Abstract
Maize (Zea mays) is a leading cereal crop in the world. The maize kernel is the storage organ and the harvest portion of this crop and is closely related to its yield and quality. The development of maize kernel is initiated by the double fertilization event, leading to the formation of a diploid embryo and a triploid endosperm. The embryo and endosperm are then undergone independent developmental programs, resulting in a mature maize kernel which is comprised of a persistent endosperm, a large embryo, and a maternal pericarp. Due to the well-characterized morphogenesis and powerful genetics, maize kernel has long been an excellent model for the study of cereal kernel development. In recent years, with the release of the maize reference genome and the development of new genomic technologies, there has been an explosive expansion of new knowledge for maize kernel development. In this review, we overviewed recent progress in the study of maize kernel development, with an emphasis on genetic mapping of kernel traits, transcriptome analysis during kernel development, functional gene cloning of kernel mutants, and genetic engineering of kernel traits.
Keywords: Maize, Kernel development, QTL, Transcriptome, Gene cloning, Transgene
Introduction
Maize (Zea mays) is one of the world’s leading cereal crops along with rice and wheat, serving as a staple food, animal feed, and industrial raw materials (Troyer 2006). Since 2001, maize has become the highest tonnage crop worldwide, with the total production surpassing that of rice and wheat (UN/FAO, 2002). In 2018, the production of maize is about 1147 M tons, while 782 M and 734 M tons that of rice and wheat in the world (http://www.fao.org). Maize kernels, like in other cereal crops, are storage organ that contains essential components for plant growth and reproduction. The kernel is comprised of three distinct compartments contributing to overall energy density, including a persistent endosperm (83%), a large embryo (11%), and a maternal pericarp (6%). The kernel contains about 72% starch, 10% protein, 4% lipid, and micronutrients such as vitamins and minerals (Nuss and Tanumihardjo 2010). Maize provides an estimated 15% of the world’s protein and 20% of the world’s calories (Council 1988; Shiferaw et al. 2011), indicating its status as a paramount crop in the context of global nutrition.
The maize kernel development is initiated by the double fertilization event, leading to the formation of a diploid embryo and a triploid endosperm (Russell 1992). Early embryo development is marked by three major events corresponding to the acquisition of apicobasal polarity, the differentiation of epidermis, and the formation of the shoot and root meristem (Goldberg et al. 1994). Then, the embryo enters a maturation phase (Dumas and Rogowsky 2008). The development of endosperm starts with the fertilized central cells (Olsen 2001), followed by cellularization and differentiation into four main cell types, including basal endosperm transfer layer (BETL), aleurone layer (AL), starchy endosperm (SE), and embryo-surrounding region (ESR) (Consonni et al. 2005). During endosperm differentiation, mitotic cell proliferation and endoreduplication occur in endosperm cells, following by maturation (cell death, dormancy, and desiccation) (Sabelli and Larkins 2009). Although embryo and endosperm are clonally and functionally separated, genetic analyses reveal that they interact extensively throughout their development (Scanlon and Takacs 2009). With its well-characterized morphogenesis and powerful genetics, the maize kernel offers an exquisite experimental system.
The investigation of maize kernel development has a long history, starting with mutant collections in the early 1900s (Demerec 1923; Wentz 1930; Jones 1920; Mangelsdorf 1923). With the development of chemical mutagens, such as ethyl methane sulfona (EMS), mutagenesis via pollen greatly expanded the collections of maize kernel mutants (Neuffer and Coe 1978). Over 100 defective kernel (dek) mutants were reported with genetic, morphological, lethality, and embryo rescue studies (Neuffer and Sheridan 1980). These mutation loci are located throughout the maize chromosomes, and most of these mutants are lethal due to the failure of germination. A recent collection of EMS-mutagenized lines, which covered more than 80% of the annotated protein-coding genes in the maize genome, was generated and sequenced (Lu et al. 2018). It greatly expanded EMS induced mutant collections for potential functional analysis of genes and desirable allelic variants associated with maize kernel development.
Apart from chemically induced mutations, DNA transposons, such as Ac/Ds (McClintock 1948), En/Spm (McClintock 1953; Pereira et al. 1986), and Mutator (Robertson 1978), were also commonly used for mutagenesis in maize. The Mutator system, with the advantage of a high mutation efficiency and no apparent insertion site bias (Bennetzen 1996; Walbot 2000), has been widely used for the construction of genome-wide mutation libraries in maize (May et al. 2003; McCarty et al. 2005; Liang et al. 2019; Settles et al. 2007; Clark and Sheridan 1991; Sheridan and Clark 1993b; Scanlon et al. 1994; Marcon et al. 2020). Regional transposon mutation systems were also developed using Ac/Ds system, based on its preferential local transposition property (Cowperthwaite et al. 2002; Vollbrecht et al. 2010). Using active Robertson’s Mutator maize stocks (Robertson 1978), 51 embryo-specific (emb) mutants (Clark and Sheridan 1991; Sheridan and Clark 1993a) and 63 kernel mutants were reported (Scanlon et al. 1994). Several sequence-indexed Mutator-induced libraries, such as UniformMu (McCarty et al. 2005), ChinaMu (Liang et al. 2019), and BonnMu (Marcon et al. 2020), and Photosynthetic Mutant Library (PML) (http://pml.uoregon.edu/photosyntheticml.html) implemented a novel strategy for harnessing the power of high-copy transposons for functional analysis of maize genome. These libraries tagged more than 50% of the annotated maize genes, providing important resources for further genetic, biochemical, and molecular analysis of genes affecting kernel development.
Maize has been a leading system for molecular cloning of functional genes in plants. Among the earliest list of genetic loci in maize, storage protein genes (Geraghty et al. 1981; Burr et al. 1982; Pedersen et al. 1982), starch biosynthesis genes (Shure et al. 1983), and pigment biosynthesis genes (Fedoroff et al. 1984; O’Reilly et al. 1985) were the first batches to be molecularly cloned. With well-characterized endogenous transposon systems and the advent of transposon tagging (Wienand et al. 1982), maize led the way in gene isolation for several years. Dozens of genes affecting kernel development have been cloned from mutants isolated from different resources. For example, the first molecularly identified transcription factor (TF), OPAQUE2 (O2), was isolated by transposon tagging in 1987 (Schmidt et al. 1987). With the release of the maize reference genome (Schnable et al. 2009), as well as the success using a variety of gene cloning methods, such as positional cloning (Bortiri et al. 2006; Gallavotti and Whipple 2015), bulked-segregant analysis (BSA) (Klein et al. 2018; Dong et al. 2019; Michelmore et al. 1991), and transcription profiling (Jansen and Nap 2001; Cheung and Spielman 2002; Swanson-Wagner et al. 2006; Stupar and Springer 2006; Pea et al. 2008), the gene cloning in maize achieved a great leap in recent year. To date, a great number of maize kernel mutant genes have been cloned (Table 1), representing one of the largest mutant categories with cloned genes in maize. These mutants can be classified into several major kernel mutant types according to their mutant phenotypes. For example, the defective kernel (dek) mutants refer to those with affected development in both embryo and endosperm (Neuffer and Sheridan 1980), the empty pericarp (emp) mutants refer to those with empty pericarp or papery kernel (Scanlon et al. 1994), and the embryo specific (emb) mutants refer to those with morphogenic effects specific to the embryo (Clark and Sheridan 1991), etc. The cloning and functional analysis of numerous kernel development related genes greatly expanded our understanding of the molecular mechanisms during maize kernel development.
Table 1.
List of cloned genes essential for maize kernel development
| Mutant phenotype | Gene name | B73 v4 ID | Functional annotation | References |
|---|---|---|---|---|
| dek | Dek1 | Zm00001d028818 | Animal calpains homolog, plant signal transduction | (Lid et al. 2002; Becraft et al. 2002) |
| Dek2 | Zm00001d034882 | PPR protein, RNA splicing | (Qi et al. 2017b) | |
| Dek5 | Zm00001d039612 | Bacterial TamB homolog, chloroplast envelope biogenesis | (Zhang et al. 2019a) | |
| Dek10 | Zm00001d053802 | PPR protein, RNA editing | (Qi et al. 2017a) | |
| Dek15 | Zm00001d052197 | SCC4, chromosome segregation | (He et al. 2019) | |
| Dek19 | Zm00001d038257 | PPR protein, unknown function | (Dong et al. 2019) | |
| Dek33 | Zm00001d016475 | Pyrimidine reductase, riboflavin biosynthesis | (Dai et al. 2019) | |
| Dek35 | Zm00001d033749 | PPR protein, RNA splicing | (Chen et al. 2017b) | |
| Dek36 | Zm00001d013136 | PPR protein, RNA editing | (Wang et al. 2017) | |
| Dek37 | Zm00001d003543 | PPR protein, RNA splicing | (Dai et al. 2018) | |
| Dek38 | Zm00001d014595 | TTI2 cochaperone, male reproductive cell development | (Garcia et al. 2017) | |
| Dek39 | Zm00001d047013 | PPR protein, RNA editing | (Li et al. 2018c) | |
| Dek40 | Zm00001d011478 | PBAC4 chaperone, 20S CP biogenesis | (Wang et al. 2019a) | |
| Dek41/Dek43 | Zm00001d021053 | PPR protein, RNA splicing | (Ren et al. 2020; Zhu et al. 2019) | |
| Dek42/Rbm48 | Zm00001d054077 | RNA-binding protein, U12-type intron splicing | (Bai et al. 2019; Zuo et al. 2019) | |
| Dek44 | Zm00001d052865 | Mitochondrial ribosomal protein L9, respiratory genes expression | (Qi et al. 2019) | |
| Dek45 | Zm00001d023331 | PPR protein, RNA editing | (Ren et al. 2019a) | |
| Dek46 | Zm00001d043107 | PPR protein, RNA editing | (Xu et al. 2020) | |
| Dek53 | Zm00001d041326 | PPR protein, RNA editing | (Dai et al. 2020) | |
| Dek605 | Zm00001d016798 | PPR protein, RNA editing | (Fan et al. 2020) | |
| Dek*/ZmReas1 | Zm00001d038475 | AAA-ATPase60S, ribosome exporting | (Qi et al. 2016) | |
| smk | Smk1 | Zm00001d007100 | PPR protein, RNA editing | (Li et al. 2014b) |
| Smk2 | Zm00001d053981 | Glutaminase, Vitamin B6 Biosynthesis | (Yang et al. 2017b) | |
| Smk3 | Zm00001d041537 | Mitochondrial transcription termination factor, RNA splicing | (Pan et al. 2019a) | |
| Smk4 | Zm00001d049196 | PPR protein, RNA editing | (Wang et al. 2019b) | |
| Smk6 | Zm00001d025446 | PPR protein, RNA editing | (Ding et al. 2019) | |
| Smk7 | Zm00001d035960 | Subunit of RNA polymerase III, expression of tRNAs and 5S rRNA | (Zhao et al. 2020) | |
| Smk9 | Zm00001d000137 | PPR protein, RNA splicing | (Pan et al. 2019b) | |
| MPPR6 | Zm00001d034111 | PPR protein, translation | (Manavski et al. 2012) | |
| Ppr78 | Zm00001d034428 | PPR protein, RNA stabilization | (Zhang et al. 2017c) | |
| emp | Emp2 | Zm00001d005675 | HEAT SHOCK BINDING PROTEIN1, Heat Shock Response | (Fu et al. 2002) |
| Emp4 | Zm00001d033869 | PPR protein, expression of mitochondrial transcripts | (Gutierrez-Marcos et al. 2007) | |
| Emp5 | Zm00001d042039 | PPR protein, RNA editing | (Liu et al. 2013) | |
| Emp6 | Zm00001d005959 | Plant organelle RNA recognition protein, BETL cell differentiation | (Chettoor et al. 2015) | |
| Emp7 | Zm00001d008298 | PPR protein, RNA editing | (Sun et al. 2015a) | |
| Emp8 | Zm00001d049796 | PPR protein, RNA splicing | (Sun et al. 2018) | |
| Emp9 | Zm00001d022480 | PPR protein, RNA editing | (Yang et al. 2017a) | |
| Emp10 | Zm00001d033992 | PPR protein, RNA splicing | (Cai et al. 2017) | |
| Emp11 | Zm00001d052450 | PPR protein, RNA splicing | (Ren et al. 2017) | |
| Emp12 | Zm00001d002098 | PPR protein, RNA splicing | (Sun et al. 2019) | |
| Emp16 | Zm00001d011559 | PPR protein, RNA splicing | (Xiu et al. 2016) | |
| Emp18 | Zm00001d034253 | PPR protein, RNA editing | (Li et al. 2019b) | |
| Emp21 | Zm00001d033495 | PPR protein, RNA editing | (Wang et al. 2019c) | |
| Emp32 | Zm00001d040363 | PPR protein, RNA splicing | (Yang et al. 2020c) | |
| Emp602 | Zm00001d028046 | PPR protein, RNA splicing | (Ren et al. 2019b) | |
| Ppr14 | Zm00001d002157 | PPR protein, RNA splicing | (Wang et al. 2020) | |
| Ppr18 | Zm00001d007927 | PPR protein, RNA splicing | (Liu et al. 2020c) | |
| Ppr27 | Zm00001d029061 | RNA editing, multiple sites | (Liu et al. 2020d) | |
| Ppr101 | Zm00001d010942 | PPR protein, RNA splicing | (Yang et al. 2020a) | |
| Ppr231 | Zm00001d018219 | PPR protein, RNA splicing | (Yang et al. 2020a) | |
| Ppr-smr | Zm00001d002345 | PPR protein, RNA splicing | (Chen et al. 2019) | |
| emb | Emb-7L | Zm00001d021871 | Plastid PPR protein, RNA splicing | (Yuan et al. 2019) |
| Emb12 | Zm00001d018366 | Plastid initiation factor 3, plastid protein synthesis | (Shen et al. 2013) | |
| Emb14 | Zm00001d054079 | Plastid-targeted cGTPase, 30S ribosome formation | (Li et al. 2015b) | |
| Emb16/Why1 | Zm00001d036148 | WHIRLY1 (WHY1), genome stability and ribosome formation | (Zhang et al. 2013) | |
| Lem1 | Zm00001d034192 | Plastid 30S ribosomal protein S9 (PRPS9) | (Ma and Dooner 2004) | |
| PPR8522 | Zm00001d034962 | Plastid PPR protein, chloroplast transcription | (Sosso et al. 2012a) | |
| ZmPRPL35-1 | Zm00001d046555 | L35 of the large subunit of plastid ribosomes | (Magnard et al. 2004) | |
| opaque/floury | DeB30 | N/A | 19-kD alpha-zein protein | (Kim et al. 2004) |
| Fl1 | Zm00001d003398 | Endoplasmic reticulum protein, zein protein body formation | (Holding et al. 2007) | |
| Fl2 | Zm00001d049243 | 22-kD alpha-zein protein | (Coleman et al. 1995) | |
| Fl3 | Zm00001d009292 | PLATZ TF, tRNA and 5S rRNA transcription | (Li et al. 2017b) | |
| Fl4 | Zm00001d048851 | z1A 19-kD alpha-zein, protein body assembly | (Wang et al. 2014a) | |
| Mc | Zm00001d005793 | 16-kDa gamma-zein protein | (Kim et al. 2006) | |
| O1 | Zm00001d052110 | Myosin XI protein, endoplasmic reticulum motility and protein body formation | (Wang et al. 2012) | |
| O2 | Zm00001d018971 | bZIP TF, regulator of diverse processes in endosperm | (Schmidt et al. 1987) | |
| O5 | Zm00001d020537 | Monogalactosyldiacylglycerol synthase MGD1, galactolipids abundance | (Myers et al. 2011) | |
| O6/Pro1 | Zm00001d010056 | Δ1-Pyrroline-5- carboxylate synthetase, biosynthesis of proline | (Wang et al. 2014b) | |
| O7 | Zm00001d026649 | Acyl-activating enzyme, storage protein synthesis | (Wang et al. 2011; Miclaus et al. 2011) | |
| O10 | Zm00001d033654 | Cereal-specific PB protein, distribution of zeins | (Yao et al. 2016) | |
| O11/ZmZHOU | Zm00001d003677 | bHLH TF, regulator of endosperm development and nutrient metabolism | (Feng et al. 2018) | |
| Pbf1 | Zm00001d005100 | Dof TF, regulator of storage protein accumulation | (Vicente-Carbajosa et al. 1997) | |
| Ocd1 | Zm00001d008739 | Oxalyl-CoA Decarboxylase 1, catalyzes oxalyl-CoA into formyl-CoA and CO2 | (Yang et al. 2018) | |
| Os1/Shai1 | Zm00001d002661 | RWP-RK domain TF, regulator of nutrient allocation and embryonic patterning | (Song et al. 2019; Mimura et al. 2018) | |
| Pdk1 | Zm00001d038163 | Pyruvate phosphate dikinase (PPDK), energy charge and storage protein gene expression | (Lappe et al. 2018) | |
| Pdk2 | Zm00001d010321 | Pyruvate phosphate dikinase (PPDK), energy charge and storage protein gene expression | (Lappe et al. 2018) | |
| ZmNAC128 | Zm00001d040189 | NAC TF, starch and zein accumulation | (Zhang et al. 2019d) | |
| ZmNAC130 | Zm00001d008403 | NAC TF, starch and zein accumulation | (Zhang et al. 2019d) | |
| shrunken (Starch related genes) | Ae1 | Zm00001d016684 | Starch branching enzyme IIB, starch biosynthesis | (Fisher et al. 1996; Kim et al. 1998) |
| Bt2 | Zm00001d050032 | ADP-glucose pyrophosphorylase (AGPase), starch biosynthesis | (Preiss et al. 1990) | |
| Se1 | Zm00001d007657 | FANTASTIC FOUR (FAF) domain protein, starch biosynthesis | (Zhang et al. 2019c) | |
| Sh1 | Zm00001d045042 | Sucrose synthase, starch biosynthesis | (Chourey and Nelson 1976) | |
| Sh2 | Zm00001d044129 | Large subunit of AGPase, starch biosynthesis | (Bhave et al. 1990) | |
| Su1 | Zm00001d049753 | Isoamylase-type DBE, starch biosynthesis | (James et al. 1995) | |
| Wx1 | Zm00001d045462 | Granule-bound starch synthase, starch biosynthesis | (Shure et al. 1983) | |
| Others | Bige1 | Zm00001d012883 | MATE transporter | (Suzuki et al. 2015) |
| Cr4 | Zm00001d023425 | TNFR-like receptor kinase, BETL differentiation | (Becraft et al. 1996) | |
| de18 | Zm00001d023718 | Yucca1, IAA biosynthesis | (Bernardi et al. 2012) | |
| hda101 | Zm00001d053595 | Histone deacetylase | (Rossi et al. 2007) | |
| Mn1 | Zm00001d003776 | Cell Wall Invertase CWI-2, BETL differentiation | (Cheng et al. 1996) | |
| MRP-1 | Zm00001d010889 | Myb-related protein-1, regulator of the differentiation of transfer cells | (Gómez et al. 2002) | |
| Nkd1 | Zm00001d002654 | INDETERMINATE DOMAIN (IDD) TF, regulator of endosperm development | (Gontarek et al. 2016) | |
| Nkd2 | Zm00001d026113 | INDETERMINATE DOMAIN (IDD) TF, regulator of endosperm development | (Gontarek et al. 2016) | |
| Ppr20 | Zm00001d039548 | PPR protein, RNA splicing | (Yang et al. 2020b) | |
| Ppr2263 | Zm00001d045089 | PPR protein, RNA editing | (Sosso et al. 2012b) | |
| qKW9 | Zm00001d048451 | PPR protein, RNA editing | (Huang et al. 2020) | |
| qVE5/ZmPORB2 | Zm00001d013937 | Chloroplast protochlorophyllideoxidoreductase, chlorophyll metabolism | (Zhan et al. 2019) | |
| RBR1 | Zm00001d007407 | Retinoblastoma-related (RBR) genes, inhibit the cell cycle | (Sabelli et al. 2005; Sabelli et al. 2009) | |
| RBR3 | Zm00001d031678 | Retinoblastoma-related (RBR) genes, inhibit the cell cycle | (Sabelli et al. 2005; Sabelli et al. 2009) | |
| Rgh3 | Zm00001d016836 | U2AF(35) Related Protein, U2-, and U12-type intron splicing | (Fouquet et al. 2011; Gault et al. 2017) | |
| Sal1 | Zm00001d046599 | Human Chmp1 homolog, BETL differentiation | (Shen et al. 2003) | |
| SWEET4c | Zm00001d015912 | Sucrose-transporting homologs, hexose transport | (Sosso et al. 2015) | |
| thk1 | Zm00001d027278 | NOT1 subunit of the CCR4-NOT complex, cell division, signaling, differentiation and metabolism | (Wu et al. 2020) | |
| Ubl1 | Zm00001d017432 | U6 biogenesis-like 1, pre-mRNA splicing | (Li et al. 2017a) | |
| Urb2 | Zm00001d028096 | Urb2 domain-containing protein, pre-ribosomal RNA processing | (Wang et al. 2018b) | |
| Vks1 | Zm00001d018624 | ZmKIN11, regulates mitosis and cytokinesis | (Huang et al. 2019b) | |
| Ysl2 | Zm00001d017427 | Metal-nicotianamine (NA) transporter, Fe distribution | (Zang et al. 2020) | |
| ZmAFL1 | Zm00001d021790 | B3 domain TF, kernel filling | (Grimault et al. 2015) | |
| ZmAFL2 | Zm00001d011712 | B3 domain TF, kernel filling | (Grimault et al. 2015) | |
| Vp1 | Zm00001d042396 | B3 domain TF, regulator of AL development and embryo-endosperm protein reallocation | (McCarty et al. 1989; Zheng et al. 2019) | |
| ZmAFL4 | Zm00001d001838 | B3 domain TF, kernel filling | (Grimault et al. 2015) | |
| ZmAFL5 | Zm00001d034965 | B3 domain TF, kernel filling | (Grimault et al. 2015) | |
| ZmAFL6 | Zm00001d052750 | B3 domain TF, kernel filling | (Grimault et al. 2015) | |
| Mdh4 | Zm00001d032695 | Cytosolic malate dehydrogenase, catalyzes the conversion from OAA to malate | (Chen et al. 2020b) | |
| ZmDof3 | Zm00001d035651 | Dof TF, starch accumulation and aleurone development | (Qi et al. 2017c) | |
| ZmGE2 | Zm00001d029526 | Cytochrome p450 protein, embryo to endosperm ratio (EER) | (Zhang et al. 2012) | |
| Zmsmu2 | Zm00001d023239 | RNA-splicing factor, protein synthesis and RNA processing | (Chung et al. 2007) | |
| ZmSMR4 | Zm00001d047159 | CKI, regulates the transition between the mitotic cycle and endoreduplication | (Li et al. 2019a) | |
| ZmTar1 | Zm00001d037498 | ZmTA-Related1, IAA biosynthesis | (Chourey et al. 2010) | |
| ZmPIN1a | Zm00001d044812 | PIN-FORMED (PIN) family protein, auxin transport | (Carraro et al. 2006; Forestan et al. 2010) | |
| ZmPIN1b | Zm00001d018024 | PIN-FORMED (PIN) family protein, auxin transport | (Carraro et al. 2006; Forestan et al. 2010) | |
| ZmPIN1c | Zm00001d052269 | PIN-FORMED (PIN) family protein, auxin transport | (Carraro et al. 2006; Forestan et al. 2010) | |
| ZmVPS29 | Zm00001d053371 | Retromer complex component, kernel morphology | (Chen et al. 2020a) |
Together with recent advancements in high informative genetic mapping technology, such as GWAS, natural genetic variations related to maize kernel traits had been analyzed and captured (Xiao et al. 2017; Yan et al. 2011; Mir et al. 2019). The quick development of high-throughput sequencing technologies, such as RNA-seq, provided massive gene expression profiles during maize kernel development. Here, we provide an overview of recent progress in maize kernel development, with an emphasis on genetic mapping of kernel traits, transcriptome analysis during kernel development, functional gene identification of kernel mutants, and genetic engineering of kernel traits.
Genetic mapping of maize kernel traits
In crops, many quantitative agronomical traits, such as grain yield and plant architecture, are governed by quantitative trait loci (QTL). Genetic mapping and molecular characterization of these functional loci facilitate molecular marker assisted breeding in crop improvement. Linkage mapping is a well-established tool for studying the genetic basis of quantitative traits in plants. During the last three decades, numerous studies were conducted and thousands of QTLs associated with various traits have been identified using molecular markers in maize (reviewed in Xiao et al. 2017; Yan et al. 2011; Mir et al. 2019).
Of particular interests, hundreds of QTLs regulating kernel-related traits, such as kernel weight and kernel size, were identified under multiple kernel developmental stages and environments (Zhang et al. 2014; Zhang et al. 2016; Hao et al. 2019; Li et al. 2013b; Raihan et al. 2016; Yang et al. 2019; Peng et al. 2011; Jiang et al. 2015; Martinez et al. 2016; Chen et al. 2016a, b; Liu et al. 2017; Chen et al. 2017a; Zhang et al. 2017b). For example, Zhang et al. (2014) collected a total of 54 unconditional main QTLs for five kernel-related traits, including kernel weight (KW), volume (KV), length (KL), thickness (KT), and width (KWI) from an immortalized F2 (IF2) maize population. Using conditional mapping analysis, they found that KWI and KV had the strongest influence on KW at the individual QTL level, followed by KT and KL; KV was mostly strongly influenced by KT, followed by KWI and KL. Chen et al. (2016a) identified 56 main-effect QTLs for yield per plant (YPP), seven ear-related traits, and seven kernel-related traits, based on the genetic linkage map constructed using 2091 bins as markers. In particular, GRMZM2G168229, which encodes an SBP-box domain protein, was identified as the candidate gene for qKRN4-3 involving in the patterning of kernel row number (Chen et al. 2016a). Liu et al. (2017) identified a total of 729 QTLs regarding KL, KWI, KT, hundred KW, and kernel test weight in 10 recombinant inbred line populations. They identified 30 candidate genes that are orthologs of 18 rice genes associated with kernel size and weight, and confirmed the effects of five genes on maize kernel size/weight in an independent association mapping panel with 540 lines by candidate gene association analysis (Liu et al. 2017). For example, overexpression of ZmINCW1, an ortholog of the rice seed weight gene GRAIN INCOMPLETE FILLING1 (GIF1), can rescue the reduced weight of the Arabidopsis homozygous mutant line in AtcwINV2 (Arabidopsis ortholog of ZmINCW1), suggesting that these genes are conserved in both monocots and dicots.
These QTLs potentially contain major genes associated with the kernel development process and can be used to improve kernel yield and quality through marker-assisted selection. However, only few kernel-related QTLs have been cloned and characterized so far. For example, with the advantage of the reference genome of small-kernel inbred line, BARELY ANY MERISTEM1d (ZmBAM1d) was identified as the QTL responsible for kernel weight variation in maize (Yang et al. 2019). Other studies reported the cloning and characterization of previously identified major QTLs, qKM4.08 (Li et al. 2013b), qVE5 (Wang et al. 2018b), and qKW9 (Raihan et al. 2016; Yang et al. 2019), implementing the list of characterized kernel-related QTLs in maize (Zhan et al. 2019; Huang et al. 2020; Chen et al. 2020a). qVE5 encodes a chloroplast protochlorophyllide oxidoreductase (ZmPORB2) that is involved in chlorophyll metabolism enabling the production of phytol (Zhan et al. 2019). Overexpression of ZmPORB2 increased tocopherol content in both leaves and kernels. Interestingly, the tocopherol content was mainly determined by maternal effect (Zhan et al. 2019). The kernel size-related QTL qKW9 encodes a pentatricopeptide repeat (PPR) protein that affects photosynthesis and grain filling (Huang et al. 2020), highlighting the importance of optimizing photosynthesis for maize grain yield production. qKM4.08 encodes a retromer complex component ZmVPS29, overexpression of which confers a slender kernel morphology and increases the yield per plant in different maize genetic backgrounds (Chen et al. 2020a).
Association mapping has recently emerged as a tool to resolve complex trait variation by exploiting historical and evolutionary recombination events at the population level (Risch and Merikangas 1996; Nordborg and Tavaré 2002), providing a powerful tool for the dissection of complex agronomic traits in plants and animals (Altshuler et al. 2008; Hunter and Crawford 2008; Zhu et al. 2008; Rafalski 2010). With the development of next-generation sequencing technologies, and the release of the maize B73 reference genome (Schnable et al. 2009), great progress has been made in QTL mapping using genome-wide association analysis (GWAS) in maize. A great number of traits including molecular and cellular, developmental and agronomic, yield, and stress resistance have been comprehensively investigated using association analysis, along with a number of cloned and candidate genes for corresponding traits (reviewed in Xiao et al. 2017; Yan et al. 2011). Several studies identified more loci associated with kernel-related traits using association analysis, revealing the genetic structure of complex quantitative traits during maize kernel development (Li et al. 2013c; Li et al. 2018a; Liu et al. 2020b; Zhang et al. 2020b; Zhang et al. 2017a). For example, kernel components-related traits, such as oil/fatty acid (Li et al. 2013c) and amylose (Li et al. 2018a), were identified using 9 million single nucleotide polymorphisms (SNPs) from 464 inbred maize lines, and 1.03 million SNPs characterized in 368 maize inbred lines, respectively. Li et al. (2013c) identified 74 loci significantly associated with kernel oil concentration and fatty acid composition, and the 26 loci associated with oil concentration could explain up to 83% of the phenotypic variation using a simple additive model. Li et al. (2018a) identified 27 associated loci involving 39 candidate genes that were linked to amylose content including transcription factors, glycosyltransferases, glycosidases, and hydrolases. A recent study reported the investigation of the genetic basis of three kernel-related traits, KL, KWI, and KT, in an association panel and a biparental population (Liu et al. 2020b). Fifty QTLs controlling these traits were detected with a total of 73 candidate genes in seven environments in the intermated B73 × Mo17 (IBM) Syn10 doubled haploid (DH) population. These studies provide insights into the mechanism of maize kernel development and the improvement of molecular marker-assisted selection for high-yield breeding in maize.
Transcriptome analysis of developing maize kernels
With the emergence of RNA-seq (Marioni et al. 2008), many transcriptome profiling studies have been conducted to uncover kernel development in different plant species, such as Arabidopsis thaliana (Le et al. 2010; Belmonte et al. 2013) and Oryza sativa (Gao et al. 2013; Xu et al. 2012). A detailed transcriptome during maize kernel development can greatly enhance our understanding of mechanisms of gene functions and cellular processes during maize kernel development.
The first transcriptome profiling investigation of maize kernel development was conducted on 9 DAP embryo and endosperm (Lu et al. 2013). The RNA-seq generated about 11 million paired-end reads from both the endosperm and the embryo. About 50.7% (8556 of 16,878) of multiexonic genes were found to be alternatively spliced, among which some transcript isoforms were specifically expressed either in the endosperm or in the embryo. In addition, many metabolic activities were specifically assigned to the endosperm or the embryo, and a number of TFs and imprinting genes were found to be specifically expressed in the endosperm or the embryo (Lu et al. 2013). A preliminary atlas of temporal and spatial gene expression patterns for early kernel development was established using RNA-seq analysis at five stages of whole kernels (0, 2, 3, 4, and 6 DAP) and three stages of isolated endosperms (8, 10, and 12 DAP) of the B73 inbred line (Li et al. 2014a). In this study, the RNA-seq generated nearly 34,000 mRNAs in the 0–6 DAP kernel and 33,000 mRNAs in endosperm at 8–12 DAP. A total of 7629 temporally regulated genes were identified, and several of the temporal patterns correlate with key developmental transitions associated with kernel and endosperm development (Li et al. 2014a). This atlas revealed a correlation between the major temporal programs and specific spatial expression programs in different compartments or tissue types of the developing endosperm. In another RNA-seq analysis of maize endosperm development at 5, 10, 15, and 20 DAP (Qu et al. 2016), more than 11,000 alternative spliced protein-coding genes and 7633 differentially expressed genes were detected during the four developmental stages. A comprehensive study of gene regulatory networks (GRNs) using 78 maize seed transcriptome profiles identified highly interwoven network communities (Xiong et al. 2017). For example, the kernel phenotype contributing community is composed of mostly unknown genes interacting with Opaque2, Brittle endosperm1, and Shrunken2. This study predicted important candidate genes in interwoven network communities that may be crucial to maize kernel development. These studies provided comprehensive insights into the transcriptome dynamics during maize kernel development.
A comprehensive study of endosperm cell differentiation was conducted in major endosperm cell types (AL, BETL, ESR, SE, central starchy endosperm (CSE), and conducting zone (CZ)), the embryo, and four maternal compartments (nucellus (NU), placento-chalazal region (PC), pericarp (PE), and pedicel (PED)) using a coupled laser-capture microdissection and RNA-Seq strategy (Zhan et al. 2015). A total of 13,009 compartment-specific genes were identified for all captured compartments at 8 DAP. Coexpression modules associated with single or multiple kernel compartments were also identified using gene coexpression network analysis (Zhan et al. 2015). For example, a detailed analysis of a coexpression module highly correlated with the BETL identified a regulatory module activated by a previously characterized BETL regulator MRP-1 (Gómez et al. 2002). This study revealed the diverged gene expression programs between filial and maternal compartments, as well as an unexpected close correlation between the embryo and the endosperm. During early kernel development, the interface between the endosperm and the embryo is developmentally dynamic (Nowack et al. 2010; Bommert and Werr 2001; Ingram and Gutierrez-Marcos 2015). Indeed, the endosperm adjacent to scutellum (EAS) was recently identified through RNA-seq analysis, representing a developmentally dynamic interface influenced by the neighboring growing embryo (Doll et al. 2020). Further phenotypic analysis of loss-of-function mutants of genes enriched in the EAS will elucidate the biological role of this newly discovered endosperm subdomain.
On the other hand, two high-resolution studies provided highly valuable temporal transcriptome landscapes of maize kernel development (Chen et al. 2014; Yi et al. 2019). Using RNA-seq data generated from 53 samples at an interval of 2 days from 0 to 38 DAP kernels, Chen et al. (2014) detected a total of 26,105 seed-expressed genes, which includes 1614 TFs and 1258 kernel-specific genes. They clearly classified the detected genes into 16, 14, and 10 coexpression modules for the embryo, endosperm, and the whole kernel, respectively. For example, the coexpression modules C5 to C8 are the active storage accumulation phase which exhibits high expression of carbohydrate metabolism genes (Chen et al. 2014). This study provides a valuable resource for the in-depth understanding of the dynamics of gene expression throughout maize kernel development. A recent study from the same group reported a high temporal-resolution investigation of transcriptomes using 31 samples collected at an interval of 4 or 6 h within the first 6 days of maize kernel development (Yi et al. 2019). In this study, they detected a total of 22,790 expressed genes in the early stages of maize kernel development, including 1415 TFs and 1093 kernel-specific genes. In the first 16 h after pollination, coenocyte formation, cellularization, and differentiation stage, 160, 22, 112, and 569 kernel-specific genes were identified to have predominant expression, respectively. Using network analysis, they also predicted 31,256 interactions among 1317 TFs and 14,540 genes, uncovering major signaling such as calcium signaling, nucleosome, auxin response, and mitosis pathways in early developmental stage.
Gene cloning analysis of maize kernel mutants
PPR genes
A surprising outcome from the gene cloning of maize kernel mutants is that an overwhelming portion of cloned genes encodes PPR proteins (Table 2). The PPR proteins are recognized to be members of the alpha-solenoid superfamily proteins (Kobe and Kajava 2000; Small and Peeters 2000), which are found in all eukaryotes and function universally in organellar gene expression (Barkan and Small 2014). In angiosperms, the PPR family is one of the largest gene families accounting for 1–2% of nearly all genomes sequenced so far (Lurin et al. 2004; Wei and Han 2016; Chen et al. 2018a; Xing et al. 2018; Liu et al. 2016; Chen et al. 2018b; O'Toole et al. 2008). PPR proteins serve as sequence-specific RNA-binding proteins inside organelles (Barkan et al. 2012; Takenaka et al. 2013; Yagi et al. 2013; Yin et al. 2013; Cheng et al. 2016), and function in every step of organellar gene expression, including RNA stabilization, RNA cleavage, RNA translation, RNA splicing, and RNA editing (Schmitz-Linneweber and Small 2008; Fujii and Small 2011; Shikanai and Fujii 2013; Dahan and Mireau 2013). The plant PPR family consists of two major subfamilies, defined as P and PLS (Lurin et al. 2004), which exhibit diverse repertories of molecular functions, mostly in mitochondria and chloroplast (Colcombet et al. 2013).
Table 2.
Mitochondrial PPR proteins characterized in maize
| PPR type | Gene name | Mutant phenotype | B73 v4 ID | Functional annotation | References |
|---|---|---|---|---|---|
| PLS-type | Dek10 | dek | Zm00001d053802 | RNA editing, nad3-61, 62, and cox2-550 | (Qi et al. 2017a) |
| Dek36 | dek | Zm00001d013136 | RNA editing, atp4-59, nad7-383, and ccmFN-302 | (Wang et al. 2017) | |
| Dek39 | dek | Zm00001d047013 | RNA editing, nad3-247 and nad3-275 | (Li et al. 2018c) | |
| Dek45 | dek | Zm00001d023331 | RNA editing, cox3-314, nad2-26, and nad5-1916 | (Ren et al. 2019a) | |
| Dek46 | dek | Zm00001d043107 | RNA editing, D5-C22 of nad7 intron 3 and 4 | (Xu et al. 2020) | |
| Dek53 | dek | Zm00001d041326 | RNA editing, multiple sites | (Dai et al. 2020) | |
| Dek605 | dek | Zm00001d016798 | RNA editing, nad1-608 | (Fan et al. 2020) | |
| Emp5 | emp | Zm00001d042039 | RNA editing, multiple sites | (Liu et al. 2013) | |
| Emp7 | emp | Zm00001d008298 | RNA editing, ccmFN-1553 | (Sun et al. 2015a) | |
| Emp9 | emp | Zm00001d022480 | RNA editing, ccmB-43 and rps4-335 | (Yang et al. 2017a) | |
| Emp18 | emp | Zm00001d034253 | RNA editing, atp6-635 and cox2-449 | (Li et al. 2019b) | |
| Emp21 | emp | Zm00001d033495 | RNA editing, multiple sites | (Wang et al. 2019b) | |
| Ppr27 | emp | Zm00001d029061 | RNA editing, ccmFN-1357, rps3-707, rps12-221, rps13-100, 256, 287, nad2-355, and cox2-482 | (Liu et al. 2020d) | |
| Ppr2263 | smk | Zm00001d045089 | RNA editing, nad5-1550 and cob-908 | (Sosso et al. 2012b) | |
| Smk1 | smk | Zm00001d007100 | RNA editing, nad7-836 | (Li et al. 2014b) | |
| Smk4 | smk | Zm00001d049196 | RNA editing, cox1-1489 | (Wang et al. 2019a) | |
| Smk6 | smk | Zm00001d025446 | RNA editing, nad1-740, nad4L-110, nad7-739, and mttB-138,139 | (Ding et al. 2019) | |
| P-type | Dek2 | dek | Zm00001d034882 | RNA splicing, nad1 intron 1 | (Qi et al. 2017b) |
| Dek35 | dek | Zm00001d033749 | RNA splicing, nad4 intron 1 | (Chen et al. 2017b) | |
| Dek37 | dek | Zm00001d003543 | RNA splicing, nad2 intron 1 | (Dai et al. 2018) | |
| Dek41/Dek43 | dek | Zm00001d021053 | RNA splicing, nad4 intron 1 and 3 | (Zhu et al. 2019; Ren et al. 2020) | |
| Emp8 | emp | Zm00001d049796 | RNA splicing, nad1 intron 4, nad2 intron 1, and nad4 intron 1 | (Sun et al. 2018) | |
| Emp10 | emp | Zm00001d033992 | RNA splicing, nad2 intron 1 | (Cai et al. 2017) | |
| Emp11 | emp | Zm00001d052450 | RNA splicing, nad1 intron 1, 2, 3, and 4 | (Ren et al. 2017) | |
| Emp12 | emp | Zm00001d002098 | RNA splicing, nad2 intron 1, 2, and 4 | (Sun et al. 2019) | |
| Emp16 | emp | Zm00001d011559 | RNA splicing, nad2 intron 4 | (Xiu et al. 2016) | |
| Emp32 | emp | Zm00001d040363 | PPR protein, RNA splicing | (Yang et al. 2020c) | |
| Emp602 | emp | Zm00001d028046 | RNA splicing, nad4 intron 1 and 3 | (Ren et al. 2019b) | |
| MPPR6 | smk | Zm00001d034111 | Translation, rps3 mRNA | (Manavski et al. 2012) | |
| Ppr14 | emp | Zm00001d002157 | RNA splicing, nad2 intron 3, nad7 intron 1 and 2 | (Wang et al. 2020) | |
| Ppr18 | emp | Zm00001d007927 | RNA splicing, nad4 intron1 | (Liu et al. 2020c) | |
| Ppr20 | dek/smk | Zm00001d039548 | RNA splicing, nad2 intron3 | (Yang et al. 2020b) | |
| Ppr78 | smk | Zm00001d034428 | RNA stabilization, nad5 mature mRNA | (Zhang et al. 2017c) | |
| Ppr101 | emp | Zm00001d010942 | RNA splicing, nad5 introns 1 and 2 | (Yang et al. 2020a) | |
| Ppr231 | emp | Zm00001d018219 | RNA splicing, nad5 introns 1, 2, 3, and nad2 intron 3 | (Yang et al. 2020a) | |
| Ppr-smr1 | emp | Zm00001d002345 | RNA splicing, multiple introns | (Chen et al. 2019) |
Early studies of PPR mutants characterized a number of chloroplast-localized PPR genes involved in translation, RNA stability, and RNA splicing, such as chloroplast RNA processing 1 (crp1) (Barkan et al. 1994; Fisk et al. 1999), ppr2 (Williams and Barkan 2003), ppr4 (Schmitz-Linneweber et al. 2006), ppr5 (Williams-Carrier et al. 2008; Beick et al. 2008), and ppr10 (Pfalz et al. 2009). Homozygous kernels of these mutants can still germinate and survive as chlorophyll-deficient seedlings until kernel reserves are exhausted (Stern et al. 2004). A later study of PPR8522 reported the first case to associate the loss of a chloroplast-localized PPR gene with an embryo-lethal phenotype in maize (Sosso et al. 2012a). PPR8522 is necessary for the transcription of nearly all plastid-encoded genes, and the ppr8522 mutation caused an embryo-lethal phenotype, however, depending on the genetic background. A recently characterized PPR protein EMB-7L (Yuan et al. 2019), provided more evidence that chloroplast-localized PPR proteins play major roles in plastid translation, the mutation of which could possibly lead to embryo lethality.
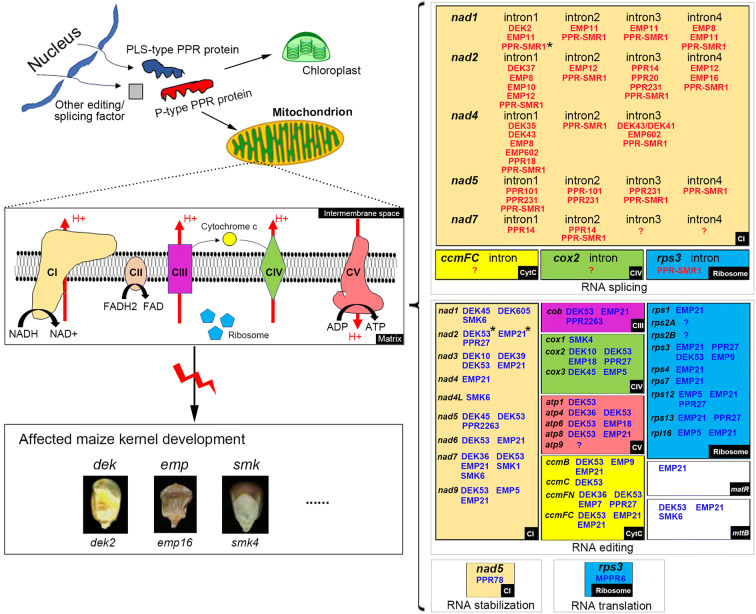
Mutations in mitochondrial PPR proteins are commonly associated with severe defects in kernel development in maize, possibly due to the disruption of respiration. The first functionally characterized maize mitochondrial PPR protein was EMP4, which was shown to be necessary for endosperm development (Gutierrez-Marcos et al. 2007). However, neither its mode of action nor its molecular target(s) is known. A study of ppr2263 kernel mutant provided the first phenotypic and molecular characterization in maize, in which PPR2263 was shown to be required for RNA editing at mitochondrial nad5-1550 and cob-908 sites, mitochondrial complex III assembly, and kernel development (Sosso et al. 2012b). In recent years, the number of characterized mitochondrial PPR proteins is expanding, with functions in almost every step of organellar gene expression, such as RNA splicing, RNA editing, RNA stabilization, and translation (Fig. 1). A summary of the detailed molecular functions of cloned mitochondrial PPR genes from maize kernel mutants is provided in Table 2.
Fig. 1.
Maize mitochondrial PPR proteins are important for kernel development. PPR proteins and other organellar editing/splicing factors are encoded in the nucleus, translated in the cytosol, and translocated into each organelle. Loss-of-function of mitochondrial PPR proteins usually causes impaired mitochondrial function and morphology, leading to defective maize kernel development with dek, emp, or smk kernel phenotypes. The dek phenotype refers to those with affected development in both embryo and endosperm, the emp phenotype refers to those with empty pericarp or papery kernel, and the smk phenotype refers to those with smaller kernels and delayed kernel development compared to the wild type. The dek and emp mutants are generally lethal, while a portion of smk mutants can germinate and develop. Shape and color variations indicate different protein families or mitochondrial respiratory components. CI to CV indicate mitochondrial complex I to complex V. PPR genes are listed by initials, targets and functions. The question marks indicate that no corresponding PPR proteins have been reported to be involved in splicing or editing of these specific mitochondrial group II introns or transcripts in maize. The asterisks mark PPR protein with multiple splicing or editing targets in maize mitochondrion
As we shall see, most reported mitochondrial PPR proteins typically function as site-specificity RNA splicing factors (P-type) or RNA editing factors (PLS-type), and each of these proteins is required for splicing or editing of only a small number of sites. The mutation of splicing factors of group II introns usually causes impaired assembly and activity of mitochondrial complexes, which results in defective mitochondrial function and morphology, leading to severe kernel phenotypes, such as dek and emp. For example, the maize Dek35 encodes a P-type PPR protein that is required for cis-spicing of mitochondrial nad4 intron 1 (Chen et al. 2017b). The dek35 mutation caused a deficiency in the complex I assembly and NADH dehydrogenase activity, producing lethal-kernel with a developmental deficiency (Chen et al. 2017b). Loss of function of maize P-type PPR protein EMP8 resulted in defects in cis-splicing of nad1 intron 4, nad4 intron 1, and nad2 intron 1, leading to severely arrested kernel development (Sun et al. 2018). However, recent studies of SMR-subgroup PPR-SMR1 revealed exceptions that the ppr-smr1 mutation affected RNA splicing of many mitochondrial group II introns (Chen et al. 2019). PPR-SMR1 is an SMR domain-containing P-type PPR protein, which is required for the splicing of 16 introns, accounting for nearly 75% of mitochondrial group II introns in maize. The failed splicing of these introns causes a deficiency of these proteins, leading to severe kernel phenotype in ppr-smr1 mutants (Chen et al. 2019). It is still unknown why PPR-SMR1 facilities splicing of a surprisingly large number of group II introns. The protein-protein interactions between the PPR-SMR1 and Zm-mCSF1 (Chen et al. 2019), as well as between the PPR-SMR1 and PPR14 (Wang et al. 2020) provided clues that these PPR proteins might be involved in the formation of large and dynamic splicing complexes.
PLS-type PPR proteins typically function as site-specific factors in RNA editing. In maize mitochondria, similar to P-type splicing factors, defects in PLS-type editing factors normally result in impaired mitochondrial function and severe kernel phenotype. The first characterized maize organellar RNA editing factor PPR2263 encoding a DYW domain-containing PPR protein that is required for RNA editing at nad5-1550 and cob-908 sites (Sosso et al. 2012b). The ppr2263 mutation caused reduced embryo and endosperm growth, resulting in small but viable kernels. Maize dek10 is a classic kernel mutant producing small kernels with delayed development (Qi et al. 2017a). Dek10 encodes an E-subgroup PPR protein required for RNA editing at nad3-61, nad3-62, and cox2-550 sites. Loss of dek10 function caused reduced assembly of complex IV and activity of NADH dehydrogenase, leading to small and shrunken kernels (Qi et al. 2017a). However, a recent study of maize EMP21, a DYW-subgroup PPR protein, showed the exception that loss of emp21 function affected RNA editing at 81 mitochondrial C targets, results in inhibited embryogenesis and delayed endosperm development (Wang et al. 2019c). Another interesting example is maize Dek53, which encodes an E-subgroup short PLS region-containing PPR protein (Dai et al. 2020). The mutation of dek53 affected RNA editing at over 60 mitochondrial C targets, resulted in the aborted assembly of mitochondrial complex III and lethal kernels (Dai et al. 2020). It was hypothesized that unique binding to specific nucleotides would not be sufficient with only seven repeats units in DEK53 to convey a tight connection within the transcriptome of the 569-kb large maize mitochondrial genome (Clifton et al. 2004). Alternatively, different genetic backgrounds might be responsible for the variation of editing extent at some sites (Kempken et al. 1995; Bentolila et al. 2005; Chu and Wei 2020).
It is intriguing that many of these reported PPR proteins have interactions with other splicing or editing factors, leading to the speculation that these proteins might function in plant organellar “group II intron spliceosome” (de Longevialle et al. 2010) or “RNA editosome” (Takenaka 2014). Indeed, PPR14 interacts with PPR-SMR1 and Zm-mCSF1 to mediate intron splicing of several mitochondrial group II introns in maize (Chen et al. 2019; Wang et al. 2020), suggesting that PPR14, PPR-SMR1, and Zm-mCSF1 may form a protein complex. In maize chloroplasts, CAF1 and CAF2 are two closely related proteins that function in concert with CRS2 to facility the splicing of group II introns (Ostheimer et al. 2006; Ostheimer et al. 2003). Several studies provided pieces of evidence that splicing factors could co-regulate the splicing of one or more specific introns by forming potential complexes (de Longevialle et al. 2010). In the case of cis-splicing event of mitochondrial nad2 intron 1, five PPR proteins have been reported to be involved, including EMP10 (Cai et al. 2017), DEK37 (Dai et al. 2018), EMP8 (Sun et al. 2018), EMP12 (Sun et al. 2019), and PPR-SMR1 (Chen et al. 2019). In Arabidopsis, several splicing factors, including MTSF1 (Haïli et al. 2013), mCSF1 (Zmudjak et al. 2013), PMH2 (Köhler et al. 2010), nMAT1 (Keren et al. 2012), and ODB1 (Samach et al. 2011), also participate in the splicing of nad2 intron 1. It is still unclear why the splicing of one specific intron needs several co-factors. It would be interesting to address whether the splicing of this specific intron was mediated by a splicing complex in mitochondria, as a high molecular weight ribonucleoprotein apparatus participating in psaA mRNA splicing has been identified in chloroplasts (Reifschneider et al. 2016).
The involvement of coordinated factors in organellar RNA editing has been studied in some detail (Sun et al. 2016), as the composition of a biochemically active editing complex has been determined in the chloroplast (Sandoval et al. 2019; Bentolila et al. 2012; Huang et al. 2019a). For example, RNA editing at ndhA C437 requires several coordinated factors, including RIPs, OZ1, ORRM1, ISE2, CLB19 and DYW2, which have all been identified in a ~ 670 kDa complex in maize chloroplast extracts (Sandoval et al. 2019), though genetic knockout data of RIPs, OZ1, ORRM1, and ISE2 were still from Arabidopsis mutant plants (Bentolila et al. 2012; Takenaka et al. 2012; Sun et al. 2013; Sun et al. 2015b; Bobik et al. 2017). Mitochondrial protein components of maize RNA editosome have also been reported in several studies, such as DEK53/ZmMORF1/ORRMs (Dai et al. 2020), PPR27/ZmMORF1 (Liu et al. 2020d), and EMP21/ZmMORF8 (Wang et al. 2019c). However, such composition of a biochemically active editing complex has not been identified in maize mitochondrial extracts, and further studies will be needed to determine the exact composition of mitochondrial editing complex.
TFs
TFs play critical roles as key regulators for gene expression during maize kernel development. The first characterized TF in maize is a bZIP family protein OPAQUE2 (O2) (Schmidt et al. 1987), regulating genes in nearly all zein families during endosperm development (Schmidt et al. 1990, 1992; Muth et al. 1996). The o2 mutation results in the opaque kernel phenotype with reduced levels of 22-kD α-zeins and increased lysine content (Mertz et al. 1964; Schmidt et al. 1987). Genome-wide identification of O2 regulated targets revealed that O2 regulated a diverse array of biological processes apart from zein biosynthesis (Frizzi et al. 2010; Hunter et al. 2002; Jia et al. 2013; Li et al. 2015a; Hartings et al. 2011; Zhan et al. 2018). Some of these diverse roles have been experimentally characterized recently. For example, an SnRK1-ZmRFWD3-O2 signaling axis that transduces source-to-sink signals and coordinates C and N assimilation was recently reported in developing maize kernels (Li et al. 2020). SUS-encoding genes (Sus1 and Sus2) can be specifically recognized and be transactivated by O2, demonstrating that O2 transcriptionally regulates the metabolic source entry for protein and starch synthesis during endosperm filling (Deng et al. 2020). These studies provide extensive evidence that O2 functions as a central player that not only transcriptionally regulates the expression of most zein genes but also, directly and indirectly, regulates starch synthesis.
Several studies subsequently indicate that O2 functions in a complex with other regulatory proteins, such as O2-heterodimerizing proteins (OHPs) (Pysh et al. 1993; Yang et al. 2016; Zhang et al. 2015), Prolamin-box binding factor1 (PBF1) (Vicente-Carbajosa et al. 1997; Zhang et al. 2015), and MADS47 (Qiao et al. 2016). OHP1 and OHP2 are O2-interacting bZIP proteins identified by screening an endosperm cDNA library with an O2 probe (Pysh et al. 1993). OHP1 and OHP2 can bind to the O2 target site in the promoters of 22-kD zein genes as a homodimer and as a heterodimer with O2 (Pysh et al. 1993) and can cotransactivate the 27-kD γ-zein promoter through protein-protein interaction with PBF1 (Zhang et al. 2015). OHPs recognize and transactivate α-zein promoters with much lower levels than O2 does, and the suppression of OHPs does not cause a significant reduction in the transcription of α-zein genes in the presence of O2 (Yang et al. 2016), indicating that OHPs function as minor TFs in this process while O2 is the primary TF. Knockdown of Ohp expression resulted in dramatically reduced RNA transcript and protein levels of 27-kD γ-zein, leading to a vitreous kernel phenotype (Zhang et al. 2015). PBF1 is a member of the Dof (DNA binding with one finger) class of plant Cys2-Cys2 zinc-finger DNA binding proteins that specifically binds to P-box in the promoters of all zein genes (Vicente-Carbajosa et al. 1997). Knockdown of Pbf1 expression resulted in opaque kernel phenotype and dramatically reduced synthesis of the 27-kDa γ- and 22-kDa α-zeins (Wu and Messing 2012; Zhang et al. 2015). O2 and PBF1 also affect starch synthesis by directly regulating Cytoplasmic pyruvate orthophosphate dikinase1 (cyPpdk1), cyPpdk2, and Starch synthase III, which are critical components in the starch biosynthetic enzyme complex (Zhang et al. 2016). A MADS-box protein ZmMADS47 is an O2-interacting protein that binds the CATGT motif in promoters of α-zein and 50-kD γ-zein genes. Transactivation of these promoters by ZmMADS47 requires the interaction with O2, thereby greatly enhances the transactivation activity (Qiao et al. 2016). A recent study identified another endosperm-specific TF, ZmbZIP22, a bZIP-type TF that binds to ACAGCTCA box in the 27-kD γ-zein promoter and activated its expression (Li et al. 2018b). The CRISPR/Cas9-generated zmbzip22 mutants showed significantly reduced accumulation of 27-kD γ-zein. Interestingly, ZmbZIP22 physically interacts with PBF1, OHP1, and OHP2, but not O2 (Li et al. 2018b), indicating that the expression of the 27-kD γ-zein gene is regulated by a complex mechanism. Two endosperm-specific NAC TFs, ZmNAC128 and ZmNAC130, are also involved in the regulation of zein genes (Zhang et al. 2019d). They specifically activated transcription of the 16-kDa γ-zein gene and Bt2, and lack of them caused reduced accumulation of protein and starch, leading to a shrunken kernel phenotype.
FLOURY3 (FL3) is an endosperm-specific TF regulated by genomic imprinting (Li et al. 2017). The fl3 mutation resulted in a semidominant negative mutant that exhibits severe defects in the endosperm. FL3 interacts with two critical factors of the RNA polymerase III, RNA polymerase III subunit 53, and transcription factor class C 1, to regulate maize endosperm development and storage reserve filling (Li et al. 2017). ZmDof3 encodes a plant-specific Dof TF that is essential for maize endosperm development (Qi et al. 2017c). Suppression of ZmDof3 resulted in a dek phenotype with reduced starch content and a partially patchy aleurone layer. ZmDOF3 directly regulates starch biosynthesis genes Du1 and Su2, and AL-associated TF Nkd1, to regulate the signaling system controlling starch accumulation and aleurone development (Qi et al. 2017c).
OPAQUE11 (O11) is an endosperm-specific bHLH TF that plays central roles in endosperm development and nutrient metabolism (Feng et al. 2018). The o11 mutation resulted in small and opaque endosperm with reduced starch and protein accumulation. O11 directly regulates key TFs in endosperm development and nutrient accumulation, such as NKD2, ZmDOF3, PBF1, and O2. O11 interacts with ZmICE1, an ortholog of Arabidopsis ICE1 functioning in cold tolerance (Chinnusamy et al. 2003) and endosperm development (Denay et al. 2014), to co-regulate genes involved in stress response during endosperm development. In addition, O11 also regulates genes specifically expressed in the ESR, such as ZmYoda, indicating that O11 might be a regulator for ESR development. Characterization of O11 highlighted an endosperm regulatory network centered around O11 in endosperm development, nutrient metabolism and stress responses (Feng et al. 2018).
VIVIPAROUS1 (VP1) is a well-documented TF that is specifically expressed in the AL and the embryo (McCarty et al. 1989; McCarty et al. 1991). VP1 is an ortholog of Arabidopsis ABA-INSENSITIVE3 (ABI3) that plays essential roles in kernel development (McCarty et al. 1989) and ABA signaling (Suzuki et al. 2003). Null alleles of vp1 and abi3 resulted in the loss of ABA sensitivity, leading to vivipary or non-dormancy in maize (McCarty et al. 1989) and Arabidopsis (Nambara et al. 1992; Nambara et al. 1994). VP1 mediates trans-repression of α-amylase expression in the AL, and the vp1 mutation repressed the germination response in developing mutant kernels (Hoecker et al. 1995). VP1 can also activate the expression of C1 (Hattori et al. 1992), a MYB family TF involved in anthocyanin biosynthesis (Cone et al. 1986; Paz-Ares et al. 1987). As the consequence, the vp1 mutation resulted in suppressed anthocyanin production in mutant kernels (McCarty et al. 1989). Interestingly, a recent study provided evidence that VP1 is also essential for scutellum development and protein reallocation from the endosperm to embryo (Zheng et al. 2019). This study used maize zein-RNAi knockdown mutants as a novel system to study nutrient allocation from the endosperm to embryo. VP1 transactivated many sulfur assimilation- and nutrient metabolism-associated genes that are differentially expressed between wild-type and zein-RNAi knockdown kernels (Zheng et al. 2019), suggesting a role for VP1 as a regulator for endosperm-embryo communication.
The maize naked endosperm1 (nkd1) nkd2 mutation produced multiple ALs with partially differentiated cells (Yi et al. 2015; Becraft and Asuncion-Crabb 2000). Nkd1 and Nkd2 encode the INDETERMINATE1 domain (IDD) containing TFs that are required for endosperm cell patterning and differentiation, possibly by regulating genes associated with cell cycle, cell growth, and division, such as Retinoblastoma-related1 and Mitotic cyclin 3B-like (Gontarek et al. 2016). nkd kernels also displayed a propensity for vivipary and anthocyanin-deficient phenotype, providing additional clues to how the NKD genes regulate endosperm development. Indeed, NKD1 and NKD2 directly activate the expression of Vp1 (Gontarek et al. 2016) and are predicted to directly activate R1. In addition, NKD1 and NKD2 can regulate O2 and PBF, indicating that NKD1 and NKD2 promote strorage protein accumulation during endosperm development. These evidences strengthened the concept that the aleurone and the starchy endosperm are not separate lineages (Becraft and Asuncion-Crabb 2000).
A prime example of BETL differentiation is ZmMRP-1, the so far only BETL-specific TF to be identified (Gómez et al. 2002). ZmMRP-1 preceded the formation of transfer cells (Gómez et al. 2002) and regulated the expression of many BETL genes, including Maternally expressed gene1 (MEG1) (Gutiérrez-Marcos et al. 2004), Transfer cell response regulator ZmTCRR-1 (Muñiz et al. 2006, 2010), and CRP-encoding genes ZmBETL1, ZmBETL2, ZmBETL9, and ZmBETL10 (Gómez et al. 2002, 2009), suggesting that ZmMRP-1 might be a key player involved in BETL differentiation. Indeed, the ectopic expression of ZmMRP-1 is sufficient to temporarily transform epidermal cells into transfer cells, and the expression of ZmMRP-1 is needed to maintain the BETL cell phenotype (Gómez et al. 2009). It is proposed that low base levels of ZmMn1 and ZmSWEET4c allow small amounts of glucose to enter the cell, which in turn induces ZmMRP-1, which then induces genes necessary for establishing the transfer cell machinery including ZmMn1 and ZmSWEET4c (Sosso et al. 2015). These properties of ZmMRP-1 had made it a determinant of BETL development.
Embryo-specific (Emb) genes
Embryo is the other important portion of fertilized maize kernel. Plant embryogenesis is a complex developmental process characterized by several major events (Kaplan and Cooke 1997). In maize, a number of embryo-specific (emb) mutants have been isolated from different genetic resources, making embryo lethality one of the most common mutant traits (Neuffer and Sheridan 1980; Clark and Sheridan 1991; Sheridan and Clark 1993c). In emb mutants, the endosperm develops normally, while the embryo shows severe developmental aberrations.
The first cloned emb mutant was lethal embryo1 (lem1), which possessed a normally developed endosperm and an aborted embryo at an early developmental stage (Ma and Dooner 2004). Lem1 encodes a plastid ribosomal protein S9 (RPS9), providing evidence that functional plastids are required for normal maize embryo development (Ma and Dooner 2004). A subsequent study of emb8516 showed a very similar kernel phenotype to lem1, in which the development of mutant embryos deviates as soon as the transition stage from that of wild-type sibling (Magnard et al. 2004). Emb8516 encodes a plastid ribosomal protein L35 (ZmPRPL35-1), which is part of 50S ribosome in plastids. The mutation caused a deficiency of protein synthesis in plastids, thereby the metabolic deficiency of embryo cells ultimately led to the abortion of the embryo. It is intriguing that several characterized emb genes, such as Ppr8522 (Sosso et al. 2012a), Emb14 (Li et al. 2015b), Emb12 (Shen et al. 2013), and Emb16 (Zhang et al. 2013), all encode plastid proteins as PPR protein, cGTPase, translation initiation factor 3, and DNA/RNA-binding protein WHIRLY1 (WHY1), respectively. Mutations in either gene caused arrested embryo development at the transition stage, suggesting that disrupts in plastid function might be linked to embryo lethality. It is generally not surprising that defects in plastid translation could produce a lethal embryo, as plastids play a fundamental role in the basic metabolism of plant cells, such as photosynthesis, fatty acid synthesis, and plant hormone biosynthesis (Bowsher and Tobin 2001). However, the characterization of some plastid PPR proteins such as PPR2 revealed exceptions, as these ppr mutant kernels displayed germinated embryo (Williams and Barkan 2003; Schmitz-Linneweber et al. 2006; Beick et al. 2008; Khrouchtchova et al. 2012). How do plastids affect embryogenesis has not been defined in maize, though genetic suppression might offer an explanation to the relationship between plastid translation and embryogenesis (Zhang et al. 2013).
Numerous efforts have been made on characterizing emb mutants in Arabidopsis, providing insights into potential mechanisms of plastid-mediated embryogenesis (Meinke 2020). A comprehensive screen for emb mutants altered in plastid proteins summarized three major types of plastid-localized proteins appearing to be most frequently associated with embryo lethality in Arabidopsis: (1) enzymes required for the biosynthesis of amino acids, vitamins, nucleotides, and fatty acids; (2) proteins required for the import, modification, and localization of essential proteins within the chloroplast; and (3) proteins required for chloroplast translation (Bryant et al. 2011). It is likely that disruption of a plastid protein should result in the lethal embryo if the function of that protein extends beyond photosynthesis. However, there is a difference in embryogenesis between maize and Arabidopsis with respect to the requirement for certain gene functions. Elucidation of this difference occurred in plastid proteins involved in posttranscriptional regulation. For example, AtPPR2 (Lu et al. 2011) and ZmPPR2 (Williams and Barkan 2003) share over 60% sequence identity at the amino acid level and are considered to be orthologous. However, the null mutation of Atppr2 and Zmppr2 result in totally different embryo fate. The mutant alleles of Atppr2 caused lethal embryo (Lu et al. 2011), while Zmppr2 null mutation did not lead to any defects in embryogenesis (Williams and Barkan 2003). Another example was from the studies of AtCAF2 (Asakura and Barkan 2006), ZmCAF2 (Ostheimer et al. 2003), and OsCAF2 (Shen et al. 2020). Loss of caf2 function results in plastid ribosome deficiency in all three species; however, embryo lethality only occurs in Arabidopsis. It will be interesting to have further evidences underline these striking differences in the developmental significance of plastid function in different plant species.
Genes related to cell cycle regulation
From about 4 to 20 DAP, the endosperm undergoes a phase of mitotic cell proliferation, followed by endoreduplication, during which a dramatic growth of the endosperm and the accumulation of storage compound emerges (Sabelli and Larkins 2009; Larkins et al. 2001). Factors affecting the cell cycle during endosperm development have been studied in maize, especially for cyclin-dependent kinases (CDKs) and CDK inhibitors. In eukaryotes, control of cell cycle progression is dependent on conserved molecular machinery consisting of protein kinases known as CDKs and their regulatory cyclin subunits (CYCs) (Vandepoele et al. 2002). In maize, reduced activity of type A CDK (CDKA), cdc2ZmA, inhibited the progression of endoreduplication cell cycles (Colasanti et al. 1991; Leiva-Neto et al. 2004). However, the lower level of endoreduplication did not affect cell size and only slightly reduced starch and storage protein accumulation (Leiva-Neto et al. 2004). CYCB2;2 is a type B2 cyclin that is sustained accumulated during maize endosperm development (Sabelli et al. 2014). The kinase activity of CYCB2;2 was observed in mitotic endosperm but was not or little observed in the immature ear and endoreduplicating endosperm, indicating that CYCB2;2 functions primarily during the mitotic cell cycle (Sabelli et al. 2014). Two CDK inhibitors, KIP-RELATED PROTEIN1 (KRP1) and KRP2, specifically inhibited cyclin A1;3- and cyclin D5;1-associated CDK activities (Coelho et al. 2005). Overexpression of KRP;1 in maize embryonic calli led to an additional round of DNA replication without nuclear division. Other CDK inhibitors, such as KRP1;1 and KRP4;2, are responsible for inhibiting kinase activity in CycD2;2-CDK, CycD4;2-CDK, and CycD5;3-CDK complexes (Godínez-Palma et al. 2017). A recent study of ZmSMR4, a member of SIAMESE-RELATED gene family, provided more evidence that CDK inhibitors play a role in plant growth and responses to abiotic stress (Li et al. 2019a).
Recent studies identified other factors regulating the cell cycle during maize kernel development (He et al. 2019; Huang et al. 2019b; Zhang et al. 2020a). During mitosis and meiosis, cohesin complexes maintain sister chromatid cohesion to ensure proper chromosome segregation (Haering et al. 2002; Nasmyth and Haering 2009). Maize Dek15 encodes a homolog of SISTER CHROMATID COHESION PROTEIN 4 (SCC4), a loader subunit of the cohesin ring. The dek15 mutation disrupted the mitotic cell cycle and endoreduplication, resulting in collapsed endosperm and embryo lethality (He et al. 2019). Another cohesin complex subunit structural maintenance of chromosome3 (SMC3) interacts with centromeric histone H3 (CENH3) during meiotic prophase I (Zhang et al. 2020a). Loss of Zmsmc3 function caused the premature loss of sister chromatid cohesion and mis-segregation of chromosomes in mitotic spreads (Zhang et al. 2020a). Maize VKS1 is a member of kinesin-14 subfamily that is essential for the migration of free nuclei in the coenocyte, as well as in mitosis and cytokinesis in early mitotic divisions (Huang et al. 2019b). The absence of vks1 caused reduced cell proliferation, resulting in varied kernel sizes (Huang et al. 2019b). With the advantage of kernel mutant libraries and transcriptome studies, more genes involved in cell cycle regulation might be identified in future studies, thereby further extending our understanding of the cell cycle regulation during maize kernel development.
Genes related with ribosome biogenesis and RNA processing
As a “house-building” function producing all cellular proteins, ribosome biogenesis is a highly regulated and coordinated multistep process (Lempiäinen and Shore 2009). It requires synthesis, processing, and modification of pre-rRNAs, assembly with ribosomal proteins, and transient interaction of numerous non-ribosomal factors with the evolving pre-ribosomal particles (Tschochner and Hurt 2003). A recent study demonstrated that “ITS1-first” and “5’ ETS-first” pathways coexist to promote the 35S pre-rRNA processing, highlighting a new 27SA pre-rRNA processing mechanism that is unique to maize and other higher plants (Liu et al. 2020a). In maize, three protein-coding genes related to rRNA and ribosome biosynthesis have been characterized (Gendra et al. 2004; Qi et al. 2016; Wang et al. 2018b). ZmDRH1 is a DEAE box RNA helicase that localizes to the nucleolus and interacts with the RNA binding protein MA6 and FIBRILLARIN (Gendra et al. 2004). MA16, ZmDRH1, and FIBRILLARIN may be forming part of a nucleolar RNP complex, and/or MA16 is an essential RNP-multifunctional protein component serving different functions in the cell. Reas1 is an AAA-ATPase that controls 60S ribosome export from the nucleus to the cytoplasm after ribosome maturation. The reas1 mutation partly repressed the maturation and export of the 60S ribosomal subunit, resulting in small kernels with delayed development (Qi et al. 2016). Maize Unhealthy Ribosome Biogenesis2 (URB2) is an ortholog of yeast URB2 that is involved in ribosome biogenesis. The urb2 kernels showed decreased ratios of 60S/40S and 80S/40S and increased ratios of polyribosomes, leading to thin kernels with delayed development (Wang et al. 2018a).
Two splicing factors, ROUGH ENDOSPERM3 (RGH3) (Fouquet et al. 2011; Gault et al. 2017) and RNA-binding motif protein 48 (RBM48)/DEK42 (Bai et al. 2019; Zuo et al. 2019), showed comprehensive evidence that the splicing of U2- and U12-type introns is critical for maize kernel development. Rgh3 encodes the maize U2AF35-related protein (URP) that is involved in both U2 and U12 splicing (Fouquet et al. 2011). The rgh3 mutation resulted in retained or miss-spliced U12-type introns, leading to a rough, etched, or pitted endosperm surface as well as a reduced kernel size (Gault et al. 2017). RBM48, also reported as DEK42 (Zuo et al. 2019), is a U12 splicing factor that functions to promote cell differentiation and repress cell proliferation. Mature rbm48 kernels had reduced endosperm size and lethal embryos (Bai et al. 2019). RBM48 can interact with RGH3, U2 auxiliary factor (U2AF), and armadillo repeat containing 7 (ARMC7), suggesting major and minor spliceosome factors required for intron recognition form complexes with RBM48. These studies greatly extended our understanding of the roles of U2- and U12-type intron splicing during maize kernel development.
Genes related with metabolism and other functions
Gene cloning of different maize kernel mutants also revealed functional genes involved in other biological processes, such as metabolic processes, ion/sugar transporting, and hormone biosynthesis. Metabolic enzymes, such as DEK33 (Dai et al. 2019) and SMK2 (Yang et al. 2017b), are involved in vitamin biosynthesis. Dek33 encodes a pyrimidine reductase in vitamin B2 biosynthesis, null mutation of dek33 caused aborted kernel development at the early developmental stage (Dai et al. 2019). SMK2 is a glutaminase in vitamin B6 biosynthesis, and the smk2 mutation resulted in arrested embryo development, but had a reduced role in endosperm development (Yang et al. 2017b). Mutation in pdg3, the third isozyme of oxPPP enzyme 6-phosphogluconate dehydrogenase (6PGDH), caused a rough endosperm kernel phenotype with reduced embryo oil (Spielbauer et al. 2013). ZmOcd1 encodes the oxalyl-CoA decarboxylase1 which catalyzes oxalyl-CoA, the product of O7 (Wang et al. 2011), into formyl-CoA and CO2 for degradation. Mutations in ocd1 caused dramatic alterations in the metabolome in the endosperm, leading to opaque endosperm phenotype (Yang et al. 2018). SEED CAROTENOID DEFICIENT (SCD) encodes an enzyme that converts 2C-methyl-d-erytrithol 2,4-cyclodiphosphate to 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate in the penultimate step of the methylerythritol phosphate (MEP) pathway in maize (Zhang et al. 2019b). The scd mutant kernels displayed pale-yellow phenotype with a reduced level of MEP-derived isoprenoids (Zhang et al. 2019b). Characterization of IAA biosynthesis genes, such as De18 (Bernardi et al. 2012; LeClere et al. 2010) and ZmTAR1 (Chourey et al. 2010), demonstrated the significance of the large abundance of IAA in developing maize kernel (Jensen and Bandurski 1994; LeClere et al. 2008). For example, the de18 mutation caused a large reduction of free IAA levels compared with De18, leading to an approximately 40% reduction of endosperm dry mass (Bernardi et al. 2012).
ZmSWEET4c mediates transepithelial hexose transport across the BETL (Sosso et al. 2015). Mutants of Zmsweet4c are defective in seed filling, indicating that a lack of hexose transport at the BETL impairs the further transfer of sugars imported from the maternal phloem (Sosso et al. 2015). A plasma membrane localized metal-nicotianamine (NA) transporter, ZmYSL2, plays an important role in Fe transport during kernel development. The Zmysl2 mutation produced smaller and collapsed kernels (Zang et al. 2020). Maize trans-Golgi associated gene Big embryo 1 (Bige1) encodes a MATE transporter BIGE1 that is required for transport of an intermediate or product associated with the CYP78A pathway. Loss of BIGE1 function caused accelerated leaf and root initiation as well as enlargement of the embryo scutellum (Suzuki et al. 2015). These studies greatly improved our understanding of the essential roles of genes involved in different biological processes during maize kernel development.
Genetic engineering
With the development of plant transformation technologies, genetic engineering became a powerful tool for crop improvement in addition to conventional breeding methods. Knowledge acquired from the identification of gene function and biochemical characterization of target proteins has been successfully applied to enhance the important agronomic traits in maize kernel (Smidansky et al. 2002; Smidansky et al. 2003; Hannah et al. 2012).
Modification of starch synthetic pathway in maize kernel is of particular interest, as the starch is the major component of yield. The synthesis of starch precursor (ADP-Glc) by the ADP-glucose pyrophosphorylase (AGPase) is a rate-limiting step in starch synthesis. AGPase is composed of two identical small and two identical large subunits. The small and large subunits of AGPase in maize endosperm are encoded by the brittle-2(bt2) and shrunken-2 (sh2) genes, respectively (Hannah 2007). The loss function of bt2 and sh2 would greatly reduce starch levels in the endosperm. Because of its allosteric properties and heat-labile characteristics, AGPase represents a suitable target for genetic manipulation. In early studies, site-specific mutagenesis by means of dissociation (Ds) transposon was used to generate a series of Sh2 revertants. Interestingly, a phosphate-insensitive mutation form of SH2 would increase seed weight 11–18% without impact the percentage of starch (Giroux et al. 1996). Heat-stable variants of AGP were also isolated by a bacterial expression system, and a single point mutation of Sh2 (Sh2hs33) was found to have increased heat stability through enhanced subunit interactions (Greene and Hannah 1998).Yield increase with enhanced AGPase transgenes was reported in different species with the use of either an engineered maize Sh2 (HS33/Rev6 Sh2) with enhanced heat stability and reduced orthophosphate inhibition or an enhanced Escherichia coli glgC-16 AGPase gene (Giroux et al. 1996; Smidansky et al. 2002; Wang et al. 2007; Smidansky et al. 2003; Lee et al. 2008; Obana et al. 2006; Sakulsingharoj et al. 2004). Surprisingly, the yield increase was due to an increased seed number rather than an increase in seed weight by expression Sh2 (Sh2hs33) driven by a native promoter in maize (Hannah et al. 2012). The extent of yield increase is also temperature-dependent. It was proposed that Sh2 not only played an important role in the endosperm but also functioned in maternal tissue to increase the frequency of seed development from the ovaries (Hannah et al. 2012). Similar to that of Sh2, the small subunit of AGPase, bt2, was found to function in maternal tissue to enhance the kernel set (Hannah et al. 2017). Nonetheless, it is showed that overexpression of wide type form of Sh2 and bt2 could increase individual maize seed weight and starch content compare to the control (Li et al. 2011).
Cell wall invertase is another example of genes which can be genetically engineered to increase maize kernel size and weight (Li et al. 2013a). It has been shown that the cell wall invertase gene plays an important role during the early grain filling (Wang et al. 2008). Mn1 encodes a cell wall invertase expressed in maize endosperm, loss function of Mn1 results in a significant reduction of seed mass (Cheng et al. 1996). When constitutively overexpression cell wall invertase gene from Arabidopsis, rice or maize, all of them can enforce the enzyme activities and promote biomass production up to ~ 145% in the transgenic maize (Li et al. 2013a). The substantially improved grain yield is due to increased starch content, enlarged kernel size, and enhanced kernel number (Li et al. 2013a).
Recently, overexpression of engineered Zmda1 and Zmdar1 in transgenic maize was also reported to enhance the sugar import into the kernels and show an improved kernel weight and kernel number (Xie et al. 2018). The ubiquitin receptor DA1/DA1R are negative regulators in seed size control, and a single base mutant DA1R358K leads to larger leaves, flowers, and seeds phenotype in Arabidopsis (Li et al. 2008). ZmDA1 and ZmDAR1 are homologous of AtDA1 and AtDAR1 in maize. The mutation forms of ZmDA1 and ZmDAR1 with the same single base change at the conserved sites (Zmda1 and Zmdar1) were transformed into maize separately. Three years of field trials confirmed that the overexpression with the mutated Zmda1 and Zmdar1 could promote basal endosperm transfer cell layer (BETL) development and the expression of starch synthase genes. On the contrary, overexpression of wild-type ZmDA1 and ZmDAR1 transgenic plants showed slowed growth and decreased yield (Xie et al. 2018).
Conclusion
In recent years, thanks to the rapid development of omics technologies, significant progress had been made in the studies of maize kernel development. Particularly, the impressive high-resolution transcriptome profiles for developing maize kernels laid a solid foundation for further studies in functional genes and gene regulatory networks. However, there is still room for improvement in gene expression analysis in the spatial dimensions. Although several studies provided valuable data based on microdissections, transcriptome at higher resolution would depend on single-cell sequencing technologies in the future. A large number of cloned maize kernel mutants greatly expanded our knowledge about functional genes and their regulation during maize kernel development. Nevertheless, compared to the model species such as Arabidopsis and rice, there is still a big gap in the number of functionally characterized genes in maize kernel development. Related to this, the available genes that can be effectively used to improving agronomic trait is very limited. In the following years, the cloning and functional analysis of more genes associated with maize kernel development is the major task. With the progress in functional genome studies and recent breakthroughs in maize stable transformation and CRISPR-Cas9 technology, modern breeding technology could further boost the yield and quality of maize kernels based on synthetic and system biology strategies.
Availability of data and materials
Not applicable.
Authors’ contributions
RS conceived and outlined the review. DD and RS performed the literature search. DD, ZM and RS wrote the paper.
Funding
This work was supported by the National Natural Science Foundation of China (grants 91935305 and 31730065 to R.S.) and the National Key Research and Development Program of China (grant 2016YFD0101003 to R.S.)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
This article is part of the Topical Collection on Maize Genetics, Genomics and Sustainable Improvement
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science (New York, NY) 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142(4):1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Corll J, Shodja DN, Davenport R, Feng G, Mudunkothge J, Brigolin CJ, Martin F, Spielbauer G, Tseung CW, Siebert AE, Barbazuk WB, Lal S, Settles AM. RNA binding motif protein 48 is required for U12 splicing and maize endosperm differentiation. Plant Cell. 2019;31(3):715–733. doi: 10.1105/tpc.18.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13(13):3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8(8):e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Asuncion-Crabb Y. Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development (Cambridge, England) 2000;127(18):4039–4048. doi: 10.1242/dev.127.18.4039. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science (New York, NY) 1996;273(5280):1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y. The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development (Cambridge, England) 2002;129(22):5217–5225. doi: 10.1242/dev.129.22.5217. [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol. 2008;28(17):5337–5347. doi: 10.1128/mcb.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci U S A. 2013;110(5):E435–E444. doi: 10.1073/pnas.1222061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. The Mutator transposable element system of maize. Curr Top Microbiol Immunol. 1996;204:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- Bentolila S, Chateigner-Boutin AL, Hanson MR. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol. 2005;139(4):2006–2016. doi: 10.1104/pp.105.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, Hanson MR. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci U S A. 2012;109(22):E1453–E1461. doi: 10.1073/pnas.1121465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi J, Lanubile A, Li QB, Kumar D, Kladnik A, Cook SD, Ross JJ, Marocco A, Chourey PS. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012;160(3):1318–1328. doi: 10.1104/pp.112.204743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave MR, Lawrence S, Barton C, Hannah LC. Identification and molecular characterization of shrunken-2 cDNA clones of maize. Plant Cell. 1990;2(6):581–588. doi: 10.1105/tpc.2.6.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik K, McCray TN, Ernest B, Fernandez JC, Howell KA, Lane T, Staton M, Burch-Smith TM. The chloroplast RNA helicase ISE2 is required for multiple chloroplast RNA processing steps in Arabidopsis thaliana. The Plant journal : for cell and molecular biology. 2017;91(1):114–131. doi: 10.1111/tpj.13550. [DOI] [PubMed] [Google Scholar]
- Bommert P, Werr W. Gene expression patterns in the maize caryopsis: clues to decisions in embryo and endosperm development. Gene. 2001;271(2):131–142. doi: 10.1016/s0378-1119(01)00503-0. [DOI] [PubMed] [Google Scholar]
- Bortiri E, Jackson D, Hake S. Advances in maize genomics: the emergence of positional cloning. Curr Opin Plant Biol. 2006;9(2):164–171. doi: 10.1016/j.pbi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Tobin AK. Compartmentation of metabolism within mitochondria and plastids. J Exp Bot. 2001;52(356):513–527. doi: 10.1093/jexbot/52.356.513. [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 2011;155(4):1678–1689. doi: 10.1104/pp.110.168120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B, Burr FA, St John TP, Thomas M, Davis RW. Zein storage protein gene family of maize. An assessment of heterogeneity with cloned messenger RNA sequences. J Mol Biol. 1982;154(1):33–49. doi: 10.1016/0022-2836(82)90415-6. [DOI] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F (2017) Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. The Plant journal : for cell and molecular biology 91(1):132–144. 10.1111/tpj.13551 [DOI] [PubMed]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142(1):254–264 [DOI] [PMC free article] [PubMed]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014;166(1):252–264. doi: 10.1104/pp.114.240689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li C, Li Y, Song Y, Zhang D, Wang T, Li Y, Shi Y. Quantitative trait loci mapping of yield and related traits using a high-density genetic map of maize. Mol Breed. 2016;36(9):134. doi: 10.1007/s11032-016-0545-0. [DOI] [Google Scholar]
- Chen L, Li YX, Li C, Wu X, Qin W, Li X, Jiao F, Zhang X, Zhang D, Shi Y, Song Y, Li Y, Wang T. Fine-mapping of qGW4.05, a major QTL for kernel weight and size in maize. BMC Plant Biol. 2016;16:81. doi: 10.1186/s12870-016-0768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, An Y, Li YX, Li C, Shi Y, Song Y, Zhang D, Wang T, Li Y. Candidate loci for yield-related traits in maize revealed by a combination of MetaQTL analysis and regional association mapping. Front Plant Sci. 2017;8:2190. doi: 10.3389/fpls.2017.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R. Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol Plant. 2017;10(3):427–441. doi: 10.1016/j.molp.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Chen G, Zou Y, Hu J, Ding Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics. 2018;19(1):720. doi: 10.1186/s12864-018-5088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Y-x L, Li C, Shi Y, Song Y, Zhang D, Li Y, Wang T. Genome-wide analysis of the pentatricopeptide repeat gene family in different maize genomes and its important role in kernel development. BMC Plant Biol. 2018;18(1):366. doi: 10.1186/s12870-018-1572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang HC, Shen J, Sun F, Wang M, Xu C, Tan BC. PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize. J Exp Bot. 2019;70(19):5245–5258. doi: 10.1093/jxb/erz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li YX, Li C, Shi Y, Song Y, Zhang D, Wang H, Li Y, Wang T. The retromer protein ZmVPS29 regulates maize kernel morphology likely through an auxin-dependent process(es) Plant Biotechnol J. 2020;18(4):1004–1014. doi: 10.1111/pbi.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fu Z, Zhang H, Tian R, Yang H, Sun C, Wang L, Zhang W, Guo Z, Zhang X, Tang J (2020b) Cytosolic malate dehydrogenase 4 modulates cellular energetics and storage reserve accumulation in maize endosperm. Plant Biotechnol J. 10.1111/pbi.13416 [DOI] [PMC free article] [PubMed]
- Cheng WH, Taliercio EW, Chourey PS. The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996;8(6):971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Gutmann B, Zhong X, Ye Y, Fisher MF, Bai F, Castleden I, Song Y, Song B, Huang J, Liu X, Xu X, Lim BL, Bond CS, Yiu SM, Small I. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. The Plant journal : for cell and molecular biology. 2016;85(4):532–547. doi: 10.1111/tpj.13121. [DOI] [PubMed] [Google Scholar]
- Chettoor AM, Yi G, Gomez E, Hueros G, Meeley RB, Becraft PW. A putative plant organelle RNA recognition protein gene is essential for maize kernel development. J Integr Plant Biol. 2015;57(3):236–246. doi: 10.1111/jipb.12234. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat Genet. 2002;32(4):522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Nelson OE. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem Genet. 1976;14(11–12):1041–1055. doi: 10.1007/bf00485135. [DOI] [PubMed] [Google Scholar]
- Chourey PS, Li QB, Kumar D. Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol Plant. 2010;3(6):1026–1036. doi: 10.1093/mp/ssq057. [DOI] [PubMed] [Google Scholar]
- Chu D, Wei L. Systematic analysis reveals cis and trans determinants affecting C-to-U RNA editing in Arabidopsis thaliana. BMC Genet. 2020;21(1):98. doi: 10.1186/s12863-020-00907-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Kim CS, Nguyen HN, Meeley RB, Larkins BA. The maize zmsmu2 gene encodes a putative RNA-splicing factor that affects protein synthesis and RNA processing during endosperm development. Plant Physiol. 2007;144(2):821–835. doi: 10.1104/pp.107.096214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JK, Sheridan WF. Isolation and characterization of 51 embryo-specific mutations of maize. Plant Cell. 1991;3(9):935–951. doi: 10.1105/tpc.3.9.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C, Meyer L, Wilson RK, Newton KJ. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136(3):3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho CM, Dante RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA. Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol. 2005;138(4):2323–2336. doi: 10.1104/pp.105.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Tyers M, Sundaresan V. Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc Natl Acad Sci U S A. 1991;88(8):3377–3381. doi: 10.1073/pnas.88.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Lopez-Obando M, Heurtevin L, Bernard C, Martin K, Berthomé R, Lurin C. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 2013;10(9):1557–1575. doi: 10.4161/rna.26128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Lopes MA, Gillikin JW, Boston RS, Larkins BA. A defective signal peptide in the maize high-lysine mutant floury 2. Proc Natl Acad Sci U S A. 1995;92(15):6828–6831. doi: 10.1073/pnas.92.15.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone KC, Burr FA, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci U S A. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni G, Gavazzi G, Dolfini S. Genetic analysis as a tool to investigate the molecular mechanisms underlying seed development in maize. Ann Bot. 2005;96(3):353–362. doi: 10.1093/aob/mci187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council NR (1988) Quality-protein maize: report of an ad hoc panel of the Advisory Committee on Technology Innovation Board on Science and Technology for International Development National Research Council, in Cooperation With the Board on Agriculture National Research Co. The National Academies Press, Washington, DC. 10.17226/18563
- Cowperthwaite M, Park W, Xu Z, Yan X, Maurais SC, Dooner HK (2002) Use of the Transposon <em>Ac</em> as a Gene-Searching Engine in the Maize Genome. Plant Cell 14 (3):713. doi:10.1105/tpc.010468 [DOI] [PMC free article] [PubMed]
- Dahan J, Mireau H. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 2013;10(9):1469–1476. doi: 10.4161/rna.25568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Luan S, Chen X, Wang Q, Feng Y, Zhu C, Qi W, Song R. Maize Dek37 encodes a P-type PPR protein that affects cis-splicing of mitochondrial nad2 intron 1 and seed development. Genetics. 2018;208(3):1069–1082. doi: 10.1534/genetics.117.300602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Tong H, Cheng L, Peng F, Zhang T, Qi W, Song R. Maize Dek33 encodes a pyrimidine reductase in riboflavin biosynthesis that is essential for oil-body formation and ABA biosynthesis during seed development. J Exp Bot. 2019;70(19):5173–5187. doi: 10.1093/jxb/erz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Jin L, Huo Z, Yan S, Ma Z, Qi W, Song R (2020) Pentatricopeptide repeat protein DEK46 is required for multi-sites mitochondrial RNA editing and maize seed development. J Exp Bot. 10.1093/jxb/eraa348 [DOI] [PubMed]
- de Longevialle AF, Small ID, Lurin C. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant. 2010;3(4):691–705. doi: 10.1093/mp/ssq025. [DOI] [PubMed] [Google Scholar]
- Demerec M. Heritable characters of maize XV—germless Seeds1. J Hered. 1923;14(7):297–300. doi: 10.1093/oxfordjournals.jhered.a102344. [DOI] [Google Scholar]
- Denay G, Creff A, Moussu S, Wagnon P, Thévenin J, Gérentes MF, Chambrier P, Dubreucq B, Ingram G. Endosperm breakdown in Arabidopsis requires heterodimers of the basic helix-loop-helix proteins ZHOUPI and INDUCER OF CBP EXPRESSION 1. Development (Cambridge, England) 2014;141(6):1222–1227. doi: 10.1242/dev.103531. [DOI] [PubMed] [Google Scholar]
- Deng Y, Wang J, Zhang Z, Wu Y (2020) Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol J. 10.1111/pbi.13349 [DOI] [PMC free article] [PubMed]
- Ding S, Liu XY, Wang HC, Wang Y, Tang JJ, Yang YZ, Tan BC. SMK6 mediates the C-to-U editing at multiple sites in maize mitochondria. J Plant Physiol. 2019;240:152992. doi: 10.1016/j.jplph.2019.152992. [DOI] [PubMed] [Google Scholar]
- Doll NM, Just J, Brunaud V, Caius J, Grimault A, Depege-Fargeix N, Esteban E, Pasha A, Provart NJ, Ingram GC, Rogowsky PM, Widiez T (2020) Transcriptomics at maize embryo/endosperm interfaces identifies a transcriptionally distinct endosperm sub-domain adjacent to the embryo scutellum. Plant Cell. 10.1105/tpc.19.00756 [DOI] [PMC free article] [PubMed]
- Dong J, Tu M, Feng Y, Zdepski A, Ge F, Kumar D, Slovin JP, Messing J. Candidate gene identification of existing or induced mutations with pipelines applicable to large genomes. The Plant journal : for cell and molecular biology. 2019;97(4):673–682. doi: 10.1111/tpj.14153. [DOI] [PubMed] [Google Scholar]
- Dumas C, Rogowsky P. Fertilization and early seed formation. Comptes rendus biologies. 2008;331(10):715–725. doi: 10.1016/j.crvi.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Fan K, Peng Y, Ren Z, Li D, Zhen S, Hey S, Cui Y, Fu J, Gu R, Wang J, Wang G, Li L (2020) Maize defective Kernel605 encodes a canonical DYW-type PPR protein that edits a conserved site of nad1 and is essential for seed nutritional quality. Plant & cell physiology. 10.1093/pcp/pcaa110 [DOI] [PubMed]
- Fedoroff NV, Furtek DB, Nelson OE. Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac) Proc Natl Acad Sci U S A. 1984;81(12):3825–3829. doi: 10.1073/pnas.81.12.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Qi W, Lv Y, Yan S, Xu L, Yang W, Yuan Y, Chen Y, Zhao H, Song R. OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell. 2018;30(2):375–396. doi: 10.1105/tpc.17.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DK, Gao M, Kim KN, Boyer CD, Guiltinan MJ. Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb. Plant Physiol. 1996;110(2):611–619. doi: 10.1104/pp.110.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18(9):2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S. ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2010;152(3):1373–1390. doi: 10.1104/pp.109.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet R, Martin F, Fajardo DS, Gault CM, Gómez E, Tseung CW, Policht T, Hueros G, Settles AM. Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development. Plant Cell. 2011;23(12):4280–4297. doi: 10.1105/tpc.111.092163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzi A, Caldo RA, Morrell JA, Wang M, Lutfiyya LL, Brown WE, Malvar TM, Huang S. Compositional and transcriptional analyses of reduced zein kernels derived from the opaque2 mutation and RNAi suppression. Plant Mol Biol. 2010;73(4–5):569–585. doi: 10.1007/s11103-010-9644-1. [DOI] [PubMed] [Google Scholar]
- Fu S, Meeley R, Scanlon MJ. Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell. 2002;14(12):3119–3132. doi: 10.1105/tpc.006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. The New phytologist. 2011;191(1):37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Whipple CJ (2015) Positional cloning in maize (Zea mays subsp. mays, Poaceae). 3(1):1400092. 10.3732/apps.1400092 [DOI] [PMC free article] [PubMed]
- Gao Y, Xu H, Shen Y, Wang J. Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA-Seq technique. Plant Mol Biol. 2013;81(4–5):363–378. doi: 10.1007/s11103-013-0009-4. [DOI] [PubMed] [Google Scholar]
- Garcia N, Li Y, Dooner HK, Messing J. Maize defective kernel mutant generated by insertion of a Ds element in a gene encoding a highly conserved TTI2 cochaperone. Proc Natl Acad Sci U S A. 2017;114(20):5165–5170. doi: 10.1073/pnas.1703498114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault CM, Martin F, Mei W, Bai F, Black JB, Barbazuk WB, Settles AM. Aberrant splicing in maize rough endosperm3 reveals a conserved role for U12 splicing in eukaryotic multicellular development. Proc Natl Acad Sci U S A. 2017;114(11):E2195–e2204. doi: 10.1073/pnas.1616173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendra E, Moreno A, Albà MM, Pages M. Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. The Plant journal : for cell and molecular biology. 2004;38(6):875–886. doi: 10.1111/j.1365-313X.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- Geraghty D, Peifer MA, Rubenstein I, Messing J. The primary structure of a plant-storage protein - ZEIN. Nucleic Acids Res. 1981;9(19):5163–5174. doi: 10.1093/nar/9.19.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux MJ, Shaw J, Barry G, Cobb BG, Greene T, Okita T, Hannah LC. A single mutation that increases maize seed weight. Proc Natl Acad Sci U S A. 1996;93(12):5824–5829. doi: 10.1073/pnas.93.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godínez-Palma SK, Rosas-Bringas FR, Rosas-Bringas OG, García-Ramírez E, Zamora-Zaragoza J, Vázquez-Ramos JM. Two maize Kip-related proteins differentially interact with, inhibit and are phosphorylated by cyclin D-cyclin-dependent kinase complexes. J Exp Bot. 2017;68(7):1585–1597. doi: 10.1093/jxb/erx054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science (New York, NY) 1994;266(5185):605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Gómez E, Royo J, Guo Y, Thompson R, Hueros G. Establishment of cereal endosperm expression domains: identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell. 2002;14(3):599–610. doi: 10.1105/tpc.010365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez E, Royo J, Muñiz LM, Sellam O, Paul W, Gerentes D, Barrero C, López M, Perez P, Hueros G. The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells. Plant Cell. 2009;21(7):2022–2035. doi: 10.1105/tpc.108.065409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontarek BC, Neelakandan AK, Wu H, Becraft PW. NKD transcription factors are central regulators of maize endosperm development. Plant Cell. 2016;28(12):2916–2936. doi: 10.1105/tpc.16.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene TW, Hannah LC. Enhanced stability of maize endosperm ADP-glucose pyrophosphorylase is gained through mutants that alter subunit interactions. Proc Natl Acad Sci U S A. 1998;95(22):13342–13347. doi: 10.1073/pnas.95.22.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimault A, Gendrot G, Chaignon S, Gilard F, Tcherkez G, Thévenin J, Dubreucq B, Depège-Fargeix N, Rogowsky PM. Role of B3 domain transcription factors of the AFL family in maize kernel filling. Plant science : an international journal of experimental plant biology. 2015;236:116–125. doi: 10.1016/j.plantsci.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O'Sullivan DM, Wormald M, Perez P, Dickinson HG. Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell. 2004;16(5):1288–1301. doi: 10.1105/tpc.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Dal Pra M, Giulini A, Costa LM, Gavazzi G, Cordelier S, Sellam O, Tatout C, Paul W, Perez P, Dickinson HG, Consonni G. Empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell. 2007;19(1):196–210. doi: 10.1105/tpc.105.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/S1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H (2013) The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res 41 (13):6650–6663. doi:10.1093/nar/gkt337 [DOI] [PMC free article] [PubMed]
- Hannah LC. Starch formation in the cereal endosperm. In: Olsen O-A, editor. Endosperm: developmental and molecular biology. Berlin Heidelberg, Berlin, Heidelberg: Springer; 2007. pp. 179–193. [Google Scholar]
- Hannah LC, Futch B, Bing J, Shaw JR, Boehlein S, Stewart JD, Beiriger R, Georgelis N, Greene T. A shrunken-2 transgene increases maize yield by acting in maternal tissues to increase the frequency of seed development. Plant Cell. 2012;24(6):2352–2363. doi: 10.1105/tpc.112.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC, Shaw JR, Clancy MA, Georgelis N, Boehlein SK. A brittle-2 transgene increases maize yield by acting in maternal tissues to increase seed number. Plant direct. 2017;1(6):e00029. doi: 10.1002/pld3.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Xue L, Zhang Z, Cheng Y, Chen G, Zhou G, Li P, Yang Z, Xu C. Combined linkage and association mapping reveal candidate loci for kernel size and weight in maize. Breed Sci. 2019;69(3):420–428. doi: 10.1270/jsbbs.18185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings H, Lauria M, Lazzaroni N, Pirona R, Motto M. The Zea mays mutants opaque-2 and opaque-7 disclose extensive changes in endosperm metabolism as revealed by protein, amino acid, and transcriptome-wide analyses. BMC Genomics. 2011;12:41. doi: 10.1186/1471-2164-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Vasil V, Rosenkrans L, Hannah LC, McCarty DR, Vasil IK. The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev. 1992;6:609–618. doi: 10.1101/gad.6.4.609. [DOI] [PubMed] [Google Scholar]
- He Y, Wang J, Qi W, Song R. Maize Dek15 encodes the Cohesin-loading complex subunit SCC4 and is essential for chromosome segregation and kernel development. Plant Cell. 2019;31(2):465–485. doi: 10.1105/tpc.18.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 1995;9(20):2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Holding DR, Otegui MS, Li B, Meeley RB, Dam T, Hunter BG, Jung R, Larkins BA. The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell. 2007;19(8):2569–2582. doi: 10.1105/tpc.107.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Li ZR, Yu QB, Ye LS, Cui YL, Molloy DP, Yang ZN. MORF2 tightly associates with MORF9 to regulate chloroplast RNA editing in Arabidopsis. Plant science : an international journal of experimental plant biology. 2019;278:64–69. doi: 10.1016/j.plantsci.2018.10.020. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang H, Huang X, Wang Q, Wang J, An D, Li J, Wang W, Wu Y. Maize VKS1 regulates mitosis and cytokinesis during early endosperm development. Plant Cell. 2019;31(6):1238–1256. doi: 10.1105/tpc.18.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lu G, Liu L, Raihan MS, Xu J, Jian L, Zhao L, Tran TM, Zhang Q, Liu J, Li W, Wei C, Braun DM, Li Q, Fernie AR, Jackson D, Yan J. The kernel size-related quantitative trait locus qKW9 encodes a pentatricopeptide repeat protein that affects photosynthesis and grain filling. Plant Physiol. 2020;183(4):1696–1709. doi: 10.1104/pp.20.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KW, Crawford NP. The future of mouse QTL mapping to diagnose disease in mice in the age of whole-genome association studies. Annu Rev Genet. 2008;42:131–141. doi: 10.1146/annurev.genet.42.110807.091659. [DOI] [PubMed] [Google Scholar]
- Hunter BG, Beatty MK, Singletary GW, Hamaker BR, Dilkes BP, Larkins BA, Jung R. Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell. 2002;14(10):2591–2612. doi: 10.1105/tpc.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G, Gutierrez-Marcos J. Peptide signalling during angiosperm seed development. J Exp Bot. 2015;66(17):5151–5159. doi: 10.1093/jxb/erv336. [DOI] [PubMed] [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7(4):417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends in genetics : TIG. 2001;17(7):388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Bandurski RS. Metabolism and synthesis of Indole-3-acetic acid (IAA) in Zea mays (levels of IAA during kernel development and the use of in vitro endosperm systems for studying IAA biosynthesis) Plant Physiol. 1994;106(1):343–351. doi: 10.1104/pp.106.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Wu H, Clay KL, Jung R, Larkins BA, Gibbon BC. Identification and characterization of lysine-rich proteins and starch biosynthesis genes in the opaque2 mutant by transcriptional and proteomic analysis. BMC Plant Biol. 2013;13:60. doi: 10.1186/1471-2229-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Ge M, Zhao H, Zhang T. Analysis of heterosis and quantitative trait loci for kernel shape related traits using triple testcross population in maize. PLoS One. 2015;10(4):e0124779. doi: 10.1371/journal.pone.0124779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DF. Heritable characters of maize: IV. A lethal factor—defective seeds. J Hered. 1920;11(4):161–168. doi: 10.1093/oxfordjournals.jhered.a101993. [DOI] [Google Scholar]
- Kaplan DR, Cooke TJ. Fundamental concepts in the embryogenesis of dicotyledons: a morphological interpretation of embryo mutants. Plant Cell. 1997;9(11):1903–1919. doi: 10.1105/tpc.9.11.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempken F, Höfken G, Pring DR. Analysis of silent RNA editing sites in atp6 transcripts of Sorghum bicolor. Curr Genet. 1995;27(6):555–558. doi: 10.1007/bf00314447. [DOI] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs Small CC, Araújo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. The Plant journal : for cell and molecular biology. 2012;71(3):413–426. doi: 10.1111/j.1365-313X.2012.04998.x. [DOI] [PubMed] [Google Scholar]
- Khrouchtchova A, Monde RA, Barkan A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA (New York, NY) 2012;18(6):1197–1209. doi: 10.1261/rna.032623.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Fisher DK, Gao M, Guiltinan MJ. Molecular cloning and characterization of the Amylose-Extender gene encoding starch branching enzyme IIB in maize. Plant Mol Biol. 1998;38(6):945–956. doi: 10.1023/a:1006057609995. [DOI] [PubMed] [Google Scholar]
- Kim CS, Hunter BG, Kraft J, Boston RS, Yans S, Jung R, Larkins BA. A defective signal peptide in a 19-kD alpha-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol. 2004;134(1):380–387. doi: 10.1104/pp.103.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Gibbon BC, Gillikin JW, Larkins BA, Boston RS, Jung R. The maize Mucronate mutation is a deletion in the 16-kDa gamma-zein gene that induces the unfolded protein response. The Plant journal : for cell and molecular biology. 2006;48(3):440–451. doi: 10.1111/j.1365-313X.2006.02884.x. [DOI] [PubMed] [Google Scholar]
- Klein H, Xiao Y, Conklin PA, Govindarajulu R, Kelly JA, Scanlon MJ, Whipple CJ, Bartlett M. Bulked-segregant analysis coupled to whole genome sequencing (BSA-Seq) for rapid gene cloning in maize. G3 (Bethesda, Md) 2018;8(11):3583–3592. doi: 10.1534/g3.118.200499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25(10):509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- Köhler D, Schmidt-Gattung S, Binder S. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol. 2010;72(4–5):459–467. doi: 10.1007/s11103-009-9584-9. [DOI] [PubMed] [Google Scholar]
- Lappe RR, Baier JW, Boehlein SK, Huffman R, Lin Q, Wattebled F, Settles AM, Hannah LC, Borisjuk L, Rolletschek H, Stewart JD, Scott MP, Hennen-Bierwagen TA, Myers AM. Functions of maize genes encoding pyruvate phosphate dikinase in developing endosperm. Proc Natl Acad Sci U S A. 2018;115(1):E24–e33. doi: 10.1073/pnas.1715668115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y. Investigating the hows and whys of DNA endoreduplication. J Exp Bot. 2001;52(355):183–192. doi: 10.1093/jexbot/52.355.183. [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, Drews GN, Fischer RL, Okamuro JK, Harada JJ, Goldberg RB. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci U S A. 2010;107(18):8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. Cell wall invertase-deficient miniature1 kernels have altered phytohormone levels. Phytochemistry. 2008;69(3):692–699. doi: 10.1016/j.phytochem.2007.09.011. [DOI] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010;153(1):306. doi: 10.1104/pp.110.155226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-M, Ryu T-H, Kim S-I, Okita T, Kim D. Kinetic and regulatory properties of plant ADP-glucose pyrophosphorylase genetically modified by heterologous expression of potato upreg mutants in vitro and in vivo. Plant Cell, Tissue and Organ Culture (PCTOC) 2008;96(2):161. doi: 10.1007/s11240-008-9472-z. [DOI] [Google Scholar]
- Leiva-Neto JT, Grafi G, Sabelli PA, Dante RA, Woo YM, Maddock S, Gordon-Kamm WJ, Larkins BA. A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell. 2004;16(7):1854–1869. doi: 10.1105/tpc.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21(6):855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008;22(10):1331–1336. doi: 10.1101/gad.463608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang S, Zhao Y, Li B, Zhang J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta. 2011;233(2):241–250. doi: 10.1007/s00425-010-1296-5. [DOI] [PubMed] [Google Scholar]
- Li B, Liu H, Zhang Y, Kang T, Zhang L, Tong J, Xiao L, Zhang H. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnol J. 2013;11(9):1080–1091. doi: 10.1111/pbi.12102. [DOI] [PubMed] [Google Scholar]
- Li C, Li Y, Sun B, Peng B, Liu C, Liu Z, Yang Z, Li Q, Tan W, Zhang Y, Wang D, Shi Y, Song Y, Wang T, Li Y. Quantitative trait loci mapping for yield components and kernel-related traits in multiple connected RIL populations in maize. Euphytica. 2013;193(3):303–316. doi: 10.1007/s10681-013-0901-7. [DOI] [Google Scholar]
- Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, Liu J, Warburton ML, Cheng Y, Hao X, Zhang P, Zhao J, Liu Y, Wang G, Li J, Yan J. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013;45(1):43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- Li G, Wang D, Yang R, Logan K, Chen H, Zhang S, Skaggs MI, Lloyd A, Burnett WJ, Laurie JD, Hunter BG, Dannenhoffer JM, Larkins BA, Drews GN, Wang X, Yadegari R. Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proc Natl Acad Sci U S A. 2014;111(21):7582–7587. doi: 10.1073/pnas.1406383111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Zhang YF, Hou M, Sun F, Shen Y, Xiu ZH, Wang X, Chen ZL, Sun SS, Small I, Tan BC. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa) The Plant journal : for cell and molecular biology. 2014;79(5):797–809. doi: 10.1111/tpj.12584. [DOI] [PubMed] [Google Scholar]
- Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, Xiao H, Song R. Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell. 2015;27(3):532–545. doi: 10.1105/tpc.114.134858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Shen Y, Meeley R, McCarty DR, Tan BC. Embryo defective 14 encodes a plastid-targeted cGTPase essential for embryogenesis in maize. The Plant journal : for cell and molecular biology. 2015;84(4):785–799. doi: 10.1111/tpj.13045. [DOI] [PubMed] [Google Scholar]
- Li J, Fu J, Chen Y, Fan K, He C, Zhang Z, Li L, Liu Y, Zheng J, Ren D, Wang G. The U6 biogenesis-like 1 plays an important role in maize kernel and seedling development by affecting the 3′ end processing of U6 snRNA. Mol Plant. 2017;10(3):470–482. doi: 10.1016/j.molp.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang J, Ye J, Zheng X, Xiang X, Li C, Fu M, Wang Q, Zhang Z, Wu Y. The maize imprinted gene Floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III. Plant Cell. 2017;29(10):2661–2675. doi: 10.1105/tpc.17.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Huang Y, Huang R, Wu Y, Wang W. The genetic architecture of amylose biosynthesis in maize kernel. Plant Biotechnol J. 2018;16(2):688–695. doi: 10.1111/pbi.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yue Y, Chen H, Qi W, Song R. The ZmbZIP22 transcription factor regulates 27-kD γ-Zein gene transcription during maize endosperm development. Plant Cell. 2018;30(10):2402–2424. doi: 10.1105/tpc.18.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gu W, Sun S, Chen Z, Chen J, Song W, Zhao H, Lai J. Defective Kernel 39 encodes a PPR protein required for seed development in maize. J Integr Plant Biol. 2018;60(1):45–64. doi: 10.1111/jipb.12602. [DOI] [PubMed] [Google Scholar]
- Li F, Wang L, Zhang Z, Li T, Feng J, Xu S, Zhang R, Guo D, Xue J. ZmSMR4, a novel cyclin-dependent kinase inhibitor (CKI) gene in maize (Zea mays L.), functions as a key player in plant growth, development and tolerance to abiotic stress. Plant science : an international journal of experimental plant biology. 2019;280:120–131. doi: 10.1016/j.plantsci.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Li XL, Huang WL, Yang HH, Jiang RC, Sun F, Wang HC, Zhao J, Xu CH, Tan BC. EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. The New phytologist. 2019;221(2):896–907. doi: 10.1111/nph.15425. [DOI] [PubMed] [Google Scholar]
- Li C, Qi W, Liang Z, Yang X, Ma Z, Song R (2020) A SnRK1-ZmRFWD3-Opaque2 signaling Axis regulates diurnal nitrogen accumulation in maize seeds. Plant Cell. 10.1105/tpc.20.00352 [DOI] [PMC free article] [PubMed]
- Liang L, Zhou L, Tang Y, Li N, Song T, Shao W, Zhang Z, Cai P, Feng F, Ma Y, Yao D, Feng Y, Ma Z, Zhao H, Song R. A sequence-indexed mutator insertional library for maize functional genomics study. Plant Physiol. 2019;181(4):1404–1414. doi: 10.1104/pp.19.00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci U S A. 2002;99(8):5460–5465. doi: 10.1073/pnas.042098799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell. 2013;25(3):868–883. doi: 10.1105/tpc.112.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-M, Xu Z-S, Lu P-P, Li W-W, Chen M, Guo C-H, Ma Y-Z. Genome-wide investigation and expression analyses of the pentatricopeptide repeat protein gene family in foxtail millet. BMC Genomics. 2016;17(1):840. doi: 10.1186/s12864-016-3184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Huang J, Guo H, Lan L, Wang H, Xu Y, Yang X, Li W, Tong H, Xiao Y, Pan Q, Qiao F, Raihan MS, Liu H, Zhang X, Yang N, Wang X, Deng M, Jin M, Zhao L, Luo X, Zhou Y, Li X, Zhan W, Liu N, Wang H, Chen G, Li Q, Yan J. The conserved and unique genetic architecture of kernel size and weight in maize and Rice. Plant Physiol. 2017;175:774–785. doi: 10.1104/pp.17.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yan P, Du Q, Wang Y, Guo Y, Fu Z, Wang H, Tang J. Pre-rRNA processing and its response to temperature stress in maize. J Exp Bot. 2020;71(4):1363–1374. doi: 10.1093/jxb/erz488. [DOI] [PubMed] [Google Scholar]
- Liu M, Tan X, Yang Y, Liu P, Zhang X, Zhang Y, Wang L, Hu Y, Ma L, Li Z, Zhang Y, Zou C, Lin H, Gao S, Lee M, Lübberstedt T, Pan G, Shen Y. Analysis of the genetic architecture of maize kernel size traits by combined linkage and association mapping. Plant Biotechnol J. 2020;18(1):207–221. doi: 10.1111/pbi.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Cao SK, Sayyed A, Xu C, Sun F, Wang X, Tan BC (2020c) The mitochondrial pentatricopeptide repeat protein PPR18 is required for the cis-splicing of nad4 intron 1 and essential to seed development in maize. Int J Mol Sci 21(11). 10.3390/ijms21114047 [DOI] [PMC free article] [PubMed]
- Liu R, Cao SK, Sayyed A, Yang HH, Zhao J, Wang X, Jia RX, Sun F, Tan BC (2020d) The DYW-subgroup pentatricopeptide repeat protein PPR27 functions on editing of multiple mitochondrial transcripts and interacts with ZmMORF1 in maize. J Exp Bot. 10.1093/jxb/eraa273 [DOI] [PubMed]
- Lu Y, Li C, Wang H, Chen H, Berg H, Xia Y. AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. The Plant journal : for cell and molecular biology. 2011;67(1):13–25. doi: 10.1111/j.1365-313X.2011.04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Chen D, Shu D, Zhang Z, Wang W, Klukas C, Chen LL, Fan Y, Chen M, Zhang C. The differential transcription network between embryo and endosperm in the early developing maize seed. Plant Physiol. 2013;162(1):440–455. doi: 10.1104/pp.113.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Liu J, Ren W, Yang Q, Chai Z, Chen R, Wang L, Zhao J, Lang Z, Wang H, Fan Y, Zhao J, Zhang C. Gene-indexed mutations in maize. Mol Plant. 2018;11(3):496–504. doi: 10.1016/j.molp.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette ML, Mireau H, Peeters N, Renou JP, Szurek B, Taconnat L, Small I. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16(8):2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Dooner HK. A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. The Plant journal : for cell and molecular biology. 2004;37(1):92–103. doi: 10.1046/j.1365-313x.2003.01942.x. [DOI] [PubMed] [Google Scholar]
- Magnard JL, Heckel T, Massonneau A, Wisniewski JP, Cordelier S, Lassagne H, Perez P, Dumas C, Rogowsky PM. Morphogenesis of maize embryos requires ZmPRPL35-1 encoding a plastid ribosomal protein. Plant Physiol. 2004;134(2):649–663. doi: 10.1104/pp.103.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N, Guyon V, Meurer J, Wienand U, Brettschneider R. An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell. 2012;24(7):3087–3105. doi: 10.1105/tpc.112.099051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf PC. The inheritance of defective seeds in Maize1. J Hered. 1923;14(3):119–125. doi: 10.1093/oxfordjournals.jhered.a102290. [DOI] [Google Scholar]
- Marcon C, Altrogge L, Win YN, Stöcker T, Gardiner JM, Portwood JL, Opitz N, Kortz A, Baldauf J, Hunter CT, McCarty DR, Koch KE, Schoof H, Hochholdinger F (2020) BonnMu: a sequence-indexed resource of transposon-induced maize mutations for functional genomics studies. Plant Physiol. 10.1104/pp.20.00478 [DOI] [PMC free article] [PubMed]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AK, Soriano JM, Tuberosa R, Koumproglou R, Jahrmann T, Salvi S. Yield QTLome distribution correlates with gene density in maize. Plant science : an international journal of experimental plant biology. 2016;242:300–309. doi: 10.1016/j.plantsci.2015.09.022. [DOI] [PubMed] [Google Scholar]
- May BP, Liu H, Vollbrecht E, Senior L, Rabinowicz PD, Roh D, Pan X, Stein L, Freeling M, Alexander D, Martienssen R. Maize-targeted mutagenesis: a knockout resource for maize. Proc Natl Acad Sci U S A. 2003;100(20):11541–11546. doi: 10.1073/pnas.1831119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Carson CB, Stinard PS, Robertson DS. Molecular analysis of viviparous-1: An abscisic acid-insensitive mutant of maize. Plant Cell. 1989;1(5):523–532. doi: 10.1105/tpc.1.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66(5):895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, Messing J, Koch KE, Hannah LC. Steady-state transposon mutagenesis in inbred maize. The Plant journal : for cell and molecular biology. 2005;44(1):52–61. doi: 10.1111/j.1365-313X.2005.02509.x. [DOI] [PubMed] [Google Scholar]
- McClintock B. Mutable loci in maize. Carnegie Institution of Washington Yearbook No. 1948;48:1948–1949. [Google Scholar]
- McClintock B (1953) Mutations in maize and chromosomal aberrations in Neurospora. Carnegie Institution of Washington. Yearbook No. 53
- Meinke DW. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. The New phytologist. 2020;226(2):306–325. doi: 10.1111/nph.16071. [DOI] [PubMed] [Google Scholar]
- Mertz ET, Bates LS, Nelson OE. Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science (New York, NY) 1964;145(3629):279–280. doi: 10.1126/science.145.3629.279. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclaus M, Wu Y, Xu JH, Dooner HK, Messing J. The maize high-lysine mutant opaque7 is defective in an acyl-CoA synthetase-like protein. Genetics. 2011;189(4):1271–1280. doi: 10.1534/genetics.111.133918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura M, Kudo T, Wu S, McCarty DR, Suzuki M (2018) Autonomous and non-autonomous functions of the maize Shohai1 gene, encoding a RWP-RK putative transcription factor, in regulation of embryo and endosperm development. The Plant journal : for cell and molecular biology. 10.1111/tpj.13996 [DOI] [PubMed]
- Mir RR, Reynolds M, Pinto F, Khan MA, Bhat MA. High-throughput phenotyping for crop improvement in the genomics era. Plant science : an international journal of experimental plant biology. 2019;282:60–72. doi: 10.1016/j.plantsci.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Muñiz LM, Royo J, Gómez E, Barrero C, Bergareche D, Hueros G. The maize transfer cell-specific type-A response regulator ZmTCRR-1 appears to be involved in intercellular signalling. The Plant journal : for cell and molecular biology. 2006;48(1):17–27. doi: 10.1111/j.1365-313X.2006.02848.x. [DOI] [PubMed] [Google Scholar]
- Muñiz LM, Royo J, Gómez E, Baudot G, Paul W, Hueros G. Atypical response regulators expressed in the maize endosperm transfer cells link canonical two component systems and seed biology. BMC Plant Biol. 2010;10:84. doi: 10.1186/1471-2229-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth JR, Müller M, Lohmer S, Salamini F, Thompson RD. The role of multiple binding sites in the activation of zein gene expression by Opaque-2. Mol Gen Genet. 1996;252(6):723–732. doi: 10.1007/bf02173979. [DOI] [PubMed] [Google Scholar]
- Myers AM, James MG, Lin Q, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW. Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell. 2011;23(6):2331–2347. doi: 10.1105/tpc.111.087205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Naito S, McCourt P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 1992;2:435–441. doi: 10.1111/j.1365-313X.1992.00435.x. [DOI] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S. Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant & cell physiology. 1994;35:509–513. [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Coe EH. Paraffin oil technique for treating mature corn pollen with chemical mutagens. Maydica. 1978;23(1):21–28. [Google Scholar]
- Neuffer MG, Sheridan WF. Defective kernel mutants of maize. I Genetic and lethality studies Genetics. 1980;95(4):929–944. doi: 10.1093/genetics/95.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Tavaré S. Linkage disequilibrium: what history has to tell us. Trends in genetics : TIG. 2002;18(2):83–90. doi: 10.1016/s0168-9525(02)02557-x. [DOI] [PubMed] [Google Scholar]
- Nowack MK, Ungru A, Bjerkan KN, Grini PE, Schnittger A. Reproductive cross-talk: seed development in flowering plants. Biochem Soc Trans. 2010;38(2):604–612. doi: 10.1042/bst0380604. [DOI] [PubMed] [Google Scholar]
- Nuss ET, Tanumihardjo SA (2010) Maize: a paramount staple crop in the context of global nutrition 9 (4):417–436. doi:10.1111/j.1541-4337.2010.00117.x [DOI] [PubMed]
- Obana Y, Omoto D, Kato C, Matsumoto K, Nagai Y, Kavakli IH, Hamada S, Edwards GE, Okita TW, Matsui H, Ito H. Enhanced turnover of transitory starch by expression of up-regulated ADP-glucose pyrophosphorylases in Arabidopsis thaliana. Plant Sci. 2006;170(1):1–11. doi: 10.1016/j.plantsci.2005.07.019. [DOI] [Google Scholar]
- Olsen OA. Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:233–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- O'Reilly C, Shepherd NS, Pereira A, Schwarz-Sommer Z, Bertram I, Robertson DS, Peterson PA, Saedler H (1985) Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1. EMBO J 4(4):877–882 [DOI] [PMC free article] [PubMed]
- Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22(15):3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer GJ, Rojas M, Hadjivassiliou H, Barkan A. Formation of the CRS2-CAF2 group II intron splicing complex is mediated by a 22-amino acid motif in the COOH-terminal region of CAF2. J Biol Chem. 2006;281(8):4732–4738. doi: 10.1074/jbc.M508921200. [DOI] [PubMed] [Google Scholar]
- O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25(6):1120–1128. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- Pan Z, Ren X, Zhao H, Liu L, Tan Z, Qiu F. A mitochondrial transcription termination factor, ZmSmk3, is required for nad1 intron4 and nad4 intron1 splicing and kernel development in maize. G3 (Bethesda, Md) 2019;9(8):2677–2686. doi: 10.1534/g3.119.400265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Liu M, Xiao Z, Ren X, Zhao H, Gong D, Liang K, Tan Z, Shao Y, Qiu F (2019b) ZmSMK9, a pentatricopeptide repeat protein, is involved in the cis-splicing of nad5, kernel development and plant architecture in maize. An international journal of experimental plant biology. Plant Sci 288, 110205. [DOI] [PubMed]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pea G, Ferron S, Gianfranceschi L, Krajewski P, Enrico Pè M. Gene expression non-additivity in immature ears of a heterotic F1 maize hybrid. Plant Sci. 2008;174(1):17–24. doi: 10.1016/j.plantsci.2007.09.005. [DOI] [Google Scholar]
- Pedersen K, Devereux J, Wilson DR, Sheldon E, Larkins BA. Cloning and sequence analysis reveal structural variation among related zein genes in maize. Cell. 1982;29(3):1015–1026. doi: 10.1016/0092-8674(82)90465-2. [DOI] [PubMed] [Google Scholar]
- Peng B, Li Y, Wang Y, Liu C, Liu Z, Tan W, Zhang Y, Wang D, Shi Y, Sun B, Song Y, Wang T, Li Y. QTL analysis for yield components and kernel-related traits in maize across multi-environments. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2011;122(7):1305–1320. doi: 10.1007/s00122-011-1532-9. [DOI] [PubMed] [Google Scholar]
- Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO J. 1986;5(5):835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3' mRNA termini in chloroplasts. EMBO J. 2009;28(14):2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J, Danner S, Summers PS, Morell M, Barton CR, Yang L, Nieder M. Molecular characterization of the Brittle-2 gene effect on maize endosperm ADPglucose pyrophosphorylase subunits. Plant Physiol. 1990;92(4):881–885. doi: 10.1104/pp.92.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Aukerman MJ, Schmidt RJ. OHP1: a maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell. 1993;5(2):227–236. doi: 10.1105/tpc.5.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Zhu J, Wu Q, Wang Q, Li X, Yao D, Jin Y, Wang G, Wang G, Song R. Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol. 2016;170(2):971–988. doi: 10.1104/pp.15.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Tian Z, Lu L, Chen X, Chen X, Zhang W, Song R. Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics. 2017;205(4):1489–1501. doi: 10.1534/genetics.116.199331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Yang Y, Feng X, Zhang M, Song R. Mitochondrial function and maize kernel development requires Dek2, a pentatricopeptide repeat protein involved in nad1 mRNA splicing. Genetics. 2017;205(1):239–249. doi: 10.1534/genetics.116.196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li S, Zhu Y, Zhao Q, Zhu D, Yu J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol Biol. 2017;93(1–2):7–20. doi: 10.1007/s11103-016-0543-y. [DOI] [PubMed] [Google Scholar]
- Qi W, Lu L, Huang S, Song R. Maize Dek44 encodes mitochondrial ribosomal protein L9 and is required for seed development. Plant Physiol. 2019;180(4):2106–2119. doi: 10.1104/pp.19.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Qi W, Wang Q, Feng Y, Yang Q, Zhang N, Wang S, Tang Y, Song R. ZmMADS47 regulates Zein gene transcription through interaction with Opaque2. PLoS Genet. 2016;12(4):e1005991. doi: 10.1371/journal.pgen.1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Ma C, Feng J, Xu S, Wang L, Li F, Li Y, Zhang R, Zhang X, Xue J, Guo D. Transcriptome dynamics during maize endosperm development. PLoS One. 2016;11(10):e0163814. doi: 10.1371/journal.pone.0163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA. Association genetics in crop improvement. Curr Opin Plant Biol. 2010;13(2):174–180. doi: 10.1016/j.pbi.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Raihan MS, Liu J, Huang J, Guo H, Pan Q, Yan J. Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 × SK maize population. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2016;129(8):1465–1477. doi: 10.1007/s00122-016-2717-z. [DOI] [PubMed] [Google Scholar]
- Reifschneider O, Marx C, Jacobs J, Kollipara L, Sickmann A, Wolters D, Kück U. A ribonucleoprotein supercomplex involved in trans-splicing of organelle group II introns. J Biol Chem. 2016;291(44):23330–23342. doi: 10.1074/jbc.M116.750570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J Exp Bot. 2017;68(16):4571–4581. doi: 10.1093/jxb/erx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren RC, Lu X, Zhao YJ, Wei YM, Wang LL, Zhang L, Zhang WT, Zhang C, Zhang XS, Zhao XY. Pentatricopeptide repeat protein DEK40 is required for mitochondrial function and kernel development in maize. J Exp Bot. 2019;70(21):6163–6179. doi: 10.1093/jxb/erz391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Fan K, Fang T, Zhang J, Yang L, Wang J, Wang G, Liu Y. Maize empty Pericarp602 encodes a P-type PPR protein that is essential for seed development. Plant & cell physiology. 2019;60(8):1734–1746. doi: 10.1093/pcp/pcz083. [DOI] [PubMed] [Google Scholar]
- Ren RC, Wang LL, Zhang L, Zhao YJ, Wu JW, Wei YM, Zhang XS, Zhao XY. DEK43 is a P-type pentatricopeptide repeat (PPR) protein responsible for the Cis-splicing of nad4 in maize mitochondria. J Integr Plant Biol. 2020;62(3):299–313. doi: 10.1111/jipb.12843. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science (New York, NY) 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Robertson DS. Characterization of a mutator system in maize. Mutat Res. 1978;51(1):21–28. doi: 10.1016/0027-5107(78)90004-0. [DOI] [Google Scholar]
- Rossi V, Locatelli S, Varotto S, Donn G, Pirona R, Henderson DA, Hartings H, Motto M. Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell. 2007;19(4):1145–1162. doi: 10.1105/tpc.106.042549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SD (1992) Double fertilization. In: Russell SD, Dumas C (eds) International review of cytology, vol 140. Academic Press, pp 357–388. 10.1016/S0074-7696(08)61102-X
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiol. 2009;149(1):14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA. RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci U S A. 2005;102(37):13005–13012. doi: 10.1073/pnas.0506160102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Hoerster G, Lizarraga LE, Brown SW, Gordon-Kamm WJ, Larkins BA. Positive regulation of minichromosome maintenance gene expression, DNA replication, and cell transformation by a plant retinoblastoma gene. Proc Natl Acad Sci U S A. 2009;106(10):4042–4047. doi: 10.1073/pnas.0813329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Nguyen HN, Gordon-Kamm WJ, Larkins BA. Expression, regulation and activity of a B2-type cyclin in mitotic and endoreduplicating maize endosperm. Front Plant Sci. 2014;5:561. doi: 10.3389/fpls.2014.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakulsingharoj C, Choi S-B, Hwang S-K, Edwards GE, Bork J, Meyer CR, Preiss J, Okita TW. Engineering starch biosynthesis for increasing rice seed weight: the role of the cytoplasmic ADP-glucose pyrophosphorylase. Plant Sci. 2004;167(6):1323–1333. doi: 10.1016/j.plantsci.2004.06.028. [DOI] [Google Scholar]
- Samach A, Melamed-Bessudo C, Avivi-Ragolski N, Pietrokovski S, Levy AA. Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell. 2011;23(12):4266–4279. doi: 10.1105/tpc.111.091744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval R, Boyd RD, Kiszter AN, Mirzakhanyan Y, Santibańez P, Gershon PD, Hayes ML. Stable native RIP9 complexes associate with C-to-U RNA editing activity, PPRs, RIPs, OZ1, ORRM1 and ISE2. The Plant journal : for cell and molecular biology. 2019;99(6):1116–1126. doi: 10.1111/tpj.14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ, Takacs EM. Kernel biology. In: Bennetzen JL, Hake SC, editors. Handbook of maize: its biology. New York, NY: Springer New York; 2009. pp. 121–143. [Google Scholar]
- Scanlon MJ, Stinard PS, James MG, Myers AM, Robertson DS. Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics. 1994;136(1):281–294. doi: 10.1093/genetics/136.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Burr B. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science (New York, NY) 1987;238(4829):960–963. doi: 10.1126/science.2823388. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci U S A. 1990;87(1):46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell. 1992;4(6):689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13(12):663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18(10):2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK. The B73 maize genome: complexity, diversity, and dynamics. Science (New York, NY) 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, Li L, O'Brien BA, Fajardo DS, Wroclawska E, Tseung CW, Lai J, Hunter CT, 3rd, Avigne WT, Baier J, Messing J, Hannah LC, Koch KE, Becraft PW, Larkins BA, McCarty DR. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics. 2007;8:116. doi: 10.1186/1471-2164-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li C, Min Z, Meeley RB, Tarczynski MC, Olsen OA. sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc Natl Acad Sci U S A. 2003;100(11):6552–6557. doi: 10.1073/pnas.0732023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Li C, McCarty DR, Meeley R, Tan BC. Embryo defective12 encodes the plastid initiation factor 3 and is essential for embryogenesis in maize. The Plant journal : for cell and molecular biology. 2013;74(5):792–804. doi: 10.1111/tpj.12161. [DOI] [PubMed] [Google Scholar]
- Shen L, Zhang Q, Wang Z, Wen H, Hu G, Ren D, Hu J, Zhu L, Gao Z, Zhang G, Guo L, Zeng D, Qian Q. OsCAF2 contains two CRM domains and is necessary for chloroplast development in rice. BMC Plant Biol. 2020;20(1):381. doi: 10.1186/s12870-020-02593-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan WF, Clark JK. Mutational analysis of morphogenesis of the maize embryo. Plant J. 1993;3(2):347–358. doi: 10.1111/j.1365-313X.1993.tb00186.x. [DOI] [Google Scholar]
- Sheridan WF, Clark JK (1993b) Mutational analysis of morphogenesis of the maize embryo. 3(2):347–358. 10.1111/j.1365-313X.1993.tb00186.x
- Sheridan WF, Clark JK. Mutational analysis of morphogenesis of the maize embryo. Plant J. 1993;3(2):347–358. doi: 10.1111/j.1365-313X.1993.tb00186.x. [DOI] [Google Scholar]
- Shiferaw B, Prasanna BM, Hellin J, Bänziger M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Security. 2011;3(3):307. doi: 10.1007/s12571-011-0140-5. [DOI] [Google Scholar]
- Shikanai T, Fujii S. Function of PPR proteins in plastid gene expression. RNA Biol. 2013;10(9):1446–1456. doi: 10.4161/rna.25207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983;35(1):225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25(2):46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci U S A. 2002;99(3):1724–1729. doi: 10.1073/pnas.022635299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidansky ED, Martin JM, Hannah LC, Fischer AM, Giroux MJ. Seed yield and plant biomass increases in rice are conferred by deregulation of endosperm ADP-glucose pyrophosphorylase. Planta. 2003;216(4):656–664. doi: 10.1007/s00425-002-0897-z. [DOI] [PubMed] [Google Scholar]
- Song W, Zhu J, Zhao H, Li Y, Liu J, Zhang X, Huang L, Lai J. OS1 functions in the allocation of nutrients between the endosperm and embryo in maize seeds. J Integr Plant Biol. 2019;61(6):706–727. doi: 10.1111/jipb.12755. [DOI] [PubMed] [Google Scholar]
- Sosso D, Canut M, Gendrot G, Dedieu A, Chambrier P, Barkan A, Consonni GM, Rogowsky P. PPR8522 encodes a chloroplast-targeted pentatricopeptide repeat protein necessary for maize embryogenesis and vegetative development. J Exp Bot. 2012;63(16):5843–5857. doi: 10.1093/jxb/ers232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D, Mbelo S, Vernoud V, Gendrot G, Dedieu A, Chambrier P, Dauzat M, Heurtevin L, Guyon V, Takenaka M, Rogowsky PM. PPR2263, a DYW-subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell. 2012;24(2):676–691. doi: 10.1105/tpc.111.091074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, Rogowsky PM, Ross-Ibarra J, Yang B, Frommer WB. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015;47(12):1489–1493. doi: 10.1038/ng.3422. [DOI] [PubMed] [Google Scholar]
- Spielbauer G, Li L, Römisch-Margl L, Do PT, Fouquet R, Fernie AR, Eisenreich W, Gierl A, Settles AM. Chloroplast-localized 6-phosphogluconate dehydrogenase is critical for maize endosperm starch accumulation. J Exp Bot. 2013;64(8):2231–2242. doi: 10.1093/jxb/ert082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 2004;9(6):293–301. doi: 10.1016/j.tplants.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Stupar RM, Springer NM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173(4):2199–2210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Germain A, Giloteaux L, Hammani K, Barkan A, Hanson MR, Bentolila S. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc Natl Acad Sci U S A. 2013;110(12):E1169–E1178. doi: 10.1073/pnas.1220162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Wang X, Bonnard G, Shen Y, Xiu Z, Li X, Gao D, Zhang Z, Tan BC. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. The Plant journal : for cell and molecular biology. 2015;84(2):283–295. doi: 10.1111/tpj.12993. [DOI] [PubMed] [Google Scholar]
- Sun T, Shi X, Friso G, Van Wijk K, Bentolila S, Hanson MR. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015;11(3):e1005028. doi: 10.1371/journal.pgen.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson MR. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016;21(11):962–973. doi: 10.1016/j.tplants.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang X, Shen Y, Wang H, Liu R, Wang X, Gao D, Yang YZ, Liu Y, Tan BC (2018) The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. The Plant journal : for cell and molecular biology. 10.1111/tpj.14030 [DOI] [PubMed]
- Sun F, Xiu Z, Jiang R, Liu Y, Zhang X, Yang YZ, Li X, Zhang X, Wang Y, Tan BC. The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. J Exp Bot. 2019;70(3):963–972. doi: 10.1093/jxb/ery432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, Li QB, McCarty DR. Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 2003;132:1664–1677. doi: 10.1104/pp.103.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Sato Y, Wu S, Kang BH, McCarty DR. Conserved functions of the MATE transporter BIG EMBRYO1 in regulation of lateral organ size and initiation rate. Plant Cell. 2015;27(8):2288–2300. doi: 10.1105/tpc.15.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci U S A. 2006;103(18):6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M. How complex are the editosomes in plant organelles? Mol Plant. 2014;7(4):582–585. doi: 10.1093/mp/sst170. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Härtel B, Brennicke A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci U S A. 2012;109(13):5104–5109. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Brennicke A, Graichen K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013;8(6):e65343. doi: 10.1371/journal.pone.0065343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer AF (2006) Adaptedness and heterosis in corn and mule hybrids 46 (2):528–543. doi:10.2135/cropsci2005.0065
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13(5):255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14(4):903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci U S A. 1997;94(14):7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, Conrad LJ, Kikuchi K, Kubinec TA, Hall BD, Weeks R, Unger-Wallace E, Muszynski M, Brendel VP, Brutnell TP. Genome-Wide Distribution of Transposed <em>Dissociation</em> Elements in Maize. Plant Cell. 2010;22(6):1667. doi: 10.1105/tpc.109.073452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V. Saturation mutagenesis using maize transposons. Curr Opin Plant Biol. 2000;3(2):103–107. doi: 10.1016/s1369-5266(99)00051-5. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen X, Wang J, Liu T, Liu Y, Zhao L, Wang G. Increasing maize seed weight by enhancing the cytoplasmic ADP-glucose pyrophosphorylase activity in transgenic maize plants. Plant Cell Tissue Org Cult. 2007;88(1):83–92. doi: 10.1007/s11240-006-9173-4. [DOI] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, Ma H, Zhang G, He Z. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40(11):1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Wang G, Sun X, Wang G, Wang F, Gao Q, Sun X, Tang Y, Chang C, Lai J, Zhu L, Xu Z, Song R. Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics. 2011;189(4):1281–1295. doi: 10.1534/genetics.111.133967. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang G, Wang F, Wang G, Wang F, Zhang X, Zhong M, Zhang J, Lin D, Tang Y, Xu Z, Song R. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell. 2012;24(8):3447–3462. doi: 10.1105/tpc.112.101360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Qi W, Wu Q, Yao D, Zhang J, Zhu J, Wang G, Wang G, Tang Y, Song R. Identification and characterization of maize floury4 as a novel semidominant opaque mutant that disrupts protein body assembly. Plant Physiol. 2014;165(2):582–594. doi: 10.1104/pp.114.238030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang J, Wang G, Fan X, Sun X, Qin H, Xu N, Zhong M, Qiao Z, Tang Y, Song R. Proline responding1 plays a critical role in regulating general protein synthesis and the cell cycle in maize. Plant Cell. 2014;26(6):2582–2600. doi: 10.1105/tpc.114.125559. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang G, Zhong M, Shuai B, Song J, Zhang J, Han L, Ling H, Tang Y, Wang G, Song R. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. The New phytologist. 2017;214(4):1563–1578. doi: 10.1111/nph.14507. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang K, Du Q, Wang Y, Fu Z, Guo Z, Kang D, Li WX, Tang J. Maize Urb2 protein is required for kernel development and vegetative growth by affecting pre-ribosomal RNA processing. The New phytologist. 2018;218(3):1233–1246. doi: 10.1111/nph.15057. [DOI] [PubMed] [Google Scholar]
- Wang H, Xu S, Fan Y, Liu N, Zhan W, Liu H, Xiao Y, Li K, Pan Q, Li W, Deng M, Liu J, Jin M, Yang X, Li J, Li Q, Yan J. Beyond pathways: genetic dissection of tocopherol content in maize kernels by combining linkage and association analyses. Plant Biotechnol J. 2018;16:1464–1475. doi: 10.1111/pbi.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fan W, Ou M, Wang X, Qin H, Feng F, Du Y, Ni J, Tang J, Song R, Wang G. Dek40 encodes a PBAC4 protein required for 20S proteasome biogenesis and seed development. Plant Physiol. 2019;180(4):2120–2132. doi: 10.1104/pp.18.01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Sayyed A, Liu XY, Yang YZ, Sun F, Wang Y, Wang M, Tan BC (2019b) SMALL KERNEL4 is required for mitochondrial cox1 transcript editing and seed development in maize. J Integr Plant Biol. 10.1111/jipb.12856 [DOI] [PubMed]
- Wang Y, Liu XY, Yang YZ, Huang J, Sun F, Lin J, Gu ZQ, Sayyed A, Xu C, Tan BC. Empty Pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PLoS Genet. 2019;15(8):e1008305. doi: 10.1371/journal.pgen.1008305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Chen Z, Yang YZ, Sun F, Ding S, Li XL, Xu C, Tan BC. PPR14 interacts with PPR-SMR1 and CRM protein Zm-mCSF1 to facilitate mitochondrial intron splicing in maize. Front Plant Sci. 2020;11:814. doi: 10.3389/fpls.2020.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Han P. Pentatricopeptide repeat proteins in maize. Mol Breed. 2016;36(12):170. doi: 10.1007/s11032-016-0596-2. [DOI] [Google Scholar]
- Wentz JB. The inheritance of germless seeds in maize. Iowa Agric Exp Sta Res Bull. 1930;121:845–379. [Google Scholar]
- Wienand U, Sommer H, Schwarz Z, Shepherd N, Saedler H, Kreuzaler F, Ragg H, Fautz E, Hahlbrock K, Harrison B, Peterson PA. A general-method to identify plant structural genes among genomic DNA clones using transposable element induced mutations. Mol Gen Genet. 1982;187(2):195–201. doi: 10.1007/bf00331117. [DOI] [Google Scholar]
- Williams PM, Barkan A. A chloroplast-localized PPR protein required for plastid ribosome accumulation. The Plant journal : for cell and molecular biology. 2003;36(5):675–686. doi: 10.1046/j.1365-313x.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA (New York, NY) 2008;14(9):1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Messing J. Rapid divergence of prolamin gene promoters of maize after gene amplification and dispersal. Genetics. 2012;192(2):507–519. doi: 10.1534/genetics.112.142372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Gontarek BC, Yi G, Beall BD, Neelakandan AK, Adhikari B, Chen R, McCarty DR, Severin A, Becraft PW (2020) The thick aleurone1 gene encodes a NOT1 subunit of the CCR4-NOT complex and regulates cell patterning in endosperm. Plant Physiol. 10.1104/pp.20.00703 [DOI] [PMC free article] [PubMed]
- Xiao Y, Liu H, Wu L, Warburton M, Yan J. Genome-wide association studies in maize: praise and stargaze. Mol Plant. 2017;10(3):359–374. doi: 10.1016/j.molp.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Xie G, Li Z, Ran Q, Wang H, Zhang J. Over-expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnol J. 2018;16(1):234–244. doi: 10.1111/pbi.12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Fu X, Yang C, Tang X, Guo L, Li C, Xu C, Luo K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci Rep. 2018;8(1):2817. doi: 10.1038/s41598-018-21269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Wang C, Zhang X, Yang Q, Shao R, Lai J, Du C. Highly interwoven communities of a gene regulatory network unveil topologically important genes for maize seed development. The Plant journal : for cell and molecular biology. 2017;92(6):1143–1156. doi: 10.1111/tpj.13750. [DOI] [PubMed] [Google Scholar]
- Xiu Z, Sun F, Shen Y, Zhang X, Jiang R, Bonnard G, Zhang J, Tan BC. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. The Plant journal : for cell and molecular biology. 2016;85(4):507–519. doi: 10.1111/tpj.13122. [DOI] [PubMed] [Google Scholar]
- Xu H, Gao Y, Wang J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS One. 2012;7(2):e30646. doi: 10.1371/journal.pone.0030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Song S, Yang YZ, Lu F, Zhang MD, Sun F, Jia R, Song R, Tan BC (2020) DEK46 performs C-to-U editing of a specific site in mitochondrial nad7 introns that is critical for intron splicing and seed development in maize. The Plant journal : for cell and molecular biology. 10.1111/tpj.14862 [DOI] [PubMed]
- Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8(3):e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Warburton M, Crouch J. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 2011;51(2):433–449. doi: 10.2135/cropsci2010.04.0233. [DOI] [Google Scholar]
- Yang J, Ji C, Wu Y. Divergent transactivation of maize storage protein Zein genes by the transcription factors Opaque2 and OHPs. Genetics. 2016;204(2):581–591. doi: 10.1534/genetics.116.192385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang HC, Sun F, Huang WL, Song S, Xu C, Tan BC. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. The New phytologist. 2017;214(2):782–795. doi: 10.1111/nph.14424. [DOI] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang Y, Li CL, Shen Y, Meeley R, McCarty DR, Tan BC. Small kernel2 encodes a glutaminase in vitamin B(6) biosynthesis essential for maize seed development. Plant Physiol. 2017;174(2):1127–1138. doi: 10.1104/pp.16.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Fu M, Ji C, Huang Y, Wu Y. Maize Oxalyl-CoA Decarboxylase1 degrades oxalate and affects the seed metabolome and nutritional quality. Plant Cell. 2018;30(10):2447–2462. doi: 10.1105/tpc.18.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Liu J, Gao Q, Gui S, Chen L, Yang L, Huang J, Deng T, Luo J, He L, Wang Y, Xu P, Peng Y, Shi Z, Lan L, Ma Z, Yang X, Zhang Q, Bai M, Li S, Li W, Liu L, Jackson D, Yan J. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat Genet. 2019;51(6):1052–1059. doi: 10.1038/s41588-019-0427-6. [DOI] [PubMed] [Google Scholar]
- Yang H, Xiu Z, Wang L, Cao SK, Li X, Sun F, Tan BC. Two pentatricopeptide repeat proteins are required for the splicing of nad5 introns in maize. Front Plant Sci. 2020;11:732. doi: 10.3389/fpls.2020.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang Y, Wang HC, Liu XY, Sun F, Xu C, Liu B, Tan BC. PPR20 is required for the cis-splicing of mitochondrial nad2 intron 3 and seed development in maize. Plant & cell physiology. 2020;61(2):370–380. doi: 10.1093/pcp/pcz204. [DOI] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Liu XY, Tang JJ, Wang Y, Sun F, Xu C, Tan BC (2020c) EMP32 is required for the cis-splicing of nad7 intron 2 and seed development in maize. RNA biology:1–11 [DOI] [PMC free article] [PubMed]
- Yao D, Qi W, Li X, Yang Q, Yan S, Ling H, Wang G, Wang G, Song R. Maize opaque10 encodes a cereal-specific protein that is essential for the proper distribution of Zeins in endosperm protein bodies. PLoS Genet. 2016;12(8):e1006270. doi: 10.1371/journal.pgen.1006270. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yi G, Neelakandan AK, Gontarek BC, Vollbrecht E, Becraft PW. The naked endosperm genes encode duplicate INDETERMINATE domain transcription factors required for maize endosperm cell patterning and differentiation. Plant Physiol. 2015;167(2):443–456. doi: 10.1104/pp.114.251413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Gu W, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, Esteban E, Pasha A, Provart NJ, Lai J. High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell. 2019;31(5):974–992. doi: 10.1105/tpc.18.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504(7478):168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- Yuan N, Wang J, Zhou Y, An D, Xiao Q, Wang W, Wu Y. EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes. Plant science : an international journal of experimental plant biology. 2019;287:110203. doi: 10.1016/j.plantsci.2019.110203. [DOI] [PubMed] [Google Scholar]
- Zang J, Huo Y, Liu J, Zhang H, Liu J, Chen H (2020) Maize ZmYSL2 is required for kernel Fe distribution and development. J Exp Bot. 10.1093/jxb/eraa332 [DOI] [PubMed]
- Zhan J, Thakare D, Ma C, Lloyd A, Nixon NM, Arakaki AM, Burnett WJ, Logan KO, Wang D, Wang X, Drews GN, Yadegari R. RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell. 2015;27(3):513–531. doi: 10.1105/tpc.114.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Li G, Ryu CH, Ma C, Zhang S, Lloyd A, Hunter BG, Larkins BA, Drews GN, Wang X, Yadegari R. Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell. 2018;30(10):2425–2446. doi: 10.1105/tpc.18.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan W, Liu J, Pan Q, Wang H, Yan S, Li K, Deng M, Li W, Liu N, Kong Q, Fernie AR, Yan J. An allele of ZmPORB2 encoding a protochlorophyllide oxidoreductase promotes tocopherol accumulation in both leaves and kernels of maize. Plant J. 2019;100:114–127. doi: 10.1111/tpj.14432. [DOI] [PubMed] [Google Scholar]
- Zhang P, Allen WB, Nagasawa N, Ching AS, Heppard EP, Li H, Hao X, Li X, Yang X, Yan J, Nagato Y, Sakai H, Shen B, Li J. A transposable element insertion within ZmGE2 gene is associated with increase in embryo to endosperm ratio in maize. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2012;125(7):1463–1471. doi: 10.1007/s00122-012-1926-3. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Hou MM, Tan BC. The requirement of WHIRLY1 for embryogenesis is dependent on genetic background in maize. PLoS One. 2013;8(6):e67369. doi: 10.1371/journal.pone.0067369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu Z, Hu Y, Li W, Fu Z, Ding D, Li H, Qiao M, Tang J. QTL analysis of Kernel-related traits in maize using an immortalized F2 population. PLoS One. 2014;9(2):e89645. doi: 10.1371/journal.pone.0089645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang J, Wu Y. Transcriptional regulation of Zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell. 2015;27(4):1162–1172. doi: 10.1105/tpc.15.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu X, Shi C, Wang R, Li S, Wang Z, Liu Z, Xue Y, Tang G, Tang J. Genetic dissection of the maize kernel development process via conditional QTL mapping for three developing kernel-related traits in an immortalized F2 population. Molecular genetics and genomics : MGG. 2016;291(1):437–454. doi: 10.1007/s00438-015-1121-8. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhou Z, Yong H, Zhang X, Hao Z, Zhang F, Li M, Zhang D, Li X, Wang Z, Weng J. Analysis of the genetic architecture of maize ear and grain morphological traits by combined linkage and association mapping. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2017;130(5):1011–1029. doi: 10.1007/s00122-017-2867-7. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang C, Wu D, Qiao F, Li W, Duan L, Wang K, Xiao Y, Chen G, Liu Q, Xiong L, Yang W, Yan J. High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol. 2017;173(3):1554. doi: 10.1104/pp.16.01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YF, Suzuki M, Sun F, Tan BC. The mitochondrion-targeted PENTATRICOPEPTIDE REPEAT78 protein is required for nad5 mature mRNA stability and seed development in maize. Mol Plant. 2017;10(10):1321–1333. doi: 10.1016/j.molp.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wu S, Boehlein SK, McCarty DR, Song G, Walley JW, Myers A, Settles AM. Maize defective kernel5 is a bacterial TamB homologue required for chloroplast envelope biogenesis. J Cell Biol. 2019;218(8):2638–2658. doi: 10.1083/jcb.201807166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang X, Wang X, Xu J, Wang M, Li L, Bai G, Fang H, Hu S, Li J, Yan J, Li J, Yang X. SEED CAROTENOID DEFICIENT functions in isoprenoid biosynthesis via the plastid MEP pathway. Plant Physiol. 2019;179(4):1723–1738. doi: 10.1104/pp.18.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mogel K, Lor VS, Hirsch CN, De Vries B, Kaeppler HF, Tracy WF, Kaeppler SM. Maize sugary enhancer1 (se1) is a gene affecting endosperm starch metabolism. Proc Natl Acad Sci U S A. 2019;116(41):20776–20785. doi: 10.1073/pnas.1902747116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci U S A. 2019;116(23):11223–11228. doi: 10.1073/pnas.1904995116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feng C, Su H, Liu Y, Liu Y, Han F. The Cohesin complex subunit ZmSMC3 participates in meiotic centromere pairing in maize. Plant Cell. 2020;32(4):1323–1336. doi: 10.1105/tpc.19.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guan Z, Wang L, Fu J, Zhang Y, Li Z, Ma L, Liu P, Zhang Y, Liu M, Li P, Zou C, He Y, Lin H, Yuan G, Gao S, Pan G, Shen Y. Combined GWAS and QTL analysis for dissecting the genetic architecture of kernel test weight in maize. Molecular genetics and genomics : MGG. 2020;295(2):409–420. doi: 10.1007/s00438-019-01631-2. [DOI] [PubMed] [Google Scholar]
- Zhao H, Qin Y, Xiao Z, Li Q, Yang N, Pan Z, Gong D, Sun Q, Yang F, Zhang Z, Wu Y, Xu C, Qiu F. Loss of function of an RNA polymerase III subunit leads to impaired maize kernel development. Plant Physiol. 2020;184(1):359–373. doi: 10.1104/pp.20.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Li Q, Li C, An D, Xiao Q, Wang W, Wu Y. Intra-kernel reallocation of proteins in maize depends on VP1-mediated scutellum development and nutrient assimilation. Plant Cell. 2019;31:2613–2635. doi: 10.1105/tpc.19.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. The Plant Genome. 2008;1(1):5–20. doi: 10.3835/plantgenome2008.02.0089. [DOI] [Google Scholar]
- Zhu C, Jin G, Fang P, Zhang Y, Feng X, Tang Y, Qi W, Song R. Maize pentatricopeptide repeat protein DEK41 affects cis-splicing of mitochondrial nad4 intron 3 and is required for normal seed development. J Exp Bot. 2019;70(15):3795–3808. doi: 10.1093/jxb/erz193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmudjak M, Colas des Francs-Small C. Keren I, Shaya F, Belausov E, Small I, Ostersetzer-Biran O. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. The New phytologist. 2013;199(2):379–394. doi: 10.1111/nph.12282. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Feng F, Qi W, Song R. Dek42 encodes an RNA-binding protein that affects alternative pre-mRNA splicing and maize kernel development. J Integr Plant Biol. 2019;61(6):728–748. doi: 10.1111/jipb.12798. [DOI] [PubMed] [Google Scholar]