Abstract
Despite renewed interest, development of chemical biology methods to study peptidoglycan metabolism has lagged in comparison to the glycobiology field in general. To address this, a panel of diamides were screened against the Gram-positive bacterium Streptococcus pneumoniae to identify inhibitors of bacterial growth. The screen identified the diamide masarimycin as a bacteriostatic inhibitor of S. pneumoniae growth with an MIC of 8 µM. The diamide inhibited detergent-induced autolysis in a concentration-dependent manner, indicating perturbation of peptidoglycan degradation as the mode-of-action. Cell based screening of masarimycin against a panel of autolysin mutants, identified a higher MIC against a ΔlytB strain lacking an endo-N-acetylglucosaminidase involved in cell division. Subsequent biochemical and phenotypic analyses suggested that the higher MIC was due to an indirect interaction with LytB. Further analysis of changes to the cell surface in masarimycin treated cells identified the overexpression of several moonlighting proteins, including elongation factor Tu which is implicated in regulating cell shape. Checkerboard assays using masarimycin in concert with additional antibiotics identified an antagonistic relationship with the cell wall targeting antibiotic fosfomycin, which further supports a cell wall mode-of-action.
Keywords: peptidoglycan, chemical biology, mode-of-action, meta-phenotype, cell wall
Introduction
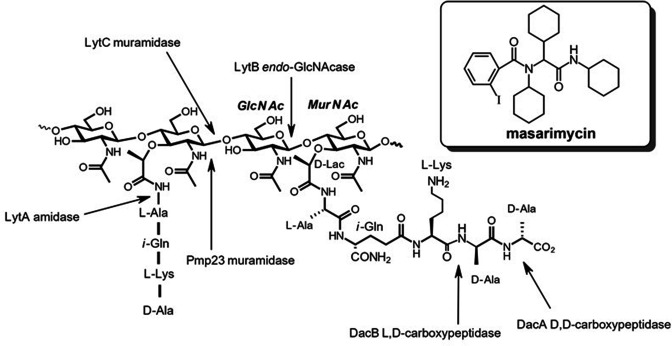
Antibiotic resistance is a growing global threat. Drug-resistant Streptococcus pneumoniae alone is estimated to cause more than two million infections with an excess of 1.3 billion USD in medical costs per annum [1, 2]. In light of this, there is need for the development of new therapeutics. Peptidoglycan (PG) is the primary structural heteropolymer conferring strength and cell shape determination in both Gram-negative and Gram-positive organisms (Fig. 1). The PG polysaccharide backbone is composed of β−1,4-linked N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc). Attached to the C-3 lactyl moiety of MurNAc is a stem peptide that is involved in cross-linking the adjacent glycan strands to form the three-dimensional structure of the cell wall. The incorporation of new material into the stress bearing layer of the existing cell wall requires the delicate homeostasis of biosynthetic and degradative enzymes to prevent lysis [3, 4]. Disruption of this interplay between degradative and biosynthetic enzymes via chemical inhibition could provide unique insights into their biological role. The degradative enzymes, collectively referred to as autolysins, are a broad class of enzymes that are differentiated based on their bond selectivity. Deciphering physiological activity of autolysins has been a formidable task as functional redundancy complicates attribution of biological activity [5]. Recent biophysical [4, 6] and computational studies [7] of bacterial autolysins have begun to unravel their roles in the release of stress in the cell wall to allow for incorporation of new material. A renaissance in PG metabolism research has started to provide new chemical biology tools to study synthesis [8–11] and the role endopeptidases play in methicillin resistance [12]. While the cell wall, and PG in particular, have provided a wealth of clinically relevant antimicrobial targets [13], our understanding of the complex interplay between degradative and synthetic steps is still developing.
Fig. 1.
Structure of PG showing the cleavage sites of several of the characterized autolysins in S. pneumoniae . Inset: structure of the antimicrobial diamide masarimycin.
Previously, we had screened a panel of 21 diamides for antibacterial activity against the Gram-positive Bacillus subtilis [14]. This screen identified the diamide masarimycin (formerly fgkc) (Fig. 1 inset) as a bacteriostatic inhibitor of B. subtilis growth that targets the major active N-acetylglucosaminidase (GlcNAcase) LytG (glycosyl hydrolase family 73 (GH73)) in vitro. Here we report on the screening of this panel of diamides against S. pneumoniae, identifying masarimycin as a bacteriostatic inhibitor of cell growth. Although an initial examination experiment implicates a related S. pneumoniae GlcNAcase (LytB, GH73) in masarimycin’s activity, it is not the direct molecular target. A series of subsequent mode-of-action studies in S. pneumoniae highlights the challenges involved in target identification.
Methods
Strains and compounds
Streptococcus pneumoniae 6305 and R6 were purchased from ATCC (Mannassas, VA), and S. pneumoniae IU1945 (ΔlytB, ΔlytC, ΔdacA, ΔdacB, Δpmp23, Δpbp1a) mutants were kindly provided by Dr Malcolm E. Winkler at the University of Indiana [15]. S. pneumoniae TIGR4 and TIGR4 ΔlytB strains were previously reported [16]. S. pneumoniae strains were grown in Mueller Hinton (MH) broth (MilliporeSigma, St. Louis, MO) supplemented with 5 % (v/v) sheep blood (Lampire Biological, Pipersville PA) or MH agar plates containing 1.5 % (m/v) Bacto agar and 5 % (v/v) sheep blood at 37 °C under anaerobic conditions. Staphylococcus aureus was grown in MH broth or solid media, Clostridiodes difficile was grown in brain heart infusion (BHI) and Escherichia coli DH5α on Luria Bertani (LB) broth or solid media. Diamide inhibitors were synthesized as described previously [14]. Other reagents, unless otherwise specified, were purchased from MilliporeSigma (St. Louis, MO).
MIC assays
MIC values were determined using the resazurin method [17]. Briefly, cells were initially grown from the freezer on MH agar plates containing 5 % defibrinated sheep blood. For all assays second passage cells of S. pneumoniae 6305, TIGR4, or R6 were used and grown overnight in MH broth, and standardized to an OD600nm = 0.4. Inhibitors were analysed via serial dilution into PBS media in microtitre plates. For masarimycin, dilutions were initially made in DMSO down to a concentration of 100 µM, further dilutions were then made into PBS. Plates containing MH broth were inoculated with a 1/20 dilution of the OD600nm = 0.4 cell culture with a final concentration of 1 % DMSO. Cultures were grown statically under anaerobic conditions for 24 h at 37 °C, followed by addition of 30 µl of a 0.01 % (m/v) solution of resazurin. The plates were incubated for 15 min to allow stabilization of colour production. MICs were read directly off the plate; MICs were recorded as the lowest concentration that completely inhibited growth. MIC assays with S. aureus were performed in MH broth, C. difficile in BHI broth, while E. coli was performed in LB.
Morphological studies of S. pneumoniae
Cultures were prepared from second passage S. pneumoniae 6305, R6 and ΔlytB [15] as previously described for MIC determination. Cells were washed in phosphate-buffered saline and chemically fixed in 20 mM HEPES pH 6.8 containing 1 % formaldehyde [18]. Samples were fixed overnight at 4 °C to limit de novo cell wall biosynthesis during fixation and stained with 0.1 % (m/v) methylene blue (solution in 20 % ethanol). Samples were gently heated to 60 °C for 15–20 min to bring cells to a common focal plane. Slides were visualized using bright-field microscopy with a Zeiss Primo Star microscope at 1000× magnification. Micrographs were acquired using an Axiocam ERc5s camera and Zen lite software.
Autolysis assays
Cellular autolysis assays were performed as previously described by Cornett and Shockman [19]. Briefly, S. pneumoniae 6305 were grown in MH broth containing 5 % (v/v) defibrinated sheep blood under anaerobic conditions. Cells were harvested by centrifugation (8000 r.p.m., 5 min) and washed with PBS. Cells were suspended in PBS and autolysis induced with the addition of Triton X-100 to a final concentration of 0.1 % (v/v) and turbidity monitored at 600 nm over 60 min. Rates were calculated using the linear portion of the autolysis curves with the rate of autolysis in the absence of inhibitor set at 100 %.
Chain dispersing assay
Dispersion of the ΔlytB chain morphology with purified LytB was carried out as previously described using the TIGR4 and associated ΔlytB strains [16]. LytB was added to the cell suspension at a final concentration of 2 µM. The final concentration of masarimycin in the assays was 40 µM.
DNA intercalation assays
To determine if masarimycin intercalates DNA, DNA mobility shift assays were performed as previously described using BamHI-linearized pUC18 plasmid [20]. The known DNA intercalator actinomycin D was used as a control.
Dnase I assays
Degradation of the pUC18 plasmid DNA was assayed using 150 ng of linearized pUC18 plasmid as described by Huang et al. [21]. Compound titrations in DMSO were added and reactions were initiated with 0.002 units of Dnase I. The reactions were incubated for 15 min at 37 °C before being subjected to agarose gel electrophoresis. EDTA was used as a control for Dnase I inhibition.
Lipotechoic acid detection by Western blot
Lipoteichoic acid profiles were analysed as previously described [22]. Briefly, S. pneumoniae R6 cells were cultured overnight, harvested (3000 g ), resuspended in 6 M urea, and incubated at 37 °C for 5 min to solubilize proteins. Samples were standardized by total protein content and separated by SDS-PAGE (16 %) and transferred to PVDF membrane. The membrane was incubated with a 1 : 5000 α-phosphocholine monoclonal antibody (SSI Biotech, Santa Cruz, CA). Blots were analysed by chemiluminescence.
Analysis of cell wall associated proteins
Changes to cell wall protein profiles were analysed as previously described [23]. Briefly, second passage S. pneumoniae R6 were inoculated 1/100 into MH broth and grown statically for 6 h anaerobically at 37 °C. Masarimycin was added to a final concentration of 0.75 × MIC, the effect of solvent was controlled by the addition of DMSO to a second flask and the cells grown overnight at 37 °C anaerobically. Cells were harvested into PBS containing sucrose (20 % w/v) and pelleted at 8000 r.p.m. for 10 min. The pellets were washed with PBS containing 20 % (w/v) sucrose and centrifuged again. The washed and pelleted bacteria were resuspended in 2 ml of 50 mM glycine-NaOH (pH 12.0) containing 20 % (w/v) sucrose and incubated for 30 min at room temperature with gentle shaking. The suspension was centrifuged (10 000 r.p.m., 20 min). The supernatant was collected, and adjusted to pH 7 with 1 M HCl, proteins precipitated with acetone and analysed by 1D-SDS PAGE and silver staining. All lanes were standardized to total A280nm loaded onto the gel. Bands were excised from the gel and sent for identification by mass spectrometry at the National Resource for Proteomics (University of Arkansas).
Proteomic analysis of SDS-PAGE gel bands
Each SDS-PAGE gel band was subjected to in-gel trypsin digestion as follows. Gel segments were destained in 50 % methanol (Fisher), 50 mM ammonium bicarbonate, (Sigma-Aldrich), followed by reduction in 10 mM Tris[2-carboxyethyl]phosphine (Pierce) and alkylation in 50 mM iodoacetamide (Sigma-Aldrich). Gel slices were then dehydrated in acetonitrile (Fisher), followed by addition of 100 ng porcine sequencing grade modified trypsin (Promega) in 50 mM ammonium bicarbonate (Sigma-Aldrich) and incubation at 37 °C for 12–16 h. Peptide products were then acidified in 0.1 % formic acid (Pierce). Tryptic peptides were separated by reverse phase Xselect CSH C18 2.5 um resin (Waters) on an in-line 150×0.075 mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 60 min gradient from 98 : 2 to 65 : 35 buffer A:B ratio. [Buffer A=0.1 % formic acid, 0.5 % acetonitrile; buffer B=0.1 % formic acid, 99.9 % acetonitrile.] Eluted peptides were ionized by electrospray (2.4 kV) followed by MS/MS analysis using higher-energy collisional dissociation (HCD) on an Orbitrap Fusion Tribrid mass spectrometer (Thermo) in top-speed data-dependent mode. MS data were acquired using the FTMS analyser in profile mode at a resolution of 240 000 over a range of 375 to 1500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyser in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0. Proteins were identified by database search using Mascot (Matrix Science) with a parent ion tolerance of 3 ppm and a fragment ion tolerance of 0.5 Da. Scaffold (Proteome Software) was used to verify MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established with less than 1.0 % false discovery by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established with less than 1.0 % false discovery and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [24].
Antagonism assay and fractional inhibitory concentration index (FICIndex) determination
Fractional inhibitory concentration index (FICIndex) was conducted to determine the interaction between masarimycin and a range of antibiotics with defined mode-of-action in a 96 well-plate microdilution broth assay. A checkerboard assay was performed with each masarimycin pair as previously described [25]. Plates were inoculated with 5 µl of a OD600nm =0.2 culture of S. pneumoniae R6 and growth monitored as previously described for the MIC assays. FICIndex was determined using the formulae:
(Eq 1) , where X is the lowest inhibitory concentration of the drug in the presence of the co-drug, and MICx is the lowest inhibitory concentration of the drug in the absence of the co-drug.
(Eq 2) FICIndex=FICmasarimycin+FICantibiotic
Drug interactions were rated as synergistic (FICIndex ≤0.5), additive (0.5<FICIndex≤1.0), indifferent (1.0<FICIndex≤4.0), and antagonistic (FICIndex >4.0), based on published standards [26].
NADP/NADP(H) ratio assays
Measurement of NADP/NADPH intracellular ratios was determined via the Amplite colorimetric NADP/NADPH ratio assay kit (Kit #15274:, AAT Bioquest Inc, Sunnyvale CA) following manufacturer protocols. S. pneumoniae R6 second passage cells grown on MH with 5 % defibrinated sheep blood under anaerobic conditions were used to inoculate 5 ml MH broth cultures (OD600nm = 0.5) containing either masarimycin (1 ×, 3 × MIC) or vehicle control. Cultures were grown under anaerobic conditions for 1.5 h and centrifuged (8000 r.p.m., 10 min). Cell pellets were resuspended in PBS and lysed via sonication. Lysed cells were then used in assays following manufacturer’s instructions. Samples were analysed in a 96 well plate format using a Molecular Devices SpectraMax190 with detection at 570 nm. Data was analysed using GraphPad Prism.
Results and discussion
We screened a previously reported [14] panel of 21 diamides against S. pneumoniae using the resazurin microtitre assay (Fig. S1, available in the online version of this article) [17]. Of the 21 compounds screened, masarimycin (formerly fgkc) was identified as a single digit micromolar bacteriostatic inhibitor (Fig. S2) of S. pneumoniae growth with an MIC of 8 µM against all three strains of S. pneumoniae that we tested – 6305, R6, and TIGR4 (Fig. S3a). Three strains were chosen for screening to account for the known genomic plasticity among S. pneumoniae isolates which can manifest as varying antibiotic sensitivity between strains [27–29]. TIGR4 was included as it is capable of causing invasive disease [30]. These results for masarimycin are comparable to those obtained against B. subtilis (MIC=3.8 µM, bacteriostatic) [14]. To further investigate masarimycin’s spectrum of activity, the compound was screened against the Gram-positive organisms Clostridiodes difficile, Staphylococcus aureus and Gram-negative Escherichia coli which possess at least one GH73. In all cases, no antimicrobial activity was observed up to 150 µM (Fig. S3b). Extrapolating from the activity of masarimycin against B. subtilis , where it inhibits LytG, a GH73 enzyme, we hypothesized that the target of masarimycin in S. pneumoniae was also a GH73 family glycosidase. S. pneumoniae possesses one glycosidase classified as a member of the GH73 family (www.cazy.org) – LytB, a cell division associated endo-β-GlcNAcase belonging to cluster 4 of GH73 [16, 31–33]. In contrast, B. subtilis LytG is an exo-acting GlcNAcase active in vegetative growth and belongs to GH73 cluster 2 [34].
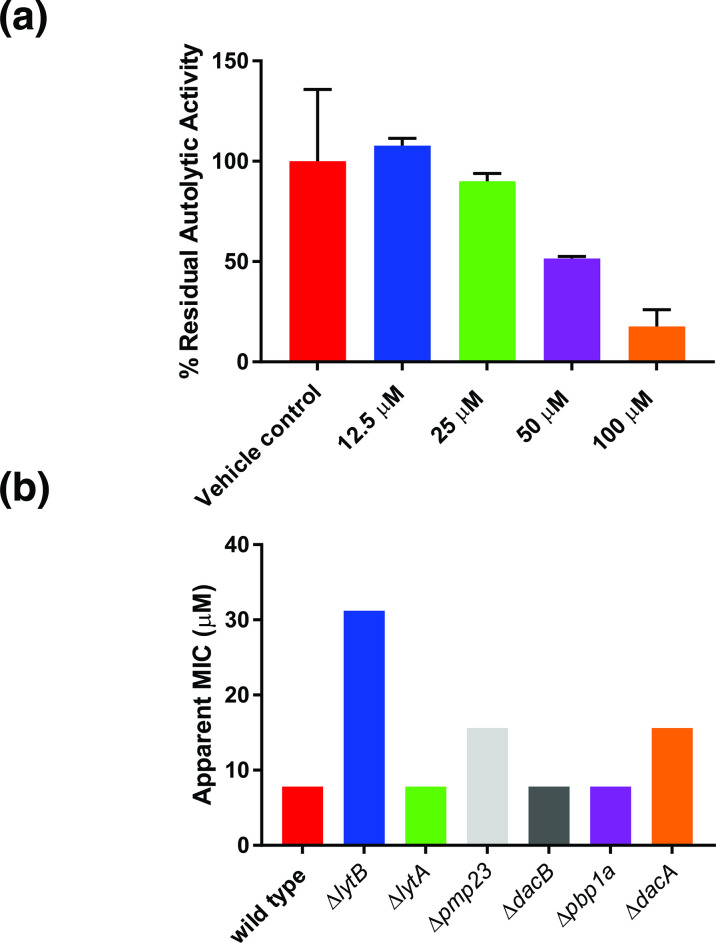
Given the potential connection to LytB and cell wall metabolism, we examined whether masarimycin inhibited autolytic activity in S. pneumoniae (Fig. 2a). Exposure of S. pneumoniae to low concentration of the non-ionic detergent (Triton X-100) induces autolysis [19]. This autolytic activity was inhibited in a concentration-dependent manner by masarimycin. The elevated concentration for near complete inhibition of autolysis in the whole cell assay is likely due to the broad dysregulation of autolysins induced by Triton X-100. Given the observed inhibition of autolytic activity, the MIC of masarimycin was determined for a series of S. pneumoniae mutant strains lacking the autolysins lytA, lytB, pmp23, dacA, or dacB, as well as the bifunctional pbp1a. It has been demonstrated that deletion of any gene that affects PG biosynthesis, stability, or regulation can make the bacterium more susceptible to compounds that target the cell wall [27]. While none of the mutants were more sensitive to masarimycin, higher MICs were observed with lytB (GH73 GlcNAcase), pmp23 (muramidase), and dacA (pbp3, d,d-carboxypeptidase) mutants. This is counter to what was observed in a screen of autolysin mutants in B. subtilis . [14] The near four-fold decrease in sensitivity to masarimycin in the ΔlytB mutant suggests that changes to the cell wall imparted by the lack LytB reduces sensitivity to masarimycin.
Fig. 2.
Screening of the diamide masarimycin against Streptococcus pneumoniae . (a) The diamide masarimycin inhibits detergent-induced autolysis in a concentration dependent manner. Percent residual activity was calculated using autolysis in the absence of inhibitor set as 100 %. Data shown is the average of experiments performed in biological and technical triplicate. Error bars denote standard deviation. (b) Activity of masarimycin against S. pneumoniae R6 autolysin and cell wall biosynthesis mutants (13) to identify changes to masarimycin sensitivity. Assays were run in biological triplicate and yielded the same MIC values.
To further explore these results, morphological changes induced by sub-MIC (0.7 ×) concentrations of masarimycin, and antibiotics with known modes-of-action were investigated (Fig. S4). Treatment of S. pneumoniae with the cell wall-acting antibiotics bacitracin, vancomycin, as well as the protein synthesis inhibitor kanamycin presented a phenotype of clumping cells. Sub-MIC treatment with masarimycin showed a change similar to these antibiotics. The clumping phenotype observed with kanamycin has been associated with antibiotics that target intracellular protein synthesis [35]. In light of this, the clumping phenotype could not be directly attributed to a cell wall mode-of-action. Comparison of the masarimycin-induced phenotype with the reported phenotypes of a ΔlytB [15], Δpmp23 [36], or ΔdacA [37] mutants, for which higher MICs with masarimycin were observed, did not correlate. Additionally, the masarimycin-induced phenotype did not correspond to phenotypes of other S. pneumoniae autolysin mutants [38–40]. The masarimycin phenotype more closely resembles a morphology in which the autolysin is still produced but is catalytically inactive, such as that reported for E61Q and D68N mutants of pmp23 [36].
To further probe alterations to the cell wall suggested by the autolysis and genetic screen assays, lipoteichoic acid (LTA) disruption was monitored by Western blot in the presence of sub-MIC masarimycin. LTA has been shown to regulate autolysin activity in several species and LytB possesses a choline binding domain, a component of S. pneumoniae LTA [41–43]. Additionally, it has been suggested that LytB function is altered when cell wall choline content is depleted [44]. Results indicated that changes to LTA and choline incorporation in the cell wall was not a contributor to the observed masarimycin-induced autolysis, genetic screen, and morphology phenotypes (Fig. S5a).
Next, changes to cell-wall-associated protein profiles were examined using high-pH extraction [23] of the S. pneumonia cell surface (Fig. S5b). Upon treatment with 0.75 × MIC masarimycin the appearance of several overexpressed proteins was observed. Proteomic analysis of SDS-PAGE gel bands identified several cell surface and moonlighting proteins that are present only in the masarimycin treated sample (Table 1, Dataset S1). Of note is the upregulation of elongation factor Tu, a known moonlighting protein that has been implicated in regulating cell shape by modulating the formation of MreB filaments in B. subtilis and E. coli [45]. The overexpression of proteins involved in complex carbohydrate catabolism (BgaA, MalX) have been previously shown to be regulated by the two-component system CiaR/H, which is also associated with sensing cell wall stress in S. pneumoniae [46]. These changes to proteins on the cell surface suggest that masarimycin is interfering with cell wall remodelling.
Table 1.
Overexpressed proteins observed in SDS-PAGE gel (Fig. S5b) of S. pneumoniae R6 cell-surface protein extracts when exposed to 0.75 x MIC masarimycin
|
Gel band |
Protein ID |
Biological function |
|---|---|---|
|
1 |
|
|
|
2 |
Spr0440, endo-β-GlcNAcase |
Surface protein, role in commencement of neuroinvasion [67] |
|
3 |
|
|
|
5 |
MalX maltooligosaccharide transporter |
Complex carbohydrate catabolism regulated by CiaR/H system which is involved in sensing cell wall stress [46, 71]. |
|
6 |
GapA glyceraldehyde 3-phosphate dehydrogenase |
Moonlighting protein [70] |
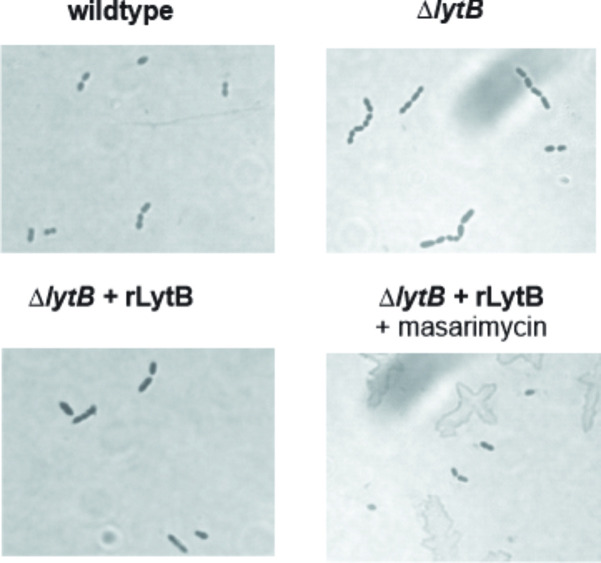
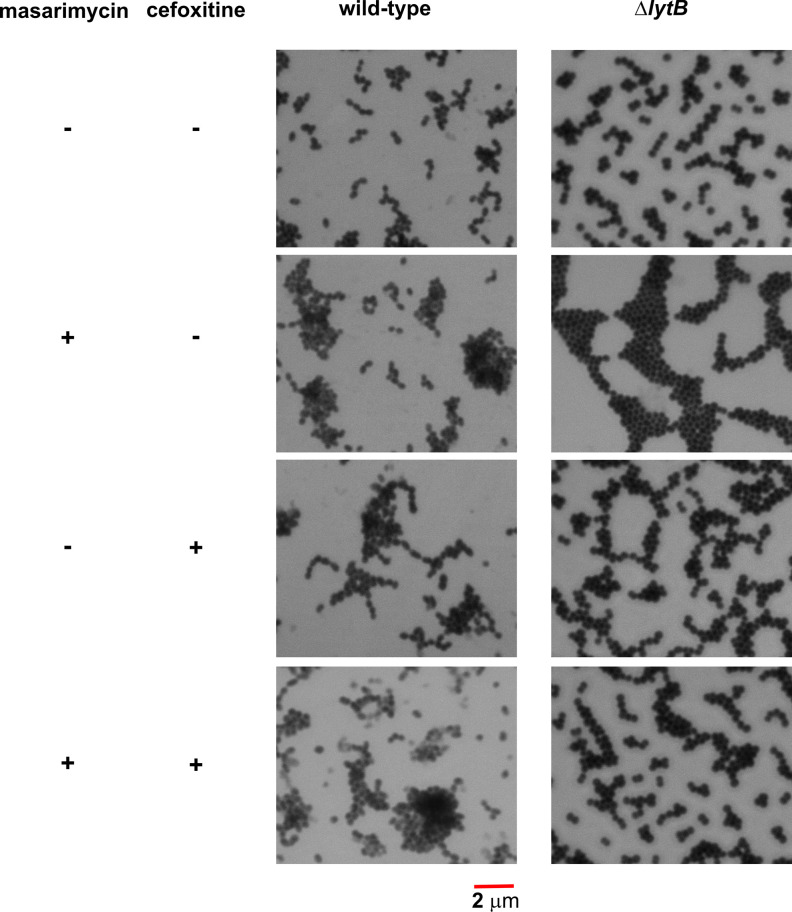
To further investigate the higher MIC observed with the ΔlytB mutant, inhibition of LytB activity was investigated in an established chain dispersing assay using the TIGR4 ΔlytB strain in the presence of exogenously added LytB [16]. When the ΔlytB mutant was treated with exogenous LytB, dispersion of the chains was observed (Fig. 3). When a five-fold MIC (40 µM) concentration of masarimycin was added, LytB catalysed chain dispersion was not inhibited. In vitro analysis with Remazol Brilliant Blue labelled PG [16, 47] and purified LytB confirmed these results. This lack of inhibition of the biochemical activity of purified LytB further suggests that the higher MIC of masarimycin observed for the ΔlytB mutant may be due to changes in cell wall structure, stability, or metabolism. These results further highlight the difficulty in target identification of small molecule inhibitors. The reduced MIC values observed in the genetic screen against the ∆lytB strain is is likely due to a more complex interplay between the deletion of LytB and the actual target(s) of masarimycin. Consistent with this hypothesis, treatment of ∆lytB with 0.7 × MIC masarimycin (Fig. 4) resulted in the conversion of the ΔlytB chaining phenotype to the clumping phenotype observed in Fig. S4. Co-administration of sub-MIC concentrations of cefoxitin, a DacA (PBP3) selective β-lactam, with masarimycin resulted in the reduction of the clumping phenotype in the ΔlytB mutant but not wild-type cells. This further suggests that masarimycin’s mode-of-action is impacted by alterations to the cell wall caused by either genetic deletion or chemical inhibition of autolysins. LytB is implicated in PG remodelling during cell division while DacA is associated with division rather than remodelling [15]. These observations suggest that masarimycin is influencing cell wall biosynthesis, turnover, stability, and/or regulation in these mutant backgrounds. Given their roles in cell wall remodelling during division (LytB) or directly involved in division or division complex (DacA, Pmp23) and the overexpression of elongation factor Tu suggests that masarimycin is either directly or indirectly impacting the cell wall during division.
Fig. 3.
Chain dispersing assay with S. pneumoniae TIGR4 ΔlytB strain and purified recombinant LytB (rLytB, 2 µM). In the presence of 40 µM masarimycin dispersion of the ΔlytB chain phenotype is not inhibited. Images were taken at 1000 x magnification.
Fig. 4.
Morphological analysis of S. pneumoniae ΔlytB mutant- [15] in the presence of sub-MIC masarimycin, the β-lactam cefoxitin (DacA/PBP3 selective) or in combination. Cells were fixed in 1 % formaldehyde, stained with methylene blue, and visualized using bright field microscopy under oil immersion.
Taken collectively, the reduction in autolysis, phenotypic changes and overexpression of surface associated proteins observed with masarimycin, along with the reduced sensitivity against multiple mutant strains, suggest that these results could be the result of meta-phenotypes - a phenotype that results from the alteration of more than one pathway [48]. For instance, the clumping meta-phenotype observed in S. pneumoniae in the presence of masarimycin could be generated via a direct mechanism (e.g. inhibition of an enzyme associated with cell wall metabolism) or an indirect one (e.g. alterations in autolysin expression levels, changes in metabolic flux through cell wall associated pathways).
To further interrogate the potential mode-of-action, masarimycin was screened in checkerboard assays with antibiotics with well-defined mechanisms (Table 2). Using pre-established guidelines [26] for interpreting FICIndex, two antibiotics, levofloxacin (DNA gyrase) and fosfomycin (MurA, first committed step of PG biosynthesis) demonstrated mild antagonism (FICIndex 4.5). Antagonistic relationships can be used to map genetic networks and reveal novel connections between pathways [25]. Antagonism with fosfomycin suggests a functional connection to the target of masarimycin. To further probe the antagonism with fosfomycin, we looked to see if masarimycin was indirectly impacting fosfomycin’s target MurA. The subsequent step, catalysed by MurB reduces the product of MurA, UDP-GlcNAc-enolpyruvate, to UDP-MurNAc utilizing NADPH [49]. Given the impact that changes to the cell wall have on masarimycin activity, we wondered if the observed antagonism with fosfomycin might be due to altered redox potential in the cell, a consequence of a destabilized cell envelope. This could impact PG metabolite flux through MurA/B by reducing the levels of NADPH in the cell. Alterations in redox potential and oxidative stress can negatively influence fosfomycin sensitivity [50]. It has been previously demonstrated in S. aureus that alterations in metabolic flux of precursors and cofactors can contribute to fosfomycin resistance [51]. Changes in NADP/NADPH levels were measured in a colorimetric assay in the presence/absence of masarimycin. The ratio of NADP/NADPH did not change when up to 3 × MIC masarimycin was present (Fig. S6) suggesting the fosfomycin antagonism is not due to an altered redox potential impacting flux through MurB.
Table 2.
Synergy and antagonism screen with masarimycin
|
Antibiotic |
MICapp antibiotic* (µM) |
FIC antibiotic |
MICapp masarimycin† (µM) |
FIC masarimycin |
FICindex |
|---|---|---|---|---|---|
|
Ampicillin |
0.010 |
1 |
2 |
0.25 |
1.25 |
|
Bacitracin |
1.66 |
0.5 |
15.6 |
2 |
2.5 |
|
Cefoxitin |
0.69 |
0.4 |
4 |
0.5 |
0.9 |
|
Cefuroxime |
0.012 |
1.59 |
2 |
0.25 |
1.75 |
|
Fosfomycin |
85.87 |
0.5 |
32 |
4 |
4.5 |
|
Kanamycin |
43 |
0.5 |
1.66 |
0.21 |
0.71 |
|
Levofloxacin |
1.08 |
0.5 |
32 |
4 |
4.5 |
|
Optochin |
7.96 |
0.5 |
8 |
1 |
1.5 |
|
Rifampicin |
0.0023 |
0.124 |
3.33 |
0.416 |
0.54 |
|
Tetracycline |
0.048 |
0.75 |
2 |
0.25 |
1 |
|
Vancomycin |
0.003 |
1 |
4 |
0.5 |
1.5 |
*MIC for antibiotics alone. ampicillin: 0.010 µM; bacitracin: 3.325 µM; cefoxitin: 1.72 µM; cefuroxime: 0.008 µM; fosfomycin: 171.5 µM; kanamycin: 86 µM; levofloxacin: 2.16 µM; optochin: 15.85 µM; rifampicin: 0.018 µM; tetracycline: 0.065 µM; vancomycin: 0.003 µM.
†MIC masarimycin 8 µM.
Antagonism with levofloxacin provides a counterpoint to the fosfomycin results. Levofloxacin introduces double-stranded DNA breaks upon inhibition of DNA gyrase, and induces the SOS response system [52]. To further evaluate this antagonistic relationship, masarimycin was evaluated for its ability to interact with DNA. Masarimycin was investigated for its ability to inhibit nuclease activity and intercalate DNA in established assays (Fig. S7) [20, 21]. Masarimycin showed no inhibition of nuclease activity or ability to intercalate DNA, suggesting the antagonism observed is likely not due to direct interaction with DNA or nucleases. Further the additive interaction with rifampicin (RNA synthesis) and the protein synthesis inhibitors tetracycline and kanamycin in Table 2 suggests a pure summation effect of the antibiotics with masarimycin. This suggests the antagonism with levofloxacin is not a product of downstream inhibition of transcription and translation. It should be noted that there is a connection between quinolone bactericidal activity and the expression of stress-induced proteins [53]. Cell wall targeting antibiotics like β-lactams can activate the SOS response system [54]. The antagonism observed with levofloxacin can be explained by the induction of the SOS system by both compounds [55].
Conclusion
Taking this data holistically, we posit that masarimycin activity is impacted by alterations to the cell wall caused by deletion of certain autolysins (LytB, DacA, Pmp23) in S. pneumoniae . These three autolysins are associated with remodelling PG during division (LytB) [31], or associated with division (DacA) [39] or with the Z-ring (Pmp23) [36] components of the cell division complex. Other PG hydrolases [56–58] might also be affected by masarimycin treatment, giving rise to the complex phenotypic results. The data presented here demonstrates that the morphological phenotypes of genetic knockouts of cell wall acting enzymes can be distinct from chemical inactivation and may more closely resemble the phenotype of catalytically inactive mutants. This distinction has previously been observed in Mycobacterium tuberculosis shikimate biosynthesis [59]. These data illustrate that the use of genetic and phenotypic screens for target identification may not always lead directly to the molecular target. The complexity involved in deciphering the underlying mechanisms associated with these meta-phenotypes obfuscates target identification. Despite these challenges, the elucidation of the molecular target of masarimycin is on-going. Based on the data provided here, masarimycin may provide a unique molecular scaffold for the development of anti- S. pneumoniae therapeutics and can play a role in furthering our understanding of PG metabolism.
The bacterial cell wall and PG biosynthesis has provided a wealth of clinically relevant antibiotic targets. While our understanding of PG biosynthetic and cross-linking steps is well established, our knowledge of the role autolytic enzymes play in the growth and maintenance of the cell wall has remained more elusive. Traditional genetic approaches to studying the biological role of autolysins are complicated by functional redundancy of these enzymes, where other autolysins can compensate for a loss in activity. The results presented here illustrate the complexity of PG metabolism and the difficulty in identifying the molecular target of small molecule inhibitors. Recent reports [4, 6, 7] have begun to elucidate the role of autolysins in relieving stress in the cell wall to allow for incorporation of new material into the stress bearing layer. The results with the diamide masarimycin demonstrate that sensitivity is impacted by alterations in the cell wall caused by the deletion of specific autolysins associated with cell division and separation. Upregulation of elongation factor Tu, a moonlighting protein known to regulate cell shape, indicates that masarimycin is impacting cell wall metabolism. Collectively our data further demonstrate that morphological, genetic, and whole cell assays (autolysis) reveal meta-phenotypes that result from the complex interaction of one or more cellular processes that appear connected to cell wall metabolism. The genetic deletion of one or more autolysins disrupts the equilibrium stoichiometry of the cell wall machinery that likely results in changes to expression levels and activity to both autolytic and biosynthetic enzymes. With interest in the development of chemical biology approaches to study PG metabolism [60–64] receiving renewed attention, the diamide inhibitor masarimycin provides a potential small molecule complement to both traditional genetic and current chemical biology approaches to studying cell wall metabolism.
Supplementary Data
Funding information
Research was partially supported by the National Science Foundation (CHE2009522) and by the Rhode Island Institutional Development Award (IDeA) Network of Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103430. NMR analysis of masarimycin was supported by the National Science Foundation major research instrumentation program award under grant number 1919644. Proteomic analysis was performed at the National Resource for Quantitative Proteomics supported through a grant from the National Institute of General Medical Sciences (R24GM137786).
Acknowledgements
We thank J. Belval for technical assistance.
Conflicts of interest
The authors A.B. and C.W.R hold patents on specific applications of masarimycin.
Footnotes
Abbreviations: BHI, brain heart infusion; DMSO, dimethylsulfoxide; FIC, fractional inhibitory concentration; GH73, glycosyl hydrolase family 73; GlcNAc, N-acetylglucosamine; GlcNAcase, N-acetylglucosaminidase; LB, Luria Bertani; MH, Mueller Hinton; MIC, minimum inhibitory concentration; MurNAc, N-acetylmuramic acid; OD, optical density; PG, peptidoglycan; PVDF, polyvinylidene fluoride; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Seven supplementary figures and supplementary data sheet are available with the online version of this article.
References
- 1.Munita JM, Bayer AS, Arias CA. Evolving resistance among Gram-positive pathogens. Clin Infect Dis. 2015;61 Suppl 2:S48–57. doi: 10.1093/cid/civ523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drug-resistant Streptococcus pneumoniae 2022. [ May 3; 2022 ]. https://www.cdc.gov/drugresistance/pdf/threats-report/strep-pneumoniae-508.pdf accessed.
- 3.Koch AL, Doyle RJ. Inside-to-outside growth and turnover of the wall of gram-positive rods. J Theor Biol. 1985;117:137–157. doi: 10.1016/s0022-5193(85)80169-7. [DOI] [PubMed] [Google Scholar]
- 4.Beeby M, Gumbart JC, Roux B, Jensen GJ. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol. 2013;88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackman SA, Smith TJ, Foster SJ. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology (Reading) 1998;144 (Pt 1):73–82. doi: 10.1099/00221287-144-1-73. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler R, Turner RD, Bailey RG, Salamaga B, Mesnage S, et al. Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. mBio. 2015;6:e00660. doi: 10.1128/mBio.00660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misra G, Rojas ER, Gopinathan A, Huang KC. Mechanical consequences of cell-wall turnover in the elongation of a Gram-positive bacterium. Biophys J. 2013;104:2342–2352. doi: 10.1016/j.bpj.2013.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taguchi A, Kahne D, Walker S. Chemical tools to characterize peptidoglycan synthases. Curr Opin Chem Biol. 2019;53:44–50. doi: 10.1016/j.cbpa.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsh MA, Schaefer K, Taguchi A, Kahne D, Walker S. Direction of chain growth and substrate preferences of shape, elongation, division, and sporulation-family peptidoglycan glycosyltransferases. J Am Chem Soc. 2019;141:12994–12997. doi: 10.1021/jacs.9b06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubino FA, Mollo A, Kumar S, Butler EK, Ruiz N, et al. Detection of transport intermediates in the peptidoglycan flippase MurJ identifies residues essential for conformational cycling. J Am Chem Soc. 2020;142:5482–5486. doi: 10.1021/jacs.9b12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjodt M, Brock K, Dobihal G, Rohs PDA, Green AG, et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature. 2018;556:118–121. doi: 10.1038/nature25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai GC, Cho H, Bernhardt TG. The mecillinam resistome reveals a role for peptidoglycan endopeptidases in stimulating cell wall synthesis in Escherichia coli . PLoS Genet. 2017;13:e1006934. doi: 10.1371/journal.pgen.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautam A, Vyas R, Tewari R. Peptidoglycan biosynthesis machinery: a rich source of drug targets. Crit Rev Biotechnol. 2011;31:295–336. doi: 10.3109/07388551.2010.525498. [DOI] [PubMed] [Google Scholar]
- 14.Nayyab S, O’Connor M, Brewster J, Gravier J, Jamieson M, et al. Diamide inhibitors of the Bacillus subtilis N-acetylglucosaminidase LytG that exhibit antibacterial activity. ACS Infect Dis. 2017;3:421–427. doi: 10.1021/acsinfecdis.7b00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boersma MJ, Kuru E, Rittichier JT, VanNieuwenhze MS, Brun YV, et al. Minimal peptidoglycan (PG) turnover in wild-type and PG hydrolase and cell division mutants of Streptococcus pneumoniae D39 growing planktonically and in host-relevant biofilms. J Bacteriol. 2015;197:3472–3485. doi: 10.1128/JB.00541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai X-H, Chen H-J, Jiang Y-L, Wen Z, Huang Y, et al. Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis. J Biol Chem. 2014;289:23403–23416. doi: 10.1074/jbc.M114.579714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palomino J-C, Martin A, Camacho M, Guerra H, Swings J, et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czarny TL, Perri AL, French S, Brown ED. Discovery of novel cell wall-active compounds using P ywaC , a sensitive reporter of cell wall stress, in the model gram-positive bacterium Bacillus subtilis . Antimicrob Agents Chemother. 2014;58:3261–3269. doi: 10.1128/AAC.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornett JB, Shockman GD. Cellular lysis of Streptococcus faecalis induced with triton X-100. J Bacteriol. 1978;135:153–160. doi: 10.1128/jb.135.1.153-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furlan RLA, Garrido LM, Brumatti G, Amarante-Mendes GP, Martins RA, et al. A rapid and sensitive method for the screening of DNA intercalating antibiotics. Biotechnol Lett. 2002;24:1807–1813. doi: 10.1023/A:1020633909762. [DOI] [Google Scholar]
- 21.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor . Mol Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman T, Ibrahim S. Choline kinase, a novel drug target for the inhibition of Streptococcus pneumoniae . Antibiotics (Basel) 2017;6:E20. doi: 10.3390/antibiotics6040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morsczeck C, Prokhorova T, Sigh J, Pfeiffer M, Bille-Nielsen M, et al. Streptococcus pneumoniae: proteomics of surface proteins for vaccine development. Clin Microbiol Infect. 2008;14:74–81. doi: 10.1111/j.1469-0691.2007.01878.x. [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 25.Farha MA, Czarny TL, Myers CL, Worrall LJ, French S, et al. Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase. Proc Natl Acad Sci U S A. 2015;112:11048–11053. doi: 10.1073/pnas.1511751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 27.van Opijnen T, Dedrick S, Bento J. Strain dependent genetic networks for antibiotic-sensitivity in a bacterial pathogen with a large pan-genome. PLoS Pathog. 2016;12:e1005869. doi: 10.1371/journal.ppat.1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, et al. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:10. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, et al. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol. 2007;189:8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aaberge IS, Eng J, Lermark G, Løvik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. doi: 10.1016/s0882-4010(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 31.García P, González MP, García E, López R, García JL. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol Microbiol. 1999;31:1275–1281. doi: 10.1046/j.1365-2958.1999.01238.x. [DOI] [PubMed] [Google Scholar]
- 32.Rico-Lastres P, Díez-Martínez R, Iglesias-Bexiga M, Bustamante N, Aldridge C, et al. Substrate recognition and catalysis by LytB, a pneumococcal peptidoglycan hydrolase involved in virulence. Sci Rep. 2015;5:16198. doi: 10.1038/srep16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipski A, Hervé M, Lombard V, Nurizzo D, Mengin-Lecreulx D, et al. Structural and biochemical characterization of the β-N-acetylglucosaminidase from Thermotoga maritima: toward rationalization of mechanistic knowledge in the GH73 family. Glycobiology. 2015;25:319–330. doi: 10.1093/glycob/cwu113. [DOI] [PubMed] [Google Scholar]
- 34.Horsburgh GJ, Atrih A, Williamson MP, Foster SJ. LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry. 2003;42:257–264. doi: 10.1021/bi020498c. [DOI] [PubMed] [Google Scholar]
- 35.Klainer AS, Perkins RL. Surface manifestations of antibiotic-induced alterations in protein synthesis in bacterial cells. Antimicrob Agents Chemother. 1972;1:164–170. doi: 10.1128/AAC.1.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacq M, Arthaud C, Manuse S, Mercy C, Bellard L, et al. The cell wall hydrolase Pmp23 is important for assembly and stability of the division ring in Streptococcus pneumoniae . Sci Rep. 2018;8:7591. doi: 10.1038/s41598-018-25882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster C, Dobrinski B, Hakenbeck R. Unusual septum formation in Streptococcus pneumoniae mutants with an alteration in the D,D-carboxypeptidase penicillin-binding protein 3. J Bacteriol. 1990;172:6499–6505. doi: 10.1128/jb.172.11.6499-6505.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barendt SM, Sham L-T, Winkler ME. Characterization of mutants deficient in the L,D-carboxypeptidase (DacB) and WalRK (VicRK) regulon, involved in peptidoglycan maturation of Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol. 2011;193:2290–2300. doi: 10.1128/JB.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morlot C, Noirclerc-Savoye M, Zapun A, Dideberg O, Vernet T. The D,D-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae . Mol Microbiol. 2004;51:1641–1648. doi: 10.1046/j.1365-2958.2003.03953.x. [DOI] [PubMed] [Google Scholar]
- 40.Severin A, Schuster C, Hakenbeck R, Tomasz A. Altered murein composition in a DD-carboxypeptidase mutant of Streptococcus pneumoniae . J Bacteriol. 1992;174:5152–5155. doi: 10.1128/jb.174.15.5152-5155.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Höltje JV, Tomasz A. Lipoteichoic acid: a specific inhibitor of autolysin activity in Pneumococcus . Proc Natl Acad Sci U S A. 1975;72:1690–1694. doi: 10.1073/pnas.72.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, et al. An autolysin ring associated with cell separation of Staphylococcus aureus . J Bacteriol. 1996;178:1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palumbo E, Deghorain M, Cocconcelli PS, Kleerebezem M, Geyer A, et al. D-alanyl ester depletion of teichoic acids in Lactobacillus plantarum results in a major modification of lipoteichoic acid composition and cell wall perforations at the septum mediated by the Acm2 autolysin. J Bacteriol. 2006;188:3709–3715. doi: 10.1128/JB.188.10.3709-3715.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakenbeck R, Madhour A, Denapaite D, Brückner R. Versatility of choline metabolism and choline-binding proteins in Streptococcus pneumoniae and commensal streptococci. FEMS Microbiol Rev. 2009;33:572–586. doi: 10.1111/j.1574-6976.2009.00172.x. [DOI] [PubMed] [Google Scholar]
- 45.Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, et al. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc Natl Acad Sci U S A. 2010;107:3163–3168. doi: 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008;16:318–325. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou R, Chen S, Recsei P. A dye release assay for determination of lysostaphin activity. Anal Biochem. 1988;171:141–144. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]
- 48.Martín-Galiano AJ, Yuste J, Cercenado MI, de la Campa AG. Inspecting the potential physiological and biomedical value of 44 conserved uncharacterised proteins of Streptococcus pneumoniae . BMC Genomics. 2014;15:652. doi: 10.1186/1471-2164-15-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, et al. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 50.Posada AC, Kolar SL, Dusi RG, Francois P, Roberts AA, et al. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus . Infect Immun. 2014;82:316–332. doi: 10.1128/IAI.01074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liebeke M, Meyer H, Donat S, Ohlsen K, Lalk M. A metabolomic view of Staphylococcus aureus and its ser/thr kinase and phosphatase deletion mutants: involvement in cell wall biosynthesis. Chem Biol. 2010;17:820–830. doi: 10.1016/j.chembiol.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 52.Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 53.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, et al. SOS response induction by ß-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 55.Baharoglu Z, Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev. 2014;38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 56.Ng W-L, Tsui H-C, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae . J Bacteriol. 2005;187:7444–7459. doi: 10.1128/JB.187.21.7444-7459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus . J Bacteriol. 2004;186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartual SG, Straume D, Stamsås GA, Muñoz IG, Alfonso C, et al. Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae . Nat Commun. 2014;5:3842. doi: 10.1038/ncomms4842. [DOI] [PubMed] [Google Scholar]
- 59.Ramachandran V, Singh R, Yang X, Tunduguru R, Mohapatra S, et al. Genetic and chemical knockdown: a complementary strategy for evaluating an anti-infective target. Adv Appl Bioinform Chem. 2013;6:1–13. doi: 10.2147/AABC.S39198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeMeester KE, Liang H, Zhou J, Wodzanowski KA, Prather BL, et al. Metabolic incorporation of N-acetyl muramic acid probes into bacterial peptidoglycan. Curr Protoc Chem Biol. 2019;11:e74. doi: 10.1002/cpch.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang H, DeMeester KE, Hou C-W, Parent MA, Caplan JL, et al. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat Commun. 2017;8:15015. doi: 10.1038/ncomms15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Lazor KM, DeMeester KE, Liang H, Heiss TK, et al. Postsynthetic modification of bacterial peptidoglycan using bioorthogonal N-acetylcysteamine analogs and peptidoglycan O-acetyltransferase B. J Am Chem Soc. 2017;139:13596–13599. doi: 10.1021/jacs.7b06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lebar MD, May JM, Meeske AJ, Leiman SA, Lupoli TJ, et al. Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J Am Chem Soc. 2014;136:10874–10877. doi: 10.1021/ja505668f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dik DA, Zhang N, Chen JS, Webb B, Schultz PG. Semisynthesis of a bacterium with non-canonical cell-wall cross-links. J Am Chem Soc. 2020;142:10910–10913. doi: 10.1021/jacs.0c02956. [DOI] [PubMed] [Google Scholar]
- 65.Singh AK, Pluvinage B, Higgins MA, Dalia AB, Woodiga SA, et al. Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA. PLoS Pathog. 2014;10:e1004364. doi: 10.1371/journal.ppat.1004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, Rahkola J, Redzic JS, Chi Y-C, Tran N, et al. Mechanism and inhibition of Streptococcus pneumoniae IgA1 protease. Nat Commun. 2020;11:6063. doi: 10.1038/s41467-020-19887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiménez-Munguía I, Pulzova L, Kanova E, Tomeckova Z, Majerova P, et al. Proteomic and bioinformatic pipeline to screen the ligands of S. pneumoniae interacting with human brain microvascular endothelial cells. Sci Rep. 2018;8:5231. doi: 10.1038/s41598-018-23485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harvey KL, Jarocki VM, Charles IG, Djordjevic SP. The diverse functional roles of elongation factor Tu (EF-Tu) in microbial pathogenesis. Front Microbiol. 2019;10:2351. doi: 10.3389/fmicb.2019.02351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ling E, Feldman G, Portnoi M, Dagan R, Overweg K, et al. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin Exp Immunol. 2004;138:290–298. doi: 10.1111/j.1365-2249.2004.02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang G, Xia Y, Cui J, Gu Z, Song Y, et al. The roles of moonlighting proteins in bacteria. Curr Issues Mol Biol. 2014;16:15–22. [PubMed] [Google Scholar]
- 71.Mascher T, Heintz M, Zähner D, Merai M, Hakenbeck R. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in beta-lactam resistance. J Bacteriol. 2006;188:1959–1968. doi: 10.1128/JB.188.5.1959-1968.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.