Abstract
Streptococcus sinensis is a recently identified member of the Mitis group of streptococci. This species has been associated with infective endocarditis; however its mechanisms of pathogenesis and virulence are not fully understood. This study aimed to investigate the influence of the competence-stimulating peptide (CSP) and the competence regulon quorum-sensing circuitry (ComABCDE) on subsequent gene transcription and expression, as well as resultant phenotypes. In this study we confirmed the native CSP identity, ascertained when endogenous CSP was produced and completed a transcriptome-wide analysis of all genes following CSP exposure. RNA sequencing analysis revealed the upregulation of genes known to be associated with competence, biofilm formation and virulence. As such, a variety of phenotypic assays were utilized to assess the correlation between increased mRNA expression and potential phenotype response, ultimately gaining insight into the effects of CSP on both gene expression and developed phenotypes. The results indicated that the addition of exogenous CSP aided in competence development and successful transformation, yielding an average transformation efficiency comparable to that of other Mitis group streptococci. Additional studies are needed to further delineate the effects of CSP exposure on biofilm formation and virulence. Overall, this study provides novel information regarding S. sinensis and provides a substantial foundation on which this species and its role in disease pathogenesis can be further investigated.
Keywords: Streptococcus sinensis, competence-stimulating peptide, quorum sensing, competence
Introduction
Bacterial competence is a physiological state bacteria enter to uptake and integrate exogenous DNA into their genomes. This process plays a vital role in both horizontal gene transfer (HGT) and genetic recombination events [1–3]. Bacterial competence also contributes to capsule polysaccharide variation and acquisition of antibiotic resistance genes to aid in bacterial survival under stressed conditions [1]. Bacterial competence has contributed to vast evolutionary development and speciation, thereby aiding the continued survival and proliferation of a multitude of bacterial species, including streptococci.
Competence has been thoroughly investigated in Streptococcus pneumoniae , 1 of 20 different species in the Mitis group of streptococci [1, 4–6]. The Mitis group of streptococci mainly comprises commensal inhabitants of the human naso-oropharyngeal tract, with some species being opportunistic pathogens [7]. Competence induction in S. pneumoniae was originally described as a quorum-sensing (QS) mechanism by which endogenous CSP production and secretion regulates the bacteria’s ability to uptake exogenous DNA [5, 8]. Further investigation of QS and the competence regulon, ComABCDE, suggests that QS also takes part in the regulation of many downstream genes and resultant proliferative phenotypes, such as biofilm formation and the production of virulence factors [9–12].
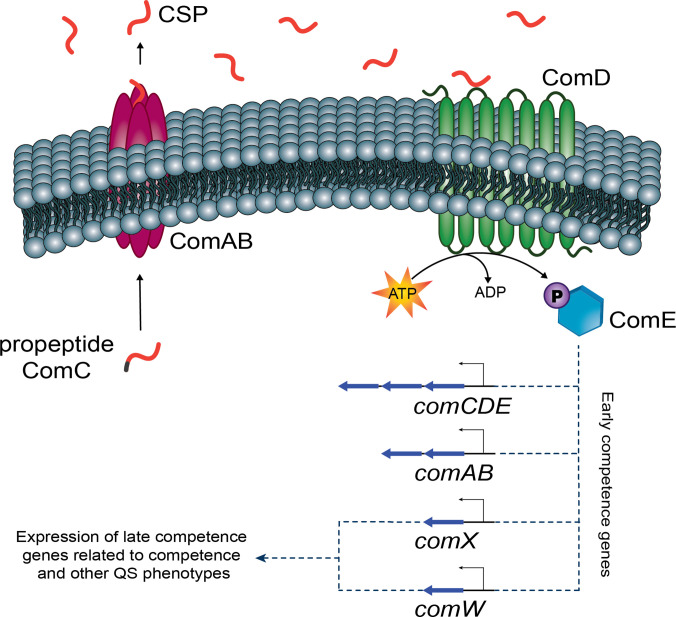
The ComABCDE QS pathway comprises the comABCDE genes, where comC encodes a precursor peptide that is cleaved and exported outside the cell by an ABC transporter, ComAB (Fig. 1). The excreted and processed peptide, now termed the competence-stimulating peptide (CSP), binds ComD, a transmembrane histidine kinase receptor, when its concentration reaches a certain threshold in the local environment [13]. The CSP : : ComD binding results in phosphorylation of a response regulator, ComE. The phosphorylation of ComE has been postulated to promote self-dimerization and the subsequent upregulation and expression of other genes, including the comABCDE regulon and an alternative sigma factor, comX [14–16]. The activation of ComX leads to a transient cascade event where multiple genes are upregulated or downregulated, giving rise to various QS phenotypes, such as competence [15, 17, 18]. Due to the energetic cost and stress associated with the competence state, this phenotype is only maintained for ~30–40 min [1, 19, 20].
Fig. 1.
Depiction of the ComABCDE QS circuit regulating competence in Mitis group streptococci.
Global transcriptome analysis during competence has been investigated in four Mitis group species: S. pneumoniae , Streptococcus gordonii , Streptococcus mitis and Streptococcus sanguinis [21–24]. Competence induction by the ComABCDE pathway has two stages: activation of early competence genes, which are controlled by phosphorylated ComE, and activation of late competence genes, which are controlled by ComX [20, 21]. Early competence genes include comABCDE, comW and comX, along with a few other genes that are species specific [25]. The function of ComW was initially unknown, but later research has clarified its role as an early competence gene and additional QS regulator [18]. Researchers confirmed that ComW aids in the stabilization of ComX and is required for the induction of late competence genes in S. pneumoniae [18]. Late competence genes, often under the direct regulation of ComX, include many genes that are involved with DNA uptake, recombination and DNA repair.
In the QS model, all of which is dependent on the initial production of CSP, early competence genes are upregulated as soon as ComE is phosphorylated, and maximal expression occurs within 10 min. Late competence genes are upregulated approximately 5 min following ComE phosphorylation, and maximal expression occurs within 15 min [20, 21].
The competence regulon in S. pneumoniae has served as a model for investigating competence in the Mitis group of streptococci. While competence has largely been attributed to QS, alternative theories also suggest that pneumococcal competence, and potentially competence in other closely related streptococcal species, may instead rely on a cell–cell contact mechanism [26]. In this model, pneumococcal competence is reliant upon paracrine CSP transmission. Additionally, the rate of competence spread is dictated by the contact between activated and latent cells, rather than the free diffusion of CSP in the extracellular environment, as is demonstrated in QS [26].
In S. pneumoniae , competence shutoff is not fully understood; however, it is well established that a separate late competence gene termed DNA-processing protein A (DprA) aids in this process [19, 27, 28]. DprA, under the control of ComX, binds phosphorylated ComE and prevents it from upregulating early competence genes [28]. Researchers also found that unabridged expression of dprA is required for complete competence shutoff [29]. An additional hypothesis for competence shutoff proposes that some unknown factor, referred to as ComZ, interacts with ComW to destabilize ComX and downregulate the expression of late competence genes [30]. Overall, the two-phase competence induction is conserved in the Mitis group of streptococci, although knowledge of other conserved key genes important for both competence induction and shutoff is lacking.
Presently, CSP-regulated competence has been shown to contribute to enhanced pathogenicity and virulence, as competent bacteria maintain the ability to uptake genes for antibiotic resistance [3, 4, 31, 32]. Such ability is well conserved among streptococcal species, although there are fine nuances between species that lend to unique phenotypes and group behaviours that have yet to be defined. Investigating streptococcal competence, as well as the effects of the competence regulon on the whole genome and proliferative phenotypes, is imperative to gain a more thorough understanding of these bacteria. Moreover, such knowledge will help establish a strong basis by which bacterial virulence and pathogenicity can be further investigated and potentially modulated. Because the QS competence regulon is known to play a significant role in bacterial competence, we set out to investigate the competence regulon and its effects in a recently reclassified species, Streptococcus sinensis .
S. sinensis , a bacterium belonging to the Mitis group of streptococci, was further examined and reclassified in 2002, where it was isolated from the blood of a Chinese woman admitted to the hospital for infective endocarditis [33]. S. sinensis naturally resides within the human oral cavity and it is currently unknown how this bacterium enters the bloodstream to cause bacteraemia and infective endocarditis [34]. Phylogenomic analysis of S. sinensis and other Mitis group streptococci revealed that S. sinensis , Streptococcus cristatus and Streptococcus oligofermentans are closely related [7, 35]. Presently, research related to S. sinensis has been limited to proper identification of this species. As such, investigation into S. sinensis Forsyth1A can serve as a model for this species. Moreover, understanding of S. sinensis Forsyth1A has the potential to aid in the investigation of closely related species and elucidate their mechanisms of pathogenesis and virulence. We therefore set out to evaluate the ComABCDE QS circuit in S. sinensis Fortsyth1A and determine its regulatory role in pathogenesis.
Methods
All chemical reagents and solvents were purchased from Sigma-Aldrich or Chem-Impex and used without further purification. Water (ddH2O) was purified using a Millipore Analyzer Feed System. All synthetic peptides were synthesized using preloaded Wang-Lysine resin (Nα-Fmoc-Nε-Boc-l-lysine 4-alkoxybenzyl alcohol resin, 0.343 meq g−1). 9-fluorenylmethoxycarbonyl (Fmoc) protected l-α-amino acids were purchased from Advanced ChemTech.
Bacterial strains
S. sinensis Forsyth1A was provided as a lyophilized powder from the Forsyth Institute by Dr Margaret Duncan [36].
Growth conditions
The lyophilized powder was resuspended in sterile Todd–Hewitt media supplemented with 0.5 % yeast extract (THY) upon arrival and incubated for 12–24 h in a CO2 incubator (37 °C with 5 % CO2). The suspension was streaked onto a THY agar plate and incubated for 12–24 h in a CO2 incubator (37 °C with 5 % CO2). Fresh colonies were picked, Gram stained and checked for purity prior to making freezer stocks using THY broth and 20 % glycerol solution. The new freezer stocks were used for all other experiments.
Species level identification
S. sinensis Forsyth1A identity was confirmed to the species level by polymerase chain reaction (PCR) amplifying a 740 base pair (bp) region unique to streptococci followed by Sanger sequencing using a method described previously [37, 38].
Isolation and characterization of competence-stimulating peptide (CSP)
Approximately 1 ml of an overnight culture of S. sinensis Forsyth1A was inoculated into 30 ml of sterile THY in a sterile 50 ml conical centrifuge tube. The culture was grown for 36 h in a CO2 incubator (37 °C with 5 % CO2). The culture was centrifuged at 3 000 g for 10 min and the supernatant was filter-sterilized using a 0.22 µM PES filter in a sterile 50 ml conical centrifuge tube. Ammonium sulfate was added to a final concentration of 40 % w/v followed by incubation at 4 °C for 2 h. The solution was centrifuged at 3 000 g for 20 min and the supernatant was discarded. The crude peptide/protein pellet was dissolved in 10 ml of DI water and lyophilized. The lyophilized powder was resuspended in 10 ml of water : ACN (90 : 10) and purified using preparative RP-HPLC. Fractions were analysed for masses corresponding to the predicted CSP sequence, DFRKVRLPRLFGK (1631 Da). Fractions that showed a mass matching the predicted CSP were lyophilized and checked for purity using analytical RP-HPLC. The identity of the fractions containing the purified natural CSP was confirmed using HRMS and sent out for peptide mapping at the Nevada Proteomics Center (Fig. S2).
Solid-phase peptide synthesis
Peptide synthesis and purification was conducted using previously established methods [39, 40]. See the Supplementary Material for full experimental details.
Development of S. sinensis reporter strain
The S. sinensis Forsyth1A luciferase-based reporter strain was constructed using previously described molecular cloning methods with some modifications [41]. Briefly, the promoter of the S. sinensis comX was ligated with the starting codon of the firefly luciferase gene in the pFW11-ffluc backbone using the Gibson Assembly Cloning kit (NEB, Ipswich, MA, USA). PCR amplification was performed with Q5 DNA polymerase (NEB, Ipswich, MA, USA) per the manufacturer’s recommendations. All constructs were verified via DNA sequencing and the observation of luminescence following exposure to both endogenous and exogenous CSP.
Temporal determination of endogenous CSP production
The temporal production and secretion of endogenous CSP in S. sinensis was determined using previously described methods with some modifications [42]. See the Supplementary Material for full experimental details.
DNA extraction and whole-genome sequencing
Genomic DNA for both Illumina sequencing and Oxford Nanopore Technology (ONT) Minion sequencing was prepared by making a 1 : 20 dilution of overnight cultures of S. sinensis Forsyth1A in 5 ml of sterile THY broth and incubating the freshly diluted culture statically for 6–8 h in a CO2 incubator (37 °C with 5 % CO2).
For Illumina sequencing, genomic DNA was extracted from pelleted cultures (4 000 g for 10 min) using the E.Z.N.A Bacterial DNA kit (Omega Bio-tek, Inc., Norcross, USA) according to the manufacturer’s instructions. The DNA was submitted to the Nevada Genomics Center (Reno, USA) for multiplexed sequencing library preparation using an Illumina Nextera XT DNA Library Preparation kit and Illumina Sequencing using an Illumina mid output v2 (2×75) sequencing kit and flowcell on an Illumina NextSeq 500 (Illumina, Inc., San Diego, CA, USA). Quality control for the genomic DNA and prepared library was conducted using an Agilent High Sensitivity DNA Chip on an Agilent 2100 BioAnalyzer System (Agilent, Inc., Santa Clara, CA, USA).
For ONT Minion sequencing, genomic DNA was extracted from pelleted cultures (4 000 g for 10 min) using a modified phenol : chloroform extraction protocol designed for obtaining high-molecular weight genomic DNA [43]. The pelleted cells were gently resuspended by pulse vortexing (5 s pulses) in 5 ml of modified buffer TLB pH 8.0 [10 mM Tris–Cl, 25 mM EDTA, 0.5 % (w/v) SDS, 20 µg ml−1 RNase A and 20 µg ml−1 lysozyme] and incubated at 37 °C for 1 h. An equal volume of Tris–EDTA (pH 8.0) saturated phenol (TE phenol) was added to the lysate and mixed by hand inversion (20 inversions min−1) for 10 min. The mixture was centrifuged at 5 000 g for 10 min. The aqueous layer was transferred to a 15 ml centrifuge tube containing 5 ml of TE phenol chloroform (1 : 1) using a cutoff P1000 pipette tip that mimics a wide-bore pipette tip and mixed by hand inversion (20 inversions min−1) for 10 min. The mixture was then poured into a new 15 ml centrifuge tube containing DOW Corning High Vacuum Grease in place of phase lock gel and centrifuged at 5 000 g for 10 min. The top aqueous layer was transferred to a new 15 ml centrifuged tube using a cutoff P1000 pipette tip that mimics a wide-bore pipette tip containing 10 ml of ice-cold 100 % ethanol and mixed by hand inversion (10 inversions) and incubated at −20 °C for 30 min. The solution was centrifuged at 5 000 g for 15 min and gently decanted to give a white glassy pellet. The pellet was gently dislodged with 0.5 ml 70 % ethanol solution and gently transferred using a cutoff P1000 pipette tip that mimics a wide-bore pipette tip into a 1.5 ml Lo-Bind microcentrifuge tube. The sample was centrifuged on a microcentrifuge at 10 000 g for 10 min. The 70 % ethanol solution was gently pipetted off the pellet without disturbing the pellet. The pellet was dried on a heat block set at 40 °C (15 min). The dried pellet was dissolved by adding 50 µl of nuclease-free water and incubating on a heat block set at 40 °C for 10 min. Isolated genomic DNA of S. sinensis Forsyth1A was barcoded for multiplexed sequencing using the Oxford Nanopore 1D ligation sequencing kit (SQK-LSK109) with a native barcoding expansion kit (EXP-NBD103). The ONT protocol library prep was performed according to the manufacturer’s instructions with the following exceptions: the Axygen AxyPrep Mag PCR Clean-Up kit was used instead of Agencourt AMPure XP beads and the QuantiFluor dsDNA System was used with a Biotek Synergy H1 multimode plate reader (BioTek Instruments, Inc., Winooski, VT, USA) to quantify DNA instead of a Qubit. The sequencing ran for 8 h on the Minion using MinKNOW (v3.1.19) for basecalling.
De novo genome assembly
Raw Illumina reads were analysed using FastQC (v0.11.8), adapter removal and quality trimming was performed using Trimmomatic software (v.0.38) [44]. Raw ONT FASTQ format files were demultiplexed using EPI2ME Agent (v2.59.1896509) and those sequences containing the appropriate barcode were collected into one FASTQ output file (specifically, ONT native barcode ‘BC12’). Unicycler software (v0.4.7) was used to create a hybrid assembly using both Illumina and ONT data [45]. Different methods of assembly were compared using bold, conservative and normal modes. Assembly using the bold mode produced the best draft genome with the fewest number of contigs. Assembly quality was assessed using QUAST (v5.0.0) [46] with S. sinensis HKU4 (GCF_000767835.1) as a reference genome. This assembly was submitted to the National Center of Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline and DFAST for annotation (BioProject: PRJNA526114; BioSample: SAMN11268212; SRA: SRX5584543 and SRX5962323) [47, 48].
Culturing and harvesting of S. sinensis Forsyth1A for RNA sequencing experiments
On the evening prior to the day of the experiment, 100 µl of S. sinensis Forsyth1A freezer stock was inoculated into 20 ml of sterile THY in a 50 ml sterile centrifuge tube and incubated statically in a CO2 incubator (37 °C with 5 % CO2). On the morning of the experiment, the overnight culture of S. sinensis Forsyth1A was transferred to 980 ml of sterile THY +1 % sterile horse serum contained in a sterile 1 l media bottle. The culture was incubated statically in a CO2 incubator (37 °C with 5 % CO2) for 2 h. After 2 h incubation (OD600 ~0.05–0.15), 25 ml aliquots were dispensed for each time point into labelled sterile 50 ml centrifuge tubes containing 20 µl of either DMSO or synthetic CSP (800 nM final concentration). The tubes were incubated for 3 and 20 min with shaking at 200 r.p.m. at 37 °C. After the specified time point had passed, 3 ml of cold stop solution (5 % phenol in absolute ethanol) was added to each tube and the tube was inverted several times before being centrifuged at 4 °C (5 000 g ) for 10 min. The supernatants were discarded, and the cell pellets were snap-freezed using dry ice and ethanol. The cell pellets were stored at −80 °C until total RNA extraction commenced. The RNA-seq experiment was conducted using two biological replicates for each condition (CSP and DMSO) for each time point (3 and 20 min) (Fig. S7).
Total RNA isolation
For each sample, the frozen cell pellet was thawed on ice. The thawed cell pellet was resuspended in 600 µl of lysis solution (sodium acetate 30 mM, sodium dodecyl sulfate 10 mM and ethylenediaminetetraacetic acid 1 mM, pH 5.5), 20 mg of sterile glass beads (0.1 mM) was added to the suspension and vortexed for 3–5 min. The solution was centrifuged (10000 g at 4 °C) for 2 min and the supernatant was transferred to a new sterile 1.5 ml microfuge tube containing 500 µl of acidic phenol (pH 5.5). Total RNA was extracted using phenol–chloroform extraction followed by DNase treatment (PureLink DNase Set, Thermo Fisher Scientific) according to the manufacturer’s specifications. Another round of phenol–chloroform extraction was performed to yield total RNA. The extracted RNA samples were resuspended in 16 µl of nuclease-free water and purity was determined using a NanoDrop.
RT-qPCR
For each condition, a volume corresponding to 500 ng of RNA (according to NanoDrop quantification values) was used for cDNA synthesis. cDNA was created using Maxima Reverse Transcriptase (Thermo Fisher Scientific), random hexamers (Thermo Fisher Scientific), RiboLock RNase Inhibitor (Thermo Fisher Scientific) and 10 mM dNTP mix (Thermo Fisher Scientific) following the manufacturer’s protocol. A 1 : 5 dilution of the generated cDNA was made using RNase-free water. The primers used are detailed in Table 1.
Table 1.
Primers used in RT-qPCR
|
Primer name |
Nucleotide sequence |
|---|---|
|
gyrB Fwd |
5′-GCTTTCCTCAATCGTGGTCTA-3′ |
|
gyrB Rev |
5′-CGTAACTAGAGATTCCGCCTTC-3′ |
|
comX5 Fwd |
5′-TATCCGCAAACAAGAGAGTCAA-3′ |
|
comX5 Rev |
5′-ATTTCACGTGCCTTCAGTTTATG-3′ |
|
cinA2 Fwd |
5′-GACCTTGCGATTGTCCACTAA-3′ |
|
cinA2 Rev |
5′-CAGATAAAGGCTGTCCCTCAAA −3′ |
|
comE2 Fwd |
5′-GTTGCTCGCTTTATTCGTCATC-3′ |
|
comE2 Rev |
5′-AATCGAGAGCGGATACCTTATATTT −3′ |
The qPCR experiments were performed using iTaq Universal SYBR Green Supermix (Bio-Rad). The total volume of each qPCR reaction was 10 µl with the following components: 5 µl of SYBR Green Supermix, 3 µl of 1 : 5 diluted cDNA, 1 µl of forward primer (10 µM) and 1 µl of reverse primer (10 µM). The qPCR assays were performed on a CFX 96 system (Bio-Rad) using the following parameters: 95 °C 2 min, 40 cycles of 95 °C for 15 s, 53 °C for 30 s and 72 °C for 60 s. A melt curve analysis was performed at the end of the amplification starting at 65 °C and increasing at 0.5 °C increments to 95 °C.
Library preparation and Illumina sequencing
The total extracted RNA samples were fluorometrically quantified using the QuantiFluor RNA System (Promega) on a Biotek Synergy H1 multimode plate reader (BioTek Instruments, Inc., Winooski, VT, USA). Library preparation was constructed with a Universal Prokaryotic RNA-seq, Prokaryotic AnyDeplete kit (Tecan Genomics, CA, USA) using 120 ng of total RNA input per sample. Library preparations were sent to the Nevada Genomics Center for library quality and quantity checking (Bioanalyzer High Sensitivity Analysis) and for Illumina sequencing on a NextSeq500 using a NextSeq 500/550 Mid Output kit (v2.5 150 cycles).
Read mapping and quantification and differential gene analysis
Raw Illumina reads were analysed using FastQC (v0.11.8), adapter removal and quality trimming was performed using Fastp (v0.20.0) and reanalysed using MultiQC (v1.8). Trimmed reads were then mapped to the S. sinensis Forsyth1A genome (RefSeq Accession: GCF_019090945.1) using Bowtie 2 (v2.3.0). Read quantification was performed using subread featurecounts (v2.0.0). Differential analysis using DESeq2 (v1.0.0) was conducted in RStudio (v. 3.6) [49]. Volcano plot was created using Enhanced Volcano (v1.4.0).
Average nucleotide identity (ANI) analysis
The following genome assemblies were downloaded from the NCBI database: GCF_000767835.1 ( S. sinensis HKU4), GCF_001578725.1 ( S. sinensis DD04) and GCF_902460385.1 ( Streptococcus sp. Marseille-P7376). ANI analysis was performed using fastANI (v1.3) [50].
Transformation assay
A single colony of S. sinensis was grown at 37 °C with 5 % CO2 in 10 ml of THY media (pH 7.3) for 18 h. The bacteria were initially incubated overnight to accomplish appropriate growth concentrations. Following incubation, the overnight S. sinensis culture was diluted 1 : 10 in fresh THY media and underwent static incubation at 37 °C with 5 % CO2 for 1 h to achieve late-lag/early-exponential phase growth. The CSP was added at final concentrations of 10 000, 1000, 100 and 10 nM, along with 300 ng pDL278 (SpecR). pDL278 is a replicative plasmid obtained from Addgene (Addgene plasmid #46882; https://www.addgene.org/46882/). Parallel assays without the CSP or without both the CSP and plasmid were used as controls to assess the indigenous competence or antibiotic resistance of S. sinensis under the tested conditions. After 6 h of incubation at 37 °C with 5 % CO2, 100 µl of the culture was plated on THY agar containing 300 µg ml−1 spectinomycin and incubated at 37 °C with 5 % CO2 for 48 h to identify positive transformants. Experiments were performed in triplicate on three separate days.
Transformation efficiency
A single colony of S. sinensis was grown at 37 °C with 5 % CO2 in 10 ml of THY media (pH 7.3) for 18 h. Following incubation, 100 µl of the overnight culture was used to inoculate 900 µl of fresh THY media and underwent static incubation at 37 °C with 5 % CO2 for 1 h . CSP was added at final concentrations of 10 000 nM, 1000 nM, 100 nM and 10 nM, along with 300 ng pDL278 (SpecR). After 6 h of incubation at 37 °C with 5 % CO2, 40 µl aliquots were spread onto either THY plates or THY plates with 300 µg ml−1 spectinomycin. Those spread onto non-selective plates were first diluted by a factor of 106. Plates were incubated at 37 °C with 5 % CO2 for 24 h to 36 h to identify positive transformants. Transformation efficiency was calculated using the following equation:
Biofilm formation assay
Biofilm formation was assessed using a previously described method with minor alterations [51, 52]. See the Supplementary Material for full experimental details.
Haemolysis assay
Virulence factor production and associated haemolysis were assessed using a previously described method with minor alterations [51]. See the Supplementary Material for full experimental details.
Results
For this work we elected to investigate a previously unidentified streptococcal clinical isolate obtained from the Forsyth Institute. This particular strain was isolated from an adult patient suffering from aggressive periodontitis in the late 1970s [36]. In that study, researchers identified the infective bacterium as a S. mitis derivative. We sequenced the RNA polymerase β subunit (rpoB) gene, a unique marker specific to different bacterial species, and confirmed that this species was in fact a strain of S. sinensis [37, 53].
A genomic search revealed that the only S. sinensis genome that is publicly available through the NCBI database is that of S. sinensis HKU4T (GenBank assembly accession: GCA_000767835.1) [35]. Evaluation of S. sinensis HKU4 genome revealed that it has the following predicted CSP sequence, DFRKVRLPRLFGK, based on the sequence preceding the double glycine motif in its ComC protein sequence (GenBank accession: KGM37872.1).
PCR amplification of the comCDE gene revealed that the S. sinensis strain received from the Forsyth Institute instead demonstrated a different comC gene with a unique CSP signal, DSRRLNFGGFIKFFGK [54]. As such, this new strain was termed S. sinensis Forsyth1A. Protein blast of the S. sinensis Forsyth1A ComC sequence matched with Streptococcus sp. DD04 and Streptococcus sp. Marseille-P7376 [55].
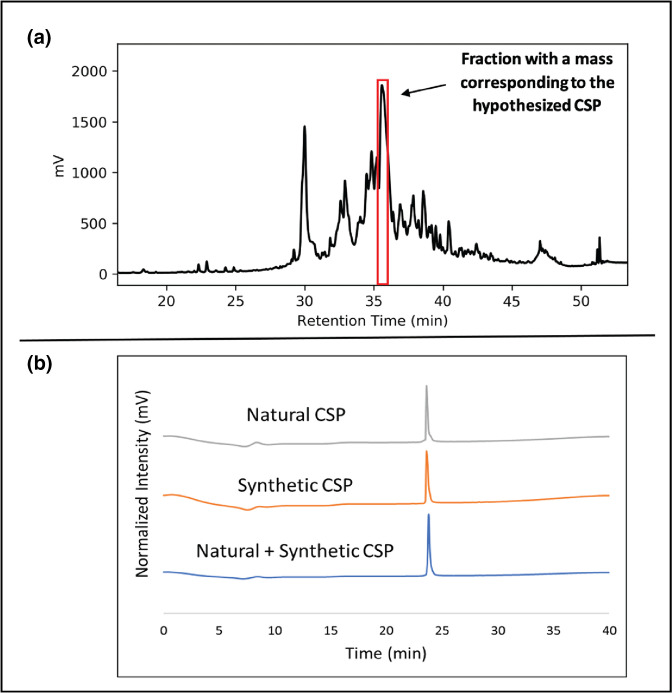
To evaluate the activity of the competence regulon in S. sinensis Forsyth1A and validate the identity of the predicted CSP sequence, the supernatant of S. sinensis Forsyth1A was screened for a peptide with a mass corresponding to our hypothesized sequence (DSRRLNFGGFIKFFGK; Fig. 2a). The endogenous CSP was isolated from the cell-free supernatant of a S. sinensis culture grown to stationary phase. Comparison of the naturally isolated peptide with a synthetic CSP synthesized via solid-phase peptide synthesis (SPPS) was used to further confirm the CSP sequence (Figs 2b and S1).Tandem mass spectometry (MS/MS) analysis of the natural CSP validated its connectivity and identity as well (Fig. S2). Together, these results confirmed that the predicted CSP sequence for S. sinensis is the 16 amino acid peptide, DSRRLNFGGFIKFFGK. This sequence was found to demonstrate homology with the CSP sequences of other, closely related streptococci (Fig. 3).
Fig. 2.
(a) Preparative RP-HPLC trace of S. sinensis Forsyth1A spent supernatant following 30 % ammonium sulfate precipitation. (b) Analytical RP-HPLC trace comparing naturally isolated S. sinensis Forsyth1A CSP to synthetic CSP.
Fig. 3.
clustalw alignment of the ComC proteins of several Mitis group streptococci. ComC is presented as a propeptide that is typically cleaved following the double-glycine motif to render the mature CSP signal.
While the detection of endogenous CSP was prevalent after the culture had reached stationary phase, it was of interest to further determine when endogenous CSP production was initiated, and thus when competence was initiated. To accomplish this, we developed a luciferase-based reporter in which pcomX was placed upstream of the gene for luciferase, thereby allowing for the quantification of comX activity via luminescence detection. Wild-type S. sinensis has one copy of comX. The wild-type strain was then transformed with this plasmid construct and monitored for luminescence. The results indicated that endogenous CSP production stimulated luminescence in S. sinensis transformed with the luciferase-based reporter system, whereas luminescence was not exhibited in the wild-type strain (Fig. S3). Furthermore, luminescence, and therewith CSP production, is initiated at an OD600 of approximately 0.230. These OD600 values correspond to the early- and mid-exponential growth phases, respectively. Luminescence, and suspected CSP production, tapers as the cultures near the late-exponential and stationary phases. Overall, our results support the notion that endogenous CSP production leads to ComE phosphorylation and the subsequent upregulation of pcomX, as visualized by luminescence.
Prior to global transcriptome analysis of the S. sinensis comABCDE regulon, whole-genome sequencing was performed using a hybrid approach combining Illumina sequencing and Oxford Nanopore sequencing data (GenBank assembly accession: GCA_019090945.2). The draft genome obtained is 2.12 Mbp and comprises 61 contigs with 1997 coding sequences (Fig. S4). Average nucleotide identity (ANI) analysis was performed comparing Forsyth1A against DD04, HKU4 and Marseille-P7376 genomes, revealing that isolates Forsyth1A, DD04 and HKU4 meet the species boundary ANI cutoff value of 94 % (Table 2). Strain Marseille-P7376 had an 87.95 ANI with Forsyth1A, which is below the suggested species boundary ANI cutoff value, suggesting that it belongs to a different species of the Mitis group of streptococci [50].
Table 2.
Average nucleotide identity results comparing the genomes of different S. sinensis strains
|
Isolate comparison |
ANI value |
|---|---|
|
Forsyth1A vs HKU4 |
96.64 % |
|
Forsyth1A vs DD04 |
95.06 % |
|
Forsyth1A vs Marseille-P7376 |
87.95 % |
Whole-genome mining of Forsyth1A revealed many genes associated with virulence and competence (Table 3) [17, 56–58]. Virulence gene comparison of Forsyth1A against HKU4 and DD04 revealed that the three strains share the same genes (Table S1). Moreover, both Forsyth1A and DD04 have the same capsular gene profile compared to HKU4.
Table 3.
List of genes found in the S. sinensis Forsyth1A genome that are associated with competence, biofilm formation and virulence
|
Gene function |
Genes |
Annotation |
Locus |
|---|---|---|---|
|
pavA |
Fibronectin-binding proteins |
FOA32_000330 |
|
|
lmb |
Laminin-binding protein |
FOA32_001121 |
|
|
Adherence |
srtA |
Sortase A |
FOA32_001697 |
|
slrA |
Streptococcal lipoprotein rotamase A |
FOA32_000620 |
|
|
plr/gapA |
Streptococcal plasmin receptor |
FOA32_001298 |
|
|
srtB |
Sortase B |
FOA32_001612 |
|
|
eno |
Streptococcal enolase |
FOA32_000730 |
|
|
Biofilm and capsule production |
cap3 |
UDP-glucose-6-dehydrogenase |
FOA32_000241 |
|
galE |
UDP-glucose-4-epimerase |
FOA32_07480 |
|
|
Manganese uptake |
psaA |
Pneumococcal surface antigen A |
FOA32_000346 |
|
htrA |
Serine protease |
FOA32_001484 |
|
|
Protease |
cppA |
Carbon catabolite protein A |
FOA32_000227 |
|
tig/ropA |
Trigger factor |
FOA32_001645 |
|
|
comA |
ABC transporter |
FOA32_001444 |
|
|
comB |
ABC transporter |
FOA32_001445 |
|
|
comC |
Competence-stimulating peptide |
FOA32_001482 |
|
|
comD |
Histidine kinase |
FOA32_001481 |
|
|
comE |
Response regulator |
FOA32_001480 |
|
|
comX |
Alternative sigma factor |
FOA32_000049 |
|
|
comW |
Competence protein |
FOA32_19470 |
|
|
comYA |
Competence protein |
FOA32_000391 |
|
|
comYB |
Competence protein |
FOA32_000390 |
|
|
comYC |
Competence protein |
FOA32_000389 |
|
|
comYD |
Competence protein |
FOA32_000388 |
|
|
Competence |
comGF |
Competence protein |
FOA32_000386 |
|
celA |
Competence protein |
FOA32_000664 |
|
|
comEC |
Competence protein |
FOA32_000665 |
|
|
coiA |
Competence protein |
FOA32_000685 |
|
|
tfoX |
Competence protein |
FOA32_000685 |
|
|
cinA |
Competence–damage inducible protein |
FOA32_001005 |
|
|
recX |
Recombination protein |
FOA32_001238 |
|
|
rmuC |
DNA recombination protein |
FOA32_001890 |
|
|
recN |
Recombination protein |
FOA32_000639 |
|
|
ydel/omp |
Bacteriocin protection protein |
FOA32_001270 |
|
|
pepF |
Oligoendopeptidase F |
FOA32_001503 |
|
|
recF |
Recombination protein |
FOA32_001471 |
|
|
recO |
Recombination protein |
FOA32_001839 |
|
|
radA |
DNA repair protein |
FOA32_001984 |
|
|
recJ |
Recombination protein |
FOA32_000543 |
|
|
mecA |
Adapter protein |
FOA32_000035 |
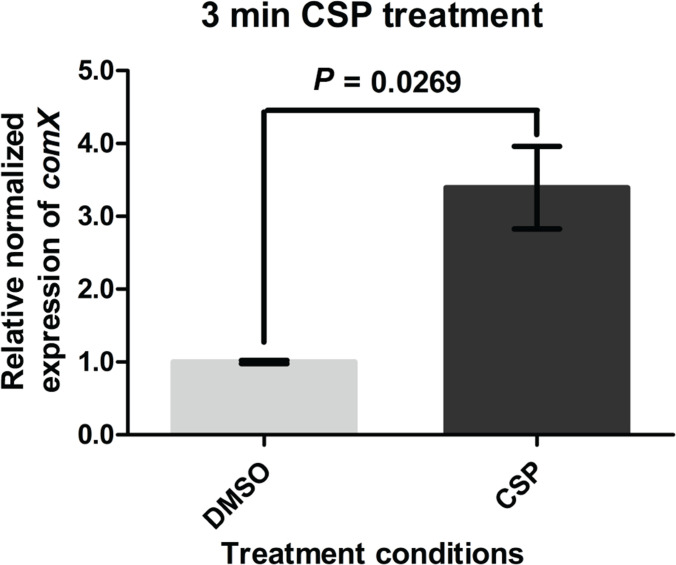
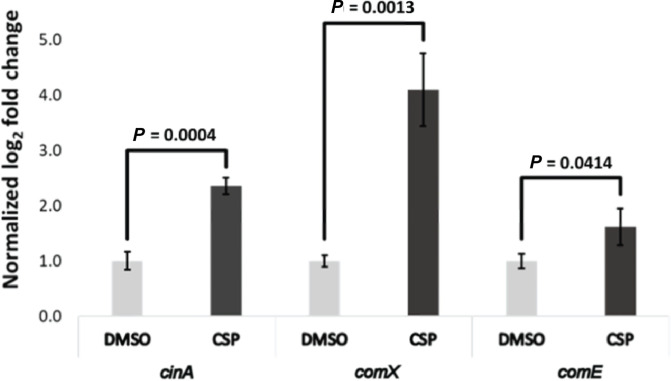
RT-qPCR analysis revealed that S. sinensis Forsyth1A follows the same pattern, common in the Mitis group of streptococci, where activation of the comX gene occurs within 5 min after addition of synthetic CSP and declines to baseline expression after 20 min (Figs 4 and S5).
Fig. 4.
RT-qPCR results after 3 min synthetic CSP treatment. Treatment with exogenous CSP for 3 min leads to activation of the comX gene. After a 3 min incubation, stop solution was added to each sample to stop transcription. Data are normalized to expression of gyrB. The RT-qPCR experiment was conducted using two biological replicates for both the CSP and DMSO conditions.
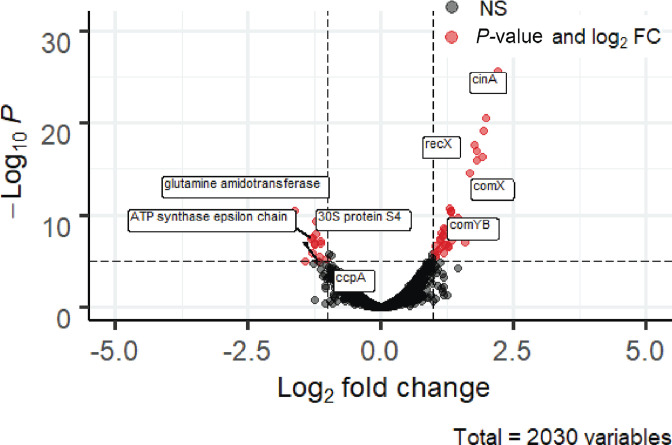
RNA-seq analysis of S. sinensis with either a 3 min treatment of synthetic CSP or DMSO control was performed. Analysis revealed that 147 genes were significantly upregulated while 127 genes were downregulated (log fold change >1, P-value <0.05, FDR value <0.05) (Fig. 5). The early competence genes, comABCDE, were upregulated, but the predicted comW gene was not significantly upregulated (Table 4). Many late competence genes associated with DNA uptake and integration were also upregulated (comYA, comYB, comYC, comYD, comGF, celA, comEC, coiA, recX, recN, rmuC), suggesting that activation of the competence regulon aids in the induction of competence in S. sinensis . An RT-qPCR analysis of comX, comE and cinA was used to validate the differential gene expression trends (Fig. 6).
Fig. 5.
Volcano plot of differentially expressed genes comparing treatment with DMSO (negative control) to synthetic CSP. Grey, non-significant genes expressed; red, genes with both a log fold change >2 and cutoff P-value >10−6 (statistically significant genes).
Table 4.
Differential expression results highlighting various genes associated with competence and virulence being upregulated when comparing DMSO (negative control) to synthetic CSP
|
Locus tag |
Gene |
Annotation |
Log2 change |
P-adj |
|---|---|---|---|---|
|
FOA32_000391 |
comYA |
Competence protein YA |
1.1362 |
1.96E-05 |
|
FOA32_000390 |
comYB |
Competence protein YB |
1.4176 |
7.27E-08 |
|
FOA32_000389 |
comYC |
Competence protein YC |
1.3002 |
1.21E-05 |
|
FOA32_000388 |
comYD |
Competence protein YD |
0.6785 |
2.42E-02 |
|
FOA32_000386 |
comGF |
Competence protein GF |
1.1461 |
2.46E-06 |
|
FOA32_000664 |
celA |
Competence protein |
1.4683 |
1.05E-03 |
|
FOA32_000665 |
comEC |
Competence protein |
1.8266 |
3.21E-15 |
|
FOA32_000685 |
coiA |
Competence protein |
0.8556 |
2.51E-03 |
|
FOA32_001444 |
comA |
ABC transporter |
1.1788 |
1.51E-06 |
|
FOA32_001445 |
comB |
ABC transporter |
0.8518 |
3.91E-04 |
|
FOA32_000049 |
comX |
Transcriptional regulator of competence-specific genes |
1.8639 |
5.05E-11 |
|
FOA32_001005 |
cinA |
Competence–damage inducible protein A |
1.9992 |
1.53E-18 |
|
FOA32_001482 |
comC |
Competence-stimulating peptide |
1.4634 |
1.08E-07 |
|
FOA32_001481 |
comD |
Histidine kinase |
0.8049 |
3.67E-03 |
|
FOA32_001480 |
comE |
Response regulator |
0.9189 |
4.93E-04 |
|
FOA32_001238 |
recX |
Recombination protein |
1.8172 |
2.60E-14 |
|
FOA32_001890 |
rmuC |
Recombination protein |
1.3208 |
6.69E-08 |
|
FOA32_000639 |
recN |
DNA repair protein |
1.0128 |
6.99E-03 |
|
FOA32_000971 |
cap3 |
UDP-glucose-6-dehyrogenase |
0.6498 |
1.74E-03 |
|
FOA32_001484 |
htrA |
Serine protease |
0.5213 |
4.87E-02 |
|
FOA32_001961 |
pbp 2A |
enicillin-binding protein 2A |
0.9618 |
8.45E-06 |
Fig. 6.
RT-qPCR analysis of original RNA samples used in the RNA sequencing experiment as validation of differential gene expression analysis. After a 3 min incubation, stop solution was added to each sample to stop transcription. Data are normalized to expression of gyrB. The RT-qPCR experiment was conducted using two biological replicates for both the CSP and DMSO conditions.
Due to the limited knowledge about S. sinensis , many of the genes that were affected by inducing the competence regulon are of unknown function; however, a few genes identified as virulence-related and metabolic-related were affected by synthetic CSP treatment (Table 4) [56]. For example, high-temperature requirement A serine protease (htrA), which is induced during competence development and has been shown to play a role in virulence phenotypes in S. pneumoniae , was upregulated [59, 60]. Another example is catabolite control protein A (ccpA), a regulatory metabolic protein that directs selective metabolism of available sugars and has been found to play a role in competence in two Mitis group streptococci: S. cristatus and S. gordonii [61, 62]. Transcriptome analysis of S. gordonii during competence revealed that ccpA was downregulated during the competence state, while a similar observation was made with S. sinensis Forsyth1A (Table 5) [23].
Table 5.
Differential expression results highlighting various genes associated with competence and virulence being downregulated when comparing DMSO (negative control) to synthetic CSP
|
Locus tag |
Gene |
Annotation |
log2 FC |
p-adj |
|---|---|---|---|---|
|
FOA32_000227 |
ccpA |
Catabolite control protein A |
−0.7463 |
6.89E-03 |
|
FOA32_001808 |
- |
PTS fructose transporter subunit IIA |
−0.9372 |
1.16E-04 |
|
FOA32_001809 |
- |
PTS fructose transporter subunit IID |
−0.9665 |
8.87E-05 |
|
FOA32_000099 |
- |
PTS glucose transporter subunit IIABC |
−0.6123 |
3.76E-02 |
|
FOA32_001341 |
- |
PTS mannose/fructose/sorbose transporter subunit IIC |
−1.1371 |
4.67E-06 |
|
FOA32_000475 |
crcB |
Putative fluoride ion transporter |
−0.8183 |
2.49E-02 |
|
FOA32_000035 |
mecA |
Adapter protein |
−0.8028 |
1.49E-02 |
|
FOA32_000761 |
amy |
Alpha-amylase |
−1.2753 |
4.96E-04 |
|
FOA32_001226 |
ftsA |
Cell division protein |
−0.6769 |
2.98E-02 |
|
FOA32_000717 |
ftsX |
Cell division protein |
−0.9199 |
1.40E-02 |
|
FOA32_001028 |
malR |
LacI family transcriptional regulator |
−0.6059 |
4.17E-02 |
|
FOA32_000006 |
rsfS |
Ribosomal silencing factor |
−1.4111 |
4.93E-04 |
|
FOA32_001645 |
tig |
Trigger factor |
−0.6221 |
4.44E-02 |
|
FOA32_001049 |
- |
Type VII secretion protein EssA |
−0.9494 |
1.12E-02 |
|
FOA32_001443 |
rpsD |
30S ribosomal protein S4 |
−1.2171 |
7.21E-08 |
|
FOA32_001630 |
rpsH |
30S ribosomal protein S8 |
−0.9610 |
2.05E-03 |
|
FOA32_001505 |
rplM |
50S ribosomal protein L13 |
−1.2907 |
6.32E-05 |
|
FOA32_001635 |
rplO |
50S ribosomal protein L15 |
−1.0335 |
7.64E-04 |
|
FOA32_000259 |
infC |
Translation initiation factor |
−1.1224 |
8.05E-06 |
|
FOA32_000143 |
- |
Membrane protein |
−0.9259 |
1.34E-02 |
|
FOA32_000586 |
- |
Membrane protein |
−0.8700 |
1.59E-02 |
|
FOA32_000860 |
scrB |
Membrane protein |
−0.6828 |
1.84E-02 |
|
FOA32_001191 |
yidC_1 |
Insertase YidC |
−1.2089 |
1.24E-05 |
Phenotypic assays following the exogenous addition of synthetic CSP were utilized to assess the correlation between the increase of mRNA transcript abundance as determined by RT-qPCR and related proliferative phenotype expression. It has been well established in several streptococcal species that the competence regulon either completely or partially aids in the regulation of specific phenotypes, including competence, biofilm formation and virulence factor production [63, 64]. As such, experiments were conducted to evaluate whether these processes are indeed regulated by the ComABCDE regulon in S. sinensis .
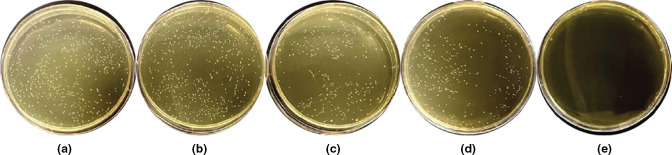
As described previously, bacterial competence is enhanced by exogenous CSP exposure in many streptococcal species [5, 65–67]. To evaluate this effect in S. sinensis Forsyth1A, transformation assays were completed in which cultures of the wild-type strain were incubated with the synthetic native peptide and plasmid-containing genes for antibiotic resistance. Conditions included final peptide concentrations of 10 000 nM, 1000 nM, 100 nM and 10 nM, as well as a control with no exogenous CSP. A correlation between the addition of exogenous CSP and bacterial competence was observed, suggesting the successful internalization of plasmid DNA via induced transformation (Fig. 7). While the addition of CSP was needed for successful transformation, there was no significant difference between the varying concentrations of added exogenous CSP.
Fig. 7.
S. sinensis transformation assay. S. sinensis demonstrates dependence on the addition of exogenous synthetic CSP for the successful uptake of added plasmid (pDL278). The synthetic CSP was added at final concentrations of 10 000 nM (a), 1000 nM (b), 100 nM (c) and 10 nM (d). The control includes no exogenous CSP (e). See the Methods section for full experimental details.
Alternatively, competence was not achieved when S. sinensis Forsyth1A was incubated with the HKU4 CSP pherotype, DFRKVRLPRLFGK, under the same experimental conditions (Fig. S6). Overall, our results suggest that the competence regulon does in fact regulate competence in S. sinensis and is specific to the given pherotype.
At this point we also sought to determine the transformation efficiency for S. sinensis to aid in additional characterization of this strain. Comparison of total transformants grown on selective media to total colonies grown on non-selective media yielded transformation efficiencies of approximately 1.14×10–4, 5.61×10–5, 3.88×10–5 and 3.70×10–5 %; these values correspond to final exogenous CSP concentrations of 10 000 nM, 1000 nM, 100 nM and 10 nM, respectively. Previous studies have indicated that the transformation efficiencies for many streptococci, including S. mitis , tend to be low, often <1 % [68–70]. The transformation efficiency of S. sinensis is comparable to these values. Further experimentation is needed to optimize this value and define conditions that allow for enhanced transformation efficiency in S. sinensis .
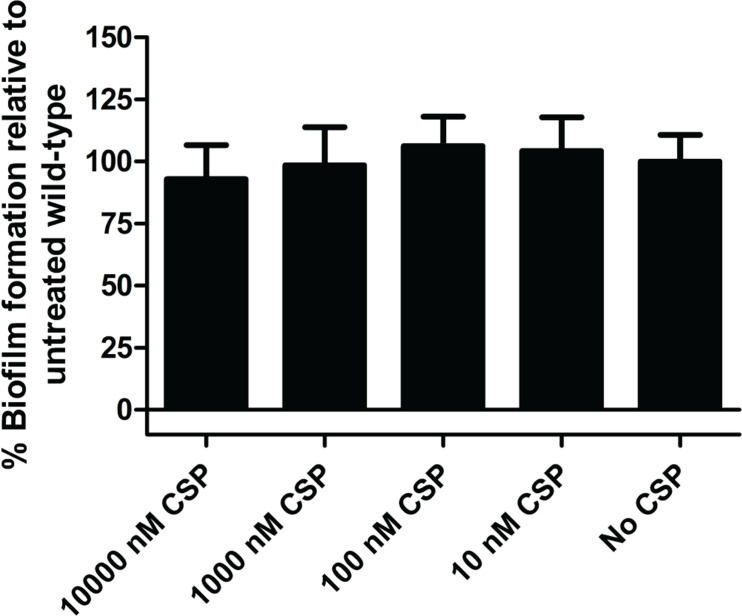
The competence regulon has also been shown to aid in the regulation of biofilm formation in many streptococcal species [63, 71]. This trait is associated with increased bacterial pathogenesis and virulence, and as such it is of great interest to delineate the relationship that exists between the competence regulon and biofilm formation. Here, a crystal violet biofilm quantification assay was used to assess the formation of bacterial biofilms [51, 52]. Cultures were exposed to exogenous CSP in a dose-dependent manner, yielding final concentrations of 10000 nM, 1000 nM, 100 nM and 10 nM synthetic CSP. The data are normalized to the amount of biofilm produced in the wild-type system (no addition of exogenous CSP). The data indicated that there is no significant difference in the quantity of biofilm formed when incubated with CSP, when compared to the untreated wild-type strain of S. sinensis Forsyth1A (Fig. 8). The OD600 and OD595 of wild-type S. sinensis cultures were used to quantify biofilm growth. Cultures of S. sinensis exhibited significant levels of biofilm formation compared to media-only samples (Table S2). This assay alone cannot fully determine the involvement of the competence regulon in the regulation of biofilm formation. However, this does indicate that the competence regulon does not affect biofilm formation under the described experimental conditions. CSP may instead influence biofilm formation in the context of polymicrobial interactions that take place in the microbiome.
Fig. 8.
Effect of exogenous CSP on S. sinensis biofilm formation. The addition of exogenous CSP at varying concentrations does not significantly affect biofilm formation in S. sinensis . The results thereby indicate that the competence regulon QS circuitry in S. sinensis does not seem to have an effect on biofilm formation. See the Supplementary Material for full experimental details.
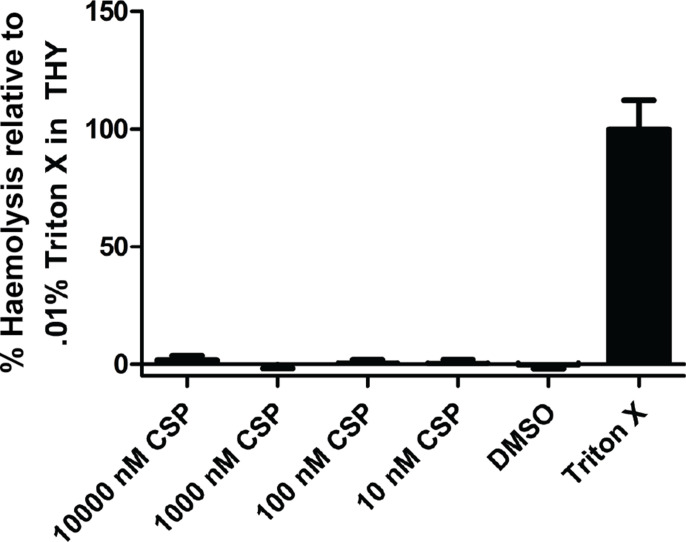
Lastly, many streptococcal species, including many closely related Mitis group species, are known to produce virulence factors to promote their own survival and pathogenesis in both human and animal models [72]. For example, it has been shown that S. pneumoniae produces a haemolytic toxin and virulence factor, pneumolysin, known to aid in the development of pneumococcal pathogenesis [73]. As such, it was of interest to determine whether S. sinensis produced similar virulence factors capable of haemolysis. This phenotype was assessed using a haemolysis assay in which the lysis of defibrinated rabbit erythrocytes was quantified. Cultures of S. sinensis Forsyth1A exposed to varying concentrations of synthetic CSP were incubated with red blood cells (RBCs). The results indicate that the presence of CSP does not directly influence haemolysis when normalized to the 0.01 % Triton X control (Fig. 9). This assay alone does not fully assess for virulence factor production and subsequent effects on RBCs. The upregulation of genes associated with virulence factor production may instead have a greater effect on interspecies interactions and potential competition that occur in the microbiome rather than direct effects on RBCs.
Fig. 9.
Effect of exogenous CSP on S. sinensis virulence factor production. The addition of exogenous CSP at varying concentrations does not significantly affect lysis of RBCs by S. sinensis . See the Supplementary Material for full experimental details.
Ultimately, our results indicated that the S. sinensis competence regulon does have an effect on competence development, but does not directly impact biofilm formation or virulence factor production under the described testing conditions. As such, further experimentation is needed to clarify the effect of CSP on subsequent S. sinensis phenotypes, both within and outside of a polymicrobial community.
Discussion
Investigation into the S. sinensis ComABCDE QS pathway revealed that this pathway is functional in strain Forsyth1A, demonstrating that this member of the Mitis group of streptococci is also naturally competent. S. sinensis likely has at least two CSP pherotypes, DFRKVRLPRLFGK (strain HKU4) and DSRRLNFGGFIKFFGK (strains DD04 and Forsyth1A). The genome of S. sinensis Forsyth1A was sequenced and whole-genome comparison with other isolates suggests that Forsyth1A is more closely related to strain DD04 compared to strain HKU4. This strain demonstrated a transformation efficiency of <0.001%, which is consistent with other streptococcal species. S. sinensis also demonstrated initial production of endogenous CSP in early- to mid-exponential phase, as detected via luminescence.
This is the first in-depth transcriptome investigation into the competence regulon in S. sinensis . Through this analysis, many competence-related genes were observed to be differentially expressed after 3 min incubation with synthetic CSP. S. sinensis competence regulation differs from S. pneumoniae and S. mitis in that it does not exhibit the upregulation of comW during competence [21, 74]. Further experiments are needed to confirm the basal expression level of comW, as well as comW expression during competence shutoff. It is probable that this gene is differentially expressed in S. sinensis while still playing a regulatory role in bacterial competence, but additional research is needed to ascertain such information.
A separate gene, htrA, demonstrated increased expression following CSP induction in S. sinensis . Previously, researchers had found that the presence of HtrA appears to be required for appropriate transformation in S. pneumoniae [60]. Furthermore, it was recently shown that in S. pneumoniae , S. gordonii and S. mitis, HrtA degrades the CSP signal and certain competence proteins involved with DNA uptake to assist with competence termination [75]. The observed increase in expression of htrA in S. sinensis suggests that it may be playing a role in regulating competence in Mitis group streptococci. Again, additional experimentation examining the expression levels and activity of HtrA throughout the process of competence induction and shutoff will be needed to further delineate its role as a potential competence regulator.
The expression of another known competence regulatory element, CcpA, was downregulated during the 3 min CSP treatment transcriptome snapshot. This result directly indicates that its expression is somewhat suppressed or reduced during early competence development. Researchers had previously found that CcpA indirectly aids in the regulation of early competence genes, including comCDE, in Mitis group streptococci [62, 76]. For example, it was found that an S. oligofermentans ΔccpA mutant had higher expression levels of phosphorylated ComE, which is known to bear great influence on other competence genes, such as comX [61]. As such, ccpA is potentially downregulated in early competence to allow for the adequate expression of early competence genes that are needed for successful transformation. Nonetheless, these findings highlight the need to perform additional competence transcriptome analyses on members of the Mitis group of streptococci to fully elucidate the complexity of this phenotype.
Lastly, the increased expression of genes related to competence, biofilm formation and virulence was of particular interest, as these have potential to influence proliferative phenotypic response. It was found that the S. sinensis Forsyth1A competence regulon does play a role in the development of bacterial competence and subsequent transformation. However, incubation with CSP does not have an effect on biofilm formation or virulence as it relates to haemolysis. These results do not fully explain the dynamic relationship that exists between the competence regulon QS circuitry and these phenotypes. Rather, these results suggest that there is no direct influence of the CSP on biofilm formation and virulence in the described tested conditions. Instead, the upregulated genes may instead have an effect on various interspecies interactions that take place in polymicrobial communities.
Future studies are needed to clarify the effects of CSP exposure on S. sinensis Forsyth1A phenotypes in the context of mixed microbial interactions, thereby providing even more insight into the role of the native CSP on the competence regulon. Overall, this study sheds light on a newly identified species, S. sinensis Forsyth1A, and provides a robust foundation for future investigation of this species, as well as its interactions and effects on other members of the oral microbiome.
Supplementary Data
Funding information
This work was supported by a grant from the National Science Foundation (CHE-1808370) and a grant from the National Institutes of Health (R35GM128651).
Acknowledgements
The S. sinensis Forsyth1A strain was a generous gift from M. Duncan (Forsyth Institute).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, Average nucleotide identity; bp, base pair; ccpA, catabolite control protein A; CSP, competence stimulating peptide; DprA, DNA-processing protein A; Fmoc, 9-Fluorenylmethoxycarbonyl; htrA, high-temperature requirement A serine protease; NCBI, National Center of Biotechnology Information; ONT, Oxford Nanopore Technology; PCR, polymerase chain reaction; QS, quorum-sensing; RBC, red blood cells; rpoB, RNA polymerase β subunit; SPPS, solid-phase peptide synthesis; THY, Todd-Hewitt media supplemented with yeast extract.
Seven supplementary figures and two supplementary tables are available with the online version of this article.
References
- 1.Salvadori G, Junges R, Morrison DA, Petersen FC. Competence in Streptococcus pneumoniae and close commensal relatives: mechanisms and implications. Front Cell Infect Microbiol. 2019;9:94. doi: 10.3389/fcimb.2019.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, et al. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol Med Microbiol. 2010;59:345–349. doi: 10.1111/j.1574-695X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhu L, Lau GW. Inhibition of competence development, horizontal gene transfer and virulence in Streptococcus pneumoniae by a modified competence stimulating peptide. PLoS Pathog. 2011;7:e1002241. doi: 10.1371/journal.ppat.1002241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gómez-Mejia A, Gámez G, Hammerschmidt S. Streptococcus pneumoniae two-component regulatory systems: the interplay of the pneumococcus with its environment. Int J Med Microbiol. 2018;308:722–737. doi: 10.1016/j.ijmm.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Lee MS, Morrison DA. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/JB.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lella M, Tal-Gan Y. Strategies to attenuate the competence regulon in Streptococcus pneumoniae . Pept Sci. 2021;113:e24222. doi: 10.1002/pep2.24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen A, Scholz CFP, Kilian M. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus . Int J Syst Evol Microbiol. 2016;66:4803–4820. doi: 10.1099/ijsem.0.001433. [DOI] [PubMed] [Google Scholar]
- 8.Pestova EV, Håvarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 9.Johnston C, Hinds J, Smith A, van der Linden M, Van Eldere J, et al. Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J Clin Microbiol. 2010;48:2762–2769. doi: 10.1128/JCM.01746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, et al. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol. 2001;40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 11.Jack AA, Daniels DE, Jepson MA, Vickerman MM, Lamont RJ, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans . Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo P, Li H, Morrison DA. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae . Mol Microbiol. 2003;50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 13.McBrayer DN, Cameron CD, Tal-Gan Y. Development and utilization of peptide-based quorum sensing modulators in gram-positive bacteria. Org Biomol Chem. 2020;18:7273–7290. doi: 10.1039/d0ob01421d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knutsen E, Ween O, Håvarstein LS. Two separate quorum-sensing systems upregulate transcription of the same ABC transporter in Streptococcus pneumoniae . J Bacteriol. 2004;186:3078–3085. doi: 10.1128/JB.186.10.3078-3085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Gámez S, Sorg RA, Domenech A, Kjos M, Weissing FJ, et al. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun. 2017;8:854. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zu Y, Li W, Wang Q, Chen J, Guo Q. ComDE two-component signal transduction systems in oral streptococci: structure and function. Curr Issues Mol Biol. 2019;32:201–258. doi: 10.21775/cimb.032.201. [DOI] [PubMed] [Google Scholar]
- 17.Luo P, Li H, Morrison DA. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae . Mol Microbiol. 2003;50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 18.Sung CK, Morrison DA. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae . J Bacteriol. 2005;187:3052–3061. doi: 10.1128/JB.187.9.3052-3061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, Lau GW. DprA-dependent exit from the competent state regulates multifaceted Streptococcus pneumoniae virulence. Infect Immun. 2019;87:e00349-19. doi: 10.1128/IAI.00349-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 21.Salvadori G, Junges R, Åmdal HA, Chen T, Morrison DA, et al. High-resolution profiles of the Streptococcus mitis CSP signaling pathway reveal core and strain-specific regulated genes. BMC Genomics. 2018;19:453. doi: 10.1186/s12864-018-4802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez AM, Callahan JE, Fawcett P, Ge X, Xu P, et al. Physiological and molecular characterization of genetic competence in Streptococcus sanguinis . Mol Oral Microbiol. 2011;26:99–116. doi: 10.1111/j.2041-1014.2011.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vickerman MM, Iobst S, Jesionowski AM, Gill SR. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol. 2007;189:7799–7807. doi: 10.1128/JB.01023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slager J, Aprianto R, Veening J-W. Refining the pneumococcal competence regulon by RNA sequencing. J Bacteriol. 2019;201:e00780-18. doi: 10.1128/JB.00780-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo P, Li H, Morrison DA. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae . Mol Microbiol. 2004;54:172–183. doi: 10.1111/j.1365-2958.2004.04254.x. [DOI] [PubMed] [Google Scholar]
- 26.Prudhomme M, Berge M, Martin B, Polard P. Pneumococcal competence coordination relies on a cell-contact sensing mechanism. PLoS Genet. 2016;12:e1006113. doi: 10.1371/journal.pgen.1006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng L, Piotrowski A, Morrison DA. Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS One. 2013;8:e64197. doi: 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirouze N, Bergé MA, Soulet A-L, Mortier-Barrière I, Quentin Y, et al. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A. 2013;110:E1035–44. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston C, Mortier-Barriere I, Khemici V, Polard P. Fine-tuning cellular levels of DprA ensures transformant fitness in the human pathogen Streptococcus pneumoniae . Mol Microbiol. 2018;109:663–675. doi: 10.1111/mmi.14068. [DOI] [PubMed] [Google Scholar]
- 30.Weyder M, Prudhomme M, Bergé M, Polard P, Fichant G. Dynamic modeling of Streptococcus pneumoniae competence provides regulatory mechanistic insights into its tight temporal regulationCompetence Provides Regulatory Mechanistic Insights Into Its Tight Temporal Regulation. Front Microbiol. 2018;9:1637. doi: 10.3389/fmicb.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin J, Zhu L, Lau GW. Disentangling competence for genetic transformation and virulence in Streptococcus pneumoniae . Curr Genet. 2016;62:97–103. doi: 10.1007/s00294-015-0520-z. [DOI] [PubMed] [Google Scholar]
- 32.Watkins RR, Bonomo RA. Overview: global and local impact of antibiotic resistance. Infect Dis Clin North Am. 2016;30:313–322. doi: 10.1016/j.idc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Woo PCY, Tam DMW, Leung K-W, Lau SKP, Teng JLL, et al. Streptococcus sinensis sp. nov., a novel species isolated from a patient with infective endocarditis. J Clin Microbiol. 2002;40:805–810. doi: 10.1128/JCM.40.3.805-810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo PC, Teng JL, Leung K-W, Lau SK, Tse H, et al. Streptococcus sinensis may react with Lancefield group F antiserum. J Med Microbiol. 2004;53:1083–1088. doi: 10.1099/jmm.0.45745-0. [DOI] [PubMed] [Google Scholar]
- 35.Teng JLL, Huang Y, Tse H, Chen JHK, Tang Y, et al. Phylogenomic and MALDI-TOF MS analysis of Streptococcus sinensis HKU4T reveals a distinct phylogenetic clade in the genus Streptococcus . Genome Biol Evol. 2014;6:2930–2943. doi: 10.1093/gbe/evu232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 37.Drancourt M, Roux V, Fournier P-E, Raoult D. rpoB gene sequence-based identification of aerobic Gram-positive cocci of the genera Streptococcus, Enterococcus, Gemella, Abiotrophia, and Granulicatella . J Clin Microbiol. 2004;42:497–504. doi: 10.1128/JCM.42.2.497-504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrington A, Tal-Gan Y. Identification of Streptococcus gallolyticus subsp. gallolyticus (Biotype I) competence-stimulating peptide pheromone. J Bacteriol. 2018;200:00709–00717. doi: 10.1128/JB.00709-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Koirala B, Sanchez LA, Phillips NR, Hamry SR, et al. Structure-activity relationships of the competence stimulating peptides (CSPs) in Streptococcus pneumoniae reveal motifs critical for intra-group and cross-group ComD receptor activation. ACS Chem Biol. 2017;12:1141–1151. doi: 10.1021/acschembio.7b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan W, White P. Fmoc Solid Phase Peptide Synthesis: A Practical Approach, vol 222. OUP Oxford; 1999. [DOI] [Google Scholar]
- 41.Guo M, Ye L, Yu T, Han L, Li Q, et al. IL-1β enhances the antiviral effect of IFN-α on HCV replication by negatively modulating ERK2 activation. ACS Infect Dis. 2020;6:1708–1718. doi: 10.1021/acsinfecdis.9b00506. [DOI] [PubMed] [Google Scholar]
- 42.Kjos M, Miller E, Slager J, Lake FB, Gericke O, et al. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog. 2016;12:e1005422. doi: 10.1371/journal.ppat.1005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol: chloroform. CSH Protoc. 2006;2006:pdb.prot4045. doi: 10.1101/pdb.prot4045. [DOI] [PubMed] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mull RW, Tal-Gan Y. Elucidating the role and structure-activity relationships of the Streptococcus oligofermentans competence-stimulating peptide. ACS Chem Biol. 2021;16:2834–2844. doi: 10.1021/acschembio.1c00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kratochvil MJ, Tal-Gan Y, Yang T, Blackwell HE, Lynn DM. Nanoporous superhydrophobic coatings that promote the extended release of water-labile quorum sensing inhibitors and enable long-term modulation of quorum sensing in Staphylococcus aureus . ACS Biomater Sci Eng. 2015;1:1039–1049. doi: 10.1021/acsbiomaterials.5b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogier J-C, Pagès S, Galan M, Barret M, Gaudriault S. rpoB, a promising marker for analyzing the diversity of bacterial communities by amplicon sequencing. BMC Microbiol. 2019;19:171. doi: 10.1186/s12866-019-1546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Håvarstein LS, Hakenbeck R, Gaustad P. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denapaite D, Rieger M, Köndgen S, Brückner R, Ochigava I, et al. Highly variable Streptococcus oralis strains are common among viridans streptococci isolated from primates. mSphere. 2016;1:e00041-15. doi: 10.1128/mSphere.00041-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, Yang J, Yu J, Yao Z, Sun L, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–8. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Saizieu A, Gardès C, Flint N, Wagner C, Kamber M, et al. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/JB.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shanker E, Federle MJ. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes (Basel) 2017;8:E15. doi: 10.3390/genes8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cassone M, Gagne AL, Spruce LA, Seeholzer SH, Sebert ME. The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J Biol Chem. 2012;287:38449–38459. doi: 10.1074/jbc.M112.391482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. Role of HtrA in the virulence and competence of Streptococcus pneumoniae . Infect Immun. 2004;72:3584–3591. doi: 10.1128/IAI.72.6.3584-3591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Cai J, Shang N, Zhu L, Shao N, et al. The carbon catabolite repressor CcpA mediates optimal competence development in Streptococcus oligofermentans through post-transcriptional regulation. Mol Microbiol. 2019;112:552–568. doi: 10.1111/mmi.14274. [DOI] [PubMed] [Google Scholar]
- 62.Zheng L, Chen Z, Itzek A, Herzberg MC, Kreth J. CcpA regulates biofilm formation and competence in Streptococcus gordonii . Mol Oral Microbiol. 2012;27:83–94. doi: 10.1111/j.2041-1014.2011.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suntharalingam P, Cvitkovitch DG. Quorum sensing in streptococcal biofilm formation. Trends Microbiol. 2005;13:3–6. doi: 10.1016/j.tim.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Brennan AA, Mehrani M, Tal-Gan Y. Modulating streptococcal phenotypes using signal peptide analogues. Open Biol. 2022;12:220143. doi: 10.1098/rsob.220143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shanker E, Federle MJ. quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes (Basel) 2017;8:E15. doi: 10.3390/genes8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler ME, Morrison DA. Competence beyond genes: filling in the details of the pneumococcal competence transcriptome by a systems approach. J Bacteriol. 2019;201:e00238-19. doi: 10.1128/JB.00238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bikash CR, Tal-Gan Y. Identification of highly potent competence stimulating peptide-based quorum sensing activators in Streptococcus mutans through the utilization of N-methyl and reverse alanine scanning. Bioorg Med Chem Lett. 2019;29:811–814. doi: 10.1016/j.bmcl.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salvadori G, Junges R, Morrison DA, Petersen FC. Overcoming the barrier of low efficiency during genetic transformation of Streptococcus mitis . Front Microbiol. 2016;7:1009. doi: 10.3389/fmicb.2016.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bensing BA, Rubens CE, Sullam PM. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect Immun. 2001;69:1373–1380. doi: 10.1128/IAI.69.3.1373-1380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rukke HV, Kalluru RS, Repnik U, Gerlini A, José RJ, et al. Protective role of the capsule and impact of serotype 4 switching on Streptococcus mitis . Infect Immun. 2014;82:3790–3801. doi: 10.1128/IAI.01840-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jack AA, Daniels DE, Jepson MA, Vickerman MM, Lamont RJ, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jedrzejas MJ. Pneumococcal virulence factors: structure and function. Microbiol Mol Biol Rev. 2001;65:187–207. doi: 10.1128/MMBR.65.2.187-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berry AM, Yother J, Briles DE, Hansman D, Paton JC. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae . Infect Immun. 1989;57:2037–2042. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tovpeko Y, Bai J, Morrison DA. Competence for genetic transformation in Streptococcus pneumoniae: mutations in σa bypass the comw requirement for late gene expression. J Bacteriol. 2016;198:2370–2378. doi: 10.1128/JB.00354-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y, Zeng Y, Huang Y, Gu L, Wang S, et al. HtrA-mediated selective degradation of DNA uptake apparatus accelerates termination of pneumococcal transformation. Mol Microbiol. 2019;112:1308–1325. doi: 10.1111/mmi.14364. [DOI] [PubMed] [Google Scholar]
- 76.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae . J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.