresed at ~96 V in 1× sodium dodecyl sulfate (SDS) running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Proteins were then transferred onto nitrocellulose membranes (Fisher Scientific) at 90 V for 1 h at 4°C in 1× Towbin transfer buffer (25 mM Tris, 192 mM glycine, 0.01% SDS, 20% methanol). Membranes were blocked for 1 h at room temperature in Tris-buffered saline (TBS)-based Intercept blocking buffer (LI-COR, Lincoln, NE, USA) and stained overnight at 4°C with the following primary antibodies diluted in blocking buffer: pIRF3 S386 (no. ab76493; 1:1,000; Abcam, Cambridge, UK), pIRF3 S396 (no. 4947; 1:1,000; Cell Signaling Technology [CST], Danvers, MA, USA), IRF3 (no. 4302; 1:1,000; CST), IκBα (no. 4814; 1:1,000; CST), IFIT1 (no. 971; 1:1,000; in-house), ZIKV NS5 (no. GTX133327; 1:2,500; GeneTex, Irvine, CA, USA), ZIKV NS1 (no. ARG65781, 1:1,000, Arigo Biolaboratories, Hsinchu City, Taiwan), ZIKV capsid (no. GTX133317, 1:1,000, GeneTex), pSTAT1 (no. 7649; 1:1,000; CST), STAT1 (no. 9172; 1:1,000; CST), STAT2 (no. 72604; 1:1,000; CST), IFITM1 (no. 60074-1-1g; 1:1,000; ProteinTech Group, Rosemont, IL, USA), OAS1 (no. 14498; 1:1,000; CST), Mx1 (no. 340B; 1:500; in-house), RIG-I (no. AG-20B-0009-C100; 1:1,000; Adipogen, San Diego, CA, USA), and actin (no. MAB 1501; 1:1,000; Sigma-Aldrich). The next day, membranes were washed with TBS containing Tween 20 (TBST), and Alexa Fluor 680-conjugated (715-625-151 or 711-625-152) or Alexa Fluor 790-conjugated secondary antibodies (711-655-152 or 715-655-150) (all from Jackson ImmunoResearch) diluted 1:10,000 in blocking buffer were added and incubated for 1 h at room temperature. Prior to imaging, membranes were washed with TBST followed by 1× TBS. Membranes were imaged on an Odyssey CLx imager (LI-COR). If necessary, membranes were stripped with 5× NewBlot IR stripping buffer (LI-COR) diluted 1:5 in dH2O and reprobed. Protein abundance was quantified in Fiji and normalized to actin abundance.
Recombinant DNA construct generation and plasmid transfections.
ZIKV/Dakar and ZIKV/Malaysia NS5 sequences were amplified from either ZIKV/Dakar or ZIKV/Malaysia cDNA and cloned into the multiple cloning site in the pcDNA3.1+ mammalian expression plasmid (Table 4) using the TaKaRa In-Fusion cloning kit. HEK293T cells were seeded at a density of 4 × 105 cells per well in a 6-well plate and transfected with 2.5 μg of the empty pcDNA3.1+, pcDNA3.1+ ZIKV/Dakar NS5, or pcDNA3.1+ ZIKV/Malaysia NS5 plasmid using Lipofectamine 3000 (Thermo Fisher-Life Technologies) according to the manufacturer’s protocol. The ratio of plasmid DNA (μg) to Lipofectamine 3000 (μL) to P3000 reagent (μL) to Opti-MEM (μL) (Fisher Scientific) was 1:3:2:50.
IFN-β treatment.
A549 NTC and RIG-I KO cells were seeded at a density of 1 × 105 cells per well on no. 1 glass coverslips (Electron Microscopy Sciences, Hatfield, PA, USA) in a 24-well plate. Cells were mock infected or infected with ZIKV variants the next day. At 18 or 40 hpi, A549 NTC and RIG-I KO cells were treated with 100 IU/mL of IFN-β (Toray, Tokyo, Japan) diluted in cDMEM for 1 h for immunofluorescence analysis. For flow cytometry analysis, A549 RIG-I KO cells were seeded at a density of 4 × 106 in 10-cm dishes and infected with ZIKV variants the next day. At various time points postinfection, cells were detached with 0.05% trypsin-EDTA, pelleted, resuspended in medium containing 100 IU/mL of IFN-β, and incubated at 37°C for 30 min with rocking. HEK293T cells were seeded and transfected as described above. The next day, cells were detached with 0.05% trypsin-EDTA, pelleted, and resuspended in medium containing 1,000 IU/mL of IFN-β for 8 h for flow cytometry analysis.
Immunofluorescence analysis.
A549 cells were seeded on no. 1 glass coverslips in a 24-well plate at a density of 1 × 105 cells per well. The next day, cells were mock infected or infected with ZIKV strains and fixed with 4% formaldehyde (Fisher Scientific) in filtered 1× TBS for 30 min at room temperature. Cells were then washed with filtered 1× TBS and permeabilized and blocked in 5% NGS in 1× TBS (5% NGS/TBS) containing 0.1% Triton X-100 for 30 min at 4°C. For pSTAT1 staining, cells were permeabilized with 100% ice-cold methanol (Fisher Scientific) for 10 min at −20°C and washed in 1× TBS for 5 min before blocking in 5% NGS/TBS for 30 min at room temperature. For all immunofluorescence staining, cells were stained for 1 h at room temperature or overnight at 4°C with the following primary antibodies diluted in 5% NGS/TBS: dsRNA (J2, no. 10010500; 1:800; Scicons, Budapest, Hungary), ZIKV NS5 (GTX133327; 1:500; GeneTex), and pSTAT1 (no. 7649; 1:400; CST). Cells were washed with filtered 1× TBS and probed for 1 h at room temperature with isotype-specific and fluorophore-conjugated secondary antibodies (A-21131, A-11036, A-11008, or A-21241; Thermo Fisher-Life Technologies) diluted 1:1,000 in 5% NGS/TBS containing 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; 1:10,000; Thermo Fisher-Life Technologies) to stain cell nuclei. After staining with secondary antibodies, cells were washed with filtered 1× TBS followed by dH2O. Excess water was carefully removed from coverslips, and coverslips were mounted onto glass slides (VWR) with ProLong gold mounting medium (Thermo Fisher-Life Technologies) and dried overnight at room temperature. Images and Z-stacks with 2.5-μm step sizes were acquired on an Eclipse Ti confocal microscope (Nikon, Tokyo, Japan) with a 60× oil immersion objective. Images were merged and processed using the Nikon confocal analysis software (NIS Elements AR 5.11.03). Further image analyses were performed in Fiji as indicated in the figure legends.
Flow cytometry.
Cells were seeded, infected, or transfected and treated with IFN-β as described above. Following IFN-β treatment, A549 RIG-I KO and HEK293T cells were pelleted and fixed with 4% formaldehyde in 1× PBS at room temperature for 15 min. Cells were then washed with excess 1× PBS, pelleted, and resuspended in 1× PBS for storage at 4°C. Fixed cells were permeabilized with 90% ice-cold methanol in 1× PBS for at least 10 min on ice. Cells were washed with excess 1× PBS and incubated for 1 h at room temperature with the following fluorescently conjugated primary antibodies prepared in 0.5% PBSA: pSTAT1-PE (no. 8062, 1:50, CST) and ZIKV NS5 AF647 (1:2,000, conjugated in-house). ZIKV NS5 was conjugated to AF647 using an Alexa Fluor 647 antibody labeling kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Following staining, cells were washed with 1× PBS and data were acquired on a Canto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Flow cytometry data were analyzed in FlowJo (BD Biosciences).
Statistical analysis.
Statistical analyses were carried out as indicated in the figure legends. Statistical analyses were performed in GraphPad Prism or in RStudio.
Illustrations.
Figures were generated using BioRender.
Data availability.
Viral consensus sequences were deposited in GenBank under the following accession numbers: OQ180929 (ZIKV/Dakar); OQ165285 (ZIKV/Malaysia). The NanoString data set can be found in File S1 in the supplemental material.
ACKNOWLEDGMENTS
We thank the members of the University of Washington and the Center for Innate Immunity and Immune Disease for helpful discussions and advice throughout the course of this study. We thank Michael Diamond (Washington University in St. Louis) and Robert Tesh (University of Texas Medical Branch) for providing hybridomas and ZIKV isolates. We are grateful to the Seattle Genomics sequencing group for sequencing ZIKV stocks and the University of Washington Cell Analysis Facility for technical assistance.
This work was supported by National Institutes of Health grants AI143265, AI145296, and AI151698.
This work is part of the thesis of A.Y.L.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael Gale, Jr., Email: mgale@uw.edu.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Dick GWA, Kitchen SF, Haddow AJ. 1952. Zika virus (I). Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Faye O, Freire CCM, Iamarino A, Faye O, de Oliveira JVC, Diallo M, Zanotto PMA, Sall AA. 2014. Molecular evolution of Zika virus during its emergence in the 20th century. PLoS Negl Trop Dis 8:e2636. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffy MR, Chen T-H, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 4.Lee JK, Shin OS. 2019. Advances in Zika virus-host cell interaction: current knowledge and future perspectives. Int J Mol Sci 20:1101. doi: 10.3390/ijms20051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musso D, Gubler DJ. 2016. Zika virus. Clin Microbiol Rev 29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pielnaa P, Al-Saadawe M, Saro A, Dama MF, Zhou M, Huang Y, Huang J, Xia Z. 2020. Zika virus-spread, epidemiology, genome, transmission cycle, clinical manifestation, associated challenges, vaccine and antiviral drug development. Virology 543:34–42. doi: 10.1016/j.virol.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilak R, Ray S, Tilak VW, Mukherji S. 2016. Dengue, chikungunya … and the missing entity—Zika fever: a new emerging threat. Med J Armed Forces India 72:157–163. doi: 10.1016/j.mjafi.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faria NR, Azevedo RDSDS, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami FGDS, Macedo FLDL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, Medeiros DBDA, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, Simith DDB, Messina JP, Abade L, Lourenço J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, Lemos PDS, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Júnior JL, da SG, Mir D, Bello G, Delatorre E, et al. 2016. Zika virus in the Americas: early epidemiological and genetic findings. Science 352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastère S, Valour F, Baudouin L, Mallet HP, Musso D, Ghawche F. 2014. Zika virus infection complicated by Guillain-Barré syndrome – case report, French Polynesia, December 2013. Eurosurveillance 19:20720. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 11.Dudley DM, Van Rompay KK, Coffey LL, Ardeshir A, Keesler RI, Bliss-Moreau E, Grigsby PL, Steinbach RJ, Hirsch AJ, MacAllister RP, Pecoraro HL, Colgin LM, Hodge T, Streblow DN, Tardif S, Patterson JL, Tamhankar M, Seferovic M, Aagaard KM, Martín CS-S, Chiu CY, Panganiban AT, Veazey RS, Wang X, Maness NJ, Gilbert MH, Bohm RP, Adams Waldorf KM, Gale M, Rajagopal L, Hotchkiss CE, Mohr EL, Capuano SV, Simmons HA, Mejia A, Friedrich TC, Golos TG, O’Connor DH. 2018. Miscarriage and stillbirth following maternal Zika virus infection in nonhuman primates. Nat Med 24:1104–1107. doi: 10.1038/s41591-018-0088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams Waldorf KM, Stencel-Baerenwald JE, Kapur RP, Studholme C, Boldenow E, Vornhagen J, Baldessari A, Dighe MK, Thiel J, Merillat S, Armistead B, Tisoncik-Go J, Green RR, Davis MA, Dewey EC, Fairgrieve MR, Gatenby JC, Richards T, Garden GA, Diamond MS, Juul SE, Grant RF, Kuller L, Shaw DWW, Ogle J, Gough GM, Lee W, English C, Hevner RF, Dobyns WB, Gale M, Rajagopal L. 2016. Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate. Nat Med 22:1256–1259. doi: 10.1038/nm.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams Waldorf KM, Nelson BR, Stencel-Baerenwald JE, Studholme C, Kapur RP, Armistead B, Walker CL, Merillat S, Vornhagen J, Tisoncik-Go J, Baldessari A, Coleman M, Dighe MK, Shaw DWW, Roby JA, Santana-Ufret V, Boldenow E, Li J, Gao X, Davis MA, Swanstrom JA, Jensen K, Widman DG, Baric RS, Medwid JT, Hanley KA, Ogle J, Gough GM, Lee W, English C, Durning WM, Thiel J, Gatenby C, Dewey EC, Fairgrieve MR, Hodge RD, Grant RF, Kuller L, Dobyns WB, Hevner RF, Gale M, Rajagopal L. 2018. Congenital Zika virus infection as a silent pathology with loss of neurogenic output in the fetal brain. Nat Med 24:368–374. doi: 10.1038/nm.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald EM, Duggal NK, Ritter JM, Brault AC. 2018. Infection of epididymal epithelial cells and leukocytes drives seminal shedding of Zika virus in a mouse model. PLoS Negl Trop Dis 12:e0006691. doi: 10.1371/journal.pntd.0006691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musso D, Nhan T, Robin E, Roche C, Bierlaire D, Zisou K, Yan AS, Cao-Lormeau VM, Broult J. 2014. Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Eurosurveillance 19:20761. doi: 10.2807/1560-7917.es2014.19.14.20761. [DOI] [PubMed] [Google Scholar]
- 16.Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau V-M. 2015. Potential sexual transmission of Zika virus. Emerg Infect Dis 21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musso D, Richard V, Teissier A, Stone M, Lanteri MC, Latoni G, Alsina J, Reik R, Busch MP. Recipient Epidemiology and Donor Evaluation Study (REDS-III) ZIKV Study Group. 2017. Detection of Zika virus RNA in semen of asymptomatic blood donors. Clin Microbiol Infect 23:1001.e1–1001.e3. doi: 10.1016/j.cmi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Ortenzio E, Matheron S, Yazdanpanah Y, de Lamballerie X, Hubert B, Piorkowski G, Maquart M, Descamps D, Damond F, Leparc-Goffart I. 2016. Evidence of sexual transmission of Zika virus. N Engl J Med 374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- 19.Driggers RW, Ho C-Y, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, Timofeev J, Rodriguez FJ, Levanov L, Razak J, Iyengar P, Hennenfent A, Kennedy R, Lanciotti R, Du Plessis A, Vapalahti O. 2016. Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med 374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 20.Motta IJF, Spencer BR, Cordeiro da Silva SG, Arruda MB, Dobbin JA, Gonzaga YBM, Arcuri IP, Tavares RCBS, Atta EH, Fernandes RFM, Costa DA, Ribeiro LJ, Limonte F, Higa LM, Voloch CM, Brindeiro RM, Tanuri A, Ferreira OC. 2016. Evidence for transmission of Zika virus by platelet transfusion. N Engl J Med 375:1101–1103. doi: 10.1056/NEJMc1607262. [DOI] [PubMed] [Google Scholar]
- 21.Kazmi SS, Ali W, Bibi N, Nouroz F. 2020. A review on Zika virus outbreak, epidemiology, transmission and infection dynamics. J Biol Res 27:5. doi: 10.1186/s40709-020-00115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill SC, Vasconcelos J, Neto Z, Jandondo D, Zé-Zé L, Aguiar RS, Xavier J, Thézé J, Mirandela M, Cândido ALM, Vaz F, Sebastião CDS, Wu C-H, Kraemer MUG, Melo A, Schamber-Reis BLF, Azevedo GSD, Tanuri A, Higa LM, Clemente C, Silva SPD, Candido DDS, Claro IM, Quibuco D, Domingos C, Pocongo B, Watts AG, Khan K, Alcantara LCJ, Sabino EC, Lackritz E, Pybus OG, Alves M-J, Afonso J, Faria NR. 2019. Emergence of the Asian lineage of Zika virus in Angola: an outbreak investigation. Lancet Infect Dis 19:1138–1147. doi: 10.1016/S1473-3099(19)30293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Valderramos SG, Wu A, Ouyang S, Li C, Brasil P, Bonaldo M, Coates T, Nielsen-Saines K, Jiang T, Aliyari R, Cheng G. 2016. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe 19:561–565. doi: 10.1016/j.chom.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikan N, Smith DR. 2016. Zika virus: history of a newly emerging arbovirus. Lancet Infect Dis 16:e119–e126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- 26.Anfasa F, Siegers JY, van der Kroeg M, Mumtaz N, Stalin Raj V, de Vrij FMS, Widagdo W, Gabriel G, Salinas S, Simonin Y, Reusken C, Kushner SA, Koopmans MPG, Haagmans B, Martina BEE, van Riel D. 2017. Phenotypic differences between Asian and African lineage Zika viruses in human neural progenitor cells. mSphere 2:e00292-17. doi: 10.1128/mSphere.00292-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esser-Nobis K, Aarreberg LD, Roby JA, Fairgrieve MR, Green R, Gale M. 2019. Comparative analysis of African and Asian lineage-derived Zika virus strains reveals differences in activation of and sensitivity to antiviral innate immunity. J Virol 93:e00640-19. doi: 10.1128/JVI.00640-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Bos S, Tsetsarkin KA, Pletnev AG, Desprès P, Gadea G, Zhao RY. 2019. The roles of prM-E proteins in historical and epidemic Zika virus-mediated infection and neurocytotoxicity. Viruses 11:157. doi: 10.3390/v11020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheridan MA, Balaraman V, Schust DJ, Ezashi T, Roberts RM, Franz AWE. 2018. African and Asian strains of Zika virus differ in their ability to infect and lyse primitive human placental trophoblast. PLoS One 13:e0200086. doi: 10.1371/journal.pone.0200086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DR, Sprague TR, Hollidge BS, Valdez SM, Padilla SL, Bellanca SA, Golden JW, Coyne SR, Kulesh DA, Miller LJ, Haddow AD, Koehler JW, Gromowski GD, Jarman RG, Alera MTP, Yoon I-K, Buathong R, Lowen RG, Kane CD, Minogue TD, Bavari S, Tesh RB, Weaver SC, Linthicum KJ, Pitt ML, Nasar F. 2018. African and Asian Zika virus isolates display phenotypic differences both in vitro and in vivo. Am J Trop Med Hyg 98:432–444. doi: 10.4269/ajtmh.17-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Liu J, Du S, Shan C, Nie K, Zhang R, Li X-F, Zhang R, Wang T, Qin C-F, Wang P, Shi P-Y, Cheng G. 2017. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature 545:482–486. doi: 10.1038/nature22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Shan C, Zhu Y, Ma E, Wang J, Wang P, Shi P-Y, Cheng G. 2021. A mutation-mediated evolutionary adaptation of Zika virus in mosquito and mammalian host. Proc Natl Acad Sci USA 118:e2113015118. doi: 10.1073/pnas.2113015118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan C, Xia H, Haller SL, Azar SR, Liu Y, Liu J, Muruato AE, Chen R, Rossi SL, Wakamiya M, Vasilakis N, Pei R, Fontes-Garfias CR, Singh SK, Xie X, Weaver SC, Shi P-Y. 2020. A Zika virus envelope mutation preceding the 2015 epidemic enhances virulence and fitness for transmission. Proc Natl Acad Sci USA 117:20190–20197. doi: 10.1073/pnas.2005722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubry F, Jacobs S, Darmuzey M, Lequime S, Delang L, Fontaine A, Jupatanakul N, Miot EF, Dabo S, Manet C, Montagutelli X, Baidaliuk A, Gámbaro F, Simon-Lorière E, Gilsoul M, Romero-Vivas CM, Cao-Lormeau V-M, Jarman RG, Diagne CT, Faye O, Faye O, Sall AA, Neyts J, Nguyen L, Kaptein SJF, Lambrechts L. 2021. Recent African strains of Zika virus display higher transmissibility and fetal pathogenicity than Asian strains. Nat Commun 12:916. doi: 10.1038/s41467-021-21199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Liu Y, Shan C, Nunes BTD, Yun R, Haller SL, Rafael GH, Azar SR, Andersen CR, Plante K, Vasilakis N, Shi P-Y, Weaver SC. 2021. Role of mutational reversions and fitness restoration in Zika virus spread to the Americas. Nat Commun 12:595. doi: 10.1038/s41467-020-20747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collette NM, Lao VHI, Weilhammer DR, Zingg B, Cohen SD, Hwang M, Coffey LL, Grady SL, Zemla AT, Borucki MK. 2020. Single amino acid mutations affect Zika virus replication in vitro and virulence in vivo. Viruses 12:1295. doi: 10.3390/v12111295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan L, Huang X-Y, Liu Z-Y, Zhang F, Zhu X-L, Yu J-Y, Ji X, Xu Y-P, Li G, Li C, Wang H-J, Deng Y-Q, Wu M, Cheng M-L, Ye Q, Xie D-Y, Li X-F, Wang X, Shi W, Hu B, Shi P-Y, Xu Z, Qin C-F. 2017. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 358:933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 38.Loo Y-M, Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dutta S, Das N, Mukherjee P. 2020. Picking up a fight: fine tuning mitochondrial innate immune defenses against RNA viruses. Front Microbiol 11:1990. doi: 10.3389/fmicb.2020.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onomoto K, Onoguchi K, Yoneyama M. 2021. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol 18:539–555. doi: 10.1038/s41423-020-00602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chow KT, Gale M, Loo Y-M. 2018. RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol 36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 42.Chazal M, Beauclair G, Gracias S, Najburg V, Simon-Lorière E, Tangy F, Komarova AV, Jouvenet N. 2018. RIG-I recognizes the 5′ region of dengue and Zika virus genomes. Cell Rep 24:320–328. doi: 10.1016/j.celrep.2018.06.047. [DOI] [PubMed] [Google Scholar]
- 43.Schilling M, Bridgeman A, Gray N, Hertzog J, Hublitz P, Kohl A, Rehwinkel J. 2020. RIG-I plays a dominant role in the induction of transcriptional changes in Zika virus-infected cells, which protect from virus-induced cell death. Cells 9:1476. doi: 10.3390/cells9061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kell AM, Gale M Jr. 2015. RIG-I in RNA virus recognition. Virology 479–480:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 46.Honda K, Yanai H, Takaoka A, Taniguchi T. 2005. Regulation of the type I IFN induction: a current view. Int Immunol 17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 47.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nan Y, Wu C, Zhang Y-J. 2017. Interplay between Janus kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front Immunol 8:1758. doi: 10.3389/fimmu.2017.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crosse KM, Monson EA, Beard MR, Helbig KJ. 2018. Interferon-stimulated genes as enhancers of antiviral innate immune signaling. J Innate Immun 10:85–93. doi: 10.1159/000484258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayashida E, Ling ZL, Ashhurst TM, Viengkhou B, Jung SR, Songkhunawej P, West PK, King NJC, Hofer MJ. 2019. Zika virus encephalitis in immunocompetent mice is dominated by innate immune cells and does not require T or B cells. J Neuroinflammation 16:177. doi: 10.1186/s12974-019-1566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. 2016. Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl Trop Dis 10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan S, Lew I, Wu F, Fritts L, Fontaine KA, Tomar S, Trapecar M, Shehata HM, Ott M, Miller CJ, Sanjabi S. 2019. Low expression of RNA sensors impacts Zika virus infection in the lower female reproductive tract. Nat Commun 10:4344. doi: 10.1038/s41467-019-12371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tripathi S, Balasubramaniam VRMT, Brown JA, Mena I, Grant A, Bardina SV, Maringer K, Schwarz MC, Maestre AM, Sourisseau M, Albrecht RA, Krammer F, Evans MJ, Fernandez-Sesma A, Lim JK, García-Sastre A. 2017. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog 13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja M-K, Diamond MS, Gale M Jr. 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suthar MS, Diamond MS, Gale M Jr. 2013. West Nile virus infection and immunity. Nat Rev Microbiol 11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 60.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M Jr. 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrows NJ, Campos RK, Liao K-C, Prasanth KR, Soto-Acosta R, Yeh S-C, Schott-Lerner G, Pompon J, Sessions OM, Bradrick SS, Garcia-Blanco MA. 2018. Biochemistry and molecular biology of flaviviruses. Chem Rev 118:4448–4482. doi: 10.1021/acs.chemrev.7b00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazeaud C, Freppel W, Chatel-Chaix L. 2018. The multiples fates of the flavivirus RNA genome during pathogenesis. Front Genet 9:595. doi: 10.3389/fgene.2018.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajah MM, Monel B, Schwartz O. 2020. The entanglement between flaviviruses and ER-shaping proteins. PLoS Pathog 16:e1008389. doi: 10.1371/journal.ppat.1008389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neufeldt CJ, Joyce MA, Van Buuren N, Levin A, Kirkegaard K, Gale M Jr, Tyrrell DLJ, Wozniak RW. 2016. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog 12:e1005428. doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, Li N, Dai S, Hou G, Guo K, Chen X, Yi C, Liu W, Deng F, Wu Y, Cao X. 2019. Zika virus circumvents host innate immunity by targeting the adaptor proteins MAVS and MITA. FASEB J 33:9929–9944. doi: 10.1096/fj.201900260R. [DOI] [PubMed] [Google Scholar]
- 66.Ma J, Ketkar H, Geng T, Lo E, Wang L, Xi J, Sun Q, Zhu Z, Cui Y, Yang L, Wang P. 2018. Zika Virus Non-structural Protein 4A Blocks the RLR-MAVS Signaling. Front Microbiol 9:1350. doi: 10.3389/fmicb.2018.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin S, Yang S, He J, Guest JD, Ma Z, Yang L, Pierce BG, Tang Q, Zhang Y-J. 2019. Zika virus NS5 protein antagonizes type I interferon production via blocking TBK1 activation. Virology 527:180–187. doi: 10.1016/j.virol.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li A, Wang W, Wang Y, Chen K, Xiao F, Hu D, Hui L, Liu W, Feng Y, Li G, Tan Q, Liu Y, Wu K, Wu J. 2020. NS5 conservative site is required for Zika virus to restrict the RIG-I signaling. Front Immunol 11:51. doi: 10.3389/fimmu.2020.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lundberg R, Melén K, Westenius V, Jiang M, Österlund P, Khan H, Vapalahti O, Julkunen I, Kakkola L. 2019. Zika virus non-structural protein NS5 inhibits the RIG-I pathway and interferon lambda 1 promoter activation by targeting IKK epsilon. Viruses 11:1024. doi: 10.3390/v11111024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ngueyen TTN, Kim S-J, Lee JY, Myoung J. 2019. Zika virus proteins NS2A and NS4A are major antagonists that reduce IFN-β promoter activity induced by the MDA5/RIG-I signaling pathway. J Microbiol Biotechnol 29:1665–1674. doi: 10.4014/jmb.1909.09017. [DOI] [PubMed] [Google Scholar]
- 71.Riedl W, Acharya D, Lee J-H, Liu G, Serman T, Chiang C, Chan YK, Diamond MS, Gack MU. 2019. Zika virus NS3 mimics a cellular 14–3-3-binding motif to antagonize RIG-I- and MDA5-mediated innate immunity. Cell Host Microbe 26:493–503.e6. doi: 10.1016/j.chom.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JY, Nguyen TTN, Myoung J. 2020. Zika virus NS2A and NS4A downregulate the promoter activity of nuclear factor kappa-light-chain-enhancer of activated B cells. J Microbiol Biotechnol 30:1651–1659. doi: 10.4014/jmb.2011.11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Airo AM, Felix-Lopez A, Mancinelli V, Evseev D, Lopez-Orozco J, Shire K, Paszkowski P, Frappier L, Magor KE, Hobman TC. 2022. Flavivirus capsid proteins inhibit the interferon response. Viruses 14:968. doi: 10.3390/v14050968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fanunza E, Carletti F, Quartu M, Grandi N, Ermellino L, Milia J, Corona A, Capobianchi MR, Ippolito G, Tramontano E. 2021. Zika virus NS2A inhibits interferon signaling by degradation of STAT1 and STAT2. Virulence 12:1580–1596. doi: 10.1080/21505594.2021.1935613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fanunza E, Grandi N, Quartu M, Carletti F, Ermellino L, Milia J, Corona A, Capobianchi MR, Ippolito G, Tramontano E. 2021. INMI1 Zika virus NS4B antagonizes the interferon signaling by suppressing STAT1 phosphorylation. Viruses 13:2448. doi: 10.3390/v13122448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roby JA, Esser-Nobis K, Dewey-Verstelle EC, Fairgrieve MR, Schwerk J, Lu AY, Soveg FW, Hemann EA, Hatfield LD, Keller BC, Shapiro A, Forero A, Stencel-Baerenwald JE, Savan R, Gale M Jr. 2020. Flavivirus nonstructural protein NS5 dysregulates HSP90 to broadly inhibit JAK/STAT signaling. Cells 9:899. doi: 10.3390/cells9040899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Best SM. 2017. The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J Virol 91:e01970-16. doi: 10.1128/JVI.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau V-M, Coulpier M, Missé D, Jouvenet N, Tabibiazar R, Gressens P, Schwartz O, Amara A. 2017. Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep 18:324–333. doi: 10.1016/j.celrep.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 80.Simonin Y, van Riel D, de Perre PV, Rockx B, Salinas S. 2017. Differential virulence between Asian and African lineages of Zika virus. PLoS Negl Trop Dis 11:e0005821. doi: 10.1371/journal.pntd.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchette NJ, Garcia R, Rudnick A. 1969. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg 18:411–415. doi: 10.4269/ajtmh.1969.18.411. [DOI] [PubMed] [Google Scholar]
- 82.Yun S-I, Song B-H, Frank JC, Julander JG, Polejaeva IA, Davies CJ, White KL, Lee Y-M. 2016. Complete genome sequences of three historically important, spatiotemporally distinct, and genetically divergent strains of Zika virus: MR-766, P6-740, and PRVABC-59. Genome Announc 4:e00800-16. doi: 10.1128/genomeA.00800-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fontes-Garfias CR, Shan C, Luo H, Muruato AE, Medeiros DBA, Mays E, Xie X, Zou J, Roundy CM, Wakamiya M, Rossi SL, Wang T, Weaver SC, Shi P-Y. 2017. Functional analysis of glycosylation of Zika virus envelope protein. Cell Rep 21:1180–1190. doi: 10.1016/j.celrep.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carbaugh DL, Baric RS, Lazear HM. 2019. Envelope protein glycosylation mediates Zika virus pathogenesis. J Virol 93:e00113-19. doi: 10.1128/JVI.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, Medeiros DBA, Zou J, Xie X, Giraldo MI, Vasconcelos PFC, Weaver SC, Wang T, Rajsbaum R, Shi P-Y. 2018. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun 9:414. doi: 10.1038/s41467-017-02816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goytain A, Ng T. 2020. NanoString nCounter technology: high-throughput RNA validation, p 125–139. In Li H, Elfman J (ed), Chimeric RNA: methods and protocols. Springer US, New York, NY. [DOI] [PubMed] [Google Scholar]
- 87.Wang B, Thurmond S, Zhou K, Sánchez-Aparicio MT, Fang J, Lu J, Gao L, Ren W, Cui Y, Veit EC, Hong H, Evans MJ, O’Leary SE, García-Sastre A, Zhou ZH, Hai R, Song J. 2020. Structural basis for STAT2 suppression by flavivirus NS5. Nat Struct Mol Biol 27:875–885. doi: 10.1038/s41594-020-0472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Willard KA, Demakovsky L, Tesla B, Goodfellow FT, Stice SL, Murdock CC, Brindley MA. 2017. Zika virus exhibits lineage-specific phenotypes in cell culture, in Aedes aegypti mosquitoes, and in an embryo model. Viruses 9:383. doi: 10.3390/v9120383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, Fedorova NB, Puri V, Shabman RS, Pulendran B, Suthar MS. 2017. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog 13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shao Q, Herrlinger S, Zhu Y-N, Yang M, Goodfellow F, Stice SL, Qi X-P, Brindley MA, Chen J-F. 2017. The African Zika virus MR-766 is more virulent and causes more severe brain damage than current Asian lineage and dengue virus. Dev Camb Engl 144:4114–4124. doi: 10.1242/dev.156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamel R, Ferraris P, Wichit S, Diop F, Talignani L, Pompon J, Garcia D, Liégeois F, Sall AA, Yssel H, Missé D. 2017. African and Asian Zika virus strains differentially induce early antiviral responses in primary human astrocytes. Infect Genet Evol 49:134–137. doi: 10.1016/j.meegid.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 92.Simonin Y, Loustalot F, Desmetz C, Foulongne V, Constant O, Fournier-Wirth C, Leon F, Molès J-P, Goubaud A, Lemaitre J-M, Maquart M, Leparc-Goffart I, Briant L, Nagot N, Van de Perre P, Salinas S. 2016. Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine 12:161–169. doi: 10.1016/j.ebiom.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramos da Silva S, Cheng F, Huang I-C, Jung JU, Gao S-J. 2019. Efficiencies and kinetics of infection in different cell types/lines by African and Asian strains of Zika virus. J Med Virol 91:179–189. doi: 10.1002/jmv.25306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guyatt KJ, Westaway EG, Khromykh AA. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods 92:37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- 95.Tan B-H, Fu J, Sugrue RJ, Yap E-H, Chan Y-C, Tan YH. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 96.Foo S-S, Chen W, Chan Y, Bowman JW, Chang L-C, Choi Y, Yoo JS, Ge J, Cheng G, Bonnin A, Nielsen-Saines K, Brasil P, Jung JU. 2017. Asian Zika virus strains target CD14+ blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nat Microbiol 2:1558–1570. doi: 10.1038/s41564-017-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol 80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thorne LG, Bouhaddou M, Reuschl A-K, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MVX, Hosmillo M, Fossati A, Ragazzini R, Jungreis I, Ummadi M, Rojc A, Turner J, Bischof ML, Obernier K, Braberg H, Soucheray M, Richards A, Chen K-H, Harjai B, Memon D, Hiatt J, Rosales R, McGovern BL, Jahun A, Fabius JM, White K, Goodfellow IG, Takeuchi Y, Bonfanti P, Shokat K, Jura N, Verba K, Noursadeghi M, Beltrao P, Kellis M, Swaney DL, García-Sastre A, Jolly C, Towers GJ, Krogan NJ. 2022. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602:487–495. doi: 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, García-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang J-J, Liao C-L, Liao J-T, Lee Y-L, Lin Y-L. 2009. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine 27:2746–2754. doi: 10.1016/j.vaccine.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 101.Slonchak A, Wang X, Aguado J, Sng JDJ, Chaggar H, Freney ME, Yan K, Torres FJ, Amarilla AA, Balea R, Setoh YX, Peng N, Watterson D, Wolvetang E, Suhrbier A, Khromykh AA. 2022. Zika virus noncoding RNA cooperates with the viral protein NS5 to inhibit STAT1 phosphorylation and facilitate viral pathogenesis. Sci Adv 8:eadd8095. doi: 10.1126/sciadv.add8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miner JJ, Diamond MS. 2017. Zika virus pathogenesis and tissue tropism. Cell Host Microbe 21:134–142. doi: 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yockey LJ, Jurado KA, Arora N, Millet A, Rakib T, Milano KM, Hastings AK, Fikrig E, Kong Y, Horvath TL, Weatherbee S, Kliman HJ, Coyne CB, Iwasaki A. 2018. Type I interferons instigate fetal demise after Zika virus infection. Sci Immunol 3:eaao1680. doi: 10.1126/sciimmunol.aao1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luo H, Li G, Wang B, Tian B, Gao J, Zou J, Shi S, Zhu S, Peng B-H, Adam A, Martinez A, Hein K, Winkelmann ER, Mahmoud Y, Zhou X, Shan C, Rossi S, Weaver S, Barrett ADT, Sun S-C, Zhang W, Shi P-Y, Wu P, Wang T. 2020. Peli1 signaling blockade attenuates congenital zika syndrome. PLoS Pathog 16:e1008538. doi: 10.1371/journal.ppat.1008538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Conde JN, Schutt W, Mladinich M, Sohn S-Y, Hearing P, Mackow ER. 2020. NS5 sumoylation directs nuclear responses that permit Zika virus to persistently infect human brain microvascular endothelial cells. J Virol 94:e01086-20. doi: 10.1128/JVI.01086-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bhatnagar J, Rabeneck DB, Martines RB, Reagan-Steiner S, Ermias Y, Estetter LBC, Suzuki T, Ritter J, Keating MK, Hale G, Gary J, Muehlenbachs A, Lambert A, Lanciotti R, Oduyebo T, Meaney-Delman D, Bolaños F, Saad EAP, Shieh W-J, Zaki SR. 2017. Zika virus RNA replication and persistence in brain and placental tissue. Emerg Infect Dis 23:405–414. doi: 10.3201/eid2303.161499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ireland DDC, Manangeeswaran M, Lewkowicz AP, Engel K, Clark SM, Laniyan A, Sykes J, Lee H-N, McWilliams IL, Kelley-Baker L, Tonelli LH, Verthelyi D. 2020. Long-term persistence of infectious Zika virus: Inflammation and behavioral sequela in mice. PLoS Pathog 16:e1008689. doi: 10.1371/journal.ppat.1008689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Strange DP, Jiyarom B, Sadri-Ardekani H, Cazares LH, Kenny TA, Ward MD, Verma S. 2021. Paracrine IFN response limits ZIKV infection in human sertoli cells. Front Microbiol 12:667146. doi: 10.3389/fmicb.2021.667146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wongsurawat T, Athipanyasilp N, Jenjaroenpun P, Jun S-R, Kaewnapan B, Wassenaar TM, Leelahakorn N, Angkasekwinai N, Kantakamalakul W, Ussery DW, Sutthent R, Nookaew I, Horthongkham N. 2018. Case of microcephaly after congenital infection with Asian lineage Zika virus, Thailand. Emerg Infect Dis 24:1758–1761. doi: 10.3201/eid2409.180416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lan PT, Quang LC, Huong VTQ, Thuong NV, Hung PC, Huong TTLN, Thao HP, Thao NTT, Mounts AW, Nolen LD. 2017. Fetal Zika virus infection in Vietnam. PLoS Curr 9. doi: 10.1371/currents.outbreaks.1c8f631e0ef8cd7777d639eba48647fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Davis MA, Voss K, Turnbull JB, Gustin AT, Knoll M, Muruato A, Hsiang T-Y, Dinnon KH, III, Leist SR, Nickel K, Baric RS, Ladiges W, Akilesh S, Smith KD, Gale M Jr. 2022. A C57BL/6 mouse model of SARS-CoV-2 infection recapitulates age- and sex-based differences in human COVID-19 disease and recovery. Vaccines 11:47. doi: 10.3390/vaccines11010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gong D, Zhang T-H, Zhao D, Du Y, Chapa TJ, Shi Y, Wang L, Contreras D, Zeng G, Shi P-Y, Wu T-T, Arumugaswami V, Sun R. 2018. High-throughput fitness profiling of Zika virus E protein reveals different roles for glycosylation during infection of mammalian and mosquito cells. iScience 1:97–111. doi: 10.1016/j.isci.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guo Y, Bao L, Xu Y, Li F, Lv Q, Fan F, Qin C. 2021. The ablation of envelope protein glycosylation enhances the neurovirulence of ZIKV and cell apoptosis in newborn mice. J Immunol Res 2021:5317662. doi: 10.1155/2021/5317662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jvi.01982-22-s0001.xlsx, XLSX file, 0.1 MB (149.6KB, xlsx)
Data Availability Statement
Viral consensus sequences were deposited in GenBank under the following accession numbers: OQ180929 (ZIKV/Dakar); OQ165285 (ZIKV/Malaysia). The NanoString data set can be found in File S1 in the supplemental material.