Abstract
Objectives
In veterinary medicine, colistin has been widely used as therapeutic and prophylactic agent, and for growth promotion. However, colistin has been re-introduced into treatment of human MDR bacterial infections. We assessed the characteristics and spread of plasmid-borne colistin resistance among healthy pigs, workers with animal-contact and their household members in Thailand.
Methods
WGS and MIC data of 146 mcr-positive isolates from a cross-sectional One Health study were analysed. Long-read sequencing and conjugation were performed for selected isolates.
Results
mcr-carrying isolates were detected in 38% of pooled-pig samples and 16% of human faecal samples. Of 143 Escherichia coli and three Escherichia fergusonii, mcr-1, mcr-3, and mcr-9 variants were identified in 96 (65.8%), 61 (41.8%) and one (0.7%) isolate, respectively. Twelve E. coli co-harboured two mcr variants (mcr-1 and mcr-3). Clonal transmission was detected in five out of 164 farms. mcr-1 was mostly harboured by epidemic IncX4 and IncHI1 plasmids (89.9%). Conversely, mcr-3 was harboured by a range of different plasmids. Comparative plasmid studies suggested IncP and IncFII plasmids as possible endemic mcr-3 plasmids in Asian countries. Moreover, mcr-3 was associated with different mobile genetic elements including TnAs2, ISKpn40 and IS26/15DI. Detected genetic signatures (DRs) indicated recent mcr-3 transpositions, underlining the mobilizable nature of the mcr-3 cassette.
Conclusions
The epidemiology of mcr and the possible evolution of successful plasmids and transposition modules should be carefully monitored. Of special concern is the growing number of different horizontal gene transferring pathways encompassing various transposable modules the mcr genes can be shared between bacteria.
Introduction
Colistin (polymyxin E), has been re-introduced into the treatment of infections caused by MDR Gram-negative bacteria, particularly carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae and other members of Enterobacteriaceae.1 However, in veterinary medicine, colistin has been widely used both as a therapeutic and prophylactic agent and for growth promotion.2 Bacteria resistant to colistin have emerged worldwide with mechanisms not only caused by mutations or interruptions in specific chromosomal genes,1 but also by plasmid-borne genes.3 Until now, 10 plasmid-borne colistin resistance genes, i.e. mcr-1 to mcr-10, have been reported.4,5mcr-1 has been extensively studied since the first description in 2015. Plasmids belonging to the incompatibility groups IncI2, IncX4 and IncHI2 are considered common dissemination vectors for mcr-1.6,7 Co-occurrence of two plasmids with different variants of mcr in a single isolate has scarcely been reported. However, some studies have recently reported such an event, co-occurrence of mcr-1 and mcr-3 genes in E. coli from patients in China8 and in New Zealand,9 in E. coli from pigs in China,10 and recently in K. pneumoniae from a healthy human in Thailand.11
The prevalence of mcr-positive bacteria in clinical isolates in Thailand have been described in E. coli and K. pneumoniae with 29.7% and 1.4%, respectively.12 Furthermore, environmental dissemination of mcr-1 has been found in 16% of investigated blowflies in a Northern province in Thailand,13 as well as in 7% of flies collected in animal farms.14 However, dissemination of plasmids containing mcr genes has never been assessed in human- and animal-originating bacteria in Thailand. Here, we investigated the distribution of mcr genes and performed an in-depth characterization of mcr plasmids in pig and human bacterial isolates collected as a part of a One Health cross-sectional study performed in Northern Thailand. Furthermore, we investigated the geographic distribution of mcr plasmids, determined the genetic environment of mcr and performed detailed characterization of strains harbouring multiple mcr gene variants.
Materials and methods
Ethics
The study was conducted according to the Helsinki Declaration for the human subjects and the EU Directive 2010/63/EU for animal experiments; the protocol involving human participants and animals was approved by the Khon Kaen University Ethics Committee (Project ID: HE612268 and 0514.1.75/66, respectively). Informed consent was obtained from all participants. Pig samples were collected with the permission of the owner of the pig herd.
Study participants
Sample collection, methods and main findings have previously been described.15–17 Briefly, rectal swabs from pigs and human faecal samples were collected from 164 pig farms between September and December 2018 in Khon Kaen, Thailand. From each farm an attempt was made to collect faecal samples from a farmworker (termed contact, C) and an individual living in the same household, however, without direct contact with pigs (termed non-contact, U). Pig samples (up to 10 individual rectal swabs per farm) were pooled and analysed as one sample per farm (termed pig, P). The samples were investigated selectively for carbapenem, extended-spectrum cephalosporin and colistin-resistant bacteria, and isolates were subjected to WGS and MIC determination as previously described.16
WGS and genetic analyses
Illumina WGS data of 146 Escherichia spp. isolates containing at least one mcr variant were further analysed. The study included one mcr-1-positive K. pneumoniae (strain 90CP1) from a human contact, from our previous study16 as this isolate might share mcr-plasmid with an E. coli isolate from pigs at the same farm (farm no. 90). Fourteen selected isolates were sequenced using Oxford Nanopore technologies sequence platform. This included 10 isolates with multiple mcr gene variants, one mcr-9 carrying isolate, one IncFIA/FIB/mcr-3.5 carrying isolate and two IncX4/mcr-1 harboured by different bacterial species (K. pneumoniae and E. coli) from pig and human hosts at the same farm (farm no. 90). Hybrid genome assembly of both short and long reads was sequentially performed. Due to budget limitations, the two remaining genomes (71PM1 and 98UM1) harbouring two mcr gene variants and initially sequenced by Illumina, were mapped to the hybrid assembled genomes (35CM1 and 98CM1) to circularize the plasmid sequences. In 16 isolates, we were able to fully close all plasmids with mcr genes. In subsequent steps, resistance genes, plasmid replicons, conjugation modules and insertion sequences were identified in all included isolates. The closed plasmids served as reference for additional analyses and mapping of mcr plasmids and contigs from other strains. Geographic distribution of the most prevalent mcr-1 and mcr-3 plasmids was created using ArcGIS (https://www.arcgis.com/index.html). The genetic surroundings of mcr-1 and mcr-3 genes were investigated and analysed. Conjugation experiments was performed with 16 strains with closed plasmids. Details of sequencing and methodological approaches are described in the Supplementary Information/Material.
Availability of data and materials
Reads (fastq files) from the study have been submitted to the European Nucleotide Archive (www.ebi.ac.uk/ena) under study accession number PRJEB38313 and PRJEB39885.
Results
mcr gene diversity, gene location and geographical distribution of mcr plasmids
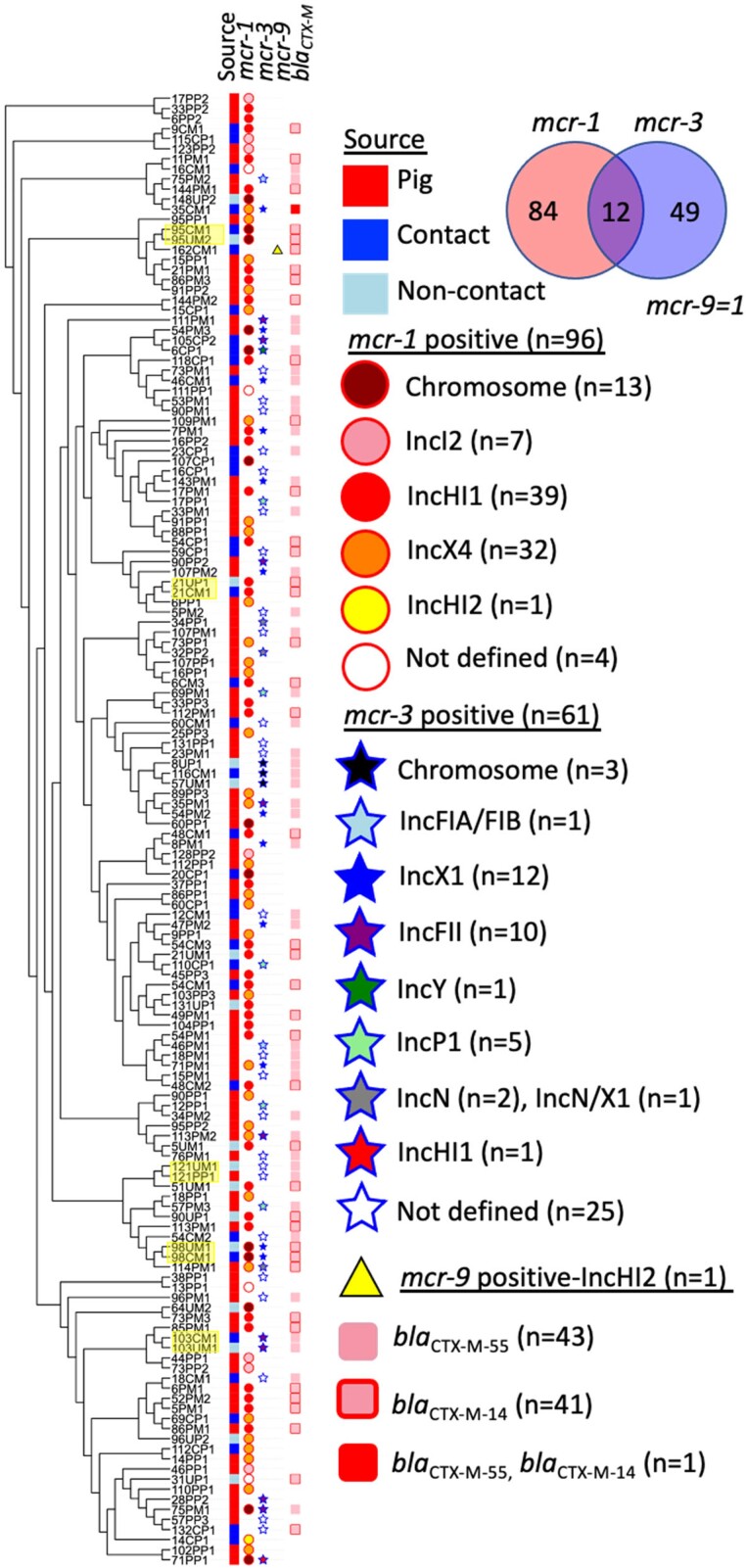
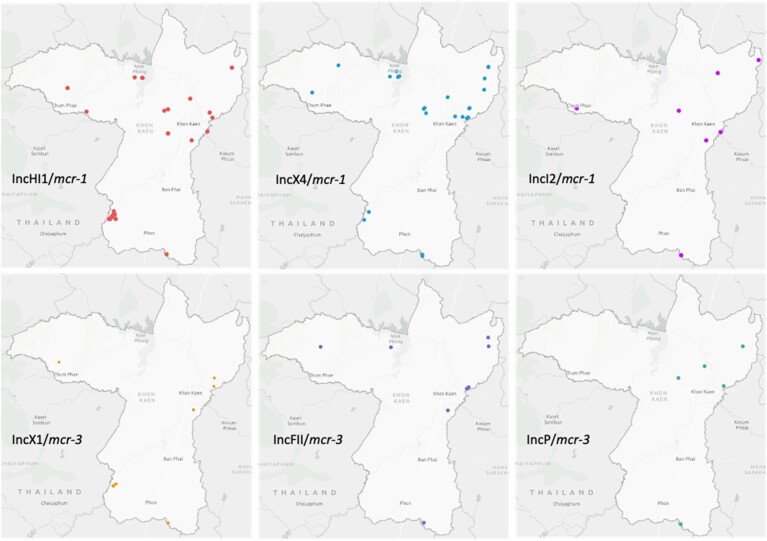
WGS data of the 146 included genomes identified several mcr variants. Most of the variants belonged to the mcr-1 (mcr-1.1, mcr-1.11, mcr-1.2) and mcr-3 group (mcr-3.1, mcr-3.19, mcr-3.2, mcr-3.39 and mcr-3.5) while only one isolate harboured mcr-9 (Figure 1, Table S1, available as Supplementary data at JAC Online). mcr-3.39 represents a new variant, available in GenBank under accession no MT872721. In total, 96 strains contained an mcr-1 gene variant, 61 contained an mcr-3 gene variant and one contained mcr-9 (strain 162CM1). Interestingly, 12 isolates co-harboured two mcr variants, mcr-1 and mcr-3. In addition, 83 isolates harboured blaCTX-M variants, blaCTX-M-55 andblaCTX-M-14. Apart from mcr variants, the associated plasmid incompatibility group or chromosomal location could be determined for 117 E. coli isolates (Figure 1). In the remaining 29 isolates (mcr-1/n = 4; mcr-3/n = 25), mcr location was uncertain due to the short contig lengths (<10 kb) and/or no replicon within the contig. These isolates were omitted from further analysis. The most common IncHI1, IncX4, IncI2 carrying mcr-1 and IncX1, IncFII, IncP carrying mcr-3 plasmids (Figure 1) were assessed for their geographical origin. The plasmids were mostly interspersed throughout the sampling area except for IncHI1/mcr-1 plasmids that clustered on several farms located in the South-West part (Figure 2).
Figure 1.
Cladogram of mcr-positive Escherichia coli/fergusonnii isolates. Maximum likelihood tree of isolates from pigs, contacts and non-contacts based on SNPs in the core genome. Isolate pairs from the same farm with zero SNPs differences were highlighted in yellow. Abbreviations: P, pigs; C, contacts; NC, non-contacts; ST, sequence type.
Figure 2.
Geographical distribution of the most common mcr-1 and mcr-3 plasmids. The map was created using ArcGIS (https://www.arcgis.com/index.html).
Co-resistance to other classes of antimicrobial agents and colistin minimum inhibitory concentrations
Most isolates were resistant to ampicillin, tetracycline, sulfamethoxazole, trimethoprim, chloramphenicol, ciprofloxacin and cefotaxime (Table 1). A majority (n = 126/146) were MDR (resistant to three or more antibiotic classes). Colistin MIC values ranged between 1–16 mg/L. Most isolates were resistant to colistin except the mcr-9 isolate (162CM1) and two mcr-3.5-positive isolates (34PM2 and 60CM1, with colistin MICs at 1 mg/L). Colistin MIC of isolates co-harbouring two mcr variants ranged between 4 and 8 mg/L. Detailed antimicrobial resistance profiles are presented in Table S1.
Table 1.
Antimicrobial susceptibility profiles of mcr-positive E. coli and E. fergusonii
| Number (%) of isolates resistant | All isolates (n = 146) | mcr-1 (n = 84) | mcr-3 (n = 49) | mcr-1 and mcr-3 (n = 12) | mcr-9 (n = 1) |
|---|---|---|---|---|---|
| Ampicillin | 140 (96) | 79 (94) | 49 (100) | 11 (92) | 1 (100) |
| Cefotaxime | 72 (49) | 23 (27) | 37 (75) | 11 (92) | 1 (100) |
| Ceftazidime | 17 (12) | 6 (7) | 9 (18) | 2 (17) | 0 (0) |
| Colistin | 143 (98) | 84 (100) | 47 (96) | 12 (100) | 0 (0) |
| Gentamicin | 23 (16) | 10 (12) | 9 (18) | 3 (25) | 1 (100) |
| Tetracycline | 118 (81) | 67 (80) | 39 (80) | 11 (92) | 1 (100) |
| Meropenem | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 80 (55) | 38 (45) | 31 (63) | 11 (92) | 0 (0) |
| Chloramphenicol | 82 (56) | 37 (44) | 35 (71) | 9 (75) | 1 (100) |
| Tigecycline | 3 (2) | 2 (2) | 1 (2) | 0 (0) | 0 (0) |
| Trimethoprim | 85 (58) | 48 (57) | 28 (57) | 8 (67) | 1 (100) |
| Sulfamethoxazole | 110 (75) | 57 (68) | 42 (86) | 10 (83) | 1 (100) |
| Azitromycin | 13 (9) | 7 (8) | 3 (6) | 2 (17) | 1 (100) |
| Nalidixic acid | 31 (21) | 14 (17) | 12 (25) | 4 (33) | 1 (100) |
Genetic relatedness among mcr-containing strains
The 143 E. coli isolates grouped into 78 different STs, including 17 new STs identified by the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (Figure 1, Table S1). ST10 clonal complex was the most prevalent ST (49/143) (Figure S1). Three E. fergusonii containing mcr-1, all from pigs, belonged to ST10568, ST10576 and ST7059. A maximum likelihood tree based on detected SNPs in the core genomes of the 146 included strains, with additional strain characteristics, is shown in Figure 1.
To investigate a possible transfer of mcr-containing strains between different hosts on the same farm we inspected the number of SNPs between isolates grouping into the same ST, obtained from the same farm. Between-host clonal transmission within farms was found on five farms, no. 21, 95, 98, 103 and 121 (Figure 1). Each pair of isolates (pig–human and/or human–human) exhibited zero SNP differences (Table S2).
Genetic analysis of mcr-1-carrying plasmids and mcr-1 genetic context
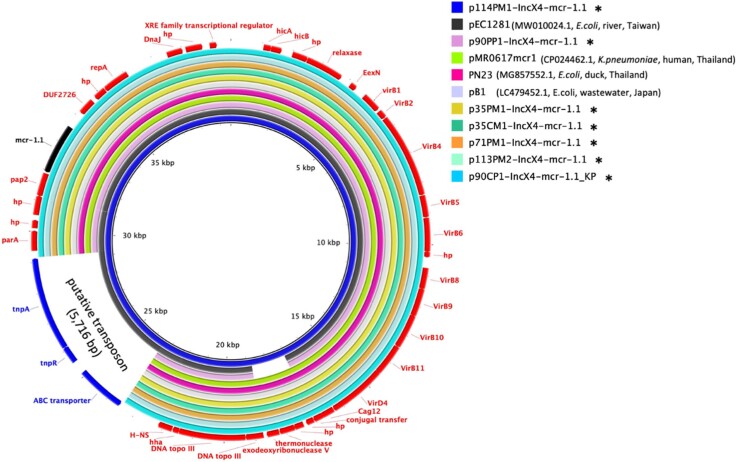
Our data enabled an in-depth analysis of 92 mcr-1 positive isolates. In 79 isolates, mcr-1 was present on a plasmid, while the 13 remaining isolates harboured mcr-1 on the chromosome. In seven E. coli and one K. pneumoniae, the mcr-1 plasmids were fully closed, plasmid-related contigs were identified in the remaining 71 isolates. Most mcr-1 plasmids belonged to incompatibility groups IncHI1 (n = 39/79) and IncX4 (n = 32/79) (Figure 1; Table 2). In addition, seven isolates contained IncI2/mcr-1 plasmids and one contained IncHI2/mcr-1. Table 2 shows an overview of mcr variants and their genome location. Characteristics of strains co-harbouring two mcr variants, mcr-1 and mcr-3, are presented in Figure 3. All detected mcr-1 plasmids and plasmid-related contigs exhibited high similarity with previously published plasmids as illustrated in Figure S2A–E. Seven complete IncX4/mcr-1 plasmid sequences were obtained, ranging between 33 309 and 39 574 bp sharing identical backbone structure with p90PP1(33 309 bp) (Figure 4). p114PM1-IncX4/mcr-1 had an additional insertion of a putative transposon (5716 bp) previously found on other plasmids (Figure 4)18. Identical IncX4/mcr-1 plasmids were shared between strains from human and animal hosts. In addition, this plasmid was present in K. pneumoniae and E. coli obtained from the same farm, suggesting inter-species plasmid transmission (Figure 4).
Table 2.
Distribution of mcr gene and number of closed plasmids
| Gene (no. of isolates) | mcr location | Number of closed plasmids (size of plasmid) |
|---|---|---|
| mcr-1 (n = 96) | IncHI1 (n = 39) | one (217 496 bp) |
| IncX4 (n = 32) | seven (33 309–39 574 bp) | |
| IncI2 (n = 7) | — | |
| IncHI2 (n = 1) | — | |
| chromosome (n = 13) | — | |
| not defined (n = 4) | — | |
| mcr-3 (n = 61) | IncX1 (n = 12) | six (32 893–53 563 bp) |
| IncFII (n = 10) | three (42 864–89 737 bp) | |
| IncP1 (n = 5) | — | |
| IncN (n = 2) | — | |
| IncX1/N (n = 1) | one (64 994 bp) | |
| IncFIA/FIB (n = 1) | one (70 757 bp) | |
| IncHI1 (n = 1) | one (302 729 bp) | |
| IncY (n = 1) | one (109 103 bp) | |
| chromosome (n = 3) | — | |
| not defined (n = 25) | — | |
| mcr-9 (n = 1) | IncHI2 (n = 1) | one (263 699 bp) |
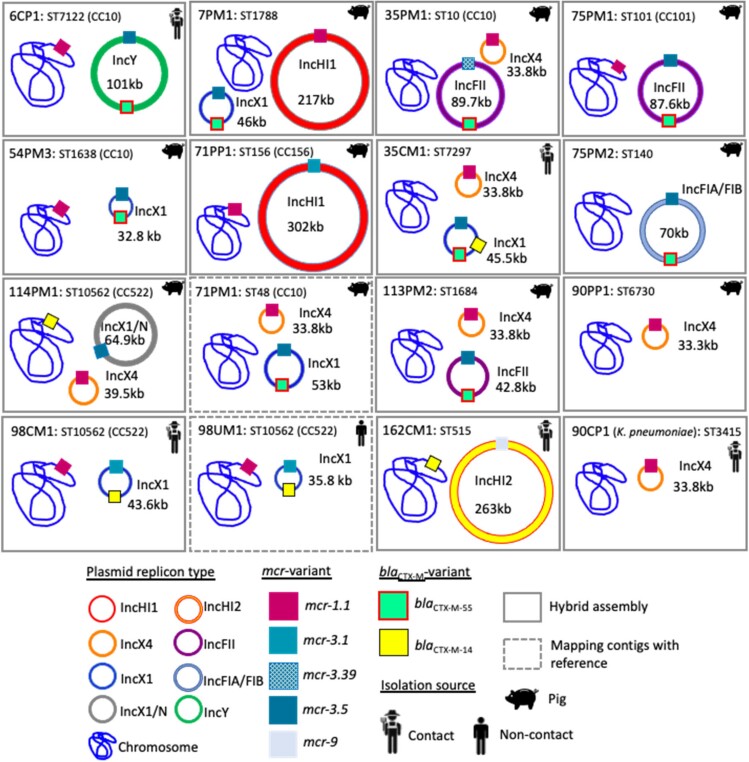
Figure 3.
Location of mcr and blaCTX-M variant in 15 E. coli and one K. pneumoniae.
Figure 4.
Circular alignments of IncX4/mcr-1 complete plasmids. Alignment of mcr-1/IncX4 complete plasmids in this study (marked with asterisk*) with plasmid sequences from Genbank: pEC1281 (accession no. MW010024.1), pMR0617mcr1 (accession no. CP024462.1), PN23 (accession no. MG557852.1) and pB1 (accession no. LC479452.1). The innermost circle represents the main reference sequence in the alignment. ARGs and mobile genetic elements are labelled in the outermost circle.
We characterized the genetic context of mcr-1 in all isolates (Figure S3). The most common mcr-1 context was mcr-1-pap2,19 presumably generated by the initial insertion of Tn6330.20 Truncated or complete Tn6330 was identified on seven chromosomes (Figure S3). Five of these isolates harboured Tn6330 encircled with a 2-bp direct repeat, AG/AC, as described previously.19,21 Moreover, an IS1294 insertion into a complete Tn6330 found on two chromosomes generated an ISApl1-mcr-1-Δpap2-IS1294-Δpap2-ISApl1 cassette, a putative circular intermediate indicating plasticity of mcr-1 mobilization.22 However, in most mcr-1-carrying isolates, we detected a truncated form of Tn6330 lacking a single copy of ISApl1 downstream or upstream of mcr-1 [Figure S3(A–C)]. Complete loss of both ISApl1 was discovered in all IncX4 plasmids (32/32), in three IncHI1, one IncHI2 and two IncI2-related contigs. The genetic surroundings of mcr-1 genes of included isolates are shown in Figure S3(A–C).
Genetic analysis of mcr-3 carrying plasmids and mcr-3 genetic context
Our data enabled an in-depth analysis of 36 mcr-3 positive isolates. mcr-3 was plasmid located in 33, and chromosomally located in three isolates. In contrast to mcr-1 plasmid types, mcr-3 plasmids were more diverse including IncFII (n = 10), IncX1 (n = 12), IncP1 (n = 5), IncN (n = 2), IncX1/N (n = 1), IncFIA/FIB (n = 1), IncHI1 (n = 1) and IncY (n = 1) incompatibility groups (Figure 1, Table 2).
In total, 13 mcr-3 plasmids were circularized (Figure 3). BLAST search with these plasmids against NCBI and PLSDB databases did not result in an exact match for IncX1, IncX1/N and IncFIA/FIB plasmids. Characteristics of these plasmids and plasmid-related contigs are shown in Table S3, Figure S4A–I. Conversely, p71PP1-IncHI1/mcr-3.1 (E. coli 71PP1; ST48) shared almost identical backbone structure with pCP131-IncHI1/mcr-3.1 identified in E. coli (ST156) isolated from a pig slaughterhouse in China (accession number CP053721.1) as illustrated in Figure S4F. Moreover, p6CP1-IncY/mcr-3.5 (E. coli 6CP1; ST7122) had a similar backbone structure with a non-conjugative phage P7-like plasmid, pHYEC7-mcr-1, hosted by an E. coli isolated from a pig farm in China (accession number: KX518745.1) (Figure S4G). Additionally, BLASTing of five almost identical mcr-3 harbouring IncP-1 related contigs (99% similarity, ranging in size from 49 345 to 52 980 bp) resulted in matching, with previously published, IncP1 plasmids hosted by E. coli from pigs and humans in Taiwan and China (100% coverage and >99.5% identity) (Figure S4H). Finally, IncFII/mcr-3 plasmids/plasmid-related contigs shared identical backbone structure with pPN42 identified in E. coli from a duck in Thailand (Figure S4I). Our findings suggest that some conserved mcr-3 containing plasmids, mainly belonging to IncP and IncFII, and to some extent also IncHI1 and IncY may be circulating among human and animal hosts in South-East Asia.
Investigations of the genetic context of mcr-3 showed a high level of variability regardless of plasmid type. Overall, an mcr-3-dgkA was found to be a core structure in all mcr-3 containing isolates. The structure was found flanked by several combinations of ISs (Figure 5, Figure S5).
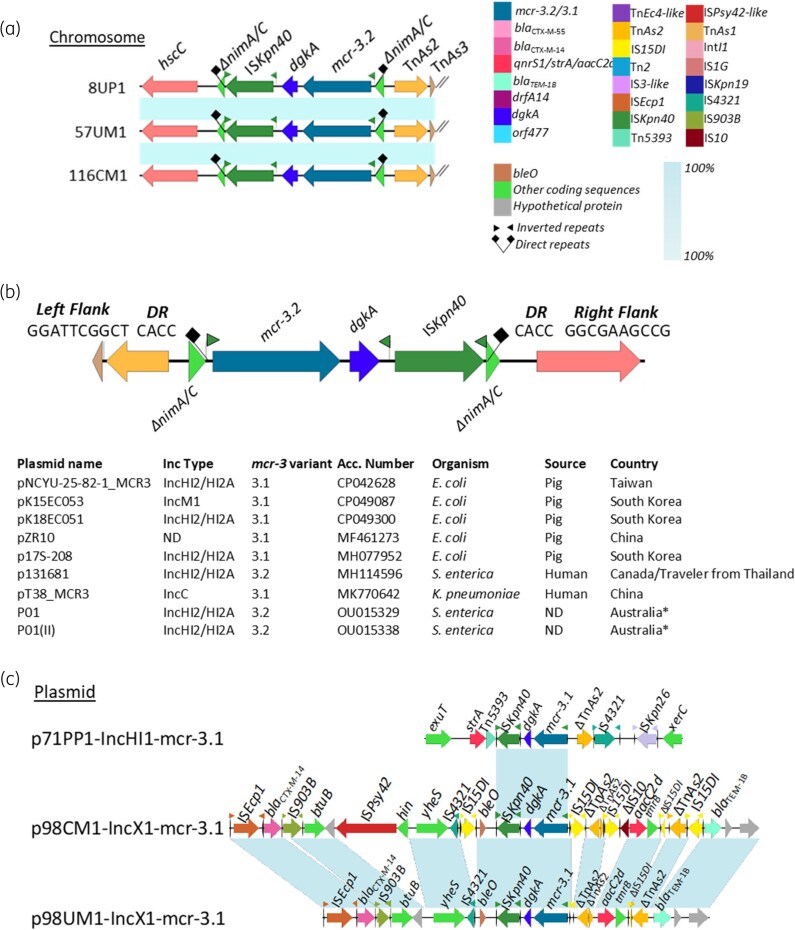
Figure 5.
Genetic environment of mcr-3.2/3.1 variants in chromosome and complete plasmids. (a) The genetic environment of mcr-3.2 on chromosomes; (b) the 4-bp direct repeats surrounding on mcr-3.2 structure found in this study and on other plasmids in GenBank and (c) the genetic environment of mcr-3.1 on plasmids.
On mcr-3.2 carrying chromosomes of three human-originating ST10 isolates, we identified a structure that might represent a transposition unit (TU)-like structure, mcr-3.2-dgkA-ISKpn40. While left and right inverted repeats (IR), IRL and IRR flanked exclusively ISKpn40, an alternative IRR1 and IRL flanked the complete structure (Figure 5A). The TU was found inserted into the nimA/C gene flanked by the 4 bp DRs, CACC (Figure 5B). As identical DRs are generated by the insertion of the composite transposon ISKpn40-mcr-3-dgkA-ISKpn40, the TU-like structure might originate from the same composite transposon.23,24 Moreover, a search in GenBank showed that nimA/C was targeted by the same TU on nine previously published plasmids of different incompatibility groups originating from various Enterobacteriaceae isolates in Asia (Figure 5B). On all these nine plasmids, the same DR repeat was found indicating a recent insertion event.
For the mcr-3.1 gene, the identical structure, mcr-3.1-dgkA-ISKpn40 was found on three plasmids characterized in this study (Figure 5C) hosted by isolates 71PP1, 98CM1 and 98UM1. Isolates 98CM1 and 98UM1 were found to be closely related (ST10562, 0 SNPs), sharing a similar plasmid. However, we were unable to detect DRs in these plasmids. The similar mcr-3.19-dgkA-ISKpn40 structure was also found on four IncFII/mcr-3.19 plasmid-related contigs; in isolates 103CM1 (ST278), 103UM1 (ST278), 111PM1 (ST48) and 28PP2 (ST101) (Figure S5). Neither an alternative IR nor DRs were found.
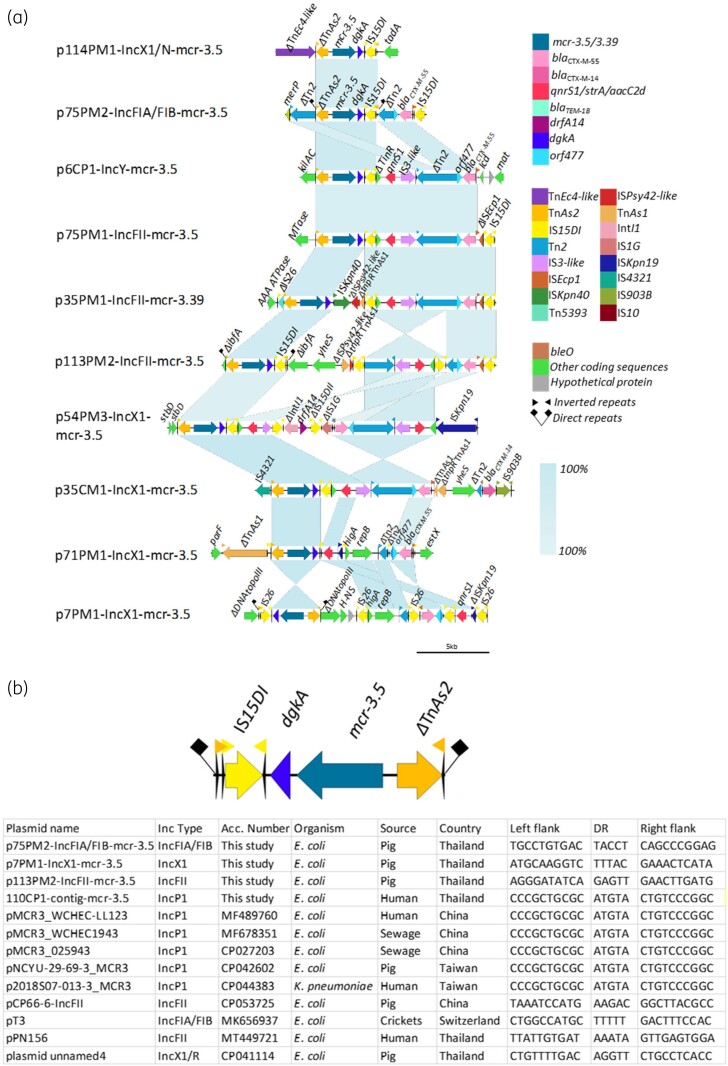
In our study, we identified a ΔTnAs2-mcr-3-dgkA-IS26/15DI structure on nine closed plasmids (IncX1, IncFIA/FIB, IncY, IncFII) and 13 contigs (IncX1, IncN, IncP1) (Figure 6A and Figure S5). Due to the end of contigs only IS26/15 remnants were detected upstream from dgkA (Figure S5). Two similar structures, ΔIS26/15DI-TnAs2-mcr-3.5-dgkA and ΔIS26-ΔTnAs2-mcr-3.1-dgkA-ISKpn40-ble identified on plasmids and harboured by two E. coli isolates (from a pig slaughterhouse, China) have been previously described.25 These structures have been shown to generate a circularized intermediate, indicating their possible mobilization.24 Identification of DRs surrounding the structure found in our study further provides evidence of mobilization. A pair of 5 bp DRs was identified on three complete plasmids: p75PM2-IncFIA/FIB (TACCT), p7PM1-IncX1 (TTTAC), p113PM2-IncFII (GAGTT) and on one IncP1/mcr-3.5-related contig 110CP1 (ATGTA) (Figure 6B). Moreover, an identical structure was also identified on GenBank deposited plasmids belonging to different Inc types, IncP1, IncFII, IncFIA/FIB and IncX1/R, hosted by various bacterial hosts, from different countries (Figure 6B). The 5 bp DR sequence on these plasmids was unique for each except for IncP1-related contig and GenBank available IncP1 plasmids that shared identical DR sequence (ATGTA) and backbone structure (Figure 6B and Figure S4H).
Figure 6.
Genetic environment of mcr-3.5/3.39 variants in complete plasmids. (a) Linear comparison of mcr-3.5 genetic environment identified in complete plasmids/contigs. (b) The 5-bp direct repeats surrounding on mcr-3.5/3.39 structure found in this study and on other plasmids in Genbank.
On 11 of 13 mcr-3 circular plasmids (mcr-3.1, n = 2; mcr-3.39/3.5, n = 9), we found several AMR genes in the vicinity of mcr-3 indicating a possible hotspot26 for accumulation of various AMR genes (Figure 5C and Figure 6A). On two IncX1/mcr-3.1 plasmids (98CM1 and 98UM1), blaCTX-M-14 was carried by ISEcp1-blaCTX-M-14-IS903 transposon (2874 bp)27 downstream of the mcr-3 cassette (Figure 5C). On nine mcr-3.5 carrying plasmids, we observed an identical pattern of qnrS1, which was almost exclusively found associated with IS3-like insertion sequence. Whereas blaCTX-M-55-orf477 was always found in close proximity to ISEcp1 remnants, a key mobilizer of blaCTX-M29,28 (Figure 6A). Different Inc-type plasmids, p6CP1-IncY/mcr-3.5 and p75PM1-IncFII/mcr-3.5 shared almost an identical structure of TnAs2-mcr-3.5-dgkA-IS15DI-TinR-qnrS1-IS3like-Tn2-orf477-blaCTX-M-55. This might indicate the involvement of a putative mobile element on these plasmids.
mcr-9 gene cassette
One mcr-9.1-positive isolate was found (162CM1), originating from a human host. The plasmid, belonging to IncHI2 group, shared identical backbone with plasmid pD610-IncHI2 (accession number MG288680.1, K. pneumoniae, China) containing an ‘IS26-wbuC-mcr-9.1-IS903B’ genetic structure (Figure S6), which has been reported in several bacterial species.30
Co-occurrence and co-transfer of mcr-1 and mcr-3 variants
The isolates co-harbouring mcr-1 and mcr-3 were analysed by long-read sequencing (n = 10) and mapping (n = 2) to resolve the location of the mcr variants within the genomes. Six isolates had chromosomally located mcr-1. mcr-3 was exclusively found on plasmids in these 12 strains. Figure 3 shows the characteristics of strains co-harbouring mcr-1 and mcr-3 genes.
We circularized 22 mcr-carrying plasmids in this study (Figure 3). Conjugation experiments were performed with all 16 isolates (Table S4 and Table S5) with fully closed plasmids as donors. Seven out of eight mcr-1 plasmids were successfully transferred including the plasmid in K. pneumoniae. Only two mcr-3 carrying plasmids were found transferable, simultaneously with mcr-1 plasmids from the pig-originating strains 7PM1 (IncHI1/mcr-1 and IncX1/mcr-3.5) and 114PM1 (IncX4/mcr-1 and IncX1/N/mcr-3.5). Among 14 out of 22 circular plasmids, transfer-related modules such as oriT, relaxase gene, T4CP and type IV secretion system cluster were found (Table S4).
Discussion
In Thailand, colistin was banned as a feed additive in March 2018. However, prolonged cumulative selective pressure of both colistin and other antimicrobial agents might have resulted in a high prevalence of bacteria resistant to these agents.31,32 Here, we conducted a comprehensive sampling and screening of plasmid-mediated colistin resistance in pig and human samples from farms in Northern Thailand collected in late 2018. A strength of our study was the sampling behind our dataset. We sampled 164 farms in the Khon Kaen Province and from most of the farms (n = 156), samples from both pigs and humans were obtained. Our sampling design and extensive genome sequencing enabled us to study mcr prevalence, transmission of strains, plasmids and mcr variants among hosts living on the same farm. To the best of our knowledge, our study includes the highest number of sampled epidemiological units (farms) to study mcr transmission between pigs and humans.
Colistin was reported to be used on only two of the 164 farms.33 However, mcr-1/mcr-3 carrying Escherichia spp. was found in high occurrence; 37.8% (62/164) and 16.1% (44/164) pig and human samples, respectively. This may be explained by the following (i) higher-than-reported use of colistin on the study farms, (ii) prolonged effects of previous colistin consumption and/or (iii) dissemination of colistin-resistant strains in the community. Since mcr-carrying isolates were more common among pigs than humans suggest the first two alternative explanations as more important factors. The percentage of mcr-positive samples on farms was significantly higher than in previous reports from other provinces, which ranged between 4.45% and 5.8%.34,35 However, the prevalence of E. coli co-harbouring mcr-1 and mcr-3 was comparable to a study of flies collected from farms in Thailand.14 Furthermore, our results showed that 86.3% of mcr-carrying E. coli were MDR. The predominant ESBL genes accompanying mcr were blaCTX-M-55 and blaCTX-M-14, the most common ESBL-resistance genes found in Enterobacteriaceae in Thailand.29,36mcr genes were present in a highly diverse set of host strains. Clonal transmission of mcr-positive E. coli within a farm was detected on five farms indicating a limited number of human-to-human or pig-to-human whole bacterium transmission events. Conversely, the evidence of plasmid spread was noted, particularly dissemination of epidemic IncX4/mcr-1 and IncHI1/mcr-1 plasmids. Similar plasmids have also been described in the Thai community previously.13,37 This may indicate that horizontal gene transfer/plasmid transfer plays the most important role in the spread of mcr. Our result contradicts previous reports of IncI2 as the major mcr-1 plasmid type in Asia.6 Furthermore, the dissemination of conserved IncP/mcr-3 and IncFII/mcr-3 plasmids over the Asian continent was suggested and should be further investigated.23,38,39
Assessment of the mcr genetic contexts showed that Tn6330 and its derivatives were common vectors of horizontal mcr-1 transfer as previously reported.40 The observation of complete loss of ISApl1 from Tn6330 renders the mcr-1-pap2 non-mobilizable indicating a stabilization of mcr-140 on IncX4, IncHI1 and IncI2 plasmids in the study. Due to the shorter assembled sequences, we were unable to retrieve the full length of mobile genetic elements surrounding mcr-3 gene for all isolates. Despite this limitation, we discovered various plasmid types harbouring mcr-3 flanked by several combinations of mobile elements including TnAs2, IS26/IS15DI and ISKpn40. These mobile elements have been previously considered key elements of mcr-3 transposition between different bacterial species.23–25,41 Additionally, the identified DRs in our study indicated a recent insertion of mcr-3-dgkA-ISKpn40 and ΔTnAs2-mcr-3-dgkA-IS15DI structures, and thereby possible mobilization and circulation in Asia.
The presence of blaCTX-M and other AMR genes in the vicinity of mcr-3 might indicate accumulation of AMR genes as reported earlier in our K. pneumoniae study.26 Evidence of co-transfer of mcr-1 and mcr-3 plasmids was confirmed in two isolates. However, the experimental conditions may have been suboptimal for transmission of all collected plasmids. Isolates co-harbouring two mcr variants might be considered an exceptional mcr gene disseminator.26
In conclusion, our One Health approach enabled us to explore the epidemiology of mcr genes, mcr genetic contexts and plasmids. We discovered that mcr-1, was mostly carried by conserved epidemic plasmids. mcr-3 was located on a more diverse set of plasmid types, some of them not previously reported as mcr-3 carriers. Our study also discovered possible endemic IncP and IncFII plasmids with mcr-3, disseminated in several Asian countries. We found genetic signatures in line with recent mcr-3 transposition events, underlining the mobilizable nature of the mcr-3 gene cassette. Of special concern is the growing number of different horizontal gene transferring pathways encompassing various transposable modules the mcr genes can be shared between bacteria. Careful monitoring and follow-up studies are needed to get more insight into the epidemiology mcr genes and the possible evolution of successful plasmids and transposition modules containing mcr and other antimicrobial resistance genes of clinical relevance.
Supplementary Material
Acknowledgements
We thank Ellen Gulbrandsen for contribution to the sampling and fieldwork. The farmers in the Khon Kaen Province are greatly acknowledged for their willingness to participate. We acknowledge Fiona Franklin and Hanna Ilag for their experimental assistance.
Contributor Information
Thongpan Leangapichart, Section for Food Safety and Animal Health Research, Norwegian Veterinary Institute, Ås, Norway.
Milan S Stosic, Section for Food Safety and Animal Health Research, Norwegian Veterinary Institute, Ås, Norway.
Rachel A Hickman, Department of Medical Biochemistry and Microbiology, Zoonosis Science Center, Uppsala University, Uppsala, Sweden; Department of Medical Sciences, Zoonosis Science Center, Uppsala University, Uppsala, Sweden.
Kamonwan Lunha, Department of Clinical Sciences, Swedish University of Agricultural Sciences, Uppsala, Sweden.
Jatesada Jiwakanon, Research Group for Animal Health Technology, Faculty of Veterinary Medicine, Khon Kaen University, Khon Kaen, Thailand.
Sunpetch Angkititrakul, Research Group for Animal Health Technology, Faculty of Veterinary Medicine, Khon Kaen University, Khon Kaen, Thailand.
Ulf Magnusson, Department of Clinical Sciences, Swedish University of Agricultural Sciences, Uppsala, Sweden.
Thomas P Van Boeckel, Health Geography and Policy Group, ETH Zurich, Zurich, Switzerland; Center for Diseases Dynamics Economics & Policy, Washington, DC, USA.
Josef D Järhult, Department of Medical Sciences, Zoonosis Science Center, Uppsala University, Uppsala, Sweden.
Marianne Sunde, Section for Food Safety and Animal Health Research, Norwegian Veterinary Institute, Ås, Norway.
Funding
This work was funded by the Fifth Call of the Joint Programming Initiative for Antimicrobial Resistance (JPIAMR) to the REDUCEAMU project (#2017-026), and the Norwegian Veterinary Institute.
Transparency declarations
None to declare.
Supplementary data
Methods, Tables S1–S5 and Figures S1–S6 are available as Supplementary data at JAC Online.
References
- 1. Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014; 5: 1–18. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rhouma M, Beaudry F, Thériault Wet al. . Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front Microbiol 2016; 7: 1789. 10.3389/fmicb.2016.01789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu YY, Wang Y, Walsh TRet al. . Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 4. Ling Z, Yin W, Shen Zet al. . Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother 2020; 75: 3087–95. 10.1093/jac/dkaa205 [DOI] [PubMed] [Google Scholar]
- 5. Wang C, Feng Y, Liu Let al. . Identification of novel mobile colistin resistance gene mcr-10. Emerging Microbes & Infections 2020; 9: 508–16. 10.1080/22221751.2020.1732231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matamoros S, van Hattem JM, Arcilla MSet al. . Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep 2017; 7:15364. 10.1038/s41598-017-15539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zelendova M, Papagiannitsis CC, Valcek Aet al. . Characterization of the complete nucleotide sequences of mcr-1-encoding plasmids from Enterobacterales isolates in retailed raw meat products from the Czech Republic. Front Microbiol 2021; 11: 3552. 10.3389/fmicb.2020.604067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu L, Feng Y, Zhang Xet al. . New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 2017; 61: e01757-17. 10.1128/AAC.01757-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Creighton J, Anderson T, Howard Jet al. . Co-occurrence of mcr-1 and mcr-3 genes in a single Escherichia coli in New Zealand. J Antimicrob Chemother 2019; 74: 3113–6. 10.1093/jac/dkz311 [DOI] [PubMed] [Google Scholar]
- 10. Li R, Zhang P, Yang Xet al. . Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J Antimicrob Chemother 2019; 74: 1517–20. 10.1093/jac/dkz058 [DOI] [PubMed] [Google Scholar]
- 11. Yu Y, Andrey DO, Yang Ret al. . A Klebsiella pneumoniae strain co-harbouring mcr-1 and mcr-3 from a human in Thailand. J Antimicrob Chemother 2020; 75: 2372–4. 10.1093/jac/dkaa133 [DOI] [PubMed] [Google Scholar]
- 12. Eiamphungporn W, Yainoy S, Jumderm Cet al. . Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J Glob Antimicrob Resist 2018; 15: 32–5. 10.1016/j.jgar.2018.06.007 [DOI] [PubMed] [Google Scholar]
- 13. Yang QE, Tansawai U, Andrey DOet al. . Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): an increasing public health risk. Environ Int 2019; 122: 281–90. 10.1016/j.envint.2018.11.021 [DOI] [PubMed] [Google Scholar]
- 14. Fukuda A, Usui M, Okubo Tet al. . Co-harboring of cephalosporin (bla)/colistin (mcr) resistance genes among Enterobacteriaceae from flies in Thailand. FEMS Microbiol Lett 2018; 365: fny178. 10.1093/femsle/fny178 [DOI] [PubMed] [Google Scholar]
- 15. Lunha K, Leangapichart T, Jiwakanon Jet al. . Antimicrobial resistance in fecal Escherichia coli from humans and pigs at farms at different levels of intensification. Antibiotics 2020; 9: 662. 10.3390/antibiotics9100662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hickman RA, Leangapichart T, Lunha Ket al. . Exploring the antibiotic resistance burden in livestock, livestock handlers and their non-livestock handling contacts: a one health perspective. Front Microbiol 2021; 12: 651461. 10.3389/fmicb.2021.651461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huber L, Hallenberg GS, Lunha Ket al. . Geographic drivers of antimicrobial use and resistance in pigs in Khon Kaen Province, Thailand. Front Vet Sci 2021; 8: 659051. 10.3389/fvets.2021.659051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teng CH, Wu PC, Tang SLet al. . A large spatial survey of colistin-resistant gene mcr-1-carrying E. coli in rivers across Taiwan. Microorganisms 2021; 9: 449–56. 10.3390/microorganisms9040722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kieffer N, Aires-de-Sousa M, Nordmann Pet al. . High rate of MCR-1–producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg Infect Dis 2017; 23: 2023–9. 10.3201/eid2312.170883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li R, Xie M, Zhang Jet al. . Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 2017; 72: 393–401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- 21. Zhao F, Feng Y, Lü Xet al. . Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol 2017; 8: 2094. 10.3389/fmicb.2017.02094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun J, Li XP, Fang LXet al. . Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: persistence of colistin resistance in Escherichia coli. Int J Antimicrob Agents 2018; 51: 842–7. 10.1016/j.ijantimicag.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 23. Xiang R, Liu BH, Zhang AYet al. . Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an Escherichia coli isolate from a chicken farm. Antimicrob Agents Chemother 2018; 62: e00501-18. 10.1128/AAC.00501-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He Y-ZZ, Long T-FF, Chen C-PPet al. . ISKpn40-mediated mobilization of the colistin resistance gene mcr-3.11 in Escherichia coli. Antimicrob Agents Chemother 2020; 64: 1–5. 10.1128/AAC.00851-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li R, Du P, Zhang Pet al. . Comprehensive genomic investigation of coevolution of mcr genes in Escherichia coli strains via nanopore sequencing. Glob Challenges 2021; 5: 2000014. 10.1002/gch2.202000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stosic MS, Leangapichart T, Lunha Ket al. . Novel mcr-3.40 variant co-located with mcr-2.3 and blaCTX-M-63 on an IncHI1B/IncFIB plasmid found in Klebsiella pneumoniae from a healthy carrier in Thailand. J Antimicrob Chemother 2021; 76: 2218–20. 10.1093/jac/dkab147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park J, Shin E, Park AKet al. . Co-infection with chromosomally-located blaCTX-M-14 and plasmid-encoding blaCTX-M-15 in pathogenic Escherichia coli in the Republic of Korea. Front Microbiol 2020; 11: 545591. 10.3389/fmicb.2020.545591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeng S, Luo J, Chen Xet al. . Molecular epidemiology and characteristics of CTX-M-55 extended-spectrum β-Lactamase-producing Escherichia coli From Guangzhou, China. Front Microbiol 2021; 12: 730012. 10.3389/fmicb.2021.730012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tansawai U, Walsh TR, Niumsup PR. Extended spectrum ß-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult Sci 2019; 98: 2622–31. 10.3382/ps/pez009 [DOI] [PubMed] [Google Scholar]
- 30. Fuentes-Castillo D, Sellera FP, Goldberg DWet al. . Colistin-resistant Enterobacter kobei carrying mcr-9.1 and blaCTX-M-15 infecting a critically endangered Franciscana dolphin (Pontoporia blainvillei), Brazil. Transbound Emerg Dis 2021; 68: 3048–54. 10.1111/tbed.13980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lekagul A, Tangcharoensathien V, Mills Aet al. . How antibiotics are used in pig farming: a mixed-methods study of pig farmers, feed mills and veterinarians in Thailand. BMJ Glob Heal 2020; 5: e001918. 10.1136/bmjgh-2019-001918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu F, Zeng X, Hinenoya Aet al. . MCR-1 confers cross-resistance to bacitracin, a widely used in-feed antibiotic. mSphere 2018; 3: e00411-18. 10.1128/mSphere.00411-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hallenberg GS, Jiwakanon J, Angkititrakul Set al. . Antibiotic use in pig farms at different levels of intensification—farmers’ practices in Northeastern Thailand. PLoS One 2020; 15: e0243099. 10.1371/journal.pone.0243099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khine NO, Lugsomya K, Kaewgun Bet al. . Multidrug resistance and virulence factors of Escherichia coli harboring plasmid-mediated colistin resistance: mcr-1 and mcr-3 genes in contracted pig farms in Thailand. Front Vet Sci 2020; 7: 582899. 10.3389/fvets.2020.582899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pungpian C, Lee S, Trongjit Set al. . Colistin resistance and plasmid-mediated mcr genes in Escherichia coli and Salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces. J Vet Sci 2021; 22: e68. 10.4142/jvs.2021.22.e68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lay KK, Jeamsripong S, Sunn KPet al. . Colistin resistance and ESBL production in Salmonella and Escherichia coli from pigs and pork in the Thailand, Cambodia, Lao PDR, and Myanmar Border Area. Antibiotics 2021; 10: 657. 10.3390/antibiotics10060657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Srijan A, Margulieux KR, Ruekit Set al. . Genomic characterization of nonclonal mcr-1-positive multidrug-resistant Klebsiella pneumoniae from clinical samples in Thailand. Microb Drug Resist 2018; 24: 403–10. 10.1089/mdr.2017.0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tansawai U, Yu Y, Kiddee Aet al. . Emergence of mcr-3-mediated IncP and IncFII plasmids in Thailand. J Glob Antimicrob Resist 2021; 24: 446–7. 10.1016/j.jgar.2021.02.006 [DOI] [PubMed] [Google Scholar]
- 39. Zhou Y, Farzana R, Sihalath Set al. . A one-health sampling strategy to explore the dissemination and relationship between colistin resistance in human, animal, and environmental sectors in Laos. Engineering 2022; 15: 45–56. 10.1016/j.eng.2022.01.013 [DOI] [Google Scholar]
- 40. Snesrud E, McGann P, Chandler M. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. MBio 2018; 9: e02381-17. 10.1128/mBio.02381-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z, Fu Y, Du X-Det al. . Potential transferability of mcr-3 via IS26-mediated homologous recombination in Escherichia coli. Emerg Microbes Infect 2018; 7: 55. 10.1038/s41426-018-0057-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reads (fastq files) from the study have been submitted to the European Nucleotide Archive (www.ebi.ac.uk/ena) under study accession number PRJEB38313 and PRJEB39885.